- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Institute of Applied Ecology, Fujian Agriculture & Forestry University, Fuzhou, China
- 3Graham Centre for Agricultural Innovation (Charles Sturt University & NSW Department of Primary Industries), Orange, NSW, Australia
- 4NSW Department of Primary Industries, Wagga Wagga Agricultural Institute, Wagga Wagga, NSW, Australia
A Corrigendum on
Phytoplasmas–The “Crouching Tiger” Threat of Australian Plant Pathology
by Liu, J., Gopurenko, D., Fletcher, M. J., Johnson, A. C., & Gurr, G. M. (2017). Front. Plant Sci. 8:599. doi: 10.3389/fpls.2017.00599
In the original article, information for phytoplasmas in Table 1 did not fully reflect recent changes in taxonomy, or showed changes only as footnotes. Corrections have been made in the sections below and in Table 1.
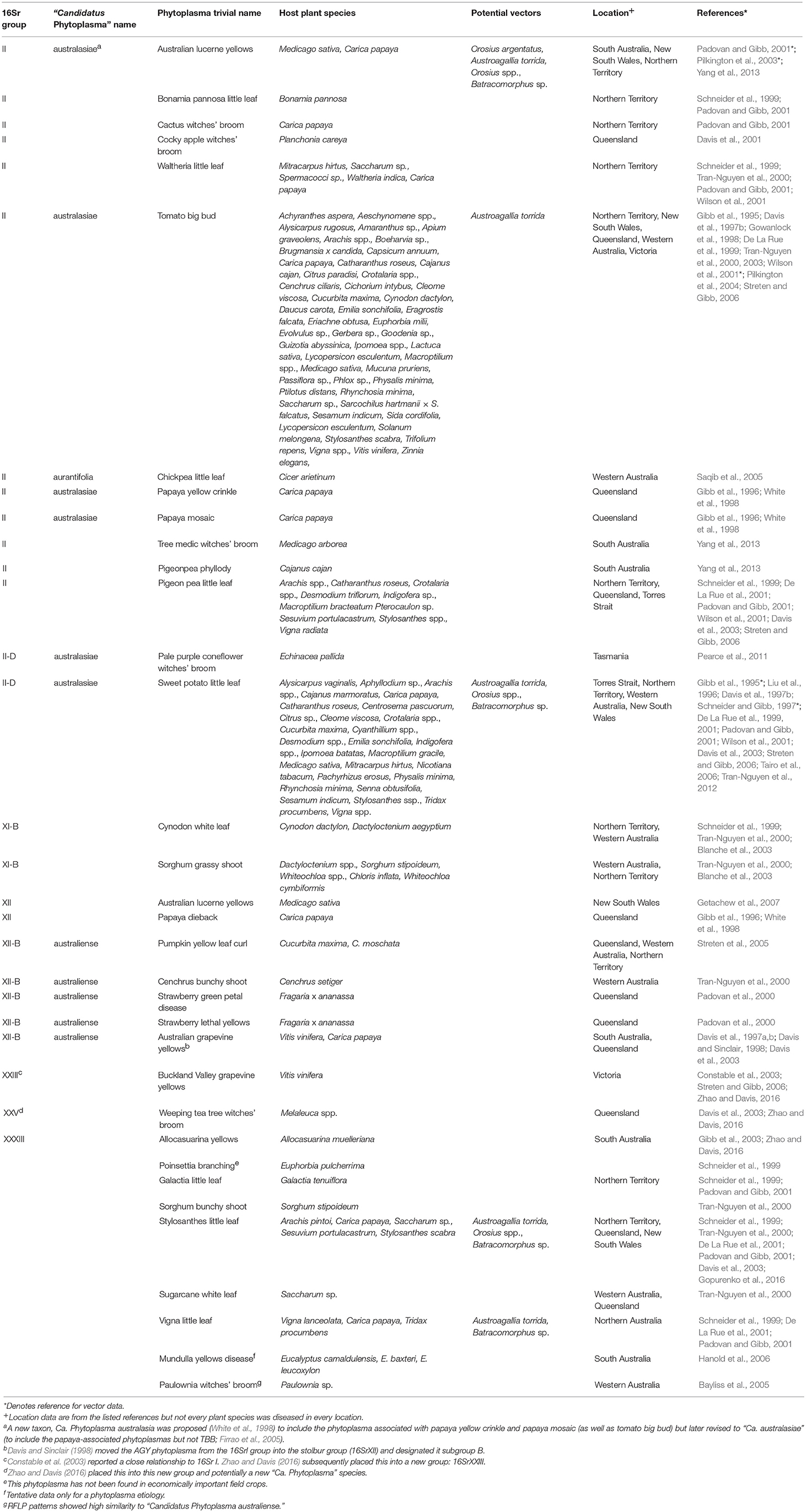
Table 1. Taxonomic and biological information on phytoplasmas in Australia (empty cells denote the absence of available information).
