
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 04 December 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01766
This article is part of the Research Topic Organ Modification for Edible Parts of Horticultural Crops View all 9 articles
Plant organ shape is determined by the spatial-temporal expression of genes that control the direction and rate of cell division and expansion, as well as the mechanical constraints provided by the rigid cell walls and surrounding cells. Despite the importance of organ morphology during the plant life cycle, the interplay of patterning genes with these mechanical constraints and the cytoskeleton is poorly understood. Shapes of harvestable plant organs such as fruits, leaves, seeds and tubers vary dramatically among, and within crop plants. Years of selection have led to the accumulation of mutations in genes regulating organ shapes, allowing us to identify new genetic and molecular components controlling morphology as well as the interactions among the proteins. Using tomato as a model, we discuss the interaction of Ovate Family Proteins (OFPs) with a subset of TONNEAU1-recruiting motif family of proteins (TRMs) as a part of the protein network that appears to be required for interactions with the microtubules leading to coordinated multicellular growth in plants. In addition, SUN and other members of the IQD family also exert their effects on organ shape by interaction with microtubules. In this review, we aim to illuminate the probable mechanistic aspects of organ growth mediated by OFP-TRM and SUN/IQD via their interactions with the cytoskeleton.
Plant organs display remarkable phenotypic diversity among and within species. Especially for cultivated crops, selection for the harvestable organs has led to greatly increased size and variable shapes of the produce. This diversity is critical for the successful marketing of a wide array of foods such as fruits, vegetables, seeds, leaves, and tubers. Recent studies have revealed many genes that control the growth form of agriculturally important organs (Zuo and Li, 2014; van der Knaap and Ostergaard, 2017). This includes a newly discovered genetic pathway, which through protein interactions and associations with microtubules is proposed to lead to changes in cell division patterns that accompany the different growth forms (Wu et al., 2018). Mechanistically, how the growth forms are controlled by these genes is largely unknown.
The classification of varieties of the same crop based on morphological descriptors is critical. Organizations such as the Union for the Protection of New Varieties of Plants1 and the International Plant Genetic Resources Institute (IPGRI)2 developed descriptors of the shape of many vegetables and fruits such that varieties can be distinguished from one another. These descriptors have become the framework for the identification of genes underlying the morphological variation in crops like tomato (Brewer et al., 2006; Rodriguez et al., 2011a). Consumers recognize the shape and size of vegetables and fruits for the different culinary purposes and/or cultural significances (Pickersgill, 2007; Daunay et al., 2008; De Haan, 2009; Monforte et al., 2014). Similarly important for grains, the slender rice grain shape is associated with improved transparent appearance and reduced undesirable grain quality and is, therefore, highly sought-after in certain cuisines (Calingacion et al., 2014; Harberd, 2015).
Lateral plant organs such as leaves and fruits typically initiate in the flanks of apical meristems. Together with the hormone auxin, AGAMOUS (for ovaries/fruits) and CUP SHAPED COTYLEDON/NO APICAL MERISTEM initiate organ primordia by setting up organ identity and boundaries (Maugarny-Cales and Laufs, 2018). To change from a meristematic cell fate to an organ fate, the down regulation of KNOXI transcription factors by ASYMMETRIC LEAVES1 and LATERAL ORGAN BOUNDARIES DOMAIN proteins is required (Maugarny-Cales and Laufs, 2018). Many hormones play important roles in the growth of organs, in particular gibberellins and brassinosteroids.
The patterns of further outgrowth occur along different axes: the proximal-distal, the medial-lateral and the abaxial-adaxial axis (Van der Knaap et al., 2014). Simply stated, isotropic growth along all three axes tends to lead to larger and round shapes whereas anisotropic growth leads to alternate shapes. For multidimensional organs such as the fruit, the different tissue types grow in an anisotropic way and together form an overall spherical or elongated shape (van der Knaap and Ostergaard, 2017). At the cellular level, the growth patterns are manifested by a combination of cell proliferation (growth and division) and cell enlargement (growth without cell division) driven by turgor pressure. The directions of cell enlargement are guided and restricted by cellulose microfibrils, which are glucose polymers bundled together by hydrogen bonds and Van der Waals forces. These polymers are deposited into the cell wall by CSCs guided by cMTs, In cells undergoing anisotropic expansion, cellulose microfibrils are deposited perpendicular to the axis of expansion and are coaligned with cMTs (Szymanski and Cosgrove, 2009; McFarlane et al., 2014). During cell proliferation, the plane of cell division is determined by the positioning of the preprophase band (PPB) (Rasmussen et al., 2011). The duration and rates of cell proliferation also affect the pattern of growth. Since plant cells are bound to surrounding cells by cell walls, once division has taken place, including formation of the phragmoplast, plant cells are positioned in the same relative location as when they were formed. Therefore, the orientation of cell division has a dramatic effect on the final shape of plant organs (Figure 1; Meyerowitz, 1997; Jenik and Irish, 2000; Van Damme et al., 2007; Schaefer et al., 2017). Mechanical forces provide direct signals leading to coordinated growth toward the final organ shape and size (von Wangenheim et al., 2016). During lateral organ initiation, a highly organized supracellular alignment of microtubule arrays forms along the maximal stress in the region between the meristematic dome and lateral primordia (Hamant et al., 2008). The microtubules guide the directional deposition of cellulose microfibrils, which reinforces the cell wall strength along the appropriate axes to separate the new organs and the undifferentiated cells. During growth, microtubule array dynamics are regulated to respond to the mechanical forces (Uyttewaal et al., 2010). The reorientation of microtubule arrays along the maximal tensile stress can control the directions of cell division and cell expansion leading to heterogeneous growth.
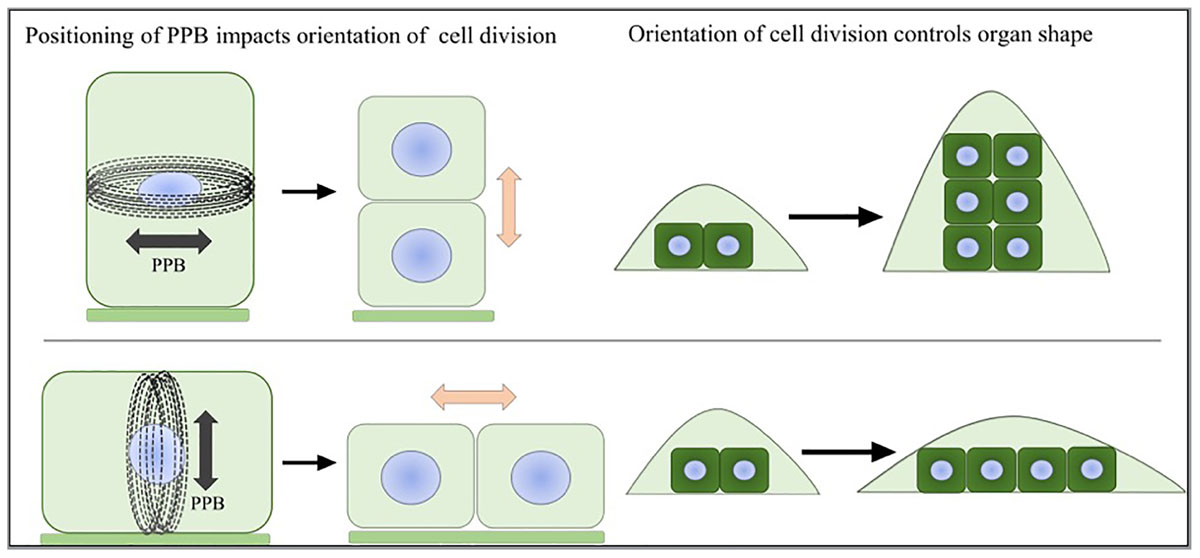
FIGURE 1. Preprophase band and organ shape. The positioning of the PPB marks the future site of cell division. The direction of cell division will greatly influence the shape of the emerging organ.
Understanding the molecular basis of shape of harvestable organs comes mostly from studies conducted in tomato and rice. The increase in rice grain size is often accompanied with altered shape, and found to be under the control of proteins involved in diverse pathways such as G-protein signaling, the ubiquitin–proteasome pathway, phytohormone signaling including brassinolides, auxin and cytokinin, as well as transcriptional regulation (Zuo and Li, 2014; Zheng et al., 2015). In the case of tomato, the identified proteins appear functionally less diverse as they seem to interact with the cytoskeleton. Specifically, a mutation in OVATE, the founding member of the OFP class, and another member named SlOFP20 result in a distinct pear shaped tomato fruit (Wu et al., 2018). OVATE and SlOFP20 interact with several members of the Tonneau1 Recruitment Motif (TRM) proteins, which are often found associated with microtubules (Hamant et al., 2008; Wu et al., 2018). SUN, a member of the IQ Domain (IQD) family, also impacts tomato fruit shape (Xiao et al., 2008). Members of the IQD family have been found to interact with calmodulin (CaM) as well as microtubule binding proteins Kinesin Light Chain-Related protein-1 (KLCR1) and SPR2 to regulate microtubule structure based on external auxin and calcium inputs (Burstenbinder et al., 2013, 2017a,b; Wendrich et al., 2018; Yang et al., 2018).
OVATE and SUN are two important genes controlling tomato fruit shape. The shape of many oval shaped varieties, including grape tomatoes, is controlled by OVATE. SUN can be found in very elongated, tapered or oxheart shaped heirloom as well as commercially grown plum tomatoes (Ku et al., 1999; Liu et al., 2003; Rodriguez et al., 2011b; Van der Knaap et al., 2014). OVATE is the founding member of the OFP class. Recently a new fruit shape gene was identified as a suppressor of ovate (sov1). This fruit shape gene is another member of the same family, SlOFP20 (Huang et al., 2009; Rodriguez et al., 2013; Wu et al., 2018). Whereas ovate is a null, the Slofp20 allele shows reduced expression and the effects of both mutations on ovary shape are already apparent at anthesis (Van der Knaap and Tanksley, 2001; Van der Knaap et al., 2014; Wu et al., 2018). This finding implies that the patterning mediated by these OFPs occurs early in the ontogeny of the ovary, perhaps immediately after organ initiation. SUN also affects ovary shape before anthesis and continues to promote fruit elongation immediately after fertilization (Van der Knaap and Tanksley, 2001; Xiao et al., 2009; Wu et al., 2011). sun is due to a transposon-mediated duplication event leading to high expression of the transposed gene during reproductive development (Xiao et al., 2008). Over-expression of SUN in both wild and cultivated tomatoes leads to evenly elongated fruit shape (Xiao et al., 2008). Interestingly, sun synergistically interacts with ovate and together the two promote growth at the proximal end to form a pear-shaped and pointed tomato fruit (Wu et al., 2015; Figure 2). ovate and sov1 also synergistically interact to form a pear-shaped tomato but with a round bottom shape (Wu et al., 2018; Figure 2). This suggests that obovoid organ shapes may be achieved by alleles from different sets of proteins or that the pathways intersect.
