
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 11 July 2018
Sec. Plant Breeding
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.01011
This article is part of the Research TopicThe Use of Systems Biology for Enhancing Crop Abiotic Stress ToleranceView all 15 articles
Salt tolerance in cotton is highly imperative for improvement in the response to decreasing farmland and soil salinization. However, little is known about the genetic basis underlying salt tolerance in cotton, especially the seedling stage. In this study, we evaluated two salt-tolerance-related traits of a natural population comprising 713 upland cotton (Gossypium hirsutum L.) accessions worldwide at the seedling stage and performed a genome-wide association study (GWAS) to identify marker-trait associations under salt stress using the Illumina Infinium CottonSNP63K array. A total of 23 single nucleotide polymorphisms (SNPs) that represented seven genomic regions on chromosomes A01, A10, D02, D08, D09, D10, and D11 were significantly associated with the two salt-tolerance-related traits, relative survival rate (RSR) and salt tolerance level (STL). Of these, the two SNPs i46598Gh and i47388Gh on D09 were simultaneously associated with the two traits. Based on all loci, we screened 280 possible candidate genes showing different expression levels under salt stress. Most of these genes were involved in transcription factors, transporters and enzymes and were previously reported as being involved in plant salt tolerance, such as NAC, MYB, NXH, WD40, CDPK, LEA, and CIPK. We further validated six putative candidate genes by qRT-PCR and found a differential expression level between salt-tolerant and salt-sensitive varieties. Our findings provide valuable information for enhancing the understanding of complicated mechanisms of salt tolerance in G. hirsutum seedlings and cotton salt tolerance breeding by molecular marker-assisted selection.
Soil salinity is a major abiotic stress that threatens crop yield, the ecological environment and agricultural sustainability (Hamwieh et al., 2011; Farooq et al., 2017). Indeed, salinity almost affected one billion hectares of arid and semi-arid areas globally (Dagar and Minhas, 2016), and soil salinity will become progressively more severe over time due to climatic changes, unscientific irrigation and excessive fertilization (Li et al., 2008; Han J. et al., 2015). Salt salinity is predicted to affect more than 50% of all arable land by 2050 (Ashraf and Wu, 1994; Demiral and Türkan, 2006).
Cotton (Gossypium hirsutum L., AADD, 2n = 52) is an economically important crop worldwide and can be used for soil reclamation as a pioneer crop of saline-alkali land, which provides improved farmland for grain crops production (Huang et al., 2013). Meanwhile, the demand for cotton fiber production will increase as the human population grows. To meet this challenge, breeders are working on developing new varieties using conventional and modern breeding methods to enhance cotton yield and tolerance under salt conditions. First, it is important to dissect the complex and dynamic mechanisms of plants under saline stress. Salinity affects plants in osmotic and ionic ways. The osmotic component of salinity leads to the reduction of water transport and partial stomatal closure. Interestingly, plants temporarily adapt to saline conditions using K+, Na+ and osmoprotectants for osmoregulation (Le et al., 1984; Munns and Tester, 2008). However, under long-term salt exposure, plants are affected by the continuing osmotic damage plus the excessive accumulation of Na+ in plant cells. High concentrations of Na+ inhibit enzyme activities and compete against K+ which is particularly important for enzyme functions (Bertorello et al., 1991; Tester and Davenport, 2003). To counter ionic toxicity, plants have mechanisms to compartmentalize Na+ in vacuoles and to remove the excess of Na+ from the cytosol to the apoplast (Maathuis and Amtmann, 1999; Munns et al., 2012).
Conventionally, Quantitative trait locus (QTL) mapping of salt-related traits has become an effective approach, which measures associations between genetic markers and traits using bi-parental populations. In the past few years, QTL mapping has been used for many plants using different statistical models, genetic markers and crossed populations (Mano and Takeda, 1997; Wang et al., 2012; Xu et al., 2012; Oyiga et al., 2017). In cotton, there have been only a few studies addressing salt tolerance traits (Tiwari et al., 2013; Oluoch et al., 2016). However, the constraint to QTL mapping is that QTLs are located in big genomic regions and contain too many genes, and genetic diversity between the parents and recombination progeny is limited. Moreover, backcrossing may take months or even years (Weigel, 2012).
Genome-wide association studies based on linkage disequilibrium (LD) are an alternative tool for studying the associations between phenotype and genotype. Compared to traditional QTL mapping, GWAS could use SNPs that are positioned across the genome as molecular markers for dissecting complex traits (Si et al., 2016). Moreover, GWAS can handle up to several million SNP markers and thousands of natural accessions as a mapping population (Huang et al., 2010; Chen et al., 2014). GWAS has been successfully applied in rice, Arabidopsis, maize, wheat, barley and other crops and contributed to identifying SNP markers that are associated with a trait of interest across diverse natural accessions (Atwell et al., 2010; Xue et al., 2013; Chen et al., 2016; McCouch et al., 2016). In cotton, association analysis had been adopted for important target traits, but all based on SSR markers (Abdurakhmonov et al., 2008; Mei et al., 2013; Cai et al., 2014; Jia et al., 2014b; Qin et al., 2015; Nie et al., 2016). Recently, with the development of SNP arrays, genotyping and sequencing technologies, some GWAS studies have been successfully applied for detecting genetic variations underlying diverse complex traits such as flowering time, yield, fiber quality and resistance to Verticillium wilt of cotton (Su et al., 2016c; Huang et al., 2017; Li et al., 2017; Sun et al., 2017). However, only few studies have applied a GWAS strategy of large-scale cotton accessions and SNP markers to unravel the molecular mechanism for salt tolerance (Diouf et al., 2017).
In the current study, we performed GWAS for the relative survival rate (RSR) and salt tolerance level (STL) with 10,511 SNPs at seedling stage under salt stress in 713 G. hirsutum accessions. The objectives of this study were to identify the associated SNPs and candidate genes, to contribute to understanding the mechanisms for salt tolerance and to develop molecular markers for accelerating breeding to enhance salt tolerance in cotton.
The association panel of 713 upland cottons accessions was used in this study derived from the recently published study (Sun et al., 2017). These germplasms were collected from different regions of China (585) and other countries (128) including the United States of America and Australia (Supplementary Table S1).
After the cotton seeds were delinted by sulfuric acid, 400 healthy and full seeds were selected from each germplasm accession. Each accession was set to three replicates and one water control treatment, and each replicate and water treatment contained 100 seeds. The seeds were placed evenly in a germination box containing 800 g dry quartz sand and covered evenly with 250 g dry quartz sand above the seeds, eventually adding 250 mL of 0.3% saline solution. The germination boxes were placed in a constant-temperature chamber at 28°C for 7 days with a 12/12-h light/dark regimen during the period without any treatment. We counted the number of surviving seedlings after sowing for 7 days and calculated the survival rate. Because the vitality of the seeds themselves will bring the error, it is necessary to calculate the RSR. Using a two-factor test design for optimal NaCl concentration, the A factor was the salt content (refers to the weight percentage of NaCl on dry quartz sand) with the above five different concentrations (0.1, 0.2, 0.3, 0.4, and 0.5%); the B factor was the control varieties containing the salt-tolerant cultivar Yu2067 and salt-sensitive cultivar Lumianyan21. The two cultivars were treated with five different NaCl concentrations. The RSR of the two cultivars had no difference when the NaCl concentration was 0.1 and 0.2% and the salt-sensitive cultivar Lumianyan21 had almost no survival when the NaCl concentrations were 0.4 and 0.5%. The results showed that the 0.3% NaCl content was suitable to evaluate the salt tolerance of the cotton seedlings.
The evaluated salt-tolerant traits included the RSR [RSR % = SR/control SR × 100, where SR % = the number of survival seedlings/total seed number used in the test × 100], and STL [STL was divided into four grades (from 1 to 4) according to the RSR; 4 represents salt sensitivity (0–49.9%), 3 represents salt tolerance (50.0–74.9%), 2 represents salt resistance (75.0–89.9%), and 1 represents high salt resistance (>90%)]. Statistical analysis of the RSR and STL was performed in SPSS 22.0. The frequency distribution of each trait and descriptive statistics were performed using all the phenotypic data from 713 cotton accessions.
Population structure, relative kinship and LD analysis had already been analyzed in a previous study (Sun et al., 2017). Here, association analysis was conducted using the Q + K model with a total of 10,511 SNPs [call rate > 85% and minor allele frequency (MAF) > 0.05] from the Illumina CottonSNP63K array (Hulse-Kemp et al., 2015) and the Q + K model, which was implemented via a mixed linear model (MLM) in TASSEL 3.0 (Bradbury et al., 2007). According to the Bonferroni correction principle, -log10 (P) > 3.97 (P = 1/n, n is the SNP numbers in this study) should be as threshold, but the Bonferroni correction is too stringent, we can’t almost identify the significant SNPs for two traits with this threshold. To obtain more associated SNPs, the significantly associated SNP markers with salt-tolerant-related traits were identified according to -log10 P > 3.0 (Hatzig et al., 2015; Zhang et al., 2015a).
The genes within 200 kb near significant SNP loci were assigned as putative candidates based on the gene annotation in the G. hirsutum TM-1 genome (Zhang et al., 2015b). The interval of 200 kb was considering the LD decay distances and in comparison with other crops (Sun et al., 2017). GO enrichment and KEGG pathway analysis were carried out for all candidate genes (Xie et al., 2011). To further screen the possible candidate genes involved in the salt response, these genes were analyzed using the expression level of the seedlings at 1, 3, 6, and 12 h under 400 mM salt concentration reported by Zhang et al. (2015b).
