- 1Department of Cell Biology, Institute of Biological Sciences, University of Brasília, Brasília, Brazil
- 2National Center for Vegetable Crops Research (CNPH), Embrapa Vegetables, Brasília, Brazil
- 3Laboratory of Virology, Wageningen University and Research Center, Wageningen, Netherlands
The Sw-5 gene cluster encodes protein receptors that are potentially able to recognize microbial products and activate signaling pathways that lead to plant cell immunity. Although there are several Sw-5 homologs in the tomato genome, only one of them, named Sw-5b, has been extensively studied due to its functionality against a wide range of (thrips-transmitted) orthotospoviruses. The Sw-5b gene is a dominant resistance gene originally from a wild Peruvian tomato that has been used in tomato breeding programs aiming to develop cultivars with resistance to these viruses. Here, we provide an overview starting from the first reports of Sw-5 resistance, positional cloning and the sequencing of the Sw-5 gene cluster from resistant tomatoes and the validation of Sw-5b as the functional protein that triggers resistance against orthotospoviruses. Moreover, molecular details of this plant–virus interaction are also described, especially concerning the roles of Sw-5b domains in the sensing of orthotospoviruses and in the signaling cascade leading to resistance and hypersensitive response.
Introduction
Orthotospoviruses (family Tospoviridae) cause substantial losses in crop production worldwide (Pappu et al., 2009). They can be transmitted mechanically, mostly associated with experimental transmission, and by a limited number of thrips species (order Thysanoptera) in nature, replicating in both plant and invertebrate hosts (Rotenberg et al., 2015). Until recently, the orthotospoviruses used to be taxonomically classified within the family Bunyaviridae (currently reclassified into the new order Bunyavirales) together with vertebrate-infecting viruses due to their similar virion shape, genomic organization and, of course, phylogenetic relationship (Adams et al., 2017).
Due to their ability to infect many crops, farmers, seed industry, and researchers have been constantly looking for natural resistance sources against orthotospoviruses. Among few that have been explored commercially (de Ronde et al., 2014), the one from tomato (Solanum lycopersicum L.) is the subject of this mini-review. Initially, this resistance source used to be referred to as Sw-5, being utilized in tomato breeding programs. Sequencing of the resistance gene locus, originally from Solanum peruvianum Mill. (a wild Peruvian tomato), revealed five paralogs, the so-called Sw-5 gene cluster (Spassova et al., 2001). Isolation of two resistance gene candidates (RGCs) and their subsequent transformation into tobacco plants helped to demonstrate that the gene copy Sw-5b is solely responsible for a broad-spectrum resistance to orthotospoviruses (Spassova et al., 2001; Hallwass et al., 2014; Leastro et al., 2017).
The first sections of this mini-review will describe historical aspects of Sw-5 resistance, from S. peruvianum to the first bred commercial tomatoes, genetic characterization of the Sw-5 gene cluster and the identification of Sw-5b as the functional gene copy against orthotospoviruses. The last sections will focus on the identification of the avirulence-determinant/effector from orthotospoviruses and the interplay with and among domains of encoded proteins by different Sw-5 alleles. The mini-review finishes with some perspectives on challenging research questions for the future that may advance our understanding of the Sw-5b-mediated resistance.
Sw-5 Resistance: From Peru to a Worldwide Scope
Solanum peruvianum varieties naturally occur in the coastal and medium elevated regions of central and southern Peru and in northern Chile (Nakazato et al., 2012). These plants have been targeted as a wild source for crop improvement because of their genetic resemblance to cultivated tomatoes (Nesbitt and Tanksley, 2002). The first reports of S. peruvianum harboring a broad resistance source against tomato spotted wilt virus (TSWV) date from the end of the 1930s (Stevens et al., 1991). Since then, efforts have been made to cross this wild tomato with commercial cultivars. One of the first tomato fresh market cultivars originated from such cross was named Stevens (Stevens et al., 1991). Subsequent inheritance studies indicated that a single, dominant gene/locus (named as Sw-5) was responsible for resistance that was initially found to be effective against a wide array of orthotospovirus isolates from United States (Stevens et al., 1991) and Brazil (Boiteux and de Giordano, 1993).
Discovery of the Sw-5 Gene Cluster
In spite of being widely used in tomato breeding programs, the genetic identity of the Sw-5 resistance remained unknown for decades. However, by gene mapping, the resistance locus was first located on the long arm of chromosome 9 of tomato cultivar Stevens (Chagué et al., 1996). Two other studies mapped the Sw-5 locus near markers CT220 and SCAR421 (Stevens et al., 1995; Brommonschenkel and Tanksley, 1997) and allowed Folkertsma et al. (1999) to physically fine map the locus and identify Sw-5 RGCs using a bacterial artificial chromosome (BAC) library made from tomato cultivar Stevens.
