- 1The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand
- 2The New Zealand Institute for Plant and Food Research Limited, Palmerston North, New Zealand
- 3The New Zealand Institute for Plant and Food Research Limited, Motueka, New Zealand
- 4School of Biological Sciences, The University of Auckland, Auckland, New Zealand
The Vaccinium genus in the family Ericaceae comprises many species, including the fruit-bearing blueberry, bilberry, cranberry, huckleberry, and lingonberry. Commercially, the most important are the blueberries (Vaccinium section Cyanococcus), such as Vaccinium corymbosum (northern highbush blueberry), Vaccinium virgatum (rabbiteye blueberry), and Vaccinium angustifolium (lowbush blueberry). The rising popularity of blueberries can partly be attributed to their “superfood” status, with an increasing body of evidence around human health benefits resulting from the fruit metabolites, particularly products of the phenylpropanoid pathway such as anthocyanins. Activation of anthocyanin production by R2R3-MYB transcription factors (TFs) has been characterized in many species, but despite recent studies on blueberry, cranberry, and bilberry, no MYB anthocyanin regulators have been reported for Vaccinium. Indeed, there has been conjecture that at least in bilberry, MYB TFs divergent to the usual type are involved. We report identification of MYBA from blueberry, and show through sequence analysis and functional studies that it is homologous to known anthocyanin-promoting R2R3-MYBs of subgroup 6 of the MYB superfamily. In transient assays, MYBA complemented an anthocyanin MYB mutant of Antirrhinum majus and, together with a heterologous bHLH anthocyanin regulator, activated anthocyanin production in Nicotiana benthamiana. Furthermore anthocyanin accumulation and anthocyanin structural gene expression (assayed by qPCR and RNA-seq analyses) correlated with MYBA expression, and MYBA was able to transactivate the DFR promoter from blueberry and other species. The RNA-seq data also revealed a range of other candidate genes involved in the regulation of anthocyanin production in blueberry fruit. The identification of MYBA will help to resolve the regulatory mechanism for anthocyanin pigmentation in the Vaccinium genus. The sequence information should also prove useful in developing tools for the accelerated breeding of new Vaccinium cultivars.
Introduction
Vaccinium (family Ericaceae) includes many species with fruit eaten by humans, including blueberries, bilberry, cranberry, huckleberry, and lingonberry. The most important commercial species are the blueberries (Vaccinium section Cyanococcus), notable species of which are Vaccinium corymbosum (northern highbush blueberry) Vaccinium virgatum (rabbiteye blueberry), and Vaccinium angustifolium (lowbush blueberry). Driving market growth of this popularly called “superfood” is the increasing body of evidence around human health benefits resulting from the fruit metabolites, particularly products of the phenylpropanoid pathway such as anthocyanins (Davies and Espley, 2013; Norberto et al., 2013; Shi et al., 2017; Govers et al., 2018), which pigment the fruit. Blueberry species and cultivars show a great variety in the amounts and types of anthocyanins produced in the berries (McGhie et al., 2003; Lohachoompol et al., 2008; Stevenson and Scalzo, 2012; Li et al., 2017).
Central to the direct control of anthocyanin production is the MBW complex, consisting of R2R3MYB and bHLH transcription factors (TFs) and a WD-repeat (WDR) protein (Ramsay and Glover, 2005; Allan et al., 2008; Hichri et al., 2011; Davies et al., 2012). It is the R2R3MYBs that are generally the key factors in determining the spatial and temporal occurrence of anthocyanins (Borevitz et al., 2000; Schwinn et al., 2006; Albert et al., 2011). These are from subgroups (SGs) 5 and 6 of the plant R2R3MYB superfamily. Based on sequences currently available, SG6 types are characteristic of dicot species, but are also found in some non-grass lineages of monocots, while SG5 types are found in grasses and the orchid lineage of monocots (Paz-Ares et al., 1987; Ithal and Reddy, 2004; Hsu et al., 2015; Schwinn et al., 2016). In a rarity, both types have been reported to have roles in peach (Prunus persica), with SG6 types regulating pigment in flowers and fruits (Zhou et al., 2016) and an SG5 type reported to be functioning in petals of an ornamental variety (Uematsu et al., 2014). SG5 also contains R2R3MYBs that activate production of proanthocyanidins (PAs) and their polymers, condensed tannins. PAs are closely related to anthocyanins, and share common precursors in the flavonoid pathway. The SG5 PA regulator is exemplified by Arabidopsis TT2 (Nesi et al., 2001) and grape MYBPA2 (Terrier et al., 2009). There is also another phylogenetically distinct clade of R2R3MYB PA regulators, exemplified by grape PA1 (Bogs et al., 2007).
In recent years, genomics resources have been growing for the Vaccinium genus, including draft genomes for V. corymbosum (Gupta et al., 2015) and the cranberry species V. macrocarpon; Polashock et al., 2014), transcriptomics of developing fruit, and studies aimed at elucidating the control of phenylpropanoid/flavonoid production in the fruit (Li et al., 2012, 2016; Zifkin et al., 2012; Lin et al., 2018). Intriguingly, despite these studies, R2R3MYB SG6/SG5 activators of the anthocyanin pathway have not been identified in blueberries (Jaakola et al., 2010; Zifkin et al., 2012). Moreover, investigations into anthocyanin production in members of the Myrtilis section of Vaccinium, which unlike highbush blueberry and other members of the Cyanococcus section, have taxa with anthocyanin-colored berry flesh in addition to the skin, have resulted in conjecture that R2R3MYBs of the PA1 type are involved (Jaakola et al., 2010; Primetta et al., 2015; Zorenc et al., 2017). This includes VmMYBPA1 (VmMYB2) from bilberry (V. myrtillus), which has an expression pattern that correlates with anthocyanin accumulation during berry development (Jaakola et al., 2010). VmMYBPA1 transcript abundance is reduced in the green sections of fruit in which a berry developmental regulatory gene has been silenced (Jaakola et al., 2010) and in fruit of white-fleshed germplasm (Zorenc et al., 2017). Similarly, MYBPA1 expression is reduced in white-fleshed berries of V. uliginosum (bog bilberry) (Primetta et al., 2015). However, a sequence from V. corymbosum, also named MYBPA1, was proposed to regulate PA and not anthocyanin production, based on the ability to activate promoters of two PA genes, but not that of an anthocyanin specific gene (Zifkin et al., 2012). Thus, there have been outstanding questions on the MYB TFs that control anthocyanin production in Vaccinium.
We present here the identification and characterization of an anthocyanin-promoting R2R3MYB SG6 sequence from blueberry that will help in resolving the regulatory mechanism for anthocyanin pigmentation in the Vaccinium genus. The sequence information should also prove useful in developing tools for the accelerated breeding of new Vaccinium cultivars.
Materials and Methods
Plant Material
Mature rabbiteye blueberry fruit skin and flesh were taken from pooled replicate samples harvested from Plant and Food Research in Motueka. Leaf anthocyanin and gene expression analysis was performed on newly emerged red or green leaves from glasshouse grown rabbiteye blueberry. Flowers from highbush blueberry were taken from the same glasshouse. For anthocyanin analysis, samples were freeze dried before extraction. For qPCR analysis, samples were flash frozen in liquid nitrogen before RNA isolation using the SpectrumTM Plant Total RNA Kit (Sigma) according to the manufacturer’s instructions.
For RNA-seq, rabbiteye blueberry fruit from Plant and Food Research in Motueka, New Zealand, was flash frozen in liquid nitrogen. Three pooled replicate samples of the separated fruit skin and fruit flesh were processed for RNA isolation, as above. RNA-seq library preparation was conducted using the SENSE Total RNA-Seq Library Prep Kit (Lexogen) according to the supplied protocol. Input RNA was poly(A) selected using Dynabeads® Oligo (dT)25 (Ambion®). RNA integrity was assessed using the Agilent RNA 6000 Nano Kit on an Agilent 2100 Bioanalyzer.
Antirrhinum majus plants were grown under standard glasshouse conditions in Palmerston North, New Zealand. The glasshouse was heated at 15°C and vented at 25°C, without supplementary lighting. The roseadorsea line was used, which carries a non-functional allele of the R2R3MYB Rosea1 (Schwinn et al., 2006).
PCR Isolation of MYBA
RNA was extracted from whole pink immature fruit of highbush blueberry using the SpectrumTM Plant Total RNA Kit (Sigma) according to the manufacturer’s instructions. The 3′-RACE PCR amplification (Frohman et al., 1988) used the primers K119 and “short primer” (SP) in the first round and the primers K115 and SP for nested PCR on the first-round products (see Supplementary Table S1 for primer sequences). This generated several cDNAs putatively for the same gene that varied in the length of the 3′-UTR. The remaining 5′ section of the sequence was obtained with 5′-RACE PCR amplification using the Invitrogen 5′-RACE system. First strand cDNA synthesis used the primer K342, which is located in the 3′-UTR. First round PCR used K345 or K346 and the Abridged Anchor Primer (from the kit), followed by two subsequent rounds with the same specific primer and the Abridged Universal Amplification Primer.
