
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 03 June 2019
Sec. Technical Advances in Plant Science
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00714
This article is part of the Research Topic Advances in High-throughput Plant Phenotyping by Multi-platform Remote Sensing Technologies View all 32 articles
Reliable, automatic, multifunctional, and high-throughput phenotypic technologies are increasingly considered important tools for rapid advancement of genetic gain in breeding programs. With the rapid development in high-throughput phenotyping technologies, research in this area is entering a new era called ‘phenomics.’ The crop phenotyping community not only needs to build a multi-domain, multi-level, and multi-scale crop phenotyping big database, but also to research technical systems for phenotypic traits identification and develop bioinformatics technologies for information extraction from the overwhelming amounts of omics data. Here, we provide an overview of crop phenomics research, focusing on two parts, from phenotypic data collection through various sensors to phenomics analysis. Finally, we discussed the challenges and prospective of crop phenomics in order to provide suggestions to develop new methods of mining genes associated with important agronomic traits, and propose new intelligent solutions for precision breeding.
Persistent food and feed supply needs, resources shortages, climate change and energy use are some of the challenges we face in our dependence on plants. Until 2050, crop production will have to double to meet the projected production demands of the global population (Ray et al., 2013). Demand for crop production is expected to grow 2.4% a year, but the average rate of increase in crop yield is only 1.3%. Moreover, production yields have stagnated in up to 40% of land under cereal production (Fischer and Edmeades, 2010). Genetic improvements in crop performance remain the key role in improving crop productivity, but the current rate of improvement cannot meet the needs of sustainability and food security.
Molecular breeding strategies pay more attention to selections based on genotypic information, but phenotypic data are still needed (Jannick et al., 2010; Araus et al., 2014). Phenotyping is necessary to improve the selection efficiency and reproducibility of results in transgenic studies (Gaudin et al., 2013; Yang et al., 2014; Feng et al., 2017) (Table 1). Considering that molecular breeding populations can include a range from at least 200 to at most 10,000 lines, the ability to accurately and high-throughput characterize hundreds of lines at the same time is still challenging (Lorenz et al., 2011; Araus et al., 2014). Apparently, compared with the vast genetic information, there is a phenotyping bottleneck hampering progress in understanding the genetic basis of complex traits. To break this bottleneck and improve the efficiency of molecular breeding, reliable, automatic, and high-throughput phenotypic technologies were urgently needed to provide breeding scientists with new insights in selecting new species to adapt to the resources shortages and global climate change.
During the past decade, plant phenomics has evolved from an emerging niche to a thriving research field, which was defined as the gathering of multi-dimensional phenotypic data at multiple levels from cell level, organ level, plant level to population level (Houle et al., 2010; Dhondt et al., 2013; Lobos et al., 2017). Crop phenotypes are extremely complicated because they are the result of interaction between genotypes (G) and a multitude of envirotypes (E) (Xu, 2016). This interaction influences not only the growth and development process of crops measured by the structural traits on cellular, tissue, organ and plant level, but also the plant functioning measured by the physiological traits. These internal phenotypes in turn determine crop external phenotypes such as morphology, biomass and yield performance (Houle et al., 2010; Dhondt et al., 2013) (Figure 1). Crop phenomics research integrates agronomy, life sciences, information science, math and engineering sciences, and combines high-performance computing and artificial intelligence technology to explore multifarious phenotypic information of crop growth in a complex environment, of which the ultimate goal is to construct an effective technical system able to phenotype crops in a high-throughput, multi-dimensional, big-data, intelligent and automatically measuring manner, and create a tool comprehensively integrating big data achieved from a multi-modality, multi-scale, phenotypic + environmental + genotypic condition, in order to develop new methods of mining genes associated with important agronomic traits, and propose new intelligent solutions for precision breeding.
Here, we provide an overview of crop phenomics research, focusing on two parts, from phenotypic data collection through various sensors to phenomics analysis. We systematically introduced the crop phenotyping approaches from cellular, tissue, organ, and plant level to field level, discussing application and practical problems in research. Based on overwhelming amounts of phenotypic data, we then discussed the phenotyping extraction, phenotype information analysis and knowledge storage. We emphasize Phenomics is entering the big-data era with multi-domain, multi-level, and multi-scale characteristics. We highlight necessity and importance of building multi-scale, multi-dimensional and trans-regional crop phenotyping big database, researching E-trait depth analysis schemas and technical systems for precise identification of crop phenotypes, realizing functional-structural plant modeling based on phenomics, and developing bioinformatics technologies that integrate genomes and phenotypes.
Currently, the plant phenotyping community seems some-what divided between high-throughput, low-resolution phenotyping and in-depth phenotyping at lower throughput and higher resolution (Dhondt et al., 2013). Phenotyping systems and tools applied in different scales are focused on different key characteristics –automated phenotyping platforms in controllable environment and high-throughput methodologies in field environment highlights high-throughput, while phenotyping covering the organ, tissue and cellular level emphasizes in-depth phenotyping and higher resolution. In this part, we systematically introduced the crop phenotyping approaches covering from cellular and tissue level to field level, and discussed the application and practical problems of which technologies in crop researches.
Compared with the whole-plant phenotying technologies, phenotyping at higher spatial and temporal resolutions of tissue and cellar scales is more difficult (Hall et al., 2016). To reveal crop micro-phenotypes, the pre-treatment of plant or organ is usually destructive when sampled in a cumbersome and involves multi-step procedure, and the high resolution imaging of samples is also inefficient in view of the output in micron level. More challenging, automated image analysis techniques are urgently needed to quantify cell and tissue traits of crop from the larger high qualified image. In recent years, many emerging algorithms and tools have been proposed to handle with microscopic images from hand-cutting slices, micro-CT, fluorescent, laser, and paraffin sections imaging (Table 2).
Root anatomical traits have important effects on plant function, including acquisition of nutrients and water from the soil and transportation to the aboveground part. Over the last years, novel micro-image acquisition technologies and computer vision have been introduced to improve our understanding of anatomical structure and function of roots. In 2011, computer-aided calculation of wheat root vascular bundle phenotypic information extraction were introduced based on sequence images of paraffin sections (Wu et al., 2011). Burton et al. (2012) (Penn State College of Agricultural Sciences Roots Lab) have developed a high-throughput, high-resolution phenotyping platform, which combines laser optics and serial imaging with three-dimensional (3D) image reconstruction (RootSlice) and quantification to understand root anatomy by semi-auto RootScan (Chimungu et al., 2015). Chopin et al. (2015) developed a fully automated RootAnalyzer software for root microscopic phenotypic information extraction to further improve image segmentation efficiency and ensure high accuracy. Pan et al. (2018) (Beijing Key Laboratory of Digital Plant) introduced X-ray micro-CT into 3D imaging of maize root tissues and developed an image processing scheme for the 3D segmentation of metaxylem vessels. Compared with traditional manual measurement of vascular bundles of maize roots, the proposed protocols significantly improved the efficiency and accuracy of the micron-scale phenotypic quantification.
Different from the root system, crop stalks have a more complex microstructure. The complexity and diversity in microscopic image data poses greater challenges for developing suitable data analysis workflows in the detection and identification of microscopic phenotypes of stalk tissue. Zhang et al. (2013) and Legland et al. (2017) presented semi- automated and automated analysis method for the stained microscopic images of stalk sections. Legland et al. (2014) and Heckwolf et al. (2015) created an image analysis tool that could operate on images of hand-cut stalk transections to measure anatomical features in high throughput. Those tools have significantly improved the measurement efficiency of vascular bundle, but the anatomical traits corresponding to the rind and the detection accuracy remained a challenge. Du et al. (2016) (Beijing Key Laboratory of Digital Plant) introduced micro-computed tomography (CT) technology for stalk imaging and developed the VesselParser 1.0 algorithm, which made it possible to automatically and accurately analyze phenotypic traits of vascular bundles within entire maize stalk cross-sections. So far, Beijing Key Laboratory of Digital Plant has built a novel method to improve the X-ray absorption contrast of maize tissue suitable for ordinary micro-CT scanning. Based on CT images, they introduced a set of image processing workflows for maize root, stalk and leaf to effectively extract microscopic phenotypes of vascular bundles (Zhang et al., 2018).
Phenotyping at tissue and cellar scales still requires complex procedures. The need to simplify the sample preparation process and explore advanced imaging techniques is crucial to accelerate microscopic phenotyping studies. Moreover, image processing is another major bottleneck in micro-phenotyping researches. Due to the specific crop organ and cell phenotypic characteristics of the huge differences, most micro-image analysis algorithms have been developed for specific biological assays currently.
