- 1Department of Biological Sciences, Purdue University, West Lafayette, IN, United States
- 2Laboratory of Crop Physiology, Research Faculty of Agriculture, Hokkaido University, Sapporo, Japan
- 3Department of Botany and Plant Pathology, Purdue University, West Lafayette, IN, United States
The bZIP transcription factor VIP1 interacts with the Agrobacterium virulence protein VirE2, but the role of VIP1 in Agrobacterium-mediated transformation remains controversial. Previously tested vip1-1 mutant plants produce a truncated protein containing the crucial bZIP DNA-binding domain. We generated the CRISPR/Cas mutant vip1-2 that lacks this domain. The transformation susceptibility of vip1-2 and wild-type plants is similar. Because of potential functional redundancy among VIP1 homologs, we tested transgenic lines expressing VIP1 fused to a SRDX repression domain. All VIP1-SRDX transgenic lines showed wild-type levels of transformation, indicating that neither VIP1 nor its homologs are required for Agrobacterium-mediated transformation. Because VIP1 is involved in innate immune response signaling, we tested the susceptibility of vip1 mutant and VIP1-SRDX plants to Pseudomonas syringae and Botrytis cinerea. vip1 mutant and VIP1-SRDX plants show increased susceptibility to B. cinerea but not to P. syringae infection, suggesting a role for VIP1 in B. cinerea, but not in P. syringae, defense signaling. B. cinerea susceptibility is dependent on abscisic acid (ABA) which is also important for abiotic stress responses. The germination of vip1 mutant and VIP1-SRDX seeds is sensitive to exogenous ABA, suggesting a role for VIP1 in response to ABA. vip1 mutant and VIP1-SRDX plants show increased tolerance to growth in salt, indicating a role for VIP1 in response to salt stress.
Introduction
Virulent strains of the soil bacterium Agrobacterium tumefaciens cause the tumorigenic disease crown gall. Agrobacterium-mediated plant genetic transformation involves mobilization of transferred-DNA (T-DNA) and five virulence proteins (VirD2, VirE2, VirE3, VirD5, and VirF) from the bacterium into a plant cell (Gelvin, 2003, 2012).
The effector protein VirE2 has non-specific single-stranded DNA-binding activity and is thought to coat single-stranded T-DNA (T-strands) after entry into the plant cell (Citovsky et al., 1992), protecting T-strands from nucleolytic degradation (Yusibov et al., 1994; Rossi et al., 1996). In addition to this structural role, VirE2 interacts with a number of plant proteins including VirE2-interacting protein 1 (VIP1; Tzfira et al., 2001) and VIP2 (Anand et al., 2007). VIP1, a bZIP transcription factor which is a target of the mitogen-activated protein kinase 3 (MPK3), is thought to be involved in plant defense responses (Djamei et al., 2007; Pitzschke et al., 2009). Phosphorylation of VIP1 on serine 79 by MPK3 results in the import of VIP1 into the plant nucleus (Djamei et al., 2007). VIP1 may subsequently bind to VIP1 response elements (VREs) to activate transcription of its target genes (Pitzschke et al., 2009). VIP1 may also be involved in sulfur utilization, starch accumulation, osmosensory signaling, and touch-induced root waving (Ishida et al., 2004; Wu et al., 2010; Tsugama et al., 2012, 2014, 2016; Chen et al., 2015).
The importance and role of VIP1 in Agrobacterium-mediated transformation are controversial (Tzfira et al., 2001; Shi et al., 2014). Previous studies using transgenic tobacco lines expressing antisense constructs targeting VIP1, and the Arabidopsis thaliana T-DNA insertion mutant vip1-1, found that these plants showed decreased stable transformation compared to that of wild-type plants (Tzfira et al., 2001; Li et al., 2005). Overexpression of VIP1 in tobacco resulted in increased transformation susceptibility, suggesting that VIP1 plays a role in transformation (Tzfira et al., 2001). However, quantitative transformation assays with the vip1-1 mutant and with 59 A. thaliana VIP1 overexpressing lines showed no effect on transformation susceptibility (Shi et al., 2014), suggesting that VIP1 is not important for Agrobacterium-mediated transformation.
Previous studies indicated that β-glucoronidase-(GUS) or YFP-tagged VirE2 localizes to the plant nucleus (Citovsky et al., 1992, 1994; Tzfira and Citovsky, 2001). However, other studies showed exclusively cytoplasmic localization of VirE2 (Bhattacharjee et al., 2008; Grange et al., 2008; Lee et al., 2008; Sakalis et al., 2013; Shi et al., 2014). VirE2 possesses a weak putative nuclear localization signal (NLS) sequence which does not bind strongly to importin α protein (Citovsky et al., 1994; Chang et al., 2014). Early work indicated that VirE2 does not interact with Arabidopsis importin alpha-1 (IMPa-1, also known as AtKAPα) in yeast (Ballas and Citovsky, 1997), although Bhattacharjee et al. (2008) subsequently detected such interactions in yeast, in planta, and in vitro. However, VirE2-IMPa-1 complexes remained cytoplasmic in plants (Lee et al., 2008). VirE2 nuclear import has been attributed to its interaction with VIP1 (Tzfira et al., 2001), a protein that localizes to both the cytoplasm and the nucleus (Djamei et al., 2007; Shi et al., 2014). Activation of VIP1 by MPK3 and subsequent binding of phosphorylated VIP1 to VirE2 may facilitate nuclear localization of VIP1-VirE2-T-strand complexes (the Trojan-horse model; Djamei et al., 2007).
The vip1-1 mutant still produces ∼80% of the VIP1 protein, including the crucial bZIP DNA-binding domain (Li et al., 2005). Because this domain may be important for function, we used CRISPR technology to generate a homozygous mutant, vip1-2, that produces a smaller protein lacking the bZIP domain. Transient and stable transformation assays indicated no effect of this mutation on Agrobacterium-mediated transformation. Furthermore, transformation assays of single and multiple null mutant lines of VIP1 homologs, and transgenic lines overexpressing VIP1 fused to a modified EAR-like motif repression domain (SRDX; Hiratsu et al., 2002), also failed to show any major effect on transformation. We therefore conclude that VIP1 and its homologs are not required for Agrobacterium-mediated transformation. However, VIP1 may be important for defense responses against the fungus Botrytis cinerea, for abscisic acid (ABA) signaling, and for growth under salt stress conditions.
Materials and Methods
Plasmids and Strain Constructions
Supplementary Tables 1, 2 list the plasmids, strains, and single-guide RNA sequences used in this study. To make VIP1 CRISPR-Cas9 constructs, we designed three sets of sgRNA constructs targeting the VIP1 gene within the first exon. For each set, two 20-nucleotide oligomers of target DNA sequences were synthesized with an additional GATC on the 5′ end of the sense-strand and AAAC on the 5′ end of the antisense-strand. After annealing, we cloned this double stranded oligomer into the BbsI site of psgR-Cas9-At. A HindIII-KpnI fragment from this plasmid (containing both sgRNA and Cas9 expression cassettes) was cloned into pCAMBIA1300 to make the plasmids pE4351, pE4352, and pE4353. These T-DNA binary vectors were introduced by electroporation into A. tumefaciens GV3101 (Van Larebeke et al., 1974) to generate A. tumefaciens At2115, At2116, and At2117, respectively.
To make the vip1-2-Venus (out of frame) fusion construct, we cloned the vip1-2 RT-PCR product into the SmaI site of pBluescript KS+ to create pE4443. BglII and BamHI sites were used to remove the vip1-2 cDNA fragment, which was ligated to the Venus gene in pE3857 to produce pE4451 (confirmed by sequencing; Supplementary Table 1).
To create the vip1-2-GUS-Venus fusion construct, the nucleotides encoding the first 145 amino acids of vip1-2 was amplified by PCR using the primers VIP1-BglII-FP1 and vip1-2 peptide, flanked by BglII and BamHI sites, respectively. The PCR product was cloned into the SmaI site of pE886 to create pE4516. The BglII-BamHI fragment was made blunt with Klenow fragment of DNA polymerase (New England Biolabs) and cloned into the BglII site of pE3835 to make pE4521. A plasmid (pE4517) containing the VIP1-GUS-Venus fusion was made by cloning the BamHI-BglII fragment from pE3857 into the BglII site of pE3835.
To create the inducible VIP1 overexpression construct, we excised an SphI-XhoI fragment, containing the LexA operator and a minimal Cauliflower Mosaic Virus 35S promoter, from pER8 (Zuo et al., 2000). The fragment, made blunt using Klenow fragment of DNA polymerase, was ligated to pE3542 digested with AgeI and XhoI and made blunt. The resulting plasmid, pE3542, is a pSAT1-derived cloning vector used to generate β-estradiol-inducible gene constructions.
To generate the T-DNA binary vector into which the inducible gene constructions were placed, we ligated a blunted SbfI-NcoI fragment containing the XVE expression cassette and a Pnos-partial hptII gene into the blunted SwaI-NcoI site of pE4145 (pPZP-RCS-hptII) to make pE4216. We then ligated a NcoI fragment containing part of the hptII gene from pER8 to the NcoI site of pE4216 to generate a complete hptII gene (pE4215). pE4215 is a T-DNA binary vector containing the XVE and hptII expression cassettes and a AscI site into which the inducible gene expression cassette can be cloned.
The SwaI-NotI fragment containing the VIP1 gene was removed from the plasmid pE4132 and cloned into the SmaI and NotI sites of the β-estradiol inducible promoter plasmid pE3542, making pE4275. pE4275 was then digested with AscI and the inducible VIP1 fragment was cloned into the AscI site of the binary vector pE4215 to create pE4288. pE4288 was introduced by electroporation into A. tumefaciens GV3101 (Van Larebeke et al., 1974) to generate A. tumefaciens At2082.
Generation and Screening of VIP1 CRISPR/Cas9 and Inducible VIP1 Transgenic A. thaliana Plants
Wild-type Col-0 ecotype A. thaliana plants were transformed by A. tumefaciens At2115, At2116, At2117, or At2082 using a flower dip protocol (Clough and Bent, 1998). T0 generation seeds harvested from transformed plants were surface sterilized for 15 min using a 50% Bleach and 0.1% sodium dodecylsulfate (SDS) before washing 5 times with sterile water. After incubation overnight at 4°C, the seeds were plated on solidified Gamborg’s B5 medium (Caisson Labs) containing 100 μg mL-1 Timentin and 20 μg mL-1 hygromycin. The seeds were incubated at 23°C using a 16/8-h light/dark cycle. Hygromycin-resistant seedlings (T1 generation) were transplanted to soil and grown under the same temperature and light conditions. Seeds were harvested from each T1 plant and T2 generation plants grown in soil. For the VIP1 CRISPR/Cas9 plants, DNA isolated from leaves of individual T2 plants was used to PCR-amplify a region surrounding the sgRNA target site using primers listed in Supplementary Table 2. The PCR products were analyzed for mutations using a T7 endonuclease I (New England Biolabs) mismatch assay (Babon et al., 2003). Mutations were confirmed by sequencing. For inducible VIP1 plants, seeds were harvested from the T2 generation plants and selected on hygromycin. Seeds from homozygous plants (100% progeny surviving on selection) were used for future experiments.
VIP1 Induction in the Presence and Absence of Agrobacterium
The T3 generation inducible VIP1 seedlings were germinated on B5 medium containing 100 μg mL-1 Timentin and 20 μg mL-1 hygromycin. After 2 weeks, the seedlings were transferred to plates containing B5 medium only which were placed vertically in racks to allow for root tissue to grow on the surface of the medium. After 7 days, B5 liquid medium containing 1 μM of β-estradiol suspended in DMSO (induction solution) or B5 with DMSO only (control solution) was pipetted onto the plates until a thin layer of liquid covered the root tissue. To determine differential gene expression in the presence of Agrobacterium, cells of A. tumefaciens A136 (lacking a Ti-plasmid) were suspended in either induction or control solution at a concentration of 108 cells mL-1. The roots were incubated in the treatment solution for either 3 or 12 h before cutting the roots from the stems using a razor blade, rinsing with sterile water, dabbing them dry with a paper towel, and freezing them in liquid nitrogen. For each treatment, the root tissue was pooled from 30 individual plants. The tissue was stored at -80°C.
Preparation of Samples for Quantitative RT-PCR
The RNA was isolated from the root tissue of untreated, non-induced, induced, non-induced in the presence of Agrobacterium, and induced in the presence of Agrobacterium after 0, 3, and 12 h of incubation. This was done for two biological replicates of inducible VIP1 A. thaliana transgenic line #12 and inducible VIP1 A. thaliana transgenic line #8.
A total of 1.45 μg of total RNA was treated with Ambion DNase I (Thermo Fisher Scientific) and SuperScriptIII reverse transcriptase (Thermo Fisher Scientific) was used to synthesize cDNA according to the manufacturer’s protocols. Quantitative RT-PCR was performed with a Roche LightCycler 96 using FastStart Essential Green Master reagents (Roche). Primers used to amplify the genes are described in Supplementary Table 2. Data were analyzed using the LightCycler 96 software, REST1 2009 software, and Microsoft Excel.
