- 1Jiangsu Key Laboratory of Oral Diseases, Nanjing Medical University, Nanjing, China
- 2Department of Orthodontics, Affiliated Stomatological Hospital, Nanjing Medical University, Nanjing, China
- 3Department of Dentistry, The Third Affiliated Hospital of Soochow University, The First People’s Hospital of Changzhou, Changzhou, China
- 4State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, China
Non-syndromic tooth agenesis (NSTA) is one of the most common dental abnormalities. MiRNAs participated in the craniofacial and tooth development. Therefore, single nucleotide polymorphisms (SNPs) in miRNA genes may contribute to the susceptibility of non-syndromic tooth agenesis. Here, a total of 625 non-syndromic tooth agenesis cases and 1,144 healthy controls were recruited, and four miRNA SNPs (miR-146a/rs2910164, miR-196a2/rs11614913, pre-miR-605/rs2043556, pre-miR-618/rs2682818) were genotyped by the TaqMan platform. Rs2043556 showed nominal associations with risk of non-syndromic tooth agenesis (PAdd = 0.021) in the overall analysis, as well as upper lateral incisor agenesis (PAdd = 0.047) and lower incisor agenesis (PAdd = 0.049) in the subgroup analysis. Notably, its significant association with upper canine agenesis was observed (PAdd = 0.0016). Rs2043556 affected the mature of miR-605-3p and miR-605-5p while dual-luciferase report analysis indicated that MDM2 was the binding target of miR-605-5p. Our study indicated that pre-miR-605 rs2043556 was associated with risk of non-syndromic tooth agenesis.
Introduction
Tooth agenesis (TA) is one of the most common developmental abnormalities in humans, defined as the absence of one or more permanent teeth, which affects the esthetic, masticatory and occlusal functions of humans. Congenital tooth deficiency affects approximately 20% of the population worldwide (Vastardis, 2000). TA can occur as a syndromic form, which is associated with other genetic diseases, such as non-lethal Raine syndrome (Acevedo et al., 2015), while the more common type is non-syndromic tooth agenesis (NSTA), which occurs as an isolated condition without other birth defects (Fauzi et al., 2018).
Most of human tooth agenesis cases are caused by genetic factors and mutations in AXIN2, EDA, LRP6, MSX1, PAX9, WNT10A, WNT10B, and BMP4 are known to cause tooth agenesis (Yu et al., 2016, 2019a,b; Wong et al., 2018). Single nucleotide polymorphisms (SNPs) are a type of DNA sequence polymorphism caused by variation in a single nucleotide at the genome level (Botstein and Risch, 2003). Previous studies indicated that genomic SNPs were associated with tooth agenesis. For instance, (Liu et al., 2013) found an association between rs929387 of GLI3 and non-syndromic tooth agenesis in Chinese Han individuals. In addition, our previous study indicated that rs17563 (Gong et al., 2015), rs15705 and rs317250 (Lu et al., 2016) of BMP4 were also associated with non-syndromic tooth agenesis.
miRNA, a variety of small non-protein-coding RNAs with a length of approximately 22 nucleotides encoded by endogenous genes, provides an efficient pathway for the regulation of gene expression at the post-transcriptional level (Sehic et al., 2017). In the past few years, the relationship between miRNA and human diseases had been extensively studied. The influence of miRNA on tooth growth and development, as well as tooth-related diseases, had also attracted the wide attention of oral biologists. Studies showed that the miR-200 family was involved in the regulation of epithelial stem cells and that miR-200c regulated tooth enamel formation by influencing signal transduction among ameloblasts (Peng et al., 2012). MiR-21 was also associated with the osteogenesis of alveolar bone (Liu et al., 2014). In addition, miR-34a interacted with notch signaling and promoted both odontogenic and osteogenic differentiation of apical papilla stem cells (SCAPs) (Sun et al., 2014). These studies collectively showed the important role of miRNA in tooth growth and development.
Single nucleotide polymorphisms in miRNA genes may alter pre-miRNA processing or maturation and affect their target specificity (Ha and Kim, 2014), thus resulting in phenotypic differences. Specifically, SNPs in pre-miRNA or pre-miRNA gene regions may affect the expression of mature miRNA, while SNPs around mature miRNA or regions that target genes bind to may affect the binding of miRNA-mRNA (Kroliczewski et al., 2018). Any of the above situations may interfere with the interaction of miRNA-target genes, thus affecting the occurrence of diseases. For instance, (Ghanbari et al., 2016) identified associations of rs897984 in miR-4519 and rs11651671 in miR-548at-5p with Parkinson’s disease. In addition, rs11614913 in miR-196a2, rs2910164 in miR-146a, and rs3746444 in miR-499 contributed to the risk of prostate cancer in Asian descendants (Mi et al., 2018). However, to the best of our knowledge, the associations between miRNA gene SNPs and TA susceptibility have not been explored.
With the aim of addressing this issue, here, we selected four miRNA gene SNPs to evaluate their associations with the risk of non-syndromic tooth agenesis in a case-control study and explored the potential underlying mechanism.
Materials and Methods
Human Subjects
This study consisted of 625 non-syndromic tooth agenesis cases (The average number of missing teeth per person is 1.62, including 6 oligodontia cases and 619 hypodontia cases. Tooth missing pattern was shown in Supplementary Table S1) and 1,144 healthy controls from the Affiliated Stomatological Hospital of Nanjing Medical University between October 2005 and June 2017. All of the participants were from Nanjing and surrounding cities in south-eastern China.
Informed consent to participate in this study was obtained from all enrolled subjects. The controls and cases were identified by two dentists according to panoramic radiographs, dental examinations, and treatment records. The inclusion criteria of healthy controls were (1) full permanent dentition from the third molar to the contralateral third molar and (2) no metal or gold crown restorations. The non-syndromic tooth agenesis cases had at least one missing tooth, excluding the third molar. Any subjects with additional non-dental abnormalities, such as cleft lip and/or cleft palate (CL/P) or other syndromes, and missing teeth caused by acquired reasons, such as trauma, extraction or orthodontic treatment, were excluded from our study. We collected 2 ml of venous blood from all participating individuals using an anticoagulant vacutainer tube with EDTA. This study was approved by the Nanjing Medical University Institution Review Board (PJ2004-030-001), and all methods and procedures followed the Declaration of Helsinki (version 2002) and additional requirements.
