
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 31 July 2018
Sec. Plant Development and EvoDevo
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00854
The C function gene AGAMOUS (AG) encodes for a MADS-box transcription factor required for floral organ identity and floral meristem (FM) determinacy in angiosperms. Unlike Arabidopsis, most legume plants possess two AG homologs arose by an ancient genome duplication event. Recently, two euAGAMOUS genes, MtAGa and MtAGb, were characterized and shown to fulfill the C function activity in the model legume Medicago truncatula. Here, we reported the isolation and characterization of a new mtaga allele by screening the Medicago Tnt1 insertion mutant collection. We found that MtAGa was not only required for controlling the stamen and carpel identity but also affected pod and seed development. Genetic analysis indicated that MtAGa and MtAGb redundantly control Medicago floral organ identity, but have minimal distinct functions in regulating stamen and carpel development in a dose-dependent manner. Interestingly, the stamens and carpels are mostly converted to numerous vexillum-like petals in the double mutant of mtaga mtagb, which is distinguished from Arabidopsis ag. Further qRT-PCR analysis in different mtag mutants revealed that MtAGa and MtAGb can repress the expression of putative A and B function genes as well as MtWUS, but promote putative D function genes expression in M. truncatula. In addition, we found that the abnormal dorsal petal phenotype observed in the mtaga mtagb double mutant is associated with the upregulation of CYCLOIDEA (CYC)-like TCP genes. Taken together, our data suggest that the redundant MtAGa and MtAGb genes of M. truncatula employ a conserved mechanism of action similar to Arabidopsis in determining floral organ identity and FM determinacy but may have evolved distinct function in regulating floral symmetry by coordinating with specific floral dorsoventral identity factors.
As the important reproductive organs in flowering plants, flowers show remarkable variation in formation and elaboration, and provide the most trustworthy external characteristics for establishing relationships among different angiosperm species. In dicots, flowers are commonly composed of four different types of floral organs arranged in concentric whorls. From outside to the center, these floral organs are sepals in the first whorl, petals in the second whorl, stamens in the third whorl and carpels in the fourth whorl. Understanding how these distinct floral organs are specified has been a long-standing question in plant development and held the fascination of scientists for centuries (Meyerowitz et al., 1989; Schwarz-Sommer et al., 1990; Coen and Meyerowitz, 1991; Theissen et al., 2016).
In the past two decades, extensive genetic and molecular analyses of a series of floral homeotic mutants in diverse species have revealed that floral organ formation is controlled by several conserved floral organ identity genes and these floral organ regulators were proposed to function in simple genetic models. Based on the studies in model dicot plants Arabidopsis thaliana and Antirrhinum majus, the most well-known “ABC” model was outlined (Schwarz-Sommer et al., 1990; Bowman et al., 1991; Coen and Meyerowitz, 1991). In Arabidopsis, the A function genes APETALA1 (AP1) (Mandel et al., 1992) and APETALA2 (AP2) (Jofuku et al., 1994) alone determine the identity of sepals in the first whorl. However, the combined activity of A function genes and B function genes including APETALA3 (AP3) (Jack et al., 1992) and PISTILLATA (PI) (Goto and Meyerowitz, 1994) is required for the formation of petals in the second whorl. Establishment of the stamen identity in the third whorl is controlled by B function genes and C function gene AGAMOUS (AG) (Yanofsky et al., 1990), whereas the C function gene AG solely determinates the termination and differentiation of the floral meristem (FM) into carpels (Wellmer et al., 2014). Later studies revealed that more genes are involved in regulating ovule identity and development inside the carpel (Angenent et al., 1995; Colombo et al., 1995). In Arabidopsis, three MADS-domain family members SHATTERPROOF1 (SHP1, formerly known as AGL1), SHATTERPROOF2 (SHP2, formerly known as AGL5) and SEEDSTICK (STK, formerly known as AGL11) have been identified as D function genes and the shp1 shp2 stk triple mutant convert ovules into carpel-like or leaf-like structures (Favaro et al., 2003; Pinyopich et al., 2003). In addition, an E function has been assigned to another class of genes, including SEPALLATA1 (SEP1, formerly known as AGL2), SEP2 (AGL4), SEP3 (AGL9), and SEP4 (AGL3), which are essential for the identity of all floral organs in combination with the A, B, C, and D function genes (Pelaz et al., 2000; Ditta et al., 2004). The expanded “ABCDE” model maintains organ identity by a refined combination as that A+E genes control sepals, A+B+E genes specify petals, B+C+E genes determine stamens, C+E genes control carpels and C+D+E genes specify ovules (Theissen et al., 2016).
In contrast to the flowers of model dicot species such as A. thaliana or A. majus, the zygomorphic flowers in Papilionoideae plants are peculiarly arranged with pentamerous whorls of sepals and petals, two whorls of five stamens each, and a single carpel (Tucker, 2003). To investigate the molecular basis of floral organ identity in Papilionoideae, several floral organ regulation genes have been identified and characterized in the model legume Medicago truncatula. The MtPIM gene, a homolog of the A function gene AP1 in M. truncatula, is required for specification of floral meristem identity. Mutation of MtPIM leads to a flower-to-inflorescence conversion and altered flowers with sepals transformed into leaves, indicating that MtPIM controls FM identity and flower development (Benlloch et al., 2006). Function analysis of the MtNMH7 and MtTM6, which are homologs of the Arabidopsis B function gene AP3, revealed that MtNMH7 appears to play a major role in determining the petal identity, whereas MtTM6 plays a more important role in the stamen identity (Roque et al., 2013). Both MtPI and MtNGL9 encode MADS-box transcription factors related to the Arabidopsis PISTILLATA gene. Mutation of MtPI leads to defects in petals and stamens, suggesting that MtPI functions as a master regulator of B-function in M. truncatula (Benlloch et al., 2009; Roque et al., 2016). Although the floral organ arrangement and flower morphogenesis between Arabidopsis and M. truncatula are apparently different, the fact that orthologs of AP1, AP3 and PI have also been identified in M. truncatula suggests that the “ABCDE” model is generally applicable to Papilionoideae plants as well.
In Arabidopsis and Antirrhinum, the C function is, respectively, represented by AG and PLENA (PLE), which is required to control the stamen, carpel, ovule identity, to prevent the mis-expression of A function genes in the third and fourth whorls, and to establish the FM determinacy by antagonizing the key regulator of stem cell homeostasis WUSCHEL (WUS) (Bowman et al., 1989, 1991; Yanofsky et al., 1990; Bradley et al., 1993; Lenhard et al., 2001). By contrast, due to an ancient genome duplication event, the presence of duplicated AG homologs has been commonly found in several Papilionoideae plants including Lotus japonicus, Pisum sativum, and soybean (Dong et al., 2005; Shoemaker et al., 2006; Serwatowska et al., 2014). Recently, two AGAMOUS homologs MtAGa and MtAGb were functionally characterized and shown to fulfill the C function activity that promote complete stamen and carpel identity and FM determinacy in M. truncatula (Serwatowska et al., 2014). However, neither mtaga and mtagb single mutant nor VIGS/RNAi lines silenced MtAGa and MtAGb both show a complete loss of C-function phenotype as observed in Arabidopsis ag mutant (Serwatowska et al., 2014). Moreover, how the duplicated AGAMOUS homologs interact with other floral organ identity genes during M. truncatula floral morphogenesis remains to be elucidated.
In this study, we reported the isolation and characterization of a new mtaga allele by screening Tnt1 retrotransposon-tagged lines of M. truncatula. We found that MtAGa was not only required for controlling the stamen and carpel identity, but also affected pod and seed development. The mtaga mtagb double mutant analysis confirmed that MtAGa and its paralog MtAGb together fulfill a full C-function activity but exhibit minimal subfunctionalization in regulating stamen and carpel identity. Further comprehensive molecular analysis revealed that the duplicated AGAMOUS homologs MtAGa and MtAGb coordinate with floral dorsoventral identity regulators to regulate flower morphogenesis in M. truncatula.
Wild-type M. truncatula ecotype R108 and mutants were grown in greenhouse with 25°C/16-h (day) and 23°C/8-h (night) photoperiods at 60–70% humidity and 150–200 μmol m2s light intensity.
Insertional mutagenesis in M. truncatula genotype R108 using Tnt1 retrotransposon and screening conditions in the greenhouse was carried out as previously described (Tadege et al., 2008; Yarce et al., 2013). Forward genetics screening of Tnt1-tagged lines under standard conditions (16 h/8 h and 24°C/20°C day/night cycles) in greenhouse for flower mutants led to identification of the mutant line NF19601 with split carpels. To identify the gene linked to the split-carpel phenotype, Tnt1 flanking sequences of NF19601 mutant were recovered using TAIL-PCR (Cheng et al., 2014) and genotyped by PCR using flanking sequence tag (FST)-specific primers (Supplementary Table S1). One FST segregated with the homozygous mutant was analyzed by BLAST search against the M. truncatula genome at the National Center for Biotechnology Information (NCBI)1 and Phytozome2. Reverse screening of Tnt1 insertion lines were performed following the standard screening protocol (Cheng et al., 2014).
Amino acid sequences of M. truncatula and other selected species were aligned using ClustalW3, and a neighbor-joining phylogenetic tree was constructed based on the amino acid sequences of the full-length proteins using the MEGA 6 software. The most parsimonious tree with bootstrap values from 1,000 trials was used. Accession numbers used in this study were listed in Supplementary Table S2.
Total RNA from different M. truncatula tissues was extracted using the Trizol method, and RNA was treated with Turbo DNase (Ambion) to remove genomic DNA. Five micrograms of total RNA was used for cDNA synthesis by using the SuperScript® III First-Strand Synthesis System for RT-PCR kit (Life Technologies). qRT-PCR was performed using the ROCHE LightCycler® 96 detect system with the TransStart Tip Green qPCR SuperMix (TransGen Biotech). qRT-PCR data were obtained using three biological replicates and transcripts were normalized to MtActin. Primers used were listed in Supplementary Table S1.
