- State Key Laboratory of Hybrid Rice, College of Life Sciences, Wuhan University, Wuhan, China
Replication factor C (RFC) is a multisubunit complex that opens the sliding clamp and loads it onto the DNA chain in an ATP-dependent manner and is thus critical for high-speed DNA synthesis. In yeast (Saccharomyces cerevisiae) and humans, biochemical studies and structural analysis revealed interaction patterns between the subunits and architectures of the clamp loaders. Mutations of ScRFC1/2/3/4/5 lead to loss of cell viability and defective replication. However, the functions of RFC subunits in higher plants are unclear, except for AtRFC1/3/4, and the interaction and arrangement of the subunits have not been studied. Here, we identified rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants in Arabidopsis, and found that embryos and endosperm arrested at the 2/4-celled embryo proper stage and 6-8 nuclei stages, respectively. Subcellular localization analysis revealed that AtRFC1 and OsRFC1/4/5 proteins were localized in the nucleus, while AtRFC2/3/4/5 and OsRFC2/3 proteins were present both in the nucleus and cytoplasm. By using yeast two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) techniques, we demonstrated the interactions of Arabidopsis and rice (Oryza sativa) RFC subunits, and proposed arrangements of the five subunits within the RFC complex, which were AtRFC5-AtRFC4-AtRFC3/2-AtRFC2/3-AtRFC1 and OsRFC5-OsRFC2-OsRFC3-OsRFC4-OsRFC1, respectively. In addition, AtRFC1 could interact with AtRFC2/3/4/5 in the presence of other subunits, while OsRFC1 directly interacted with the other four subunits. To further characterize the regions required for complex formation, truncated RFC proteins of the subunits were created. The results showed that C-termini of the RFC subunits are required for complex formation. Our studies indicate that the localization and interactions of RFCs in Arabidopsis and rice are distinctly discrepant.
Introduction
In eukaryotes, the heteropentameric replication factor C (RFC) acts as a clamp loader that can bind and open the homotrimeric proliferating cell nuclear antigen (PCNA) clamp, and then load PCNA onto a template-primer junction in an ATP-dependent reaction (Fien and Stillman, 1992; O'Donnell and Kuriyan, 2006). Coupled with ATP hydrolysis, RFC is ejected from the PCNA clamp for the next round of DNA synthesis and the clamp recruits DNA polymerases for processive elongation of DNA chains (Yao and O'Donnell, 2012). In addition to its role in DNA replication, RFC has been reported to function in DNA polymerase switching (Maga et al., 2000; Mossi et al., 2000), DNA repair (Shivji et al., 1995; Waga and Stillman, 1998), and check-point control in cell cycle progression (Sugimoto et al., 1997; Shimada et al., 1999; Kim and Brill, 2001).
The RFC complex consists of five subunits: the large subunit is named RFC1 and four small subunits are named RFC2/3/4/5 (Yao and O'Donnell, 2012). The five subunits exist in all eukaryotes and are highly conserved in both structure and function (Chen et al., 1992; Bunz et al., 1993; Luckow et al., 1994; Cullmann et al., 1995; Gray and MacNeill, 2000; Furukawa et al., 2003). All five subunits have high homology to each other (Cullmann et al., 1995). Human RFCs were first identified as an essential replication factor for simian virus 40 (SV40) DNA replication in vitro (Tsurimoto and Stillman, 1989), and are composed of five subunits–p140, p40, p38, p37, and p36–whose molecular masses are 128.3, 39, 40.5, 39.6, and 38.5 kDa, respectively (Mossi and Hübscher, 1998). In yeast (Saccharomyces cerevisiae), a functional protein complex homologous to human RFC has been identified, which also consists of five subunits with molecular masses of 94.9, 39.9, 39.7, 38.2, and 36.2 kDa (Cullmann et al., 1995; Mossi and Hübscher, 1998). The yeast RFC subunits show high identity with human RFCs (Cullmann et al., 1995). RFC subunits possess a cluster of conserved motifs that are termed RFC boxes (Mossi and Hübscher, 1998). The four small subunits contain seven conserved RFC boxes II-VIII, which mainly exist in the N-terminal region. In addition to RFC boxes II-VIII, the N-terminal extension of the large subunit RFC1 contains an additional RFC box (box I), which shows high homology to the prokaryotic DNA ligases but lacks ligase activity (Kobayashi et al., 2010). RFC box III contains the most conserved motif: a phosphate-binding loop (P loop) that is essential for the structure and function of RFC (Cullmann et al., 1995; Podust et al., 1998; Neuwald et al., 1999; Schmidt et al., 2001). The C-terminal sequences of RFC subunits are flexible and essential for mediating subunit-subunit interaction as well as forming the integrated RFC complex (Shiomi and Nishitani, 2017).
In spite of the high sequence similarity and conserved structure, each subunit is indispensable for RFC complex formation and activity. In humans, deletion mutations of hRFC1/2/3/4/5 have shown that the C-terminal regions are indispensable for RFC complex formation (Uhlmann et al., 1997a,b). In yeast, all five subunits are essential for cell viability. Cold-sensitive cdc44 (rfc1) mutants exhibited a delay in progressing through the S phase and arrested at the G2/M phase, and were sensitive to DNA damaging agent methyl methanesulfonate (MMS) and ultraviolet (UV) irradiation. Moreover, mutation of POL30 (PCNA) suppressed cold-sensitive alleles of cdc44 but could not substitute for its function. These results indicate that ScRFC1 plays important roles in both DNA replication and DNA repair (Howell et al., 1994; McAlear et al., 1994, 1996). Mutation of ScRFC2 led to defects in DNA integrity and a disordered S-phase checkpoint, indicating that ScRFC2 was required for chromosomal DNA replication and S-phase checkpoint control. On the other hand, RFC5 suppressed the phenotype of rfc2 mutation in yeast and the rfc2-1 rfc5-1 and rfc2-1 cdc44-1 double mutants were synthetically lethal, implying that ScRFC2 interacted with ScRFC1 and ScRFC5 in the RFC complex during DNA replication (Noskov et al., 1998). Mutation of ScRFC5 gave rise to incomplete DNA replication and led to defects in entering into mitosis, indicating that ScRFC5 was also involved in DNA replication and the S-phase checkpoint (Sugimoto et al., 1996; Naiki et al., 2000). In Arabidopsis, RFC1 was homologous to the large subunit p140 of human RFC and played an important role in meiotic recombination and crossover formation, and DNA double-strand break repair during meiosis (Liu et al., 2010, 2013; Wang et al., 2012). The rfc3-1 mutant exhibited hypersensitivity to salicylic acid and enhanced resistance to virulent oomycete Hyaloperonospora arabidopsidis (H. a.) Noco2, suggesting that AtRFC3 negatively regulates systemic acquired resistance and has crucial functions in cell proliferation and DNA replication (Xia et al., 2009, 2010). Mutation of AtRFC4 had severe defects in DNA replication and led to seed abortion and seedling lethality, indicating that AtRFC4 is crucial for DNA replication (Qian et al., 2018).
Studies on the interactions and arrangement of the five subunits contribute to further understanding of the structure of the RFC complex and the precise roles of individual subunits in complex formation. The crystal structure of the E. coli γ complex, the bacterial homolog of eukaryotic RFC, reveals that the γ complex is arranged in a circular manner and the C-terminal domains form a tight circular collar to mediate the subunit-subunit interactions of the complex (Jeruzalmi et al., 2001). In yeast, biochemical studies and structural analysis revealed that the model of RFC subunits was arranged in the form of ScRFC5-ScRFC2-ScRFC3-ScRFC4-ScRFC1 (Yao et al., 2003; Bowman et al., 2004). In humans, p36 (RFC3), p37 (RFC2), and p40 (RFC4) form a stable core complex, which can unload PCNA, but cannot load PCNA onto DNA. The p36/p37/p40 complex binds cooperatively to p140 (RFC1) and p38 (RFC5) subunits and forms the functional five-subunit RFC complex (Uhlmann et al., 1996; Cai et al., 1997; Ellison and Stillman, 1998). Electron microscopic studies confirm that hRFCs are also aligned in a circle, similar to the E. coli γ complex structure (Shiomi et al., 2000).
Despite the abundant literature about the RFC complex in yeast and humans, the functions of the five RFC subunits in higher plants are still unclear and the interaction and arrangement of the five subunits have not been studied. In this study, homology and structural analysis of the five subunits in Arabidopsis and rice (Oryza sativa) were carried out. The interactions of these subunits were investigated by employing the yeast two hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) techniques. To characterize the regions required for complex formation, a series of truncated RFC proteins were produced to detect interactions. In addition, the phenotypes of rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants were characterized in Arabidopsis, which showed that both embryo and endosperm development were defective. Our studies increase knowledge for understanding subunit interaction relationships of the RFC complex, and provide new clues for further studies of the biological function of each RFC subunit.
Materials and Methods
Plant Materials and Growth Conditions
Arabidopsis thaliana Columbia-0 (Col-0) was used as the wild type in this study. The rfc2-1 (CS800312 or SAIL_6_C02), rfc3-2 (SAIL_401_E05), and rfc5-1 (SALK_029291) were obtained from the ABRC (Arabidopsis Biological Resource Center). The Arabidopsis plants and wild type Nicotiana benthamiana plants were grown in a chamber at Wuhan University at 22 ± 2°C with a 16-h light and 8-h dark cycle.
Phylogenetic Analysis
The AtRFC1/2/3/4/5 and OsRFC1/2/3/4/5 protein sequences were used to search for the homologs from other species using BLASTP. Multiple sequence alignments of the box III and full-length amino acid sequences were performed using ClustalX (1.83) (Takashi et al., 2009). The phylogenetic tree with the Neighbor–Joining algorithm was constructed through the MEGA4 program (Tamura et al., 2007).
Complementation Analysis
For complementing the rfc2-1/+ and rfc5-1/+ mutants, the full length genomic fragments of AtRFC2 and AtRFC5 including the promoters and coding sequences were amplified from wild-type genome using KOD-Plus-Neo DNA polymerase (Toyobo, Japan) and cloned into pCambia1300 vector, and then introduced into rfc2-1/+ and rfc5-1/+ heterozygote mutants by the floral dip method (Clough and Bent, 1998). Primers used in the experiments were listed in Table S4.
Ovule Clearing
Fresh ovules of Arabidopsis were dissected from siliques using forceps and mounted in Hoyer's solution [chloral hydrate: glycerol: water, 8:1:2 (w/v/v)] for 30 min or 6–8 h depending on the embryo developmental stage (Chen et al., 2015). Then, the cleared ovules were observed and photographed with differential interference contrast microscopy (Olympus TH4-200 equipped with a CCD of a SPOT digital microscope camera).
