
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 April 2018
Sec. Plant Cell Biology
Volume 9 - 2018 | https://doi.org/10.3389/fpls.2018.00475
This article is part of the Research Topic Plant RNA Biology View all 15 articles
A correction has been applied to this article in:
Corrigendum: A Role of U12 Intron in Proper Pre-mRNA Splicing of Plant Cap Binding Protein 20 Genes
 Marcin Pieczynski1†
Marcin Pieczynski1† Katarzyna Kruszka1†
Katarzyna Kruszka1† Dawid Bielewicz1
Dawid Bielewicz1 Jakub Dolata1
Jakub Dolata1 Michal Szczesniak2
Michal Szczesniak2 Wojciech Karlowski3
Wojciech Karlowski3 Artur Jarmolowski1*
Artur Jarmolowski1* Zofia Szweykowska-Kulinska1*
Zofia Szweykowska-Kulinska1*The nuclear cap-binding complex (CBC) is composed of two cap-binding proteins: CBP20 and CBP80. The CBP20 gene structure is highly conserved across land plant species. All studied CBP20 genes contain eight exons and seven introns, with the fourth intron belonging to the U12 class. This highly conserved U12 intron always divides the plant CBP20 gene into two parts: one part encodes the core domain containing the RNA binding domain (RBD), and the second part encodes the tail domain with a nuclear localization signal (NLS). In this study, we investigate the importance of the U12 intron in the Arabidopsis thaliana CBP20 gene by moving it to different intron locations of the gene. Relocation of the U12 intron resulted in a significant decrease in the U12 intron splicing efficiency and the accumulation of wrongly processed transcripts. These results suggest that moving the U12 intron to any other position of the A. thaliana CBP20 gene disturbs splicing, leading to substantial downregulation of the level of properly spliced mRNA and CBP20 protein. Moreover, the replacement of the U12 intron with a U2 intron leads to undesired alternative splicing events, indicating that the proper localization of the U12 intron in the CBP20 gene secures correct CBP20 pre-mRNA maturation and CBP20 protein levels in a plant. Surprisingly, our results also show that the efficiency of U12 splicing depends on intron length. In conclusion, our study emphasizes the importance of proper U12 intron localization in plant CBP20 genes for correct pre-mRNA processing.
The cap-binding complex (CBC) is a nuclear heterodimer composed of two cap-binding proteins: cap-binding protein 20 (CBP20) and cap-binding protein 80 (CBP80, also known as ABA Hypersensitive 1, ABH1), which binds to the 5′ cap of all RNA polymerase II transcripts. In plants, CBC is important for pre-mRNA and pri-miRNA first intron splicing and regulation of pre-mRNA alternative splicing (Szarzynska et al., 2009; Raczynska et al., 2010). The CBC is also involved in miRNA biogenesis (Kim et al., 2008; Laubinger et al., 2008; Ren and Yu, 2012). Amino acid sequence comparison of Arabidopsis thaliana and Oryza sativa CBP20 and CBP80 proteins revealed high sequence conservation of the small subunit of CBC (CBP20) and a considerably lower level of conservation of its large subunit (CBP80) (Kmieciak et al., 2002). Interestingly, in comparison to animal CBP20 proteins, plant CBP20s contain an additional carboxy-terminal fragment. The structure of the CBP20 protein in higher plants can be divided into two parts. The core part, built mostly of 138 amino acids, contains the conservative RNA binding domain (RBD), which plays a crucial role in cap structure recognition and binding (Izaurralde et al., 1994), and the tail part, built of approximately 120 amino acids, contains a nuclear localization signal (NLS). In animals, this is the CBP80 protein that carries a functional NLS and is responsible for the import of CBC into the nucleus (Dias et al., 2009). In A. thaliana, two functionally independent NLSs are located in the tail part of the CBP20 protein (Kmieciak et al., 2002). In contrast, plants CBP80s do not contain any NLS; thus, the CBP20 protein targets the whole plant CBC to the nucleus. Moreover, in A. thaliana, the CBP20 protein is stabilized by CBP80 (Kierzkowski et al., 2009). The down-regulation of nuclear cap-binding proteins in Arabidopsis leads to mild developmental abnormalities, such as serrated rosette leaves and delayed development. Interestingly, loss of CBC functions in Arabidopsis plants confers hypersensitivity to abscisic acid (ABA) during seeds germination, significant reduction of stomatal conductance, and in consequence greatly enhances tolerance of the cbp20 and cbp80 mutants to drought (Hugouvieux et al., 2001; Papp et al., 2004; Jäger et al., 2011; Pieczynski et al., 2013) and salinity (Kong et al., 2014).
Arabidopsis and rice CBP20 gene structures are conserved and contain eight exons. The core part of the CBP20 protein is encoded by the first four exons of the gene, while the tail part is encoded by the last four exons. Six out of seven introns of the CBP20 gene belong to the classical and abundant U2 introns, while intron no. 4 represents the much rarer U12 introns. Computational analyses allowed the identification in the A. thaliana genome 246 U12 introns constituting 0.17% of all predicted introns in this species (Alioto, 2007). Transcriptome analyses, however, identified about eight times more U12 introns (2069 vs. 246) than previously found using the computational approach (Marquez et al., 2012). The majority of U12 introns (89.4%) contain GT-AG terminal dinucleotides; however, a small portion (10.6%) is characterized by the presence of other terminal dinucleotides, of which AT-AC comprises almost half of such non-GT-AG introns (4.8%) (Marquez et al., 2012). U12 introns contain a very characteristic and highly conserved branch point region (TTCCTTRAY), and unlike U2 introns, they do not have any polypyrimidine tract (Lewandowska et al., 2004; Simpson and Brown, 2008; Marquez et al., 2012; Turunen et al., 2013). It has been shown that several proteins of U11/U12 di-snRNP are indispensable for the correct splicing of the U12 intron-containing genes, which is crucial for the normal development of A. thaliana (Kim et al., 2010; Kwak et al., 2012; Jung and Kang, 2014; Xu et al., 2016). Moreover, the Arabidopsis quatre-quart1 (QQT1) gene is an indispensable U12 intron-containing gene whose correct splicing is necessary for the wild type phenotype and development of Arabidopsis plants (Kwak et al., 2017).
The phylogenetic distribution of U12 introns shows that the minor (U12) splicing pathway appeared very early in eukaryotic evolution, but during the course of evolution, most U12 introns systematically changed to U2 introns (Bartschat and Samuelsson, 2010; Lin et al., 2010). Despite this process, a few U12 introns were retained in selected genes and remain very stable in some taxa (Basu et al., 2008). CBP20 genes from both Arabidopsis and rice were identified as belonging to this small number of genes containing U12 introns with AT-AC terminal dinucleotides. Moreover, the U12 intron in CBP20 gene in both plant species is located between exons no. 4 and no. 5, splitting the CBP20 coding sequence into the core and tail CBP20 protein parts in both species studied. Surprisingly, the exons encoding the core part of the protein have identical lengths (18t, 224, 139, and 34 nt), but the exons encoding the tail part of CBP20s differ considerably in length.
In this paper, we show that the CBP20 gene structure is conserved across land plant species from liverworts to higher plants. All studied CBP20 genes contain eight exons and seven introns, with the fourth intron belonging to the U12 class. In addition, the length of the first four exons is conserved, while the length of the exons encoding the CBP20 tail part varies considerably in all plant species studied. Furthermore, we show that the CBP20 U12 introns in plants may be as short as 76 nt and as long as 2733 nt. Our experiments demonstrate that the efficiency of U12 splicing depends on intron length. The experiments carried out to explain the conserved localization of the CBP20 U12 intron show that the exchange of the CBP20 U12 intron with the U2 intron leads to undesired alternative splicing events and that the proper localization of the U12 intron in the CBP20 gene secures correct CBP20 pre-mRNA maturation and CBP20 protein levels in a plant.
The experiments were performed using A. thaliana (L.) Columbia-0 wt plants (Lehle Seeds, Round Rock, TX, United States) and a homozygous T-DNA insertion line cbp20 (Papp et al., 2004). Solanum tuberosum ssp. tuberosum var. Sante, Nicotiana tabacum var. Xanthi, Hordeum vulgare var. Sebastian and liverwort Pellia endiviifolia sp B were used for the CBP20 gene sequencing.
Arabidopsis plants were grown in ‘Jiffy-7 42mm’ soil (Jiffy Products International BV, Moerdijk, Nederland) in an MLR-350H Versatile Environmental Test Chamber (Sanyo, Loughborough, Leicestershire, United Kingdom) with a 16 h light/8 h dark photoperiod (approx. 80 μmol m-2s-1), constant temperature of 22°C and humidity of 70%. Potato plants were grown in sterile conditions in a greenhouse (22°C with constant light, approximately 80 μmol m-2s-1) on ½ Murashige-Skoog medium pH 5.5–5.6. N. tabacum and H. vulgare plants were grown in a greenhouse (22°C with 12 h light/12 h dark photoperiod, approx. 120 μmol m-2s-1) on soil irrigated with mineral nutrients. P. endiviifolia sp B was grown as described by Sierocka et al. (2011).
Arabidopsis thaliana transformation was performed using the floral-dip method according to Clough and Bent (1998).
Total genomic DNA, RNA and plasmid DNA were isolated using a DNeasy Plant Mini Kit (Qiagen), RNeasy Plant Mini Kit (Qiagen), and QIAprep Spin Miniprep Kit, respectively, according to protocols supplied by the manufacturers. Purity and amounts of DNA and RNA were determined using NanoDrop Spectrophotometer (Thermo Scientific).
DNA sequencing was performed with a BigDye v3.1 sequencing kit (Applied Biosystems, Foster City, CA, United States) on a ABI Prism 3130XL Analyzer (Applied Biosystems) in the Molecular Biology Techniques Laboratory, Faculty of Biology, Adam Mickiewicz University in Poznan, Poland.
Four micrograms of total RNA was reverse-transcribed using Superscript III Reverse Transcriptase (Invitrogen) and oligo(dT)15 primer (Novazym). cDNA of CBP20 genes from potato, tobacco, barley and liverwort was amplified by PCR using primers designed according to ESTs from NCBI database (Benson et al., 2012). In the case of potato, we designed primers according to the EST 706129 sequence. Tobacco CBP20 gene cDNA was amplified using primers designed according to the contig sequence assembled on the basis of nine different EST sequences (accession numbers: AM815125, CV017334, AM818189, EB679251, EB440185, AM827799, AM808999, EB444475, and EB678046). Barley CBP20 cDNA was assembled on the basis of the following EST sequences: TC111197 and TC78828 from TIGR database and BU991417, BJ480285, AJ463125, and AJ475973 from NCBI. Primers for cDNA CBP20 amplification from P. endiviifolia were designed according to the results of whole transcriptome sequencing.
