
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 08 October 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01387
 Przemysław Jagodzik1
Przemysław Jagodzik1 Małgorzata Tajdel-Zielinska2
Małgorzata Tajdel-Zielinska2 Agata Ciesla2
Agata Ciesla2 Małgorzata Marczak2
Małgorzata Marczak2 Agnieszka Ludwikow2*
Agnieszka Ludwikow2*Mitogen-activated protein kinase (MAPK) modules play key roles in the transduction of environmental and developmental signals through phosphorylation of downstream signaling targets, including other kinases, enzymes, cytoskeletal proteins or transcription factors, in all eukaryotic cells. A typical MAPK cascade consists of at least three sequentially acting serine/threonine kinases, a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK) and finally, the MAP kinase (MAPK) itself, with each phosphorylating, and hence activating, the next kinase in the cascade. Recent advances in our understanding of hormone signaling pathways have led to the discovery of new regulatory systems. In particular, this research has revealed the emerging role of crosstalk between the protein components of various signaling pathways and the involvement of this crosstalk in multiple cellular processes. Here we provide an overview of current models and mechanisms of hormone signaling with a special emphasis on the role of MAPKs in cell signaling networks.
One-sentence summary: In this review we highlight the mechanisms of crosstalk between MAPK cascades and plant hormone signaling pathways and summarize recent findings on MAPK regulation and function in various cellular processes.
Mitogen-activated protein kinases (MAPKs) are one of the largest group of transferases, catalyzing phosphorylation of appropriate protein substrates on serine or threonine residues. MAPK cascades are among the most common mechanisms by which cell functions are regulated and are evolutionarily conserved throughout the eukaryotes, including plants, fungi and mammals (Zanke et al., 1996; Ligterink and Hirt, 2001; Xu et al., 2017). In plants, they play essential roles in the transduction of environmental and developmental signals. MAPKs are present in the cytoplasm and nucleus, and take part in different cellular processes including growth, development and stress responses (Seguí-Simarro et al., 2005; Pitzschke et al., 2009; Gupta and Chakrabarty, 2013; Sheikh et al., 2013; Danquah et al., 2015; Wang Z. et al., 2015). By regulating MAPK cascades, cells are able to respond to a range of stresses caused by high or low temperature, UV radiation, ozone, reactive oxygen species, drought, high or low osmolarity, heavy metals, wounding and pathogen infections (Sinha et al., 2011; Opdenakker et al., 2012; Danquah et al., 2014; de Zelicourt et al., 2016). Importantly, hormones such as auxin (AUX), abscisic acid (ABA), jasmonic acid (JA), salicylic acid (SA), ethylene (ET), brassinosteroids (BR), and gibberellins (GA) are known to influence signaling through MAPK cascades (Mishra et al., 2006; Rodriguez et al., 2010; Smekalova et al., 2013; Hettenhausen et al., 2014; Lu et al., 2015). In this review we highlight the mechanisms of crosstalk between MAPK cascades and plant hormone signaling pathways and summarize recent findings on MAPK regulation and function in various cellular processes.
The transduction and enhancement of input signals by MAPK cascades involves three types of kinase: mitogen activated protein (MAP) kinase kinase kinases (MAPKKKs; also known as MAP3Ks or MEKKs), MAP kinase kinases (MKKs; also known as MAP2Ks or MEKs) and MAP kinases (MAPKs; also known as MPK) (Figure 1). MAP kinase kinase kinase kinases (MAPKKKKs) have also been identified in plants (Colcombet and Hirt, 2008; Raja et al., 2017). The first signal transduction step is the activation of a MAPKKKK or MAPKKK by stimulation of plasma membrane receptors. The MAPKKK then activates a downstream MAPKK by phosphorylation of two serine or threonine residues in the S/T-X5-S/T (X is any amino acid) motif of its activation loop. Once activated, the MAPKK behaves as a dual-specificity kinase, which phosphorylates a MAPK on the threonine and tyrosine residues in the T-X-Y motif of an activation loop located between subdomains VII and VIII of its catalytic domain (Rodriguez et al., 2010; Hettenhausen et al., 2014). MAPKs are serine/threonine kinases that activate various effector proteins in the cytoplasm or nucleus, including other kinases, enzymes, cytoskeletal proteins or transcription factors (Khokhlatchev et al., 1998; Rodriguez et al., 2010). Interactions between the kinases are mediated by docking sites in the enzymes themselves and/or by external scaffolding proteins. Such a series of phosphorylation events is termed a MAPK cascade.
Nearly 110 genes encoding MAPK cascade kinases have been identified in the Arabidopsis thaliana genome; these genes encode 20 MAPK, 10 MAPKK, and 80 MAPKKK proteins (Colcombet and Hirt, 2008; de Zelicourt et al., 2016; Raja et al., 2017). There are comparable numbers of MAPK cascade kinase genes in other plant species, e.g., the rice (Oryza sativa) genome contains 17 MAPK, 8 MAPKK and 75 MAPKKK genes (Xiong et al., 2001; Singh et al., 2012; Wankhede et al., 2013), the tomato (Solanum lycopersicum) genome 17 MAPK (Mohanta et al., 2015), 5 MAPKK and 89 MAPKKK genes (Wu et al., 2014) and the maize (Zea mays) genome 19 MAPK, 9 MAPKK and 74 MAPKKK genes (Kong et al., 2013) (Supplementary Tables 1, 2). Similar gene sets in other plant genomes and the presence of MAPK cascade kinases in a huge range of species indicate that they are evolutionarily conserved among higher plants (Danquah et al., 2014).
The MAPKKK family forms the largest and most heterogeneous group of MAPK cascade components (Danquah et al., 2014). Based on sequence analysis, Arabidopsis MAPKKKs can be divided into three main classes: MEKK-like (21 members), Raf-like (48 members) and ZIK-like (11 members; also known as WNK [with no lysine (K)] MAPKKKs (Danquah et al., 2014).
MEKK-like kinases fall into four subgroups (A1 – A4) (MAPK Group, 2002). Subgroup A1 in A. thaliana includes four functionally characterized protein kinases: MAP/ERK kinase kinase 1 (AtMEKK1, also called AtMAPKKK8), AtMEKK2 (AtMAPKKK9), AtMEKK3 (AtMAPKKK10), and AtMEKK4 (AtMAPKKK11) (MAPK Group, 2002). MEKK-like kinases of subgroup A1 have also been identified in other species, e.g., Nicotiana benthamiana MAPKKKβ (NbMAPKKKβ) and Brassica napus MAPKKK8 (BnaMAPKKK8, also known as BnMAP3Kβ1) (Jouannic et al., 1999; Hashimoto et al., 2012; Sun et al., 2014). Subgroup A2 consists of AtMAP3Kα (AtMAPKKK3), AtMAP3Kγ (AtMAPKKK5), AtYODA (AtMAPKKK4), BnaMAPKKK3 (also known as BnaMAP3Kα1), NbMAPKKKα and NbMAPKKKγ (Hashimoto et al., 2012; Sun et al., 2014), while subgroup A3 comprises AtANP1 (also called AtMAPKKK1), AtANP2 (AtMAPKKK2), AtANP3 (AtMAPKKK12) and their ortholog Nicotiana protein kinase 1 (NtNPK1) (MAPK Group, 2002; Sun et al., 2014). Subgroup A4, as the last functionally characterized subgroup of MEKK-like kinases, consists of AtMAP3Kε1 (AtMAPKKK7), AtMAP3Kε2 (AtMAPKKK6) and AtMAPKKK13–21 in A. thaliana, and BnaMAPKKK6 (BnMAP3Kε1) and BnaMAPKKK19-20 in Brassica napus (MAPK Group, 2002; Chaiwongsar et al., 2012; Sun et al., 2014).
Raf-like MAPKKKs have been classified into groups B and C (MAPK Group, 2002; Wu et al., 2014). Two of the best-characterized Raf-like kinases, Arabidopsis constitutive triple response 1 (AtCTR1, also known as AtRaf1) and enhanced disease resistance 1 (AtEDR1, also known as AtRaf2), together with orthologs such as O. sativa OsEDR1 (OsMAPKKK1) and drought-hypersensitive mutant 1 (OsDSM1) (OsMAPKKK6), and BnaCTR1 and BnaEDR1 (BnaRaf2), are members of group B (MAPK Group, 2002; Shen et al., 2011; Yin et al., 2013; Danquah et al., 2014; Sun et al., 2014; Virk et al., 2015). Members of group C remain mostly uncharacterized.
Although both Raf-like and ZIK-like kinases are clearly members of the MAPKKK family, they have not been confirmed to phosphorylate MAPKKs in plants (Danquah et al., 2014; Chardin et al., 2017). However, Sun et al. (2014) have recently shown that Raf-like and ZIK-like kinases interact with MAPKKs in canola (Brassica napus L.).
Plant MAPKKs, with the exception of MKK10 homologs, feature a S/T-X5-S/T motif in the activation loop (Jonak et al., 2002; MAPK Group, 2002; Doczi et al., 2007, 2012; Danquah et al., 2014; Poyraz, 2015). Some MKK10 homologs, such as ZmMKK10-2 and OsMKK10-2, show only a partial MAPKK consensus motif (R-X5-S/T), while others, such as AtMKK10, ZmMKK10-1, OsMKK10-1 and OsMKK10-3, do not have this consensus sequence at all (Supplementary Figure 1). Some MAPKKs in green algae, such as Chlamydomonas reinhardtii MKK6 (CreinMKK6) and Volvox carteri MKK3 (VcMKK3), also lack the consensus motif (Supplementary Figure 1).
The Arabidopsis genome contains ten MAPKK genes, which have been divided into four groups, A – D (Supplementary Figure 2 and Supplementary Table 1). All AtMAPKKs in group A have also been shown to interact with AtMPK6 and AtMPK11 (Supplementary Figure 3) (Meszaros et al., 2006; Lee J.S. et al., 2008). In addition, AtMKK1 activates AtMPK3 and AtMPK12 (Meszaros et al., 2006; Lee et al., 2009), AtMKK2 interacts with AtMPK5, AtMPK10 and AtMPK13 (Teige et al., 2004; Gao et al., 2008; Lee J.S. et al., 2008), while AtMKK6 besides AtMPK4 activates AtMPK13 and interacts with AtMPK12 in yeast cells (Melikant et al., 2004; Lee J.S. et al., 2008; Takahashi et al., 2010; Zeng et al., 2011).
Group A MAPKKs have also been reported in rice, alfalfa (Medicago sativa), tobacco (Nicotiana tabacum L., Nicotiana benthamiana), tomato (Solanum lycopersicum L., Lycopersicon esculentum,), green algae (Volvox carteri and Chlamydomonas reinhardtii), lycophyte (Selaginella moellendorffii), maize (Zea mays L.) and canola (Brassica napus L.) (Cardinale et al., 2002; Xie et al., 2012; Liang et al., 2013; Cai et al., 2014; Li X. et al., 2014) (Supplementary Figure 2 and Supplementary Table 1). Group B MAPKKs include AtMKK3 in A. thaliana and their homologs in O. sativa (Wankhede et al., 2013), Z. mays (Liang et al., 2013; Kong et al., 2013), Brassica napus, S. lycopersicum, N. tabacum, S. moellendorffii, C. reinhardtii, and V. carteri (Liang et al., 2013) (Supplementary Figure 2). Group C MAPKKs include AtMKK4 and AtMKK5 as well as MKK4 and/or MKK5 proteins in other species (Supplementary Figure 2) (Kong et al., 2011; Furuya et al., 2014).
The last category of MAPKKs is group D, which includes the remaining Arabidopsis MAPKKs, such as AtMKK7, AtMKK8, AtMKK9, and AtMKK10 (Supplementary Figure 2). AtMKK7 interacts with AtMPK2 (Lee J.S. et al., 2008), AtMPK12 (Lee et al., 2009) and AtMPK15 (Lee J.S. et al., 2008), while AtMKK9 interacts with AtMPK6, AtMPK10 (Lee J.S. et al., 2008), AtMPK12 (Lee et al., 2009), AtMPK17 and AtMPK20 (Lee J.S. et al., 2008). In other species, only homologs of AtMKK8 have not been identified (Kong et al., 2013; Wankhede et al., 2013). Interestingly, AtMKK10, OsMKK10-2 and ZmMKK10-1, family members that lack (partially or completely) the MAPKK consensus motif (Supplementary Figure 1), nevertheless interact with MAPKs (Lee J.S. et al., 2008; Singh et al., 2012; Wankhede et al., 2013; Kong et al., 2013) (Supplementary Figure 3).
The MAPKs themselves form the last category of MAPK cascade component. MAPKs feature the conserved T-X-Y motif, which is phosphorylated by MAPKKs during signal transduction. Based on sequence similarities, the 20 Arabidopsis MAPKs have been divided into two subtypes, TEY (12 MAPKs) and TDY (8 MAPKs). The MAPKs of the TEY subtype carry a T-E-Y (Thr-Glu-Tyr) motif and can be divided into three groups (A – C). The more evolutionarily distant group D is formed by MAPKs of the TDY subtype, which contain a T-D-Y (Thr-Asp-Tyr) motif at the phosphorylation site (Bigeard and Hirt, 2018) (Supplementary Table 2). Recent phylogenetic analysis of MAPKs from 40 plant species revealed that group A, as well as MAPKs carrying the T-E-Y motif, also contains MAPKs sharing a T-Q-Y (Thr-Gln-Tyr) motif, while group B also includes MAPKs carrying M-E-Y (Met-Glu-Tyr), T-E-C (Thr-Glu-Cys) and T-V-Y (Thr-Val-Tyr) motifs (Mohanta et al., 2015) (Supplementary Figure 4). Three MAPKs of group C also contain a motif that is different to the typical T-E-Y sequence. These are OlMPK7 from Ostreococcus lucimarinus, which carries a T-S-Y (Thr-Ser-Tyr) motif, and Picea abies MPK7-1 (PaMPK7-1) and PaMPK20, which both contain a M-S-Y (Met-Ser-Tyr) motif sequence (Supplementary Figure 4). A MAPK motif was not found in the sequence of OsMPK20-2 from group D (Supplementary Figure 4). Phylogenetic analysis of MAPKs from 40 plant species allowed two additional groups to be distinguished, i.e., E and F, which mainly contain MAPKs of lower eukaryotic and gymnosperm plants (Mohanta et al., 2015). However, our own phylogenetic analysis of MAPKs clearly identified group E MAPKs with T-E-Y, T-D-Y, T-R-M (Thr-Arg-Met), T-E-M (Thr-Ser-Met) and T-Q-M (Thr-Gln-Met) motifs (Supplementary Figure 4), but MAPKs such as CrenMPK4-1, Micromonas pusila MPK4 (MpMPK4), OlMPK6 and VcMPK4-1 assigned to group F by Mohanta et al. (2015) are actually members of group C (Supplementary Figure 5). Therefore, we suggest retaining the group E category, but MAPKs of group F should be incorporated into group C.
Group A TEY MAPKs include AtMPK3, AtMPK6 and AtMPK10 in Arabidopsis, and their homologs in canola, alfalfa, cucumber, tobacco, rice, spruce (Picea abies), tomato, and maize (MAPK Group, 2002; Liang et al., 2013; Wankhede et al., 2013; Mohanta et al., 2015; Wang J. et al., 2015) (Supplementary Table 2). Group B includes AtMPK4, AtMPK5, AtMPK11-13, whereas AtMPK1, AtMPK2, AtMPK7, AtMPK14 are members of group C. Members of group B and C are also present in others plants including gymnosperms and algae (Liang et al., 2013; Wankhede et al., 2013; Mohanta et al., 2015; Wang J. et al., 2015) (Supplementary Figure 5 and Supplementary Table 2). Group D is formed by MAPKs of the TDY subtype, such as AtMPK8-9 and MPK15-20 (MAPK Group, 2002; Liang et al., 2013; Wankhede et al., 2013; Mohanta et al., 2015; Wang J. et al., 2015) (Supplementary Table 2). Group E includes 12 MAPKs from species such as C. reinhardtii, C. subellipsoidea, M. pusila, N. tabacum, S. moellendorffii, P. abies and V. carteri (Supplementary Figures 4, 5).
The phytohormone auxin, indole 3-acetic acid (IAA), plays a crucial role in plant growth and development, including embryogenesis (Friml et al., 2003; Blilou et al., 2005; Morris et al., 2005; Leyser, 2017), organogenesis (Benkova et al., 2003; Reinhardt et al., 2003; Heisler et al., 2005; Smekalova et al., 2014; Contreras-Cornejo et al., 2015; Enders et al., 2017; Wójcikowska and Gaj, 2017; Corredoira et al., 2017; Zhao, 2018), tissue patterning, tropism and growth responses to environmental stimuli (Benkova et al., 2003; Leyser, 2003; Reinhardt et al., 2003; Morris et al., 2004; Simonini et al., 2016; Liu et al., 2017; Kamada et al., 2018). The involvement of auxin in this multiplicity of biological processes results from its regulation of cell division, expansion and differentiation (Chen and Baluska, 2013). Auxin synthesis takes place mainly in the shoot, after which it is distributed directionally throughout the plant. Auxin distribution patterns are asymmetric within tissues and they vary dynamically throughout different developmental stages (Friml et al., 2003; Tanaka et al., 2006; Béziat and Kleine-Vehn, 2018). Since Mizoguchi et al. (1994) observed that auxin can activate MAPKs in tobacco cells, several MAPK cascades have been implicated in the regulation of auxin biosynthesis, transport and signal transduction. However, published studies on the connection between auxin and MAPK signaling have given conflicting results, such that in some cases the same MAPK activities apparently mediate different functions.
The pioneering work of Mizoguchi et al. (1994) suggested a link between auxin and MAPK activity. They reported that in vitro phosphorylation of myelin basic protein (MBP) and a recombinant MAPK by extracts of tobacco BY-2 cells increased when cells were subjected to prior treatment with a high concentration of the synthetic auxin. However, using the same system, Tena and Renaudin (1998) showed that auxin at low concentrations does not induce MBP kinase activity in tobacco cell lines. Activation of MAPK was observed only after treatment with very high concentrations of synthetic auxin and was probably a consequence of cytoplasmic acidification caused by its accumulation (Tena and Renaudin, 1998; Mockaitis and Howell, 2000).
In other early studies, Kovtun et al. (1998) showed that Nicotiana protein kinase NPK1, a member of the MAPKKK family in tobacco, initiates a MAPK cascade that negatively regulates early IAA-inducible genes. Recombinant NPK1 was transiently expressed in leaf protoplasts to determine its influence on the activity of the soybean GH3 promoter, which is known to be auxin-responsive (Liu et al., 1994; Kovtun et al., 1998). Overexpression of NPK1 specifically blocked the auxin inducibility of the GH3 promoter, while a MAPK-specific phosphatase (MKP1) was able to abolish this effect (Kovtun et al., 1998). Similar results were obtained using orthologs of NPK1, i.e., Arabidopsis ANP1, ANP2, and ANP3. In an experiment with constitutively active ANPs, transiently overexpressed in protoplasts, it was shown that the ANPs selected can also suppress auxin signaling (Kovtun et al., 2000). ANP1 mediates H2O2-induced activation of the known stress MAPKs, AtMPK3 and AtMPK6, and the end result of this activation cascade is inhibition of auxin-inducible genes (Kovtun et al., 2000; Hirt, 2000). This evidence for a role of NPK1 and its orthologs in auxin signaling is consistent with their involvement in cytokinesis (Jin et al., 2002; Takahashi et al., 2010). MPK12 is another negative regulator of auxin signaling and its kinase activity increases after auxin treatment. MPK12 RNAi lines were hypersensitive to auxin in a root growth inhibition assay. Furthermore, IBR5, which mediates crosstalk between the auxin and ABA signaling pathways, has been identified as a specific MPK12 phosphatase (Monroe-Augustus et al., 2003; Lee et al., 2009; Raja et al., 2017).
In contrast to the negative role of MAPK in auxin signaling, Zhao et al. (2013) found that MAPKs positively regulate some auxin genes (e.g., OsYUCCA4) under conditions of cadmium stress in rice roots, while other genes (e.g., OsPINc) are negatively regulated. The complex relationship between MAPKs and auxin signaling was further studied with respect to cadmium and zinc stresses. Zhao et al. (2014a,b) performed a comprehensive expression analysis of 67 key genes in the auxin signaling pathway. Seven genes were positively regulated by MAPK cascades, namely OsYUCCA3, OsPIN1c, OsPIN10b, OsPID, OsARF20, OsIAA9 and OsIAA30. In addition, 14 genes were negatively regulated by MAPKs (OsYUCCA1, OsYUCCA2, OsPIN5a, OsPIN5b, OsARF7, OsARF8, OsARF12, OsARF15, OsARF16, OsARF21, OsARF22, OsARF25, OsIAA12, and OsIAA15). It should be emphasized that the combined results of Zhao et al., 2014b indicate that MAPKs function at the interface between H2O2 and auxin signaling under Cd and Zn stress conditions. This evidence suggests a model where MAPKs regulate auxin distribution through H2O2, while H2O2 in turn may act downstream of MAPKs but upstream of the auxin signaling pathway. It would be interesting to investigate the precise MAPK-dependent regulatory mechanisms that facilitate auxin/ROS (reactive oxygen species) regulation.
Polar auxin transport (PAT) is an active process whereby auxin is delivered to specific plant tissues (Tanaka et al., 2006; Petrasek and Friml, 2009; Leyser, 2017; Zhao, 2018). Interestingly, PAT is regulated by PIN proteins and reversible protein phosphorylation, mediated by protein kinases and protein phosphatases, and it can, for example, control the activity of auxin transport proteins (Muday and DeLong, 2001; Dai et al., 2006; Ganguly et al., 2014; Dory et al., 2018). Localization of the plasma membrane localized PIN proteins is also controlled by several MAP kinases including MPK4 and the MKK7/MPK6 module (Jia et al., 2016; Dory et al., 2018). The involvement of MKK7 in PAT was shown by analyses of the Arabidopsis bud1 mutant, which has significantly fewer lateral roots than wild-type (Mou et al., 2002; Dai et al., 2006) and shows disrupted PAT from shoots into roots, as well as a deficiency in auxin signaling (Reed et al., 1998; Xie et al., 2000; Rogg et al., 2001; Dai et al., 2006). Molecular genetic analysis of bud1 plants by Dai et al. (2006) revealed increased expression of the AtMKK7 gene, which results in defective auxin transport, while lowering AtMKK7 mRNA levels using antisense RNA causes an improvement in auxin transport. Together, these data suggest that AtMKK7 is a negative regulator of PAT (Dai et al., 2006; Zhang X. et al., 2008).
Recently, another module, AtMKK2/AtMPK10, has been implicated in the regulation of PAT. The results of Stanko et al. (2014) may indicate that the AtMKK2/AtMPK10 module regulates auxin transport, with consequences for venation complexity and other developmental phenomena. It seems that at least two MAPK pathways connect auxin to development, but the precise regulatory connections have not yet been fully elucidated.
