- Department of Genetics, Institute for Plant Biotechnology, University of Stellenbosch, Stellenbosch, South Africa
Starch is a plant storage polyglucan that accumulates in plastids. It is composed of two polymers, amylose and amylopectin, with different structures and plays several roles in helping to determine plant yield. In leaves, it acts as a buffer for night time carbon starvation. Genetically altered plants that cannot synthesize or degrade starch efficiently often grow poorly. There have been a number of successful approaches to manipulate leaf starch metabolism that has resulted in increased growth and yield. Its degradation is also a source of sugars that can help alleviate abiotic stress. In edible parts of plants, starch often makes up the majority of the dry weight constituting much of the calorific value of food and feed. Increasing starch in these organs can increase this as well as increasing yield. Enzymes involved in starch metabolism are well known, and there has been much research analyzing their functions in starch synthesis and degradation, as well as genetic and posttranslational regulatory mechanisms affecting them. In this mini review, we examine work on this topic and discuss future directions that could be used to manipulate this metabolite for improved yield.
Introduction
The need for improved crop yields due to an increase in world population and a decrease in available agricultural land is well known (Edgerton, 2009). This problem will likely be exacerbated through alterations in environmental conditions caused by anthropomorphic CO2 release that may lead to increases in both biotic and abiotic stresses (Fodor et al., 2017). There are many potential biotechnological methods that can lead to increased yield, and one of these involves altering starch metabolism. This mini review will examine work that has been performed to improve plant yield through manipulation of this metabolite and suggest new avenues that could be explored.
Starch is a polyglucan that is stored as granules within plastids. It consists of two polymers with differing structures, amylose and amylopectin. Amylose contains relatively long (normally composed of several hundred glucose monomers) α1,4 linked chains, while amylopectin is composed of many short (approximately 5–50 glucose monomers) α1,4 linked chains, linked together by α1,6 branch points in an ordered, crystalline array (Zeeman et al., 2010).
Manipulation of starch metabolism is important for improving plant yield for several reasons. As starch is the major form of calories within plants, increasing starch concentrations in plant tissues can mean that less food or fodder has to be consumed to supply the same energetic value (Ruckle et al., 2017). It is also a major sink within storage organs, so increasing starch here can lead to increased plant yield simply as more accumulates. In leaves, it plays two roles. Firstly, it is synthesized during the day and degraded at night, buffering the plant from night time carbon starvation (Stitt and Zeeman, 2012; Arias et al., 2014) which leads to transcriptional upregulation of stress-related genes (Stitt et al., 2007) and an inhibition of gibberellin synthesis (Paparelli et al., 2013). Secondly, it is a source of carbon skeletons for the production of compatible solutes that help plants to overcome abiotic stress (Thalmann and Santelia, 2017). Finally, starch has been shown to affect developmental processes (Matsoukas et al., 2013), and therefore, its manipulation could increase yield through altering plant or seed development.
Starch Metabolism
Due to its many roles, the pathway of starch metabolism has been studied intensively, and many enzymatic steps involved in its metabolism have been elucidated. This knowledge has been used to construct a detailed model of its metabolism, which has been described in several recent reviews (Bahaji et al., 2014; Pfister and Zeeman, 2016; MacNeill et al., 2017). The rest of this section will briefly outline the major enzymes involved to allow for further discussion of the manipulation of the pathway.
Starch polymer formation (Figure 1B) involves the synthesis of adenosine diphosphate (ADP)-glucose by ADP-glucose pyrophosphorylase (AGPase). This is used by starch synthases (SS) to form linear α1,4 glucan chains. One starch synthase isoform is responsible for amylose synthesis, while several others are involved in granule initiation and amylopectin synthesis (Pfister and Zeeman, 2016; Nazarian-Firouzabadi and Visser, 2017). The branch points within amylopectin are introduced by starch branching enzyme (SBE) isoforms, while excess α1,6 links are removed by isoamylases (Pfister and Zeeman, 2016; MacNeill et al., 2017).
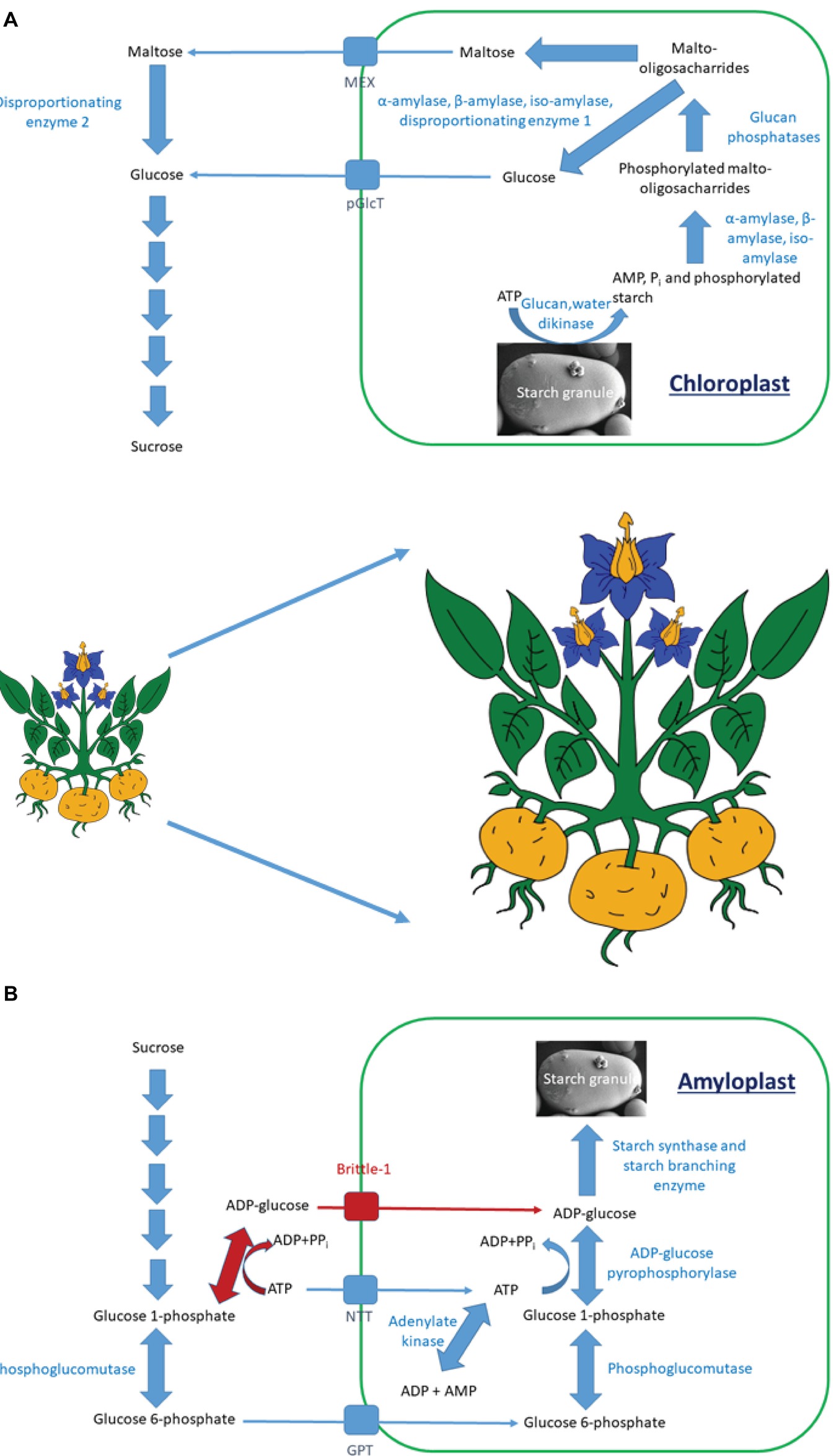
