- 1Rice and Sorghum Research Institute, Sichuan Academy of Agricultural Sciences/Key Laboratory of Southwest Rice Biology and Genetic Breeding, Ministry of Agriculture, Deyang, China
- 2Rice Research Institute, Sichuan Agricultural University, Chengdu, China
- 3Mianyang Academy of Agricultural Sciences, Mianyang, China
Rice blast caused by Magnaporthe oryzae (M. oryzae) is a major threat to global rice production. In recent years, small interference RNAs (siRNAs) and host-induced gene silencing (HIGS) has been shown to be new strategies for the development of transgenic plants to control fungal diseases and proved a useful tool to study gene function in pathogens. We here tested whether in vitro feeding artificial siRNAs (asiRNAs) could compromise M. oryzae virulence and in vivo HIGS technique could improve rice blast resistance. Our data revealed that silencing of M. oryzae MoAP1 by feeding asiRNAs targeting MoAP1 (i.e., asiR1245, asiR1362, and asiR1115) resulted in inhibited fungal growth, abnormal spores, and decreased pathogenicity. Among the asiRNAs, asiR1115 was the most inhibitory toward the rice blast fungus. Conversely, the asiRNAs targeting three other genes (i.e., MoSSADH, MoACT, and MoSOM1) had no effect on fungal growth. Transgenic rice plants expressing RNA hairpins targeting MoAP1 exhibited improved resistance to 11 tested M. oryzae strains. Confocal microscopy also revealed profoundly restricted appressoria and mycelia in rice blast-infected transgenic rice plants. Our results demonstrate that in vitro asiRNA and in vivo HIGS were useful protection approaches that may be valuable to enhance rice blast resistance.
Introduction
Rice blast, caused by the fungal pathogen Magnaporthe oryzae, is one of the most destructive rice diseases, potentially leading to considerable economic losses for the global grain industry. It is estimated that annual rice harvests are decreased by 10–30% because of this disease (Dolgin, 2009). Therefore, it is imperative that the effective disease control strategies are developed. It is now clear that, in essence, rice has two branches of innate immune system [pathogen-associated molecular pattern- (PAMP-), triggered immunity (PTI), and effector-triggered immunity (ETI)] against M. oryzae (Chen and Ronald, 2011). PTI is mediated by plant pattern recognition receptors (PRRs), such as CEBiP, LYP4, LYP6, and to recognize the pathogen-derived PAMP chitin (Chandran et al., 2019). ETI is activated upon recognition of effectors from M. oryzae by the nucleotide binding and leucine rich repeat (NBS-LRR) type resistance (R) proteins (Chen and Ronald, 2011). So far, more than 100 rice blast R genes have been identified, of which more than 30 genes were isolated (Wang et al., 2017). All of the cloned R genes encode NBS-LRR proteins, except Pid2 and the recessive pi21 mutant. Pid2 encodes a β-lectin receptor-like kinase (Chen et al., 2006). The wild type Pi21 encodes an uncharacterized proline-rich protein containing a metal-binding domain (Fukuoka et al., 2009). Notably, several recent studies have reported that blast-resistance can be trade-off with yield penalty. For example, the interconnected two NBS-LRR receptors, PigmR and PigmS, balance high rice blast disease resistance, and yield traits via an epigenetic regulatory mechanism (Deng et al., 2017). The transcription factor ideal plant architecture 1 (IPA1) switches DNA binding specificity to regulate the expression of two down-stream genes via phosphorylation upon infection of M. oryzae to improve both blast disease resistance and grain yield (Wang et al., 2018). In addition, some microRNAs have been identified to be important factors in fine-tuning the trade-off between blast disease resistance and yield-traits (Chandran et al., 2019). However, R gene-mediated resistance is easily evaded by M. oryzae due to the high race-specificity of R genes and the high frequency of race variation in M. oryzae population. Thus, new approaches leading to broad-spectrum resistance are urgently needed to control the blast disease in rice production.
RNA interference (RNAi) is a conserved integral gene regulatory process in most eukaryotes (Fire, 2008). This process typically involves the cleavage of a double-stranded RNA (dsRNA) into small-interfering RNA (siRNA) homologs to a target sequence for the subsequent silencing of the genes containing that sequence. RNAi is an important pathway for functional genomics researches in many different organisms such as worms, protists, fungi, animals, plants and human (Harborth et al., 2001; Li et al., 2010; Zhu et al., 2017). The growing evidence have indicated that RNAi technology can be used in crop protection strategies (Nunes and Dean, 2012; Vinay et al., 2016). Especially, the RNAi has been used commercially to engineer virus-resistant plants via the expression of viral sequences as transgenes (Frizzi and Huang, 2010), indicating that the host-induced gene silencing (HIGS) is an RNAi-based technology that emerged as a powerful tool for generating transgenic plants to control fungal diseases and study the function of candidate pathogenicity genes in pathogenic fungi (Nunes and Dean, 2012). This technology has been proven to be effective in the following fungal pathogens by in planta expression of RNAi constructs specifically silencing certain fungal genes, including Fusarium graminearum (Koch et al., 2013; Cheng et al., 2015), Fusarium culmorum (Chen et al., 2016), Blumeria graminis (Nowara et al., 2010; Pliego et al., 2013), Puccinia striiformis f. sp. tritici (Yin et al., 2011; Zhang et al., 2012), Puccinia triticina (Panwar et al., 2013), Fusarium oxysporum f. sp. cubense (Ghag et al., 2014), Botrytis cinerea (Wang et al., 2016), and Bremia lactucae (Govindarajulu et al., 2015). In recent researches, HIGS has been used to study the rice-M. oryzae interaction. Wang et al. (2015) cloned M. oryzae NoxI and NAC gene fragments and constructed HIGS transgenic rice plants, but they did not show whether the transgenic rice lines exhibited resistance to blast disease. Zhu et al. (2017) silenced 3 rice blast pathogenicity genes by transient BMV-HIGS system, and indicated that combining introduction of fungal gene sequences in sense and antisense orientation could significantly enhanced the efficiency of BMV-HIGS. However, they didn’t get the stable resistent transgenic rice lines.
Magnaporthe oryzae infects in host to obtain nutrients for its subsequent proliferation. The infection process involves multiple steps, including conidial germination, appressorium formation, penetration peg formation, and invasive hyphal growth (Wilson and Talbot, 2009). Each step is regulated by different genes, some of which have been functionally characterized. For example, PLS1 is involved in the regulation of appressorial function that is essential for the penetration of host cells by the fungus (Clergeot et al., 2001). PMK1 regulates appressorium formation and hyphal growth (Park et al., 2002), and GAS1, GAS2 and MST12 may function downstream of PMK1 to regulate the expression of genes involved in appressorial penetration and hyphal growth (Park et al., 2002; Xue et al., 2002). Furthermore, PDE1 (Balhadere and Talbot, 2001) and MPS1 (Xu et al., 1998) influence appressorium-mediated plant infection, whereas, RVS167 and Las17 affect appressorial penetration (Dagdas et al., 2012). Penn et al. (2015) indicated that protein kinase C is essential for growth and development of the rice blast fungus and may play key roles in spore germination, cell wall biogenesis, polarized growth, and hyphal development. The infection process of M. oryzae also involves transcription factors. For example, MoAP1 encodes an M. oryzae bZIP transcription factor essential for conidial production, appressorial development, aerial hyphal growth, and pathogenicity (Guo et al., 2011). MoAP1 regulates the expression of MoAAT, MoSSADH and MoACT, which are important for the growth, development, and pathogenicity of M. oryzae (Guo et al., 2011). Additionally, MoSOM1 and MoCDTF1 are also transcription regulators associated with cellular differentiation of invasive hyphae during infections of the rice blast fungus (Yan et al., 2011). Therefore, disrupting the infection process via HIGS of these genes may result in the weakening or loss of pathogenicity, leading to broad-spectrum rice blast resistance.
In this study, we demonstrated that silencing of the M. oryzae transcription factor MoAP1 by RNAi in vitro led to compromised M. oryzae virulence and by HIGS in transgenic rice lines led to improved blast disease resistance. Our findings may be relevant for generating new rice lines exhibiting a HIGS-based broad-spectrum resistance to rice blast disease.
Materials and Methods
Plant Material and Rice Blast Fungus
The indica rice accession Kasalath was used for transgenic analyses. The M. oryzae strain Guy11 was used as a reference strain. The GFP-tagged strain GZ8 was used in observation of the infection process. Eleven strains (Supplementary Table S1) stored in the Plant Pathology Laboratory (the Rice Research Institute, Sichuan Agricultural University, China) were used for disease assay.
asiRNA Design and Synthesis
The genomic sequences for MoAP1 (MGG_12814), MoSSADH (MGG_01230), MoACT (MGG_15157), and MoSOM1 (MGG_04708) were download from the M. oryzae database1. Nineteen-nucleotide sequences in three different sites specific to each genes were selected as the asiRNAs candidates. The specificity of the sequences was confirmed by BLASTn against the M. oryzae genomic sequences to avoid off-targeting. The complementary sequences of these candidates were then designed as double strand artificial small interference RNAs (asiRNAs) (Supplementary Table S2). Double T nucleotides were added to the 3′-terminus to stabilize the asiRNAs. Then, the TT-overhang double strand twenty-one-nucleotide sequences were synthesized by Shanghai Genepharma. Co., LTD. (China).
