
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 September 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01308
This article is part of the Research Topic Setaria as a Model Genetic System to Accelerate Yield Increases in Cereals, Forage Crops, and Bioenergy Grasses View all 12 articles
A yellow-green leaf mutant was isolated from EMS-mutagenized lines of Setaria italica variety Yugu1. Map-based cloning revealed the mutant gene is a homolog of Arabidopsis thaliana AtEGY1. EGY1 (ethylene-dependent gravitropism-deficient and yellow-green 1) is an ATP-independent metalloprotease (MP) that is required for chloroplast development, photosystem protein accumulation, hypocotyl gravitropism, leaf senescence, and ABA signal response in A. thaliana. However, the function of EGY1 in monocotyledonous C4 plants has not yet been described. The siygl2 mutant is phenotypically characterized by chlorotic organs, premature senescence, and damaged PS II function. Sequence comparisons of the AtEGY1 and SiYGL2 proteins reveals the potential for SiYGL2 to encode a partially functional protein. Phenotypic characterization and gene expression analysis suggested that SiYGL2 participates in the regulation of chlorophyll content, leaf senescence progression, and PS II function. Additionally, our research will contribute to further characterization of the mechanisms regulating leaf senescence and photosynthesis in S. italica, and in C4 plants in general.
AtEGY1 is an ATP-independent metalloprotease (MP) belonging to the M50 family (van der Hoorn, 2008). Members of the M50 family typically contain four to eight transmembrane helices (TMHs), three adjacent ones of which can form a conserved three-TMH core structure. The conversed motifs HEXXH and NPDG are located in the first TMH and the third TMH of the core structure, respectively (Kinch et al., 2006; Feng et al., 2007; Koussis et al., 2017). M50 family members contain three metal ligand-binding sites required for protease activity. Two of the ligand-binding sites reside in the HEXXH motif and the third is coordinated by the Asp residue in NPDG motif (Yu and Kroos, 2000; Feng et al., 2007). As MPs, M50 family proteins participate in multiple cellular processes by cleaving substrates such as membrane-tethered proteins, transcription factors, and signal peptides (Bölter et al., 2006; Kinch et al., 2006; Mukherjee et al., 2009; Che et al., 2010; Saito et al., 2011). AtEGY1 is targeted to chloroplasts and can be upregulated by both light and ethylene treatment in A. thaliana (Chen et al., 2005). The function of AtEGY1 has been characterized in a number of different studies. AtEGY1 has three conversed motifs: GNLR, HEXXH, and NPDG. The first is a signature motif unique to EGY family proteins (Chen et al., 2005); the function of this motif has not yet been clarified. The latter two motifs are metal-chelating motifs unique to M50 family members. AtEGY1 was shown to play a role in many biological processes including the regulation of chloroplast development, ethylene-dependent hypocotyl gravitropism, the accumulation of membrane-bound chlorophyll a/b binding (CAB) proteins, and responding to ammonium and phosphate stress (Chen et al., 2005; Guo et al., 2008; Li et al., 2012; Yu et al., 2016). In addition, AtEGY1 was reported to be involved in leaf senescence. The T-DNA insertion mutant of AtEGY1, Ategy1, had yellow rosette leaves and reduced chlorophyll content, leaf survival, and Fv/Fm. Mutants also showed decreased soluble protein content and increased ion leakage compared with WT. The expression of senescence-associated genes were increased in Ategy1. Under dark treatment, the aging phenotype in Ategy1 was more obvious than that in WT. These data indicated that the loss of AtEGY1 function accelerated leaf aging and decreased photosystem II (PS II) efficiency. In addition, exogenously applied glucose could rescue these mutant phenotypes (Chen et al., 2016).
Leaf senescence is an important plant developmental process that is associated with a range of unique physiological and biochemical characteristics. During leaf senescence, leaves turn yellow; chlorophyll is degraded; proteins, fatty acids, nucleic acids, and other macromolecules are metabolized; plastids disintegrate rapidly; and nitrogen and nutrients are efficiently transferred into growing tissues and sink organs (Yang and Ohlrogge, 2009; Sakamoto and Takami, 2014; Diaz-Mendoza et al., 2016). Concurrently, thousands of senescence-associated genes are upregulated or downregulated at the transcriptional or post-transcriptional level as senescence is a well-controlled process that transfers nutrients from source to sink (Gupta et al., 2016; Wu et al., 2016). Crop yield and quality are, therefore, closely related to the timing of senescence. Proteins in old organs are extensively degraded into amino acids, amides, and ammoniums. For cereal crops, leaf senescence provides most of the nitrogen in grains. It has been reported that delayed senescence could lead to high yields (Lu et al., 2017) because the filling period is prolonged and sugar and nitrogen accumulation is increased (Egli, 2011; Gregersen et al., 2013). Conversely, grain quality has negative correlations with senescence progression. Because delayed senescence could cause inefficient nitrogen remobilization. The grain quality parameters such as proteins and micronutrients are diluted by carbohydrates, thereby leading to a low grain quality (Gregersen et al., 2008). There is a balance between grain yield and quality. Thus, the better understanding of the senescence process could help improve crop production and seed quality (Diaz-Mendoza et al., 2016).
Metalloproteases (MPs) are one type of plant proteases, which plays important roles in leaf senescence. There are approximately 100 MPs in plants, belonging to 19 families which are classified by the similarity in amino acid sequence. Although they are involved in many biological processes, the roles of MPs in leaf senescence are poorly understood (van der Hoorn, 2008). The most studied senescence-associated MPs are FtsH (filamentation temperature sensitive H) proteases that belong to the M41 family. FtsH1, 2, 5, and 8 can form a hexameric ring in the chloroplast thylakoid to regulate the thylakoid structure and remove photodamaged D1 protein in PS II under light stress (Yoshioka-Nishimura et al., 2014). Changes in the expression of FtsH1, 2, and 5 observed in senescing and aging leaves was found to be associated with light stress, dark conditions, or nitrogen stress (Roberts et al., 2012). FtsH6 is reportedly involved in detached leaf senescence and dark-induced leaf senescence. Other proteases including members of the M10 and M17 MP families have also been reported associated with leaf senescence. Matrix MPs (MMPs) belong to the M10 family and are reportedly upregulated during senescence (Roberts et al., 2012). A mutant of At2-MMP shows a late flowering and early senescence phenotype, suggesting that MMPs are responsible for senescence regulation (Golldack et al., 2002). Leucine aminopeptidase 2 (LAP2) is a member of the M17 family. In A. thaliana, a mutant of LAP2 displays an early-senescence phenotype, suggesting that LAPs are involved in leaf longevity (Waditee-Sirisattha et al., 2011). EGY1 belongs to M50 family. To date, seven M50 family members have been identified in A. thaliana, Oryza sativa, and Zea mays. These proteases play roles in chloroplast development, hypocotyl elongation and gravitropism, stress response, nuclear plastid signaling pathway, and plant development, respectively (Nishimura et al., 2016).
Setaria italica is a new model C4 monocotyledonous species that promises to accelerate functional genomics studies in the grasses (Doust et al., 2009). In this study, a pale green mutant siygl2 was isolated from EMS-mutagenized lines of the S. italica Yugu1 cultivar. Map-based cloning indicated that the mutant gene encoded SiYGL2, a homolog of AtEGY1. Phenotypic surveys showed that siygl2, such as Ategy1, is characterized by chlorotic organs, premature senescence, and damaged PS II function. Unlike Ategy1, however, chloroplast development is not impaired in siygl2. Our study focused on the function of SiYGL1 in regulating senescence and photosynthesis in S. italica to supplement the existing functional gene knowledge available for this C4 model plant. Additionally, our research provides further insight into the mechanisms underlying the regulation of leaf senescence.
The siygl2 mutant was identified in screens of EMS mutagenized populations of S. italica cultivar Yugu1. To determine the chlorophyll content and the photosynthetic rate, plants were planted under natural conditions in the experimental field of institute of Crop Sciences, Chinese Academy of Agricultural Sciences, in Beijing (116.6°E, 40.1°N) in summer season. For dark-induced senescence, detached leaves were incubated in water at 28°C.
For analysis of photosynthetic pigments, leaves were cut into pieces and soaked in 95% alcohol for approximately 72 h until leaf pieces were completely transparent. The absorbance values of the supernatant were measured at 665 and 649 nm with UV-1800 ultraviolet/visible light. Chlorophyll a (Chl a) and chlorophyll b (Chl b) levels were then calculated as described by Lichtenthaler (1987). Statistics analysis was conducted using Welch’s two-sample t test. Multiple comparisons were made with LSD by IBM SPSS Statistics 23.0. The photosynthetic rate and chlorophyll fluorescence were measured on sunny days using a Li-6400 portable photosynthesis system (LI-COR, Lincoln, NE, United States) using mature leaves from five individuals. The light source used for measuring the photosynthetic parameters is 6400-02B LED light source, the ParIn parameter was set to 1000 μmol m-2 s1. The equations for the photosynthetic parameters are calculated as follows (Hu et al., 2014).
The second leaf and seventh leaf were obtained from plants that were at the eight-leaf stage. Leaf material was cut into 2 mm × 1 mm pieces and fixed overnight in 0.1 M phosphate buffer with 2.5% glutaraldehyde. The samples were then washed with 0.2 M phosphate buffer three times and post-fixed in 1% osmium tetroxide for 1 h. After staining with uranyl acetate, samples were further dehydrated in a gradient ethanol series and finally embedded into resin. Ultrathin sections were made and examined by JEM 1230 transmission electron microscopy (TEM). The areas of plastoglobulis are calculated by Image-Pro plus 6.0 (Media Cybernetics, Silver Spring, Georgia Avenue, United States). Statistics treatment was made with Welch’s two-sample t test.
An F2 population generated from a cross between siygl2 and the SSR41 cultivar was used for mapping of the SiYGL2 locus. Sixty-five SSR markers based on earlier studies (Jia et al., 2009; Zhang et al., 2014) were adopted for gene cross positioning. Thirteen cleaved amplified polymorphism sequences (CAPS) markers were newly designed for fine mapping based on the single-nucleotide polymorphism information between Yugu1 and SSR41 (Jia et al., 2013) (Supplementary Table S1). To identify mutant locations, 40 sequencing primers were developed that covered the whole candidate region based on the genome sequence information from the S. italica genome project V2.21 database (Supplementary Table S2).
