- 1Department of Biology, University of Padova, Padua, Italy
- 2Department of Cell Biology and Imaging, Jagiellonian University, Kraków, Poland
Sleep-like states have been described in Drosophila and the mechanisms and factors that generate and define sleep-wake profiles in this model organism are being thoroughly investigated. Sleep is controlled by both circadian and homeostatic mechanisms, and environmental factors such as light, temperature, and social stimuli are fundamental in shaping and confining sleep episodes into the correct time of the day. Among environmental cues, light seems to have a prominent function in modulating the timing of sleep during the 24 h and, in this review, we will discuss the role of light inputs in modulating the distribution of the fly sleep-wake cycles. This phenomenon is of growing interest in the modern society, where artificial light exposure during the night is a common trait, opening the possibility to study Drosophila as a model organism for investigating shift-work disorders.
Introduction
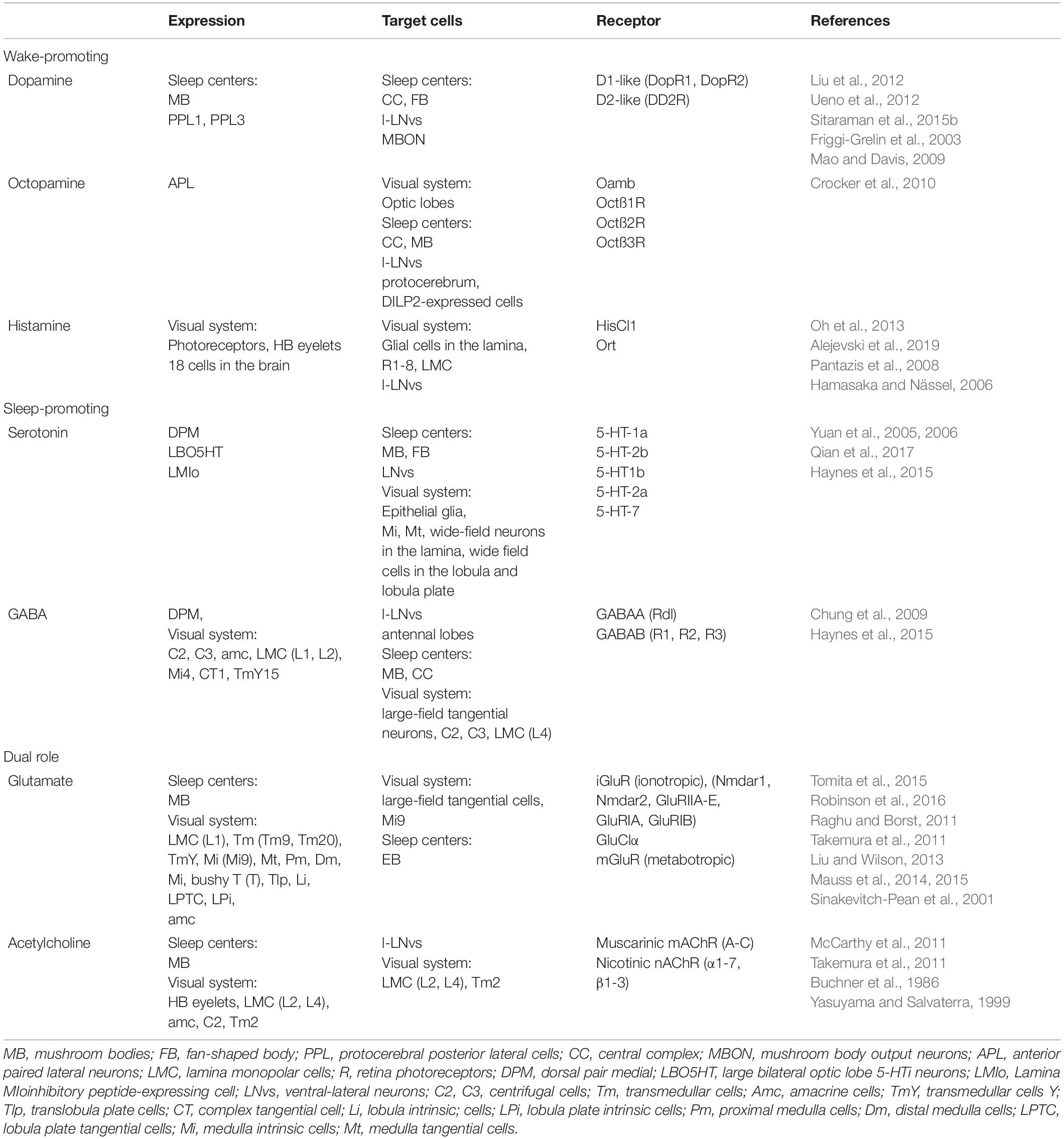
Life on Earth has been shaped by rhythmic changes of environmental cues and living organisms have evolved endogenous mechanisms to coordinate physiological and behavioural functions. For example, in humans and other diurnal animals, most activities occur during the day, contrary to nocturnal animals, mostly active during the night. Among environmental factors, light plays a major role in adjusting temporal niches of animal behaviour in relation to natural surroundings, in the sense that it acts as an arousal signal for diurnal animals and at the same time promotes sleep in nocturnal ones (Redlin, 2001). Drosophila exhibits a very well-established daily activity pattern: under 12 h Light-12 h Dark cycles (LD12:12), flies display distinct morning and evening bouts of activity, separated by a prolonged siesta in the middle of the day. This behavioural output is the result of an orchestrated activity of different clusters of clock cells and signals (Grima et al., 2004; Stoleru et al., 2004; Picot et al., 2007; Cusumano et al., 2009; Zhang et al., 2009; Yao and Shafer, 2014; Chatterjee et al., 2018; Díaz et al., 2019; Schlichting et al., 2019b). In Drosophila, the circadian oscillator is located in about 150 neurons that, based on their anatomical location, are classified as: small and large ventral-lateral neurons (s-LNvs and l-LNvs, respectively), dorsal-lateral neurons (LNds), lateral posterior neurons (LPN), and three groups of dorsal neurons (DN1s, DN2s, and DN3s) (Schubert et al., 2018; Figure 1A). Among these, the s-LNvs and LNds are specifically involved in the control of morning and evening activity, respectively (Grima et al., 2004; Stoleru et al., 2004). Daily activity has specific pattern with two peaks: just after lights-on and around lights-off (Figure 1B). Morning peak is mostly driven by light, as in constant darkness (DD) it is much weaker, while evening peak is under circadian control. In addition, morning and evening anticipation is observed, which means that activity starts to increase around 3 h before the lights-on and lights-off (Figure 1B). Moreover, bimodal pattern of activity is observed also in clock mutants, but only in light-dark conditions, in constant darkness flies are completely arrhythmic. Clock mutants do not show morning anticipation, as they need light pulse to enhance activity level.
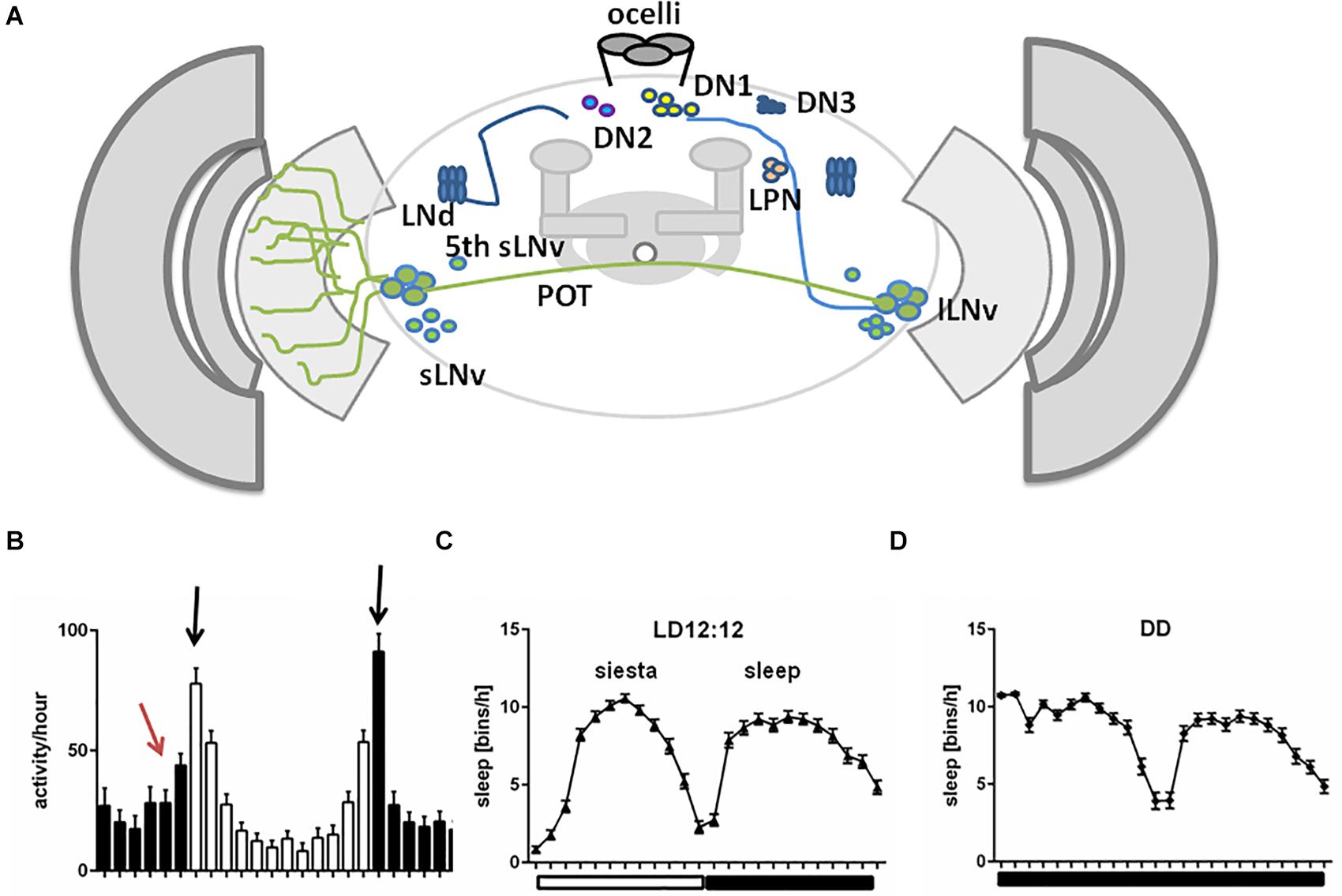
Figure 1. A schematized representation of clock network in Drosophila brain. (A) The main pacemaker cells, lateral ventral neurons, small (s-LNvs) and large (l-LNvs) are located in accessory medulla. l-LNvs send projections, called posterior optic tract (POT), to the contralateral hemisphere and form network of processes in the medulla neuropil, while s-LNvs innervate dorsal brain. Six lateral dorsal neurons (LNds) and three lateral posterior neurons (LPNs) are located above the main pacemakers. In the dorsal brain, three groups of dorsal neurons are located (DN1s, DN2s, DN3). (B) Representative activity profile in light:dark (LD12:12) conditions with pointed morning/evening peaks (black arrows) and morning anticipation (red arrow). (C,D) Representative sleep pattern observed in LD cycles (C), with siesta during the day and sleep during the night and constant darkness (DD) conditions (D).
Compelling evidence attests to the influence of light on Drosophila rest-activity rhythms (recently reviewed in Helfrich-Förster, 2019). For instance, flies kept in constant darkness are sensitive to brief light pulses: they delay or advance their activity when the light stimulus is delivered in the early or late subjective night, respectively (Stanewsky et al., 1998). Also flies lacking compound eyes, clieya mutants (Helfrich-Förster et al., 2001), or with impaired photoreceptor signal transduction, due to deficiency in norpA-encoded phospholipase C-β activity (Bloomquist et al., 1988), have a clearly advanced evening activity (Schlichting et al., 2019a) and a similar phenotype has been recently reported in flies with degenerated photoreceptors (Cusumano et al., 2018; Niu et al., 2019; Weigelt et al., 2019).
Here we will review the role of light and light input pathways in shaping the fly sleep-wake pattern. In particular, we will initially describe neurotransmitters that regulate sleep in Drosophila. We will then focus on the sleep centers and pathways (visual and not visual) mediating light signal to the brain. Then, we will review the neuronal networks involving circadian pacemaker cells and finally the influence of light (timing and intensity) on sleep architecture.
Sleep in Drosophila
As in mammals, sleep in insects is characterized by specific sleep posture and elevated sensory threshold. Although sleep patterns vary between different strains, sleep is always composed of daytime sleep, called “siesta,” with the maximum around noon, and nighttime sleep with peak at midnight (Figures 1C,D). Daytime sleep is less deep, with shorter single sleep episodes and lower arousal threshold (the level of sensory stimuli required for behavioural response), meaning that flies are more sensitive to awakening factors during the day than during the night (Hendricks et al., 2000; Huber et al., 2004). Wake/rest daily rhythms in Drosophila can be recorded by placing individuals in glass tubes and monitoring the movements using infrared beam-based activity monitors (DAMS, Trikinetics) or video recordings. Sleep in flies is defined as at least 5 min of total inactivity (Shaw et al., 2000), meaning that during this time no infrared break is recorded by the system. Recordings of local field potential in the brain suggest that Drosophila sleep can be divided to specific phases of different intensities, similar to mammalian sleep (Nitz et al., 2002; van Alphen et al., 2013; Raccuglia et al., 2019). Sleep differs according to sex: males sleep more, with comparable resting time during the day and night, while mated females sleep mostly during the night, and they are more active during the day (Huber et al., 2004). Sleep in Drosophila can be defined by the following parameters: bouts of sleep (number of sleep episodes), sleep bout length, which is useful for analysis of sleep fragmentation, and sleep latency/night offset (time between lights-off and the first sleep bout).
