- 1Department of Plant Science, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai, China
- 2Institute of Agro-Food Science and Technology, Key Laboratory of Agro-Products Processing Technology of Shandong, Shandong Academy of Agricultural Sciences, Jinan, China
ABA plays a crucial role in controlling several ripening-associated processes in grape berries. The soluble proteins named as PYR (pyrabactin resistant)/PYL (PYR-like)/RCAR (regulatory component of ABA receptor) family have been characterized as ABA receptors. Here, the function of a grape PYL1 encoding gene involved in the response to ABA was verified through heterologous expression. The expression level of VlPYL1 was highest in grape leaf and fruit tissues of the cultivar Kyoho, and the expression of VlPYL1 was increased during fruit development and showed a reduction in ripe berries. Over-expression of VlPYL1 enhances ABA sensitivity in Arabidopsis. Using the transient overexpression technique, the VlPYL1 gene was over-expressed in grape berries. Up-regulation of the VlPYL1 gene not only promoted anthocyanin accumulation but also induced a set of ABA-responsive gene transcripts, including ABF2 and BG3. Although tobacco rattle virus (TRV)-induced gene silencing (VIGS) was not successfully applied in the “Kyoho” grape, the application of the transient overexpression technique in grape fruit could be used as a novel tool for studying grape fruit development.
Introduction
Fruit development and ripening are complex plant processes. Fruits are classified into climacteric and non-climacteric depending on respiration and ethylene production during maturation and senescence (Giovannoni, 2001). From extensive studies on climacteric fruits, such as the tomato, banana, peach, apple, and melon fruit, ripening has been shown to be marked by a respiratory burst linked to the production of ethylene, which plays a pivotal role in the regulation of climacteric fruit ripening (Alexander and Grierson, 2002; Adams-Phillips et al., 2004; Gupta et al., 2006; Pech et al., 2008; Cherian et al., 2014). Contrastingly, in non-climacteric fruits, such as the strawberry, grape, orange, cucumber, bilberry, and blueberry, a respiratory burst does not occur, and ethylene may have a less central impact on ripening, while ABA appears to have a pivotal regulatory role in the process (Ji et al., 2012; Zifkin et al., 2012; Karppinen et al., 2013; Wang et al., 2013; Cherian et al., 2014; Jia et al., 2016).
Grapevines are cultivated world-wide and are an economically valuable fruit crop. The development and ripening process of grape berries have their own characteristic with a double sigmoid growth pattern, which is resulting from two rapid stages of growth separated by a lag phase of slow or no growth (Coombe and McCarthy, 2000; Xie et al., 2009). Berry acidity increases during the initial stage of rapid berry growth and declines sharply after the lag phase, while anthocyanin and sugar contents increase rapidly (Xie et al., 2009). Comprehensive studies have shown that ABA is an important hormone in the inception and color development stage of grape berries ripening (Ferrara et al., 2015; Fortes et al., 2015; Jia et al., 2017; Pilati et al., 2017). A strong rise in berry ABA content corresponds with the increase in total anthocyanin content that is recorded around the véraison and during the initial stages of ripening (Wheeler et al., 2009; Lacampagne et al., 2010; Sun et al., 2010; Wang et al., 2016). Anthocyanins keep increasing throughout the ripening phase, while ABA levels simultaneously start to decrease (Wheeler et al., 2009; Wang et al., 2016), implying that ABA triggers but does not necessarily maintain color acquisition (Kuhn et al., 2014). Additionally, treatments by ABA cause an advance in red color development including Olympia (Hiratsuka et al., 2001a), Kyoho (Ban et al., 2003), Crimson Seedless (Cantin et al., 2007; Ferrara et al., 2013), Cabernet Sauvignon (Wheeler et al., 2009), Alicante Bouschet (Castellarin et al., 2011) and Fujiminori (Jia et al., 2017).
Cells need to sense ABA and then transduceit to the cellular components for ABA function (Nakashima and Yamaguchi-Shinozaki, 2013). In 2009, independent groups simultaneously discovered the structural and functional mechanisms of ABA sensing by cytosolic PYRABACTIN RESISTANCE1(PYR1)/PYR1-like (PYL)/REGULATORY COM PONENTS OF ABA RECEPTORS (RCAR) receptor proteins (Ma et al., 2009; Park et al., 2009). PYR/PYL/RCAR proteins bind to ABA and thus inhibit the activity of PP2C proteins (Ma et al., 2009; Park et al., 2009), leading to the derepression of SnRK2 protein kinases and the activation of ABA-responsive binding factors (ABFs), which are thought to be necessary to promote ABA-induced gene expression (Kobayashi Y. et al., 2005; Umezawa et al., 2009).
The PYR/PYL/RCAR family contains 14 members (PYR1 and PYL1-13) in Arabidopsis, 12 members in rice (He et al., 2014) and 14 members in tomatoes (Gonzalez-Guzman et al., 2014). In Arabidopsis, all of the 14 members can activate ABA-responsive gene expression (Fujii and Zhu, 2009; Fuchs et al., 2014). Many studies have shown that perception of ABA by the PYR/PYL/RCARs plays a critical role in the regulation of seed germination, seedling establishment, vegetative and reproductive growth, stomata closure, and transcriptional responses to the hormone (Nakashima and Yamaguchi-Shinozaki, 2013). As ABA receptors, PYR/PYL/RCAR will be of great value in increasing ABA sensitivity (Zhang et al., 2013b).
One ABA receptor from grape, VvPYL1 can bind ABA and then inhibit the ABI1 phosphatase activity (Li et al., 2012). Eight PYR/PYL/RCARs, four of which have been identified and induced by ABA in the leaves of grapevines (Boneh et al., 2012a,b). Also, high levels of VvPYL1 (VvRCAR7) expression were induced in grape leaves by drought, salt and cold stress (Boneh et al., 2012b). Recently, the ABA receptor PYR1 was cloned from the cultivar “Fujiminori,” whose expression levels were low in the early stage of fruit development and increased rapidly in the former stage of the grape berry, being PYR1 expression pattern consistent with ABA content changes (Jia et al., 2017). However, solid evidence has not been provided to verify the function of VvPYL1 during the ripening process of grape fruits.
In this study, the data on the ripening-related expression of PYL1 in the grape cultivar “Kyoho” fruit were provided. Heterologous overexpression in Arabidopsis, transient overexpression and Tobacco rattle virus (TRV)-induced gene silencing (VIGS) assays in the grape berry were used to try to verify the function of the isolated putative PYR gene. The VlPYL1 gene played an important role in fruit development, as up-regulation of the VlPYL1 gene could promote anthocyanin accumulation in grape berry skin. Additionally, ABA-related genes, such as ABF2 and BG3, could directly respond to VlPYL1 expression levels. These results indicate that the ABA receptor VlPYL1 acts as a positive regulator to promote ripening in grapevine.
Materials and Methods
Plant Materials
The grape cultivar “Kyoho” (Vitis vinifera × Vitis labrusca) and strawberry plants (Fragaria × ananassa cv. “Hongyu”) were grown in a glasshouse at Shanghai Jiao Tong University, Shanghai, China (31°11′N, 121°29′W). Thirty vines of uniform “Kyoho” grapevines were selected. The cultivation medium was a mixture of sand, loam and perlite (1:1:1). All vines were placed at a spacing of 1.5 m × 2.0 m in north–south oriented rows, and the shoots were trained vertically with three shoots per vine. Vines were maintained under natural ventilated and light conditions throughout the growing season except on rainy days. Nutrition and irrigation management was conducted as described previously (Wang B. et al., 2012).
Strawberry plants were planted in pots (diameter, 250 mm; and depth, 250 mm) containing a mixture of soil, sand, and organic fertilizer (5:2:1, v/v/v). The seedlings were grown in growth chamber under the following conditions: 25°C, and a 12-h photoperiod with a photosynthetic photon flux density of 300 μmol m-2s-2.
The Arabidopsis thaliana (ecotype Columbia-0) seeds were sown on 1/2 Murashige and Skoog (MS) medium for 1 week, and then transferred in plastic basins filled with peat and vermiculite (1:1 by volume). The Arabidopsis plants were cultured in the controlled environment cabinets under LD (16 h light/8 h dark) conditions with a fluence rate of 200 μmol m-2s-2 of white light (produced by cool-white fluorescent lamps) at 22°C.
Spatiotemporal and Tissue-Specific Expression of VlPYL1
Different developmental stages of grape fruit were collected at 7, 21, 28, 42, 56, 74, and 96 days after anthesis (DAA). Analysis was performed on three separate pools of c. 20 fruits each. Each pool was from three different plants. After removal of seeds, the pulp and peel were cut into small cubes of 0.5–0.8 cm3. Mixed samples of grape berries were harvested at each stage, while leaves, young apical shoot internodes (stems), flowers, and roots used for organ-specific expression analysis were collected at the flowering stage with three replicates. All samples were quickly frozen in liquid nitrogen and stored at -80°C until use. Quantitative reverse transcription PCR (RT-qPCR) primers to detect VlPYL1 were designed using Primer Express 3.0.1. Primer sequences are shown in Supplementary Table S1.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from different developmental stages of grape, as well as root, stem leaf, and flower tissues using a TaKaRa MiniBEST Plant RNA Extraction Kit (Takara, Dalian, China). Genomic DNA was eliminated using a TaKaRa MiniBEST Plant Genomic DNA Extraction Kit (TaKaRa) followed by an RNA Clean Purification Kit (TaKaRa). The purity and integrity of RNA were analyzed by both agarose gel electrophoresis and NANODROP2000 (Thermo Scientific, Wilmington, DE, United States). To generate first-strand cDNA, 1 μg of total RNA was reverse-transcribed using the PrimeScriptTM RT reagent kit (TaKaRa) according to the manufacturer’s protocol.
Gene Isolation and Sequence Analysis
The cDNA obtained above was used as a template for amplifying the full encoding length of VlPYL1. PCR was performed under the following conditions: 94°C for 90 s, 35 cycles at 94°C for 20 s, 57°C for 20 s, and 72°C for 30 s using the Fast Pfu Master DNA Polymerase (Novoprotein Scientific Inc.). PCR products were ligated into a pEASY-Blunt Simple vector and subsequently transformed into Escherichia coli DH5α. Positive colonies were selected, amplified, and sequenced by Tsingke China (Shanghai, China).
For promoter isolation, 1.5 kb upstream sequence of the VlPYL1 start codon ‘ATG’ was cloned as promoter based on the grape gene library1 (Vitulo et al., 2014). Then the VlPYL1 promoter was analyzed for cis-acting elements using the Plant-CARE database2. To identify the conserved regions, multiple sequence alignments were performed using ClustalX (version 1.83) (Thompson et al., 1997) and DNAMAN 6.0 (Lynnon Biosoft, Quebec, Canada). For phylogenetic analysis, MEGA 6.0 (Tamura et al., 2013) was used to construct maximum parsimony (MP) trees with the following parameters: Poisson model, pairwise deletion and bootstrap (1000 replicates; random seed).
Quantitative Real-Time PCR Analysis
Primers used for quantitative real-time PCR (qPCR) were designed using Primer express 3.0.1 and are listed in Supplementary Table S1. The VvActin and AtACT2 genes were used as internal controls for grape and A. thaliana, respectively. qPCR was performed on a CFX connect Real Time PCR Detection System (Bio-Rad) using the following program: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 60°C for 10 s. The relative expression levels of the amplified products were analyzed using the comparative CT method based on CT values (Livak and Schmittgen, 2001).
