- Zhejiang Key Laboratory of Aquatic Germplasm Resources, College of Biological & Environmental Sciences, Zhejiang Wanli University, Ningbo, China
Bone morphogenetic proteins (BMPs) not only play essential roles in bone development but also are involved in embryonic growth, organogenesis cell proliferation and differentiation. However, the previous studies on the functions of shellfish BMPs genes are still very limited. To better understand its molecular structure and biological function, BMP7 of the razor clam Sinonovacula constricta (Sc-BMP7) was cloned and characterized in this study. The full length of Sc-BMP7 is 2252 bp, including an open reading frame (ORF) of 1257 bp encoding 418 amino acids. The protein sequence included a signal peptide (1–32 aa), a prodomain (38–270 aa) and a TGF-β domain (317–418 aa). The quantitative expression of eleven adult tissues showed that Sc-BMP7 was significantly higher expressed in the gill, foot, and mantle (P < 0.05), but lower in hemocytes and hepatopancreas. In the early development stages, low expression was detected in the stages of unfertilized mature eggs, fertilized eggs, 4-cell embryos, blastula, gastrulae, whereas it increased after the stage of trochophore and demonstrated the highest expression in umbo larvae (P < 0.01). In shell repair experiment, Sc-BMP7 showed increasing expression level after 12 h. The higher expression of Sc-BMP7 was detected while Ca2+ concentration was reduced in seawater. After inhibiting Sc-BMP7 expression using RNA interference (RNAi) technology, expression of Sc-BMP7 mRNA and protein were significantly down-regulated (P < 0.05) in the central zone of mantle (nacre formation related tissue) and the pallial zone of mantle (prismatic layer formation related tissue). Association analysis identified two shared SNPs in exon of Sc-BMP7 gene from 246 individuals of two groups. These results indicated that BMP7 might be involved in shell formation and growth. These results would contribute to clarify the role of Sc-BMP7 in the regulation of growth and shell formation, and provide growth-related markers for molecular marker assisted breeding of this species.
Introduction
Transforming growth factor-β (TGF-β) proteins comprise a family of structurally related cytokines that occur widely among various vertebrates and invertebrates. TGF-β is known to be involved in various biological processes, including bone and organ formation, cell proliferation, differentiation, apoptosis, and so on (Feng and Derynck, 2005; Massague, 2008; Ramesh et al., 2009). The TGF-β superfamily is divided into two main categories: TGF-β/activin/nodal and bone morphogenetic protein (BMP)/differentiation factor (GDF)/Müllerian inhibiting substance (MIS) (Shi and Massagué, 2003), based on sequence identity and activation of downstream pathways. All TGF-β superfamily members split from a precursor at a specific site to release a mature polypeptide, and their biological activity relies on the formation of dimers by two identical or different subunits (Kingsley, 1994). Among these proteins, BMPs constitute the largest subgroup of the TGF-β superfamily (Bragdon et al., 2011). BMPs are extensively expressed during mammalian development, with a wide range of biological activities, including development, proliferation, and extracellular matrix synthesis (Salazar et al., 2016). BMPs can distinguish and bind to serine/threonine kinase receptors, with subsequent signaling mediated by both Smad-dependent and -independent pathways (Wordinger and Clark, 2007; Sánchez-Duffhues et al., 2015).
In mollusks, the whole-body growth was determined by growth of soft parts and shell, the latter involving crystal growth regulated by the secretion of stromatin. Both shell and bone are products of biomineralization. Proteins account for <5% of the biomineralized shell but are primarily responsible for controlling the CaCO3 polymorph and texture (Marin and Luquet, 2004; Liu and Li, 2013). Most studies of BMPs in vertebrates have focused on BMP2, BMP4, and BMP7 (Salazar et al., 2016). However, other studies have shown that BMPs in mollusks also have important functions (Dong, 2012; Feng et al., 2013; Lin, 2014; Liu et al., 2014; Yan et al., 2014; Qian, 2015; Zhou, 2016; Fan et al., 2018), similar to higher animals.
BMP7 possesses the classical TGF-β domains, including a large pro-domain that facilitates protein folding and a mature signaling peptide (Xu et al., 2018), and plays important roles during skeletogenesis and postnatal bone homeostasis (Cook and Rueger, 1996). Mammalian BMP7 has been shown to induce the differentiation of primitive osteoblast progenitor cells and accelerate the healing of fractures (Schmal et al., 2012). Recombinant human BMP7 protein has been used in the clinic to promote bone regeneration (Asahina et al., 1996; Han et al., 2008; Hurtig et al., 2009). BMP7 has also been shown to play significant roles in the development of various types of tissues, such as kidney and brown adipose tissue. In mice, BMP7 was involved in sertoli cell proliferation during early postnatal development, and BMP7 gene knockout mice caused infertility (Puglisi et al., 2004; Monsivais et al., 2017). BMP7 has also been shown to participate in embryogenesis, tissue growth, and neurogenesis (Bragdon et al., 2011; Kowtharapu et al., 2018). The BMP7 gene was also identified as a candidate gene related to growth in cattle and chickens, and growth-associated single nucleotide polymorphisms (SNPs) have been identified (Chen et al., 2013; Huang et al., 2013; Wang et al., 2018). So far, BMP7 genes have been reported in some species of bivalves, including Tegillarca granosa (Dong, 2012), Pinctada martensii (Yan et al., 2014; Fan et al., 2018), and Hyriopsis cumingii (Lin, 2014). These studies also found the highest expression levels of BMP7 in the mantle, suggesting that it was related to shell formation.
The razor clam Sinonovacula constricta is an economically important maricultured bivalve species with over 800,000 metric tons of annual production in China (Xu and Zhang, 2008). However, despite recent fast developments in artificial breeding and aquaculture, new varieties of razor clams for artificial breeding are still severely lacking. At present, limited research has reported on growth-related genes in S. constricta, including IGFBP (Xie et al., 2015), MSTN (Niu et al., 2015), and GRB2 (Zhao et al., 2018). There is thus a need to study growth-related genes and carry out molecular breeding of high-yield new varieties to support the sustainable development of the clam aquaculture industry. In this study, we identified the promotor and exon of S. constricta BMP7 (Sc-BMP7) gene, and detected its expression profiles in different tissues and developmental stages. Furthermore, we also analyzed the association of Sc-BMP7 SNPs with growth traits, thus providing the basis for screening candidate genes for growth traits and for studying the molecular mechanisms of growth regulation.
Materials and Methods
Experimental Animals and Sample Collection
Adult clams (shell length 50 ± 5 mm, total weight 7.0 ± 1.0 g) were obtained from Yinzhou Danyan Aquaculture Field in Ningbo, China, for cloning and gene expression analysis of Sc-BMP7. Eleven tissues, including mantle (pallial zone, marginal zone and central zone), adductor muscle, digestive gland, foot, gill, blood, gonad, and siphon were dissected, frozen immediately in liquid nitrogen, and then stored at −80°C. Embryos/larvae were cultured in 13‰ salinity seawater, fed with golden-brown algae, and collected at 10 developmental stages (unfertilized mature egg, fertilized egg, 4-cell embryo, blastula, gastrula, trochophore, D-shaped larva, umbo larva, eyespot larva, juvenile clam) and preserved at −80°C.
A total of 246 adult clams were collected to screen for Sc-BMP7 SNPs. 122 individuals from Yongle NO 1 strain (fast-growing strain, selected for four generations by our team from Changle population, Fujian Province, China) and 124 individuals from Lianjiang population (wild population from Lianjiang county, Fujian Province, China) were randomly sampled. The two groups were cultured in the same growing environmental conditions, and the main growth traits (shell length, shell width, shell height, and total weight) were measured. The foot and mantle were dissected, frozen immediately in liquid nitrogen, and then stored at −80°C.
Cloning of Full-Length cDNA and Promoter
Total RNA was extracted from the mantle using Trizol reagent (Sangon, China). RNA integrity was determined by formaldehyde-denatured 1.2% agarose gel electrophoresis and staining, and the quality and quantity were assessed by ultraviolet spectrophotometry. First-strand cDNA was synthesized using SMART RACE reagent (Clontech, United States).
Expressed sequence tag (EST) sequences of Sc-BMP7 gene were retrieved from the razor clam transcriptome in the SRA database (NCBI) with accession number SRP2162898. Primers for 5’-RACE (Sc-BMP7-F1) and 3’-RACE (Sc-BMP7-R1) were designed (Table 1). Polymerase chain reaction (PCR) products were purified using a gel extraction kit (Tiangen, China) and then cloned into the T1 vector (TaKaRa, Japan). The vector was transformed into T1 cells (Tiangen) according to the manufacturer’s protocols, and positive clones were sequenced.
To confirm the accuracy of the cloning and sequencing, the full-length cDNA was reamplified using a pair of specific primers, Sc-BMP7-F2 and Sc-BMP7-R2 (Table 1), designed based on the Sc-BMP7 cDNA. The PCR products were cloned and sequenced following the procedures described above.