Figure 1. The pathway of starch degradation in (A) leaves and of starch synthesis in (B) storage organs. (A) Starch is degraded through a series of enzymatic steps to maltose and glucose. These are transported from the chloroplast by either the maltose export (MEX) or plastidial glucose transport (pGlcT) proteins. Within the cytosol, they are converted to sucrose for further transport. (B) There are two pathways of starch synthesis in vascular plants. One is shared in all plants (blue arrows), while the other is present only in cereal endosperm (red arrows). Sucrose is degraded to glucose 6-phosphate, which is transported into amyloplasts by the glucose 6-phosphate/phosphate transporter (GPT). The ADP-glucose pyrophosphorylase step within the amyloplast utilizes ATP, which is imported into the amyloplast by the plastidial ATP/ADP transporter (NTT). Within cereal endosperm, ADP-glucose is synthesized extraplastidially and is imported into amyloplasts by the Brittle-1 transporter.
Starch degradation (Figure 1A) is initiated by glucan water dikinase (GWD) enzymes that phosphorylate the granule, helping to solubilize it and allow access to α-, β-, and isoamylases (Streb et al., 2012; Mahlow et al., 2016). These release phosphorylated malto-oligosaccharides into the stroma, where the phosphate is removed by glucan phosphatases, allowing further degradation to maltose and glucose by the combined actions of amylases and disproportionating enzyme 1. Maltose and glucose are exported to the cytosol through specific transport proteins, where maltose becomes further mobilized by disproportionating enzyme 2 (DPE2; Lloyd and Kossmann, 2015).
The regulation of starch metabolism takes place at several levels, including both transcriptional and posttranslational mechanisms. A number of transcription factors affecting expression of genes encoding starch metabolizing enzymes have been identified, and their roles are currently being examined. At the posttranslational level, allosteric regulation (Zeeman et al., 2010) protein phosphorylation (Kötting et al., 2010) and reducing/oxidation (redox) conditions (Santelia et al., 2015; Skryhan et al., 2018) are known to influence both enzyme activities and the formation of enzyme complexes. The roles of these processes will probably differ between leaves and heterotrophic storage organs. For example, the redox status within chloroplasts varies over a day/night cycle, where reducing condition predominates during the day and oxidizing conditions at night. Within amyloplasts of heterotrophic tissues, this day/night cycle will not occur, although it has been shown that the alteration in redox status of chloroplasts can be sensed by amyloplasts (Balmer et al., 2006). Finally, nutrient sensing mechanisms would be expected to link the accumulation of soluble sugars, such as sucrose, with starch synthesis through a combination of mechanisms including 14-3-3 proteins, hexokinase (Rolland et al., 2006), SnRKs (Crozet et al., 2014; Wurzinger et al., 2018), and TOR kinases (Rolland et al., 2006; Dobrenel et al., 2016; Shi et al., 2018).
ADP-Glucose Pyrophosphorylase—A Key Enzyme for Yield in Both Storage Organs and Leaves
ADP-glucose pyrophosphorylase (Figure 1B) has been shown to be a key enzyme influencing starch accumulation in both leaves and storage organs. One complicating factor is the level of posttranslational control of the enzyme by a combination of allosteric effectors and redox, which means that overexpression of the native enzyme may not increase flux into starch synthesis (Boehlein et al., 2013a). This has been overcome through the use of mutated sequences encoding allosteric and redox insensitive isoforms (Tuncel and Okita, 2013). In cereal endosperm, another complication is that most AGPase activity is found within the cytosol, with a specific transporter localized in the outer plastid membrane importing ADP-glucose into the stroma, while in noncereal species it is found only in the plastid. Within cereal endosperm, the cytosolic pathway plays a greater role than the plastidial one (Tuncel and Okita, 2013; Tetlow and Emes, 2017).
It has been known for many years that increasing AGPase activity leads to increased starch synthesis, and as was mentioned above, increasing starch in food and fodder crops would improve their calorific value (Ruckle et al., 2017, 2018). However, interestingly, increasing starch amounts in this way can have beneficial effects on plant productivity. In Arabidopsis and rice, where leaf AGPase activity has been upregulated, leaf starch amounts are increased at the end of the day and the plants grow larger (Gibson et al., 2011; Schlosser et al., 2014; Oiestad et al., 2016), most likely caused by elevated nighttime sugar levels (Stitt and Zeeman, 2012; Arias et al., 2014).
