
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 19 September 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01395
 Linyi Qiao1†
Linyi Qiao1† Wenping Zhang2†
Wenping Zhang2† Xiaoyan Li3
Xiaoyan Li3 Lei Zhang4
Lei Zhang4 Xiaojun Zhang1
Xiaojun Zhang1 Xin Li1
Xin Li1 Huijuan Guo1
Huijuan Guo1 Yuan Ren1
Yuan Ren1 Jun Zheng1*
Jun Zheng1* Zhijian Chang1*
Zhijian Chang1*Auxin response factors (ARFs) are important transcription factors involved in both the auxin signaling pathway and the regulatory development of various plant organs. In this study, 23 TaARF members encoded by a total of 68 homeoalleles were isolated from 18 wheat chromosomes (excluding chromosome 4). The TaARFs, including their conserved domains, exon/intron structures, related microRNAs, and alternative splicing (AS) variants, were then characterized. Phylogenetic analysis revealed that members of the TaARF family share close homology with ARFs in other grass species. qRT-PCR analyses revealed that 20 TaARF members were expressed in different organs and tissues and that the expression of some members significantly differed in the roots, stems, and leaves of wheat seedlings in response to exogenous auxin treatment. Moreover, protein network analyses and co-expression results showed that TaTIR1–TaARF15/18/19–TaIAA13 may interact at both the protein and genetic levels. The results of subsequent evolutionary analyses showed that three transcripts of TaARF15 in the A subgenome of wheat exhibited high evolutionary rate and underwent positive selection. Transgenic analyses indicated that TaARF15-A.1 promoted the growth of roots and leaves of Arabidopsis thaliana and was upregulated in the overexpression plants after auxin treatment. Our results will provide reference information for subsequent research and utilization of the TaARF gene family.
Auxin signaling is key to many plant growth and developmental processes ranging from embryogenesis to senescence (Strader and Zhao, 2016; Mironova et al., 2017). Auxin response factors (ARFs), which generally consist of an amino-terminal DNA-binding domain (DBD), a middle region (MR) that functions as either an activation domain or a repression domain, and a carboxy-terminal dimerization domain (CTD), are transcription factors involved in the well-described transport inhibitor response 1/auxin signaling F-Box (TIR1/AFB) auxin signaling pathway (Boer et al., 2014; Dinesh et al., 2015). In the absence of auxin, auxin/indole acetic acid (Aux/IAA) repressor proteins bind to the CTD of ARFs and inhibit their function. When present, auxin promotes the TIR1/AFB-mediated ubiquitin–proteasome-dependent degradation of Aux/IAAs (Jing et al., 2015), thus relieving ARFs from repression and allowing them to activate or repress the expression of auxin-responsive genes; this activation or repression occurs via the DBD that binds to auxin response elements (AuxREs) within the promoters of target genes (Dinesh et al., 2015; Winkler et al., 2017).
In the model plant Arabidopsis thaliana, 23 ARF genes have been identified (Wei et al., 2006); several of these members, such as AtARF1–8 (Ellis et al., 2005; Marin et al., 2010; Pitaksaringkarn et al., 2014; Moller et al., 2017), AtARF10, AtARF16 (Yang et al., 2014), AtARF17 (Gutierrez et al., 2009), and AtARF19 (Galvan-Ampudia and Vernoux, 2014), are involved in regulating the morphology and growth of roots, stems, leaves, flowers, and fruits. Based on these AtARFs whose functions are known, homology cloning has been used to identify many ARFs in other species, including OsARF8 (Yang et al., 2006), SlARF8 (Ma et al., 2015), GmARF8 (Wang et al., 2015, NtARF8 (Zhu et al., 2013), and InARF8 (Glazinska et al., 2014), all of which are homologous to AtARF8. In addition, whole-genome sequencing studies have led to the isolation of 25 ARFs from rice (Wang et al., 2007), 24 ARFs from sorghum (Wang et al., 2010), 31 ARFs from maize (Xing et al., 2011), 51 ARFs from soybean (Ha et al., 2013), 19 ARFs from tomato (Zouine et al., 2014), and 24 ARFs from Medicago truncatula (Shen et al., 2015). However, few studies have investigated this gene family in bread wheat (Triticum aestivum L.), one of the most widely grown crops worldwide, as this species has an enormous and complex hexaploid genome. Currently, only one wheat expressed sequence tag (GenBank No. AY902381) has been reported, which responds to aluminum stress (Liu et al., 2017).
The rapid development of sequencing and assembly technologies has led to the completion (via different sequencing technologies) of the draft genome of “Chinese Spring” bread wheat (Brenchley et al., 2012; Mayer et al., 2014; Zimin et al., 2017). In addition, the physical map (IWGSC, 20171) as well as a high-quality genome (Clavijo et al., 2017) have been published, allowing the isolation and analysis of gene families on a genomic scale. In this study, 23 ARF members, with a total of 68 homeoalleles, were isolated in wheat, and the genomic location, sequence characteristics, related microRNAs, alternative splicing (AS) variants, phylogenetic relationships, and expression patterns of those ARFs were analyzed. In addition, the evolutionary rate and transgene function of TaARF15-A were verified. These results will provide reference information for subsequent research and utilization of the TaARF gene family.
The TGACv1 collection of whole-protein sequences of “Chinese Spring” wheat was downloaded from the Ensembl database2. The sequences of the predicted wheat ARF proteins were obtained by retrieving the whole-protein sequence data based on the ARF family Hidden Markov Model profiles (Pfam accession number PF00931) and checking the ARF domains using the hmmsearch and hmmscan programs of the nhmmer software (Wheeler and Eddy, 2013) program, respectively; a cutoff of E ≤1e-5 was used. Via their registration number (Supplementary Table S1), the protein sequences of ARFs from Arabidopsis, rice and other species were downloaded directly from the NCBI database (National Center for Biotechnology Information3).
Multiple sequence alignments of the ARFs were performed using Clustal X (Larkin et al., 2007). Phylogenetic trees were constructed using MEGA6.0 software (Tamura et al., 2013) with the neighbor-joining method and 1000 bootstraps. The secondary structures of the protein sequences were predicted using the NPS@ server (Network Protein Sequence Analysis4; Combet et al., 2000). The STRING database (Search Tool for the Retrieval of Interacting Genes/Proteins5; Szklarczyk et al., 2015) was used to predict interactions among ARFs, Aux/IAAs, and TIR1 proteins in wheat. The TaTIR1 protein sequence was retrieved from the Ensembl database by BLASTP queries of the OsTIR1 sequence (Xia et al., 2012), and we obtained the sequences of Aux/IAA proteins from our previous research (Qiao et al., 2015).
The coding DNA sequences (CDS), AS variants, and genomic sequences of the TaARF gene family members were extracted (via their protein accession number) from the Ensembl database. In reference to previous studies (Kim et al., 2007; Kaur et al., 2017), the AS events of TaARFs were identified by comparing the genetic structure of AS variants with the assumed wild-type sequence (generally the AS variant numbered 1) so that each variant was identified as one of the five categories: intron retention (IR), alternative 5′ splicing (A5SS), alternative 3′ splicing (A3SS), exon skipping (ES), or mutually exclusive exon (MXE). The position information of these members was determined by using the RefSeq v1.0 iteration of the wheat whole genome, which was downloaded from the IWGSC database6, after which the TaARFs were assigned to their corresponding chromosomes. The sequences of wheat microRNAs that may regulate TaARFs were obtained by retrieving tae-miR target sequence data (downloaded from the Ensembl database) using TaARF CDS as queries with a similarity >90%. Then, a professional small RNA target analysis server, psRNATarget (Dai et al., 2018), was used for the validation of bioinformatics with maximum expectation set as zero. Gene structures were determined using GSDS 2.0 (Gene Structure Display Server7; Hu et al., 2015). Putative promoter regions (2000 bp upstream of the start codon) of TaIAAs were obtained from their genomic sequences, and the AuxREs that bind ARF proteins, TGTCTC (Ulmasov et al., 1995) and TGTCGG (Boer et al., 2014), were identified by manually scanning the TaIAA promoters.
The ARF codon sequences were used for constructing phylogenetic trees and in subsequent calculations. The ratio of non-synonymous substitutions per non-synonymous site to synonymous substitutions per synonymous site (ω value) of branches was computed by using the maximum likelihood method of the branch model in the PAML 4.4 software package (Yang, 2007). If the likelihood ratio test (LRT) indicated that the ω value of a specified branch significantly differed from the constant ω value across all branches, that branch was used for the next positive selection analysis of the branch-site model using the Bayes empirical Bayes method described by Yang et al. (2005). The TuARF, AetARF, and TdARF sequences used for the evolutionary analysis were obtained by using BLASTN to query the TaARF15 CDS against the predicted gene sequences of Triticum urartu (Ling et al., 2013), Aegilops tauschii (Zhao et al., 2017), and Triticum dicoccoides (Avni et al., 2017), three ancestors of wheat.
