
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 11 July 2018
Sec. Plant Nutrition
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00922
This article is part of the Research Topic Plant Adaptations to Phosphate Deficiency View all 13 articles
 Mostafa Abdelrahman1,2
Mostafa Abdelrahman1,2 Magdi A. El-Sayed2
Magdi A. El-Sayed2 Abeer Hashem3,4
Abeer Hashem3,4 Elsayed Fathi Abd_Allah5†
Elsayed Fathi Abd_Allah5† Abdulaziz A. Alqarawi5
Abdulaziz A. Alqarawi5 David J. Burritt6
David J. Burritt6 Lam-Son Phan Tran7,8*
Lam-Son Phan Tran7,8*Phosphate (Pi) deficiency is a critical environmental constraint that affects the growth and development of several legume crops that are usually cultivated in semi-arid regions and marginal areas. Pi deficiency is known to be a significant limitation for symbiotic nitrogen (N2) fixation (SNF), and variability in SNF is strongly interlinked with the concentrations of Pi in the nodules. To deal with Pi deficiency, plants trigger various adaptive responses, including the induction and secretion of acid phosphatases, maintenance of Pi homeostasis in nodules and other organs, and improvement of oxygen (O2) consumption per unit of nodule mass. These molecular and physiological responses can be observed in terms of changes in growth, photosynthesis, and respiration. In this mini review, we provide a brief introduction to the problem of Pi deficiency in legume crops. We then summarize the current understanding of how Pi deficiency is regulated in legumes by changes in the transcriptomes and metabolomes found in different plant organs. Finally, we will provide perspectives on future directions for research in this field.
Phosphorus(P) is a crucial element required for plant growth and development, playing a pivotal role in a diverse array of cellular processes, including photosynthesis, energy production, redox reactions, symbiotic nitrogen (N2) fixation (SNF), and carbohydrate metabolism (Sulieman and Tran, 2015; Kleinert et al., 2017). However, more than 30–40% of the world’s arable soils have low P contents, and by 2050 rock phosphate (Pi) reserves, the most inexpensive form of P for fertilization of agricultural soils, may be exhausted (Vance et al., 2003; Herrera-Estrella and López-Arredondo, 2016; Kleinert et al., 2017). Uptake of Pi from some soils can be problematic for plants due to slow Pi diffusion rates and the formation of insoluble Pi complexes with cations, especially iron and aluminum in acid weathered soils (Valentine et al., 2010; Castro-Guerrero et al., 2016). In several cropping systems, Pi-containing fertilizers are applied frequently to soils to enhance Pi availability, and thus yield. With the increasing demand for food, Pi fertilizer demand has increased four- to five-fold in last few decades, and is expected to continue increasing1 (Figure 1). This fact combined with a significant increase in Pi fertilizer production will add further pressure on the limited Pi reserve in the coming years (Figure 1). The use of Pi-containing fertilizers is a short-term solution to a much greater problem, as the real challenge for scientists and farmers is to deliver food with high nutritional quality using sustainable agricultural practices (Castro-Guerrero et al., 2016).
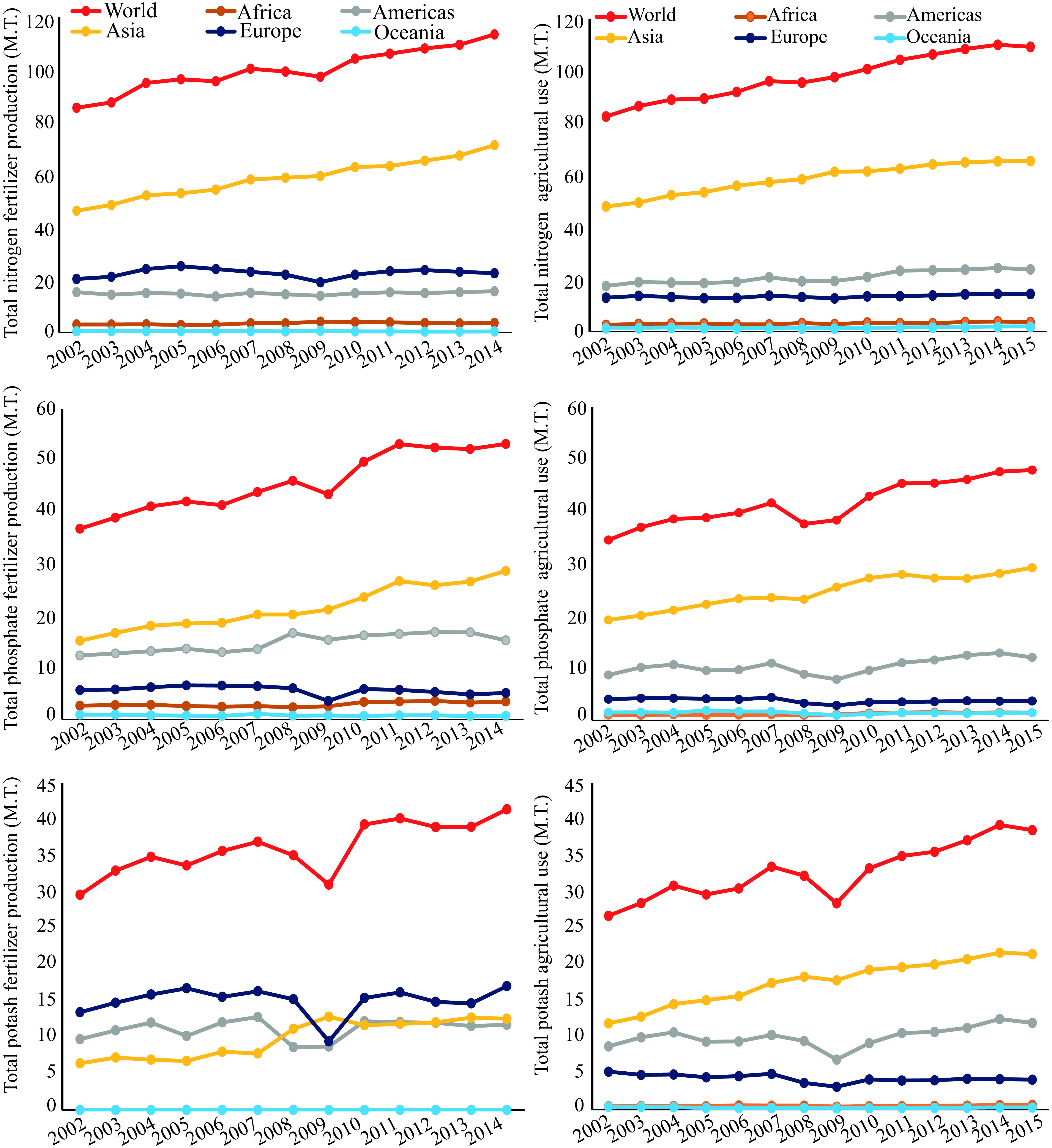
FIGURE 1. World nitrogen, phosphate (P2O5), and potash (K2O) fertilizer production from 2002 to 2014, and their agricultural use from 2002 to 2015 in millions of tonnes (M.T.), according to FAO (www.fao.org, accessed 2018).