Phenotypic Characterization of vip1-2 Plants
Homozygous vip1-2 and wild-type Col-0 plants were grown on soil at 23°C in a chamber with a 16/8-h light/dark cycle. After germination, plants were thinned to one plant per pot and photos were taken every 2–3 days throughout growth. Rosette, leaf, and flower bolt sizes were measured using image processing software and statistical analysis was performed using a Student’s t-test.
Isolation and Transfection of Arabidopsis and Tobacco BY-2 Protoplasts
Protoplasts were isolated from leaves of wild-type (ecotype Col-0) and vip1-2 A. thaliana plants and tobacco BY-2 cells and transfected as described in Lee et al. (2012). pE3170 (mRFP-nuclear marker) was co-transfected into protoplasts with the appropriate clones. Protoplasts were imaged 16 h after transfection using a Nikon A1R Confocal Laser Microscope System as described in Shi et al. (2014).
Agrobacterium-Mediated Transient and Stable Transformation Assays
Agrobacterium thaliana lines tested in this study are listed in Supplementary Table 3. Homozygous lines for the annotated T-DNA insertions were confirmed by PCR (primer sequences listed in Supplementary Table 2). Roots from 20-day-old A. thaliana plants grown in baby food jars containing sterile Gamborg’s B5 medium were cut into 3–5 mm segments. Root segments were assayed as described in Tenea et al. (2009). A. tumefaciens At849 [GV3101 containing pBISN1 (Narasimhulu et al., 1996)] was used for transient transformation assays, whereas A. tumefaciens A208 was used for stable transformation. Three replicates were performed for each experiment and root segments were pooled from 6 to 10 plants for each replicate. A total of 80 or more root segments were scored for each data point. Statistical analysis was performed using a Student’s t-test.
Quantitative RT-PCR of vip1-2
The RNA was isolated from leaf tissue harvested from 3-week-old plants grown on soil using TriZol2 reagent. For each sample, 1 μg of total RNA was treated with Ambion DNase I (Thermo Fisher Scientific) and SuperScriptIII reverse transcriptase (Thermo Fisher Scientific) was used to synthesize cDNA according to the manufacturer’s protocols. Real-time PCR was performed with a Roche LightCycler 96 using the FastStart Essential Green Master reagents (Roche). Primers used to amplify the 3′ end of the VIP1 transcript are described in Supplementary Table 2. Data were analyzed using the LightCycler 96 software, REST3 2009 software, and Microsoft Excel.
Botrytis cinerea and Pseudomonas syringae Pathogenesis Assays
Five-week-old A. thaliana wild-type (Col-0), vip1-1, vip1-2, and VIP1-SRDX Line #11 leaves (from plants grown on soil) were inoculated with 5 μL of B. cinerea at a concentration of 1.0 × 105 spores/mL. Lesion size was measured 3 days after infection and averaged over 18 leaves per genotype (36 leaves for Col-0). Standard error was calculated over two separate experiments and a Student’s t-test was used to test for significant differences.
Five-week-old A. thaliana wild-type (Col-0), vip1-1, vip1-2, and VIP1-SRDX Line #11 leaves (from plants grown on soil) were syringe-inoculated with P. syringae pv. tomato DC3000 (virulent) or P. syringae pv. tomato DC3000 hrcC (avirulent) at an optical density (A600) of 0.001 and 0.005, respectively. Bacterial growth was determined at 0 and 4 days after infection by isolating bacteria from six leaf disks for each plant and plating a dilution series to calculate the number of colony-forming units (cfu) per square centimeter of leaf material. Standard error was calculated over three replicates and a Student’s t-test was used to test for significant differences.
ABA and Hyper-Osmotic Germination and Root Growth Assays
Seeds were plated onto ½ MS 1% sucrose medium containing 0, 0.3, or 0.5 μM ABA or onto MS 2% sucrose medium containing 0, 50, 75, or 100 mM NaCl. A total of 25 seeds per genotype were placed on each plate with two (ABA) or four (NaCl) plates prepared for each treatment. The plates were incubated in a 23°C chamber with a 16/8-h light/dark cycle. Germinated seeds were scored 8 days after plating. Any seed with a radicle protruding was considered to have germinated. The number of germinated seeds was divided by the total number to calculate the percent germination and this was averaged over all the plates for each treatment. Student’s t-test was used to test for statistically significant differences.
To test the effect of exogenous ABA or hyper-osmotic conditions on root growth, seeds were germinated on ½ MS, 1% sucrose medium (ABA) or MS 2% sucrose medium (hyper-osmotic). The 5-day-old seedlings with a root length of ∼1 cm were transferred to ½ MS 1% sucrose containing 0, 2, or 20 μM of ABA or MS 2% sucrose with 0, 50, 75, or 100 mM of NaCl. These plates were placed vertically in racks in a 16/8-h light/dark cycle growth chamber at 23°C. A total of 11 plates were prepared for each treatment. Pictures were taken of each of the ABA and NaCl plates, 8 and 11 days (respectively), after the seedlings were transferred. Root length was determined using ImageJ software. Root length was averaged over 11 seedlings for each genotype for each treatment. The rate of growth was determined by subtracting the initial root length from the final root length divided by the number of days of growth. Student’s t-test was used to test for statistically significant differences.
Results
Generation of the vip1-2 Mutant
Several laboratories have utilized the T-DNA insertion mutant vip1-1 (SALK_001014.38.85.x) to study the role of VIP1 in Agrobacterium-mediated transformation and other cellular processes (Li et al., 2005; Pitzschke et al., 2009; Wu et al., 2010; Tsugama et al., 2012, 2016; Shi et al., 2014; Chen et al., 2015). However, vip1-1 is not a transcriptional null mutant and still produces the first 244 amino acids of the 341 amino acid VIP1 protein (Li et al., 2005; Shi et al., 2014). The VIP1-1 protein lacks the C-terminal domain necessary for self-dimerization and interaction with histone H2A (Li et al., 2005; Lacroix et al., 2008), but still contains the transcriptional activation domain as well as the majority of the bZIP DNA-binding domain (Figure 1A). We therefore used CRISPR-Cas9 technology (Feng et al., 2013) to generate a vip1 mutant that produces a smaller VIP1 protein (Figure 1B).
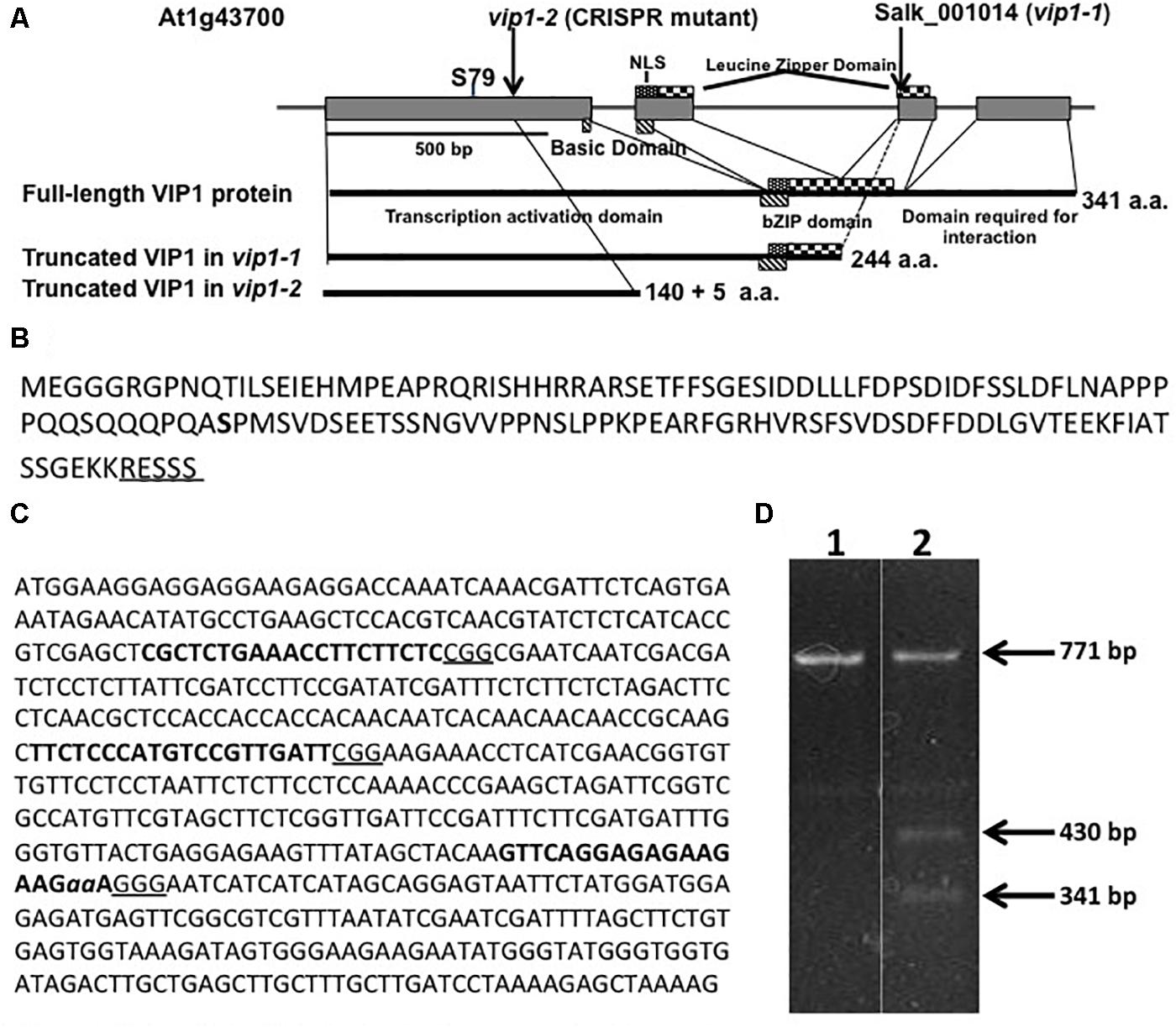
FIGURE 1. The VIP1, vip1-1, and vip1-2 genes and coding regions. (A) Map of the VIP1 coding region. Important protein domains are shown for the full-length and truncated VIP1-1 and VIP1-2 proteins. The C-terminal domain absent in both mutant proteins is required for VIP1 dimerization and VIP1-Histone H2A interactions (Li et al., 2005). Serine79, a phosphorylation site important for nuclear targeting of VIP1 (Djamei et al., 2007), is indicated. (B) Amino acid sequence of the VIP1-2 protein. The five amino acids shown underlined are not from the VIP1 protein, but result from the 2 bp deletion before a stop codon is reached. Serine79 is indicated in bold. (C) DNA sequence of the first exon of VIP1. Target sites for single-guide RNAs are highlighted in bold. The PAM sequences are underlined. The two nucleotides aa are deleted in the vip1-2 mutant. (D) T7 endonuclease I digestion of VIP1 PCR products (771 bp of gDNA surrounding the mutation in vip1-2) using wild-type gDNA as template (Lane 1) or a mixture of PCR products from both wild-type and vip1-2 mutant gDNAs (Lane 2). The mutation in vip1-2 creates a 2-bp mismatch generating two bands of 430 and 341 bp after T7 endonuclease cleavage.
We designed three guide RNAs to target multiple positions in the first exon of VIP1 (Figure 1C). T7 endonuclease analysis (Babon et al., 2003) of numerous T2 generation transgenic Arabidopsis lines expressing individual guide RNAs and Cas9 failed to identify mutations using constructs targeting the two most 5′-proximal regions of the VIP1 gene. However, the third guide RNA generated several different mutations (Figure 1D). DNA sequence analysis confirmed that one of these mutations resulted in a two base pair deletion, generating a premature stop codon. This mutant, vip1-2, encodes the first 140 amino acids of VIP1 plus five additional amino acids resulting from the frame-shift mutation. VIP1-2 lacks the bZIP DNA-binding domain, the nuclear localization signal (NLS) sequence, and the C-terminal domain important for VIP1 dimerization or interaction with histone H2A (Figure 1A).
Properties of the vip1-2 Gene and VIP1-2 Protein
RNA was isolated from homozygous vip1-2 leaves and RT-PCR was performed to determine whether VIP1 transcripts were still produced (Supplementary Figure 1A). Despite the presence of an early stop codon within the first exon of the vip1-2 gene, primers set at the 3′ end of the gene amplified a product, indicating that the VIP1 transcript was still produced. However, quantitative RT-PCR detected the VIP1 transcript at 35% of the level found in wild-type plants (Supplementary Figure 1B). The reduced level of the VIP1 transcripts may result from nonsense-mediated decay (Brogna and Wen, 2009). To verify that the vip1-2 mutant cannot make full-length VIP1 protein, we fused the vip1-2 cDNA to a Venus fluorescent protein coding sequence just before the position of the stop codon of the wild-type VIP1 cDNA. When introduced into BY-2 cells, this cDNA should not result in fluorescence because of the premature stop codon in the vip1-2 cDNA. Supplementary Figure 1C shows that a wild-type VIP1-Venus cDNA fusion construction could promote fluorescence in BY-2 cells. However, the vip1-2-Venus cDNA fusion construction could not.
