- National Institute of Plant Genome Research, New Delhi, India
Plant pathogens like virus, bacteria, and fungi incur a huge loss of global productivity. Targeting the dominant R gene resulted in the evolution of resistance in pathogens, which shifted plant pathologists’ attention toward host susceptibility factors (or S genes). Herein, the application of sequence-specific nucleases (SSNs) for targeted genome editing are gaining more importance, which utilize the use of meganucleases (MN), zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN) with the latest one namely clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9). The first generation of genome editing technologies, due to their cumbersome nature, is becoming obsolete. Owing to its simple and inexpensive nature the use of CRISPR/Cas9 system has revolutionized targeted genome editing technology. CRISPR/Cas9 system has been exploited for developing resistance against virus, bacteria, and fungi. For resistance to DNA viruses (mainly single-stranded DNA viruses), different parts of the viral genome have been targeted transiently and by the development of transgenic plants. For RNA viruses, mainly the host susceptibility factors and very recently the viral RNA genome itself have been targeted. Fungal and bacterial resistance has been achieved mainly by targeting the host susceptibility genes through the development of transgenics. In spite of these successes CRISPR/Cas9 system suffers from off-targeting. This and other problems associated with this system are being tackled by the continuous discovery/evolution of new variants. Finally, the regulatory standpoint regarding CRISPR/Cas9 will determine the fate of using this versatile tool in developing pathogen resistance in crop plants.
Introduction
Plants are continuously being exposed to various pathogens including bacteria, fungi and viruses resulting in 20–40% yield loss globally (Savary et al., 2012; Borrelli et al., 2018). Dominant R-gene-mediated breeding has been the classical approach (Dangl et al., 2013) to achieve resistance against pathogens, which, due to strong selection pressure, resulted in the evolution of resistance among pathogens (Vleeshouwers et al., 2011; Win et al., 2012). Host susceptibility factors (or S genes) came out as alternatives, which are mainly negative regulators of immunity or host proteins, which, upon manipulation by a pathogen, support their growth (Langner et al., 2018). Over the past few years, new breeding techniques (NBTs) have been developed as alternatives to classical plant breeding for crop improvement including pathogen- resistance (Lusser and Davies, 2013; Nelson et al., 2018). NBT include the usage of sequence-specific nucleases (SSNs) such as meganucleases (MNs), zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindrome repeats (CRISPR)/CRISPR-associated protein 9 (Cas9), which have revolutionized targeted modifications of genomes. The requirement of sophisticated protein engineering rendered MN, ZFN, and TALEN techniques less practicable.
