
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 August 2018
Sec. Plant Pathogen Interactions
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01153
This article is part of the Research Topic Activation and Suppression of Plant Immunity View all 27 articles
 Min Li1,2
Min Li1,2 Huan Chen1,2
Huan Chen1,2 Jian Chen1,2
Jian Chen1,2 Ming Chang1,2
Ming Chang1,2 Ian A. Palmer1
Ian A. Palmer1 Walter Gassmann3
Walter Gassmann3 Fengquan Liu2,4*
Fengquan Liu2,4* Zheng Qing Fu1*
Zheng Qing Fu1*In Arabidopsis, TEOSINTE BRANCHED 1, CYCLOIDEA, PCF1 (TCP) transcription factors (TF) play critical functions in developmental processes. Recent studies suggest they also function in plant immunity, but whether they play an important role in systemic acquired resistance (SAR) is still unknown. NON-EXPRESSER OF PR GENES 1 (NPR1), as an essential transcriptional regulatory node in SAR, exerts its regulatory role in downstream genes expression through interaction with TFs. In this work, we provide biochemical and genetic evidence that TCP8, TCP14, and TCP15 are involved in the SAR signaling pathway. TCP8, TCP14, and TCP15 physically interacted with NPR1 in yeast two-hybrid assays, and these interactions were further confirmed in vivo. SAR against the infection of virulent strain Pseudomonas syringae pv. maculicola (Psm) ES4326 in the triple T-DNA insertion mutant tcp8-1 tcp14-5 tcp15-3 was partially compromised compared with Columbia 0 (Col-0) wild type plants. The induction of SAR marker genes PR1, PR2, and PR5 in local and systemic leaves was dramatically decreased in the tcp8-1 tcp14-5 tcp15-3 mutant compared with that in Col-0 after local treatment with Psm ES4326 carrying avrRpt2. Results from yeast one-hybrid and chromatin immunoprecipitation (ChIP) assays demonstrated that TCP15 can bind to a conserved TCP binding motif, GCGGGAC, within the promoter of PR5, and this binding was enhanced by NPR1. Results from RT-qPCR assays showed that TCP15 promotes the expression of PR5 in response to salicylic acid induction. Taken together, these data reveal that TCP8, TCP14, and TCP15 physically interact with NPR1 and function redundantly to establish SAR, that TCP15 promotes the expression of PR5 through directly binding a TCP binding site within the promoter of PR5, and that this binding is enhanced by NPR1.
TCP proteins, as plant specific transcription factors (TFs), are named after the first characterized members, TEOSINTE BRANCHED1 (TB1) in maize (Zea mays), CYCLOIDEA (CYC) in snapdragon (Antirrhinum majus) and PCF in rice (Oryza sativa) (Nicolas and Cubas, 2016). Based on the sequence of the TCP specific helix-loop-helix DNA binding domain, 24 Arabidopsis TCP members are divided into class I and class II groups (Cubas et al., 1999; Martín-Trillo and Cubas, 2010). Class I proteins prefer to bind the consensus element KHGGGVC (Davière et al., 2014), whereas class II proteins bind the GTGGNCCC consensus DNA sequence (Aggarwal et al., 2010). TCP proteins govern essential functions in developmental processes, including endo-reduplication, seed germination, internode length, leaf shape, and flower development (Kieffer et al., 2011; Uberti-Manassero et al., 2012; Resentini et al., 2015; Lucero et al., 2017). In addition to regulating developmental processes, accumulating experimental evidence also implies that TCP TFs play key functions in plant immunity. Being convergently targeted by effectors from multiple pathogens suggested TCP TFs function essentially in plant immunity (Mukhtar et al., 2011; Weßling et al., 2014). TCP TFs were subsequently found to interact with SUPPRESSOR OF rps4-RLD1 (SRFR1), a negative immune regulator, and contributed redundantly to effector-triggered immunity (ETI) (Kim et al., 2014). TCP TFs can also mediate the activity of phytohormone. For instance, TCP8, TCP9, and other TCP proteins were verified to coordinately regulate the expression of ISOCHORISMATE SYNTHASE 1 (ICS1) which is responsible for pathogen-induced salicylic acid (SA) biosynthesis (Wang et al., 2015). Furthermore, Type III effector HopBB1 promotes disease susceptibility via targeting and degrading TCP14, which functions as a negative regulator of the jasmonic acid signaling pathway (Yang et al., 2017). In addition to separate functions in plant immunity or developmental processes, TCP TFs can also serve as a bridge to connect both responses. A recent study provided evidence that TCP15 connects the plant immune response with cell cycle progression by interacting with MODIFIER OF snc1-1 (MOS1) (Zhang et al., 2018).
Systemic acquired resistance (SAR) is an induced plant immunity which can be activated by pathogen infection or SA application (Fu and Dong, 2013). Pathogen infection induces the accumulation of SA which functions as an endogenous immune signal (Fu et al., 2012). SA is required for SAR, because blocking SA accumulation suppresses SAR induction (Gaffney et al., 1993). In Arabidopsis, the expression of the PATHOGENESIS-RELATED (PR) genes PR1, PR2 (encoding β-1,3-glucanase), and PR5 (encoding a thaumatin-like protein) are used as hallmarks for SAR because they maintain high expression levels during SAR (Ward et al., 1991; Uknes et al., 1992). After synthesis on the rough endoplasmic reticulum, small PR proteins (5–75 kDa) are secreted and targeted to vacuoles, or to the apoplast where bacterial pathogens are found (Dong, 2004; Edreva, 2005). PR proteins are associated with disease resistance because they exhibit anti-microbial functions both in vitro and in vivo (Ryals et al., 1996; Edreva, 2005; Breen et al., 2017).
Arabidopsis NPR1 is required for SAR and SA induced expression of PR1, PR2, and PR5 (Cao et al., 1994, 1997). As a critical transcriptional regulatory node in SAR, NPR1 regulates the expression of 2,248 out of 2,280 SA responsive genes (Wang et al., 2006). NPR1 lacks a DNA binding domain but contains two protein-protein interaction domains, suggesting that NPR1 functions as a cofactor by interacting with TFs to regulate downstream gene expression (Fan and Dong, 2002; Rochon et al., 2006; Boyle et al., 2009). Indeed, it was found that NPR1 interacts with the TGA subclass of basic Leu zipper (bZIP) family TFs to regulate the expression of PR1 (Fan and Dong, 2002). Induced SA upon pathogen infection results in cellular redox potential change, which triggers the reduction of cytosolic oligomeric NPR1 into monomeric NPR1 (Mou et al., 2003). Monomeric NPR1 proteins then enter the nucleus and interact with TGAs to facilitate the expression of PR1 (Fan and Dong, 2002; Rochon et al., 2006).
Since PR1, PR2, and PR5 are co-induced by SAR inducers, TGA TFs are thought to co-regulate their expression. However, there are no TGA binding sites (TGACGt/g, ACGTCA) (Jakoby et al., 2002) within the promoter of PR5. In addition to chemical evidence, genetic evidence also suggests separate regulators exist that regulate the expression of PR1, PR2, and PR5. First, PR2 and PR5, but not PR1, were found constitutively expressed in WRKY70 overexpression transgenic plants (Li et al., 2004). Second, only PR1 mRNA levels were found to be reduced in enhanced disease susceptibility 5-1 mutants, while the mRNA levels of PR2 and PR5 showed no apparent change (Rogers and Ausubel, 1997). All these data suggest that additional NPR1-interacting TFs are required to explain how NPR1 regulates the expression of PR5, and prompted us to screen for new NPR1-interacting TFs. Here, we show that TCP8, TCP14, and TCP15 interact with NPR1, and that they contribute redundantly to SAR establishment. TCP15 is shown to bind to the TCP binding site within the promoter of PR5 and promote its expression. In addition, the binding ability of TCP15 to the PR5 promoter was enhanced by NPR1.
