- Department of Plant Sciences, University of Saskatchewan, Saskatoon, SK, Canada
Polyphenols comprise the largest group of plant secondary metabolites and have critical roles in plant physiology and response to the biotic and abiotic environment. Changes in the content of polyphenols in the root extracts and root tissues of wild (Lens ervoides) and cultivated (Lens culinaris) lentil genotypes were examined in response to infection by Aphanomyces euteiches using liquid chromatography mass spectrometry (LC-MS). Genotype, infection and their interaction determined the composition of polyphenols in lentil roots. The levels of several polyphenols were lower in the root extract of the low-tannin genotype L. culinaris ZT-4 compared to L. ervoides L01-827A. Kaempferol derivatives including kaempferol dirutinoside and kaempferol 3-robinoside 7-rhamnoside were more concentrated in the healthy root tissues of L. ervoides L01-827A than in L. culinaris genotypes. Infection increased the concentration of kaempferol, apigenin, and naringenin in the root tissues of all genotypes, but had no effect on some polyphenols in the low-tannin genotype L. culinaris ZT-4. The concentrations of apigenin, naringenin, apigenin 4-glucoside, naringenin7-rutinoside, diosmetin, and hesperetin 7-rutinoside were higher in the infected root tissues of L. ervoides L01-827A compared with the L. culinaris genotypes. Organic acids including coumaric acid, vanillic acid, 4-aminosalicylic acid, 4-hydroxybenzoic acid, and 3,4-dihydroxybenzoic acid effectively suppressed the in-vitro hyphal growth of A. euteiches. Some of these bioactive polyphenols were more concentrated in roots of L. ervoides L01-827A but were low to undetectable in ZT-4. This study shows that genotypic differences exist in the composition of root polyphenols in lentil, and is related to the response to infection caused by A. euteiches. Polyphenols, particularly the organic acid content could be useful for selection and breeding of lentil genotypes that are resistant to Aphanomyces root rot (ARR) disease.
Introduction
Lentil (Lens culinaris Medik.) is an important cool season legume, grown in more than 70 countries around the world. Canada is the largest lentil producer, contributing nearly 41% of the global production (FAO, 2017). Lentil seeds are a rich source of proteins, carbohydrates, vitamins, minerals, fibers and antioxidants (Roy et al., 2010). They also contain non-nutritional compounds such as tannins and phytic acid that serve as a defense mechanism against pathogens, insects and parasites (Urbano et al., 2007; Constabel et al., 2014; Sánchez-Chino et al., 2015). Lens species differ in the size of seeds (5–90 mg per seed) and the color and pattern of seed coats, which vary from yellow or gray to dark brown (Ladizinsky, 1979a,b; Ferguson et al., 2000). As a legume crop, lentil provides agro-ecological services by incorporating atmospheric nitrogen into the soil through biological fixation, and promoting the diversity of soil microbial communities (Borrell et al., 2016).
Aphanomyces euteiches is a soil-borne oomycete that causes Aphanomyces root rot (ARR) in many legume crops such as lentil, pea (Pisum sativum L.), alfalfa (Medicago sativa L.), common bean (Phaseolus vulgaris L.), and red clover (Trifolium pratense; Wicker et al., 2001; Chatterton et al., 2016). This pathogen has recently been reported in western Canada (Armstrong-Cho et al., 2014; Chatterton et al., 2016). ARR can cause major crop losses especially under environmental conditions that are conducive to pathogen growth and disease development (McPhee, 2003; Gossen et al., 2016). Susceptible crop plants generally have a limited range of genetic resistance. No effective seed treatment is available for protection against A. euteiches, and its spores are long-lived in soil (Gossen et al., 2016). These characteristics make ARR the most difficult and serious root pathogen of susceptible legumes. Crop rotation and the use of cultivars with partial resistance are the only efficient ways to control ARR (Wicker et al., 2001; Moussart et al., 2013).
Polyphenols are the largest group of secondary metabolites and have very diverse structures (Cheynier, 2012) that make them unique and multifunctional natural products in plants (Quideau et al., 2011). They often have important ecological roles and are involved in a range of functions in plant growth, development, and defense. Polyphenols can attract, repel or protect plants against insects, fungi, bacteria, and viruses (Bennett and Wallsgrove, 1994; Daayf et al., 2012; Olivoto et al., 2017). Synthesis of polyphenols is a complex process that is associated with the shikimate, malonate, and phenylpropanoid pathways (Lattanzio, 2013) and is encoded by multiple genes (Vermerris and Nicholson, 2008; Olivoto et al., 2017). A detailed understanding of how these polyphenols fit into the biochemical pathways for disease resistance is necessary to develop strategies to control plant diseases.
The roots of plants are able to produce a diversity of compounds that directly or indirectly influence microbial species (Lanoue et al., 2010; Baetz and Martinoia, 2014; Bazghaleh et al., 2016). The production of antimicrobial polyphenols have been reported in various plant species (Puupponen-Pimiä et al., 2001; Deng et al., 2015; Shalaby and Horwitz, 2015; Pagliarulo et al., 2016) which could be independent or part of a plant's response to infection. Some polyphenols accumulate in plant tissues even prior to an active defense response (Nicholson and Hammerschmidt, 1992; Balmer et al., 2015). Knowledge of the components of the range of polyphenols in the root tissues and root extracts of crop plants and the potential effect of those compounds on pathogenic microbial species could be used as a first step for identifying plant genotypes that are more resistant to plant diseases (Wink, 1988; Reuveni et al., 1991; Wille et al., 2018).
Polyphenols have been detected in plants roots and their effects on soil pathogens described (Evidente et al., 2010), however, little is known about the composition of these metabolites, particularly in legume plants. This study tested the hypothesis that lentil genotypes have different compositions of polyphenols in their root tissues and root extracts. We also hypothesized that infection by A. euteiches alters the accumulation of specific polyphenols in the root tissues of lentil, and that some of these might inhibit the hyphal growth of A. euteiches. We compared the polyphenol profiles found in root tissues and root extracts of selected genotypes of cultivated (L. culinaris) and wild (Lens ervoides) lentil, including a low-tannin cultivar, using liquid chromatography-mass spectrometry. The effect of the individual polyphenols on the growth of A. euteiches was investigated in vitro. The aim of this study was to identify potential polyphenols that could be involved in providing resistance to ARR in lentil.
Materials and Methods
Composition of Polyphenols in Root Extracts of Lentil
Experimental Design and Plant Growth Conditions
Seeds of the L. culinaris cultivars ZT-4 (low-tannin), CDC Maxim (gray seed coat), Eston (green seed coat), and L. ervoides genotype L01-827A (wild type seed coat) were obtained from the Crop Development Centre at the University of Saskatchewan (Saskatoon, SK, Canada). The lentil genotypes were selected based on differences in the color and polyphenolic profile of their seed coats (Mirali et al., 2016a,b, 2017). The seeds were scarified by puncturing the seed coats using a small blade. The seeds were surface sterilized by successive immersion in 95% ethanol for 30 s, in sterile distilled water for 30 s, in 2.5% Javex® bleach solution (sodium hypochlorite) for 2 min, and then in distilled water for 5 min. The seeds were germinated on moist, sterile filter paper in Petri dishes in the dark at 25°C for 72 h prior to use. Eight germinated seeds of each genotype were transplanted into a 10 cm plastic pot filled with porous ceramic media (Profile® Greens Grade™, BrettYoung, Edmonton, AB). The planting holes in the media were treated with a peat-based Rhizobium leguminosarum inoculant (Nitragin Nitrastick GC®, Nitragin Inc., Brookfield, WI, USA). The pots were arranged as a randomized complete block design in four replicates. Plants were grown under 22/16°C d/night temperatures and 16/8 h d/night length for 30 d in a growth room (GR48, Conviron, Winnipeg, MB) located in the controlled environment facilities of the College of Agriculture and Bioresources at the University of Saskatchewan. Distilled water was added to pots as needed, and plants fertilized weekly with 100 mL of the half strength modified Hoagland's nutrient solution.