Phylogenetic Analysis
The deduced amino acid sequences were aligned using MUSCLE (Edgar, 2004) within Geneious (v 10.0.9) sequence analysis software (Kearse et al., 2012). For the maximum likelihood phylogeny, the aligned sequences were trimmed to include only the R2R3MYB region, because the C-terminal regions do not align between SGs of MYB proteins. The tree was constructed with PhyML (Guindon and Gascuel, 2003), with 1000 bootstrap replicates within Geneious. Sequences used and GenBank accessions: Actinidia chinensis AcMYB110 (AHY00342), AcMYB10 (PSS35990); Arabidopsis thaliana AtMYB4 (At4g38620), AtMYB12 (ABB03913), AtMYB75/PAP1 (AAG42001), AtMYB90/PAP2 (NP_176813.1), AtMYB113 (OAP11934.1), AtMYB114 (AEE34502.1), AtMYB5 (NP_187963.1), AtMYB123/TT2 (CAC40021); Fragaria × ananassa FaMYB1 (AAK84064.1), FaMYB10 (ABX79947.1); Lotus japonicus LjTT2a (AB300033); Malus × domestica MdMYB10 (ACQ45201); Medicago truncatula MtMYB5 (XP_003601609.3), MtLAP1 (ACN79541.1), MtMYB14 (XP_003594801.1); Petunia hybrida PhMYB27 (AHX24372), PhANTHOCYANIN2 (AAF66727.1), PhDEEP PURPLE (ADQ00393.1), PhPURPLE HAZE (ADQ00388.1), PhMYB4 (ADX33331.1); Populus tremuloides PtMYB134 (ACR83705.1); Solanum lycopersicum SlMYB12 (ACB46530.1); Trifolium arvense TaMYB14 (AFJ53053.1); Trifolium repens TrMYB4 AMB27079), TrMYB133 (AMB27081), TrMYB134 (AMB27082); V. corymbosum VcMYBA (this study: MH105054), VcMYBPA1 (JQ085966.1), VcMYB17 partial (ALP43798.1); Vaccinium myrtillus VmMYB2 partial (ADK79068.1); Vaccinium uliginosum VuMYBC2 (AKR80571), VuMYBPA1 (AKC94840.1); and Vitis vinifera VvMYBA1 (BAD18977), VvMYBA2 (BAD18978), VvMYBF1 (ACV81697), VvMYB5b (AAX51291), VvMYBC2-L1 (AFX64995.1), VvMYB-L2 (ACX50288.2), VvMYBPA1 (CAJ90831.1), VvMYBPA2 (ACK56131.1).
Transient Transformation Assays
The vector pSAK277 (Gleave, 1992) containing the 35S:VcMYBA coding sequence with/without 35S:bHLH was transformed into Agrobacterium tumefaciens strain GV3101 by electroporation followed by incubation on plate before infiltration. Nicotiana benthamiana plants were grown under glasshouse conditions using natural light with daylight extension to 16 h as previously described (Espley et al., 2007). Three leaves of 6-weeks-old N. benthamiana were used for infiltration and kept under the same growth conditions. Leaves were photographed and harvested 6 days after infiltration in liquid nitrogen and stored at -80°C until analysis.
Antirrhinum particle bombardment experiments using pKES8 were performed as described in Schwinn et al. (2016). pKES8 was formed by amplifying VcMYBA cDNA using primers K351 (AGT CGA ATT CAT GGA CAT AGT TCC ATT G), which added an EcoRI site next to the ATG that best fit the Kozak consensus sequence, and K352 (ACG TTC TAG AAG CGT AAC AAT CGA TGG A), which added an XbaI site to the end of 3′-UTR. The amplicon was digested and cloned into EcoRI/XbaI sites of pART7 (Gleave, 1992), putting it under the control of the CaMV35S promoter. The control was a CaMV35S:GFP-ER construct (Haselhoff et al., 1997), which localizes GFP to the endoplasmic reticulum. The white adaxial surface of the dorsal petals was bombarded using 300 or 400 kPa helium pressure. After bombardment, the petals were then cultured on half-strength MS medium (Murashige and Skoog, 1962) under 20–50 μmol m-2 s-1 light from Osram 36 W grolux fluorescent tubes (16 h photoperiod) at 25°C. At least two flowers were used for each construct per experiment, and each experiment was repeated at least twice.
Promoter Activation Assays in Tobacco
Promoter fragments for DFR from Arabidopsis, apple, and rabbiteye blueberry, containing 1909, 1647, and 646 bp, respectively, upstream of the start code ATG of each candidate gene were isolated by PCR. Fragments were inserted into the cloning site of pGreenII 0800-LUC (Hellens et al., 2005) and modified to introduce an NcoI site at the 3′ end of the sequence. This allowed the promoters to be cloned as a transcriptional fusion with the firefly luciferase gene (LUC). The promoter-LUC fusion in pGreenII 0800-LUC was used in transient transformation by mixing 100 μL of Agrobacterium strain GV3101 (MP90) transformed with the reporter cassette with 450 μL each of two other Agrobacterium cultures. These cultures had been transformed with cassettes containing a cDNA of MYB TF gene or a bHLH TF gene fused to the 35S promoter, respectively, in either pSAK277 or pHex2 (Hellens et al., 2005). N. benthamiana growing conditions, Agrobacterium infiltration processes and luminescent measurements were as described by Hellens et al. (2005).
Analysis of Transcript Abundance Using qPCR
Quantitative RT-PCR (qPCR) was used to measure transcript abundance for MYBA and DFR in five blueberry tissue types: berry flesh and skin (rabbiteye), green and red leaves (rabbiteye), and flowers (highbush). cDNA was synthesized according to the manufacturer’s recommendations (QuantiTect® Reverse Transcription Kit, QIAGEN). qPCR was conducted using the LightCycler® SYBR Green I Master (Roche LightCycler® 480 System). Data shown are mean expression from four technical replicates of three biological replicates for each tissue type. Expression level reported is relative to the housekeeping gene Actin. Gene specific primers are listed in Supplementary Table S1. Actin and DFR primers are from Zifkin et al. (2012). Primer efficiencies were calculated using serial dilution. Reactions contained 2.5 μL Master Mix, 0.25 μL of each primer (10 μM), 1.25 μL diluted cDNA (1:25), and nuclease-free water (Roche Diagnostics) to a total volume of 5 μL, using reaction conditions previously reported (Espley et al., 2007) Analysis was performed using LightCycler software (Roche; version 1.5.0 SP4).
HPLC Analysis of Anthocyanins
Approximately 500 mg of each sample of rabbiteye tissue (berry flesh and skin) was extracted in 5 mL ethanol: MilliQ: formic acid (80:20:1), with mixing at room temperature on a rotary shaker for 30 min, and incubation overnight at 1°C. Samples were brought to room temperature then centrifuged for 10 min at 3000 rpm; 500 μL from each sample were combined to create a composite sample for quality control. Samples were diluted either two or 10 times and 1-mL aliquots were analyzed by HPLC. The composite was diluted 10 times.
The HPLC system consisted of a Dionex Ultimate 3000 RS system: a SRD-3400 degasser, HPG-3400RS pump module, and WPS-3000 autosampler connected to a DAD-3000RS diode array detector under the control of Chromeleon software (version 7.2.3.7553, Thermo Fisher, United States). Separation was carried out on a Zorbax SB-C18 2.1 mm × 150 mm, 2.1-μm column (Agilent, United States) with the following gradient program: 0–0.5 min, 5% B; 0.5–10 min, 5–20% B; 10–15 min, 20–95% B; 15–16.5 min, 95% B; 16.5–16.8 min, 95–5% B; 16.8–20 min, 5% B. Solvent percentage was made up to 100% with A, where A = formic acid/MilliQ (5/95) and B = acetonitrile. The flow rate was 0.350 mL/min and the injection volume was 2 μL. Anthocyanin components were detected at 530 nm and peaks identified by comparison of peaks to authentic standards and previous reports of blueberry anthocyanins. The identifications were confirmed by mass spectrometry of a single sample.
Infiltrated leaf patches of N. benthamiana were harvested, weighed, and snap frozen in liquid nitrogen before freeze drying for 24 h. Leaf samples were then powdered, extracted, and analyzed by HPLC using the same protocol described for blueberry skin and flesh. Anthocyanin components were detected at 530 nm and quantified by comparison to authentic standards using a five point calibration curve.
RNA-seq Differential Expression Analysis
Next-Generation Illumina sequencing with 100 bp paired end reads was performed by The Australian Genome Research Facility (Melbourne, Australia). Raw RNA-seq read data were pre-processed by trimming the raw reads (497,170,448 reads), filtering out contaminants, adapters, and low-quality base calls (<20), and quality checking (6.28% of reads filtered out as either homopolymer or <50 bp after clipping). The cleaned data were aligned to the gene models extracted from the published V. corymbosum genome1 using the .bed file for the genes provided. These alignments were counted (by fragment) and subjected to differential expression analysis using the DESeq2 analysis software package (Love et al., 2014). The resulting 48,637 differentially expressed genes were annotated with BLAST against A. thaliana proteins. To obtain results with known annotations of transcripts and/or genes, the cleaned data were aligned to the V. corymbosum transcriptome2, which is annotated, removing the requirement for annotation in the analysis. The counts from these alignments were again analyzed for differential expression with DESeq2, and the resulting 39,461 differentially expressed transcripts were combined with the annotations for the transcriptome. Notably, some transcripts in this transcriptome, and also in the set of the differentially expressed transcripts, did not have annotation provided. These will require manual annotation if required for further analysis. All data presented for differential gene expression were with a cut-off value of 2 log2fold.
Results
Identification of a Blueberry cDNA for an R2R3MYB Subgroup 6 Factor
BLAST analysis of an in-house V. corymbosum fruit EST library did not reveal a strong candidate for a R2R3MYB anthocyanin regulator. Therefore, 3′-RACE was used on cDNA from RNA derived from immature pink-colored whole fruit, using degenerate primers sited in the conserved MYB domain of characterized anthocyanin-related sequences. This amplified a candidate anthocyanin-related MYB sequence. The sequence was extended by 5′-RACE and a full ORF amplified. The gene was named MYBA (GenBank Accession MH105054). Transcript for MYBA was also present in subsequently obtained berryfruit RNA-seq data from rabbiteye blueberry (gene model 38459, Table 2 and Supplementary Figure S1). Interestingly, our examination of the draft cranberry genome (Polashock et al., 2014) and transcriptome of its fruit (Sun et al., 2015) revealed MYBA to be present and expressed (Supplementary Figure S1), something that was not reported on in those publications.
Phylogenetic analysis was performed on MYBA and published Vaccinium R2R3MYB sequences against characterized flavonoid-related R2R3MYB sequences from other dicot species. The tree (Figure 1) was formed on an alignment of the deduced amino acid sequence for the MYB domain only. R2R3MYB sequences that activate the biosynthesis of different flavonoid classes separated into distinct clades, specifically for PAs (SG5), anthocyanins (SG6), flavonols (SG7), and R2R3MYB repressors (SG4). MYBA fell in SG6, while the previously identified Vaccinium sequences formed part of other clades: VuMYBC2 in SG4, VcMYB17 in SG5, and VmMYB2, VcMYBPA1, and VuMYBPA1 clustering with the grape sequence VvMYBPA1.