The coding sequences of MtAGa and MtAGb were PCR amplified from wild-type M. truncatula ecotype R108 and cloned into the pENTR/D-TOPO cloning vector (Invitrogen), then transferred into the pMDC83 Gateway vector using the Gateway LR reaction (Invitrogen) to generate the pMDC83-MtAGa-GFP and pMDC83-MtAGb-GFP destination vectors. The constructs were introduced into A. tumefaciens stain GV2260 by chemical transformation and the agrobacterium was infiltrated into 4-week-old Nicotiana benthamiana leaves. P19 was used to inhibit transgenic silencing. After culturing for 2–3 days, the GFP signal was visualized under the Zeiss LSM700 confocal laser-scanning microscope. Primers used for vectors construction were listed in Supplementary Table S1.
Juvenile flowers of M. truncatula from 3-month-old plants were fixed and embedded as previously described (Lin et al., 2009). The tissues were sliced into 8- to 10-μm sections with a Leica RM2265 microtome, affixed to microscope slides, and stained with Toluidine blue. Images were obtained with a digital camera mounted on the Olympus BX-51compound microscope.
For SEM, fresh M. truncatula floral buds from 5-month-old wild-type and different mutant plants were fixed by vacuum infiltration with 3.0% glutaraldehyde in 25 mM phosphate buffer (pH 7.0) for 2 days, then plant tissues were further fixed with 1.0% osmium tetroxide in the same phosphate buffer for 2 h and subsequently dehydrated in a graded ethanol series. The desiccated tissues were critical-point dried in liquid CO2, mounted on aluminum stubs, and sputter coated with gold. Specimens were then observed using a JSM-8404 microscope (S3400N; Hitachi Ltd.).
One floral development mutant NF19601 was identified from a forward genetic screening of Tnt1 retrotransposon-tagged population in M. truncatula genotype R108 (Tadege et al., 2008; Yarce et al., 2013). In contrast to the wild-type, the mutant flowers were morphologically normal in the sepals and petals (Figures 1A,B), whereas the staminal tube was slightly separated and the stamens were occasionally transformed into petal-like structures (Figures 1C–E). Besides, multiple carpels were frequently observed and trichomes were formed on the back of the main carpel in NF19601 (Figures 1F–I). A close examination of the carpels showed that the mutant had exposed ovules and some ovules were missing in the basal part of carpels (Figures 1H,J–L). Sometimes floral bud-like structures were formed at the bottom of carpels (Figures 1I,L). In this mutant, the defective carpels developed to abnormally spiny pods with reduced whirl numbers (Figures 1M–T and Supplementary Figure S1A), subsequently resulting in decreased seed number per pod (Supplementary Figure S1B). In addition, the seed size and weight in the floral mutant also significantly decreased compared to that of wild-type (Figures 1U,V and Supplementary Figures S1C,D). Taken together, these phenotypes indicated that this mutant is defective in the floral organ identity, especially in the development of stamens and carpels in M. truncatula.
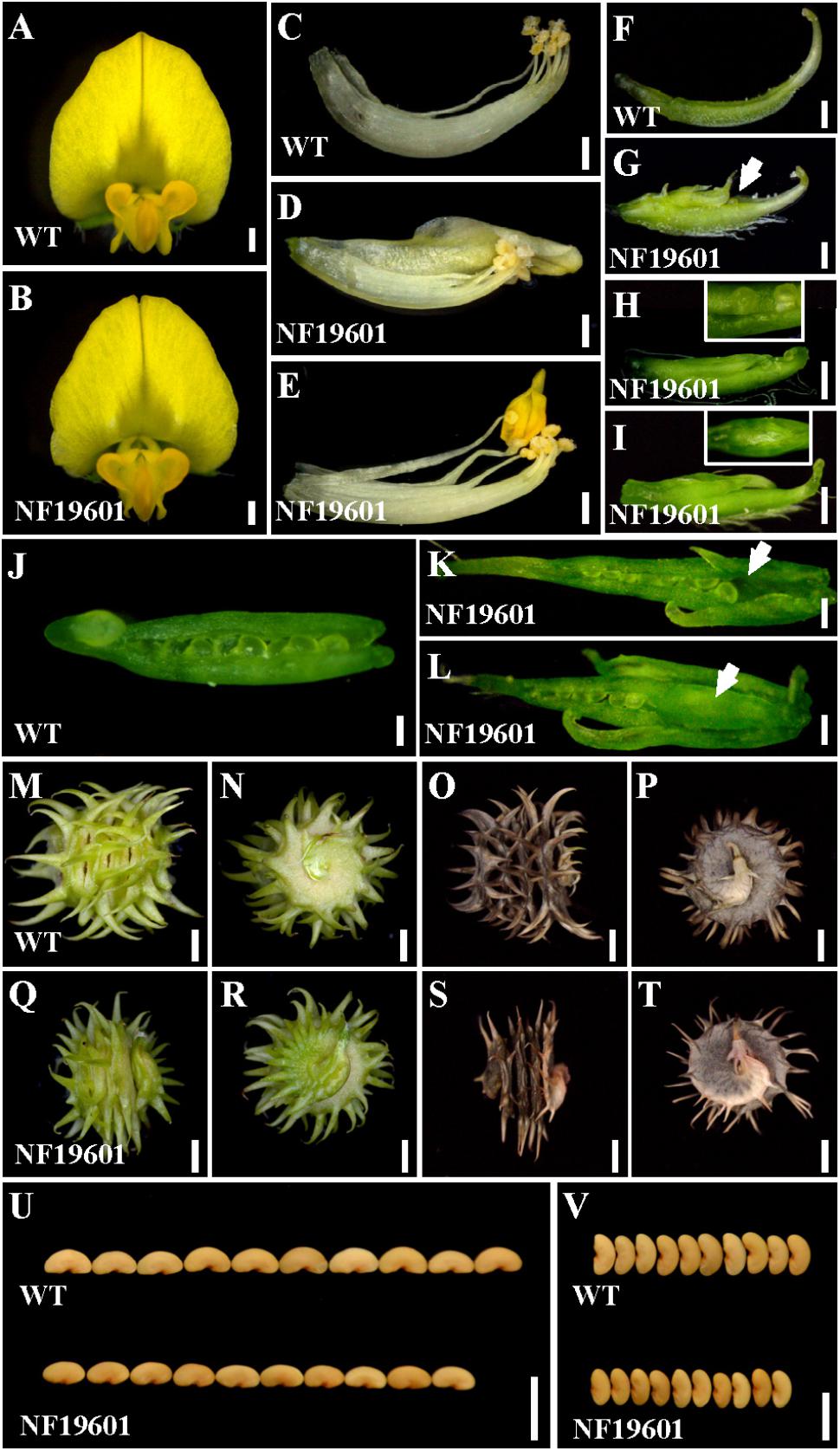
FIGURE 1. NF19601 mutant of M. truncatula shows defects in floral organ, pod and seed development. (A,B) Flowers of the wild-type and NF19601. Bars = 500 μm. (C–E) Stamens of the wild-type and NF19601. The staminal tube is slightly separated in NF19601. Bars = 500 μm. (F–I) Comparison of carpels in the wild-type and NF19601. White arrow in (G) points to the exposed ovule. The inset in (H) shows a close-up view of the distal part of the carpel with exposed ovules. The inset in (I) shows a ventral view of the paraxial part of the carpel with a floral bud-like structure formed. Bars = 500 μm. (J–L) Dissected carpels of the wild-type and NF19601. White arrow in (K) shows no ovules in the carpel base of NF19601. White arrow in (L) shows a floral bud-like structure in the carpel base of NF19601. Bars = 200 μm. (M–P) Side (M,O) and bottom (N,P) views of the wild-type immature (M,N) and mature (O,P) pods. Bars = 2 mm. (Q–T) Side (Q,S) and bottom (R,T) views of NF19601 immature (Q,R) and mature (S,T) pods. Bars = 2 mm. (U,V) Comparison of seed length (U) and width (V) between the wild type and NF19601. Bars = 5 mm.
To identify the gene associated with the mutant phenotype, thermal asymmetric interlaced-PCR was performed to recover the flanking sequences of Tnt1 from line NF19601 (Tadege et al., 2008). Based on the genotyping results, one Tnt1 insertion segregating with the mutant phenotype was identified. Genetic analysis revealed that the mutant phenotype segregates as a single-gene recessive mutant and all mutant plants harbored homozygous insertion for the particular FST. The full length gene sequence corresponding to this FST was recovered and sequence alignment showed that the candidate gene encodes a MADS-box transcription factor, which is identical to previously reported MtAGa (Serwatowska et al., 2014). Genomic PCR analysis showed that the Tnt1 was inserted in the fourth exon of MtAGa, resulting in the abolished transcription of the full-length MtAGa (Figures 2A,B). To confirm that the phenotypes of defective floral organ development as well as abnormal pods and seeds were caused by disruption of MtAGa, two additional Tnt1 insertion lines NF13380 (mtaga/mtag-2) and NF10148 (named mtaga-3 here) were obtained by reverse screening (Cheng et al., 2014; Serwatowska et al., 2014; Figures 2A,B). Phenotypic observation showed that both NF10148 and NF13380 mutants displayed same phenotypes as described in NF19601 (named mtaga-2 here), confirming that the MtAGa gene is not only required for controlling stamen, carpel identity and FM determinacy, but also functions in pod and seed development in M. truncatula.
