
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 June 2018
Sec. Plant Development and EvoDevo
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00835
One of the crucial steps in the life cycle of angiosperms is the development of carpels. They are the most complex plant organs, harbor the seeds, and, after fertilization, develop into fruits and are thus an important ecological and economic trait. CRABS CLAW (CRC), a YABBY protein and putative transcription factor, is one of the major carpel developmental regulators in A. thaliana that includes a C2C2 zinc finger and a domain with similarities to an HMG box. CRC is involved in the regulation of processes such as carpel fusion and growth, floral meristem termination, and nectary formation. While its genetic interactions with other carpel development regulators are well described, its biochemical properties and molecular way of action remain unclear. We combined Bimolecular Fluorescence Complementation, Yeast Two-Hybrid, and Yeast One-Hybrid analyzes to shed light on the molecular biology of CRC. Our results showed that CRC dimerizes, also with other YABBY proteins, via the YABBY domain, and that its DNA binding is mainly cooperative and is mediated by the YABBY domain. Further, we identified that CRC is involved in floral meristem termination via transcriptional repression while it acts as a transcriptional activator in nectary development and carpel fusion and growth control. This work increases our understanding on how YABBY transcription factors interact with other proteins and how they regulate their targets.
Floral development and especially carpel development is one of the most crucial steps in the life cycle of flowering plants, since it is the prerequisite for reproduction and, subsequently, the formation of seeds and fruits. The orchestration of carpel development requires a large number of physically and genetically interacting transcription factors that are members of a several different transcription factor families (Reyes-Olalde et al., 2013). One central regulator of carpel development genetically linked with many others is CRABS CLAW, a YABBY transcription factor. Most members of this family act in regulatory networks together with members of the HD-ZIP III family to regulate adaxial-abaxial polarity in lateral plant organs, e.g., leaves, carpels, and ovules (Sablowski, 2015; Tatematsu et al., 2015).
The plant-specific YABBY proteins with only six members in Arabidopsis thaliana [CRC, FILAMENTOUS FLOWER (FIL), INNER NO OUTER (INO), YABBY2, 3, and 5] are transcription factors with a C2C2 zinc finger domain and the family specific YABBY domain, a modified HMG box with a basic helix-loop-helix secondary structure. Identified by Bowman and Smyth (1999), it was named after the crabs claw like appearance of the apically unfused carpels of the crc-1 mutant. Han et al. (2012) showed that CRC is able to bind to DNA, in particular promoter regions of 3-KETOACYL-COA SYNTHASE 7 and 15 (KCS7 and KCS15), two genes that are involved in the synthesis of very-long-chain fatty acids which are then used as signaling molecules or in cuticular wax synthesis (Joubès et al., 2008). However, CRC’s DNA binding motif or target genes involved in carpel developmental processes remain unknown.
The crc-1 mutant shows shorter and wider gynoecia with the two carpels being unfused at the apex and flowers lacking nectaries (Bowman and Smyth, 1999). The lack of carpel fusion is intensified in double mutants of SPATULA (SPT) and CRC in which the two carpels fuse only at their base (Alvarez and Smyth, 1999, 2002). All YABBY genes are expressed in flowers but only CRC and INO’s expressions are restricted to flowers, to carpels and ovules, respectively. CRC and INO regulate, in combination with KANADI (KAN) genes and ETTIN (ETT), adaxial-abaxial patterning of the carpels and of the developing ovules by specifying the abaxial side (reviewed in Sablowski, 2015). Furthermore, the action of adaxial regulators, like members of the HD-ZIP III family is suppressed, maybe by activating the transcription of miR165/166 (Tatematsu et al., 2015).
The crc-1 mutant shows a weak meristem indeterminacy phenotype and thus a mild surplus in carpels (Alvarez and Smyth, 1999; Bowman and Smyth, 1999; Alvarez and Smyth, 2002). A more detailed analysis revealed that CRC acts redundantly with REBELOTE (RBL), ULTRAPETALA 1 (ULT1), and SQUINT (SQN) in floral meristem termination (Prunet et al., 2008). The loss of floral meristem determinacy is at least partially caused by a reduced expression of AGAMOUS (AG) in double mutants of these three genes with CRC (Prunet et al., 2008). While, RBL, ULT1, and SQN have been shown to act epistatic to AG, CRC is a direct target of AG (Gomez-Mena et al., 2005; Lee et al., 2005; Ó’Maoiléidigh et al., 2013). AG and other MADS box transcription factors, like PISTILLATA (PI), APETALA 1 (AP1), and APETALA 3 (AP3) directly bind to the CRC promoter. They either activate, as in the case of AG, or repress as in the case of PI, AP1, and AP3 CRC expression in the developing flower and restrict CRC expression to nectaries and carpels (Gomez-Mena et al., 2005; Lee et al., 2005; Ó’Maoiléidigh et al., 2013).
This regulation of CRC transcription causes a highly complex spatial and temporal expression pattern inside the developing carpels (Bowman and Smyth, 1999; Alvarez and Smyth, 2002). CRC expression starts in stage 6 (staging according to Smyth et al., 1990) and continues to stage 7 –8 in two distinct domains of the carpels: In the epidermis of the carpels, surrounding the circumference, and in four interior stripes adjacent to the developing placenta (Bowman and Smyth, 1999; Alvarez and Smyth, 2002; Lee et al., 2005). The expression in the four stripes forms a basal-apical gradient and decreases in later floral stages, whereas, the epidermal expression ceases in the future replum but is maintained in the valves until the mid of stage 12 (Bowman and Smyth, 1999; Alvarez and Smyth, 2002; Lee et al., 2005).
CRC orthologs from other angiosperms show additional functions suggesting a complex molecular evolution of the protein (Fourquin et al., 2007; Orashakova et al., 2009). The Oryza sativa CRC ortholog DROOPING LEAF (DL) regulates carpel identity, floral meristem termination, and leaf midrib formation (Yamaguchi et al., 2004; Ishikawa et al., 2009). In the basal eudicot Eschscholzia californica, a knock-down of EcCRC leads to a loss of floral meristem determinacy, impaired replum formation, and a reduced seed set, due to malformed placenta tissue (Orashakova et al., 2009). Similar to this, a knock-down of PsCRC in Pisum sativum impaired carpel fusion and the seed set was reduced due to stigma and style malformations (Fourquin et al., 2014). However, while the CRC phenotype and its genetic interactions have received considerable attention over the past decades the molecular functions of CRC protein domains are unknown as well as the molecular mechanism of its action. Here, we assign functions to CRC’s protein domains and show that CRC acts through activating regulatory processes during carpel development.
The full length coding sequence (CDS) of CRC, CRC deletion constructs CRCΔZinc Finger, (CRCΔZF) CRCΔIntermediate (CRCΔIM), CRCΔYABBY (CRCΔYD), and single domains CRC Zinc Finger (CRC-ZF), CRC Intermediate (CRC-IM), CRC YABBY (CRC-YD), CRC-YDΔnuclear localization signal (CRC-YDΔNLS), and CRC-NLS were cloned as EcoRI/BamHI fragments (for primer sequences see Supplementary Table 1, and for length of the individual fragments see Figure 1A and Supplementary Figure 2A) into the N-terminal GFP fusion vector pEGAD (Cutler et al., 2000). The borders of the CRC domains (ZF aa 26–53, IM aa 54–108, YD aa 109–155) are based on the classification of Bowman and Smyth (1999). As Bowman and Smyth (1999) predicted only the secondary structure of the YABBY domain, we performed a secondary structure prediction of full length CRC with REPPER (Jones, 1999; Gruber et al., 2005; Zimmermann et al., 2017) to identify additional secondary structures. To identify CRC’s nuclear localization signal, single amino acids in the core region of CRC’s putative nuclear localization signal were substituted via site directed mutagenesis (Hemsley et al., 1989). The CRC versions encoding for CRC K110T, P111A, P112A, E113G, K114T, K115T, Q116L, and R117T were introduced into the GreenGate system [the plasmid kit used for generation of plant transformation constructs was a gift from Jan Lohmann (Addgene kit # 1000000036)] and tagged with C-terminal GFP under the control of the CaMV 35S promoter (Lampropoulos et al., 2013). The CRC versions were transiently expressed in leaves of 4 weeks old Nicotiana benthamiana plants after infiltration with Agrobacterium tumefaciens GV3101, harboring the respective CRC version. For GreenGate based plasmids, A. tumefaciens GV3101 pSOUP+ was used. Small leaf disks were excised and stained with 1 ng/ml DAPI 2 days after infiltration. Microscopical analysis was performed with the Leica fluorescence microscope DCM5500 (Leica Microsystems GmbH, Wetzlar, Germany) using A4 filter for DAPI fluorescence and L5 filter for GFP fluorescence, or with Leica TCS SP8 confocal laser scanning microscope with excitation at 405 nm for DAPI and 488 nm for GFP, and a detection range of 413 – 460 nm for DAPI fluorescence and of 496 – 569 nm for GFP fluorescence.
