- 1Key Laboratory of Growth and Developmental Regulation for Protected Vegetable Crops of Beijing, Department of Vegetable Science, College of Horticulture, China Agricultural University, Beijing, China
- 2Changping Agricultural Technology Service Center, Beijing, China
Cucumber (Cucumis sativus L.) is threatened by substantial yield losses due to the south root-knot nematode (Meloidogyne incognita). However, understanding of the molecular mechanisms underlying the process of nematode infection is still limited. In this study, we found that M. incognita infection affected the structure of cells in cucumber roots and treatment of the cytoskeleton inhibitor (cytochalasin D) reduced root-knot nematode (RKN) parasitism. It is known that Actin-Depolymerizing Factor (ADF) affects cell structure, as well as the organization of the cytoskeleton. To address the hypothesis that nematode-induced abnormal cell structures and cytoskeletal rearrangements might be mediated by the ADF genes, we identified and characterized eight cucumber ADF (CsADF) genes. Phylogenetic analysis showed that the cucumber ADF gene family is grouped into four ancient subclasses. Expression analysis revealed that CsADF1, CsADF2-1, CsADF2-2, CsADF2-3 (Subclass I), and CsADF6 (Subclass III) have higher transcript levels than CsADF7-1, CsADF7-2 (Subclass II genes), and CsADF5 (Subclass IV) in roots. Members of subclass I genes (CsADF1, CsADF2-1, CsADF2-2, and CsADF2-3), with the exception of CsADF2-1, exhibited a induction of expression in roots 14 days after their inoculation (DAI) with nematodes. However, the expression of subclass II genes (CsADF7-1 and CsADF7-2) showed no significant change after inoculation. The transcript levels of CsADF6 (Subclass III) showed a specific induction at 21 DAI, while CsADF5 (Subclass IV) was weakly expressed in roots, but was strongly up-regulated as early as 7 DAI. In addition, treatment of roots with cytochalasin D caused an approximately 2-fold down-regulation of the CsADF genes in the treated plants. These results suggest that CsADF gene mediated actin dynamics are associated with structural changes in roots as a consequence of M. incognita infection.
Introduction
Root-knot nematodes (Meloidogyne spp., RKN) are one of the most destructive pathogens of vegetables, even low nematode levels can cause high yield losses (Mukhtar et al., 2013; Liu et al., 2015). The infective second-stage juveniles (J2s) of nematodes penetrate plant roots and migrate into the vascular cylinder toward the zone of differentiation. The J2s do not kill parasitized cells, but induce the generation of some giant cells as the sole nutritive source by expansion of parenchyma cells in the root vascular tissue (Jones et al., 2013; Molinari et al., 2014). During giant cell expansion, the organization of the actin cytoskeleton is significantly altered and permanent rearranged, showing large numbers of thick actin bundles and cables throughout the cell cortex (de Almeida Engler et al., 2004; Clément et al., 2009). These actin cables within giant cells may be required to guide the vesicle trafficking that is needed for extensive plasma membrane and cell wall biogenesis during their isotropic growth (Favery et al., 2004).
The plant actin cytoskeleton undergoes a striking reorganization in response to internal and external signals and is involved in different cellular processes essential for plant development (Clément et al., 2009). In response to multiple cellular processes, a range of actin binding proteins (ABPs) can dynamically reorganize and remodele the actin cytoskeleton (Ayscough, 1998; Hussey et al., 2006). The turnover of filamentous actins are regulated by members of the actin-depolymerizing factor (ADF) or cofilin family (Staiger et al., 1997; Carlier, 1998; Maciver and Hussey, 2002). The ADF proteins bind G-/F-actin and sever the actin filaments to increase actin turnover (Carlier et al., 1997; Maciver, 1998; Chen et al., 2000; Andrianantoandro and Pollard, 2006; Pavlov et al., 2007).
Cucumber (Cucumis sativus L.) is a good source of vitamins, minerals, fiber, and roughage (Mukhtar et al., 2013; Zhang et al., 2014b), which is one of the reasons why it is grown all over the world. However, this popular vegetable is threatened by tremendous yield losses from Meloidogyne incognita, a nematode that attacks cucumber roots, inducing giant cell formation (Wehner et al., 1991; Walters et al., 1993; Mukhtar et al., 2013). A clearer understanding of how M. incognita affects cucumber roots, the effects of infection and the associated changes in host genes expression would be of considerable value in developing strategies to prevent such attacks; however, to date such research has been limited.
The cucumber genomic sequence that provides an opportunity to study the nature of the structure, organization and expression of the constituent gene families (Huang et al., 2009). In this current study, we identified the cucumber ADF family and compared similarities with sequences to the corresponding orthologs in Arabidopsis thaliana. We found ample information on the chromosomal locations, genomic structures, and the expression of cucumber ADF genes after nematode infection or cytoskeleton inhibitor treatment, which leads us to a better understanding of the relationships between members of the cucumber ADF family and nematode infection.
Materials and Methods
Plant Material and Cytoskeleton Inhibitors Treatment
Seeds of cucumber (C. sativus) inbred line 3461 (China Agricultural University) were used in the experiments. Cucumber seeds were surface sterilized and sown in a autoclaved soil (50 nutritive soil and 50 % vermiculite) at 26 ± 2°C in a greenhouse. Single cucumber seedlings with two leaves were transplanted into a 15-cm diameter plastic pot filled with steam-sterilized river sand and randomly placed in a greenhouse, provided with a regular regime of 16 h of light/day and temperature maintained between 26 and 30°C. Plants were regularly watered with Hoagland's solution. For the cytochalasin D treatments, a previously reported method was used (de Almeida Engler et al., 2004), with minor modifications. Individual plants were pre-treated on medium with 2 μM cytochalasin D (Sigma, Beijing, China) for 1 day before being transplanted into 15-cm diameter plastic pots, after that 50 ml 0.5 μM cytochalasin D was added every day at 9 am.
