- 1State Key Laboratory of Cotton Biology, Cotton Institute of the Chinese Academy of Agricultural Sciences, Key Laboratory of Cotton Genetic Improvement, Ministry of Agriculture, Anyang, China
- 2Department of Plant and Environmental Sciences, New Mexico State University, Las Cruces, NM, United States
Cotton (Gossypium spp.) is the most important natural fiber crop and the source of cottonseed oil, a basic by-product after ginning. AtWRI1 and its orthologs in several other crop species have been previously used to increase triacylglycerols in seeds and vegetative tissues. In the present study, we identified 22, 17, 9, and 11 WRI-like genes in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii, respectively. This gene family was divided into four subgroups, and a more WRI2-like subfamily was identified compared with dicotyledonous Arabidopsis. An analysis of chromosomal distributions revealed that the 22 GhWRI genes were distributed on eight At and eight Dt subgenome chromosomes. Moreover, GhWRI1a was highly expressed in ovules 20–35 days after anthesis and was selected for further functional analysis. Ectopic expression of GhWRI1a rescued the seed phenotype of a wri1-7 mutant and increased the oil content of Arabidopsis seeds. Our comprehensive genome-wide analysis of the cotton WRI-like gene family lays a solid foundation for further studies.
Introduction
Cotton, especially upland cotton (Gossypium hirsutum L.), is the main source of renewable textile fibers and is also well known for its oil-rich seeds. After ginning, fuzzy cottonseed is processed into four major products: hulls (26%), linters (9%), oil (16%), and meal (45%), with 4% lost in processing (Liu et al., 2009). The most valuable cottonseed oil is typically composed of approximately 26% palmitic acid (C16:0), 15% oleic acid (C18:1), and 58% linoleic acid (C18:2) (Liu et al., 2002). Because cotton is the world’s sixth largest source of vegetable oil (Liu et al., 2002, 2009), increasing cottonseed oil content through classical breeding techniques and biotechnological approaches is important.
Triacylglycerols (TAGs), which accumulate in plant seeds and fruits, are major renewable sources of reduced carbon used as food, industrial feedstocks, and fuel (Bates and Browse, 2012). As reviewed previously (Bates and Browse, 2012), plants use two main pathways to produce diacylglycerol (DAG), the immediate precursor molecule to TAG synthesis. AtWRI1, an AP2 transcription factor, is involved in the regulation of seed storage metabolism in Arabidopsis (Focks and Benning, 1998; Cernac and Benning, 2004). The homozygous atwri1 mutant has a wrinkled seed phenotype and exhibits an 80% reduction in seed oil content compared with wild type (WT) Arabidopsis (Focks and Benning, 1998; Cernac and Benning, 2004). Expression of AtWRI1 cDNA under the control of the cauliflower mosaic virus 35S-promoter has been found to lead to increased seed oil content and the accumulation of TAGs in developing seedlings (Cernac and Benning, 2004). The involvement of orthologous genes of AtWRI1 in the regulation of oil content has also been reported in many other plant species (Liu et al., 2010; Shen et al., 2010; Pouvreau et al., 2011; An and Suh, 2015; Grimberg et al., 2015; Yang et al., 2015; Hofvander et al., 2016; An et al., 2017). For example, in rapeseed, overexpression of WRI1-like (BnWRI1) cDNAs driven by cauliflower mosaic virus 35S-promoter results in 10–40% increased seed oil content and enlarged seed size and mass in 51 transgenic Arabidopsis lines (Liu et al., 2010). In maize, overexpression of ZmWRI1 results in an oil increase without affecting germination, seedling growth, or grain yield in transgenic maize (Shen et al., 2010; Pouvreau et al., 2011). Eighteen putative target genes of ZmWRI1a have been identified by transcriptomic experiments, 12 of which contain the cis-element bound by AtWRI1 in their upstream regions. Interestingly, the higher seed oil content is not accompanied by a reduction of starch in ZmWRI1a transgenic lines, and could be utilized in transgenic breeding (Pouvreau et al., 2011). Recently, expression of CsWRI1A, B, or C has rescued the seed phenotype of the Arabidopsis wri1-3 loss-of-function mutant (An et al., 2017).
In cotton, WRI-like genes are found to be participated in the fiber development and oil seed content. Silencing of the expression of WRINKLED1 by TRV-VIGS (tobacco rattle virus triggers virus-induced gene silencing), corresponding to GhWRI1b in our study, has been found to increase fiber length but reduce oil seed content, suggesting the possibility of increasing fiber length by repartitioning carbon flow (Qu et al., 2012). Fiber transcriptome of G. hirsutum producing short-fiber and long-fiber is compared with the transcriptome of extra-long fiber producing G. barbadense, and find that expression pattern of a Wrinkeled1 gene shows close association with fiber length. The authors speculate that Wrinkled1 transcription factor (GenBank accession number: DW505003.1), also corresponding to GhWRI1b in the present study, is involved in the development of extra-long staples in cotton (Qaisar et al., 2017). Moreover, compared with WT, overexpression of GhWRI1 (GenBank accession number: JX270189), corresponding to GhWRI1b in our study, has been observed to increase seed lipid content and decrease protein content in transgenic upland cotton (Liu et al., 2018).
Four WRI1-like genes, named AtWRI1-4, are present in Arabidopsis (To et al., 2012). Seed-specific overexpression of AtWRI3 and AtWRI4, but not AtWRI2, can suppress the wrinkled phenotype of wri1-4 and restore normal oil accumulation (To et al., 2012). These results imply that WRI-like family genes play important roles in the developmental regulation of fatty acid and TAG production in plants. In this study, we performed a comprehensive genome-wide analysis to further understand the complexity of WRI-like family genes in cotton. In addition, a transgenic approach was used to clarify the function of GhWRI1a in TAG production.
Materials and Methods
Sequence Retrieval, Multiple Sequence Alignment, and Phylogenetic Analysis
Genome sequences of G. arboreum (A2, BGI_V1.0) (Li et al., 2014), G. raimondii (D5, BGI_V1.0) (Wang et al., 2012), G. hirsutum acc. TM-1 (AD1, NBI_V1.1) (Zhang et al., 2015), and G. barbadense acc.3-79 (AD2, SGI_V1.0) (Yuan et al., 2015) were downloaded from the CottonGen website1. AtWRI1, AtWRI2, AtWRI3, and AtWRI4 were acquired from TAIR 102. WRI-like genes in cacao were acquired from the cacao genome database3. WRI-like genes in rice were acquired from the rice annotation project database4. To identify WRI-like genes from Gossypium, AtWRI1, AtWRI2, AtWRI3, and AtWRI4 protein sequences were used as queries against the above-mentioned cotton genomes. ClustalX version 2.0 (Larkin et al., 2007) was used to perform multiple sequence alignments of all identified WRI-like genes in this study (Supplementary File 1). A phylogenetic analysis was carried out using the neighbor-joining algorithm with the pairwise deletion option, Poisson correction model, and uniform rates, with the statistical reliability of the resulting tree evaluated using 1,000 bootstrap replicates (Tamura et al., 2013). The online ExPASy tool5 was used to calculate the sequence length, theoretical molecular weight (MW), and isoelectric point (pI) of WRI-like proteins.
Chromosomal Location, Gene Duplication, and Gene Loss
MapChart6 was used to visualize the mapping of WRI-like genes (Voorrips, 2002). Gene duplication events were defined as previously described criteria (Dong et al., 2016; Cui et al., 2017). Gene loss evens were analyzed based on the best match and the syntenic blocks in the CottonGen website7. DnaSP software of phylogenetic analysis by the maximum likelihood method was used to calculate Ka and Ks of the duplicated gene pairs.
Genetic Structure Analysis and Protein Domain Detection
GhWRI gene structures were generated using the Gene Structure Display Server (GSDS)8. The SMART database9 was used for detection of GhWRI protein domains.
Expression Pattern Analysis of GhWRI Genes Based on RNA-Seq Data
FPKM values of GhWRI genes were calculated using previously reported RNA-seq data of 22 cotton tissues (SRA accession code: PRJNA248163) (Zhang et al., 2015).
Transgenic Plant Generation and Expression Analysis
The complete coding sequence of GhWRI1a (Supplementary File 2) was amplified with gene specific primers. The resulting PCR product was cloned into a digested pBI121 vector with BamH I and Sac I using ClonExpress® II One Step Cloning Kit (Vazyme, Nanjing, China). We used Agrobacterium tumefaciens strain GV3101 containing this binary construct to transform Arabidopsis plants. Transformants were selected on MS medium supplemented with kanamycin (50 mg/L). The progeny of transformants showed an approximately 3:1 segregation of live and dead phenotypes, and homozygous lines in the T3 generation were used for further analysis (Zang et al., 2017). To detect the relative expression level of GhWRI1a in the transgenic Arabidopsis lines, siliques were collected 15 days after anthesis (DPA), frozen immediately in liquid nitrogen, and stored at -80 °C for RNA isolation. Quantitative real-time PCR (qRT-PCR) was performed to determine the expression pattern of GhWRI1a, with the 2-ΔΔCt method (Livak and Schmittgen, 2001) used to quantify the expression level of GhWRI1a relative to the 18S rRNA endogenous control. Each experiment was independently repeated in triplicate. Primers are listed in Supplementary Table S1.