Most plant phenotyping platforms concentrated the high-throughput of individual plants (Chaivivatrakul et al., 2014; Cabrera-Bosquet et al., 2016). Hence the phenotypic accuracy of organs on the plants was always compromised (Dhondt et al., 2013). The most frequently used phenotyping index, such as leaf length, leaf area, and fruit volume, could be obtained in phenotyping platforms simply (Klukas et al., 2014; Zhang et al., 2017). The smartphone app platform was developed for field phenotyping by taking pictures and image analysis at organ level (Confalonieri et al., 2017). It is very convenient to acquire leaf angles and leaf length using the app in a 2D view. The plants do not need to be destructively sampled indoors. 2D cameras are low cost and were integrated into most of the phenotyping platforms, which provided effective phenotypic solution for branch structured plants, especially for tracking the dynamic growth of organs on plants (Brichet et al., 2017). However, 2D images lost another dimension data in 3D space, and some of the estimated morphological traits still need to be calibrated (Zhang et al., 2017). Multi-view stereo (MVS) approach (Duan et al., 2016; Vazquez-Arellano et al., 2016; He et al., 2017; Hui et al., 2018) is another popular low-cost alternative for organ level phenotyping. 3D point clouds were reconstructed using multi-view images through structure from motion techniques (Wu, 2011), and then phenotypic traits were extracted through segmented organs of individual plants (Thapa et al., 2018). This cost-effective 3D reconstruction method depicts an alternative to an expensive laser scanner in the studied scenarios with potential for automated procedures (Rose et al., 2015; Yin et al., 2016; Gibbs et al., 2018). There are significant differences between these organ traits within various cultivars, thus tiny errors could be ignored in large scale omics analysis (Yang et al., 2014; Zhang et al., 2017).
However, besides these measured length, area, and volume phenotypic parameters, there are lots of obvious differences that can be observed by humans of plant organs, such as blade profiles, blade folds, vein curves, and leaf colors, of which are difficult to obtain the morphological data and quantitatively describe their differences. A series of mathematical approaches (Li et al., 2018) need to be developed to quantitatively describe these differences in order to discover more detailed phenotyping traits with high precision phenotyping data of organ level. Researchers use high resolution 3D scanners to acquire the morphological structure of plant organs (Rist et al., 2018). High resolution 3D scanners are relatively expensive while the acquired morphological data are more accurate than MVS reconstructed organs, especially for non-planner surface plants. 2D LiDAR scanners (Thapa et al., 2018) and depth sensors (Hu et al., 2018) were also used, combined with turn table and translation devices to estimate phenotypic traits through 3D recovery of point clouds. Most 2D or 3D imaging could solve the 3D organ scale phenotyping data acquisition problems, except for tall plants for which it is difficult due to a limited field of vision. High resolution 3D point clouds of plant organs are quite complicated in order to extract more phenotypic traits, thus computer graphics algorithms, such as skeleton extraction (Huang et al., 2013), surface reconstruction (Yin et al., 2016; Gibbs et al., 2017), and feature-preserving remeshing (Wen et al., 2018), need to be applied to process the high-resolution morphological data for 3D phenotypic trait extraction. Because of the difficulty and complexity of detailed organ data acquisition and analysis, Wen et al. (2017) proposed a data acquisition standard and constructed a resource database of plant organs using measured in situ morphological data, to realize the integration and sharing of high quality data of plant organs.
Automation and robotics, new sensors, and imaging technologies (hardware and software) have provided an opportunity for high-throughput plant phenotyping platforms (HTPPs) development (Gennaro et al., 2017). In the past 10 years, great improvements have been made in researching and developing HTPPs (Furbank et al., 2011; Fiorani and Schurr, 2013; Virlet et al., 2016). Depending on the overall design, HTPPs in a growth chamber or greenhouse can generally be classified as sensor-to-plant or plant-to-sensor based on whether the plants occupy a fixed position during a measurement routine and an imaging setup moves to each of those positions or the plants are transported to an imaging station, respectively. Collectively, the techniques used in HTPPs in the growth chamber or greenhouse mainly include (Table 3):
(1) RGB imaging, which obtains the phenotypes of plant morphology, color, and texture.
(2) Chlorophyll fluorescence imaging, which obtains photosynthetic phenotypes.
(3) Hyperspectral imaging, which obtains phenotypes such as pigment composition, biochemical composition, nitrogen content, and moisture content.
(4) Thermal imaging, which obtains the plant surface temperature distribution, stomatal conductance and transpiration phenotype.
(5) Lidar, which obtains the three-dimensional structure phenotype of plants.
Moreover, other advanced imaging techniques widely used in medicine, such as MRI, PET, and CT, have also been introduced into HTPPs in the growth chamber or greenhouse. The following table lists the major imaging techniques used in the HTPPs in the growth chamber or greenhouse.
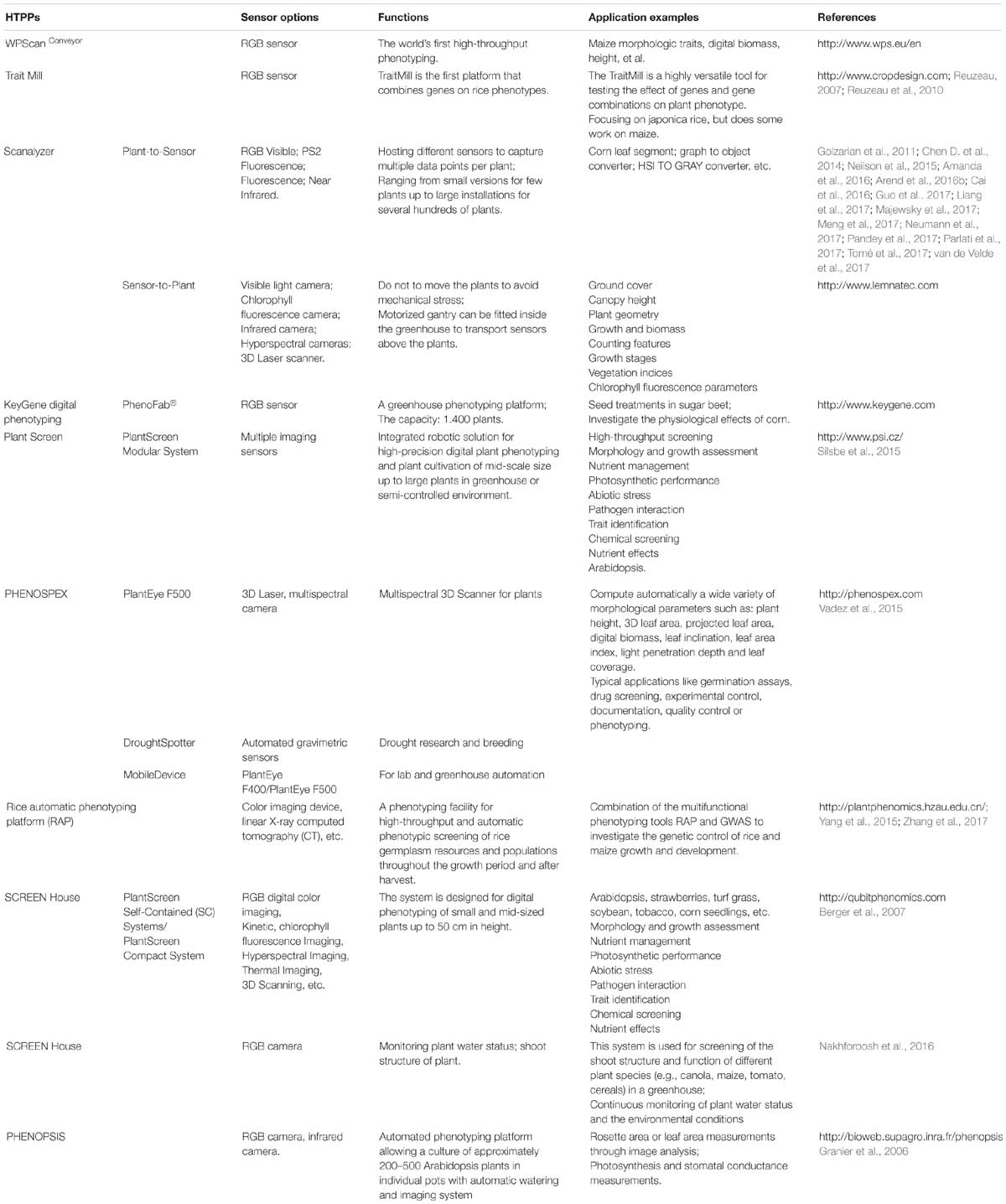
Table 4. Exhaustive list of digital technologies and platform equipments for crop phenotyping in controllable environment.
After integration of information technology, digital technology and platform equipment with plant phenotyping, several HTPPs in growth chamber or greenhouse have been listed in Table 4. Such high-throughput phenotyping platforms are characterized by automation, high-throughput, and high precision, which greatly improve plant data collection efficiency and accuracy, in order to improve the efficiency of crop breeding.
However, most HTPPs still have high construction, operating, and maintenance costs, and most academic and research institutions do not have access to these techniques as a result (Kolukisaoglu and Thurow, 2010). For example, the number of fully automatic and high-throughput phenotyping platforms is limited in China: one was developed by the Crop Phenotyping Center (CPC), http://plantphenomics.hzau.edu.cn/, and the others were introduced by LemnaTec, such as http://bri.caas.net.cn/, http://www.kenfeng.com/, and http://www.zealquest.com/. Because of this, there has been increased research into developing affordable, small-scale plant phenotyping platforms and technology. In fact, affordable phenotypic acquisition techniques or platforms are constantly being updated, such as the imaging system developed by Tsaftaris and Noutsos (2009) that uses wireless-connected consumer digital cameras, and the low-cost Glyph phenotyping platform (Pereyra-Irujo et al., 2012). Lowering the cost of these platforms could, therefore, significantly increase the scope of phenotypic research and advance the rapid expansion of phenotypic–genotypes analysis for complex traits.