Sequencing of BAC clones containing genomic DNA fragments of the cultivar “Stevens” revealed five paralogs (Folkertsma et al., 1999; Brommonschenkel et al., 2000; Spassova et al., 2001). These genes, named Sw-5a to Sw-5e, encode proteins that contain nucleotide-binding (NB) and leucine-rich repeat (LRR) domains, which are often observed in other plant resistance proteins (van der Biezen and Jones, 1998; Andolfo et al., 2014). From those genes, two highly homologous genes, named Sw-5a and Sw-5b, mapped close to the markers and exhibited significant resemblance to the tomato nematode and aphid resistance gene Mi (Brommonschenkel et al., 2000; Spassova et al., 2001). Furthermore, the presence of a prominent matrix attachment region (MAR) between those two genes (Spassova et al., 2001), and indicative of genomic regions accessed by the nuclear transcription machinery (Stief et al., 1989), supported their RGC signature. To find out which one of those two conferred resistance to TSWV, copies of Sw-5a and Sw-5b together with their own regulatory sequences were transferred to Nicotiana tabacum L. (Spassova et al., 2001). Only transgenic plants transformed with the Sw-5b gene were resistant after challenge with TSWV isolates (Spassova et al., 2001), even though the Sw-5a and Sw-5b genes share a high sequence identity of 97.7% (Spassova et al., 2001; De Oliveira et al., 2016).
In tomatoes carrying Sw-5, high levels of resistance have been observed to TSWV, tomato chlorotic spot virus (TCSV), groundnut ringspot virus (GRSV), and chrysanthemum stem necrosis virus (CSNV) (Boiteux and de Giordano, 1993; Dianese et al., 2011). This broad spectrum resistance is quite unique for a dominant NB-LRR type of resistance gene and contrasts the Tsw resistance gene from pepper (Capsicum chinense L.) which provides resistance to TSWV isolates only (Boiteux and Deavila, 1994). More recently, N. benthamiana plants transformed with the Sw-5b gene copy were challenged with six different orthotospoviruses. Besides the aforementioned four orthotospoviruses, Sw-5b provided resistance to alstroemeria necrotic streak virus (ANSV) and impatiens necrotic spot virus (INSV) as well (Leastro et al., 2017). Thus, the Sw-5b gene alone is sufficient for broad-spectrum resistance, although this applies to phylogenetically related orthotospoviruses that are clustered in the same evolutionary clade (de Oliveira et al., 2012; Lima et al., 2016). Conversely, viruses phylogenetically distant from this “TSWV clade,” e.g., melon yellow spot virus (MYSV), are not sensed and overcome the resistance when mechanically inoculated onto these Sw-5b gene-transformed N-benthamiana plants (Hallwass et al., 2014).
Hypersensitive Cell Death Response Triggered by Sw-5b
Another observation made regarding the transgenic tobacco plants involves the activation of a hypersensitive response (HR). This phenotype is usually observed as local necrotic lesions/spots and often appears (within several days) on leaves upon activation of immune protein receptors containing the NB and LRR domains (de Ronde et al., 2014). During HR, infected and neighboring cells are thought to kill themselves to halt the pathogen spread throughout the plant tissue. While the Sw-5b-transformed N. tabacum plants do not show any HR after mechanical inoculation with TSWV (Spassova et al., 2001), and strengthen the concept that the HR is not the resistance mechanism of NB-LRR proteins (Liu et al., 2010), the Sw-5b gene-transformed N. benthamiana plants show a clear, concomitant HR (Hallwass et al., 2014; Leastro et al., 2017). The main difference between these two transgenic plants is that in N. tabacum expression of the Sw-5b gene is governed by its own regulatory sequences (Spassova et al., 2001) while in N. benthamiana the Sw-5b gene is driven by constitutive expression from the strong 35S promoter of cauliflower mosaic virus (Hallwass et al., 2014). Whether this is the cause for HR induction remains speculative, but recent studies have shown auto-activation of Sw-5b when expressed from a highly translatable expression vector (Sainsbury et al., 2009) or in the additional presence of an RNA silencing suppressor (RSS) (De Oliveira et al., 2016).
Identification of the Avr-Determinant/Effector of Sw-5b
The NB-LRR proteins are also known as NLR proteins, an acronym that also stands to nucleotide-binding (NB) and LRR domains (L)-containing (R) receptors (Wilmanski et al., 2008). Many classes of such proteins have been described in both plants and animals, diverging in structure and activation mode (Ting et al., 2008; Wilmanski et al., 2008; Qi and Innes, 2013). They recognize pathogens by sensing microbial products, usually peptides/proteins (Wilmanski et al., 2008). In plants, these products are commonly referred to as avirulence (avr)-determinants or effectors (de Ronde et al., 2014). “Avirulence” since, once identified, the pathogens cannot cause infection, not being virulent (able to induce disease symptoms).
In order to identify the avr-determinant of Sw-5b, indirect approaches have been performed due to the lack of an orthotospovirus infectious clone (Hoffmann et al., 2001; Lopez et al., 2011; Hallwass et al., 2014; Peiro et al., 2014). The bottleneck in building an infectious clone resides in the fact that the orthotospovirus genetic RNA elements are of negative (non-messenger sense) polarity. Orthotospoviruses have a tripartite single-stranded RNA genome, with each segment named according to its length as Small (S), Medium (M), and Large (L). Whereas the L segment has a negative polarity, the other two present an ambisense genetic organization (King et al., 2012). In total, the orthotospoviruses express six mature proteins: (i) Nucleocapsid (N) and a non-structural protein (NSs) encoded on opposite strands of the S segment; (ii) envelope glycoproteins Gn and Gc, and a non-structural protein (NSm) encoded on opposite strands of the M segment; (iii) and L (an RNA-dependent RNA polymerase) encoded on the viral complementary strand of the L segment. While NSs protein suppresses the RNA silencing machinery (Takeda et al., 2002), NSm protein functions as a cell-to-cell movement protein in plant hosts (Kormelink et al., 1994; Storms et al., 1995; Takeda et al., 2002).
