
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci., 12 November 2018
Sec. Plant Abiotic Stress
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01618
The Arabidopsis genome annotation include 11 glyoxalase I (GLXI) genes, all encoding for protein members of the vicinal oxygen chelate (VOC) superfamily. The biochemical properties and physiological importance of three Arabidopsis GLXI proteins in the detoxification of reactive carbonyl species has been recently described. Analyses of phylogenetic relationships and conserved GLXI binding sites indicate that the other eight GLXI genes (GLXI-like) do not encode for proteins with GLXI activity. In this perspective article we analyse the structural features of GLXI and GLXI-like proteins, and explore splice forms and transcript abundance under abiotic stress conditions. Finally, we discuss future directions of research on this topic with respect to the substrate identification of GLXI and GLXI-like proteins and the need of reliable quantitative measurements of reactive carbonyl species in plant tissues.
The glyoxalase (GLX) system was biochemically characterized almost 70 years ago Racker (1951) and is one of the most important lines of defense against glycation in most organisms (Thornalley, 2003; Sousa Silva et al., 2013). The GLX system is a two-step scavenging pathway comprising two phylogenetically unrelated enzymes ultimately detoxifying the reactive dicarbonyl species methylglyoxal (MGO). In a preceding step, MGO is scavenged by reduced glutathione (GSH) forming a hemithioacetal that is the actual substrate for the first reaction catalyzed by GLXI resulting in S-D-lactoylglutathione. In a second step, S-D-lactoylglutathione is converted into D-lactate by the action of glyoxalase II (GLXII), thereby releasing GSH. The action of the GLX system prevents the reaction of free MGO with DNA, lipids, and proteins. MGO reacts preferentially with arginine or lysine residues and any protein with these residues will be prone to glycation. The modified molecules, which are hampered in their functionality are generally called advanced glycation end-products (Sousa Silva et al., 2013).
In Arabidopsis three gene loci encode for active GLXI (Kaur et al., 2013; Jain et al., 2016; Schmitz et al., 2017). These enzymes belong to the group of VOC family proteins, all featuring bidentate coordination of a substrate to a divalent metal center through vicinal oxygen atoms as a common trait (He and Moran, 2011). GLXI use either Ni2+/Mn2+ or Zn2+ for its catalytic activity. Apart from the already characterized GLXI, the remaining eight Arabidopsis proteins fall into the category of GLXI proteins (GLXI-like), due to related structural features. This structural feature, the VOC fold, is determined by two tandem βαβββ modules assigned as single VOC domain (cd06587) forming an incompletely closed, 8-stranded β-barrel containing the catalytic center (He and Moran, 2011). Intriguingly, GLXI and some other VOC family proteins like the bleomycin resistance protein assemble the VOC fold from two monomers exchanging part of their structure to form an intertwined homodimer by a process called domain swapping (Dumas et al., 1994; Bennett and Eisenberg, 2004).
Recently, we functionally characterized the predominant GLXI isoform involved in MGO detoxification in Arabidopsis, the Zn2+-dependent GLXI;3, as well as the Ni2+-dependent GLXI;1 and GLXI;2 (Schmitz et al., 2017). All three GLXI isoforms convert MGO and glyoxal using GSH. Analyses of Arabidopsis loss-of-function lines revealed that elimination of toxic reactive carbonyl species during germination and seedling establishment highly depends on the activity of the cytosolic GLXI;3 isoform (Schmitz et al., 2017).
So far, GLXI-like proteins have not been characterized at the molecular level. The ectopic expression of a GLXI-like protein ortholog from Xerophyta humilis (DSI;1; desiccation induced 1) in Escherichia coli conferred low level of MGO tolerance, leading to the conclusion that DSI;1 homologs are unlikely to have GLXI activity (Mulako et al., 2008). The substrate specificities of all GLXI-like proteins of the VOC superfamily in plants remain unknown. A simple protein blast search in the reference protein database shows that GLXI-like proteins from Arabidopsis have no significant hits (e < 10E-13) in the Animalia, Fungi or Archaean group, but are broadly distributed and diverged in Bacteria and Viridiplantae.
