- 1College of Agronomy, Northwest A&F University/State Key Laboratory of Crop Stress Biology for Arid Areas, Yangling, China
- 2Institute of Crop Science, Chinese Academy of Agricultural Sciences (CAAS)/National Key Facility for Crop Gene Resources and Genetic Improvement, Key Laboratory of Biology and Genetic Improvement of Triticeae Crops, Ministry of Agriculture, Beijing, China
- 3College of Life Sciences, Jilin Agricultural University, Changchun, China
- 4College of Agronomy, Qingdao Agricultural University, Qingdao, China
The TGA transcription factors, a subfamily of bZIP group D, play crucial roles in various biological processes, including the regulation of growth and development as well as responses to pathogens and abiotic stress. In this study, 27 TGA genes were identified in the soybean genome. The expression patterns of GmTGA genes showed that several GmTGA genes are differentially expressed under drought and salt stress conditions. Among them, GmTGA17 was strongly induced by both stress, which were verificated by the promoter-GUS fusion assay. GmTGA17 encodes a nuclear-localized protein with transcriptional activation activity. Heterologous and homologous overexpression of GmTGA17 enhanced tolerance to drought and salt stress in both transgeinc Arabidopsis plants and soybean hairy roots. However, RNAi hairy roots silenced for GmTGA17 exhibited an increased sensitivity to drought and salt stress. In response to drought or salt stress, transgenic Arabidopsis plants had an increased chlorophyll and proline contents, a higher ABA content, a decreased MDA content, a reduced water loss rate, and an altered expression of ABA- responsive marker genes compared with WT plants. In addition, transgenic Arabidopsis plants were more sensitive to ABA in stomatal closure. Similarly, measurement of physiological parameters showed an increase in chlorophyll and proline contents, with a decrease in MDA content in soybean seedlings with overexpression hairy roots after drought and salt stress treatments. The opposite results for each measurement were observed in RNAi lines. This study provides new insights for functional analysis of soybean TGA transcription factors in abiotic stress.
Introduction
Abiotic stress, such as drought and high salinity, greatly affect plant growth and development. In order to adapt to abiotic stress, plants have evolved complex signal transduction pathways and diverse response mechanisms to protect themselves against cellular damage caused by abiotic stress (Munns and Tester, 2008; Chen and Murata, 2011; Krasensky and Jonak, 2012; Mickelbart et al., 2015; Zhu, 2016; Qi et al., 2018; Yang and Guo, 2018). Under stress conditions, changes in gene expression are the earliest responses in plants, and a number of stress-responsive genes have been noted to have important functions in drought and salt resistance. Among these genes, transcription factors are very important as the proteins they typically encode control the transcription of downstream genes (Singh et al., 2002; Li et al., 2008). Among them, the basic leucine zipper (bZIP) gene family is one of the largest transcription factor families in plants. bZIP genes were classified into 10 groups (A, B, C, D, E, F, G, H, I, and S), along with another two extra groups, J and K, based on the similarity in the basic region and additional conserved motifs (Jakoby et al., 2002; Nijhawan et al., 2008). The TGA (TGACG motif-binding factor) transcription factors belong to group D, which can recognizes as-1- type cis-elements located in the promoter region of the target genes (Katagiri et al., 1989; Zhang et al., 1999; Johnson et al., 2003). Tobacco TGA1a was the first TGA transcription factor cloned from plants and is characterized by the conserved basic region/leucine zipper domain (Katagiri et al., 1989). Subsequently, more TGA transcription factors were identified in various plants (Jakoby et al., 2002; Espín et al., 2012). For TGA proteins, the primary structure of the bZIP domain is conserved, containing an invariant motif N-x7-R/K-x9-L-x6-L-x6-L in the N-terminus, and a bZIP-D box, the motif Yx2RL[RQ]ALSS[LS]W, represents the signature domain of group D in the C-terminus (Jakoby et al., 2002).
A growing body of evidence has shown that members of TGA transcription factor are known to play crucial roles in many biological processes: defense against pathogens and plant development (Johnson et al., 2001; Thurow et al., 2005; Choi et al., 2010; Zander et al., 2012; Gatz, 2013; Wang et al., 2016). Despite this body of work, little has been reported about the functions of TGA transcription factors in plant responses to abiotic stress. The Brassica juncea BjCdR15, an orthologous gene of Arabidopsis TGA3, had an induced expression in response to cadmium and stress from other heavy metals (Fusco et al., 2005). Transgenic Arabidopsis and tobacco overexpressing BjCdR15 exhibited an enhanced tolerance to cadmium through regulation of cadmium uptake and long-distance transport (Farinati et al., 2010). The crab apple MhTGA2, which showed an induced expression in response to low temperature, NaCl, and PEG, enhanced tolerance to salt and osmotic stress in transgenic apple and tobacco (Zhang et al., 2012; Du et al., 2014). Overexpression of AtTGA4 increased tolerance to drought stress by improving nitrate transport and absorption in Arabidopsis (Zhong et al., 2015). These findings suggested that TGA transcription factors may play functions in plant adaptation to various abiotic stimuli, including drought and salt.
Soybean (Glycine max L.), an economically important oil and protein crop, is considered a moderately drought- and salt-tolerant plant, though its growth and productivity is adversely affected by soil drought and salinity. Given the potential importance of TGA genes in plant responses to abiotic stress, we conducted a genome-wide analysis of the soybean TGA family and investigated the potential functions of TGA genes in plant responses to drought and salt stress.
Materials and Methods
In silico Identification of Soybean TGA Transcription Factors
The sequences of known TGA proteins from Arabidopsis were retrieved from the TAIR database. These sequences were used as queries to search against the soybean genome database using the BLASTP program with a threshold E-value cutoff of 1.0 × e-5 (Espín et al., 2012). Then, each candidate soybean TGA sequence used as a query against the Pfam database to confirm its membership in the bZIP family (Finn et al., 2016). Repeat sequences were removed manually. The ExPASy server was used to predict several physio-chemical parameters of TGA proteins such as molecular weights (Mw) and theoretical isoelectric points (pI) (Gasteiger et al., 2003).
Sequences Analysis
The exon/intron gene boundaries were analyzed using the Gene Structure Display Server 2.0 (GSDS) tool (Hu et al., 2015). The 2.0 kb sequences upstream of the start codon of soybean TGA genes were extracted from the phytozome database as the regulatory promoter region. Putative cis-acting elements were analyzed using the PlantCARE database (Lescot et al., 2002). Multiple sequence alignments for the predicted protein sequences were performed using ClustalX software (Thompson et al., 1997). MEGA6.0 software was used to construct a phylogenetic tree based on the bootstrap neighbor-joining (NJ) approach followed by 1000 bootstrap replicates (Tamura et al., 2013).
Expression Analysis of Soybean TGA Genes in Different Tissues
The analysis of gene expression patterns in different tissues, including roots, root hairs, stem, leaves, nodules, seeds, and flowers, was carried out using transcriptome data obtained from the Phytozome database, and the heatmap was produced with HemI software (Deng et al., 2014).
Plant Materials, Growth Conditions and Stress Treatments
The soybean variety Williams 82 was used for experiments in this study. Seedlings were grown in pots containing mixed soil (humus:vermiculite = 1:1) in a greenhouse with a 14-h-light/10-h-dark photoperiod, 28/20°C day/night temperatures, and 60% relative humidity. Sixteen-day-old seedlings were subjected to drought and salinity treatments. For drought stress, seedlings were removed from the soil and left to dry on filter paper. For the high-salinity treatment, the roots of seedlings were immersed in solution containing 200 mM NaCl. For treatments with exogenous ABA, leaves were sprayed with 200 μM ABA solution. Seedlings were sampled for RNA extraction at 0, 1, 3, 6, 12, and 24 h after each respective treatment.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR)
Total RNA was isolated from soybean seedlings and hairy roots, or from Arabidopsis seedlings using Trizol reagent (TaKaRa, Japan). The first strand cDNA was synthesized using the PrimeScript 1st Strand cDNA Synthesis Kit (TaKaRa, Japan) based on the manufacturer’s instructions. qRT-PCR was performed on ABI prism 7500 Real-Time PCR system (Applied Biosystem, United States) using SYBR Green Real Master Mix (Tiangen, China) with the following PCR cycles: 95°C for 15 min, followed by 40 cycles of amplification (95°C for 10 s, 58°C for 20 s, and 72°C for 32 s). The data of qRT-PCR were determined using the 2-ΔΔCt method according to the cycle threshold (Ct) values (Livak and Schmittgen, 2001). The soybean CYP2 (GmCYP2) (Glyma.12g024700) and Arabidopsis actin2 (AtACT2) (At3g18780) genes were used for normalization. Student’s t-test was used to determine significant differences. A level of 0.05 was used for statistical significance. Primer sequences for qRT-PCR analysis were designed using the software tool Primer Premier 5.0. The primer sequences are listed in Supplementary Table S1. All reactions were conducted with four biological replicates for each sample.
Subcellular Localization and Transcriptional Activation Analysis
The ORF of GmTGA17, lacking the stop codon, was amplified using custom primers and fused to the N-terminal region of GFP. The ORF of NtTGA2.2 (AAF06696), encoding a nuclear-localized protein, was cloned into the N-terminal region of RFP as the positive control (Thurow et al., 2005). The genes were driven by CaMV35S promoter. The reconstruction plasmid of NtTGA2.2-RFP and GmTGA17-GFP were co-transferred into Arabidopsis protoplasts, and NtTGA2.2-RFP and the 35S::GFP vector were co-transformed as the control. The fluorescence signal was detected by confocal microscopy (Leica Microsystem, Heidelberg, Germany) after incubating in darkness at 22°C for 16 h. The primers used are shown in Supplementary Table S1.
To perform the transcriptional activity assay in yeast cells, the ORF of GmTGA17 was amplified and cloned into the pGBKT7 vector. pGBKT7-AtDREB2A was the positive control based on previously reported data (Sakuma et al., 2006), while the pGBKT7 empty vector was the negative control. The plasmids were transformed into yeast strain AH109 according to the method described previously (Gietz and Schiestl, 2007). The YeastmakerTM Yeast Transformation System 2 (Clontech, United States) was used for yeast transformation. Transcriptional activity was analyzed using methods established in prior work (Yang et al., 2010).
The transcriptional activity was examined in Arabidopsis protoplast system. The reporter was a plasmid containing the firefly (LUC) gene fused with 5 × GAL4 binding sites under the control of CaMV35S promoter. The other plasmid with the renilla luciferase (REN) gene was used as the internal control (Liu et al., 2018). GAL4-BD and BD-AtDREB2A were used as negative and positive effector. The each effector plasmid was co-transformed with the two reporters. Firefly and Renilla luciferase was quantified at 18 h post-transformation by using the Dual-Luciferase Reporter Assay System following the manufacturer’s instructions (Promega, United States).
Generation of Transgenic Arabidopsis Plants
In order to produce transgenic Arabidopsis lines, the ORF of GmTGA17, the stop codon, was amplified and cloned into the pCAMBIA1302 vector driven by the CaMV35S promoter. The expression vector pCAMBIA1302-GmTGA17 was transformed into Agrobacterium tumefaciens strain GV3101 and transferred into Arabidopsis Col-0 plants using the floral dip method (Clough and Bent, 1998). The harvested seeds were screened by hygromycin (40 mg/L) resistance. T3 transformed plants were confirmed by qRT-PCR analysis and used for further study based on the expression level of GmTGA17.
Agrobacterium rhizogenes-Mediated Transformation of Soybean Hairy Roots
To generate GmTGA17 promoter-GUS construct, a 1.9-kb fragment upstream from the initiation codon was amplified and then used to replace the CaMV35S promoter in pCAMBIA3301. The ORF of GmTGA17 was amplified and cloned into pCAMBIA3301 under the control of the CaMV35S promoter to generate the pCAMBIA3301-GmTGA17 overexpression vector. For construction of the RNAi suppression vector, a 542 bp ligated fragment containing the sequence from CDS positions 549 to 750, the first intron sequence and the reverse complement sequence of the sequence from CDS positions 549 to 750 (Supplementary Table S1) was synthesized (Biomed, Beijing, China) and cloned into pCAMBIA3301 to generate pCAMBIA3301 -GmTGA17-RNAi vector. All recombinant vectors were transformed into soybean hairy roots by high-efficiency Agrobacterium rhizogenes-mediated transformation as described previously (Kereszt et al., 2007).
Abiotic Stress Tolerance Assessments of Transgenic Arabidopsis and Soybean Hairy Roots
Three transgenic Arabidopsis lines with a higher expression of GmTGA17 were used to evaluate the drought and salt tolerance. For the root growth assay, 5-day-old seedlings were transferred to 1/2 MS medium ( containing 6%, 9% or 12% PEG6000 and 50, 75 or 100 mM NaCl, respectively) for vertical growth under a photoperiod of 16-h-light/8-h-dark at 22°C, 40 μmol m-2 s-1 light. Seedling roots were scanned with Expression 11000XL scanner and analyzed for total root length with WinRHIZO software after 8 days of treatments. For drought stress in soil, water was withheld from 2-week-old soil-grown seedlings until differences in phenotype were observed, then plants were watered again and recovered for 1 week to count the survival rates. For high-salinity treatment, 2-week-old soil-grown seedlings were soaked in 250 mM NaCl solution for 1 week, and the control group continued to grow under normal conditions. Proline and MDA contents were measured under both stress conditions, and chlorophyll content was measured under high-salinity treatment as described previously (Arnon, 1949; Bates et al., 1973; Cakmak and Horst, 1991).
For water loss measurement, the leaves from 3-week-old plants grown in soil under normal conditions were excised and weighted immediately (initial weight, W0). Subsequently, the detached leaves were maintained in a growth chamber (40% relative humidity) and measured at designated time intervals. The fresh weights measured at each time point were used as Wn. Three replicates were done for each line. The water loss rate was calculated as (W0-Wn)∗100/W0.