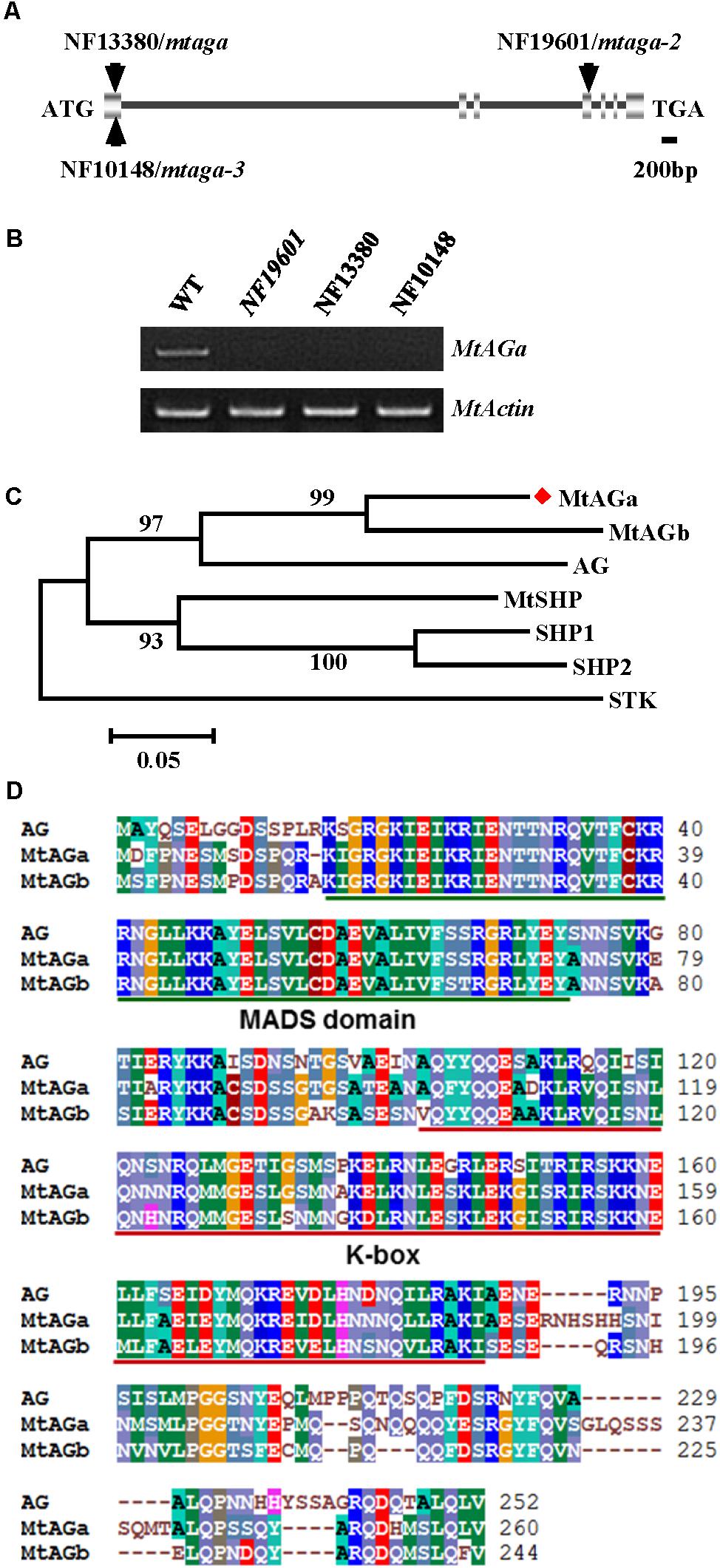
FIGURE 2. Molecular cloning of MtAGa in M. truncatula. (A) Schematic representation of the gene structure of MtAGa and the Tnt1 insertion sites of NF19601, NF13380, and NF10148. (B) RT-PCR analysis of MtAGa expression in young flowers of the wild-type, NF19601, NF13380, and NF10148. MtActin was used as the loading control. (C) Phylogenetic analysis of MtAGa and its homologs in M. truncatula and Arabidopsis based on the amino acid sequence of the full-length protein. Numbers on branches indicate bootstrap values for 1,000 replicates. (D) Sequence alignment of MtAGa, MtAGb, and Arabidopsis AG. The conserved MADS domain and K-box were highlighted with color lines, respectively.
In agreement with previous findings that MtAGa and MtAGb both show C-function activity in regulating M. truncatula floral development (Serwatowska et al., 2014), phylogenetic analysis revealed that MtAGa and its homolog MtAGb are grouped in a separate subclade of C-function family (Figure 2C) and amino acid sequence alignment indicated that MtAGa, MtAGb and Arabidopsis AG have the highly conserved MADS domain and K-box (Figure 2D). To investigate possible roles of MtAGb in M. truncatula pod and seed development, two Tnt1 insertion mutants of MtAGb were characterized. One mutant is NF4908, which has been identified as mtagb, containing a Tnt1 insertion in the first intron of MtAGb (Serwatowska et al., 2014; Figure 3A). The other mutant is NF15934 (named mtagb-2 here), which was newly identified from the Tnt1 insertional population by BLAST-searching the FST database (Cheng et al., 2014), containing a Tnt1 insertion in the first exon of MtAGb (Figure 3A). RT-PCR analysis showed that the expression of full length MtAGb were undetectable in both two alleles (Figure 3B). Consistent to the previous report, both NF15934 and NF4908 mutants exhibited normal sepals and petals, but showed an incomplete fusion of stamen tubes and weak petaloid structures developed at stamen tips (Figures 3C–F,H–K). Moreover, compared to wild-type and mtaga, the mtagb mutants showed a weak developmental defect in carpels with occasionally split carpels but no extra structure formation at the bottom (Figures 3G,L), and the pods and seeds in mtagb mutants were nearly indistinguishable from the wild-type (Figures 3M–V and Supplementary Figure S2). These data suggest that MtAGb appears to play a major role in controlling stamen fusion, whereas MtAGa contributes more to carpel development.
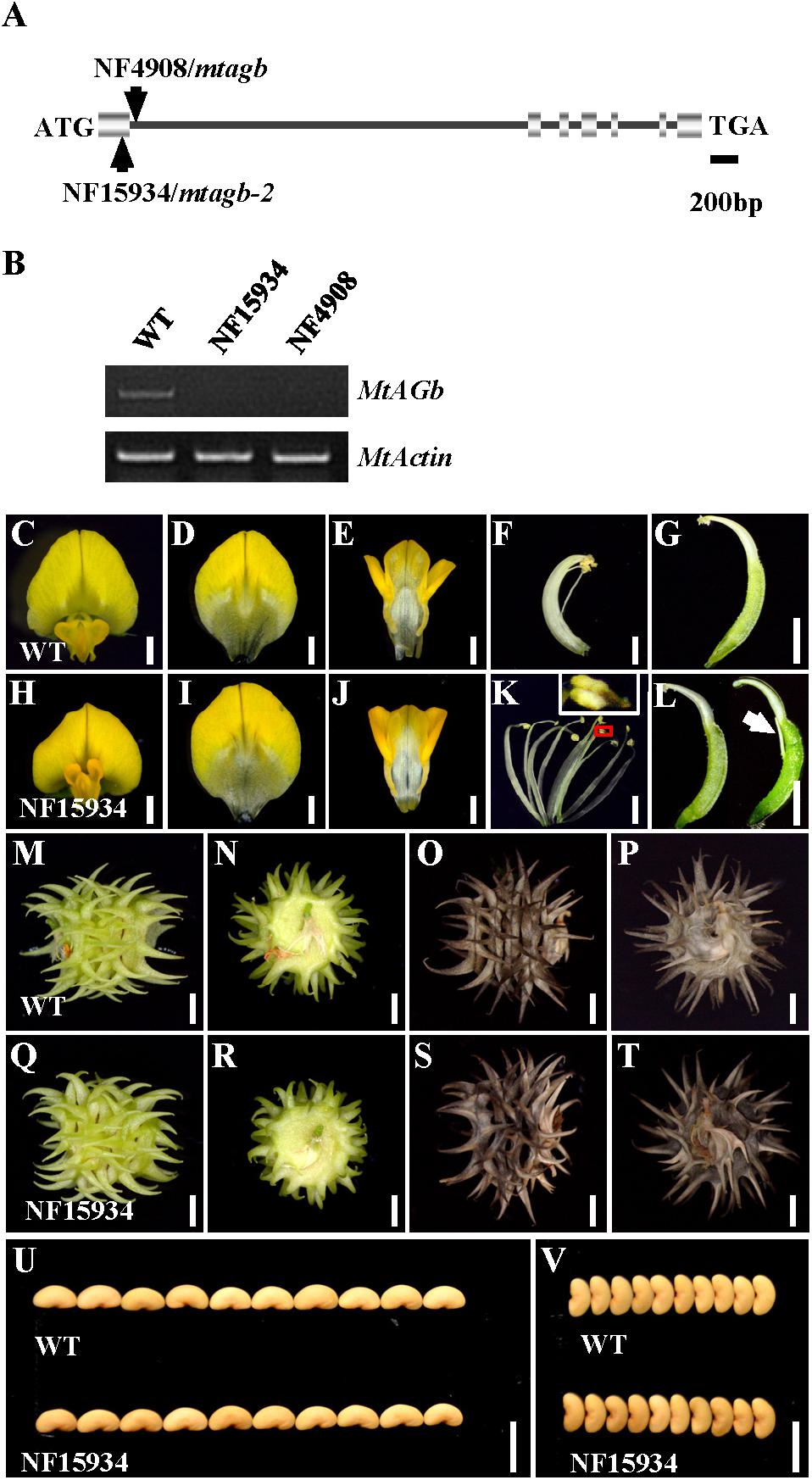
FIGURE 3. Characterization of the mtagb mutant. (A) Schematic representation of the gene structure of MtAGb and the Tnt1 insertion sites of NF15934 and NF4908 are shown. (B) RT-PCR showed the expression level of MtAGb in young flowers of the wild-type, NF15934 and NF4908. MtActin was used as the loading control. (C,H) Flowers of the wild-type and NF15934. Bars = 1 mm. (D–G) Dissected flower of the wild-type. The top view of vexillum (D), fused alae and keel petals (E), the side view of stamen (F) and carpel (G). Bars = 1 mm. (I–L) Dissected flower of NF15934. The top view of vexillum (I), fused alae and keel petals (J), the side view of stamen (K) and carpel (L). The inset in (K) shows the tip of anther changed to a petal-like structure. White arrow in (L) shows the stigmatic protuberance. Bars = 1 mm. (M–P) Side (M,O) and bottom (N,P) views of the wild-type immature (M,N) and mature (O,P) pods. Bars = 2 mm. (Q–T) Side (Q,S) and bottom (R,T) views of NF15934 immature (Q,R) and mature (S,T) pods. Bars = 2 mm. (U,V) Comparison of seed length (U) and width (V) between the wild-type and NF15934. Bars = 5 mm.
To better understand the functional specialization of MtAGa and MtAGb in controlling reproductive organ’s determination, we analyzed the expression patterns of MtAGa and MtAGb in different tissues and organs. qRT-PCR analysis revealed that both MtAGa and MtAGb are highly expressed in flowers and pods, medially expressed in floral apices, and lowly expressed in other tissues (leaf, stem, cotyledon, root, and nodule) (Figure 4A). Further qRT-PCR analysis of dissected floral organs revealed that both MtAGa and MtAGb are predominantly expressed in stamens, carpels as well as ovules with different expression levels (Figure 4B). This tissue-specific expression pattern is consistent with the proposed scenario of MtAGa and MtAGb fulfilling a C function activity during floral development in M. truncatula.