With respect to auxin treatment, the wheat cultivar “Chinese Spring” was planted under a long-day photoperiod (15 h of light, 9 h of darkness). Seedlings with three fully opened leaves were sprayed with 10 μM α-naphthylacetic acid (α-NAA) solution or distilled water (mock treatment), as described previously (Zhao et al., 2013). The roots, stems, and leaves of seedlings were sampled at 0, 0.5, 1.5, and 3 h after auxin treatment, and at each time point, samples from three plants were pooled, after which they were stored at -80°C. The total RNA was extracted using an RNA extraction kit (Tiangen Biotech, China) and reverse-transcribed into cDNA with an M-MLV reverse transcription kit (Invitrogen, United States). qRT-PCR was subsequently performed using SYBR Premix Ex Taq II (Takara Bio Inc., China), and each reaction was repeated three times; GADPH and TaAux/IAA1 (Singla et al., 2006) served as the internal and external control, respectively. The qRT-PCR primers listed in Supplementary Table S2 were designed based on the consensus sequence of homeoalleles for every TaARF member, and the specific primers for each AS variant of TaARF15-A were also designed. The qRT-PCR data were analyzed using the fold-change (Navarro et al., 2006) and the 2-ΔΔCt (Livak and Schmittgen, 2001) methods. Statistical analyses of the differences between the treatment group and the control group were performed by using t-tests (tails = 2, type = 1).
The expression data of TaARFs in different organs at different growth stages of Chinese Spring (Ramírez-González et al., 2018) were obtained from the expVIP database8 (Borrill et al., 2016) and then viewed as a heat map using the MeV tool (Multiple Experiment Viewer9).
To generate TaARF15-A1 overexpression plants, the primers 15A1-F and 15A1-R (Supplementary Table S2) were used to amplify the coding sequence of the wheat cDNA, after which the sequence under the control of the CaMV 35S promoter was inserted into a pBI121 binary vector using Gateway BP Clonase enzyme mix (Invitrogen, United States). The construct was then transformed into Arabidopsis (Col-0) by Agrobacterium tumefaciens strain GV3101 (Herrera-Estrella et al., 2005) via the floral dip method. The transformed lines were first selected on half-strength Murashige and Skoog medium that contained 50 mg L-1 kanamycin (Ahmed et al., 2012) and then screened by PCR. The resistant seedlings were subsequently transferred to a mixture of soil and vermiculite (1:1) at 22°C under a 16/8-h light/dark cycle with 70% relative humidity, after which homozygous lines were generated by self-fertilization. Plants from the F3 generation and wild-type Arabidopsis were used for morphological comparison. qRT-PCR was then used to detect the expression of TaARF15-A.1 in the transgenic plants. The total RNA was isolated from leaves of 20-day-old TaARF15-A1 overexpression plants and wild-type plants at 0, 0.5, and 2 h after 10 μM auxin treatment, and ACTIN2 served as an endogenous control. qRT-PCR for each line was based on three independent biological replicates.
By retrieving wheat protein sequence data and examining the domains, we detected 68 full-length ARF proteins, which were subsequently used to construct an unrooted tree for revealing phylogenetic relationships. The results showed that these wheat ARF proteins could be divided into 23 groups (Figure 1A); each group containing two or three homeoalleles from the wheat A, B, and D subgenomes was regarded as a member of the ARF family in wheat (TaARF family). These 23 TaARF members were distributed across all chromosomes except 4A, 4B, and 4D and were named TaARF1–TaARF23 based on their chromosome position (Supplementary Figure S1). The largest number of members (TaARF18–23) were distributed across homologous chromosome 7, while only one member (TaARF14) was found on homologous chromosome 5. With the exception of TaARF23, every TaARF member had three homeoalleles.
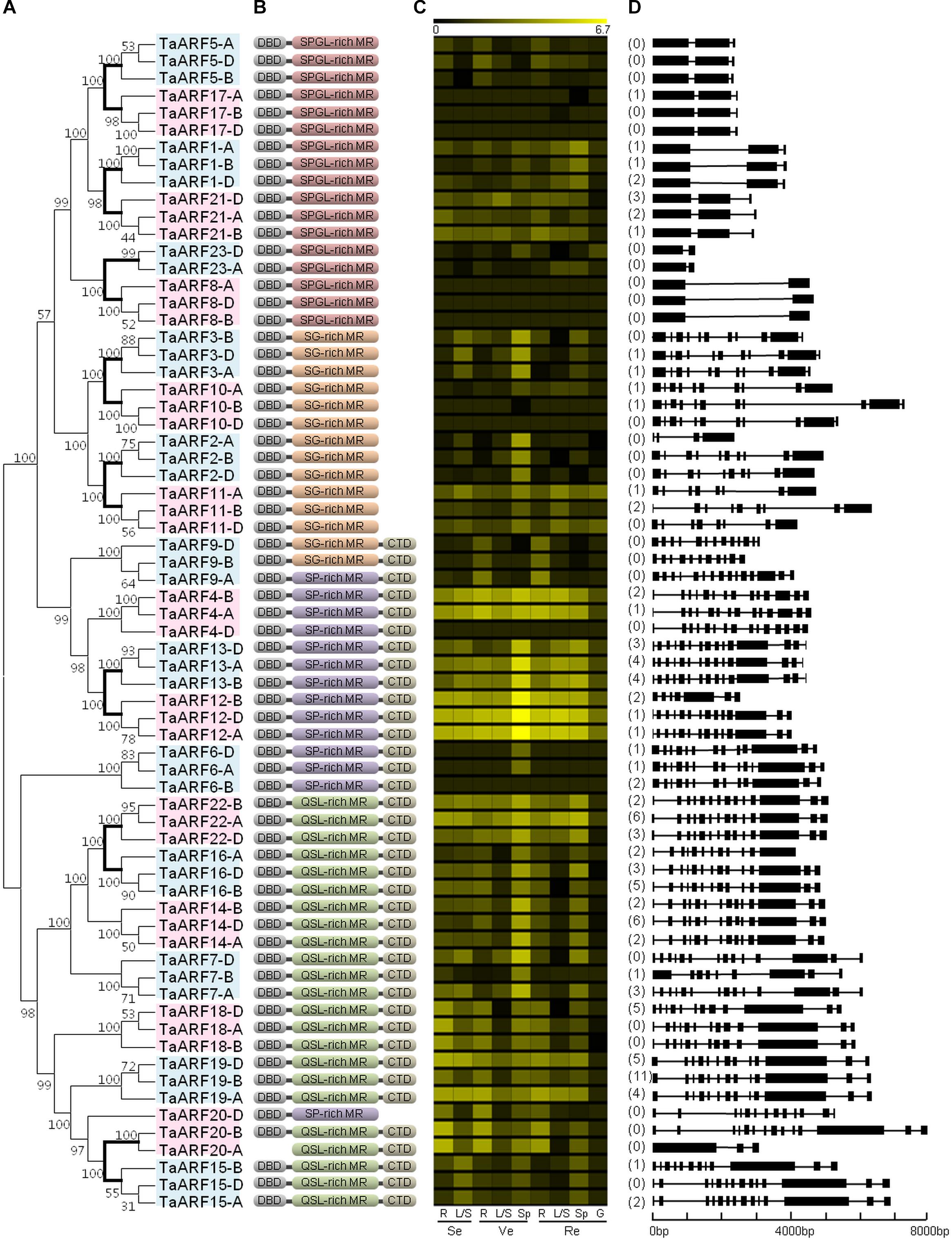
FIGURE 1. Phylogenetic relationships, domains, in silico expression profiling, and gene structure of ARFs in wheat. (A) Phylogenetic tree of TaARFs constructed from the complete alignment of 68 wheat ARF protein sequences by the neighbor-joining method with 1000 bootstrap replicates using MEGA 6.0. The bootstrap scores are indicated on the nodes, and the 23 TaARF members, all of which are coded by homeoalleles, are indicated in blue or pink boxes. The branch of paralogous TaARF members is shown in bold. (B) Conserved domains of TaARFs. DBD, DNA-binding domain; CTD, C-terminal dimerization domain; MR, middle region; S, serine; P, proline; G, glycine; L, leucine; Q, glutamine. (C) In silico expression profiling of TaARF homeoalleles in different organs at different growth stages of Chinese Spring wheat. The expression data were generated from the expVIP database (http://www.wheat-expression.com/). The color scale at the top represents the expression values: black indicates low levels of transcript abundance, and yellow indicates high levels. R, root; L/S, leaf/stem; Sp, spike; G, grain; Se, seedling stage; Ve, vegetative stage; Re, reproductive stage. (D) Exon/intron structures in the CDS of TaARF genes. The number of AS variants is listed before each gene. For genes that represent predicted AS variants, the variant with the highest in silico expression level has been selected for the gene structure analysis. Exons are represented by black boxes and introns by black lines. The size of exons and introns can be estimated using the scale at the bottom.