The transgenic hairy roots were confirmed by PCR and qRT-PCR analysis. For promoter-GUS analysis, the transgenic hairy roots were immersed in 10% PEG6000, 100 mM NaCl, or water. The GUS activity and the transcript level of GUS in soybean transgenic hairy roots were detected at 0, 3, 6, 9, 12, and 24 h post-treatments. Histochemical staining of GUS was performed as previously described (Jefferson, 1987). In addition, the expression levels of GUS were analyzed by qRT-PCR.
For abiotic stress tolerance assays, the transgenic hairy roots with higher (OE) or lower (RNAi) expression level of GmTGA17 were selected for further study. 10-day-old soil-grown plants with transgenic hairy roots were soaked in 20% (m/v) PEG6000 or 150 mM NaCl solutions for 10 days. The control group continued to grow under normal conditions. After treatments, the transgenic roots were scanned and the total root length and the total root surface were analyzed. Meanwhile, chlorophyll, proline, and MDA contents in leaves were detected as described above.
Stomatal Aperture Measurement
Stomatal aperture assay was performed as described previously (Kim and Kim, 2013). The guard cells were photographed using confocal microscopy (Leica Microsystem, Heidelberg, Germany), and the stomatal aperture (ration of width to length) was analyzed using Photoshop CS5 software (Adobe System).
Determination of ABA Content
ABA content was measured as described previously (Liu et al., 2014). The Phytodetek-ABA ELISA Kit (Agdia, United States) was used for the determination of ABA according to the manufacture’s protocol.
Statistical Analysis
All experiments above were repeated at least three replicates independently. The data were subjected to Student’s t-test analysis using functions in Excel 2007. The values are shown as mean ± standard deviation (SD). P-value cut-off of 0.05.
Primers
The primers and sequences used in this study are listed in Supplementary Table S1.
Results
Identification of TGA Transcription Factors in Soybean
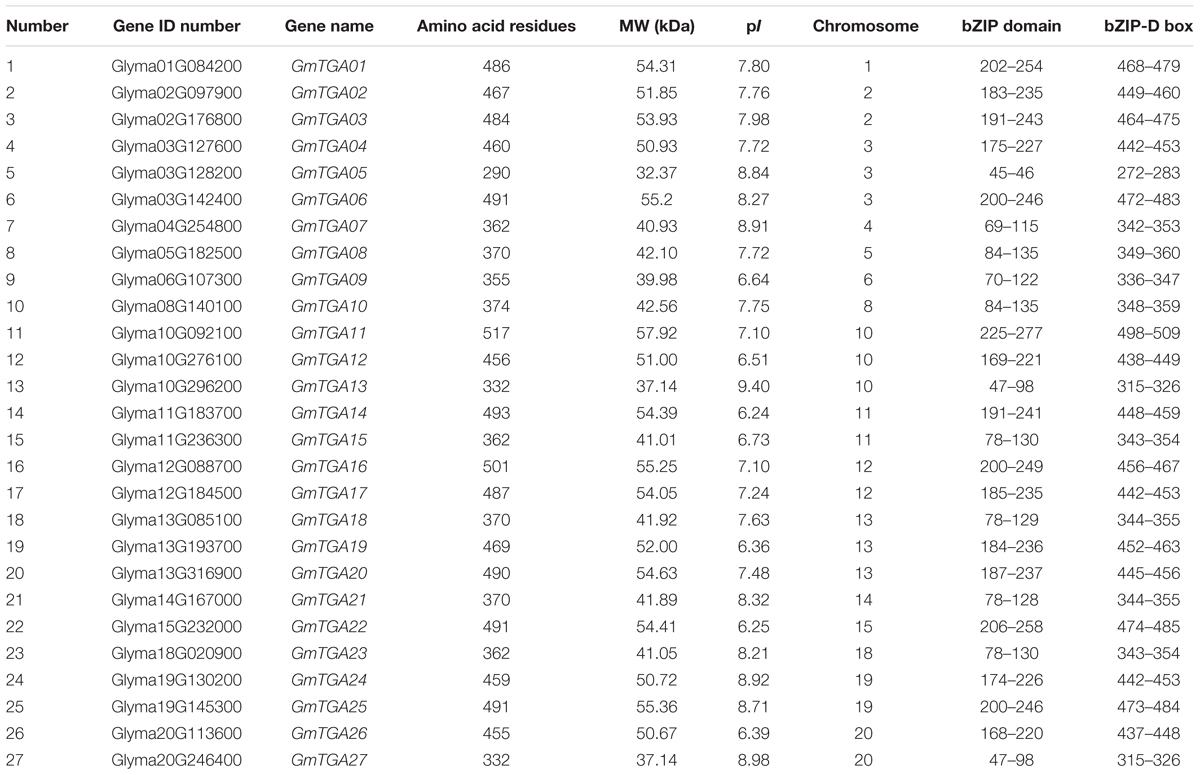
In this study, full-length proteins and conserved domains of 10 Arabidopsis TGAs were used as BLAST query sequences against the soybean genome database. A total of 27 non-redundant, putative TGA genes were identified in the soybean genome, which named GmTGA01-GmTGA27 according to the Gene ID number (Table 1). These loci were distributed across 16 chromosomes in the soybean genome, with the exceptions of chromosomes 7, 9, 16, and 17. The size range of the predicted products of these putative GmTGAs spans 290 (GmTGA05) to 517 (GmTGA17) amino acids residues, with molecular weights (Mw) ranging from 32.37 kDa (GmTGA05) to 57.92 kDa (GmTGA11), and protein isoelectric points (pIs) ranging from 6.24 (GmTGA14) to 9.4 (GmTGA13) (Table 1).
To evaluate the evolutionary history of GmTGAs and their relationships with TGAs in other plants, the TGA protein sequences were aligned and compared with TGA sequences from Arabidopsis and rice, resulting in a multiple sequence alignment and phylogeny of 53 TGA proteins. The phylogenetic tree showed that 53 TGAs were divided into three major clades: Clade I, II and III (Figure 1), which agreed with previous reports on sub-classification within TGA family proteins (Murmu et al., 2010; Espín et al., 2012). Clades I and II both contain 8 GmTGAs, while clade III has 11 members of this protein family.
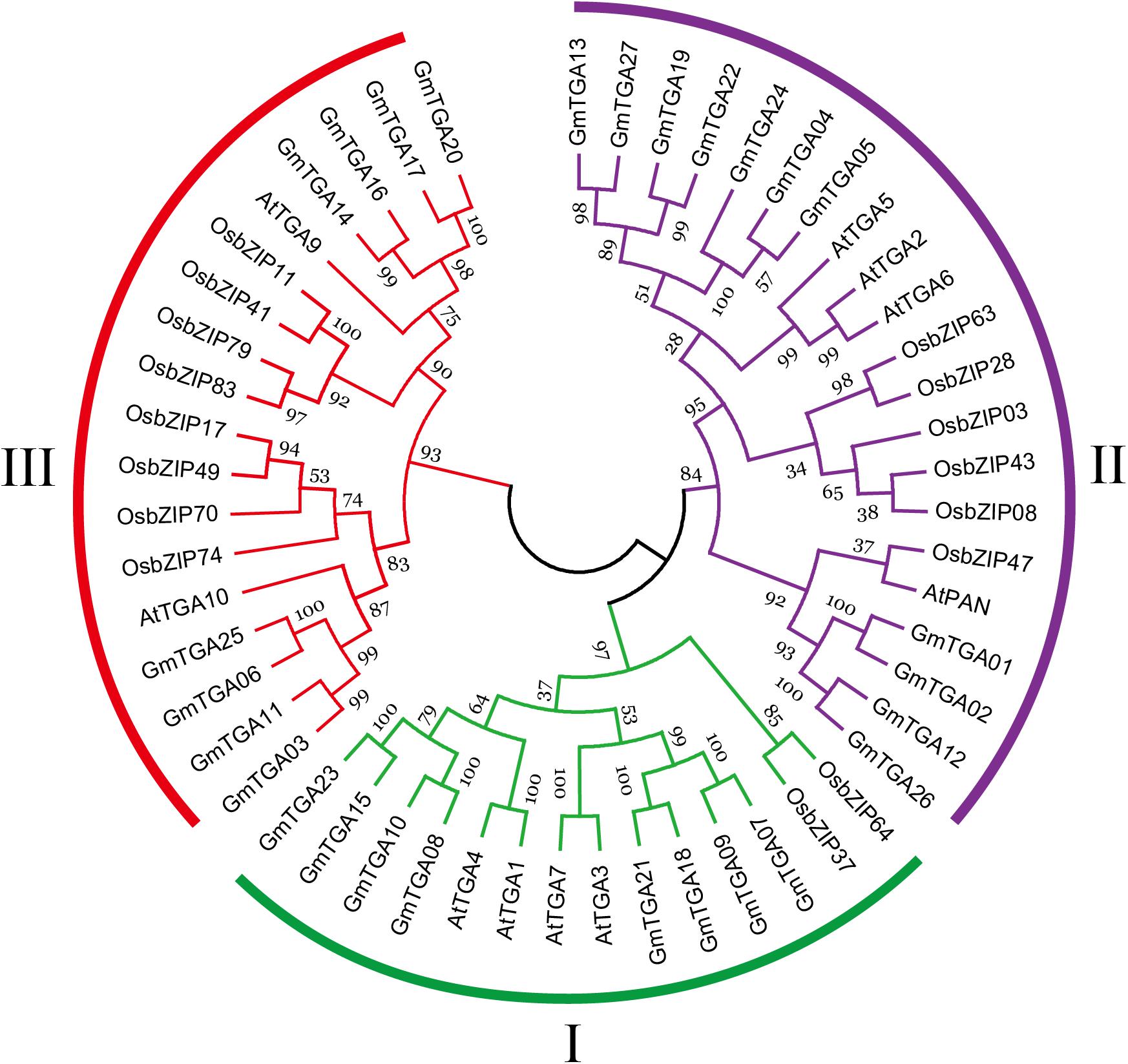
Figure 1. Phylogenetic analyses of TGA proteins from Arabidopsis, rice and soybean. Each clade is marked by a separate color.
Gene Structure and Cis-Acting Elements
To obtain some insight into the gene structure of 27 GmTGA genes, their intron-exon organization was examined. GmTGA genes were interrupted by 6–11 introns (Supplementary Figure S1). In order to better annotation and prediction of potential functions of GmTGA genes, we searched for cis-elements in their putative promoter regions. The results revealed that all GmTGA genes contained one or more of the ABRE, ARE, MYB, MYC, MBS, or G-box elements in their putative promoter regions. Only the putative promoter region of GmTGA17 carried the dehydration-responsive element (DRE). Low-temperature responsive elements (LTRE) were only observed in the putative promoter regions of GmTGA05, GmTGA25, and GmTGA27 (Supplementary Table S2).
Domain Structure of Soybean TGA Proteins
To analyze the structural characteristics of conserved domains in GmTGA proteins, a multiple sequence alignment was performed using the 27 full-length TGA amino acid sequences from soybean. Multiple alignments showed that the GmTGAs contain a typical bZIP domain, two polyglutamine domains and a unique bZIP-D box (Supplementary Figure S2). The bZIP domain consists of a DNA binding domain and a leucine zipper domain. DNA binding domains are located in the N-terminus. Highly conserved amino acid sequences in the DNA binding domains have Glutamine, Alanine and Serine residues, which are regularly spaced by three different amino acids. The leucine zipper domain is formed by a heptad repeat of leucine residues. The GmTGAs also contain the polyglutamine domains I (QI) and II (QII). All members of GmTGAs possess the bZIP-D box, located in the C-terminus (Supplementary Figure S2B). These observations are in accordance with previous reports about the structural characteristics of the TGA conserved domains (Jakoby et al., 2002; Farinati et al., 2010; Espín et al., 2012).
Expression Profiles of Soybean TGA Genes Are Different for Each Tissue
To analyze the expression patterns of GmTGA genes, the transcriptome data of seven soybean tissues and organs was extracted from the soybean genome database and made a heatmap of GmTGA genes expression profile (Figure 2). A total of 26 GmTGA genes were expressed in all evaluated tissues, whereas GmTGA01 were expressed in root, root hairs, stem, nodules, seed and flower tissue, but not in leaves. Additionally, the expression patterns were different between GmTGA genes in same tissue. For example, GmTGA04, GmTGA06, GmTGA10, GmTGA15, GmTGA17, GmTGA20, GmTGA22, and GmTGA23 were expressed at their highest levels in roots, the expressions of GmTGA07, GmTGA09, GmTGA18, and GmTGA21 were highest in stems, GmTGA01, GmTGA02, GmTGA05, GmTGA11, GmTGA14, GmTGA16, and GmTGA25 were expressed most strongly in nodules, GmTGA03, GmTGA12, and GmTGA26 transcription was most enriched in seeds, and GmTGA08, GmTGA13, GmTGA19, GmTGA22, and GmTGA24 were found most highly expressed in flowers. The expression of GmTGA01 reached its highest level in nodules, but was not expressed in leaves. These transcriptional patterns suggested that the expression of these genes might be governed by diverse and potentially tissue-dependent regulatory mechanisms.