RNA was isolated from wild-type (WT) and siygl2 stems, panicles, and 1st and 10th leaves from the top of the plant at the heading stage from fresh plant tissues using a Pure Link RNA Mini Kit (Cat no. 12183018, Invitrogen, United Kingdom). First-srtand cDNA was synthesized with a PrimerScript 1st Strand cDNA Synthesis Kit (Cat no. 6210A, TaKaRa, Otsu Shiga, Japan). Quantitative PCR was conducted using a Fast Start Universal SYBR Green Master (ROX) (Cat no. 04913914001, Roche, Mannheim, Germany) using the specific primers listed in Supplementary Table S3. Cullin was selected as the reference gene according to a previous study (Martins et al., 2016). The data were detected and analyzed using an Applied Biosystems 7300 Analyzer (Applied Biosystems, Foster City, CA, United States). Statistics treatment was made with Welch’s two-sample t test.
The sequences and structures of the candidate genes were obtained from the S. italica genome project V2.2. For phylogenetic analysis, homologs were obtained by NCBI Protein Blast2. Sequence alignments and cladograms were produced using MEGA 5.0 software. Protein TMHs were calculated using TMHMM_v.23.
The full-length cDNA of SiYGL2, excluding the stop code, was amplified from Yugu1 using the following primers: 5′TATCTCTAGAGGATCCCTATCCTCCTTCGGTCCTTCCCATT 3′ and 5′ TGCTCACCATGGATCCGAACGAAGTAACAAGCCCTACACCT 3′ [the underlined sequences are adaptors for In-fusion® PCR cloning system (Cat no. 072012, Clontech, United States) and contain BamHI cleavage sites]. The cDNA sequences were cloned into the p16318hGFP vector to form fusion proteins with the C-terminus of GFP. These vectors were then transfected into foxtail millet protoplasts by PEG-mediated transformation and detected by confocal microscopy (LSM700, Carl Zeiss, Germany).
The S. italica chlorotic mutant siygl2 was generated from the Yugu1 cultivar by EMS treatment. The siygl2 mutant showed a relatively normal phenotype in the seedling stage (Figure 1A). Throughout development, however, these plants gradually became chlorotic, and in the late developmental stages, siygl2 produced chlorotic leaves, stems, and panicles (Figures 1B–E). Additionally, the lower leaves of siygl2 had more abnormal phenotypes than the upper leaves (Figure 1F). Senescence appeared to be accelerated in siygl2 as the basal leaves of siygl2 were tip burned. Several key agronomic traits of siygl2 and WT plants were analyzed. The results showed that some yield characteristics, such as floret grain number and panicle weight, are significantly reduced compared to WT (Table 1).

FIGURE 1. Phenotypic characteristics of the siygl2 mutant. (A) Five-week-old seedling phenotypes of the wild-type (WT) cultivar Yugu1 and the siygl2 mutant. (B) Heading stage phenotypes of Yugu1 and siygl2. The red bars point out the significant abnormal phenotypes in panicles, stems, and bottom leaves. (C–E) Lower leaves, stems, and panicles of heading stage plants. (F) Leaf color comparison of the WT (left) and siygl2 (right) at the heading stage. Leaves from the top of the plant to the bottom are arranged accordingly.
To further characterize the chlorotic phenotype of the siygl2 mutant, we measured the chlorophyll content of leaf, sheath, panicle, and stem at the heading stage (Figure 2A), and found them to have 71, 35, 28, and 28% the chlorophyll content of WT tissues, respectively. Both Chl a and Chl b levels were reduced in siygl2, with the reduction in Chl b being more severe in leaves.
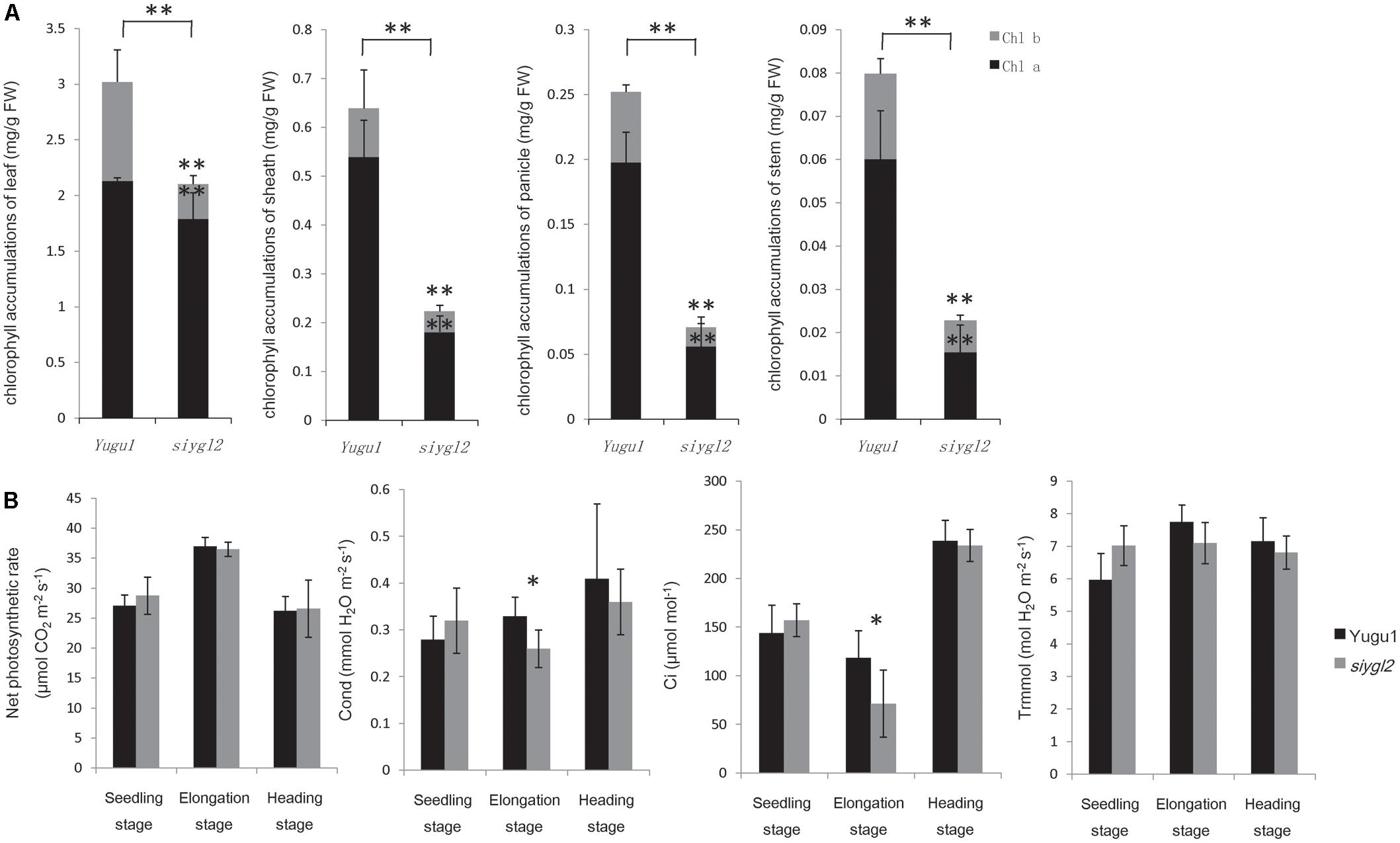
FIGURE 2. (A) Chlorophyll content of leaves, sheaths, panicles, and stems at the heading stage of wild-type cultivar Yugu1 and the siygl2 mutant. Black and gray boxes show the amount of chlorophyll a and chlorophyll b, respectively. FW, fresh weight; Chl a, chlorophyll a; Chl b, chlorophyll b. Means and standard deviations were obtained from five independent leaf samples grown under normal growth conditions. (B) Photosynthetic parameters in different developmental stages. Means and standard deviations were obtained from five independent leaf samples grown under normal growth conditions. Photo, photosynthetic rate; Cond, stomatal conductance; Ci, intercellular carbon dioxide concentration; Trmmol, transpiration rate. Means and standard deviations are obtained from five independent samples. ∗Significantly different at P = 0.05. ∗∗Significantly different at P = 0.01.
As reductions in chlorophyll may be associated with changes of the photosynthetic capacity, the photosynthetic parameters of the newly emerging leaves (from the top of the plant) were measured (Figure 2B). Surprisingly, siygl2 photosynthetic capacity was not significantly affected at the seedling stage, the elongation stage, or the heading stage. The stomatal conductance of siygl2 decreased about 22% in the elongation stage concurrent with a decrease in the intercellular carbon dioxide concentration of approximately 40%. Overall, the decrease in chlorophyll accumulation in siygl2 did not significantly affect photosynthesis in the newly developed leaves. Variations in the intercellular carbon dioxide concentration and stomatal conductance may be caused by other effects of the mutant gene or environmental factors. Further study in investigating the ACi curve of both WT and mutants would help to reveal the effects of EGY1 in photosynthesis.
As the basal leaves of siygl2 had a more obvious chlorotic phenotype than the top leaves and showed accelerated senescence (Figures 1B,F), the chlorophyll pigment levels of the 1st, 4th, 7th, and 11th leaves from the top of siygl2 and WT Yugu1 plants were examined (Figure 3A). Chlorophyll accumulation in the first leaves of siygl2 declined by approximately 30% compared with Yugu1. However, in basal leaves, siygl2 only contained one-third of the chlorophyll of the WT leaves at the same position. This suggests that chlorophyll levels decline earlier in the old leaves of siygl2 than in Yugu1. We also noticed that in the first and fourth leaves of siygl2, the Chl b levels decreased while Chl a levels did not. In the 11th leaves, both Chl a and Chl b in siygl2 are sharply decreased compared with Yugu1 (Figure 3A). Anyhow, the accelerated leaf senescence of siygl2 could be associated with changes in the function of SiYGL2.