Nine of the 11 Arabidopsis genes of the VOC family encode for single VOC domain proteins, GLXI;3 and all GLXI-like proteins, with molecular sizes of 13–22 kDa. In contrast, the two Ni2+-dependent GLXI;1 and GLXI;2 are two-domain (domain A and B) VOC proteins of 33 kDa. While these two-domain Ni2+-dependent GLXI fold and function as a monomer, the one-domain GLXI;3 and GLXI-like proteins are likely to assemble as homodimers to reconstitute the VOC fold by domain swapping (He and Moran, 2011; Turra et al., 2015). Multiple sequence alignments (MSA) as well as comparison of conserved GLXI binding sites revealed essential differences in amino acid (aa) composition among the 11 VOC superfamily members. A MAFFT MSA indicated several indels in either the GLXI or GLXI-like proteins, where the closest GLXI homolog DSI;1 shares only 25% identical aa positions with the Zn2+-dependent GLXI;3 in relation to the alignment length (Katoh et al., 2005; Sela et al., 2015). All other GLXI-like proteins share 17–21% identity with the predominant Zn2+-dependent GLXI;3 indicating a distant relationship of the VOC family members. As the MAFFT algorithm failed to align all βαβββ modules correctly, we tested various alignment programs and found that PROMALS3D (Pei et al., 2008a) performed best in detecting and aligning the secondary structure features that are responsible for the VOC fold (Figure 1A). Based on this MSA, Arabidopsis GLXI and GLXI-like proteins form three distinct clades in a phylogenetic analysis (Figure 1B). Clade I is composed of proteins with proven GLXI activity (Schmitz et al., 2017). Clade II, and Clade III are composed of GLX-like protein for which no experimental evidence of their biological function exist and no close homologs from bacteria have been characterized to date.
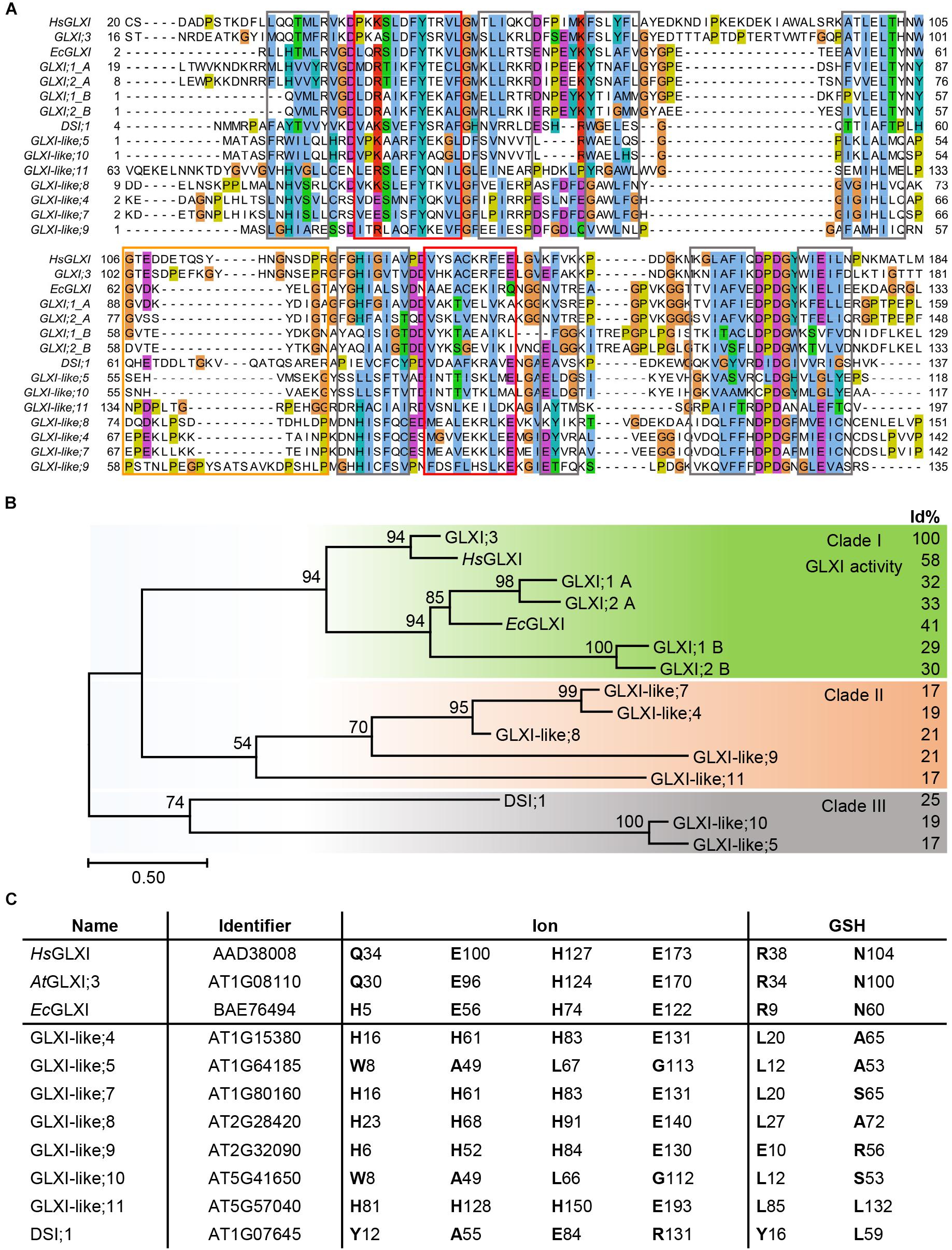
FIGURE 1. Molecular phylogenetic analysis of Arabidopsis GLXI and GLXI-like proteins. (A) Aligned amino acid sequences of GLXI- and GLXI-like proteins. PROMASL3D alignment was constructed with default settings and displayed with Jalview (Pei et al., 2008b; Waterhouse et al., 2009); non-conserved N and C-terminal parts are partially masked. highlighted: gray = β-sheet, red = α-helix, and orange = hinge region. (B) Molecular phylogenetic analysis. The evolutionary history was inferred by using the Maximum Likelihood method based on the best fitting Le_Gascuel_2008 model (Le and Gascuel, 2008). The percentage of trees in which the associated taxa clustered together is shown next to the branches based on 1000 bootstrap replicates. A discrete Gamma distribution was used to model evolutionary rate differences among sites [5 categories (+G, parameter = 3.4527)]. The rate variation model allowed for some sites to be evolutionarily invariable [(+I), 