Grain legumes are an essential source of nutrition and income for a large number of consumers and farmers worldwide (Kleinert et al., 2017; Abdelrahman et al., 2018). Legumes can create symbiotic relationships with N2-fixing rhizobia and arbuscular mycorrhizal fungi that facilitate the acquisition of nutrients; and thus, reduce the use of synthetic fertilizers, which is advantageous for sustainable agriculture (Considine et al., 2017; Valliyodan et al., 2017). SNF is an energetically expensive process, consuming ∼20 adenosine triphosphate (ATP) molecules for the production of two NH3 molecules (Thuynsma et al., 2014). Because of the requirement for large amounts of ATP for SNF, Pi deficiency is a critical constraint for efficient SNF in legumes. There is substantial evidence demonstrating that Pi deficiency can more severely affect the N:P ratio in legume tissues when compared with non-leguminous crops (Sulieman and Tran, 2015; Guo et al., 2016). Enhanced nutrient acquisition by SNF nodules formed by plants of Pi-deficient soils is crucial for the efficient fixation of N2 (Magadlela et al., 2015; Lazali et al., 2017). Legumes have evolved conserved acquisition and internal transport strategies for Pi detected in Pi-deficient soils in order to maintain nodule Pi-homeostasis and enable efficient SNF (Figure 2). These include decreased plant growth rates, modification of carbon metabolism, increased secretion of organic anions and phosphatases, changes in root architecture, expansion of root surface areas, and enhanced expression of Pi transporters (Sulieman and Tran, 2015; Considine et al., 2017; Kleinert et al., 2017; Uhde-Stone, 2017). Because of the diminishing reserves of inexpensive Pi fertilizers, plant acclimation to Pi deficiency has become a topic of considerable interest to plant researchers. Below we present recent advances in the transcriptomic and metabolomic changes that occur in legumes in response to Pi deficiency, which is essential if we are to understand the complex systemic metabolic mechanisms plants use to adapt to Pi deficiency.
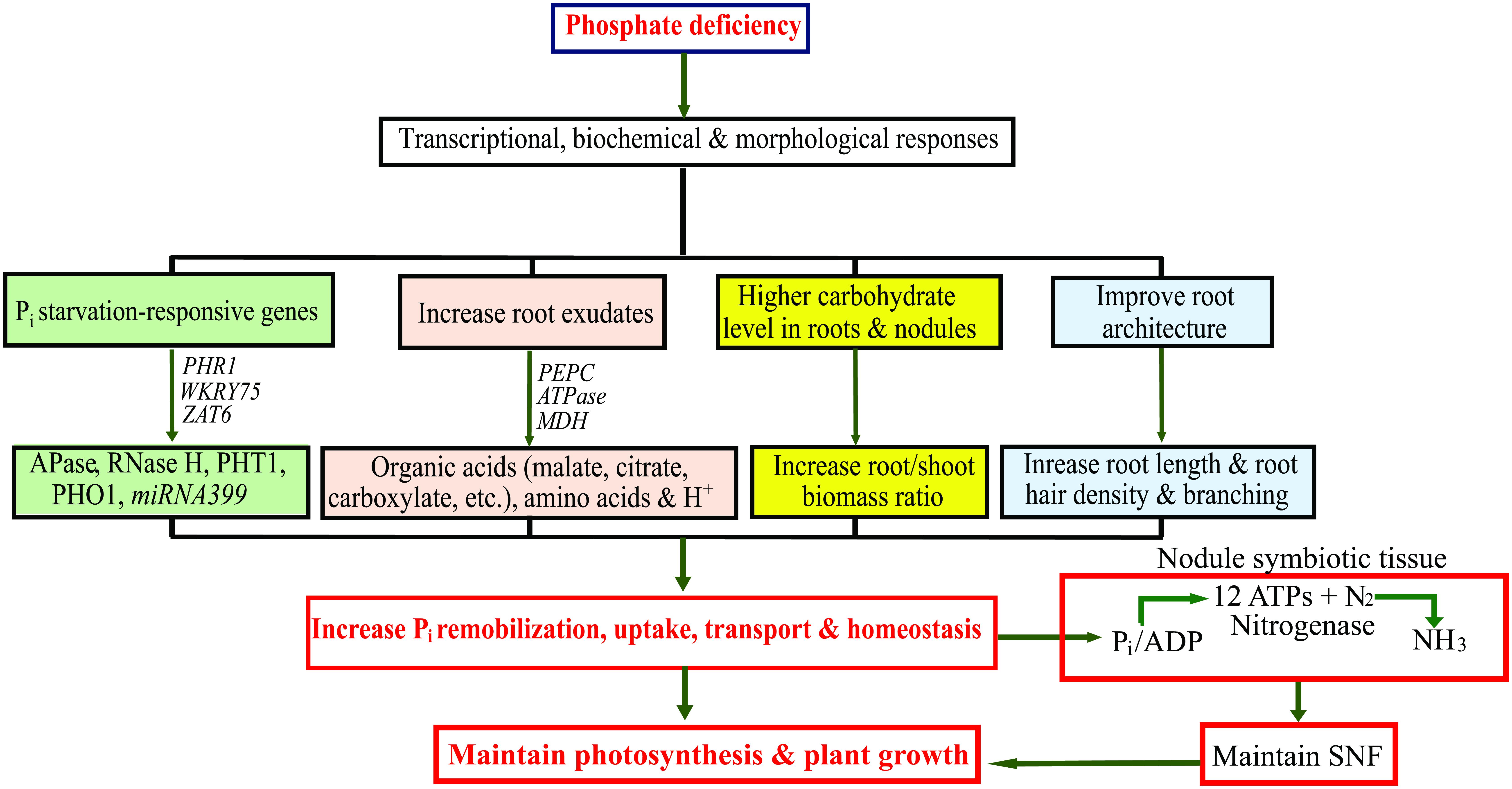
FIGURE 2. Schematic representation of transcriptional, biochemical and morphological changes in legumes under phosphate (Pi) deficiency to increase Pi uptake and optimize Pi use efficiency. APase, ACID PHOSPHATASE; ATPase, Adenosine Triphosphatase; MDH, MALATE DEHYDROGENASE; PEPC, PHOSHOENOLPYRUVATE CARBOXYLASE; PHO1, PHOSPHATE TRANSPORTER 1; PHR1, PHOSPHATE STARVATION RESPONSIVE 1; PHT1, HIGH-AFFINITY PHOSPHATE TRANSPORTER 1; RNase H, RIBONUCLEASE H; SNF, symbiotic nitrogen fixation; ZAT6, ZINC FINGER OF ARABIDOPSIS 6.