SNP Selection
Based on our previous study (Pan et al., 2018), we selected four miRNA SNPs (miR-146a/rs2910164, miR-196a2/rs11614913, pre-miR-605/rs2043556, and pre-miR-618/rs2682818) of them that were identified from miRBase (version10.0)1, dbSNP2, HapMap3, and Patrocles4, that met the following two criteria: (a) SNPs located in the pre-miRNA or mature sequences, (b) the difference in Gibbs binding free energy between the two alleles (ΔΔG) ≥ 2.60 KJ/mol, as the energy parameter ΔΔG may affect the mature miRNA products (Gong et al., 2012) and (c) minor allele frequency (MAF) ≥5% in Chinese population. Pre-miR-923/rs47960429 in our previous study was excluded in this study because miR-923 was a fragment of 28S rRNA in the latest version of miRbase (vision 22.0). Therefore, we chose four SNPs for genotyping (Supplementary Table S2).
DNA Extraction and Genotyping
DNA extraction was conducted according to the protocol of the TIANamp Genomic DNA Kit (TIANGEN, Beijing) (Fan et al., 2018). DNA samples were stored at −80°C for further manipulation after purity and concentration were measured.
Single nucleotide polymorphisms were genotyped by polymerase chain reaction with the TaqMan-MGB method performed using the ABI-Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA, United States). The reaction system includes a pair of upstream and downstream primers and two probes (Supplementary Table S3). Genotyping results were analyzed by SDS software (version 2.4.1) and reviewed by two independent investigators in a blind manner. Samples that failed to be genotyped were removed. 10% samples were randomly selected for confirmation, and the results were 100% concordant.
Predicting the Potential Binding Targets of miRNAs
To further find the target gene of miR-605, prediction was made by four databases (TargetScan, miRDB, miRTarBase, and miRWalk) according to the following criteria: (1) consistently predicted by all four databases and (2) reported to be potentially associated with tooth development.
Cell Culture
The HEK-293 (human embryonic kidney 293 cells) and COS-7 cells (Africa green monkey kidney cells) were cultured, respectively, in Eagle’s minimum essential medium (EMEM) and Dulbecco’s modified eagle’s medium (DMEM) comprised of 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml of streptomycin at 37°C in 5% CO2. The culture medium was replaced every other day.
Transfection and Quantitative Real-Time PCR
HEK-293 cells and COS-7 cells, seeded into 12-well culture plates, were instantly transfected with vector DNA containing either the rs2043556 A or G allele and miR-605-5p mimics by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). After 48 h, the cells were collected for RNA extraction using RNA extraction kit (Takara, Shiga, Japan) and reverse transcribed into single-stranded cDNA by cDNA synthesis kit (Takara, Shiga, Japan). U6 snRNA was used as corresponding internal controls for miRNA and GAPDH was for gene. Real-time quantitative PCR was performed using Power SYBR Green on a 7900 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). Data were collected and analyzed by the 2–ΔΔCt method for qualification of the relative expression levels of mature miR-605 and MDM2.
Construction of Reporter Plasmids and Dual-Luciferase Reporter Assay
The MDM2 3′-UTR wild type fragment (407 bp) was inserted at the Nhel-Xhol restriction site downstream of the luciferase gene in the pmirGLO vector (Supplementary Figure S1). HEK-293 and COS-7 cells, seeded into 24-well culture plates, were transfected with the reporter plasmids containing the MDM2 3′-UTR fragment and miR-605-5p mimic using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States). The luciferase activity in the lysates was quantified with a dual-luciferase reporter assay system after transfection for 24 h (Promega, Madison, WI, United States). The ratio of firefly luciferase to renilla luciferase activity was evaluated, representing the binding activity of miRNA to a given 3′-UTR.
Western Blot Analysis
The cells were lysed by RIPA buffer (Beyotime) and the concentration of protein was measured using BCA Kit (Beyotime). We separated the equivalent amount of protein (20 μg protein/lane) on 8% SDS-PAGE and transfer them to PVDF membrane (Millipore). The membranes were blocked with 5% non-fat milk for 2 h at room temperature and then incubated at 4°C overnight with primary antibodies including anti-MDM2 (Santa Cruz, sc-965, 1:800) and anti-GAPDH (Beyotime, AG019, 1:1000). After incubation with horseradish peroxidase-conjugated secondary antibodies (1:10000), the protein bands were visualized by chemiluminescence reagents (Merck Millipore, WBKLS0050).
In silico Analysis
We retrieved and downloaded three datasets, including GSE138180, GSE42589, and GSE48150 from the GEO database5, a public database of microarray and sequencing data that contained a large number of disease-related gene expressions. GSE138180 explored the roles of miRNAs in odontogenic differentiation of human dental pulp stem cells (DPSCs). GSE42589 showed gene expression data on dental pulp stem cells from 13 samples while GSE48150 reported gene expression of tooth germs at the cap stage from 12-week-old human embryonic oral cavity.
Statistical Analysis
Data were subsequently processed and analyzed with PLINK software (version 1.07). The chi-square test and Student’s t test were used to analyze the age and gender distribution between the case and control group. Odds ratio (OR) and confidence intervals (CIs) calculated by logistic regression analysis were used to determine whether any SNPs were preferentially associated with the risk of non-syndromic tooth agenesis under additive and allelic models. Hardy-Weinberg equilibrium (HWE) was evaluated in the controls by a goodness-of-fit χ2 test. P value ≤ 0.05 was designated as nominal association while P value ≤ 0.0025 (0.05/4/5) after the Bonferroni correction was appointed as significant association. Luciferase activity analysis and qRT-PCR results were calculated by Student’s t test and one-way analysis of variance (ANOVA).
Results
Characteristics of the Samples
A total of 625 cases (205 males and 420 females, mean age: 15.91 ± 8.34) and 1,144 controls (415 males and 729 females, mean age: 15.88 ± 7.48) were recruited, and their information is shown in Supplementary Table S1. There was no significant difference in gender distribution (P = 0.143) between the case and control groups.
The distribution of missing teeth in the maxillary (N = 219, 30.4%) and mandibular regions (N = 501, 69.6%) is shown in Supplementary Table S1. Among the samples we collected, mandibular incisors were the most common missing teeth (N = 362, 50.3%), followed by mandibular premolars (N = 139, 19.3%), maxillary lateral incisors (N = 96, 13.3%), maxillary premolar (N = 72, 10.0%) and maxillary canine (N = 51, 7.1%).