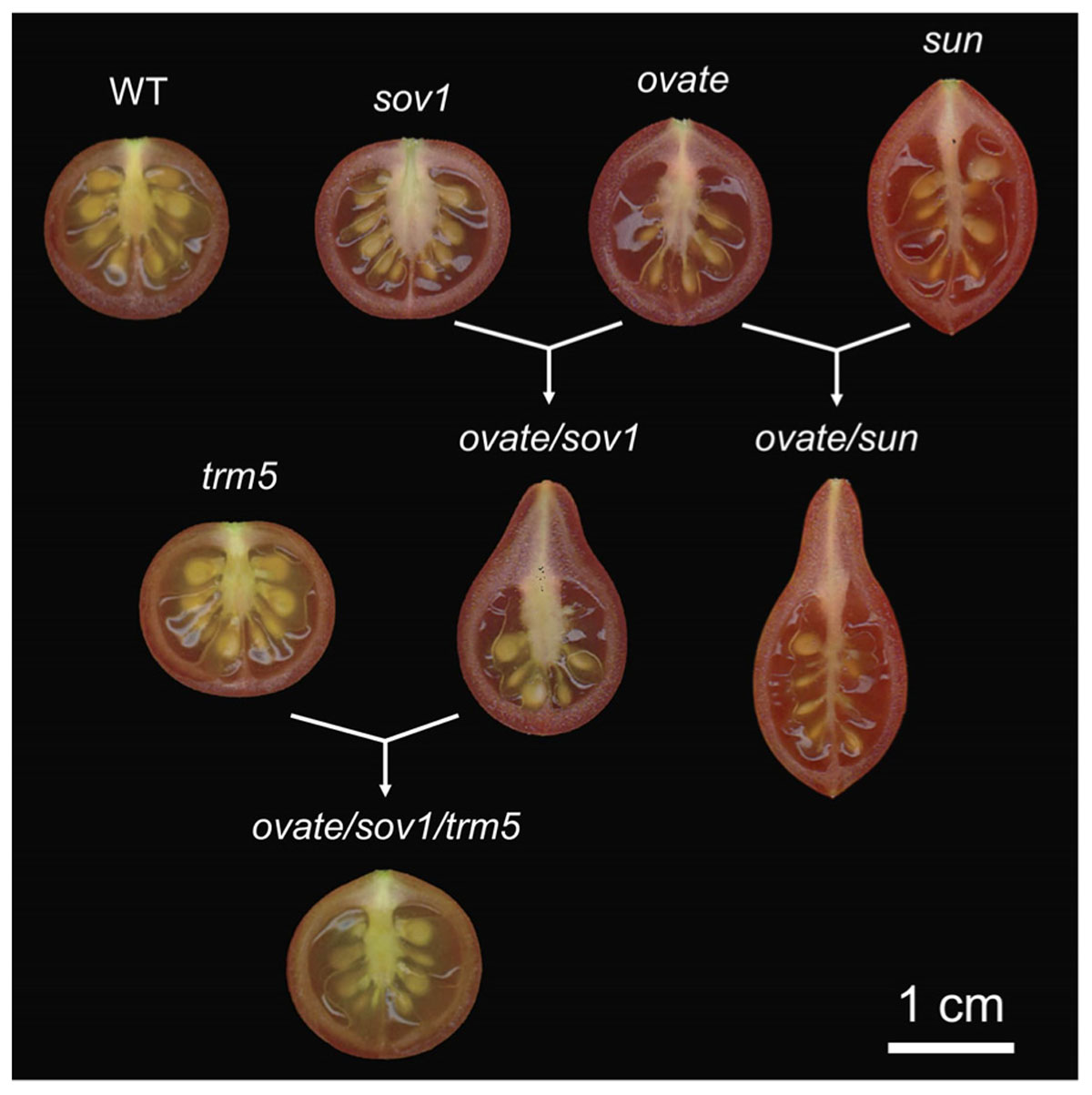
FIGURE 2. Effect of the fruit shape genes in the wild type tomato background. The loci were introgressed (sun, ovate, sov1) or edited (trm5) in the Solanum pimpinellifolium accession LA1589 background to create near isogenic lines (NILs). WT, wild type; sov1, suppressor of ovate corresponding to SlOFP20. The single natural NILs are shown on the top of the figure, whereas the double and triple NILs are shown below the single NILs.
The expression of wild type OVATE in tomato is high in the IM/FM, and its expression reduces 8 days after floral initiation (dpi) (Figure 3). In contrast, the expression of wild type SlOFP20 is relatively low in the IM/FM and increases in 6 dpi buds, with a dramatic increase at 16 dpi (Figure 3). For SUN, wild type gene expression is very low (first two time points, LA1589) whereas in the NIL with the retrotransposon-mediated duplication (sun introgressed in the LA1589 background), SUN is highly expressed during floral development (Figure 3). The initiation of the gynoecium primordia occurs at 6 dpi (Xiao et al., 2009), which is when OVATE, SlOFP20 and SUN are well expressed. At 8 and 13 dpi, the expression of OVATE and SUN respectively, is much reduced from expression levels at the earlier developmental stages coinciding with when these genes may function in development.
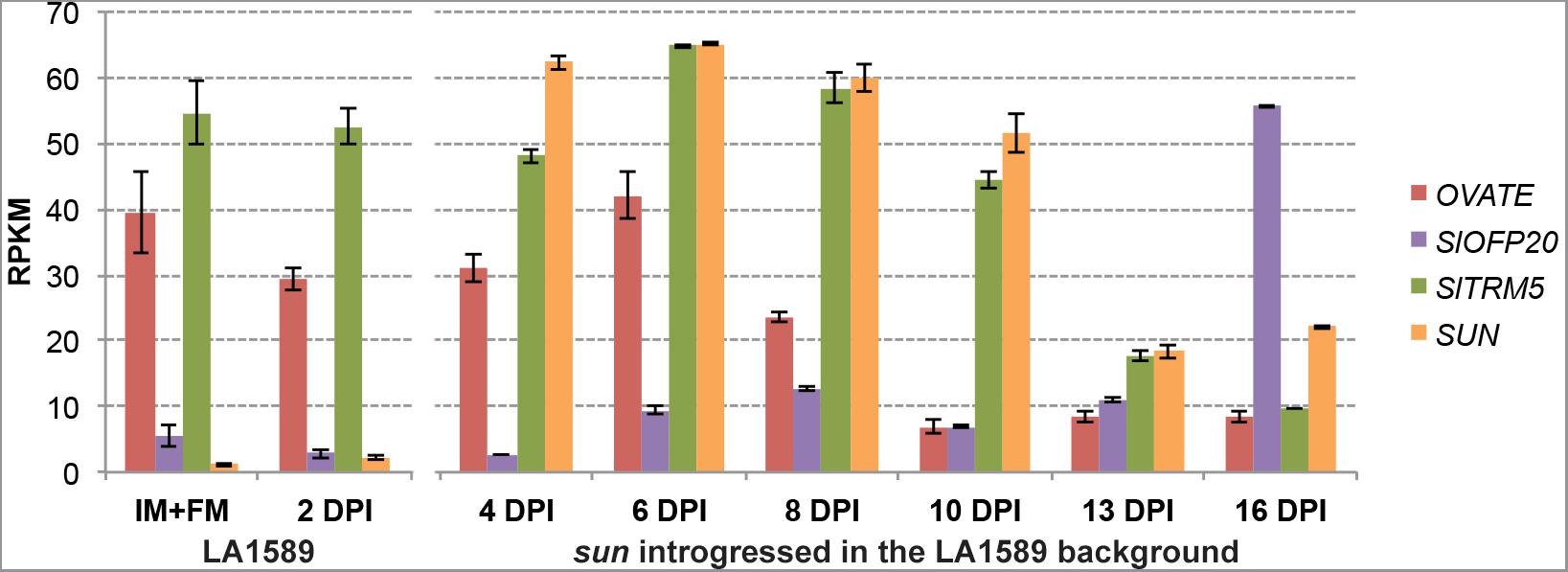
FIGURE 3. Expression patterns of OVATE, SlOFP20, SlTRM5 and SUN during floral development. Samples were collected from wild-type S. pimpinellifolium LA1589 or the sun NIL in the LA1589 background. RPKM, reads per kilobase of transcript per million mapped reads; IM, inflorescence meristem; FM, floral meristem; dpi, flower buds collected in number of days post floral initiation. Each value represents 3 to 4 biological replicates, each containing 100–150 meristems or young flower buds. The bars indicate standard errors among the four replicates. The expression data is available under BioProject number PRJNA343236 and SRP089970 as well as on the Sol Genomics Network website (https://www.sgn.cornell.edu/) at the Tomato Functional Genomics Database (http://ted.bti.cornell.edu/cgi-bin/TFGD/digital/home.cgi).
OVATE, SlOFP20 and SUN affect neither floral organ identity, nor the organization or number of floral organs (Wu, 2015). Instead, SUN, OVATE and SlOFP20 regulate organ elongation by altering the directions of cell division along the proximal-distal axis (Wu et al., 2011, 2018). Whereas SUN affects cell division along the entire proximal-distal axis, OVATE and SlOFP20 appear to have a specific role in anisotropic growth primarily at the proximal end of the ovary. In NILs that carry the ovate and sov1 mutant alleles, there is an increased number of cells in the proximal-distal direction and a reduced number of cells in the medial-lateral direction compared to wild type. Cell size is also enlarged but cell shape appears to change little in the mutant background (Wu et al., 2018). Thus, the effect of cell size and shape in fruit elongation is not clear and therefore, cell division patterns are thought to drive the shape of the ovate/sov1 fruits.