Neurotransmitters
Sleep is controlled through neurotransmitters, divided into sleep-promoting [serotonin and gamma-aminobutyric acid (GABA)], wake-promoting (dopamine, octopamine, histamine) and those playing a dual role depending on target cells (acetylcholine, glutamate) (Table 1; reviewed in Ly et al., 2018). Both sleep-promoting neurotransmitters are released by dorsal pair medial neurons (DPMs) and directly affect mushroom bodies by inhibiting their activity (Haynes et al., 2015). GABA inhibits l-LNvs activity through Rdl receptor (Chung et al., 2009), and the pharmacological administration of GABA-A agonist (Gaboxadol) induces sleep behaviour in flies (Dissel et al., 2015) and humans (Faulhaber et al., 1997), indicating conserved role of GABA receptors in promoting sleep. Among wake-promoting molecules, dopamine and octopamine regulate the activity of sleep centers, central complex (CC), and mushroom bodies (MB) (Friggi-Grelin et al., 2003; Mao and Davis, 2009; Crocker et al., 2010), while histamine links retinal and extra-retinal photoreceptors to clock neurons (Oh et al., 2013). The role of octopamine is not well defined as recent data showed that the effect of octopamine could be sleep-promoting rather than wake-promoting (Deng et al., 2019). Finally, glutamate can promote sleep (Tomita et al., 2015) or wakefulness (Zimmerman et al., 2017) depending on the postsynaptic receptors. A similar effect is described for acetylcholine which promotes wakefulness by exciting l-LNvs when released from extra-retinal photoreceptors, Hofbauer–Buchner eyelets, and L2 neurons (McCarthy et al., 2011; Muraro and Ceriani, 2015; Schlichting et al., 2016), and has a sleep-promoting effect when released from mushroom bodies (Yi et al., 2013).
Sleep Centers
Drosophila sleep centers are located in different brain regions, although the most essential ones reside in the central and dorsal region, as MB and CC, composed of dorsal fan-shaped body (FB), and ellipsoid body (EB) with the ring structure (EB-R2) (Joiner et al., 2006; Pitman et al., 2006; Donlea et al., 2011; Liu et al., 2012, 2016; Guo et al., 2016; Figures 2A,B).
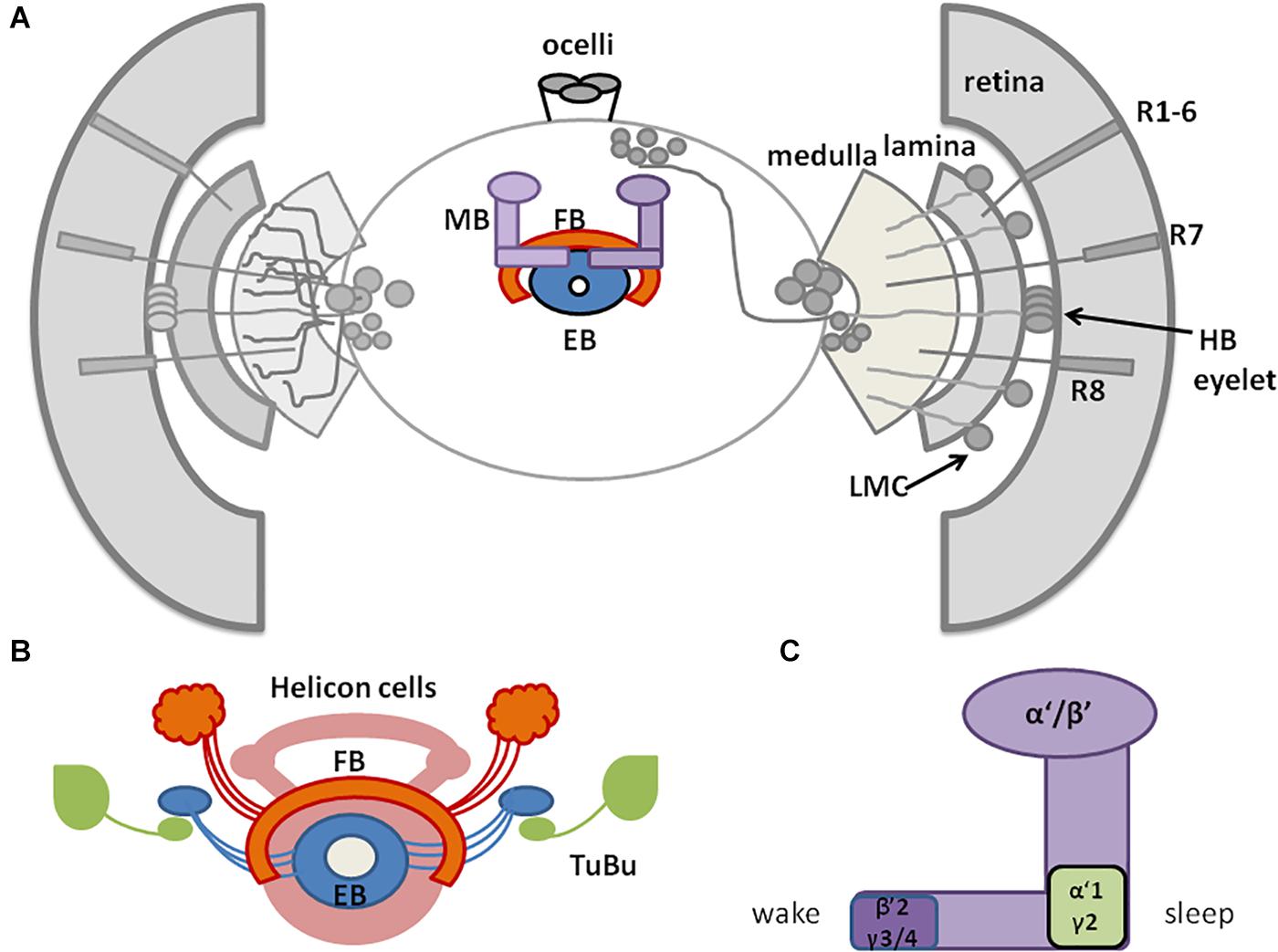
Figure 2. A schematic representation of Drosophila sleep centers and structures mediating light signals. (A) Drosophila brain with three photoreceptive structures: visual system (composed of retina and three optic neuropils: lamina, medulla, and lobula), extra-retinal Hofbauer–Buchner (HB) eyelets and ocelli, located on the top of head. Light is received by retinal photoreceptors R1–6, which terminate in the lamina, and R7–8, terminating in the medulla and transmitting through lamina monopolar cells (LMC) to the deep brain. HB eyelets, located between retina and lamina, are sensitive to high intensity light and transmit signal directly to clock neurons (ventral-lateral neurons_LNvs). In the central brain sleep centers are located with mushroom bodies (MB), ellipsoid body (EB), and fan-shaped body (FB). (B) Scheme of sleep center, composed of fan-shaped body (FB), mushroom bodies (MB), ellipsoid body (EB), and additional cells: tubercular bulbar neurons (TuBu) and helicon cells. (C) Mushroom bodies are composed of vertical (α, α’) and horizontal (β, β’, γ) lobes. Wake-promoting neurons are located in β’2, γ3/4, while sleep-promoting ones in α’1, γ2 region of MB.
Mushroom bodies are composed of neurons called Kenyon cells (Technau, 2007), whose axons form lobes: two vertical (α, α’) and three horizontal (β, β’, γ). MB contains both wake-promoting and sleep-promoting neurons, located in β’2, γ3/4 and α’1, γ2 region, respectively (Joiner et al., 2006; Sitaraman et al., 2015b; Artiushin and Sehgal, 2017; Figure 2C). MB receive inhibiting, wake-promoting signals through serotonin and GABA (Yuan et al., 2006; Haynes et al., 2015). Specific MB compartments send information to mushroom body output neurons (MBONs) via glutamate and acetylcholine, that have a wake- or sleep- promoting effect, respectively (Aso et al., 2014; Sitaraman et al., 2015a). MB express additional sleep-promoting factors. Among these, Neurocalcin (NCA) and Noktochor (Nkt) promote nighttime sleep by suppressing nocturnal arousal (Chen et al., 2019; Sengupta et al., 2019).
The CC is involved in the regulation of locomotor activity and visual processing (Liu et al., 2006; Poeck et al., 2008; Triphan et al., 2010; Seelig and Jayaraman, 2013). The upper part of CC contains the FB, with sleep-promoting ExFl2-cells, which receive signals from the protocerebral posterolateral cluster 1 (PPL1) and protocerebral posteromedial 3 (PPM3) (Liu et al., 2016). This dopaminergic pathway inhibits FB activity and suppresses sleep (Liu et al., 2012; Ueno et al., 2012; Kayser et al., 2014; Pimentel et al., 2016; Ni et al., 2019). FB can be also activated by glutamatergic input from the circadian clock, represented by Allatostatin A (AstA)-expressing LPN cells (Ni et al., 2019). Both inputs are integrated in FB to precisely control its activity and ultimately regulate shifts between sleep and wake states. In the final step, active FB releases GABA, which inhibits octopaminergic output arousal neurons (OAA), thus promoting sleep (Ni et al., 2019; Figure 3).
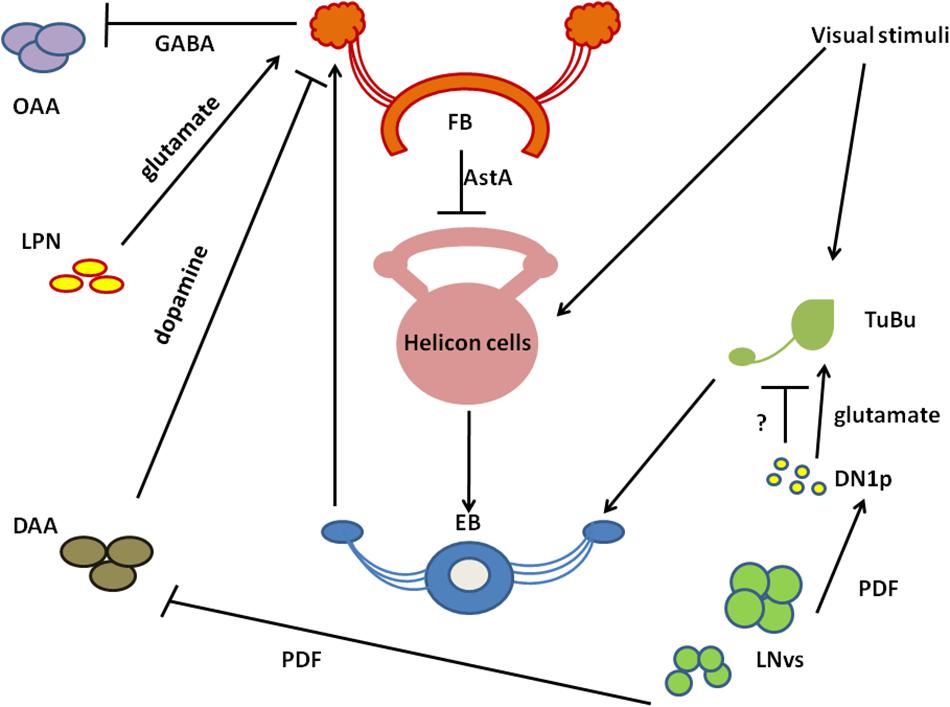
Figure 3. A schematized circuit of neurotransmission regulating sleep centers. Light affects clock neurons (ventral-lateral neurons_LNvs) activity, which stimulates pigment dispersing factor (PDF) release. In turn, PDF inhibits dopamine arousal neurons (DAAs), which normally decrease fan-shaped body (FB) activity through dopamine. Lack of dopamine signalling promotes sleep. PDF activates also Dorsal Neurons posterior (DN1sp), which can inhibit tubercular bulbar cells (TuBu), or activate them through glutamate. Visual stimuli affect Tubu and helicon cells activity, both send signals to the ellipsoid body (EB). EB coordinates FB activity and receives feedback signals from FB through helicon cells, which are inhibited by AstA released from FB. In addition, FB obtains signals from lateral posterior neurons (LPNs) through glutamate. FB processes all inputs and sends final sleep-promoting information through inhibiting GABA signalling to output arousal neurons (OAAs).
Below the FB, EB is located, which is involved in memory formation and startle response to mechanical stimulation. The major neurons composing EB are called ring neurons (R), and they receive synaptic signals from the anterior visual tract, through tubercular bulbar (TuBu) neurons (Omoto et al., 2017). Additional cells, called helicon cells, receive and integrate visual inputs and connect FB and EB: during wakefulness, helicon cells are sensitive to visual inputs and propagate signals to R2 cells, which are important for the regulation of sleep depth. R2 cells activate sleep-promoting ExFl2 neurons, thus increasing sleep need. In turn, during sleep AstA released by FB inhibits helicon cells, which causes decreased responsiveness to visual stimuli (Donlea et al., 2018; Figure 3).
How Do Light Inputs Modulate Sleep in Drosophila?
Light signals to Drosophila brain are mediated either by visual structures, such as two large compound eyes (retinal photoreceptors) and two Hofbauer–Buchner eyelets (HB eyelets, extraretinal photoreceptors), and non-visual pathways, involving three ocelli and deep brain photoreceptors CRYPTOCHROME (CRY), QUASIMODO (QSM), and Rhodopsin 7 (Rh7) (reviewed in Helfrich-Förster, 2019).
Compound Eyes
Drosophila compound eyes comprise ∼800 units, called ommatidia, organized in regular structures innervating and conveying visual signals to the four distinct neuropil regions of the optic lobe (lamina, medulla, lobula, and lobula plate). Each ommatidium houses 20 cells, eight of which are photoreceptors (R1–R8) designated in processing light inputs as function of position, spectral composition, and axonal projections. The six outer photoreceptors (R1–R6), expressing the broad spectrum rhodopsin (Rh1) (Hardie, 1979; O’Tousa et al., 1985; Zuker et al., 1985), project to the lamina (Braitenberg, 1967; Strausfeld, 1971) and are intended for motion detection and image formation (Heisenberg and Buchner, 1977; Yamaguchi et al., 2008). The two inner central photoreceptors (R7–R8) reach and innervate the distant medulla and participate in color, UV, and polarized light detection (Melamed and Trujillo-Cenóz, 1967; Montell, 2012). R7–R8 photoreceptors are clustered in 30% “pale” [R7 expressing UV-sensitive-Rh3 (331 nm) and R8 expressing blue-sensitive-Rh5 (442 nm)] and 70% “yellow” ommatidia [R7 expressing UV-sensitive-Rh4 (355 nm) and R8 expressing green-sensitive-Rh6 (515 nm)] (Fryxell and Meyerowitz, 1987; Montell et al., 1987; Zuker et al., 1987; Feiler et al., 1992; Chou et al., 1996, 1999; Huber et al., 1997; Papatsenko et al., 1997; Salcedo et al., 1999). A seventh Rhodopsin (Rh7) (Adams et al., 2000) is expressed also in the compound eyes (specifically in R8) as well as in other brain neurons (including some clock neurons) (Senthilan and Helfrich-Förster, 2016; Grebler et al., 2017; Kistenpfennig et al., 2017; Ni et al., 2017; Senthilan et al., 2019). Its role and expression pattern have been recently discussed (Senthilan et al., 2019).