First pieces of evidence already indicated that the avr-determinant of Sw-5b-mediated resistance was encoded in the M segment of TSWV. Reassorted viruses containing the S and L segments of a resistance-inducing (RI) TSWV isolate and the M segment of an RB TSWV isolate overcame the Sw-5 resistance in tomato plants (Hoffmann et al., 2001). Furthermore, comparison of genomes from several RI and RB TSWV isolates pointed toward two amino acid substitutions (C118Y or T120N) in the NSm protein that were associated with the emergence of RB isolates (Lopez et al., 2011). Final evidence for the identification of the avr-determinant came from transient expression analysis of TSWV proteins from binary clones using agrobacteria, in which only NSm triggered HR in plants harboring a functional Sw-5b gene copy (Hallwass et al., 2014; Peiro et al., 2014). Moreover, expression of NSm proteins from CSNV and TCSV isolates also triggered HR in those plants, being abrogated by the introduction of mutations C118Y or T120N (Leastro et al., 2017). All these experiments validated NSm as the avr-determinant of Sw-5b.
Sw-5b Activation Mode
Although there are other variations, most plant NLRs have at their amino (N)-terminus either a Toll and interleukin-1 receptor (TIR) domain or a coiled-coil (CC) domain, which is then followed by the central NB- and carboxy (C)-terminal LRR domains (Meyers et al., 2003). Therefore, NLRs are often also classified as TNLs (TIR-containing NLRs) or CNLs (CC-containing NLRs). The Sw-5 proteins belong to the class of CNLs but are distinct by the presence of an additional extension at their N-terminus, the so-called Solanaceae domain (SD) (De Oliveira et al., 2016). This extra domain has been reported in other Solanaceae CNLs as well (Mucyn et al., 2006). Dissection of Sw-5b has designated important functions to each domain. Using this approach, truncated versions of Sw-5b, containing one or more domains, have usually been expressed by agroinfiltration in wild-type N. benthamiana leaves (Chen et al., 2016; De Oliveira et al., 2016).
The Sw-5b protein has a nucleocytoplasmic distribution, with the SD and CC domains (covalently bound) responsible to deliver the protein to the cell nucleus (De Oliveira et al., 2016). This nuclear localization is possibly required for the signal transduction that leads to the resistance mechanism itself. The absence of SD-CC does not interfere with NSm recognition and HR triggering, but transgenic plants transformed with a truncated Sw-5b gene lacking these domains are not resistant to orthotospoviruses (Chen et al., 2016). Altogether, these results indicate that resistance and HR are uncoupled events (Chen et al., 2016; De Oliveira et al., 2016). The SD and CC domains are also involved in the activation control of Sw-5b. In the absence of NSm, CC suppresses the HR triggered by NB while SD enhances this inhibition independently (Chen et al., 2016). Interestingly, if NSm is present, SD acts as a positive regulator, relieving the inhibitory effect over NB (Chen et al., 2016).
The NB domain, which also includes the subdomains ARC 1 and ARC 2, is solely responsible for the signaling pathway that leads to HR triggering (Chen et al., 2016; De Oliveira et al., 2016). This domain switches between “off” and “on” states according to its binding to adenosine diphosphate (ADP) or adenosine triphosphate (ATP), respectively (Williams et al., 2011). Overexpression of NB in the presence of RSSs results in HR as observed for the full Sw-5b protein (De Oliveira et al., 2016). Expression of NB alone can also result in a negligible HR that is inhibited in the presence of LRR (NB-LRR covalently bound) (Chen et al., 2016).
As mentioned before, co-expression of NB-LRR (covalently bound) and NSm triggers a strong HR. If only NB is co-expressed with NSm, no HR is seen (De Oliveira et al., 2016). Thus, LRR recognizes NSm as avr-determinant and activates NB. For most studied plant NLRs, the pathogen is perceived by an indirect interaction between the LRR and the avr-determinant (Dangl and Jones, 2001; van der Hoorn and Kamoun, 2008). Using co-immunoprecipitation assays, Sw-5b NB-LRR domains and NSm were shown to directly interact in vitro and in planta (Zhu et al., 2017). A conserved 21 amino acid epitope from NSm was sufficient for the interaction and activation of Sw-5b (Zhu et al., 2017). Within the LRR domain, four polymorphic sites were critical for recognition of this NSm epitope (Zhu et al., 2017). As such, Sw-5b is one of the few CNL proteins able to perceive the avr-determinant via direct interaction. Figure 1 summarizes the functions mapped to Sw-5b domains.