The CRISPR-Cas9 system initially reported from Streptococcus pyogenes as class II bacterial adaptive immune system (Langner et al., 2018) is a two-component system consisting of the Cas9 nuclease and a customizable single guide RNA (sgRNA) (Khatodia et al., 2016). Additionally, it requires a protospacer adjacent motif (PAM) sequence (5′-NGG-3′) to induce double-stranded break (DSB) at the target site. DSBs can be repaired either by Homology-Directed Repair (HDR) or more frequently by non- homologous end joining (NHEJ) (Khatodia et al., 2016). Due to its error-prone nature, NHEJ leads to small indels (insertions/deletions) within the target region (Figure 1). This strategy of the CRISPR/Cas9 system has been exploited in plant pathology to target the S genes or the viral genomes. In this context, the mini-review will discuss the recent advances in crop protection against viral, fungal and bacterial pathogens using the CRISPR/Cas9 technology, the advantages, limitations, and possible ways for further improvement of this technology for better utilization in targeted genome editing.
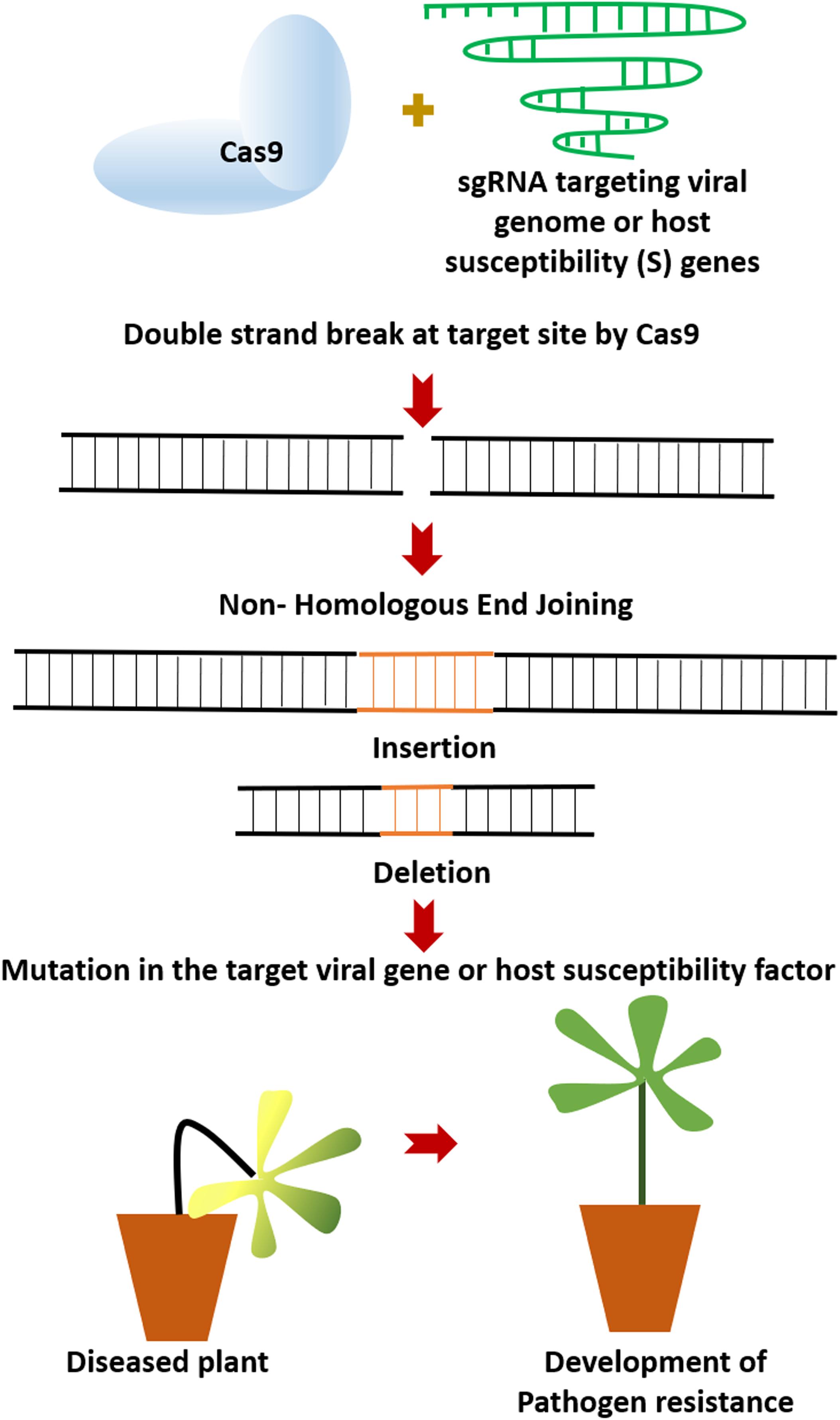
Figure 1. The CRISPR/Cas9 targeting and subsequent non- homologous end joining (NHEJ) process exploited for development of pathogen resistance in plants. SgRNA, single guide RNA.
Achievements in Plant Virus Resistance Through CRISPR/Cas9 Technology
Utilization of the CRISPR/Cas9 system for the viral resistance has been executed by targeting either the viral genome or the host susceptibility factors (Table 1). Most of the CRISPR/Cas9 mediated viral resistance have been achieved by targeting the ssDNA of the geminiviruses (Ali et al., 2015, 2016; Baltes et al., 2015; Ji et al., 2015) with mono- or bi-partite genome containing the genes encoding proteins required for viral replication, movement, and suppressor of host defense machinery (Sharma and Prasad, 2017). Majority of the reports of targeting viral genes using CRISPR/Cas9 are either of transient type or through the development of transgenics in model plants like Nicotiana benthamiana and Arabidopsis thaliana.
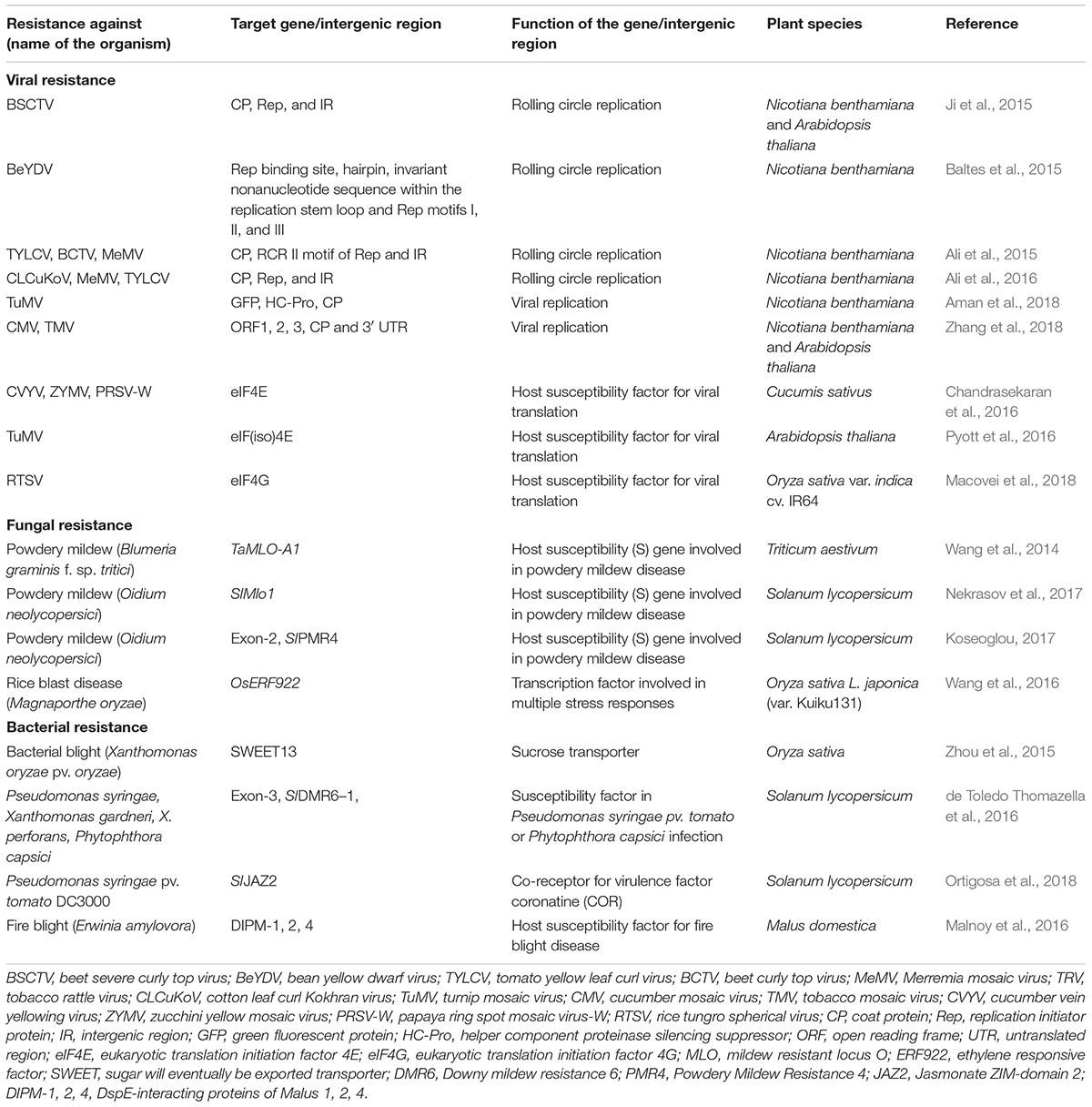
Table 1. Major applications of CRISPR/Cas9 technology for viral, fungal, and bacterial resistance in plants.
The first report of exploitation of CRISPR/Cas9 system for geminivirus resistance came from Baltes et al. (2015) and Ji et al. (2015). Ji et al. (2015) first utilized the CRISPR/Cas9 system to develop beet severe curly top virus (BSCTV) resistance in Arabidopsis and N. benthamiana plants overexpressing sgRNA-Cas9. Baltes et al. (2015) demonstrated that transgenic N. benthamiana plants constitutively expressing Cas9 and sgRNA-Cas9 exhibit enhanced resistance against bean yellow dwarf virus (BeYDV) resulting in reduced viral load and symptoms.