In vitro Culture of M. oryzae With asiRNA
Artificial siRNAs were added into solid CM medium (Choi et al., 2015) with different concentrations (i.e., 0, 50, 100, or 200 nM). Two microliter (μl) of M. oryzae spore suspension (1 × 105 spores /ml) was inoculated on the medium and incubated at 28°C with 12-h light/ 12-h darkness. The development of spores and hypha was monitored with a microscope and the aerial hyphae growth rate (AHGR) was calculated as percentage of the amount of healthy aerial hyphae divided by the sum of healthy aerial hyphae and abnormal aerial hyphae at 1 dpi in three biological replicates. Each replicate contained at least 30 spores inoculated. After sporulation, the conidia deformity rate was calculated as percentage of the amount of healthy conidia divided by the sum of healthy conidia and abnormal conidia at 7 dpi in three biological replicates. Each replicate contained at least 30 conidia inoculated.
Pathogenicity Assay for the Rice Blast Fungus Treated With asiRNA
The M. oryzae strains Guy11 were cultured on CM solid medium containing asiR1115 for 14 days at 28°C with 12-h light/ 12-h darkness. Then the spores were collected and 5 μL of spore suspension (1 × 105 spores/ml) were punch-inoculated on the third leaf of TaiPei309 (TP309) from three-leaf stage seedlings. The disease phenotypes on the leaves were observed at 7 dpi. Images were taken and the lesion area was calculated as the product of the length and width of the lesions.
qRT-PCR Assay
Three-leaves-stage transgenic rice plants were inoculated with asiR1115-treated (50 and 100 nM) M. oryzae spore suspension (1 × 105 spores/ml). After 7 dpi, disease lesions were recorded before sampling fore RNA extraction. Total RNA was extracted from the lesions using Trizol reagent (Invitrogen). The first-strand cDNA was synthesized using the Super-Script First-strand Synthesis System (Invitrogen). A qRT-PCR assay was conducted using the SYBR Green Mix (TaKaRa) to measure relative transcript abundance. MoPot 2 expression was normalized against that of OsActin1 to be the internal control (Chandran et al., 2019). The qRT-PCR primers are listed in Supplementary Table S3.
In vitro asiRNA-Spraying Assay
Four- to six-leaf stage TP309 seedlings were sprayed with 50 or 100 nM asiR1115 in 0.1% tween 20. Twelve hours after spraying, M. oryzae strain Guy11 was inoculated by punch-inoculation. The disease phenotypes on the inoculated leaves were observed at 7 dpi. Images were taken and the lesion area was measured as the product of the length and width of the lesion points.
Construction of RNAi Vectors and Genetic Transformation
Three MoAP1 exon fragments of 242, 255, and 247 bp containing asiR1115, asiR1245, and asiR1362 sequences, respectively, were amplified from M. oryzae mycelia DNA with MoAP1-specific primers (Supplementary Table S4). The PCR fragments were cloned into the pCR8-T plasmid [(Cat No.:K2500-20, Invitrogen), and then inserted into the RNAi vector pANDA using the recombination Gateway system (Cat No.:11791020, Invitrogen; Miki and Shimamoto, 2004) leading to constructs pANDA-MoAP1-1115 (1115i), pANDA-MoAP1-1245 (1245i), and pANDA-MoAP1-1362 (1362i)]. The constructs were introduced into rice accession Kasalath via Agrobacterium-mediated transformation following a previous report ( Li et al., 2017). Briefly, the pANDA-MoAP1-RNAi plasmids (i.e., 1115i, 1245i, and 1362i) were transformed into Agrobacterium strain EHA105, and then used to infect rice callus from the mature embryo of Kasalath. Positive transformants were screened by means of hygromycin resistance analysis.
Agrobacterium-Mediated Transient Expression Assay in Nicotiana benthamiana
YFP detection was assayed as previously reported (Li et al., 2017). We fused YFP with MoAP1 at its C-terminus (35S:MoAP1-YFP) and inserted into KpnI-SpeI sites of binary vector 35S-pCAMBIA1300. Then the construct was transformed into Agrobacterium strain GV3101 for agro-infiltration-mediated transient expression assay in N. benthamiana. In brief, Agrobacterium strain GV3101 harboring MoAP1-YFP constructs and Agrobacterium strain EHA105 harboring the indicated constructs (empty vector pANDA, 1115i, 1245i, and 1362i) were incubated at 28°C overnight in LB media containing kanamycin (50 mg/ mL) and carbenicillin (50 mg/ mL) on a table shaking at 250 rpm. The bacteria were collected by centrifuge at 800 × g for 5 min and resuspended in an MMA buffer (10 mM 4-Morpholineethanesulfonic acid hydrate, 10 mM MgCl2, 100 mM Acetosyringone). The indicated Agrobacteria were infiltrated into leaves of N. benthamiana. Leaves were examined at 48 h post infiltration (hpi) for image acquisition using a NikonA1 Confocal Laser Scanning Microscope (Nikon Instruments, Inc., China).
Infection Process Assay
For observing the infection process of M. oryzae, we inoculated the GFP-tagged strain GZ8 on 4-cm-long leaf sheaths as described (Kankanala et al., 2007). The inoculated epidermal layer was excised and analyzed by fluorescence microscopy (Zeiss Axio Imager A2) at 36 hpi.
Results
The Artificial siRNAs Targeting MoAP1 Effectively Suppressed the Growth of Aerial Hyphae in vitro
To screen a small RNA that could be used in HIGS transgenic analysis, we first tested whether the in vitro treatment of asiRNAs affects M. oryzae growth in an axenic culture. To this end, we designed and synthesized asiRNAs targeting M. oryzae transcription factor genes MoAP1, MoSSADH, MoACT, and MoSOM1, respectively (Supplementary Table S2). The sites targeted by asiRNAs in each of the 4 genes are shown in Figure 1A and the selected sequences for asiRNAs were BLASTn against the with M. oryzae genome sequence to make sure the asiRNAs were specific. Microscopic analyses revealed that increasing concentrations of the asiRNAs targeting MoAP1 inhibited the growth of M. oryzae aerial hyphae and resulted in abnormal hyphae consisting of degraded cytoplasm and lackng melanin. Abnormal aerial hyphae were not detected in the control M. oryzae sample (Figure 1B). By contrast, the samples treated with asiRNAs targeting the MoSSADH, MoACT, and MoSOM1 did not show obvious abnormal growth or development (Figures 1C–E). The AHGR was analyzed to quantify the inhibitory effects of asiRNAs. The AHGR of MoAP1-silenced M. oryzae was significantly decreased upon 50 nM asiRNA treatment, and further decreased when asiRNA concentrations were increased. Among the three asiRNAs targeting MoAP1, asiR1115 displayed the most inhibition on fungal aerial hyphae growth, followed by asiR1362 and asiR1245 (Figures 1B, 2A). In contrast, the asiRNAs targeting MoACT, MoSSADH, and MoSOM1 did not affect M. oryzae aerial hyphae growth (Figures 1C–E, 2B–D).