To validate the results of the GWAS, five salt-tolerant and five salt-sensitive varieties were selected for gene expression analysis of the putative candidates by qRT-PCR (Supplementary Table S6). Plants were grown in germination boxes containing quartz sand for 7 days with 0.3% NaCl content. Roots were sampled and immediately frozen in liquid nitrogen and stored at -80°C for RNA extraction. Total cDNA was synthesized with PrimeScriptTM RT reagent Kit (Perfect Real Time) (TaKaRa). The qRT-PCR reactions contained 10 μL of SYBR Premix DimerEraser (TaKaRa), 2.0 μL of cDNA, 0.6 μL of primer, and ddH2O to a final volume of 20 μL. The reactions were amplified for 30 s at 95°C, followed by 40 cycles of 95°C for 5 s, 55°C for 30 s and 72°C for 30 s. All reactions were performed in three independent biological replicates, each with three technical replicates, using the Roche LightCycler96 Real-Time PCR System. The primers are listed in Supplementary Table S7. GhUBQ14 expression was used as the internal control for qRT-PCR. Relative gene expression values were calculated using the 2-ΔCT method (Schmittgen and Livak, 2008).
A salt-tolerant cultivar (Suwu77-702) and a salt-sensitive cultivar (Shann424) were selected to grow for 7 days in a germination box, and then moved into 1/2 Hogland nutrient solution until the three-true-leaf period (about another 14 days). Under 200 mmol/L NaCl stress, leaves were sampled at 0, 3, 6, 8, 12, 24, and 48 h for measuring the content of the protein, POD, MDA, and H2O2 (Liu and Li, 2007; Cai et al., 2017). Three biological replicates were performed.
Under salt stress of 0.3% NaCl content, extensive phenotypic variations were observed for RSR and STL in 713 upland cotton accessions. The RSR ranged from 3.38 to 92.06% with an average of 35.85%, and the STL ranged from 1.00 to 4.00 with an average of 1.25. The coefficient of variation (CV) was 49.96 and 39.52% (Table 1). Of the 713 accessions, 160 were identified as salt-tolerant, and 18 were salt-resistant in which Litai8 had 92.06% for RSR and belonged to a high salt-resistant level according to the STL standard. The phenotypic distribution of RSR and STL showed continuous variation (Figure 1), while RSR was approximately a normal distribution, indicating that these two traits were quantitative traits controlled by multiple genes.
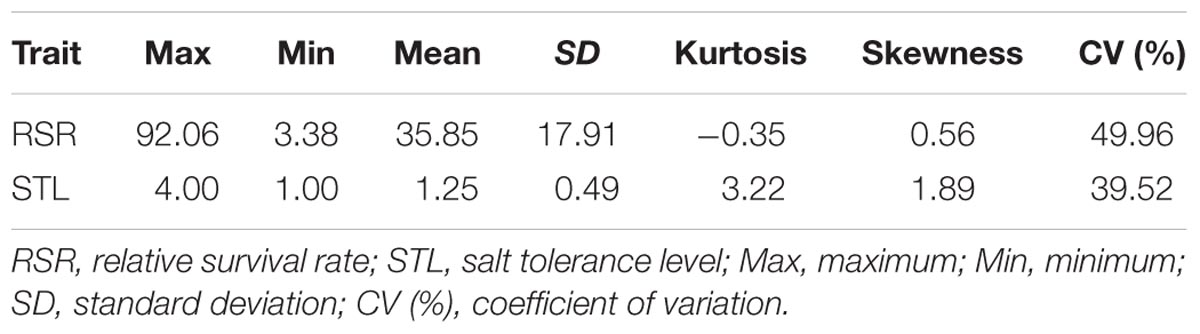
TABLE 1. Phenotypic variation for the two salt-tolerance traits under salt treatment in the panel of cotton accessions.
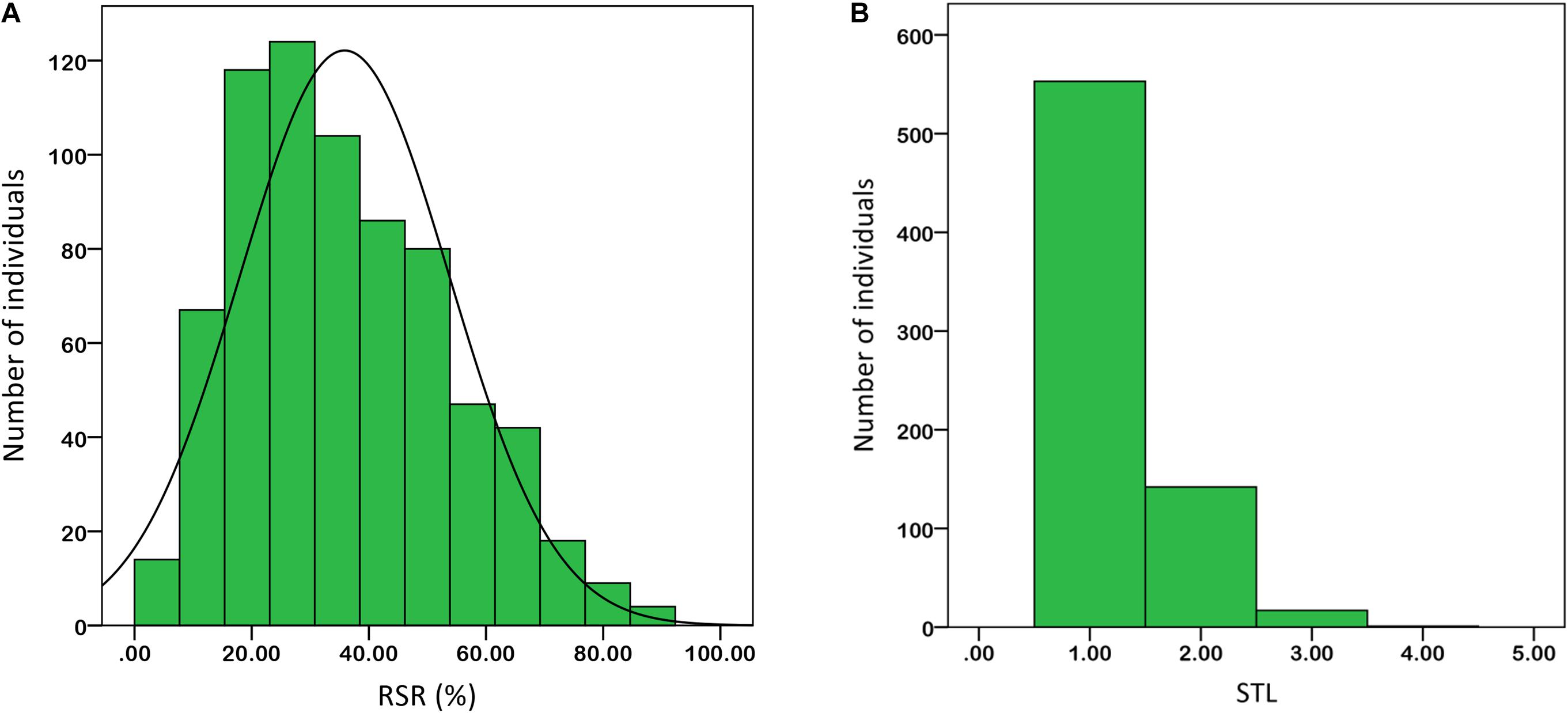
FIGURE 1. Phenotypic diversity of the two salt-tolerance-related traits in 713 cotton accessions based on 0.3% salt treatment. (A) RSR. (B) STL. Data were averaged over three replicates.
The SNP markers associated with the two salt-tolerant related traits, RSR and STL, were identified using the MLM model considering population structure (Q) and kinship (K) in TASSEL 3.0 software (Figure 2). We identified a total of 23 significantly associated SNPs for the two traits. These SNPs were distributed on chromosomes A01, A10, D02, D08, D09, D10, and D11 (Table 2).