The length of the 68 TaARF proteins varied from 354 amino acids (AAs) for TaARF23-D to 1174 AA for TaARF20-B (Supplementary Table S3). With the exception of TaARF20-A, every TaARF protein contained a DBD for binding the promoter region of target genes. Thirty-eight TaARF proteins contained a CTD, suggesting that those proteins may be inhibited by Aux/IAA proteins. In addition, all of the 68 TaARF proteins contained a MR: 17 of the proteins contained an SPGL-rich MR, 14 contained an SG-rich MR, 14 contained an SP-rich MR, and 23 contained a QSL-rich MR (Figure 1B and Supplementary Figure S2). The first three types may suppress the expression of target genes, while the last one may promote it (Guilfoyle and Hagen, 2007). Furthermore, 34 TaARF proteins contained all three domains.
The full lengths of the TaARF genes varied from 1119 bp for TaARF23-A to 7915 bp for TaARF20-B, and the number of introns ranged from zero to 14 (Supplementary Table S3). The homeoalleles for most TaARF members, including TaARF1, 3, 5, 6, 8, 10, 13, 14, 17, 19, 21, 22, and 23, showed analogous exon/intron structures (Figure 1D). Furthermore, high sequence similarities (93.76–100%) were observed between several target sequences of wheat microRNAs and the sequences of TaARF1, 5, 7, 17, 21, and 22. Finally, TaARF1-A, -B, and -D and TaARF17-B were predicted to be the targets for tae-miR160 (Supplementary Figure S1 and Supplementary Table S4).
Furthermore, 41 homeoalleles of 17 TaARF members were found to have 103 AS variants; 16 homeoalleles had only one AS variant, while TaARF19-B had the most (12) AS variants (Figure 1D and Supplementary Table S3). Furthermore, 33 of the 103 AS variants resulted from two or more AS events, resulting in a total of 149 AS events. Among these AS events, 76 were IR events, which were the most abundant (51.7%), followed by A5SS (29), A3SS (23), ES (20), and MXE (1) events (Figure 2A). On the whole, the AS variants in the wheat B subgenome presented the most AS events among all AS types (Figure 2B).
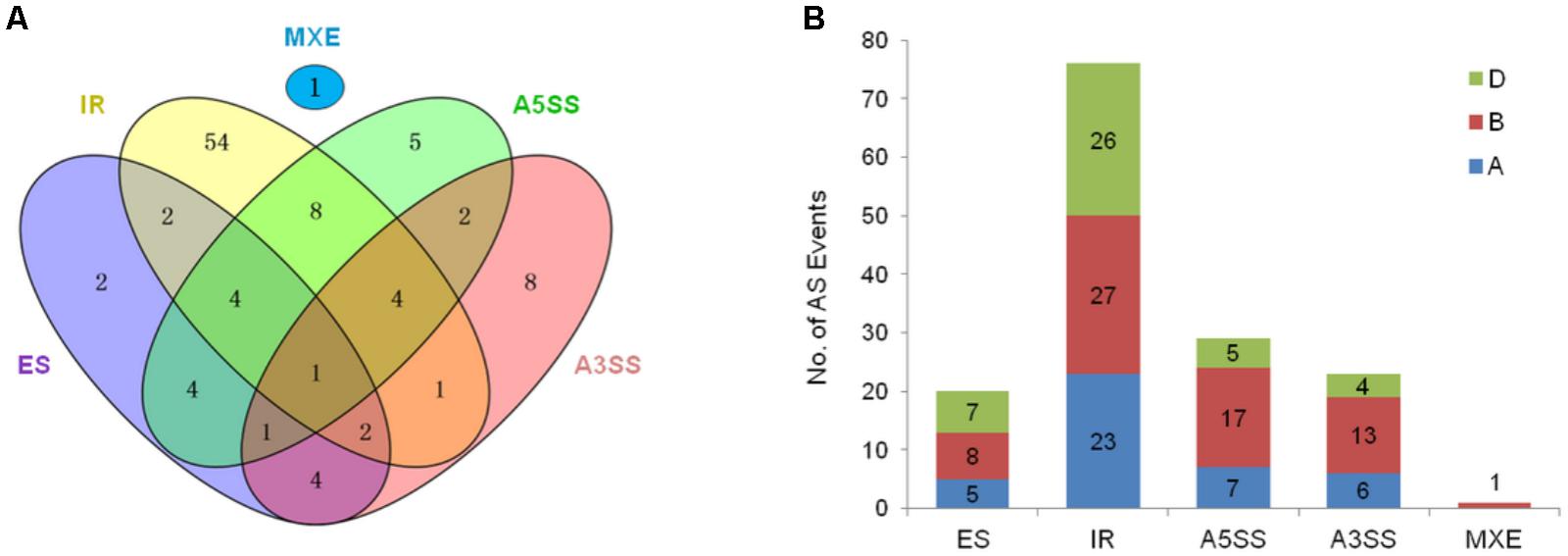
FIGURE 2. Summary of AS in TaARFs. (A) Five types of AS in 103 splice variants from TaARF homeoalleles. The number of splice variants is represented in each intersection of the Venn diagram. IR, intron retention; ES, exon skipping; A5SS, alternative 5′ splicing site; A3SS, alternative 3′ splicing site; MXE, mutually exclusive exon. (B) One hundred forty-nine AS events occurring in 103 splice variants. The AS events that occurred in the A, B, and D subgenomes are indicated in blue, red, and green, respectively.
In addition, the in silico expression data revealed that TaARF4, 12, 13, and 22 were highly expressed during seedling, vegetative, and reproductive stages in wheat (Figure 1C). The expression levels of TaARF5 and TaARF9 were highest in the root, while TaARF2 and TaARF3 were expressed highly in the spike.
We constructed a phylogenetic tree of the protein sequences of 68 TaARFs, 23 AtARFs, 25 OsARFs, and 14 ARFs with known functions from other species. These ARFs can be classified into nine groups: Groups I–IX (Supplementary Figure S3). TaARFs were distributed among all groups, and each TaARF member has an orthologous OsARF.
Based on homology cloning in previous studies, many ARFs were shown to have functions similar to those of their orthologous AtARFs, such as SlARF2 (Ren et al., 2017)–AtARF2 (Ellis et al., 2005), MtARF3 (Peng et al., 2017)–AtARF3 (Marin et al., 2010), SlARF4 (Sagar et al., 2013)–AtARF4 (Marin et al., 2010), GmARF8 (Wang et al., 2015)–AtARF8 (Pitaksaringkarn et al., 2014), SlARF9 (de Jong et al., 2015)–AtARF9, and BnARF18 (Liu et al., 2015)–AtARF18, and each of these ortholog pairs was located in the same branch (Supplementary Figure S3). Thus, TaARFs may also have functions similar to those of their orthologous OsARFs in the same branch. For example, TaARF9–OsARF1 (Shen et al., 2013) from Group V, TaARF15–OsARF5 (Indoliya et al., 2016 and TaARF20–OsARF19 (Zhang et al., 2015) from GroupVII, TaARF7–OsARF12 (Yang et al., 2006) from Group VIII, and TaARF16–OsARF6 (Meng et al., 2009) from Group IX, may have similar functions.
qRT-PCR analysis of TaARFs in the roots, stems, and leaves of wheat seedlings revealed no expression of TaARF5, TaARF7, or TaARF17. The response of the other 20 TaARF members to exogenous auxin stimuli was investigated in wheat seedlings treated with 10 μM NAA. qRT-PCR analysis revealed that seven TaARF members (TaARF7, 11, 12, 13, 15, 19, and 20) were upregulated and that two members, TaARF6 and 9, were downregulated in the roots in response to exogenous auxin treatment (Figure 3A). Among those TaARFs, the expression levels of TaARF11, 12, 13, 15, 19, and 20 increased gradually within 1.5 h after treatment (HAT) but decreased later, and the expression of TaARF6 was downregulated across all time points. Moreover, in wheat stems, the expression levels of 10 genes (TaARF1, 4, 9, 10, 12, 13, 15, 16, 19, and 20) were significantly upregulated by auxin (Figure 3B). Among those genes, the expression levels of TaARF1, 4, 10, 12, 19, and 20 were downregulated within the first 0.5 HAT but were continuously upregulated after 1.5–3 HAT, while the expression levels of TaARF9 and 13 were lower at 3 HAT than at 1.5 HAT; the expression of TaARF15 was upregulated across all time points. In addition, in wheat leaves, the expression levels of seven genes, TaARF4, 6, 9, 13, 15, 21, and 23, were significantly upregulated in response to auxin (Figure 3C), and TaARF4-21 and TaARF15-23 exhibited similar expression patterns in response to auxin. Overall, in the roots, stems, and leaves of wheat seedling, TaARF9, 13, and 15 responded significantly to auxin treatment.
