
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 28 February 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00228
This article is part of the Research TopicConstruction of Regulatory Networks in Plant Abiotic Stress ResponsesView all 4 articles
Dynamic environmental changes such as extreme temperature, water scarcity and high salinity affect plant growth, survival, and reproduction. Plants have evolved sophisticated regulatory mechanisms to adapt to these unfavorable conditions, many of which interface with plant hormone signaling pathways. Abiotic stresses alter the production and distribution of phytohormones that in turn mediate stress responses at least in part through hormone- and stress-responsive transcription factors. Among these, the APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family transcription factors (AP2/ERFs) have emerged as key regulators of various stress responses, in which they also respond to hormones with improved plant survival during stress conditions. Apart from participation in specific stresses, AP2/ERFs are involved in a wide range of stress tolerance, enabling them to form an interconnected stress regulatory network. Additionally, many AP2/ERFs respond to the plant hormones abscisic acid (ABA) and ethylene (ET) to help activate ABA and ET dependent and independent stress-responsive genes. While some AP2/ERFs are implicated in growth and developmental processes mediated by gibberellins (GAs), cytokinins (CTK), and brassinosteroids (BRs). The involvement of AP2/ERFs in hormone signaling adds the complexity of stress regulatory network. In this review, we summarize recent studies on AP2/ERF transcription factors in hormonal and abiotic stress responses with an emphasis on selected family members in Arabidopsis. In addition, we leverage publically available Arabidopsis gene networks and transcriptome data to investigate AP2/ERF regulatory networks, providing context and important clues about the roles of diverse AP2/ERFs in controlling hormone and stress responses.
Abiotic stresses such as water scarcity, extreme temperature and high salinity lead to arrested plant growth and ultimately result in massive agricultural losses (Fahad et al., 2017). Plants have developed sophisticated regulatory mechanisms to respond to external stress signals in a timely manner to ensure optimal growth and stress tolerance. Among these, hormonal signaling pathways and stress responsive transcriptional factors function together to form an interconnected network (Chen et al., 2017; Ye et al., 2017). In addition, environmental changes are often multifactorial, with several stresses occurring simultaneously. Instead of the linear stress signaling pathways, these regulatory components lead to more complex responses (Van den Broeck et al., 2017). Under abiotic stress conditions, stress hormones such as abscisic acids (ABA) and ethylene (ET) are induced, whereas the production and distribution of growth promoting hormones, such as gibberellins (GAs), brassinosteroids (BRs), and cytokinins (CTK) are also altered to enable optimal responses (Verma et al., 2016). The regulation of plant hormone signaling during abiotic stresses is partially mediated by hormone- and stress-responsive transcription factors (Nolan T. et al., 2017; Bechtold and Field, 2018).
APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family transcription factors (AP2/ERFs) have emerged as key regulators of several abiotic stresses and respond to multiple hormones (Dietz et al., 2010; Mizoi et al., 2012; Chandler, 2018). Numerous AP2/ERFs mutants with altered abiotic stress responses and hormone sensitivity have been identified, positioning this family of transcription factors as promising candidates to study the interactions between abiotic stresses and hormones. Several properties of AP2/ERFs, such as induction upon specific stresses and diverse DNA binding preferences, enable these transcription factors to integrate responses of multiple stimuli and participate in different regulatory processes. In this review, we focus on Arabidopsis thaliana to summarize the regulation and function of AP2/ERFs in hormone and abiotic stress responses. The function of AP2/ERFs in crops has been extensively reviewed elsewhere (Abiri et al., 2017; Kulkarni et al., 2017; Phukan et al., 2017). Thus, only a few examples of AP2/ERFs in other species will be discussed. Finally, we investigate AP2/ERFs regulatory networks using publicly available transcriptome data to verify known and uncover novel roles of AP2/ERFs in hormone and stress responses.
AP2/ERFs are characterized by an APETALA2 (AP2)/Ethylene Responsive Element Binding Factor (EREB) domain, which consists of 40–70 conserved amino acids involved in DNA binding (Sakuma et al., 2002; Feng et al., 2005; Nakano et al., 2006). AP2/ERFs contain the four major subfamilies: APETALA2 (AP2), RELATED TO ABSCISIC ACID INSENSITIVE 3/VIVIPAROUS 1 (RAV), DEHYDRATION-RESPONSIVE ELEMENT BINDING proteins (DREBs) (subgroup A1–A6) and ETHYLENE RESPONSIVE FACTORS (ERFs) (subgroup V-X) (Sakuma et al., 2002; Nakano et al., 2006). As transcription factors, AP2/ERFs regulate genes involved in diverse biological processes such as growth, development, hormone and stress responses through several mechanisms including transcriptional and post-translational control (Dietz et al., 2010; Mizoi et al., 2012; Licausi et al., 2013; Gibbs et al., 2015; Chandler, 2018).
AP2/ERFs expression is tightly regulated to enable proper stress responses. Gene expression profiling studies have shown that most AP2/ERFs are expressed at low levels under normal conditions, whereas the expression can be induced or repressed at certain growth stages, by hormones and stress stimuli (Feng et al., 2005; Li et al., 2017a; Owji et al., 2017). In many cases, AP2/ERFs expression is regulated by the conserved cis-elements present in their promoter regions, or a combination of multiple responsive elements (Figure 1 left). For example, Arabidopsis DREB2A is highly induced under dehydration and heat conditions (Liu et al., 1998). The expression of DREB2A is controlled by HEAT SHOCK FACTOR1 (HSFA1) and ABRE-BINDING PROTEINs/ABRE-BINDING FACTOR 3 (AREB1/AREB2/ABF3) through binding to Heat Shock Element (HSE) and ABA Response Element (ABRE) motifs on its promoter, respectively (Kim et al., 2011; Liu et al., 2011). Moreover, by aligning 20 Arabidopsis DREB gene promoters to the motifs related to abiotic stresses, it was found that DREB1A/CBF3, DREB1C/CBF2, DREB2C, DREB2G, and DEAR3 have the most types of these motifs including HSE motif to which heat shock factors bind, Low-Temperature Responsive element (LTR) that is important for the induction of cold regulated genes, and ABRE motif that responds to ABA (Sazegari et al., 2015). AP2/ERFs whose promoters contain ethylene-responsive (EBS) motifs are also likely to respond to ET signaling (Zhang et al., 2011).
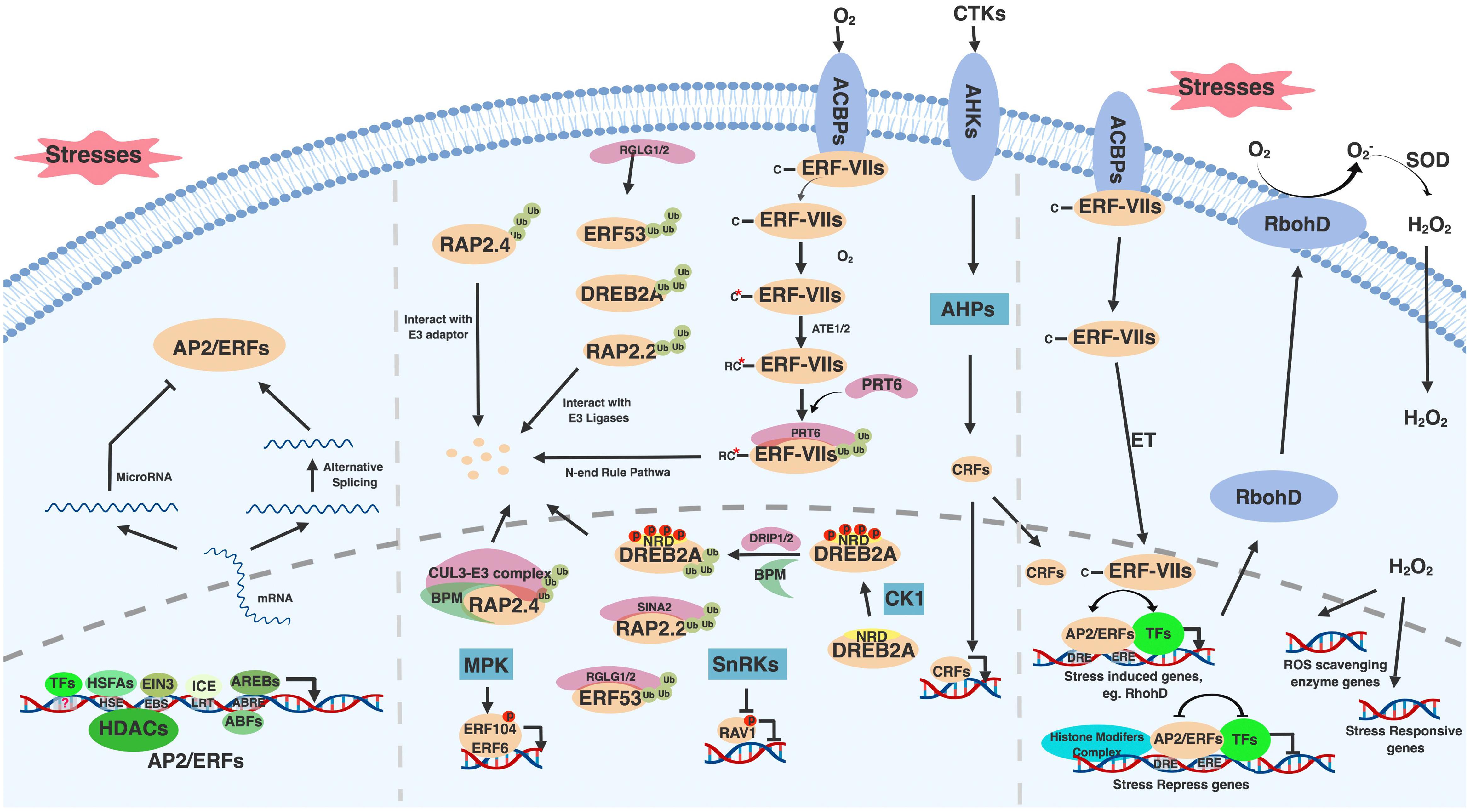
Figure 1. General regulatory mechanisms of AP2/ERF family transcription factors. AP2/ERFs are regulated by multiple stresses and stimuli at transcription, translation and protein modification levels. Upon stresses, AP2/ERFs are induced though cis-elements presented in their promoter regions (left bottom). These cis-elements include HSE, EBS, LRT, ABRE and many other unknown binding sites that respond to HSFAs, EIN3, ICE, AREBs and other transcription factors, respectively. The transcription of AP2/ERFs is also regulated by HDACs. Alternative splicing helps generate AP2/ERFs functional mRNA (left). Under normal condition, AP2/ERFs have adverse effect on plant growth and development and thus need to be eliminated. miroRNA mediated AP2/ERF silence is one of the ways to inhibit AP2/ERFs translation (left). E3 ubiquitin ligases involved proteasome degradation and phosphorylation mediated by kinases provide additional ways to regulate AP2/ERFs protein levels and activity (middle). These E3 ligases include DRIP1/2, RGLG1/2, SINA2, and PRT6, which mediate DREB2A, ERF53, RAP2.2, and ERF-VIIs degradation, respectively. ERF-VIIs undergo N-end rule mediated degradation under normal condition. BPM provides RAP2.4 docking adaptor for CUL3-E3 complex. MPKs and SnRKs mediated phosphorylation activates and represses ERF104 and RAV1 function, respectively. Under stresses, AP2/ERFs including CRFs and ERF-VIIs translocate into nucleus, bind to conserved or diverged DNA binding sites, interact with many other transcription factors and histone modification complex to either activate or repress stress responsive genes expression (right). Among stress induced genes, RbohD is responsive for ROS and H2O2 generation. Induced H2O2 serves as a signal messenger to active ROS scavenging enzyme genes and stress responsive genes expression. Figure is created with BioRender.