Ali et al. (2015) delivered guide RNAs in Cas9 expressing N. benthamiana via tobacco rattle virus (TRV) vector targeting the viral capsid protein (CP), the RCRII motif of the replication protein (Rep) and the intergenic region (IR) of tomato yellow leaf curl virus (TYLCV). Guide RNA targeting the stem-loop sequence within the origin of replication in the IR was found to be the most effective. As the stem-loop sequence of the origin of replication in the IR is conserved in all geminiviruses, this system also provided resistance to other geminiviruses like a monopartite beet curly top virus (BCTV) and the bipartite Merremia mosaic virus (MeMV). Ali et al. (2016) further extended this work to show that targeting the non-coding IR results in durable resistance as it restricted the generation of virus variants capable of replication and movement, which was not achievable by targeting the coding sequences of geminiviruses. This observation is of great importance for future researchers while targeting the viral genome for long-term, durable resistance against viruses. This work also demonstrated successful utilization of the CRISPR/Cas9 system to develop resistance against Cotton leaf curl Kokhran virus (CLCuKoV).
As Cas9 from S. pyogenes can only edit double-stranded DNA, its initial application was limited to target the DNA viruses alone. Search for RNA editing nucleases led to the discovery of FnCas9 from Francisella novicida (Hirano et al., 2016; Green and Hu, 2017) and LwaCas13a (previously known as C2c2) from Leptotrichia wadei (Abudayyeh et al., 2017; Green and Hu, 2017). The first report of direct targeting of the genome of RNA viruses came from Zhang et al. (2018). The N. benthamiana and Arabidopsis plants expressing FnCas9 and sgRNA specific for cucumber mosaic virus (CMV) or tobacco mosaic virus (TMV) showed a significant reduction in virus accumulation and weakened symptom development. More importantly, the resistance was heritable, and the resultant progenies exhibited significantly reduced virus accumulation.
The other RNA endonuclease Cas13a has dual functions: processing of pre- CRISPR RNAs (crRNAs) and sequence-directed endonucleolysis of the target single-stranded RNA (Khan et al., 2018). Aman et al. (2018) employed CRISPR/Cas13a system in N. benthamiana to interfere with the RNA genome of turnip mosaic virus (TuMV). Targeting the HC-Pro and GFP sequences provided improved resistance than targeting the coat protein (CP) sequence. Successful multiplex targeting of the viral genome by utilization of the inherent ability of Cas13a to process the poly crRNA into individual crRNAs has also been exhibited (Aman et al., 2018). Although the RNA knockdown efficiency of another Cas13 family, namely Cas13b is greater than that of the Cas13a (Cox et al., 2017; Ying, 2018), there is no report yet regarding its utilization in a plant system.