The visual cascade complex is located in the rhabdomeres of photoreceptor cells: the G-protein (Gq) activates the phospholipase C (PLC), [encoded by norpA (Bloomquist et al., 1988; Scott et al., 1995)] which hydrolyzes phosphatidylinositol 4,5-bisphosphate (PIP2) and promotes the opening of the TRP and the TRP-like (TRPL) cation channels (reviewed in Montell, 2012). The triggered calcium current is then balanced by the Na2 + /Ca2+ exchanger, Calx (Wang et al., 2005).
Each outer photoreceptor cell forms a tetrad synapse with L1 and L2 laminar neurons and L3 or amacrine cell or epithelial glia. Under certain conditions (bright light and impaired synaptic transmission) lamina interneurons feed back to photoreceptor cells, modulating their output (Zheng et al., 2006). In the lamina, projections from R1–6 are organized in synaptic modules called cartridges, in which three epithelial glial cells surround six photoreceptor terminals with invaginating capitate projections (Stuart et al., 2007). On the other hand, inner photoreceptors and lamina neurons form synaptic modules (columns) with medulla interneurons and neurons that convey visual information to the lobula and lobula plate neuropils (Stark and Carlson, 1986; Meinertzhagen and O’Neil, 1991; Perez and Steller, 1996; Rivera-Alba et al., 2011; Millard and Pecot, 2018). Recently, a new class of Drosophila interneurons has been discovered: the Allatostatin C (AstC)/crustacean cardioactive peptide receptor (CcapR) expressing neurons, which convey light input from the compound eyes directly to the circadian pacemaker neurons, through the accessory medulla (aMe) (Li et al., 2018). The medulla thus receives and processes both motion and colour information coming from different retinotopic maps (Rister et al., 2007; Gao et al., 2008).
The epithelial glial cells surrounding lamina cartridges express the Mesencephalic Astrocyte-derived Neurotrophic Factor DmMANF, orthologue of mammalian MANF and CDNF (cerebral dopamine neurotrophic factor), involved in supporting the survival of dopaminergic neurons (Lindholm and Saarma, 2010). DmMANF is also involved in the maintenance of dopaminergic neurons, as DmMANF null mutants display extremely low levels of dopamine and decreased dopaminergic neurites (Palgi et al., 2009). In the adult fly, DmMANF is also expressed in the retina, specifically in the photoreceptor cell bodies, and in the lamina (lamina cortex and synaptic neuropil) (Stratoulias and Heino, 2015). At structural level, the silencing of DmMANF in glial cells induces degeneration of the lamina, in particular in the lamina epithelial glial cells, which exhibits holes and/or tightly packed membranes and also a decrease of capitate projections in the cartridges (Walkowicz et al., 2017). In glial cells, DmMANF is also involved in controlling the levels of dopamine and other neurotransmitters responsible for Drosophila behaviour. In fact, downregulation of DmMANF in glia alters the sleep/activity pattern of flies in LD, with decreased activity in the light phase and increased activity in the dark phase of the cycle (Walkowicz et al., 2017). Conversely, these flies display a reduction of nightime sleep and a slight increase of sleep in the early day (Figure 4). The sleep modulating role of DmMANF is supported by the significant upregulation of transcripts involved in the dopamine synthesis pathway observed in hypomorphic DmMANF mutant embryos (Palgi et al., 2012).
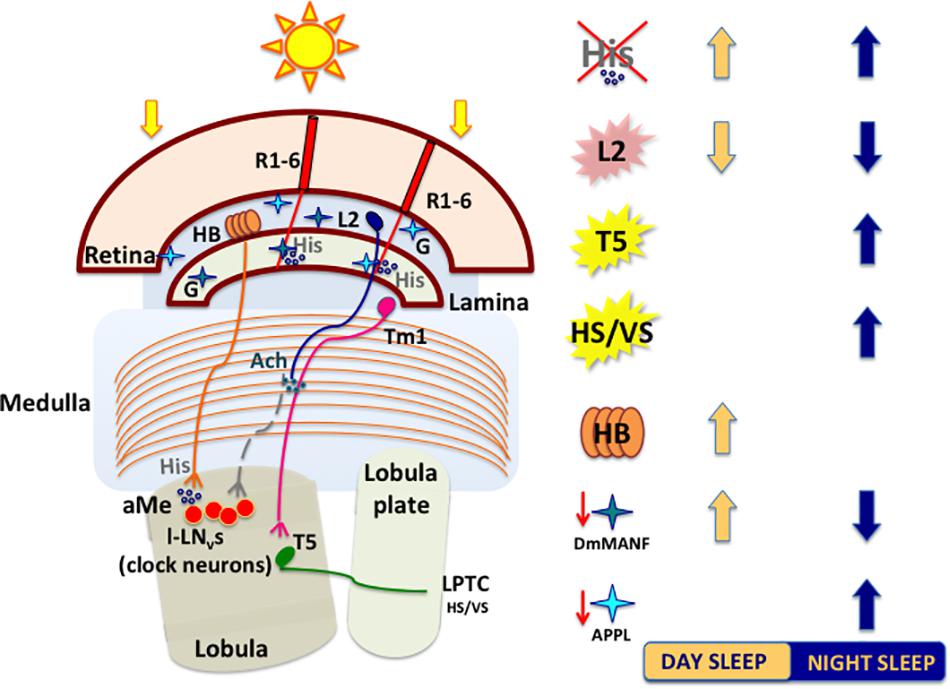
Figure 4. A schematized and simplified circuit showing how light perceived by retinal and extra-retinal structures impacts sleep. Light-activated retinal photoreceptors release histamine (His). Mutations affecting the histamine biosynthesis pathway (hdc mutants) lead to increase in sleep. R1–R6 photoreceptors convey signals to cholinergic large monopolar laminar cells (L2) that connect indirectly both clock neurons (l-LNvs) and neurons involved in motion pathways (T5). Temperature-Sensing TRP Channel activation in L2 neurons strongly suppresses sleep by releasing ACh that, in turn, promotes the bursting of l-LNvs. T5 cells axons transfer motion information to various types of lobula plate tangential neurons (LPTCs), like HS (horizontal motion) and VS (vertical motion) neurons, to further reach the central brain. Optogenetic activation of motion processing neurons (T5 or HS/VS) increases nighttime sleep. The extraretinal photoreceptors (HB eyelets) release histamine in the accessory Medulla (aMe), preventing l-LNvs from firing and thus increasing siesta during the day. Glia cells in lamina interact with photoreceptors and shape the wake/sleep pattern of flies. Downregulation of DmMANF (mesencephalic astrocytes-derived neurotrophic factor) in epithelial glia induces lamina neurodegeneration and leads to increase in daytime sleep. Downregulation of Appl (amyloid precursor protein like) in glia cells increases nighttime sleep. Light blue (APPL) and dark blue (DmMANF) stars indicate cortex and epithelial lamina glia, respectively. L2: heat-activated (pink); T5 and HS/VS: optogenetically activated (yellow). G, glia cell; His, histamine; Ach, acetylcholine.
Glial cells express the Amyloid Precursor Protein-Like (APPL), known for its crucial role in neuronal physiology and cellular biology and its involvement in age-dependent behavioural deficits and neurodegeneration, as consequence of production and deposition of toxic β-amyloid peptides, in both mammals and flies (Carmine-Simmen et al., 2009). In Drosophila, this is true also for the glial cells in the subretinal layer of lamina cortex, where the correct cleavage of APPL is fundamental for their survival. Indeed, loss of function mutation or knock-down of the beta-site Amyloid Precursor Cleaving Enzyme (dBACE) in photoreceptor neurons result in glial cell death and progressive lamina degeneration (Bolkan et al., 2012).
The role of APPL in glia is not only related to its neurotoxic effects, as it has been recently shown to be involved in the physiology and regulation of sleep/wake cycles (Farca Luna et al., 2017). The downregulation of Appl in cortex and astrocyte-like glia significantly increases nighttime sleep, which exhibits longer sleep-bouts (Figure 4) and, conversely, the overexpression of Appl in these cells results in reduced sleep amount and increased sleep latency (Farca Luna et al., 2017). This effect on sleep/wake regulation is due to an altered glutamate recycling, as the downregulation of Appl increases the expression of genes involved in reuptake and recycling of the neurotransmitter, such as the glutamate transporter excitatory amino acid transporter 1 (dEaat1) and the glutamine synthetase (Gs) (Farca Luna et al., 2017). Moreover, the downregulation of Appl changes also the cellular distribution of Innexin 2 (Inx2), highly expressed in the layers of laminar pseudocartridge and satellite glia, where it plays a fundamental role in modulating the level of carcinine, and therefore histamine, essential for a proper visual synaptic transmission (Chaturvedi et al., 2014).
As previously mentioned, R1–6 photoreceptors are also involved in visual motion, that is the detection of direction-selective signals, fundamental for fly survival. The luminance information from R1–6 is integrated by some large motion-sensitive neurons in the lobula plate, called lobula plate tangential cells (LPTCs), specific for vertical or horizontal motion (VS and HS, respectively) and responding by selective hyperpolarization or depolarization (reviewed in Borst et al., 2020). Each lamina cartridge specifically conveys brightness increments or decrements information to subsets of downstream motion detecting neurons, via a specific set of cells in the medulla, called trans-medulla Y (TmY). In particular, L1 pathway conveys luminance increments to specific layers of the lobula (T4 cells-ON channels), while the L2–4 pathway transmits information about brightness decrements to the lobula plate (T5 cells-OFF channels) (Borst et al., 2020). Axon terminals from T4 and T5 neurons then connect to the dendrites of LPTCs (HS and VS) in the lobula plate (Borst et al., 2020), from where the information is further transmitted to the central brain likely through descending neurons (Suver et al., 2016). LPTCs also receive direction information from another source. Indeed, T4 and T5 cells contact and send an inhibitory glutamatergic signal to a group of neurons in the lobula plate, the bi-stratified lobula plate intrinsic (LPi) cells, that convey this signal to the tangential cells expressing glutamatergic Cl– channel α (reviewed in Borst et al., 2020).
Visual information processed by motion circuits play an important role in sleep regulation. Flies lacking HS and VS neurons (ombH31 mutants) display a reduced and fragmented sleep compared to wild-type (wt), while the optogenetic activation of these cells results in an increase of nighttime sleep (Kirszenblat et al., 2019). Moreover, the optogenetic activation of T5 neurons leads to a consolidation of nighttime sleep, with increased bout duration and lower bouts number (Kirszenblat et al., 2019; Figure 4).
Histamine, the Major Neurotransmitter in the Compound Eyes
Histamine is the most important neurotransmitter released by the compound eyes (Hardie, 1987, 1989), and histamine-immunoreactivity has been detected in the optic lobes, in neurons adjacent to LNs and DNs, ocelli, in the eyelets axons, in 18 cell bodies in protocerebrum (HP1–4) and 2 cell bodies in the subesophageal ganglion (Nässel, 1999; Hamasaka and Nässel, 2006; Hong et al., 2006; Oh et al., 2013). The biogenic amine is synthetized in photoreceptors, from L-histidine, by the histidine decarboxylase (Hdc) and flies deficient for this enzyme activity (hdcP218) have disrupted photoreceptor synaptic transmission (Burg et al., 1993). Light-depolarization of retinal photoreceptors triggers the fast release of histamine to the downstream lamina monopolar neurons; this, in turn, opens the histamine-gated chloride channels and leads to hyperpolarization (Wang and Montell, 2007; Pantazis et al., 2008). Electroretinograms in postsynaptic lamina neurons record ON and OFF transient peaks as a function of light (Alawi and Pak, 1971; Heisenberg, 1971). The epithelial glia cells surrounding synaptic cartridges work in coordinating photoreceptor-glia communication in the lamina: in fact, in capitate projections histamine is conjugated to β-alanine by Ebony, to form β-alanylhistamine (carcinine) (Stark and Carlson, 1986; Meinertzhagen and O’Neil, 1991; Borycz et al., 2002; Richardt et al., 2002, 2003; Hartwig et al., 2014). Carcinine is then transported back to photoreceptors by the transporter CarT (Stenesen et al., 2015; Xu et al., 2015; Chaturvedi et al., 2016) and cleaved again into histamine and β-alanine by Tan (Borycz et al., 2002; Wagner et al., 2007). Interruption of this cycle results in the loss of visual transmission (Rahman et al., 2012).
In Drosophila, histamine gates two chloride channels: the outer rhabdomeres transientless (ort) and histamine-gated chloride channel subunit 1 (HisCl1) (Gengs et al., 2002; Gisselmann et al., 2002; Witte et al., 2002; Zheng et al., 2002). Ort is expressed in lamina (L1–L3 cells), medulla, lobula neuropils, ocellar postsynaptic interneurons, Pars Intercerebralis (PI), FB, cells in the lateral and central brain and thoracic ganglia (Hong et al., 2006; Gao et al., 2008; Pantazis et al., 2008; Lin et al., 2016; Schnaitmann et al., 2018). In lamina interneurons, it plays a key role in transmitting motion detection inputs coming from retina photoreceptors: its overexpression in L1 and L2 can restore the ON and OFF transients in electroretinograms and motion detection responses lost in ort-null mutants (Gengs et al., 2002; Rister et al., 2007; Gao et al., 2008; Pantazis et al., 2008). HisCl1 receptor is strongly expressed in lamina epithelial glial cells surrounding cartridges, in neurons in the medulla (Gao et al., 2008; Pantazis et al., 2008), in R7 and R8 photoreceptors (Tan et al., 2015; Schnaitmann et al., 2018; Alejevski et al., 2019; Davis et al., 2020) and many other cell types, including the large LNvs (Hamasaka and Nässel, 2006; Hong et al., 2006).
Histamine released by light-activated photoreceptors likely acts in at least two different pathways directly involved in sleep regulation: the visual (photic) input and the motion detection pathways, distinct signalling dynamics both relying on activation of lamina interneurons L2 (Meinertzhagen and O’Neil, 1991; Meinertzhagen and Sorra, 2001; Shinomiya et al., 2014, 2019; Muraro and Ceriani, 2015; Kirszenblat et al., 2019; Borst et al., 2020; Figure 4).
In mammals, histamine is known to play a wake-promoting role (Thakkar, 2011), that seems to be conserved in insect. In Drosophila, histamine treatment causes sleep time reduction (Oh et al., 2013), while administration of its receptor antagonist increases sleep (Shaw et al., 2000). Moreover, mutations in the hdc gene [hdcP211 and hdcP218 (Burg et al., 1993)], lead to a significant increase of daytime sleep duration and number of sleep episodes in comparison to wt (Oh et al., 2013; Figure 4). Similar data obtained in constant darkness indicate that the observed wake-promoting effect depends on histamine, and it is not connected with defects in photoreception in the eye (Oh et al., 2013). Of the two histamine receptors, only HisCl1 located on the surface of l-LNvs is involved in sleep regulation (Oh et al., 2013).
