- 1Key Laboratory of the Ministry of Education for Medicinal Plant Resources and Natural Pharmaceutical Chemistry, College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 2National Engineering Laboratory for Resource Development of Endangered Crude Drugs in the Northwest of China, College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 3Zhengzhou Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
Cotton is the main source of renewable fiber in the world and is primarily used for textile production. Cotton fibers are single cells differentiated from the ovule epidermis and are an excellent model system for studying cell elongation, polyploidization, and cell wall biosynthesis. Plant hormones, which are present in relatively low concentrations, play important roles in various developmental processes, and recently, multiple reports have revealed the pivotal roles of hormones in regulating cotton fiber development. For example, exogenous application of hormones has been shown to promote the initiation and growth of fiber cells. However, a comprehensive understanding about phytohormone regulating fiber development is still unknown. Here, we focus on recent advances in elucidating the roles of multiple phytohormones in the control of fiber development, namely auxin, gibberellin, brassinosteroid, ethylene, cytokinin, abscisic acid, and strigolactones. We not only review the identification of genes involved in hormone biosynthetic and signaling pathways but also discuss the mechanisms of these phytohormones in regulating the initiation and elongation of fiber cells in cotton. Auxin, gibberellin, brassinosteroid, ethylene, jasmonic acid, and strigolactones play positive roles in fiber development, whereas cytokinin and abscisic acid inhibit fiber growth. Our aim is to provide a comprehensive review of the role of phytohormones in cotton fiber development that will serve as the basis for further elucidation of the mechanisms by which plant hormones regulate fiber growth.
Introduction
Upland cotton (Gossypium hirsutum) is one of the most important economic crops, and the fiber produced by this crop is the raw material for the textile industry worldwide. Naturally grown cotton plants are perennial woody shrubs and trees, yet cultivated cotton is mostly an annual crop. Each cotton fiber is a single, phenomenally elongated cotton seed coat epidermal cell. The differentiation and development of cotton fiber cells is a very complicated process and is divided into four overlapping stages: fiber initiation (3 days before anthesis to 2 days post anthesis (DPA), 2–3 DPA), fiber cell elongation (0–25 DPA), cell wall thickening (20–45 DPA) and fiber cell maturation (45–50 days post anthesis) (Basra and Malik, 1984). In recent years, genomics, functional genomics, and proteomics studies have been done to show that ethylene and pectin as well as genes involved in synthesis of secondary cell wall promote fiber growth in cotton (Ji et al., 2003; Shi et al., 2006; Gou et al., 2007; Pang et al., 2010).
Phytohormones, namely gibberellin acid (GA), auxin, cytokinin, brassinosteroid (BR), abscisic acid (ABA), ethylene, jasmonic acid (JA), salicylic acid, and strigolactone (SL), are small endogenous signaling molecules in plants (Davière and Achard, 2016). Many of these hormones have been shown to be directly involved in plant growth, cell elongation, and cell expansion. The analysis of endogenous hormone levels in bolls and fibers and the effects of treating ovules with exogenous hormones indicate that phytohormones are responsible for the development of fiber cells in cotton (Kim and Triplett, 2001; Ahmed et al., 2018). Although the receptors and signaling pathways of these hormones have been identified in Arabidopsis in the past two decades, it is still unknown how the components of phytohormone signaling pathways participate in fiber cell development. Taking advantage of the whole genome sequence of cotton (Paterson et al., 2012; Wang et al., 2012; Li et al., 2014, 2015a,b; Zhang et al., 2015), the study of cotton fiber growth has progressed rapidly and has led to new discoveries about the hormones involved in the development of fiber cells. Our manuscript comprehensively and specifically reviews the vital roles of various phytohormones in regulating cotton fiber development, including gibberellin acid, auxin, cytokinin, brassinosteroid, abscisic acid, ethylene, jasmonic acid, and strigolactone. Compared with the review paper (Ahmed et al., 2018), our manuscript also discusses the important role of jasmonic acid and strigolactone during fiber cell development.
Auxin is Responsible for Both Fiber Cell Initiation and Elongation
Auxin plays a crucial role in various developmental processes, such as plant root development, apical dominance, embryogenesis, vascular differentiation, and the response to internal and external stimuli (Dubrovsky et al., 2008; Mashiguchi et al., 2011; Rakusova et al., 2015; Wang et al., 2015a,b; Xi et al., 2016; Liu et al., 2012). There is also evidence that auxin plays an important role in cotton fiber development. For example, supplementation of exogenous indole-3-acetic acid (IAA) can compensate for defects in fiber elongation (Jasdanwala et al., 1977). Furthermore, exogenous application of IAA, one of the most important natural auxins, significantly increases the total fiber volume (Beasley and Ting, 1974; Guinn and Brummett, 1988; Chen and Guan, 2011). Consistent with its role in fiber development, auxin begins to accumulate before flowering, peaking around 2–3 DPA, and gradually decreases to the initial level at 10 DPA (Chen and Guan, 2011). Furthermore, driving expression of the IAA biosynthetic gene iaaM under the control of the petunia MADS box gene FBP7 (Floral Binding protein 7) promoter in the epidermis from −2 DPA to 10 DPA cotton ovules was found to significantly enhance fiber cell initiation at the ovule epidermis and greatly increase the number of lint fibers, which finally resulted in a greater than 15% increase in fiber yield (Zhang et al., 2011).
Recent studies have shown that auxin in fiber cells is mainly derived from outside of the ovule rather than from in situ synthesis. Auxin is transported from the ovules into the fibroblasts by the PIN-FORMED polar auxin transporters (GhPINs). The flow of auxin in fiber cells and the establishment of hormone gradients in epidermal cells of the ovule are primarily mediated by the GhPIN3a protein. GhPIN3a gene is highly expressed in the outer cells of 0 DPA ovules. Ovule-specific inhibition of multiple GhPIN genes significantly inhibits fiber cell initiation and elongation (Zhang et al., 2016). The GhPIN1a_Dt, GhPIN6_At, and GhPIN8_At genes were preferentially expressed in the fiber initiation and elongation stages; these genes also increase the density and length of leaf trichomes, which are organs similar to fiber cells (Zhang et al., 2017a,b). These results indicate that GhPIN-mediated auxin transport plays an important role in fiber-specific auxin accumulation in cotton, which should be further verified by genetic evidences.
Besides the biosynthesis and transport pathways, the auxin signaling pathway also contributes to cotton fiber cell development. Five putative auxin response genes are highly expressed during fiber cell elongation (6–12 DPA) (Gou et al., 2007). GhARF2 and GhARF18, two auxin response factors, are highly expressed during fiber cell initiation, and overexpression of these two genes significantly promotes trichome initiation in Arabidopsis leaves (Xiao et al., 2018). These data imply that GhARF2 and GhARF18 are positive regulators of cotton fiber cell initiation. In contrast to GhARF2 and GhARF18, INDOLEACETIC ACID-INDUCED PROTEIN 16 (GhIAA16) may play a negative role in fiber initiation and elongation. GhIAA16 is relatively lowly expressed in wild-type ovules. However, in the fuzzless-lintless (fl) mutant, GhIAA16 transcripts are the most highly expressed immediately after flowering (Han et al., 2012).