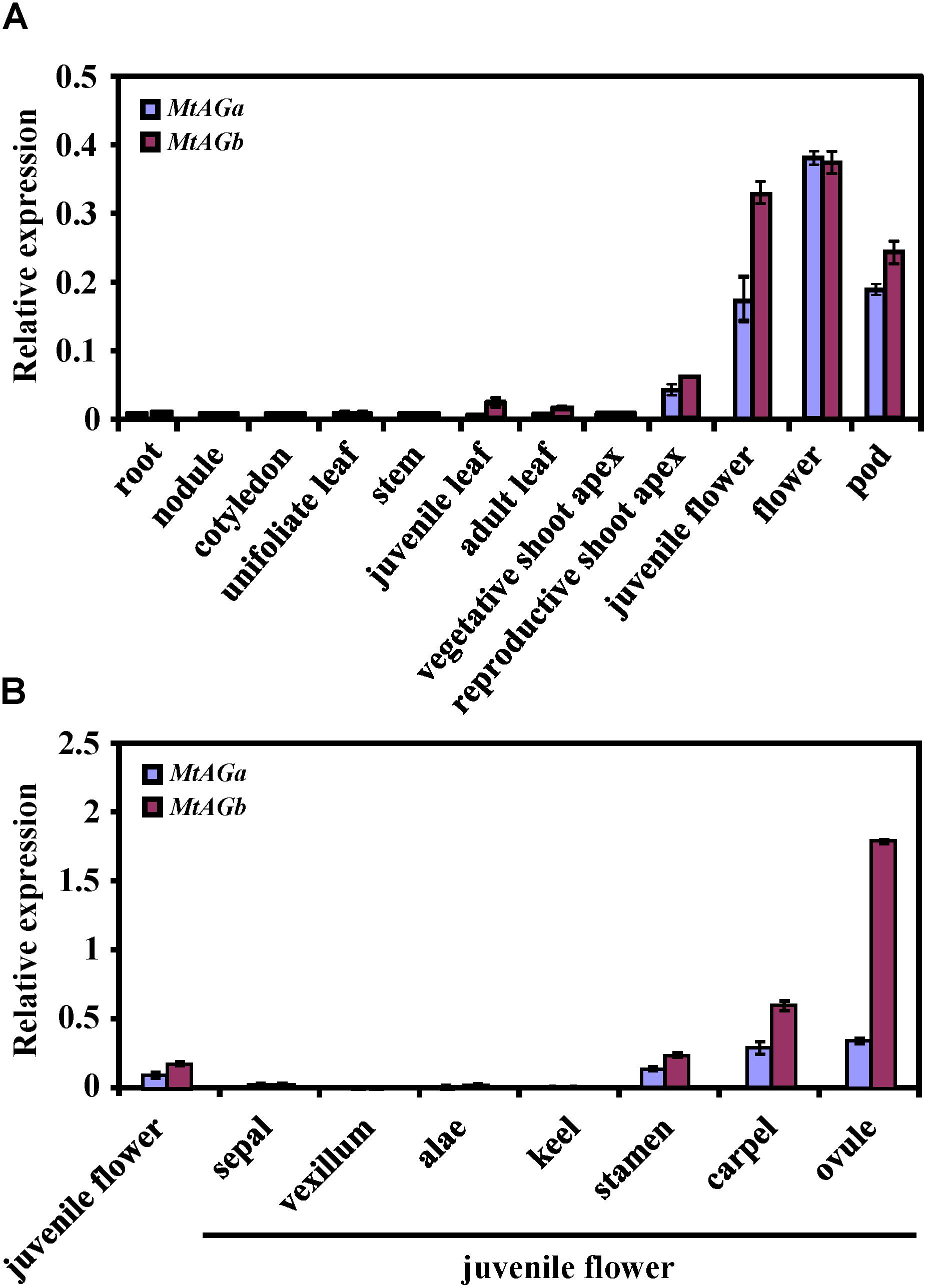
FIGURE 4. Comparison of expression pattern between MtAGa and MtAGb. (A) MtAGa and MtAGb expression levels revealed by qRT-PCR in different tissues. Values are means ± SE of three biological replicates. (B) MtAGa and MtAGb expression levels revealed by qRT-PCR analysis in different floral organs. The flower organs were dissected from juvenile flowers. Values are means ± SE of three biological replicates.
To determine the subcellular localization of the MtAGa and MtAGb proteins, we performed a transient expression experiment in leaf epidermal cells of N. benthamiana. The C terminus of MtAGa and MtAGb were individually fused with green fluorescent protein (GFP) under the control of Cauliflower mosaic virus (CaMV) 35S promoter. In contrast to the control, which was ubiquitous in leaf epidermal cells, the MtAGa-GFP and MtAGb-GFP fusion proteins were localized in both cytoplasm and nucleus (Supplementary Figure S3).
In Arabidopsis, AGAMOUS is the only C function gene, which is required for stamen and carpel identity and FM determinacy. Mutation of AG in Arabidopsis causes lack of pistils and stamens, and the flowers continue to produce sepals and petals (Bowman et al., 1989). However, in M. truncatula, either mtaga and mtagb single mutants or VIGS/RNAi lines silenced MtAGa and MtAGb did not show a complete loss of C-function phenotype as observed in Arabidopsis ag mutant (Serwatowska et al., 2014). To explore whether MtAGa and MtAGb fulfill a full C-function activity, we generated the mtaga mtagb-2 double mutant by crossing NF13380 and NF15934 and the resulting double mutant was analyzed for morphological phenotypes. Compared with wild-type and single mutants, the mtaga mtagb-2 double mutant showed additive effect on stamen and carpel development (Figures 5, 6). In the mtaga flower, the central carpel was frequently separated and was not able to fully enclose the ovules (Figures 5B, 6C), while in the mtagb-2 flower, the staminal tube was poorly fused and the stamen tips were frequently transformed into petal-like structures (Figures 5C, 6E). However, in the mtaga mtagb-2 double mutant, the stamens and carpels were mostly converted to numerous vexillum-like petals, which may be derived from multiple FMs (Figures 5G, 6M,N). These results clearly indicate that MtAGa and MtAGb together fulfill a full C-function activity but also suggest that MtAGa and MtAGb may affect petal shape somehow.

FIGURE 5. Floral phenotypes of different mtag mutants. (A) Dissected flowers of the wild-type showing normal four whorls (W1–W4). Bar = 1 mm. (B) Dissected flowers of the mtaga mutant. The white arrow indicates anther turned into petal-like structure and the inset is magnification of indicated region. The green arrow in W4 points to the split carpels and the red arrow indicates the floral bud-like structure formed at the bottom of the carpel. Bar = 1 mm. (C) Dissected flowers of the mtagb mutant. The white arrow in W3 indicates the unfused staminal tube. The inset view shows the anther tips which were changed into petal-like structure. Occasionally, the split carpel can be found in W4 (green arrow). Bar = 1 mm. (D) Dissected flowers of the mtaga/+ mtagb-2/+ plants. The floral phenotype of mtaga/+ mtagb-2/+ is similar to that of the wild-type, but occasionally the split carpel could be found in W4. The white arrow in W4 indicates the filamentous structure and the inset is magnification of indicated region. Bar = 1 mm. (E) Dissected flowers of the mtaga mtagb-2/+ plants. The white arrow indicates the wrapped stamen in the center of the W4, the red arrow shows the floral bud-like structure formed at the base of the carpel. The inset is magnification of indicated region by green arrow showing existed ovules. Bar = 1 mm. (F) Dissected flowers of the mtaga/+ mtagb-2 plants. The white arrows indicate multiple carpels, red arrow shows the floral bud-like structure at the carpel base. The inset is magnification of indicated region by green arrow showing existed ovules. Bar = 1 mm. (G) Dissected flowers of the mtaga mtagb-2 double mutant. The W2–W4 whorls mostly changed into vexillum-like petals. Bar = 1 mm.
