
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 17 December 2018
Sec. Plant Biotechnology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01873
This article is part of the Research TopicForest Genomics and BiotechnologyView all 20 articles
Boreal and temperate woody perennials are highly adapted to their local climate, which delimits the length of the growing period. Moreover, seasonal control of growth-dormancy cycles impacts tree productivity and geographical distribution. Therefore, traits related to phenology are of great interest to tree breeders and particularly relevant in the context of global warming. The recent application of transcriptional profiling and genetic association studies to poplar species has provided a robust molecular framework for investigating molecules with potential links to phenology. The environment dictates phenology by modulating the expression of endogenous molecular switches, the identities of which are currently under investigation. This review outlines the current knowledge of these molecular switches in poplar and covers several perspectives concerning the environmental control of growth-dormancy cycles. In the process, we highlight certain genetic pathways which are affected by short days, low temperatures and cold-induced signaling.
When the photoperiod falls below the critical day length, poplars undergo growth cessation, culminating in bud set and the acquisition of cold hardiness. The fact that components of light signaling, the circadian clock and orthologs of Arabidopsis flowering time regulators have all been implicated in this stage suggests the interplay between environmental signals and diurnal gene expression in dormancy switch. Thereafter, the sequential induction of ethylene and abscisic acid signaling pathways promotes bud maturation, cessation of meristematic activity and the establishment of dormancy. Once dormant, meristem becomes insensitive to growth-promoting signals. Release from dormancy strongly depends on the accumulation of a defined number of chilling hours, a mechanism for which the molecular basis is unknown. Under spring conditions, the low temperature (LT)-mediated activation of growth promoters reactivates meristem growth. Thus, poplar phenology is controlled by the orchestrated activity of molecular switches following exposure to environmental cues. This mini-review provides insight into the genetic network underlying poplar bud set, dormancy establishment and bud break with a focus on photoperiod- and temperature-dependent signaling. Moreover, we show evidence that bud set is highly sensitive to low ambient temperatures and identify candidate genes that may participate in this response.
Irrefutable experimental evidence for the short day (SD) requirement to cold acclimation comes from studies of photoperiod-insensitive oat PHYA overexpressing poplars Junttila and Kaurin, 1990; Olsen et al., 1997; Welling et al., 2002). When transgenic and wild type plants grown during SDs were subjected to freezing conditions, poplars overexpressing oat PHYA did not develop cold hardiness (Olsen et al., 1997; Welling et al., 2002). In contrast, neither transgenic nor wild type poplars grown during long days were able to survive freezing temperatures Junttila and Kaurin, 1990. This indicates that exposure to SDs acclimates poplars to cold weather conditions, and possibly other environmental stresses, which suggests that SDs may activate adaptation pathways.
The transcriptional and metabolomic profiling of poplar shoot apices and stem tissues grown under LD and SD conditions has been used to investigate the molecular signatures underlying SD-driven adaptive responses. The results revealed that SD conditions significantly alter the transcription of certain genes (Rohde et al., 2007; Ruttink et al., 2007; Karlberg et al., 2010; Hoffman et al., 2010; Resman et al., 2010). For example, a reduction in day length affects the transcription of several light signaling- and circadian clock-regulated genes (Ruttink et al., 2007). Moreover, SD also induces genes related to dehydration and cold adaptation, such as LATE EMBRYOGENESIS-ABUNDANT (LEA), DEHYDRIN (DHN), COLD REGULATED GENES (COR), and a set of transcription factors that mediate responses to abiotic stimuli, among others (Ruttink et al., 2007; Karlberg et al., 2010). Continued SD exposure stimulates ethylene signaling and abscisic acid (ABA) hormonal burst (Rohde et al., 2002; Ruttink et al., 2007; Karlberg et al., 2010). Ethylene and ABA are abiotic stress-related phytohormones required for adaptive responses in plants and are also functionally associated to the regulation of many aspects of bud development in poplar and birch (Rohde et al., 2002; Ruonala et al., 2006; Shi and Yang, 2014; Müller and Munné-Bosch, 2015; Tylewicz et al., 2015; Trivedi et al., 2016; Tylewicz et al., 2018). Thus, acclimation to winter conditions is mediated by the temporal orchestration of SD-induced transcriptional responses that partly originate from light signaling and circadian clock dependent pathways. As a result, poplars with modified circadian clock sensitivity showed different tolerance to freezing temperatures (Ibañez et al., 2010).
The effect of temperature on bud set has been poorly investigated at the molecular level. Earlier physiological studies have shown that bud set is a thermosensitive process in trees (reviewed in Tanino et al., 2010). Furthermore, recent phenotyping studies using clonally replicated poplars grown under natural conditions show that the timing of bud set depends on the local climate (Rohde et al., 2011; Evans et al., 2014). Rohde et al. (2011) showed that sub-optimal growth temperatures delay the timing of bud set. Accordingly, we observed that the timing of bud set in hybrid aspen (Populus tremula x P. alba) grown under controlled conditions is highly sensitivity to small changes in ambient temperature (Figures 1A,B). Bud set was delayed by approximately 7 days in plants grown at 18°C relative to plants grown at 21°C, and this difference was amplified when the temperature was further reduced to 15°C (Figures 1A,B). This indicates that ambient temperature can impact the SD responses controlling bud set timing. Thus, it can be hypothesized that the activation or repression of thermosensitive genes underlies the regulation of pathways that control bud set. Numerous biological processes are governed by the endogenous clock (Greenham and McClung, 2015); thus, bud set in response to low ambient temperature may additionally be mediated by the modulation of clock-related pathways. For instance, the transcription of cold tolerance genes, e.g., C-REPEAT BINDING FACTOR (CBF) genes, follow circadian rhythms in Arabidopsis (Fowler et al., 2005; Dong et al., 2011).