Nematode Inoculation and Determination of Infestation Levels
An M. incognita race 3 from Institute of Plant Protection, China Academy of Agricultural Sciences population had been reared previously in a glasshouse on susceptible cucumber plants (Liu et al., 2015). The progression of nematode infection is shown in Supplementary Figure 1. Nematode inoculation and determination of infestation levels were as previously described (Bybd et al., 1983; Molinari et al., 2014). Nematodes were distinguished as motile vermiform individuals (J2s), swollen individuals that had become sedentary (third and fourth stages, SJs) and adult females (AFs) (Molinari et al., 2014).
Paraffin Section of Roots and Galls
Paraffin embedding technique was used to achieved the paraffin section of roots and galls. Cucumber roots and galls at different time were fixed, embedded, and sectioned as previously described (Kerk et al., 2003; Zhang et al., 2013).
Transmission Electron Microscopy
Cucumber roots and galls were fixed, embedded, and sectioned as previously described (Liu et al., 2016). Thin cross-sections were made with a UC6I microtome (Leica, Germany) and examined with a JEM-123O scanning transmission electron microscope (JEOL, Japan).
Sequence Alignment and Molecular Phylogenetic Analysis
Sequence information for 11 A. thaliana ADF genes were retrieved from previous study (Ruzicka et al., 2007). To assess the corresponding cucumber orthologs, a basic local alignment search tool (BLAST) was performed for each one of the A.thaliana ADF protein sequences against the Cucumber Genome Database (http://cucumber.genomics.org.cn/page/cucumber/index.jsp). Those with the highest amino acid percentage identity were selected. Cucumber ADFs were named according to the annotation of the Cucumber Genome Database. The multiple sequence alignment of CsGID1s and related CsGID1 proteins was performed using ClustalW within the MEGA5 software package (Tamura et al., 2011), and the highlighting conserved sequences were drawn by using BoxShade (http://www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method (Saitou and Nei, 1987) with the Poisson model and 1000 bootstrap replicates test by MEGA5 (Liu et al., 2016). The gene numbers corresponding to every CsADF gene in the Cucumber Genome Database are shown in Supplementary Table 1. The PCR primers used for cloning CsADF genes are listed in Supplementary Table 2.
Subcellular Localization
The fusion 35S::CsADF2-3::GFP, 35S:: CsADF7-1::GFP, 35S::CsADF6::GFP, 35S::CsADF5:GFP constructs and the control plasmid of 35S::GFP were transformed into onion epidermal cells as previously described (Zhang et al., 2014a). The PCR primers used for cloning CsADF genes are listed in Supplementary Table 2.
RNA Isolation and qRT-PCR
Total RNA was extracted from roots or galls using the SV Total RNA Isolation System (Promega, Beijing, China), and cDNA was synthesized using the MultiScribe™ reverse transcriptase kit (Applied Biosystems, Beijing, China). Quantitative real-time RT-PCR (qRT-PCR) was performed as previously described (Liu et al., 2016), The gene specific primers used are listed in Supplementary Table 2.
Results
Cloning of ADF Genes in Cucumber
Using A.thaliana ADFs protein sequences, we identified and named eight candidates ADF genes based on a BLAST search of the Cucumber Genome Database (http://cucumber.genomics.org.cn/page/cucumber/index.jsp). All ADF genes from the cucumber were anchored to cucumber chromosomes, the chromosomal locations and the scaffold sequence directions of the 8 CsADF genes were analyzed using BLASTN. Further analysis of the cucumber ADF genes revealed that they were distributed on five of the seven cucumber chromosomes and none of the CsADF gene was found on chromosomes 6 and 7 (Figure 1A). We observed homologous CsADF genes located on different chromosomes in cucumber, suggesting that duplicated events were potentially involved in the evolution of cucumber CsADFs.