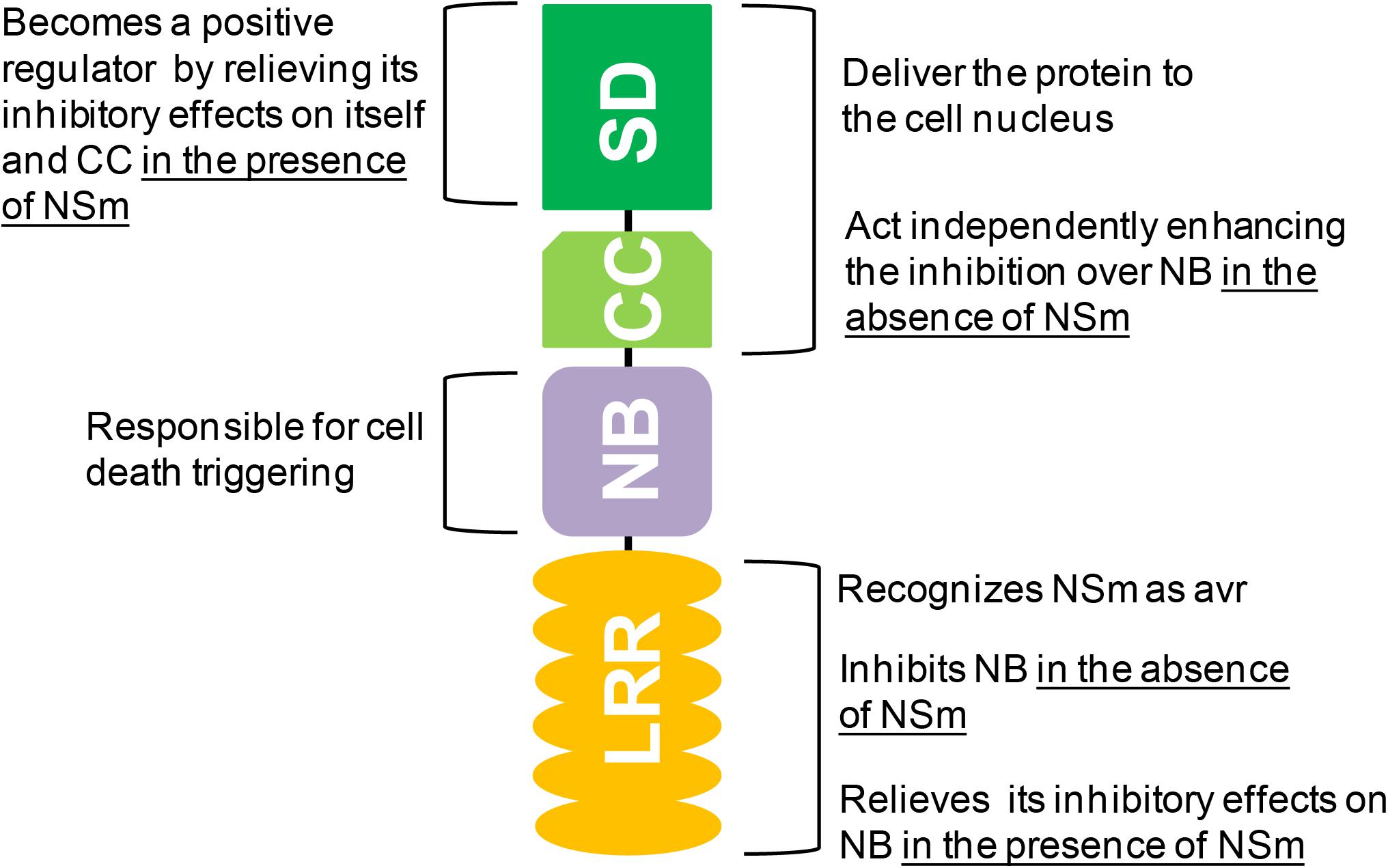
FIGURE 1. Overview of the (modulating) roles of the amino-terminal Solanaceae (SD) and coiled-coil (CC), central nucleotide-binding (NB), and carboxy-terminal leucine-rich repeat (LRR) domains in Sw-5b activation. Detailed information is found in the manuscript text.
Tomato Paralogs and Orthologs of Sw-5b
In contrast to the findings with Sw-5b, co-expression of Sw-5a and TSWV NSm does not result in HR triggering (De Oliveira et al., 2016), which could explain why Sw-5a-transformed tobacco plants are susceptible to TSWV (Spassova et al., 2001). Dissection analysis of Sw-5a, in analogy to Sw-5b, showed that overexpression of the entire Sw-5a protein or its NB domain only leads to auto-activation (HR triggering in the absence of an avr-determinant) (De Oliveira et al., 2016). Considering that Sw-5a is the highest conserved paralog of Sw-5b (95.1% aa identity), it is likely that some point mutations within the LRR domain disrupted the avr-determinant sensing of Sw-5a. Proof for this could come from testing a chimeric Sw-5a NB-LRR construct, in which its LRR domain has been swapped for the functional one from Sw-5b. So far, no studies have yet been performed with the proteins Sw-5c, Sw-5d, and Sw-5e.
In contrast to Sw-5a and Sw-5b, the protein product of the most conserved ortholog from susceptible tomato isolines (not bred with S. peruvianum), named Sw-5aS, is not able to trigger auto-HR at all, even though these three proteins share about 94% aa identity (De Oliveira et al., 2016). Analysis of the Sw-5aS NB domain revealed that a point mutation (Q599R) abolished the ability for HR-induction. A reversion of this single amino acid into the one found in the NB domains of Sw-5a and Sw-5b, restored the ability of triggering HR by the Sw-5aS NB domain (De Oliveira et al., 2016).