We designed the reverse primer using Primer 5 software based on the Sc-BMP7 cDNA. The PCR products were cloned and sequenced following the procedures described above, using a genome walker kit (TaKaRa). The possible core promoter region and potential transcription factor binding sites were predicted by the online software BDGP1 and Alibaba22. The CpG island was predicted using the online analysis software Meth primer3.
Sequence and Phylogenetic Analysis
Sequences were spliced using the National Center for Biotechnology Information database BLAST algorithm4. The deduced amino acid sequence was analyzed using the simple modular architecture research tool (SMART)5 to predict conserved domains. The presence and location of the signal peptide and cleavage sites in the amino acid sequence were predicted by SignalP 4.0 server6. Multiple alignments of BMP7 proteins between S. constricta and other species were performed using the ClustalW2 multiple alignment program7. A phylogenetic tree was constructed by the neighbor-joining method with MEGA 6.0.
Quantitative Analysis
The expression profiles of Sc-BMP7 during different developmental stages (n > 500, three sets of samples per stage) and in different adult tissues (n = 4, four sets of samples per tissue) were analyzed using real time quantitative reverse transcription PCR (qRT-PCR). Total RNA was extracted from the samples as described above. Primers RBF and RBR (Table 1) were designed using Primer 5 and 18S rRNA was used as an internal reference. PCR was conducted using a 7500 Fast Real-Time PCR machine (ABI, United States). The relative value of 2–ΔΔCt was adopted for data processing. Quantitative differences in fluorescence results were analyzed by SPSS 20.0. One way ANOVA was adopted to compare the difference of these groups. A p-value less than 0.05 (P < 0.05) was considered as statistical significance.
Shell Repair Experiment
48 clams (shell length 40 ± 5 mm) were divided randomly into three treatment groups (n = 8 per group) and three control groups (n = 8 per group). Shell incision was carried out after holding for 3 days. In the treatment groups, a V-shape cut was performed on both sides of the clam’s shells using scissors, but the mantle was not damaged during the operation. Clams in the control groups remained untreated. One treatment group and one control group were then cultured together in the same tank used sterilized seawater, with a total of three tanks. All clams were fed with the microalgae Isochrysis galbana in the morning and evening. Three shells were then sampled from the treatment group and control group at 2, 4, 8, 12, 24, 48, and 96 h, and at 7 days, respectively. The outer mantle tissue was dissected at the V-shaped notch and total RNA was extracted for qRT-PCR, as described above.
Effect of Ca2+ on BMP7 Gene Expression
A total of 60 clams (shell length 40 ± 5 mm) were divided randomly into six groups. The control group was NSG (normal seawater group). CFG (calcium free group), Ca2+ levels was lower than normal levels (seawater plus EDTA-Na2 to a final concentration of 250 mg/L), and LCG (low calcium free group), MCG (middle calcium group), HCG (high calcium group), and VHCG (very high calcium free group) groups, were levels higher than normal seawater (CaCl2 added to final concentrations of 100, 200, 300, and 400 mg/L, respectively). The clams were cultured in the same water environment (salinity 25‰, pH 8.0 ± 0.3 and temperature 25°C) for 3 weeks in 50 L barrels and fed I. galbana in the morning and evening. The seawater was changed every day. The mantle and gill tissues were then dissected, and total RNA was extracted for qRT-PCR analysis, as described above.
RNA Interference (RNAi) and Enzyme-Linked Immunosorbent Assay (ELISA)
RNAi was performed to examine the role of Sc-BMP7 in shell growth. The specific primers (Table 1) were designed based on the Sc-BMP7 cDNA and used to amplify the specific sequences. The T. granosa Hb gene (which is not expressed in S. constricta), and DEPC H2O was used as a control. Ten individuals were used in each treatment. RNAi was conducted according to Yan et al. (2014). Sc-BMP7 double-stranded (ds)RNA was diluted to 1 μg/μL with DEPC H2O, and 80 μL was injected into the adductor muscle of S. constricta (1-year-old, shell length 48 ± 5 mm), followed by the same dose 3 days after the first injection. Clams in the control groups were injected with the same volume of DEPC H2O or 1 μg/μL of hemoglobin-dsRNA (Hb-dsRNA) in DEPC H2O. Total RNA was extracted from the mantle pallium and edge 3 days after the third injection. The expression levels of the Sc-BMP7 gene after RNAi were measured by qRT-PCR using 18sRNA as the internal reference.
Sc-BMP7 proteins of ten samples from each treatment group above were detected by ELISA using the shellfish BMP-7 ELISA kit (Jianglai, China). Statistical analysis of results between RNAi group and control group were used SPSS 20.0. ANOVA was adopted to compare the difference of these groups. A p-value less than 0.05 (P < 0.05) was considered as statistical significance.
Exon SNPs
The Sc-BMP7 cDNA was used to design the primers SBMP7-F1, SBMP7-R1, SBMP7-F2, and SBMP7-R2 (Table 1). Total RNA was extracted from the samples as described above. The PCR products were verified by direct sequencing and sequence alignment was conducted using MEGA6.0 software. The relationships between SNPs and growth traits were analyzed using SPSS20.0. ANOVA was adopted to compare the difference of these genotypes. Linkage disequilibrium (LD) and haplotype were analyzed using SHEsis analysis software8.
Results
Sequence Analysis of Sc-BMP7 Gene
The deduced Sc-BMP7 protein contained 418 amino acid residues encoded by 2,247 nucleotides. The cDNA also contained a 5’ untranslated region (UTR) of 638 nucleotides and a 3’ UTR of 117 nucleotides (Figure 1A). The calculated molecular mass of the deduced mature Sc-BMP7 was 47.70 kDa, and the theoretical isoelectric point was 7.59. Amino acid sequence analysis showed that Sc-BMP7 contained a signal peptide (1–32 aa), a prodomain (33–283 aa), and a mature peptide (284–418 aa). The mature protein, which was produced by cleaving off the prodomain in the putative maturation site Arg-X-X-Arg, consisted of 135 amino acids, and a TGF-β family domain (318–429 aa) with seven conserved cysteine residues (Figure 1C). PCR amplification was carried out using two pairs of primers. After assembly, a proximal promoter of 2,252 bp was obtained using the genomic walking method. The promoter contained a CpG island, 203 transcription factor binding sites, and four potential transcription initiation sites.
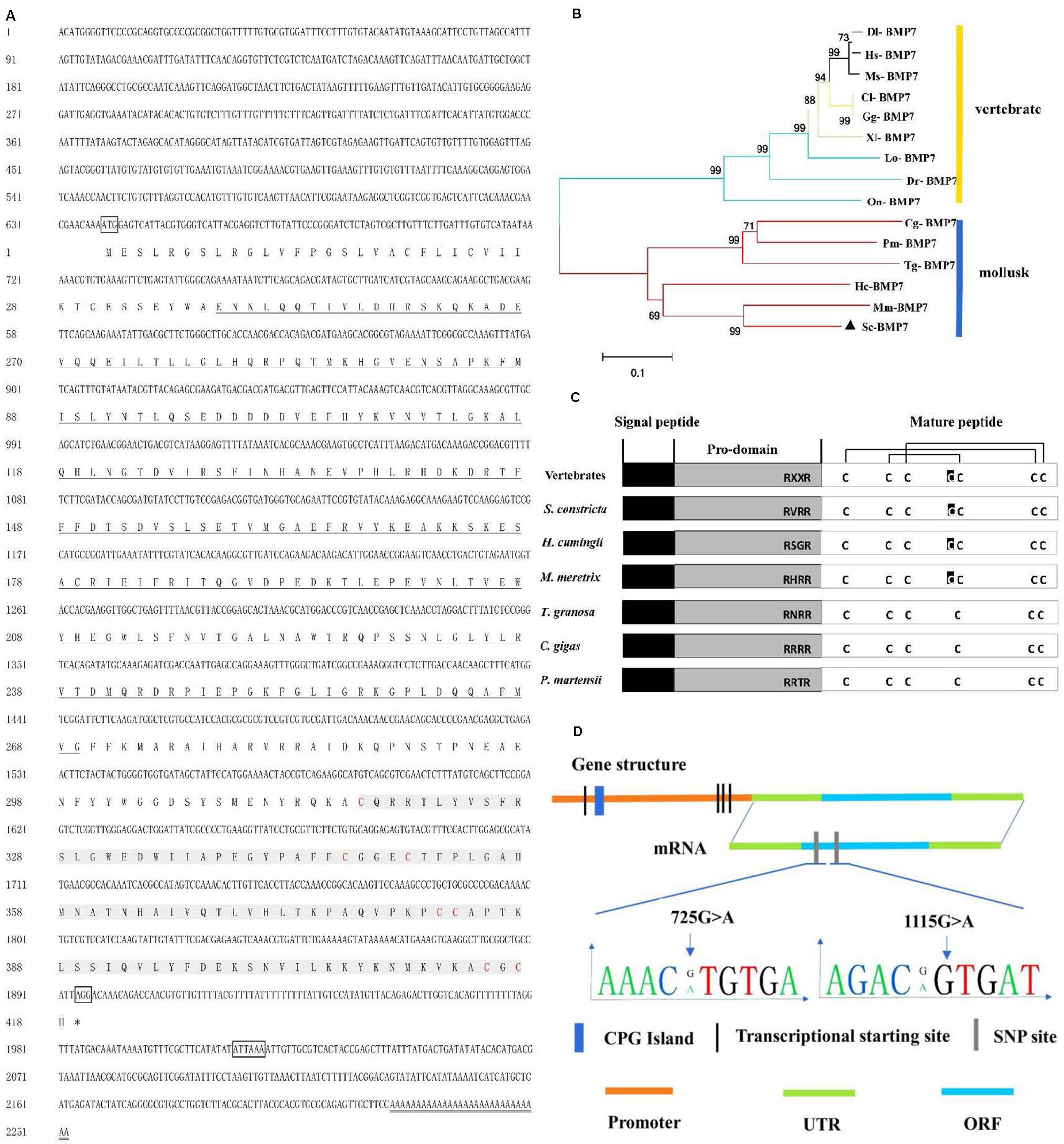
Figure 1. Analysis of sequence and structure of Sc-BMP7 gene. (A) The full-length cDNA and deduced amino acid sequence of Sc-BMP7. The letters boxes are the start codon, the stop codon and the polyadenylation signal sequence, the ∗ represents the end of the protein translation, the double line is the poly A. the bold underlined parts are the propeptide, shaded gray part is the signal peptide, red letters are cysteine residues. (B) Neighbor-joining phylogenetic tree of BMP7 between S. constricta and other species using MEGA6.0 software. The abbreviation of BMP7 and the GenBank accession numbers used to construct phylogenetic tree are given in Table 2. (C) Structure of Sc-BMP7 and selected BMP7 genes from mollusks and vertebrates. (D) Gene structure of Sc-BMP7 gene.