Expression profiling has provided further evidence that auxin plays an essential role in fiber cell elongation. The expression level of the auxin binding protein GhABP was found to increase by about 59-fold from 0 to 10 DPA. Further analyses have shown that GhABP is only expressed in elongated fibroblasts but not in villous mutants and undifferentiated epidermal cells (Chen et al., 2001). These results suggest that GhABP may be involved in the elongation of cotton fiber cells. Although ABP is essential for growth and development in Arabidopsis, the biological function of GhABP in cotton is still unknown. Overexpression and knockout of GhABP gene in cotton should be performed to study the function of GhABP gene in the future. Rac, a small G protein, mediates intracellular auxin signal transduction (Wu et al., 2011). To date, several Rac genes have been identified in cotton. GhRac1 is highly expressed during fiber elongation, and expression gradually decreases when secondary wall biosynthesis in fiber cells begins (Kim and Triplett, 2004). GhRacA and GhRacB were found to be widely expressed in roots, stems, leaves, hypocotyls, and fiber cells, with the highest transcription level observed in fiber cells at the initiation and elongation stages (Li et al., 2005). GhMAPK, a member of MAPK family involved in auxin signal pathway, is predominately expressed in elongated fiber cells (Chen et al., 2001). All members of AGAMOUS subfamily were significantly induced by auxin (Moura et al., 2016). The expression levels of most GhSAUR genes are responsive to exogenous application of IAA treatment (Li et al., 2017).
Auxin also plays an important role in fiber cell wall thickening stage. Based on the activities of IAA oxidase and superoxide enzymes, the level of IAA at the secondary wall thickening stage was four times than that in fiber cells during elongation (Singh et al., 2009), suggesting that auxin may regulate the expansion of the primary fiber cell wall and the synthesis of the secondary wall. The exogenous application of 1-naphthaleneacetic acid (NAA), an analogue of auxin, further confirmed that auxin is involved in the synthesis of secondary wall cellulose (Zhang et al., 2011).
Taken together, these findings suggest that auxin plays critical roles in fiber cell initiation and elongation in cotton (Figure 1). More genetic experiments need to be done to further confirm this conclusion by overexpressing and knocking out key genes involved in auxin synthesis and signaling pathways.
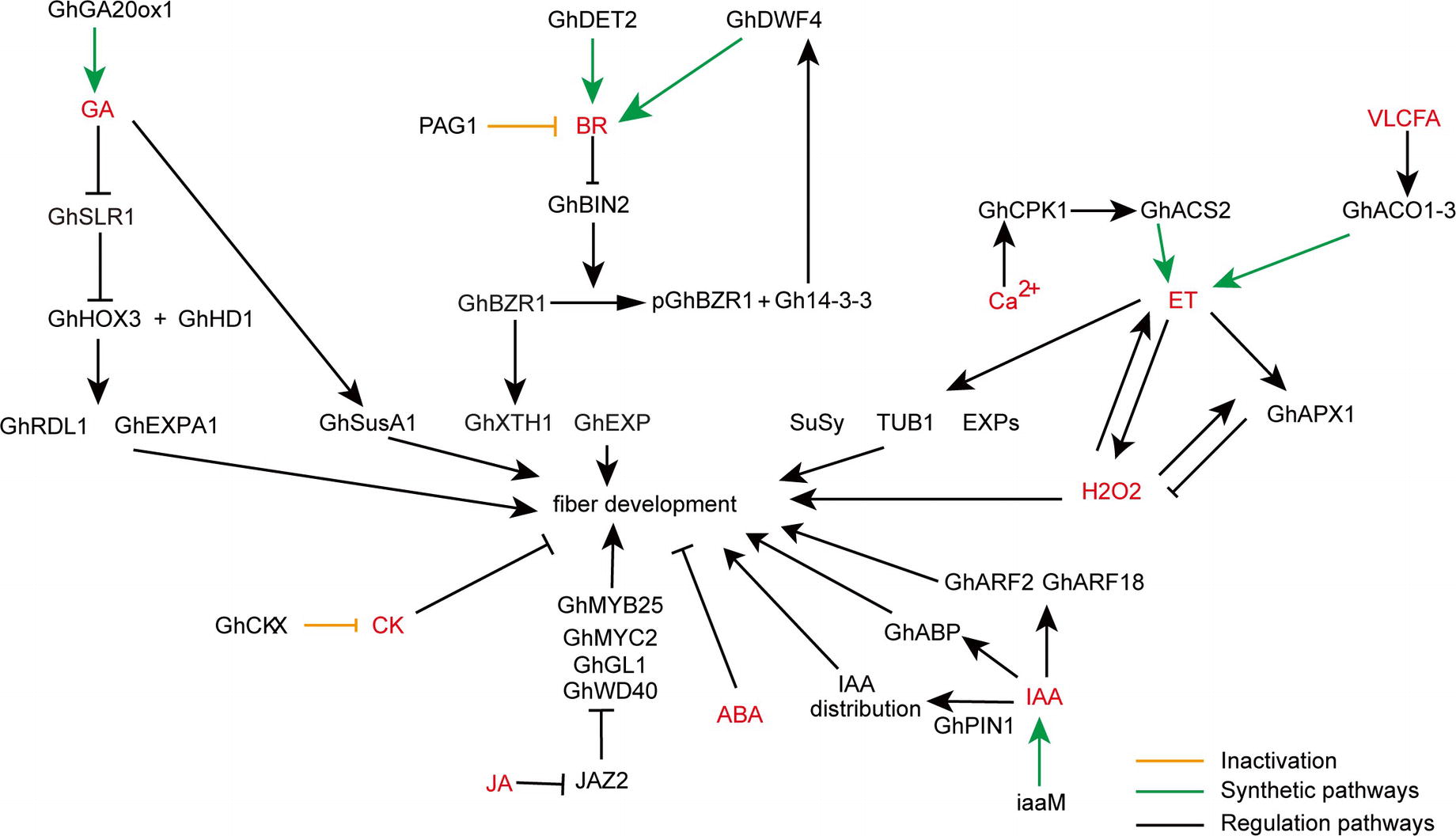
Figure 1. A schematic model showing the roles of various phytohormones during cotton fiber development. Arrows show promotion actions and bars ends show inhibitory actions. Yellow lines indicate inactivation pathway, green lines indicate synthetic pathway, and black lines indicate regulation pathway. GA, gibberellic acid; BR, brassinosteroids; VLCFA, very long-chain fatty acid; ET, ethylene; H2O2, hydrogen peroxide; CK, cytokinin; ABA, abscisic acid; JA, jasmonic acid; IAA, indoleacetic acid.
Gibberellic Acid Contributes to The Elongation of Cotton Fibers
GA is a biologically active diterpene hormone that regulates seed germination, stem elongation, epidermal hair development, and fruit development (Huang et al., 2015; David et al., 2016). Previous studies have shown that exogenous application of GA significantly promotes cotton fiber elongation (Kim and Triplett, 2001). However, when the GA synthesis inhibitor paclobutrazol was added into the ovule culture medium, there were fewer and shorter fibers than in the absence of treatment (Liao et al., 2009). The application of gibberellic acid increased the IAA and ABA content during fiber development stage and also improved fiber strength, micronaire, and maturation in natural-colored cottons (Zhang et al., 2017a,b). GA accumulation is also correlated with cotton fiber elongation. The content of GA increases rapidly after flowering, reaching a peak in 10 DPA fiber cells, and then decreases quickly (Xiao et al., 2010). The endogenous level of GA3, a bioactive form of GA, in long-staple cotton varieties is significantly higher than that in medium- and short-fiber varieties (Aleman et al., 2008). In addition, overexpression of the gibberellin 20-oxidase1 (GhGA20ox1) in transgenic cotton, which significantly increased the content of two bioactive forms of GA, GA3 and GA4, resulted in the production of more and longer fibers (Xiao et al., 2010).
