
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci., 28 May 2018
Sec. Plant Physiology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00705
This article is part of the Research TopicPost-Translational Modifications in Plant Nuclear Signaling: Novel Insights into Responses to Environmental ChangesView all 9 articles
Reactive oxygen species (ROS) are well-described by-products of cellular metabolic activities, acting as signaling molecules and regulating the redox state of proteins. Solvent exposed thiol residues like cysteines are particularly sensitive to oxidation and their redox state affects structural and biochemical capacities of many proteins. While thiol redox regulation has been largely studied in several cell compartments like in the plant chloroplast, little is known about redox sensitive proteins in the nucleus. Recent works have revealed that proteins with oxidizable thiols are important for the regulation of many nuclear functions, including gene expression, transcription, epigenetics, and chromatin remodeling. Moreover, thiol reducing molecules like glutathione and specific isoforms of thiols reductases, thioredoxins and glutaredoxins were found in different nuclear subcompartments, further supporting that thiol-dependent systems are active in the nucleus. This mini-review aims to discuss recent progress in plant thiol redox field, taking examples of redox regulated nuclear proteins and focusing on major thiol redox systems acting in the nucleus.
Oxygen is one of the most important molecules for aerobic organisms. It is necessary for cell metabolism, but it also generates reactive oxygen species (ROS) as by-products of oxidoreduction pathways. ROS include free radical species like superoxides (), hydroxyl radicals (OH•), or nitric oxide (NO•), and non-radical species like hydrogen peroxide (H2O2) and peroxynitrite (ONOO-) (Sies et al., 2017). In plants, major sources of ROS are photosynthetic and respiratory chains in chloroplasts and mitochondria. ROS are also generated by plasma membrane NADPH oxidases and peroxisomal xanthine oxidases. Oxidative eustress is playing important signaling functions by inducing post-translational modifications (PTM) and by regulating protein redox state. ROS can also trigger oxidative distress which can damage the cell (Foyer and Noctor, 2016; Choudhury et al., 2017). Plant cells display a large panel of ROS scavenging enzymes like catalases, peroxidases, and superoxide dismutases. They also generate compounds that reverse ROS-induced oxidations. Among these compounds are antioxidant molecules like glutathione and ascorbate, which both play important roles as cofactors for thiol reduction enzymes like peroxidases and reductases (Noctor, 2017; Rahantaniaina et al., 2017). Glutathione and ascorbate are themselves reduced by glutathione reductases (GRs) and dehydroascorbate reductases (DHARs). Thioredoxins (TRXs) and glutaredoxins (GRXs) are key thiol reduction enzymes. They act as reducing power of metabolic enzymes and ROS scavenging systems but they also regulate thiol-based post-transcriptional redox modifications in proteins (Meyer et al., 2012). Oxidized TRXs are generally reduced by NADPH-dependent thioredoxin reductases (NTRs), whereas the reduction of GRXs is dependent on glutathione. Due to their multifunctional thiol reduction capacities, TRXs and GRXs have been involved in many metabolic functions, controlling plant developmental programs and acting as key signaling molecules in response to abiotic and biotic stresses (Meyer et al., 2009; Rouhier et al., 2015). In this mini-review, we aim to give an updated overview of nuclear thiol-based ROS signaling in plants.
Some data suggest that ROS are actively generated in the nucleus (Ashtamker et al., 2007), but they principally accumulate in the nucleus through transfer from other cell compartments. Genetically encoded fluorescent H2O2 sensors (e.g., HyPer) have consistently shown that cytosolic H2O2 freely diffuses in the nucleus through nuclear pores (Møller et al., 2007; Rodrigues et al., 2017). It is also transferred from the chloroplasts to the nucleus under pathogen and high light (HL) conditions (Caplan et al., 2015; Exposito-Rodriguez et al., 2017).
The chemical characteristics of the sulfur atom make Cys and Met residues major sites of oxidation within proteins (Davies, 2005). Depending on their pKa and on the pH of the medium, thiol residues are deprotonated into a thiolate residue (R-S-) which is prone to oxidation. This is leading to successive oxidations to sulfenic (R-SOH), sulfinic (R-SO2H), and sulfonic (R-SO3H) acids (Davies, 2005). Thiol groups can also form a disulfide bridge (S-S) or react with reactive nitrogen species (RNS) or oxidized glutathione (GSSG) resulting in S-nitrosylation (R-SNO) or S-glutathionylation (R-S-SG). Depending on their nature, most of these thiol modifications can be reversed by dedicated thiol reduction systems (TRX, GRX, and GSNO Reductase) which exhibit disulfide bond, deglutathionylation or denitrosylation activities (Figure 1). Thiol modifications can alter the structure and/or the activity of many proteins like transcription factors, MAP kinases, and chromatin modification proteins (see below). Proteomic approaches aiming to identify oxidized thiol targets have been developed in plants. Hundreds of nuclear candidate proteins were found sulfenylated, nitrosylated, or glutathionylated (Supplementary Table 1; Zaffagnini et al., 2012, 2016; Morisse et al., 2014; Waszczak et al., 2014; Chaki et al., 2015; Pérez-Pérez et al., 2017). These approaches also revealed the complexity of the thiol redox modification networks in plants (Pérez-Pérez et al., 2017). However, among all these candidates, redox regulation has been validated only in a few cases (see below).
