
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 19 March 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00267
This article is part of the Research Topic Somatic Embryogenesis: 60 Years of Research Applied to Plant Cloning to Unravel Plant Totipotency View all 27 articles
Medicago truncatula is now widely regarded as a legume model where there is an increasing range of genomic resources. Highly regenerable lines have been developed from the wild-type Jemalong cultivar, most likely due to epigenetic changes. These lines with high rates of somatic embryogenesis (SE) can be compared with wild-type where SE is rare. Much of the research has been with the high SE genotype Jemalong 2HA (2HA). SE can be induced from leaf tissue explants or isolated mesophyll protoplasts. In 2HA, the exogenous phytohormones 1-naphthaleneacetic acid (NAA) and 6-benzylaminopurine (BAP) are central to SE. However, there are interactions with ethylene, abscisic acid (ABA), and gibberellic acid (GA) which produce maximum SE. In the main, somatic embryos are derived from dedifferentiated cells, undergo organellar changes, and produce stem-like cells. There is evidence that the SE is induced as a result of a stress and hormone interaction and this is discussed. In M. truncatula, there are connections between stress and specific up-regulated genes and specific hormones and up-regulated genes during the SE induction phase. Some of the transcription factors have been knocked down using RNAi to show they are critical for SE induction (MtWUSCHEL, MtSERF1). SE research in M. truncatula has utilized high throughput transcriptomic and proteomic studies and the more detailed investigation of some individual genes. In this review, these studies are integrated to suggest a framework and timeline for some of the key events of SE induction in M. truncatula.
Medicago truncatula is a genetic and genomic model for legumes (Cook, 1999; Rose, 2008). M. truncatula has a small diploid genome, which is sequenced and annotated (Young et al., 2011; Tang et al., 2014), and a range of genetic and genomic resources are available (Benedito et al., 2008; Li et al., 2012; Rose, 2013; Garmier et al., 2017) as well as being represented in major data bases such as NCBI and Phytozome.
M. truncatula was first regenerated by somatic embryogenesis (SE) in 1989 (Nolan et al., 1989) and required a special seed line (Rose et al., 1999) called Jemalong 2HA (2HA). SE in wild-type Jemalong is rare. Subsequently, two other M. truncatula lines were developed that could also be regenerated by SE: R108 (Hoffmann et al., 1997) and M9-10a (Araújo et al., 2004). M. truncatula has predominantly been used to study nodulation but now is also used to study a wide range of plant biology, including the regulation of SE (Rose and Nolan, 2006). Unlike Arabidopsis (Mordhorst et al., 1998; Ikeda-Iwai et al., 2002; Harding et al., 2003; Kurczyńska et al., 2007; Kadokura et al., 2018) where primary somatic embryos come from immature embryos or seedling SAMs (shoot apical meristems), simple leaf explants can be used (Nolan et al., 1989; Nolan and Rose, 1998). In M. truncatula, an auxin plus cytokinin, rather than auxin alone, is required in the medium. Auxin alone in M. truncatula produces roots from procambium cells in the leaf explant veins (Rose et al., 2006). The specific auxin and cytokinin used vary between laboratories (Table 1). The absence of an exogenous cytokinin requirement for Arabidopsis represents a significant difference to M. truncatula. Interestingly, in the perennial Medicago sativa, SE can be produced by a pulse of 2,4-D in callus induced by an auxin and cytokinin (Dudits et al., 1991). In the 2HA line, abscisic acid (ABA) and gibberellic acid (GA) at appropriate concentrations and timing can stimulate SE over and above auxin + cytokinin (Table 1).
In this review on SE in M. truncatula, the explant, the stress response, and the hormonal response, and how they are integrated in the generation of somatic embryos are considered (Figure 1).
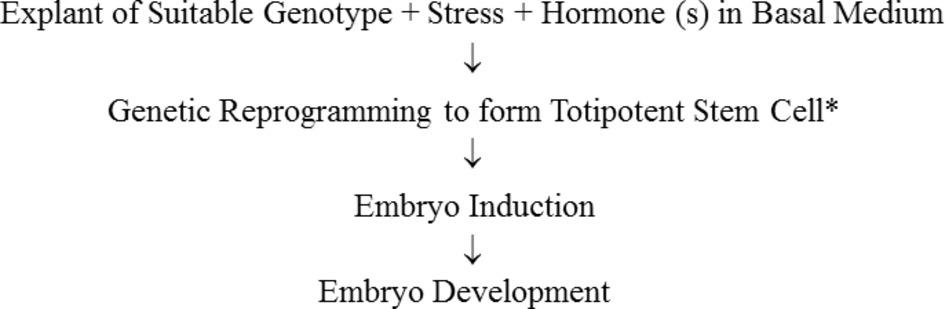
Figure 1. Sequence of steps in somatic embryogenesis. *It has been shown by tracking of a single labeled cell that embryos can develop from single somatic cells (Schmidt et al., 1997). A multicellular origin proposed by Williams and Maheswarin (1986) to occur in some cases has not been unequivocally demonstrated. Haccius (1978) concluded that somatic embryos can derive from a single cell or proembryonal cell complexes which are derived from a single segmenting cell.
For successful SE in M. truncatula, a special genotype is required. In the case of the three genotypes, all have been derived in a similar way, by selection after a cycle or cycles of tissue culture (Nolan et al., 1989; Hoffmann et al., 1997; Araújo et al., 2004). The ability of a cycle of tissue culture to consistently generate regenerable genotypes, which is heritable, is suggestive of epigenetic change as a result of the culture process. The 2HA genotype was developed from one of three rare Jemalong regenerates (Nolan et al., 1989). Each regenerate showed highly enhanced SE. Seed from the highest regenerator was selected for four generations to produce the 2HA line (Nolan et al., 1989; Rose et al., 1999). Earlier work on M. sativa had shown that regeneration capacity is genotype-specific, inherited, and could be enhanced by selection (Bingham et al., 1975; Reisch and Bingham, 1980). There is evidence that 2HA is an epigenetic variant of wild-type Jemalong. Amplified methylation polymorphism (AMP), an arbitrarily primed, methylation-sensitive PCR, showed many DNA methylation changes in 2HA, without detectable genome sequence change (Irwanto and Rose, 2008; Kurdyukov et al., 2014a). There are no obvious karyotypic differences between 2HA and wild-type Jemalong (Kurdyukov et al., 2014a). MtEIL1, an EIN3-like gene, is down-regulated and is methylated in the coding region. This methylation correlates with a small RNA that is antisense to the 3′ region. This gene is discussed further below. Another point of interest in the latter study is that two putative transposase genes, BEDHAT1 and BEDHAT2, are up-regulated. These genes likely became hypomethylated (Kooke et al., 2015). It is not known if the other regenerable genotypes have similar DNA methylation changes. However, using 5-azacytidine with the highly regenerable M9-10a line to inhibit DNA methylation stopped somatic embryogenesis (Santos and Fevereiro, 2002).
Leaf explants contain a number of different cell types in addition to the mesophyll cells. In the case of 2HA, the question of what cells are involved in SE has been examined (Wang et al., 2011). Most somatic embryos are derived from dedifferentiating mesophyll cells near the cut surface while some are derived from the vascular procambium. This was confirmed by changing the orientation of the explants. Vascularization can be greatly reduced by plating the leaf explant adaxial side down rather than abaxial side down and there is little difference in somatic embryo formation. While it is not possible to be unequivocal about why these two different cell types are involved, there are reasonable explanations based on previous literature with other species. The cells near the cut surface have ready access to wound stress molecules and stress can induce dedifferentiation (Grafi and Barak, 2015) and SE (Zavattieri et al., 2010; Rose et al., 2013; Fehér, 2015). The vascular procambium cells are stem-like cells and these types of cells are dedifferentiated and only require an SE-specific signal (Wang et al., 2011). A report in peach has also shown a procambial origin of somatic embryos (De Almeida et al., 2012).
The explants commonly used for investigation of SE in Arabidopsis are from immature zygotic embryos or the seedling SAM (Mordhorst et al., 1998; Gaj, 2001; Harding et al., 2003; Kurczyńska et al., 2007; Kadokura et al., 2018). Ikeda-Iwai et al. (2002) and Su et al. (2009) have used embryonic callus derived from primary embryos from immature zygotic embryos. Embryos develop from the edge of the callus. In general terms, the message from both Arabidopsis and M. truncatula and the wider literature is that SE can have an origin from uncommitted stem-like cells (Mordhorst et al., 1998; Rose, 2016) and cells that require dedifferentiation (Rose, 2016).
M. truncatula can form somatic embryos from isolated mesophyll protoplasts (Rose and Nolan, 1995). The isolated protoplasts form colonies that develop into callus. While embryos can initiate throughout the callus, it appears that embryos only develop fully when they approach the surface (Wang et al., 2011). This is consistent with the idea that it is peripheral cells of an explant, that may also be close to wounded cells as in leaf explants, that produce somatic embryos. Further, suitable auxin gradients may be easier to obtain near the surface of the callus, given auxin’s role in embryogenesis (Jenik and Barton, 2005).
With confocal microscopy and tracking organelles with fluorescent proteins, it is possible to visualize what happens to the organelles as the protoplasts form colonies. This has predominantly been carried out with Nicotiana and Arabidopsis, with some work on M. truncatula. There are three points of interest to emerge. Very early in culture, there is massive mitochondrial fusion in all three species (Sheahan et al., 2005), and in Nicotiana and Arabidopsis (not studied in M. truncatula), there are increases in peroxisomes (Tiew et al., 2015) and increases in P-bodies which are RNA processing bodies (Bhullar et al., 2017).
Massive mitochondrial fusion is indicative of preparation for a new generation (Rose and McCurdy, 2017) and is a response to the stress and hormones in the protoplast culture medium which ultimately leads to regeneration. The peroxisome proliferation is part of a stress response which is discussed further below. It can be argued that the increase in P-bodies reflects the degradation of transcripts characteristic of the differentiated cell, as it transits into cell division (Bhullar et al., 2017).
In analyzing the very first events in SE, it is important to distinguish between direct SE from stem-like cells and dedifferentiating cells developing embryonic callus as these early changes are different (Rose, 2016; Horstman et al., 2017a).
The M. truncatula system predominantly involves the dedifferentiation of cells and the formation of proembryogenic masses (PEMs) that have embryonic stem cells (Rose, 2016). Overall, the working hypothesis is that it is certain cells within the PEMs that have the right hormone environment to transit to SE formation. There is evidence for auxin gradients in the Arabidopsis callus, Su et al. (2009).
