
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 06 November 2018
Sec. Virology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01604
This article is part of the Research Topic Plant Responses to Phytophagous Mites/Thrips and Search for Resistance View all 25 articles
 Rajagopalbabu Srinivasan1*
Rajagopalbabu Srinivasan1* Mark R. Abney2
Mark R. Abney2 Pin-Chu Lai1
Pin-Chu Lai1 Albert K. Culbreath3
Albert K. Culbreath3 Shyam Tallury4
Shyam Tallury4 Soraya C. M. Leal-Bertioli5
Soraya C. M. Leal-Bertioli5Thrips are major pests of peanut (Arachis hypogaea L.) worldwide, and they serve as vectors of devastating orthotospoviruses such as Tomato spotted wilt virus (TSWV) and Groundnut bud necrosis virus (GBNV). A tremendous effort has been devoted to developing peanut cultivars with resistance to orthotospoviruses. Consequently, cultivars with moderate field resistance to viruses exist, but not much is known about host resistance to thrips. Integrating host plant resistance to thrips in peanut could suppress thrips feeding damage and reduce virus transmission, will decrease insecticide usage, and enhance sustainability in the production system. This review focuses on details of thrips resistance in peanut and identifies future directions for incorporating thrips resistance in peanut cultivars. Research on thrips–host interactions in peanut is predominantly limited to field evaluations of feeding damage, though, laboratory studies have revealed that peanut cultivars could differentially affect thrips feeding and thrips biology. Many runner type cultivars, field resistant to TSWV, representing diverse pedigrees evaluated against thrips in the greenhouse revealed that thrips preferred some cultivars over others, suggesting that antixenosis “non-preference” could contribute to thrips resistance in peanut. In other crops, morphological traits such as leaf architecture and waxiness and spectral reflectance have been associated with thrips non-preference. It is not clear if foliar morphological traits in peanut are associated with reduced preference or non-preference of thrips and need to be evaluated. Besides thrips non-preference, thrips larval survival to adulthood and median developmental time were negatively affected in some peanut cultivars and in a diploid peanut species Arachis diogoi (Hoehne) and its hybrids with a Virginia type cultivar, indicating that antibiosis (negative effects on biology) could also be a factor influencing thrips resistance in peanut. Available field resistance to orthotospoviruses in peanut is not complete, and cultivars can suffer substantial yield loss under high thrips and virus pressure. Integrating thrips resistance with available virus resistance would be ideal to limit losses. A discussion of modern technologies such as transgenic resistance, marker assisted selection and RNA interference, and future directions that could be undertaken to integrate resistance to thrips and to orthotospoviruses in peanut cultivars is included in this article.
Peanut (Arachis hypogaea L.) is a major food and oil seed crop that provides high quality human nutrition and is severely affected by thrips and viruses transmitted by them in many parts of the world including the Southern United States, South/Southeastern Asia, and South America (Pappu et al., 2009; Riley et al., 2011; Mandal et al., 2012). Thrips are small (<2 mm in length) and slender insects with fringed wings belonging to the order Thysanoptera. They are hemimetabolous insects with egg, larvae, prepupal (quasi pupal stage), and adult stages. The adults and larvae are the two mobile stages, with adults alone possessing wings (Lewis, 1973, 1997). The two common wing morphs include the brachypterous (short-winged) and macropterous (long-winged) forms. Depending upon seasonal environmental parameters and host availability, thrips alternate wing forms to aid their dispersal. In the United States, two thrips species, Western flower thrips, Frankliniella occidentalis (Pergande), tobacco thrips, Frankliniella fusca (Hinds), occur in most peanut producing areas (Figure 1; Sakimura, 1963; Todd et al., 1995; Riley et al., 2011). In Southeastern United States, where more than half of United States peanuts are grown, F. fusca is commonly found on peanut foliage and flowers, and is responsible for almost all the early season feeding injury in peanut (Todd et al., 1995). Western flower thrips is predominantly a flower feeder, and is often found later in the growing season. In other peanut growing areas, such as South and Southeast Asia, thrips species viz., common blossom thrips, Frankliniella schultzei (Trybom), chili thrips, Scirtothrips dorsalis (Hood), melon thrips, Thrips palmi (Karny), bean flower thrips, Megalurothrips usitatus (Bagnall), and groundnut thrips, Caliothrips indicus (Bagnall) are known to infest peanut (Amin et al., 1985; Ekvised et al., 2006a). In South America, besides Frankliniella sp. others such as Enneothrips flavens (Moulton) are commonly found on peanuts (de Souza et al., 2010; Michelotto et al., 2017).

FIGURE 1. Two common thrips species in peanuts in Southeastern United States, tobacco thrips, Frankliniella fusca (Hinds), on the left, and the western flower thrips, Frankliniella occidentalis (Pergande), on the right.
Thrips feeding in peanut is a concern from the time of seedling emergence to a few weeks following emergence. Under severe thrips pressure, thrips feeding injury early in the season can result in yield loss and/or delayed maturity (Todd et al., 1995; de Moraes et al., 2005; Funderburk et al., 2007). Thrips possess asymmetrical mouthparts, due to an atrophied mandible, and generally feed by sucking plant cell contents. In the process, thrips feeding is often characterized by “silvering” of leaves. The silvering appearance is caused by empty epidermal cells following thrips feeding on the cells’ contents (Figure 2A). Larvae and adults can feed on the peanut foliage. When thrips populations are high early in the growing season, a situation characterized by extensive larval colonization, it is common to find leaf-tip yellowing and necrosis and curling of newly developing leaflets (terminals) at the shoot tip (Figures 2B,C). Heavily infested peanut seedlings are often stunted and in severe cases can die (Figure 2D). Thrips feeding also results in transmission of viruses. Orthotospoviruses are a major peanut-infecting virus group that is of concern worldwide (Jones, 2005; Pappu et al., 2009; Riley et al., 2011). The main orthotospoviruses (Family Tospoviridae; Order Bunyavirales) include the Tomato spotted wilt virus (TSWV) in Asia and in North America, Groundnut ring spot virus (GRSV) and Groundnut bud necrosis virus (GBNV) in Asia, and GRSV in South America (Pappu et al., 2009). Increased thrips populations are often correlated with increased virus incidence (Garcia et al., 2000; Culbreath et al., 2003; Sharma et al., 2003).
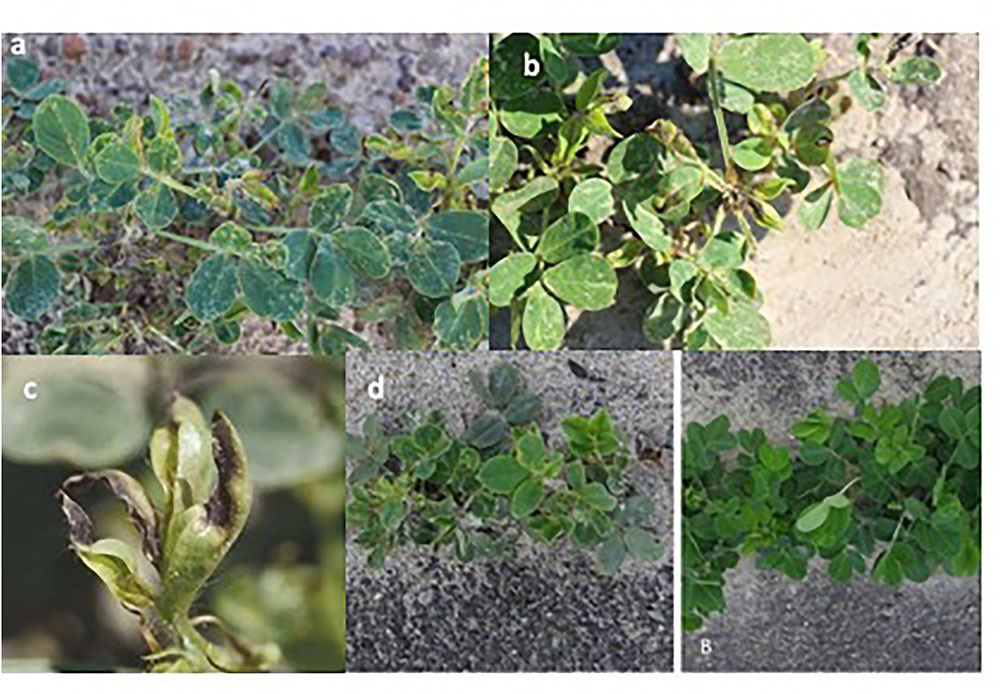
FIGURE 2. Thrips induced feeding symptoms on peanut. Silvering appearance due to thrips feeding on the epidermal cells’ contents (A), leaf-tip yellowing and necrosis and curling of newly developing leaflets (terminals) at the shoot tip (B), a close-up view of tip burning in terminal leaflets (C), and stunting of thrips-infested non-treated peanut plant on the left and treated peanut plant on the right (D).
Thrips employ haplodiploid sex determination, wherein the fertilized eggs produce diploid females, and the non-fertilized eggs result in haploid males (Moritz, 1997). This mode of reproduction and rapid life cycle (dependent on temperature) allows them to build up in large numbers. Their populations are usually characterized by one or two peaks in a typical peanut growing season. Their reproductive capacity, short lifecycles, broad host range, and thigmotactic behavior (seeking refuge in tight spaces such as unfolded peanut terminals), make thrips difficult to manage in peanut. Cultural practices can significantly affect thrips populations in peanut. Manipulating planting date to avoid coincidence of peak thrips dispersal and the susceptible seedling stage results in lower thrips densities and reduces the risk of feeding injury and virus transmission (Brown et al., 1996; McKeown et al., 2001; Culbreath et al., 2010). Likewise, seeding into heavy plant residue in conservation tillage systems reduces thrips abundance on peanut compared with conventional tillage systems with bare soil (Brown et al., 1996; Monfort et al., 2007). Increased plant density and twin row planting have also been shown to reduce thrips infestation and virus incidence, though the mechanism(s) responsible are not well understood (Culbreath et al., 2008; Tubbs et al., 2011). Unfortunately, all the management tactics discussed here have potential negative consequences. Planting dates that minimize risk of thrips infestation are not always optimal for maximizing yield. While conservation tillage offers several recognized agronomic and environmental benefits in addition to thrips management, it may have negative effects on weed management programs compared with conventional tillage systems (Johnson et al., 2001). The additional seed and specialized planting equipment needed to increase plant densities and achieve twin row patterns ultimately increase production costs for farmers.