PCR was carried out as described in Szarzynska et al. (2009) and Sierocka et al. (2011). Genomic sequence of the CBP20 gene from potato and tobacco was amplified using the Expand Long Template PCR System (Roche). PCR products were separated on a 1% agarose gel, purified using a QIAquick PCR Purification Kit (Qiagen), cloned into the pGEM-T Easy vector (Promega) and sequenced. Primers used for amplification of genomic and cDNA sequences of the CBP20 gene from different plant species are shown in Supplementary Table S1. Amplification of CBP20 cDNAs as well as genes from potato, tobacco, barley and liverwort is shown in Supplementary Figure S1.
Mini- and midi-gene constructs numbers 1, 8, and 9 were prepared by PCR amplification of appropriate A. thaliana and Physcomitrella patens CBP20 gene fragments. Mini- and midi-gene constructs numbers 2–7 were prepared as presented in Supplementary Figure S2. Each construct contains exons no. 4 and no. 5 from the A. thaliana CBP20 gene. Depending on the construct, these two exons are separated by a U12 or U2 intron originating from different plant species. Mini-gene constructs were prepared in 3- or 4-step PCR. In PCR-1, the U2 or U12 intron was amplified using a forward primer containing on its 5′ end 20 nucleotides complementary to exon no. 4. The reverse primer used in PCR-1 contained at its 5′ end 20 nucleotides complementary to exon no. 5. In PCR-2, the sequence of exon no. 5 was amplified using a forward primer complementary to the last 20 nucleotides of the U2 or U12 intron, depending on the individual mini-gene construct. The final mini-gene sequence was amplified in PCR-3, in which PCR-1 and PCR-2 products were used as templates. In mini-gene constructs containing the U12 intron derived from the S. tuberosum or Vitis vinifera CBP20 gene, an additional PCR-0 was performed in which the U12 intron sequence was amplified.
Each mini-gene construct was cloned into the pDH515 vector at a unique BamHI restriction site within an intronless zein gene encoding a maize seed storage protein. Additionally, the vector contains the 35S promoter of Cauliflower mosaic virus (CaMV 35S) and terminator sequence (Lewandowska et al., 2004).
The maxi-gene wt transgene construct was prepared by PCR amplification of the Arabidopsis CBP20 gene sequence encompassing its native promoter (1228 bp) (Kmieciak et al., 2002). The PCR product was cloned into the NotI restriction site in the pENTR vector. The other five maxi-gene constructs were obtained performing a series of PCRs using the wt transgene construct or A. thaliana genomic DNA as a template. Specific sub-fragments of each maxi-gene construct were obtained in the PCR, followed by the consecutive cohesive ends annealing and final PCR amplification (Supplementary Figure S3). The individual maxi-gene constructs were transferred from the pENTR cloning vector to the pEarleyGate302 expression vector using Gateway LR Clonase II Enzyme Mix (Thermo Fisher Scientific). All constructs were confirmed by DNA sequencing. Primers used for construct preparation are listed in Supplementary Table S1.
Splicing of mini- and midi-gene constructs was analyzed in protoplasts isolated from leaves of N. tabacum var. Xanthi (Lewandowska et al., 2004). Protoplast suspension was transfected with each mini- or midi-gene construct as described by Lewandowska et al. (2004). After overnight incubation, total RNA from protoplasts was isolated. cDNA template was synthesized using the zein3′-R reverse primer complementary to the zein sequence flanking mini- and midi-gene constructs in the pDH515 vector (Simpson and Filipowicz, 1996). Splicing analyses were carried out by RT-PCR using following primers: zeinF-FAM labeled with fluorescent phosphoramidite 6-FAM at 5′ end and zeinR (see Supplementary Table S1).
6-FAM-labeled RT-PCR products were quantitatively analyzed using capillary electrophoresis on an ABI 3130xl Genetic Analyzer. The length of the labeled products was calculated using Peak Scanner Software v1.0 (Applied Biosystems) by comparison with the GeneScan-350 TAMRA size standards (Applied Biosystems). Quantification of RT-PCR products was carried out after 23rd amplification cycle by measurement of the fluorescent peak areas of the detected fragments. RT-PCR was carried out in three biological replicates. The same RT-PCR products were separated in parallel on a 1% agarose gel. 6-FAM-labeled fragments were eluted from the gel, cloned into the pGEM T-Easy (Promega) vector and sequenced to confirm specific splicing products.
Splicing analysis of maxi-gene transcripts that were introduced into the A. thaliana cbp20 mutant was also carried out using capillary electrophoresis, as described above. Primers used for RT-PCR amplification hybridized to the sequence of exons no. 3 and no. 6 of the CBP20 gene transcript (Supplementary Table S1). The forward primer Splice-FAM-F was labeled at its 5′ end with the fluorescent phosphoramidite 6-FAM.
Real-time RT-qPCR was performed with Power SYBR® Green PCR Master MIX (Applied Biosystems, Warrington, United Kingdom) on a 7900HT Fast Real-Time PCR System (Applied Biosystems) in 10-μl reaction volumes in 384-well plates. To assess the splicing efficiency of mini-genes, an absolute quantification approach was applied. Standard curves were prepared using plasmids containing individual mini-genes. Splicing efficiency was calculated by comparison of spliced and unspliced transcript levels to the total transcript amount. To estimate the total level of the CBP20 transcript in maxi-gene transgenic lines, the fold change was calculated using the 2-ΔΔCt method. The mRNA fragment of elongation factor 1-alpha (EF1-alpha, TAIR locus: At1g07930) was amplified as a reference gene in A. thaliana.
Protein extracts were separated by 10% SDS-PAGE, transferred to a polyvinylidene fluoride membrane (PVDF; Immobilon®-P, Millipore), and analyzed by western blot using antibodies at the indicated dilutions: anti-Actin (691001; MP Biomedicals) at 1:5000 and anti-CBP20 (AS09 530; Agrisera) at 1:1000.
Comparison between DNA and protein sequences was carried out using bioinformatics tools: ClustalW21 and BoxShade2. Sequencing results were assembled together using the ContigExpress program from Vector NTI (Invitrogen).
Arabidopsis thaliana, Gene ID: 834443; Populus trichocarpa, Gene ID: 7469563; V. vinifera, Gene ID: 100265980; S. tuberosum, GU046516.1; N. tabacum, GU058037.1; O. sativa, Gene ID: 4329963; H. vulgare, FJ548567.1; S. moellendorffii – Selaginella moellendorffii v1.0, Scaffold 3333196:13; P. patens – P. patens V1.2_genome scaffold_1054.
The analysis of CBP20 genes from A. thaliana and O. sativa revealed high sequence and structural conservation. To gain deeper insight into the evolutionary conservation of the CBP20 gene, we decided to compare CBP20 sequences from various plant species across the plant kingdom. The CBP20 gene and cDNA sequences from P. trichocarpa, V. vinifera, Selaginella moellendorffii, and P. patens were found in publicly available databases (Figure 1). In addition, full-length CBP20 cDNA sequences from S. tuberosum var. Sante, N. tabacum var. Xanthi, H. vulgare var. Sebastian and the liverwort P. endiviifolia subspecies B were established in our laboratory. Based on the obtained CBP20 cDNA sequences, we determined the full-length CBP20 gene sequences in potato, tobacco, barley and liverwort genomes.
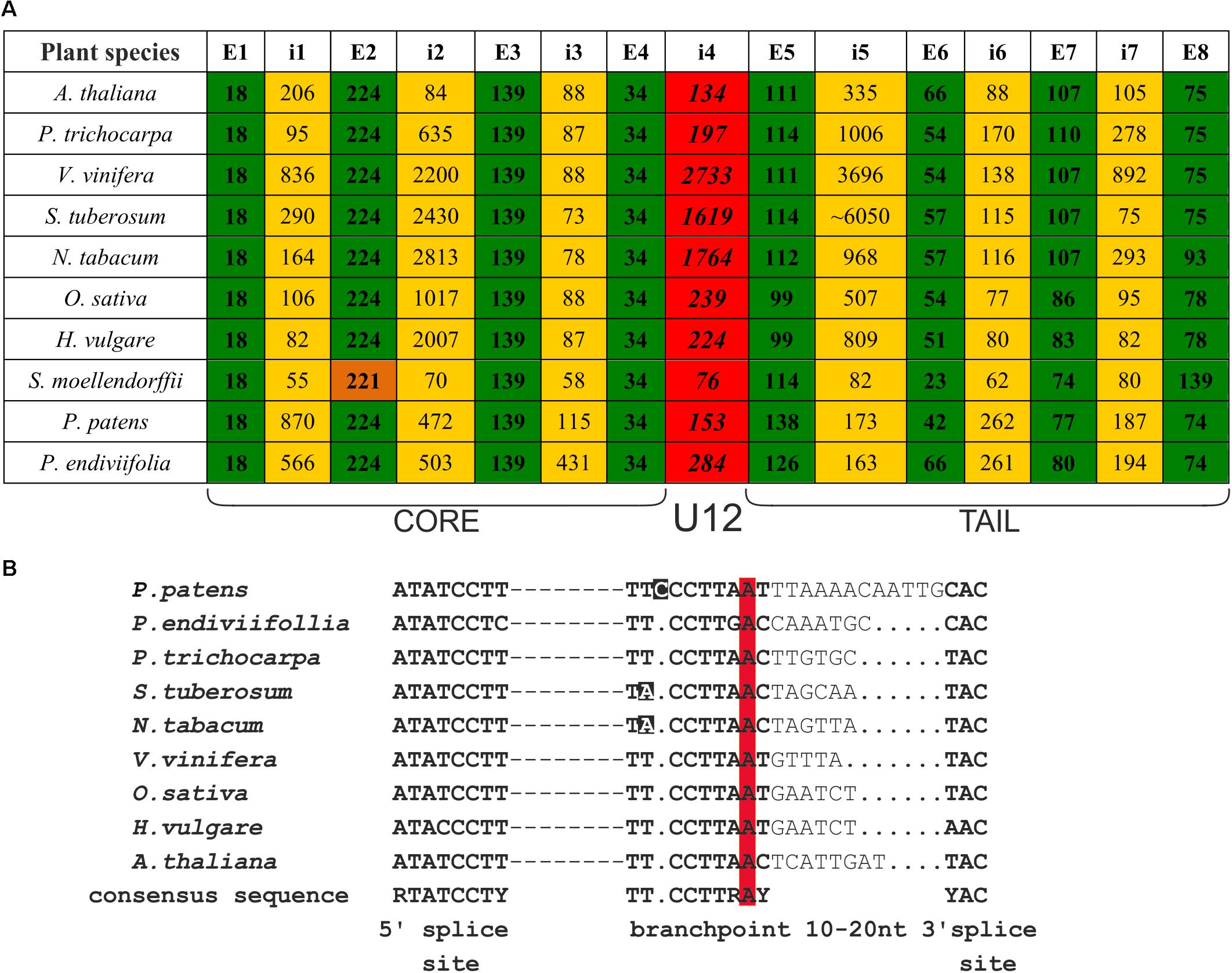
FIGURE 1. CBP20 genes derived from different plant species present very similar gene structures and conserved localization of U12 introns. (A) Comparison of CBP20 gene structures. Green color depicts exons, yellow color marks U2 introns, red color indicates U12 introns, and orange color depicts the second exon of the CBP20 gene from S. moellendorffii, which is shorter than other exons in the same position in other plants studied by 3 nucleotides. Note that with the exception of S. moellendorffii, the exon lengths are the same in all plant species studied until exon no. 5 is reached, which is separated from the protein core-encoding part of the gene by the U12 intron. E – exon, i – intron. (B) A comparison of consensus elements of CBP20 U12 introns derived from the plant species studied (Simpson and Brown, 2008). Dashed lines represent individual nucleotide sequences between the 5′ splice site and branch point. Dots mark alignment gaps. Nucleotides that differ from the consensus sequence in the branch point region are marked in black. The conserved adenine nucleotides in the branch point are marked in red.