Moreover, AP2/ERFs expression might be affected by histone modifications. Histone modifications such as phosphorylation, ubiquitination, methylation, and acetylation can either activate or repress transcription by creating more “open” or “closed” chromatin configurations (Pfluger and Wagner, 2007). A study in peanut (Arachis hypogaea L.) showed that the general inhibition of histone deacetylases (HDACs) and Polyethylene Glycol (PEG) treatment induced the acetylation around peanut DREB1 (AhDREB1) promoter region and resulted in increased AhDREB1 expression (Zhang B. et al., 2018). Given that chromatin features such as histone variants and post-translational histone modifications are altered by abiotic stresses, and these influences could be inherited to the next generation (Asensi-Fabado et al., 2017), the study of epigenetic regulation of AP2/ERFs will advance our understating the mechanisms controlling AP2/ERFs.
Another point of AP2/ERF regulation occurs at the level of splicing. Alternative splicing is required to create a functional isoform for some AP2/ERFs in rice (Oryza sativa) (OsDREB2A/2B) (Matsukura et al., 2010), maize (Zea mays) (ZmDREB2A) (Qin et al., 2007), wheat (Triticum aestivum) (WDREB2) (Egawa et al., 2006), and barley (Hordeum vulgare) (HvDRF1) (Xue and Loveridge, 2004). In these cases, plants produce an inactive AP2/ERF form containing stop codons before the DNA binding domain during normal conditions, while under stress conditions, the exon with a premature stop codons is excluded to generate a functional transcription factor. In addition, microRNA (miRNA)-mediated RNA silencing and translation repression was shown to regulate AP2/ERFs. miRNA172 targets Arabidopsis AP2 messenger RNA to inhibit its translation (Chen, 2004). Several other miRNAs such as miRNA156 and miRNA838 have also been shown to regulate AP2/ERFs in common bean (Phaseolus vulgaris) (Kavas et al., 2015).
In addition to transcriptional control, post-transcriptional regulation modulates the activity and abundance of AP2/ERF proteins (Figure 1 middle). One of the modifications, phosphorylation, affects AP2/ERF protein transactivity and stability. In Arabidopsis, SNF1-RELATED PROTEIN KINASES (SnRKs), a positive regulator in ABA signaling pathway, interacts with and phosphorylates RAV1 to inhibit its transcriptional repression function (Feng et al., 2014), while ERF104 and ERF6 are phosphorylated by mitogen-activated protein kinases (MPKs) to positively regulate pathogen responses (Bethke et al., 2009; Meng et al., 2013). The stability of Arabidopsis DREB2A is also affected by phosphorylation. DREB2A is destabilized by phosphorylation on its negative regulatory domain (NRD), which is probably mediated by Casein Kinase 1 (CK1) (Mizoi et al., 2018). Indeed, many kinases such as GSK3-like kinase BR-INSENSITIVE 2 (BIN2) and SnRKs have been reported to mediate abiotic stresses (Nolan T. et al., 2017). However, information about how these kinases regulate AP2/ERFs is limited. Therefore, future studies such as phosphoproteomics under abiotic stresses and upon perturbation of specific kinases could help identify AP2/ERF protein modifications, leading to potential strategies to modify the protein activity and stability of AP2/ERFs to confer abiotic stress tolerance.
The protein stability of AP2/ERFs is also regulated by ubiquitin-mediated protein degradation through the 26S proteasome pathway as revealed in Arabidopsis (Figure 1 middle). Under non-stress conditions, DREB2A, ERF53, and ERF75/RAP2.2 proteins are directly ubiquitinated by RING domain E3 ligases DREB2A-INTERACTING PROTEIN1/2 (DRIP1/2), RAlGDS-LIKE (RGLG1/2), and SEVEN IN ABSENTIA OF ARABIDOPSIS2 (SINAT2), respectively (Qin et al., 2008; Cheng et al., 2012; Papdi et al., 2015). A CUL3-based E3 ligase adaptor BTB/POZ AND MATH DOMAIN proteins (BPMs) also mediates DREB2A and ERF59/RAP2.4 degradation (Weber and Hellmann, 2009; Morimoto et al., 2017). Additionally, the stability of several Arabidopsis ERF-VIIs is controlled by the N-end rule pathway, where the N terminal Met (Nt-Met) of ERF71/HRE2, ERF72/RAP2.3, and ERF74/RAP2.12 is removed and the second amino acid Cys (Nt-Cys) is oxidized into cysteine sulfinic/sulfonic acid in an oxygen-dependent manner (Gibbs et al., 2015). This process triggers ubiquitination mediated by the E3 ligase PROTEOLYSIS 6 (PRT6) (Gibbs et al., 2011; Licausi et al., 2011; Abbas et al., 2015). Particularly, RAP2.12 interactes with acyl-CoA binding protein 1/2 (ACBP1/2) and is localized on the plasma membrane under normal conditions. During the limited oxygen conditions, RAP2.12 de-associates from ACBP1/2 and moves into the nucleus by an unknown mechanism (Gibbs et al., 2015). ET also promotes RAP2.3 nuclear localization (Kim et al., 2018). Overall, a number of components have been identified that regulate AP2/ERF stability, localization and activity. Since many AP2/ERFs are involved in stress responses, this tight regulation of AP2/ERF protein likely ensures that plants effectively respond to environmental stimuli without ectopic activation of AP2/ERF-mediated stress responses.
To regulate target genes, AP2/ERFs have conserved DNA binding preferences (Nakano et al., 2006). Typically, DREBs recognize Dehydration-Responsive or C-Repeat Element (DRE/CRT) with A/GCCGAC core sequence on stress-responsive genes to confer resistance to drought, cold and heat abiotic stresses. ERFs bind to Ethylene-Response Element (ERE) with AGCCGCC core sequence (also known as GCC-box) to confer resistance to biotic stresses (Shinozaki and Yamaguchi-Shinozaki, 2000; Guo and Ecker, 2004; Franco-Zorrilla et al., 2014). However, many Arabidpsis DREBs and ERFs have been reported to bind to both DRE/CRT and ERE elements, implying their potential roles in both abiotic and biotic stress. For instance, DREBs (including TINY, CBF1, ERF53, RAP2.4, and TG/RAP2.4A) and ERFs (including ERF1, ERF4, and ERF71) bind to both DRE and ERE elements (Lin et al., 2008; Sun et al., 2008; Yang et al., 2009; Cheng et al., 2012; Zhu et al., 2014; Lee et al., 2015). Similarly, the conserved DNA binding preferences of AP2/ERF are also expanded to other species such as rice (Wan et al., 2011), wheat (Gao et al., 2018), maize (Liu et al., 2013), soybean (Glycine max) (Zhang et al., 2009), and tobacco (Nicotiana tabacum) (Park et al., 2001). Recently, the combination of high-throughput protein-binding microarray and relevant transcriptome data demonstrated that transcription factors with high structure identity share similar DNA binding sites, which also enables them to share some biological relevance and explains their functional redundancy (Franco-Zorrilla et al., 2014). It provides a way to predict unknown AP2/ERFs function. Additionally, AP2/ERFs recognize cis-elements that diverge significantly from these motifs (Table 1). For example, Coupling Element 1 (CE1: TGCCACCG), Coupling Element 3-like (CE3-like: CGCG), Hypoxia-Responsive Promoter Element (HRPE) and non-specific sequences are recognized by AP2/ERFs (Kagaya et al., 1999; Welsch et al., 2007; Shaikhali et al., 2008; Bossi et al., 2009; Zhu et al., 2010; Dinh et al., 2012; Chen et al., 2016; Gasch et al., 2016; Park et al., 2016). Through different partners upon binding to different sites, the diversity of AP2/ERFs DNA binding broadens the scope of target genes and might enable them to participate in different regulatory processes. To date, both in vivo (ChIP-chip and ChIP-seq) and in vitro (DAP-seq, SELEX-seq and Protein-Binding Microarrays (PBMs)) techniques are helpful to decipher the AP2/ERFs transcriptional regulatory code (Franco-Zorrilla et al., 2014; O’Malley et al., 2016; Bartlett et al., 2017).
Through directly binding to target gene promoters, AP2/ERFs can either activate or repress target gene expression (Figure 1 right). Besides an N-terminal DNA binding domain, the C-terminal activation domain of AP2/ERFs mediates the activation of target gene expression in Arabidopsis and rice (Nakano et al., 2006). Recently, an activation EDLL motif was identified from Arabidopsis ERF98 and AP2 subfamily. The EDLL motif is strong enough to override the repression effect mediated by ERF-associated amphiphilic repression (EAR) motif, which emphasizes the transactivation activity of AP2/ERFs (Tiwari et al., 2012). However, AP2/ERFs containing an EAR motif with the consensus sequence LxLxL or DLNxxP, or B3 repression domain (BRD) with R/KLFGV sequence exhibit a repressive effect on target genes (Ikeda and Ohme-Takagi, 2009; Kagale and Rozwadowski, 2011). The EAR motif containing AP2/ERFs also interact and recruit transcription co-repressors like TOPLESS (TPL) and TOPLESS-RELATED (TPR) (Causier et al., 2012) or histone modifiers to suppress target gene expression (Song et al., 2005; Song and Galbraith, 2006). In Arabidopsis, ERF7 interacts with a human global co-repressor SIN3 homolog (ATSIN3) that in turn interacts with Histone Deacetylase 19 (HDA19) (Song et al., 2005). Similarly, ERF3 interacts with SIN3 Associated Polypeptide P18 (SAP18) and then recruits HDA19 to repress gene expression (Song and Galbraith, 2006). The BRD motif containing Arabidopsis RAV1 and RAV2 also display repressive activities (Ikeda and Ohme-Takagi, 2009). There is a report demonstrating that genes co-regulated with corresponding AP2/ERFs were enriched with the AP2/ERFs targets. Therefore, by analyzing the enrichment of cognate motifs in AP2/ERFs co-regulated genes, it is possible to identify putative target genes of transcription factors and predict their biological functions (Franco-Zorrilla et al., 2014).
AP2/ERFs regulate numerous abiotic stresses such as cold, drought, heat, salt, and freezing (Lata and Prasad, 2011; Mizoi et al., 2012; Licausi et al., 2013; Phukan et al., 2017). Although many AP2/ERFs are proposed to form an abiotic stress-specific regulatory network, the ability of AP2/ERFs to respond to multiple stimuli and regulate different stresses enable them to form a more complex stress response network. In this network, AP2/ERFs also respond to abiotic stresses with varying dynamic patterns: some AP2/ERFs are induced quickly and continuously, whereas others are regulated by prolonged stress, which indicates they might have mutual influence on each other’s function (Van den Broeck et al., 2017). However, the detailed mechanisms of how different AP2/ERFs cooperate or antagonize with each other are yet to be established. Therefore, in addition to studying the function of individual transcription factors in this family, it is also necessary to study the relationship between different AP2/ERFs in abiotic stress responses. Here we summarize recent works on the DREBs and ERFs regulation in cold, drought, heat and salt stress responses particularly in Arabidopsis, and we also discuss the mutual regulation of different AP2/ERFs.
DREBs have been extensively examined in abiotic stresses, where they respond to and positively regulate cold, drought, heat and salt tolerance by directly regulating stress-responsive genes (Figure 2). Among these, DREB1s (DREB-A1 subgroup) containing several C-Repeat-Binding Factors (CBFs) play major roles in acquisition of freezing tolerance (Chinnusamy et al., 2003). CBFs together with another major cold responsive transcription factor, Inducer of CBF Expression (ICE), establish a central cold response pathway to activate a majority of DRE containing Cold Responsive Genes (CORs) in Arabidopsis (Zhao et al., 2016; Liu J. et al., 2018). CORs encode Late Embryogenesis Abundant (LEA) proteins and enzymes for sugar metabolism and fatty acid desaturation that provide the protection for plants from cold stress (Maruyama et al., 2009). The roles of Arabidopsis CBFs in stress responses have been characterized by genetics using cbf mutants generated by CRISPR/Cas9 genome editing techniques and CBFs overexpression lines, as well as transcriptome analysis (Park et al., 2015; Zhao et al., 2016). CBFs are also reported to positively regulate plant drought and salt tolerance, which might due to a common set of stress responsive genes (Kasuga et al., 1999; Zhao and Zhu, 2016). Despite having a clear function in cold response pathways, how CBFs regulate different stresses and the mechanisms by which they confer stress tolerance are still unknown. Environmental changes are usually multifactorial and several stresses often occur simultaneously. Therefore, the multiple roles of CBFs in abiotic stresses might be necessary for plants to overcome stresses and it will be interesting to examine how CBFs regulate genes under different stress conditions.