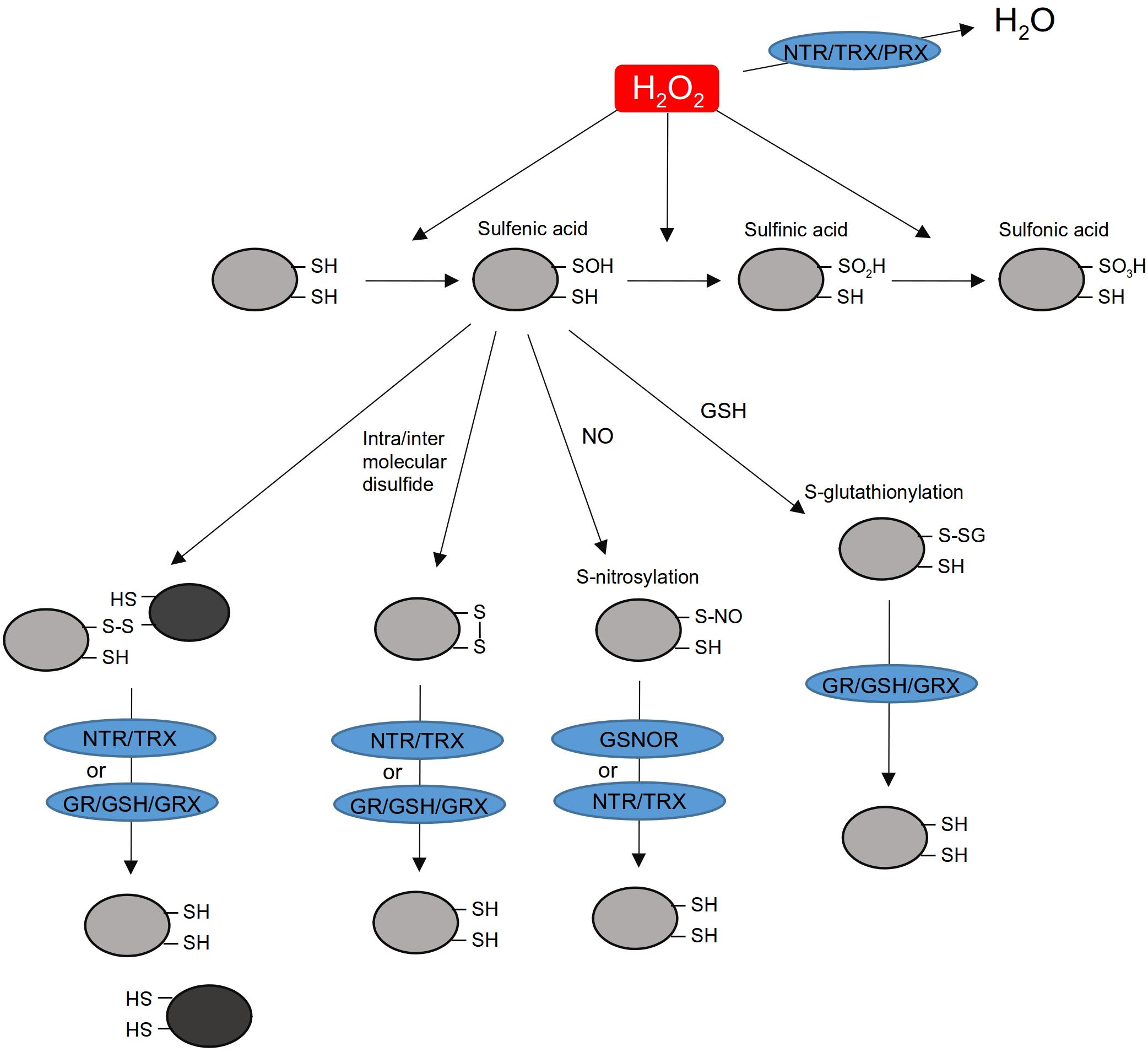
FIGURE 1. H2O2-induced thiol modifications and scavenging activities in the nucleus. H2O2 accumulating in the nucleus can be detoxified by a NTR/TRX/PRX system. While many other H2O2 detoxification enzymes are active in plants (e.g., catalases and ascorbate peroxidases), their presence in the nucleus is not demonstrated yet. They are not represented here. H2O2 can oxidize thiol residues in protein. Sulfenic acid reacts with GSH, NO or with adjacent thiol residues. Putative nuclear proteins prone to S-glutathionylated, S-nitrosylated, or disulfide bonds formation have been identified by proteomic and biochemical approaches (see Supplementary Table 1; Delorme-Hinoux et al., 2016; Pérez-Pérez et al., 2017). S-glutathionylated, S-nitrosylated, or intra/intermolecular disulfide bonds can be reduced by NTR/TRX, GR/GSH/GRX, or GSNOR. NTR, NADPH-dependent thioredoxin reductase; TRX, thioredoxin; GR, glutathione reductase; GRX, glutaredoxin; PRX, peroxiredoxin; GSNOR, S-nitrosoglutathione reductase; GSH, glutathione; NO, nitric oxide; H2O2, hydrogen peroxide.
Plants exhibit a large panel of thiol reduction systems (Meyer et al., 2012). Among them is glutathione, a low molecular weight thiol-containing tripeptide (γ-glutamyl-cysteinyl-glycine). Glutathione biosynthesis is performed in chloroplasts and in the cytosol but is found in almost all cell compartments, including the nucleus. Nuclear pores are generally assumed to allow unrestricted bidirectional diffusion of glutathione across the nuclear envelope. Therefore, nuclear glutathione translocation to the nucleus can be passive. Glutathione quantification at the subcellular level is technically challenging due to its highly dynamic compartmentation (Delorme-Hinoux et al., 2016). Thiol-specific dyes and genetically encoded probes such as reduction-oxidation-sensitive green fluorescent proteins (roGFPs) have consistently detected glutathione in the nucleus. Immunocytochemistry (ICC) coupled to electronic microscopy were also used to address its location at a subcompartment level (Zechmann et al., 2008; Zechmann and Müller, 2010). This study found glutathione uniformly spread in the nucleoplasm, without distinction between euchromatin and heterochromatin. In Arabidopsis thaliana, a glutathione reductase (GR1) is found in the nucleus, suggesting that oxidized glutathione (GSSG) is actively reduced in the nucleus (Delorme-Hinoux et al., 2016).
The functions of glutathione in the nucleus are still poorly understood. Thiol-labeling experiments using the 5-chloromethylfluorescein diacetate (CMFDA) dye and glutathione redox state measured by roGFP, suggest that a redox cycle is occurring during the cell cycle progression and is critical for cell cycle progression (Diaz Vivancos et al., 2010; Vivancos et al., 2010; de Simone et al., 2017). Consistently, sustained mild oxidation observed in ascorbate mutants also restricts nuclear functions and impairs progression through the cell cycle (de Simone et al., 2017). More than acting as a general redox buffer, glutathione could also provide reducing moiety for anti-oxidant enzymes like GRXs (i.e., GRXC1, GRXC2, GRXS17, GRXS13, and ROXY1/2/4) and Glutathione S-Transferases (i.e., GSTF5-10, GSTU19/20, and GSTT19/20), several of them having been identified in the nucleus (Rouhier et al., 2015; Palm et al., 2016).
Consistent levels of ascorbate have also been found in the nucleus but little evidence for its nuclear functions has been described (Zechmann et al., 2011; Considine and Foyer, 2014; de Simone et al., 2017; Zechmann, 2017).
Thioredoxins (TRXs) and Glutaredoxins (GRXs) are major classes of thiols reductases. Plants harbor a complex TRX and GRX network (Meyer et al., 2012). Among the 40 TRXs and 50 GRXs isoforms were found in Arabidopsis, at least 4–8 of them have been assigned to the nucleus, although often in association with a cytosolic localization (Delorme-Hinoux et al., 2016). Moreover, NTR isoforms were also found in the nucleus, where they reduce TRXh, TRXo1, and Nucleoredoxin 1 (NRX1) (Serrato et al., 2001; Serrato and Cejudo, 2003; Marty et al., 2009; Marchal et al., 2014). Some thiol reductases are constitutively located in the nucleus, but others were found to shuttle between the cytosol and nucleus. In tomato subjected to heat stress, the predominant cytosolic GRXS17 was found to relocate in the nucleus (Wu et al., 2012). In wheat and Chlamydomonas, TRXh isoforms accumulate in the nucleus upon oxidative or genotoxic stress (Serrato and Cejudo, 2003; Sarkar et al., 2005). Little is known about the subnuclear localization of these respective proteins. This is due to the low resolution of localization techniques like ICC and GFP-fusion coupled with confocal microscopy analyses. In most cases, these proteins are detected in the nucleoplasm, without distinction between heterochromatin and euchromatin, and are apparently excluded from the nucleolus. Recently, ICC analyses have detected NRX1 and NTRA in the nucleolar cavity, but the functional significance of this localization is still unknown (Marchal et al., 2014). Major thiol redox components found in the nucleus are presented in Figure 2. ROS scavenging enzymes and thiol-containing target proteins are shown as well. Most of these components have been recently reviewed by Delorme-Hinoux et al. (2016) and will not be further discussed here.