Subcellular Localization of VlPYL1
For subcellular localization of VlPYL1, the coding region of VlPYL1 lacking its stop codon was cloned into the pHB vector containing the GFP reporter gene to produce a fusion construct under the control of the CaMV35S promoter (Li et al., 2016). The fusion construct was transferred into Agrobacterium tumefaciens strain GV3101 by heat shock. Agrobacterium mediated transient transformation of tobacco (Nicotiana benthamiana) leaves was done according to Sparkes et al. (2006). The cells of transformed tobacco leaves were observed with a laser scanning confocal microscope (Leica TCS SP5-II, Germany).
Construction of Expression Vectors and Agroinfiltration
To construct a VlPYL1 over-expression (VlPYL1-OE) vector, full-length cDNAs of the VlPYL1 gene were amplified. The resultant PCR product was inserted into the pHB vector under the control of the double 35S-CaMV promoter.
For construction of silencing vectors, the tobacco rattle virus (TRV)-based pTRV1 and pTRV2 vectors used for RNAi (Dong et al., 2007) were kindly donated by Dr. Liu Yu-le, Qinghua University. A 420-bp cDNA fragment of VlPYL1 (from 137 to 556 bp) and a 390 bp fragment of VvPDS (from 166 to 556 bp) were cloned into pTRV2, respectively (Dong et al., 2007).
Agrobacterium tumefaciens strain GV3101 containing pHB-VlPYL1, pTRV1, pTRV2, or the pTRV2 derivatives pTRV2-VlPYL1 and pTRV2-VvPDS was grown at 28°C in Luria–Bertani medium supplemented with kanamycin and rifampin. When the culture reached an OD600 of approximately 1.0, Agrobacterium cells were harvested and resuspended in infection buffer (10 mM MgCl2, 10 mM MES, pH 5.8, and 100 μM acetosyringone) and shaken for 4 h at 28°C before being used for infiltration. Agrobacterium-mediated infiltration by syringe injection into strawberry and tobacco was performed as described by Dong et al. (2007), Chai et al. (2011), and Li Q. et al. (2013). Large green grape berries (45 DAA) were used and infected with a syringe until the whole fruit became hygrophanous. Approximately 2, 4, 6, and 8 days after treatment, the fruits infiltrated with the overexpression or the silencing constructs were harvested and used to detect the vector by reverse transcription PCR (RT-PCR). For analysis of phenotypes, 200 fruits were injected, with 50 fruits being transformed with VlPYL1-OE or VlPYL1-RNAi and the other 50 used as the control (i.e., injected with the pHB or TRV empty vector). For the expression analysis of ripening-related genes, three fruits were combined as an individual sample. The pulp and peels without seeds were frozen in liquid nitrogen and stored at -80°C until use.
Color Measurement
The anthocyanin content in whole grape skins was measured after inoculation for 8 days and different growth periods at 20, 30, 40, 50, 60, 70, 80, and 90 DAA; Grape skin (50 mg) was pulverized with liquid nitrogen and then each sample was homogenized in 1% (v/v) hydrochloric acid in methanol and shaken at 4°C overnight. Anthocyanin concentrations were determined by measuring the absorbance of the extract at 530 nm using an ultraviolet–visible (UV-Vis) spectrophotometer (Pirie and Mullins, 1976; Lurie et al., 2009).
The color of grape berry skin was measured after inoculation for 8 days with a tristimulus reflectance colorimeter (S-6017, Chentaike, Beijing, China). Color parameters were recorded as L∗ (lightness), a∗ (redness) and b∗ (yellowness).
ABA and Drought Tolerance Assay in Transgenic Arabidopsis
The sense vector was genetically introduced into Arabidopsis using Agrobacterium-mediated transformation as described in a previous report (Zhang et al., 2006). Transformants were selected based on their resistance to hygromycin (Hyg). Positive transgenic seedlings were grown in pots containing a mixture of soil and vermiculite (1:2, v/v) to select for T2 and T3 seeds. After surface sterilization of the seeds, approximately 100 seeds of each genotype were sowed on 1/2 MS medium supplemented with 1.5% sucrose and 0.7% agar. Seeds were vernalized at 4°C for 3 days and then incubated in a growth chamber at 22°C with a 16 h light and 8 h dark photoperiod.
Surface-sterilized seeds were sown on 1/2 MS without or with ABA (1 and 3 μM) or mannitol (100 mM). Seeds were scored as germinated after 48 h and 4 days. The germination rate was determined as the radicle emerges from the seed coat.
For ABA and mannitol treatment, 5-day-old seedlings were plated on 1/2 MS medium and 1/2 MS medium supplemented with ABA (1 and 3 μM) or mannitol (200 mM) for 8 days, and then their root lengths and leaf areas were measured using PC-aided image processing software (Bakr, 2005). The treated plants were harvested at the indicated time points, immediately frozen in liquid nitrogen, and stored at -80°C until used for RNA extractions.
Statistical Analysis
Data were analyzed using variance (ANOVA), and the averages were compared by the Duncan’s New Multiple Range test or T-test (P < 0.05) using SPSS 17.0 (SPSS, United States).
Results
Phylogenetic and Identification Analysis of VlPYL1
Three genes, named VvPYL1, VvPYL2 and VvPYL3 were cloned from Vitis vinifera cv. Muscat of Hamburg (Li et al., 2012). To clone the VlPYL1 gene in “Kyoho” (Vitis vinifera × Vitis labrusca), the nucleotide and protein sequences of PYR1 (At4G17870) and PYL1 (At5g46790) in Arabidopsis, PYR1 in strawberry (No. JF268669) and VvPYL1 (LOC100267793) (Li et al., 2012) were used to BLAST to a grape gene library3 (Vitulo et al., 2014), and 8 significantly highly similar proteins with conserved amino acid regions were obtained. In agreement with Boneh et al. (2012b), the phylogenetic analysis results showed that these proteins from the three species could be classified into three large groups: in one group was the RCAR7 protein, which was most closely related to AtPYR1, AtPYL1 and FaPYR1, followed by gene VIT 204s0008g00890.1, which is annotated as “abscisic acid receptor PYL2/RCAR8” (Supplementary Figure S1B). Although the RCAR7 showed higher homology to AtPYR1 and FaPYR1 than AtPYL1, the name VlPYL1 was used in consistency with naming by Li et al. (2012).
Based on the nucleotide sequence, gene-specific primers (Supplementary Table S1) were designed to amplify a coding sequence of the VlPYL1 gene from “Kyoho” grape fruit. A 645 bp cDNA was isolated from grape fruit through RT-PCR (Supplementary Figure S2). The cDNA included an open reading frame encoding a deduced protein of 215 amino acids (Supplementary Figure S3), in which the putative conserved domains were detected by homology analysis (protein BLAST on the NCBI website4) (Supplementary Figure S1A), which suggested that the putative grape ABA receptor gene, VlPYL1, was isolated successfully. VlPYL1 protein exhibited 74.4% sequence identity with the Arabidopsis AtPYR1 protein. VlPYL1 and strawberry PYR1 (FaPYR1) shared 72.35% sequence similarity (Supplementary Figure S3).
VlPYL1 promoter sequence was cloned and sequenced by sanger method (GenBank Accession Number MG917083). To define cis-acting elements in the VlPYL1 promoter, a motif search was conducted using the Plant-CARE databases. As illustrated in Supplementary Table S2, two ABRE (ABA-responsive element) motifs with the core sequences GCAACGTGTC and TACGTG could be identified. The sequence analysis suggested that several cis-elements, including defense and stress responsiveness (TC-rich repeats), fungal elicitor responsive element (W-box, which is recognized by the WRKY protein), cold response (LTR), and element essential for the anaerobic induction (TGGTTT) are also present in the VlPYL1 promoter sequence (Supplementary Table S2). The sequence also contains hormone responsive cis-acting elements such as methyl jasmonate (MeJA) responsive TGACG -motif, ethylene responsive element, and salicylic acid responsive elements.
Subcellular Localization and Expression Pattern of VlPYL1
To provide more insights into the function of VlPYL1, the subcellular localization of the protein was investigated using a full-length VlPYL1 N-terminal GFP fusion protein. The fusion gene of VlPYL1-GFP was transformed into tobacco (Nicotiana benthamiana) leaves using A. tumefaciens infiltration. As shown in Supplementary Figure S4, the VlPYL1-GFP fusion protein localized in both the nucleus and cytosol.
To determine the expression profile of VlPYL1 in grapes, total RNA was extracted from roots, stems, leaves, flowers, and fruits and reverse-transcribed into cDNA. Based on the VlPYL1 gene sequence, a 152 bp VlPYL1 gene specific fragment was used to carry out the RT-qPCR to identify expression patterns. The results showed that VlPYL1 was abundant in grape leaves and fruits, followed by stems and it showed a low level in roots and flowers (Figure 1A).
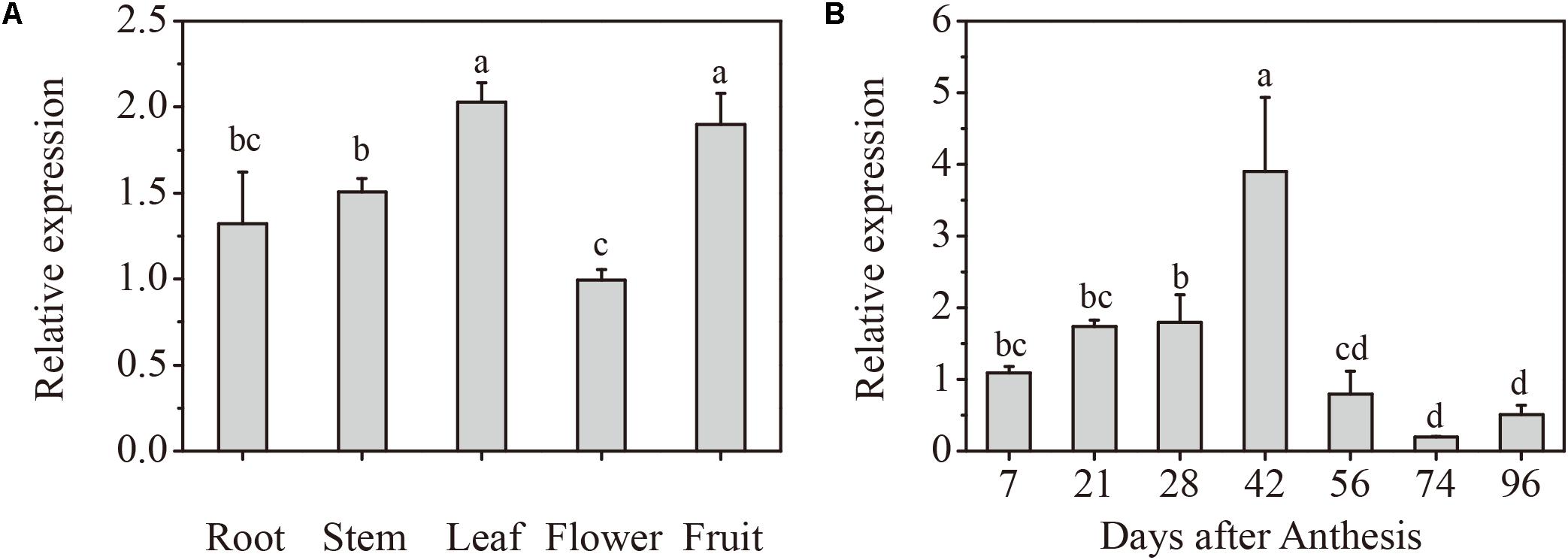
FIGURE 1. Spatiotemporal pattern of VlPYL1 expression in grape fruits. (A) RT-qPCR analysis of VlPYL1 expression in different organs of the “Kyoho” grape plant. (B) Changes in the mRNA expression level of the VlPYL1 gene in “Kyoho” grape fruits. VlPYL1 transcripts were detected by RT-qPCR in the pulp and peel during grape fruit development. Actin mRNA was used as an internal control. The error bars represent the SD (n = 3). Different letters indicated a statistical difference at P < 0.05 as determined by Duncan’s multiple range test.