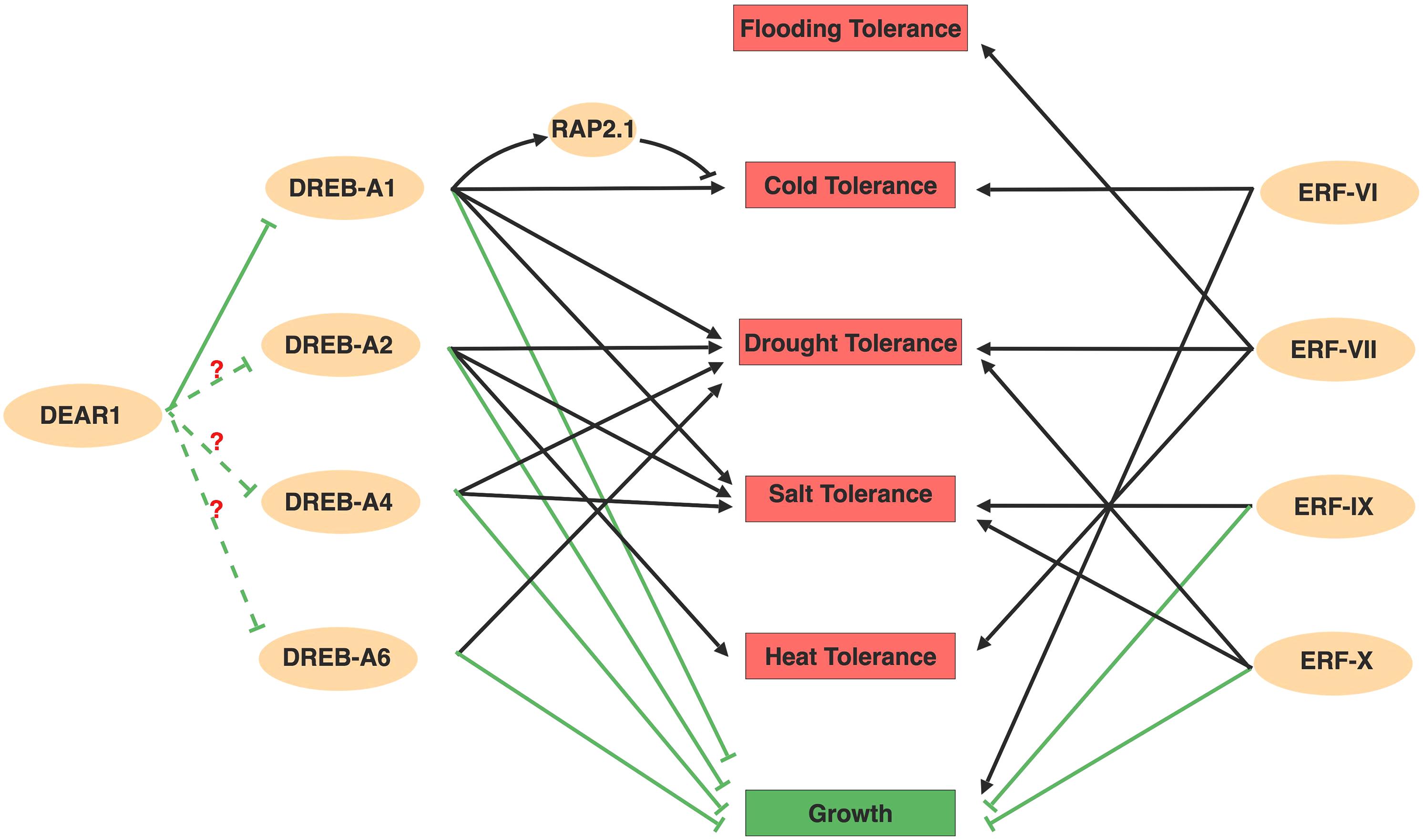
Figure 2. Overview about AP2/ERFs mediated abiotic stresses. Members in DREBs and ERFs subfamily positively regulate numerous abiotic stresses, but negatively regulate plant growth. The ability to regulate several stresses simultaneously form a comprehensive regulatory network. Among the network, DREB-A1 family might act as repressors at the upstream of DREBs with question mark. Arrows and bar ends indicate activation and repression effect, respectively. Figure is created with BioRender.
In addition to CBFs, transcriptome profiling in Arabidopsis also identified several other regulators that are activated during cold acclimation, including members in the DREB-A5 group (Fowler and Thomashow, 2002). The DREB-A5 group contains six members with EAR motifs (DEARs) acting as transcriptional repressors on DRE motif containing genes (Nakano et al., 2006), which provides a repression effect in the DREB-regulated abiotic stress network. Among these, DEAR1 likely acts upstream of CBFs, while DEAR6/RAP2.1 acts downstream of CBFs (Tsutsui et al., 2009; Dong and Liu, 2010). Overexpressed DEAR1 suppressed the cold induced expression of CBFs and displayed reduced plant freezing tolerance (Tsutsui et al., 2009). RAP2.1 is induced by cold, as well as in plants constitutively overexpressing CBFs, but the induction of RAP2.1 by cold has a later onset than the induction of CBF2 (Dong and Liu, 2010). RAP2.1 was first identified as a downstream CBFs subregulon and negatively modifies plants cold tolerance (Fowler and Thomashow, 2002). The presence of the DRE motif in the RAP2.1 promoter region also suggests that RAP2.1 might be a direct target of CBFs (Dong and Liu, 2010). Although the integration of DEAR1 and RAP2.1 in the CBF pathway still needs to be examined using genetics, this negative regulation mechanism might provide checks and balances that minimize the adverse effects of prolonged stress responses. In addition to the negative role of RAP2.1 in cold stress, it also negatively regulates drought tolerance, in that overexpression of RAP2.1 resulted in sensitivity to drought (Dong and Liu, 2010). However, how RAP2.1 regulates drought response, whether it regulates other abiotic stresses and what other DEARs function in abiotic stresses are interesting questions to answer in the future. The checks and balances of DREBs’ function in stress were also revealed in Brassica napus, where two groups DREBs (Group I and Group II) regulate cold stress responsive genes sequentially. The early induced Group I DREBs activate cold stress response pathways, whereas the Group II DREBs which are expressed later competitively inhibited Group I DREBs function (Zhao et al., 2006).
DREB2s from DREB-A2 are mostly involved in plant drought and heat tolerance, which has been reviewed in detail elsewhere (Mizoi et al., 2012). Briefly, DREB2s are induced upon drought and heat, and positively regulate DRE containing drought responsive genes such as LEAs, and heat responsive genes such as heat chaperons (Maruyama et al., 2009). Moreover, members in DREB-A4 family such as HARDY (HRD) and in DREB-A6 family such as ERF53, RAP2.4, and TG/RAP2.4A also positively regulate drought and salt tolerance (Karaba et al., 2007; Lin et al., 2008; Cheng et al., 2012; Zhu et al., 2014). Overexpression of HRD in Arabidopsis or rice remarkably improved plant drought and salt tolerance (Karaba et al., 2007). Overexpression of TRANSLUCENT GREEN (TG) resulted in vitrified leaves with increased water content in cells, leading to increased drought tolerance (Zhu et al., 2014). TG regulates cell water homeostasis mainly by directly activating several aquaporin genes (Rae et al., 2011; Zhu et al., 2014), as well as Ascorbate Peroxidases (APx) genes encoding chloroplast peroxidases that functions to protect against photo-oxidative stresses caused by Reactive Oxygen Species (ROS) (Rudnik et al., 2017).
Beside Arabidopsis, DREBs’ improvement in stress tolerance has been extensively studied in various crops such as OsDREB1s and OsDREB2s in rice (Dubouzet et al., 2003; Matsukura et al., 2010), ZmDREB2A in maize (Qin et al., 2007), TaDREB1 in wheat (Shen et al., 2003), and HvDRF1 in barley (Xue and Loveridge, 2004). These DREBs provide abundant candidate genes for the engineering of stress tolerance in crops. However, given that DREBs are a major family of AP2/ERF transcription factors that integrate multiple abiotic stress signals, cooperate or antagonize one another, and modulate downstream stress responsive genes, studying the DREB gene regulatory network will provide a platform for a more comprehensive understanding of abiotic stress responses and guide the genetic engineering of crops.
Members in ERF subfamily also contribute to abiotic stress responses (Licausi et al., 2013). Recently, two groups of ERFs have emerged as central players of abiotic stress regulation in Arabidopsis (Figure 2). CYTOKININ RESPONSE FACTORS (CRFs) in ERF-VI subfamily are induced by CTK as well as multiple abiotic stresses to positively regulate osmotic and freezing tolerance (Rashotte et al., 2006; Rashotte and Goertzen, 2010). CRF6, whose induction is dependent on the perception of CTK, alleviated the H2O2 damage on plants to positively regulate oxidative response (Zwack et al., 2016b). CRF4, one of several CRFs not transcriptionally regulated by CTK, positively regulates freezing tolerance by promoting CORs expression (Zwack et al., 2016a). However, the mechanisms by which CRFs confer stress tolerance remain to be determined.
Members of ERF-VII subfamily in Arabidopsis as well as rice have been demonstrated to play major roles in flooding, low oxygen (hypoxia) and submergence tolerance and their redundant function in hypoxia responses has been reviewed (Bailey-Serres et al., 2012; Bui et al., 2015; Gibbs et al., 2015). For example, rice SUBMERGENCE 1A (SUB1A), and SNORKEL1/2 (SK1/SK2) positively regulate flooding tolerance by two opposite mechanisms: SUB1A mediates a quiescence strategy associated with reduced growth and respiration whereas SK1/SK2 promote a deep-water escape strategy allowing rapid growth of petioles, stems, and vascular changes (Hattori et al., 2009; Locke et al., 2018).
In Arabidopsis, ERF-VIIs have conserved N-terminal domains that allow them to be degraded under anoxia conditions though oxygen-dependent N-end rule pathway (Gibbs et al., 2015). Five members including ERF71/HRE2, ERF72/RAP2.3, ERF73/HRE1, ERF74/RAP2.12, and ERF75/ RAP2.2 are induced by limited oxygen. With limited oxygen, these ERF-VIIs accumulate and positively regulate hypoxia responsive genes involved in sugar metabolism, fermentation and ET biosynthesis to achieve hypoxia tolerance. Apart from hypoxia responses, ERF-VIIs also regulate oxidative and osmotic stresses. Overexpression of RAP2.2, RAP2.3 and RAP2.12 (RAPs) results in a higher survival rate from low oxygen, oxidative and osmotic stresses, while rap2.12-2 rap2.3-1 double mutants are sensitive to these stresses (Papdi et al., 2015; Yao et al., 2017b).
RESPIRATORY BURST OXIDASE HOMOLOG D (RbohD), a NADPH oxidase, helps to generate ROS burst (Yao et al., 2017b). It is reported that RAPs regulate abiotic stresses via an RbohD-dependent mechanism. Apart from being a toxic by-product of biochemical processes, ROS serve as signaling molecules to trigger stress responses and transduce signals crossing cells according to its lower molecular weight and fast cell diffusion (Qi et al., 2018). First it was found that the ROS production and RbohD expression were compromised in single erf74 and double erf74 erf75 mutants at an early stage, which resulted in compromised stress responsive gene expression and stress tolerance. Given that ERF74 binds to RbohD promoter to activate its expression, the RbohD dependent ROS activation was essential for ERF74 and ERF75 mediated hypoxia resistance. However, too much ROS can cause cell injury and cell death. To overcome adverse effects of ROS, overexpression of ERF74 promoted increased ROS scavenging enzymes and stress responsive genes at later stage. Therefore, ERF74 acts as an on-off switch to control RbohD-dependent ROS burst in response to different stresses in Arabidopsis (Yao et al., 2017b). This newly identified mechanism provides more details and divides the stress response into early and later stages, as well as ROS balance. One example that rice ERFs OsLG3 induced ROS scavenging to positively regulate stress tolerance was reported recently, suggesting the similar mechanism exists in rice (Xiong et al., 2018).