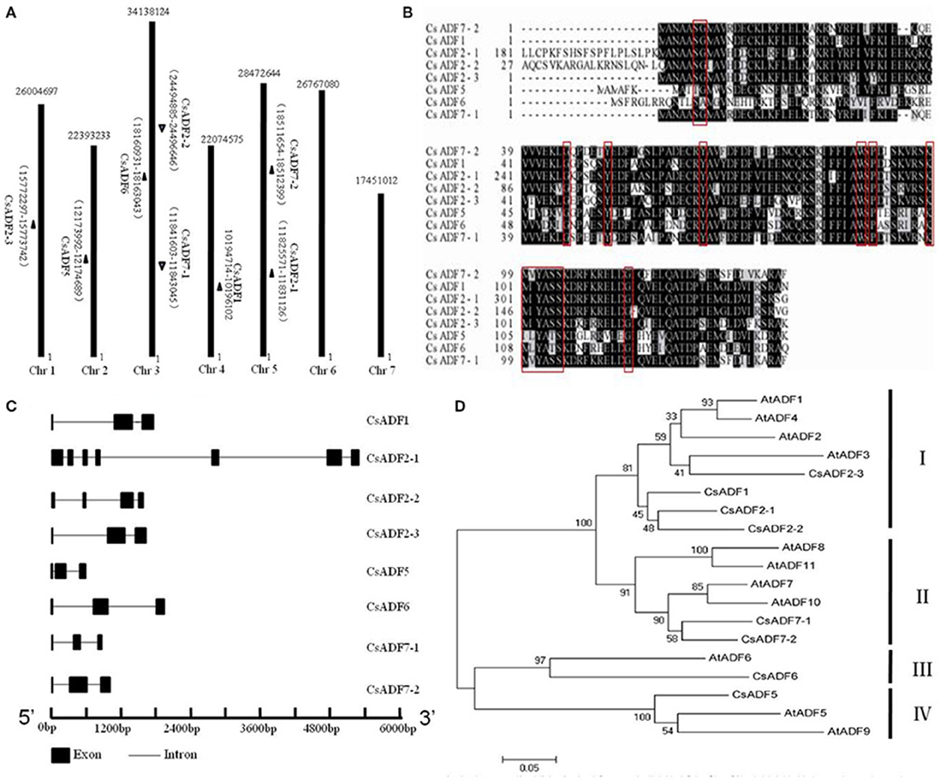
Figure 1. Cloning of cucumber ADF (CsADF) genes. (A) Genomic localization of CsADF genes. The arrows next to the gene names show the direction of the scaffold. (B) Multiple sequence alignment of eight CsADF proteins. The red boxes indicate the ADF-H domains. (C) Genomic organization of CsADF genes with exons (bars) and introns (lines). (D) Phylogenetic tree of ADF protein sequences from Arabidopsis thaliana (AtADF proteins) and cucumber CsADF proteins. Neighbor-joining protein sequence phylogeny showing all 8 ADF proteins from cucumber (Cs) and Arabidopsis (At). Bootstrap values are shown on the branch points of the tree. Subclasses I to IV are indicated by vertical bars on the right.
Previous studies have shown that ADF genes contain a conserved region called the H-domain (Lappalainen et al., 1998), In cucumber, it is present in all ADF genes (Figure 1B). As was reported to be the case with A. thaliana ADF orthologous genes, all CsADFs was predicted to contain three exons and two introns, with the exception of CsADF2-1 (having seven exons and six introns) and CsADF2-2 (having four exons and three introns; Figure 1C). Subsequently we further analyzed the phylogenetic relationship with A. thaliana. The results revealed that the cucumber ADF gene family is grouped phylogenetically into four ancient subclasses supported by the existence of homologous ADF protein sequences from each subclass in the A. thaliana. (Figure 1D). For example, CsADF6 are significantly more related in protein sequence to Arabidopsis ADF6 in subclass III than other cucumber ADF genes.
Subcellular Localization of CsADF Proteins
To further determine the subcellular localization of CsADF proteins, sequences corresponding to 35S::CsADF2-3::GFP (subclass I), 35S::CsADF7-1::GFP (subclass II), 35S::CsADF6:GFP (subclass III) and 35S::CsADF5:GFP (subclass IV) fusion proteins were constructed and transiently expressed in onion epidermal using particle bombardment (Zhang et al., 2014a). Transformation with the 35S:GFP control construct resulted in GFP localization throughout the cells (Figures 2A–C), while green fluorescence resulting from expression of each of the four ADF-GFP fusion proteins was localized in both the cytoplasm and the nucleus, with a characteristic filamentous appearance (Figures 2D–O), consistent with a previous report (Tang et al., 2016).
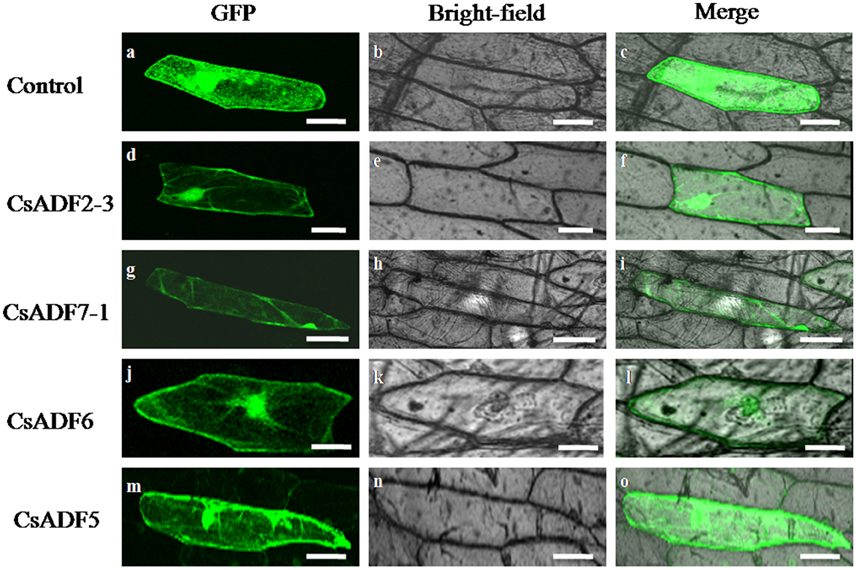
Figure 2. Subcellular localization of CsADF2-3, CsADF7-1, CsADF5 and CsADF6 in onion epidermal cells. Green fluorescent protein (GFP) (A–C), CsADF2-3:GFP (D–F), CsADF7-1:GFP (G–I), CsADF5:GFP (J-L) or CsADF6:GFP (M-O) fusion proteins were transiently expressed in onion epidermal cells under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Bar = 50 μm.