Chemical management options for thrips in peanut, like many other row crops, are limited to a few insecticide active ingredients (Reddy et al., 1995; Todd et al., 1995, 1996; Herbert et al., 2007; Culbreath et al., 2008; Marasigan et al., 2016). The most commonly used insecticide classes include organophosphates, carbamates, phenylpyrazole, pyrethroids, and neonicotinoids (Todd et al., 1996; Culbreath et al., 2003, 2016; Mandal et al., 2012; Marasigan et al., 2016; Srinivasan et al., 2017). Newer classes of insecticides such as spinosyns and diamides, though effective, are too expensive to justify their use in peanut (Marasigan et al., 2016, 2018). These products are generally reserved for high value crops such as fruits and vegetables. In addition, there is increased concern over environmental and non-target issues associated with older broad-spectrum insecticides such as organophosphates, and carbamates. Even neonicotinoids, long considered to be reduced risk options, are being scrutinized due to their presumed role in pollinator decline (Mullin et al., 2010; Nicodemo et al., 2014). In lieu of the perceived effects, the United States Environmental Agency in 2015 issued a temporary moratorium on new neonicotinoid registrations1. This moratorium has not affected any existing registrations. Neonicotinoids applied as liquid in-furrow at peanut planting offer ease of application, are relatively less expensive, and in general provide good efficacy. For these reasons, the neonicotinoid imidacloprid is now used increasingly in Georgia in Southern United States (Marasigan et al., 2016, 2018; Srinivasan et al., 2017). Thrips, in general, have effective pesticide detoxification abilities (Espinosa et al., 2005; Bielza et al., 2007, 2008; Bielza, 2008), and they have developed resistance to several insecticide classes. Increased neonicotinoid usage in cotton has already led to widespread resistance development in the Southeastern United States (Huseth et al., 2016). Preliminary research conducted in Georgia in the United States indicated no evidence of resistance to neonicotinoids in thrips populations collected from peanut (Lai, 2015). The usefulness of these insecticides in the long run remains questionable. Of course, the main concern in peanut production in many parts of the world is thrips-transmitted orthotospoviruses. No insecticide, except phorate, has been found to be effective in suppressing virus transmission significantly in the United States, and the effect of phorate is not consistent (Culbreath et al., 2003, 2008, 2016; Marasigan et al., 2018).
Because of the difficulty associated with managing thrips and the significant economic loss that accompanies virus infection, a tremendous amount of effort has been invested into breeding cultivars with resistance against orthotospoviruses. Much of the information on breeding for virus resistance comes with research conducted with TSWV in peanut in the United States and GBNV in Asia. The cultivars grown at the advent of TSWV in Southern United States, in the 1980s and in early 1990s, such as Florunner and Southern runner, were extremely susceptible to TSWV-induced spotted wilt disease (Culbreath et al., 1992, 1996). Screening and breeding efforts led to incremental increases in resistance, most of which was derived from a single genotype (PI 203363) introduced from Brazil in 1953 (Culbreath et al., 2003). The introduction of this unique genotype had a rapid and profound effect on the United States peanut breeding, as the main runner peanut cultivars have a significant proportion of PI203363 alleles (Clevenger et al., 2018). Current “third generation TSWV-resistant” peanut cultivars are highly field resistant to TSWV, and losses due to the disease have been minimized. Breeding efforts also have led to identification of moderate resistance to GBNV in Asia (Amin et al., 1985; Dwivedi et al., 1995; Reddy et al., 2000; Kesmala et al., 2004; Mandal et al., 2012). In all these instances, resistance to the virus is not complete, and often other management options are integrated. For instance, insecticides are still being employed to reduce thrips feeding injury in early season peanut (Mandal et al., 2012; Marasigan et al., 2016). Identifying and incorporating effective thrips resistance in high yielding peanut cultivars will provide significant economic benefit to producers and result in reduced environmental impact associated with pesticide use.
Thrips resistance in peanut was more actively pursued in the 1980s and early 1990s in Asia and in the United States until thrips-transmitted viruses became a more pressing issue (Amin and Mohammed, 1980; Campbell and Wynne, 1980; Amin et al., 1985; Mulder and Seuhs, 2002). Most of those early examinations were based on field screening (Young et al., 1972; Stalker and Campbell, 1983; Lynch, 1990). Consequently, thrips resistance contributing factors in peanut are not well understood. Information about thrips resistance stems mostly from work on other crops, where resistance seems to be imparted by morphological as well as biochemical traits. Each trait category is discussed in detail.
Early on increased leaf pubescence in crops such as cassava was associated with thrips resistance (Schoonhoven, 1974). Similarly, increased foliar pubescence in diploid cotton such as Gossypium arboreum L., Gossypium thurberi (Todd), Gossypium trilobum (DC.) Skovst resulted in reduced western flower thrips infestation compared with other commonly grown Gossypium hirsutum L. cv. Sicot 71 (Miyazaki et al., 2017). Another study found that thrips infestation was less in glandless cotton than in glandular cotton (Zhang et al., 2014). Increased leaf waxiness was associated with resistance against thrips in cabbage (Voorrips et al., 2008a,b). On the contrary, in onion cultivars, glossy (less wax) yellow green foliage provided more protection against thrips than cultivars with non-glossy or waxy blue green foliage (Coudriet et al., 1979; Molenaar, 1984; Diaz-Montano et al., 2010; Damon et al., 2014). Traits such as leaf angle and leaf toughness were also more influential than waxiness on host plant susceptibility to onion thrips (Njau et al., 2017). Onion foliage that was more open and round, allowed more thrips exposure to natural enemies, and thus had fewer thrips compared with onion foliage that was tight and had short angle of deviation. In addition to leaves, floral, and fruiting structures are also vulnerable to thrips. It has been shown in at least two cases that the size of floral structures can be associated with thrips resistance. Smaller flowers in both cowpea and chrysanthemum resulted in reduced incidences of Megalurothrips sjostedti (Trybom) and F. occidentalis, respectively (de Jager et al., 1995; Abudulai et al., 2006; Omo-Ikerodah et al., 2009).
Field screening of peanut genotypes revealed differences in thrips feeding injury consistently, and it was speculated that foliage color could be influencing thrips host selection patterns (Amin et al., 1985). A relatively recent study found differences in normalized vegetation index among peanut cultivars, which in turn might affect light reflectance off peanut foliage (Navia-Gine, 2012). It is not clear if these differences in light reflectance affect thrips host plant utilization. The hypothesis that reflectance contributes to host plant resistance should be evaluated, and the role of light reflectance in thrips host selection and host utilization in peanut needs to be examined in greater depth. Such differences in foliar reflectance in light could be due to differences in profiles of cuticular waxes. Peanut cultivars’ and wild species foliage hues differ substantially from light green to bluish green. Increased cuticular wax in various wild peanut species such as Arachis batizocoi Krapov. & W. C. Gregory, Arachis glandulifera Stalker, Arachis ipaensis Krapov. & W. C. Gregory, Arachis Chacoense Krapov., and Arachis paraguariensis Chodat & Hassl., is believed to be responsible for suppressing thrips feeding compared with commonly grown peanut cultivars (Yang et al., 1993; de Souza et al., 2010). Other morphological traits discussed previously such as leaf hairiness, and leaf toughness are also believed to be involved in conferring resistance to insects including thrips in peanut (Campbell and Wynne, 1980; Yang et al., 1993), but their role in suppressing thrips needs to be experimentally demonstrated.
Alkaloids and other secondary metabolites seem to be contributing to thrips resistance either in the presence or absence of morphological traits. In a study with wild tomato species viz., Solanum pennellii Correll and Solanum hirsutum Dunal, acyl sugars were implicated as being involved in conferring resistance to thrips (Mirnezhad et al., 2010; Romero González, 2011). An alkaloid, pyrrolizidine, was associated with thrips resistance in the case of Jacobaea aquatica (Hill) G. Gaertn., (Cheng et al., 2011). Similarly, glycoalkaloids provided resistance against thrips in potato, Solanum tuberosum L. (Galvez et al., 2005). Evidence for involvement of biochemical compounds besides alkaloids in thrips resistance is found in numerous crops. Isobutylamides of unsaturated fatty acids and chlorogenic acid from chrysanthemum, Chrysanthemum indicum L., conferred resistance against western flower thrips (Tsao et al., 2003; Leiss et al., 2009, 2011). A flavonoid luteolin and phenylpropanoid sinapic acid derived from carrot, Daucus carota (Hoffm.) Schübl., negatively impacted Western flower thrips development (Leiss et al., 2013). In some instances, biochemicals are thought to be involved in imparting resistance against thrips; however, they have not been identified. For instance, steam distillates from certain resistant cultivars of rice (Oryza indica L.) were toxic to Stenchaetothrips biformis (Bagnall), but it is not clear which active compound(s) in that distillate was responsible for thrips mortality (Velusamy and Saxena, 1991). Similarly, extensive studies in pepper, Capsicum sp., led to identification of resistance believed to be caused by biochemical traits, but the causal agents have not been identified (Maris et al., 2003a,b; Maharijaya, 2013). Also, some odors from rose, Rosa sp., that repelled western flower thrips, and are yet to be characterized (Gaum et al., 1994). de Souza et al. (2010) found that certain n-alkanes from wild diploid peanut species could be responsible for E. flavens resistance in Brazil. A polyphenolic compound, 2,3-Di-(E)-caffeoyl-(2B,3R)-(+)-tartaric acid, found in peanut terminals was associated with F. fusca resistance in the Southeastern United States (Snook et al., 1994). However, this study merely reported a correlation, and did not present a conclusive evidence of the compound’s involvement in resistance against thrips. Another study showed a strong negative correlation between phenols and tannins in peanut germplasm with thrips feeding damage (Kandakoor et al., 2014). The correlative roles of these compounds in thrips resistance should further be functionally characterized by isolating these compounds and conducting bioassays with thrips.
Peanut cultivars have been screened for F. fusca and F. Schultzei feeding injury in numerous studies in the Southern United States, Asia, and in South America (Young et al., 1972; Kinzer et al., 1973; Amin et al., 1985; Sharma et al., 2003). In the United States, several studies identified runner type and Virginia type peanut germplasm plant introductions with resistance to thrips in Georgia and North Carolina (Lynch, 1990). Results from these studies showed differences in thrips injury rating. However, the peanut plants generally recovered from thrips injury and yield losses only occurred under certain conditions (Funderburk et al., 2007). The losses that were observed were more often associated with Virginia type peanut cultivars than runner type (Amin and Mohammed, 1980; Campbell and Wynne, 1980; Mulder, 1999; Mulder and Seuhs, 2002; Herbert et al., 2005; Whalen et al., 2014). Choice experiments conducted with peanut cultivars and tobacco thrips suggest that cultivars may differently affect thrips density as well as severity of thrips feeding injury (Figure 3). Thrips feeding was reduced in some cultivars such as “Tifguard” and “Georganic” when compared with others such as Georgia Green and Georgia 06G (Sundaraj et al., 2014). Thrips feeding was measured by using a thrips feeding damage index, which is a measurement of silvering area on the plant as a proportion of the undamaged area (Maris et al., 2003a). The evaluated peanut cultivars were actually released with increased resistance to TSWV, but none were specifically bred for thrips resistance. These results suggest that there could be non-preference or antixenosis effects present in peanut cultivars that impact the behavior of thrips. The term non-preference was defined by Painter (1951); Kogan and Ortman (1978) later described it as antixenosis, these terms describe the inability of the insect to effectively use a host plant and instead select an alternate host plant (Smith, 2005). It is not clear how these preference patterns would influence thrips in peanut agroecosystems with relatively low genotype diversity such as those in the Southern United States. For instance, in Georgia, more than 80% of the peanut acreage (>600 K acres, NASS, 2017) is often planted with a single cultivar. More research needs to be conducted to evaluate the significance of antixenosis against thrips in commercial peanut production.