Additionally, many other Arabidopsis ERFs also regulate abiotic stresses. ERF1 and Ethylene- and Salt-inducible ERF genes (ESEs) in ERF-IX group positively regulate plant salinity tolerance by promoting salt responsive gene expression (Zhang et al., 2011). ERF6, another member in ERF-IX group, triggers growth inhibition to confer long-term osmotic stress tolerance (Dubois et al., 2013). RAP2.6L from ERF-X subgroup improves drought and salt tolerance (Yang et al., 2009; Liu et al., 2012). Additionally, Arabidopsis RAVs, especially AP2s, play central roles in developmental processes, such as organ number and size control, shoot and root meristem maintenance, flower initiation and growth (Osnato et al., 2012; Horstman et al., 2014). Members in these subfamilies are also reported to mediate diverse abiotic stress responses. AINTEGUMENTA (ANT) controls organ cell number and size throughout shoot development. ANT also negatively regulates salt tolerance by repressing SOS3-LIKE CALCIUM BINDING PROTEIN 8 (SCABP8/CBL10), a putative Ca2+ sensor that protects Arabidopsis shoots against salt stress and maintains ion homeostasis (Meng et al., 2015a). Overexpression of Arabidopsis RAV1 and RAV2 in cotton increased fiber length and even obtained the same yield under drought stress compared with control conditions (Mittal et al., 2015).
Similar as DREBs, ERFs in other plant species like rice, wheat and tomato are also involved in a broad range of abiotic stresses (Abiri et al., 2017; Phukan et al., 2017). Overall, these findings provide the potential of engineering high-efficiency crops under stress conditions. In summary, ERFs function to receive multiple stress signals and control a diverse set of stress responsive genes, where many ERFs have cooperative or antagonistic regulation on stress responses. Therefore, constructing ERF-specific gene regulatory networks would be interesting to provide insight as to how ERFs function as a unit to regulate common downstream genes.
In addition to directly regulating abiotic stresses, AP2/ERFs are also involved in hormone signaling and hormone mediated-stress responses. Plant hormones affect abiotic stresses by triggering a wide range of physiological processes (Kazan, 2013, 2015; Colebrook et al., 2014; Khan et al., 2015; Muller and Munne-Bosch, 2015; Tao et al., 2015; Sah et al., 2016; Nolan T. et al., 2017). ABA and ET are major stress hormones that are induced under abiotic stress conditions and regulate stress responses associated with AP2/ERFs (Kazan, 2015; Sah et al., 2016). GAs, CTK, and BRs are growth-related hormones that promote cell growth, proliferation and differentiation. It is becoming increasingly evident that these growth-related hormones also have direct and/or indirect effects on abiotic stresses (Kazan, 2013; Colebrook et al., 2014; Nolan T. et al., 2017). Because stress tolerance is often associated with trade-offs between growth and stress, abiotic stress-tolerant plants usually have lower growth rates and productivity (Bechtold and Field, 2018). Numerous studies in Arabidopsis have shown AP2/ERFs confer plant stresses tolerance associated with growth losses (Figure 2), such as CBFs, DREB2A, HARDY, TG, ERF6, and RAP2.6 (Kasuga et al., 1999; Sakuma et al., 2006; Karaba et al., 2007; Sharabi-Schwager et al., 2010; Krishnaswamy et al., 2011; Dubois et al., 2013; Zhu et al., 2014). However, the growth loss can be minimized by driving expression via the stress-induced RD29A promoter (Kasuga et al., 1999; Sakuma et al., 2006; Matsukura et al., 2010). Thus, in addition to understanding the basis of AP2/ERFs in abiotic stresses, it is also important to explore their roles in the hormone-regulated stresses responses. AP2/ERFs are involved in plant hormone-mediated abiotic stresses through the following aspects: (1) AP2/ERFs transcripts are regulated by hormones; (2) AP2/ERFs alter hormone sensitivity and gene expression by cooperating or antagonizing various hormone signaling components; (3) AP2/ERFs regulate hormone biosynthesis or metabolism via feedback regulation (Figure 3). In the following sections we emphasize how abiotic stress-induced (ABA and ET) and growth-promoting (GAs, CTK, and BRs) hormones are regulated by AP2/ERFs in Arabidopsis through these mechanisms.
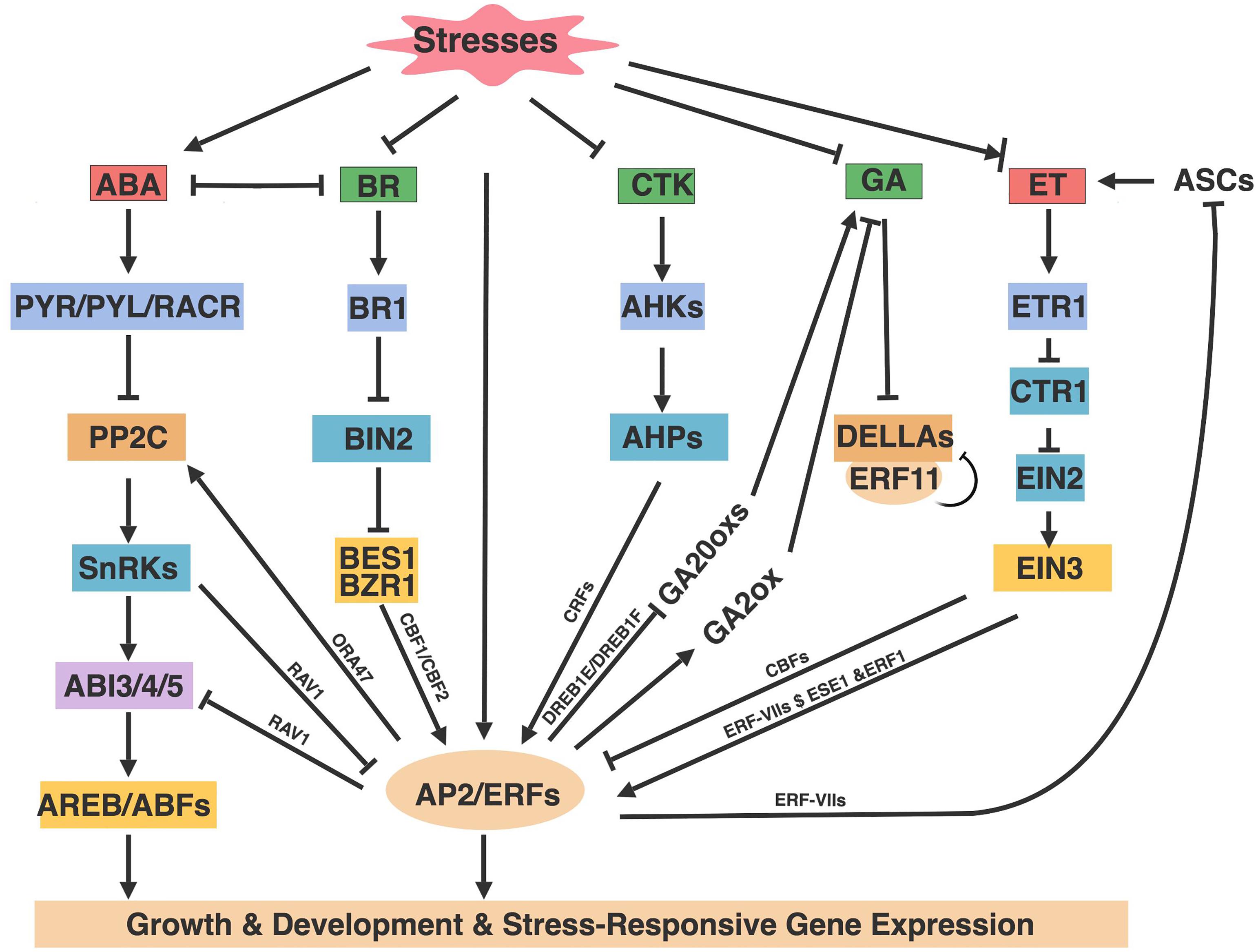
Figure 3. AP2/ERFs roles in hormone pathways. Abiotic stresses alter the production and distribution of phytohormones that in turn mediate stresses responses through hormone signaling components and AP2/ERFs. Arrows and bar ends indicate activation and repression effect, respectively. Figure is created with BioRender.
The plant hormone ABA is a pivotal hormone that regulates abiotic stress responses including drought, salinity, cold and heat stresses. ABA exerts a protective function through inducing stomata closure, modulating root architecture, and promoting the synthesis of osmolytes (Cutler et al., 2010; Sah et al., 2016). During stress conditions such as water deprivation and osmotic pressure, the rate-limiting ABA biosynthetic enzyme Nine-cis-Epoxycarotenoid Dioxygenase (NCED) is rapidly up-regulated to promote ABA biosynthesis. ABA is then sensed by a large family of PYRABACTIN RESISTANCE1 /PYR1-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS (PYR/PLY/RCAR) receptors that interact with Protein Phosphatase 2C (PP2Cs) as a ternary complex to release PP2Cs’ inhibition on SnRK2 kinases (SnRK2s). The active SnRK2s phosphorylate downstream substrate proteins, including AREBs/ABFs, ion channels, and enzymes such as NADPH oxidases, thereby inducing ABA responses (Finkelstein, 2013; Sah et al., 2016). Generally, transcription factors regulate abiotic stresses through ABA-dependent or ABA-independent pathways.
A great deal of studies have shown that DREBs regulate ABA-independent abiotic stresses by directly binding to DRE/CRT motifs on stress responsive genes (Gilmour et al., 2004; Matsukura et al., 2010; Lata and Prasad, 2011; Mizoi et al., 2012; Zhu et al., 2014). However, AP2/ERFs are indispensable for ABA-dependent stress responses as well. ANT (Meng et al., 2015b), ERF53 (Hsieh et al., 2013), ERF-VIIs (Papdi et al., 2015; Yao et al., 2017a), RAP2.6L (Liu et al., 2012), and RAP2.6 (Zhu et al., 2010) in Arabidopsis are induced by ABA to up-regulate DRE- and ABRE- element containing genes. Rice OsERF71 positively regulates ABA signaling to alter root architecture and confer drought tolerance (Lee et al., 2017; Li et al., 2018). The combination of abiotic stresses and ABA also led to the further activation of DREB2s and stress inducible genes (Lee et al., 2016). ABA INSENSITIVE 4 (ABI4), a unique one in the DREB-A3 group is a key component of the ABA signaling pathway. Upon ABA and ROS accumulation under stress conditions, ABI4 represses CCAAT Binding Factor A (CBFA) (Zhang et al., 2013). CBFA is a subunit of the trimeric transcription complex of Heme Activator Proteins (HAPs). Repression of CBFA then allows other transcription factors to enter the transcription complex and improves the efficiency of stress responsive gene transcription (Zhang et al., 2013).
Apart from the positive effect of AP2/ERFs in ABA-mediated stress responses, many Arabidopsis studies have shown that AP2/ERFs also interrupt ABA signaling, resulting in reduced sensitivity on root growth inhibition and stomata closure (Figure 3). ERF18/ ORA47 activated the PP2C family phosphatase gene ABI2. At the same time ABI1 acted upstream of ORA47 to activate ORA47, leading to an ABI1-ORA47-ABI2 regulation loop that inhibits ABA signaling as well as drought tolerance (Chen et al., 2016). RAV1 inhibites ABA sensitivity on root growth by repressing ABI3, ABI4 and ABI5 expression (Feng et al., 2014). SnRK2.2, SnRK2.3, SnRK2.6 also interact with and phosphorylate RAV1 to inhibit RAV1’s transcriptional repression of targets genes (Feng et al., 2014).