Certain OFPs and SUNs are likely to be involved in conserved mechanisms of morphology regulation across plant species. Genetic evidence indicates that the same subclade of OFPs, represented by Arabidopsis OFP1 and tomato OFP20, controls tomato fruit shape as well as aerial organ shapes in Arabidopsis (Wang et al., 2011), tuber shape in potato, and fruit shape in melon (Wu et al., 2018). Specifically, the potato (Solanum tuberosum L.) tuber shape QTL Ro has been fine-mapped in an outcrossing diploid F1 population to a region on chromosome 10 that contains the potato ortholog of SlOFP20. There is also strong association between tuber shape and StOFP20 in a separate inbred diploid F2 population. Very elongated tubers lack the StOFP20 gene, consistent with its role in the regulation of organ shape as found in tomato. In melon (Cucumis melo), fine mapping within the fruit shape QTL fsqs8.1 has identified CmOFP13 in a cross of Piel de Sapo and PI124112 (Wu et al., 2018). For SUN and other members of the IQD family, natural mutations affecting organ shape have been found in rice and species in the Cucurbitaceae family. Specifically, the rice gene GSE5 at the GW5/qSW5 locus encodes a SUN member closely related to Arabidopsis IQD25-27 (Duan et al., 2017). The change in grain shape is due to increased cell proliferation in spikelet hulls. Interestingly, in cucumber and watermelon, a SUN member that is also most similar to Arabidopsis IQD25-27 likely controls fruit shape in these two species (Pan et al., 2017; Dou et al., 2018). Another rice SUN-like gene, OsIQD14 has been shown to affect rice grain shape and this member is most closely related to another subclade of the SUN/IQD family (Yang et al., 2018). Arabidopsis, overexpression of several IQD members leads to altered organ shapes. The overexpression of microtubular localized AtIQD16 and AtIQD11 resulted in elongated aerial organs with left-handed helical growth abnormalities similar to the phenotype of the tomato SUN overexpressors (Wu et al., 2011; Burstenbinder et al., 2017b). Overexpression of AtIQD14 results in organ twisting but not cell elongation as observed in AtIQD11 and AtIQD16 (Burstenbinder et al., 2017b), a phenotype that resembles that of tortifolia/spiral mutants (Furutani et al., 2000; Buschmann et al., 2004; Shoji et al., 2004). Furthermore, overexpression of plasma membrane localized IQD25 resulted in rounder leaves and larger cells, the opposite phenotype from that observed in overexpression of microtubule localized IQDs suggesting that IQD proteins can have diverse functions in regulating the cytoskeleton and cell elongation (Burstenbinder et al., 2017b). Thus, in addition to SUN in tomato, several members of this family have been associated with changing plant organ shape.
A knockout mutation in tomato’s TONNEAU1 Recruiting Motif 5 (SlTRM5) results in a slightly flatter fruit yet its effect is most strongly noticeable in the ovate/sov1 mutant background (Figure 2). The expression of wild type SlTRM5 is high in IM/FM throughout floral development until 13 dpi (Figure 3), following similar expression dynamics as OVATE and SUN. SlTRM5 is a member of the Arabidopsis TRM1-5 subclade in which tomato carries only two TRM paralogs. At the cellular level, SlTRM5 controls the number of cells in the proximal-distal and medial-lateral direction such that the mutant allele Sltrm5 rescues the tomato fruit shape phenotype of ovate/sov1. TRM5 orthologs and close paralogs also appear to regulate organ shape in other crops. For example, the cucumber ortholog of TRM5 underlies the fs2.1 QTL controlling fruit shape (Wu et al., 2018). In rice, a major QTL for grain length encodes a TRM member in the TRM1-5 subclade. The discovery was made in three independent studies as GRAIN LENGTH ON CHROMOSOME 7 (GL7)/GRAIN WIDTH 7 (GW7)/SLENDER GRAIN ON CHROMOSOME 7 (SLG7) loci (Wang Y. et al., 2015; Wang S. et al., 2015; Zhou et al., 2015). Copy number variants at the GL7 locus contribute to grain size diversity (Wang Y. et al., 2015) and the increased expression of GW7/SLG7 increases grain length (Wang S. et al., 2015; Zhou et al., 2015). However, these studies show contrasting effects on the cellular mechanisms of grain shape changes. Higher expression of GW7 increased cell division in the proximal-distal direction and decreased cell division in the medial-lateral direction (Wang S. et al., 2015), which is similar to the effect of SlTRM5 on tomato fruit shape. On the other hand, increased expression of SLG7 increased cell length and decreased cell width with no changes in cell division (Zhou et al., 2015). In Arabidopsis, certain members of the TRM1-5 subclade control the elongation of various aerial organs. Overexpression of AtTRM1 (LONGIFOLIA2) or AtTRM2 (LONGIFOLIA1) leads to extremely long cotyledons, leaves, floral organs and siliques (Lee et al., 2006). On the other hand, loss-of-function mutations in AtTRM1 or AtTRM2 cause shortened siliques and cotyledons (Lee et al., 2006; Drevensek et al., 2012), which intriguingly mimic the phenotypes of AtOFP overexpressors (Wang et al., 2007). The more elongated leaf blades seen in the AtTRM1 and AtTRM2 overexpressors are due to increased cell expansion along the proximal-distal axis rather than an altered cell proliferation pattern (Lee et al., 2006).
As mentioned in the previous section and based on several studies, TRMs play a critical role in regulating organ shape. In tomato, TRMs were first discovered in a Yeast 2-Hybrid (Y2H) experiment using OVATE as bait. The goal of the experiment was to identify molecular interactants of OVATE to learn about cell division patterning mediated by OFP family members. A total of 11 out of 26 members of the TRM superfamily were identified in the screen. What set these OVATE-interactants apart from the other members of the TRM family was the conserved M8 motif (Van der Knaap et al., 2014; Wu et al., 2018). These findings suggest that the genetic interaction of TRM5 is through protein-protein interactions with OVATE via the TRM M8 motif. To validate the findings from Y2H, the interaction motifs were mapped in OVATE, SlOFP20, and several Ovate-interacting TRMs (Wu et al., 2018). OVATE and SlOFP20 both interact through highly conserved negatively charged amino acids in the OFP domain with TRMs via the highly conserved basic residue (K or R) in the TRM M8 motif. It is reasonable to conclude that the electrostatic interactions in the OFP domain and the M8 motif enable the interactions between these proteins (Wu et al., 2018).
The Y2H protein interactions have also been validated in a plant system. OVATE, SlOFP20 and several Ovate-interacting TRMs were expressed as fusion proteins in Nicotiana benthamiana leaf epidermal cells (Wu et al., 2018). When expressed alone, OVATE localizes in the cytoplasm and SlOFP20 is in the nucleus and cytoplasm. When SlTRM3/4 or SlTRM5 (members of the AtTRM1-5 subclade) are expressed alone, they localize to microtubules. Co-expression of OVATE and SlTRM5 dissociates SlTRM5 from microtubules and both proteins are found in the cytoplasm. On the other hand, co-expression of SlOFP20 and SlTRM5 causes the localization of SlOFP20 to microtubules coincident with SlTRM5. Co-expressions of OVATE or SlOFP20 with SlTRM3/4 both lead to a nearly complete dissociation of SlTRM3/4 from microtubules to the cytoplasm (Wu et al., 2018). These re-localizations are much reduced when mutants of OVATE, SlOFP20, SlTRM5, and SlTRM3/4 lacking the interacting charged amino acid residues are co-expressed in N. benthamiana cells. These findings imply that relocalization occurs through physical protein interactions. Bifluorescence complementation assays further demonstrate that the charged amino acid residues of OVATE and SlTRM5, or OVATE and SlTRM3/4, are responsible for their interactions as well as relocalizations (Wu et al., 2018). The relocalization of OFPs and TRMs to different subcellular compartments upon interaction suggests that a dynamic balance between cytoplasmic- and microtubular-localized OFP-TRM protein complexes regulates cell division and organ growth. A mechanistic model describing the function of OFPs and TRMs to control organ shape is shown in Figure 4.
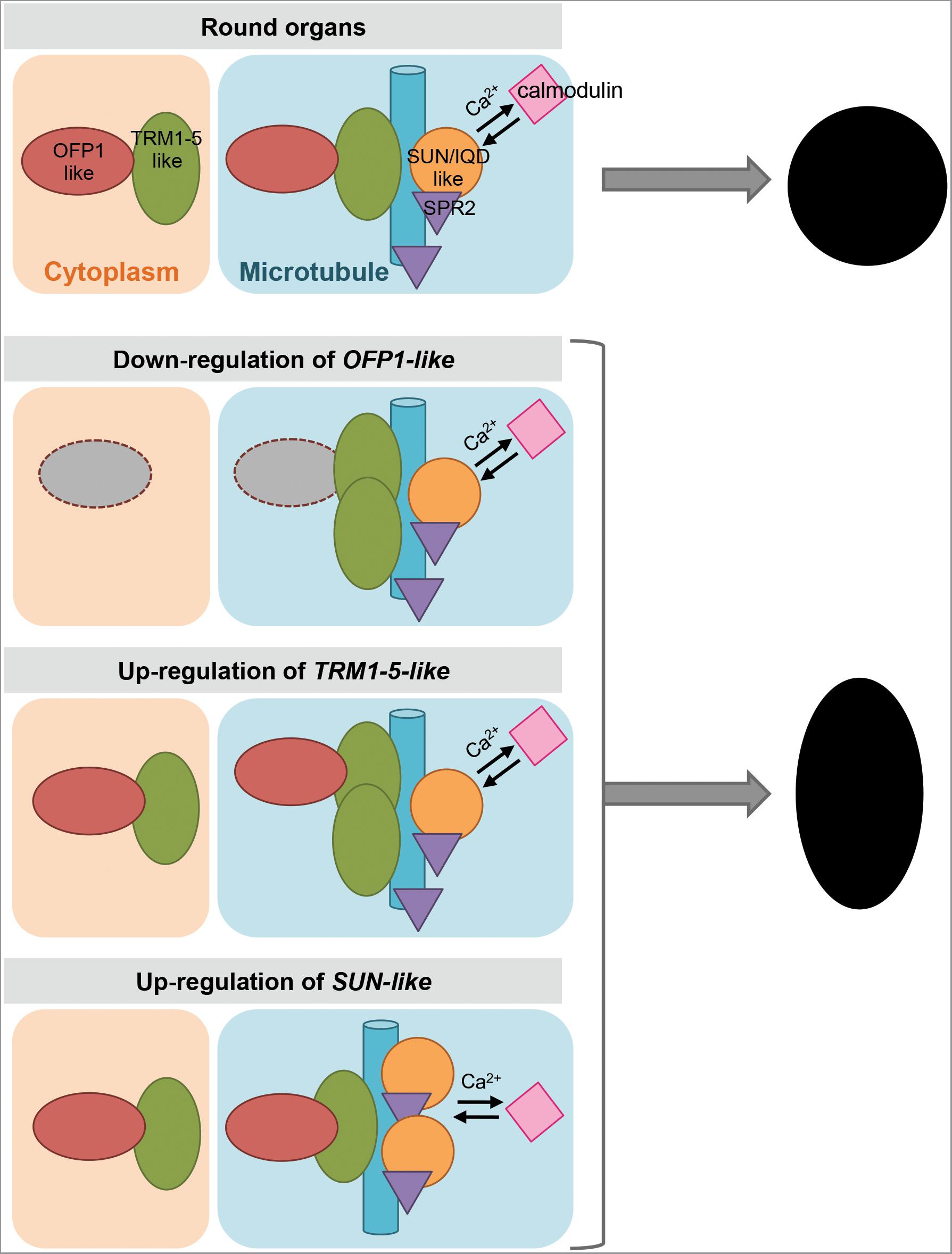
FIGURE 4. A model of the regulation of plant organ shapes. Expression levels of OFP1-like, TRM1-5-like and SUN/IQD-like lead to more or less association with microtubules to determine organ shape. Red oval shape, OFP1-like; green oval shape, TRM1-5-like; blue bar, microtubule; orange circle, SUN/IQD-like; purple triangle, SPR2; pink square, calmodulin.
