
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 January 2019
Sec. Crop and Product Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01956
This article is part of the Research Topic Biostimulants in Agriculture View all 51 articles
 Monica Agnolucci1
Monica Agnolucci1 Luciano Avio1
Luciano Avio1 Alessandra Pepe1
Alessandra Pepe1 Alessandra Turrini1
Alessandra Turrini1 Caterina Cristani2
Caterina Cristani2 Paolo Bonini3
Paolo Bonini3 Veronica Cirino4
Veronica Cirino4 Fabrizio Colosimo4
Fabrizio Colosimo4 Maurizio Ruzzi5
Maurizio Ruzzi5 Manuela Giovannetti1*
Manuela Giovannetti1*The implementation of sustainable agriculture encompasses practices enhancing the activity of beneficial soil microorganisms, able to modulate biogeochemical soil cycles and to affect soil fertility. Among them, arbuscular mycorrhizal fungi (AMF) establish symbioses with the roots of most food crops and play a key role in nutrient uptake and plant protection from biotic and abiotic stresses. Such beneficial services, encompassing improved crop performances, and soil resources availability, are the outcome of the synergistic action of AMF and the vast communities of mycorrhizospheric bacteria living strictly associated with their mycelium and spores, most of which showing plant growth promoting (PGP) activities, such as the ability to solubilize phosphate and produce siderophores and indole acetic acid (IAA). One of the strategies devised to exploit AMF benefits is represented by the inoculation of selected isolates, either as single species or in a mixture. Here, for the first time, the microbiota associated with a commercial AMF inoculum was identified and characterized, using a polyphasic approach, i.e., a combination of culture-dependent analyses and metagenomic sequencing. Overall, 276 bacterial genera were identified by Illumina high-throughput sequencing, belonging to 165 families, 107 orders, and 23 phyla, mostly represented by Proteobacteria and Bacteroidetes. The commercial inoculum harbored a rich culturable heterotrophic bacterial community, whose populations ranged from 2.5 to 6.1 × 106 CFU/mL. The isolation of functional groups allowed the selection of 36 bacterial strains showing PGP activities. Among them, 14 strains showed strong IAA and/or siderophores production and were affiliated with Actinomycetales (Microbacterium trichotecenolyticum, Streptomyces deccanensis/scabiei), Bacillales (Bacillus litoralis, Bacillus megaterium), Enterobacteriales (Enterobacter), Rhizobiales (Rhizobium radiobacter). This work demonstrates for the first time that an AMF inoculum, obtained following industrial production processes, is home of a large and diverse community of bacteria with important functional PGP traits, possibly acting in synergy with AMF and providing additional services and benefits. Such bacteria, available in pure culture, could be utilized, individually and/or in multispecies consortia with AMF, as biofertilizers and bioenhancers in sustainable agroecosystems, aimed at minimizing the use of chemical fertilizers and pesticides, promoting primary production, and maintaining soil health and fertility.
Worldwide, a major shift is taking place in agriculture, in order to meet the growing global demand for a safe production of high-quality food, able to maintain or enhance environmental quality and to conserve natural resources for future generations. The implementation of sustainable agriculture encompasses practices enhancing the activity of soil biogeochemical cycles, at the basis of long-term soil productivity and health. The most important players of soil biological fertility are represented by beneficial soil microorganisms, able to modulate biochemical and physiological soil processes, and to affect its biological and nutritional characteristics (Barea et al., 2005). Among them, arbuscular mycorrhizal fungi (AMF, Glomeromycota) are recognized as ecologically and economically important elements of sustainable food production systems, given the key role played in plant nutrition and health, by reducing the input of chemical fertilizers and pesticides (Smith and Read, 2008).
AMF are obligate mutualistic biotrophs, establishing symbioses with the roots of most land plants, including the major food and feed crops, from cereals and legumes to fruits and vegetables, including also important industrial plants, such as sunflower, tobacco, cotton, and medicinal plants (Smith and Read, 2008). AMF symbionts facilitate plant nutrient uptake, mainly phosphorus (P), nitrogen (N), sulfur (S) potassium (K), calcium (Ca), copper (Cu), and zinc (Zn), by means of a large network of extraradical hyphae spreading from colonized roots to the surrounding soil and functioning as a supplementary absorbing system (Giovannetti et al., 2001; Avio et al., 2006). Moreover, they protect plants from biotic and abiotic stresses (Augé, 2001; Evelin et al., 2009; Sikes et al., 2009), provide essential ecosystem services (Gianinazzi et al., 2010), and affect the biosynthesis of beneficial plant secondary metabolites, contributing to the production of safe and high quality food (Sbrana et al., 2014; Avio et al., 2018). However, such beneficial services, encompassing improved crop performances and soil resources availability, are the outcome of the synergistic action of AMF and the vast communities of mycorrhizospheric bacteria living strictly associated with their mycelium and spores (Hildebrandt et al., 2006; Agnolucci et al., 2015). AMF-associated microbiota has been reported to promote mycorrhizal activity (Mayo et al., 1986; Xavier and Germida, 2003; Horii and Ishii, 2006; Giovannetti et al., 2010), to protect plants from soilborne pathogens (Citernesi et al., 1996; Budi et al., 1999; Li et al., 2007; Bharadwaj et al., 2008a,b) and to provide nutrients and growth factors (Barea et al., 2002; Xavier and Germida, 2003), thus being considered as plant growth promoting (PGP) bacteria (PGPB) (Philippot et al., 2013). Molecular investigations allowed the description of the complexity and diversity of bacterial communities associated to AMF spores belonging to different species and isolates, suggesting that their differential occurrence may affect the performance of the relevant taxa in terms of infectivity and efficiency, given their important functional roles as PGPB (Roesti et al., 2005; Long et al., 2008; Agnolucci et al., 2015). Other studies, aimed at isolating and functionally characterizing spore associated bacteria, reported the occurrence of bacteria showing antagonistic activity against plant pathogens (Budi et al., 1999; Bharadwaj et al., 2008a), phosphate-solubilizing and nitrogenase activity (Cruz et al., 2008; Cruz and Ishii, 2011), and indole acetic acid (IAA) production (Bharadwaj et al., 2008a). A recent work, using a culture-dependent approach, showed that bacterial strains isolated in pure culture from Rhizophagus intraradices spores were able to solubilize P from phytate and inorganic sources (69.7 and 49.2%, respectively), produce siderophores (65.6%), and IAA (42.6%) (Battini et al., 2016). The last two molecules are very important for plant growth and nutrition. Actually, IAA, a phytohormone of the auxin class, affects the morphology and physiology of roots, enhancing cell division and elongation, and the formation of lateral roots, thus improving water and nutrient uptake and playing a key role in the regulation of plant development (Khalid et al., 2004; Aloni et al., 2006; Duca et al., 2014). Siderophores are low molecular weight, high-affinity iron-chelating compounds able to bind soluble Fe3, even at high pH when Fe solubility decreases (Mimmo et al., 2014), thus making it available to bacteria and plants (Colombo et al., 2014). Given the essential role played by iron in plant biochemical processes, such as photosynthesis and respiration (Kobayashi and Nishizawa, 2012), bacterial siderophores, facilitating plant Fe acquisition, represent important factors of plant growth and development (Crowley et al., 1988; Duijff et al., 1994a,b; Walter et al., 1994; Yehuda et al., 1996; Siebner-Freibach et al., 2003; Jin et al., 2006; Vansuyt et al., 2007; Robin et al., 2008). Moreover, siderophores have been reported to possess biocontrol activity against soilborne diseases, by means of iron competition (Thomashow et al., 1990; Glick, 1995; Whipps, 2001), inhibiting the development of deleterious plant pathogens (Davison, 1988; Arora et al., 2001).
Although the individual roles of AMF and their associated bacteria in optimizing plant performance are still to be completely dissected, AMF are progressively more considered among the main factors of sustainable food (primary) production (Philippot et al., 2013; Rouphael et al., 2015). Two main strategies have been devised to exploit the benefits deriving from the mycorrhizal symbionts: the adoption of specific management practices and the use of AMF inoculation. The first one focuses on the improvement of the activity of native AMF, pursued by using crop rotation and mycotrophic cover crops, able to raise soil mycorrhizal potential and to shape native AMF communities (Kabir and Koide, 2002; Karasawa and Takebe, 2012; Lehman et al., 2012; Njeru et al., 2014, 2015; Turrini et al., 2016, 2017), and by reducing tillage intensity or chemical fertilizations, which affect AMF species composition, spore abundance and mycorrhizal colonization (Douds et al., 1995; Jansa et al., 2003; Oehl et al., 2004; Castillo et al., 2006; Brito et al., 2012; Avio et al., 2013). The second strategy focuses on the inoculation of selected AMF, either as single species or in a mixture, reported as efficient root colonizers and plant nutrition enhancers (Jeffries et al., 2003; Gianinazzi and Vosatka, 2004; Lekberg and Koide, 2005; Rouphael et al., 2015).