Increasing AGPase in storage organs can also increase starch amounts (Zeeman et al., 2010; Sonnewald and Kossmann, 2013; Tuncel and Okita, 2013). Initial work utilized a bacterial gene that was insensitive to allosteric regulation (Stark et al., 1992); however, more recent work has used plant genes engineered to encode proteins with improved properties (Tuncel and Okita, 2013). Due to the dual cytosolic and plastidial localization of AGPase in cereal endosperm (Figure 1B) compared to its plastidial targeting in other organs and species, the subcellular targeting of AGPase is critical for successful upregulation of starch synthesis. In noncereal species, transgenic plants with increased plastidial AGPase activity accumulate increased amounts of starch, while in cereal endosperm the enzyme has to be localized to the cytosol to have an effect (Zeeman et al., 2010; Sonnewald and Kossmann, 2013; Tuncel and Okita, 2013).
It is clear that AGPase plays a critical role in determining starch yield. It is important to understand, therefore, how its properties could be altered to increase yield, especially in a changing environment. Protein engineering using plant genes has led to much knowledge about reducing redox or allosteric inhibition of its activity as well as improving its activity under heat stress (Georgelis and Hannah, 2008; Haedrich et al., 2012; Boehlein et al., 2013a,b, 2015), and many of these engineered proteins have been transferred into plants through transgenic technology (Tuncel and Okita, 2013). A recent TILLING population targeting this enzyme in Arabidopsis has been developed and may help in developing nontransgenic routes to increase starch synthesis in vivo using gene editing (Haedrich et al., 2011). Although increasing AGPase activity will increase flux into the pathway, this is not the only way that it increases yield. In several cereals, for example, over-expression of AGPase can increase seed number, which in maize has been shown to occur through an effect on maternal tissues (Smidansky et al., 2002, 2003; Hannah et al., 2012, 2017).
Another strategy used to influence the AGPase step in storage organs has been to increase supply of its substrates, adenosine triphosphate (ATP) and/or glucose 6-phosphate. In dicots, ATP limitation of AGPase within plastids has been suggested (Geigenberger, 2001), meaning that increasing supply would be a necessity to increase starch contents. This has been achieved in potato through overexpression of a plastidial adenylate translocator (NTT; which counter-exchanges ATP for ADP; Figure 1B). However, reports on the effect of increasing NTT expression on starch amounts are mixed. Two studies in potato found that this increased starch amounts (Tjaden et al., 1998; Geigenberger, 2001) while another described no alteration (Zhang et al., 2008). That later study did, however, find that combined overexpression of the NTT and a plastidial glucose 6-phosphate translocator leads to increased starch and tuber yield (Zhang et al., 2008).
A second strategy to influence ATP supply occurred through manipulation of a plastidial isoform of adenylate kinase, an enzyme that interconverts ATP with ADP and adenosine monophosphate (AMP) (Figure 1B). Repression of this enzyme led to increased ADP-glucose, tuber starch content, and yield (Regierer et al., 2002), presumably due to an increase in the plastidial ATP pool. Indeed, manipulation of nucleotide metabolism generally may be a profitable way to increase starch contents. In addition to the adenylate kinase study described above, repression of UMP synthase has been demonstrated to lead in uridine nucleotides accompanied by an increase in both cell wall and starch (Geigenberger et al., 2005). This is most likely caused by increases of flux into both components through increased sucrose degradation.
The work performed in altering substrate supply for AGPase has generally been performed in dicotyledonous plants. A recent study (Cakir et al., 2016) has examined rice plants where an increase in extra-plastidial AGPase activity was combined with overexpression of the plastidial ADP-glucose transporter (Figure 1B). They found that, although they could identify increased plastidial ADP-glucose amounts, this did not lead to an increase in starch, indicating additional stromal barriers affecting this pathway. If these could be identified, then they could be manipulated to allow increased starch accumulation.
Can Alterations in Other Starch Biosynthetic Enzymes Increase Yield?
Although most work has examined influencing AGPase as a method of increasing starch amounts, it may not be the only protein that can do this. The polymerizing enzymes SS and SBE (Figure 1B) are present as multiple isoforms, which often play differing roles in determining the structure of the amylopectin molecule. It has been reported that increased expression of SS encoding genes is associated with increased starch accumulation and grain weight caused by expression of a mutated ubiquitin receptor in maize (Xie et al., 2018), while overexpression of one SS isoform increased potato tuber starch content (Gámez-Arjona et al., 2011). The data of Gámez-Arjona et al. (2011) have, however, been questioned (Sonnewald and Kossmann, 2013) due to the lack of dry matter increase that accompanied the reported starch elevation. Nevertheless, these data indicate that manipulation of SS isoforms may be a profitable way in increasing starch content.
There are fewer reports of SBE overexpression; however, one study in potatoes demonstrated that this led to synthesis of starch with altered structure but did not report on an effect on yield (Brummell et al., 2015). Interestingly, manipulation of SBE activity in Arabidopsis leaves has revealed a potential method for improving growth. Replacement of endogenous Arabidopsis activities with two from maize led to plants with increased starch in their leaves and improved seed yield, most likely also due to increased night-time sugar levels (Liu et al., 2016).
Are Starch Degradative Enzymes Useful for Yield Increases?
Plant biotechnologists have generally attempted to increase starch yield through altering activities of enzymes involved in its synthesis. Degradative enzymes have often been ignored as targets for improving yield as mutations in them are often associated with reduced plant growth (Stitt and Zeeman, 2012; Paparelli et al., 2013). Recent work, however, has demonstrated that repression of the starch phosphorylating GWD enzyme (Figure 1A) in wheat endosperm improved both growth and seed production. Unfortunately, although this was demonstrated convincingly in glasshouse trials (Ral et al., 2012; Bowerman et al., 2016), when the same lines were examined in the field a reduction in yield was observed (Whan et al., 2017). Although it may not be a valuable way to increase starch yield in storage organs, repressing genes involved in this process could improve forage and silage crops by improving their calorific value for animal feed (Weise et al., 2012; Ruckle et al., 2018).
Manipulating starch degradation (Figure 1A) may have a greater role in helping plants overcome abiotic stress (Thalmann and Santelia, 2017). One of the main metabolites produced during starch catabolism is maltose, and it has been demonstrated that this can help stabilize photosynthetic membranes (Kaplan and Guy, 2004). dpe2 mutant plants, which accumulate maltose, demonstrate reduced freezing damage (Li et al., 2011); however, they also grow smaller than wild-type plants (Chia et al., 2004). One strategy to overcome this may be the use of stress inducible promoters to drive repression constructs that reduce Dpe2 expression only at times when increased maltose would be advantageous. Simultaneously, β-amylases that produce maltose during starch degradation (Kaplan and Guy, 2004; Kaplan et al., 2006) could be upregulated to increase levels of this metabolite further.