It is worth mentioning that other interesting links between MAPKs and auxins exist. YODA kinase (MAPKKK4) and MPK6 have been shown to be involved in an auxin-dependent regulation of cell division during post-embryogenic root development. Smekalova et al. (2014), showed that both loss-of-function (yda1) and gain-of-function (ΔNyda1) plants exhibit pronounced root phenotypes that result from visibly disorientated cell divisions. Both mutants have elevated endogenous auxin (IAA) levels, and this might be related to their phenotypes. Indeed, because the IAA level is particularly upregulated in ΔNyda1 plants, it is tempting to hypothesize that the role of YODA in the elongation of the zygote is to promote auxin signaling. It is known that YODA acts upstream of MPK3/6 in stomatal development (Bergmann et al., 2004; Lukowitz et al., 2004; Kim J.M. et al., 2012) and embryogenesis (Wang et al., 2007). Interestingly, a mpk6 mutant transformed with a kinase-dead form of MPK6 has a very similar root phenotype to yda1 plants. This indicates that MPK6 acts downstream of YODA in an auxin-dependent manner to control cell division in post-embryonic root development (Smekalova et al., 2014).
The recent work Enders et al. (2017) provided evidence that the AtMKK3-MAPK1-RBK1 (ROP binding protein kinase 1) module regulates auxin dependent cell expansion in Arabidopsis via modulation of the Rho-like GTPase (ROP4 and ROP6) activity. Both mpk1 and mkk3-1 mutants display similar phenotypes to the effects of the auxins on the inhibition of root elongation and cotyledon expansion, suggesting that the MKK3-MPK1 pathway negatively regulates auxin-dependent cell growth. Strikingly, the known upstream MKK3 activators, MAPKKK17/18, which are clearly involved in ABA signaling (Danquah et al., 2015; Matsuoka et al., 2015; Mitula et al., 2015), were not investigated. Future studies should consider following up on this preliminary result in order to investigate the role of the ABA-activated MAPKKK17/18-MKK3-MPK1/2/7/14 module in the crosstalk between ABA and auxin signaling.
Overall, this section of the review highlights exciting results that suggest a connection between MAPK modules and the auxin signaling pathway. Disappointingly, though, current knowledge of MAPK cascade involvement in auxin-mediated processes is still fragmentary and no complete MAPK module has been confirmed as having a role in auxin signaling. Thus, multiple challenges and unanswered questions remain to be addressed.
Jasmonic acid and salicylic acid are plant hormones that participate in plant growth and development. JA plays essential roles in both biotic and abiotic stress responses (Heil et al., 2012; Hou et al., 2013; Wasternack and Hause, 2013). Plant defenses against pathogens are also mediated by SA, a type of phenolic acid, which similarly to JA also plays a role in plant growth and development (Hayat and Ahmad, 2007). In addition to its involvement in the response to wounding, SA participates in systemic acquired resistance (SAR) and the responses to abiotic stresses such as water, salinity and cold stress (Miura and Tada, 2014; Wendehenne et al., 2014). MAPKs are clearly involved in both signaling pathways as both positive and negative regulators. However, much work is still needed to elucidate downstream MAPK targets involved in SA- and JA-dependent processes.
Plant hormone JA is an important regulator of plant growth and development, but it plays a more important role in the wounding response and SAR (Heil et al., 2012; Hou et al., 2013; Wasternack and Hause, 2013). Despite the fact that crosstalk between JA and MAPK signaling has been reported, only a few studies have summarized this interaction. Nevertheless, MAPKs are reported to regulate JA biosynthesis and the expression of JA-dependent genes. For example, tomato SlMPK6-1 (also known as SlMAPK2, LeMPK2 and SlMPK2) and SlMPK6-2 (SlMAPK1, LeMPK1, SlMPK1) function as positive regulators of JA biosynthesis and signaling pathways (Kandoth et al., 2007). Simultaneous silencing of SlMPK6-1 and SlMPK6-2 has been shown to reduce JA biosynthesis and the expression of JA-dependent defense genes (Figure 2). On the other hand, JA regulates both MAPK activity and MAPK gene expression. In Arabidopsis, induction of AtMPK1/2 kinase activity is observed in leaves 1 h after JA treatment (Figure 2) (Ortiz-Masia et al., 2007). Furthermore AtMPK9 and AtMPK12 together are involved in JA-induced stomatal closure (Khokon et al., 2015; de Zelicourt et al., 2016; Lee et al., 2016) (Figure 2). Induction by JA treatment, albeit only at the transcript level, has also been demonstrated in rice for OsMPK7, OsMPK20-5 and OsMPK16 (Reyna and Yang, 2006; Singh and Jwa, 2013). Increased transcript levels after JA treatment have been observed for BnaRaf30 in canola (Sun et al., 2014), for Cucumis sativus MPK6 (CsMPK6), CsMPK9-1, CsMPK20-1, CsMPK20-2, CsMKK4, CsMKK6, and CsMEKK21-1 in cucumber (Wang J. et al., 2015), for NtMPK1, NtMPK7, NtMPK22-1 (also known as NtMPK16) and NtMPK22-2 (also known as NtMPK17) in tobacco (Zhang et al., 2013), for 23 MAPKs in cotton (Gossypium raimondii) including GrMPK2/3/5-1/18-20/22-25/27-28 (Zhang et al., 2014e), and for Brachypodium distachyon MPK7-1 (BdMPK7-1) and BdMPK20-5 in purple false brome (Jiang et al., 2015). Undoubtedly much work is still needed to fully resolve the potential roles of MAPKs in JA biosynthesis, JA signaling and JA-mediated responses and these pathways are attractive targets for future research.
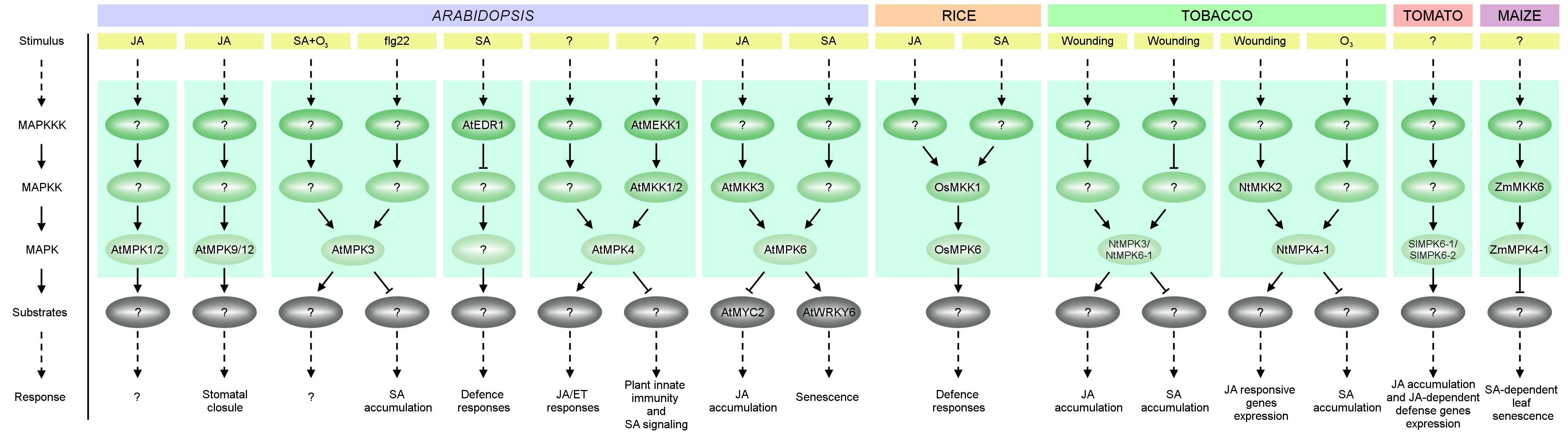
FIGURE 2. A simplified overview of MAPK cascades involved in JA and SA signaling in plant species such as: A. thaliana (At), O. sativa (Os), Nicotiana tabacum (Nt), S. lycopersicum (Sl) and Z. mays (Zm). Activation of MAPKs by various stimuli causes phosphorylation of MAPK effectors (usually transcription factors) further triggering cellular responses. See text for details.
Alongside JA, SA – a type of phenolic acid – also plays an important role in plant growth, development and defense (Hayat and Ahmad, 2007). In addition to its involvement in the response to wounding, SA participates in SAR and responses to abiotic stresses such as water, salinity, and cold stress (Miura and Tada, 2014; Wendehenne et al., 2014). The activity of Arabidopsis AtMPK3 and AtEDR1, and maize ZmMKK6-ZmMPK4-1, are known to be regulated by SA (Figure 2). Thus, AtMPK3 has been shown to be induced by ozone stress in an SA-dependent manner (Ahlfors et al., 2004; Samajova et al., 2013). In disease resistance, MPK3 seems to be an important crosstalk regulator of late immune responses (Ichimura et al., 2006; Han et al., 2010; Mao et al., 2011; Meng and Zhang, 2013). Besides its well-known role in repressing the constitutive and flg22-induced expression of defense genes (Asai et al., 2002; Galletti et al., 2011; Montillet et al., 2013), AtMPK3 also appears to be a negative regulator of flg22-induced SA accumulation (Frei Dit et al., 2014).
Other studies have revealed that SA-inducible defense responses are also negatively regulated by the Raf-like MAPKKK, AtEDR1, indicating that AtEDR1 is involved in SA signaling, but not in JA/ET signaling (Frye and Innes, 1998; Frye et al., 2001; Virk et al., 2015). Recent studies also suggest the participation of ZmMKK6 in SA signaling: expression of inactive ZmMKK6 in Arabidopsis transgenic plants induced SA accumulation and SA-dependent leaf senescence. ZmMKK6 also activates both ZmMPK4-1 (also called ZmMPK4, ZmSIMK) and AtMPK4 in vitro. These data indicate that the ZmMKK6-ZmMPK4-1 cascade may play an important role in the regulation of SA-dependent leaf senescence (Li et al., 2016).
Analysis of expression profiles of MAPK cascade kinases after treatment with SA also led to the identification of kinases that might be involved in SA signaling in other species. In tomato, three out of five known MAPKK genes (SlMAPKK1/2/4), almost half of the MEKK subfamily genes, nearly half of the RAF subfamily genes and nearly all the ZIK subfamily genes were significantly upregulated by SA treatment (Wu et al., 2014). Transcription of the genes encoding AtRaf43 in Arabidopsis (Virk et al., 2015), BdMPK3 and BdMPK17 in purple false brome (Jiang et al., 2015), BnaMAPKKK18, BnaRaf28, BnaMKK1-2, BnaMKK4, BnaMKK9, BnaMPK1, BnaMPK3, BnaMPK5, BnaMPK6, BnaMPK19 in canola (Liang et al., 2013; Sun et al., 2014), GrMPK2/3/5/6/7/8/9/12/13/16/18/22/23/25/28 in cotton (Zhang et al., 2014e), NtMPK9-2 and NtMPK15 in tobacco (Zhang et al., 2013), OsMPK17-1 and OsMPK17-2 in rice (Hamel et al., 2006; Singh and Jwa, 2013), SlMKK3 in tomato (Li X. et al., 2014), and PsMAPK3 in pea (Barba-Espín et al., 2011) has also been shown to be significantly increased after treatment with SA. Future research is needed to identify novel components and effectors of these SA-dependent MAPK pathways.
MAPKs are clearly involved in plant defense signaling. Certain MAPKs such as AtMPK4 and AtMPK6 in Arabidopsis are involved in both JA and SA signaling (Figure 2). AtMPK4 positively regulates JA/ET responses (Brodersen et al., 2006), whereas in the AtMEKK1–AtMKK1/2–AtMPK4 cascade appears to function as a negative regulator of plant innate immunity and SA signaling (Petersen et al., 2000; Andreasson et al., 2005; Brader et al., 2007; Pitzschke et al., 2009). AtMPK6 participates in SA-induced detached leaf senescence by promotion of AtNPR1 activation (Chai et al., 2014). On the other hand, activation of the AtMKK3–AtMPK6 cascade in Arabidopsis plants by JA represses a positive regulator of JA biosynthesis genes (AtMYC2), leading to suppression of JA production (Takahashi et al., 2007). In tobacco, NtMPK3 (also known as NtWIPK, NtMPK5) and NtMPK6-1 (NtSIPK, NtMPK6) appear to play an important role in wound-induced biosynthesis of JA and they function as repressors of SA accumulation in response to wounding (Seo et al., 2007; Oka et al., 2013; Hettenhausen et al., 2014). In addition to NtMPK3 and NtMPK6-1, another wounding-activated MAPK, NtMPK4-1 (NtMPK4, NtMPK1) appears to positively regulate JA signaling pathways and is also involved in SA signaling by affecting SA biosynthesis and signaling in response to ozone exposure (Gomi et al., 2005) (Figure 2). The induction of other Nicotiana MAPKs, such as NtMPK16 (NtMPK10) and NtMPK20 (NtMPK8), at the transcript level in response to MeJA and SA treatment may suggest that these kinases also play a role in JA and SA signaling, but further studies are needed to confirm this (Zhang et al., 2013). The putative involvement of MAPKs in both JA and SA signaling has also been reported in other species, such as O. sativa, Z. mays, and S. lycopersicum. In rice, the kinase activity of OsMPK17-1 (OsMPK12, OsBWMK1) is activated by both JA and SA treatment (Cheong et al., 2003; Singh and Jwa, 2013). Overexpression of OsMPK17-1 in tobacco causes SA and H2O2 accumulation and elevated PR gene expression, leading to hypersensitive response (HR)-like cell death (Cheong et al., 2003; Bigeard and Hirt, 2018). OsMKK1 (OsMEK2), as well as OsMPK6 (also called OsMPK1, OsMAPK6 and O. sativa SA-induced protein kinase – OsSIPK), OsMPK17-1 and OsMPK3 (also called OsMPK5, OsMAP1, OsMAPK2, OsMSRMK2 and OsBIMK1), are transcriptionally induced by both JA and SA treatment in rice. In addition, overexpression of OsMPK6 results in JA and SA accumulation when challenged by pathogens, indicating that the OsMKK1-OsMPK6 cascade may be involved in JA- and SA-inducible defense responses (Singh and Jwa, 2013) (Figure 2). In maize, the MAPK ZmMPK3-2 (ZmMPK3) is sensitive to various signaling molecules, including JA or SA (Wang X.J. et al., 2010; Smekalova et al., 2013). In tomato, SlMKK4 (SlMKK2) and SlMKK9 (SlMKK4) seem to be involved in both JA and SA signaling pathways (Li X. et al., 2014). SA and JA signaling are crucial to plant defense against pathogens. Many examples of MAPK cascade kinases involved more or less directly in JA, SA or both JA and SA signaling demonstrate the importance of this cooperation for plants in response to wounding. However, there is still very little known about the details of this cooperation and further studies are needed to understand how this leads to improved resistance of plants to pathogens.
Relatively recent studies have shown that crosstalk also exists between Arabidopsis MAPK cascade kinases and a class of polyhydroxylated steroid hormones, the BR (Kim T.W. et al., 2012; Kang et al., 2015). In particular, BRs participate in cell division and cell elongation, but also take part in cellular patterning (Tang et al., 2011; Khan et al., 2013) (Figure 3). It was demonstrated that BRs repress stomatal development in cotyledons, but in an AtBZR1-independent fashion (Kim T.W. et al., 2012; Serna, 2013). The signal transduction required for correct stomatal patterning is mediated by the Arabidopsis ERECTA family (AtERf) of receptor-like kinases and the AtYODA-AtMKK4/5-AtMPK3/6 cascade, which results in phosphorylation and thereby inactivation of transcription factors such as Arabidopsis speechless (AtSPCH), AtMUTE and AtFAMA (Lau and Bergmann, 2012; Kim T.W. et al., 2012; Le et al., 2014). Recent studies have shown that phosphorylation of AtSPCH Serine 186 (one of three primary phosphorylation targets) plays a crucial role in stomatal formation (Yang et al., 2015). To summarize, when the BR level is high, BR signal transduction through plasma-membrane receptor brassinosteroid-insensitive 1 (AtBRI1), BR-signaling kinase 1 (AtBSK1) and phosphatase AtBRI1 suppressor 1 (AtBSU1) inactivates the glycogen synthase kinase 3 (GSK3)-like kinase BR insensitive 2 (AtBIN2), making AtBES1 (BRI1-EMS-suppressor 1; also called brassinazole-resistant 2 – BZR2)/AtBZR1, and the MAPK cascade (repressing AtSPCH) active, which in turn leads to promotion of plant growth and inhibition of cell division and stomatal formation in cotyledons, respectively (Kim T.W. et al., 2012; Le et al., 2014; Zhang et al., 2014a). On the other hand, when BR levels are low, AtBIN2 remains active, inhibiting AtBES1/AtBZR1 and the AtYODA-AtMKK4/5-AtMPK3/6 module, and leading to inhibition of plant growth and promotion of stomatal development in cotyledons (Figure 3) (Kim T.W. et al., 2012). It has also been demonstrated in two different studies by in vitro and/or yeast two-hybrid assays that AtBIN2 seems to inhibit the AtYODA-AtMKK4/5-AtMPK3/6 cascade by direct suppression of both AtYODA (MAPKKK) and AtMKK4/5 (Kim T.W. et al., 2012; Khan et al., 2013; Le et al., 2014; Xu and Zhang, 2015).
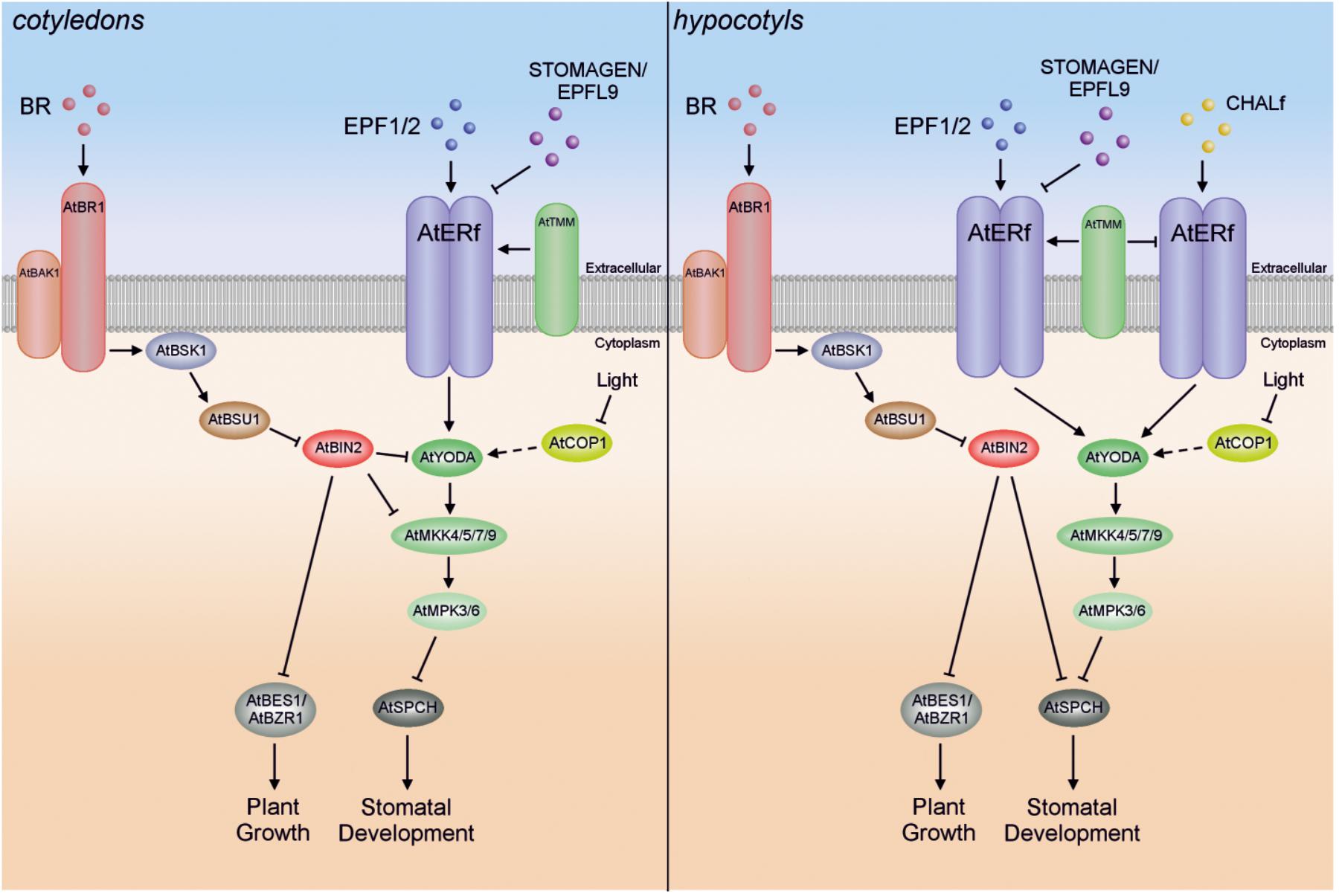
FIGURE 3. Schematic illustration of the GSK3-like kinase AtBIN2-mediated crosstalk between the AtYODA-AtMKK4/5-AtMPK3/6 cascade and BR signaling in cotyledons and in hypocotyls.
In contrast to their action in cotyledons, BRs seem to promote stomatal development in the hypocotyl, but in a BES1- and BZR1-independent manner (Serna, 2013). Interestingly, AtSPCH seems also to be controlled by AtBIN2, which phosphorylates the same AtSPCH residues as AtMPK3/6 (Gudesblat et al., 2012). Since AtYODA-AtMKK4/5-AtMPK3/6 cascade activity is likely reduced by CHALLAH family (CHALf) signaling in the hypocotyl, meaning that AtSPCH is not inhibited by this pathway, inactivation of AtSPCH by AtBIN2 might be the predominant pathway in hypocotyls (Le et al., 2014). Thus, in the hypocotyl, BR signaling inactivates AtBIN2 at high BR levels, whereas AtBES1/AtBZR1 and AtSPCH remain active, leading to promotion of plant growth, cell division and stomatal formation. Conversely, AtBIN2 remains active at low BR levels and then inhibits AtBES1/AtBZR1 and AtSPCH, resulting in inhibition of plant growth and promotion of stomatal development in hypocotyls (Figure 3).
Recent studies have also identified crosstalk between MAPK cascade kinases and BR signaling pathways in other species. OsMKK4 seems to be involved in BR signaling pathways (Duan et al., 2014) and might be involved in BR signaling in a similar manner to AtMKK4, because one of the rice orthologs of AtBIN2, OsGSK2, is involved in BR signaling (Tong et al., 2012). In tomato, all the MAPKs of group A are involved in BR-induced nematode resistance (Song et al., 2018), two of which, SlMPK6-1 (SlMPK2) and SlMPK6-2 (SlMPK1), positively regulate BR-induced pesticide metabolism (Yin et al., 2016). Only SlMPK6-1 plays a role in the regulation of BR-induced H2O2 accumulation and tolerance to oxidative and heat stress (Nie et al., 2013). In maize, the homolog of AtMPK6, ZmMPK6-2 (also called ZmMPK5), is involved in BR signaling. ZmMPK6-2 is activated by BR-induced H2O2 accumulation and in turn enhances apoplastic H2O2 accumulation via gene expression of NADPH, leading to up-regulation of antioxidant defense systems in leaves (Zhang et al., 2010). The significant involvement of MPK3 or/and MPK6 in BR signaling in different species might suggest that the crosstalk between MAPK and BR is evolutionarily conserved. However, in chinese cabbage (Brassica rapa), has the investigation of MAPK expression profiles revealed that five other genes are induced after BR treatment (Lu et al., 2015): Brassica rapa MAPK5 (BraMAPK5), BraMAPK17-1, BraMAPK17-2, BraMAPK18-1 and BraMAPK19-1.