Multiple comparisons with the BMP7 of mollusks and model animals showed that Sc-BMP7 shared the highest identity (68%) with BMP7 of Meretrix meretrix, and 39%–55% similarity with others. The abbreviations of BMP7 and the GenBank accession numbers used to construct the phylogenetic tree are shown in Table 2. The phylogenetic tree, constructed using the neighbor-joining method, showed that BMP7 protein could be divided into two groups (Figure 1B), one containing all shellfish, and another comprising mammals, reptiles, and fish. M. meretrix BMP7 was firstly clustered with Sc-BMP7 in the former group.
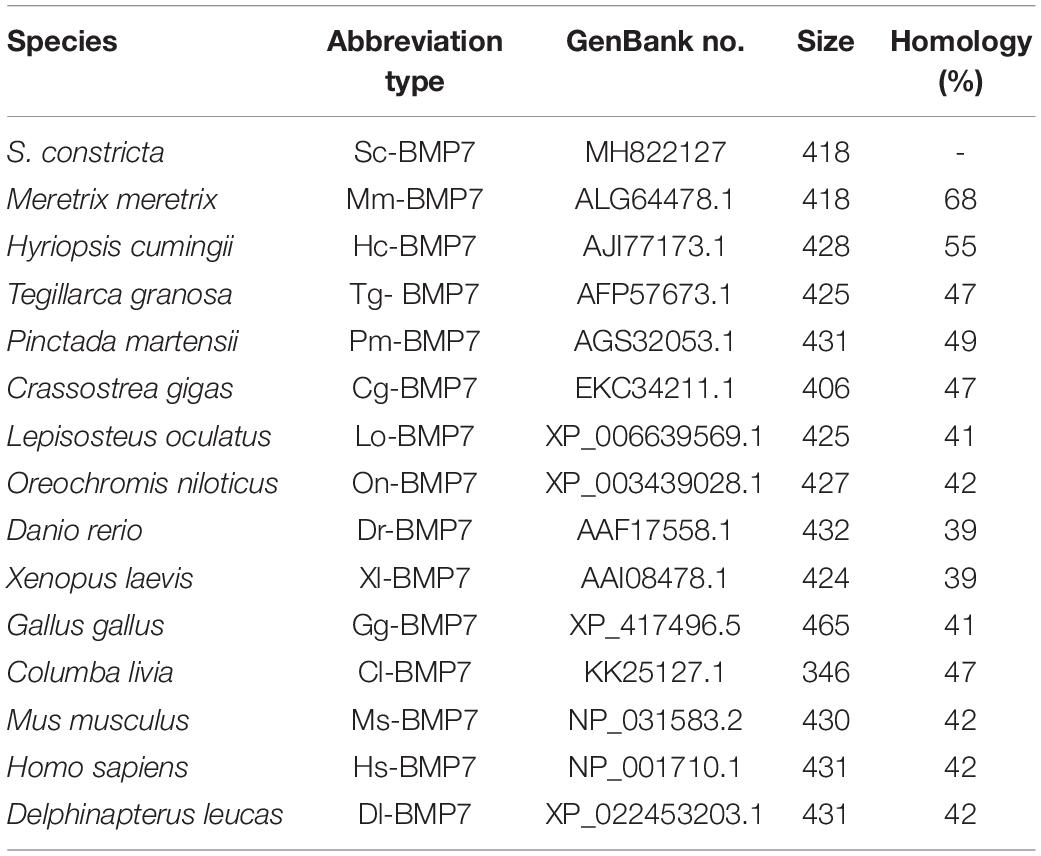
Table 2. Species and GenBank accession numbers of BMP7s sequence used for multiple alignment and phylogenetic analysis.
Quantitative Expression Analysis of Sc-BMP7
Tissue and developmental stage-specific expression of Sc-BMP7 were determined by qRT-PCR. Sc-BMP7 gene expression levels were high in the gills and mantle (P < 0.05), especially in the central zone of mantle (Figure 2A). In terms of developmental stage, Sc-BMP7 expression levels were very low before the trochophore stage, including unfertilized mature eggs, fertilized eggs, 4-cell embryos, blastulae, and gastrulae, but gradually increased in the subsequent developmental stages, with the highest levels in D-shaped larvae (P < 0.01) (Figure 2B).
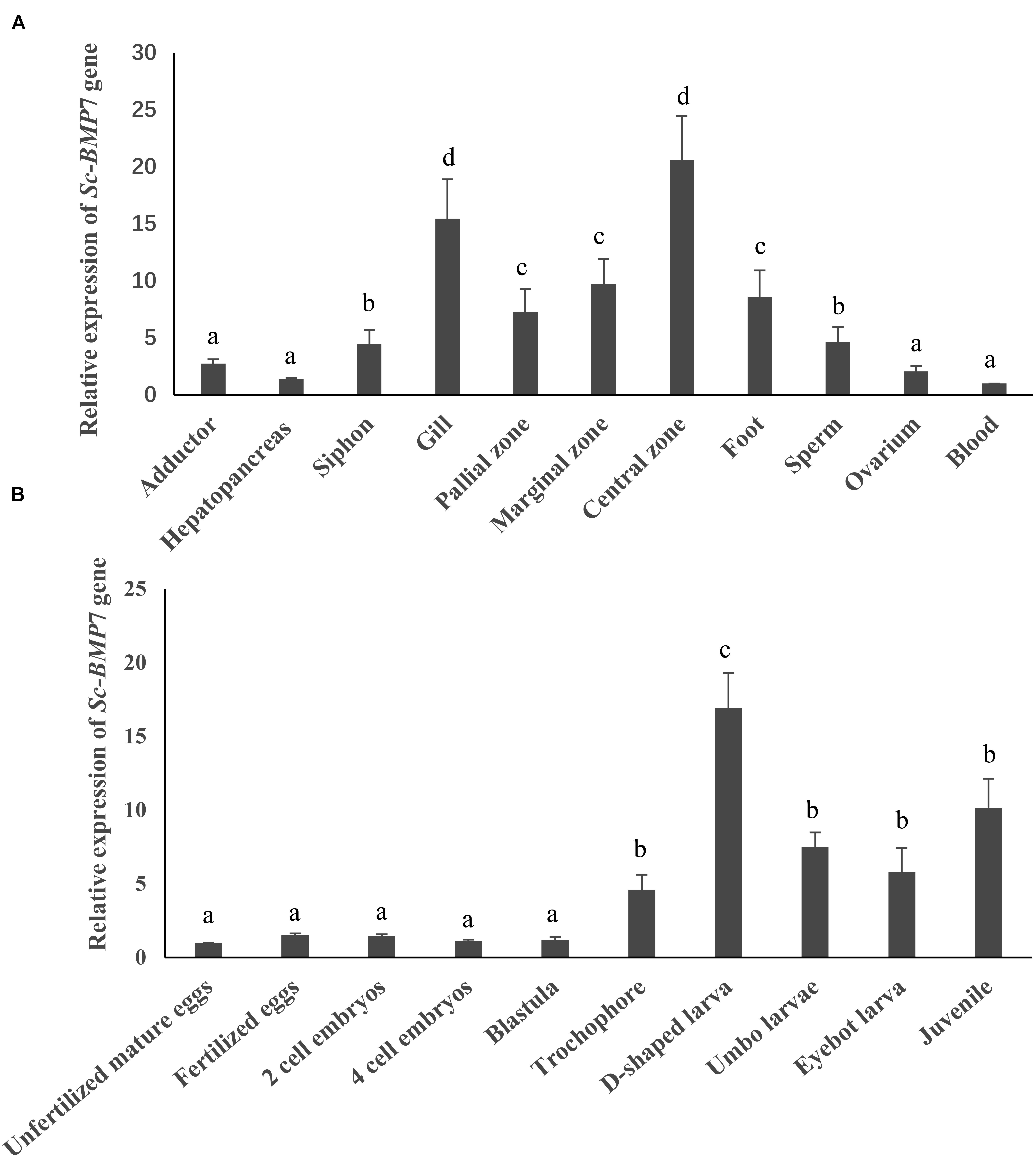
Figure 2. Quantitative expression analysis of Sc-BMP7, (A) Gene expression difference analysis of Sc-BMP7 in different tissues. The same letters indicate no difference in the level of expression, whereas different letters indicate significant differences in expression levels among various tissues (P < 0.05, based on ANOVA, n = 4). (B) Gene expression difference analysis of Sc-BMP7 in different developmental stages. The same letters indicate no difference in the level of expression, whereas different letters indicate significant differences in expression levels among various developmental stages (P < 0.05, based on ANOVA, n > 500).