Expression of CsADF Genes in M. incognita Induced Galls
Transcript expression analysis by a quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) experiment revealed that all CsADF genes are expressed in roots. Among them, CsADF1, CsADF2-1, CsADF2-2, CsADF2-3 (Subclass I genes), and CsADF6 (Subclass III gene) have a higher expression, while CsADF7-1, CsADF7-2 (Subclass II genes), and CsADF5 (Subclass IV gene) have a lower transcript (Figure 3A). To investigate whether M. incognita infection correlates with changes in the transcription of ADF genes, we quantified the expression of eight ADF isotypes using qRT-PCR in roots and galls 7, 14, and 21 days after inoculation with 300 M. incognita active J2s (DAI; Figure 3B). The transcript levels of five isotypes (CsADF1, CsADF2-2, CsADF2-3, CsADF5, and CsADF6) were higher in galls of the infected roots compared with those of the control. Members of subclass I genes (CsADF1, CsADF2-1, CsADF2-2, and CsADF2-3) except CsADF2-1 exhibited a specific induction at 14 DAI. The expression of subclass II genes (CsADF7-1 and CsADF7-2) showed no significant change. CsADF6 (Subclass III gene) showed a specific induction at 21 DAI. It's interesting that CsADF5 (Subclass IV gene) was weakly expressed in root, but its transcript level was remarkably up-regulated as early as 7 DAI.
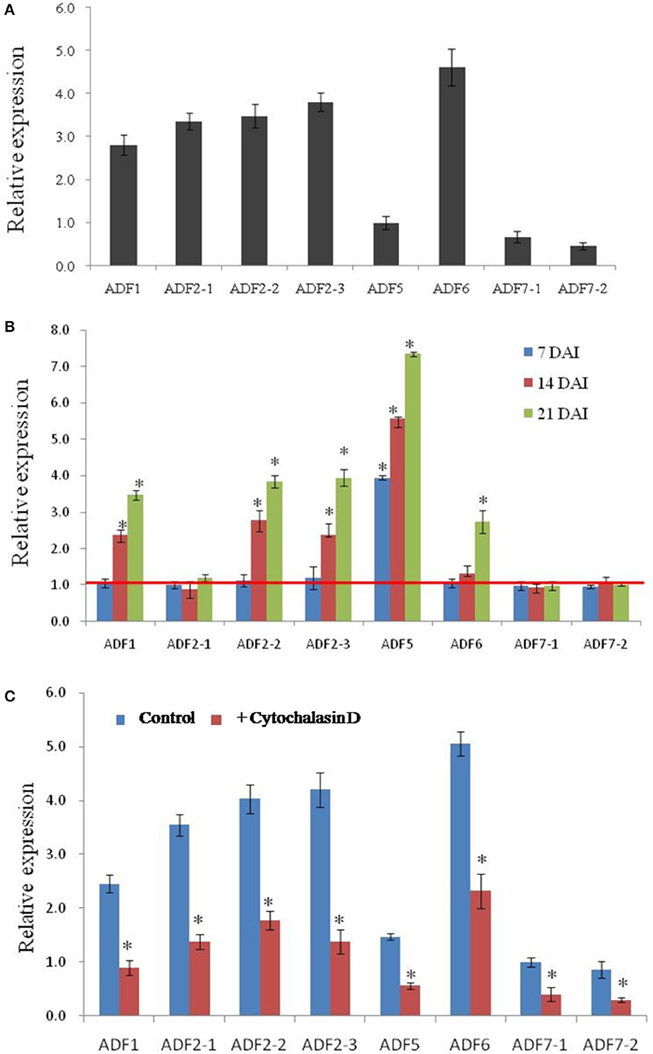
Figure 3. Analysis of CsADF gene expression levels in uninfected roots and galls, as well as in roots, after cytochalasin D treatment. (A) Quantitative real time-PCR analysis of the transcript abundance of CsADF genes in cucumber uninfected roots. (B) Relative amount of transcripts of the CsADF1, CsADF2-1, CsADF2-2, CsADF2-3, CsADF5, CsADF6, CsADF7-1, and CsADF7-2 genes in cucumber galls, 7 (blue bars), 14 (red bars), and 21 (green bars) days after infection (DAI) with the nematode, M. incognita, compared with uninfected conditions. The ratio was calculated as 2 −(ΔCT gene of interest (CT uninfected−CT infected)/Δ CT reference gene(CT uninfected−CT infected), where the gene of interest corresponds to one of the analyzed cucumber ADF genes and the reference gene is TUA (Csa021066). The ratio equals 1, meaning that the ADF gene expression is not affected by nematode infection. Three biological replicates, represented by three independent quantitative RT-PCR reactions, were performed per sample. The bars (SE) represent mean values of three independent experiments. Statistical significance of the differences between dissected galls and controls (non-meristematic root fragments from non-infected plants) were determined by the Wilcoxon test (*P < 0.05). (C) The bars represent ±SD (n = 3). Asterisks indicate that the transcripts are significantly down-regulated by cytochalasin D (Wilcoxon test, P < 0.05).
The Expression of CsADF Genes were Reduced after Cytoskeletal-Inhibitor Application
Previous study showed that microtubules and actin filaments appeared partially depolymerized throughout the feeding site development (de Almeida Engler et al., 2004), and our results indicated that nematode infection induced the expression of five CsADF genes (Figure 3B). Given that cytoskeletal-inhibitor treatments result in the cessation of the development of normal giant cells in Arabidopsis (de Almeida Engler et al., 2004), we hypothesized that cytoskeleton inhibitors are likely to reduce the expression of CsADF genes. This idea was supported by the qRT-PCR experiment revealing that an approximately 2-fold down-regulation of the CsADF genes occurred in roots of treated plants (Figure 3C).