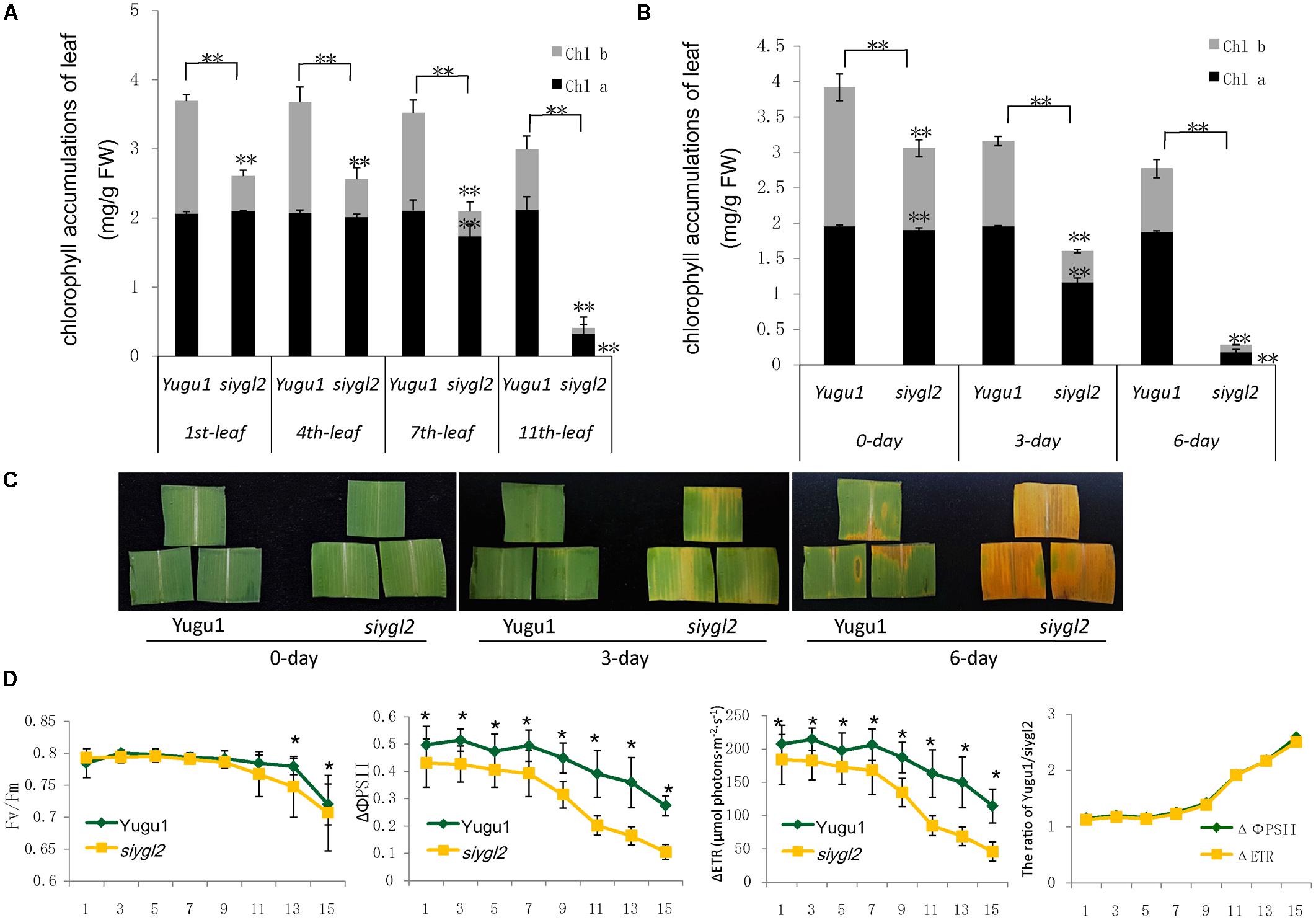
FIGURE 3. (A) Chlorophyll accumulation in different leaves at the heading stage (mg/g FW). Black and gray boxes represent Chl a and Chl b, respectively. (B) Chlorophyll accumulations after different durations of dark treatment. (C) Leaves of Yugu1 and siygl2 after different durations of dark treatment. (D) Chlorophyll fluorescence in different leaves of Yugu1 and siygl2. The X axis shows the leaf position from the top of plants. For measuring the chlorophyll content and chlorophyll fluorescence, means and standard deviations are obtained from five independent plants for each genotype. ∗Significantly different at P = 0.05. ∗∗Significantly different at P = 0.01.
To verify the changes in leaf aging in siygl2, we first conducted a detached leaf senescence assay using dark treatments (Figures 3B,C). After 3 days of dark treatment, first leaves of WT and siygl2 were compared. The leaves of siygl2 began turning yellow, while the leaves of the WT remained green (Figure 3C). After 6 d of dark treatment, siygl2 leaves were completely yellow, whereas only the cut ends of the WT leaves showed yellowing (Figure 3C). The chlorophyll content also reflected the changes induced by dark treatment (Figure 3B). These results suggest that SiYGL2 helps to negatively regulate dark-induced leaf senescence, with senescence promoted by the loss of SiYGL2 function. In addition, we noticed that at the beginning of dark treatment, the effect on the reduction of Chl b levels are stronger than on the reduction of Chl a levels (Figure 3B). This is similar with the phenomenon showed in Figure 3A.
The maximum photochemical efficiency (Fv/Fm) is a senescence-associated index. We measured the Fv/Fm values of the 1st, 3rd, 5th, 7th, 11th, 13th, and 15th leaves of WT and siygl2 plants. The Fv/Fm values of the WT Yugu1 plants decreased gradually from the upper to the lower leaves. The Fv/Fm values of the mutant siygl2 leaves displayed a similar decreasing tendency, with levels declining more sharply than in the WT in the 13th and 15th leaves (Figure 3D). This suggests that the basal leaves of siygl2 enter senescence earlier than Yugu1. We further tested effective PSII quantum yield (ΔΦPS II) and photosynthetic electron transport rate (ΔETR) to describe the changes PS II light-use efficiency (Figure 3D). Both ΔΦPS II and ΔETR declined generally in leaves when moving from the top to the bottom of the plant, indicating that PS II photosynthesis capacity decreases with aging. In the whole-plant leaves, both ΔΦPS II and ΔETR of siygl2 were lower than in Yugu1. From the 11th leaf, the ratio of both ΔΦPS II and ΔETR of Yugu1/siygl2 significantly increased compared with that in the upper leaves, indicating that these two indices drop more acutely than in siygl2 (Figure 3D). These results verify that PS II photosynthesis capacity is decreased and senescence is premature in siygl2.
Leaf senescence is always accompanied by changes in chloroplast ultrastructure. We obtained entirely expanded young leaves and basal leaves of plants at the elongating stage (eight leaf-stage) and surveyed the mesophyll cell-chloroplasts of these leaves. TEM observations showed that in the young leaves of siygl2, the chloroplasts were similar to those of Yugu1 (Figures 4A,C), indicating that thylakoid and chloroplast development were not impaired. However, in the basal leaves of siygl2, the chloroplasts were seriously disintegrated. The size of the plastoglobuli increased by 57.4% in siygl2 (Supplementary Figure S1), and irregular grana thylakoid stacks were also be observed in the chloroplasts of the mutant (Figure 4D). In siygl2 leaves at the same position as in Yugu1, chloroplasts retained their normal state (Figure 4B). This result indicates that chloroplast degradation of siygl2 is advanced.
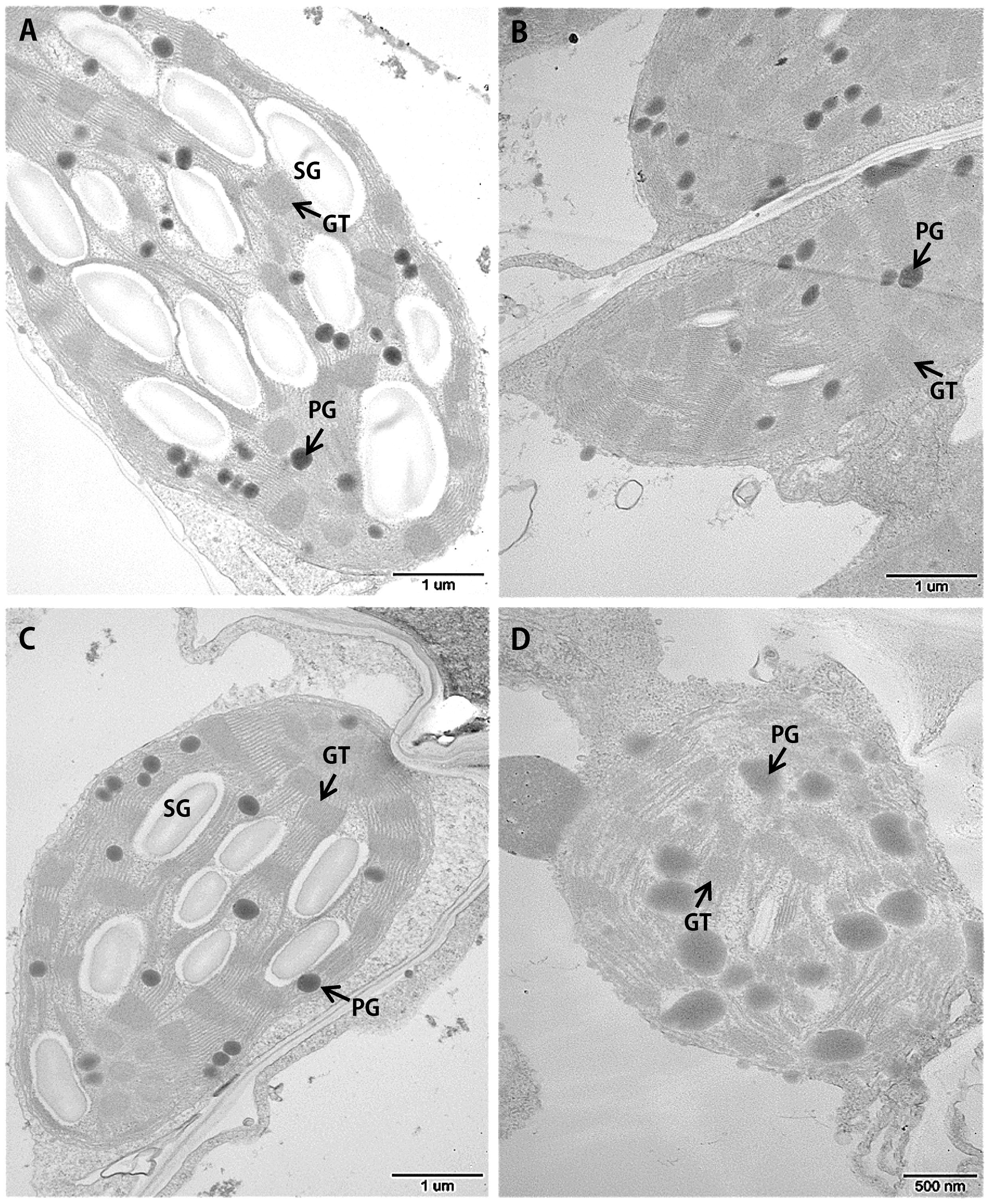
FIGURE 4. Transmission electron microscopic images of chloroplasts in the WT Yugu1 cultivar (A,B) and siygl2 mutant (C,D). (A,C) Chloroplasts in tender leaves. (B,D) Chloroplasts in tender aging leaves. SG, starch granules; PG, plastoglobuli; GT, grana thylakoid stacks.
