- Global Wheat Program, International Maize and Wheat Improvement Center (CIMMYT), Texcoco, Mexico
Aegilops species have significantly contributed to wheat breeding despite the difficulties involved in the handling of wild species, such as crossability and incompatibility. A number of biotic resistance genes have been identified and incorporated into wheat varieties from Aegilops species, and this genus is also contributing toward improvement of complex traits such as yield and abiotic tolerance for drought and heat. The D genome diploid species of Aegilops tauschii has been utilized most often in wheat breeding programs. Other Aegilops species are more difficult to utilize in the breeding because of lower meiotic recombination frequencies; generally they can be utilized only after extensive and time-consuming procedures in the form of translocation/introgression lines. After the emergence of Ug99 stem rust and wheat blast threats, Aegilops species gathered more attention as a form of new resistance sources. This article aims to update recent progress on Aegilops species, as well as to cover new topics around their use in wheat breeding.
Introduction
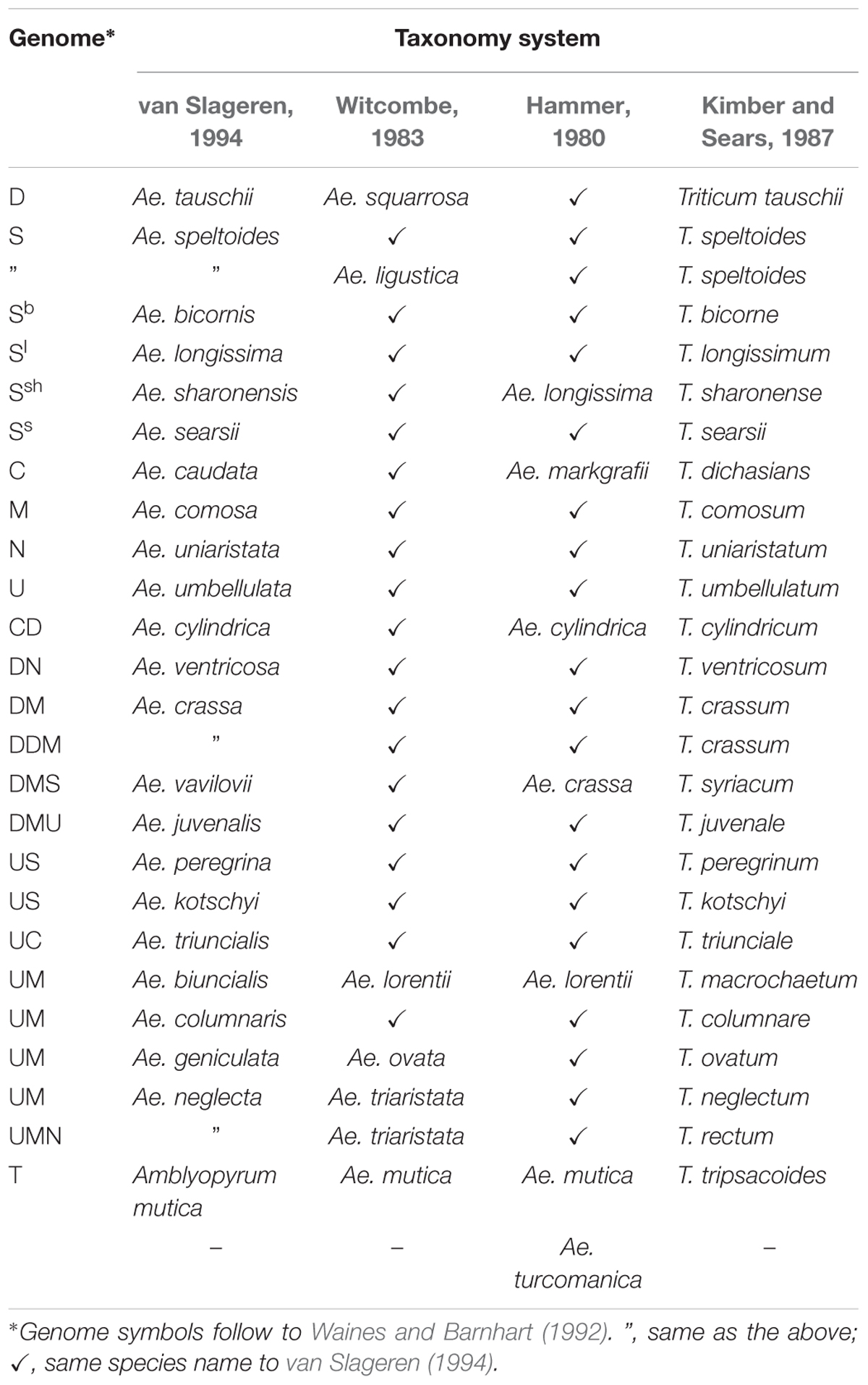
According to the latest revision of Aegilops L. taxonomy, (van Slageren system) which is used by most researchers (including this article), Aegilops consists of 23 species, having the D, S, U, C, N, and M genomes (van Slageren, 1994). Since the taxonomy has frequently changed (Kihara, 1954; Hammer, 1980; Witcombe, 1983; Kimber and Sears, 1987; van Slageren, 1994) this has led to some confusion about species names, and so a list of Aegilops species is provided below (Table 1). The biggest change from the previous taxonomy system is that Ae. mutica Boiss. has been removed and assigned a new species name: Ambylopyrum muticum (Boiss.) Eig. In the future, it will also be possible to make further modifications to reflect molecular findings (Edet et al., 2018).
One of the most important aspects of Aegilops is that it is closely related to bread wheat Triticum aestivum L. (AABBDD), which is one of the most important calorie sources for human nutrition. The D genome originated from the diploid species of Aegilops tauschii Coss. ( = Ae. squarrosa L.) (Kihara, 1946; McFadden and Sears, 1946), and the B genome was derived from a closely related species to Ae. speltoides Tausch (Riley et al., 1958; Sasanuma et al., 1996; Petersen et al., 2006; Kilian et al., 2007; Zhang W. et al., 2018) which has the S genome. Aegilops species are distributed from Europe to western China in a species-specific manner (van Slageren, 1994), adapted to many different climatic zones including drought/heat environments, different disease hot spots and nutrient-poor areas. It has been reported that Aegilops possesses useful traits for wheat breeding (For review to see; Kilian et al., 2011) including drought tolerance (Damania et al., 1992; Waines et al., 1993; Rekika et al., 1998; Monneveux et al., 2000; Farooq and Azam, 2001), heat tolerance (Waines, 1994), salinity (Colmer et al., 2006), aluminum toxicity tolerance (Miller et al., 1995) and resistance to several pests and diseases such as rust (Mihova, 1988; Anikster et al., 2005; Liu et al., 2010; Rouse et al., 2011; Vikas et al., 2014; Huang S. et al., 2018; Olivera et al., 2018), powdery mildew (Lutz et al., 1994; Buloichik et al., 2008), Hessian fly (El Bouhssini et al., 2008), cereal aphid (Holubec and Havlıckova, 1994) and barley yellow dwarf virus (BYDV) (Makkouk et al., 1994). In addition, the species can adapt to low phosphorous environments (Liu et al., 2015) and can contribute to higher iron and zinc content in wheat grain (Rawat et al., 2009).
In order to effectively exploit these useful traits in wheat, it is necessary to overcome extra difficulties with the introgression process, including a hybridization barriers, incompatibilities/hybrid abnormalities, sterility of F1s and, reduced meiotic chromosome pairings. Despite these obstacles, many Aegilops genes have been transferred to wheat and have been heavily utilized over the last 60 years (For review to see; Schneider et al., 2008; Kilian et al., 2011). Aegilops is also contributing to abate two recent threats to the global wheat production: Ug99 stem rust race derivatives and wheat blast (Magnaporthe oryzae Triticum). When Ug99 (original pathotype TTKSK) appeared in the early 2000s (Pretorius et al., 2000), more than 80% of wheat varieties did not have resistance against the race (Pretorius et al., 2000) and as such, wheat breeders sought resistance traits in Aegilops. When Wheat blast disease emerged in Bangladesh in 2016 (Ceresini et al., 2018), resistant wheat varieties were non-existent in the country, as well as neighboring India. Yet, a resistant variety was released within 2 years because of a resistance gene from Aegilops that was previously introgressed and ready for use (Cruz et al., 2016; Velu et al., 2018a; Mahmud, 2019).
In this paper, I will first review some difficulties relating to the use of Aegilops species (Supplementary Figure S1). Then, I will provide information on the contribution of Aegilops to wheat breeding in terms of identified genes in Aegilops, as well as some recent information on how Aegilops has contributed to the crisis prevention of Ug99 stem rust and wheat blast disease, which may change perspectives of Aegilops species as important sources for wheat breeding.
Hybridization Barriers Between Wheat and Aegilops Species and Crossability Genes
To utilize the genetic resources in the Aegilops genus, it is necessary to first produce hybrids between wheat and Aegilops species. Wheat can be either a female or male parent of the F1s, depending on species and specific cross combinations.
In wheat × Aegilops crosses, crossability genes on the wheat side have been highlighted for their significant role on the success rate of obtaining F1 hybrids with Aegilops species (Figure 1). This is a key point considering it is very difficult to produce F1s using low crossable wheat parents. While East Asian wheat landraces generally have higher crossability success rates with Aegilops and other alien species (e.g., rye), European ones have lower rates of success (Zeven, 1987), presumably because European wheat has had greater chances to cross-pollinate with rye histroically. Even though crossability is a QTL trait and controlled by several genes (Alfares et al., 2009), two dominant genes Kr1 (5BL) and Kr2 (5AL) were two major genes (Lein, 1943) affecting pollen tube growth (Riley and Chapman, 1967). These two genes have effects across different species including Hordeum and Aegilops (Snape et al., 1979; Koba and Shimada, 1993). Kr1 has a stronger effect than Kr2, and dominant alleles (Kr1 and Kr2) have inhibition effects. Plants with Kr1Kr2 show less than 10% crossability, Kr1kr2 showed between 10 and 25% crossability, kr1Kr2 between 25 and 50% and plants with the kr1kr2 genotype more than 50% crossability (Lein, 1943). Additionally, crossability genes were also reported as Kr3 on 5D (homoeologous of Kr1 and Kr2) and Kr4 on 1A (Krolow, 1970; Zheng et al., 1992). More recently, SKr on 5BS was reported to have a stronger crossable effect than Kr1 (Tixier et al., 1998; Alfares et al., 2009; Mishina et al., 2009).

Figure 1. Effects of crossability on seed setting. (A) A high crossable durum line. (B) A low crossable durum line. The durum spikes show seed setting 2 weeks after pollination with Ae. tasuchii. The high crossable line (A) sets six grains, while the low crossable line (B) sets zero grains.
If it is too difficult to produce F1 hybrids in wheat × certain Aegilops species (wheat as females), pollination in the opposite cross direction (Aegilops as females) may be more successful. Dale et al. (2017) reported 0% seed setting in bread wheat × Ae. tauschii crosses (probably due to a crossability problem of the bread wheat parents), while it was 30% in Ae. tauschii × bread wheat. The seed-setting rate with Aegilops as female parents is variable across these species. Yuan et al. (2017) reported the rate was about 0.2% in Ae. speltoides × bread wheat, 2–9% in Ae. cylindrica, 12–15% in Ae. ovata and 22–47% in Ae. tauschii. It must be cautioned that the seed setting does not always mean success in obtaining F1 plants.
Endosperm and Embryo Development Deficiency and Embryo Rescue
Gill et al. (1981) observed endosperm abortion and embryo lethality or semi-lethality and seedling death in crosses between Ae. tauschii and three diploid Triticum species. While the reaction types were different in each three Triticum species, the same thing is common in Ae. tauschii × bread wheat crosses. Even though the initial seed-setting rate was a 47% (Yuan et al., 2017), the seedling formation rate dropped to 1%. Sehgal et al. (2011) reported that an average of 35% initial embryo formation ended in an average of 7% F1 plants.
The degree of endosperm development deficiency is cross-combination specific. However, high polyploidy Aegilops tend to set endosperm more when crossed with wheat, while diploid Aegilops species set less (data not shown). To overcome endosperm abortion, embryo rescue is necessary to recover hybrid seedlings. In this procedure, embryos are dissected from developing grains and transferred to an agar medium with nutrients such as sugar and salts for proper development (Miller et al., 1987). While some wheat lines such as Langdon (durum wheat) or various East Asian landrace lines tend to develop enough endosperm for the embryo to form seeds (Koba and Shimada, 1993), the amount of endosperm sometimes will be lower than normal “wheat × wheat crosses” (Figure 2). It is possible to skip embryo rescue if using these lines. The genetic background of forming unreduced gametes in wheat is not known yet.
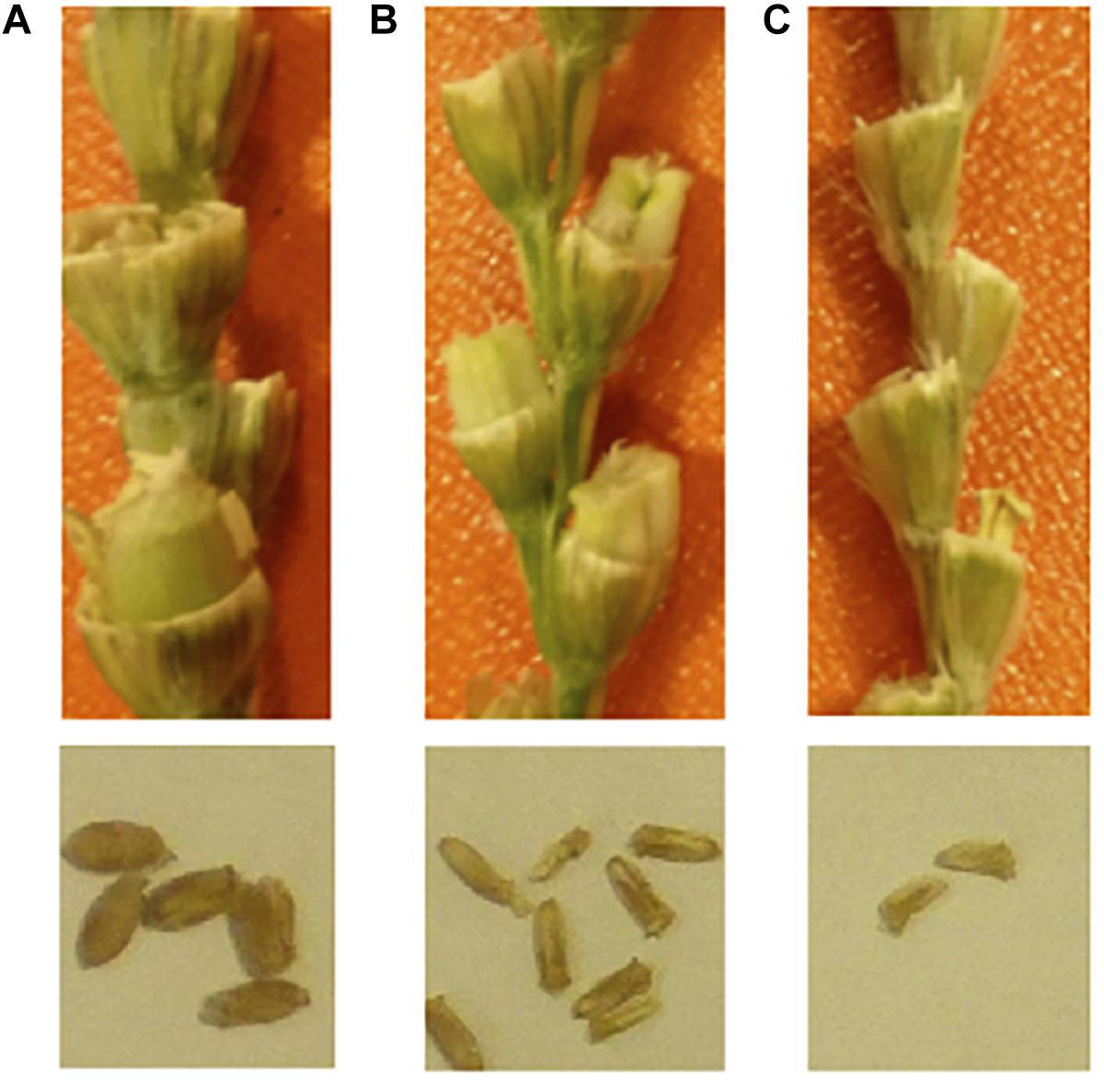
Figure 2. Endosperm development deficiency in F1 grains. (A) normal selfed-seeds of durum wheat, (B) durum wheat cv. LANGDON × Ae. tauschii; (C) durum wheat cv. CIRNO × Ae. tauschii. The seed sizes are smaller in durum × Ae. tauschii. The panel (B) has some amount of endosperm, and the seeds can germinate. The panel (C) has no endosperm, and the seeds will not germinate. Embryo rescue is necessary on the right.
Overcoming Sterility of F1S and Unreduced Gametes
The genome of F1s between wheat and Aegilops in haploids causes sterility until doubling the chromosome numbers. One option is to conduct direct backcrossing of F1s with wheat as a pollen donor. Even though the rate of seed set is extremely low, it is possible to obtain BC1 plants (Cox et al., 1990; Fritz et al., 1995; Zemetra et al., 1998; Olson et al., 2013). The alternative is through chemical treatments such as colchicine (Blakeslee and Avery, 1937; Tang and Loo, 1940; Bennett and Smith, 1979) and N2O gas (Hansen et al., 1988). Some wheat lines such as Langdon produce unreduced gametes, which is a gamete with a 2n nucleus resulting from abnormal meiosis (Fukuda and Sakamoto, 1992; Cai et al., 2010) that leads to spontaneous amphidiploid formation. The formation of unreduced gametes have been reported in durum × Ae. tauschii, Ae. speltoides, Ae. longissima, Ae. umbellulata, Ae. comosa, Ae. ovata, ( = Ae. geniculata), Ae. ventricosa, Ae. crassa and Ae. triuncialis (Xu and Dong, 1992; Matsuoka and Nasuda, 2004; Tiwari et al., 2008; Fakhri et al., 2016). The rate of formation is different among Aegilops species and prevented by the presence of a shared homologous subgenomes (Fakhri et al., 2016). Additionally, it depends on the genotype of the Aegilops parents (Matsuoka and Nasuda, 2004; Fakhri et al., 2016).