Next-generation sequencing (NGS) technologies have become essential tools to help understand the regulation of gene expression and the molecular basis of cellular responses that occur in plants exposed to biotic and/or abiotic stressors (Abdelrahman et al., 2015, 2017a,b; Miao et al., 2015; Liese et al., 2017; Nasr Esfahani et al., 2017). A number of transcriptome studies of leguminous plant species, including white lupin (Lupinus albus), common bean (Phaseolus vulgaris), soybean (Glycine max), chickpea (Cicer arietinum), and Medicago truncatula, grown under Pi deficiency have been conducted in the last several years (Hernández et al., 2007; O’Rourke et al., 2013; Liese et al., 2017; Nasr Esfahani et al., 2017; Zhang et al., 2017). RNAseq-based transcriptome profiling of nodules of Sinorhizobium meliloti-inoculated M. truncatula plants grown under Pi deficiency has shown a strong down-regulation in the expression of genes encoding NODULE-SPECIFIC CYSTEINE-RICH peptides, LEGHEMOGLOBIN and NICOTIANAMINE SYNTHASE-LIKE PROTEIN, compared with nodules of control plants grown under Pi-replete conditions (Liese et al., 2017). The down-regulation of these genes disturbs normal cellular iron distribution, restricts the supply of oxygen for respiration and eventually lowers nitrogenase activity in nodules (Liese et al., 2017). This potential disruption of normal nodule metabolism caused by Pi deficiency greatly reduces SNF efficiency in legumes (Liese et al., 2017). In addition, a reduction in shoot and nodule dry matter, and tissue Pi levels was observed in Pi-deficient M. truncatula plants relative to Pi-replete control plants, indicating that Pi deficiency can severely limit legume growth and potential crop yields (Liese et al., 2017). However, while S. meliloti-inoculated M. truncatula plants grown under Pi deficiency had much lower stem and root tissue Pi concentrations compared with control (Pi-sufficient) plants, nodule Pi levels in Pi-deficient plants were maintained at relatively high levels and did not show the considerable loss of Pi as observed for stems and roots (Liese et al., 2017). Nasr Esfahani et al. (2017) examined transcriptome changes in the nodules of Pi-deficiency-more-susceptible Mesorhizobium mediterraneum SWRI9-(MmSWRI9)-chickpea and Pi-deficiency-less-susceptible M. ciceri CP-31-(McCP-31)-chickpea associations under Pi-deficient and -sufficient conditions. The transcriptome profiles of these interactions showed that many genes related to several key cellular processes and metabolic pathways namely transcriptional regulation, detoxification, nodulation, ion/nutrient transport, and Pi signaling and remobilization were differentially expressed in MmSWRI9-induced nodules relative to McCP-31-induced nodules (Nasr Esfahani et al., 2017). Changes in the expression of Pi starvation-related genes are likely to help improve acquisition and transport of Pi in the MmSWRI9-chickpea association; and thus maintenance of the sufficient SNF capacity under Pi-deficient conditions (Nasr Esfahani et al., 2017). The above observations indicated that changes in legume transcriptomes under Pi starvation are mostly associated with facilitating Pi solubilization, acquisition and transportation into nodules, which are significant sinks for Pi, contributing to more efficient SNF and therefore higher plant productivity.