Collection of Roots and Extraction of Polyphenols
All plants in each pot were gently removed 30 d after transplanting. The intact roots were collected, weighed, and transferred into 50 mL tubes containing 10 mL 80% MeOH. Polyphenols that had been released from root tissues were extracted by soaking roots in 80% MeOH for 72 h. The tubes were stored at −20°C until further use. After 3 d, the tube was thawed at room temperature, vortexed, and 1.5 mL was transferred into a 2 mL centrifuge tube, which was centrifuged for 5 min at 12,000 rpm. Then 1 mL of the supernatant was removed and dried using a benchtop centrifugal vacuum concentrator (Labconco, Kansas City, Mo) for approximately 2 h and immediately reconstituted in 200 μL of the recon solution (90:10 H2O:MeOH v/v) to reconstitute the extract.
Composition of Polyphenols in Root Tissues of Healthy and Diseased Lentil Plants
Experimental Design and Plant Growth Conditions
A factorial pot experiment with four lentil genotypes and two infection levels was conducted in a controlled temperature environment. Seeds of the lentil genotypes ZT-4, CDC Maxim, Eston, and L. ervoides L01-827A were scarified, surface sterilized, and germinated as described for experiment 1. Eight germinated seeds of each genotype were transplanted into 10 cm plastic pots filled with peat and vermiculite-based media, Sunshine Mix 3® (Sun Gro Horticulture Canada Ltd.). The planting holes in the media were treated with a peat-based R. leguminosarum inoculant (Nitragin Nitrastick GC®, Nitragin Inc., Brookfield, WI, USA). The pots were arranged in four replicates. Plants were grown under 22/16°C d/night temperatures and 16/8 h d/night length for 30 d in the growth room (GR48, Conviron, Winnipeg, MB). Distilled water was added to pots as needed, and plants were fertilized weekly with 100 mL of the half strength modified Hoagland's nutrient solution. Roots of the infected plants were inoculated with zoospores of A. euteiches. The experiment was repeated two times.
Preparation of Inoculum of Aphanomyces euteiches and Root Infection
Zoospores of A. euteiches were produced following a protocol developed in the Plant Pathology Laboratory, University of Saskatchewan, Canada (Banniza, pers. comm.). After counting the zoospores using a haemocytometer, the final concentration was adjusted to 5 × 103 zoospores mL−1 with sterile deionized water. Ten days after planting, a 5-mL aliquot of the zoospore suspension was injected into the soil near the base of the lentil plants.
Collection of Roots and Extraction of Root Polyphenols
Fourteen days after inoculation, the plants were uprooted and the adhering media (Profile® Greens Grade™, Winnipeg, MB) were removed from the roots by soaking the roots in the tap water. The roots were then visually evaluated for infection on the basis of discoloration and root decay. The roots were stored at −80°C until use.
Polyphenols were extracted from roots of healthy and diseased plants using a procedure similar to that of Mirali et al. (2014) with a few modifications. One hundred fifty milligram of fresh root tissue was placed in 2 mL Sarstedt microtube and then freeze dried over 24 h. The freeze-dried samples were weighed, and then one ¼- inch ceramic bead (MP Bio, Cat. No. 6540-412) was placed in each tube along with 1 mL of an extraction solvent containing 70:30 v/v acetone:water plus internal standards. The root tissues were pulverized using a Mini-Beadbeater-16 (BioSpec Products, Inc. US) for 2 × 2.5 min followed by shaking using an Eppendorf mixer for 1 h at 1,400 rpm and 23°C. After centrifuging the tubes at 12,000 rpm for 5 min, 500 μL of the supernatant was pipetted into 1.5 mL Eppendorf tubes and centrifuged again at 12,000 rpm for 5 min. From the Eppendorf tube, 200 μL of the supernatant was pipetted into a new 1.5 mL Eppendorf tube and dried down for approximately 2 h using a benchtop centrifugal vacuum concentrator (Labconco, Kansas City, MO). The supernatant was then reconstituted by adding 200 μL of 90:10 MiliQ-water:MeOH to each tube followed by vortexing for 20 sec and shaking on an Eppendorf mixer for 30 min at 1,400 rpm at 23°C. The tubes were centrifuged at 12,000 rpm for 5 min and then 150 μL of the supernatant was transferred into LC vials and used for LC-MS analysis.
LC-MS Analysis
Polyphenols were purchased from Sigma-Aldrich (Missouri, USA), and Extrasynthese (Genay, France) except for the deuterated compounds, which were purchased from Toronto Research Compounds (Toronto, Canada).
Root tissues and root extracts of the lentil plants were analyzed for polyphenols using a diode-array detector (DAD) for UV-vis detection and a targeted LC-MS method. Since all polyphenols absorb UV-vis, the DAD detector detects all polyphenols in the sample and the MS gives the m/z of the ion(s) within each peak. The targeted method (list of polyphenols analyzed is given in Table 1) uses LC-selective reaction monitoring (SRM) and is based on the method initially developed by Mirali et al. (2014), that has been more recently updated (Mirali et al., 2016a; Purves et al., 2016). Note that catechin-13C3, 4-hydroxybenzoic acid-13C7, ferulic acid-d3, resveratrol 4-hydroxyphenyl-13C6, vanillin ring-13C3, and quercetin-d3 were used as internal standards. The m/z values used for the parent and fragment ions for each of the polyphenols are given in Table 1.
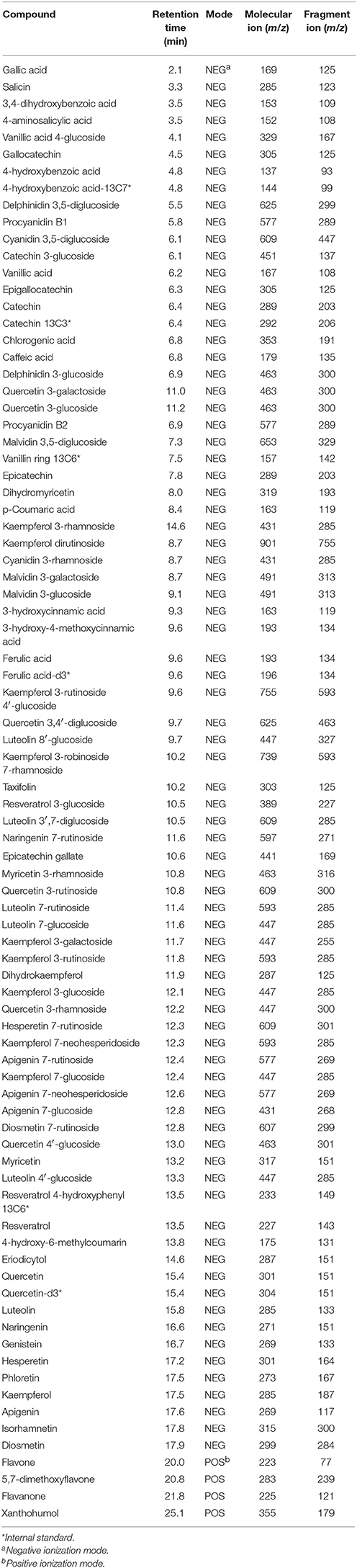
Table 1. Polyphenols analyzed in the root extracts and root tissues of cultivated lentil genotypes ZT-4, CDC Maxim, Eston, and Lens ervoides L01-827A using liquid chromatography-mass spectrometry.
