- 1Plant Cell Biology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 2Plant Physiology, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
- 3Tasmanian Institute of Agriculture, University of Tasmania, Hobart, TAS, Australia
- 4Gene Discovery Research Group, RIKEN Plant Science Center, Tsukuba, Japan
- 5Department of Biology, Healthcare and the Environment, Faculty of Pharmacy, University of Barcelona, Barcelona, Spain
Polyamines, such as putrescine (Put), spermidine (Spd), and spermine (Spm), are low-molecular-weight polycationic molecules found in all living organisms. Despite the fact that they have been implicated in various important developmental and adaptative processes, their mode of action is still largely unclear. Here, we report that Put, Spd, and Spm trigger a rapid increase in the signaling lipid, phosphatidic acid (PA) in Arabidopsis seedlings but also mature leaves. Using time-course and dose-response experiments, Spm was found to be the most effective; promoting PA responses at physiological (low μM) concentrations. In seedlings, the increase of PA occurred mainly in the root and partly involved the plasma membrane polyamine-uptake transporter (PUT), RMV1. Using a differential 32Pi-labeling strategy combined with transphosphatidylation assays and T-DNA insertion mutants, we found that phospholipase D (PLD), and in particular PLDδ was the main contributor of the increase in PA. Measuring non-invasive ion fluxes (MIFE) across the root plasma membrane of wild type and pldδ-mutant seedlings, revealed that the formation of PA is linked to a gradual- and transient efflux of K+. Potential mechanisms of how PLDδ and the increase of PA are involved in polyamine function is discussed.
Introduction
Polyamines are small polycationic molecules present in all living organisms (Galston and Kaur-Sawhney, 1995). In plants, putrescine (Put), spermidine (Spd), and spermine (Spm) are the major polyamines, where they have been implicated in a broad range of cellular events, including embryogenesis, cell division, morphogenesis, senescence, and in various biotic- and abiotic stress responses (Bagni and Pistocchi, 1988; Wallace, 2009; Tiburcio et al., 2014; Michael, 2016). Despite the fact that polyamines were discovered nearly 350 years ago, and have been intensively studied during the last decades, the molecular mechanism by which these molecules regulate such a wide range of cellular functions remains a big mystery (Bachrach, 2010; Alcázar and Tiburcio, 2014). Nonetheless, polyamines have been shown to interact with components of the nucleus and cellular membranes, including transcription factors, protein kinases and phospholipases (Miller-Fleming et al., 2015) as well as ion transporting proteins (Pottosin and Shabala, 2014; Pottosin et al., 2014). The multifaceted relationship between polyamine-mediated effects and the activation of different signaling systems adds another layer of complexity to the experimental determination of direct polyamine targets (Alcázar et al., 2010; Tiburcio et al., 2014; Miller-Fleming et al., 2015).
While most studies have focused on the interaction of endogenous polyamines with immediate subcellular targets, plants are also exposed to extracellular polyamines. In the soil, plants encounter a high degree of polyamines through decomposition of organic material by microorganisms (Young and Chen, 1997; Zandonadi et al., 2013). In addition, there are several environmental cues, such as salinity stress and abscisic acid (ABA), which trigger an efflux of polyamines into the apoplast (Moschou et al., 2008; Toumi et al., 2010). There, polyamines can be oxidized by diamine- and polyamine oxidases, producing H2O2 that in turn triggers downstream effects that eventually affect the plant’s development and/or responses to stress (Takahashi et al., 2003; Moschou et al., 2008; Toumi et al., 2010; Pottosin and Shabala, 2014). However, not all apoplastic polyamines are oxidized, as intercellular transport and local internalization of a substantial part of these compounds also takes place (Friedman et al., 1986; Pistocchi et al., 1987; Ditomaso et al., 1992a; Yokota et al., 1994; Sood and Nagar, 2005; Pommerrenig et al., 2011).
The study of polyamine uptake and transport in plant cells remains scarce. However, with the recent characterization of several polyamine-uptake transporters (PUTs), an important new area is emerging, providing interesting genetic tools to explore its potential in plant function and signaling (Fujita et al., 2012; Mulangi et al., 2012; Li et al., 2013; Strohm et al., 2015; Martinis et al., 2016; Tong et al., 2016).
Phosphatidic acid (PA) represents a minor class of membrane lipids, constituting 1–3% of total phospholipids in most plant tissues. As a precursor of glycerolipids, PA is involved in lipid biosynthesis at the ER and plastids. Over the last decade, however, PA has also emerged as a signaling molecule, playing key roles in regulating plant development and stress responses (Munnik, 2001; Testerink and Munnik, 2005, 2011; Yao and Xue, 2018). This PA is typically formed at the plasma membrane and along the endosomal membrane system, where it recruits and modulates target proteins involved in membrane trafficking, organization of the cytoskeleton and ion transport (Testerink and Munnik, 2005, 2011; Kooijman et al., 2007; Raghu et al., 2009; McLoughlin et al., 2013; Pleskot et al., 2013; Putta et al., 2016; Yao and Xue, 2018). A local increase of PA may also induce biophysical effects, affecting membrane curvature and surface charge, which facilitate membrane fission and fusion (Kooijman et al., 2003; Wang et al., 2006; Roth, 2008), also in cooperation with other lipid signals (Testerink and Munnik, 2011).
The accumulation of PA in response to stimuli is in general relatively fast, taking place within minutes after stimulation, and is generated via two pathways, i.e., via phosphorylation of diacylglycerol (DAG) by DAG kinase (DGK) and by hydrolysis of structural phospholipids by phospholipase D (PLD). DAG itself can be produced via non-specific phospholipase C (NPC), which hydrolyses structural phospholipids, or by phosphoinositide- (PI-) specific phospholipase C (PLC), which hydrolyses inositol-containing phospholipids (Munnik, 2014).
Both PLC- and PLD activities are known to be affected by polyamines. In vitro studies on isolated enzymes from animal cells and tissues have shown that polyamines can inhibit (Kimura et al., 1986; Smith and Snyderman, 1988; Wojcikiewicz and Fain, 1988; Sjöholm et al., 1993; Pawelczyk and Matecki, 1998) and stimulate PLC activity (Sagawa et al., 1983; Haber et al., 1991; Späth et al., 1991; Periyasamy et al., 1994; Pawelczyk and Lowenstein, 1997) and PLD activity (Jurkowska et al., 1997; Madesh and Balasubramanian, 1997). In plants, polyamines have been found to activate PLC in Catharanthus roseus roots (Echevarría-Machado et al., 2002, 2004), but to inhibit it in Coffea arabica cells, where an increase in PLD activity was observed (Echevarría-Machado et al., 2005).
Here, we show that polyamines trigger a rapid (minutes) PA response in Arabidopsis seedlings, with Spm being the most potent. Using differential 32Pi-labeling techniques and a PLD-specific transphosphatidylation assay (Arisz and Munnik, 2013; Munnik and Laxalt, 2013), we provide evidence that the PLD pathway is the most important contributor. Using T-DNA-insertion PLD mutants, we identified PLDδ as the main contributor of the Spm induced-PA response. Using Microelectrode Ion Flux Estimation (MIFE), we found a differential Spm induced-K+ efflux response in the pldδ KO mutant, highlighting a potential role for PA downstream of Spm signaling.
Materials and Methods
Plant Material and Growth Conditions
Arabidopsis thaliana pldα1, pldα3, pldδ, pldε, pldα1/δ, pldα1/δ/α3, pldα1/δ/ε, pldζ1, pldζ2, gapc1-1/gapc2-1, gapc1-1/gapc2-2, rmv1, spms-2 mutant null alleles and the Pro35S::RMV1 and Pro35S::SPMS-9 transgenic lines were described previously (Hong et al., 2008, 2009; Bargmann et al., 2009; Gonzalez et al., 2011; Fujita et al., 2012; Guo et al., 2012; Galvan-Ampudia et al., 2013). The lat1/2/3/5 and lat1/2/4/5 quadruple null mutant were generated by Dr. M. Fujita (unpublished; RIKEN Plant Science Center, Japan), while pldα1/δ/α3 and pldα1/δ/ε triple knock-out mutants were kindly provided by Prof. Dr. D. Bartels (University of Bonn, Germany). In most cases Arabidopsis thaliana ecotype Col-0 was used as wild type, except for rmv1 and the Pro35S::RMV1 lines, in which Ler ecotype and Col-0 empty vector, Ve-1, were used as wild type, respectively.
Seeds were surface-sterilized with chlorine gas and sown under sterile conditions on square petri dishes containing standard growth medium consisting of ½ Murashige and Skoog (MS) medium with Gamborg B5 vitamins (pH 5.7; KOH), 1% (w/v) sucrose, and 1% (w/v) agar. Plates were vernalized at 4°C for 48 h and then placed vertically under the angle of 70°, in a growth chamber (16/8 light/dark cycle, 110–130 μmol m–2 s–1) at 22°C. Five days-old seedlings were then transferred to either 2 mL Eppendorf safe-lock tubes for 32Pi labeling O/N, or to treatment plates for phenotypic analyses.
Chemicals
All chemicals were obtained from Sigma-Aldrich except 32Pi (orthophosphate, 32PO43–), which was purchased from PerkinElmer. Arabidopsis incubations with polyamines and chemicals were performed in incubation buffer, consisting of 2.5 mM MES buffer [2-(N-morpholino) ethanesulfuric acid], pH 5.7 (KOH), 1 mM KCl.
32Pi-Phospholipid Labeling, Extraction and Analysis
Phospholipid levels were measured as described earlier (Munnik and Zarza, 2013). Briefly, three seedlings per sample were metabolically labeled overnight by flotation in continuous light in 2 ml safe-lock Eppendorf tubes containing 200 μl incubation buffer (2.5 mM MES-KOH, pH 5.7, 1 mM KCl) and 2.5–10 μCi 32PO43– (stock 32Pi; carrier-free, 2.5–10 μCi/μL). For mature plants, Arabidopsis leaf disks (∅ 5 mm) were taken from 3-week-old plants and labeled using the same conditions. Treatments were performed by adding 1:1 (v/v) of a 2× solution and incubations were stopped at indicated times by adding perchloric acid (Munnik and Zarza, 2013). Lipids were extracted and analyzed by thin-layer chromatography (TLC) using an ethyl acetate solvent system (Munnik and Laxalt, 2013). Radioactivity was visualized by autoradiography and individual spots were quantified by phosphoimaging (Typhoon FLA 7000; GE Healthcare).
For certain experiments, the protocol was slightly modified, i.e.: (1) In short-labeling experiments, 32Pi was added 30 min prior treatment. (2) In transphosphatidylation assays, treatments were performed in the presence of 0.5% n-butanol (Munnik and Laxalt, 2013). (3) For tissue-dissection experiments, seedlings were labeled, treated and fixed as described above, but then carefully cut into sections with a scalpel, and every section processed separately.
For statistical analysis, letters indicate values significantly different according to Student–Newman–Keuls test at P-value <0.05, and asterisks indicate significant differences with respect to control treatments, using the Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.005. Data represent the mean ± SD. The results obtained were confirmed by at least 2 independent experiments.
Detection of ROS and NO in Arabidopsis Root
ROS production in the root tip of 5-day-old seedlings was detected by DCF fluorescence as described previously (Pei et al., 2000; Zhang et al., 2009). Briefly, seedlings were treated for the indicated times and then transferred to 10 μM H2DCFDA for 10 min followed by two washes in buffer. For ROS scavenging, seedlings were pre-treated with 5 mM N,N’-Dimethylthiourea (DMTU; Lu et al., 2009) for 60 min, before the different treatments with or without Spm 120 μM for 30 min. For NO detection, seedlings were co-incubated with the corresponding treatment and 10 μM DAR-4M for 30 min, and then washed two times with buffer. For cPTIO treatment, 0.1 mM cPTIO was applied for 60 min prior to treatments in order to scavenge NO. All incubations were performed in dark conditions. The localization of the DCF and DAR signal was done using the AMG Evos FL digital inverted microscope equipped with transmitted light GFP (470/22 to 510/42 nm). Images were converted to 8-bit using Image-J, and data was quantified as mean pixel intensity per region of interest (ROI).