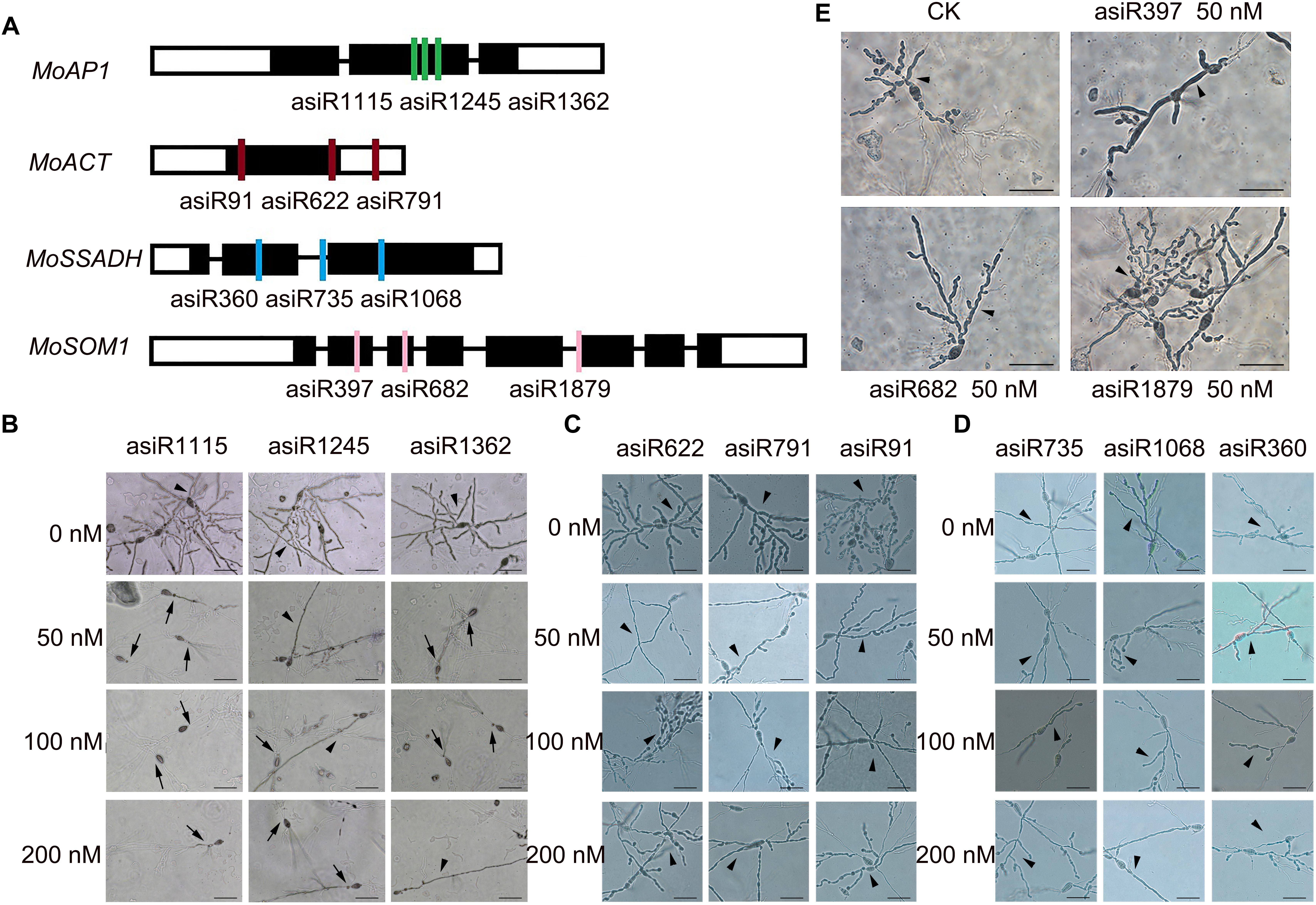
Figure 1. Growth of aerial hyphae of rice blast fungus on media containing artificial siRNAs (asiRNAs). (A) Diagrams show the target sites of the indicated artificial small interference RNAs. Black boxes denote exons. (B) The indicated asiRNAs targeting MoAP1 inhibited fungal aerial hyphae growth. The culture medium was supplemented with the indicated asiRNAs targeting MoAP1 at the indicated concentration. Control (CK) samples with 0 nM asiRNAs. Note that abnormal hyphae consisted of degraded hyphal cytoplasm and lacked melanin (arrows). Normal hyphae exhibited robust growth and had relatively high cytoplasmic melanin levels in the CK media (arrowheads). (C–E) The indicated asiRNAs targeting MoACT (C), MoSSADH (D), and MoSOM1 (E) did not affect Magnaporthe oryzae aerial hyphae growth. Images were taken at 1 day post conidial culture. Bars = 50 μm. Similar results were obtained in at least three independent experiments.
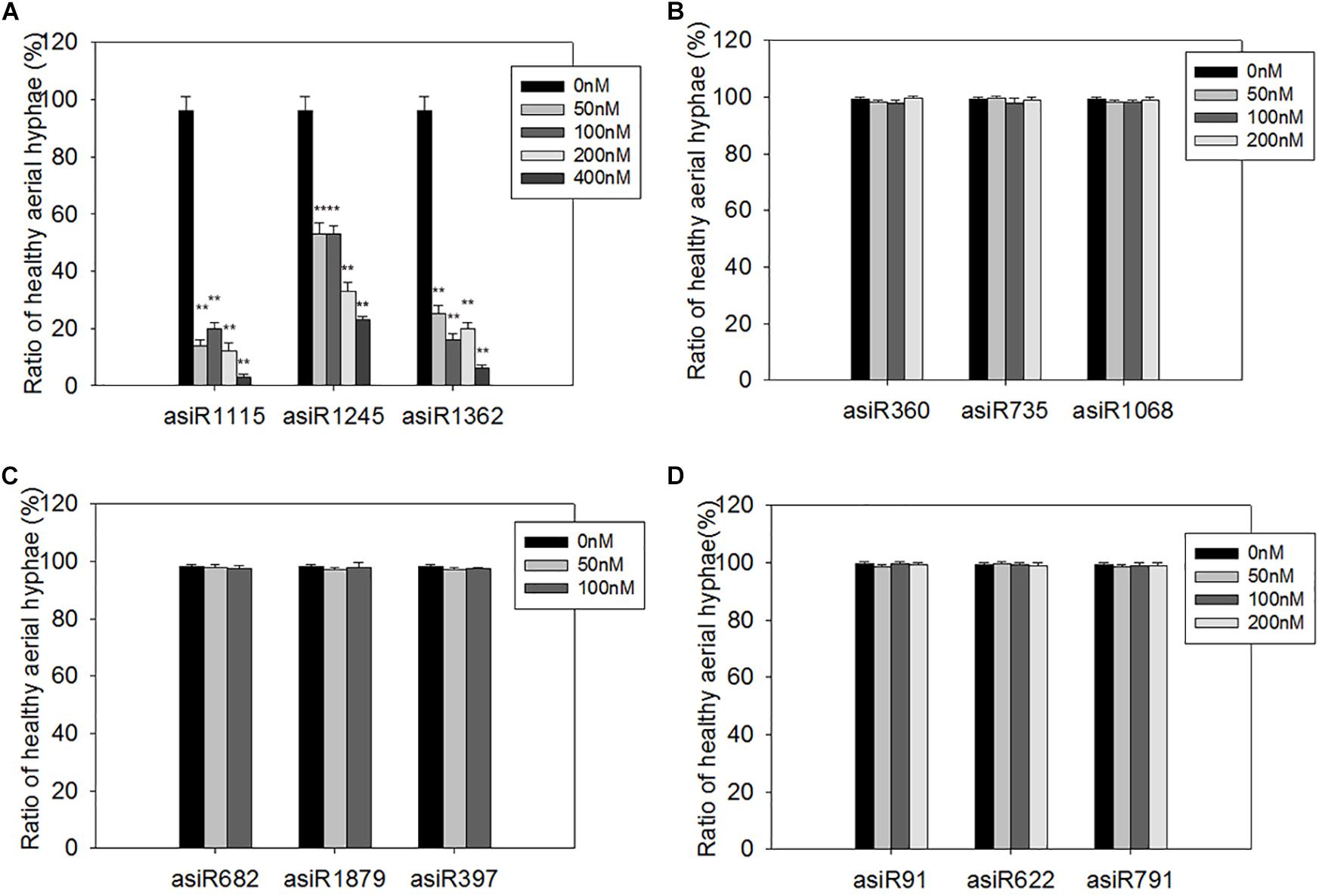
Figure 2. Quantification of healthy aerial hyphae on media containing different asiRNAs. (A–D) Quantification of healthy aerial hyphae ratio on the media containing the indicated asiRNAs targeting MoAP1 (A), MoSSADH (B), MoSOM1 (C), and MoACT (D). Note that the ratio of healthy aerial hyphae was substantially declined in the media containing asiRNAs targeting MoAP1 (A). While the indicated asiRNAs targeting MoSSADH (B), MoSOM1 (C), and MoACT (D) had no inhibitory effect on fungal aerial hyphae growth. Values were means ± SD from three replicates, in each replicate, more than 30 spores of hyphae were evaluated at 1 day post conidial culture. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 between the indicated treatment and control as determined by student’s t-test. All the experiments were repeated at least three times with similar results.
These results indicated that the asiRNAs targeting MoAP1 effectively suppressed the growth of aerial hyphae, which is consistent with the function of MoAP1 reported previously (Guo et al., 2011).
asiRNA Treatment Results in Conidial Deformity
To further test whether development of M. oryzae conidia was affected by asiRNAs, we checked the fungal conidia treated with the asiRNAs at 7 days post conidial culture. Microscopic analyses revealed that the in vitro treatment with asiRNA targeting MoAP1 resulted in malformed conidia, including elongated, and spindly morphology (Figures 3A,B). These results indicates that the three asiRNAs targeting MoAP1 influence the conidial development.
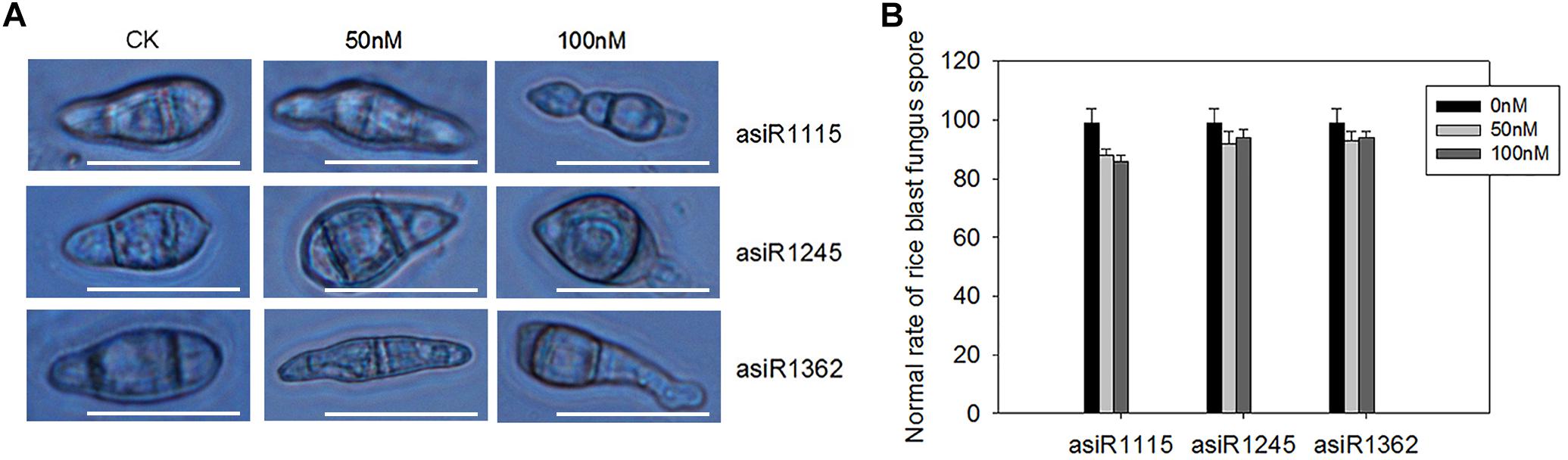
Figure 3. Abnormal conidial morphology formed on media containing asiRNA targeting MoAP1. (A) The abnormal conidial morphology with the indicated asiRNA targeting MoAP1 at the indicated concentration. Bars = 25 μm. (B) Quantitative analysis on ratio of normal fungal conidia treated with asiRNAs. Values were means ± SD from three replicates. In each replicate, more than 30 conidia were evaluated at 7 days post conidial culture. All the experiments were repeated at least three times with similar results.