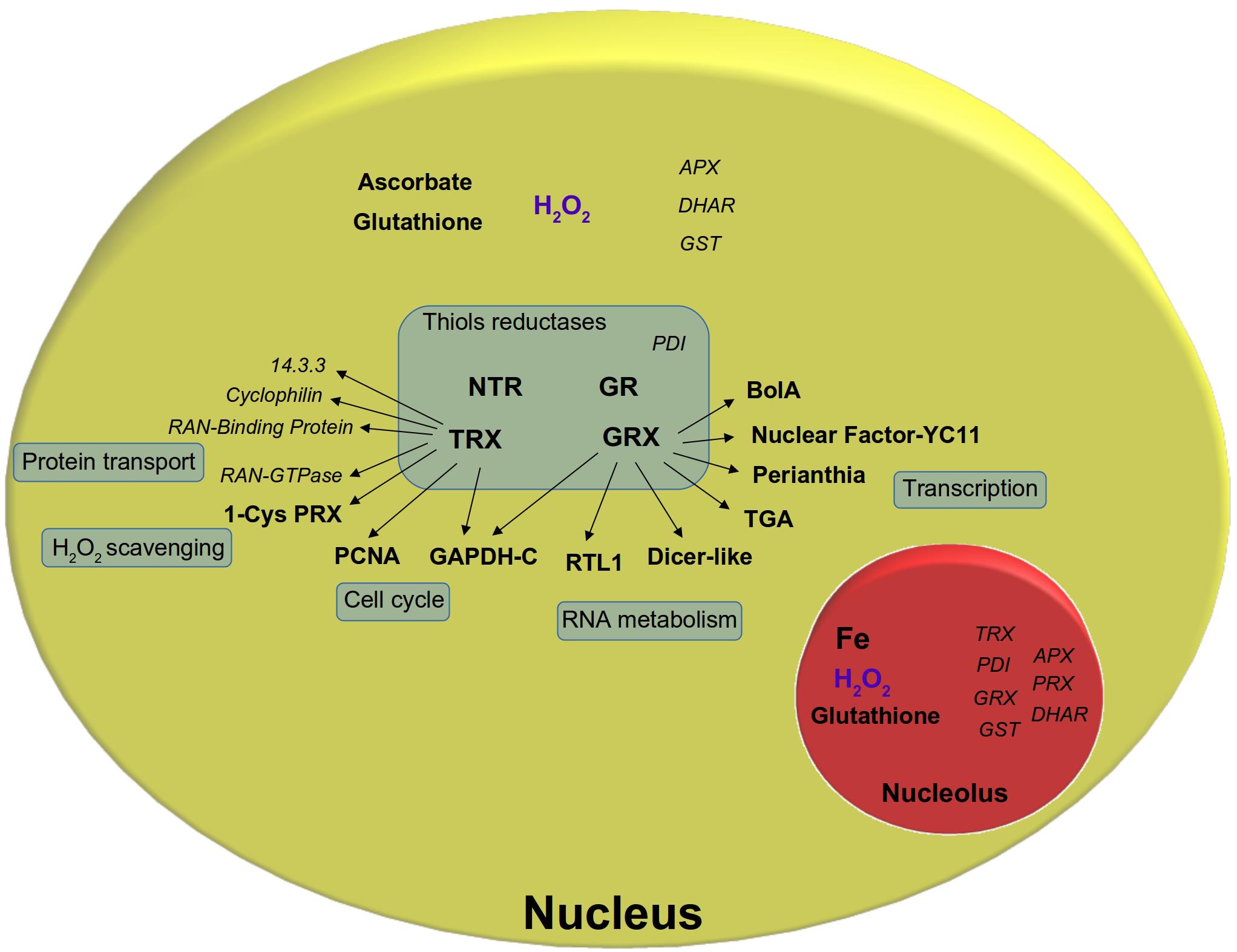
FIGURE 2. Major thiol redox components found in the nucleus. Thiol reduction systems, thiol-containing target proteins were represented as well as ROS scavenging enzymes. Proteins which were only found in proteomic data (according to Montrichard et al., 2009; Waszczak et al., 2014; Delorme-Hinoux et al., 2016; Palm et al., 2016; Montacié et al., 2017; Pérez-Pérez et al., 2017) but not further validated in the nucleus were represented in italic. NTR, NADPH-dependent thioredoxin reductase; TRX, thioredoxin; GR, glutathione reductase; GRX, glutaredoxin; PRX, peroxiredoxin; PDI, protein disulfide isomerase; GST, glutathione S-transferase; APX, ascorbate peroxidase; DHAR, dehydroascorbate reductase; Ran-GTPase, Ran-guanosine triphosphatase; PCNA, proliferating cell nuclear antigen; GAPDH-C, glyceraldehyde 3-phosphate dehydrogenase-C; RTL1, RNAse III-like 1; TGA, TGA-transcription factor; NF-YC11, nuclear factor-YC11; GSH, glutathione; NO, nitric oxide; H2O2, hydrogen peroxide.
The nucleolus, a nuclear subcompartment responsible for rRNA biosynthesis, might also be subjected to redox regulation (Saez-Vasquez and Medina, 2009). Significant accumulation of H2O2 has been detected in the nucleolus in tobacco cell suspension subjected to elicitor treatments (Ashtamker et al., 2007). Intriguingly, the nucleolus also accumulates high amounts of iron, which might provide substrates for ROS generation by Fenton reactions (Roschzttardtz et al., 2011). In addition, glutathione and several isoforms of PRXs, DHARs, APXs, TRXs, and GRX-like proteins were enriched in this compartment (Zechmann et al., 2008; Zechmann and Müller, 2010; Palm et al., 2016; Montacié et al., 2017). Whether redox activities are occurring in the nucleolus will need further investigations.
Reactive oxygen species causes drastic changes in nuclear gene expression (Gadjev et al., 2006; Willems et al., 2016; Shaikhali and Wingsle, 2017). Oxidative stress affects many pathways involved in RNA processing, including splicing, polyadenylation, exporting, and editing. It is also involved in RNA degradation and protein translation (Van Ruyskensvelde et al., 2018). Under High-light (HL) conditions, ROS originated in chloroplasts are associated with chloroplast-to-nucleus (retrograde) signaling. Among the molecules involved in the retrograde signaling (Suzuki et al., 2012; Vogel et al., 2014; Dietz et al., 2016), singlet oxygen (1O2) induces expression of subsets of 1O2-responsive genes and enhances tolerance to HL and to other abiotic and biotic stress (Wagner et al., 2004; Carmody et al., 2016). H2O2 originating by dismutation of superoxide in the chloroplast has also been recently shown to be involved in retrograde signaling upon HL exposure (Exposito-Rodriguez et al., 2017). Another retrograde signaling was suggested to involve a redox regulation of the chloroplastic cyclophilin Cyp20.3, leading to stimulation of Cys synthesis, accumulation of non-protein thiols and activation of defense gene expression (Dominguez-Solis et al., 2008; Park et al., 2013). Presumably, ROS generated in other cell compartments (mitochondria, peroxisomes, and apoplast) can also exert similar retrograde signaling (Noctor and Foyer, 2016; Rodríguez-Serrano et al., 2016).
Photorespiration produces H2O2 in peroxisomes. In this compartment, catalases play an important role in removing H2O2. The cat2 mutant inactivated in the major peroxisomal catalase accumulates a high level of peroxisomal H2O2 and impacts nuclear gene expression extensively, rapidly inducing subsets of stress and hormonal response genes (Queval et al., 2007). In this case, regulation of gene expression involves glutathione signaling also, as the transcriptomic response is partly abolished in a glutathione-defective (cad2) cat2 cad2 double mutant (Han et al., 2013a,b). A signaling role of glutathione on nuclear gene expression was also suggested by transcriptomic data in genetically or pharmacologically manipulated glutathione backgrounds (Xiang and Oliver, 1998; Ball et al., 2004; Schnaubelt et al., 2015). Other transcriptomic analyses also showed involvement of thiol reduction systems in modulating nuclear gene expression (Bashandy et al., 2009; Martins et al., unpublished data). Whether these actors are directly involved in gene expression needs further investigations.