Ion Flux Measurement
Net K+ fluxes were measured using MIFE technique (UTas Innovation, Hobart, TAS, Australia) (Newman, 2001; Shabala et al., 2006). Five days-old Arabidopsis seedlings were placed into a 30 mL measuring chamber, containing 0.5 mM KCl, 0.2 mM CaCl2, 5 mM MES, 2 mM Tris base; pH 6.0. Roots were immobilized in a horizontal position (Bose et al., 2014) and preincubated in the above buffer for at least 30 min. Electrodes were positioned near the root surface at the elongation zone (less than 2 mm from the root cap junction). First, steady-state ion fluxes were recorded over a period of 5 min, after which different concentrations of Spm were applied and net ion fluxes measured.
Root Phenotyping Assay on Plates
Arabidopsis seedlings were grown on vertical plates containing standard growth medium for 5 days. Then, seedlings were transferred to plates supplemented with or without Spm. Plates were scanned at indicated days after transfer (DAT) using an Epson Perfection V700 Scanner at 300 dpi. For root measurements EZ-Rhizo software was used (Armengaud et al., 2009). Main root (MR) growth was expressed as growth ratio (MR length divided by MR length at 0 DAT). A paired t-test in SPSS was used for statistical analysis.
Results
Polyamines Trigger the Formation of PA in Arabidopsis Seedlings
To investigate whether polyamines could affect PA signaling, Arabidopsis seedlings were 32Pi-prelabeled O/N and treated with physiological concentrations (60 μM) of Put, Spd, or Spm for 30 min. As shown in Figure 1A, in particular Spm but also Spd, was found to induce an increase in PA. Put did not show any effect until millimolar concentrations were used (Figure 1B). Spm already induced significant PA responses at ≥15 μM, while 30–60 μM was required for Spd. Thermospermine (tSpm), a minor structural isomer of Spm also present in Arabidopsis (Kakehi et al., 2008), was found to induce PA responses at a similar concentrations as Spm (Figure 1B). Diaminopropane (Dap), a diamine product of polyamine oxidation, exhibited a similar low potency like Put (Figure 1B). Together these results show that polyamines can trigger a PA response, with its potency depending on charge, i.e., Spm4+ = tSpm4+ > Spd3+>>Put2+≈Dap2+.
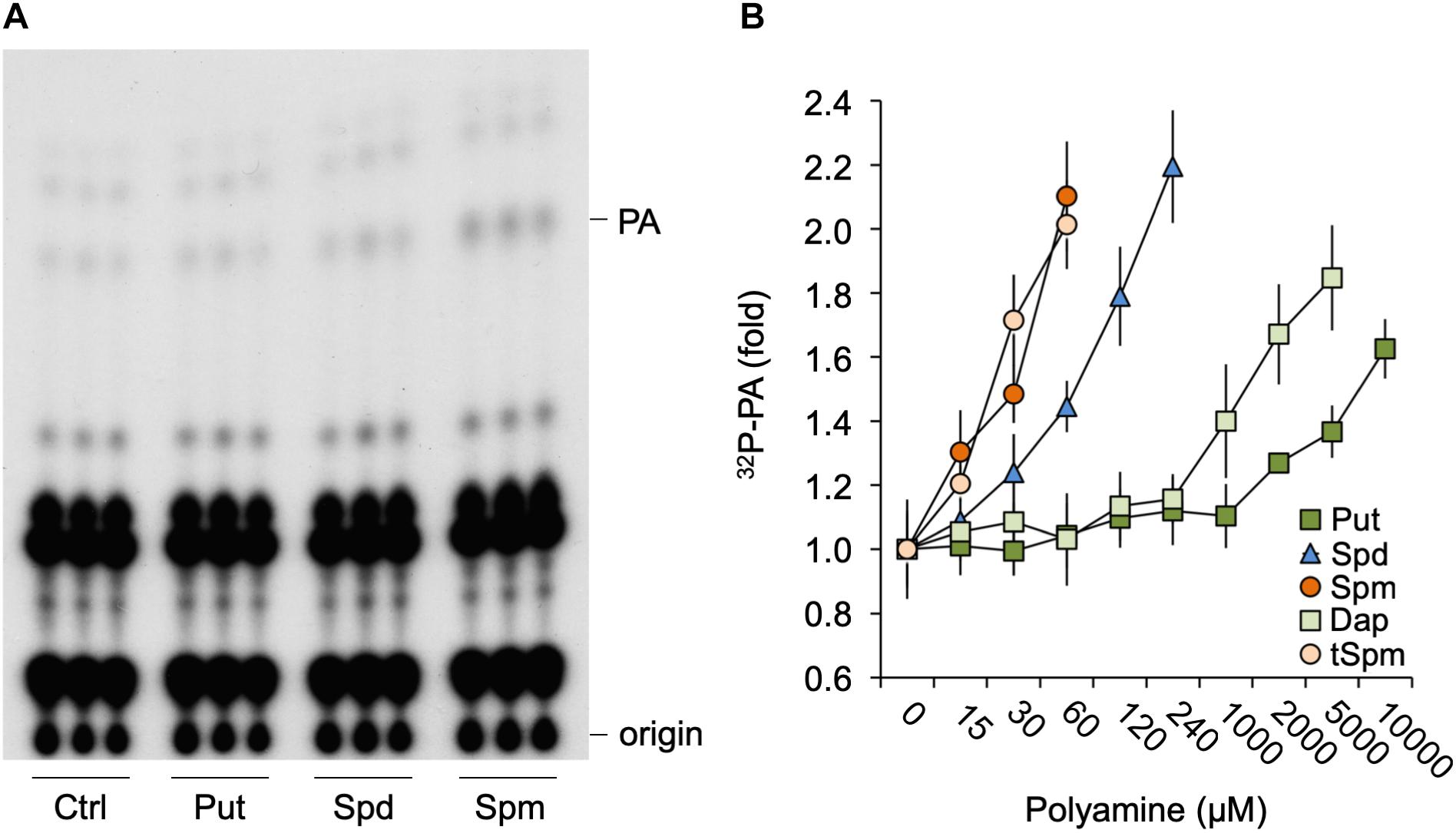
Figure 1. Polyamines trigger a PA response in Arabidopsis seedlings. (A) Six-days old seedlings that had been 32Pi-labeled O/N were treated for 30 min with 60 μM of putrescine (Put), spermidine (Spd) or spermine (Spm), or with buffer alone (control, Ctrl), after which lipids were extracted, separated by TLC, and visualized by autoradiography. Each sample represents the extract of three seedlings. (B) 32P-labeled PA fold response to 30 min Put, Spd, Spm or diaminopropane (Dap), and thermospermine (tSpm) at the indicated concentrations.
To further investigate the Spm-induced PA, detailed dose-response and time-course analyses were performed. As shown in Figure 2, Spm induced a clear dose-dependent Michaelis–Menten PA curve when treated for 30 min; starting at low μM levels and reaching a maximum 2.5-fold increase at ∼250 μM (Figures 2A,B). The response was relatively fast, starting between 8 and 15 min when using 60 μM of Spm, and PA linearly increasing (Figures 2C,D).
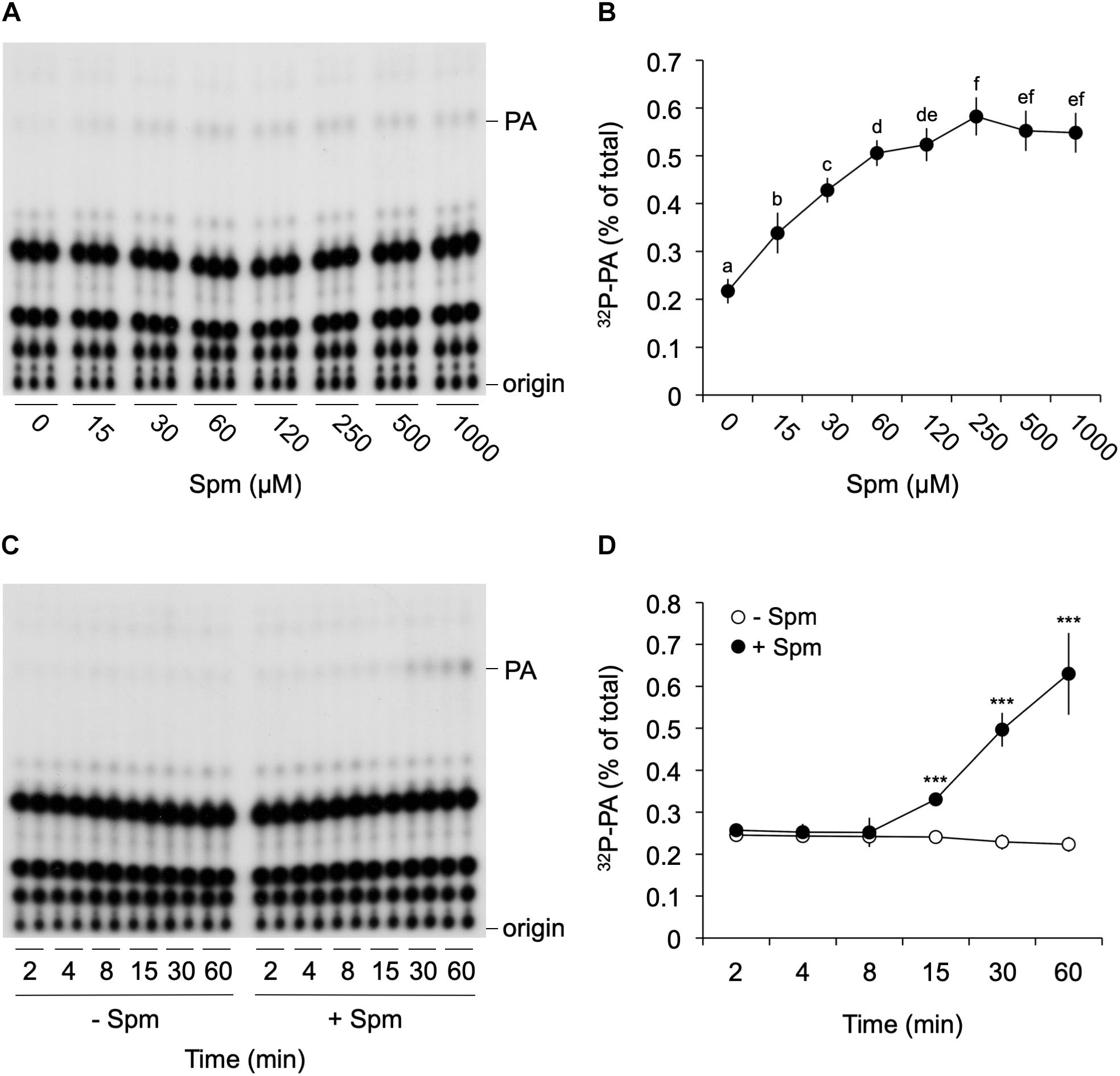
Figure 2. Dose-response and time-course analysis of the Spm induced-PA response. Six-days old 32Pi-prelabeled seedlings were treated for 30 min with different concentrations of Spm (A,B) or with 60 μM Spm or buffer alone (control) for different times (C,D) and their lipids analyzed. (A,C) Autoradiographs of a representative experiment. (B,D) Quantifications of the PA response, showing the percentage of 32P-PA with respect to the total amount of 32P-labeled phospholipids. Data are the mean ± SD of three independent experiments (n = 9 for B; n = 6 for D). Different letters indicate values significantly different according to Student-Newman-Keuls test at P-value < 0.05. Asterisks indicate significant differences with respect to control treatments, using the Student’s t-test: *P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005.
Spm Triggers PA Formation in Roots and Requires Transport Across the Plasma Membrane via RMV1
To obtain more information as to where in the seedling the PA accumulation took place, we performed the same 32Pi-labeling and treatments, but now dissected root- and shoot tissues prior to lipid extraction. Interestingly, the Spm induced-PA increase was only found in the root, not in the shoot or hypocotyl (Figure 3A). Within the root, the PA accumulation was equally distributed along the different root sections (Figure 3A). A repetition of this experiment with 120 μM Spm gave similar results (data not shown). In contrast to seedlings, we did observe a Spm induced-PA response in 32Pi-labeled leaf disks of mature, 3-weeks old plants (Supplementary Figure S1).