DELLA proteins, which are negative regulators of the GA signaling pathway, interact with transcription factors or key regulatory proteins to inhibit their binding to target genes or their transcriptional activation activities (Davière and Achard, 2013). When GA is present, it binds to the receptor GID1 to initiate the ubiquitin-mediated degradation of DELLA proteins, releasing key transcription factors that trigger the expression of GA-responsive genes (Sun, 2008; Davière and Achard, 2016). The cotton DELLA proteins GhGID1 and GhSLR1 are specifically expressed in fiber cells, and GhGID1 interacts with GhSLR1 in response to GA. Ectopic expression of GhSLR1 in Arabidopsis results in a dwarf plant phenotype and enhances the transcription of GA-responsive genes (Aleman et al., 2008; Dong et al., 2009).
In order to reveal the mechanisms by which GA promotes cell elongation, more genes responsive to GA have been identified. Xyloglucan endotransglycosylase/hydrolase (XTH) and expansin (EXP) genes involved in promoting cell elongation are induced by GA (Aleman et al., 2008). The transcript levels of the sucrose synthase gene GhSusA1 and sucrose synthase activity are significantly higher in transgenic fiber cells overexpressing GA 20-oxidase than in wild-type fiber cells (Bai et al., 2014). In addition, exogenous application of bioactive GA enhances the transcription of GhSusA1 in both fiber cells and hypocotyls (Bai et al., 2014). These results suggest that GA promotes secondary cell wall development in cotton fiber cells by regulating the expression of sucrose synthase genes (Figure 1). Recently, a new mechanism for the promotion of cotton fiber elongation by GA was identified. The transcription factor GhHOX3 is a core regulator in the GA signaling pathway (Shan et al., 2014). When the level of GA is low, the DELLA protein GhSLR1 specifically interacts with GhHOX3 and prevents GhHOX3 from regulating its target genes. However, when the level of GA is high, the GhSLR1 protein is degraded and releases GhHOX3. GhHOX3 then interacts with GhHD1 and promotes the expression of the two cell wall genes, GhRDL1 and GhEXPA1.
Brassinosteroids Play an Important Role in Fiber Initiation and Elongation
BRs are a class of plant steroid hormones involved in a variety of physiological and developmental processes, including cell division and elongation, vascular tissue differentiation, reproductive development, senescence, and tolerance to biotic and abiotic stress (Anuradha and Rao, 2001; Haubrick and Assmann, 2006; Ming et al., 2008). BRs are also important for cotton fiber cell development. In cotton, application of a low concentration of brassinolide, a bioactive BR isolated from plants, significantly promotes fiber cell elongation, whereas brassinazole (BRZ), a BR biosynthesis inhibitor, significantly inhibits fiber cell development in vitro (Sun et al., 2005). Exogenous application of BRZ to cotton floral buds leads to severe defects in fiber cell differentiation. Steroid reduction, catalyzed by steroid 5α-reductase (DET), is considered a major rate-limiting step in BR biosynthesis. GhDET2, a cotton steroid 5α-reductase, is highly expressed during fiber cell initiation and elongation, and silencing of GhDET2 transcription inhibits both fiber cell initiation and elongation (Ming et al., 2008). Application of finasteride, a steroid 5α-reductase inhibitor, also significantly inhibits fiber elongation, which can be restored by applying BR. Overexpression of GhDET2 driven by the seed coat-specific promoter pFBP7, increases both fiber number and length. GhSMT1, another gene important for sterol biosynthesis, is highly expressed in 10 DPA fiber cells (Shi et al., 2006). GhPAG1, a homolog of Arabidopsis CYP734A1 involved in BR biosynthesis, controls cotton fiber development by regulating the level of endogenous BRs (Yang et al., 2014). The cotton pag1 mutant has reduced fiber length, and fiber elongation can be restored by BR treatment. Taken together, these findings suggest that BR is required for both fiber cell initiation and elongation (Figure 1). Importantly, the biological functions of GhBZR1 and GhBES1 genes need to be further clarified during cotton fiber development.
The components in BR signaling pathway are found to be involved in fiber cell development. BRASSINOSTEROID INSENSITIVE 1 (BRI1), which encodes a plasma membrane-localized leucine-rich repeat receptor kinase, is a BR receptor (Peng et al., 2018). The cotton, GhBRI1 gene is predominantly expressed in the fiber cells at the elongation stage and completely complements the phenotype of the Arabidopsis bri1–5 mutant (Sun et al., 2004, 2005). In Arabidopsis, BRASSINOSTEROID-INSENSITIVE 2 (BIN2), a key negative regulator in BR signaling pathways, regulates root epidermal cell fate specification by phosphorylating ENHANCER OF GLABRA 3 (EGL3) and TRANSPARENT TESTA GLABRA 1 (Guo et al., 2013; Dubrovsky et al., 2008; Mashiguchi et al., 2011; Rakusova et al., 2015; Wang et al., 2015a,b; Xi et al., 2016; Liu et al., 2012). Arabidopsis plants overexpressing GhBIN2 rescue the reduced growth resulted from the loss of function of BIN2 gene (Sun and Allen, 2005). BRASSINAZOLE-RESISTANT 1 (BZR1), a core transcription factor in the BR signaling pathway, regulates the transcription of its target genes by binding to a BRRE box in the promoter region (Guo et al., 2013). 14-3-3 proteins interact with BZR1 and affect its nucleocytoplasmic shuttling in response to BR signaling (Gampala et al., 2007). In cotton, overexpression of Gh14-3-3 L promotes fiber elongation, resulting in an increase in the length of mature fibers, and silencing of Gh14-3-3 L significantly reduces the initiation and elongation of fiber cells (Zhou et al., 2015). This short-fiber phenotype can be partially restored by exogenous application of BR. Further analyses have shown that Gh14-3-3 L can interact with GhBZR1 and that the 14-3-3-regulated GhBZR1 protein can directly bind the GhXTH1 and GhEXP promoters to regulate gene expression during the fiber cell elongation stage (Zhou et al., 2015). Recently, a newly identified gene, GhbHLH282, is found to not only regulate fiber development but also involve in BR signaling (Lu et al., 2018). Interestingly, there is no report about GhBZR1 regulation of cotton fiber growth due to the severe sterility resulting from the overexpression of GhBZR1 gene in cotton.
Ethylene Regulates Fiber Cell Elongation
The phytohormone ethylene plays a pivotal role in plant growth, regulating processes such as root hair development, hypocotyl growth, and apical hook formation (Dubois and Van den Broeck Inzé, 2018). Ethylene biosynthesis consists of two steps: S-adenosylmethionine is converted into 1-aminocyclopropane-1-carboxylic acid (ACC), which is catalyzed by ACC synthases (ACS). ACC is further converted into ethylene, which is catalyzed by ACC oxidase (ACO) (Yang et al., 2015). Ethylene promotes root hair growth by regulating ethylene insensitive 3 (EIN3) and root hair defective (RHD6) activity in Arabidopsis (Feng et al., 2017). Recently, cellulose synthase-like D3 (AtCSLD3) in Arabidopsis and GhCSLD3 in cotton were found to act downstream of ethylene to mediate root growth and cell elongation (Hu et al., 2018).