Overall Analysis Between SNPs and NSTA Risk
A total of 614 cases and 1,112 healthy controls were successfully genotyped in the present study. The genotype and allele distributions of each SNP among non-syndromic tooth agenesis cases and controls were calculated. The observed genotype frequencies in the control group were consistent with HWE (P > 0.01). We evaluated the association between non-syndromic tooth agenesis risk and SNPs under additive and allelic models.
As shown in Table 1, nominal associations were detected between rs2043556 and non-syndromic tooth agenesis risk in the overall analysis under additive (P = 0.021, OR = 1.20, 95% CI = 1.03–1.40) and allelic (P = 0.021, OR = 1.20, 95% CI = 1.03–1.40) models. None of any association was observed for the other three SNPs (PAdd = 0.776 for rs2910164, PAdd = 0.557 for rs11614913, PAdd = 0.553 for rs2682818).
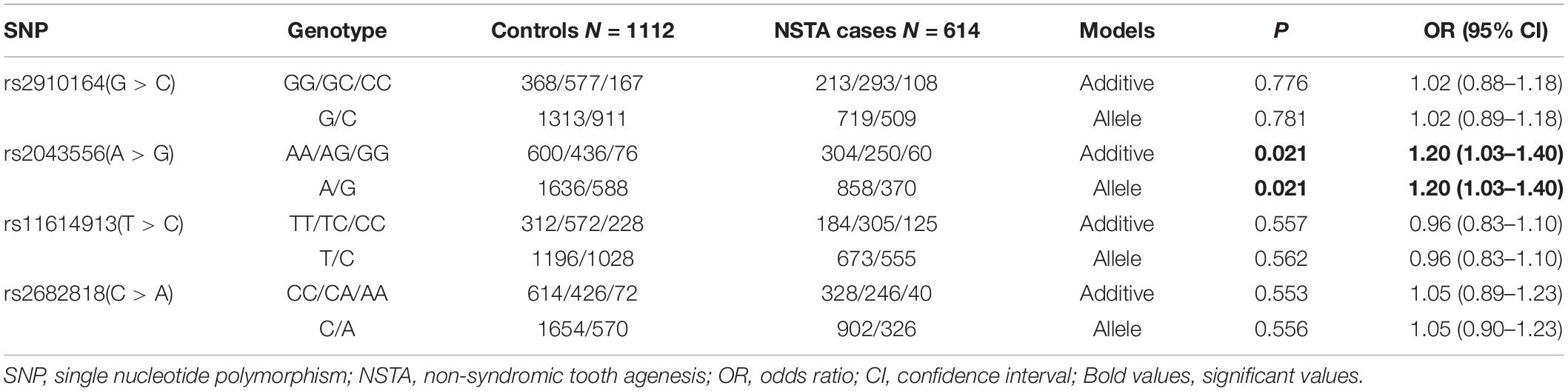
Table 1. Overall associations between the four SNPs and non-syndromic tooth agenesis (NSTA) susceptibility.
Subgroup Analysis With the Position of Missing Teeth
All cases were divided into five subgroups according to different positions of missing teeth: upper lateral incisor agenesis (N = 95, 13.4%), upper canine agenesis (N = 50, 7.1%), upper premolar agenesis (N = 72, 10.2%), lower incisor agenesis (N = 354, 49.9%) and lower premolar agenesis (N = 138, 19.5%). Taking upper lateral incisor agenesis for example, it refers to the subjects with congenitally missing upper lateral incisor with or without other types of missing teeth.
As shown in Table 2, rs2043556 was nominally associated with upper lateral incisor agenesis (PAdd = 0.047, OR = 1.38, 95% CI = 1.01–1.89; PAllelic = 0.046, OR = 1.38, 95% CI = 1.01–1.89) and lower incisor agenesis (PAdd = 0.049, OR = 1.21, 95% CI = 1.00–1.4; PAllelic = 0.049, OR = 1.21, 95% CI = 1.00–1.45). Furthermore, its significant association with upper canine agenesis was detected (PAdd = 0.0016, OR = 1.95, 95% CI = 1.29–2.95; PAllelic = 0.0016, OR = 1.93, 95% CI = 1.28–2.91). However, the subgroup analysis did not reveal any significant associations between rs2910164, rs11614913 and rs2682818 and susceptibility of non-syndromic tooth agenesis.
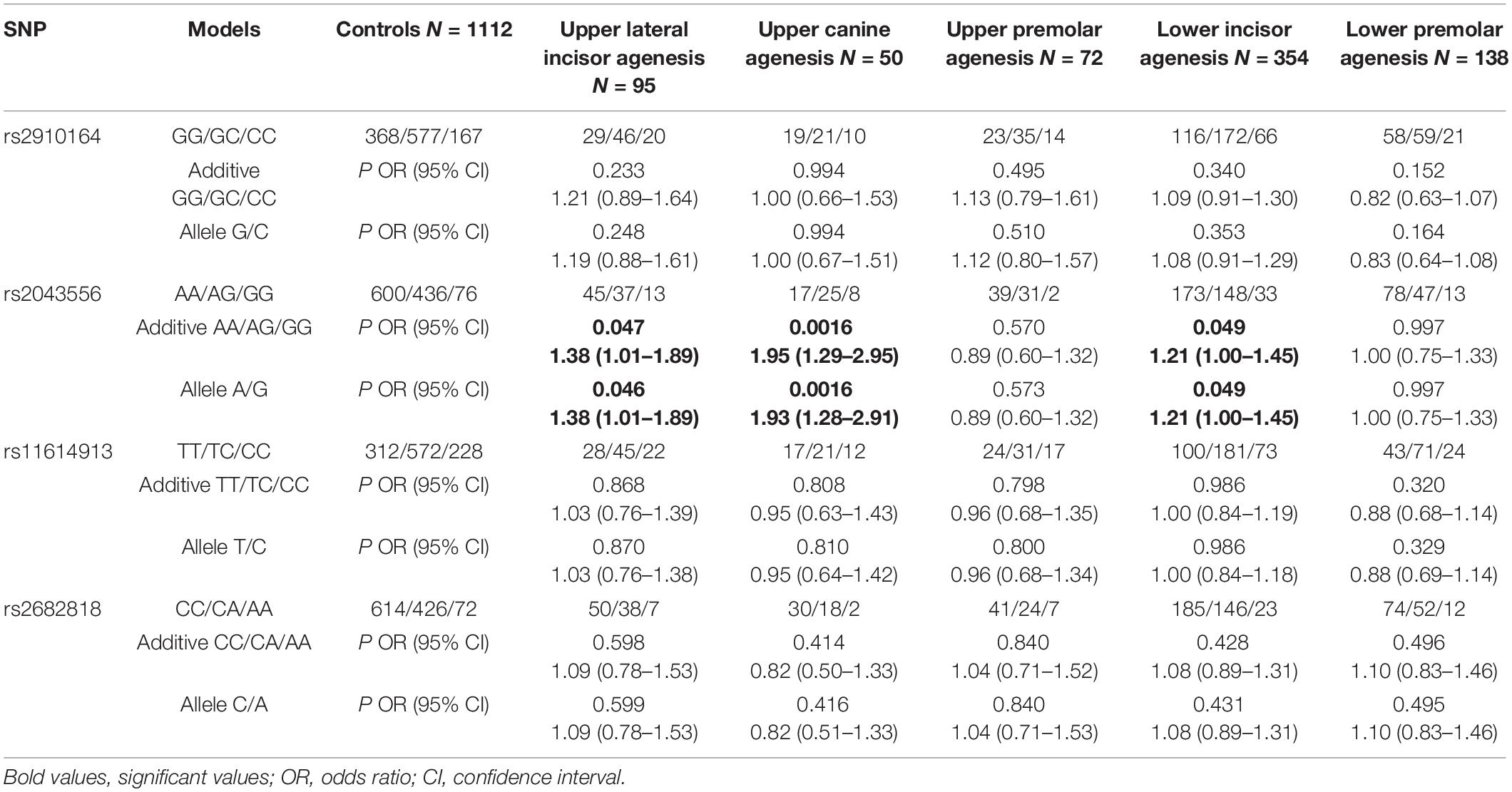
Table 2. Association of the four miRNA gene SNPs with the risk of different types of teeth agenesis.
Rs2043556 Affects the Mature of miR-605-3p and miR-605-5p
MiRNA plasmids containing pre-miR-605-A and pre-miR-605-G were transiently transfected into HEK-293 and COS-7 cells. The expression levels of mature miR-605, including miR-605-3p and miR-605-5p, were, respectively, tested with qRT-PCR. As shown in Figures 1A,B, the processing efficiency of the pre-miR-605-G plasmid was significantly lower than that of the pre-miR-605-A plasmid in HEK-293 and COS-7 cells (miR-605-3P: PCOS–7 = 8.9E-4, PHEK–293 = 1.0E-4; miR-605-5P: PCOS–7 = 7.4E-3, PHEK–293 = 3.6E-7), consistent with the study by Zhou et al. (2019).
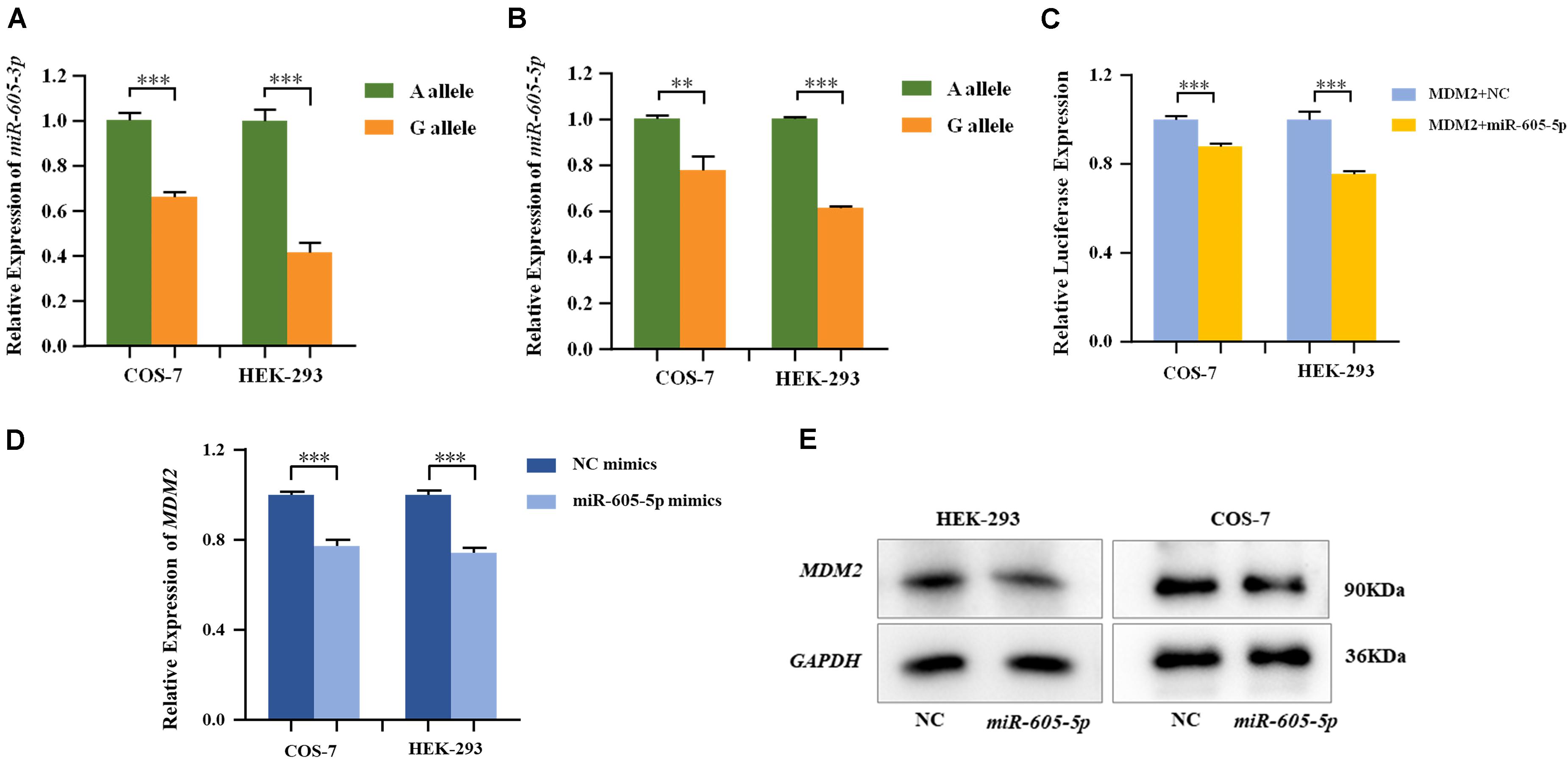
Figure 1. Expression of miR-605-3p (A) and miR-605-5p (B) in human cell lines. COS-7 and HEK-293 cells were transfected to express either the miR-605-3p A/G allele. The miR-605-3p expression levels were estimated by qRT-PCR; n = 3 (colonies) for each group and results are shown as mean values with the SD normalized to U6. (C) Dual luciferase reporter assays in COS-7 cells and HEK-293 cells demonstrated that MDM2 was direct target of miR-605-5p. MDM2 mRNA (D) and protein (E) expression in COS-7 and HEK-293 cells after transfection with miR-605-5p mimics. Transcript levels were analyzed by qPCR and normalized to GAPDH levels (**P < 0.01; ***P < 0.001).