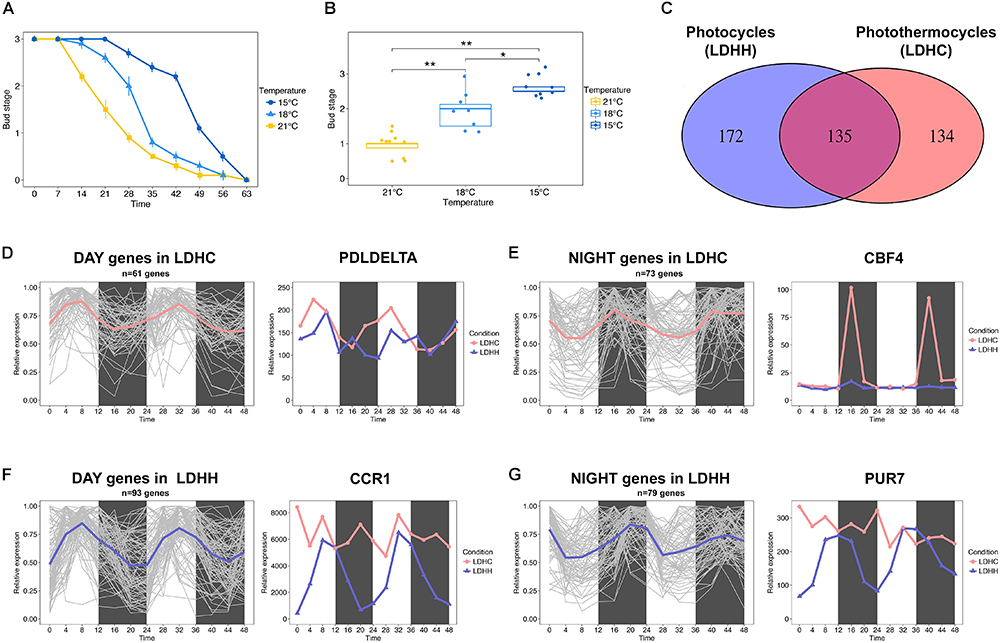
Figure 1. Bud set-associated genes in poplar are sensitive to low ambient temperature. (A) Bud set scores for hybrid poplar (Populus tremula x P. alba) plants grown under SD and three different temperatures (21, 18, and 15°C) for 9 weeks. (B) Comparative analysis of hybrid poplar bud stage progression in plants grown under SD and three different temperatures (21, 18, and 15°C) over 4 weeks. Asterisks denote significant differences between hybrid poplar plant groups (n = 8) grown at different temperatures (Kruskal–Wallis test followed by pairwise Wilcox test; ∗P < 0.05, ∗∗P < 0.01). (C) Venn diagram representing the intersection between genes with SNPs associated with bud set and that show robust diurnal oscillations (cut-off = 0.8) under light/dark cycles (LD) and constant temperature (25°C, HH) or photothermocycles (25°C during the day, 12°C during the night, HC). 172 genes (lilac) show robust diurnal rhythms only under LDHH, 134 genes (pink) show diurnal rhythms only under LDHC and 135 genes (fuchsia) oscillate under both conditions, respectively. (D,E) Clusters of genes that show diurnal expression within 0–11 h under LDHC (D, mean = pink) and within 12–23 h under LDHC (E, mean = pink). The line chart demonstrates how photothermocycles (LDHC) promote diurnal oscillation of poplar PDLDELTA (D) and CBF4 genes (E). (F,G) Clusters of genes that show diurnal expression within 0–11 h under LDHH (F, mean = lilac) and within 12–23 h under LDHH (G, mean = lilac). The line chart demonstrates how photothermocycles (LDHC) disrupt the diurnal oscillation of poplar CCR1 (F) and PUR7 genes (G).
To identify poplar candidate genes that could affect the timing of bud set under low ambient temperature, we investigated how diurnal photothermocycles affect bud set-associated single nucleotide polymorphism (SNP) genes (Mockler et al., 2007; Filichkin et al., 2011; Evans et al., 2014). Particularly, we analyzed the diurnal oscillation (cut-off 0.8) of bud set-associated SNP genes in two different conditions, photothermocycles (LDHC; Day 25°C/Night 12°C) and photocycles (LDHH; Day 25°C/Night 25°C), in Populus thricocarpa using the Diurnal database1 (Mockler et al., 2007; Filichkin et al., 2011). A total of 134 genes associated with bud set showed robust diurnal transcription patterns over a 48 h period under LDHC conditions but not under constant temperature (LDHH), which suggests that the lower night temperature promotes their rhythmic expression (Figures 1C–E). Within the clusters of genes affected by photothermal cycles, we identified diurnal expression in the poplar ortholog of Arabidopsis PHOSPHOLIPASE D DELTA (PLDDELTA; Potri.007G060300), which is involved in phospholipid metabolism, freezing tolerance and stomatal closure (Figure 1D; Chen et al., 2008a,b; Distéfano et al., 2012; Uraji et al., 2012). Moreover, the poplar ortholog of C-REPEAT-BINDING FACTOR 4 (CBF4; Potri.012G134100), which mediates the response to decreased temperatures in Arabidopsis (Wang and Hua, 2009), is also induced by cold temperatures in poplar and birch (Figure 1E; Benedict et al., 2006; Welling and Palva, 2008). These examples indicate that the rhythmic expression of genes associated with bud set – stimulated by diurnal oscillations in temperature – may be required for cold acclimation. In contrast, we identified 172 genes with bud set-associated SNPs that showed diurnal transcription patterns under constant temperatures (LDHH) but not under photothermocycles (LDHC). This suggests that reduced night-time temperatures can undermine the rhythmic expression of certain genes (Figures 1C,F,G). Within the clusters of genes with diurnal expression disrupted by photothermocycles, we highlight the poplar ortholog of Arabidopsis COLD, CIRCADIAN RHYTHM, AND RNA BINDING 1 (CCR1; Potri.009G116400) gene (Figure 1F). CCR1 contains an RNA recognition motif (RRM), which promotes alternative splicing that is coupled to degradation by nonsense-mediated decay (NMD) (Schöning et al., 2008). CCR1 is also regulated by both cold conditions and circadian rhythms in Arabidopsis (Carpenter et al., 1994). Furthermore, we found that the diurnal expression of the poplar ortholog of Arabidopsis PURIN 7 (PUR7; Potri.017G051500), which is required to generate purine dependent cofactors in tissues under high rates of cell division, was disrupted by low night-time temperatures (Senecoff et al., 1996). These examples indicate that the impairment of key, diurnally regulated nucleic acid metabolism processes by decreased night-time temperatures could be important to timely bud set in poplar. Future functional studies will hopefully elucidate the genetic network involved in bud set regulation when poplars are exposed to LTs.