Ocelli
Ocelli complex is composed of three ocellar cells, interocellar cuticle and bristles (Haynie and Bryant, 1986). They contain 80–100 photoreceptors expressing the UV-sensitive Rhodopsin2 (Mismer and Rubin, 1987; Feiler et al., 1988; Pollock and Benzer, 1988). The role of ocelli is to adjust sensitivity of the compound eyes (Hu and Stark, 1980) and to collect information about the horizontal position (Krapp, 2009). They also contribute to entrainment to long and short days (Rieger et al., 2003), via a norpA-independent pathway (Saint-Charles et al., 2016). They use histamine as neurotransmitter, and they do not contact directly with clock neuron processes (Hamasaka and Nässel, 2006). A specific role for this structure in sleep has not been reported yet.
Hofbauer–Buchner Eyelets: Direct Light Signalling to the Pacemaker
Hofbauer–Buchner eyelets originate from the larval visual system, called Bolwig organs (BO), involved in the regulation of many light-dependent behaviours (Busto et al., 1999; Hassan et al., 2000). Larval BO is cholinergic (Yasuyama and Salvaterra, 1999), but it uses norpA-dependent phototransduction pathway, similar to retinal photoreceptors (Busto et al., 1999; Hassan et al., 2000). It is composed of 12 cells: eight of them express Rh6 and 4 of them express Rh5 (Sprecher and Desplan, 2008), and their projections terminate in the area of LNvs (Kaneko et al., 1997). Rh6-expressing cells die during development, and four others switch expression from Rh5 to Rh6 (Sprecher and Desplan, 2008). Adult HB express Rh6 and are sensitive to 480 nm wavelength (Helfrich-Förster et al., 2002), yet there are evidences that they may use an alternative mechanism of phototransduction, norpA-independent, like cascade described by Chang and Ready (2000). Although Rh5 expression in HB could not be detected by immunostaining (Yasuyama and Meinertzhagen, 1999; Malpel et al., 2002), the expression of GFP under the Rh5-Gal4 driver was revealed as a weak signal (Malpel et al., 2002). In the adult, HB act as circadian photoreceptive organs (Hofbauer and Buchner, 1989; Yasuyama and Meinertzhagen, 1999) and contribute to the synchronisation of circadian clock, in terms of entrainment to long and short days (Helfrich-Förster et al., 2001, 2002; Rieger et al., 2003). At the molecular level, they are involved in synchronisation of TIM and PER expression in s-LNvs (Helfrich-Förster et al., 2001), l-LNvs and DN1s (Mealey-Ferrara et al., 2003; Veleri et al., 2007).
Hofbauer–Buchner axons terminate in the accessory medulla and they can directly contact with pigment dispersing factor (PDF)-expressing LNvs in aMe (Helfrich-Förster et al., 2002; Malpel et al., 2002). Eyelets express both histamine and acetylcholine as neurotransmitters (Hofbauer and Buchner, 1989; Pollack and Hofbauer, 1991; Yasuyama and Meinertzhagen, 1999; Damulewicz et al., 2020; Figures 4, 5).
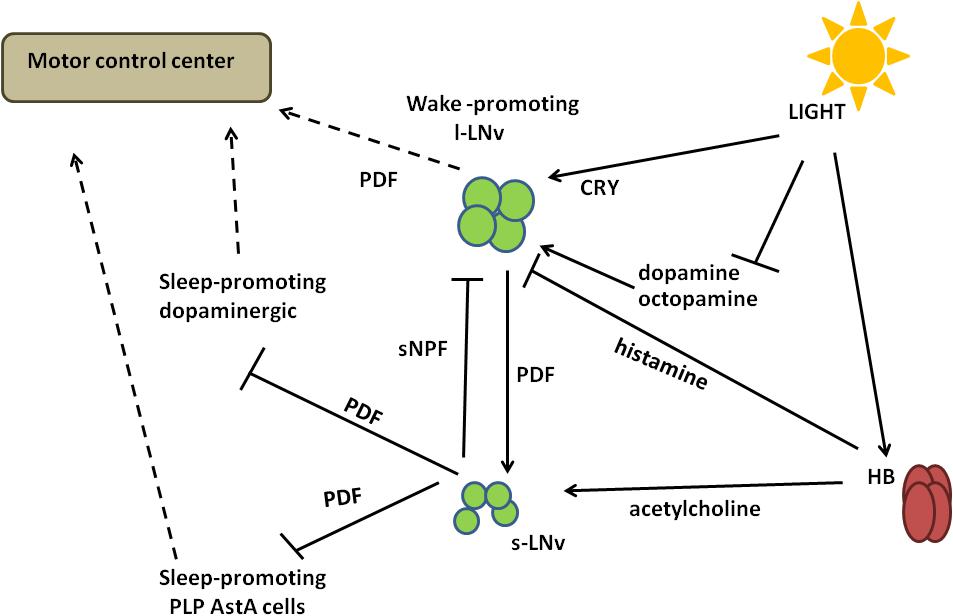
Figure 5. A schematized circuit of circadian regulation of sleep. Light signals received by Hofbauer–Buchner eyelets (HB) are transmitted through acetylcholine to small ventral-lateral neurons (s-LNvs) and through histamine to large ventral-lateral neurons (l-LNvs). Activated s-LNvs release pigment dispersing factor (PDF), which inhibits dopaminergic and AstA-expressing sleep-promoting cells. The regulation of l-LNvs activity is more complex, as they express light-sensitive Cryptochrome (CRY) as well as dopamine and octopamine receptors. Light activates CRY, but at the same time it inhibits the response to dopamine and octopamine, decreasing signal inputs to l-LNvs. In addition, l-LNvs receive inhibiting inputs from s-LNvs through short Neuropeptide F (sNPF). Different signalling inputs are processed in l-LNvs and transmitted to the motor control center through PDF.
Light signals received by HB eyelets in the morning are transmitted via acetylcholine and excite s-LNvs via nicotinic receptors (Wegener et al., 2004; McCarthy et al., 2011; Schlichting et al., 2016), causing increased cAMP levels (Lelito and Shafer, 2012) and a wake-promoting effect. At the same time, histamine released from HB inhibits l-LNvs (Schlichting et al., 2016). In the morning l-LNvs are less active, with decreased firing observed. Then they start to accumulate Ca2+ that reaches its maximal level around midday (Liang et al., 2016), when they could receive input from other cells, that is, from L2 cells in the medulla (Muraro and Ceriani, 2015). l-LNvs increase firing and release PDF to activate evening cells, ultimately increasing the evening activity.
Deep Brain Photopigments
Cryptochrome (CRY) is a blue-light-sensitive protein (VanVickle-Chavez and Van Gelder, 2007) playing many different roles, ranging from photoreceptor to magnetoreception and metabolism regulation (for review see Damulewicz and Mazzotta, 2020). It is expressed in a broad range of cells in the brain: in circadian pacemaker neurons (all five s-LNvs, l-LNvs, three of the six LNds, and some of the DN1s), but also in non-clock neurons, glia and visual system (Benito et al., 2008; Yoshii et al., 2008; Damulewicz and Pyza, 2011; Fogle et al., 2011). Its photoreceptive role allows the entrainment of molecular clock to environmental light conditions through conformational changes that expose specific domains and promote binding TIM or PER (Ceriani et al., 1999; Koh et al., 2006; Peschel et al., 2009; Rosato et al., 2001), targeting TIM to ubiquitination and degradation in proteasomes (Peschel et al., 2009). The role of CRY in the visual system is more complex, as it plays a role of circadian transcriptional repressor (Collins et al., 2006), in maintaining the proper localization of phototransduction cascade complex (Mazzotta et al., 2013; Schlichting et al., 2018) and in enhancing photosensitivity during the night (Damulewicz et al., 2017; Mazzotta et al., 2018).
Quasimodo (QSM) is a light-sensitive protein, belonging to the extracellular membrane-anchored Zona pellucida (ZP) domain family. It is expressed in all clock neuronal groups, except for LPNs, however not in every cell within the cluster. Most of the clock cells co-express both, QSM and CRY, but some of them, like DN2s and DN3s, do not express CRY, suggesting that QSM works in a CRY-independent pathway. In addition, QSM is expressed in non-clock cells, located in close proximity to the pacemaker (Chen et al., 2011). Light exposure increases QSM levels inside the cell, via a post-translational mechanism that involves the extracellular ZP domain light-dependent cleavage (Plaza et al., 2010; Buhl et al., 2016). Active QSM was proposed to change membrane conductance following interaction with the Na+, K+, Cl– co-transporter (NKCC) and the Shaw K+ channel (dKV3.1) (Buhl et al., 2016). QSM regulates electrical excitability also in clock neurons: it modulates l-LNvs daily changes of activity, as its downregulation results in a constitutively more active state, similar to that observed during the day time, while qsm overexpression leads to a constitutive less active, night-like state (Buhl et al., 2016). Moreover, QSM is involved in the light-dependent TIM degradation process and it can affect TIM stability in a CRY-independent pathway (Chen et al., 2011).
Circadian Pacemaker Neurons
Sleep timing and duration are highly influenced by the circadian clock, which promotes the consolidation of sleep during the night in diurnal species, such as Drosophila and human, and during the day in nocturnal animals, such as rodents (Kunst et al., 2014; Liu et al., 2014). Indeed, in flies lacking the main clock genes period and timeless, the sleep episodes are randomly distributed across the 24 h, although the mean rest levels do not differ from wt (Hendricks et al., 2000). Moreover, flies mutants for both Clock and cycle show a significant decrease in daily consolidated rest in DD conditions, with brief rest and prolonged activity bouts (Shaw et al., 2002; Hendricks, 2003), and cyc01 mutants show also an excessive response to sleep deprivation, with a persistent large increase in sleep (Shaw et al., 2002).
PDF Expressing LNvs
LNvs and serotoninergic signalling: modulation of the circadian light sensitivity
The serotoninergic pathway regulates many aspects of behaviour, including sleep/wake cycles (Ursin, 2002). In Drosophila, it positively controls sleep and negatively modulates circadian photosensitivity: treatment with the serotonin precursor 5-hydroxyl-L-tryptophan (5-HTP) results in significant increase of sleep amount and reduction of the light-induced phase shift, especially in response to high intensity light pulses (Yuan et al., 2005, 2006). This dual role is mediated by two distinct receptors, d5-HT1A and d5-HT1B, sharing high homology with the mammalian counterpart (5-HT1A), that controls many aspects of animal behaviour, including sleep (Boutrel et al., 2002; Yuan et al., 2005).
d5-HT1A is highly expressed in the MB, at levels that remain constant during the day (Yuan et al., 2006). d5-HT1A is specifically involved in regulating sleep amount and consolidation, as flies carrying a deleted form of d5-HT1A exhibit a significant reduction and fragmentation in sleep, with nighttime sleep bouts reduced in length but increased in number (Yuan et al., 2006). This behaviour is specifically dependent on d5-HT1A in MB, since it can be completely rescued by overexpression of the receptor in these neurons (Yuan et al., 2006). Treatment with 5-HTP increases sleep in d5-HT1A mutant flies, indicating that other unidentified serotonin receptors are also involved in sleep regulation (Yuan et al., 2006).
d5-HT1B is expressed in different brain structures, including LNvs (Yuan et al., 2005). The expression of d5-HT1B in the adult fly brain does not show circadian oscillation, neither as mRNA nor as protein, but its levels are influenced by the clock, as they appear to be upregulated in tim01 and downregulated in cyc0 mutants (Yuan et al., 2005).
In clock cells, d5-HT1B is involved in modulating the circadian light sensitivity: flies overexpressing this receptor in the clock neurons exhibit a reduced magnitude of the response to phase shift following light pulse, mirrored by a reduced light-dependent TIM degradation (more evident in s-LNvs compared to l-LNvs). Conversely, the downregulation of d5-HT1B results in an increased phase shift, also toward low light intensities (Yuan et al., 2005).
Serotonin and d5-HT1B effects on circadian light sensitivity are related to the CRY signalling pathway: while the overexpression of d5-HT1B in a wt background induces increased levels of rhythmicity in constant light, the overexpression of d5-HT1B in a cry mutant background (cryb) has no effect on the rhythmicity exhibited by cryb flies (Yuan et al., 2005).
LNvs and Neuronal Structural Remodeling
Remodeling of neuronal connections is fundamental for the neuronal circuits to detect environmental changes and drive complex behaviour. In Drosophila, the circadian behaviour also results from a clock-controlled structural plasticity that contributes to the transmission of information downstream of pacemaker neurons (Fernández et al., 2008).
PDF positive LNvs rhythmically express the miRNA miR-210 (Chen and Rosbash, 2017), that plays an important role in the phasing of the circadian locomotor activity (Cusumano et al., 2018; Niu et al., 2019). miR-210 is also involved in the regulation of sleep levels and temporal distribution, and this role is likely correlated to the morphology remodeling of l-LNvs: in fact the miR-210 overexpression in clock cells results in a significant increase in daytime sleep and a dramatic alteration of l-LNvs morphology and projections (Cusumano et al., 2018).
Small ventral-lateral neurons express dTau, a protein with microtubule-binding properties, homolog to mammalian Tau, known to be involved in the maturation and establishment of synaptic networks regulating complex behaviours (Abruzzi et al., 2017; Tracy and Gan, 2018). dTau plays an important role in shaping behavioural rhythms and sleep patterns: in either LD cycles or DD, dTau mutant flies exhibit an increased activity during the day/subjective day, more pronounced in the middle of the day, when wt flies have a “siesta” (Arnes et al., 2019). This altered locomotor phenotype is mirrored by pronounced sleep alterations: dTau null flies exhibit a significant alteration of daytime sleep, while nocturnal sleep is not affected: the total daytime sleep is significantly decreased, including the “siesta,” the sleep episodes are shorter, that is, sleep is more fragmented, and the sleep latency is significantly longer (Arnes et al., 2019).
At the neuronal level, dTau plays an essential role in modulating the structural plasticity of s-LNvs terminals: in wt flies the dorsal projections of s-LNvs neurons display a rhythmic remodeling, with significantly higher degree of axonal arborisation in the early day (ZT2) compared to early night (ZT14) (Fernández et al., 2008). dTau null flies exhibit a significant reduction in the structural morphology of the s-LNv at ZT2 compared to wt, in line with the behavioural defects (increased activity and decreased sleep) displayed in the early day (Arnes et al., 2019). Furthermore, in s-LNvs, dTau shows rhythmic expression at both mRNA and protein levels, with significantly higher levels in the early morning (ZT2) than in the early night (Abruzzi et al., 2017; Arnes et al., 2019). This temporal rhythmic pattern perfectly matches with its role in modulating the structural plasticity of s-LNvs terminals (Arnes et al., 2019).