In cotton, application of ethylene in vitro significantly promotes the elongation of fiber cells, and the ethylene synthesis inhibitor L-(2-aminoethoxyvinyl)-glycine (AVG) inhibits fiber cell elongation (Shi et al., 2006). The ethylene biosynthesis pathway is one of the most significant biochemical pathways during fiber elongation stage. GhACO1-3 is significantly expressed during this stage, which is consistent with the high ethylene production in elongating fiber cells. The transcripts of three GhACO genes were found to specifically accumulate in 10 DPA fibers. Recently, excessive levels of ACO1 and ACO3 transcripts and ethylene were found to accumulate in Gossypium raimondii, resulting in the abortion of fiber cells (Li et al., 2015a,b). Ca2+-dependent protein kinase 1 interacts with and phosphorylates cotton ACS2, which increases its activity and results in a significant increase in ethylene production (Wang et al., 2011). Conversely, the production of an appropriate amount of ethylene in Gossypium hirsutum was found to significantly promote fiber development. Interestingly, ethylene can restore the inhibition of fiber growth resulting from the application of BRZ, whereas BR does not restore the shortening of fibers caused by application of AVG. In addition, ethylene promotes the expression of genes encoding inositol synthase, tubulin, and expansion, which are all involved in cotton fiber development (Shi et al., 2006).
Ethylene may also promote fiber elongation by promoting the production of hydrogen peroxide (H2O2), a type of reactive oxygen species (ROS) that significantly promotes fiber cell elongation in vitro (Qin et al., 2008). Ascorbate peroxidase (APX) is an ROS-scavenging enzyme that is involved in the regulation of intracellular ROS levels (Jiang et al., 2017). Compared with ovules in the fl mutant, GhAPX1 is highly expressed in five DPA fiber cells in wild-type cotton (Li et al., 2007). Exogenous application of H2O2 significantly induces the transcription of GhAPX1 and enhances APX activity (Qin et al., 2008). Exogenous ethylene promotes the production of H2O2 during fiber elongation, indicating that the H2O2-induced cotton fiber elongation may occur downstream of the ethylene signaling pathway. Overexpression of the calmodulin GhCaM7 also promotes fiber elongation, whereas GhCaM7 RNAi plants have delayed fiber initiation and inhibited fiber elongation (Tang et al., 2014). In contrast to wild type, GhCaM7-overexpressing fiber cells have increased ROS content and GhCaM7 RNAi fibers have significantly reduced ROS accumulation, suggesting that GhCaM7 promotes cotton fiber cell elongation by regulating ROS production. Interestingly, H2O2 can increase the influx of Ca2+ into fiber cells.
Saturated very long-chain fatty acids (VLCFAs) also promote the production of ethylene and thus regulate cotton fiber growth (Qin et al., 2007). Ethylene effectively eliminates the inhibition of fiber cell elongation resulted from the application of 2-chloro-N-[ethoxymethyl]-N-[2-ethyl-6-methyl-phenyl]-acetamide (ACE), the inhibitor of VLCFA biosynthesis, whereas VLCFA cannot overcome this inhibition caused by AVG. Application of C24:0 fatty acids in vitro results in a significant increase in ACO transcripts, resulting in significant ethylene production. Therefore, ROS regulates the accumulation of Ca2+, which then promotes fiber elongation, likely by promoting ethylene production. Besides, VLCFA can promote ethylene biosynthesis to enhance fiber cell elongation (Qin and Zhu, 2011).
Cytokinin Contributes to the Development of Cotton Ovules, but not Fiber Cells
Cytokinin regulates many aspects of plant development, such as cell division, senescence of plant tissues and organs, and apical dominance (Liu et al., 2017). There is evidence that cytokinin plays a role in ovule development. The addition of exogenous cytokinins into ovule culture medium significantly promotes the growth of ovules but inhibits the elongation of fiber cells (Beasley et al., 1974). This may be due to the fact that rapid cell division occurs during embryo development, but only single cells expand during fiber cell elongation. The cytokinin compound kinetin (6-furfurylaminopurine) significantly increases the yield of cotton seed in vitro (Sawan et al., 2000). However, large-scale commercial applications of cytokinins are not practically feasible in crops because of high costs and the time required (Li et al., 2004). The accumulation of endogenous cytokinin is mainly in wild-type ovules, but not in fiber cells (Chen et al., 1997). Cytokinin is present at a relatively low concentration in unfertilized ovules, but levels continue to increase after anthesis (Beasley et al., 1974).
Genetic modification is a good strategy for manipulating the concentration of hormones such as cytokinin and investigating the resulting phenotypes. Cytokinin levels can be increased by overexpressing isopentenyl transferase (IPT), a rate-limiting enzyme involved in cytokinin biosynthesis (Spallek et al., 2017). Overexpression of the GhIPT gene in cotton under the control of the CaMV35S promoter or a seed-specific promoter significantly increases the accumulation of cytokinin but has no impact on fiber yield and quality (Zhu et al., 2018). Cytokinin oxidase/dehydrogenase (CKX), which catalyzes the cleavage of the unsaturated side chain of cytokinin N6 and leads to the loss of cytokinin activity, is an important negative regulator of cytokinin metabolism (Niemann et al., 2018). Inhibition of CKX expression can increase endogenous levels of cytokinin in plants. Silencing of GhCKX transcripts in transgenic cotton plants using RNAi technology resulted in a significant increase in seed number and slightly enhanced the yield of fibers (Jones and Schreiber, 1997; Zhao et al., 2015). In conclusion, we suggest that cytokinin is mainly required for seed development and plays a negative role in fiber cell growth in cotton (Figure 1). Both seed development and fiber growth need to be investigated in the transgenic cottons overexpressing the GhCKX gene.
Abscisic Acid Inhibits the Initiation of Cotton Fibers
ABA is mainly involved in seed dormancy and stress response (Tuan et al., 2018). Previous studies have revealed that in vitro application of ABA not only inhibits fiber cell elongation but also inhibits the initiation of cotton fibers (Beasley et al., 1974). This inhibition is highly correlated with an increase in ABA levels. When measuring the content of endogenous ABA in different fiber cells, it was found that the accumulation of ABA gradually increased during the fiber cell initiation and elongation stages (0–10 DPA), decreased during the period of rapid elongation (10–20 DPA), and then returned to the original low level during the maturation stage (30–50 DPA) (Davis and Addicott, 1972). The ABA in fibers around 16 DPA will undergo a transition, which is consistent with the formation of the secondary wall of fiber cells (Yang et al., 2001). This indicates that ABA may be involved in secondary wall biosynthesis. The ABA content in short-staple fibers is much higher than that in long-staple fibers throughout development (Nayyar et al., 1989). Moreover, levels of endogenous ABA in cotton ovules are positively correlated with the short fiber production (Ma et al., 2011). Much higher accumulation of ABA in 0 DPA ovules was observed in the short fiber cotton mutant Ligon-lintless 1 than in wild type (Gilbert et al., 2013). The promoter of GbEXPA2, a cotton fiber-preferential promoter, can be regulated by both GA and ABA (Li et al., 2015a,b). Taken together, these results indicate that ABA seems to be a negative regulator of cotton fiber initiation (Figure 1), which should be further confirmed by investigating the number of fibers per ovule in the transgenic cottons overexpressing ABA synthetic genes or in cotton with high ABA content.