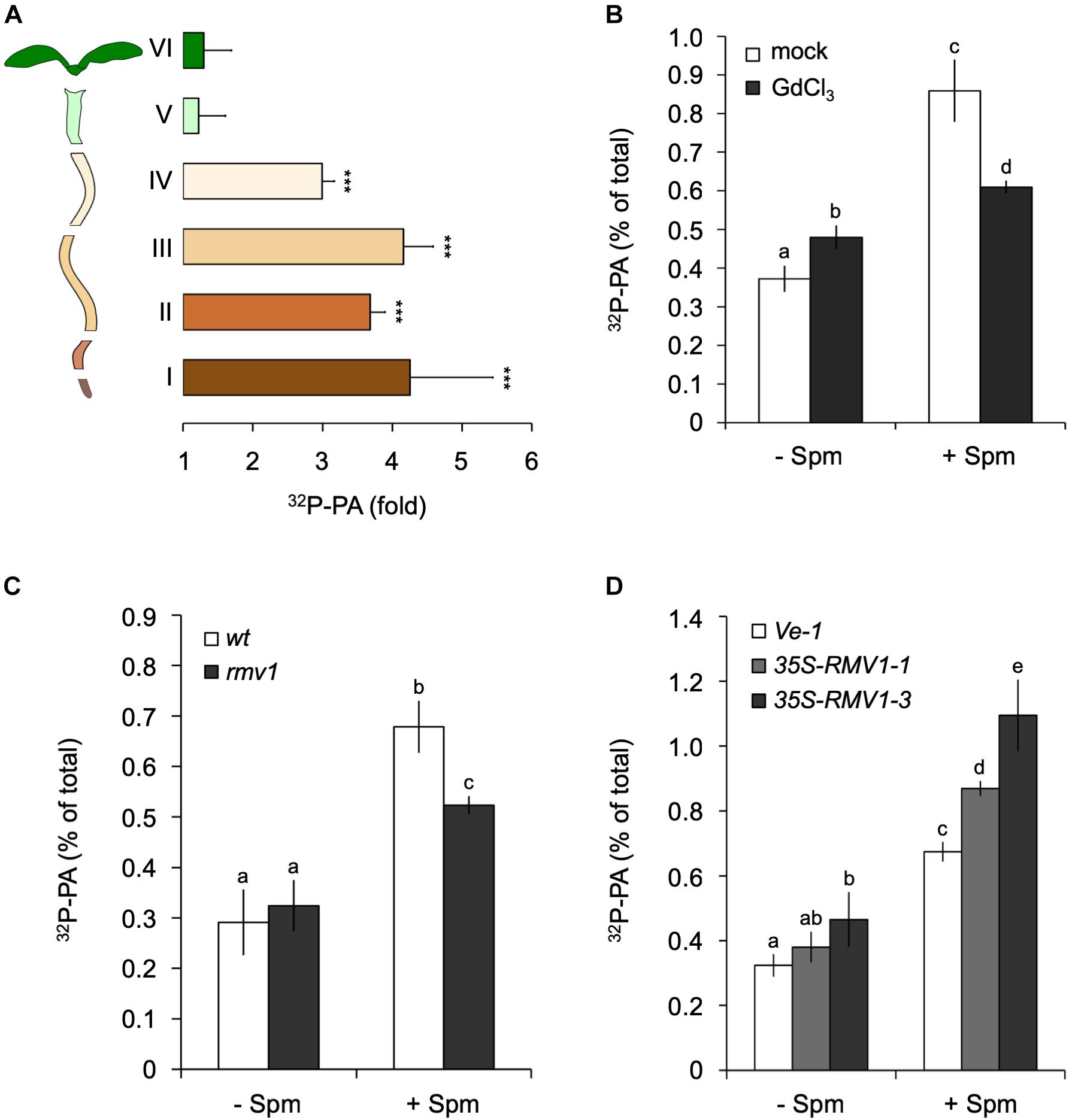
Figure 3. Spermine (Spm) induces PA responses in the root and involves polyamine-uptake transporter, RMV1. (A) 32P-PA response in different sections of 32Pi-prelabeled Arabidopsis seedlings. Length/type of section: root tip, 2 mm (I), 3 mm (II), 5 mm (III), 5–7 mm (IV), hypocotyl (V), and cotyledons (VI). Results are expressed as fold-increase of PA with respect to control treatment of each section. (B) Gadolinium blocks Spm induced-PA response. O/N 32P-labeled seedlings were pre-treated with buffer containing KCl (mock) or 100 μM GdCl3 for 60 min after which they were treated with or without 60 μM Spm for 30 min. (C,D) 32P-PA responses in rmv1 knock-out and two independent Pro35S::RMV1 over-expressor lines treated for 30 min ± 60 μM Spm. Wild-type Landsberg (Ler) and a wild-type Col-0 line containing the empty vector (Ve-1) were used as control lines, respectively. Different letters indicate values significantly different according to Student-Newman-Keuls test at P-value < 0.05. Asterisks indicate significant differences with respect to control treatments, using the Student’s t-test: *P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005.
The non-permeant cation transport blocker, gadolinium (Gd3+) is known to inhibit the uptake of Spm across the plasma membrane (Pistocchi et al., 1988; Ditomaso et al., 1992b; Pottosin et al., 2014). Incubation of seedlings with GdCl3 prior to the application of Spm triggered a small PA response itself, but significantly reduced the Spm induced-PA response to approximately 70% of the control response (Figure 3B). This may indicate that most of the PA response observed is caused intracellularly.
To further characterize Spm uptake, we analyzed the Arabidopsis PUT/L-type amino acid transporter (LAT), called Resistant to Methyl Viologen 1 (RMV1, PUT3, LAT1), which is localized in the plasma membrane and responsible for the high-affinity uptake of Spm (Fujita et al., 2012). Using the knock-out T-DNA insertion mutant rmv1 and two independent over-expressing Pro35S::RMV1 lines, we found a 35% decrease and ∼20–40% increase in PA, respectively (Figures 3C,D). These results indicate that cellular uptake of Spm is required for the PA response, and that RMV1 is one of the proteins involved in internalizing Spm across the plasma membrane.
To functionally analyze the involvement of the rest of the PUT/LAT family members, of which Arabidopsis contains five homologs (Mulangi et al., 2012), two quadruple knock-out mutants were used, i.e., lat1/2/3/5 and lat1/2/4/5, because the quintuple mutant was lethal (Fujita M., unpublished). Both lines, however, showed Spm induced-PA responses similar to wild type (Supplementary Figure S2). This discrepancy could be due to the fact that the single rmv1-KO allele is different from the quadruple mutants and belongs to a different ecotype (i.e., Ler vs. Col-0, respectively). While at least three LAT proteins exhibit polyamine transport activity (i.e., LAT1, LAT3, LAT4; Fujita et al., 2012; Mulangi et al., 2012), only LAT1 (RMV1, PUT3) is localized to the plasma membrane; LAT3 and LAT4 are localized to the ER and Golgi, respectively (Li et al., 2013; Fujita and Shinozaki, 2014). Hence, the results obtained may reflect distinct plasma membrane activity as well as genetic redundancy for Spm uptake, involving other members from the amino acid-polyamine-choline (APC) transporter family to which LAT/PUT transporters belong (Verrey et al., 2004; Rentsch et al., 2007).
Spm-Triggered PA Is Predominantly, but Not Solely Generated via PLD
A rapid PA response has traditionally been associated with increased DGK- and/or PLD activity. DGK produces PA through phosphorylation of DAG that originates from the hydrolysis of phosphoinositides or structural phospholipids by PLC or NPC, respectively (Munnik, 2014). PLD hydrolyses structural phospholipids, like PE and PC, to form PA directly. To distinguish between these two routes, a differential 32P-labeling protocol was used that highlights the DGK kinase-dependent reaction (Arisz and Munnik, 2013). This method is based on the premise that the 32Pi added to seedlings is rapidly taken-up and incorporated into ATP and subsequently into lipids that are synthesized via kinase activity (e.g., DGK), which is in huge contrast to the relatively slow incorporation of 32P into structural phospholipids via de novo synthesis (Munnik et al., 1994, 1998; Arisz and Munnik, 2013). Under short labeling conditions, PLD would hardly generate 32P-labeled PA whereas the contribution of DGK would be augmented. As shown in Figure 4, Spm was still able to trigger an increase in 32P-PA when seedlings were only prelabeled for 30 min rather than 16 hrs O/N, indicating that at least part of the Spm induced-PA response is generated via DGK.
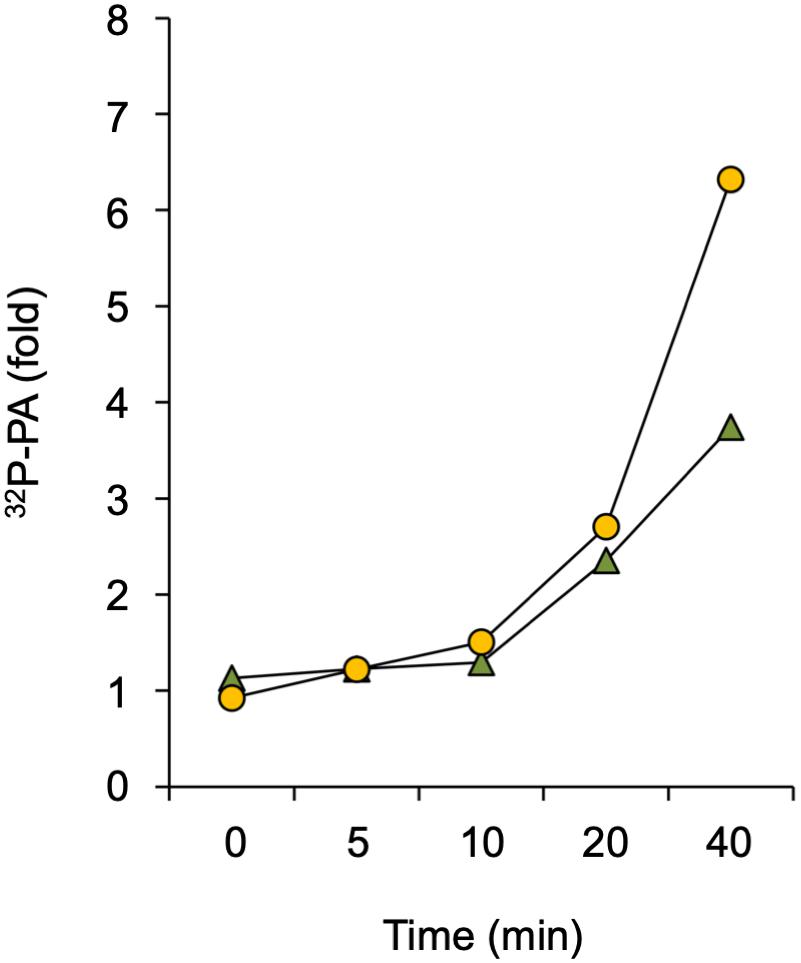
Figure 4. Analysis of DGK involvement in Spm triggered-PA response. Seedlings were pulse-labeled with 32Pi for 30 min and then treated with 60 μM Spm or buffer alone (Ctrl) for the indicated times. The fold-PA response of two independent experiments is shown (squares and triangles, respectively). Values were normalized to the 32P-labeling of phosphatidylinositol and to Ctrl, without Spm.
To analyze the potential involvement of PLD, its unique ability to catalyze a transphosphatidylation reaction was used, which produces phosphatidylbutanol (PBut) in vivo in the presence of a low concentration of n-butanol (Munnik et al., 1995, 1998; Arisz and Munnik, 2013; Munnik and Laxalt, 2013). To get a substantial proportion (though not all) of the structural phospholipids (i.e., PLD’s substrate) 32P-labeled, seedlings were incubated with 32Pi O/N and the next day treated with Spm in the presence of 0.5% n-butanol. The subsequent formation of 32P-PBut is an in vivo marker for PLD activity that can be quantified. Again dose-response and time-course experiments were performed, but this time in the presence of n-butanol. As shown in Figures 5A,B, Spm clearly triggered PLD activity, with the PBut following a similar pattern as PA (Figure 2).