Generation of CRISPR/Cas9 Transgenic Plant
For AtWRI1 gene editing, two single-guide RNAs (sgRNAs) were designed to target the first and fifth exons, namely Target1 and Target2 (Supplementary Figure S1). The two integrated targets were ligated to BsaI-digested pRGEB32-GhU6.9 as previously reported (Wang et al., 2017). This construct was introduced into Agrobacterium tumefaciens strain GV3101, which was used to transform Arabidopsis Col-0 as described above. The resulting CRISPR/Cas9 transgenic lines were genotyped for mutations using a pair of primers spanning the two target sequences (Supplementary Table S1). The homozygous T3 generation was used for further analysis.
Oil Content Analysis
We determined total oil content using an NMI20-Analyst nuclear magnetic resonance spectrometer (Niumag, Shanghai, China).
Results
Genome-Wide Identification and Phylogenetic Analysis of WRI-Like Genes in Gossypium
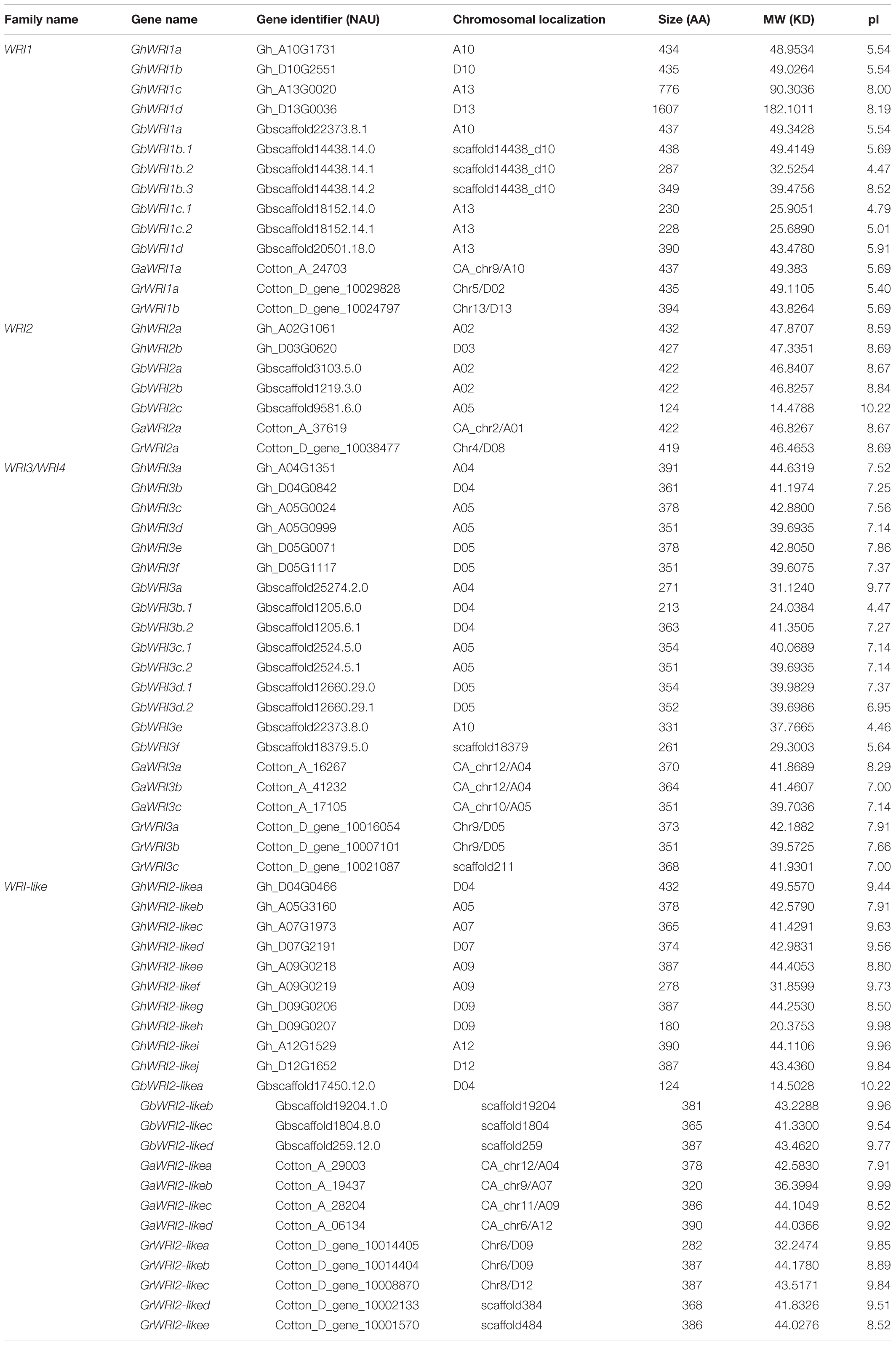
Two diploid cottons, G. arboreum (AA genome) and G. raimondii (DD genome), evolved from a common ancestor (Zhang et al., 2017). The most widely cultivated tetraploid cotton species are G. hirsutum (AADD, AD1 genome) and G. barbadense (AADD, AD2 genome), both of which originated from inter-genomic hybridization of two A- and D-genome progenitor species (Paterson et al., 2012). To identify all WRI-like proteins in AD1, AD2, AA and DD genomes, Arabidopsis WRI1-4 protein sequences (AtWRI1/AT3g54320, AtWRI2/AT2g41710, AtWRI3/AT1g16060, and AtWRI4/AT1g79700) were queried against reference genomes of the above-mentioned four species. All WRI-like candidates were further screened based on the conserved AP2 domain using the SMART database. A total of 59 WRI-like genes were identified: 11 in G. raimondii, 9 in G. arboreum, 22 in G. hirsutum, and 17 in G. barbadense (Table 1). WRI-like gene names and identifiers, gene pairs, and predicted properties of WRI-like proteins are listed in Table 1.
A phylogenetic tree was constructed to reveal the relationships of WRI-like proteins in Arabidopsis, cacao, rice, and cotton (Figure 1). This phylogenetic analysis classified WRI-like genes into WRI1, WRI2, WRI3/WRI4, and WRI2-like subfamilies. In comparison with dicotyledonous Arabidopsis, a more WRI2-like subfamily was identified interestingly. The WRI1 subfamily contained 11 members: 4 from G. hirsutum, 4 from G. barbadense, 1 from G. arboreum, and 2 from G. raimondii. The WRI2 subfamily consisted of seven members: two from G. hirsutum, three from G. barbadense, and one each from G. arboreum and G. raimondii. The WRI3/WRI4 subfamily included 18 members: 6, 6, 3, and 3 from G. hirsutum, G. barbadense, G. arboreum, and G. raimondii, respectively. Finally, the WRI2-like subfamily comprised 23 members: 10, 4, 4, and 5 in G. hirsutum, G. barbadense, G. arboreum, and G. raimondii, respectively.
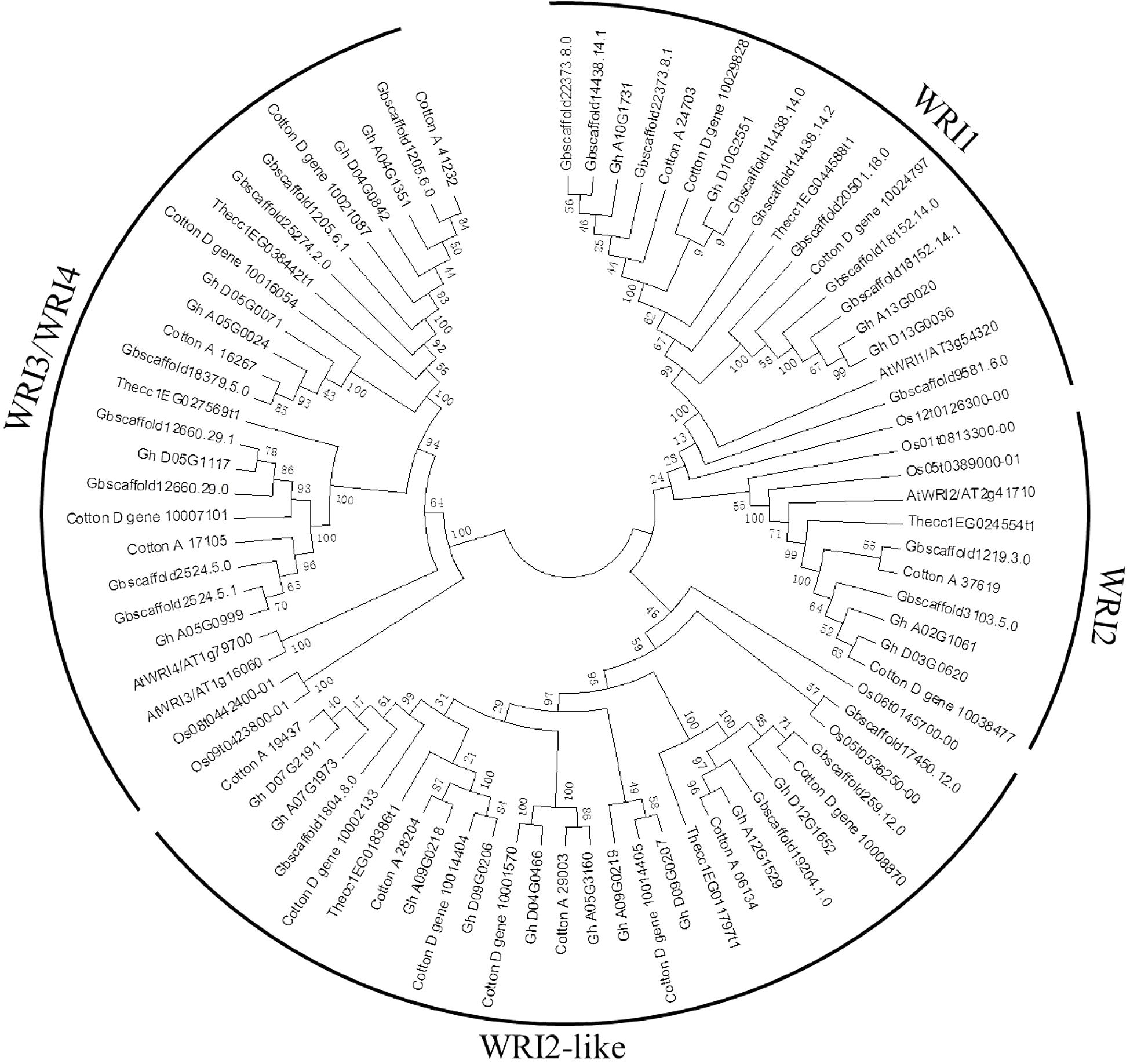
FIGURE 1. Phylogenetic tree of WRI-like gene family in Gossypium and Arabidopsis. The phylogenetic tree of all WRI-like proteins in four Gossypium species and Arabidopsis was constructed using the neighbor-joining method with 1,000 bootstrap replicates.