Jasmonic Acid Positively Controls Fiber Cell Initiation in Cotton
JA, which is an important endogenous hormone, is biosynthesized from linolenic acid via the octadecanoid pathway (Jia et al., 2015). Linolenic acid is oxygenated by lipoxygenase (LOX) to generate a peroxide, which is then converted into oxophytodienoic acid, a precursor of JA, by allene oxide synthase (AOS). Although its roles in the response to abiotic and biotic stress are well known, JA also plays a pivotal role in regulating aspects of plant growth and development, including growth inhibition, trichome development, leaf abscission, and senescence (Delker et al., 2006). For example, JA significantly enhances the number of trichomes in leaves (Traw and Bergelson, 2003). In contrast to wild-type plants, aos mutant plants have leaves with significantly fewer trichomes, and this defect can be rescued by exogenous application of JA (Yoshida et al., 2009). JA significantly promotes the expression of GL3, a key bHLH transcription factor involved in trichome formation, and increases trichome initiation. The Jasmonate-ZIM domain (JAZ) proteins are negative regulators of JA signaling (Song et al., 2011). JAZ interacts with multiple members of the WD-repeat/bHLH/MYB transcriptional complex, including GL1, GL3, and EGL3, which are key components involved in trichome growth, to repress trichome initiation (Qi et al., 2011). In the absence of JA, JAZ inhibits the expression of GL1, GL3, and EGL3, and in the presence of JA, JAZ is degraded by the 26S proteasome, thus releasing these transcription factors and promoting trichome initiation.
In cotton, JA-associated metabolism contributes to cotton fiber initiation (Wang et al., 2015a,b). GhJAZ2 is highly expressed in ovules at the fiber initiation stage, and overexpression of GhJAZ2 inhibits fiber initiation and reduces the length of fiber cells (Hu et al., 2016). GhJAZ2 can interact with the GhMYB25-like, GhGL1, GhMYC2, and GhWD40 proteins, which are the core components of the WD-repeat/bHLH/MYB transcriptional complex and are reported to be involved in fiber development (Figure 1).
Taken together, a number of genes related to phytohormone biosynthesis and signaling pathway have been found to be involved in cotton fiber cell development (Table 1). Other functions of these genes should be the focus of future research in cotton.

Table 1. Genes involved in phytohormone synthesis as well as signaling pathway and cotton fiber development.
Perspectives
Although auxin, GA, BR, ethylene, and JA have been shown to promote fiber cell development in cotton and a number of genes have been reported to be involved in cotton fiber cell development (Ahmed et al., 2018), a large number of questions still remain to be answered. Auxin polar transporters (PINs) and the core regulators of the auxin signaling pathway (ARFs) play important roles in regulating cotton fiber development (Zhang et al., 2017a,b), but the mechanisms of auxin transport between the ovule and fibers and the targets of ARF proteins are still a mystery. In the future, transgenic technology can be used to generate transgenic cotton with PIN overexpression and knockout and to study the distribution of auxin in ovules and fibers of transgenic cotton. Exploring and identifying target genes of GhARF2 and GhARF18 is also a very effective way to study the molecular mechanism of auxin signaling pathway regulating fiber development in cotton. Exogenous application of BR promotes cotton fiber development, and overexpression of the BR biosynthetic gene GhDET2 significantly increases the length of fiber cells (Sun et al., 2005; Ming et al., 2008). However, how the BR signaling pathway regulates cotton fiber development and the roles of BZR1 and BRI1-EMS-SUPPRESSOR 1 (BES1), two core factors in BR signaling pathway, are still unknown. We have too little understanding of how JA regulates fiber growth. Identification of downstream target genes of GhMYB25-like, GhGL1, GhMYC2, and GhWD40 to elucidate JA signaling pathway will help us better understand the molecular mechanism of JA regulation of fiber development. The interactions between key regulators of different hormone signaling pathways will reveal the cross talk between various phytohormones involved in fiber initiation and elongation.
Recently, another class of plant hormones, SL, has attracted scientists’ attention, and the SL signaling pathway is becoming more and more clear (Khosla and Nelson, 2016). SL was first isolated from cotton roots in 1966 (Cook et al., 1966), and more recent studies have revealed that SL mainly functions as an inhibitor of shoot branching (Jiang et al., 2013; Zhou et al., 2013; Yao et al., 2016). Interestingly, SL was found to regulate root hair elongation in Arabidopsis (Kapulnik et al., 2011a,b). The cross talk between SL, ethylene, and auxin may be essential for the elongation of root hairs (Kapulnik et al., 2011a,b), which are similar to cotton fiber cells. Although the biological function of SLs and the SL signaling pathway have been extensively studied in Arabidopsis, the roles of SLs during cotton development, especially during fiber cell growth, are still unknown. Whether SL is involved in cotton fiber development is key question and will be a topic of research in the future. The relationship between SL and fiber development can be investigated by overexpressing or knocking out SL biosynthetic and signaling pathway genes and then examining the lengths of cotton fiber cells.
The key genes of biosynthesis and signaling pathways of these hormones have been known in Arabidopsis. Our review summarizes some important members of these pathways, which contribute to cotton fiber development, and also prompts us to investigate whether the remaining key components of these pathways regulate fiber cell growth to further characterize the mechanisms of plant hormones regulating fiber growth.
Author Contributions
GX and ZP write the manuscript, ZY revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31600223), the Fundamental Research Funds for the Central Universities (Grant No. GK201803041), and State Key Laboratory of Cotton Biology Open Fund (Grant No. CB2018A03).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmed, M., Shahid, A. A., Din, S. U., Akhtar, S., Ahad, A., Rao, A. Q., et al. (2018). An overview of genetic and hormonal control of cotton fiber development. Pak. J. Bot. 50, 433–443.
Aleman, L., Kitamura, J., Abdel-Mageed, H., Lee, J., Sun, Y., Nakajima, M., et al. (2008). Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol. Biol. 68, 1–16. doi: 10.1007/s11103-008-9347-z
Anuradha, S., and Rao, S. S. S. (2001). Effect of brassinosteroids on salinity stress induced inhibition of seed germination and seedling growth of rice (Oryza sativa L.). Plant Growth Regul. 33, 151–153. doi: 10.1023/A:1017590108484
Bai, W. Q., Xiao, Y. H., Zhao, J., Song, S. Q., Hu, L., Zeng, J. Y., et al. (2014). Gibberellin overproduction promotes sucrose synthase expression and secondary cell wall deposition in cotton fibers. PLoS One 9:e96537. doi: 10.1371/journal.pone.0096537
Beasley, C., Bimbaum, E., Dugger, W., and Ting, I. P. (1974). A quantitative procedure for estimating cotton fiber growth. Biotech. Histochem. 49, 85–92.
Beasley, C. A., and Ting, I. P. (1974). Effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am. J. Bot. 188–194.