Field-based phenotyping (FBP) is a critical component of crop improvement through genetics, as it is the ultimate expression of the relative effects of genetic factors, environmental factors, and their interaction on critical production traits, such as yield potential and tolerance to abiotic/biotic stresses (Araus and Cairns, 2014; Neilson et al., 2015). Currently, the most commonly field-based phenotyping platforms (FBPPs) use ground wheeled, rigid motorized gantry or aerial vehicles, combined with a wide range of cameras, sensors and high-performance computing, to capture deep phenotyping data in time (throughout the crop cycle) and space (at the canopy level) in field environments (Fritsche-Neto and Borém, 2015) (Table 5). It cannot be denied that the efficiency of ground wheeled phenotyping system is quite low if the plot area is too large (White et al., 2012; Zhang and Kovacs, 2012). In 2016, FIELD SCANALYZERS, with rigid motorized gantry supporting a weather proof measuring platform that incorporates a wide range of cameras, sensors and illumination systems, were developed. The facility, equipped with high-resolution visible, chlorophyll fluorescence and thermal infrared camera, hyperspectral imager, and 3D laser scanner. Crops within a 10–20 m × 110–200 m area can be monitored, which realizes the continuous, automatic, and high-throughput detection of crop phenotyping detection in field (Virlet et al., 2016; Sadeghi-Tehran et al., 2017). Meanwhile, the cable-suspended field phenotyping platform covering an area of ∼1 ha was also developed for rapid and non-destructive monitoring of crop traits (Kirchgessner et al., 2016). However, these large ground-based field phenotyping platforms also have high construction, operating, and maintenance costs, and it has to be located at certain sites limiting the scale at which it can be used. Furthermore, the ground-based FBPPs are not very suitable for large crops, such as maize, except in the early stages (Montes et al., 2011).
In recent years, manned aircraft and unmanned aerial vehicle remote sensing platforms (UAV-RSPs) are becoming a high-throughput tool for crop phenotyping in the field environment (Berni et al., 2009; Liebisch et al., 2015), which meet the demands of spatial, spectral, and temporal resolutions (Yang et al., 2017) (Table 5). For example, thermal sensors fitted to manned aircraft were used to measure canopy temperature (Deery et al., 2016; Rutkoski et al., 2016). The sensors that UAV-RSPs carried typically included digital cameras, infrared thermal imagers, light detection and ranging (LIDAR), multispectral cameras, and hyperspectral sensors, which are applied to: canopy surface modeling and crop biomass estimation based on visible imaging; crop physiological status monitor, such as chlorophyll fluorescence and N levels, based on visible–near-infrared spectroscopy and high-resolution hyperspectral imaging; plant water status detection based on thermal imaging; and crop fine-scale geometric traits analysis based on LIDAR point clouds (Sugiura et al., 2005; Overgaard et al., 2010; Swain et al., 2010; Wallace et al., 2012; Gonzalez-Dugo et al.; Gonzalez-Dugo et al., 2014; Mathews and Jensen, 2013; Diaz-Varela et al., 2014; Gonzalez-Dugo et al., 2015; Nigon et al., 2015; Gómez-Candón et al., 2016; Camino et al., 2018; Roitsch et al., 2019). There are definite advantages for UAV-RSPs, including portable, high monitoring efficiency, low-cost, and suitability for field environments. On the other hand, some limiting factors for UAV-RSPs also exist, including the lack of methods for fast and automatic data processing and modeling, the strict airspace regulations, and vulnerable to different weather conditions. Recently, combining ground-based platforms and aerial platforms for phenotyping offers flexibility. For example, the tractor-based proximal crop-sensing platform, combined with UAV-based platform, was used to target complex traits such as growth and RUE in sorghum (Potgieter et al., 2018; Furbank et al., 2019).
Phenomic experiments are not directly reproducible because of the multi-model of sensors (structure, morphology, color, and physiology information), multi-scales phenotypic data (from cellular to population level), and variability of environmental conditions. Crop phenotypic data collection is only the first step in Phenomic research. How to extract phenotypic traits from raw data? How to realize phenotype data standardization and storage? How to make cross-scale, cross-dimensionality meta-analyses? Finally, based on the phenotypic big data, how to realize the model-assisted phenotyping and phenotypic-genomic selection? Hereby this part focuses on hot topics in crop phenomics analysis raised above. We then suggest that research in this area is entering a new stage of development using artificial intelligence technology, such as deep learning methods, in which can help researchers transform large numbers of omic-data into knowledge.
Image-based phenotyping, as an important and all-purpose technique, has been applied to measure and quantify vision-based and model-based traits of plant in the laboratory, greenhouse and field environments. So far, lots of image analysis methods and softwares have been developed to perform image-based plant phenotyping (Fahlgren et al., 2015b). From image analysis perspective, phenotypic traits of plant can be classified into 4 categories, i.e., quantity, geometry, color and texture, and are also classified into linear and non-linear features related to the pixel representation. Moreover, some valuable agronomic and physiological traits can be derived from image features. Generally, image techniques are specially designed according to specific crop varieties and phenotypic traits of interest, and always require with prior knowledge of research objects, as well as more or less man-machine interaction. Under highly controlled conditions, the classic image processing pipeline can provide acceptable phenotyping results, such as biomass (Leister et al., 1999), NDVI (Walter et al., 2015), chlorophyll responses (Walter et al., 2015), and compactness (Vylder et al., 2012) etc. However, the simple image processing pipeline is still very difficult to handle with non-linear, non-geometric phenotyping tasks (Pape and Klukas, 2015). Facing species and environment diversities, fully automated and intelligent image analysis will remain a long-term challenge.
Machine learning techniques, such as support vector machines, clustering algorithms, and neural networks, have been widely utilized to image-based phenotyping, which not only improve the robustness of image analysis tasks, also relieve tedious manual intervention (Singh et al., 2016). There is no doubt that machine learning techniques will have a prominent role in breaking through the bottlenecks of plant phenotyping (Tsaftaris et al., 2016). In a broad category of machine learning techniques (Ubbens and Stavness, 2017), deep learning demonstrates impressive advantages in many image-based tasks, such as object detection and localization, semantic segmentation, image classification, and others (Lecun et al., 2015). In essence, deep convolutional neural networks (CNNs) are well suited to many vision-based computer problems, e.g., recognition (Lecun, 1990), classification (He et al., 2015), and instance detection and segmentation (Girshick, 2015). Compared with the traditional image analysis methods, CNNs are simultaneously trained from end to end without image feature description and extraction procedures. As far as plant phenotyping, CNNs have been effectively applied to detect and diagnosis (Mohanty et al., 2016), classify fruits and flowers (Pawara et al., 2017), and count leaf number (Ubbens and Stavness, 2017). It is worth noting that those vision-based phenotyping tasks were driven by the massive captured and annotation plant images. From the view of machine vision perspective, deep learning has been a fundamental technique framework in image-based plant phenotyping (Tsaftaris et al., 2016).
A huge amount of complex data and the integration of a wide range of image, spectral and environmental data can be generated through by the above phenotypic technologies, usually up to GB or even petabytes, unstructured “Big Data.” Thus, the efficient storage, management and retrieval of phenotypic data have become the important issues to be considered (Wilkinson et al., 2016). The current universally accepted principle of information standardization includes three aspects: (i) the ‘minimum information’ (MI) approach is recommended to define the content of the data set, (ii) ontology terms is applied for the unique and repeatable annotation of data, and in the form of data sharing and meta-analyses (Krajewski et al., 2015; Coppens et al., 2017), (iii) and proper data formats, such as CSV, XML, RDF, MAGE-TAB, etc., are chosen for the construction of data sets. Up until now, a number of phenotyping resources have been built ranging from phenotypic data of one species to multi-data types (Coppens et al., 2017). In 2010, PODD was developed for capturing, managing, annotating and distributing the data to support both Australian and international biological research communities (Li et al., 2010); Fabre et al. (2011) built a PHENOPSIS DB information system for Arabidopsis thaliana phenotypic data acquired by the PHENOPSIS phenotyping platform; Bisque is the first web based, cross-platform, developed into a repository to store, visualize, organize and analyze images in the cloud (Kvilekval Das et al., 2010; Goff et al., 2011). In 2014, ClearedLeaves DB, an on open online database, was built to store, manage and access leaf images and phenotypic data (Das et al., 2014); AraPheno1 was the first comprehensive, central repository of population-scale phenotypes (it integrated more than 250 publicly available phenotypes from six independent studies) for A. thaliana inbred lines (Seren et al., 2017); PhenoFront was a publicly available dataset of above-ground plant tissue to the LemnaTec Phenotyper platform (Fahlgren et al., 2015a); in 2016, the plant genomics and phenomics research data repository (PGP) were developed by the Leibniz institute of plant genetics and crop plant research and the German plant phenotyping network to comprehensively publish plant phenotypic and genotypic data (Arend et al., 2016a). Obviously, from the perspective of database data standardization and storage, the storage scheme based on “cloud technology” is becoming the trend for the development of plant phenotype data storage. Cloud storage system can optimize the design of the system architecture, file structure, high-speed cache, etc., for the plant phenotype platform. At present, all kinds of phenotypic data collection platforms are still relatively independent, and have not been established at the level of regions, countries or continents. Through the advanced technology of artificial intelligence, establishing a typical crop phenotype database based on the multi-layer phenotypic information, for example GDB Human Genome Database, will of interest to a range of stakeholders.