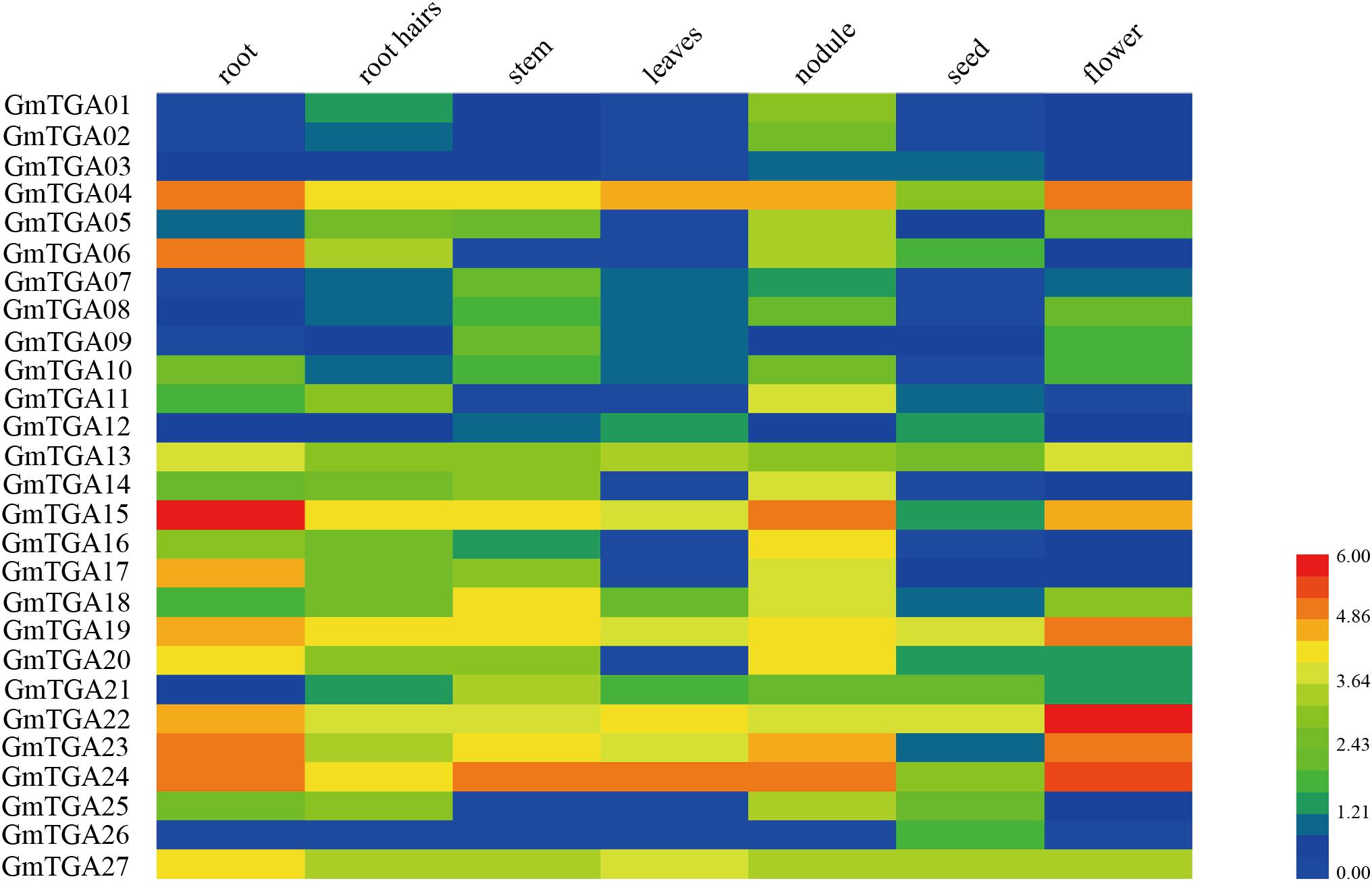
Figure 2. Expression profiles of soybean TGA genes in seven different tissues of soybean. Different colors in map represent gene transcript abundance values. The color scale is shown in the bar at bottom of figure.
GmTGA Genes Are Involved in Response to Drought and Salt
To investigate the potential roles of GmTGAs in response to multiple abiotic stresses, the expression profiles of GmTGA genes in soybean plants treated with drought, salt or water were examined by qRT-PCR. The data revealed that the transcript levels of GmTGA genes showed no obvious difference under non-stress conditions (data not show). Under drought treatment, twelve GmTGA genes had a significantly different transcriptional response to drought (Figure 3). Among them, GmTGA15, GmTGA17, GmTGA24, GmTGA26, and GmTGA27 had significant increases in transcript level after drought treatment, especially GmTGA15 (10-fold) and GmTGA17 (22-fold), which reached a peak at 24 and 12 h post-treatment, respectively. GmTGA05, GmTGA07, GmTGA10, GmTGA14, GmTGA16, GmTGA20, and GmTGA22 had significant decreases in transcript level after drought treatment. The other GmTGA genes showed no response to drought (Figure 3). Following salt treatment,; seven GmTGA genes had a significantly different transcriptional response to salt (Figure 4). Among them, GmTGA10, GmTGA13, GmTGA14, GmTGA17, and GmTGA26 had significant increases in transcript level after salt treatment, especially GmTGA13 (10-fold), GmTGA17 (26-fold), and GmTGA26 (8-fold), which had the highest transcript levels at 12 and 3 h post-treatment, respectively. GmTGA05 and GmTGA07 had significant decreases in transcript level after salt treatment. The other GmTGA genes had no significant changes under salt treatment (Figure 4). In light of its dramatic up-regulation in stress conditions, GmTGA17 was selected for further study of the roles of GmTGAs in abiotic stress tolerance.
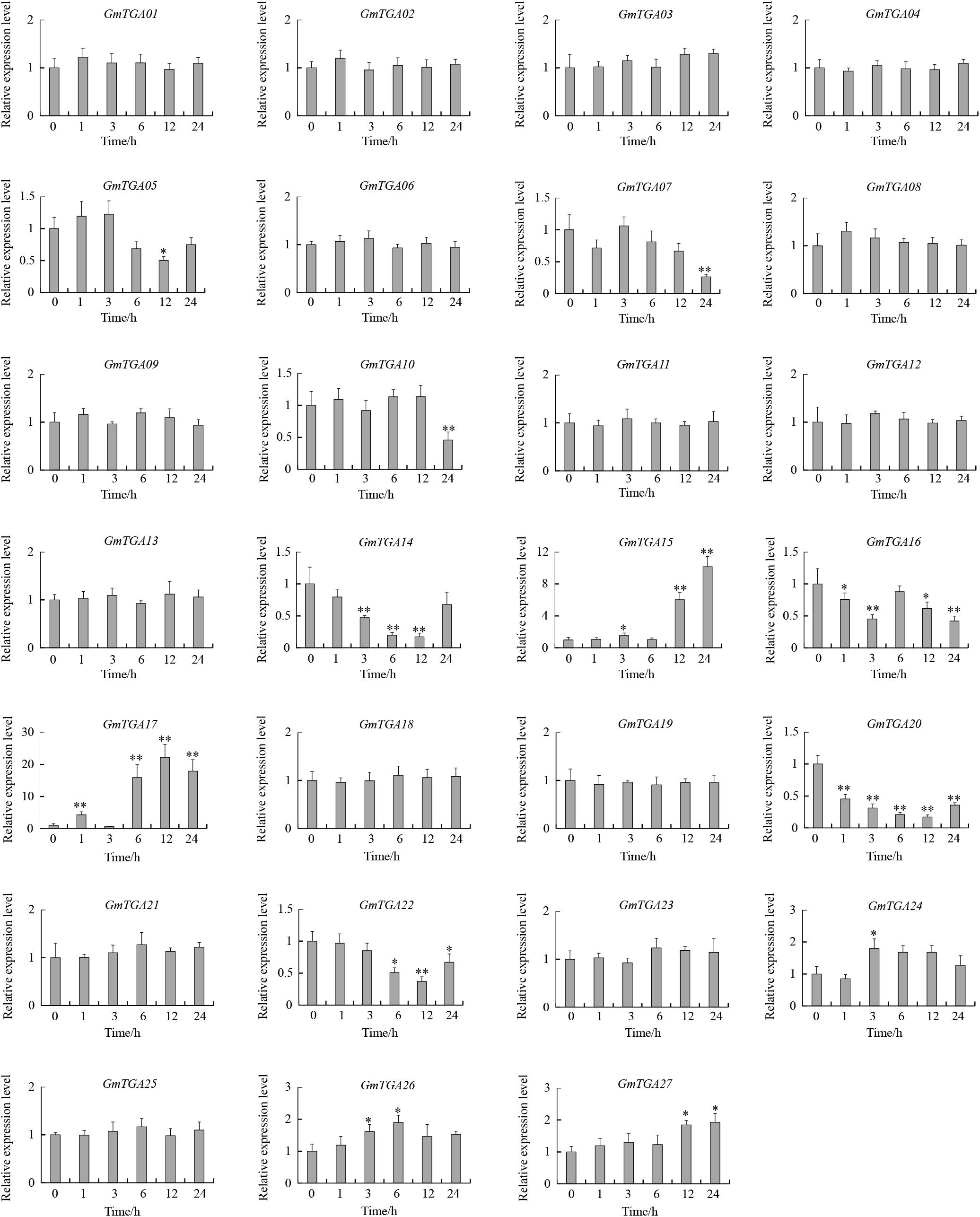
Figure 3. Expression profiles of soybean TGA genes under drought stress conditions. The expression levels were normalized to that of CYP2. Values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
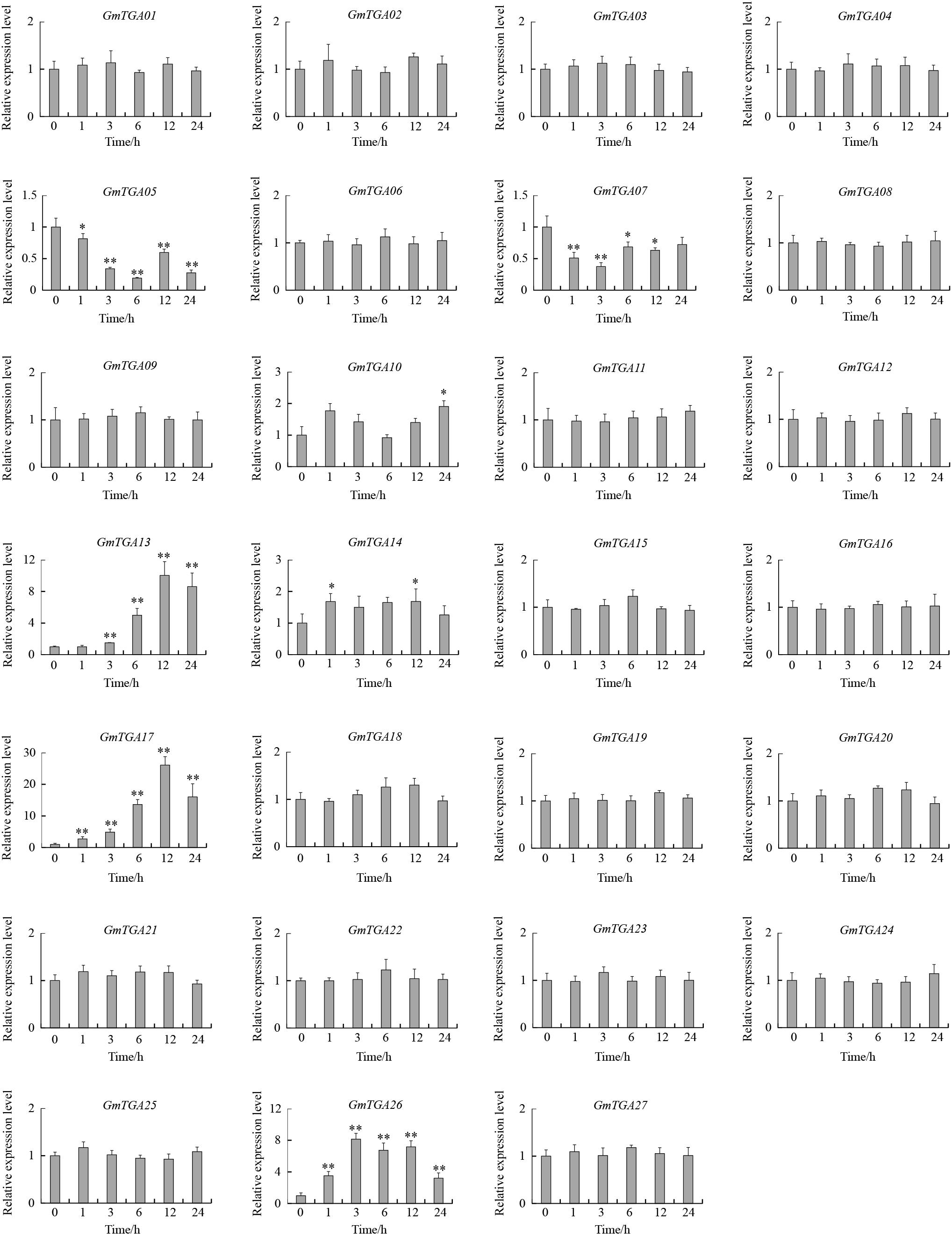
Figure 4. Expression profiles of soybean TGA genes under salt stress conditions. The expression levels were normalized to that of CYP2. Values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
GmTGA17 Is a Nuclear Protein With Transcriptional Activation Activity
To follow the subcellular localization of GmTGA17 proteins, the ORF of GmTGA17 was fused to GFP, as well as NtTGA2.2 fused to RFP as a positive control, The fusion genes were subsequently co-transformed into Arabidopsis protoplasts. The visible fluorescence showed that GmTGA17 was located in the nucleus (Figure 5A). This finding was in accordance with its putative function as a transcription factor.