MAPKs are also involved in ET biosynthesis and signaling. ET is a gaseous hormone involved in many aspects of plant biology, such as germination, plant growth, organ senescence and fruit ripening (Yang and Hoffman, 1984; Bleecker and Kende, 2000; Skottke et al., 2011; Dubois et al., 2018). Furthermore, it integrates external and internal signals to provide a dynamic response to diverse stress conditions (Yoo et al., 2009). ET sensing and signal transduction in plants are complex processes (Figure 4).
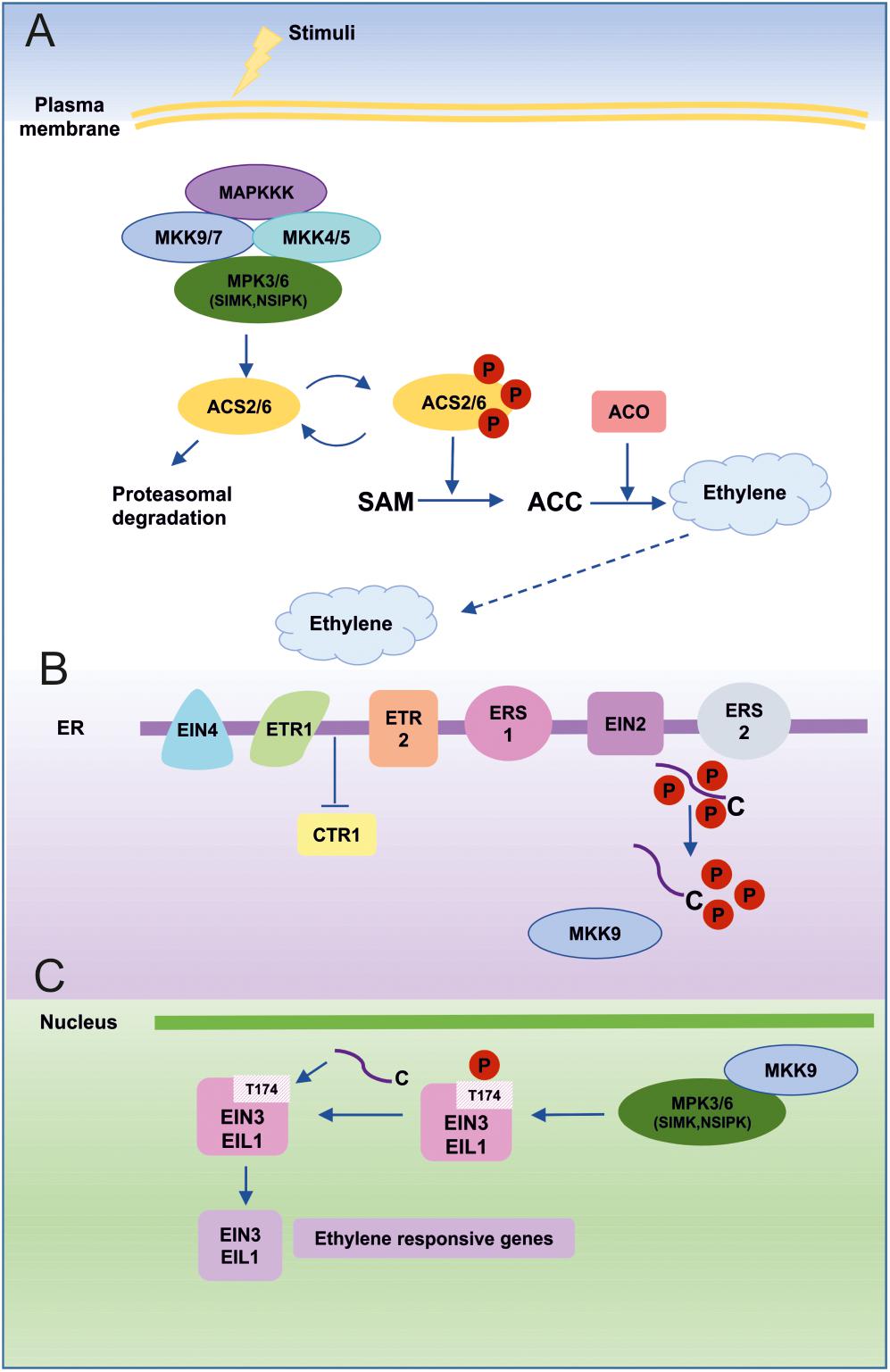
FIGURE 4. MAPKs in ET biosynthesis and signaling. (A) An external stimulus leads to activation of ET biosynthesis predominantly through the MKK9-MPK3/MPK6 cascade. Alternatively, MKK7 may also be involved in MPK3/MPK6 activation due to similarities with the MKK9 sequence and its activation mechanisms. MPK3 and MPK6 can also be activated by MKK4 and MKK5, which act in a redundant fashion upstream of MPK3/MPK6, especially after wounding-induced ET biosynthesis. SIMK (MsMPK6) and NSIPK(NtMPK6-1) are homologs of AtMPK6 from alfalfa and tobacco, respectively. Active MPK6 phosphorylates ACS2/ACS6, which initiates ET biosynthesis. (B) ET is perceived by five different receptors (ETR1, ETR2, ERS1, ERS2, EIN4) localized in the endoplasmic reticulum (ER) membrane and this leads to inhibition of CTR1 kinase activity, which is the primary negative regulator of ET signaling. As a consequence, MKK9 is released from CTR1 inhibition and translocates to the nucleus, where it activates MPK3 and MPK6. Moreover, inactive CTR1 is no longer able to phosphorylate the C-terminal domain (CEND) of EIN2. Dephosphorylated CEND moves to the nucleus and takes part in EIN3 stabilization. (C) In the nucleus, active MPK3/MK6 promotes the stability of the main plant-specific ET-dependent transcription factors (EIN3 and EIL1). Phosphorylation of EIN3 at the T174 position blocks its proteasomal degradation and enables it to activate ET-responsive genes.
As shown in many studies, the MKK9-MPK3/MPK6 cascade is involved in the regulation of ET biosynthesis (Liu and Zhang, 2004; Joo et al., 2008; Xu J. et al., 2008; Skottke et al., 2011). The basal ET level is very low, but under special conditions (abiotic stress, wounding, pathogen infection, nutrient availability) ET production increases dramatically (Zarembinski and Theologis, 1994; Wang et al., 2002; De Paepe and Van der Straeten, 2005; Stepanova and Alonso, 2009; Iqbal et al., 2013; Ludwików et al., 2014; Tao et al., 2015; Chardin et al., 2017). Key enzymes in ET biosynthesis are ACC synthases (ACS; 1-aminocyclopropane-1-carboxylate synthases), which are strictly regulated at both the transcriptional and post-translational levels, (Kende, 1993).
Kim et al. (2003) showed that NtMPK6-1 is able to induce ET biosynthesis. The authors constructed transgenic plants overexpressing a constitutively active mutant of NtMKK4DD (NtMEK2DD T227D/S233D; kinase upstream of NtMPK6-1; Yang et al., 2001) under the control of a steroid-inducible promoter. Dexamethasone treatment resulted in immediate NtMPK6-1 activation and significant elevation of ET production. In experiments with transgenic Arabidopsis plants that overexpress NtMKK4DD, AtMKK4DD, and AtMKK5DD under the control of the same steroid-inducible promoter, Liu and Zhang (2004) showed that MAPK activation mechanisms are conserved between species. Thus, NtMKK4DD is able to activate MPK6/MPK3 in Arabidopsis. Analogously, Arabidopsis MKK4DD and MKK5DD (AtMKK4 and AtMKK5 are functional orthologs of NtMKK4DD) can activate the endogenous NtMPK3/NtMPK6-1 in tobacco plants. Further experiments demonstrated that MPK6 is essential for NtMKK4DD-dependent ET biosynthesis.
Analysis of known Arabidopsis ACS protein sequences revealed potential MAPK phosphorylation sites in ACS1, ACS2, and ACS6 (Figure 4). All three ACC synthases cluster together on a phylogenetic tree (Chae et al., 2003; Yamagami et al., 2003). An in-gel kinase assay confirmed that MPK6 is responsible for ACS6 phosphorylation. Site-directed mutagenesis showed that MPK6 is able to phosphorylate three serines (S480, S483, S488) in the ACS6 sequence. However, experiments with wild-type and mutated forms of ACS6 in which single, double and triple serine (S) residues were converted to alanine (A) or aspartic acid (D) revealed that changes in phosphorylation state do not alter its enzyme activity. Instead, it was suggested by Liu and Zhang (2004) that MPK6-mediated phosphorylation may influence ACS6 and ACS2 stability. Indeed, it turns out that mutated ACS6DDD, which mimics the phosphorylated state, is much more stable in transgenic plants than wild-type ACS6 and ACS6AAA. MPK6 phosphorylation sites are localized within the C-terminal domain of ACS6 and ACS2, which is the regulatory domain responsible for their stability. Lack of MPK6-mediated phosphorylation results in decreased ACS6 and ACS2 stability and immediate targeting of both proteins for proteasomal degradation. Dephosphorylation by ABI1, a member of the protein phosphatase 2C (PP2C) family is also involved in regulating the proteasomal degradation of ACS6 (Ludwików et al., 2014; Ludwików, 2015).
MAPKs regulate ET biosynthesis by controlling transcription of ACS. Recent studies showed that wounding-induced ET biosynthesis in Arabidopis is also under the control of MAPKs (Li et al., 2018). Analysis of ET accumulation after wounding in single mpk3 and mpk6 mutants and in a double mpk3 mpk6 mutant rescued by MPK3TA or MPK6YG (chemically synthesized MPK3 and MPK6) revealed that MPK6 is the dominant kinase in this process (Xu et al., 2014, 2016). Li C.H. et al. (2014) observed a 50% reduction in wounding-triggered ET accumulation in the mkk6 mutant compared to control plants. In many developmental processes, kinases MKK4 and MKK5 are redundant and function upstream of MPK3/MPK6 (Xu and Zhang, 2015), as seen in the case of wounding-induced ET accumulation. Thus, in a single mkk4 mutant, ET biosynthesis was reduced by about 10% compared to wild-type plants, while in a mkk5 single mutant, the reduction in ET accumulation was about 50%. However, in a double mutant (mkk4 mkk5) strain, ET production was reduced by 80%. Therefore, MKK4 and MKK5 act upstream of MPK3 and MPK6 after wounding and are required for wounding-dependent ET accumulation. Among the family of ACC synthase genes, only four are induced after wounding, namely ACS2, ACS6, ACS7, and ACS8. Genetic studies confirmed that changes in expression of these four ACS genes are under the control of MKK4 and MKK5. In double mkk4 mkk5 mutant plants, ACS2, ACS6, ACS7, and ACS8 expression was reduced. Analysis of the role of downstream elements of the MAPK cascade showed that, in mpk6 single mutant plants, the expression level of all four ACC synthase genes was markedly reduced, but in mpk3 single mutant plants was unaffected. Moreover, ACS2, ACS6, ACS7, and ACS8 are activated at different times after wounding stimuli. ACS6 and ACS7 were induced very quickly, about 30 min post-wounding, while ACS2 and ACS8 expression reached a maximum around 2–6 h after wounding (Li et al., 2018). These data show that MAPKs are indeed involved in ET biosynthesis under wounding conditions. MAPKs likely influence the expression level of a subset of ACS genes, and thereby modulate ET biosynthesis, by activation of WRKY33 TFs (Li et al., 2012).
Extensive research in Arabidopsis led to the identification of key elements of the ET signaling cascade (Figure 4). After revealing that CTR1 does not function as a MAPK cascade element, but instead inactivates EIN2 by direct phosphorylation of specific residues, extensive efforts have been made to identify MAPK cascades involved in ET signaling (Ju et al., 2012; Qiao et al., 2012; Wen et al., 2012; Cho and Yoo, 2015). Novikova et al. (2000) showed that a protein extract prepared from wild-type Arabidopsis plants treated with ET contained MAPK activity and that this activity was higher in ctr1 (knockout) plants and lower in etr1 (ET-insensitive) mutant plants than in wild-type. Immunoprecipitation experiments with the Arabidopsis extracts, using antibodies specific for the mammalian MAPK ERK1, identified a putative MAPK with molecular mass of 47 kDa (Novikova et al., 2000). Later, in 2003, Hirt’s group found the MAPKs MPK6 and MPK13 to be involved in ET signaling (Ouaked et al., 2003). They isolated protein extracts from Medicago and Arabidopsis cells before and after 1-aminocyclopropane-1-carboxylic acid (ACC) treatment and used these to perform in-gel kinase assays. As a result, they discovered two protein kinases 46 and 44 kDa in size in Medicago. These experiments led to the identification of strong kinase activity associated with SIMK (46 kDa) and MMK3 (44 kDa). The researchers also noticed that an increase in kinase activity did not correlate with an increased amount of these proteins, suggesting that ET induced MAPK activation by post-translational modification. SIMK from Medicago was most similar to MPK6 from Arabidopsis, while MMK3 corresponded to MPK13. Moreover, Ouaked and coworkers showed that MPK6 is constitutively active in a ctr1 mutant and ET-dependent activation is not connected with EIN2 or EIN3. In Arabidopsis plants overexpressing Medicago MKK4 (MsSIMKK, MsSIMK kinase), they observed a ctr1-like phenotype in etiolated seedlings (Ouaked et al., 2003).
Yoo et al. (2008) demonstrated that the MKK9-MPK3/MPK6 cascade is involved in not only in ET biosynthesis, but also in ET signaling, acting downstream of CTR1 (Figure 4). It was also shown that MKK7 and MKK9 are able to activate both MPK3 and MPK6, which play a similar role in ET signaling (Novikova et al., 2000; Ouaked et al., 2003). What is more, ET-dependent activation of MPK3/6 by MKK9 is abolished in a mkk9 mutant. The same authors also showed that overexpression of MKK7 and MKK9 in ctr1 protoplasts results in specific activation of MPK3 and MPK6. These observations led to placement of the MKK9-MPK3/MPK6 cascade downstream of the key negative regulator of ET signaling, CTR1. Despite the fact that MKK7 and MKK9 are very similar in both sequence and mechanism of activation, the basal transcript level of MKK9 in protoplasts and in leaves is significantly higher than that of MKK7. Thus, MKK9 is considered predominant in ET signaling. Yoo et al. (2008) also investigated a positive role of MKK9 in ET signaling. Under most of the conditions examined, mkk9 mutants present phenotypes similar to those of ein3 mutants. Furthermore, in mkk9 and ein3 mutants, the expression of early ET signaling genes (ERF1 and ERF5) in leaves is abolished. ERF1 and ERF5 are indirect targets of EIN3, the main TF regulating ET-inducible genes (Solano et al., 1998; Yanagisawa et al., 2003). Overexpression of permanently active MKK9 (MKK9a) results in constitutive ET signaling, which cannot be blocked by ET receptor mutants (etr1) or treatment with Ag+, an inhibitor of ET perception. These data support the assumption that MKK9-MPK3/6 functions downstream of CTR1 (Yoo et al., 2008). Perhaps even more interestingly, MKK9 acts a linker between CTR1, which is located in the ER, and other components of the ET signaling cascade located in the nucleus. After ACC treatment, MKK9 is able to move to the nucleus and activate MPK3 and MPK6, which are localized in both the nucleus and cytoplasm. After activation by MKK9, MPK3 and/or MPK6 phosphorylate(s) EIN3 in the nucleus. Computational analysis predicts two MAPK phosphorylation sites in the EIN3 protein. Mutation experiments reveal that phosphorylation at T174 is mediated by MPK6 and results in enhanced EIN3 stability (Alonso et al., 2003; Binder et al., 2007). However, after phosphorylation at the second MAPK phosphorylation site (T592), the stability of EIN3 is reduced. These findings show that the MKK9-MPK3/MPK6 cascade is the key module responsible for EIN3 stability and ET signaling (Yoo et al., 2008).
Results from different studies provide evidence for ET-dependent MKK9-MAPK3/6 activation, but there is still some controversy about the proposed model. It is worth noting that the MKK9-MAPK3/6 cascade is readily activated by environmental stresses (such as wounding and touch) (Alzwiy and Morris, 2007). It is even possible to activate the MAPK cascade mechanically by spraying “treatment” instead of ET or ACC treatment (Colcombet and Hirt, 2008). The involvement of the MKK9-MAPK3/6 cascade in ET signaling therefore needs to be scrutinized by precisely controlled experiments.
It is well known that ET signaling is an indispensable element of the response to various stimuli (salt stress, pathogen attack, iron deficiency or dehydration) (Kende, 1993; Xu J. et al., 2008; Han et al., 2010; Kazan, 2015; Tao et al., 2015; Ye et al., 2015; Chen J. et al., 2017; Khan et al., 2017). Salt stress stimulates ET biosynthesis which in turn activates other internal signals (Wang et al., 2002; Xu J. et al., 2008; Dong et al., 2011). However, the mechanisms by which the external signals relating to salinity stress stimulate ET biosynthesis remain unknown. Recent studies in O. sativa report that one of the receptor-like kinases (RLKs) involved in salt stress tolerance is able to phosphorylate both MPK3 and MPK6 (Ouyang et al., 2010; Li C.H. et al., 2014). RLKs are thought to be involved in transducing external signals into the cell, and some of the large number of known RLKs in O. sativa and A. thaliana are important in plant development (Osakabe et al., 2013) and the responses to drought and salinity stress (Marshall et al., 2012; Ouyang et al., 2010; Vaid et al., 2013). Salt Intolerance 1 (SIT1) is an active RLK that plays a significant role in drought and salt stress tolerance in O. sativa. SIT1 is mainly expressed in root epidermal cells and its expression is induced immediately by NaCl. As a consequence, SIT1 activates MPK3 and MPK6 (Li C.H. et al., 2014). A co-immunoprecipitation assay showed that rice MPK3 and MPK6 are components of the SIT1 complex. What is more, in vitro phosphorylation experiments revealed that SIT1 is able to phosphorylate MPK3 and MPK6. A genetic approach confirmed that SIT1 acts upstream of MPK3 and MPK6 in O. sativa. It was also shown that SIT1 is involved in activation of antioxidant systems. ET signaling during plant stress responses is regulated by ROS production (Jung et al., 2009; Mergemann and Sauter, 2000). Li C.H. et al. (2014) demonstrated that SIT1-induced ROS accumulation requires ET production and signaling. Furthermore, they showed that after salinity-dependent activation, SIT1 is able to phosphorylate MPK3 and MPK6, resulting in salt sensitivity in rice. These findings are in line with other results showing that, when MPK3 and MPK6 are activated by MKK9, they increase salt sensitivity in Arabidopsis (Xu J. et al., 2008). There is a marked similarity between the rice SIT1-MPK3/MPK6 and Arabidopsis MKK9-MPK3/MPK6 cascades. However, whether SIT1 is involved in the rice MKK9-MPK3/MPK6 cascade needs to be examined.
Plants under attack by the necrotrophic fungal pathogen Botrytis cinerea produce high level of ET (Broekaert et al., 2006; van Loon et al., 2006), although precisely how ET biosynthesis is triggered by pathogen infection is still unclear. Han et al. (2010), using a double mpk3 mpk6 mutant rescued by a DEX-inducible MPK6 cDNA construct, were able to show that, in response to Botrytis cinerea, the MPK3/MPK6 cascade is crucial for activating ET biosynthesis. These authors also identified ACC synthase 6 (ACS6) as the main enzyme contributing to Botrytis cinerea-induced ET production.
MPK3 and MPK6 are also involved in the regulation of ET biosynthesis during iron deficiency in Arabidopsis (Ye et al., 2015). Iron (Fe) is a vital microelement because Fe ions are a component of many of the enzymes controlling basic physiological processes including photosynthesis and chlorophyll biosynthesis (Kobayashi and Nishizawa, 2012). Ye et al. (2015) showed that lack of iron induces transcription of the MPK3 and MPK6 genes, as well as increasing MPK3 and MPK6 kinase activity. Moreover, the transcript levels of some ACC synthases are also increased in Fe-deficient plants. Although the regulation of Fe-induced ET biosynthesis needs further analysis, the work of Ye et al. (2015) highlights another mechanism involving the MPK3/MPK6 cascade.
MAPKs can be activated by many different stimuli. For example, in Arabidopsis seedlings, MPK6 may be activated by drought and rapidly inactivated during rehydration (MAPK Group, 2002; Tsugama et al., 2012; Xu and Chua, 2012). Interplay between dehydration and rehydration in plants is especially important for the cut-flower industry. Thus, rehydration after dehydration induces rapid ET production for a short duration in rose (Rosa hybrida) flowers (Tsugama et al., 2012). Further research on rose flowers showed that, particularly in the gynoecia, protein levels of RhMPK6 are high during both dehydration and rehydration, but RhMPK6 kinase activity was observed only within the first hour of rehydration. Active RhMPK6 is able to phosphorylate and stabilize RhACS1, stimulating ET production (Meng et al., 2014). The RhMPK6-RhACS1 module seems to be crucial for transduction of the rehydration signal and triggering of ET biosynthesis, which controls flower opening and senescence in rose. Rehydration-induced ET biosynthesis also seems to involve RhMKK9. RhMKK9 is expressed 30 min after rehydration, but after 12 h the expression is almost undetectable. Chen and co-workers correlated these results for RhMKK9 with the expression and activity of RhMPK6 and RhACS1, and proposed that RhMKK9 functions as an activator of RhMPK6-RhACS1 (Chen J. et al., 2017). However, whether RhMKK9 is actually an upstream activator of RhMPK6 in dehydration-dependent ET biosynthesis in rose gynoecia must be confirmed by further experiments.
Abscisic acid signaling has been intensively studied and comprises multiple components including MAPKs. The plant hormone ABA functions as a key regulator in many developmental and physiological processes in plants, including seed dormancy and germination (Finkelstein et al., 2002; Nambara and Marion-Poll, 2003; Gutierrez et al., 2007; Chen M. et al., 2017; Née et al., 2017), seedling growth (Leon-Kloosterziel et al., 1996; Chen M. et al., 2017; Trupkin et al., 2017) and also adaptation to various biotic and abiotic stress conditions (Lee and Luan, 2012; Wang et al., 2018). Interestingly, the application of exogenous ABA to plant structures initiates the effect of stress conditions and results in transcriptional regulation, protein accumulation and activation of MAPKs, suggesting an important role for MAPK pathways in ABA signaling (Fujita et al., 2006; Xing et al., 2008; Zhang et al., 2014c,d; Li Y. et al., 2017, Li K. et al., 2017; Li Q. et al., 2017). Subsequent to binding of the hormone by different cellular receptors, ABA functions through a complex network of signal transduction pathways, which activate responses including the regulation of stomatal aperture and the expression of stress-responsive genes (Himmelbach et al., 2003; Leung and Giraudat, 1998; Finkelstein, 2013; Mitula et al., 2015; Albert et al., 2017; Eisenach et al., 2017). The core components of the ABA signaling pathway have been identified and characterized relatively recently (Fujii et al., 2009; Ma et al., 2009; Park et al., 2009). Initial steps in ABA signal transduction involve the PYR/PYL/RCAR ABA receptors and also the phosphatase/kinase enzyme pairs, PP2Cs and SnRK2s, respectively, which have antagonistic functions. The outcome of ABA signaling is the activation of gene expression by transcription factors under the control of SnRK2s (Figure 5) (Cutler et al., 2010; Klingler et al., 2010; Finkelstein, 2013; Nakashima and Yamaguchi-Shinozaki, 2013; Tan et al., 2018). These findings have certainly contributed to a more rapid understanding of the protein complexes that perceive and transmit ABA signals. Many previous studies indicate the participation of MAPK cascades in ABA-mediated responses, including antioxidant defense, guard cell signaling and seed germination (for reviews see also Liu, 2012; Danquah et al., 2014; Colcombet et al., 2016; de Zelicourt et al., 2016). Thus, the interactions between ABA signaling and other signaling pathways, including MAPK pathways, are beginning to be deciphered.