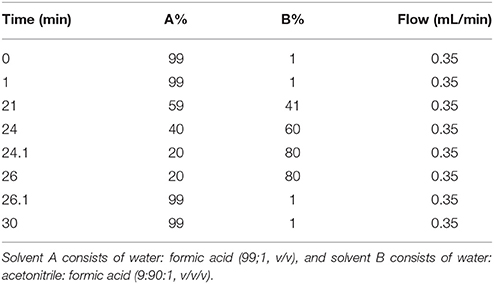
Previous chromatographic conditions optimized by Mirali et al. (2016a) were applied with some modifications. An Agilent 1290 UPLC (Agilent Technologies, Santa Clara, CA) equipped with an auto sampler (G4226A), a binary pump (G4220), a thermostatted column compartment (G1316), and a diode array detector (G4212) was used. Compounds were separated using a Phenomenex (Torrance, CA) core-shell Kinetex pentafluorophenyl (PFP) column (100 mm × 2.1 mm, 2.6 μm particle size). The mobile phase employed a 30 min binary gradient shown in Table 2; solvent A consisted of H2O: CH2O2 99:1, and solvent B was ACN:H2O: CH2O2 90:9:1 (v/v). The injection volume was 5 μL and the flow rate was 0.35 mL/min. Polyphenols exiting the column were detected by the DAD and then subsequently with a Thermo Fisher TSQ Vantage triple quadrupole mass spectrometer equipped with a heated electrospray ionization (HESI) interface.
Data were processed by comparing peak areas of polyphenols normalized to the peak area of an internal standard (IS) using Thermo Xcalibur 2.2 software. Each compound (Table 1) was quantified using a 4-point calibration curve that was obtained using serial dilutions. Kaempferol dirutinoside and catechin-3-glucoside were not commercially available, however, their presence has been reported in lentil seeds, and they were examined here based on previous reports (Aguilera et al., 2010; Mirali et al., 2016a). The standard curves of kaempferol 3-rutinoside and catechin were used to estimate the quantities of these compounds, respectively. Concentration values were converted to ng/g based on dry sample weight.
Bioassay of the Effect of Polyphenols on Mycelial Growth of Aphanomyces euteiches
The polyphenols found in the root tissues and root extracts were evaluated for their effect on mycelial growth of the pathogen A. euteiches (AE1) isolated from Saskatchewan soils. Commercially available polyphenols were used to prepare the stock solutions. A 1 mm well was created in the center of 5% corn meal agar (CMA) plates. An aliquot of 20 μL of 1 g L−1 of each polyphenol was added to the well.
Control plates received 20 μL of sterilized deionized water and a 1 mm plug of A. euteiches was cut from the margins of an actively growing culture plate and placed in the well. Inoculated plates were incubated in the dark at 22°C and radial growth was measured daily for up to 5 d. Percentage inhibition or stimulation of radial growth was calculated using the formula:
Where C = growth in the control plate and T = growth in the treated plate (Dickinson and Skidmore, 1976). The experiment was repeated three times with three replicated plates per treatment.
Polyphenols found to be active against the pathogen A. euteiches were selected for further examination of their minimum inhibitory concentration. Using the same protocol described above, three concentrations of the selected polyphenols were examined for their effect on radial growth of the pathogen. Each concentration included a 10 times dilution level. We used a wide range of concentrations for each polyphenol to detect the possible inhibitory effects. The experiment was repeated three times with three replicated plates per treatment.
Statistical Analysis
Analysis of variance (ANOVA) was performed to test the significance of the effect of genotype, treatments and the interactions between genotype and treatment on the concentration of polyphenols in roots tissues. The least significant difference (LSD) test was used to determine differences in the concentration of the compounds among genotypes and between the treatments. LSD was also used to test differences among genotypes in the concentration of the compounds in the root extract. A p-value of 0.05 was used as the threshold below which the null hypothesis was rejected. Comparisons of mean values and principal component analysis (PCA) were completed using R package (R, v.3.3.2) (www.r-project.org). A Dunnett's test was used to compare the A. euteiches mycelial inhibition of each polyphenol/concentration with control (P < 0.05).
Results
Since all polyphenols absorb in the UV-vis region, the combination of LC-DAD with LC-MS is a powerful approach for investigating polyphenols. Figure 1 shows a comparison of LC-DAD spectra (UV-vis 250–600 nm) obtained using extracts of (A) healthy roots and (B) diseased root tissues of lentil genotype ZT-4. The figure shows that the most significant changes in the spectra are observed at 9.75 and 12.41 min, with less significant changes being observed at other retention times. The compound at 9.75 min has an m/z of 755 (negative mode MS) and no standard has this m/z and retention time in our targeted LC-SRM method. This compound was observed to have a similar product ion spectrum compared with kaempferol 3-O-rutinoside-4′-glucoside (m/z 755, tr = 9.6 min included in the targeted method) and therefore is consistent with being a structural isomer of kaempferol 3-O-rutinoside-4′-glucoside. Similarly, the compound at 12.41 min has an m/z of 901 (negative mode) and no standard has this m/z and retention time in our targeted method. This unknown compound was observed to have many of the same product ions as kaempferol dirutinoside (m/z 901, tr = 8.69 min, included in the targeted method) and therefore is consistent with being a structural isomer of kaempferol dirutinoside. Thus, the UV-vis spectra not only detected large changes in the profile but also the presence of polyphenols not included in the targeted SRM method. For this study, we are focusing on quantifying polyphenols already present in our library.
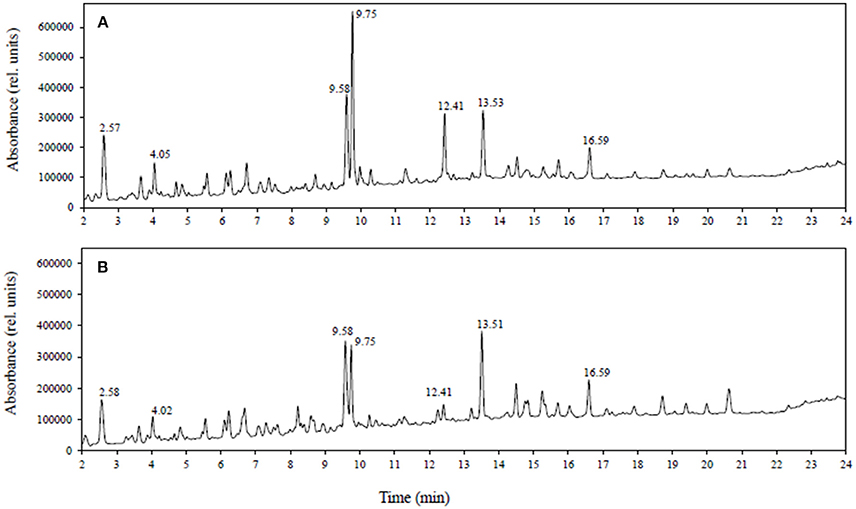
Figure 1. LC-DAD spectra (UV-vis: 250–600 nm) obtained from tissue extracts of (A) healthy roots and (B) diseased roots of lentil genotype ZT-4.
Biochemical Analysis of Root Extracts
Fourteen polyphenols detected by LC-SRM were common to the root extracts of all 4 genotypes of lentil. Variation in the level of these polyphenols among the 4 genotypes of lentil was shown in Figure 2. Lens ervoides L01-827A had elevated amounts of most of the polyphenols in its root extract (Table 3).