miR-605-3p and miR-605-5p Expressions in Human Dental Pulp Stem Cells (DPSCs)
GSE138180 contained miRNAs expression of DPSCs in control groups and differentiated groups were cultured with an odontogenic differentiation medium (Supplementary Table S4). Here, we found miR-605-5p expression was significantly higher than miR-605-3p in both two groups (Pcontrol = 1.27E-7, Pdifferentiation = 1.62E-5). In addition, miR-605-5p expression increased after odontogenic differentiation (P = 1.12E-3) while miR-605-3p showed no significant change (P = 3.67E-1). Taken together, miR-605-5p played a more important role in tooth development and warranted the further research.
MDM2 Was Identified as the Target Gene of miR-605-5p
MDM2, highly expressed in the odontoblasts in the dental papilla cells of mouse incisors and molars (Zheng et al., 2019), was consistently predicted by four databases to be a favorable target of miR-605-5p (Supplementary Figure S2). In addition, P53, the canonical substrate of MDM2, was expressed in all layers of the human tooth buds (Muica Nagy-Bota et al., 2014). We therefore cloned luciferase reporter plasmids with the MDM2 3′-UTR and co-transfected with miRNA in HEK-293 and COS-7 cells. As presented in Figure 1C, luciferase reporter gene analysis showed that the activity of the MDM2 3’-UTR reporter gene was significantly decreased by miR-605-5p (PCOS–7=6.5E-4, PHEK–293 = 4.0E-4). In addition, when transfecting miR-605-5p mimics into these cell lines, the expression of MDM2 mRNA and protein were decreased significantly (Figures 1D,E).
MDM2 and P53 Are Expressed During the Cap Stage of Tooth Germs
As the gene expression data (GSE48150) in GEO datasets showed, tooth germs of molar, incisor, and canine at the cap stage were, respectively, dissected from 12-week-old human embryonic oral cavity for RNA extraction. We found that both MDM2 and P53 were expressed in the dental tissues by multiple transcripts (Figure 2A). Furthermore, P53 exhibited higher expression in canine germs than in molar (P = 0.09) and incisor germs (P = 0.04) compared by the paired t test (Supplementary Table S5).
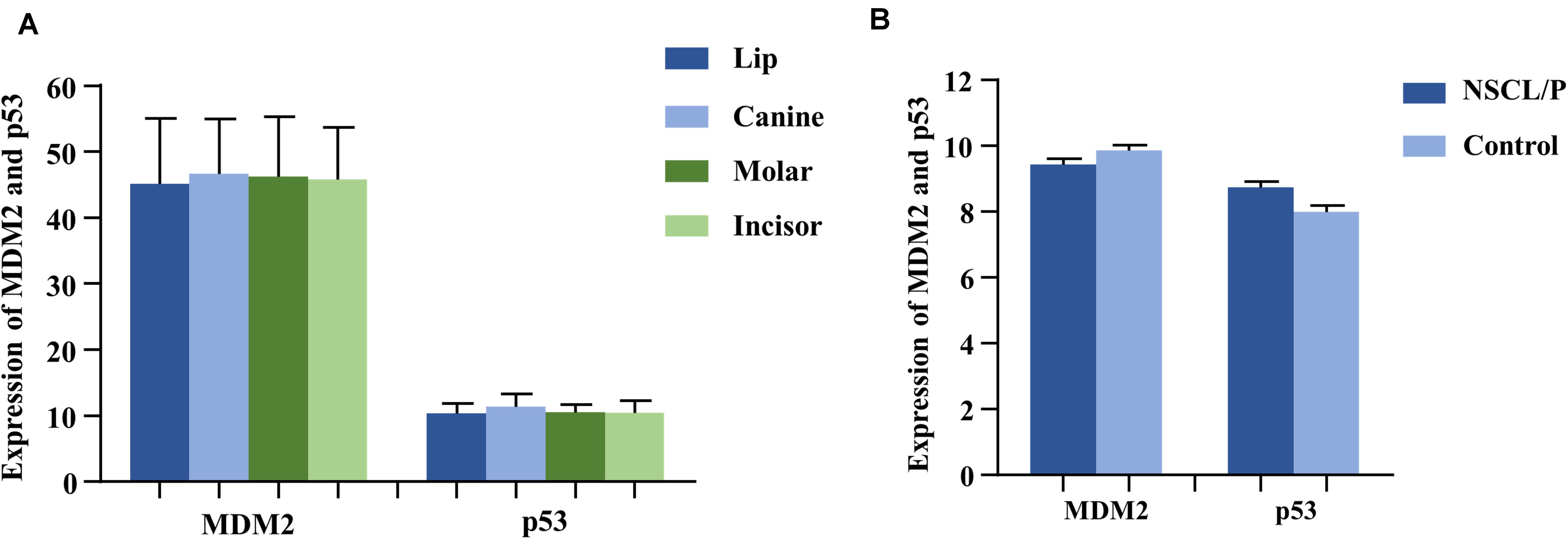
Figure 2. The expression of MDM2 and p53 in human tooth germs (A) from GEO datasets (GSE48150) and dental pulp stem cells (B) from GEO datasets (GSE42589). NSCL/P, non-syndromic cleft lip with or without cleft palate.