Because wild-type VIP1 protein dimerizes (Li et al., 2005), we were concerned that Venus-tagged VIP1 may interact with untagged VIP1 present in cells, and that untagged full-length VIP1 may direct the subcellular localization of the dimer complex. We therefore conducted VIP1 subcellular localization experiments in both wild-type and vip1-2 mutant protoplasts. Wild-type VIP1-Venus fusion protein localized to both the plant cytoplasm and nucleoplasm, but not the nucleolus, of wild-type and vip1-2 mutant Arabidopsis protoplasts (Figures 2A,B; Djamei et al., 2007; Shi et al., 2014). The VIP1-2 protein is small (16,016 Da), and even when fused to Venus would produce a protein below the nuclear exclusion limit (< 60 kDa; Dingwall and Laskey, 1991), permitting nuclear entry of a VIP1-2-Venus fusion protein by diffusion. We therefore fused the VIP1-2 protein in-frame with a GUS-Venus protein, creating a protein (111.77 kDa) that exceeds the nuclear size exclusion limit. Transfection of a plasmid containing a VIP1-2-GUS-Venus expression cassette, together with a plasmid encoding a red fluorescence protein (RFP) nuclear marker, revealed exclusive cytoplasmic yellow fluorescence (Figure 2C), indicating that the VIP1-2 protein does not possess strong nuclear targeting capabilities. This result is consistent with deletion of the putative NLS from the VIP1-2 protein (Tsugama et al., 2012). Somewhat surprisingly, wild-type VIP1, when fused to GUS-Venus, also remains exclusively in the cytoplasm of both Arabidopsis and tobacco BY-2 protoplasts (Figures 2D,E). This result suggests either that the VIP1 nuclear localization signal sequence is not strong enough to target this large fusion protein to the nucleus, or that this fusion prevents phosphorylation of VIP1 serine-79 or some other aspect of VIP1 nuclear targeting.
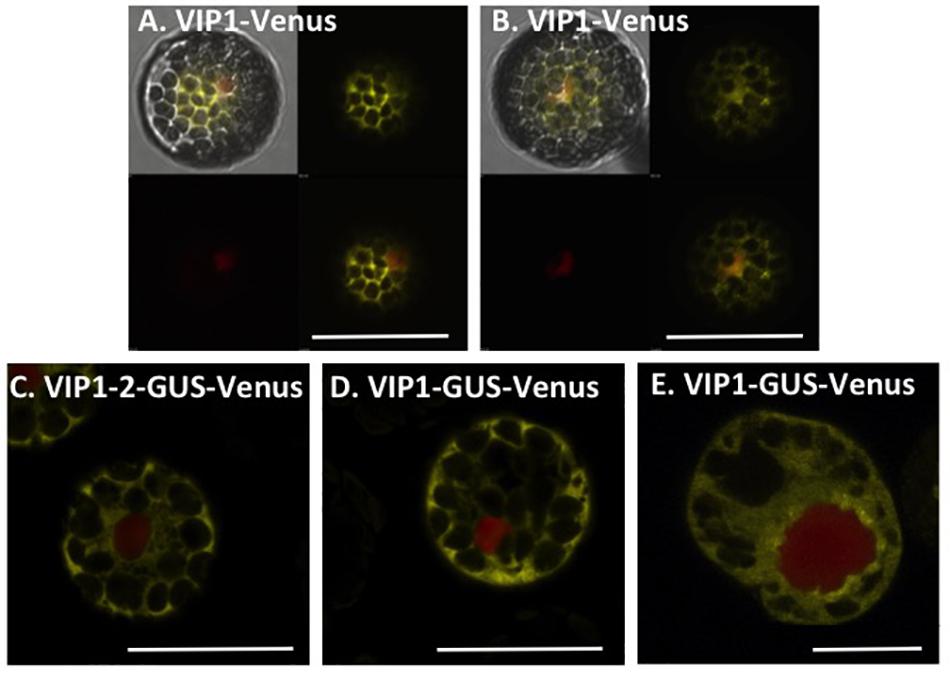
FIGURE 2. Subcellular localization of VIP1 and VIP1-2 proteins in protoplasts. Protoplasts were co-transfected with the indicated Venus-tagged constructs and a mRFP-NLS construct that marks the nucleus. A total of 16 h after transfection, the cells were imaged by confocal microscopy. VIP1-Venus localizes in both the cytoplasm and the nucleus of Col-0 (A) and vip1-2 (B) protoplasts; VIP1-2-GUS-Venus localizes exclusively in the cytoplasm of Col-0 protoplasts (C); localization of VIP1-GUS-Venus is limited to the cytoplasm of Col-0 (D) and tobacco BY-2 (E) protoplasts. In (A,B), four images of the same cell are presented (clockwise from top left: merged YFP, mRFP, and DIC; YFP; YFP + mRFP; mRFP). In (C–E), only the merged YFP and mRFP images are presented. Bars indicate 20 μm.
vip1-2 Plants Show Altered Growth Characteristics
We examined vip1-2 plants for abnormal growth or developmental phenotypes. vip1-2 plants exhibited increased rosette and leaf size compared to wild-type plants (Figures 3A–C). This growth phenotype suggests a role for VIP1 in the regulation of rosette leaf development. However, flowering time did not differ significantly from that of wild-type plants (flowering occurred 26 days after seed sowing).
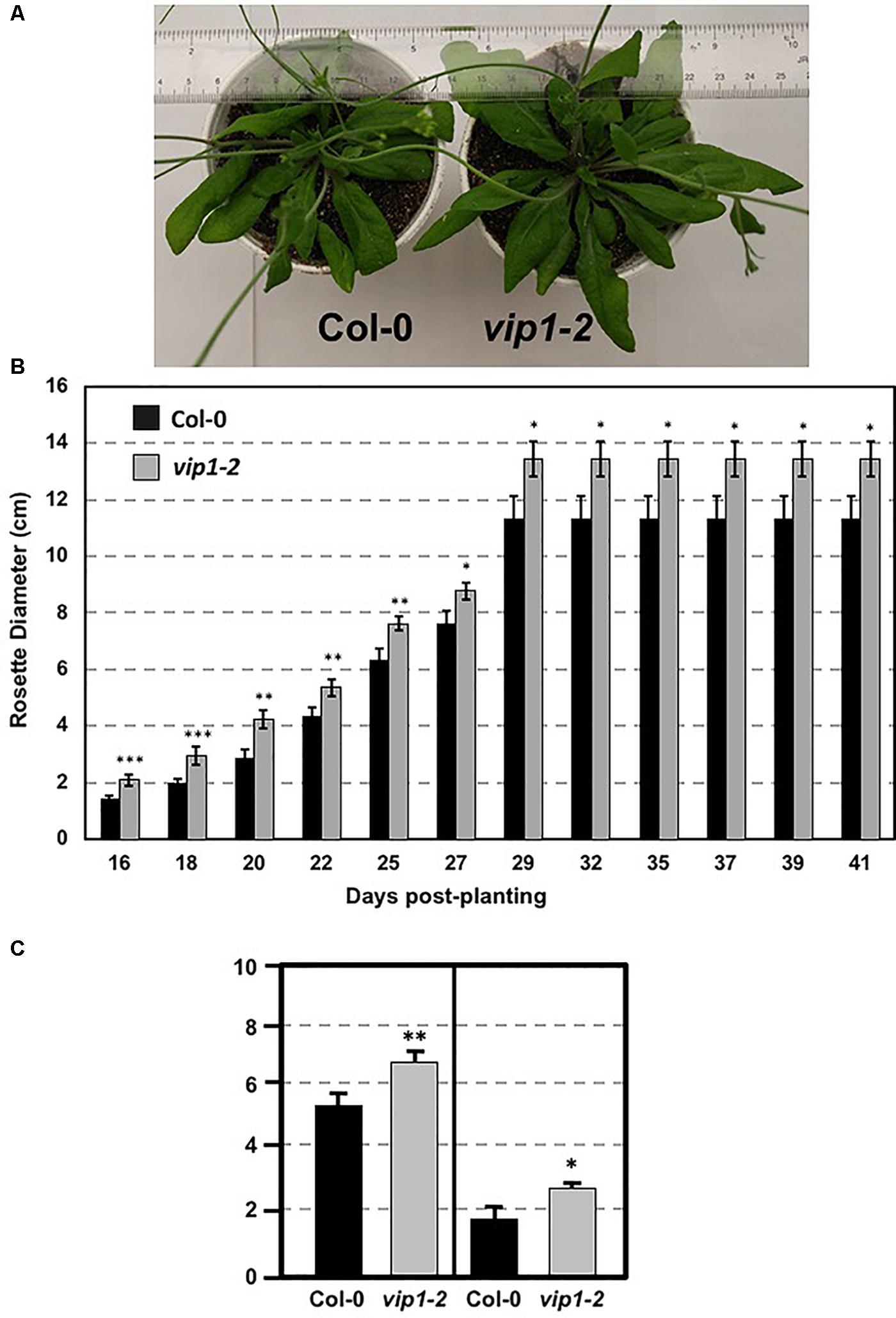
FIGURE 3. Growth of wild-type and vip1-2 mutant Arabidopsis plants. (A) Mature (29-day-old) wild-type (Col-0, left) and vip1-2 mutant (right) plants. (B) Bars represent the average diameter, ± SE, of leaf rosettes on 5–7 plants grown for the indicated number of days. (C) Bars represent the average leaf length (left) and width (right), ± SE, of the three largest leaves on five plants of each genotype grown for 29 days. Student’s t-test ∗P-value < 0.1, ∗∗P-value < 0.05, ∗∗∗P-value < 0.01.
vip1-2 Plants Show Wild-Type Susceptibility to Agrobacterium-Mediated Transformation
We tested transient and stable Agrobacterium-mediated transformation susceptibility of root segments from wild-type and vip1-2 plants. Root segments were infected with a non-tumorigenic Agrobacterium strain carrying a GUS reporter, At849 (transient transformation), or the tumorigenic strain A. tumefaciens A208 (stable transformation; Nam et al., 1997, 1999; Zhu et al., 2003; Shi et al., 2014) at several bacterial concentrations. Root segments of wild-type and vip1-2 plants had similar susceptibility to both transient and stable transformation at all bacterial concentrations tested (Figure 4; Supplementary Figure 2). These results correspond to our previous observations (Shi et al., 2014) that the vip1-1 mutant is not deficient in transformation susceptibility.
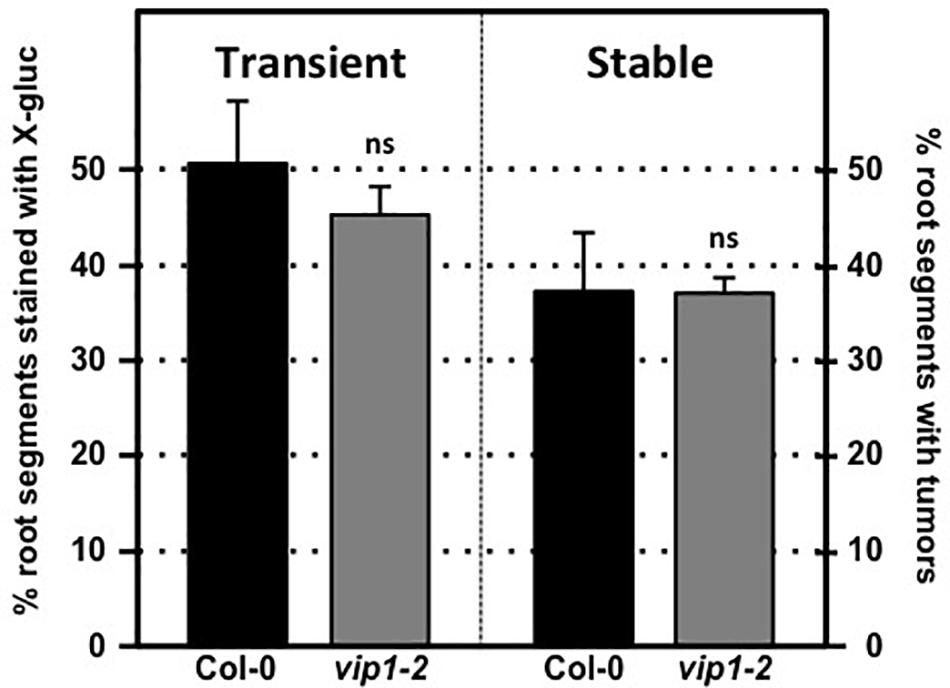
FIGURE 4. Transformation susceptibility of Arabidopsis wild-type and vip1-2 mutant plants. Agrobacterium-mediated transient (left) or stable (right) transformation assays were conducted on wild-type and vip1-2 mutant plants. Root segments were inoculated with 107 cfu/ml of the A. tumefaciens strains At849 (transient) or A208 (stable). For the transient assay, the root segments were stained with X-gluc 6 days after infection. For stable transformation, tumors were scored 30 days after infection. Numbers represent an average of three biological replicates (each replicate containing > 60 root segments) ± SE. Student’s t-test. ns: not significant.