The circadian clock creates endogenous, 24 h rhythms to help plants and animals anticipate daily and seasonal environmental changes (Schultz and Kay, 2003). The circadian clock controls physiology, growth and development, as temporal transcriptional profiles have revealed that more than 30% of genes in Arabidopsis and poplar show circadian rhythms (Harmer et al., 2000; Covington et al., 2008; Michael et al., 2008; Hoffman et al., 2010). Recent research has revealed a role for the circadian clock in the genetic network regulating growth-dormancy cycles. It is widely accepted that circadian rhythms play a central role in the photoperiodic mechanism that controls poplar shoot apical growth (reviewed in Triozzi et al., 2018). However, the implication of the biological clock in the control of growth cessation and bud set has recently emerged (Ibañez et al., 2010; Kozarewa et al., 2010; Ding et al., 2018). The circadian rhythms of several pathways involved in growth cessation and bud set can persist 8–10 weeks after exposure to continuous SDs (Triozzi et al., personal communication). This is supported by the finding that the downregulation of poplar clock-related genes LATE ELONGATED HYPOCOTYL (LHY) and TIMING OF CAB EXPRESSION 1 (TOC1) delays growth cessation and bud set (Ibañez et al., 2010). Furthermore, it has been firstly shown that chestnut clock-related genes display arrhythmic expression under winter conditions (Ramos et al., 2005; Ibañez et al., 2008). Moreover, chestnut and poplar clock-related genes respond to cold temperatures (4°C), showing high and constant expression irrespective of photoperiod (Ramos et al., 2005; Ibañez et al., 2010). It has been suggested that circadian clock disruption may facilitate bud set under cold temperatures (Johansson et al., 2015). Accordingly, cold-induced disruption of circadian rhythms caused wide transcriptional rearrangement of cold response genes in Arabidopsis (Bieniawska et al., 2008). Additionally, when exposed to freezing temperatures, LHY-RNAi poplars showed far more severe stem injuries than control plants. This indicates that the circadian clock is pivotal in the development of cold hardiness during bud set (Ibañez et al., 2010). Nevertheless, the functional implications of cold-induced disruption of clock-regulated pathways during bud set needs further investigation.
Prolonged SD exposure after growth cessation and bud set results in dormancy establishment in various plants (Heide, 1974; Espinosa-Ruiz et al., 2004; Ruttink et al., 2007; Maurya and Bhalerao, 2017). However, until recently, our knowledge of the molecular basis of bud dormancy was rudimentary. Conclusive evidences for dormancy establishment has come from studies on plant hormones ethylene and ABA, both of which are required for apical bud formation (Figure 2). For example, ethylene-insensitive dominant mutant etr1-1 birch plants failed to develop apical buds under SD conditions (Ruonala et al., 2006). Ethylene has also been suggested to participate in dormancy induction in plants such as leafy spurge and Chrysanthemum (Sumitomo et al., 2008; Doğramacı et al., 2013. Nevertheless, genetic evidences for how ethylene affects bud dormancy are missing.
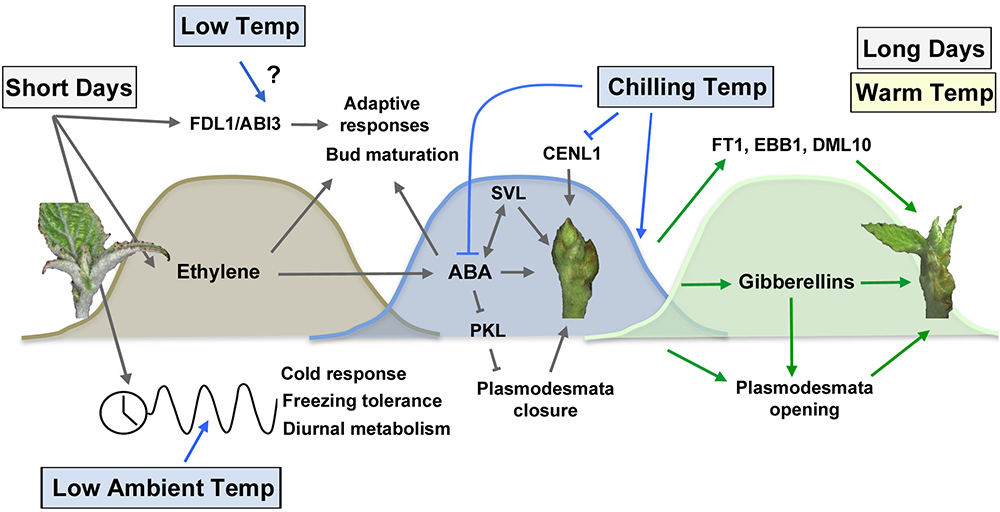
Figure 2. Photoperiodic and temperature control of bud set, dormancy establishment and bud break. Short days promote bud set via activation of Flowering locus D1 (FLD1) and Abscisic acid insensitive 3 (ABI3) (Tylewicz et al., 2015), the ethylene and ABA pathways (Rohde et al., 2002; Ruonala et al., 2006) as well as the circadian clock pathway (Hoffman et al., 2010; Ibañez et al., 2010). The timing of bud set is sensitive to low ambient temperature, which alters the expression of the circadian pathway (Figure 1). ABA plays an essential role in dormancy establishment by promoting plasmodesmata closure via PKL repression (Tylewicz et al., 2018) and activating SVL growth repressive gene (Singh et al., 2018). Increased exposure to chilling temperatures is necessary to the downregulation of ABA and degradation of growth repressors such as CENL1 (Karlberg et al., 2010; Mohamed et al., 2010; Tylewicz et al., 2018). Dormancy release increases gibberellin levels (Rinne et al., 2011), mediates plasmodesmata opening (Tylewicz et al., 2018) and activates bud break-promoting factors such as FT1 (Hsu et al., 2011; Rinne et al., 2011), EBB1 (Yordanov et al., 2014), and DML10 (Conde et al., 2017). Long days and warm temperatures restore shoot apical growth.