Hybrid Necrosis/Weakness Abnormality
Hybrid necrosis, chlorosis and bushy plant formation is very common in “normal” wheat × wheat cross (Hermsen, 1963; Hermsen and Waninge, 1972; Pukhalskiy et al., 2000; Chu et al., 2006). The Ne1-Ne2 necrosis system is the best known hybrid necrosis system in wheat, which is caused when two complementary genes of Ne1 (5BL) and Ne2 (2BS) are found in the same plant (Tsunewaki, 1960; Nishikawa et al., 1974; Chu et al., 2006). However, this phenomenon is more frequent and complex in wheat × Aegilops crosses. Necrosis in T. turgidum × Ae. tauschii was first reported in the 1960s (Nishikawa, 1960, 1962a,b; Figure 3). Mizuno et al. (2010) did further analysis using a set of synthetic wheat lines that had one common durum wheat parent “Langdon” and different Ae. tauschii accessions. They found four different types of hybrid abnormality and responsible genes Net1 (7DS), Net2 (2DS), and Hybrid chlorosis1 (Hch1; 7DS) in Ae. tauschii (Mizuno et al., 2010, 2011; Nakano et al., 2015). The mode of action of these genes should be complementary with genes on the durum side, because the hybrid abnormalities take place only when Ae. tauschii is crossed with durum wheat. Hypersensitive response-like reactions were observed for Net1 necrosis, indicating that it is a kind of disease response reaction (Jeuken et al., 2009; Mizuno et al., 2010). Okada et al. (2017) also reported growth abortion and grass-clump dwarf phenotype in durum × Ae. umbellulata. They also showed a repressed expression of the shoot meristem maintenance-related and cell cycle-related genes in the plants with the grass-clump dwarf phenotype. To avoid a problem with hybrid seedling death, Dhaliwal et al. (1986) reported the suppression of Ne1-Ne2 necrosis at high temperatures. The author also confirmed that incubation at 28°C suppressed necrosis in F1s between emmer × Ae. tauschii (Supplementary Figure S2). However, the high temperature causes pollen sterility.
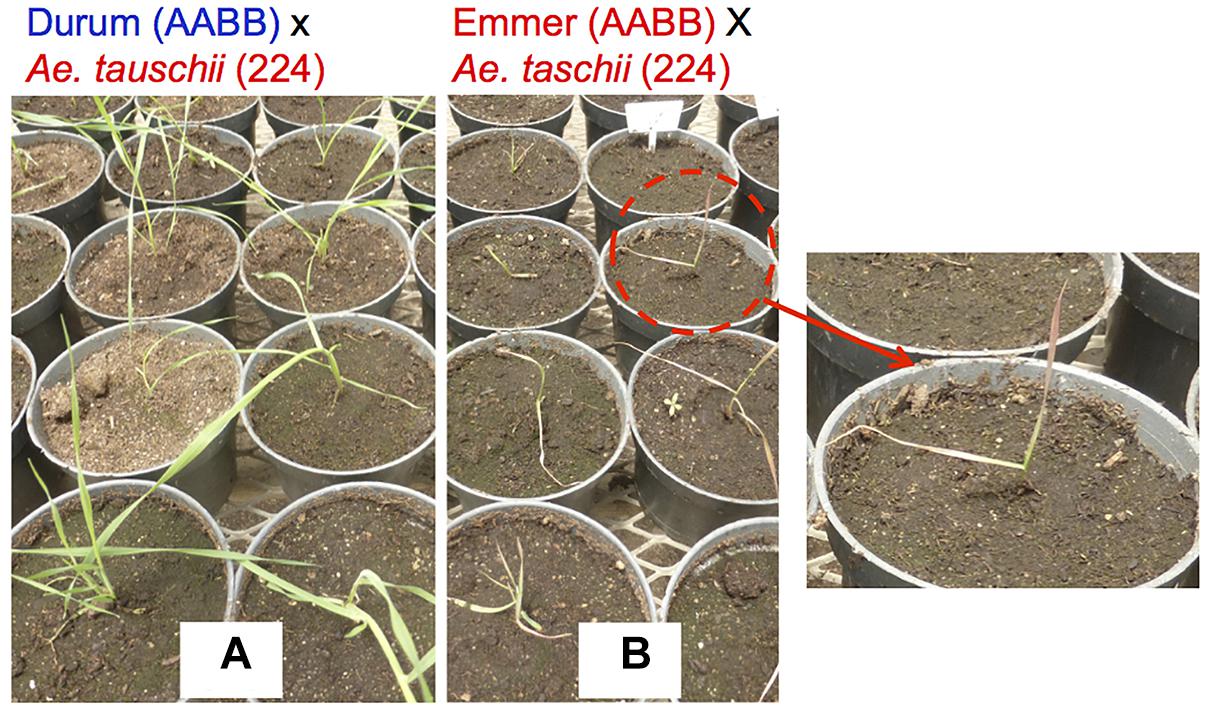
Figure 3. Hybrid necrosis observed in tetraploid wheat × Ae. tauschii. (A) F1 from Triticum turgidum ssp. durum cv. CIRNO C 2008 × Ae. tauschii WX (224); all plants of this group are growing normally. (B) F1 from Triticum turgidum ssp. dicoccum (PI 233433) × Ae. tauschii WX (224); all plants are showing a necrosis symptom of reddish leaf color.
Gametocidal Genes
A group of gametocidal genes (Gc), sometimes considered as selfish genes, is another type of obstacle in which the genes cause chromosome breakages in gametes without Gc (Endo and Tsunewaki, 1975; Maan, 1975; Endo and Katayama, 1978). This happens when a plant becomes heterozygous in Gc —half of the gametes will have Gc and the other half will have no Gc. Gametes without Gc show reduce fitness, which is to the advantage of gametes with Gc for the transmission to the next generation (For review, see Tsujimoto, 2005; Endo, 2007; Niranjana, 2017). Gc genes have been identified in accessions of certain species that have C, S, Sl, or M genomes and mostly confined to three different homoeologous groups: 2, 3, and 4 (Endo, 2007). The identified genes include chromosome 3C of Ae. markgrafii ( = Ae. caudata) and Ae. triuncialis (Endo and Tsunewaki, 1975), 2C of Ae cylindrica (Endo, 1979), 2Sl and 4Sl of Ae. longissima, 2Ssh and 4Ssh of Ae. sharonensis (Maan, 1975; Endo, 1985), 2S and 6S of Ae. speltoides (Tsujimoto and Tsunewaki, 1984, 1988; Kota and Dvorak, 1988) and 4M of Ae. geniculata (Kynast et al., 2000). The effects of Gc genes are variable; some cause lethality to gametes, while others are mild, allowing incorporation of the gamete into progenies. King et al. (2018) reported the presence of a 2S chromosome segment in all of the developed wheat-Ae. speltoides introgression lines due to the gametocidal effect. When researchers use these species, it is better to keep in mind that extra difficulties may arise from Gc genes. The suppression of Gc genes was reported in Norin 26, which inhibits Ae. triuncialis Gc3-C1 action and is designated as Igc1 (Tsujimoto and Tsunewaki, 1985). The presence of additional suppressor genes can also be predicted because the effect of a Gc gene is different in various wheat backgrounds. The Gc of chromosome 3C is usually lethal but when found in “Chinese Spring” background, it is mild. In addition, Friebe et al. (2003) produced a mutant of the Ae. sharonensis Gc2 gene (designated as Gc2mut) which has a suppression effect on Gc2, which will be useful to reduce problems of Gc genes in wheat breeding scheme.
The Use of Ae. tauschii For Wheat Breeding
Aegilops tauschii is the easiest species in this genus to utilize in wheat breeding, because there is little to no inhibition to meiotic chromosome pairing with the D genome chromosomes of bread wheat. According to several sources, bread wheat originated about 10,000 years ago (Wang et al., 2013; Matsuoka and Takumi, 2017), which is relatively recent and not long enough for genomic differentiation. Furthermore, Ae. tauschii contrasts with diploid A genome ancestors. Luo et al. (2000) reported about a 1/6 recombination-rate reduction between Triticum monococcum 5A and bread wheat 5A chromosomes when compared to the recombination rate between two T. monococcum 5A homologous chromosomes. Even though the A genome of bread wheat and that of the diploid ancestor can form perfect bivalents during meiosis in the F1s of AAB (Gill et al., 1988), there are likely to be significant differences in base sequences and chromosome structures (such as inversion, translocations, deletion/duplications, or heterochromatin structures) after the tetraploid wheat formation – i.e., 100,000–500,000 years ago (Huang et al., 2002).
The spontaneous formation of bread wheat in nature was a rare event during which only a very limited number of Ae. tauschii plants were involved, based on molecular data and field observations (Dvorak et al., 1998; Matsuoka, 2011; Wang et al., 2013). The genetic diversity of Ae. tauschii is far greater in comparison to bread wheat’s D genome diversity (Dvorak et al., 1998; Wang et al., 2013). Matsuoka et al. (2013) proposed sub-dividing Ae. tauschii into three groups, TauL1, L2, and L3, and found that bread wheat is close to TauL2 but distinct from TuL1. Even though it is not obvious as in the case of T. monococcum, crosses of bread wheat with Ae. tauschii accessions of TauL1 may show a reduction in chromosome recombination rates of the A-genome chromosomes.
Figure 4 represents two ways to utilize Ae. tauschii in wheat breeding, either through direct crossing or indirect crossing (synthetic wheat). With indirect crossing, tetraploid wheat (AABB) will be crossed with Ae. tauschii (DD) to produce an F1 (ABD), and subsequently this F1 will have its chromosome number doubled naturally or artificially to produce so-called synthetic wheat (AABBDD). Synthetic wheat can then be used in wheat breeding by crossing with bread wheat. Synthetic wheat lines were first developed in the United States and Japan in 1940s (Kihara, 1944, 1946; McFadden and Sears, 1944). During the next few decades, a number of synthetic wheat lines were developed by various groups (Kihara and Lilienfeld, 1949; Tanaka, 1961; Dyck and Kerber, 1970; Kerber and Dyck, 1979; Hatchett et al., 1981; Chèvre et al., 1989; Valkoun et al., 1990; Lange and Jochemsen, 1992; Lutz et al., 1994; Wang et al., 2006). Later in the 1980s, CIMMYT started a large-scale production of synthetic wheat, developing more than 1,000 lines (Das et al., 2016; Li et al., 2018). Matsuoka et al. (2007) also reported another set of “Langdon” synthetic wheat lines, and Zeng et al. (2016) produced synthetic wheat using local Chinese land races that were more adaptable to China.
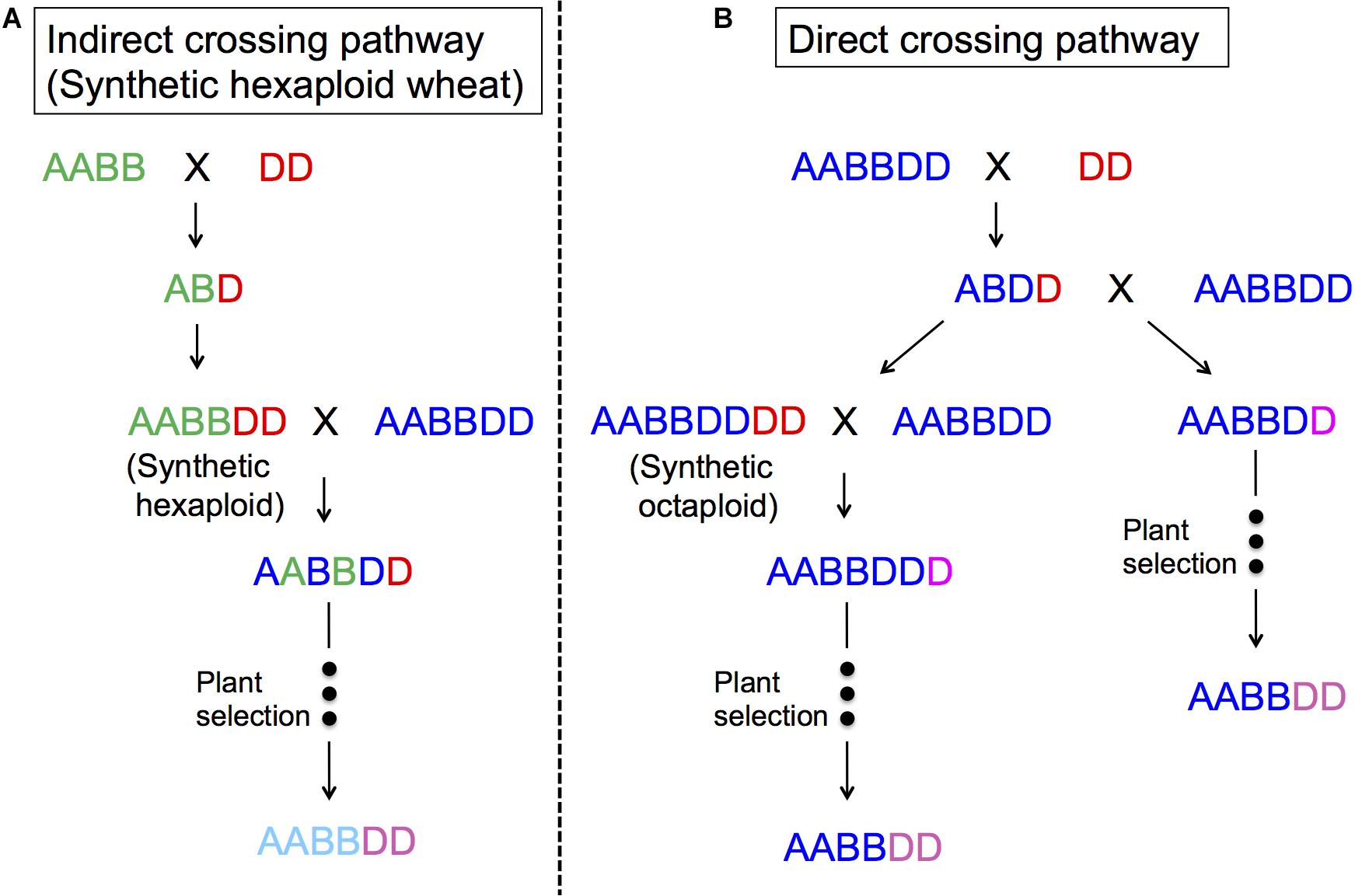
Figure 4. The use of Ae. tauschii in wheat breeding. (A) indirect crossing pathway (synthetic hexaploid pathway) through durum × Ae. tauschii crosses; (B) direct crossing pathway through bread wheat × Ae. tauschii crosses.
In the direct crossing pathway, Ae. tauschii (DD) is crossed with bread wheat (AABBDD) to make an F1 (ABDD). These F1s are then backcrossed with the same bread wheat (AABBDD) to generate BC1, where the plant selection process begins. Gill and Raupp (1987) and Cox et al. (1992) reported this method as successful for transferring Hessian fly and rust resistance. The merit of this method is that it will only change the D genome, making it easy to perform some analyses, as well as directly improving the “best” line without contribution from durum wheat. One of disadvantage of this method may be sterility of the F1 plants even as females, and as such, it is necessary to backcross a large number of spikes to have enough BC1 seeds to introgress the whole genome (Cox et al., 1990; Fritz et al., 1995; Olson et al., 2013). It is important to note that the seed setting rates in F1 plants also depend on Ae. tauschii accessions (Matsuoka and Takumi, 2017).
Octaploid synthetic wheat is another way to utilize Ae. tauschii in wheat breeding, in which an F1 (ABDD) from bread wheat (AABBDD) × Ae. tauschii (DD) has its chromosome number doubled to produce an octaploid synthetic wheat (AABBDDDD) (Chèvre et al., 1989). Sehgal et al. (2011) and Zhang D. et al. (2018) reported the production of five and one AABBDDDD lines, respectively. CIMMYT has also produced a few hundred octaploid synthetic wheat lines (Supplementary Figure S3). This research resulted in the successful transfer of a dormancy QTL (Dale et al., 2017) and Septoria tritici Blotch resistance (Figure 5).
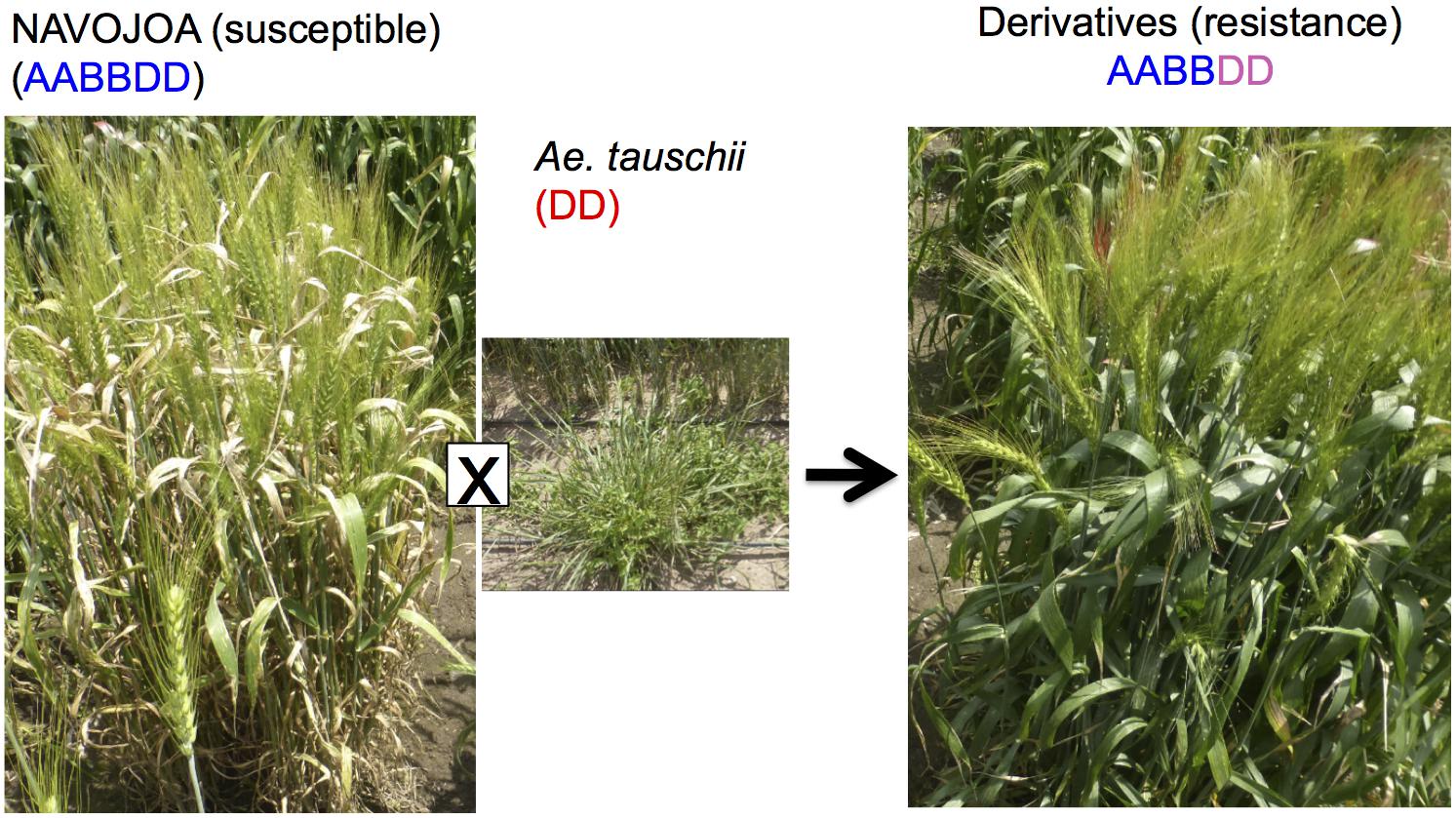
Figure 5. Improvement of Septoria disease resistance through synthetic octaploid wheat. The derivative shows resistance while wheat parent line “NAVOJOA” is susceptible.