Shell Repair Experiment
Sc-BMP7 expression showed no significant increase to 8 h after incision but increased significantly after 12 h (P < 0.05) and peaked after 48 h (Figure 3). Sc-BMP7 expression then began to decline until the seventh day, with extremely significant difference between two groups (P < 0.01).
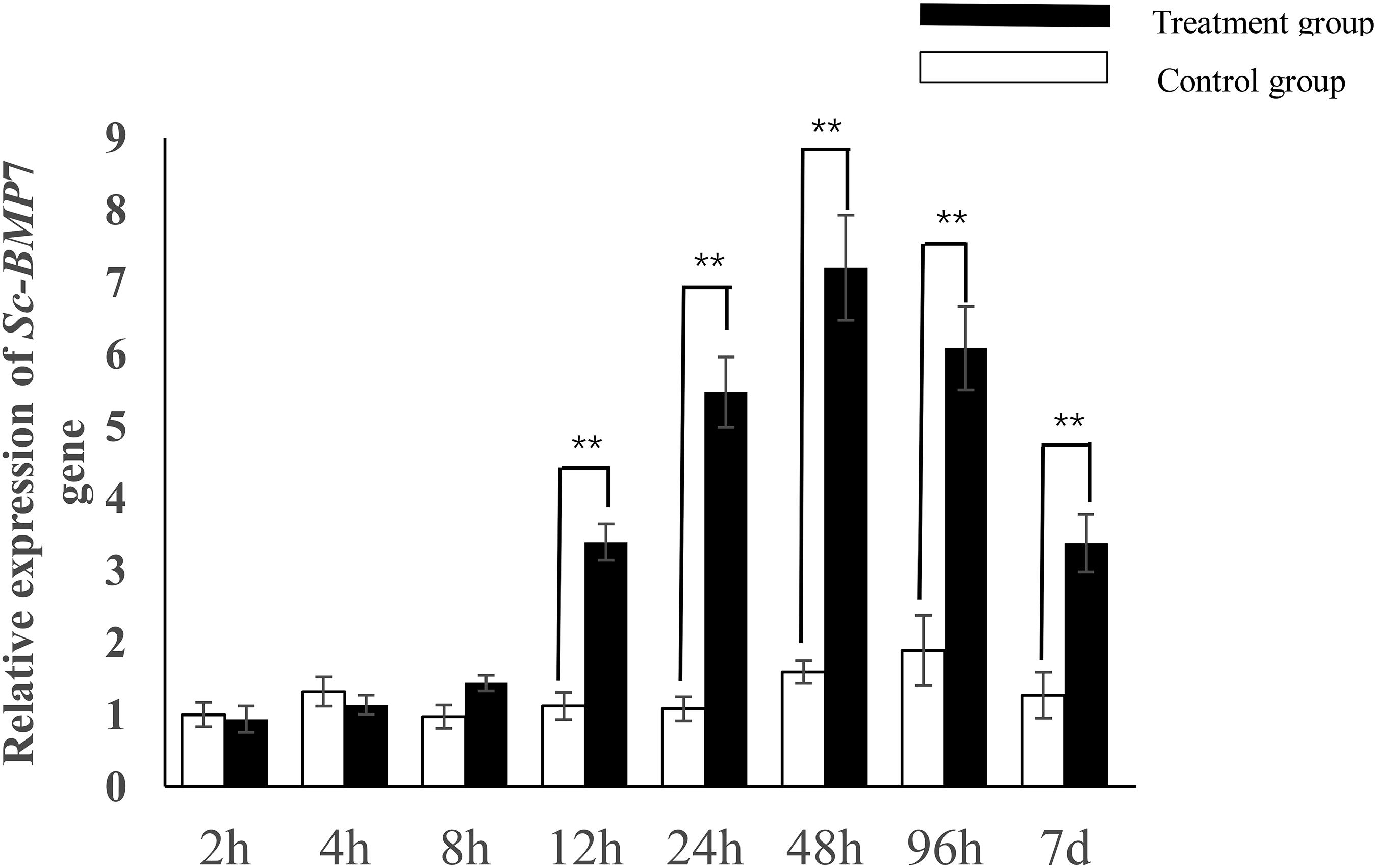
Figure 3. Quantitative expression analysis of Sc-BMP7 after shell notching. ∗∗ represents extremely significant differences, P < 0.01.
Effect of Ca2+ on BMP7 Gene Expression
Sc-BMP7 expression in the mantle was significantly higher in the CFG group compared with the NSG group. Sc-BMP7 expression in the mantle was increased by the addition of 100 mg/L CaCl2, and was unaffected by the addition of 200 mg/L or 300 mg/L CaCl2 to the seawater. However, the expression levels were reduced after the addition of 400 mg/L CaCl2 compared with normal seawater (Figure 4).
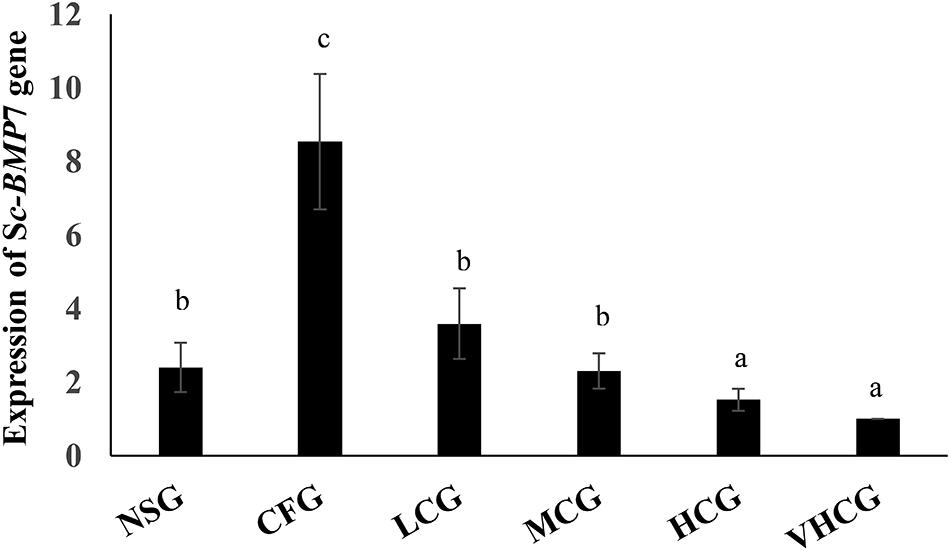
Figure 4. Quantitative expression analysis of Sc-BMP7 in seawater with different Ca2+ concentration. Different letters represent significant differences, P < 0.05. NSG: normal seawater group; CFG: calcium free group, add EDTA-Na2 to a final concentration of 250 mg/L; LCG, MCG, HCG, and VHCG represent CaCl2 added to final Ca2+ concentrations of 100, 200, 300, and 400 mg/L, respectively.
Role of Sc-BMP7 in Shell Growth
We further investigated the function of Sc-BMP7 in shell biomineralization in vivo using RNAi to inhibit the expression of Sc-BMP7 gene. We measured Sc-BMP7 mRNA levels in the mantle pallium and the mantle edge using qRT-PCR. Sc-BMP7 gene expression levels in the RNAi group were downregulated to approximately 38% in the mantle pallium and 29% in the mantle edge compared with control levels (Figure 5A). Sc-BMP7 protein levels were significantly lower in the Sc-BMP7 RNAi group (64.31 pg/mL) compared with the control group (165.37 pg/mL) (Figure 5B).