Abscisic acid levels increase following SD exposure (Ruttink et al., 2007; Karlberg et al., 2010). Interestingly, the apical buds of etr1-1 mutant birches failed to accumulate ABA under SD conditions, which may suggest that defects in these plants may stem from the inability to increase ABA levels. Moreover, under SD conditions, ABI3oe plants display apical buds with defects during bud maturation (Rohde et al., 2002; Ruttink et al., 2007). Although poplar ABI3 has not been demonstrated to be ABA responsive or contribute to the ABA response per se, this finding may suggest a role for ABA, or at least ABI3, in bud maturation. Other research suggests that molecules associated with the opening and closing of plasmodesmata are involved in dormancy establishment (Rinne et al., 2011). It has been proposed that plasmodesmata closure in shoot apical meristem (SAM) is important for dormancy establishment (Rinne et al., 2011; Singh et al., 2017). Growth-promoting signals such a florigen move symplastically through plasmodesmata to SAM to promote growth under appropriate conditions, yet molecular and genetic evidence for this mechanism was only recently published (Singh et al., 2017; Tylewicz et al., 2018).
Abscisic acid-insensitive hybrid aspen plants overexpressing the dominant negative abi1-1 allele failed to establish dormancy under short photoperiods (SPs) (Tylewicz et al., 2018). Both WT and abi1-1 plants underwent growth cessation and apical bud formation when grown under SPs. However, when transferred from 11 weeks of SP to growth-promoting long photoperiods (LPs), WT plants did not start growing, which suggests that these plants were in a dormant state. However, abi1-1 lines restarted growth 11–15 days after being transferred to LPs. The transcriptomic analysis of WT and abi1-1 plants after 0, 6, and 10 weeks of SPs suggests that 1000s of genes are differentially regulated. Interestingly, many of these genes are associated with plasmodesmata closure and opening. Plasmodesmata closure-related genes such as GERMIN-LIKE 10, REMORIN-LIKE 1 and 2 and CALLOSE SYNTHASE 1 were upregulated in the apices of WT plants whereas genes related to plasmodesmata opening, such as GH17_39, were downregulated. The transcript levels of these genes differed between abi1-1 and WT plants. Microscopic analyses of WT and abi1-1 plants suggest that 83 and less than 3% of plasmodesmata in these plants, respectively, were closed after 10W of SPs (Tylewicz et al., 2018). Flowering Locus T (FT) protein acts as a mobile signal that can move from leaves to apex (Corbesier et al., 2007) and promotes growth of poplar trees even under SP (Böhlenius et al., 2006). Furthermore, when rootstocks of Flowering Locus T1 (FT1) plants were grafted to the scions of 10W SP grown WT and abi1-1 plants, it was able to activate the growth of abi1-1 plants even in SP conditions but not in WT scions.
Many of the transcripts associated with the plasmodesmata closure and opening also responded to the SPs (Tylewicz et al., 2018). Overexpression of plasmodesmata-located protein 1 (PDLP1) in abi1-1 plants leads to dormancy establishment, as abi1-1/PDLP1oe plants transferred from SPs to LPs did not show growth reactivation. Certain genes involved in chromatin remodeling, such as FERTILIZATION INDEPENDENT ENDOSPERM (FIE) and PICKLE (PKL), have been shown to be upregulated after SD exposure (Ruttink et al., 2007). FIE is a component of the polycomb repressive complex 2 (PRC2), while PKL is its antagonist. PRC2 plays important roles in keeping the genes in the repressed state (Margueron and Reinberg, 2011). Transcript level of PKL was upregulated in abi1-1 plants, and its downregulation in abi1-1 background developed dormancy by closing plasmodesmata (Tylewicz et al., 2018).
Taken together, these results suggest that SP influences ABA levels, which then differentially regulate plasmodesmata opening and closure-related genes to induce dormancy. Once the plasmodesmata are closed, growth-promoting signals cannot enter the SAM due to the formation of dormancy sphincters through callose deposition in plasmodesmata.