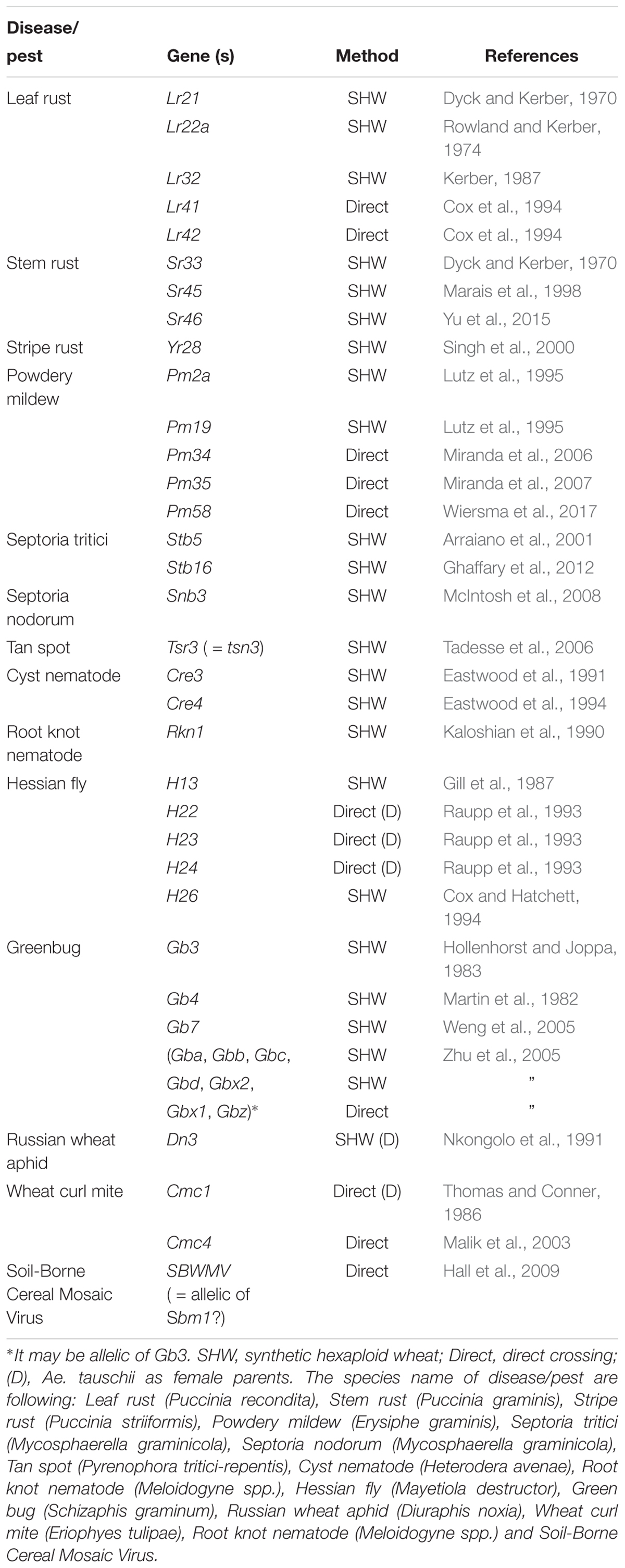
Table 2 summarizes the resistance genes identified and/or transferred into wheat, including leaf rust, stem rust, stripe rust, powdery mildew, and Hessian fly resistance. It is difficult to identify genes related to abiotic stress (drought and heat) and yield potential, as these traits are not obvious by sight. However, synthetically derived lines have shown up to a 30% yield increase under rain-fed conditions, and a 45% yield increase under drought condition over their wheat parents (Narasimhamoorthy et al., 2006; Dreccer et al., 2007; Trethowan and Mujeeb-Kazi, 2008; Li et al., 2014) and better performance under heat (Iehisa and Takumi, 2012; Jafarzadeh et al., 2016). The percentage of synthetic derivative lines (SDLs) in the Semiarid Wheat Yield Trial reached 52% in 2010, with a five-year average (2010–2015) of 35%. At least, 62 wheat varieties were released using CIMMYT synthetic wheat in their pedigree around the world since 2003 (Li et al., 2018). Elbashir et al. (2017) reported that synthetic derivative lines are promissive for improving heat tolerance in the analysis of multiple synthetic derivative (MSD) lines that cover the whole diversity of Ae. tauschii.
The Use of Tetraploid/Hexaploid Aegilops Species With a D Genome
Hybrids between tetraploid Aegilops species with the D genome can show meiotic pairing with the D genome chromosomes of bread wheat (Kimber and Zhao, 1983). These species include Ae. cylindrica, Ae. crassa, and Ae. ventricosa, Ae. juvenalis and Ae. vavilovii. Amphiploids of wheat with Ae. crassa have been reported (Jovkova et al., 1977; Xu and Dong, 1992), and a number of amphiploids of Ae. ventricosa × durum wheat were produced (Delibes et al., 1987). CIMMYT has also developed 20 amphiploid lines of these species using durum and bread wheat (Figure 6) for bread wheat D genome improvement (Supplementary Figure S4).

Figure 6. Amphidiploid lines derived from between durum and diploid or tetraploid Aegilops with a D genome. (A) durum wheat cv. CIRNO C 2008, (AABB); (B) durum cv. CIRNO C 2008 × Ae. tauchii (895), (AABBDD); (C) durum cv. CIRNO C 2008 × Ae. ventricosa (PI 542385), (AABBDDNN); (D) durum cv. ACONCHI 89 × Ae. crassa (PI 542385), (AABBDDMM); (E) durum cv. CIRNO C 2008 × Ae. crassa (PI 542385), (AABBDDMM); (F) durum cv. CIRNO C 2008 × Ae. cylindrica (PI 639294), (AABBDDCC). All amphidiploids were produced and maintained at CIMMYT.
The eye spot resistance gene Pch1 (one of two strong seedling resistance genes) was transferred from an amphiploid (AABBDDNN) between tetraploid wheat (AABB) × Ae. ventricosa (DDNN) (Table 2). This amphiploid was crossed with bread wheat (AABBDD) to have a derivative line named “VPM1” (Maia, 1967; Doussinault et al., 1983). It was later determined that the location of the transferred Pch1 is chromosome 7D (Mena et al., 1992).
The Use of Other Aegilops Species and Chromosome Pairing
For the use of other Aegilops species, a reduced chromosome pairing frequency is more problematic. Ae. cylindrica, Ae. crassa, Ae. ventricosa, Ae. Juvenalis, and Ae. vavilovii are also categorized in this group due to the presence of non-D genomes.
Because of the lack of recombination, it is common to produce so-called alien chromosome addition or substitution lines in which one pair of Aegilops (or alien) chromosomes is added to or substituted for a pair of wheat chromosomes, respectively (Supplementary Figure S5). A number of addition lines have been produced from 14 different Aegilops species (For a review see; Schneider et al., 2008; Kilian et al., 2011). These addition lines are very useful for analysis and locating useful genes at the chromosomal level. However, addition lines have less breeding value, because they have many negative factors and the presence of an extra alien chromosome disrupts the genetic harmony of a genome. To be more appropriate for breeding, it is necessary to produce introgression lines (small Aegilops chromosome segment transfers) or Robertsonian/centromeric translocation lines (Robertson, 1916), in which one of the Aegilops chromosome arms is translocated to a wheat chromosome, replacing an arm of that wheat chromosome (Supplementary Figure S5). These translocations can be obtained spontaneously from addition/substitution lines or amphiploids in backcrossing populations, or using wheat monosomic lines (2n = 41; with one of the homoeologous chromosomes is missing). All of these can lead to the occurrence of univalent chromosomes during meiosis. Then, the meiotic spindle fiber will attach to the both sides of univalent chromosomes, which then causes chromosome breakages through the centromeric regions at high frequency. The broken chromosome arms are sticky and may fuse to other broken chromosomes to produce centromeric translocations (Supplementary Figure S6).
Homoeologous meiotic pairing between chromosomes of wheat and Aegilops species is inhibited mostly by the Ph1 gene (5BL) (Okamoto, 1957; Riley and Chapman, 1958; Sears and Okamoto, 1958; Riley, 1960). Therefore, the meiotic barrier can be overcome by suppressing of Ph1 activity. Sears (1977) produced the ph1b mutant in which the Ph1 locus is missing and this is the most widely used Ph1 gene mutant in wheat breeding. Another gene which affects homoeologous chromosome pairing was identified as Ph2 (3D) (Mello-Sampayo, 1971) and has a mild inhibition effect on Ph1 (Sears, 1982). Additional mutants, ph1c (Giorgi, 1983) and ph2 (Sears, 1982) are also available, even though they have been rarely used in the breeding. It is also known that the presence of Ph1 suppressors or promotors of homoeologous chromosome paring are present in some accessions of Aegilops species: Ae. speltoides (Feldman and Mello-Sampayo, 1967; Dover and Riley, 1977; Dvorak et al., 2006), Ae. longissima (7), Ae. mutica (Dover and Riley, 1972), Ae. umbellulata (Riley et al., 1973), Ae. Peregrina, and Ae. kotschyi (Fernandez-Calvin and Orellana, 1991) and Ae. geniculata (Koo et al., 2017). Therefore, the transfer of traits may be easier in accessions that have the suppressive effects. The Ae. speltoides genes are considered to be suppressants, because they can promote more meiotic pairing in the presence of the Ph1 gene (Dover and Riley, 1972). A couple of suppressor genes of Ae. speltoides has been transferred into wheat and designated as PhI (Chen et al., 1994) and Su1-Ph1 (7S) and Su2-Ph1 (3S) (Dvorak et al., 2006; Li et al., 2017). Since these genes are dominant, they can be faster and easier to utilize in breeding. Yet the effects of PhI have been shown to be lower than that of ph1b (Aghaee-Sarbarzeh et al., 2000).
Sometimes it is difficult to induce homoeologous recombination due to different homoeologous co-linearity between wheat and Aegilops chromosomes (Molnár et al., 2013, 2016). It is also known that centromeric and other chromosomal regions may have very low recombination rates, even in wheat × wheat crosses (Saintenac et al., 2009). In these cases, other methodologies become an alternatives. Sears (1956) demonstrated a successful transfer of Ae. umbellulata Lr9 gene into wheat using irradiation. Yet this is the only success story using irradiation for introgression of Aegilops chromatin for wheat breeding until recently. Singh et al. (2016) and Verma et al. (2016) recently reported the production of a translocation by irradiation of Ae. kotschyi hybrids. Mild effect Gc genes and some chemicals can also induce random translocations, much like irradiation. Even though it is not for breeding purposes, the Gc system has been used for producing translocations of wheat-rye and wheat-barley (Joshi et al., 2013; Li et al., 2013; Ishihara et al., 2014).
Useful Genes of Aegilops Transferred to Wheat
Through the use of the various techniques described above, a number of genes have been transferred from Aegilops (including Ae. tauschii) to wheat (Tables 3, 4). In terms of total number, leaf rust resistance genes are the most numerous (20), followed by powdery mildew (15), and green bug (12). Since more than 75 resistance gene loci have been identified and permanently designated as resistance genes by 2018 (Ponce-Molina et al., 2018). Aegilops provided more than 20% of them. For powdery mildew, 54 resistance loci were found by 2018 (Tang et al., 2018), and Aegilops contributed about 20%. For Cereal Cyst Nematodes (CCN) resistance genes, a total of 12 genes have been identified, including Cre1-8, CreR, CreV, CreX, CreY (Ali et al., 2019). Of them, two (Cre1 and Cre8) are indigenous to the wheat genepool. The others are from Ae. tasuchii (Cre3 and Cre4), Ae. ventricosa (Zhuk.) (Cre2, Cre5, and Cre6), Ae. triuncialis L. (Cre7); Ae. peregrina (CreX and CreY), Secale cereale (CreR) and Dasypyrum villosum (CreV) (Zhang et al., 2016), showing that two thirds of them are from Aegilops. In terms of actual species of origin, Ae. tauschii has provided the most number of genes, followed by Ae. speltoides and then Ae. ventricosa. It is worth noting that most of the disease resistances from Ae. ventricosa are provided by a single 2NS-2AS translocation, including Lr37, Sr38, Yr17, Cre5, Rkn3 (Bariana and McIntosh, 1993, 1994; Jahier et al., 1996, 2001; Helguera et al., 2003; Tanguy et al., 2005; Williamson et al., 2013); this translocation has originated from VPM1 (Maia, 1967) that also has Pch1 resistance on 7D (Mena et al., 1992).
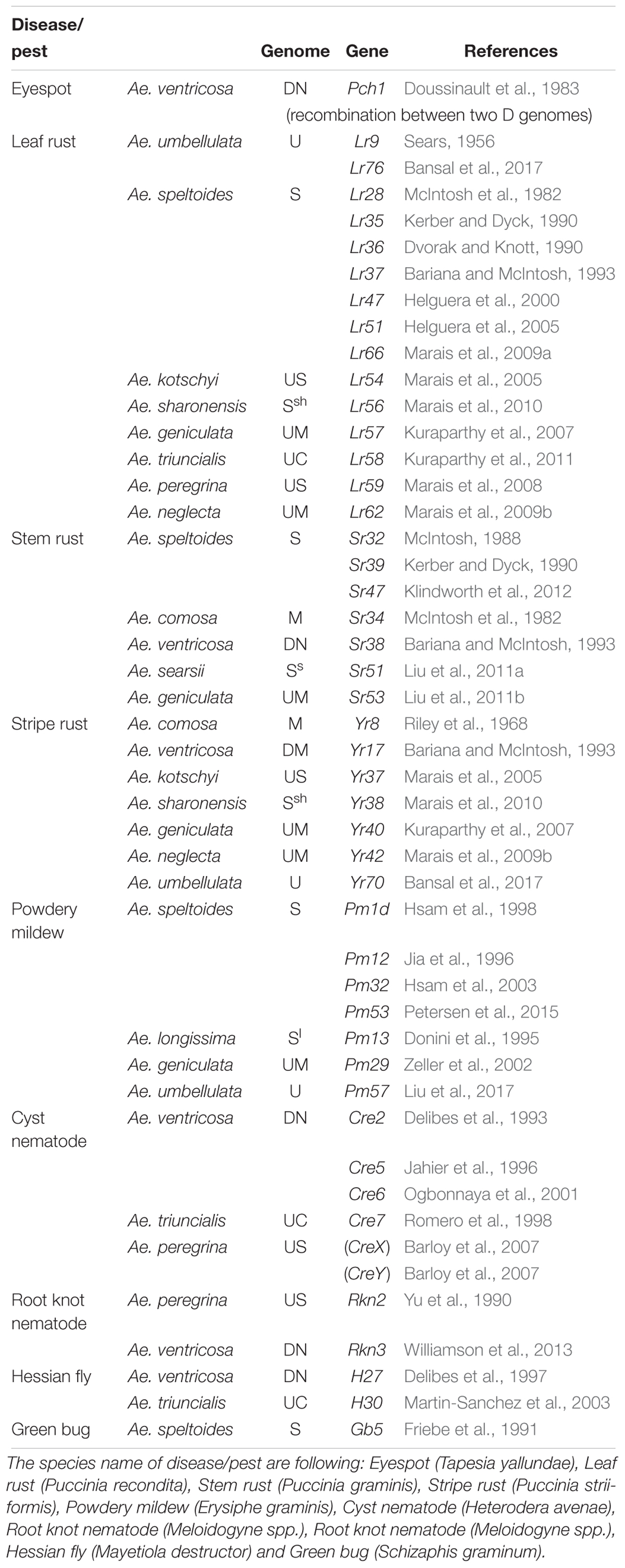
Table 3. Identified or transferred biotic resistance genes in Aegilops (other than from Ae. tauschii) into wheat.
Recently, Aegilops has gathered more attention for improving micro-nutrient content (such as Fe and Zn) in wheat grains. Zn deficiency affects 17.3% of the world’s population across Asia and Africa, leading to the deaths of more than 400,000 children each year (Cakmak, 2007; Black et al., 2013; Velu et al., 2018b). Micro-nutrient rich wheat, i.e., bio-fortified wheat, can improve the lives of these people. It is difficult to find high Zn and Fe content germplasm in the wheat genepool (Cakmak et al., 2010), even though some Aegilops species show three to four-fold higher Zn and Fe grain content, including Ae. longissima (Sl), Ae. kotschyi (US), Ae. peregrina (US), Ae. cylindrica (CD), Ae. ventricosa (DN), Ae. geniculata (UM) (Rawat et al., 2009). Amphiploid durum- Ae. longissima and partial amphiploids of wheat – Ae. kotschyi show two to three times higher levels of Zn and Fe grain content than the parental wheat line (Tiwari et al., 2008, 2010). Rawat et al. (2011) further reported Zn grain content three times higher in wheat- Ae. kotschyi addition/substitution lines than the wheat parent.
In addition to the benefit for wheat breeding mentioned above, it is also important to highlight that Aegilops introgression lines have a level of diversity and unique traits that wheat lacks. Even though these are of no immediate benefit at this moment, their value could be seen in the future, as exemplified by two recent global wheat production threats.