Chen, J. G., Du, X. M., Zhou, X., and Zhao, H. Y. (1997). Levels of cytokinins in the ovules of cotton mutants with altered fiber development. J. Plant Growth Regul. 16, 181–185. doi: 10.1007/PL00006994
Chen, J. G., Ullah, H., Young, J. C., Sussman, M. R., and Jones, A. M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15, 902–911. doi: 10.1101/gad.866201
Chen, Z. J., and Guan, X. (2011). Auxin boost for cotton. Nat. Biotechnol. 29, 407–409. doi: 10.1038/nbt.1858
Cook, C. E., Whichard, L. P., Turner, B., Wall, M. E., and Egley, G. H. (1966). Germination of witchweed (Striga lutea Lour): isolation and properties of a potent stimulant. Science 154, 1189–1190. doi: 10.1126/science.154.3753.1189
David, V. D., Seifi, S., Filipe, O., Haeck, A., Huu, S. N., Demeestere, K., et al. (2016). The DELLA protein SLR1 integrates and amplifies salicylic acid- and jasmonic acid-dependent innate immunity in rice. Plant Physiol. 170, 1831–1847.
Davière, J. M., and Achard, P. (2013). Gibberellin signaling in plants. Development 140, 1147–1151. doi: 10.1242/dev.087650
Davière, J. M., and Achard, P. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9, 10–20. doi: 10.1016/j.molp.2015.09.011
Davis, L. A., and Addicott, F. T. (1972). Abscisic acid: correlations with abscission and with development in the cotton fruit. Plant Physiol. 49, 644–648. doi: 10.1104/pp.49.4.644
Delker, C., Stenzel, I., Hause, B., Miersch, O., Feussner, I., and Wasternack, C. (2006). Jasmonate biosynthesis in Arabidopsis thaliana-enzymes, products, regulation. Plant Biol. 8, 297–306. doi: 10.1055/s-2006-923935
Dong, J., Yin, M. H., Yang, F., Zhao, J., Qin, S., Hou, L., et al. (2009). Cloning and expression profiling of gibberellin insensitive dwarf GID1 homologous genes from cotton. Acta Agron. Sin. 35, 1822–1830.
Dubois, M., and Van den Broeck Inzé, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. doi: 10.1016/j.tplants.2018.01.003
Dubrovsky, J. G., Sauer, M., Napsucialy-Mendivil, S., Ivanchenko, M. G., Friml, J., Shishkova, S., et al. (2008). Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 105, 8790–8794.
Feng, Y., Xu, P., Li, B., Li, P., Wen, X., An, F., et al. (2017). Ethylene promotes root hair growth through coordinated EIN3/EIL1 and RHD6/RSL1 activity in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, 13834–13839. doi: 10.1073/pnas.1711723115
Gampala, S. S., Kim, T. W., He, J. X., Tang, W., Deng, Z., Bai, M. Y., et al. (2007). An essential role for 14-3-3 proteins in brassinoste oid signal transduction in Arabidopsis. Dev. Cell 13, 177–178. doi: 10.1016/j.devcel.2007.06.009
Gilbert, M. K., Bland, J. M., Shockey, J. M., Cao, H., Hinchliffe, D. J., Fang, D. D., et al. (2013). A transcript profiling approach reveals an abscisic acid-specific glycosyltransferase (UGT73C14) induced in developing fiber of Ligon lintless-2 mutant of cotton (Gossypium hirsutum L.). PLoS One 8:e75268. doi: 10.1371/journal.pone.0075268
Gou, J. Y., Wang, L. J., Chen, S. P., Hu, W. L., and Chen, X. Y. (2007). Gene expression and metabolite profiles of cotton fiber during cell elongation and secondary cell wall synthesis. Cell Res. 17, 422–434. doi: 10.1038/sj.cr.7310150
Guinn, G., and Brummett, D. L. (1988). Changes in abscisic acid and indoleacetic acid before and after anthesis relative to changes in abscission rates of cotton fruiting forms. Plant Physiol. 87, 629–631. doi: 10.1104/pp.87.3.629
Guo, H., Li, L., Aluru, M., Aluru, S., and Yin, Y. (2013). Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 16, 545–553. doi: 10.1016/j.pbi.2013.08.002
Han, X. Y., Xu, X. Y., Fang, D. D., Zhang, T. Z., and Guo, W. Z. (2012). Cloning and expression analysis of novel Aux/IAA family genes in Gossypium hirsutum. Gene 503, 83–91. doi: 10.1016/j.gene.2012.03.069
Haubrick, L. L., and Assmann, S. M. (2006). Brassinosteroids and plant function: some clues, more puzzles. Plant Cell Environ. 29, 446–457. doi: 10.1111/j.1365-3040.2005.01481.x
Hu, H., He, X., Tu, L., Zhu, L., Zhu, S., Ge, Z., et al. (2016). GhJAZ2 negatively regulates cotton fiber initiation by interacting with the R2R3-MYB transcription factor GhMYB25-like. Plant J. 88, 921–935. doi: 10.1111/tpj.13273
Hu, H., Zhang, R., Dong, S., Li, Y., Fan, C., Wang, Y., et al. (2018). AtCSLD3 and GhCSLD3 mediate root growth and cell elongation downstream of the ethylene response pathway in Arabidopsis. J. Exp. Bot. Oxf. Acad. 69, 1065–1080.
Huang, D., Wang, S., Zhang, B., Shang-Guan, K., Shi, Y., Zhang, D., et al. (2015). A gibberellin-mediated DELLA-NAC signaling cascade regulates cellulose synthesis in rice. Plant Cell 27, 1681–1696. doi: 10.1105/tpc.15.00015
Jasdanwala, R. T., Sing, Y. D., and Chinoy, J. J. (1977). Auxin metabolism in developing cotton hairs. J. Exp. Bot. 28, 1111–1116. doi: 10.1093/jxb/28.5.1111
Ji, S. J., Lu, Y. C., Feng, J. X., Wei, G., Li, J., Shi, Y. H., et al. (2003). Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 31, 2534–2543. doi: 10.1093/nar/gkg358
Jia, H. F., Zhang, C., Pervaiz, T., Zhao, P. C., Liu, Z. J., Wang, B. J., et al. (2015). Jasmonic acid involves in grape fruit ripening and resistant against Botrytis cinerea. Funct Integr Genomics 16, 79–94.
Jiang, L., Wang, W., Chen, Z., Gao, Q., Xu, Q., and Cao, H. (2017). A role for APX1 gene in lead tolerance in Arabidopsis thaliana. Plant Sci. 256, 94–102. doi: 10.1016/j.plantsci.2016.11.015
Jiang, L. J., Liu, X., Xiong, G. S., Liu, H., Chen, F., Wang, L., et al. (2013). DWARF 53 acts as a repressor of strigolactone signaling in rice. Nature 504, 401–405. doi: 10.1038/nature12870
Jones, R. J., and Schreiber, B. M. N. (1997). Role and function of cytokinin oxidase in plants. Plant Growth Regul. 23, 123–134. doi: 10.1023/A:1005913311266
Kapulnik, Y., Delaux, P. M., Resnick, N., Mayzlish-Gati, E., Wininger, S., Bhattacharya, C., et al. (2011a). Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216.