Treatment of Cucumber Roots with the Cytoskeleton Inhibitor Cytochalasin D Affects Cell Structure and Reduces RKN Parasitism
As shown in Figure 4A, infection with M. incognita resulted in cucumber root galls; however, in cytochalasin D-treated plantlets, the galls were smaller than in untreated controls. A detailed morphological comparison was performed of nematode infected roots and controls (Figures 4B–I). Cells in both longitudinal and cross sections of control roots were regularly arranged (Figures 4B,D), but in nematode infected root galls, most of the cells were less organized and giant cells were induced at feeding sites (Figures 4C,E). To provide higher resolution of cellular structures, root cells were examined by transmission electron microscopy (TEM) (Figures 4F,G). We focused on variation in the appearance of the cell walls and plasma membrane as several studies have shown that their morphology is particularly important for understanding giant cell development (Jones and Dropkin, 1975, 1976; Jones and Payne, 1978). In the control roots, each cell had a distinct and normal cell wall (Figure 4F), while in the infected roots, ingrowths and degradation occurred on the walls of the giant cells, such that the wall itself was sparse (Figure 4G). The numerous ingrowths greatly increased the surface area of the plasma membrane and were most pronounced adjacent to the vascular tissue, which is a potential site of nutrient influx into giant cells (Baldwin, 1978). Treatment of uninfected roots with cytochalasin D caused cellular defects (Figure 4D), such as a wider root central cylinder and larger cell files. Giant cells in roots at the same stage of infected seedling treated with cytochalasin D were much smaller than those of uninfected and treated roots (Figure 4I) and there were fewer of them than in untreated plants (Figure 4E).
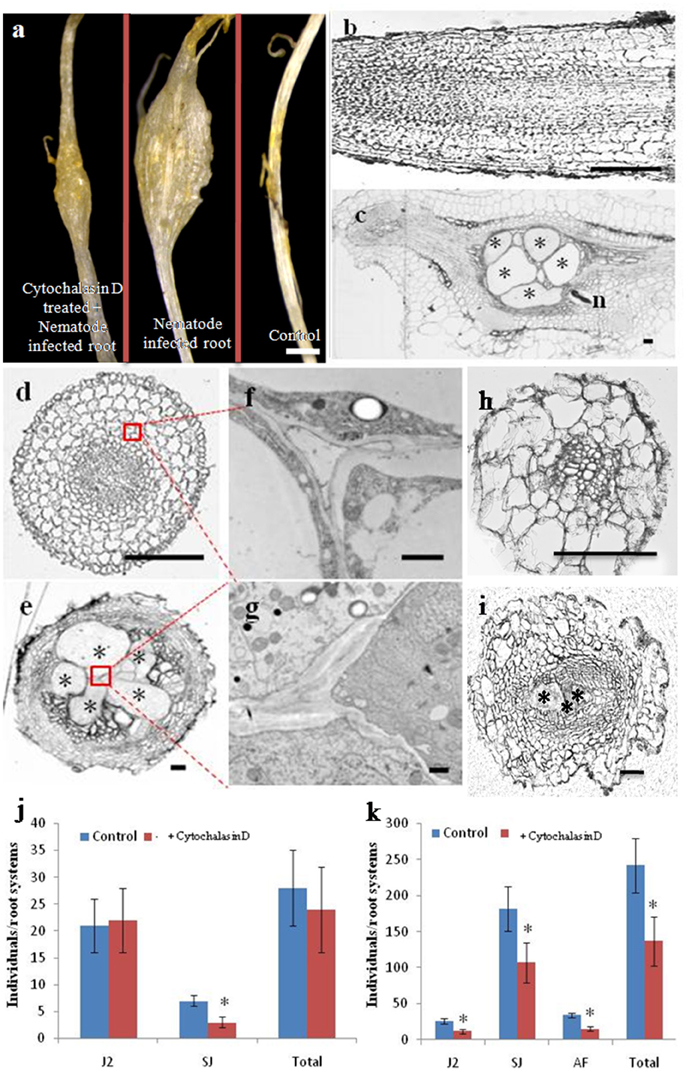
Figure 4. Cytochalasin D treatment reduces the number of giant cells, as well as RKN parasitism. Morphological analysis of cucumber roots infected by the nematode M. incognita. (A) Morphology of cytochalasin D treated and nematode infected, nematode infected, or control roots, Bar = 1 mm. (B,C) Longitudinal sections of control and nematode infected roots, Bar = 200 um. (D,E) Cross sections of control and nematode infected roots, Bar = 200 um. (F,G) Enlarged transmission electron micrographs of the boxes in (D,E), respectively, Bar = 1 um. (H,I) Cross sections of control and nematode infected roots in the presence of cytochalasin D, Bar = 200 μm. Samples in (A–I) were collected 21 days after infection. Asterisk indicates giant cells. n indicates nematode. (J,K) Number of nematodes that penetrated cucumber roots of untreated (Control) and cytochalasin D -treated (+ cytochalasin D) plants, 7 days (J) and 21 days (K) after inoculation with 300 M. incognita invasive juveniles (J2s). Motile juveniles (J2s), sedentary juveniles (SJs) and adult females (AFs) were determined. Values are expressed as averages (obtained from three biological replicates) of the numbers of individuals per root system ±SD (n = 20). Asterisks indicate that the means are significantly different, as determined by a t-test (P < 0.05).