Chromosomal Distribution of WRI-Like Genes
The identified WRI-like genes were physically mapped to the chromosomes of cotton using the reference genome sequences (Figure 2 and Table 1). In the G. arboretum genome, nine GaWRIs were evenly distributed on seven chromosomes (A01, A04, A05, A07, A09, A10, and A12) (Figure 2A). One GaWRI gene each was located on chromosomes A01, A05, A07, A09, A10, and A12, and three GaWRI genes were found on chromosome A04. Nine of the 11 GrWRI genes in the G. raimondii genome were uniformly distributed on six chromosomes (D02, D05, D08, D09, D12, and D13), with one each positioned on chromosomes D05 and D09 (Figure 2B). The other three GrWRI genes were only located on the scaffolds. Among the 22 GhWRI genes identified in the G. hirsutum genome, 11 originated from the eight At subgenome chromosomes (A02, A04, A05, A07, A09, A10, A12, and A13), while 11 were derived from the eight Dt subgenome chromosomes (D03, D04, D05, D07, D09, D10, D12, and D13) (Figure 2C). Two genes each were located on chromosomes D04, D05, A09, and D09, while chromosome A05 harbored three GhWRIs. Each of the remaining chromosomes contained one GhWRI gene each. Among the 17 GbWRI genes identified in the G. barbadense genome, nine were located on the five At subgenome chromosomes (two on A02, one on A04, two on A05, two on A10, and two on A13), four were mapped to the three Dt subgenome chromosomes (two on D04, one on D05, and one on D10), and four were located on scaffolds (Figure 2D). Most of WRI-like genes were distributed evenly on the chromosomes (Figure 2 and Table 1), which provided a clue to their evolution.
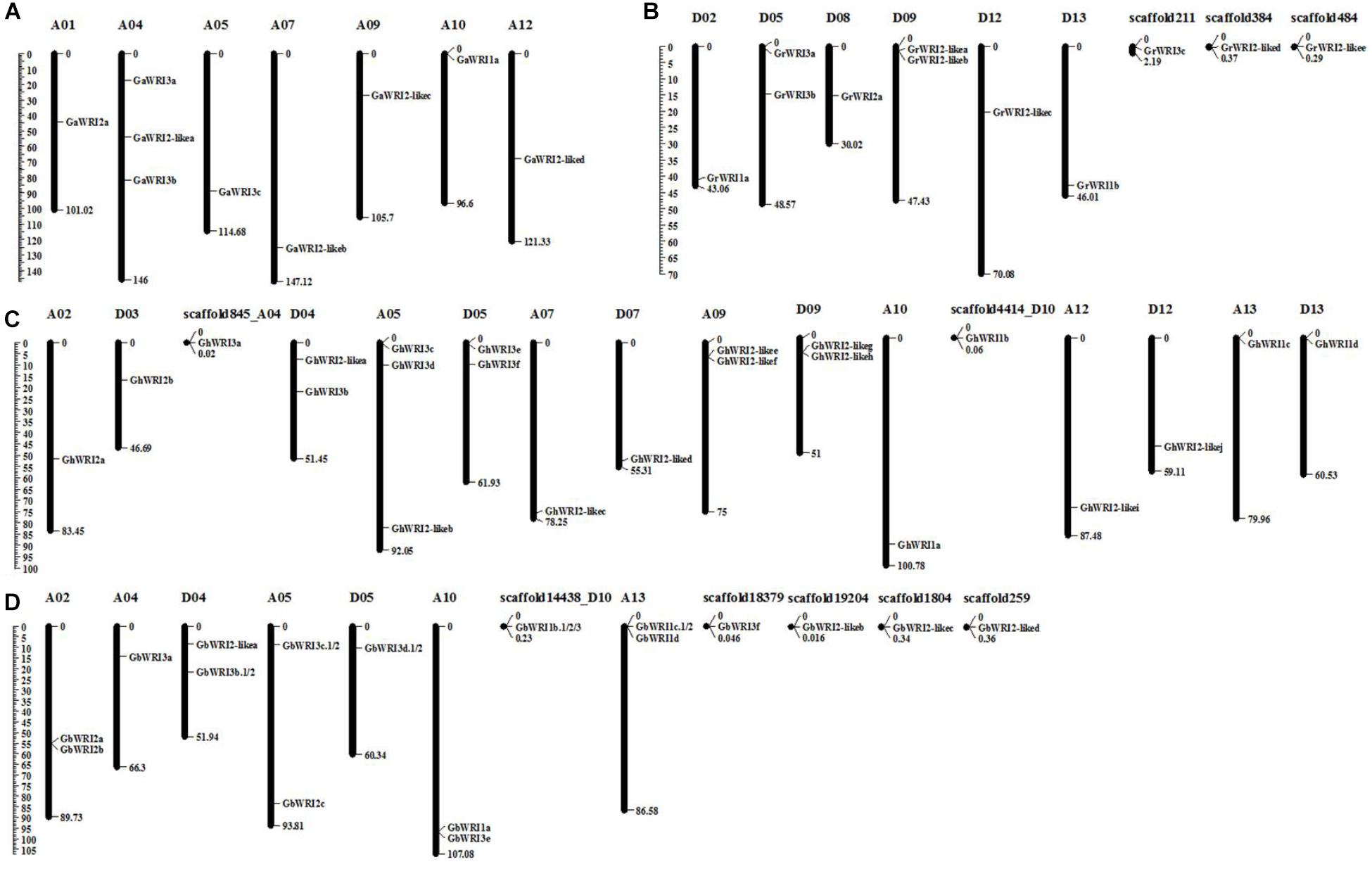
FIGURE 2. Chromosomal localization of WRI-like genes in four Gossypium species. A total of 69 WRI-like genes were mapped onto different chromosomes or scaffolds of G. arboreum (A), G. raimondii (B), G. hirsutum (C), and G. barbadense (D). The scale represents the megabases (Mb).
