
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 June 2019
Sec. Plant Nutrition
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00700
This article is part of the Research Topic Iron Nutrition and Interactions in Plants View all 28 articles
Ferric reductase oxidase (FRO), the enzyme that reduced ferric iron [Fe (III)] into ferrous iron [Fe (II)], is known to play important roles in Fe absorption and homeostasis in plants that utilize a strategy I mechanism to obtain iron. Rice can use both strategies I and II for Fe uptake depending on the growth conditions. FRO is encoded by two genes in rice genome. Amino acid sequence alignment shows that OsFRO1 contains all necessary predicted motifs for a functional FRO enzyme, whereas OsFRO2 lacks a complete transmembrane domain at the N-terminal. Transient expression of OsFRO1: GFP protein fusion revealed that OsFRO1 is localized to the vacuolar membrane in rice protoplast. OsFRO1 is primarily expressed in leaves and transcript abundance was decreased under excess Fe conditions. Transgenic plants overexpressing OsFRO1 were more sensitive to Fe toxicity, in contrast RNA interference lines showed more tolerance to Fe excess stress. Furthermore, RNAi lines showed decreased Fe concentrations compared to wild type plants under Fe excess condition. Together these data show that OsFRO1 is involved in reducing ferric Fe into ferrous Fe in the vacuole, and makes the vacuolar stored Fe available to the cytoplasm through Fe (II) or chelated Fe (II) transporters. Under Fe excess condition, the downregulation of OsFRO1 in the RNAi plants reduced the amount of Fe (II) available for cytoplasm, to alleviate Fe excess toxicity. This indicates that OsFRO1 plays an important role to maintain Fe homeostasis between the cytoplasm and vacuole in rice.
Iron (Fe) is an essential micronutrient for most living organisms. The transition between two oxidation states, the Fe (II) and Fe (III), makes it an indispensable element for multiple biological processes for plant growth (Connorton et al., 2017). Fe participates in various metabolic pathways including photosynthesis, respiration, chlorophyll biosynthesis, and nitrogen fixation (Kobayashi and Nishizawa, 2012; Briat et al., 2015). The redox properties also allow Fe to function as the catalytic component, in the form of heme or Fe–S clusters, in a wide variety of proteins (Guerinot and Yi, 1994).
Although abundant in the Earth’s crust Fe bioavailability is strongly restricted by the insolubility of ferric oxide present in well-aerated or alkaline soils, which is not easily accessible for uptake by plants (Mori, 1999). Fe deficiency retards plant growth and reduces crop yields, but excess Fe also cause toxicity to the plants (Halliwell and Gutteridge, 1992). Plants have two distinct Fe acquisition strategies to obtain Fe from the soil (Römheld and Marschner, 1986; Connorton et al., 2017). Non-grass plants, such as the model plant Arabidopsis thaliana, utilize strategy I, which is also known as the reducing strategy. When exposed to Fe-limiting conditions, plants secrete protons and phenolic compounds into the rhizosphere that lower the pH and enhance Fe (III) solubility (Santi and Schmidt, 2009). Fe (III) is reduced to Fe (II) by Ferric Reduction Oxidase 2 (FRO2) localized on the plasma membrane (Robinson et al., 1999). Afterward, Fe (II) is transported across the plasma membrane into the root epidermal cells by the iron-regulated transporter 1 (IRT1) (Eide et al., 1996; Varotto et al., 2002; Vert et al., 2002). Grass plants, such as barley and maize, utilize strategy II which is also called chelating strategy to take up Fe from the soil. In strategy II plants phytosiderophores (PS) are released into the rhizosphere (Kobayashi et al., 2010). After binding Fe (III) in the soil, Fe (III)-PS complexes are taken up by specific transporters belonging to the yellow stripe (YS) or YS like (YSL) family of proteins (Curie et al., 2001; Inoue et al., 2009). Rice has functional IRT1 and IRT2 that allow it to take up Fe (II) directly under submerge conditions (Ishimaru et al., 2006). In addition, rice can secrete phenolic compounds to chelate Fe (III) and solubilize apoplasmic Fe (Bashir et al., 2011; Ishimaru et al., 2011). Thus, rice is considered to use a mixed model for Fe uptake.
Under anaerobic conditions and low pH in flooded soils Fe is present mainly as soluble Fe (II) due to the low redox potential (Becker and Asch, 2005). Fe (II) can be taken up excessively by plant roots, resulting in cellular Fe overloaded, which can be toxic for plants. Fe toxicity has become a widespread nutrient stress affecting the growth of wetland rice, especially in Asia and West Africa (Sahrawat, 2004; Audebert and Fofana, 2009). In plant cells excess amount of Fe (II) contributes to the formation of hydroxyl radicals (⋅OH) and other reactive oxygen species (ROS) through the Fenton reaction (Fenton, 1984; Becana et al., 1998). These hydroxyl radicals can react with lipids, DNA and proteins, causing irreversible damage and oxidative stress (Briat et al., 1995; Fang et al., 2001). In rice the most obvious symptoms of Fe-overloading stress are leaf bronzing (Asch et al., 2005; Becker and Asch, 2005). Moreover, excess Fe in the soil can damage the root uptake system and adversely affect the acquisition of other nutrients, such as phosphorus, zinc and magnesium, leading to reduced growth and yield loss and even death of plants (Sahrawat, 2004). Thus, the cellular Fe level must be tightly controlled to maintain Fe homeostasis in plants.
The mechanisms for Fe excess tolerance can be divided into three aspects: (1) Avoiding excess Fe accumulation – Fe deposits at the root surface as the Fe plaque and forms a physical barrier to exclude excess Fe uptake (Deng et al., 2010). Reduced Fe translocation from roots to shoots could be important to prevent oxidative stress in leaves. (2) Storing high Fe levels – Fe partitioning into different sub-cellular compartments, mainly apoplast and vacuoles, are crucial to alleviate Fe toxicity (Moore et al., 2014). Ferritins can store up to 4,000 Fe atoms and are considered essential for tolerance to excess Fe (Briat et al., 2010). (3) Scavenging of ROS by antioxidants – Plants may also utilize antioxidants such as ascorbate, glutathione, phenolics or antioxidant enzymes such as superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (CAT) to detoxify oxidative molecules (Fang et al., 2001; Wu et al., 2017).
A significantly amount of work has been done on Fe-deficiency in plants. In contrast, the molecular aspects of Fe excess responses remain largely unknown. Several quantitative trait loci (QTL) co-localized on chromosome 1 were reported to be associated with tolerance to Fe excess stress in rice (Wu et al., 2014; Dufey et al., 2015). These results were also confirmed by genome-wide association study (GWAS) (Matthus et al., 2015). A transcriptomic study compared the expression profiling of rice seedlings after short- and long-term exposure to Fe excess (Quinet et al., 2012). It was found that more genes were up- or down-regulated after 3 days than after 3 weeks of Fe excess treatment. Transcriptomic analysis of rice in response to Fe deficiency and excess revealed that there is crosstalk between the two treatments and small open reading frames might play an important role in the response of plants to Fe deficiency and excess (Bashir et al., 2014). OsWRKY80 is the first reported transcription factor induced by Fe excess in plants (Ricachenevsky et al., 2010). However, the biological function of this gene is still under investigation.
Ferric reductase oxidase (FRO), the enzyme to reduce Fe (III) into Fe (II), is known to play important roles in Fe absorption and homeostasis in strategy I plants. There are two FRO genes in rice genome, but their function has not yet been characterized. In this study, the predicted amino acid sequence alignment shows that OsFRO1 contains all necessary motifs for a functional FRO enzyme. OsFRO1 is localized in the vacuolar membrane, which is different from other plant FRO proteins previously reported. Under Fe excess conditions, the transcript abundance of OsFRO1 was decreased in abundance. Knockdown of OsFRO1 by RNAi alleviated Fe toxicity in transgenic rice. This work indicates that OsFRO1 plays an important role to maintain Fe homeostasis in rice.
The rice cultivar (Oryza sativa L. ssp. japonica cv. Dongjin) was used for physiological experiments and rice transformations. Seeds were germinated in water for 3 days. After germination uniform seedlings were transferred to nutrient solution containing 1.425 mM NH4NO3, 0.323 mM NaH2PO4, 0.513 mM K2SO4, 0.998 mM CaCl2, 1.643 mM MgSO4, 0.009 mM MnCl2, 0.075 mM (NH4)6Mo7O24, 0.019 mM H3BO3, 0.155 mM CuSO4, 0.152 mM ZnSO4, 125 μM Fe (II)-EDTA, with pH 5.5 (Yoshida et al., 1976). For gene expression assay, 2-week old seedlings were transferred to normal, Fe deficient or excess nutrient solutions. The solution contains 0, 0.125 mM Fe or 1 mM Fe (II)-EDTA, respectively. Plants were harvested at 7 days after Fe treatment. For phenotypic analysis, 2-week-old seedlings were grown in nutrient solution with 7 or 0.125 mM Fe (II)-EDTA for 3 days.