The asiRNA Targeting MoAP1 Affects Fungal Colony Morphology
Magnaporthe oryzae colony morphology was affected by different concentrations of asiR1115. Treatments with the asiRNA targeting MoAP1 resulted in significantly decreased growth of aerial hyphae (arrows), and an obviously delayed development of outer circle of the fungal colony, which differed from that of the controls (Figure 4B). This result indicated that the asiRNAs targeting MoAP1 effectively suppressed the growth of aerial hyphae leading to change in colony morphology. However, the asiRNAs targeting MoACT, MoSSADH, and MoSOM1 did not affect fungal colony morphology (Figures 4C,D).
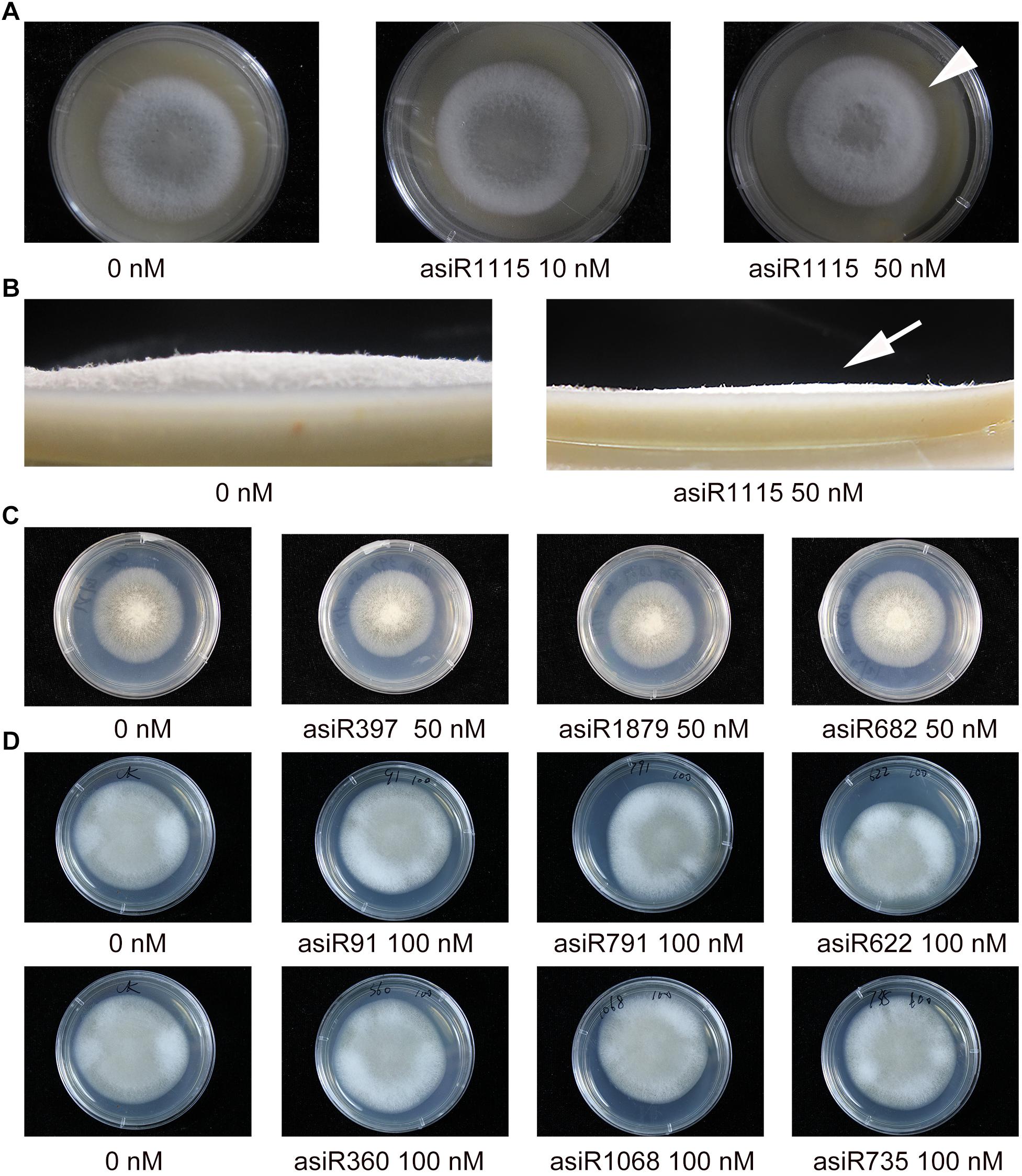
Figure 4. The effect of asiRNAs targeting MoAP1 on fungal colony morphology. (A) The colony morphology of M. oryzae growing on media containing the indicated asiRNA targeting MoAP1. Note that the asiRNA treatments resulted in short and sparse aerial hyphae, with an obviously delayed development of outer circle of the fungal colony (arrowhead), which differed from that of the control. (B) Side view of the colony morphology of M. oryzae growing on media containing the indicated asiRNA targeting MoAP1. It showed decreased growth of aerial hyphae on media containing 50 nM of asiR1115 targeting MoAP1 in comparison with that of 0 nM (arrow). (C,D) Colony morphology in media supplemented with different concentrations of asiRNAs targeting MoSOM1, MoACT and MoSSADH, respectively. Note that these asiRNAs did not affect the colony morphology. Images were taken at 7 days post mycelia colony culture. Similar results were obtained in three independent experiments.
In vitro Silencing of MoAP1 by asiRNAs Resulted in Lower Virulence of M. oryzae
Because asiRNA targeting MoAP1 influenced the growth and development of M. oryzae, we asked the question whether the affected fungal conidia were virulent to rice. To this end, we inoculated susceptible rice cultivar TP309 plants with spores of the M. oryzae strain Guy11 cultured on the media containing asiRNAs targeting MoAP1. Intriguingly, all three asiRNAs-treated M. oryzae Guy11 resulted in compromised symptom with smaller lesion area (Figures 5A,B). Furthermore, asiR1115-treated Guy11 developed the slightest symptom and smallest lesion area (Figures 5A,B). These data indicated that all three asiRNAs targeting MoAP1 effectively compromised the virulence of the rice blast fungus. These results were consistent with that loss-of function in MoAP1 led to loss virulence of the fungus (Guo et al., 2011).
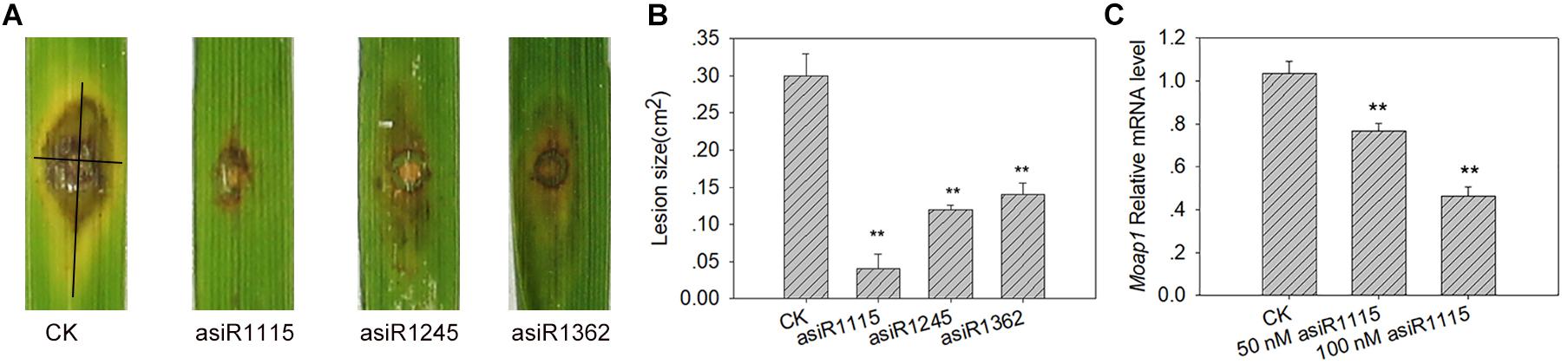
Figure 5. Silence of MoAP1 by asiRNAs resulted lower virulence of M. oryzae. (A) Disease phenotypes of TP309 at 7 days post-inoculation (dpi) by asiRNA-treated M. oryzae strains Guy11. Rice cultivar TP309 was punch-inoculated with Guy11 (1 × 105 spores/ml) cultured on media containing the indicated asiRNAs targeting MoAP1. (B) Quantitative analysis on lesion area at 7 dpi. The length and width of the symptom were measured, and the product of the length and width were designed as the area of the symptom. Black lines in lesion (A) were used to represent length and width. Values were means ± standard deviations (SD) of 15 disease spots. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 as determined by student’s t-test analysis. (C) The relative mRNA level of MoAP1 in the mycelium from the lesions of (A) generated by asiR1115-treated Guy11 and control. The expression of MoAP1 was determined by qRT-PCR. The relative MoAP1 mRNA levels were measured by using the mRNA level of M. oryzae MoAP1. Values are means ± standard deviations (SD) of three replicates. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 as determined by student’s t-test. All experiments repeated three times with similar results.