A Story of Aegilops Translocations on Stem Rust UG99 Race
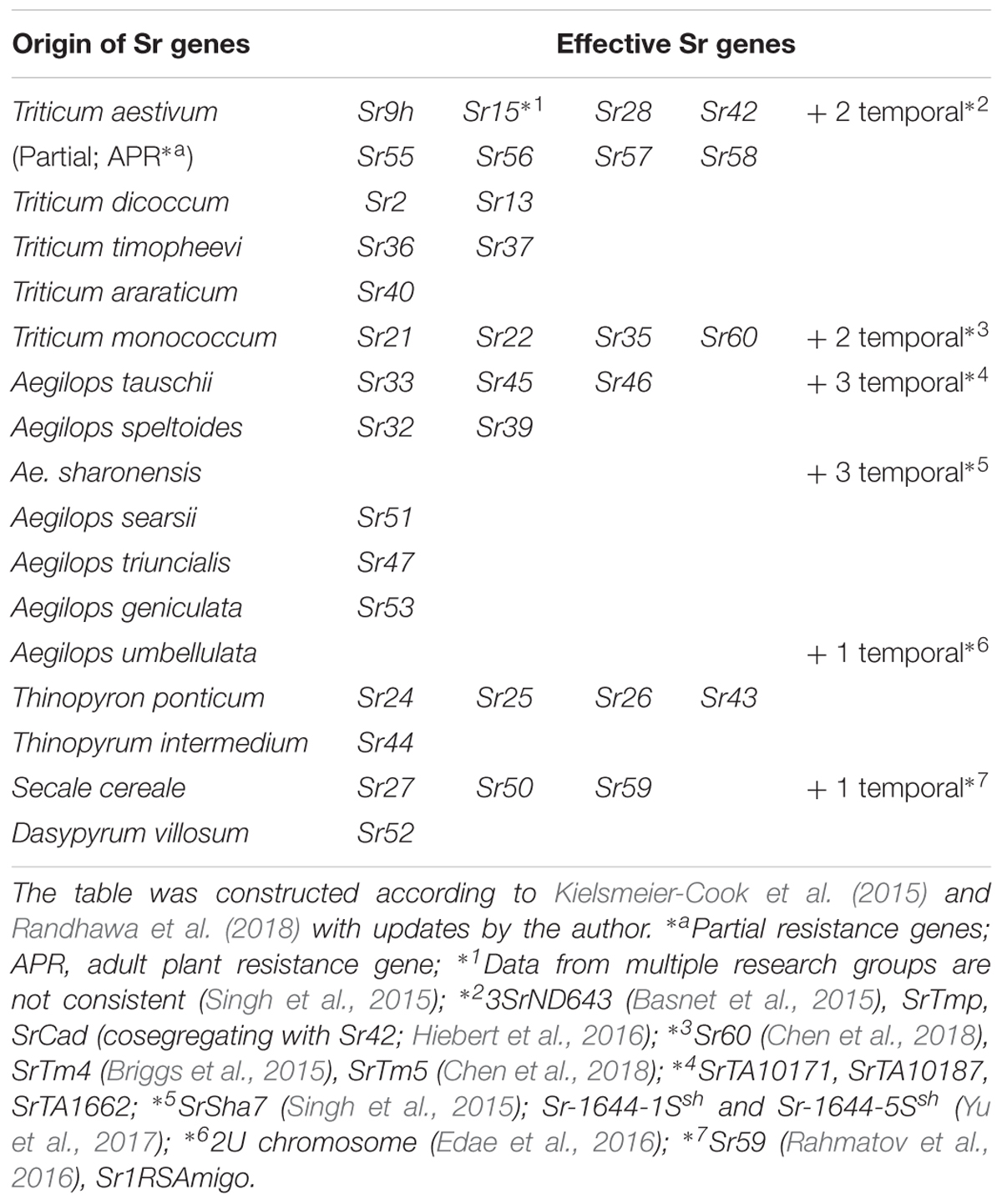
A serious threat to global wheat production is the emergence of stem rust Ug99 race, which was recognized in Uganda in 1999 (Pretorius et al., 2000). This disease had the potential to develop into a global catastrophe, as more than 70% of wheat varieties around the world did not have resistance against Ug99 in the early 2000s (Singh et al., 2015). Many wheat breeders and pathologists, who had thought stem rust was no longer a problem, were caught unprepared and were then spurred to search for new resistant sources. The researchers realized that while the bread and durum wheat gene pools do not have many resistant sources, resistance is available outside the genepool from ancestral and alien species including many in Aegilops (Table 4, based on Yu et al., 2014; Kielsmeier-Cook et al., 2015; Randhawa et al., 2018 with updates by the author). This has also promoted various studies to identify new stem rust resistance genes, which led the identification of Sr46 (Ae. tauschii; Yu et al., 2015), Sr47 (Ae. triuncialis; Klindworth et al., 2012), Sr51 (Ae. searsii; Liu et al., 2011a), Sr53 (Ae. geniculate; Liu et al., 2011b) and three additional genes in Ae. tauschii (Rouse et al., 2011), three genes in Ae. sharonensis (Singh et al., 2015; Yu et al., 2017) and one gene in Ae. umbellulata (Edae et al., 2016). In addition, it has been reported that 81% of Ae. longissima (out of 394 accessions), 94% of Ae. neglecta (189 out of 202 accessions tested), 88% of Ae. cylindrica (DDCC) and Ae. peregrina (SSUU) were Ug99 resistant (Huang S. et al., 2018; Olivera et al., 2018).
Even though introgression lines of two Ug99 resistance genes (Sr32 and Sr39) from Ae. speltoides were available, they were not used in wheat breeding program due to the presence of large Ae. speltoides segments and associated negative factors on agronomy (Friebe et al., 1996). Fortunately, researchers in Australia and the United States started preparing for the possible appearance of dangerous new stem rust pathogen races back in the early 1990s and the reports of Ug99 just confirmed their expectations. Based on that work, shortened introgressions of chromosome 2S segments with Sr32 and Sr39 were already developed using the ph1b mutant and have been quickly distributed around the world (Mago et al., 2009, 2013; Niu et al., 2011).
It is notable that it has eight resistance genes (+ three temporary assigned genes) in the bread wheat gene pool are effective to Ug99, but four of them (Sr55, Sr56, Sr57, and Sr58) are partial or adult plant resistance genes (APR), so it is necessary to combine them with other genes to exert a higher level of resistance (Gustafson and Shaner, 1982; McIntosh et al., 1995, 1998, 2012).
A Story of the 2NS Translocation in Relation to Wheat Blast Disease
Wheat blast caused by Pyricularia oryzae (Magnaporthe oryzae) is an emerging disease that was first recognized in Brazil in the 1980s (Igarashi et al., 1986). The pathogen gained an ability to infect the new host plant wheat through a mutation of an avirulence gene (Inoue et al., 2017). Since then, it has been a serious obstacle for wheat production in central and south Brazil, south-east Paraguay and eastern Argentina, affecting 300 million ha of wheat fields and reducing the yield of infected areas 100–10% (Kohli et al., 2011; Perello et al., 2015; Duveiller et al., 2016). The disease jumped to Bangladesh in 2016 and spread to 15,000 ha (Malaker et al., 2016). Because of this serious threat to the wheat production of South Asia, quick remedial action was required to prevent a devastating epiphytotic (Mottaleb et al., 2018). Eight different resistance genes against wheat blast (Rmg1-8) have been reported, and only two of them (Rmg7 and Rmg8) are effective in the field in Bangladesh (Anh et al., 2017). Since Rmg7 and Rmg8 recognize the same avirulence gene peptide of the pathogen, both resistance genes are functionally equivalent to a single gene for resistance (Anh et al., 2017). Despite of lacking resistance sources, a new resistance wheat variety, “BARI com” was released in Bangladesh within 2 years in 2018. This happened because of the existence of the 2NS-2AS translocation (Cruz et al., 2016; Velu et al., 2018a; Mahmud, 2019). This translocation has been utilized in wheat breeding programs because of rust resistances (Juliana et al., 2017), but it also happens to have a strong wheat blast resistance. If 2NS-2AS had not have been produced, the wheat blast issue would have been a much more serious problem in the last few years. Another amazing finding with the 2NS-2AS translocation is that nearly 90% of advanced lines of the CIMMYT bread wheat program have this translocation (Juliana et al., 2017; Philomin Juliana, Personal communication). As in the case of the T1BL.1RS translocation that dominated wheat cultivars for decades, a beneficial translocation can have a huge impact on wheat breeding and production.
The Use of Aegilops in the Genomic Age for Breeding and Pre-Breeding
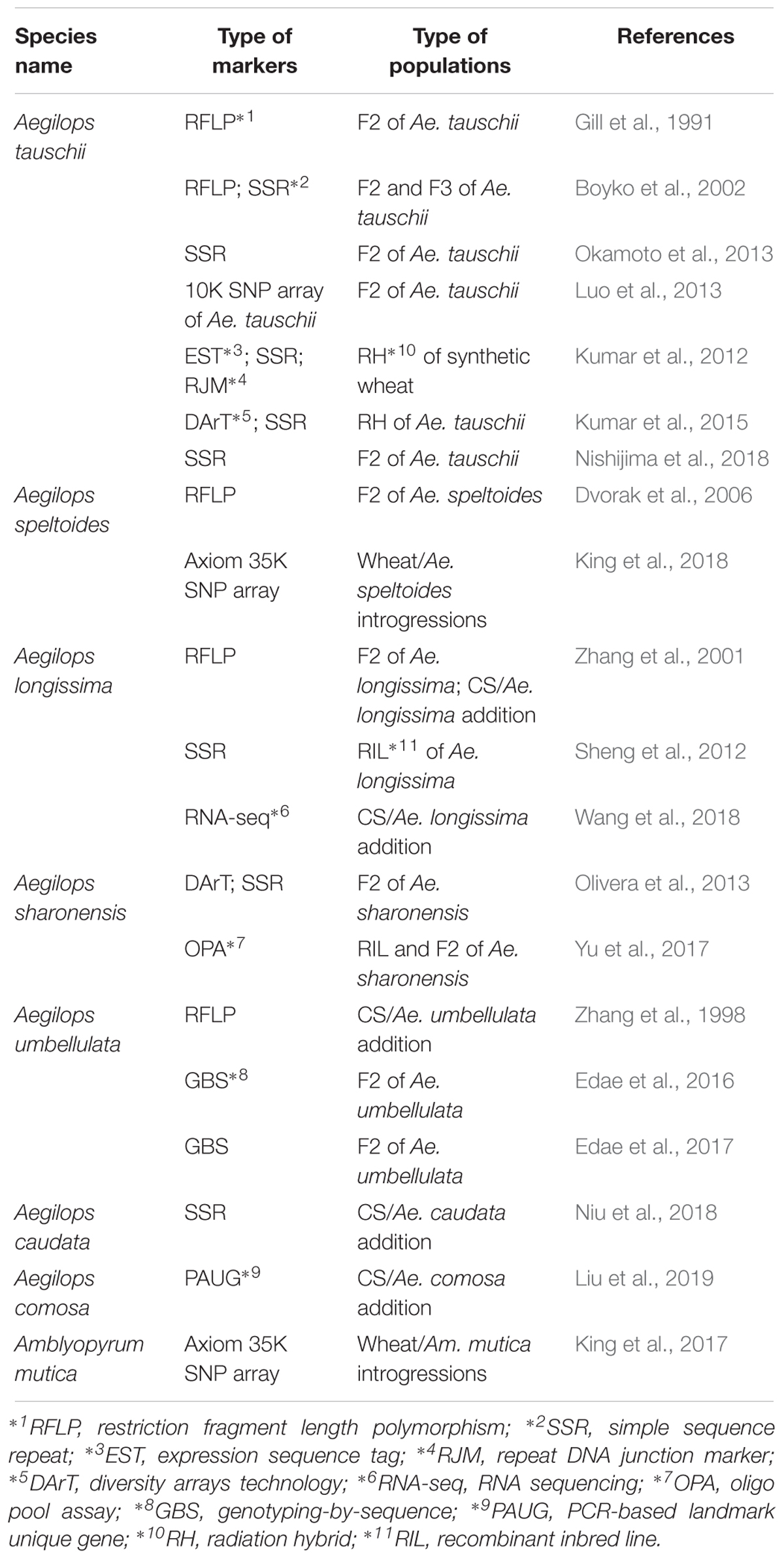
During the last several decades, cytogenetic methods not only have been essential tools for screening and understanding the nature of translocations and alien introgressions from a number of progenies, but also possess the major constraint in handling large numbers of samples. But new cytogenetic FISH/GISH technology using oligo probes expands the capacity, proving a valuable tool in cytogenetics (Du et al., 2017; Huang X. et al., 2018). More importantly, recent progress in high through-put genotyping technology and availability of molecular methods makes it possible to detect alien segments very easily, which has been promoting the production of alien segment introgressions. Niu et al. (2011) screened about 1,000 plants and found 40 smaller alien recombinants of Ae. speltoides 2S chromosome using the ph1b mutant. The development of translocations which cover a whole genome have been demonstrated in Ambylopyrum mutica ( = Ae. mutica) (King et al., 2017) and Ae. speltoides (King et al., 2018). The number of estimated introgression segments obtained are about 200 of Am. mutica (King et al., 2017), and a map of about 600 cM was made with 540 plants in the case of Ae. speltoides (King et al., 2018), which allowed the construction of linkage maps even using wheat- Aegilops introgression lines and the Axiom 35K SNP array that was constructed on a wheat sequence based Axiom 820K SNP array by optimizing for finding polymorphism between wheat and Aegilops species. An increased number of whole genome linkage or physical maps in Aegilops species have been available (Table 5). A 4-gigabase physical map based on BAC clones of Ae. tauschii led the construction of a 10 K Ae. tauschii Infinium SNP array (Luo et al., 2013). Moreover, the draft sequence of Ae. tauschii has been recently reported (Luo et al., 2017), and a TILLING population of Ae. tauschii was reported (Rawat et al., 2018). It will be possible to have additional physical maps and draft sequences in another Aegilops species in near future that will facilitate their use in wheat breeding and gene identifications.
Yet the biggest limitation and challenge for the use of Aegilops is still reduced recombination rates between wheat and Aegilops chromosomes that is sometimes prohibitive in producing an Aegilops introgression segment. The new technologies such as MutChromSeq (mutant chromosome sequencing), MutRenSeq (Mutagenesis Resistance gene enrichment and sequencing) and AgRenSeq (Association Genetics R gene enrichment Sequencing) may provide an alternative to overcome gene identification obstacles. These techniques allow a rapid isolation of mutated genes with mutagenesis by sequencing sorted chromosomes (MutChromSeq) or enriching target gene families by exome capture (MutRenSeq) or a rapid isolation of natural variants by enriching target gene family (AgRenSeq) and sequencing for resistance gene homologs. Steuernagel et al. (2016) reported the cloning of Sr22 and Sr45 from bread wheat using MutRenSeq, Sánchez-Martín et al. (2016) reported the cloning of Pm2 using MutChromSeq, and, Arora et al. (2019) demonstrated the discovering and cloning of Sr33, Sr45, Sr46, and SrTA1662 from a panel of about 200 Ae. tauschii accessions using AgRenSeq. Development of new methodologies which can compensate the reduced recombination rate may overcome the biggest constrains of the use of Aegilops. Alternatively, we may be able to find new variations or genes to increase the recombination rate from Aegilops like PhI genes (Chen et al., 1994; Dvorak et al., 2006; Li et al., 2017).
As we can see from the stories of Ug99 and wheat blast, Aegilops species are important not only for pre-breeding but also for a proactive main-stream breeding. It is still necessary to induce a certain level of recombination between wheat and alien chromosomes for the use of Aegilops, but the new technologies are opening up a new era of Aegilops for wheat breeding.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by the CGIAR Research Program on Wheat (CRP-Wheat) and Japan ODA from Ministry of Foreign Affairs.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Special thanks to reviewers for the meaningful comments to improve the quality of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00585/full#supplementary-material
FIGURE S1 | Various constrains in obtaining F1/amphiploids between wheat and Aegilops species. In this case, the cross is between bread wheat (AABBDD) and Ae. speltoides (SS). Measures corresponds to the constraints of the left.
FIGURE S2 | Suppression of hybrid necrosis in F1s between emmer x Ae. tauschii by incubation at 28°C. (A): incubation at 28°C; (B): incubation at 22°C.
FIGURE S3 | Synthetic octaploid and hexaploid wheat lines. (A) bread wheat cv. BORLAUG 100 × Ae. tauchii (WX 700), (AABBDDDD); (B) bread wheat cv. BORLAUG 100 × Ae. tauchii (WX 1195), (AABBDDDD); (C): bread wheat cv. BORLAUG 100 × Ae. tauchii (KU 2096), (AABBDDDD); (D): bread wheat cv. BORLAUG 100 × Ae. tauchii (KU 2811), (AABBDDDD); (E): durum cv. ACONCHI 89 × Ae. tauchii (KU 2811), (AABBDD); (F): durum cv. CIRNO C 2008 × Ae. tauchii (KU 2811), (AABBDD). DD, Ae. tauschii. All amphiploids were produced and maintained at CIMMYT.
FIGURE S4 | The use of the D genome in tetraploid Aegilops species.
FIGURE S5 | The use of Aegilops species (except Ae. tauschii) for wheat breeding. Introgression lines can be produced from any part of F1 haploid, amphiploid, addition/substitution lines, and centromeric translocation line.
FIGURE S6 | The mechanism of forming Robertsonian (centromeric) translocation. During meiosis, spindle fibers will attach to the both side of univalent chromosomes, which leads chromosome breakage at the centromeric region at high frequency (the right one). Broken chromosomes may fuse with other broken chromosome arm, forming centromeric translocation. The 3A, 3B, and 3U are 3A, 3B, and 3U chromosomes. S, short arm; L, long arm.
Abbreviations
BC1, backcrossed generation 1; CIMMYT, International Maize and Wheat Improvement Center; SHW, synthetic hexaploid wheat.
References
Aghaee-Sarbarzeh, M., Singh, H., and Dhaliwal, H. S. (2000). Ph1 gene derived from Aegilops speltoides induces homoeologous chromosome pairing in wide crosses of Triticum aestivum. J. Hered. 91, 417–421. doi: 10.1093/jhered/91.5.417
Alfares, W., Bouguennec, A., Balfourier, F., Gay, G., Bergès, H., Vautrin, S., et al. (2009). Fine mapping and marker development for the crossability gene skr on chromosome 5bs of hexaploid wheat (Triticum aestivum L.). Genetics 183, 469–481. doi: 10.1534/genetics.109.107706
Ali, M. A., Shahzadi, M., Zahoor, A., Dababat, A. A., Toktay, H., Bakhsh, A., et al. (2019). Resistance to cereal cyst nematodes in wheat and barley: an emphasis on classical and modern approaches. Int. J. Mol. Sci. 20:E432. doi: 10.3390/ijms20020432
Anh, V. L., Inoue, Y., Asuke, S., Vy, T. T. P., Anh, N. T., Wang, S., et al. (2017). Rmg8 and Rmg7, wheat genes for resistance to the wheat blast fungus, recognize the same avirulence gene AVR-Rmg8. Mol. Plant Pathol. 19, 1252–1256. doi: 10.1111/mpp.12609
Anikster, Y., Manisterski, J., Long, D. L., and Leonard, K. J. (2005). Resistance to leaf rust, stripe rust, and stem rust in Aegilops species in Israel. Plant Dis. 89, 303–308. doi: 10.1094/PD-89-0303
Arora, S., Steuernagel, B., Gaurav, K., Chandramohan, S., Long, Y., Matny, O., et al. (2019). Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 37, 139–143. doi: 10.1038/s41587-018-0007-9
Arraiano, L. S., Worland, A. J., Ellerbrook, C., and Brown, J. K. M. (2001). Chromosomal location of a gene for resistance to septoria tritici blotch (Mycosphaerella graminicola) in a hexaploid wheat ‘synthetic 6X’. Theor. Appl. Genet. 103, 758–764. doi: 10.1007/s001220100668
Bansal, M., Kaur, S., Dhaliwal, H. S., Baines, N. S., Bariana, H. S., Chhuneja, P., et al. (2017). Mapping of Aegilops umbellulata - derived leaf rust and stripe rust loci in wheat. Plant Pathol. 66, 38–44. doi: 10.1111/ppa.12549
Bariana, H. S., and McIntosh, R. A. (1993). Cytogenetic studies in wheat. XV. location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome 36, 476–482. doi: 10.1139/g93-065
Bariana, H. S., and McIntosh, R. A. (1994). Characterization and origin of rust and powdery mildew resistance genes in VPM1 wheat. Euphytica 76, 53–61. doi: 10.1007/BF00024020
Barloy, D., Lemoine, J., Abelard, P., Tanguy, A. M., Rivoal, R., and Jahier, J. (2007). Marker-assisted pyramiding of two cereal cyst nematode resistance genes from Aegilops variabilis in wheat. Mol. Breed. 20, 31–40. doi: 10.1007/s11032-006-9070-x
Basnet, B. R., Singh, S., Lopez-Vera, E. E., Huerta-Espino, J., Bhavani, S., Jin, Y., et al. (2015). Molecular mapping and validation of SrND643: a new wheat gene for resistance to the stem rust pathogen Ug99 race group. Phytopathology 105, 470–476. doi: 10.1094/PHYTO-01-14-0016-R
Bennett, M. D., and Smith, J. B. (1979). The effect of colchicine on fibrillar material in wheat meiocytes. J. Cell Sci. 38, 33–47.