Plants are highly plastic to genotypes, environment and management via changing morphological traits and adjusting their physiological behavior. Complex interactions between genotypes, environment and managements at different scales determine the development of plants, but their separate contribution to the phenotype remains unclear. Dynamic models have been proven to be an efficient tool in dissection of abiotic and biotic effects on plant phenotypes (Tardieu and Tuberosa, 2010). Functional–structural plant (FSP) (Vos et al., 2010) models simulate plant growth and development in time and three-dimension (3D) space, and quantify complex interactions between architecture and physiological processes. The combination of FSP modeling and phenotyping have been used as a facile technique to address research questions in two ways.
Firstly, FSP models offer a tool to dissect phenotype governed by a set of mechanisms. For example, Zhu et al. (2015) used a FSP model to dissect net biodiversity effect into the effect induced by interspecific trait differences, and the effect induced by phenotypic plasticity by simulating whole-vegetation light capture for scenarios with and without phenotypic plasticity based on experimental plant trait data. The separate effect of each architectural trait (leaf angle, leaf curvature and internode length etc.) on dry mass production and light interception was quantified by simulating canopy growth using a dynamic FSP model for tomato (Chen T.W. et al., 2014). Radiation interception and radiation use efficiency were dissected into an environmental and a genetic term via conducting virtual multi-genotype canopies, in which the FSP model for maize was applied to calculate light interception for each plant (Chen et al., 2018). FSP modeling has proven to be highly effective for disentangling the relative contribution of each underlying process.
Secondly, high-throughput phenotyping techniques facilitate the automate and precise calibration of FSP models. Image-based analysis was performed daily to reconstruct individual maize architecture, in order to calculate light interception using a FSP model and to estimate leaf area and the fresh plant weight of individual plants (Cabrera-Bosquet et al., 2016). Terrestrial LiDAR scanning was used to reconstruct complex tree canopy for predicting three-dimensional distribution of microclimate-related quantities in terms of net radiation, surface temperature and evapotranspiration (Bailey et al., 2016). OPENSIMROOT integrated a root model that can simulate growth of a root system with 3D phenotyping techniques, such as magnetic resonance imaging (MRI) and X-ray computed tomography (CT) (Postma et al., 2017). Phenotyping techniques not only provide an efficient method to evaluate the ability of the model to simulate plant architecture and geometry but also help researchers to understand functional responses based on images.
Although genomic data has a major role in crop genetic improvements and breeding programs, with the advent of the era of omics, considerable gain can only be achieved by tightly coupling genomic discovery to plant phenomics (Cobb et al., 2013; Bolger et al., 2017). In recent years, phenomic researches that combine genomic data with data on quantitative variation in phenotypes have been initiated in many species, which rapidly decoded the function of a mass of unknown genes and improved understanding the G-P map (Campbell et al., 2015, 2017; Neumann et al., 2017).
Many agronomic traits are complex and controlled by many genes, each with a small effect. Identifying the molecular basis of such complex traits requires genotyping and phenotyping of suitable mapping populations, enabling quantitative trait locus (QTL) mapping and genome-wide association studies (GWAS), which have been widely carried out in crop plants (Salvi and Tuberosa, 2015; Muraya et al., 2017). Busemeyer et al. (2013) associated phenotypic traits of small grain cereals with genome information to dissect the genetic architecture of biomass accumulation. In 2014, based on 13 traditional agronomic traits and 2 newly defined traits of rice, Yang et al. (2014) identified 141 associated loci by GWAS. In 2015, Combining GWAS with 29 leaf traits at three growth stages using high-throughput leaf scoring (HLS), 73 new loci with leaf size, 123 of leaf color, and 177 of leaf shape were detected (Yang et al., 2015). In 2017, large-scale quantitative trait locus (QTL) mapping was performed, combined with 106 agronomic traits of maize inbred line from seedling to tasselling stage, and a total of 988 QTLs were identified (Zhang et al., 2017). Also, plant sizes of 252 diverse maize inbred lines were monitored at 11 different developmental time points, and 12 main-effect marker-trait associations were identified (Muraya et al., 2017).
Obviously, combining the high-throughput phenotyping technology and large-scale QTL or GWAS analysis not only greatly expanded our knowledge of the crop dynamic development process but also provided a novel tool for studies of crop genomics, gene characterization and breeding. We believe that with a complete system of genetic information, combined with crop high-throughput phenotyping technology, phenotypic-genomic analysis will revolutionize how we deal with complex traits and underpin a new era of crop improvement.
Phenomics is entering the era of ‘Big Data,’ thus the crop science community need to combine artificial intelligence technology and collaborative research at the national and international levels, to build a new theory for analyzing crop phenotypic information, construct an effective technical system able to phenotype crops in a high-throughput, multi-dimensional, big-data, intelligent and automatically measuring manner, and create a tool comprehensively integrating big data achieved from a multi-modality, multi-scale, phenotypic + environmental + genotypic condition. There is no denying that there are many challenges that crop phenomics need to address in the next 5–10 years, such as:
(1) Phenomics is entering the big-data era with high-throughput, multi-dimensionality, and multi-scale. We emphasize various phenotyping approaches for crop morphology, structure, and physiological data with three multi- characteristics: multi-domain (phenomics, genomics etc.), multi-level (traditional small to medium scale up to large-scale omics), and multi-scale (crop morphology, structure, and physiological data from cell to whole-plant). The single and individual phenotypic information cannot satisfy the association analysis in the new era called ‘-omics,’ and the systematic and complete phenomics information will be the foundation of future research.
(2) In response to emerging challenges, new methods and techniques based on artificial intelligence shall be introduced to advance image-based phenotyping. An automated phenotyping system and platform result in lots of digital features, which need to prove their values throughout large sample statistics and relationship analysis with traditional agronomic traits. How to precisely and efficiently evaluate, understand and interpret these digital image-based features, and dig out valuable quantitative traits for functional genomes are key problems in the development and application of plant phenotyping.
(3) With the multi-domain, multi-level, and multi-scale phenotypic information, we urgently need to make use of the latest achievements of artificial intelligence in depth learning, data fusion, hybrid intelligence and swarm intelligence to develop big-data management producers for supporting data integration, interoperability, ontologies, shareability and globality.
(4) Modeling is a powerful tool to understand G × E × M interactions, identify key traits of interest for target environments. Nevertheless, several scientific and technical challenges need to be overcome. For example the validity and practicality of the models in terms of modeling processes and their interactions need further verification, and the interaction and feedbacks of multi-scale phenotypes between modeling processes also need to be solved. Only then will we be able to streamline and speed up the tortuous gene-to-phenotype journey through modeling to develop the required agricultural outputs and sustainable environments for everybody.
(5) Crop genotype (G) -phenotype (p) -envirotype (E) information comprehensive analysis and utilization. In short, as Coppens et al. (2017) said “the future of plant phenotyping lies in synergism at the national and international levels.” We need to seek novel solutions to the grand challenges of multi-omics data, such as intelligent data-mining analytics, which offers a powerful tool to unravel the biological processes governing plant growth and development, and to advance plant breeding for much-needed climate-resilient and high-yielding crops.
CZ, XG, and YZ conducted the literature survey and drafted the article. JD, WW, SG, JW, and JF provided the data and tables about the crop phenotyping researches by NERCITA. All authors read and approved the final manuscript.