It has been reported that cytochalasin D treatment blocks cytoskeleton dynamics, resulting in the cessation of development of normal giant cell in Arabidopsis (de Almeida Engler et al., 2004). We investigated whether cytochalasin D treatment similarly affected RKN parasitism during a compatible interaction with cucumber. Individual plants were pre-treated on media with high concentrations of cytochalasin D (2 μM) for 1 day, after which they were transferred to soil and inoculated with 300 active J2s. Next, 50 ml 0.5 μM cytochalasin D was added every day. Seven and 21 days after inoculation, we counted the number of nematodes that had penetrated the roots (Figures 4J,K). At day 7 after inoculation, there was no significant difference in total nematode number and the numbers of motile invasive forms (J2s) between untreated and cytochalasin D-treated plants (Figure 4J). We observed very few invasive juveniles as they had become sedentary in the roots of untreated plants. However, even fewer sedentary juveniles (SJs) were present in the cytochalasin D treated plants (Figure 4J). Twenty-one days after inoculation, the number of juveniles developing in the roots increased substantially, whereas the presence of motile invasive forms was reduced to a small fraction. However, the roots of cytochalasin D treated plants exhibited a significant reduction in the number of both J2s and SJs compared with untreated plants (Figure 4K). Although very few AFs were present at this stage, no differences were evident between treated and untreated plants. In general, the number of juveniles that penetrated the roots 21 days after inoculation was approximately two-thirds of the number of inoculated juveniles in the untreated plants and one-half of the number of inoculated juveniles in the treated plants.
Discussion
Although breeders looked for RKN -resistant cucumber species from many years ago, none of cucumber species was found immune or highly resistant to M. incognita (Wehner et al., 1991; Walters et al., 1993; Mukhtar et al., 2013). Moreover, molecular mechanism on how nematodes inducing giant cell formation and expansion in cucumber root is still unclear.
Gene expression patterns and protein subcellular localization and can provide important clues as to their function, and so we investigated the subcellular localization of the CsADF proteins. As shown in Figure 2, heterologously expressed GFP proteins fused to CsADF2-3 (subclass I), CsADF7-1 (subclass II), CsADF6 (subclass III) and CsADF5 (subclass IV) were detected in both the cytoplasm and nucleus, in the form of filaments, which is consistent with previous studies of ADF proteins (Tang et al., 2016).
It has been reported that cytoskeletal changes in giant cells are triggered by the depolymerization of F-actin and microtubules (MTs) (Caillaud et al., 2008; Clément et al., 2009), and that this depolymerization may cause a decrease in the viscosity of the cytoplasm (Gross et al., 1993; de Almeida Engler et al., 2004). The actin depolymerizing factor (ADF)/cofilin family has emerged as a central regulator of actin turnover (Augustine et al., 2008). Biochemical studies show that ADF/cofilin is capable of severing and depolymerizing actin filaments from their pointed ends (Carlier et al., 1997; Andrianantoandro and Pollard, 2006)., and the relaxation of the cytoskeleton in nematode feeding sites may open the way for a successful infection (de Almeida Engler et al., 2004). Analysis of the transcription profiles of eight cucumber ADF genes revealed that in the course of infection over 21 days, a specific ADF family expression signature was observed. Quantitative RT-PCR results showed that the transcription of five genes (ADF1, 2–2, 2–3, 5, and 6) was significantly increased in nematode-induced galls (Figure 3B). Out of these five, ADF5 was induced from early stages of infection (7 DAI); ADF1, ADF2–2, and ADF2–3 were induced from the middle stages of infection (14 DAI); ADF6 was induced from the later stages of infection (14–21 DAI). Transcriptional activation of ADF genes suggests that more ADF or cofilin family may be needed to rearrangement the cytoskeleton in the large galls. These results above revealed that ADF genes possibly facilitate nematodes feeding in cucumber root.
Cytochalasin D is an alkaloid compound that inhibits globular actin addition and loss from actin filaments, resulting in a fragmented ACT cytoskeleton (Sampath and Pollard, 1991). Studies with A. thaliana indicated that extended treatments with low concentrations (0.5 μM) of cytochalasin D do not prevent root growth, and that nematodes are able to complete their life cycle in the treated roots (Sampath and Pollard, 1991; de Almeida Engler et al., 2004). Moreover, it was reported that microtubules and actin filaments appeared to be partially depolymerized throughout the feeding site development (de Almeida Engler et al., 2004), which likely reflects the up-regulated expression of ADF genes. Our results indicate that the transcript levels of the ADF genes were reduced after cytochalasin D application (Figure 3), suggesting that the application of cytochalasin D and nematode infection had opposite effects on the expression of ADF genes.
Giant cells induced by root knot nematodes are large multinucleated feeding cells containing a dense cytoplasm generated during a complex host-parasite association in plant roots (de Almeida Engler et al., 2004). Our morphological analyses showed that giant cells were induced and most of the cells were less organized in nematodes infected root galls (Figure 4), suggesting that the actin cytoskeleton may be dynamically reorganized and remodeled by M. incognita in cucumber roots. In Arabidopsis, nematode feeding involves the retrieval of large cytoplasm volumes (Muller et al., 1981), and a degree of cytoskeleton fragmentation may facilitate the suction and ingestion of cytoplasm during the feeding process of nematodes (de Almeida Engler et al., 2004). In addition, de Almeida Engler et al. (2004) also demonstrated that the cytoskeleton rearrangements and depolymerization induced by parasitic nematodes may be essential for a successful feeding process and the development of galls. Moreover, the expression levels of cytoskeletal components were up-regulated to allow the assembly of a new cytoskeleton in expanding feeding cells (de Almeida Engler et al., 2004). The results presented in Figure 4 indicate that the number of giant cells and RKN parasitism were reduced after cytochalasin D application, from which we infer that the application of cytochalasin D reduces RKN parasitism by affecting the giant cell development.
To conclude, we identified eight cucumber ADF genes that are homologs of A. thaliana ADF genes, and determined that the transcript levels of several ADF genes were affected by M. incognita infection. In addition, we found that the expression of CsADF genes and RKN parasitism were reduced by the application of cytochalasin D. Our results suggest that CsADF gene-mediated actin dynamics may be involved in M. incognita infection of cucumber roots.