Control Mechanisms Affecting Starch Metabolism
Although alterations of individual enzymatic steps within the starch pathway can have a beneficial effect on yield, alterations in transcriptional control mechanisms allow the possibility to influence these in a beneficial manner through altering multiple steps simultaneously. A few transcriptional regulators affecting starch metabolism have been identified (Zhang et al., 2005; Fu and Xue, 2010; Guan et al., 2011; Wang et al., 2013; Gontarek et al., 2016; Xiao et al., 2017), and manipulation of one of these led to increased seed size and yield in rice (Fu and Xue, 2010). More recently, transcriptional analysis has led to the identification of genes putatively involved in regulating starch metabolic genes (see for example Van Harsselaar et al., 2017), but their roles have often not been studied in detail. Such functional analyses would help in identification of factors that could be used to improve yield.
Post-transcriptional regulation will also be influential in controlling starch metabolism. Many sugar sensing mechanisms involving 14-3-3- and SnRK proteins, trehalose metabolism, TOR kinases, and hexokinase are known to affect starch metabolism. Alterations in expression of some 14-3-3 and SnRK proteins can lead to improved starch accumulation or to the upregulation of enzymes involved in starch synthesis, in several species (Sehnke et al., 2001; McKibbin et al., 2006; Wang et al., 2016, 2018). Trehalose 6-phosphate (T6P) has been proposed to activate AGPase through a post-translational redox mechanism (Kolbe et al., 2005), while genetic manipulation of T6P amounts can increase starch in leaves through repression of starch degradation (Martins et al., 2013) as well as increase yield in maize (Nuccio et al., 2015). More recently, it has been demonstrated that application of plant permeable analogues of T6P increases endosperm starch content and yield in wheat. The reasons for this are not entirely clear as many transcriptional and metabolic changes were identified; however, application of the same analog to Arabidopsis increased AGPase activity which would provide a direct explanation for the increased starch (Griffiths et al., 2016).
Post-translational alteration of proteins involved in starch metabolism can involve protein phosphorylation (Kötting et al., 2010) or reduction/oxidation mechanisms (Glaring et al., 2012; Santelia et al., 2015). Little is known about protein phosphorylation influencing starch metabolism, although a recent paper has identified plastidially localized protein kinases and phosphatases that may interact with starch metabolic enzymes (White-Gloria et al., 2018), which is a first step in the study of this process. More is known about redox control as several genes involved in starch metabolism are known to be redox regulated (Fu et al., 1998; Ballicora et al., 2000; Tiessen et al., 2002; Sokolov et al., 2006; Valerio et al., 2011; Glaring et al., 2012; Seung et al., 2013; Shaik et al., 2014), and expression of thioredoxin-f increased starch amounts in tobacco leaves (Sanz-Barrio et al., 2013), although it is not clear which enzymes were affected. The best characterization of the role of redox on starch metabolizing enzymes in vivo has been the examination of AGPase and GWD, where redox insensitive proteins have been expressed in plants (Haedrich et al., 2012; Skeffington et al., 2014). Redox insensitive AGPase led to increased leaf starch, although this was dependent on day length (Haedrich et al., 2012). On the other hand, constitutive expression of a redox insensitive GWD had little effect on leaf starch degradation (Skeffington et al., 2014), which agrees with the observation that reducing conditions (which would be expected to be present in chloroplasts during the day) activate the wild-type enzyme (Mikkelsen et al., 2005).
Future Prospects
Although much has been achieved over the past decades in the manipulation of starch metabolism, there are still improvements that can be made. Detailed analysis of both transcriptional and post-translational control mechanisms will help fine tune current attempts at manipulating the pathway. Perhaps most importantly is the rational integration of metabolic engineering simultaneously in leaves and storage organs. One attempt to accomplish this involved reducing starch synthesis in potato leaves in order to increase soluble sugar export, while simultaneously increasing substrate supply to starch synthesis in tubers through overexpression of two plastidial transport proteins (Jonik et al., 2012). This approach led to an increase over and above the amounts found when either leaf or tuber metabolism was altered alone, doubling starch yield. We believe that such integrated approaches will lead to the greatest benefit for crop improvement.
Author Contributions
JL wrote the first draft and edited the manuscript alongside JK.