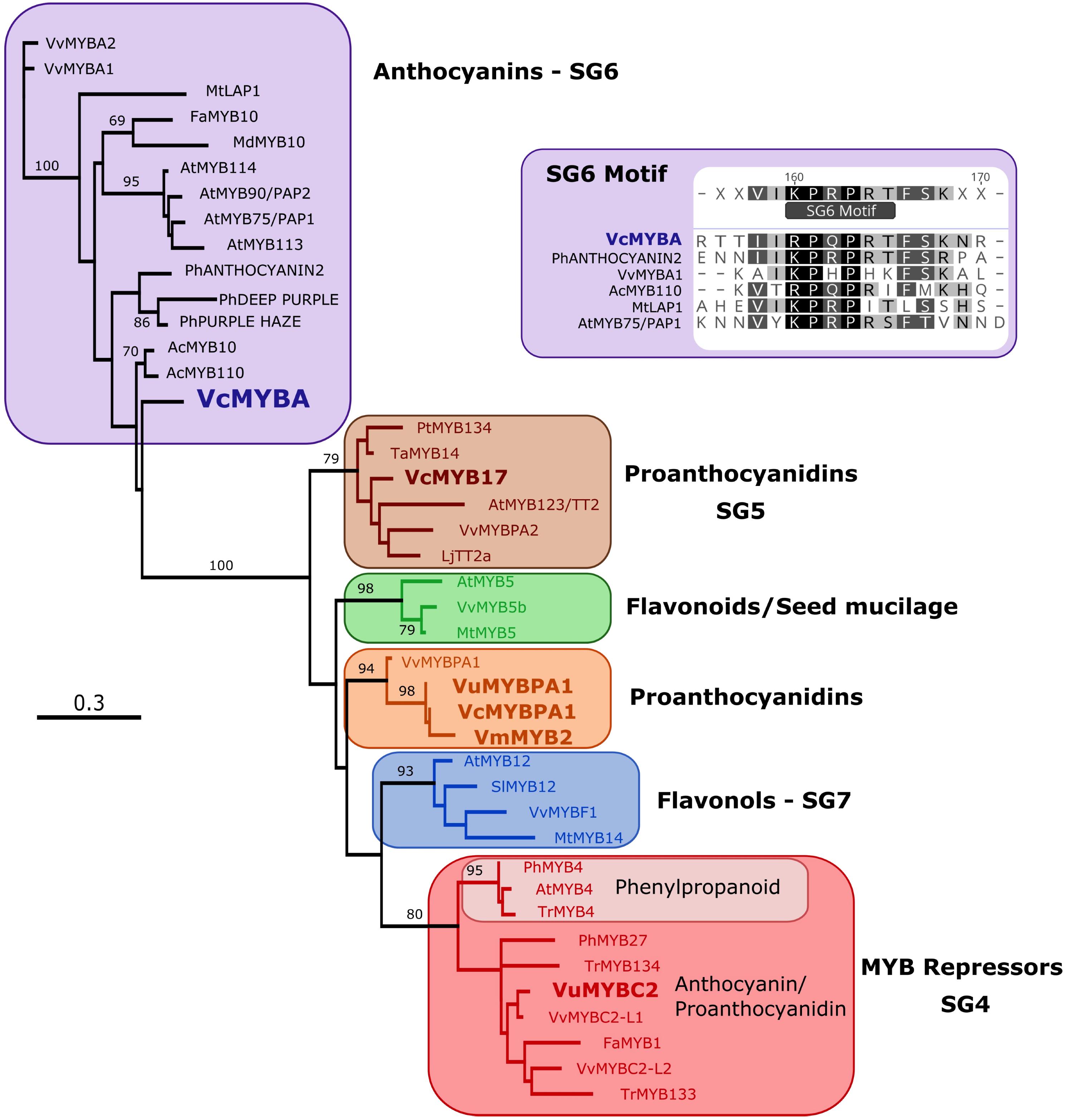
FIGURE 1. MYBA is a subgroup-6 R2R3MYB protein. A maximum likelihood phylogenetic tree of R2R3MYB sequences was generated using an alignment of the deduced amino acid sequences for the MYB domain. Sequences from Vaccinium species are in bold type. Node support > 65% from 1000 bootstrap replicates is shown. The inset shows that MYBA has the motif that is present in the variable C-terminal region of subgroup-6 anthocyanin regulators (Stracke et al., 2001; Schwinn et al., 2016).
The MYB domain of MYBA contains the residues identified as being key for R2R3MYB interaction with bHLH proteins, including the motif [D/E]Lx2[R/K]x3Lx6Lx3R (Zimmermann et al., 2004; Supplementary Figure S2). Of the amino acid residues within the MYB domain that distinguish between R2R3MYB of SG5 and 6 (Lin-Wang et al., 2010; Schwinn et al., 2016), VcMYBA contained five of the six SG6 residues (Supplementary Figure S2), the exception being an N at position 82, which is also an N in CmAN1. Of the SG5/6 distinguishing amino acid residues within the variable C-terminal region (Stracke et al., 2001; Yamagishi et al., 2010; Schwinn et al., 2016), MYBA contained the SG6 residues (Figure 1 and Supplementary Figure S2). In contrast, the published flavonoid-related R2R3MYB sequences from Vaccinium, VcMYBPA1, VmMYBPA1, and VuMYBPA1, have the SG5 characteristic residues in the MYB domain (Supplementary Figure S2).
A search of all non-redundant GenBank protein sequences gave a 97% identity score to a partial V. corymbosum sequence (ALP43799) that may be an allele of MYBA or a member of a gene family with MYBA.
MYBA Activates Anthocyanin Synthesis in planta
The phylogenetic analysis suggested that MYBA was associated with anthocyanin regulation. We tested the ability of MYBA to complement the A. majus roseadorsea mutant (Schwinn et al., 2006), which has been used previously to test the activity of a range of SG6 anthocyanin MYB genes from diverse plant species (Albert et al., 2011, 2015; Schwinn et al., 2016). Biolistic transformation of roseadorsea petals with GFP-ER alone was not capable of inducing pigmentation in transformed cells (Figure 2). Conversely, MYBA restored pink anthocyanin pigmentation to the transformed cells, which were identified by the GFP internal control (Figure 2). These assays were extended to a cultivar of highbush blueberry with white flowers. The transformation efficiency of petal cells was very low compared with that of Antirrhinum, as determined by the relatively infrequently occurring GFP-fluorescing foci. Nevertheless, while transformation with GFP-ER alone did not induce anthocyanin pigmentation, co-transforming with 35S:VcMYBA resulted in some pink pigmentation (Supplementary Figure S3), suggesting that MYBA does regulate anthocyanin biosynthesis in highbush blueberry.
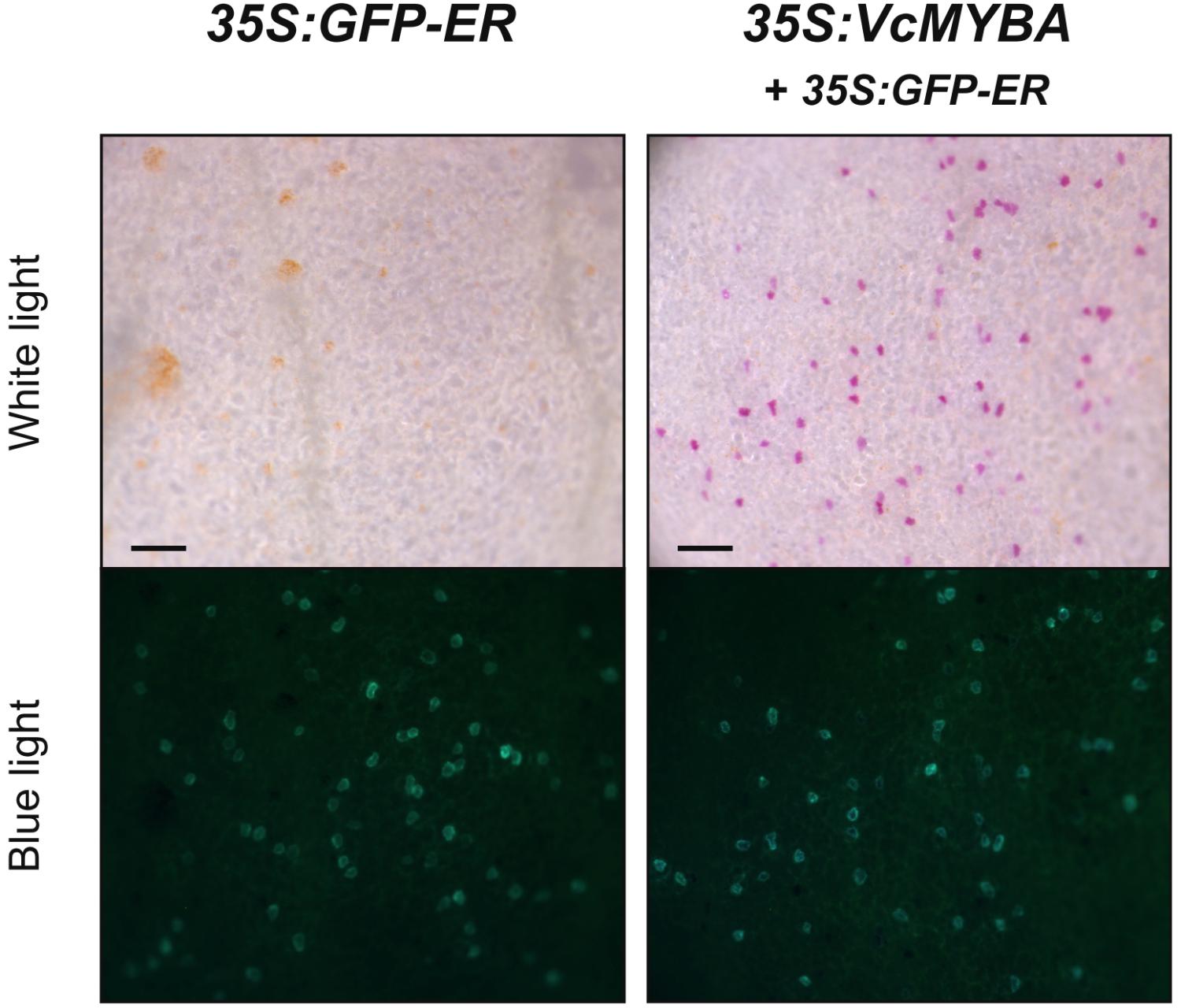
FIGURE 2. MYBA restores anthocyanin biosynthesis in Antirrhinum roseadorsea. VcMYBA under the control of the CaMV35S promoter was biolistically introduced into white adaxial petal tissue of the A. majus line roseadorsea, which carries a mutation in the main anthocyanin-promoting R2R3MYB Rosea1. Complementation is seen as restoration of pigmentation to single cells. GFP-ER was co-transformed as an internal control and viewed with blue light. Scale bar = 200 μm.