We compared the CBP20 gene structure from ten plant species, including representatives of Bryophyta (liverworts, mosses), Leucophyta (lycophyte), and Spermatophyta (monocots and dicots) (Figure 1A). In all species analyzed, the CBP20 genes contain 8 exons and 7 introns. Interestingly, their first four exons, coding for the core part of the protein, are of the same length with one exception, S. moellendorffii, whose second exon is three nucleotides shorter than that in the other CBP20 genes studied. In all CBP20 genes, a U12 intron was found separating the evolutionarily conserved core part that encodes a canonical RBD from the carboxy-terminal fragment (tail) that encodes the NLS and generally exhibits a much lower degree of evolutionary conservation. All these U12 introns contain the canonical AT-AC terminal dinucleotides and a branch point sequence typical of U12 introns (Figure 1B). The CBP20 U12 intron length varies from 76 nt in S. moellendorffii to 2733 nt in V. vinifera. In general, plant species can be divided into two classes: those carrying short U12 introns (from 76 to 284 nt) and those carrying long U12 introns (from 1619 to 2733 nt) (Figure 1A).
There are considerable differences between different plant CBP20 gene lengths. The S. moellendorffii CBP20 gene (1245 bp) is 10 times shorter than that from S. tuberosum (11430 bp). In the potato CBP20 gene, the fifth intron is extremely long, approximately 6050 bp, which largely accounts for the unusual length of this gene. Computational analysis of this intron sequence has revealed that it contains characteristic signatures of the LINE1 retrotransposon. Since we could not identify any full open reading frame for reverse transcriptase, we assumed that this retroelement is defective.
Comparison of all plant CBP20 amino acid sequences revealed the presence of conserved residues within the core domain that were reported to be responsible for binding to the cap structure and for recognition of RNA (Supplementary Figure S4) (Mazza et al., 2002). In the tail fragment of all CBP20 proteins, we identified potential NLS sequences. In the majority of plant species, the CBP20 gene contains two NLS motives, as was shown before for A. thaliana, while in P. trichocarpa and P. patens, we identified only one motif: the proximal NLS. Generally, sequence homology of full-length CBP20 proteins in plants ranges from 61 to 79% identity, and that of the highly conserved core part ranges from 80 to 89% identity. The homology within the tail part is rather low (39–69% identity), but the distal part of this tail region shows again a relatively high degree of conservation, suggesting that this part of the protein may play a structural or functional role.
It was shown that U12 introns over the course of evolution were usually exchanged with U2 introns (Burge et al., 1998; Bartschat and Samuelsson, 2010). The evolutionarily conserved localization of the plant CBP20 U12 intron separating the conserved core domain from the tail part of the protein in all CBP20 plant genes characterized encouraged us to investigate the role of this intron in CBP20 pre-mRNA maturation.
As we have shown, the length of CBP20 U12 introns in the studied plant species varies considerably. To test whether U12 intron length may influence U12 intron splicing efficiency, we prepared five mini-gene constructs. These mini-genes consist of A. thaliana CBP20 exon no. 4 (34 bp) and exon no. 5 (111 bp) separated by the following: construct no. 1 – the original A. thaliana CBP20 U12 intron (134 bp), construct no. 2 – the P. patens CBP20 U12 intron (153 bp), construct no. 3 – the O. sativa CBP20 U12 intron (239 bp), construct no. 4 –the S. tuberosum CBP20 U12 intron (1619 bp) and construct no. 5 – the V. vinifera CBP20 U12 intron (2733 bp) (Figure 2A). The mini-genes were cloned into the pDH515 expression vector (Lewandowska et al., 2004) and sequenced. These recombinant plasmids were then used for tobacco mesophyll protoplasts transfection. Total RNA was isolated from the transfected protoplasts incubated overnight, and cDNA derived from the mini-gene transcripts was prepared. Splicing analyses were carried out applying RT-PCR with fluorescent-labeled primers. The splicing efficiency of each construct was determined as the mean of three independent experiments. All U12 introns were spliced correctly; no alternative splicing events were observed (Figure 2B). However, the splicing efficiency of different mini-genes varied considerably. We found a correlation between the U12 intron length and splicing efficiency as revealed by RT-qPCR using the absolute quantification method (Figure 2C and Supplementary Table S2). The splicing efficiency of the 134 nt long U12 intron from A. thaliana was assumed in this comparison to be 1. The U12 introns derived from O. sativa, S. tuberosum and V. vinifera CBP20 genes were spliced 1.28, 7.17, and 34.65 times more efficiently than that from A. thaliana, respectively (Figure 2C).
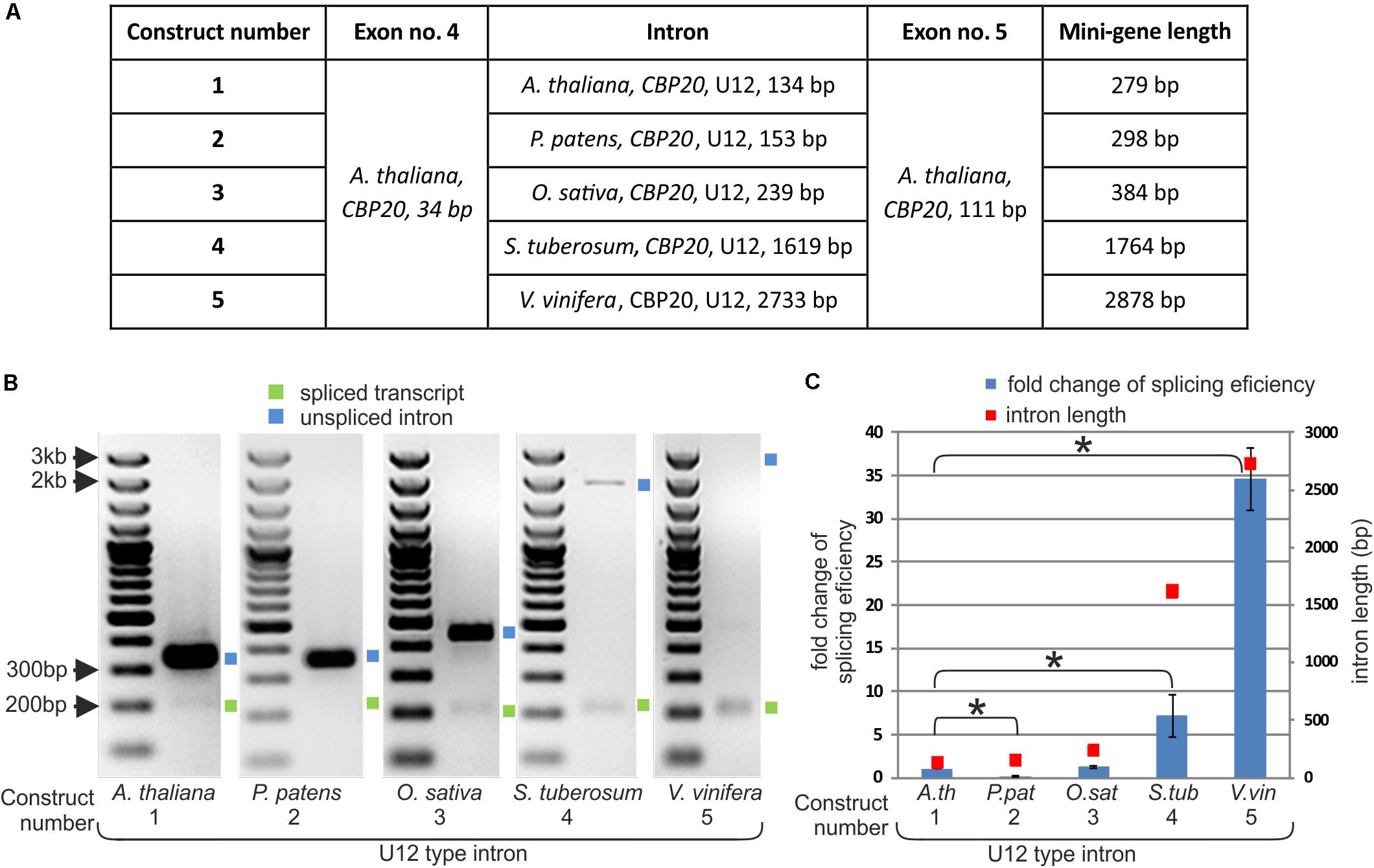
FIGURE 2. The efficiency of CBP20 U12 intron splicing depends on intron length. (A) A table showing structures of five mini-gene constructs containing exons no. 4 and no. 5 from the A. thaliana CBP20 gene separated by one of the U12 introns differing in length and derived from different plant species. (B) Qualitative RT-PCR analysis of splicing of mini-gene transcripts in tobacco protoplasts. Blue and green squares represent unspliced and spliced transcripts, respectively. (C) Quantitative Real-time PCR analysis of mini-gene transcript splicing efficiency. Splicing efficiency of the mini-gene no. 1 transcript carrying the U12 intron from the Arabidopsis CBP20 gene was taken as 1. Blue bars represent fold change in splicing efficiency in comparison to the mini-gene no. 1 transcript. Red squares show the length of U12 introns derived from various plant species studied. Values are shown as the mean ± SD (n = 3) from three independent experiments. Asterisk – P < 0.001, Student’s t-tests.