A likely impact of ROS on nuclear gene expression relies on the regulation of redox-sensitive transcription factors (Considine and Foyer, 2014; Dietz, 2014; Rouhier et al., 2015; Waszczak et al., 2015). In most cases, redox regulation induces conformation changes in transcription factors or associated proteins. Such modifications can occur in the cytosol and trigger nuclear translocation, e.g., by uncovering of a nuclear localization sequence (NLS). A well-documented example is the thiol redox-dependent nuclear translocation of the glycolytic enzyme Glucose 6-Phosphate Dehydrogenase C (GAPDH-C) which impacts both its metabolic activity and its moonlighting function as a transcriptional activator of glycolytic genes (Holtgrefe et al., 2008; Vescovi et al., 2013; Zaffagnini et al., 2013; Testard et al., 2016; Zhang et al., 2017). The HL- and H2O2-dependent nuclear translocation of Heat-Shock Factors (HSFA1D and HSFA8) is also dependent on specific Cys residues (Miller and Mittler, 2006; Jung et al., 2013; Giesguth et al., 2015; Dickinson et al., 2018). The pathogen-induced Salicylic Acid (SA)-dependent transcriptional response is mediated by redox-dependent nuclear translocation of NON-EXPRESSOR OF PR GENES1 (NPR1). In this particular case, NPR1 is kept in the cytosol in a disulfide-bound oligomeric homocomplex. Upon pathogen attack, SA induces TRXh5 expression which counteracts NPR1 oligomer formation by reducing NPR1 disulfides. Moreover, through its denitrosylase activity, TRXh5 also suppresses the stimulatory effect of Cys156 S-nitrosylation on formation of disulfide-linked NPR1 oligomer (Tada et al., 2008; Kneeshaw et al., 2014). NPR1 is translocated in the nucleus where it promotes PR gene expression through interaction with TGA transcription factors such as TGA1 (Després et al., 2003; Mou et al., 2003; Tada et al., 2008; Kneeshaw et al., 2014).
Indeed, other members of the TGA transcription factors are likely redox regulated in the nucleus. Among the 10 TGA factors found in Arabidopsis, several of them (i.e., TGA1, TGA2, TGA3, TGA7, and Perianthia) interact with type III GRXs (ROXY1 and 2) and are involved in the development of petals, anthers and microspores. Although the redox dependent control of these TGA is not fully established, ROXY/TGA interactions are occurring in the nucleus and affect TGA-regulated gene expression (Xing et al., 2005; Xing and Zachgo, 2008; Li et al., 2009, 2011; Murmu et al., 2010; reviewed by Dietz, 2014 and Delorme-Hinoux et al., 2016).
R2R3-type MYB transcription factors from maize require reducing conditions for DNA binding. Under non-reducing conditions, Cys49 and Cys53 form a disulfide bond that prevents the R2R3 MYB domain from binding DNA (Williams and Grotewold, 1997; Heine et al., 2004). More recently, the structure and the DNA binding activity of a AtMYB30 transcription factor were shown to be influenced by S-nitrosylation (Tavares et al., 2014).
AP2/ethylene response factor (ERF) is another class of transcription factors which undergoes redox regulation (Welsch et al., 2007; Shaikhali et al., 2008; Vogel et al., 2014). One of the most striking examples was described for the Rap2.12-dependent regulation of hypoxia response genes. Under aerobic conditions, Rap2.12 is bound to the plasma membrane within an acyl-CoA binding protein 1 or 2 (ACBP1/2) complex. In low oxygen, Rap2.12 is released from the plasma membrane by a mechanism involving a N-terminal Cys2 residue, and is translocated to the nucleus where it activates hypoxia response genes (Gibbs et al., 2011; Licausi et al., 2011; Licausi, 2013).
Comelli and Gonzalez (2007) also reported a redox regulation of conserved Cys in the homeodomain (HD) DNA of plant class III HD-Zip proteins. Here, DNA binding capacities are only maintained when an intramolecular disulfide bond is reduced by a thioredoxin (Comelli and Gonzalez, 2007). A Cys-dependent redox regulation of the DNA binding activity of basic region leucine zipper (bZIP) transcription factors has also been reported (Shaikhali et al., 2012).
Finally, a subunit of the Nuclear Factor-Y (NF-Y) transcription factor complex (NY-YC11) physically interacts with the iron-sulfur cluster glutaredoxin GRXS17 in the nucleus. It is not known yet if this interaction is redox-dependent (Knuesting et al., 2015). In the cytosol and the nucleus, GRXS17 also interacts with and reduces BolA2, a factor involved in iron metabolism (Couturier et al., 2014; Qin et al., 2015).
Redox regulation of epigenetic processes has mostly been addressed in mammals (García-Giménez et al., 2017), but this field is poorly explored in plants (Delorme-Hinoux et al., 2016; Shen et al., 2016). Nevertheless, due to the ubiquity of these basic mechanisms in living organisms, it is likely that such regulation occurs in plants as well. Different enzymes involved in histone methylation are prone to redox regulation, affecting both positive and negative histone marks (e.g., H3K4me2, H3K4me3, H3K79me3, H3K27me2, and H3K9me2) (Chen et al., 2006; Zhou et al., 2008, 2010; Niu et al., 2015). In mammals, nuclear histone acetylation activities are redox sensitive, affecting chromatin conformation and transcription (Ito et al., 2004; Ago et al., 2008; Nott et al., 2008; Doyle and Fitzpatrick, 2010). During brain development, neurotrophic factors induce S-nitrosylation at conserved Cys of HDAC2 in neurons, resulting in changes of histone modification and gene expression (Nott et al., 2008). Upon cardiac hypertrophy, a ROS/TRX-dependent redox switch of key Cys residues affects nuclear trafficking of a class II HDAC and subsequent gene expression (Ago et al., 2008). Within the large family of HDAC identified in plants (Pandey et al., 2002), members of the class I RPD-3 like HDAC (HDAC9, 19) have been shown to be sensitive to oxidation (Liu et al., 2015; Mengel et al., 2017), but the physiological significance of those modifications is still poorly understood. NO-induced HDAC inhibition is proposed to operate in plant stress response by facilitating the stress-induced transcription of genes (Mengel et al., 2017).
In addition to methylation and acetylation, mammalian histone H3 has been shown to be glutathionylated on a conserved and unique Cys residue (García-Giménez et al., 2014). Histone H3 glutathionylation increases during cell proliferation and decreases during aging. This produces structural changes affecting nucleosome stability and leading to a more open chromatin structure (García-Giménez et al., 2013, 2014, 2017; Xu et al., 2014).
Small RNAs (siRNA and miRNA) are key regulators of gene expression, involved in most developmental and stress response processes in eukaryotic cells (Leisegang et al., 2017). Biogenesis of small RNAs is orchestrated by DICER-LIKE (DCL) and RNASE THREE-LIKE (RTL) endonucleases that process almost every class of double-stranded RNA precursors. Charbonnel et al. (2017) have recently demonstrated that members of DCL and RTL families in Arabidopsis are glutathionylated on a conserved Cys which affects their RNase III activity. R-S-SG of RTL1 is reversed by type I GRXs, suggesting that small RNA biogenesis and subsequent gene expression responses are under the control of the cell redox environment (Charbonnel et al., 2017). Indeed, the RNase activity of another member of the family (RTL2) was previously shown to be regulated by its dimerization state through an intermolecular disulfide bond (Comella et al., 2008), showing that a redox switch might regulate small RNA biogenesis.