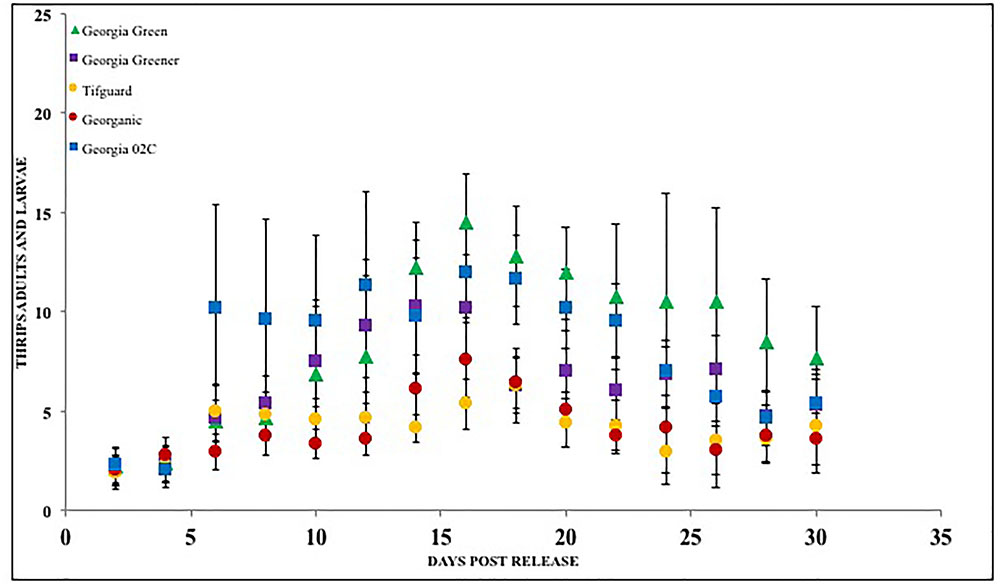
FIGURE 3. Thrips larvae and adults counted on peanut foliage of various cultivars that are resistant and susceptible to Tomato spotted wilt virus under a choice situation. Georgia Green is considered as a TSWV-susceptible cultivar. Thrips counts were taken at two-day intervals since release, and counted for 3 weeks post initial thrips release.
Microcosmic “Munger”(45 cm3 thrips-proof cages) studies indicate that peanut cultivar differences could differentially influence tobacco thrips, F. fusca fitness. No-choice tests to monitor thrips development and thrips survival were conducted with commonly grown cultivars in Georgia. Results revealed that cultivar differences significantly influenced tobacco thrips survival (Figure 4; Sundaraj et al., 2014). Thrips fitness was consistently reduced in some of the “second-generation TSWV resistant” peanut cultivars compared to ones released earlier such as “Georgia Green.” Antibiosis is a resistance mechanism by which a host plant adversely affects the biology of the insect, often resulting in increased mortality or reduced longevity and fecundity (Teetes, 1996). The Munger cage studies revealed that thrips fecundity (adults recovered per adult released) and longevity was reduced in some cultivars such as Georganic and Tifguard. The innate factors in these cultivars that contribute to antibiosis against thrips are yet to be identified. Unique parentage of these cultivars could be influencing resistance to thrips. For instance, “Tifguard,” which possesses resistance to nematodes seems to be the most resistant against thrips (Holbrook et al., 2008; Shrestha et al., 2013). Nematode resistance in Tifguard is derived from a germplasm accession line “COAN”; it is not clear if the nematode resistance and thrips resistance are interlinked (Holbrook et al., 2008). Fitness experiments also revealed that the median thrips developmental time from egg to adult in Tifguard was lower than in any other cultivar evaluated (Figure 5; Shrestha et al., 2013). The reduced developmental time (by 2 days) of F. fusca on a resistant cultivar could be a strategy used by thrips to overcome unfavorable characteristics in that resistant genotype. This strategy is not unique to F. fusca and peanut; variations in thrips developmental time have been associated with F. occidentalis on thrips-resistant pepper (Maris et al., 2003b; Maharijaya et al., 2012). This phenomenon has also been observed in other insects (Leather et al., 1998).
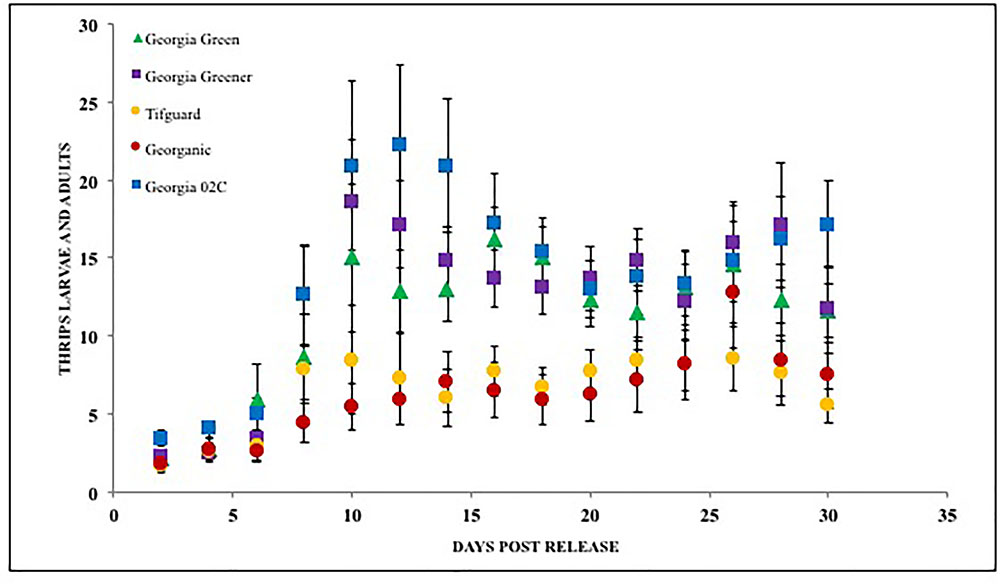
FIGURE 4. Thrips larvae and adults counted on peanut foliage of various cultivars that are resistant and susceptible to Tomato spotted wilt virus under a no-choice situation. Georgia Green is considered as a TSWV-susceptible cultivar. Thrips counts were taken at two-day intervals since release, and counted for 3 weeks post initial thrips release.
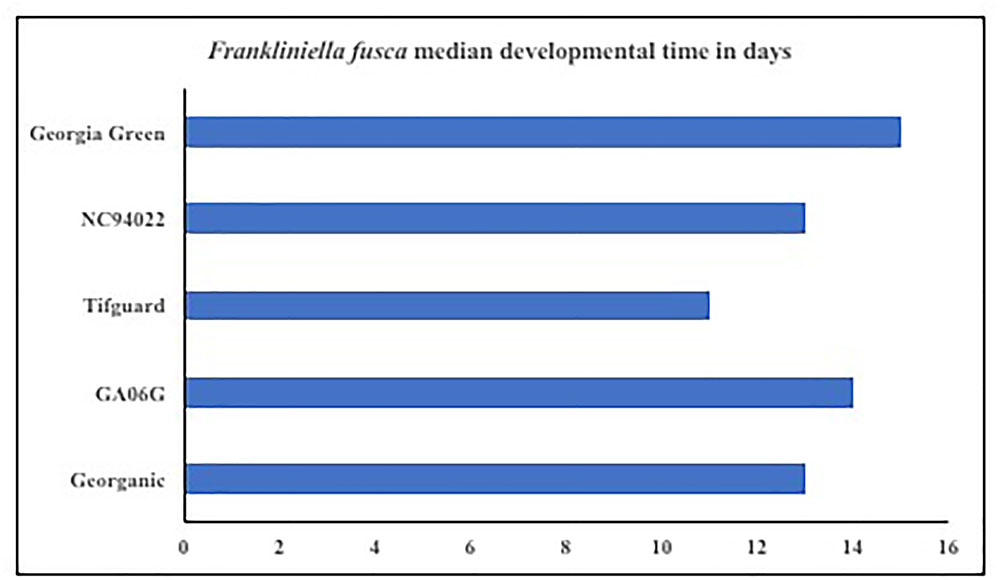
FIGURE 5. Thrips developmental time on Tomato spotted wilt virus resistant and susceptible cultivars. Developmental time refers to the time take from egg to adulthood, and counts were taken once every 2 days. Thrips developmental time was monitored using Plexiglass arenas called Munger cages.
Breeding efforts for thrips resistance in peanut has dwindled in the United States and continues on a minor scale in Asia Culbreath et al., 2003; Pappu et al., 2009; Culbreath and Srinivasan, 2011; Mandal et al., 2012; Srinivasan et al., 2017). A study on heritability of thrips resistance in Thailand found that there was a weak correlation between thrips resistance parameters and agronomic traits and predicted that these characters are independently inherited (Ekvised et al., 2006b). With increased adoption of TSWV and other virus-resistant cultivars in most peanut production areas, there is heightened concern about evolution of resistance breaking virus strains (Sundaraj et al., 2014). Prior evidence of TSWV resistance breakdown in solanaceous crop hosts should serve as a warning (Qiu and Moyer, 1999), and all protections should be taken to prevent virus-resistance breakdown in peanut cultivars. Evidence presented above suggests that peanut cultivar-thrips interactions could influence virus transmission by thrips (Sundaraj et al., 2014). Results from Sundaraj et al. (2014) revealed that some of the TSWV-resistant cultivars such as Tifguard and Georganic accumulated less viral copies (up to 1/3rd less) and were infected at a lower percentage (up to 20%) than other TSWV resistant cultivars such as Georgia Green and Georgia Greener. These results suggest that cultivars that negatively affect thrips preference and/or fitness further suppress virus incidence and accumulation in cultivars that are already moderately resistant to TSWV, thereby providing additive effects. Therefore, it is logical to assume that stacking thrips resistance with virus resistance would reduce the amount of selection pressure imparted against the virus itself, delay the development of resistance breaking strains, and prolong the usefulness of these resistant cultivars that are heavily relied upon. In the United States in Georgia and North Carolina, more than 80% of the peanut acreage is planted with TSWV-resistant cultivars. Losing TSWV resistance in these cultivars would be devastating to peanut production. One way to prevent such a resistance breakdown would be to identify and incorporate thrips resistance in conjunction with TSWV resistance.
Wild species of peanut have been sources of resistance for many pests and diseases in peanut. Resistance to thrips is no exception. The most remarkable being the production of varieties with resistance to root-knot nematode, leaf spot, and to rust, all derived from wild species (Simpson, 1991; Simpson et al., 2003; Khedikar et al., 2010; Chu et al., 2011). Several wild species have been associated with thrips resistance in peanut. Stalker and Campbell (1983) evaluated more than 30 wild species of Arachis over 3 years through field screening and identified several species that possess resistance to thrips. The number of thrips damaged leaves per plot on A. batizocoi, Arachis correntina (Benth.), Arachis villosa Benth., Arachis spegazzini Greg., A. chacoense, Arachis cardenasii Krapov., A. stenosperma Krapov. & W. C. Greg., Arachis duranensis Krapov, Arachis rigonii Krapov., Arachis paraguariensis Chodat & Hassl., Arachis pusilla Benth., and Arachis repens Handro, were two to ∼100 times less than on peanut cultivars. Similar evaluations were conducted in other places in the Southern United States as well as in Asia, and several genotypes were found to possess thrips resistance (Amin and Mohammed, 1980; Lynch, 1990). Diploid wild species such as Arachis vallsii Krapov. & W. C. Greg., Arachis kempff-mercadoi Krapov., W. C. Greg. & C. E. Simpson, Arachis williamsii Krapov. & W. C. Greg., A. duranensis, and amphidiploids such as A. batizocoi × A. kempff-mercadoi, A. gregoryi × A. stenosperma, A. magna × A. cardenasii also exhibited substantial levels of resistance to E. flavens in South America (Michelotto et al., 2017). Most of these evaluations were based on field screening, and the mechanism by which resistance is conferred in some of these wild species is not known. Few studies have examined the resistance contributing factors in these wild species. Epicuticular waxes containing n-alkanes from A. batizocoi, A. chacoense, A. paraguariensis, A. glandulifera, and A. ipaensis were speculated to confer resistance to thrips and other sucking insects, probably through non-preference/antixenosis (Yang et al., 1993). The same group of compounds, were also identified in Arachis monticola Krapov. & Rigoni and A. stenosperma, through gas chromatography (de Souza et al., 2010). In Brazil, another study showed that various wild species exhibited field resistance to E. flavens (Michelotto et al., 2017). For a more comprehensive list of wild species with resistance to thrips and TSWV, please refer to review by Stalker (2017).