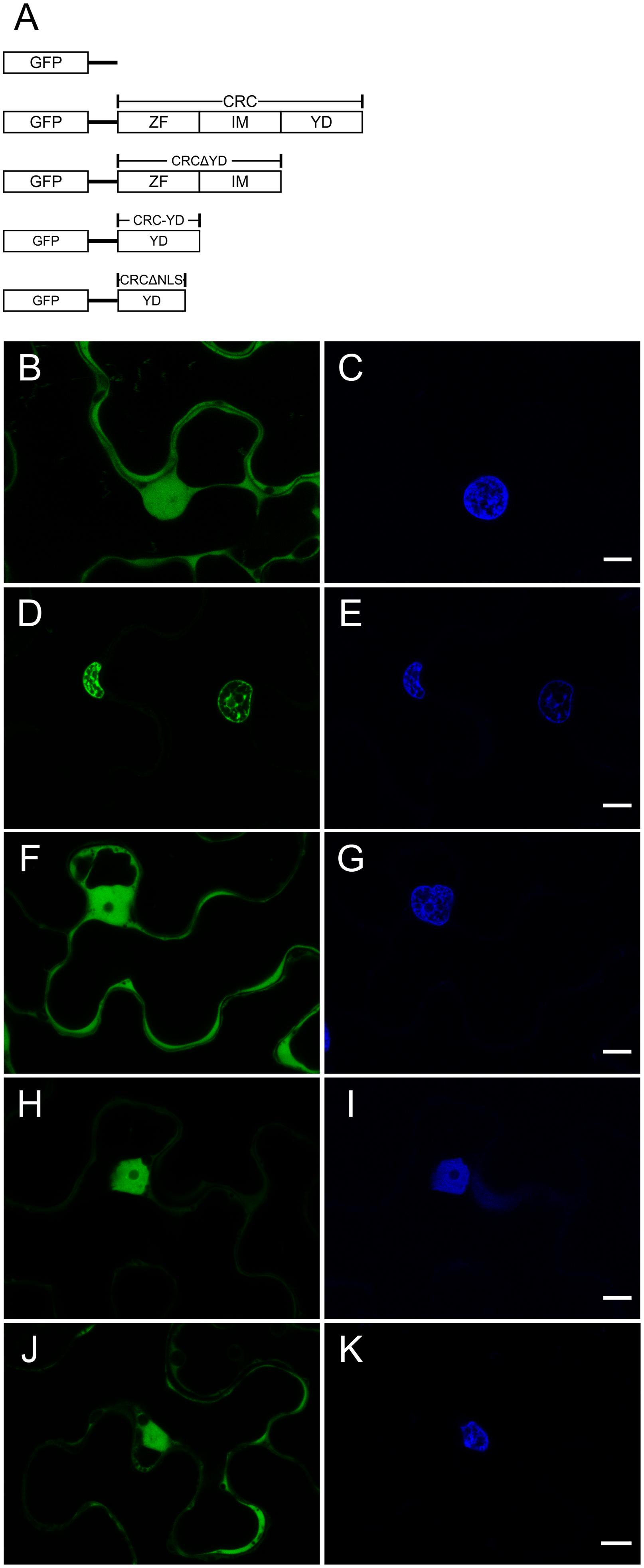
FIGURE 1. Analysis of the intracellular localization of GFP::CRC employing different CRC deletion variants. GFP fusion proteins were detected by CLSM. False colors were assigned to GFP (green, left panels) and DAPI (blue, right panels) which stains DNA. (A) Schematic representation of the GFP-CRC constructs used in this study. (B,C) 35S::GFP (29.26 kDa), (D,E) 35S::GFP::CRC (48.97 kDa). (F,G) 35S::GFP::CRCΔYD (40.75 kDa); (H,I) 35S::GFP::CRC-YD (34.75 kDa); (J,K) 35S::GFP::CRCΔNLS (47.97 kDa). ZF, zinc finger domain; IM, intermediate domain; YD, YABBY domain; NLS, nuclear localization signal. All scale bars represent 10 μm.
Parts of the promoter regions of KCS7 and KCS15 from A. thaliana Ler-0 (Han et al., 2012) were cloned into the Y1H vector pAbAi (Takara Clontech, Saint-Germain-en-Laye, France) using HindIII and SacI restriction sites. The assembled pAbAi proKCS7 and pAbAi proKCS15 constructs were linearized with BstBI and transformed into Saccharomyces cerevisiae Y1HGold according to the Yeast Transformation System 2 manual (Takara Clontech). Clones carrying the desired DNA fragment were screened for auto activation on synthetic uracil dropout medium, supplemented with 100 ng/ml, 150 ng/ml, 200 ng/ml, 500 ng/ml, and 1000 ng/ml Aureobasidin A (Takara Clontech). Bait strain colonies that showed no auto activation were selected and were transformed with prey plasmids: full-length CRC, deletion constructs, and single domains in pGADT7 (Takara Clontech), according to the modified Yeast Transformation System 2 manual. Additionally, three CRC mutant versions were generated via site directed mutagenesis, in which either the zinc finger or the YABBY domain was non-functional, and a combination of both: CRC zinc finger mutated (CRC ZFm), with C26W and C29W; CRC YDm with A121P, N123P, M126P, E129P, R132P, F146P, A149P, N152P, and K155P; and the combination: CRC ZFm/YDm.
Protein interaction tests via Y2H were carried out as described in Lange et al. (2013). The CDS of YABBY 3 (YAB3), INO, and CRC were translationally fused to the GAL4 activation domain (AD) or GAL4 DNA binding domain (BD) by cloning them into pGADT7 and pGBKT7, respectively. The yeast 2-hybrid vectors of FILAMENTOUS FLOWER (FIL), YABBY 2 (YAB2), and YABBY 5 (YAB5) were a kind gift of John Golz (Stahle et al., 2009). Transformation of S. cerevisiae AH109 was carried out as described (Kaufmann et al., 2005). Growth assays on synthetic leucine, tryptophan, and histidine dropout medium (SD-Leu/-Trp/-His), supplemented with 3 mM 3-Amino-1,2,4-triazole (3-AT), and MEL1 based α-galactosidase assay was carried out in triplicates. As positive control, a combination of AD-EcSEIRENA/BD-EcDEFICIENS2 (Lange et al., 2013) was used and a combination of the empty vectors pGADT7/pGBKT7 as negative control.
Bimolecular fluorescence complementation assays to test for homodimerization of CRC were performed as described in Lange et al. (2013). The different CRC variants (CRC, CRCΔZF, CRCΔIM, CRCΔYD, CRC-ZF, CRC-IM, and CRC-YD) were cloned into the respective pNBV vector as described above (Walter et al., 2004; Lange et al., 2013) and the expression cassettes were transferred to the plant transformation vector pMLBART and transformed into A. tumefaciens GV3101. Leaves of 4 weeks old N. benthamiana plants were inoculated with the A. tumefaciens strains, expressing the proteins to be tested, according to Lange et al. (2013). Small leaf disks were excised and stained with 1 μg/ml DAPI 2 days after infiltration. Microscopical analysis was performed with Leica fluorescence microscope DCM5500 using A4 filter for DAPI fluorescence and L5 filter for GFP fluorescence.
The CDS encoding the EDLL domain (Tiwari et al., 2012) and the SRDX domain (Hiratsu et al., 2003) were synthesized as oligonucleotides and introduced into the GreenGate system. Using the GreenGate system, proUBQ10:N-Dummy:CRC:C-Dummy:tUBQ10; proMAS:BASTA:tMAS, proUBQ10:N-Dummy: CRC:SRDX:tUBQ10; proMAS:BASTA:tMAS, and proUBQ10:N-Dummy:CRC:EDLL:tUBQ10; proMAS:BASTA:tMAS were assembled according to Lampropoulos et al. (2013). Plasmids verified by sequencing were transformed into A. tumefaciens GV3101 pSOUP+. All constructs were introduced into A. thaliana Ler-0 plants via floral dip (Davis et al., 2009). Putatively transgenic seeds were grown on soil and selected by spraying with 300 μM Basta (Bayer CropScience Deutschland GmbH, Langenfeld, Germany) 7 days after germination with repetitions on every second day. Basta resistant seedlings were further genotyped by PCR. Expression strength was analyzed via qRT-PCR. Total RNA from leaves and of buds of representative wild type, crc-1, CRC over expression (CRCoe), CRC-SRDX, and CRC-EDLL was isolated in triplicates using the NucleoSpin RNA Plant kit (Macherey-Nagel GmbH & Co., KG, Düren, Germany) and transcribed into cDNA using RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific Inc., Schwerte, Germany) with random hexamer primer. The cDNA was diluted 1:50 and added to Luna master mix (NEB Inc., Frankfurt am Main, Germany), followed by a PCR run using Lightcycler 480 II (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) to detect CRC expression in leaves and KCS7 and KCS15 expression in buds. Afterward, the obtained data was analyzed according to Pfaffl (2001) and Taylor et al. (2010).
The promoter regions of BLADE ON PETIOLE1 (BOP1, AT3G57130), BOP2 (AT2G41370) POLYUBIQUITIN10 (UBQ10, AT4G05320), GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C-2 (GAPDH, AT1G13440), and ELONGATION FACTOR 1 α (EF-1α, AT1G07920) were screened for the presence of the four YABBY binding motifs (Shamimuzzaman and Vodkin, 2013; Franco-Zorrilla et al., 2014), using PlantPAN (Chow et al., 2016).