A proposed relationship between stress and hormones for M. truncatula is shown in Figure 2. In this context, the gene Mt STRESS KINASE1 (MtSK1) has been investigated (Nolan et al., 2006). This gene is first expressed in the callus induction phase and is expressed in explants cultured in the presence or absence of the hormones auxin and cytokinin. MtSK1 is a stress-related kinase, responding to wounding and salt stress. Its close relationship to SnRK2.4 of Arabidopsis and with no obvious responses to ABA (Nolan et al., 2006) like SnRK2 class 2 and 3 genes (Ng et al., 2014) suggests it is a class 1 SnRK2. How MtSK1 is connected to the SE response is not clear. However, there is an interesting relationship between class1 SnRK2 genes and the VARICOSE (VCS) mRNA decapping activator. VCS is the substrate for SnRK2 genes which subsequently causes decay of mRNA transcripts (Soma et al., 2017; Chantarachot and Bailey-Serres, 2018). In Nicotiana mesophyll protoplasts, VCS-containing P-bodies increase in the initial dedifferentiation phase of protoplast culture (first 48 h). It is plausible that MtSK1 and VCS are linked in dedifferentiation to remove transcripts associated with the dedifferentiation of the mesophyll explant cells.
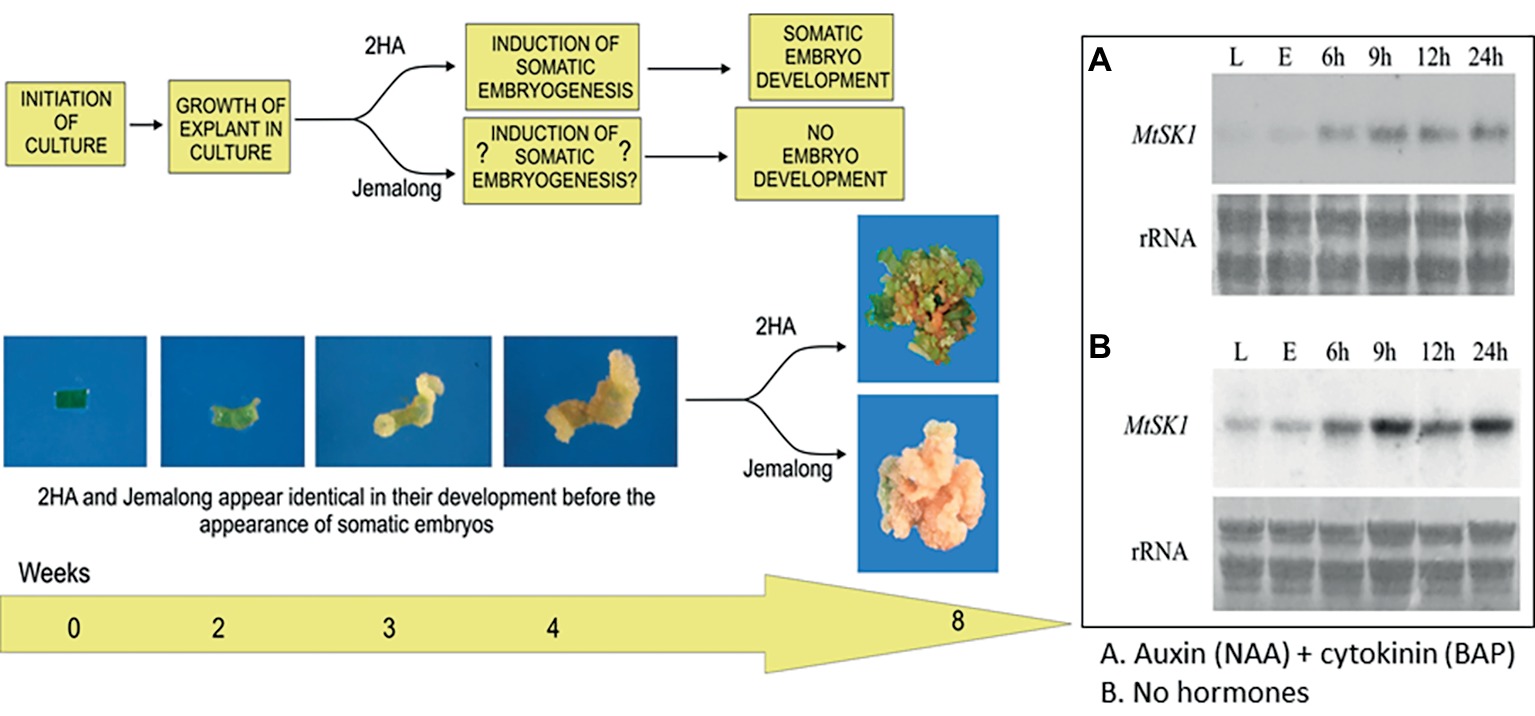
Figure 2. Interactions between stress and hormones in somatic embryogenesis. Development of explants from 2HA and wild-type Jemalong. Jemalong leaf explants develop into calli while 2HA explants develop into embryogenic calli capable of producing regenerated plants. Inset shows MtSK1 expression (northern blots) in 2HA explants (E) from leaves (L) cultured for 24 h with and without hormones. Component figures reproduced from Nolan et al. (2006) with permission.
The increased expression of MtSK1 occurs prior to 6 h after excision and plating, there are no data prior to 6 h. The very first change in the explant (Wang et al., 2011) is the production of reactive oxygen species (ROS), which occurs in seconds (Figure 3A). Soares et al. (2009) showed that after wounding M. truncatula leaves, there is an initial burst of O2 − for 0–30 min, which converts to H2O2 by superoxide dismutase. DAB (3,3′-diaminobenzidine) staining shows that the ROS is associated predominantly with the wound surface (Figure 3B), where most SE derives, with less staining associated with the vasculature (Wang et al., 2011). ROS can act as a signal (Mittler et al., 2011) but excessive ROS can be toxic (Foyer and Noctor, 2011). In quantitative proteomic studies of embryogenic 2HA versus wild-type Jemalong, enzymes involved in ROS detoxification were up-regulated: ascorbate peroxidase, thioredoxin h (TrnH), and peroxidoredoxin (Imin et al., 2005). This is consistent with ROS modulation in M. truncatula SE by up-regulation of redox genes and is supported by proteomic studies in other species (Heringer et al., 2018).
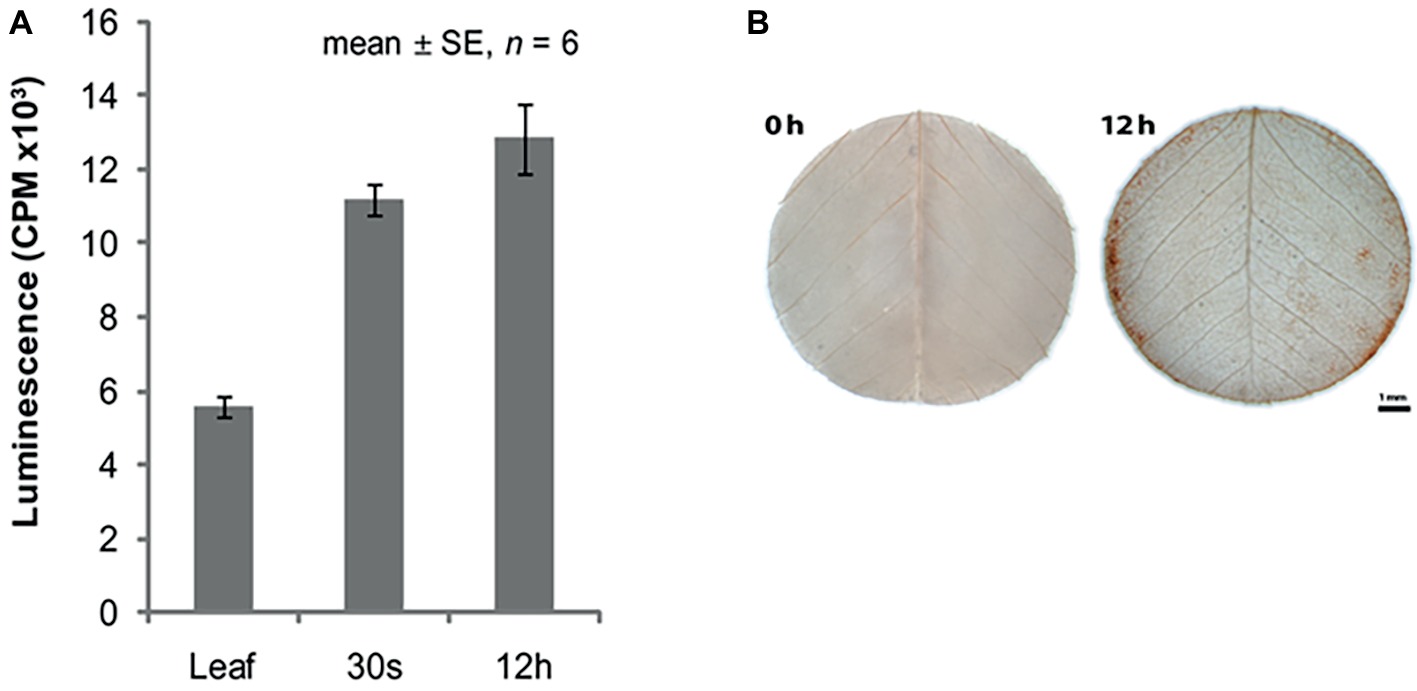
Figure 3. ROS in relation to somatic embryogenesis induction. (A) ROS production in leaf explants. (B) DAB (3,3'-diaminobenzidine) staining for H2O2. Bar = 1 mm. (A,B) reproduced with permission from Wang et al., 2011.
ROS homeostasis is essential for dedifferentiation and cell division induction, the beginning of callus formation (Fehér et al., 2008; Tiew et al., 2015). Zhang et al. (2018) have shown that thioredoxin regulates ROS homeostasis and de novo shoot regeneration in Arabidopsis. Excessive ROS is mitochondrially produced and inhibits shoot regeneration.
ROS inhibitors prevent SE induction in M. truncatula (Tiew, 2015). Whether there is a connection to MtSK1 is not known but it is an area that requires investigation. Mitochondria clearly produce a lot of ROS in the culture process that needs to be regulated (Tiew et al., 2015), but it is not the only source of ROS. ROS are produced by NADPH oxidases, encoded by respiratory burst oxidase homologs (RBOHs) in a plasma membrane complex. The MtRBOHA gene expression is up-regulated within the first week of culture, and is reduced by ABA+GA (Nolan et al., 2014), which increases SE, again suggesting the importance of modulation of ROS. In M. sativa protoplasts, ROS has been linked to auxin action and cell division induction in culture (Fehér et al., 2008). In M. truncatula, there is a link between ROS activity and ethylene production, which is discussed further in the sections below.
In understanding the SE process, it is ultimately necessary to understand the signaling processes involved and how this relates to both the hormones in the medium as well as endogenous hormones. While no doubt a number of parts of the process are similar across species, the detailed operation of the gene networks involved is likely to be species-specific. In the case of M. truncatula, the expression of some genes has been linked to specific hormones.