Susceptible and resistant tomato plants can be easily differentiated by using a co-dominant molecular marker that detects a series of conserved deletions (of variable sizes depending upon the cultivar) upstream of the coding sequence of Sw-5aS (Dianese et al., 2010). Apart from these deletions, the regulatory sequences of Sw-5a, Sw-5b, and Sw-5aS are highly conserved. With the availability of the tomato genome sequence (Sato et al., 2012), all Sw-5b orthologs from the susceptible tomato cultivar “Heinz 1706” could be mapped. Including Sw-5aS, three complete Sw-5 genes (containing the SD-CC-NB-LRR domains) and a truncated gene are found in chromosome 9 of tomato cultivar “Heinz 1706” (Turina et al., 2016). One of these Sw-5 genes encodes a protein referred to as Sw-5f, although it is an ortholog and not a paralog of Sw-5b. The Sw-5f protein interacts with the effector protein SPRYSEC-19 from nematode Globodera rostochiensis (Rehman et al., 2009). This effector inhibits HR triggered by many CNL proteins, including an auto-active mutant of Sw-5b (Postma et al., 2012). Figure 2 schematizes some Sw-5-related proteins found in resistant and susceptible tomatoes to orthotospoviruses.
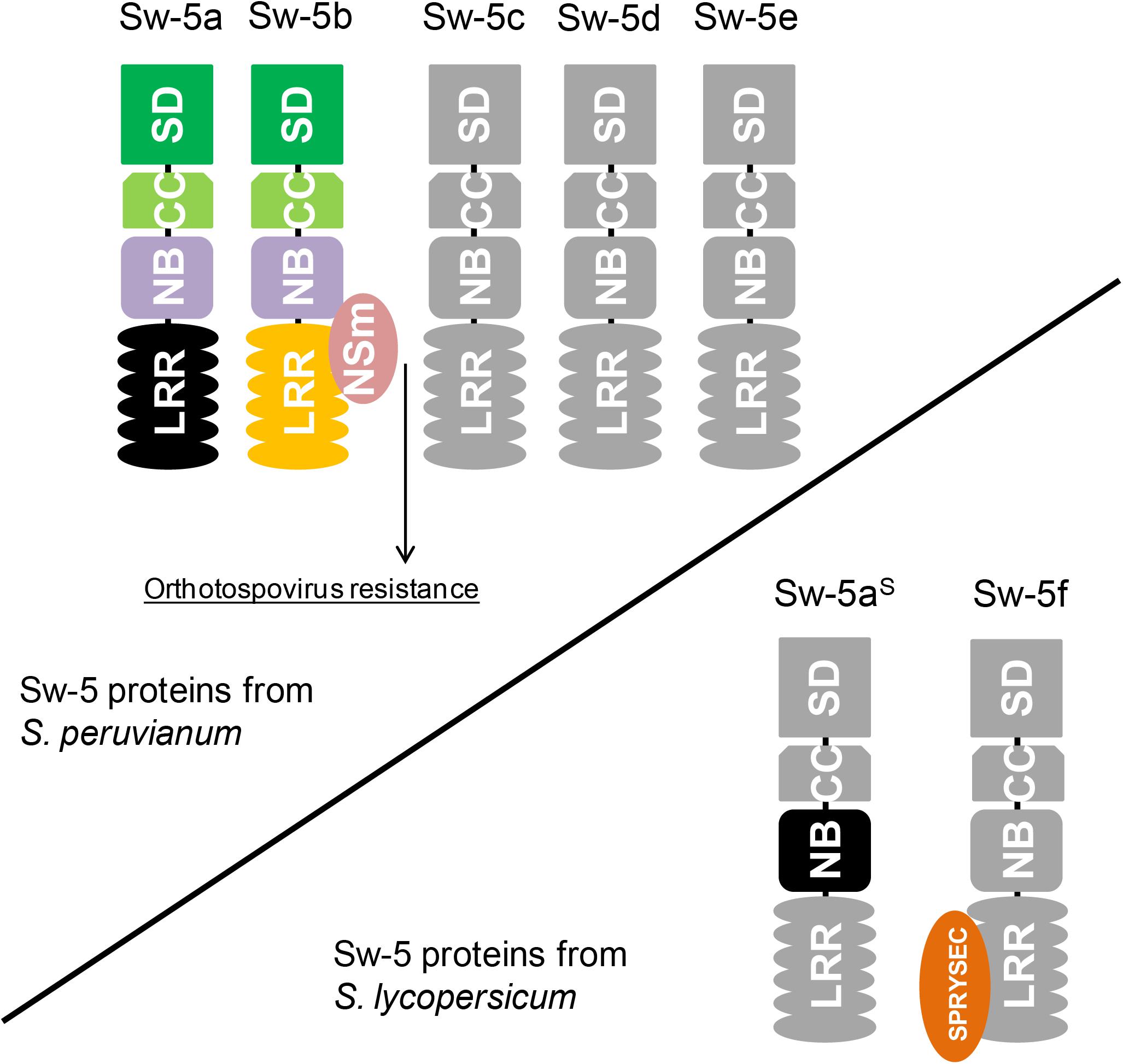
FIGURE 2. Topology of Sw-5 proteins encoded by paralogs from the Sw-5 gene cluster and orthologs collected from tomatoes susceptible to orthotospoviruses. The upper schematic encompasses the Sw-5 proteins of S. peruvianum. So far, there are only functional studies on Sw-5a and Sw-5b. The latter directly interacts with NSm (orthotospovirus cell-to-cell movement protein), which activates orthotospovirus resistance. On the contrary, Sw-5a is unable to recognize NSm as avirulence (avr)-determinant. The lower schematic shows two Sw-5 proteins found in tomato cultivars susceptible to orthotospoviruses. The most conserved homolog of Sw-5a and Sw-5b, named Sw-5aS, contains a mutation in its nucleotide-binding (NB) domain that halts hypersensitive response (HR) triggering. Apart from this, Sw-5aS cannot recognize NSm as avr-determinant. Sw-5f has been first reported interacting with an effector (SPRYSEC-19) of nematode Globodera rostochiensis that inhibits HR triggered by several resistance proteins.