An exception to this rule was observed in the splicing efficiency of the U12 intron derived from P. patens. Despite its almost identical length with the U12 intron from A. thaliana, the splicing efficiency of this U12 intron was five times lower.
To learn more about the role of the U12 intron in CBP20 plant genes, we constructed a series of mini-genes containing A. thaliana exons no. 4 and no. 5 separated by the original CBP20 U12 intron (134 bp – construct no. 1), the U2 intron derived from the A. thaliana CBP80 gene (intron no. 11, 146 bp – construct no. 6) and the U2 intron derived from the P. sativum legumin gene (intron no. 1, 138 bp – construct no. 7) (Figure 3A). Both U2 introns were chosen because of their similar length to that of the original A. thaliana CBP20 U12 intron. The selected U2 introns have canonical GT-AG splice site dinucleotides and a polypyrimidine tract near the 3′ splice site and show a low GC content, 26% and 29% for the pea legumin and Arabidopsis CBP80 genes, respectively. U12 intron from the A. thaliana CBP20 gene has also a low GG content – 36%.
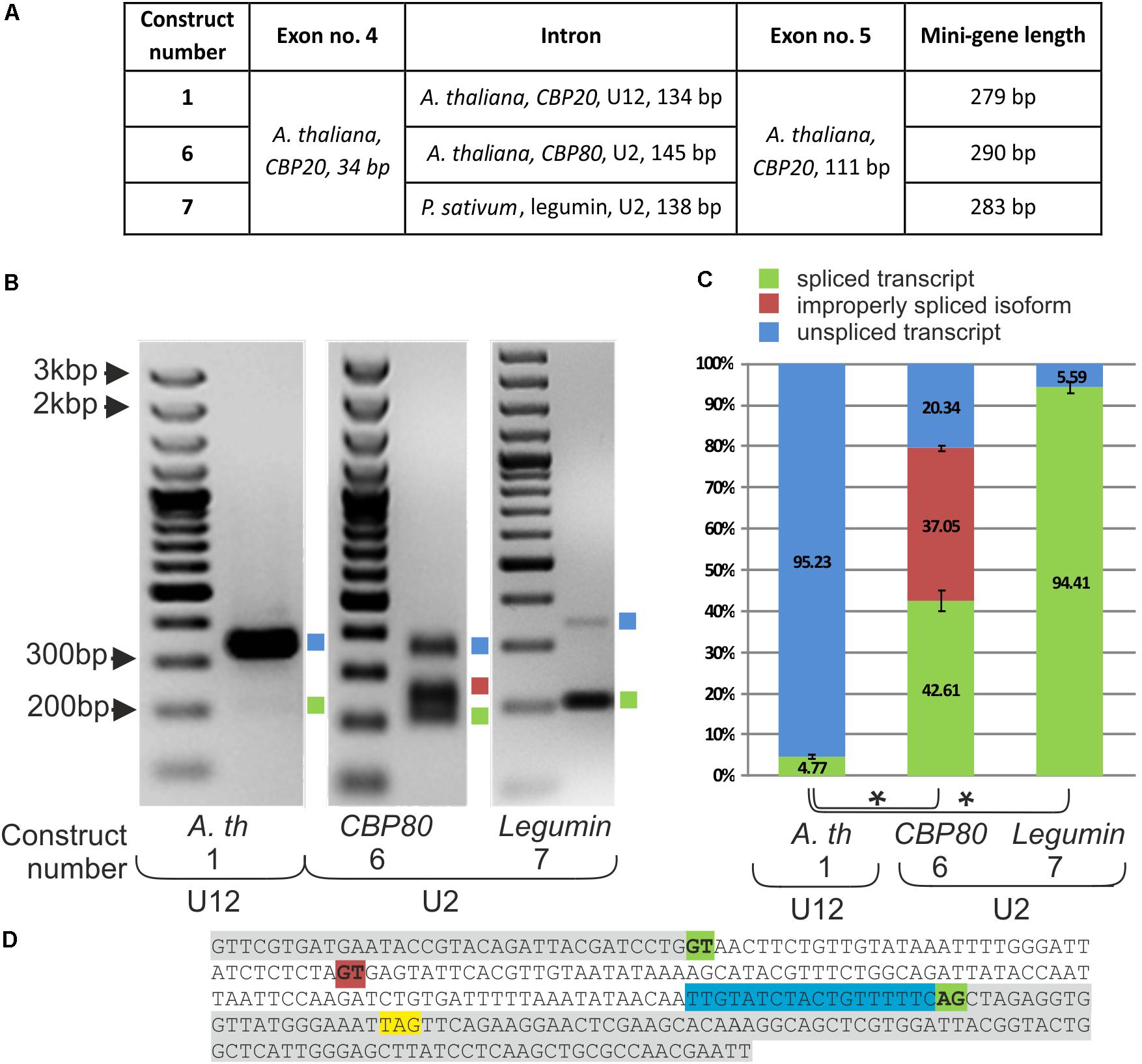
FIGURE 3. Replacement of the original CBP20 U12 intron by a U2 intron may lead to deleterious splicing events. (A) A table showing structures of three mini-gene constructs containing exons no. 4 and no. 5 from the A. thaliana CBP20 gene separated by the original U12 intron (construct no. 1), by the U2 intron from the Arabidopsis CBP80 gene (construct no. 6), and by the U2 intron from the P. sativum legumin gene (construct no. 7). (B) Qualitative RT-PCR analysis of splicing of mini-gene transcripts in tobacco protoplasts. Blue and green squares represent unspliced and spliced transcripts, respectively. The red square represents the alternatively spliced CBP80 U2 intron. (C) Quantitative analysis of mini-gene transcripts RT-PCR products using capillary electrophoresis. Splicing efficiency is calculated as a percentage of the sum of all detectable products. Values are shown as the mean ± SD (n = 3) from three independent experiments. (D) Nucleotide sequence of the mini-gene no. 6 containing the Arabidopsis CBP80 gene U2 intron. Green color depicts constitutive U2 5′ and 3′ splice site dinucleotides, red color represents an alternative 5′ splice site, the STOP codon is marked in yellow, and blue color stands for the polypyrimidine tract. Asterisk – P < 0.005, Student’s t-tests.
Splicing of the mini-gene transcripts was studied in the tobacco protoplast system, as described in the case of mini-genes containing various U12 introns. The original U12 intron was spliced with only 4.77% efficiency, whereas both studied U2 introns, from the Arabidopsis CBP80 gene and pea legumin gene, were spliced, reaching 79.66 and 94.4% efficiency, respectively (Figure 3B). These results are supported by data that have been previously obtained in our laboratory (Lewandowska et al., 2004), showing that A. thaliana U12 introns are spliced less efficiently than are U2 introns in tobacco protoplasts. However, in the mini-gene containing the U2 intron derived from the A. thaliana CBP80 gene, an alternative splicing event was observed: two alternative 5′ splice sites were recognized by the tobacco U2 splicing machinery, leading to the proper splicing of the CBP20 mini-gene transcript (42.61%) or to the inclusion of an additional 38 nt from the 5′ intron end into the spliced mRNA (37.05%) (Figures 3C,D). The 38 nt long insertion introduces a premature stop codon resulting in premature translation termination. To test whether this alternative splicing event also occurs naturally in the CBP80 gene, we analyzed the splicing of this intron in the CBP80 transcript. We were able to detect only constitutively, properly spliced CBP80 mRNA, without any additional alternative splicing events (Supplementary Figure S5).
It has been shown that the extension of mini-genes composed of two exons from GSH2 interrupted by a U12 intron with neighboring exons and U2 introns improves U12 splicing efficiency (Lewandowska et al., 2004). To test whether this is also true for other genes containing U12 introns, we prepared two midi-genes that were based on A. thaliana and P. patens CBP20 genes. A. thaliana (construct no. 8) and P. patens (construct no. 9) midi-genes consist of E3-i3(U2)-E4-i4(U12)-E5-i5(U2)-E6 from the A. thaliana and P. patens CBP20 genes, respectively (Figure 4A). The midi-genes were studied in tobacco protoplasts as described before in the case of mini-genes.
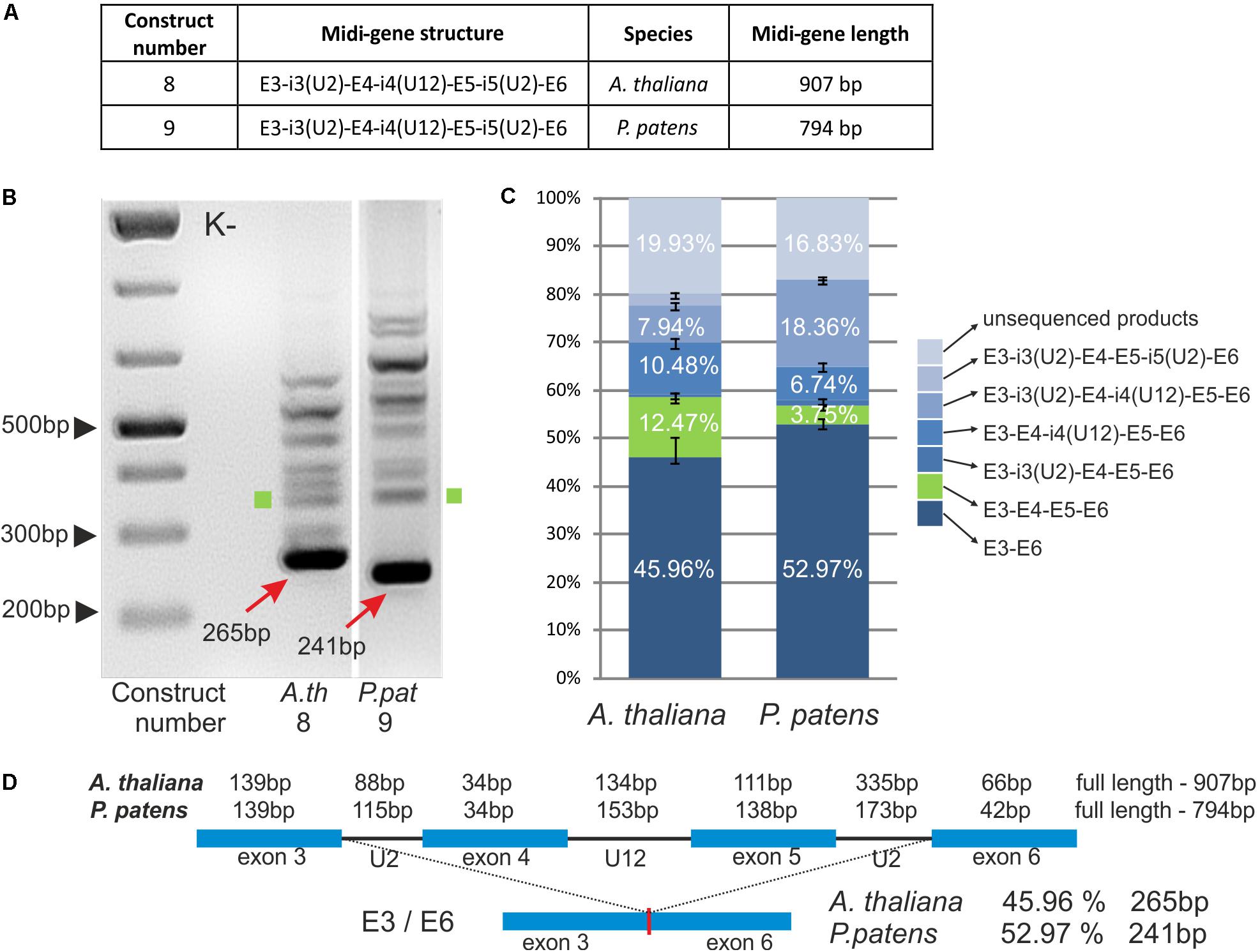
FIGURE 4. Plant CBP20 U12 intron surrounded by four exons and two U2 introns is not efficiently recognized by the U12 splicing machinery. (A) A table showing structures of two midi-gene constructs containing A. thaliana or P. patens CBP20 gene fragment of the following composition: exon 3 – U2 intron 3 – exon 4 – U12 intron 4 – exon 5 – U2 intron 5 – exon 6. (B) Qualitative RT-PCR analysis of splicing of midi-gene transcripts in tobacco protoplasts. Arrows point to the improperly spliced products in which exons no. 4 and no. 5 are skipped. Green squares represent properly spliced midi-gene CBP20 transcripts. The rest of the bands above the main amplification products represent a variety of partially spliced transcripts. (C) Quantitative analysis of midi-gene transcript RT-PCR products using capillary electrophoresis. Splicing efficiency is calculated as a percentage of the sum of all detectable products. Values are shown as the mean ± SD (n = 3) from three independent experiments. Green color depicts properly spliced CBP20 mRNA isoform. (D) Structure of midi-genes and fully spliced transcript representing the most abundant splicing event.