ET is also reported to regulate abiotic stress responses including salt, cold, and flooding (Kazan, 2015). ET is synthesized from the rate limiting enzymes ACC Synthases (ACSs), a major target for the regulation for ET production under stresses (Tao et al., 2015). The binding of ET with its receptor ETHYLENE INSENSITIVE 1 (ETR1) deactivates CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) kinase activity to release CTR1’s inhibition on ETHYLENE INSENSITIVE 2 (EIN2). Then the C-terminal of EIN2 translocates into the nucleus to activate ETHYLENE INSENSITIVE 3 (EIN3) as well as the transcriptional cascade of ethylene-regulated genes (Qiao et al., 2012; Muller and Munne-Bosch, 2015).
In stress responses, AP2/ERFs, especially the ERF subfamily, are the major downstream regulatory factors of the ET signaling pathway (Licausi et al., 2013; Gibbs et al., 2015; Kazan, 2015; Muller and Munne-Bosch, 2015). ET regulates several aspects of Arabidopsis abiotic stress responses, including inhibiting CBFs to negatively regulate cold stress (Figure 3). Conversely, ET positively regulates flooding and submergence mediated by ERF-VIIs, and improves salt tolerance by activating ERF1 and ESEs.
For ET-regulated cold response, the production of ET is inhibited after exposure to cold, which results in compromised cold tolerance (Shi et al., 2012). Consistently, ET insensitive mutants etr1-1, ein2-5, and ein3-1 displayed increased freezing tolerance. EIN3 inhibits the expression of CBFs by directly binding to their promoters. In addition, ET plays crucial roles on plant survival and recovery from flooding, especially in rice. Flooding causes oxygen deficiency, which promotes ET production (Yang et al., 2011) and activates the expression of a set of ERF-VIIs, whose function in hypoxia is discussed above. ET also promotes RAP2.3 nuclear localization and advances ORA59 mediated ethylene responses which is dependent on RAP2.3 (Kim et al., 2018). However, ERF-VIIs regulate hypoxia response partially through ET-independent pathways. The induction of ERF73/HRE1 under hypoxia was not completely abolished in ethylene-insensitive mutants or in the presence of ethylene biosynthesis inhibitors. ERF-VIIs also negatively regulate ET signaling and homeostasis probably via feedback regulations (Hinz et al., 2010; Yang et al., 2011). For instance, HRE1-RNAi seedlings displayed exaggerated triple responses; ACSs was decreased in RAP2.2 overexpression plants, but up-regulated in rap2.2-2 knockout mutant (Hinz et al., 2010). However, how ERF-VIIs control ET homeostasis via negative feedback mechanism under stresses needs further investigation.
ET also has complex regulation in salt stress, which has been extensively discussed (Kazan, 2015; Muller and Munne-Bosch, 2015; Tao et al., 2015). In Arabidopsis, ET signaling is required for plant tolerance to salinity stress as EIN3 activates ERF1 and ESEs to activate downstream stress-related genes and promote salinity tolerance. However, knockout mutants of ACSs also led to salt tolerance, leading to an opposite conclusion in terms of ET signaling and salt tolerance. These different conclusions might be due to the different mutants, growth conditions or experimental setups used. Future investigation is necessary to further our understanding of the role of ET in plant salinity response.
The plant hormone GAs is known to promote plant growth and development. GAs have also been shown to regulate abiotic stresses, as reduced GA content slows down plant growth upon exposure to several abiotic stresses including cold, salt, and osmotic stresses (Claeys et al., 2012; Colebrook et al., 2014). GAs are synthesized through several key oxidases including GA 20-oxidases (GA20oxs) and GA 3-oxidases (GA3oxs), and catabolized by GA 2-oxidase (GA2ox) that depletes pools of GA precursors to maintain GA homeostasis (Phillips et al., 1995; Rieu et al., 2008). In the absence of GAs, a group of DELLA proteins (DELLAs) inhibit GA response, and this inhibition can be released by the degradation of DELLAs in the presence of GAs (Claeys et al., 2012). Generally, abiotic stresses cause reduction of GA content and signaling through the inhibition of GA biosynthesis enzymes mediated by Arabidopsis AP2/ERFs (Figure 3). DREB1E and DREB1F confer salt stress-induced growth retardation mostly through the repression of GA20oxs (Magome et al., 2004). CBF1 and ERF6 overexpression plants were sensitive to stress-induced growth retardation because of increased GA2oxs expression as well as the accumulation of DELLAs (Achard et al., 2008; Dubois et al., 2013). Conversely, ERF11 promotes plant internode elongation by activating GA biosynthesis, and expression of GA3ox1 and GA20oxs are increased in ERF11 overexpression plants (Zhou et al., 2016). Nevertheless, ERF11 and ERF6 show antagonistic regulation on stress-induced growth inhibition. ERF11 suppresses the extreme dwarf phenotype of ERF6 overexpression plants and represses ERF6-induced gene expression (Dubois et al., 2015). The opposite regulation by ERF6 and ERF11 reveals that dynamic mechanisms must exist in plants to fine-tune and maintain the balance between plant growth and stress responses.
In addition to GA regulation in Arabidopsis abiotic stresses, rice regulates flooding coping submergence tolerance by two opposite GA regulations. The first quiescence strategy was that SUB1A increased the accumulation of SLENDER RICE1 (SLR1) and SLENDER RICE1 LIKE1 (SLRL1) (DELLA like proteins in rice) to restrict GA signaling and sensitivity, which resulted in inhibition of plant internode elongation and reduced carbohydrate consumption (Fukao and Bailey-Serres, 2008; Locke et al., 2018; Perata, 2018). The second deep-water escape strategy involves SK1/SK2, which lead to up-regulated GA20oxs and promoted internode elongation to escape submergence in water (Hattori et al., 2009; Ayano et al., 2014).
The plant hormone CTK not only plays diverse roles in plant growth and development, but also has been reported to regulate plant abiotic stresses (Zwack and Rashotte, 2015), one of which is mediate by CRFs (Figure 3). CRFs are essential for CTK-mediated embryo, cotyledon, and leaf development, as both single and multiple CRF1/2/3/5/6 mutants displayed cell proliferation deficient phenotypes (Rashotte et al., 2006). The roles of CRF’s regulation on CTK-mediated development were further confirmed by the transcriptome analysis of crf 1,2,5 and crf2,3,6 mutants, with or without CTK treatment in Arabidopsis. About 60% of the CTK responsive genes were regulated by both CRFs and type-B ARRs (the typical cytokinin-responsive transcription factors), suggesting a model that CRFs acted tandemly with type-B ARRs to mediate CTK response. CRF6 also cooperated with CTK signaling to inhibit stress-induced leaf senescence through a common subset of CTK-regulated genes (Zwack et al., 2013). Apart from the CRFs positive effect in the CTK pathway, CRF6 also represses CTK-associated target genes involved in CTK biosynthesis, signaling and transport, to alleviate the adverse effect of CTK on abiotic stress (Zwack et al., 2016b). The opposite regulation between CRF6 and CTK on stresses and similar regulation on senescence suggest that CRF6 regulates CTK signaling through two subsets of genes: one set of genes alleviate the negative effect of CTK on abiotic stresses, while the other set helps CTK to promote plant development. The detailed mechanisms of CRF regulation in these processes remain to be determined. Identification of CRF target genes and the upstream signaling could allow for a better understanding about ERF-VIs function and how CTK regulates abiotic stresses.
The plant hormone BRs play important roles throughout plant development, such as cell elongation, leaf development, pollen tube growth, xylem differentiation, senescence, and photomorphogenesis as well as stress response (Clouse et al., 1996; Ye et al., 2017). BRs are sensed by plasma membrane located receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) to inhibit negative regulator BRASSINOSTEROID INSENSITIVE 2 (BIN2), leading to accumulation of transcription factors BRASSINAZOLE-RESISTANT 2/BRI1-EMS-SUPPRESSOR 1 (BES1/BZR1) to regulate 1000s of BR responsive genes involved in plant growth and stresses responses (Guo et al., 2013).
BR regulates cold and drought responses through several pathways. For example, BR positively regulates cold tolerance partially though CBF-mediated cold response pathway, where BZR1 binds and promotes the expression of CBF1/CBF2 in response to cold. Cold stress also promotes accumulation of the unphosphorylated and active form of BZR1 by unknown mechanisms (Li et al., 2017b; Figure 3). However, BR negatively regulates drought tolerance via antagonizing with drought induced transcription factor RD26 on drought responsive genes (Ye et al., 2017). BR also antagonizes with ABA pathway from receptors to transcription factors and regulates the trade-off of plants growth under stress conditions (Nolan T. et al., 2017). Although there are no reports of AP2/ERFs in BR mediated drought response, BES1 and BZR1 target genes include numerous AP2/ERFs, implying that AP2/ERFs have a potential function to integrate the BR pathway with abiotic stresses (Sun et al., 2010; Yu et al., 2011). Future studies in this area will shed light on the mechanisms and roles of AP2/ERFs in BR and stress responses.
In addition, AP2/ERFs regulate the BR pathway through different mechanisms. ERF72/RAP2.3 antagonizes BZR1 and AUXIN RESPONSIVE FACTOR 6 (ARF6) to inhibit hypocotyl elongation, while its role in BR regulated stresses response is unknown (Liu K. et al., 2018). The role of ERF72 in controlling growth implies that ERF72 might be a candidate for the study of the integration of BR and stresses. In fact, rice SUB1A mediates GA and BR cross-talk to control submergence tolerance. SUB1A activates BR biosynthesis and signaling, which in turn induces GA catabolic gene GA2ox7 to lower GAs content, and ultimately promotes rice DELLA protein accumulation (Schmitz et al., 2013).
Based on AP2/ERFs roles in abiotic stresses and hormone signaling, AP2/ERFs function through complicated regulatory networks. These networks are influenced by diverse environment stimuli and plant hormones. Regulatory mechanisms such as protein–protein interactions and cooperative or antagonistic regulation of target genes are involved in dictating the output of AP2/ERF networks for plant growth, development and abiotic stresses. The large number of the AP2/ERF transcription factors coupled with functional redundancy and their diverse roles have made it difficult to fully understand AP2/ERFs networks. A promising approach is to apply computational tools to dissect AP2/ERFs function. These include analyzing promoter sequences for abiotic stress-related motifs (Sazegari et al., 2015), and constructing stress response gene regulatory networks under different stress conditions (Dubois et al., 2017; Van den Broeck et al., 2017). For instance, analysis of 20 Arabidopsis DREB genes promoters demonstrated that they contained most types of HSE LTR and ABRE promoter elements. The multiple stress responsive motifs on one hand help explain the induction of DREBs under various environment stimuli, and on the other hand, they imply that these DREBs might form a central response network to control diverse abiotic stresses (Sazegari et al., 2015).
In addition to promoter analysis, Van den Broeck et al. examined a complex, highly interconnected network of 20 Arabidopsis transcription factors (more than half are AP2/ERFs) to illustrate how stress inhibits plant growth (Van den Broeck et al., 2017). Under stresses condition, plants limit growth to promote survival (Claeys et al., 2014; Bechtold and Field, 2018). In this case, members from ERF-VIIIs (ERF8, ERF9, ERF11), ERF-IXs (ERF-1, ERF2, ERF5, ERF6, ERF59, ERF98) and ERF-X (RAP2.6L) were significantly up-regulated in proliferating and expanding tissues upon short-term mannitol exposure. Among these, ERF6 (Dubois et al., 2013) and RAP2.6L (Krishnaswamy et al., 2011; Liu et al., 2012) have already been shown to positively regulate stress tolerance and inhibit growth, implying they have potential role in growth and stress trade-offs. This work also illustrated that the activation of these ERFs was sequential. ERF5, ERF6, ERF11, and ERF98 showed a fast, strong and continuous induction, while other ERFs were regulated mildly and slowly. Transcriptomic profiling of each ERF using inducible overexpression plants demonstrated that they formed a highly interconnected gene regulation network consisting of redundant regulation mediated via interaction, activation, repression, and internal regulation. By combining osmotic stress and each individual ERFs transcriptiomic data it was possible to generate a network that simplifies the signaling cascades into linear pathways, although further genetic confirmation is needed.