To further characterize the activity of MYBA, we performed transient Agrobacterium infiltration assays on N. benthamiana leaves (Figure 3). This landrace of N. benthamiana was used as it is capable of producing anthocyanin pigments when anthocyanin regulatory genes are expressed (Bally et al., 2015). In positive control experiments infiltration with the characterized apple anthocyanin regulator MdMYB10 together with MdbHLH3 resulted in purple pigmentation, and the intensity of the pigmentation was greater with the peach regulators, PpMYB10 and PpbHLH3, than with the apple orthologs (Figure 3A). Infiltration of blueberry MYBA with either MdbHLH3 or PpbHLH3 resulted in even more intense purple pigmentation than in the positive control experiments. The intensity of coloration observed in the infiltrated leaves (Figure 3A) correlated with the amounts of anthocyanin pigments present in the tissue (Figure 3B). Over 95% of the anthocyanin in the leaves was identified as delphinidin 3-O-glucoside; however, a minor peak (λmax 536 nm) was also present on the chromatogram that may represent a second anthocyanin species, which could not be identified by comparison to the reference standards.
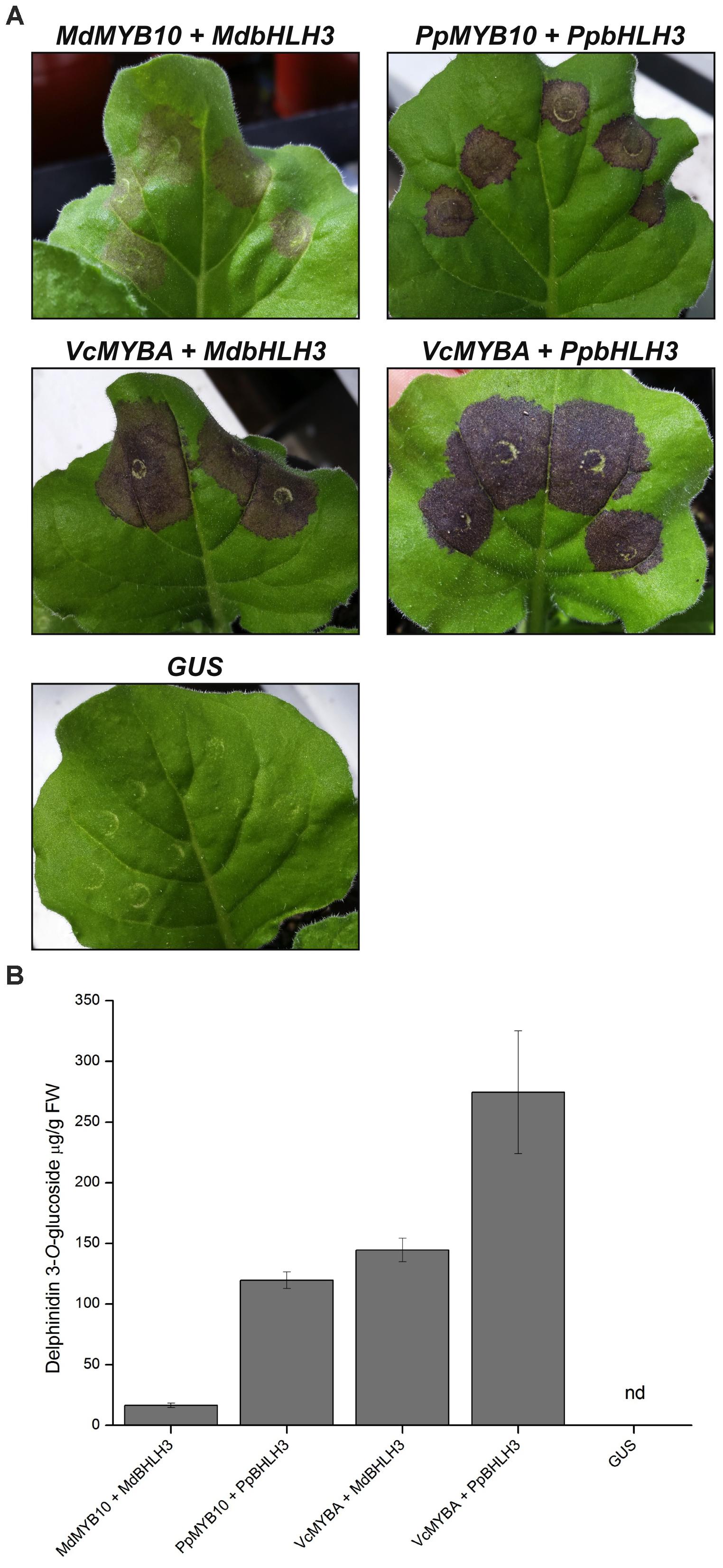
FIGURE 3. Heterologous expression of MYBA in Nicotiana benthaminana induces anthocyanin accumulation. (A) Agrobacterium infiltration of N. benthamiana leaves with SG6 R2R3MYB transcription factors MdMYB10 (apple), PpMYB10 (peach), or VcMYBA (blueberry) co-infiltrated with bHLH partners MdbHLH3 or PpbHLH3 induces pigmentation. 35S:GUS was included as a negative control. (B) Anthocyanin quantification from infiltrated tissues. Nd = not detected. Mean of three biological replicates. Error bars represent ± SEM.
MYBA Activates Promoters of a Key Anthocyanin Biosynthetic Gene
To test the functionality of MYBA, we used the dual luciferase promoter activation assay in N. benthamiana. We chose a key gene in the anthocyanin pathway, dihydroflavonol 4-reductase (DFR), and isolated promoters from Arabidopsis, apple, and blueberry, which were each fused to a luciferase reporter. Promoters alone were infiltrated into N. benthamiana to check for background activity and also co-infiltrated with MYBA alone or with known anthocyanin-associated bHLH factors from apple (MdbHLH3) or peach (PpbHLH3) (Figure 4). MYBA alone led to strong activation of all three DFR promoter fusions. The addition of MdbHLH3 further increased this activation of the Arabidopsis promoter. However, the addition of bHLH did not appear to have an effect on the apple promoter (with PpbHLH3) or on the blueberry promoter, (with PpbHLH3 or MdbHLH3). This suggests that MYBA is able to activate the DFR promoter from all three species without the dependence on a co-infiltrated bHLH cofactor. This is in contrast to apple MdMYB10, which is reliant on a co-infiltrated bHLH for DFR promoter activation (Espley et al., 2007). However, it is likely that endogenous N. benthamiana bHLH TFs (Montefiori et al., 2015) act as the co-factor with MYBA to drive DFR activation. These results demonstrate that MYBA is able to strongly activate the DFR promoter from blueberry and is capable of recognizing the promoters from heterologous hosts.
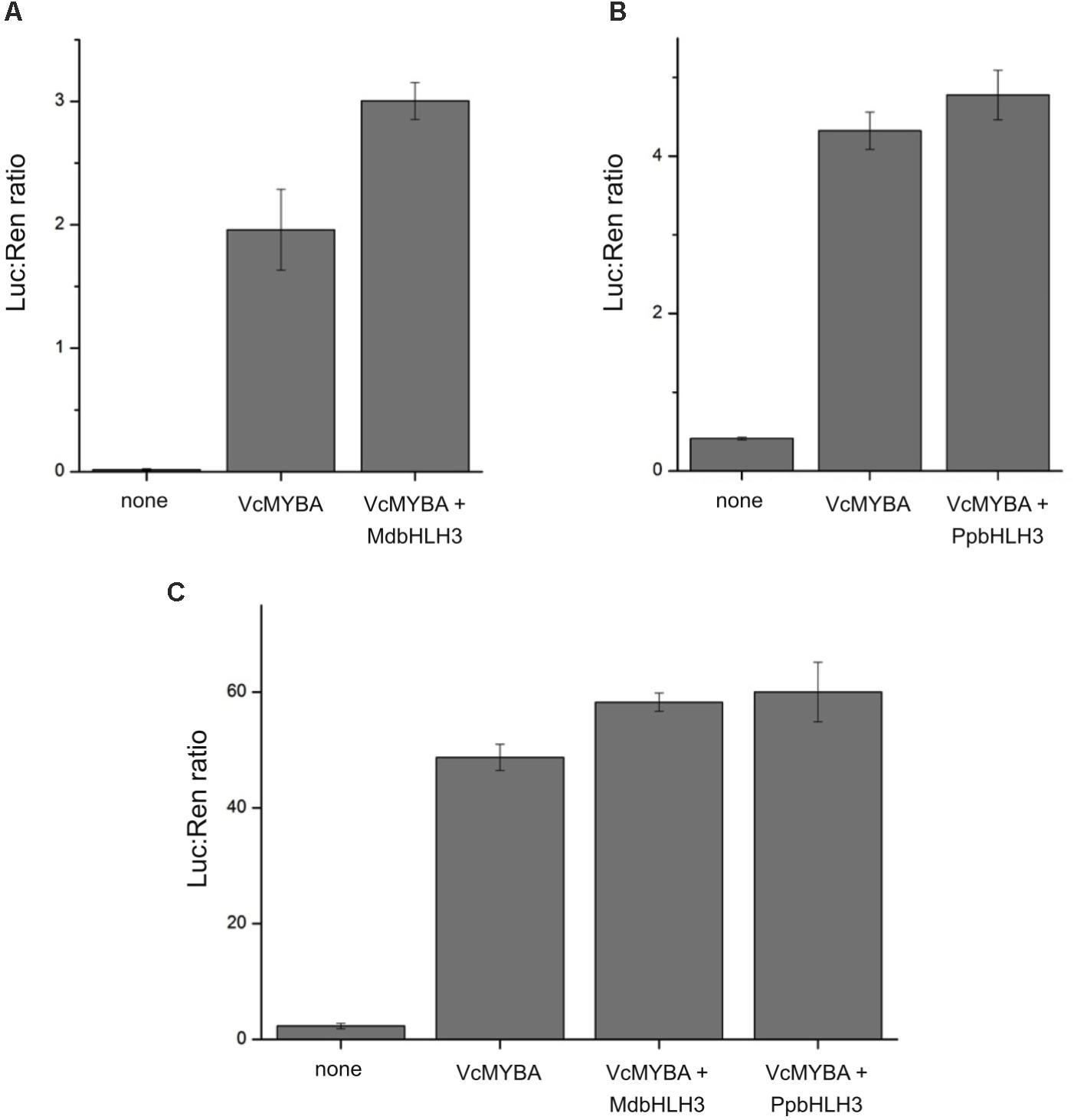
FIGURE 4. MYBA activates the promoter of dihydroflavonol 4-reductase. The dihydroflavonol 4-reductase (DFR) promoter sequences from (A) Arabidopsis (pAtDFR), (B) apple (pMdDFR), and (C) blueberry (pVcDFR) were isolated and cloned into dual luciferase reporter constructs. Promoter activation assays were performed by Agrobacterium infiltration of effector and reporter constructs into Nicotiana benthamiana leaves. Combinations of VcMYBA together with either MdbHLH3 from apple or PpbHLH3 from peach were tested. Mean of three biological replicates. Error bars represent ± SEM.