MtWUSCHEL is an ortholog of AtWUSCHEL hybridizing to the SAM stem cell niche (Chen et al., 2009) and in zygotic embryogenesis has the same time course expression as for Arabidopsis (Kurdyukov et al., 2014b). WUSCHEL (WUS) expression in M. truncatula is cytokinin dependent (Figure 4A), its expression increases a few days after excision, and RNAi studies have shown it is essential for SE (Chen et al., 2009). Similar results have also been shown in Arabidopsis (Su et al., 2009). However, in Arabidopsis, this early WUS expression in relation to SE is auxin dependent. Cytokinin in Arabidopsis cultures induces WUS expression and shoots (Gordon et al., 2007). In the case of M. truncatula, auxin alone produces numerous root primordia, from procambial cells, forming adventitious roots (Rose et al., 2006). Early WUS expression is characteristic of SE induction in Arabidopsis, M. truncatula, and Brassica (Chen et al., 2009; Su et al., 2009; Elhiti et al., 2010), consistent with the model of Fehér (2015). The well-established SAM expression occurs later as the bipolar embryo is formed (Mayer et al., 1998). This suggests that WUS in relation to SE can initiate an embryonic stem cell that progresses into embryogenesis. In MtWUS::GUS studies in M. truncatula, there are three expression stages: an initial phase throughout the early callus, then, when the explant is more fully callused, there are clusters of expression, and then embryo-associated expression. In M. truncatula zygotic embryogenesis, WUS expression also occurs in the ovule and early cell divisions of the embryo (Kurdyukov et al., 2014b), as it does in Arabidopsis (Groß-Hardt et al., 2002). It is feasible that the earliest expression found in 2HA, and also found in M9-10a but where there were no GUS studies (Orłowska and Kępczyńska, 2018), is more analogous to the ovule stage, the patches to PEMs, and subsequently the classic embryo WUS expression (Mayer et al., 1998). How then do these WUS expressing patches in the callus occur? The assumption is that this reflects parts of the callus where PEMs form as a result of suitable auxin and cytokinin concentrations. There are no data on this in M. truncatula but in the Su et al. (2009) study dealing with SE in Arabidopsis callus, WUS induction in patches is associated with PIN-FORMED (PIN1) expression and the setting up of appropriate auxin gradients. In M. truncatula 2HA, there is an initial peak expression of WUS at 7 d and then declines. CLAVATA3 (CLV3) expression starts as the somatic embryo forms, establishing the WUS, CLV3 feedback loop characteristic of the SAM. A question that arises is given the role of WUS in the stem cells of the SAM, how can it be involved as an embryonic stem cell destined to form shoot and root meristems. Sarkar et al. (2007) have shown that WUS can act interchangeably with WOX5, the root stem cell maintenance gene. It was shown that a WOX5-WUScDNA transgene restored stem cells in the root meristem of a wox5 mutant.
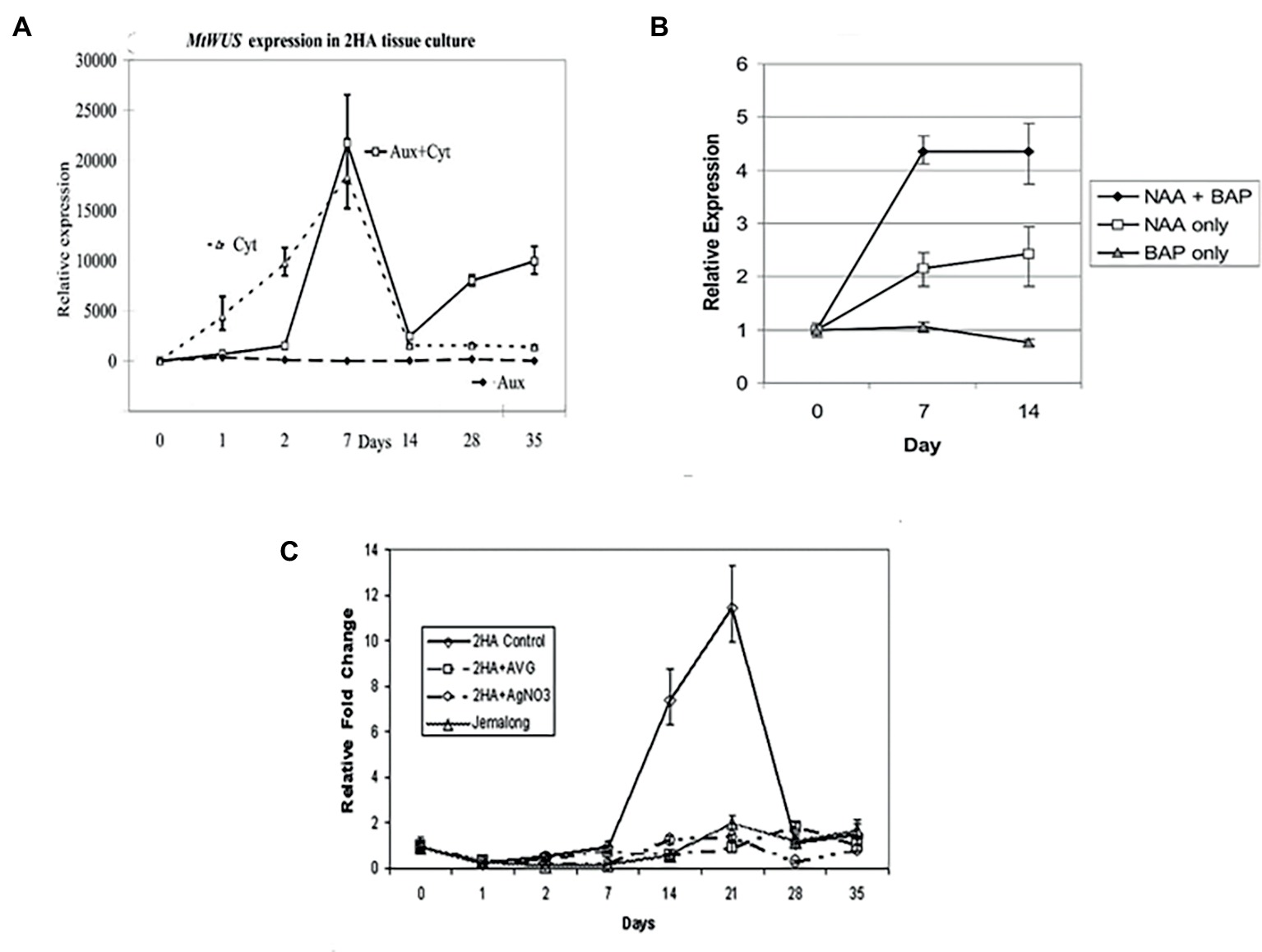
Figure 4. Effects of different hormones on somatic embryo induction. (A) MtWUSCHEL expression; (B) MtSERK1 expression; (C) MtSERF1 expression. AVG (10 μM aminoethoxyvinylglycine) and AgNO3 (10 μM) are ethylene biosynthesis and ethylene perception inhibitors, respectively. Aux = NAA = 10 μM 1-naphthalene acetic acid, Cyt = BAP = 4 μM 6-benzylaminopurine. (A) From Chen et al. (2009), authors’ copyright. (B) From Nolan et al. (2003), www.plantphysiol.org, Copyright American Society of Plant Biologists. (C) From Mantiri et al. (2008a,b), www.plantphysiol.org, Copyright American Society of Plant Biologists.
The SOMATIC EMBRYO RECEPTOR-LIKE KINASE1 (SERK1) has been implicated in the induction of SE since its discovery by Schmidt et al. (1997) in carrot, where it is auxin-induced and expressed in cells destined to form somatic embryos. In Arabidopsis, overexpression of SERK1 stimulates SE (Hecht et al., 2001) and is expressed to the early globular stage. SERK1 is also expressed in ovules and early zygotic embryos. In M. truncatula, SERK1 is induced in both somatic embryo and root forming cultures (Nolan et al., 2003) in response to auxin. There is no response with cytokinin alone (Figure 4B). As noted earlier, auxin alone induces root formation. With auxin plus cytokinin, somatic embryos are induced and cytokinin acts synergistically with auxin to increase MtSERK1 expression above auxin alone (Figure 4B). The M. truncatula results suggest that SERK1 is not specific to SE or embryogenesis. In follow-up studies with MtSERK1 with promoter-GUS analysis, MtSERK1 expression was found to be associated with developmental change (Nolan et al., 2009). There is expression of MtSERK1 when callus is initiated and when somatic embryos are initiated. Expression is also associated with primary meristems of the shoot and root. The M. truncatula data do not indicate that SERK1 is not important in SE induction, rather that it is a gene connected to reprogramming of cells associated with developmental change. Studies with a range of species show SERK1 expression is characteristic of early SE (Pandey and Chaudhary, 2014; Rocha et al., 2016). What is of interest in the case of M. truncatula is that cytokinin is key for WUSCHEL expression and auxin for SERK1 expression.
The MtSERF1 gene (SOMATIC EMBRYO RELATED FACTOR1) was discovered in the context of a cDNA microarray study in M. truncatula protoplasts at the transition stage, between callus and SE induction (Mantiri et al., 2008a). The microarray study showed up-regulation of ethylene biosynthesis and ethylene response genes. The MtSERF1 gene is a member of the ERF sub-family of the AP2/ERF super family. It is up-regulated in 2HA but not wild-type Jemalong (Figure 4C). The expression of this gene peaks at 21d, the transition period between callus and somatic embryo production. The expression of the gene is ethylene dependent. The expression of this gene not only requires ethylene but is dependent on the presence of both auxin plus cytokinin (Rose and Song, 2018). This is a link between induced endogenous hormones and hormones supplied in the medium. What is the function of MtSERF1? There is some evidence that it is related to the action of WUS. This is based on the promoter region sequence of MtSERF1 having WUS binding sites and the localization of MtSERF1 expression to the apical region of the heart stage somatic embryo (Mantiri et al., 2008a).
As for auxin and cytokinin, the exact role for ethylene is species dependent, though it is clear that it is involved in SE (Kępczyńska and Zielińska, 2011; Fehér, 2015). The MADS box transcription factor AGAMOUS-LIKE15 (AGL15) stimulates SE when overexpressed in Arabidopsis and soybean (Zheng et al., 2013). AGL15 is able to stimulate ethylene production and SERF1 expression (Zheng et al., 2013, 2016) suggesting that AGL15 could be associated with the stress response as well as modulating the auxin and GA response. There has been speculation on the targets of SERF1 in M. truncatula (Mantiri et al., 2008b) where the HD-Zip III genes PHABULOSA, PHAVOLUTA, and REVOLUTA were suggested, but there is no hard evidence on targets.
In M. truncatula, there is evidence that ethylene signaling is modified. Microarray studies by Imin et al. (2008) and more specific studies by Kurdyukov et al. (2014a) have shown that one of the two EIN3-like genes is down-regulated. This could possibly represent a necessary modulation of the ethylene response, preventing excess stress.