All the experiments were carried out in green house with a day/night temperature of 30°C/28°C and a 12 h photoperiod. The nutrient solution was changed every 3 days.
To investigate the subcellular localization of OsFRO1 and OsFRO2, the coding sequences of OsFRO1 and OsFRO2 without a stop codon was amplified and cloned into pDEST/CGFP using the gateway system. Rice protoplasts were isolated from 2-week-old seedlings using cellulose R10 and macerozyme R10 as previously described (Ying et al., 2017). Two hundred micro liter of protoplast suspension was transformed with 8∼10 μg plasmid DNA using the PEG (polyethylene glycol)-mediated transformation method (Chen et al., 2011). After incubation at 25°C in the dark for 12 to 14 h, fluorescence images were captured using confocal microscopy.
For over-expression the full-length cDNA of OsFRO1 was cloned into entry vector pENTR-D-TOPO. After sequencing the LR recombination reaction was performed between an attL-containing entry clone and an attR-containing destination vector pEarlyGate 303 to generate the overexpression (OsFRO1-Oe) vector. For RNA-interference vector, a 300 bp fragment from the OsFRO1 coding region was amplified and subcloned into entry vector pDONR201 via BR reaction. Then the fragment was inserted into the destination vector pH7GWIWG2(I) in both a sense and anti-sense orientation to get the interference (OsFRO1-Ri) vector (Karimi et al., 2002). For expression pattern analysis, the 1800 bp upstream of the ATG start codon of OsFRO1 was amplified and cloned into pBI101.3 to generate the POsFRO1: GUS vector. All the corresponding primers are listed in Supplementary Table S1.
The OsFRO1-Oe, OsFRO1-Ri and POsFRO1: GUS vectors were transformed into Agrobacterium strain EHA101 or EHA105. Transgenic rice plants for the three constructs were regenerated via Agrobacterium-mediated transformation as described (Chen et al., 2003).
T1 transgenic seeds were germinated and grown in nutrient solution with 125 μM Fe (II)-EDTA. Tissues were immersed immediately in staining solution with 1 mM X-Gluc (5-bromo-4-chloro-3-indolyl-b-D-glucuronidase) overnight at 37°C. Afterward, roots were imbedded in 3% (w/v) agarose (Biowest, Spain). Samples were sectioned to a thickness of 30 μm by vibrating microtome (VT 1000 S, Leica, Bensheim, Germany). The Spur’s resin was used to imbed leaves. Sections of 10 μm were cut and the images were observed under a microscope (Eclipse 90i, Nikon, Tokyo, Japan).
For assaying Fe (III) reduction activity of OsFRO1 and OsFRO2, the full-length cDNA of OsFRO1, OsFRO2 and AtFRO2 were subcloned into pYES2.0 vector to generate pYES2.0-OsFRO1, pYES2.0-OsFRO2 and pYES2.0-AtFRO2, respectively. Constructs were transformed into S. cerevisiae wild-type strain BJ2168 using the lithium acetate method according to the manufacturer’s manual. Yeast transformants were grown following the protocol described previously (Li et al., 2004). Cells were harvested in the mid-log phase, and Fe (III) chelate reductase activity was quantified based on the absorbance at 562 nm (Yi and Guerinot, 1996).
Total RNA was extracted from the roots and shoots using TRIzol reagent (Invitrogen, Shanghai, China), according to the manufacturer’s instructions. cDNA was synthesized from 2 μg of total RNA using the PrimeScriptTM RT reagent Kit (Takara Bio, Inc., Dalian, China). qRT-PCR were conducted using LightCycler 480 SYBR Green I Master Kit (Roche Diagnostics) on a LightCycler480 machine. The relative expression levels were normalized to the housekeeping gene OsACTIN1 by the formula 2−ΔΔCt. The primers for qRT-PCR analysis are given in Supplementary Table S1.
To determine the metal concentration in plants, root or shoot samples were dried for 3 days at 80°C. About 0.1 g dry weight was used for digestion. Metal concentration analysis was performed using inductively coupled plasma spectroscopy (ICP-MS, Agilent 7500ce, Santa Clara, CA, United States).
In situ detection of superoxide (O2−) in leaves was conducted using NBT staining as described (Höller et al., 2014). In brief, the expanded leaves were excised and immersed in staining solution containing 0.1% (w/v) NBT, 10 mM sodium azide, 50 mM potassium phosphate (pH 6.4). Vacuum infiltrate the leaves for 30 min until they were completely infiltrated. Afterward, incubate the leaves in 10 mL of staining solution (0.1% NBT) for 15 min. ROS formation was visualized by a scanner (CanoScan 9000F MarkII, Tokyo, Japan).
In Arabidopsis thaliana there are predicted to be eight FRO genes in the genome based on high sequence similarity with AtFRO2 (Ling et al., 2005). In order to identify FRO genes of rice, BLAST searches against the rice genome database using the amino acid sequences of AtFROs were performed. Two putative proteins, named OsFRO1 (LOC_Os04g36720) and OsFRO2 (LOC_Os04g48930) were found as ortyhologs of AtFROs. Phylogenic analysis of FROs in higher plants showed that OsFRO1 displayed higher similarity to AtFRO7, while OsFRO2 clustered together with three characterized FROs, PsFRO1 (Waters et al., 2002), LeFRO1 (Li et al., 2004), CsFRO1 (Waters et al., 2007) in a small group (Figure 1A). OsFRO1 and OsFRO2 have the highest similarity with AtFRO6 (60.9%) and AtFRO2 (58.5%) among the Arabidopsis FROs, respectively (Figure 1B).
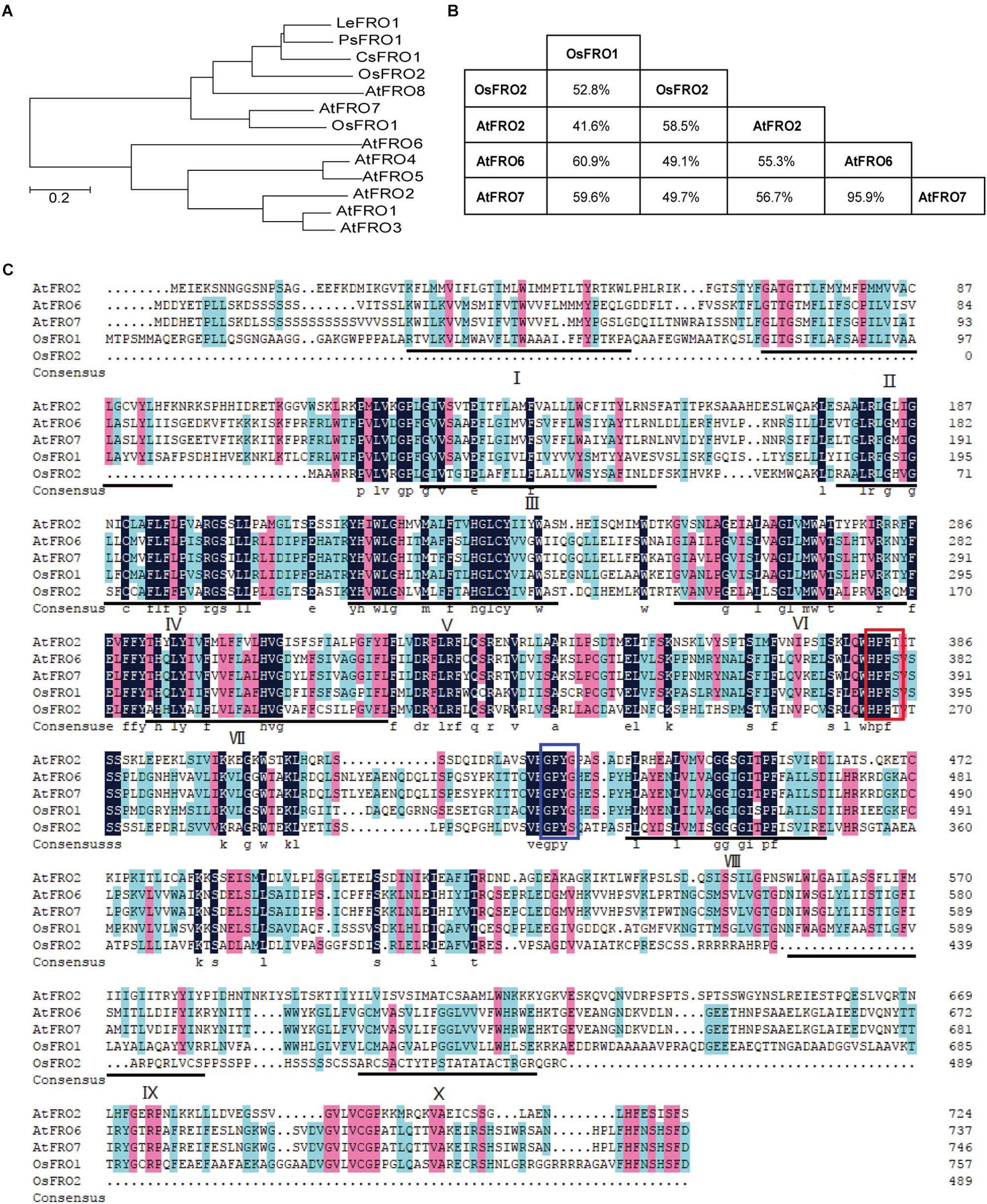
Figure 1. Phylogenetic analysis and alignments of FROs in plants. (A) Phylogenetic analysis of FRO proteins in plants. Amino acid sequences were used and the evolutionary history was conducted in MEGA6 via neighbor-joining method. (B) The similarity matrix for FRO proteins from rice and Arabidopsis. (C) Amino acids sequences alignment of FRO proteins from rice and Arabidopsis. The FAD and NADPH binding site were highlighted by red and blue boxes, respectively. The locations of the 10 potential transmembrane domains were underlined and numbered I-X.