As mentioned earlier, dormancy establishment makes the SAM impermeable to growth-promoting signals, even under LD conditions. Release from dormancy requires that buds are exposed to LT for a prolonged period (Saure, 1985; Hannerz et al., 2003; Brunner et al., 2014; Fu et al., 2015). How temperature regulates dormancy release remains a poorly studied topic. Interestingly, the exposure of chilled buds to warm temperature is sufficient to induce dormancy release and subsequent bud burst. By monitoring the dormancy release time in plants with altered basal level of certain genes, we can anticipate their role in this process. Since plasmodesmata closure is required for dormancy establishment (Tylewicz et al., 2018), their opening could be expected to result in dormancy release. It has been suggested that LT treatment leads to the opening of plasmodesmata (Rinne et al., 2011), but this does not have proper experimental proof until now. However, some of the genes needed for the removal of plasmodesmatal dormancy sphincters by degrading the deposited callose, such as 1,3-β-glucanase (glucan hydrolase family 17 [GH17]), are upregulated after chilling treatment (Rinne et al., 2011).
Gibberellic acids (GAs) and FT are positive regulators of growth; as such, LT induces the transcription of FT1 and genes implicated in GA metabolism, those that encode members of the GA3 and GA20 oxidases (Rinne et al., 2011). GAs may promote GH17s expression to reopen the plasmodesmata and thus restart the symplastic, growth-promoting cell-to-cell signaling within the SAM (Figure 2). However, genetic and experimental proof of this mechanism is still lacking. CONSTANS (CO) expression was shown to be upregulated almost threefold after 2 weeks of chilling. This suggests that the CO/FT module may be initiated by LT, yet CO expression remains at a similar level throughout the cold period and is only further enhanced when chilled buds sense LDs and warmer temperatures (Rinne et al., 2011). However, the expression of CENL1/TFL1 (CENTRORADIALIS-LIKE1/TERMINAL FLOWER 1), a negative regulator of growth, remains at low levels in chilled buds throughout the LT period (Figure 2; Rinne et al., 2011). Experiments with CENL1oe and CENL-RNAi lines showed delayed and early bud burst, respectively, relative to wild type plants (Mohamed et al., 2010). This suggests that CENL1 represses dormancy release and bud burst. In chilled buds, CENL1 expression is only upregulated once the buds are transferred to warmer temperatures.
The process of dormancy release in trees is comparable to vernalization in Arabidopsis (Chouard, 1960). Many of the MAD-box transcription factors – termed DORMANCY ASSOCIATED MADS-BOX (DAM) – are known to be induced in the dormant buds of many plants, and are similar to short vegetative protein (SVP) and AGAMOUS-LIKE (AGL) transcription factors which control flowering in Arabidopsis (Ríos et al., 2014). Similar to Flowering Locus C (FLC) in Arabidopsis, these DAM genes are also epigenetically regulated by cold conditions. Cold-induced epigenetic silencing of floral repressor FLC is required during vernalization to induce flowering in Arabidopsis (Amasino, 2004; Bastow et al., 2004). The finding that expression of DAM genes goes down after cold treatment suggests that these genes are repressors of dormancy release (Shim et al., 2014; Howe et al., 2015). However, the exact roles of DAM genes can only be elucidated through future genetic studies.
Like dormancy release, our knowledge about bud burst is also very limited in perennial plants. A large number of genes are differentially regulated during dormancy release (Shim et al., 2014; Howe et al., 2015), but we do not have enough genetic evidence to conclusively describe their roles in dormancy release. Very recently, a genetic network mediating the dormancy release and bud break has been described (Singh et al., 2018). This study demonstrated that short vegetative protein-like (SVL), a tree ortholog of Arabidopsis SVP, acts as a central regulator of dormancy release and bud burst in poplar. It negatively regulates dormancy release and bud burst by promoting and repressing the expressions of negative and positive regulators of growth, respectively, after cold treatment (Singh et al., 2018). The expression of Early Bud-Break 1 (EBB1), a putative APETALA2/Ethylene responsive factor, also increases after cold periods during dormancy release and before bud burst. However, EBB1 transcripts are undetectable during the dormancy period. Transgenic lines with EBB1 overexpression and downregulation show early and delayed bud burst relative to wild type plants, respectively. This suggests that EBB1 is a positive regulator of bud burst (Figure 2; Yordanov et al., 2014).
The effect of temperature on tree phenology is an important topic in the context of climate change. Extended seasonal growth and shifts in latitudinal distribution demonstrate how plants are already adapting to increasing global temperatures. Although previous research has attempted to elucidate how temperature influences growth cessation and bud set in trees, very little is still known about the molecular mechanisms involved in these phenomena. For this reason, understanding how environmentally sensitive molecular switches regulate the perception and transduction of temperature signals in woody plants will be crucial to predicting how plants will adapt to warmer environments and designing appropriate breeding programs for this scenario.
PT and MP analyzed the data. JM, PT, RB, and MP contributed to the writing of the manuscript.
The work of MP was supported by the Ramón y Cajal Programme of MINECO (RYC-2012-10194) to PT. The Ph.D. project of PT was supported by a program at the CEI campus of the Universidad Politécnica de Madrid (L1UF00-47-JX9FYF). The work in the lab of RB was funded by grants from Vetenskapsrådet and Knut and Alice Wallenberg Foundation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Allona for the critical reading of the manuscript. We apologize to all colleagues whose work has not been cited because of space limitations.