Quantitative Real-Time PCR Analysis
Total RNA from various tissues was extracted by RNAiso Plus (TaKaRa, Japan). RNA was used to transcribe into cDNA using ReverTra Ace qPCR RT Kit (TOYOBO). Quantitative Real-Time PCR was carried out as described (Li et al., 2017) and the relative expression levels were analyzed according to the reported method (Ma and Zhao, 2010). The expression level of the GAPDH gene was used as reference for the mRNA level. Three or more independent biological replicates and three technical replicates of each sample were performed for quantitative PCR analysis. Primers used in the experiments were listed in Table S4.
Subcellular Localizations
To construct GFP fusion vectors with OsRFC1/2/3/4/5 and Venus fusion vectors with AtRFC1/2/3/4/5, the coding sequence of the 10 proteins were fused in-frame to the N terminus of the enhanced GFP and Venus coding sequence under the control of the CaMV 35S promoter in the pCAMBIA1300-EGFP and pCAMBIA1300-Venus vector, respectively. The constructs were introduced into A. tumefaciens strain GV3101 and transformed into tobacco (Nicotiana benthamiana) leaves simultaneously by the agrobacterium-mediated transformation. After 2 days of incubation, fluorescence imaging of the tobacco epidermal cell was observed under an Olympus FluoView FV1000 confocal microscopy. Meanwhile, the 35S::Venus, 35S::AtRFC1/2/3/4/5-Venus transgenic plants and 35S::GFP, 35S::OsRFC2/3-GFP transgenic callus cells were obtained for subcellular localization analysis. The excitation and emission wavelength for GFP and Venus were 488 and 505–530 nm as well as 514 and 526–600 nm, respectively. Primers used were listed in Table S4.
Yeast Two-Hybrid
The full-length open reading frames (ORFs) of AtRFC1/2/3/4/5 and OsRFC1/2/3/4/5 were cloned into the pGADT7 and pGBKT7 vector separately. Y2H assay was performed as described in our lab (Deng et al., 2014). The results were repeated at least three times. Primers used were listed in Table S4.
Construction of Tandem Expression Vectors
To construct the tandem expression vectors of Arabidopsis RFC complex, the CDS (coding sequence) of AtRFC2 (SalI+KpnI), AtRFC3 (SalI+SacI), AtRFC4 (SalI+KpnI), and AtRFC5 (SalI+KpnI) were amplified from the cDNA sample of 2-weeks seedlings using KOD-Plus-Neo DNA polymerase (Toyobo, Japan). The above CDS were then inserted into mpCAMBIA1300 vector to build 35S::AtRFC2-NOST, 35S::AtRFC3-NOST, 35S::AtRFC4-NOST, and 35S::AtRFC5-NOST single vectors, respectively. Afterwards, primer pairs of 35S-FP-Hind III and NOST-RP-HindIII, 35S-FP-EcoRI and NOST-RP-EcoRI were used to obtain 35S::AtRFC3-NOST-35S::AtRFC2-NOST, 35S::AtRFC4-NOST-35S::AtRFC2-NOST, 35S::AtRFC4-NOST-35S::AtRFC3-NOST, 35S::AtRFC2-NOST-35S::AtRFC5-NOST, 35S::AtRFC3-NOST-35S::AtRFC5-NOST, and 35S::AtRFC4-NOST-35S::AtRFC5-NOST dual vectors. Similarly, the 35S:: AtRFC3-NOST-35S::AtRFC2-NOST-35S::AtRFC5-NOST, 35S:: AtRFC4-NOST-35S::AtRFC2-NOST-35S::AtRFC5-NOST, 35S:: AtRFC4-NOST-35S::AtRFC2-NOST-35S::AtRFC3-NOST, and 35S:: AtRFC4-NOST-35S::AtRFC3-NOST-35S::AtRFC5-NOST triple vectors were constructed. Primers used were listed in Table S4.
BiFC Assays
The full-length ORFs of AtRFC1/2/3/4/5 and OsRFC1/2/3/4/5 and their truncated cDNA were inserted into the pCAMBIA-SPYNE and pCAMBIA-SPYCE vectors, respectively. The constructs were transferred into Agrobacterium tumefaciens strain GV3101 by a freeze–thaw method. The experimental procedure was performed as described previously (Sparkes et al., 2006; Strzalka et al., 2015). Fluorescent images of YFP were taken with an Olympus FluoView FV1000 confocal microscope to determine whether the two proteins interact with each other. The excitation and emission wavelength for YFP was 515 and 505–530 nm, respectively. Empty vectors of BiFC constructs were used as a negative control. The results were repeated at least three times. Primers used in this test were listed in Table S4.
Accession Numbers
The accession numbers used in the article are AtRFC1 (At5g22010), AtRFC2 (At1g63160), AtRFC3 (At1g77470), AtRFC4 (At1g21690), AtRFC5 (At5g27740), OsRFC1 (Os11g0572100), OsRFC2 (Os12g0176500), OsRFC3 (Os02g0775200), OsRFC4 (Os04g0569000), and OsRFC5 (Os03g0792600).
Results
RFC Subunits Are Conserved in Eukaryotes
To determine the identity of RFC subunits between Arabidopsis and rice, we performed full alignment of the amino acid sequences. As shown in Figure 1A, the amino acid sequence of rice RFC subunits exhibited high identity with Arabidopsis ranging from 59 to 85%. Moreover, the RFC subunits shared highly conserved regions, defined as boxes I–VIII. The small RFC2/3/4/5 subunits contained RFC boxes II–VIII, while AtRFC1 and OsRFC1 had an additional box I in their N-terminal sequences. To gain insights toward the evolutionary relationships of RFC subunits in yeast, humans, C. elegans, mice, Arabidopsis, and rice, phylogenetic analyses were performed. The results indicated that the RFC complex was conserved in eukaryotes, and the Arabidopsis RFC proteins were closely related to the rice RFC subunits (Figure S1). To further study the conservation of RFC subunits of Arabidopsis and rice at the structure level, three-dimensional structures were constructed based on their homologs in yeast (Figure 1B). The models suggested that the rice RFC subunits exhibited extremely high similarity with Arabidopsis RFC subunits, providing further evidence that RFC structures were conserved in eukaryotes. As one of the most conserved motifs in RFC subunits, box III plays an essential role in ATP-binding in yeast and humans (Kelch, 2016). Sequence alignment of box III of the five subunits from Arabidopsis, rice and other eukaryotes revealed high sequence similarities and the conserved motif GxxxxGK (S/T) (Figure 1C).
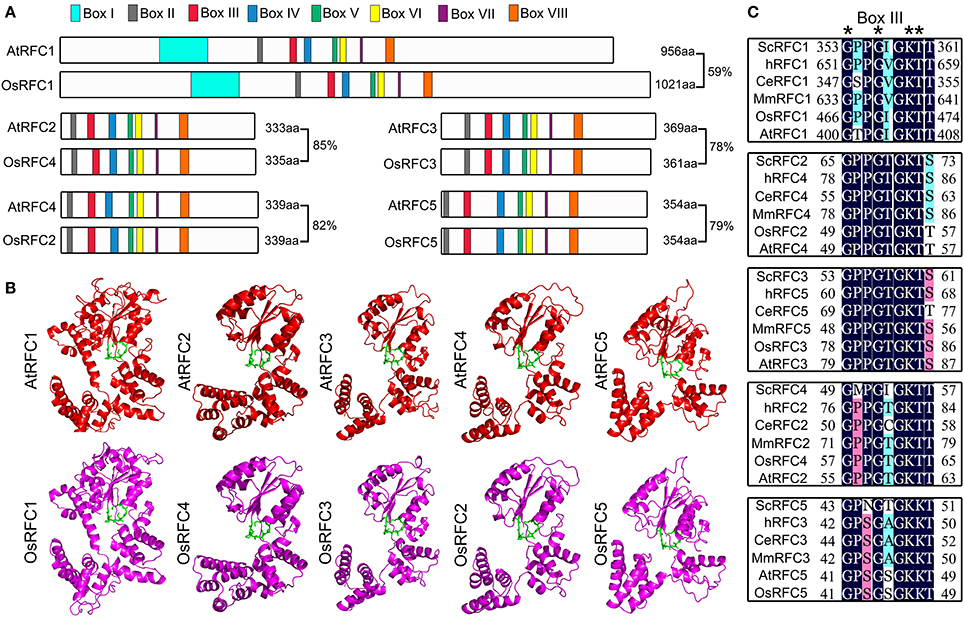
Figure 1. The conservation analysis of the five RFC subunits in Arabidopsis and rice. (A) Schematic diagram of the Arabidopsis and rice RFC subunits. AtRFC1 and OsRFC1 contain eight boxes I–VIII, while AtRFC2/3/4/5 and OsRFC2/3/4/5 contain seven boxes II-VIII. The length of the amino acid sequence is indicated at the end of each gene. (B) The three-dimensional structures of RFC subunits are modeled according to the crystal structures of their yeast homologs through SWISS-MODEL (https://www.swissmodel.expasy.org/). The potential ATP-binding domains (also known as box III) are colored in green. (C) Sequence alignment of the box III of the five subunits in different organisms. Asterisks indicate the conserved residues. At, Arabidopsis thaliana; Os, Oryza sativa; Sc, Saccharomyces cerevisiae; Ce, Caenorhabditis elegans; H, Homo sapiens; Mm, Mus musculus.
Knock-Out of AtRFC2/3/5 Seriously Inhibits the Division of Embryo Cells and Endosperm Free Nuclei
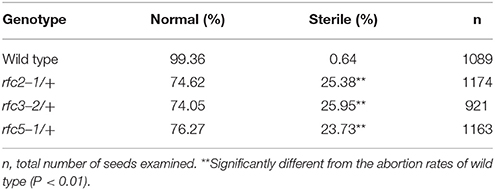
In Arabidopsis, the expression and function of RFC1 and RFC4 have been reported (Liu et al., 2013; Qian et al., 2018). To assess the functions of AtRFC2/3/5, we first detected their expressions in various Arabidopsis tissues using quantitative real-time PCR (qRT-PCR) assays. The results showed that AtRFC2/3/5 were expressed in nearly all of the vegetative and reproductive tissues, and the expression levels were higher in the flowers and siliques (Figure S2). Meanwhile three T-DNA insertion mutants rfc2-1, rfc3-2, and rfc5-1 were obtained from the Arabidopsis Biological Resource Center (ABRC). Primers were designed to identify the precise positions of the T-DNA insertions and sequencing showed that rfc2-1 possessed a T-DNA in the second exon of AtRFC2, rfc3-2 in the third intron of AtRFC3 and rfc5-1 in the second exon of AtRFC5 (Figure 2A). We found that viable homozygotes in the progenies of the three mutant plants could not be obtained. Though no developmental defects were observed in heterozygous plants of the mutants during vegetative growth, their mature siliques contained white abnormal ovules at a frequency of about 25% (Figure 2B; Table 1). All of these results showed that the mutations in AtRFC2/3/5 were lethal in homozygous progenies, similar to the mutant rfc4 (Qian et al., 2018).