Many types of commercial AMF inoculum are available on the market, including sterile products obtained in vitro using genetically modified Ri T-DNA roots and the species Rhizoglomus irregulare (synonym Rhizophagus irregularis, basionym Glomus irregulare). However, most of the commercial products are obtained from greenhouse multiplication on mycotrophic trap plants and represent a multipartite symbiosis, where a rich community of bacteria may thrive, associated with AMF propagules, and exert important functional activities, as PGPB. Here, for the first time, we explored the bacterial metagenome of a commercially available AMF inoculum by Illumina high-throughput sequencing, a method able to provide information about culturable and unculturable members of the inoculum microbiota. Moreover, we isolated and functionally selected culturable bacteria showing important PGP traits, as the ability to produce IAA and siderophores, to be utilized, individually and/or in multispecies consortia with AMF as beneficial biofertilizers/bioenhancers in sustainable agroecosystems.
The commercial inoculum utilized consisted of the substrate where trap plants (Allium ampeloprasum var. porrum L.) were grown and of mycorrhizal root fragments, AMF spores, and extraradical mycelium of Rhizoglomus irregulare BEG72 (synonym Rhizophagus irregularis, basionym Glomus irregulare). The substrate (vermiculite) and the seeds utilized for the inoculum production were sterilized prior to their utilization. The only microbial input in the commercial product arised from the AM fungus and its associated microbiota. The corresponding AMF inoculum is available on the market under the name “AEGIS” (Atens, Agrotecnologias Naturales S.L.). The percentage of mycorrhizal colonization of the roots contained within the inoculum was assessed on three 5 g samples by the gridline intersect method, after clearing with 10% KOH and staining with 0.05% Trypan blue in lactic acid (Giovannetti and Mosse, 1980). The mycorrhizal potential of the commercial inoculum was assessed by using the Mycorrhizal Inoculum Potential (MIP) bioassay, as described in Njeru et al. (2014). Briefly, three replicate inoculum samples were sown with Cichorium intybus L. cv. Zuccherina di Trieste, put in sun-transparent bags and maintained in a growth chamber at 27°C and 16/8 h light/dark daily cycle until harvest. Roots were harvested 30 days after sowing, cleaned with tap water and cleared, stained, and examined for AMF colonization assessment, as described above.
The composition of the bacterial community of three commercial inoculum samples was determined by Next-generation high-throughput DNA sequencing (NGS; Ansorge, 2009). Total community DNA was extracted from each sample using DNeasy PowerSoil Kit (Qiagen, Hilden, Germany). In brief, 50 g of sample and 0.1 mL Tween 20 were suspended in saline phosphate buffer (100 mL) and homogenized in a paddle blender (BagMixer® 400, Interscience, Saint Nom, France) for three min at maximum speed. Substrate soil and root fragments were removed by low speed (1,000 g) centrifugation, then, for DNA extraction, cells were collected after centrifugation and lysed using the DNeasy PowerSoil reagents and Qiagen spin columns on a QIAcube automated station (Qiagen, Hilden, Germany).
Three 16S rRNA gene amplicon libraries were prepared by PCR amplification of an approximate 630 bp region within the hypervariable (V3-V4) region of the 16S rRNA gene according to the Illumina 16S metagenomic sequencing library protocol. PCR amplification was performed with broad spectrum 16S rRNA primers (forward primer: 5′-TCGTCGGCAGCGTCAGATGTGTAT AAGAGACAGCCTACGGGNGGCWGCAG-3′, reverse primer: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC-3′) (Klindworth et al., 2013), using Kapa HiFi HotStart 2 × ReadyMix DNA polymerase (Kapa Biosystems Ltd., London, UK). Cycle conditions were: an initial step at 95°C for 3 min; 25 cycles of 95°C (30 s), 55°C (30 s), 72°C (30 s); a final extension of 5 min at 72°C. Libraries were purified using AMPure XP beads (LABPLAN; Naas, Ireland) according to the Illumina 16S metagenomic sequencing library protocol. Dual indices and Illumina sequencing adapters from the Illumina Nextera XT index kits v2 B and C (Illumina, San Diego, USA) were added to the target amplicons in a second Index PCR step using Kapa HotStart HiFi 2 × ReadyMix DNA polymerase (Kapa Biosystems Ltd.). Cycle conditions were: 95°C (3 min); 9 cycles of 95°C (30 s), 55°C (30 s), 72°C (30 s); a final extension of 5 min at 72°C. Libraries were again purified using AMPure XP beads (LABPLAN; Naas, Ireland) according to the Illumina 16S metagenomic sequencing library protocol. Libraries were quantified using a Qubit fluorometer (Life Technologies, Paisley, UK) and pooled in equal concentrations (4 nM) into a single pool, according to their Qubit quantification measurement. The library pool was diluted and denatured according to the Illumina MiSeq library preparation guide. The amplicon library (8 pM) was spiked with 10% denatured and diluted PhiX Illumina control library (12.5 pM). The sequencing run was conducted on the Illumina MiSeq using the 600 cycle MiSeq reagent kit (version 3) with paired 300 bp reads.
Illumina sequencing was performed using MiSeq (Illumina, San Diego, CA). Paired-end sequencing used custom primers and a 600-cycle sequencing kit (V3), according to manufacturer instructions. Amplicon sequencing was carried out in the presence of 10% PhiX control (Illumina, San Diego, CA) to allow proper focusing and matrix calculations.
Raw data processing, run de-multiplexing and operational taxonomic unit (OTU) analysis were performed using the CLC Genomics Workbench (Version 11.0.1) with CLC Microbial Genomics Module (Version 3.5) (Qiagen Bioinformatics, Hilden, Germany). Such programme was used also for the estimation of alpha diversity (total richness in OTUs). Taxonomy attribution was performed against SILVA 16S v132 at the identity level of 97%.
Three 40 g samples of the commercial inoculum were suspended in 360 mL of sterile physiological solution added with Tween 80 (0.36 μL). The suspension was shaken for 30 min using a multi wrist shaker (Labline Instruments, Illinois, USA). Hundred microliter suspension for each sample were plated in triplicate onto Petri dishes containing different agar media. Culturable heterotrophic bacteria were isolated on Tryptic Soil Agar (TSA, 30 g L−1 tryptic soy broth, 20 g L−1 bacteriological agar, Oxoid, Milan, Italy), a medium which, given its non-selectivity, allows the recovery of a wide range of aerobic and facultative anaerobic gram-negative and gram-positive bacteria. In order to isolate specific functional bacterial groups, two additional selective media were used. The selective N-free Winogradsky medium (N-free W) (Tchan, 1984) was utilized for the isolation of putative nitrogen-fixing bacteria, able to grow on N-free medium. For the isolation of bacteria able to solubilize inorganic phosphate the National Botanical Research Institute's Phosphate growth (NBRIP) medium was used (Nautiyal, 1999). The three culture media were supplemented with 100 mg L−1 of cyclohexymide and 500 UI L−1 of nystatin (Sigma–Aldrich, Milan, Italy) to inhibit possible fungal development. The number of colony forming units (CFU) was assessed after 2 and 7 days of incubation at 28°C for TSA and the other two media, respectively. Bacteria grown on N-free W and those showing halo zones formation on NBRIP, were selected and purified by streaking four times onto the same medium used for the isolation. In addition, bacteria grown in TSA medium were randomly selected on the basis of phenotypic colony characteristics, i.e., shape, size, edge morphology, surface and pigment and inoculated onto N-free W and NBRIP media and then purified by streaking four times onto the same medium. The purified strains were maintained at −80°C in cryovials with 20% (v/v) of glycerol in the collection IMA (International Microbial Archives) of the Department of Agriculture, Food and Environment, University of Pisa.
All the bacterial strains isolated and selected as described above, were screened in vitro for two functional traits linked to the promotion of plant growth and performance, i. e., the ability to produce IAA and siderophores. The production of IAA was assessed using Luria–Bertani Broth (LBB) (Bharadwaj et al., 2008a) and following the method described by Battini et al. (2016). Briefly, strains were inoculated in 4 mL of LBB amended with 1 mg mL−1 of l-tryptophan (Sigma–Aldrich, Milan, Italy), incubated at 20°C until exponential growth phase was reached. They were centrifugated (7500 rpm for 10 min) and 1mL of supernantant was transferred in a 24-well plate, mixed with 2 mL of Salkowski reagent (1.2% FeCl3 in 37% sulfuric acid). The non-inoculated medium represented the negative control, and the medium amended with pure IAA the positive one. Development of red–purple color after 3 min incubation in the dark indicated positive strains for IAA production. Strains were classified using a rating scale as follows (Figure 1): –, no production (no color development); +/–, low production (pale pink); +, production (light purple); ++, moderate production (bright purple); +++, high production (dark purple), considering color intensity of the positive controls, IAA (66 μg/mL) representing the maximum value (10+) and IAA 1:2 the half (5+). The test was replicated three times. The ability to produce siderophores was investigated using the overlay Chrome Azurol S assay (CAS) described by Pérez-Miranda et al. (2007). CAS agar was prepared following the procedure described by Louden et al. (2011). Siderophore-producing bacterial strains showed a change in color, from blue to yellow/orange, in the overlaid medium around the colonies. After 7 days the radius of the halo was measured (mm) from the colony edge to the edge of the colored halo. Strains were classified using a rating scale as follows: no production (halo = 0 mm), +/– = low production (halo < 2 mm), + = production (3 mm ≤ halo ≤ 8 mm), ++ = moderate production (9 mm < halo < 14 mm), +++ = high production (halo > 15 mm).