Although most studies to date have focused on nodule transcriptomes, exploring the transcriptomes of other plant tissues is also important, as these may provide valuable insight into Pi deficiency acclimation mechanisms in legumes. For example, transcriptome analysis of leaves and roots (combined cluster and normal roots) of white lupine plants grown under Pi-deficient or -sufficient conditions identified 1,342 and 904 differentially expressed genes, respectively, in response to Pi deficiency (O’Rourke et al., 2013). In leaves, the most highly expressed transcripts were involved in amino acid metabolism, tetrapyrrole synthesis, photosynthesis, carbohydrate catabolism, and flavonoid biosynthesis; whereas in roots the most highly expressed transcripts were involved in sugar/nutrient signaling and transport, lignin biosynthesis, phospholipid and carbohydrate metabolism, and amino acid synthesis. Interestingly, 12 transcripts identified in the above study were commonly induced by low-Pi stress across three species, white lupine, Arabidopsis, and potato (Solanum tuberosum), making them excellent candidates to investigate responses to Pi starvation in plants (O’Rourke et al., 2013). Among these 12 transcripts, three transcripts that were highly up-regulated in Pi-deficient lupine plants compared with Pi-sufficient ones (O’Rourke et al., 2013) encode SPX domain-containing proteins [RECOMBINANT Saccharomyces cerevisiae PROTEIN (SYG1)/PHOSPHATASE(PHO81)/XENOTROPIC and POLYTROPICRETROVIRUS RECEPTOR 1(XPR1)] that are essential regulators involved in Pi homeostasis and signaling responses to Pi deficiency (Chiou and Lin, 2011; Secco et al., 2012). In addition, Pi solubilization- and transport-related genes, encoding PHOSPHATE TRANSPORTER 1, PHOSPHOLIPASE, PYROPHOSPHATASE, PURPLE ACID PHOSPHATASE, and MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE, were also up-regulated in lupine Pi-deficient relative to Pi-sufficient plants (O’Rourke et al., 2013). A recent study by Zhang et al. (2017) provided transcriptome datasets obtained from the roots and leaves of soybean plants grown under Pi-deficient and -sufficient conditions, and showed a significant role for the acid phosphatase-encoding gene GmACP1 in regulating Pi use efficiency in soybean. These results were in agreement with a previous finding by the same research group (Zhang et al., 2014), who used a genome-wide association study of 192 soybean accessions to identify a quantitative trait locus (QTL) on soybean chromosome 8, namely qPE8, which was associated with improvement of soybean Pi use efficiency under Pi starvation. This qPE8 QTL contained the candidate genes Glyma08g20700, Glyma08g20710, and Glyma08g20800, Glyma08g20820/GmACP1 and Glyma08g20830, which encode CALCINEURIN B, PHOSPHOLIPASE D and putative PHOSPHATASES, respectively. Glyma08g20820/GmACP1 was up-regulated under Pi deficiency; however, the transcript levels of the remaining genes were not changed (Zhang et al., 2014). In addition, overexpression study of GmACP1 using hairy-root transformation showed that the transgenic hairy roots displayed a 2.3-fold increase in acid phosphatase activity and an 11.2–20.0% higher Pi use efficiency relative to wild-type plants under Pi starvation (Zhang et al., 2014).
Transcriptome analysis of wild legumes is also critical to aid in understanding the differences that exist between domesticated legumes and their wild progenitors (Abdelrahman et al., 2018). This could also help provide a better understanding of the Pi stress adaptation mechanisms present in wild legumes. In addition, transcriptome correlation analyses between different legume species under Pi deficiency may provide crucial information about the conserved Pi deficiency-responsive genes, which could be used as molecular markers for screening for low Pi- tolerant/susceptible cultivars or genetic engineering to enhance the growth and productivity of crop plants grown on low Pi soils.
Plants adapt to Pi starvation by an array of molecular responses in which transcription factors (TFs) are key components in the regulation of these processes (Jain et al., 2012). The transcriptional regulations of the Pi starvation responses have been extensively studied in other plant species; however, these important processes have not much investigated yet in legumes. Four TF-encoding genes BASIC HELIX-LOOP-HELIX 32 (BHLH32), WRKY75, PHOSPHATE STARVATION RESPONSIVE 1 (PHR1), and ZINC FINGER OF ARABIDOPSIS 6 (ZAT6) involved in Pi starvation signaling have been identified in Arabidopsis (Valdés-López and Hernández, 2008). AtPHR1 and its orthologs from rice (Oryza sativa, OsPHR1 and OsPHR2) are regarded as the key positive regulators controlling plant transcriptional responses to Pi deficiency (Rubio et al., 2001; Zhou et al., 2008). The overexpression of AtPHR1 induced Pi-responsive genes involved in Pi remobilization (ACID PHOSPHATASES and RIBONUCLEASE H), transport [HIGH-AFFINITY PHOSPHATE TRANSPORTER 1 (PHT1), PHOSPHATE TRANSPORTER 1(PHO1)] and homeostasis (miRNA399 and At4), in addition to genes involved in anthocyanin biosynthesis (Valdés-López and Hernández, 2008). Hernández et al. (2007) reported 17 TF-encoding genes differentially expressed in common bean roots under Pi deficiency. Of these genes, TC2883 MYB gene was highly induced under Pi starvation and exhibited 63% homology to AtPHR1, suggesting an important role of PHR1 in common bean response to Pi deficiency. Likewise, the Arabidopsis At4 plays a significant role in translocation of Pi from roots to shoots, and its ortholog from M. truncatula, the Mt4, showed strongly induced expression in roots under Pi deficiency (Valdés-López and Hernández, 2008). WKRY75 and ZAT6 are also up-regulated under Pi starvation, and both two genes are implicated in Pi remobilization, transport, and homeostasis as well as root architecture. In contrast, BHLH32 is down-regulated under Pi deficiency, and its role in modification of root architecture has been proposed (Chen et al., 2007).