Taken together, the above results suggest that the SiYGL2 gene may play a role in maintaining chlorophyll accumulation and chloroplast structure in aging leaves, and may delay the onset of leaf senescence. Suppression of SiYGL2 function could, therefore, lead to early senescence of basal leaves.
For genetic analysis of the siygl2 mutant, we constructed an F2 population by hybridizing siygl2 with the Yugu1 cultivar. The F2 progeny showed a segregation ratio of 3:1 (266:80, χ2 = 0.56 < χ20.05 = 3.84), suggesting that this chlorotic phenotype was controlled by a single recessive gene.
To map the siygl2 locus, an F2 mapping population was generated from a cross between siygl2 and the cultivar SSR41. Using this F2 population (883 yellow leaf plants), we generated a DNA pool of 40 individual mutant plants and preliminarily mapped the siygl2 gene to a 948.3-kb genomic region on chromosome 9 by bulked segregation analysis. On this basis, a 31.3-kb genomic region between the SSR marker P44 and the CAPS marker zs912 was then defined. Within this region, eight open reading frames (ORFs) were predicted from the data on phytozome4 (Figure 5A). These ORFs were amplified and sequenced and only the fifth ORF (Seita.9G060500) was found to carry a 17-bp deletion (AATGTTTGACATATCAA) at the position 3,478,889–3,478,905 of chromosome 9. The gene structure of Seita.9G060500 is predicted to contain 11 exons and 10 introns. The identified deletion in this gene leads to a frameshift that results in a termination codon at the fifth exon (Figure 5A).
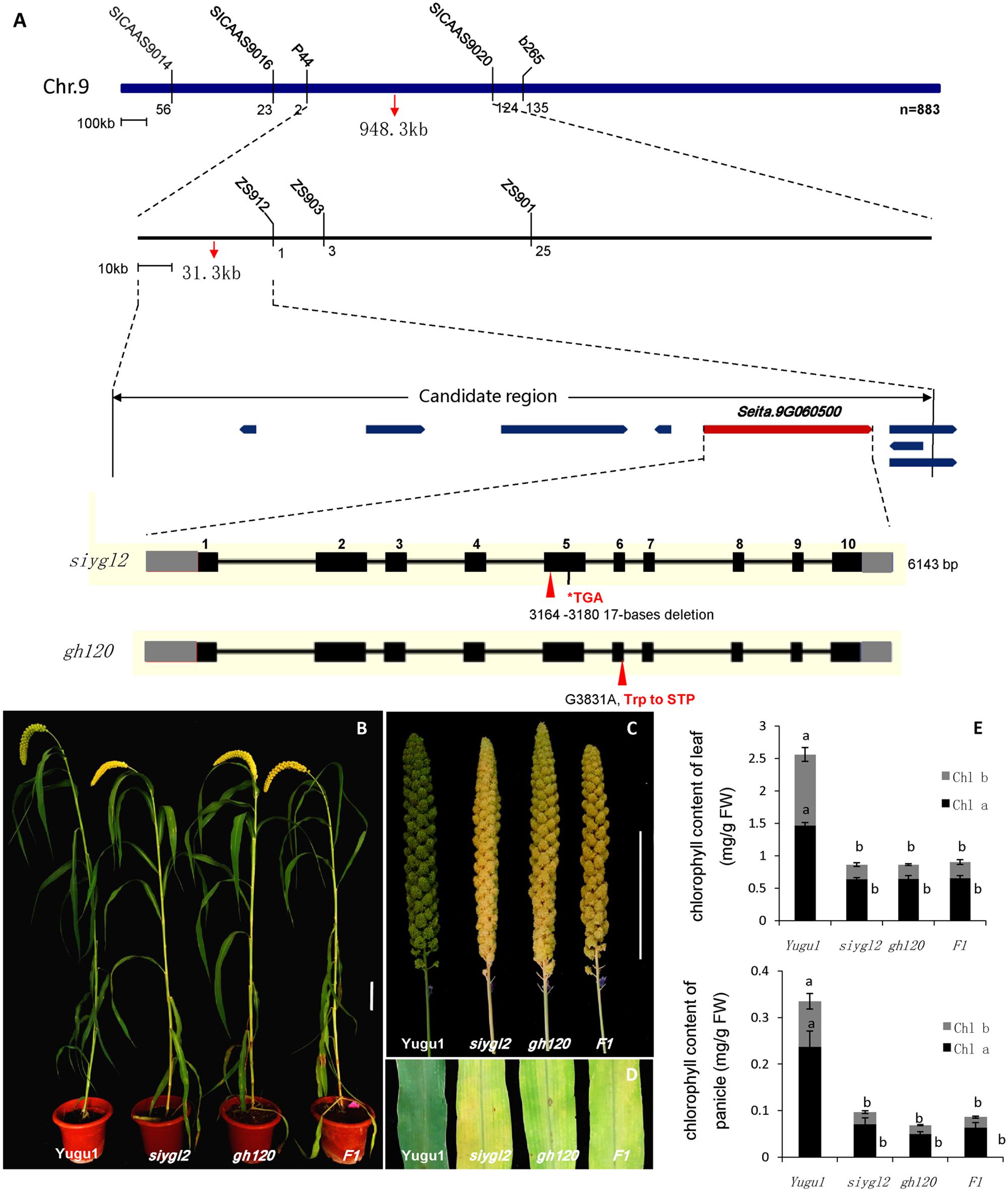
FIGURE 5. (A) Map-based cloning, gene structure of Seita.9G060500, and the mutant positions of siygl2 and gh120. (B–D) are phenotypes of WT Yugu1 plants and mutants. (E) Chlorophyll content of siygl2, gh120, and F1 individuals. Means and standard deviations are obtained from five independent samples. Values connected by same letters are not statistically different; values not connected by same letters are statistically different at P = 0.05.
We also identified another leaf chlorotic mutant, gh120, that shows the same phenotypes as siygl2 (Figures 5B–E). Genome sequencing indicated that there is a single base change (G 3831 A) at the sixth exon of Seita.9G060500 that leads to the alteration of Trp to a termination codon. The F1 individuals of a cross between siygl2 and gh120 are hemizygous at both the mutation sites of their parents (Supplementary Figure S2) and display similar phenotypes to their parents (Figures 5B,D). The chlorophyll content of F1 plants is consistent with that of siygl2 and gh120 mutant (Figure 5E). These results suggest that the mutations in Seita.9G060500 indeed cause the abnormal phenotype in the siygl2 and gh120.
Amino acid sequence comparison indicated that SiYGL2 is most closely related to AtEGY1, with these proteins sharing 77.3% amino acid sequence identity. SiYGL2 was, therefore, proposed to be a homolog of AtEGY1, a S2P-like chloroplast membrane-located MP. Phylogenetic analysis further confirmed this relationship (Figure 6). In Arabidopsis, there are three EGY proteins: AtEGY1, AtEGY2, and AtEGY3 (Chen et al., 2005). Similarly, two homologs of SiYGL2, Seita.5G097600 (SiEGY2) and Seita.9G108100 (SiEGY3), were identified in S. italica (Figure 6).
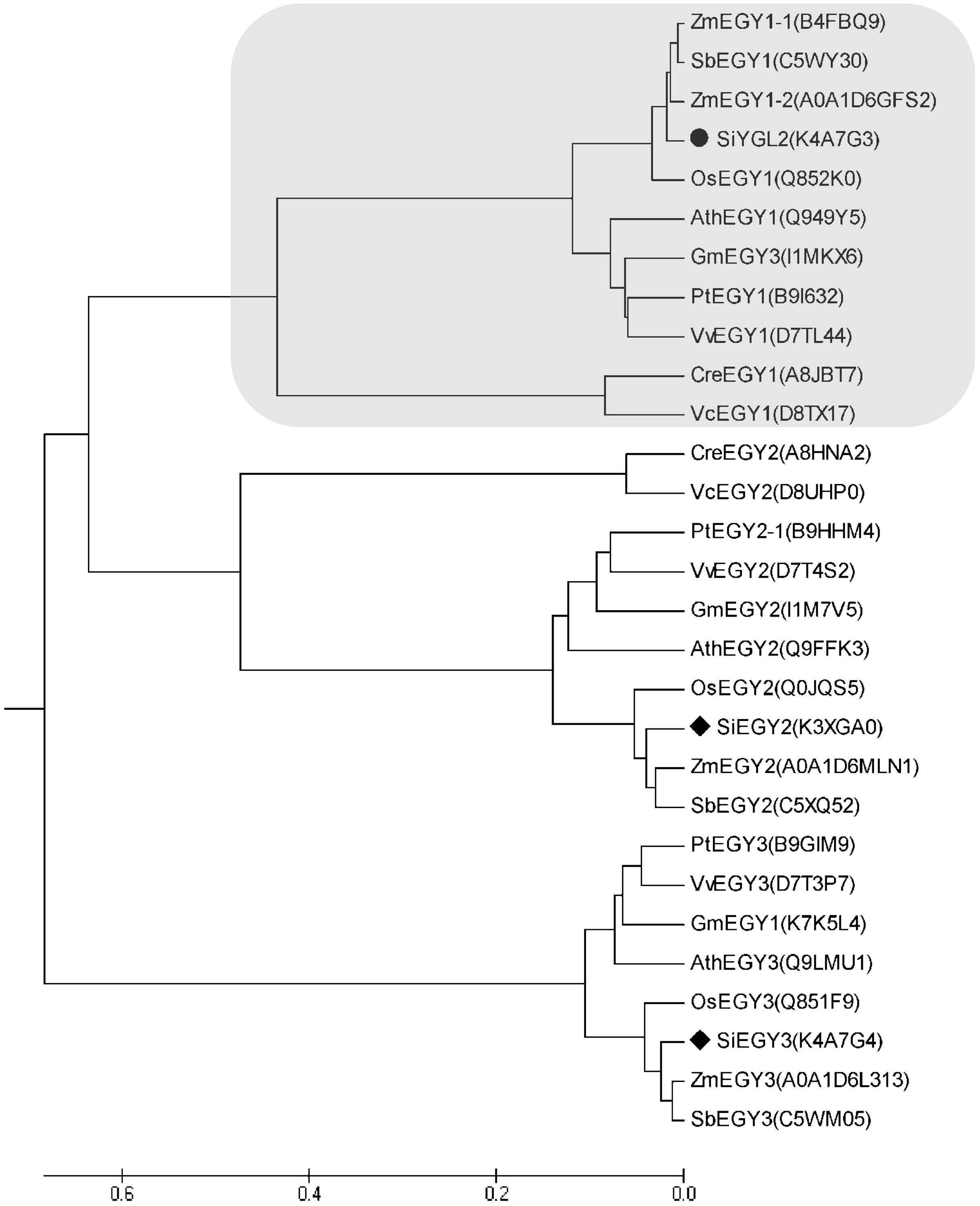
FIGURE 6. Phylogenetic analysis of EGY proteins in diverse species. Ath, A. thaliana; Cre, Chlamydomonas reinhardtii; Gm, Glycine max; Os, Oryza sativa; Pt, Populus trichocarpa; Sb, Sorghum bicolor; Si, S. italica; Vv, Vitis vinifera; Zm, Zea mays. Protein IDs are listed in brackets and are archived in the UNIPROT database (http://www.uniprot.org/). SiEGY2, Seita.5G097600. SiEGY3, Seita.9G108100.