To investigate whether VlPYL1 is involved in grape fruit development, the levels of its transcript during berry ripening were observed. The results showed that VlPYL1 was expressed in both green and purple fruits. The transcripts of the VlPYL1 gene increased gradually coupled with green fruit growth and enlargement (from 7 to 42 DAA). However, transcripts decreased rapidly at the veraison stage (56 DAA), and reached their lowest level in ripe fruits (74 and 96 DAA) (Figure 1B).
Overexpression of VlPYL1 in Arabidopsis Leads to the ABA and Osmotic Stress Hypersensitive Phenotype During Germination
The VlPYL1 overexpression (VlPYL1-OE) vector was introduced into Arabidopsis plants, and four transgenic lines were obtained. The T3 homozygous 1-1 and 2-4 lines with high and moderate levels of expression of VlPYL1, respectively, were chosen for functional analysis (Supplementary Figure S5).
We treated the 1-1 and 2-4 lines and wild type with 1 and 3 μM ABA and then investigated their seed germination phenotypes. No significant differences in seed germination rate between VlPYL1-OE plants and wild type were found under control conditions. However, the seed germination rates of the two VlPYL1-OE transgenic Arabidopsis lines were less than the wild type in the presence of 1 μM ABA and 3 μM ABA (Figures 2A,B).
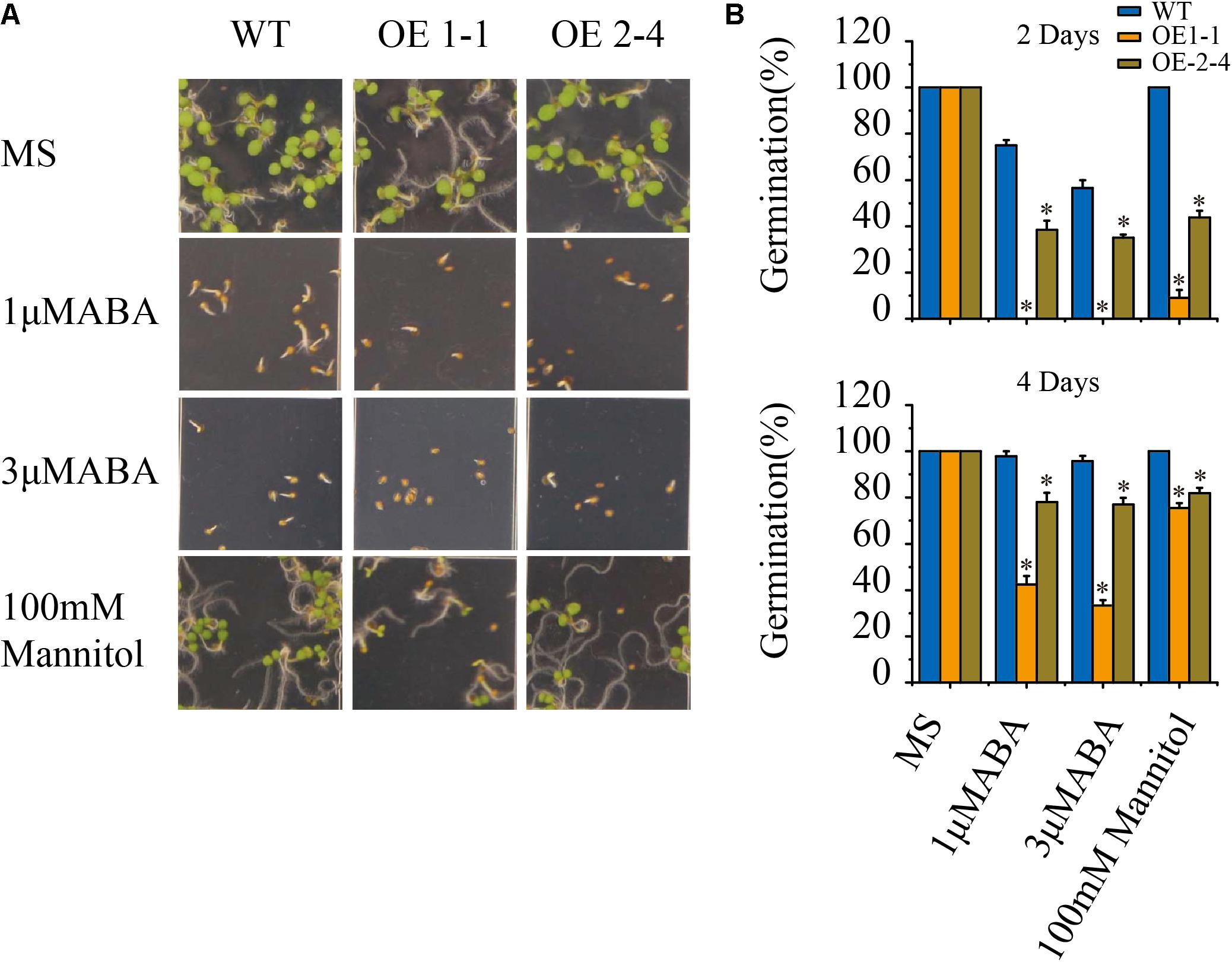
FIGURE 2. Seed germination phenotypes of 2 × 35S:: VlPYL1 transgenic Arabidopsis plants. (A) ABA and osmotic stress hypersensitive inhibition of germination in 2 × 35S:: VlPYL1 transgenic Arabidopsis lines compared with wild-type plants. Seeds were grown on MS medium with or without 1 μmol ABA, or 3 μmol ABA, or 100 mM mannitol. Photographs were taken 4 days after sowing. (B) Percentage of seed germination in the presence of the stress treatment. Germination percentage is determined at 2 (upper graph) and 4 days (lower graph) after seeds are scored on MS, 1 μmol ABA, 3 μmol ABA, or 100 mM mannitol. Data are averages ± SD from three independent experiments (n∼50 seeds per experiment). ∗p-value < 0.05 when comparing data for each genotype versus the wild-type under the same conditions.
Since the ABA sensitivity at the seed germination stage of VlPYL1-OE transgenic Arabidopsis plants have been enhanced, we speculated that overexpression of VlPYL1 may also affect plant osmotic stress tolerance. To test this, the seed germination of VlPYL1-OE transgenic Arabidopsis lines under 100 mM mannitol treatment was analyzed. Similar to the ABA treatment, the germination greening ratio and germination rate of VlPYL1 transgenic lines were significantly inhibited by 100 mM mannitol, but the inhibition in the wild type was less severe (Figures 2A,B).
These results demonstrate that VlPYL1-OE seeds exhibit a sensitivity to inhibition of germination by exogenous ABA and osmotic stress.
Overexpression of VlPYL1 in Arabidopsis Confers ABA Hypersensitive Phenotype and Promotes Plant Root Growth to Osmotic Stress During Seedling Growth
ABA acts as a repressor in root growth, and root growth under ABA treatment is important indicator to evaluate plant ABA sensitivity (Zhang et al., 2013a). To fully examine the plant phenotype under ABA treatment, we sowed wild type and the two transgenic lines on MS medium. After 5 days, plants were transferred to vertical plates and supplemented with 0, 1, or 3 μM ABA for 8 days (Figure 3A). When grown on plates without ABA, the plants displayed no visible root development phenotype. However, the two transgenic lines showed significantly reduced root length and leaf area comparing with the wild-type plants on the plates with the supplement 1 or 3 μM ABA (Figures 3A,B). These results indicated that VlPYL1 overexpression lines were hypersensitive to ABA-mediated inhibition of root growth in Arabidopsis.
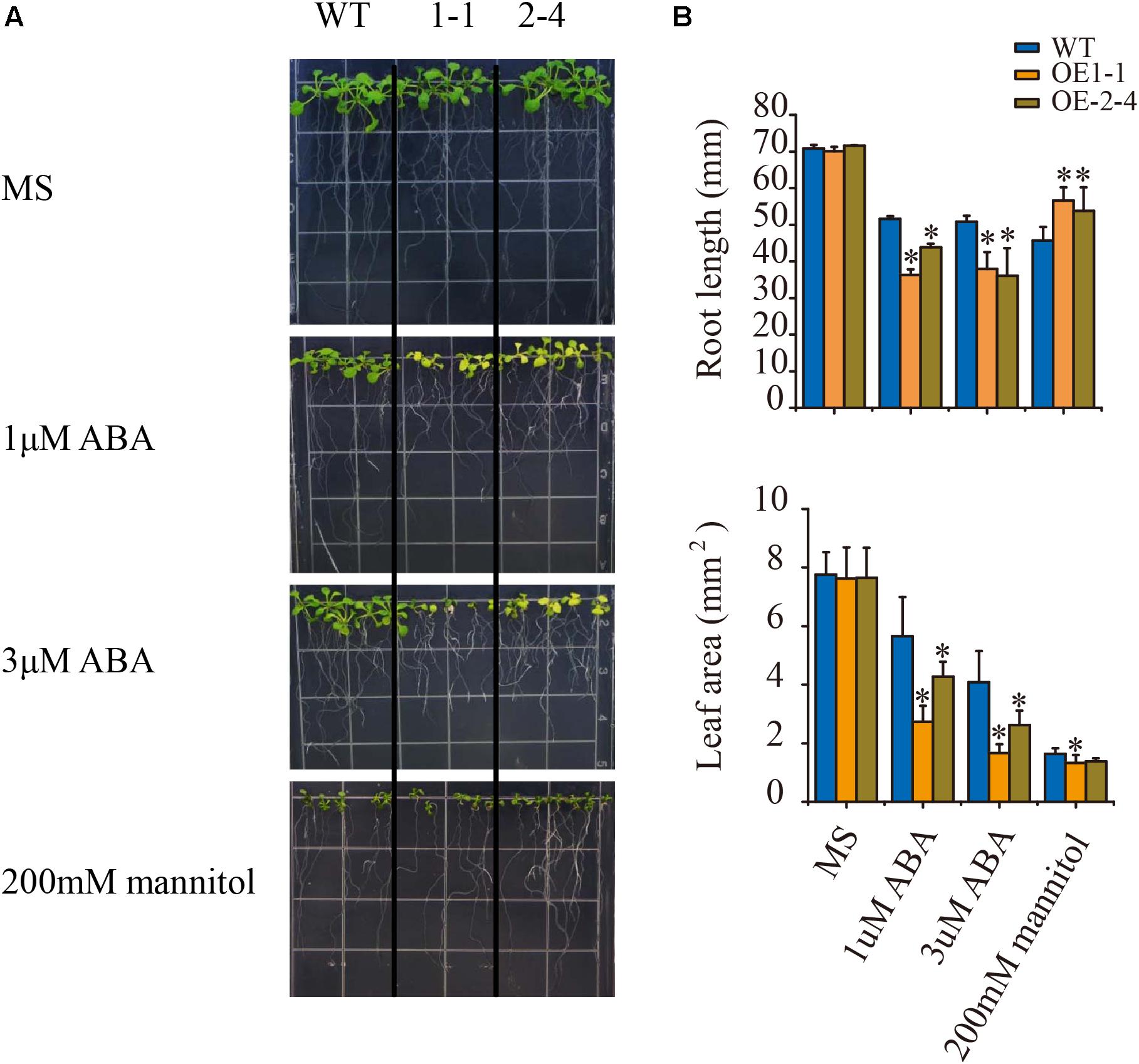
FIGURE 3. ABA hypersensitive inhibition of root growth in 2 × 35S:: VlPYL1 transgenic Arabidopsis. (A) Growth of Col-0, 2 × 35S:: VlPYL1, two independent 2 × 35S:: VlPYL1 lines (1-1, 2-4) in medium (MS) supplemented with 1 and 3 μM ABA. Photographs of representative seedlings 8 days after the transfer of 5-day-old seedlings from MS to plates supplemented with ABA. (B) Root length and leaf area of WT and two transgenic lines on 1 μM ABA, 3 μM ABA, or 100 mM mannitol.