Overexpression of the rice osa-miR827 and Arabidopsis miR399/miRNA399 that target the SPX-MAJOR FACILITATOR SUPERFAMILY (MFS) protein-encoding genes and the Pi transporter genes, respectively, drastically impacts Pi homeostasis and accumulation in transgenic plants (Wang et al., 2012; Chen et al., 2017). During Pi starvation, miR399/miRNA399 supresses its target gene PHO2 and allows sufficient transcript of PHT1 accumulated in the membrane of Pi-starved transgenic plants, thereby increasing Pi acquisition (Franco-Zorrilla et al., 2007). Recently, Chen et al. (2017) demonstrated a crucial role of the TamiR167a in mediating tobacco (Nicotiana tabacum) growth and adaptation to Pi starvation via regulation of various biological processes, including Pi acquisition and reactive oxygen species homeostasis. Thus, distinct miRNAs are also important regulators in mediating the plant response to Pi stress as well.
The development of crop plants that are able to produce good yield on nutrient-deficient soils requires an in-depth knowledge of physiological and biochemical processes that allow plants to survive under these stressful conditions. Integrated transcriptomic and metabolomic studies can aid in obtaining this knowledge (Hirai et al., 2004; Last et al., 2007; Hernández et al., 2009; Saito, 2013; Abdelrahman et al., 2014, 2015, 2017c,d; Jin et al., 2017). Plant metabolites are synthesized by numerous proteins/enzymes encoded in the plant genome, and integration of gene expression atlas with targeted/non-targeted metabolite profile is an innovative approach to identify gene-to-metabolite associations/networks (Hirai et al., 2004; Saito, 2013; Abdelrahman et al., 2017d). The use of metabolic profiling has been quite limited for legume crops, but this approach has recently been applied to help understand the metabolic changes associated with legume-rhizobial symbiosis. Symbiotic N2-fixing bacteria secrete lipo-chitooligosaccharide signaling molecules, also known as Nod factors, upon perception of isoflavonoids and flavonoids secreted by legume roots (Zhang et al., 2012). The Nod factors are perceived by their receptors on the plasma membranes of root cells of leguminous plants, which then activate signaling processes within the nucleus and cytoplasm of target cells (Zhang et al., 2012). Untargeted metabolite profiling of the extracts of M. truncatula seedlings treated with rhizobial lipo-chitooligosaccharide molecules has shown a significant decrease in oxylipin-related compounds in M. truncatula. Oxylipins are precursors of the jasmonic acid biosynthesis pathway, and both oxylipins and jasmonic acids inhibit Nod factor signaling, suggesting that these oxylipin-related compounds act as negative regulators of the early stages of symbiosis (Zhang et al., 2012).
In an early study, Hernández et al. (2009) used integrated non-targeted metabolite profiling and transcriptome analysis to identify changes in the roots and nodules of common bean plants inoculated with Rhizobium tropici and grown under Pi-deficient and -sufficient conditions. They showed clear metabolic differences between plants grown under these two contrasting conditions. Integrative analysis of nodule transcriptome and metabolome allowed the authors to identify 13 metabolites that could be assigned to repressed or induced pathways in response to Pi deficiency. Of these 13 Pi starvation-responsive metabolites, a reduction in N metabolism-related metabolites, including spermidine, putrescine, urea, glycine, serine, glutamine, and threonine, was detected in nodules of Pi-deficient common bean plants relative to that of Pi-sufficient ones, and this change might contribute to a decrease in SNF efficiency (Hernández et al., 2009). In addition, Pi-deficient common bean roots show reduced levels of organic acids like tartaric acid and 2,4-dihydroxybutanoic acid, due to the secretion of these organic acids into the rhizosphere (Hernández et al., 2009). In contrast, alteration of carbon (C) metabolism in Pi-deficient common bean results in lower and higher carbohydrate levels in the shoots and roots, respectively, thereby contributing to the increased root/shoot biomass ratio and altering root morphology (Hernández et al., 2009). Similarly, Nasr Esfahani et al. (2016) showed lower SNF efficiency and decreased Pi level in the Pi-deficiency-more-susceptible MmSWRI9-chickpea nodules than the Pi-deficiency-less-susceptible McCP-31-chickpea nodules under Pi deficiency, which was evident by significant differences in C and N metabolism-related metabolites. For example, in McCP-31-inoculated plants, Pi deficiency increased total level of identified sugars by 68.8%, whereas that remained unchanged in MmSWRI9-induced nodules (Nasr Esfahani et al., 2016). In addition, Pi deficiency induced a remarkable increase in total level of organic acids in McCP-31-nodulated roots, whereas it decreased that in MmSWRI9-nodulated roots (Nasr Esfahani et al., 2016). These results revealed the existence of crosstalk among various signaling pathways involved in regulation of Mesorhizobium-chickpea adaptation to Pi deficiency, in-depth understanding of which at genetic level will be useful for genetic engineering of chickpea cultivars and other leguminous crops that can sustain efficient SNF under Pi deficiency. C and N metabolism is essential for SNF, and is a significant determinant of plant and nodule responses to Pi starvation (Kleinert et al., 2017). Some studies have shown that even under Pi deficiency, plant nodules continue to act as very strong nutrient sinks for C in order to maintain SNF, and underground biomass often continues to increase even at the expense of whole plant growth (Thuynsma et al., 2014; Magadlela et al., 2015). Interestingly, for white lupine plants grown under Pi-deficient and -sufficient conditions, no significant differences in the above and below ground biomass between Pi-deficient and -sufficient plants were observed, nor were any large differences in resource allocation (N and Pi) between the shoot and root/nodule systems (Thuynsma et al., 2014). However, white lupine plants produced more cluster root biomass, up to 24% of the root system under Pi deficiency; relative to approximately 5% increase of the root system with sufficient Pi supply. In contrast, less nodule biomass (up to 14% of the root system) was detected under Pi deficiency than (up to 20% of the root system) sufficient Pi supply. In addition, cluster roots exhibited a significant increase in Pi acquisition rates under deficient Pi than sufficient Pi conditions (Thuynsma et al., 2014). These results suggest that underground adaptations, rather than large changes in shoot/root biomass ratio, may underpin the ability of lupine plants to grow well on Pi-deficient soils, as more cluster root biomass would result in an improved Pi uptake rate; and hence help maintain high Pi level in the nodules, consequently efficient SNF under Pi deficiency (Thuynsma et al., 2014).