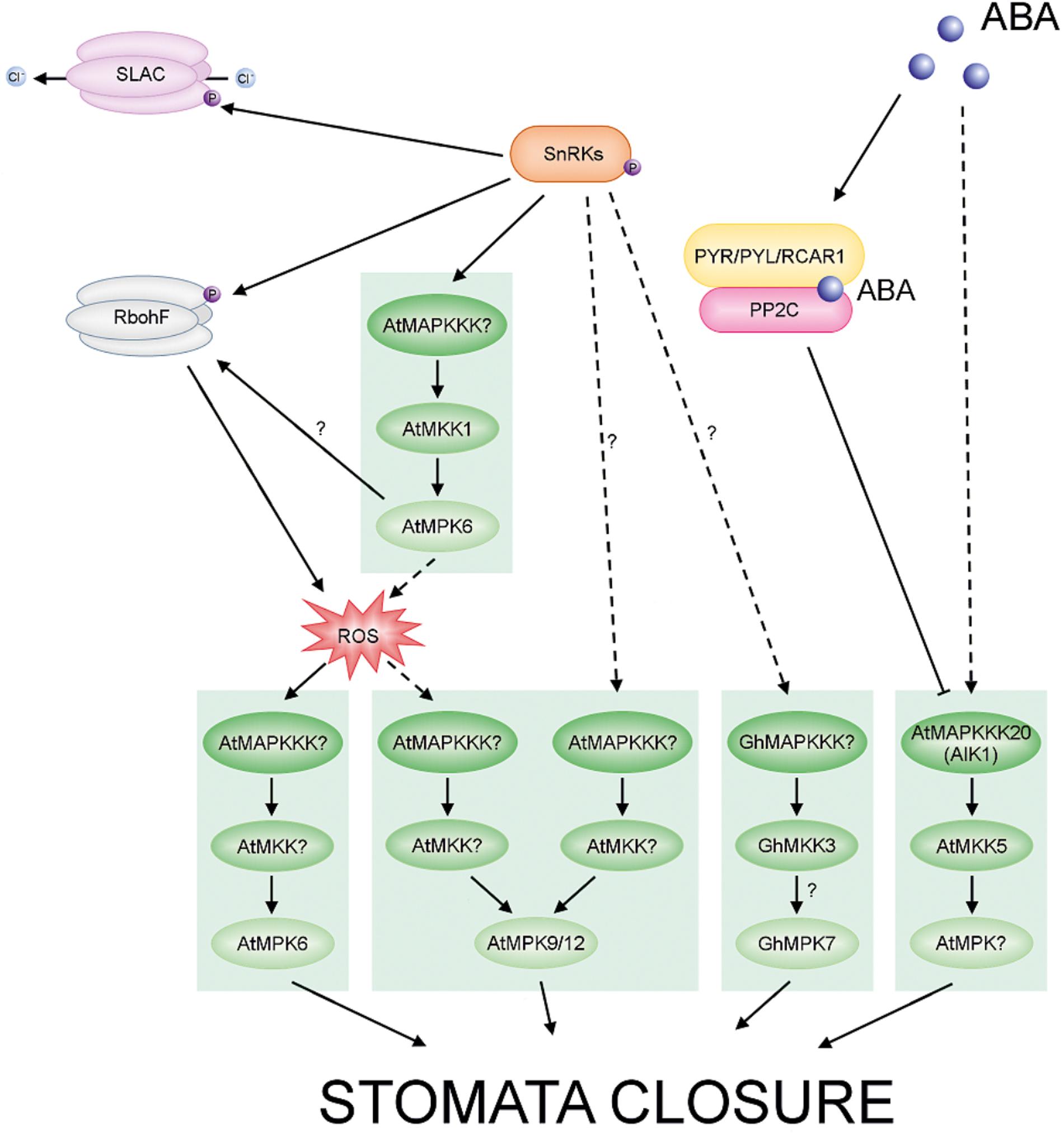
FIGURE 5. ABA-regulated MAPKs in Arabidopsis and cotton. ABA promotes stomatal closing. The different cascades are distinguished by different colors in the scheme. Arrows with solid lines represent established signaling pathways, while arrows with dashed lines represent putative signaling pathways. In the presence of ABA, PYR/PYL/RCAR receptors bind the phytohormone and inhibit group A PP2Cs. These events result in activation of SnRK2s. Activated SnRK2s phosphorylate and activate downstream targets, including MAPKs, Respiratory Burst Oxidase Homolog (RBOH) and Slowly Activating Anion Conductance (SLAC S-type). Active RBOH mediates ROS production. Note that in guard cells crosstalk between ABA signaling and ROS signaling may coincide at the MAPK level and regulates stomatal closure.
In recent years, there has been a significant increase in research on ABA-mediated gene expression in a variety of plant species. ABA signal transduction pathways modulate gene expression, including changes in transcription levels, transcript processing and stability (Cutler et al., 2010). The regulation of ABA-responsive gene activity involves TFs, which recognize and bind to cis-elements in the promoter regions upstream of their target genes (Zhang et al., 2014b). Importantly, in addition to the action of TFs, ABA-responsive gene expression is mediated by receptors, secondary messengers and protein kinase/phosphatase cascades (Fujita et al., 2011). Nearly 10% of the protein-coding genes in A. thaliana are regulated by ABA, a far greater percentage than for other hormones (Shinozaki et al., 2003; Nakashima et al., 2009; Cutler et al., 2010; Fujita et al., 2011). Several Arabidopsis genes encoding particular members of the MAPK family have been reported to be transcriptionally regulated by ABA. These include AtMPK1, AtMPK2 (Ortiz-Masia et al., 2007; Hwa and Yang, 2008; Umezawa et al., 2013), AtMPK3 (Lu et al., 2002; Wang et al., 2011), AtMPK5, AtMPK7 (Menges et al., 2008), AtMPK18, AtMPK20 (Wang et al., 2011), AtMKK9 (Menges et al., 2008), AtMAPKKK1 (ANP1), AtMAPKKK5 (Menges et al., 2008), AtMAPKKK15 (Wang et al., 2011), AtMAPKKK16 (Wang et al., 2011), AtMAPKKK17, AtMAPKKK18 (Menges et al., 2008; Wang et al., 2011), AtMAPKKK19 (Wang et al., 2011), AtMAPKKK20 (Li K. et al., 2017), and AtRaf6, AtRaf12, and AtRaf35 (Menges et al., 2008), all of which are regulated at the transcriptional level, indicating possible participation of these kinases in ABA signaling. It is worth mentioning that, despite the large number of ABA-regulated genes, the roles of most of them in ABA signaling have not been characterized. In searches for rice (O. sativa) MAPK genes transcriptionally activated by ABA, many genes were identified (Supplementary Table 3). It is worth mentioning that OsMPK3 (OsMPK5) is the best characterized of all the rice MAPKs, having been studied independently by at least six research groups and shown to be regulated by a variety of biotic and abiotic stresses (Agrawal et al., 2002; Huang et al., 2002; Song and Goodman, 2002; Wen et al., 2002; Reyna and Yang, 2006; Chen and Ronald, 2011; Nautiyal et al., 2013; Sharma et al., 2013; Jaemsaeng et al., 2018). Suppression of OsMPK3 (OsMPK5) by RNAi on the one hand results in reduced sensitivity to ABA, and on the other hand causes an increase in levels of endogenous ET (Xiong and Yang, 2003; Sharma et al., 2013). Many ABA-regulated genes have also been characterized in other plant species and these can be classified into two groups, upregulated and downregulated (Supplementary Table 3). The response of MAPK genes to ABA treatment suggest the involvement of these genes in ABA signaling. So far, the role of only a few of the kinases listed in Supplementary Table 3 has been investigated in detail, and in the following sections the functional characterization of these kinase modules and the downstream responses they control is reviewed.
Abscisic acid is the main regulator of stomatal movement (Burnett et al., 2000; Dodd et al., 2003; Jiang and Song, 2008; Albert et al., 2017; Qu et al., 2018). The phytohormone may also cause the production of ROS in various plant cells or tissues (Hu et al., 2005; Zhang et al., 2011; Shang et al., 2016; Qi et al., 2017) and ABA signaling in guard cells is mediated by ROS (Jammes et al., 2009). Studies showing that MAPKs can be activated by ROS may indicate that ABA signaling and ROS signaling coincide at the MAPK level (Zhang et al., 2007), and crosstalk between these pathways could regulate stomatal closure. H2O2 is an another important signaling molecule in ABA-induced stomatal closure (Pei et al., 2000; Li Q. et al., 2017; Rodrigues et al., 2017). Thus, the generation of H2O2 in response to ABA results in a reduction in size of the stomatal aperture (Wang and Song, 2008; Li Q. et al., 2017). In A. thaliana MPK3 is involved in the perception of ABA and H2O2 in guard cells. The results of Gudesblat et al. (2007) indicate that MPK3 functions downstream of ROS in ABA inhibition of stomatal opening, but not in ABA-induced stomatal closure. Another study showed that the atmkk1 and atmpk6 mutants block ABA-dependent H2O2 production in guard cells (Xing et al., 2008) (Figure 5). In apparent contradiction of these results, Montillet et al. (2013) recently found that AtMPK3 and AtMPK6 are not involved in ABA-induced stomatal closure, but instead are involved in stomatal closure induced by biotic stress. The same authors confirmed, however, that ABA-induced stomatal closure is mediated by MPK9 and MPK12: atmpk9 atmpk12 double mutants, but not single mutants, are impaired in ABA-induced stomatal closure, in ABA inhibition of stomatal opening, and in inhibition of the promotion of stomatal closure by H2O2 (Figure 5) (Jammes et al., 2009; Salam et al., 2012; de Zelicourt et al., 2016). Recently, Mitula et al. (2015) found an ABA-activated kinase MAPKKK18 to be involved in stomatal development and function. Under normal growth conditions, mapkkk18 mutant plants show increased stomatal aperture and decreased abaxial stomatal index, compared to the wild-type. Moreover, Li Y. et al. (2017) demonstrated that the mapkkk18 mutant displays impaired ABA-induced stomatal closure. The authors hypothesized that MAPKKK18 is probably involved in drought stress resistance by accelerating stomatal closing when drought stress occurs (Li Y. et al., 2017). Consistent with this, studies of the transcriptional regulation of the MKKK18 promotor revealed high promoter activity following ABA stimulation in guard cells (Mitula et al., 2015). Importantly, experimental results indicate that MAPKKK18 interacts directly with two of the key proteins of the ABA core signaling module, PP2C phosphatase ABI1 (Mitula et al., 2015) and kinase SnRK2.6 (Tajdel et al., 2016). ABI1, in the absence of ABA, not only inhibits MAPKKK18 kinase activity by dephosphorylation, but also targets MAPKKK18 for degradation by the ubiquitin-proteasome pathway (UPS) (Ludwików, 2015; Mitula et al., 2015). However, when ABA binds to PYR/PYL receptors, MAPKKK18 degradation is blocked, and this stabilization allows the kinase to activate downstream components of the signaling module (Mitula et al., 2015). It is worth mentioning that recently two independent research groups reconstructed a complete MAPK cascade initiated by MAPKKK18 and regulated by ABA (Danquah et al., 2015; Matsuoka et al., 2015). These authors showed the ABA-regulated MAP3K17/18-MKK3-MPK1/2/7/14 cascade to be involved in stress signaling (Danquah et al., 2015) and timing of senescence (Matsuoka et al., 2015), and as previously mentioned it is also known to have a role in drought stress resistance (Li Y. et al., 2017) (Figure 6). Importantly, a close paralogue of MAPKKK18, MAPKKK17, was found in the Arabidopsis genome and was included in the study of Danquah et al. (2015). The kinase activity of both MAPKKK17 and MAPKKK18 is significantly increased after ABA treatment (Danquah et al., 2015; Mitula et al., 2015). In addition, there is a positive correlation between the transcription levels of ABA core signaling genes and the MAPKKK17/MAPKKK18 genes (Danquah et al., 2015).
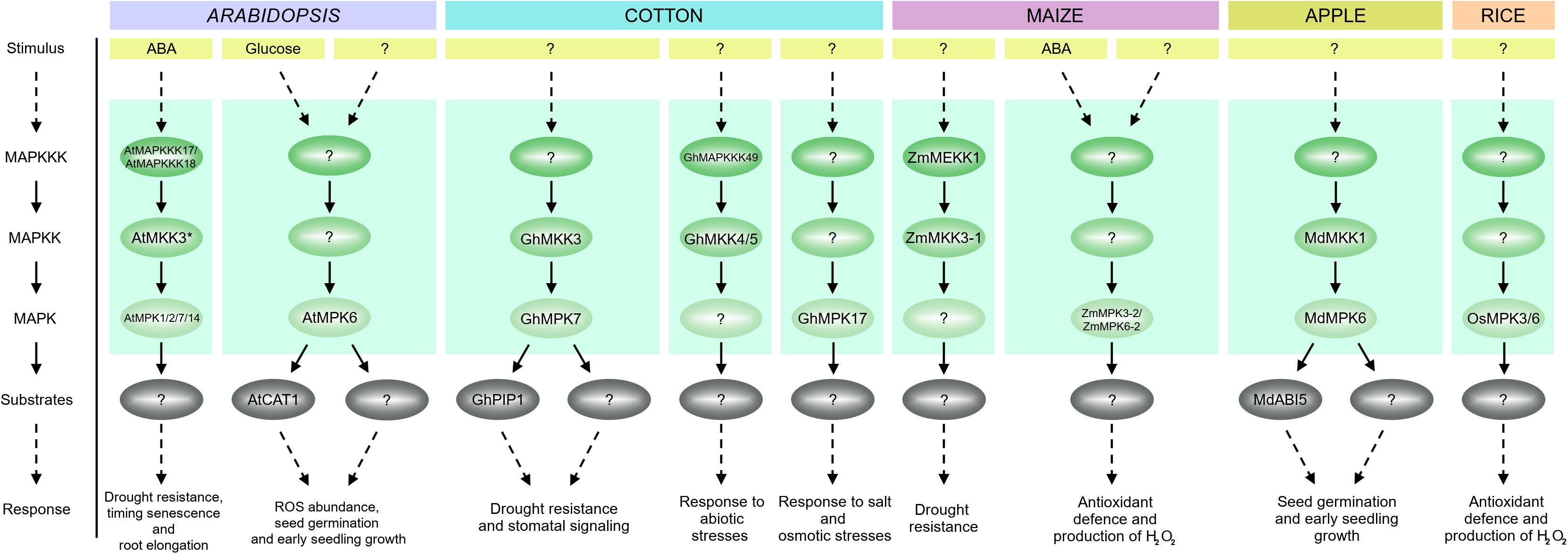
FIGURE 6. Overview of MAPKs regulated by ABA in different plant species. A single ABA-activated MAPK cascade MAPKKK17/18-MKK3-MPK1/2/7/14 has been identified in Arabidopsis. This pathway is involved in drought resistance, senescence, stomatal development and signaling. In addition, MKK3 in both maize and cotton has been shown to function in response to ABA. In maize, MKK3 acts downstream of MEKK1 and transcripts for both kinases are upregulated on ABA treatment. In cotton, ABA and drought induce activation of a MAPK cascade composed of MKK3, MPK7 and PIP1. These two pathways, MEKK1–MKK3 in maize and MKK3–MPK7–PIP1 in cotton, are associated with drought resistance and stomatal signaling. Another module in cotton, MAPKKK49-MKK4/MKK5, is involved in the ABA-mediated response to abiotic stress. MPK17 is another well-characterized MAPK in cotton, which regulates the response to salt and osmotic stresses. ABA-inducible genes encoding cotton MAPK cascade components presented in the scheme are MKK3, MAP3K49 and MPK17, respectively. Some MAPK cascades have a similar function in different plant species. In Arabidopsis and apple, the MKK1-MPK6 module affects seed germination and early seedling growth. ABA treatment induces transcription of the genes encoding MKK1 and MPK6 in both plant species. In Arabidopsis, MKK1 mediates activation of MPK6, thereby regulating CATALASE1 expression in ROS homeostasis. Additionally, glucose treatment significantly increases MKK1 and MPK6 activities. In apple, ABA-responsive transcription factor ABI5 may act as a downstream target of this MAPK cascade. MPK5 and MPK3 in maize and MPK1 and MPK5 in rice are required for ABA-induced antioxidant defense and play a similar role to Arabidopsis MPK6. In maize, ABA treatment significantly increases MPK5 and MPK3 activities. In rice, ABA treatment induces MPK1 and MPK5 expression.
Arabidopsis ABA-insensitive protein kinase 1 (AIK) is another MAPKKK, MAPKKK20, involved in the regulation of ABA-induced responses. Very recently, Li K. et al. (2017) documented that MAPKKK20 is a positive regulator of ABA-induced stomatal closure and also regulates the effect of ABA on root architecture. Arabidopsis AIK insertion mutants are insensitive to ABA and do not display stomatal closure and root elongation in response to ABA treatment. Moreover, the number of stomata in aik1 mutants is greater than in wild-type plants. The authors also showed that, as in the case of MAPKKK18 (Mitula et al., 2015), MAPKKK20 is regulated by ABA at both transcript and protein levels. ABA induces AIK1 activity in Arabidopsis and tobacco and, significantly, this kinase activity is inhibited by ABI1, which dephosphorylates AIK. Finally, analysis of mpk6 and mkk5 single mutant plants showed them to have a similar phenotype to aik1 single mutant plants and experiments using bimolecular fluorescence complementation demonstrated that AIK1 works upstream of MKK5-MPK6: MKK5 is phosphorylated and is thus activated by AIK1 in an ABA-activated process. From these findings, it is tempting to hypothesize that sequential phosphorylations of the AIK1 (MKKK20)-MKK5-MPK6 module are involved in ABA- mediated regulation of both the stomatal response and primary root growth (Li K. et al., 2017) (Figure 5).
In addition to stomatal closure, ABA has other important physiological effects relating to seed maturation and the inhibition of seed germination (Koornneef et al., 2002; Xing et al., 2009; Chiu et al., 2016; Devic and Roscoe, 2016; Huang et al., 2017; Leprince et al., 2017). Using ABA-mediated inhibition of germination as a selection criterion, a number of important players in ABA signaling, including MAPKs, have been discovered through genetic screens (Joseph et al., 2014). Other evidence indicates that MAPK cascades are positive regulators of ABA signaling during seed germination, when plants overexpressing AtMPK1 and AtMPK2 display hypersensitivity to ABA (Hwa and Yang, 2008). Interestingly, a phosphoproteomic study showed that SnRK2 promotes activation of AtMPK1 and AtMPK2 in an ABA-dependent manner (Umezawa et al., 2013). AtMKK3 has been suggested as the upstream activator of AtMPK1 and AtMPK2 (Hwa and Yang, 2008). Indeed, Danquah et al. (2015) reported that the MKK3-MPK1/2/7/14 module mediates ABA signaling during germination and root elongation. Thus, mkk3-1 plants are hypersensitive to ABA during germination and root elongation, while the seeds of this mutant are hypersensitive to increasing ABA concentrations. Correspondingly, MKK3-overexpressing seeds were less sensitive to increasing ABA concentrations (Danquah et al., 2015). Importantly, MAPKKK18, which functions upstream of MKK3, is associated with the control of seed development and dormancy. Mitula et al. (2015) demonstrated that the germination of mkkk18 knockout plant lines is inhibited in medium supplied with ABA. Taken together, these results suggest that the MAPKKK18-MKK3 module mediates ABA signaling during germination and root elongation.
The Raf10 and Raf11 kinases are also involved in regulating seed dormancy and the response to ABA, as they affect the expression of ABA-regulated genes (including ABI3, ABI5) (Lee et al., 2015). The above mentioned AtMKK1–AtMPK6 cascade is also involved in ABA signaling during seed germination. The single mutants mkk1 and mpk6, as well as the mkk1 mpk6 double mutant, all show insensitivity to ABA during germination, while plants overexpressing MKK1 and MPK6 are hypersensitive to ABA (Xing et al., 2007, 2008). Interestingly, in apple, the MdMKK1–MdMPK1 cascade has a similar function to AtMKK1–AtMPK6 in Arabidopsis. Expression of MdMKK1 and MdMPK1 results in ABA hypersensitivity during seed germination, implicating MdMKK1 and MdMPK1 in the positive regulation of ABA signaling during seed germination and early seedling growth (Wang X.J. et al., 2010) (Figure 6).
So far, only a few members of the MAPK family have been identified and well documented in Z. mays. These include ZmMPK3-2 and ZmMPK6-2 (ZmMPK5), which are both activated by ABA-induced production of H2O2 and increase the tolerance of plants to drought, salt stress and oxidative stress (Wang J. et al., 2010). ZmMPK3 and ZmMPK6-2 play a similar role in ABA-induced antioxidant defense as AtMPK6 in Arabidopsis (Xing et al., 2008), and OsMPK3 (OsMPK5) and OsMPK6 (OsMPK1) in rice (Zhang H. et al., 2012; Shi et al., 2014). Interestingly, ZmCPK11, one of the calcium-dependent protein kinases (CDPKs), has been shown to act upstream of ZmMPK6-2 in ABA signaling in maize (Ding et al., 2013). Moreover, very recently the underlying molecular mechanisms have been elucidated. Ma et al. (2016), identified ZmABA2 as a protein interacting with ZmMPK6-2. ZmMPK6-2 phosphorylates ZmABA2, which results in an increase in ABA content. These findings show that ZmABA2 is a direct target of ZmMPK6-2 and participates in ABA biosynthesis and function.
Another study implicates the maize gene ZmMKK3-1 (ZmMKK3), which encodes a MAPKK, in the ABA signal transduction pathway, since ZmMKK3-1 is upregulated by ABA. Its overexpression on the one hand results in increased tolerance to osmotic and oxidative stresses, but on the other hand causes a decrease in ABA sensitivity in transgenic tobacco plants (Zhang M. et al., 2012). In maize root the expression of another MAPKK, ZmMKK1, is also induced by ABA. Overexpression of ZmMKK1 confers tolerance to salt and drought in Arabidopsis and yeast. ZmMKK1 interacts with ZmMEKK1 in vitro, and this, importantly, represents the first characterized MAPK cascade in maize (Cai et al., 2014). ABA has also been shown to induce transcription of other MAPKs in maize, including ZmMPK4-1 (Wang et al., 2014), ZmMPK7 (Zong et al., 2009), and ZmMPK17 (Pan et al., 2012). ZmMPK7 together with ZmMPK3 is activated by ZmMKK10 (Chang et al., 2017).
The participation of MAPKs in ABA signaling has been best characterized in A. thaliana. Nevertheless, MAPKs are known to be involved in this signaling pathway in other species, as recent research has shown. In mulberry (Morus L.) expression of MnMPK1 is upregulated by ABA (Liu et al., 2017). In pea (P. sativum L.), using a kinase activity assay, Ortiz-Masia et al. (2008) showed that ABA can activate PsMPK2. Furthermore, the activation profile of PsMPK2 is similar to that described above for AtMPK1 and AtMPK2. JA and H2O2 also cause an increase in activity of this kinase, which in turn suggests that MAPKs may have the same functions across species in this context (Ortiz-Masia et al., 2008). In wild tobacco, Nicotiana attenuata, NaMPK4 plays a critical role in ABA-induced stomatal closure responses. NaMPK4-silenced plants (irNaMPK4) are impaired in their response to ABA- and H2O2-mediated stomatal closure. NPK4 is also involved in defense against aphids, invading pathogenic bacteria and Alternaria alternata (tobacco pathotype) (Slavov et al., 2004; Hettenhausen et al., 2012; Sun et al., 2014; Guo et al., 2017).