1.49% sites]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (Scale bar). The analysis involved 15 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 101 positions in the final dataset. Evolutionaryanalyses were conducted in MEGA7 (Kumar et al., 2016). Percent of identical amino acid positions relative to Arabidopsis GLXI;3 (AtGLXI;3) in relation to the length of a pairwise alignment are displayed to the right (Id%). (C) Conserved amino acid positions for GLXI function. Amino acid positions involved in ion or glutathione binding were interfered from the structure-based sequence alignment of Human GLXI (HsGLXI), Escherichia coli (EcGLXI), AtGLXI;3, and compared to the Arabidopsis GLXI-like proteins.
Several studies have identified the aa positions responsible for either metal ion or substrate binding and hence for the catalytic activity of the GLXI homologs. In Zn2+-dependent GLXI the metal ion binding center within a β-barrel is formed by four essential aa, a glutamine, two glutamic acids, and a histidine (in Human GLXI: Q34, E100, H127, and E173) (Cameron et al., 1997). In Ni2+-dependent GLXI the glutamine is exchanged for a histidine (in E. coli GLXI: H5, E56, H74, and E122) (He et al., 2000). Even though not the aa but rather an α-structural component determines GLXI metal selectivity (Clugston et al., 2004; Suttisansanee et al., 2015), mutation studies on the ion binding aa clearly confirmed their importance for the catalytic activity (Ridderstrom et al., 1998; Frickel et al., 2001). The aa of the ion binding site are strictly conserved among active Ni2+- or Zn2+-dependent GLXI proteins but are different within the GLXI-like proteins. In GLXI-like; 4, 7, 8, 9, and 11 the aa at these specific positions are all changed to H, H, H, E, suggesting a similar biochemical property for the GLXI-like proteins of Clade II (Figures 1B,C).
Two conserved arginine and asparagine residues lying in close proximity to the catalytic site in the tertiary structure are responsible for glutathione binding and are highly conserved among GLXI proteins (in Human GLXI: R38, and N104). Furthermore, Cameron et al. (1997) postulated five important conserved aa within a hinge region involved in domain swapping (in Human HsGLXI: G106, Y115, G118, N119, and G124). Interestingly, position G106 is conserved in all GLXI but not in GLXI-like proteins. Actually, the putative hinge region is quite diverse in all VOC family proteins and predicted to have a disordered structure explaining why the other aa Y115, G118, N119, and G124 are not found at the exact positions within the MSA. Notably, none of the Arabidopsis GLXI-like proteins have conserved Zn2+ or Ni2+ ion, and GSH binding sites suggesting a different biochemical function for the GLXI-like proteins (Figure 1C).