While there is evidence that wild species exhibit resistance against thrips, their ploidy level, makes it difficult to incorporate their resistance into cultivated tetraploid peanuts. Most wild species are diploids. Resistance introgression from wild species is achieved by crossing and subsequent backcrossing. This process is time consuming, and a major concern has been the reduction of level of resistance while backcrossing. Arachis diogoi is a diploid wild species that is known to possess resistance to numerous pests and pathogens including TSWV (Lyerly et al., 2002). Attempts were made to introgress TSWV resistance from A. diogoi to the Virginia type cultivar Gregory, using the hexaploid route (Stalker HT et al., 1979; Figure 6). Several progenies from those crosses accumulated fewer copies of the virus when compared with Gregory (Lai, 2015). Subsequently evaluations for thrips resistance were done using microcosmic Munger cages (Lai, 2015). Results reiterated that A. diogoi was highly resistant to thrips when compared with the cultivar Gregory; however, the genotypes from the intraspecific crosses were not anymore resistant than the recurrent parent Gregory (Lai, 2015). Thrips fitness parameters such as developmental time and fecundity were evaluated, and thrips developmental time was (20%) longer and its fecundity was lower on A. diogoi than on tetraploid Gregory. These results revealed direct effects on thrips biology suggesting that there could be antibiosis-based resistance against thrips.

FIGURE 6. Leaf phenotype of a diploid wild species, Arachis diogoi (A), cultivated tetraploid Arachis hypogaea cultivar Gregory (C), and a cross between the two (B).
A different route of introgression, called the tetraploid route involves crossing two wild species of complementary genomes and doubling the resultant sterile diploid hybrid with colchicine (Simpson, 1991; Leal-Bertioli et al., 2012, 2015). The resultant synthetic allotetraploid possesses all genes from both wild species and is compatible with peanut, being, therefore, useful for breeding. In Brazil, synthetics developed using this strategy were evaluated for field resistance to thrips, and three of them [(A. batizocoi × A. kempff-mercadoi)4x, (A. gregoryi × A. stenosperma)4x, and (A. magna × A. cardenasii)4x] were found to have superior resistance than the cultivars tested, and are, therefore potentially useful for breeding (Michelotto et al., 2017).
Availability of novel tools and resources could facilitate a renewed interest in breeding for thrips resistant peanut cultivars, a few of them are discussed below.
Improvement in traditional and novel breeding approaches will result in hundreds and hundreds of peanut genotypes that need to be screened for thrips resistance. Until recently, most phenotypic evaluations for thrips resistance have focused on field screening for foliar feeding damage. Recently, a few studies have performed laboratory experiments to examine resistance to thrips in peanut (Shrestha et al., 2013; Sundaraj et al., 2014). These assays were used to evaluate both behavior such as preference and end-point parameters such as thrips feeding damage and biological fitness. These tests need to be conducted for weeks if not months, and could be laborious especially if there are many genotypes involved. Until recently, there was no automated high-throughput screening tool available for thrips, but have been available for hemipteran insects such as aphids. Recently, one such automated thrips-resistance screening tool has become available (Thoen et al., 2016). Thoen et al. (2016) used an automated video monitoring parallel choice test platform to screen ∼350 Arabidopsis accessions within a week. By using this assay, they could measure parameters that are obtained in traditional choice tests such as leaf damage and number of nymphs produced within a certain time. In addition to these usual parameters, they were also able to get information pertaining to thrips behavior on the host plant using a behavior analyzing software Ethovision® XT 10. The parameters estimated included time spent on the foliage, time not moving on the foliage, time not moving, distance moved, and movement velocity. Assessing these parameters for peanut genotype screening against thrips will provide more insights into thrips-peanut plant interactions facilitate faster and better screening for resistance. Electronic nose is another tool that could be used for quick and efficient screening for thrips resistance in peanut. This technique was found to be useful to discriminate Western flower thrips resistant genotypes from susceptible chrysanthemum genotypes effectively using headspace volatile profiles following simulated thrips feeding and thrips feeding bioassays (McKellar et al., 2005). The usefulness of such a technique needs to be explored in screening for thrips resistance in peanut.
Genomic tools have been used for crop breeding to develop improved varieties. In particular, the application of trait-linked DNA markers to facilitate trait selection (Marker Assisted Selection – MAS) for crop improvement have proved successful for major crops. MAS uses trait-linked markers, instead of trait itself, has the advantage of eliminating plants with undesirable gene combinations, allowing the breeder to concentrate on a lower number of better lines. It has proven successful for cultivar improvement on many major crops. Peanut, however, has lagged behind the major crops due to the intrinsic narrow genetic variability delaying the identification of markers useful for selection. With a concerted effort by the Peanut Genome Initiative, an international group of scientists, genetic and genomic tools became available to the community and made marker assisted selection a reality in for peanut breeding (Stalker and Mozingo, 2001; Wang et al., 2018). Various marker systems have been developed throughout the years, following technology development (for a comprehensive review, see Wang et al., 2018). Recently, the most significant achievement was the sequence of the genome of the two wild progenitors of peanut, A. duranensis and A. ipaensis (Bertioli et al., 2016) and the development of a publicly available genotyping tool: a chip with nearly 60,000 SNPs (Clevenger et al., 2017; Pandey et al., 2017).
To date, only one study was conducted that showed correlation of a molecular marker with a peanut virus vector, Aphis craccivora, which transmits Groundnut rosette virus (Genus Umbravirus; Family Tombusviridae) (Herselman et al., 2004). In other crops (but not peanuts), markers have been associated with resistance to thrips (e.g., cowpea Lucas et al., 2012; pepper Maharijaya et al., 2015; tomato, Escobar-Bravo et al., 2017). For peanut, we still need to understand better the nature of thrips resistance and develop molecular markers.
Research on Orthotospovirus resistance, on the other hand, have seen more advances. For instance, TSWV host resistance has been established as a major factor to reduce disease risk, and therefore breeding for resistance has become a major goal in the breeding programs in the United States. Phenotypic selection for TSWV is inaccurate as field resistance expression varies significantly from year to year, depending on the environment (Culbreath et al., 2010; Tseng et al., 2016). In vitro transmission is also not considered reliable as it bypasses some possible plant defense responses (Zhao et al., 2018). Therefore, breeding for resistance to orthotospoviruses such as TSWV could greatly be facilitated by implementation of MAS in breeding programs. The first population, major QTLs were found in LG A01, and in the second study, 11 minor QTLs were found in different regions of the genome (Khera et al., 2016; Pandey et al., 2017). Work with another population, Florida EPTM13 x Georgia Valencia revealed major QTLs on LG01, and markers tightly linked to TSWV resistance (Tseng et al., 2016; Zhao et al., 2018). Association genetics using these markers on the United States minicore collection, however, did not show association between TSWV resistance and the markers (Li et al., 2018; Zhao et al., 2018). This is probably because the resistance allelic region to which the marker is associated is from a unique source, which is not present in the United States peanut mini core gene pool. This well documented study flags caution on the use of correct and validated molecular markers on genotypes with the same allele variants.
All the studies described above were conducted mainly using Simple Sequence Repeat (SSR) markers, which are costly, time consuming and not very abundant. New genotyping methods using Single Nucleotide Polymorphism (SNPs) are currently available (Bertioli et al., 2014; Clevenger et al., 2017; Pandey et al., 2017) and, together with more precise genotyping, are likely to speed the discovery of tightly linked markers and consequently, the implementation of MAS for in peanut breeding against thrips and viruses. MAS is not the only, or the main one, but it is undoubtedly, a very useful tool in the breeder’s tool box.
Peanut transformation was first reported by Brar et al. (1994) but no resistance tests were performed. Peanut has been transformed with cry genes derived from Bacillus thuringiensis (Berliner) that confer resistance to several insects, especially of the order Lepidoptera (Krishna et al., 2015). However, to our knowledge, no transgenic peanut has been tested or found to be resistant to thrips. In other crops, transgenesis has been successful to transfer resistance to thrips: resistance to western flower thrips has been engineered in potato plants (Outchkourov et al., 2004). Members of inhibitors of cysteine and aspartic proteases viz., stefin, potato cystatin, equistatin, and cystatin were fused and made into a functional unit and expressed in potato plants. The plants containing these multidomain proteins had fewer larvae and adults when compared with non-transgenic control plants (Outchkourov et al., 2004).
Resistance to orthotospoviruses in transgenic peanut has been achieved multiple times. In the United States and in India, both Valencia and runner genotypes were transformed with TSWV/GBNV N-gene or coat protein-based constructs, and pathogen-derived resistance was achieved in these transformants. Transformed peanut seedlings provided substantial field or in vitro resistance against TSWV and GBNV (Li et al., 1997; Magbanua et al., 2000; Yang et al., 2004; Rao et al., 2013). Mehta et al. (2013) also obtained transgenic peanut resistant to Peanut stem necrosis virus. So, the potential for viral resistance is very significant.
RNA interference has been deployed to provide resistance against a wide range of organisms including viruses and insects (Whyard et al., 2009; Gan et al., 2010; Zhang et al., 2013). In this process, the dsRNA pertaining to an invading organism is degraded into short interfering (si) RNAs (20–23 nucleotides long) with the help of an enzyme complex, and subsequently the siRNAs block the mRNA translation (Fire et al., 1998; Fire, 2007). RNAi and its usefulness have not been demonstrated in peanuts against viruses and/or thrips. Nevertheless, the usefulness of RNAi has been demonstrated with F. occidentalis, wherein double stranded RNA (dsRNA) pertaining to an important enzyme (vacuolar ATP-Synthase or V-ATPase) was silenced, following which a significant reproductive fitness was observed in F. occidentalis (Badillo-Vargas et al., 2015). Advancements in thrips genomics and transcriptomics has led to identification of various developmental genes associated with western flower thrips and tobacco thrips in recent years (Schneweis et al., 2017; Shrestha et al., 2017). The usefulness of these genes should first be validated through in vitro assays, and further their in planta expression and effectiveness against thrips should be attempted to examine the usefulness of RNAi as a management option. Though consumer preference will keep transgenic peanut from coming to the marketplace anytime soon, available transgenic technology does offer some very interesting research possibilities. Technologies such as RNAi could be modified in certain ways to circumvent plant-based transgene delivery. In fact, the versatility of RNAi has been demonstrated in other cropping systems by mere exogenous application. For instance, Gogoi et al. (2017) observed that exogenous application of Zucchini yellow mosaic (Genus Potyvirus; Family Potyviridae) virus specific dsRNA molecules when applied to tomato plants were detected in the plants up to 14 days post application, and they were also detected in phloem feeding insects such as aphids and whiteflies up to 10 days post application (Gogoi et al., 2017). In another study, exogenous application of dsRNA specific to Tobacco mosaic virus (TMV) (Genus Tobamovirus; Family Virgaviridae) limited the systemic movement of TMV and conferred resistance to TMV inoculation (Konakalla et al., 2016). RNAi is very specific to the target organism and should greatly minimize non-target effects, and therefore could be highly useful and acceptable especially when an exogenous application strategy is considered. This RNAi strategy (exogenous application) will help alleviate the regulatory hurdles and consumer concerns associated with peanut or any other crop. However, this technology is just emerging, its suitability and its commercial viability for peanut production need to be developed and explored in greater depth. Nevertheless, it is promising and reassuring that thrips management in peanut may not have to exclusively rely on spraying insecticides in the long run. Stacking thrips resistance with pathogen resistance could have multifaceted benefits to peanut growers worldwide.