Large Ventral-Lateral Neurons: The Heart of the Sleep Circuit
Large ventral-lateral neurons are among the first clock neurons that have been identified (Zerr et al., 1990) and they have a predominant role in detecting light and transferring the photic information to the circadian clock (reviewed in Helfrich-Förster, 2019). By using different signalling pathways l-LNvs integrate light stimuli and produce appropriate behavioural responses (Figure 5).
l-LNvs are directly activated by light
Large ventral-lateral neurons display an acute increase in their firing rate in response to light, and this altered electrical activity influences locomotor behaviour, sleep and arousal (Sheeba et al., 2008a). l-LNvs hyperexcited flies exhibit an increase in nocturnal activity compared to controls, mirrored by a disruption in the quantity and quality of nocturnal sleep (Sheeba et al., 2008a). Moreover, the increased nocturnal behaviour of l-LNvs hyperexcited flies is mediated by a PDF-dependent mechanism, as Pdf mutants exhibit a nocturnal activity significantly lower compared to wt (Sheeba et al., 2008a). The light-induced firing rate of l-LNvs is dependent on the presence of the circadian photoreceptor CRY, highly expressed in these clock cells (Emery et al., 2000). Indeed, in cryb hypomorphic mutants, the electrophysiological response is attenuated (Sheeba et al., 2008b), while it is completely abolished in cry-null flies (Fogle et al., 2011). Conversely, the light-induced firing of l-LNvs is functionally rescued by targeted expression of CRY in the l-LNvs.
Large ventral-lateral neurons are part of the peptidergic arousal pathways in Drosophila. The hyperactivation of these cells by overexpression of NaChBacGFP, a bacterial-derived voltage-gated sodium channel (Nitabach et al., 2005), results in a dramatic increase of nighttime activity and, by a genetic manipulation, it has been also shown that the stimulation of l-LNvs is sufficient to promote arousal at night (Shang et al., 2008). Moreover, l-LNvs-mediated arousal is light-dependent: flies in which this subset of clock cells is genetically ablated exhibit an increased sleep in LD, even more evident in LL, a phenotype completely lost when flies are moved to DD (Shang et al., 2008). Another important feature of l-LNvs is that they signal light information to the circadian clock at dawn: indeed, l-LNvs-deficient flies exhibit no phase advance response to light at ZT21 compared to control, while no differences between the two genotypes are observed for light pulse at ZT15 (Shang et al., 2008).
Pigment dispersing factor is specifically involved in increasing flies’ activity in the late night: Pdf01 mutants, as well as flies with null mutation in the receptor for PDF (Pdfrhan5304), exhibit an increased sleep during the late night, while flies in which the PDF-expressing neurons are genetically ablated, show a prolonged sleep (Chung et al., 2009). The lack of PDF-mediated signalling is partially compensated by light: in DD, Pdf01, Pdfrhan5304, and PDF-ablated flies exhibit a significant increase in total sleep during the subjective day, which is not visible in LD (Chung et al., 2009; Figure 5).
Light negatively regulates dopamine and octopamine signalling in l-LNvs
Large ventral-lateral neurons express high levels of dopamine receptors (DopR, DopR2, and D2R) as well as the two major octopamine GPC receptors, OA2 and OAMB (Kula-Eversole et al., 2010). By GRASP (GFP Reconstitution Across Synaptic Partners) analysis (Feinberg et al., 2008) it has been shown that they form membrane contacts with dopaminergic and octopaminergic neurons (Shang et al., 2011). Both dopamine and octopamine represent arousal signals in l-LNvs (Shafer et al., 2008). The response to dopamine is negatively regulated by light and it is time of day-independent, with no significant difference between day/subjective day versus the night/subjective night. However, responses in DD are much stronger during both the subjective day and subjective night, in comparison to those at the same circadian times in LD cycles. The effects of octopamine on l-LNvs are both light and time dependent: the responses from subjective day are similar to those of daytime in LD while during subjective night they are far stronger compared to daytime, nighttime, or subjective day (Shang et al., 2011). The time-sensitivity of l-LNvs response to octopamine is a clock-controlled feature, since in per01 mutants the responsiveness during the night is much weaker compared to controls (Shang et al., 2011; Figure 5).
Dopamine signalling in l-LNvs also involves the circadian photoreceptor CRY, expressed at high levels in ClkJrk flies, that display a nocturnal behaviour and a reduction in total sleep (Kim et al., 2002; Lu et al., 2008). This CRY-driven nighttime activity of Clk mutants is suppressed when dopamine signalling is blocked either pharmacologically or genetically (Kumar et al., 2012).
l-LNvs mediate histamine wake-promoting signals
Pigment dispersing factor neurons can receive histaminergic wake-activation signals. Loss-of-function mutations in the HisCl1 and hdc genes result in increased sleep duration, especially during the day (Oh et al., 2013). l-LNvs play an important role in mediating these histaminergic wake-promoting signals: the targeted downregulation of HisCl1 in PDF cells increases both the daytime and nighttime sleep duration, while the targeted overexpression of HisCl1 with either tim-Gal4 or Pdf-Gal4 is able to restore the increased sleep duration of HisCl1 mutant (Oh et al., 2013; Figure 5).
l-LNvs activity is modulated by potassium channels
During sleep, neuronal activity undergoes large-scale changes, and different types of potassium channels are required for normal wake–sleep cycles (Cirelli et al., 2005; Bushey et al., 2007; Allebrandt et al., 2013, 2017). In l-LNvs, the Shal/Kv4, voltage-gated K+ channel plays an important role in controlling wake–sleep transition at dusk (Feng et al., 2018). Kv4 acts as sleep-promoter, since flies with a pan-neuronal expression of a dominant-negative form of Kv4 (DNKv4) exhibit a reduced nighttime sleep, as consequence of a decrease in sleep-bout duration. The expression of DNKv4 limited to all PDF positive neurons induces a marked increase in sleep latency and decrease in nighttime sleep, even more evident when the expression is further restricted to l-LNvs (Feng et al., 2018). In l-LNvs, both the frequency of the action potential (AP) currents and the resting membrane potential (RMP) exhibit a strong rhythmicity, with a higher firing rate during daytime and more RMPs significantly depolarized at dawn (ZT1) compared to dusk (ZT13). Both features are dependent on Kv4, since the expression of DNKv4 results in the increase of either the frequency of AP currents or the firing rate during dusk (Feng et al., 2018).
l-LNvs and modulation of sleep/wake behaviour at transcriptional level
Many brain neurons, including PDF-positive LNvs, express apterous (ap), a well-known LIM-homeodomain transcription factor involved in development and neuropeptide expression (Hobert and Westphal, 2000; Shimada et al., 2016). ap levels are particularly high in l-LNvs, where they also exhibit a daily oscillation generated by a light-dependent mechanism. In LD both mRNA and protein show a rhythmic expression with a peak during the night (ZT16 and ZT18, respectively), while this oscillation is lost in DD, at least at protein level (Claridge-Chang et al., 2001; Shimada et al., 2016). This transcription factor is involved in buffering light-driven arousal: specific knock-down of ap in these PDF neurons results in promoting arousal (reduction in sleep amount and increase in waking time) under LD conditions, whereas the sleep/wake pattern is not affected in DD (Shimada et al., 2016). ap knock-down does not significantly affect PDF, neither its expression nor its release; therefore, other neuropeptides or signalling inputs/synaptic output are involved. ap acts in cooperation with the transcription factor Chip (Chi), to drive the expression of developmental genes (Van Meyel et al., 1999). In PDF neurons, the two transcription factors act as a complex playing a key role in transcriptional modulation of sleep/wake behaviour. In fact, while the knock-down of ap only results in a general decrease of sleep, regardless of the time of the day, when both proteins are inactive only the daytime sleep amount is decreased. This indicates that this complex modulates mechanisms that act specifically in regulating sleep/wake at different times of the day (Shimada et al., 2016).
Small Ventral-Lateral Neurons: A Secondary Role in the Arousal Circuit
s-LNvs and PDFR signalling
The arousal activity of l-LNvs is mediated by PDF and a functional PDFR signalling is required for a proper sleep/wake regulation. In fact, Pdfr mutants display an increased sleep, specifically during the day (Parisky et al., 2008; Chung et al., 2009; Potdar and Sheeba, 2018; Sheeba et al., 2008a). The PDFR signalling pathway targets the dopaminergic neurons (i.e., PPM3) and plays a crucial role in regulating daytime wakefulness. The downregulation of Pdfr in these neurons results in a significant increase of daytime sleep, with longer sleep bouts, while Pdfr overexpression suppresses daytime sleep and delays sleep onset (Potdar and Sheeba, 2018).
PDF and dopaminergic neurons are synaptically connected, specifically in the region of s-LNvs axonal projections (Potdar and Sheeba, 2018). Importantly, this observation confirms not only that dopaminergic neurons are a downstream target of PDFR signalling, but also that s-LNvs contribute to the wake-promoting activity of l-LNvs. The involvement of s-LNvs in the arousal circuit was already suggested: (1) the downregulation of Pdfr in s-LNvs results in the increase of total sleep (both daytime and nighttime) (Parisky et al., 2008); and (2) the electrical activity of s-LNvs contributes in modulating the phase of evening activity under long photoperiods (Potdar and Sheeba, 2018).
A PDFR signalling originating from the s-LNvs targets also a group of neurons, posterior lateral protocerebrum (PLP) cells, that express the neuropeptide AstA and are involved in sleep promotion (Chen et al., 2016). The thermogenic activation of AstA-PLP neurons causes a significant decrease in locomotor activity and an increase of sleep, either in LD or DD and LL; conversely, the silencing of AstA cells results in a significant reduction of sleep, especially during the midday siesta time, either in LD or DD (Chen et al., 2016).
PLP cells represent downstream target of PDF signalling; they are post-synaptically connected to s-LNvs and express functional PDF receptors and, furthermore, constitutive activation of PDF signalling in AstA-expressing neurons significantly increases the amount of sleep (Chen et al., 2016; Figure 5).
s-LNvs and short neuropeptide F (sNPF) signalling
sNPF is broadly expressed in various brain regions, including MB, PI, and CC neurons (Nässel et al., 2008; Johard et al., 2009), and known to regulate different aspects of fly physiology and behaviour (Kahsai et al., 2010; Nagy et al., 2019). sNPF has an important role in promoting and maintaining normal sleep: flies carrying a hypomorphic mutation in sNPF or with a knock-down of sNPF in adult brain, exhibit a reduced and fragmented sleep compared to control. Moreover, the silencing of sNPF neurons results in a significant reduction of the sleep levels during the daytime (Shang et al., 2013).
The sleep-promoting activity of sNPF neurons is normally suppressed by GABAA signalling during the daytime, as the downregulation of the GABAA receptor Rdl in these neurons leads to a significant increase of both daytime and total sleep time and to a lengthening of sleep bouts (Shang et al., 2013). sNPF is also involved in the response to sleep deprivation: the hyperactivation of sNPF neurons during mechanical sleep deprivation causes a partial sleep-like state and induces less sleep rebound or recovery sleep (Shang et al., 2013).
In s-LNvs sNPF acts in promoting normal nighttime sleep. sNPF mRNA levels exhibit a robust oscillation in s-LNvs, while in l-LNvs it is barely expressed (Kula-Eversole et al., 2010). Flies with downregulation of sNPF in PDF neurons exhibit a decreases in nighttime sleep, while daytime sleep is not affected (Shang et al., 2013). These sNPF sleep-promoting signals from s-LNvs are transmitted to l-LNvs (Figure 5); the downregulation of sNPFR (sNPF receptor) in l-LNvs, where it is normally expressed at high levels (Kula-Eversole et al., 2010), results in a significant fragmentation of nighttime sleep (Shang et al., 2013).
The sleep-promoting role of sNPF is essentially exerted by an inhibitory effect on arousal neurons activity, as it is the result of a balance between the sNPF and the dopamine (DA) signallings in the l-LNvs: in fact, the co-application of DA and sNPF suppresses the cAMP response in the l-LNvs, strongly elicited by DA alone (Shang et al., 2013).
In this section we have focused on those signalling pathways involving PDF neurons specifically related to both light and sleep. Nevertheless, LNvs participate in many other signalling pathways that act in synchronising their activity and regulating sleep/wake behaviour. Among these, (1) glutamatergic transmission mediated by the metabotropic glutamate receptor DmGluRA is important for inhibiting activity in the dark (Hamasaka et al., 2007); (2) GABAergic signalling in the l-LNvs, mediated by GABAA receptor Rdl and modulated by the ankyrin repeats domain containing protein WIDE AWAKE (WAKE), plays a role in either the initiation or the maintenance of sleep (Agosto et al., 2008; Parisky et al., 2008; Chung et al., 2009; Liu et al., 2014); and (3) cholinergic inputs to the l-LNvs, mediated by nicotinic acetylcholine receptors (nAChRs) and modulated by glutamate-gated Cl– channels, ensure a highly synchronized rhythmic membrane activity with a simultaneous occurrence of depolarized and hyperpolarized phases (McCarthy et al., 2011).
Lateral Posterior Neurons: Connection Between the Clock Network and the Sleep Center
The sleep promoting function of LPNs is modulated by the circadian clock, as the expression of a dominant negative form of Clock in these cells reduces the sleep during the daytime (Ni et al., 2019).
The LPN express the neuropeptide AstA and form synaptic connections with the FB; moreover, the inhibition of neurotransmission from the LPNs results in a reduction of sleep (Ni et al., 2019). The excitatory neurotransmitter in LPN that activates FB neurons and promotes sleep is glutamate: in fact, the inhibition of glutamate transport results in a significant reduction of nighttime sleep bouts length and therefore increases sleep fragmentation (Ni et al., 2019).
Fan-shaped body cells are synaptically connected and receive inhibitory input from dopamine arousal (DAA) neurons, as hyperactivation of DAAs antagonizes the effect on sleep promotion observed when LPNs are hyperactivated; moreover, hyperactivation of both LPNs and DAAs significantly fragments sleep (Ni et al., 2019). FB cells promote sleep via GABAergic signalling, as the inhibition of GABA synthesis in these neurons eliminates the sleep promoted by their activation (Ni et al., 2019). FB sleep promoting neurons negatively regulate the activity of OAA neurons: they are closely connected with the FB axon terminals and their neuronal activity is dramatically reduced by the hyperactivation of FB cells (Ni et al., 2019; Figure 3).
Dorsal Neurons: A Major Role in Shaping and Maintaining the Sleep/Wake Pattern
Dorsal neurons (DN1s) are at the same time sleep- and wake-promoting cells, as a result of different signalling pathways that either act on different subsets of neurons or are predominant at different times of day, to promote activity in the morning or sleep at siesta and during the night.