In order to gain additional insight into AP2/ERF function in abiotic stresses and hormone response pathways, we investigated Arabidopsis AP2/ERFs in gene co-expression networks (Chockalingam et al., 2016; Chockalingam et al., 2017) that were generated by processing and classifying 1000s of public microarray datasets into tissue and process specific categories. If AP2/ERFs are involved in hormone- and stress- responsive gene regulation, then these transcription factors would be expected to connect to hormone/stress regulated genes in the network. We used transcriptome data from various stress and hormone treatments (Maruyama et al., 2009; Bai et al., 2012; Park et al., 2015; Song et al., 2016; Hickman et al., 2017; Albihlal et al., 2018; de Zelicourt et al., 2018; Guo et al., 2018; Xie et al., 2018; Zhang F. et al., 2018) to test this idea and found that many AP2/ERFs connect to more hormone/stress regulated genes in the network than would be expected by chance (Figure 4A, Fisher’s exact test). We further divided AP2/ERFs into four clusters based on enrichment for hormone or stress responsive genes in the network. The role of CBFs in stress and hormone responses was investigated as an example. CBFs are distributed into four clusters that include AP2/ERFs enriched for cold, drought, salt and heat responsive genes. These clusters also include AP2/ERFs enriched for ABA, ET, and BR responsive genes. In line with these network predictions, the function of CBFs in ET and cold stress (including those mediated by BRs) has been confirmed by genetics or transcriptomic studies (Shi et al., 2012; Li et al., 2017b). We further compared genes differentially expressed in cbf123 triple mutants after cold treatment, BR and ET responsive genes (Figure 4B; Zhao et al., 2016; Nolan T. M. et al., 2017; de Zelicourt et al., 2018; Zhang F. et al., 2018). The comparison indicates that CBFs share a significant amount of genes with cold, ET and BR. For example, cold regulates ∼4000 genes, ∼68% of which are regulated by CBFs, supporting CBFs’ major contribution in cold tolerance. Among these, ∼20% of CBF regulated cold responsive genes are regulated by ET and BR, indicating that ET and BR regulated cold stress are dependent on the CBF pathway. ET and BR negatively and positively regulated CBFs through their specific transcription factors EIN3 (Shi et al., 2012) and BES1/BZR1 (Li et al., 2017b), respectively (Figure 4C). Taken together, these examples of network and transcriptome comparisons can (1) predict AP2/ERFs functions, (2) study AP2/ERFs function in specific stress/hormone responses; (3) examine the cross-talk of hormones in controlling growth and stress tolerance; (4) help clarify their upstream regulators and downstream targets.
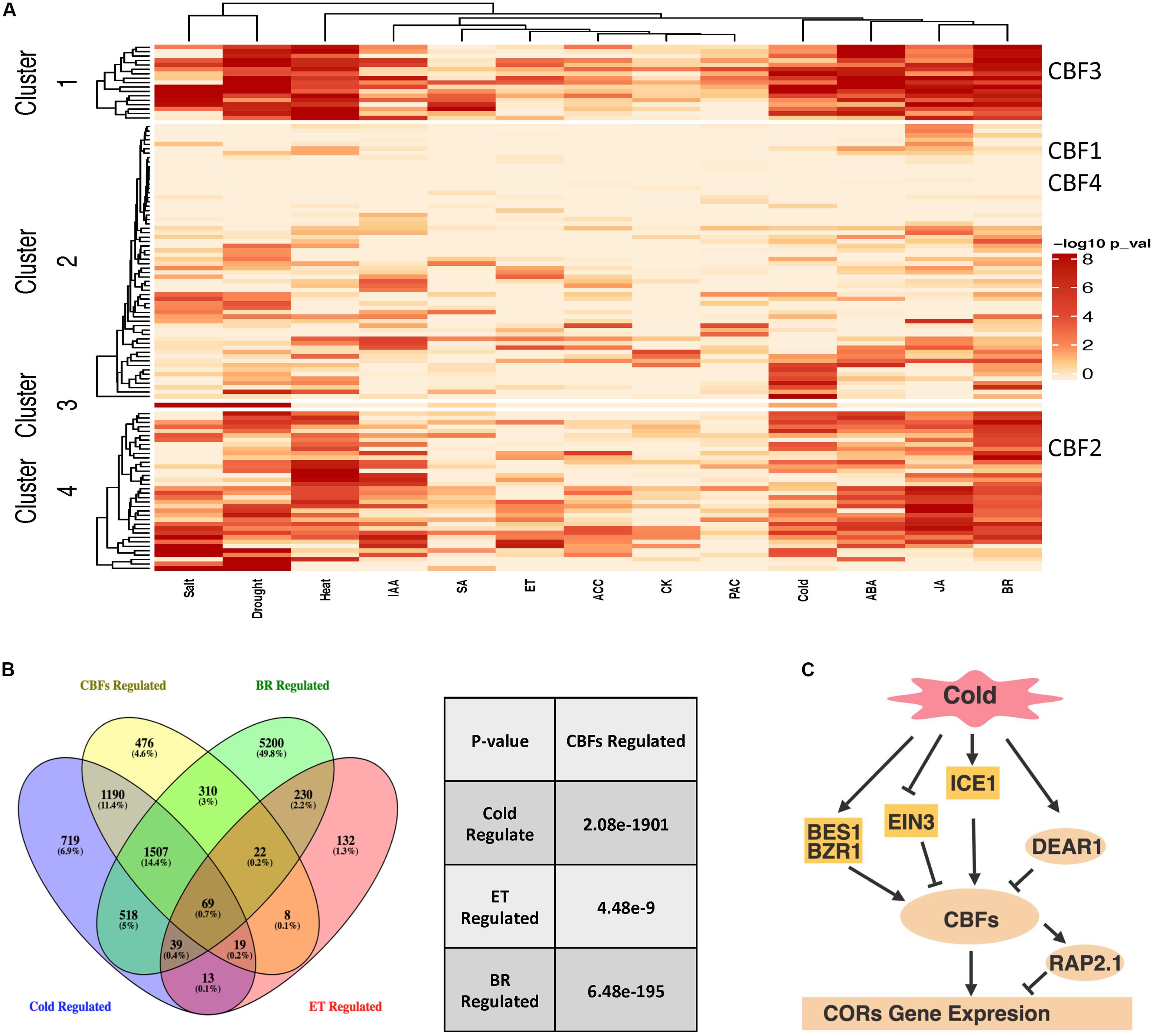
Figure 4. AP2/ERFs regulatory network in abiotic stresses and hormones. (A) Arabidopsis AP2/ERFs in abiotic stresses and hormone responsive gene co-expression networks. Networks from Chockalingam et al. (2016) and Chockalingam et al. (2017) were used to investigate the functions of AP2/ERFs. The color legend indicates normalized p-value for the enrichment of the indicated hormone- or stress-responsive gene set by Fisher’s exact test. (B) Venn diagram showing overlaps among CBFs-regulated genes compared with cold-regulated, ET-regulated and BR-regulated genes (left). Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to perform the comparisons. The significance of overlapping genes regulated by CBFs versus cold, ET and BR is calculated by hypergeometric test (right). (C) CBFs regulated cold response pathway with integration of ET and BR signaling components. Panel (C) is created with BioRender.
In summary, AP2/ERFs are regulated by stresses and control multiple plant stress responses to coordinate plant growth under stress conditions. AP2/ERFs are both regulated by plant hormone pathways and modulate plant hormone biosynthesis and signaling. Considering the complex roles of AP2/ERFs in abiotic stress and hormonal responses, future studies are required to fully understand this unique family of transcription factors. The regulation of AP2/ERFs by various hormone signaling pathways should be examined at transcriptional and post-transcriptional levels. The genome-wide identification of AP2/ERF target genes should help understand their functions as well as gauge the scope of their actions. Establishing gene regulatory networks using gene expression data and computational tools will be crucial to understand the large family of genes with 100s–1000s of target genes. Considering the complexity, the functions of AP2/ERFs should be examined in different tissues/cell types and in a temporal manner. Many AP2/ERFs are potential candidates for crops stress tolerance engineering. Thus, a full understanding of AP2/ERFs can guide crop engineering to achieve optimal plant growth and crop production under changing environmental conditions.
Publicly available datasets were analyzed in this study. This data can be found here: http://www.plantphysiol.org/content/171/4/2744/tab-figures-data.
ZX, TN, HJ, and YY conceived the topic. TN and HJ collected the gene regulation data. TN performed the network analysis. ZX wrote the manuscript with edits from other co-authors.
The work was supported by grants from NSF (MCB-1818160) and Plant Science Institute at Iowa State University. ZX was partially supported by a China Scholarship Council fellowship.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Hongqing Guo at Iowa State University for critical editing of the manuscript.