To extend the osmotic stress-tolerance analysis, 5-day-old seedlings were transferred to 1/2 MS plates without (control) or with 200 mM mannitol for 8 days. In the presence of 200 mM mannitol, the shoot growth of VlPYL1-OE plants was inhibited slightly compared to that of the control, but transgenic Arabidopsis plants displayed longer primary roots than wild type (Figures 3A,B).
Analysis of ABA-Responsive Gene Expression in Transgenic Arabidopsis Plants
To examine whether the enhancement of ABA sensitivity in transgenic plants was accompanied by altered mRNA levels of the stress-responsive genes, the abiotic stress-responsive genes RD29A, RD29B, RAB18 and KIN1 mRNA levels were determined by RT-qPCR analysis using total RNAs isolated from mock-treated Arabidopsis or ABA-treated plants for 8 days (Zhang et al., 2013a). As shown in Figure 4, the transcript levels of most of these stress marker genes were higher in ABA stress plants compared with plants grown in the MS medium without ABA treatment. Compared with the WT lines, both AtRD29B and RAB18 exhibited a significantly higher level of expression in transgenic lines following ABA treatment. Notably, the expression level of AtRD29A was slightly higher in WT plants than in transgenic plants under ABA stress. Expression of KIN1 was significantly higher in 1-1 lines under 1 μM ABA treatment, but both transgenic plants expressed similar levels under 3 μM ABA treatment (Figure 4).
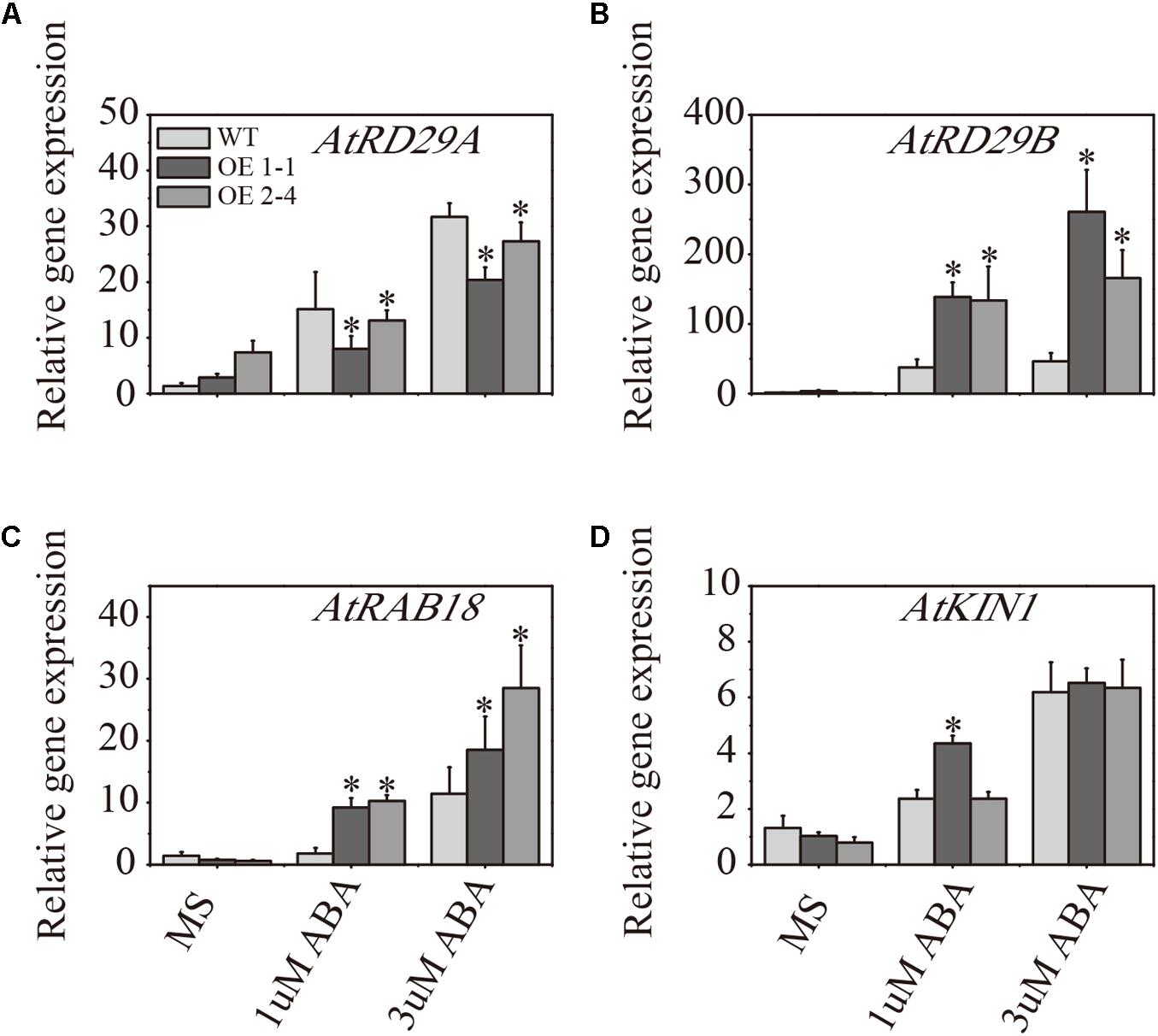
FIGURE 4. Expression analysis of ABA-regulated genes in 2 × 35S::VlPYL1 transgenic Arabidopsis compared to wild-type. The mRNA levels of the indicated genes, including RD29A (A), RD29B (B), RAB18 (C), and KIN1 (D) are determined by RT-qPCR analysis using total RNAs isolated from mock-treated Arabidopsis or ABA-treated plants. Data are averages ± SD from three independent experiments. Expression of β-ACTIN2 was used to normalize data. ∗p-value < 0.05 indicate statistically significant expression change of transgenic lines w.r.t. respective WT in different treatments.
Over-Expression of VlPYL1 Alters the Developmental Processes of Grape Fruits
Developmental and ripening parameters were characterized (Supplementary Figure S6). The earliest sample stage corresponded with the green-hard stage characterized by berries at least 11 mm in diameter with a minimum 6° Brix. An increasing trend in weight, size, soluble solids, and anthocyanin were consistent with the phase transition from green-hard (0 DAA) to green-soft (50 DAA), pink-soft (60 DAA), and red-soft (80 DAA) developmental stages (Supplementary Figure S6).
Big green grape berries (45 days after anthesis) were selected for injection with the empty vector pHB and VlPYL1-OE with a 1-ml syringe. After being infiltrated for 8 days, the VlPYL1-OE-infiltrated berries showed faster color development compared with the empty vector pHB infiltrated berries (Figure 5A). A significant increase in the red color (a∗), a decrease in the yellowness (b∗) and similar lightness (L∗) values were obtained for OE fruits as compared to the control fruits. Fruit anthocyanin contents also increased in OE fruits compared with the control fruits (Figures 5C,D). The number of VlPYL1-OE berries with the observed red spot was higher than control berries 8 days after injection (Supplementary Table S3). However, the accumulation began to converge in the VlPYL1-OE and control berries 16 days after infiltration (Supplementary Tables S3, S4).
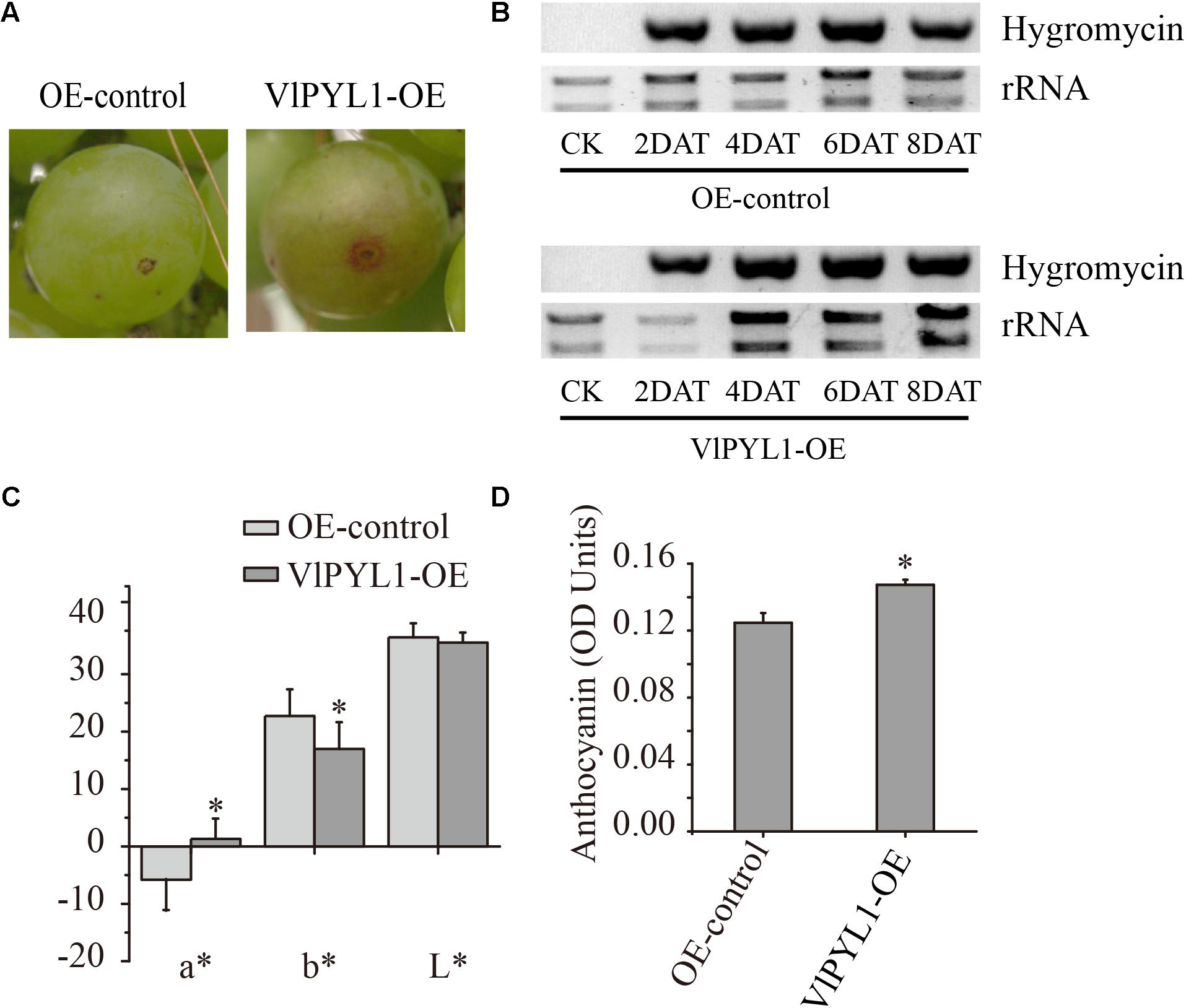
FIGURE 5. (A) “Kyoho” fruits agroinfiltrated with empty vector (EV) and agroinfiltrated fruit with the VlPYL1-OE construct (overexpression) at 6 days after injection. (B) RT-PCR analysis of OE vector expression in fruits. Hygromycin gene (resistance marker gene) expression was detected in fruits infiltrated with Agrobacterium containing the OE vector (harvested on day 2, 4, 6, and 8 post-injection) but was not detected in fruits infiltrated with Agrobacterium alone (lane 1). (C) Color parameters (L∗, a∗, b∗) of the control and overexpression fruits. (D) The total anthocyanin content was measured in the control and overexpression fruits. Asterisks indicated statistically significant differences at P < 0.05 as determined by Student’s t-test.