Anthocyanin and Phenylpropanoid Biosynthetic Genes Are Differentially Expressed in the Skin and Flesh of Blueberry Fruit
The fruit of blueberries (highbush and rabbiteye) are rich in anthocyanins, but this is restricted to the fruit skin, with the flesh tissues containing only trace amounts (Figure 5A). To gain additional information on anthocyanin production in the fruit and to identify other genes possibly involved in anthocyanin regulation, RNA-seq analysis was conducted on skin and flesh samples of rabbiteye blueberries, and differentially expressed genes were identified using DESeq2. Phenylpropanoid structural genes that provide precursor for anthocyanin production along with all the core structural genes in the anthocyanin biosynthesis pathway showed greater than 2 log2fold differences (Table 1). This was apparent from the first precursor step of phenylalanine ammonia-lyase (PAL) onward, with a number of gene models showing between 4 and 5.8 log2fold increase in fruit skin compared with flesh. These increases included cinnamate 4-hydroxylase (C4H) and 4-coumarateCoA ligase (4CL), representing the next steps after PAL and which together provide the precursor products for use by the first committed enzyme in the flavonoid pathway, chalcone synthase (CHS). CHS produces naringenin chalcone and showed as much as 5.7 log2fold difference. The subsequent isomerization of naringenin chalcone by chalcone isomerase (CHI) to produce naringenin showed a 3.4 log2fold difference. The hydroxylation steps performed by flavanone 3-hydroxylase (F3H), which converts naringenin to dihydroflavonol, and the B-ring hydroxylases F3′H and F3′5′H, which determine the type of dihydroflavonol formed, were also well represented in the upregulated gene set. Two gene models for DFR, which reduces dihydroflavonol to leucoanthocyanidin, were also shown to be strongly upregulated in the skin, as was leucoanthocyanidin dioxygenase (LDOX), which produces anthocyanidin. One gene model for the glycosylation step to anthocyanin, uridine diphosphate (UDP)-glucose:flavonoid 3-O-glucosyltransferase (UFGT), was upregulated by 2.3 log2fold in fruit skin.
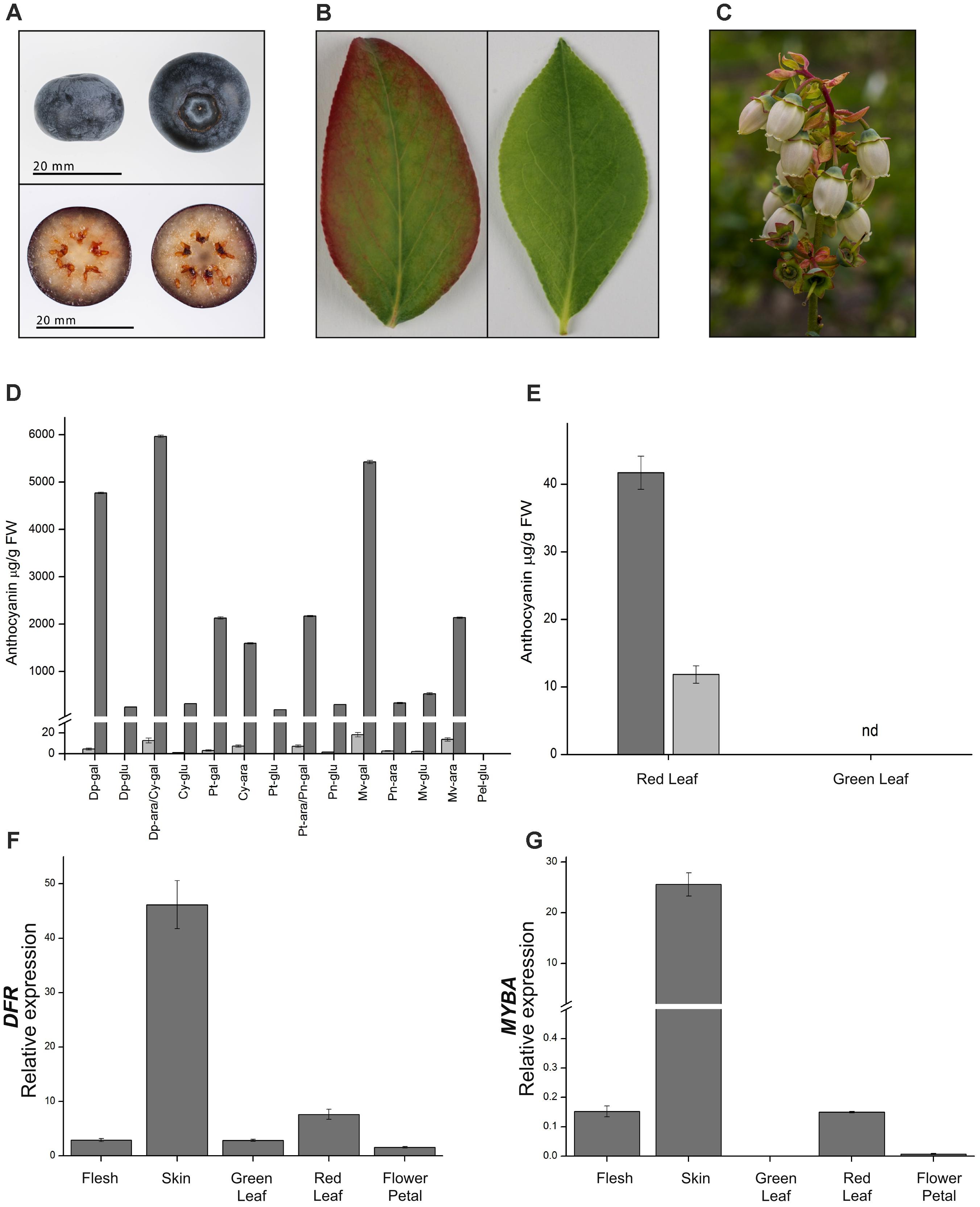
FIGURE 5. Anthocyanin accumulation and gene expression in blueberry fruits, leaves, and flowers. (A) Representative berry as used at mature stage. (B) Representative leaves at the same developmental stage showing presence (left) or absence (right) of anthocyanin pigmentation. (C) Flowers showing little, if any, anthocyanin. (D) Anthocyanin composition and concentration for fruit flesh (light gray bars) and skin (dark gray bars). Cy, cyanidin; Pn, peonidin; Dp, delphinidin; Pt, petunidin; Mv, malvidin; ara, arabinoside; gal, galactoside; glu, glucoside; nd, not detectable. Compounds Dp-ara/Cy-gal and Pt-ara/Pn-gal were not distinguishable at the resolution used for this analysis. (E) Anthocyanin composition in red and green leaves, cyanidin 3-galactoside (dark gray bars), and cyanidin 3-O-glucoside (light gray bars). Relative gene expression of (F) DFR or (G) MYBA in fruit flesh and skin, green and red leaves, and flower petals. Mean of three biological replicates. Error bars represent ± SEM. Rabbiteye and highbush blueberry used for A/B and C, respectively.