A recent metabolite profiling study of common bean root exudates grown in liquid culture media supplemented with Pi concentrations ranged from 0 to 8 mg L-1 showed that the levels of some organic acids, nucleic acids, and amino acids were much higher in common bean root exudates under Pi-deficient conditions than Pi-sufficient ones (Tawaraya et al., 2014). On the other hand, levels of phosphate esters, including glucose-6-phosphate, fructose-6-phosphate, and fructose-1, 6-phosphate, were lower in Pi-deficient relative to Pi-sufficient conditions (Tawaraya et al., 2014). The increase in amino acid and organic acid levels in the root exudates changed the respiration rate and influenced microsymbiont community in the root nodules, improving SNF under Pi deficiency (Tawaraya et al., 2014). While relatively few metabolomic profiling studies of legume crops under Pi deficiency have been conducted to date, future studies on legumes using an integrated metabolomic-transcriptomic approach may provide valuable information on the metabolic reprogramming at molecular level, which is required by plants for better adaptation to Pi deficiency.
Legume crops are widely cultivated in many semi-arid and tropical parts of the world where Pi deficiency poses severe threats to crop productivity. To sustain legume cultivation under deficient Pi conditions, crop improvement programs require innovative methods, such as an integrated approach of transcriptomics and metabolomics to gain in-depth understanding of how plants respond to Pi deficiency at the molecular level. This mini review provides an overview of several transcriptomic and metabolomic studies conducted for legumes grown under Pi deficiency and their potential to help understand how legume crops respond to Pi deficiency (Figure 2). Transcriptomics and metabolomics have generated gigabyte-size data sets that require specialized computational software and bioinformatic tools to analyze them. To aid with this, several transcriptomic and metabolomic databases have been created; e.g., the MedicCyc for M. truncatula2, which includes more than 250 pathways with related metabolites, enzymes and associated genes. Another database, the Soybean Knowledge Base (SoyKB)3 has also been constructed. This resource is not only useful for soybean translational genomics, but also for legume proteomics and metabolomics. Identification of candidate genes and metabolic pathways important for the adaptation of legumes to Pi deficiency could be used in future for the marker-assisted selection of Pi-efficient genotypes. Characterizing the proteomes of legumes under Pi deficiency is also a significant task for the future. In addition, information generated from transcriptomics and metabolomics combined with information from other types of analyses, including reverse and forward genetic analyses, could lead to the long-elusive goal of improvement of N2 fixation in agronomically essential grain legumes grown under Pi deficiency.
MA and L-SPT conceived the idea. MA, ME-S, AH, EA_A, AA, DB, and L-SPT wrote the manuscript. All authors read and approved the final manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer AK and handling Editor declared their shared affiliation.
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding to the Research Group number (RGP-271).
Abdelrahman, M., Burritt, D. J., and Tran, L. P. (2017a). The use of metabolomic quantitative trait locus mapping and osmotic adjustment traits for the improvement of crop yields under environmental stresses. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2017.06.020 [Epub ahead of print].
Abdelrahman, M., El-Sayed, M., Jogaiah, S., Burritt, D. J., and Tran, L. P. (2017b). The “STAY-GREEN” trait and phytohormone signaling networks in plants under heat stress. Plant Cell Rep. 36, 1009–1025. doi: 10.1007/s00299-017-2119-y
Abdelrahman, M., El-Sayed, M., Sato, S., Hirakawa, H., Ito, S.-I., Tanaka, K., et al. (2017c). RNA-sequencing-based transcriptome and biochemical analyses of steroidal saponin pathway in a complete set of Allium fistulosum-A. cepa monosomic addition lines. PLoS One 12:e0181784. doi: 10.1371/journal.pone.0181784
Abdelrahman, M., Suzumura, N., Mitoma, M., Matsuo, S., Ikeuchi, T., Mori, M., et al. (2017d). Comparative de novo transcriptome profiles in Asparagus officinalis and A. kiusianus during the early stage of Phomopsis asparagi infection. Sci. Rep. 7:2608. doi: 10.1038/s41598-017-02566-7
Abdelrahman, M., Hirata, S., Ito, S.-I., Yamauchi, N., and Shigyo, M. (2014). Compartmentation and localization of bioactive metabolites in different organs of Allium roylei. Biosci. Biotechnol. Biochem. 78, 1112–1122. doi: 10.1080/09168451.2014.915722
Abdelrahman, M., Jogaiah, S., Burritt, D. J., and Tran, L.-P. (2018). Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. doi: 10.1111/pce.13123 [Epub ahead of print].