In cotton (Gossypium hirsutum), GhMPK17 expression is upregulated by ABA and also by NaCl. Notably, overexpression of GhMPK17 in A. thaliana results in increased tolerance to salt and osmotic stresses, as well as in changes in H2O2 levels and in the expression of stress-related genes (Zhang et al., 2014c). In recent work by Liu et al. (2016), a novel cotton MAPKKK gene, GhMAPKKK49, was isolated and shown to be significantly induced by exogenous treatment with ABA or H2O2. As GhMAPKKK49 also interacts with GhMKK4 and GhMKK9, it is tempting to hypothesize that a GhMAPKKK49–GhMKK4 or GhMAPKKK49–MKK9 cascade participates in ABA- and H2O2-mediated responses to abiotic stress (Figure 6). Recently, experimental work by Wang et al. (2016) showed that GhMKK3 plays an important role in drought tolerance by controlling the rate of water loss. Overexpression of GhMKK3 in N. benthamiana results in more efficient ABA-induced stomatal closure and a decrease in the number of stomata (Figure 5). Intriguingly, both GhMKK3 and GhPIP1 interact with GhMPK7 to form a functional ABA- and drought-activated MAPK module. In support of this result, previous studies in Arabidopsis demonstrated that group C MAPKs, including AtMPK7, are activated by ABA in an MKK3-dependent manner (Danquah et al., 2015).
In Brassica napus, MKK1 appears to be involved in ABA signaling (Supplementary Table 3). Interestingly, overexpression of BnMKK1 in transgenic tobacco plants causes rapid water loss, resulting in increased sensitivity to drought stress (Yu et al., 2014).
In this review, we provide a comprehensive picture of our current understanding of the function of MAPKs and their interaction networks in plants. MAPK cascades are responsible for protein phosphorylation and signal transduction events associated with plant hormone signaling and therefore they play an essential role in the regulation of development, senescence, stress signaling and acclimation. Many cases of MAPK involvement in AUX, ABA, JA, SA, ET, and BR signaling have been identified and these demonstrate the complex structure, extensive crosstalk and dynamics of the signaling network. For example, analysis of MAPK function in ABA and ET signaling highlights MAPK regulation of target protein stability and the control of MAPK pathways by UPS degradation. However, despite the impressive current knowledge of MAPK cascades, their participation in BR and AUX signaling remains relatively unexplored. The very limited amount of data available on crosstalk between MAPK and GA signaling pathways (Huttly and Phillips, 1995; Marcote and Carbonell, 2000; Li Y. et al., 2014; Lu et al., 2015) highlights significant opportunities for further study in this area. In oat Aspk9 (also known as AsMAP1) seems to be negatively regulated by GA (Huttly and Phillips, 1995). The expression of the PsMAPK3 gene is induced by GA in unpollinated pea ovary after fruit set (Marcote and Carbonell, 2000). Overexpression of GhMKK4 (homolog of AtMKK4 and AtMKK5) from G. hirsutum in transgenic N. benthamiana increases sensitivity of the plant to GA (as well as ABA) and significantly reduces GA levels after infection with R. solanacearum (Li Y. et al., 2014). In Chinese cabbage, BraMAPK17-2 and BraMAPK19-1 are upregulated by GA3, whereas transcript levels of most BraMAPKs, such as BraMAPK3, BraMAPK10-2/3, BraMAPK10-4/5, BraMAPK5, BraMAPK13, BraMAPK1. BraMAPK7-1, BraMAPK7-2, BraMAPK8-1 and BraMAPK16-2 are significantly lower after treatment with GA3 (Lu et al., 2015). Hypocotyl elongation in Arabidpopsis mkk3-1 knockout seedlings is hypersensitive to exogenous GA3, and is proportional to GA3 concentration. However, no significant activation of MAPK cascade kinases has been observed in the Arabidopsis protoplast system after treatment with exogenous GA3, indicating that crosstalk may occur between AtMKK3 and GA in Arabidopsis, but is probably mediated through common downstream targets (Lee, 2015).
The molecular mechanisms that regulate MAPK assembly, activity (both activation and inactivation) and substrate binding require further elucidation. Nevertheless, several distinctive features of the mode of action of MAPKs have already been identified. The specificity of the MAPK module is achieved by coordinated expression of its components, by protein complex assembly and by subcellular localization (Danquah et al., 2014; Mitula et al., 2015; Bigeard and Hirt, 2018). In general, MAPK, MAPKK and MAPKKKs are localized in either nucleus or cytoplasm or both compartments. Localization in other cellular compartments is also evident. For example, MEKK1, besides being present in the nucleus and cytoplasm, can also be observed at the plasma membrane and in endosomes (Yang et al., 2010; Bigeard and Hirt, 2018). In addition, MAPK localization can be associated with its enzymatic activity. The active form of MAPKKK18 is localized in the nucleus, while the kinase-inactive isoform is found in the cytoplasm (Mitula et al., 2015). This suggests that not only MAPKKK18 activity but also the concentration of the protein is tightly controlled within the target compartment.
The activation of a typical MAPK module is rapid but transient. Subsequently, MAPK inactivation is achieved via dephosphorylation by dedicated protein phosphatases that function as part of a negative feedback loop to control the hormonal response. For example, MPK1 protein phosphatase regulates MPK3, MPK4 and MPK6 (Ulm et al., 2002; Bartels et al., 2009). Another MAPK phosphatase, MKP2, interacts with and controls the activity of MPK3 and MPK6 (Lumbreras et al., 2010). Furthermore, following ABA treatment, MPK3 and MPK6 kinase activities are inhibited by AP2C1 and PP2C5 phosphatases (Brock et al., 2010). Finally, MPK6 and MAPKKK18 are also regulated by the ABI1 protein phosphatase, a negative regulator of ABA signaling (Mitula et al., 2015). Besides protein phosphatases, activated MAPKs can be also controlled by the UPS dependent proteolytic pathway. For example ABA-regulated MAPKKK18 is regulated in this way (Mitula et al., 2015), demonstrating that MAPKKK18 downregulation by UPS is a significant factor in determining the nature of the MAPK signal output.
A consistent feature of MAPK function in hormone signaling is the existence of central MAPK-dependent hubs allowing extensive crosstalk between hormonal pathways, which leads to precisely regulated cellular functions. In principle, these MAPKs may use more sophisticated mechanisms to diversify signal outputs determined by different stimuli, such as tissue distribution and the formation of acontext-specific signalosome. The most prominent examples are AtMPK3/4/6. MPK3/6 are partially redundant in their activities and are crucial for coordinated response in scope of JA, SA, BR and ABA signaling pathway (Figures 2, 5, 6). AtMPK4 regulates crosstalk between the JA/ET/SA and AUX responses (Figure 2). All three kinases can autophosphorylate (Huang et al., 2000; Pecher et al., 2014) suggesting that they may be regulated by alternative mechanisms or potentially can even escape from the canonical model of MAPK activation. Another example is the AtMKK4/5-MPK6 module which precisely regulates ET and BR signaling and the corresponding responses (Figures 3, 4).
The very complexity of MAPK cascades means that it is often difficult to define them in detail and to assign them a specific role in a particular biological process. Thus, to date, no MAPK cascade, together with its downstream substrates, has been defined in its entirety in any plant system. We also need a better characterization of the functional diversity and redundancy of MAPK complexes. MAPK cascades share many of their components, but nevertheless are still able to deliver hormonal signals to the cell interior precisely and specifically. Importantly, we also need to understand the consequences of phosphorylation by MAPKs for the function, localization or stability of their protein targets. Therefore, many questions remain, some of which are listed below:
• Which cellular elements function as molecular switches to support precise crosstalk and interaction outcomes between MAPK cascades? How do plants discriminate between hormone signaling pathways? How do MAPK cascades maintain specificity?
• What governs MAPK distribution within the cell? What post-transcriptional and translational mechanisms are employed to regulate this distribution?
• Which signaling systems are responsible for MAPK inactivation? How do these work? Which ligands target MAPK pathways to regulate their activity?
• What is the relative importance of different MAPK pathways in hormonal responses?
We believe that the answers to these questions will provide exciting discoveries and establish further the crucial role of MAPKs in plants. MAPK cascades, like other signaling networks, display a wide range of regulatory properties and the extension of MAPK research to all economically important crops is particularly relevant for ensuring sustainable food production globally.
PJ and AL designed the paper. All the authors wrote and revised the review. PJ, MT-Z, AC, and MM prepared the figures. PJ and MT-Z prepared the Supplemental Figures and Supplementary Tables.
This work was supported by grants from the National Science Centre (2012/05/B/NZ3/00352 and 2014/15/B/NZ3/00358 to AL and 2015/17/N/NZ3/00532 to MT-Z.); the Dean of the Faculty of Biology (GDBWB-08/15 to MT-Z and GDWB-02/2017 to MM); and KNOW Poznań RNA Centre (01/KNOW2/2014).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank the reviewers for their useful suggestions.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01387/full#supplementary-material
Agrawal, G. K., Agrawal, S. K., Shibato, J., Iwahashi, H., and Rakwal, R. (2003a). Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem. Biophys. Res. Commun. 300, 775–783. doi: 10.1016/S0006-291X(02)02868-1
Agrawal, G. K., Tamogami, S., Iwahashi, H., Agrawal, V. P., and Rakwal, R. (2003b). Transient regulation of jasmonic acid-inducible rice MAP kinase gene OsBWMK1 by diverse biotic and abiotic stresses. Plant Physiol. Biochem. 41, 355–361. doi: 10.1016/S0981-9428(03)00030-5
Agrawal, G. K., Rakwal, R., and Iwahashi, H. (2002). Isolation of novel rice (Oryza sativa L.) multiple stress responsive MAP kinase gene, OsMSRMK2, whose mRNA accumulates rapidly in response to environmental cues. Biochem. Biophys. Res. Commun. 294, 1009–1016. doi: 10.1016/S0006-291X(02)00571-5
Ahlfors, R., Macioszek, V., Rudd, J., Brosche, M., Schlichting, R., Scheel, D., et al. (2004). Stress hormone-independent activation and nuclear translocation of mitogen-activated protein kinases in Arabidopsis thaliana during ozone exposure. Plant J. 40, 512–522. doi: 10.1111/j.1365-313X.2004.02229.x
Albert, R., Acharya, B. R., Jeon, B. W., Zañudo, J. G. T., Zhu, M., Osman, K., et al. (2017). A new discrete dynamic model of ABA-induced stomatal closure predicts key feedback loops. PLoS Biol. 15:e2003451. doi: 10.1371/journal.pbio.2003451
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Alzwiy, I. A., and Morris, P. C. (2007). A mutation in the Arabidopsis MAP kinase kinase 9 gene results in enhanced seedling stress tolerance. Plant Sci. 173, 302–308. doi: 10.1016/j.plantsci.2007.06.007
Andreasson, E., Jenkins, T., Brodersen, P., Thorgrimsen, S., Petersen, N. H., Zhu, S., et al. (2005). The MAP kinase substrate MKS1 is a regulator of plant defense responses. EMBO J. 24, 2579–2589. doi: 10.1038/sj.emboj.7600737
Asai, T., Tena, G., Plotnikova, J., Willmann, M. R., Chiu, W. L., Gomez-Gomez, L., et al. (2002). MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983. doi: 10.1038/415977a
Barba-Espín, G., Clemente-Moreno, M. J., Alvarez, S., Garcia-Legaz, M. F., Hernandez, J. A., and Diaz-Vivancos, P. (2011). Salicylic acid negatively affects the response to salt stress in pea plants. Plant Biol. 13, 909–917. doi: 10.1111/j.1438-8677.2011.00461.x
Bartels, S., Anderson, J. C., González Besteiro, M. A., Carreri, A., Hirt, H., Buchala, A., et al. (2009). MAP KINASE PHOSPHATASE1 and PROTEIN TYROSINE PHOSPHATASE1 are repressors of salicylic acid synthesis and SNC1-mediated responses in Arabidopsis. Plant Cell 21, 2884–2897. doi: 10.1105/tpc.109.067678
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/S0092-8674(03)00924-3
Berberich, T., Sano, H., and Kusano, T. (1999). Involvement of a MAP kinase, ZmMPK5, in senescence and recovery from low-temperature stress in maize. Mol. Gen. Genet. 262, 534–542. doi: 10.1007/s004380051115
Bergmann, D. C., Lukowitz, W., and Somerville, C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. doi: 10.1126/science.1096014
Béziat, C., and Kleine-Vehn, J. (2018). The road to auxin-dependent growth repression and promotion in apical hooks. Curr. Biol. 28, R519–R525. doi: 10.1016/j.cub.2018.01.069
Bigeard, J., and Hirt, H. (2018). Nuclear signaling of plant MAPKs. Front. Plant Sci. 9:469. doi: 10.3389/fpls.2018.00469
Binder, B. M., Walker, J. M., Gagne, J. M., Emborg, T. J., Hemmann, G., Bleecker, A. B., et al. (2007). The Arabidopsis EIN3 binding F-Box proteins EBF1 and EBF2 have distinct but overlapping roles in ethylene signaling. Plant Cell 19, 509–523. doi: 10.1105/tpc.106.048140
Blanco, F. A., Zanetti, M. E., Casalongué, C. A., and Daleo, G. R. (2006). Molecular characterization of a potato MAP kinase transcriptionally regulated by multiple environmental stresses. Plant Physiol. Biochem. 44, 315–322. doi: 10.1016/j.plaphy.2006.05.005
Bleecker, A. B., and Kende, H. (2000). Ethylene, a gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 16, 1–18. doi: 10.1146/annurev.cellbio.16.1.1
Blilou, I., Xu, J., Wildwater, M., Willemsen, V., Paponov, I., Friml, J., et al. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44. doi: 10.1038/nature03184
Brader, G., Djamei, A., Teige, M., Palva, E. T., and Hirt, H. (2007). The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol. Plant Microbe Interact. 20, 589–596. doi: 10.1094/MPMI-20-5-0589
Brock, A. K., Willmann, R., Kolb, D., Grefen, L., Lajunen, H. M., Bethke, G., et al. (2010). The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol. 153, 1098–1111. doi: 10.1104/pp.110.156109
Brodersen, P., Petersen, M., Bjorn, N. H., Zhu, S., Newman, M. A., Shokat, K. M., et al. (2006). Arabidopsis MAP kinase 4 regulates salicylic acid- and jasmonic acid/ethylene-dependent responses via EDS1 and PAD4. Plant J. 47, 532–546. doi: 10.1111/j.1365-313X.2006.02806.x
Broekaert, W. F., Delaure, S. L., De Bolle, M. F., and Cammue, B. P. (2006). The role of ethylene in host-pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416. doi: 10.1146/annurev.phyto.44.070505.143440
Burnett, E. C., Desikan, R., Moser, R. C., and Neill, S. J. (2000). ABA activation of an MBP kinase in Pisum sativum epidermal peels correlates with stomatal responses to ABA. J. Exp. Bot. 51, 197–205. doi: 10.1093/jexbot/51.343.197
Cai, G., Wang, G., Wang, L., Pan, J., Liu, Y., and Li, D. (2014). ZmMKK1, a novel group A mitogen-activated protein kinase kinase gene in maize, conferred chilling stress tolerance and was involved in pathogen defense in transgenic tobacco. Plant Sci. 214, 57–73. doi: 10.1016/j.plantsci.2013.09.014
Cardinale, F., Meskiene, I., Ouaked, F., and Hirt, H. (2002). Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases. Plant Cell 14, 703–711. doi: 10.1105/tpc.010256
Chae, H. S., Faure, F., and Kieber, J. J. (2003). The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15, 545–559. doi: 10.1105/tpc.006882
Chai, J., Liu, J., Zhou, J., and Xing, D. (2014). Mitogen-activated protein kinase 6 regulates NPR1 gene expression and activation during leaf senescence induced by salicylic acid. J. Exp. Bot. 65, 6513–6528. doi: 10.1093/jxb/eru369
Chaiwongsar, S., Strohm, A. K., Su, S. H., and Krysan, P. J. (2012). Genetic analysis of the Arabidopsis protein kinases MAP3Kepsilon1 and MAP3Kepsilon2 indicates roles in cell expansion and embryo development. Front. Plant Sci. 3:228. doi: 10.3389/fpls.2012.00228
Chang, Y., Yang, H., Ren, D., and Li, Y. (2017). Activation of ZmMKK10, a maize mitogen-activated protein kinase kinase, induces ethylene-dependent cell death. Plant Sci. 264, 129–137. doi: 10.1016/j.plantsci.2017.09.012
Chardin, C., Schenk, S. T., Hirt, H., Colcombet, J., and Krapp, A. (2017). Review: mitogen-activated protein kinases in nutritional signaling in Arabidopsis. Plant Sci. 260, 101–108. doi: 10.1016/j.plantsci.2017.04.006
Chen, L., Hu, W., Tan, S., Wang, M., Ma, Z., Zhou, S., et al. (2012). Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS One 7:e46744. doi: 10.1371/journal.pone.0046744
Chen, J., Zhang, Q., Wang, Q., Feng, M., Li, Y., Meng, Y., et al. (2017). RhMKK9, a rose MAP KINASE KINASE gene, is involved in rehydration-triggered ethylene production in rose gynoecia. BMC Plant Biol. 17:51. doi: 10.1186/s12870-017-0999-1
Chen, M., Xie, S., Ouyang, Y., and Yao, J. (2017). Rice PcG gene OsEMF2b controls seed dormancy and seedling growth by regulating the expression of OsVP1. Plant Sci. 260, 80–89. doi: 10.1016/j.plantsci.2017.04.005
Chen, R., and Baluska, F. (2013). “Polar auxin transport,” in Series, Signaling and Communication in Plants, Vol. 17, eds R. Chen and F. Baluška (Berlin: Springer-Verlag), 330. doi: 10.1007/978-3-642-35299-7
Chen, X., and Ronald, P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16, 451–459. doi: 10.1016/j.tplants.2011.04.003
Cheong, Y. H., Moon, B. C., Kim, J. K., Kim, C. Y., Kim, M. C., Kim, I. H., et al. (2003). BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972. doi: 10.1104/pp.103.023176
Chiu, R. S., Pan, S., Zhao, R., and Gazzarrini, S. (2016). ABA-dependent inhibition of the ubiquitin proteasome system during germination at high temperature in Arabidopsis. Plant J. 88, 749–761. doi: 10.1111/tpj.13293
Cho, Y. H., and Yoo, S. D. (2015). Novel connections and gaps in ethylene signaling from the ER membrane to the nucleus. Front. Plant Sci. 5:733. doi: 10.3389/fpls.2014.00733
Colcombet, J., and Hirt, H. (2008). Arabidopsis MAPKs, a complex signalling network involved in multiple biological processes. Biochem. J. 413, 217–226. doi: 10.1042/BJ20080625
Colcombet, J., Sözen, C., and Hirt, H. (2016). Convergence colcombet of multiple MAP3Ks on MKK3 identifies a set of novel stress MAPK modules. Front. Plant Sci. 7:1941. doi: 10.3389/fpls.2016.01941
Contreras-Cornejo, H. A., Lopez-Bucio, J. S., Mendez-Bravo, A., Macias-Rodriguez, L., Ramos-Vega, M., Guevara-Garcia, A. A., et al. (2015). Mitogen-activated protein kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root-system architecture alterations by Trichoderma atroviride. Mol. Plant Microbe Interact. 28, 701–710. doi: 10.1094/MPMI-01-15-0005-R
Corredoira, E., Cano, V., Bárány, I., Solís, M. T., Rodríguez, H., Vieitez, A. M., et al. (2017). Initiation of leaf somatic embryogenesis involves high pectin esterification, auxin accumulation and DNA demethylation in Quercus alba. J. Plant Physiol. 213, 42–54. doi: 10.1016/j.jplph.2017.02.012