All mutants and transgenic lines were derived from Arabidopsis [(Arabidopsis thaliana (L.) Heynh.)] ecotype Columbia-0 (Col-0). Single T-DNA insertion mutants tcp8-1 (CS875709), tcp14-5 (CS458588), tcp15-3 (CS68533) (Kim et al., 2014), and tcp15-1 (CS875923) (Kieffer et al., 2011) were purchased from Arabidopsis Biological Resource Center (ABRC). Double mutants tcp8-1 tcp14-5, tcp8-1 tcp15-3, tcp14-5 tcp15-3 and triple mutant tcp8-1 tcp14-5 tcp15-3 were described before (Kim et al., 2014). The mutant npr1-2 was described before (Cao et al., 1997). Agrobacterium tumefaciens (strain GV3101) mediated transformation was used to construct transgenic plants through the floral dipping method. Transgenic lines pTA:TCP15-EYFP and 35S:GFP (Wang et al., 2015) were provided by Dr. Ai-wu Dong. The Dex:Flag-TCP15 constructs were transformed into the Col-0 and npr1-2 background to generate Dex:Flag-TCP15/Col-0 and Dex:Flag-TCP15/npr1-2 transgenic plants. T3 homozygous transgenic lines were screened on 1/2 Murashige, and Skoog (MS) medium with 10 μM hygromycin B. Inducible FLAG-TCP15 protein expression level was verified by immunoblot. The point mutation in the npr1-2 mutant was confirmed by restriction enzyme digestion (Cao et al., 1997).
Seeds were vernalized at 4°C for 3 days before growth. Soil-grown plants were placed in a growth chamber at 22°C with 60% humidity under 12 h light. For in vitro growth, surface sterilized seeds were grown on MS plates at 22°C with 50% humidity under 16 h light. The bacterial strains of Pseudomonas syringae pv. maculicola (Psm) ES4326 and Psm ES4326 carrying avrRpt2 were grown on King's B (KB) medium under streptomycin and both streptomycin and tetracycline selection, respectively at 28°C. SA solutions were diluted from a 100 mM sodium salicylate (Sigma Aldrich) stock solution. Dexamethasone (DEX) solutions were diluted from a 30 mM stock solution dissolved in ethanol.
Primers used to amplify gene-specific sequences are listed in Supplementary Table 1. Fragments used in all constructs were validated by DNA sequencing. The Arabidopsis transcription factor library was purchased form ABRC (CD4-89). The entire coding regions of NPR1, TCP8, TCP14, and TCP15 were amplified by PCR with Phusion® DNA Polymerase (NEB). PCR products were subsequently introduced into the Gateway® (GW) entry vector pDONR207 (Clontech) using GW BP Clonase II Enzyme Mix (Invitrogen). Resulting entry clones were cloned into GW destination vector pDEST22 or pDEST32 using LR Clonase II Enzyme Mix (Clontech) to generate pDEST22-TCP8, pDEST22-TCP14, pDEST22-TCP15, and pDEST32-NPR1. The promoter sequences of PR1 (2,380 bp), PR2 (1,513 bp), PR5 (1,000 and 500 bp), and NPR1 (1,000 bp) were amplified by PCR and also introduced into pDONR207 by BP reactions. Resulting entry clones were remobilized into GW destination vector pLacZi (Pruneda-Paz et al., 2014) using the LR reaction to generate pPR1:lacZ, pPR2:lacZ, pPR5:lacZ, and pNPR1:lacZ constructs. To introduce site-specific mutations within the promoter of PR5, the pPR5: lacZ plasmid DNA was amplified with a pair of complementary primers with GCGGGAC to ATAAACT mutations. After digestion with Dpnl enzyme (NEB), the resulting PCR products were transformed into Escherichia coli (E. coli) strain TOP10 by electroporation. TCP15 from pDONR207-TCP15 was introduced into the GW compatible vector pTA7002_Flag-GW (Chen et al., 2017) to generate Dex:Flag-TCP15 constructs. TCP14 and TCP15 from pDONR207-TCP14 and pDONR207-TCP15, respectively, were cloned into the pEarlyGate201 destination vector to make transient expression constructs 35S:HA-TCP14 and 35S:HA-TCP15 by LR reaction. TCP8 from pDONR207-TCP8 was cloned into the pEarlyGate202 destination vector to make a transient expression construct 35S:Flag-TCP8. The 35S:NPR1-GFP construct generated with pCB302 binary vector was described before (Mou et al., 2003).
Bait plasmid pDEST32-NPR1 was transformed into the Saccharomyces cerevisiae strain AH109 (MATa). Prey plasmids pDEST22-TFs were transformed into the S. cerevisiae strain Y187 (MATα). Y2H library screening was described before Ou et al., 2011). The pDEST22 and pDEST32 empty vectors were served as negative controls. After mating, healthy diploid yeast cells growing on the double dropout medium without leucine and tryptophan (control plates) were selected. The triple dropout medium lacking leucine, tryptophan, and histidine with 1 mM 3-amino-1,2,4-triazole (3-AT) was used as selective plates. Aliquots of 10 μl diploid yeast cell suspension were spotted on control and selective plates at a concentration of OD600 = 1, 0.1, and 0.01. The yeast transformation, mating, plasmid isolation and interaction test processes described in the Yeast Protocols Handbook (Clontech) were followed.
The promoter DNA:lacZ constructs were first digested with restriction enzyme NcoI (NEB), and resulting linearized constructs were subsequently integrated into the chromosome of S. cerevisiae strain YM4271 (MATa). The constructs of pDEST22-TCP8, pDEST22-TCP14, and pDEST22-TCP15 were transformed into the yeast strain YU (MATα) (Pruneda-Paz et al., 2014). The pDEST22 empty vector was also included to serve as a negative control. After mating, healthy diploid yeast cells growing on the double dropout medium lacking tryptophan and uracil were selected. The binding ability of the prey transcription factors to the bait promoter was calculated with the β-galactosidase activity assay described previously (Zheng et al., 2015).
The constructs 35S:HA-TCP14, 35S:HA-TCP15, 35S:HA-EV, 35S:Flag-TCP8, 35S:Flag-EV, 35S:NPR1-GFP, 35S:EV-GFP and p19 were transformed into the A. tumefaciens strain GV3101. Resulting strains were grown in YEB culture with gentamicin, rifampicin, and kanamycin A at 28°C overnight. Bacterial cells were collected by centrifugation at 5,000 g for 5 min. Precipitated cells were washed twice and resuspended in induction buffer (10 mM MES, pH 5.7, 10 mM MgCl2, and 200 μM acetosyringone). A final concentration of resuspended cells at OD600 = 0.5 was used to infiltrate into young leaves of 4-week-old Nicotiana benthamiana. Infiltrated leaves were harvested at 48 h after infiltration and stored at −80°C for subsequent analysis.