This study was supported by the Construction of Collaborative Innovation Center of Beijing Academy and Forestry Science (Collaborative Innovation Center of Crop Phenomics), the National Natural Science Foundation of China (Nos. 31801254 and 31671577), Research Development Program of China (No. 2016YFD0300605-01), and Beijing Academy of Agricultural and Forestry Sciences Grant (Nos. KJCX20170404 and KJCK20180423).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Amanda, D., Doblin, M. S., Galletti, R., Bacic, A., Ingram, G. C., and Johnson, K. L. (2016). Defective kernel1 (DEK1) regulates cell walls in the leaf epidermis. Plant Physiol. 172, 2204–2218. doi: 10.1104/pp.16.01401
Araus, J. L., and Cairns, J. E. (2014). Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61. doi: 10.1016/j.tplants.2013.09.008
Arend, D., Junker, A., Scholz, U., Schüler, D., Wylie, J., and Lange, M. (2016a). PGP repository: a plant phenomics and genomics data publication infrastructure. Database 2016:baw033. doi: 10.1093/database/baw033
Arend, D., Lange, M., Pape, J. M., Weigelt-Fischer, K., Arana-Ceballos, F., Mücke, I., et al. (2016b). Quantitative monitoring of Arabidopsis thaliana growth and development using high-throughput plant phenotyping. Sci. Data 3:160055. doi: 10.1038/sdata.2016.55
Bailey, B. N., Stoll, R., Pardyjak, E. R., and Miller, N. E. (2016). A new three-dimensional energy balance model for complex plant canopy geometries: model development and improved validation strategies. Agric. For. Meteorol. 218, 146–160. doi: 10.1016/j.agrformet.2015.11.021
Berger, S., Benediktyová, Z., Matous, K., Bonfig, K., Mueller, M. J., Nedbal, L., et al. (2007). Visualization of dynamics of plant-pathogen interaction by novel combination of chlorophyll fluorescence imaging and statistical analysis: differential effects of virulent and avirulent strains of P. Syringae and of Oxylipins on A. thaliana. J. Exp. Bot. 58, 797–806. doi: 10.1093/jxb/erl208
Berni, J. A. J., Zarco-Tejada, P. J., Suarez, L., and Fereres, E. (2009). Thermal and narrowband multispectral remote sensing for vegetation monitoring from an unmanned aerial vehicle. IEEE Trans. Geosci. Remote Sens. 47, 722–738. doi: 10.1109/tgrs.2008.2010457
Bolger, M., Schwacke, R., Gundlach, H., Schmutzer, T., Chen, J., Arend, D., et al. (2017). From plant genomes to phenotypes. J. Biotechnol. 261, 46–52. doi: 10.1016/j.jbiotec.2017.06.003
Brichet, N., Fournier, C., Turc, O., Strauss, O., Artzet, S., Pradal, C., et al. (2017). A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 13:12. doi: 10.1186/s13007-017-0246-7
Burton, A. L., Williams, M., Lynch, J. P., and Brown, K. M. (2012). Rootscan: software for high-throughput analysis of root anatomical traits. Plant Soil 357, 189–203. doi: 10.1007/s11104-012-1138-2
Busemeyer, L., Ruckelshausen, A., Möller, K., Melchinger, A. E., Alheit, K. V., Maurer, H. P., et al. (2013). Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci. Rep. 3:2442. doi: 10.1038/srep02442
Cabrera-Bosquet, L., Fournier, C., Brichet, N., Welcker, C., Suard, B., and Tardieu, F. (2016). High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol. 212, 269–281. doi: 10.1111/nph.14027
Cai, J., Okamoto, M., Atieno, J., Sutton, T., Li, Y., and Miklavcic, S. J. (2016). Quantifying the onset and progression of plant senescence by color image analysis for high throughput applications. PLoS One 11:e0157102. doi: 10.1371/journal.pone.0157102
Camino, C., González-Dugo, V., Hernández, P., Sillero, J. C., and Zarco-Tejada, P. J. (2018). Improved nitrogen retrievals with airborne-derived fluorescence and plant traits quantified from VNIR-SWIR hyperspectral imagery in the context of precision agriculture. Int. J. Appl. Earth Obs. Geoinf. 70, 105–117. doi: 10.1016/j.jag.2018.04.013
Campbell, M. T., Du, Q., Liub, K., Brien, C. J., Berger, B., Zhang, C., et al. (2017). A comprehensive image-based phenomic analysis reveals the complex genetic architecture of shoot growth dynamics in rice (Oryza sativa). Plant Genome 10, 1–14. doi: 10.3835/plantgenome2016.07.0064
Campbell, M. T., Knecht, A. C., Berger, B., Brien, C. J., Wang, D., and Walia, H. (2015). Integrating image-based phenomics and association analysis to dissect the genetic architecture of temporal salinity responses in rice. Plant Physiol. 168, 1476–1489. doi: 10.1104/pp.15.00450
Carrolla, A. A., Clarke, J., Fahlgrenc, N. A., Gehanc, M. J., Lawrence-Dilld, C., and Lorencee, L. (2019). NAPPN: who we ae, where we are going, and why you should join us! Plant Phenome J. 2:180006.
Chaivivatrakul, S., Tang, L., Dailey, M. N., and Nakarmi, A. D. (2014). Automatic morphological trait characterization for corn plants via 3d holographic reconstruction. Comput. Electron. Agric. 109, 109–123. doi: 10.1016/j.compag.2014.09.005
Chapman, S. C., Merz, T., Chan, A., Jackway, P., Hrabar, S., Dreccer, M. F., et al. (2014). Pheno-copter: a low-altitude, autonomous remote-sensing robotic helicopter for high-throughput field-based phenotyping. Agronomy 4, 279–301. doi: 10.3390/agronomy4020279
Chen, D., Neumann, K., Friedel, S., Kilian, B., Chen, M., Altmann, T., et al. (2014). Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26, 4636–4655. doi: 10.1105/tpc.114.129601
Chen, T. W., Nguyen, T. M. N., Kahlen, K., and Stützel, H. (2014). Quantification of the effects of architectural traits on dry mass production and light interception of tomato canopy under different temperature regimes using a dynamic functional–structural plant model. J. Exp. Bot. 65, 6399–6410. doi: 10.1093/jxb/eru356
Chen, T.-W., Cabrera-Bosquet, L., Prado, S. A., Perez, R., Artzet, S., Pradal, C., et al. (2018). Genetic and environmental dissection of biomass accumulation in multi-genotype maize canopies. J. Exp. Bot. 70, 2523–2534. doi: 10.1093/jxb/ery309
Chimungu, J. G., Loades, K. W., and Lynch, J. P. (2015). Root anatomical phenes predict root penetration ability and biomechanical properties in maize. J Exp. Bot. 66, 3151–3162. doi: 10.1093/jxb/erv121
Chopin, J., Laga, H., Huang, C. Y., Heuer, S., and Miklavcic, S. J. (2015). Rootanalyzer: a cross-section image analysis tool for automated characterization of root cells and tissues. PLoS One 10:e0137655. doi: 10.1371/journal.pone.0137655
Cobb, J. N., DeClerck, G., Greenberg, A., Clark, R., and McCouch, S. (2013). Next-generation phenotyping: requirements and strategies for enhancing our understanding of genotype–phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 126, 867–887. doi: 10.1007/s00122-013-2066-0
Confalonieri, R., Paleari, L., Foi, M., Movedi, E., Vesely, F. M., Thoelke, W., et al. (2017). Pocketplant3d: analysing canopy structure using a smartphone. Biosys. Eng. 164, 1–12. doi: 10.1016/j.biosystemseng.2017.09.014
Coppens, F., Wuyts, N., Inzé, D., and Dhondt, S. (2017). Unlocking the potential of plant phenotyping data through integration and data-driven approaches. Curr. Opin. Syst. Biol. 4, 58–63. doi: 10.1016/j.coisb.2017.07.002
Das, A., Bucksch, A., Price, C. A., and Weitz, J. S. (2014). Clearedleavesdb: an online database of cleared plant leaf images. Plant Methods 10:8. doi: 10.1186/1746-4811-10-8
Deery, D. M., Rebetzke, G. J., Jimenez-Berni, J. A., James, R. A., Condon, A. G., Bovill, W. D., et al. (2016). Methodology for high-throughput field phenotyping of canopy temperature using airborne thermography. Front. Plant Sci. 7:e1808. doi: 10.3389/fpls.2016.01808
Dhondt, S., Wuyts, N., and Inzé, D. (2013). Cell to whole-plant phenotyping: the best is yet to come. Trends Plant Sci. 18, 428–439. doi: 10.1016/j.tplants.2013.04.008
Diaz-Varela, R. A., Zarco-Tejada, P. J., Angileri, V., and Loudjani, P. (2014). Automatic identification of agricultural terraces through object-oriented analysis of very high resolution DSMs and multispectral imagery obtained from an unmanned aerial vehicle. J. Environ. Manag. 134, 117–126. doi: 10.1016/j.jenvman.2014.01.006
Du, J., Zhang, Y., Guo, X., Ma, L., Shao, M., Pan, X., et al. (2016). Micron-scale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stems based on micro-CT scanning. Funct. Plant Biol. 44, 10–22.
Duan, T., Chapman, S. C., Holland, E., Rebetzke, G. J., Guo, Y., and Zheng, B. (2016). Dynamic quantification of canopy structure to characterize early plant vigour in wheat genotypes. J. Exp. Bot. 67, 4523–4534. doi: 10.1093/jxb/erw227
Fabre, J., Dauzat, M., Nègre, V., Wuyts, N., Tireau, A., Gennari, E., et al. (2011). Phenopsis DB: an information system for Arabidopsis thaliana phenotypic data in an environmental context. BMC Plant Biol. 11:77. doi: 10.1186/1471-2229-11-77
Fahlgren, N., Feldman, M., Gehan, M. A., Wilson, M. S., Shyu, C., Bryant, D. W., et al. (2015a). A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria. Mol. Plant 8, 1520–1535. doi: 10.1016/j.molp.2015.06.005
Fahlgren, N., Gehan, M. A., and Baxter, I. (2015b). Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 24:93. doi: 10.1016/j.pbi.2015.02.006
Feng, H., Guo, Z., Yang, W., Huang, C., Chen, G., Fang, W., et al. (2017). An integrated hyperspectral imaging and genome-wide association analysis platform provides spectral and genetic insights into the natural variation in rice. Sci. Rep. 7:4401. doi: 10.1038/s41598-017-04668-8
Fiorani, F., and Schurr, U. (2013). Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 64, 267–291. doi: 10.1146/annurev-arplant-050312-120137
Fischer, R. A. T., and Edmeades, G. O. (2010). Breeding and cereal yield progress. Crop Sci. 50, S85–S98.
Fritsche-Neto, R., and Borém, A. (2015). Phenomics How Next-Generation Phenotyping is Revolutionizing Plant Breeding. Switzerland: Springer Press.