The splicing patterns of A. thaliana and P. patens midi-transcripts were similar (Figure 4B). We obtained one main and many additional minor products. Sequencing of these products has shown that the main band (265 bp in A. thaliana and 241 bp in P. patens) represents a spliced product consisting of exons no. 3 and no. 6. Thus, splicing in that case leads to the exon skipping event by the removal of all three introns and exons no. 4 and no. 5. The remaining minor bands represent various spliced products listed in Figure 4C, among which a fully and properly spliced transcript containing all four exons (E3-E4-E5-E6) was also detected. Quantitative analysis shows that the main improperly spliced product was present in approximately 50% of all other differentially spliced RNA molecules, in both Arabidopsis and Physcomitrella midi-genes (Figures 4C,D). The fully spliced transcript containing all four exons (E3-E4-E5-E6) was present only in approximately 12.47% of all splicing products in Arabidopsis and 3.75% in Physcomitrella. Thus, in contrast to the GSH2 midi-gene, the neighboring U2 introns and exons do not enhance the recognition of the U12 intron in the case of Arabidopsis and moss CBP20 midi-genes. Instead, the U12 intron signals are mainly not recognized properly in these constructs.
To study the influence of the U12 intron on the splicing efficiency of the full CBP20 pre-mRNA in plants, we prepared constructs composed of the natural A. thaliana CBP20 gene promoter and the whole wild-type (wt) gene sequence or its variants (the maxi-genes series) (Figure 5B). In addition to the wt maxi-gene (wt transgene), we constructed CBP20 gene variants in which (i) the U12 intron was replaced with the U2 intron derived from the A. thaliana CBP80 gene (intron no. 11; U12→U2), (ii) the U12 intron was removed (ΔU12), and (iii) exons no. 4 and no. 5 were swapped (exon swap). Additionally, we prepared two constructs based on the CBP20 gene variant U12→U2. In the first construct, the U12 intron was moved upstream in the CBP20 gene and replaced the U2 intron no. 3 (U12 in core); by this, we moved the U12 intron into the core-encoding part of the CBP20 gene. In the second construct, the U12 intron replaced the original U2 intron localized between exons no. 5 and no. 6 (U12 in tail); in this construct, the U12 intron was moved into the tail-encoding part of the CBP20 gene. A. thaliana cbp20 mutant plants were transformed with these constructs, and three independent homozygous lines of each gene variant were further tested.
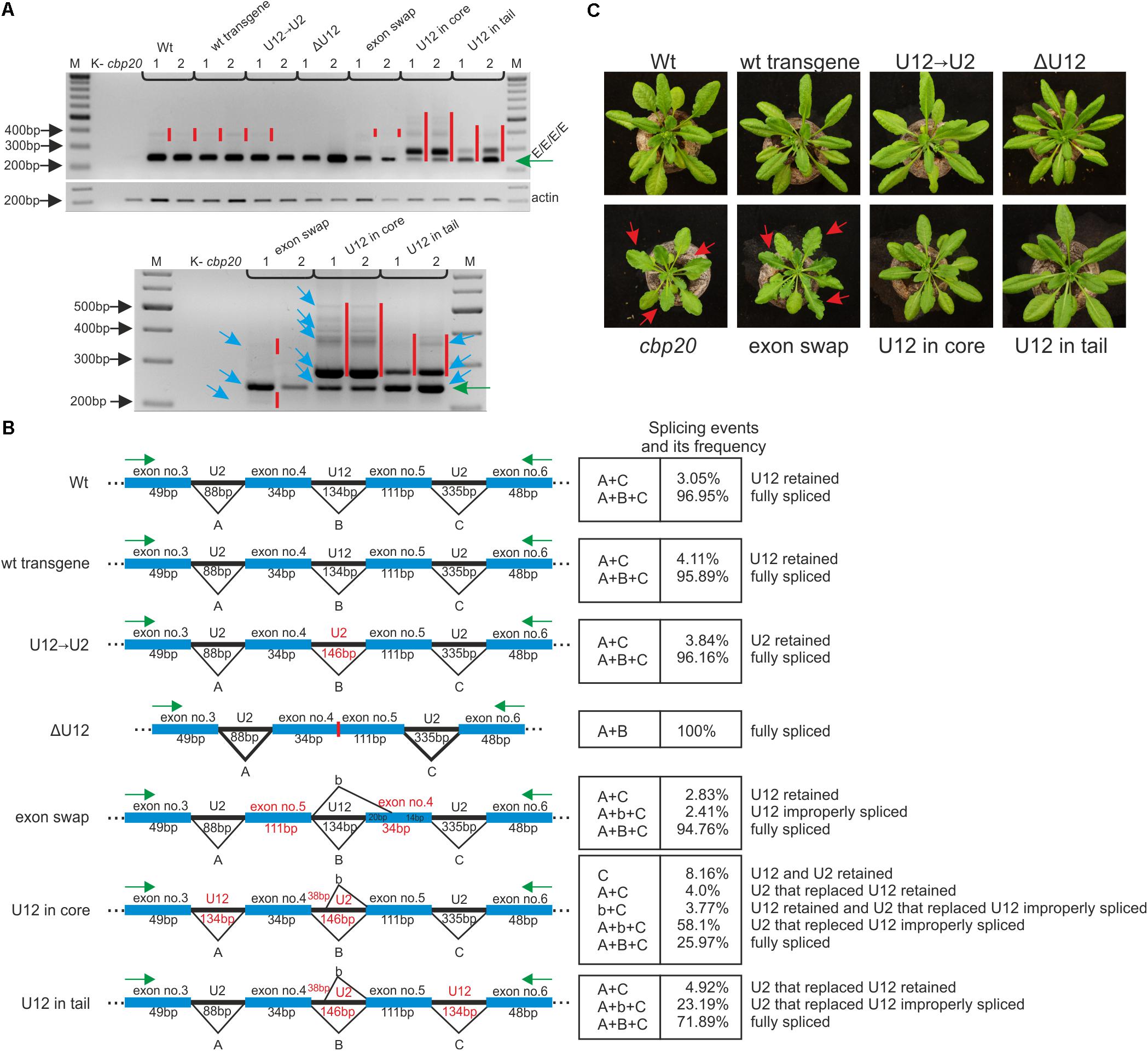
FIGURE 5. The proper localization of the U12 intron in the CBP20 gene secures correct pre-mRNA maturation in plant. (A) Qualitative RT-PCR analysis of splicing of CBP20 full-length maxi-gene transcripts. For each construct, two independent transgenic lines were analyzed. Electrophoretic separation of RT-PCR products was carried out in a 1.2% agarose gel. Abbreviations: K- – negative control (no template); cbp20 – mutant line; Wt – wild-type plants; wt transgene – depicts wild-type CBP20 gene; U12→U2 – the CBP20 gene in which the original U12 intron was exchanged by the U2 intron derived from the Arabidopsis CBP80 gene; ΔU12 – the CBP20 gene with U12 intron deletion; exon swap – the CBP20 gene mutant in which exons no. 4 and no. 5 flanking the U12 intron have been exchanged; U12 in core – construct derived from the U12→U2 construct in which the U12 intron has been moved between exons no. 3 and no. 4 (core part of the CBP20 gene); and U12 in tail – construct derived from the U12→U2 construct in which the U12 intron has been moved between exons no. 5 and no. 6 (tail part of the CBP20 gene). All constructs are under a native promoter. Green arrow points the properly spliced CBP20 transcript (E/E/E/E). Red bars show additional detectable partially spliced or improperly spliced products. A panel below shows a zoomed-in fragment of the above gel; blue arrows depict sequenced and quantitatively analyzed products. Actin – loading control. M – molecular DNA marker. (B) A scheme presenting all detected CBP20-gene derived mutant splicing isoforms. Capital A, B, and C letters represent constitutive splicing events for introns no. 3, no. 4, and no. 5, respectively. b – depicts improper splicing events of intron no. 4. Quantitative analysis of mini-gene transcript RT-PCR products was performed using capillary electrophoresis. Splicing efficiency is calculated as a percentage of the sum of all detectable products. Values are shown as the mean ± SD (n = 3) from three independent experiments. (C) Arabidopsis vegetative rosettes of wt and CBP20 maxi-gene lines after 30 days of growth.