Aliquots of 0.15 g plant sample were homogenized with 150 μl protein extraction buffer [PEB, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 0.1% TritonTM X-100 (Sigma-Aldrich), 0.2% IGEPAL CA-630 (Sigma-Aldrich), 50 μM MG115 (Signa-Aldrich), 1 mM PMSF, 10 mM DTT, and 1 × protease inhibitor cocktail (Sigma-Aldrich)] using 2010 Geno/Grinder (SPEX). Total protein extracts were obtained by centrifuging homogenized culture at 15,000 g for 15 min twice at 4°C. Protein concentration was quantified with Bradford reagent (Bio-Rad) using a spectrophotometer (Eppendorf). Protein samples in 5 × sample buffer (250 mM Tris-HCl, pH 6.8, 500 mM DTT, 6% SDS, 0.08% bromophenol blue, and 30% glycerol) were denatured at 70°C for 15 min. After separation on a precast ExpressTM PAGE gel (GeneScript) by electrophoresis, denatured protein samples were transferred onto a nitrocellulose membrane (GE Healthcare). Total protein was stained with Ponceau S solution (0.1% Ponceau S (Abcam) and 5% acetic acid) to verify equal protein loading. The membrane was first incubated with a primary antibody [anti-GFP (Clontech), anti-HA-peroxidase (3F10, Roche), or anti-FLAG M2 (Sigma-Aldrich)], then incubated with a secondary antibody [goat-anti-rabbit lgG-HRP (Agrisera) or goat-anti-mouse lgG-HRP (Santa Cruz Biotech)]. After incubation with chemiluminescent agent SuperSignal™ West Pico or Dura substrate (ThermoFisher), targeted proteins were visualized on photographic film using an SRX-101A Medical Film Processor (Konica).
NPR1-GFP with FLAG-TCP8, HA-TCP14, or HA-TCP15 were transiently co-expressed in N. benthamiana. After 48 h, 1.5 g N. benthamiana leaves were homogenized with PEB. Homogenized samples were centrifuged at 20,000 g for 30 min twice at 4°C. For the interaction of NPR1 with TCP14 or TCP15, protein extracts were incubated with 20 μl GFP-Trap®_ MA Beads (Chromotek) with gentle rotation at 4°C overnight. The conjugated beads were separated by a magnetic stand (Promega) and subsequently washed three times with cold wash buffer (10 mM Tris/HCl pH 7.5, 150 mM NaCl, and 0.5 mM EDTA). Beads were resuspended in 2 × Laemmli Sample Buffer (Bio-Rad). Immunoprecipitated proteins were eluted by boiling beads for 10 min. The bound HA-TCP14 and HA-TCP15 were detected by immunoblots with the anti-HA antibody. For the interaction between NPR1 and TCP8, protein extracts were incubated with 20 μl anti-FLAG M2 Magnetic Beads (Sigma-Aldrich) with gentle rotation at 4°C overnight. The bound NPR1-GFP proteins were detected by immunoblot with the anti-GFP antibody. No DTT was added to any buffer to prevent disrupting disulfide linkages in the anti-FLAG beads.
Two grams of 12-day-old seedlings were used in each experiment. After treatment with 30 μM DEX for 24 h, samples were harvested at 24 h after treatment with 0.5 mM SA. The ChIP assays were performed according to a previously described protocol (Komar et al., 2016). For pTA:TCP15-EYFP and 35S:GFP transgenic lines, chromatin was immunoprecipitated by PierceTM Protein A/G Magnetic Beads (ThermoFisher) bound with the anti-GFP antibody. For Dex:Flag-TCP15/Col-0 and Dex:Flag-TCP15/npr1-2 transgenic lines, chromatin was immunoprecipitated by beads bound with the anti-FLAG M2 antibody. Primers used in the ChIP assays are listed in Supplementary Table 1.
RNA was extracted with TRIzol™ (Invitrogen) according to its protocol. cDNA was synthesized using reverse transcriptase (Quanta). Real-time quantitative polymerase chain reaction (RT-qPCR) was performed with SYBR® Green (Quanta). The expression level of UBIQUITIN 5 (UBQ5) was used as an internal control. Three biological replicates were assayed. Primers for RT-qPCR are listed in Supplementary Table 1.
Two lower leaves of 3-week-old plants were hand-infiltrated with 10 mM MgSO4 or avirulent pathogen Psm ES4326 carrying avrRpt2 (OD600 = 0.02). Three days later, three upper leaves were hand-infiltrated with virulent pathogen Psm ES4326 (OD600 = 0.001). Leaf samples for bacterial growth were collected at 3 days after the secondary infection. Six plants were used in each treatment.
To test our hypothesis that NPR1 can interact with TFs other than TGAs, we performed Y2H screens (Ou et al., 2011) using Arabidopsis NPR1 as a bait and an Arabidopsis transcription factor library (Pruneda-Paz et al., 2014) as prey. Fifteen interactors were identified. To eliminate false positive interactions, we performed Y2H assays using individual candidate TFs as preys. Interactions were tested by the growth of diploid yeast cells on triple dropout medium lacking leucine, tryptophan, and histidine with 1 mM 3-AT. Supporting our initial assays, diploids containing NPR1 fused with a GAL4 DNA binding domain (BD-NPR1) and empty vector with a GAL4 activation domain (AD-EV) did not grow on selective plates (Figure 1A). Yeast diploids with BD-EV and AD-TFs were included as negative controls to exclude self-activation activity of TFs (Figure 1A). For this study, we focused on transcription factors in the TCP family. Yeast diploids containing BD-NPR1 and AD-TCP15 grew on selective plates (Figure 1A), indicating TCP15 was a true positive interactor of NPR1. We also found that yeast diploids containing BD-NPR1 with AD-TCP8 and AD-TCP14 grew on selective plates (Figure 1A), indicating NPR1 interacts with TCP8 and TCP14 in Y2H assays.
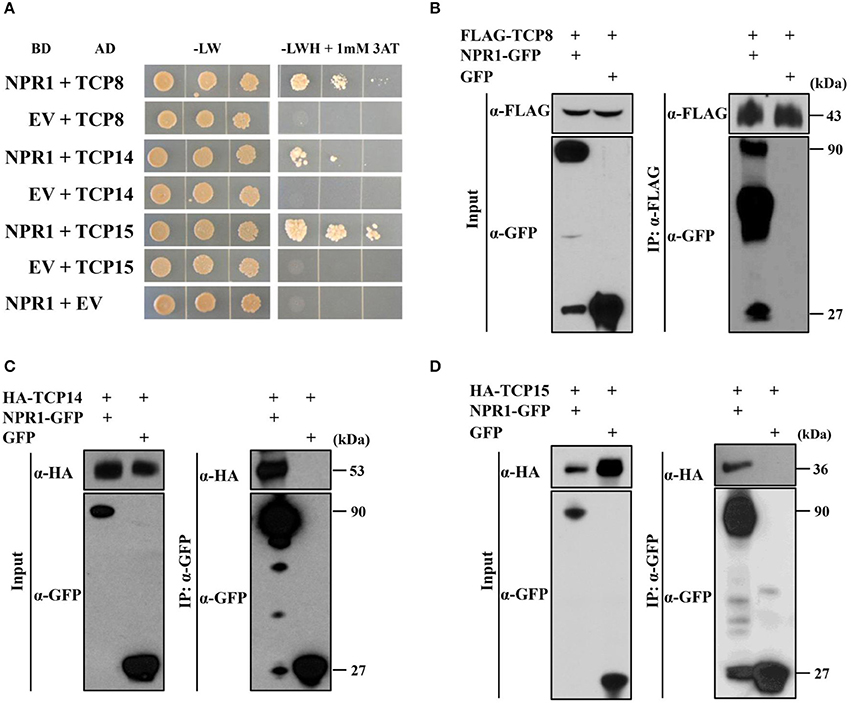
Figure 1. TCP8, TCP14, and TCP15 interact with NPR1. (A) Interactions between NPR1 with TCP8, TCP14, and TCP15 in yeast two-hybrid assays. Diploid yeast cells were serially diluted at OD600 = 1.0, 0.1, and 0.01. Aliquots of 10 μL of each dilution were plated on double synthetic dropout control plates without leucine and tryptophan (-LW) and selective triple synthetic dropout plates without leucine, tryptophan, and histidine with 1 mM 3-amino-1,2,4-triazole (–LWH + 1 mM 3AT). Photographs were taken 5 days after plating. EV, Empty vector; BD, GAL4 DNA-binding domain; AD, GAL4 activation domain. (B) Co-immunoprecipitation (Co-IP) of NPR1 with TCP8 in Nicotiana benthamiana. NPR1-GFP and FLAG-TCP8 under control of the cauliflower mosaic virus (CMV) 35S promoter were transiently co-expressed in N. benthamiana by agroinfiltration. Total protein extracts were immunoprecipitated with anti-FLAG M2 magnetic beads. The input and immunoprecipitated protein were analyzed by immunoblot using the anti-FLAG and anti-GFP antibodies. kDa, kilodaltons. (C,D) Co-IP of TCP14 and TCP15 with NPR1 in N. benthamiana. Constitutively expressed NPR1-GFP was transiently co-expressed with HA-TCP14 (C) and HA-TCP15 (D) under control of the CMV 35S promoter in N. benthamiana by agroinfiltration. Total protein extracts were immunoprecipitated using anti-GFP trap beads. The input and immuno-precipitated proteins were analyzed by immunoblot using anti-HA and anti-GFP antibodies.