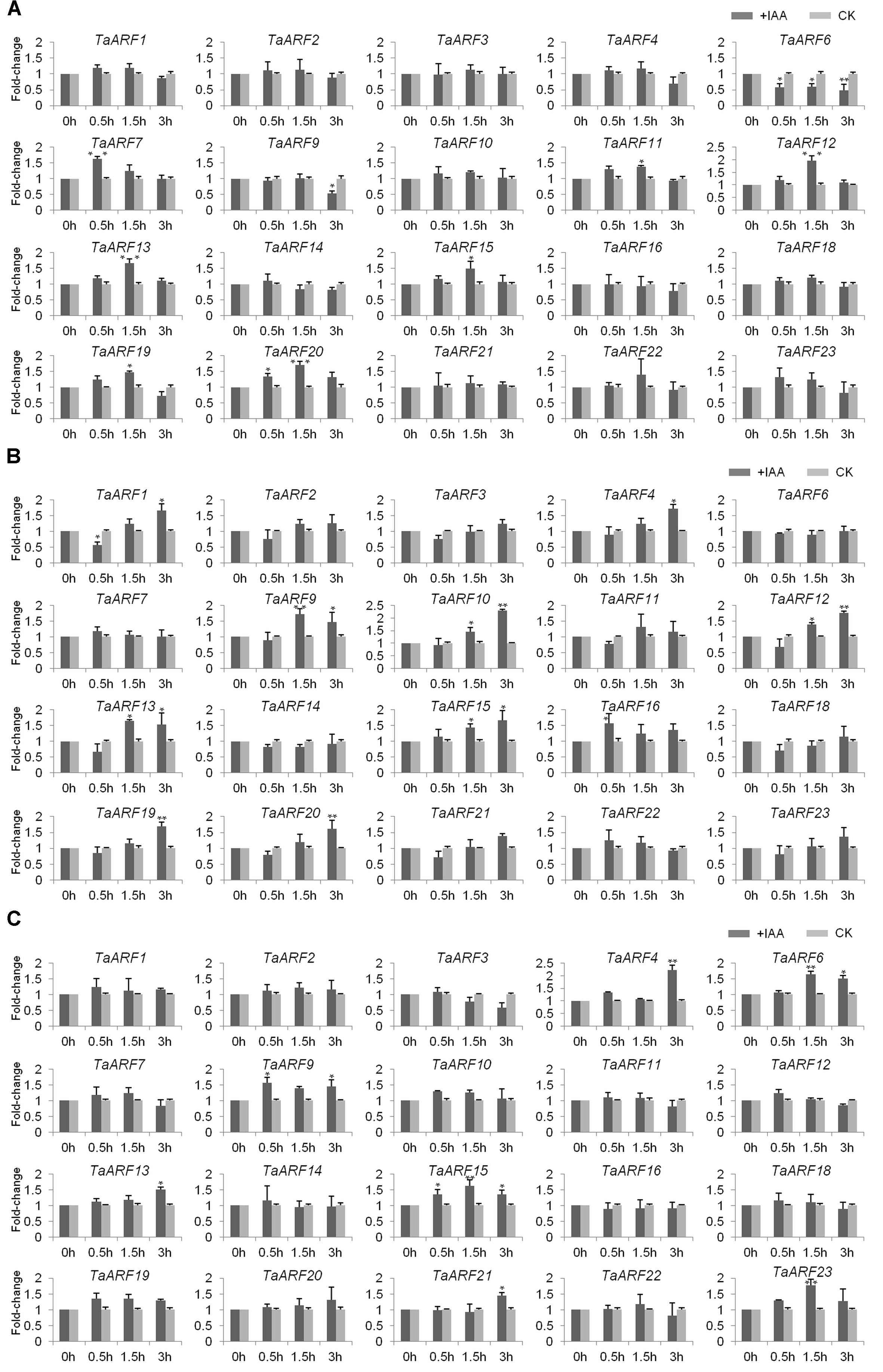
FIGURE 3. Induction (fold change) of TaARF genes in response to exogenous auxin stimuli in the roots (A), stems (B), and leaves (C) of wheat seedlings. The roots, stems and leaves of wheat cultivar “Chinese Spring” at the three-leaf stage were harvested at 0, 0.5, 1.5, and 3 HAT; the plants were treated with 10 μM α-NAA solution or distilled water (mock treatment). The relative expression level of each gene was measured three times and then normalized to that of the GADPH gene, after which the levels were analyzed using the fold-change method. Paired t-tests were used to detect significant differences in relative expression levels of genes between the auxin treatment and the mock treatment at each time point. The asterisks indicate significant differences, and the error bars indicate the SD.
In addition, the expression level of external control TaAux/IAA1 was similar to that reported in a previous study (Singla et al., 2006), indicating that the results are reliable (Supplementary Figure S4).
Based on the Arabidopsis and rice interaction network models, 46 proteins expressed in wheat, including 20 TaARF members, 25 TaIAA members (Qiao et al., 2015), and one TaTIR1, were subjected to an interaction analysis. The results showed that four closely related TaARF members (TaARF15, 18, 19, and 20; Figure 1A) could bind to TaIAA12, 13, and 19 in both models and that TaTIR1 could also bind to TaIAA proteins and inhibit their function (Figures 4A,B); these results are consistent with the TIR1/AFB–IAA–ARF interaction model in the auxin signaling pathway (Boer et al., 2014; Dinesh et al., 2015). The clustering results showed that the common domain III and domain IV of the CTD of TaARF15, 18, 19, and 20 as well as those of TaIAA12, 13, and 19 showed high sequence similarity, which suggests the occurrence of a similar secondary structure that facilitates protein interactions (Figure 4C).
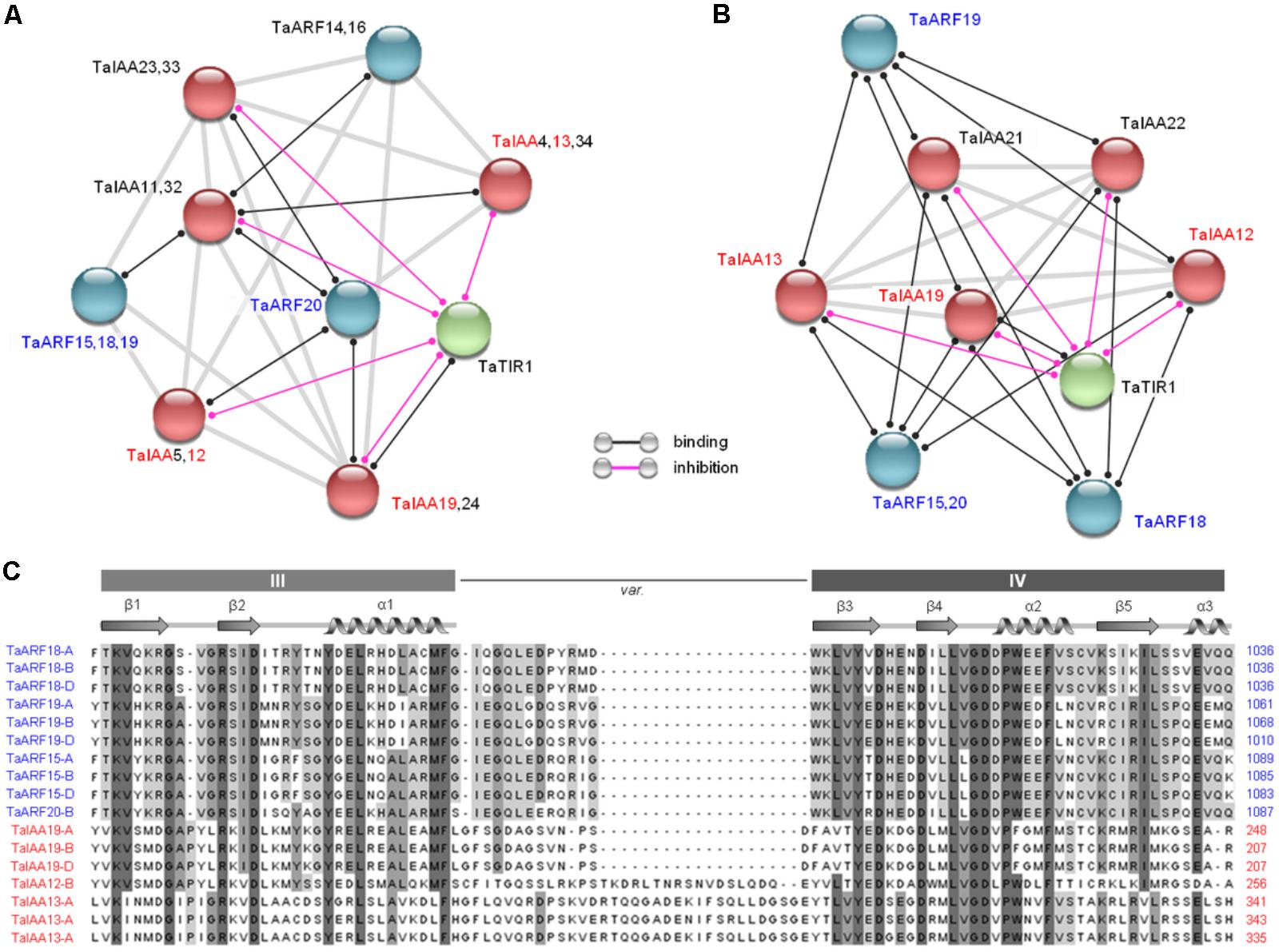
FIGURE 4. Prediction of protein interactions among TaARFs, TaIAAs, and TaTIR1. (A,B) Interaction networks of 20 TaARF members, 25 TaIAA members, and TaTIR1 expressed in wheat based on the interaction models of Arabidopsis and rice. Proteins for which interactions were not detected are not shown. The line colors indicate the types of interactions, which are listed in the bottom. (C) Residues predicted to participate in ARF and IAA protein interactions. The sequence alignment of TaARF and TaIAA proteins revealed canonical CTD domain features, including the conserved domain III and domain IV. The variable linker region is indicated by var. The α structures are shown by a spiral, and the β structures are shown by arrows.