Analysis of Gene Duplication and Loss of WRI-Like Genes
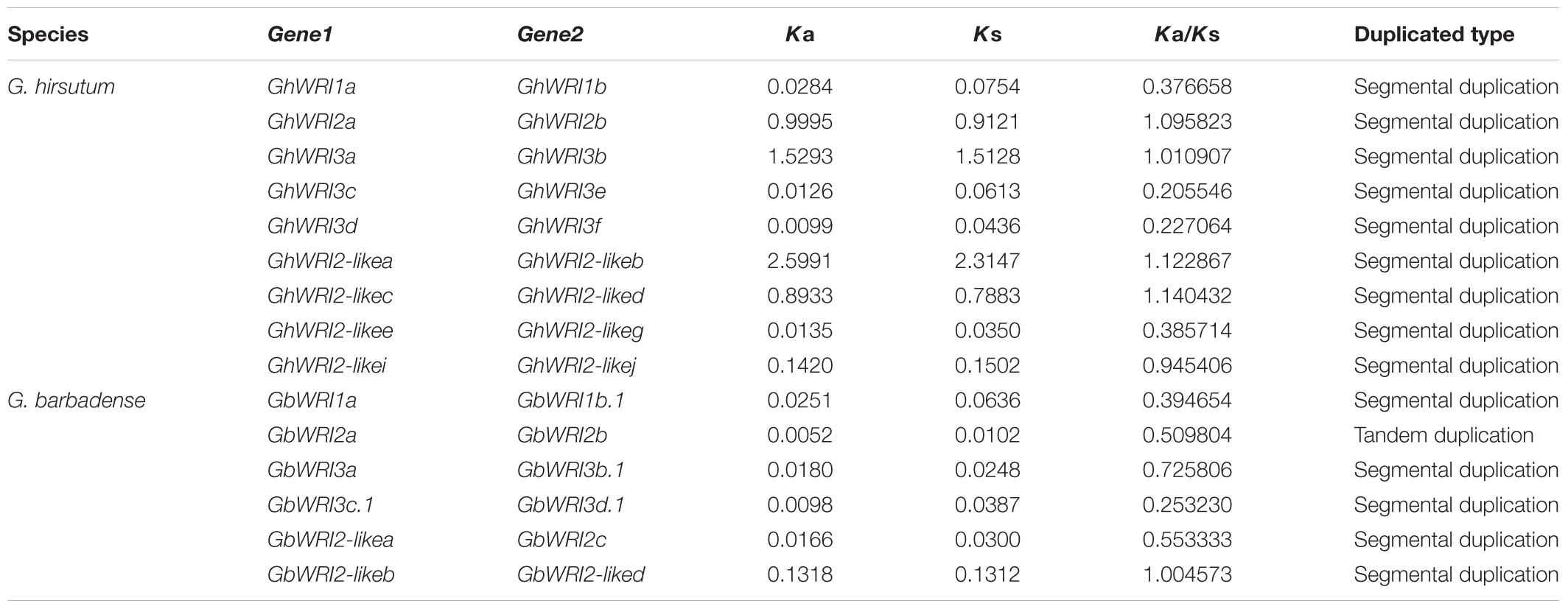
Large-scale duplication events have occurred during Gossypium evolution progress (Paterson et al., 2012). Gene duplication events, including tandem and segmental duplications, are considered as the major forces for expansion of gene families. In contrast to Arabidopsis, cacao, and rice, the WRI-like genes were expanded in cotton (Figure 1). We investigated the possible tandem and segmental duplication events of WRI-like genes in the four cotton species, respectively (Table 2 and Supplementary Table S2). Among them, no duplicated gene pairs were found in genome of G. raimondii and G. arboretum. In G. hirsutum, nine duplicated gene pairs were found to be segmental duplication events. In G. barbadense, six duplicated gene pairs were found, containing five segmental duplication events and one tandem event. These results indicated that segmental duplication were the main driving forces of the in the expansion of the WRI-like gene family.
During the process of evolution, gene pairs are subject to three alternative fates, i.e., non-functionalization, subfunctionalization, and neofunctionalization (Lynch and Conery, 2000). In this study, the Ka/Ks ratios for 15 duplicated WRI-like gene pairs were calculated (Table 2). The Ka/Ks ratios of ten pairs were less than 1, which suggests that these duplicated WRI-like genes have mainly experienced purifying selection pressure. The Ka/Ks ratios of other five pairs were more than 1, indicating positive selection pressure in the progress of evolution.
Then, WRI-like gene conservation and loss were analyzed based on the best match and the syntenic blocks in the CottonGen website (Figure 3 and Supplementary Table S3). Four homologous WRI-like clusters were ultra-conserved in four cotton species (Figure 3A and Supplementary Table S3). Ten homologous WRI-like genes were lost from the At, Dt or both subgenomes of G. barbadense and two were lost from G. arboretum (Figure 3B and Supplementary Table S3). Additionally, two genes were only present in G. barbadense (Figure 3C and Supplementary Table S3). This indicated that the GbWRIs and GaWRIs experienced a higher frequency of genic sequence losses than GhWRIs and GrWRIs.
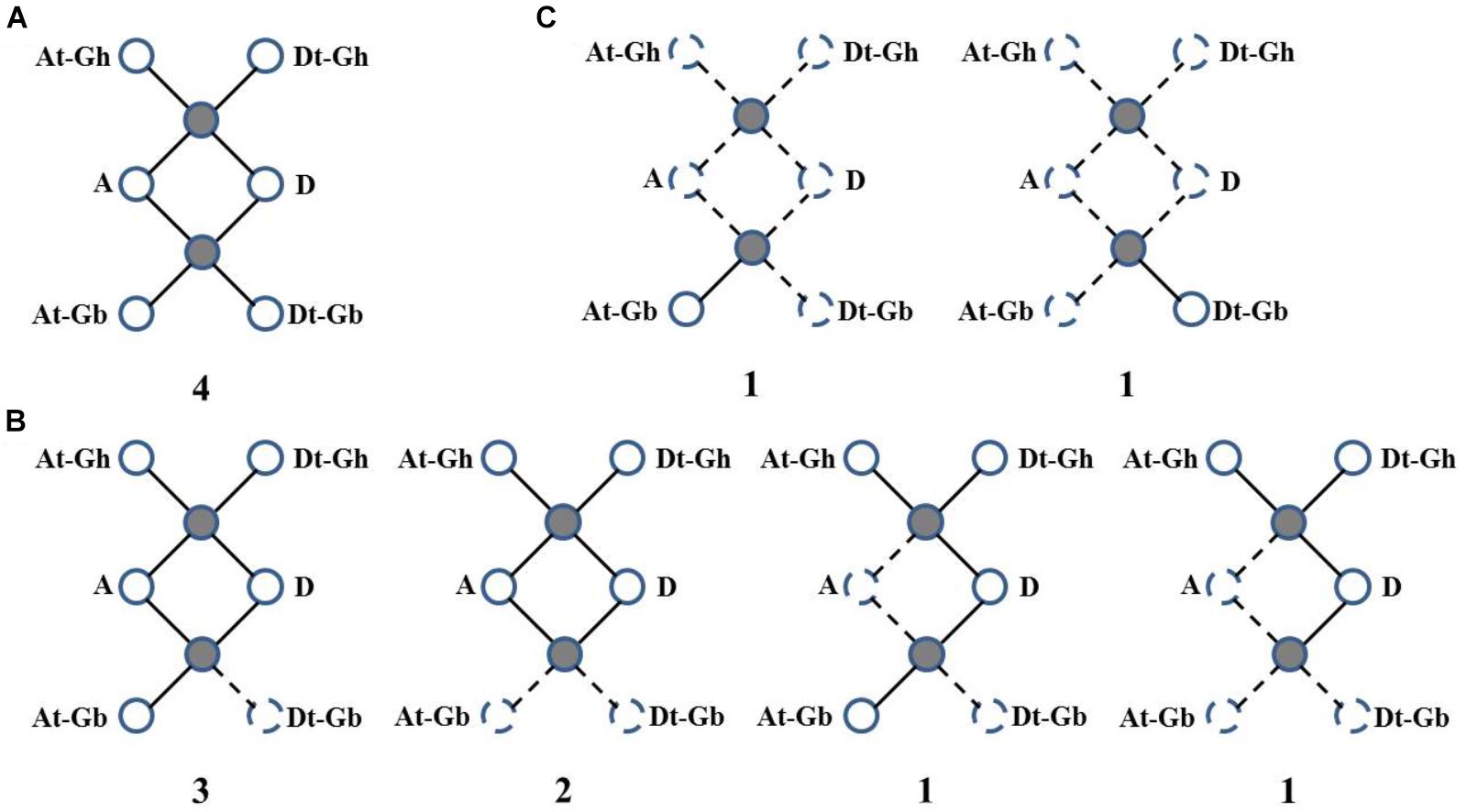
FIGURE 3. Gene conservation and loss analysis of the WRI-like genes in Gossypium. (A) Ultra-conserved homologous WRI-like clusters in four cotton species. (B) Homologous WRI-like genes lost from one and/or two cotton species. (C) WRI-like genes only present in G. barbadense. Statistics and scenarios of gene conservation and loss analysis in the four cotton species. The observed genes were indicated as solid lines and balls, and the lost genes were indicated as dotted lines and balls. The number of gene clusters found in the four cotton species is provided below each graphic.
Gene Structure and Protein Domain Analyses of WRI-Likes in G. hirsutum
Generic Feature Format files of the four Gossypium species and a phylogenetic tree of deduced amino acids of GhWRIs were used to analyze the similarity and diversity of their exon–intron structures (Figure 4). The AtWRI2 gene contained 10 introns and 11 exons, whereas WRI2 subfamily genes GhWRI2a (Gh_A02G1061) and GhWRI2b (Gh_D03G0620) harbored seven introns and eight exons. AtWRI1, AtWRI3, and AtWRI4 genes contained seven introns and eight exons. In contrast, most GhWRI1, GhWRI3/GhWRI4, and GhWRI2-like family genes fell into two categories: those containing five introns and six exons, and those having six introns and seven exons. GhWRI3b (Gh_D04G0842), GhWRI3c (Gh_A05G0024), GhWRI3d (Gh_A05G0999), GhWRI3f (Gh_D05G1117), GhWRI2-likeb (Gh_A05G3160), GhWRI2-likec (Gh_A07G1973), GhWRI2-liked (Gh_D07G2191), GhWRI2-likef (Gh_A09G0219), GhWRI2-likei (Gh_A12G1529), and GhWRI2-likej (Gh_D12G1652) contained six introns and seven exons. GhWRI1a (Gh_A10G1731), GhWRI1b (Gh_D10G2551), GhWRI3a (Gh_A04G1351), GhWRI3e (Gh_D05G0071), GhWRI2-likee (Gh_A09G0218), and GhWRI2-likeg (Gh_D09G0206) contained five introns and six exons. Four genes had unique intron–exon compositions: GhWRI1c (Gh_A13G0020) with 20 introns and 21 exons, GhWRI1d (Gh_D13G0036) with 24 introns and 25 exons, GhWRI2-likea (Gh_D04G0466) with four introns and five exons, and GhWRI2-likeh (Gh_D09G0207) containing three introns and four exons.