Before the discovery of RNA-guided RNA editing systems, the only way to combat the RNA viruses was to target the host susceptibility factors for viral infection such as the eukaryotic translation initiation factor 4E (eIF4E), eIF(iso)4E, and eIF4G (Sanfacon, 2015). The eIF4E from plants was reported as a crucial host susceptibility component for viral infection and forms the largest group of recessive virus resistance genes in monocots and dicots (Ruffel et al., 2006; Hofinger et al., 2011). The eIF4E and eIF(iso)4E from tomato and melon exhibited recessive resistance against viruses (Mazier et al., 2011; Rodriguez-Hernandez et al., 2012). Targeting eIF4E of cucumber within non-homologous regions of exons 1 and 3 resulted in enhanced resistance against potyviruses such as cucumber vein yellowing virus, zucchini yellow mosaic virus and papaya ringspot mosaic virus-W in homozygous T3 lines (Chandrasekaran et al., 2016).
In a similar genome editing approach using CRISPR/Cas9 technology, Pyott et al. (2016) targeted the eIF(iso)4E locus in Arabidopsis thaliana. The resulting genome edited plants were resistant toward TuMV. Segregation of CRISPR/Cas9 transgene was observed in the T2 generation, and the resulting T3 homozygous lines exhibited morphologically normal phenotype. Macovei et al. (2018) developed tungro disease resistance [causal agent: rice tungro spherical virus (RTSV)] in susceptible rice cultivar IR64 by targeting translation initiation factor 4 gamma (eIF4G) gene.
The advantage of targeting host susceptibility genes is that it results in recessive resistance, which is more durable than dominant R gene-mediated resistance. The probable reason for this is that viruses endure a lower selection pressure impeding their evolution (Borrelli et al., 2018). A problem in targeting the susceptibility genes is that they are also required for translation of the host cells. Hence, although editing of eIF4E results in potyvirus resistance in lettuce, it also impairs with the physiology of the plant (Abdul-Razzak et al., 2009). In another study, disruption of OsSEC3A by CRISPR/Cas9 in rice resulted in enhanced resistance to Magnaporthe oryzae, but it also impaired the normal growth of the plant resulting in a dwarf phenotype (Ma et al., 2018).
Development of Fungal Resistance Via CRISPR/Cas9 Technology
Fungal resistance via CRISPR/Cas9 were mainly achieved till date by targeting the S genes like mildew resistance locus O (MLO), rice Ethylene Response Factor 922 (Table 1). The mildew resistance locus O (MLO) is the most widely known S gene locus. Since its identification in 1942 (Jørgensen, 1992), several mutants have been generated to provide resistance against powdery mildew in barley. MLO encodes a seven transmembrane domain-containing calmodulin binding protein located at the plasma membrane (Kim et al., 2002). Its role in susceptibility toward powdery mildew disease in monocot and dicot plants has also been confirmed (Kusch and Panstruga, 2017). Wang et al. (2014) targeted all three homoeoalleles- MLO-A1, MLO-B1, and MLO-D1 by TALEN and TaMLO-A1 allele of exon 2 of bread wheat using CRISPR/Cas9 technology. Both the approaches were successful in generation of heritable resistance against powdery mildew caused by Blumeria graminis f. sp. tritici.
Powdery mildew in tomato is caused by Ascomycete fungus Oidium neolycopersici (Jones et al., 2001). Tomelo, a non-transgenic tomato variety resistant to O. neolycopersici has been developed by Nekrasov et al. (2017) using CRISPR/Cas9 technology. Targeting the SlMlo1 locus by two sgRNAs resulted in the deletion of 48 bp in the said locus. Segregation of T-DNA was achieved by selfing of the T0 transformants, which was further confirmed by whole-genome Illumina sequencing. Besides MLO, other S genes associated with powdery mildew have also been identified from Arabidopsis. One such example is Powdery Mildew Resistance 4 (PMR4) which encodes for a callose synthase (Huibers et al., 2013). Koseoglou (2017) targeted its ortholog in tomato SlPMR4 using CRISPR/Cas9 technology- deletion and rare inversion mutation were observed in the targeted exon-2. The resulting T2 progenies exhibited partial resistance against O. neolycopersici.
Rice OsERF922 encodes an APETELA2/ethylene response factor (AP2/ERF) type transcription factor, which is strongly induced by M. oryzae (Liu et al., 2012). The identification of specific ERFs as negative regulators of plant immunity made them potential targets for genome editing (Langner et al., 2018). Targeting the OsERF922 gene using CRISPR/Cas9 technology in rice showed resistance to blast disease. The T2 mutant lines were similar to the wild-type rice plants with regard to several agronomic traits (Wang et al., 2016).
Development of Resistance Against Bacteria Using CRISPR/Cas9 Technology
Compared to viral and fungal resistance few reports are available for utilization of CRISPR/Cas9 to combat bacterial diseases of crops (Table 1). The γ-proteobacterium, Xanthomonas oryzae pv. oryzae utilizes type III transcription-activator-like effectors (TALEs) to induce host gene expression resulting in host susceptibility. OsSWEET13, a sucrose transporter gene was identified as a susceptibility gene for X. oryzae pv. oryzae effector protein, PthXo2. Transfer of the OsSWEET13 allele from indica rice IR24 to japonica rice Kitaake conferred disease susceptibility, whereas, mutations in the allele via CRISPR/Cas9 conferred resistance to bacterial blight (Zhou et al., 2015).