Individual VIP1 Homologs Are Not Essential for Agrobacterium-Mediated Transformation
VIP1 is a single copy gene, but 11 close VIP1 homologs are present in Arabidopsis. VIP1 and these VIP1 homologs comprise the group I bZIP protein family. Their C-terminal regions, which include the bZIP domain, are highly similar to each other, whereas their N-terminal regions are variable (Jakoby et al., 2002; Tsugama et al., 2014). Of the 12 genes encoding the group I bZIP proteins, VIP1, and six other genes (bZIP18, bZIP29, bZIP30, bZIP52, bZIP69, and PosF21) are expressed at moderate levels in seedlings, roots, shoots, and flowers, whereas the other five genes (UNE4, bZIP31, bZIP33, bZIP71, and bZIP74) are hardly expressed in any of these tissues (Tsugama et al., 2014; Supplementary Figure 3). Many of these family members have similar subcellular localization, form homo- and heterodimers, and can similarly bind DNA fragments with the AGCTGT/G motif (Pitzschke et al., 2009; Tsugama et al., 2014, 2016; O’Malley et al., 2016). To test the importance of individual family members for transformation susceptibility, we obtained and confirmed homozygous mutants for six of the more highly expressed VIP1 homologs (bZIP18, bZIP29, bZIP30, bZIP33, bZIP52, and posF21). No aberrant phenotypes were observed in these single knockout mutants under normal growth conditions. Transient and stable root transformation assays indicated that each mutant had transformation susceptibility similar to that of wild-type plants (Figures 5A,B). Thus, in addition to VIP1, none of these six transcription factors is essential for Agrobacterium-mediated transformation. We additionally tested a triple mutant (vip1-1/posf21/bzip29) for transient and stable transformation susceptibility. Using two concentrations of bacterial inoculum, the vip1-1/posf21/bzip29 mutant had a slight (1.5-fold) reduction in both transient and stable transformation efficiency (Figures 5C,D).
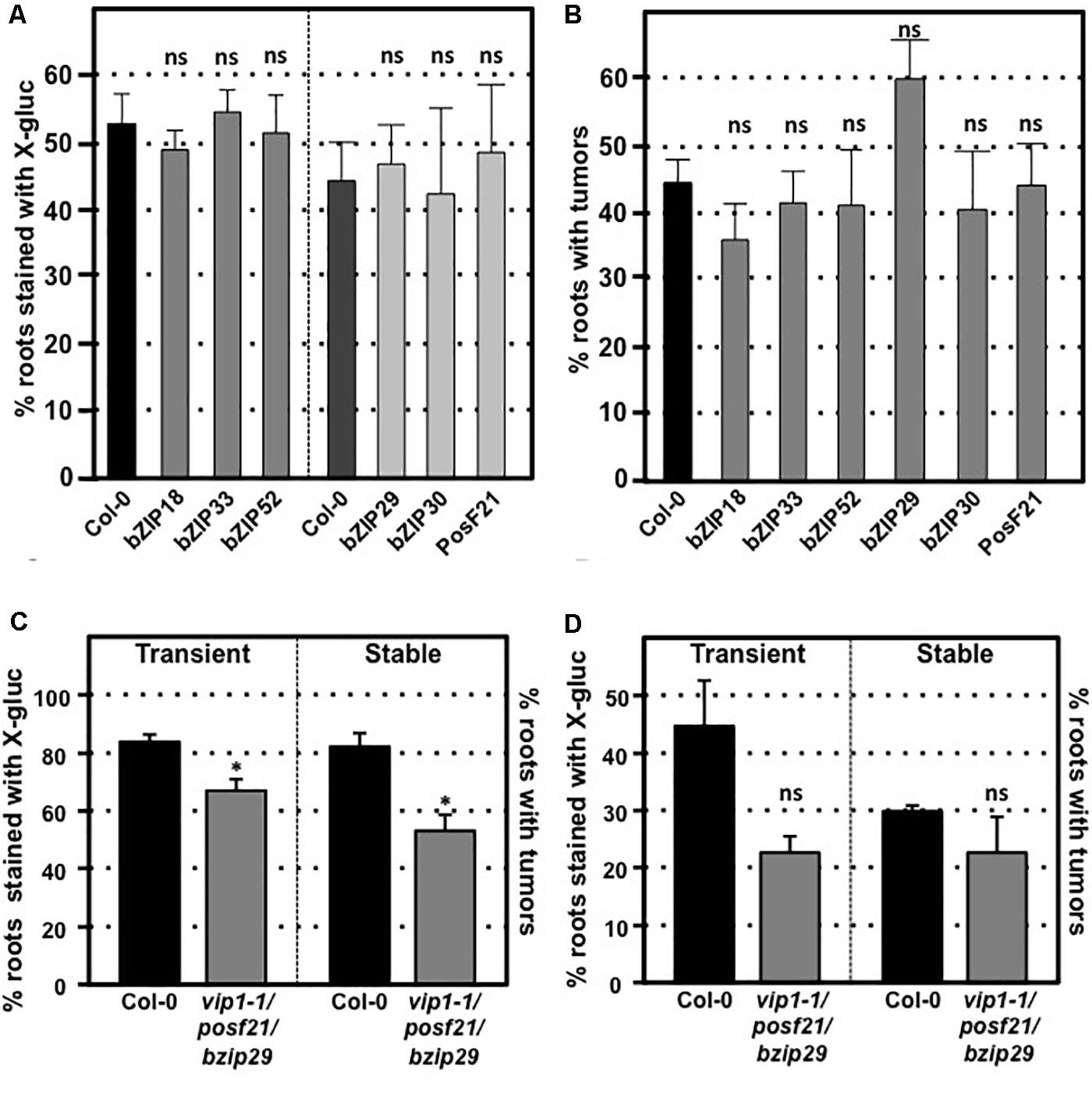
FIGURE 5. Transformation susceptibility of Arabidopsis VIP1 homolog mutant roots. Agrobacterium-mediated transient or stable transformation assays were conducted on wild-type and VIP1 homolog mutant plants. Root segments of VIP1 homolog single gene mutants and one triple gene mutant were infected with A. tumefaciens At849 (transient) or A208 (stable) at the concentration of 106 cfu/ml. For the transient assay, the root segments were stained with X-gluc 6 days after infection. For stable transformation, the tumors were scored 30 days after infection. Transient and stable transformation efficiencies of six VIP1 homolog mutants are shown in (A,B), respectively. The transformation efficiencies of the triple gene mutant with an inoculum at 107 cfu/ml and at 106 cfu/ml are shown in (C,D), respectively. Numbers represent an average of three biological replicates (each replicate containing > 60 root segments) ± SE. Student’s t-test ∗P-value < 0.05, ns: not significant.
Dominant Repression of VIP1 Family Function by a VIP1-SRDX Fusion Does Not Affect Transformation Susceptibility
To circumvent potential redundant roles among VIP1 family members, we assayed the transformation susceptibility of root segments from three transgenic Arabidopsis lines expressing VIP1 fused to the EAR motif repression domain SRDX (Mitsuda et al., 2006; Tsugama et al., 2016). The three independent lines of VIP1-SRDX plants used in this study all showed high expression levels of VIP1-SRDX and root waving phenotypes in a previous study (Tsugama et al., 2016), indicating the efficacy of the EAR motif in repressing expression of genes regulated by VIP1 family members. However, they showed transient and stable transformation susceptibility similar to that of wild-type plants (Figures 6A,B). Tumor size and morphology also did not change on roots of these lines. We conclude that VIP1 and its homologs are not essential for Agrobacterium-mediated transformation.
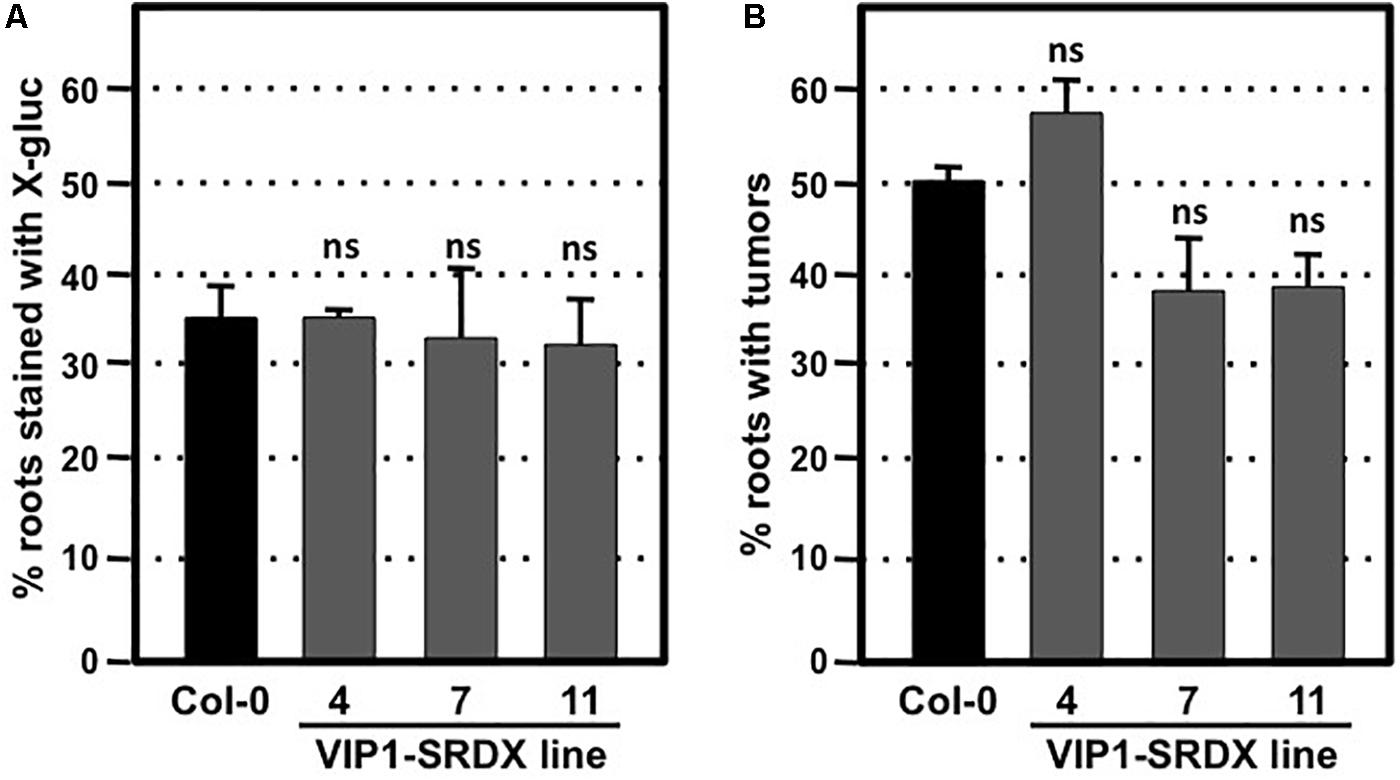
FIGURE 6. Transformation susceptibility of Arabidopsis wild-type and VIP1-SRDX mutant roots. Agrobacterium-mediated transient or stable transformation assays were conducted on wild-type and VIP1-SRDX plants. Root segments were inoculated with the strains A. tumefaciens At849 (106 cfu/ml for transient) or A208 (107 cfu/ml for stable). For transient transformation (A), root segments were stained with X-gluc 6 days after infection; for stable transformation (B), tumors were scored 30 days after infection. Numbers represent an average of three biological replicates (each replicate containing > 60 root segments) ± SE. Student’s t-test. ns: not significant.
Subcellular Localization of VIP1 Homologs and Their Interactions With VirE2
We transfected tobacco BY-2 protoplasts using constructs encoding GFP-tagged VIP1, bZIP52, PosF21, bZIP29, bZIP31, UNE4, and bZIP33 expressed from a Cauliflower Mosaic Virus (CaMV) 35S promoter (Figure 7). The subcellular localization of VIP1, bZIP52, bZIP31, and UNE4 is in both the cytoplasm and the nucleus, except that VIP1 localizes predominantly to the nucleoplasm (Figure 7A), whereas the other transcription factors also localized to the nucleolus (Figures 7B,E,F). PosF21 and bZIP33 localized predominantly to the cytoplasm (Figures 7C,G), with bZIP33 showing perinuclear aggregates (Figure 7G). bZIP29 showed exclusively nucleoplasmic localization (Figure 7D). Free GFP localized throughout the cytoplasm and nucleus (Figure 7H). Thus, although these related transcription factors showed overlapping subcellular localization patterns, none of these patterns is identical to that of VIP1.