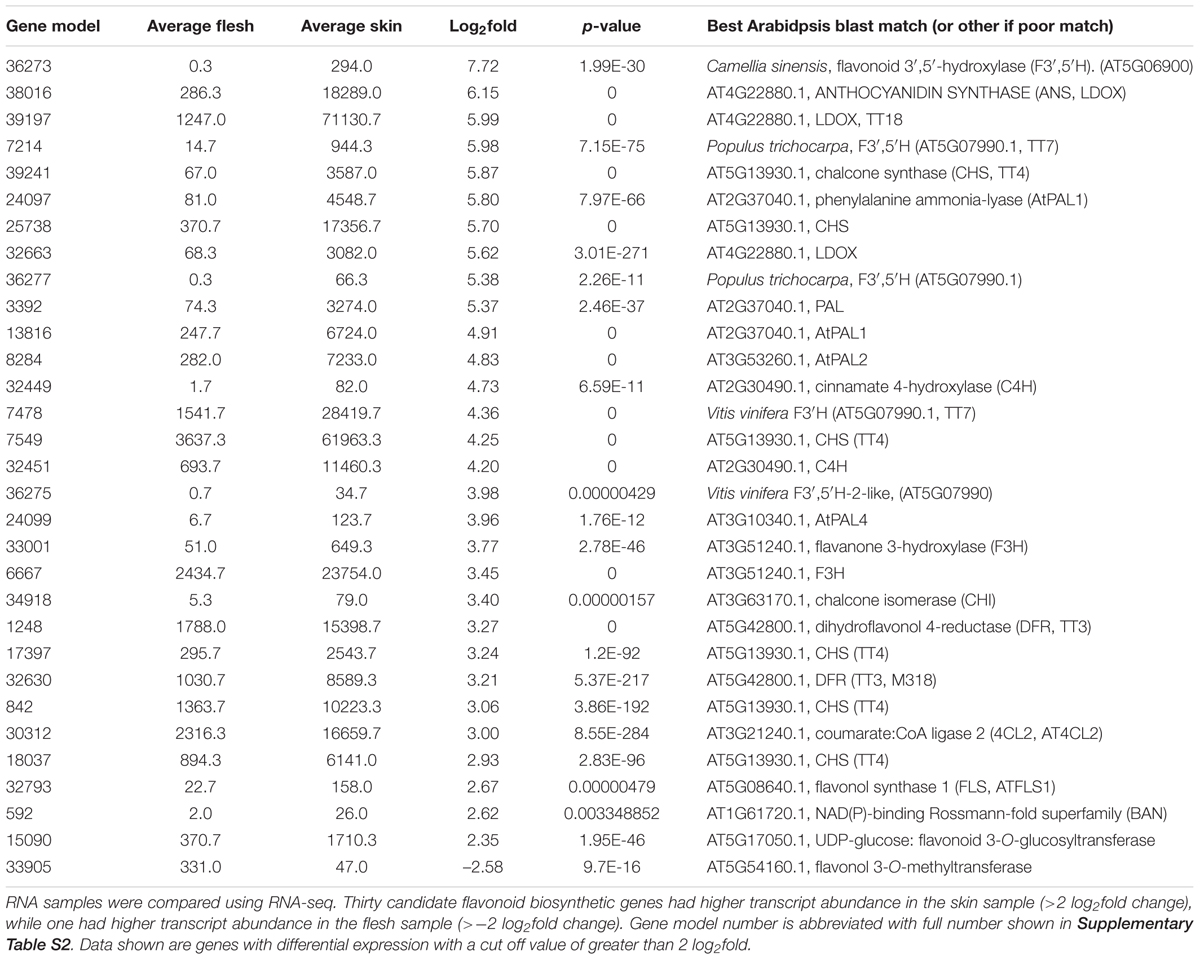
TABLE 1. Comparative abundance of transcripts encoding flavonoid and phenylpropanoid biosynthetic genes between skin and flesh of blueberry fruit.
RNA-seq Identifies Additional Candidates for Anthocyanin Regulation
Results are presented in Table 2. The most differentially expressed MYB TF in the dataset was MYBA (gene model 38459; Supplementary Figure S1), which was present at 97 fold higher in skin (a 6.6 log2fold differential). This further supports a role for MYBA in anthocyanin production. A gene ontology search for other TFs differentially expressed in the fruit skin and flesh revealed a number of transcripts corresponding to different TF families. Of these, 38 were more highly expressed in skin than in flesh and 17 higher in flesh. Of interest was the highly differentiated gene model 17527, which showed closest BLAST match to At1G22640, which encodes AtMYB3. In Arabidopsis, this SG4 repressor motif-containing R2R3MYB has been shown to be an active repressor of phenylpropanoid biosynthesis, acting via direct repression of the C4H gene, and its overexpression reduces anthocyanin accumulation (Zhou et al., 2017). Other highly differential MYBs included genes probably homologous to AtMYB94, involved in wax biosynthesis regulation; AtMYB5, a repressor of trichome branching; AtMYB36 and AtMYB58, both associated with lignin regulation; and AtMYB60, involved in the light-induced opening of stomata.
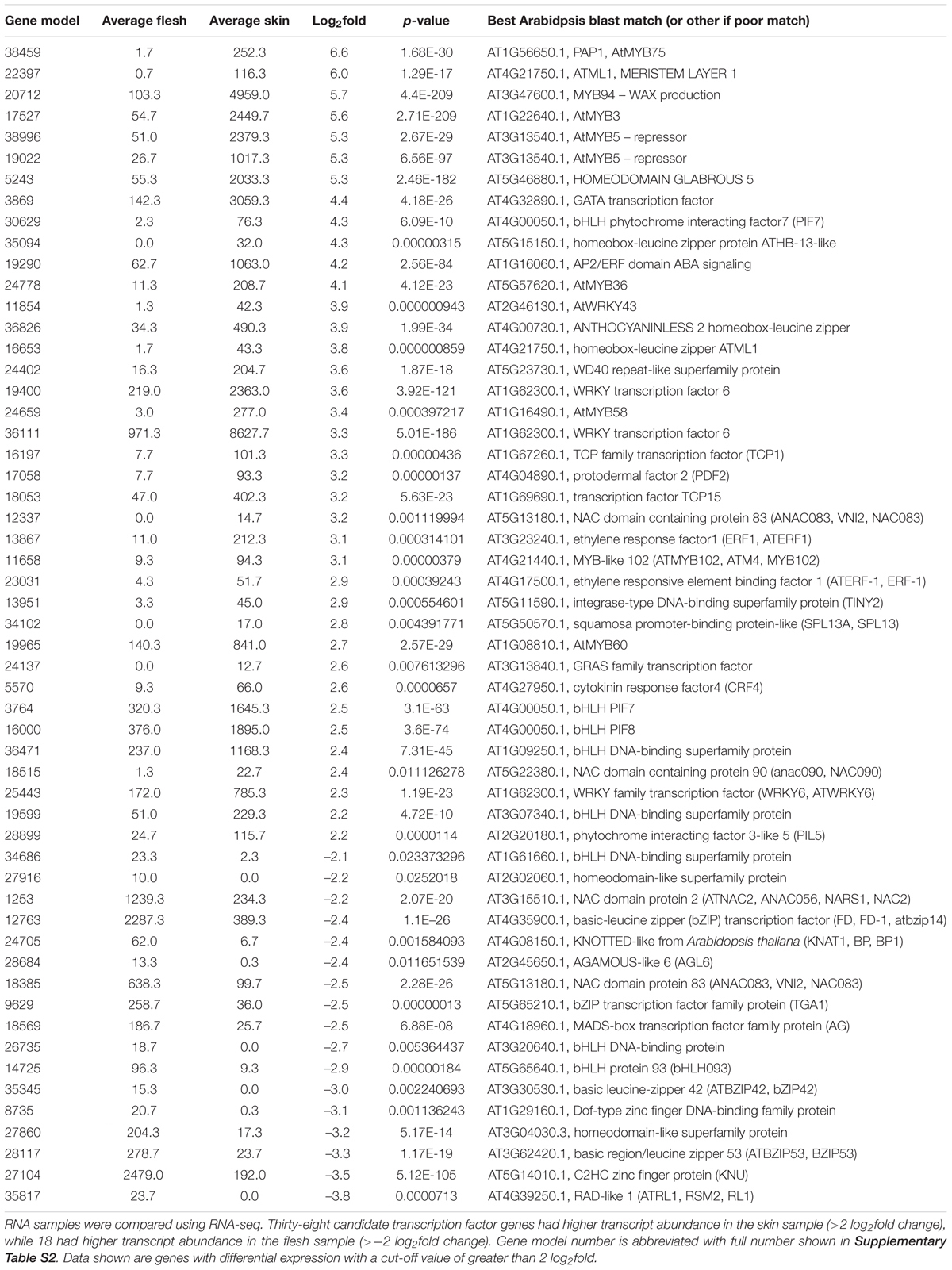
TABLE 2. Comparative abundance of transcripts corresponding to transcription factors between skin and flesh of blueberry fruit.
A number of bHLH TF gene models were strongly differential in the two tissues, including phytochrome-interacting factor 7 and phytochrome-interacting factor 3-like 5 (PIL5) which were in greater abundance in skin. Two bHLH gene models were more highly represented in flesh. Other TF classes represented included NACs, WRKYs, ERFs, and bZIPs, all classes with examples of potential association with anthocyanin regulation. Also of note was the homeodomain Anthocyaninless2 (ANL2)-like gene model (36826), with a 3.9 log2fold differential. This gene controls anthocyanin accumulation in sub-epidermal tissues in Arabidopsis (Kubo et al., 1999) and may act upstream of the MBW complex (Kubo et al., 2008), although this remains unresolved.
MYBA Transcript Abundance Correlates With Anthocyanin Production
Blueberries (highbush and rabbiteye) produce anthocyanins in a range of tissues. In fruit, anthocyanins are produced during ripening and are limited to the skin, while the flesh is essentially acyanic (Figure 5A). Mature rabbiteye blueberry leaves are uniformly green in color; however, newly formed leaves sometimes have a pinkish red blush around the leaf margins (Figure 5B). Blueberry flowers commonly have white or weakly blushed petals (Figure 5C). Analysis of anthocyanin content by HPLC identified a variety of anthocyanins in fruit skin, which were predominantly purple-blue trihydroxylated delphinidin-derived (Figure 5D). In contrast, the anthocyanin profile of red leaves was much simpler, consisting of cyanidin 3-O-galactoside and cyanidin 3-O-glucoside, which were not detected in green leaves (Figure 5E).
The RNA-seq data showed that MYBA expression was highly differential between the pigmented skin and acyanic flesh of the berry. To more widely examine the association of MYBA expression with anthocyanin accumulation patterns in blueberry, we conducted qPCR analysis and included the key biosynthetic gene DFR (Figures 5F,G). The anthocyanin-rich berry skin had the highest relative expression of DFR and MYBA, and anthocyanin-containing red leaves also displayed higher DFR and MYBA expression than green leaves. Expression levels of the genes in red leaves were over 6 (DFR) and 150 fold (MYBA) lower than in the skin, possibly reflecting the considerably lower concentrations of total anthocyanins. Whilst the relative expression of MYBA was significantly higher in red leaves than in green leaves, its relative amount was similar to that in the acyanic berry flesh. This might indicate that MYBA does not regulate anthocyanin accumulation in the leaves, or might suggest that this relatively low level of MYBA expression is not sufficient to drive the pathway flux toward anthocyanin accumulation in blueberry flesh.
Discussion
In dicot species characterized to date, R2R3MYBs of SG6 are the key activators of the flavonoid biosynthetic pathway for the production of anthocyanin pigments in fruit. Nevertheless, previous studies on regulation of the anthocyanin pathway in the Myrtilis section of Vaccinium have suggested that R2R3MYBs of a clade of PA regulators (separate to either SG5 or SG6) fulfill this role in these species. However, our collective findings presented here on blueberry taxa (Vaccinium section Cyanococcus) agree with the data for other dicots, and identify the SG6 R2R3MYB MYBA as an anthocyanin regulator and the probable central activator of berry skin pigmentation.