To confirm the results caused by VlPYL1-OE in grape berries, a pHB-specific primer for a 752 bp amplicon of the vector were designed. As shown in Figure 5B, the PCR products could be detected accompanying the color development in the infiltrated fruits. These results confirmed that over-expression of VlPYL1 could alter the developmental processes of grape fruits.
Overexpression of the VlPYL1 Gene Alters a Set of ABA-Responsive and Flavonoid Biosynthesis-Related Gene Transcripts
Here, the mRNA expression levels of the ABA-responsive genes, including ABA synthesis-associated genes NCED1, NCED2, BG1, BG2 and BG3, ABA degradation-related gene CYP1, ABA signal transduction associated genes PP2C9, SnRK2.1, SnRK2.6, ABF1 and ABF2 were analyzed by RT-qPCR (Figure 6A). The results showed that PYL1, NCED1, CYP1, BG3 and ABF2 were significantly up-regulated in the VlPYL1-OE grape fruits.
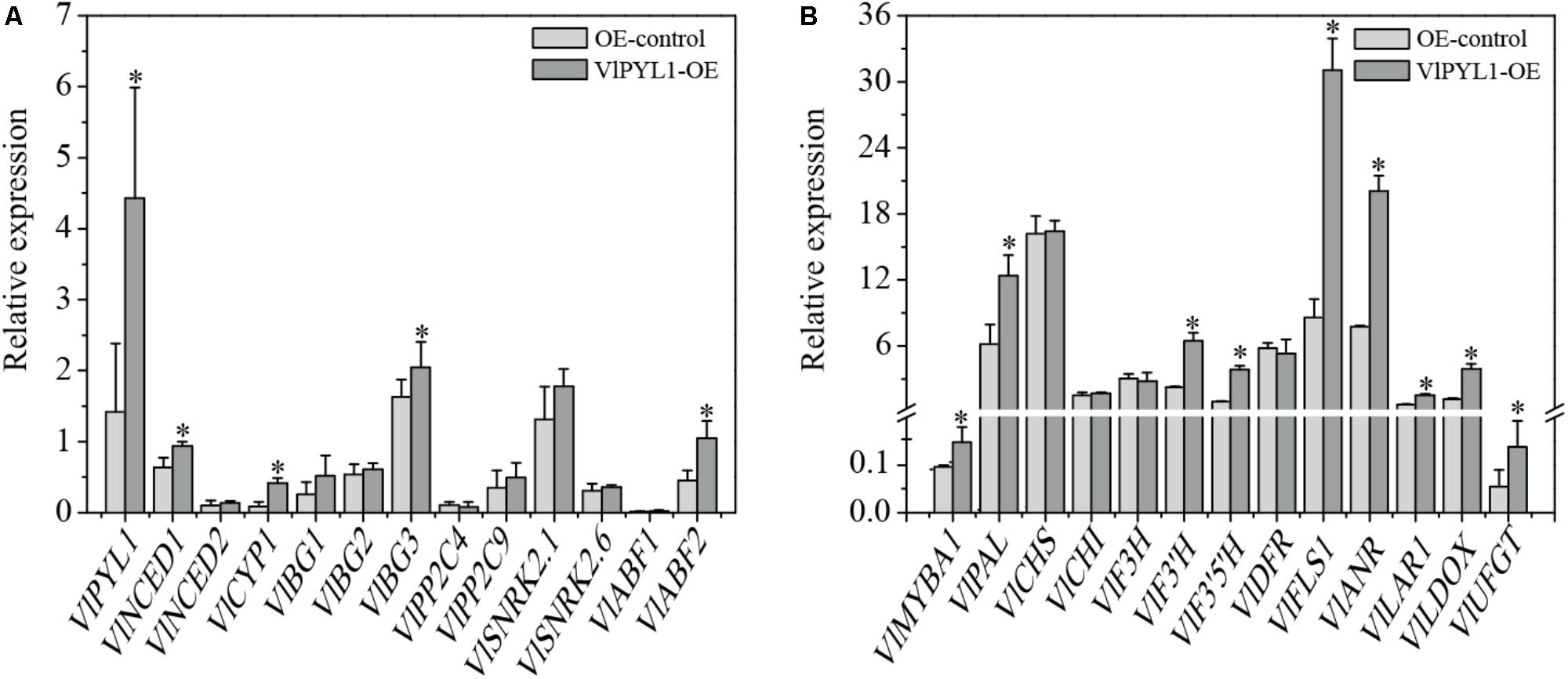
FIGURE 6. Transcriptional changes of the ABA signaling genes and flavonoid biosynthesis-related genes in VlPYL1-OE fruits. (A) The mRNA expression levels of ABA signaling genes in VlPYL1 fruit and control fruit were detected by RT-qPCR. (B) Relative expression by RT-qPCR of flavonoid biosynthetic pathway genes in the grape fruits of VlPYL1 overexpression fruits or control solution. Data are averages ± SD from three independent experiments. Expression of VlActin was used to normalize data. Asterisks indicate significant differences by t-test analysis: ∗P < 0.05.
To explore the relationship of anthocyanin biosynthesis and VlPYL1, the mRNA levels of the flavonoid upstream pathway gene (PAL), the flavonoid biosynthesis-related genes (CHS, CHI, F3H, F3′H, F3′5′H, DFR, LDOX, UFGT, FLS1, LAR1, and ANR) and a regulatory gene (MybA1) were also analyzed by RT-qPCR (Figure 6B). As shown in Figure 6B, the mRNA expression levels of MYBA1, PAL, F3′H, F3′5′H, FLS1, ANR, LAR1, LDOX and UFGT were significantly up-regulated in the VlPYL1-OE grape berries.
Transient Expression of VlPYL1- RNAi Vector Mediated by Agrobacterium
After being infiltrated for 12 days, the pTRV2-VlPYL1 infiltrated grape berries and controls were both beginning to turn into the veraison stage (Supplementary Figure S7A). However, the strawberry fruits infiltrated with pTRV1 and pTRV2-VlPYL1 showed an arrest of color development (Chai et al., 2011) compared to the control strawberry fruits (Supplementary Figure S8A). Further analysis of FaPYR1 transcripts by RT-qPCR was performed in the receptacle of independent fruits injected with either the silencing construct or the empty vector. As shown in Supplementary Figure S8B, VlPYL1-RNAi resulted in a dramatic decrease in FaPYR1 transcript level. To test whether the TRV-vector can directly infect grape berries, a TRV-RNA1-specific primer for a 647 bp amplicon and a TRV-RNA2-specific primer for a 367 bp amplicon were designed. RT-PCR analysis of the vector transcripts were performed in the control and RNAi grape berries. As shown in Supplementary Figure S7B, PCR products were only detected in 2-day-old and 4-day-old TRV-infiltrated berries, but could not be detected 6 days after infiltration. In addition, we first cloned the CDS sequence of PDS (PHYTOENE DESATURASE) in grape and then silenced it. Photo-bleaching was shown on the newly developed leaves 2 weeks after agroinoculation in tobacco, but not in grape leaves, also indicating that TRV vector cannot be used to silence genes in “Kyoho” grapes (Supplementary Figures S7C,D).
Discussion
PYL family genes encode receptors that are involved in ABA signal transduction (Ma et al., 2009; Park et al., 2009). Most studies on ABA receptors have focused on Arabidopsis, whereas only a few of these receptors have been characterized in other plants. Here, we report an in planta functional characterization of a grape PYL which is functional and translates the ABA signaling during osmotic stress or ABA treatment in Arabidopsis. Consistent with findings from Boneh et al. (2012b), phylogenetic and sequence analysis revealed that RCAR7/VlPYL1 amino acid sequence was highly similar to the members of the well-established ABA-related Arabidopsis PYR/PYL/RCAR family, belonging to group III of grape PYL (Supplementary Figures S1B, S3). Two different bases did not lead to the change in amino acid between VlPYL1 from “Kyoho” grape and VvPYL1 from Vitis vinifera cv. Muscat of Hamburg (Supplementary Figure S2). It seemed that the expression level of VvPYL1 of Muscat of Hamburg in leaf and stem was higher than in root (Li et al., 2012). Similarly, “Kyoho” VlPYL1 showed high expression in the leaf and fruit, which seems to support a relevant role for ABA perception. Moreover, subcellular localization suggested that the VlPYL1 protein resided in both the nucleus and the cytosol in agreement with previous result (Li et al., 2012). This outcome is similar to the pattern seen in Arabidopsis (Park et al., 2009) and gives the evidence to support the cellular function of VlPYL1 involves signaling components in both the nucleus and cytosol. Moreover, in silico promoter analysis revealed the presence of two ABA responsive elements (ABRE) that were also found within 1.5 kb upstream sequence of VlPYL1 (Supplementary Table S2). These observations indicated a functional role of this grape PYL1 in the ABA and abiotic stress signaling network. Two independent homozygous Arabidopsis transgenic lines constitutively expressing VlPYL1 were analyzed for ABA sensitivity and stress tolerance on MS-based media. It showed that VlPYL1 overexpression in Arabidopsis results in ABA and osmotic stress hypersensitive seed germination. Moreover, when grown on plates supplemented with 1 and 3 μM ABA, transgenic lines showed statistically significant differences compared with WT, and both transgenic lines showed significantly reduced root length and leaf area (Figure 4B). The genetic evidence suggested that VlPYL1-OE plants were sensitive to ABA similar to the PYLs in Arabidopsis (Ma et al., 2009; Santiago et al., 2009; Lee et al., 2013) and that it also acts as positive regulator of ABA signaling. Moreover, in this study, the stress marker genes such as RD29B and RAB18 were significantly up-regulated in VlPYL1-OE lines in comparison to WT under exogenous ABA treatment. It has been reported that these stress marker genes were regulated through ABA signaling during the abiotic stress response (Lang and Palva, 1992; Jakab et al., 2005; Li F. et al., 2013). And AtRD29B was function in the ABA-dependent pathway, whereas AtRD29A was function in the ABA-independent pathway (Shinozaki and Yamaguchi-Shinozaki, 2000). Also, the RD29B promoter was highly responsive to salt stress, whereas the RD29A promoter was more sensitive to drought and cold stresses (Msanne et al., 2011). The variable expression patterns of stress markers such as RD29A and RD29B (Figure 4C) could be attributed to different ABA responsive element in promoter and multiple layers of gene expression regulation. Key transcription factors such as DREB and AREB, which ultimately control the expression of stress genes such as RD29A and RD29B, are controlled by VlPYL1 or other components such as PP2C (since PP2C is downstream of the PYLs) or SnRK2s, which are known to regulate the stress-responsive transcription factors in dose-dependent way (Gonzalez-Guzman et al., 2012). It may be speculated that VlPYL1 might interact with and regulate some critical components of the ABA signal transduction pathway such as Ser/Thr kinases, especially PP2C (Park et al., 2009; Li W. et al., 2013; Pizzio et al., 2013); these kinases have been implicated in ABA stress and could influence the expression of stress-responsive genes to regulate the physiological process.