Black, R. E., Victora, C. G., Walker, S. P., Bhutta, Z. A., Christian, P., de Onis, M., et al. (2013). Maternal and child undernutrition and overweight in low-income and middle-income countries. Lancet 382, 427–451. doi: 10.1016/s0140-6736(13)60937-x
Blakeslee, A. F., and Avery, A. G. (1937). Methods of inducing doubling of chromosomes in plants by treatment with colchicine. J. Hered. 28, 393–411. doi: 10.1093/oxfordjournals.jhered.a104294
Boyko, E., Kalendar, R., Korzun, V., Fellers, J., Korol, A., Schulman, A. H., et al. (2002). A high-density cytogenetic map of the Aegilops tauschii genome incorporating retrotransposons and defense-related genes: insights into cereal chromosome structure and function. Plant Mol. Biol. 48, 767–790. doi: 10.1023/A:1014831511810
Briggs, J., Chen, S. S., Zheng, W. J., Nelson, S., Dubcovsky, J., and Rouse, M. N. (2015). Mapping of SrTm4, a recessive stem rust resistance gene from diploid wheat effective to UG99. Phytopathology 105, 1347–1354. doi: 10.1094/PHYTO-12-14-0382-R
Buloichik, A. A., Borzyak, V. S., and Voluevich, E. A. (2008). Influence of alien chromosomes on the resistance of soft wheat to biotrophic fungal pathogens. Cytol. Genet. 42, 9–15. doi: 10.1007/s11956-008-1002-8
Cai, X., Xu, S. S., and Zhu, X. W. (2010). Mechanism of haploidy-dependent unreductional meiotic cell division in polyploid wheat. Chromosoma 119, 275–285. doi: 10.1007/s00412-010-0256-y
Cakmak, I. (2007). Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 302, 1–17. doi: 10.1007/s11104-007-9466-3
Cakmak, I., Pfeiffer, W. H., and Mcclafferty, B. (2010). Biofortification of durum wheat with zinc and iron. Cereal Chem. J. 87, 10–20. doi: 10.1094/cchem-87-1-0010
Ceresini, P. C., Castroagudín, L. V., Rodrigues, A. F., Rios, J. A., Aucique-Pérez, C. E., Moreira, S. I., et al. (2018). Wheat blast: from its origins in South America to its emergence as a global threat. Mol. Plant. Pathol. 20, 155–172. doi: 10.1111/mpp.12747
Chen, P. D., Tsujimoto, H., and Gill, B. S. (1994). Transfer of PhI genes promoting homoeologous pairing from Triticum speltoides to common wheat. Theor. Appl. Genet. 88, 97–101. doi: 10.1007/BF00222400
Chen, S., Guo, Y., Briggs, J., Dubach, F., Chao, S., Zhang, W., et al. (2018). Mapping and characterization of wheat stem rust resistance genes SrTm5 and Sr60 from Triticum monococcum. Theor. Appl. Genet. 131, 625–635. doi: 10.1007/s00122-017-3024-z
Chèvre, A. M., Jahier, J., and Trottet, M. (1989). Expression of disease resistance genes in amphiploid wheats-Triticum tauschii (Coss.) Schmal. Cer. Res. Comm. 17, 23–29.
Chu, C. G., Faris, J. D., Friesen, T. L., and Xu, S. S. (2006). Molecular mapping of hybrid necrosis genes Ne1 and Ne2 in hexaploid wheat using microsatellite markers. Theor. Appl. Genet. 112, 1374–1381. doi: 10.1007/s00122-006-0239-9
Colmer, T. D., Flowers, T. J., and Munns, R. (2006). Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 57, 1059–1078. doi: 10.1093/jxb/erj124
Cox, T. S., Hatcher, J. H., Gill, B. S., Raupp, W. J., and Sears, R. G. (1990). Agronomic performance of hexaploid wheat lines derived from direct crosses between wheat and Aegilops squarrosa. Plant Breed. 105, 271–277. doi: 10.1111/j.1439-0523.1990.tb01285.x
Cox, T. S., and Hatchett, J. H. (1994). Resistance gene H26 transferred from dipoid goatgrass to common wheat. Crop Sci. 34, 958–960. doi: 10.2135/cropsci1994.0011183X003400040023x
Cox, T. S., Raupp, W. J., and Gill, B. S. (1994). Leaf rust-resistance genes Lr41, Lr42 and Lr43 transferred from Triticum tauschii to common wheat. Crop Sci. 34, 339–343. doi: 10.2135/cropsci1994.0011183X003400020005x
Cox, T. S., Sears, R. G., and Gill, B. S. (1992). Registration of KS90WGRC10 leaf rust-resistant hard red winter wheat germplasm. Crop Sci. 32:506. doi: 10.2135/cropsci1992.0011183X003200020060x
Cruz, C. D., Peterson, G. L., Bockus, W. W., Kankanala, P., Dubcovsky, J., Jordan, K. W., et al. (2016). The 2NS translocation from Aegilops ventricosa confers resistance to the Triticum pathotype of Magnaporthe oryzae. Crop Sci. 56, 990–1000. doi: 10.2135/cropsci2015.07.0410
Dale, Z., Jie, H., Luyu, H., Cancan, Z., Yun, Z., Yarui, S., et al. (2017). An advanced backcross population through synthetic octaploid wheat as a “bridge”: development and QTL detection for seed dormancy. Front. Plant Sci. 8:2123. doi: 10.3389/fpls.2017.02123
Damania, A. B., Altunji, H., and Dhaliwal, H. S. (1992). Evaluation of Aegilops spp. for Drought and Frost Tolerance. Genetics Research Unit Annual Report 1992. Aleppo: ICARDA, 45–46.
Das, M. K., Bai, G. H., Mujeeb-Kazi, A., and Rajaram, S. (2016). Genetic diversity among synthetic hexaploid wheat accessions (Triticum aestivum) with resistance to several fungal diseases. Genet. Resour. Crop Evol. 63, 1285–1296. doi: 10.1007/s10722-015-0312-9
Delibes, A., Del Morala, J., Martin-Sanchez, J. A., Mejias, A., Gallego, M., Casado, D., et al. (1997). Hessian fly-resistance gene transferred from chromosome 4Mv of Aegilops ventricosa to Triticum aestivum. Theor. Appl. Genet. 94, 858–864. doi: 10.1007/s001220050487
Delibes, A., Lopez-Brafia, I., Mena, M., and Garcia-Olmedo, F. (1987). Genetic transfer of resistance to powdery mildew and of an associated biochemical marker from Aegilops ventricosa to hexaploid wheat. Theor. Appl. Genet. 73, 605–608. doi: 10.1007/BF00289201
Delibes, A., Romero, D., Aguaded, S., Duce, A., Mena, M., López-Braña, I., et al. (1993). Resistance to the cereal cyst nematode (Heterodera avenae Woll.) transferred from the wild grass Aegilops ventricosa to hexaploid wheat by a “stepping-stone” procedure. Theor. Appl. Genet. 87, 402–408. doi: 10.1007/BF01184930
Dhaliwal, H. S., Sharma, S. K., and Randhawa, A. S. (1986). How to overcome hybrid necrosis in wheat? Wheat Inf. Serv. 61, 27–28.
Donini, P., Koebner, R. M., and Ceoloni, C. (1995). Cytogenetic and molecular mapping of the wheat-Aegilops longissima chromatin breakpoints in powdery mildew-resistant introgression lines. Theor. Appl. Genet. 91, 738–743. doi: 10.1007/BF00220952
Doussinault, G., Delibes, A., Sanchez-Monge, R., and Garcia-Olmedo, F. (1983). Transfer of a dominant gene for resistance to eyespot disease from a wild grass to hexaploid wheat. Nature 303, 698–700. doi: 10.1038/303698a0
Dover, G. A., and Riley, R. (1972). Variation at two loci affecting homeologous meiotic chromosome pairing in Triticum aestivum × Aegilops mutica hybrids. Nat. New Biol. 235, 1–6. doi: 10.1038/newbio235061a0
Dover, G. A., and Riley, R. (1977). Inferences from genetical evidence on the course of meiotic chromosome pairing in plants. Philos. Trans. R. Soc. Lond. B. 277, 313–328. doi: 10.1098/rstb.1977.0020
Dreccer, A. F., Borgognone, A. G., Ogbonnaya, F. C., Trethowan, R. M., and Winter, B. (2007). CIMMYT-selected derived synthetic bread wheats for rainfed environments: yield evaluation in Mexico and Australia. Field Crops Res. 100, 218–228. doi: 10.1016/j.fcr.2006.07.005
Du, P., Zhuang, L., Wang, Y., Yuan, L., Wang, Q., Wang, D., et al. (2017). Development of oligonucleotides and multiplex probes for quick and accurate identification of wheat and Thinopyrum bessarabicum chromosomes. Genome 60, 93–103. doi: 10.1139/gen-2016-0095
Duveiller, E., He, X., and Singh, P. K. (2016). “Wheat blast: An emerging disease in South America potentially threatening wheat production,” in World Wheat Book, A History of Wheat, eds A. Bonjean and M. van Ginkel (Paris: Lavoisier), 1107–1122.
Dvorak, J., Deal, K. R., and Luo, M. C. (2006). Discovery and mapping of wheat Ph1 suppressors. Genetics 174, 17–27. doi: 10.1534/genetics.106.058115
Dvorak, J., and Knott, D. R. (1990). Location of a Triticum speltoides chromosome segment conferring resistance to leaf rust in Triticum aestivum. Genome 33, 892–897. doi: 10.1139/g90-134
Dvorak, J., Luo, M. C., Yang, Z. L., and Zhang, H. B. (1998). The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 97, 657–670. doi: 10.1007/s001220050942
Dyck, P. L., and Kerber, E. R. (1970). Inheritance in hexaploid wheat of adult-plant leaf rust resistance derived from Aegilops squarrosa. Can. J. Genet. Cytol. 13, 480–483. doi: 10.1139/g70-025
Eastwood, R. F., Lagudah, E. S., and Appels, R. (1994). A directed search for DNA sequences tightly linked to cereal cyst nematode resistance genes in Triticum tauschii. Genome 37, 311–319. doi: 10.1139/g94-043
Eastwood, R. F., Lagudah, E. S., Appels, R., Hannah, M., and Kollmorgen, J. (1991). Triticum tauschii: a novel source of resistance to cereal cyst nematode (Heterodera avenae). Aust. J. Agric. Res. 42, 69–77. doi: 10.1270/jsbbs.15158
Edae, E. A., Olivera, P. D., Jin, Y., Poland, J. A., and Rouse, M. N. (2016). Genotype-by-sequencing facilitates genetic mapping of a stem rust resistance locus in Aegilops umbellulata, a wild relative of cultivated wheat. BMC Genomics 17:1039. doi: 10.1186/s12864-016-3370-2
Edae, E. A., Olivera, P. D., Jin, Y., and Rouse, M. N. (2017). Genotyping-by-sequencing facilitates a high-density consensus linkage map for Aegilops umbellulata, a wild relative of cultivated wheat. G3 7, 1551–1561. doi: 10.1534/g3.117.039966
Edet, O. U., Gorafi, Y. S. A., Nasuda, S., and Tsujimoto, H. (2018). DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 8:16397. doi: 10.1038/s41598-018-34811-y
El Bouhssini, M., Nachit, M. M., Valkoun, J., Abdalla, O., and Rihawi, F. (2008). Sources of resistance to hessian fly (Diptera: Cecidomyiidae) in Syria identified among Aegilops species and synthetic derived bread wheat lines. Genet. Resour. Crop Evol. 55, 1215–1219. doi: 10.1007/s10722-008-9321-2
Elbashir, A. A. E., Gorafi, Y. S. A., Tahir, I. S. A., Elhashimi, A. M. A., Abdalla, M. G. A., and Tsujimoto, H. (2017). Genetic variation in heat tolerance-related traits in a population of wheat multiple synthetic derivatives. Breed Sci. 67, 483–492. doi: 10.1270/jsbbs.17048
Endo, T. R. (1979). Selective gametocidal action of a chromosome of Aegilops cylindrica in a cultivar of common wheat. Wheat Info. Serv. 50, 24–28.
Endo, T. R. (1985). Two types of gametocidal chromosomes of Ae. sharonensis and Ae. longissima. Jpn. J. Genet. 60, 125–135. doi: 10.1266/jjg.60.125
Endo, T. R. (2007). The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res. 15, 67–75. doi: 10.1007/s10577-006-1100-3
Endo, T. R., and Katayama, Y. (1978). Finding of a selectively retained chromosome of Aegilops caudata L. in common wheat. Wheat Info. Serv. 48, 32–35.
Endo, T. R., and Tsunewaki, K. (1975). Sterility of common wheat with Aegilops triuncialis cytoplasm. J. Hered. 66, 13–18. doi: 10.1093/oxfordjournals.jhered.a108562
Fakhri, Z., Mirzaghaderi, G., Ahmadian, S., and Mason, A. S. (2016). Unreduced gamete formation in wheat × Aegilops spp. hybrids is genotype specific and prevented by shared homologous subGenomes. Plant Cell. Rep. 35, 1143–1154. doi: 10.1007/s00299-016-1951-9
Farooq, S., and Azam, F. E. (2001). Co existence of salt and drought tolerance in Triticeae. Hereditas 135, 205–210. doi: 10.1111/j.1601-5223.2001.00205.x
Feldman, M., and Mello-Sampayo, T. (1967). Suppression of homeologous pairing in hybrids of polyploid wheats × Triticum speltoides. Can. J. Genet. Cytol. 9, 307–313. doi: 10.1139/g67-029
Fernandez-Calvin, B., and Orellana, J. (1991). Metaphase I bound arms frequency and Genome analysis in wheat-Aegilops hybrids. 1. Ae. variabilis-wheat and Ae. kotschyi-wheat hybrids with low and high homoeologous pairing. Theor. Appl. Genet. 83, 264–272. doi: 10.1007/BF00226261
Friebe, B., Jiang, J., Raupp, W. J., McIntosh, R. A., and Gill, B. S. (1996). Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91, 59–87. doi: 10.1007/BF00035277
Friebe, B., Mukai, Y., Dhaliwal, H. S., Martin, T. J., and Gill, B. S. (1991). Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germplasm by C-banding and in situ hybridization. Theor. Appl. Genet. 81, 381–389. doi: 10.1007/BF00228680
Friebe, B., Zhang, P., Nasuda, S., and Gill, B. S. (2003). Characterization of a knock-out mutation at the Gc2 locus in wheat. Chromosoma 111, 509–517. doi: 10.1007/s00412-003-0234-8
Fritz, A. K., Cox, T. S., Gill, B. S., and Sears, R. G. (1995). Molecular marker-facilitated analysis of introgression in winter wheat × Triticum tauschii populations. Crop Sci. 35, 1691–1695. doi: 10.2135/cropsci1995.0011183X003500060030x
Fukuda, K., and Sakamoto, S. (1992). Cytological studies on unreduced male gamete formation in hybrids between tetraploid emmer wheats and Aegilops squarrosa L. Jpn. J. Breed. 42, 255–266. doi: 10.1111/j.1601-5223.1992.tb00150.x
Ghaffary, S. M. T., Faris, J. D., Friesen, T. L., Visser, R. G. F., van der Lee, T. A. G., Robert, O., et al. (2012). New broad-spectrum resistance to Septoria tritici blotch derived from synthetic hexaploid wheat. Theor. Appl. Genet. 124, 125–142. doi: 10.1007/s00122-011-1692-7
Gill, B. S., Dhaliwal, H. S., and Multani, D. S. (1988). Synthesis and evaluation of Triticum durum — T. monococcum amphiploids. Theor. Appl. Genet. 75, 912–916. doi: 10.1007/BF00258053
Gill, B. S., Hatchett, J. H., and Raupp, W. J. (1987). Chromosomal mapping of Hessian fly resistance gene H13 in the D Genome of wheat. J. Hered. 78, 97–100. doi: 10.1093/oxfordjournals.jhered.a110344
Gill, B. S., and Raupp, W. J. (1987). Direct genetic transfers from Aegilops squarrosa L. to hexaploid wheat. Crop Sci. 27, 445–450. doi: 10.2135/cropsci1987.0011183X002700030004x
Gill, B. S., Waines, J. G., and Sharma, H. C. (1981). Endosperm abortion and the production of viable Aegilops squarrosa x triticum boeoticum hybrids by embryo culture. Plant Sci. Lett. 23, 181–187. doi: 10.1016/0304-4211(81)90010-9
Gill, K. S., Lubbers, E. L., Gill, B. S., Raupp, W. J., and Cox, T. S. (1991). A genetic linkage map of Triticum tauschii (DD) and its relationship to the D-genome of bread wheat (AABBDD). Genome 14, 362–374. doi: 10.1139/g91-058
Giorgi, B. (1983). “Origin, behaviour and utilization of a Ph1 mutant of durum wheat, Triticum turgidum (L.) var. durum,” in Proceedings of the 6th International Wheat Genetics Symposium Kyoto, ed. S. Sakamoto Japan,1033–1040.
Gustafson, G. D., and Shaner, G. (1982). Influence of plant age on the expression of slow-mildewing resistance in wheat. Phytopathology 72, 746–749. doi: 10.1094/Phyto-72-746
Hall, M. D., Brown-Guedira, G., Klatt, A., and Fritz, A. K. (2009). Genetic analysis of resistance to soil-borne wheat mosaic virus derived from Aegilops tauschii. Euphytica 169, 169–176. doi: 10.1007/s10681-009-9910-y
Hammer, K. (1980). Vorarbeiten zur monographischen darstellung von wildpflanzen sortimenten: Aegilops L. Kulturpflanze 28, 33–180.