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid, emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dai, Y., Wang, H., Li, B., Huang, J., Liu, X., Zhou, Y., et al. (2006). Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell 18, 308–320. doi: 10.1105/tpc.105.037846
Danquah, A., Zélicourt, A., Boudsocq, M., Neubauer, J., Frei Dit, F. N., Leonhardt, N., et al. (2015). Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J. 82, 232–244. doi: 10.1111/tpj.12808
Danquah, A., Zélicourt, A., Colcombet, J., and Hirt, H. (2014). The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol. Adv. 32, 40–52. doi: 10.1016/j.biotechadv.2013.09.006
Daxberger, A., Nemak, A., Mithöfer, A., Fliegmann, J., Ligterink, W., Hirt, H., et al. (2007). Activation of members of a MAPK module in beta-glucan elicitor-mediated non-host resistance of soybean. Planta 225, 1559–1571. doi: 10.1007/s00425-006-0442-6
De Paepe, A., and Van der Straeten, D. (2005). Ethylene biosynthesis and signaling, an overview. Vitam. Horm. 72, 399–430. doi: 10.1016/S0083-6729(05)72011-2
de Zelicourt, A., Colcombet, J., and Hirt, H. (2016). The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 21, 677–685. doi: 10.1016/j.tplants.2016.04.004
Devic, M., and Roscoe, T. (2016). Seed maturation: simplification of control networks in plants. Plant Sci. 252, 335–346. doi: 10.1016/j.plantsci.2016.08.012
Ding, Y., Cao, J., Ni, L., Zhu, Y., Zhang, A., Tan, M., et al. (2013). ZmCPK11 is involved in abscisic acid-induced antioxidant defence and functions upstream of ZmMPK5 in abscisic acid signalling in maize. J. Exp. Bot. 64, 871–884. doi: 10.1093/jxb/ers366
Doczi, R., Brader, G., Pettko-Szandtner, A., Rajh, I., Djamei, A., Pitzschke, A., et al. (2007). The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19, 3266–3279. doi: 10.1105/tpc.106.050039
Doczi, R., Okrész, L., Romero, A. E., Paccanaro, A., and Bögre, L. (2012). Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 17, 518–525. doi: 10.1016/j.tplants.2012.05.009
Dodd, I. C., Tan, L. P., and He, J. (2003). Do increases in xylem sap pH and/or ABA concentration mediate stomatal closure following nitrate deprivation? J. Exp. Bot. 54, 1281–1288. doi: 10.1093/jxb/erg122
Dong, H., Zhen, Z., Peng, J., Chang, L., Gong, Q., and Wang, N. N. (2011). Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J. Exp. Bot. 62, 4875–4887. doi: 10.1093/jxb/err143
Dory, M., Hatzimasoura, E., Kállai, B. M., Nagy, S. K., Jäger, K., Darula, Z., et al. (2018). Coevolving MAPK and PID phosphosites indicate an ancient environmental control of PIN auxin transporters in land plants. FEBS Lett. 592, 89–102. doi: 10.1002/1873-3468.12929
Droillard, M., Boudsocq, M., Barbier-Brygoo, H., and Laurière, C. (2002). Different protein kinase families are activated by osmotic stresses in Arabidopsis thaliana cell suspensions. Involvement of the MAP kinases AtMPK3 and AtMPK6. FEBS Lett. 527, 43–50. doi: 10.1016/S0014-5793(02)03162-9
Duan, P., Rao, Y., Zeng, D., Yang, Y., Xu, R., Zhang, B., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77, 547–557. doi: 10.1111/tpj.12405
Dubois, M., Van den Broeck, L., and Inzé, D. (2018). The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. doi: 10.1016/j.tplants.2018.01.003
Eisenach, C., Baetz, U., Huck, N. V., Zhang, J., De Angeli, A., Beckers, G. J. M., et al. (2017). ABA-induced stomatal closure involves ALMT4, a phosphorylation-dependent vacuolar anion channel of Arabidopsis. Plant Cell 29, 2552–2569. doi: 10.1105/tpc.17.00452
Enders, T. A., Frick, E. M., and Strader, L. C. (2017). An Arabidopsis kinase cascade influences auxin-responsive cell expansion. Plant J. 92, 68–81. doi: 10.1111/tpj.13635
Finkelstein, R. (2013). Abscisic Acid synthesis and response. Arabidopsis Book 11:e0166. doi: 10.1199/tab.0166
Finkelstein, R. R., Gampala, S. S. L., and Rock, C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.), s15–s45. doi: 10.1105/tpc.010441
Frei Dit, F. N., Garcia, A. V., Bigeard, J., Zaag, R., Bueso, E., Garmier, M., et al. (2014). Functional analysis of Arabidopsis immune-related MAPKs uncovers a role for MPK3 as negative regulator of inducible defences. Genome Biol. 15:R87. doi: 10.1186/gb-2014-15-6-r87
Friml, J., Vieten, A., Sauer, M., Weijers, D., Schwarz, H., Hamann, T., et al. (2003). Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426, 147–153. doi: 10.1038/nature02085
Frye, C. A., and Innes, R. W. (1998). An Arabidopsis mutant with enhanced resistance to powdery mildew. Plant Cell 10, 947–956. doi: 10.1105/tpc.10.6.947
Frye, C. A., Tang, D., and Innes, R. W. (2001). Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl. Acad. Sci. U.S.A. 98, 373–378. doi: 10.1073/pnas.98.1.373
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Fujita, M., Fujita, Y., Noutoshi, Y., Takahashi, F., Narusaka, Y., Yamaguchi-Shinozaki, K., et al. (2006). Crosstalk between abiotic and biotic stress responses, a current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9, 436–442. doi: 10.1016/j.pbi.2006.05.014
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
Furuya, T., Matsuoka, D., and Nanmori, T. (2014). Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett. 588, 2025–2030. doi: 10.1016/j.febslet.2014.04.032
Galletti, R., Ferrari, S., and De, L. G. (2011). Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 157, 804–814. doi: 10.1104/pp.111.174003
Ganguly, A., Park, M., Kesawat, M. S., and Cho, H. T. (2014). Functional analysis of the hydrophilic loop in intracellular trafficking of Arabidopsis PIN-FORMED proteins. Plant Cell 26, 1570–1585. doi: 10.1105/tpc.113.118422
Gao, M., Liu, J., Bi, D., Zhang, Z., Cheng, F., Chen, S., et al. (2008). MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 18, 1190–1198. doi: 10.1038/cr.2008.300
Ghawana, S., Kumar, S., and Ahuja, P. S. (2010). Early low-temperature responsive mitogen activated protein kinases RaMPK1 and RaMPK2 from Rheum australe D. Don respond differentially to diverse stresses. Mol. Biol. Rep. 37, 933–938. doi: 10.1007/s11033-009-9726-9
Gomi, K., Ogawa, D., Katou, S., Kamada, H., Nakajima, N., Saji, H., et al. (2005). A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol. 46, 1902–1914. doi: 10.1093/pcp/pci211
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., et al. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38, W695–W699. doi: 10.1093/nar/gkq313
Gu, L., Liu, Y., Zong, X., Liu, L., Li, D. P., and Li, D. Q. (2010). Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol. Biol. Rep. 37, 4067–4073. doi: 10.1007/s11033-010-0066-6
Gudesblat, G. E., Iusem, N. D., and Morris, P. C. (2007). Guard cell-specific inhibition of Arabidopsis MPK3 expression causes abnormal stomatal responses to abscisic acid and hydrogen peroxide. New Phytol. 173, 713–721. doi: 10.1007/s11033-010-0066-6
Gudesblat, G. E., Schneider-Pizon, J., Betti, C., Mayerhofer, J., Vanhoutte, I., van Dongen, W., et al. (2012). SPEECHLESS integrates brassinosteroid and stomata signalling pathways. Nat. Cell Biol. 14, 548–554. doi: 10.1038/ncb2471
Guo, H., Peng, X., Gu, L., Wu, J., Ge, F., and Sun, Y. (2017). Up-regulation of MPK4 increases the feeding efficiency of the green peach aphid under elevated CO2 in Nicotiana attenuata. J. Exp. Bot. 68, 5923–5935. doi: 10.1093/jxb/erx394
Gupta, R., and Chakrabarty, S. K. (2013). Gibberellic acid in plant, still a mystery unresolved. Plant Signal. Behav. 8:e25504. doi: 10.4161/psb.25504
Gutierrez, L., Van, W. O., Castelain, M., and Bellini, C. (2007). Combined networks regulating seed maturation. Trends Plant Sci. 12, 294–300. doi: 10.1016/j.tplants.2007.06.003
Hadiarto, T., Nanmori, T., Matsuoka, D., Iwasaki, T., Sato, K., Fukami, Y., et al. (2006). Activation of Arabidopsis MAPK kinase kinase (AtMEKK1) and induction of AtMEKK1-AtMEK1 pathway by wounding. Planta 223, 708–713. doi: 10.1007/s00425-005-0126-7
Hamel, L. P., Nicole, M. C., Sritubtim, S., Morency, M. J., Ellis, M., Ehlting, J., et al. (2006). Ancient signals, comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 11, 192–198. doi: 10.1016/j.tplants.2006.02.007
Han, L., Li, G. J., Yang, K. Y., Mao, G., Wang, R., Liu, Y., et al. (2010). Mitogen-activated protein kinase 3 and 6 regulate Botrytis cinerea-induced ethylene production in Arabidopsis. Plant J. 64, 114–127. doi: 10.1111/j.1365-313X.2010.04318.x
Hashimoto, M., Komatsu, K., Maejima, K., Okano, Y., Shiraishi, T., Ishikawa, K., et al. (2012). Identification of three MAPKKKs forming a linear signaling pathway leading to programmed cell death in Nicotiana benthamiana. BMC Plant Biol. 12:103. doi: 10.1186/1471-2229-12-103
Hayat, S., and Ahmad, A. (2007). Salicylic Acid - A Plant Hormone. Berlin: Springer. doi: 10.1007/1-4020-5184-0
Heil, M., Ibarra-Laclette, E., Adame-Alvarez, R. M., Martinez, O., Ramirez-Chavez, E., Molina-Torres, J., et al. (2012). How plants sense wounds, damaged-self recognition is based on plant-derived elicitors and induces octadecanoid signaling. PLoS One 7:e30537. doi: 10.1371/journal.pone.0030537
Heisler, M. G., Ohno, C., Das, P., Sieber, P., Reddy, G. V., Long, J. A., et al. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911. doi: 10.1016/j.cub.2005.09.052
Hettenhausen, C., Baldwin, I. T., and Wu, J. (2012). Silencing MPK4 in Nicotiana attenuata enhances photosynthesis and seed production but compromises abscisic acid-induced stomatal closure and guard cell-mediated resistance to Pseudomonas syringae pv tomato DC3000. Plant Physiol. 158, 759–776. doi: 10.1104/pp.111.190074
Hettenhausen, C., Schuman, M. C., and Wu, J. (2014). MAPK signaling - a key element in plant defense response to insects. Insect Sci. 22, 157–164. doi: 10.1111/1744-7917.12128
Himmelbach, A., Yang, Y., and Grill, E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6, 470–479. doi: 10.1016/S1369-5266(03)00090-6
Hirt, H. (2000). MAP kinases in plant signal transduction. Results Probl. Cell Differ. 27, 1–9. doi: 10.1007/978-3-540-49166-8_1
Hou, X., Ding, L., and Yu, H. (2013). Crosstalk between GA and JA signaling mediates plant growth and defense. Plant Cell Rep. 32, 1067–1074. doi: 10.1007/s00299-013-1423-4
Hu, X., Jiang, M., Zhang, A., and Lu, J. (2005). Abscisic acid-induced apoplastic H2O2 accumulation up-regulates the activities of chloroplastic and cytosolic antioxidant enzymes in maize leaves. Planta 223, 57–68. doi: 10.1007/s00425-005-0068-0
Huang, H. J., Fu, S. F., Tai, Y. H., Chou, W. C., and Huang, D. D. (2002). Expression of Oryza sativa MAP kinase gene is developmentally regulated and stress-responsive. Physiol. Plant. 114, 572–580. doi: 10.1034/j.1399-3054.2002.1140410.x
Huang, X., Zhang, X., Gong, Z., Yang, S., and Shi, Y. (2017). ABI4 represses the expression of type-A ARRs to inhibit seed germination in Arabidopsis. Plant J. 89, 354–365. doi: 10.1111/tpj.13389
Huang, Y., Li, H., Gupta, R., Morris, P. C., Luan, S., and Kieber, J. J. (2000). ATMPK4, an Arabidopsis homolog of mitogen-activated protein kinase, is activated in vitro by AtMEK1 through threonine phosphorylation. Plant Physiol. 122, 1301–1310. doi: 10.1104/pp.122.4.1301
Huttly, A. K., and Phillips, A. L. (1995). Gibberellin-regulated expression in oat aleurone cells of two kinases that show homology to MAP kinase and a ribosomal protein kinase. Plant Mol. Biol. 27, 1043–1052. doi: 10.1007/BF00037031
Hwa, C.-M., and Yang, X.-C. (2008). The AtMKK3 pathway mediates ABA and salt signaling in Arabidopsis. Acta Physiol. Plant. 30, 277–286. doi: 10.1007/s11738-007-0117-3
Ichimura, K., Casais, C., Peck, S. C., Shinozaki, K., and Shirasu, K. (2006). MEKK1 is required for MPK4 activation and regulates tissue-specific and temperature-dependent cell death in Arabidopsis. J. Biol. Chem. 281, 36969–36976. doi: 10.1074/jbc.M605319200
Iqbal, N., Trivellini, A., Masood, A., Ferrante, A., and Khan, N. A. (2013). Current understanding on ethylene signaling in plants: the influence of nutrient availability. Plant Physiol. Biochem. 73, 128–138. doi: 10.1016/j.plaphy.2013.09.011
Jaemsaeng, R., Jantasuriyarat, C., and Thamchaipenet, A. (2018). Molecular interaction of 1-aminocyclopropane-1-carboxylate deaminase (ACCD)-producing endophytic Streptomyces sp. GMKU 336 towards salt-stress resistance of Oryza sativa L. cv. KDML105. Sci. Rep. 8:1950. doi: 10.1038/s41598-018-19799-9
Jalmi, S. K., and Sinha, A. K. (2016). Functional involvement of a mitogen activated protein kinase module, OsMKK3-OsMPK7-OsWRK30 in mediating resistance against Xanthomonas oryzae in rice. Sci. Rep. 6:37974. doi: 10.1038/srep37974
Jammes, F., Song, C., Shin, D., Munemasa, S., Takeda, K., Gu, D., et al. (2009). MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 20520–20525. doi: 10.1073/pnas.0907205106
Jeong, M. J., Lee, S. K., Kim, B. G., Kwon, T. R., Cho, W. S., Park, Y. T., et al. (2006). A rice (Oryza sativa L.) MAP kinase gene, OsMAPK44, is involved in response to abiotic stresses. Plant Cell Tissue Organ Cult. 85, 151–160. doi: 10.1007/s11240-005-9064-0
Jia, W., Li, B., Li, S., Liang, Y., Wu, X., Ma, M., et al. (2016). Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 14:e1002550. doi: 10.1371/journal.pbio.1002550
Jiang, J., and Song, C. P. (2008). MEK1/2 and p38-like MAP kinase successively mediate H(2)O(2) signaling in Vicia guard cell. Plant Signal. Behav. 3, 996–998. doi: 10.1007/s00299-007-0449-x
Jiang, M., Wen, F., Cao, J., Li, P., She, J., and Chu, Z. (2015). Genome-wide exploration of the molecular evolution and regulatory network of mitogen-activated protein kinase cascades upon multiple stresses in Brachypodium distachyon. BMC Genomics 16:228. doi: 10.1186/s12864-015-1452-1
Jin, H., Axtell, M. J., Dahlbeck, D., Ekwenna, O., Zhang, S., Staskawicz, B., et al. (2002). NPK1, an MEKK1-like mitogen-activated protein kinase kinase kinase, regulates innate immunity and development in plants. Dev. Cell 3, 291–297. doi: 10.1016/S1534-5807(02)00205-8
Jonak, C., Okresz, L., Bogre, L., and Hirt, H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5, 415–424. doi: 10.1016/S1369-5266(02)00285-6
Jonak, C., Pay, A., Bogre, L., Hirt, H., and Herberle-Bors, E. (1993). The plant homologue of MAPK is expressed in a cell cycle-dependent and organ specific manner. Plant J. 3, 611–617. doi: 10.1046/j.1365-313X.1993.03040611.x
Joo, S., Liu, Y., Lueth, A., and Zhang, S. (2008). MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 54, 129–140. doi: 10.1111/j.1365-313X.2008.03404.x
Joseph, M. P., Papdi, C., Kozma-Bognar, L., Nagy, I., Lopez-Carbonell, M., Rigo, G., et al. (2014). The Arabidopsis ZINC FINGER PROTEIN3 interferes with abscisic acid and light signaling in seed germination and plant development. Plant Physiol. 165, 1203–1220. doi: 10.1104/pp.113.234294
Jouannic, S., Hamal, A., Leprince, A. S., Tregear, J. W., Kreis, M., and Henry, Y. (1999). Characterisation of novel plant genes encoding MEKK/STE11 and RAF-related protein kinases. Gene 229, 171–181. doi: 10.1016/S0378-1119(99)00012-8
Ju, C., Yoon, G. M., Shemansky, J. M., Lin, D. Y., Ying, Z. I., Chang, J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signaling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 19486–19491. doi: 10.1073/pnas.1214848109
Jung, J. Y., Shin, R., and Schachtman, D. P. (2009). Ethylene mediates response and tolerance to potassium deprivation in Arabidopsis. Plant Cell 21, 607–621. doi: 10.1105/tpc.108.063099
Kamada, M., Miyamoto, K., Oka, M., Ueda, J., and Higashibata, A. (2018). Regulation of asymmetric polar auxin transport by PsPIN1 in endodermal tissues of etiolated Pisum sativum epicotyls, focus on immunohistochemical analyses. J. Plant Res. 131, 681–692. doi: 10.1007/s10265-018-1031-z
Kandoth, P. K., Ranf, S., Pancholi, S. S., Jayanty, S., Walla, M. D., Miller, W., et al. (2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. U.S.A. 104, 12205–12210. doi: 10.1073/pnas.0700344104
Kang, S., Yang, F., Li, L., Chen, H., Chen, S., and Zhang, J. (2015). The Arabidopsis transcription factor BRASSINOSTEROID INSENSITIVE1-ETHYL METHANESULFONATE-SUPPRESSOR1 is a direct substrate of MITOGEN-ACTIVATED PROTEIN KINASE6 and regulates immunity. Plant Physiol. 167, 1076–1086. doi: 10.1104/pp.114.250985
Kazan, K. (2015). Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20, 219–229. doi: 10.1016/j.tplants.2015.02.001
Kende, H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Biol. 44, 283–307. doi: 10.1146/annurev.pp.44.060193.001435
Khan, M., Rozhon, W., Bigeard, J., Pflieger, D., Husar, S., Pitzschke, A., et al. (2013). Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 288, 7519–7527. doi: 10.1074/jbc.M112.384453
Khan, N. A., Khan, M. I. R., Ferrante, A., and Poor, P. (2017). Ethylene: a key regulatory molecule in plants. Front. Plant Sci. 8:1782. doi: 10.3389/fpls.2017.01782
Khokhlatchev, A. V., Canagarajah, B., Wilsbacher, J., Robinson, M., Atkinson, M., Goldsmith, E., et al. (1998). Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell 93, 605–615. doi: 10.1016/S0092-8674(00)81189-7
Khokon, M. A., Salam, M. A., Jammes, F., Ye, W., Hossain, M. A., Uraji, M., et al. (2015). Two guard cell mitogen-activated protein kinases, MPK9 and MPK12, function in methyl jasmonate-induced stomatal closure in Arabidopsis thaliana. Plant Biol. 17, 946–952. doi: 10.1111/plb.12321
Kim, H. S., Park, S. C., Ji, C. Y., Park, S., Jeong, J. C., Le, H. S., et al. (2016). Molecular characterization of biotic and abiotic stress-responsive MAP kinase genes, IbMPK3 and IbMPK6, in sweet potato. Plant Physiol. Biochem. 108, 37–48. doi: 10.1016/j.plaphy.2016.06.036
Kim, J. A., Agrawal, G. K., Rakwal, R., Han, K. S., Kim, K. N., Yun, C. H., et al. (2003). Molecular cloning and mRNA expression analysis of a novel rice (Oryzasativa L.) MAPK kinase kinase, OsEDR1, an ortholog of Arabidopsis AtEDR1, reveal its role in defense/stress signalling pathways and development. Biochem. Biophys. Res. Commun. 300, 868–876. doi: 10.1016/S0006-291X(02)02944-3
Kim, J. M., Woo, D. H., Kim, S. H., Lee, S. Y., Park, H. Y., Seok, H. Y., et al. (2012). Arabidopsis MKKK20 is involved in osmotic stress response via regulation of MPK6 activity. Plant Cell Rep. 31, 217–224. doi: 10.1007/s00299-011-1157-0
Kim, T. W., Michniewicz, M., Bergmann, D. C., and Wang, Z. Y. (2012). Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422. doi: 10.1038/nature10794
Klingler, J. P., Batelli, G., and Zhu, J. K. (2010). ABA receptors, the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61, 3199–3210. doi: 10.1093/jxb/erq151
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kong, Q., Qu, N., Gao, M., Zhang, Z., Ding, X., Yang, F., et al. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24, 2225–2236. doi: 10.1105/tpc.112.097253
Kong, X., Pan, J., Zhang, D., Jiang, S., Cai, G., Wang, L., et al. (2013). Identification of mitogen-activated protein kinase kinase gene family and MKK-MAPK interaction network in maize. Biochem. Biophys. Res. Commun. 441, 964–969. doi: 10.1016/j.bbrc.2013.11.008
Kong, X., Pan, J., Zhang, M., Xing, X., Zhou, Y., Liu, Y., et al. (2011). ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 34, 1291–1303. doi: 10.1111/j.1365-3040.2011.02329.x
Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36.