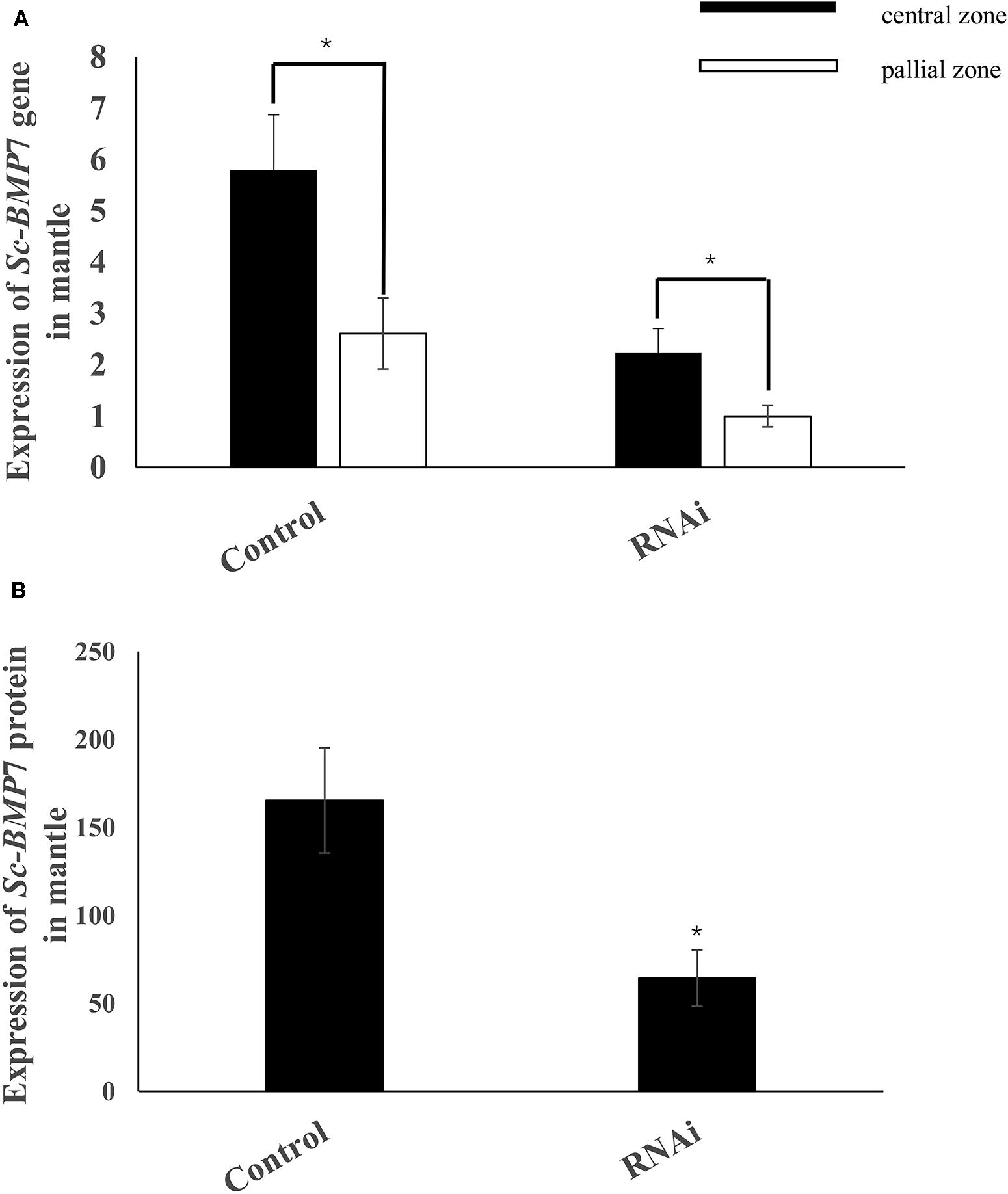
Figure 5. Expression analysis of Sc-BMP7. (A) Expression analysis of Sc-BMP7 gene in the central zone and pallial zone of mantle; (B) Expression analysis of Sc-BMP7 protein in mantle. ∗ represents significant differences (P < 0.05, based on ANOVA).
Growth-Related SNPs in Sc-BMP7 Exon
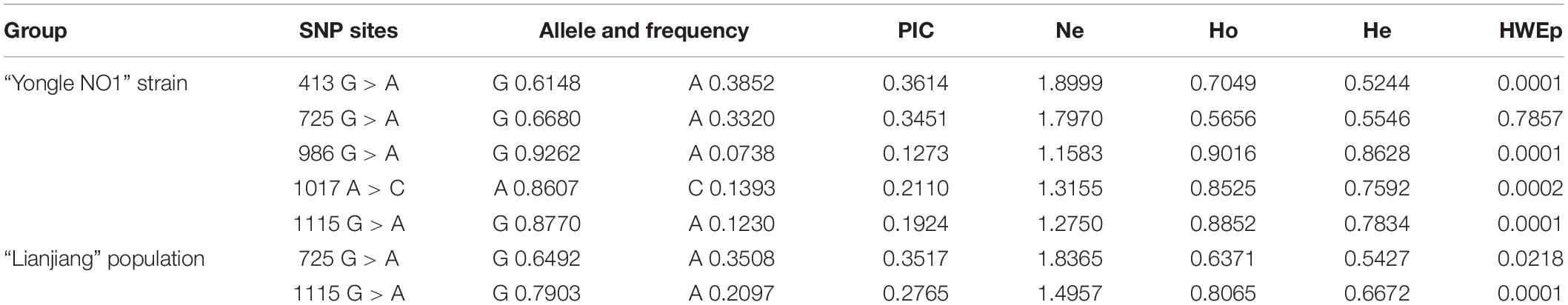
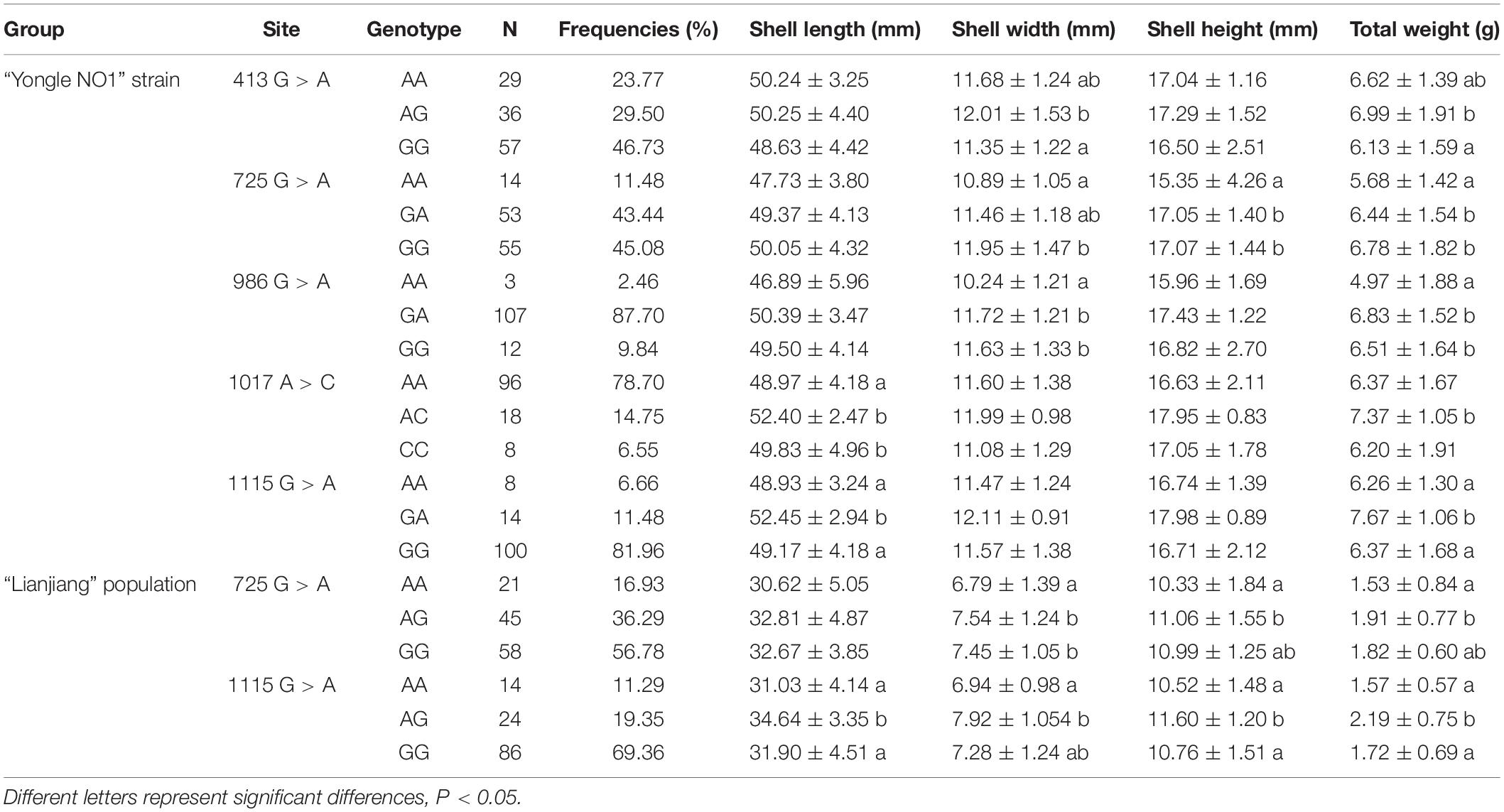
Sequence comparisons detected 20 SNPs in Sc-BMP7. All the SNPs were synonymous mutations, with A/G transversions accounting for 50%. Of these, nine SNPs were in the 5’ UTR or 3’ UTR, and others in the ORF. The associations between its SNPs and growth traits are shown in Table 3. Five SNPs (413G > A, 725G > A, 986G > A, 1017A > C, 1115G > A) were associated with growth traits in the Yongle NO1 strain. The genotype, allele frequencies and polymorphism information content (PIC) values in Yongle NO1 strain are shown in Table 4. Furthermore, analysis of polymorphic parameters indicated that all these SNPs were moderately polymorphic (0.25 < PIC < 0.5).