A subgroup of DN1 neurons, the posterior DN1s (DN1ps), express the narrow abdomen (na), involved in light-mediated control of diurnal behaviour (morning activity and lights-on response) (Lear et al., 2005). Under LD cycles, DN1ps promote morning activity and their contribution to circadian behaviour is strongly influenced by light intensity (Zhang et al., 2010). They also express PDF receptor that, in these cells, is necessary for periodicity in DD (Zhang et al., 2010).
DN1s and DH31 wake-promoting signalling
DN1s secrete the neuropeptide diuretic hormone 31 (DH31) and express its receptor DH31-R1, homologous to vertebrate calcitonin gene-related peptide (CGRP) and its receptor (Mertens et al., 2005). DH31/DH31R are involved in sleep regulation: flies with a loss-of-function mutation in DH31 exhibit a significant increase of sleep, especially during the night, while the pan-neuronal overexpression of DH31 significantly decreases nighttime sleep (Kunst et al., 2014). More precisely, DH31 acts as negative regulator of sleep maintenance and awakens flies in anticipation of dawn; indeed the increased sleep of DH31 mutants is more prominent in the second half of the night and immediately before lights-on, and the overexpressing DH31 flies exhibit a decreased sleep and increase of sleep fragmentation, which is more pronounced in the late night (Kunst et al., 2014). These altered sleep features can be completely rescued by restoring expression of DH31 specifically in DN1s, that can also re-establish the anticipation of the lights-on (Kunst et al., 2014). DN1s are a direct target of PDF signalling from s-LNvs, that modulates sleep by controlling the time of DH31 secretion: PDF specifically activates PDFR late at night, and the consequent secretion of DH31 results in a reduced nighttime sleep and an awakening of flies at dawn (Kunst et al., 2014).
DN1s and glutamatergic signalling to clock cells
DN1s neuronal activity is also fundamental in promoting sleep: blocking the synaptic neurotransmission of these cells results in a marked increase of the flies’ activity and a decrease of siesta and nighttime total sleep, due to reduced sleep episodes duration (Guo et al., 2016).
DN1s directly contact core pacemaker cells: GRASP assay identifies a functional direct interaction of DN1s presynaptic regions with either dendritic regions of the Evening cells (the CRY-positive LNds and the 5th s-LNv) or dorsal axon regions of Morning cells (the s-LNvs) (Guo et al., 2016). This neuronal transmission is mediated by glutamatergic signalling, with inhibitory effect. DN1s express the vesicular glutamate transporter, DvGluT, while E cells express the metabotropic glutamate receptor DmGluRA, whose mRNA exhibits cycling levels with a peak in the middle of the day. This support the predominant inhibitory role of DN1s on E cell-derived locomotor activity, which is then confined in the late day-early night (Guo et al., 2016).
DN1s signals to sleep center
DN1s can be both wake- and sleep- promoting, according to synaptic types and targets. A CRY-positive subset of DN1s (anterior-projecting DNs, APDNs or a-DN1ps) sends post-synaptic projections also to the anterior region of adult brain, the superior lateral protocerebrum, target of the AstA sleep-promoting signalling from PLPs (Guo et al., 2016), and innervates the anterior optic tubercle (AOTU) (Guo et al., 2018; Lamaze et al., 2018). In particular, they target a small subset of neurons within the AOTU (TuBu), that receive visual inputs from medullo-tubercular neurons and transmit this information to EB-R neurons (Guo et al., 2018; Lamaze et al., 2018). However, DN1ps connect TuBu using both excitatory and inhibitory synapses. They suppress the sleep-promoting activity of TuBu neurons, as their acute inhibition results in an increase of TuBu electrical activity, while thermo-genetic excitation of TuBu neurons profoundly induces sleep throughout both day and night (Lamaze et al., 2018). On the other hand, DN1ps activate TuBu using glutamate, giving sleep-promoting effect (Guo et al., 2018).
Moreover, a CRY-negative subset of DN1s (ventro-contralateral-projecting DN1p neurons, vc-DN1ps) send ventral and contralateral projections to the PI region (Cavanaugh et al., 2014), that represents an activity-promoting output of DN1s, since the activation of these cells promotes wakefulness and inhibits sleep (Guo et al., 2018).
Light Exposure: Timing and Intensity
The quality and architecture of sleep is also influenced by the characteristics of the light stimulus, such as the intensity of light and the timing of light exposure (morning/daytime versus evening/nighttime).
Nocturnal Light Affects Daytime Sleep
Natural pattern of light availability assumes dark nighttime, therefore a different administration of light disrupts the sleep pattern. Sleep analysis of flies exposed to 4 days of discontinuous nocturnal light stimulation (DLS) showed a reduction of sleep episode duration (but not the sleep bouts number) specifically during the day, while it does not affect nighttime sleep. Moreover, during recovery time after light disruption, the quality of nighttime sleep is increased, opposite to daytime sleep. On the molecular level, discontinuous light stimulation disrupts CRY daily oscillation at both mRNA and protein levels, and decreases TIM levels during the night (Liu and Zhao, 2014).
Light Intensity Impacts on Sleep Timing
In the natural environment, light intensity changes between 0 and 100,000 lux, depending on the time of the day, time of the year, weather, etc. Insects are sensitive to a broad range of light, and they can adjust their behaviour according to light exposure, through an adaptative mechanism that allows avoiding of bright light in the middle of the day, especially during summertime (Lazopulo et al., 2019).
Flies are able to detect low light intensity by using CRY (Vinayak et al., 2013) and the four rhodopsins expressed in photoreceptors cells in the compound eyes (Rh1, Rh3, Rh4, Rh6) (Saint-Charles et al., 2016). In natural conditions, dim light appears during full moon nights and around dawn and dusk. Moonlight causes the phase shift of molecular clock in pacemaker cells, resulting in the advance of morning activity and the delay of the evening one, with the overall flies’ activity becoming more nocturnal. Experiments performed with clock mutants have clearly demonstrated that the effect on nighttime activity is light-dependent (Kempinger et al., 2009) and the response to moonlight is mediated by R1–6 and Rh6-expressing R8 photoreceptors (Schlichting et al., 2014), while CRY is not involved (Bachleitner et al., 2007). Moreover, dim light does not affect clock protein expression pattern in peripheral oscillators in the retina (Bachleitner et al., 2007). Periodogram analysis of wt flies under L:ML conditions (Light:MoonLight) reveals that sleep time is also affected: relative level of activity is increased compared with flies reared in light:dark conditions, and activity is continuous during the whole night, with flies sleeping mostly during the day (Schlichting et al., 2014).
The effect of twilight is opposite to moonlight: morning peak of activity is delayed and evening peak is advanced, while nocturnal activity in reduced (Rieger et al., 2007). R7 and R8 photoreceptors are involved in the response to twilight (Schlichting et al., 2014). When flies are exposed to both dim light at dawn/dusk and moonlight, the twilight effect dominates in terms of shifted morning and evening peak of activity and reduced nocturnal locomotor activity. This more composite light exposure has an effect which is more similar to natural conditions: fly activity is not shifted to the nighttime during full moon nights (Schlichting et al., 2014; Vanin et al., 2012).
In the middle of the day, flies are exposed to high intensity light (HI). Response to HI is independent from CRY, ocelli, and compound eyes, and most probably it is mediated by HB eyelets (Schlichting et al., 2016), which communicate with s-LNvs through acetylcholine. The possible mechanism at the basis of activity and sleep regulation mediated by HI assumes that activation of acetylcholine receptor on s-LNvs increases Ca2+ level, which, in turn, causes delayed PER degradation during the day. Then, s-LNvs propagate signal through PDF pathway, affecting PER cycling in downstream neurons DN1s, known to be sleep regulators (Guo et al., 2016). Indeed, flies exposed to HI show delayed evening peak of activity and lengthened siesta time (Schlichting et al., 2019a), thus avoiding bright light in the middle of the day during hot summer time.
Light Influences Temperature-Dependent Regulation of Sleep
Light plays a role in the regulation of temperature-dependent sleep pattern. Nighttime sleep is decreased by high temperature, but this effect is influenced by light presence during the preceding day, as it was shown in experiments performed in DD conditions. The mechanism of this process requires CRY, as cryout mutants do not show decreased nighttime sleep in response to heat (Parisky et al., 2016). This effect seems to be connected with the wake-promoting role of dopamine on nighttime sleep, as it was shown that light increases expression of inhibitory dopamine receptors (Shang et al., 2011). The involvement of light in temperature-dependent sleep control was comprehensively reviewed elsewhere (Lamaze and Stanewsky, 2020).
Conclusion
Understanding the mechanisms underlying the relations between light exposure and sleep disturbances has become a challenge. Modern society no longer relies on the day-night differences in external conditions that have shaped life on Earth: from shift-work schedules to the current widespread use of digital technology until late at night, we are more and more exposed to stimuli that not only are not coordinated with our body’s internal time but can also stimulate alertness and extend sleep latency. Taking advantage of the fruit fly Drosophila melanogaster, we have tried to address the contribution of the different light signalling pathways involved in promoting, consolidating, or preventing sleep. The picture derived is complex: the architecture of sleep is regulated by an intricate set of structures, neurotransmitters, and networks that integrate environmental signals.
However, as most of the essential sleep features are shared between flies and mammals, the knowledge of how light regulates this complex behaviour in Drosophila can be fundamental for future research in humans, addressing how light can promote high wakefulness during the day and good sleep during the night.
Author Contributions
GM, MD, and PC equally contributed to writing the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by grants from Department of Biology, University of Padua (Italy) PRID-SEED 2018 to PC and PRID-SEED 2019 to GM, the Polish National Science Centre (Narodowe Centrum Nauki, NCN_Grant UMO-2017/27/B/NZ3/00859), and Polish National Agency for Academic Exchange to MD.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sraboni Ghose (Ph.D.) for her comments with regard to this manuscript.
References
Abruzzi, K. C., Zadina, A., Luo, W., Wiyanto, E., Rahman, R., Guo, F., et al. (2017). RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet. 13:e1006613. doi: 10.1371/journal.pgen.1006613
Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. doi: 10.1126/science.287.5461.2185
Agosto, J., Choi, J. C., Parisky, K. M., Stilwell, G., Rosbash, M., and Griffith, L. C. (2008). Modulation of GABAA receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat. Neurosci. 11, 354–359. doi: 10.1038/nn2046
Alawi, A. A., and Pak, W. L. (1971). On-transient of insect electroretinogram: its cellular origin. Science 172, 1057–1057. doi: 10.1126/science.172.3987.1055
Alejevski, F., Saint-Charles, A., Michard-Vanhée, C., Martin, B., Galant, S., Vasiliauskas, D., et al. (2019). The HisCl1 histamine receptor acts in photoreceptors to synchronize Drosophila behavioural rhythms with light-dark cycles. Nat. Commun. 10:252.
Allebrandt, K. V., Amin, N., Müller-Myhsok, B., Esko, T., Teder-Laving, M., Azevedo, R. V. D. M., et al. (2013). A KATP channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol. Psychiatry 18, 122–132. doi: 10.1038/mp.2011.142
Allebrandt, K. V., Teder-Laving, M., Cusumano, P., Frishman, G., Levandovski, R., Ruepp, A., et al. (2017). Identifying pathways modulating sleep duration: from genomics to transcriptomics. Sci. Rep. 7:4555.