Next, we examined the expression of MoAP1 in the mycelium developed on the leaf lesions which were inoculated with Guy11 cultured on the media containing asiR1115 targeting MoAP1. The qRT-PCR results indicated that MoAP1 expression was significantly lower in the lesions generated by asiR1115-treated Guy11 than in the control lesions (Figure 5C), indicating that asiR1115 effectively suppressed MoAP1 expression in vitro and such suppression could maintain in the subsequent fungal mass grown in rice leaves.
AsiRNA-Treated Leaves Shows Increased Resistance to Rice Blast Fungus
To test whether asiRNAs can be used on rice to improve rice resistance against M. oryzae, we pre-treated rice leaves with 50 or 100 nM asiR1115. Then, we inoculated the pre-treated leaves with Guy11 by punch-inoculation (Li et al., 2017). The control plants were pre-treated with water. After 7 dpi, the lesion area were measured. Our results showed that the asiR1115-pretreated leaves formed lesions much smaller than the control leaves (Figures 6A,B), indicating that asiRNA may be used as an in vitro treatment to enhance rice blast resistance.
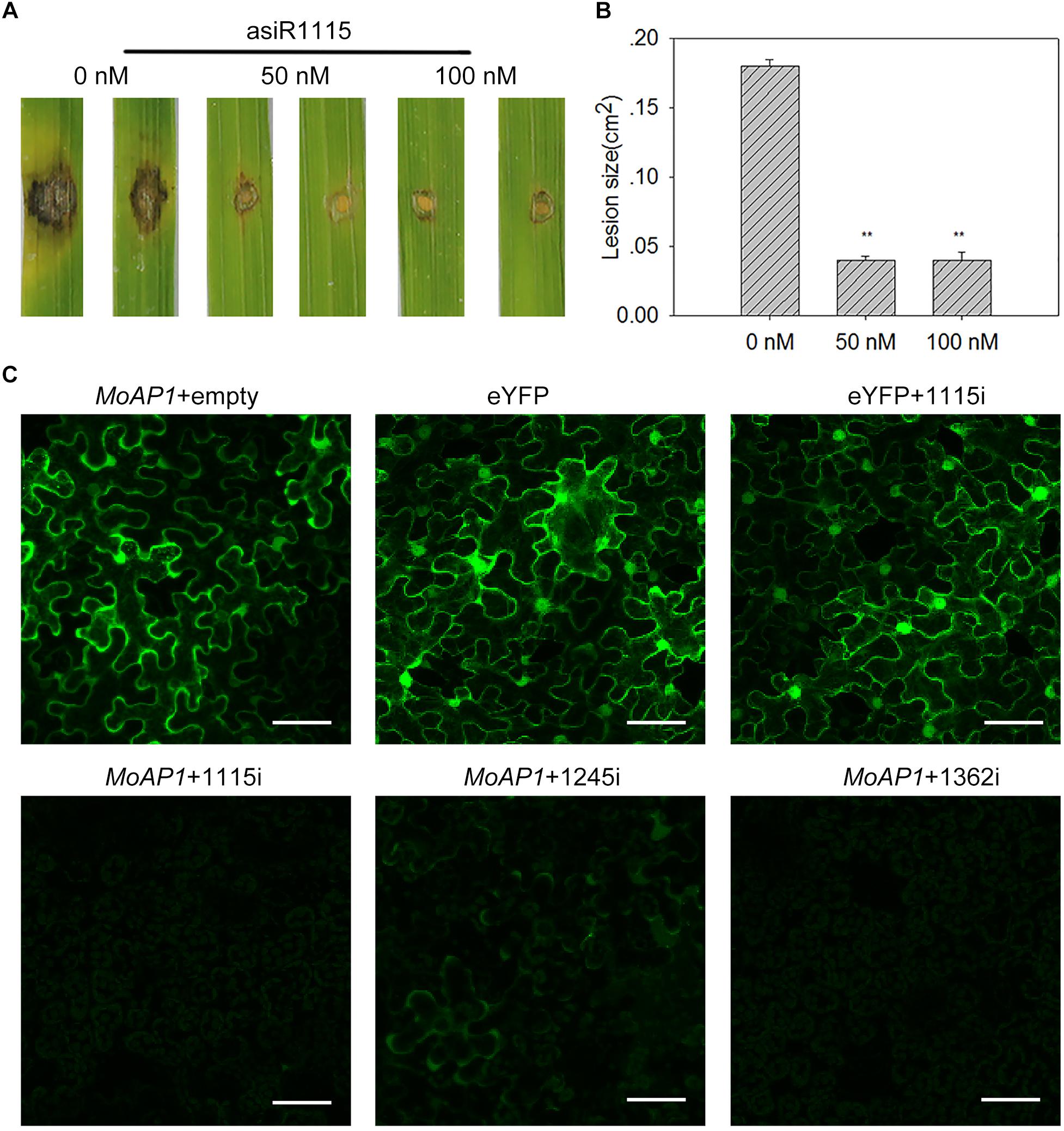
Figure 6. Artificial siRNAs targeting MoAP1 inhibits the expression of MoAP1 in vivo and compromises M. oryzae virulence. (A) Disease phenotypes of TP309 leaves pre-treated with the indicated concentration of asiRNA1115. TP309 third leaves of seedlings were smeared by a brush soaked in 50 or 100 nM asiR1115 solution before inoculation with 5 μL of spore suspension (5 × 105 spores/ml) of M. oryzae strain Guy11. Disease phenotype was recorded at 7 dpi (B) Quantitative analysis of lesion area at 7 dpi. The length and width of the symptom were measured, and the product of the length, and width were designed as the area of the symptom. Values are means ± standard deviations (SD) of 15 disease spots. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 as determined by student’s t-test. This experiment was repeated three times with similar results. (C) Confocal images show the inhibition of asiRNAs on the expression of MoAP1-eYFP in N. benthamiana. Transient expression of 35:MoAP1-eYFP (MoAP1) plasmid with an empty vector (no asiRNA) in tobacco as positive control (Supplementary Figure S2). 35:MoAP1-eYFP co-expressed with the asiRNAs (Supplementary Figure S1) as the test group. The eYFP and eYFP co-expressed with asiRNA1115 as the negative control. The images were taken at 2 days post infiltration. Bars = 50 μm. All experiments were repeated three times with similar results.
In vivo Suppression of MoAP1 Gene Expression by asiRNAs in a Heterologous System
The in vitro data indicated that asiRNA-treatment could suppress MoAP1 expression to decrease M. oryzae virulence. We then tested whether asiRNAs could silence MoAP1 in planta. To this end, we made constructs expressing eYFP fused with the MoAP1 at its 5′-terminus (35S:MoAP1-eYFP) (Supplementary Figure S1B) and constructs expressing asiRNAs containing asiRNA1115, asiRNA1245, and asiRNA1362, respectively (Supplementary Figure S1A). Then, 35S:MoAP1-eYFP was transiently expressed in N. benthamiana alone or together with asiRNA1115, asiRNA1245, or asiRNA1362. The YFP intensity expressed from 35S:MoAP1-eYFP was obviously lower when co-expressed with either one of the asiRNAs than 35S:MoAP1-eYFP alone (Figure 6C). In contrast, the YFP intensity expressed from 35S:eYFP was unchanged when co-expressed with asiRNA1115 (Figure 6C), indicating the tested three asiRNAs could suppress MoAP1 expression in vivo.
HIGS of MoAP1 in Transgenic Rice Results in Compromised Susceptibility
Next, we tried to make rice transgenic lines overexpressing the three asiRNAs and successfully got transgenic lines containing asiR1362 (MoAP1-1362, Supplementary Figure S2). Then, these transgenic lines were subjected to disease assay. Positive T1 transgenic plants were punch-inoculated with M. oryzae strain Guy11. Disease phenotype was recorded at 7 dpi. As our expectation, the transgenic lines displayed infection area obviously smaller than the control plants (Figures 7A,B). Then, we examined the expression of MoAP1 in the fungal mass collected at the infection area. Consistently with the smaller infection area, MoAP1 expression was significantly decreased in the fungal mass collected from transgenic lines (Figure 7C), indicating HIGS of MoAP1. These results indicated that HIGS of MoAP1 could improve rice resistance against M.oryzae.