The grape berry ripening process is non-climacteric and does not rely on a sole ethylene signal. Numerous studies have highlighted ABA as an important hormone in the inception and color development stages of fruit ripening (Peppi et al., 2006; Cantin et al., 2007; Ferrara et al., 2015). However, the molecular mechanism of ABA in the regulation of grape fruit development, especially fruit maturity, is still under question. The PYR/PYL/RCARs have been suggested to play a role in grape fruit ripening (Jia et al., 2017), but substantial evidence has been lacking. Here, we also provided evidence to determine the role of PYLs on anthocyanin accumulation in grape. A previous study reported that downregulation of FaPYR1 transcripts by RNA interference (RNAi) inhibited fruit abnormal reddening. This loss of red coloring in FaPYR1 RNAi fruits could not be rescued by exogenously applied ABA (Chai et al., 2011), indicating that PYL was a necessary regulator of fruit ripening. Similar result with the previous reports of FaPYR1 (Chai et al., 2011), CsPYL1 and CsPYL2 (Wang Y. et al., 2012) and all of the MaPYLs (except for MaPYL1) (Zhu et al., 2017), the expression of VlPYL1 kept higher level during the early stages of fruit development suggested that it might participate in the regulation of fruit development. However, the expression changes of PYR1 in grape cv. Fujiminori were consistent with ABA content which increased rapidly in the former phase and stay high level at 14 weeks post flowering (Jia et al., 2017). Genetic characteristics of the plants may lead to the different expression patterns of PYL1 between “Fujiminori” and “Kyoho” grapes. Other PYR/PYL/RCAR family members in Kyoho may contribute to the ABA signal transduction after veraison stage. Obviously, VlPYL1 transcripts decreased rapidly just before the veraison (Figure 1B), whereas anthocyanin content began rapidly increasing after the veraison stage (Supplementary Figure S6D). It suggested that the VlPYL1 might be mainly involved in “Kyoho” early fruit development and stimulation of ripening initiation.
Stable transformation is a method of assessing the functional significance of transgenes, but the stable transformation system is time-consuming and requires great care in handling of the experiment. On the other hand, the transient gene expression system allows for rapid in vivo analysis of genes in plants such as strawberry (Guidarelli and Baraldi, 2015). Many studies have successfully applied transient gene expression system using agroinfiltration to grape leaves (Jelly et al., 2014), but no similar technique has been reported in grape berries. In this study, we applied a transient gene expression assay to analyze the effects of gene expression due to overexpression of ABA receptor gene in grape fruits. Over-expression of the VlPYL1 gene in grapes led to more PYL1 transcript (Figure 6A), and the VlPYL1-OE fruit exhibited an increase of anthocyanin accumulation compared to the control fruit. Figure 3C also shows that over-expression of the VlPYL1 gene improved color of the grape skin due to higher values of a∗. Furthermore, the transcripts of the up-regulated ABA-dependent genes, including NCED, BG3 and ABF2, and pigment-related genes, including PAL, UFGT and MYBA1, were up-regulated in the VlPYL1-OE berries, in which the mRNA expression level of VlPYL1 was up-regulated threefold (Figure 6A). A previous study demonstrated that VvABF2 was involved in the regulation of grape fruit ripening (Nicolas et al., 2014), and high expression of VvBG1 suggested a regulatory role in grape berry ripening (Sun et al., 2015). It is also interesting to note that FaBG3 was a positive regulator of strawberry fruit ripening (Li Q. et al., 2013). These results were consistent with a positive role of VlPYL1 in ABA signaling during fruit ripening.
The transcript levels of VlPAL, VlF3’H, VlF3’5’H, VlFLS1, VlANR, VlLAR1, VlLDOX, VlUFGT and VlMYBA1 were significantly up-regulated in the VlPYL1-OE berries, suggesting that VlPYL1 may affect anthocyanin biosynthesis through regulating the expression of these flavonoid biosynthetic pathway genes. In the phenylpropanoid biosynthesis pathway, PAL is the first and rate-limited enzyme that contributes to the biosynthesis of anthocyanins, stilbenes and tannins. In addition, similar to other plants (such as strawberry), in both the field grown grapes and the in vitro experiments, VvPAL could be induced by exogenous ABA and directly influenced the accumulation of flavonols and anthocyanins (Hiratsuka et al., 2001a,b; Gagne et al., 2006). It is likely that the synthesis of more flavonols was also beneficial to VlPYL1-OE berries resulting from ∼3-fold higher expression levels of the FLS1 gene (Downey et al., 2003). Higher expression levels of LAR1 and ANR also indicated that these genes were favored in the enhanced synthesis of proanthocyanidins in VlPYL1-OE berries (Xie and Dixon, 2005). UFGT encodes a crucial enzyme that mediates carbon flow to anthocyanins in grapes (Boss et al., 1996). UFGT has been implicated in grape berry coloration (Ageorges et al., 2006; Castellarin et al., 2007), and overexpression of the VVMYBA1-2 gene could increase the transcript level of UFGT (Cutanda-Perez et al., 2009). In grapes, MYB1 was reported to regulate the anthocyanin synthesis pathway (Kobayashi S. et al., 2005; Walker et al., 2007; Azuma et al., 2008), and ABA has been reported to induce VVMYBA1 expression (Jeong et al., 2004; Azuma et al., 2015). Since F3′H regulates the synthesis of cyanidin-type anthocyanins and F3′5′H, diverting the flux to synthesis of delphinidin derivatives, expression profiles of these two genes in concert with increased expression of LDOX and UFGT and MybA1 would ensure the flux of flavonoid intermediates toward the synthesis of five main anthocyanidin aglycones including delphinidin, cyanidin, peonidin, petunidin and malvidin in VlPYL1-OE berries (Wang B. et al., 2012). These results suggested that the ABA signaling pathway initiated and controlled by PYL1 could activate the expression of the anthocyanin biosynthetic genes and accelerate the process of grape skin coloring.
Conclusion
In this study, the VlPYL1 gene was isolated from “Kyoho” grape fruits, and its biological role was assessed in fruits attached to the plant. Overexpression of VlPYL1 significantly promoted anthocyanin accumulation in grape berry skins. Transient overexpression affected expression of the ABA pathway genes, increasing the transcript level of genes closely related to anthocyanin biosynthesis. The results showed that VlPYL1 exerts a regulatory effect at the transcriptional level in a regulatory effect of ABA pathway aimed at promoting anthocyanin production in “Kyoho” grape berry skins. These data provide experimental information that supports the biological role of VlPYL1 on the regulation of anthocyanin accumulation in grape berry skin as previously reported for FaPYR1 in strawberry. In addition, we have demonstrated that TRV VIGS vectors could not be used to suppress functional genes in “Kyoho” grapes.
Author Contributions
SS, WX, CZ, SW, and CM contributed to the project design. ZG performed the experiments and conducted statistical analysis. QL, YC, ML, HL, YW, JW, SD, and LW took part in the experimental work. ZG and JL wrote the manuscript. SW and CM revised the manuscript. All the authors read and approved the final manuscript.
Funding
This research was supported by the Shanghai Pujiang Program (Grant No. 16PJ1404900), National Natural Science Foundation of China (Grant No. 31471836), and the Special Funds of Modern Industrial Technology System for Agriculture (Grant No. CARS-29-zp-7).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00592/full#supplementary-material
FIGURE S1 | Sequence analysis of VlPYL1. (A) Finding conserved domains in the VlPYL1 protein. Conserved domains were determined for the VlPYL1 protein via a 215-amino acid polypeptide BLAST in the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The PYR/PYL/RCAR-like family belongs to the SRPBCC (START/RHO-alpha-C/PITP/Bet-v1/CoxG/CalC) domain superfamily of proteins that binds hydrophobic ligands. SRPBCC domains have a deep hydrophobic ligand-binding pocket. (B) Phylogenetic tree showing the relationship between VlPYL1 and other PYR/PYL/RCAR proteins. The tree presented here is a Neighbor–Joining tree based on an amino acid sequence alignment. The numbers next to each node give bootstrap values for 1000 replicates. Genes and accession numbers are as follows: AtPYR1, At4g17870; At PYL1, At5g46790; At PYL2, At2g26040; At PYL3, At1g73000; At PYL4, At2g38310; At PYL5, At5g05440; At PYL6, At2g40330; At PYL7, At4g01026; At PYL8, At5g53160; At PYL9, At1g01360; At PYL10, At4g27920; At PYL11, At5g45860; At PYL12, At5g45870; At PYL13, At4g18620; FaPYR1, JF268669; RCAR1, VIT 215s0046g01050; RCAR2, VIT 216s0050g02620; RCAR3, VIT 202s0025g01340; RCAR4, VIT 210s0003g01335; RCAR5, VIT 208s0058g00470; RCAR6, VIT 213s0067g01940; VlPYL1/RCAR7, VIT 202s0012g01270 and RCAR8, VIT 204s0008g00890.
FIGURE S2 | Coding-sequence analysis of VlPYL1 from “Kyoho” grape and VvPYL1 from Vitis vinifera cv. Muscat of Hamburg reported by Li et al. (2012).
FIGURE S3 | Amino acid sequence analysis of VlPYL1. Sequence alignment of the VlPYL1 protein with PYR1 and PYL1 in Arabidopsis and PYR1 in strawberries. Positions with identical amino acid residues are highlighted in blue, while similar amino acid residues are colored in pink and cyan.
FIGURE S4 | Subcelluar localization of VlPYL1 in tobacco leaves. Nicotiana benthamiana leaves were transiently infiltrated with A. tumefaciens GV3101 containing vectors expressing 2 × 35S::eGFP and 2 × 35S:: VlPYL1-eGFP. Confocal images were captured 72 h after inoculation. GFP image fluorescences are shown in the left panels, bright field images are shown in the middle panels, and overlap images are shown in the right panels.
FIGURE S5 | Expression analysis of the VlPYL1 gene in transgenic A. thaliana plants and WT. The A. thaliana actin gene was used as an internal control. ∗∗p-value < 0.01 when comparing data for each genotype versus the wild-type.
FIGURE S6 | Morphological and physiological changes in “Kyoho” fruits during development. (A) Changes in fruit size and color, (B) berry weight (means ± SD, n = 30), (C) total soluble solids (means ± SD, n = 30), (D) anthocyanin content. Vertical bars represent SD of the values. Different letters indicated a statistical difference at P < 0.05 as determined by Duncan’s multiple range test.