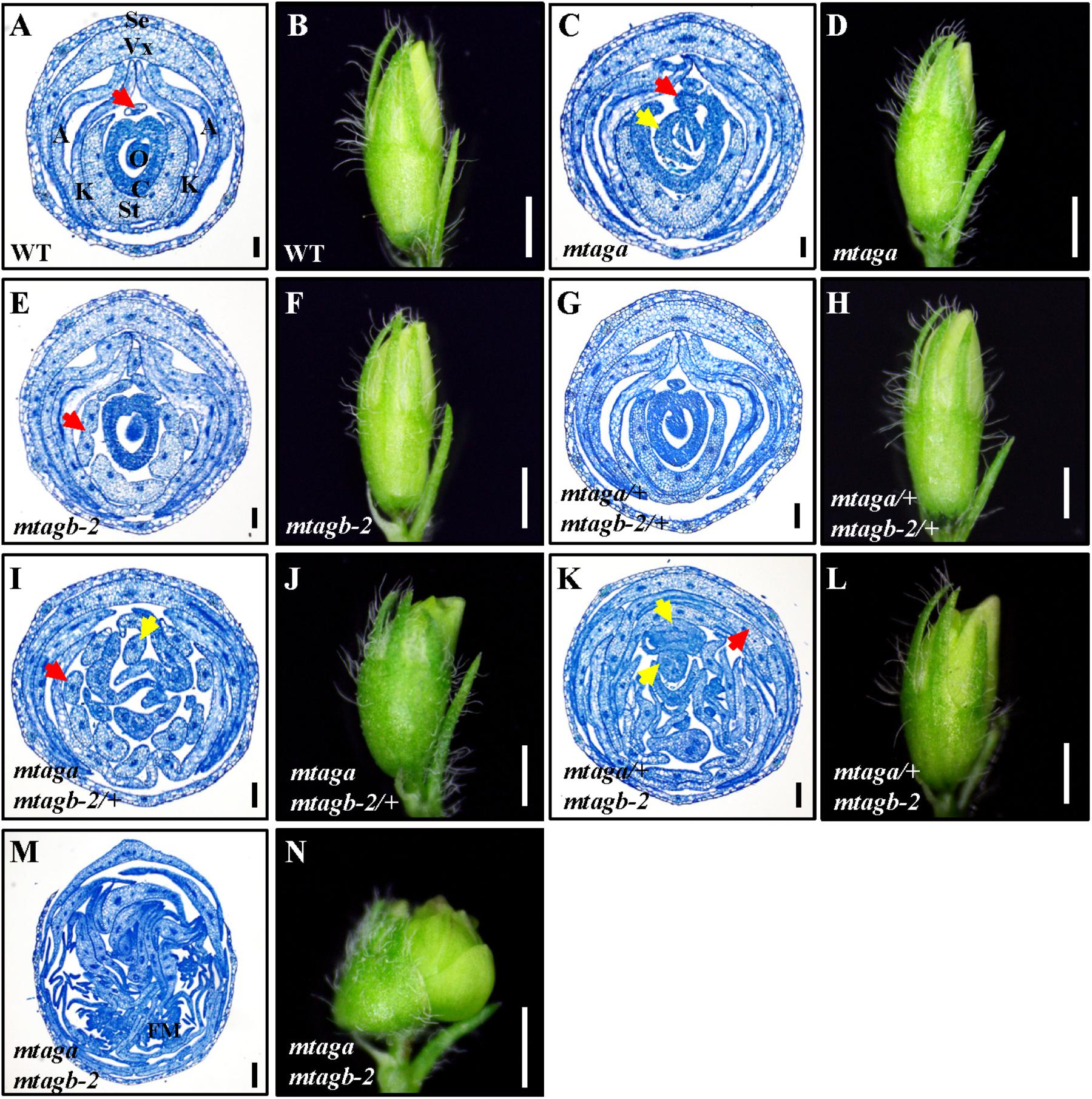
FIGURE 6. The histological analysis of juvenile flowers in the wild-type and different mtag mutants. (A,B) Cross section and morphology of juvenile flower in the wild-type. The red arrow points to the stamen filament separated from the fused nine. Se, sepal; Vx, vexillum; A, alae; K, keel; St, staminal tube; C, carpel; O, ovule. Bar = 100 μm in (A), 1 mm in (B). (C,D) Cross section and morphology of juvenile flower in mtaga mutant. Red arrow indicates exposed ovule and yellow arrow points to the extra floral bud-like structure, respectively. Bar = 100 μm in (C), 1 mm in (D). (E,F) Cross section and morphology of juvenile flower in mtagb mutant. The red arrow indicates the incompletely fused staminal tube. Bar = 100 μm in (E), 1 mm in (F). (G,H) Cross section and morphology of juvenile flower in mtaga/+ mtagb-2/+. Bar = 100 μm in (G), 1 mm in (H). (I,J) Cross section and morphology of juvenile flower in mtaga mtagb-2/+. The red arrow indicates the separated staminal tube, the yellow arrow points to the extra stamen. Bar = 100 μm in (I), 1 mm in (J). (K,L) Cross section and morphology of juvenile flower in mtaga/+ mtagb-2. Red arrow points to petaloid organ and yellow arrows indicate the extra floral bud-like structures. Bar = 100 μm in (K), 1 mm in (L). (M,N) Cross section and morphology of juvenile flower in the mtaga mtagb-2 double mutant. FM, floral meristem. Bar = 100 μm in (M), 1 mm in (N).
Interestingly, during the screening of the mtaga mtagb-2 double mutant, we found that some of the F2 progeny from the NF13380 × NF15934 crossing showed peculiar floral phenotypes, which are different from the single and double mutants. Genotyping analysis revealed that these peculiar floral phenotypes are associated with particular genotypes. Compared with the wild-type and single mutants, the mtaga mtagb-2/+ (/+ means the heterozygous mutant) flowers displayed normal sepals and petals in whorl 1 and 2, but in whorl 3 stamens usually changed into irregular petals or petaloid structures, and extra organs such as filament-like structure and petaloid extensions were often observed (Figures 5E, 6I and Supplementary Figure S4A). Whorl 4 was usually composed of mosaic organs of sepaloid and petaloid structures and/or leaf-like structures in place of carpels and ovules (Figures 5E, 6I and Supplementary Figures S4B,C). Moreover, a new layer of floral organs like stamen or floral bud were frequently formed in the center of whorl 4, suggesting FM determinacy was disrupted in mtaga mtagb-2/+ (Figures 5E, 6I and Supplementary Figure S4B). By contrast, in the mtaga/+ mtagb plants, flowers also displayed normal whorl 1 and 2, but in whorl 3, three of 10 stamens were usually converted to one vexillum and two alae-like petals, the other seven stamens showed petaloid structures (Figures 5F, 6K). Besides, extra petal and petaloid tissues were often observed in whorl 3 (Figure 6K and Supplementary Figure S5A). Whorl 4 was frequently composed of multiple carpels similar to mtaga, but leaf-like tissues and new floral buds were often found in the center of whorl 4 (Figures 5F, 6K and Supplementary Figure S5B), suggesting mtaga/+ mtagb-2 mutant flowers were indeterminate as well. However, in double heterozygote mtaga/+ mtagb-2/+ plants, whorl 1–3 of flowers did not exhibit any obvious phenotypic defects compared to that of wild-type (Figures 5A,D, 6A,G), but in whorl 4, a filamentous structure was observed at the carpel base (Figure 5D). These observations indicated that MtAGa and MtAGb redundantly control floral organs identity in whorl 3 and 4 of M. truncatula, but have minimal distinct functions in regulating stamen and carpel development in a dose-dependent manner.
To further understand the effects of mutation of the duplicated MtAGa and MtAGb on early stage floral development in M. truncatula, we used SEM to compare flowers of mtag mutants with that of wild-type. All mtag mutants, except the homozygous mtaga mtagb-2 double mutant, displayed similar floral ontogeny as wild-type (Figures 7A,E,I,M,Q,U). In the double mutant, the sepals were indistinguishable from that of wild-type flowers, but there were no obvious petals, stamens, as well as carpel primordia in the inner three whorls, and several FMs could be observed (Figures 7A,Y), which continued to generate vexillum-like petals as well as new FMs. Differences between mtag mutants and wild-type flowers became evident when the primordia of the third- and fourth-whorl organs started to differentiate (Figures 7B–D,F–H,J–L,N–P,R–T,V–X,Z,A1). In wild-type, the filaments and the anther locules appeared after stage 7 (Benloch et al., 2003) and then the central carpel became closed (Figures 7B–D). However, at the same stage, trichomes formed on the abaxial side of the carpel and bulges were observed at the base of the carpel margin in mtaga (Figures 7F–H). This observation may explain why mature mtaga flowers contain split carpels. Meanwhile, some weak petaloids in the anther tips were observed in mtagb-2 flowers (Figure 7K). In the double heterozygote mtaga/+ mtagb-2/+ plants, no obvious developmental defects exhibited in whorls 1-3 (Figures 7M–O), but there was one bulge at carpel base (Figure 7P).
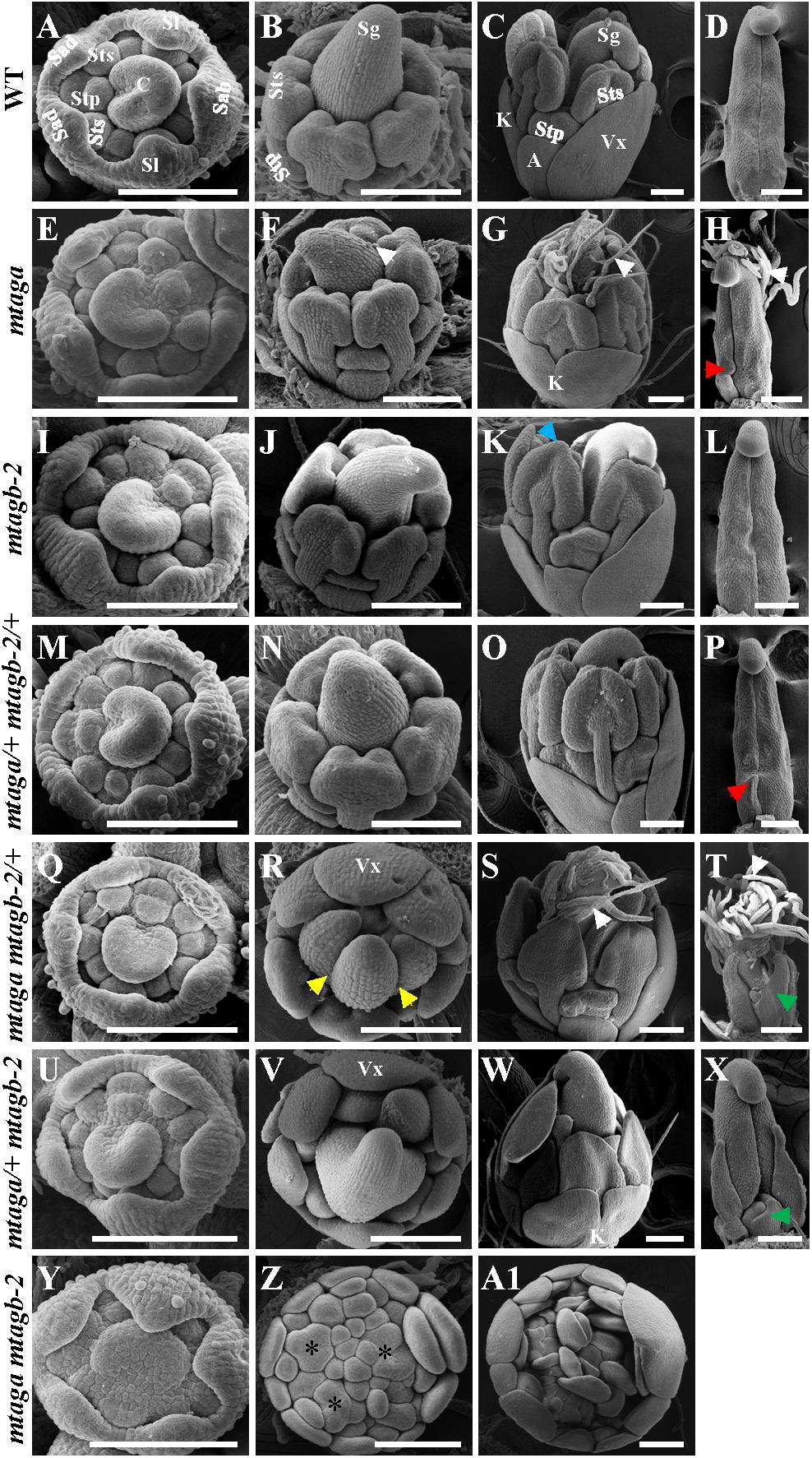
FIGURE 7. The scanning electron microscopy (SEM) analysis of floral development in the wild-type and mtag mutants. (A–D) Different developmental stage flowers (A–C) and carpel (D) of the wild-type. Sab, abaxial sepal; Sl, lateral sepal; Sad, adaxial sepal; Stp, inner antepetal stamen; Sts, outer antesepal stamen; C, carpel primordium; A, alae; K, keel; Vx, vexillum; Sg, stigma. Bars = 100 μm. (E–H) Different developmental stage of flowers (E–G) and carpel (H) in mtaga mutant. Red arrow points to the bulge structure. White arrows indicate trichomes. Bars = 100 μm. (I–L) Different developmental stage flowers (I–K) and carpel (L) of mtagb mutant. Blue arrow points to the petaloid structure of anther. Bars = 100 μm. (M–P) Different developmental stage flowers (M–O) and carpel (P) of mtaga/+ mtagb-2/+. Red arrow indicates a bulge at the base of carpel. Bars = 100 μm. (Q–T) Different developmental stage flowers (Q–S) and carpel (T) of mtaga mtagb-2/+. Yellow arrows point to split carpels, green arrow points to extra floral bud-like structure and white arrows indicate trichomes. Vx, vexillum. Bars = 100 μm. (U–X) Different developmental stage flowers (U–W) and carpel (X) of mtaga/+ mtagb-2. Green arrow points to extra floral bud-like structure at the carpel base. Vx, vexillum; K, keel. Bars = 100 μm. (Y–A1) Different developmental stage flowers of the mtaga mtagb-2 double mutant. Asterisks indicate abnormal floral meristems which will develop to petals. Bars = 100 μm.