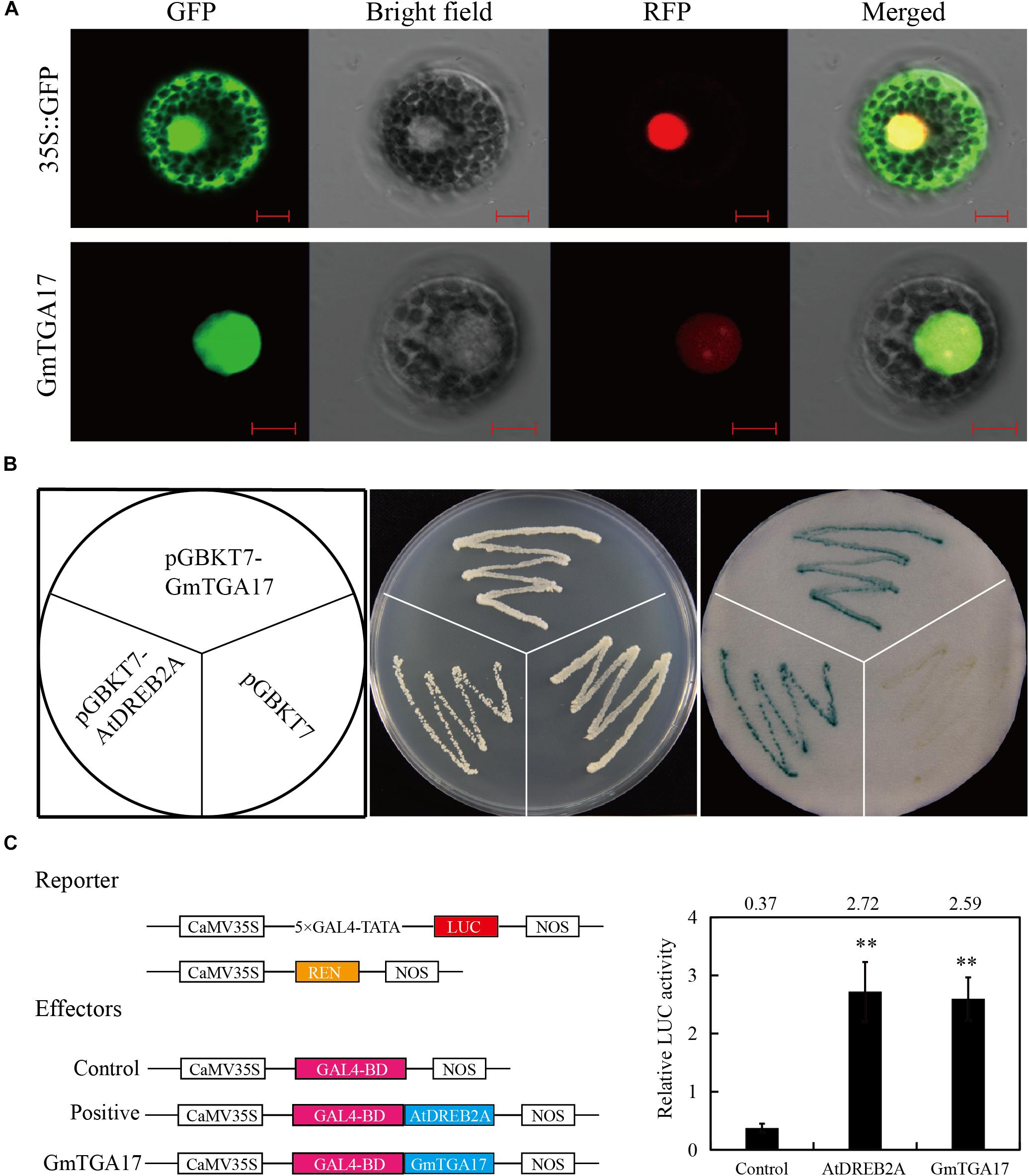
Figure 5. Subcellular localization and transcription activation analysis of GmTGA17. (A) Co-localization of GmTGA17. The recombinant plasmids of GmTGA17-GFP and NtTGA2.2-RFP were co-transformed into Arabidopsis protoplasts. Results were detected with confocal microscopy. Scale bars = 10 μm. (B) Transcriptional activity analysis of GmTGA17 in yeast cells. pGBKT7-AtDREB2A and pGBKT7 were used as positive and negative controls, respectively. (C) Transient expression assay in Arabidopsis protoplasts investigating the transcriptional activation activity of GmTGA17. The relative LUC activities in each sample was normalized relative to that of the internal control. Values are the mean ± SD of three independent replicates and the asterisks indicate a significant difference (∗∗P < 0.01) compared with the corresponding controls.
In order to test for transcriptional activity of GmTGA17, the fusion plasmids pGBKT7-GmTGA17, the positive control pGBKT7-AtDREB2A, and the negative control pGBKT7 were separately transformed into yeast strain AH109. The transformed yeast strain grew well in non-selective medium SD/–Trp and, using X-α-Gal, it was observed that both the positive control and the cells harboring pGBKT7-GmTGA17 displayed β-galactosidase activity, whereas the negative control exhibited no β-galactosidase activity (Figure 5B), suggesting that GmTGA17 possesses transcriptional activation activity in yeast cells.
To further confirm whether GmTGA17 has transcriptional activation activity, we used a dual reporter system for a transient expression assays in Arabidopsis protoplasts (Liu et al., 2018). In this system, the reporter plasmid was constructed by fusing the firefly luciferase (LUC) gene to a 5 × GAL4 binding site. The renilla luciferase (REN) gene under the control of the CaMV35S promoter was used as the internal control (Figure 5C). The ORF of GmTGA17 was fused to the GAL4 binding domain (GAL4-BD) as effector plasmid (Figure 5C). After co-transformed three plasmids into Arabidopsis protoplasts, the relative LUC activity was determined. The results showed that the relative LUC activity was significantly upregulated when GmTGA17 or the positive control AtDREB2A expressed, compared with the empty vector control (Figure 5C). The above findings suggested that GmTGA17 potentially acts as transcriptional activation in plant cell nucleus.
Promoter Activity of GmTGA17 During Drought and Salt Stress
Analysis by qRT-PCR showed that the expression of GmTGA17 is responsive to drought and salt treatments. We used an established promoter-GUS construct in conjunction with a 1.9 kb GmTGA17 promoter upstream of the transcription start site, to further study the activity of the GmTGA17 promoter under drought and salt treatments. The construct was then transformed into soybean hairy roots. Staining of transgenic hairy roots showed that GUS activity was significantly increased in hairy roots after 6 h of drought or salt treatments compared with the hairy roots treated with water (Figure 6A). qRT-PCR date, agreed with the observations of GUS histochemical staining, showed that GUS was also up-regulated (Figure 6B). The above results suggested that the promoter activity of GmTGA17 was enhanced by drought and salt treatments.
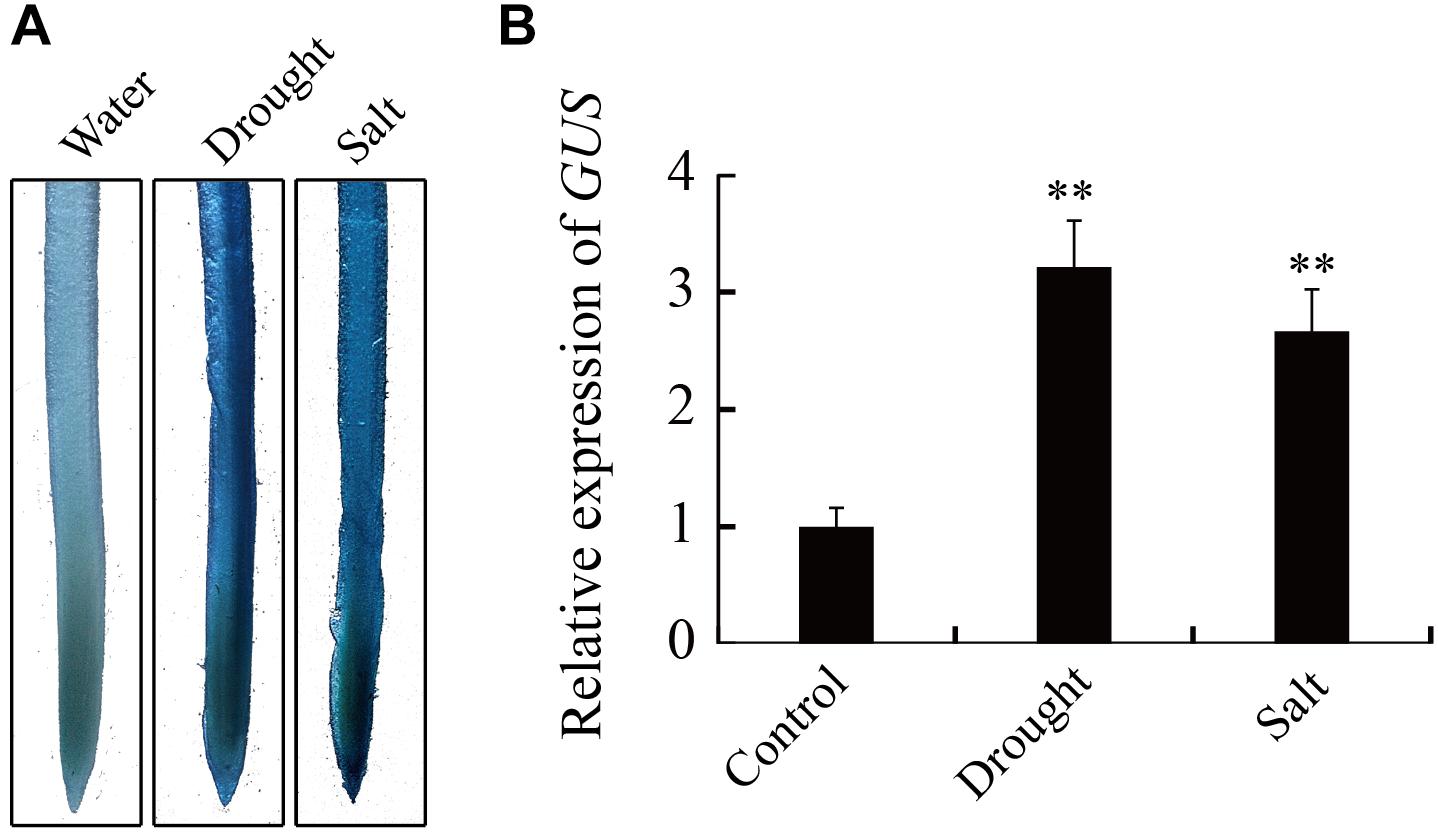
Figure 6. Expression of the GUS reporter gene under the control of GmTGA17 promoter in transgenic soybean hairy roots. (A) Histochemical assay and (B) relative expression of GUS. The GmTGA17pro-GUS transgenic soybean hairy roots were treated with distilled water, 10% (m/v) PEG6000, or 100 mM NaCl for 6 h before being subjected to histochemical and expression analysis. The expression level of CYP2 was used as quantitative control. Values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗∗P < 0.01) compared with the corresponding controls.
Overexpression of GmTGA17 Improved Arabidopsis Tolerance to Drought
Since GmTGA17 was identified as a predicted stress-tolerance regulator, the drought and salt tolerance phenotypes of transgenic Arabidopsis lines grown in medium and soil were examined. The semi-qRT-PCR result was shown in Figure 7A, and T3 transgenic Arabidopsis lines (OE-1, OE-4, and OE-6) with the high expression of GmTGA17 were selected for further experiments (Supplementary Figure S3). PEG6000 was used to simulate drought stress when grown in medium. In the absence of PEG, the total root length and fresh weight showed no difference between transgenic and WT plants. In contrast, when exposed to 1/2 MS medium with 6% PEG6000 for 8 days, the growth of transgenic and WT plants was strongly inhibited, though the degree of inhibition in the transgenic plants was much lower than that of WT plants. The total root length and fresh weight of transgenic plants were significantly longer and greater than those of WT lines (Figures 7B,C). The total root length and fresh weight were not significantly different between transgenic lines and WT plants when grown on 1/2 MS medium supplemented with 9% and 12% PEG (Figures 7B,C).