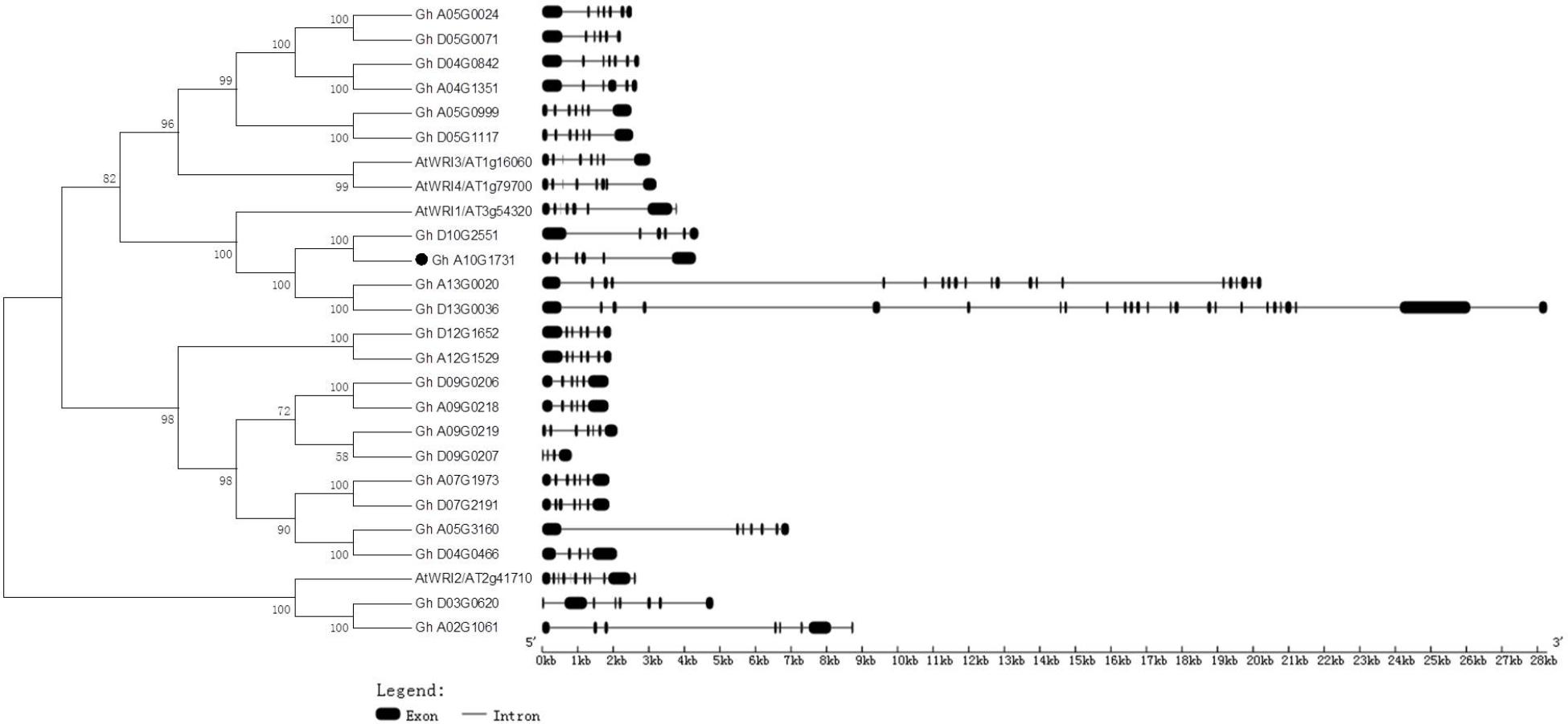
FIGURE 4. Phylogenetic relationships and gene structures of GhWRIs. Phylogeny of 22 GhWRI proteins in G. hirsutum and Arabidopsis. The phylogenetic tree (Left) was constructed using the neighbor-joining (NJ) method with 1,000 bootstrap replicates. The exon–intron structure (Right) of GhWRI genes was drawn using the GSDS server. Black boxes show exons, and lines show introns. The scale bar is shown at the bottom.
To better understand the similarity and diversity of GhWRI protein structures, their putative protein domains were predicted using the SMART database. The WRI-like proteins belonged to the AP2-EREPB family of transcription factors (Cernac and Benning, 2004; To et al., 2012). As shown in Figure 5, most GhWRIs contained two AP2 domains; the exceptions were GhWRI1d, GhWRI2a, GhWRI2b, and GhWRI2-likeh, all having only one each. Interestingly, we also found many other putative protein domains in GhWRI1c and GhWRI1d that need to be further verified.
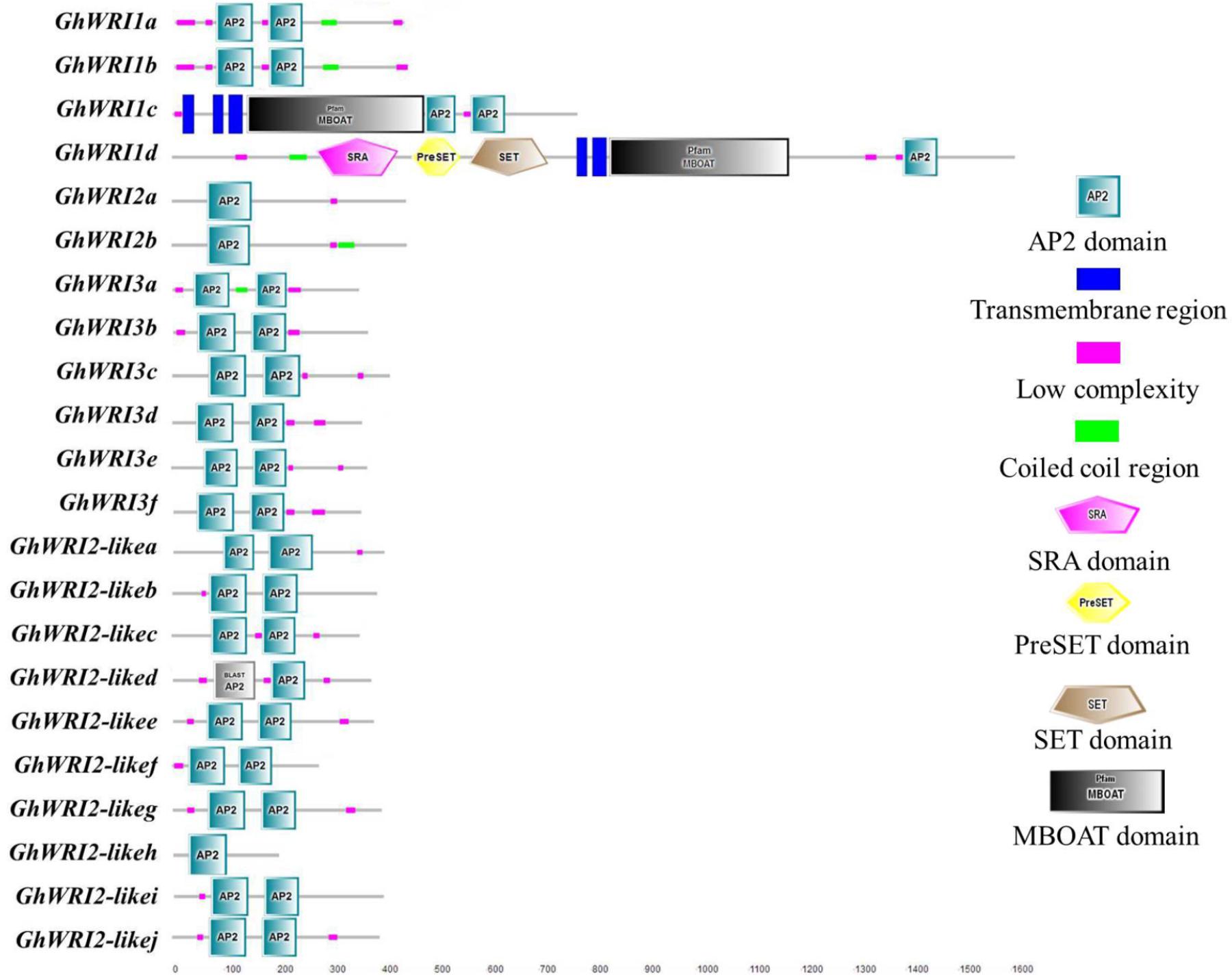
FIGURE 5. GhWRI protein domain prediction. Potential AP2 domains of GhWRI proteins were identified using the SMART database.
Tissue-Specific Expression Profiles of GhWRI Genes
The expression pattern of a gene can be a direct indication of its involvement in developmental or differential events (Zang et al., 2017). To reveal the tissue-specific expression profiles of the 22 GhWRI genes identified in this study, published TM-1 expression data (Zhang et al., 2015) were used to analyze the transcript profiles of GhWRI genes in 22 cotton tissues (Supplementary Figure S2). GhWRI genes from WRI1, WRI2, and WRI3/WRI4 subfamilies were widely detected in different tissues, whereas GhWRI genes from the WRI2-like subfamily exhibited very low expression levels in most tissues. Interestingly, we found GhWRI1a and GhWRI1b (gene pairs from the corresponding At and Dt subgenome) were highly expressed in 20–35 DPA ovules (Figure 6 and Supplementary Figure S2). GhWRI1a was thus selected for further functional analysis.