Plant FROs were identified based on its sequence similarity to human phagocytic NADPH oxidase gp91phox and yeast ferric-chelate reductases such as FREs (Dancis et al., 1990; Babior, 1995). Similar to FREs and gp91phox, AtFRO2 contains the two conserved motifs HPFT and GPYG that are associated with FAD and NADPH binding sites of the cofactor (Robinson et al., 1999). Amino acid sequence alignment of OsFROs with the AtFROs revealed that both OsFRO1 and OsFRO2 contained the two conservative motifs. The FAD binding site of OsFRO1 shared 100% identity with AtFRO6/7 (HPFS), whereas the FAD motif of OsFRO2 is “HPFT,” identical to AtFRO2. The NADPH binding site of OsFRO1 is GPYS, identical to the other FROs, whereas that of OsFRO2 is GPYT, different from the functional Arabidopsis FROs analyzed (Figure 1C).
The THTMM program was used to predict the membrane topology of two OsFRO proteins, resulting in predictions of 10 transmembrane domains in OsFRO1 and 6 transmembrane domains in OsFRO2 (Supplementary Figure S1). OsFRO2 lacks complete transmembrane domains in its N-terminal and might not be functional. Thus, the study is focused on OsFRO1 for further investigation.
To determine the subcellular localization of OsFRO1 and OsFRO2 GFP was fused to the C terminus of OsFRO1 and OsFRO2 coding regions. The GFP-fusion constructs were transformed into rice protoplasts isolated from stems and leaves of etiolated seedlings. The green fluorescence signal of OsFRO1 was detected in the vacuolar membrane, whereas that of OsFRO2 located mainly in the cytoplasm (Figure 2). To further confirm the subcellular localization of OsFRO1, a vacuolar membrane marker, OsSPX-MFS3 (Wang et al., 2015), was co-expressed with OsFRO1 in rice protoplasts. The GFP signal mostly overlapped with the mCherry signal of the vacuolar membrane marker (Supplementary Figure S2). Noticeably, the subcellular localization of OsFRO1 was completely different with that of other FROs in plants, since it is the only FRO family member localized to tonoplast to our knowledge. This result indicated that OsFRO1 may play a novel role in Fe homeostasis.
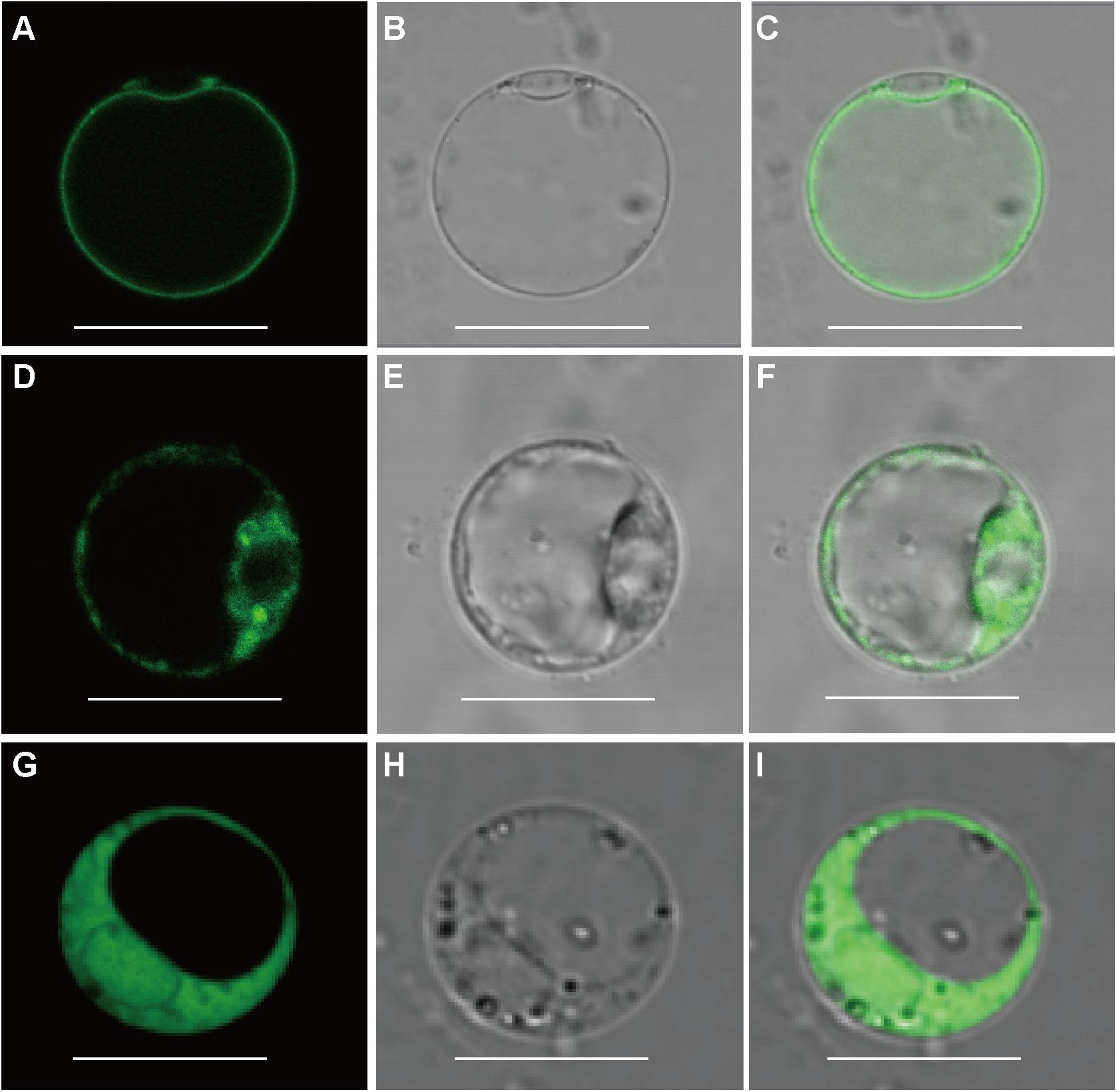
Figure 2. Subcellular localization of OsFRO1 and OsFRO2. (A) OsFRO1-GFP expression. (B) Bright field image of a protoplast transformed with OsFRO1-GFP. (C) Overlay image of (A,B). (D) OsFRO2-GFP expression. (E) Bright field image of a protoplast transformed with OsFRO2-GFP. (F) Overlay image of (D,E). (G) 35s-GFP expression. (H) Bright field image of a protoplast transformed with 35s-GFP. (I) Overlay image of (G,H). Bars = 20 μm.
To investigate the spatial expression of OsFRO1, transgenic plants carrying the promoter region of OsFRO1 fused to the GUS gene (POsFRO1: GUS) were generated. GUS expression was observed in all tissues, including roots, stems, leaves, flowers and seeds (Figure 3A). Cross-sections of leaves showed that no staining was observed in bulliform cells and part of epidermal cells (Figure 3B). In roots, GUS expression was mainly observed in the mature roots, but not at the young roots. Little staining was detected in root tips (Figure 3C). Cross-sections of roots exhibited that GUS expression was observed throughout root except the epidermal cells (Figure 3D). In the floral organs, GUS was expressed in stigma and pollen (Figures 3E–G). In mature seeds, GUS expression was detected in the glumes (Figure 3H). One day after germination, GUS expression was also detected in the endosperm (Figures 3I,J).