DMR6 (Downy mildew resistance 6) functions as a negative regulator of plant defense (Zeilmaker et al., 2015; Langner et al., 2018). de Toledo Thomazella et al. (2016) demonstrated that DMR6 ortholog SlDMR6–1 is upregulated in tomato during infection with Pseudomonas syringae pv. tomato or Phytophthora capsici (Langner et al., 2018). Targeting exon-3 of SlDMR6–1 resulted in mutated plants with a truncated version of SlDMR6 showing broad-spectrum resistance against Xanthomonas gardneri, X. perforans, P. syringae, and P. capsici (de Toledo Thomazella et al., 2016; Langner et al., 2018).
Pseudomonas syringae pv. tomato (Pto) DC3000 is the causal agent of tomato bacterial speck disease. It produces coronatine (COR) which induces stomatal opening ensuing invasion of bacteria. In Arabidopsis, this stomatal response to COR is dependent on COR co-receptor AtJAZ2 (Jasmonate ZIM-domain-2). The truncated form of JAZ2 lacking the C-terminal Jas domain (JAZ2Δjas) prevent stomatal opening by COR (Gimenez-Ibanez et al., 2017). Ortigosa et al. (2018) identified ortholog of AtJAZ2 in tomato (SlJAZ2), and it was targeted by CRISPR/Cas9 to generate dominant JAZ2 repressor- SlJAZ2Δjas which prevented COR induced stomatal opening and provided resistance to biotrophic microbe Pto DC3000. Their experiment is also an example of successful uncoupling between the jasmonate (JA) and salicylate (SA) mediated defense pathways toward necrotroph and biotroph respectively. Effectual defense against biotrophs generally leads to increased susceptibility to necrotrophs and vice versa (Gimenez-Ibanez et al., 2017). As in this experiment, JA-signaling outside the stomata remained unaffected, Sljaz2Δjas plants were also resistant to the necrotrophic fungi Botrytis cinerea which causes tomato gray mold.
The enterobacterium Erwinia amylovora causes fire blight disease in apple and other commercially important Rosaceae plants (Malnoy et al., 2016). The pathogenicity effector (DspE) of E. amylovora interacts with four leucine-rich-repeat, receptor-like serine/theonine kinases produced by DspE-interacting proteins of Malus (DIPM) genes- DIPM 1, 2, 3, 4 (Borejsza-Wysocka et al., 2006). Malnoy et al. (2016) utilized the CRISPR/Cas9 system to target DIPM 1, 2, and 4 genes in apple protoplast to develop resistance against fire blight disease. The experiment by Malnoy et al. (2016) also demonstrates successful direct delivery of CRISPR/Cas9 ribonucleoproteins (RNPs) (pre-assembled sgRNA/Cas9 complex) into plant protoplasts which has several benefits like rapid targeting efficiency, improved on-target and reduced off-target activity (Malnoy et al., 2016; Borrelli et al., 2018).
Advantages and Limitations of CRISPR/Cas9 Technology
The main advantage of CRISPR/Cas9 technology is its inexpensiveness and ease of use. Unlike ZFN and TALEN, which is dependent on protein engineering, synthesis and validation (Joung and Sander, 2013; Voytas, 2013; Puchta and Fauser, 2014), here only the guide RNA needs to be designed. This single guide RNA (sgRNA) provides target-site specificity in CRISPR/Cas9 system (Jinek et al., 2012). Another advantage of CRISPR/Cas9 system compared to the first-generation genome editing techniques is the ability of multiplex genome editing, i.e., targeting multiple genes using a single construct (Murugan et al., 2017; Borrelli et al., 2018). Additionally, using the CRISPR/Cas9 system transgene-free genome edited plants can be obtained in very few generations (Khatodia et al., 2016; Langner et al., 2018). The efficiency of CRISPR/Cas9 system relies on the method of transformation. In plants generally routinely used methods are Agrobacterium-mediated transformation, biolistic transformation, and protoplast transformation. To perform CRISPR based homology-directed repair, the biolistic method is preferred over the other two methods (Baysal et al., 2016; Shi et al., 2017). Several factors such as the type of promoter used for driving the expression of Cas9 (e.g., 35S, rice ubiquitin promoter) and promoter driving sgRNA (e.g., rice snoRNA U3 promoter, Arabidopsis U6 promoter) also determines the targeted genome editing efficiency (Ma et al., 2016).
Like every other technique CRISPR/Cas9 system also has its limitations. Although less frequent in plants, CRISPR/Cas9 system suffers from off-target mutations (Langner et al., 2018). This can be tackled by using paired nickases where the RuvC domain of Cas9 is inactivated. As a result, it creates a nick instead of a double-strand break at the target site (Khatodia et al., 2016; Langner et al., 2018). Two nicks induced in close proximity ultimately produces a double-strand break (Hsu et al., 2014). In addition, the paired nickase system is also useful in high-efficiency HDR (Khatodia et al., 2016). The problem of off-targeting can also be tackled by the use of recently discovered CRISPR/Cpf1 from Prevotella and Francisella 1 (Cpf1) (Zetsche et al., 2015; Puchta, 2017; Zaidi et al., 2017) which creates a staggered double-strand break at the target site. Recent reports of genome editing by Cpf1 exhibited little to no off-target effects in rice (Tang et al., 2017; Xu et al., 2017). Additionally, Woo et al. (2015) reported that when ribonucleoproteins complexes or RNPs, instead of DNA were transfected into lettuce protoplasts, no off-target mutations were detected in the genome.