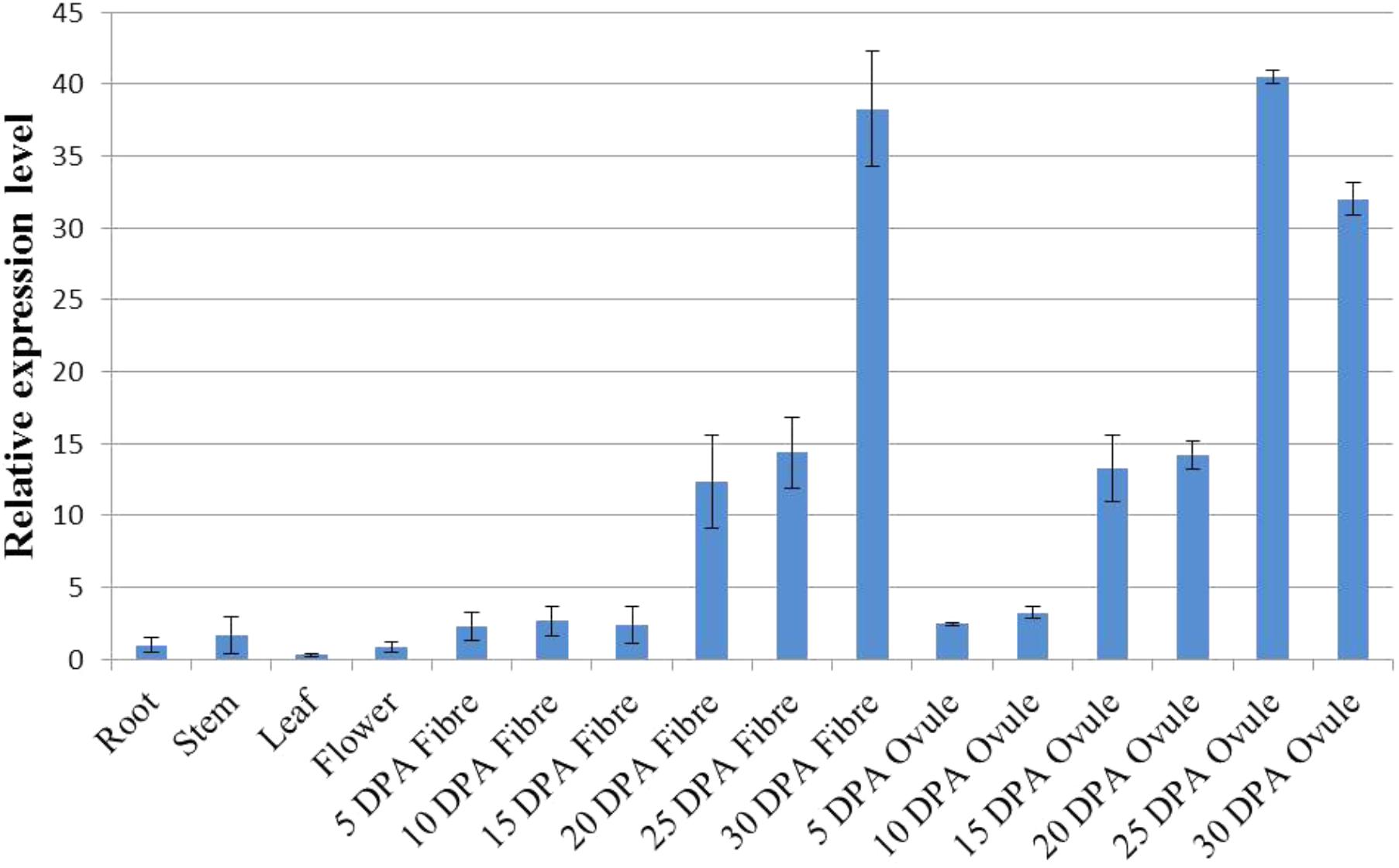
FIGURE 6. Tissue-specific expression profile of GhWRI1a in different tissues of G. hirsutum accession TM-1. The ΔCt value of GhWRI1a in root was set as the control. The data presented are the means ± SD of three replicates.
Ectopic Expression of GhWRI1a Rescued the Seed Phenotype of the wri1-7 Mutant and Increased the Oil Content of Arabidopsis Seeds
To characterize the biological functions of GhWRI1a in regard to oil content, we generated transgenic Arabidopsis plants overexpressing GhWRI1a. qRT-PCR was performed to analyze relative expression levels of GhWRI1a in transgenic Arabidopsis using cDNA from three different transgenic lines and WT as templates (Figure 7A). GhWRI1a was highly expressed in the transgenic lines. To evaluate the applicability of GhWRI1a in transgenic breeding for oil content, we characterized the phenotypes of GhWRI1a transgenic Arabidopsis at different developmental stages. No visible difference between transgenic and WT plants was observed (data not shown). To determine whether GhWRI1a had increased the oil content, we compared the oil contents of transgenic and WT plants. Significantly increased oil content, 6.96–14.24% higher, was observed in the transgenic plants (Figure 7B).
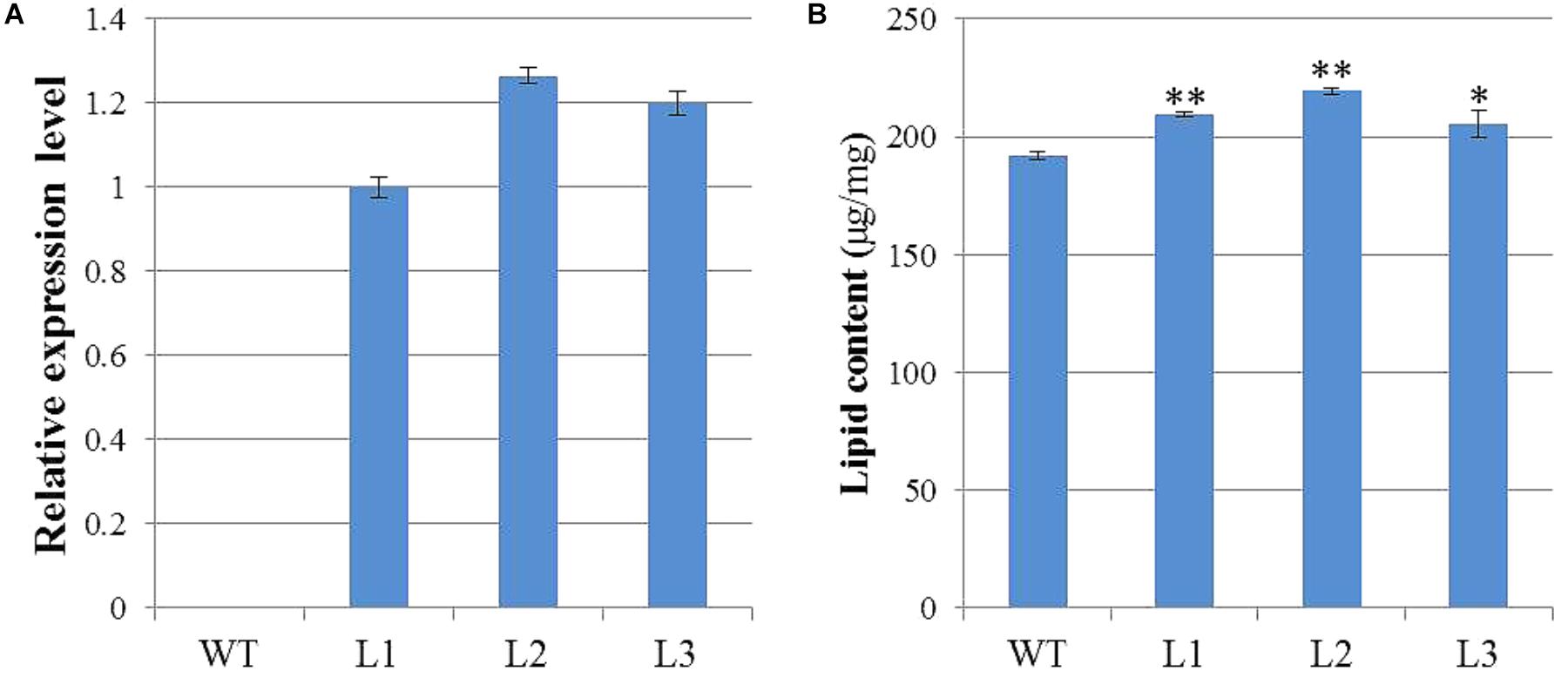
FIGURE 7. Improved oil content of GhWRI1a transgenic plants. (A) Relative expression level of GhWRI1a in three transgenic Arabidopsis lines (L1, L2, and L3). The ΔCt value of GhWRI1a in transgenic line L1 was set as the control. The data presented are the means ± SD of three replicates. (B) Seed oil content of GhWRI1a transgenic lines (L1, L2, and L3) and WT. The data presented are the means ± SD of three replicates; ∗P < 0.05; ∗∗P < 0.01 (Student’s t-test).