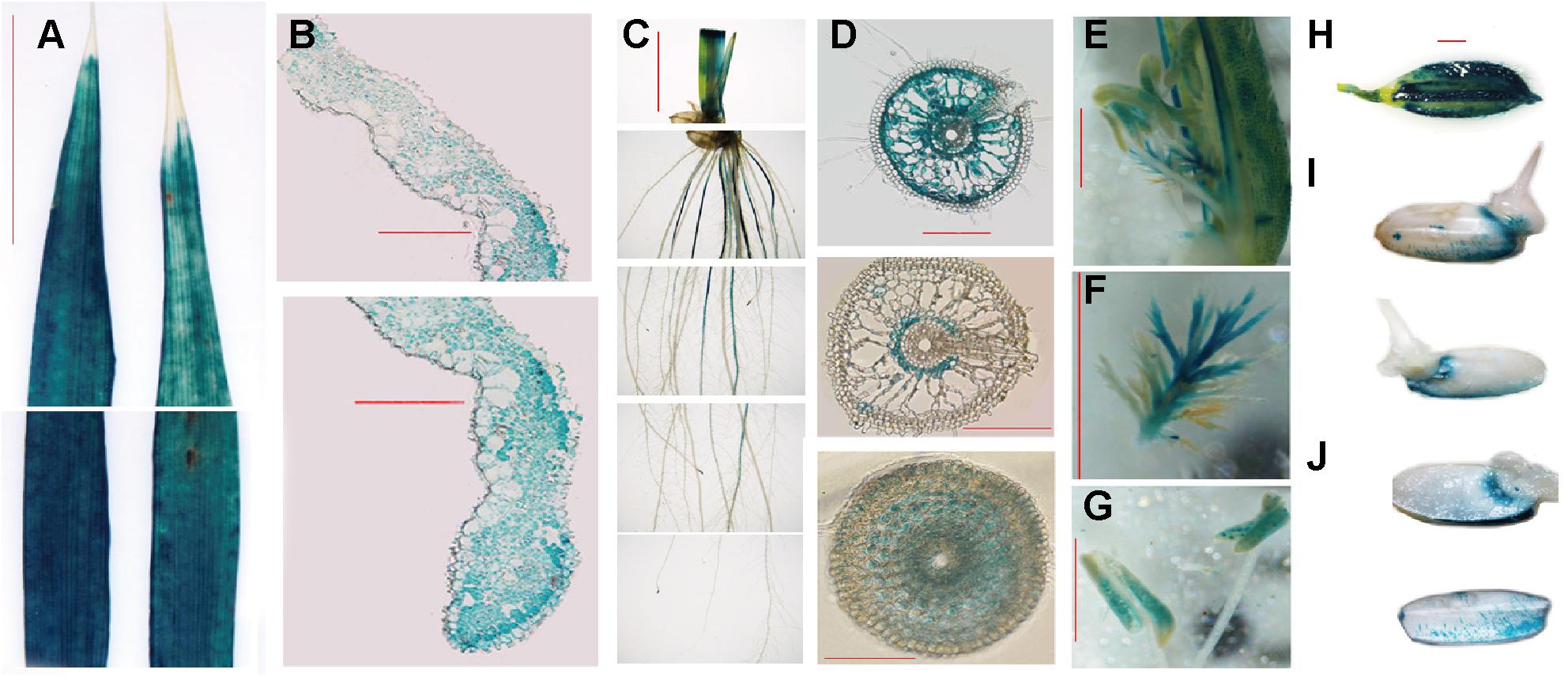
Figure 3. OsFRO1 promoter-driven GUS expression pattern. (A) Leaves from 2-week-old seedlings. (B) Cross-sections of (A). (C) Roots from -week-old seedling. (D) Cross-sections of (C). Images of the sections that were close to the root–stem junction (upper panel), 5 cm away from the root tip (middle panel) and 0.5 cm away from the root tip (lower panel). (E) GUS staining of the mature flowers. (F) Pistil of (E). (G) Stamen of (E). (H) Seeds before germination. (I) Seeds geminated after 1 day. (J) Cross-sections of (I). Bar = 1 cm (A,C), Bar = 100 μm (BD), Bar = 1 mm (E–J).
To gain further insight into the expression pattern of OsFRO1 the transcript abundance of OsFRO1 was analyzed in leaves and roots. Results showed that the basal level of OsFRO1 in leaves was 1000 times as high as that in roots (Figure 4A). Similar spatial expression pattern can be found from the microarray data retrieved from GENEVESTIGATOR (Supplementary Figure S3). While the mRNA abundance of OsFRO1 was not affected by Fe deficiency, it is greatly inhibited when plants exposed to solution culture containing excess Fe (Figure 4B). In addition, GUS staining of the PFRO1: GUS transgenic plants showed that excess Fe supply significantly suppressed the GUS activity in old leaves, but not in the young leaves (Figure 4C).
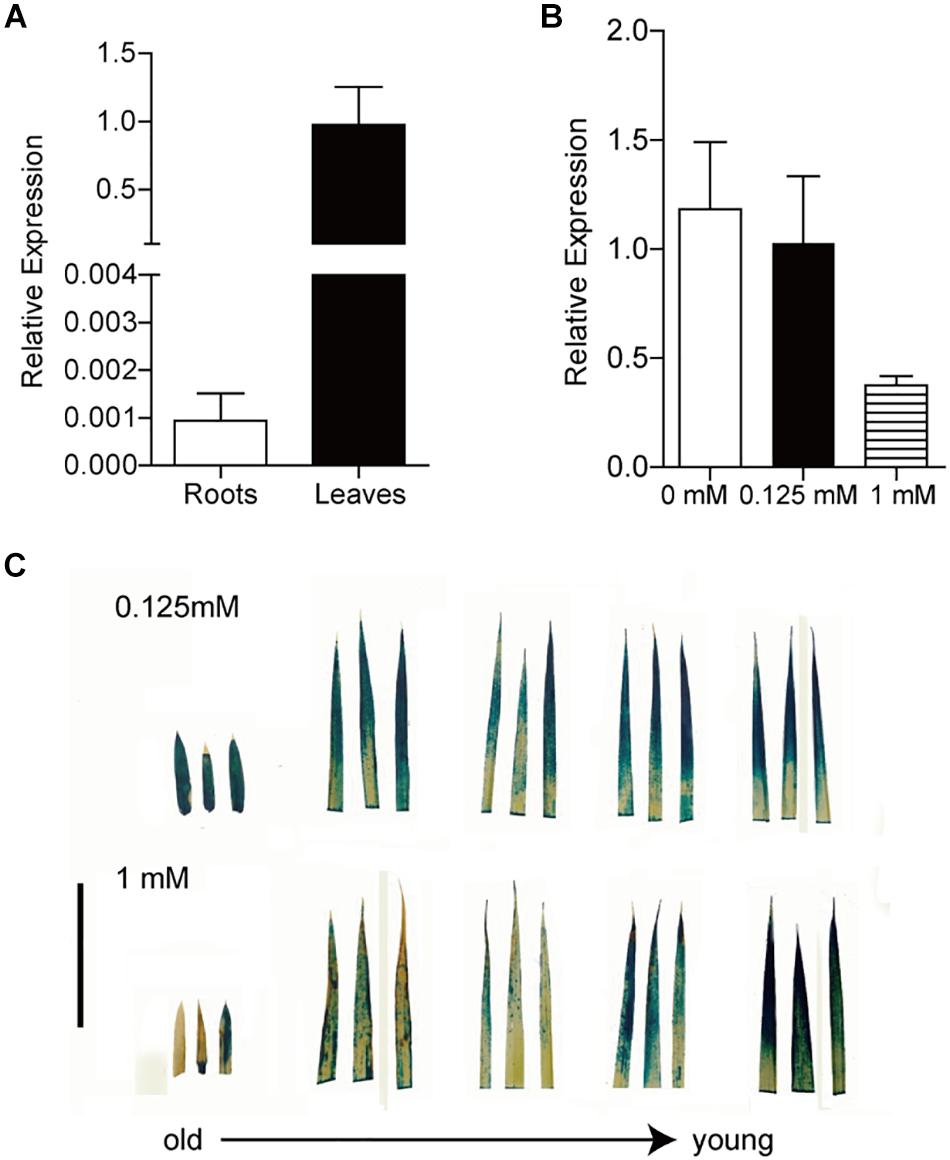
Figure 4. Expression analysis of OsFRO1 transcript abundance in Rice. (A) Transcript abundance of OsFRO1 in leaves and roots (the expression in leaves was set to 1.0). Seedlings were grown in normal nutrient solution for 3 weeks. Leaves and roots were sampled for RNA extraction. (B) Transcript levels of OsFRO1 under different Fe supply. Two-week-old seedlings cultured in normal nutrient solution (0.125 mM Fe) were transferred to 0 or 1 mM Fe solution for 7 days. Leaves were sampled for RNA extraction. (C) GUS staining of POsFRO1: GUS transgenic seedlings under 1 mM Fe treatment. Bar = 3 cm. Data are shown as the mean ± SD (n = 3).