To test the association of NPR1 with TCP8, TCP14, and TCP15 in planta, we performed co-immunoprecipitation (Co-IP) assays in N. benthamiana. TCP8 fused with an N-terminal FLAG tag (FLAG-TCP8) and NPR1 fused with a C-terminal GFP tag (NPR1-GFP) were transiently co-expressed in N. benthamiana by agroinfiltration. NPR1-GFP proteins were co-immunoprecipitated with FLAG-TCP8 bound to the anti-FLAG magnetic beads (Figure 1B), indicating NPR1 associates with TCP8 in planta. NPR1-GFP and TCP14 fused with an N-terminal HA tag (HA-TCP14) (Figure 1C) or NPR1-GFP and HA-TCP15 (Figure 1D) were also transiently co-expressed in N. benthamiana by agroinfiltration. HA-TCP14 (Figure 1C) and HA-TCP15 (Figure 1D) proteins were co-immunoprecipitated with NPR1-GFP bound to the anti-GFP beads, indicating TCP14 and TCP15 are associated with NPR1 in planta. Together, results from Y2H and Co-IP assays indicate that TCP8, TCP14, and TCP15 physically interact with NPR1.
To investigate whether NPR1 affects the transcription of TCP8, TCP14, and TCP15, their mRNA levels in wild-type Col-0 and the npr1-2 mutant after 24 h with or without SA application were examined by RT-qPCR. The SA-induced increase of TCP8 mRNA levels was abolished in npr1-2, while the mRNA levels of TCP14 and TCP15 did not show a significant difference between npr1-2 and Col-0 with or without SA for 24 h (Supplementary Figure 1). Our results indicate that NPR1 does not affect the transcription of TCP14 and TCP15 at this later time point, and an increase in SA-induced TCP8 mRNA levels is dependent on NPR1.
To investigate whether NPR1 interactors TCP8, TCP14, and TCP15 also function in SAR, we carried out SAR tests in the tcp8-1, tcp14-5, and tcp15-3 single, corresponding double, and triple mutants (Kim et al., 2014). Two lower leaves were infected with Psm ES4326 carrying avrRpt2 to induce SAR. Three days later, two upper leaves were infiltrated with the virulent pathogen Psm ES4326. The bacterial growth of Psm ES4326 in the tcp8-1, tcp14-5, and tcp15-3 single and corresponding double mutants decreased 8.6- to 12.8-fold which was similar to that reduction (11.7-fold) in Col-0 after SAR induction (Figure 2A). These results demonstrated that the deletion of one or two of TCP8, TCP14, or TCP15 genes is not enough to disrupt SAR. However, the Psm ES4326 population in tcp8-1 tcp14-5 tcp15-3 (tcp8/14/15) only decreased 2.5-fold after SAR induction (Figure 2A), indicating that SAR induction was partially compromised in tcp8/14/15. Consistent with bacterial growth results, Psm ES4326 infected systemic leaves of the single and double mutants showed less chlorosis compared with that of tcp8/14/15 after SAR induction (Supplementary Figure 2).
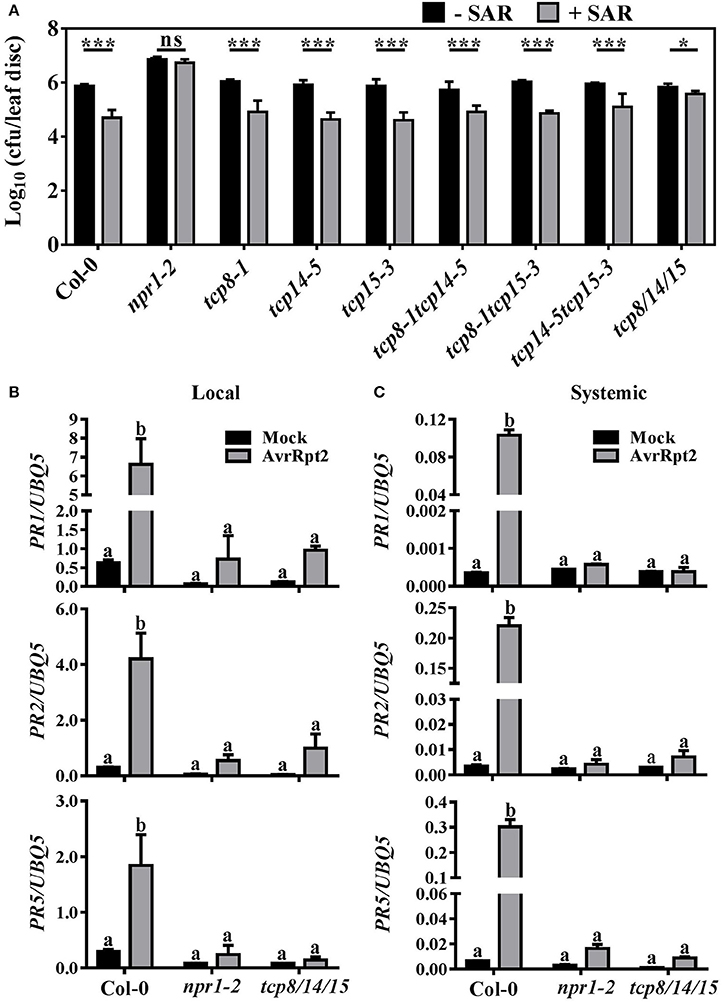
Figure 2. TCP8, TCP14, and TCP15 contribute redundantly to SAR establishment. (A) Bacterial population in Col-0, npr1-2, and the tcp mutants. Two lower leaves of 3-week-old plants were infiltrated with 10 mM MgCl2 (-SAR) or Pseudomonas syringae pv. maculicola (Psm) ES4326 carrying avrRpt2 at OD600 = 0.02 (+SAR). Three days later, two upper leaves were infiltrated with Psm ES4326 at OD600 = 0.001. Bacterial growth in infected systemic leaves was calculated 3 days after the second infection. Error bars represent SD of six biological repeats. Statistical analysis was studied by t-test (*p < 0.05, ***p < 0.001) using Excel 2016. (B,C) The expression of PR1, PR2, and PR5 in Col-0, npr1-2, and tcp8/14/15. Two local leaves were inoculated with 10 mM MgCl2 (Mock) or Psm ES4326 with avrRpt2 at OD600 = 0.02 (AvrRpt2). The mRNA levels of PR1, PR2, and PR5 in local leaves at 12 h post inoculation (hpi) (B) and in systemic leaves at 48 hpi (C) were analyzed by RT-qPCR. Values were normalized to the UBQ5 mRNA levels. Error bars represent SD of three biological repeats. Statistical analysis was studied by two-way ANOVA following multiple comparisons with turkey test (95% confidence interval) using GraphPad Prism 7. Different small letters above the bars mean significant differences.