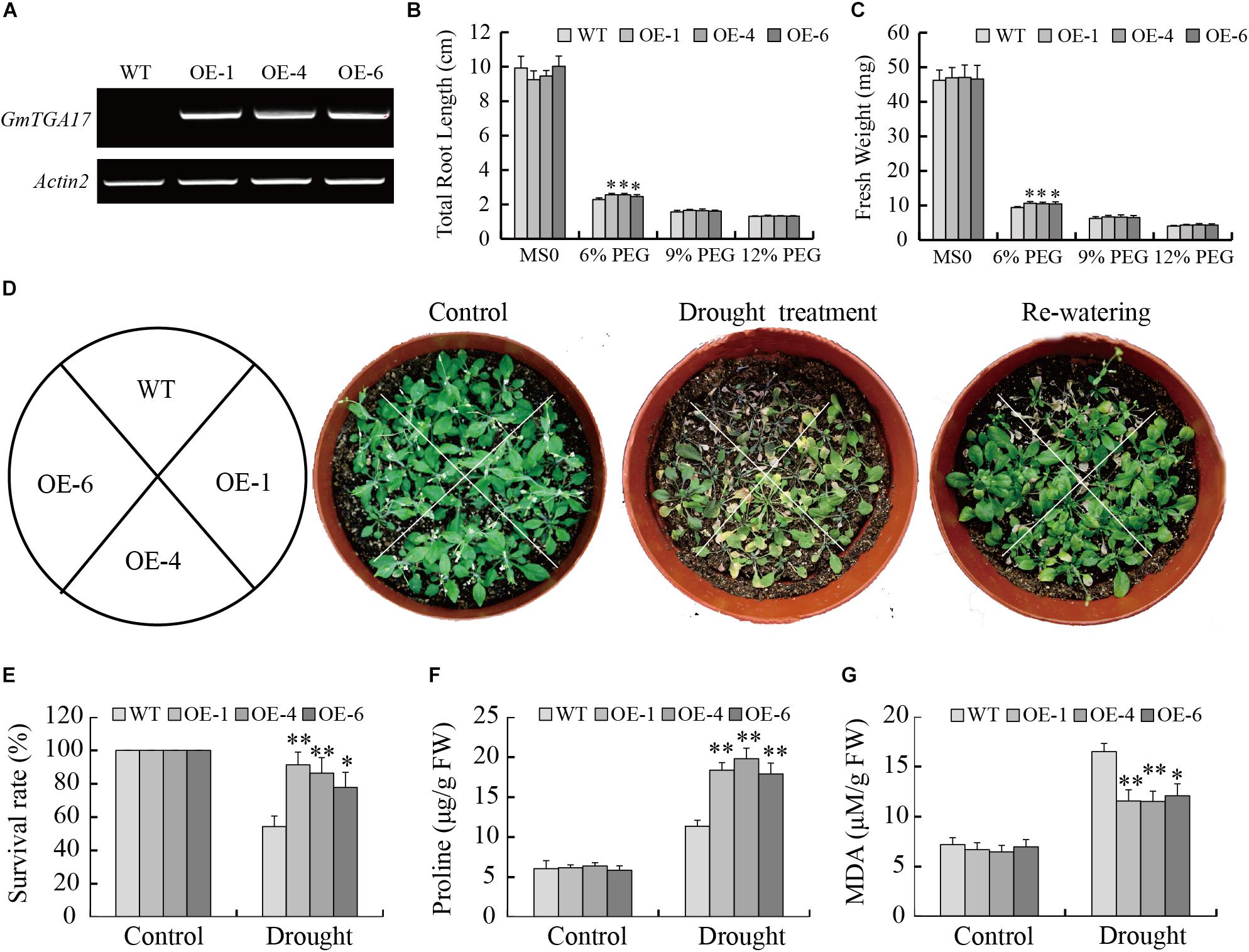
Figure 7. Heterologous overexpression of GmTGA17 in Arabidopsis enhanced drought tolerance. (A) Semi-qRT-PCR analysis of GmTGA17 expression levels in T3 transgenic Arabidopsis lines. (B) Total root length and (C) fresh weight of WT and transgenic lines exposed to 0, 6, 9, and 12% (m/v) PEG6000 for 8 days. (D) Phenotypes of WT and transgenic plants grown in soil under drought stress: control, treated for 2 weeks and re-watered for 1 week. (E) Survival rates was counted after 1 week of recovery. (F) Proline and (G) MDA contents were measured 10 days after treatment. All values are presented as means of three independent replicates (n = 40). The error bars indicate SD. The asterisks denote a significant difference (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
To compare with the above results, we also tested the tolerance of transgenic Arabidopsis lines growing in the soil under drought stress conditions. After a 2-week water-deficit regimen, the leaves of WT plants became shriveled or died, while transgenic lines were only slightly shriveled and showed lower mortality than WT plants (Figure 7D). Compared to a 54.32% survival rate of WT plants, the survival rate of transgenic plants was 77.78–91.35% after 1 week of re-watering following treatment (Figure 7E). After a 10-day water-withholding treatment, two stress-related parameters, proline and MDA contents, were compared between the transgenic and WT seedlings. The results showed that proline and MDA contents in transgenic seedlings (17.90–18.37 μg/g FW and 11.52–12.07 μM/g FW) were significantly higher and lower than in WT plants (11.37 μg/g FW and 16.49 μM/g FW) under drought stress conditions, respectively (Figures 7F,G). Collectively, these results indicated that GmTGA17 may have a potential function in enhancement of the transgenic plants tolerance to drought stress.
Overexpression of GmTGA17 Improved Arabidopsis Tolerance to Salinity
To determine if GmTGA17 is in involved in responses to salt stress, we tested the salt tolerance of the transgenic Arabidopsis lines. When grown on 1/2 MS medium, the no difference in phenotype was observed between the transgenic and WT seedlings (Figure 8A). When grown on 1/2 MS medium containing 75 mM NaCl for 1 week, the total root length of transgenic plants was significantly longer than those of WT plants (Figure 8B). Similarly, the fresh weight of transgenic plants was significantly greater than those of WT plants (Figure 8C). In response to 50 or 100 mM NaCl treatments, the total root length and fresh weight were different between the transgenic and WT seedlings, though not significant at the 0.05 level (Figures 8B,C).
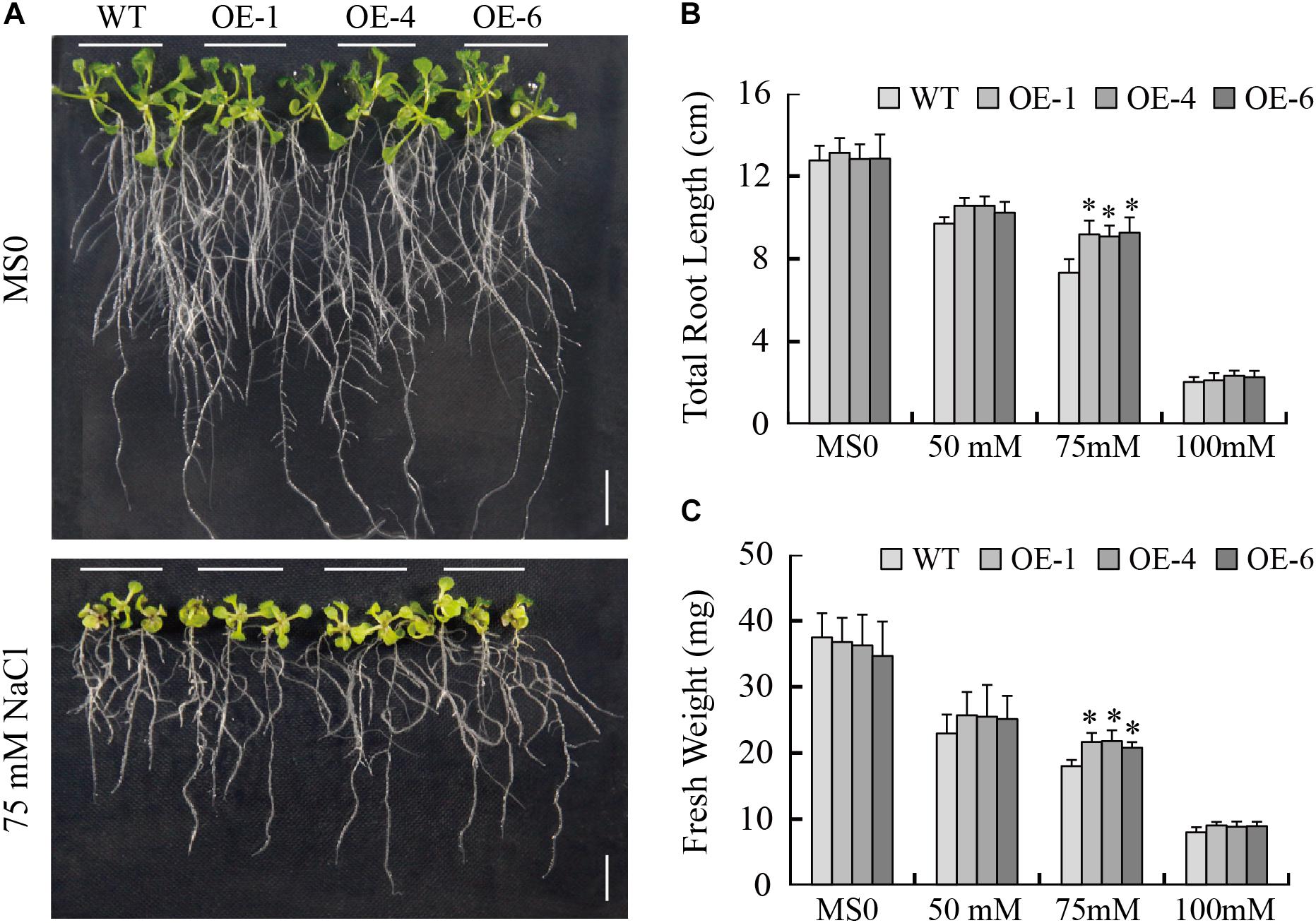
Figure 8. Response of WT and transgenic Arabidopsis plants to NaCl treatment. (A) Phenotypes of WT and transgenic seedlings under NaCl treatment. The photographs were taken after 8 days of treatment. (B) Total root length and (C) fresh weight of WT and transgenic lines exposed to 0, 50, 75, and 100 mM NaCl for 8 days. Scale bars = 1 cm. All values are presented as means of three independent replicates (n = 30). The error bars indicate SD. The asterisks denote a significant difference (∗P < 0.05) compared with the corresponding controls.
The capacity of GmTGA17 transgenic lines to respond to high salinity stress was also assessed. The leaves of WT plants were severely wilted, whereas, the leaves of transgenic plants were slightly damaged but still remained green after treated for 1 week (Figure 9A). The transgenic plants displayed a significantly higher survival rate compared with WT plants under salinity conditions (Figure 9B). We also analyzed the chlorophyll content of all experimental plants and observed a significantly higher chlorophyll content in the transgenic plants compared with WT plants (Figure 9C). Together, these results demonstrated that overexpression of GmTGA17 enhanced tolerance to salt stress in transgenic Arabidopsis plants.
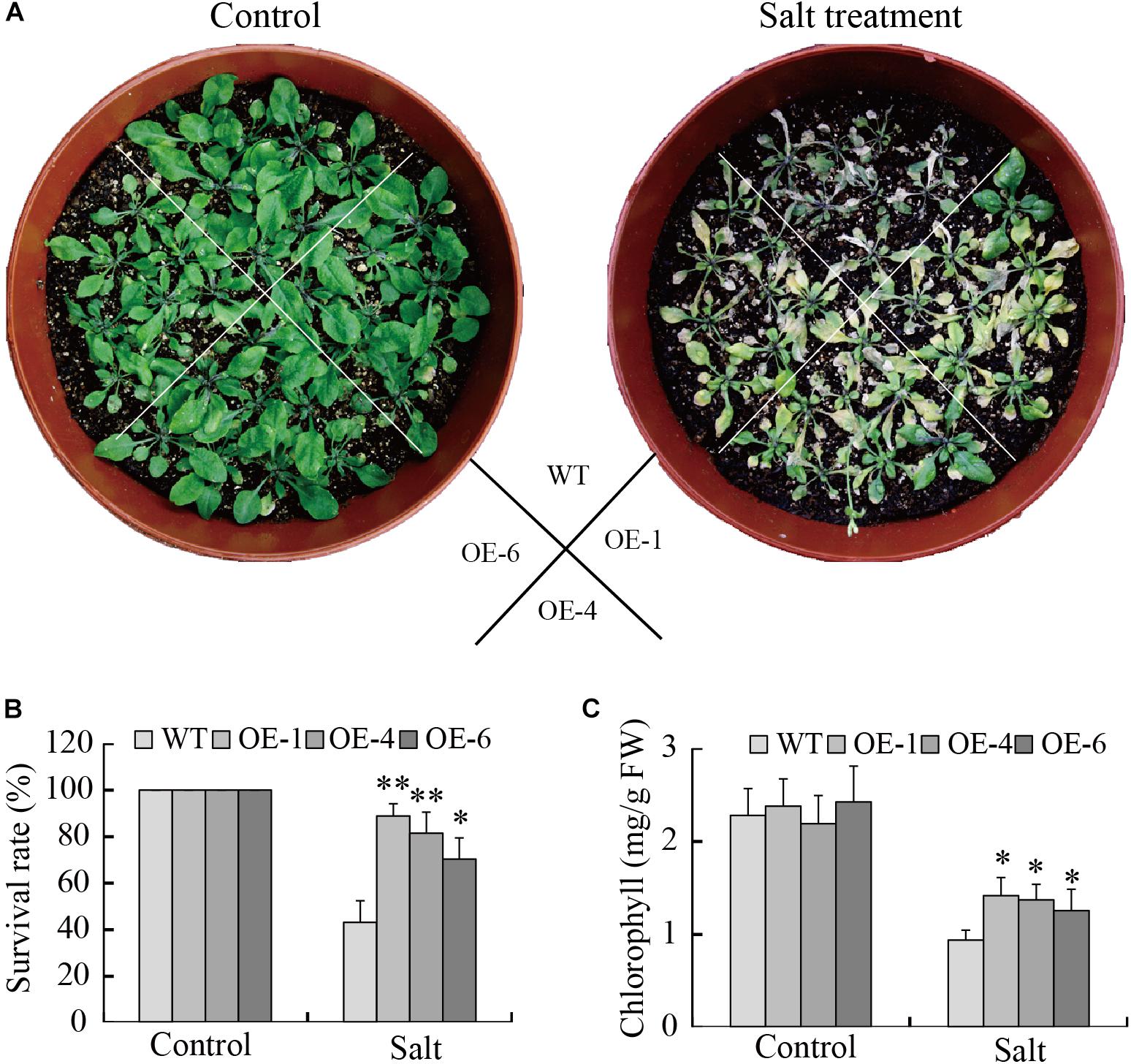
Figure 9. Enhance salt tolerance mediated by heterologous overexpression of GmTGA17 in Arabidopsis. (A) Phenotypes of WT and transgenic plants grown in soil under high salinity treatment conditions. The photographs were taken after 1 week of treatment. (B) Survival rates was counted after 1 week of treatment. (C) Chlorophyll content was measured 5 days after treatment. All values are presented as means of three independent replicates (n = 40). The error bars indicate SD. The asterisks denote a significant difference (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
GmTGA17 Promoted ABA-Induced Stomatal Closure
A reduced water loss rate is a major factor contributing to the maintenance of moisture under drought stress. In the present study, data showed that the detached leaves from transgenic Arabidopsis lines lost water more slowly than those of WT plants (Figure 10A). Since the stomatal aperture has a close link with water loss, we examined the stomata of transgenic Arabidopsis and WT plants with or without ABA treatment. Under normal growth conditions, there was no obvious difference in stomatal aperture between transgenic Arabidopsis and WT plants. In the presence of 10 or 15 μM ABA, the stomatal aperture of transgenic Arabidopsis plants was significantly smaller than that of WT plants (Figures 10C,D). Therefore, transgenic Arabidopsis lines showed more rapid ABA-induced stomatal closure than did WT plants.