The CRC protein includes a single C2C2 type zinc finger and the YABBY domain which shows weak similarities to protein domains found in HMG (high mobility group) proteins (Bowman and Smyth, 1999). Secondary structure prediction (Supplementary Figure 1) by REPPER (Gruber et al., 2005; Zimmermann et al., 2017) suggests that the zinc finger (between amino acid position 26–53) is formed by two neighboring α-helices (red) flanked by two β-strands (green). The intermediate segment (between position 54–108) is predicted to contain two β-strands that are followed by an unstructured stretch of amino acids that may serve as flexible linker region and the YABBY domain (between position 109–155) is predicted to include one short and two long α-helices.
The presence of a zinc finger, which may bind DNA, RNA, or proteins (as reviewed in Burdach et al., 2012), suggests that CRC is a transcription factor, a hypothesis supported also by its DNA binding capacity (Han et al., 2012). We aimed to support this hypothesis by studying CRC’s intracellular localization (Figure 1). The full-length open reading frame (ORF) of CRC, several deletion constructs, and the individual domains were translationally fused to GFP and driven by the CaMV 35S (35S) promoter (Figure 1A and Supplementary Figure 2A). These constructs were transiently expressed in N. benthamiana epidermal cells to observe the intracellular localization of the encoded protein fusions by confocal-laser-scanning (Figure 1) and fluorescence microscopy (Supplementary Figure 2). As a control, native GFP driven by the 35S promoter was found to be present in both, the cytoplasm and nuclei (Figure 1B), as a comparison with the DAPI staining indicates (Figure 1C) but was absent in other organelles like the vacuole. In contrast, GFP::CRC was localized exclusively to the nuclei of the leaf epidermal cells (Figures 1D,E), supporting the hypothesis that CRC acts in the nucleus.
To identify the protein domain required for nuclear localization of CRC, we truncated the protein and used both, individual domains and deletion constructs for further localization studies (Figure 1A and Supplementary Figure 2A). The deletion of the YABBY domain led to an accumulation of GFP::CRCΔYD in the cytoplasm, similar to native GFP (Figures 1F,G). The removal of the zinc finger or of the serine-proline rich intermediate domain from the full-length protein instead resulted in the respective fusion protein being confined to the nucleus with no fluorescence detected in the cytoplasm (Supplementary Figures 2B–E). The localization of the individual zinc finger and of the intermediate domains showed the same subcellular localization as native GFP, throughout the cytoplasm and the nuclei (Supplementary Figures 2F–I).
Since the deletion of the YABBY domain increased the amount of GFP::CRCΔYD in the cytoplasm, we further analyzed the YABBY domain. When GFP was fused to only the YABBY domain, the fusion protein showed a restored nuclear localization (Figures 1H,I). This suggests that CRC’s NLS is located in the YABBY domain. With the position of the NLS restricted to the YABBY domain, we compared the predictions about the position of the NLS of Bowman and Smyth (1999) with known NLS consensus sequences (Kosugi et al., 2008) and identified eight possible amino acid residues in the N-terminal part of the YABBY domain that might be part of the NLS. When we removed these eight residues from the YABBY domain (GFP::CRCΔNLS), the exclusive nuclear transport stopped and the fusion protein accumulated in the cytoplasm like the native GFP (Figures 1J,K). However, if only these eight amino acid residues were fused to GFP (Supplementary Figures 2J,K), the distribution of the fusion protein was identical with native GFP, and thus, the eight residues alone, were insufficient to induce exclusive nuclear import. We were then interested in analyzing the NLS sequence in more detail and produced single amino acid substitutions of these eight amino acids. The eight CRC versions with single substitutions (CRC K110T, P111A, P112A, E113G, K114T, K115T, Q116L, and R117T) were fused to GFP and transiently expressed in N. benthamiana leaves. However, none of the single amino acid substitutions changed the localization of the respective CRC fusion variants and they all remained inside the nuclei (Supplementary Figure 3).
In summary we find that the nuclear localization of the CRC protein depends on the presence of the YABBY domain. However, a conventional NLS as predicted by Kosugi et al. (2008) which can be abolished by single amino acid exchanges does not reside in the YABBY domain.
As other transcription factors with a HMG box or with only one zinc finger domain are known to form homo- and heterodimers (Yanagisawa, 1997; Sanchez-Giraldo et al., 2015), we hypothesize that CRC also forms dimeric complexes. We thus performed a BiFC with full-length CRC, CRC deletion constructs, and CRC single domains to elucidate the formation of homodimers and to identify the necessary domain for this protein–protein interaction. Additionally, an Y2H analysis was carried out in which we combined full-length AD-CRC/BD-CRC. A restoration of YFP fluorescence in the nuclei of N. benthamiana leaf cells by the formation of CRC homodimers was only observed when YFPC and YFPN tag were fused to CRC in the same orientation (Figures 2A,B). The same restriction in tag orientation was found in the other constructs such that homodimer formation was also observed when the deletion constructs CRCΔZF (Figures 2C,D) and CRCΔIM (Figures 2E,F) were combined, respectively. Here, the reconstituted YFP was localized to the nuclei, similar to the full-length CRC combinations, as the comparison with DAPI staining indicates. In contrast, combinations with the other single domains (Supplementary Figure 4) or with CRCΔYD (Figures 2G,H) failed to restore YFP fluorescence. Vice versa, only YABBY domain combinations were able to reconstitute the YFP signal when the individual domains were tested (Figures 2I,J). This indicates that the YABBY domain harbors the protein-protein interaction site. However, this YABBY domain mediated homodimerization did not occur in the Y2H analysis, where the combination of AD-CRC/BD-CRC was neither able to grow on histidine drop-out medium (SD- Leu/Trp/His), nor produced a blue staining after incubation with X-α-Gluc (Supplementary Figure 5 and Supplementary Table 2).
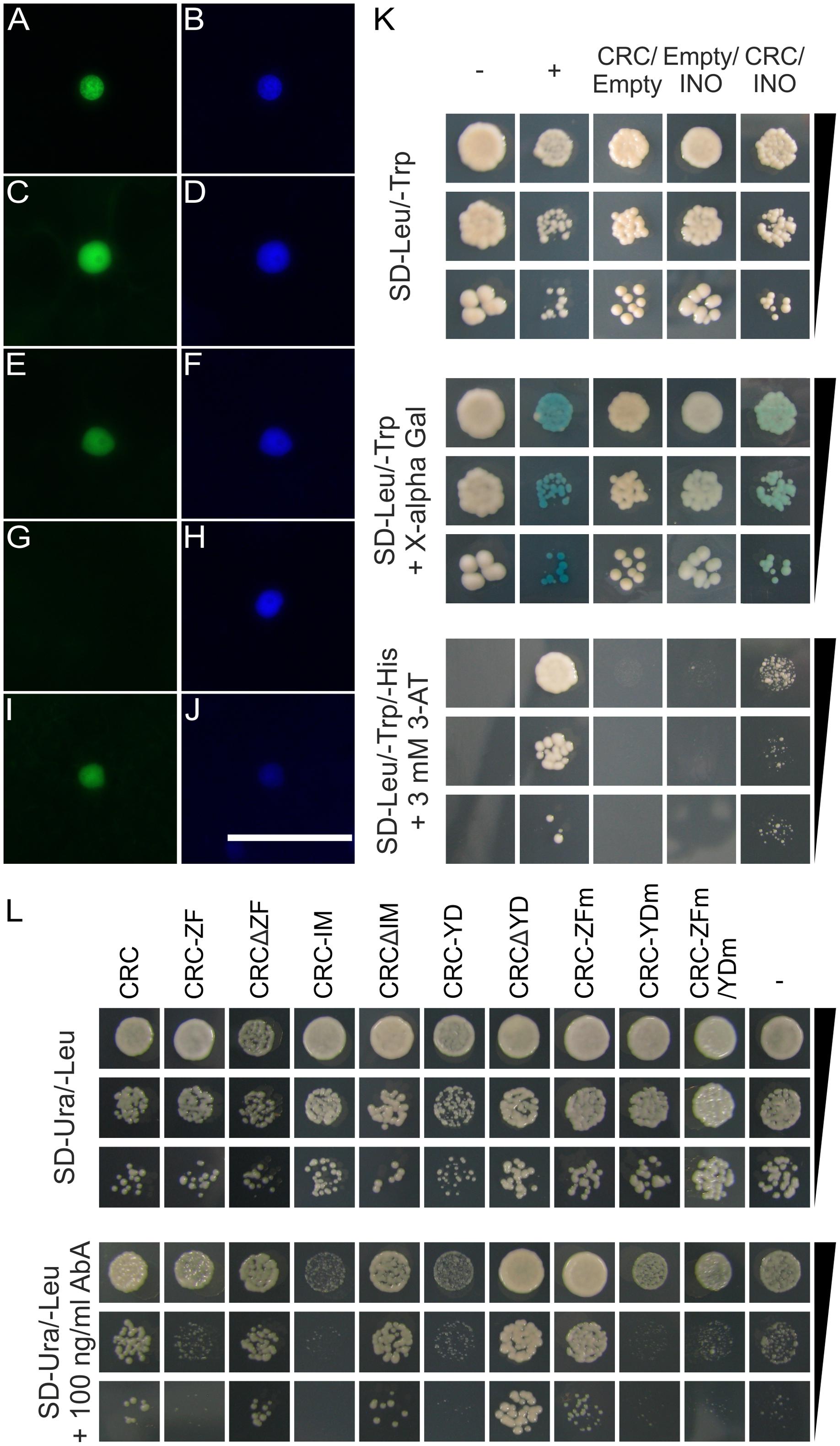
FIGURE 2. Protein interaction analysis of CRC and analysis of interacting domains. (A–J) Protein interaction analysis by BiFC using multiple CRC versions. YFPC and YFPN tagged full-length CRC, deletion version of CRC, and single domains were detected by fluorescence microscopy. (A) Combination CRC-YFPC-/CRC-YFPN visualizing the YFP signal, (B) DAPI staining of the cell shown in (A). (C) Combination YFPC-CRCΔZF/YFPN-CRCΔZF visualizing the YFP signal, (D), DAPI staining of the cell shown in (C). (E) Combination YFPC-CRCΔIM/YFPN-CRCΔIM visualizing the YFP signal. (F) DAPI staining of the cell shown in (E). (G) Combination YFPC-CRCΔYD/YFPN-CRCΔYD visualizing the YFP signal. (H) DAPI staining of the cell shown in (G). (I) Combination YFPC-CRC-YD/YFPN-CRC-YD visualizing the YFP signal. (J) DAPI staining of the cell shown in (I). ZF, zinc finger domain; IM, intermediate domain; YD, YABBY domain. Scale bar represents 50 μm. (K) Y2H analysis of CRCs interaction with INO. Yeast cell suspensions of the respective test strains with an OD600 of 0.1, 0.01, and 0.001 plated on SD-Leu/-Trp medium and stained with X-α-Gluc after 5 days of incubation and on SD-Leu/-Trp/-His + 3mM 3-AT. As positive control, a combination of AD-EcSEI/BD-EcDEF2 (Lange et al., 2013) was used and a combination of the empty vectors pGADT7/pGBKT7 as negative control. (L) CRC’s DNA binding capabilities in an Y1H analysis. The S. cerevisiae proKCS15 reporter strain was transformed with full length CRC, single domains, deletion constructs, and mutant versions, fused to the activation domain of GAL4. Yeast cell suspensions of the respective test strains with an OD600 of 0.1, 0.01, and 0.001 plated on SD-Ura/-Leu and on SD-Ura/-Leu + 100 ng/ml AbA. As negative controls, the proKCS15 bait strain was transformed with an empty pGADT7 vector.