SiYGL2 is predicted to encode a 548-aa protein, with a peptide chain that contains three conversed motifs, GNLR (aa 169–178), HEXXH (aa 311–315), and NPDG (aa 442–454) (Figure 7A), that reportedly also occur in AtEGY1 (Chen et al., 2005). However, the mutant SiYGL2 protein (ΔSiYGL2) was predicted to lack 165 amino acid residues from the carboxyl terminus. Furthermore, frameshift mutation caused changes in the sequence from 362–383 aa, leading to the loss of NPDG motif. This suggests that the protein structure and function of ΔSiYGL2 is likely to differ significantly from that of SiYGL2.
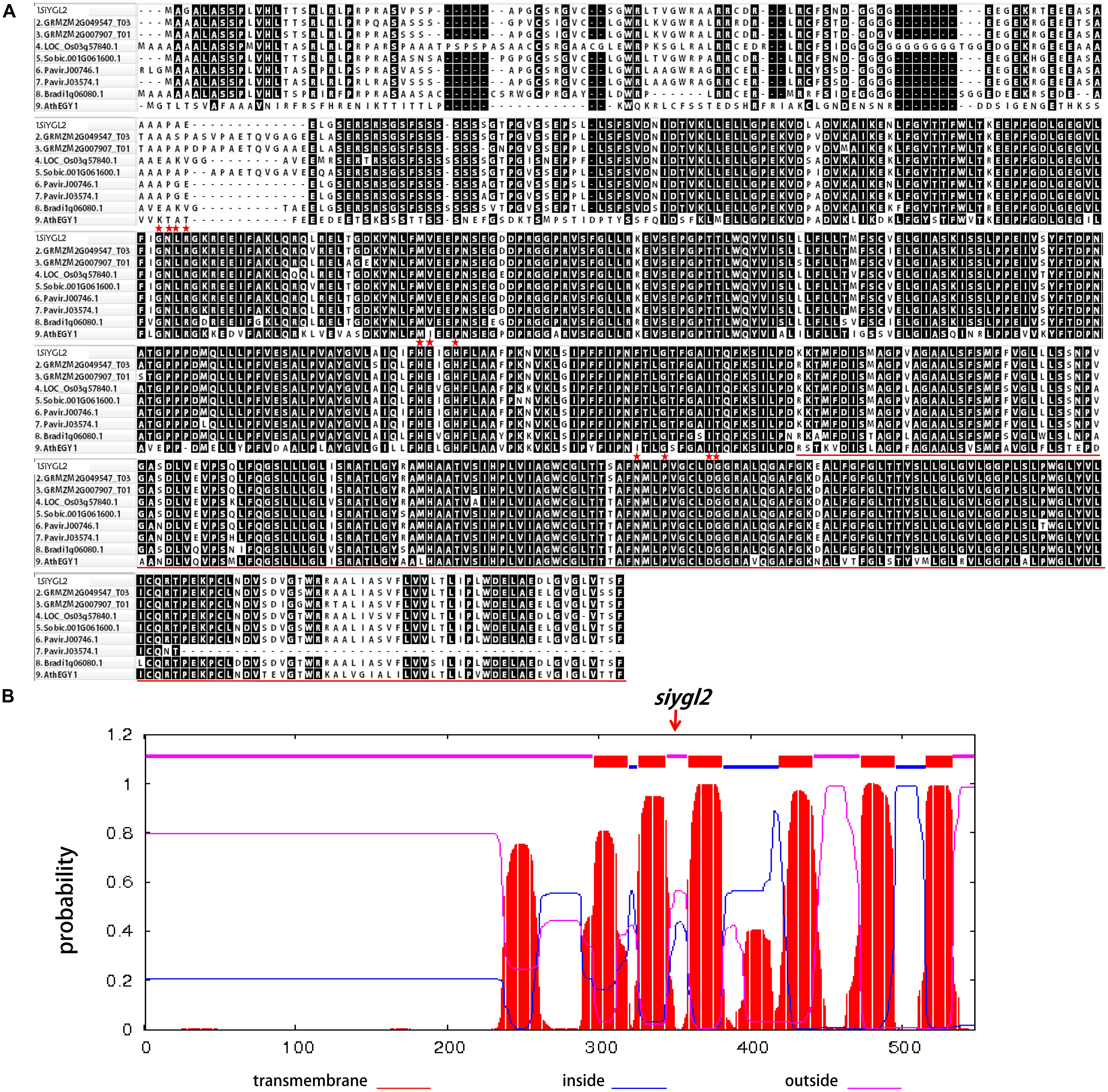
FIGURE 7. (A) Amino acid sequence alignment of the homologous proteins. Red stars show the position of the conserved motifs GNLR, HEXXH, and NPDG, respectively. The red lines show lost amino acid sequence of ΔSiYGL2. (B) Hydropathy plot of SiYGL2 and ΔSiYGL2. Red boxes show the transmembrane helices. Blue lines and pink lines at the top of the figure indicate the inside and outside peptide chains, respectively. The arrow indicates the mutated position of siygl2.
Hydropathy analysis revealed that the AtEGY1 protein is highly hydrophobic as it contains six predicted transmembrane (tm) helices in its C-terminus (tm1, aa 340–362; tm2, aa 369–387; tm3, aa 402–424; tm4, aa 462–484; tm5, aa 516–538; tm6, aa 559–576). Conversely, ΔSiYGL2 lost four of these tm helices (Figure 7B).
As the siygl2 mutant showed chlorotic leaves, stems, and panicles, with this phenotype more apparent in older leaves, the expression pattern of SiYGL2 was investigated in different organs and leaves at different developmental stages by qRT-PCR (Figure 8). SiYGL2 was more highly expressed in panicles and young leaves (1st leaves) than in stems and old leaves (10th leaves), suggesting that SiYGL2 is typically more active in young organs. However, SiYGL2 expression in siygl2 organs differed significantly; in panicle, stem, and the 10th leaf in siygl2, SiYGL2 expression decreased to 27.7, 43.5, and 20.0% of those in WT, respectively. In the 10th leaf, in particular, transcript accumulation was very low. However, the expression SiYGL2 in the first leaf of siygl2 was higher (1.25-fold) than in the WT. These results suggest that the expression of SiYGL2 in siygl2 is not dependent on the function of the SiYGL2 protein.
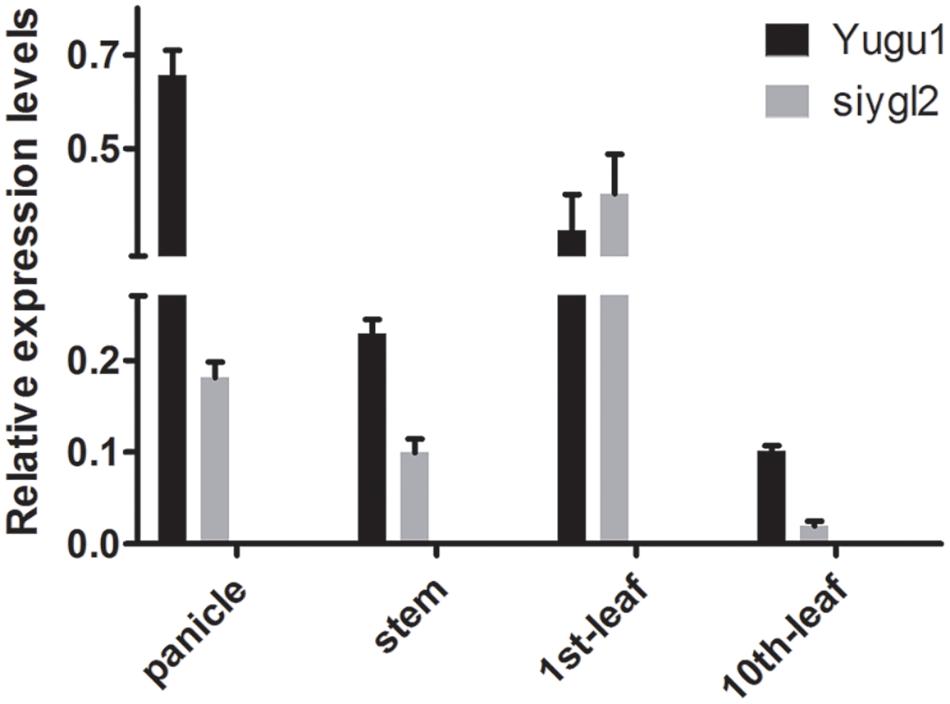
FIGURE 8. Transcript accumulation of SiYGL2 in panicle, stem, and leaves. Black boxes, Yugu1; gray boxes, siygl2. Means and standard deviations are obtained from three independent samples and three independent assays. Statistics treatment was made with Welch’s two-sample t test. ∗Significantly different at P = 0.05.
To explore the subcellular location of SiYGL2, we generated a SiYGL2-GFP fusion gene. The fusion gene was placed under the control of the CaMV 35S promoter and introduced into protoplasts of Yugu1. The results obtained from confocal laser microscopy show that SiYGL2 is localized to the chloroplast (Figure 9A). This result is similar to that of AtEGY1 (Chen et al., 2005). In the GFP transgenic control line, GFP fluorescence was found in the cytomembrane, the plasma, and the nucleus (Figure 9B).