Funding
This work was supported by the NRF SARCHI chair “Genetic Tailoring of Biopolymers.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Arias, M. C., Pelletier, S., Hilliou, F., Wattebled, F., Renou, J.-P., and D’Hulst, C. (2014). From dusk till dawn: the Arabidopsis thaliana sugar starving responsive network. Front. Plant Sci. 5:482. doi: 10.3389/fpls.2014.00482
Bahaji, A., Li, J., Sánchez-López, Á. M., Baroja-Fernández, E., Muñoz, F. J., Ovecka, M., et al. (2014). Starch biosynthesis, its regulation and biotechnological approaches to improve crop yields. Biotechnol. Adv. 32, 87–106. doi: 10.1016/j.biotechadv.2013.06.006
Ballicora, M. A., Frueauf, J. B., Fu, Y., Schurmann, P., and Preiss, J. (2000). Activation of the potato tuber ADP-glucose pyrophosphorylase by thioredoxin. J. Biol. Chem. 275, 1315–1320. doi: 10.1074/jbc.275.2.1315
Balmer, Y., Vensel, W. H., Cai, N., Manieri, W., Schurmann, P., Hurkman, W. J., et al. (2006). A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc. Natl. Acad. Sci. U. S. A. 103, 2988–2993. doi: 10.1073/pnas.0511040103
Boehlein, S. K., Shaw, J. R., Hwang, S. K., Stewart, J. D., and Curtis Hannah, L. (2013a). Deciphering the kinetic mechanisms controlling selected plant ADP-glucose pyrophosphorylases. Arch. Biochem. Biophys. 535, 215–226. doi: 10.1016/j.abb.2013.04.003
Boehlein, S. K., Shaw, J. R., McCarty, D. R., Hwang, S.-K., Stewart, J. D., and Hannah, L. C. (2013b). The potato tuber, maize endosperm and a chimeric maize-potato ADP-glucose pyrophosphorylase exhibit fundamental differences in Pi inhibition. Arch. Biochem. Biophys. 537, 210–216. doi: 10.1016/j.abb.2013.07.019
Boehlein, S. K., Shaw, J. R., Stewart, J. D., Sullivan, B., and Hannah, L. C. (2015). Enhancing the heat stability and kinetic parameters of the maize endosperm ADP-glucose pyrophosphorylase using iterative saturation mutagenesis. Arch. Biochem. Biophys. 568, 28–37. doi: 10.1016/j.abb.2015.01.008
Bowerman, A. F., Newberry, M., Dielen, A.-S., Whan, A., Larroque, O., Pritchard, J., et al. (2016). Suppression of glucan, water dikinase in the endosperm alters wheat grain properties, germination and coleoptile growth. Plant Biotechnol. J. 14, 398–408. doi: 10.1111/pbi.12394
Brummell, D. A., Watson, L. M., Zhou, J., McKenzie, M. J., Hallett, I. C., Simmons, L., et al. (2015). Overexpression of STARCH BRANCHING ENZYME II increases short-chain branching of amylopectin and alters the physicochemical properties of starch from potato tuber. BMC Biotechnol. 15:28. doi: 10.1186/s12896-015-0143-y
Cakir, B., Shiraishi, S., Tuncel, A., Matsusaka, H., Satoh, R., Singh, S., et al. (2016). Analysis of the rice ADPglucose transporter (OsBT1) indicates the presence of regulatory processes in the amyloplast stroma that control ADPglucose flux into starch. Plant Physiol. 170, 1271–1283. doi: 10.1104/pp.15.01911
Chia, T., Thorneycroft, D., Chapple, A., Messerli, G., Chen, J., Zeeman, S. C., et al. (2004). A cytosolic glucosyltransferase is required for conversion of starch to sucrose in Arabidopsis leaves at night. Plant J. 37, 853–863. doi: 10.1111/j.1365-313X.2003.02012.x
Crozet, P., Margalha, L., Confraria, A., Rodrigues, A., Martinho, C., Adamo, M., et al. (2014). Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00190
Dobrenel, T., Caldana, C., Hanson, J., Robaglia, C., Vincentz, M., Veit, B., et al. (2016). TOR signaling and nutrient sensing. Annu. Rev. Plant Biol. 67, 261–285. doi: 10.1146/annurev-arplant-043014-114648
Edgerton, M. D. (2009). Increasing crop productivity to meet global needs for feed, food, and fuel. Plant Physiol. 149, 7–13. doi: 10.1104/pp.108.130195
Fodor, N., Challinor, A., Droutsas, I., Ramirez-Villegas, J., Zabel, F., Koehler, A.-K., et al. (2017). Integrating plant science and crop modeling: assessment of the impact of climate change on soybean and maize production. Plant Cell Physiol. 58, 1833–1847. doi: 10.1093/pcp/pcx141
Fu, F.-F., and Xue, H.-W. (2010). Coexpression analysis identifies rice starch regulator 1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol. 154, 927–938. doi: 10.1104/pp.110.159517
Fu, Y., Ballicora, M. A., Leykam, J. F., and Preiss, J. (1998). Mechanism of reductive activation of potato tuber ADP-glucose pyrophosphorylase. J. Biol. Chem. 273, 25045–25052. doi: 10.1074/jbc.273.39.25045
Gámez-Arjona, F. M., Li, J., Raynaud, S., Baroja-Fernández, E., Muñoz, F. J., Ovecka, M., et al. (2011). Enhancing the expression of starch synthase class IV results in increased levels of both transitory and long-term storage starch. Plant Biotechnol. J. 9, 1049–1060. doi: 10.1111/j.1467-7652.2011.00626.x
Geigenberger, P. (2001). Tuber physiology and properties of starch from tubers of transgenic potato plants with altered plastidic adenylate transporter activity. Plant Physiol. 125, 1667–1678. doi: 10.1104/pp.125.4.1667
Geigenberger, P., Regierer, B., Nunes-Nesi, A., Leisse, A., Urbanczyk-Wochniak, E., Springer, F., et al. (2005). Inhibition of de novo pyrimidine synthesis in growing potato tubers leads to a compensatory stimulation of the pyrimidine salvage pathway and a subsequent increase in biosynthetic performance. Plant Cell 17, 2077–2088. doi: 10.1105/tpc.105.033548
Georgelis, N., and Hannah, L. C. (2008). Isolation of a heat-stable maize endosperm ADP-glucose pyrophosphorylase variant. Plant Sci. 175, 247–254. doi: 10.1016/j.plantsci.2008.04.005