To determine the biological function of OsFRO1 in Fe homeostasis, OsFRO1 overexpression (Oe) or the RNA interference (Ri) transgenic lines were generated and the transcript abundance of OsFRO1 in the Oe and Ri plants was determined by quantitative real-time PCR (qRT-PCR) analyses. The OsFRO1 transcript abundance were 10–40 times higher in the Oe plants, or 2–5 times lower in the Ri plants, compared to that in the wild type (WT) plants (Supplementary Figure S4). Two independent transgenic lines of each construct were selected for further experimental analysis, i.e., Oe-1, Oe-3, Ri-1, and Ri-3.
To evaluate the effect of OsFRO1 on Fe homeostasis, the transgenic plants were grown under Fe deficiency or Fe excess conditions. Under normal or Fe deficient conditions, there was no significant difference in growth performance and Fe concentrations between the transgenic plants and WT control (Supplementary Figure S5). When treated with excess Fe for 3 days, all the plants showed bronzing in the leaves, and Fe plaques in roots, which are common symptoms of Fe overaccumulation. The 4th and 5th leaves of the WT and transgenic plants were detached and compared. As shown in Figure 5A, OsFRO1-Ri lines displayed less bronzing symptom in the leaves compared to WT and Oe plants. No significant differences in plant length or fresh weight were observed between the transgenic plants and WT control (Figure 5B).
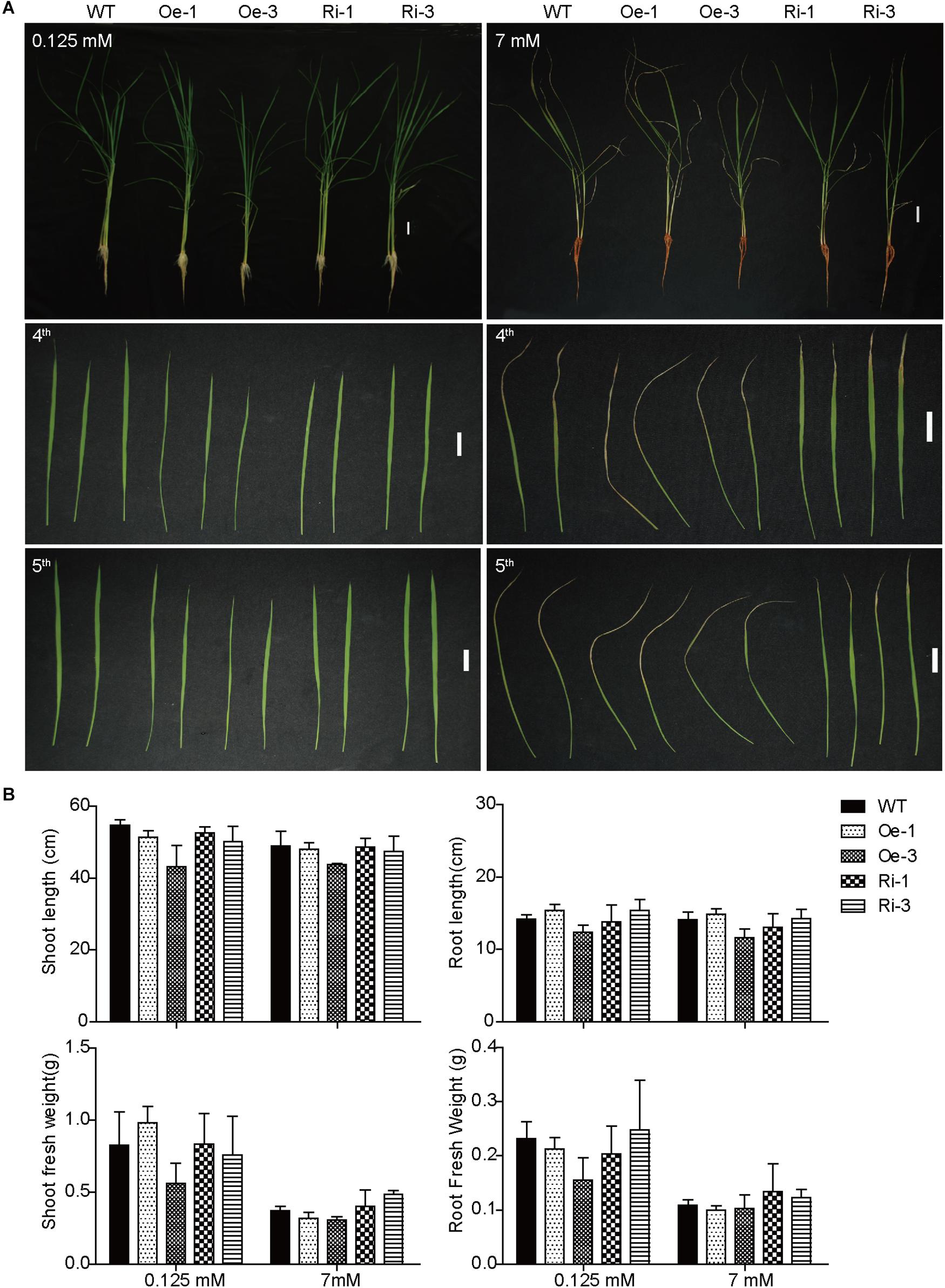
Figure 5. Phenotype analysis of WT and transgenic plants under treatment of excess Fe. (A) Growth performance of WT and transgenic plants under normal and Fe excess conditions. Two-week-old plants were grown on nutrient solution supplied with 0.125 or 7 mM Fe (II)-EDTA for 3 days. The fourth and fifth leaves were detached to display their phenotypes. (B) Plant length and biomass of WT and transgenic plants under normal and HFe conditions. Bar = 2 cm. Data are shown as the mean and standard deviation (n = 3).
No significant difference was found in leaf and root Fe concentrations among the WT, Ri, and Oe plants when grown in normal Fe supply condition (Figure 6A). Under excess Fe supply condition, the leaf Fe concentrations of the Ri-1 and Ri-3 plants were significantly (p < 0.05) or marginally (p < 0.1) lower than WT (Figure 6A). In addition, the root Fe concentrations of the Ri-1 and Ri-3 plants were significantly lower than that of WT (Figure 6B). There was no significant difference in the Zn, Mn, Cu concentrations among the leaves and roots of OsFRO1-Oe and Ri lines compared to WT (Supplementary Figure S6).
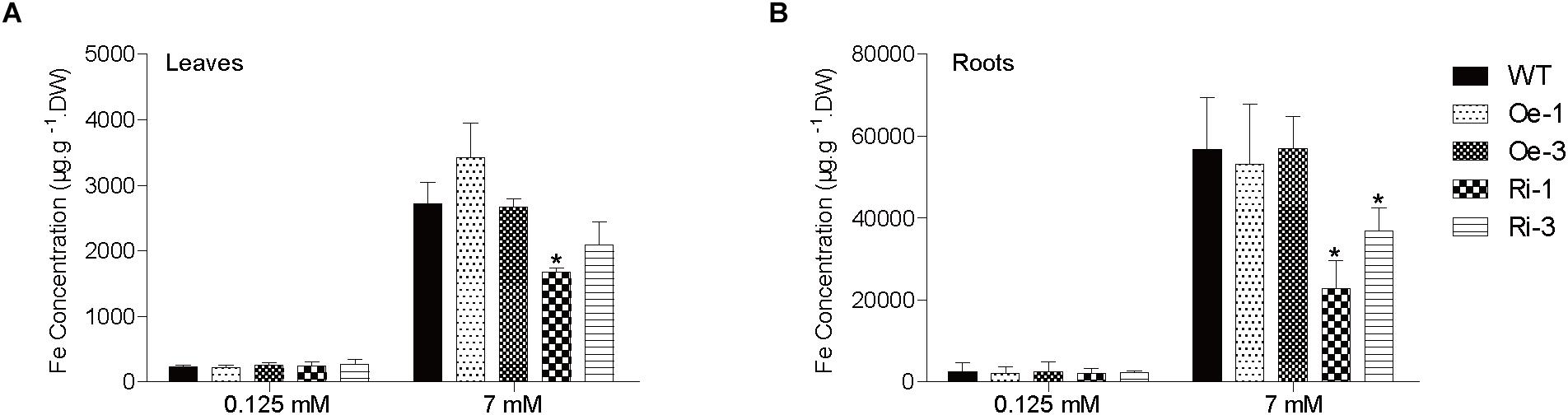
Figure 6. Analysis of Fe concentrations in WT and transgenic plants. Two-week-old plants were grown on nutrient solution supplied with 0.125 or 7 mM Fe (II)-EDTA for 3 days. Fe concentrations in leaves (A) and roots (B) were measured by ICP-OES. Data are shown as the mean and standard deviation (n = 3). Significance of differences compared to WT is indicated by asterisks (Tukey’s ANOVA test; ∗p < 0.05).