Amasino, R. (2004). Vernalization, competence, and the epigenetic memory of winter. Plant Cell 16, 2553–2559. doi: 10.1105/tpc.104.161070
Bastow, R., Mylne, J. S., Lister, C., Lippman, Z., Martienssen, R. A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. doi: 10.1038/nature02269
Benedict, C., Skinner, J. S., Meng, R., Chang, Y., Bhalerao, R., Huner, N. P. A., et al. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant Cell Environ. 29, 1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x
Bieniawska, Z., Espinoza, C., Schlereth, A., Sulpice, R., Hincha, D. K., and Hannah, M. A. (2008). Disruption of the Arabidopsis circadian clock is responsible for extensive variation in the cold-responsive transcriptome. Plant Physiol. 147, 263–279. doi: 10.1104/pp.108.118059
Böhlenius, H., Huang, T., Charbonnel-Campaa, L., Brunner, A. M., Jansson, S., Strauss, S. H., et al. (2006). CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. doi: 10.1126/science.1126038
Brunner, A. M., Evans, L. M., Hsu, C. Y., and Sheng, X. (2014). Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front. Plant Sci. 5:732. doi: 10.3389/fpls.2014.00732
Carpenter, C. D., Kreps, J. A., and Simon, A. E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104, 1015–1025. doi: 10.1104/pp.104.3.1015
Chen, Q. F., Xiao, S., and Chye, M. L. (2008a). Arabidopsis ACBP6 is an acyl-CoA-binding protein associated with phospholipid metabolism. Plant Signal. Behav. 3, 1019–1020. doi: 10.4161/psb.6762
Chen, Q. F., Xiao, S., and Chye, M. L. (2008b). Overexpression of the Arabidopsis 10-kilodalton Acyl-coenzyme a-binding protein acbp6 enhances freezing tolerance. Plant Physiol. 148, 304–315. doi: 10.1104/pp.108.123331
Chouard, P. (1960). Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 11, 191–238. doi: 10.1146/annurev.pp.11.060160.001203
Conde, D., Le Gac, A. L., Perales, M., Dervinis, C., Kirst, M., Maury, S., et al. (2017). Chilling-responsive demeter-like DNA demethylase mediates in poplar bud break. Plant Cell Environ. 40, 2236–2249. doi: 10.1111/pce.13019
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Covington, M. F., Maloof, J. N., Straume, M., Kay, S. A., and Harmer, S. L. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 9:R130. doi: 10.1186/gb-2008-9-8-130
Ding, J., Böhlenius, H., Rühl, M. G., Chen, P., Sane, S., Zambrano, J. A., et al. (2018). Gigantea-like genes control seasonal growth cessation in populus. New Phytol. 218, 1491–1503. doi: 10.1111/nph.15087
Distéfano, A. M., Scuffi, D., García-Mata, C., Lamattina, L., and Laxalt, A. M. (2012). Phospholipase Dδ is involved in nitric oxide-induced stomatal closure. Planta 236, 1899–1907. doi: 10.1007/s00425-012-1745-4
Doğramacı, M., Foley, M. E., Chao, W. S., Christoffers, M. J., and Anderson, J. V. (2013). Induction of endodormancy in crown buds of leafy spurge (Euphorbia esula L.) implicates a role for ethylene and cross-talk between photoperiod and temperature. Plant Mol. Biol. 81, 577–593. doi: 10.1007/s11103-013-0026-3
Dong, M. A., Farre, E. M., and Thomashow, M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the c-repeat binding factor (cbf) pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 7241–7246. doi: 10.1073/pnas.1103741108
Espinosa-Ruiz, A., Saxena, S., Schmidt, J., Mellerowicz, E., Miskolczi, P., Bako, L., et al. (2004). Differential stage-specific regulation of cyclin-dependent kinases during cambial dormancy in hybrid aspen. Plant J. 38, 603–615. doi: 10.1111/j.1365-313X.2004.02070.x
Evans, L. M., Slavov, G. T., Rodgers-Melnick, E., Martin, J., Ranjan, P., Muchero, W., et al. (2014). Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat. Genet. 46, 1089–1096. doi: 10.1038/ng.3075
Filichkin, S. A., Breton, G., Priest, H. D., Dharmawardhana, P., Jaiswal, P., Fox, S. E., et al. (2011). Global profiling of rice and poplar transcriptomes highlights key conserved Circadian-controlled pathways and cis-regulatory modules. PLoS One 6:e16907. doi: 10.1371/journal.pone.0016907
Fowler, S. G., Cook, D., and Thomashow, M. F. (2005). Low temperature induction of Arabidopsis CBF1,2, and 3 is gated by the circadian clock. Plant Physiol. 137, 961–968. doi: 10.1104/pp.104.058354
Fu, Y. H., Piao, S., Vitasse, Y., Zhao, H., Boeck, H. J. D., Liu, Q., et al. (2015). Increased heat requirement for leaf flushing in temperate woody species over 1980–2012: effects of chilling, precipitation and insolation. Global Change Biol. 21, 2687–2697. doi: 10.1111/gcb.12863
Greenham, K., and McClung, C. R. (2015). Integrating circadian dynamics with physiological processes in plants. Nat. Rev. Genet. 16, 598–610. doi: 10.1038/nrg3976
Hannerz, M., Ekberg, I., and Norell, L. (2003). Variation in chilling requirements for completing bud rest between provenances of Norway spruce. Silvae Genet. 52, 161–168.
Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Heide, O. M. (1974). Growth and dormancy in Norway spruce ecotypes (Picea abies) I. Interaction of photoperiod and temperature. Physiol. Plant. 30, 1–12. doi: 10.1111/j.1399-3054.1974.tb04983.x
Hoffman, D. E., Jonsson, P., Bylesjö, M., Trygg, J., Antti, H., Eriksson, M. E., et al. (2010). Changes in diurnal patterns within the Populus transcriptome and metabolome in response to photoperiod variation. Plant Cell Environ. 33, 1298–1313. doi: 10.1111/j.1365-3040.2010.02148.x
Howe, G. T., Horvath, D. P., Dharmawardhana, P., Priest, H. D., Mockler, T. C., and Strauss, S. H. (2015). Extensive transcriptome changes during natural onset and release of vegetative bud dormancy in Populus. Front. Plant Sci. 6:989. doi: 10.3389/fpls.2015.00989
Hsu, C. Y., Adams, J. P., Kim, H., No, K., Ma, C., Strauss, S. H., et al. (2011). Flowering locus t duplication coordinates reproductive and vegetative growth in perennial poplar. Proc. Natl. Acad. Sci. U.S.A. 108, 10756–10761. doi: 10.1073/pnas.1104713108
Ibañez, C., Kozarewa, I., Johansson, M., Ogren, E., Rohde, A., and Eriksson, M. E. (2010). Circadian clock components regulate entry and affect exit of seasonal dormancy as well as winter hardiness in Populus trees. Plant Physiol. 153, 1823–1833. doi: 10.1104/pp.110.158220
Ibañez, C., Ramos, A., Acebo, P., Contreras, A., Casado, R., Allona, I., et al. (2008). Overall alteration of circadian clock gene expression in the chestnut cold response. PLoS One 3:e3567. doi: 10.1371/journal.pone.0003567
Johansson, M., Ramos-Saìnchez, J. M., Conde, D., IbaìnÞez, C., Takata, N., and Allona, I. (2015). “Role of the circadian clock in cold acclimation and winter dormancy in perennial plants,” in Advances in Plant Dormancy, ed. J. V. Anderson (Basel: Springer International Publishing), 51–74.