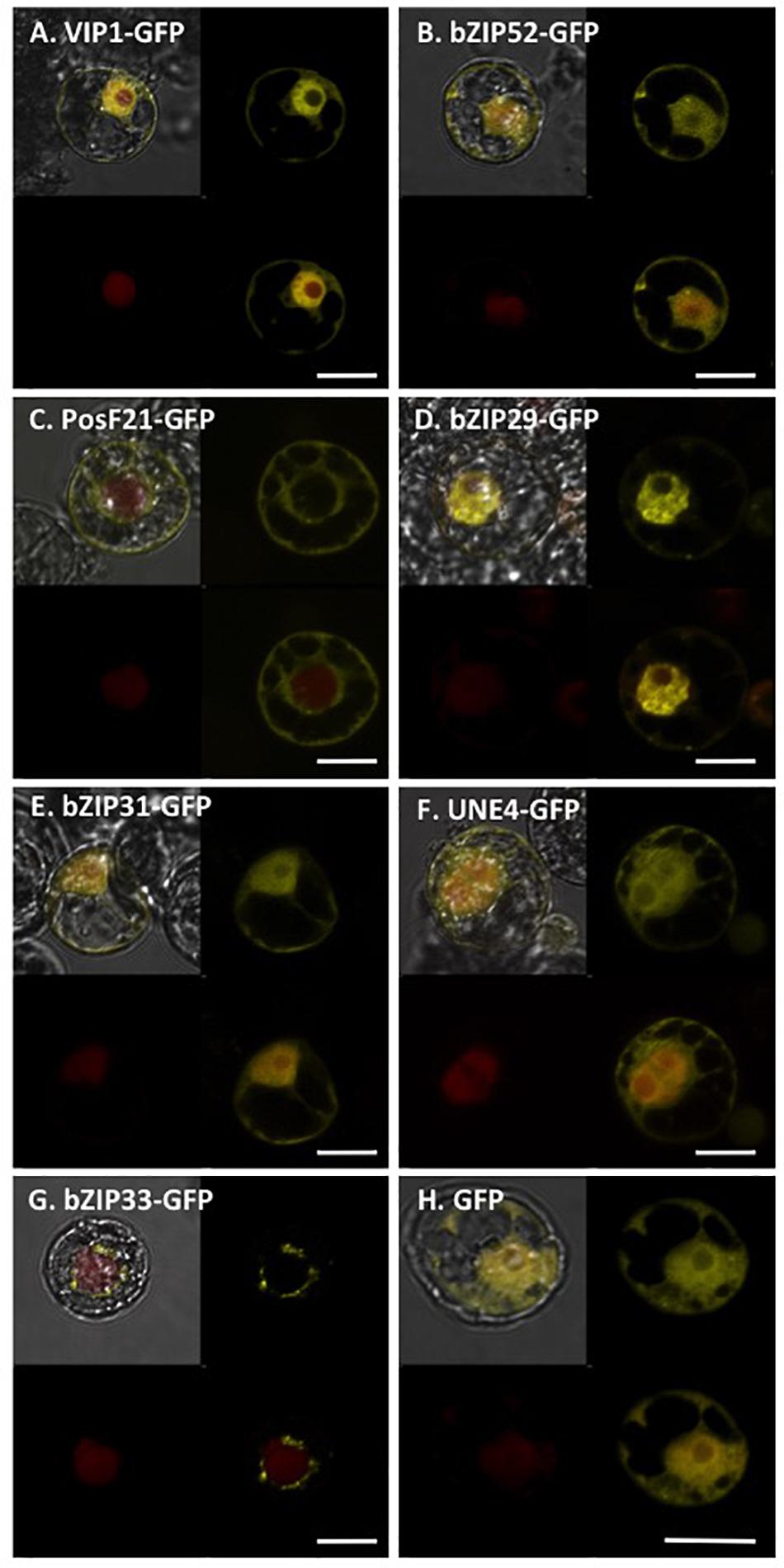
FIGURE 7. Subcellular localization of VIP1 and its homologs in tobacco BY-2 protoplasts. DNA of Venus-tagged VIP1 or its homologs were co-transfected with a nuclear marker mRFP-NLS into tobacco BY-2 protoplasts. Cells were imaged by confocal microscopy 16 h after transfection. Four images of each cell are presented (clockwise from top left: merged YFP, mRFP, and DIC; YFP; YFP + mRFP; mRFP). Bars indicate 20 μm.
We examined the interaction of VirE2 with VIP1, bZIP52, and PosF21 using BiFC. VIP1-VirE2 complexes localized to the perinuclear area and formed aggregates (Figure 8A; Shi et al., 2014). The interaction and co-localization patterns of bZIP52 and PosF21 with VirE2 (Figures 8B,C) resemble the pattern of VirE2 localization (Figure 8D). These data suggest that through interaction, VirE2 relocalizes these transcription factors in plant cells (compare Figures 7, 8). In our control, we did not detect interaction of VirE2-nYFP with cCFP (Figure 8E).
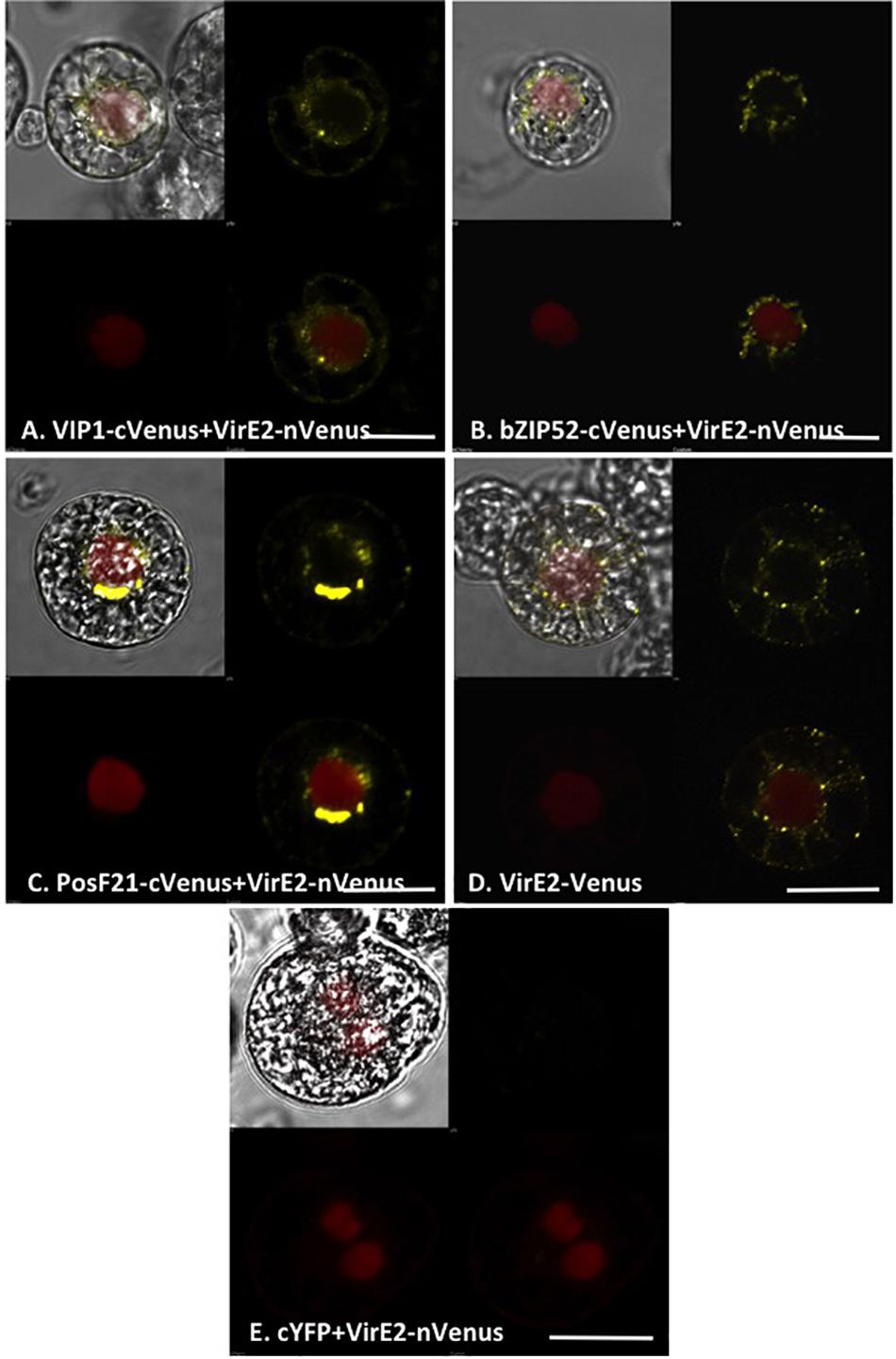
FIGURE 8. Subcellular localization of complexes formed by VirE2 with VIP1 homologs in tobacco BY-2 protoplasts. Tobacco BY-2 protoplasts were co-transfected with constructs comprised of the indicated cVenus-tagged VIP1 homologs and VirE2-nVenus (A–C); a construct encoding VirE2-Venus (D), or constructs encoding VirE2-nVenus and cCFP (E). A nuclear marker encoding mRFP-NLS was also included in all transfection experiments. The cells were imaged by confocal microscopy after 16 h. Four images of each cell are presented (clockwise from top left: merged YFP, mRFP, and DIC; YFP; YFP + mRFP; mRFP). Bars indicate 20 μm.
VIP1 Target Gene Expression in the Absence and Presence of Agrobacterium
To elucidate the expression of VIP1 target genes in the presence of Agrobacterium, we generated transgenic A. thaliana expressing VIP1 under the control of an inducible promoter. We incubated roots of these plants in induction or control solutions for 0, 3, or 12 h in the absence or presence of the avirulent strain A. tumefaciens A136 lacking a Ti plasmid. Incubation with bacteria induces plant PAMP (pattern associated molecular pattern) defense responses. After various times, we harvested root tissue and isolated total RNA. We performed quantitative RT-PCR analysis to measure the expression of previously identified VIP1 target genes (Pitzschke et al., 2009; Tsugama et al., 2012, 2014; Andrea Pitzschke, personal communication). These experiments were performed as three technical replicates each of three biological replicates. Representative data are shown in Figures 9A–E, and the full analysis is shown in Supplementary Table 4. The VIP1 transgene was strongly expressed in the induced but not the non-induced samples, both in the absence and in the presence of Agrobacterium (Figure 9A). The VIP1 target gene MYB44 (At5g67300) showed slightly elevated expression to similar levels in all of the induced samples compared to the non-induced samples, both in the presence and in the absence of Agrobacterium (Figure 9B). The putative VIP1 target gene PHI-1 (At1g35140) showed the highest expression 12 h after induction in the presence of Agrobacterium (Figure 9C). Although CYP707A1 (At4g19230) showed a modest two-fold response to induction of VIP1, CYP707A3 (At5g45340) showed an even greater increase in expression, especially 12 h after induction in the presence of Agrobacterium. However, the expression of CYP707A3 in the presence of Agrobacterium was at a level similar to that found in the 3-h samples (Figure 9E). CYP707A3 is involved in the inactivation of ABA signaling, suggesting that VIP1 may play a role in modulating ABA responses during stress responses (Tsugama et al., 2012). ABA is a key hormone involved in defense responses against fungal pathogens such as Botrytis cinerea (Audenaert et al., 2002; Fan et al., 2009; Sivakumaran et al., 2016). Therefore, we tested the susceptibility of vip1 mutant plants to Botrytis cinerea (Figure 10).