Excess Fe resulted an oxidative stress in plants through the formation of ROS by the Fenton reaction (Becana et al., 1998). In order to evaluate the Fe-induced oxidative stress, superoxide (⋅O2−) contents representing the major ROS was estimated using NBT staining. Under Fe excess conditions, all the plants showed typical symptoms of Fe overaccumulation (Figure 7A). While the excess Fe supply resulted in significant accumulation of ROS in leaves of WT, Oe and Ri plants, the NBT staining in the OsFRO1-Oe leaves was at the highest level (Figure 7B). As contrast, the blue staining in Ri leaves were significantly lower than that in WT and Oe leaves. This NBT staining result is in accordance with the bronzing phenotypes described (Figure 5A).
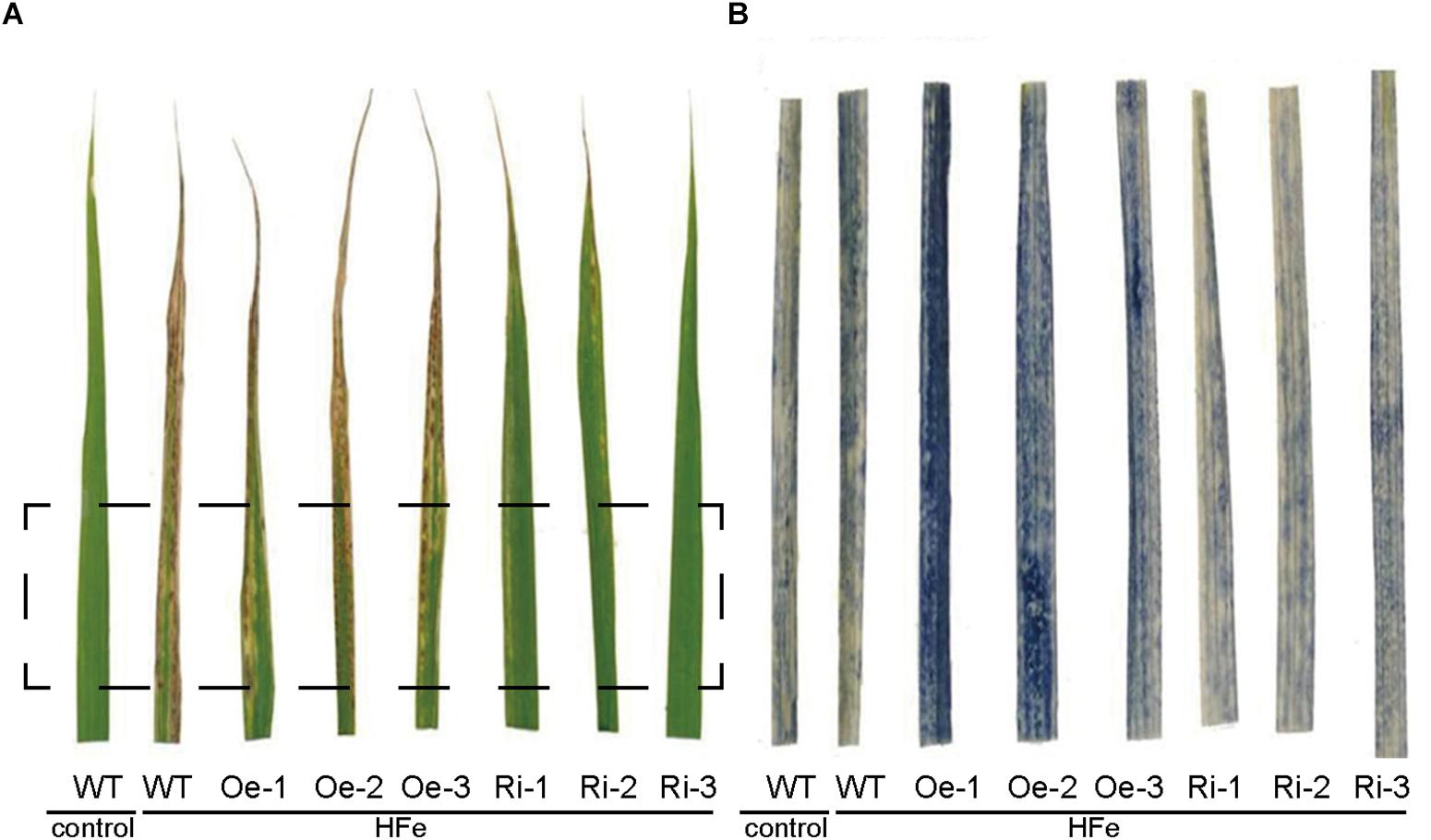
Figure 7. NBT staining of superoxide (O2−) in leaves of WT and transgenic rice plants. Three-week-old seedlings cultured in normal nutrient solution were transferred to nutrient solution containing 1 mM Fe and grown for 7 days. Leaves were detached for visualization. (A) The leaf-bronzing symptom of WT and transgenic plants under Fe excess conditions. (B) The zoom-in region that correspond to rectangle in (A) after NBT staining.
A rice vacuolar membrane ferric-chelate reductase, OsFRO1, which was down-regulated in shoots under Fe excess conditions was identified in this study. Suppression of the expression of OsFRO1 by RNAi resulted in tolerant to Fe excess stress in transgenic plants, suggesting that OsFRO1 might be involved in maintaining the Fe balance between the cytoplasm and vacuoles. We propose that OsFRO1 is involved in reducing ferric Fe into ferrous Fe in vacuole, and makes the vacuolar stored Fe available to cytoplasm through Fe (II) or chelated Fe (II) transporters. Under Fe excess condition, the decrease in transcript abundance of OsFRO1 in the RNAi plants reduced the amount of Fe (II) available for cytoplasm, and therefore alleviates Fe excess toxicity.
Previously, rice FROs were predicted to be non-functional because OsFRO1 and OsFRO2 do not contain perfect conserved HPFT or GPYG motifs compared to Arabidopsis FRO2, which are essential for the binding to the FAD and NADPH in cofactors, respectively (Ishimaru et al., 2006). In the study, we performed a multiple sequence alignment of OsFRO1, OsFRO2, AtFRO2, AtFRO6, and AtFRO7. Results showed that both HPFT and GPYG motifs of OsFRO1 were identical to that of AtFRO6/7 (Figure 1), while the GPYG motif of OsFRO2 is not conserved with the four analyzed Arabidopsis FROs. AtFRO6 was demonstrated to be a functional ferric reductase, which could facilitate the reduction of Fe in tobacco leaves (Li et al., 2011). AtFRO7 was reported to be a chloroplast ferric chelate reductase (Jeong et al., 2008). In this regard, OsFRO1 is likely a functional ferric reductase. To test the reductase activity of OsFRO1 and OsFRO2, we performed a Fe (III)-chelate reductase assay in yeast. Unfortunately, we were not able to detect significant Fe (III)-chelate reductase activity in OsFRO1-expressing or OsFRO2-expressing cells, while AtFRO2-expressing cells showed up to fivefold more Fe (III)-chelate reductase activity than cells transformed with an empty vector (Supplementary Figure S7). The failure of detection of Fe (III)-chelate reductase activity could be caused by the non-plasma membrane localization of OsFRO1 and OsFRO2 in the yeast (data not shown). Because Fe (III)-chelate reductase activity assay requires the membrane impermeant property of bathophenanthroline disulfonic acid, it is difficult to measure the Fe (III) chelate reductase activity of a non-plasma membrane localized FRO. Similar observation was reported in AtFRO7 (Jeong et al., 2008). AtFRO7 is localized in chloroplasts, whose Fe (III) chelate reductase activity could not be detected in the yeast. Further ferric Fe reductase activity assay in yeast may first fuse the OsFRO1 to a protein targeting signal, for instance, the COOH-terminal domain of Ist2p (Jüschke et al., 2005), to facilitate the trafficking of OsFRO1 protein to the plasma membrane in yeast.