The subcellular relocalization when co-expressing OVATE or SlOFP20 on the one hand and SlTRM5 or SlTRM3/4 on the other hand suggests that certain OFPs play a critical role in localizing protein complexes. The discovery of the OFP-TRM module may provide an explanation for how other OFPs would serve as regulators in various distinct developmental processes by their effect on subcellular localization. AtOFP5 negatively affects the function of BLH1-KNAT3 complex in early embryo sac development (Pagnussat et al., 2007). The authors show that this is due to abnormal migration and positioning of embryo sac nuclei during megagametogenesis. They propose that this could be due to a change in the behavior of microtubules (Pagnussat et al., 2007) that serve as tracks for nuclear movement in plant cells (Vogelmann et al., 1981; Meindl, 1983; Mineyuki and Furuya, 1985). However, no evidence was available at that time to link the function of AtOFP5 with microtubule dynamics. Another example is offered by AtOFP4 that interacts with KNAT7 to regulate secondary cell wall formation (Li et al., 2011). cMTs participate in secondary cell wall development by directing the deposition of cell wall matrix components (Oda et al., 2005). The defects in secondary cell wall formation could be caused by an abnormal microtubule behavior or due to mislocalization of the CSCs due to the loss-of-function of AtOFP4. Again, this suggests that subcellular localization of CSCs may be disrupted by an OFP. Interestingly, SUN-like protein, AtIQD13 is also associated with positioning the CSCs by influencing the organization of the cMT arrays that guide them (Sugiyama et al., 2017). Other research has shown that the BEL1-like homeodomain 1 (BLH1) protein is shuttled from the nucleus to the cytoplasm when interacting with AtOFP1 or AtOFP5. It was therefore proposed that AtOFPs affect the activities of TALE transcription factors by altering their subcellular localization (Hackbusch et al., 2005). AtOFP1 was also identified as a protein partner of AtKu70, which plays a role in non-homologous end-joining DNA repair (Wang et al., 2010). Interestingly, the centromeric function of Ku70 depends on the presence of microtubules (Cabrero et al., 2013). Therefore, it is reasonable to propose that AtOFP1 may function in DNA-repair by affecting the anchoring of AtKu70 to microtubules. Thus, the OFP-TRM module could explain these seemingly unrelated pathways where OFP controls the subcellular localization of protein complexes. This idea is in stark contrast with the notion in the literature that AtOFPs are transcriptional repressors (Wang et al., 2007, 2011). However, these conclusions were primarily made based on protoplast expression assays as well as expression correlation when overexpressing AtOFP1 (Hackbusch et al., 2005; Wang et al., 2007) and thus transcriptional repression was not validated in intact plants.
A broader function of the TRMs is their role in assembling the TTP (TON1-TRM-PP2A) complex. Before members of this family were associated with the TTP complex, two members namely LONGIFOLIA (LNG) 1 and LNG2, were identified to control organ shape in Arabidopsis (Lee et al., 2006). The entire TRM family, however, was identified in Arabidopsis in a Y2H study using TON1 as bait (Drevensek et al., 2012). The Arabidopsis TRMs consists of 34 members and all contain the TON1-interacting M2 motif at the C terminus. A typical TRM in Arabidopsis is AtTRM1, a microtubule-associated protein that localizes to cMTs in vivo and binds microtubules in vitro. AtTRM1 recruits TON1 to microtubule arrays in N. benthamiana leaf cells. A subset of TRMs target the TTP complex to microtubules (Drevensek et al., 2012). The TTP complex has been proposed to regulate the organization of microtubule arrays and PPB formation, and thus cell division patterns and cell growth (Camilleri et al., 2002; Azimzadeh et al., 2008; Drevensek et al., 2012; Spinner et al., 2013; Schaefer et al., 2017). Throughout interphase in plant cells, microtubules are found just beneath the plasma membrane in the cell cortex. These cMTs determine cell shape as they form patterns in the absence of focused nucleation centers like centrosomes in animal and fungal cells. The nucleation of new cMTs is geometrically constrained (Fishel and Dixit, 2013). Most initiate from a nucleation site on the side of a parent microtubule. These nucleation sites contain γ-tubulin and associated γ-tubulin complex proteins (Nakamura et al., 2010; Murata and Hasebe, 2011) to form the γ-TuRC. After nucleation, new microtubules elongate at about 40o from the parent microtubule (Chan et al., 2009; Nakamura et al., 2010; Murata and Hasebe, 2011). New microtubules also grow parallel to the parent microtubule and move alongside existing microtubules by polymer treadmilling (Chan et al., 2009; Nakamura et al., 2010). The branched form of nucleation is the dominant pattern, while parallel nucleation occurs about half as frequently. About 1–2% of nucleation events occur de novo, where a new microtubule elongates independently of an existing microtubule (Shaw et al., 2003; Chan et al., 2009; Nakamura et al., 2010). As a component of the TTP complex, the Arabidopsis thaliana B” subunit of protein phosphatase 2A is encoded by the TONNEAU2/FASS (TON2) gene (Camilleri et al., 2002), and microtubule branching nucleation is specifically promoted by this regulatory subunit (Kirik et al., 2012). In ton2-15 mutants, the frequency of microtubule branching nucleation is reduced 4-fold while the frequency of parallel nucleation is increased 2.4-fold. The branching angle of new microtubules is unchanged. In hypocotyl cells, loss of TON2 function also results in the inability of microtubule arrays to reorient in response to light, suggesting an essential role for TON2 and microtubule branching nucleation in the reorganization of microtubule arrays (Kirik et al., 2012). It has been postulated that TON2 may influence the orientation of initial polymerization through a direct interaction with or phosphorylation of a component of the γ-TuRC (Fishel and Dixit, 2013). Microtubule assembly at the γ-TuRC is also partly regulated by the microtubule severing protein katanin (Nakamura et al., 2010) and katanin1-mediated microtubule rearrangement is proposed to play a role in regulating rice grain shape controlled by a SUN-like gene, OsIQD14 (Yang et al., 2018). This finding supports the notion that the regulation of organ shape might be functionally linked by the OFP-TRM and SUN/IQD pathways.
The formation and function of different microtubule arrays are regulated by microtubule nucleation, dynamics and stability. Microtubule assembly is a polarized process starting from one or several MTOCs (Murata and Hasebe, 2011). The centrosome is the major MTOC in animal cells to recruit and modify cell cycle proteins (Wu and Akhmanova, 2017). Even though vascular plant cells lack centrosomes, TTP components have sequence similarity to animal centrosomal proteins. For example, the N-terminus of TON1 has sequence similarity to FOP and OFD1, proteins required for microtubule anchoring and stability within the centrosome, respectively (Yan et al., 2006; Azimzadeh et al., 2008; Singla et al., 2010). In addition, three TRM motifs (i.e., M3-M4-M2) are found in the human centrosomal protein CAP350, which interacts with FOP. The C-terminal M2 motif in CAP350 is responsible for FOP recruitment to the human centrosome and facilitates microtubule anchoring within the centrosome (Yan et al., 2006). Similarly, TON1 and the TTP complex bind to the PPB, which marks the future division plane by promoting spindle bipolarity and limiting spindle rotation to ensure properly patterned cell division. The consequence of the sequence similarity and overlapping motifs between TTP complex proteins and animal centrosomal proteins may be the functional similarities among the complexes in plant and animal cell division (Schaefer et al., 2017). The TTP complex is required for proper PPB assembly and division plane establishment (Spinner et al., 2013). The PPB is an array of microtubules and actin filaments that forms a ring at the cell periphery during G2 and persists throughout prophase. Although the PPB is disassembled as the nuclear envelope breaks down and the mitotic spindle forms, its position precisely correlates with the position of the future division plane. The spatial information of the PPB is preserved by selective recruitment and depletion of proteins that lead to the generation of the cortical division zone and the precise positioning of the cell plate during cytokinesis (Rasmussen et al., 2011; Rasmussen and Bellinger, 2018). TON1A, TON1B and the PP2A subunit FASS/TON2 (in Arabidopsis) or DISCORDIA1/ALTERNATIVE DISCORDIA1 (FASS/TON2 orthologs in monocots) are required for PPB formation. Knockout mutants lack PPBs and have incorrectly positioned division planes (Camilleri et al., 2002; Azimzadeh et al., 2008; Wright et al., 2009; Spinner et al., 2010). Thus, a potential mechanistic link between OFPs, OVATE-interacting TRMs and cell patterning is established through interactions with the TTP complex thereby regulating organ shape.