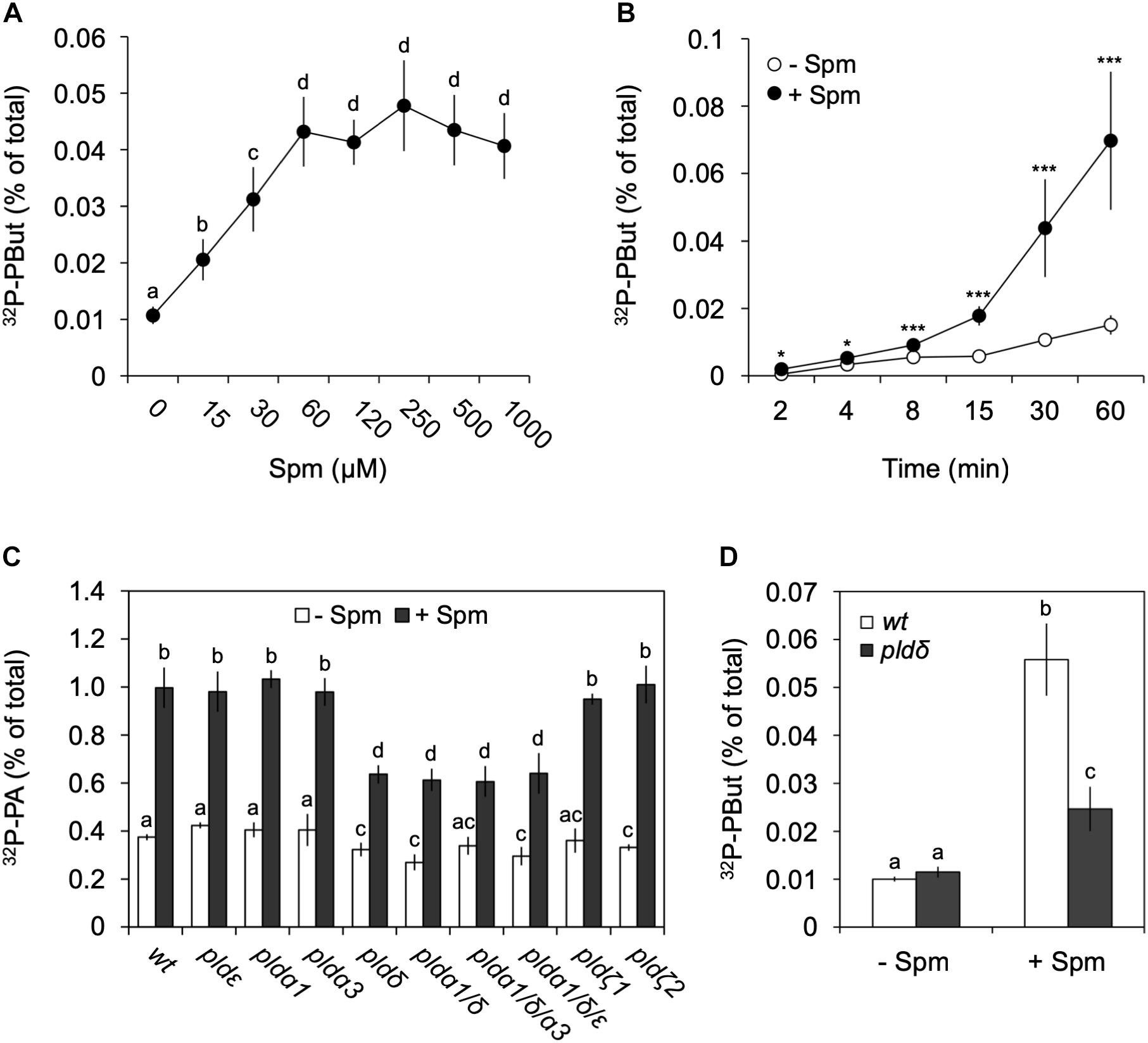
Figure 5. Analysis of Spm-induced PLD activity and gene identification. Dose-response (A) and time-course (B) experiments were performed as described earlier but this time in the presence of 0.5% n-butanol, to quantify the production of the PLD-dependent transphosphatidylation product, 32P-PBut. (A,B) Quantification of 32P-PBut. Data are the means ± SD of three independent experiments (n = 6–9). (C) 32P-PA responses in PLD T-DNA insertion mutants treated with (+) or without (–) 60 μM Spm for 30 min. Mutants analyzed: plda1, plda3, pldδ, pldε, pldζ1, pldζ2, and the mutant combinations plda1/δ, plda1/δ/a3 and plda1/δ/ε. (D) 32P-PBut response in pldδ with respect to wt. Different letters indicate values significantly different according to Student-Newman-Keuls test at P-value < 0.05. Asterisks indicate significant differences with respect to control treatments, using the Student’s t-test: *P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.005.
Arabidopsis contains 12 PLDs, i.e., 3 PLDαs, 2 PLDβs, 3 PLDγs, 1 PLDδ, 1 PLDε, and 2 PLDζs (Zhang et al., 2005). Validating the 32P-labeled PBut- and PA response in various T-DNA KO mutants, we identified PLDδ as the main contributor, with the pldδ-KO mutant alone or in combination with other KOs, showing a ∼55% reduction in PA and ∼70% reduction in PBut accumulation (Figures 5C,D). Interestingly, in Arabidopsis this isoform is located in the plasma membrane (Wang and Wang, 2001; Pinosa et al., 2013), while promoter-GUS analyses suggests it is mainly expressed in roots (Katagiri et al., 2001), which is in agreement with the PA response observed here.
H2O2 or NO Are Not Involved in the Spm Induced-PA Response
Spermine is known to cause an accumulation of NO and H2O2 (Cona et al., 2006; Tun, 2006; Moschou et al., 2008), which is likely mediated by polyamine oxidase (PAO) and diamine oxidase (DAO) activities (Tun, 2006; Wimalasekera et al., 2011). We confirmed that Spm was able to trigger an increase in H2O2 and NO under our conditions, as evidenced by the increase in fluorescence of their reporters, i.e., 2′,7′-dichlorofluorescein diacetate (H2DCFDA; Figure 6A) and diaminorhodamine-4M acetoxymethyl ester (DAR-4M AM; Figure 6B).
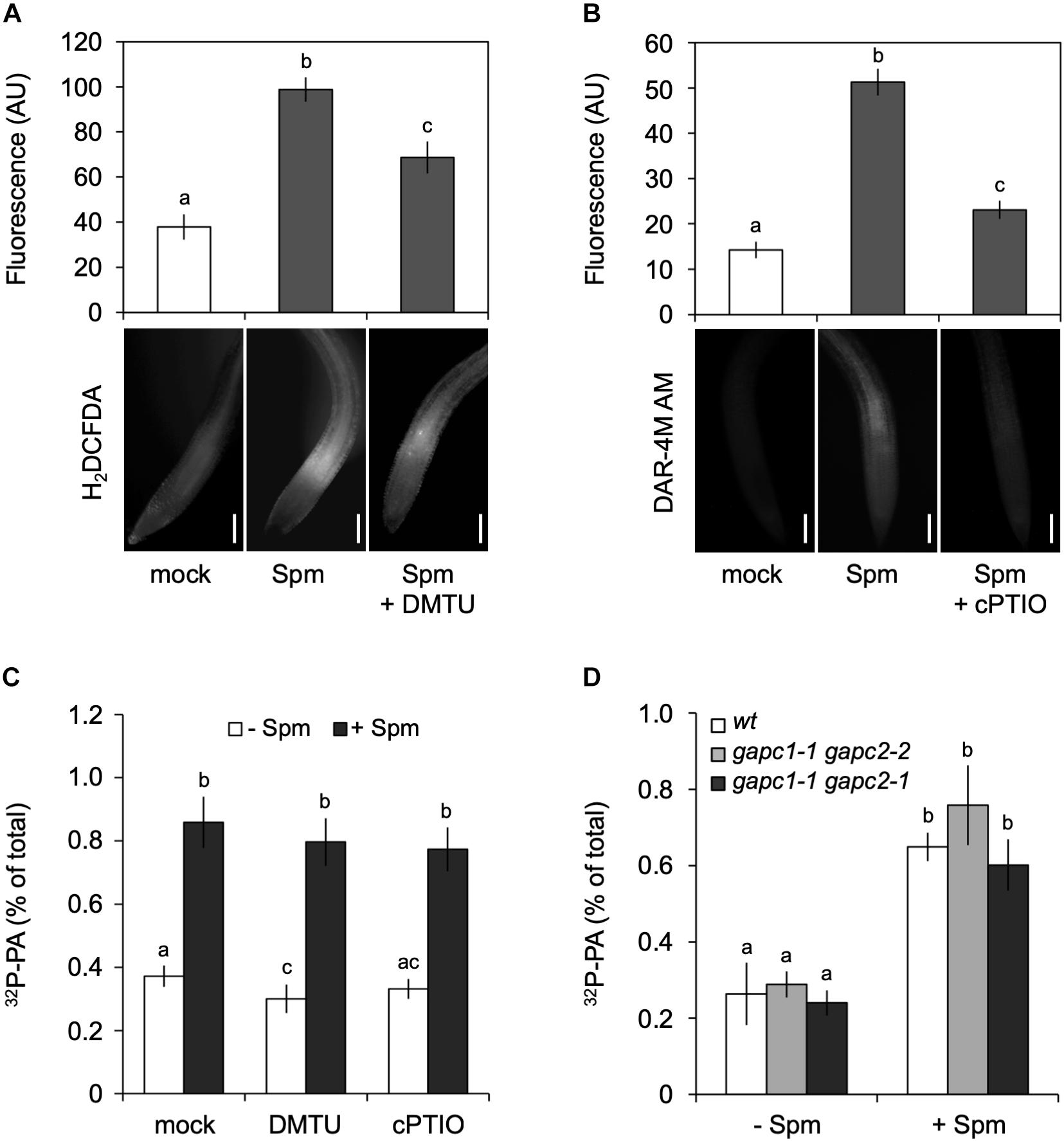
Figure 6. Spermine (Spm) induced-PA response is not generated through the production of H2O2 or NO. Visualization and quantification of Spm induced-H2O2 (A) or -NO production (B) using corresponding dyes and scavengers, respectively (H2DCFDA and DMTU for H2O2; DAR-4M AM and cPTIO for NO). Bars represent 100 μm. The fluorescence in the root tip is expressed as arbitrary units (AU; Mean ± SD, n = 10). (C) 32P-PA levels in seedlings incubated in presence of buffer (Ctrl) or scavengers ± 120 μM Spm. (D) 32P-PA response in gapc1-1/gapc2-2 and gapc1-1/gapc2-1 double knock-out mutants respect to wt. Mean ± SD (n = 3). All experiments were repeated twice with similar results. Different letters indicate values significantly different according to Student-Newman-Keuls test at P-value < 0.05.
Since PLD activity can be activated by H2O2 (Wang and Wang, 2001; Zhang et al., 2003, 2009) or by NO (Distéfano et al., 2008; Lanteri et al., 2008; Raho et al., 2011), this could be a potential mechanism by which the production of PA was stimulated, especially since H2O2 and NO have been found to act upstream of PLDδ in response to ABA induced-stomatal closure (Distéfano et al., 2012). Similarly, H2O2 has been found to promote the binding of cytosolic glyceraldehyde-3-phosphate dehydrogenase (GAPC) to PLDδ and increase its activity (Guo et al., 2012).
To investigate whether H2O2 and NO were responsible for the Spm induced-PA response, the effects of ROS scavenger, DMTU and NO scavenger, carboxy-PTIO (cPTIO) were analyzed. While able to significantly reduce the accumulation of Spm-derived H2O2 and NO (Figures 6A,B), the scavengers had no effect on the Spm induced-PA response (Figure 6C), suggesting that the increase in PA was independent of these secondary metabolites. Moreover, double gapc1-1 gapc2-1 or gapc1-1 gapc2-2 knock-out mutants, revealed a PA response similar to wild type (Figure 6D). These results indicate that the Spm-induced PA is not caused via ROS- or NO induction.