The majority of the YABBY proteins is able to form homo- and heterodimers with other YABBY proteins (Stahle et al., 2009) and we were interested to see if this is also the case for CRC. We performed an Y2H analysis of CRC and the remaining five A. thaliana YABBY proteins (FIL, YAB2, YAB3, INO, and YAB5) (Figure 2K, Supplementary Figure 5, and Supplementary Table 2), by combining AD-CRC with the respective BD-YABBY constructs and vice versa. Interestingly, only the combination AD-CRC/BD-INO showed an interaction indicated by weak growth on the SD-Leu/Trp/His + 3-AT plates and on the blue staining in the α-Gal assay (Figure 2K).
In addition to the nuclear localization of a protein, the ability to bind DNA is a requirement for its action as a transcription factor. In silico analysis by Bowman and Smyth (1999), predicted that CRC exhibits two putative DNA binding domains: The N-terminal C2C2 zinc finger domain and the C-terminal YABBY domain, which has a helix-loop-helix structure with similarities to a HMG box. We used partial promoter regions of KCS7 and KCS15, identified by Han et al. (2012), as direct targets of CRC, in an Y1H experiment to analyze which protein domain mediates the DNA binding. However, the proKCS7 bait strain was discarded due to a high degree of autoactivation.
Full-length CRC, translationally fused to the activation domain of the yeast transcription factor GAL4, was able to bind to the promoter region of KCS15 and enabled colony growth (colonies with > 1 mm) (Figure 2L) on SD-Ura/-Leu medium, supplemented with 100 ng/ml Aureobasidin A. When transforming the proKCS15 bait strain with the respective single domains of CRC in this Y1H assay, well grown colonies (>1 mm) were observed only in the following combinations of the deletion constructs and proKCS15 (Figure 2L): intermediate domain plus YABBY domain (CRCΔZF); zinc finger plus YABBY domain (CRCΔIM): and zinc finger plus intermediate (CRCΔYD). In contrast, no interaction could be shown when only the single domains (proKCS15/CRC-ZF, proKCS15/CRC-IM, and proKCS15/CRC-YD) are used in the analysis, suggesting that single domains alone are unable to mediate DNA binding. Interestingly, we did not observe differences in KCS7 or KCS15 gene expression in buds between wild type and crc-1 plants (Supplementary Figure 8).
To further analyze this effect, we mutated CRC with site directed mutagenesis and rendered either the zinc finger (CRC-ZFm) or the YABBY domain (CRC-YDm) non-functional by changing two and nine amino acids, respectively. A third CRC version comprised a fusion of the two non-functional domains (CRC-ZFm/YDm). When these CRC versions were introduced into the proKCS15 bait strain, only the CRC version with a mutagenized zinc finger (CRC-ZFm) grew well on the selection medium. In summary, we show that the intermediate domain does not participate in DNA binding. For sequence specific DNA binding, the YABBY and ZF domains are required, but if the ZF three-dimensional structure is abolished, DNA binding remains unaffected. However, if the YABBY domain’s 3D structure is destroyed, DNA binding is not possible anymore suggesting a more prominent role for the YABBY domain in CRC-DNA binding.
CRC’s role as a transcription factor relies not only on its ability to bind to DNA but also on the effect of this interaction. We were interested to answer the question if CRC acts as an activator or repressor of transcription or if CRC has a dual role in which it represses some developmental processes but activates others. Thus, CRC was transformed into a constitutive repressor of transcription by translational fusion of the SRDX domain to the CRC CDS. Consequently, molecular functions requiring transcriptional activation of target genes cannot be carried out by the fusion protein resulting in a mutant phenotype (Figure 3). Conversely, the translational fusion of the EDLL domain converted CRC in a constitutive activator of transcription, abolishing repressive function of CRC. The respective fusion constructs, driven by A. thaliana UBQ10 promoter were transformed into A. thaliana Ler-0 and the plants were genotyped. For phenotypical analysis, 10 gynoecia of each of the 10 independent transgenic lines, except for CRCoe with only three transgenic lines and 30 analyzed flowers, were compared to equal numbers of Ler-0 and crc-1 gynoecia (Figure 3). Wild type gynoecia had an average length of 2.40 ± 0.44 mm (Figure 3C), similar to CRCoe gynoecia (2.45 ± 0.21), whereas crc-1 gynoecia were significantly shorter (1.92 ± 0.28 mm). Both, CRC-SRDX and CRC-EDLL, were significantly shorter than wild type gynoecia, with 0.45 ± 0.04 mm and 2.03 ± 0.24 mm respectively. Interestingly, the CRC-SRDX lines developed gynoecia even shorter than the crc-1 plants. Gynoecia of crc-1 (0.48 ± 0.04 mm), CRC-SRDX (0.45 ± 0.04 mm), and CRCoe (0.44 ± 0.02) plants were wider than gynoecia of wild type plants (0.41 ± 0.04 mm) and of CRC-EDLL plants with only two carpels (0.42 ± 0.05 mm) (Figure 3D). Additional to these growth parameters, the presence or absence of other known crc-1 phenotypes (carpel fusion, nectary formation, and the presence of additional carpels) was analyzed in the three transgenic lines (Figure 3E). All analyzed gynoecia of wild type plants were completely fused in their apical region, exhibited nectaries at their base and had two carpels. In contrast, all gynoecia of crc-1 plants were apically unfused, and failed to develop nectaries. Additionally, 4 % of the analyzed crc-1 gynoecia consisted of more than two carpels. CRCoe gynoecia were identical with wild type gynoecia in terms of carpel number, apical fusion, and presence of nectaries. CRC-SRDX gynoecia failed in 93% to complete the apical fusion, their septa were partially unfused, and they showed a reduced amount of style and stigma tissue, similar to the crc-1 phenotype. Moreover, ovules protruded from the inside of the carpels (Figure 3G) and nectaries were present in only 6% of the analyzed gynoecia but additional carpels were not observed.