Methylglyoxal can be produced in several reactions, such as lipid peroxidation, oxidation of aa, and the enzymatic oxidation of ketone bodies, but its main source is the action of triosephosphate isomerase during glycolysis (Semchyshyn, 2014). The enediol phosphate intermediate of the isomerisation can escape from the catalytic center and decompose spontaneously into inorganic phosphate, and MGO, linking in this way, central sugar metabolism and MGO formation. In line with the GLXI function in glycation defense, Arabidopsis GLXI isoforms are highly expressed in heterotrophic as well as autotrophic tissues (Figure 2A) and the transcription of the Ni2+-dependent GLXI;2 shows regulation upon alterations in cellular sugar levels (Schmitz et al., 2017). In silico analyses of transcriptional responses of GLX to abiotic stress in Medicago, Glycine, Arabidopsis, and Oryza consistently indicated that the expression of GLXI-like homologs is highly modulated by abiotic stresses, while Ni2+- and Zn2+-dependent GLXI homologs are expressed at a high constitutive level, and show no, or low transcriptional regulation under abiotic stress (Mustafiz et al., 2011; Ghosh and Islam, 2016; Ghosh, 2017).
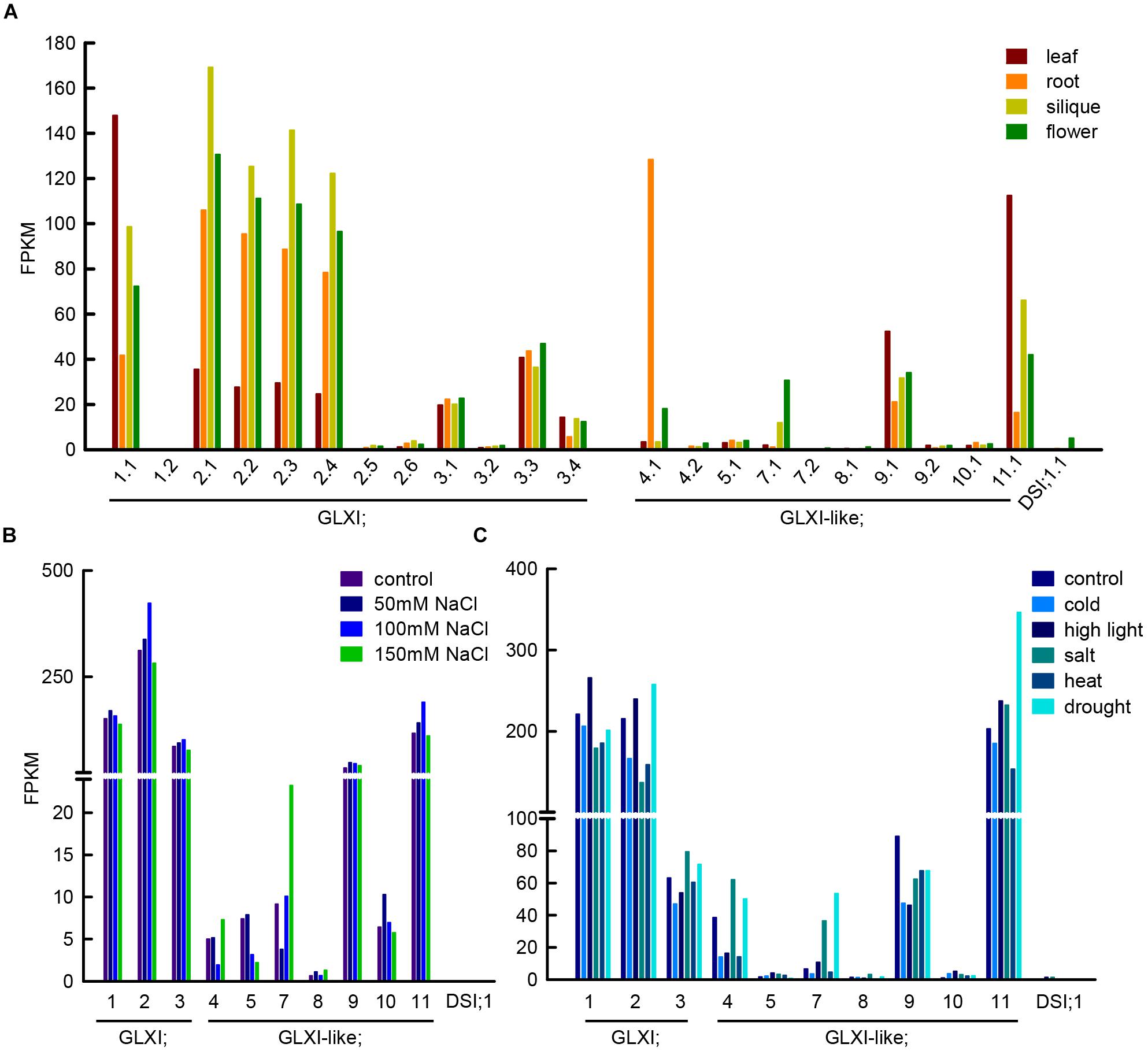
FIGURE 2. Transcript abundance of GLXI and GLXI-like splice forms and gene expression under abiotic stress conditions. (A) FPKM (fragments per kilo base of exon per million fragments mapped) values of GLXI and GLXI-like splice forms in Arabidopsis leaf, root, siliques, and flowers. Raw data was taken from Liu et al. (2012) (PRJNA168212). (B) GLXI, and GLXI-like transcript abundance in FPKM in 21-days-old Arabidopsis plants after 24 h treatment with 50, 100, and 150 mM NaCl. Raw data taken from Shafi et al. (2015) (PRJNA217812). (C) GLXI, and GLXI-like transcripts in FPKM in whole 12-days-old Arabidopsis seedlings after 24 h of abiotic stress exposure. Raw data taken from Filichkin et al. (2010) (SRA009031). Raw data was re-mapped with high stringency to the TAIR 10 annotation as described in Schmitz et al. (2017).