Abstract
Phytoplasmas are insect-vectored bacteria that cause disease in a wide range of plant species. The increasing availability of molecular DNA analyses, expertise, and additional methods in recent years has led to a proliferation of discoveries of phytoplasma-plant host associations and in the numbers of taxonomic groupings for phytoplasmas. The widespread use of common names based on the diseases with which they are associated, as well as separate phenetic and taxonomic systems for classifying phytoplasmas based on variation at the 16S rRNA-encoding gene, complicates interpretation of the literature. We explore this issue and related trends through a focus on Australian pathosystems, providing the first comprehensive compilation of information for this continent, covering the phytoplasmas, host plants, vectors, and diseases. Of the 33 16Sr groups reported internationally, only groups II, XI, XII, XXIII, XXV, and XXXIII have been recorded in Australia and this highlights the need for ongoing biosecurity measures to prevent the introduction of additional pathogen groups. Many of the phytoplasmas reported in Australia have not been sufficiently well-studied to assign them to 16Sr groups so it is likely that unrecognized groups and sub-groups are present. Wide host plant ranges are apparent among well studied phytoplasmas, with multiple crop and non-crop species infected by some. Disease management is further complicated by the fact that putative vectors have been identified for few phytoplasmas, especially in Australia. Despite rapid progress in recent years using molecular approaches, phytoplasmas remain the least well-studied group of plant pathogens, making them a “crouching tiger” disease threat.
Issue 2: Complex taxonomic nomenclature, paragraphs 2 and 3
Second, as molecular methods became available, workers were able to group and phenetically classify phytoplasmas using restricted fragment length polymorphism (RFLP) analysis of a PCR amplified portion of the 16S rRNA gene with a defined set of restriction enzymes (Lee et al., 1998). The RFLP profiles generated for different phytoplasmas are generally consistent with sequence-based phylogenetic analyses of the 16S rRNA gene, particularly in the co-identification and grouping of related strains. The 33 16Sr groups currently defined each have a similarity of less than 85% compared with any representative phytoplasma from within an established 16Sr group (Zhao and Davis, 2016). Table 1 summarizes available information on the 16Sr groups reported in Australian studies. Of the 33 16Sr groups reported internationally, only groups II, XI, XII, XXIII, XXV, and XXXIII have been recorded in Australia and this highlights the need for ongoing biosecurity measures to prevent the introduction of additional pathogen groups.