Arnes, M., Alaniz, M. E., Karam, C. S., Cho, J. D., Lopez, G., Javitch, J. A., et al. (2019). Role of tau protein in remodeling of circadian neuronal circuits and sleep. Front. Aging Neurosci. 11:320. doi: 10.3389/fnagi.2019.00320
Artiushin, G., and Sehgal, A. (2017). The Drosophila circuitry of sleep–wake regulation. Curr. Opin. Neurobiol. 44, 243–250. doi: 10.1016/j.conb.2017.03.004
Aso, Y., Sitaraman, D., Ichinose, T., Kaun, K. R., Vogt, K., Belliart-Guérin, G., et al. (2014). Mushroom body output neurons encode valence and guide memory-based action selection in Drosophila. eLife 3:e04580. doi: 10.7554/eLife.04580
Bachleitner, W., Kempinger, L., Wülbeck, C., Rieger, D., and Helfrich-Förster, C. (2007). Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 104, 3538–3543. doi: 10.1073/pnas.0606870104
Benito, J., Houl, J. H., Roman, G. W., and Hardin, P. E. (2008). The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS. J. Biol. Rhythms 23, 296–307. doi: 10.1177/0748730408318588
Bloomquist, B. T., Shortridge, R. D., Schneuwly, S., Perdew, M., Montell, C., Steller, H., et al. (1988). Isolation of a putative phospholipase c gene of drosophila, norpA, and its role in phototransduction. Cell 54, 723–733. doi: 10.1016/s0092-8674(88)80017-5
Bolkan, B. J., Triphan, T., and Kretzschmar, D. (2012). β-secretase cleavage of the fly amyloid precursor protein is required for glial survival. J. Neurosci. 32, 16181–16192. doi: 10.1523/JNEUROSCI.0228-12.2012
Borst, A., Haag, J., and Mauss, A. S. (2020). How fly neurons compute the direction of visual motion. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 206, 109–124. doi: 10.1007/s00359-019-01375-9
Borycz, J., Borycz, J. A., Loubani, M., and Meinertzhagen, I. A. (2002). Tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. J. Neurosci. 22, 10549–10557. doi: 10.1523/jneurosci.22-24-10549.2002
Boutrel, B., Monaca, C., Hen, R., Hamon, M., and Adrien, J. (2002). Involvement of 5-HT1A receptors in homeostatic and stress-induced adaptive regulations of paradoxical sleep: studies in 5-HT 1A Knock-Out Mice. J. Neurosci. 22, 4686–4692. doi: 10.1523/jneurosci.22-11-04686.2002
Braitenberg, V. (1967). Patterns of projection in the visual system of the fly. I. Retina-lamina projections. Exp. Brain Res. 3, 271–298. doi: 10.1007/BF00235589
Buchner, E., Buchner, S., Crawford, G., Mason, W. T., Salvaterra, P. M., and Sattelle, D. B. (1986). Choline acetyltransferase-like immunoreactivity in the brain of Drosophila melanogaster. Cell Tissue Res. 246, 57–62. doi: 10.1007/BF00218999
Buhl, E., Bradlaugh, A., Ogueta, M., Chen, K. F., Stanewsky, R., and Hodge, J. J. L. (2016). Quasimodo mediates daily and acute light effects on Drosophila clock neuron excitability. Proc. Natl. Acad. Sci. U.S.A. 113, 13486–13491. doi: 10.1073/pnas.1606547113
Burg, M. G., Sarthy, P. V., Koliantz, G., and Pak, W. L. (1993). Genetic and molecular identification of a Drosophila histidine decarboxylase gene required in photoreceptor transmitter synthesis. EMBO J. 12, 911–919. doi: 10.1002/j.1460-2075.1993.tb05732.x
Bushey, D., Huber, R., Tononi, G., and Cirelli, C. (2007). Drosophila Hyperkinetic mutants have reduced sleep and impaired memory. J. Neurosci. 27, 5384–5393. doi: 10.1523/JNEUROSCI.0108-07.2007
Busto, M., Iyengar, B., and Campos, A. R. (1999). Genetic dissection of behaviour: modulation of locomotion by light in the Drosophila melanogaster larva requires genetically distinct visual system functions. J. Neurosci. 19, 3337–3344. doi: 10.1523/jneurosci.19-09-03337.1999
Carmine-Simmen, K., Proctor, T., Tschäpe, J., Poeck, B., Triphan, T., Strauss, R., et al. (2009). Neurotoxic effects induced by the Drosophila amyloid-β peptide suggest a conserved toxic function. Neurobiol. Dis. 33, 274–281. doi: 10.1016/j.nbd.2008.10.014
Cavanaugh, D. J., Geratowski, J. D., Wooltorton, J. R. A., Spaethling, J. M., Hector, C. E., Zheng, X., et al. (2014). Identification of a circadian output circuit for rest: activity rhythms in drosophila. Cell 157, 689–701. doi: 10.1016/j.cell.2014.02.024
Ceriani, M. F., Darlington, T. K., Staknis, D., Más, P., Petti, A. A., Weitz, C. J., et al. (1999). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556. doi: 10.1126/science.285.5427.553
Chang, H. Y., and Ready, D. F. (2000). Rescue of photoreceptor degeneration in rhodopsin-null Drosophila mutants by activated RAC1. Science 290, 1978–1980. doi: 10.1126/science.290.5498.1978
Chatterjee, A., Lamaze, A., De, J., Mena, W., Chélot, E., Martin, B., et al. (2018). Reconfiguration of a multi-oscillator network by light in the Drosophila circadian clock. Curr. Biol. 28, 2007–2017.e4. doi: 10.1016/j.cub.2018.04.064
Chaturvedi, R., Luan, Z., Guo, P., and Li, H. S. (2016). Drosophila vision depends on carcinine uptake by an organic cation transporter. Cell Rep. 14, 2076–2083. doi: 10.1016/j.celrep.2016.02.009
Chaturvedi, R., Reddig, K., and Li, H. S. (2014). Long-distance mechanism of neurotransmitter recycling mediated by glial network facilitates visual function in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 117, 2812–2817. doi: 10.1073/pnas.1323714111
Chen, J., Reiher, W., Hermann-Luibl, C., Sellami, A., Cognigni, P., Kondo, S., et al. (2016). Allatostatin a signalling in drosophila regulates feeding and sleep and is modulated by PDF. PLoS Genet. 12:e1006346. doi: 10.1371/journal.pgen.1006346
Chen, K. F., Lowe, S., Lamaze, A., Krätschmer, P., and Jepson, J. (2019). Neurocalcin regulates nighttime sleep and arousal in Drosophila. eLife 8:e38114. doi: 10.7554/eLife.38114
Chen, K. F., Peschel, N., Zavodska, R., Sehadova, H., and Stanewsky, R. (2011). QUASIMODO, a novel GPI-anchored Zona Pellucida protein involved in light input to the drosophila circadian clock. Curr. Biol. 21, 719–729. doi: 10.1016/j.cub.2011.03.049
Chen, X., and Rosbash, M. (2017). MicroRNA-92a is a circadian modulator of neuronal excitability in Drosophila. Nat. Commun. 8:14707. doi: 10.1038/ncomms14707
Chou, W. H., Hall, K. J., Wilson, D. B., Wideman, C. L., Townson, S. M., Chadwell, L. V., et al. (1996). Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17, 1101–1115. doi: 10.1016/s0896-6273(00)80243-3
Chou, W. H., Huber, A., Bentrop, J., Schulz, S., Schwab, K., Chadwell, L. V., et al. (1999). Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126, 607–616.
Chung, B. Y., Kilman, V. L., Keath, J. R., Pitman, J. L., and Allada, R. (2009). The GABAA receptor RDL Acts in Peptidergic PDF neurons to promote sleep in Drosophila. Curr. Biol. 19, 386–390. doi: 10.1016/j.cub.2009.01.040
Cirelli, C., Bushey, D., Hill, S., Huber, R., Kreber, R., Ganetzky, B., et al. (2005). Reduced sleep in Drosophila Shaker mutants. Nature 434, 1087–1092. doi: 10.1038/nature03486
Claridge-Chang, A., Wijnen, H., Naef, F., Boothroyd, C., Rajewsky, N., and Young, M. W. (2001). Circadian regulation of gene expression systems in the Drosophila head. Neuron 32, 657–671. doi: 10.1016/s0896-6273(01)00515-3
Collins, B., Mazzoni, E. O., Stanewsky, R., and Blau, J. (2006). Drosophila CRYPTOCHROME is a circadian transcriptional repressor. Curr. Biol. 16, 441–449. doi: 10.1016/j.cub.2006.01.034
Crocker, A., Shahidullah, M., Levitan, I. B., and Sehgal, A. (2010). Identification of a neural circuit that underlies the effects of octopamine on sleep: wake behaviour. Neuron 65, 670–681. doi: 10.1016/j.neuron.2010.01.032
Cusumano, P., Biscontin, A., Sandrelli, F., Mazzotta, G. M., Tregnago, C., De Pittà, C., et al. (2018). Modulation of miR-210 alters phasing of circadian locomotor activity and impairs projections of PDF clock neurons in Drosophila melanogaster. PLoS Genet. 14:e1007500. doi: 10.1371/journal.pgen.1007500
Cusumano, P., Klarsfeld, A., Chélot, E., Picot, M., Richier, B., and Rouyer, F. (2009). PDF-modulated visual inputs and cryptochrome define diurnal behaviour in Drosophila. Nat. Neurosci. 12, 1431–1437. doi: 10.1038/nn.2429
Damulewicz, M., and Mazzotta, G. M. (2020). One actor, multiple roles: the performances of cryptochrome in Drosophila. Front. Physiol. 11:99. doi: 10.3389/fphys.2020.00099
Damulewicz, M., Ispizua, J. I., Ceriani, M. F., and Pyza, E. M. (2020). Communication among photoreceptors and the central clock affects sleep profile. Front. Physiol. 11. doi: 10.3389/fphys.2020.00993
Damulewicz, M., Mazzotta, G. M., Sartori, E., Rosato, E., Costa, R., and Pyza, E. M. (2017). Cryptochrome is a regulator of synaptic plasticity in the visual system of Drosophila melanogaster. Front. Mol. Neurosci. 10:165. doi: 10.3389/fnmol.2017.00165
Damulewicz, M., and Pyza, E. (2011). The clock input to the first optic neuropil of Drosophila melanogaster expressing neuronal circadian plasticity. PLoS One 6:e21258. doi: 10.1371/journal.pone.0021258
Davis, F. P., Nern, A., Picard, S., Reiser, M. B., Rubin, G. M., Eddy, S. R., et al. (2020). A genetic, genomic, and computational resource for exploring neural circuit function. eLife 9:50901. doi: 10.7554/eLife.50901
Deng, B., Li, Q., Liu, X., Cao, Y., Li, B., Qian, Y., et al. (2019). Chemoconnectomics: mapping chemical transmission in Drosophila. Neuron 101, 876–893.e4. doi: 10.1016/j.neuron.2019.01.045
Díaz, M. M., Schlichting, M., Abruzzi, K. C., Long, X., and Rosbash, M. (2019). Allatostatin-C/AstC-R2 is a novel pathway to modulate the circadian activity pattern in Drosophila. Curr. Biol. 29, 13–22.e3. doi: 10.1016/j.cub.2018.11.005
Dissel, S., Angadi, V., Kirszenblat, L., Suzuki, Y., Donlea, J., Klose, M., et al. (2015). Sleep restores behavioural plasticity to drosophila mutants. Curr. Biol. 25, 1270–1281. doi: 10.1016/j.cub.2015.03.027
Donlea, J. M., Pimentel, D., Talbot, C. B., Kempf, A., Omoto, J. J., Hartenstein, V., et al. (2018). Recurrent circuitry for balancing sleep need and sleep. Neuron 97, 378–389.e4. doi: 10.1016/j.neuron.2017.12.016
Donlea, J. M., Thimgan, M. S., Suzuki, Y., Gottschalk, L., and Shaw, P. J. (2011). Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576. doi: 10.1126/science.1202249
Emery, P., Stanewsky, R., Helfrich-Förster, C., Emery-Le, M., Hall, J. C., and Rosbash, M. (2000). Drosophila CRY is a deep brain circadian photoreceptor. Neuron 26, 493–504. doi: 10.1016/s0896-6273(00)81181-2
Farca Luna, A. J., Perier, M., and Seugnet, L. (2017). Amyloid precursor protein in Drosophila glia regulates sleep and genes involved in glutamate recycling. J. Neurosci. 37, 4289–4300. doi: 10.1523/JNEUROSCI.2826-16.2017
Faulhaber, J., Steiger, A., and Lancel, M. (1997). The GABA(A) agonist THIP produces slow wave sleep and reduces spindling activity in NREM sleep in humans. Psychopharmacology 130, 285–291. doi: 10.1007/s002130050241
Feiler, R., Bjornson, R., Kirschfeld, K., Mismer, D., Rubin, G. M., Smith, D. P., et al. (1992). Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: visual physiology and photochemistry of transgenic animals. J. Neurosci. 12, 3862–3868. doi: 10.1523/jneurosci.12-10-03862.1992
Feiler, R., Harris, W. A., Kirschfeld, K., Wehrhahn, C., and Zuker, C. S. (1988). Targeted misexpression of a Drosophila opsin gene leads to altered visual function. Nature 333, 737–741. doi: 10.1038/333737a0
Feinberg, E. H., VanHoven, M. K., Bendesky, A., Wang, G., Fetter, R. D., Shen, K., et al. (2008). GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron 57, 353–363. doi: 10.1016/j.neuron.2007.11.030
Feng, G., Zhang, J., Li, M., Shao, L., Yang, L., Song, Q., et al. (2018). Control of sleep onset by Shal/Kv4 channels in Drosophila circadian neurons. J. Neurosci. 38, 9059–9071. doi: 10.1523/JNEUROSCI.0777-18.2018
Fernández, M. P., Berni, J., and Ceriani, M. F. (2008). Circadian remodeling of neuronal circuits involved in rhythmic behaviour. PLoS Biol. 6:e69. doi: 10.1371/journal.pbio.0060069
Fogle, K. J., Parson, K. G., Dahm, N. A., Holmes, T. C., Sheeba, V., Parisky, K. M., et al. (2011). CRYPTOCHROME is a blue-light sensor that regulates neuronal firing rate. Science 331, 1409–1413. doi: 10.1126/science.1199702
Friggi-Grelin, F., Coulom, H., Meller, M., Gomez, D., Hirsh, J., and Birman, S. (2003). Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J. Neurobiol. 54, 618–627. doi: 10.1002/neu.10185
Fryxell, K. J., and Meyerowitz, E. M. (1987). An opsin gene that is expressed only in the R7 photoreceptor cell of Drosophila. EMBO J. 6, 443–451. doi: 10.1002/j.1460-2075.1987.tb04774.x
Gao, S., Takemura, S.-Y., Ting, C. Y., Huang, S., Lu, Z., Luan, H., et al. (2008). The neural substrate of spectral preference in Drosophila. Neuron 60, 328–342. doi: 10.1016/j.neuron.2008.08.010
Gengs, C., Leung, H. T., Skingsley, D. R., Iovchev, M. I., Yin, Z., Semenov, E. P., et al. (2002). The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA). J. Biol. Chem. 277, 42113–42120. doi: 10.1074/jbc.M207133200
Gisselmann, G., Pusch, H., Hovemann, B. T., and Hatt, H. (2002). Two cDNAS coding for histamine-gated ion channels in D. melanogaster. Nat. Neurosci. 5, 11–12. doi: 10.1038/nn787
Grebler, R., Kistenpfennig, C., Rieger, D., Bentrop, J., Schneuwly, S., Senthilan, P. R., et al. (2017). Drosophila Rhodopsin 7 can partially replace the structural role of Rhodopsin 1, but not its physiological function. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 203, 649–659. doi: 10.1007/s00359-017-1182-8
Grima, B., Chélot, E., Xia, R., and Rouyer, F. (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. doi: 10.1038/nature02935
Guo, F., Holla, M., Díaz, M. M., and Rosbash, M. (2018). A circadian output circuit controls sleep-wake arousal in Drosophila. Neuron 100, 624–635.e4. doi: 10.1016/j.neuron.2018.09.002
Guo, F., Yu, J., Jung, H. J., Abruzzi, K. C., Luo, W., Griffith, L. C., et al. (2016). Circadian neuron feedback controls the Drosophila sleep-activity profile. Nature 536, 292–297. doi: 10.1038/nature19097
Hamasaka, Y., and Nässel, D. R. (2006). Mapping of serotonin, dopamine, and histamine in relation to different clock neurons in the brain of Drosophila. J. Comp. Neurol. 494, 314–330. doi: 10.1002/cne.20807
Hamasaka, Y., Rieger, D., Parmentier, M. L., Grau, Y., Helfrich-Förster, C., and Nässel, D. R. (2007). Glutamate and its metabotropic receptor in Drosophila clock neuron circuits. J. Comp. Neurol. 505, 32–45. doi: 10.1002/cne.21471
Hardie, R. C. (1979). Electrophysiological analysis of fly retina. I: Comparative properties of R1-6 and R 7 and 8. J. Comp. Physiol. A 129, 19–33. doi: 10.1007/BF00679908
Hardie, R. C. (1987). Is histamine a neurotransmitter in insect photoreceptors? J. Comp. Physiol. A 161, 201–213. doi: 10.1007/BF00615241
Hardie, R. C. (1989). A histamine-activated chloride channel involved in neurotransmission at a photoreceptor synapse. Nature 339, 704–706. doi: 10.1038/339704a0
Hartwig, S., Dovengerds, C., Herrmann, C., and Hovemann, B. T. (2014). Drosophila Ebony: a novel type of nonribosomal peptide synthetase related enzyme with unusually fast peptide bond formation kinetics. FEBS J. 281, 5147–5158. doi: 10.1111/febs.13054
Hassan, J., Busto, M., Iyengar, B., and Campos, A. R. (2000). Behavioural characterization and genetic analysis of the Drosophila melanogaster larval response to light as revealed by a novel individual assay. Behav. Genet. 30, 59–69. doi: 10.1023/A:1002090627601
Haynes, P. R., Christmann, B. L., and Griffith, L. C. (2015). A single pair of neurons links sleep to memory consolidation in Drosophila melanogaster. eLife 7:e03868. doi: 10.7554/eLife.03868
Haynie, J. L., and Bryant, P. J. (1986). Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 237, 293–308. doi: 10.1002/jez.1402370302
Heisenberg, M. (1971). Separation of receptor and lamina potentials in the electroretinogram of normal and mutant Drosophila. J. Exp. Biol. 55, 85–100.