Epigenetic regulation of gene expression is performed by DNA methylation. Some key metabolic enzymes involved in DNA methylation are suspected to be redox regulated. Among them are enzymes of the S-Adenosyl Methionine (SAM) cycle which provide precursors for DNA and histone methylation (Shen et al., 2016). Other nuclear candidates are the DNA demethylases Repressor of Silencing1 (ROS1) and Demeter-like (DME, DML2, and DML3) enzymes which remove methylated bases from the DNA backbone (Zhu, 2009). All these enzymes contain an iron-sulfur (Fe-S) cluster which might be susceptible to oxidation by ROS. Moreover, different members of the cytosolic Fe-S cluster assembly machinery (i.e., MET18 and AE7) are involved in DNA methylation, likely because they affect the nuclear DNA demethylases Fe-S cluster metabolism (Luo et al., 2012; Duan et al., 2015). Therefore, all these examples show an emerging link between redox regulation and epigenetic regulation.
Data supporting the role of redox regulation in nuclear functions are rapidly increasing. ROS are key actors of this regulation, influencing gene expression at multiple levels (transcription and post-transcription), notably by modulating activities of transcription regulators. While H2O2 has been detected in the nucleus, little is known about ROS metabolism and dynamics in this cell compartment. The recent discovery of a H2O2 flux from the chloroplast to the nucleus opens new perspectives to decipher the role of ROS in gene expression. In this way, the identification of a redox regulation of key transcriptional regulators (e.g., transcription factors, HDAC) will shed light on the ways ROS are acting on gene expression. Key for these questions are the proteomic approaches aiming to identify nuclear PTM occurring on redox-sensitive residues, and the structure biology techniques designed to visualize redox-based modifications on protein structure (Waszczak et al., 2014; Parker et al., 2015; Zaffagnini et al., 2016; Pérez-Pérez et al., 2017). While those redox proteome approaches have identified hundreds of nuclear proteins which could be prone to redox modifications, biochemical and functional evidence is missing to support the biological significance of these redox switches. This will be a major challenge for future research in redox biology.
LM, JT-H and J-PR wrote the paper. LM and JT-H performed the figures.
This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (ANR-Blanc Cynthiol 12-BSV6-0011). LM was supported by a Ph.D. grant from the Université de Perpignan Via Domitia (Ecole Doctorale Energie et Environnement ED305). JT-H was supported by a Ph.D. grant from Consejo Nacional de Ciencia y Tecnología (CONACYT: 254639/411761).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00705/full#supplementary-material
Ago, T., Liu, T., Zhai, P., Chen, W., Li, H., Molkentin, J. D., et al. (2008). A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy. Cell 133, 978–993. doi: 10.1016/j.cell.2008.04.041
Ashtamker, C., Kiss, V., Sagi, M., Davydov, O., and Fluhr, R. (2007). Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco Bright Yellow-2 cells. Plant Physiol. 143, 1817–1826. doi: 10.1104/pp.106.090902
Ball, L., Accotto, G.-P., Bechtold, U., Creissen, G., Funck, D., Jimenez, A., et al. (2004). Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell 16, 2448–2462. doi: 10.1105/tpc.104.022608
Bashandy, T., Taconnat, L., Renou, J.-P., Meyer, Y., and Reichheld, J.-P. (2009). Accumulation of flavonoids in an ntra ntrb mutant leads to tolerance to UV-C. Mol. Plant 2, 249–258. doi: 10.1093/mp/ssn065
Caplan, J. L., Kumar, A. S., Park, E., Padmanabhan, M. S., Hoban, K., Modla, S., et al. (2015). Chloroplast stromules function during innate immunity. Dev. Cell 34, 45–57. doi: 10.1016/j.devcel.2015.05.011
Carmody, M., Crisp, P. A., d’Alessandro, S., Ganguly, D., Gordon, M., Havaux, M., et al. (2016). Uncoupling high light responses from singlet oxygen retrograde signaling and spatial-temporal systemic acquired acclimation. Plant Physiol. 171, 1734–1749. doi: 10.1104/pp.16.00404
Chaki, M., Shekariesfahlan, A., Ageeva, A., Mengel, A., von Toerne, C., Durner, J., et al. (2015). Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 238, 115–126. doi: 10.1016/j.plantsci.2015.06.011
Charbonnel, C., Niazi, A. K., Elvira-Matelot, E., Nowak, E., Zytnicki, M., de Bures, A., et al. (2017). The siRNA suppressor RTL1 is redox-regulated through glutathionylation of a conserved cysteine in the double-stranded-RNA-binding domain. Nucleic Acids Res. 45, 11891–11907. doi: 10.1093/nar/gkx820
Chen, H., Ke, Q., Kluz, T., Yan, Y., and Costa, M. (2006). Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol. Cell. Biol. 26, 3728–3737. doi: 10.1128/MCB.26.10.3728-3737.2006
Choudhury, F. K., Rivero, R. M., Blumwald, E., and Mittler, R. (2017). Reactive oxygen species, abiotic stress and stress combination. Plant J. 90, 856–867. doi: 10.1111/tpj.13299
Comella, P., Pontvianne, F., Lahmy, S., Vignols, F., Barbezier, N., Debures, A., et al. (2008). Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3’ETS in Arabidopsis. Nucleic Acids Res. 36, 1163–1175. doi: 10.1093/nar/gkm1130
Comelli, R. N., and Gonzalez, D. H. (2007). Conserved homeodomain cysteines confer redox sensitivity and influence the DNA binding properties of plant class III HD-Zip proteins. Arch. Biochem. Biophys. 467, 41–47. doi: 10.1016/j.abb.2007.08.003
Considine, M. J., and Foyer, C. H. (2014). Redox regulation of plant development. Antioxid. Redox Signal. 21, 1305–1326. doi: 10.1089/ars.2013.5665
Couturier, J., Wu, H.-C., Dhalleine, T., Pégeot, H., Sudre, D., Gualberto, J. M., et al. (2014). Monothiol glutaredoxin-BolA interactions: redox control of Arabidopsis thaliana BolA2 and SufE1. Mol. Plant 7, 187–205. doi: 10.1093/mp/sst156
Davies, M. J. (2005). The oxidative environment and protein damage. Biochim. Biophys. Acta 1703, 93–109. doi: 10.1016/j.bbapap.2004.08.007
de Simone, A., Hubbard, R., de la Torre, N. V., Velappan, Y., Wilson, M., Considine, M. J., et al. (2017). Redox changes during the cell cycle in the embryonic root meristem of Arabidopsis thaliana. Antioxid. Redox Signal. 27, 1505–1519. doi: 10.1089/ars.2016.6959
Delorme-Hinoux, V., Bangash, S. A. K., Meyer, A. J., and Reichheld, J.-P. (2016). Nuclear thiol redox systems in plants. Plant Sci. 243, 84–95. doi: 10.1016/j.plantsci.2015.12.002
Després, C., Chubak, C., Rochon, A., Clark, R., Bethune, T., Desveaux, D., et al. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15, 2181–2191. doi: 10.1105/tpc.012849
Diaz Vivancos, P., Wolff, T., Markovic, J., Pallardó, F. V., and Foyer, C. H. (2010). A nuclear glutathione cycle within the cell cycle. Biochem. J. 431, 169–178. doi: 10.1042/BJ20100409
Dickinson, P. J., Kumar, M., Martinho, C., Yoo, S. J., Lan, H., Artavanis, G., et al. (2018). Chloroplast signaling gates thermotolerance in Arabidopsis. Cell Rep. 22, 1657–1665. doi: 10.1016/j.celrep.2018.01.054
Dietz, K.-J. (2014). Redox regulation of transcription factors in plant stress acclimation and development. Antioxid. Redox Signal. 21, 1356–1372. doi: 10.1089/ars.2013.5672
Dietz, K.-J., Mittler, R., and Noctor, G. (2016). Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 171, 1535–1539. doi: 10.1104/pp.16.00938
Dominguez-Solis, J. R., He, Z., Lima, A., Ting, J., Buchanan, B. B., and Luan, S. (2008). A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 105, 16386–16391. doi: 10.1073/pnas.0808204105