Abbas, M., Berckhan, S., Rooney, D. J., Gibbs, D. J., Vicente Conde, J., Sousa Correia, C., et al. (2015). Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25, 1483–1488. doi: 10.1016/j.cub.2015.03.060
Abiri, R., Shaharuddin, N. A., Maziah, M., Yusof, Z. N. B., Atabaki, N., Sahebi, M., et al. (2017). Role of ethylene and the APETALA 2/ethylene response factor superfamily in rice under various abiotic and biotic stress conditions. Environ. Exp. Bot. 134, 33–44. doi: 10.1016/j.envexpbot.2016.10.015
Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P., and Genschik, P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. doi: 10.1105/tpc.108.058941
Albihlal, W. S., Obomighie, I., Blein, T., Persad, R., Chernukhin, I., Crespi, M., et al. (2018). Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b regulates multiple developmental genes under benign and stress conditions. J. Exp. Bot. 69, 2847–2862. doi: 10.1093/jxb/ery142
Asensi-Fabado, M. A., Amtmann, A., and Perrella, G. (2017). Plant responses to abiotic stress: the chromatin context of transcriptional regulation. Biochim. Biophys. Acta 1860, 106–122. doi: 10.1016/j.bbagrm.2016.07.015
Ayano, M., Kani, T., Kojima, M., Sakakibara, H., Kitaoka, T., Kuroha, T., et al. (2014). Gibberellin biosynthesis and signal transduction is essential for internode elongation in deepwater rice. Plant Cell Environ. 37, 2313–2324. doi: 10.1111/pce.12377
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Bailey-Serres, J., Fukao, T., Gibbs, D. J., Holdsworth, M. J., Lee, S. C., Licausi, F., et al. (2012). Making sense of low oxygen sensing. Trends Plant Sci. 17, 129–138. doi: 10.1016/j.tplants.2011.12.004
Bartlett, A., O’Malley, R. C., Huang, S. C., Galli, M., Nery, J. R., Gallavotti, A., et al. (2017). Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat. Protoc. 12, 1659–1672. doi: 10.1038/nprot.2017.055
Bechtold, U., and Field, B. (2018). Molecular mechanisms controlling plant growth during abiotic stress. J. Exp. Bot. 69, 2753–2758. doi: 10.1093/jxb/ery157
Bethke, G., Unthan, T., Uhrig, J. F., Poschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Bossi, F., Cordoba, E., Dupre, P., Mendoza, M. S., Roman, C. S., and Leon, P. (2009). The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J. 59, 359–374. doi: 10.1111/j.1365-313X.2009.03877.x
Bui, L. T., Giuntoli, B., Kosmacz, M., Parlanti, S., and Licausi, F. (2015). Constitutively expressed ERF-VII transcription factors redundantly activate the core anaerobic response in Arabidopsis thaliana. Plant Sci. 236, 37–43. doi: 10.1016/j.plantsci.2015.03.008
Causier, B., Ashworth, M., Guo, W., and Davies, B. (2012). The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Chandler, J. W. (2018). Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci. 23, 151–162. doi: 10.1016/j.tplants.2017.09.016
Chen, H. Y., Hsieh, E. J., Cheng, M. C., Chen, C. Y., Hwang, S. Y., and Lin, T. P. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613. doi: 10.1111/nph.13914
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Cheng, M. C., Hsieh, E. J., Chen, J. H., Chen, H. Y., and Lin, T. P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158, 363–375. doi: 10.1104/pp.111.189738
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Chockalingam, S., Aluru, M., and Aluru, S. (2016). Microarray data processing techniques for genome-scale network inference from large public repositories. Microarrays 5:E23. doi: 10.3390/microarrays5030023
Chockalingam, S. P., Aluru, M., Guo, H., Yin, Y., and Aluru, S. (2017). “Reverse engineering gene networks: a comparative study at genome-scale,” in Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics (Boston, MA: ACM). doi: 10.1145/3107411.3107428
Claeys, H., Skirycz, A., Maleux, K., and Inze, D. (2012). DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol. 159, 739–747. doi: 10.1104/pp.112.195032
Claeys, H., Van Landeghem, S., Dubois, M., Maleux, K., and Inze, D. (2014). What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol. 165, 519–527. doi: 10.1104/pp.113.234641
Clouse, S. D., Langford, M., and McMorris, T. C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111, 671–678. doi: 10.1104/pp.111.3.671
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
de Zelicourt, A., Synek, L., Saad, M. M., Alzubaidy, H., Jalal, R., Xie, Y., et al. (2018). Ethylene induced plant stress tolerance by Enterobacter sp. SA187 is mediated by 2-keto-4-methylthiobutyric acid production. PLoS Genet. 14:e1007273. doi: 10.1371/journal.pgen.1007273
Dietz, K. J., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. doi: 10.1007/s00709-010-0142-8
Dinh, T. T., Girke, T., Liu, X., Yant, L., Schmid, M., and Chen, X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139, 1978–1986. doi: 10.1242/dev.077073
Dong, C. J., and Liu, J. Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10:47. doi: 10.1186/1471-2229-10-47
Dubois, M., Claeys, H., Van den Broeck, L., and Inze, D. (2017). Time of day determines Arabidopsis transcriptome and growth dynamics under mild drought. Plant Cell Environ. 40, 180–189. doi: 10.1111/pce.12809
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Dubois, M., Van den Broeck, L., Claeys, H., Van Vlierberghe, K., Matsui, M., and Inze, D. (2015). The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 169, 166–179. doi: 10.1104/pp.15.00335
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., and Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81, 77–91. doi: 10.1266/ggs.81.77
Fahad, S., Bajwa, A. A., Nazir, U., Anjum, S. A., Farooq, A., Zohaib, A., et al. (2017). Crop Production under drought and heat stress: plant responses and management options. Front. Plant Sci. 8:1147. doi: 10.3389/fpls.2017.01147
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., and Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. doi: 10.1111/tpj.12670
Feng, J. X., Liu, D., Pan, Y., Gong, W., Ma, L. G., Luo, J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59, 853–868. doi: 10.1007/s11103-005-1511-0
Finkelstein, R. (2013). Abscisic Acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14, 1675–1690. doi: 10.1105/tpc.003483
Franco-Zorrilla, J. M., Lopez-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Fukao, T., and Bailey-Serres, J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 16814–16819. doi: 10.1073/pnas.0807821105
Gao, T., Li, G. Z., Wang, C. R., Dong, J., Yuan, S. S., Wang, Y. H., et al. (2018). Function of the ERFL1a Transcription Factor in Wheat Responses to Water Deficiency. Int. J. Mol. Sci. 19:E1465. doi: 10.3390/ijms19051465
Gasch, P., Fundinger, M., Muller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Gibbs, D. J., Conde, J. V., Berckhan, S., Prasad, G., Mendiondo, G. M., and Holdsworth, M. J. (2015). Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169, 23–31. doi: 10.1104/pp.15.00338
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Guo, H., Li, L., Aluru, M., Aluru, S., and Yin, Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16, 545–553. doi: 10.1016/j.pbi.2013.08.002
Guo, H., Nolan, T. M., Song, G., Liu, S., Xie, Z., Chen, J., et al. (2018). FERONIA receptor kinase contributes to plant immunity by suppressing jasmonic acid signaling in Arabidopsis thaliana. Curr. Biol. 28, 3316.e–3324.e. doi: 10.1016/j.cub.2018.07.078
Hattori, Y., Nagai, K., Furukawa, S., Song, X. J., Kawano, R., Sakakibara, H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. doi: 10.1038/nature08258
Hickman, R., Van Verk, M. C., Van Dijken, A. J. H., Mendes, M. P., Vroegop-Vos, I. A., Caarls, L., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 29, 2086–2105. doi: 10.1105/tpc.16.00958
Hinz, M., Wilson, I. W., Yang, J., Buerstenbinder, K., Llewellyn, D., Dennis, E. S., et al. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772. doi: 10.1104/pp.110.155077
Horstman, A., Willemsen, V., Boutilier, K., and Heidstra, R. (2014). AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends Plant Sci. 19, 146–157. doi: 10.1016/j.tplants.2013.10.010
Hsieh, E. J., Cheng, M. C., and Lin, T. P. (2013). Functional characterization of an abiotic stress-inducible transcription factor AtERF53 in Arabidopsis thaliana. Plant Mol. Biol. 82, 223–237. doi: 10.1007/s11103-013-0054-z
Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. doi: 10.1093/pcp/pcp048
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. doi: 10.1093/nar/27.2.470
Karaba, A., Dixit, S., Greco, R., Aharoni, A., Trijatmiko, K. R., Marsch-Martinez, N., et al. (2007). Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. U.S.A. 104, 15270–15275. doi: 10.1073/pnas.0707294104
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. doi: 10.1038/7036
Kavas, M., Kizildogan, A., Gokdemir, G., and Baloglu, M. C. (2015). Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J. 14, 1187–1206. doi: 10.17179/excli2015-600
Kazan, K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 1655–1665. doi: 10.1093/aob/mct229
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Khan, M. I., Fatma, M., Per, T. S., Anjum, N. A., and Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6:462. doi: 10.3389/fpls.2015.00462
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Kim, N. Y., Jang, Y. J., and Park, O. K. (2018). AP2/ERF family transcription factors ORA59 and RAP2.3 interact in the nucleus and function together in ethylene responses. Front. Plant Sci. 9:1675. doi: 10.3389/fpls.2018.01675
Krishnaswamy, S., Verma, S., Rahman, M. H., and Kav, N. N. (2011). Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol. Biol. 75, 107–127. doi: 10.1007/s11103-010-9711-7
Kulkarni, M., Soolanayakanahally, R., Ogawa, S., Uga, Y., Selvaraj, M. G., and Kagale, S. (2017). Drought response in wheat: key genes and regulatory mechanisms controlling root system architecture and transpiration efficiency. Front. Chem. 5:106. doi: 10.3389/fchem.2017.00106
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lee, D. K., Yoon, S., Kim, Y. S., and Kim, J. K. (2017). Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 12:e1268311. doi: 10.1080/15592324.2016.1268311
Lee, S. Y., Boon, N. J., Webb, A. A., and Tanaka, R. J. (2016). Synergistic Activation of RD29A via integration of salinity stress and abscisic acid in Arabidopsis thaliana. Plant Cell Physiol. 57, 2147–2160. doi: 10.1093/pcp/pcw132
Lee, S. Y., Hwang, E. Y., Seok, H. Y., Tarte, V. N., Jeong, M. S., Jang, S. B., et al. (2015). Arabidopsis AtERF71/HRE2 functions as transcriptional activator via cis-acting GCC box or DRE/CRT element and is involved in root development through regulation of root cell expansion. Plant Cell Rep. 34, 223–231. doi: 10.1007/s00299-014-1701-9
Li, H., Wang, Y., Wu, M., Li, L., Li, C., Han, Z., et al. (2017a). Genome-Wide Identification of AP2/ERF transcription factors in cauliflower and expression profiling of the ERF family under salt and drought stresses. Front. Plant Sci. 8:946. doi: 10.3389/fpls.2017.00946
Li, H., Ye, K., Shi, Y., Cheng, J., Zhang, X., and Yang, S. (2017b). BZR1 positively regulates freezing tolerance via CBF-Dependent and CBF-independent pathways in Arabidopsis. Mol. Plant 10, 545–559. doi: 10.1016/j.molp.2017.01.004
Li, J., Guo, X., Zhang, M., Wang, X., Zhao, Y., Yin, Z., et al. (2018). OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 270, 131–139. doi: 10.1016/j.plantsci.2018.01.017
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lin, R. C., Park, H. J., and Wang, H. Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1, 42–57. doi: 10.1093/mp/ssm004
Liu, H. C., Liao, H. T., and Charng, Y. Y. (2011). The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ. 34, 738–751. doi: 10.1111/j.1365-3040.2011.02278.x
Liu, J., Shi, Y., and Yang, S. (2018). Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795. doi: 10.1111/jipb.12657
Liu, K., Li, Y., Chen, X., Li, L., Liu, K., Zhao, H., et al. (2018). ERF72 interacts with ARF6 and BZR1 to regulate hypocotyl elongation in Arabidopsis. J. Exp. Bot. 69, 3933–3947. doi: 10.1093/jxb/ery220
Liu, P., Sun, F., Gao, R., and Dong, H. (2012). RAP2.6L overexpression delays waterlogging induced premature senescence by increasing stomatal closure more than antioxidant enzyme activity. Plant Mol. Biol. 79, 609–622. doi: 10.1007/s11103-012-9936-8
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Liu, S., Wang, X., Wang, H., Xin, H., Yang, X., Yan, J., et al. (2013). Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 9:e1003790. doi: 10.1371/journal.pgen.1003790
Liu, W., Karemera, N. J. U., Wu, T., Yang, Y., Zhang, X., Xu, X., et al. (2017). The ethylene response factor AtERF4 negatively regulates the iron deficiency response in Arabidopsis thaliana. PLoS One 12:e0186580. doi: 10.1371/journal.pone.0186580
Locke, A. M., Barding, G. A. Jr., Sathnur, S., Larive, C. K., and Bailey-Serres, J. (2018). Rice SUB1A constrains remodelling of the transcriptome and metabolome during submergence to facilitate post-submergence recovery. Plant Cell Environ. 41, 721–736. doi: 10.1111/pce.13094
Magome, H., Yamaguchi, S., Hanada, A., Kamiya, Y., and Oda, K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37, 720–729. doi: 10.1111/j.1365-313X.2003.01998.x