Compared to wild type, the mtaga mtagb-2/+ and mtaga/+ mtagb-2 mutants displayed serious developmental defects in whorl 3 and 4. In mtaga mtagb-2/+, anthers were transformed into petaloid structures (Figures 7R,S) and the carpel clearly changed into compound leaf-like structure with trichomes and a floral bud-like organ was observed at its base (Figures 7R–T). Similarly, anthers also changed into petal-like structures in mtaga/+ mtagb-2 (Figures 7V,W) and floral bud-like organs formed at the base of carpels (Figure 7X). In the mtaga mtagb-2 double mutant, there were no gynoecium primordia formed but replaced by several meristem-like tissues, which continue to produce petal primordia (Figures 7Z,A1).
To further understand the molecular basis of abnormal floral organs in diverse mtag mutants, we compared the expression of M. truncatula homologs of A, B, C, D, and E function genes in flowers of wild-type and different mtag mutants. First, we searched for potential A, B, D, and E function genes in M. truncatula database using the Arabidopsis A, B, D and E function proteins as a BLAST query in the National Center for Biotechnology Information (NCBI) and Phytozome (Supplementary Figure S6). Besides the previously identified AP1-like gene MtPIM (Benlloch et al., 2006), PI homologs MtPI and MtNGL9 (Benlloch et al., 2009), AP3-like genes MtNMH7 and MtTM6 (Roque et al., 2013), AG homologs MtAGa and MtAGb (Serwatowska et al., 2014), and the PLENA-like gene MtSHP (Serwatowska et al., 2014), one additional AP1-like gene Medtr5g046790 (named MtAP1b), two AP2-like genes Medtr4g094868 and Medtr5g016810 (named MtAP2a and MtAP2b, respectively), one STK-like gene Medtr3g005530 (named MtSTK) and five SEPALLATA-like genes Medtr6g015975, Medtr7g016600, Medtr3g084980, Medtr8g097090, and Medtr4g109810 (named, respectively, MtSEP1/2a, MtSEP1/2b, MtSEP3a, MtSEP3b, MtSEP4) were identified. Phylogenetic analysis showed that MtPIM and MtAP1b were closely related to Arabidopsis AP1. Amino acid sequence alignment indicated that MtPIM and MtAP1b have the highly conserved MADS-box and K-box (Vandenbussche et al., 2003) (Supplementary Figure S7). The MtAP2a and MtAP2b sequences are also very close to Arabidopsis AP2. Sequence alignment indicated that both sequences have the highly conserved YRG element and RAYD element that are characteristic of the subfamily (Supplementary Figure S8). The MtSTK shows highest level of sequence identity with the STK protein from Arabidopsis (75% amino acid identity). MtSTK contains the conserved MADS-box and K-box (Supplementary Figure S9). Amino acid sequence alignment of MtSEPs with homologs from Arabidopsis showed that all five MtSEPs have the conserved MADS-box, I region, K-box and C region (Supplementary Figure S10). Phylogenetic analysis of MtSEP1-5 with other MADS-box homologs showed that MtSEPs belong to the SEP-like clade specifically (Supplementary Figure S6).
After identifying these putative M. truncatula “ABCDE” model homolog genes, we then analyzed their expression levels by qRT-PCR in juvenile flowers of different mtag mutants. We found that the expression of three putative A function genes MtPIM, MtAP2a and MtAP2b, was strongly upregulated in the mtaga mtagb-2 double mutant but slightly upregulated in mtaga mtagb-2/+, while only MtAP2a was weakly upregulated in mtaga/+ mtagb-2 (Figure 8A). This result coincides with the scenario that the A and C function genes negatively regulate each other. Loss of C function genes relieved the expression of A function genes (Figures 8A,C). Accordingly, the expression of two identified B function genes MtPI and MtTM6, which are required for petal and stamen identify, significantly increased in mtaga mtagb-2, and only MtTM6 weakly increased in mtaga/+ mtagb-2. The expression of MtNGL9 and MtNMH7 was indistinguishable in wild-type and mtag mutants (Figure 8B). The expressions of these putative M. truncatula B function genes strongly diversified in mtag mutants, suggesting the existence of function differentiation. Consistent with the serious defects in ovule formation in mtaga mtagb-2, mtaga mtagb-2/+ and mtaga/+ mtagb-2 mutants, the expression of putative D function genes MtSTK and MtSHP was downregulated in above three mutants (Figure 8D). The decreased level of MtSTK and MtSHP appeared to be correlated with the different ovule defect phenotypes observed in mtaga mtagb-2, mtaga mtagb-2/+ and mtaga/+ mtagb-2 mutants, that is, mtaga mtagb-2 had no ovule with the lowest MtSTK and MtSHP expression and mtaga/+ mtagb-2 had the highest amount of ovule with the highest MtSTK and MtSHP expression. These results suggested that the ovule formation in M. truncatula is dependent on the level of MtSTK and MtSHP expression. However, compared to wild-type, the expression of five putative E function genes MtSEP1/2a, MtSEP1/2b, MtSEP3a, MtSEP3b and MtSEP4 was not altered in all mtag mutants (Figure 8E), suggesting that loss of MtAGs does not affect the expression of MtSEPs, although SEP genes genetically interact with AG in controlling stamens and carpels identity.
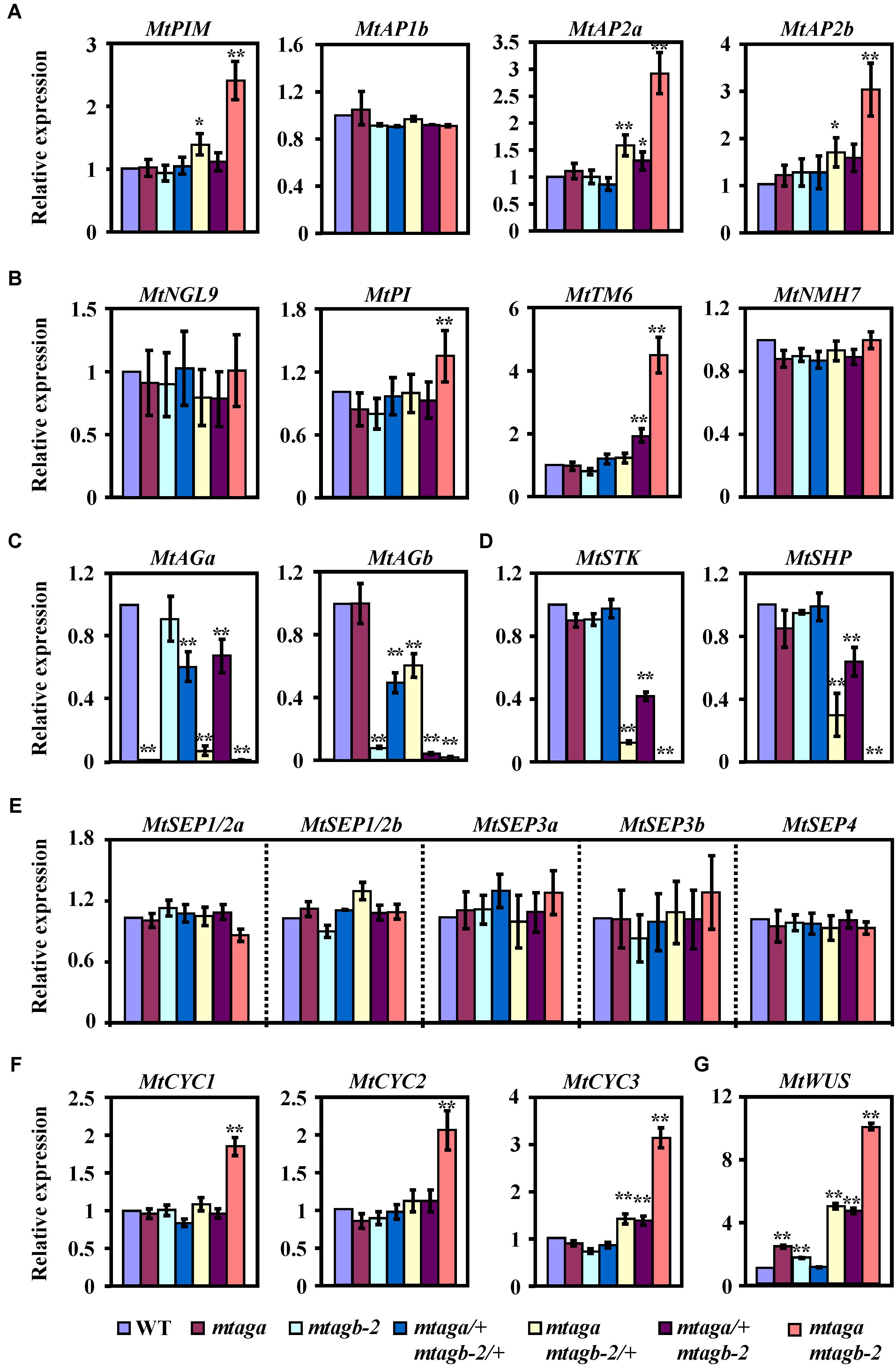
FIGURE 8. The expression levels of floral development related genes in the juvenile flowers of wild-type and mtag mutants. (A) Comparison of putative A function genes expression in the wild-type and different mtag mutants. (B) Expression of putative B function genes in the wild-type and different mtag mutants. (C) Expression of MtAGa and MtAGb in the wild-type and different mtag mutants. (D) Comparison of putative D function genes expression in the wild-type and different mtag mutants. (E) Expression of M. truncatula SEP homologs genes in the wild-type and different mtag mutants. (F) Comparison of TCP family genes MtCYC1-3 expression in wild-type and different mtag mutants. (G) Expression of the MtWUS gene in wild-type and different mtag mutants. Values are means ± SE of three biological replicates. ∗P < 0.05, ∗∗P < 0.01 (Student’s t-test).