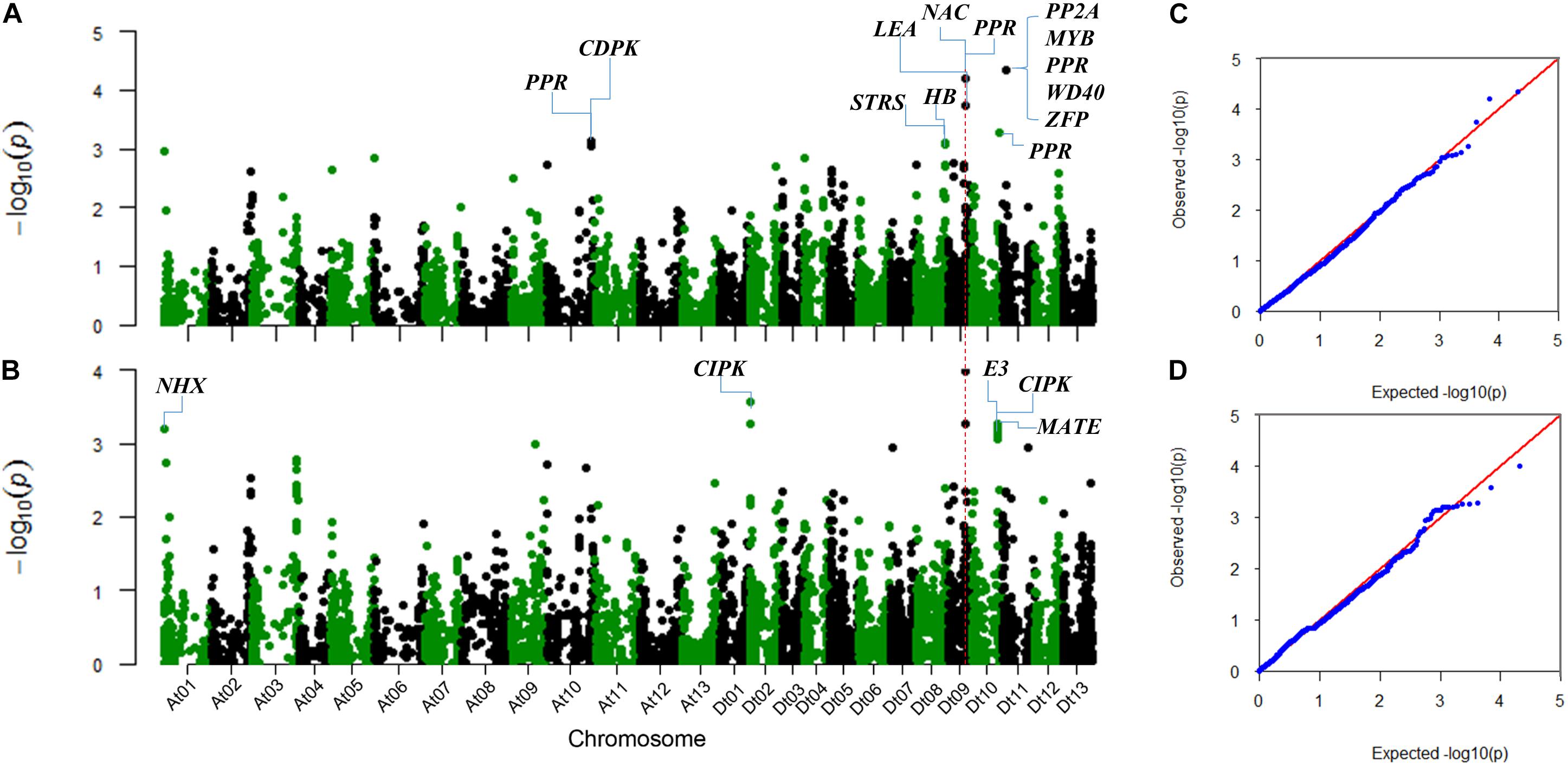
FIGURE 2. Manhattan and QQ plots for the two salt-tolerance-related traits. (A) Manhattan plot for RSR. (B) Manhattan plot for STL. (C) QQ plot for RSR. (D) QQ plot for STL. The corresponding Arabidopsis thaliana homologous genes linked to the significant SNPs are shown.
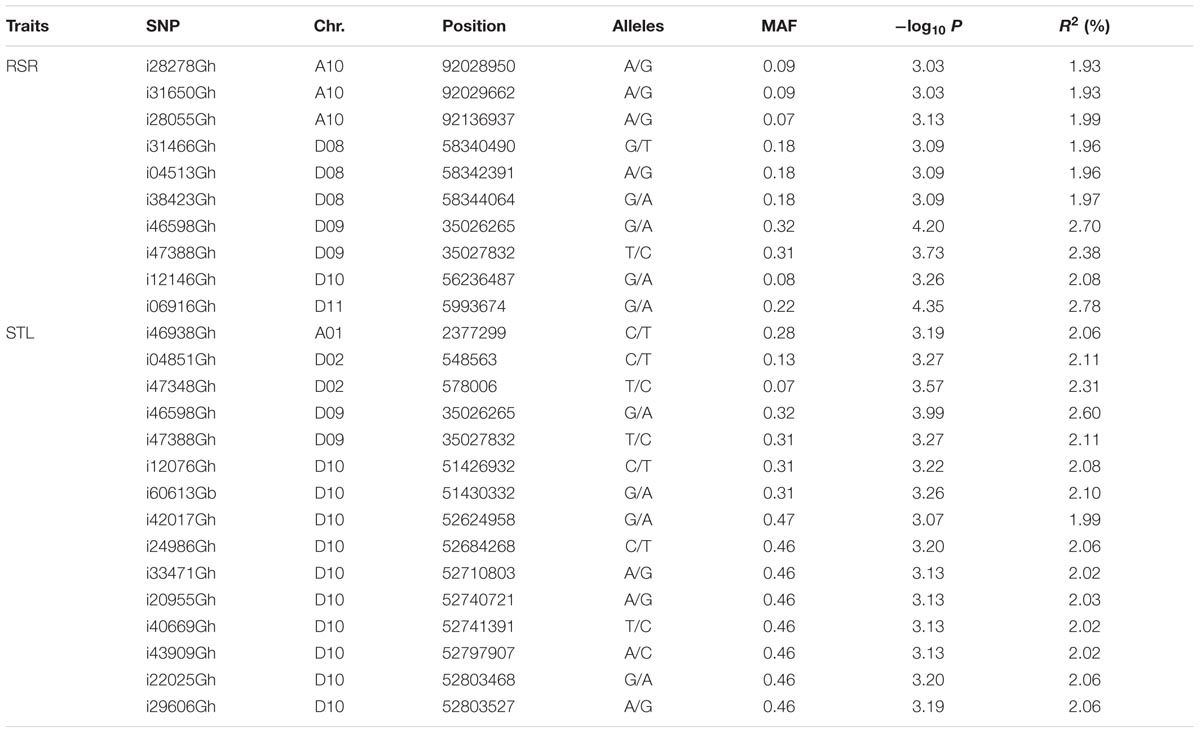
TABLE 2. List of SNP markers significantly associated with RSR and STL in the upland cotton association panel.
For RSR, 10 associated SNPs were identified on chromosomes A10, D08, D09, D10, and D11 (Table 2). The phenotypic variation explained by a single SNP ranged from 1.93 to 2.78%, and the strongest association locus (i06916Gh) was found at 5.99 Mb on D11 (Figure 2A).
For STL, a total of 15 association signals were detected and distributed on chromosomes A01, D02, D09, and D10, explaining 1.99–2.60% of the phenotypic variation (Table 2). Among them, the strongest association signal (i46598Gh) was found at 35.02 Mb on D09. Notably, the SNP i46598Gh and i47388Gh on D09 were significantly associated with both STL and RSR. In addition, a GWAS peak containing 10 SNPs was detected on chromosome D10 (Figure 2B), of which eight SNPs were closed in the chromosome (Table 2).
To predict the potential candidate genes related to RSR and STL, we identified genes located within 200 kb in the genome according to the physical position of each significant locus and the functional annotation of the homologous genes in Arabidopsis. Based on the G. hirsutum TM-1 reference genome, we obtained 280 candidate genes associated with the two traits (Supplementary Table S2). GO enrichment and KEGG pathway analysis were conducted for all candidate genes. Tetrahydrofolate interconversion (GO: 0035999) with four genes and the regulation of the phenylpropanoid metabolic process (GO: 2000762) with four genes were the categories most significantly enriched (Supplementary Table S3). These genes were in the amino acid dehydrogenase family protein and peroxidase superfamily protein. Meanwhile, there were three genes enriched significantly in oxidoreductase activity (GO: 0016646). For the KEGG pathway, the top significantly enriched pathway was monoterpenoid biosynthesis with six genes, which was part of the NAD (P)-binding Rossmann-fold superfamily of proteins (Supplementary Table S3).
In chromosome D09, 35 common candidate genes were identified for the RSR and STL surrounding peak SNPs (i46598Gh and i47388Gh) with high LD (Figure 3A). These candidate genes were involved in different transcription factors, binding proteins, membrane transport proteins and other proteins of unknown function. Further, these candidate genes were screened using transcriptome sequencing data1. The results showed that the most of the genes had varying degrees of expression at four time points (1, 3, 6, and 12 h) under salt stress (Figure 3B). Among them, Gh_D09G0943 and Gh_D09G0950 are orthologous to the genes NAC061 and NAC089, respectively, and Gh_D09G0958 and Gh_D09G0959 are LEA genes (Supplementary Table S4). These genes could be promising candidates according to previous reports of salt-related genes in other plants (Puranik et al., 2012; Hu et al., 2016).