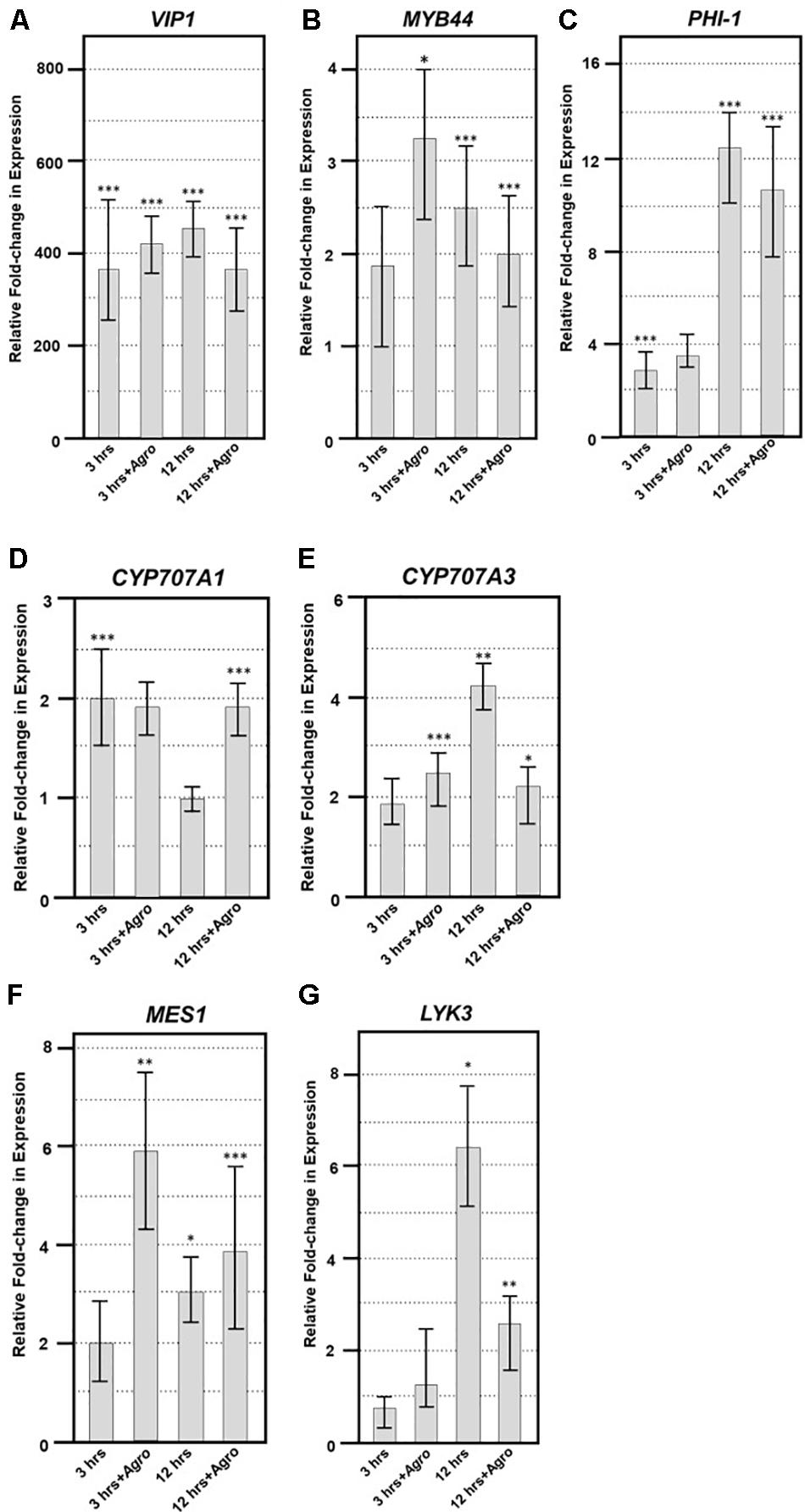
FIGURE 9. Quantitative RT-PCR of VIP1 target and fungal defense genes. Quantitative RT-PCR of (A) VIP1 transgene, (B) MYB44, (C) PHI-1, (D) CYP707A1, (E) CYP707A3, (F) MES1, and (G) LYK3 gene expression in induced relative to that of non-induced roots (Y-axis). Results represent an average of three replicates ± SE. Relative expression is shown after 3 and 12 h of induction in the absence or presence of Agrobacterium (+Agro) on the X-axis. Asterisks indicate SE according to Student’s t-test: ∗P-value < 0.05, ∗∗P-value < 0.01, ∗∗∗P-value < 0.001.
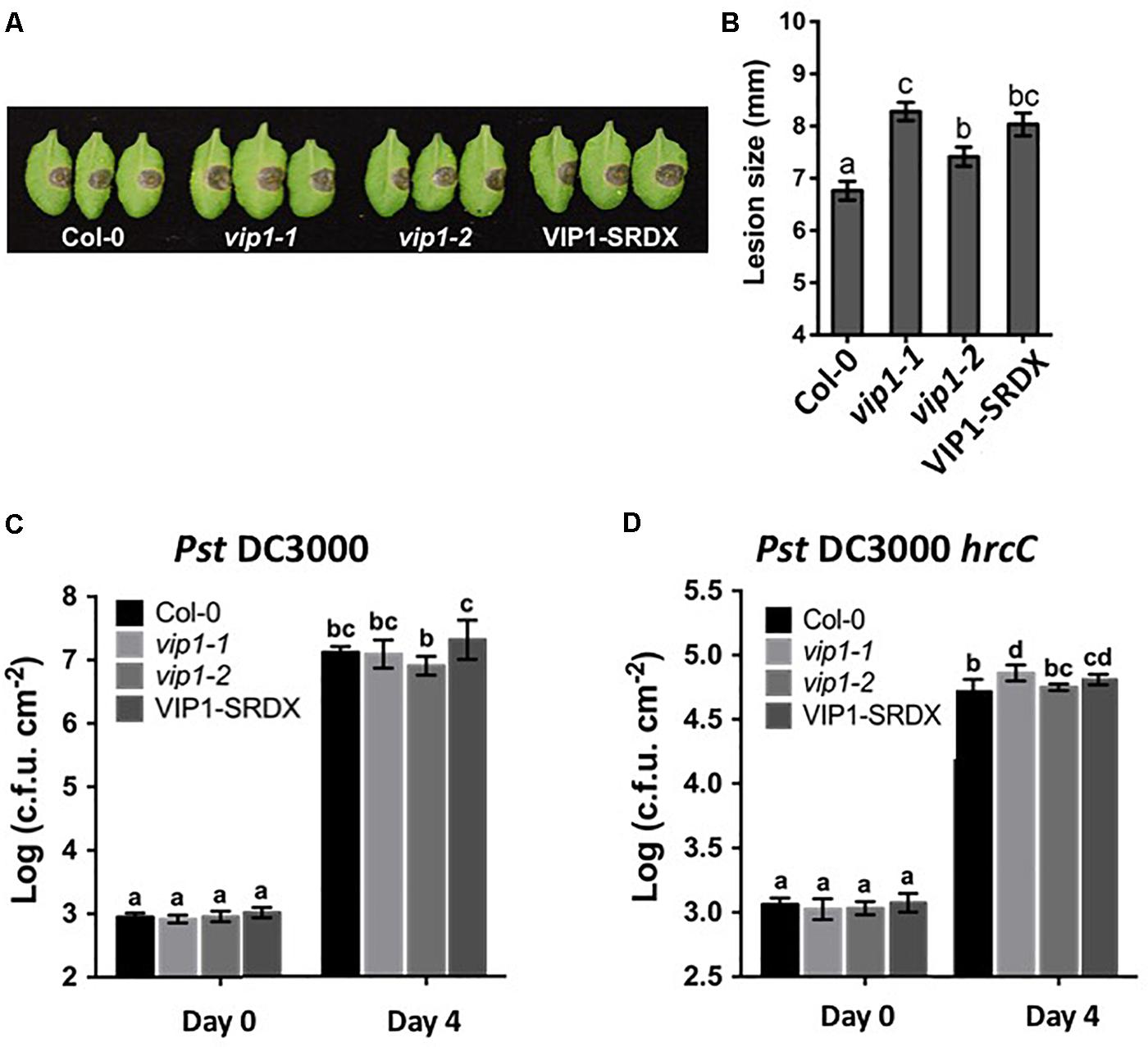
FIGURE 10. VIP1 is important for fungal but not bacterial infection of Arabidopsis. (A) Disease symptoms on leaves of various plants 3 days after inoculation with 5 μl of B. cinerea at the concentration of 1.0 × 105 spores/ml; (B) Average lesion size of 18 leaves of each genotype (36 leaves for Col-0) after B. cinerea inoculation; (C) Leaves of 5-week-old plants were syringe inoculated with Pseudomonas syringe c.v. tomato Pst DC3000 (A600 = 0.001) or (D) Pst DC3000 hrcC (A600 = 0.005). Growth of bacteria on leaves was measured at 0 and 4 dpi. Average numbers of bacteria and SDs were obtained from three replicates, each consisting of six leaf disks. Different letters indicate significant differences (P < 0.05, Student’s t-test).
vip1 Mutant and VIP1-SRDX Lines Show Increased Susceptibility to Botrytis cinerea, but Not to Pseudomonas syringae Infection
VIP1 is a target of the MAPK cascade and has been proposed to be involved in defense responses (Pitzschke et al., 2009). Although we were unable to find a role for VIP1 and its homologs in defense against Agrobacterium, we considered that VIP1 may play a role in defense against other pathogens. We therefore conducted pathogenesis assays, using Pseudomonas syringae and Botrytis cinerea, on wild-type, vip1-1, vip1-2, and VIP1-SRDX plants. Leaf lesion size was significantly larger on B. cinerea infected vip1-1, vip1-2, and VIP1-SRDX leaves than on wild-type leaves, indicating that VIP1, and perhaps additionally its paralogs, are involved in defense against Botrytis infection (Figures 10A,B).
The vip1 mutants and VIP1-SRDX lines responded similarly as did wild-type plants to treatment with both a virulent (Pst DC3000) and avirulent (Pst DC3000 hrcC) P. syringae strains (Figures 10C,D). These results suggest that VIP1 plays a role in B. cinerea, but not P. syringae and A. tumefaciens, defense. We also found that expression of the fungal defense genes MES1 (At2g23620) and LYK3 (At1g51940) were elevated after induction of VIP1 transgene expression, suggesting a role for VIP1 in fungal defense (Figures 9F,G; Vlot et al., 2008; Paparella et al., 2014).
vip1 Mutant and VIP1-SRDX Lines Are Sensitive to Exogenous ABA During, but Not After, Germination
Botrytis cinerea produces exogenous ABA to suppress plant defense responses (Audenaert et al., 2002; Fan et al., 2009; Sivakumaran et al., 2016). VIP1 may play a role in ABA signaling (Tsugama et al., 2012, 2013, 2014). Under hypo-osmotic conditions, VIP1 re-localizes to the nucleus and activates transcription of CYP707A1 and CYP707A3 (Tsugama et al., 2012) which encode proteins that degrade ABA and are therefore involved in osmosensory regulation of plant growth (Kushiro et al., 2004; Umezawa et al., 2006; Supplementary Figure 4). In the absence of VIP1, plants may be less able to degrade exogenous ABA, which may explain the increased susceptibility of vip1 mutant plants and VIP1-SRDX lines to B. cinerea infection. Because ABA is also a negative regulator of germination (Gimeno-Gilles et al., 2009; Supplementary Figure 4), we hypothesized that vip1 mutant and VIP1-SRDX lines may display altered germination in the presence of exogenous ABA. We germinated seeds of wild-type, vip1-1, vip1-2, and VIP1-SRDX lines 7–1 and 11 on medium containing either 0, 0.3, or 0.5 μM ABA. In the presence of ABA, almost all the wild-type seeds germinated within 8 days after imbibition. Seeds of the vip1-1 and vip1-2 mutants, and two VIP1-SRDX lines, showed reduced germination in the presence of ABA (Figure 11).
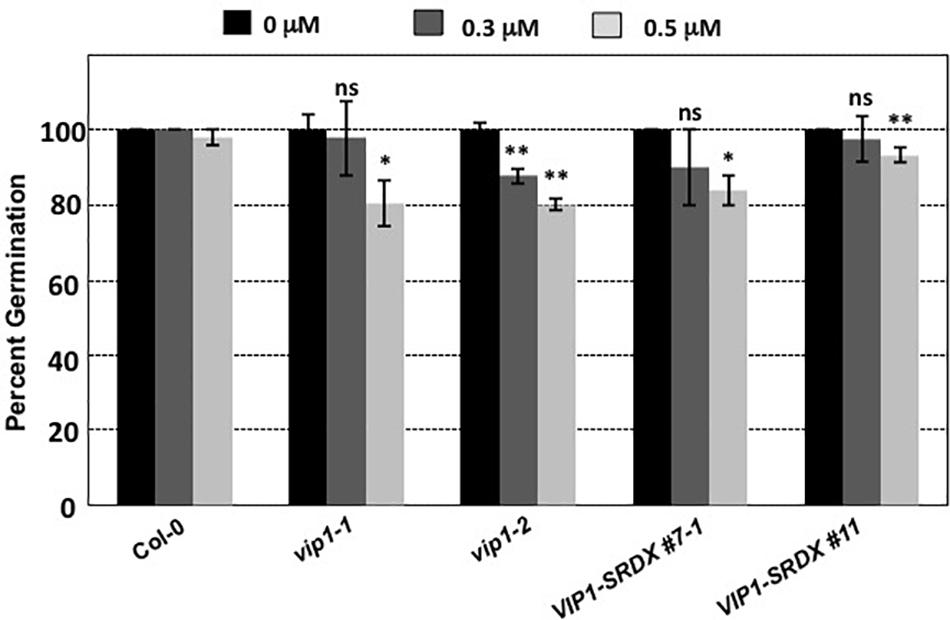
FIGURE 11. Germination of wild-type, vip1 mutants, and a VIP1-SRDX line on medium containing ABA. Seeds of the indicated lines were germinated on B5 medium containing 0, 0.3, or 0.5 μM ABA. Data represent the average percent germination ± SE. Student’s t-test ∗P-value < 0.1, ∗∗P-value < 0.05, ns: not significant.