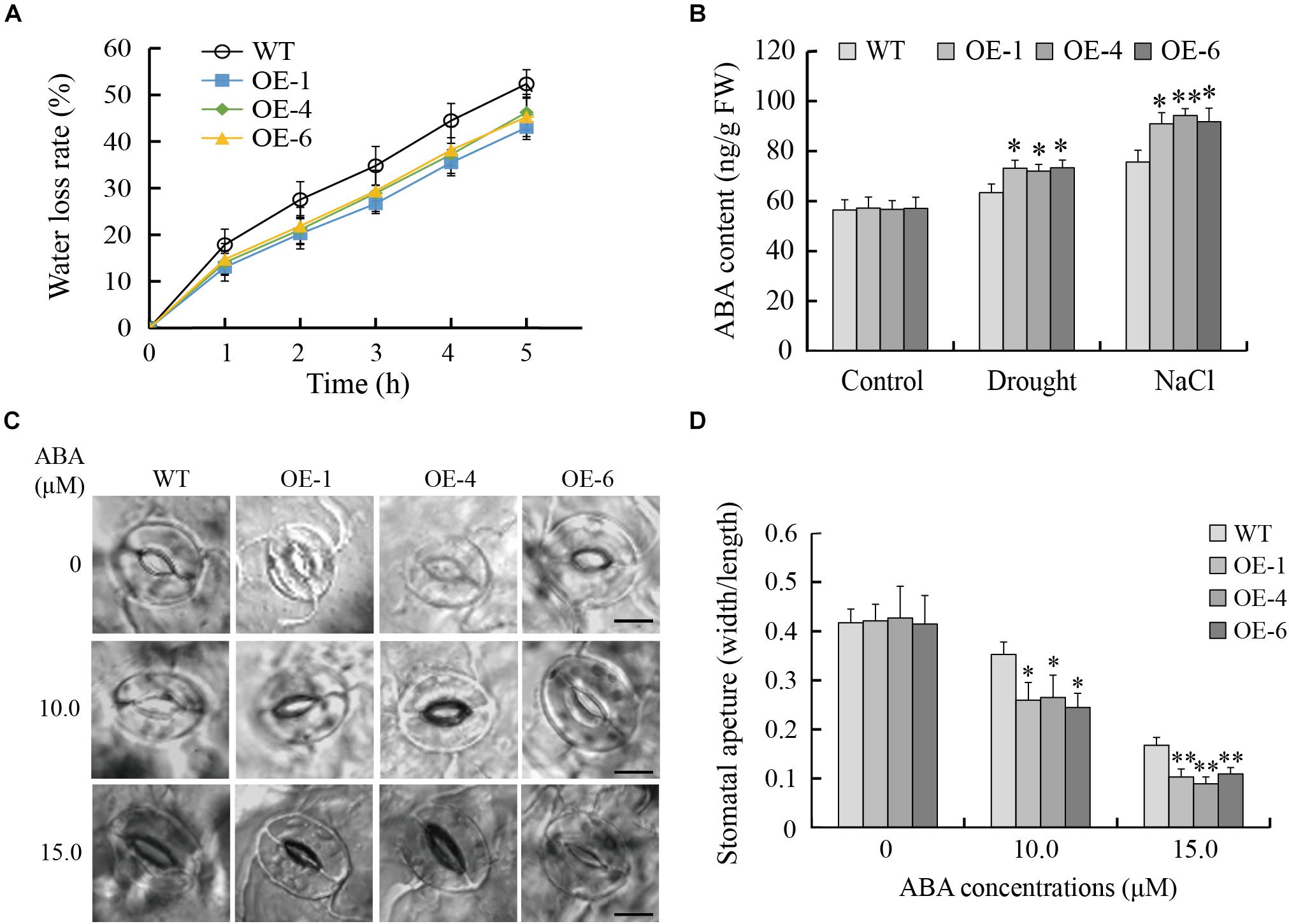
Figure 10. GmTGA17 promotes ABA-induced stomatal closure. (A) Water loss from detached leaves of transgenic Arabidopsis lines and WT. The water loss of 1.0 g detached leaves from different lines was expressed as the percentage of initial fresh weight. Values are presented as means of three independent replicates. The error bars indicate SD. (B) Endogenous ABA content in leaves of transgenic Arabidopsis lines and WT plants before and after different abiotic stress treatments. ELISA was used to measure ABA content in leaves of transgenic Arabidopsis lines and WT plants with drought treatment for 10 days or salt treatment for 5 days. Values are presented as means ± SD of three independent replicates. The asterisks denote a significant difference (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls. (C,D) ABA-mediated stomatal closure. Rosette leaves from 3-week-old transgenic Arabidopsis lines and WT plants were treated with stomatal opening solution and in ABA solution (0, 10 or 15 μM) for another 3 h. Scale bars = 10 μm. Data are means of three independent replicates (n = 40). The error bars indicate SD. The asterisks denote a significant difference (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
In addition, endogenous ABA content were measured in transgenic Arabidopsis lines and WT plants before and after stress treatments. As shown in Figure 10B, there was no significant difference in ABA content in leaves between transgenic Arabidopsis and WT plants under normal growth conditions. However, in response to drought or salt stress, the leaves of transgenic Arabidopsis lines accumulated significantly higher content of ABA than WT plants, suggesting that drought- and salt-tolerance phenotypes of transgenic Arabidopsis lines are at least partially derived from higher endogenous ABA content.
GmTGA17 Upregulated ABA-Responsive Genes in Transgenic Arabidopsis
To further elucidate the possible mechanism of GmTGA17 during the abiotic stress response, the transcript levels of eight known ABA-responsive genes, i.e., RD29A, RD29B, RD22, KIN1, COR15A, NCED3, COR47, and RAB18, were analyzed in transgenic Arabidopsis lines and WT plants under drought or salt stress treatment. The qRT-PCR data showed that the expressions of RD29A, RD29B, and RAB18 were significantly up-regulated in transgenic Arabidopsis lines compared with WT plants under both non-stress conditions and stress conditions (Figure 11). Under non-stress conditions, no obvious differences in the expressions of RD22, KIN1, COR15A, NCED3, and COR47 were observed between transgenic Arabidopsis lines and WT plants. However, the transcript levels of RD22, KIN1, COR15A, NCED3, and COR47 were significant higher in transgenic Arabidopsis lines than those in WT plants under both drought and salt treatments (Figure 11).
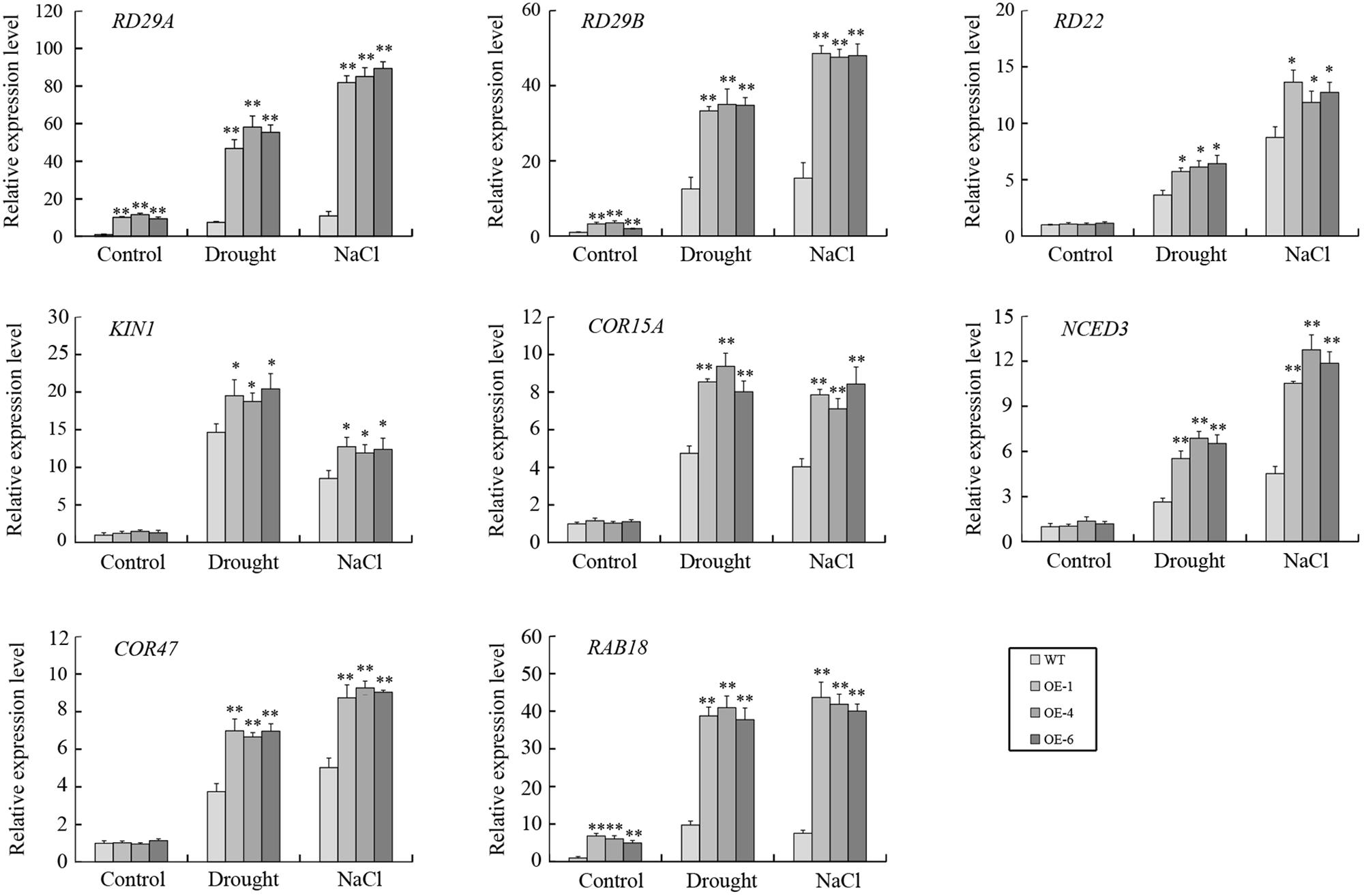
Figure 11. The expressions of several ABA-responsive genes in transgenic Arabidopsis lines and WT plants following drought and salt stress. qRT-PCR was used to detect expression levels of stress-related genes in transgenic Arabidopsis lines and WT plants with drought treatment for 10 days or salt treatment for 5 days. Values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
GmTGA17 Improves Drought and Salt Stress Tolerance in Transgenic Soybean Hairy Roots
The participation of GmTGA17 in drought and salt tolerance was further investigated by performing similar abiotic stress assays in Agrobacterium rhizogenes-mediated soybean hairy roots (Collier et al., 2005; Kereszt et al., 2007; Sun et al., 2015; Wang et al., 2015). qRT-PCR analysis showed that the expression level of GmTGA17 was significantly higher in hairy roots overexpressing GmTGA17, and significantly lower in RNAi hairy roots suppressing GmTGA17 compared with the empty vector control (Figure 12C).
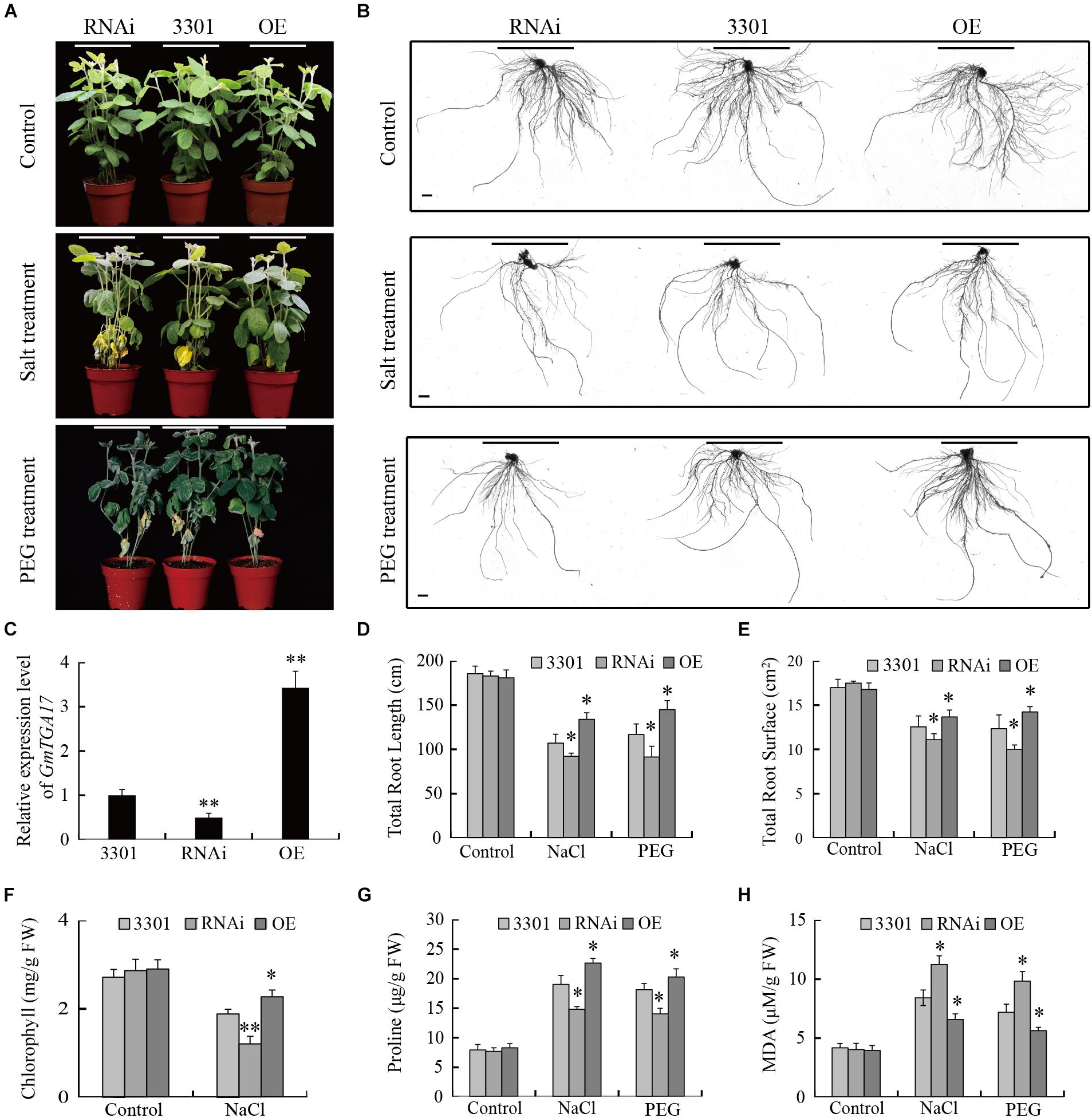
Figure 12. Performance of GmTGA17 transgenic soybean hairy roots under drought and salt stress. (A) Phenotypes of plants with transgenic soybean hairy roots under PEG and high salinity treatments. (B) Transgenic hairy root images: control and after treatment. Scale bars = 1 cm. (C) Relative GmTGA17 expression in roots overexpressing GmTGA17, RNA interference hairy roots, and control roots as shown by qRT-PCR. qRT-PCR quantifications were normalized to the expression of CYP2. Values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗∗P < 0.01) compared with the corresponding controls. (D,E) Total root length and total root surface were measured 10 days after treatments (F–H) Chlorophyll, proline, and MDA contents in leaves of plants with transgenic hairy roots were measured 1 week after treatments. All values are presented as means of three independent replicates (n > 45). The error bars indicate SD. The asterisks denote a significant difference (∗P < 0.05 and ∗∗P < 0.01) compared with the corresponding controls.