In phylogenetic analysis, MYBA separated clearly in the SG6 clade, while the previously identified Vaccinium sequences were placed in other clades. Moreover, the deduced amino acid sequence of MYBA had the conserved motifs identified for SG6 proteins. The MYBA cDNA sequence was able to functionally substitute for the endogenous R2R3MYB anthocyanin regulator of the model species A. majus. Additionally, MYBA expression correlated with anthocyanin production and MYBA activated promoters of the anthocyanin biosynthetic gene DFR, both from blueberry and from other species (Arabidopsis and apple). This activation was (variably) increased when MYBA was co-introduced with a heterologous bHLH partner, and MYBA together with a heterologous bHLH partner gave effective induction of anthocyanin production in N. benthamiana leaves. These data also provide strong evidence for MYBA acting within a MYB-bHLH-WDR complex as shown for similar anthocyanin regulators of other species.
Confirming that MYBA is the central, coordinating activator of blueberry fruit skin pigmentation will require additional genetic evidence. However, we did not find evidence supporting a role for other types of R2R3MYB in anthocyanin pathway activation. There were no SG5 candidate sequences with significant differential expression in the skin. The other R2R3MYB sequences that did have higher abundance in the skin transcriptome are candidate sequences for flavonoid pathway repressors, which in other species are known to interact with the MBW complex to modulate anthocyanin production. Other transcripts corresponding to TFs with a characterized role in negatively regulating flavonoid biosynthesis were also more abundant in the skin, such as for squamosa promoter binding protein-like sequences.
We do not know why SG6 R2R3MYB anthocyanin regulators have not been identified previously in Vaccinium species. One possibility is that there is variation in the presence of SG6 genes across the genus. However, MYBA is present in the draft cranberry genome (Polashock et al., 2014) and is expressed (Sun et al., 2015; Supplementary Figure S1). As cranberry is in a different lineage to blueberry, being part of the Oxycoccus section of Vaccinium, this suggests widespread occurrence of MYBA-like genes in the genus. However, this does not preclude the Myrtillis lineage from having divergent regulatory factors. Alternatively, the SG6 genes could have narrow windows of active expression during berry development, so that previous transcriptome studies have not been at the appropriate developmental stage.
All dicot species studied in detail to date have a small multi-gene family for the SG6 anthocyanin regulatory genes, with different family members having distinct expression profiles to generate tissue-specific or developmental variations in pigmentation (Schwinn et al., 2006; Walker et al., 2007; Albert et al., 2011, 2015). In grape, berry color is primarily controlled by VvMYBA1 and VvMYBA2, which exist within a complex locus on chr2 that contains 4 SG6 R2R3MYB genes (Walker et al., 2007), while vegetative pigmentation patterns are determined by a locus on chr14 that contains three SG6 MYB genes, with VvMYBA7 linked to bud coloration and VvMYBA6 to pigmented leaves and tendrils (Matus et al., 2017). Our PCR and transcriptome analyses for blueberry identified only a single transcript type encoding a functional SG6 protein. However, as MYBA was relatively lowly expressed in blueberry leaf tissues (Figure 5E), it is expected that another MYB gene family member is responsible for controlling pigmentation in vegetative tissues. In petunia for example, a specific gene family member PURPLE HAZE is light-regulated in vegetative tissues and in immature flower buds, conferring pigmentation to stems and leaves and a blush phenotype to flowers (Albert et al., 2011), with additional MYB genes responsible for controlling full flower color during development (Quattrocchio et al., 1999). In blueberry, young or stressed leaves produce anthocyanins (Figure 5B) and immature berries are also blushed with anthocyanin on the sun-exposed side, and only later develop fully pigmented fruit during maturation (Zifkin et al., 2012). Interestingly, the anthocyanins detected in blueberry leaves were cyanidin-based (Figure 5E), while berry skin has predominantly trihydroxylated anthocyanins (Figure 5D). In grape, the vegetative MYB genes were not effective at regulating F3′5′H, resulting in cyanidin-based pigments, while the berry VvMYBA1 gene strongly activated this gene, resulting in trihydroxylated anthocyanins (Matus et al., 2017). Thus, it seems likely that a small family of R2R3-MYB genes controls anthocyanin biosynthesis in blueberry.
Whether the skin, flesh, or both skin and flesh are anthocyanin pigmented, and the strength of any such pigmentation, is a trait that varies across the Vaccinium genus, and that is of interest to plant breeding programs targeting novel commercial characters. In this regard, MYBA is a good candidate sequence for marker development for accelerated breeding approaches. The RNA-seq data will provide an additional resource for a comparison of gene expression between skin and flesh in blueberry. The other TFs identified with strongly enhanced expression in the skin could also be useful candidates for further examination for an association with differing pigmentation intensities.
Author Contributions
KS, BP, RE, KD, AA, and NA contributed to project planning and conducted sequence analysis. KS cloned MYBA. JT provided germplasm and expertise in Vaccinium sp. KS and EG conducted biolistic transformations. BP performed the Nicotiana transformations and promoter activation assays. SC and AD conducted metabolite assays and analysis. BW carried out bioinformatics. KS, BP, KD, RE, and NA wrote the manuscript. All authors contributed to manuscript editing.
Funding
This work was supported by The New Zealand Ministry of Business, Innovation, and Employment (MBIE) contract C11X1704 “Filling the void: boosting the nutritional content of NZ fruit.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Prof. Peter Waterhouse at Queensland University of Technology for the N. benthamiana seeds. We thank Monica Dragulescu, Ian King, and Julie Ryan for care of plants. We also thank Rebecca Henry-Kirk for project support. We acknowledge the services of the Australian Genome Research Facility.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01300/full#supplementary-material
FIGURE S1 |MYBA orthologs from rabbiteye blueberry and cranberry. Nucleotide alignment of the coding sequence of MYBA from highbush blueberry (Vaccinium corymbosum; Vc) and orthologs from rabbiteye blueberry (V. ashei; Va) and cranberry (V. macrocarpon; Vm). Sequence identity between VcMYBA and the orthologs from rabbiteye blueberry and cranberry are 97 and 95%, respectively. The putative start codon that has a strong Kozak sequence is indicated; the first ATG has a weak Kozak sequence and is not in-frame in rabbiteye blueberry.
FIGURE S2 | Alignment of the deduced amino acid sequence of VcMYBA with those of selected flavonoid-related R2R3MYBs. The deduced amino acid sequence of VcMYBA was aligned with representative R2R3MYB genes from SGs 4, 5, 6 and the PA1-type clade using MUSCLE. Sequences from Vaccinium species are in bold. The C-terminal motifs that define each SG (Stracke et al., 2001) are indicated, as are the bHLH interaction motif (Zimmermann et al., 2004), the “ANDV” motif present within the MYB domain of SG6 genes, and the TLLLFR repression domain present in some SG4 MYB repressors. At, Arabidopsis thaliana; Vc, Vaccinium corymbosum; Vv, Vitis vinifera; Vu, Vaccinium ulignosum.
FIGURE S3 | MYBA induces anthocyanin biosynthesis in blueberry petals. Petals from V. corymbosum were biolistically transformed with either 35S:GFP-ER alone or 35S:VcMYBA + 35S:GFP-ER. Transformed cells were identified by GFP fluorescence, observed with blue light. Transformation with VcMYBA induced pink pigmentation in transformed cells, while GFP-ER alone did not.
TABLE S1 | Sequences of DNA primers used.
TABLE S2 | Full list of differentially expressed genes in the skin and flesh of mature-stage blueberry fruit, with a differential greater than 2 log2fold.