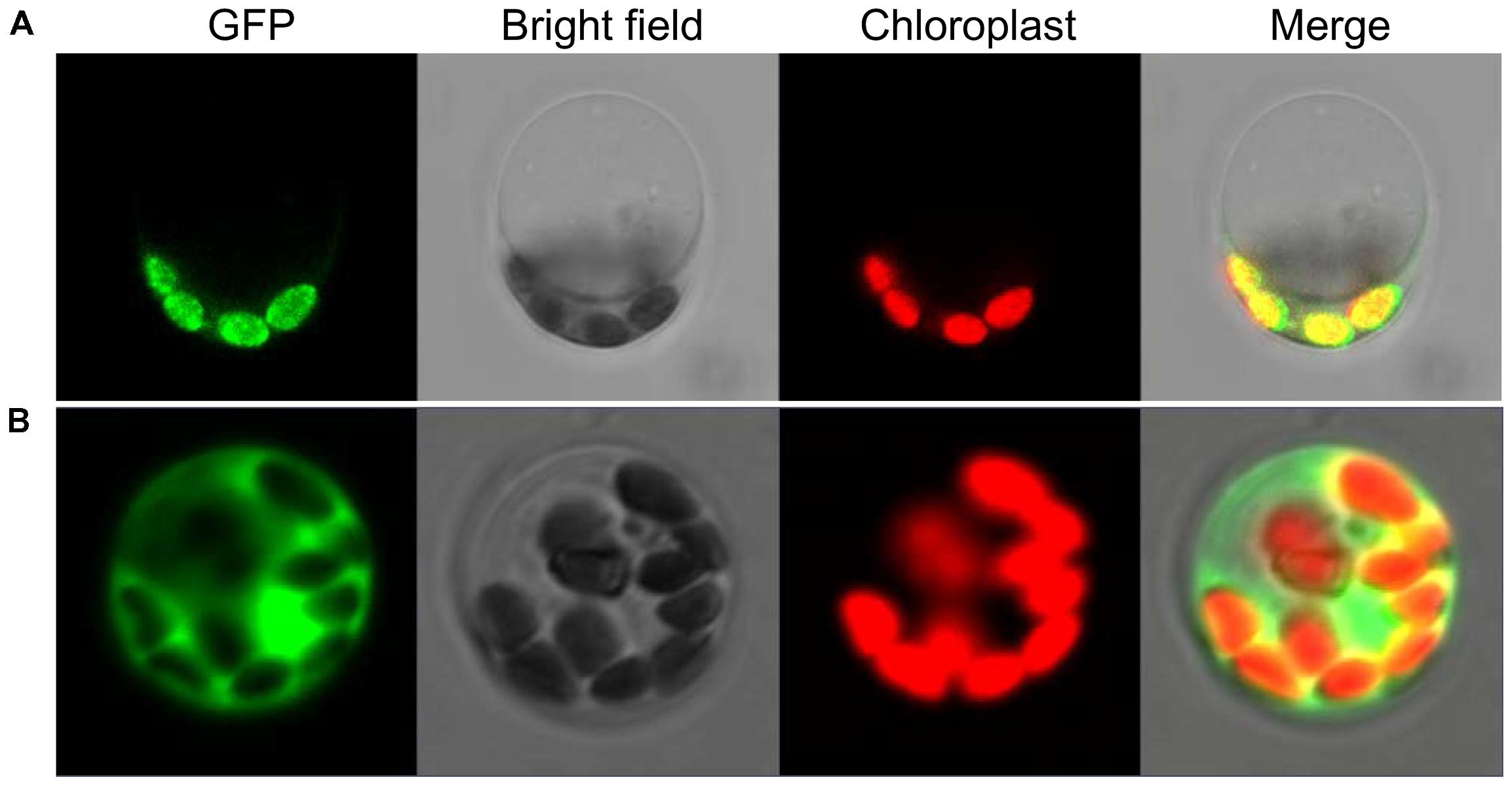
FIGURE 9. Subcellular localization of SiYGL2. (A) SiYGL2:GFP signal. (B) Empty GFP vector as a control.
To further verify the influence of the SiYGL2 mutation on leaf senescence, several senescence-related genes examined using qRT-PCR (Figure 10). AtSAG12, encoding a putative cysteine protease, is a negative regulator of senescence. In A. thaliana, expression of AtSAG12 is strictly associated with senescence progression, with expression levels increasing throughout leaf aging before decreasing at the end of the aging process (Weaver et al., 1998). We used SiSAG12, a homolog of AtSAG12 as a reporter to follow the progression of leaf senescence. In old 10th leaves of Yugu1, the expression level of SiSAG12 was upregulated dramatically (approximately sevenfold), consistent with the reported expression of AtSAG12. While SiSAG12 expression levels in both young and old leaves of siygl2 were lower than those in the corresponding leaves of Yugu1. SiSAG12 expression did increase by approximately threefold in 10th leaf than that in young leaf of siygl2 (Figure 10). These results suggest that the expression of senescence-associated genes in siygl2 is impaired, reflecting the variations in aging processes between siygl2 and WT.
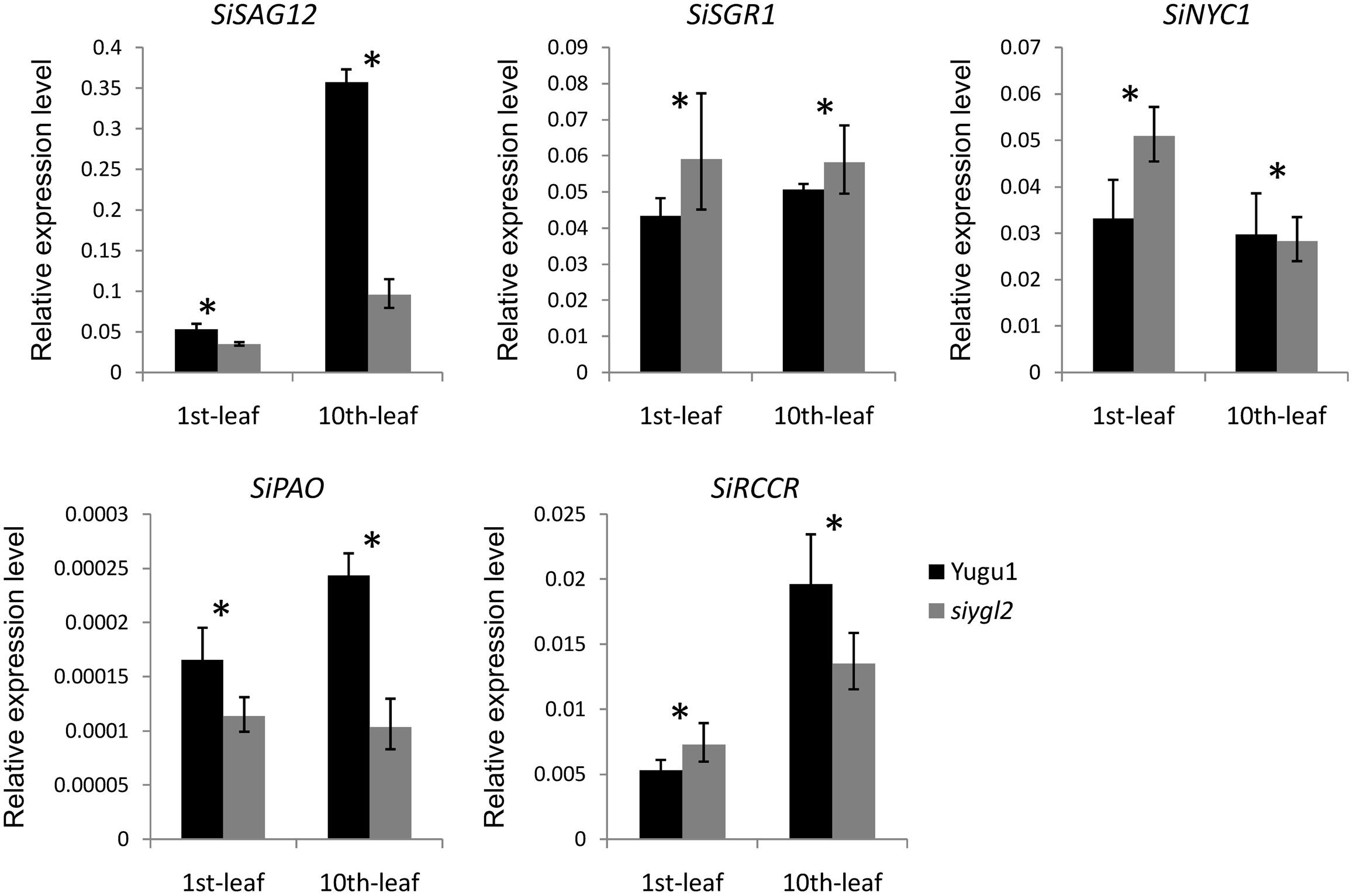
FIGURE 10. Relative expression levels of chlorophyll degradation- and leaf senescence-associated genes.
SGR1 and NYC1 are reportedly involved in chlorophyll degradation and disassembly of the light-harvesting complex of PS II (LHC II) during leaf senescence (Kusaba et al., 2007; Sakuraba et al., 2012). In the young leaves of siygl2, the expression of these two genes was higher than in the young WT leaves. Furthermore, the transcript accumulation of these two genes in mutant old leaves also differed significantly from their expression in WT. These results indicate that chlorophyll metabolism and LHC II stability during the aging process may be affected in the mutant. Two other senescence-associated genes, RCCR and PAO, also reportedly participate in chlorophyll degradation (Sakuraba et al., 2016). These two genes were upregulated in old WT leaves. In siygl2, however, the expression of these two genes differed significantly from the WT in both the 1st and 10th leaves. This suggests that chlorophyll degradation in siygl2 is affected.
In summary, we conclude that changes in SiYGL2 function affects the regulation of several senescence-associated genes, with chlorophyll degradation and leaf senescence also affected.