Whether the PPB is absolutely required for division plane patterning is not clear as cells in certain tissues appear to divide without TON1a and PPB formation in other tissues (Zhang et al., 2016a; Costa, 2017). Further insights about the function of the PPB and certain TRMs show that TRM7 is a specific PPB marker whereas TRM6 and TRM8 are constitutively expressed throughout the cell cycle (Schaefer et al., 2017). The frequency of normal PPB formation is reduced in trm7 mutants and no PPBs are found in the trm678 triple mutants. Cells with disrupted PPB formation retain the capacity to define a cortical division zone but lose precision in the orientation of this division zone. Intriguingly, mutant trm678 plants are fertile with normal organs that do not exhibit aberrant cell division patterns. The phragmoplast-orienting kinesin 1 (POK1) is a factor controlling the timing and efficiency of the cortical division zone (Lipka et al., 2014). In trm678 mutant cells, even though POK1 targeting to the cell cortex is altered in the absence of the PPB, POK1 still forms a cortical ring that corresponds with the division zone (Schaefer et al., 2017). These results suggest that the PPB may be less of a causal determinant of the cell division plane and more of a regulator to ensure the fidelity of a division plane defined by another mechanism. Regardless, the position of the cell division plane has profound impact on the shape of plant organs, and therefore, much remains to be discovered of how plane positioning is regulated.
The tomato SUN/AtIQD12 is a member of the IQ67 domain (IQD) protein family (Xiao et al., 2008; Huang et al., 2013). The IQ67 domain of IQD proteins is a conserved region of 67 amino acids and contains up to three regularly spaced IQ motifs which promote calmodulin (CaM) binding in the presence of Ca2+ (Rhoads and Friedberg, 1997; Abel et al., 2005, 2013). Ca2+ is a common secondary messenger in all eukaryotes and is used to regulate many cellular processes in response to both cellular and environmental stimuli, including cell division and shape (Cardenas, 2009; Steinhorst and Kudla, 2013; Burstenbinder et al., 2017b). The IQ67 domain of several IQD proteins interacts with CaM demonstrating that this family of proteins may serve as a large class of CaM binding proteins in plants (Burstenbinder et al., 2013, 2017b; Yang et al., 2018). The founding member of the IQD family, IQD1, localizes to microtubules with CaM2 in Arabidopsis and both IQD1 and IQD20 were found to interact with CaMs by Y2H, suggesting that IQDs may integrate Ca2+ sensing in regulation of the cytoskeleton (Levy et al., 2005; Burstenbinder et al., 2013).
Expression analyses in N. benthamiana showed that the N-terminus of most IQD proteins localizes to microtubules and that half of the IQDs localize to the plasma membrane (Burstenbinder et al., 2017b). There is also evidence that certain IQDs have differential subcellular localization dependent on the cell cycle stage (Wendrich et al., 2018). Cells in plants overexpressing AtIQD16 had altered orientation of cMTs with more oblique aligned microtubules and significantly elongated cells. Colocalization of IQDs with CaM also suggests that IQD proteins are capable of sequestering or recruiting CaM to specific subcellular domains. Subcellular localization of several Arabidopsis IQDs (IQD12, IQD22, IQD24, and IQD25) showed punctate structures that are reminiscent of regions within the plasma membrane, which may act as signaling centers in the cell. IQD14 is the rice ortholog to the Arabidopsis IQD15-18 subclade and loss of function alleles result in shorter and wider grains than wild type rice (Yang et al., 2018).
OsIQD14 was found to localize to the nucleus and cytoplasm and also in punctate locations along the microtubules, suggesting that the protein may function at specific points in microtubule regulation or on particular microtubule structures. Interestingly, expression of rice IQD14-GFP N- or C-terminal regions in N. benthamiana showed that the C-terminal region localized to the microtubules while the N-terminal region localized to the nucleus. This result is similar to the localization observed in the Arabidopsis IQD15-18 clade where the full length IQD protein was found on both microtubules and in the nucleus (Burstenbinder et al., 2017b; Yang et al., 2018). Rice IQD14 was also found to interact with Arabidopsis SPR2 by Y2H, and the orthologous Arabidopsis IQD15-18 subclade members were found to interact with SPR2 and CaM as well (Wendrich et al., 2018; Yang et al., 2018). SPR2 is a microtubule binding protein involved in protecting the minus end of microtubules and promoting severing and reorientation of the cMT arrays. SPR2 generally localizes to microtubules and does not distinguish dynamic from stable microtubules. However, IQD proteins may serve to direct the location of SPR2 function to specific regions to regulate reorganization the cytoskeleton in response to a certain signal (Buschmann et al., 2004; Shoji et al., 2004; Yao et al., 2008; Nakamura et al., 2018).
Auxin has been suggested to influence MT dynamics, but the mechanism is unclear. However, recent studies suggest that auxin-mediated cytoskeletal changes may involve IQD proteins (Chen et al., 2014; Wendrich et al., 2018) and IQDs are likely downstream targets of AUXIN RESPONSE FACTOR5/ MONOPTEROS (Boer et al., 2014; Moller et al., 2017). It has been proposed that IQDs in low auxin/ Ca2+ environments do not bind to CaM and instead bind to SPR2, inhibiting its function. This results in stabilized microtubules and a less dynamic cytoskeleton (Leong et al., 2018; Nakamura et al., 2018). Auxin leads to an increase in Ca2+, which promotes CaM binding to the IQD, and CaM binding then prevents IQDs from binding to SPR2. Unbound SPR2 can then bind the minus end of microtubules and promote microtubule branching and changes to cytoskeletal architecture in response to auxin (Wendrich et al., 2018). IQD proteins may function to resolve these signals and changes in Ca2+ levels within developing tissues in response to the environment, thereby directing cell elongation and expansion to ultimately drive organ shape.
As organ shape is controlled by both cell division and directed cell expansion, the regulation of microtubule dynamics is important in both of these processes to determine morphology. Some IQD proteins regulate cytoskeletal architecture by guiding the formation of ROP domains in the plasma membrane. ROPs are plant specific Rho GTPases with diverse functions (Yalovsky, 2015). One function of ROPs is in organizing the microtubule and actin cytoskeleton to determine a cell’s final shape. Some ROPs have been shown to promote aggregation of fine actin filaments and inhibit the assembly of organized microtubule arrays (ROP2 and ROP4), while ROP6 has been shown to have the opposite role and promotes accumulation of organized microtubule arrays (Fu et al., 2005, 2009; Ivakov and Persson, 2013). Furthermore, downstream of ROP6, the microtubule severing protein katanin is activated and promotes microtubule reorganization (Hamant, 2013; Lin et al., 2013). Formation of distinct subcellular domains of ROPs with these opposing functions can alter cytoskeleton composition and fine-tune the overall cell shape (Fu et al., 2002, 2005, 2009; Lin et al., 2013). The plasma membrane localization of Arabidopsis IQD13 has been shown to regulate ROP function in the xylem by promoting cMT growth and interaction with the membrane surface, thus restricting the formation of ROP11 domains (Oda and Fukuda, 2012; Sugiyama et al., 2017). In the presence of IQD13, the active ROP11 is restricted within narrow plasma membrane domains where it can recruit additional proteins to ultimately deplete the region of cMTs and form narrow pits in the secondary cell wall. In the absence of IQD13 or in the presence of truncated IQD13 lacking the plasma membrane associated domain, ROP11 forms circular domains that are independent of the cMTs and ultimately forms round pits in the secondary cell wall (Sugiyama et al., 2017). Secondary cell wall pit architecture is further refined by an interplay between the restriction of ROP11 to narrow domains by IQD13 and the impairment of ROP11 restriction and the resulting delineation of cMTs by CORTICAL MICROTUBULE DISORDERING1 protein (CORD) (Sasaki et al., 2017). AtIQD5 has recently been shown to regulate microtubule dynamics that affect cMT organization and subsequent cell shape formation in leaf pavement cells. IQD5 is enriched in lobed regions of pavement cells where cMTs are organized in parallel arrays and cell expansion is restricted (Liang et al., 2018; Mitra et al., 2018). In iqd5 mutant cells, regions lacking IQD5 expression no longer form lobes and have altered cellulose deposition (Mitra et al., 2018). In summary, these results suggest that IQDs may regulate microtubule organization in distinct subcellular regions through interactions with ROPs in order to impact a cells final shape and ultimately organ shape.
Since organ shape is also influenced by anisotropic cell expansion (Maugarny-Cales and Laufs, 2018), the regulation of this expansion is an important factor controlling morphology. While cell expansion is driven by isotropic turgor pressure, the direction of expansion is controlled by the pattern of cellulose microfibrils, which are generally deposited perpendicular to the axis of expansion. This pattern resists turgor driven expansion in the direction parallel with the cellulose microfibrils, thus promoting expansion in the perpendicular direction (Szymanski and Cosgrove, 2009; McFarlane et al., 2014). cMTs regulate the deposition pattern of cellulose microfibrils within the cell wall by interacting with cellulose synthase complexes in the plasma membrane (Paredez et al., 2006; McFarlane et al., 2014). Therefore, the organization of microtubules, in part controlled by IQDs also impacts the arrangement of cellulose microfibrils which will impact how the cell wall expands and the ultimate shape of the cell. While randomly aligned cMTs are located in the cytoplasm away from the plasma membrane, cMTs closely anchored to the plasma membrane are organized in parallel bundles to preferentially serve as tracks for cellulose synthase movement (Barton et al., 2008). These cMTs rarely display lateral displacement from their parallel organization due to their tight association with the plasma membrane (Shaw et al., 2003). Cellulose microfibrils are synthesized by large CSCs, composed of 18–36 cellulose synthase subunits and their accessory proteins (McFarlane et al., 2014). Cellulose synthases are assembled and matured in the Golgi and sorted by the Trans Golgi network into small cellulose synthase compartments/ microtubule-associated cellulose synthase compartment vesicles for its secretion to the plasma membrane (Crowell et al., 2009; Gutierrez et al., 2009; Sampathkumar et al., 2013; Zhang et al., 2016b). CSCs are tethered to cMTs through cellulose synthase interactive protein 1 (CSI1) (Gu et al., 2010; Bringmann et al., 2012; Li et al., 2012; Mei et al., 2012), which determines their trajectory along the cMTs (Paredez et al., 2006). Delivery to the plasma membrane is also mediated by CSI1 and fusion with the plasma membrane is mediated by the plant specific protein PATROL1 and the exocyst complex (Zhu et al., 2018). AtIQD1 interacts with KLCR1 in Y2H and in planta where its recruitment to microtubules is dependent on IQD1 (Abel et al., 2013; Burstenbinder et al., 2013). KLCR and cellulose-microtubule uncoupling (CMU) are in the same protein family (Burstenbinder et al., 2017a) and the same proteins in Arabidopsis where At4g10840 encodes KLCR1 and CMU1 and At3g27960 encodes KLCR2 and CMU2 (Burstenbinder et al., 2013; Liu et al., 2016). CMU1 and CMU2 are localized as static puncta along microtubules. Disruption of CMU function causes lateral microtubule displacement. This compromises microtubule-based guidance of CSCs leading to cell twisting and altered growth (Liu et al., 2016). Together these results indicate that IQD proteins regulate how microtubules direct CSCs and the pattern of cellulose microfibril deposition, which influences cell expansion and organ shape. It is intriguing that AtOFP4 also regulates cell wall formation through its interaction with KNAT7 (Li et al., 2011). Thus, both IQDs and OFPs may influence organ shape at the cellular level through their regulation of cell wall formation.