Third, phytoplasmas are classified in the provisional genus “Candidatus Phytoplasma” (IRPCM, 2004). To date, there are 42 formally described species and ten potentially novel phytoplasma species (Davis et al., 2015). This number exceeds the current number of 16s rRNA groups because some of these groups contain several “Candidatus Phytoplasma” species. At least 100 subgroups are known (Dickinson and Hodgetts, 2013). According to Phytoplasma/Spiroplasma Working Team-Phytoplasma Taxonomy Group, a novel “Ca. Phytoplasma” species description should refer to a single, unique 16S rRNA gene sequence (>1,200 bp), and a strain can be recognized as a novel “Ca. Phytoplasma” species if its 16S rRNA gene sequence has <97.5% similarity to that of any previously described “Ca. Phytoplasma” species (Duduk and Bertaccini, 2011). Additional biological characters such as antibody specificity, host range and vector transmission specificity as well as genetic markers can also be used in an integrative taxonomy approach for species differentiation. Of the 42 recognized “Ca. Phytoplasma” species, only Ca. Phytoplasma aurantifolia, Ca. Phytoplasma australasiae and Ca. Phytoplasma australiense are reported in Australia (Table 1) but uncertainty exists because many papers appear without Ca. Phytoplasma names which are used consistently only in the case of the GenBank database.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Bayliss, K. L., Saqib, M., Dell, B., Jones, M. G. K., and Hardy, G. E. S. J. (2005). First record of “Candidatus Phytoplasma australiense” in Paulownia trees. Australas. Plant Pathol. 34, 123–124. doi: 10.1071/AP04089
Blanche, K. R., Tran-Nguyen, L. T. T., and Gibb, K. S. (2003). Detection, identification and significance of phytoplasmas in grasses in northern Australia. Plant Pathol. 52, 505–512. doi: 10.1046/j.1365-3059.2003.00871.x
Constable, F. E., Whiting, J. R., Jones, J., Gibb, K. S., and Symons, R. H. (2003). The distribution of grapevine yellows disease associated with the buckland valley grapevine yellows phytoplasma. J. Phytopathol. 151, 65–73. doi: 10.1046/j.1439-0434.2003.00681.x
Davis, R., Dally, E. L., Gundersen, D. E., Lee, I. M., and Habili, N. (1997a). “Candidatus Phytoplasma australiense,” a New Phytoplasma Taxon Associated with Australian Grapevine Yellows. Int. J. Syst. Evol. Microbiol. 47, 262–269. doi: 10.1099/00207713-47-2-262
Davis, R. I., Henderson, J., Jones, L. M., McTaggart, A. R., O'Dwyer, C., Tsatsia, F., et al. (2015). First record of a wilt disease of banana plants associated with phytoplasmas in Solomon Islands. Australas. Plant Dis. Notes 10:14. doi: 10.1007/s13314-015-0163-4
Davis, R., Schneider, B., and Gibb, K. (1997b). Detection and differentiation of phytoplasmas in Australia. Aust. J. Agric. Res. 48, 535–544. doi: 10.1071/A96114
Davis, R. E., and Sinclair, W. A. (1998). Phytoplasma identity and disease etiology. Phytopathology 88, 1372–1376. doi: 10.1094/PHYTO.1998.88.12.1372
Davis, R. I., Jacobson, S. C., Rue, S. J., Tran-Nguyen, L., Gunua, T. G., and Rahamma, S. (2003). Phytoplasma disease surveys in the extreme north of Queensland, Australia, and the island of New Guinea. Australas. Plant Pathol. 32, 269–277. doi: 10.1071/AP03020
Davis, R. I., Jacobson, S. C., Waldeck, G. J., De La Rue, S. J., and Gibb, K. S. (2001). A witches' broom of cocky apple (Planchonia careya) in north Queensland. Australas. Plant Pathol. 30, 179–179. doi: 10.1071/AP01016
De La Rue, S., Padovan, A., and Gibb, K. (2001). Stylosanthes is a Host for Several Phytoplasmas, One of which Shows Unique 16S-23S Intergenic Spacer Region Heterogeneity. J. Phytopathol. 149, 613–619. doi: 10.1046/j.1439-0434.2001.00683.x
De La Rue, S. J., Schneider, B., and Gibb, K. S. (1999). Genetic variability in phytoplasmas associated with papaya yellow crinkle and papaya mosaic diseases in Queensland and the Northern Territory. Australas. Plant Pathol. 28, 108–114. doi: 10.1071/AP99019
Dickinson, M., and Hodgetts, J. (eds) (2013). Phytoplasma: Methods and Protocols, Dordrecht. Netherlands: Science+Business Media.