Hansen, F. L., Andersen, S. B., Due, I. K., and Olesen, A. (1988). Nitrous oxide as a possible alternative agent for chromosome doubling of wheat haploids. Plant Sci. 54, 219–222. doi: 10.1016/0168-9452(88)90116-1
Hatchett, J. H., Martin, T. J., and Livers, R. W. (1981). Expression and inheritance of resistance to hessian fly in synthetic hexaploid wheats derived from Triticum tauschii (coss) schmal.1. Crop Sci. 21, 731–734. doi: 10.2135/cropsci1981.0011183X002100050025x
Helguera, M., Khan, A. I., Kolmer, J., Lijavetzky, D., Zhongqi, L., and Dubcovsky, J. (2003). PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Sci. 43, 1839–1847. doi: 10.2135/cropsci2003.1839
Helguera, M., Khan, I. A., and Dubcovsky, J. (2000). Development of PCR markers for the wheat leaf rust gene Lr47. Theor. Appl. Genet. 101, 625–631. doi: 10.1007/s001220051397
Helguera, M., Vanzetti, L., Soria, M., Khan, I. A., Kolmer, J., and Dubcovsky, J. (2005). PCR markers for Triticum speltoides leaf rust resistance gene Lr51 and their use to develop isogenic hard red spring wheat lines. Crop Sci. 45, 728–734. doi: 10.2135/cropsci2005.0728
Hermsen, J. G. T. (1963). Hybrid necrosis as a problem for the wheat breeder. Euphytica 12, 1–16. doi: 10.1007/BF00033587
Hermsen, J. G. T., and Waninge, J. (1972). Attempts to localize the gene Ch1 for hybrid chlorosis in wheat. Euphytica 21, 204–208. doi: 10.1007/BF00036760
Hiebert, C. W., Kassa, M. T., McCartney, C. A., You, F. M., Rouse, M. N., Fobert, P., et al. (2016). Genetics and mapping of seedling resistance to Ug99 stem rust in winter wheat cultivar Triumph 64 and differentiation of SrTmp, SrCad, and Sr42. Theor. Appl. Genet. 129, 2171–2177. doi: 10.1007/s00122-016-2765-4
Hollenhorst, M. M., and Joppa, L. R. (1983). Chromosomal location of genes for resistance to greenbug in ‘Largo’ and ‘Amigo’ wheats. Crop Sci. 23, 91–93. doi: 10.2135/cropsci1983.0011183X002300010026x
Holubec, V., and Havlıckova, A. (1994). Interspecific differences in cereal aphid infestation of 20 Aegilops species. Genet. Slecht 30, 81–87.
Hsam, S. L. K., Huang, X. Q., Ernst, F., Hartl, L., and Zeller, F. J. (1998). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 5. Alleles at the Pm1 locus. Theor. Appl. Genet. 96, 1129–1134. doi: 10.1007/s001220050848
Hsam, S. L. K., Lapochkina, I. F., and Zeller, F. J. (2003). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 8. gene Pm32 in a wheat-Aegilops speltoides translocation line. Euphytica 133, 367–370. doi: 10.1023/A:1025738513638
Huang, S., Sirikhachornkit, A., Su, X. J., Faris, J., Gill, B., Haselkorn, R., et al. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. U.S.A. 99, 8133–8138. doi: 10.1073/pnas.072223799
Huang, S., Steffenson, B. J., Sela, H., and Stinebaugh, K. (2018). Resistance of Aegilops longissima to the rusts of wheat. Plant Dis. 102, 1124–1135. doi: 10.1094/PDIS-06-17-0880-RE
Huang, X., Zhu, M., Zhuang, L., Zhang, S., Wang, J., Chen, X., et al. (2018). Structural chromosome rearrangements and polymorphisms identified in Chinese wheat cultivars by high-resolution multiplex oligonucleotide FISH. Theor. Appl. Genet. 131, 1967–1986. doi: 10.1007/s00122-018-3126-2
Iehisa, J. C. M., and Takumi, S. (2012). Variation in abscisic acid responsiveness of Aegilops tauschii and hexaploid wheat synthetics due to the D-Genome diversity. Genes Genet. Syst. 87, 9–18. doi: 10.1266/ggs.87.9
Igarashi, S., Utiamada, C. M., Igarashi, L. C., Kazuma, A. H., and Lopes, R. S. (1986). Pyricularia in wheat. 1. occurrence of Pyricularia sp. in Parana State. (in Portuguese). Fitopatol. Bras. 11, 351–352.
Inoue, Y., Vy, T. T. P., Yoshida, K., Asano, H., Mitsuoka, C., Asuke, S., et al. (2017). Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science 357, 80–83. doi: 10.1126/science.aam9654
Ishihara, A., Mizuno, N., Islam, R. A., Doležel, J., Endo, T. R., and Nasuda, S. (2014). Dissection of barley chromosomes 1H and 6H by the gametocidal system. Genes Genet. Syst. 89, 203–214. doi: 10.1266/ggs.89.203
Jafarzadeh, J., Bonnett, D., Jannink, J. L., Akdemir, D., Dreisigacker, S., and Sorrells, M. E. (2016). Breeding value of primary synthetic wheat genotypes for grain yield. PLoS One 11:e0162860. doi: 10.1371/journal.pone.0162860
Jahier, J., Abelard, P., Tanguy, A. M., Dedryver, F., Rivoal, R., Khatkar, S., et al. (2001). The Aegilops ventricosa segment on chromosome 2AS of the wheat cultivar ‘VPM1’ carries the cereal cyst nematode resistance gene Cre5. Plant Breed. 120, 125–128. doi: 10.1046/j.1439-0523.2001.00585.x
Jahier, J., Tanguy, A. M., Abelard, P., and Rivoal, R. (1996). Utilization of deletions to localize a gene for resistance to the cereal cyst nematode, Heterodera avenae, on an Aegilops ventricosa chromosome. Plant Breed. 115, 282–284. doi: 10.1111/j.1439-0523.1996.tb00919.x
Jeuken, M. J. W., Zhang, N. W., McHale, L. K., Pelgrom, K., den Boer, E., Lindhout, P., et al. (2009). Rin4 causes hybrid necrosis and race-specific resistance in an interspecific lettuce hybrid. Plant Cell 21, 3368–3378. doi: 10.1105/tpc.109.070334
Jia, J., Devos, K. M., Chao, S., Miller, T. E., Reader, S. M., and Gale, M. D. (1996). RFLP-based maps of the homoeologous group-6 chromosomes of wheat and their application in the tagging of Pm12, a powdery mildew resistance gene transferred from Aegilops speltoides to wheat. Theor. Appl. Genet. 92, 559–565. doi: 10.1007/BF00224558
Joshi, G. P., Endo, T. R., and Nasuda, S. (2013). PCR and sequence analysis of barley chromosome 2H subjected to the gametocidal action of chromosome 2C. Theor. Appl. Genet. 126, 2381–2390. doi: 10.1007/s00122-013-2142-5
Jovkova, M. E., Kondeva, E., and Kostova, R. (1977). Biochemical investigations on Aegilops crassa x Triticum aestivum hybrids. Genet. Sel. 10, 91–98.
Juliana, P., Singh, R. P., Singh, P. K., Crossa, J., Huerta-Espino, J., Lan, C., et al. (2017). Genomic and pedigree-based prediction for leaf, stem, and stripe rust resistance in wheat. Theor. Appl. Genet. 130, 1415–1430. doi: 10.1007/s00122-017-2897-1
Kaloshian, I., Roberts, P. A., Waines, J. G., and Thomason, I. J. (1990). Inheritance of resistance to root-knot nematodes in Aegilops squarrosa L. J. Hered. 81, 170–172. doi: 10.1093/oxfordjournals.jhered.a110956
Kerber, E. R. (1987). Resistance to leaf rust in hexaploid wheat: Lr32 a third gene derived from Triticum tauschii. Crop Sci. 27, 204–206. doi: 10.2135/cropsci1987.0011183X002700020013x
Kerber, E. R., and Dyck, P. L. (1979). “Resistance to stem rust and leaf rust of wheat in Aegilops squarrosa and transfer of a gene for stem rust resistance to hexaploid wheat,” in Proceedings of the 5th international wheat genetics symposium, ed. S. Ramanujam (New Delhi: Indian Society of Genetics and Plant Breeding), 358–364.
Kerber, E. R., and Dyck, P. L. (1990). Transfer to hexaploid wheat of linked genes for adult-plant leaf rust and seedling stem rust resistance from an amphiploid of Aegilops speltoides x Triticum monococcum. Genome 33, 530–537. doi: 10.1139/g90-079
Kielsmeier-Cook, J., Danilova, T. V., Friebe, B., and Rouse, M. N. (2015). Resistance to the Ug99 race group of Puccinia graminis f. sp. tritici in wheat-intra/intergeneric hybrid derivatives. Plant Dis. 99, 1317–1325. doi: 10.1094/PDIS-09-14-0922-RE
Kihara, H. (1944). Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hortic. 19, 889–890. doi: 10.1016/j.jgg.2011.07.002
Kihara, H. (1946). Maturation division in F1 hybrids between Triticum dicoccoides × Aegilops squarrosa. La Kromosoma 1, 6–11.
Kihara, H. (1954). Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 19, 336–357. doi: 10.1508/cytologia.19.336
Kihara, H., and Lilienfeld, F. A. (1949). “New synthesized 6x-wheat,” in Proceedings of the 8th International Congress of Genetics, ed. G. B. A. R. Larsson Hereditas Stockholm, 307–319.
Kilian, B., Mammen, K., Millet, E., Sharma, R., Graner, A., Salamini, F., et al. (2011). “Aegilops,” in Wild Crop Relatives: Genomic and Breeding Resources, ed. C. Kole (New York, NY: Spring-Verlag), 1–76.
Kilian, B., Özkan, H., Deusch, O., Effgen, S., Brandolini, A., Kohl, J., et al. (2007). Independent wheat B and G Genome origins in outcrossing Aegilops progenitor haplotypes. Mol. Biol. Evol. 24, 217–227. doi: 10.1093/molbev/msl151
Kimber, G., and Sears, E. R. (1987). “Evolution in the genus Triticum and the origin of cultivated wheat,” in Wheat and Wheat Improvement, 2nd Edn, ed. E. G. Heyne (Madison, WI: American Society of Agronomy), 154–164.
Kimber, G., and Zhao, Y. H. (1983). The D genome of the Triticeae. Can. J. Genet. Cytol. 25, 581–589. doi: 10.1139/g83-088
King, J., Grewal, S., Yang, C. Y., Hubbart, S., Scholefield, D., Ashling, S., et al. (2017). A step change in the transfer of interspecific variation into wheat from Amblyopyrum muticum. Plant Biotech. J. 15, 217–226. doi: 10.1111/pbi.12606
King, J., Grewal, S., Yang, C. Y., Hubbart, S., Scholefield, D., Ashling, S., et al. (2018). Introgression of Aegilops speltoides segments in Triticum aestivum and the effect of the gametocidal genes. Ann. Bot. 121, 229–240. doi: 10.1093/aob/mcx149
Klindworth, D. L., Niu, Z., Chao, S., Friesen, T. L., Jin, Y., Faris, J. D., et al. (2012). Introgression and characterization of a goatgrass gene for a high level of resistance to ug99 stem rust in tetraploid wheat. G3 2, 665–673. doi: 10.1534/g3.112.002386
Koba, T., and Shimada, T. (1993). Crossability of common wheat with Aegilops squarrosa. Wheat Inf. Serv. 77, 7–12.
Kohli, M. M., Mehta, Y. R., Guzman, E., De Viedma, L., and Cubilla, L. E. (2011). Pyricularia blast – A threat to wheat cultivation. Czech J. Genet. Plant Breed. 47, S130–S134. doi: 10.17221/3267-CJGPB
Koo, D. H., Liu, W., Friebe, B., and Gill, B. S. (2017). Homoeologous recombination in the presence of Ph1 gene in wheat. Chromosoma 126, 531–540. doi: 10.1007/s00412-016-0622-5
Kota, R. S., and Dvorak, J. (1988). Genomic instability in wheat induced by chromosome 6Bs of Triticum speltoides. Genetics 120, 1085–1094.
Krolow, K. D. (1970). Investigations on compatibility between wheat and rye. Z PXanzenzuchtg 64, 44–72.
Kumar, A., Seetan, R., Mergoum, M., Tiwari, V. K., Iqbal, M. J., Wang, Y., et al. (2015). Radiation hybrid maps of the D-genome of Aegilops tauschii and their application in sequence assembly of large and complex plant genomes. BMC Genomics 16:800. doi: 10.1186/s12864-015-2030-2
Kumar, A., Simons, K., Iqbal, M. J., de Jiménez, M. M., Bassi, F. M., Ghavami, F., et al. (2012). Physical mapping resources for large plant genomes: radiation hybrids for wheat D-genome progenitor Aegilops tauschii. BMC Genomics 13:597. doi: 10.1186/1471-2164-13-597
Kuraparthy, V., Chhuneja, P., Dhaliwal, H. S., Kaur, S., Bowden, R. L., and Gill, B. S. (2007). Characterization and mapping of cryptic alien introgression from Aegilops geniculata with new leaf rust and stripe rust resistance genes Lr57 and Yr40 in wheat. Theor. Appl. Genet. 114, 1379–1389. doi: 10.1007/s00122-007-0524-2
Kuraparthy, V., Sood, S., Guedira, G.-B., and Gill, B. S. (2011). Development of a PCR assay and marker-assisted transfer of leaf rust resistance gene Lr58 into adapted winter wheats. Euphytica 180, 227–234. doi: 10.1007/s10681-011-0383-4
Kynast, R. G., Friebe, B., and Gill, B. S. (2000). Fate of multicentric and ring chromosomes induced by a new gametocidal factor located on chromosome 4Mg of Aegilops geniculata. Chrom. Res. 8, 133–139. doi: 10.1023/A:1009294519798
Lange, W., and Jochemsen, G. (1992). Use of the gene pools of Triticum turgidum ssp. dicoccoides and Aegilops squarrosa for the breeding of common wheat (T. aestivum), through chromosome-doubled hybrids. II. Morphology and meiosis of the amphiploids. Euphytica 59, 213–220. doi: 10.1007/BF00041273
Lein, A. (1943). The genetical basis of the crossability between wheat and rye. Z. Indukt. Abstamm. Vererbungsl. 81, 28–59.
Li, A., Liu, D., Yang, W., Kishii, M., and Mao, L. (2018). Synthetic hexaploid wheat: yesterday, today, and tomorrow. Engineering 4, 552–558. doi: 10.1016/j.eng.2018.07.001
Li, H., Deal, K. R., Luo, M. C., Ji, W., Distelfeld, A., and Dvorak, J. (2017). Introgression of the Aegilops speltoides Su1-Ph1 suppressor into wheat. Front Plant Sci. 8:2163. doi: 10.3389/fpls.2017.02163
Li, J., Nasuda, S., and Endo, T. R. (2013). Dissection of rye chromosomes by the gametocidal system. Genes Genet. Syst. 88, 321–327. doi: 10.1266/ggs.88.321
Li, J., Wan, H. S., and Yang, W. Y. (2014). Synthetic hexaploid wheat enhances variation and adaptive evolution of bread wheat in breeding processes. J. Syst. Evol. 52, 735–742. doi: 10.1111/jse.12110
Liu, C., Gong, W., Han, R., Guo, J., Li, G., Li, H., et al. (2019). Characterization, identification and evaluation of a set of wheat-Aegilops comosa chromosome lines. Sci Rep. 9:4773. doi: 10.1038/s41598-019-41219-9
Liu, D. C., Zhang, L. Q., Yan, Z. H., Lan, X. J., and Zheng, Y. L. (2010). Stripe rust resistance in Aegilops tauschii and its genetic analysis. Genet. Resour. Grop Evol. 57, 325–328. doi: 10.1007/s10722-009-9510-7
Liu, W., Jin, Y., Rouse, M., Friebe, B., Gill, B. S., and Pumphrey, M. O. (2011a). Development and characterization of wheat-Ae. searsii robertsonian translocations and a recombinant chromosome conferring resistance to stem rust. Theor. Appl. Genet. 122, 1537–1545. doi: 10.1007/s00122-011-1553-4
Liu, W., Koo, D. H., Xia, Q., Li, C., Bai, F., Song, Y., et al. (2017). Homoeologous recombination-based transfer and molecular cytogenetic mapping of powdery mildew-resistant gene Pm57 from Aegilops searsii into wheat. Theor. Appl. Genet. 130, 841–848. doi: 10.1007/s00122-017-2855-y
Liu, W., Rouse, M., Friebe, B., Jin, Y., Gill, B. S., and Pumphrey, M. O. (2011b). Discovery and molecular mapping of a new gene conferring resistance to stem rust, Sr53, derived from Aegilops geniculata and characterization of spontaneous translocation stocks with reduced alien chromatin. Chromosome Res. 19, 669–682. doi: 10.1007/s10577-011-9226-3
Liu, Y., Wang, L., Deng, M., Li, Z., Lu, Y., Wang, J., et al. (2015). Genome-wide association study of phosphorus-deficiency-tolerance traits in Aegilops tauschii. Theor. Appl. Genet. 128, 2203–2212. doi: 10.1007/s00122-015-2578-x
Luo, M. C., Gu, Y. Q., Puiu, D., Wang, H., Twardziok, S. O., Deal, K. R., et al. (2017). Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature 551, 498–502. doi: 10.1038/nature24486
Luo, M. C., Gu, Y. Q., You, F. M., Deal, K. R., Ma, Y., Hu, Y., et al. (2013). A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. U.S.A. 110, 7940–7945. doi: 10.1073/pnas.1219082110
Luo, M. C., Yang, Z. L., Kota, R. S., and Dvorák, J. (2000). Recombination of chromosomes 3A(m) and 5A(m) of Triticum monococcum with homeologous chromosomes 3A and 5A of wheat: the distribution of recombination across chromosomes. Genetics 154, 1301–1308.
Lutz, J., Hsam, S. L. K., Limpert, E., and Zeller, F. J. (1994). Powdery mildew resistance in Aegilops tauschii coss. and synthetic hexaploid wheats. Genet. Resour. Crop Evol. 41, 151–158. doi: 10.1007/BF00051631
Lutz, J., Hsam, S. L. K., Limpert, E., and Zeller, F. J. (1995). Chromosomal location of powdery mildew resistance genes in Triticum aestivum L. (common wheat) 2. genes Pm2 and Pm19 from Aegilops squarrosa L. Heredity 74, 152–156.
Maan, S. S. (1975). Exclusive preferential transmission of an alien chromosome in common wheat. Crop Sci. 15, 287–292. doi: 10.2135/cropsci1975.0011183X001500030002x
Mago, R., Verlin, D., Zhang, P., Bansal, U., Bariana, H., Jin, Y., et al. (2013). Development of wheat-Aegilops speltoides recombinants and simple PCR-based markers for Sr32 and a new stem rust resistance gene on the 2S#1 chromosome. Theor. Appl. Genet. 126, 2943–2955. doi: 10.1007/s00122-013-2184-8
Mago, R., Zhang, P., Bariana, H. S., Verlin, D. C., Bansal, U. K., Ellis, D. G., et al. (2009). Development of wheat lines carrying stem rust resistance gene Sr39 with reduced Aegilops speltoides chromatin and simple PCR markers for marker-assisted selection. Theor. Appl. Genet. 119, 1441–1450. doi: 10.1007/s00122-009-1146-7
Mahmud, H. (2019). Wheat blast in Bangladesh threatening South Asia wheat production. Acta Sci. Microbiol. 2, 08–09. doi: 10.1371/journal.pone.0197555
Maia, N. (1967). Obtention des bles tendres resistants au pietin-verse par croisements interspecifiques bles_Aegilops. C. R. Acad. Agric. 53, 149–154.