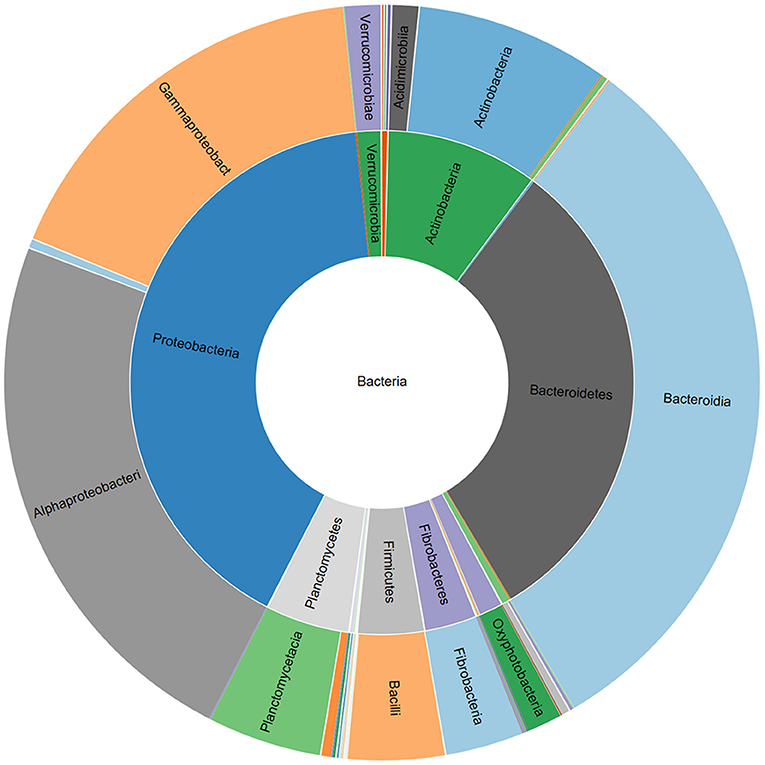
Figure 1. Relative abundance of bacterial phyla and classes associated with a commercial AMF inoculum.
The purified bacterial strains showing the best ability to produce IAA and siderophores were identified based on 16S rDNA sequencing. Genomic DNA was extracted from bacterial liquid cultures grown overnight at 28°C using “MasterPure™ Yeast DNA Purification Kit” (Epicenter®), following the manufacturer's protocol. Bacterial 16S rRNA gene was amplified using the primers 27F (5′-GAGAGTTTGATC CTG GCT CAG-3′) e 1495R (5′-CTA CGG CTA CCT TGT TAC GA-3′) (Lane, 1991; Weisburg et al., 1991). The amplification reaction was carried out in a final volume of 25 μL, containing: 5 μL of DyNAzyme buffer 10X (Finnzymes), 0.2 μM of each primer, 0.2 mM of each dNTPs (EuroClone), 0.625 U of Taq DyNAzyme II DNA polymerase (Finnzymes) and 10–20 ng of DNA. The samples were amplified using an iCycleriQ Multicolor Real-Time PCR Detection System (BIORAD), with the following PCR protocol: 95°C 2 min; 94°C 1 min and 20 s, 54°C 1 min, 72°C 1 min, and 30 s for 35 cycles; 72°C 5 min. PCR amplicons were analyzed by 1.5% agarose gel electrophoresis, stained with ethidium bromide, visualized and captured as TIFF format files by the UVITEC UV1-1D program for UVITEC Gel Documentation system Essential V6 (Cambridge, UK). The amplification products were purified by the Clean PCR CleanUp kit” (CABRU), quantified and 5′ sequenced by Eurofins Genomics (Ebersberg, Germany), as reported in Palla et al. (2017). Sequences were analyzed using BLAST on the NCBI web (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences were aligned using MUSCLE, and phylogenetic trees were constructed using the Neighbor-Joining method based on Tamura 3-parameter method in MEGA10 (Kumar et al., 2018) software with 1,000 bootstrap replicates. The sequences were submitted to the European Nucleotide Archive under the accession numbers from LS999506 to LS999519.
The percentage of colonized length of the root fragments contained within the inoculum was 77±0.7%. The mycorrhizal potential of the inoculum ranged from 20 to 30%.
The V3-V4 region of 16S rRNA gene was sequenced to analyze the composition of the bacterial microbiota associated with three different lots of AMF inoculum. NGS analysis allowed us to generate a number of reads per sample comprised between 3.1 and 3.9 million (Supplementary Material 1). Approximately 88% of raw reads per sample passed merging, trimming and chimera filtering steps and were analyzed for OTU search. The clustering produced a mean of reads in OTUs of 386,899 ± 25,087 with an average read length after trim of 232 bp. Alpha diversity (OTUs richness) value was 1485 ± 14 (Supplementary Material 1). In total, 23 phyla, 107 orders, 165 families, and 276 bacterial genera were identified in the samples. Nine phyla accounted for 95.8% of the sequence reads across all samples with the majority being Proteobacteria (36.9%) and Bacteroidetes (29.3%; Figure 1). Other phyla that comprised ≥2.5% of the bacterial communities were: Actinobacteria (8.4%), Planctomycetes (6.3%), Verrucomicrobia (3.7%), Firmicutes (3.3%), Patescibacteria (3.1%), Deinococcus-Thermus (2.6%), and Fibrobacteres (2.5%). The predominant orders were: Rhizobiales (23.6%), Caulobacterales (12.9%), Sphingomonadales (12.1%), and Cellvibrionales (9.0%) among Proteobacteria; Sphingobacteriales (45.1%) and Flavobacteriales (34.7%) among Bacteroidetes. A deeper phylogenetic classification of the reads revealed that the most represented genera were Sphingobacterium (10% of total bacteria), Flavobacterium (6.6%), Brevundimonas (3.4%), Allorhizobium/Neorhizobium/Pararhizobium/Rhizobium group (3.4%), Stenotrophomonas (3.4%), Cellvibrio (2.9%), and Devosia (2.7%) (Figure 2).
Microbiological analyses allowed the determination of the bacterial cells associated with the inoculum. The CFU/ml number of heterotrophic bacteria ranged from 2.5 ± 0.2 to 6.1 ± 1.4 × 106, while putative N-fixers and P-solubilizers ranged from 9.7 ± 0.8 × 105 to 2.2 ± 0.4 × 106 and from 9.2 ± 0.3 × 105 to 2.4 ± 0.9 × 106, respectively (Table1). A total of 26 putative N-fixers and 9 P-solubilizers were obtained in pure culture. As an additional strain showed both characteristics, the total strains successively tested for their PGP traits were 36.
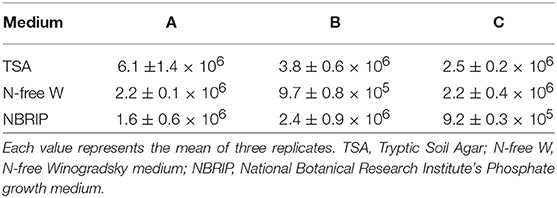
Table 1. Number of culturable bacteria isolated from three 40 g samples (A, B, C) of the AMF commercial inoculum (mean CFU/mL ± SE) isolated from three different microbiological substrates.
Among the 36 strains analyzed for IAA production, 6 showed the red/orange color similar to the positive controls. Such IAA producers were the isolates N-67 and N-92 within the putative N-fixers, and P-30, P-36, P-42, and P-57 within the P-solubilizers (Supplementary Material 2). The other isolates produced lower levels of IAA, as indicated by the golden yellow color of the substrate (Supplementary Table S1).
As to siderophores production, most strains showed the indicative clarification halo around the colonies, 5 of them producing a halo with a diameter higher than 5 mm, i.e. isolates N-21, N-75, N-78, and N-87 within the putative N-fixers and the isolate P-24 within P-solubilizers (Supplementary Material 2).
Three additional strains, P-23, N-P-27 and N-64, showed a moderate siderophore production, together with IAA production.The 14 bacterial isolates showing the best combination of PGP traits (production of IAA and siderophores) were 16S rDNA sequenced and affiliated to bacterial genera and species. Sequences were affiliated with Actinomycetales (Microbacterium trichotecenolyticum, Streptomyces deccanensis/scabiei), Bacillales (Bacillus litoralis, Bacillus megaterium), Enterobacteriales (Enterobacter), Rhizobiales (Rhizobium radiobacter, syn Agrobacterium radiobacter/tumefaciens) (Table 2, Figure 3).
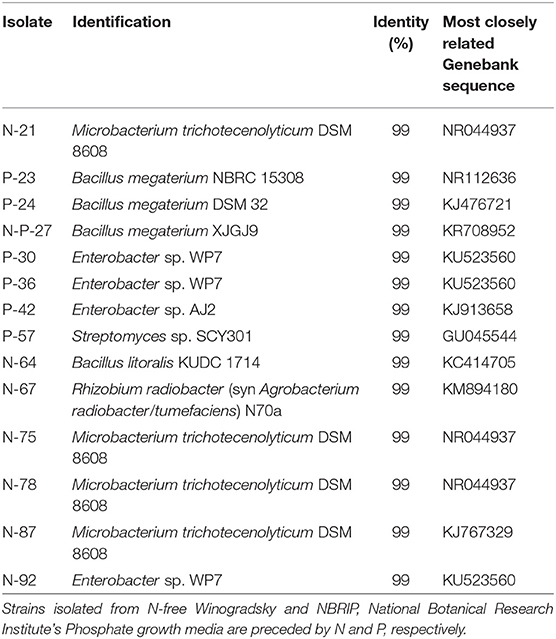
Table 2. Phylogenetic identification of the 14 best performing plant growth promoting bacteria isolated from the mycorrhizal commercial inoculum (sequence accession numbers from LS999506 to LS999519).