Conclusions and Perspectives
Few plant immune receptors against viruses have been deeply studied to date. Apart from tomato Sw-5b, most of the efforts concentrated on the resistance mechanisms orchestrated by potato Rx against potato virus X (PVX) and tobacco N against tobacco mosaic virus (TMV) (de Ronde et al., 2014). The findings obtained from studying these proteins, especially concerning their activation mode, can be extrapolated to other plant NLR proteins with a similar topology (domain organization). Since HR is not the resistance mechanism itself, the actual downstream signaling cascade and cellular events leading to Sw-5b-mediated resistance remain to be elucidated. In this context, identifying plant proteins that directly or indirectly interact with Sw-5b as well as identifying the genes that are up- or down-regulated during the resistance response may provide further clues for this.
We do not rule out that in addition to Sw-5b, the other Sw-5-related proteins trigger resistance to different pathogens. This has been observed for NLRs from potato, in which the paralogs Rx and GPa2 trigger resistance to PVX and nematode G. pallida, respectively (van der Vossen et al., 2000). Taking into consideration that Sw-5a and Sw-5aS share high identity with Sw-5b, these two homologs could still be reverted to functional genes against orthotospoviruses. However, as we discussed in previous sections, point mutations in single domains, e.g., only in LRR that recognizes NSm as avr-determinant, may not be sufficient. Structural and modeling studies need to be performed to identify (a combination of) modifications that have to be implemented to turn non-functional copies into one that resembles the functional Sw-5b. The insertion of these modifications in the tomato genome, especially in those containing only Sw-5aS, could be done by genome editing tools such as CRISPR and TALENs (Gaj et al., 2013).
Author Contributions
AdO organized and wrote most of this manuscript. LB, RK, and RR revised and added complementary information.
Funding
The authors acknowledge the Brazilian institutions CNPq (Conselho Nacional de Desenvolvimento Cientfico e Tecnológico), CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), and FAPDF (Fundação de Apoio à Pesquisa do Distrito Federal), and the Dutch Technology Foundation (STW), Applied Science Division of the Netherlands Organization for Scientific Research (NWO) for financial support.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, M. J., Lefkowitz, E. J., King, A. M. Q., Harrach, B., Harrison, R. L., Knowles, N. J., et al. (2017). Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch. Virol. 162, 2505–2538. doi: 10.1007/s00705-017-3358-5
Andolfo, G., Jupe, F., Witek, K., Etherington, G. J., Ercolano, M. R., and Jones, J. D. (2014). Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14:120. doi: 10.1186/1471-2229-14-120
Boiteux, L. S., and de Giordano, B. L. (1993). Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon esculentum). Euphytica 71, 151–154. doi: 10.1007/BF00023478
Boiteux, L. S., and Deavila, A. C. (1994). Inheritance of a resistance specific to tomato spotted wilt Tospovirus in Capsicum-chinense PI159236. Euphytica 75, 139–142. doi: 10.1007/BF00221009
Brommonschenkel, S. H., Frary, A., and Tanksley, S. D. (2000). The broad-spectrum tospovirus resistance gene Sw-5 of tomato is a homolog of the root-knot nematode resistance gene Mi. Mol. Plant Microbe Interact. 13, 1130–1138. doi: 10.1094/MPMI.2000.13.10.1130
Brommonschenkel, S. H., and Tanksley, S. D. (1997). Map-based cloning of the tomato genomic region that spans the Sw-5 tospovirus resistance gene in tomato. Mol. Gen. Genet. 256, 121–126. doi: 10.1007/s004380050553
Chagué, V., Mercier, J. C., Guénard, M., de Courcel, A., and Vedel, F. (1996). Identification and mapping on chromosome 9 of RAPD markers linked to Sw-5 in tomato by bulked segregant analysis. Theor. Appl. Genet. 92, 1045–1051. doi: 10.1007/BF00224047
Chen, X., Zhu, M., Jiang, L., Zhao, W., Li, J., Wu, J., et al. (2016). A multilayered regulatory mechanism for the autoinhibition and activation of a plant CC-NB-LRR resistance protein with an extra N-terminal domain. New Phytol. 212, 161–175. doi: 10.1111/nph.14013
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