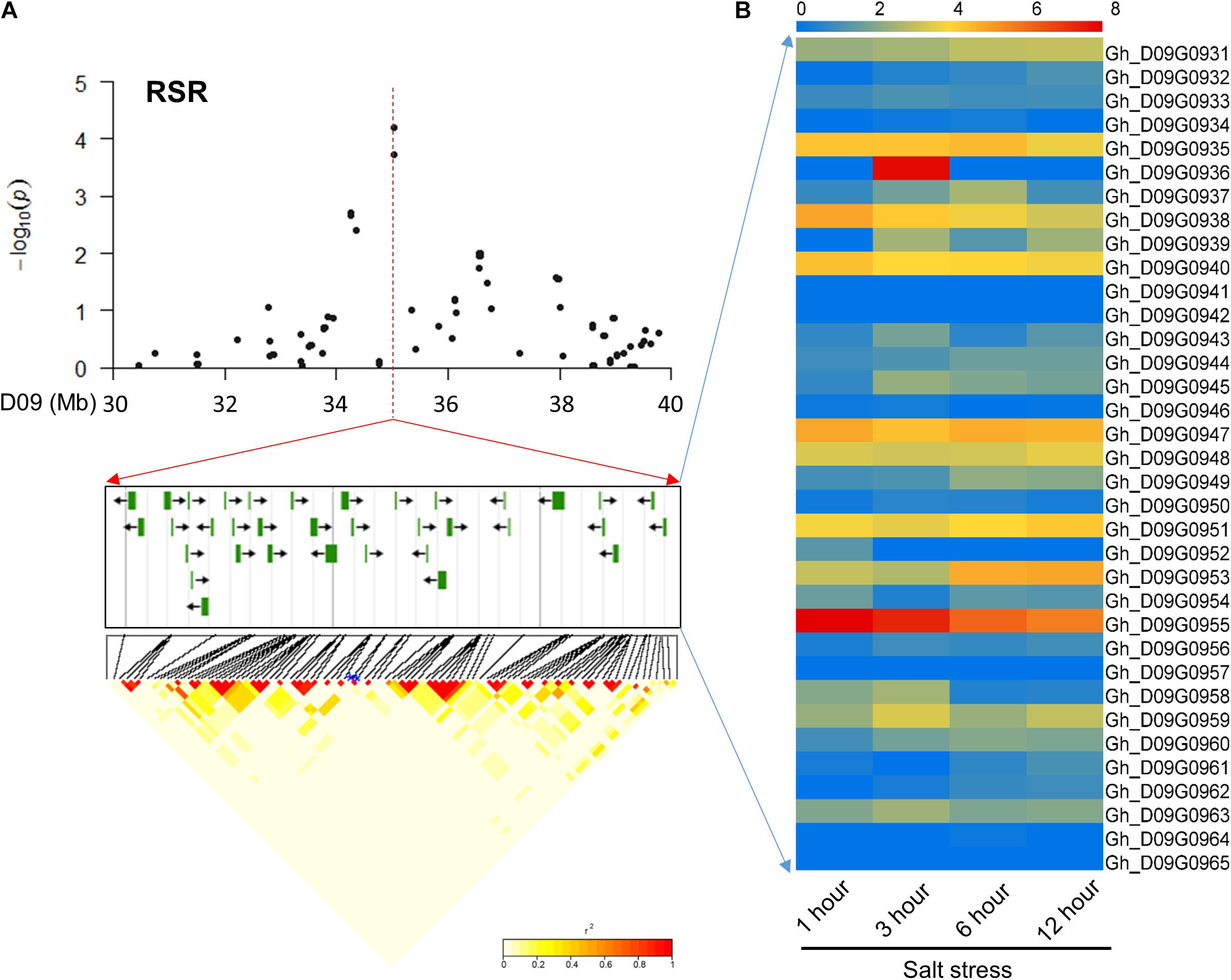
FIGURE 3. Identification of the candidate genes for RSR and STL on the peak of chromosome D09. (A) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak. The 35 candidate genes were presented in the middle. (B) Heat map of the expression of the 35 candidate genes at four time points. The samples were collected at 1, 3, 6, and 12 h under salt stress.
For STL, 26 candidate genes surrounding the eight significant SNP loci with high LD were found on chromosome D10 (Figure 4A). Most of these genes had preferential expression at different time points with salt treatment (Supplementary Table S5). Among these genes, Gh_D10G1882 and Gh_D10G1888 were preferentially expressed and Gh_D10G1874 had higher expression at 3 h (Figure 4B). The above three genes were annotated as peroxidase, E3 ubiquitin-protein ligase gene and CBL-interacting protein kinase, respectively, which were involved and functioned in plant responses to salt stress (Yee and Goring, 2009; Jin et al., 2016).
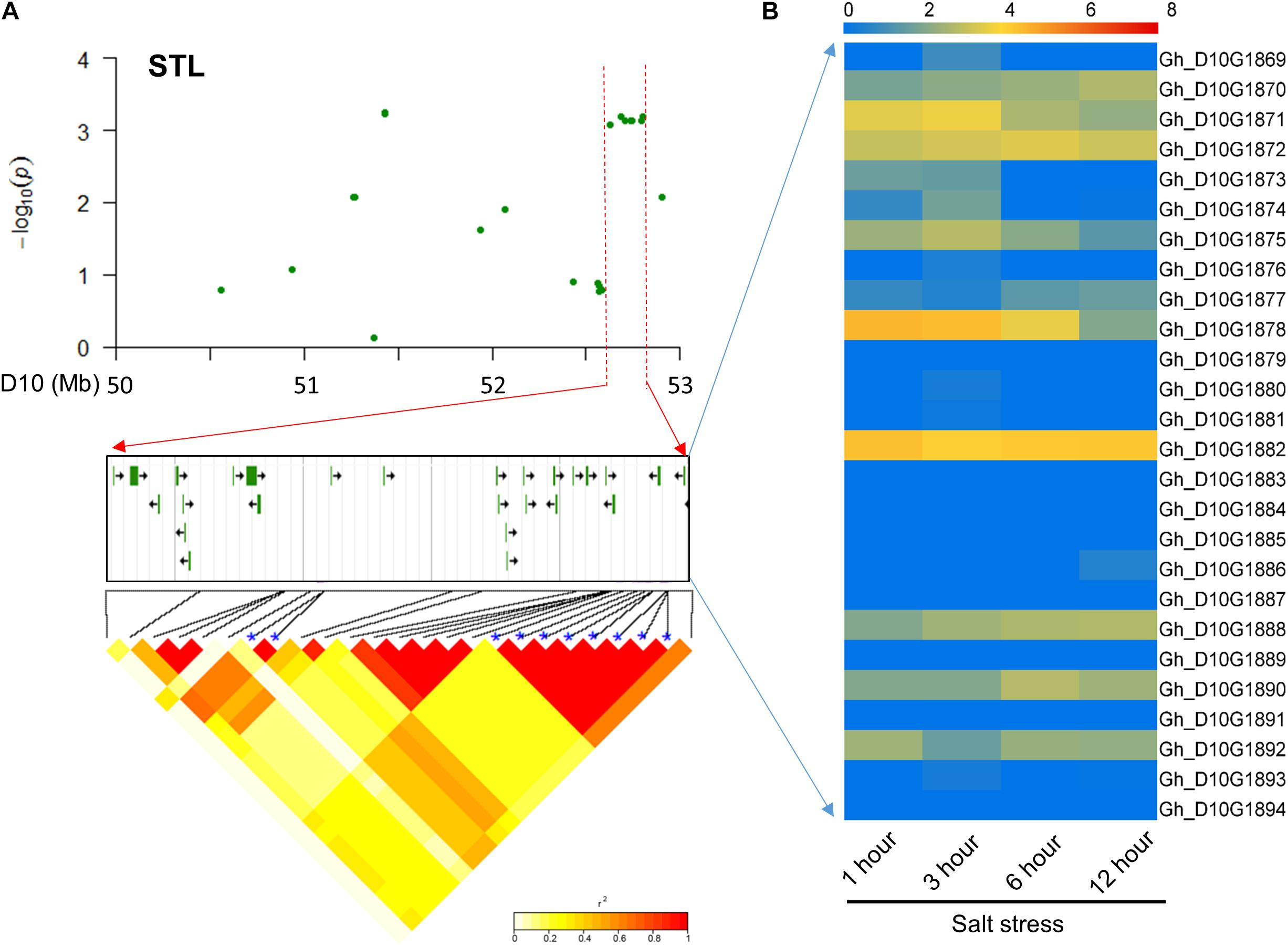
FIGURE 4. Identification of the candidate genes for STL on the peak of chromosome D10. (A) Local Manhattan plot (top) and LD heatmap (bottom) surrounding the peak. The 26 candidate genes were presented in the middle. (B) Heat map of the expression of the 26 candidate genes at four time points. The samples were collected at 1, 3, 6, and 12 h under salt stress.
A total of 35 and 26 candidate genes were within the LD decay region on D09 and D10, respectively (Supplementary Tables S3, S4). Gh_D09G0943 and Gh_D09G0958 were close to the peak SNP i46598Gh at 34.83–35.22 Mb. Gh_D10G1888 was located near the peak SNP i20955Gh at 52.62–52.80 Mb. Another three genes (Gh_A10G1756, Gh_D02G0060, and Gh_D10G1821) were located near the association peak of other chromosomes. The expression levels of these six genes were measured using five salt-tolerant varieties and five salt-sensitive varieties by qRT-PCR. In comparison to the salt-sensitive varieties, Gh_D10G1888, Gh_D02G0060, Gh_D09G0943, and Gh_D10G1821 had a higher relative expression in the salt-tolerant varieties. Notably, the relative expression of Gh_D10G1821 was high in the salt-tolerant varieties, but almost no expression was found in the salt-sensitive varieties. However, the other two genes, Gh_D09G0958 and Gh_A10G1756, had higher expression in the salt-sensitive varieties (Figure 5). This provides further evidence that the six putative genes were closely associated with salt tolerance.