MDM2 and P53 Expressions in Human Dental Pulp Stem Cells (DPSCs)
Based on the analysis of gene expression data (GSE42589) in DPSCS from GEO datasets, both MDM2 and P53 mRNA were detected in DPSCs from 13 samples, including 6 control samples and 7 non-syndromic cleft lip with or without cleft palate (NSCL/P) samples (Figure 2), which often accompanied by tooth agenesis. Interestingly, a significant reverse correlation between MDM2 and P53 was observed by Spearman’s correlation (r = −0.709, P = 0.007).
Discussion
Single nucleotide polymorphisms in miRNA genes may affect the expression of miRNA and its binding efficiency with target genes, leading to the occurrence of common human diseases (Ryan et al., 2010; Schoen et al., 2017). Existing evidence shows that SNPs in miRNA can be useful markers for a variety of diseases (Hui et al., 2016; Pan et al., 2018). The four SNPs in the present study had been reported in various human diseases. For example, pre-miR-605/rs2043556 had previously been related to the lung cancer (Xu et al., 2018) and oral squamous cell carcinoma (Miao et al., 2016). miR-146a/rs2910164 was associated with the non-syndromic orofacial cleft (Pan et al., 2018) and bladder cancer (Wang et al., 2012). miR-196a2/rs11614913 showed an association with the hepatocellular carcinoma (Zheng et al., 2017) and type 1 diabetes mellitus (Ibrahim et al., 2019). pre-miR-618/rs2682818 had been reported in the colorectal cancer (Chen et al., 2018) and lymphomagenesis (Fu et al., 2014). However, their roles in tooth development remain to be explored.
Here, we selected four common variants in miRNA genes and found that rs2043556 was nominally associated with non-syndromic tooth agenesis risk in the overall analysis. The type of non-syndromic tooth agenesis may be associated with specific types of mutation according to the previous study (Williams and Letra, 2018). Jonsson et al. (2018) found two variants, located in EDA and FOXP1, were associated with agenesis of the maxillary lateral incisors. EDA was also identified as a genetic risk factor for maxillary lateral incisor agenesis in another research (Alves-Ferreira et al., 2014). In the present study, rs2043556 showed significant association with the risk of upper canine agenesis and nominal association with the risk of upper lateral incisor and lower incisor agenesis. To the best of our knowledge, this is the first finding of genetic variants contributing to susceptibility of canine agenesis.
Rs2043556 was located in pre-miR-605 and our study showed that this SNP was associated with differential expression of mature miR-605 (including miR-605-3p and miR-605-5p), consistent with the study by Zhou et al. (2017) Pre-miR-605 had previously been linked to a number of malignancies, such as prostate cancer, breast cancer (Morales et al., 2018), non-small-cell lung cancer (Zhou and Li, 2019), and oral squamous cell carcinoma (Miao et al., 2016), suggesting its potential as a biomarker for cancer. Interestingly, tooth agenesis and cancer development share common molecular pathways and previous study found that almost all types of tumors were more common in families with tooth agenesis (Kuchler et al., 2013).
MiR-605, a component in the P53 gene network, was the first miRNA identified as upregulating P53 by repressing the translation of MDM2 (Xiao et al., 2011). One of the core functions of P53 was to activate apoptosis (Fridman and Lowe, 2003), which existed in all stages of tooth development and played an important role in the development and formation of the final form of the tooth crown. Matalova et al. (2004) found strong expressions of P53 in the dental buds of mouse embryos. Furthermore, It had been reported that P53 was expressed in all layers of the human tooth buds (Muica Nagy-Bota et al., 2014). Recently, (Zheng et al., 2019) found that MDM2 promoted the odontoblast-like differentiation of mouse dental papilla cells (mDPCs) by ubiquitinating both DLX3 and P53. Here, we confirmed that MDM2 was the target gene of miR-605-5p and found a negative correlation between MDM2 and P53 in dental pulp stem cells. Thus, we hypothesized that the MDM2: miR-605: P53 feedback loop may also exist during tooth development and rs2043556, significantly associated with differential expression of miR-605, contributed to the risk of tooth agenesis probably by influencing this loop (Supplementary Figure S3).
Taken together, this is the first study to evaluate SNPs in miRNA and non-syndromic tooth agenesis susceptibility in a relatively large sample size. Our findings indicated that miR-605 rs2043556 was associated with risk of non-syndromic tooth agenesis probably by affecting MDM2: miR-605: P53 feedback loop.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved by Nanjing Medical University Institution Review Board (PJ2004-030-001). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin.
Author Contributions
LW and YP directed the study, obtained the financial support and were responsible for the study design, interpretation of results, and manuscript writing. MG performed the overall project management with LF. XY performed the statistical analyses with SL and FY, and drafted the initial manuscript with GZ. LM directed each participating study and jointly organized the study. FY and SL were responsible for the sample processing and managed the genotyping data. LF and XY were responsible for the subject recruitment and sample collection. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81830031, 81970969, and 81570959), the State Key Lab of Reproductive Medicine of Nanjing Medical University (JX116GSP 20171416), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD-2018-87), the Natural Science Foundation of Jiangsu Province (BL2014073 and 15KJA320002 to LW), the Qing Lan Project (YP), Six Distinguished Talent (2016-WSW-008) and Jiangsu Provincial Medical Talent (ZDRCC2016023 to YP), and the Jiangsu Provincial Key Medical Discipline (zdxka2016026).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all the study participants, research staff, and students who contributed to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2020.01052/full#supplementary-material