With respect to the regulation of organ shape by SUN/IQDs and the role in cell division patterning, these proteins may directly influence this process. AtIQD5 is localized to the PPB, spindle, and phragmoplast of dividing root cells (Liang et al., 2018). ROPs may also be involved in PPB formation (Oda, 2018) because two ROP GTPase activating proteins interact with POK1 and are required for accurate orientation of the PPB, phragmoplast, and cell plate (Stockle et al., 2016). POK1 is required for division plane maintenance (Lipka et al., 2014; Stockle et al., 2016) and its function is also influenced by TRMs (Schaefer et al., 2017). It is plausible that SUN/IQD and TRM proteins coordinate cell division planes, which contribute to organ shape.
A model to describe organ shape in the context of interactions of SUN/IQD, OFP and TRM, and associations with microtubules is shown in Figure 4. Assuming round shape as the default, the down regulation of OFP would lead to the association of more TRMs to the microtubules and hence elongated shape. This organ shape might also be attained via a similar mechanism when up-regulating TRM (Figure 4). Conversely, up-regulation of OFP and down regulation of TRM would result in rounder or even flat shapes due to less microtubular association of TRMs. SUN/IQD proteins are often found at the microtubules where their interaction with SPR2 and CaM might lead to altered cytoskeleton activities. Higher expression of SUN/IQD would lead to more association with the microtubules and hence elongated shape. Together with findings in Arabidopsis and crop plants, further information has shown that OFPs, TRMs and SUN/IQDs impact microtubular activities, offering mechanistic insights into how the different shapes of plant organs are realized.
ML and EvdK wrote and edited the review with significant contributions and further edits from SW, AS, and YW.
This research was supported by National Science Foundation IOS 0922661, NSF IOS 1564366, and AFRI 2017-67013-26199 of the USDA National Institute of Food and Agriculture.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
CAP350, centrosome associated protein 350; cMT, cortical microtubules; CMU, cellulose microtubule uncoupling; CSC, cellulose synthase complex; FOP, fibroblast growth factor receptor 1 oncogene partner; IQD, isoleucine glutamine domain; KLCR, kinesin light chain related; MTOC, microtubule organizing center; NIL, nearly isogenic line; OFD1, orofaciodigital syndrome 1; OFP, ovate family protein; POK, phragmoplast-orienting kinesin; PPB, preprophase band; ROP, rho-GTPase of plants; SPR, spiral; TRM, tonneau1 recruiting motif; TTP, tonneau1-TRM-phosphatase 2C; γ-TuRC, tubulin ring complex.
Abel, S., Burstenbinder, K., and Muller, J. (2013). The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal. Behav. 8:e24369. doi: 10.4161/psb.24369
Abel, S., Savchenko, T., and Levy, M. (2005). Genome-wide comparative analysis of the IQD gene families in Arabidopsis thaliana and Oryza sativa. BMC Evol. Biol. 5:72. doi: 10.1186/1471-2148-5-72
Azimzadeh, J., Nacry, P., Christodoulidou, A., Drevensek, S., Camilleri, C., Amiour, N., et al. (2008). Arabidopsis TONNEAU1 proteins are essential for preprophase band formation and interact with centrin. Plant Cell 20, 2146–2159. doi: 10.1105/tpc.107.056812
Barton, D. A., Vantard, M., and Overall, R. L. (2008). Analysis of cortical arrays from Tradescantia virginiana at high resolution reveals discrete microtubule subpopulations and demonstrates that confocal images of arrays can be misleading. Plant Cell 20, 982–994. doi: 10.1105/tpc.108.058503
Boer, D. R., Freire-Rios, A., van den Berg, W. A., Saaki, M. T., Manfield, I. W., Kepinski, S., et al. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. doi: 10.1016/j.cell.2013.12.027
Brewer, M. T., Lang, L., Fujimura, K., Dujmovic, N., Gray, S., and van der Knaap, E. (2006). Development of a controlled vocabulary and software application to analyze fruit shape variation in tomato and other plant species. Plant Physiol. 141, 15–25. doi: 10.1104/pp.106.077867
Bringmann, M., Landrein, B., Schudoma, C., Hamant, O., Hauser, M.-T., Persson, S. (2012). Cracking the elusive alignment hypothesis: the microtubule–cellulose synthase nexus unraveled. Trends Plant Sci. 17, 666–674. doi: 10.1016/j.tplants.2012.06.003
Burstenbinder, K., Mitra, D., and Quegwer, J. (2017a). Functions of IQD proteins as hubs in cellular calcium and auxin signaling: a toolbox for shape formation and tissue-specification in plants? Plant Signal. Behav. 12:e1331198. doi: 10.1080/15592324.2017.1331198
Burstenbinder, K., Moller, B., Plotner, R., Stamm, G., Hause, G., Mitra, D., et al. (2017b). The IQD family of calmodulin-binding proteins links calcium signaling to microtubules, membrane subdomains, and the nucleus. Plant Physiol. 173, 1692–1708. doi: 10.1104/pp.16.01743
Burstenbinder, K., Savchenko, T., Muller, J., Adamson, A. W., Stamm, G., Kwong, R., et al. (2013). Arabidopsis calmodulin-binding protein IQ67-domain 1 localizes to microtubules and interacts with kinesin light chain-related protein-1. J. Biol. Chem. 288, 1871–1882. doi: 10.1074/jbc.M112.396200
Buschmann, H., Fabri, C. O., Hauptmann, M., Hutzler, P., Laux, T., Lloyd, C. W., et al. (2004). Helical growth of the Arabidopsis mutant tortifolia1 reveals a plant-specific microtubule-associated protein. Curr. Biol. 14, 1515–1521. doi: 10.1016/j.cub.2004.08.033
Cabrero, J., Bakkali, M., Navarro-Domínguez, B., Ruíz-Ruano, F. J., Martín-Blázquez, R., López-León, M. D., et al. (2013). The Ku70 DNA-repair protein is involved in centromere function in a grasshopper species. Chromosome Res. 21, 393–406. doi: 10.1007/s10577-013-9367-7
Calingacion, M., Laborte, A., Nelson, A., Resurreccion, A., Concepcion, J. C., Daygon, V. D., et al. (2014). Diversity of global rice markets and the science required for consumer-targeted rice breeding. PLoS One 9:e85106. doi: 10.1371/journal.pone.0085106
Camilleri, C., Azimzadeh, J., Pastuglia, M., Bellini, C., Grandjean, O., and Bouchez, D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14, 833–845. doi: 10.1105/tpc.010402
Cardenas, L. (2009). New findings in the mechanisms regulating polar growth in root hair cells. Plant Signal. Behav. 4, 4–8. doi: 10.4161/psb.4.1.7341
Chan, J., Sambade, A., Calder, G., and Lloyd, C. (2009). Arabidopsis cortical microtubules are initiated along, as well as branching from, existing microtubules. Plant Cell 21, 2298–2306. doi: 10.1105/tpc.109.069716
Chen, X., Grandont, L., Li, H., Hauschild, R., Paque, S., Abuzeineh, A., et al. (2014). Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516, 90–93. doi: 10.1038/nature13889
Costa, S. (2017). Are division plane determination and cell-cycle progression coordinated? New Phytol. 213, 16–21. doi: 10.1111/nph.14261
Crowell, E. F., Bischoff, V., Desprez, T., Rolland, A., Stierhof, Y. D., Schumacher, K., et al. (2009). Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21, 1141–1154. doi: 10.1105/tpc.108.065334
Daunay, M. C., Laterrot, H., and Janick, J. (2008). Iconography and history of solanaceae: antiquity to the 17th century. Horticult. Rev. 34, 1–111.
De Haan, S. (2009). Potato Diversity at Height: Multiple Dimensions of Farmer-Driven In-Situ Conservation in the Andes. PhD, Wageningen University, Wageningen.
Dou, J., Zhao, S., Lu, X., He, N., Zhang, L., Ali, A., et al. (2018). Genetic mapping reveals a candidate gene (ClFS1) for fruit shape in watermelon (Citrullus lanatus L.). Theor. Appl. Genet. 131, 947–958. doi: 10.1007/s00122-018-3050-5
Drevensek, S., Goussot, M., Duroc, Y., Christodoulidou, A., Steyaert, S., Schaefer, E., et al. (2012). The Arabidopsis TRM1-TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell 24, 178–191. doi: 10.1105/tpc.111.089748
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Fishel, E. A., and Dixit, R. (2013). Role of nucleation in cortical microtubule array organization: variations on a theme. Plant J. 75, 270–277. doi: 10.1111/tpj.12166
Fu, Y., Gu, Y., Zheng, Z., Wasteneys, G., and Yang, Z. (2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687–700. doi: 10.1016/j.cell.2004.12.026
Fu, Y., Li, H., and Yang, Z. (2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14, 777–794. doi: 10.1105/tpc.001537
Fu, Y., Xu, T., Zhu, L., Wen, M., and Yang, Z. (2009). A ROP GTPase signaling pathway controls cortical microtubule ordering and cell expansion in Arabidopsis. Curr. Biol. 19, 1827–1832. doi: 10.1016/j.cub.2009.08.052
Furutani, I., Watanabe, Y., Prieto, R., Masukawa, M., Suzuki, K., Naoi, K., et al. (2000). The SPIRAL genes are required for directional control of cell elongation in Aarabidopsis thaliana. Development 127, 4443–4453.