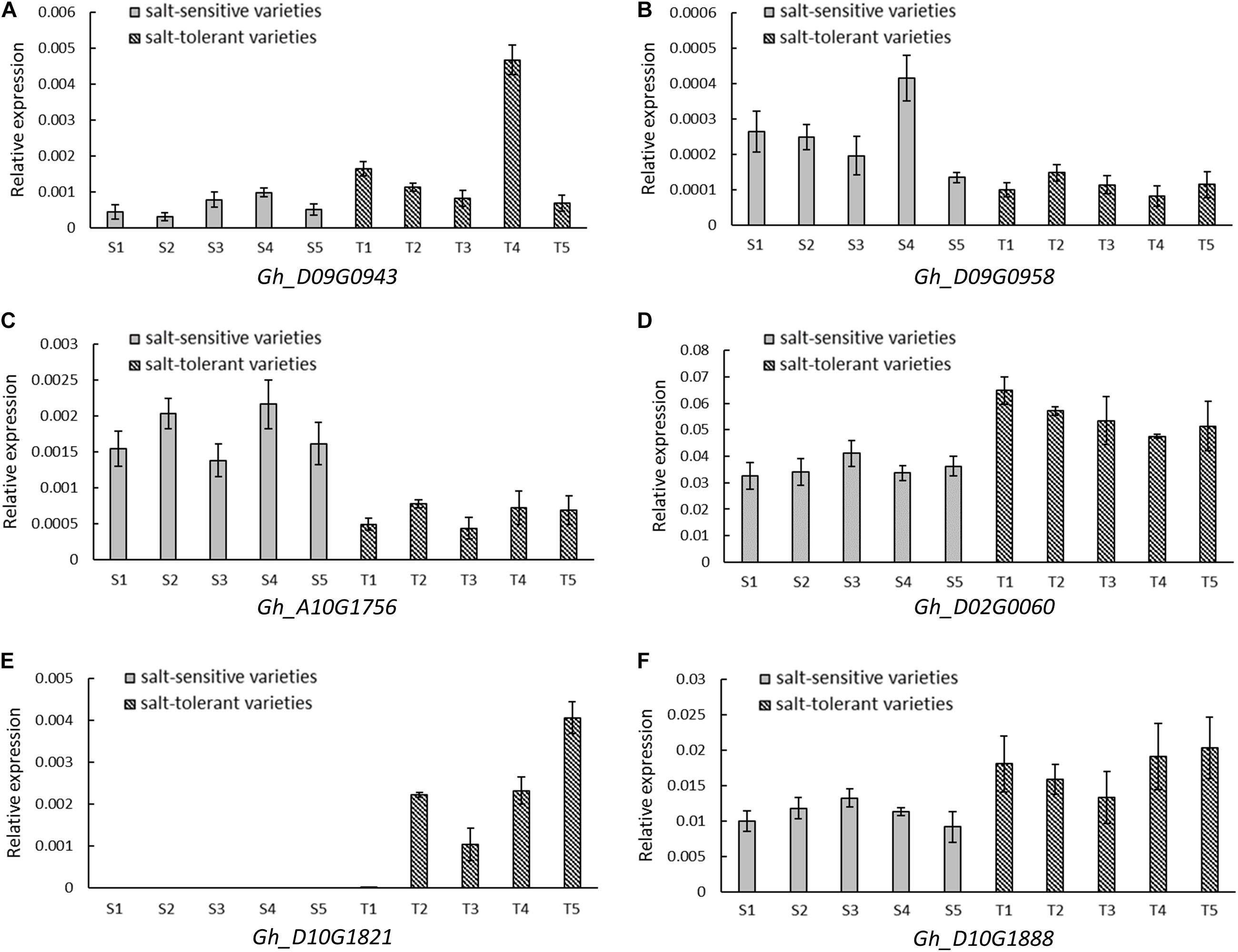
FIGURE 5. Relative expression by quantitative RT-PCR of six candidate genes significantly associated with RSR and STL. (A) Gh_D09G0943 (NAC090). (B) Gh_D09G0958 (LEA). (C) Gh_A10G1756 (CPK1). (D) Gh_D02G0060 (CIPK2). (E) Gh_D10G1821 (UPL6). (F) Gh_D10G1888 (SINAT2). Five salt-sensitive varieties, S1: Pengze70, S2: Shannmian10, S3: Lumianyan35, S4: Jiwu2833, S5: Xinluzao38; five salt-tolerant varieties, T1: Zhongwu1038, T2: CCRI20, T3: Zhongkang5, T4: Litai8, T5: Zhongwu3385.
In addition, we selected the two cultivars with significant difference of morphological characteristics under salt treatment for subsequent analysis. The contents of protein, POD, MDA, and H2O2 were used as physiological and biochemical indexes to characterize the tolerance to salt stress. Protein content increased gradually in the salt-tolerant cultivar and was significantly higher in the salt-tolerant cultivar than in the salt-sensitive cultivar at 48 h. POD content reached a peak at 3 h under salt treatment in the salt-tolerant cultivar; however, the POD content showed an upward tendency and continued to 24 h, where it reached a peak in the salt-sensitive cultivar. The MDA content totally decreased in the two types of cultivars, but showed a peak at 12 and 24 h. The H2O2 content increased in the salt-sensitive cultivar, however, it was basically unchanged in the salt-tolerant cultivar (Figure 6). These results indicated that there were differences in salt tolerance mechanisms between the two types of cultivars.
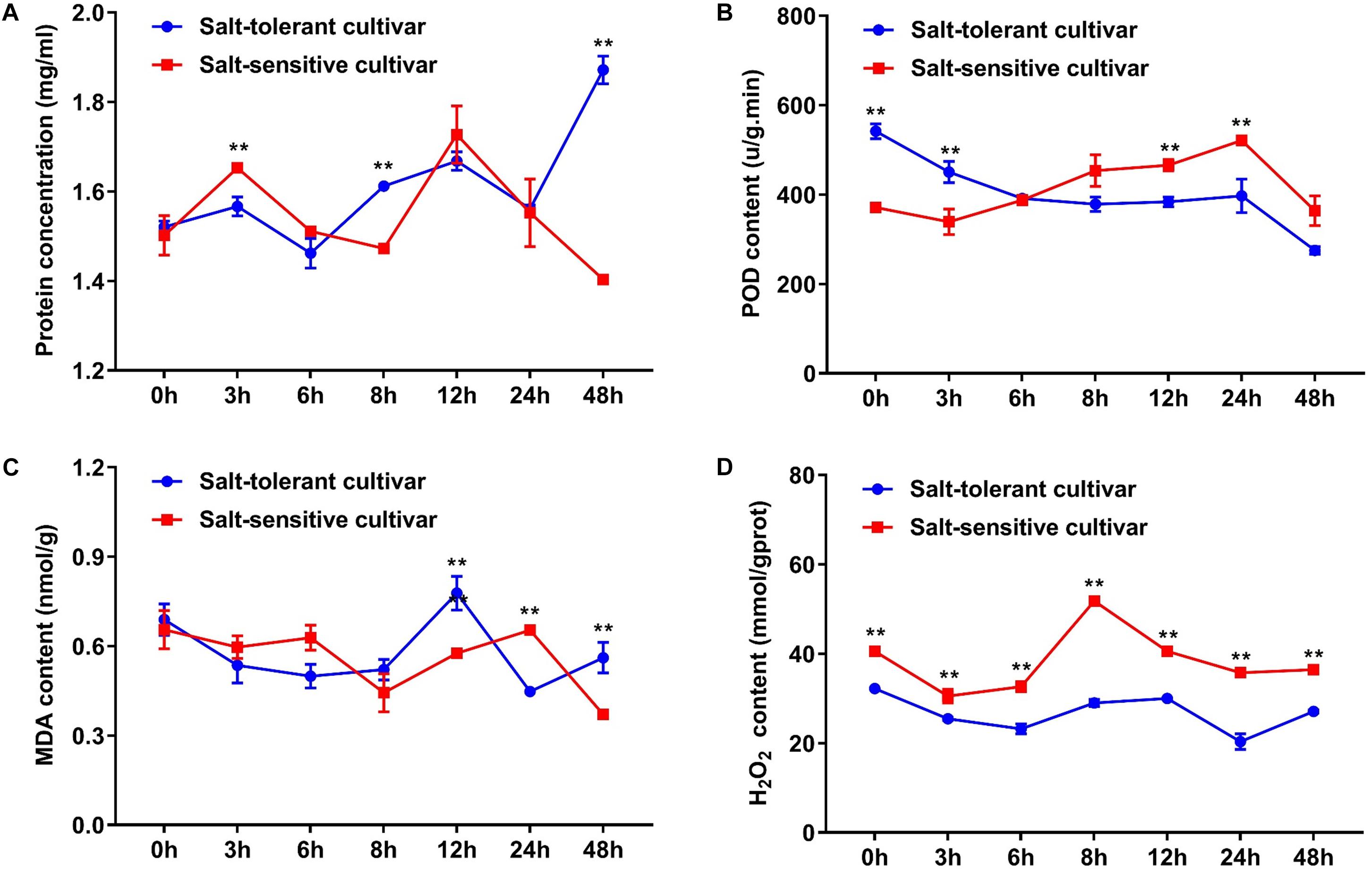
FIGURE 6. Comparison of the protein, POD, MDA, and H2O2 contents between the salt-tolerant cultivar and the salt-sensitive cultivar under salt stress. (A) Protein concentration. (B) POD content. (C) MDA content. (D) H2O2 content. ∗∗ indicates significance at P < 0.01 level (two-tailed t-test). Error bars indicate ± SD; n = 3 independent biological replicates.