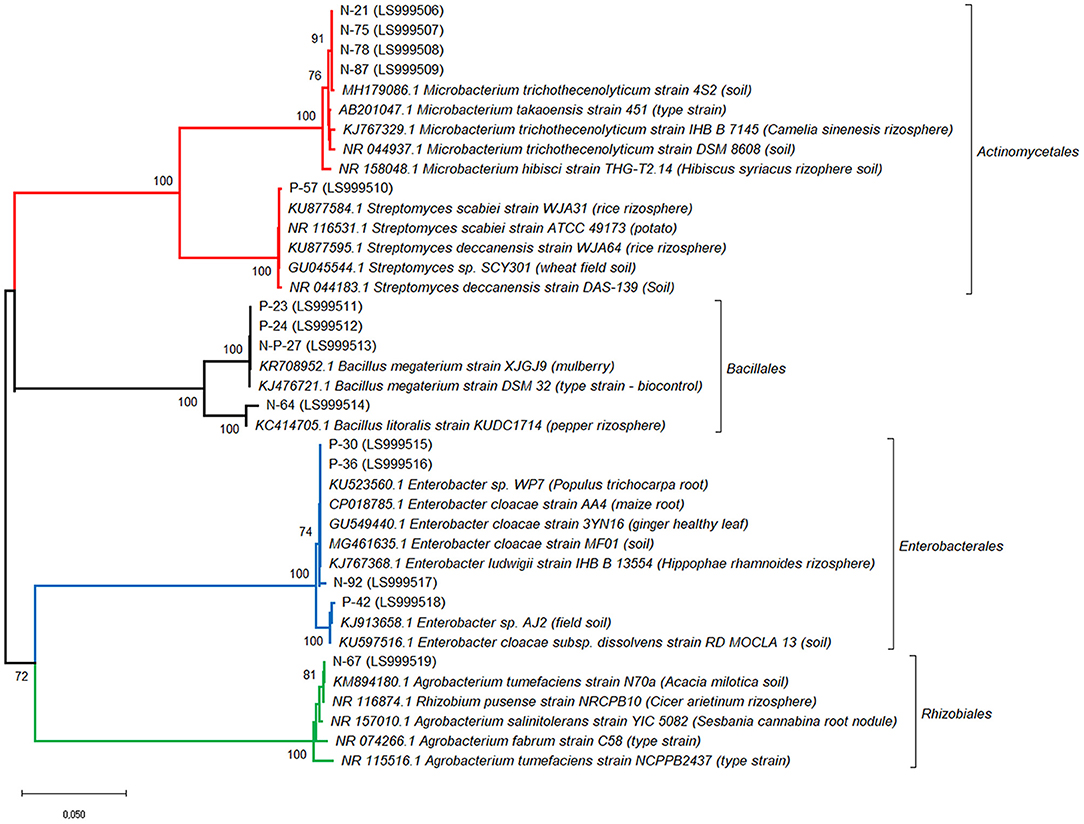
Figure 3. Affiliation of the sequences of the 14 bacterial strains showing the best PGP traits with the existing 16S rRNA gene sequences. Phylogenetic analysis was inferred by using the Neighbor-Joining method. The evolutionary distances were computed using the Tamura 3-parameter method. The rate variation among sites was modeled with a gamma distribution (shape parameter = 1). Bootstrap (1,000 replicates) values below 70 are not shown. Evolutionary analyses were conducted in MEGA10.
In this work, for the first time, the microbiota associated with a commercial AMF inoculum was identified and characterized, using a polyphasic approach, i.e., a combination of traditional microbiological culture-dependent analyses and metagenomic sequencing. A complex and highly diverse bacterial community was identified by Illumina high-throughput sequencing and several bacteria showing important PGP traits, as the ability to produce IAA and siderophores, were isolated and identified.
The assessment of mycorrhizal colonization of the roots contained in the inoculum and of the MIP was the necessary prerequisite for the feasibility of our study, given the recent data on the poor colonization of plant roots by a commercial AMF inoculum (Berruti et al., 2013). In our material, both roots contained in commercial inoculum and those of the plants used for the MIP bioassay were well colonized, showing that the commercial inoculum was highly infective and able to rapidly establish the mycorrhizal symbiosis.
The crude inoculum analyzed, consisting of the substrate where trap plants were grown (mycorrhizal root fragments, AMF spores and mycelium) harbored a rich culturable heterotrophic bacterial community, whose populations ranged from 2.5 to 6.1 × 106 CFU/mL. Such values are high, when considering the origin of the sampled material, which did not derive from living roots, but from a dry inoculum, and show that the rich bacterial community thriving in the particular ecological niche, rich in nutrients and exudates, represented by trap plants during AMF inoculum production, is able to maintain its vitality and activity through the different phases leading to the production of the commercial AMF inoculum, from plant harvest to substrate drying. Moreover, present data confirm previous molecular findings which detected large and complex bacterial communities associated with AMF spores (Roesti et al., 2005; Long et al., 2008; Agnolucci et al., 2015).
The culture-independent approach revealed the occurrence of 7 most represented bacterial genera known to include species isolated from a variety of environments that can be subjected to different environmental stresses. For example, bacteria belonging to Sphingobacterium, the most represented genus in the commercial inoculum, can survive at temperatures lower than 5°C (Shivaji et al., 1992) and higher than 65°C (Yoo et al., 2007), or can survive in soil contaminated with herbicides (Lü et al., 2006) or solvents (Mohammad et al., 2006). Some species of this genus have been reported to have PGP activities, such as inorganic phosphate solubilization, surfactant and IAA production (Marques et al., 2010; Ahmad et al., 2014; Ali et al., 2017), that can improve the efficacy of AMF inocula. Plant growth-promoting traits were also reported in bacteria belonging to other genera associated with the inoculum, including Flavobacterium (phosphate solubilization, production of phytohormones and antimicrobial compounds, Nishioka et al., 2016), Brevundimonas (production of IAA and ammonia, Kumar and Gera, 2014), Stenotrophomonas (production of antibiotics and plant growth regulators, Messiha et al., 2007) and Devosia (development of a nitrogen-fixing root-nodule symbiosis, Rivas et al., 2002). The potential contribution of these bacteria to the efficacy of AMF inocula is supported by recent findings reporting that inoculation with PGPB Flavobacterium and Stenotrophomonas can be effective in promoting plant growth under draft (Gontia-Mishra et al., 2016) or salinity stress (Singh and Jha, 2017). Interestingly, several sequences (2.9%) were assigned to Cellvibrio, a genus known for its cellulose and complex carbohydrate degradation potential, which was previously retrieved from AMF spores, where it was supposed to feed on components of the spore walls, thus facilitating AMF spore germination (Roesti et al., 2005). Many other genera were represented in the bacterial community associated with the commercial inoculum (Supplementary Material 1). Among them, several sequences occurring at low frequencies were ascribed to Streptomyces (0.22%), Enterobacter (0.24%), Bacillus (0.66%), Microbacterium (0.83%), genera to which our selected strains belonged.
Here, the inoculation and successive purification on selective media allowed the initial isolation of 36 bacterial strains, and their subsequent screening allowed the selection of the 14 best performing strains showing important PGP traits. Six and five strains were strong producers of IAA and siderophores, respectively, while two of them (N-67 and N-92), displayed at high levels the two PGP traits. The occurrence of such bacterial functional groups in the commercial inoculum further supports our previous evidence that the beneficial microbiota associated with AMF maintains not only its vitality and activity, but also its functional properties during the different phases of the life cycle (Battini et al., 2016). The ability of such strains to produce IAA, a hormone enhancing cell division and boosting the development of plant root systems (Patten and Glick, 2002) and siderophores, able to facilitate plant acquisition of Fe, thus acting as potential biocontrol agents against soilborne plant pathogens (Glick, 1995; Arora et al., 2001; Whipps, 2001; Battini et al., 2016), confirms the need and utility of adopting culture-dependent methods in order to gain knowledge on functional traits of AMF-associated bacteria. The availability of such beneficial bacteria in pure culture allows their use in ecological studies aimed at investigating their mycorrhizospheric competence and role in plant growth promotion.