At the protein level, by interacting with CTDs, Aux/IAAs inhibit ARFs. However, at the gene expression level, Aux/IAAs are regulated by ARFs because Aux/IAAs are the target genes of ARFs. Previous transcriptomic research in rice has shown that the expression levels of TIR1 and most ARF genes increased in response to exogenous auxin stimulation, which subsequently regulated the expression of downstream Aux/IAAs (Indoliya et al., 2016). In this study, in response to auxin treatment, the expression levels of seven of eight proteins that may interact were analyzed; TaARF18 was not chosen because its response to auxin was not significant (Figure 3). The results showed that the expression trends of TaTIR1, TaIAA13, TaARF15, TaARF19, and TaARF20 were consistent in wheat roots and that the expression patterns of TaTIR1, TaIAA13, and TaARF15 were similar not only in the stems but also in the leaves (Figure 5A). In addition, the TaIAA13 promoter contains AuxRE elements that are the binding sites for the DBD of ARF proteins (Supplementary Figure S5). Therefore, we inferred that TaTIR1–TaARF15/19/20–TaIAA13 could also interact at the gene expression level.
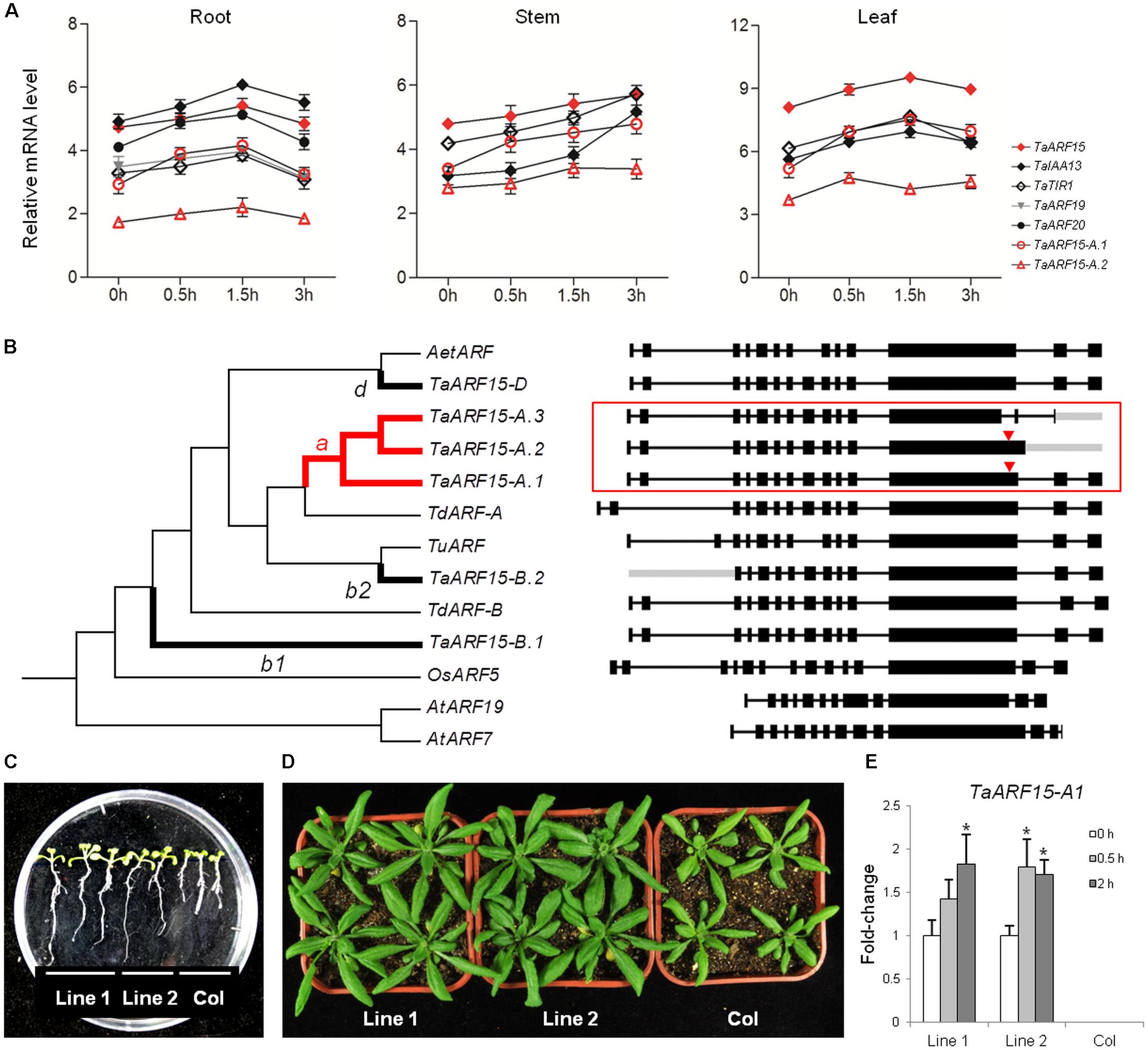
FIGURE 5. Co-expression, evolutionary, and functional analysis of TaARF15. (A) Auxin-related genes expressed simultaneously with TaARF15 in the roots, stems, and leaves of wheat cultivar “Chinese Spring” at the seedling stage. The qRT-PCR data were processed by the 2-ΔΔCt method, and GADPH served as the control. (B) Phylogenetic analyses of the six transcribed sequences of TaARF15 as well as those of its homologs in T. urartu (TuARF, TRIUR3_19224), Aegilops speltoides (AetARF, AEGTA09095), T. dicoccoides (TdARF-A, TRIDC6AG014880.2; TdARF-B, TRIDC6BG020370.14), and rice (OsARF5, Os02g04810); AtARF7 (AT5G20730.3) and AtARF19 (AT1G19220.1) served as out branches. The exon/intron structures are listed on the right. The branches with TaARF15-A, B, and D are shown as thick lines and labeled with letters a, b1/b2, or c, respectively, and the ω values were calculated in accordance with the two-ratio model. The branch that underwent positive selection is labeled in red, and the positive selection sites of sequences are marked with red triangles. (C,D) Morphology of the roots and leaves of 35S::TaARF15-A.1 lines and Col, the latter of which represents wild-type Arabidopsis plants. (E) Relative expression levels of TaARF15-A.1 in both Col and homozygous F3 transgenic lines. The total RNA was isolated from 20-day-old leaves of Arabidopsis plants at 0, 0.5, and 2 h after 10 μM auxin solution treatment. ACTIN2 served as an endogenous control.
On the whole, in response to auxin treatment, TaTIR1–TaARF15–TaIAA13 showed the same expression trends in the roots, stems and leaves. In addition to those of TaARF15-A, TaARF15-B, and TaARF15-D, the transcripts of TaARF15 can exist as two or one additional AS variant in the A and B subgenomes, respectively. These six transcripts of TaARF15 and their orthologs in both wheat ancestral species and rice were used to construct a phylogenetic tree. The result showed that ARF genes from wheat and its ancestors clustered into the same group (Figure 5B). Among those genes, TaARF15-D is located in the same branch as its ortholog AetARF in A. tauschii, which is the donor of the wheat D subgenome. Furthermore, TaARF15-A is located in the same branch as its orthologs TdARF-A and TuARF from the ancestral species of the A subgenome. However, TaARF15-B is distantly related to its ancestral ortholog TdARF-B.
Based on the two-ratio model, the evolutionary rates of TaARF15-A.1–3 (ωa = 0.66) and TaARF15-B.1 (ωb1 = 0.99) were significantly higher than those of the background branch (ω0 = 0.17–0.18, Supplementary Table S5). Additional analyses revealed that only branch-an underwent positive selection; the specific site was the 717L–736S segment of the TaARF15-A.1 protein-coding sequence, which is located on the exon 12 of the gene. TaARF15-A.2 also contains a positive selection site, but its translation is terminated prematurely because of an IR event within intron 12. In addition, TaARF15-A.3 lost a positive selection site in exon 12 because of an A5SS event. These examples may represent techniques used by wheat to regulate genes in different organs or stages.
qRT-PCR analysis for three AS variants of TaARF15 showed that TaARF15-A.1 was more highly expressed than TaARF15-A.2, while the expression of TaARF15-A.3 was not detected. Because of the response to exogenous auxin and the more complete gene structure among TaARF15-A transcripts, TaARF15-A.1 was transformed into Arabidopsis for functional verification. The results showed that the root length of the overexpression lines were significantly higher than those of the Col control lines (Figure 5C). Additionally, the leaf area of the overexpression lines exceeded those of the control lines (Figure 5D). Furthermore, qRT-PCR showed that TaARF15-A.1 was upregulated in the overexpression plants in response to exogenous auxin treatment. Thus, it can be inferred that TaARF15-A.1 participates in the development of roots and leaves during the vegetative growth stage of the plants (Figure 5E).