Furbank, R. T., Jimenez-Berni, J. A., George-Jaeggli, B., Potgieter, A. B., and Deery, D. M. (2019). Field crop phenomics: enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. [Epub ahead of print]
Furbank, R. T., and Tester, M. (2011). Phenomics-technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 16, 635–644. doi: 10.1016/j.tplants.2011.09.005
Gaudin, A. C. M., Henry, A., Sparks, A. H., and Slamet-Loedin, I. H. (2013). Taking transgenic rice drought screening to the field. J. Exp. Bot. 64, 109–117. doi: 10.1093/jxb/ers313
Gennaro, S. F. D., Rizza, F., Badeck, F. W., Berton, A., Delbono, S., Gioli, B., et al. (2017). UAV-based high-throughput phenotyping to discriminate barley vigour with visible and near-infrared vegetation indices. Int. J. Remote Sens. 39, 5330–5344. doi: 10.1080/01431161.2017.1395974
Gibbs, J. A., Pound, M., French, A. P., Wells, D. M., Murchie, E., and Pridmore, T. (2017). Approaches to three-dimensional reconstruction of plant shoot topology and geometry. Funct. Plant Biol. 44, 62–75.
Gibbs, J. A., Pound, M., French, A. P., Wells, D. M., Murchie, E., and Pridmore, T. (2018). Plant phenotyping: an active vision cell for three-dimensional plant shoot reconstruction. Plant Physiol. 178, 524–534. doi: 10.1104/pp.18.00664
Girshick, R. (2015). “Fast R-CNN,” in Proceedings of the IEEE International Conference on Computer Vision, Piscataway, NJ.
Goff, S. A., Vaughn, M., McKay, S., Lyons, E., Stapleton, A. E., Gessler, D., et al. (2011). The iPlant collaborative: cyberinfrastructure for plant biology. Front. Plant Sci. 2:34. doi: 10.3389/fpls.2011.00034
Golzarian, M. R., Frick, R. A., Rajendran, K., Berger, B., Roy, S., Tester, M., et al. (2011). Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 7:2. doi: 10.1186/1746-4811-7-2
Gómez-Candón, D., Virlet, N., Labbé, S., Jolivot, A., and Regnard, J. L. (2016). Field phenotyping of water stress at tree scale by UAV-sensed imagery: new insights for thermal acquisition and calibration. Precis. Agric. 17, 786–800. doi: 10.1007/s11119-016-9449-6
Gonzalez-Dugo, V., Hernandez, P., Solis, I., and Zarco-Tejada, P. J. (2015). Using high-resolution hyperspectral and thermal airborne imagery to assess p-physiological condition in the context of wheat phenotyping. Remote Sens. 7, 13586–13605. doi: 10.3390/rs71013586
Gonzalez-Dugo, V., Zarco-Tejada, P. J., and Fereres, E. (2014). Applicability and limitations of using the crop water stress index as an indicator of water deficits in citrus orchards. Agric. Forest Meteorol. 198, 94–104. doi: 10.1016/j.agrformet.2014.08.003
Gonzalez-Dugo, V., Zarco-Tejada, P., Nicolás, E., Nortes, P. A., Alarcón, J. J., Intrigliolo, D. S., et al. (2013). Using high resolution UAV thermal imagery to assess the variability in the water status of five fruit tree species within a commercial orchard. Precis. Agric. 14, 660–678. doi: 10.1007/s11119-013-9322-9
Granier, C., Aguirrezabal, L., Chenu, K., Cookson, S. J., Dauzat, M., Hamard, P., et al. (2006). PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 169, 623–635. doi: 10.1111/j.1469-8137.2005.01609.x
Guo, D., Juan, J., Chang, L., Zhang, J., and Huang, D. (2017). Discrimination of plant root zone water status in greenhouse production based on phenotyping and machine learning techniques. Sci. Rep. 7:8303. doi: 10.1038/s41598-017-08235-z
Hall, H. C., Fakhrzadeh, A., Luengo Hendriks, C. L., and Fischer, U. (2016). Precision automation of cell type classification and sub-cellular fluorescence quantification from laser scanning confocal images. Front. Plant Sci. 7:119. doi: 10.3389/fpls.2016.00119
He, J. Q., Harrison, R. J., and Li, B. (2017). A novel 3d imaging system for strawberry phenotyping. Plant Methods 13:8. doi: 10.1186/s13007-017-0243-x
He, K., Zhang, X., Ren, S., and Sun, J. (2015). Deep residual learning for image recognition. computer vision and pattern recognition (cs.CV). arXiv
Heckwolf, S., Heckwolf, M., Kaeppler, S. M., de Leon, N., and Spalding, E. P. (2015). Image analysis of anatomical traits in stem transections of maize and other grasses. Plant Methods 11:26. doi: 10.1186/s13007-015-0070-x
Herwitz, S. R., Johnson, L. F., Dunagan, S. E., Higgins, R. G., Sullivan, D. V., Zheng, J., et al. (2004). Imaging from an unmanned aerial vehicle: agricultural surveillance and decision support. Comp. Electron. Agric. 44, 49–61. doi: 10.1016/j.compag.2004.02.006
Houle, D., Govindaraju, D. R., and Omholt, S. (2010). Phenomics: the next challenge. Nat. Rev. Genet. 11, 855–866. doi: 10.1038/nrg2897
Hu, Y., Wang, L., Xiang, L., Wu, Q., and Jiang, H. (2018). Automatic non-destructive growth measurement of leafy vegetables based on kinect. Sensors 18:806. doi: 10.3390/s18030806
Huang, H., Wu, S. H., Cohen-Or, D., Gong, M. L., Zhang, H., Li, G., et al. (2013). L1-medial skeleton of point cloud. ACM Trans. Graph. 32:8.
Hui, F., Zhu, J., Hu, P., Meng, L., Zhu, B., Guo, Y., et al. (2018). Image-based dynamic quantification and high-accuracy 3d evaluation of canopy structure of plant populations. Ann. Bot. 121, 1079–1088. doi: 10.1093/aob/mcy016
Jannick, J. L., Lorenz, A. J., and Iwata, H. (2010). Genomic selection in plant breeding: from theory to practice. Brief. Funct. Genomics 9, 166–177. doi: 10.1093/bfgp/elq001
Jimenez-Berni, J. A., Deery, D. M., Rozas-Larraondo, P., Condon, A. T. G., Rebetzke, G. J., James, R. A., et al. (2018). High throughput determination of plant height, ground cover, and above-ground biomass in wheat with lidar. Front. Plant Sci. 9:237. doi: 10.3389/fpls.2018.00237
Johannsen, W. (2014). The genotype conception of heredity. Int. J. Epidemiol. 43, 989–1000. doi: 10.1093/ije/dyu063
Kirchgessner, N., Liebisch, F., Yu, K., Pfeifer, J., Friedli, M., Hund, A., et al. (2016). The ETH field phenotyping platform FIP: a cable-suspended multi-sensor system. Funct. Plant Biol. 44, 154–168.
Klukas, C., Chen, D. J., and Pape, J. M. (2014). Integrated analysis platform: an open-source information system for high-throughput plant phenotyping. Plant Physiol. 165, 506–518. doi: 10.1104/pp.113.233932
Kolukisaoglu, U., and Thurow, K. (2010). Future and frontiers of automated screening in plant sciences. Plant Sci. 178, 476–484. doi: 10.1016/j.plantsci.2010.03.006
Krajewski, P., Chen, D., Æwiek, H., van Dijk, A. D., Fiorani, F., Kersey, P., et al. (2015). Towards recommendations for metadata and data handling in plant phenotyping. J. Exp. Bot. 66, 5417–5427. doi: 10.1093/jxb/erv271
Kvilekval Das, K., Fedorov, D., Obara, B., Singh, A., and Manjunath, B. S. (2010). Bisque: a platform for bioimage analysis and management. Bioinformatics 26, 544–552. doi: 10.1093/bioinformatics/btp699
Lecun, Y. (1990). Handwritten digit recognition with a back-propagation network. Adv. Neural Inform. Process. Sys. 2, 396–404.
Lecun, Y., Bengio, Y., and Hinton, G. (2015). Deep learning. Nature 521:436. doi: 10.1038/nature14539
Legland, D., Devaux, M. F., and Guillon, F. (2014). Statistical mapping of maize bundle intensity at the stem scale using spatial normalisation of replicated images. PLoS One 9:e90673. doi: 10.1371/journal.pone.0090673
Legland, D., El-Hage, F., Méchin, V., and Reymond, M. (2017). Histological quantification of maize stem sections from FASGA-stained images. Plant Methods 13:84. doi: 10.1186/s13007-017-0225-z
Leister, D., Varotto, C., Pesaresi, P., Niwergall, A., and Salamini, F. (1999). vLarge-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol. Biochem. 37, 671–678. doi: 10.3389/fpls.2014.00770
Li, M., Frank, M., Coneva, V., Mio, W., Chitwood, D. H., and Topp, C. N. (2018). The persistent homology mathematical framework provides enhanced genotype-to-phenotype associations for plant morphology. Plant Physiol. 177, 1382–1395. doi: 10.1104/pp.18.00104
Li, Y.-F., Kennedy, G., Davies, F., and Hunter, J. (2010). PODD: an ontology-driven data repository for collaborative phenomics research. Lect. Notes Comput. Sci. 6102, 179–188. doi: 10.1007/978-3-642-13654-2_22
Li, Z., Chen, Z., Wang, L., Liu, J., and Zhou, Q. (2014). Area extraction of maize lodging based on remote sensing by small unmanned aerial vehicle. Trans. Chin. Soc. Agric. Eng. 30, 207–213.