Gibson, K., Park, J.-S., Nagai, Y., Hwang, S.-K., Cho, Y.-C., Roh, K.-H., et al. (2011). Exploiting leaf starch synthesis as a transient sink to elevate photosynthesis, plant productivity and yields. Plant Sci. 181, 275–281. doi: 10.1016/j.plantsci.2011.06.001
Glaring, M. A., Skryhan, K., Kötting, O., Zeeman, S. C., and Blennow, A. (2012). Comprehensive survey of redox sensitive starch metabolising enzymes in Arabidopsis thaliana. Plant Physiol. Biochem. 58, 89–97. doi: 10.1016/j.plaphy.2012.06.017
Gontarek, B. C., Neelakandan, A. K., Wu, H., and Becraft, P. W. (2016). NKD transcrition factors are central regulators of maize endosperm development. Plant Cell 28, 2916–2936. doi: 10.1105/tpc.16.00609
Griffiths, C. A., Sagar, R., Geng, Y., Primavesi, L. F., Patel, M. K., Passarelli, M. K., et al. (2016). Chemical intervention in plant sugar signalling increases yield and resilience. Nature 540, 574–578. doi: 10.1038/nature20591
Guan, S., Wang, P., Liu, H., Liu, G., Ma, Y., and Zhao, L. (2011). Production of high-amylose maize lines using RNA interference in sbe2a. African J. Biotechnol. 10, 15229–15237. doi: 10.5897/AJB11.943
Haedrich, N., Gibon, Y., Schudoma, C., Altmann, T., Lunn, J. E., and Stitt, M. (2011). Use of TILLING and robotised enzyme assays to generate an allelic series of Arabidopsis thaliana mutants with altered ADP-glucose pyrophosphorylase activity. J. Plant Physiol. 168, 1395–1405. doi: 10.1016/j.jplph.2011.01.013
Haedrich, N., Hendriks, J. H. M., Kötting, O., Arrivault, S., Feil, R., Zeeman, S. C., et al. (2012). Mutagenesis of cysteine 81 prevents dimerization of the APS1 subunit of ADP-glucose pyrophosphorylase and alters diurnal starch turnover in Arabidopsis thaliana leaves. Plant J. 70, 231–242. doi: 10.1111/j.1365-313X.2011.04860.x
Hannah, L. C., Futch, B., Bing, J., Shaw, J. R., Boehlein, S., Stewart, J. D., et al. (2012). A shrunken-2 transgene increases maize yield by acting in maternal tissues to increase the frequency of seed development. Plant Cell 24, 2352–2363. doi: 10.1105/tpc.112.100602
Hannah, L. C., Shaw, J. R., Clancy, M. A., Georgelis, N., and Boehlein, S. K. (2017). A brittle-2 transgene increases maize yield by acting in maternal tissues to increase seed number. Plant Direct 1–9. doi: 10.1002/pld3.29
Jonik, C., Sonnewald, U., Hajirezaei, M.-R., Flügge, U.-I., and Ludewig, F. (2012). Simultaneous boosting of source and sink capacities doubles tuber starch yield of potato plants. Plant Biotechnol. J. 10, 1088–1098. doi: 10.1111/j.1467-7652.2012.00736.x
Kaplan, F., and Guy, C. L. (2004). Beta-amylase induction and the protective role of maltose during temperature shock. Plant Physiol. 135, 1674–1684. doi: 10.1104/pp.104.040808
Kaplan, F., Sung, D. Y., and Guy, C. L. (2006). Roles of beta-amylase and starch breakdown during temperatures stress. Physiol. Plant. 126, 120–128. doi: 10.1111/j.1399-3054.2006.00604.x
Kolbe, A., Tiessen, A., Schluepmann, H., Paul, M., Ulrich, S., and Geigenberger, P. (2005). Trehalose 6-phosphate regulates starch synthesis via posttranslational redox activation of ADP-glucose pyrophosphorylase. Proc. Natl. Acad. Sci. U. S. A. 102, 11118–11123. doi: 10.1073/pnas.0503410102
Kötting, O., Kossmann, J., Zeeman, S. C., and Lloyd, J. R. (2010). Regulation of starch metabolism: the age of enlightenment? Curr. Opin. Plant Biol. 13, 321–329. doi: 10.1016/j.pbi.2010.01.003
Li, T., Xu, S.-L., Oses-Prieto, J. A., Putil, S., Xu, P., Wang, R.-J., et al. (2011). Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis. Mol. Plant 4, 361–374. doi: 10.1093/mp/ssq078
Liu, F., Zhao, Q., Mano, N., Ahmed, Z., Nitschke, F., Steup, M., et al. (2016). Modification of starch metabolism in transgenic Arabidopsis thaliana increases plant biomass and doubles oil seed production. Plant Biotechnol. J. 14, 976–985. doi: 10.1111/pbi.12453
Lloyd, J. R., and Kossmann, J. (2015). Transitory and storage starch metabolism: two sides of the same coin? Curr. Opin. Biotechnol. 32, 143–148. doi: 10.1016/j.copbio.2014.11.026
MacNeill, G. J., Mehrpouyan, S., Minow, M. A. A., Patterson, J. A., Tetlow, I. J., and Emes, M. J. (2017). Starch as a source, starch as a sink: the bifunctional role of starch in carbon allocation. J. Exp. Bot. 68, 4433–4453. doi: 10.1093/jxb/erx291
Mahlow, S., Orzechowski, S., and Fettke, J. (2016). Starch phosphorylation: insights and perspectives. Cell. Mol. Life Sci. 73, 2753–2764. doi: 10.1007/s00018-016-2248-4
Martins, M. C. M., Hejazi, M., Fettke, J., Steup, M., Feil, R., Krause, U., et al. (2013). Feedback inhibition of starch degradation in Arabidopsis leaves mediated by trehalose 6-phosphate. Plant Physiol. 163, 1142–1163. doi: 10.1104/pp.113.226787
Matsoukas, I. G., Massiah, A. J., and Thomas, B. (2013). Starch metabolism and antiflorigenic signals modulate the juvenile-to-adult phase transition in Arabidopsis. Plant. Cell Environ. 36, 1802–1811. doi: 10.1111/pce.12088
McKibbin, R. S., Muttucumaru, N., Paul, M. J., Powers, S. J., Burrell, M. M., Coates, S., et al. (2006). Production of high-starch, low-glucose potatoes through over-expression of the metabolic regulator SnRK1. Plant Biotechnol. J. 4, 409–418. doi: 10.1111/j.1467-7652.2006.00190.x
Mikkelsen, R., Mutenda, K. E., Mant, A., Schurmann, P., and Blennow, A. (2005). Alpha-glucan, water dikinase (GWD): a plastidic enzyme with redox-regulated and coordinated catalytic activity and binding affinity. Proc. Natl. Acad. Sci. U. S. A. 102, 1785–1790. doi: 10.1073/pnas.0406674102