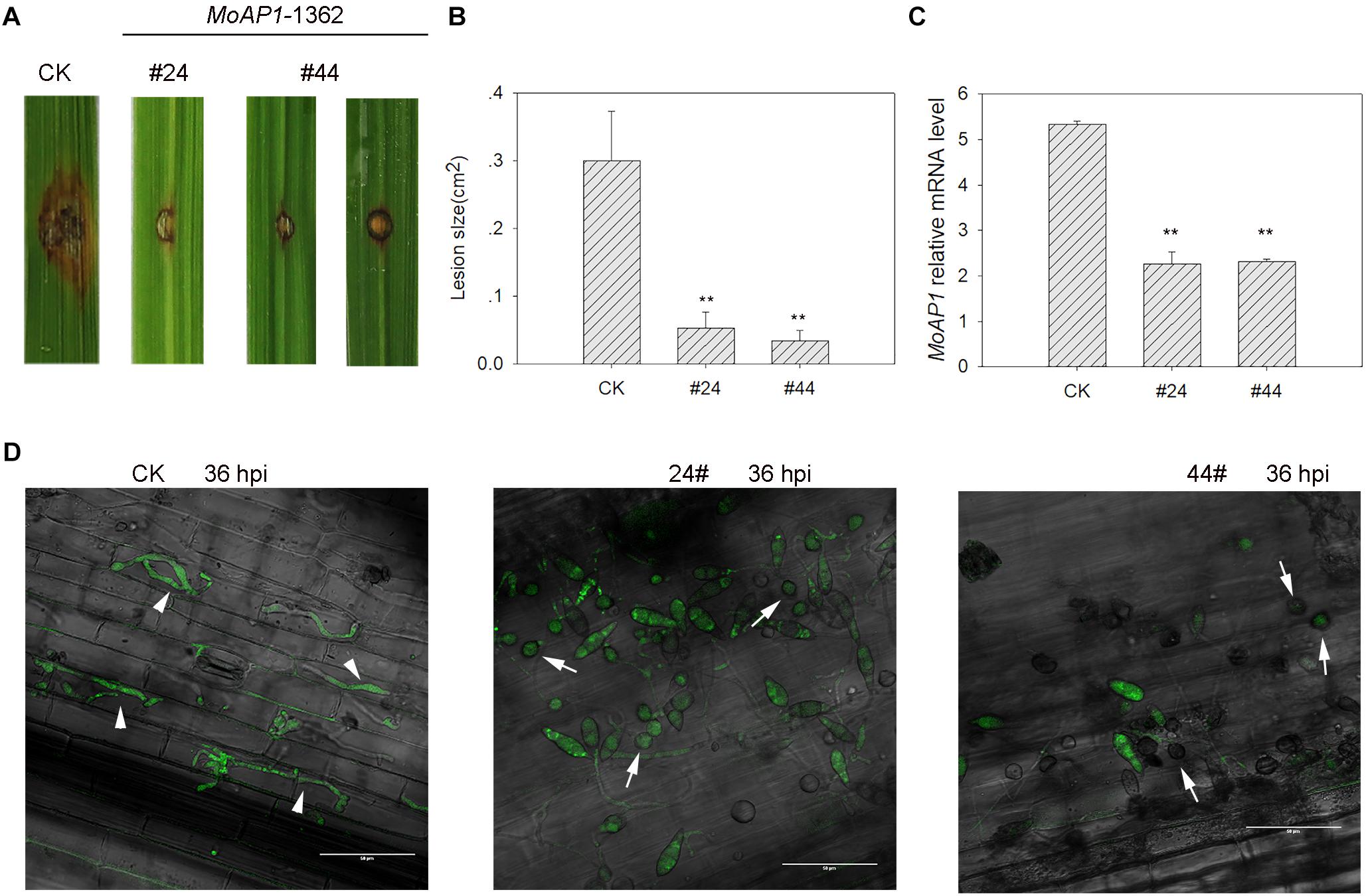
Figure 7. Analyses of the resistance of MoAP1-RNAi transgenic plants to the rice blast fungus and the MoAP1 expression in inoculated T1 transgenic plants. (A) Blast disease assay on the indicated T1 transgenic lines. The control plants (CK) and MoAP1-1362 T1 transgenic lines were punch-inoculated with 5 μL of spore suspension (5 × 105 spores mL−1) of Magnaporthe oryzae Guy11. Images were taken at 7 dpi. CK: the rice cultivar Kasalath expressing empty vector; #24 and #44: MoAP1-1362 T1 generation transgenic lines. (B) Quantitative analysis of lesion area at 7 dpi. The length and width of the lesions were measured, and the product of the length and width was designed as the lesion size. Values are means ± standard deviations (SD) of 15 disease spots. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 between the indicated lines and control as determined by student’s t-test. This experiment was repeated three times with similar results. (C) qRT-PCR showing that MoAP1 expression in MoAP1-1362 T1 generation transgenic lines in comparison with control at 7 dpi. Asterisks (∗∗) above the bars indicate significant differences at P < 0.01 between the indicated lines and control as determined by student’s t-test. (D) Confocal images show the infection status of M. oryzae GZ8 in the indicated lines at 36 hpi. Note that appressoria (arrows) were formed and delayed to 36 hpi on MoAP1-1362 transgenic plants, by contrast, the invasive hyphae (arrowheads) were formed and extended to the neighboring cell on empty vector plants at 36 hpi. Bars = 20 μm. All experiments repeated three times with similar results.
Cellular Responses of Transgenic Rice Lines to M. oryzae
To understand how HIGS of MoAP1 leading to enhanced blast resistance, we examined the infection process of the GFP-tagged strain GZ8 by laser scanning confocal microscopy. To this end, GZ8 was inoculated on sheath of MoAP1-1362 transgenic plants and control plants. Then, the infection stage was recorded under fluorescence microscopy. At 36 hpi, the invasive hyphae were formed and extended to the neighboring cells on control plants. By contrast, the conidia on the transgenic lines mostly formed long germ tubes outside the host cells and stayed at appressoria-forming stage (Figure 7D), indicating failure in penetration.
HIGS of MoAP1 Coffers Broad-Spectrum Resistance to M. oryzae
Next, we asked whether the MoAP1-1362 transgenic plants also were resistant to other M. oryzae isolates. To address this question, we inoculated MoAP1-1362 transgenic lines with 11 M. oryzae strains derived from rice fields in Ya’an, Sichuan Province, China (Supplementary Table S1). We found that the transgenic lines displayed increased resistance or compromised susceptibility to all 11 tested M. oryzae strains (Figure 8 and Supplementary Table S1), indicating that the in vivo expression of siRNAs triggered HIGS of MoAP1 leading to increased rice resistance against rice blast disease.
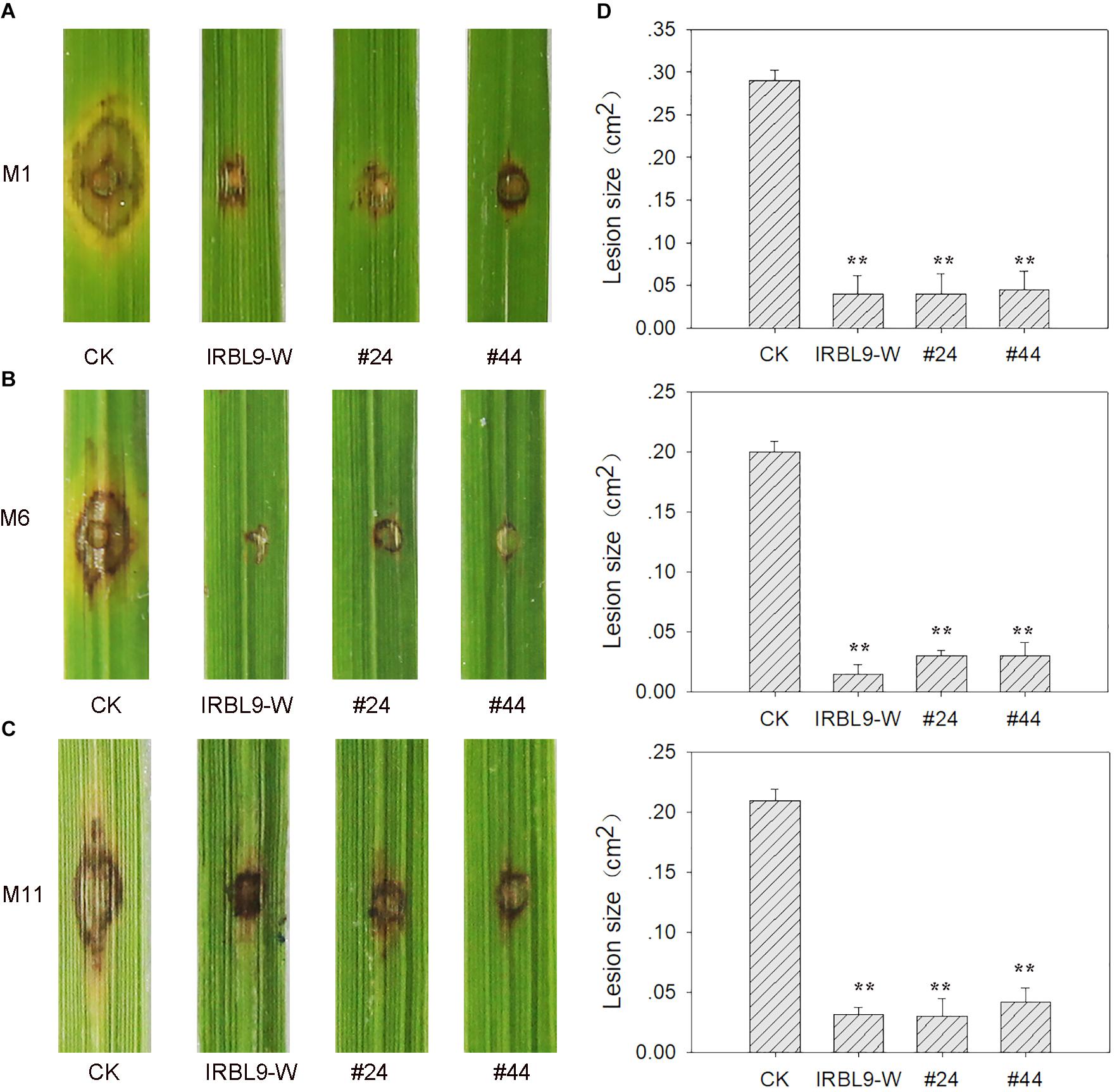
Figure 8. Disease resistance spectrum of transgenic plants. (A–C) Disease phenotype of the indicated lines punch-inoculated with M. oryzae strains M1 (Tepep) (A), M6 (Zhong1) (B), and M11 (089) (C). Two T3 lines (i.e., MoAP1-1362 transgenic lines #44 and #24) were punch-inoculated with 5 μL of spore suspension (5 × 105 spores mL−1) of the indicated M. oryzae strains. CK, the rice cultivar Kasalath expressing empty vector. IRBL9-W, a monogenetic rice line containing the blast resistance gene Pi9. Images were taken at 7 dpi. (D) Quantitative analysis of lesion area at 7 dpi. The length and width of the lesions were measured, and the product of the length and width was designed as the lesion size. Values are means ± standard deviations (SD) of 15 disease spots. Asterisks (∗∗) above the bars indicate significant differences between the indicated lines and control at P < 0.01 as determined by student’s t-test. All experiments repeated three times with similar results.