The relationship of the PICKLE (PKL) gene to SE was discovered by Ogas et al. (1997). In the pkl mutant, cultured roots on basal medium could undergo SE without the application of plant hormones. SE in the pkl mutant is inhibited by GA. PKL, then, is a negative regulator of SE. This is conceptually important as the capacity for embryogenesis has to be switched off in somatic cells. If this gene is repressed, then the capacity for SE is enabled. The study by Zhang et al. (2008) shows that PKL and GA can act synergistically via separate pathways to repress expression of seed-associated genes. PKL is an ATP-dependent CHD3 chromatin remodeler which is part of complex that promotes the trimethylation of histone H3 lysine 27 (H3K27me3), a negative histone mark (Zhang et al., 2008). PKL represses expression of the embryo-specific transcriptional program, including the master regulators of embryogenesis LEAFY COTYLEDON1 (LEC1) and LEAFY COTYLEDON2 (LEC2), extensively studied in relation to SE (Zhang et al., 2012). It is not clear whether GA acts upstream or downstream of LEC genes in PKL repression of embryogenesis (Braybrook and Harada, 2008). In any event, models based on Arabidopsis data show that there is a nexus between PKL, LEC1, LEC2, FUSCA3 (FUS3), AGL15, GA, and ABA (Braybrook and Harada, 2008). Essentially, high ABA/GA ratios promote SE. However, in the case of M. truncatula, it is low ABA/GA ratios that promote SE and inhibit PKL (Nolan et al., 2014). Though ABA/GA ratios have not been investigated in M. sativa, the ABA and GA data alone are more similar to M. truncatula (Nolan and Rose, 1998; Ruduś et al., 2002; Ruduś et al., 2009; Nolan et al., 2014). This again shows how genes can be common to SE across species, but behave differently to plant hormones.
Some aspects of GA metabolism in relation to SE have been studied in M. truncatula. GA3 is synthesized in the SE induction period in the M9-10A embryogenic line (Igielski and Kępczyńska, 2017). GA in the medium inhibited SE in m9-10A, but at lower concentrations than in 2HA. In m9-10A, the GA biosynthesis inhibitor paclobutrazol also inhibited SE. This again suggests there are differences to Arabidopsis where this inhibitor promotes SE (Wang et al., 2004).
In general terms, stress in the acquisition of SE has implicated 2,4-dichlorophenoxyacetic acid (2,4-D) at high concentrations, ABA, and ethylene (Karami and Saidi, 2010). Stress is involved in dedifferentiation (Grafi and Barak, 2015; Zhou et al., 2016) as well as in the activation of embryonic cell division (Pasternak et al., 2002; Rose et al., 2013). ROS are important in a plant’s response to stress (Chamnongpol et al., 1998; Podgórska et al., 2017). In the M. truncatula system (Tiew, 2015), as in M. sativa, ROS inhibition prevents SE (Fehér et al., 2008). In M. sativa protoplasts, ROS interact with auxin to initiate the cell cycle (Fehér et al., 2008). In cotton SE, there is an interplay between ROS and auxin to modulate SE (Zhou et al., 2016). Qu et al. (2017) have shown that H2O2 can regulate auxin distribution in lateral root development in Arabidopsis by regulation of PIN2. As shown in Figure 3B, ROS accumulate at sites where somatic embryos ultimately form.
Ethylene can be induced by ROS (Chamnongpol et al., 1998; Song et al., 2007) and is ROS dependent in M. truncatula (Tiew, 2015). In incubated excised mung bean hypocotyls, ROS promotes auxin-induced ethylene production (Song et al., 2007). Ethylene, together with auxin and cytokinin, is required for MtSERF1 expression (Rose and Song, 2018). It is plausible that MtSERF1, with its requirement for ethylene, is a nexus between stress and auxin and cytokinin action (Mantiri et al., 2008a,b). In Arabidopsis and soybean studies, it has been shown that AGL15 stimulates expression of SERF1 (Zheng et al., 2013), as well as LEC2 (not tested in soybean), FUS3, and ABSCISIC ACID INSENSITIVE3 (ABI3) genes which encode a B3 domain (Zheng et al., 2009; Zheng and Perry, 2014). AGL15 expression has not been examined in M. truncatula, nor have the targets of SERF1. It would be expected that there would be some similarity to soybean where AGL15 not only stimulates SERF1 but the FUS3 and ABI3 genes that are influenced by GA:ABA ratios (Braybrook and Harada, 2008; Zheng et al., 2009) and are required for embryogenesis. It is possible that ethylene influences GA action as well as auxin as discussed by Zheng et al. (2016). The conclusion from these latter studies in both soybean and Arabidopsis is that ethylene accumulation and response reduce the GA response facilitating SE. DELLA proteins may be significant in these interactions, where at least some ethylene response factors have been shown to interact with DELLA (Marín-de la Rosa et al., 2014). Ethylene biosynthesis and action have been shown to be important in the proliferation of embryogenic suspensions and embryo development in M. sativa L.cv. Rangelander (Kępczyńska et al., 2009, Kępczyńska and Zielińska, 2011) but not the initial induction (Kępczyńska et al., 2009).
It is known from high throughput gene expression studies in legumes (Thibaud-Nissen et al., 2003; Imin et al., 2008; Mantiri et al., 2008a) that there are large numbers of gene expression changes associated with SE. However, there are a number of major genes that need to be put in perspective before the complexity of the integration of all the metabolic events associated with the developmental changes can be assembled. Genes or metabolites related to SE studied in M. truncatula and their approximate timeline are shown in Table 2. The different molecules and genes set out in Table 2 for M. truncatula can be discussed in the following framework.
In cultured M. truncatula, ROS is the first signal as a result of the stress from the excision and plating of the explant. ROS is an important signal, but needs to be modulated as excess ROS can be toxic (Liu and He, 2016; Podgórska et al., 2017). Redox control then becomes essential and this is consistent with up-regulation of TrnH and ascorbate oxidase (Table 2, Imin et al., 2005). Increased number of peroxisomes also assists in redox homeostasis in this phase in Arabidopsis (Tiew et al., 2015). ROS can be generated by both NADPH oxidase and the electron transfer chain of mitochondria. There is some evidence that the initial ROS signal is due to NADPH oxidase (Soares et al., 2009) and MtRBOHA expression occurs before 7 d of culture (Nolan et al., 2014).
The transduction of the ROS signal together with the plant hormones in the medium initiate the chromatin changes leading to dedifferentiation and cell division initiation. The work of Zhao et al. (2001) shows isolated protoplasts undergo chromatin decondensation and there is increased DNA accessibility with propidium iodide. How this is linked to the stress/ROS/hormone interaction is unclear. Chromatin remodeling is influenced by PKL, POLYCOMB REPRESSIVE COMPLEX1 and 2 (PRC1 and PRC2), and the TRITHORAX GROUP proteins (TrxG) shown in Table 2. PKL contributes to H3K27me3 enrichment of loci, which is a repressive mark (Zhang et al., 2012), so down-regulation of PKL can facilitate derepression of genes required for SE. The PRC2 complex also increases H3K27me3 levels and is required for callus formation from leaf tissue where it represses the genes encoding leaf characteristics (He et al., 2012). The PRC1 complex ubiquinates histone H2A lysine 119 to compact chromatin (Schuettengruber et al., 2011; He et al., 2012) and needs to be down-regulated for SE. The up-regulation of TRITHORAX genes facilitates increased gene expression (Schuettengruber et al., 2011). Chromatin remodeling is clearly important in the transition to the dedifferentiated state where some genes need to be repressed and during SE where a number of genes need to be activated.
The specific role of MtSK1, characteristic of the excision and plating of the explant, in SE is not yet clear. However, given that it is a class 1 SnRK2 gene, implicated with RNA processing bodies, suggests a role in the degradation of transcripts from the original explant cells by interaction with P-bodies as the cells become meristematic (Soma et al., 2017; Chantarachot and Bailey-Serres, 2018). This degradation of existing transcripts characteristic of the explant leaf cells is critical to cell fate. The fusion of mitochondria appears to be important in ensuring that the integrity of the mitochondrial genome is maintained, to ensure cells have the capacity for regeneration (Rose and McCurdy, 2017).
Callus proliferation itself has not been studied in any detail in M. truncatula where the focus in the callus phase has been on the transition of callus cells to stem-like cells that will initiate embryos. In Arabidopsis WOUND INDUCED DEDIFFERENTIATION (WIND) transcription factors, members of the AP2/ERF transcription factor family have been shown to be important regulators of wound-induced callus (Iwase et al., 2011; Ikeuchi et al., 2017). The WUSCHEL-RELATED HOMEOBOX5 (WOX5) root meristem transcription factor is important in callus induction from pericycle cells in response to 2,4-D and kinetin in Arabidopsis (Sugimoto et al., 2010). In M. truncatula, WOX5 expression is high in explants cultured with auxin where callus and root primordia come from procambium cells (Chen et al., 2009). Less WOX5 expression occurs in the auxin + cytokinin medium (Chen et al., 2009; Orłowska and Kępczyńska, 2018), with massive callus coming from dedifferentiation of mesophyll cells rather than the procambium (Chen et al., 2009), and is where SE mainly occurs.
Early expression of WUS is characteristic of SE (Fehér 2015; Mahdavi-Darvari et al., 2015) and there is a good case that it is a critical gene required for the production of embryonic stem cells (Zuo et al., 2002; Chen et al., 2009; Su et al., 2009; Elhiti et al., 2010). In M. truncatula SE from leaf explants, the very early expression of WUS is more analogous to the ovule expression. Subsequently, the induction of CLV3 is important in forming patches of WUS expressing cells (Mantiri et al., 2008a) in the callus, similar to patches of expression in Arabidopsis embryogenic callus (Su et al., 2009). These patches of WUS expression could correspond to the densely cytoplasmic cells of the PEMs. Su et al. (2009), in Arabidopsis, have shown that auxin gradients and PIN genes are required for WUS expression in SE, but the location of PIN gene expression has not been studied in M. truncatula SE. In de novo shoot regeneration investigations in Arabidopsis in response to cytokinin, there was reduced DNA methylation, increased levels of histone H3K4me3 and H3k9ac, and reduced levels of H3Kme2 at the WUS sequences (Li et al., 2011).
Expressing slightly later than WUS is the KNOX gene SHOOT MERISTEMLESS (STM) (Orłowska and Kępczyńska, 2018) which in zygotic embryogenesis in M. truncatula has a similar time course of expression to WUS (Kurdyukov et al., 2014b). In Arabidopsis, STM is required for continued stem cell function in the SAM by sustaining expression of WUS (Scofield et al., 2014).
For the embryonic stem cells to develop into embryos, appropriate hormone and transcription interactions are required (Table 2, Figure 5). Essentially upstream of the LEC gene transcription, there are the SERK1, AGL15, SERF1, and BBM genes.