Doyle, K., and Fitzpatrick, F. A. (2010). Redox signaling, alkylation (carbonylation) of conserved cysteines inactivates class I histone deacetylases 1, 2, and 3 and antagonizes their transcriptional repressor function. J. Biol. Chem. 285, 17417–17424. doi: 10.1074/jbc.M109.089250
Duan, C.-G., Wang, X., Tang, K., Zhang, H., Mangrauthia, S. K., Lei, M., et al. (2015). MET18 connects the cytosolic iron-sulfur cluster assembly pathway to active DNA demethylation in Arabidopsis. PLoS Genet. 11:e1005559. doi: 10.1371/journal.pgen.1005559
Exposito-Rodriguez, M., Laissue, P. P., Yvon-Durocher, G., Smirnoff, N., and Mullineaux, P. M. (2017). Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signalling mechanism. Nat. Commun. 8:49. doi: 10.1038/s41467-017-00074-w
Foyer, C. H., and Noctor, G. (2016). Stress-triggered redox signalling: what’s in pROSpect? Plant Cell Environ. 39, 951–964. doi: 10.1111/pce.12621
Gadjev, I., Vanderauwera, S., Gechev, T. S., Laloi, C., Minkov, I. N., Shulaev, V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445. doi: 10.1104/pp.106.078717
García-Giménez, J. L., Ibañez-Cabellos, J. S., Seco-Cervera, M., and Pallardó, F. V. (2014). Glutathione and cellular redox control in epigenetic regulation. Free Radic. Biol. Med. 75(Suppl. 1):S3. doi: 10.1016/j.freeradbiomed.2014.10.828
García-Giménez, J. L., Òlaso, G., Hake, S. B., Bönisch, C., Wiedemann, S. M., Markovic, J., et al. (2013). Histone h3 glutathionylation in proliferating mammalian cells destabilizes nucleosomal structure. Antioxid. Redox Signal. 19, 1305–1320. doi: 10.1089/ars.2012.5021
García-Giménez, J. L., Romá-Mateo, C., Pérez-Machado, G., Peiró-Chova, L., and Pallardó, F. V. (2017). Role of glutathione in the regulation of epigenetic mechanisms in disease. Free Radic. Biol. Med. 112, 36–48. doi: 10.1016/j.freeradbiomed.2017.07.008
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Giesguth, M., Sahm, A., Simon, S., and Dietz, K.-J. (2015). Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett. 589, 718–725. doi: 10.1016/j.febslet.2015.01.039
Han, Y., Chaouch, S., Mhamdi, A., Queval, G., Zechmann, B., and Noctor, G. (2013a). Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox Signal. 18, 2106–2121. doi: 10.1089/ars.2012.5052
Han, Y., Mhamdi, A., Chaouch, S., and Noctor, G. (2013b). Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 36, 1135–1146. doi: 10.1111/pce.12048
Heine, G. F., Hernandez, J. M., and Grotewold, E. (2004). Two cysteines in plant R2R3 MYB domains participate in REDOX-dependent DNA binding. J. Biol. Chem. 279, 37878–37885. doi: 10.1074/jbc.M405166200
Holtgrefe, S., Gohlke, J., Starmann, J., Druce, S., Klocke, S., Altmann, B., et al. (2008). Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133, 211–228. doi: 10.1111/j.1399-3054.2008.01066.x
Ito, K., Hanazawa, T., Tomita, K., Barnes, P. J., and Adcock, I. M. (2004). Oxidative stress reduces histone deacetylase 2 activity and enhances IL-8 gene expression: role of tyrosine nitration. Biochem. Biophys. Res. Commun. 315, 240–245. doi: 10.1016/j.bbrc.2004.01.046
Jung, H.-S., Crisp, P. A., Estavillo, G. M., Cole, B., Hong, F., Mockler, T. C., et al. (2013). Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc. Natl. Acad. Sci. U.S.A. 110, 14474–14479. doi: 10.1073/pnas.1311632110
Kneeshaw, S., Gelineau, S., Tada, Y., Loake, G. J., and Spoel, S. H. (2014). Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant Immunity. Mol. Cell 56, 153–162. doi: 10.1016/j.molcel.2014.08.003
Knuesting, J., Riondet, C., Maria, C., Kruse, I., Bécuwe, N., König, N., et al. (2015). Arabidopsis glutaredoxin S17 and its partner, the nuclear factor Y subunit C11/negative cofactor 2α, contribute to maintenance of the shoot apical meristem under long-day photoperiod. Plant Physiol. 167, 1643–1658. doi: 10.1104/pp.15.00049
Leisegang, M. S., Schröder, K., and Brandes, R. P. (2017). Redox regulation and noncoding RNAs. Antioxid. Redox Signal. doi: 10.1089/ars.2017.7276 [Epub ahead of print].
Li, S., Gutsche, N., and Zachgo, S. (2011). The ROXY1 C-terminal L∗∗LL motif is essential for the interaction with TGA transcription factors. Plant Physiol. 157, 2056–2068. doi: 10.1104/pp.111.185199
Li, S., Lauri, A., Ziemann, M., Busch, A., Bhave, M., and Zachgo, S. (2009). Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell 21, 429–441. doi: 10.1105/tpc.108.064477
Licausi, F. (2013). Molecular elements of low-oxygen signaling in plants. Physiol. Plant. 148, 1–8. doi: 10.1111/ppl.12011
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Liu, P., Zhang, H., Yu, B., Xiong, L., and Xia, Y. (2015). Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci. Rep. 5:8625. doi: 10.1038/srep08625
Luo, D., Bernard, D. G., Balk, J., Hai, H., and Cui, X. (2012). The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24, 4135–4148. doi: 10.1105/tpc.112.102608
Marchal, C., Delorme-Hinoux, V., Bariat, L., Siala, W., Belin, C., Saez-Vasquez, J., et al. (2014). NTR/NRX define a new thioredoxin system in the nucleus of Arabidopsis thaliana cells. Mol. Plant 7, 30–44. doi: 10.1093/mp/sst162
Marty, L., Siala, W., Schwarzländer, M., Fricker, M. D., Wirtz, M., Sweetlove, L. J., et al. (2009). The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 9109–9114. doi: 10.1073/pnas.0900206106
Mengel, A., Ageeva, A., Georgii, E., Bernhardt, J., Wu, K., Durner, J., et al. (2017). Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 173, 1434–1452. doi: 10.1104/pp.16.01734
Meyer, Y., Belin, C., Delorme-Hinoux, V., Reichheld, J.-P., and Riondet, C. (2012). Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxid. Redox Signal. 17, 1124–1160. doi: 10.1089/ars.2011.4327
Meyer, Y., Buchanan, B. B., Vignols, F., and Reichheld, J.-P. (2009). Thioredoxins and glutaredoxins: unifying elements in redox biology. Annu. Rev. Genet. 43, 335–367. doi: 10.1146/annurev-genet-102108-134201
Miller, G., and Mittler, R. (2006). Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 98, 279–288. doi: 10.1093/aob/mcl107
Møller, I. M., Jensen, P. E., and Hansson, A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. doi: 10.1146/annurev.arplant.58.032806.103946
Montacié, C., Durut, N., Opsomer, A., Palm, D., Comella, P., Picart, C., et al. (2017). Nucleolar proteome analysis and proteasomal activity assays reveal a link between nucleolus and 26S proteasome in A. thaliana. Front. Plant Sci. 8:1815. doi: 10.3389/fpls.2017.01815
Montrichard, F., Alkhalfioui, F., Yano, H., Vensel, W. H., Hurkman, W. J., and Buchanan, B. B. (2009). Thioredoxin targets in plants: the first 30 years. J. Proteomics 72, 452–474. doi: 10.1016/j.jprot.2008.12.002
Morisse, S., Zaffagnini, M., Gao, X.-H., Lemaire, S. D., and Marchand, C. H. (2014). Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid. Redox Signal. 21, 1271–1284. doi: 10.1089/ars.2013.5632
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Murmu, J., Bush, M. J., DeLong, C., Li, S., Xu, M., Khan, M., et al. (2010). Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 154, 1492–1504. doi: 10.1104/pp.110.159111
Niu, Y., DesMarais, T. L., Tong, Z., Yao, Y., and Costa, M. (2015). Oxidative stress alters global histone modification and DNA methylation. Free Radic. Biol. Med. 82, 22–28. doi: 10.1016/j.freeradbiomed.2015.01.028
Noctor, G. (2017). ROS-related redox regulation and signaling in plants. Semin. Cell Dev. Biol. doi: 10.1016/j.semcdb.2017.07.013 [Epub ahead of print].