Makkouk, K. M., Comeau, A., and Ghulam, W. (1994). Resistance to barley yellow dwarf luteovirus in Aegilops species. Can. J. Plant Sci. 74, 631–634. doi: 10.4141/cjps94-113
Malaker, P. K., Barma, N. C. D., Tiwari, T. P., Collis, W. J., Duveiller, E., Singh, P. K., et al. (2016). First report of wheat blast caused by Magnaporthe oryzae pathotype Triticum in Bangladesh. Plant Dis. 100:2330. doi: 10.1094/PDIS-05-16-0666-PDN
Malik, R., Brown-Guerdira, G. L., Smith, C. M., Harvey, T. L., and Gill, B. S. (2003). Genetic mapping of wheat curl mite resistance genes Cmc3 and Cmc4 in common wheat. Crop Sci. 43, 644–650. doi: 10.2135/cropsci2003.0644
Marais, G. F., Badenhorst, P. E., Eksteen, A., and Pretorius, Z. A. (2010). Reduction of Aegilops sharonensis chromatin associated with resistance genes Lr56 and Yr38 in wheat. Euphytica 171, 15–22. doi: 10.1007/s10681-009-9973-9
Marais, G. F., Bekker, T. A., Eksteen, A., McCallum, B., Fetch, T., and Marais, A. S. (2009a). Attempts to remove gametocidal genes co-transferred to wheat with rust resistance from Aegilops speltoides. Euphytica 171, 71–85. doi: 10.1007/s10681-009-9996-2
Marais, G. F., Marais, A. S., McCallum, B., and Pretorius, Z. (2009b). Transfer of leaf rust and stripe rust resistance genes Lr62 and and Yr42 from Aegilops neglecta Req. ex Bertol. to common wheat. Crop Sci. 49, 871–879. doi: 10.2135/cropsci2008.06.0317
Marais, G. F., McCallum, B., and Marais, A. S. (2008). Wheat leaf rust resistance gene Lr59 derived from Aegilops peregrina. Plant Breed. 127, 340–345. doi: 10.1111/j.1439-0523.2008.01513.x
Marais, G. F., McCallum, B., Snyman, J. E., Pretorius, Z. A., and Marais, A. S. (2005). Leaf rust and stripe rust resistance genes Lr54 and Yr37 transferred to wheat from Aegilops kotschyi. Plant Breed. 124, 538–541. doi: 10.1111/j.1439-0523.2005.01116.x
Marais, G. F., Wessels, W. G., Horn, M., and du Toit, F. (1998). Association of a stem rust resistance gene (Sr45) and two Russian wheat aphid resistance genes (Dn5 and Dn7) with mapped structural loci in common wheat. S. Afr. J. Plant Soil 15, 67–71. doi: 10.1080/02571862.1998.10635119
Martin, T. J., Harvey, T. L., and Hatchett, J. H. (1982). Registration of greenbug and Hessian fly resistant wheat germplasm. Crop Sci. 22:1089. doi: 10.2135/cropsci1982.0011183X002200050061x
Martin-Sanchez, J. A., Gomez-Colmenarejo, M., Del Morel, J., Sin, E., Montes, M. J., Gonzalez-Belinchon, C., et al. (2003). A new Hessian fly resistance gene (H30) transferred from wild grass Aegilops triuncialis to hexaploid wheat. Theor. Appl. Genet. 106, 1248–1255. doi: 10.1007/s00122-002-1182-z
Matsuoka, Y. (2011). Evolution of polyploid Triticum wheats under cultivation: the role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52, 750–764. doi: 10.1093/pcp/pcr018
Matsuoka, Y., and Nasuda, S. (2004). Durum wheat as a candidate for the unknown female progenitor of bread wheat: an empirical study with a highly fertile F1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 109, 1710–1717. doi: 10.1007/s00122-004-1806-6
Matsuoka, Y., Nasuda, S., Ashida, Y., Nitta, M., Tsujimoto, H., Takumi, S., et al. (2013). Genetic basis for spontaneous hybrid Genome doubling during allopolyploid speciation of common wheat shown by natural variation analyses of the paternal species. PLoS One 8:e68310. doi: 10.1371/journal.pone.0068310
Matsuoka, Y., and Takumi, S. (2017). The role of reproductive isolation in allopolyploid speciation patterns: empirical insights from the progenitors of common wheat. Sci. Rep. 7:16004. doi: 10.1038/s41598-017-15919-z
Matsuoka, Y., Takumi, S., and Kawahara, T. (2007). Natural variation for fertile triploid F1 hybrid formation in allohexaploid wheat speciation. Theor. Appl. Genet. 115, 509–518. doi: 10.1007/s00122-007-0584-3
McFadden, E. S., and Sears, E. R. (1944). The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 13, 26–27.
McFadden, E. S., and Sears, E. R. (1946). The origin of Triticum spelta and its free-threshing hexaploid relatives. J. Hered. 37, 107–116. doi: 10.1093/oxfordjournals.jhered.a105594
McIntosh, R. A. (1988). “Catalogue of gene symbols for wheat,” in Proceedings of the 7th international wheat Genetics Symposium, eds T. E. Miller and R. M. D. Koebner (Bath: Bath Press), 1273.
McIntosh, R. A., Dubcovsky, J., Rogers, W. J., Morris, C. F., Appels, R., and Xia, X. C. (2012). Catalogue of gene symbols for wheat: 2012 supplement. Ann. Wheat Newsl. 58, 259–279.
McIntosh, R. A., Hart, G. E., and Gale, M. D. (1995). “Catalogue of gene symbols for wheat,” in Proceedings of the 8th International Wheat genetics symposium, eds Z. S. Li and Z. Y. Xin (Beijing: China Agricultural Scientech), 1333–1500.
McIntosh, R. A., Hart, G. E., Gale, M. D., and Rogers, W. J. (1998). “Catalogue of gene symbols for wheat,” in Proceedings of the 9th International Wheat Genet Symposium, Vol. 5, ed. A. E. Slinkard (Saskatoon: University Extension Press, University of Saskatchewan), 1–235.
McIntosh, R. A., Miller, T. E., and Chapman, V. (1982). Cytogenetical studies in wheat XII. Lr28 for resistance to Puccinia recondita and Sr34 for resistance to P. graminis tritici. Z. Pflanzenzucht 89, 295–306.
McIntosh, R. A., Yamazaki, Y., Dubcovsky, J., Rogers, W. J., Morris, C. F., Sommers, D. J., et al. (2008). “Catalogue of gene symbols for wheat: 2008,” in Proceedings of the 11th International Wheat Genetics, eds R. Appels, R. Eastwood, E. Lagudah, P. Langridge, M. Mackay, L. McIntyre, et al. (Sydney: Sydney University Press).
Mello-Sampayo, T. (1971). Genetic regulation of meiotic chromosome pairing by chromosome 3D of Triticum aestivum. Nat. New Biol. 230, 22–23. doi: 10.1038/newbio230022a0
Mena, M., Doussinault, G., Lopez-Braña, I., Aguaded, S., García-Olmedo, F., and Delibes, A. (1992). Eyespot resistance gene Pch-1 in H-93 wheat lines. Evidence of linkage to markers of chromosome group 7 and resolution from the endopeptidase locus Ep-D1b. Theor. Appl. Genet. 83, 1044–1047. doi: 10.1007/BF00232970
Mihova, S. (1988). The resistance of Aegilops species to Puccinia striiformis West f. sp. tritici in relation with their ploidy and Genome composition. Genet. Selekt. 21, 20–25.
Miller, T. E., Reader, S. M., Ainsworth, C. C., and Summers, R. W. (1987). “The introduction of a major gene for resistance to powdery mildew of wheat, Erysiphe graminis f. sp. tritici, from Aegilops speltoides into wheat,” in Proceedings of the EUCARPIA Conference Cereal Breeding Related to Integrated Cereal Production, eds M. L. Jorna and L. A. J. Shootmaker Wageningen, 179–183.
Miller, T. E., Reader, S. M., Mahmood, A., Purdie, K. A., and King, I. P. (1995). “Chromosome 3N of Aegilops uniaristata – a source of tolerance to high levels of aluminium for wheat,” in Proceedings of the 8th International Wheat Genetics Symposium, eds Z. S. Li and Z. Y. Xin (Beijing: China Agricultural Scientech), 1037–1042.
Miranda, L. M., Murphy, J. P., Marshall, D., Cowger, C., and Leath, S. (2007). Chromosomal location of Pm35, a novel Aegilops tauschii derived powdery mildew resistance gene introgressed into common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 1451–1456. doi: 10.1007/s00122-007-0530-4
Miranda, L. M., Murphy, J. P., Marshall, D., and Leath, S. (2006). Pm34: a new powdery mildew resistance gene transferred from Aegilops tauschii Coss. to common wheat (Triticum aestivum L.). Theor. Appl. Genet. 113, 1497–1504. doi: 10.1007/s00122-006-0397-9
Mishina, K., Sato, H., Manickavelu, A., Sassa, H., and Koba, T. (2009). Molecular mapping of Skr for crossabiltiy in common wheat. Breed. Sci. 59, 679–684. doi: 10.1270/jsbbs.59.679
Mizuno, N., Hosogi, N., Park, P., and Takumi, S. (2010). Hypersensitive response-like reaction is associated with hybrid necrosis in interspecific crosses between tetraploid wheat and Aegilops tauschii coss. PLoS One 5:e11326. doi: 10.1371/journal.pone.0011326
Mizuno, N., Shitsukawa, N., Hosogi, N., Park, P., and Takumi, S. (2011). Autoimmune response and repression of mitotic cell division occur in inter-specific crosses between tetraploid wheat and Aegilops tauschii Coss. that show low temperature-induced hybrid necrosis. Plant J. 68, 114–128. doi: 10.1111/j.1365-313X.2011.04667.x
Molnár, I., Šimková, H., Leverington-Waite, M., Goram, R., Cseh, A., Vrána, J., et al. (2013). Syntenic relationships between the U and M Genomes of Aegilops, wheat and the model species Brachypodium and rice as revealed by COS markers. PLoS One 8:e70844. doi: 10.1371/journal.pone.0070844
Molnár, I., Vrána, J., Burešová, V., Cápal, P., Farkas, A., Darkó, É, et al. (2016). Dissecting the U, M, S and C Genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J. 88, 452–467. doi: 10.1111/tpj.13266
Monneveux, P., Zaharieva, M., and Rekika, D. (2000). “The utilisation of Triticum and Aegilops species for the improvement of durum wheat,” in Durum Wheat Improvement in the Mediterranean Region: New Challenges, eds C. Royo, M. N. Nachit, N. Di Fonzo, and J. L. Araus (Zaragoza: CIHEAM), 71–81.
Mottaleb, K. A., Singh, P. K., Sonder, K., Kruseman, G., Tiwari, T. P., Barma, N. C. D., et al. (2018). Threat of wheat blast to South Asia’s food security: an ex-ante analysis. PLoS One 13:e0197555. doi: 10.1371/journal.pone.0197555
Nakano, H., Mizuno, N., Tosa, Y., Yoshida, K., Park, P., and Takumi, S. (2015). Accelerated senescence and enhanced disease resistance in hybrid chlorosis lines derived from interspecific crosses between tetraploid wheat and Aegilops tauschii. PLoS One 10:e0121583. doi: 10.1371/journal.pone.0121583
Narasimhamoorthy, B., Gill, B. S., Fritz, A. K., Nelson, J. C., and Brown-Guedira, G. L. (2006). Advanced backcross QTL analysis of a hard winter wheat × synthetic wheat population. Theor. Appl. Genet. 112, 787–796. doi: 10.1007/s00122-005-0159-0
Niranjana, M. (2017). Gametocidal genes of Aegilops: segregation distorters in wheat-Aegilops wide hybridization. Genome 60, 639–647. doi: 10.3389/fpls.2018.01756
Nishijima, R., Ikeda, T. M., and Takumi, S. (2018). Genetic mapping reveals a dominant awn-inhibiting gene related to differentiation of the variety anathera in the wild diploid wheat Aegilops tauschii. Genetica 146, 75–84. doi: 10.1007/s10709-017-9998-2
Nishikawa, K. (1960). Hybrid lethality in crosses between emmer wheats and Aegilops squarrosa, I. Vitality of F1 hybrids between emmer wheats and Ae. squarrosa var. typica. Seiken Ziho 11, 21–28.
Nishikawa, K. (1962a). Hybrid lethality in crosses between Emmer wheats and Aegilops squarrosa, II. Synthesized 6x wheats employed as test varieties. Jpn. J. Genet. 37, 227–236. doi: 10.1266/jjg.37.227
Nishikawa, K. (1962b). Hybrid lethality in crosses between emmer wheats and Aegilops squarrosa, III. Gene analysis of type-2 necrosis. Seiken Ziho 14, 45–50.
Nishikawa, K., Mori, T., Takami, N., and Furuta, Y. (1974). Mapping of progressive necrosis gene Ne1 and Ne2 of common wheat by the telocentric method. Jpn. J. Breed. 24, 277–281. doi: 10.1270/jsbbs1951.24.277
Niu, Z., Chao, S., Cai, X., Whetten, R. B., Breiland, M., Cowger, C., et al. (2018). Molecular and cytogenetic characterization of six wheat-Aegilops markgrafii disomic addition lines and their resistance to rusts and powdery mildew. Front. Plant Sci. 9:1616. doi: 10.3389/fpls.2018.01616
Niu, Z., Klindworth, D. L., Friesen, T. L., Chao, S., Jin, Y., Cai, X., et al. (2011). Targeted introgression of a wheat stem rust resistance gene by DNA marker-assisted chromosome engineering. Genetics 187, 1011–1021. doi: 10.1534/genetics.110.123588
Nkongolo, K. K., Quick, J. S., Limin, A. E., and Fowler, D. B. (1991). Sources and inheritance of resistance to Russian wheat aphid in Triticum species, amphiploids and Triticum tauschii. Can. J. Plant Sci. 71, 703–708. doi: 10.4141/cjps91-103
Ogbonnaya, F. C., Seah, S., Delibes, A., Jahier, J., López-Braña, I., Eastwood, R. F., et al. (2001). Molecular-genetic characterisation of a new nematode resistance gene in wheat. Theor. Appl. Genet. 102, 623–629. doi: 10.1007/s001220051689
Okada, M., Yoshida, K., and Takumi, S. (2017). Hybrid incompatibilities in interspecific crosses between tetraploid wheat and its wild diploid relative Aegilops umbellulata. Plant Mol. Biol. 95, 625–645. doi: 10.1007/s11103-017-0677-6
Okamoto, Y., Nguyen, A. T., Yoshioka, M., Iehisa, J. C., and Takumi, S. (2013). Identification of quantitative trait loci controlling grain size and shape in the D genome of synthetic hexaploid wheat lines. Breed Sci. 63, 423–429. doi: 10.1270/jsbbs.63.423
Olivera, P. D., Kilian, A., Wenzl, P., and Steffenson, B. J. (2013). Development of a genetic linkage map for Sharon goatgrass (Aegilops sharonensis) and mapping of a leaf rust resistance gene. Genome 56, 367–376. doi: 10.1139/gen-2013-0065
Olivera, P. D., Rouse, M. N., and Jin, Y. (2018). Identification of new sources of resistance to wheat stem rust in Aegilops spp. in the tertiary Genepool of Wheat. Front. Plant Sci. 9:1719. doi: 10.3389/fpls.2018.01719
Olson, E. L., Rouse, M. N., Pumphrey, M. O., Bowden, R. L., Gill, B. S., and Poland, J. A. (2013). Simultaneous transfer, introgression, and genomic localization of genes for resistance to stem rust race TTKSK (Ug99) from Aegilops tauschii to wheat. Theor. Appl. Genet. 126, 1179–1188. doi: 10.1007/s00122-013-2045-5
Perello, A., Martinez, L., and Molina, M. (2015). First report of virulence and effects of Magnaporthe oryzae isolates causing wheat blast in Argentina. Plant Dis. 99, 1177–1178. doi: 10.1094/PDIS-11-14-1182-PDN
Petersen, G., Seberg, O., Yde, M., and Berthelsen, K. (2006). Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D Genomes of common wheat (Triticum aestivum). Mol. Phylogenet. Evol. 39, 70–82. doi: 10.1016/j.ympev.2006.01.023
Petersen, S., Lyerly, J. H., Worthington, M. L., Parks, W. R., Cowger, C., Marshall, D. S., et al. (2015). Mapping of powdery mildew resistance gene Pm53 introgressed from Aegilops speltoides into soft red winter wheat. Theor. Appl. Genet. 128, 303–312. doi: 10.1007/s00122-014-2430-8
Ponce-Molina, L. J., Huerta-Espino, J., Singh, R. P., Basnet, B. R., Alvarado, G., Randhawa, M. S., et al. (2018). Characterization of leaf rust and stripe rust resistance in spring wheat ‘Chilero’. Plant Dis. 102, 421–427. doi: 10.1094/PDIS-11-16-1545-RE
Pretorius, Z. A., Singh, R. P., Wagoire, W. W., and Payne, T. S. (2000). Detection of virulence to wheat stem rust resistance gene Sr31 in Puccinia graminis f. sp. tritici in Uganda. Plant Dis. 84:203. doi: 10.1094/PDIS.2000.84.2.203B
Pukhalskiy, V. A., Martynov, S. P., and Dobrotvorskaya, T. V. (2000). Anal-ysis of geographical and breeding-related distribution of hybridnecrosis genes in bread wheat (Triticum aestivum L.). Euphytica 114, 233–240. doi: 10.1023/A:1003915708448
Rahmatov, M., Rouse, M. N., Nirmala, J., Danilova, T., Friebe, B., Steffenson, B. J., et al. (2016). A new 2DS⋅2RL Robertsonian translocation transfers stem rust resistance gene Sr59 into wheat. Theor. Appl. Genet. 129, 1383–1392. doi: 10.1007/s00122-016-2710-6
Randhawa, M. S., Singh, R. P., Dreisigacker, S., Bhavani, S., Huerta-Espino, J., Rouse, M. N., et al. (2018). Identification and validation of a common stem rust resistance locus in two bi-parental populations. Front. Plant Sci. 9:1788. doi: 10.3389/fpls.2018.01788
Raupp, W. J., Amri, A., Hatchett, J. H., Gill, B. S., Wilson, D. L., and Cox, T. S. (1993). Chromosomal location of hessian fly-resistance genes H22, H23, and H24 derived from Tricum tauschii in the D Genome of wheat. J. Hered. 84, 142–145. doi: 10.1093/oxfordjournals.jhered.a111300
Rawat, N., Neelam, K., Tiwari, V. K., Randhawa, G. S., Friebe, B., Gill, B. S., et al. (2011). Development and molecular characterization of wheat-Aegilops kotschyi addition and substitution lines with high grain protein, iron, and zinc. Genome 54, 943–953. doi: 10.1139/g11-059
Rawat, N., Schoen, A., Singh, L., Mahlandt, A., Wilson, D. L., Liu, S., et al. (2018). TILL-D: An Aegilops tauschii TILLING resource for wheat improvement. Front. Plant Sci. 9:1665. doi: 10.3389/fpls.2018.01665
Rawat, N., Tiwari, V. K., Singh, N., Randhawa, G. S., Singh, K., Chhuneja, P., et al. (2009). Evaluation and utilization of Aegilops and wild Triticum species for enhancing iron and zinc content in wheat. Genet. Resour. Crop Evol. 56, 53–64. doi: 10.1007/s10722-008-9344-8
Rekika, D., Zaharieva, M., Stankova, P., Xu, X., Souyris, I., and Monneveux, P. (1998). “Abiotic stress tolerance in Aegilops species,” in Proceedings of the Durum Research Network, SEWANA, South Europe, West Asia and North Africa, eds M. N. Nachit, M. Baum, E. Porceddu, P. Monneveux, and E. Picard (Aleppo: ICARDA), 113–128.