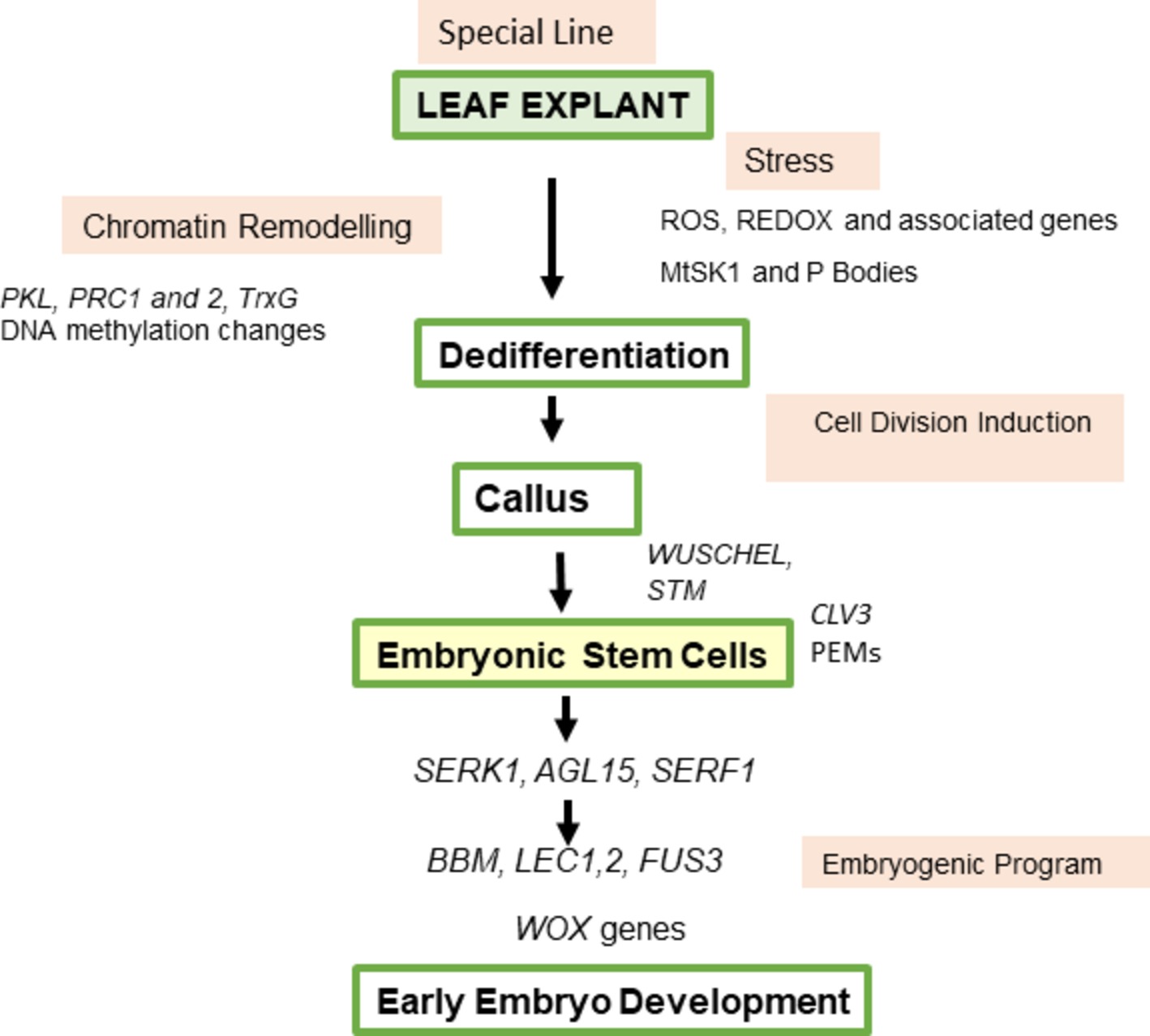
Figure 5. Model for sequence of events in somatic embryogenesis from M. trunctula leaf explants. Excision and plating of the explant produce ROS that probably in interaction with hormones cause chromatin remodeling involving PKL, PRC complexes, and TRITHORAX genes, and a stress kinase that maybe linked to transcript degradation via RNA processing bodies. Cell divisions follow and callus is produced. Cytokinin-dependent WUS expression is essential for SE and there is an initial expression analogous to that occurring in the ovule followed by stem cell development linked to PEMs in patches on the callus, correlating with CLV3 and STM expression. AGL15, SERK, and SERF are part of the correct hormone milieu leading to expression of BBM and LEC genes and the embryogenic program. The changes diagramed go hand in hand with hormonal changes. Auxin is initially high then lowers as differentiation starts, followed by auxin regulation associated with embryo patterning in the formation of the bipolar embryo. Cytokinin is important for specific genes and the cell cycle. Endogenous ethylene is produced and endogenous ABA/GA ratios specific to M. truncatula influence SE. Ethylene, ABA, and GA can be involved in regulating auxin responses. Abbreviations are in the text.
There are interesting interactions between SERK1, AGL15, and SERF1. MtSERF1 is ethylene dependent, responding to increased ethylene and possibly WUS (Mantiri et al., 2008a). In soybean, AGL15 stimulates LEC2, FUS3, ABI3, and SERF1 expression (Zheng et al., 2009, 2013). AGL15 also represses the auxin response and interacts with GA metabolism to influence LEC genes (Zheng et al., 2016). In relation to the auxin response, a common classic response to auxin and SE is that high auxin is required for the initiation of SE and then auxin removal for embryo development (Halperin, 1964; Rose, 2004). In the model proposed by Fehér (2015), removal of 2,4-D blocks cell proliferation and triggers differentiation. Later, endogenous auxin is produced as part of normal embryogenesis paralleling zygotic embryogenesis. Ethylene and ERF genes are potentially capable of reducing the GA response through DELLA interactions (Achard et al., 2007; Marín-de la Rosa, 2014; Zheng et al., 2016). There is also evidence for a connection between AGL15 and SERK1. AGL15 has been found in the same complex with SERK1, supporting an involvement in the same signaling pathway (Karlova et al., 2006). It is possible that these interactions with SERK1, AGL15, and SERF1, and their relationship to hormone effects, provide the milieu for the activation of SE genes.
The key roles of the leafy cotyledon genes in SE are well established (Lotan et al., 1998; Stone et al., 2001; Gaj et al., 2005; Braybrook and Harada, 2008). As master regulators of embryogenesis (Santos-Mendoza et al., 2008), it is the LEC1, LEC2, and FUS3 genes that finally need to be switched on to set in train the embryogenic program. As indicated here, and in other reviews, the leafy cotyledon genes are expressed downstream of WUS (Fehér, 2015). An argument can be made that 35S/LEC1 and 35S/LEC2 seedlings produce somatic embryos (Lotan et al., 1998; Stone et al., 2001) because of the presence of pre-existing stem cells that exist in the vascular and apical meristems. It has been shown that LEC2 can stimulate local auxin synthesis via YUCCA genes in Arabidopsis (Stone et al., 2008; Wójcikowska et al., 2013) and this is consistent with regulating hormone auxin levels required for embryo development (Friml et al., 2003). Overexpression of BABY BOOM (BBM), like LEC genes, can induce SE (Boutilier et al., 2002) and recent investigations place BBM upstream of LEC genes and part of the same SE pathway (Horstman et al., 2017b). This is consistent with the Table 2 timeline of M. truncatula gene expression. That the cell context is important was shown in the BBM studies where overexpression at different stages of germination caused differences in the way embryos were produced, with and without a callus phase.
The overexpression of WUS can also induce SE in Arabidopsis (Zuo et al., 2002). Again, pre-existing cells and the degree of stemness could be key as to the cells that respond. WUS expression is upstream of BBM in M. truncatula (Chen et al., 2009; Orłowska et al., 2017; Orłowska and Kępczyńska, 2018). Ectopic expression of AtWUS produced embryogenic callus in cotton (Zheng et al., 2014), but not regeneration, and resulted in up-regulation of GhLEC1, GhLEC2, and GhFUS3. Overexpression of WUS in Coffea canephora also increased SE (Arroyo-Herrera et al., 2008). In a number of monocotyledons, overexpression of both WUS and BBM initiates high rates of SE (Lowe et al., 2016). In addition to their established roles in the induction of SE in Arabidopsis and M. truncatula, WUS (Mayer et al., 1998; Chen et al., 2009) and BBM (Imin et al., 2007; ten Hove et al., 2015) have pivotal roles in the SAM and RAM (root apical meristem) respectively, which suggests that SE is able to co-ordinate these zygotic embryogenesis roles in the SE induction phase. Some years ago, work on M. sativa based on intercrossings indicated that two genes were important in SE determination (Reisch and Bingham, 1980; Hernandez-Fernandez and Christie, 1989; Kielly and Bowley, 1992). This suggests that in recalcitrant legume varieties, including the Medicago genus, overexpression of key genes is worth further investigation.
Once embryos start to develop, then WOX genes become important and the controls characteristic of zygotic embryogenesis follow (Kurdyukov et al., 2014b; ten Hove et al., 2015).
One aspect that also requires further investigation comes from microarray data in the study by Kurdyukov et al. (2014a) where 28 d cultures showed up-regulation of a number of genes that are also linked to nodulation.
In the case of indirect callus-based SE in M. truncatula, the following model is suggested, based on current understanding (Figure 5).
The hormonology and stress responses for SE are characteristic of different species and cultivars, but the principles illustrated in M. truncatula provide a basis for understanding indirect callus-based SE from this legume model (Figure 5). Current data (Kurdyukov et al., 2014a) with M. truncatula indicate that the special lines required for SE have epigenetic changes but at this stage which genes are critical have not been ascertained. This is an important question for further work, as are epigenetic change and stress (Grafi and Barak, 2015). The M. truncatula studies provide some insights into how exogenous hormones (auxin and cytokinin) and endogenous hormones (ABA, GA, and ethylene) contribute to different SE components and their integration. Some of the work with PRC1, PRC2, and TRX genes and MtSK1 and P-bodies suggest ways to explore chromatin remodeling and dedifferentiation on the way to cell division initiation. Overexpression and gene knockdown studies to assist in more fully defining the sequence of events and networks responsible for SE in M. truncatula are required. The very earliest changes involving ROS suggest that what happens at the cell membrane is also an area requiring detailed exploration.