Gu, Y., Kaplinsky, N., Bringmann, M., Cobb, A., Carroll, A., Sampathkumar, A., et al. (2010). Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 12866–12871. doi: 10.1073/pnas.1007092107
Gutierrez, R., Lindeboom, J. J., Paredez, A. R., Emons, A. M., and Ehrhardt, D. W. (2009). Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat. Cell Biol. 11, 797–806. doi: 10.1038/ncb1886
Hackbusch, J., Richter, K., Muller, J., Salamini, F., and Uhrig, J. F. (2005). A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 4908–4912. doi: 10.1073/pnas.0501181102
Hamant, O. (2013). Integrative cell biology: katanin at the crossroads. Curr. Biol. 23, R206–R208. doi: 10.1016/j.cub.2013.01.031
Hamant, O., Heisler, M. G., Jonsson, H., Krupinski, P., Uyttewaal, M., Bokov, P., et al. (2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322, 1650–1655. doi: 10.1126/science.1165594
Harberd, N. P. (2015). Shaping taste: the molecular discovery of rice genes improving grain size, shape and quality. J. Genet. Genomics 42, 597–599. doi: 10.1016/j.jgg.2015.09.008
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Huang, Z., Van Houten, J., Gonzalez, G., Xiao, H., and van der Knaap, E. (2013). Genome-wide identification, phylogeny and expression analysis of SUN, OFP and YABBY gene family in tomato. Mol. Genet. Genomics 288, 111–129. doi: 10.1007/s00438-013-0733-0
Ivakov, A., and Persson, S. (2013). Plant cell shape: modulators and measurements. Front. Plant Sci. 4:439. doi: 10.3389/fpls.2013.00439
Jenik, P. D., and Irish, V. F. (2000). Regulation of cell proliferation patterns by homeotic genes during Arabidopsis floral development. Development 127, 1267–1276.
Kirik, A., Ehrhardt, D. W., and Kirik, V. (2012). TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24, 1158–1170. doi: 10.1105/tpc.111.094367
Ku, H. M., Doganlar, S., Chen, K. Y., and Tanksley, S. D. (1999). The genetic basis of pear-shaped tomato fruit. Theor. Appl. Genet. 99, 844–850. doi: 10.1007/s001220051304
Lee, Y. K., Kim, G. T., Kim, I. J., Park, J., Kwak, S. S., Choi, G., et al. (2006). LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133, 4305–4314. doi: 10.1242/dev.02604
Leong, S. Y., Yamada, M., Yanagisawa, N., and Goshima, G. (2018). SPIRAL2 stabilises endoplasmic microtubule minus ends in the moss physcomitrella patens. Cell Struct. Funct. 43, 53–60. doi: 10.1247/csf.18001
Levy, M., Wang, Q., Kaspi, R., Parrella, M. P., and Abel, S. (2005). Arabidopsis IQD1, a novel calmodulin-binding nuclear protein, stimulates glucosinolate accumulation and plant defense. Plant J. 43, 79–96. doi: 10.1111/j.1365-313X.2005.02435.x
Li, E., Wang, S., Liu, Y., Chen, J. G., and Douglas, C. J. (2011). OVATE FAMILY PROTEIN4 (OFP4) interaction with KNAT7 regulates secondary cell wall formation in Arabidopsis thaliana. Plant J. 67, 328–341. doi: 10.1111/j.1365-313X.2011.04595.x
Li, S., Lei, L., Somerville, C. R., and Gu, Y. (2012). Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc. Natl. Acad. Sci. U.S.A. 109, 185–190. doi: 10.1073/pnas.1118560109
Liang, H., Zhang, Y., Martinez, P., Rasmussen, C. G., Xu, T., and Yang, Z. (2018). The microtubule-associated protein IQ67 DOMAIN5 modulates microtubule dynamics and pavement cell shape. Plant Physiol. 177, 1555–1568. doi: 10.1104/pp.18.00558
Lin, D., Cao, L., Zhou, Z., Zhu, L., Ehrhardt, D., Yang, Z., et al. (2013). Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr. Biol. 23, 290–297. doi: 10.1016/j.cub.2013.01.022
Lipka, E., Gadeyne, A., Stockle, D., Zimmermann, S., De Jaeger, G., Ehrhardt, D. W., et al. (2014). The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell 26, 2617–2632. doi: 10.1105/tpc.114.124933
Liu, J., Cong, B., and Tanksley, S. D. (2003). Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiol. 132, 292–299. doi: 10.1104/pp.102.018143
Liu, Z., Schneider, R., Kesten, C., Zhang, Y., Somssich, M., Zhang, Y., et al. (2016). Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev. Cell 38, 305–315. doi: 10.1016/j.devcel.2016.06.032
Maugarny-Cales, A., and Laufs, P. (2018). Getting leaves into shape: a molecular, cellular, environmental and evolutionary view. Development 145:dev161646. doi: 10.1242/dev.161646
McFarlane, H. E., Doring, A., and Persson, S. (2014). The cell biology of cellulose synthesis. Annu. Rev. Plant Biol. 65, 69–94. doi: 10.1146/annurev-arplant-050213-040240
Mei, Y., Gao, H. B., Yuan, M., and Xue, H. W. (2012). The Arabidopsis ARCP protein, CSI1, which is required for microtubule stability, is necessary for root and anther development. Plant Cell 24, 1066–1080. doi: 10.1105/tpc.111.095059
Meindl, U. (1983). Cytoskeletal control of nuclear migration and anchoring in developing cells of Micrasterias denticulata and the change caused by the anti-microtubular herbicide amiprophos-methyl (APM). Protoplasma 118, 75–90. doi: 10.1007/bf01284749
Meyerowitz, E. M. (1997). Genetic control of cell division patterns in developing plants. Cell 88, 299–308. doi: 10.1016/S0092-8674(00)81868-1
Mineyuki, Y., and Furuya, M. (1985). Involvement of microtubules on nuclear positioning during apical growth in Adiantum protonemata. Plant Cell Physiol. 26, 627–634. doi: 10.1093/oxfordjournals.pcp.a076951
Mitra, D., Kumari, P., Quegwer, J., Klemm, S., Moeller, B., Poeschl, Y., et al. (2018). Microtubule-associated protein IQ67 DOMAIN5 regulates interdigitation of leaf pavement cells in Arabidopsis thaliana. bioRxiv [Preprint]. doi: 10.1101/268466
Moller, B. K., Ten Hove, C. A., Xiang, D., Williams, N., Lopez, L. G., Yoshida, S., et al. (2017). Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl. Acad. Sci. U.S.A. 114, E2533–E2539. doi: 10.1073/pnas.1616493114
Monforte, A. J., Diaz, A. I., Cano-Delgado, A., and van der Knaap, E. (2014). The genetic basis of fruit morphology in horticultural crops: lessons from tomato and melon. J. Exp. Bot. 65, 4625–4637. doi: 10.1093/jxb/eru017
Murata, T., and Hasebe, M. (2011). Microtubule Nucleation and Organization in Plant Cells. The Plant Cytoskeleton. B. Liu. New York, NY: Springer New York, 81–94.
Nakamura, M. D., Ehrhardt, W., and Hashimoto, T. (2010). Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12, 1064–1070. doi: 10.1038/ncb2110
Nakamura, M., Lindeboom, J. J., Saltini, M., Mulder, B. M., and Ehrhardt, D. W. (2018). SPR2 protects minus ends to promote severing and reorientation of plant cortical microtubule arrays. J. Cell Biol. 217, 915–927. doi: 10.1083/jcb.201708130
Oda, Y. (2018). Emerging roles of cortical microtubule-membrane interactions. J. Plant Res. 131, 5–14. doi: 10.1007/s10265-017-0995-4
Oda, Y., and Fukuda, H. (2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337, 1333–1336. doi: 10.1126/science.1222597
Oda, Y., Mimura, T., and Hasezawa, S. (2005). Regulation of secondary cell wall development by cortical microtubules during tracheary element differentiation in Arabidopsis cell suspensions. Plant Physiol. 137, 1027–1036. doi: 10.1104/pp.104.052613
Pagnussat, G. C., Yu, H. J., and Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592. doi: 10.1105/tpc.107.054890
Pan, Y., Liang, X., Gao, M., Liu, H., Meng, H., Weng, Y., et al. (2017). Round fruit shape in WI7239 cucumber is controlled by two interacting quantitative trait loci with one putatively encoding a tomato SUN homolog. Theor. Appl. Genet. 130, 573–586. doi: 10.1007/s00122-016-2836-6
Paredez, A. R., Somerville, C. R., and Ehrhardt, D. W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312, 1491–1495. doi: 10.1126/science.1126551
Pickersgill, B. (2007). Domestication of plants in the Americas: insights from mendelian and molecular genetics. Ann. Bot. 100, 925–940. doi: 10.1093/aob/mcm193
Rasmussen, C. G., and Bellinger, M. (2018). An overview of plant division-plane orientation. New Phytol. 219, 505–512. doi: 10.1111/nph.15183
Rasmussen, C. G., Humphries, J. A., and Smith, L. G. (2011). Determination of symmetric and asymmetric division planes in plant cells. Annu. Rev. Plant Biol. 62, 387–409. doi: 10.1146/annurev-arplant-042110-103802
Rhoads, A. R., and Friedberg, F. (1997). Sequence motifs for calmodulin recognition. FASEB J. 11, 331–340. doi: 10.1096/fasebj.11.5.9141499
Rodriguez, G. R., Francis, D., van der Knaap, E., Strecker, J., Njanji, I., Thomas, J., et al. (2011a). “New features and many improvements to analyze morphology and color of digitalized plant organs are available in tomato analyzer 3.0,” in Proceedings of the 22nd Midwest Artificial Intelligence and Cognitive Science Conference 2011, Cincinnati, Ohio.