Author Contributions
HR conceived the project, BL designed the research; BL, XL, YL, MD, YZ, HL, BZ, CQ, and NZ performed the experiments; HR, BL, and YL analyzed data; BL drafted the manuscript, XL and HR revised the manuscript, all authors approved the manuscript.
Funding
This work was supported by National Public Service Sectors (Agriculture) Project of China (201203003), Beijing Agricultural Innovation Consortium (BAIC01-2016) and Agricultural Scientific and Technological Project (20160415) to HR.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Atrouli Chatterjee and Nathan Lindberg at Cornell University English Language Support Office for the language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01393
Supplementary Table 1. Sequence accession numbers of GID1 genes.
Supplementary Table 2. List of primers and their uses.
Supplementary Figure 1. The developmental of nematodes in infected tissues. (A) Second stage juvenile (J2s) 3 days after inoculation. (B) Sedentary juvenile (SJs) nematode 7 days after inoculation. (C) Sedentary juvenile (SJs) nematode 14 days after inoculation. (D) Adult female nematode 21 days after inoculation. (E) Egg masses 30 days after inoculation. Arrows indicate nematodes or egg masses. Bars = 500 μm.
References
Andrianantoandro, E., and Pollard, T. D. (2006). Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23. doi: 10.1016/j.molcel.2006.08.006
Augustine, R. C., Vidali, L., Kleinman, K. P., and Bezanilla, M. (2008). Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 54, 863–875. doi: 10.1111/j.1365-313X.2008.03451.x
Ayscough, K. R. (1998). In vivo functions of actin-binding proteins. Curr. Opin. Cell Biol. 10, 102–111. doi: 10.1016/S0955-0674(98)80092-6
Baldwin, J. (1978). Giant cells Induced by Nematodes of the Heteroderidae. Nematology Circular, Division of Plant Industry, Florida Department of Agriculture and Consumer Service. Avon Park, FL.
Bybd, D. Jr., Kirkpatrick, T., and Barker, K. (1983). An improved technique for clearing and staining plant tissues for detection of nematodes. J. Nematol. 15, 142.
Caillaud, M.-C., Lecomte, P., Jammes, F., Quentin, M., Pagnotta, S., Andrio, E., et al. (2008). MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20, 423–437. doi: 10.1105/tpc.107.057422
Carlier, M.-F. (1998). Control of actin dynamics. Curr. Opin. Cell Biol. 10, 45–51. doi: 10.1016/S0955-0674(98)80085-9
Carlier, M.-F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G.-X., et al. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322. doi: 10.1083/jcb.136.6.1307
Chen, H., Bernstein, B. W., and Bamburg, J. R. (2000). Regulating actin-filament dynamics in vivo. Trends Biochem. Sci. 25, 19–23. doi: 10.1016/S0968-0004(99)01511-X
Clément, M., Ketelaar, T., Rodiuc, N., Banora, M. Y., Smertenko, A., Engler, G., et al. (2009). Actin-depolymerizing factor2-mediated actin dynamics are essential for root-knot nematode infection of Arabidopsis. Plant Cell 21, 2963–2979. doi: 10. 1105/tpc.109.069104
de Almeida Engler, J., Van Poucke, K., Karimi, M., De Groodt, R., Gheysen, G., Engler, G., et al. (2004). Dynamic cytoskeleton rearrangements in giant cells and syncytia of nematode-infected roots. Plant J. 38, 12–26. doi: 10.1105/tpc.109.069104
Favery, B., Chelysheva, L. A., Lebris, M., Jammes, F., Marmagne, A., De Almeida-Engler, J., et al. (2004). Arabidopsis formin AtFH6 is a plasma membrane–associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16, 2529–2540. doi: 10.1105/tpc.104.024372
Gross, P., Julius, C., Schmelzer, E., and Hahlbrock, K. (1993). Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defence gene activation in infected, cultured parsley cells. EMBO J. 12, 1735.
Huang, S., Li, R., Zhang, Z., Li, L., Gu, X., Fan, W., et al. (2009). The genome of the cucumber, Cucumis sativus L. Nat. Genet. 41, 1275–1281. doi: 10.1038/ng.475
Hussey, P. J., Ketelaar, T., and Deeks, M. J. (2006). Control of the actin cytoskeleton in plant cell growth. Annu. Rev. Plant Biol. 57, 109–125. doi: 10.1146/annurev.arplant.57.032905.105206
Jones, J. T., Haegeman, A., Danchin, E. G., Gaur, H. S., Helder, J., Jones, M. G., et al. (2013). Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Jones, M., and Dropkin, V. (1975). Cellular alterations induced in soybean roots by three endoparasitic nematodes. Physiol. Plant Pathol. 5, 119–124. doi: 10.1016/0048-4059(75)90015-6
Jones, M., and Dropkin, V. (1976). Scanning electron microscopy of nematode-induced giant transfer cells. Cytobios 15, 149–161.
Jones, M., and Payne, H. (1978). Early stages of nematode-induced giant-cell formation in roots of Impatiens balsamina. J. Nematol. 10:70.