Abdelrahman, M., Sawada, Y., Nakabayashi, R., Sato, S., Hirakawa, H., El-Sayed, M., et al. (2015). Integrating transcriptome and target metabolome variability in doubled haploids of Allium cepa for abiotic stress protection. Mol. Breed. 35:195. doi: 10.1007/s11032-015-0378-2
Castro-Guerrero, N. A., Isidra-Arellano, M. C., Mendoza-Cozatl, D. G., and Valdés-López, O. (2016). Common bean: a legume model on the rise for unraveling responses and adaptations to iron, zinc, and phosphate deficiencies. Front. Plant Sci. 7:600. doi: 10.3389/fpls.2016.00600
Chen, X., Liu, Z., Shi, G., Bai, Q., Guo, C., and Xiao, K. (2017). MIR167a transcriptionally regulates ARF6 and ARF8 and mediates drastically plant Pi-starvation response via modulation of various biological processes. Plant Cell Tissue Org. Cult. 133, 177–191. doi: 10.1007/s11240-017-1371-8
Chen, Z. H., Nimmo, G. A., Jenkins, G., and Nimmo, H. G. (2007). BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 405, 191–198. doi: 10.1042/BJ20070102
Chiou, T.-J., and Lin, S.-I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Considine, M. J., Siddique, K. H. M., and Foyer, C. H. (2017). Nature’s pulse power: legumes, food security and climate change. J. Exp. Bot. 68, 1815–1818. doi: 10.1093/jxb/erx099
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Samoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Guo, Y., Yang, X., Schöb, C., Jiang, Y., and Tang, Z. (2016). Legume shrubs are more nitrogen-homeostatic than non-legume shrubs. Front. Plant Sci. 8:1662. doi: 10.3389/fpls.2017.01662
Hernández, G., Ramírez, M., Valdés-López, O., Tesfaye, M., Graham, M. A., Czechowski, T., et al. (2007). Phosphorus stress in common bean: root transcript and metabolic responses. Plant Physiol. 144, 752–767. doi: 10.1104/pp.107.096958
Hernández, G., Valdés-López, O., Ramírez, M., Goffard, N., Weiller, G., Aparicio-Fabre, R., et al. (2009). Global changes in the transcript and metabolic profiles during symbiotic nitrogen fixation in phosphorus-stressed common bean plants. Plant Physiol. 151, 1221–1238. doi: 10.1104/pp.109.143842
Herrera-Estrella, L., and López-Arredondo, D. L. (2016). Phosphorus: the underrated element for feeding the world. Trends Plant Sci. 21, 461–463. doi: 10.1016/j.tplants.2016.04.010
Hirai, M. Y., Yano, M., Goodenowe, D. B., Kanaya, S., Kimura, T., Awazuhara, M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 1010, 10205–10210. doi: 10.1073/pnas.0403218101
Jain, A., Nagarajan, V. K., and Raghothama, K. G. (2012). Transcriptional regulation of phosphate acquisition by higher plants. Cell. Mol. Life Sci. 69, 3207–3224. doi: 10.1007/s00018-012-1090-6
Jin, J., Zhang, H., Zhang, J., Liu, P., Chen, X., Li, Z., et al. (2017). Integrated transcriptomics and metabolomics analysis to characterize cold stress responses in Nicotiana tabacum. BMC Genomics 18:496. doi: 10.1186/s12864-017-3871-7
Kleinert, A., Thuynsma, R., Magadlela, A., Benedito, V. A., and Valentine, A. J. (2017). “Metabolism and transport of carbon in legume nodules under phosphorus deficiency,” in Legume Nitrogen Fixation in Soils with Low Phosphorus Availability, eds S. Sulieman and L. P. Tran (Cham: Springer International Publishing), 77–95.