Another factor that limits the utilization of Cas9 is the PAM specificity. The stringent requirement of NGG motif immediately after the protospacer element limits targeting of the high AT-rich genome (Zetsche et al., 2015). The CRISPR/Cpf1 system mentioned earlier is also useful in this regard. Cpf1 recognizes a T-rich PAM sequence 5′-TTTN-3′ (or 5′-TTTV-3′; V = A, C, or G, in some cases) instead of 5′-NGG-3′. Cpf1 generates a staggered double-strand break with cohesive ends, which can also be useful for increasing the HDR efficiency (Zaidi et al., 2017). To tackle the problem of PAM specificity mutations have also been generated in the PAM-interacting domain of wild-type SpCas9 (Kleinstiver et al., 2015) which recognize alternative PAM sequences like NGCG, NGAG (Anders et al., 2016; Langner et al., 2018).
Future Perspectives
Targeted genome editing by CRISPR/Cas9 can yield desired disease resistant traits within a very short period which cannot be achievable by traditional breeding methods (Borrelli et al., 2018; Langner et al., 2018). When resistance achieved via dominant R genes is amenable to be overcome by the adaptive potential of the pathogens, targeting the host susceptibility factors seems to be a smarter alternative. The availability of the genome sequences of economically important crops and their transcriptomics datasets can be useful in the identification of new S genes (Zaidi et al., 2018). However, the CRISPR/Cas9 technology is still in its juvenile phase- field trial of the genome edited crops is limited (Shi et al., 2017) which will be essential to check the durability of the incurred pathogen resistance. The regulatory issues regarding genome edited crops will play an important role in this regard. The United States Department of Agriculture (USDA) does not regulate genome edited plants which could otherwise have been developed through traditional breeding techniques (Jaganathan et al., 2018). On the other hand, in the European Union, genome edited crops are currently subject to regulations as genetically modified (GM) organisms (Callaway, 2018). Although safety issues regarding the application of CRISPR/Cas9 technology must be examined by scientific means and considering the associated practical and societal aspects (Bechtold, 2018), regulatory standpoint in favor of this promising technology will assist in its proper dissemination leading to better crop management.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The authors’ work on plant molecular virology is funded by the research grant of J.C. Bose National Fellowship Award from DST-SERB, Ministry of Science & Technology, Government of India (File No. JCB/2018/000001/SSC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AD acknowledges DST-SERB, Ministry of Science & Technology, Government of India for N-PDF Award (File No. PDF/2016/003148). The authors are thankful to DBT-eLibrary Consortium (DeLCON) for providing access to the e-resources.
References
Abdul-Razzak, A., Guiraud, T., Peypelut, M., Walter, J., Houvenaghel, M. C., Candresse, T., et al. (2009). Involvement of the cylindrical inclusion (CI) protein in the overcoming of an eIF4E- mediated resistance against Lettuce mosaic potyvirus. Mol. Plant Pathol. 10, 109–113. doi: 10.1111/j.1364-3703.2008.00513.x
Abudayyeh, O. O., Gootenberg, J. S., Essletzbichler, P., Han, S., Joung, J., Belanto, J. J., et al. (2017). RNA targeting with CRISPR-Cas13. Nature 550, 280–284. doi: 10.1038/nature24049
Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16:238. doi: 10.1186/s13059-015-0799-6
Ali, Z., Ali, S., Tashkandi, M., Shan, S., Zaidi, A., and Mahfouz, M. M. (2016). CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci. Rep. 6:26912. doi: 10.1038/srep26912
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19:1. doi: 10.1186/s13059-017-1381-1
Anders, C., Bargsten, K., and Jinek, M. (2016). Structural plasticity of PAM recognition by engineered variants of the RNA-guided endonuclease Cas9. Mol. Cell. 61, 895–902. doi: 10.1016/j.molcel.2016.02.020
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 1:15145. doi: 10.1038/nplants.2015.145
Baysal, C., Bortesi, L., Zhu, C., Farré, G., Schillberg, S., and Christou, P. (2016). CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa. Mol. Breed. 36:108. doi: 10.1007/s11032-016-0533-4
Bechtold, S. (2018). Beyond risk considerations: where and how can a debate about non-safety related issues of genome editing in agriculture take place? Front. Plant Sci. 9:1724. doi: 10.3389/fpls.2018.01724
Borejsza-Wysocka, E. E., Malnoy, M., Aldwinckle, H. S., Meng, X., Bonasera, J. M., Nissinen, R. M., et al. (2006). The fire blight resistance of apple clones in which DspE-interacting proteins are silenced. Acta Hortic. 704, 509–514. doi: 10.17660/ActaHortic.2006.704.80
Borrelli, V. M. G., Brambilla, V., Rogowsky, P., Marocco, A., and Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9:1245. doi: 10.3389/fpls.2018.01245
Callaway, E. (2018). CRISPR plants now subject to tough GM laws in European Union. Nature 560:16. doi: 10.1038/d41586-018-05814-6