Subsequently, we evaluated the drought and salt tolerance in plants with transgenic hairy root grown in soil watered with 20% (m/v) PEG or 150 mM NaCl solutions for 10 days. To avoid damaging the transgenic hairy roots, PEG6000 was also used for simulating drought stress instead of withholding water in this assay. All plants grew well and displayed a similar phenotype under non-stress conditions (Figure 12A). Under high-salinity, the leaves of all experimental plants gradually yellowed and shriveled, while under PEG treatments, the leaves of all experimental plants wilted and bleached from bottom to top. Moreover, the plants carrying the GmTGA17 RNAi hairy roots were the most sensitive to high salinity and PEG stress, while less susceptible were the empty vector control hairy roots, and least susceptible were the GmTGA17 overexpression hairy roots (Figures 12A,B). Under high-salinity stress, we separately observed a significantly higher and lower chlorophyll content in leaves of plants with overexpression hairy roots and RNAi hairy roots compared with that of the empty vector control (Figure 12F). Under both stress conditions, proline content was higher in soybean plants with overexpression hairy roots but lower in RNAi lines relative to the empty vector control. Similarly, MDA content in plants with overexpression hairy roots were lower but higher in RNAi lines relative to the empty vector control (Figures 12G,H). Additionally, compared with the empty vector control, the total root length and root surface of the overexpression hairy roots were longer and greater, but the total root length of the RNAi hairy roots were shorter with decreased surface area under high-salinity and PEG stress conditions (Figures 12D,E). These results indicated that GmTGA17 positively regulated tolerance to drought and salt stress in transgenic soybean hairy roots.
Discussion
TGA transcription factors have been reported to function in various biological processes in plants. However, there is no comprehensive analysis of TGA transcription factors in soybean, and especially concerning their participation in abiotic stress tolerance. In the present study, a total of 27 TGA members were identified in the soybean genome. The GmTGAs could be clustered together with TGAs from Arabidopsis and rice in the same clade (Figure 1), suggesting that the evolution of TGA genes are conserved between monocots and dicots. However, the number of TGAs in soybean is expanded compared to that in Arabidopsis, rice, maize, and papaya (Jakoby et al., 2002; Nijhawan et al., 2008; Espín et al., 2012; Wei et al., 2012). Gene structure analysis showed that putative GmTGA genes within the same clade share a similar intron-exon organization (Supplementary Figure S1), and similar patterns were observed in Arabidopsis and maize (Murmu et al., 2010; Wei et al., 2012). This indicates that the diverse status of intron and exon splicing might thus be meaningful for the evolution of TGA genes. All predicted GmTGA proteins possessed QI and QII domains, which is reported to function as a transcriptional activation domain (Schindler et al., 1992; Schwechheimer et al., 1998; Chuang et al., 1999). Additionally, we found that GmTGA17 showed transcriptional activation activity (Figures 5A,C). Taken together, we speculate that GmTGAs may act as transcription activator. TGA proteins have a conserved bZIP-D box in addition to the common domains of bZIP proteins (Jakoby et al., 2002; Farinati et al., 2010). Although the bZIP-D box is a characteristic domain for TGA family, there are currently no articles of which we are aware reporting the association of this feature with any particular biological function.
It is well known that cis-elements in the promoter region of a given gene are closely related to the biological functions of gene product (Cheng et al., 2013). In the present study, the cis-acting elements analysis revealed that the putative promoter regions of all GmTGA genes possess ERE, W-box, P-box or WUN-motif pathogen-related cis-elements (Raventós et al., 1995; Choi et al., 2004), suggesting involvement of GmTGA genes in defense against pathogens. This observation was in accordance with the fact that TGA genes involved in response to pathogen attack (Jakoby et al., 2002; Wei et al., 2012). Moreover, a number of stress-related cis-elements have been identified in the putative promoter regions of GmTGA genes (Supplementary Table S2), with evidence demonstrating participation of these cis-elements in responses to drought, salt, ABA, and low temperature (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Busk and Pagès, 1998; Gómez-Cadenas et al., 2001; Maestrini et al., 2009). The expression profiles of these genes in different plant tissues showed that 26 deduced GmTGA genes were expressed in roots, root hairs, stems, leaves, nodules, seeds, and flowers, with the exception of GmTGA01, which had no expression in leaves (Figure 2). It is thus evident that GmTGA genes likely play a broad role in soybean development.
We hypothesized that the existence of abundant stress-related cis-elements suggests that GmTGA genes may participate in plant responses to a range of abiotic stresses. Our qRT-PCR data confirmed this hypothesis, with results showing that GmTGA genes are involved in drought and salt stress responses, though the response mechanisms are diverse. For example, GmTGA15, GmTGA17 and GmTGA25 were up-regulated, while GmTGA05 and GmTGA07 were down-regulated by drought and salt treatments. GmTGA10 and GmTGA14 were up-regulated by drought treatment, but down-regulated by salt treatment. Similar stress response mechanisms were observed in bZIP family genes of rice and Brachypodium distachyon (Nijhawan et al., 2008; Liu and Chu, 2015).
Using transgenic Arabidopsis and soybean hairy root assays, we further characterized the roles of GmTGA17 in abiotic stress tolerance. Our data showed that GmTGA17 could confer tolerance to drought and salinity in transgenic plants (Figures 7–9, 12). Plant tolerance to abiotic stresses are modulated by complicated plant hormone signal transduction pathways and metabolism, especially the main stress hormone ABA (Huang et al., 2008). Drought and salt stress can induce the accumulation of ABA, and this in turn regulates the expression of stress-responsive genes to improve the plant adaption to stress (Xiong et al., 2002). In this study, endogenous ABA content was significantly higher in transgenic Arabidopsis seedlings than WT plants under drought and salt stress treatments (Figure 10B). Coincidentally, the transcript level of a key ABA-biosynthesis gene, AtNCED3, was significantly up-regulated in drought- and salt-treated transgenic Arabidopsis lines compared with WT plants (Figure 11). Based on our stomatal aperture assay data, we speculated that transgenic Arabidopsis plants exhibited reduced leaves water loss due to induced ABA accumulation. Additionally, qRT-PCR data revealed that GmTGA17 also had an induced expression response to ABA treatment (Supplementary Figure S4), and upregulated several ABA-responsive genes in transgenic Arabidopsis lines under drought and salt stress treatments (Figure 11). The above results suggest that GmTGA17 may participate in plant response to drought and salinity in an ABA-dependent pathway.
Previously, AtTGA1-AtTGA7 were grouped within three clades associated with plant defense, based on sequence homology (Xiang et al., 1997; Kesarwani et al., 2007). However, the other three AtTGA members were not included in this classification. Subsequently, TGAs from Arabidopsis and papaya were divided into three main clades (I, II and III) according to the similarity of their protein sequences (Murmu et al., 2010; Espín et al., 2012). Clade I contains AtTGA1, AtTGA3, AtTGA4, AtTGA7, CpTGA1, and CpTGA3. AtTGA2, AtTGA5, AtTGA6, AtPAN, CpTGA2, and CpTGA4 belong to clade II. AtTGA9, AtTGA10 clustered together with CpTGA5 and CpTGA6 in the clade III.
Our phylogenetic analysis revealed that the 27 predicted GmTGAs could also be divided into clades I, II, and III, mentioned above (Figure 1). In clade I, AtTGA3 and AtTGA4 participate in plant pathogen response and root development, as well as playing a vital role in the regulation of response various abiotic stresses (Kesarwani et al., 2007; Alvarez et al., 2014; Du et al., 2014; Zhong et al., 2015; Canales et al., 2017). CpTGA3 was shown to be responsive to Salicylic Acid, suggesting its potential involvement in the defense response (Espín et al., 2012). qRT-PCR data generated in this study showed that GmTGA07, GmTGA10, and GmTGA15 are involved in responses to drought or salt stress.
In clade II, AtPAN is a key regulator in the control of floral patterning (Running and Meyerowitz, 1996; Chuang et al., 1999). AtTGA2, AtTGA5 and AtTGA6 play important roles in disease resistance and development (Zhang et al., 2003; Mueller et al., 2008; Stotz et al., 2013). Our results revealed that the expression of GmTGA05, GmTGA13, GmTGA22, GmTGA24, GmTGA26, and GmTGA27 were up- or down-regulated by drought and salt stress. Interestingly, GmTGA13, GmTGA22, and GmTGA24 were expressed most strongly in flowers (Figure 2), this indicates that they might be also involved in floral development processes in soybean.
Of the TGAs assigned to clade III, AtTGA9 and AtTGA10 were shown to participate in anther development as well as ROS-mediated responses to pathogens (Murmu et al., 2010; Noshi et al., 2016). CpTGA5 was strongly expressed, but only in petals, suggesting an associated role in floral development (Espín et al., 2012). OsbZIP41 was found to be up-regulated in seedlings under blue light treatment while OsbZIP79 and OsbZIP83 were down-regulated under abiotic stress conditions (Nijhawan et al., 2008). Miyamoto et al. (2015) found that OsbZIP79 suppressed production of the diterpenoid phytoalexin, an antimicrobial metabolite. Among the GmTGA genes, the expression of GmTGA14, GmTGA16, GmTGA17, and GmTGA20 were activated or repressed under drought and salt stress. Moreover, GmTGA17 is a positive regulator of plant tolerance to drought and salt stress. The above findings suggested the functional diversification of a certain TGA gene or the TGA members in the same clade. However, further studies are needed to determine the specific functions of the GmTGA family genes by additional experiments.
Conclusion
Twenty-seven soybean TGA genes were identified in the soybean genome. Expression analysis showed that soybean TGA family genes may participate in responses to drought and salt stress. GmTGA17 conferred tolerance to drought and salt stress in both transgenic Arabidopsis plants and soybean hairy roots. However, RNAi hairy roots silenced for GmTGA17 exhibited an increased sensitivity to drought and salt stress. Taken together, soybean TGA genes may act as important components of abiotic stress tolerance in plants.
Author Contributions
Z-SX coordinated the project, conceived and designed the experiments, and edited the manuscript. BL performed the experiments and wrote the first draft. YL and J-DF conducted the bioinformatic work and performed the experiments. W-JZ, J-HL, L-GJ, Y-BZ, MC and D-HM contributed with valuable discussions. X-YC revised and edited the manuscript. Y-ZM coordinated the team. All authors read and approved the final manuscript.
Funding
This research was financially supported the National Transgenic Key Project of the Chinese Ministry of Agriculture (2018ZX0800909B and 2016ZX08002-002), the National Natural Science Foundation of China (31871624 and 31871611), and the Program of Introducing Talents of Innovative Discipline to Universities (Project 111) from the State Administration of Foreign Experts Affairs (#B18042) “Crop Breeding for Disease Resistance and Genetic Improvement.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Lijuan Qiu of the Institute of Crop Science, Chinese Academy of Agricultural Sciences for kindly providing soybean seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00549/full#supplementary-material
FIGURE S1 | Phylogenetic analysis and intron/exon structures of TGA genes in soybean. The phylogenetic tree (left panel) was created using MEGA6.0 software. Exon/intron structures of TGA genes are shown in the right panel. Gene models were drawn to scale as indicated on the bottom.
FIGURE S2 | Structure and alignment of GmTGA proteins. (A) Primary structure of TGA (Jakoby et al., 2002). (B) Alignment of GmTGA amino acid sequences. Black arrows indicate Glutamine, Alanine, and Serine residues. Asterisks indicate the leucine zipper domain.
FIGURE S3 | PCR detection and the expression levels of GmTGA17 in transgenic Arabidopsis lines overexpressing GmTGA17. (A) Genomic DNA PCR detection of GmTGA17 in transgenic Arabidopsis lines (OE-1, OE-4, and OE-6). “+” represents positive control; “–” represents negative control (WT plant). The size of target band is 671 bp. (B) Relative expression levels of GmTGA17 in transgenic Arabidopsis lines. Actin2 was used as a control.