Testing polyamines on seedling growth, we observed a significant reduction in primary root growth (Supplementary Figure S3). Previous reports have shown that this root growth inhibition is associated to H2O2 accumulation, derived from PAO activity (de Agazio et al., 1995; Couée et al., 2004; Tisi et al., 2011). However, loss-of function pldδ seedlings did not show any apparent root phenotype when transferred to agar plates containing μM concentrations of Spm. Only a slight increase in root growth inhibition with respect to wt was observed at higher Spm concentrations, i.e., 150 μM (Supplementary Figure S3). Those results are consistent with previous reports indicating that pldδ is more sensitive to H2O2-induced stress (Zhang et al., 2003).
PLDδ Is Involved in the Spm-Induced K+-Efflux Response in the Root-Elongation Zone
Application of exogenous polyamines has been shown to trigger a K+ efflux in pea- (Zepeda-Jazo et al., 2011) and maize roots (Pandolfi et al., 2010), which has consequences for the membrane potential, inducing the plasma membrane to depolarize (Ozawa et al., 2010; Pottosin et al., 2014). To study this in our context, we performed MIFE ion-flux analyses at the root elongation zone of Arabidopsis seedlings using different concentrations of Spm. As shown in Figures 7A,B, a clear dose-dependent efflux of K+ was detected, which correlated with the response in PA (Figure 2B). While the Spm induced-K+ efflux slowly restored to pre-treatment values after 50 min with 60–200 μM, the efflux persisted when only 10–20 μM Spm was used.
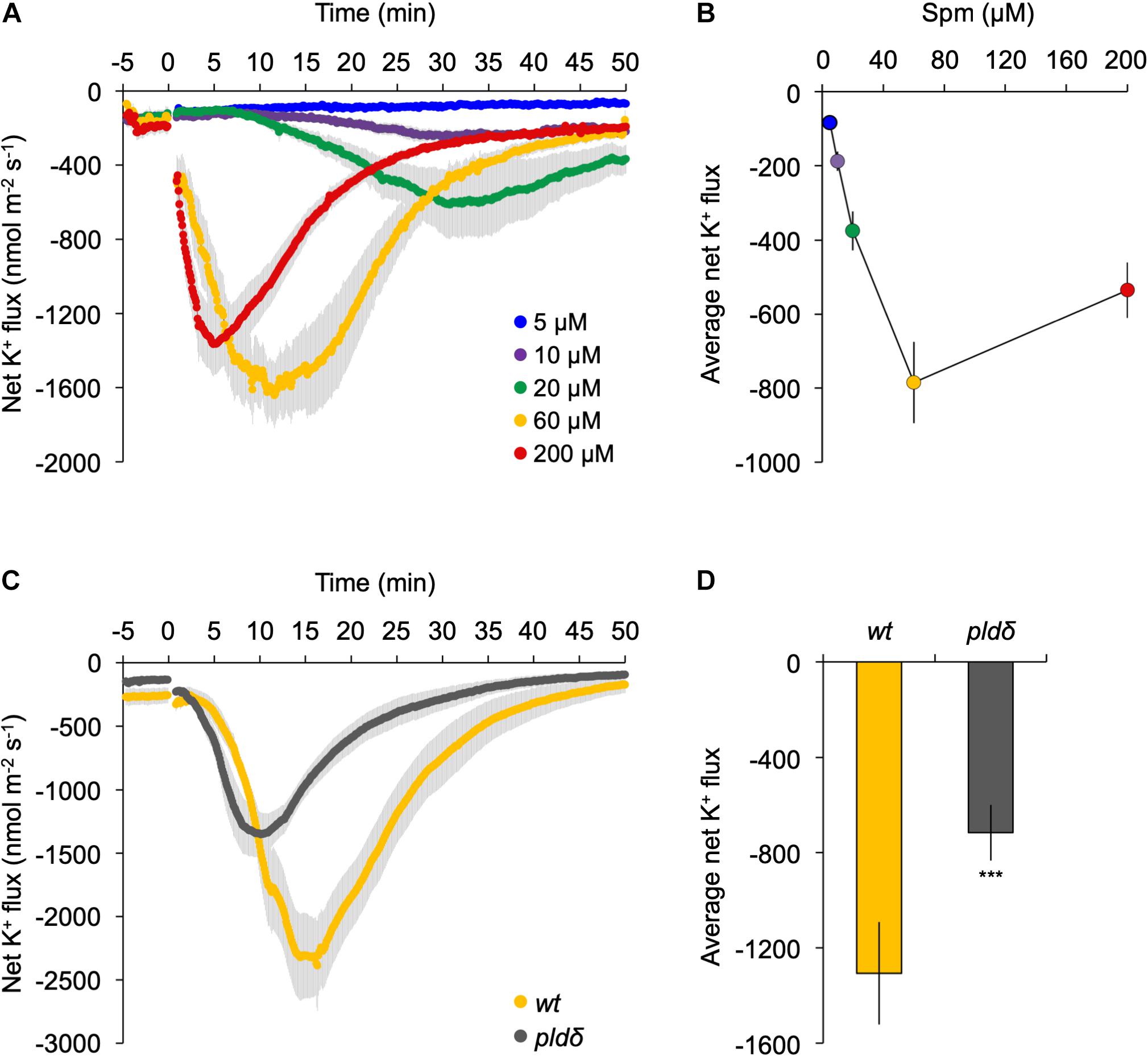
Figure 7. K+ flux at the root elongation zone is strongly reduced in pldδ mutant. (A) Dose-response analyses of K+ flux kinetics measured by MIFE in the root tip. Spm was added at the indicated concentrations (t = 0). (B) Quantification of the average K+ flux during the 50 min period of treatment. (C) Transient K+-flux kinetics in wt- and pldδ seedlings upon addition of 60 μM Spm at t = 0. (D) Quantification of the average K+ flux over 30 min of Spm treatment (means ± SE, n = 6–7). Asterisks indicate significant differences using Student’s t-test: *P < 0.05, **P < 0.01, ***P < 0.005. For all MIFE data, negative values represent net efflux of ions across the PM into the apoplast.
To investigate potential involvement of PLDδ, the response of wild type was compared with that of the pldδ knock-out mutant in the same root zone. Prior to Spm application, roots showed a small net K+ efflux of 150–250 nmol m–2 s–1, likely due to transferring seedlings from nutrient-rich MS medium (∼20 mM K+) to poorer, basic-salt medium (BSM; 0.2 mM K+; Figure 7). Upon 60 μM Spm application to wild-type seedlings, the K+ efflux increased gradually, reaching a peak around 15 min, and returning to basal levels after ∼50 min (Figure 7C). In pldδ, the response was significantly different, showing an faster K+ efflux peak at 10 min, and recovery (Figure 7C). Overall, pldδ showed ∼60% reduction in net K+ loss compared to wt (Figure 7D), placing PLDδ and derived PA upstream of the Spm induced-K+ efflux. Since the Spm induced-K+ efflux is completely abolished by gadolinium (Supplementary Figure S4), these results indicate that PLDδ is likely activated by Spm from the inside of the cell, after its uptake.
Discussion
Polyamines Trigger a Charge-Dependent PA Response
Polyamines are naturally occurring polycationic molecules involved in a plethora of cellular events (Tiburcio et al., 2014), yet it is still largely unknown how this works at the molecular level. Here, a link to the formation of the lipid second messenger PA is reported, which is predominantly generated by PLDδ and plays a role in Spm induced-K+ efflux.
In this paper, we show that low μM concentrations of Spm trigger a rapid (minutes) PA response in the roots of Arabidopsis seedlings (Figure 2). In older plant material, Spm also triggered a PA response in leaves (Supplementary Figure S1), so multiple tissues are sensitive to Spm. Other polyamines, like Spd, Put and Dap were also able to trigger an accumulation in PA but this required higher concentrations, especially the diamines. In contrast, tSpm was as effective as Spm (Figure 1). These results indicate that the capacity of polyamines to activate the formation of PA is charge-dependent, with Spm4+ = tSpm4+>Spd3+>>Put2+, which may indicate an electrostatic interaction between the positive charges of the polyamine and a negatively charged target (Bertoluzza et al., 1988; Kurata et al., 2004; DeRouchey et al., 2010; Rudolphi-Skórska et al., 2014).
Triggering PA Response Requires Uptake of Polyamines
The saturation of the response at relatively low concentrations (Figure 2) may reflect a saturation of polyamine uptake (Pistocchi et al., 1987; Ditomaso et al., 1992a). In that regard, the Spm induced-PA responses were found to be strongly reduced in the plasma membrane localized PUT, RMV1-KO, also known as LAT1 or PUT3 (Fujita et al., 2012; Fujita and Shinozaki, 2014), while overexpression of RMV1 resulted in a much stronger PA response upon Spm treatment (Figure 3). The affinity of this transporter for Spm (Km = 0.6 μM), Spd (Km = 2.2 μM), and Put (Km = 56.5 μM), respectively (Fujita et al., 2012; Fujita and Shinozaki, 2014), is consistent with their potency to activate a PA response and indicates, together with the inhibition of the PA response by gadolinium (Figure 3), that polyamines are taken up before triggering a PA increase.
Link Between Polyamine Synthesis and PLDδ
Differential 32P-labeling experiments combined with transphosphatidylation assays revealed that part of the PA response was generated via the PLD pathway. Using T-DNA insertion PLD-KO mutants, we found that the majority of the PA is generated through PLDδ. Arabidopsis contains 3α-, 2β-, 3γ-, 1δ-, 1ε-, and 2ζ PLDs, which differ in amino acid-sequence conservation and lipid-binding domains (Bargmann and Munnik, 2006; Hong et al., 2016; Hou et al., 2016). PLDδ is typically localized in plasma membrane facing the cytosol (Wang and Wang, 2001; Pinosa et al., 2013) while all others PLDs are cytosolic though can transiently bind to various microsomal membranes (Wang, 2002). PLDδ has been implicated in drought and salinity stress (Katagiri et al., 2001; Bargmann et al., 2009; Distéfano et al., 2015) and in freezing tolerance (Li et al., 2004), which are stress responses in which polyamines have also been implicated to play a role (Alcázar et al., 2010). The specific involvement of PLDδ further implies that the polyamine induced-PA response predominantly occurs at the plasma membrane.
Both Spm and tSpm activated PLDδ equally well. In Arabidopsis, Spm is synthesized by spermine synthase (SPMS) while tSpm by ACAULIS5 (Panicot et al., 2002; Kakehi et al., 2008). Both are encoded by single genes, SPMS and ACL5, which are predominantly expressed in the phloem and xylem, respectively, in both roots and leaves (Supplementary Figure S5; Brady et al., 2007; Winter et al., 2007; Sagor et al., 2011; Yoshimoto et al., 2016). Interestingly, their expression strongly overlaps with that of PLDδ, especially SPMS, which are both strongly induced upon salt- and osmotic stress (Supplementary Figures S5, S6; Katagiri et al., 2001; Brady et al., 2007; Winter et al., 2007). So potentially, the local synthesis and/or transport of spermine could activate PLDδ to generate PA. In agreement, significantly increased PA levels were found in an SPMS-overexpressor line (35S::SPMS-9; Gonzalez et al., 2011) at control conditions (Supplementary Figure S7A). An spms-2 KO line exhibited normal PA levels, however, in response to salt stress, a strongly reduced PA response was found while the SPMS-overexpressor line revealed a much higher PA response than wt (Supplementary Figures 7A,B). These results support the idea of a novel, interesting link between PA and Spm in stress responses. Alternatively, since polyamines are rich in soil and produced by various microbes (Young and Chen, 1997; Chibucos and Morris, 2006; Zandonadi et al., 2013; Zhou et al., 2016), our observation that extracellular polyamines trigger intracellular PA responses may also reflect the action of natural, exogenous polyamines.