The author confirms being the sole contributor of this work and has approved it for publication.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Achard, P., Baghour, M., Chapple, A., Hedden, P., Van Der Straeten, D., Genschik, P., et al. (2007). The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 104, 6484–6489. doi: 10.1073/pnas.0610717104
Araújo, S. S., Duque, A. S. R. L. A., Santos, D. M. M. F., and Fevereiro, M. P. S. (2004). An efficient transformation method to regenerate a high number of transgenic plants using a new embryogenic line of M. truncatula cv. Jemalong. Plant Cell Tissue Organ Cult. 78, 123–131. doi: 10.1023/B:TICU.0000022540.98231.f8
Arroyo-Herrera, A., Gonzalez, A. K., Moo, R. C., Quiroz-Figueroa, F. R., Loyola-Vargas, V. M., Rodriguez-Zapata, L. C., et al. (2008). Expression of WUSCHEL in Coffea canephora causes ectopic morphogenesis and increases somatic embryogenesis. Plant Cell Tissue Organ Cult. 94, 171–180. doi: 10.1007/s11240-008-9401-1
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume M. truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Bhullar, D. S., Sheahan, M. B., and Rose, R. J. (2017). RNA processing body (P-body) dynamics in mesophyll protoplasts re-initiating cell division. Protoplasma 254, 1627–1637. doi: 10.1007/s00709-016-1053-0
Bingham, E. T., Hurley, L. V., Kaatz, D. M., and Saunders, J. W. (1975). Breeding alfalfa which regenerates from callus tissue in culture. Crop Sci. 15, 719–721. doi: 10.2135/cropsci1975.0011183X001500050032x
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Braybrook, S. A., and Harada, J. J. (2008). LECs go crazy in embryo development. Trends Plant Sci. 13, 624–630. doi: 10.1016/j.tplants.2008.09.008
Chabaud, M., Larsonneau, C., Marmouget, C., and Huguet, T. (1996). Transformation of barrel medic (M. truncatula Gaertn.) by Agrobacterium tumefaciens and regeneration via somatic embryogenesis of transgenic plants with the MtENOD12 nodulin promoter fused to the gus reporter gene. Plant Cell Rep. 15, 305–310. doi: 10.1007/BF00232361
Chamnongpol, S., Willekens, H., Moeder, W., Langebartels, C., Sandermann, H., Van Montagu, M., et al. (1998). Defense activation and enhanced pathogen tolerance induced by H2O2 in transgenic tobacco. Proc. Natl. Acad. Sci. USA 95, 5818–5823. doi: 10.1073/pnas.95.10.5818
Chantarachot, T., and Bailey-Serres, J. (2018). Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol. 176, 254–269. doi: 10.1104/pp.17.01468
Chen, S. K., Kurdyukov, S., Kereszt, A., Wang, X. D., Gresshoff, P. M., and Rose, R. J. (2009). The association of homeobox gene expression with stem cell formation and morphogenesis in cultured M. truncatula. Planta 230, 827–840. doi: 10.1007/s00425-009-0988-1
Cook, D. R. (1999). M. truncatula–a model in the making! Curr. Opin. Plant Biol. 2, 301–304. doi: 10.1016/S1369-5266(99)80053-3
De Almeida, M., De Almeida, C. V., Graner, E. M., Brondani, G. E., and de Abreu-Tarazi, M. F. (2012). Pre-procambial cells are niches for pluripotent and totipotent stem-like cells for organogenesis and somatic embryogenesis in the peach palm: a histological study. Plant Cell Rep. 31, 1495–1515. doi: 10.1007/s00299-012-1264-6
Dudits, D. N., Bogre, L. A., and Gyorgyey, J. A. (1991). Molecular and cellular approaches to the analysis of plant embryo development from somatic cells in vitro. J. Cell Sci. 99, 473–482.
Elhiti, M., Tahir, M., Gulden, R. H., Khamiss, K., and Stasolla, C. (2010). Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. J. Exp. Bot. 61, 4069–4085. doi: 10.1093/jxb/erq222
Fehér, A. (2015). Somatic embryogenesis—stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta Gene Regul. Mech. 1849, 385–402. doi: 10.1016/j.bbagrm.2014.07.005
Fehér, A., Ötvös, K., Pasternak, T. P., and Pettkó-Szandtner, A. (2008). The involvement of reactive oxygen species (ROS) in the cell cycle activation (G0-to-G1 transition) of plant cells. Plant Signal. Behav. 3, 823–826. doi: 10.4161/psb.3.10.5908
Foyer, C. H., and Noctor, G. (2011). Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18. doi: 10.1104/pp.110.167569
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., et al. (2003). Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Garmier, M., Gentzbittel, L., Wen, J., Mysore, K. S., and Ratet, P. (2017). M. truncatula: genetic and genomic resources. Curr. Protoc. Plant Biol. 2, 318–349. doi: 10.1002/cppb.20058
Gaj, M. D. (2001). Direct somatic embryogenesis as a rapid and efficient system for in vitro regeneration of Arabidopsis thaliana. Plant Cell Tissue Organ Cult. 64, 39–46. doi: 10.1023/A:1010679614721
Gaj, M. D., Zhang, S., Harada, J. J., and Lemaux, P. G. (2005). Leafy cotyledon genes are essential for induction of somatic embryogenesis of Arabidopsis. Planta 222, 977–988. doi: 10.1007/s00425-005-0041-y
Gordon, S. P., Heisler, M. G., Reddy, G. V., Ohno, C., Das, P., and Meyerowitz, E. M. (2007). Pattern formation during de novo assembly of the Arabidopsis shoot meristem. Development 134, 3539–3548. doi: 10.1242/dev.010298
Grafi, G., and Barak, S. (2015). Stress induces cell dedifferentiation in plants. Biochim. Biophys. Acta, Gene Regul. Mech. 1849, 378–384. doi: 10.1016/j.bbagrm.2014.07.015
Groß-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. doi: 10.1101/gad.225202
Haccius, B. (1978). Question of unicellular origin of nonzygotic embryos in callus cultures. Phytomorphology 28, 74–81.
Halperin, W. (1964). Morphogenetic studies with partially synchronized cultures of carrot embryos. Science 146, 408–410. doi: 10.1126/science.146.3642.408
Harding, E. W., Tang, W., Nichols, K. W., Fernandez, D. E., and Perry, S. E. (2003). Expression and maintenance of embryogenic potential is enhanced through constitutive expression of AGAMOUS-Like 15. Plant Physiol. 133, 653–663. doi: 10.1104/pp.103.023499
He, C., Chen, X., Huang, H., and Xu, L. (2012). Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet. 8:e1002911. doi: 10.1371/journal.pgen.1002911
Hecht, V., Vielle-Calzada, J. P., Hartog, M. V., Schmidt, E. D., Boutilier, K., Grossniklaus, U., et al. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. doi: 10.1104/pp.010324
Heringer, A. S., Santa-Catarina, C., and Silveira, V. (2018). Insights from proteomic studies into plant somatic embryogenesis. Proteomics 18:1700265. doi: 10.1002/pmic.201700265
Hernandez-Fernandez, M. M., and Christie, B. R. (1989). Inheritance of somatic embryogenesis in alfalfa (M. sativa L.). Genome 32, 318–321. doi: 10.1139/g89-447
Hoffmann, B., Trinh, T. H., Leung, J., Kondorosi, A., and Kondorosi, E. (1997). A new M. truncatula line with superior in vitro regeneration, transformation, and symbiotic properties isolated through cell culture selection. Mol. Plant-Microbe Interact. 10, 307–315. doi: 10.1094/MPMI.1997.10.3.307
Horstman, A., Bemer, M., and Boutilier, K. (2017a). A transcriptional view on somatic embryogenesis. Regeneration 4, 201–216. doi: 10.1002/reg2.91
Horstman, A., Li, M., Heidmann, I., Weemen, M., Chen, B., Muino, J. M., et al. (2017b). The BABY BOOM transcription factor activates the LEC1-ABI3-FUS3-, network to induce somatic embryogenesis. Plant Physiol. 175, 848–857. doi: 10.1104/pp.17.00232
Iantcheva, A., Revalska, M., Zehirov, G., and Vassileva, V. (2014). Agrobacterium-mediated transformation of M. truncatula cell suspension culture provides a system for functional analysis. In Vitro Cell. Dev. Biol. Plant 50, 149–157. doi: 10.1007/s11627-013-9554-4
Igielski, R., and Kępczyńska, E. (2017). Gene expression and metabolite profiling of gibberellin biosynthesis during induction of somatic embryogenesis in M. truncatula Gaertn. PLoS One 12:e0182055. doi: 10.1371/journal.pone.018205
Ikeda-Iwai, M., Satoh, S., and Kamada, H. (2002). Establishment of a reproducible tissue culture system for the induction of Arabidopsis somatic embryos. J. Exp. Bot. 53, 1575–1580. doi: 10.1093/jxb/erf006
Ikeuchi, M., Iwase, A., Rymen, B., Lambolez, A., Kojima, M., Takebayashi, Y., et al. (2017). Wounding triggers callus formation via dynamic hormonal and transcriptional changes. Plant Physiol. 175, 1158–1174. doi: 10.1104/pp.17.01035
Imin, N., Goffard, N., Nizamidin, M., and Rolfe, B. G. (2008). Genome-wide transcriptional analysis of super-embryogenic M. truncatula explant cultures. BMC Plant Biol. 8:110. doi: 10.1186/1471-2229-8-110
Imin, N., Nizamidin, M., Daniher, D., Nolan, K. E., Rose, R. J., and Rolfe, B. G. (2005). Proteomic analysis of somatic embryogenesis in M. truncatula explant cultures grown under 6-benzylaminopurine and 1-naphthaleneacetic acid treatments. Plant Physiol. 137, 1250–1260. doi: 10.1104/pp.104.055277
Imin, N., Nizamidin, M., Wu, T., and Rolfe, B. G. (2007). Factors involved in root formation in M. truncatula. J. Exp. Bot. 58, 439–451. doi: 10.1093/jxb/erl224
Irwanto, R. R., and Rose, R. J. (2008). AFLP and AMP fingerprints as markers to evaluate genetic differences between M. truncatula line Jemalong and 2HA, a new line produced by in vitro culture selection. J. Math. Fundam. Sci. 40, 138–150. doi: 10.5614%2Fitbj.sci.2008.40.2.4
Iwase, A., Mitsuda, N., Koyama, T., Hiratsu, K., Kojima, M., Arai, T., et al. (2011). The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr. Biol. 21, 508–514. doi: 10.1016/j.cub.2011.02.020
Jenik, P. D., and Barton, M. K. (2005). Surge and destroy: the role of auxin in plant embryogenesis. Development 132, 3577–3585. doi: 10.1242/dev.01952
Kadokura, S., Sugimoto, K., Tarr, P., Suzuki, T., and Matsunaga, S. (2018). Characterization of somatic embryogenesis initiated from the Arabidopsis shoot apex. Dev. Biol. 442, 13–27. doi: 10.1016/j.ydbio.2018.04.023
Karami, O., and Saidi, A. (2010). The molecular basis for stress-induced acquisition of somatic embryogenesis. Mol. Biol. Rep. 7, 2493–2507. doi: 10.1007/s11033-009-9764-3