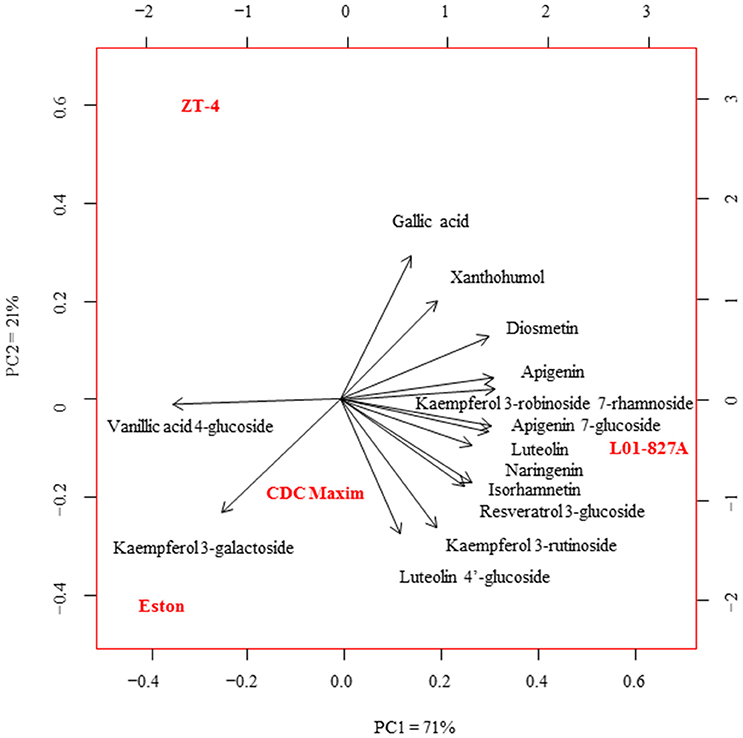
Figure 2. Principal component analysis of the concentration of phenolic compounds in the root extracts of cultivated lentil genotypes ZT-4, CDC Maxim and Eston and Lens ervoides L01-827A using liquid chromatography-mass spectrometry (n = 4). Phenolic compounds were extracted from root extracts using 80% MeOH, and analyzed using LC-SRM.
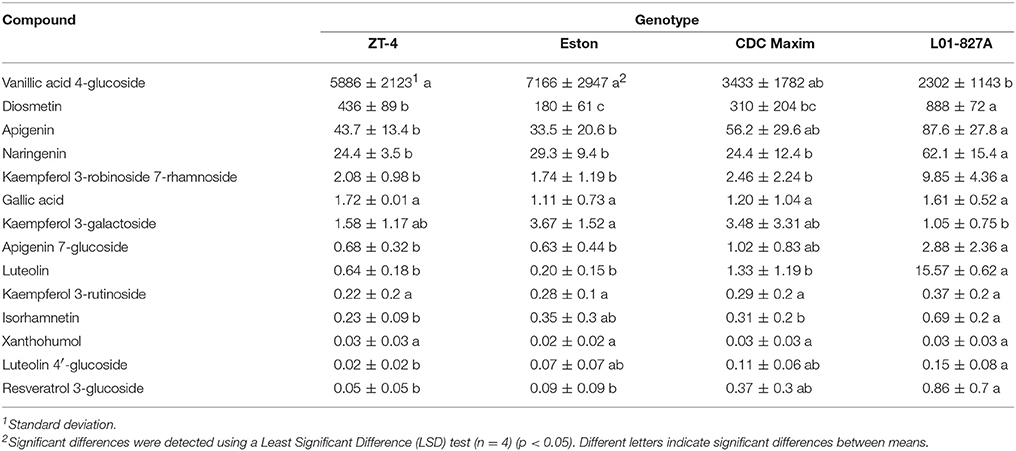
Table 3. Concentration of phenolic compounds (ng/g root) detected in the root extracts of cultivated lentil genotypes ZT-4, CDC Maxim and Eston and Lens ervoides L01-827A using liquid chromatography-mass spectrometry.
Biochemical Analysis of Healthy and Diseased Root Tissues
The LC-SRM analysis detected 17 polyphenols in the root tissues of all 4 lentil genotypes. The effect of genotype, infection and the interactions between genotype and infection on the concentration of each of these polyphenols was described in Table 4. Concentrations of some of the polyphenols varied among the genotypes in healthy plants, whereas in diseased plants, the concentration of most of the polyphenols varied among the genotypes (Table 5). Concentration of several compounds varied between diseased and healthy root tissues of the same genotype. The variation observed in the concentration of polyphenols between healthy and diseased roots was larger in L. ervoides L01-827A whereas it was smaller in ZT-4 and CDC Maxim (Figure 3). The concentration of apigenin, kaempferol, and naringenin was higher in diseased root tissues for all four lentil genotypes (Table S1). Diosmetin, hesperetin 7-rutinoside, kaempferol 3-galactoside, kaempferol 3-rutinoside, naringenin 7-rutinoside, and vanillic acid were only significantly more abundant in diseased roots of L. ervoides L01-827A (Table S1). Coumaric acid was only detected in diseased roots of L. ervoides L01-827A and Eston. 3,4-dihydroxybenzoic acid was detected in diseased roots of all and in healthy roots of L. ervoides L01-827A (Table S2).
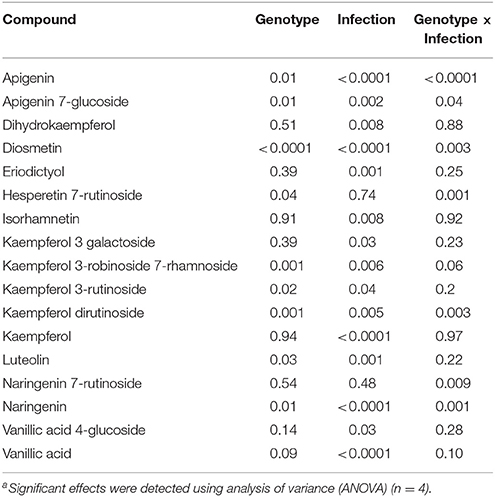
Table 4. Statistical significance of the effecta of genotype and infection on the composition of some phenolic compounds in the root tissues of cultivated lentil genotypes ZT-4, CDC Maxim, Eston, and Lens ervoides L01-827A determined by liquid chromatography-mass spectrometry (n = 4).
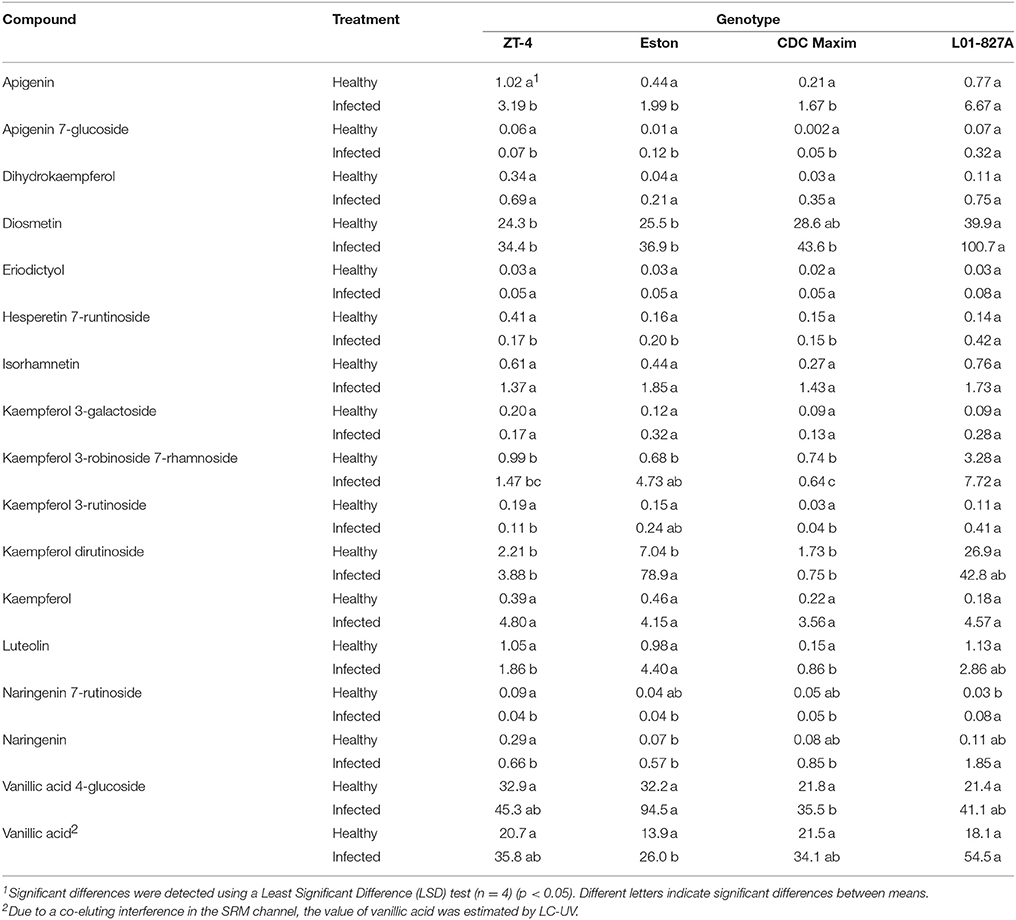
Table 5. Concentration of phenolic compounds (μg g−1 root) detected in the root tissues of cultivated lentil genotypes ZT-4, CDC Maxim and Eston and Lens ervoides L01-827A using liquid chromatography-mass spectrometry.