Last, R. L., Jones, A. D., and Shachar-Hill, Y. (2007). Innovations: towards the plant metabolome and beyond. Nat. Rev. Mol. Cell Biol. 8, 167–174. doi: 10.1038/nrm2098
Lazali, M., Blavet, D., Pernot, C., Desclaux, D., and Drevon, J. J. (2017). Efficiency of phosphorus use for dinitrogen fixation varies between common bean genotypes under phosphorus limitation. Agron. J. 109, 283–290. doi: 10.2134/agronj2016.01.0034
Liese, R., Schulze, J., and Cabeza, R. A. (2017). Nitrate application or P deficiency induce a decline in Medicago truncatula N2-fixation by similar changes in the nodule transcriptome. Sci. Rep. 7:46264. doi: 10.1038/srep46264
Magadlela, A., Vardien, W., Kleinert, A., Dreyer, L. L., and Valentine, A. J. (2015). The role of phosphorus deficiency in nodule microbial composition, and carbon and nitrogen nutrition of a native legume tree in the Cape fynbos ecosystem. Aust. J. Bot. 63, 379–386. doi: 10.1071/BT14216
Miao, Z., Xu, W., Li, D., Hu, X., Liu, J., Zhang, R., et al. (2015). De novo transcriptome analysis of Medicago falcata reveals novel insights about the mechanisms underlying abiotic stress responsive pathway. BMC Genomics 16:818. doi: 10.1186/s12864-015-2019-x
Nasr Esfahani, M., Inoue, K., Chu, H. D., Nguyen, K. H., Ha, C. V., Watanabe, Y., et al. (2017). Comparative transcriptome analysis of nodules of two Mesorhizobium-chickpea associations with differential symbiotic efficiency under phosphate deficiency. Plant J. 95, 911–926. doi: 10.1111/tpj.13616
Nasr Esfahani, M., Kusano, M., Nguyen, K. H., Watanabe, Y., Ha, C. V., Saito, K., et al. (2016). Adaptation of the symbiotic Mesorhizobium–chickpea relationship to phosphate deficiency relies on reprogramming of whole-plant metabolism. Proc. Natl. Acad. Sci. U.S.A. 113, E4610–E4619. doi: 10.1073/pnas.1609440113
O’Rourke, J. A., Yang, S. S., Miller, S. S., Bucciarelli, B., Liu, J., Rydeen, A., et al. (2013). An RNA-Seq transcriptome analysis of orthophosphate-deficient white lupin reveals novel insights into phosphorus acclimation in plants. Plant Physiol. 161, 705–724. doi: 10.1104/pp.112.209254
Rubio, V., Linhares, F., Solano, R., Martın, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plant and unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Saito, K. (2013). Phytochemical genomics — a new trend. Curr. Opin. Plant Biol. 16, 373–380. doi: 10.1016/j.pbi.2013.04.001
Secco, D., Wang, C., Arpat, B. A., Wang, Z., Poirier, Y., Tyerman, S. D., et al. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193, 842–851. doi: 10.1111/j.1469-8137.2011.04002.x
Sulieman, S., and Tran, L. P. (2015). Phosphorus homeostasis in legume nodules as an adaptive strategy to phosphorus deficiency. Plant Sci. 239, 36–43. doi: 10.1016/j.plantsci.2015.06.018
Tawaraya, K., Horie, R., Saito, S., Wagatsuma, T., Saito, K., and Oikawa, A. (2014). Metabolite profiling of root exudates of common bean under phosphorus deficiency. Metabolites 4, 599–611. doi: 10.3390/metabo4030599
Thuynsma, R., Valentine, A., and Kleinert, A. (2014). Phosphorus deficiency affects the allocation of below-ground resources to combined cluster roots and nodules in Lupinus albus. J. Plant Physiol. 171, 285–291. doi: 10.1016/j.jplph.2013.09.001
Uhde-Stone, C. (2017). “White lupin: a model system for understanding plant adaptation to low phosphorus availability,” in Legume Nitrogen Fixation in Soils with Low Phosphorus Availability, eds S. Sulieman and L. P. Tran (Cham: Springer International Publishing), 77–95.
Valdés-López, O., and Hernández, G. (2008). Transcriptional regulation and signaling in phosphorus starvation: what about legumes? J. Integr. Plant Biol. 50, 1213–1222. doi: 10.1111/j.1744-7909.2008.00758.x
Valentine, A. J., Benedito, V. A., and Kang, Y. (2010). “Legume nitrogen fixation and soil abiotic stress: from physiology to genomics and beyond,” in Nitrogen Metabolism in Plants in the Post-Genomic Era: Annual Plant Reviews, Vol. 42, eds C. H. Foyer and H. Zhang (Oxford: Wiley-Blackwell).
Valliyodan, B., Ye, H., Song, L., Murphy, M., Shannon, J. G., and Nguyen, H. T. (2017). Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 68, 1835–1849. doi: 10.1093/jxb/erw433
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Wang, C., Huang, W., Ying, Y., Li, S., Secco, D., Tyerman, S., et al. (2012). Functional characterization of the rice SPX–MFS family reveals a key role of OsSPX–MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 196, 139–148. doi: 10.1111/j.1469-8137.2012.04227.x
Zhang, D., Song, H., Cheng, H., Hao, D., Wang, H., Kan, G., et al. (2014). The acid phosphatase-encoding gene GmACP1 contributes to soybean tolerance to low-phosphorus stress. PLoS Genet. 10:e1004061. doi: 10.1371/journal.pgen.1004061
Zhang, H., Chu, S., and Zhang, D. (2017). Transcriptome dataset of soybean (Glycine max) grown under phosphorus-deficient and -sufficient conditions. Data 2:17. doi: 10.3390/data2020017
Zhang, N., Venkateshwaran, M., Boersma, M., Harms, A., Howes-Podoll, M., den Os, D., et al. (2012). Metabolomic profiling reveals suppression of oxylipin biosynthesis during the early stages of legume–rhizobia symbiosis. FEBS Lett. 586, 3150–3158. doi: 10.1016/j.febslet.2012.06.046
Keywords: legumes, metabolomics, transcriptomics, phosphate deficiency, nitrogen fixation
Citation: Abdelrahman M, El -Sayed MA, Hashem A, Abd_Allah EF, Alqarawi AA, Burritt DJ and Tran L-SP (2018) Metabolomics and Transcriptomics in Legumes Under Phosphate Deficiency in Relation to Nitrogen Fixation by Root Nodules. Front. Plant Sci. 9:922. doi: 10.3389/fpls.2018.00922
Received: 22 January 2018; Accepted: 11 June 2018;
Published: 11 July 2018.
Edited by:
Alex Joseph Valentine, Stellenbosch University, South AfricaReviewed by:
Oswaldo Valdes-Lopez, Universidad Nacional Autónoma de México, MexicoCopyright © 2018 Abdelrahman, El-Sayed, Hashem, Abd_Allah, Alqarawi, Burritt and Tran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lam-Son Phan Tran, c29uLnRyYW5AcmlrZW4uanA=
†orcid.org/0000-0002-8509-8953
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.