Even though thrips-transmitted viruses have garnered substantial attention in the last two decades, it is abundantly clear that thrips continue to cause damage by direct feeding in peanut production systems throughout the world. Host-resistance to orthotospoviruses is the main management option adopted in peanut, but it is used in conjunction with cultural practices and insecticide applications targeted at thrips. Unlike other crops such as tomato and pepper where the resistance is governed by a major gene, neither is resistance to orthotospoviruses complete nor is the mechanism of resistance known in peanut (Shrestha et al., 2013; Sundaraj et al., 2014). Studies suggest that virus resistance in peanut may not be an exclusive result of host–virus interactions, but could also be influenced by peanut–thrips interactions (do Nascimento et al., 2006; Sundaraj et al., 2014; Shrestha et al., 2015). A. hypogaea, an allotetraploid, has a very narrow genetic base and has little to no inherent resistance to thrips and/or viruses transmitted by them (Ratnaparkhe et al., 2011). Numerous studies have documented that wild species source of resistance for thrips and orthotospoviruses. Therefore, it may be possible to incorporate resistance to both thrips and the virus simultaneously. Differences in ploidy level and the subsequent dilution effect in resistance introgression due to backcrossing represent the biggest hurdle for using wild species in breeding for resistance. This obstacle could be overcome by using novel strategies such as development of synthetic tetraploids (Leal-Bertioli et al., 2015). Molecular markers could also be potentially used to link thrips and virus resistance (Bertioli et al., 2014). Stacking resistance against thrips and orthotospoviruses could reduce selection pressure on the virus itself, delay or prevent evolution of highly virulent strains, prolong the usefulness of resistant cultivars, reduce insecticide usage and allied non-target effects, and promote sustainability in peanut production.
RS developed the idea and contributed to writing. MA co-developed the idea and contributed to writing. P-CL conducted the experiments included in manuscript and contributed to writing. AC contributed to writing. ST was a collaborator on Arachis diogoi work and contributed to writing. SL-B contributed to writing.
The authors appreciate the funding support received from grower-funded National Peanut Board and the Georgia Peanut Commission to pursue peanut-thrips research outlined in the Southeastern United States. Funding from other agencies including United States Department of Agriculture National Institute of Food and Agriculture and Georgia Seed Foundation area also acknowledged.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The technical help rendered by Mr. Simmy Mckeown and Ms. Sheran Thompson as part of the peanut entomology program at the University of Georgia Tifton campus is well appreciated.
Abudulai, M., Salifu, A., and Haruna, M. (2006). Screening of cowpeas for resistance to the flower bud thrips, Megalurothrips sjostedti Trybom (Thysanoptera: Thripidae). J. Appl. Sci. 6, 1621–1624. doi: 10.3923/jas.2006.1621.1624
Amin, P. W., and Mohammed, A. B. (1980). “Groundnut pest research at ICRISAT,” in Proceedings of the International Workshop on Groundnuts, ICRISAT Center, Patancheru, 157–168.
Amin, P. W., Singh, K. N., Dwivedi, S. L., and Rao, V. R. (1985). Sources of resistance to the jassid (Empoasca kerri Pruthi), thrips (Frankliniella schultzei (Trybom)) and termites (Odontotermes sp.) in groundnut (Arachis hypogaea L.). Peanut Sci. 12, 58–60. doi: 10.3146/pnut.12.2.0002
Badillo-Vargas, I. E., Rotenberg, D., Schneweis, B. A., and Whitfield, A. E. (2015). RNA interference tools for the western flower thrips, Frankliniella occidentalis. J. Insect Physiol. 76, 36–46. doi: 10.1016/j.jinsphys.2015.03.009
Bertioli, D. J., Cannon, S. B., Froenicke, L., Huang, G., Farmer, A. D., Cannon, E. K., et al. (2016). The genome sequences of Arachis duranensis and Arachis ipaensis, the diploid ancestors of cultivated peanut. Nat. Genet. 48, 438–446. doi: 10.1038/ng.3517
Bertioli, D. J., Ozias-Akins, P., Chu, Y., Dantas, K. M., Santos, S. P., Gouvea, E., et al. (2014). The use of SNP markers for linkage mapping in diploid and tetraploid peanuts. G3 4:89. doi: 10.1534/g3.113.007617
Bielza, P. (2008). Insecticide resistance management strategies against the western flower thrips, Frankliniella occidentalis. Pest. Manag. Sci. 64, 1131–1138. doi: 10.1002/ps.1620
Bielza, P., Espinosa, P. J., Quinto, V., Abellán, J., and Contreras, J. (2007). Synergism studies with binary mixtures of pyrethroid, carbamate and organophosphate insecticides on Frankliniella occidentalis (Pergande). Pest. Manag. Sci. 63, 84–89. doi: 10.1002/ps.1328
Bielza, P., Quinto, V., Grávalos, C., Fernández, E., and Abellán, J. (2008). Impact of production system on development of insecticide resistance in Frankliniella occidentalis (Thysanoptera: Thripidae). J. Econ. Entomol. 101, 1685–1690. doi: 10.1093/jee/101.5.1685
Brar, G. S., Cohen, B. A., Vick, C. L., and Johnson, G. W. (1994). Recovery of transgenic peanut (Arachis hypogaea L.) plants from elite cultivars utilizing ACCELL technology. Plant J. 5, 745–753. doi: 10.1111/j.1365-313X.1994.00745.x
Brown, S., Todd, J., and Culbreath, A. (1996). Effect of selected cultural practices on incidence of Tomato spotted wilt virus and populations of thrips vectors in peanuts. Tospoviruses Thrips Floral Veg. Crops 431, 491–498. doi: 10.17660/ActaHortic.1996.431.45
Campbell, W. V., and Wynne, J. C. (1980). “Resistance to groundnuts to insects and mites,” in Proceedings of the International Workshop on Groundnuts. (International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Patancheru, 149–157.
Cheng, D., Kirk, H., Vrieling, K., Mulder, P. P., and Klinkhamer, P. G. (2011). The relationship between structurally different pyrrolizidine alkaloids and western flower thrips resistance in F2 hybrids of Jacobaea vulgaris and Jacobaea aquatica. J. Chem. Ecol. 37:1071. doi: 10.1007/s10886-011-0021-6
Chu, Y., Wu, C. L., Holbrook, C. C., Tillman, B. L., Person, G., and Ozias-Akins, P. (2011). Marker-assisted selection to pyramid nematode resistance and the high oleic trait in peanut. Plant Genome 4, 110–117. doi: 10.3835/plantgenome2011.01.0001
Clevenger, J., Chu, Y., Chavarro, C., Agarwal, G., Bertioli, D. J., Leal-Bertioli, S. C. M., et al. (2017). Genome-wide SNP Genotyping Resolves signatures of selection and tetrasomic recombination in peanut. Mol. Plant. 10, 309–322. doi: 10.1016/j.molp.2016.11.015
Clevenger, J., Chu, Y., Chavarro, C., Botton, S., Culbreath, A., Isleib, T. G., et al. (2018). Mapping late leaf spot resistance in peanut (Arachis hypogaea) using QTL-seq reveals markers for marker-assisted selection. Front. Plant Sci. 9:83. doi: 10.3389/fpls.2018.00083
Coudriet, D. L., Kishaba, A. N., McCreight, J. D., and Bohn, G. W. (1979). Varietial resistance in onions to thrips. J. Econ. Entomol. 72, 614–615.
Culbreath, A., Todd, J., and Brown, S. (2003). Epidemiology and management of tomato spotted wilt in peanut. Annu. Rev. Phytopathol. 41, 53–75. doi: 10.1146/annurev.phyto.41.052002.095522
Culbreath, A., Todd, J., Demski, J., and Chamberlin, J. (1992). Disease progress of spotted wilt in peanut cultivars Florunner and Southern Runner. Phytopathology 82, 766–771. doi: 10.1094/Phyto-82-766
Culbreath, A. K., Todd, J. W., Gorbet, D. W., Branch, W. D., Sprenkel, R. K., Shokes, F. M., et al. (1996). Disease progress of Tomato spotted wilt virus in selected peanut cultivars and advanced breeding lines. Plant Dis. 80, 70–73. doi: 10.1094/PD-80-0070
Culbreath, A. K., Selph, A. C., Williams, B. W., Kemerait, R. C., Srinivasan, R., Abney, M. R., et al. (2016). Effects of new field resistant cultivars and in-furrow applications of phorate insecticide on tomato spotted wilt of peanut. Crop Protect. 81, 70–75. doi: 10.1016/j.cropro.2015.12.002
Culbreath, A. K., and Srinivasan, R. (2011). Epidemiology of spotted wilt disease of peanut caused by Tomato spotted wilt virus in the southeastern U.S. Virus Res. 159, 101–109. doi: 10.1016/j.virusres.2011.04.014
Culbreath, A. K., Tillman, B. L., Gorbet, D. W., Holbrook, C. C., and Nischwitz, C. (2008). Response of new field-resistant peanut cultivars to twin-row pattern or in-furrow applications of phorate for management of spotted wilt. Plant Dis. 92, 1307–1312.