To examine whether the expression of SAR marker genes was affected, the mRNA levels of PR1, PR2, and PR5 in local and systemic leaves of Col-0, npr1-2, and tcp8/14/15 were examined by RT-qPCR assays. After locally treated with 10 mM MgCl2, the mRNA levels of PR1, PR2, and PR5 in both local and systemic leaves of Col-0, npr1-2, and tcp8/14/15 did not show a significant difference (Figure 2B), indicating TCP8, TCP14, and TCP15 did not affect the basal expression of PR1, PR2, and PR5. Whereas, after locally infected with Psm ES4326 with avrRpt2, the local and systemic induction of PR1, PR2, and PR5 were all obviously decreased in tcp8/14/15 compared with Col-0 (Figures 2B,C), consistent with the increased local susceptibility of tcp8/14/15 to avrRpt2-expressing DC3000 (Kim et al., 2014). Together, these results reveal that TCP8, TCP14, and TCP15 contribute redundantly to establish SAR either directly or indirectly.
Since the reduction of mRNA levels of PR1, PR2, and PR5 in tcp8/14/15 may be a direct or indirect effect, we subsequently investigated whether TCP8, TCP14, and TCP15 can directly bind to the promoters of PR1, PR2, and PR5 through yeast one-hybrid (Y1H) assays. Constructs of pDEST22-TCPs were transformed into the yeast strain YU, and PR promoters constructed in the pLacZi vector were integrated into the yeast strain YM4271 (Pruneda-Paz et al., 2014). Healthy yeast diploids growing on double dropout medium lacking tryptophan and uracil were selected. The binding affinity of TFs to promoters was quantified by β-galactosidase activity shown as fold change over empty vector control. In addition, 3-fold induction was set as the cut-off (Zheng et al., 2015). Expression of any TCP8, TCP14, or TCP15 with PR1 and PR2 promoters fused with the LacZ reporter gene did not activate more than a 3-fold change of β-galactosidase activity (Figure 3A), demonstrating that these TCP TFs did not bind to the promoter of PR1 and PR2; however, expression of AD-TCP15 with the LacZ reporter gene fused PR5 promoter resulted in a 4.13-fold change of β-galactosidase activity compared with vector control (Figure 3A), demonstrating that TCP15 physically bound to the PR5 promoter and functioned as a transcriptional activator in yeast.
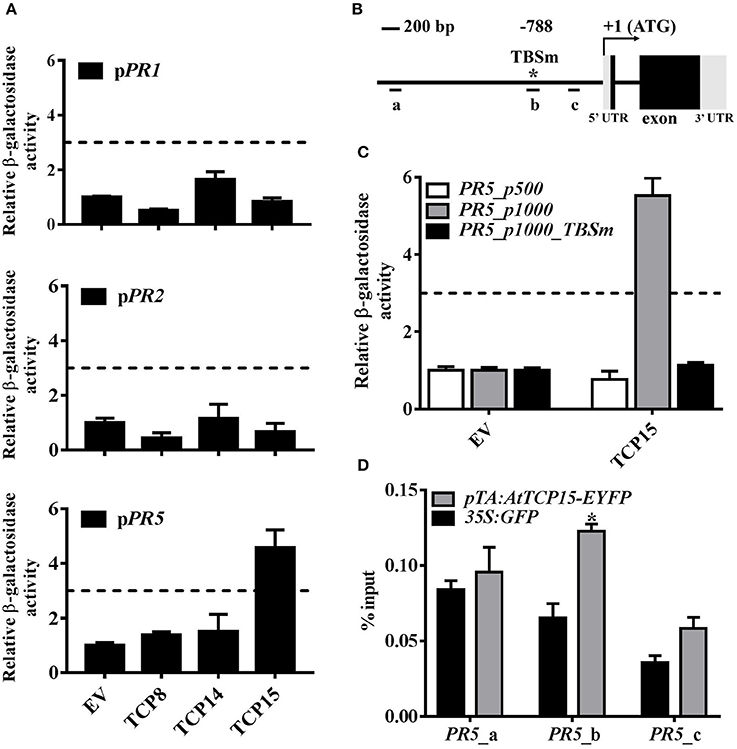
Figure 3. TCP15 binds to the PR5 promoter at the TCP binding site. (A) Interactions between the TCP transcription factors and the PR promoters in yeast one-hybrid system (Y1H). Constructs expressing pDEST22-TCPs and empty pDEST22 (EV) were transformed into the yeast strain YU. The PR promoters fused with LacZ reporter gene were integrated into the yeast strain YM4271. Healthy diploids grew on dropout medium without tryptophan and uracil were selected. Binding ability between TCPs and the PR promoters was quantified by β-galactosidase activity which was calculated in fold change over EV control. Three-fold induction was set as the cut-off. The binding ability of TCP8, TCP14, and TCP15 to the PR1 promoter (upper panel), the PR2 promoter (middle panel), and the PR5 promoter (lower panel) is shown. Error bars represent SD of three repeats. (B) Diagram of the PR5 gene structure. (+1) means transcription start site; long line indicates the promoter of PR5; short lines marked by a, b, and c represent the fragments amplified by RT-PCR in (D); star shows TCP binding site located at −788 bp; TCPm represents TCP binding site with mutations. (C) Interactions between TCP15 with the PR5 promoter containing a TCPm and the PR5 promoter (500 bp) in Y1H assays. Error bars represent SD of three repeats. (D) Association between TCP15 and the PR5 promoter was studied by chromatin immunoprecipitation (ChIP) assays combined with RT-PCR analysis. Twelve-day-old seedlings of pTA:AtTCP15-EYFP and 35S:GFP transgenic plants were treated with 30 μM DEX for 24 h. Samples were collected at another 24 h after 0.5 mM SA induction. The enrichment of PR5 promoter DNA in immunoprecipitated samples was shown as % input. Error bars represent SD of three technical repeats. Statistical analysis was studied by t-test (*p < 0.05) using Excel 2016. Another independent repeat showed similar results.
As a member of the class I TCP TFs, TCP15 was shown to bind to the consensus element KHGGGVC (Davière et al., 2014). Such a motif, GCGGGAC, was found in the promoter of PR5 at −788 bp from the transcription start site (TSS) (Figure 3B). To investigate whether TCP15 targets the PR5 promoter specifically at the TCP binding site (TBS), site mutations (GCGGGAC to ATAAACT) were introduced into the TBS (TBSm). The presence of TCP15 did not activate the expression of the LacZ reporter gene fused after the PR5 promoter with TBSm (Figure 3C). To further confirm the binding specificity, a shorter PR5 promoter (500 bp) without TBS was used. The presence of TCP15 did not activate reporter gene expression either (Figure 3C). These results indicate that the TBS within the promoter of PR5 is required for the binding of TCP15 in yeast.