Junttila, O., and Kaurin, Å (1990). Environmental control of cold acclimation in salix pentandra. Scand. J. For. Res. 5, 195–204. doi: 10.1080/02827589009382605
Karlberg, A., Englund, M., Petterle, A., Molnar, G., Sjödin, A., and Bako, L. (2010). Analysis of global changes in gene expression during activity-dormancy cycle in hybrid aspen apex. Plant Biotechnol. 27, 1–16. doi: 10.5511/plantbiotechnology.27.1
Kozarewa, I., Ibanez, C., and Johansson, M. (2010). Alteration of PHYA expression change circadian rhythms and timing of bud set in Populus. Plant Mol. Biol. 73, 143–156. doi: 10.1007/s11103-010-9619-2
Margueron, R., and Reinberg, D. (2011). The polycomb complex PRC2 and its mark in life. Nature 469, 343–349. doi: 10.1038/nature09784
Maurya, J. P., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective. Ann. Bot. 120, 351–360. doi: 10.1093/aob/mcx061
Michael, T. P., Mockler, T. C., Breton, G., McEntee, C., Byer, A., and Trout, J. D. (2008). Network discovery pipeline elucidates conserved time-of-day-specific cis-regulatory modules. PLoS Genet. 4:e14. doi: 10.1371/journal.pgen.0040014
Mockler, T. C., Michael, T. P., Priest, H. D., Shen, R., Sullivan, C. M., and Givan, S. A. (2007). The diurnal project: diurnal and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb. Symp. Quant. Biol. 72, 353–363. doi: 10.1101/sqb.2007.72.006
Mohamed, R., Wang, C. T., Ma, C., Shevchenko, O., Dye, S. J., Puzey, J. R., et al. (2010). Populus CEN/TFL1 regulates first onset of flowering, axillary meristem identity and dormancy release in Populus. Plant J. 62, 674–688. doi: 10.1111/j.1365-313X.2010.04185.x
Müller, M., and Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Olsen, J. E., Junttila, O., Nilsen, J., Eriksson, M. E., Martinussen, I., Olsson, O., et al. (1997). Ectopic expression of oat phytochrome A in hybrid aspen changes critical daylength for growth and prevents cold acclimatization. Plant J. 12, 1339–1350. doi: 10.1046/j.1365-313x.1997.12061339.x
Ramos, A., Pérez-Solís, E., Ibáñez, C., Casado, R., Collada, C., Gómez, L., et al. (2005). Winter disruption of the circadian clock in chestnut. Proc. Natl. Acad. Sci. U.S.A. 102, 7037–7042. doi: 10.1073/pnas.0408549102
Resman, L., Howe, G., Jonsen, D., Englund, M., Druart, N., Schrader, J., et al. (2010). Components acting downstream of short day perception regulate differential cessation of cambial activity and associated responses in early and late clones of hybrid poplar. Plant Physiol. 154, 1294–1303. doi: 10.1104/pp.110.163907
Rinne, P. L., Welling, A., Vahala, J., Ripel, L., Ruonala, R., Kangasjarvi, J., et al. (2011). Chilling of dormant buds hyperinduces flowering locus t and recruits GA-inducible 1,3-betaglucanases to reopen signal conduits and release dormancy in Populus. Plant Cell 23, 130–146. doi: 10.1105/tpc.110.081307
Ríos, G., Leida, C., Conejero, A., and Badenes, M. L. (2014). Epigenetic regulation of bud dormancy events in perennial plants. Front. Plant Sci. 5:247. doi: 10.3389/fpls.2014.00247
Rohde, A., Bastien, C., and Boerjan, W. (2011). Temperature signals contribute to the timing of photoperiodic growth cessation and bud set in poplar. Tree Physiol. 31, 472–482. doi: 10.1093/treephys/tpr038
Rohde, A., Prinsen, E., De Rycke, R., Engler, G., Van Montagu, M., and Boerjan, W. (2002). PtABI3 impinges on the growth and differentiation of embryonic leaves during bud set in poplar. Plant Cell 14, 1885–1901. doi: 10.1105/tpc.003186
Rohde, A., Ruttink, T., Hostyn, V., Sterck, L., Van Driessche, K., and Boerjan, W. (2007). Gene expression during the induction, maintenance, and release of dormancy in apical buds of poplar. J. Exp. Bot. 58, 4047–4060. doi: 10.1093/jxb/erm261
Ruonala, R., Rinne, P. L. H., Baghour, M., Moritz, T., Tuominen, H., and Kangasjärvi, J. (2006). Transitions in the functioning of the shoot apical meristem in birch (Betula pendula) involve ethylene. Plant J. 46, 628–640. doi: 10.1111/j.1365-313X.2006.02722.x
Ruttink, T., Arend, M., Morreel, K., Storme, V., Rombauts, S., Fromm, J., et al. (2007). A molecular timetable for apical bud formation and dormancy induction in poplar. Plant Cell 19, 2370–2390. doi: 10.1105/tpc.107.052811
Saure, M. (1985). Dormancy release in deciduous fruit trees. Hortic. Rev. 7, 239–300. doi: 10.1002/9781118060735.ch6
Schöning, J. C., Streitner, C., Meyer, I. M., Gao, Y., and Staiger, D. (2008). Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 36, 6977–6987. doi: 10.1093/nar/gkn847