Our study has shown that SiYGL2 gene expression levels are higher in panicles and first leaves than in stems and old leaves (Figure 8). The flag leaf and panicle are the most important source and sink organs, respectively, and should retain high chlorophyll levels and photosynthetic capacities to avoid premature aging and ensure their continued output. The high expression of SiYGL2 in these organs suggests that SiYGL2 may play a role in maintaining the physiological state of the leaf and its photosynthetic capacity. Mutation of SiYGL2 results in premature chlorophyll degradation, chloroplast disintegration, and leaf senescence in the basal leaves (Figures 3A, 4). Furthermore, aging of the leaves of the siygl2 mutant can be induced more easily by dark than in the WT (Figures 3B,C). Taken together, these results suggest that SiYGL2 may be involved in delaying the onset of leaf senescence. This is consistent with the results reported for the SiYGL2 homolog AtEGY1 in Arabidopsis (Chen et al., 2016).
The chlorophyll content of siygl2 was lower than in WT in both the top and basal leaves (Figure 3A), indicating that SiYGL2 may be related to chlorophyll metabolism. The chlorophyll degradation-associated genes SiNYC1, SiSGR1, and SiRCCR were expressed more highly in young leaves of siygl2 than in Yugu1 (Figure 10), suggesting that chlorophyll degradation is mobilized early in young siygl2 leaves. We also find that the effects on Chl b caused by SiYGL2 mutant, natural senescence, and dark treatment are stronger than that on Chl a (Figures 2A, 3A,B). First, this may be explained by that the degradation of Chl b is prior to Chl a. And Chl b can converted into Chl a when undergoing degradation (Schelbert et al., 2009). Thus, at the preliminary stage of senescence, Chl b is sharply decreased while Chl a seems to be not impaired. With the aging process, Chl a is also degraded. Therefore, both the Chl a and Chl b contents are reduced in later period of senescence. Second, the reduction in Chl b content but not Chl a suggesting that the light harvesting is altered. The mutant may not grow well under low light condition, but would be fine under moderately high light. Shading and light deficiency may be one of the reasons the bottom leaf had earlier senescence. This hypothesis could be tested with a light response curve in further study.
In all leaves at different ages, both ΔΦPS II and ΔETR were lower in siygl2 than in WT (Figure 3D), indicating that the PS II function was impaired in siygl2. Thus, we propose that SiYGL2 plays a role in the regulation of PS II function. In conclusion, in monocotyledonous S. italica, SiYGL2 is associated with the regulation of leaf senescence, chlorophyll metabolism, and PS II efficiency. As described in previous studies, AtEGY1 participates in chlorophyll accumulation, leaf senescence, and PS II function (Chen et al., 2005, 2016). This indicates that the function of SiYGL2 in monocotyledonous S. italica is quite similar to that its homolog AtEGY1 in dicotyledonous Arabidopsis.
The Arabidopsis mutant egy1-1 displayed a defective chloroplast development phenotype (Chen et al., 2005). However, the S. italica EGY1 mutant siygl2 has relatively normal chloroplasts (Figure 4C). Further analysis of the function of SiYGL2 in chloroplast development is therefore required. We compared the peptide chains of these two mutants and found that the mutant AtEGY1 protein lacks the HEXXH and NPDG motifs, while the mutant SiYGL2 protein lacks only the NPDG motif. Thus, we propose two hypotheses for the differences in chloroplast development between the egy1-1 and siygl2 mutants. First, it has been reported that for S2P family members in Bacillus subtilis, HEXXH and NPDG together form the catalytic center required for protease function (Rudner et al., 1999). Furthermore, two other MPs, AtVAR2 and AtVIR3, which have the HEXXH motif but lack the NPDG motif, could regulate chloroplast development (Chen et al., 1999; Qi et al., 2016). We, therefore, speculate that HEXXH is essential for chloroplast development-related function of EGY1 and alone can guarantee normal chloroplast development, but requires NPDG to regulate leaf senescence. Second, SiYGL2 and AtEGY1 share 77.3% amino acid sequence similarity, so the differences between remaining amino acids may lead to differences in function. Homologous proteins commonly show different functions in different species; EGY1 may potentially have changed its chloroplast development-related function in dicotyledonous C3 Arabidopsis relative to the monocotyledonous C4 plant S. italica during evolution. These two hypotheses could be verified in future by removing the HEXXH motif of SiYGL2 and observing the chloroplast structure of the resulting plants.
Chlorophylls functions as the antenna pigment in the photosynthesis and as such is an essential component to PS II function (Stirbet et al., 2014; Kato et al., 2016). However, while chlorophyll content, ΔΦPS II, and ΔETR were lower in siygl2 than in WT (Figures 10B,C), net photosynthetic rate of siygl2 remained the same as in the WT (Figure 2B). Similar results have been reported in the S. italica ygl1 mutant and rice ygl7 mutant, which are two mutants of D submit of Mg-chelatase encoding gene, that show decreased chlorophyll levels and defective chloroplasts and, yet display increased photosynthetic rates and photosynthetic reaction center activity (Deng et al., 2014; Li et al., 2016). A previous report has stated that plants contain an abundance of light-harvesting pigments and absorb more light than they can use (Ort et al., 2015). Following this, we speculate that in the siygl2 mutant leaves, even though chlorophyll accumulation is reduced, photosynthesis can still be maintained at the same level as in the WT because sufficient light is still harvested by the chlorophyll present. The fact that a reduction in PS II efficiency does not impair the photosynthetic rate of the mutant may suggest that the siygl2 mutant may have improvements in other photosynthesis-related components that compensate for the decrease in PS II efficiency. In addition, the induction of alternative electron transport (water–water cycle, etc.) may be also a reason leading to the alteration in ΔETR but not photosynthetic rate. These should be confirmed by further experiments.
Despite the conserved photosynthetic rate, the yield of siygl2 was significantly reduced when compared with WT plants. We propose two explanations for this phenomenon. On the one hand, yield is closely related to leaf senescence, so accelerated leaf senescence could influence the mobilization of nutrients to reproductive organs and reduce the grain yield. Alternatively, the panicle has photosynthetic carbon assimilation capabilities and plays an important role in grain formation. Glumes can ensure remobilization of nutrients to grains as a vital part of crop source–sink translocation. Compared with the flag leaf, glumes are more crucial in the later period of grain filling (Kong et al., 2015). In our study, the siygl2 mutant panicle contained only 28% of the chlorophyll contained in the WT panicle at the heading stage (Figure 2). This suggests that the low chlorophyll content and impaired photosynthetic capability of the glume may be associated with the reduced yield in the siygl2 mutant.
XD conceived the project. SZ, WL, JS, and CT carried out the experimental work. HZ provided the materials and did the field trials. SZ did the data analysis and wrote the manuscript. XD, GJ, and ST guided the experimental work. All authors read and approved the final manuscript.
This work was supported by Fundamental Research Funds of CAAS (CAAS-XTCX2016002), China Agricultural Research System (CARS06-13.5-A04), the National Natural Science Foundation of China (31501324 and 31522040), and the Agricultural Science and Technology Innovation Program of the Chinese Academy of Agricultural Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer RZ and handling editor declared their shared affiliation at the time of the review.
We thank Emma Tacken, Ph.D., from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01308/full#supplementary-material
FIGURE S1 | The plastoglobulis areas of the young leaves and old leaves in Yugu1 and siygl2. Means and standard deviations are obtained from 10 plastoglobulis. Statistics treatment was made with Welch’s two-sample t test. ∗∗Significantly different at P = 0.01.
FIGURE S2 | Sequences at the mutation sites of Yugu1, siygl2, gh120, and F1 individuals. The red boxes and arrow show the mutant sites.
TABLE S1 | CAPS markers for fine mapping.
TABLE S2 | Sequencing primers for the candidate region.
TABLE S3 | Primers used for qRT-PCR analysis.