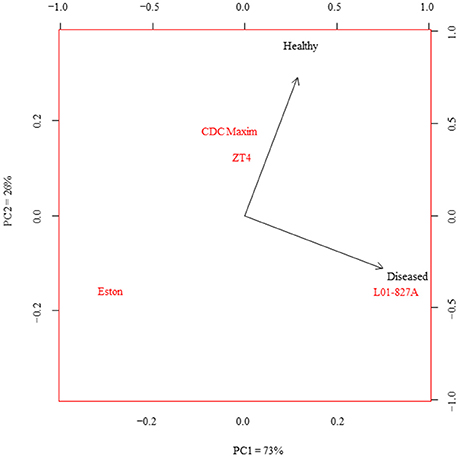
Figure 3. Principal component analysis of the concentration of phenolic compounds in the healthy and diseased root tissues of cultivated lentil genotypes ZT-4, CDC Maxim and Eston and Lens ervoides L01-827A using liquid chromatography-mass spectrometry (n = 4). Phenolic compounds were extracted from root tissues, and analyzed using LC-SRM.
In vitro Assay Testing the Effect of Polyphenols on Mycelial Growth of Aphanomyces euteiches
Seven polyphenols including vanillic acid, 4-hydroxybenzoic acid, 3,4-dihydroxybenzoic acid, 4-aminosalicylic acid, coumaric acid, delphinidin 3-glucoside and phloretin inhibited the mycelial growth of A. euteiches at 1 mg mL−1 in vitro (p < 0.0001). Coumaric acid and phloretin also inhibited A. euteiches at 0.1 g L−1 (10x dilution). No inhibition was observed for other compounds at 0.1 g L−1 (10x dilution), or at lower concentrations of any of these polyphenols (Table 6).
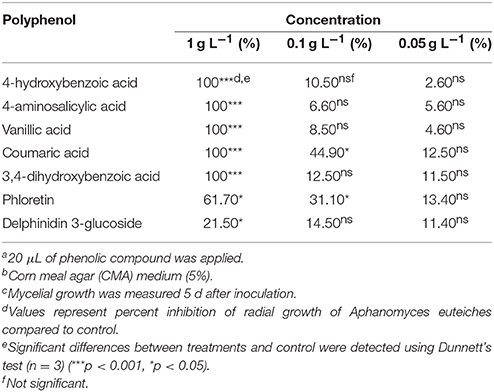
Table 6. Inhibitory effect of bioactive polyphenolsa detected in lentil root tissues on the in vitrob radial growth of Aphanomyces euteichesc.
Discussion
We used semi-quantitative mass spectrometry to investigate genotypic variations within the composition of polyphenols that naturally exist in the root tissues and root extracts of lentil. We analyzed a wide range of polyphenols and demonstrated that the qualitative and quantitative composition of polyphenols not only differs among the lentil genotypes, but also between the healthy and A. euteiches infected roots.
The higher concentration of polyphenols in the root extracts of the L. ervoides genotype (tertiary gene pool) compared to the cultivated L. culinaris genotypes (primary gene pool) could be an evolutionary divergence or a consequence of selection for larger plants and seeds during the domestication process (Alo et al., 2011; Wong et al., 2015). It was shown that the selection of new cultivars for specific traits or under optimal growth conditions may inadvertently lead to the loss of specific genes and phytochemicals (Hättenschwiler and Vitousek, 2000; Dixon, 2001) including those involved in the synthesis of polyphenols.
Lentil genotype ZT-4 is characterized by the expression of the single recessive gene, tan, that affects the color and thickness of the seed coat (Vaillancourt et al., 1986). The seed coats of low-tannin lentils lack many of the polyphenols found in normal seed coats, causing them to be thinner and more fragile compared to the seed coat of normal genotypes (Matus et al., 1993; Mirali et al., 2016a). This study shows that the tan gene is also expressed in the root tissues and results in a lower concentration of most of the polyphenols.
Variations observed in the levels of specific polyphenols between healthy and diseased root tissues may be due to defensive responses through the inactivation of microbial enzymes, reinforcement of plant structural components or creation of strategic barriers at points of entry (Sylvia and Sinclair, 1983; Beckman, 2000; Vermerris and Nicholson, 2008). In this study, the elevated concentration of apigenin, kaempferol, and naringenin in the diseased roots of all four lentil genotypes (Table S1) may suggest that these three polyphenols are associated with a general response against A. euteiches. Support for this hypothesis can be found in previous studies in which naringenin and apigenin were shown to have inhibitory effects on the bacterial pathogens and nematodes (Baidez et al., 2006; Treutter, 2006). Since such inhibitory effect was not detected against A. euteiches in our bioassay, these polyphenols might have other potential roles in a response against A. euteiches. Similar levels of several compounds in healthy and diseased roots of the low-tannin genotype ZT-4 may suggest that the expression of the gene tan is one of the factors influencing the composition of polyphenols in responses to phytopathogens. The differences observed in the concentration of polyphenols between healthy and diseased roots in different genotypes (Tables 3, 4) may indicate that the synthesis of defensive metabolites is genotype dependent. The variation was larger in L. ervoides L01-827A (Figure 3), suggesting that this wild genotype might have a wider capacity for a defensive response to plant pathogens.
In this study, the lentil genotypes were treated by the endosymbiont R. leguminosarum. It is notable that some polyphenols act as chemoattractants for rhizobium, improving the root colonization (Fisher and Long, 1992; Begum et al., 2001; Cooper, 2004) and, rhizobium may aid the plant to resist against pathogen by mediating the polyphenolic content of root tissues (Mishra et al., 2006). However, rhizobium may interact differently with the lentil genotypes resulting in different degrees of root colonization, and different responses to plant pathogens.