Schultz, T. F., and Kay, S. A. (2003). Circadian clocks in daily and seasonal control of development. Science 301, 326–328. doi: 10.1126/science.1085935
Senecoff, J. F., McKinney, E. C., and Meagher, R. B. (1996). De novo purine synthesis in Arabidopsis thaliana. II. The PUR7 gene encoding 5’-phosphoribosyl-4-(N-succinocarboxamide)-5-aminoimidazole synthetase is expressed in rapidly dividing tissues. Plant Physiol. 112, 905–917. doi: 10.1104/pp.112.3.905
Shi, Y., and Yang, S. (2014). “ABA regulation of the cold stress response in plants,” in Abscisic Acid: Metabolism, Transport and Signaling, ed. D. P. Zhang (Dordrecht: Springer), 337–363. doi: 10.1007/978-94-017-9424-4_17
Shim, D., Ko, J. H., Kim, W. C., Wang, Q., Keathley, D. E., and Han, K. H. (2014). A molecular framework for seasonal growth–dormancy regulation in perennial plants. Hortic. Res. 1:14059. doi: 10.1038/hortres.2014.59
Singh, R. K., Maurya, J. P., Azeez, A., Miskolczi, P., Tylewicz, S., Stojkoviè, K., et al. (2018). A genetic network mediating the control of bud break in hybrid aspen. Nat. Commun. 9:4173. doi: 10.1038/s41467-018-06696-y
Singh, R. K., Svystun, T., AlDahmash, B., Jönsson, A. M., and Bhalerao, R. P. (2017). Photoperiod- and temperature-mediated control of phenology in trees – a molecular perspective. New Phytol. 213, 511–524. doi: 10.1111/nph.14346
Sumitomo, K., Narumi, T., Satoh, S., and Hisamatsu, T. (2008). Involvement of the ethylene response pathway in dormancy induction in chrysanthemum. J. Exp. Bot. 59, 4075–4082. doi: 10.1093/jxb/ern247
Tanino, K. K., Kalcsits, L., Silim, S., Kendall, E., and Gray, G. R. (2010). Temperature-driven plasticity in growth cessation and dormancy development in deciduous woody plants: a working hypothesis suggesting how molecular and cellular function is affected by temperature during dormancy induction. Plant Mol. Biol. 73, 49–65. doi: 10.1007/s11103-010-9610-y
Triozzi, P. M., Ramos-Sánchez, J. M., Hernández-Verdeja, T., Moreno-Cortés, A., Allona, I., and Perales, M. (2018). Photoperiodic control of shoot apical growth in poplar. Front. Plant Sci. 9:1030. doi: 10.3389/fpls.2018.01030
Trivedi, D. K., Gill, S. S., and Tuteja, N. (2016). “Abscisic acid (ABA): biosynthesis, regulation, and role in abiotic stress tolerance,” in Abiotic Stress Response in Plants, eds N. Tuteja and S. S. Gill (Hoboken, NJ: Wiley), doi: 10.1002/9783527694570.ch15
Tylewicz, S., Petterle, A., Marttila, S., Miskolczi, P., Azeez, A., Singh, R. K., et al. (2018). Photoperiodic control of seasonal growth is mediated by ABA acting on cell-cell communication. Science 360, 212–215. doi: 10.1126/science.aan8576
Tylewicz, S., Tsuji, H., Miskolczi, P., Petterle, A., Azeez, A., Jonsson, K., et al. (2015). Dual role of tree florigen activation complex component FD in photoperiodic growth control and adaptive response pathways. Proc. Natl. Acad. Sci. U.S.A. 112, 3140–3145. doi: 10.1073/pnas.1423440112
Uraji, M., Katagiri, T., Okuma, E., Ye, W., Hossain, M. A., Masuda, C., et al. (2012). Cooperative function of pld and pld 1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 159, 450–460. doi: 10.1104/pp.112.195578
Wang, Y., and Hua, J. (2009). A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. Plant J. 60, 340–349. doi: 10.1111/j.1365-313X.2009.03959.x
Welling, A., Moritz, T., Palva, E. T., and Junttila, O. (2002). Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiol. 129, 1633–1641. doi: 10.1104/pp.003814
Welling, A., and Palva, E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147, 1199–1211. doi: 10.1104/pp.108.117812
Keywords: poplar, adaptive response, cold response, circadian clock, short day, low ambient temperature, bud set, winter dormancy
Citation: Maurya JP, Triozzi PM, Bhalerao RP and Perales M (2018) Environmentally Sensitive Molecular Switches Drive Poplar Phenology. Front. Plant Sci. 9:1873. doi: 10.3389/fpls.2018.01873
Received: 30 July 2018; Accepted: 04 December 2018;
Published: 17 December 2018.
Edited by:
Ronald Ross Sederoff, North Carolina State University, United StatesReviewed by:
Hairong Wei, Michigan Technological University, United StatesCopyright © 2018 Maurya, Triozzi, Bhalerao and Perales. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rishikesh P. Bhalerao, UmlzaGkuQmhhbGVyYW9Ac2x1LnNl Mariano Perales, bWFyaWFuby5wZXJhbGVzQHVwbS5lcw==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.