We stringently re-mapped RNAseq data from Liu et al. (2012) from leaf, root, flower, and siliques, and discriminated between the different splice forms of the GLXI-like proteins in Arabidopsis. We found that only the first of the predicted splice forms is preferentially transcribed (Figure 2A). GLXI-like;4 transcripts are highly abundant in roots, GLXI-like;7 is moderately expressed in flowers, and GLXI-like;9, and 11 are expressed in all tissues tested, with higher expression in leaf. GLXI-like;5, 8, 10, and DSI;1 are extremely low expressed (FPKM < 5) in all organs tested (Figure 2A). Mulako et al. (2008) demonstrated that Arabidopsis DSI;1 mRNA transcripts are found at high levels in mature seeds and are not induced upon desiccation stress in other vegetative tissues. The analysis of transcriptional regulation of GLXI, and GLXI-like proteins in Arabidopsis leaves upon 24 h exposure to different NaCl concentrations (Shafi et al., 2015) shows that the Ni2+-dependent GLXI;2 is moderately induced at 100 mM NaCl (Figure 2B). Among the GLXI-like proteins, the homologs 7, 10, and 11 respond to NaCl stress (Figure 2B). However, the induction of expression is not correlated with increasing NaCl concentrations. The analysis of an alternative RNAseq study on 12-day-old seedlings treated during 24 h with different stresses (Filichkin et al., 2010), indicates increase of transcript abundance of the Ni2+-dependent GLXI;1, GLXI;2 and of GLXI-like;11 under high light. In this study, the Zn2+-dependent GLXI;3, GLXI-like;4, 7, and 11 show induction by 100 mM NaCl. This analysis also shows the induction of the Ni2+-dependent GLXI;2, GLXI-like;4, 7, and 11 by drought stress (Figure 2C). A directed and elaborate qPCR-based approach discriminating between the GLXI and GLXI-like gene expression in correlation with abiotic stresses in different plant tissues and stages is needed. This will help dissecting the role of GLXI and GLXI-like proteins in housekeeping and abiotic stress responses.
Determination of the substrate specificities is fundamental to dissect the biological functions and to understand the importance of the GLXI and GLXI-like proteins. The actual substrate of GLXI is the hemithioacetal formed by the spontaneous reaction of MGO and GSH. Alternative substrate activities have been shown for glyoxal and phenylglyoxal in the presence of GSH (Vander Jagt et al., 1972; Schmitz et al., 2017). Whether the GLXI-like proteins utilize GSH is unclear. From our present analysis we hypothesize that GLXI-like proteins do not use GSH, as the GSH binding sites are not conserved in these proteins. GLXI use bivalent metal ions as cofactors, these are important for the catalytic activity and in the case of Ni2+-dependent GLXI also for substrate specificity (Schmitz et al., 2017). The specific aa involved in metal ion binding in the GLXI proteins are not conserved in GLXI-like proteins, but the majority of the VOC family members need bound metal ions for their catalytic activity (Fe2+, Mn2+, Zn2+, Ni2+, or Mg2+) (He and Moran, 2011). It is tempting to speculate that the GLXI-like proteins convert other α-keto aldehydes, that might be produced during abiotic stress without using GSH, as in the case of the VOC family member 4-hydroxyphenylpyruvate dioxygenase (Moran, 2014). Developing enzymatic assays with purified proteins for substrate screening should clarify the role of the still uncharacterized GLXI-like proteins (Hüdig et al., 2018).