Bölter, B., Nada, A., Fulgosi, H., and Soll, J. (2006). A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett. 580, 789–794. doi: 10.1016/j.febslet.2005.12.098
Che, P., Bussell, J. D., Zhou, W., Estavillo, G. M., Pogson, B. J., and Smith, S. M. (2010). Signaling from the endoplasmic reticulum activates brassinosteroid signaling and promotes acclimation to stress in Arabidopsis. Sci. Signal. 3:ra69. doi: 10.1126/scisignal.2001140
Chen, C. Y., Wang, J., and Zhao, X. (2016). Leaf senescence induced by EGY1 defection was partially restored by glucose in Arabidopsis thaliana. Bot. Stud. 57:5. doi: 10.1186/s40529-016-0120-3
Chen, G., Bi, Y. R., and Li, N. (2005). EGY1 encodes a membrane-associated and ATP-independent metalloprotease that is required for chloroplast development. Plant J. 41, 364–375. doi: 10.1111/j.1365-313X.2004.02308.x
Chen, M., Jensen, M., and Rodermal, S. (1999). The yellow variegated mutant of Arabidopsis is plastid autonomous and delayed in chloroplast biogenesis. J. Heredity 90, 207–214. doi: 10.1093/jhered/90.1.207
Deng, X. J., Zhang, H. Q., Wang, Y., He, F., Liu, J. L., and Xiao, X. (2014). Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS One 9:e99564. doi: 10.1371/journal.pone.00
Diaz-Mendoza, M., Velasco-Arroyo, B., Santamaria, M. E., Gonzalez-Melendi, P., Martinez, M., and Diaz, I. (2016). Plant senescence and proteolysis: two processes with one destiny. Genet. Mol. Biol. 39, 329–338. doi: 10.1590/1678-4685-GMB-2016-0015
Doust, A. N., Kellogg, E. A., Devos, K. M., and Bennetzen, J. L. (2009). Foxtail millet: a sequence-driven grass model system. Plant Physiol. 149, 137–141. doi: 10.1104/pp.108.129627
Egli, D. B. (2011). Time and the productivity of agronomic crops and cropping systems. Agron. J. 103, 743–750. doi: 10.2134/agronj2010.0508
Feng, L., Yan, H., Wu, Z., Yan, N., Wang, Z., and Jeffrey, P. D. (2007). Structure of a site-2 protease family intramembrane metalloprotease. Science 38, 1608–1612. doi: 10.1126/science.1150755
Golldack, D., Popova, O. V., and Dietz, K. J. (2002). Mutation of the matrix metalloproteinase At2-MMP inhibits growth and causes late flowering and early senescence in Arabidopsis. J. Biol. Chem. 277, 5541–5547. doi: 10.1074/jbc.M106197200
Gregersen, P. L., Culetic, A., Boschian, L., and Krupinska, K. (2013). Plant senescence and crop productivity. Plant Mol. Biol. 82, 603–622. doi: 10.1007/s11103-013-0013-8
Gregersen, P. L., Holm, P. B., and Krupinska, K. (2008). Leaf senescence and nutrient remobilization in barley and wheat. Plant Biol 10, 37–49. doi: 10.1111/j.1438-8677.2008.00114.x
Guo, D., Gao, X. R., Li, H., Zhang, T., Chen, G., Huang, P. B., et al. (2008). EGY1 plays a role in regulation of endodermal plastid size and number that are involved in ethylene-dependent gravitropism of light-grown Arabidopsis hypocotyls. Plant Mol. Biol. 66, 345–360. doi: 10.1007/s11103-007-9273-5
Gupta, R., Lee, S. J., Min, C. W., Kim, S. W., Park, K. H., and Bae, D. W. (2016). Coupling of gel-based 2-DE and 1-DE shotgun proteomics approaches to dig deep into the leaf senescence proteome of Glycine max. J. Proteom. 148, 65–74. doi: 10.1016/j.jprot.2016.07.025
Hu, H. Q., Wang, L. H., Wang, Q. Q., Jiao, L. Y., Hua, W. Q., and Zhou, Q. (2014). Photosynthesis, chlorophyll fluorescence, and chlorophyll content of soybean seedlings unde combined stress of bisphenol a and cadmium. Environ. Toxicol. Chem. 33, 2455–2462. doi: 10.1002/etc.2720
Jia, G. Q., Huang, X. H., Zhi, H., Zhao, Y., Zhao, Q., and Li, W. J. (2013). A haplotype map of genomic variations and genome-wide association studies of agronomic traits in foxtail millet (Setaria italica). Nat. Genet. 45, 957–U167. doi: 10.1038/ng.2673
Jia, X. P., Zhang, Z. H., Liu, Y. H., Zhang, C. W., Shi, Y. S., and Song, Y. C. (2009). Development and genetic mapping of SSR markers in foxtail millet [Setaria italica (L.) P. Beauv.]. Theor. Appl. Genet. 118, 821–829. doi: 10.1007/s00122-008-0942-9
Kato, Y., Nagao, R., and Noguchi, T. (2016). Redox potential of the terminal quinone electron acceptor QB in photosystem II reveals the mechanism of electron transfer regulation. Proc. Natl. Acad. Sci. U.S.A. 113, 620–625. doi: 10.1073/pnas.1520211113
Kinch, L. N., Ginalski, K., and Grishin, N. V. (2006). Site-2 protease regulated intramembrane proteolysis: sequence homologs suggest an ancient signaling cascade. Protein Sci. 15, 84–93. doi: 10.1110/ps.051766506
Kong, L. G., Sun, M. Z., Xie, Y., Wang, F. H., and Zhao, Z. D. (2015). Photochemical and antioxidative responses of the glume and flag leaf to seasonal senescence in wheat. Front. Plant Sci. 6:358. doi: 10.3389/fpls.2015.00358
Koussis, K., Goulielmaki, E., Chaliri, A., Withers-Martinez, C., Siden-Kiamos, I., and Matuschewski, K. (2017). Targeted deletion of a plasmodium site-2 Protease impairs life cycle progression in the mammalian host. PLoS One 12:e0170260. doi: 10.1371/journal.pone.0170260
Kusaba, M., Ito, H., Morita, R., Iida, S., Sato, Y., and Fujimoto, M. (2007). Rice non-yellow coloring1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19, 1362–1375. doi: 10.1105/tpc.106.042911
Li, B., Li, Q., Xiong, L., Kronzucker, H. J., Kramer, U., and Shi, W. (2012). Arabidopsis plastid AMOS1/EGY1 integrates abscisic acid signaling to regulate global gene expression response to ammonium stress. Plant Physiol. 160, 2040–2051. doi: 10.1104/pp.112.206508
Li, W., Tang, S., Zhang, S., Shan, J. G., Tang, C. J., and Chen, Q. N. (2016). Gene mapping and functional analysis of the novel leaf color gene siygl1 in foxtail millet [Setaria italica (L.) P. Beauv]. Physiol. Plant. 157, 24–37. doi: 10.1111/ppl.12405
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lu, G., Casaretto, J. A., Ying, S., Mahmood, K., Liu, F., Bi, Y. M., et al. (2017). Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 94, 215–227. doi: 10.1007/s11103-017-0604-x
Martins, P. K., Mafra, V., De Souza, W. R., Ribeiro, A. P., Vinecky, F., and Basso, M. F. (2016). Selection of reliable reference genes for RT-qPCR analysis during developmental stages and abiotic stress in Setaria viridis. Sci. Rep. 6:28348. doi: 10.1038/srep28348
Mukherjee, P., Sureka, K., Datta, P., Hossain, T., Barik, S., and Das, K. P. (2009). Novel role of Wag31 in protection of mycobacteria under oxidative stress. Mol. Microbiol. 73, 103–119. doi: 10.1111/j.1365-2958.2009.06750.x
Nishimura, K., Kato, Y., and Sakamoto, W. (2016). Chloroplast Proteases: updates on proteolysis within and across suborganellar compartments. Plant Physiol. 171, 2280–2293. doi: 10.1104/pp.16.00330
Ort, D. R., Merchant, S. S., Alric, J., Barkan, A., Blankenship, R. E., and Bock, R. (2015). Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc. Natl. Acad. Sci. U.S.A. 112, 8529–8536. doi: 10.1073/pnas.1424031112
Qi, Y. F., Liu, X. Y., Liang, S., Wang, R., Li, Y. F., and Zhao, J. (2016). A putative chloroplast thylakoid metalloprotease VIRESCENT3 regulates chloroplast development in Arabidopsis thaliana. J. Biol. Chem. 291, 3319–3332. doi: 10.1074/jbc.M115.681601
Roberts, I. N., Caputo, C., Criado, M. V., and Funk, C. (2012). Senescence-associated proteases in plants. Physiol. Plant. 145, 130–139. doi: 10.1111/j.1399-3054.2012.01574.x
Rudner, D., Fawcett, P., and Losick, R. (1999). A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. U.S.A. 96, 14765–14770. doi: 10.1073/pnas.96.26.14765
Saito, A., Hizukuri, Y., Matsuo, E., Chiba, S., Mori, H., and Nishimura, O. (2011). Post-liberation cleavage of signal peptides is catalyzed by the site-2 protease (S2P) in bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 13740–13745. doi: 10.1073/pnas.1108376108
Sakamoto, W., and Takami, T. (2014). Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J. Exp. Bot. 65, 3835–3843. doi: 10.1093/jxb/eru091
Sakuraba, Y., Han, S.-H., Lee, S.-H., Hörtensteiner, S., and Paek, N.-C. (2016). Arabidopsis NAC016 promotes chlorophyll breakdown by directly upregulating STAYGREEN1 transcription. Plant Cell Rep. 35, 155–166. doi: 10.1007/s00299-015-1876-8
Sakuraba, Y., Schelbert, S., Park, S. Y., Han, S. H., Lee, B. D., and Andres, C. B. (2012). STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 21, 767–785. doi: 10.1105/tpc.111.089474
Schelbert, S., Aubry, S., Burla, B., Agne, B., Kessler, F., and Krupinska, N.-C. (2009). Pheophytin pheophorbide hydrolase (Pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 24, 507–518. doi: 10.1105/tpc.108.064089
Stirbet, A., Riznienko, G. Y., Rubin, A. B., and Goindjee, P. (2014). Modeling chlorophyll a fluorescence transient: relation to photosynthesis. Biochem. Moscow 79, 291–323. doi: 10.1134/S0006297914040014
van der Hoorn, R. A. (2008). Plant proteases: from phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 59, 191–223. doi: 10.1146/annurev.arplant.59.032607.092835
Waditee-Sirisattha, R., Shibato, J., Rakwal, R., Sirisattha, S., Hattori, A., and Nakano, T. (2011). The Arabidopsis aminopeptidase LAP2 regulates plant growth, leaf longevity and stress response. New Phytol. 191, 958–969. doi: 10.1111/j.1469-8137.2011.03758.x
Weaver, L. M., Gan, S. S., Quirino, B., and Amasino, R. M. (1998). A comparison of the expression patterns of several senescence-associated genes in response to stress and hormone treatment. Plant Mol. Biol. 37, 455–469. doi: 10.1023/A:1005934428906
Wu, X. Y., Hu, W. J., Luo, H., Xia, Y., Zhao, Y., and Wang, L. D. (2016). Transcriptome profiling of developmental leaf senescence in sorghum (Sorghum bicolor). Plant Mol. Biol 92, 555–580. doi: 10.1007/s11103-016-0532-1
Yang, Z., and Ohlrogge, J. B. (2009). Turnover of fatty acids during natural senescence of Arabidopsis, Brachypodium, and switchgrass and in Arabidopsis beta-oxidation mutants. Plant Physiol. 150, 1981–1989. doi: 10.1104/pp.109.140491
Yoshioka-Nishimura, M., Nanba, D., Takaki, T., Ohba, C., Tsumura, N., and Morita, N. (2014). Quality control of photosystem II: direct imaging of the changes in the thylakoid structure and distribution of FtsH proteases in spinach chloroplasts under light stress. Plant Cell Physiol. 55, 1255–1265. doi: 10.1093/pcp/pcu079
Yu, F. W., Zhu, X. F., Li, G. J., Kronzucker, H. J., and Shi, W. M. (2016). The chloroplast protease AMOS1/EGY1 affects phosphate homeostasis under phosphate stress. Plant Physiol. 172, 1200–1208. doi: 10.1104/pp.16.00786
Yu, Y. T., and Kroos, L. (2000). Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J. Bacteriol. 182, 3305–3309. doi: 10.1128/JB.182.11.3305-3309.2000
Keywords: EGY1, Setaria italica, chlorotic mutant, leaf senescence, PS II efficiency
Citation: Zhang S, Zhi H, Li W, Shan J, Tang C, Jia G, Tang S and Diao X (2018) SiYGL2 Is Involved in the Regulation of Leaf Senescence and Photosystem II Efficiency in Setaria italica (L.) P. Beauv. Front. Plant Sci. 9:1308. doi: 10.3389/fpls.2018.01308
Received: 29 November 2017; Accepted: 20 August 2018;
Published: 04 September 2018.
Edited by:
Thomas P. Brutnell, Shandong Agricultural University, ChinaReviewed by:
Ru Zhang, Donald Danforth Plant Science Center, United StatesCopyright © 2018 Zhang, Zhi, Li, Shan, Tang, Jia, Tang and Diao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianmin Diao, ZGlhb3hpYW5taW5AY2Fhcy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.