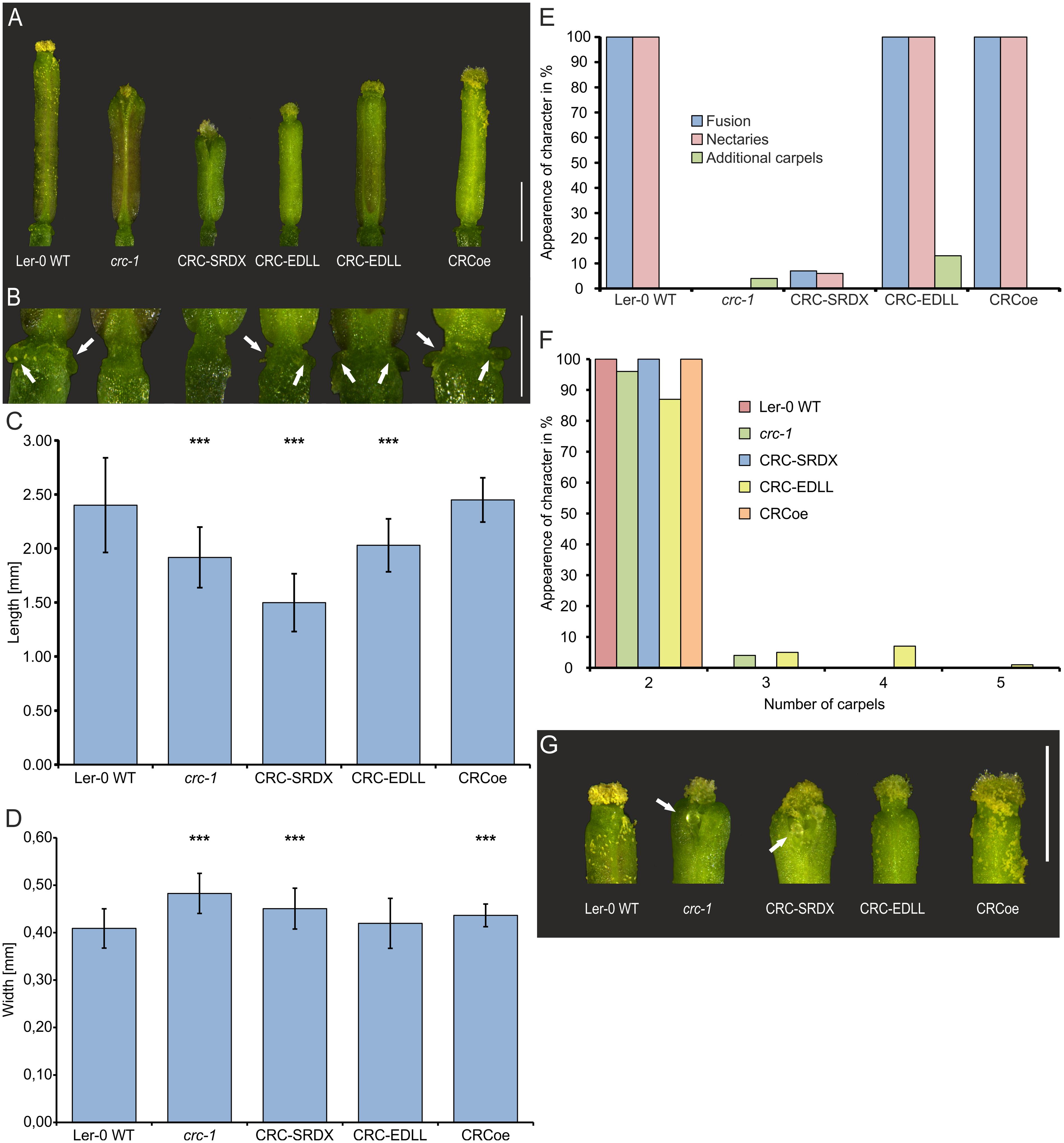
FIGURE 3. Phenotypic analysis of CRCoe, CRC-SRDX, and CRC-EDLL expressing A. thaliana Ler-0 plants. (A) Representative gynoecia of Ler-0 wild type, crc-1, proUBQ10:CRC (CRCoe), proUBQ10:CRC:SRDX, and proUBQ10:CRC:EDLL plants. Scale bar represents 1 mm. (B) Magnification of the gynophore region of the exemplary gynoecia with arrows highlighting the nectaries. Scale bar represents 500 μm. Statistical analysis of gynoecium length (C), width (D), a summary of other described defects of the crc-1 phenotype (E), and the number of carpels in the analyzed gyneocia of the four plant lines. In each line, except for CRCoe (n = 30), 100 randomly picked gynoecia were analyzed. Both, length and width comparisons (C,D) are mean values with their respective standard deviation. Percent values are shown in (E,F). Student’s t-test was applied to compare the wild type gynoecia with the other lines and significant differences were marked with up to three asterisks (p < 0.001). (G) Magnification of the apical region of representative gynoecia of the respective lines showing protruding ovules (arrows). Scale bar represents 1 mm.
CRC-EDLL gynoecia showed no defects in carpel or septum fusion and nectaries were present in all gynoecia. However, 13% of the analyzed gynoecia were composed of more than two, in most cases four carpels (Figure 3F). We then hypothesized that this phenotype may be dosage dependent and carried out qRT-PCR on plants over expressing CRC-EDLL with CRC-specific primers (Supplementary Figure 6). Interestingly, expression of CRC-EDLL does not significantly differ in the transgenic lines between plants developing a normal carpel number vs. plants with a surplus in carpels, suggesting that the EDLL domain has a stronger effect on downstream targets than the absence of CRC protein.
Taken together, these data suggest that CRC acts as a transcriptional activator in the regulation of carpel fusion and nectary development, in promoting longitudinal growth and restricting the width of the gynoecium. However, repression of target genes by CRC is required for floral meristem termination.
CRC is one of the major carpel developmental regulators in Arabidopsis. In this study, we have further characterized the protein by assigning functions to the different protein domains of CRC and showed that CRC acts as both, an activator and a repressor of transcription.
The localization studies of GFP::CRC and different CRC variants showed that CRC is a nuclear localized protein and is almost completely absent from the cytoplasm, which is similar to the subcellular localization of e.g., FIL and INO (Sawa et al., 1999; Meister et al., 2002). Required for this nuclear localization is the C-terminal YABBY domain (Figures 1B–K) but remarkably, the single amino acid mutagenesis of the predicted NLS, did not abolish the nuclear localization (Supplementary Figure 3). Successive deletion of the eight amino acids in the NLS only led to an accumulation of the GFP fusion protein in the cytoplasm suggesting an additive effect of these amino acids toward nuclear localization. Further, this finding may suggest the presence of NLS supporting amino acids in the YABBY domain allowing more efficient binding to certain IMPORTINα isoforms (Sankhala et al., 2017). However, some proteins such as the floral homeotic regulators AP3 and PI (McGonigle et al., 1996) or the mismatch repair system proteins MLH1 and PMS2 (Wu et al., 2003) must enter the nucleus as dimer, as only their dimerization leads to the reconstitution of a functional NLS. In our experiment, the construct with only the short NLS sequence (8 amino acids) is missing the largest part of the YABBY domain, and thus the GFP::CRC-NLS protein is most likely unable to homodimerize such that a functional NLS cannot be reconstituted.
At present, we cannot conclude from our data if the lack in dimerization, or missing NLS supportive amino acids are the reason for the failing nuclear import of CRC-NLS. Interestingly, an influx of the single domain GFP::CRC-ZF, GFP::CRC-IM, and the truncated GFP::CRCΔYD, GFP::CRC-YDΔNLS fusion proteins into the nuclei could be observed in combination with strong residual cytoplasmic localization (Figure 1F and Supplementary Figure 2). This is most likely due to the small size of the fusion proteins and the fact that the GFP fusion construct concentration is higher than the endogenous CRC concentration in wild type plants would be. Generally, nuclear pore complexes allow non-specific influx of proteins up to a size of ∼60 kDa (Haasen et al., 1999), hence, the fusion proteins are small enough (∼28–47 kDa, Figure 1A and Supplementary Figure 2A) to diffuse into nuclei. However, this diffusional influx yields to an active nuclear import if the entire YABBY domain is present in the respective fusion protein.
Two putative DNA binding domains, the N-terminal zinc finger and the C-terminal YABBY domain were suggested previously (Bowman and Smyth, 1999). Interestingly, our yeast-1-hybrid analysis using the KCS15 promoter (Han et al., 2012) demonstrated that neither the zinc finger nor the YABBY domain alone binds sufficiently enough to cause transcription of the selective marker. Conversely, all deletion constructs mildly induced colony growth on selection medium, suggesting that one single domain alone is not able to bind to DNA, but cooperatively with other parts of CRC DNA binding is achieved (Figure 2L). The YABBY domain is most likely the main interacting domain, as we could show that mutations in the YABBY domain drastically reduce DNA binding (CRC-YDm and CRC-ZFm/YDm). These mutations disrupted the two long α-helices in the YABBY domain (Supplementary Figure 1), which are responsible for DNA bending and binding in HMG box proteins (reviewed in Malarkey and Churchill, 2012). Furthermore, the YABBY domain shows a high degree of conservation of those amino acid residues that are necessary for DNA binding in HMG boxes (Bowman and Smyth, 1999). In contrast, mutations in the zinc finger domain did not result in a loss of DNA binding of CRC, which is in line with observations from experiments using another YABBY protein (Smyth et al., 1990; Yang et al., 2016). These findings from YABBY proteins are similar to the DNA binding behavior of major mammalian cell cycle regulator and tumor suppressor protein p53 whose DNA binding capability of the central DNA binding domain additionally requires the C-terminal domain (Laptenko et al., 2015), hence, cooperative DNA binding seems a more generally applicable concept.