Rodriguez, G. R., Munos, S., Anderson, C., Sim, S. C., Michel, A., Causse, M., et al. (2011b). Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156, 275–285. doi: 10.1104/pp.110.167577
Rodriguez, G. R., Kim, H. J., and van der Knaap, E. (2013). Mapping of two suppressors of OVATE (sov) loci in tomato. Heredity 111, 256–264. doi: 10.1038/hdy.2013.45
Sampathkumar, A., Gutierrez, R., McFarlane, H. E., Bringmann, M., Lindeboom, J., Emons, A. M., et al. (2013). Patterning and lifetime of plasma membrane-localized cellulose synthase is dependent on actin organization in Arabidopsis interphase cells. Plant Physiol. 162, 675–688. doi: 10.1104/pp.113.215277
Sasaki, T., Fukuda, H., and Oda, Y. (2017). Cortical microtubule disordering1 is required for secondary cell wall patterning in xylem vessels. Plant Cell 29, 3123–3139. doi: 10.1105/tpc.17.00663
Schaefer, E., Belcram, K., Uyttewaal, M., Duroc, Y., Goussot, M., Legland, D., et al. (2017). The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356, 186–189. doi: 10.1126/science.aal3016
Shaw, S. L., Kamyar, R., and Ehrhardt, D. W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300, 1715–1718. doi: 10.1126/science.1083529
Shoji, T., Narita, N. N., Hayashi, K., Asada, J., Hamada, T., Sonobe, S., et al. (2004). Plant-specific microtubule-associated protein SPIRAL2 is required for anisotropic growth in Arabidopsis. Plant Physiol. 136, 3933–3944. doi: 10.1104/pp.104.051748
Singla, V., Romaguera-Ros, M., Garcia-Verdugo, J. M., and Reiter, J. F. (2010). Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell 18, 410–424. doi: 10.1016/j.devcel.2009.12.022
Spinner, L., Gadeyne, A., Belcram, K., Goussot, M., Moison, M., Duroc, Y., et al. (2013). A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 4:1863. doi: 10.1038/ncomms2831
Spinner, L., Pastuglia, M., Belcram, K., Pegoraro, M., Goussot, M., Bouchez, D., et al. (2010). The function of TONNEAU1 in moss reveals ancient mechanisms of division plane specification and cell elongation in land plants. Development 137, 2733–2742. doi: 10.1242/dev.043810
Steinhorst, L., and Kudla, J. (2013). Calcium – A central regulator of pollen germination and tube growth. Biochim. Biophys. Acta 1833, 1573–1581. doi: 10.1016/j.bbamcr.2012.10.009
Stockle, D., Herrmann, A., Lipka, E., Lauster, T., Gavidia, R., Zimmermann, S., et al. (2016). Putative RopGAPs impact division plane selection and interact with kinesin-12 POK1. Nat. Plants 2:16120. doi: 10.1038/nplants.2016.120
Sugiyama, Y., Wakazaki, M., Toyooka, K., Fukuda, H., and Oda, Y. (2017). A novel plasma membrane-anchored protein regulates xylem cell-wall deposition through microtubule-dependent lateral inhibition of Rho GTPase domains. Curr. Biol. 27:e2524. doi: 10.1016/j.cub.2017.06.059
Szymanski, D. B., and Cosgrove, D. J. (2009). Dynamic coordination of cytoskeletal and cell wall systems during plant cell morphogenesis. Curr. Biol. 19, R800–R811. doi: 10.1016/j.cub.2009.07.056
Uyttewaal, M., Traas, J., and Hamant, O. (2010). Integrating physical stress, growth, and development. Curr. Opin. Plant Biol. 13, 46–52. doi: 10.1016/j.pbi.2009.10.004
Van Damme, D., Vanstraelen, M., and Geelen, D. (2007). Cortical division zone establishment in plant cells. Trends Plant Sci. 12, 458–464. doi: 10.1016/j.tplants.2007.08.011
Van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5:227. doi: 10.3389/fpls.2014.00227
van der Knaap, E., and Ostergaard, L. (2017). Shaping a fruit: developmental pathways that impact growth patterns. Semin. Cell Dev. Biol. 79, 27–36. doi: 10.1016/j.semcdb.2017.10.028
Van der Knaap, E., and Tanksley, S. D. (2001). Identification and characterization of a novel locus controlling early fruit development in tomato. Theor. Appl. Genet. 103, 353–358. doi: 10.1007/s001220100623
Vogelmann, T. C., Bassel, A. R., and Miller, J. H. (1981). Effects of microtubule-inhibitors on nuclear migration and rhizoid differentiation in germinating fern spores (Onoclea sensibilis). Protoplasma 109, 295–316. doi: 10.1007/bf01287449
von Wangenheim, D., Fangerau, J., Schmitz, A., Smith, R. S., Leitte, H., Stelzer, E. H., et al. (2016). Rules and self-organizing properties of post-embryonic plant organ cell division patterns. Curr. Biol. 26, 439–449. doi: 10.1016/j.cub.2015.12.047
Wang, S., Chang, Y., Guo, J., and Chen, J. G. (2007). Arabidopsis ovate family protein 1 is a transcriptional repressor that suppresses cell elongation. Plant J. 50, 858–872. doi: 10.1111/j.1365-313X.2007.03096.x
Wang, S., Chang, Y., Guo, J., Zeng, Q., Ellis, B. E., and Chen, J. G. (2011). Arabidopsis ovate family proteins, a novel transcriptional repressor family, control multiple aspects of plant growth and development. PLoS One 6:23. doi: 10.1371/journal.pone.0023896
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, Y., Xiong, G., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Wang, Y. K., Chang, W. C., Liu, P. F., Hsiao, M. K., Lin, C. T., Lin, S. M., et al. (2010). Ovate family protein 1 as a plant Ku70 interacting protein involving in DNA double-strand break repair. Plant Mol. Biol. 74, 453–466. doi: 10.1007/s11103-010-9685-5
Wendrich, J., Yang, B.-J., Mijnhout, P., Xue, H.-W., De Rybel, B., and Weijers, D. (2018). IQD proteins integrate auxin and calcium signaling to regulate microtubule dynamics during Arabidopsis development. bioRxiv [Preprint]. doi: 10.1101/275560
Wright, A. J., Gallagher, K., and Smith, L. G. (2009). Discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21, 234–247. doi: 10.1105/tpc.108.062810
Wu, J., and Akhmanova, A. (2017). Microtubule-organizing centers. Annu. Rev. Cell Dev. Biol. 33, 51–75. doi: 10.1146/annurev-cellbio-100616-060615
Wu, S. (2015). The Roles of OVATE and Other Elongation Genes in Regulating Proximal-Distal Patterning of Tomato Fruit. PhD, The Ohio State University, Columbus, OH.
Wu, S., Clevenger, J. P., Sun, L., Visa, S., Kamiya, Y., Jikumaru, Y., et al. (2015). The control of tomato fruit elongation orchestrated by sun, ovate and fs8.1 in a wild relative of tomato. Plant Sci. 238, 95–104. doi: 10.1016/j.plantsci.2015.05.019
Wu, S., Xiao, H., Cabrera, A., Meulia, T., and van der Knaap, E. (2011). SUN regulates vegetative and reproductive organ shape by changing cell division patterns. Plant Physiol. 157, 1175–1186. doi: 10.1104/pp.111.181065
Wu, S., Zhang, B., Keyhaninejad, N., Rodriguez, G. R., Kim, H. J., Chakrabarti, M., et al. (2018). A common genetic mechanism underlies morphological diversity in plants. Nat. Commun. 9–4734. doi: 10.1038/s41467-018-07216-8
Xiao, H., Jiang, N., Schaffner, E. K., Stockinger, E. J., and Van der Knaap, E. (2008). A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science 319, 1527–1530. doi: 10.1126/science.1153040
Xiao, H., Radovich, C., Welty, N., Hsu, J., Li, D., Meulia, T., et al. (2009). Integration of tomato reproductive developmental landmarks and expression profiles, and the effect of SUN on fruit shape. BMC Plant Biol. 9:49. doi: 10.1186/1471-2229-9-49
Yalovsky, S. (2015). Protein lipid modifications and the regulation of ROP GTPase function. J. Exp. Bot. 66, 1617–1624. doi: 10.1093/jxb/erv057
Yan, X., Habedanck, R., and Nigg, E. A. (2006). A complex of two centrosomal proteins, CAP350 and FOP, cooperates with EB1 in microtubule anchoring. Mol. Biol. Cell 17, 634–644. doi: 10.1091/mbc.e05-08-0810
Yang, B.-J., Wendrich, J. R., De Rybel, B., Weijers D., and Xue, H.-W. (2018). OsIQD14 regulates rice grain shape through modulating the microtubule cytoskeleton. bioRxiv [Preprint]. doi: 10.1101/275552
Yao, M., Wakamatsu, Y., Itoh, T. J., Shoji, T., and Hashimoto, T. (2008). Arabidopsis SPIRAL2 promotes uninterrupted microtubule growth by suppressing the pause state of microtubule dynamics. J. Cell Sci. 121, 2372–2381. doi: 10.1242/jcs.030221
Zhang, Y., Iakovidis, M., and Costa, S. (2016a). Control of patterns of symmetric cell division in the epidermal and cortical tissues of the Arabidopsis root. Development 143, 978–982. doi: 10.1242/dev.129502
Zhang, Y., Nikolovski, N., Sorieul, M., Vellosillo, T., McFarlane, H. E., Dupree, R., et al. (2016b). Golgi-localized STELLO proteins regulate the assembly and trafficking of cellulose synthase complexes in Arabidopsis. Nat. Commun. 7:11656. doi: 10.1038/ncomms11656
Zheng, J., Zhang, Y., and Wang, C. (2015). Molecular functions of genes related to grain shape in rice. Breed Sci. 65, 120–126. doi: 10.1270/jsbbs.65.120
Zhou, Y., Miao, J., et al. (2015). Natural variations in SLG7 regulate grain shape in rice. Genetics 201, 1591–1599. doi: 10.1534/genetics.115.181115
Zhu, X., Li, S., Pan, S., Xin, X., and Gu, Y. (2018). CSI1, PATROL1, and exocyst complex cooperate in delivery of cellulose synthase complexes to the plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 115, E3578–E3587. doi: 10.1073/pnas.1800182115
Keywords: OFP, TRM, SUN, IQD, microtubules, organ shape
Citation: Lazzaro MD, Wu S, Snouffer A, Wang Y and van der Knaap E (2018) Plant Organ Shapes Are Regulated by Protein Interactions and Associations With Microtubules. Front. Plant Sci. 9:1766. doi: 10.3389/fpls.2018.01766
Received: 05 August 2018; Accepted: 14 November 2018;
Published: 04 December 2018.
Edited by:
Han Xiao, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Li Zhengguo, Chongqing University, ChinaCopyright © 2018 Lazzaro, Wu, Snouffer, Wang and van der Knaap. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Esther van der Knaap, dmFuZGVya25AdWdhLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.