FIGURE S4 | Expression profile of GmTGA17 under ABA treatment. Expression pattern of GmTGA17 in soybean seedlings under ABA treatment. The expression levels were normalized to that of CYP2. All values are means and SD obtained from four biological replicates. The asterisks indicate a statistical significance (∗∗P < 0.01) compared with the corresponding controls.
TABLE S1 | Primers and sequences used in this study.
TABLE S2 |Cis-elements in promoters of GmTGA genes.
Abbreviations
ABA, abscisic acid; ABRE, ABA-responsive element; ARE, anaerobic responsive element; ERE, elicitor-response element; FW, fresh weight; GFP, green fluorescent protein; MS, Murashige and Skoog; ORF, open reading frame; P-box, pathogen-inducible box; qRT-PCR, quantitative real-time-PCR; W-box, wound-inducible box; WT, wild type.
References
Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-Lopez, O., Tamayo, K. P., et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factor as important regulatory components of the nitrate response of Arabidopsis thaliana. Plant J. 80, 1–13. doi: 10.1111/tpj.12618
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. doi: 10.1104/pp.24.1.1
Baker, S. S.,and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Busk, P. K., and Pagès, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. doi: 10.1023/A:1006058700720
Cakmak, I., and Horst, W. J. (1991). Effect of aluminium on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol. Plant 83, 463–468. doi: 10.1111/j.1399-3054.1991.tb00121.x
Canales, J., Contreras-López, O., Álvarez, J. M., and Gutiérrez, R. A. (2017). Nitrate induction of root hair density is mediated by TGA1/TGA4 and CPC transcription factors in Arabidopsis thaliana. Plant J. 92, 305–316. doi: 10.1111/tpj.13656
Chen, T. H. H., and Murata, N. (2011). Glycinebetaine protects plants against abiotic stress: mechanisms and biotechnological applications. Plant Cell Environ. 34, 1–20. doi: 10.1111/j.1365-3040.2010.02232.x
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Choi, J., Huh, S. U., Kojima, M., Sakakibara, H., Paek, K. H., and Hwang, I. (2010). The cytokinin-activated transcription factor ARP2 promoters plant immunity via TGA3/NPR1-development salicylic acid signaling in Arabidopsis. Dev. Cell 19, 284–295. doi: 10.1016/j.devcel.2010.07.011
Choi, J. J., Klosterman, S. J., and Hadwiger, L. A. (2004). A promoter from pea gene DRR206 is suitable to regulate and elicitor-coding gene and develop disease resistance. Phytopathology 94, 651–660. doi: 10.1094/PHYTO.2004.94.6.651
Chuang, C. F., Running, M. P., Williams, R. W., and Meyerowitz, E. M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344. doi: 10.1101/gad.13.3.334
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j1365-313x.1998.00343x
Collier, R., Fuchs, B., Walter, N., Lutke, W. K., and Taylor, C. G. (2005). Ex vitro composite plants an inexpensive, rapid method for root biology. Plant J. 43, 449–457. doi: 10.1111/j.1365-313X.2005.02454.x
Deng, W. K., Wang, Y. B., Liu, Z. X., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Du, X., Du, B., Chen, X., Zhang, S., Zhang, Z., and Qu, S. (2014). Overexpression of the MhTGA2 gene from crab apple (Malus hupehensis) confers increased tolerance to salt stress in transgenic apple (Malus domestica). J. Agric. Sci. 152, 634–641. doi: 10.1017/S0021859613000130
Espín, F. M. I., Peraza-Echeverria, S., Fuentes, G., and Santamaría, J. M. (2012). In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol. Biochem. 54, 113–122. doi: 10.1016/j.plaphy.2012.02.011
Farinati, S., DalCorso, G., Varotto, S., and Furini, A. (2010). The brassica juncea BjCdR15, an ortholog of Arabidopsis TGA3, is a regulator of cadmium uptake, transport and accumulation in shoots and confers cadmium tolerance in transgenic plants. New Phytol. 185, 964–978. doi: 10.1111/j.1469-8137.2009.03132.x
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fusco, N., Micheletto, L., DalCorso, G., Borgato, L., and Furini, A. (2005). Identification of cadmium-regulated genes by cDNA-AFLP in the heavy metal accumulator Brassica juncea L. J. Exp. Bot. 56, 3017–3027. doi: 10.1093/jxb/eri299
Gasteiger, E., Gattiker, A., Hoogland, C., Ivanyi, I., Appel, R. D., and Bairoch, A. (2003). ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788. doi: 10.1093/nar/gkg563
Gatz, C. (2013). From pioneers to team players: TGA transcription factors provide a molecular link between different stress pathways. Mol. Plant Microbe Interact. 26, 151–159. doi: 10.1094/MPMI-04-12-0078-IA
Gietz, R. D., and Schiestl, R. H. (2007). Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 35–37. doi: 10.1038/nprot.2007.14
Gómez-Cadenas, A., Zentella, R., Sutliff, T. D., and Ho, T. H. (2001). Involvement of multiple cis-elements in the regulation of GA responsive promoters: definition of a new cis-element in the Amy32b gene promoter of barley (Hordeum vulgare). Physiol. Plant 112, 211–216. doi: 10.1034/j.1399-3054.2001.1120209.x
Hu, B., Jin, J. P., Guo, A. Y., Zhang, H., Luo, J. C., and Gao, G. (2015). GSDS2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Huang, D. Q., Wu, W. R., Abrams, S. R., and Cutler, A. J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. doi: 10.1093/jxb/ern155
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jefferson, R. (1987). Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. doi: 10.1007/BF02667740
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-acting TGA factor to defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. doi: 10.1105/tpc.012211
Johnson, C., Boden, E., Desai, M., Pascuzzi, P., and Arias, J. (2001). In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 28, 237–243. doi: 10.1046/j.1365-313X.2001.01147.x
Katagiri, F., Lam, E., and Chua, N. H. (1989). Two tobacco DNA-binding proteins with homology to the nuclear factor CREB. Nature 340, 727–730. doi: 10.1038/340727a0
Kereszt, A., Li, D. X., Indrasumunar, A., Nguyen, C. D. T., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kesarwani, M., Yoo, J., and Dong, X. (2007). Genetic interactions of TGA transcription factors in the regulation of pathogenesis related genes and disease resistance in Arabidopsis. Plant Physiol. 144, 336–346. doi: 10.1104/pp.106.095299
Kim, J. K., and Kim, W. T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162, 1733–1749. doi: 10.1104/pp.113.220103
Krasensky, J., and Jonak, C. (2012). Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 63, 1593–1608. doi: 10.1093/jxb/err460
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, W. X., Oono, Y., Zhu, J. H., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopisi NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promoter drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Liu, N., Fromm, M., and Avramova, Z. (2014). H3K27me3 and H3K4me3 chromatin environment at super-induced dehydration stress memory genes of Arabidopsis thaliana. Mol. Plant 7, 502–513. doi: 10.1093/mp/ssu001
Liu, P., Liu, J., Dong, H. X., and Sun, J. Q. (2018). Functional regulation of Q by microRNA172 and transcriptional co-repressor TOPLESS in controlling breed wheat spikelet density. Plant Biotechnol. J. 16, 495–506. doi: 10.1111/pbi.12790
Liu, X., and Chu, Z. Q. (2015). Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genomics 16:227. doi: 10.1186/s12864-015-1457-9
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Maestrini, P., Cavallini, A., Rizzo, M., Giordani, T., Bernardi, R., Durante, M., et al. (2009). Isolation and expression analysis of low temperature-induced genes in white poplar (Populus alba). J. Plant Physiol. 166, 1544–1556. doi: 10.1016/j.jplph.2009.03.014
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Miyamoto, K., Nishizawa, Y., Minami, E., Nojiri, H., Yamane, H., and Okada, K. (2015). Overexpression of the bZIP transcription factor OsbZIP suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 173, 19–27. doi: 10.1016/j.jplph.2014.09.001
Mueller, S., Hibert, B., Dueckershoff, K., Roitsch, T., Krischke, M., Mueller, M. J., et al. (2008). General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell 20, 768–785. doi: 10.1105/tpc.107.054809
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and redundantly required for anther development. Plant Physiol. 154, 1492–1504. doi: 10.1104/pp.110.159111
Nijhawan, A., Jain, M., Tyagi, A. K., and Khurana, J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol. 146, 333–350. doi: 10.1104/pp.107.112821
Noshi, M., Mori, D., Tanabe, N., and Maruta, T. (2016). Arabidopsis clade IV TGA transcription factors, TGA10 and TGA9, are involved in ROS-mediated responses to bacterial PAMP flag22. Plant Sci. 252, 12–21. doi: 10.1016/j.plantsci.2016.06.019
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Raventós, D., Jensen, A. B., Rask, M. B., Casacuberta, J. M., Mundy, J., and Segundo, B. S. (1995). A 20bp cis-acting element is both necessary and sufficient to mediate elicitor response of a maize PRms gene. Plant J. 7, 147–155. doi: 10.1046/j.1365-313X.1995.07010147.x
Running, M. P., and Meyerowitz, E. M. (1996). Mutations in the PERIANTHIA gene of Arabidopsis specifically alter floral organ number and initiation pattern. Development 122, 1261–1269.
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Schindler, U., Beckmann, H., and Cashmore, A. R. (1992). TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4, 1309–1319. doi: 10.1105/tpc.4.10.1309
Schwechheimer, C., Smith, C., and Bevan, M. W. (1998). The activities of acidic and glutamine-rich transcriptional activation domains in plant cells: design of modular transcription factors for high-level expression. Plant Mol. Biol. 36, 195–204. doi: 10.1023/A:1005990321918
Singh, K. B., Foley, R. C., and Oñate-Sánchèz, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/S1369-5266(02)00289-3
Stotz, H. U., Mueller, S., Zoeller, M., Mueller, M. J., and Berger, S. (2013). TGA transcription factors and jasmonate-independent col1 signalling regulate specific plant responses to reactive oxylipins. J. Exp. Bot. 64, 963–975. doi: 10.1093/jxb/ers389
Sun, X. J., Hu, Z., Chen, R., Jiang, Q. Y., Song, G. H., Zhang, H., et al. (2015). Target mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5:10342. doi: 10.1038/srep10342
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kuma, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tool. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolor, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113. doi: 10.1111/j.1365-313X.2005.02513.x
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, X., Yang, B., Li, K., Kang, Z., Cantu, D., and Dubcovsky, J. (2016). A conserved Puccinia striiformis protein interacts with wheat NPR1 and reduces induction of pathogenesis-related genes in response to pathogens. Mol. Plant Microbe Interact. 29, 977–989. doi: 10.1094/MPMI-10-16-0207-R
Wei, K., Chen, J., Wang, Y., Chen, S., Lin, Y., Pan, S., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi: 10.1093/dnares/dss026
Xiang, C. B., Miao, Z. H., and Lam, E. (1997). DNA-binding properties, genomic organization and expression pattern of TGA6, a new member of the TGA family of bZIP transcription factors in Arabidopsis thaliana. Plant Mol. Biol. 34, 403–415. doi: 10.1023/A:1005873500238
Xiong, L. M., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi: 10.1105/tpc.000596
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in a Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.1105/tpc.6.2.251
Yang, L., Ji, W., Zhu, Y. M., Gao, P., Li, Y., Cai, H., et al. (2010). GsCBRLK, a calcium/calmodulin-binding receptor-like kinase, is a positive regulator of plant tolerance to salt and ABA stress. J. Exp. Bot. 61, 2519–2533. doi: 10.1093/jxb/erq084
Yang, Y. Q., and Guo, Y. (2018). Unraveling salt stress signaling in plants. J. Integr. Plant Biol. 60, 796–804. doi: 10.1111/jipb.12689
Zander, M., Chen, S. X., Imkampe, J., Thurow, C., and Gatz, C. (2012). Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol. Plant 5, 831–840. doi: 10.1093/mp/ssr113
Zhang, J. Y., Qu, S. C., Du, X. L., Qiao, Y. S., Cai, B. H., Guo, Z. R., et al. (2012). Overexpression of the Malus hupehensis MhTGA2 gene, a novel bZIP transcription factor for increased tolerance to salt and osmotic stress in transgenic tobacco. Int. J. Plant Sci. 173, 441–453. doi: 10.1086/665262
Zhang, Y., Fan, W., Kinkema, M., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y. L., Tessaro, M. J., Lassner, M., and Li, X. (2003). Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
Zhong, L., Chen, D. D., Min, D. H., Li, W. W., Xu, Z. S., Zhou, Y. B., et al. (2015). AtTGA4, a bZIP transcription factor, confers drought resistance by enhancing nitrate transport and assimilation in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 457, 433–439. doi: 10.106/j.bbrc.2015.01.009
Keywords: soybean, TGA transcription factor, molecular characterization, abiotic stress response, drought and salt tolerance
Citation: Li B, Liu Y, Cui X-Y, Fu J-D, Zhou Y-B, Zheng W-J, Lan J-H, Jin L-G, Chen M, Ma Y-Z, Xu Z-S and Min D-H (2019) Genome-Wide Characterization and Expression Analysis of Soybean TGA Transcription Factors Identified a Novel TGA Gene Involved in Drought and Salt Tolerance. Front. Plant Sci. 10:549. doi: 10.3389/fpls.2019.00549
Received: 06 November 2018; Accepted: 10 April 2019;
Published: 16 May 2019.
Edited by:
Yan Guo, China Agricultural University, ChinaReviewed by:
Yong Hwa Cheong, Sunchon National University, South KoreaSoumitra Paul, University of Calcutta, India
Copyright © 2019 Li, Liu, Cui, Fu, Zhou, Zheng, Lan, Jin, Chen, Ma, Xu and Min. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhao-Shi Xu, eHV6aGFvc2hpQGNhYXMuY24= Dong-Hong Min, bWRoMjQ5M0AxMjYuY29t
†These authors have contributed equally to this work
 Bo Li1,2†
Bo Li1,2† Ming Chen
Ming Chen Zhao-Shi Xu
Zhao-Shi Xu