Culbreath, A. K., Tillman, B. L., Tubbs, R. S., Beasley, J. P., Kemerait, R. C., and Brenneman, T. B. (2010). Interactive effects of planting date and cultivar on tomato spotted wilt of peanut. Plant Dis. 94, 898–904. doi: 10.1094/PDIS-94-7-0898
Damon, S. J., Groves, R. L., and Havey, M. J. (2014). Variation for epicuticular waxes on onion foliage and impacts on numbers of onion thrips. J. Am. Soc. Hortic. Sci. 139, 495–501.
de Jager, C. M., Butôt, R. P. T., Klinkhamer, P. G. L., Jong, T. J., Wolff, K., and Meijden, E. (1995). Genetic variation in chrysanthemum for resistance to Frankliniella occidentalis. Entomologia Experimentalis et Applicata 77, 277–287. doi: 10.1111/j.1570-7458.1995.tb02325.x
de Moraes, A. R. A., Lourenção, A. L., de Godoy, I. J., and Teixeira, G. D. C. (2005). Infestation by Enneothrips flavens Moulton and yield of peanut cultivars. Sci. Agric. 62, 469–472. doi: 10.1590/S0103-90162005000500010
de Souza R.J.C., Silva, S. I., and de Oliveira, A. F. M. (2010). Chemical similarity among domesticated and wild genotypes of peanut based on n-alkanes profiles. Pesquisa Agropecuária Brasileira 45, 1321–1323. doi: 10.1590/S0100-204X2010001100013
Diaz-Montano, J., Fuchs, M., Nault, B. A., and Shelton, A. M. (2010). Evaluation of onion cultivars for resistance to onion thrips (Thysanoptera: Thripidae) and Iris yellow spot virus. J. Econ. Entomol. 103, 925–937. doi: 10.1603/EC09263
do Nascimento, L. C., Pensuk, V., da Costa, N. P., de Assis Filho, F. M., Pio-Ribeiro, G., Deom, C. M., et al. (2006). Evaluation of peanut genotypes for resistance to Tomato spotted wilt virus by mechanical and thrips inoculation. Pesquisa Agropecuária Brasileira 41, 937-942. doi: 10.1590/S0100-204X2006000600006.
Dwivedi, S. L., Nigam, S. N., Reddy, D. V. R., Reddy, A. S., and Ranga Rao, G. V. (1995). “Progress in breeding groundnut varieties resistant to peanut bud necrosis virus and its vector,” in Recent Studies on Peanut Bud Necrosis Disease: Proceedings of a Meeting, eds A. A. M. Buiel, J. E. Parlevliet, and J. M. Lenne (Nairobi: ICRISAT Asia Center), 35–40.
Ekvised, S., Jogloy, S., Akkasaeng, C., Keerati-Kasikorn, M., Kesmala, T., Buddhasimma, I., et al. (2006a). Field evaluation of screening procedures for thrips resistance in peanut. Asian J. Plant Sci. 5, 838–846. doi: 10.3923/ajps.2006.838.846
Ekvised, S., Jogloy, S., Akkasaeng, C., Keerati-Kasikorn, M., Kesmala, T., Buddhasimma, I., et al. (2006b). Heritability and correlation of thrips resistance and agronomic traits in peanut. Asian J. Plant Sci. 5, 923–931. doi: 10.3923/ajps.2006.923.931
Escobar-Bravo, R., Klinkhamer, P. G. L., and Leiss, K. A. (2017). Induction of jasmonic acid-associated defenses by thrips alters host suitability for conspecifics and correlates with increased trichome densities in tomato. Plant Cell Physiol. 58, 622–634. doi: 10.1093/pcp/pcx014
Espinosa, P. J., Contreras, J., Quinto, V., Grávalos, C., Fernández, E., and Bielza, P. (2005). Metabolic mechanisms of insecticide resistance in the western flower thrips, Frankliniella occidentalis (Pergande). Pest. Manag. Sci. 61, 1009–1015. doi: 10.1002/ps.1069
Fire, A. Z. (2007). Gene silencing by double-stranded RNA (Nobel lecture). Cell Death. Differ. 14, 1998–2012. doi: 10.1038/sj.cdd.4402253
Fire, A. Z., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic inter- ference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Funderburk, J. E., Gorbet, D. W., Teare, I. D., and Stavisky, J. (2007). Thrips injury can reduce peanut yield and quality under conditions of multiple stress. Agron. J. 90, 563–566. doi: 10.2134/agronj1998.00021962009000040020x
Galvez, H. F., Fernandez, E. C., and Hautea, D. M. (2005). Molecular mapping of resistance to thrips in potato. Philipp. Agric. 88, 268–280.
Gan, D., Zhang, J., Jiang, H., Jiang, T., Zhu, S., and Cheng, B. (2010). Bacterially expressed dsRNA protects maize against SCMVinfection. Plant Cell Rep. 29, 1261–1268. doi: 10.1007/s00299-010-0911-z
Garcia, L. E., Brandenburg, R. L., and Bailey, J. E. (2000). Incidence of Tomato spotted wilt virus (Bunyaviridae) and tobacco thrips in Virginia-type peanuts in North Carolina. Plant Dis. 84, 459–464. doi: 10.1094/PDIS.2000.84.4.459
Gaum, W. G., Giliomee, J. H., and Pringle, K. L. (1994). Resistance of some rose cultivars to the western flower thrips, Frankliniella occidentalis (Thysanoptera: Thripidae). Bull. Entomol. Res. 84, 487–492. doi: 10.1017/S0007485300032715
Gogoi, A., Sarmah, N., Kaldis, A., Perdikis, D., and Voloudakis, A. (2017). Plant insects and mites uptake double-stranded RNA upon its exogenous application on tomato leaves. Planta 246, 1233–1241. doi: 10.1007/s00425-017-2776-7
Herbert, D. A. Jr., Malone, S., Aref, S., Brandenburg, R. L., Jordan, D. L., Royals, B. M., et al. (2007). Role of insecticides in reducing thrips injury to plants and incidence of tomato spotted wilt virus in virginia market-type peanut. J. Econ. Entomol. 100, 1241–1247. doi: 10.1603/0022-0493(2007)100[1241:ROIIRT]2.0.CO;2
Herbert, D. A., Malone, S., and Goerger, C. (2005). Evaluation of selected in-furrow and foliar applied insecticides for control of thrips in peanut. Arthropod. Manag. Tests 30:F57.
Herselman, L., Thwaites, R., Kimmins, F. M., Courtois, B., van der Merwe, P. J., and Seal, S. E. (2004). Identification and mapping of AFLP markers linked to peanut (Arachis hypogaea L.) resistance to the aphid vector of groundnut rosette disease. Theor. Appl. Genet. 109, 1426–1433. doi: 10.1007/s00122-004-1756-z
Holbrook, C. C., Timper, P., Culbreath, A. K., and Kvien, C. K. (2008). Registration of ‘Tifguard’ Peanut. J. Plant Registr. 2, 92–94. doi: 10.3198/jpr2007.12.0662crc
Huseth, A. S., Chappell, T. M., Langdon, K., Morsello, S. C., Martin, S., Greene, J. K., et al. (2016). Frankliniella fusca resistance to neonicotinoid insecticides: an emerging challenge for cotton pest management in the eastern United States. Pest Manag. Sci. 72, 1934–1945. doi: 10.1002/ps.4232
Johnson, W. C., Brenneman, T. B., Baker, S. H., Johnson, A. W., Sumner, D. R., and Mullinix, B. G. (2001). Tillage and pest management considerations in a peanut–cotton rotation in the southeastern coastal plain. Agron. J. 93, 570–576. doi: 10.2134/agronj2001.933570x
Jones, D. R. (2005). Plant viruses transmitted by thrips. Eur. J. Plant Pathol. 113, 119–157. doi: 10.1007/s10658-005-2334-1
Kandakoor, B. S., Khan, K., Chakravarthy, A. K., Ashok Kumar, C. T., and Venkataravana, P. (2014). Biochemical constituents influencing thrips resistance in groundnut germplasm. J. Environ. Biol. 35, 675–681.
Kesmala, T., Jogloy, S., Wongkaew, S., Akkasaeng, C., Vorasoot, N., and Patanothai, A. (2004). Heritability and phenotypic correlation of resistance to Peanut bud necrosis virus (PBNV) and agronomic traits in peanut Songklanakarin. J. Sci. Technol. 26, 129–138.
Khedikar, Y. P., Gowda, M. V. C., Sarvamangala, C., Patgar, K. V., Upadhyaya, H. D., and Varshney, R. V. (2010). A QTL study on late leaf spot and rust revealed one major QTL for molecularbreeding for rust resistance in groundnut (Arachis hypogaea L.). Theor. Appl. Genet. 121, 971–984. doi: 10.1007/s00122-010-1366-x
Khera, P., Pandey, M. K., Wang, H., Feng, S., Qiao, L., Culbreath, A. K., et al. (2016). Mapping Quantitative Trait Loci of resistance to tomato spotted wilt virus and leaf spots in a recombinant inbred line population of peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022. PLoS One 11:e0158452. doi: 10.1371/journal.pone.0158452
Kinzer, D. R., Pitts, J. T., Walton, R. R., and Kirby, J. S. (1973). Thrips resistance in plant introductions and in selections made for peanut improvement in Oklahoma. J. Econ. Entomol. 66, 91–95. doi: 10.1093/jee/66.1.91
Kogan, M., and Ortman, E. E. (1978). Antixenosis - a new term proposed to replace Painter’s “nonpreference” modality of resistance. Bull. Entomol. Soc. Am. 24, 175–176. doi: 10.1093/besa/24.2.175
Konakalla, N. C., Kaldis, A., Berbati, M., Masarapu, H., and Voloudakis, A. E. (2016). Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 244, 961–969. doi: 10.1007/s00425-016-2567-6
Krishna, G., Singh, B. K., Kim, E.-K, Morya, V. K., and Ramteke, P. W. (2015). Progress in genetic engineering of peanut (Arachis hypogaea L.)—a review. Plant Biotechnol. J. 13, 147–162. doi: 10.1111/pbi.12339
Lai, P. (2015). Evaluation of Cultural Tactics, Insecticides, and Peanut Genotypes for Thrips and Spotted Wilt Disease Management in Peanut. Doctoral dissertation, University of Georgia, Athens, GA.
Leal-Bertioli, S. C. M., Bertioli, D. J., Guimarães, P. M., Pereira, T. D., Galhardo, I., Silva, J. P., et al. (2012). The effect of tetraploidization of wild Arachis on leaf morphology and other drought-related traits. Environ. Exp. Bot. 84, 17–24. doi: 10.1016/j.envexpbot.2012.04.005
Leal-Bertioli, S. C. M., Santos, S. P., Dantas, K. M., Inglis, P. W., Nielen, S., Araujo, A. C. G., et al. (2015). Arachis batizocoi: a study of its relationship to cultivated peanut (A. hypogaea) and its potential for introgression of wild genes into the peanut crop using induced allotetraploids. Ann. Bot. 115, 237–249. doi: 10.1093/aob/mcu237
Leather, S. R., Beare, J. A., Cooke, R. C. A., and Fellowes, M. D. E. (1998). Are differences in life history parameters of the Pine Beauty Moth Panolis flammea modified by host plant quality or gender? Entomol. Expt. Appl. 87, 237–243. doi: 10.1046/j.1570-7458.1998.00327.x
Leiss, K. A., Choi, Y. H., Verpoorte, R., and Klinkhamer, P. G. L. (2011). An overview of NMR-based metabolomics to identify secondary plant compounds involved in host plant resistance. Phytochem. Rev. 10, 205–216. doi: 10.1007/s11101-010-9175-z
Leiss, K. A., Cristofori, G., van Steenis, R., Verpoorte, R., and Klinkhamer, P. G. L. (2013). An eco-metabolomic study of host plant resistance to western flower thrips in cultivated, biofortified and wild carrots. Phytochemistry 93, 63–70. doi: 10.1016/j.phytochem.2013.03.011
Leiss, K. A., Maltese, F., Choi, Y. H., Verpoorte, R., and Klinkhamer, P. G. L. (2009). Identification of chlorogenic acid as a resistance factor for thrips in chrysanthemum. Plant Physiol. 150, 1567–1575. doi: 10.1104/pp.109.138131
Lewis, T. (1997). “Pest thrips in perspective,” in Thrips as Crop Pests, ed. T. Lewis (Wallingford: CAB International), 1–13.