Noctor, G., and Foyer, C. H. (2016). Intracellular redox compartmentation and ROS-related communication in regulation and signaling. Plant Physiol. 171, 1581–1592. doi: 10.1104/pp.16.00346
Nott, A., Watson, P. M., Robinson, J. D., Crepaldi, L., and Riccio, A. (2008). S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455, 411–415. doi: 10.1038/nature07238
Palm, D., Simm, S., Darm, K., Weis, B. L., Ruprecht, M., Schleiff, E., et al. (2016). Proteome distribution between nucleoplasm and nucleolus and its relation to ribosome biogenesis in Arabidopsis thaliana. RNA Biol. 13, 441–454. doi: 10.1080/15476286.2016.1154252
Pandey, R., Müller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055.
Park, S.-W., Li, W., Viehhauser, A., He, B., Kim, S., Nilsson, A. K., et al. (2013). Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc. Natl. Acad. Sci. U.S.A. 110, 9559–9564. doi: 10.1073/pnas.1218872110
Parker, J., Balmant, K., Zhu, F., Zhu, N., and Chen, S. (2015). cysTMTRAQ-An integrative method for unbiased thiol-based redox proteomics. Mol. Cell. Proteomics 14, 237–242. doi: 10.1074/mcp.O114.041772
Pérez-Pérez, M. E., Mauriès, A., Maes, A., Tourasse, N. J., Hamon, M., Lemaire, S. D., et al. (2017). The deep thioredoxome in Chlamydomonas reinhardtii: new insights into redox regulation. Mol. Plant 10, 1107–1125. doi: 10.1016/j.molp.2017.07.009
Qin, L., Wang, M., Zuo, J., Feng, X., Liang, X., Wu, Z., et al. (2015). Cytosolic BolA plays a repressive role in the tolerance against excess iron and MV-induced oxidative stress in plants. PLoS One 10:e0124887. doi: 10.1371/journal.pone.0124887
Queval, G., Issakidis-Bourguet, E., Hoeberichts, F. A., Vandorpe, M., Gakière, B., Vanacker, H., et al. (2007). Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. doi: 10.1111/j.1365-313X.2007.03263.x
Rahantaniaina, M.-S., Li, S., Chatel-Innocenti, G., Tuzet, A., Issakidis-Bourguet, E., Mhamdi, A., et al. (2017). Cytosolic and chloroplastic DHARs cooperate in oxidative stress-driven activation of the salicylic acid pathway. Plant Physiol. 174, 956–971. doi: 10.1104/pp.17.00317
Rodrigues, O., Reshetnyak, G., Grondin, A., Saijo, Y., Leonhardt, N., Maurel, C., et al. (2017). Aquaporins facilitate hydrogen peroxide entry into guard cells to mediate ABA- and pathogen-triggered stomatal closure. Proc. Natl. Acad. Sci. U.S.A. 114, 9200–9205. doi: 10.1073/pnas.1704754114
Rodríguez-Serrano, M., Romero-Puertas, M. C., Sanz-Fernández, M., Hu, J., and Sandalio, L. M. (2016). Peroxisomes extend peroxules in a fast response to stress via a reactive oxygen species-mediated induction of the peroxin PEX11a. Plant Physiol. 171, 1665–1674. doi: 10.1104/pp.16.00648
Roschzttardtz, H., Grillet, L., Isaure, M.-P., Conéjéro, G., Ortega, R., Curie, C., et al. (2011). Plant cell nucleolus as a hot spot for iron. J. Biol. Chem. 286, 27863–27866. doi: 10.1074/jbc.C111.269720
Rouhier, N., Cerveau, D., Couturier, J., Reichheld, J.-P., and Rey, P. (2015). Involvement of thiol-based mechanisms in plant development. Biochim. Biophys. Acta 1850, 1479–1496. doi: 10.1016/j.bbagen.2015.01.023
Sarkar, N., Lemaire, S., Wu-Scharf, D., Issakidis-Bourguet, E., and Cerutti, H. (2005). Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage. Eukaryot. Cell 4, 262–273. doi: 10.1128/EC.4.2.262-273.2005
Schnaubelt, D., Queval, G., Dong, Y., Diaz-Vivancos, P., Makgopa, M. E., Howell, G., et al. (2015). Low glutathione regulates gene expression and the redox potentials of the nucleus and cytosol in Arabidopsis thaliana. Plant Cell Environ. 38, 266–279. doi: 10.1111/pce.12252
Serrato, A. J., and Cejudo, F. J. (2003). Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217, 392–399. doi: 10.1007/s00425-003-1009-4
Serrato, A. J., Crespo, J. L., Florencio, F. J., and Cejudo, F. J. (2001). Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol. Biol. 46, 361–371. doi: 10.1023/A:1010697331184
Shaikhali, J., Heiber, I., Seidel, T., Ströher, E., Hiltscher, H., Birkmann, S., et al. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8:48. doi: 10.1186/1471-2229-8-48
Shaikhali, J., Norén, L., de Dios Barajas-López, J., Srivastava, V., König, J., Sauer, U. H., et al. (2012). Redox-mediated mechanisms regulate DNA binding activity of the G-group of basic region leucine zipper (bZIP) transcription factors in Arabidopsis. J. Biol. Chem. 287, 27510–27525. doi: 10.1074/jbc.M112.361394
Shaikhali, J., and Wingsle, G. (2017). Redox-regulated transcription in plants: emerging concepts. AIMS Mol. Sci. 4, 301–338. doi: 10.3934/molsci.2017.3.301
Shen, Y., Issakidis-Bourguet, E., and Zhou, D.-X. (2016). Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 67, 5291–5300. doi: 10.1093/jxb/erw310
Sies, H., Berndt, C., and Jones, D. P. (2017). Oxidative stress. Annu. Rev. Biochem. 86, 715–748. doi: 10.1146/annurev-biochem-061516-045037
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321, 952–956. doi: 10.1126/science.1156970
Tavares, C. P., Vernal, J., Delena, R. A., Lamattina, L., Cassia, R., and Terenzi, H. (2014). S-nitrosylation influences the structure and DNA binding activity of AtMYB30 transcription factor from Arabidopsis thaliana. Biochim. Biophys. Acta 1844, 810–817. doi: 10.1016/j.bbapap.2014.02.015
Testard, A., Da Silva, D., Ormancey, M., Pichereaux, C., Pouzet, C., Jauneau, A., et al. (2016). Calcium- and nitric oxide-dependent nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in response to long chain bases in tobacco BY-2 cells. Plant Cell Physiol. 57, 2221–2231. doi: 10.1093/pcp/pcw137
Van Ruyskensvelde, V., Van Breusegem, F., and Van Der Kelen, K. (2018). Post-transcriptional regulation of the oxidative stress response in plants. Free Radic. Biol. Med. doi: 10.1016/j.freeradbiomed.2018.02.032 [Epub ahead of print].