Unraveling the physiological significance of GLXI and GLXI-like proteins requires a detailed and precise knowledge of steady state concentrations of reactive carbonyl species, such as MGO, in different plant organs and cell compartments under physiological as well as adverse environmental conditions. Measurement of MGO is hampered by the high reactivity of this compound, the need for a derivatization reaction, and its rather low accumulation levels. Several studies have used different methods for extraction, derivatization and detection in different plant species and conditions. This might explain why quantitative reports on MGO range between 3 nmol⋅gFW-1 (Rabbani and Thornalley, 2014) and 100 μmol⋅gFW-1 (Yadav et al., 2005a). Due to its reactive nature, MGO extraction procedures might impact on the results and lead to an overestimation of the levels (Sousa Silva et al., 2013; Rabbani and Thornalley, 2014). Taking into account that triose phosphates and glucose are the major sources of MGO in physiological metabolism and that their steady state concentrations in Arabidopsis leaves are ∼50 nmol⋅gFW-1 in the case of triose phosphates and ∼1 μmol⋅gFW-1 in the case of glucose (Arrivault et al., 2009), free MGO levels far beyond these levels are only possible with a completely abolished GLXI function. Considering a steady state MGO concentration of 3 nmol⋅gFW-1, it can be deduced that under physiological conditions the GSH:MGO ratio would be around 100:1 (Krueger et al., 2009; Noctor et al., 2011). Thus, conditions that induce a depletion of the GSH pools would imply a fundamental increase in MGO content.
Each abiotic stress factor, like high light, heat, cold, salt or drought has particular as well as overlapping effects and will perturb metabolism by formation of reactive oxygen species, alteration in osmotic potential or disruption of enzymatic functions. Thalmann et al. (2016) demonstrated the importance of starch degradation during the day to regulate osmotic adjustment and growth upon short term osmotic stress. Hence, increase in soluble sugar content may also increase steady state fluxes through glycolysis and with it MGO formation. Under abiotic stress conditions other sources of MGO, such as lipid peroxidation induced by reactive oxygen species, may become important and would explain the vast number of publications reporting that overexpression of Zn2+-dependent GLXI or its expression together with a GLXII can confer tolerance toward general stresses in plants (Veena et al., 1999; Yadav et al., 2005b; Gupta et al., 2018). A reproducible and standardized MGO quantification method as that established by Rabbani and Thornalley (2014) should be used to determine the in vivo concentrations of MGO in different stress conditions.
Even though all GLXI and GLXI-like proteins share the structural features of VOC superfamily proteins, they belong to three distinct clades in a phylogenetic analysis. Through analysis of homology and aa conservation, we found that Arabidopsis GLXI-like proteins do not have GLXI conserved substrate and metal binding sites, and in contrast to GLXI proteins their phylogenetic occurrence seems to be restricted to Bacteria and the green lineage. GLXI expression is high and rather constitutive in different plant organs, whereas expression of GLXI-like;4, 7, and 11 mainly respond to abiotic stresses in our analyses. Quantitative measurements of MGO and other reactive carbonyl species from different plant tissues, in different physiological and abiotic stress conditions using loss-of function mutant lines will support the characterization of GLXI-like proteins and will pinpoint their physiological significance. It seems plausible that GLXI-like proteins diverged in plants to fulfill a different function other than MGO detoxification.
JS and VGM analyzed data and wrote the manuscript. JS the performed the phylogenetic analysis. AWR performed the re-mapping of RNAseq data.