Fourteen bacterial strains showing the best combination of PGP traits were identified by 16S rDNA sequencing. Interestingly, 5 out of 14 strains (36%) belonged to Actinomycetales: among them, the Microbacterium trichotecenolyticum strains N-21, N-75, N-78, N-87, and the Streptomyces sp. strain P-57 were strong siderophores and IAA producers, respectively. Actinobacteria are ubiquitous in the soil and able to produce many biologically active secondary metabolites, including antibacterial, antifungal, antiparasitic, anticancer and immunosuppressant drugs (Wolf and Zähner, 1972; Weitnauer et al., 2001; Ritacco and Eveleigh, 2008; Qin et al., 2014) and/or to utilize a wide range of complex compounds (Vandera et al., 2015). They were previously reported to live in strict association with spores and hyphae of different AMF, including F. coronatum, F. mosseae, and R. intraradices (Walley and Germida, 1996; Andrade et al., 1997; Bharadwaj et al., 2008b; Agnolucci et al., 2015; Battini et al., 2016). Many Actinobacteria showed PGP traits, acting as antagonists against plant pathogens, and mycorrhizal helper traits, enhancing mycorrhizal colonization and AMF functionality (Bharadwaj et al., 2008a; Hamdali et al., 2008; Giovannetti et al., 2010).
Members of the genus Microbacterium are ubiquitous in many environments and considered important players of biogeochemical cycles, due to their diazotrophic properties and endophytic behavior (Miliute et al., 2015). Consistent with our findings a M. trichotecenolyticum strain isolated from roots of wild Dodonaea viscosa L. was reported to possess multiple plant growth promoting activities, such as siderophore and IAA production (Afzal et al., 2017).
The genus Streptomyces is one of the main component of soil bacterial communities and is considered within the promising taxa to be investigated for PGP activity, given its ability to solubilize phosphates and produce growth regulators (Mohandas et al., 2013; Hamedi and Mohammadipanah, 2015), two activities shown also by our strain P-57. Actually, two Streptomyces strains, W94 and W77, isolated from the spores of the AM fungus R. irregularis IMA6, significantly increased the uptake and translocation of 33P in maize plants, and hyphal length specific 33P uptake, respectively, compared with control plants (Battini et al., 2017). On the other hand, other IAA-producing bacteria isolated from AMF propagules were able to increase AMF development (Bidondo et al., 2011), in agreement with previous data reporting that Streptomyces spp. boosted AMF spore germination and hyphal growth (Mugnier and Mosse, 1987; Tylka et al., 1991; Carpenter-Boggs et al., 1995), thus showing mycorrhizal helper traits.
Four out of 14 strains (28%) were affiliated with Bacillales, and belonged to the species Bacillus megaterium and Bacillus litoralis. All of them produced siderophores, activity previously reported in other members of the order (Battini et al., 2016), known for their ability to control soilborne pathogens (Jeong et al., 2014) and to act as PGP and mycorrhizal helper bacteria, facilitating mycorrhizal establishment and improving plant growth (Budi et al., 2013; Pérez-Montaño et al., 2014; Zhao et al., 2014). The isolation of Bacillus species from our commercial inoculum represents a further confirmation of previous data obtained by culture-independent methods (Agnolucci et al., 2015).
One strain, Rhizobium radiobacter (syn. Agrobacterium radiobacter/tumefaciens) N-67, was affiliated to the Rhizobiales, an order thoroughly investigated for the ability of its members to fix nitrogen. This isolate was one of the two only strains able to produce both IAA and siderophores, confirming previous data on PGP ability of some rhizobia to boost plant nutritional status by producing phytohormones (Zahir et al., 2003; Chandra et al., 2007; Dodd et al., 2010). Its persistence in the AMF inoculum may be ascribed to the formation of biofilms containing exopolysaccharides which allow an efficient colonization of roots and mycorrhizal hyphae (Bianciotto et al., 1996; Toljander et al., 2006).
A very interesting finding is represented by the isolation of 4 strains, P-30, P-36, P-42, N-92, affiliated with Enterobacteriales (Enterobacter cloacae/ludwigii), which were strong producers of IAA, confirming previous data on the capacity of a strain of E. cloacae to produce as much IAA as a Pseudomonas strain (Imen et al., 2013). Recent works reported that a few strains of the genus Enterobacter, isolated from legume plants, possessed multiple plant-growth promoting characteristics, such as phosphate solubilisation activity and IAA production, thus affecting plant growth and development (Ghosh et al., 2015; Khalifa et al., 2016). On the other hand, one of our isolates, N-92, produced also siderophores, activity already reported for members of the genus Enterobacter (Tian et al., 2009).
In conclusion, this work demonstrates for the first time that an AMF inoculum, produced following industrial production processes, is home of a large and diverse community of bacteria with important functional PGP properties, possibly acting in synergy with AMF and providing new services and benefits. The commercial AMF product could be enriched with the selected beneficial bacterial isolates utilized as an additional inoculum, further boosting plant growth, nutrition and health, in order to optimize plant performance in sustainable food production systems. Indeed, our findings imply a new perspective of AM symbiosis, that of a multipartite association - host plants, AMF and bacteria - where different microbial functional groups are active: for example, specific mycorrhizospheric bacteria, by solubilizing P and fixing N, may improve the availability of key mineral nutrients, then absorbed and translocated to the host plant by AMF extraradical hyphae, while other bacteria, by producing siderophores and IAA, may control plant pathogens and promote plant growth. Notwithstanding, so far only few works have been carried out either on the isolation and functional characterization of mycorrhizospheric microbiota, or on their occurrence and significance in AMF inocula. Yet, these studies are necessary and urgent, in the perspective of developing new strategies for sustainable intensification in agriculture, aimed at minimizing the use of chemical fertilizers and pesticides, promoting primary production and maintaining soil health and fertility. To this aim, the most diverse combinations of AMF and bacteria should be studied, in model experimental systems and in the field, to discover possible synergistic effects on different host plants, in order to select the best performing ones for their targeted use in sustainable food production systems in the years to come.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The authors gratefully acknowledge the financial support by the University of Pisa (Fondi di Ateneo) and by ATENS-Agrotecnologias Naturales SL, La Riera de Gaia, Tarragona, Spain.
MG received funding from and VC and FC are employees of Atens - Agrotecnologias Naturales SL, La Riera de Gaia, Tarragona, Spain.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01956/full#supplementary-material
Afzal, I., Iqrar, I., Shinwari, Z. K., and Yasmin, A. (2017). Plant growth-promoting potential of endophytic bacteria isolated from roots of wild Dodonaea viscosa L. Plant Growth Regul. 81, 399–408. doi: 10.1007/s10725-016-0216-5
Agnolucci, M., Battini, F., Cristani, C., and Giovannetti, M. (2015). Diverse bacterial communities are recruited on spores of different arbuscular mycorrhizal fungal isolates. Biol. Fertil. Soils 51, 379–389. doi: 10.1007/s00374-014-0989-5
Ahmad, M., Zahir, Z. A., Jamil, M., Nazli, F., Latif, M., and Akhtar, M. F. (2014). Integrated use of plant growth promoting rhizobacteria, biogas slurry and chemical nitrogen for sustainable production of maize under salt–affected conditions. Pak. J. Bot 46, 375–382.
Ali, L., Khalid, M., Asghar, H. N., and Asgher, M. (2017). Scrutinizing of rhizobacterial isolates for improving drought resilience in maize (Zea mays). Int. J. Agric. Biol. 19, 1054–1064. doi: 10.17957/IJAB/15.0387
Aloni, R., Aloni, E., Langhans, M., and Ullrich, C. I. (2006). Role of cytokinin and auxin in shaping root architecture: regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 97, 883–893. doi: 10.1093/aob/mcl027
Andrade, G., Mihara, K. L., Linderman, R. G., and Bethlenfalvay, G. J. (1997). Bacteria from rhizosphere and hyphosphere soils of different arbuscular-mycorrhizal fungi. Plant Soil 192, 71–79. doi: 10.1023/A:1004249629643
Ansorge, W. J. (2009). Next-generation DNA sequencing techniques. New Biotechnol. 25, 195–203. doi: 10.1016/j.nbt.2008.12.009
Arora, N. K., Kang, S. C., and Maheshwari, D. K. (2001). Isolation of siderophore-producing strains of Rhizobium meliloti and their biocontrol potential against Macrophomina phaseolina that causes charcoal rot of groundnut. Curr. Sci. 81, 673–677.