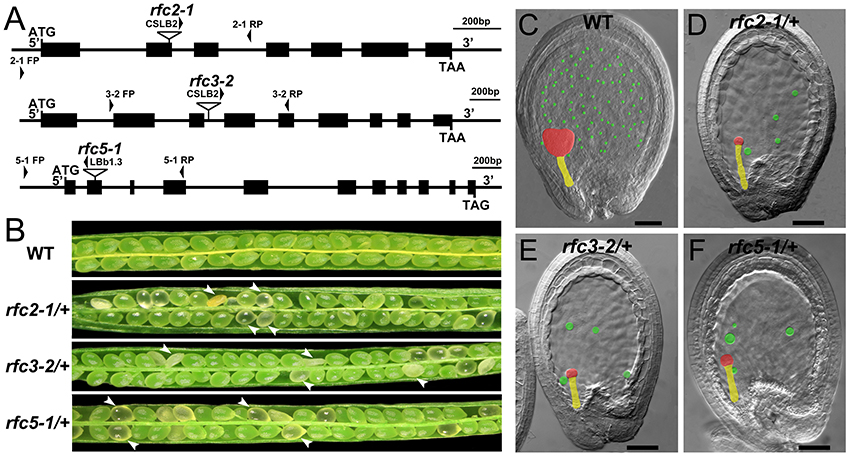
Figure 2. Characterization of Arabidopsis rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants. (A) Schematic diagrams of the AtRFC2/3/5 genes and the positions of T-DNA insertions of their mutants. Exons are shown as black boxes, and 5′ regions, 3′ regions and introns as lines. Arrowheads indicate the positions of FP/RP primers used for genotyping. (B) Silique phenotypes of Arabidopsis wild type and rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants. The white arrowheads show the aborted white ovules. (C–F) Phenotypes of embryo and endosperm free nuclei in wild type, rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants. Red, yellow and green marks indicate the embryo proper, the suspensor and the endosperm free nuclei, respectively.
To clarify whether the null mutation in RFC2/3/5 affected gametophyte activity, segregation of the three self-fertilized mutant progenies were analyzed. The T-DNA insertion in rfc2-1/+ and rfc3-2/+ mutants harbors the Basta (Bas) resistance gene; segregation analysis of the mutant alleles was performed with resistance screening. Due to the lack of kanamycin resistance of the rfc5-1/+ mutant, its segregation was determined by PCR. All of the segregation ratios were about 2:1 (resistant:sensitive), instead of the expected 3:1 (Table S2). The results showed that the rfc2-1/+, rfc3-2/+, and rfc5-1/+ mutants contained a single copy T-DNA insertion in their respective genomes, and which led to aborted seeds in the mature siliques. We performed further reciprocal crosses with rfc2-1/+, rfc3-2/+, and rfc5-1/+ to wild-type plants, respectively, and the results showed that the transmission capacity of both females and males in the three mutants was normal (Table S3), indicating that knock out of RFC2/3/5 genes did not affect the viability of gametophytes.
Detailed ovule phenotypes of rfc2-1/+, rfc3-2/+, and rfc5-1/+ were also investigated. At 4 days after pollination (DAP), all embryos in wild-type ovules had developed into late globular or heart stage (Figure 2C). However, in aborted ovules of the three mutants, all embryos arrested at the 2/4-celled embryo proper stage and the number of endosperm free nuclei decreased dramatically (Figures 2D–F; Table 2), indicating that embryo and endosperm development were severely delayed and repressed in the mutants. The result showed that endosperm proliferation was suppressed as early as the elongated zygote stage and finally reached 6-8 nuclei stages (Table 2). This finding suggested that AtRFC2/3/5 played a crucial role in maintaining mitosis in early embryo cells and endosperm free nuclei.
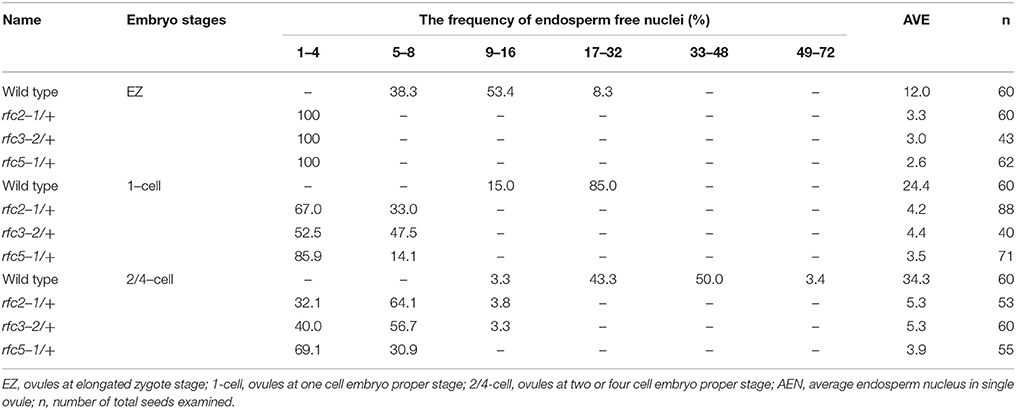
Table 2. Distribution of endosperm free nuclei in the rfc2-1/+, rfc3-2/+, and rfc5-1/+ aborted ovules at sequential development stages.
To verify that the rfc2-1/+ and rfc5-1/+ phenotypes were caused by mutation in the AtRFC2 and AtRFC5 gene, respectively, we performed complementation of the two mutants by transforming the full-length genomic sequence of AtRFC2 and AtRFC5 into the rfc2-1/+ and rfc5-1/+ plants. Homozygous mutants were obtained from the progenies of the transgenic plants through screening, showing that their seed abortion phenotypes were rescued (Figures S3A,B). The results indicated that mutation of AtRFC2/5 genes is responsible for the seed defective phenotype of rfc2-1/+ and rfc5-1/+ mutants.
Subcellular Localization of the Arabidopsis and Rice RFC Subunits
To characterize the subcellular localization of AtRFCs and OsRFCs, vectors expressing fusion proteins of AtRFC1/2/3/4/5 Venus and OsRFC1/2/3/4/5-GFP under the control of the CaMV 35S promoter were generated and expressed transiently in tobacco epidermal cells. The 35S::Venus and 35S::GFP vectors were used as controls, and their fluorescent signals were evenly distributed in the cytoplasm and the nucleus (Figures 3A,G). As previously reported (Liu et al., 2010), AtRFC1-Venus was preferentially localized in the nucleus (Figure 3B), whereas the fluorescent signals of AtRFC2/3/4/5-Venus fusion proteins accumulated in both the cytoplasm and the nucleus (Figures 3C–F). Consistent with AtRFC1-Venus, OsRFC1-GFP was also mainly localized in the nucleus (Figure 3H), while OsRFC2-GFP and OsRFC3-GFP were expressed ubiquitously (Figures 3I,J). Like OsRFC1, OsRFC4-GFP, and OsRFC5-GFP were only detected in the nucleus (Figures 3K,L). To further analyze the subcellular localization of AtRFCs and OsRFCs, we obtained the 35S::Venus, 35S::AtRFC1/2/3/4/5-Venus transgenic plants and 35S::OsRFC2/3-GFP transgenic callus cells, and observed their fluorescent signals. The results showed that the fluorescent signals of 35S::Venus and 35S::GFP were distributed in cytoplasm and nucleus (Figure 3M; Figure S4A). AtRFC1 was only localized in the nucleus of the root tip cells (Figure 3N). However, AtRFC2/3/4/5-Venus were mainly located in the nucleus, and a small amount in the cytoplasm of the root tip cells (Figures 3O–R). In rice callus cells, OsRFC2-GFP and OsRFC3-GFP were also mainly localized in the nucleus (Figures S4B,C). All of these results indicate that the RFC subunits were preferentially localized in the nucleus in Arabidopsis and rice.
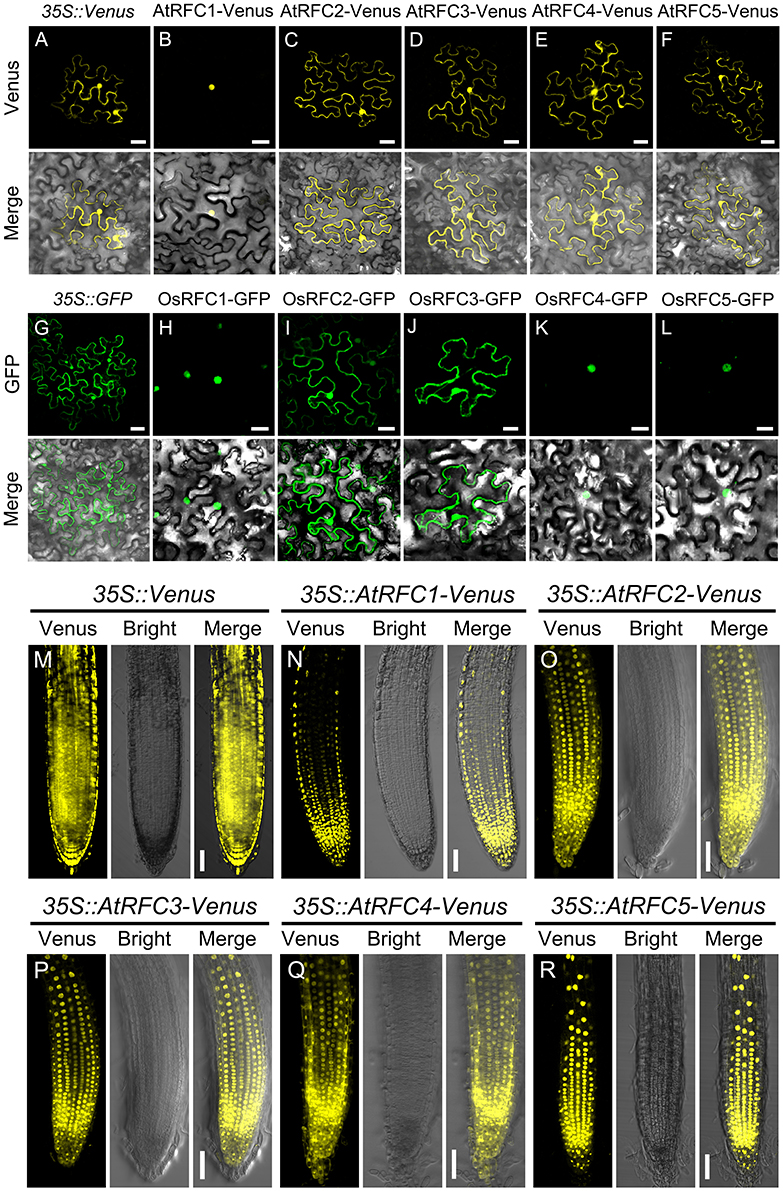
Figure 3. Subcellular localization of AtRFC1/2/3/4/5 and OsRFC1/2/3/4/5. (A–F) The subcellular localization of AtRFC1/2/3/4/5 in epidermic cells of the transiently-expressed Nicotiana Benthamiana. Bars = 30 μm. (G–L) The subcellular localization of OsRFC1/2/3/4/5 in epidermic cells of transiently-expressed Nicotiana Benthamiana. Bars = 30 μm. (M–R) The subcellular localization of AtRFC1/2/3/4/5 in root tip cells of the stably transformed Arabidopsis plants. Bars = 50 μm.