Certain phytochemicals were shown to attract the zoospores or inhibit the hyphal growth of A. euteiches (Yokosawa et al., 1986; Smolinska et al., 1997; Cannesan et al., 2012). In our study, the first involving lentil, some of the root polyphenols that inhibited the hyphal growth of the A. euteiches were more concentrated in specific lentil genotypes. For instance, the concentration of vanillic acid and 3,4-dihydroxybenzoic acid were higher in L. ervoides L01-827A which has a marbled gray seed coat. Coumaric acid was detected only in diseased roots of L. ervoides L01-827A, and in the green seed coat of the cultivar Eston (Mirali et al., 2016b). Differences in the quantity and quality of the phenylpropanoid metabolites in different genotypes of lentil suggests there could be useful genotypic variation in the level of resistance to A. euteiches in lentil. Since the majority of these A. euteiches inhibitors were organic acids, the organic acid content of root tissues and extracts appears to be associated with some level of resistance to ARR. Organic acids comprise a major portion of root extracts in most plant species (Neumann and Römheld, 2001; Cawthray, 2003; Sandnes et al., 2005). Organic acids and their derivatives were detected in both root tissue and root extracts of lentil. The observation that some organic acids, including caffeic acid, gallic acid, and 3-hydroxy-4-methoxycinnamic acid, did not affect the hyphal growth of A. euteiches suggests that the inhibitory effect on A. euteiches is limited to specific organic acids. The concentrations of root polyphenols could be variable in real conditions depending on various factors. However, an accumulative concentration of the bioactive polyphenols could be achieved and be effective to inhibit the root rot pathogen. Polyphenols, particularly those released from roots, are involved in a wide range of agro-ecological processes, and could affect specific ecological niches in the rhizosphere (Aira et al., 2010; Doornbos et al., 2012). The selective release of organic acids and other polyphenols that inhibit A. euteiches may improve soil health by reducing the abundance of the pathogen in soil, and the severity of ARR in subsequent susceptible crops in rotation.
To date there is no report of detection of effective resistance to A. euteiches in lentil. Among the genotypes used in this study, ZT4 was more susceptible to A. euteiches (Knowpulse Report, 2015), while L01-827A and Eston have shown a moderate level of resistance to A. euteiches (Banniza, pers. comm.; Saskpulse, 2015). One reason for their better tolerance to A. euteiches could be the elevated levels of specific of polyphenols. Such variations could increase the understanding of pathways of biochemical resistance to plant diseases and could be used as a starting point for identification of lentil genotypes that are resistant to root rot diseases in breeding programs.
Our study revealed that genotypic variations for the composition of polyphenols exist in lentil, and that there is a relationship between the composition of polyphenols and tolerance to A. euteiches. The tan gene in lentil influences the composition of polyphenols in root extracts. A number of studies have described the potential role of polyphenols in providing resistance against root rot diseases in different plant species (Farkas and Kiraaly, 1962; Lagrimini et al., 1993; Daayf et al., 2012; Aksoya et al., 2017). Such studies usually focus on a small number of compounds and thus cannot provide a clear image of the function of polyphenols. Here, we analyzed a wide range of polyphenols in response to root rot disease. Understanding of the role of polyphenols in providing resistance against root rot diseases, and their potential application in breeding programs is still not well defined. In lentil breeding programs, the exploitation of polyphenol content has mainly targeted the seed coats in order to improve seed quality, storage, marketability and nutritional values (Vaillancourt et al., 1986; Mirali et al., 2016b, 2017; Ganesan and Xu, 2017). Further research is required to identify and quantify all the polyphenols that are differentially expressed in uninfected and infected root tissues of lentil species. It is necessary to detect correlations between the concentration of specific polyphenols and other metabolites in the root tissues in order to identify mechanisms of resistance to A. euteiches. The induction of resistance in host plants can be influenced by a number of factors, including plant genotype, nutrition and environmental conditions (Walters et al., 2013). Therefore, it is necessary to understand the influence of plant nutrition on the polyphenolic content of roots, since this may override the influence of plant genetics on the composition of polyphenols. We are currently investigating unknown peaks in the UV spectra to identify new compounds of importance for roots that will be added to the polyphenol library in future studies.
Author Contributions
NB grew plants and collected the root materials. NB and RP contributed to LCMS analyses. PP prepared the inoculum and conducted the in-vitro plate assay. All authors contributed to experimental design and to preparation of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge financial assistance from the NSERC Industrial Research Chair program, and Saskatchewan Pulse Growers. We also acknowledge the assistance provided by Thermo Fisher Scientific (San Jose, CA) through instrumentation made available as part of a collaboration between Thermo Fisher and the University of Saskatchewan. Additional support was provided by the University of Saskatchewan Health Sciences core mass spectrometry facility, and by the Pulse Research Crew at the Crop Development Centre, University of Saskatchewan.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01131/full#supplementary-material
Abbreviations
ARR, Aphanomyces Root Rot; CDC, Crop Development Centre; LC, Liquid Chromatography; MS, Mass Spectrometry; SRM, Single Reaction Monitoring; ZT, Zero Tannin.
References
Aguilera, Y., Dueñas, M., Estrella, I., Hernández, T., Benitez, V., Esteban, R. M., et al. (2010). Evaluation of phenolic profile and antioxidant properties of Pardina lentil as affected by industrial dehydration. J. Agric. Food Chem. 58, 10101–10108. doi: 10.1021/jf102222t
Aira, M., Gomez-Brandon, M., Lazcano, C., Baath, E., and Domínguez, J. (2010). Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 42, 2276–2281. doi: 10.1016/j.soilbio.2010.08.029
Aksoya, H. M., Kaya, Y., Ozturk, M., Secgin, Z., Onder, H., and Okumus, A. (2017). Pseudomonas putida – Induced response in phenolic profile of tomato seedlings (Solanum lycopersicum L.) infected by Clavibacter michiganensis subsp. michiganensis. Biol. Control 105, 6–12. doi: 10.1016/j.biocontrol.2016.11.001
Alo, F., Furman, B. J., Akhunov, E., Dvorak, J., and Gepts, P. (2011). Leveraging genomic resources of model species for the assessment of diversity and phylogeny in wild and domesticated lentil. J Hered. 102, 315–329. doi: 10.1093/jhered/esr015
Armstrong-Cho, C., Tetreault, M., Banniza, S., Bhadauria, V., and Morrall, R. A. A. (2014). Reports of Aphanomyces euteiches in Saskatchewan. The Canadian Phytopathological Society. Canadian Plant Disease Survey. Disease Highlights.