Augé, R. M. (2001). Water relations, drought and vesicular–arbuscular mycorrhizal symbiosis. Mycorrhiza 11, 3–42. doi: 10.1007/s005720100097
Avio, L., Castaldini, M., Fabiani, A., Bedini, S., Sbrana, C., Turrini, A., et al. (2013). Impact of nitrogen fertilization and soil tillage on arbuscular mycorrhizal fungal communities in a Mediterranean agroecosystem. Soil Biol. Biochem. 67, 285–294. doi: 10.1016/j.soilbio.2013.09.005
Avio, L., Pellegrino, E., Bonari, E., and Giovannetti, M. (2006). Functional diversity of arbuscular mycorrhizal fungal isolates in relation to extraradical mycelial networks. New Phytol. 172, 347–357. doi: 10.1111/j.1469-8137.2006.01839.x
Avio, L., Turrini, A., Giovannetti, M., and Sbrana, C. (2018). Designing the ideotype mycorrhizal symbionts for the production of healthy food. Front. Plant Sci. 9:1089. doi: 10.3389/fpls.2018.01089
Barea, J. M., Azcón, R., and Azcón-Aguilar, C. (2002). Mycorrhizosphere interactions to improve plant fitness and soil quality. Anton. Van Leeuw. 81, 343–351. doi: 10.1023/A:1020588701325
Barea, J. M., Pozo, M. J., Azcón, R., and Aczón-Aguilar, C. (2005). Microbial cooperation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. doi: 10.1093/jxb/eri197
Battini, F., Cristani, C., Giovannetti, M., and Agnolucci, M. (2016). Multifunctionality and diversity of culturable bacterial communities strictly associated with spores of the plant beneficial symbiont Rhizophagus intraradices. Microb. Res. 183, 68–79. doi: 10.1016/j.micres.2015.11.012
Battini, F., Grønlund, M., Agnolucci, M., Giovannetti, M., and Jakobsen, I. (2017). Facilitation of phosphorus uptake in maize plants by mycorrhizosphere bacteria. Sci. Rep. 7:4686. doi: 10.1038/s41598-017-04959-0
Berruti, A., Borriello, R., Della Beffa, M. T., Scariot, V., and Bianciotto, V. (2013). Application of nonspecific commercial AMF inocula results in poor mycorrhization in Camellia japonica L. Symbiosis 61, 63–76. doi: 10.1007/s13199-013-0258-7
Bharadwaj, D. P., Lundquist, P. O., and Alström, S. (2008a). Arbuscular mycorrhizal fungal spore-associated bacteria affect mycorrhizal colonization, plant growth and potato pathogens. Soil Biol. Biochem. 40, 2494–2501. doi: 10.1016/j.soilbio.2008.06.012
Bharadwaj, D. P., Lundquist, P. O., Persson, P., and Alström, S. (2008b). Evidence for specificity of cultivable bacteria associated with arbuscular mycorrhizal fungal spores. FEMS Microbiol. Ecol. 65, 310–322. doi: 10.1111/j.1574-6941.2008.00515.x
Bianciotto, V., Bandi, C. D., Minerdi, M., Sironi, H., Tichy, V., and Bonfante, P. (1996). An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62, 3005–3010.
Bidondo, L. F., Silvani, V., Colombo, R., Pérgola, M., Bompadre, J., and Godeas, A. (2011). Pre-symbiotic and symbiotic interactions between Glomus intraradices and two Paenibacillus species isolated from AM propagules. In vitro and in vivo assays with soybean (AG043RG) as plant host. Soil Biol. Biochem. 43, 1866–1872. doi: 10.1016/j.soilbio.2011.05.004
Brito, I., Goss, M. J., De Carvalho, M., Chatagnier, O., and van Tuinen, D. (2012). Impact of tillage system on arbuscular mycorrhiza fungal communities in the soil under Mediterranean conditions. Soil Tillage Res. 121, 63–67. doi: 10.1016/j.still.2012.01.012
Budi, S. W., Bakhtiar, Y., and May, N. L. (2013). Bacteria associated with arbuscula mycorrhizal spores Gigaspora margarita and their potential for stimulating root mycorrhizal colonization and neem (Melia azedarach Linn) seedling growth. Microbiol. Indones. 6, 180–188. doi: 10.5454/mi.6.4.6
Budi, S. W., van Tuinen, D., Martinotti, G., and Gianinazzi, S. (1999). Isolation from Sorghum bicolor mycorrhizosphere of a bacterium compatible with arbuscular mycorrhiza development and antagonistic towardssoil-borne fungal pathogens. Appl. Environ. Microbiol. 65, 5148–5150.
Carpenter-Boggs, L., Loynachan, T. E., and Stahl, P. D. (1995). Spore germination of Gigaspora margarita stimulated by volatiles of soil-isolated actinomycetes. Soil Boil. Biochem. 27, 1445–1451. doi: 10.1016/0038-0717(95)00075-P
Castillo, C. G., Rubio, R., Rouanet, J. L., and Borie, F. (2006). Early effects of tillage and crop rotation on arbuscular mycorrhizal fungal propagules in an ultisol. Biol. Fertil. Soils 43, 83–92. doi: 10.1007/s00374-005-0067-0
Chandra, S., Choure, K., Dubey, R. C., and Maheshwari, D. K. (2007). Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of Indian mustard (Brassica campestris). Braz. J. Microbiol. 38, 124–130. doi: 10.1590/S1517-83822007000100026
Citernesi, A. S., Fortuna, P., Filippi, C., Bagnoli, G., and Giovannetti, M. (1996). The occurrence of antagonistic bacteria in Glomus mosseae pot cultures. Agronomie 16, 671–677.
Colombo, C., Palumbo, G., He, J. Z., Pinton, R., and Cesco, S. (2014). Review on iron availability in soil: interaction of Fe minerals, plants, and microbes. J. Soil Sed. 14, 538–548. doi: 10.1007/s11368-013-0814-z
Crowley, D. E., Reid, C. P. P., and Szaniszlo, P. J. (1988). Utilization of microbial siderophores in iron acquisition by oat. Plant Physiol. 87, 680–685. doi: 10.1104/pp.87.3.680
Cruz, A. F., Horii, S., Ochiai, S., Yasuda, A., and Ishii, T. (2008). Isolation and analysis of bacteria associated with spores of Gigaspora margarita. J. Appl. Microbiol. 104, 1711–1717. doi: 10.1111/j.1365-2672.2007.03695.x
Cruz, A. F., and Ishii, T. (2011). Arbuscular mycorrhizal fungal spores host bacteria that affect nutrient biodynamics and biocontrol of soil-borne plant pathogens. Biol. Open 1, 52–57. doi: 10.1242/bio.2011014
Davison, J. (1988). Plant beneficial bacteria. Nat. Biotechnol. 6, 282–286. doi: 10.1038/nbt0388-282
Dodd, I. C., Zinovkina, N. Y., Safronova, V. I., and Belimov, A. A. (2010). Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. doi: 10.1111/j.1744-7348.2010.00439.x
Douds, D. D., Galvez, L., Janke, R. R., and Wagoner, P. (1995). Effect of tillage and farming system upon populations and distribution of vesicular-arbuscular mycorrhizal fungi. Agric. Ecosyst. Environ. 52, 111–118. doi: 10.1016/0167-8809(94)00550-X
Duca, D., Lorv, J., Patten, C. L., Rose, D., and Glick, B. R. (2014). Indole-3-acetic acid in plant–microbe interactions. Anton. Van Leeuw. 106, 85–125. doi: 10.1007/s10482-013-0095-y
Duijff, B., Bakker, P. A. H. M., and Schippers, B. (1994a). Ferric pseudobactin 358 as an iron source for carnation. J. Plant Nutr. 17, 2069–2078. doi: 10.1080/01904169409364866
Duijff, B., De Kogel, W. J., Bakker, P. A. H. M., and Schippers, B. (1994b). Influence of pseudobactin 358 on the iron nutrition of barley. Soil Biol. Biochem. 26, 1681–1688. doi: 10.1016/0038-0717(94)90321-2
Evelin, H., Kapoor, R., and Giri, B. (2009). Arbuscular mycorrhizal fungi in alleviation of salt stress: a review. Ann. Bot. 104, 1263–1280. doi: 10.1093/aob/mcp251
Ghosh, P. K., Sen, S. K., and Maiti, T. K. (2015). Production and metabolism of IAA by Enterobacter spp. (Gammaproteobacteria) isolated from root nodules of a legume Abrus precatorius L. Biocat. Agric. Biotech. 4, 296–303. doi: 10.1016/j.bcab.2015.04.002
Gianinazzi, S., Gollotte, A., Binet, M. N., van Tuinen, D., Redecker, D., and Wipf, D. (2010). Agroecology the key role of arbuscular mycorrhizas in ecosystem services. Mycorrhiza 20, 519–530. doi: 10.1007/s00572-010-0333-3
Gianinazzi, S., and Vosatka, M. (2004). Inoculum of arbuscular mycorrhizal fungi for production systems, science meets business. Can. J. Bot. 82, 1264–1271. doi: 10.1139/b04-072
Giovannetti, M., Avio, L., and Sbrana, C. (2010). “Fungal spore germination and pre-symbiotic mycelial growth–physiological and genetic aspects,” in Arbuscular Mycorrhizas: Physiology and Function, eds H. Koltai and Y. Kapulnik (Dordrecht: Springer), 3–32.
Giovannetti, M., Fortuna, P., Citernesi, A. S., Morini, S., and Nuti, M. P. (2001). The occurrence of anastomosis formation and nuclear exchange in intact arbuscular mycorrhizal networks. New Phytol. 151, 717–724. doi: 10.1046/j.0028-646x.2001.00216.x
Giovannetti, M., and Mosse, B. (1980). An evaluation of techniques for measuring vesicular-arbuscular infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Glick, B. R. (1995). The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Gontia-Mishra, I., Sapre, S., Sharma, A., and Tiwari, S. (2016). Amelioration of drought tolerance in wheat by the interaction of plant growth-promoting rhizobacteria. Plant Biol. 18, 992–1000. doi: 10.1111/plb.12505
Hamdali, H., Hafidi, M., Virolle, M. J., and Ouhdouch, Y. (2008). Growth promotion and protection against damping-off of wheat by two rock phosphate solubilizing actinomycetes in a P-deficient soil under greenhouse conditions. Appl. Soil Ecol. 40, 510–517. doi: 10.1016/j.apsoil.2008.08.001
Hamedi, J., and Mohammadipanah, F. (2015). Biotechnological application and taxonomical distribution of plant growth promoting actinobacteria. J. Ind. Microbiol. Biotechnol. 42, 157–171. doi: 10.1007/s10295-014-1537-x
Hildebrandt, U., Ouziad, F., Marner, F.-J. J., and Bothe, H. (2006). The bacterium Paenibacillus validus stimulates growth of the arbuscular mycorrhizal fungus Glomus intraradices up to the formation of fertile spores. FEMS Microbiol. Lett. 254, 258–267. doi: 10.1111/j.1574-6968.2005.00027.x
Horii, S., and Ishii, T. (2006). Identification and function of Gigaspora margarita growth-promoting microorganisms. Symbiosis 41, 135–141.