Low concentrations of ABA promote root growth, whereas high concentrations inhibit growth (Pilet and Saugy, 1987; Sharp and LeNoble, 2002). To elucidate whether VIP1 plays a role in ABA signaling during root growth, we first germinated vip1 mutant and VIP1-SRDX plants on MS medium, then transferred the seedlings to plates containing 0, 2, or 20 μM ABA to continue growth. The rate of root growth did not significantly differ from that of wild-type for any of the vip1 mutant or VIP1-SRDX lines (Supplementary Figure 5). These results suggest that although VIP1 appears important for ABA defense signaling and germination, it does not play a role in ABA-dependent regulation of root growth.
vip1 Mutant and VIP1-SRDX Roots Are More Tolerant to Growth in High Salt
To determine whether VIP1 also plays a role under hyper-osmotic conditions, we performed seed germination and root growth assays on wild-type Col-0, vip1-2, vip1-1/posf21/bzip29 mutants, and VIP1-SRDX lines. All seeds germinated well on medium containing elevated concentrations of NaCl (Supplementary Figure 6). However, roots of all vip1 mutant lines, and two VIP1-SRDX lines, grew better on medium containing salt than did wild-type roots (Figure 12). These results indicate that VIP1 plays a role in root growth under salt stress conditions.
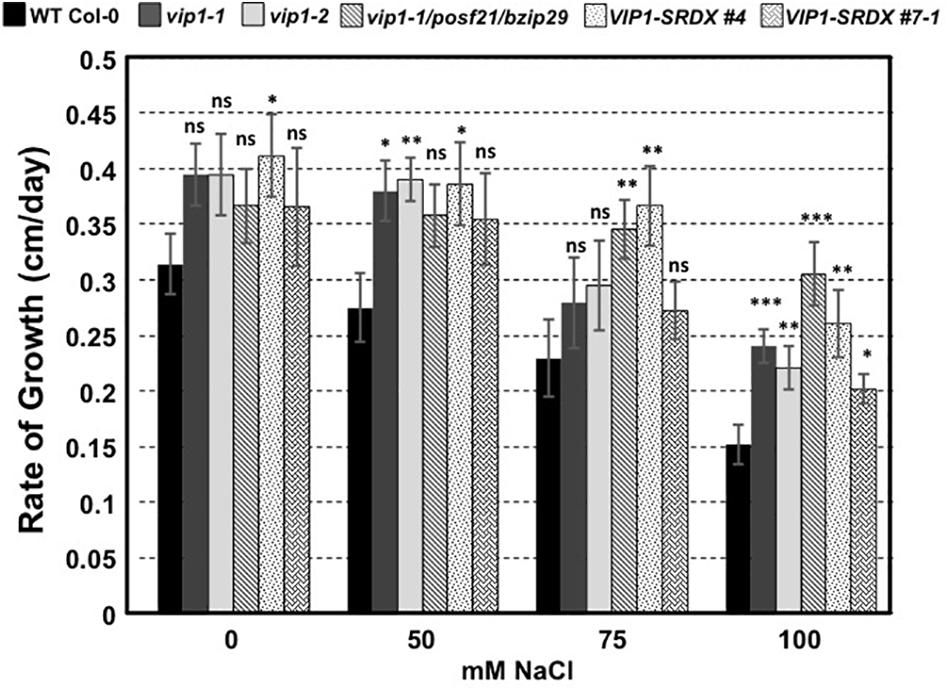
FIGURE 12. Root growth rates of wild-type, vip1 mutant, and VIP1-SRDX lines on various concentrations of NaCl. Data represent the average rate of root growth ± SE. Student’s t-test ∗P-value < 0.05, ∗∗P-value < 0.01, ∗∗∗P-value < 0.001, ns: not significant.
Discussion
Previous reports indicated that, compared to wild-type plants, vip1-1 A. thaliana and VIP1 antisense tobacco plants showed reduced stable transformation susceptibility, suggesting an important role for VIP1 in Agrobacterium-mediated transformation (Tzfira et al., 2001, 2002; Li et al., 2005). As a consequence of these reports, and observations that plant-generated VirE2 localizes to the nucleus (Zupan et al., 1996) but cannot interact with importin α-1 (AtKapα; Ballas and Citovsky, 1997), VIP1 was proposed to act as an adaptor molecule between importin α-1 and VirE2 for nuclear entry of VirE2-bound T-DNA (the Trojan-horse model; Djamei et al., 2007). However, we have observed that VirE2 can interact with all tested Arabidopsis importin α isoforms in vitro, in yeast, and in plants (Bhattacharjee et al., 2008). We and others have observed that VirE2 and VIP1-VirE2 complexes synthesized in planta localize to the cytoplasm (Bhattacharjee et al., 2008; Lee et al., 2008; Sakalis et al., 2013; Shi et al., 2014; this study), indicating that VIP1 does not act as an adaptor to localize VirE2 to the nucleus. Recent reports, however, suggest that some VirE2 molecules delivered from Agrobacterium may reach the nucleus (Li et al., 2014; Yang et al., 2017). Thus, the role of VirE2 in helping deliver T-strands to the plant nucleus remains controversial. Regardless of the role of VirE2 in nuclear import of T-strands, this current, and a previous, study (Shi et al., 2014) found no significant change in Agrobacterium-mediated transformation susceptibility in any VIP1 mutant background or in VIP1 overexpressing transgenic lines. Therefore, our data do not support the Trojan Horse model (Djamei et al., 2007).
Although we could not find a role for VIP1 in Agrobacterium-mediated plant transformation, we were concerned that other group I bZIP transcription factors related to Arabidopsis VIP1 could mask the effect of VIP1 on transformation. VIP1 is one of a 12-gene family whose members may have redundant functions. Analysis of null mutants of six individual VIP1 homologs did not reveal any transformation phenotypes, and the vip1-1/posf21/bzip29 triple mutant showed only a modest reduction in transformation susceptibility, suggesting that some VIP1 family members may slightly potentiate transformation. We therefore analyzed transgenic lines overexpressing VIP1 fused to a SRDX repression domain. The binding of other transcription factors to the promoters of VIP1 target genes is blocked in these lines (Mitsuda et al., 2006; Tsugama et al., 2016). VIP1-SRDX lines also displayed transformation characteristics similar to those of wild-type plants. We therefore conclude that full expression of VIP1 and its homologs is not required for Agrobacterium-mediated transformation. It is possible, however, that residual expression of VIP1 target genes may facilitate transformation.
While testing inducible VIP1 plants in the absence and presence of Agrobacterium, we observed differential expression of the VIP1 target genes MYB44 (At5g67300) and CYP707A3 (At5g45340) previously identified in the literature (Figures 9B,E; Pitzschke et al., 2009; Tsugama et al., 2012, 2014). Pitzschke et al. (2009) found that MYB44 was upregulated in wild-type plants after treatment with flg22, and that the gene contains multiple copies of a VIP1 responsive element (VRE) in its promoter. We were also able to detect upregulation of the putative VIP1 target gene PHI-1 (At1g35140) in response to VIP1 induction (Figure 9C; Andrea Pitzschke, personal communication). CYP707A3 expression also increases upon VIP1 induction, as well as after tissue rehydration and in the presence of mannitol in VIP1 overexpressing plants (Figure 9E; Tsugama et al., 2012). Although Tsugama et al. (2012, 2014) showed that CYP707A1 (At4g19230) is differentially expressed under the same conditions as is CYP707A3, we only detected a modest change in CYP707A1 expression in our experiments (Figure 9D), suggesting that CYP707A1 upregulation requires conditions not present in our protocol.
VIP1 is a phosphorylation target of MPK3 and has been proposed to be involved in plant defense responses (Pitzschke et al., 2009). Increased susceptibility of vip1-1, vip1-2, and VIP1-SRDX A. thaliana plants to Botrytis cinerea, but not to P. syringae, suggests that VIP1 may play a role in fungal but not bacterial defense responses. Resistance to broad host necrotrophic fungi such as B. cinerea is mediated by quantitative resistance mechanisms involving the contributions of many genes (Poland et al., 2009; Lai and Mengiste, 2013). The VIP1 gene contributes to this resistance, as indicated by the increase in disease lesion size when VIP1 is debilitated (Figure 10A). This model is supported by our observation that the LYK3 gene, involved in response to chitin, is upregulated in inducible VIP1 plants (Figure 9G). VIP1 may be phosphorylated by MPK3 in response to B. cinerea, leading to the activation of its target genes CYP707A1 and CYP707A3, which are ABA degradation enzymes (Kushiro et al., 2004; Umezawa et al., 2006). CYP707A1 and CYP707A3 may be important for the degradation of exogenous ABA produced by B. cinerea, preventing the suppression of defense responses (Audenaert et al., 2002; Fan et al., 2009; Sivakumaran et al., 2016). The role of ABA in fungal infection is consistent with the observation that ABA-deficient tomato plants are highly resistant to B. cinerea infection (Asselbergh et al., 2007). The precise role of VIP1 in defense signaling during B. cinerea infection remains unknown. Measuring the expression of VIP1 target genes throughout infection should provide clues as to how VIP1 contributes to defense against B. cinerea during early and late stages of infection.
The altered growth of vip1-2 leaves (Figures 3A,B) suggests a role for VIP1 in plant growth and development. This role is supported by the observation of increased touch-induced root waving in VIP1-SRDX plants (Tsugama et al., 2016) and is consistent with previous findings that two other group I bZIP proteins, bZIP29 and bZIP30, also regulate leaf growth (Lozano-Sotomayor et al., 2016; Van Leene et al., 2016). This and previous studies also suggest a role for VIP1 in the regulation of abiotic stress responses, specifically to hypo- and hyperosmotic conditions (Figure 12; Tsugama et al., 2012). A previous study did not observe any major growth differences of VIP1-SRDX compared to wild-type plants in the presence of ABA or under hyperosmotic conditions (mannitol) after 3 weeks of growth (Tsugama et al., 2016). Our study, however, measured seed germination and the rate of root growth of plants at earlier times (less than 12 days after plating). This assay allowed us to quantify better the sensitivity of these plants to ABA and hyperosmotic conditions. VIP1 enters the nucleus and binds to the promoters of its target genes, CYP707A1 and CYP707A3, upon rehydration of plant roots, leading to an increase in their expression (Tsugama et al., 2012). It is unknown whether the increase in the expression of CYP707A1 and CYP707A3 by VIP1 after rehydration leads to the degradation of ABA or contributes to some other signaling pathway. Whether VIP1 affects plant growth and development under various osmotic conditions via CYP707A1/CYP707A3 degradation of ABA or by other ABA-dependent or -independent signaling mechanisms will require further investigation.
Author Contributions
RL and SG designed the experiments. RL, L-YL, SL, and SG conducted the experiments. DT contributed to the reagents. RL and SG wrote the article with contributions from L-YL, TM, and DT.
Funding
RL was funded by an Andrews Fellowship awarded by the Purdue University Graduate School, and by a graduate research grant from the Purdue Research Foundation. Work in the laboratory of SG was supported by the National Science Foundation and a P30 grant to the Purdue University Cancer Center.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank our undergraduate researchers Eder Xhako, Laurel Jahn, Lauren Huemmer, Emily Traxler, and Erica Wolfe for their help performing transformation assays and genotyping mutant A. thaliana lines. We thank Dr. Xiao-Hong Zhu and Ms. Wen-Shan Liu for their help constructing the sgRNA vectors. We also acknowledge the use of the facilities of the Bindley Bioscience Center at Purdue University, a core facility of the NIH-funded Indiana Clinical and Translational Sciences Institute, and the Purdue Genomics Core Facility.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00749/full#supplementary-material
Footnotes
- ^ http://www.gene-quantification.de/rest-2009.html
- ^ https://www.thermofisher.com
- ^ http://www.gene-quantification.de/rest-2009.html
References
Anand, A., Krichevsky, A., Schomack, S., Lahaye, T., Tzfira, T., Tang, Y., et al. (2007). Arabidopsis VIRE2 INTERACTING PROTEIN 2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19, 1695–1708. doi: 10.1105/tpc.106.042903
Asselbergh, B., Curvers, K., França, S. C., Audenaert, K., Vuylsteke, M., Van Breusegem, F., et al. (2007). Resistance to Botrytis cinerea in sitiens, an abscisic acid-deficient tomato mutant, involves timely production of hydrogen peroxide and cell wall modifications in the epidermis. Plant Physiol. 144, 1863–1877. doi: 10.1104/pp.107.099226
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Babon, J. J., McKenzie, M., and Cotton, R. G. H. (2003). The use of resolvases T4 endonuclease VII and T7 endonuclease I in mutation detection. Mol. Biotechnol. 23, 73–81. doi: 10.1385/MB:23:1:73
Ballas, N., and Citovsky, V. (1997). Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. U.S.A. 94, 10723–10728. doi: 10.1073/pnas.94.20.10723
Bhattacharjee, S., Lee, L.-Y., Oltmanns, H., Cao, H., Veena, Cuperus, J., et al. (2008). AtImpα-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 20, 2661–2680. doi: 10.1105/tpc.108.060467
Brogna, S., and Wen, J. (2009). Nonsense-mediated mRNA decay (n.d.) mechanisms. Nat. Struct. Mol. Biol. 16, 107–113. doi: 10.1038/nsmb.1550
Chang, C.-W., Williams, S. J., Cunago, R. M., and Kobe, B. (2014). Structural basis of interaction of bipartite nuclear localization signal from Agrobacterium VirD2 with rice importin-α. Mol. Plant 7, 1061–1064. doi: 10.1093/mp/ssu014
Chen, J., Yi, Q., Cao, Y., Wei, B., Zheng, L., Xiao, Q., et al. (2015). ZmbZIP91 regulates expression of starch synthesis-related genes by binding to ACTCAT elements in their promoters. J. Exp. Bot. 67, 1327–1338. doi: 10.1093/jxb/erv527
Citovsky, V., Warnick, D., and Zambryski, P. (1994). Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl. Acad. Sci. U.S.A. 91, 3210–3214. doi: 10.1073/pnas.91.8.3210
Citovsky, V., Zupan, J., Warnick, D., and Zambryski, P. (1992). Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256, 1802–1805. doi: 10.1126/science.1615325
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dingwall, C., and Laskey, R. A. (1991). Nuclear targeting sequences—a consensus? Trends Biochem. Sci. 16, 478–481.