SNPs 725G > A and 1115G > A were significantly associated with growth traits in Lianjiang population (Figure 1D). Shell width, shell height, and total weight were significantly higher in clams with the GA compared with the AA genotype of 725G > A in the two groups. Shell length and total weight were significantly higher in clams with heterozygous GA type 1115G > A, compared with homozygous AA or GG in both groups (P < 0.05; Table 3). Meanwhile, the frequency of the dominant AG genotype of 725G > A was significantly higher in Yongle NO1 strain than in Lianjiang population, while the AA genotype was less common in Yongle NO1 strain compared with Lianjiang population, possibly related to the breeding of Yongle NO 1 strain.
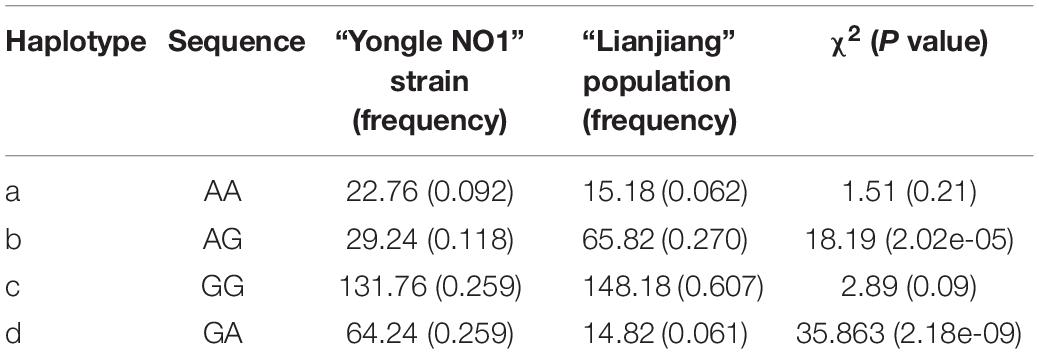
LD analysis using SHEsis online software showed linkage in 725G > A and 1115G > A in both strains (Table 5). LD and haplotypes across SNPs are shown in Table 5. There were two haplotypes of the BMP7 gene. The frequencies of the AG and GA haplotypes in Yongle NO1 strain were significantly higher than in Lianjiang population, but there was no significant difference between the two groups in terms of the AA and GG haplotypes.
Discussion
The results of our current study showed that the Sc-BMP7 gene was highly similar in sequence size to BMP7 genes from some species (Dong, 2012; Xu et al., 2018), but differed from those of P. martensii (Yan et al., 2014) and M. meretrix. The full-length cDNA may have differed from the two homologous genes because of the existence of the additional homologous gene, BMP7b (Shawi and Serluca, 2008; Fan et al., 2018). Previous research showed that all BMPs were synthesized as large precursors (Xiao et al., 2007; Nelsen and Christian, 2009). Mature BMPs in vertebrates share seven conserved cysteines, which can build a cystine knot, active hetero- or homodimers by forming intrachain disulfide bonds or interchain disulfide bonds (Fairlie et al., 2001; Wozney, 2002). This phenomenon was also found in the Sc-BMP7 protein, but only six conserved cysteines were found in C. gigas and T. granosa (Dong, 2012). This may indicate that Sc-BMP7 may possess certain different functions in mollusks.
BMP7 plays an important role in early embryonic development, organ formation, and development (Cook and Rueger, 1996; Bragdon et al., 2011). Bivalves have similar shell-formation processes, and shells occur at an early stage of embryonic development. Development of the shell can be divided into five stages (Kin et al., 2009). The shell begins to form prodissoconch I during the larval stage, followed by prodissoconch II (Miyazaki et al., 2010) in D-larvae. The prodissoconch I is completely formed during gastrula formation, and the prodissoconch II begins to form with formation of the velum (Kakoi et al., 2008). In this study, Sc-BMP7 expression was lower before the trochophore stage, probably because cell division is the main event in early embryonic development. However, BMP7 expression began to increase in D-shaped larva, with the beginning of shell formation (P < 0.05), like the situation in T. granosa (Dong, 2012). Higher expression of Sc-BMP7 in the juvenile stage then reflects the fast growth of razor clams, and the rapid development of organs and shells at this stage (Wang and Wang, 2008).
Previous studies on mollusks found higher expression levels of BMP7 in the gills and mantle (Dong, 2012; Lin, 2014; Yan et al., 2014; Fan et al., 2018). In this study, Sc-BMP7 was expressed in all tissues, especially showed higher expression in the gills and mantle. Gills are the respiratory and filter-feeding organs in bivalves and consume large amounts of energy during the processes of food filtration and gas exchange (Gajaraville et al., 1990; Cui et al., 2006), associated with metabolic vigor and rapid cell proliferation, potentially requiring more BMP7 to regulate cell function. The mantle is the main organ for shell formation and regulates the extracellular growth of crystals, and secretes matrix proteins (Awaji and Machii, 2011; Liu and Li, 2013). The mantle can be divided into marginal, pallial, and central zones according to its different functions (Daisuke et al., 2014; Fan et al., 2018). The Sc-BMP7 gene expression is higher in the central zone than in the other zones. Formation of the prismatic layer of the shell mainly depends on the marginal zone of the mantle, while the central zone secretes the pearl layer because the calcium ion channels (Barry and Diamond, 1971; Shi et al., 2012). Sc-BMP7 may play a crucial role in nacre formation of the shell, as well as being involved in formation of the prismatic layer. The results of the gene expression by calcium concentration showed that the Sc-BMP7 expression in low calcium environment was increased compared with normal seawater. Probably because biomineralization requires a certain amount of Ca2+. Ca2+ was absorbed mainly through the digestive organs and gills, and then transported to the mantle to participate in shell formation (Luo et al., 2010). Sc-BMP7 gene expression increased to maintain mineralization with a small amount of calcium ion when the Ca2+ concentration in seawater decreased. In addition, mollusks have a stable calcium metabolism system, the secretion of calcium regulated by matrix protein, as an essential factor of shell formation. Excessive calcium ions may affect normal shell growth. When the concentration of Ca2+ in seawater increases substantially, Sc-BMP7 gene was low expressed to stabilize the biomineralization in razor clam. The result is similar to the study of BMP3 in P. martensii (Zhou, 2016).
We further elucidated the role of Sc-BMP7 in shell formation in razor clams by RNAi and ELISA analysis. Sc-BMP7 expression decreased significantly following RNAi, by approximately 62% in the pallial zone and 71% in the central zone of the mantle. Meanwhile, Sc-BMP7 protein also decreased significantly by approximately 52%. BMPs are not only a bone-inducing factor, and it is also a major component of bone (Urist, 1965; Wozney, 2002). In mollusks, CaCO3 polymorph, and size and shape of the crystals are controlled by matrix proteins secreted by mantle epidermal cells (Marin and Luquet, 2004; Liu and Li, 2013). In addition, preosteoblast differentiation can be induced by nacre, especially the water soluble matrix fraction of nacre, finally leading to bone formation (Silve et al., 1992; Mouriès et al., 2002), and it is also involved in the activation of mantle cells in mollusks (Sud et al., 2001). The BMP7 gene has been shown to be highly expressed in the mantle in various shellfish. Furthermore, inhibition of the BMP7 gene by RNAi disrupted the growth of aragonite tablets and resulted in holes in the calcite crystals in the mantle of P. martensii, indicating that BMP7 participated in the formation of nacre and the prismatic layer (Yan et al., 2014). In the current study, RNAi treatment decreased the gene and protein expression of Sc-BMP7, potentially resulting in decreased activation and secretion of matrix proteins by the mantle epidermal cells.