Karlova, R., Boeren, S., Russinova, E., Aker, J., Vervoort, J., and de Vries, S. (2006). The Arabidopsis somatic embryogenesis receptor-like kinase1 protein complex includes brassinosteroid-insensitive1. Plant Cell 18, 626–638. doi: 10.1105/tpc.105.039412
Kępczyńska, E., Ruduś, I., and Kępczyński, J. (2009). Endogenous ethylene in indirect somatic embryogenesis of M. sativa L. Plant Growth Regul. 59, 63–73. doi: 10.1007/s10725-009-9388-6
Kępczyńska, E., and Zielińska, S. (2011). Disturbance of ethylene biosynthesis and perception during somatic embryogenesis in M. sativa L. reduces embryos’ ability to regenerate. Acta Physiol. Plant. 33, 1969–1980. doi: 10.1007/s11738-011-0745-5
Kielly, G. A., and Bowley, S. R. (1992). Genetic control of somatic embryogenesis in alfalfa. Genome 35, 474–477. doi: 10.1139/g92-070
Kooke, R., Johannes, F., Wardenaar, R., Becker, F., Etcheverry, M., Colot, V., et al. (2015). Epigenetic basis of morphological variation and phenotypic plasticity in Arabidopsis thaliana. Plant Cell 27, 337–348. doi: 10.1105/tpc.114.133025
Kurdyukov, S., Mathesius, U., Nolan, K. E., Sheahan, M. B., Goffard, N., Carroll, B. J., et al. (2014a). The 2HA line of M. truncatula has characteristics of an epigenetic mutant that is weakly ethylene insensitive. BMC Plant Biol. 14:174. doi: 10.1186/1471-2229-14-174
Kurdyukov, S., Song, Y., Sheahan, M. B., and Rose, R. J. (2014b). Transcriptional regulation of early embryo development in the model legume M. truncatula. Plant Cell Rep. 33, 349–362. doi: 10.1007/s00299-013-1535-x
Kurczyńska, E. U., Gaj, M. D., Ujczak, A., and Mazur, E. (2007). Histological analysis of direct somatic embryogenesis in Arabidopsis thaliana (L.) Heynh. Planta 226, 619–628. doi: 10.1007/s00425-007-0510-6
Li, J., Dai, X., Liu, T., and Zhao, P. X. (2012). LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 40, D1221–D1229. doi: 10.1093/nar/gkr939
Li, W., Liu, H., Cheng, Z. J., Su, Y. H., Han, H. N., Zhang, Y., et al. (2011). DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7:e1002243. doi: 10.1371/journal.pgen.1002243
Liu, Y., and He, C. (2016). Regulation of plant reactive oxygen species (ROS) in stress responses: learning from AtRBOHD. Plant Cell Rep. 35, 995–1007. doi: 10.1007/s00299-016-1950-x
Lotan, T., Ohto, M. A., Yee, K. M., West, M. A. L., Lo, R., Kwong, R. W., et al. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in somatic cells. Cell 93, 1195–1205. doi: 10.1016/S0092-8674(00)81463-4
Lowe, K., Wu, E., Wang, N., Hoerster, G., Hastings, C., Cho, M. J., et al. (2016). Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 28, 1998–2015. doi: 10.1105/tpc.16.00124
Mahdavi-Darvari, F., Noor, N. M., and Ismanizan, I. (2015). Epigenetic regulation and gene markers as signals of early somatic embryogenesis. Plant Cell Tissue Organ Cult. 120, 407–422. doi: 10.1007/s11240-014-0615-0
Mantiri, F. R., Kurdyukov, S., Lohar, D. P., Sharopova, N., Saeed, N. A., Wang, X. D., et al. (2008a). The transcription factor MtSERF1 of the ERF subfamily identified by transcriptional profiling is required for somatic embryogenesis induced by auxin plus cytokinin in M. truncatula. Plant Physiol. 146, 1622–1636. doi: 10.1104/pp.107.110379
Mantiri, F. R., Kurdyukov, S., Chen, S. K., and Rose, R. J. (2008b). The transcription factor MtSERF1 may function as a nexus between stress and development in somatic embryogenesis in M. truncatula. Plant Signal. Behav. 3, 498–500. doi: 10.4161/psb.3.7.6049
Marín-de la Rosa, N., Sotillo, B., Miskolczi, P., Gibbs, D. J., Vicente, J., Carbonero, P., et al. (2014). Large-scale identification of gibberellin-related transcription factors defines group VII ETHYLENE RESPONSE FACTORS as functional DELLA partners. Plant Physiol. 166, 1022–1032. doi: 10.1104/pp.114.244723
Mayer, K. F., Schoof, H., Haecker, A., Lenhard, M., Jürgens, G., and Laux, T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815. doi: 10.1016/S0092-8674(00)81703-1
Mordhorst, A. P., Voerman, K. J., Hartog, M. V., Meijer, E. A., van Went, J., Koornneef, M., et al. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563.
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Ng, L. M., Melcher, K., Teh, B. T., and Xu, H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35, 567–584. doi: 10.1038/aps.2014.5
Nolan, K. E., and Rose, R. J. (1998). Plant regeneration from cultured M. truncatula with particular reference to abscisic acid and light treatments. Aust. J. Bot. 46, 151–160. doi: 10.1071/BT96138
Nolan, K. E., Irwanto, R. R., and Rose, R. J. (2003). Auxin up-regulates MtSERK1 expression in both M. truncatula root-forming and embryogenic cultures. Plant Physiol. 133, 218–230. doi: 10.1104/pp.103.020917
Nolan, K. E., Kurdyukov, S., and Rose, R. J. (2009). Expression of the SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 (SERK1) gene is associated with developmental change in the life cycle of the model legume M. truncatula. J. Exp. Bot. 60, 1759–1771. doi: 10.1093/jxb/erp046
Nolan, K. E., Rose, R. J., and Gorst, J. R. (1989). Regeneration of M. truncatula from tissue culture: increased somatic embryogenesis using explants from regenerated plants. Plant Cell Rep. 8, 278–281. doi: 10.1007/BF00274129
Nolan, K. E., Saeed, N. A., and Rose, R. J. (2006). The stress kinase gene MtSK1 in M. truncatula with particular reference to somatic embryogenesis. Plant Cell Rep. 25, 711–722. doi: 10.1007/s00299-006-0135-4
Nolan, K. E., Song, Y., Liao, S., Saeed, N. A., Zhang, X., and Rose, R. J. (2014). An unusual abscisic acid and gibberellic acid synergism increases somatic embryogenesis, facilitates its genetic analysis and improves transformation in M. truncatula. PLoS One 9:e99908. doi: 10.1371/journal.pone.0099908
Ogas, J., Cheng, J. C., Sung, Z. R., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. doi: 10.1126/science.277.5322.91
Orłowska, A., Igielski, R., Łagowska, K., and Kępczyńska, E. (2017). Identification of LEC1, L1L and Polycomb Repressive Complex 2 genes and their expression during the induction phase of M. truncatula Gaertn. somatic embryogenesis. Plant Cell Tissue Organ Cult. 129, 119–132. doi: 10.1007/s11240-016-1161-8
Orłowska, A., and Kępczyńska, E. (2018). Identification of Polycomb Repressive Complex1, Trithorax group genes and their simultaneous expression with WUSCHEL, WUSCHEL-related Homeobox5 and SHOOT MERISTEMLESS during the induction phase of somatic embryogenesis in M. truncatula Gaertn. Plant Cell Tissue Organ Cult. 134, 345–356. doi: 10.1007/s11240-018-1425-6
Pandey, D. K., and Chaudhary, B. (2014). Oxidative stress responsive SERK1 gene directs the progression of somatic embryogenesis in cotton (Gossypium hirsutum L. cv. Coker 310). Am. J. Plant Sci. 5, 80–102. doi: 10.4236/ajps.2014.51012
Pasternak, T. P., Prinsen, E., Ayaydin, F., Miskolczi, P., Potters, G., Asard, H., et al. (2002). The role of auxin, pH, and stress in the activation of embryogenic cell division in leaf protoplast-derived cells of alfalfa. Plant Physiol. 129, 1807–1819. doi: 10.1104/pp.000810
Podgórska, A., Burian, M., and Szal, B. (2017). Extra-cellular but extra-ordinarily important for cells: apoplastic reactive oxygen species metabolism. Front. Plant Sci. 8:1353. doi: 10.3389/fpls.2017.01353
Qu, Y., Wang, Q., Guo, J., Wang, P., Song, P., Jia, Q., et al. (2017). Peroxisomal CuAOζ and its product H2O2 regulate the distribution of auxin and IBA-dependent lateral root development in Arabidopsis. J. Exp. Bot. 68, 4851–4867. doi: 10.1093/jxb/erx290
Reisch, B., and Bingham, E. T. (1980). The genetic control of bud formation from callus cultures of diploid alfalfa. Plant Sci. Lett. 20, 71–77. doi: 10.1016/0304-4211(80)90071-1
Rocha, D. I., Pinto, D. L. P., Vieira, L. M., Tanaka, F. A. O., Dornelas, M. C., and Otoni, W. C. (2016). Cellular and molecular changes associated with competence acquisition during passion fruit somatic embryogenesis: ultrastructural characterization and analysis of SERK gene expression. Protoplasma 253, 595–609. doi: 10.1007/s00709-015-0837-y
Rose, R. J. (2004). “Somatic embryogenesis in plants” in Encyclopedia of plant and crop science. ed. R. M. Goodman (New York: Marcel Dekker), 1165–1168.
Rose, R. J. (2008). M. truncatula as a model for understanding plant interactions with other organisms, plant development and stress biology: past, present and future. Funct. Plant Biol. 35, 253–264. doi: 10.1071/FP07297
R. J. Rose (ed.) (2013). Legume genomics, methods and protocols. Methods in molecular biology, Vol. 1069. New York: Humana Press, Springer.
Rose, R. J. (2016). “Genetic reprogramming of plant cells in vitro via dedifferentiation or pre-existing stem cells” in Molecular cell biology of the growth and differentiation of plant cells. ed. R. J. Rose (Boca Raton, London, NY: CRC Press), 320–339.
Rose, R. J., and McCurdy, D. W. (2017). New beginnings: mitochondrial renewal by massive mitochondrial fusion. Trends Plant Sci. 22, 641–643. doi: 10.1016/j.tplants.2017.06.005
Rose, R. J., and Nolan, K. E. (1995). Regeneration of M. truncatula from protoplasts isolated from kanamycin-sensitive and kanamycin-resistant plants. Plant Cell Rep. 14, 349–353. doi: 10.1007/BF00238595
Rose, R. J., and Nolan, K. E. (2006). Genetic regulation of somatic embryogenesis with particular reference to Arabidopsis thaliana and M. truncatula. In Vitro Cell. Dev. Biol. Plant 42, 473–481. doi: 10.1079/ivp2006806
Rose, R. J., Nolan, K. E., and Bicego, L. (1999). The development of the highly regenerable seed line Jemalong 2HA for transformation of M. truncatula - implications for regenerability via somatic embryogenesis. J. Plant Physiol. 155, 788–791. doi: 10.1016/S0176-1617(99)80097-2
Rose, R. J., Sheahan, M. B., and Tiew, T. W.-Y. (2013). “Connecting stress to development in the induction of somatic embryogenesis” in Somatic embryogenesis and gene expression. eds. J. Aslam, P. S. Srivastava, and M. P. Sharma (NewDelhi: Narosa Publishing House), 146–165.