Discussion
Recent studies have shown that HIGS technology was a great strategy for protecting rice plants against the pathogenic fungi M. oryzae, but they didn’t get the stable resistent transgenic rice lines (Zhu et al., 2017). In this study, we demonstrated that HIGS of M. oryzae transcription factor gene MoAP1 is efficient for controlling the infection of rice blast fungus. Transcription factor genes, such as MoAP1, MoSSADH, MoACT, and MoSOM1 in the rice blast fungus, are important regulators of mycelial development, sporulation, and pathogenicity. we designed three asiRNAs for each gene and tested their role on fungal growth by adding them in the medium. The three asiRNAs (i.e., asiR1245, asiR1362, and asiR1115) targeting MoAP1 all resulted in abnormal aerial hyphae, deformed spores, and decreased virulence with different inhibitory effects (Figures 1–5). AsiR1115 had the greatest inhibitory effect in vitro, with a minimal effective concentration of 50 nM (Figure 6). Therefore, artificial siRNA in vitro assay could serve as a quick prescreening to identify efficient HIGS target for stable transformation. Interestingly, the persistence of the silencing effect was found in inoculum grown on medium with MoAP1 asiRNA. The three asiRNAs-treated M. oryzae Guy11 resulted in compromised symptom with smaller lesion area on rice. The possible explanation is that, the fungus cultured on medium may absorb asiRNA from the medium, which resulted in the suppressed hyphae growth and malformed conidia, including elongated, and spindly morphology, indicating that the silencing effect is hereditary and persistence. Next, we focused on asiRNAs targeting MoAP1 to verify whether HIGS could provide resistance against the rice blast fungus in vivo. We constructed transgenic lines expressing siRNAs that targeted MoAP1 based on HIGS technology. These transgenic lines exhibited enhanced resistance or compromised susceptibility when different M. oryzae strains were inoculated (Figures 7, 8). During infection, the fungus may be able to take up siRNAs from the plant cells, resulting in decreased MoAP1 gene expression via an RNAi mechanism. As a result, the virulence of the fungus may be compromised due to the lower expression of MoAP1.
The HIGS system represents a potentially powerful reverse genetics tool for analyses of fungal gene functions. The effect of HIGS may differ on different genes and on different target sites of one gene. Here we found that asiRNAs targeting MoAP1 were effective in vitro and HIGS of MoAP1 worked in vivo (Figures 5–8). Nevertheless, the three asiRNAs targeted at different sites of MoAP1 and the site on the most up-stream of MoAP1 seems the best in silencing of MoAP1 (Figure 2). However, the asiRNAs targeting the other three genes (i.e., MoSSADH, MoACT, and MoSOM1) did not result in any in vitro phenotypes (Figures 1, 2). Further investigation is needed to make out whether the asiRNA sites we selected were not work or silencing these genes did not lead to any obvious phenotypes. In fact, some genes are not suitable for HIGS because knocking-down there expression may not have effect. For example, Yin et al. (2011) chose twelve pathogenic-related genes of P. striiformis f. sp. tritici or P. graminis f. sp. tritici to determine whether silencing signals can be delivered to the pathogen and suppress expression of the fungal genes by HIGS. The results indicated that some candidate genes are not suitable as targets for HIGS. Govindarajulu et al. (2015) verified this conclusion. HIGS efficiency of different target genes may be related to the function of target genes and their expression patterns. Selection of certain pathogenic genes transcribed highly in the infectious hyphae may be beneficial to control rice blast disease effectively (Zhu et al., 2017). In addition, the size, sequence specificity, and position of the dsRNA are also important in considerations for HIGS (Thomas et al., 2001; Holen et al., 2002; Burch-Smith et al., 2004). The higher the homology between siRNA and the target mRNA, the better the effect of gene silencing. several siRNAs synthesized against different sites on the same target mRNA (human Tissue Factor) demonstrated striking differences in silencing efficiency. And previous studies have indicated that, if the gene fragment selection was not suitable, it may cause off-target silencing, so that no effective silent phenotype can be obtained (Xu et al., 2006; Senthil-Kumar and Mysore, 2011), and several siRNAs synthesized against different sites on the same target mRNA demonstrated striking differences in silencing efficiency (Holen et al., 2002). To achieve a better silencing effect, the length of the gene fragment inserted into the carrier should be limited to 200–350 bp (Burch-Smith et al., 2006), and using a hairpin RNA expression pattern will significant increase efficiency of gene silencing (Pflieger et al., 2008; Zhu et al., 2017). And some strategies are feasible to exploit HIGS for durable resistance. (i) using a silencing construct that targets entire gene families, (ii) multiple lines deployed in rotation, (iii) a HIGS line conferring resistance to single or multiple fungal pathogens or (iv) a combination of both classical R genes and HIGS (Nunes and Dean, 2012).
We observed phenotypes on silencing of MoAP1. The hyphae can grow and appressoria can form on the surface of transgenic plant leaves (Figure 7D), but are strongly inhibited in vitro (Figure 1B). We think the possible reason is that in vitro experiments, the fungus cultured on medium may absorb asiRNA from the medium, and as a result, the growth of the fungus is suppressed. In the transgenic lines expressing the siRNAs, the fungus can temporarily grow forming germ tubes and appressoria before penetration but could not invads into the rice cells (Figure 7). It is possibly that before the fungus invaded into the rice cells, the nutrients required for aerial hyphae and appressorium growth is provided by the spores. As a result, the hyphae and appressorium develop well. When the appressorium begins to invade the rice cells, the siRNAs generated from the hairpin constructs in the transgenic rice cells suppress MoAP1 expression, leading to suppressed or delayed invasion. This conjecture needs to be further verified.
The asiRNAs targeting MoAP1 may be used as an in vitro reagent to enhance rice blast resistance. This is consistent with the effects of asiRNAs in the other pathogens. For example, Koch et al. (2016) showed that the growth of F. graminearum was severely inhibited after spraying of target long dsRNAs on the surface of barley leaves. Wang et al. (2016) also showed that significant inhibition of gray mold disease could be obtained by applying asiRNAs or dsRNAs targeting Botrytis DCL1 and DCL2 genes on the surface of fruits, vegetables and flowers. Consequently, given the ease of design and applicability to diverse pathogens, the use of target-specific asiRNA as an anti-fungal agent offers unprecedented potential as a new plant protection strategy.
Taken together, we obtained a target gene and a target site that can be used in HIGS. Thus, this work may be relevant for the development of a strategy to engineer broad-spectrum blast-disease resistance in rice.