Molecular markers can be used to allow selection at the molecular level during assisted breeding, thus greatly improving the breeding efficiency. SNP is the most widely distributed molecular marker in genome and has been used in molecular breeding. BMP7 has previously been identified as a candidate growth-related gene in SNP studies in cattle (Wang, 2009) and chickens (Chen et al., 2013; Wang et al., 2018), but not in mollusks. The current study detected 20 SNPs in the Sc-BMP7 cDNA sequence in Yongle NO1 strain and Lianjiang population, implying a high frequency of SNPs. All the SNPs showed moderate (0.25 < PIC < 0.5) or low polymorphism (PIC < 0.25), presumably because SNP markers are typically biallelic making it difficult to show high polymorphism, as seen for simple sequence repeat (SSR) markers (Hubert et al., 2009).
Analysis of the SNPs identified two (725A > G and 1115A > G) that were associated with growth traits in both groups. GA at 725A > G was significantly associated with shell height and total weight compared with AA, while GA at 1115G > A was significantly associated with shell length and total weight compared with GG and AA in both Yongle NO1 and Lianjiang groups. Furthermore, the two SNPs were located within the coding region of the Sc-BMP7 gene with no amino acid changes. Various studies have shown that synonymous mutations can regulate gene transcription and translation by affecting transcriptional efficiency or changing the mRNA molecules and spatial structure of the protein (Greenwood and Kelsoe, 2003; Capon et al., 2004; Kimchi-Sarfaty et al., 2007).
In conclusion, we got the cDNA and promoter of Sc-BMP7 gene and then analyzed the sequence characteristics and phylogenetic relationship. Analysis of tissue- and development-specific expression demonstrated Sc-BMP7 mRNA was the highest in the central zone of mantle (P < 0.05) and D-shaped larva (P < 0.05), suggesting that it may be involved in the formation and growth of shells. Further results of shell repair experiment and RNAi indicated that Sc-BMP7 gene plays a vital role in repairing shell damage and its function is affected by the Ca2+ concentration in seawater. Moreover, association analysis identified two shared SNPs in exon of Sc-BMP7 gene from 246 individuals of two groups. These results of the present study would contribute to clarify the role of Sc-BMP7 in the regulation of growth and shell formation, and provide growth-related markers for molecular marker assisted breeding in S. constricta.
Data Availability Statement
The BMP7 SNP data has been deposited into the European Variation Archive (EVA) with the project accession PRJEB39579 and analysis accession ERZ1468016.
Author Contributions
YD and ZL conceived and designed the project. HY collected the samples and contributed reagents. JZ and BC performed the experiments and data analysis. YD and JZ wrote and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Key Research and Development Program of China (2018YFD0901405), Zhejiang Major Program of Science and Technology (2016C02055-9), Ningbo Major Project of Science and Technology (2019B10005), and National Marine Genetic Resource Center Program.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^ http://www.fruitfly.org/seq_tools/promoter.html
- ^ http://www.gene-regulation.com/pub/programs/alibaba2/index.html
- ^ http://www.urogene.org/methprimer/index1.html
- ^ http://www.ncbi.nlm.nih.gov/blast/
- ^ http://smart.embl-heidelberg.de
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://www.ebi.ac.uk/Tools/msa/clustalw2/
- ^ http://analysis.bio-x.cn/myAnalysis.php
References
Asahina, I., Sampath, T., and Hauschka, P. (1996). Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp. Cell Res. 222, 38–47. doi: 10.1006/excr.1996.0005
Awaji, M., and Machii, A. (2011). Fundamental studies on in vivo and in vitro pearl formation—contribution of outer epithelial cells of pearl oyster mantle and pearl sacs. Terrapub 4, 1–39. doi: 10.5047/absm.2011.00401.0001
Barry, P., and Diamond, J. (1971). A theory of ion permeation through membranes with fixed neutral sites. J. Membr. Biol. 4, 295–330. doi: 10.1007/BF02431977
Bragdon, B., Moseychuk, O., Saldanha, S., King, D., Julian, J., and Nohe, A. (2011). Bone morphogenetic proteins: a critical review. Cell. Signal. 23, 609–620. doi: 10.1016/j.cellsig.2010.10.003
Capon, F., Allen, M., Ameen, M., Burden, A., Tillman, D., Barker, J., et al. (2004). A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum. Mol. Genet. 13, 2361–2368. doi: 10.1093/hmg/ddh273
Chen, B., Li, L., Shouzhi, W., Xi, C., and Hui, L. (2013). Association between polymorphism of BMP7 gene and growth and body composition traits in broiler chickens. China Poultry 35, 6–10. doi: 10.3969/j.issn.1004-6364.2013.03.003
Cook, S., and Rueger, D. (1996). Osteogenic protein-1: biology and applications. Clin. Orthopaedics Relat. Res. 324, 29–38. doi: 10.1097/00003086-199603000-00005
Cui, L., Hou, Z., and Zhou, X. (2006). Light and electron microscopic observation of gill in Sinonvaculina constricta. Fish. Sci. 25, 129–132. doi: 10.3969/j.issn.1003-1111.2006.03.006
Daisuke, F., Fumito, O., Shigeharu, K., Hiroki, K., Saeri, M., Ayaka, O., et al. (2014). Novel genes participating in the formation of prismatic and nacreous layers in the pearl oyster as revealed by their tissue distribution and RNA interference knockdown. PLoS One 9:e84706. doi: 10.1371/journal.pone.0084706
Dong, Y. (2012). Transcriptome Analysis Using 454 Pyrosequencing and Cloning and Expression of Growth-Related Genes for the Blood Clam Tegillarca granosa (Linnaeus, 1758). dissertation thesis, Ocean University of China, Shangdong.
Fairlie, W., Zhang, H., Wu, W., Pankhurst, S., Bauskin, A., Russell, P., et al. (2001). The propeptide of the transforming growth factor-beta superfamily member, macrophage inhibitory cytokine-1 (MIC-1), is a multifunctional domain that can facilitate protein folding and secretion. J. Biol. Chem. 276, 16911–16918. doi: 10.1074/jbc.M010000200
Fan, S., Zhou, D., Liu, B., Deng, Z., Guo, Y., and Yu, D. (2018). Molecular cloning and expression analysis of BMP 7b from Pinctada fucata. South China Fish. Sci. 14, 121–126. doi: 10.3969/j.issn.2095-0780.2018.01.016
Feng, L., Guo, H., Li, X., Yu, Q., Hu, X., Zhang, L., et al. (2013). Cloning and expression analysis of bone morphogenetic protein 2 gene of Chlamys farreri. Period. Ocean Univ. China 43, 48–55. doi: 10.16441/j.cnki.hdxb.2013.12.008
Feng, X., and Derynck, R. (2005). Specificity and versatility in TGF-beta signaling through Smads. Annu. Rev. Cell. Dev. Biol. 21, 659–693. doi: 10.1146/annurev.cellbio.21.022404.142018
Gajaraville, M., Marigomez, J., and Angulo, E. (1990). Light and electron microscopic study of the gill epithelium of Littorina littorea (Gastropoda, prosobranch). Biol. Struct. Morp. 3, 1–12.
Greenwood, T., and Kelsoe, J. (2003). Promoter and intronic variants affect the transcriptional regulation of the human dopamine transporter gene. Genomics 82, 511–520. doi: 10.1016/S0888-7543(03)00142-3
Han, Q., Gou, S., and Wang, L. (2008). In vivo bone morphogenetic protein 7 gene transfection mediated by polyethyleneimine for femoral fracture healing in old rats. J. Clin. Rehabil. Tissue Eng. Res. 12, 1205–1208. doi: 10.3321/j.issn:1673-8225.2008.07.002
Huang, Y., Wang, X., He, H., Lan, X., Lei, C., Zhang, C., et al. (2013). Identification and genetic effect of haplotype in the bovine BMP7 gene. Gene 532, 281–287. doi: 10.1016/j.gene.2013.03.009
Hubert, S., Bussey, J., Higgins, B., Curtis, B., and Bowman, S. (2009). Development of single nucleotide polymorphism markers for Atlantic cod (Gadus morhua) using expressed sequences. Aquaculture 296, 7–14. doi: 10.1016/j.aquaculture.2009.07.027
Hurtig, M., Chubinskaya, S., Dickey, J., and Rueger, D. (2009). BMP7 protects against progression of cartilage degeneration after impact injury. J. Orthopaedic Res. 27, 602–611. doi: 10.1002/jor.20787
Kakoi, S., Kin, K., Miyazaki, K., and Wada, H. (2008). Early development of the Japanese spiny oyster (Saccostrea kegaki): characterization of some genetic markers. Zool. Sci. 25, 455–464. doi: 10.2108/zsj.25.455
Kimchi-Sarfaty, C., Oh, J., Kim, I., Sauna, Z., Calcagno, A., Ambudkar, S., et al. (2007). A” silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 315, 525–528. doi: 10.1126/science.1135308
Kin, K., Kakoi, S., and Wada, H. (2009). A novel role for dpp in the shaping of bivalve shells revealed in a conserved molluscan developmental program. Dev. Biol. 329, 152–166. doi: 10.1016/j.ydbio.2009.01.021
Kingsley, D. (1994). The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 8, 133–146. doi: 10.1101/gad.8.2.133
Kowtharapu, B., Prakasam, R., Murín, R., Koczan, D., Stahnke, T., Wree, A., et al. (2018). Role of bone morphogenetic protein 7 (BMP7) in the modulation of corneal stromal and epithelial cell functions. Int. J. Mol. Sci. 19:1415. doi: 10.3390/ijms19051415
Lin, J. (2014). Cloning and Expression Analysis of Genes Involved in the Pearl Formation of Hyriopsis cumingii. master’s thesis, Shanghai Ocean University, Shanghai.