Liang, Z., Pandey, P., Stoerger, V., Xu, Y., Qiu, Y., Ge, Y., et al. (2017). Conventional and hyperspectral time-series imaging of maize lines widely used in field trials. Gigascience 7, 1–11. doi: 10.1093/gigascience/gix117
Liebisch, F., Kirchgessner, N., Schneider, D., Walter, A., and Hund, A. (2015). Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 11:9. doi: 10.1186/s13007-015-0048-8
Lobos, G. A., Camargo, A. V., Del Pozo, A., Araus, J. L., Ortiz, R., and Doonan, J. H. (2017). Editorial: plant phenotyping and phenomics for plant breeding. Front. Plant Sci. 8:2181. doi: 10.3389/fpls.2017.02181
Lorenz, A. J., Chao, S., Asoro, F. G., Heffner, E. L., Hayashi, T., Iwata, H., et al. (2011). Genomic selection in plant breeding: knowledge and prospects. Adv. Agron. 110, 77–123.
Majewsky, V., Scherr, C., Schneider, C., Sebastian, P. A., and Stephan, B. (2017). Reproducibility of the effects of homeopathically potentised argentum nitricum on the growth of Lemna gibba L. in a randomised and blinded bioassay. Homeopathy. 106, 145–154. doi: 10.1016/j.homp.2017.04.001
Mathews, A. J., and Jensen, J. L. R. (2013). Visualizing and quantifying vineyard canopy LAI using an unmanned aerial vehicle (UAV) collected high density structure from motion point cloud. Remote Sens. 5, 2164–2183. doi: 10.3390/rs5052164
Meng, R., Saade, S., Kurtek, S., Berger, B., Brien, C., Pillen, K., et al. (2017). Growth curve registration for evaluating salinity tolerance in barley. Plant Methods 13:18. doi: 10.1186/s13007-017-0165-7
Mohanty, S. P., Hughes, D. P., and Salathe, M. (2016). Using deep learning for image-based plant disease detection. Front. Plant Sci. 7:1419. doi: 10.3389/fpls.2016.01419
Montes, J. M., Technow, F., Dhillon, B., Mauch, F., and Melchinger, A. (2011). High-throughput non-destructive biomass determination during early plant development in maize under field conditions. Field Crops Res. 121, 268–273. doi: 10.1016/j.fcr.2010.12.017
Muraya, M. M., Chu, J., Zhao, Y., Junker, A., Klukas, C., Reif, J. C., et al. (2017). Genetic variation of growth dynamics in maize (Zea mays L.). revealed through automated non-invasive phenotyping. Plant J. 89, 366–380. doi: 10.1111/tpj.13390
Nakhforoosh, A., Bodewein, T., Fiorani, F., and Bodner, G. (2016). Identification of water use strategies at early growth stages in durum wheat from shoot phenotyping and physiological measurements. Front. Plant Sci. 7:1155. doi: 10.3389/fpls.2016.01155
Neilson, E. H., Edwards, A. M., Blomstedt, C. K., Berger, B., Moller, B. L., and Gleadow, R. M. (2015). Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C-4 cereal crop plant to nitrogen and water deficiency over time. J. Exp. Bot. 66, 1817–1832. doi: 10.1093/jxb/eru526
Neumann, K., Zhao, Y., Chu, J., Keilwagen, J., Reif, J. C., Kilian, B., et al. (2017). Genetic architecture and temporal patterns of biomass accumulation in spring barley revealed by image analysis. BMC Plant Biol. 17:137. doi: 10.1186/s12870-017-1085-4
Nigon, T. J., Mulla, D. J., Rosen, C. J., Cohen, Y., Alchanatis, V., Knight, J., et al. (2015). Hyperspectral aerial imagery for detecting nitrogen stress in two potatocultivars. Comput. Electron. Agric. 112, 36–46. doi: 10.1016/j.compag.2014.12.018
Overgaard, S. I., Isaksson, T., Kvaal, K., and Korsaeth, A. (2010). Comparisons of two hand-held, multispectral field radiometers and a hyperspectral airborne imager in terms of predicting spring wheat grain yield and quality by means of powered partial least squares regression. J. Near Infrared Spectrosc. 18, 247–261. doi: 10.1255/jnirs.892
Pan, X., Ma, L., Zhang, Y., Wang, J., Du, J., and Guo, X. (2018). Reconstruction of maize roots and quantitative analysis of metaxylem vessels based on X-ray micro-computed tomography. Can. J. Plant Sci. 98, 457–466.
Pandey, P., Ge, Y., Stoerger, V., and Schnable, J. C. (2017). High throughput in vivo analysis of plant leaf chemical properties using hyperspectral imaging. Front. Plant Sci. 8:1348. doi: 10.3389/fpls.2017.01348
Pape, J. M., and Klukas, C. (2015). 3-D histogram-based segmentation and leaf detection for rosette plants. Asia Pac. Conf. Concept. Model. 43,107–114.
Parlati, A., Valkov, V. T., D’Apuzzo, E., Alves, L. M., Petrozza, A., Summerer, S., et al. (2017). Ectopic expression of PII induces stomatal closure in Lotus japonicus. Front. Plant Sci. 8:1299. doi: 10.3389/fpls.2017.01299
Pawara, P., Okafor, E., Surinta, O., Schomaker, L., and Wiering, M. (2017). “Comparing local descriptors and bags of visual words to deep convolutional neural networks for plant recognition,” in Proceedings of the International Conference on Pattern Recognition Applications and Methods, Lisbon.
Pereyra-Irujo, G. A., Gasco, E. D., Peirone, L. S., and Aguirrezábal, L. A. N. (2012). GlyPh: a low-cost platform for phenotyping plant growth and water use. Funct. Plant Biol. 39, 905–913.
Postma, J. A., Kuppe, C., Owen, M. R., Mellor, N., Griffiths, M., Bennett, M. J., et al. (2017). OpenSimRoot: widening the scope and application of root architectural models. New Phytol. 215, 1274–1286. doi: 10.1111/nph.14641
Potgieter, A. B., Watson, J., Eldridge, M., Laws, K., George-Jaeggli, B., Hunt, C., et al. (2018). “Determining crop growth dynamics in sorghum breeding trials through remote and proximal sensing technologies,” in Proceedings of the IGARSS 2018-2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia.
Ray, D. K., Mueller, N. D., West, P. C., and Foley, J. A. (2013). Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428. doi: 10.1371/journal.pone.0066428
Reuzeau, C. (2007). TraitMill (TM): a high throughput functional genomics platform for the phenotypic analysis of cereals. Vitro Cell. Dev. Biol. Anim. 43:S4.
Reuzeau, C., Pen, J., Frankard, V., de Wolf, J., Peerbolte, R., Broekaert, W., et al. (2005). TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Mol. Plant Breed. 1, 1–6.
Reuzeau, C., Pen, J., Frankard, V., de Wolf, J., Peerbolte, R., Broekaert, W., et al. (2006). TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Plant Genet. Res. 4, 20–24.
Reuzeau, C., Pen, J., Frankard, V., Wolf, J., Peerbolte, R., Broekaert, W., et al. (2010). TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Plant Gene Trait. 1, 1–6. doi: 10.5376/pgt.2010.01.0001
Rist, F., Herzog, K., Mack, J., Richter, R., Steinhage, V., and Töpfer, R. (2018). High-precision phenotyping of grape bunch architecture using fast 3d sensor and automation. Sensors 18:763. doi: 10.3390/s18030763
Roitsch, T., Cabrera-Bosquet, L., Fournier, A., Ghamkhar, K., Jiménez-Berni, J., Pinto, F., et al. (2019). Review: new sensors and data-driven approaches-A path to next generation phenomics. Plant Sci. 282, 2–10. doi: 10.1016/j.plantsci.2019.01.011
Rose, J. C., Paulus, S., and Kuhlmann, H. (2015). Accuracy analysis of a multi-view stereo approach for phenotyping of tomato plants at the organ level. Sensors 15, 9651–9665. doi: 10.3390/s150509651
Rutkoski, J., Poland, J., Mondal, S., Autrique, E., Pérez, L. G., Crossa, J., et al. (2016). Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat.Genes Genom. Genet. 6, 2799–2808. doi: 10.1534/g3.116.032888
Sadeghi-Tehran, P., Sabermanesh, K., Virlet, N., and Hawkesford, M. J. (2017). Automated method to determine two critical growth stages of wheat: heading and flowering. Front. Plant Sci. 8:252. doi: 10.3389/fpls.2017.00252
Salvi, S., and Tuberosa, R. (2015). The crop QTLome comes of age. Curr. Opin. Biotechnol. 32, 179–185. doi: 10.1016/j.copbio.2015.01.001
Schork, N. J. (1997). Genetics of complex disease: approaches, problem, and solutions. Am. J. Respir. Crit. Care Med. 156, S103–S109.