Kerk, N. M., Ceserani, T., Tausta, S. L., Sussex, I. M., and Nelson, T. M. (2003). Laser capture microdissection of cells from plant tissues. Plant Physiol. 132, 27–35. doi: 10.1104/pp.102.018127
Lappalainen, P., Kessels, M. M., Cope, M. J. T., and Drubin, D. G. (1998). The ADF homology (ADF-H) domain: a highly exploited actin-binding module. Mol. Biol. Cell 9, 1951–1959. doi: 10.1091/mbc.9.8.1951
Liu, B., Liu, X., Yang, S., Chen, C., Xue, S., Cai, Y., et al. (2016). Silencing of the gibberellin receptor homolog, CsGID1a, affects locule formation in cucumber (Cucumis sativus) fruit. New Phytol. 210, 551–563. doi: 10.1111/nph.13801
Liu, B., Ren, J., Zhang, Y., An, J., Chen, M., Chen, H., et al. (2015). A new grafted rootstock against root-knot nematode for cucumber, melon, and watermelon. Agron. Sus. Dev. 35, 251–259. doi: 10.1007/s13593-014-0234-5
Maciver, S. K. (1998). How ADF/cofilin depolymerizes actin filaments. Curr. Opin. Cell Biol. 10, 140–144. doi: 10.1016/S0955-0674(98)80097-5
Maciver, S. K., and Hussey, P. J. (2002). The ADF/cofilin family: actin-remodeling proteins. Genome Biol. 3, reviews3007.1–reviews3007.12. doi: 10.1186/gb-2002-3-5-reviews3007
Molinari, S., Fanelli, E., and Leonetti, P. (2014). Expression of tomato salicylic acid (SA)-responsive pathogenesis-related genes in Mi-1-mediated and SA-induced resistance to root-knot nematodes. Mol. Plant Pathol. 15, 255–264. doi: 10.1111/mpp.12085
Mukhtar, T., Kayani, M. Z., and Hussain, M. A. (2013). Response of selected cucumber cultivars to Meloidogyne incognita. Crop Prot. 44, 13–17. doi: 10.1016/j.cropro.2012.10.015
Muller, J., Rehbock, K., and Wyss, U. (1981). Growth of Heterodera schachtii with remarks on amounts of food consumed. Revue de Nématologie 4, 227–234.
Pavlov, D., Muhlrad, A., Cooper, J., Wear, M., and Reisler, E. (2007). Actin filament severing by cofilin. J. Mol. Biol. 365, 1350–1358. doi: 10.1016/j.jmb.2006.10.102
Ruzicka, D. R., Kandasamy, M. K., McKinney, E. C., Burgos-Rivera, B., and Meagher, R. B. (2007). The ancient subclasses of Arabidopsis Actin Depolymerizing Factor genes exhibit novel and differential expression. Plant J. 52, 460–472. doi: 10.1111/j.1365-313X.2007.03257.x
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sampath, P., and Pollard, T. D. (1991). Effects of cytochalasin, phalloidin and pH on the elongation of actin filaments. Biochemistry 30, 1973–1980. doi: 10.1021/bi00221a034
Staiger, C. J., Gibbon, B. C., Kovar, D. R., and Zonia, L. E. (1997). Profilin and actin-depolymerizing factor: modulators of actin organization in plants. Trends Plant Sci. 2, 275–281. doi: 10.1016/S1360-1385(97)86350-9
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, C., Deng, L., Chang, D., Chen, S., Wang, X., and Kang, Z. (2016). TaADF3, an Actin-Depolymerizing Factor, negatively modulates wheat resistance against Puccinia striiformis. Front. Plant Sci. 6:1214. doi: 10.3389/fpls.2015.01214
Walters, S. A., Wehner, T. C., and Barkel, K. R. (1993). Root-knot nematode resistance in cucumber and horned cucumber. HortScience 28, 151–154.
Wehner, T. C., Walters, S. A., and Barker, K. R. (1991). Resistance to root-knot nematodes in cucumber and horned cucumber. J. Nematol. 23, 611.
Zhang, Y., Liu, B., Yang, S., An, J., Chen, C., Zhang, X., et al. (2014a). A cucumber DELLA homolog CsGAIP may inhibit staminate development through transcriptional repression of B class floral homeotic genes. PLoS ONE 9:e91804. doi: 10.1371/journal.pone.0091804
Zhang, Y., Zhang, X., Liu, B., Wang, W., Liu, X., Chen, C., et al. (2014b). A GAMYB homologue CsGAMYB1 regulates sex expression of cucumber via an ethylene-independent pathway. J. Exp. Bot. 65, 3201–3213. doi: 10.1093/jxb/eru176
Zhang, X., Zhou, Y., Ding, L., Wu, Z., Liu, R., and Meyerowitz, E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101. doi: 10.1105/tpc.112.107854
Keywords: cucumber, meloidogyne incognita, actin-depolymerizing factor (ADF) genes, giant cells, cytoskeleton
Citation: Liu B, Liu X, Liu Y, Xue S, Cai Y, Yang S, Dong M, Zhang Y, Liu H, Zhao B, Qi C, Zhu N and Ren H (2016) The Infection of Cucumber (Cucumis sativus L.) Roots by Meloidogyne incognita Alters the Expression of Actin-Depolymerizing Factor (ADF) Genes, Particularly in Association with Giant Cell Formation. Front. Plant Sci. 7:1393. doi: 10.3389/fpls.2016.01393
Received: 16 May 2016; Accepted: 01 September 2016;
Published: 16 September 2016.
Edited by:
Christopher J. Staiger, Purdue University, USAReviewed by:
Brad Day, Michigan State University, USANoriko Inada, Nara Institute of Science and Technology, Japan
Copyright © 2016 Liu, Liu, Liu, Xue, Cai, Yang, Dong, Zhang, Liu, Zhao, Qi, Zhu and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huazhong Ren, cmVuaHVhemhvbmdAY2F1LmVkdS5jbg==
†These authors have contributed equally to this work.
 Bin Liu1†
Bin Liu1† Huazhong Ren
Huazhong Ren