The Subunit Interactions of RFC in Arabidopsis and Rice
Structural analyses of E. coli γ complex and yeast RFC complex revealed that the five subunits are arranged in a circular fashion (Jeruzalmi et al., 2001; Yao et al., 2003). However, it remains unclear how RFC subunits connect with each other to form an integrated RFC complex in higher plants. To investigate the subunit interactions among RFC subunits in Arabidopsis and rice, yeast two-hybrid assays were conducted. As shown in Figure 4A, OsRFC1 could interact with OsRFC2/3/4/5, OsRFC2 with OsRFC3/5, and OsRFC3 with OsRFC4, but no interaction was detected between OsRFC2 and OsRFC4. Meanwhile, it was found that AtRFC2, AtRFC3, and AtRFC4 could interact with each other, while AtRFC5 only interacted with AtRFC4 (Figure 4B). Unlike OsRFC1, AtRFC1 could not interact with AtRFC2/3/4/5 subunits.
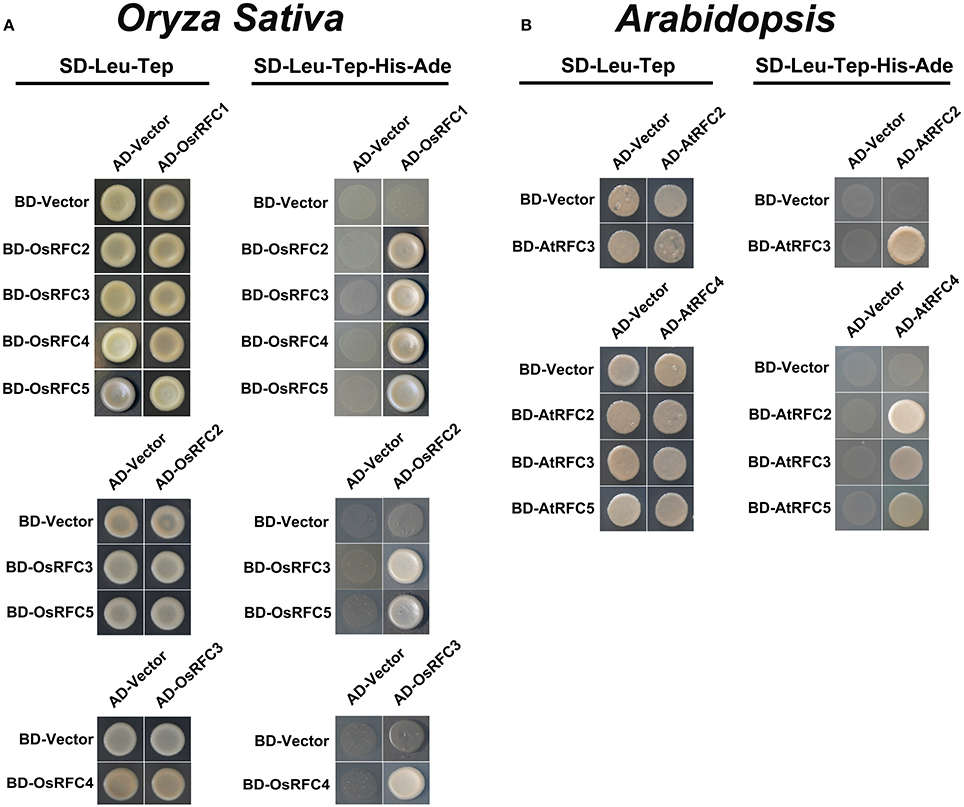
Figure 4. Yeast two-hybrid assay showing interactions among AtRFC2/3/4/5 subunits or OsRFC1/2/3/4/5 subunits. (A) Interactions between OsRFC1/2/3/4/5 subunits in rice. (B) Interactions between AtRFC2/3/4/5 subunits in Arabidopsis. Yeast two-hybrid assays were carried out and the co-transformed strains were spotted on SD-L-T/SD-L-T-H-A plates to test the direct interaction between the expressed proteins. Yeast strains co-transformed with the “empty” AD or BD plasmids were used as negative controls. AD, pGADT7 vector; BK, pGBKT7 vector; SD, synthetic dextrose; L, Leu; T, Trp; H, His; A, adenine.
Following the Y2H analysis, to further confirm the interactions of RFC subunits in Arabidopsis and rice, BiFC technique was employed. The N- and C-terminal ends of YFP protein were fused with AtRFCs and OsRFCs, respectively, and then the fusion proteins were co-expressed in tobacco leaves. As shown in Figure 5, no YFP signal was accumulated in epidermal cells co-transfected with YFPN and AtRFC2-YFPC or YFPN and AtRFC4-YFPC (Figures 5A,B,E,F). Obvious YFP signals were detected, however, in cells co-transfected with AtRFC3-YFPN and AtRFC2-YFPC, AtRFC2/3/5- YFPN and AtRFC4-YFPC (Figures 5C,D,G–L), consistent with the results of Y2H assays, indicating that AtRFC2, AtRFC3, and AtRFC4 could interact with each other, while AtRFC5 interacted only with AtRFC4. On the other hand, YFP signals of OsRFC2-YFPC and OsRFC3/5-YFPN, OsRFC3-YFPC and OsRFC4-YFPN were observed in the nucleus and cytoplasm of the transformed epidermal cells (Figures 5O–R,U,V), but there were no YFP signals between YFPN and OsRFC2-YFPC or YFPN and OsRFC3-YFPC (Figures 5M,N,S,T). Because AtRFC1 did not interact with any AtRFC2/3/4/5 subunits, it was speculated that the large subunit might be assembled into the complex with the assistance of other RFC subunits. Consistent with this presumption, YFP signals were observed in the nucleus of cells co-expressing AtRFC1-YFPC and AtRFC2/3/4/5-YFPN in the presence of all four AtRFC2/3/4/5 subunits (Figures 6C–J; Table S1), whereas YFP signals of OsRFC1-YFPC and OsRFC2/3/4/5-YFPN were only detected in the nucleus of the epidermal cells in the absence of RFC2/3/4/5 subunits (Figures 6M–T). No YFP signals were detected between YFPN and AtRFC1-YFPC or YFPN and OsRFC1-YFPC (Figures 6A,B,K,L). These results showed that the interaction patterns of AtRFCs were different from that of OsRFCs.
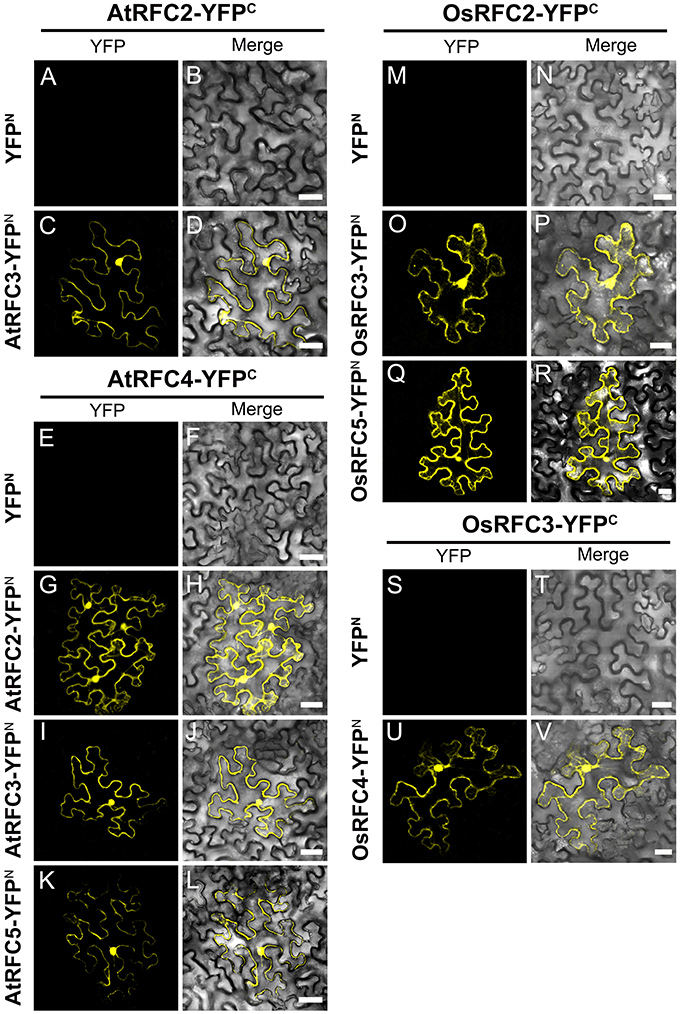
Figure 5. Analysis of interactions between AtRFC2/3/4/5 subunits or between OsRFC2/3/4/5 subunits using the BiFC assay in tobacco leaf epidermis cells. (A–D) AtRFC2 and AtRFC3 can interact with each other. (E–L) AtRFC4 can interact with AtRFC2/3/5, respectively. (M–R) OsRFC2 interacts with OsRFC3/5. (S–V) OsRFC3 interacts with OsRFC4. The tobacco epidermal cells were co-transfected with constructs encoding the indicated fusion proteins. YFPC, YFP C-terminal fragment (aa 156–239); YFPN, YFP N-terminal fragment (aa 1–155). Bars = 30 μm.
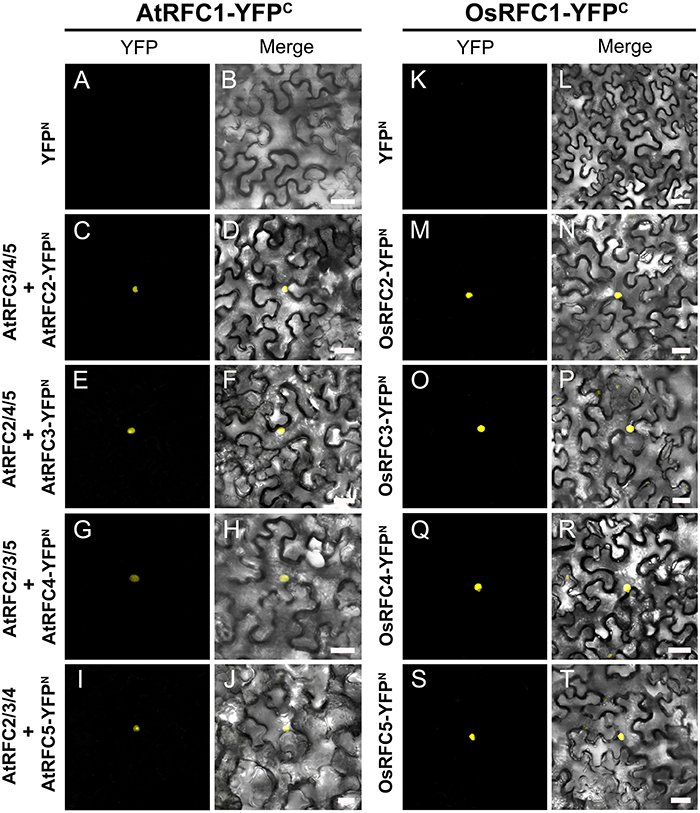
Figure 6. Analysis of the interactions between AtRFC1 and AtRFC2/3/4/5 as well as between OsRFC1 and OsRFC2/3/4/5 using the BiFC assay. (A–J) AtRFC1 can interact with AtRFC2/3/4/5 in the presence of all four AtRFC2/3/4/5 subunits in tobacco leaf cells. (K–T) OsRFC1 can directly interact with OsRFC2/3/4/5 in tobacco leaf cells. YFPC, YFP C-terminal fragment (aa 156–239); YFPN, YFP N-terminal fragment (aa 1–155). Bars = 30 μm.