Baetz, U., and Martinoia, E. (2014). Root exudates: the hidden part of plant defense. Trends Plant Sci. 19, 90–98. doi: 10.1016/j.tplants.2013.11.006
Baidez, A. G., Gomez, P., Del Rio, J. A., and Ortuno, A. (2006). Antifungal capacity of major phenolic compounds of Olea europaea L. against Phytophthora megasperma Drechsler and Cylindrocarpon destructans (Zinssm.) Scholten. Physiol. Mol. Plant Pathol. 69, 224–229. doi: 10.1016/j.pmpp.2007.05.001
Balmer, A., Pastor, V., Gamir, J., Flors, V., and Mauch-Mani, B. (2015). The “prime-ome”: towards a holistic approach to priming. Trends Plant Sci. 20, 443–452. doi: 10.1016/j.tplants.2015.04.002
Banniza, S. (2015). BRE1519: Developing Rapid Generation Technology Involving Wild Lentil Crosses to Produce Aphanomyces-Resistant Lentil Varieties–Proof of Concept. Saskpulse Report. Available online at: http://saskpulse.com/files/general/BRE1519_%28Banniza%29.pdf
Bazghaleh, N., Hamel, C., Gan, Y., Knight, J. D., Vujanovic, V., Cruz, A. F., et al. (2016). Phytochemicals induced in chickpea roots selectively and non-selectively stimulate and suppress fungal endophytes and pathogens. Plant Soil 409, 479–493. doi: 10.1007/s11104-016-2977-z
Beckman, C. H. (2000). Phenolic-storing cells: keys to programmed cell death and periderm formation in wilt disease resistance and in general defence responses in plants? Physiol. Mol. Plant Pathol. 57, 101–110. doi: 10.1006/pmpp.2000.0287
Begum, A. A., Leibovitch, S., Migner, P., and Zhang, F. (2001). Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 52, 1537–1543. doi: 10.1093/jexbot/52.360.1537
Bennett, R., and Wallsgrove, R. (1994). Secondary metabolites in plant defence mechanisms. New Phytol. 127, 617–633. doi: 10.1111/j.1469-8137.1994.tb02968.x
Borrell, A. N., Shi, Y., Gan, Y., Bainard, L. D., Germida, J. J., and Hamel, C. (2016). Fungal diversity associated with pulses and its influence on the subsequent wheat crop in the Canadian prairies. Plant Soil 414, 13–31. doi: 10.1007/s11104-016-3075-y
Cannesan, M. A., Durand, C., Burel, C., Gangneux, C., Lerouge, P., Ishii, T., et al. (2012). Effect of arabinogalactan proteins from the root caps of pea and brassica napus on Aphanomyces euteiches zoospore chemotaxis and germination. Plant Physiol. 159, 1658–1670. doi: 10.1104/pp.112.198507
Cawthray, G. R. (2003). An improved reversed-phase liquid chromatographic method for the analysis of low-molecular mass organic acids in plant root exudates. J. Chromatogr. A 1011, 233–240. doi: 10.1016/S0021-9673(03)01129-4
Chatterton, S., Bowness, R., and Harding, M. W. (2016). First report of root rot of field pea caused by Aphanomyces euteiches in Alberta, Canada. Plant Dis. 99:288. doi: 10.1094/PDIS-09-14-0905-PDN
Cheynier, V. (2012). Phenolic compounds: from plants to foods. Phytochem. Rev. 11, 153–177. doi: 10.1007/s11101-012-9242-8
Constabel, C. P., Yoshida, K., and Walker, V. (2014). Diverse ecological roles of plant tannins: plant defense and beyond. Recent Adv. Polyphen. Res. 4, 115–142. doi: 10.1002/9781118329634.ch5
Cooper, J. E. (2004). Multiple responses of rhizobia to flavonoids during legume root infection. Adv. Bot. Res. 41, 1–62. doi: 10.1016/S0065-2296(04)41001-5
Daayf, F., El Hadrami, A., El-Bebany, A. F., Henriquez, M. A., Yao, Z., Derksen, H., et al. (2012). Phenolic compounds in plant defense and pathogen counter-defense mechanisms. Recent Adv. Polyphen. Res. 3, 191–208. doi: 10.1002/9781118299753.ch8
Deng, Y., Zhao, Y., Padilla-Zakour, O., and Yang, G. (2015). Polyphenols, antioxidant and antimicrobial activities of leaf and bark extracts of Solidago canadensis L. Ind. Crops Prod. 74, 803–809. doi: 10.1016/j.indcrop.2015.06.014
Dickinson, C. H., and Skidmore, A. M. (1976). Interactions between germinating spores of Septoria nodorum and phylloplane fungi. Trans. Br. Mycol. Soc. 66, 45–56. doi: 10.1016/S0007-1536(76)80091-5
Dixon, R. A. (2001). Natural products and plant disease resistance. Nature 411, 843–847. doi: 10.1038/35081178
Doornbos, R. F., Van Loon, L. C., and Bakker, P. A. H. M. (2012). Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32, 227–243. doi: 10.1007/s13593-011-0028-y
Evidente, A., Cimmino, A., Fernandez-Aparicio, M., Andolfi, A., Rubiales, D., and Motta, A. (2010). Polyphenols, including the new peapolyphenols A–C, from pea root exudates stimulate Orobanche foetida seed germination. J. Agric. Food Chem. 58, 2902–2907. doi: 10.1021/jf904247k
FAO (2017). FAOSTAT. Food and Agriculture Organization of the United Nations. Rome. Available online at: http://faostat.fao.org
Farkas, G. L., and Kiraaly, Z. (1962). Role of phenolic compounds in the physiology of plant diseases and disease resistance. J. Phytopathol. 44, 105–150. doi: 10.1111/j.1439-0434.1962.tb02005.x
Ferguson, M. E., Maxted, N., Van Slageren, M., and Robertson, L. D. (2000). A re-assessment of the taxonomy of Lens Mill. (Leguminosae, Papilionaceae, Vicieae). Bot. J. Linn. Soc. 133, 41–59. doi: 10.1111/j.1095-8339.2000.tb01536.x
Fisher, R. F., and Long, S. R. (1992). Rhizobium–plant signal exchange. Nature 357, 655–660. doi: 10.1038/357655a0
Ganesan, K., and Xu, B. (2017). Polyphenol-rich lentils and their health promoting effects. Int. J. Mol. Sci. 18, 1–23. doi: 10.3390/ijms18112390
Gossen, B. D., Conner, R. L., Chang, K. F., Pasche, J. S., McLaren, D. L., Henriquez, M. A., et al. (2016). Identifying and managing root rot of pulses on the northern great plains. Plant Dis. 100, 1965–1978. doi: 10.1094/PDIS-02-16-0184-FE
Hättenschwiler, S., and Vitousek, P. M. (2000). The role of polyphenols in terrestrial ecosystems nutrient cycling. Trends Ecol. Evol. 15, 238–243. doi: 10.1016/S0169-5347(00)01861-9
Knowpulse Report (2015). Trichoderma: A Possible Solution for Aphanomyces? Root rot report. Available online at: http://knowpulse.usask.ca/portal/sites/default/files/NAPIARootRot_Prashar.pdf
Ladizinsky, G. (1979a). The genetics of several morphological traits in the lentil. J. Hered. 70, 135–137.
Lagrimini, L. M., Vaughn, J., Erb, W. A., and Miller, S. A. (1993). Peroxidase overproduction in tomato: wound-induced polyphenol deposition and disease resistance. Hortscience 28, 218–221.
Lanoue, A., Burlat, V., Henkes, G. J., Koch, I., Schurr, U., and Röse, U. S. R. (2010). De novo biosynthesis of defense root exudates in response to fusarium attack in barley. New Phytol. 185, 577–588. doi: 10.1111/j.1469-8137.2009.03066.x
Lattanzio, V. (2013). “Phenolic compounds,” in Natural Products, eds K. G. Ramawat and J. M. Merillon (Berlin; Heidelberg: Springer-Verlag), 1543–1580.
Matus, A., Slinkard, A. E., and Vandenberg, A. (1993). “The potential of zero tannin lentil,” in New Crops, eds J. Janick and J. E. Simon (New York, NY: Wiley), 279–282.
Mirali, M., Ambrose, S. J., Wood, S. A., Vandenberg, A., and Purves, R. W. (2014). Development of a fast extraction method and optimization of liquid chromatography-mass spectrometry for the analysis of phenolic compounds in lentil seed coats. J. Chromatogr. B 969, 149–161. doi: 10.1016/j.jchromb.2014.08.007
Mirali, M., Purves, R. W., Stonehouse, R., Song, R., Bett, K., and Vandenberg, A. (2016a). Genetics and biochemistry of zero-tannin lentils. PLoS ONE 11:e0164624. doi: 10.1371/journal.pone.0164624
Mirali, M., Purves, R. W., and Vandenberg, A. (2016b). Phenolic profiling of green lentil (Lens culinaris Medic.) seeds subjected to long-term storage. Eur. Food Res. Technol. 242, 2161–2170. doi: 10.1007/s00217-016-2713-1
Mirali, M., Purves, R. W., and Vandenberg, A. (2017). Profiling the phenolic compounds of the four major seed coat types and their relation to color genes in lentil. J. Nat. Prod. 80, 1310–1317. doi: 10.1021/acs.jnatprod.6b00872
Mishra, R. P., Singh, R. K., Jaiswal, H. K., Kumar, V., and Maurya, S. (2006). Rhizobium-mediated induction of phenolics and plant growth promotion in rice (Oryza sativa L.). Curr. Microbiol. 52, 383–389. doi: 10.1007/s00284-005-0296-3
Moussart, A., Even, M. N., Lesne, A., and Tivoli, B. (2013). Successive legumes tested in a greenhouse crop rotation experiment modify the inoculum potential of soils naturally infested by Aphanomyces euteiches. Plant Pathol. 62, 545–551. doi: 10.1111/j.1365-3059.2012.02679.x
Neumann, G., and Römheld, V. (2001). “The release of root exudates as affected by the plant's physiological status,” in The Rhizosphere: Biochemistry and Organic Substances at the Soil–Plant Interface, eds R. Pinton, Z. Varanini, and P. Nannipieri (New York, NY: Dekker), 23–72.