Riley, R., and Chapman, V. (1958). Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182, 713–715. doi: 10.1038/182713a0
Riley, R., and Chapman, V. (1967). The inheritance in wheat of crossability with rye. Genet. Res. 9, 259–267.
Riley, R., Chapman, V., and Johnson, R. (1968). The incorporation of alien disease resistance to wheat by genetic interference with regulation of meiotic chromosome synapsis. Genet. Res. Camb. 12, 199–219. doi: 10.1017/S0016672300011800
Riley, R., Chapman, V., and Miller, T. E. (1973). “The determination of meiotic chromosome pairing,” in Proceedings of the 4th International Wheat Genetics Symposium, eds E. R. Sears and L. M. S. Sears (Columbia), 731–738.
Riley, R., Unrau, J., and Chapman, V. (1958). Evidence on the origin of the B Genome of wheat. J. Hered. 49, 90–98. doi: 10.1093/oxfordjournals.jhered.a106784
Robertson, W. R. B. (1916). Chromosome studies. I. taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae. V-shaped chromosomes and their significance in Acrididae, Locustidae and Gryllidae: chromosome and variation. J. Morph. 27, 179–331. doi: 10.1002/jmor.1050270202
Romero, M. D., Montes, M. J., Sin, E., López-Braña, I., Duce, A., Martn-Sanchez, J. A., et al. (1998). A cereal cyst nematode (Heterodera avenae Woll.) resistance gene transferred from Aegilops triuncialis to hexaploid wheat. Theor. Appl. Genet. 96, 1135–1140. doi: 10.1007/s001220050849
Rouse, M. N., Olson, E. L., Gill, B. S., Pumphrey, M. O., and Jin, Y. (2011). Stem rust resistance in Aegilops tauschii germplasm. Crop Sci. 51, 2074–2078. doi: 10.2135/cropsci2010.12.0719
Rowland, G. G., and Kerber, E. R. (1974). Telocentric mapping in hexaploid wheat of genes for leaf rust resistance and other characters derived from Aegilops squarrosa. Can. J. Genet. Cytol. 16, 137–144. doi: 10.1139/g74-013
Saintenac, C., Falque, M., Martin, O. C., Paux, E., Feuillet, C., and Sourdille, P. (2009). Detailed recombination studies along chromosome 3B provide new insights on crossover distribution in wheat (Triticum aestivum L.). Genetics 181, 393–403. doi: 10.1534/genetics.108.097469
Sánchez-Martín, J., Steuernagel, B., Ghosh, S., Herren, G., Hurni, S., Adamski, N., et al. (2016). Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 17:221. doi: 10.1186/s13059-016-1082-1
Sasanuma, T., Miyashita, N. T., and Tsunewaki, K. (1996). Wheat phylogeny determined by RFLP analysis of nuclear DNA. 3. Intra- and interspecific variations of five Aegilops sitopsis species. Theor. Appl. Genet. 92, 928–934. doi: 10.1007/BF00224032
Schneider, A., Molnár, I., and Molnár-Láng, M. (2008). Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica 163, 1–19. doi: 10.1007/s10681-007-9624-y
Sears, E. R. (1956). “The transfer of leaf rust resistance from Aegilops umbellulata to wheat,” in Genetics in Plant Breeding. Brook-Haven Symposia in Biology, ed. Brookhaven National Laboratory (Upton: Biology Dept), 1–22.
Sears, E. R. (1977). An induced mutant with homoeologous pairing in common wheat. Can. J. Genet. Cytol. 19, 585–593. doi: 10.1139/g77-063
Sears, E. R. (1982). A wheat mutant conditioning an intermediate level of homoeologous chromosome pairing. Can. J. Genet. Cytol. 24, 715–719. doi: 10.1139/g82-076
Sears, E. R., and Okamoto, M. (1958). “Intergenomic chromosome relationship in hexaploid wheat,” in Proceedings of the 10th International Congress of Genetics, Montreal, 258–259.
Sehgal, S. K., Kaur, S., Gupta, S., Sharma, A., Kaur, R., and Bains, N. S. (2011). A direct hybridization approach to gene transfer from Aegilops tauschii Coss. to Triticum aestivum L. Plant Breed. 130, 98–100. doi: 10.1111/j.1439-0523.2010.01817.x
Sheng, H., See, D. R., and Murray, T. D. (2012). Mapping QTL for resistance to eyespot of wheat in Aegilops longissima. Theor. Appl. Genet. 125, 355–366. doi: 10.1007/s00122-012-1838-2
Singh, J., Sheikh, I., Sharma, P., Kumar, S., Verma, S. K., Kumar, R., et al. (2016). Transfer of HMW glutenin subunits from Aegilops kotschyi to wheat through radiation hybridization. J. Food Sci. Technol. 53, 3543–3549. doi: 10.1007/s13197-016-2333-6
Singh, R. P., Hodson, D. P., Jin, Y., Lagudah, E. S., Ayliffe, M. A., Bhavani, S., et al. (2015). Emergence and spread of new races of wheat stem rust fungus: continued threat to food security and prospects of genetic control. Phytopathology 105, 872–884. doi: 10.1094/PHYTO-01-15-0030-FI
Singh, R. P., Nelson, J. C., and Sorrells, M. E. (2000). Mapping Yr28 and other genes for resistance to stripe rust in wheat. Crop Sci. 40, 1148–1155. doi: 10.2135/cropsci2000.4041148x
Snape, J. W., Chapman, V., Moss, J., Blanchard, C. E., and Miller, T. E. (1979). The crossabilities of wheat varieties with Hordeum bulbosum. Heredity 42, 291–298. doi: 10.1038/hdy.1979.32
Steuernagel, B., Periyannan, S. K., Hernández-Pinzón, I., Witek, K., Rouse, M. N., Yu, G., et al. (2016). Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat. Biotechnol. 34, 652–655. doi: 10.1038/nbt.3543
Tadesse, W., Hsam, S. L. K., Wenzel, G., and Zeller, F. J. (2006). Identification and monosomic analysis of tan spot resistance genes in synthetic wheat lines (Triticum turgidum L. x Aegilops tauschii Coss.). Crop Sci. 46, 1212–1217. doi: 10.2135/cropsci2005.10-0396
Tanaka, M. (1961). New amphidiploids, synthesized 6x-wheats, derived from Emmer wheat × Aegilops squarrosa. Wheat Inf. Serv. 12:11.
Tang, P. S., and Loo, W. S. (1940). Polyploidy in soybean, pea, wheat and rice, induced by colchicine treatment. Science 91:222. doi: 10.1126/science.91.2357.222
Tang, S., Hu, Y., Zhong, S., and Luo, P. (2018). The potential role of powdery mildew-resistance gene Pm40 in Chinese wheat-breeding programs in the post-Pm21 Era. Engineering 4, 500–506. doi: 10.1016/j.eng.2018.06.004
Tanguy, A. M., Coriton, O., Abélard, P., Dedryver, F., and Jahier, J. (2005). Structure of Aegilops ventricosa chromosome 6Nv, the donor of wheat genes Yr17, Lr37, Sr38, and Cre5. Genome 48, 541–546. doi: 10.1139/g05-001
Thomas, J. B., and Conner, R. I. (1986). Resistance to colonization by the wheat curl mite in Aegilops squarrosa and its inheritance after transfer to common wheat. Crop Sci. 26, 527–530. doi: 10.2135/cropsci1986.0011183X002600030019x
Tiwari, V. K., Rawat, N., Neelam, K., Kumar, S., Randhawa, G. S., and Dhaliwal, H. S. (2010). Random chromosome elimination in synthetic Triticum-Aegilops amphiploids leads to development of a stable partial amphiploid with high grain micro- and macronutrient content and powdery mildew resistance. Genome 53, 1053–1065. doi: 10.1139/G10-083
Tiwari, V. K., Rawat, N., Neelam, K., Randhawa, G. S., Singh, K., Chhuneja, P., et al. (2008). Development of Triticum turgidum subsp. durum–Aegilops longissima amphiploids with high iron and zinc content through unreduced gamete formation in F1 hybrids. Genome 51, 757–766. doi: 10.1139/G08-057
Tixier, M. H., Sourdille, P., Charmet, G., Gay, G., Jaby, C., Cadalen, T., et al. (1998). Detection of QTLs for crossability in wheat using a doubled haploid population. Theor. Appl. Genet. 97, 1076–1082. doi: 10.1007/s001220050994
Trethowan, R. M., and Mujeeb-Kazi, A. (2008). Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Sci. 48, 1255–1265. doi: 10.2135/cropsci2007.08.0477
Tsujimoto, H. (2005). “Gametocidal genes in wheat as the inducer of chromosome breakage,” in Frontiers of Wheat Bioscience, Memorial Issue, Wheat Information Service No.100, ed. K. Tsunewaki (Minato: Yokohama), 33–48.
Tsujimoto, H., and Tsunewaki, K. (1984). Gametocidal genes in wheat and its relatives. I. Genetic analyses in common wheat of a gametocidal gene derived from Aegilops speltoides. Can. J. Genet. Cytol. 26, 78–84. doi: 10.1139/g84-013
Tsujimoto, H., and Tsunewaki, K. (1985). Gametocidal genes in wheat and its relatives. II. Suppressor of the chromosome 3C gametocidal gene of Aegilops triuncialis. Can. J. Genet. Cytol. 27, 178–185. doi: 10.1139/g85-027
Tsujimoto, H., and Tsunewaki, K. (1988). Gametocidal genes in wheat and its relatives. III. Chromosome location and effects of two Aegilops speltoides-derived gametocidal genes in common wheat. Genome 30, 239–244. doi: 10.1139/g88-041
Tsunewaki, K. (1960). Monosomic and conventional gene analysis in common wheat. III. Lethality. Jpn. J. Genet. 35, 71–75. doi: 10.1266/jjg.35.71
Valkoun, J., Dostal, J., and Kucerova, D. (1990). “Triticum × Aegilops hybrids through embryo culture,” in Biotechnology in Agriculture and Forestry 13, ed. Y. P. S. Bajaj (Berlin: Springer Verlag), 152–166.
van Slageren, M. W. (1994). Wild Wheats: a Monograph of Aegilops L. and Amblyopyrum (Jaub. & Spach) Eig (Poaceae). Wageningen: Agricultural University Papers.
Velu, G., Crespo-Herrera, L., Guzman, C., Huerta, J., Payne, T., and Singh, R. P. (2018a). Assessing genetic diversity to breed competitive biofortified wheat with enhanced grain Zn and Fe concentrations. Front. Plant Sci. 9:1971. doi: 10.3389/fpls.2018.01971
Velu, G., Singh, R. P., Crespo-Herrera, L., Juliana, P., Dreisigacker, S., Valluru, R., et al. (2018b). Genetic dissection of grain zinc concentration in spring wheat for mainstreaming biofortification in CIMMYT wheat breeding. Sci. Rep. 8:13526. doi: 10.1038/s41598-018-31951-z
Verma, S. K., Kumar, S., Sheikh, I., Malik, S., Mathpal, P., Chugh, V., et al. (2016). Transfer of useful variability of high grain iron and zinc from Aegilops kotschyi into wheat through seed irradiation approach. Int. J. Radiat. Biol. 92, 132–139. doi: 10.3109/09553002.2016.1135263
Vikas, V. K., Sivasamy, M., Kumar, J., Jayaprakash, P., Kumar, S., Parimalan, R., et al. (2014). Stem and leaf rust resistance in wild relatives of wheat with D Genome (Aegilops spp.). Genet. Resour. Crop Evol. 61, 861–874. doi: 10.1007/s10722-014-0085-6
Waines, J. G. (1994). High temperature in wild wheats and spring wheats. Aust. J. Plant Physiol. 21, 705–715.
Waines, J. G., and Barnhart, D. (1992). Biosystematic research in Aegilops and Triticum. Hereditas 116, 207–212. doi: 10.1111/j.1601-5223.1992.tb00825.x
Waines, J. G., Rafi, M. M., and Ehdaie, B. (1993). “Yield components and transpiration efficiency in wild wheats,” in Biodiversity and Wheat Improvement, ed. A. B. Damania (Chichester: Wiley), 173–186.
Wang, J., Luo, M. C., Chen, Z., You, F. M., Wei, Y., Zheng, Y., et al. (2013). Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-Genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 198, 925–937. doi: 10.1111/nph.12164
Wang, K., Lin, Z., Wang, L., Wang, K., Shi, Q., Du, L., et al. (2018). Development of a set of PCR markers specific to Aegilops longissima chromosome arms and application in breeding a translocation line. Theor. Appl. Genet. 131, 13–25. doi: 10.1007/s00122-017-2982-5
Wang, T., Xu, S. S., Harris, M. O., Hu, J., Liu, L., and Cai, X. (2006). Genetic characterization and molecular mapping of Hessian fly resistance genes derived from I in synthetic wheat. Theor. Appl. Genet. 113, 611–618. doi: 10.1007/s00122-006-0325-z
Weng, Y., Li, W., Devkota, R. N., and Rudd, J. C. (2005). Microsatellite markers associated with two Aegilops tauschii-derived greenbug resistance loci in wheat. Theor. Appl. Genet. 110, 462–469. doi: 10.1007/s00122-004-1853-z
Wiersma, A. T., Pulman, J. A., Brown, L. K., Cowger, C., and Olson, E. L. (2017). Identification of Pm58 from Aegilops tauschii. Theor. Appl. Genet. 130, 1123–1133. doi: 10.1007/s00122-017-2874-8
Williamson, V. M., Thomas, V., Ferris, H., and Dubcovsky, J. (2013). An Aegilops ventricosa translocation confers resistance against root-knot nematodes to common wheat. Crop Sci. 53, 1412–1418. doi: 10.2135/cropsci2012.12.0681
Witcombe, J. R. (1983). A Guide to the Species of Aegilops L.: Their Taxonomy, Morphology, and Distribution. Rome: International Board for Plant Genetic Resources (IPGRI).
Xu, S., and Dong, Y. (1992). Fertility and meiotic mechanisms of hybrids between chromosome autoduplication tetraploid wheats and Aegilops species. Genome 35, 379–384. doi: 10.1139/g92-057
Yu, G., Champouret, N., Steuernagel, B., Olivera, P. D., Simmons, J., Williams, C., et al. (2017). Discovery and characterization of two new stem rust resistance genes in Aegilops sharonensis. Theor. Appl. Genet. 130, 1207–1222. doi: 10.1007/s00122-017-2882-8
Yu, G., Zhang, Q., Friesen, T. L., Rouse, M. N., Jin, Y., Zhong, S., et al. (2015). Identification and mapping of Sr46 from Aegilops tauschii accession CIae 25 conferring resistance to race TTKSK (Ug99) of wheat stem rust pathogen. Theor. Appl. Genet. 128, 431–443. doi: 10.1007/s00122-014-2442-4
Yu, L. Y., Barbier, H., Rouse, M. N., Singh, S., Singh, R. P., Bhavani, S., et al. (2014). A consensus map for Ug99 stem rust resistance loci in wheat. Theor. Appl. Genet. 127, 1561–1581. doi: 10.1007/s00122-014-2326-7
Yu, M. Q., Person-Dedrywer, F., and Jahier, J. (1990). Resistance to root knot nematode, Meloidogyne naasi (Franklin) transferred from Aegilops variabilis Eig to bread wheat. Agronomie 6, 451–456. doi: 10.1051/agro:19900603
Yuan, B., Cao, X., and Lvb, A. (2017). Gene introgression from common wheat into Aegilops L. Saudi J. Biol. Sci. 24, 813–816. doi: 10.1016/j.sjbs.2016.05.016
Zeller, F. J., Kong, L., Hartl, L., Mohler, V., and Hsam, S. L. K. (2002). Chromosomal location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L. em Thell.). 7. Gene Pm29 in line Pova. Euphytica 123, 187–194. doi: 10.1023/A:1014944619304
Zemetra, R. S., Hansen, J., and Mallory-Smith, C. A. (1998). Potential for gene transfer between wheat (Triticum aestivum) and jointed goatgrass (Aegilops cylindrica). Weed Sci. 46, 313–317. doi: 10.1007/s00122-009-1009-2
Zeng, D., Luo, J., Li, Z., Chen, G., Zhang, L., Ning, S., et al. (2016). High transferability of homoeolog-specific markers between bread wheat and newly synthesized hexaploid wheat lines. PLoS One 11:e0162847. doi: 10.1371/journal.pone.0162847
Zeven, A. C. (1987). Crossability percentages of some 1400 bread wheat varieties and lines with rye. Euphytica 36, 299–319. doi: 10.1007/BF00730677
Zhang, D., Zhou, Y., Zhao, X., Lv, L., Zhang, C., Li, J., et al. (2018). Development and utilization of introgression lines using synthetic octaploid wheat (Aegilops tauschii × hexaploid wheat) as donor. Front. Plant Sci. 9:1113. doi: 10.3389/fpls.2018.01113
Zhang, H., Jia, J., Gale, M. D., and Devos, K. M. (1998). Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor. Appl. Genet. 96, 69–75. doi: 10.1007/s001220050710
Zhang, H., Reader, S. M., Liu, X., Jia, J. Z., Gale, M. D., and Devos, K. M. (2001). Comparative genetic analysis of the Aegilops longissimi and Ae. sharonensis genomes with common wheat. Theor. Appl. Genet. 103, 518–525. doi: 10.1007/s001220100656
Zhang, R. Q., Feng, Y. X., Li, H. F., Yuan, H. X., Dai, J. L., Cao, A. Z., et al. (2016). Cereal cyst nematode resistance gene CreV effective against Heterodera filipjevi transferred from chromosome 6VL of Dasypyrum villosum to bread wheat. Mol. Breed. 2016:122. doi: 10.1007/s11032-016-0549-9
Zhang, W., Zhang, M., Zhu, X., Sun, Q., Ma, G., Chao, S., et al. (2018). Molecular cytogenetic and genomic analyses reveal new insights into the origin of the wheat B Genome. Theor. Appl. Genet. 131, 365–375. doi: 10.1007/s00122-017-3007-0
Zheng, Y. L., Luo, M. C., Yen, C., and Yang, J. L. (1992). Chromosome location of a new crossability gene in common wheat. Wheat Inf. Serv. 75, 36–40. doi: 10.1186/1471-2229-10-266
Keywords: Aegilops, introgression, translocation, wheat breeding, stress tolerance, wild species, genetic resources
Citation: Kishii M (2019) An Update of Recent Use of Aegilops Species in Wheat Breeding. Front. Plant Sci. 10:585. doi: 10.3389/fpls.2019.00585
Received: 20 February 2019; Accepted: 18 April 2019;
Published: 09 May 2019.
Edited by:
Peter Shewry, Rothamsted Research (BBSRC), United KingdomReviewed by:
Matthew Rouse, United States Department of Agriculture, United StatesIan Stewart Dundas, The University of Adelaide, Australia
Copyright © 2019 Kishii. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Masahiro Kishii, bS5raXNoaWlAY2dpYXIub3Jn
 Masahiro Kishii
Masahiro Kishii