Conserved Substitution in the Arabidopsis and Rice RFC Subunits
Sequence alignment revealed that the RFC subunits in Arabidopsis and rice are highly conserved. To gain insights on the conserved nature of AtRFCs and OsRFCs, interaction assays between RFC subunits of Arabidopsis and rice were performed via BiFC in tobacco epidermal cells. According to the phylogenetic analysis (Figure S1), we exchanged AtRFC1 with OsRFC1, AtRFC2 with OsRFC4, AtRFC3 with OsRFC3, AtRFC4 with OsRFC2, and AtRFC5 with OsRFC5. The results showed that AtRFC1 could bind OsRFC2/3/4/5 when all four small RFC subunits existed simultaneously (Figures S5A–J), while OsRFC1 could directly interact with AtRFC2/3/4/5 in the absence of other AtRFCs subunits (Figures S5K–T), which was different from the interactions between AtRFC1 and AtRFC2/3/4/5. Likewise, OsRFC2 could interact with AtRFC2/3/5, respectively (Figures S6A–C). YFP signals of OsRFC3-YFPN and AtRFC2/4-YFPC were detected in the nucleus and cytoplasm of the tobacco epidermal cells (Figures S6D,E). YFP signals could also be observed in cells co-expressing OsRFC4-YFPN and AtRFC3/4-YFPC (Figures S6F,G). Moreover, OsRFC5 could directly interact with AtRFC4 (Figure S6H). Taken together, these results suggested that Arabidopsis and rice RFC proteins were highly conserved.
The Regions Required for Complex Formation of the Five Arabidopsis RFC Subunits
In humans, deletion analysis of the large subunit p140 has shown that a region between amino acids 822 and 1,142 is required for the formation of the RFC complex (Uhlmann et al., 1997a). In the same way, sequences close to the C terminus of each of the small subunits were required for formation of the five-subunit complex (Uhlmann et al., 1997b). To identify the regions that were responsible for subunit interactions and complex formation of the five RFC subunits in Arabidopsis and rice, a series of truncated RFC proteins were fused with N- or C-terminus of the YFP and used in the BiFC assay.
As shown in Figure 7A and Figure S7, the YFP signals of AtRFC1 Δ1-334-YFPN and AtRFC2/3/4/5 YFPC were observed in the nucleus of the transformed tobacco epidermal cells (Figures S7A–D), indicating that the region of amino acids 1–334 in AtRFC1 was not involved in the formation of the five-subunit RFC complex. However, no fluorescence signal was detected in cells co-expressing AtRFC1 Δ1-457-YFPN and AtRFC2/3/4/5-YFPC (Figures S7E–H), suggesting that the region of amino acids 335–457 of AtRFC1 was required for assembly of the RFC complex. On the other hand, we found that a deletion of 40 C-terminal amino acids (AtRFC1 Δ917-956-YFPN) did not affect interactions with AtRFC2/3/4/5 (Figures S7I–L), while an additional deletion of 20 C-terminal amino acids (AtRFC1 Δ897-956-YFPN) did not support the RFC complex formation (Figures S7M–P). These results indicated that the region within amino acids 335–457 and 898–917 of AtRFC1 mediated its interaction with AtRFC2/3/4/5 to form the five-subunit complex.
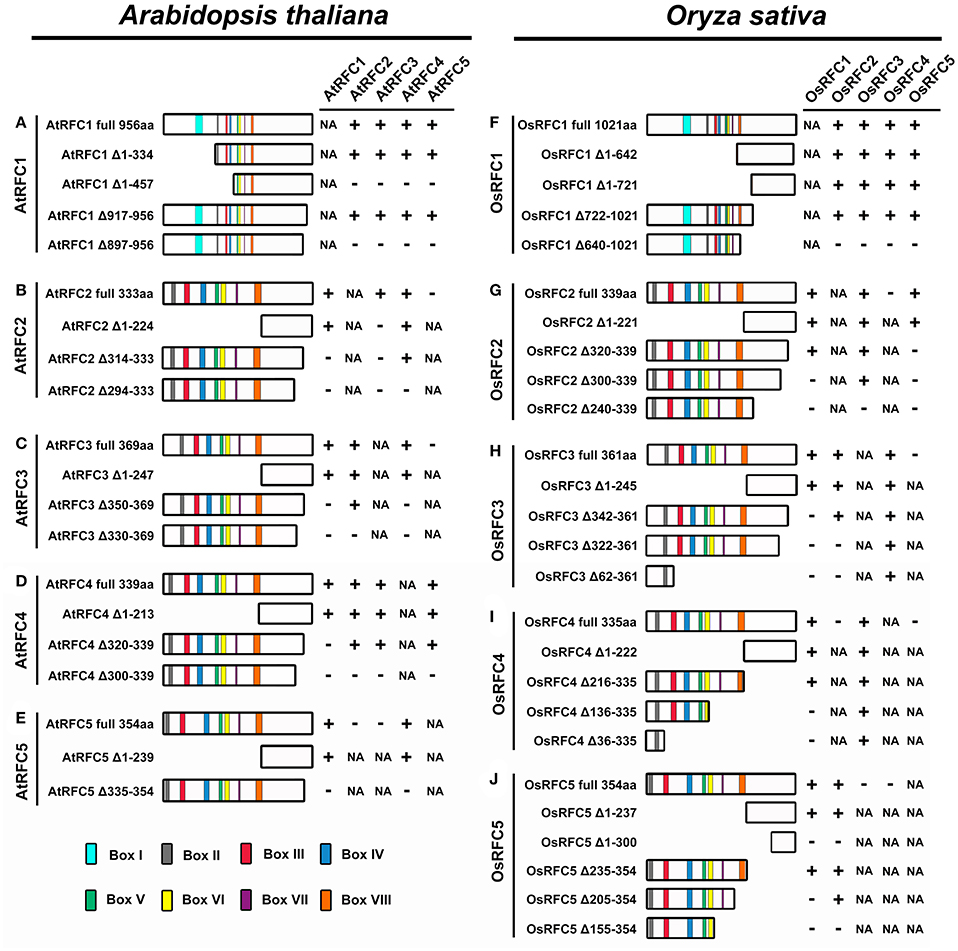
Figure 7. Summary of the truncated RFC subunits and their effects on RFC complex formation in Arabidopsis and rice. (A–E) Schematic diagrams of the regions required for RFC complex formation in Arabidopsis. (F–J) Schematic diagrams of the regions required for RFC complex formation in rice. The symbol “+” indicates that these proteins can interact; the symbol “–” indicates that these proteins cannot interact; the abbreviation NA indicates not applicable.
Deletion analysis of the AtRFC2 subunit is shown in Figure 7B and Figure S8. Deletion from the N terminus of AtRFC2 to amino acid 224 (AtRFC2 Δ1–224) did not affect the interactions between AtRFC2 and AtRFC1/4 (Figures S8A,C). AtRFC2 Δ314–333, with 20 C-terminal amino acids deleted, could interact with AtRFC4 (Figure S8F), but could not interact with AtRFC1 and AtRFC3 (Figures S8D,E). Deletion of an additional 20 amino acids at the C-terminus (AtRFC2 Δ294–333) led to a subunit variant that could not interact with AtRFC1/4 (Figures S8G,I). Moreover, the interactions between AtRFC2 and AtRFC3 disappeared no matter whether the N- or C-terminal sequence of AtRFC2 was truncated (Figures S8B,E,H). These results indicated that the C-terminal region between amino acids 314-333 of AtRFC2 mediated its interaction with AtRFC1, while the C-terminal region between amino acids 294–314 of AtRFC2 mediated its interaction with AtRFC4. In contrast to truncated AtRFC2, only the entire AtRFC2 protein could interact with AtRFC3.
Similar experiments were performed on deletion variants of the AtRFC3 subunit (Figure 7C; Figure S9). Full-length AtRFC3, as well as N-terminal deletion of AtRFC3 (AtRFC3 Δ1–247), supported interactions between AtRFC3 and AtRFC1/2/4 (Figures S9A–C). AtRFC3 Δ350–369, which lacked the 20 C-terminal amino acids no longer supported interactions between AtRFC3 and AtRFC1/4 (Figures S9D,F), but this did not affect its connection with AtRFC2 (Figure S9E). AtRFC3 ΔC330–369, which possessed an additional deletion of 20 C-terminal amino acids, did not interact with AtRFC1/2/4 (Figures S9G–I). These findings suggested that the C-terminal region of AtRFC3 between amino acids 350–369 was required for the interactions between AtRFC3 and AtRFC1/4, and the C-terminal region of AtRFC3 between amino acids 330–350 was required for interaction between AtRFC2 and AtRFC3.
Consistent with the results of AtRFC2/3, as a deletion variant of the AtRFC4 subunit, AtRFC4 Δ1–213 did not affect complex formation with AtRFC1/2/3/5, as the full-length AtRFC4 (amino acids 1–339) did (Figure 7D; Figures S10A–D). AtRFC4 Δ320–339, with 20 C-terminal amino acids deleted, supported interactions with AtRFC2/3/5, but did not support interaction with AtRFC1 (Figures S10E–H). When the full-length RFC4 subunit was replaced by RFC4 Δ300–339, a deletion protein lacking 40 C-terminal amino acids, its ability to interact with AtRFC1/2/3/5 was lost (Figures S10I–L). These results indicated that a C-terminal region of AtRFC4 between amino acids 320 and 339 was required for connection with AtRFC1, while the C-terminal region between 300 and 320 mediated the interactions between AtRFC4 and AtRFC2/3/5.
In the same way, we identified the regions responsible for complex formation of AtRFC5 (Figure 7E; Figure S11). The truncated protein lacking 239 N-terminal amino acids (AtRFC5 Δ1–239) did not affect its interactions with AtRFC1 and AtRFC4 (Figures S11A,B). However, when the 20 C-terminal amino acids of AtRFC5 (AtRFC5 Δ335–354) were deleted, the above interactions between AtRFC5 and AtRFC1/4 disappeared (Figures S11C,D). These results demonstrated that sequences from 335 to 354 of the C termini of AtRFC5 were required for the interactions with AtRFC1 and AtRFC4.