To further confirm this specific binding in planta, we performed chromatin immunoprecipitation (ChIP) assays using DEX inducible TCP15 transgenic seedlings pTA:AtCP15-EYFP (Li et al., 2012). The relative enrichment of selected PR5 promoter regions was tested by RT-PCR. Primers were designed to amplify three PR5 promoter fragments a, b, and c, in which the b region contained the TBS (Figure 3B). Our data show that fragments a and c did not show obvious enrichment, while fragment b showed significant enrichment in the immunoprecipitated samples from pTA:AtCP15-EYFP compared with 35S:GFP (Figure 3D), demonstrating that TCP15 binds in vivo to the TBS within the promoter of PR5. Our results from Y1H assays and ChIP assays indicate that TCP15 activates the transcription of PR5 via binding to the TBS within its promoter.
To investigate whether TCP15 affects the expression of PR5, the mRNA levels of PR5 in Col-0, npr1-2, and the two TCP15 T-DNA insertion lines tcp15-1 (Kieffer et al., 2011) and tcp15-3 were analyzed after SA treatment using RT-qPCR assays. The SA-induced PR5 transcript level was significantly compromised in tcp15-1 and tcp15-3 compared with Col-0 (Figure 4A), indicating that TCP15 is required for SA-induced PR5 expression.
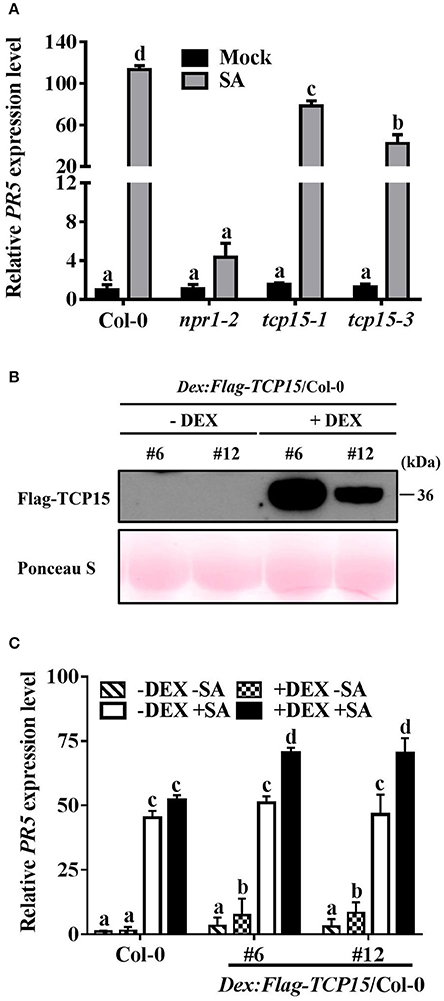
Figure 4. TCP15 regulates the expression of PR5. (A) The expression of PR5 in Col-0, npr1-2, tcp15-1, and tcp15-3. Twelve-day-old seedlings were treated with water (Mock) or 0.5 mM SA (SA) for 24 h. The mRNA levels of PR5 were analyzed by RT-qPCR. Values were normalized to the UBQ5 mRNA levels, then to the value of Col-0 (mock), which was arbitrarily set at 1. Statistical analysis was studied by two-way ANOVA following multiple comparisons with turkey test (95% confidence interval) using GraphPad Prism 7. (B) FLAG-TCP15 protein levels in DEX inducible transgenic lines. Two T3 homozygous lines expressing FLAG-TCP15 under control of a DEX-inducible promoter in the Col-0 background (DEX:FLAG-TCP15/Col-0) were used. Twelve-day-old seedlings were treated with either 0.01% ethanol (–DEX) or 30 μM DEX (+DEX) for 24 h. Protein levels were measured by immunoblot with the anti-FLAG antibody. Rubisco large subunits stained with Ponceau S were used to show equal total protein loading. (C) The expression of PR5 in Col-0 and two DEX:FLAG-TCP15/Col-0 lines. Twelve-day-old seedlings described in (B) were treated with either 0.5 mM SA (+SA) or water (–SA) for 24 h after being treated with 0.01% alcohol (-DEX) or 30 μM DEX (+DEX) for 24 h. The PR5 mRNA levels were examined by RT-qPCR. Values were normalized to the UBQ5 mRNA levels, then to the value of Col-0 (–DEX –SA) which was arbitrarily set at 1. Error bars represent SD of three biological replicates. Statistical analysis was studied by two-way ANOVA following multiple comparisons with turkey test (95% confidence interval) using GraphPad Prism 7. Different small letters above the bars mean significant differences.
To further confirm that TCP15 promotes the expression of PR5, the mRNA levels of PR5 in Col-0, npr1-2, and two DEX inducible TCP15 transgenic lines were analyzed after SA treatment using RT-qPCR assays. Since most constitutive overexpression TCP15 transgenic lines showed growth arrest (Li et al., 2012), Arabidopsis transgenic lines expressing FLAG-TCP15 under the control of a DEX-inducible promoter in the Col-0 background (DEX:Flag-TCP15/Col-0) were generated. Two T3 homozygous transgenic lines, #6 and #10, which exhibited different FLAG-TCP15 protein levels after DEX treatment, were used (Figure 4B). Before SA treatment, the presence of TCP15 in both transgenic lines significantly increased the mRNA levels of PR5 (Figure 4C). With SA treatment, the presence of TCP15 increased the PR5 mRNA levels to a dramatically higher level (Figure 4C), indicating that the up-regulation of PR5 by TCP15 was promoted by SA. Taken together, these results indicate that TCP15 positively regulates the expression of PR5.
Without a DNA binding domain, NPR1 functions as a transcriptional co-activator to facilitate TGA TFs binding to the promoter of PR1 by interacting with TGA TFs (Zhang et al., 1999; Fan and Dong, 2002; Johnson et al., 2003). To test whether NPR1 can enhance TCP15 binding to the PR5 promoter, we performed ChIP assays using Arabidopsis transgenic lines expressing FLAG-TCP15 under the control of a DEX-inducible promoter in the Col-0 background and npr1-2 background (DEX:FLAG-TCP15/npr1-2). The DEX:FLAG-TCP15/Col-0 transgenic lines #6 and #10 and DEX:FLAG-TCP15/npr1-2 transgenic lines #13 and #17, which expressed similar Flag-TCP15 protein levels after DEX application (Figure 5A), were used. FLAG-TCP15 proteins and their cross-linked DNA were immuno-precipitated from chromatin extracts using beads bound with the anti-FLAG antibody. Immuno-precipitated DNA was analyzed by RT-PCR using primers amplifying fragment b described in Figure 3B. Fragment b showed significantly higher enrichment in DEX:FLAG-TCP15/Col-0 compared with DEX:FLAG-TCP15/npr1-2 (Figure 5B), demonstrating that NPR1 facilitates TCP15 binding to the promoter of PR5 in Arabidopsis.