DNA binding activities of other YABBY proteins were analyzed previously with partially conflicting results: Whereas OsYABBY1 from Oryza sativa, specifically binds to a GA-responsive element in the promoter of GIBBERELLIN 3-BETA-DIOXYGENASE 2 (2GA3ox2) (Dai et al., 2007), FIL has been described to bind unspecifically to DNA via its YABBY domain (Kanaya et al., 2002). However, a binding motif analysis via protein-binding microarrays (PBM) identified the DNA binding motifs of FIL and YABBY5 (Franco-Zorrilla et al., 2014). Interestingly, typical zinc finger DNA binding motifs were identified by chromatin immunoprecipitation DNA-sequencing (ChIP-Seq) of YABBY proteins from Glycine max (Shamimuzzaman and Vodkin, 2013). And, even more confusing, a recent study identified the YABBY domain as DNA interaction domain by binding to a typical zinc finger motif (Shamimuzzaman and Vodkin, 2013; Yang et al., 2016).
Hence, we propose a model in which CRC forms dimers mediated by the YABBY domain. This dimerization enables the YABBY domain and the zinc finger to individually bind to DNA, most likely to different DNA motifs as both, typical zinc finger DNA binding motifs and non-zinc finger DNA binding motifs have been identified for YABBY proteins (Shamimuzzaman and Vodkin, 2013; Franco-Zorrilla et al., 2014).
Our analyzes did not show a clear function for the intermediate domain, and phylogenetic analysis of different YABBY orthologs show a high variability of the domain (Yamada et al., 2011). Hence, we hypothesize that the intermediate domain, even though it is not necessary for homodimerization and the interaction with DNA, may act as a flexible linker (Supplementary Figure 1) between die DNA-binding YABBY domain and the zinc finger.
Our protein interaction analysis (Figures 2A–K) has shown that CRC forms homodimers and interacts with INO mediated by the YABBY domain. The YABBY domain is predicted to form two amphipathic helices (Supplementary Figure 1) with similarities to HMG boxes, the latter are also involved in dimerization between HMG box containing proteins (Roemer et al., 2008; Sanchez-Giraldo et al., 2015), suggesting conservation on sequence and two-dimensional structure level between classical HMB boxes and YABBY domains.
Our analysis with the functionally modified CRC protein (Figure 3) shows that CRC exhibits characteristics of both, activator and repressor: It serves as an activator of target gene transcription required for carpel fusion, septum formation, and nectary development. Conversely, CRC represses transcription of target genes involved in the termination of the floral meristem. Furthermore, the transgenic lines showed enhanced phenotypes compared to the crc-1 mutant, especially in terms of supernumerous carpels. The crc-1 mutant exhibits up to three carpels at a low frequency. Carpel number can be increased by crossing crc-1 with mutants of the redundant acting regulators of floral meristem termination (RBL, SQN, ULT1, and AG). We thus think that CRC-SRDX and CRC-EDLL may superimpose these regulatory pathways and increase the strength of their respective phenotypes [as shown before for other YABBY proteins by Bonaccorso et al. (2012), and as a general phenomenon by Tiwari et al. (2012), and Figueroa and Browse (2015)]. Interestingly, mild over expression of CRC via the UBQ10 promoter resulted only in a minor increase in the width of the respective gynoecia compared to wild type gynoecia, but the gynoecia were still significantly thinner than gynoecia of crc-1 or CRC-SRDX gynoecia. Hence, the overexpression of CRC via proUBQ10 shows no adverse effects related to over expression like transgene silencing or co-suppression (Elmayan and Vaucheret, 1996; Elmayan et al., 1998; Mishiba et al., 2005) which might inhibit phenotypical analysis.
There is a growing number of examples for bifunctional transcription factors in animals (Willy, 2000; Bernadt et al., 2005) and in plants. E.g., the homeobox gene WUSCHEL activates early AG expression but otherwise acts as a repressor for CLV3 expression to maintain stem cell identity and homeostasis in the shoot apical meristem and floral meristems (Lohmann et al., 2001; Leibfried et al., 2005; Ikeda et al., 2009) and APETALA 2 that activates floral repressors like AGAMOUS-LIKE 15 and represses floral activators such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (Yant et al., 2010). So far, the A. thaliana members of the YABBY family, except for FIL and YAB3, show mainly repressive effects, as they form regulatory complexes with the corepressors LEUNIG and LEUNIG HOMOLOG and repress the transcription of e.g., EXPANSIN 11 (Navarro et al., 2004; Stahle et al., 2009; Bonaccorso et al., 2012). Also in rice, OsYAB1 and OsYAB4 are involved in the regulation of gibberellin levels by repressing GA3ox2 expression (Dai et al., 2007; Yang et al., 2016). Additionally, Yamaguchi et al. (2017) found that CRC represses the expression of TORNADO 2, a plasma membrane localized protein involved in auxin homeostasis in the gynoecium during early flower development. Interestingly, FIL and YAB3 can also act as activators on the expression of AUXIN RESPONSE FACTOR 4 and KAN1. Furthermore, BLADE ON PETIOLE 1 and 2 are, as CRC, necessary for nectary development and as CRC was reported to act epistatically on these two genes (McKim et al., 2008). Both genes may be direct target genes of CRC as their promoter regions exhibit multiple predicted YABBY binding sites in close proximity to their respective start codons (Supplementary Figure 7). Especially the putative YABBY binding motif (YBM) 4 (WATNATW, Franco-Zorrilla et al., 2014) appears frequently in proBOP1 and proBOP2 (28 times and 33 times, respectively). To rule out the possibility of random occurrence of this motif in any given stretch of DNA, we analyzed the promoter regions of the established qRT-PCR reference genes POLYUBIQUITIN 10 (UBQ10), GLYCERALDEHYDE – 3 - PHOSPHATE DEHYDRO-GENASE C – 2 (GAPDH2), and ELONGATION FACTOR 1 α (EF1α) (Czechowski et al., 2005) and found YABBY binding motifs completely absent. However, further studies are required to experimentally corroborate these in silico predictions.
Siggers et al. (2014) showed that the conserved DNA binding motifs of a C2H2 zinc finger transcription factors can change in reaction to different protein interaction partners. Apparently, protein changes mediate binding to new target sites while not affecting the binding to a core set of common target sites. In the light of our observation combined with previously published results we hypothesize that CRC also binds to multiple DNA motifs via the zinc finger and/or the YABBY domain. However, all experiments carried out so far fail to show affinity to these binding sites in a quantitative way and it could well be that some experimentally identified binding sites require the interaction of CRC with specific co-factors not present in our yeast-based experiments. Because of the multitude of binding motifs, we hypothesize that CRC may use different DNA binding motifs in dependence of its interaction partners when activating vs. repressing transcription of target genes.
These two possibilities might explain the duality of YABBY proteins and especially of CRC, as certain functions might be dependent on the presence or absence of different protein interactors. Repression of transcription could be achieved through the interaction with the co-repressors LEU and LUG which are part of the regulatory network of abaxial-adaxial determination of lateral organs (Stahle et al., 2009). Additional interaction partners were identified in a high-throughput protein-protein interaction screen of CRC by Trigg et al. (2017): NGATHA4 or TEOSINTE BRANCHED1-CYCLOIDEA-PCF15 (TCP15), transcription factors involved in style development via regulation of the of auxin biosynthesis (Trigueros et al., 2009; Lucero et al., 2015).
Activation of transcription may be mediated by the interaction of CRC with INDETERMINATE DOMAIN15, which is involved in auxin biosynthesis and auxin homeostasis during lateral organ development (Cui et al., 2013; Trigg et al., 2017). It forms complexes with the co-activators ANGUSTIFOLIA3 and GROWTH-REGULATING FACTOR2, which are both involved in carpel development (Lee et al., 2018; Trigg et al., 2017) and auxin homeostasis (Vercruyssen et al., 2014; Schuster et al., 2015). A tight link between CRC function and auxin has been established previously as the apical defects in the crc-1 gynoecium can be rescued by exogenous auxin application (Ståldal et al., 2008).
Taken together, CRC is a bifunctional transcription factor that regulates carpel fusion and nectary formation by activating the expression of target genes and terminates the meristematic activity of the floral meristem by repression of target gene expression. To fulfill its functions, it forms homo- and heterodimers via the YABBY domain which is also the main DNA interacting domain and harbors also the NLS. CRC is directly regulated by multiple MADS box proteins (Lee et al., 2005; Ó’Maoiléidigh et al., 2013) and is tightly integrated in the carpel developmental network by interactions with many auxin regulating proteins (Ståldal et al., 2008; Yamaguchi et al., 2017). Previously, it was shown that the function of CRC’s orthologs from only distantly related species can be very diverse, such that the rice ortholog DL is also involved in leaf midrib formation and carpel organ identity (Yamaguchi et al., 2004). EcCRC from E. californica is essential for placental development, ovule initiation and lateral carpel margin formation (for a more comprehensive overview of CRC ortholog functions, see Orashakova et al., 2009). It is astonishing how a comparatively small and highly conserved transcription factor can take up many different functions in diverse plant lineages and one may wonder how this is achieved. We and others have shown that the A. thaliana CRC protein has multiple target sequences and our work indicates that it represses transcription of some while activates transcription of other target genes, most likely depending on its protein interaction partners. CRC orthologs from other angiosperm species may also have these flexible properties and may interact with a different set of proteins. This may, at least in part, explain the functional versatility of these transcription factors in the angiosperms analyzed to date.