Regions Required for Complex Formation of the Five Rice RFC Subunits
To investigate the regions that were required for complex formation in OsRFC1, deletion variants were used in the BiFC assay. The truncated proteins OsRFC1 Δ1–642 and OsRFC1 Δ1–721 that lacked 642 and 721 aa at the N-terminal end did not affect the interactions with the OsRFC2/3/4/5 (Figure 7F; Figures S12A–H). Similar results were observed when the region between 722 and 1,021 aa in the C-terminal of OsRFC1 was deleted (Figures S12I–L). However, OsRFC1 Δ640–1021, with another 82 C-terminus amino acids deleted, did not support RFC complex formation (Figures S12M–P). This indicated that a region between amino acids 722 and 1021 of OsRFC1 was sufficient for RFC complex formation and interactions between OsRFC1 and OsRFC2/3/4/5.
OsRFC2 Δ1–221, a variant containing a deletion of the boxes II-VIII was able to interact with OsRFC1/3/5 (Figure 7G; Figures S13A–C). Consistent with this, a deletion of 20 amino acids from the C terminus (OsRFC2 Δ320–339) did not affect the interaction with OsRFC1 or OsRFC3 (Figures S13D,E), but the connection with OsRFC5 disappeared (Figure S13F). Deletion of 40 C-terminal amino acids of OsRFC2 (OsRFC2 Δ300–339) resulted in a subunit variant incapable of supporting interaction with OsRFC1 or OsRFC5 (Figures S13G,I), while the deletion did not affect its ability to interact with OsRFC3 (Figure S13H). OsRFC2 Δ240–339, which lacked 100 C-terminal amino acids was unable to interact with OsRFC1/3/5 (Figures S13J–L). These results demonstrated that in the C terminus of OsRFC2, the region 320–339 mediated interaction with OsRFC5, the region 300–320 mediated interaction with OsRFC1, and the region 240–300 mediated interaction with OsRFC3.
Similar to OsRFC2, the truncated OsRFC3 Δ1–245 that lacked 245 aa N-terminal amino acids did not affect interactions with the other three subunits of OsRFC1/2/4 (Figure 7H; Figures S14A–C). OsRFC3 Δ342–361,with 20 C-terminal amino acids deleted, could interact with OsRFC2 and OsRFC4 but did not interact with OsRFC1 (Figures S14D–F). OsRFC3 Δ322–361, which possessed an additional deletion of 20 C-terminal amino acids, was able to interact with OsRFC4, but could not interact with OsRFC1 and OsRFC2 (Figures S14G–I). OsRFC3 Δ62–361, lacking 300 C-terminal amino acids, still interacted with OsRFC4, but not with OsRFC1 and OsRFC2 (Figures S14J–L). These results indicated that the regions between amino acids 342–361 and 322–342 were necessary for the interactions with OsRFC1 and OsRFC2, respectively. Different from the results of OsRFC1 and OsRFC2, the N-terminus and C-terminus of OsRFC3 were required for the interaction with OsRFC4.
Next, to identify the regions required for interactions with OsRFC1 or OsRFC3, deletion variants of OsRFC4 were prepared. OsRFC4 Δ1–222 that lacked 222 N-terminal amino acids, supported the interactions with OsRFC1 or OsRFC3 (Figure 7I; Figures S15A,B). Consistent with this, OsRFC4, with 120 amino acids deleted from the C terminus (OsRFC4 Δ216–335) was also able to interact with OsRFC1 or OsRFC3 (Figures S15C,D). However, OsRFC4 Δ136–335, with an additional 80 C-terminal amino acids deleted, did not support interaction with OsRFC1, but still interacted with OsRFC3 (Figures S15E,F). This indicated that a region between amino acids 136 and 216 within the OsRFC4 C-terminus was indispensable for interaction with OsRFC1, which was similar with the results of other small RFC subunits. OsRFC4 Δ36–335, with 300 C-terminal amino acids deleted, retained its ability to interact with OsRFC3 (Figure S15H), suggesting that the interaction between OsRFC4 and OsRFC3 might be very tight and thus was not affected by deleting N-terminal or C-terminal regions of OsRFC4.
Furthermore, N-terminal deletion of OsRFC5 that removed the 237 amino acids of its N-terminal end (OsRFC5Δ1–237) did not affect complex formation with OsRFC1 and OsRFC2 (Figure 7J; Figures S15I,J). However, when 300 N-terminal amino acids were deleted from of OsRFC5 (OsRFC5 Δ1–300), its ability to interact with OsRFC1 and OsRFC2 was lost (Figures S15K,L). A C-terminal deletion of the OsRFC5 that ended at amino acid 235 (OsRFC5 Δ235–354) did not affect its interactions with the OsRFC1 and OsRFC2 (Figures S15M,N). OsRFC5 Δ205–354, with an additional 30 C-terminal amino acids deleted, did not support interaction with OsRFC1 (Figure S15O), but still interacted with OsRFC2 (Figure S15P). OsRFC5 Δ155–354, with an additional 50 C-terminal amino acids deleted, did not support interaction with OsRFC1 and OsRFC2 (Figures S15Q,R). These results demonstrated that the region between amino acids 205 and 235 within the OsRFC5 C-terminus was indispensable for interaction with OsRFC1, while the amino acid sequences from 155 to 205 in its C terminus were required for interaction with OsRFC2.
A Model for Subunit Arrangement in RFCs
Interaction models for the organization of the five subunits within the RFC complex were summarized on the basis of our studies in Arabidopsis and rice as well as the reported data in yeast and humans (Shiomi et al., 2000; Yao et al., 2003; Bowman et al., 2004). In humans, subunits p36, p37, and p40 form a three-subunit core complex where any two of the three subunits interact with each other; the p38 and p140 subunits cooperatively bind to the core complex in assembling the RFC complex (Figure 8A). The yeast RFC subunits assemble into a ring-shape structure with the arrangement ScRFC5-ScRFC2-ScRFC3-ScRFC4-ScRFC1 (Figure 8B). In Arabidopsis, as shown in Figure 8C, the subunits AtRFC2, AtRFC3, and AtRFC4 interacted with each other and formed a three-subunit sub-complex, AtRFC5 interacted with AtRFC4, and then AtRFC1 bound to the four-subunit complex assembling into a heteropentamer. The organization pattern of AtRFCs was AtRFC5-AtRFC4-AtRFC3/2-AtRFC2/3-AtRFC1, which was similar to those of humans, but the interactions between RFC1 and the other subunits were different. In rice, the interaction results provided a model where the subunits were arranged within the circular pentamer like OsRFC5-OsRFC2-OsRFC3-OsRFC4-OsRFC1 which was similar to yeast RFCs (Figure 8D), except that OsRFC1 could interact with the other four subunits. These results indicated that the organizations of the five RFC subunits were discrepant in different species, implying that the structure and function of different subunit interactions were diverse between human, yeast, and higher plants.
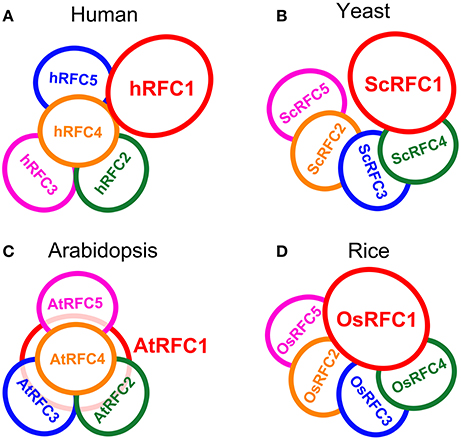
Figure 8. The models of RFC subunits arrangement in human (A), yeast (B), Arabidopsis (C), and rice (D).
Discussion
The Complex of Five RFC Subunits in Arabidopsis and Rice May Assemble in Different Pentameric Forms
The clamp loaders composed of five distinct subunits are arranged in a circular shape in eukaryotes (Bowman et al., 2004). Biochemical analysis, electron microscopy and atomic force microscopy studies, and crystal structure of the RFC complex have provided detailed views of subunit interdigitates and the architecture of RFC in yeast and humans (Shiomi et al., 2000; Jeruzalmi et al., 2001; O'Donnell et al., 2001; Yao et al., 2003; Bowman et al., 2004). We summarized the interaction models of the RFC subunits within the complexes on the basis of the reported data in yeast and human as well as our studies in Arabidopsis and rice (Figure 8). In Arabidopsis, the arrangement of the subunits within the circular RFC complex was AtRFC5-AtRFC4-AtRFC3/2-AtRFC2/3-AtRFC1 (Figure 8C). The assembly of the five subunits may be initiated by formation of the RFC 2/3/4/5 tetramer, followed by recruitment of the large RFC1 to form a pentamer. The interaction pattern of the Arabidopsis RFC subunits was similar to that of human RFC (Figure 8A), but AtRFC1 can interact with the other four subunits in the presence of other subunits, while hRFC1 (p140) only interacts with hRFC4 (p40) and hRFC5 (p38) (Uhlmann et al., 1996; Cai et al., 1997; Ellison and Stillman, 1998). In yeast and rice, the models showed that the arrangements of the subunits were similar, namely RFC5-RFC2-RFC3-RFC4-RFC1 (Figures 8B,D). However, the difference was that OsRFC1 can directly interact with the other four subunits in rice, while ScRFC1 can interact only with ScRFC4 and ScRFC5 in yeast. Although the RFC complex forms a circular pentamer like in eukaryotes, the organizations of the five subunits are discrepant in different species, indicating that the structure and arrangement of different subunit interactions are diverse from yeast and humans to higher plants.
The C-Terminal Regions of the RFC Subunits Are Required for Complex Formation
In humans, the five individual subunits were expressed though transcription/translation system and interacted to reconstitute a stable and bioactive complex with a three-subunit core complex. The large p140 (RFC1) interacted with the core complex in the presence of the p38 subunit (Cai et al., 1996, 1997; Podust and Fanning, 1997). Protein-protein interaction studies of yeast RFC showed that RFC2 interacted with RFC3/RFC5 and RFC3 interacted with RFC4 (Yao et al., 2003). In this study, we examined the interactions in the Arabidopsis and rice RFC subunits using Y2H and BiFC techniques. Our characterizations of the interactions between the Arabidopsis subunits were similar to the interaction pattern of the human subunits. AtRFC2, AtRFC3, and AtRFC4, like the p36p37p40 complex, interacted with each other and formed a three-subunit core complex. AtRFC1, like p140, binds to the core complex with the help of the other subunits. In rice, consistent with the interactions of the yeast subunits, the small subunits formed three sub-complexes –RFC2/3, RFC2/5 and RFC3/4–while OsRFC1 interacted with each of the other subunits.