Author Contributions
W-MW and HX have designed the research. X-YG performed the research with help from J-QZ. X-YG, YL, JF, and F-XX analyzed the experimental results. Y-FW and X-LC assisted with the inoculation test. J-QZ, JS, and YS assisted with the rice transgenic work. X-YG and W-MW wrote the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31430072 and 31672090 to W-MW), Sichuan Academy of Agricultural Sciences Youth Fund (2015QNJJ-018) awarded to X-YG, and The National Key R&D Program of China (2017YFD0300400).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Drs. Shunyuan Xiao, Yinong Yang, and Xian-Feng Ma for their helpful discussions and suggestions. We also thank Rong Xie for providing rice blast fungus strains.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00433/full#supplementary-material
Footnotes
References
Balhadere, P. V., and Talbot, N. (2001). PDE1 a P-ATPase involved in appressorium- mediated plant infection by the rice blast fungus Magnaporthe grisea. Plant Cell 13, 1987–2004. doi: 10.1105/tpc.13.9.1987
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., and Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313x.2004.02158.x
Burch-Smith, T. M., Schiff, M., Liu, Y., and Dinesh-Kumar, S. P. (2006). Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 142, 21–27. doi: 10.1104/pp.106.084624
Chandran, V., Wang, H., Gao, F., Cao, X. L., Chen, Y. P., Li, G. B., et al. (2019). miR396-OsGRFs module balances growth and rice blast disease-resistance. Front. Plant Sci. 9:9999. doi: 10.3389/fpls.2018.01999
Chen, W. X., Christine, K., Nowara, D., Oliveira, G. E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
Chen, X., Shang, J., Chen, D., Lei, C., Zou, Y., Zhai, W., et al. (2006). A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 46, 794–804. doi: 10.1111/j.1365-313x.2006.02739.x
Cheng, W., Song, X. S., Li, H. P., Cao, L. H., Sun, K., Qiu, X. L., et al. (2015). Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 13, 1335–1345. doi: 10.1111/pbi.12352
Choi, J., Chung, H., Lee, G. W., Koh, S. K., Chae, S. K., and Lee, Y. H. (2015). Genome-wide analysis of hypoxia-responsive genes in the rice blast fungus, Magnaporthe oryzae. PLoS One 10:e0134939. doi: 10.1371/journal.pone.0134939
Clergeot, P. H. G., Ourgues, M., Cots, J., and Lebrun, M. (2001). PLS1,a gene encoding a tetraspanin-like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea. PNAS 98, 6963–6968. doi: 10.1073/pnas.111132998
Dagdas, Y. F., Yoshino, K., Dagdas, G., Ryder, L. S., Bielska, E., Steinberg, G., et al. (2012). Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336, 1590–1595. doi: 10.1126/science.1222934
Deng, Y. W., Zhai, K. R., Xie, Z., Yang, D. Y., Zhu, X. D., Liu, J. Z., et al. (2017). Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science 355, 962–965. doi: 10.1126/science.aai8898
Frizzi, A., and Huang, S. (2010). Tapping RNA silencing pathways for plant biotechnology. Plant Biotechnol. J. 8, 655–677. doi: 10.1111/j.1467-7652.2010.00505.x
Fukuoka, S., Saka, N., Koga, H., Ono, K., Shimizu, T., Ebana, K., et al. (2009). Loss of function of a proline-containing protein confers durable disease resistance in rice. Science 325, 998–1001. doi: 10.1126/science.1175550
Ghag, S. B., Shekhawat, U. K., and Ganapathi, T. R. (2014). Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 12, 541–553. doi: 10.1111/pbi.12158
Govindarajulu, M., Epstein, L., Wroblewski, T., and Michelmore, R. W. (2015). Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. doi: 10.1111/pbi.12307
Guo, M., Chen, Y., Du, Y., Dong, Y. H., Guo, W., Zhai, S., et al. (2011). The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1001302. doi: 10.1371/journal.ppat.1001302
Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T., and Weber, K. (2001). Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114, 4557–4565.
Holen, T., Amarzguioui, M., Wiiger, M. T., Babaie, E., and Prydz, H. (2002). Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 30, 1757–1766. doi: 10.1093/nar/30.8.1757
Kankanala, P., Czymmek, K., and Valent, B. (2007). Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell 19, 706–724. doi: 10.1105/tpc.106.046300
Koch, A., Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., et al. (2016). An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLos Pathog. 12:e1005901. doi: 10.1371/journal.ppat.1005901
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Li, L., Chang, S. S., and Liu, Y. (2010). RNA interference pathways in filamentous fungi. Cell. Mol. Life Sci. 67, 3849–3863. doi: 10.1007/s00018-010-0471-y
Li, W. T., Zhu, Z. W., Chern, M. S., Yin, J. J., Yang, C., Ran, L., et al. (2017). A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170:114-126.e15.
Miki, D., and Shimamoto, K. (2004). Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45, 490–495. doi: 10.1093/pcp/pch048
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS:host induced gene silencing in the obligate biotrophic fungal pathogen. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Nunes, C. C., and Dean, R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. doi: 10.1111/j.1364-3703.2011.00766.x
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Park, G., Xue, C., Zheng, L., Stephen, L., and Xu, J. R. (2002). MST12 regulates infections growth but not appressorium formation in the rice blast fungus Magnaporthe grisea. Mol. Plant Microbe Interact. 15, 183–192.
Penn, T. J., Wood, M. E., Soanes, D. M., Csukai, M., Corran, A. J., and Talbot, N. J. (2015). Protein kinase C is essential for viability of the rice blast fungus Magnaporthe oryzae. Mol. Microbiol. 98, 403–419. doi: 10.1111/mmi.13132
Pflieger, S., Blanchet, S., Camborde, L., Drugeon, G., Rousseau, A., Noizet, M., et al. (2008). Efficient virus-induced gene silencing in Arabidopsis using a‘one-step’TYMV-derived vector. Plant J. 56, 678–690. doi: 10.1111/j.1365-313X.2008.03620.x
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew.reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Senthil-Kumar, M., and Mysore, K. S. (2011). Caveat of RNAi in Plants: The Off-Target Effect//RNAi and Plant Gene Function Analysis. New York, NY: Humana Press, 13–25.
Thomas, C. L., Jones, L., Baulcombe, D. C., and Maule, A. J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana benthamiana using a potato virus X vector. Plant J. 25, 417–425. doi: 10.1046/j.1365-313x.2001.00976.x
Vinay, P., McCallum, B., Jordan, M., Loewen, M., Fobert, P., McCartney, C., et al. (2016). RNA silencing approaches for identifying pathogenicity and virulence elements towards engineering crop resistance to plant pathogenic fungi. CAB Rev. 11, 1–13.
Wang, B. H., Ebbole, D. J., and Wang, Z. H. (2017). The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J. Integr. Agric. 16, 2746–2760. doi: 10.1016/j.fgb.2018.04.005
Wang, J., Zhou, L., Shi, H., Chern, M., Yu, H., Yi, H., et al. (2018). A single transcription factor promotes both yield and immunity in rice. Science 361, 1026–1028. doi: 10.1126/science.aat7675
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., and Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 9:16151.
Wang, M. Y., Liu, J., Qu, S. H., Wang, X. L., and Liu, G. L. (2015). Construction of rice blast resistant HIGS expression vectors for rice genetic transformation. Plant Protect. 41, 73–78.
Wilson, R. A., and Talbot, N. (2009). Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 7, 185–195. doi: 10.1038/nrmicro2032
Xu, J. R., Staiger, C., and Hamer, J. E. (1998). Inactivation of the mitogen –activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci.U.S.A. 1998, 12713–12718. doi: 10.1073/pnas.95.21.12713
Xu, P., Zhang, Y., Kang, L., Roossinck, M. J., and Mysore, K. S. (2006). Computational estimation and experimental verification of off-target silencing during post transcriptional gene silencing in plants. Plant Physiol. 142, 429–440. doi: 10.1104/pp.106.083295
Xue, C. Y., Park, G., Choi, W., and Xu, J. R. (2002). Two novel fungal virulence genes specifically expressed in appressoria of the rice blast fungus. Plant Cell 14, 2107–2119. doi: 10.1105/tpc.003426
Yan, X., Li, Y., Yue, X. F., Wang, C. C., Que, Y. W., Kong, D. D., et al. (2011). Two novel transcriptional regulators are essential for infection-related morphogenesis and pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 7:e1002385. doi: 10.1371/journal.ppat.1002385
Yin, C., Jurgenson, J., and Hulbert, S. H. (2011). Development of a host-induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24, 554–561. doi: 10.1094/mpmi-10-10-0229
Zhang, H., Guo, J., Voegele, R. T., and Kang, Z. S. (2012). Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp.tritici using a host-induced RNAi system. PLoS One 7:e49262. doi: 10.1371/journal.pone.0049262
Keywords: host-induced gene silencing, rice blast, small interfering RNA, MoAP1, resistance
Citation: Guo X-Y, Li Y, Fan J, Xiong H, Xu F-X, Shi J, Shi Y, Zhao J-Q Wang Y-F, Cao X-L and Wang W-M (2019) Host-Induced Gene Silencing of MoAP1 Confers Broad-Spectrum Resistance to Magnaporthe oryzae. Front. Plant Sci. 10:433. doi: 10.3389/fpls.2019.00433
Received: 20 October 2018; Accepted: 21 March 2019;
Published: 09 April 2019.
Edited by:
Kostya Kanyuka, Rothamsted Research (BBSRC), United KingdomReviewed by:
Wanxin Chen, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyGuus Bakkeren, Summerland Research and Development Centre, Canada
Copyright © 2019 Guo, Li, Fan, Xiong, Xu, Shi, Shi, Zhao, Wang, Cao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Ming Wang, ajMxNndlbm1pbmd3YW5nQHNpY2F1LmVkdS5jbg==; ajMxNndlbm1pbmd3YW5nQDE2My5jb20=
 Xiao-Yi Guo
Xiao-Yi Guo Yan Li
Yan Li Jing Fan2
Jing Fan2 Wen-Ming Wang
Wen-Ming Wang