Duduk, B., and Bertaccini, A. (2011). Phytoplasma classification: taxonomy based on 16S ribosomal gene, is it enough? Phytopathogenic Mollicutes 1, 3–13. doi: 10.5958/j.2249-4669.1.1.001
Firrao, G., Gibb, K., and Streten, C. (2005). Short taxonomic guide to the genus “Candidatus Phytoplasma.” J. Plant Pathol. 87, 249–263. doi: 10.4454/jpp.v87i4.926
Getachew, M. A., Mitchell, A., Gurr, G. M., Fletcher, M. J., Pilkington, L. J., and Nikandrow, A. (2007). First report of a “Candidatus phytoplasma australiense”-related strain in lucerne (Medicago sativa) in Australia. Plant Dis. 91, 111–111. doi: 10.1094/PD-91-0111A
Gibb, K., Padovan, A., and Mogen, B. (1995). Studies on sweet potato little-leaf phytoplasma detected in sweet potato and other plant species growing in Northern Australia. Phytopathology 85, 169–174. doi: 10.1094/Phyto-85-169
Gibb, K., Persley, D., Schneider, B., and Thomas, J. (1996). Phytoplasmas associated with papaya diseases in Australia. Plant Dis. 80, 174–178. doi: 10.1094/PD-80-0174
Gibb, K. S., Tran-Nguyen, L. T. T., and Randles, J. W. (2003). A new phytoplasma detected in the South Australian native perennial shrub, Allocasuarina muelleriana. Ann. Appl. Biol. 142, 357–364. doi: 10.1111/j.1744-7348.2003.tb00261.x
Gopurenko, D., Fletcher, M. J., Liu, J., and Gurr, G. M. (2016). Expanding and exploring the diversity of phytoplasmas from lucerne (Medicago sativa). Sci. Rep. 6:37746. doi: 10.1038/srep37746
Gowanlock, D. H., Ogle, H. J., and Gibb, K. S. (1998). Phytoplasma associated with virescence in an epiphytic orchid in Australia. Australas. Plant Pathol. 27, 265–268. doi: 10.1071/AP98031
Hanold, D., Gowanlock, D., Stukely, M. J. C., Habili, N., and Randles, J. W. (2006). Mundulla Yellows disease of eucalypts: descriptors and preliminary studies on distribution and etiology. Australas. Plant Pathol. 35, 199–215. doi: 10.1071/AP06013
IRPCM (2004). “Candidatus Phytoplasma,” a taxon for the wall-less, non-helical prokaryotes that colonize plant phloem and insects. Int. J. Syst. Evol. Microbiol. 54, 1243–1255. doi: 10.1099/ijs.0.02854-0
Lee, I.-M., Gundersen-Rindal, D. E., Davis, R. E., and Bartoszyk, I. M. (1998). Revised classification scheme of phytoplasmas based on RFLP Analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 48, 1153–1169. doi: 10.1099/00207713-48-4-1153
Liu, B., White, D., Walsh, K., and Scott, P. (1996). Detection of phytoplasmas in dieback, yellow crinkle, and mosaic diseases of papaya using polymerase chain reaction techniques. Aust. J. Agric. Res. 47, 387–394. doi: 10.1071/AR9960387
Padovan, A., Gibb, K., and Persley, D. (2000). Association of “Candidatus Phytoplasma australiense” with green petal and lethal yellows diseases in strawberry. Plant Pathol. 49, 362–369. doi: 10.1046/j.1365-3059.2000.00461.x
Padovan, A. C., and Gibb, K. S. (2001). Epidemiology of phytoplasma diseases in papaya in Northern Australia. J. Phytopathol. 149, 649–658. doi: 10.1046/j.1439-0434.2001.00688.x
Pearce, T. L., Scott, J. B., and Pethybridge, S. J. (2011). First report of a 16SrII-D subgroup phytoplasma associated with pale purple coneflower Witches'-Broom disease in Australia. Plant Dis. 95, 773–773. doi: 10.1094/PDIS-03-11-0155
Pilkington, L. J., Gibb, K. S., Gurr, G. M., Fletcher, M. J., Nikandrow, A., Elliott, E., et al. (2003). Detection and identification of a phytoplasma from lucerne with Australian lucerne yellows disease. Plant Pathol. 52, 754–762. doi: 10.1111/j.1365-3059.2003.00934.x
Pilkington, L. J., Gurr, G. M., Fletcher, M. J., Nikandrow, A., and Elliott, E. (2004). Vector status of three leafhopper species for Australian lucerne yellows phytoplasma. Aust. J. Entomol. 43, 366–373. doi: 10.1111/j.1440-6055.2004.00419.x