Imen, C. F., Manel, C., Omar, S., Moez, J., and Salwa, H. J. (2013). Phytostabilization of moderate copper contaminated soils using co-inoculation of Vicia faba with plant-growth-promoting bacteria. J. Basic Microbiol. 53, 1–9. doi: 10.1002/jobm.201300323
Jansa, J., Mozafar, A., Kuhn, G., Anken, T., Ruh, R., Sanders, I. R., et al. (2003). Soil tillage affects the community structure of mycorrhizal fungi in maize roots. Ecol. Appl. 13, 1164–1176. doi: 10.1890/1051-0761(2003)13[1164:STATCS]2.0.CO;2
Jeffries, P., Gianinazzi, S., Perotto, S., Turnau, K., and Barea, J. M. (2003). The contribution of arbuscular mycorrhizal fungi in sustainable maintenance of plant health and soil fertility. Biol. Fertil. Soils 37, 1–16. doi: 10.1007/s00374-002-0546-5
Jeong, H., Choi, S. K., Kloepper, J. W., and Ryu, C. M. (2014). Genome sequence of the plant endophyte Bacillus pumilus INR7, triggering induced systemic resistance in field crops. Genome Announc. 2:e01093–1114. doi: 10.1128/genomeA.01093-14
Jin, C. W., He, Y. F., Tang, C. X., Wu, P., and Zheng, S. J. (2006). Mechanisms of microbially enhanced Fe acquisition in red clover (Trifolium pratense L.). Plant Cell Environ. 29, 888–897. doi: 10.1111/j.1365-3040.2005.01468.x
Kabir, Z., and Koide, R. T. (2002). Effect of autumn and winter mycorrhizal cover crops on soil properties, nutrient uptake and yield of sweet corn in Pennsylvania, USA. Plant Soil 238, 205–215. doi: 10.1023/A:1014408723664
Karasawa, T., and Takebe, M. (2012). Temporal or spatial arrangements of cover crops to promote arbuscular mycorrhizal colonization and P uptake of upland crops grown after nonmycorrhizal crops. Plant Soil 353, 355–366. doi: 10.1007/s11104-011-1036-z
Khalid, A., Arshad, M., and Kahir, Z. A. (2004). Screening plant growth promoting rhizobacteria for improving growth and yield of wheat. J. Appl. Microbiol. 96, 473–480. doi: 10.1046/j.1365-2672.2003.02161.x
Khalifa, A. Y., Alsyeeh, A. M., Almalki, M. A., and Saleh, F. A. (2016). Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J Biol. Sci. 23, 79–86. doi: 10.1016/j.sjbs.2015.06.008
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:e1. doi: 10.1093/nar/gks8
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Ann. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molec. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kumar, V., and Gera, R. (2014). Isolation of a multi-trait plant growth promoting Brevundimonas sp. and its effect on the growth of Bt-cotton. 3 Biotech 4, 97–101. doi: 10.1007/s13205-013-0126-4
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematic, eds E. Stackebrandt and M. Goodfellow (New York, NY: Wiley), 115–175.
Lehman, R. M., Taheri, W. I., Osborne, S. L., Buyer, J. S., and Douds, D. D. Jr (2012). Fall cover cropping can increase arbuscular mycorrhizae in soils supporting intensive agricultural production. Appl. Soil Ecol. 61, 300–304. doi: 10.1016/j.apsoil.2011.11.008
Lekberg, Y., and Koide, R. T. (2005). Is plant performance limited by abundance of arbuscular mycorrhizal fungi? A meta-analysis of studies published between 1988 and 2003. New Phytol. 168, 189–204. doi: 10.1111/j.1469-8137.2005.01490.x
Li, B., Ravnskov, S., Xie, G., and Larsen, J. (2007). Biocontrol of Pythium damping-off in cucumber by arbuscular mycorrhiza-associated bacteria from the genus Paenibacillus. Biocontrol 52, 863–875. doi: 10.1007/s10526-007-9076-2
Long, L., Zhu, H., Yao, Q., and Ai, Y. (2008). Analysis of bacterial communities associated with spores of Gigaspora margarita and Gigaspora rosea. Plant Soil 310, 1–9. doi: 10.1007/s11104-008-9611-7
Louden, B. C., Haarmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Ed. 12, 51–53. doi: 10.1128/jmbe.v12i1249
Lü, Z., Min, H., Li, N., Shao, T., and Ye, Y. (2006). Variations of bacterial community structure in flooded paddy soil contaminated with herbicide quinclorac. J. Environ. Sci. Health Part B 41, 821–832. doi: 10.1080/03601230600805873
Marques, A. P., Pires, C., Moreira, H., Rangel, A. O., and Castro, P. M. (2010). Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol. Biochem. 42, 1229–1235. doi: 10.1016/j.soilbio.2010.04.014
Mayo, K., Davis, R. E., and Motta, J. (1986). Stimulation of germination of spores of Glomus versiforme by spore-associated bacteria. Mycologia 78, 426–431. doi: 10.2307/3793046
Messiha, N. A. S., Van Diepeningen, A. D., Farag, N. S., Abdallah, S. A., Janse, J. D., and Van Bruggen, A. H. C. (2007). Stenotrophomonas maltophilia: a new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur. J. Plant Phatol. 118, 211–225. doi: 10.1007/s10658-007-9136-6
Miliute, I., Buzaite, O., Baniulis, D., and Stanys, V. (2015). Bacterial endophytes in agricultural crops and their role in stress tolerance: a review. Zemdirbyste 102, 465–478. doi: 10.13080/z-a.2015.102.060
Mimmo, T., Del Buono, D., Terzano, R., Tomasi, N., Vigani, G., Crecchio, C., et al. (2014). Rhizospheric organic compounds in the soil microorganism-plant system: their role in iron availability. Eur. J. Soil Sci. 65, 629–642. doi: 10.1111/ejss.12158
Mohammad, B. T., Wright, P. C., and Bustard, M. T. (2006). Bioconversion of isopropanol by a solvent tolerant Sphingobacterium mizutae strain. J. Industr. Microbiol. Biotechnol. 33, 975–983. doi: 10.1007/s10295-006-0143-y
Mohandas, S., Poovarasan, S., Panneerselvam, P., Saritha, B., Upreti, K. K., Kamal, R., et al. (2013). Guava (Psidium guajava L.) rhizosphere Glomus mosseae spores harbor actinomycetes with growth promoting and antifungal attributes. Sci. Hortic. 150, 371–376. doi: 10.1016/j.scienta.2012.11.019
Mugnier, J., and Mosse, B. (1987). Spore germination and viability of a vesicular arbuscular mycorrhizal fungus, Glomus mosseae. Trans. Br. Mycol. Soc. 88, 411–413. doi: 10.1016/S0007-1536(87)80018-9
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Nishioka, T., Elsharkawy, M. M., Suga, H., Kageyama, K., Hyakumachi, M., and Shimizu, M. (2016). Development of culture medium for the Isolation of Flavobacterium and Chryseobacterium from rhizosphere soil. Microb. Environ. 31, 104–110. doi: 10.1264/jsme2.ME15144
Njeru, E. M., Avio, L., Bocci, G., Sbrana, C., Turrini, A., Bàrberi, P., et al. (2015). Contrasting effects of cover crops on ‘hot spotș arbuscular mycorrhizal fungal communities in organic tomato. Biol. Fertil. Soils 51, 151–166. doi: 10.1007/s00374-014-0958-z
Njeru, E. M., Avio, L., Sbrana, C., Turrini, A., Bocci, G., Bàrberi, P., et al. (2014). First evidence for a major cover crop effect on arbuscular mycorrhizal fungi and organic maize growth. Agron. Sustain. Dev. 34, 841–848. doi: 10.1007/s13593-013-0197-y
Oehl, F., Sieverding, E., Mäder, P., Dubois, D., Ineichen, K., Boller, T., et al. (2004). Impact of long-term conventional and organic farming on the diversity of arbuscular mycorrhizal fungi. Oecologia 138, 574–583. doi: 10.1007/s00442-003-1458-2
Palla, M., Cristani, C., Giovannetti, M., and Agnolucci, M. (2017). Identification and characterization of lactic acid bacteria and yeasts of PDO Tuscan bread sourdough by culture dependent and independent methods. Int. J. Food Microbiol. 250, 19–26. doi: 10.1016/j.ijfoodmicro.2017.03.015
Patten, C. L., and Glick, B. R. (2002). Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. doi: 10.1128/AEM.68.8.3795-3801.2002
Pérez-Miranda, S., Cabirol, N., George-Téllez, R., Zamudio-Rivera, L. S., and Fernández, F. J. (2007). O-CAS, a fast and universal method for siderophore detection. J. Microbiol. Method 70, 127–131. doi: 10.1016/j.mimet.2007.03.023