Qualitative RT-PCR-based analyses of the CBP20 mRNA levels revealed the presence of one dominant product representing fully spliced transcripts in the case of wt plants as well as transgenic plants carrying the wt CBP20 gene construct (wt transgene) and the U12→U2 construct. A small amount of partially unspliced products was also detected (Figure 5A, upper panel). In transgenic plants carrying mutated CBP20 genes in which the U12 intron was deleted (ΔU12), we also observed the presence of fully and properly spliced CBP20 mRNA. Moreover, the transgenic plants exhibited a wt phenotype, indicating that the CBP20 protein is produced in the transgenic lines analyzed (Figure 5C, upper panel).
Only transgenic plants carrying the CBP20 gene with swapped exons (exon swap) gave mRNAs in which the CBP20 coding sequence was disrupted. This impaired mRNA accumulated at a lower level than in wt and wt transgene plants containing a wt copy of the CBP20 gene (Figure 5A, upper panel). The exon swapping mRNA contains a premature stop codon (in exon no. 5) that might cause the synthesis of putative shorter proteins with a disturbed core fragment. As expected, this mutant plants’ phenotype is very similar to that of the null cbp20 mutant, exhibiting serrated rosette leaves and growth retardation (Figure 5C, lower panel), confirming that CBP20 is indeed not produced in these plants. In the case of mutants in which the U12 intron was moved into the core- or tail-encoding parts of the gene, additional splice products were observed (Figure 5A, upper panel; the lower panel represents a zoomed in part of the upper one).
Quantitative RT-qPCR analyses using fluorescent-labeled primers and capillary electrophoresis were carried out to measure the levels of individual spliced products. Cloning and sequencing of these products were performed to identify particular splicing isoforms. In the case of ΔU12, apart from the fully spliced product, we did not detect any additional mRNA isoforms. In the wt, wt transgene and U12→U2 plants, the fully spliced mRNA represented 96–97% of all spliced products, while U12- or U2-containing mRNA isoforms were present in 3–4% of all spliced products (Figure 5B). The exchange of exons no. 4 and no. 5 within the CBP20 gene resulted in the presence of properly spliced mRNA (approximately 95% of all isoforms), the isoform still containing the U12 intron (approximately 3% of all splicing isoforms) and an additional mRNA isoform in which the 3′ alternative U12 splice site was selected within exon no. 4 (approximately 2.5% of all splicing isoforms).
Two A. thaliana lines in which the U12 intron was relocated into the core- or tail-encoding part of the CBP20 gene (U12 in core and U12 in tail) produced complex patterns of mRNA isoforms. In the case of the U12 in core variant, we identified and quantitatively measured the levels of five mRNA isoforms. The fully and correctly spliced CBP20 mRNA isoform represented only 26% of all splicing products (Figure 5B). Surprisingly, the most abundant mRNA isoform showed an improperly spliced U2 intron replacing the original U12 intron. The alternative 5′ splice site was selected, leading to the inclusion of an additional 38 nt from the 5′ intron end into the processed mRNA (58.1%) (Figure 5B). However, these transgenic plants exhibit the wt phenotype, suggesting that the lower level of properly spliced CBP20 mRNA is sufficient to fulfill plant requirements for the CBP20 protein (Figure 5C). Interestingly, an identical improper splicing event was observed in the mini-gene containing the same U2 intron instead of the original U12 intron (Figure 3B). Three additional mRNA isoforms were observed in small amounts: (1) the isoform in which the U12 intron as well as the U2 intron that replaced the original U12 intron were retained (8.16%), (2) the isoform in which the U2 intron that replaced the original U12 intron was retained (4.0%), and (3) the isoform in which the U12 intron was retained and the U2 intron located between exons no. 3 and no. 4 was incorrectly spliced (3.8%).
In the U12 in tail maxi-gene variant we identified and measured the levels of three mRNA isoforms. The fully and correctly spliced CBP20 mRNA isoform represented almost 72% of all mRNA isoforms (Figure 5B). A minor portion of spliced products were identified as an isoform still containing the U2 intron that replaced the original U12 intron (4.9%). We also detected an mRNA isoform identical to that in the U12 in core maxi-gene, which contained the improperly spliced U2 intron that replaced the original U12 intron (23.2%) (Figure 5B). As expected, all these plants exhibit a wt phenotype (Figure 5C). These results suggest that shifting the U12 intron into other intron positions in the Arabidopsis CBP20 gene disturbs splicing events, leading to substantial downregulation of the properly spliced mRNA level.
To quantify the total level of CBP20 transcript in the wt and maxi-gene transgenic lines analyzed, we performed real-time RT-PCR using primers designed to recognize the 3′ ends of the transcripts. No significant differences in the CBP20 mRNA levels between wt and transgenic plants were observed (Figure 6A). However, we did detect changes between maxi-gene transgenic lines when the CBP20 protein level was analyzed by western blotting (Figure 6B). A similar protein level was observed for the wt transgene and ΔU12 lines. As expected, no CBP20 protein was detected in the exon swapping maxi-gene line due to the premature STOP codon existing in exon no. 5 of this construct. Interestingly, the decreased level of CBP20 was observed in the U12→U2 transgenic line as well as in the U12 in core and U12 in tail transgenic lines.
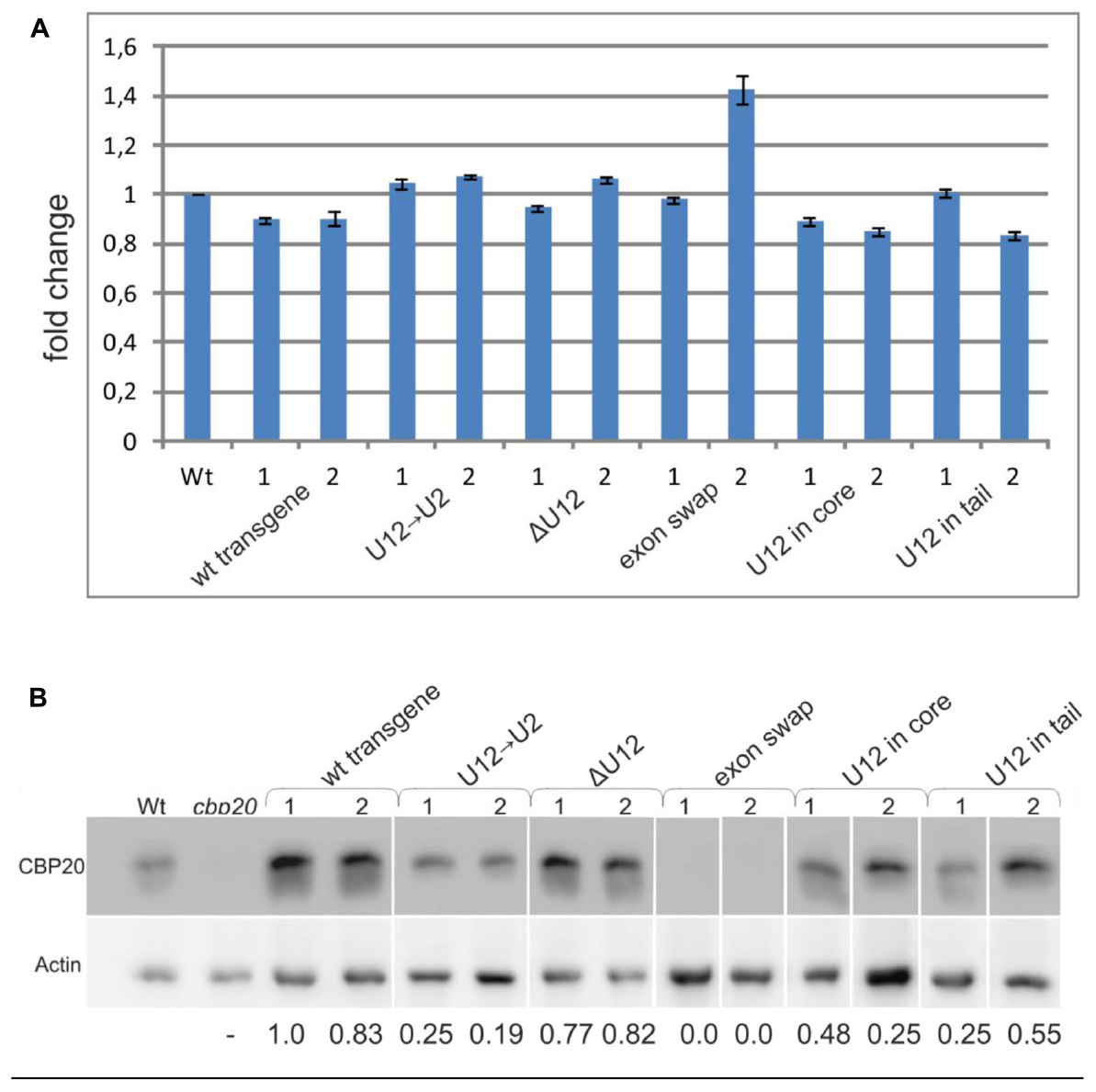
FIGURE 6. The localization, lack or replacement of the U12 intron in the CBP20 gene does not influence the total level of the CBP20 transcript but impacts the level of CBP20 protein in plants. (A) Real-time qPCR analysis of the total level of the CBP20 transcript in maxi-gene transgenic lines. For each construct, two independent transgenic lines were analyzed. Wt – wild-type plants; cbp20 – cbp20 mutant line; wt transgene – wild-type CBP20 gene structure; U12→U2 – CBP20 gene in which the original U12 intron was replaced with the U2 intron derived from the Arabidopsis CBP80 gene; ΔU12 – CBP20 gene with U12 intron deletion; exon swap – the CBP20 gene in which exons no. 4 and no. 5 flanking the U12 intron have been exchanged; U12 in core - a derivative of the U12→U2 construct in which the U12 intron has been introduced between exons no. 3 and no. 4; and U12 in tail - derivative of the U12→U2 construct in which the U12 intron has been introduced between exons no. 5 and no. 6. The CBP20 mRNA level in Wt was taken as 1. Values are shown as the mean ± SD (n = 3) from three independent experiments. (B) Western blot analysis of CBP20 protein levels in transgenic Arabidopsis plants. For each construct, two independent transgenic lines were analyzed. Upper panel – immunoblot using antibodies against CBP20 protein; lower panel – immunoblot with antibodies against actin used as a loading control. Numbers below the western blot image are relative intensities of CBP20 bands calculated using the wt transgene (line 1) CBP20 level as 1. Lines are described as previously. The western blot representing recognition of CBP20 in wild type and various mutants of this gene in A. thaliana is fused because the original blots contained data from additional transgenic lines, not included in the paper. The data are from two independent western blots. All calculations concerning the amount of the CBP20 protein were carried out separately for each western.