In this study, 23 members of the ARF family were isolated from the hexaploid wheat genome, which is closely related to that of its ancestral species, which include T. urartu (21), A. tauschii (23), and Triticum turgidum (25); gramineous grasses such as barley (25), Brachypodium distachyon (28), and rice (25, Wang et al., 2007); and the dicotyledon A. thaliana (23, Wei et al., 2006), and each ARF member exhibited good homology among grass species (Supplementary Figure S6). Phylogenetic analyses (Supplementary Figures S3, S6) revealed the poor sequence homology between TaARFs and AtARFs, which indicated that the ARF family differentiated after the divergence of mono- and dicotyledons (∼130 MYA, D’Hont et al., 2012) and that further specific differentiation occurred in Arabidopsis and gramineous plants. Interestingly, there are no ARF members on chromosome 4 in wheat, T. urartu, A. tauschii, T. turgidum, or barley. Therefore, the ARF family evolutionary progress at the stage during which the Triceae diverged from their grass ancestors (10–95 MYA, Pont et al., 2013) warrants further study.
There are eight pairs of paralogs in the TaARF family (Figure 1A). Among those paralogs, TaARF12-13 are both located on the long arm of wheat chromosome 3 at a close genomic distance, suggesting that those genes may have undergone a tandem duplication event. TaARF12-13 had homologous gene pairs in allied Triceae species but only one common ortholog in A. thaliana, rice, and B. distachyon, indicating that the tandem duplication occurred after the divergence of the Triceae ancestor (∼10 MYA, Pont et al., 2013). The remaining seven paralog pairs may have undergone segmental duplication events. Among these paralogs, TaARF1-21 has a homologous gene-pair, AtARF10-16, in A. thaliana, suggesting that this duplication event occurred the earliest, before the monocot–dicot divergence (∼130 MYA, D’Hont et al., 2012); however, the origins of TaARF2-11, TaARF3-10, TaARF5-17, TaARF8-23, TaARF15-20, and TaARF16-22 may occurred after the divergence. In addition, some redundant genes produced during genome doubling and duplication events may be recombined or modified, causing loss of function; these modifications include changes in structural variation, domains, and gene expression regulation as well as gradual loss (Chen, 2007; Otto, 2007). In this study, TaARF2-A and TaARF9-B lacked some exons, while TaARF20-A and TaARF20-D lacked a DBD and a CTD, respectively. Thus, these genes may suffer from the same lost fate as did TaARF23-B.
Alternative splicing is important for increasing the diversity and adaptability of plants (Matlin et al., 2005). In the hexaploid wheat genome, approximately 31% of the predicted coding genes have AS variants, among which IR events were the prevalent AS event (34%), followed by A3SS (27%), ES (20%), A5SS (19%), and MXE (0.04%) events (Clavijo et al., 2017). In this study, 103 AS variants were identified from 41 sequences of 17 TaARF members, and a total of 149 AS events, including IR (51.0%), A3SS (15.4%), ES (13.4%), A5SS (19.5%), and MXE (0.7%) events, occurred. IRs still represented the most common AS event in the TaARF splicing variants, and 18 of 77 IR events resulted in premature termination codons (PTCs). These PTC transcripts are often recognized and degraded by the nonsense-mediated mRNA decay (NMD) mechanism to avoid cell toxicity resulting from the accumulation of truncated protein products (Lykke-Andersen and Jensen, 2015); occasionally, these PTC transcripts encode shortened protein products that have new structures and functions (Romero et al., 2006). In addition, 17 of the 149 AS events occurred in the 5′-untranslated (UTR) region; if located in the regulatory protein binding site, the AS sites in this region can lead to altered expression of the TaARF gene. Similarly, 10 AS events occurred in the 3′-UTR region; these occurrences may also affect the microRNA- or long non-coding RNA (lncRNA)-based regulation of TaARF expression (Hughes, 2006).
Since the homeoalleles of most tri-genes exhibited similar expression levels (Pfeifer et al., 2014), we used universal genomic primers to analyze the expression of each TaARF member. The results showed that three TaARF members, TaARF5, 7, and 17, were not expressed, but the remaining members were all expressed in the roots, stems and leaves of wheat seedlings. Among the TaARF members, TaARF9 and 13 in Group IV and TaARF15 in Group VI both significantly responded to exogenous auxin. Since other ARFs in the same group, including AtARF2, SlARF2, OsARF1, and OsARF24 in Group IV as well as AtARF7, AtARF19, OsARF19, SlARF19, and OsARF5 in Group VI, were confirmed to be involved in the regulatory development of plant roots, stems, and leaves (Ellis et al., 2005; Inukai et al., 2005; Sakamoto and Inukai, 2013; Galvan-Ampudia and Vernoux, 2014; Ma et al., 2015; Zhang et al., 2015; Indoliya et al., 2016; Ren et al., 2017), it is speculated that TaARF9, 13, and 15 also have similar functions. In addition, the in silico expression data revealed that in Group IV, TaARF9 showed a high expression level in roots and TaARF12 as well as TaARF13 were highly expressed during seedlings, vegetative and reproductive stages in wheat. Moreover, OsARF1, the homologous gene of TaARF9, promoted the response of primary roots, lateral roots, and root hair to auxin (Shen et al., 2013; numbered as OsARF16); AtARF2 (Ellis et al., 2005; Okushima et al., 2005; Lim et al., 2010), SlARF2 (Xu et al., 2016; Ren et al., 2017), and ZmARF25 (von Behrens et al., 2011; Li et al., 2014), in the same subgroup as TaARF12 and TaARF13, were all confirmed to be involved in the regulation various growth stages. Therefore, TaARF9, 12, and 13 may also have similar functions. Furthermore, TaARF13 has a very close relationship with the expressed sequence tag AY902381 (Supplementary Figure S7), the only ARF sequence reported in wheat (Liu et al., 2017).
In addition to the temporal and spatial specificity of TaARF members and their induction by exogenous hormones, the expression of TaARF members may also be regulated by microRNA, which is a complex processes. For example, TaARF17 is predicted to be the target gene of tae-miR160; thus, the lack of expression of TaARF17 in the roots, stems, and leaves of wheat seedlings in this study is most likely due to the inhibition of miRNA. Next, we will focus on the impact of AS and microRNAs on the expression levels of some TaARF members.
TaARF15 is an ortholog of OsARF5, which regulates the development of rice at different stages (Indoliya et al., 2016). In this study, TaARF15 was expressed in the roots, stems, and leaves of wheat seedling, and its expression levels significantly differed in response to exogenous auxin treatment. Because stems are the major organ involved in polar transport and are relatively insensitive to auxin, ARFs could be constantly upregulated by auxin stimuli. Therefore, the expression level of TaARF15 continuously increased in wheat stems. In the roots and leaves, however, the expression level of TaARF15 continuously increased during the first 1.5 HAT but decreased at 3 HAT, suggesting that a negative autoregulatory feedback loop (de Jong et al., 2009) occurs in the roots and leaves sensitive to auxin treatment, causing the expression of TaARF15 to gradually return to its initial level. In the above process, the expression patterns of the genes TaARF15 and TaTIR1 and the downstream gene TaIAA13 were consistent. Because the TaARF15 protein has a QSL-MR, which may promote downstream gene expression, as well as a DBD that can bind with the AuxRE of the TaIAA13 promoter, we speculated that TaIAA13 is the target gene of TaARF19. The network prediction indicated that there is also a protein-level interaction involving TaTIR1–TaIAA13–TaARF15. Furthermore, TaARF15 can express a total of six transcripts, in which TaARF15-A.1–3 from the A subgenome exhibited a high evolutionary rate, and positive selection sites were detected in TaARF15. In addition, TaARF15-A.1, which has a more complete gene structure and can better respond to exogenous auxin than TaARF15-A.2 and -A.3, was transferred to A. thaliana. Compared with the wild-type plants, the transgenic plants had longer roots and greater leaf area, and TaARF15-A.1 could respond to exogenous auxin, which meant that TaARF15-A.1 may participate in the regulatory development of the roots and leaves. The function of the remaining five transcripts needs further study.
The TaARF family has a total of 23 members, and each member except two TaARF members may be the targets of tae-miR160. Seventeen TaARF members have extra transcripts that undergo 149 AS events, including IR (76), A5SS (29), A3SS (23), ES (20), and MXE (1) events. Thirty-seven (54%) TaARF protein sequences have a DBD, MR, and CTD. Twenty TaARF members are expressed among different organs and tissues. In response to auxin treatment, the expression of nine, 10, and seven TaARF members significantly differed in the roots, stems, and leaves of wheat seedlings, respectively. Overall, TaARF9, 13, and 15 responded significantly to auxin treatment in all the three organs. In addition, TaTIR1-TaARF15, 18, 19, and 20-TaIAA12, 13, and 19 were predicted to be interactive proteins, and TaTIR1–TaARF15/19/20–TaIAA13 exhibited similar expression patterns at the genetic level. TaARF15-A.1 is likely involved in the regulation of roots and leaves of A. thaliana.