FIGURE S7 |(A) Effect of TRV-VlPYL1 on grape fruit ripening. Note that both the control fruit and the RNAi fruit became red. The total anthocyanin content was measured in the control and TRV-RNAi fruits. (B) RT-PCR analysis of TRV expression in fruits. After infiltration, virus vector gene expression of both 647 bp pTRV1 and 367 bp pTRV2 was detected in fruits infiltrated with Agrobacterium containing TRV (lane 2 and lane 3: TRV-VlPYL1-treated grape fruits 2 and 4 days after infiltration) but were not detected in fruits infiltrated with Agrobacterium alone (lane 1) nor in TRV-VlPYL1-treated grape fruits 6 days (lane 4) and 8 days (lane 5) after infiltration. (C) TRV-VvPDS-treated grape plants 30 days after infiltration. (D) Mock-treated and TRV-VvPDS-treated tobacco plants 16 days after infiltration. The photobleached phenotype was observed in the plant (left) infiltrated with TRV-VvPDS, while the plant treated with TRV alone (right) remained green.
FIGURE S8 |(A) Strawberry fruits agroinfiltrated with empty vector (EV) and agroinfiltrated fruit with VlPYL1-RNAi construct (RNAi) at 7 days after injection. (B) FaPYR1 expression by RT-qPCR of strawberry receptacle infiltrated with empty vector (Control) and VlPYL1-RNAi vector, 7 days after injection. Error bars indicate + SD of three biological replicates. The asterisk indicates a significant difference by t-test analysis: ∗∗P < 0.01.
TABLE S1 | Primers for quantitative real-time PCR (qPCR) and constructs.
TABLE S2 | Important cis-elements found in the VlPYL1 promoter using PLANT-CARE database analysis.
TABLE S3 | The number of red spots 8 days after injection.
TABLE S4 | The number of coloring fruits 16 days after injection.
Footnotes
- ^http://genomes.cribi.unipd.it/grape/
- ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^http://genomes.cribi.unipd.it/grape/
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Adams-Phillips, L., Barry, C., and Giovannoni, J. (2004). Signal transduction systems regulating fruit ripening. Trends Plant Sci. 9, 331–338. doi: 10.1016/j.tplants.2004.05.004
Ageorges, A., Fernandez, L., Vialet, S., Merdinoglu, D., Terrier, N., and Romieu, C. (2006). Four specific isogenes of the anthocyanin metabolic pathway are systematically co-expressed with the red colour of grape berries. Plant Sci. 170, 372–383. doi: 10.1016/j.plantsci.2005.09.007
Alexander, L., and Grierson, D. (2002). Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J. Exp. Bot. 53, 2039–2055. doi: 10.1093/jxb/erf072
Azuma, A., Fujii, H., Shimada, T., and Yakushiji, H. (2015). Microarray analysis for the screening of genes inducible by light or low temperature in post-veraison grape berries. Hort. J. 84, 214–226. doi: 10.2503/hortj.MI-041
Azuma, A., Kobayashi, S., Mitani, N., Shiraishi, M., Yamada, M., Ueno, T., et al. (2008). Genomic and genetic analysis of Myb-related genes that regulate anthocyanin biosynthesis in grape berry skin. Theor. Appl. Genet. 117, 1009–1019. doi: 10.1007/s00122-008-0840-1
Bakr, E. M. (2005). A new software for measuring leaf area, and area damaged by Tetranychus urticae Koch. J. Appl. Entomol. 129, 173-175. doi: 10.1111/j.1439-0418.2005.00948.x
Ban, T., Ishimaru, M., Kobayashi, S., Shiozaki, S., Goto-Yamamoto, N., and Horiuchi, S. (2003). Abscisic acid and 2,4-dichlorophenoxyacetic acid affect the expression of anthocyanin biosynthetic pathway genes in ‘Kyoho’ grape berries. J. Hort. Sci. Biotechnol. 78, 586–589. doi: 10.1080/14620316.2003.11511668
Boneh, U., Biton, I., Schwartz, A., and Ben-Ari, G. (2012a). Characterization of the ABA signal transduction pathway in Vitis vinifera. Plant Sci. 187, 89–96. doi: 10.1016/j.plantsci.2012.01.015
Boneh, U., Biton, I., Zheng, C. L., Schwartz, A., and Ben-Ari, G. (2012b). Characterization of potential ABA receptors in Vitis vinifera. Plant Cell Rep. 31, 311–321. doi: 10.1007/s00299-011-1166-z
Boss, P. K., Davies, C., and Robinson, S. P. (1996). Expression of anthocyanin biosynthesis pathway genes in red and white grapes. Plant Mol. Biol. 32, 565–569. doi: 10.1007/BF00019111
Cantin, C. M., Fidelibus, M. W., and Crisostoc, C. H. (2007). Application of abscisic acid (ABA) at veraison advanced red color development and maintained postharvest quality of ‘Crimson Seedless’ grapes. Postharvest Biol. Technol. 46, 237–241. doi: 10.1016/j.postharvbio.2007.05.017
Castellarin, S. D., Gambetta, G. A., Wada, H., Shackel, K. A., and Matthews, M. A. (2011). Fruit ripening in Vitis vinifera: spatiotemporal relationships among turgor, sugar accumulation, and anthocyanin biosynthesis. J. Exp. Bot. 62, 4345–4354. doi: 10.1093/jxb/err150
Castellarin, S. D., Matthews, M. A., Di Gaspero, G., and Gambetta, G. A. (2007). Water deficits accelerate ripening and induce changes in gene expression regulating flavonoid biosynthesis in grape berries. Planta 227, 101–112. doi: 10.1007/s00425-007-0598-8
Chai, Y.-M., Jia, H.-F., Li, C.-L., Dong, Q.-H., and Shen, Y.-Y. (2011). FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 62, 5079–5089. doi: 10.1093/jxb/err207
Cherian, S., Figueroa, C. R., and Nair, H. (2014). ‘Movers and shakers’ in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 65, 4705–4722. doi: 10.1093/jxb/eru280
Coombe, B. G., and McCarthy, M. G. (2000). Dynamics of grape berry growth and physiology of ripening. Aust. J. Grape Wine Res. 6, 131–135. doi: 10.1111/j.1755-0238.2000.tb00171.x
Cutanda-Perez, M. C., Ageorges, A., Gomez, C., Vialet, S., Terrier, N., Romieu, C., et al. (2009). Ectopic expression of VlmybA1 in grapevine activates a narrow set of genes involved in anthocyanin synthesis and transport. Plant Mol. Biol. 69, 633–648. doi: 10.1007/s11103-008-9446-x
Dong, Y., Burch-Smith, T. M., Liu, Y., Mamillapalli, P., and Dinesh-Kumar, S. P. (2007). A ligation-independent cloning tobacco rattle virus vector for high-throughput virus-induced gene silencing identifies roles for NbMADS4-1 and -2 in floral development. Plant Physiol. 145, 1161–1170. doi: 10.1104/pp.107.107391
Downey, M. O., Harvey, J. S., and Robinson, S. P. (2003). Analysis of tannins in seeds and skins of Shiraz grapes throughout berry development. Aust. J. Grape Wine Res. 9, 15–27. doi: 10.1111/j.1755-0238.2003.tb00228.x
Ferrara, G., Mazzeo, A., Matarrese, A. M. S., Pacucci, C., Pacifico, A., Gambacorta, G., et al. (2013). Application of abscisic acid (S-ABA) to ‘Crimson Seedless’ grape berries in a mediterranean climate: effects on color, chemical characteristics, metabolic profile, and S-ABA concentration. J. Plant Growth Regul. 32, 491–505. doi: 10.1007/s00344-012-9316-2
Ferrara, G., Mazzeo, A., Matarrese, A. M. S., Pacucci, C., Punzi, R., Faccia, M., et al. (2015). Application of abscisic acid (S-ABA) and sucrose to improve colour, anthocyanin content and antioxidant activity of cv. Crimson Seedless grape berries. Aust. J. Grape Wine Res. 21, 18–29. doi: 10.1111/ajgw.12112
Fortes, A. M., Teixeira, R. T., and Agudelo-Romero, P. (2015). Complex interplay of hormonal signals during grape berry ripening. Molecules 20, 9326–9343. doi: 10.3390/molecules20059326
Fuchs, S., Tischer, S. V., Wunschel, C., Christmann, A., and Grill, E. (2014). Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. U.S.A. 111, 5741–5746. doi: 10.1073/pnas.1322085111
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Gagne, S., Esteve, K., Deytieux, C., Saucier, C., and Geny, L. (2006). Influence of abscisic acid in triggering “veraison” in grape berry skins of Vitis vinifera L. cv. Cabernet-Sauvignon. J. Int. Sci. Vigne Vin 40, 7–14. doi: 10.20870/oeno-one.2006.40.1.882
Giovannoni, J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 725–749. doi: 10.1146/annurev.arplant.52.1.725
Gonzalez-Guzman, M., Pizzio, G. A., Antoni, R., Vera-Sirera, F., Merilo, E., Bassel, G. W., et al. (2012). Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 24, 2483–2496. doi: 10.1105/tpc.112.098574
Gonzalez-Guzman, M., Rodriguez, L., Lorenzo-Orts, L., Pons, C., Sarrion-Perdigones, A., Fernandez, M. A., et al. (2014). Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 65, 4451–4464. doi: 10.1093/jxb/eru219
Guidarelli, M., and Baraldi, E. (2015). Transient transformation meets gene function discovery: the strawberry fruit case. Front. Plant Sci. 6:444. doi: 10.3389/fpls.2015.00444
Gupta, S. M., Srivastava, S., Sane, A. P., and Nath, P. (2006). Differential expression of genes during banana fruit development, ripening and 1-MCP treatment: presence of distinct fruit specific, ethylene induced and ethylene repressed expression. Postharvest Biol. Technol. 42, 16–22. doi: 10.1016/j.postharvbio.2006.05.002
He, Y., Hao, Q., Li, W. Q., Yan, C. Y., Yan, N., and Yin, P. (2014). Identification and characterization of ABA receptors in Oryza sativa. PLoS One 9:e95246. doi: 10.1371/journal.pone.0095246
Hiratsuka, S., Onodera, H., Kawai, Y., Kubo, T., Itoh, H., and Wada, R. (2001a). ABA and sugar effects on anthocyanin formation in grape berry cultured in vitro. Sci. Hort. 90, 121–130. doi: 10.1016/S0304-4238(00)00264-8
Hiratsuka, S., Onodera, H., Kawai, Y., Kubo, T., Itoh, H., and Wada, R. (2001b). Enzyme activity changes during anthocyanin synthesis in ‘Olympia’ grape berries. Sci. Hort. 90, 255–264. doi: 10.1016/S0304-4238(00)00266-1
Jakab, G., Ton, J., Flors, V., Zimmerli, L., Metraux, J. P., and Mauch-Mani, B. (2005). Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 139, 267–274. doi: 10.1104/pp.105.065698
Jelly, N. S., Valat, L., Walter, B., and Maillot, P. (2014). Transient expression assays in grapevine: a step towards genetic improvement. Plant Biotechnol. J. 12, 1231–1245. doi: 10.1111/pbi.12294
Jeong, S. T., Goto-Yamamoto, N., Kobayashi, S., and Esaka, A. (2004). Effects of plant hormones and shading on the accumulation of anthocyanins and the expression of anthocyanin biosynthetic genes in grape berry skins. Plant Sci. 167, 247–252. doi: 10.1016/j.plantsci.2004.03.021
Ji, K., Chen, P., Sun, L., Wang, Y. P., Dai, S. J., Li, Q., et al. (2012). Non-climacteric ripening in strawberry fruit is linked to ABA, FaNCED2 and FaCYP707A1. Funct. Plant Biol. 39, 351–357. doi: 10.1071/FP11293