PA Function
Phosphatidic acid is an important plant phospholipid. Besides its role as precursor for all glycerolipids at the ER, PA has emerged as important lipid second messenger, generated through the PLC/DGK- and/or PLD pathways in response to various (a)biotic stresses, including plant defense, wounding, salt, drought, cold, and heat stress, where it is linked to various cellular processes, like vesicular trafficking, membrane fission and -fusion, and transport (Munnik et al., 2000; Testerink and Munnik, 2005, 2011; Vergnolle et al., 2005; Zhao, 2015; Hong et al., 2016; Hou et al., 2016). A local accumulation of PA in cellular membranes may affect the enzymatic- or structural properties of protein targets in that membrane or, alternatively, recruit cytosolic protein targets via PA-binding domains. PA targets include protein kinases, phosphatases, ion transporters, PEPC, GAPDH, NADPH oxidases (Rboh) (Kim et al., 2013; McLoughlin et al., 2013; Pleskot et al., 2013; Putta et al., 2016; Ufer et al., 2017). Here, evidence is provided for a role of PA in ion transport. MIFE analyses showed that Spm triggers a rapid efflux of K+ ions, which was strongly reduced in the pldδ mutant (Figure 7), indicating a direct or indirect role for PA in K+ gating. Associated to Spm uptake, we observed a fast net H+ influx (cytosolic acidification) followed by a gradual increase of H+ efflux (cytosolic alkalinisation), which correlated with the K+ efflux peak and its gradual recovery (Supplementary Figure S8). The cytosolic alkalinisation is related to the opening of voltage-gated inward-rectifying K+ channels (Kin) to compensate the K+ efflux (Dreyer and Uozumi, 2011; Karnik et al., 2016). In animal cells, PA has been shown to regulate voltage-gated potassium (Kv) channels and has been proposed to stabilize K+-inward channels (Kin) in its closed conformation, thus reducing K+ inward currents (Hite et al., 2014). In plants, PA has been shown to inactivate Kin channels in guard cells of Vicia faba and Arabidopsis (Jacob et al., 1999; Uraji et al., 2012), in which PLDδ has been implicated (Uraji et al., 2012). These observations are in agreement with a role for PA in regulating K+ fluxes of which the precise mechanism requires further investigation.
In summary, we provide molecular evidence that polyamines functionally require PLD and PA for their mode of action. This knowledge and the use of PLD- and polyamine synthesis mutants may shed new light on this phenomenon in other studies.
Data Availability
All datasets for this study are included in the manuscript and the Supplementary Files.
Author Contributions
XZ, SS, and TM designed the experiments. LS performed the MIFE experiments while XZ performed the rest. MF, AT, and MH added materials, ideas and discussions. XZ and TM wrote the manuscript.
Funding
This work was supported by the Spanish Ministerio de Ciencia e Innovación (BIO2011-29683 and CSD2007-00036), the Generalitat de Catalunya (SGR2009-1060 and BE DGR 2011), and the Netherlands Organisation for Scientific Research (NWO 867.15.020).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank L. Tikovsky and H. Lemereis for the assistance in the greenhouse, Dr. T. Takahashi (Okayama University, Japan) for supplying tSpm and Dr. I. Pottosin (University of Colima, Mexico) for discussion and critical review of the MIFE experiments.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00601/full#supplementary-material
References
Alcázar, R., Altabella, T., Marco, F., Bortolotti, C., Reymond, M., Koncz, C., et al. (2010). Polyamines: molecules with regulatory functions in plant abiotic stress tolerance. Planta 231, 1237–1249. doi: 10.1007/s00425-010-1130-1130
Alcázar, R., and Tiburcio, A. F. (2014). Plant polyamines in stress and development: an emerging area of research in plant sciences. Front. Plant Sci. 5:319. doi: 10.3389/fpls.2014.00319
Arisz, S. A., and Munnik, T. (2013). Distinguishing phosphatidic acid pools from de novo synthesis, PLD, and DGK. Methods Mol. Biol. 1009, 55–62. doi: 10.1007/978-1-62703-401-2_6
Armengaud, P., Zambaux, K., Hills, A., Sulpice, R., Pattison, R. J., Blatt, M. R., et al. (2009). EZ-Rhizo: integrated software for the fast and accurate measurement of root system architecture. Plant J. 57, 945–956. doi: 10.1111/j.1365-313X.2008.03739.x
Bachrach, U. (2010). The early history of polyamine research. Plant Physiol. Biochem. 48, 490–495. doi: 10.1016/j.plaphy.2010.02.003
Bagni, N., and Pistocchi, R. (1988). Polyamines as growth substances in higher plants. Adv. Exp. Med. Biol. 250, 547–558. doi: 10.1007/978-1-4684-5637-0_49
Bargmann, B. O. R., Laxalt, A. M., ter Riet, B., van Schooten, B., Merquiol, E., Testerink, C., et al. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50, 78–89. doi: 10.1093/pcp/pcn173
Bargmann, B. O. R., and Munnik, T. (2006). The role of phospholipase D in plant stress responses. Curr. Opin. Plant Biol. 9, 515–522. doi: 10.1016/j.pbi.2006.07.011
Bertoluzza, A., Bonora, S., Fini, G., and Morelli, M. A. (1988). Spectroscopic and calorimetric studies of phospholipid-polyamine molecular interactions. J. Raman Spectrosc. 19, 369–373. doi: 10.1002/jrs.1250190512
Bose, J., Shabala, L., Pottosin, I., Zeng, F., Velarde-Buendía, A.-M., Massart, A., et al. (2014). Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K(+) -permeable channels to reactive oxygen species: physiological traits that differentiate salinity tolerance between pea and barley. Plant Cell Environ. 37, 589–600. doi: 10.1111/pce.12180
Brady, S. M., Orlando, D. A., Lee, J.-Y., Wang, J. Y., Koch, J., Dinneny, J. R., et al. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806. doi: 10.1126/science.1146265
Chibucos, M. C., and Morris, P. F. (2006). Levels of polyamines and kinetic characterization of their uptake in the soybean pathogen Phytophthora sojae. Appl. Environ. Microbiol. 72, 3350–3356. doi: 10.1128/AEM.72.5.3350-3356.2006
Cona, A., Rea, G., Angelini, R., Federico, R., and Tavladoraki, P. (2006). Functions of amine oxidases in plant development and defence. Trends Plant Sci. 11, 80–88. doi: 10.1016/j.tplants.2005.12.009
Couée, I., Hummel, I., Sulmon, C., Gouesbet, G., and El Amrani, A. (2004). Involvement of polyamines in root development. Plant Cell. Tissue Organ. Cult. 76, 1–10. doi: 10.1023/A:1025895731017
de Agazio, M., Grego, S., Ciofi-Luzzatto, A., Rea, E., Zaccaria, M. L., and Federico, R. (1995). Inhibition of maize primary root elongation by spermidine: effect on cell shape and mitotic index. J. Plant Growth Regul. 14, 85–89. doi: 10.1007/BF00203118
DeRouchey, J., Parsegian, V. A., and Rau, D. C. (2010). Cation charge dependence of the forces driving DNA assembly. Biophys. J. 99, 2608–2615. doi: 10.1016/j.bpj.2010.08.028
Dinneny, J. R., Long, T. A., Wang, J. Y., Jung, J. W., Mace, D., Pointer, S., et al. (2008). Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945. doi: 10.1126/science.1153795
Distéfano, A. M., García-Mata, C., Lamattina, L., and Laxalt, A. M. (2008). Nitric oxide-induced phosphatidic acid accumulation: a role for phospholipases C and D in stomatal closure. Plant. Cell Environ. 31, 187–194. doi: 10.1111/j.1365-3040.2007.01756.x
Distéfano, A. M., Scuffi, D., García-Mata, C., Lamattina, L., and Laxalt, A. M. (2012). Phospholipase Dδ is involved in nitric oxide-induced stomatal closure. Planta 236, 1899–1907. doi: 10.1007/s00425-012-1745-1744
Distéfano, A. M., Valiñas, M. A., Scuffi, D., Lamattina, L., ten Have, A., García-Mata, C., et al. (2015). Phospholipase Dδ knock-out mutants are tolerant to severe drought stress. Plant Signal. Behav. 10:e1089371. doi: 10.1080/15592324.2015.1089371
Ditomaso, J. M., Hart, J. J., and Kochian, L. V. (1992a). Transport kinetics and metabolism of exogenously applied putrescine in roots of intact maize seedlings. Plant Physiol. 98, 611–620. doi: 10.1104/pp.98.2.611
Ditomaso, J. M., Hart, J. J., Linscott, D. L., and Kochian, L. V. (1992b). Effect of inorganic cations and metabolic inhibitors on putrescine transport in roots of intact maize seedlings. Plant Physiol. 99, 508–514. doi: 10.1104/pp.99.2.508
Dreyer, I., and Uozumi, N. (2011). Potassium channels in plant cells. FEBS J. 278, 4293–4303. doi: 10.1111/j.1742-4658.2011.08371.x
Echevarría-Machado, I., Ku-González, A., Loyola-Vargas, V. M., and Hernández-Sotomayor, S. M. T. (2004). Interaction of spermine with a signal transduction pathway involving phospholipase C, during the growth of Catharanthus roseus transformed roots. Physiol. Plant 120, 140–151. doi: 10.1111/j.0031-9317.2004.0212.x
Echevarría-Machado, I., Muñoz-Sánchez, A., Loyola-Vargas, V. M., and Hernández-Sotomayor, S. M. T. (2002). Spermine stimulation of phospholipase C from Catharanthus roseus transformed roots. J. Plant Physiol. 159, 1179–1188. doi: 10.1078/0176-1617-1893
Echevarría-Machado, I., Ramos-Díaz, A., Brito-Argáez, L., Racagni-Di Palma, G., Loyola-Vargas, V. M., and Hernández-Sotomayor, S. M. T. (2005). Polyamines modify the components of phospholipids-based signal transduction pathway in Coffea arabica L. cells. Plant Physiol. Biochem. 43, 874–881. doi: 10.1016/j.plaphy.2005.08.013
Friedman, R., Levin, N., and Altman, A. (1986). Presence and identification of polyamines in xylem and Phloem exudates of plants. Plant Physiol. 82, 1154–1157. doi: 10.1104/pp.82.4.1154
Fujita, M., Fujita, Y., Iuchi, S., Yamada, K., Kobayashi, Y., Urano, K., et al. (2012). Natural variation in a polyamine transporter determines paraquat tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 6343–6347. doi: 10.1073/pnas.1121406109
Fujita, M., and Shinozaki, K. (2014). Identification of polyamine transporters in plants: paraquat transport provides crucial clues. Plant Cell Physiol. 55, 855–861. doi: 10.1093/pcp/pcu032
Galston, A. W., and Kaur-Sawhney, R. (1995). “Polyamines as endogenous growth regulators,” in Plant Hormones: Physiology, Biochemistry and Molecular Biology, 2nd Edn, ed. P. J. Davis (Dordrecht: Springer), 158–178. doi: 10.1007/978-94-011-0473-9_8
Galvan-Ampudia, C. S., Julkowska, M. M., Darwish, E., Gandullo, J., Korver, R. A., Brunoud, G., et al. (2013). Halotropism is a response of plant roots to avoid a saline environment. Curr. Biol. 23, 2044–2050. doi: 10.1016/j.cub.2013.08.042
Gonzalez, M. E., Marco, F., Minguet, E. G., Carrasco-Sorli, P., Blazquez, M. A., Carbonell, J., et al. (2011). Perturbation of spermine synthase gene expression and transcript profiling provide new insights on the role of the tetraamine spermine in Arabidopsis defense against Pseudomonas viridiflava. Plant Physiol. 156, 2266–2277. doi: 10.1104/pp.110.171413
Guo, L., Devaiah, S. P., Narasimhan, R., Pan, X., Zhang, Y., Zhang, W., et al. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24, 2200–2212. doi: 10.1105/tpc.111.094946
Haber, M. T., Fukui, T., Lebowitz, M. S., and Lowenstein, J. M. (1991). Activation of phosphoinositide-specific phospholipase Cδ from rat liver by polyamines and basic proteins. Arch. Biochem. Biophys. 288, 243–249. doi: 10.1016/0003-9861(91)90191-K
Hite, R. K., Butterwick, J. A., MacKinnon, R., Dart, C., Dickson, E., Falkenburger, B., et al. (2014). Phosphatidic acid modulation of Kv channel voltage sensor function. eLife 3, 3169–3178. doi: 10.7554/eLife.04366
Hong, Y., Devaiah, S. P., Bahn, S. C., Thamasandra, B. N., Li, M., Welti, R., et al. (2009). Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58, 376–387. doi: 10.1111/j.1365-313X.2009.03788.x
Hong, Y., Pan, X., Welti, R., and Wang, X. (2008). Phospholipase Da3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20, 803–816. doi: 10.1105/tpc.107.056390
Hong, Y., Zhao, J., Guo, L., Kim, S.-C., Deng, X., Wang, G., et al. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress. Plant. Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Jacob, T., Ritchie, S., Assmann, S. M., and Gilroy, S. (1999). Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Natl. Acad. Sci. U.S.A. 96, 12192–12197. doi: 10.1073/pnas.96.21.12192
Jurkowska, G., Rydzewska, G., and Andrzejewska, A. (1997). The influence of polyamines synthesis inhibition on pancreas regeneration and phospholipase D activity after acute caerulein induced pancreatitis in rats. Biochemical and ultrastructural study. J. Physiol. Pharmacol. 48, 789–804.