Vescovi, M., Zaffagnini, M., Festa, M., Trost, P., Lo Schiavo, F., and Costa, A. (2013). Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 162, 333–346. doi: 10.1104/pp.113.215194
Vivancos, P. D., Dong, Y., Ziegler, K., Markovic, J., Pallardó, F. V., Pellny, T. K., et al. (2010). Recruitment of glutathione into the nucleus during cell proliferation adjusts whole-cell redox homeostasis in Arabidopsis thaliana and lowers the oxidative defence shield. Plant J. 64, 825–838. doi: 10.1111/j.1365-313X.2010.04371.x
Vogel, M. O., Moore, M., König, K., Pecher, P., Alsharafa, K., Lee, J., et al. (2014). Fast retrograde signaling in response to high light involves metabolite export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF transcription factors in Arabidopsis. Plant Cell 26, 1151–1165. doi: 10.1105/tpc.113.121061
Wagner, D., Przybyla, D., Op den Camp, R., Kim, C., Landgraf, F., Lee, K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185. doi: 10.1126/science.1103178
Waszczak, C., Akter, S., Eeckhout, D., Persiau, G., Wahni, K., Bodra, N., et al. (2014). Sulfenome mining in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 111, 11545–11550. doi: 10.1073/pnas.1411607111
Waszczak, C., Akter, S., Jacques, S., Huang, J., Messens, J., and Van Breusegem, F. (2015). Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 66, 2923–2934. doi: 10.1093/jxb/erv084
Welsch, R., Maass, D., Voegel, T., Dellapenna, D., and Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145, 1073–1085. doi: 10.1104/pp.107.104828
Willems, P., Mhamdi, A., Stael, S., Storme, V., Kerchev, P., Noctor, G., et al. (2016). The ROS wheel: refining ROS transcriptional footprints. Plant Physiol. 171, 1720–1733. doi: 10.1104/pp.16.00420
Williams, C. E., and Grotewold, E. (1997). Differences between plant and animal Myb domains are fundamental for DNA binding activity, and chimeric Myb domains have novel DNA binding specificities. J. Biol. Chem. 272, 563–571. doi: 10.1074/jbc.272.1.563
Wu, Q., Lin, J., Liu, J.-Z., Wang, X., Lim, W., Oh, M., et al. (2012). Ectopic expression of Arabidopsis glutaredoxin AtGRXS17 enhances thermotolerance in tomato. Plant Biotechnol. J. 10, 945–955. doi: 10.1111/j.1467-7652.2012.00723.x
Xiang, C., and Oliver, D. J. (1998). Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell 10, 1539–1550. doi: 10.1105/tpc.10.9.1539
Xing, S., Rosso, M. G., and Zachgo, S. (2005). ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development 132, 1555–1565. doi: 10.1242/dev.01725
Xing, S., and Zachgo, S. (2008). ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 53, 790–801. doi: 10.1111/j.1365-313X.2007.03375.x
Xu, Y.-M., Du, J.-Y., and Lau, A. T. Y. (2014). Posttranslational modifications of human histone H3: an update. Proteomics 14, 2047–2060. doi: 10.1002/pmic.201300435
Zaffagnini, M., Bedhomme, M., Marchand, C. H., Morisse, S., Trost, P., and Lemaire, S. D. (2012). Redox regulation in photosynthetic organisms: focus on glutathionylation. Antioxid. Redox Signal. 16, 567–586. doi: 10.1089/ars.2011.4255
Zaffagnini, M., De Mia, M., Morisse, S., Di Giacinto, N., Marchand, C. H., Maes, A., et al. (2016). Protein S-nitrosylation in photosynthetic organisms: a comprehensive overview with future perspectives. Biochim. Biophys. Acta 1864, 952–966. doi: 10.1016/j.bbapap.2016.02.006
Zaffagnini, M., Fermani, S., Costa, A., Lemaire, S. D., and Trost, P. (2013). Plant cytoplasmic GAPDH: redox post-translational modifications and moonlighting properties. Front. Plant Sci. 4:450. doi: 10.3389/fpls.2013.00450
Zechmann, B. (2017). Compartment-specific importance of ascorbate during environmental stress in plants. Antioxid. Redox Signal. doi: 10.1089/ars.2017.7232 [Epub ahead of print].
Zechmann, B., Mauch, F., Sticher, L., and Müller, M. (2008). Subcellular immunocytochemical analysis detects the highest concentrations of glutathione in mitochondria and not in plastids. J. Exp. Bot. 59, 4017–4027. doi: 10.1093/jxb/ern243
Zechmann, B., and Müller, M. (2010). Subcellular compartmentation of glutathione in dicotyledonous plants. Protoplasma 246, 15–24. doi: 10.1007/s00709-010-0111-2
Zechmann, B., Stumpe, M., and Mauch, F. (2011). Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 233, 1–12. doi: 10.1007/s00425-010-1275-x
Zhang, H., Zhao, Y., and Zhou, D.-X. (2017). Rice NAD+-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 45, 12241–12255. doi: 10.1093/nar/gkx825
Zhou, X., Sun, H., Chen, H., Zavadil, J., Kluz, T., Arita, A., et al. (2010). Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 70, 4214–4221. doi: 10.1158/0008-5472.CAN-09-2942
Zhou, X., Sun, H., Ellen, T. P., Chen, H., and Costa, M. (2008). Arsenite alters global histone H3 methylation. Carcinogenesis 29, 1831–1836. doi: 10.1093/carcin/bgn063
Keywords: nucleus, thiol, glutathione, thioredoxin, glutaredoxin, ROS
Citation: Martins L, Trujillo-Hernandez JA and Reichheld J-P (2018) Thiol Based Redox Signaling in Plant Nucleus. Front. Plant Sci. 9:705. doi: 10.3389/fpls.2018.00705
Received: 09 February 2018; Accepted: 09 May 2018;
Published: 28 May 2018.
Edited by:
Christian Lindermayr, Helmholtz Zentrum München – Deutsches Forschungszentrum für Gesundheit und Umwelt, GermanyReviewed by:
Renu Deswal, University of Delhi, IndiaCopyright © 2018 Martins, Trujillo-Hernandez and Reichheld. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean-Philippe Reichheld, anByQHVuaXYtcGVycC5mcg==
†These authors should be considered as co-first authors.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.