TG carried out the all the experiments, except for the Y1H analysis, in which SB was heavily involved. TG and AB analyzed the data. TG and AB drafted the manuscript. AB developed the project.
This work was funded by the Justus Liebig University Giessen, Germany and the DAAD (German Academic Exchange Service Grant No. 57162231 to AB).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Kai Pfannebecker for valuable discussions on the manuscript, Martin Hardt for supervising the CLSM work, and Paula Elomaa for granting the opportunity to learn the Y1H technique in Helsinki and her helpful discussions. Also, we would like to thank Andrea Weisert and Claudia Jung-Blasini for excellent technical support in the lab, Dietmar Haffer for valuable advice on plant growth and Jan Lohmann for generously providing vectors. We thank John Golz for his kind contribution of pGADT7/pGBKT7 FIL, YAB2 and YAB5 and we thank Kai Pfannebecker for cloning pGADT7/pGBKT7 CRC, YAB3 and INO. Furthermore, we would like to thank the students Evgenia Diehl, Stefanie Weisgerber, Verena Deutschmeyer, Dominik Lotz, Vanessa Kramer, and David Lonergan for assisting in the various cloning steps and localization experiments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00835/full#supplementary-material
Alvarez, J., and Smyth, D. R. (1999). CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 126, 2377–2386.
Alvarez, J., and Smyth, D. R. (2002). CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 163, 17–41. doi: 10.1086/324178
Bernadt, C. T., Nowling, T., Wiebe, M. S., and Rizzino, A. (2005). NF-Y behaves as a bifunctional transcription factor that can stimulate or repress the FGF-4 promoter in an enhancer-dependent manner. Gene Expr. 12, 193–212. doi: 10.3727/000000005783992052
Bonaccorso, O., Lee, J. E., Puah, L., Scutt, C. P., and Golz, J. F. (2012). FILAMENTOUS FLOWER controls lateral organ development by acting as both an activator and a repressor. BMC Plant Biol. 12:176. doi: 10.1186/1471-2229-12-176
Bowman, J. L., and Smyth, D. R. (1999). CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 126, 2387–2396.
Burdach, J., O’Connell, M. R., Mackay, J. P., and Crossley, M. (2012). Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem. Sci. 37, 199–205. doi: 10.1016/j.tibs.2012.02.001
Chow, C.-N., Zheng, H.-Q., Wu, N.-Y., Chien, C.-H., Huang, H.-D., Lee, T.-Y., et al. (2016). PlantPAN 2.0: an update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 44, 1154–1160. doi: 10.1093/nar/gkv1035
Cui, D., Zhao, J., Jing, Y., Fan, M., Liu, J., Wang, Z., et al. (2013). The Arabidopsis IDD14, IDD15, and IDD16 cooperatively regulate lateral organ morphogenesis and gravitropism by promoting auxin biosynthesis and transport. PLoS Genet. 9:e1003759. doi: 10.1371/journal.pgen.1003759
Cutler, S. R., Ehrhardt, D. W., Griffitts, J. S., and Somerville, C. R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. U.S.A. 97, 3718–3723. doi: 10.1073/pnas.97.7.3718
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W.-R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Dai, M., Zhao, Y., Ma, Q., Hu, Y., Hedden, P., Zhang, Q., et al. (2007). The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 144, 121–133. doi: 10.1104/pp.107.096586
Davis, A. M., Hall, A., Millar, A. J., Darrah, C., and Davis, S. J. (2009). Protocol: streamlined sub-protocols for floral-dip transformation and selection of transformants in Arabidopsis thaliana. Plant Methods 5:3. doi: 10.1186/1746-4811-5-3
Elmayan, T., Balzergue, S., Béon, F., Bourdon, V., Daubremet, J., Guénet, Y., et al. (1998). Arabidopsis mutants impaired in cosuppression. Plant Cell 10, 1747–1758. doi: 10.1105/tpc.10.10.1747
Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. doi: 10.1046/j.1365-313X.1996.9060787.x
Figueroa, P., and Browse, J. (2015). Male sterility in Arabidopsis induced by overexpression of a MYC5-SRDX chimeric repressor. Plant J. 81, 849–860. doi: 10.1111/tpj.12776
Fourquin, C., Primo, A., Martínez-Fernández, I., Huet-Trujillo, E., and Ferrándiz, C. (2014). The CRC orthologue from Pisum sativum shows conserved functions in carpel morphogenesis and vascular development. Ann. Bot. 114, 1535–1544. doi: 10.1093/aob/mcu129
Fourquin, C., Vinauger-Douard, M., Chambrier, P., Berne-Dedieu, A., and Scutt, C. P. (2007). Functional conservation between CRABS CLAW orthologues from widely diverged angiosperms. Ann. Bot. 100, 651–657. doi: 10.1093/aob/mcm136
Franco-Zorrilla, J. M., López-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Gomez-Mena, C., Folter, S., de Costa, M. M. R., Angenent, G. C., and Sablowski, R. (2005). Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 132, 429–438. doi: 10.1242/dev.01600
Gruber, M., Söding, J., and Lupas, A. N. (2005). REPPER–repeats and their periodicities in fibrous proteins. Nucleic Acids Res. 33, W239–W243. doi: 10.1093/nar/gki405
Haasen, D., Kohler, C., Neuhaus, G., and Merkle, T. (1999). Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 20, 695–705. doi: 10.1046/j.1365-313X.1999.00644.x
Han, X., Yin, L., and Xue, H. (2012). Co-expression analysis identifies CRC and AP1 the regulator of Arabidopsis fatty acid biosynthesis. J. Integr. Plant Biol. 54, 486–499. doi: 10.1111/j.1744-7909.2012.01132.x
Hemsley, A., Arnheim, N., Toney, M. D., Cortopassi, G., and Galas, D. J. (1989). A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 17, 6545–6551. doi: 10.1093/nar/17.16.6545
Hiratsu, K., Matsui, K., Koyama, T., and Ohme-Takagi, M. (2003). Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 34, 733–739. doi: 10.1046/j.1365-313X.2003.01759.x
Ikeda, M., Mitsuda, N., and Ohme-Takagi, M. (2009). Arabidopsis WUSCHEL is a bifunctional transcription factor that acts as a repressor in stem cell regulation and as an activator in floral patterning. Plant Cell 21, 3493–3505. doi: 10.1105/tpc.109.069997
Ishikawa, M., Ohmori, Y., Tanaka, W., Hirabayashi, C., Murai, K., Ogihara, Y., et al. (2009). The spatial expression patterns of DROOPING LEAF orthologs suggest a conserved function in grasses. Genes Genet. Syst. 84, 137–146. doi: 10.1266/ggs.84.137
Jones, D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202. doi: 10.1006/jmbi.1999.3091
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67, 547–566. doi: 10.1007/s11103-008-9339-z
Kanaya, E., Nakajima, N., and Okada, K. (2002). Non-sequence-specific DNA binding by the FILAMENTOUS FLOWER protein from Arabidopsis thaliana is reduced by EDTA. J. Biol. Chem. 277, 11957–11964. doi: 10.1074/jbc.M108889200
Kaufmann, K., Anfang, N., Saedler, H., and Theissen, G. (2005). Mutant analysis, protein-protein interactions and subcellular localization of the Arabidopsis Bsister (ABS) protein. Mol. Genet. Genomics 274, 103–118. doi: 10.1007/s00438-005-0010-y
Kosugi, S., Hasebe, M., Matsumura, N., Takashima, H., Miyamoto-Sato, E., Tomita, M., et al. (2008). Six classes of nuclear localization signals specific to different binding grooves of Importin α. J. Biol. Chem. 284, 478–485. doi: 10.1074/jbc.M807017200
Lampropoulos, A., Sutikovic, Z., Wenzl, C., Maegele, I., Lohmann, J. U., and Forner, J. (2013). GreenGate – a novel, versatile, and efficient cloning system for plant transgenesis. PLoS One 8:e83043. doi: 10.1371/journal.pone.0083043
Lange, M., Orashakova, S., Lange, S., Melzer, R., Theissen, G., Smyth, D. R., et al. (2013). The seirena B class floral homeotic mutant of California Poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. Plant Cell 25, 438–453. doi: 10.1105/tpc.112.105809
Laptenko, O., Shiff, I., Freed-Pastor, W., Zupnick, A., Mattia, M., Freulich, E., et al. (2015). The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol. Cell. 57, 1034–1046. doi: 10.1016/j.molcel.2015.02.015
Lee, J.-Y., Baum, S. F., Alvarez, J., Patel, A., Chitwood, D. H., and Bowman, J. L. (2005). Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 17, 25–36. doi: 10.1105/tpc.104.026666
Lee, S.-J., Lee, B. H., Jung, J.-H., Park, S. K., Song, J. T., and Kim, J. H. (2018). GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR specify meristematic cells of gynoecia and anthers. Plant Physiol. 176, 717–729. doi: 10.1104/pp.17.00960
Leibfried, A., To, J. P., Busch, W., Stehling, S., Kehle, A., Demar, M., et al. (2005). WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175. doi: 10.1038/nature04270
Lohmann, J. U., Hong, R. L., Hobe, M., Busch, M. A., Parcy, F., Simon, R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. doi: 10.1016/S0092-8674(01)00384-1
Lucero, L. E., Uberti-Manassero, N. G., Arce, A. L., Colombatti, F., Alemano, S. G., and Gonzalez, D. H. (2015). TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 84, 267–282. doi: 10.1111/tpj.12992
Malarkey, C. S., and Churchill, M. E. A. (2012). The high mobility group box: the ultimate utility player of a cell. Trends Biochem. Sci. 37, 553–562. doi: 10.1016/j.tibs.2012.09.003
McGonigle, B., Bouhidel, K., and Irish, V. F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10, 1812–1821. doi: 10.1101/gad.10.14.1812
McKim, S. M., Stenvik, G.-E., Butenko, M. A., Kristiansen, W., Cho, S. K., Hepworth, S. R., et al. (2008). The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 135, 1537–1546. doi: 10.1242/dev.012807
Meister, R. J., Kotow, L. M., and Gasser, C. S. (2002). SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development 129, 4281–4289.