To further investigate the regions within the five subunits responsible for complex formation, Y2H and BiFC assays were carried out using constructs of truncated RFC proteins. The ability of the truncated proteins of each subunit to form the RFC complex is summarized in Figure 7. The results showed that each subunit was required to form the RFC complex. The C-terminal regions of all Arabidopsis and rice RFC subunits were indispensable for subunit interactions. This was consistent with previous biochemical studies in humans and yeast as well as structural analysis of the E. coli and yeast clamp loaders (Uhlmann et al., 1996, 1997a,b; Jeruzalmi et al., 2001; Yao et al., 2003; Bowman et al., 2004). Different from the other subunits, in addition to the C-terminus, the N-terminal sequence of AtRFC2 was also required for the interaction with AtRFC3 (Figure 7B). Basically, the C-terminal regions mediated the strong interactions between individual subunits. In contrast, the truncated proteins lacking the conserved boxes close to their N-terminal regions did not affect the interactions with other subunits. This might explain why the C-terminal sequences of all five subunits are unique.
Conservation and Divergence of RFC Subunits in Arabidopsis and Rice
The clamp loading mechanism is functionally conserved across the domains of life (Trakselis and Benkovic, 2001; O'Donnell and Kuriyan, 2006). RFC subunits are evolutionarily conserved in both structure and function and exhibited significant sequence similarity to each other and to γ and δ′ of the E. coli γ complex (O'Donnell et al., 1993; Cullmann et al., 1995; Guenther et al., 1997). In particular, the RFC boxes II to VIII, which are mainly localized to the N-terminal half shared by the clamp loader, are highly conserved (Cullmann et al., 1995; Mossi and Hübscher, 1998). In this study, we analyzed and compared the structures and sequences of Arabidopsis and rice RFC complex (Figure 1). The molecular masses of AtRFC1 and OsRFC1 were 104.3 and 110.8 kDa, respectively. Similarly, the other four small subunits of Arabidopsis and rice possessed similar molecular mass, ranging from 36 to 42 kDa (Furukawa et al., 2003). In addition, the amino acid sequences of the five subunits in Arabidopsis showed a high degree of homology to the subunits of rice, up to 85%. The amino acid sequences of box III in the small subunits are similar as in their Arabidopsis counterpart. Although these subunits were homologous, it was not clear whether the rice subunits were the functional equivalent of the Arabidopsis orthologs. Thus, a substitution assay between Arabidopsis and rice was performed, and the results showed that every subunit played a similar role during the interaction among the subunits, implying that this complex was conserved in higher plants (Figures S5, S6).
Although most of the amino acids are highly conserved between Arabidopsis and rice RFC subunits, there are many variations in three-dimensional structure and sequence (Figure 1). For example, OsRFC1 has a longer N terminus compared with AtRFC1, and thus it is larger. The N-terminal extension of RFC1 is not required for cell viability and clamp loading activity but is related to DNA damage in vivo (Uhlmann et al., 1997a; Gomes et al., 2000). The P loop within box III is a general Walker-type ATPase motif with the consensus sequence GXXGXGKT. The residue Pro467 in box III of OsRFC1 corresponds to Thr401 in box III of AtRFC1. This subtle change may result in the differences in function. Consistent with the difference in structure and sequence, AtRFC1 can interact with AtRFC2/3/4/5 in the presence of the other subunits, while OsRFC1 can directly bind OsRFC2/3/4/5. For example, the mutation of four subunits in humans (not including p38) results in the Lys of the P-loop motif being replaced by Glu, which affects complex assembly and RFC function (Podust et al., 1998). The C-terminal regions of the RFC subunits in Arabidopsis required for subunit-subunit interactions were different from their counterparts in rice. Taken together, these differences may result in structural changes and may lead to divergence of functions between Arabidopsis and rice.
RFC Subunits Are Essential for Cell Viability in Arabidopsis and Rice
Compared to the eukaryotic RFC clamp loader possessing four distinct small subunits, the gp44/62 complex in T4 bacteriophages possesses one large and four identical small subunits to load the clamp onto DNA (Moarefi et al., 2000). This indicates that there was some divergence between the clamp loaders in different organisms over evolutionary time, and the distinct small subunits of the RFC complex may have evolved diverse functions. In yeast, ScRFC1 plays an essential role in DNA replication and DNA repair. Mutations in ScRFC2, ScRFC3, ScRFC4, and ScRFC5 had defects in DNA replication and checkpoint controls, indicating that the subunits play crucial roles in DNA replication and the cell cycle checkpoint (Sugimoto et al., 1996, 1997; Noskov et al., 1998; Shimada et al., 1999; Gray and MacNeill, 2000; Kim and Brill, 2001). In Drosophila, DmRfc4 mutants had reduced numbers of replicating cells and had defects in mitotic chromosomes and the cell cycle checkpoint, demonstrating that DmRFC4 is essential for checkpoint control (Krause et al., 2001). In Arabidopsis, AtRFC1 plays important roles in meiotic recombination and CO (crossover) formation, and DNA double-strand break repair during meiosis (Liu et al., 2010, 2013; Wang et al., 2012). AtRFC3 plays a crucial role in systemic acquired resistance, cell proliferation and DNA replication (Xia et al., 2009, 2010). AtRFC4 is crucial for DNA replication (Qian et al., 2018). In this study, we found that the rfc2-1, rfc3-2, and rfc5-1 of the T-DNA insertion lines caused seed lethality (Figure 2A). Ovule clearing showed that all embryos arrested at the 2/4-celled embryo proper stage and the number of endosperm free nuclei decreased dramatically and finally endosperm reached 6–8 nuclei stages (Figures 2D–F; Table 2). The results suggested that AtRFC2/3/5 were required for maintaining mitosis in early embryo cells and endosperm free nuclei. Our study and the previous reports suggested that RFCs have a variety of functions in addition to DNA replication in higher plants. Based on the conservation of the RFC complex in sequence, structure and function in loading the clamps onto DNA in different species, in particular Arabidopsis, we speculated that the rice RFC subunits also played important roles in DNA replication and DNA repair. In rice, the five RFC subunits are highly expressed in tissues where cell division is active and cell cycle inhibitors significantly reduced the expression of OsRFC5 and slightly affected the expression of OsRFC1/2/3/4 (Furukawa et al., 2003). These findings indicate that RFCs may be involved in cell proliferation and cell cycle progression. However, the specific functions of the Arabidopsis RFC2/3/5 subunits and rice RFC subunits are still being studied.
Author Contributions
YC performed most of the experiments, analyzed the research results and wrote the paper; JQ performed most of the experiments and analyzed the data; LY participated in Y2H and BiFC assays; XZ and JJ participated in BiFC assays; YL participated in temporal and spatial expressions of AtRFC2/3/5; JZ conceived the research plans, guided the whole study, and modified the paper.
Funding
This research was supported by National Natural Science Foundation of China (31370348, 31670312).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00779/full#supplementary-material
References
Bowman, G. D., O'Donnell, M., and Kuriyan, J. (2004). Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429, 724–730. doi: 10.1038/nature02585
Bunz, F., Kobayashi, R., and Stillman, B. (1993). cDNAs encoding the large subunit of human replication factor C. Proc. Natl. Acad. Sci. U.S.A. 90, 11014–11018. doi: 10.1073/pnas.90.23.11014
Cai, J., Gibbs, E., Uhlmann, F., Phillips, B., Yao, N., O'Donnell, M., et al. (1997). A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA dependent ATPase and an intermediate in the assembly of the holoenzyme. J. Biol. Chem. 272, 18974–18981.
Cai, J., Uhlmann, F., Gibbs, E., Flores-Rozas, H., Lee, C. G., Phillips, B., et al. (1996). Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc. Natl. Acad. Sci. U.S.A. 93, 12896–12901. doi: 10.1073/pnas.93.23.12896
Chen, H., Zou, W., and Zhao, J. (2015). Ribonuclease J is required for chloroplast and embryo development in Arabidopsis. J. Exp. Bot. 66, 2079–2091. doi: 10.1093/jxb/erv010
Chen, M., Pan, Z. Q., and Hurwitz, J. (1992). Studies of the cloned 37 kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 89, 5211–5215. doi: 10.1073/pnas.89.12.5211
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743.
Cullmann, G., Fien, K., Kobayashi, R., and Stillman, B. (1995). Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 4661–4671. doi: 10.1128/MCB.15.9.4661
Deng, Y., Zou, W., Li, G., and Zhao, J. (2014). TRANSLOCASE OF THE INNER MEMBRANE9 and 10 are essential for maintaining mitochondrial function during early embryo cell and endosperm free nucleus divisions in Arabidopsis. Plant Physiol. 166, 853–868. doi: 10.1104/pp.114.242560
Ellison, V., and Stillman, B. (1998). Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J. Biol. Chem. 273, 5979–5987. doi: 10.1074/jbc.273.10.5979
Fien, K., and Stillman, B. (1992). Identification of replication factor C from Saccharomyces cerevisiae: a component of the leading-strand DNA replication complex. Mol. Cell Biol. 12, 155–163. doi: 10.1128/MCB.12.1.155
Furukawa, T., Ishibashi, T., Kimura, S., Tanaka, H., Hashimoto, J., and Sakaguchi, K. (2003). Characterization of all the subunits of replication factor C from a higher plant, rice (Oryza sativa L.), and their relation to development. Plant Mol. Biol. 53, 15–25. doi: 10.1023/B:PLAN.0000009258.04711.62
Gomes, X. V., Gary, S. L., and Burgers, P. M. (2000). Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 275, 14541–14549. doi: 10.1074/jbc.275.19.14541
Gray, F. C., and MacNeill, S. A. (2000). The Schizosaccharomyces pombe rfc3+ genes encodes a homologue of the human hRFC36 and Saccharomyces cerevisiae RFC3 subunits of replication factor C. Curr. Genet. 37, 159–167. doi: 10.1007/s002940050514
Guenther, B., Onrust, R., Sali, A., O'Donnell, M., and Kuriyan, J. (1997). Crystal structure of the delta' subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91, 335–345.