Kapulnik, Y., Resnick, N., Mayzlish Gati, E., Kaplan, Y., Wininger, S., Hershenhorn, J., et al. (2011b). Strigolactones interact with ethylene and auxin in regulating root-hair elongation in Arabidopsis. J. Exp. Bot. 62, 2915–2924.
Khosla, A., and Nelson, D. C. (2016). Strigolactones, super hormones in the fight against Striga. Curr. Opin. Plant Biol. 33, 57–63. doi: 10.1016/j.pbi.2016.06.001
Kim, H. J., and Triplett, B. A. (2001). Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 127, 1361–1366. doi: 10.1104/pp.010724
Kim, H. J., and Triplett, B. A. (2004). Characterization of GhRac1 and GTPase expressed in developing cotton (Gossypium hirsutum L.) fibers. Biochim. Biophys. Acta 1679, 214–221.
Li, F. G., Fan, G. Y., Lu, C. R., Sun, F., Yuan, Y., Song, G., et al. (2015a). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530.
Li, F. G., Fan, G. Y., Wang, K. B., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, H. B., Qin, Y. M., Pang, Y., Song, W. Q., Mei, W. Q., and Zhu, Y. X. (2007). A cotton ascorbate peroxidase is involved in hydrogen peroxide homeostasis during fibre cell development. New Phytol. 175, 462–471. doi: 10.1111/j.1469-8137.2007.02120.x
Li, X., Liu, G., Geng, Y., Wu, M., Pei, W., Zhai, H., et al. (2017). A genome-wide analysis of the small auxin-up RNA (SAUR) gene family in cotton. BMC Genomics 18:815. doi: 10.1186/s12864-017-4224-2
Li, X. B., Xiao, Y. H., Luo, M., et al. (2005). Cloning and expression analysis of two Rac genes from cotton (Gossypium hirsutum L.). Acta Genet. Sin. 32, 72–78.
Li, Y., Duan, H., Wu, Y., McAvoy, R., Pei, Y., Zhao, D. G., et al. (2004). “Transgenics of plant hormones and their potential application in horticultural crops” in Genetically modified crops, their development, uses, and risks. eds. Liang, G. H. Liang and D. Z. Skinner (New York: The Haworth Press), 101–117.
Li, Y., Tu, L., Ye, Z., Wang, M., Gao, W., and Zhang, X. (2015b). A cotton fiber-preferential promoter, PGbEXPA2, is regulated by GA and ABA in Arabidopsis. Plant Cell Rep. 34, 1539–1549.
Liao, W. B., Ruan, M. B., Cui, B. M., Xu, N. F., Lu, J. J., and Peng, M. (2009). Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum. Plant Growth Regul. 58, 35–45. doi: 10.1007/s10725-008-9350-z
Liu, J., Moore, S., Chen, C., and Lindsey, K. (2017). Crosstalk complexities between auxin, cytokinin, and ethylene in Arabidopsis root development: from experiments to systems modeling, and back again. Mol. Plant 10, 1480–1496. doi: 10.1016/j.molp.2017.11.002
Liu, K., Sun, J., Yao, L., Hu, Y., Feng, H., Chang, L., et al. (2012). Transcriptome analysis reveals critical genes and key pathways for early cotton fiber elongation in Ligon lintless-1 mutant. Genomics 100, 42–50. doi: 10.1016/j.ygeno.2012.04.007
Lu, R., Zhang, J., Liu, D., Wei, Y. L., Wang, Y., and Li, X. B. (2018). Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum). BMC Plant Biol . 18:304. doi: 10.1186/s12870-018-1523-y
Luo, M., Xiao, Y. H., Li, X. B., Li, D. M., Hou, L., Hu, M. Y., et al. (2008). GhDET2, a steroid 5alpha-reductase, plays an important role in cotton fiber cell initiation and elongation. Cotton Sci. 20:130.
Ma, Q., Hedden, P., and Zhang, Q. (2011). Heterosis in rice seedlings: its relationship to gibberellin content and expression of gibberellin metabolism and signaling genes. Plant Physiol. 156, 1905–1920. doi: 10.1104/pp.111.178046
Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H., Natsume, M., et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 108, 18512–18517.
Moura, S. M., Artico, S., Lima, C., Nardeli, S. M., Berbel, A., Oliveira-Neto, O. B., et al. (2016). Functional characterization of AGAMOUS-subfamily members from cotton during reproductive development and in response to plant hormones. Plant Reproduct. 30, 19–39.
Nayyar, H., Kaur, K., Basra, A. S., and Malik, C. P. (1989). Hormonal regulation of cotton fibre elongation in Gossypium arboreum L. in vitro and in vivo. Biochem. Physiol. Pflanz. 185, 415–421. doi: 10.1016/S0015-3796(89)80065-4
Niemann, M. E., Weber, H., Hluska, T., Leonte, G., Anderson, S. M., Novák, O., et al. (2018). The cytokinin oxidase/dehydrogenase CKX1 is a membrane-bound protein requiring homooligomerization in the endoplasmic reticulum for its cellular activity. Plant Physiol. 176, 2024–2039. doi: 10.1104/pp.17.00925
Pang, C. Y., Wang, H., Pang, Y., Xu, C., Jiao, Y., Qin, Y. M., et al. (2010). Comparative proteomics indicates that biosynthesis of pectin precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol. Cell. Proteomics 9, 2019–2033. doi: 10.1074/mcp.M110.000349
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Peng, Y., Chen, L., Li, S., Zhang, Y., Xu, R., Liu, Z., et al. (2018). BRI1 and BAK1 interact with G proteins and regulate sugar-responsive growth and development in Arabidopsis. Nat. Commun. 9:1522.
Qi, T. C., Song, S. S., Ren, Q. C., Wu, D. W., Huang, H., Chen, Y., et al. (2011). The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate Jasmonate –mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814.
Qin, Y. M., Hu, C. Y., Pang, Y., Kastaniotis, A. J., Hiltunen, J. K., and Zhu, Y. X. (2007). Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19, 3692–3704. doi: 10.1105/tpc.107.054437
Qin, Y. M., Hu, C. Y., and Zhu, Y. X. (2008). The ascorbate peroxidase regulated by H2O2 and ethylene is involved in cotton fiber cell elongation by modulating ROS homeostasis. Plant Signal. Behav. 3, 194–196.
Qin, Y. M., and Zhu, Y. X. (2011). How cotton fibers elongate: a tale of linear cell-growth mode. Curr. Opin. Plant Biol. 14, 106–111. doi: 10.1016/j.pbi.2010.09.010
Rakusova, H., Fendrych, M., and Friml, J. (2015). Intracellular trafficking and PIN mediated cell polarity during tropic responses in plants. Curr. Opin. Plant Biol. 23, 116–123. doi: 10.1016/j.pbi.2014.12.002
Sawan, Z. M., Mohamed, A. A., Sakr, R. A., and Tarrad, A. M. (2000). Effect of kinetin concentration and methods of application on seed germination, yield components, yield and fiber properties of the Egyptian cotton (Gossypium barbadense). Environ. Exp. Bot. 44, 59–68. doi: 10.1016/S0098-8472(00)00054-X
Shan, C. M., Shangguan, X. X., Zhao, B., Zhang, X. F., Chao, L. M., Yang, C. Q., et al. (2014). Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat. Commun. 5:5519.