De Oliveira, A. S., Koolhaas, I., Boiteux, L. S., Caldararu, O. F., Petrescu, A. J., Oliveira Resende, R., et al. (2016). Cell death triggering and effector recognition by Sw-5 SD-CNL proteins from resistant and susceptible tomato isolines to Tomato spotted wilt virus. Mol. Plant Pathol. 17, 1442–1454. doi: 10.1111/mpp.12439
de Oliveira, A. S., Melo, F. L., Inoue-Nagata, A. K., Nagata, T., Kitajima, E. W., and Resende, R. O. (2012). Characterization of bean necrotic mosaic virus: a member of a novel evolutionary lineage within the Genus Tospovirus. PLoS One 7:e38634. doi: 10.1371/journal.pone.0038634
de Ronde, D., Butterbach, P., and Kormelink, R. (2014). Dominant resistance against plant viruses. Front. Plant Sci. 5:307. doi: 10.3389/fpls.2014.00307
Dianese, E. C., de Fonseca, M. E. N., Goldbach, R., Kormelink, R., Inoue-Nagata, A. K., Resende, R. O., et al. (2010). Development of a locus-specific, co-dominant SCAR marker for assisted-selection of the Sw-5 (Tospovirus resistance) gene cluster in a wide range of tomato accessions. Mol. Breed. 25, 133–142. doi: 10.1007/s11032-009-9313-8
Dianese, E. C., Fonseca, M. E. N., Inoue-Nagata, A. K., Resende, R. O., and Boiteux, L. S. (2011). Search in Solanum (section Lycopersicon) germplasm for sources of broad-spectrum resistance to four Tospovirus species. Euphytica 180, 307–319. doi: 10.1007/s10681-011-0355-8
Folkertsma, R. T., Spassova, M. I., Prins, M., Stevens, M. R., Hille, J., and Goldbach, R. W. (1999). Construction of a bacterial artificial chromosome (BAC) library of Lycopersicon esculentum cv. stevens and its application to physically map the Sw-5 locus. Mol. Breed. 5, 197–207. doi: 10.1023/A:1009650424891
Gaj, T., Gersbach, C. A., and Barbas, C. F. III. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Hallwass, M., de Oliveira, A. S., de Campos Dianese, E., Lohuis, D., Boiteux, L. S., Inoue-Nagata, A. K., et al. (2014). The Tomato spotted wilt virus cell-to-cell movement protein (NSm) triggers a hypersensitive response in Sw-5-containing resistant tomato lines and in Nicotiana benthamiana transformed with the functional Sw-5b resistance gene copy. Mol. Plant Pathol. 15, 871–880. doi: 10.1111/mpp.12144
Hoffmann, K., Qiu, W. P., and Moyer, J. W. (2001). Overcoming host- and pathogen-mediated resistance in tomato and tobacco maps to the M RNA of Tomato spotted wilt virus. Mol. Plant Microbe Interact. 14, 242–249. doi: 10.1094/MPMI.2001.14.2.242
King, A. M. Q., Adams, M. J., Carstens, E. B., and Lefkowitz, E. J. (eds) (2012). Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press.
Kormelink, R., Storms, M., Vanlent, J., Peters, D., and Goldbach, R. (1994). Expression and subcellular location of the nsm protein of tomato spotted wilt virus (tswv), a putative viral movement protein. Virology 200, 56–65. doi: 10.1006/viro.1994.1162
Leastro, M. O., De Oliveira, A. S., Pallás, V., Sánchez-Navarro, J. A., Kormelink, R., and Resende, R. O. (2017). The NSm proteins of phylogenetically related tospoviruses trigger Sw-5b-mediated resistance dissociated of their cell-to-cell movement function. Virus Res. 240, 25–34. doi: 10.1016/j.virusres.2017.07.019
Lima, R. N., De Oliveira, A. S., Leastro, M. O., Blawid, R., Nagata, T., Resende, R. O., et al. (2016). The complete genome of the tospovirus Zucchini lethal chlorosis virus. Virol. J. 13:123. doi: 10.1186/s12985-016-0577-4
Liu, P. P., Bhattacharjee, S., Klessig, D. F., and Moffett, P. (2010). Systemic acquired resistance is induced by R gene-mediated responses independent of cell death. Mol. Plant Pathol. 11, 155–160. doi: 10.1111/j.1364-3703.2009.00564.x
Lopez, C., Aramburu, J., Galipienso, L., Soler, S., Nuez, F., and Rubio, L. (2011). Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 92, 210–215. doi: 10.1099/vir.0.026708-0
Meyers, B. C., Kozik, A., Griego, A., Kuang, H., and Michelmore, R. W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. doi: 10.1105/tpc.009308
Mucyn, T. S., Clemente, A., Andriotis, V. M., Balmuth, A. L., Oldroyd, G. E., Staskawicz, B. J., et al. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18, 2792–2806. doi: 10.1105/tpc.106.044016
Nakazato, T., Franklin, R. A., Kirk, B. C., and Housworth, E. A. (2012). Population structure, demographic history, and evolutionary patterns of a green-fruited tomato, Solanum peruvianum (Solanaceae), revealed by spatial genetics analyses. Am. J. Bot. 99, 1207–1216. doi: 10.3732/ajb.1100210
Nesbitt, T. C., and Tanksley, S. D. (2002). Comparative sequencing in the genus Lycopersicon. Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 162, 365–379.