Footnotes
- ^ http://microrna.sanger.ac.uk/
- ^ http://ncbi.nlm.nih.gov/SNP
- ^ http://www.hapmap.org
- ^ http://www.patrocles.org/
- ^ http://www.ncbi.nlm.nih.gov/geo
References
Acevedo, A. C., Poulter, J. A., Alves, P. G., de Lima, C. L., Castro, L. C., Yamaguti, P. M., et al. (2015). Variability of systemic and oro-dental phenotype in two families with non-lethal Raine syndrome with FAM20C mutations. BMC Med. Genet. 16:8. doi: 10.1186/s12881-015-0154-5
Alves-Ferreira, M., Pinho, T., Sousa, A., Sequeiros, J., Lemos, C., and Alonso, I. (2014). Identification of genetic risk factors for maxillary lateral incisor agenesis. J. Dent. Res. 93, 452–458. doi: 10.1177/0022034514523986
Botstein, D., and Risch, N. (2003). Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33(Suppl.), 228–237. doi: 10.1038/ng1090
Chen, Y., Du, M., Chen, W., Zhu, L., Wu, C., Zhang, Z., et al. (2018). Polymorphism rs2682818 in miR-618 is associated with colorectal cancer susceptibility in a Han Chinese population. Cancer Med. 7, 1194–1200. doi: 10.1002/cam4.1409
Fan, L., Kan, S., Yang, F., Xu, H., Li, H., Zhu, G., et al. (2018). Non-syndromic cleft lip with or without palate susceptible loci is associated with tooth agenesis. Oral Dis. 25, 803–811. doi: 10.1111/odi.13024
Fauzi, N. H., Ardini, Y. D., Zainuddin, Z., and Lestari, W. (2018). A review on non-syndromic tooth agenesis associated with PAX9 mutations. Jpn. Dent. Sci. Rev. 54, 30–36. doi: 10.1016/j.jdsr.2017.08.001
Fridman, J. S., and Lowe, S. W. (2003). Control of apoptosis by p53. Oncogene 22, 9030–9040. doi: 10.1038/sj.onc.1207116
Fu, A., Hoffman, A. E., Liu, R., Jacobs, D. I., Zheng, T., and Zhu, Y. (2014). Targetome profiling and functional genetics implicate miR-618 in lymphomagenesis. Epigenetics 9, 730–737. doi: 10.4161/epi.27996
Ghanbari, M., Darweesh, S. K., de Looper, H. W., van Luijn, M. M., Hofman, A., Ikram, M. A., et al. (2016). Genetic variants in MicroRNAs and their binding sites are associated with the risk of parkinson disease. Hum. Mutat. 37, 292–300. doi: 10.1002/humu.22943
Gong, J., Tong, Y., Zhang, H. M., Wang, K., Hu, T., Shan, G., et al. (2012). Genome-wide identification of SNPs in microRNA genes and the SNP effects on microRNA target binding and biogenesis. Hum. Mutat. 33, 254–263. doi: 10.1002/humu.21641
Gong, M., Qian, Y. J., Gu, N., Wang, W., Wang, H., Ma, L., et al. (2015). Association of BMP4 polymorphisms with isolated tooth agenesis in a Chinese Han population: a case-control study. Eur. Rev. Med. Pharmacol. Sci. 19, 2188–2194.
Ha, M., and Kim, V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell. Biol. 15, 509–524. doi: 10.1038/nrm3838
Hui, L., Wu, H. U. A., Yang, N., Guo, X., and Jang, X. (2016). Identification of prognostic microRNA candidates for head and neck squamous cell carcinoma. Oncol. Rep. 35, 3321–3330. doi: 10.3892/or.2016.4698
Ibrahim, A. A., Ramadan, A., Wahby, A. A., Hassan, M., Soliman, H. M., and Abdel Hamid, T. A. (2019). Micro-RNA 196a2 expression and miR-196a2 (rs11614913) polymorphism in T1DM: a pilot study. J. Pediatr. Endocrinol. Metab. 32, 1171–1179. doi: 10.1515/jpem-2019-0226
Jonsson, L., Magnusson, T. E., Thordarson, A., Jonsson, T., Geller, F., Feenstra, B., et al. (2018). Rare and common variants conferring risk of tooth agenesis. J. Dent. Res. 97, 515–522. doi: 10.1177/0022034517750109
Kroliczewski, J., Sobolewska, A., Lejnowski, D., Collawn, J. F., and Bartoszewski, R. (2018). microRNA single polynucleotide polymorphism influences on microRNA biogenesis and mRNA target specificity. Gene 640, 66–72. doi: 10.1016/j.gene.2017.10.021
Kuchler, E. C., Lips, A., Tannure, P. N., Ho, B., Costa, M. C., Granjeiro, J. M., et al. (2013). Tooth agenesis association with self-reported family history of cancer. J. Dent. Res. 92, 149–155. doi: 10.1177/0022034512468750
Liu, H., Han, D., Wong, S., Nan, X., Zhao, H., and Feng, H. (2013). rs929387 of GLI3 is involved in tooth agenesis in Chinese Han population. PLoS One 8:e80860. doi: 10.1371/journal.pone.0080860
Liu, W., Konermann, A., Guo, T., Jager, A., Zhang, L., and Jin, Y. (2014). Canonical Wnt signaling differently modulates osteogenic differentiation of mesenchymal stem cells derived from bone marrow and from periodontal ligament under inflammatory conditions. Biochim. Biophys. Acta 1840, 1125–1134. doi: 10.1016/j.bbagen.2013.11.003
Lu, Y., Qian, Y., Zhang, J., Gong, M., Wang, Y., Gu, N., et al. (2016). Genetic variants of BMP2 and their association with the risk of non-syndromic tooth agenesis. PLoS One 11:e0158273. doi: 10.1371/journal.pone.0158273
Matalova, E., Tucker, A. S., and Sharpe, P. T. (2004). Death in the life of a tooth. J. Dent. Res. 83, 11–16. doi: 10.1177/154405910408300103
Mi, Y., Ren, K., Zou, J., Bai, Y., Zhang, L., Zuo, L., et al. (2018). The association between three genetic variants in MicroRNAs (Rs11614913, Rs2910164, Rs3746444) and prostate cancer risk. Cell Physiol. Biochem. 48, 149–157. doi: 10.1159/000491671
Miao, L., Wang, L., Zhu, L., Du, J., Zhu, X., Niu, Y., et al. (2016). Association of microRNA polymorphisms with the risk of head and neck squamous cell carcinoma in a Chinese population: a case-control study. Chin. J. Cancer 35:77. doi: 10.1186/s40880-016-0136-9
Morales, S., De Mayo, T., Gulppi, F. A., Gonzalez-Hormazabal, P., Carrasco, V., Reyes, J. M., et al. (2018). Genetic variants in pre-miR-146a, pre-miR-499, pre-miR-125a, pre-miR-605, and pri-miR-182 are associated with breast cancer susceptibility in a South American population. Genes 9:427. doi: 10.3390/genes9090427
Muica Nagy-Bota, M. C., Pap, Z., Denes, L., Ghizdavat, A., Brinzaniuc, K., Lup Cosarca, A. S., et al. (2014). Immunohistochemical study of Ki67, CD34 and p53 expression in human tooth buds. Rom. J. Morphol. Embryol. 55, 43–48.