Kovtun, Y., Chiu, W. L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 97, 2940–2945. doi: 10.1073/pnas.97.6.2940
Kovtun, Y., Chiu, W. L., Zeng, W., and Sheen, J. (1998). Suppression of auxin signal transduction by a MAPK cascade in higher plants. Nature 395, 716–720. doi: 10.1038/27240
Kumar, K. R. R., Srinivasan, T., and Kirti, P. B. (2009). A mitogen-activated protein kinase gene, AhMPK3 of peanut: molecular cloning, genomic organization, and heterologous expression conferring resistance against Spodoptera litura in tobacco. Mol. Genet. Genomics 282, 65–81. doi: 10.1007/s00438-009-0446-6
Lau, O. S., and Bergmann, D. C. (2012). Stomatal development, a plant’s perspective on cell polarity, cell fate transitions and intercellular communication. Development 139, 3683–3692. doi: 10.1242/dev.080523
Le, J., Zou, J., Yang, K., and Wang, M. (2014). Signaling to stomatal initiation and cell division. Front. Plant Sci. 5:297. doi: 10.3389/fpls.2014.00297
Lee, H. (2015). Mitogen-activated protein kinase kinase 3 is required for regulation during dark-light transition. Mol. Cells 38, 651–656. doi: 10.14348/molcells.2015.0055
Lee, J. S., Huh, K. W., Bhargava, A., and Ellis, B. E. (2008). Comprehensive analysis of protein-protein interactions between Arabidopsis MAPKs and MAPK kinases helps define potential MAPK signalling modules. Plant Signal. Behav. 3, 1037–1041. doi: 10.4161/psb.3.12.6848
Lee, M. O., Cho, K., Kim, S. H., Jeong, S. H., Kim, J. A., Jung, Y. H., et al. (2008). Novel rice OsSIPK is a multiple stress responsive MAPK family member showing rhythmic expression at mRNA level. Planta 227, 981–990. doi: 10.1007/s00425-007-0672-2
Lee, J. S., Wang, S., Sritubtim, S., Chen, J. G., and Ellis, B. E. (2009). Arabidopsis mitogen-activated protein kinase MPK12 interacts with the MAPK phosphatase IBR5 and regulates auxin signaling. Plant J. 57, 975–985. doi: 10.1111/j.1365-313X.2008.03741.x
Lee, S. C., and Luan, S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35, 53–60. doi: 10.1111/j.1365-3040.2011.02426.x
Lee, S. J., Lee, M. H., Kim, J. I., and Kim, S. Y. (2015). Arabidopsis putative MAP kinase kinase kinases Raf10 and Raf11 are positive regulators of seed dormancy and ABA response. Plant Cell Physiol. 56, 84–97. doi: 10.1093/pcp/pcu148
Lee, S. K., Kim, B. G., Kwon, T. R., Jeong, M. J., Park, S. R., Lee, J. W., et al. (2011). Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.)1. J. Biosci. 36, 139–151. doi: 10.1007/s12038-011-9002-8
Lee, Y., Kim, Y. J., Kim, M.-H., and Kwak, J. M. (2016). MAPK cascades in guard cell signal transduction. Front. Plant Sci. 7:80. doi: 10.3389/fpls.2016.00080
Leon-Kloosterziel, K. M., van de Bunt, G. A., Zeevaart, J. A., and Koornneef, M. (1996). Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 110, 233–240. doi: 10.1104/pp.110.1.233
Leprince, O., Pellizzaro, A., Berriri, S., and Buitink, J. (2017). Late seed maturation, drying without dying. J. Exp. Bot. 68, 827–841. doi: 10.1093/jxb/erw363
Leung, J., and Giraudat, J. (1998). ABSCISIC ACID SIGNAL TRANSDUCTION. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Leyser, O. (2003). Regulation of shoot branching by auxin. Trends Plant Sci. 8, 541–545. doi: 10.1016/j.tplants.2003.09.008
Li, C. H., Wang, G., Zhao, J. L., Zhang, L. Q., Ai, L. F., Han, Y. F., et al. (2014). The receptor-like kinase SIT1 mediates salt sensitivity by activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell 26, 2538–2553. doi: 10.1105/tpc.114.125187
Li, G., Meng, X., Wang, R., Mao, G., Han, L., Liu, Y., et al. (2012). Dual-level regulation of ACC synthase activity by MPK3/MPK6 cascade and its downstream WRKY transcription factor during ethylene induction in Arabidopsis. PLoS Genet. 8:e1002767. doi: 10.1371/journal.pgen.1002767
Li, K., Yang, F., Zhang, G., Song, S., Li, Y., Ren, D., et al. (2017). AIK1, a mitogen-activated protein kinase, modulates abscisic acid responses through the MKK5-MPK6 kinase cascade. Plant Physiol. 173, 1391–1408. doi: 10.1104/pp.16.01386
Li, Q., Wang, Y., Liu, C., Pei, Z. M., and Shi, W.-L. (2017). The crosstalk between ABA, nitric oxide, hydrogen peroxide, and calcium in stomatal closing of Arabidopsis thaliana. Biologia 72, 1140–1146. doi: 10.1515/biolog-2017-0126
Li, S., Han, X., Yang, L., Deng, X., Wu, H., Zhang, M., et al. (2018). Mitogen-activated protein kinases and calcium-dependent protein kinases are involved in wounding-induced ethylene biosynthesis in Arabidopsis thaliana. Plant Cell Environ. 41, 134–147. doi: 10.1111/pce.12984
Li, X., Zhang, Y., Huang, L., Ouyang, Z., Hong, Y., Zhang, H., et al. (2014). Tomato SlMKK2 and SlMKK4 contribute to disease resistance against Botrytis cinerea. BMC Plant Biol. 14:166. doi: 10.1186/1471-2229-14-166
Li, Y., Cai, H., Liu, P., Wang, C., Gao, H., Wu, C., et al. (2017). Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem. Biophys. Res. Commun. 484, 292–297. doi: 10.1016/j.bbrc.2017.01.104
Li, Y., Chang, Y., Zhao, C., Yang, H., and Ren, D. (2016). Expression of the inactive ZmMEK1 induces salicylic acid accumulation and salicylic acid-dependent leaf senescence. J. Integr. Plant Biol. 58, 724–736. doi: 10.1111/jipb.12465
Li, Y., Qin, L., Zhao, J., Muhammad, T., Cao, H., Li, H., et al. (2017). SlMAPK3 enhances tolerance to tomato yellow leaf curl virus (TYLCV) by regulating salicylic acid and jasmonic acid signaling in tomato (Solanum lycopersicum). PLoS One 12:e0172466. doi: 10.1371/journal.pone.0172466
Li, Y., Zhang, L., Lu, W., Wang, X., Wu, C. A., and Guo, X. (2014). Overexpression of cotton GhMKK4 enhances disease susceptibility and affects abscisic acid, gibberellin and hydrogen peroxide signalling in transgenic Nicotiana benthamiana. Mol. Plant Pathol. 15, 94–108. doi: 10.1111/mpp.12067
Liang, W., Yang, B., Yu, B. J., Zhou, Z., Li, C., Jia, M., et al. (2013). Identification and analysis of MKK and MPK gene families in canola (Brassica napus L.). BMC Genomics 14:392. doi: 10.1186/1471-2164-14-392
Lieberherr, D., Thao, N. P., Nakashima, A., Umemura, K., Kawasaki, T., and Shimamoto, K. (2005). A sphingolipid elicitor-inducible mitogen-activated protein kinase is regulated by the small GTPase OsRac1 and heterotrimeric G-protein in rice 1. Plant Physiol. 138, 1644–1652. doi: 10.1104/pp.104.057414
Ligterink, W., and Hirt, H. (2001). Mitogen-activated protein [MAP] kinase pathways in plants, versatile signaling tools. Int. Rev. Cytol. 201, 209–275. doi: 10.1016/S0074-7696(01)01004-X
Lin, F., Ding, H., Wang, J., Zhang, H., Zhang, A., Zhang, Y., et al. (2009). Positive feedback regulation of maize NADPH oxidase by mitogen-activated protein kinase cascade in abscisic acid signalling. J. Exp. Bot. 60, 3221–3238. doi: 10.1093/jxb/erp157
Liu, D. D., Zhu, M., Hao, L. L., Chen, X. B., Gao, Y., Guo, X. Q., et al. (2016). GhMAPKKK49, a novel cotton (Gossypium hirsutum L.) MAPKKK gene, is involved in diverse stress responses. Acta Physiol. Plant. 38, 1–12. doi: 10.1007/s1173
Liu, H., Wang, Y., Xu, J., Su, T., Liu, G., and Ren, D. (2008). Ethylene signaling is required for the acceleration of cell death induced by the activation of AtMEK5 in Arabidopsis. Cell Res. 18, 422–432. doi: 10.1038/cr.2008.29
Liu, X., Lin, Y., Liu, D., Wang, C. H., Zhao, Z., Cui, X., et al. (2017). MAPK-mediated auxin signal transduction pathways regulate the malic acid secretion under aluminum stress in wheat (Triticum aestivum L.). Sci. Rep. 7:1620. doi: 10.1104/pp.103.3.695
Liu, Y. (2012). Roles of mitogen-activated protein kinase cascades in ABA signaling. Plant Cell Rep. 31, 1–12. doi: 10.1007/s00299-011-1130-y
Liu, Y., Li, X., Tan, H., Liu, M., Zhao, X., and Wang, J. (2010). Molecular characterization of RsMPK2, a C1 subgroup mitogen-activated protein kinase in the desert plant Reaumuria soongorica. Plant Physiol. Biochem. 48, 836–844. doi: 10.1016/j.plaphy.2010.07.001
Liu, Y., Zhang, D., Wang, L., and Li, D. (2013). Genome-wide analysis of mitogen-activated protein kinase gene family in maize. Plant Mol. Biol. Rep. 31, 1446–1460. doi: 10.1007/s11105-013-0623-y
Liu, Y., and Zhang, S. (2004). Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 16, 3386–3399. doi: 10.1105/tpc.104.026609
Liu, Z., Shi, L., Liu, Y., Tang, Q., Shen, L., Yang, S., et al. (2015). Genome-wide identification and transcriptional expression analysis of mitogen-activated protein kinase and mitogen-activated protein kinase kinase genes in Capsicum annuum. Front. Plant Sci. 6:780. doi: 10.3389/fpls.2015.00780
Liu, Z. B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T. J. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. doi: 10.1105/tpc.6.5.645
Lu, C., Han, M. H., Guevara-Garcia, A., and Fedoroff, N. V. (2002). Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 99, 15812–15817. doi: 10.1073/pnas.242607499
Lu, K., Guo, W., Lu, J., Yu, H., Qu, C., Tang, Z., et al. (2015). Genome-wide survey and expression profile analysis of the mitogen-activated protein kinase (MAPK) gene family in Brassica rapa. PLoS One 10:e0132051. doi: 10.1371/journal.pone.0132051
Ludwików, A. (2015). Targeting proteins for proteasomal degradation-a new function of Arabidopsis ABI1 protein phosphatase 2C. Front. Plant Sci. 6:310. doi: 10.3389/fpls.2015.00310
Ludwików, A., Ciesla, A., Kasprowicz-Maluski, A., Mitula, F., Tajdel, M., Galganski, L., et al. (2014). Arabidopsis protein phosphatase 2C ABI1 interacts with type I ACC synthases and is involved in the regulation of ozone-induced ethylene biosynthesis. Mol. Plant 7, 960–976. doi: 10.1093/mp/ssu025
Lukowitz, W., Roeder, A., Parmenter, D., and Somerville, C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109–119. doi: 10.1016/S0092-8674(03)01067-5
Lumbreras, V., Vilela, B., Irar, S., Solé, M., Capellades, M., Valls, M., et al. (2010). MAPK phosphatase MKP2 mediates disease responses in Arabidopsis and functionally interacts with MPK3 and MPK6. Plant J. 63, 1017–1030. doi: 10.1111/j.1365-313X.2010.04297.x
Ma, F., Ni, L., Liu, L., Li, X., Zhang, H., Zhang, A., et al. (2016). ZmABA2, an interacting protein of ZmMPK5, is involved in abscisic acid biosynthesis and functions. Plant Biotechnol. J. 14, 771–782. doi: 10.1111/pbi.12427
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
MAPK Group (2002). Mitogen-activated protein kinase cascades in plants, A new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/S1360-1385(02)02302-6
Marcote, M. J., and Carbonell, J. (2000). Transient expression of a pea MAP kinase gene induced by gibberellic acid and 6-benzyladenine in unpollinated pea ovaries. Plant Mol. Biol. 44, 177–186. doi: 10.1023/A:100643433
Marshall, A., Aalen, R. B., Audenaert, D., Beeckman, T., Broadley, M. R., Butenko, M. A., et al. (2012). Tackling drought stress, receptor-like kinases present new approaches. Plant Cell 24, 2262–2278. doi: 10.1105/tpc.112.096677
Matsuoka, D., Yasufuku, T., Furuya, T., and Nanmori, T. (2015). An abscisic acid inducible Arabidopsis MAPKKK, MAPKKK18 regulates leaf senescence via its kinase activity. Plant Mol. Biol. 87, 565–575. doi: 10.1007/s11103-015-0295-0
Melikant, B., Giuliani, C., Halbmayer-Watzina, S., Limmongkon, A., Heberle-Bors, E., and Wilson, C. (2004). The Arabidopsis thaliana MEK AtMKK6 activates the MAP kinase AtMPK13. FEBS Lett. 576, 5–8. doi: 10.1016/j.febslet.2004.08.051
Meng, X., and Zhang, S. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Meng, Y., Ma, N., Zhang, Q., You, Q., Li, N., Ali, K. M., et al. (2014). Precise spatio-temporal modulation of ACC synthase by MPK6 cascade mediates the response of rose flowers to rehydration. Plant J. 79, 941–950. doi: 10.1111/tpj.12594
Menges, M., Doczi, R., Okresz, L., Morandini, P., Mizzi, L., Soloviev, M., et al. (2008). Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 179, 643–662. doi: 10.1111/j.1469-8137.2008.02552.x
Mergemann, H., and Sauter, M. (2000). Ethylene induces epidermal cell death at the site of adventitious root emergence in rice. Plant Physiol. 124, 609–614. doi: 10.1111/j.1469-8137.2008.02552.x
Meszaros, T., Helfer, A., Hatzimasoura, E., Magyar, Z., Serazetdinova, L., Rios, G., et al. (2006). The Arabidopsis MAP kinase kinase MKK1 participates in defence responses to the bacterial elicitor flagellin. Plant J. 48, 485–498. doi: 10.1111/j.1365-313X.2006.02888.x
Mishra, N. S., Tuteja, R., and Tuteja, N. (2006). Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68. doi: 10.1016/j.abb.2006.05.001
Mitula, F., Tajdel, M., Ciesla, A., Kasprowicz-Maluski, A., Kulik, A., Babula-Skowronska, D., et al. (2015). Arabidopsis ABA-activated kinase MAPKKK18 is regulated by protein phosphatase 2C ABI1 and the ubiquitin-proteasome pathway. Plant Cell Physiol. 56, 2351–2367. doi: 10.1093/pcp/pcv146
Miura, K., and Tada, Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5:4. doi: 10.3389/fpls.2014.00004
Mizoguchi, T., Gotoh, Y., Nishida, E., Yamaguchi-Shinozaki, K., Hayashida, N., Iwasaki, T., et al. (1994). Characterization of two cDNAs that encode MAP kinase homologues in Arabidopsis thaliana and analysis of the possible role of auxin in activating such kinase activities in cultured cells. Plant J. 5, 111–122. doi: 10.1046/j.1365-313X.1994.5010111.x
Mockaitis, K., and Howell, S. H. (2000). Auxin induces mitogenic activated protein kinase (MAPK) activation in roots of Arabidopsis seedlings. Plant J. 24, 785–796. doi: 10.1111/j.1365-313X.2000.00921.x
Mohanta, T. K., Arora, P. K., Mohanta, N., Parida, P., and Bae, H. (2015). Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genomics 16:58. doi: 10.1186/s12864-015-1244-7
Monroe-Augustus, M., Zolman, B. K., and Bartel, B. (2003). IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15, 2979–2991. doi: 10.1105/tpc.017046
Montillet, J. L., Leonhardt, N., Mondy, S., Tranchimand, S., Rumeau, D., Boudsocq, M., et al. (2013). An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11:e1001513. doi: 10.1371/journal.pbio.1001513
Morris, D. A., Friml, J., and Zazimalova, E. (2004). “The transport of auxins,” in Plant 36 Hormones, Biosynthesis, Signal Transduction, Action!, Vol. 37, ed. P. J. Davies (Dordrecht: Kluwer Academic Publishers), 437–469.
Morris, S. E., Cox, M. C., Ross, J. J., Krisantini, S., and Beveridge, C. A. (2005). Auxin dynamics after decapitation are not correlated with the initial growth of axillary buds. Plant Physiol. 138, 1665–1672. doi: 10.1104/pp.104.058743
Mou, Z., Wang, X., Fu, Z., Dai, Y., Han, C., Ouyang, J., et al. (2002). Silencing of phosphoethanolamine N-methyltransferase results in temperature-sensitive male sterility and salt hypersensitivity in Arabidopsis. Plant Cell 14, 2031–2043. doi: 10.1105/tpc.001701
Muday, G. K., and DeLong, A. (2001). Polar auxin transport, controlling where and how much. Trends Plant Sci. 6, 535–542. doi: 10.1016/S1360-1385(01)02101-X
Nakashima, K., Ito, Y., and Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nambara, E., and Marion-Poll, A. (2003). ABA action and interactions in seeds. Trends Plant Sci. 8, 213–217. doi: 10.1016/S1360-1385(03)00060-8
Nautiyal, C. S., Srivastava, S., Chauhan, P. S., Seem, K., Mishra, A., and Sopory, S. K. (2013). Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Biochem. 66, 1–9. doi: 10.1016/j.plaphy.2013.01.020
Née, G., Kramer, K., Nakabayashi, K., Yuan, B., Xiang, Y., Miatton, E., et al. (2017). DELAY OF GERMINATION1 requires PP2C phosphatases of the ABA signalling pathway to control seed dormancy. Nat. Commun. 8:72. doi: 10.1038/s41467-017-00113-6
Nie, W. F., Wang, M. M., Xia, X. J., Zhou, Y. H., Shi, K., Chen, Z., et al. (2013). Silencing of tomato RBOH1 and MPK2 abolishes brassinosteroid-induced H2O2 generation and stress tolerance. Plant Cell Environ. 36, 789–803. doi: 10.1111/pce.12014
Ning, N., Li, X., Hicks, L. M., and Xiong, L. (2010). A raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. doi: 10.1104/pp.109.149856
Novikova, G. V., Moshkov, I. E., Smith, A. R., and Hall, M. A. (2000). The effect of ethylene on MAPKinase-like activity in Arabidopsis thaliana. FEBS Lett. 474, 29–32. doi: 10.1016/S0014-5793(00)01565-9
Oka, K., Amano, Y., Katou, S., Seo, S., Kawazu, K., Mochizuki, A., et al. (2013). Tobacco MAP kinase phosphatase (NtMKP1) negatively regulates wound response and induced resistance against necrotrophic pathogens and lepidopteran herbivores. Mol. Plant Microbe Interact. 26, 668–675. doi: 10.1094/MPMI-11-12-0272-R
Opdenakker, K., Remans, T., Vangronsveld, J., and Cuypers, A. (2012). Mitogen-Activated Protein (MAP) kinases in plant metal stress, regulation and responses in comparison to other biotic and abiotic stresses. Int. J. Mol. Sci. 13, 7828–7853. doi: 10.3390/ijms13067828
Ortiz-Masia, D., Perez-Amador, M. A., Carbonell, J., and Marcote, M. J. (2007). Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 581, 1834–1840. doi: 10.1016/j.febslet.2007.03.075
Ortiz-Masia, D., Perez-Amador, M. A., Carbonell, P., Aniento, F., Carbonell, J., and Marcote, M. J. (2008). Characterization of PsMPK2, the first C1 subgroup MAP kinase from pea (Pisum sativum L.). Planta 227, 1333–1342. doi: 10.1007/s00425-008-0705-5
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2013). Sensing the environment, key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64, 445–458. doi: 10.1093/jxb/ers354
Ouaked, F., Rozhon, W., Lecourieux, D., and Hirt, H. (2003). A MAPK pathway mediates ethylene signaling in plants. EMBO J. 22, 1282–1288. doi: 10.1093/emboj/cdg131
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Pan, J., Zhang, M., Kong, X., Xing, X., Liu, Y., Zhou, Y., et al. (2012). ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 235, 661–676. doi: 10.1007/s00425-011-1510-0
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Pecher, P., Eschen-Lippold, L., Herklotz, S., Kuhle, K., Naumann, K., Bethke, G., et al. (2014). The Arabidopsis thaliana mitogen-activated protein kinases MPK3 and MPK6 target a subclass of ‘VQ-motif’-contai proteins to regulate immune responses. New Phytol. 203, 592–606. doi: 10.1111/nph.12817
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klûsener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. doi: 10.1016/S0092-8674(00)00213-0
Petrasek, J., and Friml, J. (2009). Auxin transport routes in plant development. Development 136, 2675–2688. doi: 10.1242/dev.030353
Pitzschke, A., Djamei, A., Bitton, F., and Hirt, H. (2009). A major role of the MEKK1-MKK1/2-MPK4 pathway in ROS signalling. Mol. Plant 2, 120–137. doi: 10.1093/mp/ssn079
Poyraz, I. (2015). Molecular cloning and characterization of a mitogen-activated protein kinase kinase (OoMAPKK1) in Origanum onites L. (Lamiaceae). J. Plant Biochem. Biotechnol. 24, 75–83. doi: 10.1021/jf001494m
Qi, J., Wang, J., Gong, Z., and Zhou, J. M. (2017). Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 38, 92–100. doi: 10.1016/j.pbi.2017.04.022
Qiao, H., Shen, Z., Huang, S. S., Schmitz, R. J., Urich, M. A., Briggs, S. P., et al. (2012). Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science 338, 390–393. doi: 10.1126/science.1225974
Qu, Y., Song, P., Hu, Y., Jin, X., Jia, Q., Zhang, X., et al. (2018). Regulation of stomatal movement by cortical microtubule organization in response to darkness and ABA signaling in Arabidopsis. Plant Growth Regul. 84, 467–479. doi: 10.1007/s10725-017-0353-5
Raja, V., Majeed, U., Kang, H., Andrabi, K. I., and John, R. (2017). Abiotic stress: interplay between ROS, hormones and MAPKs. Environ. Exp. Bot. 137, 142–157. doi: 10.1016/j.envexpbot.2017.02.010
Reed, R. C., Brady, S. R., and Muday, G. K. (1998). Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol. 118, 1369–1378. doi: 10.1104/pp.118.4.1369
Reinhardt, D., Pesce, E. R., Stieger, P., Mandel, T., Baltensperger, K., Bennett, M., et al. (2003). Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. doi: 10.1038/nature02081
Reyna, N. S., and Yang, Y. (2006). Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 19, 530–540. doi: 10.1094/MPMI-19-0530
Rodrigues, O., Reshetnyak, G., Grondin, A., Saijo, Y., Leonhardt, N., Maurel, C., et al. (2017). Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. U.S.A. 114, 9200–9205. doi: 10.1073/pnas.1704754114
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252
Rogg, L. E., Lasswell, J., and Bartel, B. (2001). A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13, 465–480. doi: 10.1105/tpc.13.3.465
Salam, M. A., Jammes, F., Hossain, M. A., Ye, W., Nakamura, Y., Mori, I. C., et al. (2012). MAP kinases, MPK9 and MPK12, regulate chitosan-induced stomatal closure. Biosci. Biotechnol. Biochem. 76, 1785–1787. doi: 10.1271/bbb.120228
Samajova, O., Plihal, O., Al-Yousif, M., Hirt, H., and Samaj, J. (2013). Improvement of stress tolerance in plants by genetic manipulation of mitogen-activated protein kinases. Biotechnol. Adv. 31, 118–128. doi: 10.1016/j.biotechadv.2011.12.002
Seguí-Simarro, J. M., Testillano, P. S., Jouannic, S., Henry, Y., and Risueño, M. C. (2005). Mitogen-activated protein kinases are developmentally regulated during stress-induced microspore embryogenesis in Brassica napus L. Histochem. Cell Biol. 123, 541–551. doi: 10.1007/s00418-004-0749-y
Seo, S., Katou, S., Seto, H., Gomi, K., and Ohashi, Y. (2007). The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J. 49, 899–909. doi: 10.1111/j.1365-313X.2006.03003.x
Serna, L. (2013). What causes opposing actions of brassinosteroids on stomatal development? Plant Physiol. 162, 3–8. doi: 10.1104/pp.112.213058
Shang, Y., Dai, C., Lee, M. M., Kwak, J. M., and Nam, K. H. (2016). BRI1-associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Mol. Plant 9, 447–460. doi: 10.1016/j.molp.2015.12.014
Sharma, R., De, V. D., Sharma, M. K., and Ronald, P. C. (2013). Recent advances in dissecting stress-regulatory crosstalk in rice. Mol. Plant 6, 250–260. doi: 10.1093/mp/sss14
Sheikh, A. H., Raghuram, B., Jalmi, S. K., Wankhede, D. P., Singh, P., and Sinha, A. K. (2013). Interaction between two rice mitogen activated protein kinases and its possible role in plant defense. BMC Plant Biol. 13:121. doi: 10.1186/1471-2229-13-121
Shen, X., Liu, H., Yuan, B., Li, X., Xu, C., and Wang, S. (2011). OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ. 34, 179–191. doi: 10.1111/j.1365-3040.2010.02219.x
Shi, B., Ni, L., Liu, Y., Zhang, A., Tan, M., and Jiang, M. (2014). OsDMI3-mediated activation of OsMPK1 regulates the activities of antioxidant enzymes in abscisic acid signalling in rice. Plant Cell Environ. 37, 341–352. doi: 10.1111/pce.12154
Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417. doi: 10.1016/S1369-5266(03)00092-X