In the present study, we conducted a GWAS of salt-tolerant related traits at the seedling stage with a great many upland cotton accessions and SNPs from the CottonSNP63K array. This study provides new insights into the genetic basis of salt tolerance and the identification of the novel alleles underlying the variation in the salt-tolerant traits and candidate genes, allowing for accelerating the progress of cotton tolerance breeding.
The seedling stage is a very important stage in the cotton growth period. The RSR is the percentage of surviving seedlings divided by the total number of seeds used in the test, which is a critical feature to evaluate the seedling quality and viability. STL is considered a reliable index for evaluating salt tolerance on the corresponding seedling survival rate. In our study, therefore, both RSR and STL were selected to evaluate the upland cotton seedlings’ ability to salt stress. The seedling stage of cotton is about 10 days in the field and slightly shorter in the chamber; thus, we chose 7 days to investigate the two traits (Wang et al., 2011). Du et al. (2016) performed the evaluation of salt tolerance at the germination and seedling stages of 304 upland cotton cultivars and identified 43 advanced salt-tolerant cultivars by cluster analysis of 10 salt-tolerance related traits. Of the 713 accessions in this study, 142 were identified as salt-tolerant cultivars, 17 were salt-resistant cultivars and one accession had a high salt-resistance level. In addition, we found that the obsolete cultivars or lines were more salt-tolerant than the newly bred ones (Supplementary Table S1). This is perhaps because salt tolerance was gradually lost when breeders focused on high yield and improved fiber quality. In China, the planting area for the cotton already reduced greatly when agricultural industrial restructuring, and breeders are trying to develop and utilize saline-alkali land suitable for farming. However, because of the lack of excellent improved salt-tolerant cotton varieties, salinity is a serious constraint to cotton productivity in such areas. Therefore, these better salt-tolerant accessions could be selected as parents to accelerate the progress of cotton tolerance breeding by molecular design (Borsani et al., 2003; Rai et al., 2011).
Salt tolerance is a genetically and physiologically complex trait controlled by multiple small effect genes (Flowers, 2004). With the fast development of SNP arrays and next-generation sequencing technology, GWAS is becoming a novel and effective method for determining useful genes in crop plants. Association analysis has been successfully used in mining candidate genes of important agronomic traits in cotton, such as fiber quality, yield and its components, and Verticillium wilt resistance (Su et al., 2016a,b; Fang et al., 2017; Li et al., 2017; Sun et al., 2017; Wang et al., 2017). However, only a few studies related to salt tolerance were reported with natural populations and genome-wide molecular markers using the GWAS strategy. Jia et al. (2014a) only detected three SSR markers associated with salt tolerance in MLM using 106 SSRs in 323 G. hirsutum germplasms. Cai et al. (2017) found 9 intron length polymorphisms (ILPs) markers for 10 salt stress traits with 535 ILP markers in 264 G. hirsutum accessions. In this study, 713 upland cotton accessions were used to conduct a GWAS of two salt-tolerance related traits with 10,511 SNPs, and we identified 10 and 15 loci for RSR and STL, respectively, of which two common SNP loci (i46598Gh and i47388Gh) on D09 were significantly associated with the two traits. Perhaps this chromosome has important genomic hotspots controlling salt tolerance similar to other traits (Said et al., 2015). Furthermore, we screened 280 possible candidate genes that were involved in several kinds of functional proteins including transcription factors, transporters and enzymes. However, little is known about the genetic architecture of salt tolerance in cotton. These associated SNPs and candidates remain to be further verified and discussed in future research. Currently, many studies have dissected the genetic mechanisms of salt tolerance in Arabidopsis and other crop species, and many important candidate genes and pathways for salinity tolerance have been identified in Arabidopsis thaliana (Zhu, 2001; Ashraf et al., 2013; Deinlein et al., 2014).
On chromosome D09, the two genes Gh_D09G0943 and Gh_D09G0950 encode NAC domain-containing proteins. NAC proteins have received much attention as regulators in various stress signaling pathways, which may include the interplay of phytohormones (Puranik et al., 2012). It was demonstrated that ONAC022 improved drought and salt stress tolerance through modulating an ABA-mediated pathway in rice (Hong et al., 2016). Two NAC transcription factors from Caragana intermedia altered ABA signaling during seed germination and enhanced salt tolerance of transgenic Arabidopsis (Han X. et al., 2015). Late embryogenesis abundant (LEA) proteins participated in tolerance to salinity and drought in many different organisms. We identified two candidate genes Gh_D09G0958 and Gh_D09G0959, encoding LEA proteins. The LEA proteins could act as membrane stabilizers to prevent cellular collapse (Wang et al., 1996). In rice, the overexpression of OsLEA4 in transgenic plants conferred increased resistance to salt, drought and heavy metal stresses (Hu et al., 2016). In addition, five homogenous genes located on different chromosomes belonged to pentatricopeptide repeat (PPR) proteins family, of which two (Gh_D09G0932 and Gh_D09G0961) were located on D09. PPR proteins are considered to play important role in photosynthesis, plant development, and environmental responses (Barkan and Small, 2014). It was reported that the overexpression of a PPR gene improved salt tolerance in Arabidopsis (Zsigmond et al., 2012).
For RSR, the gene Gh_A10G1756 encodes a calcium-dependent protein kinase (CPK or CDPK). In rice, the overexpression of OsCPK4 positively regulated salt and drought tolerance via protection against membrane lipid peroxidation (Campo et al., 2014). On chromosome D08, the gene Gh_D08G1976 coded for a homeobox-leucine zipper protein (HB protein). The ectopic expression of HaHB1 from sunflowers in Arabidopsis conferred tolerance to drought and salinity stresses by the induction of proteins that stabilize membranes (Cabello and Chan, 2012). There is another gene, Gh_D08G1982, encoding a DEAD-box helicase protein. A DEAD-box helicase, PDH45, conferred salinity tolerance to rice in both the seedling and reproductive stage (Amin et al., 2011). Additionally, the four genes Gh_D11G0691, Gh_D11G0694, Gh_D11G0702, and Gh_D11G0706 on chromosome D11 are homologous genes of PP2A, MYB40, WD40, and ZFP in Arabidopsis, respectively. Protein phosphatase 2A (PP2A) was one of the major serine/threonine protein phosphatases and played important roles in cellular processes in plants (Farkas et al., 2007). In Arabidopsis, PP2A-C5 increased salt tolerance in a pathway different from the SOS signaling pathway (Hu et al., 2017). Plant MYB transcription factors control diverse biological processes. In birch, BplMYB46 improved salt and osmotic stress tolerance and mediated secondary cell wall deposition (Guo et al., 2017). A wheat WD40 repeat-containing protein increased plant tolerance to ABA, salt stress and osmotic stress during seed germination and seedling development (Kong et al., 2015). In cotton, GhZFP1, a novel CCCH-type zinc finger protein from cotton, acted as a novel positive regulator to confer salt tolerance and fungal pathogen resistance to plants (Guo et al., 2009).
For STL, the three genes Gh_D10G1821, Gh_D10G1842, and Gh_D10G1888 encode the E3 ubiquitin-protein ligase. E3 ubiquitin was involved in the response to dehydration stress and the regulation of proline biosynthesis in Arabidopsis (Yee and Goring, 2009). The Arabidopsis U-box E3 ubiquitin ligase AtPUB30 participated in the salt stress tolerance as a negative factor during the germination stage (Hwang et al., 2015). There are two genes, Gh_D02G0060 and Gh_D10G1874, that encode CBL-interacting protein kinase (CIPK). CaCIPK6 increased auxin transport and hypersensitivity to auxin and promoted salt tolerance in transgenic tobacco (Tripathi et al., 2009). The overexpression of TaCIPK25 impaired salt tolerance, which was mediated by a WRKY transcription factor in an ABA-dependent pathway under saline conditions (Jin et al., 2016). The gene Gh_D10G1824 encoded a MATE efflux family protein that modulates ABA sensitivity and increases tolerance to drought with lower stomatal conductance (Zhang et al., 2014). Another gene Gh_A01G0243, encodes an Na+/H+ antiporter. It was responsible for the regulation of internal pH, cell volume and sodium level in the cytoplasm (Bassil et al., 2011). CottonGhNHX1 functioned as a tonoplast Na+/H+ antiporter and played an important role in the salt tolerance of cotton (Wu et al., 2004).
The cotton accessions consisting of diverse germplasms worldwide showed large variations in RSR and STL under salt stress. A total of 10 and 15 SNPs significantly associated with RSR and STL were identified, respectively, of which the two SNPs i46598Gh and i47388Gh on D09 were simultaneously associated with the two traits. We further screened 280 candidate genes, many of which were related to plant salt tolerance. In addition, we validated the expression level of six putative genes using five salt-tolerant and five salt-sensitive varieties under salt stress by qRT-PCR. Our results provided a valuable reference for the study of salt tolerance in G. hirsutum. These SNP loci and candidate genes would be useful for future salt tolerance breeding programs in cotton.
ZM, GZ, and XW conceived and designed the research. ZS, HL, YZ, ZL, HK, and LW performed the experiments. ZS and HL conducted statistical analysis of data. ZS and XW wrote the manuscript. ZM revised the manuscript.