Pan, Y., Li, D., Lou, S., Zhang, C., Du, Y., Jiang, H., et al. (2018). A functional polymorphism in the pre-miR-146a gene is associated with the risk of nonsyndromic orofacial cleft. Hum. Mutat. 39, 742–750. doi: 10.1002/humu.23415
Peng, C. G., Li, N., Ng, Y. K., Zhang, J. Z., Meier, F., Theis, F. J., et al. (2012). A unilateral negative feedback loop between miR-200 microRNAs and Sox2/E2F3 controls neural progenitor cell-cycle exit and differentiation. J. Neurosci. 32, 13292–13308. doi: 10.1523/Jneurosci.2124-12.2012
Ryan, B. M., Robles, A. I., and Harris, C. C. (2010). Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer 10, 389–402. doi: 10.1038/nrc2867
Schoen, C., Aschrafi, A., Thonissen, M., Poelmans, G., Von den Hoff, J. W., and Carels, C. E. L. (2017). MicroRNAs in palatogenesis and cleft palate. Front. Physiol. 8:165. doi: 10.3389/fphys.2017.00165
Sehic, A., Tulek, A., Khuu, C., Nirvani, M., Sand, L. P., and Utheim, T. P. (2017). Regulatory roles of microRNAs in human dental tissues. Gene 596, 9–18. doi: 10.1016/j.gene.2016.10.009
Sun, F., Wan, M., Xu, X., Gao, B., Zhou, Y., Sun, J., et al. (2014). Crosstalk between miR-34a and notch signaling promotes differentiation in apical papilla stem cells (SCAPs). J. Dent. Res. 93, 589–595. doi: 10.1177/0022034514531146
Vastardis, H. (2000). The genetics of human tooth agenesis: new discoveries for understanding dental anomalies. Am. J. Orthod. Dentofacial Orthop 117, 650–656.
Wang, M., Chu, H., Li, P., Yuan, L., Fu, G., Ma, L., et al. (2012). Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer Res. 72, 6173–6182. doi: 10.1158/0008-5472.can-12-0688
Williams, M. A., and Letra, A. (2018). The changing landscape in the genetic etiology of human tooth agenesis. Genes 9:255. doi: 10.3390/genes9050255
Wong, S. W., Han, D., Zhang, H., Liu, Y., Zhang, X., Miao, M. Z., et al. (2018). Nine novel PAX9 mutations and a distinct tooth agenesis genotype-phenotype. J. Dent. Res. 97, 155–162. doi: 10.1177/0022034517729322
Xiao, J., Lin, H., Luo, X., Luo, X., and Wang, Z. (2011). miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress. EMBO J. 30, 524–532. doi: 10.1038/emboj.2010.347
Xu, X., Yin, Z., Ren, Y., Guan, P., and Zhou, B. (2018). Associations of miR-605 rs2043556 polymorphism with the susceptibility and overall survival of lung cancer in Chinese non-smoking females. Int. J. Clin. Exp. Pathol. 11, 438–447.
Yu, M., Wang, H., Fan, Z., Xie, C., Liu, H., Liu, Y., et al. (2019a). BMP4 mutations in tooth agenesis and low bone mass. Arch. Oral Biol. 103, 40–46. doi: 10.1016/j.archoralbio.2019.05.012
Yu, M., Wong, S. W., Han, D., and Cai, T. (2019b). Genetic analysis: wnt and other pathways in nonsyndromic tooth agenesis. Oral Dis. 25, 646–651. doi: 10.1111/odi.12931
Yu, P., Yang, W., Han, D., Wang, X., Guo, S., Li, J., et al. (2016). Mutations in WNT10B are identified in individuals with oligodontia. Am. J. Hum. Genet. 99, 195–201. doi: 10.1016/j.ajhg.2016.05.012
Zheng, H., Yang, G., Fu, J., Chen, Z., and Yuan, G. (2019). Mdm2 promotes odontoblast-like differentiation by ubiquitinating Dlx3 and p53. J. Dent. Res. 99, 320–328. doi: 10.1177/0022034519893672
Zheng, L., Zhuang, C., Zhao, J., and Ming, L. (2017). Functional miR-146a, miR-149, miR-196a2 and miR-499 polymorphisms and the susceptibility to hepatocellular carcinoma: an updated meta-analysis. Clin. Res. Hepatol. Gastroenterol. 41, 664–676. doi: 10.1016/j.clinre.2017.03.005
Zhou, W., and Li, R. (2019). microRNA-605 inhibits the oncogenicity of non-small-cell lung cancer by directly targeting Forkhead Box P1. Onco Targets Ther. 12, 3765–3777. doi: 10.2147/OTT.S193675
Zhou, W. L., Mo, Z. Z., Xiao, F. Y., Dai, W., Wang, G., Zhou, G., et al. (2019). microRNA-605 rs2043556 polymorphisms affect clopidogrel therapy through modulation of CYP2B6 and P2RY12 in acute coronary syndrome patients. Platelets [Epub ahead of print]. doi: 10.1080/09537104.2019.1696455
Keywords: miRNA, single nucleotide polymorphism, tooth agenesis, miR-605-5p, MDM2, p53
Citation: Gu M, Yu X, Fan L, Zhu G, Yang F, Lou S, Ma L, Pan Y and Wang L (2020) Genetic Variants in miRNAs Are Associated With Risk of Non-syndromic Tooth Agenesis. Front. Physiol. 11:1052. doi: 10.3389/fphys.2020.01052
Received: 17 April 2020; Accepted: 31 July 2020;
Published: 21 August 2020.
Edited by:
Pierfrancesco Pagella, University of Zurich, SwitzerlandReviewed by:
Sing-Wai Wong, University of North Carolina at Chapel Hill, United StatesAlexandre Rezende Vieira, University of Pittsburgh, United States
Maria Gazouli, National and Kapodistrian University of Athens, Greece
Hailan Feng, Peking University, China
Copyright © 2020 Gu, Yu, Fan, Zhu, Yang, Lou, Ma, Pan and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lin Wang, bHc2MDNAbmptdS5lZHUuY24=; Yongchu Pan, cGFueW9uZ2NodUBuam11LmVkdS5jbg==
†These authors have contributed equally to this work
 Min Gu1,2,3†
Min Gu1,2,3† Xin Yu
Xin Yu Shu Lou
Shu Lou Lan Ma
Lan Ma Yongchu Pan
Yongchu Pan