Saqib, M., Bayliss, K. L., Dell, B., Hardf, G. E. S., and Jones, M. G. K. (2005). First record of a phytoplasma-associated disease of chickpea (Cicer arietinum) in Australia. Australas. Plant Pathol. 34, 425–426. doi: 10.1071/AP05047
Schneider, B., and Gibb, K. S. (1997). Detection of phytoplasmas in declining pears in Southern Australia. Plant Dis. 81, 254–258. doi: 10.1094/PDIS.1997.81.3.254
Schneider, B., Padovan, A., Rue, S., Eichner, R., Davis, R., Bernuetz, A., et al. (1999). Detection and differentiation of phytoplasmas in Australia: an update. Aust. J. Agric. Res. 50, 333–342. doi: 10.1071/A98106
Streten, C., Conde, B., Herrington, M., Moulder, J., and Gibb, K. (2005). Candidatus Phytoplasma australiense is associated with pumpkin yellow leaf curl disease in Queensland, Western Australia and the Northern Territory. Australas. Plant Pathol. 34, 103–105. doi: 10.1071/AP04077
Streten, C., and Gibb, K. S. (2006). Phytoplasma diseases in sub-tropical and tropical Australia. Australas. Plant Pathol. 35, 129–146. doi: 10.1071/AP06004
Tairo, F., Jones, R. A. C., and Valkonen, J. P. T. (2006). Phytoplasma from little leaf disease affected sweetpotato in Western Australia: detection and phylogeny. Ann. Appl. Biol. 149, 9–14. doi: 10.1111/j.1744-7348.2006.00065.x
Tran-Nguyen, L., Blanche, K. R., Egan, B., and Gibb, K. S. (2000). Diversity of phytoplasmas in northern Australian sugarcane and other grasses. Plant Pathol. 49, 666–679. doi: 10.1046/j.1365-3059.2000.00498.x
Tran-Nguyen, L. T. T., Persley, D. M., and Gibb, K. S. (2003). First report of phytoplasma disease in capsicum, celery and chicory in Queensland, Australia. Australas. Plant Pathol. 32, 559–560. doi: 10.1071/AP03055
Tran-Nguyen, L. T. T., Smith, S. H., and Liberato, J. R. (2012). Sweet potato little leaf strain V4 phytoplasma associated with snake bean in the Northern Territory, Australia. Australas. Plant Dis. Notes 7, 147–150. doi: 10.1007/s13314-012-0071-9
White, D. T., Blackall, L. L., Scott, P. T., and Walsh, K. B. (1998). Phylogenetic positions of phytoplasmas associated with dieback, yellow crinkle and mosaic diseases of papaya, and their proposed inclusion in “Candidatus Phytoplasma australiense” and a new taxon, “Candidatus Phytoplasma australasia.” Int. J. Syst. Evol. Microbiol. 48, 941–951. doi: 10.1099/00207713-48-3-941
Wilson, D., Blanche, K. R., and Gibb, K. S. (2001). Phytoplasmas and disease symptoms of crops and weeds in the semi-arid tropics of the Northern Territory, Australia. Australas. Plant Pathol. 30, 159–163. doi: 10.1071/AP01015
Yang, S., Habili, N., Aoda, A., Dundas, I., Paull, J., and Randles, J. (2013). Three group 16SrII phytoplasma variants detected in co-located pigeonpea, lucerne and tree medic in South Australia. Australas. Plant Dis. Notes 8, 125–129. doi: 10.1007/s13314-013-0113-y
Keywords: “Candidatus Phytoplasma”, 16S rRNA, biosecurity, taxonomy, biodiversity, vector, seed transmission, host range
Citation: Liu J, Gopurenko D, Fletcher MJ, Johnson AC and Gurr GM (2018) Corrigendum: Phytoplasmas—The “Crouching Tiger” Threat of Australian Plant Pathology. Front. Plant Sci. 9:1298. doi: 10.3389/fpls.2018.01298
Received: 21 June 2018; Accepted: 17 August 2018;
Published: 26 October 2018.
Edited and reviewed by: Brigitte Mauch-Mani, University of Neuchâtel, Switzerland
Copyright © 2018 Liu, Gopurenko, Fletcher, Johnson and Gurr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Geoff M. Gurr, Z2d1cnJAY3N1LmVkdS5hdQ==
 Jian Liu
Jian Liu David Gopurenko
David Gopurenko Murray J. Fletcher3
Murray J. Fletcher3 Anne C. Johnson
Anne C. Johnson Geoff M. Gurr
Geoff M. Gurr