LQ, ZC, and JZ conceived and designed the experiments. LQ, WZ, XZ, XL, HG, and YR performed the experiments. LQ analyzed the data. XyL and LQ contributed reagents, materials, and analysis tools. LQ and LZ wrote the manuscript.
This study was funded by the National Key Research and Development Plan of China (2017YFD0100600), the Key Project of Science and Technology Innovation Platform of Shanxi Province (201605D151002), the Natural Science Foundation of Shanxi Province (201601D102051), the Special Project for Talents of Shanxi Province (201705D211025), and the Foundation for Youths of Institute of Crop Science, Shanxi Academy of Agricultural Sciences (ZZQ1701).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01395/full#supplementary-material
Ahmed, S. S., Gong, Z. H., Ji, J. J., Yin, Y. X., Xiao, H. J., Khan, M. A., et al. (2012). Construction of the intermediate vector pVBG2307 by incorporating vital elements of expression vectors pBI121 and pBI221. Genet. Mol. Res. 11, 3091–3104. doi: 10.4238/2012.August.31.7
Avni, R., Nave, M., Barad, O., Baruch, K., Twardziok, S. O., Gundlach, H., et al. (2017). Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357, 93–97. doi: 10.1126/science.aan0032
Boer, D. R., Freire-Rios, A., van den Berg, W. A., Saaki, T., Manfield, I. W., Kepinski, S., et al. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. doi: 10.1016/j.cell.2013.12.027
Borrill, P., Ramirez-Gonzalez, R., and Uauy, C. (2016). expVIP: a customisable RNA-seq data analysis and visualisation platform. Plant Physiol. 170, 2172–2186. doi: 10.1104/pp.15.01667
Brenchley, R., Spannagl, M., Pfeifer, M., Barker, G. L., D’Amore, R., Allen, A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. doi: 10.1038/nature11650
Chen, Z. J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58, 377–406. doi: 10.1146/annurev.arplant.58.032806.103835
Clavijo, B. J., Venturini, L., Schudoma, C., Accinelli, G. G., Kaithakottil, G., Wright, J., et al. (2017). An improved assembly and annotation of the allohexaploid wheat genome identifies complete families of agronomic genes and provides genomic evidence for chromosomal translocations. Genome Res. 27, 885–896. doi: 10.1101/gr.217117.116
Combet, C., Blanchet, C., Geourjon, C., and Deleage, G. (2000). NPS@: network protein sequence analysis. Trends Biochem. Sci 25, 147–150. doi: 10.1016/S0968-0004(99)01540-6
Dai, X., Zhuang, Z., and Zhao, P. X. (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46, W49–W54. doi: 10.1093/nar/gky316
de Jong, M., Wolters-Arts, M., Feron, R., Mariani, C., and Vriezen, W. H. (2009). The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170. doi: 10.1111/j.1365-313X.2008.03671.x
de Jong, M., Wolters-Arts, M., Schimmel, B. C., Stultiens, C. L., de Groot, P. F., Powers, S. J., et al. (2015). Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 66, 3405–3416. doi: 10.1093/jxb/erv152
D’Hont, A., Denoeud, F., Aury, J. M., Baurens, F. C., Carreel, F., Garsmeur, O., et al. (2012). The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. doi: 10.1038/nature11241
Dinesh, D. C., Kovermann, M., Gopalswamy, M., Hellmuth, A., Calderón Villalobos, L. I., Lilie, H., et al. (2015). Solution structure of the PsIAA4 oligomerization domain reveals interaction modes for transcription factors in early auxin response. Proc. Natl. Acad. Sci. U.S.A. 112, 6230–6235. doi: 10.1073/pnas.1424077112
Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J., and Reed, J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574. doi: 10.1242/dev.02012
Galvan-Ampudia, C. S., and Vernoux, T. (2014). Signal integration by GSK3 kinases in the root. Nat. Cell Biol. 16, 21–23. doi: 10.1038/ncb2898
Glazinska, P., Wojciechowski, W., Wilmowicz, E., Zienkiewicz, A., Frankowski, K., and Kopcewicz, J. (2014). The involvement of InMIR167 in the regulation of expression of its target gene InARF8, and their participation in the vegetative and generative development of ipomoea nil plants. J. Plant Physiol. 171, 225–234. doi: 10.1016/j.jplph.2013.07.011
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Gutierrez, L., Bussell, J. D., Pacurar, D. I., Schwambach, J., Pacurar, M., and Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Ha, C. V., Le, D. T., Nishiyama, R., Watanabe, Y., Sulieman, S., Tran, U. T., et al. (2013). The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Res. 20, 511–524. doi: 10.1093/dnares/dst027
Herrera-Estrella, L., Simpson, J., and Martinez-Trujillo, M. (2005). Transgenic plants: an historical perspective. Methods Mol. Biol. 286, 3–32. doi: 10.1385/1-59259-827-7:003
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hughes, T. A. (2006). Regulation of gene expression by alternative untranslated regions. Trends Genet. 22, 119–122. doi: 10.1016/j.tig.2006.01.001
Indoliya, Y., Tiwari, P., Chauhan, A. S., Goel, R., Shri, M., Bag, S. K., et al. (2016). Decoding regulatory landscape of somatic embryogenesis reveals differential regulatory networks between japonica and indica rice subspecies. Sci. Rep. 6:23050. doi: 10.1038/srep23050
Inukai, Y., Sakamoto, T., Ueguchi-Tanaka, M., Shibata, Y., Gomi, K., Umemura, I., et al. (2005). Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell 17, 1387–1396. doi: 10.1105/tpc.105.030981
Jing, H., Yang, X., Zhang, J., Liu, X., Zheng, H., Dong, G., et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nat. Commun. 6:7395. doi: 10.1038/ncomms8395
Kaur, S., Dhugga, K. S., Beech, R., and Singh, J. (2017). Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol. 17:193. doi: 10.1186/s12870-017-1142-z
Kim, E., Magen, A., and Ast, G. (2007). Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35, 125–131. doi: 10.1093/nar/gkl924
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Li, C., Wang, C., Meng, L., Xing, J., Wang, T., Yang, H., et al. (2014). Ectopic expression of a maize hybrid down-regulated gene ZmARF25 decreases organ size by affecting cellular proliferation in Arabidopsis. PLoS One 9:e94830. doi: 10.1371/journal.pone.0094830
Lim, P. O., Lee, I. C., Kim, J., Kim, H. J., Ryu, J. S., Woo, H. R., et al. (2010). Auxin response factor 2 (ARF2) plays a major role in regulating auxin-mediated leaf longevity. J. Exp. Bot. 61, 1419–1430. doi: 10.1093/jxb/erq010
Ling, H. Q., Zhao, S., Liu, D., Wang, J., Sun, H., Zhang, C., et al. (2013). Draft genome of the wheat A-genome progenitor Triticum urartu. Nature 496, 87–90. doi: 10.1038/nature11997
Liu, J., Hua, W., Hu, Z., Yang, H., Zhang, L., Li, R., et al. (2015). Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. U.S.A. 112, E5123–E5132. doi: 10.1073/pnas.1502160112
Liu, X., Lin, Y., Liu, D., Wang, C., Zhao, Z., Cui, X., et al. (2017). MAPK-mediated auxin signal transduction pathways regulate the malic acid secretion under aluminum stress in wheat (Triticum aestivum L.). Sci. Rep. 7:1620. doi: 10.1038/s41598-017-01803-3
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lykke-Andersen, S., and Jensen, T. H. (2015). Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat. Rev. Mol. Cell Biol. 16, 665–677. doi: 10.1038/nrm4063
Ma, C., Meir, S., Xiao, L., Tong, J., Liu, Q., Reid, M. S., et al. (2015). A knotted1-like homeobox protein regulates abscission in tomato by modulating the auxin pathway. Plant Physiol. 167, 844–853. doi: 10.1104/pp.114.253815
Marin, E., Jouannet, V., Herz, A., Lokerse, A. S., Weijers, D., Vaucheret, H., et al. (2010). miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell 22, 1104–1117. doi: 10.1105/tpc.109.072553
Matlin, A. J., Clark, F., and Smith, C. W. (2005). Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6, 386–398. doi: 10.1038/nrm1645
Mayer, K. F., Rogers, J., Doležel, J., Pozniak, C., Eversole, K., Feuillet, C., et al. (2014). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788. doi: 10.1126/science.1251788
Meng, Y., Huang, F., Shi, Q., Cao, J., Chen, D., Zhang, J., et al. (2009). Genome-wide survey of rice microRNAs and microRNA-target pairs in the root of a novel auxin-resistant mutant. Planta 230, 883–898. doi: 10.1007/s00425-009-0994-3