Pappu, H. R., Jones, R. A. C., and Jain, R. K. (2009). Global status of tospovirus epidemics in diverse cropping systems: successes achieved and challenges ahead. Virus Res. 141, 219–236. doi: 10.1016/j.virusres.2009.01.009
Peiro, A., Canizares, M. C., Rubio, L., Lopez, C., Moriones, E., Aramburu, J., et al. (2014). The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol. Plant Pathol. 15, 802–813. doi: 10.1111/mpp.12142
Postma, W. J., Slootweg, E. J., Rehman, S., Finkers-Tomczak, A., Tytgat, T. O. G., van Gelderen, K., et al. (2012). The effector SPRYSEC-19 of Globodera rostochiensis suppresses CC-NB-LRR-mediated disease resistance in plants. Plant Physiol. 160, 944–954. doi: 10.1104/pp.112.200188
Qi, D., and Innes, R. W. (2013). Recent advances in plant nlr structure, function, localization, and signaling. Front. Immunol. 4:348. doi: 10.3389/fimmu.2013.00348
Rehman, S., Postma, W., Tytgat, T., Prins, P., Qin, L., Overmars, H., et al. (2009). A secreted SPRY domain-containing protein (SPRYSEC) from the plant-parasitic nematode Globodera rostochiensis interacts with a CC-NB-LRR protein from a susceptible tomato. Mol. Plant Microbe Interact. 22, 330–340. doi: 10.1094/MPMI-22-3-0330
Rotenberg, D., Jacobson, A. L., Schneweis, D. J., and Whitfield, A. E. (2015). Thrips transmission of tospoviruses. Curr. Opin. Virol. 15, 80–89. doi: 10.1016/j.coviro.2015.08.003
Sainsbury, F., Thuenemann, E. C., and Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7, 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Spassova, M. I., Prins, T. W., Folkertsma, R. T., Klein-Lankhorst, R. M., Hille, J., Goldbach, R. W., et al. (2001). The tomato gene Sw5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol. Breed. 7, 151–161. doi: 10.1023/A:1011363119763
Stevens, M. R., Lamb, E. M., and Rhoads, D. D. (1995). Mapping the sw-5 locus for tomato spotted wilt virus-resistance in tomatoes using rapd and rflp analyses. Theor. Appl. Genet. 90, 451–456. doi: 10.1007/BF00221989
Stevens, M. R., Scott, S. J., and Gergerich, R. C. (1991). Inheritance of a gene for resistance to tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica 59, 9–17.
Stief, A., Winter, D. M., Strätling, W. H., and Sippel, A. E. (1989). A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature 341, 343–345. doi: 10.1038/341343a0
Storms, M. M., Kormelink, R., Peters, D., Van Lent, J. W., and Goldbach, R. W. (1995). The nonstructural NSm protein of tomato spotted wilt virus induces tubular structures in plant and insect cells. Virology 214, 485–493. doi: 10.1006/viro.1995.0059
Takeda, A., Sugiyama, K., Nagano, H., Mori, M., Kaido, M., Mise, K., et al. (2002). Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 532, 75–79. doi: 10.1016/S0014-5793(02)03632-3
Ting, J. P., Willingham, S. B., and Bergstralh, D. T. (2008). NLRs at the intersection of cell death and immunity. Nat. Rev. Immunol. 8, 372–379. doi: 10.1038/nri2296
Turina, M., Kormelink, R., and Resende, R. O. (2016). Resistance to tospoviruses in vegetable crops: epidemiological and molecular aspects. Annu. Rev. Phytopathol. 54, 347–371. doi: 10.1146/annurev-phyto-080615-095843
van der Biezen, E. A., and Jones, J. D. (1998). The NB-ARC domain: a novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, 226–227. doi: 10.1016/S0960-9822(98)70145-9
van der Hoorn, R. A., and Kamoun, S. (2008). From guard to decoy: a new model for perception of plant pathogen effectors. Plant Cell 20, 2009–2017. doi: 10.1105/tpc.108.060194
van der Vossen, E. A., van der Voort, J. N., Kanyuka, K., Bendahmane, A., Sandbrink, H., Baulcombe, D. C., et al. (2000). Homologues of a single resistance-gene cluster in potato confer resistance to distinct pathogens: a virus and a nematode. Plant J. 23, 567–576. doi: 10.1046/j.1365-313x.2000.00814.x
Williams, S. J., Sornaraj, P., deCourcy-Ireland, E., Menz, R. I., Kobe, B., Ellis, J. G., et al. (2011). An autoactive mutant of the M flax rust resistance protein has a preference for binding ATP, whereas wild-type M protein binds ADP. Mol. Plant Microbe Interact. 24, 897–906. doi: 10.1094/MPMI-03-11-0052
Wilmanski, J. M., Petnicki-Ocwieja, T., and Kobayashi, K. S. (2008). NLR proteins: integral members of innate immunity and mediators of inflammatory diseases. J. Leukoc. Biol. 83, 13–30. doi: 10.1189/jlb.0607402
Keywords: tomato, orthotospovirus, resistance, Sw-5, Sw-5b, NLR, NB-LRR
Citation: de Oliveira AS, Boiteux LS, Kormelink R and Resende RO (2018) The Sw-5 Gene Cluster: Tomato Breeding and Research Toward Orthotospovirus Disease Control. Front. Plant Sci. 9:1055. doi: 10.3389/fpls.2018.01055
Received: 08 May 2018; Accepted: 28 June 2018;
Published: 19 July 2018.
Edited by:
Ralf Georg Dietzgen, The University of Queensland, AustraliaReviewed by:
Gian Paolo Accotto, Istituto per la Protezione Sostenibile delle Piante (IPSP), ItalyDenis Michael Persley, Queensland Government, Australia
Copyright © 2018 de Oliveira, Boiteux, Kormelink and Resende. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Athos S. de Oliveira, YXRob3Muc29AZ21haWwuY29t
 Athos S. de Oliveira
Athos S. de Oliveira Leonardo S. Boiteux2
Leonardo S. Boiteux2 Richard Kormelink
Richard Kormelink Renato O. Resende
Renato O. Resende