It has been reported that the TCP transcription factor CYCLOIDEA (CYC) is involved in the control of petal shape and floral zygomorphy in Fabaceae (Luo et al., 1996; Citerne et al., 2006; Feng et al., 2006; Wang et al., 2008; Xu et al., 2013). Because of presence of multiple vexillum-like petals in flowers of the mtaga mtagb-2 double mutant (Figure 5G), we wondered whether this abnormal petal phenotype is related to the CYC genes and wanted to test the expression levels of the M. truncatula homologs of CYC in juvenile flowers of mtag mutants. Three potential CYC homologs (named here MtCYC1, MtCYC2, and MtCYC3) were found using the Lotus japonicas LjCYC1, LjCYC2 and LjCYC3 proteins as a BLAST query in the NCBI and Phytozome database. Phylogenetic analysis showed that MtCYC1, MtCYC2 and MtCYC3 are closely related to L. japonicas LjCYC1-3 (Supplementary Figure S11) and sequence alignment indicated that MtCYC1-3 sequences have the highly conserved TCP domain and R domain (Supplementary Figure S12), which are characteristic of the TCP transcription factors. qRT-PCR analysis revealed that the expression of MtCYC1, MtCYC2 and MtCYC3 were significantly upregulated in either the whole juvenile flowers or dissected petals of the mtaga mtagb-2 double mutant, while only the expression of MtCYC3 was slightly increased in mtaga mtagb-2/+ and mtaga/+ mtagb-2 (Figure 8F and Supplementary Figure S13). This result suggested that the abnormal dorsal petal shape observed in the mtaga mtagb-2 double mutant might be associated with the ectopic expression of MtCYC genes. Taken together, our data demonstrated that the duplicated AGAMOUS homologs MtAGa and MtAGb redundantly fulfill a C-function activity in determining floral organ identity and coordinate with other floral homeotic genes and dorsoventral identity factors to control flower morphogenesis in M. truncatula.
The C function genes, belonging to the AGAMOUS (AG) lineage and encoding MADS-box transcription factors, are required for stamen and carpel identity and FM determinacy in angiosperms (Theissen, 2001; Theissen et al., 2016). Unlike Arabidopsis in which the AGAMOUS gene is represented by a single genomic sequence and essentially confers the C-function in the FM (Yanofsky et al., 1990; Wellmer et al., 2014), most extant legume plants are the products of an ancient genome duplication event and the presence of duplicated AGAMOUS homologs has been found in several legume species (Shoemaker et al., 2006; Serwatowska et al., 2014). Recent studies of the AG lineage genes in the model legume M. truncatula revealed that two AG homologs MtAGa and MtAGb are present in the M. truncatula genome and redundantly control the C-function activity in the third and fourth floral whorls during floral development (Serwatowska et al., 2014). In this study, we further investigated the genetic interaction of the duplicated MtAG homologs and uncovered interesting aspects of the regulatory control of floral organ identity as well as floral zygomorphy in M. truncatula.
By characterizing the MtAGa and MtAGb loss-of-function insertional (Tnt1) mutants, we confirmed that both MtAGa and MtAGb shows a conserved C function activity, but with a little functional differentiation in determining the floral organ identity and FM determinacy. In consistent with previous description (Serwatowska et al., 2014), the mtaga mutants show partial conversion of stamens to petaloid structures, whereas serious defects in carpel fusion and ovule formation were observed at the early stage, leading to split carpels and producing abnormal pods with reduced seed number (Figures 1, 5B and Supplementary Figures S1A,B). In contrast, the mtagb mutants produce flowers that display homeotic transformations predominantly in the third whorl (Figures 3, 5C). Loss-of-function of MtAGb leads to an incomplete fusion of staminal tubes with weak stamen-petaloid conversion and occasionally resulting in split carpels (Figures 3K,L, 5C, 6E). These phenotypic differences between mtaga and mtagb mutants indicated that MtAGa may contribute more to carpel and ovule identity than MtAGb, whereas MtAGb appears to play a more important role in stamen identity than MtAGa. Besides, mutation of MtAGa causes extra structure formation at the base of carpels, which was not observed in the mtagb mutant (Figures 1I,L, 5B), suggesting that MtAGa may play an important role in FM determinacy of M. truncatula.
The conserved function and subfunctionalization of MtAGa and MtAGb in controlling floral organ development may be partially explained by their spatial and temporal expression profile. qRT-PCR analysis revealed that both MtAGa and MtAGb predominantly express in floral tissues of stamens, carpels and ovules although MtAGa and MtAGb exhibit different expression levels (Figures 4A,B). In agreement with this result, previous northern blot and in situ hybridization analysis also indicated that the expression of both paralogs is distributed uniformly in whorls 3 and 4, while MtAGb shows a stronger signal than MtAGa (Serwatowska et al., 2014). Moreover, compared to the throughout expression of MtAGa in the whole FM, MtAGb transcript was observed in the region of the common primordia that will give rise to stamens at early development stage (Serwatowska et al., 2014). This specific expression is consistent with the notion that MtAGa and MtAGb are redundantly implicated in specification of the third and fourth whorls while MtAGb contributes more to stamen identity.
In Arabidopsis, AGAMOUS establishes the FM determinacy by repressing the key regulator of stem cell homeostasis WUSCHEL (Lenhard et al., 2001; Ming and Ma, 2009; Liu et al., 2011). Because mtag mutants not only show defects in stamen and carpel development but also exhibit serious indeterminate flower phenotypes, we wondered whether this would be the case in M. truncatula as well. qRT-PCR analysis showed that the transcript level of MtWUS (Chen et al., 2009) is significantly upregulated in all mtag mutants excepting the double heterozygous mtaga/+ mtagb-2/+ (Figure 8G). Consistent with the FM termination determinacy phenotypes, the mtaga mtagb-2 double mutant has the highest transcript levels of MtWUS, whereas the mtaga mtagb-2/+ and mtaga/+ mtagb-2 have intermediate levels, and the mtaga and mtagb-2 single mutants have low levels of MtWUS transcript (Figure 8G). Although the function of MtWUS in meristem maintenance is unclear at this point, our data suggested that the molecular mechanism of FM determinacy through AG-mediated repression of WUS might be also conserved in M. truncatula.
In agreement with previous results that MtAGa and MtAGb show a C-function activity, the mtaga mtagb-2 double mutant exhibits a complete loss of C function phenotype. The third whorl (stamens) and the fourth whorl (carpel) are entirely replaced by petals in the mtaga mtagb-2 double mutant (Figures 5G, 6M). However, in contrast to the single and double mutants, mtaga mtagb-2/+ and mtaga/+ mtagb-2 plants show an intermediate but distinct floral phenotypes (Figures 5E,F, 6I–L), indicating a clear additive effect. Interestingly, despite the floral phenotypes of the double heterozygote mtaga/+ mtagb-2/+ mutant are generally similar to that of wild-type, occasional split carpels were observed in mtaga/+ mtagb-2/+, suggesting a fine tuning expression of MtAGa and MtAGb in regulating carpel and stamen development. These results support the quartet model of floral organ specification (FQM) that floral homeotic gene function varies with respect to the amount of gene product required for different organ-specific tetrameric complexes (Theissen et al., 2016). The correlation between the level of MtAGa and MtAGb accumulation and the alteration of floral organ development suggests that the duplicated AGAMOUS homologs control M. truncatula C-function in a dose-depend manner.
In Arabidopsis, the A function genes (AP1 and AP2) and C function gene (AG) antagonize each other, enforcing proper domains of activity (Bowman et al., 1989, 1991; Yanofsky et al., 1990). In consistency with this scenario, molecular analysis of mtag mutants revealed that the transcript levels of three putative A-function genes MtPIM, MtAP2a and MtAP2b are significantly upregulated in the mtaga mtagb-2 double mutant (Figure 8A). Similarly, we also found that loss-of-function of MtAGa and MtAGb leads to significantly increased expression of a subset of floral homeotic B genes MtPI and MtTM6 (Figure 8B), which is in agreement with previous report that floral homeotic C-function genes can repress the expression of specific B-function genes in California poppy (Yellina et al., 2010). These results indicated that this type of C-function-dependent regulation of A- and B- function genes may also be conserved in M. truncatula. In contrast to the upregulation of A- and B- function genes, the putative D-function genes MtSTK and MtSHP are significantly downregulated in mtaga mtagb-2/+, mtaga/+ mtagb-2 and mtaga mtagb-2 mutants (Figure 8D), which may explain the serious defects of ovule formation in above mutants. However, compared to wild-type, the expression of putative E function genes MtSEP1-5 is not altered in the mtaga mtagb-2 double mutant (Figure 8E), suggesting no direct regulation between MtAGs and MtSEPs at the transcriptional level. This is conceivable because the expression pattern of C function genes is not altered in the Arabidopsis sep1 sep2 sep3 triple mutants as well, although the C and E function proteins form a protein complex postulated for stamen and carpel identity in FQM (Pelaz et al., 2000; Theissen et al., 2016).