Maruyama, K., Takeda, M., Kidokoro, S., Yamada, K., Sakuma, Y., Urano, K., et al. (2009). Metabolic pathways involved in cold acclimation identified by integrated analysis of metabolites and transcripts regulated by DREB1A and DREB2A. Plant Physiol. 150, 1972–1980. doi: 10.1104/pp.109.135327
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Meng, L. S., Wang, Y. B., Yao, S. Q., and Liu, A. (2015a). Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J. Cell Sci. 128, 2919–2927. doi: 10.1242/jcs.172072
Meng, L. S., Wang, Z. B., Yao, S. Q., and Liu, A. (2015b). The ARF2-ANT-COR15A gene cascade regulates ABA-signaling-mediated resistance of large seeds to drought in Arabidopsis. J. Cell Sci. 128, 3922–3932. doi: 10.1242/jcs.171207
Meng, X., Xu, J., He, Y., Yang, K. Y., Mordorski, B., Liu, Y., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Mittal, A., Jiang, Y., Ritchie, G. L., Burke, J. J., and Rock, C. D. (2015). AtRAV1 and AtRAV2 overexpression in cotton increases fiber length differentially under drought stress and delays flowering. Plant Sci. 241, 78–95. doi: 10.1016/j.plantsci.2015.09.013
Mizoi, J., Kanazawa, N., Kidokoro, S., Takahashi, F., Qin, F., Morimoto, K., et al. (2018). Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 294, 902–917. doi: 10.1074/jbc.RA118.002662
Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 86–96. doi: 10.1016/j.bbagrm.2011.08.004
Morimoto, K., Ohama, N., Kidokoro, S., Mizoi, J., Takahashi, F., Todaka, D., et al. (2017). BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, E8528–E8536. doi: 10.1073/pnas.1704189114
Muller, M., and Munne-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nolan, T., Chen, J., and Yin, Y. (2017). Cross-talk of Brassinosteroid signaling in controlling growth and stress responses. Biochem. J. 474, 2641–2661. doi: 10.1042/BCJ20160633
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 Mediated by DSK2 balances plant growth and survival. Dev. Cell 41:e37. doi: 10.1016/j.devcel.2017.03.013
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Osnato, M., Castillejo, C., Matias-Hernandez, L., and Pelaz, S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3:808. doi: 10.1038/ncomms1810
Owji, H., Hajiebrahimi, A., Seradj, H., and Hemmati, S. (2017). Identification and functional prediction of stress responsive AP2/ERF transcription factors in Brassica napus by genome-wide analysis. Comput. Biol. Chem. 71, 32–56. doi: 10.1016/j.compbiolchem.2017.09.004
Papdi, C., Perez-Salamo, I., Joseph, M. P., Giuntoli, B., Bogre, L., Koncz, C., et al. (2015). The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12. RAP2.2 and RAP2.3. Plant J. 82, 772–784. doi: 10.1111/tpj.12848
Park, C. S., Go, Y. S., and Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88, 257–270. doi: 10.1111/tpj.13248
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., and Paek, K. H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Park, S., Lee, C. M., Doherty, C. J., Gilmour, S. J., Kim, Y., and Thomashow, M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796
Perata, P. (2018). The rice SUB1A gene: making adaptation to submergence and post-submergence possible. Plant Cell Environ. 41, 717–720. doi: 10.1111/pce.13122
Pfluger, J., and Wagner, D. (2007). Histone modifications and dynamic regulation of genome accessibility in plants. Curr. Opin. Plant Biol. 10, 645–652. doi: 10.1016/j.pbi.2007.07.013
Phillips, A. L., Ward, D. A., Uknes, S., Appleford, N. E., Lange, T., Huttly, A. K., et al. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108, 1049–1057. doi: 10.1104/pp.108.3.1049
Phukan, U. J., Jeena, G. S., Tripathi, V., and Shukla, R. K. (2017). Regulation of Apetala2/ethylene response factors in plants. Front. Plant Sci. 8:150. doi: 10.3389/fpls.2017.00150
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjarvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Qiao, H., Shen, Z., Huang, S. S., Schmitz, R. J., Urich, M. A., Briggs, S. P., et al. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338, 390–393. doi: 10.1126/science.1225974
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Rae, L., Lao, N. T., and Kavanagh, T. A. (2011). Regulation of multiple aquaporin genes in Arabidopsis by a pair of recently duplicated DREB transcription factors. Planta 234, 429–444. doi: 10.1007/s00425-011-1414-z
Rashotte, A. M., and Goertzen, L. R. (2010). The CRF domain defines cytokinin response factor proteins in plants. BMC Plant Biol. 10:74. doi: 10.1186/1471-2229-10-74
Rashotte, A. M., Mason, M. G., Hutchison, C. E., Ferreira, F. J., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. U.S.A. 103, 11081–11085. doi: 10.1073/pnas.0602038103
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20, 2420–2436. doi: 10.1105/tpc.108.058818
Rudnik, R., Bulcha, J. T., Reifschneider, E., Ellersiek, U., and Baier, M. (2017). Specificity versus redundancy in the RAP2.4 transcription factor family of Arabidopsis thaliana: transcriptional regulation of genes for chloroplast peroxidases. BMC Plant Biol. 17:144. doi: 10.1186/s12870-017-1092-5
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sazegari, S., Niazi, A., and Ahmadi, F. S. (2015). A study on the regulatory network with promoter analysis for Arabidopsis DREB-genes. Bioinformation 11, 101–106. doi: 10.6026/97320630011101
Schmitz, A. J., Folsom, J. J., Jikamaru, Y., Ronald, P., and Walia, H. (2013). SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 198, 1060–1070. doi: 10.1111/nph.12202
Shaikhali, J., Heiber, I., Seidel, T., Stroher, E., Hiltscher, H., Birkmann, S., et al. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8:48. doi: 10.1186/1471-2229-8-48
Sharabi-Schwager, M., Samach, A., and Porat, R. (2010). Overexpression of the CBF2 transcriptional activator in Arabidopsis counteracts hormone activation of leaf senescence. Plant Signal. Behav. 5, 296–299. doi: 10.4161/psb.5.3.10739
Shen, Y. G., Zhang, W. K., He, S. J., Zhang, J. S., Liu, Q., and Chen, S. Y. (2003). An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 106, 923–930. doi: 10.1007/s00122-002-1131-x
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Song, C. P., and Galbraith, D. W. (2006). AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257. doi: 10.1007/s11103-005-3880-9
Song, L., Huang, S. C., Wise, A., Castanon, R., Nery, J. R., Chen, H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354, aag1550. doi: 10.1126/science.aag1550
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.M706800200
Sun, Y., Fan, X. Y., Cao, D. M., Tang, W., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Tao, J. J., Chen, H. W., Ma, B., Zhang, W. K., Chen, S. Y., and Zhang, J. S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6:1059. doi: 10.3389/fpls.2015.01059
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Tsutsui, T., Kato, W., Asada, Y., Sako, K., Sato, T., Sonoda, Y., et al. (2009). DEAR1, a transcriptional repressor of DREB protein that mediates plant defense and freezing stress responses in Arabidopsis. J. Plant Res. 122, 633–643. doi: 10.1007/s10265-009-0252-6
Van den Broeck, L., Dubois, M., Vermeersch, M., Storme, V., Matsui, M., and Inze, D. (2017). From network to phenotype: the dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 13:961. doi: 10.15252/msb.20177840
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Wan, L., Zhang, J., Zhang, H., Zhang, Z., Quan, R., Zhou, S., et al. (2011). Transcriptional activation of OsDERF1 in OsERF3 and OsAP2-39 negatively modulates ethylene synthesis and drought tolerance in rice. PLoS One 6:e25216. doi: 10.1371/journal.pone.0025216
Weber, H., and Hellmann, H. (2009). Arabidopsis thaliana BTB/ POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276, 6624–6635. doi: 10.1111/j.1742-4658.2009.07373.x
Welsch, R., Maass, D., Voegel, T., Dellapenna, D., and Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145, 1073–1085. doi: 10.1104/pp.107.104828
Woo, H. R., Kim, J. H., Kim, J., Kim, J., Lee, U., Song, I. J., et al. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61, 3947–3957. doi: 10.1093/jxb/erq206
Xie, M., Chen, H., Huang, L., O’Neil, R. C., Shokhirev, M. N., and Ecker, J. R. (2018). A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 9:1604. doi: 10.1038/s41467-018-03921-6
Xiong, H., Yu, J., Miao, J., Li, J., Zhang, H., Wang, X., et al. (2018). Natural variation in OsLG3 increases drought tolerance in rice by inducing ROS scavenging. Plant Physiol. 178, 451–467. doi: 10.1104/pp.17.01492
Xue, G. P., and Loveridge, C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37, 326–339. doi: 10.1046/j.1365-313X.2003.01963.x
Yang, C. Y., Hsu, F. C., Li, J. P., Wang, N. N., and Shih, M. C. (2011). The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 156, 202–212. doi: 10.1104/pp.111.172486
Yang, S., Wang, S., Liu, X., Yu, Y., Yue, L., Wang, X., et al. (2009). Four divergent Arabidopsis ethylene-responsive element-binding factor domains bind to a target DNA motif with a universal CG step core recognition and different flanking bases preference. FEBS J. 276, 7177–7186. doi: 10.1111/j.1742-4658.2009.07428.x
Yao, Y., Chen, X., and Wu, A. M. (2017a). ERF-VII members exhibit synergistic and separate roles in Arabidopsis. Plant Signal. Behav. 12:e1329073. doi: 10.1080/15592324.2017.1329073
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017b). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Ye, H., Liu, S., Tang, B., Chen, J., Xie, Z., Nolan, T. M., et al. (2017). RD26 mediates crosstalk between drought and brassinosteroid signalling pathways. Nat. Commun. 8:14573. doi: 10.1038/ncomms14573
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Zhang, B., Su, L., Hu, B., and Li, L. (2018). Expression of AhDREB1, an AP2/ERF transcription factor gene from peanut, is affected by histone acetylation and increases abscisic acid sensitivity and tolerance to osmotic stress in Arabidopsis. Int. J. Mol. Sci. 19:E1441. doi: 10.3390/ijms19051441
Zhang, F., Wang, L., Ko, E. E., Shao, K., and Qiao, H. (2018). Histone deacetylases SRT1 and SRT2 Interact with ENAP1 to mediate ethylene-induced transcriptional repression. Plant Cell 30, 153–166. doi: 10.1105/tpc.17.00671
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, L., Li, Z., Quan, R., Li, G., Wang, R., and Huang, R. (2011). An AP2 domain-containing gene, ESE1, targeted by the ethylene signaling component EIN3 is important for the salt response in Arabidopsis. Plant Physiol. 157, 854–865. doi: 10.1104/pp.111.179028
Zhang, Z. W., Feng, L. Y., Cheng, J., Tang, H., Xu, F., Zhu, F., et al. (2013). The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Mol. Biol. 83, 445–458. doi: 10.1007/s11103-013-0102-8
Zhao, C., Zhang, Z., Xie, S., Si, T., Li, Y., and Zhu, J. K. (2016). Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 171, 2744–2759. doi: 10.1104/pp.16.00533
Zhao, C., and Zhu, J. K. (2016). The broad roles of CBF genes: from development to abiotic stress. Plant Signal. Behav. 11:e1215794. doi: 10.1080/15592324.2016.1215794
Zhao, T. J., Sun, S., Liu, Y., Liu, J. M., Liu, Q., Yan, Y. B., et al. (2006). Regulating the drought-responsive element (DRE)-mediated signaling pathway by synergic functions of trans-active and trans-inactive DRE binding factors in Brassica napus. J. Biol. Chem. 281, 10752–10759. doi: 10.1074/jbc.M510535200
Zhou, X., Zhang, Z. L., Park, J., Tyler, L., Yusuke, J., Qiu, K., et al. (2016). The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 171, 2760–2770. doi: 10.1104/pp.16.00154
Zhu, D., Wu, Z., Cao, G., Li, J., Wei, J., Tsuge, T., et al. (2014). TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol. Plant 7, 601–615. doi: 10.1093/mp/sst152
Zhu, Q., Zhang, J., Gao, X., Tong, J., Xiao, L., Li, W., et al. (2010). The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457, 1–12. doi: 10.1016/j.gene.2010.02.011
Zwack, P. J., Compton, M. A., Adams, C. I., and Rashotte, A. M. (2016a). Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 35, 573–584. doi: 10.1007/s00299-015-1904-8
Zwack, P. J., De Clercq, I., Howton, T. C., Hallmark, H. T., Hurny, A., Keshishian, E. A., et al. (2016b). Cytokinin response factor 6 represses cytokinin-associated genes during oxidative stress. Plant Physiol. 172, 1249–1258.
Zwack, P. J., and Rashotte, A. M. (2015). Interactions between cytokinin signalling and abiotic stress responses. J. Exp. Bot. 66, 4863–4871. doi: 10.1093/jxb/erv172
Keywords: AP2/ERF, plant stress, plant growth, plant hormones, gene regulatory network
Citation: Xie Z, Nolan TM, Jiang H and Yin Y (2019) AP2/ERF Transcription Factor Regulatory Networks in Hormone and Abiotic Stress Responses in Arabidopsis. Front. Plant Sci. 10:228. doi: 10.3389/fpls.2019.00228
Received: 20 November 2018; Accepted: 11 February 2019;
Published: 28 February 2019.
Edited by:
Henrik Aronsson, University of Gothenburg, SwedenReviewed by:
Sameer Hassan, University of Gothenburg, SwedenCopyright © 2019 Xie, Nolan, Jiang and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanhai Yin, eWluQGlhc3RhdGUuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.