This work was supported by the Deutsche Forschungsgemeinschaft under Grants MA2379/11–2, and EXC 1028 to VGM.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arrivault, S., Guenther, M., Ivakov, A., Feil, R., Vosloh, D., Van Dongen, J. T., et al. (2009). Use of reverse-phase liquid chromatography, linked to tandem mass spectrometry, to profile the Calvin cycle and other metabolic intermediates in Arabidopsis rosettes at different carbon dioxide concentrations. Plant J. 59, 824–839. doi: 10.1111/j.1365-313X.2009.03902.x doi: 10.1111/j.1365-313X.2009.03902.x
Bennett, M. J., and Eisenberg, D. (2004). The evolving role of 3D domain swapping in proteins. Structure 12, 1339–1341. doi: 10.1016/j.str.2004.07.004
Cameron, A. D., Olin, B., Ridderström, M., Mannervik, B., and Jones, T. A. (1997). Crystal structure of human glyoxalase I–evidence for gene duplication and 3D domain swapping. EMBO J. 16, 3386–3395. doi: 10.1093/emboj/16.12.3386
Clugston, S. L., Yajima, R., and Honek, J. F. (2004). Investigation of metal binding and activation of Escherichia coli glyoxalase I: kinetic, thermodynamic and mutagenesis studies. Biochem. J. 377, 309–316. doi: 10.1042/bj20030271
Dumas, P., Bergdoll, M., Cagnon, C., and Masson, J. M. (1994). Crystal structure and site-directed mutagenesis of a bleomycin resistance protein and their significance for drug sequestering. EMBO J. 13, 2483–2492. doi: 10.1002/j.1460-2075.1994.tb06535.x
Filichkin, S. A., Priest, H. D., Givan, S. A., Shen, R., Bryant, D. W., Fox, S. E., et al. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20, 45–58. doi: 10.1101/gr.093302.109
Frickel, E. M., Jemth, P., Widersten, M., and Mannervik, B. (2001). Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 276, 1845–1849. doi: 10.1074/jbc.M005760200
Ghosh, A. (2017). Genome-wide identification of glyoxalase genes in medicago truncatula and their expression profiling in response to various developmental and environmental stimuli. Front. Plant Sci. 8:836. doi: 10.3389/fpls.2017.00836
Ghosh, A., and Islam, T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16:87. doi: 10.1186/s12870-016-0773-9
Gupta, B. K., Sahoo, K. K., Ghosh, A., Tripathi, A. K., Anwar, K., Das, P., et al. (2018). Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant Cell Environ. 41, 1186–1200. doi: 10.1111/pce.12968
He, M. M., Clugston, S. L., Honek, J. F., and Matthews, B. W. (2000). Determination of the structure of Escherichia coli glyoxalase I suggests a structural basis for differential metal activation. Biochemistry 39, 8719–8727. doi: 10.1021/bi000856g
He, P., and Moran, G. R. (2011). Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J. Inorg. Biochem. 105, 1259–1272. doi: 10.1016/j.jinorgbio.2011.06.006
Hüdig, M., Schmitz, J., Engqvist, M. K. M., and Maurino, V. G. (2018). Biochemical control systems for small molecule damage in plants. Plant Signal. Behav. 13:e1477906. doi: 10.1080/15592324.2018.1477906
Jain, M., Batth, R., Kumari, S., and Mustafiz, A. (2016). Arabidopsis thaliana contains both Ni2 + and Zn2 + dependent glyoxalase I enzymes and ectopic expression of the latter contributes more towards abiotic stress tolerance in E. coli. PLoS One 11:e0159348. doi: 10.1371/journal.pone.0159348
Katoh, K., Kuma, K., Toh, H., and Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518. doi: 10.1093/nar/gki198
Kaur, C., Vishnoi, A., Ariyadasa, T. U., Bhattacharya, A., Singla-Pareek, S. L., and Sopory, S. K. (2013). Episodes of horizontal gene-transfer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 3:3076. doi: 10.1038/srep03076
Krueger, S., Niehl, A., Lopez Martin, M. C., Steinhauser, D., Donath, A., Hildebrandt, T., et al. (2009). Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant Cell Environ. 32, 349–367. doi: 10.1111/j.1365-3040.2008.01928.x
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345. doi: 10.1105/tpc.112.102855
Moran, G. R. (2014). 4-Hydroxyphenylpyruvate dioxygenase and hydroxymandelate synthase: exemplars of the alpha-keto acid dependent oxygenases. Arch. Biochem. Biophys. 544, 58–68. doi: 10.1016/j.abb.2013.10.022
Mulako, I., Farrant, J. M., Collett, H., and Illing, N. (2008). Expression of Xhdsi-1VOC, a novel member of the vicinal oxygen chelate (VOC) metalloenzyme superfamily, is up-regulated in leaves and roots during desiccation in the resurrection plant Xerophyta humilis (Bak) dur and schinz. J. Exp. Bot. 59, 3885–3901. doi: 10.1093/jxb/ern226
Mustafiz, A., Singh, A. K., Pareek, A., Sopory, S. K., and Singla-Pareek, S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11, 293–305. doi: 10.1007/s10142-010-0203-2