Nazarian-Firouzabadi, F., and Visser, R. G. F. (2017). Potato starch synthases: functions and relationships. Biochem. Biophys. Rep. 10, 7–16. doi: 10.1016/j.bbrep.2017.02.004
Nuccio, M. L., Wu, J., Mowers, R., Zhou, H.-P., Meghji, M., Primavesi, L. F., et al. (2015). Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 33, 862–869. doi: 10.1038/nbt.3277
Oiestad, A. J., Martin, J. M., and Giroux, M. J. (2016). Overexpression of ADP-glucose pyrophosphorylase in both leaf and seed tissue synergistically increase biomass and seed number in rice (Oryza sativa ssp. japonica). Funct. Plant Biol. 43, 1194–1204. doi: 10.1071/FP16218
Paparelli, E., Parlanti, S., Gonzali, S., Novi, G., Mariotti, L., Ceccarelli, N., et al. (2013). Nighttime sugar starvation orchestrates gibberellin biosynthesis and plant growth in Arabidopsis. Plant Cell 25, 3760–3769. doi: 10.1105/tpc.113.115519
Pfister, B., and Zeeman, S. C. (2016). Formation of starch in plant cells. Cell. Mol. Life Sci. 73, 2781–2807. doi: 10.1007/s00018-016-2250-x
Ral, J.-P., Bowerman, A. F., Li, Z., Sirault, X., Furbank, R., Pritchard, J. R., et al. (2012). Down-regulation of glucan, water-dikinase activity in wheat endosperm increases vegetative biomass and yield. Plant Biotechnol. J. 10, 871–882. doi: 10.1111/j.1467-7652.2012.00711.x
Regierer, B., Fernie, A. R., Springer, F., Perez-Melis, A., Leisse, A., Koehl, K., et al. (2002). Starch content and yield increase as a result of altering adenylate pools in transgenic plants. Nat. Biotechnol. 20, 1256–1260. doi: 10.1038/nbt760
Rolland, F., Baena-Gonzalez, E., and Sheen, J. (2006). Sugar sensing and signaling in plants conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Ruckle, M., Bernasconi, L., Kölliker, R., Zeeman, S., and Studer, B. (2018). Genetic diversity of diurnal carbohydrate accumulation in white clover (Trifolium repens L.). Agronomy 8:47. doi: 10.3390/agronomy8040047
Ruckle, M., Meier, M., Frey, L., Eicke, S., Kölliker, R., Zeeman, S., et al. (2017). Diurnal leaf starch content: an orphan trait in forage legumes. Agronomy 7:16. doi: 10.3390/agronomy7010016
Santelia, D., Trost, P., and Sparla, F. (2015). New insights into redox control of starch degradation. Curr. Opin. Plant Biol. 25, 1–9. doi: 10.1016/j.pbi.2015.04.003
Sanz-Barrio, R., Corral-Martinez, P., Ancin, M., Segui-Simarro, J. M., and Farran, I. (2013). Overexpression of plastidial thioredoxin f leads to enhanced starch accumulation in tobacco leaves. Plant Biotechnol. J. 11, 618–627. doi: 10.1111/pbi.12052
Schlosser, A. J., Martin, J. M., Beecher, B. S., and Giroux, M. J. (2014). Enhanced rice growth is conferred by increased leaf ADP-glucose pyrophosphorylase activity. J. Plant Physiol. Pathol. 2:4. doi: 10.4172/2329-955X.1000136
Sehnke, P. C., Chung, H.-J., Wu, K., and Ferl, R. J. (2001). Regulation of starch accumulation by granule-associated plant 14-3-3 proteins. Proc. Natl. Acad. Sci. U. S. A. 98, 765–770. doi: 10.1073/pnas.021304198
Seung, D., Thalmann, M., Sparla, F., Abou Hachem, M., Lee, S. K., Issakidis-Bourguet, E., et al. (2013). Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic alpha-amylase. J. Biol. Chem. 288, 33620–33633. doi: 10.1074/jbc.M113.514794
Shaik, S. S., Carciofi, M., Martens, H. J., Hebelstrup, K. H., and Blennow, A. (2014). Starch bioengineering affects cereal grain germination and seedling establishment. J. Exp. Bot. 65, 2257–2270. doi: 10.1093/jxb/eru107
Shi, L., Wu, Y., and Sheen, J. (2018). TOR signaling in plants: conservation and innovation. Development 145:dev160887. doi: 10.1242/dev.160887
Skeffington, A. W., Graf, A., Duxbury, Z., Gruissem, W., and Smith, A. M. (2014). Glucan, water dikinase exerts little control over starch degradation in Arabidopsis leaves at night. Plant Physiol. 165, 866–879. doi: 10.1104/pp.114.237016
Skryhan, K., Gurrieri, L., Sparla, F., Trost, P., and Blennow, A. (2018). Redox regulation of starch metabolism. Front. Plant Sci. 9:1344. doi: 10.3389/fpls.2018.01344
Smidansky, E. D., Clancy, M., Meyer, F. D., Lanning, S. P., Blake, N. K., Talbert, L. E., et al. (2002). Enhanced ADP-glucose pyrophosphorylase activity in wheat endosperm increases seed yield. Proc. Natl. Acad. Sci. U. S. A. 99, 1724–1729. doi: 10.1073/pnas.022635299
Smidansky, E. D., Martin, J. M., Hannah, L. C., Fischer, A. M., and Giroux, M. J. (2003). Seed yield and plant biomass increases in rice are conferred by deregulation of endosperm ADP-glucose pyrophosphorylase. Planta 216, 656–664. doi: 10.1007/s00425-002-0897-z
Sokolov, L. N., Dominguez-Solis, J. R., Allary, A. L., Buchanan, B. B., and Luan, S. (2006). A redox-regulated chloroplast protein phosphatase binds to starch diurnally and functions in its accumulation. Proc. Natl. Acad. Sci. U. S. A. 103, 9732–9737. doi: 10.1073/pnas.0603329103
Sonnewald, U., and Kossmann, J. (2013). Starches-from current models to genetic engineering. Plant Biotechnol. J. 11, 223–232. doi: 10.1111/pbi.12029
Stark, D. M., Timmerman, K. P., Barry, G. F., Preiss, J., and Kishore, G. M. (1992). Regulation of the amount of starch in plant tissues by ADP glucose pyrophosphorylase. Science 258, 287–292. doi: 10.1126/science.258.5080.287
Stitt, M., Gibon, Y., Lunn, J. E., and Piques, M. (2007). Multilevel genomics analysis of carbon signalling during low carbon availability: coordinating the supply and utilisation of carbon in a fluctuating environment. Funct. Plant Biol. 34, 526–549. doi: 10.1071/FP06249