Shi, Y. H., Zhu, S. W., Mao, X. Z., Feng, J. X., Qin, Y. M., Zhang, L., et al. (2006). Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18, 651–664. doi: 10.1105/tpc.105.040303
Singh, B., Cheek, H. D., and Haigler, C. H. (2009). A synthetic auxin (NAA) suppresses secondary wall cellulose synthesis and enhances elongation in cultured cotton fiber. Plant Cell Rep. 28, 1023–1032. doi: 10.1049/iet-syb.2009.0002
Song, S., Qi, T., Huang, H., Ren, Q., Wu, D., Chang, C., et al. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23, 1000–1013. doi: 10.1105/tpc.111.083089
Spallek, T., Melnyk, C. W., Wakatake, T., Zhang, J., Sakamoto, Y., Kiba, T., et al. (2017). Interspecies hormonal control of host root morphology by parasitic plants. Proc. Natl. Acad. Sci. USA 114, 5283–5288. doi: 10.1073/pnas.1619078114
Sun, T. (2008). Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 6:e0103. doi: 10.1199/tab.0103
Sun, Y., and Allen, R. D. (2005). Functional analysis of the BIN2 genes of cotton. Mol. Gen. Genom. 274, 51–59. doi: 10.1007/s00438-005-1122-0
Sun, Y., Fokar, M., Asami, T., Yoshida, S., and Allen, R. D. (2004). Characterization of the brassinosteroid insensitive 1 genes of cotton. Plant Mol. Biol. 54, 221–232. doi: 10.1023/B:PLAN.0000028788.96381.47
Sun, Y., Veerabomma, S., Abdel-Mageed, H. A., Fokar, M., Asami, T., Yoshida, S., et al. (2005). Brassinosteroid regulates fiber development on cultured cotton ovules. Plant Cell Physiol. 46, 1384–1391. doi: 10.1093/pcp/pci150
Tang, W., Tu, L., Yang, X., Tan, J., Deng, F., Hao, J., et al. (2014). The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 202, 509–520.
Traw, M. B., and Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375.
Tuan, P. A., Kumar, R., Rehal, P. K., Toora, P. K., and Ayele, B. T. (2018). Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 9:668.
Wang, H., Mei, W., Qin, Y., and Zhu, Y. (2011). 1-Aminocyclopropane-1-carboxylic acid synthase 2 is phosphorylated by calcium-dependent protein kinase 1 during cotton fiber elongation. Acta Biochim. Biophys. Sin. 43, 654–661. doi: 10.1093/abbs/gmr056
Wang, K. B., Wang, Z. W., Li, F. G., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wang, L. M., Zhu, Y. M., Hu, W. J., Zhang, X. Y., Cai, C. P., and Guo, W. Z. (2015b). Comparative transcriptomics reveals jasmonic acid-associated metabolism related to cotton fiber initiation. PLoS One 10:e0129854.
Wang, Y. Q., Chai, C. L., Valliyodan, B., Maupin, C., Annen, B., and Nguyen, H. T. (2015a). Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max). BMC Genomics 16:951.
Wu, H. M., Hazak, O., Cheung, A. Y., and Yalovsky, S. (2011). RAC/ROP GTPases and auxin signaling. Plant Cell 23, 1208–1218.
Xi, W. Y., Gong, X. M., Yang, Q. Y., Yu, H., and Liou, Y. C. (2016). Pin1At regulates PIN1 polar localization and root gravitropism. Nat. Commun. 7:10430.
Xiao, G. H., He, P., Zhao, P., Liu, H., Zhang, L., Pang, C. Y., et al. (2018). Genome-wide identification of the GhARF gene family reveals that GhARF2 and GhARF18 are involved in cotton fibre cell initiation. J. Exp. Bot. 69, 4323–4337. doi: 10.1093/jxb/ery219
Xiao, Y. H., Li, D. M., Yin, M. H., Li, X. B., Zhang, M., Wang, Y. J., et al. (2010). Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J. Plant Physiol. 167, 829–837. doi: 10.1016/j.jplph.2010.01.003
Yang, C., Lu, X., Ma, B., Chen, S. Y., and Zhang, J. S. (2015). Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol. Plant 8, 495–505. doi: 10.1016/j.molp.2015.01.003
Yang, Y. M., Xu, C. N., Wang, B. M., and Jia, J. Z. (2001). Effects of plant growth regulators on secondary wall thickening of cotton fibres. Plant Growth Regul. 35, 233–237.
Yang, Z., Zhang, C., Yang, X., Liu, K., Wu, Z., Zhang, X., et al. (2014). PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 203, 437–448. doi: 10.1111/nph.12824
Yao, R., Ming, Z., Yan, L., Li, S., Wang, F., Ma, S., et al. (2016). DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. doi: 10.1038/nature19073
Yoshida, Y., Sano, R., Wada, T., Takabayashi, J., and Okada, K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136, 1039–1048. doi: 10.1242/dev.030585
Zhang, M., Zeng, J. Y., Long, H., Xiao, Y. H., Yan, X. Y., and Pei, Y. (2016). Auxin regulates cotton fiber initiation via GhPIN-mediated auxin transport. Plant Cell Physiol. 58, 385–397.
Zhang, M., Zheng, X., Song, S., Zeng, Q., Hou, L., Li, D., et al. (2011). Spatiotemporal manipulation of auxin biosynthesis in cotton ovule epidermal cells enhances fiber yield and quality. Nat. Biotechnol. 29, 453–458. doi: 10.1038/nbt.1843
Zhang, T. Z., Hu, Y., Jiang, W. K., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, X., Hu, D. P., Li, Y., Chen, Y., Eltayib, A., Dong, Z. D., et al. (2017b). Developmental and hormonal regulation of fiber quality in two natural-colored cotton cultivars. J. Integr. Agric. 16, 1720–1729.
Zhang, Y., He, P., Yang, Z., Huang, G., Wang, L., Pang, C., et al. (2017a). A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front. Plant Sci. 8:461.
Zhao, J., Bai, W., Zeng, Q., Song, S., Zhang, M., Li, X., et al. (2015). Moderately enhancing cytokinin level by down-regulation of GhCKX expression in cotton concurrently increases fiber and seed yield. Mol. Breed. 35:60.
Zhou, F., Lin, Q., Zhu, L., Ren, Y., Zhou, K., Shabek, N., et al. (2013). D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. doi: 10.1038/nature12878
Zhou, Y., Zhang, Z. T., Li, M., Wei, X. Z., Li, X. J., Li, B. Y., et al. (2015). Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol. J. 13, 269–280. doi: 10.1111/pbi.12275
Keywords: gene expression, signaling pathway, cotton, fiber initiation, phytohormone, fiber elongation
Citation: Xiao G, Zhao P and Zhang Y (2019) A Pivotal Role of Hormones in Regulating Cotton Fiber Development. Front. Plant Sci. 10:87. doi: 10.3389/fpls.2019.00087
Edited by:
Prem Bhalla, The University of Melbourne, AustraliaReviewed by:
Karl Kunert, University of Pretoria, South AfricaAndrzej Bajguz, University of Białystok, Poland
Copyright © 2019 Xiao, Zhao and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guanghui Xiao, Z3VhbmdodWl4QHNubnUuZWR1LmNu
 Guanghui Xiao
Guanghui Xiao Peng Zhao1,2,3
Peng Zhao1,2,3