Noctor, G., Queval, G., Mhamdi, A., Chaouch, S., and Foyer, C. H. (2011). Glutathione. Arabidopsis Book 9, 1–32. doi: 10.1199/tab.0142
Pei, J. M., Kim, B. H., and Grishin, N. V. (2008a). PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 36, 2295–2300. doi: 10.1093/nar/gkn072
Pei, J. M., Tang, M., and Grishin, N. V. (2008b). PROMALS3D web server for accurate multiple protein sequence and structure alignments. Nucleic Acids Res. 36, W30–W34. doi: 10.1093/nar/gkn322
Rabbani, N., and Thornalley, P. J. (2014). Measurement of methylglyoxal by stable isotopic dilution analysis LC-MS/MS with corroborative prediction in physiological samples. Nat. Protoc. 9, 1969–1979. doi: 10.1038/nprot.2014.129
Ridderstrom, M., Cameron, A. D., Jones, T. A., and Mannervik, B. (1998). Involvement of an active-site Zn2 + ligand in the catalytic mechanism of human glyoxalase I. J. Biol. Chem. 273, 21623–21628. doi: 10.1074/jbc.273.34.21623
Schmitz, J., Dittmar, I. C., Brockmann, J. D., Schmidt, M., Hudig, M., Rossoni, A. W., et al. (2017). Defense against reactive carbonyl species involves at least three subcellular compartments where individual components of the system respond to cellular sugar Status. Plant Cell 29, 3234–3254. doi: 10.1105/tpc.17.00258
Sela, I., Ashkenazy, H., Katoh, K., and Pupko, T. (2015). GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. doi: 10.1093/nar/gkv318
Semchyshyn, H. M. (2014). Reactive carbonyl species in vivo: generation and dual biological effects. Sci. World J. 2014, 27–31. doi: 10.1155/2014/417842
Shafi, A., Chauhan, R., Gill, T., Swarnkar, M. K., Sreenivasulu, Y., Kumar, S., et al. (2015). Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol. Biol. 87, 615–631. doi: 10.1007/s11103-015-0301-6
Sousa Silva, M., Gomes, R. A., Ferreira, A. E. N., Ponces Freire, A., and Cordeiro, C. (2013). The glyoxalase pathway: the first hundred years.. and beyond. Biochem. J. 453, 1–15. doi: 10.1042/BJ20121743
Suttisansanee, U., Ran, Y., Mullings, K. Y., Sukdeo, N., and Honek, J. F. (2015). Modulating glyoxalase I metal selectivity by deletional mutagenesis: underlying structural factors contributing to nickel activation profiles. Metallomics 7, 605–612. doi: 10.1039/c4mt00299g
Thalmann, M., Pazmino, D., Seung, D., Horrer, D., Nigro, A., Meier, T., et al. (2016). Regulation of Leaf starch degradation by abscisic acid is important for osmotic stress tolerance in plants. Plant Cell 28, 1860–1878. doi: 10.1105/tpc.16.00143
Thornalley, P. J. (2003). Glyoxalase I - structure, function and a critical role in the enzymatic defense against glycation. Biochem. Soc. Trans. 31, 1343–1348. doi: 10.1042/bst0311343
Turra, G. L., Agostini, R. B., Fauguel, C. M., Presello, D. A., Andreo, C. S., Gonzalez, J. M., et al. (2015). Structure of the novel monomeric glyoxalase I from Zea mays. Acta Crystallogr. D. Biol. Crystallogr. 71, 2009–2020. doi: 10.1107/S1399004715015205
Vander Jagt, D. L., Han, L. P. B., and Lehman, C. H. (1972). Kinetic evaluation of substrate specificity in the glyoxalase-I-catalyzed disproportionation of alpha-ketoaldehydes. Biochemistry 11, 3735–3740. doi: 10.1021/bi00770a011
Veena, Reddy, V. S., and Sopory, S. K. (1999). Glyoxalase I from Brassica juncea: molecular cloning, regulation and its over-expression confer tolerance in transgenic tobacco under stress. Plant J. 17, 385–395. doi: 10.1046/j.1365-313X.1999.00390.x
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M., and Barton, G. J. (2009). Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Yadav, S. K., Singla-Pareek, S. L., Ray, M., Reddy, M. K., and Sopory, S. K. (2005a). Methylglyoxal levels in plants under salinity stress are deaendent on glyoxalase I and glutathione. Biochem. Biophys. Res. Commun. 337, 61–67. doi: 10.1016/j.bbrc.2005.08.263
Keywords: glyoxalase I, methylglyoxal, scavenging, abiotic stress, reactive carbonyl species, glyoxalase system
Citation: Schmitz J, Rossoni AW and Maurino VG (2018) Dissecting the Physiological Function of Plant Glyoxalase I and Glyoxalase I-Like Proteins. Front. Plant Sci. 9:1618. doi: 10.3389/fpls.2018.01618
Received: 15 September 2018; Accepted: 18 October 2018;
Published: 12 November 2018.
Edited by:
Paula Casati, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
Kazuo Nakashima, Japan International Research Center for Agricultural Sciences, JapanCopyright © 2018 Schmitz, Rossoni and Maurino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jessica Schmitz, amVzc2ljYS5zY2htaXR6QHVuaS1kdWVzc2VsZG9yZi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.