The members of FRO family has been well-documented and characterized in fungi, plants, and mammals. Many of them, such as AtFRO2, AtFRO6 in Arabidopsis and FRE1 in yeast, localize to the plasma membrane (Jeong and Connolly, 2009). There are also a subset of FRO family members localizes to internal membranes, such as Arabidopsis AtFRO7 in chloroplast, AtFRO3 and AtFRO8 in mitochondria, and yeast FRE6 in vacuolar membrane, suggesting functional roles in several subcellular compartments (Singh et al., 2007; Jeong et al., 2008; Jain et al., 2014). Vacuoles are considered as the main compartment for excess Fe storage in rice and Arabidopsis (Roschzttardtz et al., 2009). Since all known vacuolar transporters, including AtVIT1, AtNRAMP3 and AtNRAMP4, transport Fe across vacuolar membranes in the form of Fe(II) (Lanquar et al., 2005; Kim et al., 2006), it was postulated that Fe reduction might be involved in the vacuolar membrane (Jeong and Connolly, 2009). In yeast, reduction of Fe (III) in vacuoles is carried out by FRE6, which is critical for remobilization of Fe from vacuoles into the cytosol (Raguzzi et al., 1988; Singh et al., 2007). While none of the eight Arabidopsis FROs was reported or predicted to reside on the vacuolar membrane, OsFRO1 was located in the vacuolar membrane in this study. OsFRO1 is likely to reduce Fe (III) into Fe (II) in the vacuolar membrane similar to FRE6 in yeast. The reduced Fe (II) can be transported across the vacuolar membrane into the cytosol by other Fe (II) transporters. Knock down of OsFRO1 resulted in more tolerance to Fe toxicity in rice plants. The similar phenotype was observed in the rice mutant MuFRO1 that had a mutation in the OsFRO1 gene, isolated from a fast neutron mutant library (Ruengphayak et al., 2015). In the OsFRO1 mutant or knockdown plants, the vacuolar ferric reductase activities are reduced, which might lead to the reduction of the amount of Fe (II) being transported into the cytosol. When exposed to excess Fe, the mutant plants should have less Fe accumulation in the cytosol, and thus showed enhanced tolerance to the excess Fe. The NBT staining showed a significant reduction of ROS accumulation in the OsFRO1-Ri leaves, supports this working model for OsFRO1 (Figure 5A).
OsFRO1-Ri and OsFRO1 mutant had a similar decrease in root and shoot Fe concentrations (Figure 6; Ruengphayak et al., 2015). qRT-PCR was carried out to determine the transcript abundance of genes involving in Fe acquisition and translocation, including IRT1, FRDL1, YSL15, and no significant difference between WT and OsFRO1-Ri plants was detected. This indicates the mechanism by which the Fe concentrations were reduced in the OsFRO1-Ri or mutant plants merits more investigation to give insight into Fe sensing and response in rice.
In summary, rice FRO, OsFRO1 was shown to function in the vacuolar membrane to converting Fe (III) into Fe (II). Knockdown of OsFRO1 could alleviate the Fe excess toxicity.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
LL carried out most experiments and drafted the manuscript. QK performed the phenotypic analyses and helped in results interpretation. LY helped to conceive the study and its design, and participated in the critical review of the manuscript. HS conceived the study, its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
This work was supported by the 111 project (B14027) and National Natural Science Foundation of China (Grant No. 31401934).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Shelong Zhang for the technical support on confocal microscope manipulation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00700/full#supplementary-material
FIGURE S1 | The predicted membrane topology of FROs in Rice and Arabidopsis.
FIGURE S2 | Subcellular localization of OsFRO1.
FIGURE S3 | Expression levels of OsFRO1 in different tissues (A) and different development stages (B) based on microarray data retrieved from GENEVESTIGATOR (https://genevestigator.com/gv/).
FIGURE S4 | Identification of OsFRO1 overexpression and RNAi lines.
FIGURE S5 | Growth performance of WT and transgenic plants.
FIGURE S6 | Analysis of Cu, Zn, Mn concentrations in WT and transgenic plants.
FIGURE S7 | Fe (III) chelate reductase assay in yeast cells.
TABLE S1 | Primers used in this work.
Asch, F., Becker, M., and Kpongor, D. S. (2005). A quick and efficient screen for resistance to iron toxicity in lowland rice. J. Plant Nutr. Soil Sci. 168, 764–773. doi: 10.1002/jpln.200520540
Audebert, A., and Fofana, M. (2009). Rice yield gap due to iron toxicity in West Africa. J. Agron. Crop Sci. 195, 66–76. doi: 10.1111/j.1439-037X.2008.00339
Bashir, K., Hanada, K., Shimizu, M., Seki, M., Nakanishi, H., and Nishizawa, N. K. J. R. (2014). Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 7:18. doi: 10.1186/s12284-014-0018-1
Bashir, K., Ishimaru, Y., Shimo, H., Kakei, Y., Senoura, T., Takahashi, R., et al. (2011). Rice phenolics efflux transporter 2 (PEZ2) plays an important role in solubilizing apoplasmic iron. Soil Sci. Plant Nutr. 57, 803–812. doi: 10.1080/00380768.2011.637305
Becana, M., Moran, J. F., and Iturbe-Ormaetxe, I. (1998). Iron-dependent oxygen free radical generation in plants subjected to environmental stress: toxicity and antioxidant protection. Plant Soil 201, 137–147. doi: 10.1023/a:1004375732137
Becker, M., and Asch, F. (2005). Iron toxicity in rice—conditions and management concepts. J. Plant Nutr. Soil Sci. 168, 558–573. doi: 10.1002/jpln.200520504
Briat, J.-F., Dubos, C., and Gaymard, F. (2015). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20, 33–40. doi: 10.1016/j.tplants.2014.07.005
Briat, J.-F., Duc, C., Ravet, K., and Gaymard, F. (2010). Ferritins and iron storage in plants. Biochim. Biophys. Acta 1800, 806–814. doi: 10.1016/j.bbagen.2009.12.003
Briat, J.-F., Fobis-Loisy, I., Grignon, N., Lobréaux, S., Pascal, N., Savino, G., et al. (1995). Cellular and molecular aspects of iron metabolism in plants. Biol. Cell 84, 69–81. doi: 10.1016/0248-4900(96)81320-7
Chen, J., Liu, Y., Ni, J., Wang, Y., Bai, Y., Shi, J., et al. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157, 269–278. doi: 10.1104/pp.111.181669
Chen, S. Y., Jin, W. Z., Wang, M. Y., Zhang, F., Zhou, J., Jia, Q. J., et al. (2003). Distribution and characterization of over 1000 T-DNA tags in rice genome. Plant J. 36, 105–113. doi: 10.1046/j.1365-313X.2003.01860.x
Connorton, J. M., Balk, J., and Rodríguez-Celma, J. (2017). Iron homeostasis in plants – a brief overview. Metallomics 9, 813–823. doi: 10.1039/c7mt00136c
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J.-F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Dancis, A., Klausner, R. D., Hinnebusch, A. G., and Barriocanal, J. G. (1990). Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol. Cell. Biol. 10, 2294–2301. doi: 10.1128/mcb.10.5.2294
Deng, D., Wu, S.-C., Wu, F.-Y., Deng, H., and Wong, M.-H. (2010). Effects of root anatomy and Fe plaque on arsenic uptake by rice seedlings grown in solution culture. Environ. Pollut. 158, 2589–2595. doi: 10.1016/j.envpol.2010.05.015
Dufey, I., Mathieu, A.-S., Draye, X., Lutts, S., and Bertin, P. (2015). Construction of an integrated map through comparative studies allows the identification of candidate regions for resistance to ferrous iron toxicity in rice. Euphytica 203, 59–69. doi: 10.1007/s10681-014-1255-5
Eide, D., Broderius, M., Fett, J., and Guerinot, M. L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 5624–5628. doi: 10.1073/pnas.93.11.5624
Fang, W.-C., Wang, J.-W., Lin, C. C., and Kao, C. H. (2001). Iron induction of lipid peroxidation and effects on antioxidative enzyme activities in rice leaves. Plant Growth Regul. 35, 75–80. doi: 10.1023/a:1013879019368
Fenton, M. A. (1984). Oxidation of tartaric acid in presence of iron. J. Chem. Soc. Trans. 65, 899–910. doi: 10.1039/ct8946500899
Guerinot, M. L., and Yi, Y. (1994). Iron: nutritious, noxious, and not readily available. Plant Physiol. 104, 815–820. doi: 10.1104/pp.104.3.815
Halliwell, B., and Gutteridge, J. M. C. (1992). Biologically relevant metal ion-dependent hydroxyl radical generation an update. FEBS Lett. 307, 108–112. doi: 10.1016/0014-5793(92)80911-Y