Jia, H., Xie, Z., Wang, C., Shangguan, L., Qian, N., Cui, M., et al. (2017). Abscisic acid, sucrose, and auxin coordinately regulate berry ripening process of the Fujiminori grape. Funct. Integr. Genomics 17, 441–457. doi: 10.1007/s10142-017-0546-z
Jia, H. F., Jiu, S. T., Zhang, C., Wang, C., Tariq, P., Liu, Z. J., et al. (2016). Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol. J. 14, 2045–2065. doi: 10.1111/pbi.12563
Karppinen, K., Hirvela, E., Nevala, T., Sipari, N., Suokas, M., and Jaakola, L. (2013). Changes in the abscisic acid levels and related gene. expression during fruit development and ripening in bilberry (Vaccinium myrtillus L.). Phytochemistry 95, 127–134. doi: 10.1016/j.phytochem.2013.06.023
Kobayashi, S., Yamamoto, N. G., and Hirochika, H. (2005). Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin - color mutants. J. Jpn. Soc. Hort. Sci. 74, 196–203. doi: 10.2503/jjshs.74.196
Kobayashi, Y., Murata, M., Minami, H., Yamamoto, S., Kagaya, Y., Hobo, T., et al. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949. doi: 10.1111/j.1365-313X.2005.02583.x
Kuhn, N., Guan, L., Dai, Z. W., Wu, B. H., Lauvergeat, V., Gomes, E., et al. (2014). Berry ripening: recently heard through the grapevine. J. Exp. Bot. 65, 4543–4559. doi: 10.1093/jxb/ert395
Lacampagne, S., Gagne, S., and Geny, L. (2010). Involvement of abscisic acid in controlling the proanthocyanidin biosynthesis pathway in grape skin: new elements regarding the regulation of tannin composition and leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) activities and expression. J. Plant Growth Regul. 29, 81–90. doi: 10.1007/s00344-009-9115-6
Lang, V., and Palva, E. T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. doi: 10.1007/bf00027165
Lee, S. C., Lim, C. W., Lan, W., He, K., and Luan, S. (2013). ABA signaling in guard cells entails a dynamic protein-protein interaction relay from the PYL-RCAR family receptors to ion channels. Mol. Plant 6, 528–538. doi: 10.1093/mp/sss078
Li, F., Han, Y., Feng, Y., Xing, S., Zhao, M., Chen, Y., et al. (2013). Expression of wheat expansin driven by the RD29 promoter in tobacco confers water-stress tolerance without impacting growth and development. J. Biotechnol. 163, 281–291. doi: 10.1016/j.jbiotec.2012.11.008
Li, Q., Ji, K., Sun, Y., Luo, H., Wang, H., and Leng, P. (2013). The role of FaBG3 in fruit ripening and B. cinerea fungal infection of strawberry. Plant J. 76, 24–35. doi: 10.1111/tpj.12272
Li, W., Wang, L., Sheng, X., Yan, C., Zhou, R., Hang, J., et al. (2013). Molecular basis for the selective and ABA-independent inhibition of PP2CA by PYL13. Cell Res. 23, 1369–1379. doi: 10.1038/cr.2013.143
Li, G., Xin, H., Zheng, X. F., Li, S., and Hu, Z. (2012). Identification of the abscisic acid receptor VvPYL1 in Vitis vinifera. Plant Biol. 14, 244–248. doi: 10.1111/j.1438-8677.2011.00504.x
Li, J., Jiang, M. M., Ren, L., Liu, Y., and Chen, H. Y. (2016). Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 291(4), 1769-1781. doi: 10.1007/s00438-016-1218-8
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lurie, S., Ovadia, R., Nissim-Levi, A., Oren-Shamir, M., Kaplunov, T., Zutahy, Y., et al. (2009). Abscisic acid improves colour development in ‘Crimson Seedless’ grapes in the vineyard and on detached berries. J. Hort. Sci. Biotechnol. 84, 639–644. doi: 10.1080/14620316.2009.11512579
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Msanne, J., Lin, J., Stone, J. M., and Awada, T. (2011). Characterization of abiotic stress-responsive Arabidopsis thaliana RD29A and RD29B genes and evaluation of transgenes. Planta 234, 97–107. doi: 10.1007/s00425-011-1387-y
Nakashima, K., and Yamaguchi-Shinozaki, K. (2013). ABA signaling in stress-response and seed development. Plant Cell Rep. 32, 959–970. doi: 10.1007/s00299-013-1418-1
Nicolas, P., Lecourieux, D., Kappel, C., Cluzet, S., Cramer, G., Delrot, S., et al. (2014). The basic leucine zipper transcription factor Abscisic Acid Response Element-Binding Factor2 is an important transcriptional regulator of abscisic acid-dependent grape berry ripening processes. Plant Physiol. 164, 365–383. doi: 10.1104/pp.113.231977
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Pech, J. C., Bouzayen, M., and Latche, A. (2008). Climacteric fruit ripening: ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Sci. 175(1-2), 114–120. doi: 10.1016/j.plantsci.2008.01.003
Peppi, M. C., Fidelibus, M. W., and Dokoozlian, N. (2006). Abscisic acid application timing and concentration affect firmness, pigmentation, and color of ‘flame seedless’ grapes. HortScience 41, 1440–1445.
Pilati, S., Bagagli, G., Sonego, P., Moretto, M., Brazzale, D., Castorina, G., et al. (2017). Abscisic acid is a major regulator of grape berry ripening onset: new insights into ABA signaling network. Front. Plant Sci. 8:1093. doi: 10.3389/fpls.2017.01093
Pirie, A., and Mullins, M. G. (1976). Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic Acid. Plant Physiol. 58, 468–472. doi: 10.1104/pp.58.4.468
Pizzio, G. A., Rodriguez, L., Antoni, R., Gonzalez-Guzman, M., Yunta, C., Merilo, E., et al. (2013). The PYL4 A194T mutant uncovers a key role of PYR1-LIKE4/PROTEIN PHOSPHATASE 2CA interaction for abscisic acid signaling and plant drought resistance. Plant Physiol. 163, 441–455. doi: 10.1104/pp.113.224162
Santiago, J., Dupeux, F., Round, A., Antoni, R., Park, S. Y., Jamin, M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. doi: 10.1038/nature08591
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)00067-4
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Sun, J. J., Dong, Y. H., Li, C. L., and Shen, Y. Y. (2015). Transcription and enzymatic analysis of beta-glucosidase VvBG1 in grape berry ripening. Plant Growth Regul. 75, 67–73. doi: 10.1007/s10725-014-9932-x
Sun, L. A., Zhang, M., Ren, J., Qi, J. X., Zhang, G. J., and Leng, P. (2010). Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biol. 10:257. doi: 10.1186/1471-2229-10-257
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Vitulo, N., Forcato, C., Carpinelli, E. C., Telatin, A., Campagna, D., D’Angelo, M., et al. (2014). A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 14:99. doi: 10.1186/1471-2229-14-99
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A. J., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wang, B., He, J. J., Duan, C. Q., Yu, X. M., Zhu, L. N., Xie, Z. S., et al. (2012). Root restriction affects anthocyanin accumulation and composition in berry skin of ‘Kyoho’ grape (Vitis vinifera L. x Vitis labrusca L.) during ripening. Sci. Hort. 137, 20–28. doi: 10.1016/j.scienta.2011.10.006
Wang, J. F., Wang, S. Q., Liu, G. T., Edwards, E. J., Duan, W., Li, S. H., et al. (2016). The synthesis and accumulation of resveratrol are associated with veraison and abscisic acid concentration in beihong (Vitis vinifera x Vitis amurensis) berry skin. Front. Plant Sci. 7:1605. doi: 10.3389/fpls.2016.01605
Wang, Y., Wang, Y., Ji, K., Dai, S., Hu, Y., Sun, L., et al. (2013). The role of abscisic acid in regulating cucumber fruit development and ripening and its transcriptional regulation. Plant Physiol. Biochem. 64, 70–79. doi: 10.1016/j.plaphy.2012.12.015
Wang, Y., Wu, Y., Duan, C., Chen, P., Li, Q., Dai, S., et al. (2012). The expression profiling of the CsPYL, CsPP2C and CsSnRK2 gene families during fruit development and drought stress in cucumber. J. Plant Physiol. 169, 1874–1882. doi: 10.1016/j.jplph.2012.07.017
Wheeler, S., Loveys, B., Ford, C., and Davies, C. (2009). The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Aust. J. Grape Wine Res. 15, 195–204. doi: 10.1111/j.1755-0238.2008.00045.x
Xie, D. Y., and Dixon, R. A. (2005). Proanthocyanidin biosynthesis - still more questions than answers? Phytochemistry 66, 2127–2144. doi: 10.1016/j.phytochem.2005.01.008
Xie, Z. S., Li, B., Forney, C. F., Xu, W. P., and Wang, S. P. (2009). Changes in sugar content and relative enzyme activity in grape berry in response to root restriction. Sci. Hort. 123, 39–45. doi: 10.1016/j.scienta.2009.07.017
Zhang, F., Lu, X., Lv, Z., Zhang, L., Zhu, M., Jiang, W., et al. (2013a). Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin content in Artemisia annua L. PLoS One 8:e56697. doi: 10.1371/journal.pone.0056697
Zhang, F., Lu, X., Lv, Z. Y., Zhang, L., Zhu, M. M., Jiang, W. M., et al. (2013b). Overexpression of the Artemisia orthologue of ABA receptor, AaPYL9, enhances ABA sensitivity and improves artemisinin content in Artemisia annua L. PLoS One 8:e56697. doi: 10.1371/journal.pone.0056697
Zhang, X. R., Henriques, R., Lin, S. S., Niu, Q. W., and Chua, N. H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1, 641–646. doi: 10.1038/nprot.2006.97
Zhu, P., Cai, Y., Yu, J., Zhao, A., Liang, Y., Liu, C., et al. (2017). Characterization and expression of abscisic acid signal transduction genes during mulberry fruit ripening. Acta Physiol. Plant. 39:149. doi: 10.1007/s11738-017-2442-5
Zifkin, M., Jin, A., Ozga, J. A., Zaharia, L. I., Schernthaner, J. P., Gesell, A., et al. (2012). Gene expression and metabolite profiling of developing highbush blueberry fruit indicates transcriptional regulation of flavonoid metabolism and activation of abscisic acid metabolism. Plant Physiol. 158, 200–224. doi: 10.1104/pp.111.180950
Keywords: grape, anthocyanin, PYL1, ABA sensitivity, gene over-expression
Citation: Gao Z, Li Q, Li J, Chen Y, Luo M, Li H, Wang J, Wu Y, Duan S, Wang L, Song S, Xu W, Zhang C, Wang S and Ma C (2018) Characterization of the ABA Receptor VlPYL1 That Regulates Anthocyanin Accumulation in Grape Berry Skin. Front. Plant Sci. 9:592. doi: 10.3389/fpls.2018.00592
Received: 01 September 2017; Accepted: 16 April 2018;
Published: 18 May 2018.
Edited by:
Vicent Arbona, Jaume I University, SpainReviewed by:
Miguel Gonzalez-Guzman, Consejo Superior de Investigaciones Científicas (CSIC), SpainCarlos R. Figueroa, University of Talca, Chile
Copyright © 2018 Gao, Li, Li, Chen, Luo, Li, Wang, Wu, Duan, Wang, Song, Xu, Zhang, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiping Wang, ZnJ1aXRAc2p0dS5lZHUuY24= Chao Ma, Y2hhb21hMjAxNUBzanR1LmVkdS5jbg==
 Zhen Gao
Zhen Gao Qin Li1
Qin Li1 Chao Ma
Chao Ma