The evolutionary conservation from liverworts to higher plants of the U12 intron localization within the plant CBP20 genes encouraged us to study the role of this intron in the CBP20 pre-mRNA maturation. It has been reported that U12 intron positions are more strongly conserved between animals and plants than are the positions of U2 introns (Basu et al., 2008). Similar to plant CBP20 genes, human, chicken, and starlet sea anemone CBP20 genes also contain U12 introns. However, the animal CBP20 gene structures and positions of U12 introns differ significantly from those of plant CBP20 genes (Supplementary Figure S6). In contrast to their animal orthologs, plant CBP20 genes contain an additional coding sequence representing the so-called tail part of CBP20. In each plant CBP20 gene, a U12 intron separates the core part of the protein from the tail fragment. The tail part of plant CBP20s usually contains two NLSs, which is characteristic of plant small CBC subunits. Our in silico analyses revealed, however, that the Chlamydomonas reinhardtii CBP20 gene does not have the tail-encoding part (GenBank: EDP01640.1). Interestingly, the Chlamydomonas CBP20 gene also does not contain a U12 intron. Moreover, we have not found U12 introns in fungal CBP20 genes that also lack the plant-specific CBP20 tail. Therefore, it can be speculated that the U12 intron in CBP20 genes appeared during land plant evolution and was introduced into the gene coding sequence together with the tail-encoding part of CBP20.
Comparison of the CBP20 U12 introns of various plants revealed their broad spectrum of length: they can be as short as 76 bp (S. moellendorffii) and as long as 2733 bp (V. vinifera). Browsing the U12 database (U12DB) for U12 introns present in the A. thaliana genome revealed the presence of 246 U12 introns (Alioto, 2007). The length of these introns varies between 68 and 3731 bp. However, the number of U12 introns exceeding 1000 bp in length is rather low (only 6 such introns were described). The number of Arabidopsis U12 introns presented in the U12DB is underestimated, because more recent studies based on transcriptomic data revealed 2069 U12 introns in Arabidopsis (Marquez et al., 2012). Our data show that plant CBP20 U12 introns derived from different plant species, when inserted into the mini-genes containing two flanking A. thaliana exons, exhibit differential impact on their splicing efficiency. Surprisingly, the longer the U12 intron was, the higher splicing efficiency was observed. We have previously reported that the splicing efficiency of plant U12 introns depends on a combination of factors, including TA content, exon splicing enhancer sequences (ESEs), and the presence of an adenosine at the upstream purine position in the plant U12 branch point consensus TCCTTRATY (Lewandowska et al., 2004). All CBP20 U12 introns studied in this paper exhibit a similar high TA content (approximately 65%), and almost all (except P. endiviifolia) contain an adenosine at the upstream position in their branch point sequences (Figure 1). Since the flanking exons are identical in all constructs, one can suggest that putative ESEs should influence all studied U12 intron splicing in the same way. Thus, the only evident difference between short and long CBP20 U12 introns is that the longer U12 introns contain more TA-rich sequences, which may help in recognition of these introns by the U12 splicing machinery. Therefore, it is possible that one of the yet-unidentified hnRNP proteins recognizing TA-rich sequences within U12 introns plays an important role in plant minor spliceosome recruitment.
We noticed that the CBP20 U12 intron from P. patens was extremely inefficiently spliced in tobacco protoplasts (Figure 2). Previously we also observed very low splicing efficiency of mini-genes containing U12 introns originated from different plant genes (Lewandowska et al., 2004). We showed that U12 intron splicing was enhanced by increased TA-richness of the intron. In this study we found that the branch point of the P. patens CBP20 U12 intron has an additional C nucleotide in the 5′ part of the consensus sequence compared to the CBP20 U12 introns from other plant species (Figure 1). We analyzed all U12 introns from the P. patens genome and compared their branch points to the consensus sequence. Only four P. patens U12 introns containing the additional C nucleotide in the 5′ part of the branch point were found (124 P. patens U12 introns were analyzed). Therefore, the low efficiency of P. patens CBP20 U12 intron splicing could be due to the specific sequence of its branch point. Searching the U12DB for this additional C in the A. thaliana 5′ part of all U12 branch point sequences revealed the presence of only six such examples (Alioto, 2007). This shows that the presence of an additional cytidine in the branch point sequence is acceptable for both Physcomitrella and Arabidopsis U12 splicing machineries but may be a reason for the inefficient splicing of introns containing such branch point sequence. Further investigations are needed to uncover the role of this additional C in plant U12 intron splicing. On the other hand, the low splicing efficiency of the P. patens CBP20 U12 intron may also be associated with the compatibility of specific intron/exon sequences as well as ESEs that are present or absent in different exonic sequences. Comparison of P. patens and other plant species exon sequences that surround CBP20 U12 introns revealed a high similarity between exon no. 4, but in the case of exon no. 5, four insertions (3, 6, 14, and 5 bp long) and a single nucleotide deletion were found in P. patents (Supplementary Figure S7). Therefore, we cannot exclude the possibility that these exon differences between Arabidopsis and Physcomitrella may cause the incompatibility in the proper recognition of P. patens U12 intron-containing mini-gene transcripts by the tobacco U12 minor spliceosome.
To explain a role of the U12 intron in CBP20 pre-mRNA maturation, several mini-, midi- and maxi-gene constructs were prepared. This approach allowed us to dissect the effect of given CBP20 transcript fragments on U12 intron splicing. The shortest constructs (the mini-genes series) contained two CBP20 exons that originally flank the U12 intron. The U12-containing mini-transcripts were correctly but poorly spliced (4.77%). In a similar assay, the pea U2 legumin intron was spliced correctly and very efficiently (94.41%). The CBP80 U2 intron was spliced more efficiently (42.61%) than the original U12 intron but less efficiently than the legumin gene-derived U2 intron. In addition, alternatively spliced products were frequently observed (37.05%) (Figure 3C). These results have shown that the replacement of the U12 intron with a U2 intron increases splicing efficiency but may lead to undesired improper splicing events. Our observation agrees with the published data showing that U12 splicing slows down pre-mRNA processing (Lewandowska et al., 2004; Simpson and Brown, 2008). We demonstrated, however, that U2 introns when inserted between exons normally flanking a U12 intron are spliced improperly, which may lead to the downregulation of CBP20 protein expression (Figure 4). It has been shown in animal cells that U12 intron splicing is a limiting step in pre-mRNA processing (Patel et al., 2002; Niemelä and Frilander, 2014).
Since the CBP20 U12 intron splicing efficiency of the Arabidopsis mini-gene transcript (construct no. 1, Figure 2) was very low (0.34%), we decided to extend the construct, and we modified it by the introduction of two U2 introns and two flanking exons of the Arabidopsis CBP20 gene (the midi-gene series). Indeed, we improved the proper splicing efficiency almost three times (from 4.77 to 12.47%, Figure 4). Unexpectedly, this correct splicing was predominated by an extensive skipping event resulting in the production of fused exons no. 3 and no. 6 (approximately 50% of all spliced products). In addition, a plethora of minor alternatively spliced products were observed. A similar effect was observed in the case of midi-gene transcripts containing the Physcomitrella CBP20 intron, suggesting the general character of the results obtained (Figure 4). In our earlier studies, we used a similar approach to study analogous mini- and midi-gene constructs of the Arabidopsis GSH2 gene containing a U12 intron (Lewandowska et al., 2004). Unlike the results of the experiment presented in this paper, in the case of GSH2, we detected only constitutive splicing events, and as a result, we obtained the fully spliced products consisting of two or four exons when the mini- and midi-genes were analyzed, respectively. In addition, the transcript derived from the midi-gene (containing additional exon and intron sequences that flanked the U12 intron) was spliced more efficiently. This suggested that the U2 introns located upstream and downstream of U12 participated in the mechanism of definition of the U12 intron (Lewandowska et al., 2004). Interestingly, overexpression of the RBP45 protein, an hnRNP-like RNA binding protein with affinity for U-rich sequences (Lorković et al., 2000), resulted in a complete abolishment of the normal splicing pattern of the GSH2 midi-gene transcript. RBP45 increased the overall splicing efficiency of the transcript, but caused an extensive skipping event resulting in the accumulation of RNA molecules that were composed of fused two terminal exons (Lewandowska et al., 2004). In the majority of CBP20 midi-gene constructs analyzed in this paper, the presence of the U12 intron was ignored, and the most proximal 5′ as well as distal 3′ U2 splice sites were predominantly selected, without overexpression of RBP45. This can suggest that within the CBP20 midi-gene transcripts are sequences with high affinity to RBP45 or other yet-unidentified tobacco RBP45-like proteins that can stimulate selection of the most external 5′ and 3′ splice sites.
Interestingly, the skipping effects that were observed during splicing of the midi-gene constructs were not detected when full-length CBP20 gene variants (the maxi-genes series) were tested in transgenic plants. The wt transgene transcripts were correctly spliced, and removing the U12 intron (the ΔU12 CBP20 gene version) did not change the correct splicing pattern. The splicing pattern was changed, however, when the position of the U12 was altered (moved to the core or tail part of the CBP20 gene), and a U2 intron replaced the original Arabidopsis U12 intron. In transgenic lines expressing these two constructs, incorrect splicing of the U2 intron, which served in the tested gene variants as intron no. 4, was detected. This is interesting since the same intron surrounded only by two U2 introns (a midi-gene) was spliced correctly, without any alternative events. Thus, in context of the whole CBP20 gene, the U12 intron present upstream or downstream of its natural location induced selection of alternative splice sites. Moreover, the exon-swapping version of the CBP20 gene (exons no. 4 and no. 5 were swapped) caused alterations in splice site selection of the U12 intron. These results show that sequences of both exons no. 4 and no. 5, as well as location of the U12 intron, are necessary for the proper maturation of A. thaliana CBP20 pre-mRNA. However, in all maxi-gene constructs analyzed, the levels of their transcripts were comparable to those of the wt (Figure 6A), suggesting that the alternative splicing events detected in some CBP20 gene variants do not dramatically influence the levels of primary transcripts produced from the transgenes used in this study. Surprisingly, the protein level of CBP20 originating from the U12→U2 construct was lower than that expressed from the wt transgene. This low level of CBP20 was observed in two independent transgenic lines. Less CBP20 was also observed in the U12 in core and U12 in tail transgenes, and as expected, no CBP20 was detected in the exon swapping. These differences in the levels of CBP20, which are not supported by alterations in CBP20 gene expression at the mRNA level, suggest that mature mRNAs originating from some of our constructs are partially retained in the nucleus and are not exported to the cytoplasm to be used as templates in translation. Further studies must be carried out to fully uncover the connection between U12 intron splicing, mRNA export and translation in the cytoplasm.