Höller, S., Meyer, A., and Frei, M. (2014). Zinc deficiency differentially affects redox homeostasis of rice genotypes contrasting in ascorbate level. J. Plant Physiol. 171, 1748–1756. doi: 10.1016/j.jplph.2014.08.012
Inoue, H., Kobayashi, T., Nozoye, T., Takahashi, M., Kakei, Y., Suzuki, K., et al. (2009). Rice OsYSL15 is an iron-regulated iron (III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479. doi: 10.1074/jbc.M806042200
Ishimaru, Y., Kakei, Y., Shimo, H., Bashir, K., Sato, Y., Sato, Y., et al. (2011). A rice phenolic efflux transporter is essential for solubilizing precipitated apoplasmic iron in the plant stele. J. Biol. Chem. 286, 24649–24655. doi: 10.1074/jbc.M111.221168
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Jain, A., Wilson, G., and Connolly, E. (2014). The diverse roles of FRO family metalloreductases in iron and copper homeostasis. Front. Plant Sci. 5:100. doi: 10.3389/fpls.2014.00100
Jeong, J., Cohu, C., Kerkeb, L., Pilon, M., Connolly, E. L., and Guerinot, M. L. (2008). Chloroplast Fe (III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 10619–10624. doi: 10.1073/pnas.0708367105
Jeong, J., and Connolly, E. L. (2009). Iron uptake mechanisms in plants: functions of the FRO family of ferric reductases. Plant Sci. 176, 709–714. doi: 10.1016/j.plantsci.2009.02.011
Jüschke, C., Wächter, A., Schwappach, B., and Seedorf, M. (2005). SEC18/NSF- independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J. Cell Biol. 169, 613–622. doi: 10.1083/jcb.200503033
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kim, S. A., Punshon, T., Lanzirotti, A., Li, L., Alonso, J. M., Ecker, J. R., et al. (2006). Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314, 1295–1298. doi: 10.1126/science.1132563
Kobayashi, T., Nakanishi, H., and Nishizawa, N. K. (2010). Recent insights into iron homeostasis and their application in graminaceous crops. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 900–913. doi: 10.2183/pjab.86.900
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Lanquar, V., Lelièvre, F., Bolte, S., Hamès, C., Alcon, C., Neumann, D., et al. (2005). Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 24, 4041–4051. doi: 10.1038/sj.emboj.7600864
Li, L., Cheng, X., and Ling, H.-Q. (2004). Isolation and characterization of Fe (III)-chelate reductase gene LeFRO1 in tomato. Plant Mol. Biol. 54, 125–136. doi: 10.1023/b:plan.0000028774.82782.16
Li, L.-Y., Cai, Q.-Y., Yu, D.-S., and Guo, C.-H. (2011). Overexpression of AtFRO6 in transgenic tobacco enhances ferric chelate reductase activity in leaves and increases tolerance to iron-deficiency chlorosis. Mol. Biol. Rep. 38, 3605–3613. doi: 10.1007/s11033-010-0472-9
Ling, H.-Q., Wu, H., Li, L., Du, J., Cheng, X., and Yuan, Y. (2005). Molecular and biochemical characterization of the Fe (III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 46, 1505–1514. doi: 10.1093/pcp/pci163
Matthus, E., Wu, L.-B., Ueda, Y., Höller, S., Becker, M., and Frei, M. (2015). Loci, genes, and mechanisms associated with tolerance to ferrous iron toxicity in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 2085–2098. doi: 10.1007/s00122-015-2569-y
Moore, K. L., Chen, Y., van de Meene, A. M. L., Hughes, L., Liu, W., Geraki, T., et al. (2014). Combined NanoSIMS and synchrotron X-ray fluorescence reveal distinct cellular and subcellular distribution patterns of trace elements in rice tissues. New Phytol. 201, 104–115. doi: 10.1111/nph.12497
Mori, S. (1999). Iron acquisition by plants. Curr. Opin. Plant Biol. 2, 250–253. doi: 10.1016/S1369-5266(99)80043-0
Quinet, M., Vromman, D., Clippe, A., Bertin, P., Lequeux, H., Dufey, I., et al. (2012). Combined transcriptomic and physiological approaches reveal strong differences between short- and long-term response of rice (Oryza sativa) to iron toxicity. Plant Cell Environ. 35, 1837–1859. doi: 10.1111/j.1365-3040.2012.02521.x
Raguzzi, F., Lesuisse, E., and Crichton, R. R. (1988). Iron storage in Saccharomyces cerevisiae. FEBS Lett. 231, 253–258. doi: 10.1016/0014-5793(88)80742-7
Ricachenevsky, F. K., Sperotto, R. A., Menguer, P. K., and Fett, J. P. (2010). Identification of Fe-excess-induced genes in rice shoots reveals a WRKY transcription factor responsive to Fe, drought and senescence. Mol. Biol. Rep. 37, 3735–3745. doi: 10.1007/s11033-010-0027-0
Robinson, N. J., Procter, C. M., Connolly, E. L., and Guerinot, M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Römheld, V., and Marschner, H. (1986). Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol. 80, 175–180. doi: 10.1104/pp.80.1.175
Roschzttardtz, H., Conéjéro, G., Curie, C., and Mari, S. (2009). Identification of the endodermal vacuole as the iron storage compartment in the arabidopsis embryo. Plant Physiol. 151, 1329–1338. doi: 10.1104/pp.109.144444
Ruengphayak, S., Ruanjaichon, V., Saensuk, C., Phromphan, S., Tragoonrung, S., Kongkachuichai, R., et al. (2015). Forward screening for seedling tolerance to Fe toxicity reveals a polymorphic mutation in ferric chelate reductase in rice. Rice 8:36. doi: 10.1186/s12284-014-0036-z
Sahrawat, K. L. (2004). Iron toxicity in wetland rice and the role of other nutrients. J. Plant Nutr. 27, 1471–1504. doi: 10.1081/pln-200025869
Santi, S., and Schmidt, W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183, 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
Singh, A., Kaur, N., and Kosman, D. J. (2007). The Metalloreductase Fre6p in Fe-efflux from the yeast vacuole. J. Biol. Chem. 282, 28619–28626. doi: 10.1074/jbc.M703398200
Varotto, C., Maiwald, D., Pesaresi, P., Jahns, P., Salamini, F., and Leister, D. (2002). The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31, 589–599. doi: 10.1046/j.1365-313X.2002.01381
Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M. L., Briat, J.-F., et al. (2002). IRT1, an arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.001388
Wang, C., Yue, W., Ying, Y., Wang, S., Secco, D., Liu, Y., et al. (2015). Rice SPX-major facility superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. J. Plant Physiol. 169, 2822–2831. doi: 10.1104/pp.15.01005
Waters, B. M., Blevins, D. G., and Eide, D. J. (2002). Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 129, 85–94. doi: 10.1104/pp.010829
Waters, B. M., Lucena, C., Romera, F. J., Jester, G. G., Wynn, A. N., Rojas, C. L., et al. (2007). Ethylene involvement in the regulation of the H+-ATPase CsHA1 gene and of the new isolated ferric reductase CsFRO1 and iron transporter CsIRT1 genes in cucumber plants. Plant Physiol. Biochem. 45, 293–301. doi: 10.1016/j.plaphy.2007.03.011
Wu, L. B., Shhadi, M. Y., Gregorio, G., Matthus, E., Becker, M., and Frei, M. (2014). Genetic and physiological analysis of tolerance to acute iron toxicity in rice. Rice 7:8. doi: 10.1186/s12284-014-0008-3
Wu, L. B., Ueda, Y., Lai, S.-K., and Frei, M. (2017). Shoot tolerance mechanisms to iron toxicity in rice (Oryza sativa L.). Plant Cell Environ. 40, 570–584. doi: 10.1111/pce.12733
Yi, Y., and Guerinot, M. L. (1996). Genetic evidence that induction of root Fe (III) chelate reductase activity is necessary for iron uptake under iron deficiency†. Plant J. 10, 835–844. doi: 10.1046/j.1365-313x.1996.10050835.x
Ying, Y., Yue, W., Wang, S., Li, S., Wang, M., Zhao, Y., et al. (2017). Two h-Type thioredoxins interact with the E2 ubiquitin conjugase PHO2 to fine-tune phosphate homeostasis in rice. Plant Physiol. 173, 812–824. doi: 10.1104/pp.16.01639
Keywords: rice, ferric reductase oxidase, iron excess, vacuole, iron homeostasis
Citation: Li L, Ye L, Kong Q and Shou H (2019) A Vacuolar Membrane Ferric-Chelate Reductase, OsFRO1, Alleviates Fe Toxicity in Rice (Oryza sativa L.). Front. Plant Sci. 10:700. doi: 10.3389/fpls.2019.00700
Received: 16 February 2019; Accepted: 13 May 2019;
Published: 04 June 2019.
Edited by:
Thomas J. Buckhout, Humboldt University of Berlin, GermanyReviewed by:
Khurram Bashir, RIKEN, JapanCopyright © 2019 Li, Ye, Kong and Shou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huixia Shou, aHVpeGlhQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.