Footnotes
- ^ http://www.igbquickload.org/blueberry/V_corymbosum_Aug_2015/
- ^ https://www.vaccinium.org/analysis/36
References
Albert, N. W., Griffiths, A. G., Cousins, G. R., Verry, I. M., and Williams, W. M. (2015). Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytol. 205, 882–893. doi: 10.1111/nph.13100
Albert, N. W., Lewis, D. H., Zhang, H., Schwinn, K. E., Jameson, P. E., and Davies, K. M. (2011). Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 65, 771–784. doi: 10.1111/j.1365-313X.2010.04465.x
Allan, A. C., Hellens, R. P., and Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
Bally, J., Nakasugi, K., Jia, F. Z., Jung, H. T., Ho, S. Y. W., Wong, M., et al. (2015). The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 1:15165. doi: 10.1038/nplants.2015.165
Bogs, J., Jaffé, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Borevitz, J. O., Xia, Y. J., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2393. doi: 10.1105/tpc.12.12.2383
Davies, K. M., Albert, N. W., and Schwinn, K. E. (2012). From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 39, 619–638. doi: 10.1071/fp12195
Davies, K. M., and Espley, R. V. (2013). Opportunities and challenges for the metabolic engineering of plant secondary metabolite pathways for improved human health characters in fruit and vegetable crops. N. Z. J. Crop Hort. Sci. 41, 154–177. doi: 10.1080/01140671.2013.793730
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Espley, R. V., Hellens, R. P., Putterill, J., Stevenson, D. E., Kutty-Amma, S., and Allan, A. C. (2007). Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 49, 414–427. doi: 10.1111/j.1365-313X.2006.02964.x
Frohman, M. A., Dush, M. K., and Martin, G. R. (1988). Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Nat. Acad. Sci. U.S.A. 85, 8998–9002. doi: 10.1073/pnas.85.23.8998
Gleave, A. P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. doi: 10.1007/BF00028910
Govers, C., Berkel Kasikci, M., Van Der Sluis, A. A., and Mes, J. J. (2018). Review of the health effects of berries and their phytochemicals on the digestive and immune systems. Nutr. Rev. 76, 29–46. doi: 10.1093/nutrit/nux039
Guindon, S., and Gascuel, O. (2003). PhyML: a simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. System. Biol. 52, 696–704. doi: 10.1080/10635150390235520
Gupta, V., Estrada, A. D., Blakley, I., Reid, R., Patel, K., Meyer, M. D., et al. (2015). RNA-Seq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. Gigascience 4:5. doi: 10.1186/s13742-015-0046-9
Haselhoff, J., Siemering, K. R., Prasher, D. C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U.S.A. 94, 2122–2127. doi: 10.1073/pnas.94.6.2122
Hellens, R. P., Allan, A. C., Friel, E. N., Bolitho, K., Grafton, K., Templeton, M. D., et al. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1:13. doi: 10.1186/1746-4811-1-13
Hichri, I., Barrieu, F., Bogs, J., Kappel, C., Delrot, S., and Lauvergeat, V. (2011). Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 62, 2465–2483. doi: 10.1093/jxb/erq442
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., and Chen, H. H. (2015). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Ithal, N., and Reddy, A. R. (2004). Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 166, 1505–1513. doi: 10.1016/j.plantsci.2004.02.002
Jaakola, L., Poole, M., Jones, M. O., Kämäräinen-Karppinen, T., Koskimäki, J. J., Hohtola, A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153, 1619–1629. doi: 10.1104/pp.110.158279
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kubo, H., Kishi, M., and Goto, K. (2008). Expression analysis of ANTHOCYANINLESS2 gene in Arabidopsis. Plant Sci. 175, 853–857. doi: 10.1016/j.plantsci.2008.08.006
Kubo, H., Peeters, A. J. M., Aarts, M. G. M., Pereira, A., and Koornneef, M. (1999). ANTHOCYANINLESS2, a homeobox gene affecting anthocyanin distribution and root development in Arabidopsis. Plant Cell 11, 1217–1226. doi: 10.1105/tpc.11.7.1217
Li, D., Li, B., Ma, Y., Sun, X., Lin, Y., and Meng, X. (2017). Polyphenols, anthocyanins, and flavonoids contents and the antioxidant capacity of various cultivars of highbush and half-high blueberries. J. Food Comp. Anal. 62, 84–93. doi: 10.1016/j.jfca.2017.03.006
Li, L., Zhang, H., Liu, Z., Cui, X., Zhang, T., Li, Y., et al. (2016). Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 16:223. doi: 10.1186/s12870-016-0866-5
Li, X., Sun, H., Pei, J., Dong, Y., Wang, F., Chen, H., et al. (2012). De novo sequencing and comparative analysis of the blueberry transcriptome to discover putative genes related to antioxidants. Gene 511, 54–61. doi: 10.1016/j.gene.2012.09.021
Lin, Y., Wang, Y., Li, B., Tan, H., Li, D., Li, L., et al. (2018). Comparative transcriptome analysis of genes involved in anthocyanin synthesis in blueberry. Plant Physiol. Biochem. 127, 561–572. doi: 10.1016/j.plaphy.2018.04.034
Lin-Wang, K., Bolitho, K., Grafton, K., Kortstee, A., Karunairetnam, S., Mcghie, T. K., et al. (2010). An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 10:50. doi: 10.1186/1471-2229-10-50
Lohachoompol, V., Mulholland, M., Srzednicki, G., and Craske, J. (2008). Determination of anthocyanins in various cultivars of highbush and rabbiteye blueberries. Food Chem. 111, 249–254. doi: 10.1016/j.foodchem.2008.03.067
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Matus, J. T., Cavallini, E., Loyola, R., Höll, J., Finezzo, L., Dal Santo, S., et al. (2017). A group of grapevine MYBA transcription factors located in chromosome 14 control anthocyanin synthesis in vegetative organs with different specificities compared with the berry color locus. Plant J. 91, 220–236. doi: 10.1111/tpj.13558
McGhie, T., Ainge, G., Barnett, L., Cooney, J., and Jensen, D. (2003). Anthocyanin glycosides from berry fruit are absorbed and excreted unmetabolized by both humans and rats. J. Agric. Food Chem. 51, 4539–4548. doi: 10.1021/jf026206w
Montefiori, M., Brendolise, C., Dare, A. P., Lin-Wang, K., Davies, K. M., Hellens, R. P., et al. (2015). In the Solanaceae, a hierarchy of bHLHs confer distinct target specificity to the anthocyanin regulatory complex. J. Exp. Bot. 66, 1427–1436. doi: 10.1093/jxb/eru494
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nesi, N., Jond, C., Debeaujon, I., Caboche, M., and Lepiniec, L. (2001). The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13, 2099–2114. doi: 10.1105/tpc.13.9.2099
Norberto, S., Silva, S., Meireles, M., Faria, A., Pintado, M., and Calhau, C. (2013). Blueberry anthocyanins in health promotion: a metabolic overview. J. Funct. Foods 5, 1518–1528. doi: 10.1016/j.jff.2013.08.015
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558.
Polashock, J., Zelzion, E., Fajardo, D., Zalapa, J., Georgi, L., Bhattacharya, D., et al. (2014). The American cranberry: first insights into the whole genome of a species adapted to bog habitat. BMC Plant Biol. 14:165. doi: 10.1186/1471-2229-14-165
Primetta, A. K., Karppinen, K., Riihinen, K. R., and Jaakola, L. (2015). Metabolic and molecular analyses of white mutant Vaccinium berries show down-regulation of MYBPA1-type R2R3 MYB regulatory factor. Planta 242, 631–643. doi: 10.1007/s00425-015-2363-8
Quattrocchio, F., Wing, J., Van Der Woude, K., Souer, E., De Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.1105/tpc.11.8.1433
Ramsay, N. A., and Glover, B. J. (2005). MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 10, 63–70. doi: 10.1016/j.tplants.2004.12.011
Schwinn, K., Venail, J., Shang, Y. J., Mackay, S., Alm, V., Butelli, E., et al. (2006). A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. Plant Cell 18, 831–851. doi: 10.1105/tpc.105.039255
Schwinn, K. E., Ngo, H., Kenel, F., Brummell, D. A., Albert, N. W., Mccallum, J. A., et al. (2016). The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 7:1865. doi: 10.3389/fpls.2016.01865
Shi, M., Loftus, H., Mcainch, A. J., and Su, X. Q. (2017). Blueberry as a source of bioactive compounds for the treatment of obesity, type 2 diabetes and chronic inflammation. J. Funct. Foods 30, 16–29. doi: 10.1016/j.jff.2016.12.036
Stevenson, D., and Scalzo, J. (2012). Anthocyanin composition and content of blueberries from around the world. J. Berry Res. 2, 179–189.
Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. doi: 10.1016/S1369-5266(00)00199-0
Sun, H., Liu, Y., Gai, Y., Geng, J., Chen, L., Liu, H., et al. (2015). De novo sequencing and analysis of the cranberry fruit transcriptome to identify putative genes involved in flavonoid biosynthesis, transport and regulation. BMC Genomics 16:652. doi: 10.1186/s12864-015-1842-4
Terrier, N., Torregrosa, L., Ageorges, A., Vialet, S., Verries, C., Cheynier, V., et al. (2009). Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 149, 1028–1041. doi: 10.1104/pp.108.131862
Uematsu, C., Katayama, H., Makino, I., Inagaki, A., Arakawa, O., and Martin, C. (2014). Peace, a MYB-like transcription factor, regulates petal pigmentation in flowering peach ‘Genpei’ bearing variegated and fully pigmented flowers. J. Exp. Bot. 65, 1081–1094. doi: 10.1093/jxb/ert456
Walker, A. R., Lee, E., Bogs, J., Mcdavid, D. A., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Yamagishi, M., Shimoyamada, Y., Nakatsuka, T., and Masuda, K. (2010). Two R2R3-MYB genes, homologs of petunia AN2, regulate anthocyanin biosyntheses in flower tepals, tepal spots and leaves of Asiatic hybrid lily. Plant Cell Physiol. 51, 463–474. doi: 10.1093/pcp/pcq011
Zhou, H., Peng, Q., Zhao, J., Owiti, A., Ren, F., Liao, L., et al. (2016). Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 7:1557. doi: 10.3389/fpls.2016.01557
Zhou, M., Zhang, K., Sun, Z., Yan, M., Chen, C., Zhang, X., et al. (2017). LNK1 and LNK2 corepressors interact with the MYB3 transcription factor in phenylpropanoid biosynthesis. Plant Physiol. 174, 1348–1358. doi: 10.1104/pp.17.00160
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Zimmermann, I. M., Heim, M. A., Weisshaar, B., and Uhrig, J. F. (2004). Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 40, 22–34. doi: 10.1111/j.1365-313X.2004.02183.x
Zorenc, Z., Veberic, R., Slatnar, A., Koron, D., Miosic, S., Chen, M.-H., et al. (2017). A wild ‘albino’ bilberry (Vaccinium myrtillus L.) from Slovenia shows three bottlenecks in the anthocyanin pathway and significant differences in the expression of several regulatory genes compared to the common blue berry type. PLoS One 12:e0190246. doi: 10.1371/journal.pone.0190246
Keywords: blueberry, flavonoid, Vaccinium, anthocyanin, phenylpropanoid, MYB transcription factor, regulation, fruit
Citation: Plunkett BJ, Espley RV, Dare AP, Warren BAW, Grierson ERP, Cordiner S, Turner JL, Allan AC, Albert NW, Davies KM and Schwinn KE (2018) MYBA From Blueberry (Vaccinium Section Cyanococcus) Is a Subgroup 6 Type R2R3MYB Transcription Factor That Activates Anthocyanin Production. Front. Plant Sci. 9:1300. doi: 10.3389/fpls.2018.01300
Received: 13 June 2018; Accepted: 17 August 2018;
Published: 11 September 2018.
Edited by:
Stefan Martens, Fondazione Edmund Mach, ItalyReviewed by:
Laura Jaakola, UiT The Arctic University of Norway, NorwayHiroshi Noguchi, Nihon Pharmaceutical University, Japan
Copyright © 2018 Plunkett, Espley, Dare, Warren, Grierson, Cordiner, Turner, Allan, Albert, Davies and Schwinn. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kathy E. Schwinn, kathy.schwinn@plantandfood.co.nz
 Blue J. Plunkett
Blue J. Plunkett Richard V. Espley
Richard V. Espley Andrew P. Dare
Andrew P. Dare Ben A. W. Warren
Ben A. W. Warren Ella R. P. Grierson2
Ella R. P. Grierson2 Andrew C. Allan
Andrew C. Allan Nick W. Albert
Nick W. Albert Kevin M. Davies
Kevin M. Davies Kathy E. Schwinn
Kathy E. Schwinn