In order to determine whether the GhWRI1a transcription factor is involved in the activation of the whole fatty acid biosynthetic pathway, we created an atwri1 mutant named wri1-7 by the CRISPR method. DNA sequence comparison revealed the presence of a 722 bp deletion and a single adenine (A) insertion from the first to the fifth exon in the wri1-7 mutant (Figure 8A and Supplementary Figure S1). Microscopic observation of mature dry seeds of the wri1-7 mutant also revealed a wrinkled phenotype (Figure 8B), similar to previously reported wri1 mutant seeds (Cernac and Benning, 2004; To et al., 2012). The ability of the overexpression constructs to complement the seed phenotype of the wri1-7 mutant was confirmed by crossing L1, L2, and L3 transgenic plants with the wri1-7 mutant. Over accumulation of GhWRI1a RNA in the transgenic lines was verified by qRT-PCR (Figure 8B). Microscopic observation of mature dry seeds revealed a reversion to the wrinkled phenotype in wri1-7 seeds overexpressing GhWRI1a (Figure 8C). An analysis of total oil content of the dry seeds confirmed the ability of GhWRI1a to efficiently activate fatty acid biosynthesis and to thus complement the oil accumulation of wri1-7 seeds (Figure 8D).
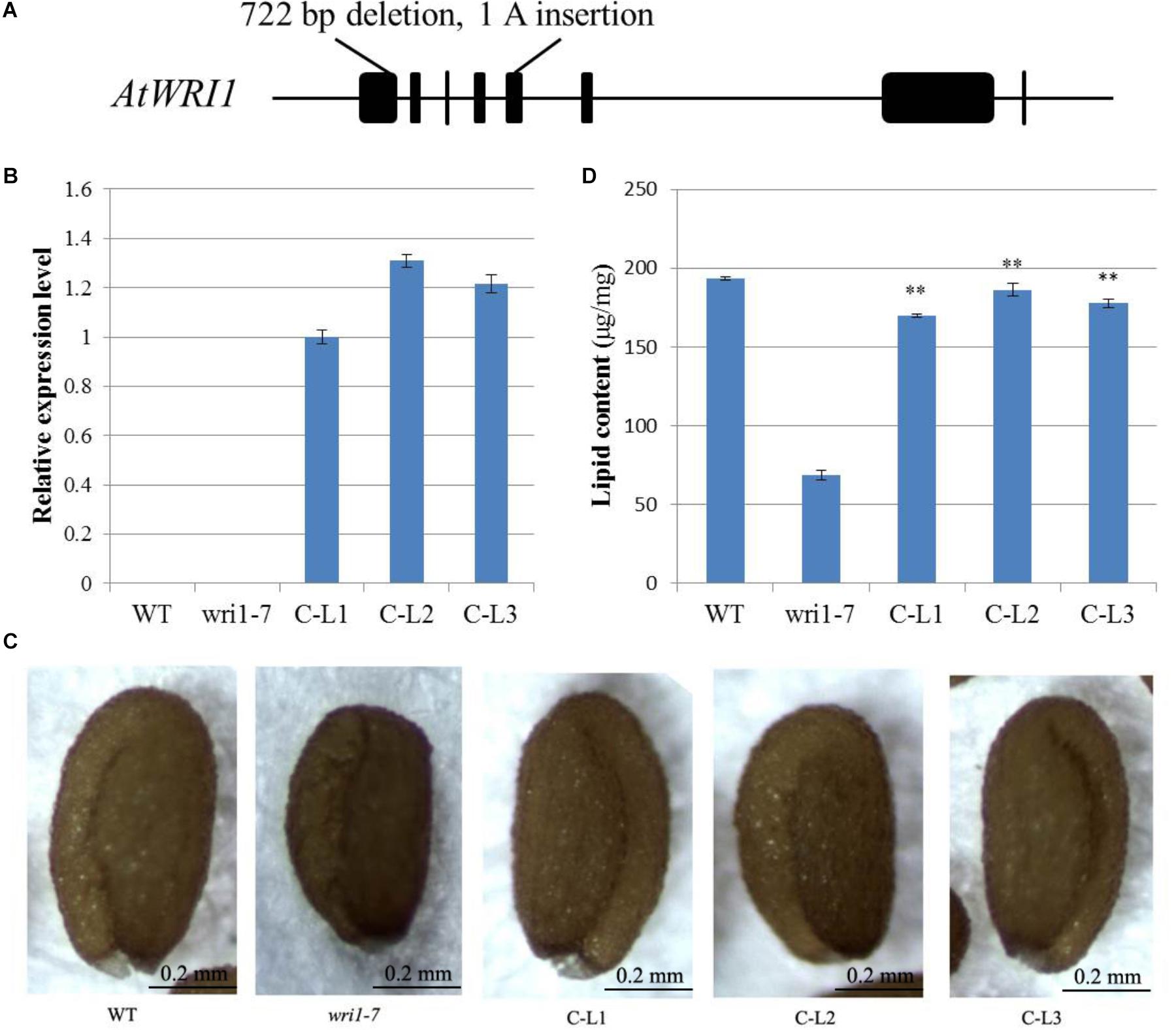
FIGURE 8. Arabidopsis wri1-7 mutant displaying a wrinkled phenotype rescued by overexpression of GhWRI1a. (A) Isolation and molecular characterization of the wri1-7 mutant. (B) Relative expression level of GhWRI1a in three GhWRI1a-complemented lines (C-L1, C-L2, and C-L3). The ΔCt value of GhWRI1a in complemented line C-L1 was set as the control. The data presented are the means ± SD of three replicates. (C) Rescue of the wrinkled phenotype of mature wri1-4 seeds by overexpression of GhWRI1a. Dry seeds from complemented lines were observed by stereoscopic microscopy. Representative seeds are shown. Bar = 0.2 mm. (D) Seed oil content of complemented lines (C-L1, C-L2, and C-L3), wri1-7, and WT. The data presented are the means ± SD of three replicates; ∗∗P < 0.01 (Student’s t-test).
Discussion
Numerous studies have revealed a crucial role for WRI-like genes in TAG biosynthesis, including GhWRI1 corresponding to GhWRI1b (Liu et al., 2018). Nevertheless, the naming of WRI-like family genes in cotton is confusing, and their systematic exposition is incomplete. In this study, we have accomplished the first-ever identification of WRI-like genes in four representative types of cotton, i.e., allotetraploid cotton species G. hirsutum and G. barbadense and their diploid ancestors G. arboreum, and G. raimondii. Our findings provide significant insights into the sequence variation, adaptive evolution, protein domains, expression profiles, co-localization with QTLs and GhWRI1a functions in cotton.
Our analysis revealed details of 22 deduced GhWRIs, most of which contain two AP2 domains, with only four GhWRIs (GhWRI1d, GhWRI2a, GhWRI2b, and GhWRI2-likeh) having just one AP2 domain (Figure 5). The WRI-like gene family is a branch of the AP2/EREBP (APETALA2/ethylene responsive element binding protein) transcription factor family. The AP2/EREBP family is one of the largest plant transcription factor families and plays an important role in plant growth and development (Okamuro et al., 1997; Riechmann et al., 2000; Zhou et al., 2013). This superfamily, comprising AP2, EREBP, and RAV subfamilies, is defined by the AP2/ERF DNA binding domain. AP2 family proteins contain two repeated AP2/ERF domains, EREBP family proteins have a single AP2/ERF domain, and RAV family proteins possess a B3 DNA-binding domain in addition to a single AP2/ERF domain (Sakuma et al., 2002; Feng et al., 2005; Nakano et al., 2006). AtWRI1, AtWRI2, AtWRI3, and AtWRI4 proteins all belong to the AP2 subfamily (Feng et al., 2005). These proteins generally contain two repeated AP2/ERF domains, the exception is AtWRI2, which was found to possess only one AP2/ERF domain (Supplementary Figure S3), consistent with previous reports (Nakano et al., 2006). Consequently, GhWRI1d, GhWRI2a, GhWRI2b, and GhWRI2-likeh, which contain only one AP2 domain, are typical representatives of the AP2 subfamily. The WRI-like genes identified in this study belong to the AP2 subfamily of the AP2/EREBP family.
Cottonseed oil accumulates in ovules after 15 DPA. At this stage, most GhWRIs were found to be expressed in our study. GhWRI1a and GhWRI1b had the highest expression levels (Supplementary Figure S2), indicating that these two genes play important roles in TAG biosynthesis in developing cotton seeds. In this study, we demonstrated that ectopic expression of GhWRI1a could rescue the seed phenotype of the wri1-7 mutant and increase the oil content of Arabidopsis seeds. In addition, four WRI-like genes were localized in cottonseed oil QTL intervals, which suggests their association with natural variation in cottonseed oil content.
We further discovered that GhWRIs were expressed in developing fibers. GhWRI1a and GhWRI1b, in particular, were highly expressed in 25-DPA developing fibers (Figure 6 and Supplementary Figure S2), suggesting their additional involvement in fiber development. Other studies of upland cotton have also indicated the involvement of GhWRIs in fiber length (Qu et al., 2012; Qaisar et al., 2017). The regulatory relationship between GhWRIs and fiber development needs to be further verified.
In short, we have performed a comprehensive genome-wide analysis of the WRI-like gene family in G. hirsutum, G. barbadense, G. raimondii, and G. arboreum. A total of 69 WRI-like genes grouped into four distinct subfamilies were identified in four sequenced Gossypium species. Our detailed analysis has established a solid foundation for further studies of WRI-like genes in cotton.