Mironova, V., Teale, W., Shahriari, M., Dawson, J., and Palme, K. (2017). The systems biology of auxin in developing embryos. Trends Plant Sci. 22, 225–235. doi: 10.1016/j.tplants.2016.11.010
Moller, B. K., Ten Hove, C. A., Xiang, D., Williams, N., Lopez, L. G., Yoshida, S., et al. (2017). Auxin response cell-autonomously controls ground tissue initiation in the early Arabidopsis embryo. Proc. Natl. Acad. Sci. U.S.A. 114, E2533–E2539. doi: 10.1073/pnas.1616493114
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Okushima, Y., Mitina, I., Quach, H. L., and Theologis, A. (2005). AUXIN RESPONSE FACTOR 2 (ARF2): a pleiotropic developmental regulator. Plant J. 43, 29–46. doi: 10.1111/j.1365-313X.2005.02426.x
Otto, S. P. (2007). The evolutionary consequences of polyploidy. Cell 131, 452–462. doi: 10.1016/j.cell.2007.10.022
Peng, J., Berbel, A., Madueno, F., and Chen, R. (2017). AUXIN RESPONSE FACTOR3 regulates compound leaf patterning by directly repressing PALMATE-LIKE PENTAFOLIATA1 expression in Medicago truncatula. Front. Plant Sci. 8:1630. doi: 10.3389/fpls.2017.01630
Pfeifer, M., Kugler, K. G., Sandve, S. R., Zhan, B., Rudi, H., Hvidsten, T. R., et al. (2014). Genome interplay in the grain transcriptome of hexaploid bread wheat. Science 345:1250091. doi: 10.1126/science.1250091
Pitaksaringkarn, W., Ishiguro, S., Asahina, M., and Satoh, S. (2014). ARF6 and ARF8 contribute to tissue reunion in incised Arabidopsis inflorescence stems. Plant Biotechnol. 31, 49–53. doi: 10.5511/plantbiotechnology.13.1028b
Pont, C., Murat, F., Guizard, S., Flores, R., Foucrier, S., Bidet, Y., et al. (2013). Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J. 76, 1030–1044. doi: 10.1111/tpj.12366
Qiao, L., Zhang, X., Han, X., Zhang, L., Li, X., Zhan, H., et al. (2015). A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front. Plant Sci. 6:770. doi: 10.3389/fpls.2015.00770
Ramírez-González, R. H., Borrill, P., Lang, D., Harrington, S. A., Brinton, J., Venturini, L., et al. (2018). The transcriptional landscape of polyploid wheat. Science 361:eaar6089. doi: 10.1126/science.aar6089
Ren, Z., Liu, R., Gu, W., and Dong, X. (2017). The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 256, 103–111. doi: 10.1016/j.plantsci.2016.12.008
Romero, P. R., Zaidi, S., Fang, Y. Y., Uversky, V. N., Radivojac, P., Oldfield, C. J., et al. (2006). Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc. Natl. Acad. Sci. U.S.A. 103, 8390–8395. doi: 10.1073/pnas.0507916103
Sagar, M., Chervin, C., Mila, I., Hao, Y., Roustan, J. P., Benichou, M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374. doi: 10.1104/pp.113.213843
Sakamoto, T., and Inukai, Y. (2013). Characterization of a Tos17 insertion mutant of rice auxin signal transcription factor gene, OsARF24. Am. J. Plant Sci. 4, 84–91. doi: 10.4236/ajps.2013.41013
Shen, C., Wang, S., Zhang, S., Xu, Y., Qian, Q., Qi, Y., et al. (2013). OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 36, 607–620. doi: 10.1111/pce.12001
Shen, C., Yue, R., Sun, T., Zhang, L., Xu, L., Tie, S., et al. (2015). Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front. Plant Sci. 6:73. doi: 10.3389/fpls.2015.00073
Singla, B., Chugh, A., Khurana, J. P., and Khurana, P. (2006). An early auxin-responsive Aux/IAA gene from wheat (Triticum aestivum) is induced by epibrassinolide and differentially regulated by light and calcium. J. Exp. Bot. 57, 4059–4070. doi: 10.1093/jxb/erl182
Strader, L. C., and Zhao, Y. (2016). Auxin perception and downstream events. Curr. Opin. Plant Biol. 33, 8–14. doi: 10.1016/j.pbi.2016.04.004
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. doi: 10.1093/nar/gku1003
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Ulmasov, T., Liu, Z. B., Hagen, G., and Guilfoyle, T. J. (1995). Composite structure of auxin response elements. Plant Cell 7, 1611–1623. doi: 10.1105/tpc.7.10.1611
von Behrens, I., Komatsu, M., Zhang, Y., Berendzen, K. W., Niu, X., Sakai, H., et al. (2011). Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J. 66, 341–353. doi: 10.1111/j.1365-313X.2011.04495.x
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, S., Bai, Y., Shen, C., Wu, Y., Zhang, S., Jiang, D., et al. (2010). Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genomics 10, 533–546. doi: 10.1007/s10142-010-0174-3
Wang, Y., Li, K., Chen, L., Zou, Y., Liu, H., Tian, Y., et al. (2015). MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 168, 984–999. doi: 10.1104/pp.15.00265
Wei, H., Cui, B., Ren, Y., Li, J., Liao, W., Xu, N., et al. (2006). Research progresses on auxin response factors. J Integr. Plant Biol. 48, 622–627. doi: 10.1111/j.1744-7909.2006.00280.x
Wheeler, T. J., and Eddy, S. R. (2013). nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489. doi: 10.1093/bioinformatics/btt403
Winkler, M., Niemeyer, M., Hellmuth, A., Janitza, P., Christ, G., Samodelov, S. L., et al. (2017). Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 8:15706. doi: 10.1038/ncomms15706
Xia, K., Wang, R., Ou, X., Fang, Z., Tian, C., Duan, J., et al. (2012). OsTIR1 and OsAFB2 downregulation via OsMIR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One 7:e30039. doi: 10.1371/journal.pone.0030039
Xing, H., Pudake, R. N., Guo, G., Xing, G., Hu, Z., Zhang, Y., et al. (2011). Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 12:178. doi: 10.1186/1471-2164-12-178
Xu, T., Liu, X., Wang, R., Dong, X., Guan, X., Wang, Y., et al. (2016). SlARF2a plays a negative role in mediating axillary shoot formation. Sci. Rep. 6:33728. doi: 10.1038/srep33728
Yang, J. H., Han, S. J., Yoon, E. K., and Lee, W. S. (2006). Evidence of an auxin signal pathway, microRNA167-ARF8-GH3, and its response to exogenous auxin in cultured rice cells. Nucleic Acids Res. 34, 1892–1899. doi: 10.1093/nar/gkl118
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yang, Z., Wong, W. S., and Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Yang, Z. B., Geng, X., He, C., Zhang, F., Wang, R., Horst, W. J., et al. (2014). TAA1-regulated local auxin biosynthesis in the root-apex transition zone mediates the aluminum-induced inhibition of root growth in Arabidopsis. Plant Cell 26, 2889–2904. doi: 10.1105/tpc.114.127993
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhao, G., Zou, C., Li, K., Wang, K., Li, T., Gao, L., et al. (2017). The Aegilops tauschii genome reveals multiple impacts of transposons. Nat. Plants 3, 946–955. doi: 10.1038/s41477-017-0067-8
Zhao, Z., Zhang, Y., Liu, X., Zhang, X., Liu, S., Yu, X., et al. (2013). A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev. Cell 27, 113–122. doi: 10.1016/j.devcel.2013.09.005
Zhu, Q., Li, B., Mu, S., Han, B., Cui, R., Xu, M., et al. (2013). TTG2-regulated development is related to expression of putative AUXIN RESPONSE FACTOR genes in tobacco. BMC Genomics 14:806. doi: 10.1186/1471-2164-14-806
Zimin, A. V., Puiu, D., Hall, R., Kingan, S., Clavijo, B. J., and Salzberg, S. L. (2017). The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. Gigascience 6, 1–7. doi: 10.1093/gigascience/gix097
Keywords: genomewide, ARFs, alternative splicing, expression pattern, PAML, transgenic functional verification
Citation: Qiao L, Zhang W, Li X, Zhang L, Zhang X, Li X, Guo H, Ren Y, Zheng J and Chang Z (2018) Characterization and Expression Patterns of Auxin Response Factors in Wheat. Front. Plant Sci. 9:1395. doi: 10.3389/fpls.2018.01395
Received: 01 April 2018; Accepted: 03 September 2018;
Published: 19 September 2018.
Edited by:
Zhong-Nan Yang, Shanghai Normal University, ChinaReviewed by:
Han Xiao, Shanghai Institutes for Biological Sciences (CAS), ChinaCopyright © 2018 Qiao, Zhang, Li, Zhang, Zhang, Li, Guo, Ren, Zheng and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Zheng, emhlbmdqc3hhYXNAMTI2LmNvbQ== Zhijian Chang, Y3pqc3hhYXNAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.