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Cox, D. B. T., Gootenberg, J. S., Abudayyeh, O. O., Franklin, B., Kellner, M. J., Joung, J., et al. (2017). RNA editing with CRISPR-Cas13. Science 358, 1019–1027. doi: 10.1126/science.aaq0180
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
de Toledo Thomazella, D. P., Brail, Q., Dahlbeck, D., and Staskawicz, B. J. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv [Preprint]. doi: 10.1101/064824
Gimenez-Ibanez, S., Boter, M., Ortigosa, A., García-Casado, G., Chini, A., Lewsey, M. G., et al. (2017). JAZ2 controls stomata dynamics during bacterial invasion. New Phytol. 213, 1378–1392. doi: 10.1111/nph.14354
Green, J. C., and Hu, J. S. (2017). Editing plants for virus resistance using CRISPR-Cas. Acta Virol. 61, 138–142. doi: 10.4149/av_2017_02_02
Hirano, H., Gootenberg, J. S., Horii, T., Abudayyeh, O. O., Kimura, M., Hsu, P. D., et al. (2016). Structure and engineering of Francisella novicida Cas9. Cell 164, 950–961. doi: 10.1016/j.cell.2016.01.039
Hofinger, B. J., Russell, J. R., Bass, C. G., Baldwin, T., dos Reis, M., Hedley, P. E., et al. (2011). An exceptionally high nucleotide and haplotype diversity and a signature of positive selection for the eIF4E resistance gene in barley are revealed by allele mining and phylogenetic analyses of natural populations. Mol. Ecol. 20, 3653–3668. doi: 10.1111/j.1365-294X.2011.05201.x
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Huibers, R. P., Loonen, A. E., Gao, D., Van den Ackerveken, G., Visser, R. G., and Bai, Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One 8:e67467. doi: 10.1371/journal.pone.0067467
Jaganathan, D., Ramasamy, K., Sellamuthu, G., Jayabalan, S., and Venkataraman, G. (2018). CRISPR for crop improvement: an update review. Front. Plant Sci. 9:985. doi: 10.3389/fpls.2018.00985
Ji, X., Zhang, H., Zhang, Y., Wang, Y., and Gao, C. (2015). Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1:15144. doi: 10.1038/nplants.2015.144
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, H., Whipps, J. M., and Gurr, S. J. (2001). The tomato powdery mildew fungus Oidium neolycopersici. Mol. Plant Pathol. 2, 303–309. doi: 10.1046/j.1464-6722.2001.00084.x
Jørgensen, I. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63, 141–152. doi: 10.1007/BF00023919
Joung, J. K., and Sander, J. D. (2013). TALENs: a widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 14, 49–55. doi: 10.1038/nrm3486
Khan, M. Z., Amin, I., Hameed, A., and Mansoor, S. (2018). CRISPR-Cas13a: prospects for plant virus resistance. Trends Biotechnol. 36, 1207–1210. doi: 10.1016/j.tibtech.2018.05.005
Khatodia, S., Bhatotia, K., Passricha, N., Khurana, S. M., and Tuteja, N. (2016). The CRISPR/Cas genome-editing tool: application in improvement of crops. Front. Plant Sci. 7:506. doi: 10.3389/fpls.2016.00506
Kim, M. C., Panstruga, R., Elliott, C., Müller, J., Devoto, A., Yoon, H. W., et al. (2002). Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 416, 447–451. doi: 10.1038/416447a
Kleinstiver, B. P., Prew, M. S., Tsai, S. Q., Topkar, V. V., Nguyen, N. T., Zheng, Z., et al. (2015). Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 523, 481–485. doi: 10.1038/nature14592
Koseoglou, E. (2017). The Study of SlPMR4 CRISPR/Cas9-Mediated Tomato Allelic Series for Resistance Against Powdery Mildew. Master’s thesis, Wageningen University and Research, Wageningen.
Kusch, S., and Panstruga, R. (2017). mlo-Based resistance: an apparently universal “weapon” to defeat powdery mildew disease. Mol. Plant Microbe Interact. 30, 179–189. doi: 10.1094/MPMI-12-16-0255-CR
Langner, T., Kamoun, S., and Belhaj, K. (2018). CRISPR Crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56, 479–512. doi: 10.1146/annurev-phyto-080417-050158
Liu, D., Chen, X., Liu, J., Ye, J., and Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Lusser, M., and Davies, H. V. (2013). Comparative regulatory approaches for groups of new plant breeding techniques. New Biotechnol. 30, 437–446. doi: 10.1016/j.nbt.2013.02.004
Ma, J., Chen, J., Wang, M., Ren, Y., Wang, S., Lei, C., et al. (2018). Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 69, 1051–1064. doi: 10.1093/jxb/erx458
Ma, X., Zhu, Q., Chen, Y., and Liu, Y. G. (2016). CRISPR/Cas9 platforms for genome editing in plants: developments and applications. Mol. Plant. 9, 961–974. doi: 10.1016/j.molp.2016.04.009
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Èermák, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Mazier, M., Flamain, F., Nicolai, M., Sarnette, V., and Caranta, C. (2011). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS One 6:e29595. doi: 10.1371/journal.pone.0029595
Murugan, K., Babu, K., Sundaresan, R., Rajan, R., and Sashital, D. G. (2017). The revolution continues: newly discovered systems expand the CRISPR-Cas toolkit. Mol. Cell. 68, 15–25. doi: 10.1016/j.molcel.2017.09.007
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7:482. doi: 10.1038/s41598-017-00578-x
Nelson, R., Wiesner-Hanks, T., Wisser, R., and Balint-Kurti, P. (2018). Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 19, 21–33. doi: 10.1038/nrg.2017.82
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2018). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. doi: 10.1111/pbi.13006 [Epub ahead of print].