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Simonini, S., Deb, J., Moubayidin, L., Stephenson, P., Valluru, M., Freire-Rios, A., et al. (2016). A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 30, 2286–2296. doi: 10.1101/gad.285361.116
Singh, R., and Jwa, N. S. (2013). The rice MAPKK-MAPK interactome, the biological significance of MAPK components in hormone signal transduction. Plant Cell Rep. 32, 923–931. doi: 10.1007/s00299-013-1437-y
Singh, R., Lee, M. O., Lee, J. E., Choi, J., Park, J. H., Kim, E. H., et al. (2012). Rice mitogen-activated protein kinase interactome analysis using the yeast two-hybrid system. Plant Physiol. 160, 477–487. doi: 10.1104/pp.112.200071
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
Skottke, K. R., Yoon, G. M., Kieber, J. J., and Delong, A. (2011). Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 7:e1001370. doi: 10.1371/journal.pgen.1001370
Slavov, S., Mayama, S., and Atanassov, A. (2004). Some aspects of epidemiology of Alternaria alternata tobacco pathotype. Biotechnol. Biotechnol. Equip. 18, 28–33. doi: 10.1080/13102818.2004.10817083
Smekalova, V., Doskocilova, A., Komis, G., and Samaj, J. (2013). Crosstalk between secondary messengers, hormones and MAPK modules during abiotic stress signalling in plants. Biotechnol. Adv. 32, 2–11. doi: 10.1016/j.biotechadv.2013.07.009
Smekalova, V., Luptovciak, I., Komis, G., Samajova, O., Ovecka, M., Doskocilova, A., et al. (2014). Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203, 1175–1193. doi: 10.1111/nph.12880
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling, a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Song, D., Chen, J., Song, F., and Zheng, Z. (2006). A novel rice MAPK gene, OsBIMK2, is involved in disease-resistance responses. Plant Biol. 8, 587–596. doi: 10.1055/s-2006-924149
Song, F., and Goodman, R. M. (2002). OsBIMK1, a rice MAP kinase gene involved in disease resistance responses. Planta 215, 997–1005. doi: 10.1007/s00425-002-0794-5
Song, L. X., Xu, X. C., Wang, F. N., Wang, Y., Xia, X. J., Shi, K., et al. (2018). Brassinosteroids act as a positive regulator for resistance against root-knot nematode involving RESPIRATORY BURST OXIDASE HOMOLOG-dependent activation of MAPKs in tomato. Plant Cell Environ. 41, 1113–1125. doi: 10.1111/pce.12952
Soyano, T., Nishihama, R., Morikiyo, K., Ishikawa, M., and Machida, Y. (2003). NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev. 17, 1055–1067. doi: 10.1101/gad.1071103
Stanko, V., Giuliani, C., Retzer, K., Djamei, A., Wahl, V., Wurzinger, B., et al. (2014). Timing is everything, highly specific and transient expression of a MAP kinase determines auxin-induced leaf venation patterns in Arabidopsis. Mol. Plant 7, 1637–1652. doi: 10.1093/mp/ssu080
Stepanova, A. N., and Alonso, J. M. (2009). Ethylene signaling and response, where different regulatory modules meet. Curr. Opin. Plant Biol. 12, 548–555. doi: 10.1016/j.pbi.2009.07.009
Sun, J., Zhou, R., Li, Y., Ding, W. H., Xiatian, Q., Qiong, W., et al. (2016). A Brachypodium distachyon MAPKK gene BdMKK6.2 negatively regulates drought stress tolerance in transgenic tobacco plants. J. Plant Growth Regul. 35, 121–134. doi: 10.1007/s00344-015-9512-y
Sun, W., Chen, H., Wang, J., Sun, H. W., Yang, S. K., Sang, Y. L., et al. (2015). Expression analysis of genes encoding mitogen-activated protein kinases in maize provides a key link between abiotic stress signaling and plant reproduction. Funct. Integr. Genomics 15, 107–120. doi: 10.1007/s10142-014-0410-3
Sun, Y., Wang, C., Yang, B., Wu, F., Hao, X., Liang, W., et al. (2014). Identification and functional analysis of mitogen-activated protein kinase kinase kinase (MAPKKK) genes in canola (Brassica napus L.). J. Exp. Bot. 65, 2171–2188. doi: 10.1093/jxb/eru092
Tajdel, M., Mitula, F., and Ludwikow, A. (2016). Regulation of Arabidopsis MAPKKK18 by ABI1 and SnRK2, components of the ABA signaling pathway. Plant Signal. Behav. 11:e1139277. doi: 10.1080/15592324.2016.1139277
Takahashi, F., Yoshida, R., Ichimura, K., Mizoguchi, T., Seo, S., Yonezawa, M., et al. (2007). The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19, 805–818. doi: 10.1105/tpc.106.046581
Takahashi, Y., Soyano, T., Kosetsu, K., Sasabe, M., and Machida, Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51, 1766–1776. doi: 10.1093/pcp/pcq135
Tan, W., Zhang, D., Zhou, H., Zheng, T., Yin, Y., and Lin, H. (2018). Transcription factor HAT1 is a substrate of SnRK2.3 kinase and negatively regulates ABA synthesis and signaling in Arabidopsis responding to drought. PLoS Genet. 14:e1007336. doi: 10.1371/journal.pgen.1007336
Tanaka, H., Dhonukshe, P., Brewer, P. B., and Friml, J. (2006). Spatiotemporal asymmetric auxin distribution, a means to coordinate plant development. Cell. Mol. Life Sci. 63, 2738–2754. doi: 10.1007/s00018-006-6116-5
Tang, W., Yuan, M., Wang, R., Yang, Y., Wang, C., Oses-Prieto, J. A., et al. (2011). PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 13, 124–131. doi: 10.1038/ncb2151
Tao, J. J., Chen, H. W., Ma, B., Zhang, W. K., Chen, S. Y., and Zhang, J. S. (2015). The role of ethylene in plants under salinity stress. Front. Plant Sci. 6:1059. doi: 10.3389/fpls.2015.01059
Teige, M., Scheikl, E., Eulgem, T., Doczi, R., Ichimura, K., Shinozaki, K., et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell. 15, 141–152. doi: 10.1016/j.molcel.2004.06.023
Tena, G., and Renaudin, J. P. (1998). Cytosolic acidification but not auxin at physiological concentration is an activator of MAP kinases in tobacco cells. Plant J. 16, 173–182. doi: 10.1046/j.1365-313x.1998.00283.x
Tong, H., Liu, L., Jin, Y., Du, L., Yin, Y., Qian, Q., et al. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24, 2562–2577. doi: 10.1105/tpc.112.097394
Trupkin, S. A., Auge, G. A., Zhu, J.-K., Sánchez, R. A., and Botto, J. F. (2017). SALT OVERLY SENSITIVE 2 (SOS2) and interacting partners SOS3 and ABSCISIC ACID-INSENSITIVE 2 (ABI2) promote red-light-dependent germination and seedling deetiolation in Arabidopsis. Int. J. Plant Sci. 178, 485–493. doi: 10.1086/692097
Tsugama, D., Liu, S., and Takano, T. (2012). Drought-induced activation and rehydration-induced inactivation of MPK6 in Arabidopsis. Biochem. Biophys. Res. Commun. 426, 626–629. doi: 10.1016/j.bbrc.2012.08.141
Ulm, R., Ichimura, K., Mizoguchi, T., Peck, S. C., Zhu, T., Wang, X., et al. (2002). Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. doi: 10.1093/emboj/cdf646
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8. doi: 10.1126/scisignal.2003509
Vaid, N., Macovei, A., and Tuteja, N. (2013). Knights in action, lectin receptor-like kinases in plant development and stress responses. Mol. Plant 6, 1405–1418. doi: 10.1093/mp/sst033
van Loon, L. C., Geraats, B. P., and Linthorst, H. J. (2006). Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 11, 184–191. doi: 10.1016/j.tplants.2006.02.005
Virk, N., Li, D., Tian, L., Huang, L., Hong, Y., Li, X., et al. (2015). Arabidopsis raf-like mitogen-activated protein kinase kinase kinase gene Raf43 is required for tolerance to multiple abiotic stresses. PLoS One 10:e0133975. doi: 10.1371/journal.pone.0133975
Wang, C., Lu, W., He, X., Wang, F., Zhou, Y., Guo, X., et al. (2016). The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol. 57, 1629–1642. doi: 10.1093/pcp/pcw090
Wang, H., Ngwenyama, N., Liu, Y., Walker, J. C., and Zhang, S. (2007). Stomatal development and patterning are regulated by environmentally responsive mitogen-activated protein kinases in Arabidopsis. Plant Cell 19, 63–73. doi: 10.1105/tpc.106.048298
Wang, J., Ding, H., Zhang, A., Ma, F., Cao, J., and Jiang, M. (2010). A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 52, 442–452. doi: 10.1111/j.1744-7909.2010.00906.x
Wang, X. J., Zhu, S. Y., Lu, Y. F., Zhao, R., Xin, Q., Wang, X. F., et al. (2010). Two coupled components of the mitogen-activated protein kinase cascade MdMPK1 and MdMKK1 from apple function in ABA signal transduction. Plant Cell Physiol. 51, 754–766. doi: 10.1093/pcp/pcq037
Wang, J., Pan, C., Wang, Y., Ye, L., Wu, J., Chen, L., et al. (2015). Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genomics 16:386. doi: 10.1186/s12864-015-1621-2
Wang, Z., Jia, C., Li, J., Xu, B., and Jin, Z. (2015). Identification of six mitogen-activated protein kinase (MAPK) genes in banana (Musa acuminata L. AAA group, cv. Cavendish) under infection of Fusarium oxysporum f. sp cubense tropical race. Acta Physiol. Plant. 37:115. doi: 10.1007/s11738-015-1868-x
Wang, K. L., Li, H., and Ecker, J. R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14(Suppl.), S131–S151. doi: 10.1105/tpc.001768
Wang, L., Liu, Y., Cai, G., Jiang, S., Pan, J., and Li, D. (2014). Ectopic expression of ZmSIMK1 leads to improved drought tolerance and activation of systematic acquired resistance in transgenic tobacco. J. Biotechnol. 172, 18–29. doi: 10.1016/j.jbiotec.2013.11.006
Wang, P., and Song, C. P. (2008). Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 178, 703–718. doi: 10.1111/j.1469-8137.2008.02431.x
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell. 69, 100–112. doi: 10.1016/j.molcel.2017.12.002
Wang, R. S., Pandey, S., Li, S., Gookin, T. E., Zhao, Z., Albert, R., et al. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12:216. doi: 10.1186/1471-2164-12-216
Wankhede, D. P., Misra, M., Singh, P., and Sinha, A. K. (2013). Rice mitogen activated protein kinase kinase and mitogen activated protein kinase interaction network revealed by in-silico docking and yeast two-hybrid approaches. PLoS One 8:e65011. doi: 10.1371/journal.pone.0065011
Wasternack, C., and Hause, B. (2013). Jasmonates, biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct06
Wen, J. Q., Oono, K., and Imai, R. (2002). Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 129, 1880–1891. doi: 10.1104/pp.006072
Wen, X., Zhang, C., Ji, Y., Zhao, Q., He, W., An, F., et al. (2012). Activation of ethylene signaling is mediated by nuclear translocation of the cleaved EIN2 carboxyl terminus. Cell Res. 22, 1613–1616. doi: 10.1038/cr.2012.145
Wendehenne, D., Gao, Q. M., Kachroo, A., and Kachroo, P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20, 127–134. doi: 10.1016/j.pbi.2014.05.012
Wójcikowska, B., and Gaj, M. D. (2017). Expression profiling of AUXIN RESPONSE FACTOR genes during somatic embryogenesis induction in Arabidopsis. Plant Cell Rep. 36, 843–858. doi: 10.1007/s00299-017-2114-3
Wu, J., Wang, J., Pan, C., Guan, X., Wang, Y., Liu, S., et al. (2014). Genome-wide identification of MAPKK and MAPKKK gene families in tomato and transcriptional profiling analysis during development and stress response. PLoS One 9:e103032. doi: 10.1371/journal.pone.0103032
Xie, G., Kato, H., and Imai, R. (2012). Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem. J. 443, 95–102. doi: 10.1042/BJ20111792
Xie, K., Chen, J., Wang, Q., and Yang, Y. (2014). Direct phosphorylation and activation of a mitogen-activated protein kinase by a calcium-dependent protein kinase in rice. Plant Cell 26, 3077–3089. doi: 10.1105/tpc.114.126441
Xie, Q., Frugis, G., Colgan, D., and Chua, N. H. (2000). Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 14, 3024–3036. doi: 10.1101/gad.852200
Xing, Y., Jia, W., and Zhang, J. (2007). AtMEK1 mediates stress-induced gene expression of CAT1 catalase by triggering H2O2 production in Arabidopsis. J. Exp. Bot. 58, 2969–2981. doi: 10.1093/jxb/erm144
Xing, Y., Jia, W., and Zhang, J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x
Xing, Y., Jia, W., and Zhang, J. (2009). AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol. Biol. 70, 725–736. doi: 10.1007/s11103-009-9503-0
Xiong, L., Lee, M. W., Qi, M., and Yang, Y. (2001). Identification of defense-related rice genes by suppression subtractive hybridization and differential screening. Mol. Plant Microbe Interact. 14, 685–692. doi: 10.1094/MPMI.2001.14.5.685
Xiong, L., and Yang, Y. (2003). Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15, 745–759. doi: 10.1105/tpc.008714
Xu, C., Liu, R., Zhang, Q., Chen, X., Qian, Y., and Fang, W. (2017). The diversification of evolutionarily conserved MAPK cascades correlates with the evolution of fungal species and development of lifestyles. Genome Biol. Evol. 9, 311–322. doi: 10.1093/gbe/evw051
Xu, H., Wang, X., Sun, X., Shi, Q., Yang, F., and Donliang, D. (2008). Molecular cloning and characterization of a cucumber MAP kinase gene in response to excess NO. Sci. Hortic. 117, 1–8. doi: 10.1016/j.scienta.2008.01.012
Xu, J., and Chua, N. H. (2012). Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J. 31, 1975–1984. doi: 10.1038/emboj.2012.56
Xu, J., Li, Y., Wang, Y., Liu, H., Lei, L., Yang, H., et al. (2008). Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 283, 26996–27006. doi: 10.1074/jbc.M801392200
Xu, J., Meng, J., Meng, X., Zhao, Y., Liu, J., Sun, T., et al. (2016). Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28, 1144–1162. doi: 10.1105/tpc.15.00871
Xu, J., Xie, J., Yan, C., Zou, X., Ren, D., and Zhang, S. (2014). A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234. doi: 10.1111/tpj.12382
Xu, J., and Zhang, S. (2015). Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 20, 56–64. doi: 10.1016/j.tplants.2014.10.001
Yamagami, T., Tsuchisaka, A., Yamada, K., Haddon, W. F., Harden, L. A., and Theologis, A. (2003). Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J. Biol. Chem. 278, 49102–49112. doi: 10.1074/jbc.M308297200
Yan, Y., Wang, L., Ding, Z., Tie, W., Ding, X., Zeng, C., et al. (2016). Genome-wide identification and expression analysis of the mitogen-activated protein kinase gene family in cassava. Front. Plant Sci. 7:1294. doi: 10.3389/fpls.2016.01294
Yanagisawa, S., Yoo, S. D., and Sheen, J. (2003). Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425, 521–525. doi: 10.1038/nature01984
Yang, K. Y., Liu, Y., and Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. U.S.A. 98, 741–746. doi: 10.1073/pnas.98.2.741
Yang, K. Z., Jiang, M., Wang, M., Xue, S., Zhu, L. L., Wang, H. Z., et al. (2015). Phosphorylation of serine 186 of bHLH transcription factor SPEECHLESS promotes stomatal development in Arabidopsis. Mol. Plant 8, 783–795. doi: 10.1016/j.molp.2014.12.014
Yang, S. F., and Hoffman, N. E. (1984). Ethylene biosynthesis and its regulation in higher plants. Annu. Rev. Plant Physiol. 35, 155–189. doi: 10.1146/annurev.pp.35.060184.001103
Yang, T., Shad Ali, G., Yang, L., Du, L., Reddy, A. S., and Poovaiah, B. W. (2010). Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 5, 991–994. doi: 10.1074/jbc.M109.035659
Ye, L., Li, L., Wang, L., Wang, S., Li, S., Du, J., et al. (2015). MPK3/MPK6 are involved in iron deficiency-induced ethylene production in Arabidopsis. Front. Plant Sci. 6:953. doi: 10.3389/fpls.2015.00953
Yin, H., Zhao, X., Bai, X., and Du, Y. (2010). Molecular cloning and characterization of a Brassica napus L. MAP kinase involved in oligochitosan-induced defense signaling. Plant Mol. Biol. Rep. 28, 292–301. doi: 10.1007/s11105-009-0152-x
Yin, Y. L., Zhou, Y., Zhou, Y. H., Shi, K., Zhou, J., Yu, Y., et al. (2016). Interplay between mitogen-activated protein kinase and nitric oxide in brassinosteroid-induced pesticide metabolism in Solanum lycopersicum. J. Hazard. Mater. 316, 221–231. doi: 10.1016/j.jhazmat.2016.04.070
Yin, Z., Wang, J., Wang, D., Fan, W., Wang, S., and Ye, W. (2013). The MAPKKK gene family in Gossypium raimondii, genome-wide identification, classification and expression analysis. Int. J. Mol. Sci. 14, 18740–18757. doi: 10.3390/ijms140918740
Yoo, S. D., Cho, Y., and Sheen, J. (2009). Emerging connections in the ethylene signaling network. Trends Plant Sci. 14, 270–279. doi: 10.1016/j.tplants.2009.02.007
Yoo, S. D., Cho, Y. H., Tena, G., Xiong, Y., and Sheen, J. (2008). Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451, 789–795. doi: 10.1038/nature06543
Yoo, S. J., Kim, S. H., Kim, M. J., Ryu, C. M., Kim, Y. C., Cho, B. H., et al. (2014). Involvement of the OsMKK4-OsMPK1 cascade and its downstream transcription factor OsWRKY53 in the wounding response in rice. Plant Pathol. J. 30, 168–177. doi: 10.5423/PPJ.OA.10.2013.0106
You, M. K., Oh, S. I., Ok, S. H., Cho, S. K., Shin, H. Y., Jeung, J. U., et al. (2007). Identification of putative MAPK kinases in Oryza minuta and O. sativa responsive to biotic stresses. Mol. Cells 23, 108–114.
Yu, S., Zhang, L., Chen, C., Li, J., Ye, S., Liu, G., et al. (2014). Isolation and characterization of BnMKK1 responsive to multiple stresses and affecting plant architecture in tobacco. Acta Physiol. Plant 36, 1313–1324. doi: 10.1007/s11738-014-1510-3
Zanke, B. W., Boudreau, K., Rubie, E., Winnett, E., Tibbles, L. A., Zon, L., et al. (1996). The stress-activated protein kinase pathway mediates cell death following injury induced by cis-platinum, UV irradiation or heat. Curr. Biol. 6, 606–613. doi: 10.1016/S0960-9822(02)00547-X
Zarembinski, T. I., and Theologis, A. (1994). Ethylene biosynthesis and action, a case of conservation. Plant Mol. Biol. 26, 1579–1597. doi: 10.1007/BF00016491
Zeng, Q., Sritubtim, S., and Ellis, B. E. (2011). AtMKK6 and AtMPK13 are required for lateral root formation in Arabidopsis. Plant Signal. Behav. 6, 1436–1439. doi: 10.4161/psb.6.10.17089
Zhang, A., Jiang, M., Zhang, J., Ding, H., Xu, S., Hu, X., et al. (2007). Nitric oxide induced by hydrogen peroxide mediates abscisic acid-induced activation of the mitogen-activated protein kinase cascade involved in antioxidant defense in maize leaves. New Phytol. 175, 36–50. doi: 10.1111/j.1469-8137.2007.02071.x
Zhang, A., Jiang, M., Zhang, J., Tan, M., and Hu, X. (2006). Mitogen-activated protein kinase is involved in abscisic acid-induced antioxidant defense and acts downstream of reactive oxygen species production in leaves of maize plants. Plant Physiol. 141, 475–487. doi: 10.1104/pp.105.075416
Zhang, A., Zhang, J., Ye, N., Cao, J., Tan, M., Zhang, J., et al. (2010). ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassinosteroid-induced antioxidant defence in leaves of maize. J. Exp. Bot. 61, 4399–4411. doi: 10.1093/jxb/erq243
Zhang, D., Ye, H., Guo, H., Johnson, A., Lin, H., and Yin, Y. (2014a). Transcription factors involved in brassinosteroid repressed gene expression and their regulation by BIN2 kinase. Plant Signal. Behav. 9:e27849. doi: 10.4161/psb.27849
Zhang, H., Liu, Y., Wen, F., Yao, D., Wang, L., Guo, J., et al. (2014b). A novel rice C2H2-type zinc finger protein, ZFP36, is a key player involved in abscisic acid-induced antioxidant defence and oxidative stress tolerance in rice. J. Exp. Bot. 65, 5795–5809. doi: 10.1093/jxb/eru313
Zhang, H., Ni, L., Liu, Y., Wang, Y., Zhang, A., Tan, M., et al. (2012). The C2H2-type zinc finger protein ZFP182 is involved in abscisic acid-induced antioxidant defense in rice. J. Integr. Plant Biol. 54, 500–510. doi: 10.1111/j.1744-7909.2012.01135.x
Zhang, H., Zhu, H., Pan, Y., Yu, Y., Luan, S., and Li, L. (2014c). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532. doi: 10.1093/mp/ssu063
Zhang, J., Zou, D., Li, Y., Sun, X., Wang, N. N., Gong, S. Y., et al. (2014d). GhMPK17, a cotton mitogen-activated protein kinase, is involved in plant response to high salinity and osmotic stresses and ABA signaling. PLoS One 9:e95642. doi: 10.1371/journal.pone.0095642
Zhang, M., Pan, J., Kong, X., Zhou, Y., Liu, Y., Sun, L., et al. (2012). ZmMKK3, a novel maize group B mitogen-activated protein kinase kinase gene, mediates osmotic stress and ABA signal responses. J. Plant Physiol. 169, 1501–1510. doi: 10.1016/j.jplph.2012.06.008
Zhang, W., Jeon, B. W., and Assmann, S. M. (2011). Heterotrimeric G-protein regulation of ROS signaling and calcium currents in Arabidopsis guard cells. J. Exp. Bot. 62, 2371–2379. doi: 10.1093/jxb/erq424
Zhang, X., Cheng, T., Wang, G., Yan, Y., and Xia, Q. (2013). Cloning and evolutionary analysis of tobacco MAPK gene family. Mol. Biol. Rep. 40, 1407–1415. doi: 10.1007/s11033-012-2184-9
Zhang, X., Wang, L., Xu, X., Cai, C., and Guo, W. (2014e). Genome-wide identification of mitogen-activated protein kinase gene family in Gossypium raimondii and the function of their corresponding orthologs in tetraploid cultivated cotton. BMC Plant Biol. 14:345. doi: 10.1186/s12870-014-0345-9
Zhang, X., Xiong, Y., Defraia, C., Schmelz, E., and Mou, Z. (2008). The Arabidopsis MAP kinase kinase 7: a crosstalk point between auxin signaling and defense responses? Plant Signal. Behav. 3, 272–274. doi: 10.1111/j.1365-313X.2007.03294.x
Zhang, X., Xu, X., Yu, Y., Chen, C., Wang, J., Cai, C., et al. (2016). Integration analysis of MKK and MAPK family members highlights potential MAPK signaling modules in cotton. Sci. Rep. 6:29781. doi: 10.1038/srep29781
Zhao, C., Nie, H., Shen, Q., Zhang, S., Lukowitz, W., and Tang, D. (2014a). EDR1 physically interacts with MKK4/MKK5 and negatively regulates a MAP kinase cascade to modulate plant innate immunity. PLoS Genet. 10:e1004389. doi: 10.1371/journal.pgen.1004389
Zhao, F. Y., Han, M. M., Zhang, S. Y., Ren, J., Hu, F., and Wang, X. (2014b). MAPKs as a cross point in H2O2 and auxin signaling under combined cadmium and zinc stress in rice roots. Russ. J. Plant Physiol. 61, 608–618. doi: 10.1134/S1021443714040232
Zhao, F. Y., Hu, F., Zhang, S. Y., Wang, K., Zhang, C. R., and Liu, T. (2013). MAPKs regulate root growth by influencing auxin signaling and cell cycle-related gene expression in cadmium-stressed rice. Environ. Sci. Pollut. Res. Int. 20, 5449–5460. doi: 10.1007/s11356-013-1559-3
Zhao, Y. (2018). Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 69, 417–435. doi: 10.1146/annurev-arplant-042817-040226
Keywords: MAP kinase cascade, auxin, abscisic acid, jasmonic acid, salicilic acid, ethylene, brassinosteroids, gibberellin
Citation: Jagodzik P, Tajdel-Zielinska M, Ciesla A, Marczak M and Ludwikow A (2018) Mitogen-Activated Protein Kinase Cascades in Plant Hormone Signaling. Front. Plant Sci. 9:1387. doi: 10.3389/fpls.2018.01387
Received: 26 June 2018; Accepted: 31 August 2018;
Published: 08 October 2018.
Edited by:
Nigel G. Halford, Rothamsted Research (BBSRC), United KingdomReviewed by:
Mingyi Jiang, Nanjing Agricultural University, ChinaCopyright © 2018 Jagodzik, Tajdel-Zielinska, Ciesla, Marczak and Ludwikow. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnieszka Ludwikow, bHVkd2lrYUBhbXUuZWR1LnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.