Rose, R. J., and Song, Y. (2018). “The Jemalong 2HA line used for M. truncatula transformation: hormonology and epigenetics” in The model legume M. truncatula. ed. F. J. de Bruijn (New Jersey: Wiley). In press.
Rose, R. J., Wang, X.-D., Nolan, K. E., and Rolfe, B. G. (2006). Root meristems in M. truncatula tissue culture arise from vascular-derived procambial-like cells in a process regulated by ethylene. J. Exp. Bot. 57, 2227–2235. doi: 10.1093/jxb/erj187
Ruduś, I., Kępczyńska, E., and Kępczyński, J. (2002). Regulation of M. sativa L. somatic embryogenesis by gibberellins. Plant Growth Regul. 36, 91–95. doi: 10.1023/A:1014751125297
Ruduś, I., Weiler, E. W., and Kępczyńska, E. (2009). Do stress-related phytohormones, abscisic acid and jasmonic acid play a role in the regulation of M. sativa L. somatic embryogenesis? Plant Growth Regul. 59, 159–169. doi: 10.1007/s10725-009-9399-3
Santos, D., and Fevereiro, P. (2002). Loss of DNA methylation affects somatic embryogenesis in M. truncatula. Plant Cell Tissue Organ Cult. 70, 155–161. doi: 10.1023/A:1016369921067
Santos-Mendoza, M., Dubreucq, B., Baud, S., Parcy, F., Caboche, M., and Lepiniec, L. (2008). Deciphering gene regulatory networks that control seed development and maturation in Arabidopsis. Plant J. 54, 608–620. doi: 10.1111/j.1365-313X.2008.03461.x
Sarkar, A. K., Luijten, M., Miyashima, S., Lenhard, M., Hashimoto, T., Nakajima, K., et al. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814. doi: 10.1038/nature05703
Schmidt, E. D. L., Guzzo, F., Toonen, M. A. J., and DeVries, S. C. (1997). A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124, 2049–2062.
Schuettengruber, B., Martinez, A. M., Iovino, N., and Cavalli, G. (2011). Trithorax group proteins: switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 12, 799–814. doi: 10.1038/nrm3230
Scofield, S., Dewitte, W., and Murray, J. A. (2014). STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal. Behav. 9:e28934. doi: 10.4161/psb.28934
Sheahan, M. B., McCurdy, D. W., and Rose, R. J. (2005). Mitochondria as a connected population: ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 44, 744–755. doi: 10.1111/j.1365-313X.2005.02561.x
Soares, N. C., Francisco, R., Vielba, J. M., Ricardo, C. P., and Jackson, P. A. (2009). Associating wound-related changes in the apoplast proteome of Medicago with early steps in the ROS signal-transduction pathway. J. Proteome Res. 8, 2298–2309. doi: 10.1021/pr8009353
Soma, F., Mogami, J., Yoshida, T., Abekura, M., Takahashi, F., Kidokoro, S., et al. 2017. ABA-unresponsive SnRK2 protein kinases regulate mRNA decay under osmotic stress in plants. Nat. Plants 3:16204. doi: 10.1038/nplants.2016.204
Song, Y. J., Joo, J. H., Ryu, H. Y., Lee, J. S., Bae, Y. S., and Nam, K. H. (2007). Reactive oxygen species mediate IAA-induced ethylene production in mungbean (Vigna radiata L.) hypocotyls. J. Plant Biol. 50, 18–23. doi: 10.1007/BF03030595
Stone, S. L., Braybrook, S. A., Paula, S. L., Kwong, L. W., Meuser, J., Pelletier, J., et al. (2008). Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. Proc. Natl. Acad. Sci. USA 105, 3151–3156. doi: 10.1073/pnas.0712364105
Stone, S. L., Kwong, L. W., Yee, K. M., Pelletier, J., Lepiniec, L., Fischer, R. L., et al. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98, 11806–11811. doi: 10.1073/pnas/201413498
Su, Y. H., Zhao, X. Y., Liu, Y. B., Zhang, C. L., O’Neill, S. D., and Zhang, X. S. (2009). Auxin-induced WUS expression is essential for embryonic stem cell renewal during somatic embryogenesis in Arabidopsis. Plant J. 59, 448–460. doi: 10.1111/j.1365-313X.2009.03880.x
Sugimoto, K., Jiao, Y., and Meyerowitz, E. M. (2010). Arabidopsis regeneration from multiple tissues occurs via a root development pathway. Dev. Cell 18, 463–471. doi: 10.1016/j.devcel.2010.02.004
Tang, H., Krishnakumar, V., Bidwell, S., Rosen, B., Chan, A., Zhou, S., et al. (2014). An improved genome release (version Mt4.0) for the model legume M. truncatula. BMC Genomics 15:312. doi: 10.1186/1471-2164-15-312
ten Hove, C. A., Lu, K.-J., and Weijers, D. (2015). Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142, 420–430. doi: 10.1242/dev.111500
Thibaud-Nissen, F., Shealy, R. T., Khanna, A., and Vodkin, L. O. (2003). Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol. 132, 118–136. doi: 10.1104/pp.103.019968
Tiew, T. W.-Y. (2015). The influence of peroxisomes on reactive oxygen species regulation and regeneration induction in plants. Ph.D. thesis. Newcastle (Australia): The University of Newcastle.
Tiew, T. W.-Y., Sheahan, M. B., and Rose, R. J. (2015). Peroxisomes contribute to reactive oxygen species homeostasis and cell division induction in Arabidopsis protoplasts. Front. Plant Sci. 6:658. doi: 10.3389/fpls.2015.00658
Wang, H., Caruso, L. V., Downie, A. B., and Perry, S. E. (2004). The embryo MADS domain protein AGAMOUS-Like 15 directly regulates expression of a gene encoding an enzyme involved in gibberellin metabolism. Plant Cell 16, 1206–1219. doi: 10.1105/tpc.021261
Wang, X. D., Nolan, K. E., Irwanto, R. R., Sheahan, M. B., and Rose, R. J. (2011). Ontogeny of embryogenic callus in M. truncatula: the fate of the pluripotent and totipotent stem cells. Ann. Bot. 107, 599–609. doi: 10.1093/aob/mcq269
Williams, E. G., and Maheswaran, G. (1986). Somatic embryogenesis: factors influencing coordinated behaviour of cells as an embryogenic group. Ann. Bot. 57, 443–462. doi: 10.1093/oxfordjournals.aob.a087127
Wójcikowska, B., Jaskóła, K., Gąsiorek, P., Meus, M., Nowak, K., and Gaj, M. D. (2013). LEAFY COTYLEDON2 (LEC2) promotes embryogenic induction in somatic tissues of Arabidopsis, via YUCCA-mediated auxin biosynthesis. Planta 238, 425–440. doi: 10.1007/s00425-013-1892-2
Young, N. D., Debellé, F., Oldroyd, G. E., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Zavattieri, M. A., Frederico, A. M., Lima, M., Sabino, R., and Arnholdt-Schmitt, B. (2010). Induction of somatic embryogenesis as an example of stress-related plant reactions. Electron J. Biotechnol. 13, 1–9. doi: 10.2225/vol13-issue1-fulltext-4
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., and Ogas, J. (2012). The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, H., Rider, S. D., Henderson, J. T., Fountain, M., Chuang, K., Kandachar, V., et al. (2008). The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J. Biol. Chem. 283, 22637–22648. doi: 10.1074/jbc.M802129200
Zhang, H., Zhang, T. T., Liu, H., Shi, D. Y., Wang, M., Bie, X. M., et al. (2018). Thioredoxin-mediated ROS homeostasis explains natural variation in plant regeneration. Plant Physiol. 176, 2231–2259. doi: 10.1104/pp.17.00633
Zhao, J., Morozova, N., Williams, L., Libs, L., Avivi, Y., and Grafi, G. (2001). Two phases of chromatin decondensation during dedifferentiation of plant cells: distinction between competence for cell fate switch and a commitment for S phase. J. Biol. Chem. 276, 22772–22778. doi: 10.1074/jbc.M101756200
Zheng, Q., and Perry, S. E. (2014). Alterations in the transcriptome of soybean in response to enhanced somatic embryogenesis promoted by orthologs of AGAMOUS-Like15 and AGAMOUS-Like18. Plant Physiol. 164, 1365–1377. doi: 10.1104/pp.113.234062
Zheng, W., Zhang, X., Yang, Z., Wu, J., Li, F., Duan, L., et al. (2014). AtWuschel promotes formation of the embryogenic callus in Gossypium hirsutum. PLoS One 9:e87502. doi: 10.1371/journal.pone.0087502
Zheng, Y., Ren, N., Wang, H., Stromberg, A. J., and Perry, S. E. (2009). Global identification of targets of the Arabidopsis MADS domain protein AGAMOUS-Like15. Plant Cell 21, 2563–2577. doi: 10.1105/tpc.109.068890
Zheng, Q., Zheng, Y., and Perry, S. E. (2013). AGAMOUS-Like15 promotes somatic embryogenesis in Arabidopsis and soybean in part by the control of ethylene biosynthesis and response. Plant Physiol. 161, 2113–2127. doi: 10.1104/pp.113.216275
Zheng, Q., Zheng, Y., Ji, H., Burnie, W., and Perry, S. E. (2016). Gene regulation by the AGL15 transcription factor reveals hormone interactions in somatic embryogenesis. Plant Physiol. 172, 2374–2387. doi: 10.1104/pp.16.00564
Zhou, T., Yang, X., Guo, K., Deng, J., Xu, J., Gao, W., et al. (2016). ROS homeostasis regulates somatic embryogenesis via the regulation of auxin signaling in cotton. Mol. Cell. Proteomics 15, 2108–2124. doi: 10.1074/mcp.M115.049338
Keywords: Auxin, cytokinin, ethylene, kinases, M. truncatula, somatic embryogenesis, totipotency, transcription factors
Citation: Rose RJ (2019) Somatic Embryogenesis in the Medicago truncatula Model: Cellular and Molecular Mechanisms. Front. Plant Sci. 10:267. doi: 10.3389/fpls.2019.00267
Received: 14 June 2018; Accepted: 19 February 2019;
Published: 19 March 2019.
Edited by:
Sandra Isabel Correia, University of Coimbra, PortugalReviewed by:
Atanas Ivanov Atanassov, Joint Genomic Center, BulgariaCopyright © 2019 Rose. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this�journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ray J. Rose, UmF5LlJvc2VAbmV3Y2FzdGxlLmVkdS5hdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.