Li, J., Tang, Y., Jacobson, A. L., Dang, P. M., Li, X., Wang, M. L., et al. (2018). Population structure and association mapping to detect QTL controlling tomato spotted wilt virus resistance in cultivated peanuts. Crop J. 6, 516–526. doi: 10.1016/j.cj.2018.04.001
Li, Z., Jarret, R. L., and Demski, J. W. (1997). Engineered resistance to tomato spotted wilt virus in transgenic peanut expressing the viral nucleocapsid gene. Transgenic Res. 6, 297–305. doi: 10.1023/A:1018462729127
Lucas, M. R., Ehlers, J. D., Roberts, P. A., and Close, T. J. (2012). Markers for Quantitative Inheritance of resistance to foliar thrips in cowpea. Crop Sci. 52, 2075–2081. doi: 10.2135/cropsci2011.12.0684
Lyerly, J. H., Stalker, H. T., Moyer, J. W., and Hoffman, K. (2002). Evaluation of Arachis species for resistance to Tomato spotted wilt virus. Peanut Sci. 29, 79–84. doi: 10.3146/pnut.29.2.0001
Lynch, R. E. (1990). Resistance in peanut to major arthropod pests. Florida Entomol. 73:422. doi: 10.2307/3495460
Magbanua, Z. V., Wilde, H. D., Roberts, J. K., Chowdhury, K., Abad, J., Moyer, J. W., et al. (2000). Field resistance to Tomato spotted wilt virus in transgenic peanut (Arachis hypogaea L.) expressing an antisense nucleocapsid gene sequence. Mol. Breed. 6, 227–236. doi: 10.1023/A:1009649408157
Maharijaya, A. (2013) Resistance to Thrips in Pepper. Ph.D. thesis, Wageningen University, Wageningen.
Maharijaya, A., Vosman, B., Steenhuis-Broers, G., Pelgrom, K., Purwito, A., Visser, R. G. F., et al. (2015). QTL mapping of thrips resistance in pepper. Theor. Appl. Genet. 128, 1945–1956. doi: 10.1007/s00122-015-2558-1
Maharijaya, A., Vosman, B., Verstappen, F., Steenhuis-Broers, G., Mumm, R., Purwito, A., et al. (2012). Resistance factors in pepper inhibit larval development of thrips (Frankliniella occidentalis). Entomol. Exp. Appl. 145, 62–71. doi: 10.1111/j.1570-7458.2012.01304.x
Mandal, B., Jain, R. K., Krishnareddy, M., Krishna Kumar, N. K., Ravi, K. S., and Pappu, H. R. (2012). Emerging problems of Tospoviruses (Bunyaviridae) and their management in the Indian Sub-continent. Plant Dis. 96, 468–479. doi: 10.1094/PDIS-06-11-0520
Marasigan, K., Toews, M., Kemerait, J. R., Abney, M. R., Culbreath, A., and Srinivasan, R. (2016). Evaluation of alternatives to carbamate and organophosphate insecticides against thrips and Tomato spotted wilt virus in peanut production. J. Econ. Entomol. 109, 544–557. doi: 10.1093/jee/tov336
Marasigan, K., Toews, M., Kemerait, J. R., Abney, M. R., Culbreath, A., and Srinivasan, R. (2018). Evaluation of alternatives to an organophosphate insecticide with selected cultural practices: effects on thrips, Frankliniella fusca, and incidence of spotted wilt in peanut farmscapes. J. Econ. Entomol 111, 1030–1041. doi: 10.1093/jee/toy079
Maris, P. C., Joosten, N. N., Peters, D., and Goldbach, R. W. (2003a). Thrips resistance in pepper and its consequences for the acquisition and inoculation of Tomato spotted wilt virus by the western flower thrips. Phytopathology 93, 96–101. doi: 10.1094/PHYTO.2003.93.1.96
Maris, P. C., Joosten, N. N., Goldbach, R. W., and Peters, D. (2003b). Restricted spread of Tomato spotted wilt virus in thrips-resistant pepper. Phytopathology 93, 1223–1227. doi: 10.1094/PHYTO.2003.93.10.1223
McKellar, R. C., Mcgarvey, B. D., Tsao, R., Lu, X., and Knight, K. P. (2005). Application of the electronic nose to the classification of resistance to western flower thrips in chrysanthemums. J. Chem. Ecol. 31, 2439–2450. doi: 10.1007/s10886-005-7111-2
McKeown, S., Todd, J., Culbreath, A., Gorbet, D., and Weeks, J. (2001). Planting date effects on tomato spotted wilt in resistant and susceptible peanut cultivars. Phytopathology 91:S60.
Mehta, R., Radhakrishnan, T., Kumar, A., Yadav, R., Dobaria, J. R., Thirumalaisamy, P. P., et al. (2013). Coat protein-mediated transgenic resistance of peanut (Arachis hypogaea L.) to peanut stem necrosis disease through Agrobacterium-mediated genetic transformation. Indian J. Virol. 24, 205–213. doi: 10.1007/s13337-013-0157-9
Michelotto, M. D., de Godoy, I. J., Pirotta, M. Z., dos Santos, J. F., Finoto, E. L., and Pereira Fávero, A. (2017). Resistance to thrips (Enneothrips flavens) in wild and amphidiploid Arachis species. PLoS One 12:e0176811. doi: 10.1371/journal.pone.0176811
Mirnezhad, M., Romero-González, R. R., Leiss, K. A., Choi, H. K., Verpoorte, R., and Klinkhamer, P. G. L. (2010), Metabolomic analysis of host plant resistance to thrips in wild and cultivated tomatoes. Phytochem. Anal. 21, 110–117. doi: 10.1002/pca.1182
Miyazaki, J., Stiller, W. N., and Wilson, L. J. (2017). Sources of plant resistance to thrips: a potential core component in cotton IPM. Entomologia Experimentalis et Applicata 162, 30–40. doi: 10.1111/eea.12501
Molenaar, N. D. (1984). Genetics, Thrips (Thrips tabaci L.) Resistance and Epicuticular Wax Characteristics of Nonglossy and Glossy Onions (Allium cepa L.). Doctoral dissertation, Wisconsin University, Madison, WI.
Monfort, W. S., Culbreath, A. K., Stevenson, K. L., Brenneman, T. B., and Perry, C. D. (2007). Use of resistant peanut cultivars and reduced fungicide inputs for disease management in strip-tillage and conventional tillage systems. Plant Health Prog. 8. doi: 10.1094/PHP-2007-0614-01-RS
Moritz, G. (1997). “Structure, growth and development,” in Thrips as Crop Pests, ed. T. Lewis (New York, NY: CAB International), 15–63.
Mulder, P. (1999). Effects of insecticides on thrips population, peanut injury, growth and yield. Arthropod. Manag. Tests 24:F84.
Mulder, P. G., and Seuhs, S. K. (2002). Control of thrips with various insecticide formulations and methods of application on peanut. Arthropod. Manag. Tests 27:F81.
Mullin, C. A., Frazier, M., Frazier, J. L., Ashcraft, S., Simonds, R., vanEngelsdorp, D., et al. (2010). High levels of miticides and agrochemicals in North American apiaries: implications for honey bee health. PLoS One 5:e9754. doi: 10.1371/journal.pone.0009754
NASS (2017). United States Department of Agriculture National Agricultural Statistics Service. Available at: https://quickstats.nass.usda.gov/
Navia-Gine, P. A., (2012). Characterization of the Relationship between Leaf Spot Severity and Yield in New Peanut Runner-Type Cultivars and Effects of New Peanut Genotypes on Leaf Spot Epidemics. Doctoral dissertation, University of Georgia, Atlanta.
Nicodemo, D., Maioli, M. A., Medeiros, H. C. D., Guelfi, M., Balieira, K. V. B., Jong, D. D., et al. (2014). Fipronil and imidacloprid reduce honeybee mitochondrial activity. Environ. Toxicol. Chem. 33, 2070–2075. doi: 10.1002/etc.2655
Njau, G. M., Nyomora, A. M., Dinssa, F. F., Chang, J. C., Malini, P., Subramanian, S., et al. (2017). Evaluation of onion (Allium cepa) germplasm entries for resistance to onion thrips, Thrips tabaci (Lindeman) in Tanzania. Int. J. Trop. Insect Sci. 37, 98–113. doi: 10.1017/S1742758417000078
Omo-Ikerodah, E. E., Fatokun, C. A., and Fawole, I. (2009). Genetic analysis of resistance to flower bud thrips (Megalurothrips sjostedti) in cowpea (Vigna unguiculata (L.) Walp). Euphytica 165, 145–154. doi: 10.1007/s10681-008-9776-4
Outchkourov, N. S., Kogel, W. J. D., Wiegers, G. L., Abrahamson, M., and Jongsma, M. A. (2004). Engineered multidomain cysteine protease inhibitors yield resistance against western flower thrips (Frankliniella occidentalis) in greenhouse trials. Plant Biotechnol. J. 2, 449–458. doi: 10.1111/j.1467-7652.2004.00089.x
Pandey, M., Gaurav, A., Kale, S., Clevenger J., Nayak S., Sriswathi M., et al. (2017). Development and evaluation of a high density genotyping ‘Axiom_Arachis’ array with 58K SNPs for accelerating genetics and breeding in groundnut (Arachis species). Sci. Rep. 7:40577. doi: 10.1038/srep40577
Pappu, H. R., Jones, R. A. C., and Jain, R. K. (2009). Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141, 219–236. doi: 10.1016/j.virusres.2009.01.009
Qiu, W., and Moyer, J. W. (1999). Tomato spotted wilt tospovirus adapts to the TSWV N gene-derived resistance by genome reassortment. Phytopathology 89, 575–582. doi: 10.1094/PHYTO.1999.89.7.575
Rao, S. C., Bhatnagar-Mathur, P., Lava Kumar, P., Sudarshan Reddy, A., and Sharma, K. K. (2013). Pathogen-derived resistance using a viral nucleocapsid gene confers only partial non-durable protection in peanut against peanut bud necrosis virus. Arch. Virol. 158, 133–143. doi: 10.1007/s00705-012-1483-8
Ratnaparkhe, M. B., Wang, X., Li, J., Compton, R. O., Rainville, L. K., Lemke, C., et al. (2011). Comparative analysis of peanut NBS-LRR gene clusters suggests evolutionary innovation among duplicated domains and erosion of gene microsynteny. New Phytol. 192, 164–178. doi: 10.1111/j.1469-8137.2011.03800.x
Reddy, A. S., Reddy, L. J., Mallikarjuna, N., Abdurahaman, M. D., Reddy, Y. V., Bramel, P. J., et al. (2000). Identification of resistance sources to Peanut bud necrosis virus (PBNV) in wild Arachis germplasm. Ann. Appl. Biol. 137, 135–139. doi: 10.1111/j.1744-7348.2000.tb00045.x
Reddy, D. V. R., Buiel, A. A. M., Satyanarayana, T., Dwivedi, S. L., Reddy, A. S., Ratna, A. S., et al. (1995). “Peanut bud necrosis disease: an overview,” in Recent Studies on Peanut Bud Necrosis Disease: Proceedings of A Meeting, eds A. A. M. Buiel, J. E. Parlevliet, and J. M. Lenne (Nairobi: ICRISAT Asia Center), 3–7.