Mishiba, K., Nishihara, M., Nakatsuka, T., Abe, Y., Hirano, H., Yokoi, T., et al. (2005). Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 44, 541–556. doi: 10.1111/j.1365-313X.2005.02556.x
Navarro, C., Efremova, N., Golz, J. F., Rubiera, R., Kuckenberg, M., Castillo, R., et al. (2004). Molecular and genetic interactions between STYLOSA and GRAMINIFOLIA in the control of Antirrhinum vegetative and reproductive development. Development 131, 3649–3659. doi: 10.1242/dev.01205
Ó’Maoiléidigh, D. S., Wuest, S. E., Rae, L., Raganelli, A., Ryan, P. T., Kwasniewska, K., et al. (2013). Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. Plant Cell 25, 2482–2503. doi: 10.1105/tpc.113.113209
Orashakova, S., Lange, M., Lange, S., Wege, S., and Becker, A. (2009). The CRABS CLAW ortholog from California poppy (Eschscholzia californica, Papaveraceae), EcCRC, is involved in floral meristem termination, gynoecium differentiation and ovule initiation. Plant J. 58, 682–693. doi: 10.1111/j.1365-313X.2009.03807.x
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Prunet, N., Morel, P., Thierry, A.-M., Eshed, Y., Bowman, J. L., Negrutiu, I., et al. (2008). REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. Plant Cell 20, 901–919. doi: 10.1105/tpc.107.053306
Reyes-Olalde, J. I., Zuniga-Mayo, V. M., Chavez Montes, R. A., Marsch-Martinez, N., and de Folter, S. (2013). Inside the gynoecium: at the carpel margin. Trends Plant Sci. 18, 644–655. doi: 10.1016/j.tplants.2013.08.002
Roemer, S. C., Adelman, J., Churchill, M. E. A., and Edwards, D. P. (2008). Mechanism of high-mobility group protein B enhancement of progesterone receptor sequence-specific DNA binding. Nucleic Acids Res. 36, 3655–3666. doi: 10.1093/nar/gkn249
Sablowski, R. (2015). Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 66, 1065–1073. doi: 10.1093/jxb/eru514
Sanchez-Giraldo, R., Acosta-Reyes, F. J., Malarkey, C. S., Saperas, N., Churchill, M. E. A., and Campos, J. L. (2015). Two high-mobility group box domains act together to underwind and kink DNA. Acta Crystallogr. D Biol. Crystallogr. 71, 1423–1432. doi: 10.1107/S1399004715007452
Sankhala, R. S., Lokareddy, R. K., Begum, S., Pumroy, R. A., Gillilan, R. E., and Cingolani, G. (2017). Three-dimensional context rather than NLS amino acid sequence determines importin α subtype specificity for RCC1. Nat. Commun. 8:979. doi: 10.1038/s41467-017-01057-7
Sawa, S., Watanabe, K., Goto, K., Kanaya, E., Morita, E. H., and Okada, K. (1999). FILAMENTOUS FLOWER, a meristem and organ identiy gene of Arabidopsis encodes a protein witch a zinc finger and HMG-related domains. Genes Dev. 13, 1079–1088. doi: 10.1101/gad.13.9.1079
Schuster, C., Gaillochet, C., and Lohmann, J. U. (2015). Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 142, 3343–3350. doi: 10.1242/dev.120444
Shamimuzzaman, M., and Vodkin, L. (2013). Genome-wide identification of binding sites for NAC and YABBY transcription factors and co-regulated genes during soybean seedling development by ChIP-Seq and RNA-Seq. BMC Genomics 14:477. doi: 10.1186/1471-2164-14-477
Siggers, T., Reddy, J., Barron, B., and Bulyk, M. L. (2014). Diversification of transcription factor paralogs via noncanonical modularity in C2H2 zinc finger DNA binding. Mol. Cell. 55, 640–648. doi: 10.1016/j.molcel.2014.06.019
Smyth, D. R., Bowman, J. L., and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2, 755–767. doi: 10.1105/tpc.2.8.755
Stahle, M. I., Kuehlich, J., Staron, L., Arnim, A. G., and von Golz, J. F. (2009). YABBYs and the transcriptional corepressors LEUNIG and LEUNIG HOMOLOG maintain leaf polarity and meristem activity in Arabidopsis. Plant Cell 21, 3105–3118. doi: 10.1105/tpc.109.070458
Ståldal, V., Sohlberg, J. J., Eklund, D. M., Ljung, K., and Sundberg, E. (2008). Auxin can act independently of CRC, LUG, SEU, SPT and STY1 in style development but not apical-basal patterning of the Arabidopsis gynoecium. New Phytol. 180, 798–808. doi: 10.1111/j.1469-8137.2008.02625.x
Tatematsu, K., Toyokura, K., Miyashima, S., Nakajima, K., and Okada, K. (2015). A molecular mechanism that confines the activity pattern of miR165 in Arabidopsis leaf primordia. Plant J. 82, 596–608. doi: 10.1111/tpj.12834
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M., and Nguyen, M. (2010). A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–S5. doi: 10.1016/j.ymeth.2010.01.005
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Trigg, S. A., Garza, R. M., MacWilliams, A., Nery, J. R., Bartlett, A., Castanon, R., et al. (2017). CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods 14, 819–825. doi: 10.1038/nmeth.4343
Trigueros, M., Navarrete-Gómez, M., Sato, S., Christensen, S. K., Pelaz, S., Weigel, D., et al. (2009). The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21, 1394–1409. doi: 10.1105/tpc.109.065508
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S.-K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Willy, P. J. (2000). A basal transcription factor that activates or represses transcription. Science 290, 982–984. doi: 10.1126/science.290.5493.982
Wu, X., Platt, J. L., and Cascalho, M. (2003). Dimerization of MLH1 and PMS2 limits nuclear localization of MutL. Mol. Cell. Biol. 23, 3320–3328. doi: 10.1128/MCB.23.9.3320-3328.2003
Yamada, T., Yokota, S., Hirayama, Y., Imaichi, R., Kato, M., and Gasser, C. S. (2011). Ancestral expression patterns and evolutionary diversification of YABBY genes in angiosperms. Plant J. 67, 26–36. doi: 10.1111/j.1365-313X.2011.04570.x
Yamaguchi, N., Huang, J., Xu, Y., Tanoi, K., and Ito, T. (2017). Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 8:1125. doi: 10.1038/s41467-017-01252-6
Yamaguchi, T., Nagasawa, N., Kawasaki, S., Matsuoka, M., Nagato, Y., and Hirano, H.-Y. (2004). The YABBY gene DROOPING LEAF regulates carpel specification and midrib development in Oryza sativa. Plant Cell 16, 500–509. doi: 10.1105/tpc.018044
Yanagisawa, S. (1997). Dof DNA-binding domains of plant transcription factors contribute to multiple protein-protein interactions. Eur. J. Biochem. 250, 403–410. doi: 10.1111/j.1432-1033.1997.0403a.x
Yang, C., Ma, Y., and Li, J. (2016). The rice YABBY4 gene regulates plant growth and development through modulating the gibberellin pathway. J. Exp. Bot. 67, 5545–5556. doi: 10.1093/jxb/erw319
Yant, L., Mathieu, J., Dinh, T. T., Ott, F., Lanz, C., Wollmann, H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156–2170. doi: 10.1105/tpc.110.075606
Keywords: CRABS CLAW, carpel, protein domains, Arabidopsis thaliana, YABBY
Citation: Gross T, Broholm S and Becker A (2018) CRABS CLAW Acts as a Bifunctional Transcription Factor in Flower Development. Front. Plant Sci. 9:835. doi: 10.3389/fpls.2018.00835
Received: 13 February 2018; Accepted: 29 May 2018;
Published: 20 June 2018.
Edited by:
Cristina Ferrandiz, Instituto de Biología Molecular y Celular de Plantas (IBMCP), SpainReviewed by:
Adriana Garay, Universidad Nacional Autónoma de México, MexicoCopyright © 2018 Gross, Broholm and Becker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thomas Gross, dGhvbWFzLmdyb3NzQGJvdDEuYmlvLnVuaS1naWVzc2VuLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.