Nicholson, R. L., and Hammerschmidt, R. (1992). Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. doi: 10.1146/annurev.py.30.090192.002101
Olivoto, T., Nardino, M., Carvalho, I. R., Follmann, D. N., Szareski, V. J., Ferrari, M., et al. (2017). Plant secondary metabolites and its dynamical systems of induction in response to environmental factors: a review. Afr. J. Agric. Res. 12, 71–84. doi: 10.5897/AJAR2016.11677
Pagliarulo, C., De Vito, V., Picariello, G., Colicchio, R., Pastore, G., Salvatore, P., et al. (2016). Inhibitory effect of pomegranate (Punica granatum L.) polyphenol extracts on the bacterial growth and survival of clinical isolates of pathogenic Staphylococcus aureus and Escherichia coli. Food chem. 190, 824–831. doi: 10.1016/j.foodchem.2015.06.028
Purves, R. W., Mirali, M., Bazghaleh, N., and Vandenberg, A. (2016). “Semi quantitative LC-MRM workflow for the analysis of over 100 polyphenols in lentil crops,” in 29th Workshop on Tandem Mass Spectrometry (Lake Louise, AB), 1.
Puupponen-Pimiä, R., Nohynek, L., Meier, C., Kähkönen, M., Heinonen, M., Hopia, A., et al. (2001). Antimicrobial properties of phenolic compounds from berries. J. Appl. Microbiol. 90, 494–507. doi: 10.1046/j.1365-2672.2001.01271.x
Quideau, S., Deffieux, D., Douat-Casassus, C., and Pouységu, L. (2011). Plant polyphenols: chemical properties, biological activities, and synthesis. Angew. Chemie. Int. Ed. 50, 586–621. doi: 10.1002/anie.201000044
Reuveni, R., Shimoni, M., Karchi, Z., and Kuc, J. (1991). Peroxidase activity as a biochemical marker for resistance of muskmelon (Cucumis melo) to Pseudoperonospora cubensis. Resistance 82, 749–753.
Roy, F., Boye, J. I., and Simpson, B. K. (2010). Bioactive proteins and peptides in pulse crops: pea, chickpea and lentil. Food Res. Int. 43, 432–442. doi: 10.1016/j.foodres.2009.09.002
Sánchez-Chino, X., Jiménez-Martínez, C., Dávila-Ortiz, G., Álvarez-González, I., and Madrigal-Bujaidar, E. (2015). Nutrient and nonnutrient components of legumes, and its chemopreventive activity: a review. Nutr. Cancer 67, 401–410. doi: 10.1080/01635581.2015.1004729
Sandnes, A., Eldhuset, T. D., and Wollebaek, G. (2005). Organic acids in root exudates and soil solution of Norway spruce and silver birch. Soil Biol. Biochem. 37, 259–269. doi: 10.1016/j.soilbio.2004.07.036
Shalaby, S., and Horwitz, B. A. (2015). Plant phenolic compounds and oxidative stress: integrated signals in fungal–plant interactions. Curr. Genet. 61, 347–357. doi: 10.1007/s00294-014-0458-6
Smolinska, U., Knudsen, G. R., Morra, M. J., and Borek, V. (1997). Inhibition of Aphanomyces euteiches f. sp. pisi by volatiles produced by hydrolysis of Brassica napus seed meal. Plant Dis. 81, 288–292.
Sylvia, D. M., and Sinclair, W. A. (1983). Phenolic compounds and resistance to fungal pathogens induced in primary roots of douglas fir seedlings by the ectomycorrhizal fungus Laccaria laccata. Phytopathology 73, 390–397. doi: 10.1094/Phyto-73-390
Treutter, D. (2006). Significance of flavonoids in plant resistance: a review. Environ. Chem. Lett. 4, 147–157. doi: 10.1007/s10311-006-0068-8
Urbano, G., Porres, J. M., Frías, J., and Vidal-valverde, C. (2007). “Nutritional value,” in Lentil: An Ancient Crop for Modern Times, eds S. S. Yadav, D. McNeil, and P. C. Stevenson (Dordrecht: Springer), 47–93.
Vaillancourt, R., Slinkard, A. E., and Reichert, R. D. (1986). The inheritance of condensed tannin concentration in lentil. Can. J. Plant Sci. 66, 241–246. doi: 10.4141/cjps86-038
Vermerris, W., and Nicholson, R. (eds.). (2008). “The role of phenols in plant defense,” in Phenolic Compound Biochemistry (Dordrecht: Springer), 211–234.
Walters, D. R., Ratsep, J., and Havis, N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280. doi: 10.1093/jxb/ert026
Wicker, E., Hullé, M., and Rouxel, F. (2001). Pathogenic characteristics of isolates of Aphanomyces euteiches from pea in France. Plant Pathol. 50, 433–442. doi: 10.1046/j.1365-3059.2001.00590.x
Wille, L., Messmer, M. M., Studer, B., and Hohmann, P. (2018). Insights to plant-microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. doi: 10.1111/pce.13214. [Epub ahead of print].
Wink, M. (1988). Plant breeding: importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 75, 225–233. doi: 10.1007/BF00303957
Wong, M. M., Gujaria-Verma, N., Ramsay, L., Yuan, H. Y., Caron, C., Diapari, M., et al. (2015). Classification and characterization of species within the genus lens using genotyping-by-sequencing (GBS). PLoS ONE 10:e0122025. doi: 10.1371/journal.pone.0122025
Keywords: lentil, Lens ervoides, Lens culinaris, polyphenols, Aphanomyces euteiches, root rot, liquid chromatography, mass spectrometry
Citation: Bazghaleh N, Prashar P, Purves RW and Vandenberg A (2018) Polyphenolic Composition of Lentil Roots in Response to Infection by Aphanomyces euteiches. Front. Plant Sci. 9:1131. doi: 10.3389/fpls.2018.01131
Received: 13 April 2018; Accepted: 13 July 2018;
Published: 03 August 2018.
Edited by:
Andreia Figueiredo, Universidade de Lisboa, PortugalReviewed by:
Alessandra Durazzo, Consiglio per la Ricerca in Agricoltura e l'Analisi dell'Economia Agraria (CREA), ItalyGalina Suvorova, All-Russia Research Institute of Legumes and Groat Crops, Russia
Marisa Porrini, Università degli Studi di Milano, Italy
Copyright © 2018 Bazghaleh, Prashar, Purves and Vandenberg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Navid Bazghaleh, bmF2aWQuYmF6Z2hhbGVoQHVzYXNrLmNh
 Navid Bazghaleh
Navid Bazghaleh Pratibha Prashar
Pratibha Prashar Randy W. Purves
Randy W. Purves Albert Vandenberg
Albert Vandenberg