Puchta, H. (2017). Applying CRISPR/Cas for genome engineering in plants: the best is yet to come. Curr. Opin. Plant Biol. 36, 1–8. doi: 10.1016/j.pbi.2016.11.011
Puchta, H., and Fauser, F. (2014). Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J. 78, 727–741. doi: 10.1111/tpj.12338
Pyott, D. E., Sheehan, E., and Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Rodriguez-Hernandez, A. M., Gosalvez, B., Sempere, R. N., Burgos, L., Aranda, M. A., and Truniger, V. (2012). Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. doi: 10.1111/j.1364-3703.2012.00785.x
Ruffel, S., Gallois, J. L., Moury, B., Robaglia, C., Palloix, A., and Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089–2098. doi: 10.1099/vir.0.81817-0
Sanfacon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Savary, S., Ficke, A., Aubertot, J. N., and Hollier, C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4, 519–537. doi: 10.1007/s00203-017-1426-6
Sharma, N., and Prasad, M. (2017). An insight into plant–tomato leaf curl New Delhi virus interaction. Nucleus 60, 335–348.
Shi, J., Gao, H., Wang, H., Lafitte, H. R., Archibald, R. L., Yang, M., et al. (2017). ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 15, 207–216. doi: 10.1111/pbi.12603
Tang, X., Lowder, L. G., Zhang, T., Malzahn, A. A., Zheng, X., Voytas, D. F., et al. (2017). A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat. Plants. 3:17018. doi: 10.1038/nplants.2017.18
Vleeshouwers, V. G., Raffaele, S., Vossen, J. H., Champouret, N., Oliva, R., Segretin, M. E., et al. (2011). Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49, 507–531. doi: 10.1146/annurev-phyto-072910-095326
Voytas, D. F. (2013). Plant genome engineering with sequence-specific nucleases. Annu. Rev. Plant Biol. 64, 327–350. doi: 10.1146/annurev-arplant-042811-105552
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., and Gao, Y. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Win, J., Chaparro-Garcia, A., Belhaj, K., Saunders, D. G., Yoshida, K., Dong, S., et al. (2012). Effector biology of plant associated organisms: concepts and perspectives. Cold Spring Harb. Symp. Quant. Biol. 77, 235–247. doi: 10.1101/sqb.2012.77.015933
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Xu, R., Qin, R., Li, H., Li, D., Li, L., Wei, P., et al. (2017). Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol. J. 15, 713–717. doi: 10.1111/pbi.12669
Ying, X. (2018). CRISPR, a new powerful weapon for plant to combat viruses. Virol. Mycol. 7:e111. doi: 10.4172/2161-0517.1000e111
Zaidi, S. S., Mahfouz, M. M., and Mansoor, S. (2017). CRISPR-Cpf1: a new tool for plant genome editing. Trends Plant Sci. 22, 550–553. doi: 10.1016/j.tplants.2017.05.001
Zaidi, S. S., Mukhtar, M. S., and Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zeilmaker, T., Ludwig, N. R., Elberse, J., Seidl, M. F., Berke, L., Van Doorn, A., et al. (2015). Downy mildew resistant 6 and dmr6-like oxygenase 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 81, 210–222. doi: 10.1111/tpj.12719
Zetsche, B., Gootenberg, J. S., Abudayyeh, O. O., Slaymaker, I. M., Makarova, K. S., Essletzbichler, P., et al. (2015). Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163, 759–771. doi: 10.1016/j.cell.2015.09.038
Zhang, T., Zheng, Q., Yi, X., An, H., Zhao, Y., Ma, S., et al. (2018). Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol. J. 16, 1415–1423. doi: 10.1111/pbi.12881
Keywords: CRISPR/Cas9, guide RNA, protospacer adjacent motif, genome editing, pathogen- resistance, host susceptibility factor
Citation: Das A, Sharma N and Prasad M (2019) CRISPR/Cas9: A Novel Weapon in the Arsenal to Combat Plant Diseases. Front. Plant Sci. 9:2008. doi: 10.3389/fpls.2018.02008
Received: 30 November 2018; Accepted: 31 December 2018;
Published: 15 January 2019.
Edited by:
Giorgio Gambino, Institute for Sustainable Plant Protection, Italian National Research Council (IPSP-CNR), ItalyReviewed by:
John Hu, University of Hawaii at Manoa, United StatesYuanhuai Han, Shanxi Agricultural University, China
Copyright © 2019 Das, Sharma and Prasad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manoj Prasad, bWFub2pfcHJhc2FkQG5pcGdyLmFjLmlu
 Ayan Das
Ayan Das Namisha Sharma
Namisha Sharma Manoj Prasad
Manoj Prasad