Liu, G., Huang, P., and Liu, B. (2014). Clone of dpp gene and its functions in larval shell formation of the Pacific oyster Crassostrea gigas. Mar. Sci. 38, 7–12. doi: 10.11759/hykx20130205002
Liu, X., and Li, J. (2013). Matrix proteins in the shell of cultured pearl bivalves. J. Shanghai Ocean Univ. 22, 200–205.
Luo, W., Yang, S., Ding, Y., Mi, Z., and Zheng, D. (2010). Calcium metabolism in Hyriopsis cumingii during early pearl-forming stages. Oceanol. Limnol. Sin. 41, 895–900. doi: 10.3724/SP.J.1238.2010.00453
Marin, F., and Luquet, G. (2004). Molluscan shell proteins. Comptes Rendus Palevol. 3, 469–492. doi: 10.1016/j.crpv.2004.07.009
Miyazaki, Y., Nishida, T., Aoki, H., and Samata, T. (2010). Expression of genes responsible for biomineralization of Pinctada fucata during development. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 155, 241–248. doi: 10.1016/j.cbpb.2009.11.009
Monsivais, D., Clementi, C., Peng, J., Fullerton, P. Jr., and Prunskaite-Hyyryläinen, R. (2017). BMP7 induces uterine receptivity and blastocyst attachment. Endocrinology 158, 979–992. doi: 10.1210/en.2016-1629
Mouriès, L., Almeida, M., Milet, C., Berland, S., and Lopez, E. (2002). Bioactivity of nacre water-soluble organic matrix from the bivalve mollusk Pinctada maxima, in three mammalian cell types: fibroblasts, bone marrow stromal cells and osteoblasts. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 132, 217–229. doi: 10.1016/S1096-4959(01)00524-3
Nelsen, S., and Christian, J. (2009). Site-specific cleavage of BMP4 by furin, PC6, and PC7. J. Biol. Chem. 284, 27157–27166. doi: 10.1074/jbc.m109.028506
Niu, D., Wang, L., Bai, Z., Xie, S., Zhao, H., and Li, J. (2015). Identification and expression characterization of the myostatin (MSTN) gene and association analysis with growth traits in the razor clam Sinonovacula constricta. Gene 555, 297–304. doi: 10.1016/j.gene.2014.11.020
Puglisi, R., Montanari, M., Chiarella, P., Stefanini, M., and Boitani, C. (2004). Regulatory role of BMP2 and BMP7 in spermatogonia and Sertoli cell proliferation in the immature mouse. Eur. J. Endocrinol. 151, 511–520. doi: 10.1530/eje.0.1510511
Qian, X. (2015). Cloning and Spatiotemporal Expression Analysis of Smad1/5, BMP2/4 Gene and Growth Traits Related SNP Screening in the Tegillarca granosa. master’s thesis, Shanghai Ocean University, Shanghai.
Ramesh, S., Wildey, G., and Howe, P. (2009). Transforming growth factor β (TGFβ)-induced apoptosis: the rise and fall of Bim. Cell Cycle 8, 11–17. doi: 10.4161/cc.8.1.7291
Salazar, V., Gamer, L., and Rosen, V. (2016). BMP signaling in skeletal development, disease and repair. Nat. Rev. Endocrinol. 12:203. doi: 10.1038/nrendo.2016.12
Sánchez-Duffhues, G., Hiepen, C., Knaus, P., and ten Dijke, P. (2015). Bone morphogenetic protein signaling in bone homeostasis. Bone 80, 43–59. doi: 10.1016/j.bone.2015.05.025
Schmal, H., Mehlhorn, A., Pilz, I., Doviakue, D., and Kirchhoff, C. (2012). Immunohistological localization of BMP-2, BMP-7, and their receptors in knee joints with focal cartilage lesions. Sci. World J. 2012:467892. doi: 10.1100/2012/467892
Shawi, M., and Serluca, F. (2008). Identification of a BMP7 homolog in zebrafish expressed in developing organ systems. Gene Express. Patterns 8, 369–375. doi: 10.1016/j.gep.2008.05.004
Shi, S., Jiao, Y., Du, X., and Zhang, Y. (2012). Ultrastructure of mantle epithelial cells in Pinctada martensii. Guangdong Agric. Sci. 39, 135–137. doi: 10.3969/j.issn.1004-874X.2012.08.042
Shi, Y., and Massagué, J. (2003). Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700. doi: 10.1016/S0092-8674(03)00432-X
Silve, C., Lopez, E., Vidal, B., Smith, D., Camprasse, S., Camprasse, G., et al. (1992). Nacre initiates biomineralization by human osteoblasts maintained in vitro. Calcif. Tissue Int. 51, 363–369. doi: 10.1007/BF00316881
Sud, D., Doumenc, D., Lopez, E., and Milet, C. (2001). Role of water-soluble matrix fraction, extracted from the nacre of Pinctada maxima, in the regulation of cell activity in abalone mantle cell culture (Haliotis tuberculate). Tissue Cell 33, 154–160. doi: 10.1054/tice.2000.0166
Urist, M. (1965). Bone: formation by autoinduction. Science 150, 893–899. doi: 10.1126/science.150.3698.893
Wang, R., and Wang, Z. (2008). Science of Marine Shellfish Culture. Qingdao: China Ocean University of China Press.
Wang, X. (2009). Studies on Polymorphisms of GLI2 and BMP7 Genes and their Associations with Growth Traits in Cattle. Dissertation thesis, Northwest Science-Forestry University, Yangling.
Wang, Y., Guo, F., Qu, H., Luo, C., Wang, J., and Shu, D. (2018). Associations between variants of bone morphogenetic protein 7 gene and growth traits in chickens. Br. Poultry Sci. 59, 264–269. doi: 10.1080/00071668.2018.1454586
Wordinger, R., and Clark, A. (2007). Bone morphogenetic proteins and their receptors in the eye. Exp. Biol. Med. 232, 979–992. doi: 10.3181/0510-mr-345
Wozney, J. M. (2002). Overview of bone morphogenetic proteins. Spine 27, 2–8. doi: 10.1097/00007632-200208151-00002
Xiao, Y., Xiang, L., and Shao, J. (2007). Bone morphogenetic protein. Biochem. Biophys. Res. Commun. 362, 550–553. doi: 10.1016/j.bbrc.2007.08.045
Xie, S., Niu, D., Ruan, H., Wang, Z., Wang, F., Chen, F., et al. (2015). Molecular characterization of IGFBP and association analysis with growth traits in the razor clam Sinonovacula constricta[J]. J. Fish. China 39, 799–809. doi: 10.11964/jfc.20141209622
Xu, F., and Zhang, S. (2008). An Illustrated Bivalvia Mollusca Fauna of China Seas. Beijing: Science Press, 211–213.
Xu, Y., Wang, G., Zhou, Y., and Yang, W. (2018). The characterization and potential roles of bone morphogenetic protein 7 during spermatogenesis in Chinese mitten crab Eriocheir sinensis. Gene 673, 119–129. doi: 10.1016/j.gene.2018.06.020
Yan, F., Luo, S., Jiao, Y., Deng, Y., Du, X., Huang, R., et al. (2014). Molecular characterization of the BMP7 gene and its potential role in shell formation in Pinctada martensii. Int. J. Mol. Sci. 15, 21215–21228. doi: 10.3390/ijms151121215
Zhao, J., Cui, B., Dong, Y., and Yao, H. (2018). Cloning, spatiotemporal expression and SNPs identification of GRB2 gene in Sinonovacula constricta. Haiyang Xuebao 40, 87–94. doi: 10.11964/jfc.20141209592
Keywords: Sinonovacula constricta, BMP7, SNP, growth traits, association, RNAi
Citation: Zhao J, Cui B, Yao H, Lin Z and Dong Y (2020) A Potential Role of Bone Morphogenetic Protein 7 in Shell Formation and Growth in the Razor Clam Sinonovacula constricta. Front. Physiol. 11:1059. doi: 10.3389/fphys.2020.01059
Received: 18 March 2020; Accepted: 31 July 2020;
Published: 28 August 2020.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Zhenhua Ma, South China Sea Fisheries Research Institute, ChinaHaihui Ye, Xiamen University, China
Copyright © 2020 Zhao, Cui, Yao, Lin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinghui Dong, ZG9uZ3lpbmdodWkxMThAMTI2LmNvbQ==
 Jiaxi Zhao
Jiaxi Zhao Baoyue Cui
Baoyue Cui Yinghui Dong
Yinghui Dong