Kakehi, J.-I., Kuwashiro, Y., Niitsu, M., and Takahashi, T. (2008). Thermospermine is required for stem elongation in Arabidopsis thaliana. Plant Cell Physiol. 49, 1342–1349. doi: 10.1093/pcp/pcn109
Karnik, R., Waghmare, S., Zhang, B., Larson, E., Lefoulon, C., Gonzalez, W., et al. (2016). Commandeering channel voltage sensors for secretion, cell turgor, and volume control. Trends Plant Sci. 22, 81–95. doi: 10.1016/j.tplants.2016.10.006
Katagiri, T., Takahashi, S., and Shinozaki, K. (2001). Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. 26, 595–605. doi: 10.1046/j.1365-313x.2001.01060.x
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, S.-C., Guo, L., and Wang, X. (2013). Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J. Biol. Chem. 288, 11834–11844. doi: 10.1074/jbc.M112.427229
Kimura, T., Imamura, K., Eckhardt, L., and Schulz, I. (1986). Ca2+-, phorbol ester-, and cAMP-stimulated enzyme secretion from permeabilized rat pancreatic acini. Am. J. Physiol. 250, G698–G708.
Kooijman, E. E., Chupin, V., de Kruijff, B., and Burger, K. N. J. (2003). Modulation of membrane curvature by phosphatidic acid and lysophosphatidic acid. Traffic 4, 162–174. doi: 10.1034/j.1600-0854.2003.00086.x
Kooijman, E. E., Tieleman, D. P., Testerink, C., Munnik, T., Rijkers, D. T. S., Burger, K. N. J., et al. (2007). An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 282, 11356–11364. doi: 10.1074/jbc.M609737200
Kurata, H. T., Phillips, L. R., Rose, T., Loussouarn, G., Herlitze, S., Fritzenschaft, H., et al. (2004). Molecular basis of inward rectification: polyamine interaction sites located by combined channel and ligand mutagenesis. J. Gen. Physiol. 124, 541–554. doi: 10.1085/jgp.200409159
Lanteri, M. L., Laxalt, A. M., and Lamattina, L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase D during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147, 188–198. doi: 10.1104/pp.107.111815
Li, J., Mu, J., Bai, J., Fu, F., Zou, T., An, F., et al. (2013). Paraquat resistant1, a golgi-localized putative transporter protein, is involved in intracellular transport of paraquat. Plant Physiol. 162, 470–483. doi: 10.1104/pp.113.213892
Li, W., Li, M., Zhang, W., Welti, R., and Wang, X. (2004). The plasma membrane–bound phospholipase Dδ enhances freezing tolerance in Arabidopsis thaliana. Nat. Biotechnol. 22, 427–433. doi: 10.1038/nbt949
Lu, S., Su, W., Li, H., and Guo, Z. (2009). Abscisic acid improves drought tolerance of triploid bermudagrass and involves H2O2- and NO-induced antioxidant enzyme activities. Plant Physiol. Biochem. 47, 132–138. doi: 10.1016/j.plaphy.2008.10.006
Madesh, M., and Balasubramanian, K. A. (1997). Activation of intestinal mitochondrial phospholipase D by polyamines and monoamines. Biochim. Biophys. Acta 1348, 324–330. doi: 10.1016/s0005-2760(97)00074-x
Martinis, J., Gas-Pascual, E., Szydlowski, N., Crèvecoeur, M., Gisler, A., Bürkle, L., et al. (2016). Long-distance transport of thiamine (vitamin B1) is concomitant with that of polyamines. Plant Physiol. 171, 542–553. doi: 10.1104/pp.16.00009
McLoughlin, F., Arisz, S. A., Dekker, H. L., Kramer, G., de Koster, C. G., Haring, M. A., et al. (2013). Identification of novel candidate phosphatidic acid-binding proteins involved in the salt-stress response of Arabidopsis thaliana roots. Biochem. J. 450, 573–581. doi: 10.1042/BJ20121639
Michael, A. J. (2016). Polyamines in eukaryotes, bacteria, and archaea. J. Biol. Chem. 291, 14896–14903. doi: 10.1074/jbc.R116.734780
Miller-Fleming, L., Olin-Sandoval, V., Campbell, K., and Ralser, M. (2015). Remaining mysteries of molecular biology: the role of polyamines in the cell. J. Mol. Biol. 427, 3389–3406. doi: 10.1016/j.jmb.2015.06.020
Moschou, P. N., Paschalidis, K. A., Delis, I. D., Andriopoulou, A. H., Lagiotis, G. D., Yakoumakis, D. I., et al. (2008). Spermidine exodus and oxidation in the apoplast induced by abiotic stress is responsible for H2O2 signatures that direct tolerance responses in tobacco. Plant Cell 20, 1708–1724. doi: 10.1105/tpc.108.059733
Mulangi, V., Chibucos, M. C., Phuntumart, V., and Morris, P. F. (2012). Kinetic and phylogenetic analysis of plant polyamine uptake transporters. Planta 236, 1261–1273. doi: 10.1007/s00425-012-1668-1660
Munnik, T. (2001). Phosphatidic acid – an emerging plant lipid second messenger. Trends Plant Sci. 6, 227–233.
Munnik, T. (2014). “PI-PLC: phosphoinositide-phospholipase C in plant signaling,” in Phospholipases in Plant Signaling. Signaling and Communication in Plants, ed. X. Wang (Heidelberg: Springer), 27–54. doi: 10.1007/978-3-642-42011-5_2
Munnik, T., Arisz, S. A., De Vrije, T., and Musgrave, A. (1995). G protein activation stimulates phospholipase D signaling in plants. Plant Cell 7, 2197–2210. doi: 10.1105/tpc.7.12.2197
Munnik, T., and Laxalt, A. M. (2013). Measuring PLD activity in vivo. Methods Mol. Biol. 1009, 219–231. doi: 10.1007/978-1-62703-401-2_20
Munnik, T., Meijer, H. J., Ter Riet, B., Hirt, H., Frank, W., Bartels, D., et al. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. doi: 10.1046/j.1365-313x.2000.00725.x
Munnik, T., Musgrave, A., and de Vrije, T. (1994). Rapid turnover of polyphosphoinositides in carnation flower petals. Planta 193, 89–98. doi: 10.1007/BF00191611
Munnik, T., Van Himbergen, J. A. J., Ter Riet, B., Braun, F. J., Irvine, R. F., Van Den Ende, H., et al. (1998). Detailed analysis of the turnover of polyphosphoinositides and phosphatidic acid upon activation of phospholipases C and D in Chlamydomonas cells treated with non-permeabilizing concentrations of mastoparan. Planta 207, 133–145. doi: 10.1007/s004250050465
Munnik, T., and Zarza, X. (2013). Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol. Biol. 1009, 3–15. doi: 10.1007/978-1-62703-401-2_1
Newman, I. A. (2001). Ion transport in roots: measurement of fluxes using ion-selective microelectrodes to characterize transporter function. Plant Cell Environ. 24, 1–14. doi: 10.1046/j.1365-3040.2001.00661.x
Ozawa, R., Bertea, C. M., Foti, M., Narayana, R., Arimura, G.-I., Muroi, A., et al. (2010). Polyamines and jasmonic acid induce plasma membrane potential variations in Lima bean. Plant Signal. Behav. 5, 308–310. doi: 10.4161/psb.5.3.10848
Pandolfi, C., Pottosin, I., Cuin, T., Mancuso, S., and Shabala, S. (2010). Specificity of polyamine effects on NaCl-induced ion flux kinetics and salt stress amelioration in plants. Plant Cell Physiol. 51, 422–434. doi: 10.1093/pcp/pcq007
Panicot, M., Minguet, E. G., Ferrando, A., Alcázar, R., Blázquez, M. A., Carbonell, J., et al. (2002). A polyamine metabolon involving aminopropyl transferase complexes in Arabidopsis. Plant Cell 14, 2539–2551. doi: 10.1105/TPC.004077
Pawelczyk, T., and Lowenstein, J. M. (1997). The effect of different molecular species of sphingomyelin on phospholipase Cδ1 activity. Biochimie 79, 741–748. doi: 10.1016/s0300-9084(97)86932-5
Pawelczyk, T., and Matecki, A. (1998). Localization of phospholipase Cδ3 in the cell and regulation of its activity by phospholipids and calcium. Eur. J. Biochem. 257, 169–177. doi: 10.1046/j.1432-1327.1998.2570169.x
Pei, Z.-M., Murata, Y., Benning, G., Thomine, S., Klüsener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Periyasamy, S., Kothapalli, M. R., and Hoss, W. (1994). Regulation of the phosphoinositide cascade by polyamines in brain. J. Neurochem. 63, 1319–1327. doi: 10.1046/j.1471-4159.1994.63041319.x
Pinosa, F., Buhot, N., Kwaaitaal, M., Fahlberg, P., Thordal-Christensen, H., Ellerstrom, M., et al. (2013). Arabidopsis phospholipase Dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 163, 896–906. doi: 10.1104/pp.113.223503
Pistocchi, R., Bagni, N., and Creus, J. A. (1987). Polyamine uptake in carrot cell cultures. Plant Physiol. 84, 374–380. doi: 10.1104/pp.84.2.374
Pistocchi, R., Keller, F., Bagni, N., and Matile, P. (1988). Transport and subcellular localization of polyamines in carrot protoplasts and vacuoles. Plant Physiol. 87, 514–518. doi: 10.1104/pp.87.2.514
Pleskot, R., Li, J., Žárskı, V., Potockı, M., and Staiger, C. J. (2013). Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 18, 496–504. doi: 10.1016/j.tplants.2013.04.005
Pommerrenig, B., Feussner, K., Zierer, W., Rabinovych, V., Klebl, F., Feussner, I., et al. (2011). Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis. Plant Cell 23, 1904–1919. doi: 10.1105/tpc.110.079657
Pottosin, I., and Shabala, S. (2014). Polyamines control of cation transport across plant membranes: implications for ion homeostasis and abiotic stress signaling. Front. Plant Sci. 5:154. doi: 10.3389/fpls.2014.00154
Pottosin, I., Velarde-Buendía, A. M., Bose, J., Fuglsang, A. T., and Shabala, S. (2014). Polyamines cause plasma membrane depolarization, activate Ca2+-, and modulate H+-ATPase pump activity in pea roots. J. Exp. Bot. 65, 2463–2472. doi: 10.1093/jxb/eru133
Putta, P., Rankenberg, J., Korver, R. A., van Wijk, R., Munnik, T., Testerink, C., et al. (2016). Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim. Biophys. Acta Biomembr. 1858, 2709–2716. doi: 10.1016/j.bbamem.2016.07.014
Raghu, P., Manifava, M., Coadwell, J., and Ktistakis, N. T. (2009). Emerging findings from studies of phospholipase D in model organisms (and a short update on phosphatidic acid effectors). Biochim. Biophys. Acta 1791, 889–897. doi: 10.1016/j.bbalip.2009.03.013
Raho, N., Ramirez, L., Lanteri, M. L., Gonorazky, G., Lamattina, L., ten Have, A., et al. (2011). Phosphatidic acid production in chitosan-elicited tomato cells, via both phospholipase D and phospholipase C/diacylglycerol kinase, requires nitric oxide. J. Plant Physiol. 168, 534–539. doi: 10.1016/j.jplph.2010.09.004