This work was supported by the National Key Research and Development Program of China (2016YFD0101006), the Science and Technology Support Program of Hebei Province (16226307D), and Young Talents Support Program of Hebei Province.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Tianzhen Zhang (Zhejiang University) for releasing RNA-seq data.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01011/full#supplementary-material
Abdurakhmonov, I. Y., Kohel, R. J., Yu, J. Z., Pepper, A. E., Abdullaev, A. A., Kushanov, F. N., et al. (2008). Molecular diversity and association mapping of fiber quality traits in exotic G. hirsutum L. germplasm. Genomics 92, 478–487. doi: 10.1016/j.ygeno.2008.07.013
Amin, M., Elias, S. M., Hossain, A., Ferdousi, A., Rahman, M. S., Tuteja, N., et al. (2011). Over-expression of a DEAD-box helicase, PDH45, confers both seedling and reproductive stage salinity tolerance to rice (Oryza sativa L.). Mol. Breed. 30, 345–354. doi: 10.1007/s11032-011-9625-3
Ashraf, M., Foolad, M. R., and Tuberosa, R. (2013). Crop breeding for salt tolerance in the era of molecular markers and marker-assisted selection. Plant Breed. 132, 10–20. doi: 10.1111/pbr.12000
Ashraf, M., and Wu, L. (1994). Breeding for salinity tolerance in plants. Crit. Rev. Plant Sci. 13, 17–42. doi: 10.1080/07352689409701906
Atwell, S., Huang, Y. S., Vilhjalmsson, B. J., Willems, G., Horton, M., Li, Y., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. doi: 10.1038/nature08800
Barkan, A., and Small, I. (2014). Pentatricopeptide repeat proteins in plants. Annu. Rev. Plant Biol. 65, 415–442. doi: 10.1146/annurev-arplant-050213-040159
Bassil, E., Tajima, H., Liang, Y. C., Ohto, M. A., Ushijima, K., Nakano, R., et al. (2011). The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell 23, 3482–3497. doi: 10.1105/tpc.111.089581
Bertorello, A. M., Aperia, A., Walaas, S. I., Nairn, A. C., and Greengard, P. (1991). Phosphorylation of the catalytic subunit of Na+,K+-ATPase inhibits the activity of the enzyme. Proc. Natl. Acad. Sci. U.S.A. 88, 11359–11362. doi: 10.1073/pnas.88.24.11359
Borsani, O., Valpuesta, V., and Botella, M. A. (2003). Developing salt tolerant plants in a new century: a molecular biology approach. Plant Cell Tissue Organ Cult. 73, 101–115. doi: 10.1023/a:1022849200433
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Cabello, J. V., and Chan, R. L. (2012). The homologous homeodomain-leucine zipper transcription factors HaHB1 and AtHB13 confer tolerance to drought and salinity stresses via the induction of proteins that stabilize membranes. Plant Biotechnol. J. 10, 815–825. doi: 10.1111/j.1467-7652.2012.00701.x
Cai, C., Wu, S., Niu, E., Cheng, C., and Guo, W. (2017). Identification of genes related to salt stress tolerance using intron-length polymorphic markers, association mapping and virus-induced gene silencing in cotton. Sci. Rep. 7:528. doi: 10.1038/s41598-017-00617-7
Cai, C., Ye, W., Zhang, T., and Guo, W. (2014). Association analysis of fiber quality traits and exploration of elite alleles in Upland cotton cultivars/accessions (Gossypium hirsutum L.). J. Integr. Plant Biol. 56, 51–62. doi: 10.1111/jipb.12124
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., and San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Chen, G., Zhang, H., Deng, Z., Wu, R., Li, D., Wang, M., et al. (2016). Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica 212, 173–185. doi: 10.1007/s10681-016-1750-y
Chen, W., Gao, Y., Xie, W., Gong, L., Lu, K., Wang, W., et al. (2014). Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 46, 714–721. doi: 10.1038/ng.3007
Dagar, J. C., and Minhas, P. S. (2016). “Global perspectives on agroforestry for the management of salt-affected soils,” in Agroforestry for the Management of Waterlogged Saline Soils and Poor-Quality Waters, J. C. Dagar and P. S. Minhas (New Delhi: Springer), 5–32. doi: 10.1007/978-81-322-2659-8_2
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Demiral, T., and Türkan, I. (2006). Exogenous glycinebetaine affects growth and proline accumulation and retards senescence in two rice cultivars under NaCl stress. Environ. Exp. Bot. 56, 72–79. doi: 10.1016/j.envexpbot.2005.01.005
Diouf, L., Pan, Z., He, S. P., Gong, W. F., Jia, Y. H., Magwanga, R. O., et al. (2017). High-density linkage map construction and mapping of salt-tolerant qtls at seedling stage in upland cotton using genotyping by sequencing (GBS). Int. J. Mol. Sci. 18:E2622. doi: 10.3390/ijms18122622
Du, L., Cai, C., Wu, S., Zhang, F., Hou, S., and Guo, W. (2016). Evaluation and exploration of favorable QTL alleles for salt stress related traits in cotton cultivars (G. hirsutum L.). PLoS One 11:e0151076. doi: 10.1371/journal.pone.0151076
Fang, L., Wang, Q., Hu, Y., Jia, Y., Chen, J., Liu, B., et al. (2017). Genomic analyses in cotton identify signatures of selection and loci associated with fiber quality and yield traits. Nat. Genet. 49, 1089–1098. doi: 10.1038/ng.3887
Farkas, I., Dombradi, V., Miskei, M., Szabados, L., and Koncz, C. (2007). Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12, 169–176. doi: 10.1016/j.tplants.2007.03.003
Farooq, M., Gogoi, N., Barthakur, S., Baroowa, B., Bharadwaj, N., Alghamdi, S. S., et al. (2017). Drought stress in grain legumes during reproduction and grain filling. J. Agron. Crop Sci. 203, 81–102. doi: 10.1111/jac.12169
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55, 307–319. doi: 10.1093/jxb/erh003
Guo, H., Wang, Y., Wang, L., Hu, P., Wang, Y., Jia, Y., et al. (2017). Expression of the MYB transcription factor gene BplMYB46 affects abiotic stress tolerance and secondary cell wall deposition in Betula platyphylla. Plant Biotechnol. J. 15, 107–121. doi: 10.1111/pbi.12595
Guo, Y. H., Yu, Y. P., Wang, D., Wu, C. A., Yang, G. D., Huang, J. G., et al. (2009). GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 183, 62–75. doi: 10.1111/j.1469-8137.2009.02838.x
Hamwieh, A., Tuyen, D. D., Cong, H., Benitez, E. R., Takahashi, R., and Xu, D. H. (2011). Identification and validation of a major QTL for salt tolerance in soybean. Euphytica 179, 451–459. doi: 10.1007/s10681-011-0347-8
Han, J., Shi, J., Zeng, L., Xu, J., and Wu, L. (2015). Effects of nitrogen fertilization on the acidity and salinity of greenhouse soils. Environ. Sci. Pollut. Res. Int. 22, 2976–2986. doi: 10.1007/s11356-014-3542-z
Han, X., Feng, Z., Xing, D., Yang, Q., Wang, R., Qi, L., et al. (2015). Two NAC transcription factors from Caragana intermedia altered salt tolerance of the transgenic Arabidopsis. BMC Plant Biol. 15:208. doi: 10.1186/s12870-015-0591-5
Hatzig, S. V., Frisch, M., Breuer, F., Nesi, N., Ducournau, S., Wagner, M. H., et al. (2015). Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front. Plant Sci. 6:221. doi: 10.3389/fpls.2015.00221
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Hu, R., Zhu, Y., Wei, J., Chen, J., Shi, H., Shen, G., et al. (2017). Overexpression of PP2A-C5 that encodes the catalytic subunit 5 of protein phosphatase 2A in Arabidopsis confers better root and shoot development under salt conditions. Plant Cell Environ. 40, 150–164. doi: 10.1111/pce.12837
Hu, T., Zhu, S., Tan, L., Qi, W., He, S., and Wang, G. (2016). Overexpression of OsLEA4 enhances drought, high salt and heavy metal stress tolerance in transgenic rice (Oryza sativa L.). Environ. Exp. Bot. 123, 68–77. doi: 10.1016/j.envexpbot.2015.10.002
Huang, C., Nie, X., Shen, C., You, C., Li, W., Zhao, W., et al. (2017). Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol. J. 15, 1374–1386. doi: 10.1111/pbi.12722
Huang, X., Wei, X., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Huang, Z. B., Sun, Z. J., and Lu, Z. H. (2013). Effects of soil amendments on coastal saline-alkali soil improvement and the growth of plants. Adv. Mater. Res. 634-638, 152–159. doi: 10.4028/www.scientific.net/AMR.634-638.152
Hulse-Kemp, A. M., Lemm, J., Plieske, J., Ashrafi, H., Buyyarapu, R., Fang, D. D., et al. (2015). Development of a 63K SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3 5, 1187–1209. doi: 10.1534/g3.115.018416
Hwang, J. H., Seo, D. H., Kang, B. G., Kwak, J. M., and Kim, W. T. (2015). Suppression of Arabidopsis AtPUB30 resulted in increased tolerance to salt stress during germination. Plant Cell Rep. 34, 277–289. doi: 10.1007/s00299-014-1706-4
Jia, Y., Sun, J., Wang, X., Zhou, Z., Pan, Z., He, S., et al. (2014a). Molecular diversity and association analysis of drought and salt tolerance in Gossypium hirsutum L. germplasm. J. Integr. Agric. 13, 1845–1853. doi: 10.1016/s2095-3119(13)60668-1