Riley, D. G., Joseph, S. V., Srinivasan, R., and Diffie, S. (2011). Thrips vectors of tospoviruses. J. Integ. Pest. Manag. 2, I1–I10. doi: 10.1603/IPM10020
Romero González, R. R. (2011). A Metabolomics Approach to Thrips Resistance in Peanut. Doctoral dissertation, Leiden University, Leiden.
Sakimura, K. (1963). Frankliniella fusca and additional vector for the Tomato spotted wilt virus, with notes on Thrips tabaci, another vector. Phytopathology 53, 412–415.
Schneweis, D. J., Whitfield, A. E., and Rotenberg, D. (2017). Thrips developmental stage-specific transcriptome response to tomato spotted wilt virus during the virus infection cycle in Frankliniella occidentalis, the primary vector. Virology 500, 226–237. doi: 10.1016/j.virol.2016.10.009
Schoonhoven, A. V. (1974). Resistance to thrips damage in cassava. J. Econ. Ent. 67, 728–730. doi: 10.1093/jee/67.6.728
Sharma, H. C., Pampapathy, G., Dwivedi, S. L., and Reddy, L. J. (2003). Mechanisms and diversity of resistance to insect pests in wild relatives of groundnut. J. Econ. Ent. 96, 1886–1897. doi: 10.1093/jee/96.6.1886
Shrestha, A., Champagne, D. E., Culbreath, A. K., Rotenberg, D., Whitfield, A. E., and Srinivasan, R. (2017). Transcriptome changes associated with Tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (Hinds). J. Gen. Virol. 98, 2156–2170. doi: 10.1099/jgv.0.000874
Shrestha, A., Srinivasan, R., Sundaraj, S., Culbreath, A. K., and Riley, D. G. (2013). Second generation peanut genotypes resistant to thrips-transmitted Tomato spotted wilt virus exhibit tolerance rather than true resistance and differentially affect thrips fitness. J. Econ. Entomol. 106, 587–596. doi: 10.1603/EC12430
Shrestha, A., Sundaraj, S., Culbreath, A. K., Riley, D. G., Abney, M. R., and Srinivasan, R. (2015). Effects of thrips density, mode of inoculation, and plant age on Tomato spotted wilt virus transmission in peanut plants. Environ. Entomol. 44, 136–143. doi: 10.1093/ee/nvu013
Simpson, C. E. (1991). Pathways for introgression of pest resistance into Arachis hypogaea L. Peanut Science 18, 22–26. doi: 10.3146/i0095-3679-18-1-8
Simpson, C. E., Starr, J. L., Church, G. T., Burow, M. D., and Paterson, A. H. (2003). Registration of ‘NemaTAM’ Peanut. Crop Sci. 43:1561. doi: 10.2135/cropsci2003.1561
Smith, C. M. (2005). Plant Resistance to Arthropods: Molecular and Conventional Approaches. Dordrecht: Springer. doi: 10.1007/1-4020-3702-3
Snook, M. E., Lynch, R. E., Culbreath, A. K., and Costello, C. E. (1994). 2,3-Di-(E)-caffeoyl-(2R,3R)-( + )-tartaric acid in terminals of peanut (Arachis hypogaea) varieties with different resistances to late leaf spot disease (Cercosporidium personatum) and the insects tobacco thrips (Frankliniella fusca) and potato leafhopper (Empoasca fabae). J. Agric. Food Chem. 42, 1572–1574. doi: 10.1021/jf00043a035
Srinivasan, R., Abney, M. R., Culbreath, A. K., Kemerait, R. C., Tubbs, R. S., Monfort, W. S., et al. (2017). Three decades of managing Tomato spotted wilt virus in peanut in southeastern United States. Virus Res. 241, 203–212. doi: 10.1016/j.virusres.2017.05.016
Stalker, H. T. (2017). Utilizing wild species for peanut improvement. Crop Sci. 57, 1102–1120. doi: 10.1007/BF00266988
Stalker, H. T., and Campbell, W. V. (1983). Resistance of wild species of peanut to an insect complex. Peanut Sci. 10, 30–33. doi: 10.3146/i0095-3679-10-1-9
Stalker, H. T., and Mozingo, L. G. (2001). Molecular markers of Arachis and marker-assisted selection. Peanut Sci. 28, 117–123 doi: 10.3146/i0095-3679-28-2-13
Stalker HT, Wynne JC, and Company M. (1979). Variation in progenies of an Arachis hypogaea x diploid wild species hybrid. Euphytica 28, 675–684. doi: 10.1007/BF00038934
Sundaraj, S., Srinivasan, R., Culbreath, A. K., Riley, D. G., and Pappu, H. R. (2014). Host plant resistance against Tomato spotted wilt virus in peanut (Arachis hypogaea) and its impact on susceptibility to the virus, virus population genetics, and vector feeding behavior and survival. Phytopathology 104, 202–210. doi: 10.1094/PHYTO-04-13-0107-R
Teetes, G. L. (1996). “Plant resistance to insects: a fundamental component of IPM,” in Radcliffe’s IPM World Textbook, eds Radcliffe and W. D. Hutchison (Minneapolis, MN: University of Minnesota).
Thoen, M. P. M., Kloth, K. J., Wiegers, G. L., Krips, O. E., Noldus, L. P. J. J., Dicke, M., et al. (2016). Automated video tracking of thrips behavior to assess host-plant resistance in multiple parallel two-choice setups. Plant Methods 12:1. doi: 10.1186/s13007-016-0102-1
Todd, J. W., Culbreath, A. K., Chamberlin, J. R., Beshear, R. J., and Mullinix, B. G. (1995). “Colonization and population dynamics of thrips in peanuts in the southern United States,” in Thrips Biological Management, B. Parker, M. Skinner and T. Lewis (New York, NY: Plenum Press), 453–460.
Todd, J. W., Culbreath, A. K., and Brown, M. R. (1996). Dynamics of vector populations and progress of spotted wilt disease relative to insecticide use in peanuts. Acta Hortic. 431, 483–490. doi: 10.17660/ActaHortic.1996.431.44
Tsao, R., Attygalle, A. B., Schroeder, F. C., Marvin, C. H., and McGarvey, B. D. (2003). Isobutylamides of unsaturated fatty acids from Chrysanthemum morifolium associated with host-plant resistance against the western flower thrips. J. Nat. Prod. 66, 1229–1231. doi: 10.1021/np0301745
Tseng, Y. C., Tillman, B. L., Peng, Z., and Wang, J. (2016). Identification of major QTLs underlying tomato spotted wilt virus resistance in peanut cultivar Florida-EPTM ‘113. BMC Genet. 17:128. doi: 10.1186/s12863-016-0435-9
Tubbs, R. S., John, P. Beasley, J., Culbreath, A. K., Kemerait, R. C., Smith, N. B., and Smith, A. R. (2011). Row pattern and seeding rate effects on agronomic, disease, and economic factors in large-seeded runner peanut. Peanut Sci. 38, 93–100. doi: 10.3146/ps10-19.1
Velusamy, R., and Saxena, R. C. (1991). Genetic evaluation for resistance to rice thrips (Thysanoptera: Thripidae) in leafhopper- and planthopper- resistant rice varieties. J. Econ. Entomol. 84, 664–668. doi: 10.1093/jee/84.2.664
Voorrips, R. E., Steenhuis-Broers, G., Tiemens-Hulscher, M., and Lammerts van Bueren, E. T. (2008a). “Plant traits affecting thrips resistance in cabbage,” in Proceedings of the Cultivating the Future Based on Science: 2nd Conference of the International Society of Organic Agriculture Research ISOFAR,Modena.
Voorrips, R. E., Steenhuis-Broers, G., Tiemens-Hulscher, M., and van Bueren, E. T. L. (2008b). Plant traits associated with resistance to Thrips tabaci in cabbage (Brassica oleracea var capitata). Euphytica 163:409. doi: 10.1007/s10681-008-9704-7
Wang, H., Lei, Y., Yan, Y., Wan, L., Cai, Y., Yang, Z. et al. (2018). Development and validation of simple sequence repeat markers from Arachis hypogaea transcript sequences. Crop J. 6, 172–180. doi: 10.1007/s00438-015-1115-6
Whalen, R., Herbert, D. A., and Malone, S. (2014). Influence of seed treatments and granular insecticide on two peanut cultivars for thrips management. Arthropod. Manag. Tests 39:F57.
Whyard, S., Singh, A. D., and Wong, S. (2009). Ingested double-stranded RNAs can act as species specific insecticides. Insect Biochem. Mol. Biol. 39, 824–832. doi: 10.1016/j.ibmb.2009.09.007
Yang, G., Espelie, K. E., Todd, J. W., Culbreath, A. K., Pittman, R. N., and Demski, J. W. (1993). Cuticular lipids from wild and cultivated peanuts and the relative resistance of these peanut species to fall armyworm and thrips. J. Agric. Food Chem. 41, 814–818. doi: 10.1021/jf00029a026
Yang, H., Ozias-Akins, P., Culbreath, A. K., Gorbet, D. W., Weeks, J. R., Mandal, B., et al. (2004). Field evaluation of Tomato spotted wilt virus resistance in transgenic peanut (Arachis hypogaea). Plant Dis. 88, 259–264. doi: 10.1094/PDIS.2004.88.3.259
Young, S., Kinzer, R. E., Walton, R. R., and Matlock, R. S. (1972). Field screening for tobacco thrips resistance in peanuts. J. Econ. Entomol. 65, 828–832. doi: 10.1093/jee/65.3.828
Zhang, H., Li, H. C., and Miao, X. X. (2013). Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci. 20, 15–30. doi: 10.1111/j.1744-7917.2012.01513.x
Zhang, J., Idowu, O. J., Wedegaertner, T., and Hughs, S. E. (2014). Genetic variation and comparative analysis of thrips resistance in glandless and glanded cotton under field conditions. Euphytica 199, 373–383. doi: 10.1007/s10681-014-1137-x
Zhao, Z., Tseng, Y. C., Peng, Z., Lopez, Y., Chen, C. Y., Tillman, B. L., et al. (2018). Refining a major QTL controlling spotted wilt disease resistance in cultivated peanut (Arachis hypogaea L.) and evaluating its contribution to the resistance variations in peanut germplasm. BMC Genetics 19:17. doi: 10.1186/s12863-018-0601-3
Keywords: Frankliniella fusca, peanut, resistance, wild species, vector, Orthotospovirus
Citation: Srinivasan R, Abney MR, Lai P-C, Culbreath AK, Tallury S and Leal-Bertioli SCM (2018) Resistance to Thrips in Peanut and Implications for Management of Thrips and Thrips-Transmitted Orthotospoviruses in Peanut. Front. Plant Sci. 9:1604. doi: 10.3389/fpls.2018.01604
Received: 01 June 2018; Accepted: 17 October 2018;
Published: 06 November 2018.
Edited by:
Kirsten Leiss, Wageningen University & Research, NetherlandsReviewed by:
Maria Balota, Virginia Tech, United StatesCopyright © 2018 Srinivasan, Abney, Lai, Culbreath, Tallury and Leal-Bertioli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rajagopalbabu Srinivasan, YmFidXNyaUB1Z2EuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.