It has been reported that CYCLOIDEA (CYC)-like TCP genes play key roles in dorsoventral differentiation of zygomorphic flowers in Papilionoideae legumes (Luo et al., 1996; Citerne et al., 2006; Feng et al., 2006; Wang et al., 2008; Xu et al., 2013). Considering the phenotype of multiple vexillum petals in flowers of the mtaga mtagb-2 double mutant, we postulated that the expression of M. truncatula CYC homologs may be altered in mtaga mtagb-2. qRT-PCR analysis revealed that the expression of all three MtCYC homologs is significantly upregulated in the mtaga mtagb-2 double mutant (Figure 8F), suggesting that the abnormal petal shape variation observed in mtag mutants may be caused by the ectopic expression of MtCYC genes. The hypothesis is further supported by findings in Lotus japonicas that ectopic expression of CYC-like genes LjCYC1 and LjCYC2 leads to the transformation of lateral and ventral petals to vexilla (Xu et al., 2013). These results indicated that loss-of-function of C function genes in M. truncatula may affect the flower dorsoventral differentiation by ectopically expressing MtCYC genes, although the direct/indirect regulation between MtAG and MtCYC genes remains to be elucidated. Further investigation of the genetic interaction between MtAG and MtCYC genes may add new insights into understanding the molecular mechanism underlying the floral morphogenesis in M. truncatula.
Taken together, our study analyze the functional conservation and diversification of the M. truncatula duplicated AGAMOUS homolog genes in regulating floral development and provide information for understanding the coordination of floral organ identity factors and floral dorsoventral identity regulators in determining flower morphogenesis of Papilionoideae legumes.
BZ, HuL, LN, and HaL designed the research. BZ, HuL, and LN performed the experiments. JW and KM contributed analytical tools. BZ, HuL, XW, YP, LN, and HaL analyzed the data. BZ, LN, and HaL wrote the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31570309 and 31770347), Fundamental Research Funds for Central Non-profit Scientific Institution (Y2018LM04), Xinjiang Key R&D Program (2018B01006-3 and 2018E02053), and the Capital Science and Beijing Municipal Science and Technology Commission (lj201812). Generation and screening of M. truncatula Tnt1 lines were supported by National Science Foundation, United States (DBI 0703285 and IOS 1127155) and in part by the Noble Research Institute.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would prefer to thank Lin and Niu lab members for their assistance and support throughout this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00854/full#supplementary-material
Angenent, G. C., Franken, J., Busscher, M., van Dijken, A., van Went, J. L., Dons, H. J., et al. (1995). A novel class of MADS box genes is involved in ovule development in petunia. Plant Cell 7, 1569–1582. doi: 10.1105/tpc.7.10.1569
Benlloch, R., d’Erfurth, I., Ferrandiz, C., Cosson, V., Beltran, J. P., Canas, L. A., et al. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142, 972–983. doi: 10.1104/pp.106.083543
Benlloch, R., Roque, E., Ferrandiz, C., Cosson, V., Caballero, T., Penmetsa, R. V., et al. (2009). Analysis of B function in legumes: PISTILLATA proteins do not require the PI motif for floral organ development in Medicago truncatula. Plant J. 60, 102–111. doi: 10.1111/j.1365-313X.2009.03939.x
Benloch, R., Navarro, C., Beltran, J. P., and Canas, L. A. (2003). Floral development of the model legume Medicago truncatula: ontogeny studies as a tool to better characterize homeotic mutations. Sex. Plant Reprod. 15, 231–241.
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell 1, 37–52. doi: 10.1105/tpc.1.1.37
Bowman, J. L., Smyth, D. R., and Meyerowitz, E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20.
Bradley, D., Carpenter, R., Sommer, H., Hartley, N., and Coen, E. (1993). Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena locus of Antirrhinum. Cell 72, 85–95. doi: 10.1016/0092-8674(93)90052-R
Chen, S. K., Kurdyukov, S., Kereszt, A., Wang, X. D., Gresshoff, P. M., and Rose, R. J. (2009). The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta 230, 827–840. doi: 10.1007/s00425-009-0988-1
Cheng, X., Wang, M., Lee, H. K., Tadege, M., Ratet, P., Udvardi, M., et al. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 201, 1065–1076. doi: 10.1111/nph.12575
Citerne, H. L., Pennington, R. T., and Cronk, Q. C. (2006). An apparent reversal in floral symmetry in the legume Cadia is a homeotic transformation. Proc. Natl. Acad. Sci. U.S.A. 103, 12017–12020. doi: 10.1073/pnas.0600986103
Coen, E. S., and Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Colombo, L., Franken, J., Koetje, E., van Went, J., Dons, H. J., Angenent, G. C., et al. (1995). The petunia MADS box gene FBP11 determines ovule identity. Plant Cell 7, 1859–1868. doi: 10.1105/tpc.7.11.1859
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., and Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Dong, Z. C., Zhao, Z., Liu, C. W., Luo, J. H., Yang, J., Huang, W. H., et al. (2005). Floral patterning in Lotus japonicus. Plant Physiol. 137, 1272–1282. doi: 10.1104/pp.104.054288
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15, 2603–2611. doi: 10.1105/tpc.015123
Feng, X., Zhao, Z., Tian, Z., Xu, S., Luo, Y., Cai, Z., et al. (2006). Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. U.S.A. 103, 4970–4975. doi: 10.1073/pnas.0600681103
Goto, K., and Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Jack, T., Brockman, L. L., and Meyerowitz, E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. doi: 10.1016/0092-8674(92)90144-2
Jofuku, K. D., den Boer, B. G., Van Montagu, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Lenhard, M., Bohnert, A., Jurgens, G., and Laux, T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. doi: 10.1016/S0092-8674(01)00390-7
Lin, H., Wang, R., Qian, Q., Yan, M., Meng, X., Fu, Z., et al. (2009). DWARF27, an iron-containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21, 1512–1525. doi: 10.1105/tpc.109.065987
Liu, X., Kim, Y. J., Muller, R., Yumul, R. E., Liu, C., Pan, Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23, 3654–3670. doi: 10.1105/tpc.111.091538
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. doi: 10.1038/383794a0
Mandel, M. A., Gustafson-Brown, C., Savidge, B., and Yanofsky, M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. doi: 10.1038/360273a0
Meyerowitz, E. M., Smyth, D. R., and Bowman, J. L. (1989). Abnormal flowers and pattern formation in floral development. Development 106, 209–217.
Ming, F., and Ma, H. (2009). A terminator of floral stem cells. Genes Dev. 23, 1705–1708. doi: 10.1101/gad.1834409
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., and Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Roque, E., Fares, M. A., Yenush, L., Rochina, M. C., Wen, J., Mysore, K. S., et al. (2016). Evolution by gene duplication of Medicago truncatula PISTILLATA-like transcription factors. J. Exp. Bot. 67, 1805–1817. doi: 10.1093/jxb/erv571
Roque, E., Serwatowska, J., Cruz Rochina, M., Wen, J., Mysore, K. S., Yenush, L., et al. (2013). Functional specialization of duplicated AP3-like genes in Medicago truncatula. Plant J. 73, 663–675. doi: 10.1111/tpj.12068
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., and Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931–936. doi: 10.1126/science.250.4983.931
Serwatowska, J., Roque, E., Gomez-Mena, C., Constantin, G. D., Wen, J., Mysore, K. S., et al. (2014). Two euAGAMOUS genes control C-function in Medicago truncatula. PLoS One 9:e103770. doi: 10.1371/journal.pone.0103770
Shoemaker, R. C., Schlueter, J., and Doyle, J. J. (2006). Paleopolyploidy and gene duplication in soybean and other legumes. Curr. Opin. Plant Biol. 9, 104–109. doi: 10.1016/j.pbi.2006.01.007
Tadege, M., Wen, J., He, J., Tu, H., Kwak, Y., Eschstruth, A., et al. (2008). Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 54, 335–347. doi: 10.1111/j.1365-313X.2008.03418.x
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/S1369-5266(00)00139-4
Theissen, G., Melzer, R., and Rumpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Tucker, S. C. (2003). Floral development in legumes. Plant Physiol. 131, 911–926. doi: 10.1104/pp.102.017459
Vandenbussche, M., Zethof, J., Souer, E., Koes, R., Tornielli, G. B., Pezzotti, M., et al. (2003). Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15, 2680–2693. doi: 10.1105/tpc.017376
Wang, Z., Luo, Y., Li, X., Wang, L., Xu, S., Yang, J., et al. (2008). Genetic control of floral zygomorphy in pea (Pisum sativum L.). Proc. Natl. Acad. Sci. U.S.A. 105, 10414–10419. doi: 10.1073/pnas.0803291105
Wellmer, F., Graciet, E., and Riechmann, J. L. (2014). Specification of floral organs in Arabidopsis. J. Exp. Bot. 65, 1–9. doi: 10.1093/jxb/ert385
Xu, S., Luo, Y., Cai, Z., Cao, X., Hu, X., Yang, J., et al. (2013). Functional diversity of CYCLOIDEA-like TCP genes in the control of zygomorphic flower development in Lotus japonicus. J. Integr. Plant Biol. 55, 221–231. doi: 10.1111/j.1744-7909.2012.01169.x
Yanofsky, M. F., Ma, H., Bowman, J. L., Drews, G. N., Feldmann, K. A., and Meyerowitz, E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Yarce, J. C., Lee, H. K., Tadege, M., Ratet, P., and Mysore, K. S. (2013). Forward genetics screening of Medicago truncatula Tnt1 insertion lines. Methods Mol. Biol. 1069, 93–100. doi: 10.1007/978-1-62703-613-9_8
Keywords: AGAMOUS homologs, C function genes, floral morphogenesis, functional diversification, Medicago truncatula, Papilionoideae
Citation: Zhu B, Li H, Wen J, Mysore KS, Wang X, Pei Y, Niu L and Lin H (2018) Functional Specialization of Duplicated AGAMOUS Homologs in Regulating Floral Organ Development of Medicago truncatula. Front. Plant Sci. 9:854. doi: 10.3389/fpls.2018.00854
Received: 14 February 2018; Accepted: 01 June 2018;
Published: 31 July 2018.
Edited by:
Amy Litt, University of California, Riverside, United StatesReviewed by:
Zhipeng Liu, Lanzhou University, ChinaCopyright © 2018 Zhu, Li, Wen, Mysore, Wang, Pei, Niu and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lifang Niu, bml1bGlmYW5nQGNhYXMuY24= Hao Lin, bGluaGFvQGNhYXMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.