Howell, E. A., McAlear, M. A., Rose, D., and Holm, C. (1994). CDC44: a putative nucleotide-binding protein required for cell cycle progression that has homology to subunits of replication factor C. Mol. Cell. Biol. 14, 255–267. doi: 10.1128/MCB.14.1.255
Jeruzalmi, D., O'Donnell, M., and Kuriyan, J. (2001). Crystal structure of the processivity clamp loader γ complex of E. coli DNA polymerase III. Cell 106, 429–441. doi: 10.1016/S0092-8674(01)00463-9
Kelch, B. A. (2016). The lord of the rings: structure and mechanism of the sliding clamp loader. Biopolymers 105, 532–546. doi: 10.1002/bip.22827
Kim, H. S., and Brill, S. J. (2001). Rfc4 interacts with Rpa1 and is required for both DNA replication and DNA damage checkpoints in Saccharomyces cerevisiae. Mol. Cell Biol. 21, 3725–3737. doi: 10.1128/MCB.21.11.3725-3737.2001
Kobayashi, M., Ab, E., Bonvin, A. M., and Siegal, G. (2010). Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex. J. Biol. Chem. 285, 10087–10097. doi: 10.1074/jbc.M109.054106
Krause, S. A., Loupart, M. L., Vass, S., Schoenfelder, S., Harrison, S., and Heck, M. M. (2001). Loss of cell cycle checkpoint control in Drosophila Rfc4 mutants. Mol. Cell Biol. 21, 5156–5168. doi: 10.1128/MCB.21.15.5156-5168.2001
Li, G., Zou, W., Jian, L., Qian, J., Deng, Y., and Zhao, J. (2017). Non-SMC elements 1 and 3 are required for early embryo and seedling development in Arabidopsis. J. Exp. Bot. 68, 1039–1054. doi: 10.1093/jxb/erx016
Liu, Q., Wang, J., Miki, D., Xia, R., Yu, W., He, J., et al. (2010). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22, 2336–2352. doi: 10.1105/tpc.110.076349
Liu, Y., Deng, Y., Li, G., and Zhao, J. (2013). Replication factor C1 (RFC1) is required for double-strand break repair during meiotic homologous recombination in Arabidopsis. Plant J. 73, 154–165. doi: 10.1111/tpj.12024
Luckow, B., Bunz, F., Stillman, B., Lichter, P., and Schutz, G. (1994). Cloning, expression, and chromosomal localization of the 140-kilodalton subunit of replication factor C from mice and human. Mol. Cell Biol. 14, 1626–1634. doi: 10.1128/MCB.14.3.1626
Ma, H., and Zhao, J. (2010). Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J. Exp. Bot. 61, 2647–2668. doi: 10.1093/jxb/erq104
Maga, G., Stucki, M., Spadari, S., and Hübscher, U. (2000). DNA polymerase switching: I. Replication factor C displaces DNA polymerase alpha prior to PCNA loading. J. Mol. Biol. 295, 791–801. doi: 10.1006/jmbi.1999.3394
McAlear, M. A., Howell, E. A., Espenshade, K. K., and Holm, C. (1994). Proliferating cell nuclear antigen (pol30). mutations suppress cdc44 mutations and identify potential regions of interaction between the two encoded proteins. Mol. Cell Biol. 14, 4390–4397.
McAlear, M. A., Tuffo, K. M., and Holm, C. (1996). The large subunit of replication factor C (Rfc1p/Cdc44p). is required for DNA replication and DNA repair in Saccharomyces cerevisiae. Genetics 142, 65–78.
Moarefi, I., Jeruzalmi, D., Turner, J., O'Donnell, M., and Kuriyan, J. (2000). Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J. Mol. Biol. 296, 1215–1223. doi: 10.1006/jmbi.1999.3511
Mossi, R., and Hübscher, U. (1998). Clamping down on clamps and clamp loaders–the eukaryotic replication factor C. Eur. J. Biochem. 254, 209–216.
Mossi, R., Keller, R. C., Ferrari, E., and Hübscher, U. (2000). DNA polymerase switching: II. Replication factor C abrogates primer synthesis by DNA polymerase alpha at a critical length. J. Mol. Biol. 295, 803–814. doi: 10.1006/jmbi.1999.3395
Naiki, T., Shimomura, T., Kondo, T., Matsumoto, K., and Sugimoto, K. (2000). Rfc5, in cooperation with rad24, controls DNA damage checkpoints throughout the cell cycle in Saccharomyces cerevisiae. Mol. Cell Biol. 20, 5888–5896. doi: 10.1128/MCB.20.16.5888-5896.2000
Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. (1999). AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43.
Noskov, V. N., Araki, H., and Sugino, A. (1998). The RFC2 gene, encoding the third-largest subunit of the replication factor C complex, is required for an S-phase checkpoint in Saccharomyces cerevisiae. Mol. Cell Biol. 18, 4914–4923. doi: 10.1128/MCB.18.8.4914
O'Donnell, M., Jeruzalmi, D., and Kuriyan, J. (2001). Clamp loader structure predicts the architecture of DNA polymerase III holoenzyme and RFC. Curr. Biol. 11, R935–946. doi: 10.1016/S0960-9822(01)00559-0
O'Donnell, M., and Kuriyan, J. (2006). Clamp loaders and replication initiation. Curr. Opin. Struct. Biol. 16, 35–41. doi: 10.1016/j.sbi.2005.12.004
O'Donnell, M., Onrust, R., Dean, F. B., Chen, M., and Hurwitz, J. (1993). Homology in accessory proteins of replicative polymerases–E. coli to humans. Nucleic Acids Res. 21, 1–3. doi: 10.1093/nar/21.1.1
Podust, V. N., and Fanning, E. (1997). Assembly of functional replication factor C expressed using recombinant baculoviruses. J. Biol. Chem. 272, 6303–6310. doi: 10.1074/jbc.272.10.6303
Podust, V. N., Tiwari, N., Ott, R., and Fanning, E. (1998). Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J. Biol. Chem. 273, 12935–12942. doi: 10.1074/jbc.273.21.12935
Qian, J., Chen, Y., Hu, Y., Deng, Y., Liu, Y., Li, G., et al. (2018). Arabidopsis replication factor C4 is critical for DNA replication during mitotic cell cycle. Plant J. 94, 288–303. doi: 10.1111/tpj.13855
Schmidt, S. L., Pautz, A. L., and Burgers, P. M. (2001). ATP utilization by yeast replication factor C. IV. RFC ATP-binding mutants show defects in DNA replication, DNA repair, and checkpoint regulation. J. Biol. Chem. 276, 34792–34800. doi: 10.1074/jbc.M011671200
Shimada, M., Okuzaki, S., Tanaka, T., Tougan, K. K., Tamai, C., Shimoda, C., et al. (1999). Replication factor C3 of Schizosaccharomyces pombe, a small subunit of replication factor C complex, plays a role in both replication and damage checkpoints. Mol. Biol. Cell 10, 3991–4003. doi: 10.1091/mbc.10.12.3991
Shiomi, Y., and Nishitani, H. (2017). Control of genome integrity by RFC complexes; Conductors of PCNA loading onto and unloading from chromatin during DNA replication. Genes 8:52. doi: 10.3390/genes8020052
Shiomi, Y., Usukura, J., Masamura, Y., Takeyasu, K., Nakayama, Y., Obuse, C., et al. (2000). ATP-dependent structural change of the eukaryotic clamp-loader protein, replication factor C. Proc. Natl. Acad. Sci. U.S.A. 97, 14127–14132. doi: 10.1073/pnas.97.26.14127
Shivji, M. K., Podust, V. N., Hübscher, U., and Wood, R. D. (1995). Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34, 5011–5017.
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Strzalka, W. K., Aggarwal, C., Krzeszowiec, W., Jakubowska, A., Sztatelman, O., and Banas, A. K. (2015). Arabidopsis PCNAs form complexes with selected D-type cyclins. Front. Plant Sci. 6:516. doi: 10.3389/fpls.2015.00516
Sugimoto, K., Ando, S., Shimomura, T., and Matsumoto, K. (1997). Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol. 17, 5905–5914. doi: 10.1128/MCB.17.10.5905
Sugimoto, K., Shimomura, T., Hashimoto, K., Araki, H., Sugino, A., and Matsumoto, K. (1996). Rfc5, a small subunit of replication factor C complex, couples DNA replication and mitois in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 7048–7052. doi: 10.1073/pnas.93.14.7048
Takashi, Y., Kobayashi, Y., Tanaka, K., and Tamura, K. (2009). Arabidopsis replication protein A 70a is required for DNA damage response and telomere length homeostasis. Plant Cell Physiol. 50, 1965–1976. doi: 10.1093/pcp/pcp140
Tamura, K., Dudley, J., Nei, M., and Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA). software version 4.0. Mol. Biol. Evol. 24, 1596–1599. doi: 10.1093/molbev/msm092
Trakselis, M. A., and Benkovic, S. J. (2001). Intricacies in ATP-dependent clamp loading: variations across replication systems. Structure 9, 999–1004. doi: 10.1016/S0969-2126(01)00676-1
Tsurimoto, T., and Stillman, B. (1989). Purification of a cellular replication factor, RF-C, that is required for coordinated synthesis of leading and lagging strands during simian virus 40 DNA replication in vitro. Mol. Cell Biol. 9, 609–619. doi: 10.1128/MCB.9.2.609
Uhlmann, F., Cai, J., Flores-Rozas, H., Dean, F. B., Finkelstein, J., O'Donnell, M., et al. (1996). In vitro reconstitution of human replication factor C from its five subunits. Proc. Natl. Acad. Sci. U.S.A. 93, 6521–6526.
Uhlmann, F., Cai, J., Gibbs, E., O'Donnell, M., and Hurwitz, J. (1997a). Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J. Biol. Chem. 272, 10058–10064.
Uhlmann, F., Gibbs, E., Cai, J., O'Donnell, M., and Hurwitz, J. (1997b). Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J. Biol. Chem. 272, 10065–10071.
Waga, S., and Stillman, B. (1998). The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751. doi: 10.1146/annurev.biochem.67.1.721
Wang, Y., Cheng, Z., Huang, J., Shi, Q., Hong, Y., Copenhaver, G. P., et al. (2012). The DNA replication factor RFC1 is required for interference-sensitive meiotic crossovers in Arabidopsis thaliana. PLoS Genet. 8:e1003039. doi: 10.1371/journal.pgen.1003039
Xia, S., Xiao, L., Gannon, P., and Li, X. (2010). RFC3 regulates cell proliferation and pathogen resistance in Arabidopsis. Plant Signal. Behav. 5, 168–170. doi: 10.4161/psb.5.2.10526
Xia, S., Zhu, Z., Hao, L., Chen, J. G., Xiao, L., Zhang, Y., et al. (2009). Negative regulation of systemic acquired resistance by replication factor C subunit 3 in Arabidopsis. Plant Physiol. 150, 2009–2017. doi: 10.1104/pp.109.138321
Yao, N., Coryell, L., Zhang, D., Georgescu, R. E., Finkelstein, J., Coman, M. M., et al. (2003). Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J. Biol. Chem. 278, 50744–50753. doi: 10.1074/jbc.M309206200
Keywords: Arabidopsis, DNA replication, protein interaction, rice, replication factor C
Citation: Chen Y, Qian J, You L, Zhang X, Jiao J, Liu Y and Zhao J (2018) Subunit Interaction Differences Between the Replication Factor C Complexes in Arabidopsis and Rice. Front. Plant Sci. 9:779. doi: 10.3389/fpls.2018.00779
Received: 02 March 2018; Accepted: 22 May 2018;
Published: 19 June 2018.
Edited by:
Michael James Considine, University of Western Australia, AustraliaReviewed by:
Cécile Raynaud, Centre National de la Recherche Scientifique (CNRS), FranceJie Le, Institute of Botany (CAS), China
Copyright © 2018 Chen, Qian, You, Zhang, Jiao, Liu and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jie Zhao, anpoYW9Ad2h1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Yueyue Chen
Yueyue Chen Jie Qian†
Jie Qian† Jie Zhao
Jie Zhao