Rentsch, D., Schmidt, S., and Tegeder, M. (2007). Transporters for uptake and allocation of organic nitrogen compounds in plants. FEBS Lett. 581, 2281–2289. doi: 10.1016/j.febslet.2007.04.013
Roth, M. G. (2008). Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x
Rudolphi-Skórska, E., Zembala, M., and Filek, M. (2014). Mechanical and electrokinetic effects of polyamines/phospholipid interactions in model membranes. J. Membr. Biol. 247, 81–92. doi: 10.1007/s00232-013-9614-z
Sagawa, N., Bleasdale, J. E., and Di Renzo, G. C. (1983). The effects of polyamines and aminoglycosides on phosphatidylinositol-specific phospholipase C from human amnion. Biochim. Biophys. Acta 752, 153–161. doi: 10.1016/0005-2760(83)90243-90246
Sagor, G. H. M., Yamaguchi, K., Watanabe, K., Berberich, T., Kusano, T., and Takahashi, Y. (2011). Spatio-temporal expression analysis of Arabidopsis thaliana spermine synthase gene promoter Gene Note. Plant Biotechnol. 28, 407–411. doi: 10.5511/plantbiotechnology.11.0704a
Shabala, L., Ross, T., McMeekin, T., and Shabala, S. (2006). Non-invasive microelectrode ion flux measurements to study adaptive responses of microorganisms to the environment. FEMS Microbiol. Rev. 30, 472–486. doi: 10.1111/j.1574-6976.2006.00019.x
Sjöholm, A., Arkhammar, P., Welsh, N., Bokvist, K., Rorsman, P., Hallberg, A., et al. (1993). Enhanced stimulus-secretion coupling in polyamine-depleted rat insulinoma cells. An effect involving increased cytoplasmic Ca2+, inositol phosphate generation, and phorbol ester sensitivity. J. Clin. Invest. 92, 1910–1917. doi: 10.1172/JCI116784
Smith, C. D., and Snyderman, R. (1988). Modulation of inositol phospholipid metabolism by polyamines. Biochem. J. 256, 125–130. doi: 10.1042/bj2560125
Sood, S., and Nagar, P. K. (2005). Xylem and phloem derived polyamines during flowering in two diverse rose species. J. Plant Growth Regul. 24, 36–40. doi: 10.1007/s00344-004-0026-22
Späth, M., Woscholski, R., and Schächtele, C. (1991). Characterization of multiple forms of phosphoinositide-specific phospholipase C from bovine aorta. Cell. Signal. 3, 305–310. doi: 10.1016/0898-6568(91)90059-90054
Strohm, A. K., Vaughn, L. M., and Masson, P. H. (2015). Natural variation in the expression of ORGANIC CATION TRANSPORTER 1 affects root length responses to cadaverine in Arabidopsis. J. Exp. Bot. 66, 853–862. doi: 10.1093/jxb/eru444
Takahashi, Y., Berberich, T., Miyazaki, A., Seo, S., Ohashi, Y., and Kusano, T. (2003). Spermine signalling in tobacco: activation of mitogen-activated protein kinases by spermine is mediated through mitochondrial dysfunction. Plant J. 36, 820–829. doi: 10.1046/j.1365-313X.2003.01923.x
Testerink, C., and Munnik, T. (2005). Phosphatidic acid: a multifunctional stress signaling lipid in plants. Trends Plant Sci. 10, 368–375. doi: 10.1016/j.tplants.2005.06.002
Testerink, C., and Munnik, T. (2011). Molecular, cellular, and physiological responses to phosphatidic acid formation in plants. J. Exp. Bot. 62, 2349–2361. doi: 10.1093/jxb/err079
Tiburcio, A. F., Altabella, T., Bitrián, M., and Alcázar, R. (2014). The roles of polyamines during the lifespan of plants: from development to stress. Planta 240, 1–18. doi: 10.1007/s00425-014-2055-2059
Tisi, A., Federico, R., Moreno, S., Lucretti, S., Moschou, P. N., Roubelakis-Angelakis, K. A., et al. (2011). Perturbation of polyamine catabolism can strongly affect root development and xylem differentiation. Plant Physiol. 157, 200–215. doi: 10.1104/pp.111.173153
Tong, W., Imai, A., Tabata, R., Shigenobu, S., Yamaguchi, K., Yamada, M., et al. (2016). Polyamine resistance is increased by mutations in a nitrate transporter gene NRT1.3 (AtNPF6.4) in Arabidopsis thaliana. Front. Plant Sci. 7:834. doi: 10.3389/fpls.2016.00834
Toumi, I., Moschou, P. N., Paschalidis, K. A., Bouamama, B., Ben Salem-Fnayou, A., Ghorbel, A. W., et al. (2010). Abscisic acid signals reorientation of polyamine metabolism to orchestrate stress responses via the polyamine exodus pathway in grapevine. J. Plant Physiol. 167, 519–525. doi: 10.1016/j.jplph.2009.10.022
Tun, N. N. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47, 346–354. doi: 10.1093/pcp/pci252
Ufer, G., Gertzmann, A., Gasulla, F., Röhrig, H., and Bartels, D. (2017). Identification and characterization of the phosphatidic acid-binding A. thaliana phosphoprotein PLDrp1 that is regulated by PLDα1 in a stress-dependent manner. Plant J. 92, 276–290. doi: 10.1111/tpj.13651
Uraji, M., Katagiri, T., Okuma, E., Ye, W., Hossain, M. A., Masuda, C., et al. (2012). Cooperative function of PLD and PLD 1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 159, 450–460. doi: 10.1104/pp.112.195578
Vergnolle, C., Vaultier, M.-N., Taconnat, L., Renou, J.-P., Kader, J.-C., Zachowski, A., et al. (2005). The cold-induced early activation of phospholipase C and D pathways determines the response of two distinct clusters of genes in Arabidopsis cell suspensions. Plant Physiol. 139, 1217–1233. doi: 10.1104/pp.105.068171
Verrey, F., Closs, E. I., Wagner, C. A., Palacin, M., Endou, H., and Kanai, Y. (2004). CATs and HATs: the SLC7 family of amino acid transporters. Pflugers Arch. Eur. J. Physiol. 447, 532–542. doi: 10.1007/s00424-003-1086-z
Wallace, H. M. (2009). The polyamines: past, present and future. Essays Biochem. 46, 1–9. doi: 10.1042/bse0460001
Wang, C., and Wang, X. (2001). A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiol. 127, 1102–1112. doi: 10.1104/pp.127.3.1102
Wang, X. (2002). Phospholipase D in hormonal and stress signaling. Curr. Opin. Plant Biol. 5, 408–414. doi: 10.1016/S1369-5266(02)00283-282
Wang, X., Devaiah, S. P., Zhang, W., and Welti, R. (2006). Signaling functions of phosphatidic acid. Prog. Lipid Res. 45, 250–278. doi: 10.1016/j.plipres.2006.01.005
Wimalasekera, R., Tebartz, F., and Scherer, G. F. E. (2011). Polyamines, polyamine oxidases and nitric oxide in development, abiotic and biotic stresses. Plant Sci. 181, 593–603. doi: 10.1016/j.plantsci.2011.04.002
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2:e718. doi: 10.1371/journal.pone.0000718
Wojcikiewicz, R. J., and Fain, J. N. (1988). Polyamines inhibit phospholipase C-catalysed polyphosphoinositide hydrolysis. Studies with permeabilized GH3 cells. Biochem. J. 255, 1015–1021. doi: 10.1042/bj2551015
Yao, H.-Y., and Xue, H.-W. (2018). Phosphatidic acid plays key roles regulating plant development and stress responses. J. Integr. Plant Biol. 60, 851–863. doi: 10.1111/jipb.12655
Yokota, T., Nakayama, M., Harasawa, I., Sato, M., Katsuhara, M., and Kawabe, S. (1994). Polyamines, indole-3-acetic acid and abscisic acid in rice phloem sap. Plant Growth Regul. 15, 125–128. doi: 10.1007/BF00024101
Yoshimoto, K., Takamura, H., Kadota, I., Motose, H., Takahashi, T., Igarashi, K., et al. (2016). Chemical control of xylem differentiation by thermospermine, xylemin, and auxin. Sci. Rep. 6:21487. doi: 10.1038/srep21487
Young, C. C., and Chen, L. F. (1997). Polyamines in humic acid and their effect on radical growth of lettuce seedlings. Plant Soil 195, 143–149. doi: 10.1023/A:1004247302388
Zandonadi, D. B., Santos, M. P., Busato, J. G., Peres, L. E. P., and Façanha, A. R. (2013). Plant physiology as affected by humified organic matter. Theor. Exp. Plant Physiol. 25, 13–25. doi: 10.1590/S2197-00252013000100003
Zepeda-Jazo, I., Velarde-Buendía, A. M., Enríquez-Figueroa, R., Bose, J., Shabala, S., Muñiz-Murguía, J., et al. (2011). Polyamines interact with hydroxyl radicals in activating Ca(2+) and K(+) transport across the root epidermal plasma membranes. Plant Physiol. 157, 2167–2180. doi: 10.1104/pp.111.179671
Zhang, W., Wang, C., Qin, C., Wood, T., Olafsdottir, G., Welti, R., et al. (2003). The oleate-stimulated phospholipase D, PLDdelta, and phosphatidic acid decrease H2O2-induced cell death in Arabidopsis. Plant Cell 15, 2285–2295. doi: 10.1105/tpc.013961
Zhang, W., Yu, L., Zhang, Y., and Wang, X. (2005). Phospholipase D in the signaling networks of plant response to abscisic acid and reactive oxygen species. Biochim. Biophys. Acta 1736, 1–9. doi: 10.1016/j.bbalip.2005.07.004
Zhang, Y., Zhu, H., Zhang, Q., Li, M., Yan, M., Wang, R., et al. (2009). Phospholipase da1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. Plant Cell 21, 2357–2377. doi: 10.1105/tpc.108.062992
Zhao, J. (2015). Phospholipase D and phosphatidic acid in plant defence response: from protein–protein and lipid–protein interactions to hormone signalling. J. Exp. Bot. 66, 1721–1736. doi: 10.1093/jxb/eru540
Keywords: phospholipase D (PLD), phosphatidic acid (PA), lipid signaling, polyamines (putrescine, spermidine, spermine), phospholipids, MIFE
Citation: Zarza X, Shabala L, Fujita M, Shabala S, Haring MA, Tiburcio AF and Munnik T (2019) Extracellular Spermine Triggers a Rapid Intracellular Phosphatidic Acid Response in Arabidopsis, Involving PLDδ Activation and Stimulating Ion Flux. Front. Plant Sci. 10:601. doi: 10.3389/fpls.2019.00601
Received: 05 March 2019; Accepted: 24 April 2019;
Published: 21 May 2019.
Edited by:
Luo Jie, Huazhong Agricultural University, ChinaReviewed by:
Rebecca L. Roston, University of Nebraska–Lincoln, United StatesThomas Berberich, Senckenberg Nature Research Society, Germany
Copyright © 2019 Zarza, Shabala, Fujita, Shabala, Haring, Tiburcio and Munnik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teun Munnik, dC5tdW5uaWtAdXZhLm5s
 Xavier Zarza
Xavier Zarza Lana Shabala
Lana Shabala Miki Fujita
Miki Fujita Sergey Shabala
Sergey Shabala Michel A. Haring
Michel A. Haring Antonio F. Tiburcio
Antonio F. Tiburcio Teun Munnik
Teun Munnik