MP and KK designed and performed the experiments, analyzed the data, and wrote the manuscript. DB and JD performed the experiments and contributed to data analysis. MS and WK conducted the bioinformatic analyses. AJ advised on experiments and data analysis and assisted in drafting the manuscript. ZS-K conceived the study contributed to data interpretation and participated in the manuscript writing. All authors read and approved the final manuscript.
This work was supported by the National Science Centre project UMO-2013/11/B/NZ1/02099 and the Faculty of Biology at Adam Mickiewicz University in Poznan, Poland (Badania Statutowe). Funding for open access charge: Polish Ministry of Science and Higher Education [01/KNOW2/2014].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00475/full#supplementary-material
Alioto, T. S. (2007). U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 35, D110–D115. doi: 10.1093/nar/gkl796
Bartschat, S., and Samuelsson, T. (2010). U12 type introns were lost at multiple occasions during evolution. BMC Genomics 11:106. doi: 10.1186/1471-2164-11-106
Basu, M. K., Makalowski, W., Rogozin, I. B., and Koonin, E. V. (2008). U12 intron positions are more strongly conserved between animals and plants than U2 intron positions. Biol. Direct. 3:19. doi: 10.1186/1745-6150-3-19
Benson, D. A., Karsch-Mizrachi, I., Clark, K., Lipman, D. J., Ostell, J., and Sayers, E. W. (2012). GenBank. Nucleic Acids Res. 40, D48–D53. doi: 10.1093/nar/gkr1202
Burge, C. B., Padgett, R. A., and Sharp, P. A. (1998). Evolutionary fates and origins of U12-type introns. Mol. Cell. 2, 773–785. doi: 10.1016/S1097-2765(00)80292-0
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Dias, S. M., Wilson, K. F., Rojas, K. S., Ambrosio, A. L., and Cerione, R. A. (2009). The molecular basis for the regulation of the cap-binding complex by the importins. Nat. Struct. Mol. Biol. 16, 930–937. doi: 10.1038/nsmb.1649
Hugouvieux, V., Kwak, J. M., and Schroeder, J. I. (2001). An mRNA cap binding protein, ABH1, modulates early abscisic acid signal transduction in Arabidopsis. Cell 106, 477–487. doi: 10.1016/S0092-8674(01)00460-3
Izaurralde, E., Lewis, J., McGuigan, C., Jankowska, M., Darzynkiewicz, E., and Mattaj, I. W. (1994). A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell 78, 657–668. doi: 10.1016/0092-8674(94)90530-4
Jäger, K., Fábián, A., Tompa, G., Deák, C., Höhn, M., Olmedilla, A., et al. (2011). New phenotypes of the drought-tolerant cbp20 Arabidopsis thaliana mutant have changed epidermal morphology. Plant Biol. 13, 78–84. doi: 10.1111/j.1438-8677.2010.00343.x
Jung, H. J., and Kang, H. (2014). The Arabidopsis U11/U12-65K is an indispensible component of minor spliceosome and plays a crucial role in U12 intron splicing and plant development. Plant J. 78, 799–810. doi: 10.1111/tpj.12498
Kierzkowski, D., Kmieciak, M., Piontek, P., Wojtaszek, P., Szweykowska-Kulinska, Z., and Jarmolowski, A. (2009). The Arabidopsis CBP20 targets the cap-binding complex to the nucleus, and is stabilized by CBP80. Plant J. 59, 814–825. doi: 10.1111/j.1365-313X.2009.03915.x
Kim, S., Yang, J. Y., Xu, J., Jang, I. C., Prigge, M. J., and Chua, N. H. (2008). Two cap-binding proteins CBP20 and CBP80 are involved in processing primary MicroRNAs. Plant Cell Physiol. 49, 1634–1644. doi: 10.1093/pcp/pcn146
Kim, W. Y., Jung, H. J., Kwak, K. J., Kim, M. K., Oh, S. H., Han, Y. S., et al. (2010). The Arabidopsis U12-type spliceosomal protein U11/U12-31K is involved in U12 intron splicing via RNA chaperone activity and affects plant development. Plant Cell 22, 3951–3962. doi: 10.1105/tpc.110.079103
Kmieciak, M., Simpson, C. G., Lewandowska, D., Brown, J. W., and Jarmolowski, A. (2002). Cloning and characterization of two subunits of Arabidopsis thaliana nuclear cap-binding complex. Gene 283, 171–183. doi: 10.1016/S0378-1119(01)00859-9
Kong, X., Ma, L., Yang, L., Chen, Q., Xiang, N., Yang, Y., et al. (2014). Quantitative proteomics analysis reveals that the nuclear cap-binding complex proteins arabidopsis CBP20 and CBP80 modulate the salt stress response. J. Proteome Res. 1, 2495–2510. doi: 10.1021/pr4012624
Kwak, K. J., Jung, H. J., Lee, K. H., Kim, Y. S., Kim, W. Y., and Ahn, S. J. (2012). The minor spliceosomal protein U11/U12-31K is an RNA chaperone crucial for U12 intron splicing and the development of dicot and monocot plants. PLoS One 7:e43707. doi: 10.1371/journal.pone.0043707
Kwak, K. J., Kim, B. M., Lee, K., and Kang, H. (2017). quatre-quart1 is an indispensable U12 intron-containing gene that plays a crucial role in Arabidopsis development. J. Exp. Bot. 68, 2731–2739. doi: 10.1093/jxb/erx138
Laubinger, S., Sachsenberg, T., Zeller, G., Busch, W., Lohmann, J. U., Rätsch, G., et al. (2008). Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 8795–8800. doi: 10.1073/pnas.0802493105
Lewandowska, D., Simpson, C. G., Clark, G. P., Jennings, N. S., Barciszewska-Pacak, M., Lin, C. F., et al. (2004). Determinants of plant U12-dependent intron splicing efficiency. Plant Cell 16, 1340–1352. doi: 10.1105/tpc.020743
Lin, C. F., Mount, S. M., Jarmołowski, A., and Makałowski, W. (2010). Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol. Biol. 10:47. doi: 10.1186/1471-2148-10-47
Lorković, Z. J., Wieczorek Kirk, D. A., Klahre, U., Hemmings-Mieszczak, M., and Filipowicz, W. (2000). RBP45 and RBP47, two oligouridylate-specific hnRNP-like proteins interacting with poly(A)+ RNA in nuclei of plant cells. RNA 6, 1610–1624. doi: 10.1017/S1355838200001163
Marquez, Y., Brown, J. W., Simpson, C., Barta, A., and Kalyna, M. (2012). Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 22, 1184–1195. doi: 10.1101/gr.134106.111
Mazza, C., Segref, A., Mattaj, I. W., and Cusack, S. (2002). Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21, 5548–5557. doi: 10.1093/emboj/cdf538
Niemelä, E. H., and Frilander, M. J. (2014). Regulation of gene expression through inefficient splicing of U12-type introns. RNA Biol. 11, 1325–1329. doi: 10.1080/15476286.2014.996454
Papp, I., Mur, L. A., Dalmadi, A., Dulai, S., and Koncz, C. (2004). A mutation in the cap binding protein 20 gene confers drought tolerance to Arabidopsis. Plant Mol. Biol. 55, 679–686. doi: 10.1007/s11103-004-1680-2
Patel, A. A., McCarthy, M., and Steitz, J. A. (2002). The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J. 21, 3804–3815. doi: 10.1093/emboj/cdf297
Pieczynski, M., Marczewski, W., Hennig, J., Dolata, J., Bielewicz, D., Piontek, P., et al. (2013). Down-regulation of CBP80 gene expression as a strategy to engineer a drought-tolerant potato. Plant Biotechnol. J. 11, 459–469. doi: 10.1111/pbi.12032
Raczynska, K. D., Simpson, C. G., Ciesiolka, A., Szewc, L., Lewandowska, D., McNicol, J., et al. (2010). Involvement of the nuclear cap-binding protein complex in alternative splicing in Arabidopsis thaliana. Nucleic Acids Res. 38, 265–278. doi: 10.1093/nar/gkp869
Ren, G., and Yu, B. (2012). Critical roles of RNA-binding proteins in miRNA biogenesis in Arabidopsis. RNA Biol. 9, 1424–1428. doi: 10.4161/rna.22740
Sierocka, I., Rojek, A., Bielewicz, D., Karlowski, W., Jarmolowski, A., and Szweykowska-Kulinska, Z. (2011). Novel genes specifically expressed during the development of the male thalli and antheridia in the dioecious liverwort Pellia endiviifolia. Gene 485, 53–62. doi: 10.1016/j.gene.2011.06.012
Simpson, C. G., and Brown, J. W. (2008). U12-dependent intron splicing in plants. Curr. Top. Microbiol. Immunol. 326, 61–82. doi: 10.1007/978-3-540-76776-3_4
Simpson, G. G., and Filipowicz, W. (1996). Splicing of precursors to mRNA in higher plants: mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 32, 1–41. doi: 10.1007/BF00039375
Szarzynska, B., Sobkowiak, L., Pant, B. D., Balazadeh, S., Scheible, W. R., Mueller-Roeber, B., et al. (2009). Gene structures and processing of Arabidopsis thaliana HYL1-dependent pri-miRNAs. Nucleic Acids Res. 37, 3083–3093. doi: 10.1093/nar/gkp189
Turunen, J. J., Niemelä, E. H., Verma, B., and Frilander, M. J. (2013). The significant other: splicing by the minor spliceosome. Wiley Interdiscip. Rev. RNA 4, 61–76. doi: 10.1002/wrna.1141
Keywords: U12 introns, U2 introns, mRNA splicing, CBP20, Arabidopsis thaliana
Citation: Pieczynski M, Kruszka K, Bielewicz D, Dolata J, Szczesniak M, Karlowski W, Jarmolowski A and Szweykowska-Kulinska Z (2018) A Role of U12 Intron in Proper Pre-mRNA Splicing of Plant Cap Binding Protein 20 Genes. Front. Plant Sci. 9:475. doi: 10.3389/fpls.2018.00475
Received: 12 January 2018; Accepted: 27 March 2018;
Published: 16 April 2018.
Edited by:
Dora Szakonyi, Instituto Gulbenkian de Ciência (IGC), PortugalReviewed by:
Song Li, Duke University, United StatesCopyright © 2018 Pieczynski, Kruszka, Bielewicz, Dolata, Szczesniak, Karlowski, Jarmolowski and Szweykowska-Kulinska. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Artur Jarmolowski, YXJ0amFybW9AYW11LmVkdS5wbA== Zofia Szweykowska-Kulinska, em9mc3p3ZXlAYW11LmVkdS5wbA==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.