Djamei, A., Pitzschke, A., Nakagami, H., Rajh, I., and Hirt, H. (2007). Trojan horse strategy in Agrobacterium transformation: abusing MAPK defense signaling. Science 318, 453–456. doi: 10.1126/science.1148110
Fan, J., Hill, L., Crooks, C., Doerner, P., and Lamb, C. (2009). Abscisic acid has a key role in modulating diverse plant-pathogen interactions. Plant Physiol. 150, 1750–1761. doi: 10.1104/pp.109.137943
Feng, Z., Zhang, B., Ding, W., Liu, X., Yang, D.-L., Wei, P., et al. (2013). Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23, 1229–1232. doi: 10.1038/cr.2013.114
Gelvin, S. B. (2003). Agrobacterium-mediated plant transformation: the biology behind the “gene- jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37. doi: 10.1128/MMBR.67.1.16-37.2003
Gelvin, S. B. (2012). Traversing the cell: Agrobacterium T-DNA’s journey to the host genome. Front. Plant Sci. 3:52. doi: 10.3389/fpls.2012.00052
Gimeno-Gilles, C., Lelièvre, E., Viau, L., Malik-Ghulam, M., Ricoult, C., Niebel, A., et al. (2009). ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol. Plant 2, 108–119. doi: 10.1093/mp/ssn092
Grange, W., Duckely, M., Husale, S., Jacob, S., Engel, A., and Hegner, M. (2008). VirE2: A unique ssDNA-compacting molecular machine. PLoS Biol. 6:0343–0351. doi: 10.1371/journal.pbio.0060044
Hiratsu, K., Ohta, M., Matsui, K., and Ohme-Takagi, M. (2002). The SUPERMAN is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Lett. 514, 351–354. doi: 10.1016/S0014-5793(02)02435-3
Ishida, S., Fukazawa, J., Yuasa, T., and Takahashi, Y. (2004). Involvement of 14-3-3 signaling protein binding in the functional regulation of the transcriptional activator REPRESSION OF SHOOT GROWTH by gibberellins. Plant Cell 16, 2641–2651. doi: 10.1105/tpc.104.024604
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Kushiro, T., Okamoto, M., Nakabayashi, K., Yamagishi, K., Kitamura, S., Asami, T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8’-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121
Lacroix, B., Loyter, A., and Citovsky, V. (2008). Association of the Agrobacterium T-DNA-protein complex with plant nucleosomes. Proc. Natl. Acad. Sci. U.S.A. 105, 15429–15434. doi: 10.1073/pnas.0805641105
Lai, Z., and Mengiste, T. (2013). Genetic and cellular mechanisms regulating plant responses to necrotrophic pathogens. Curr. Opin. Plant Biol. 16, 505–512. doi: 10.1016/j.pbi.2013.06.014
Lee, L.-Y., Fang, M.-J., Kuang, L.-Y., and Gelvin, S. B. (2008). Vectors for multi-color bimolecular fluorescence complementation to investigate protein-protein interactions in living plant cells. Plant Methods 4:24. doi: 10.1186/1746-4811-4-24
Lee, L.-Y., Wu, F.-H., Hsu, C.-T., Shen, S.-C., Yeh, H.-Y., Liao, D.-C., et al. (2012). Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24, 1746–1759. doi: 10.1105/tpc.112.097998
Li, J., Krichevsky, A., Vaidya, M., Tzifa, T., and Citovsky, V. (2005). Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738. doi: 10.1073/pnas.0404118102
Li, X., Yang, Q., Tu, H., Lim, Z., and Pan, S. Q. (2014). Direct visualization of Agrobacterium-delivered VirE2 in recipient cells. Plant J. 77, 487–495. doi: 10.1111/tpj.12397
Lozano-Sotomayor, P., Chávez Montes, R. A., Silvestre-Vañó, M., Herrera-Ubaldo, H., Greco, R., Pablo-Villa, J., et al. (2016). Altered expression of the bZIP transcription factor DRINK ME affects growth and reproductive development in Arabidopsis thaliana. Plant J. 88, 437–451. doi: 10.1111/tpj.13264
Mitsuda, N., Hiratsu, K., Todaka, D., Nakashima, K., Yamaguchi-Shinozaki, K., and Ohme-Takagi, M. (2006). Efficient production of male and female sterile plants by expression of a chimeric repressor in Arabidopsis and rice. Plant Biotechnol. J. 4, 325–332. doi: 10.1111/j.1467-7652.2006.00184.x
Nam, J., Matthysse, A. G., and Gelvin, S. B. (1997). Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9, 317–333. doi: 10.1105/tpc.9.3.317
Nam, J., Mysore, K. S., Zheng, C., Knue, M. K., Matthysse, A. G., and Gelvin, S. B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261, 429–438. doi: 10.1007/s004380050985
Narasimhulu, S. B., Deng, X.-B., Sarria, R., and Gelvin, S. B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8, 873–886. doi: 10.1105/tpc.8.5.873
O’Malley, R. C., Huang, S. C., Song, L., Lewsey, M. G., Bartlett, A., Nery, J. R., et al. (2016). Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292. doi: 10.1016/j.cell.2016.04.038
Paparella, C., Savatin, D. V., Marti, L., De Lorenzo, G., and Ferrari, S. (2014). The Arabidopsis LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 165, 262–276. doi: 10.1104/pp.113.233759
Pilet, P.-E., and Saugy, M. (1987). Effect of root growth on endogenous and applied IAA and ABA. Plant Physiol. 83, 33–38. doi: 10.1104/pp.83.1.33
Pitzschke, A., Djamei, A., Teige, M., and Hirt, H. (2009). VIP1 response elements mediate mitogen-activated protein kinase 3-induced stress gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 18414–18419. doi: 10.1073/pnas.0905599106
Poland, J. A., Balint-Kurti, P. J., Wisser, R. J., Pratt, R. C., and Nelson, R. J. (2009). Shades of gray: the world of quantitative disease resistance. Trends Plant Sci. 14, 21–29. doi: 10.1016/j.tplants.2008.10.006
Rossi, L., Hohn, B., and Tinland, B. (1996). Integration of complete transferred DNA is units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 93, 126–130. doi: 10.1073/pnas.93.1.126
Sakalis, P. A., van Heusden, G. P. H., and Hooykaas, P. J. J. (2013). Visualization of VirE2 protein translocation by the Agrobacterium type IV secretion system into host cells. Microbiol. Open 3, 104–117. doi: 10.1002/mbo3.152
Sharp, R. E., and LeNoble, M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53, 33–37. doi: 10.1093/jexbot/53.366.33
Shi, Y., Lee, L.-Y., and Gelvin, S. B. (2014). Is VIP1 important for Agrobacterium-mediated plant transformation? Plant J. 79, 848–860. doi: 10.1111/tpj.12596
Sivakumaran, A., Akinyemi, A., Mandon, J., Cristescu, S. M., Hall, M. A., Harren, F. J. M., et al. (2016). ABA suppresses Botrytis cinerea elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. doi: 10.3389/fpls.2016.00709
Tenea, G. N., Spantzel, J., Lee, L.-Y., Zhu, Y., Lin, K., Johnson, S. J., et al. (2009). Overexpression of several Arabidopsis histone genes increases Agrobacterium-mediated transformation and transgene expression in plants. Plant Cell 21, 3350–3367. doi: 10.1105/tpc.109.070607
Tsugama, D., Liu, S., and Takano, T. (2012). A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol. 159, 144–155. doi: 10.1104/pp.112.197020
Tsugama, D., Liu, S., and Takano, T. (2013). A bZIP protein, VIP1, interacts with Arabidopsis heterotrimeric G protein β subunit, AGB1. Plant Physiol. Biochem. 71, 240–246. doi: 10.1016/j.plaphy.2013.07.024
Tsugama, D., Liu, S., and Takano, T. (2014). Analysis of functions of VIP1 and its close homologs in osmosensory responses of Arabidopsis thaliana. PLoS One 9:e103930. doi: 10.1371/journal.pone.0103930
Tsugama, D., Liu, S., and Takano, T. (2016). The bZIP protein VIP1 is involved in touch responses in Arabidopsis roots. Plant Physiol. 171, 1355–1365. doi: 10.1104/pp.16.00256
Tzfira, T., and Citovsky, V. (2001). Comparison between nuclear localization of nopaline- and octopine-specific Agrobacterium VirE2 proteins in plant, yeast, and mammalian cells. Mol. Plant Pathol. 2, 171–176. doi: 10.1046/j.1364-3703.2001.00065.x
Tzfira, T., Vaidya, M., and Citovsky, V. (2001). VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607. doi: 10.1093/emboj/20.13.3596
Tzfira, T., Vaidya, M., and Citovsky, V. (2002). Increasing plant susceptibility to Agrobacterium infection by over-expression of the Arabidopsis nuclear protein VIP1. Proc. Natl. Acad. Sci. U.S.A. 99, 10435–10440. doi: 10.1073/pnas.162304099
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8’-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. 4, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Van Larebeke, N., Engler, G., Holsters, M., Van Den Elsacker, S., Zaenen, I., Schilperoort, R. A., et al. (1974). Large plasmid in Agrobacterium tumefaciens essential for crown gall- inducing ability. Nature 252, 169–170. doi: 10.1038/252169a0
Van Leene, J., Blomme, J., Kulkarni, S. R., Cannoot, B., De Winne, N., Eeckhout, D., et al. (2016). Functional characterization of the Arabidopsis transcription factor bZIP29 reveals its role in leaf and root development. J. Exp. Bot. 67, 5825–5840. doi: 10.1093/jxb/erw347
Vlot, A. C., Liu, P. P., Cameron, R. K., Park, S. W., Yang, Y., Kumar, D., et al. (2008). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56, 445–456. doi: 10.1111/j.1365-313X.2008.03618.x
Wu, Y., Zhao, Q., Gao, L., Yu, X.-M., Fang, P., Oliver, D. J., et al. (2010). Isolation and characterization of low-sulfur-tolerant mutants in Arabidopsis. J. Exp. Bot. 61, 3407–3422. doi: 10.1093/jxb/erq161
Yang, Q., Li, X., Tu, H., and Pan, S. Q. (2017). Agrobacterium-delivered virulence protein VirE2 is trafficked inside host cells via a myosin XI-K-powered ER/actin network. Proc. Natl. Acad. Sci. U.S.A. 114, 2982–2287. doi: 10.1073/pnas.1612098114
Yusibov, V. M., Steck, T. R., Gupta, V., and Gelvin, S. B. (1994). Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 91, 2994–2998. doi: 10.1073/pnas.91.8.2994
Zhu, Y., Nam, J., Humara, J. M., Mysore, K. S., Lee, L.-Y., Cao, H., et al. (2003). Identification of Arabidopsis rat mutants. Plant Physiol. 132, 494–505. doi: 10.1104/pp.103.020420
Zuo, J., Niu, Q.-W., and Chua, N.-H. (2000). An estrogen receptor-based system transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24, 265–273. doi: 10.1046/j.1365-313x.2000.00868.x
Keywords: Agrobacterium-mediated transformation, fungal tolerance, protein localization, protein–protein interactions, salt tolerance, VIP1 transcriptome, VirE2
Citation: Lapham R, Lee L-Y, Tsugama D, Lee S, Mengiste T and Gelvin SB (2018) VIP1 and Its Homologs Are Not Required for Agrobacterium-Mediated Transformation, but Play a Role in Botrytis and Salt Stress Responses. Front. Plant Sci. 9:749. doi: 10.3389/fpls.2018.00749
Received: 14 December 2017; Accepted: 15 May 2018;
Published: 12 June 2018.
Edited by:
Hua Lu, University of Maryland, Baltimore County, United StatesReviewed by:
Yuanhu Xuan, Shenyang Agricultural University, ChinaArun Jagannath, University of Delhi, India
Copyright © 2018 Lapham, Lee, Tsugama, Lee, Mengiste and Gelvin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stanton B. Gelvin, Z2VsdmluQHB1cmR1ZS5lZHU=; Z2VsdmluQGJpbGJvLmJpby5wdXJkdWUuZWR1
 Rachelle Lapham
Rachelle Lapham Lan-Ying Lee
Lan-Ying Lee Daisuke Tsugama
Daisuke Tsugama Sanghun Lee
Sanghun Lee Tesfaye Mengiste
Tesfaye Mengiste Stanton B. Gelvin
Stanton B. Gelvin