Pérez-Montaño, F., Alías-Villegas, C., Bellogín, R. A., Del Cerro, P., Espuny, M. R., Jiménez-Guerrero, I., et al. (2014). Plant growth promotion in cereal and leguminous agricultural important plants: from microorganism capacities to crop production. Microbiol. Res. 169, 325–336. doi: 10.1016/j.micres.2013.09.011
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Qin, Z., Wang, X., Rateb, M. E., Ass'ad, L. A., Jaspars, M., Deng, Z., et al. (2014). Disruption of a methyltransferase gene in actinomycin G gene cluster in Streptomyces iakyrus increases the production of phenazinomycin. FEMS Microbiol. Lett. 352, 62–68. doi: 10.1111/1574-6968.12370
Ritacco, F. V., and Eveleigh, D. E. (2008). Molecular and phenotypic comparison of phaeochromycin-producing strains of Streptomyces phaeochromogenes and Streptomyces ederensis. J. Ind. Microbiol. Biotechnol. 35, 931–945. doi: 10.1007/s10295-008-0367-0
Rivas, R., Velázquez, E., Willems, A., Vizcaíno, N., Subba-Rao, N. S., Mateos, P. F., et al. (2002). A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (Lf) Druce. Appl. Environ. Microbiol. 68, 5217–5222. doi: 10.1128/AEM.68.11.5217-5222.2002
Robin, A., Vansuyt, G., Hinsinger, P., Meyer, J. M., Briat, J. F., and Lemanceau, P. (2008). Iron dynamics in the rhizosphere: consequences for plant health and nutrition. Adv. Agron. 99, 183–225. doi: 10.1016/S0065-2113(08)00404-5
Roesti, D., Ineichen, K., Braissant, O., Redecker, D., Wiemken, A., and Aragno, M. (2005). Bacteria associated with spores of the arbuscular mycorrhizal fungi Glomus geosporum and Glomus constrictum. Appl. Environ. Microbiol. 71, 6673–6679. doi: 10.1128/AEM.71.11.6673-6679.2005
Rouphael, Y., Franken, P., Schneider, C., Schwarz, D., Giovannetti, M., Agnolucci, M., et al. (2015). Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Sci. Hortic. 196, 91–108. doi: 10.1016/j.scienta.2015.09.002
Sbrana, C., Avio, L., and Giovannetti, M. (2014). Beneficial mycorrhizal symbionts affecting the production of health-promoting phytochemicals. Electrophoresis 35, 1535–1546. doi: 10.1002/elps.201300568
Shivaji, S., Ray, M. K., Rao, N. S., Saisree, L., Jagannadham, M. V., Kumar, G. S., et al. (1992). Sphingobacterium antarcticus sp. nov., a psychrotrophic bacterium from the soils of Schirmacher Oasis, Antarctica. Int. J. Sys. Evol.Microbiol. 42, 102–106. doi: 10.1099/00207713-42-1-102
Siebner-Freibach, H., Hadar, Y., and Chen, Y. (2003). Siderophores sorbed on Ca-montmorillonite as an iron source for plants. Plant Soil 251, 115–124. doi: 10.1023/A:1022984431626
Sikes, B. A., Kottenie, K., and Klironomos, J. N. (2009). Plant and fungal identity determines pathogen protection of plant roots by arbuscular mycorrhizas. J. Ecol. 97, 1274–1280. doi: 10.1111/j.1365-2745.2009.01557.x
Singh, R. P., and Jha, P. N. (2017). The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 8:1945. doi: 10.3389/fmicb.2017.01945
Tchan, Y. T. (1984). “Azotobacteraceae,” in Bergey's Manual of Systematic Bacteriology, vol. 1, eds N. Krieg and J. G. Holt (London: Williams and Wikins), 219–225.
Thomashow, L. S., Weller, D. M., Bonsall, R. F., and Pierson, L. S. (1990). Production of the antibiotic phenazine-1-carbox-ylic acid by fluorescent Pseudomonas species in the rhizosphere of wheat. Appl. Environ. Microbiol. 56, 908–912.
Tian, F., Ding, Y., Zhu, H., Yao, L., and Du, B. (2009). Genetic diversity of siderophore-producing bacteria of tobacco rhizosphere. Braz. J. Microbiol. 40, 276–284. doi: 10.1590/S1517-83822009000200013
Toljander, J. F., Artursson, V., Paul, L. R., Jansson, J. K., and Finlay, R. D. (2006). Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol. Lett. 254, 34–40. doi: 10.1111/j.1574-6968.2005.00003.x
Turrini, A., Caruso, G., Avio, L., Gennai, C., Palla, M., Agnolucci, M., et al. (2017). Protective green cover enhances soil respiration and native mycorrhizal potential compared with soil tillage in a high-density olive orchard in a long term study. Appl. Soil Ecol. 116, 70–78. doi: 10.1016/j.apsoil.2017.04.001
Turrini, A., Sbrana, C., Avio, L., Njeru, E. M., Bocci, G., Bàrberi, P., et al. (2016). Changes in the composition of native root arbuscular mycorrhizal fungal communities during a short-term cover crop-maize succession. Biol. Fertil. Soils 52, 643–653. doi: 10.1007/s00374-016-1106-8
Tylka, G. L., Hussey, R. S., and Roncadori, R. W. (1991). Axenic germination of vesicular–arbuscular mycorrhizal fungi: effects of selected Streptomyces species. Phytopathology 81, 754–759.
Vandera, E., Samiotaki, M., Parapouli, M., Panayotou, G., and Koukkou, A. I. (2015). Comparative proteomic analysis of Arthrobacter phenanthrenivorans Sphe3 on phenanthrene, phthalate and glucose. J. Proteomics 113, 73–89. doi: 10.1016/j.jprot.2014.08.018
Vansuyt, G., Robin, A., Briat, J., Curie, C., and Lemanceau, P. (2007). Iron acquisition from Fe-pyoverdine by Arabidopsis thaliana. Mol. Plant-Micr. Interac. 20, 441–447. doi: 10.1094/MPMI-20-4-0441
Walley, F. L., and Germida, J. J. (1996). Failure to decontaminate Glomus clarum NT4spores is due to spore wall-associated bacteria. Mycorrhiza 6, 43–49. doi: 10.1007/s005720050104
Walter, A., Römheld, V., Marschner, H., and Crowley, D. E. (1994). Iron nutrition of cucumber and maize: effect of Pseudomonas putida YC3 and its siderophore. Soil Biol. Biochem. 26, 1023–1031. doi: 10.1016/0038-0717(94)90117-1
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Weitnauer, G., Mühlenweg, A., Trefzer, A., Hoffmeister, D., Süssmuth, R. D, Jung, G., et al. (2001). Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tü57 and production of new antibiotics. Chem. Biol. 8, 569–581. doi: 10.1016/S1074-5521(01)00040-0
Whipps, J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. doi: 10.1093/jexbot/52.suppl_1.487
Xavier, L. J. C., and Germida, J. J. (2003). Bacteria associated with Glomus clarum spores influence mycorrhizal activity. Soil Biol. Biochem. 35, 471–478. doi: 10.1016/S0038-0717(03)00003-8
Yehuda, Z., Shenker, M., Romheld, V., Marschner, H., Hadar, Y., and Chen, Y. (1996). The role of ligand exchange in the uptake of iron from microbial siderophores by gramineous plants. Plant Physiol. 112, 1273–1280. doi: 10.1104/pp.112.3.1273
Yoo, S. H., Weon, H. Y., Jang, H. B., Kim, B. Y., Kwon, S. W., Go, S. J., et al. (2007). Sphingobacterium composti sp. nov., isolated from cotton-waste composts. Int. J. Sys. Evol.Microbiol. 57, 1590–1593. doi: 10.1099/ijs.0.64948-0
Zahir, Z. A., Arshad, M., and Frankenberger, W. T. Jr. (2003). “Plant growth promoting rhizobacteria: applications and perspectives in agriculture,” in Advances in Agrononomy vol. 81, ed D. L. Sparks (Newark, NY: Academic Press), 97–168.
Keywords: arbuscular mycorrhizal symbionts, mycorrhizosphere, plant-growth promoting bacteria, siderophores production, indole acetic acid production, metagenomics
Citation: Agnolucci M, Avio L, Pepe A, Turrini A, Cristani C, Bonini P, Cirino V, Colosimo F, Ruzzi M and Giovannetti M (2019) Bacteria Associated With a Commercial Mycorrhizal Inoculum: Community Composition and Multifunctional Activity as Assessed by Illumina Sequencing and Culture-Dependent Tools. Front. Plant Sci. 9:1956. doi: 10.3389/fpls.2018.01956
Received: 27 September 2018; Accepted: 17 December 2018;
Published: 14 January 2019.
Edited by:
Youssef Rouphael, University of Naples Federico II, ItalyReviewed by:
Alessandra Salvioli, Università degli Studi di Torino, ItalyCopyright © 2019 Agnolucci, Avio, Pepe, Turrini, Cristani, Bonini, Cirino, Colosimo, Ruzzi and Giovannetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manuela Giovannetti, bWFudWVsYS5naW92YW5uZXR0aUB1bmlwaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.