Seren, Ü, Grimm, D., Fitz, I., Weigel, D., Nordborg, M., Borgwardt, K., et al. (2017). AraPheno: a public database for Arabidopsis thaliana phenotypes. Nucleic Acids Res. 4, 1054–1059. doi: 10.1093/nar/gkw986
Silsbe, G. M., Oxborough, K., Suggett, D. J., Forster, R. M., Ihnken, S., Komárek, O., et al. (2015). Toward autonomous measurements of photosynthetic electron transport rates: an evaluation of active fluorescence-based measurements of photochemistry. Limnol. Oceanogr. Methods 13, 138–155.
Singh, A., Ganapathysubramanian, B., Singh, A. K., and Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21:110. doi: 10.1016/j.tplants.2015.10.015
Sugiura, R., Noguchi, N., and Ishii, K. (2005). Remote-sensing technology for vegetation monitoring using an unmanned helicopter. Biosys. Eng. 90, 369–379. doi: 10.1016/j.biosystemseng.2004.12.011
Swain, K. C., Thomson, S. J., and Jayasuriya, H. P. W. (2010). Adoption of an unmanned helicopter for low-altitude remote sensing to estimate yield and total biomass of a rice crop. Trans. Asabe. 53, 21–27. doi: 10.13031/2013.29493
Tardieu, F., Cabrera-Bosquet, L., Pridmore, T., and Bennett, M. (2017). Plant phenomics, from sensors to knowledge. Curr. Biol. 27, R770–R783. doi: 10.1016/j.cub.2017.05.055
Tardieu, F., and Tuberosa, R. (2010). Dissection and modelling of abiotic stress tolerance in plants. Curr. Opin. Plant Biol. 13, 206–212. doi: 10.1016/j.pbi.2009.12.012
Tattaris, M., Reynolds, M. P., and Chapman, S. C. (2016). A direct comparison of remote sensing approaches for high-throughput phenotyping in plant breeding. Front. Plant Sci. 7:1131. doi: 10.3389/fpls.2016.01131
Thapa, S., Zhu, F., Walia, H., Yu, H., and Ge, Y. (2018). A novel lidar-based instrument for high-throughput, 3d measurement of morphological traits in maize and sorghum. Sensors 18:1187. doi: 10.3390/s18041187
Tomé, F., Jansseune, K., Saey, B., Grundy, J., Vandenbroucke, K., Hannah, M. A., et al. (2017). rosettR: protocol and software for seedling area and growth analysis. Plant Methods. 13:13. doi: 10.1186/s13007-017-0163-9
Tsaftaris, S., and Noutsos, C. (2009). Plant Phenotyping with Low Cost Digital Cameras and Image Analytics. Berlin: Springer Press.
Tsaftaris, S. A., Minervini, M., and Scharr, H. (2016). Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 21, 989–991. doi: 10.1016/j.tplants.2016.10.002
Tuberosa, R. (2012). Phenotyping for drought tolerance of crops in the genomics era. Front. Physiol. 3:347. doi: 10.3389/fphys.2012.00347
Ubbens, J. R., and Stavness, I. (2017). Deep plant phenomics: a deep learning platform for complex plant phenotyping tasks. Front. Plant Sci. 8:1190. doi: 10.3389/fpls.2017.01190
Vadez, V., Kholová, J., Hummel, G., Zhokhavets, U., Gupta, S. K., and Hash, C. T. (2015). LeasyScan: a novel concept combining 3D imaging and lysimetry for high-throughput phenotyping of traits controlling plant water budget. J. Exp. Bot. 66, 5581–5593. doi: 10.1093/jxb/erv251
van de Velde, K., Chandler, P. M., van der Straeten, D., and Rohde, A. (2017). Differential coupling of gibberellin responses by Rht-B1c suppressor alleles and Rht-B1b in wheat highlights a unique role for the DELLA N-terminus in dormancy. J. Exp. Bot. 68, 443–455. doi: 10.1093/jxb/erw471
Vazquez-Arellano, M., Griepentrog, H. W., Reiser, D., and Paraforos, D. S. (2016). 3-d imaging systems for agricultural applications-a review. Sensors 16:24.
Virlet, N., Sabermanesh, K., Sadeghi-Tehran, P., and Hawkesford, M. J. (2016). Field scanalyzer: an automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 44, 143–153.
Vos, J., Evers, J. B., Buck-Sorlin, G. H., Andrieu, B., Chelle, M., and de Visser, P. H. (2010). Functional-structural plant modelling: a new versatile tool in crop science. J. Exp. Bot. 61, 2101–2115. doi: 10.1093/jxb/erp345
Vylder, J. D., Vandenbussche, F., Hu, Y., Philips, W., and Straeten, D. V. D. (2012). Rosette tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiol. 160:1149. doi: 10.1104/pp.112.202762
Wallace, L., Lucieer, A., Watson, C., and Turner, D. (2012). Development of a UAV-LiDAR system with application to forest inventory. Remote Sens. 4, 1519–1543. doi: 10.3390/rs4061519
Walter, A., Liebisch, F., and Hund, A. (2015). Plant phenotyping: from bean weighing to image analysis. Plant Methods 11:14. doi: 10.1186/s13007-015-0056-8
Wen, W., Guo, X., Wang, Y., Zhao, C., and Liao, W. (2017). Constructing a three-dimensional resource database of plants using measured in situ morphological data. Appl. Eng. Agric. 33, 747–756. doi: 10.13031/aea.12135
Wen, W., Li, B., Li, B.-J., and Guo, X. (2018). A leaf modeling and multi-scale remeshing method for visual computation via hierarchical parametric vein and margin representation. Front. Plant Sci. 9:783. doi: 10.3389/fpls.2018.00783
White, J. W., Andrade-Sanchez, P., Gore, M. A., Bronson, K. F., Coffelt, T. A., Conley, M. M., et al. (2012). Field-based phenomics for plant genetics research. Field Crops Res. 133, 101–112. doi: 10.1016/j.fcr.2012.04.003
Wilkinson, M. D., Dumonter, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., et al. (2016). The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3:160018. doi: 10.1038/sdata.2016.18
Wu, C. (2011). Visualsfm: A Visual Structure from Motion System. Available at: http://ccwu.me/vsfm/ (accessed June 11, 2014).
Wu, H., Jaeger, M., Wang, M., Li, B., and Zhang, B. G. (2011). Three-dimensional distribution of vessels, passage cells and lateral roots along the root axis of winter wheat (Triticum aestivum). Ann. Bot. 107, 843–853. doi: 10.1093/aob/mcr005
Xu, Y. (2016). Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 129, 653–673. doi: 10.1007/s00122-016-2691-5
Yang, G., Liu, J., Zhao, C., Li, Z., Huang, Y., Yu, H., et al. (2017). Unmanned aerial vehicle remote sensing for field-based crop phenotyping: current status and perspectives. Front. Plant Sci. 8:1111. doi: 10.3389/fpls.2017.01111
Yang, W., Guo, Z., Huang, C., Duan, L., Chen, G., Jiang, N., et al. (2014). Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 5:5087. doi: 10.1038/ncomms6087
Yang, W., Guo, Z., Huang, C., Wang, K., Jiang, N., Feng, H., et al. (2015). Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J. Exp. Bot. 66, 5605–5615. doi: 10.1093/jxb/erv100
Yin, K. X., Huang, H., Long, P. X., Gaissinski, A., Gong, M. L., Sharf, A., et al. (2016). Full 3d plant reconstruction via intrusive acquisition. Comput. Graph. Forum 35, 272–284. doi: 10.1111/cgf.12724
Zarco-Tejada, P. J., Diaz-Varela, R., Angileri, V., and Loudjani, P. (2014). Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. Eur. J. Agron. 55, 89–99. doi: 10.1016/j.eja.2014.01.004
Zhang, C. H., and Kovacs, J. M. (2012). The application of small unmanned aerial systems for precision agriculture: a review. Precis. Agric. 13, 693–712. doi: 10.1007/s11119-012-9274-5
Zhang, X., Huang, C., Wu, D., Qiao, F., Li, W., Duan, L., et al. (2017). High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize plant growth. Plant Physiol. 173, 1554–1564. doi: 10.1104/pp.16.01516
Zhang, Y., Legay, S., Barrière, Y., Méchin, V., and Legland, D. (2013). Color quantification of stained maize stem section describes lignin spatial distribution within the whole stem. J. Sci. Food Agric. 61, 3186–3192. doi: 10.1021/jf400912s
Zhang, Y., Ma, L., Pan, X., Wang, J., Guo, X., and Du, J. (2018). Micron-scale phenotyping techniques of maize vascular bundles based on X-ray microcomputed tomography. J. Vis. Exp. 140:e58501. doi: 10.3791/58501
Keywords: crop phenomics, phenotyping extraction, data storage, functional–structural plant modeling, phenotype–genotype association analysis
Citation: Zhao C, Zhang Y, Du J, Guo X, Wen W, Gu S, Wang J and Fan J (2019) Crop Phenomics: Current Status and Perspectives. Front. Plant Sci. 10:714. doi: 10.3389/fpls.2019.00714
Received: 30 October 2018; Accepted: 14 May 2019;
Published: 03 June 2019.
Edited by:
Jose Antonio Jimenez-Berni, Spanish National Research Council (CSIC), SpainReviewed by:
Young-Su Seo, Pusan National University, South KoreaCopyright © 2019 Zhao, Zhang, Du, Guo, Wen, Gu, Wang and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunjiang Zhao, emhhb2NqQG5lcmNpdGEub3JnLmNu
†Co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.