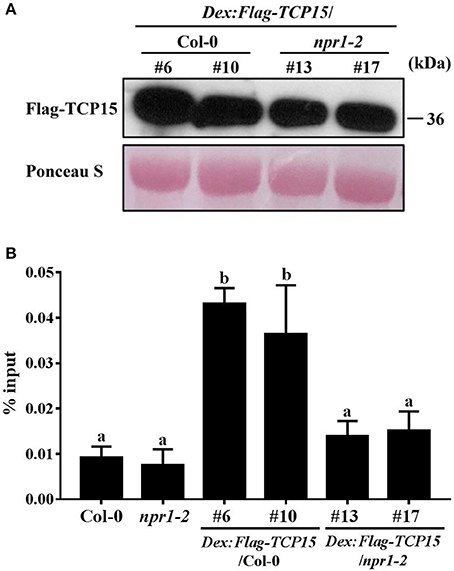
Figure 5. NPR1 enhances TCP15 binding to the PR5 promoter. (A) FLAG-TCP15 protein levels in DEX inducible transgenic lines. Twelve-day-old seedlings of T3 homozygous lines expressing FLAG-TCP15 under the control of a DEX-inducible promoter in the Col-0 and npr1-2 background (DEX:FLAG-TCP15/npr1-2) were treated with 30 μM DEX for 24 h. Protein levels were measured by immunoblot with the anti-FLAG antibody. (B) The binding ability of TCP15 to the PR5 promoter in transgenic plants described in (A) were examined with ChIP assays combined with RT-PCR analysis. Twelve-day-old seedlings were treated with 30 μM DEX, then treated with 0.5 mM SA after 24 h. Samples were collected 24 h after SA treatment. Primers amplifying fragment b described in Figure 3B were used in RT-PCR. The enrichment of PR5 promoter DNA in immunoprecipitated samples was shown as % input. Error bars represent SD of three technical replicates. Statistical analysis was studied by two-way ANOVA following multiple comparisons with turkey test (95% confidence interval) using GraphPad Prism 7. Different small letters above the bars mean significant differences. Another independent replication demonstrated similar results.
In this work, we provide clear evidence that TCP8, TCP14, and TCP15 physically interact with NPR1 and coordinately contribute to SAR establishment, that TCP15 positively regulates the expression of PR5 by directly binding to the TBS within the PR5 promoter, and that NPR1 can enhance this binding. Our results suggest that in addition to acting as mediators of SA biosynthesis (Wang et al., 2015), TCP TFs are also essential players in the SA signaling pathway, and that TCP proteins not only contribute to ETI (Kim et al., 2014), but also function in SAR.
Taking advantage of the comprehensive Arabidopsis transcription factor library (Pruneda-Paz et al., 2014), TCP15 was identified as a novel NPR1 interactor through Y2H screens. Interactions between NPR1 and all the 24 Arabidopsis TCP proteins were also tested using Y2H assays, and more interactors were identified (unpublished data). As a critical transcriptional regulatory node, NPR1 directly regulates the expression of 2,248 SA-responsive genes (Wang et al., 2006). The absence of a DNA binding domain and the presence of two protein-protein interaction domains suggest NPR1 regulates the expression of downstream genes through interactions with TFs (Fu and Dong, 2013). Apparently, the known interactions between NPR1 and TGAs are not sufficient to explain the expression of all the genes regulated by NPR1. TCP proteins, as novel NPR1 interactors, will be excellent candidates to fill these missing links.
TCP proteins were reported to mediate SA biosynthesis by coordinately regulating the expression of ICS1, which encodes an enzyme responsible for pathogen-induced SA biosynthesis (Wang et al., 2015). This suggests that TCP proteins can affect the activity of NPR1. Although one conserved TCP binding motif was located at −169 bp from the TSS within the promoter of NPR1, no direct interaction was found between the NPR1 promoter and TCP proteins in yeasts (Supplementary Figure 3), suggesting that TCP proteins could affect NPR1 activity indirectly. Because TCP8, TCP14, and TCP15 can interact with each other, and one characteristic of TCP TFs is their functional redundancy (Kim et al., 2014), it is not surprising that SAR could still be induced in the tcp8-1, tcp14-5, and tcp15-3 single or corresponding double mutants (Figure 2A). SAR was abolished in the npr1-2 mutants, while it was partially compromised in the tcp8/14/15 triple mutants (Figure 2A), suggesting that other TFs are required in NPR1-mediated SAR signaling pathway. These TFs could be TGA proteins or other NPR1-interacting TCP proteins.
In addition, failed SAR induction could be caused by decreased initial immunity signal production and/or blocked mobile signal transduction. The expression of SAR marker genes (PR1, PR2, and PR5) in both local and systemic leaves were all significantly decreased after local infection with an avirulent pathogen (Figures 2B,C), consistent with a reduced initial immune signal production in tcp8/14/15 plants (Kim et al., 2014). Our finding that TCP15 directly promotes the expression of PR5 (Figures 4A,B) supports this explanation. As TCP8 and TCP14 did not bind the promoter of PR5 in yeast, they may facilitate TCP15 binding to the promoter of PR5 through a complex formation. Since TCP8, TCP14, and TCP15 did not show direct interactions with the promoter of PR1 and PR2 in Y1H assays (Figure 3A), the decrease of PR1 and PR2 may be indirectly regulated by these TCP TFs; however, whether TCP8, TCP14, and TCP15 are involved in SAR by mediating the activity of a mobile signal is unknown and will be an exciting topic to study in the future. Because PR1 and PR2 promoters both have TGA binding sites, it is possible that either TCP8, TCP14, and TCP15 interact with TGA transcription factors or the interactions between NPR1 and TCP8/14/15 promote the interactions between NPR1 and TGAs, to facilitate the expression of PR1 and PR2 to establish SAR.
How TCP15 responses to SA signaling and bind to the promoter of PR5 is unclear. Based on the activity of TCP15 is dependent on redox modulation (Viola et al., 2013, 2016), the molecular mechanism of how SA induces the TCP15-dependent PR5 expression is proposed. Under oxidizing conditions, the DNA binding activity of TCP15 is inhibited by dimers formed with disulfide bonds, while under reducing conditions induced by SA accumulation, the inhibition is reversed, resulting in extensively upregulated expression of PR5.
Although NPR1 could enhance TCP15 binding to the promoter of PR5 (Figure 5B), the exact molecular mechanism through which NPR1 facilitates this association is unclear and will be an interesting topic to investigate. It is perhaps significant in this context that NPR1 activity is also regulated by SUMOylation-induced complex formation (Saleh et al., 2015), and TCP8, TCP14, and TCP15 were recently shown to associate with the nuclear SUMOylation machinery and to be SUMOylated as well (Mazur et al., 2017).
ML, HC, JC, MC, WG, FL, and ZF contributed conception and design of the study. ML collected the data. ML, HC, WG, JC, and ZF organized the data. ML performed the statistical analyses. ML, IP, WG, and ZF wrote the manuscript. All authors contributed to manuscript revision, and have read and approved the submitted version.
This work is supported by a SPARC Graduate Research Grant (2017-2018) sponsored by the University of South Carolina (to ML) and the National Science Foundation (grant IOS-1758994 to ZF and grant IOS-1456181 to WG).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ai-wu Dong for providing the seeds of pTA:TCP15-EYFP and 35:GFP transgenic lines; Li-jia Qu for providing the pDEST22 empty vector; and Steve A. Kay for providing the pLacZi plasmid, yeast strain YM4271 and YU. We thank Johannes Stratmann, Beth Krizek, Hexin Chen, and Kim E. Creek for working as the committee members of ML, and for providing suggestions about the experimental designs used in this study. We thank the Chinese Scholarship Council for providing a fellowship to ML.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01153/full#supplementary-material
Aggarwal, P., Das Gupta, M., Joseph, A. P., Chatterjee, N., Srinivasan, N., and Nath, U. (2010). Identification of specific DNA binding residues in the TCP family of transcription factors in Arabidopsis. Plant Cell 22, 1174–1189. doi: 10.1105/tpc.109.066647
Boyle, P., Le Su, E., Rochon, A., Shearer, H. L., Murmu, J., Chu, J. Y., et al. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21, 3700–3713. doi: 10.1105/tpc.109.069971
Breen, S., Williams, S. J., Outram, M., Kobe, B., and Solomon, P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Chen, H., Chen, J., Li, M., Chang, M., Xu, K., Shang, Z., et al. (2017). A bacterial Type III effector targets the master regulator of salicylic acid signaling, NPR1, to subvert plant immunity. Cell Host Microbe 22, 777–788. doi: 10.1016/j.chom.2017.10.019
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222. doi: 10.1046/j.1365-313X.1999.00444.x
Davière, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Edreva, A. (2005). Pathogenesis-related proteins: research progress in the last 15 years. Gen. Appl. Plant Physiol. 31, 105–124.