Jia, Y., Sun, X., Sun, J., Pan, Z., Wang, X., He, S., et al. (2014b). Association mapping for epistasis and environmental interaction of yield traits in 323 cotton cultivars under 9 different environments. PLoS One 9:e95882. doi: 10.1371/journal.pone.0095882
Jin, X., Sun, T., Wang, X., Su, P., Ma, J., He, G., et al. (2016). Wheat CBL-interacting protein kinase 25 negatively regulates salt tolerance in transgenic wheat. Sci. Rep. 6:28884. doi: 10.1038/srep28884
Kong, D., Li, M., Dong, Z., Ji, H., and Li, X. (2015). Identification of TaWD40D, a wheat WD40 repeat-containing protein that is associated with plant tolerance to abiotic stresses. Plant Cell Rep. 34, 395–410. doi: 10.1007/s00299-014-1717-1
Le, R. D., Strom, A. R., Dandekar, A. M., Smith, L. T., and Valentine, R. C. (1984). Molecular biology of osmoregulation. Science 224, 1064–1068. doi: 10.1126/science.224.4653.1064
Li, T., Ma, X., Li, N., Zhou, L., Liu, Z., Han, H., et al. (2017). Genome-wide association study discovered candidate genes of Verticillium wilt resistance in upland cotton (Gossypium hirsutum L.). Plant Biotechnol. J. 15, 1520–1532. doi: 10.1111/pbi.12734
Li, Y., Pang, H., Zhang, H., and Chen, F. (2008). Effect of irrigation management on soil salinization in Manas River Valley, Xinjiang, China. Front. Agric. China 2, 216–223. doi: 10.1007/s11703-008-0028-0
Maathuis, F. J. M., and Amtmann, A. (1999). K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann. Bot. 84, 123–133. doi: 10.1006/anbo.1999.0912
Mano, Y., and Takeda, K. (1997). Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94, 263–272. doi: 10.1023/A:1002968207362
McCouch, S. R., Wright, M. H., Tung, C. W., Maron, L. G., Mcnally, K. L., Fitzgerald, M., et al. (2016). Open access resources for genome-wide association mapping in rice. Nat. Commun. 7:10532. doi: 10.1038/ncomms10532
Mei, H., Zhu, X., and Zhang, T. (2013). Favorable QTL alleles for yield and its components identified by association mapping in Chinese Upland cotton cultivars. PLoS One 8:e82193. doi: 10.1371/journal.pone.0082193
Munns, R., James, R. A., Xu, B., Athman, A., Conn, S. J., Jordans, C., et al. (2012). Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 30, 360–364. doi: 10.1038/nbt.2120
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nie, X., Huang, C., You, C., Li, W., Zhao, W., Shen, C., et al. (2016). Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genomics 17:352. doi: 10.1186/s12864-016-2662-x
Oluoch, G., Zheng, J., Wang, X., Khan, M. K. R., Zhou, Z., Cai, X., et al. (2016). QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum. Euphytica 209, 223–235. doi: 10.1007/s10681-016-1674-6
Oyiga, B. C., Sharma, R. C., Baum, M., Ogbonnaya, F. C., Léon, J., and Ballvora, A. (2017). Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ. 41, 919–935. doi: 10.1111/pce
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qin, H., Chen, M., Yi, X., Bie, S., Zhang, C., Zhang, Y., et al. (2015). Identification of associated SSR markers for yield component and fiber quality traits based on frame map and Upland cotton collections. PLoS One 10:e0118073. doi: 10.1371/journal.pone.0118073
Rai, M. K., Kalia, R. K., Singh, R., Gangola, M. P., and Dhawan, A. K. (2011). Developing stress tolerant plants through in vitro selection—An overview of the recent progress. Environ. Exp. Bot. 71, 89–98. doi: 10.1016/j.envexpbot.2010.10.021
Said, J. I., Song, M., Wang, H., Lin, Z., Zhang, X., Fang, D. D., et al. (2015). A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G. hirsutum x G. barbadense populations. Mol. Genet. Genomics 290, 1003–1025. doi: 10.1007/s00438-014-0963-9
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Su, J., Fan, S., Li, L., Wei, H., Wang, C., Wang, H., et al. (2016a). Detection of favorable QTL alleles and candidate genes for lint percentage by gwas in Chinese upland cotton. Front. Plant Sci. 7:1576. doi: 10.3389/fpls.2016.01576
Su, J., Li, L., Pang, C., Wei, H., Wang, C., Song, M., et al. (2016b). Two genomic regions associated with fiber quality traits in Chinese upland cotton under apparent breeding selection. Sci. Rep. 6:38496. doi: 10.1038/srep38496
Su, J., Pang, C., Wei, H., Li, L., Liang, B., Wang, C., et al. (2016c). Identification of favorable SNP alleles and candidate genes for traits related to early maturity via GWAS in upland cotton. BMC Genomics 17:687. doi: 10.1186/s12864-016-2875-z
Sun, Z., Wang, X., Liu, Z., Gu, Q., Zhang, Y., Li, Z., et al. (2017). Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol. J. 15, 982–996. doi: 10.1111/pbi.12693
Tester, M., and Davenport, R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91, 503–527. doi: 10.1093/aob/mcg058
Tiwari, R. S., Picchioni, G. A., Steiner, R. L., Jones, D. C., Hughs, S. E., and Zhang, J. (2013). Genetic variation in salt tolerance at the seedling stage in an interspecific backcross inbred line population of cultivated tetraploid cotton. Euphytica 194, 1–11. doi: 10.1007/s10681-013-0927-x
Tripathi, V., Parasuraman, B., Laxmi, A., and Chattopadhyay, D. (2009). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58, 778–790. doi: 10.1111/j.1365-313X.2009.03812.x
Wang, B., Ho, T. H. D., and Wu, R. (1996). Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 110, 249–257. doi: 10.1104/pp.110.1.249
Wang, J., Wang, D., Fan, W., Song, G., Wang, S., and Ye, W. (2011). The characters of salt-tolerance at different growth stages in cotton. Acta Ecol. Sin. 31, 3720–3727.
Wang, M., Tu, L., Lin, M., Lin, Z., Wang, P., Yang, Q., et al. (2017). Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet. 49, 579–587. doi: 10.1038/ng.3807
Wang, Z., Cheng, J., Chen, Z., Huang, J., Bao, Y., Wang, J., et al. (2012). Identification of QTLs with main, epistatic and QTL × environment interaction effects for salt tolerance in rice seedlings under different salinity conditions. Theor. Appl. Genet. 125, 807–815. doi: 10.1007/s00122-012-1873-z
Weigel, D. (2012). Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol. 158, 2–22. doi: 10.1104/pp.111.189845
Wu, C. A., Yang, G. D., Meng, Q. W., and Zheng, C. C. (2004). The cotton GhNHX1 gene encoding a novel putative tonoplast Na+/H+ antiporter plays an important role in salt stress. Plant Cell Physiol. 45, 600–607. doi: 10.1093/pcp/pch071
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322. doi: 10.1093/nar/gkr483
Xu, Y. F., An, D. G., Liu, D. C., Zhang, A. M., Xu, H. X., and Li, B. (2012). Mapping QTLs with epistatic effects and QTL × treatment interactions for salt tolerance at seedling stage of wheat. Euphytica 186, 233–245. doi: 10.1007/s10681-012-0647-7
Xue, Y., Warburton, M. L., Sawkins, M., Zhang, X., Setter, T., Xu, Y., et al. (2013). Genome-wide association analysis for nine agronomic traits in maize under well-watered and water-stressed conditions. Theor. Appl. Genet. 126, 2587–2596. doi: 10.1007/s00122-013-2158-x
Yee, D., and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60, 1109–1121. doi: 10.1093/jxb/ern369
Zhang, H., Zhu, H., Pan, Y., Yu, Y., Luan, S., and Li, L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532. doi: 10.1093/mp/ssu063
Zhang, J., Song, Q., Cregan, P. B., Nelson, R. L., Wang, X., Wu, J., et al. (2015a). Genome-wide association study for flowering time, maturity dates and plant height in early maturing soybean (Glycine max) germplasm. BMC Genomics 16:217. doi: 10.1186/s12864-015-1441-4
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015b). Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Keywords: cotton, salt tolerance, seedling, genome-wide association study, SNP, candidate genes
Citation: Sun Z, Li H, Zhang Y, Li Z, Ke H, Wu L, Zhang G, Wang X and Ma Z (2018) Identification of SNPs and Candidate Genes Associated With Salt Tolerance at the Seedling Stage in Cotton (Gossypium hirsutum L.). Front. Plant Sci. 9:1011. doi: 10.3389/fpls.2018.01011
Received: 31 August 2017; Accepted: 21 June 2018;
Published: 11 July 2018.
Edited by:
Hernâni Gerós, University of Minho, PortugalReviewed by:
Mehboob-ur- Rahman, National Institute for Biotechnology and Genetic Engineering, PakistanCopyright © 2018 Sun, Li, Zhang, Li, Ke, Wu, Zhang, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xingfen Wang, Y290dG9uQGhlYmF1LmVkdS5jbg== Zhiying Ma, bXpoeUBoZWJhdS5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.