Stitt, M., and Zeeman, S. C. (2012). Starch turnover: pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15, 282–292. doi: 10.1016/j.pbi.2012.03.016
Streb, S., Eicke, S., and Zeeman, S. C. (2012). The simultaneous abolition of three starch hydrolases blocks transient starch breakdown in Arabidopsis. J. Biol. Chem. 287, 41745–41756. doi: 10.1074/jbc.M112.395244
Tetlow, I. J., and Emes, M. J. (2017). Starch biosynthesis in the developing endosperms of grasses and cereals. Agronomy 7:81. doi: 10.3390/agronomy7040081
Thalmann, M., and Santelia, D. (2017). Starch as a determinant of plant fitness under abiotic stress. New Phytol. 214, 943–951. doi: 10.1111/nph.14491
Tiessen, A., Hendriks, J. H. M., Stitt, M., Branscheid, A., Gibon, Y., Farre, E. M., et al. (2002). Starch synthesis in potato tubers is regulated by post-translational redox modification of ADP-glucose pyrophosphorylase: a novel regulatory mechanism linking starch synthesis to the sucrose supply. Plant Cell 14, 2191–2213. doi: 10.1105/tpc.003640
Tjaden, J., Mohlmann, T., Kampfenkel, K., Neuhaus, G. H., and Ekkehard Neuhaus, H. (1998). Altered plastidic ATP/ADP-transporter activity influences potato (Solanum tuberosum L.) tuber morphology, yield and composition of tuber starch. Plant J. 16, 531–540. doi: 10.1046/j.1365-313x.1998.00317.x
Tuncel, A., and Okita, T. W. (2013). Improving starch yield in cereals by over-expression of ADPglucose pyrophosphorylase: expectations and unanticipated outcomes. Plant Sci. 211, 52–60. doi: 10.1016/j.plantsci.2013.06.009
Valerio, C., Costa, A., Marri, L., Issakidis-Bourguet, E., Pupillo, P., Trost, P., et al. (2011). Thioredoxin-regulated β-amylase (BAM1) triggers diurnal starch degradation in guard cells, and in mesophyll cells under osmotic stress. J. Exp. Bot. 62, 545–555. doi: 10.1093/jxb/erq288
Van Harsselaar, J. K., Lorenz, J., Senning, M., Sonnewald, U., and Sonnewald, S. (2017). Genome-wide analysis of starch metabolism genes in potato (Solanum tuberosum L.). BMC Genom. 18:37. doi: 10.1186/s12864-016-3381-z
Wang, F., Ren, G., Li, F., Wang, B., Yang, Y., Ma, X., et al. (2018). Overexpression of GmSnRK1, a sorbean sucrose non-fermenting-1 related protein kinase 1 gene, results in directional alteration tof carbohydrate metabolism in transgenic Arabidopsis. Biotechnol. Biotechnol. Equip. doi: 10.1080/13102818.2018.1469431
Wang, J.-C., Xu, H., Zhu, Y., Liu, Q.-Q., and Cai, X.-L. (2013). OsbZIP58, a basic leucine xipper transcrition factor, regulates starch biosynthesis in rice endosperm. J. Exp. Bot. 64, 3453–3466, doi: 10.1093/jxb/ert187
Wang, X., Chang, L., Tong, Z., Wang, D., Yin, Q., Wang, D., et al. (2016). Proteomics profiling reveals carbohydrate metabolic enzymes and 14-3-3 proteins play important roles for starch accumulation during Cassava root tuberization. Sci. Rep. 6:19643. doi: 10.1038/srep19643
Weise, S. E., Aung, K., Jarou, Z. J., Mehrshahi, P., Li, Z., Hardy, A. C., et al. (2012). Engineering starch accumulation by manipulation of phosphate metabolism of starch. Plant Biotechnol. J. 10, 545–554. doi: 10.1111/j.1467-7652.2012.00684.x
Whan, A., Verbyla, A., Mieog, J., Howitt, C., and Ral, J.-P. (2017). Transferring a biomass enhancement biotechnology from glasshouse to field: a case study on wheat GWD RNAi. Agronomy 7:82. doi: 10.3390/agronomy7040082
White-Gloria, C., Johnson, J. J., Marritt, K., Kataya, A., Vahab, A., and Moorhead, G. B. (2018). Protein kinases and phosphatases of the plastid and their potential role in starch metabolism. Front. Plant Sci. 9:1032. doi: 10.3389/fpls.2018.01032
Wurzinger, B., Nukarinen, E., Nägele, T., Weckwerth, W., and Teige, M. (2018). The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 176, 1085–1094. doi: 10.1104/pp.17.01404
Xiao, Q., Wang, Y., Du, J., Li, H., Wei, B., Wang, Y., et al. (2017). ZmMYB14 is an important transcription factor involved in the regulation of the activity of the ZmBT 1 promoter in starch biosynthesis in maize. FEBS J. 284, 3079–3099. doi: 10.1111/febs.14179
Xie, G., Li, Z., Ran, Q., Wang, H., and Zhang, J. (2018). Over-expression of mutated ZmDA1 or ZmDAR1 gene improves maize kernel yield by enhancing starch synthesis. Plant Biotechnol. J. 16, 234–244. doi: 10.1111/pbi.12763
Zeeman, S. C., Kossmann, J., and Smith, A. M. (2010). Starch: its metabolism, evolution, and biotechnological modification in plants. Annu. Rev. Plant Biol. 61, 209–234. doi: 10.1146/annurev-arplant-042809-112301
Zhang, L., Häusler, R. E., Greiten, C., Hajirezaei, M.-R., Haferkamp, I., Neuhaus, H. E., et al. (2008). Overriding the co-limiting import of carbon and energy into tuber amyloplasts increases the starch content and yield of transgenic potato plants. Plant Biotechnol. J. 6, 453–464. doi: 10.1111/j.1467-7652.2008.00332.x
Keywords: starch, sucrose, carbohydrate partitioning, photosynthesis, metabolism
Citation: Lloyd JR and Kossmann J (2019) Starch Trek: The Search for Yield. Front. Plant Sci. 9:1930. doi: 10.3389/fpls.2018.01930
Edited by:
Miguel Angel Ballicora, Loyola University Chicago, United StatesReviewed by:
Robert D. Hancock, James Hutton Institute, United KingdomNishikant Wase, University of Nebraska-Lincoln, United States
Copyright © 2019 Lloyd and Kossmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Kossmann, a29zc21hbm5Ac3VuLmFjLnph
 James R. Lloyd
James R. Lloyd Jens Kossmann
Jens Kossmann