Fan, W., and Dong, X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Johnson, C., Boden, E., and Arias, J. (2003). Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. Plant Cell 15, 1846–1858. doi: 10.1105/tpc.012211
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 68, 147–158. doi: 10.1111/j.1365-313x.2011.04674.x
Kim, S. H., Son, G. H., Bhattacharjee, S., Kim, H. J., Nam, J. C., Nguyen, P. D., et al. (2014). The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 78, 978–989. doi: 10.1111/tpj.12527
Komar, D. N., Mouriz, A., Jarillo, J. A., and Piñeiro, M. (2016). Chromatin immunoprecipitation assay for the identification of Arabidopsis protein-DNA interactions in vivo. J. Vis. Exp. 107:e53422. doi: 10.3791/53422
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, Z. Y., Li, B., and Dong, A. W. (2012). The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 5, 270–280. doi: 10.1093/mp/ssr086
Lucero, L. E., Manavella, P. A., Gras, D. E., Ariel, F. D., and Gonzalez, D. H. (2017). Class I and class II TCP transcription factors modulate SOC1-dependent flowering at multiple levels. Mol. Plant 10, 1571–1574. doi: 10.1016/j.molp.2017.09.001
Martín-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Mazur, M. J., Spears, B. J., Djajasaputra, A., van der Gragt, M., Vlachakis, G., Beerens, B., et al. (2017). Arabidopsis TCP transcription factors interact with the SUMO conjugating machinery in nuclear foci. Front. Plant Sci. 8:2043. doi: 10.3389/fpls.2017.02043
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Mukhtar, M. S., Carvunis, A. R., Dreze, M., Epple, P., Steinbrenner, J., Moore, J., et al. (2011). Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601. doi: 10.1126/science.1203659
Nicolas, M., and Cubas, P. (2016). TCP factors: new kids on the signaling block. Curr. Opin. Plant Biol. 33, 33–41. doi: 10.1016/j.pbi.2016.05.006
Ou, B., Yin, K. Q., Liu, S. N., Yang, Y., Gu, T., Wing Hui, J. M. et al. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4, 546–555. doi: 10.1093/mp/ssr002
Pruneda-Paz, J. L., Breton, G., Nagel, D. H., Kang, S. E., Bonaldi, K., Doherty, C. J., et al. (2014). A genome-scale resource for the functional characterization of Arabidopsis transcription factors. Cell Rep. 8, 622–632. doi: 10.1016/j.celrep.2014.06.033
Resentini, F., Felipo-Benavent, A., Colombo, L., Blázquez, M. A., Alabad,í, D., and Masiero, S. (2015). TCP14 and TCP15 mediate the promotion of seed germination by gibberellins in Arabidopsis thaliana. Mol. Plant 8, 482–485. doi: 10.1016/j.molp.2014.11.018
Rochon, A., Boyle, P., Wignes, T., Fobert, P. R., and Després, C. (2006). The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 18, 3670–3685. doi: 10.1105/tpc.106.046953
Rogers, E. E., and Ausubel, F. M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alterations in PR-1 gene expression. Plant Cell 9, 305–316. doi: 10.1105/tpc.9.3.305
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H. Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. doi: 10.1105/tpc.8.10.1809
Saleh, A., Withers, J., Mohan, R., Marqués, J., Gu, Y., Yan, S., et al. (2015). Posttranslational modifications of the master transcriptional regulator NPR1 enable dynamic but tight control of plant immune responses. Cell Host Microbe 18, 169–182. doi: 10.1016/j.chom.2015.07.005
Uberti-Manassero, N. G., Lucero, L. E., Viola, I. L., Vegetti, A. C., and Gonzalez, D. H. (2012). The class I protein AtTCP15 modulates plant development through a pathway that overlaps with the one affected by CIN-like TCP proteins. J. Exp. Bot. 63, 809–823. doi: 10.1093/jxb/err305
Uknes, S., Mauch-Mani, B., Moyer, M., Potter, S., Williams, S., Dincher, S., et al. (1992). Acquired resistance in Arabidopsis. Plant Cell 4, 645–656. doi: 10.1105/tpc.4.6.645
Viola, I. L., Camoirano, A., and Gonzalez, D. H. (2016). Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 170, 74–85. doi: 10.1104/pp.15.01016
Viola, I. L., Güttlein, L. N., and Gonzalez, D. H. (2013). Redox modulation of plant developmental regulators from the class I TCP transcription factor family. Plant Physiol. 162, 1434–1447. doi: 10.1104/pp.113.216416
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wang, X., Gao, J., Zhu, Z., Dong, X., Wang, X., Ren, G., et al. (2015). TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J. 82, 151–162. doi: 10.1111/tpj.12803
Ward, E. R., Uknes, S. J., Williams, S. C., Dincher, S. S., Wiederhold, D. L., Alexander, D. C., et al. (1991). Coordinate gene activity in response to agents that induce systemic acquired resistance. Plant Cell 3, 1085–1094. doi: 10.1105/tpc.3.10.1085
Weßling, R., Epple, P., Altmann, S., He, Y., Yang, L., Henz, S. R., et al. (2014). Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host Microbe 16, 364–375. doi: 10.1016/j.chom.2014.08.004
Yang, L., Teixeira, P. J., Biswas, S., Finkel, O. M., He, Y., Salas-Gonzalez, I., et al. (2017). Pseudomonas syringae Type III effector HopBB1 promotes host transcriptional repressor degradation to regulate phytohormone responses and virulence. Cell Host Microbe 21, 156–168. doi: 10.1016/j.chom.2017.01.003
Zhang, N., Wang, Z., Bao, Z., Yang, L., Wu, D., Shu, X., et al. (2018). MOS1 functions closely with TCP transcription factors to modulate immunity and cell cycle in Arabidopsis. Plant J. 93, 66–78. doi: 10.1111/tpj.13757
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Keywords: plant immunity, systemic acquired resistance, transcriptional regulation, NON-EXPRESSER OF PR GENES 1, TCP transcription factors, PATHOGENESIS-RELATED genes
Citation: Li M, Chen H, Chen J, Chang M, Palmer IA, Gassmann W, Liu F and Fu ZQ (2018) TCP Transcription Factors Interact With NPR1 and Contribute Redundantly to Systemic Acquired Resistance. Front. Plant Sci. 9:1153. doi: 10.3389/fpls.2018.01153
Received: 14 May 2018; Accepted: 19 July 2018;
Published: 14 August 2018.
Edited by:
Hua Lu, University of Maryland, Baltimore County, United StatesReviewed by:
Zhilong Bao, Shandong Agricultural University, ChinaCopyright © 2018 Li, Chen, Chen, Chang, Palmer, Gassmann, Liu and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fengquan Liu, ZnFsaXUyMDAxMUBzaW5hLmNvbQ==
Zheng Qing Fu, emZ1QG1haWxib3guc2MuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.