Author Contributions
JY and XZ directed the experiments. WP, MW, YG, NW, GL, JM, DL, YC, XL, and JZ participated in the study. XZ conceived the study, performed the experiments and wrote the manuscript. JY and JZ revised the manuscript. All authors read and approved the final manuscript.
Funding
The research was supported by grants from the National Natural Science Foundation of China (Grant No. 31621005), the National Key Research and Development Program of China (Grant No. 2016YFD0101400), and the National Research and Development Project of Transgenic Crops of China (Grant No. 2016ZX08005005).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01516/full#supplementary-material
FIGURE S1 | Sequence alignment of AtWRI1 in the wri1-7 mutant and WT. Target1 and Target2 are the two designed single-guide RNAs.
FIGURE S2 | Expression analysis of GhWRIs in 22 tissues of G. hirsutum accession TM-1 (Zhang et al., 2015). The RNA-seq profiles of TM-1 were used to identify GhWRI gene expression levels. FPKM, fragments per kilobase of exon model per million mapped reads.
FIGURE S3 | Protein domain prediction for AtWRIs. The potential AP2 domains of AtWRI proteins were identified using the SMART database.
TABLE S1 | Primers used in this paper.
TABLE S2 | Duplicated genes of WRI-like family in Gossypium.
TABLE S3 | Gene conservation and loss analysis of WRI-like family in Gossypium.
FILE S1 | Phylogenetic data of Figure 1.
FILE S2 | Coding sequence of GhWRI1a.
Footnotes
- ^ https://www.cottongen.org/
- ^ http://www.arabidopsis.org/
- ^ https://www.cacaogenomedb.org/
- ^ https://rapdb.dna.affrc.go.jp/
- ^ http://web.expasy.org/
- ^ http://www.earthatlas.mapchart.com/
- ^ http://www.cottonqtldb.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://smart.embl-heidelberg.de/
References
An, D., and Suh, M. C. (2015). Overexpression of Arabidopsis WRI1 enhanced seed mass and storage oil content in Camelina sativa. Plant Biotechnol. Rep. 9, 137–148. doi: 10.1007/s11816-015-0351-x
An, D., Kim, H., Ju, S., Go, Y. S., Kim, H. U., and Suh, M. C. (2017). Expression of Camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front. Plant Sci. 8:34. doi: 10.3389/fpls.2017.00034
Bates, P. D., and Browse, J. (2012). The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3:147. doi: 10.3389/Fpls.2012.00147
Cernac, A., and Benning, C. (2004). WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J. 40, 575–585. doi: 10.1111/j.1365-313X.2004.02235.x
Cui, Y. P., Liu, Z. J., Zhao, Y. P., Wang, Y. M., Huang, Y., Li, L., et al. (2017). Overexpression of heteromeric GhACCase subunits enhanced oil accumulation in upland cotton. Plant Mol. Biol. Rep. 35, 287–297. doi: 10.1007/s11105-016-1022-y
Dong, Y., Li, C., Zhang, Y., He, Q., Daud, M. K., Chen, J., et al. (2016). Glutathione S-transferase gene family in Gossypium raimondii and G. arboreum: comparative genomic study and their expression under salt stress. Front. Plant Sci. 7:139. doi: 10.3389/Fpls.2016.00139
Feng, J. X., Liu, D., Pan, Y., Gong, W., Ma, L. G., Luo, J. C., et al. (2005). An annotation update via cDNA sequence analysis and comprehensive profiling of developmental, hormonal or environmental responsiveness of the Arabidopsis AP2/EREBP transcription factor gene family. Plant Mol. Biol. 59, 853–868. doi: 10.1007/s11103-005-1511-0
Focks, N., and Benning, C. (1998). wrinkled1: a novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 118, 91–101. doi: 10.1104/pp.118.1.91
Grimberg, A., Carlsson, A. S., Marttila, S., Bhalerao, R., and Hofvander, P. (2015). Transcriptional transitions in Nicotiana benthamiana leaves upon induction of oil synthesis by WRINKLED1 homologs from diverse species and tissues. BMC Plant Biol. 15:192. doi: 10.1186/s12870-015-0579-1
Hofvander, P., Ischebeck, T., Turesson, H., Kushwaha, S. K., Feussner, I., Carlsson, A. S., et al. (2016). Potato tuber expression of Arabidopsis WRINKLED1 increase triacylglycerol and membrane lipids while affecting central carbohydrate metabolism. Plant Biotechnol. J. 14, 1883–1898. doi: 10.1111/pbi.12550
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Liu, Q., Singh, S. P., and Green, A. G. (2002). High-stearic and high-oleic cottonseed oils produced by hairpin RNA-mediated post-transcriptional gene silencing. Plant Physiol. 129, 1732–1743. doi: 10.1104/pp.001933
Liu, Q., Surinder, S., Chapman, K., and Green, A. (2009). “Bridging traditional and molecular genetics in modifying cottonseed oil,” in Genetics and Genomics of Cotton. Plant Genetics and Genomics: Crops and Models, Vol. 3, ed. A. H. Paterson (London: Springer Science + Business Media),353–382.
Liu, Z. J., Zhao, Y. P., Liang, W., Cui, Y. P., Wang, Y. M., and Hua, J. P. (2018). Over-expression of transcription factor GhWRI1 in upland cotton. Biol. Plantarum. 62, 335–342. doi: 10.1007/s10535-018-0777-4
Liu, J., Hua, W., Zhan, G., Wei, F., Wang, X., Liu, G., et al. (2010). Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol. Biochem. 48, 9–15. doi: 10.1016/j.plaphy.2009.09.007
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Okamuro, J. K., Caster, B., Villarroel, R., Van Montagu, M., and Jofuku, K. D. (1997). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 94, 7076–7081. doi: 10.1073/pnas.94.13.7076
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Pouvreau, B., Baud, S., Vernoud, V., Morin, V., Py, C., Gendrot, G., et al. (2011). Duplicate maize Wrinkled1 transcription factors activate target genes involved in seed oil biosynthesis. Plant Physiol. 156, 674–686. doi: 10.1104/pp.111.173641
Qaisar, U., Akhtar, F., Azeem, M., and Yousaf, S. (2017). Studies on involvement of Wrinkled1 transcription factor in the development of extra-long staple in cotton. Indian J. Genet. Plant Breed. 77, 298–303. doi: 10.5958/0975-6906.2017.00040.2
Qu, J., Ye, J., Geng, Y. F., Sun, Y. W., Gao, S. Q., Zhang, B. P., et al. (2012). Dissecting functions of KATANIN and WRINKLED1 in cotton fiber development by virus-induced gene silencing. Plant Physiol. 160, 738–748. doi: 10.1104/pp.112.198564
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Shen, B., Allen, W. B., Zheng, P., Li, C., Glassman, K., Ranch, J., et al. (2010). Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 153, 980–987. doi: 10.1104/pp.110.157537
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
To, A., Joubès, J., Barthole, G., Lécureuil, A., Scagnelli, A., Jasinski, S., et al. (2012). WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24, 5007–5023. doi: 10.1105/tpc.112.106120
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, P., Zhang, J., Sun, L., Ma, Y., Xu, J., Liang, S., et al. (2017). High efficient multi-sites genome editing in allotetraploid cotton (Gossypium hirsutum) using CRISPR/Cas9 system. Plant Biotechnol. J. 16, 137–150. doi: 10.1111/pbi.12755
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Yang, Y., Munz, J., Cass, C., Zienkiewicz, A., Kong, Q., Ma, W., et al. (2015). Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol. 169, 1836–1847. doi: 10.1104/pp.15.01236
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Jin, X., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Zang, X., Geng, X., Liu, K., Wang, F., Liu, Z., Zhang, L., et al. (2017). Ectopic expression of TaOEP16-2-5B, a wheat plastid outer envelope protein gene, enhances heat and drought stress tolerance in transgenic Arabidopsis plants. Plant Sci. 258, 1–11. doi: 10.1016/j.plantsci.2017.01.011
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhang, Y., He, P., Yang, Z., Huang, G., Wang, L., Pang, C., et al. (2017). A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development. Front. Plant Sci. 8:461. doi: 10.3389/fpls.2017.00461
Keywords: cotton, WRI-like, expression pattern, cottonseed oil, GhWRI1a
Citation: Zang X, Pei W, Wu M, Geng Y, Wang N, Liu G, Ma J, Li D, Cui Y, Li X, Zhang J and Yu J (2018) Genome-Scale Analysis of the WRI-Like Family in Gossypium and Functional Characterization of GhWRI1a Controlling Triacylglycerol Content. Front. Plant Sci. 9:1516. doi: 10.3389/fpls.2018.01516
Received: 06 July 2018; Accepted: 27 September 2018;
Published: 16 October 2018.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Qing Liu, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaNan Lu, University of North Texas, United States
Copyright © 2018 Zang, Pei, Wu, Geng, Wang, Liu, Ma, Li, Cui, Li, Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiwen Yu, eXVqdzY2NkBob3RtYWlsLmNvbQ==
 Xinshan Zang
Xinshan Zang Wenfeng Pei1
Wenfeng Pei1 Man Wu
Man Wu Nuohan Wang
Nuohan Wang Jianjiang Ma
Jianjiang Ma Yupeng Cui
Yupeng Cui Jinfa Zhang
Jinfa Zhang