Heisenberg, M., and Buchner, E. (1977). The rôle of retinula cell types in visual behaviour of Drosophila melanogaster. J. Comp. Physiol. 117, 127–162. doi: 10.1007/BF00612784
Helfrich-Förster, C. (2019). Light input pathways to the circadian clock of insects with an emphasis on the fruit fly Drosophila melanogaster. J. Comp. Physiol. A. 206, 259–272. doi: 10.1007/s00359-019-01379-5
Helfrich-Förster, C., Edwards, T., Yasuyama, K., Wisotzki, B., Schneuwly, S., Stanewsky, R., et al. (2002). The extraretinal eyelet of Drosophila: development, ultrastructure, and putative circadian function. J. Neurosci. 22, 9255–9266. doi: 10.1523/jneurosci.22-21-09255.2002
Helfrich-Förster, C., Winter, C., Hofbauer, A., Hall, J. C., and Stanewsky, R. (2001). The circadian clock of fruit flies is blind after elimination of all known photoreceptors. Neuron 30, 249–261. doi: 10.1016/s0896-6273(01)00277-x
Hendricks, J. C. (2003). Invited review: sleeping flies don’t lie: the use of Drosophila melanogaster to study sleep and circadian rhythms. J. Appl. Physiol. 94, 1660–1672. doi: 10.1152/japplphysiol.00904.2002
Hendricks, J. C., Finn, S. M., Panckeri, K. A., Chavkin, J., Williams, J. A., Sehgal, A., et al. (2000). Rest in Drosophila is a sleep-like state. Neuron 25, 129–138. doi: 10.1016/s0896-6273(00)80877-6
Hobert, O., and Westphal, H. (2000). Functions of LIM-homeobox genes. Trends Genet. 16, 75–83. doi: 10.1016/s0168-9525(99)01883-1
Hofbauer, A., and Buchner, E. (1989). Does Drosophila have seven eyes? Naturwissenschaften 76, 335–336. doi: 10.1007/BF00368438
Hong, S. T., Bang, S., Paik, D., Kang, J., Hwang, S., Jeon, K., et al. (2006). Histamine and its receptors modulate temperature-preference behaviours in Drosophila. J. Neurosci. 26, 7245–7256. doi: 10.1523/JNEUROSCI.5426-05.2006
Hu, K. G., and Stark, W. S. (1980). The roles of Drosophila ocelli and compound eyes in phototaxis. J. Comp. Physiol. A 135, 85–95. doi: 10.1007/BF00660183
Huber, A., Schulz, S., Bentrop, J., Groell, C., Wolfrum, U., and Paulsen, R. (1997). Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells. FEBS Lett. 406, 6–10. doi: 10.1016/S0014-5793(97)00210-X
Huber, R., Hill, S. L., Holladay, C., Biesiadecki, M., Tononi, G., and Cirelli, C. (2004). Sleep homeostasis in Drosophila melanogaster. Sleep 27, 628–639. doi: 10.1093/sleep/27.4.628
Johard, H. A. D., Yoishii, T., Dircksen, H., Cusumano, P., Rouyer, F., Helfrich-Förster, C., et al. (2009). Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J. Comp. Neurol. 516, 59–73. doi: 10.1002/cne.22099
Joiner, W. J., Crocker, A., White, B. H., and Sehgal, A. (2006). Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760. doi: 10.1038/nature04811
Kahsai, L., Martin, J.-R., and Winther, A. M. E. (2010). Neuropeptides in the Drosophila central complex in modulation of locomotor behaviour. J. Exp. Biol. 213, 2256–2265. doi: 10.1242/jeb.043190
Kaneko, M., Helfrich-Förster, C., and Hall, J. C. (1997). Spatial and temporal expression of the period and timeless genes in the developing nervous system of drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. J. Neurosci. 17, 6745–6760. doi: 10.1523/jneurosci.17-17-06745.1997
Kayser, M. S., Yue, Z., and Sehgal, A. (2014). A critical period of sleep for development of courtship circuitry and behaviour in Drosophila. Science 344, 269–274. doi: 10.1126/science.1250553
Kempinger, L., Dittmann, R., Rieger, D., and Helfrich-Förster, C. (2009). The nocturnal activity of fruit flies exposed to artificial moonlight is partly caused by direct light effects on the activity level that bypass the endogenous clock. Chronobiol. Int. 26, 151–166. doi: 10.1080/07420520902747124
Kim, E. Y., Bae, K., Ng, F. S., Glossop, N. R. J., Hardin, P. E., and Edery, I. (2002). Drosophila clock protein is under posttranscriptional control and influences light-induced activity. Neuron 34, 69–81. doi: 10.1016/s0896-6273(02)00639-6
Kirszenblat, L., Yaun, R., and Van Swinderen, B. (2019). Visual experience drives sleep need in Drosophila. Sleep. 42:zsz102. doi: 10.1093/sleep/zsz102
Kistenpfennig, C., Grebler, R., Ogueta, M., Hermann-Luibl, C., Schlichting, M., Stanewsky, R., et al. (2017). A new Rhodopsin influences light-dependent daily activity patterns of fruit flies. J. Biol. Rhythms 32, 406–422. doi: 10.1177/0748730417721826
Koh, K., Zheng, X., and Sehgal, A. (2006). JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science 312, 1809–1812. doi: 10.1126/science.1124951
Kula-Eversole, E., Nagoshi, E., Shang, Y., Rodriguez, J., Allada, R., and Rosbash, M. (2010). Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 107, 13497–13502. doi: 10.1073/pnas.1002081107
Kumar, S., Chen, D., and Sehgal, A. (2012). Dopamine acts through Cryptochrome to promote acute arousal in Drosophila. Genes Dev. 26, 1224–1234. doi: 10.1101/gad.186338.111
Kunst, M., Hughes, M. E., Raccuglia, D., Felix, M., Li, M., Barnett, G., et al. (2014). Calcitonin gene-related peptide neurons mediate sleep-specific circadian output in Drosophila. Curr. Biol. 24, 2652–2664. doi: 10.1016/j.cub.2014.09.077
Lamaze, A., Krätschmer, P., Chen, K. F., Lowe, S., and Jepson, J. E. C. (2018). A wake-promoting circadian output circuit in Drosophila. Curr. Biol. 28, 3098–3105.e3. doi: 10.1016/j.cub.2018.07.024
Lamaze, A., and Stanewsky, R. (2020). DN1p or the “Fluffy”. Cerberus of Clock Outputs. Front. Physiol. 10:1540. doi: 10.3389/fphys.2019.01540
Lazopulo, S., Lazopulo, A., Baker, J. D., and Syed, S. (2019). Daytime colour preference in Drosophila depends on the circadian clock and TRP channels. Nature 574, 108–111. doi: 10.1038/s41586-019-1571-y
Lear, B. C., Lin, J. M., Keath, J. R., McGill, J. J., Raman, I. M., and Allada, R. (2005). The ion channel narrow abdomen is critical for neural output of the Drosophila circadian pacemaker. Neuron 48, 965–976. doi: 10.1016/j.neuron.2005.10.030
Lelito, K. R., and Shafer, O. T. (2012). Reciprocal cholinergic and GABAergic modulation of the small ventrolateral pacemaker neurons of Drosophila’s circadian clock neuron network. J. Neurophysiol. 107, 2096–2108. doi: 10.1152/jn.00931.2011
Li, M. T., Cao, L. H., Xiao, N., Tang, M., Deng, B., Yang, T., et al. (2018). Hub-organized parallel circuits of central circadian pacemaker neurons for visual photoentrainment in Drosophila. Nat. Commun. 9:4247.
Liang, X., Holy, T. E., and Taghert, P. H. (2016). Synchronous drosophila circadian pacemakers display nonsynchronous Ca2+ rhythms in vivo. Science 351, 976–981. doi: 10.1126/science.aad3997
Lin, T. Y., Luo, J., Shinomiya, K., Ting, C. Y., Lu, Z., Meinertzhagen, I. A., et al. (2016). Mapping chromatic pathways in the Drosophila visual system. J. Comp. Neurol. 524, 213–227. doi: 10.1002/cne.23857
Lindholm, P., and Saarma, M. (2010). Novel CDNF/MANF family of neurotrophic factors. Dev. Neurobiol. 70, 360–371. doi: 10.1002/dneu.20760
Liu, G., Seiler, H., Wen, A., Zars, T., Ito, K., Wolf, R., et al. (2006). Distinct memory traces for two visual features in the Drosophila brain. Nature 439, 551–556. doi: 10.1038/nature04381
Liu, Q., Liu, S., Kodama, L., Driscoll, M. R., and Wu, M. N. (2012). Two dopaminergic neurons signal to the dorsal fan-shaped body to promote wakefulness in Drosophila. Curr. Biol. 22, 2114–2123. doi: 10.1016/j.cub.2012.09.008
Liu, S., Lamaze, A., Liu, Q., Tabuchi, M., Yang, Y., Fowler, M., et al. (2014). WIDE AWAKE mediates the circadian timing of sleep onset. Neuron 82, 151–166. doi: 10.1016/j.neuron.2014.01.040
Liu, S., Liu, Q., Tabuchi, M., and Wu, M. N. (2016). Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360. doi: 10.1016/j.cell.2016.04.013
Liu, W. W., and Wilson, R. I. (2013). Glutamate is an inhibitory neurotransmitter in the Drosophila olfactory system. Proc. Natl. Acad. Sci. U.S.A. 110, 10294–10299. doi: 10.1073/pnas.1220560110
Liu, Z., and Zhao, Z. (2014). Effects of light interruption on sleep and viability of Drosophila melanogaster. PLoS One 9:e105678. doi: 10.1371/journal.pone.0105678
Lu, B., Liu, W., Guo, F., and Guo, A. (2008). Circadian modulation of light-induced locomotion responses in Drosophila melanogaster. Genes Brain Behav. 7, 730–739. doi: 10.1111/j.1601-183X.2008.00411.x
Ly, S., Pack, A. I., and Naidoo, N. (2018). The neurobiological basis of sleep: insights from Drosophila. Neurosci. Biobehav. Rev. 87, 67–86. doi: 10.1016/j.neubiorev.2018.01.015
Malpel, S., Klarsfeld, A., and Rouyer, F. (2002). Larval optic nerve and adult extra-retinal photoreceptors sequentially associate with clock neurons during Drosophila brain development. Development 129, 1443–1453.
Mao, Z., and Davis, R. L. (2009). Eight different types of dopaminergic neurons innervate the Drosophila mushroom body neuropil: anatomical and physiological heterogeneity. Front. Neural Circuits 3:5. doi: 10.3389/neuro.04.005.2009
Mauss, A. S., Meier, M., Serbe, E., and Borst, A. (2014). Optogenetic and pharmacologic dissection of feedforward inhibition in Drosophila motion vision. J. Neurosci. 34, 2254–2263. doi: 10.1523/JNEUROSCI.3938-13.2014
Mauss, A. S., Pankova, K., Arenz, A., Nern, A., Rubin, G. M., and Borst, A. (2015). Neural circuit to integrate opposing motions in the visual field. Cell 162, 351–362. doi: 10.1016/j.cell.2015.06.035
Mazzotta, G., Rossi, A., Leonardi, E., Mason, M., Bertolucci, C., Caccin, L., et al. (2013). Fly cryptochrome and the visual system. Proc. Natl. Acad. Sci. U.S.A. 110, 6163–6168. doi: 10.1073/pnas.1212317110
Mazzotta, G. M., Bellanda, M., Minervini, G., Damulewicz, M., Cusumano, P., Aufiero, S., et al. (2018). Calmodulin enhances cryptochrome binding to INAD in Drosophila photoreceptors. Front. Mol. Neurosci. 11:280. doi: 10.3389/fnmol.2018.00280
McCarthy, E. V., Wu, Y., deCarvalho, T., Brandt, C., Cao, G., and Nitabach, M. N. (2011). Synchronized bilateral synaptic inputs to Drosophila melanogaster neuropeptidergic rest/arousal neurons. J. Neurosci. 31, 8181–8193. doi: 10.1523/JNEUROSCI.2017-10.2011
Mealey-Ferrara, M. L., Montalvo, A. G., and Hall, J. C. (2003). Effects of combining a cryptochrome mutation with other visual-system variants on entrainment of locomotor and adult-emergence rhythms in Drosophila. J. Neurogenet. 17, 171–221. doi: 10.1080/neg.17.2-3.171.221
Meinertzhagen, I. A., and O’Neil, S. D. (1991). Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. J. Comp. Neurol. 305, 232–263. doi: 10.1002/cne.903050206
Meinertzhagen, I. A., and Sorra, K. E. (2001). Synaptic organization in the fly’s optic lamina: few cells, many synapses and divergent microcircuits. Prog. Brain Res. 131, 53–69. doi: 10.1016/S0079-6123(01)31007-5
Melamed, J., and Trujillo-Cenóz, O. (1967). The fine structure of the central cells in the ommatidia of dipterans. J. Ultrasruct. Res. 21, 313–334. doi: 10.1016/s0022-5320(67)80098-4
Mertens, I., Vandingenen, A., Johnson, E. C., Shafer, O. T., Li, W., Trigg, J. S., et al. (2005). PDF receptor signalling in Drosophila contributes to both circadian and geotactic behaviours. Neuron 48, 213–219. doi: 10.1016/j.neuron.2005.09.009
Millard, S. S., and Pecot, M. Y. (2018). Strategies for assembling columns and layers in the Drosophila visual system. Neural Dev. 13:11.
Mismer, D., and Rubin, G. M. (1987). Analysis of the promoter of the ninaE opsin gene in Drosophila melanogaster. Genetics 116, 565–578.
Montell, C. (2012). Drosophila visual transduction. Trends Neurosci. 35, 356–363. doi: 10.1016/j.tins.2012.03.004
Montell, C., Jones, K., Zuker, C., and Rubin, G. (1987). A second opsin gene expressed in the ultraviolet-sensitive R7 photoreceptor cells of Drosophila melanogaster. J. Neurosci. 7, 1558–1566. doi: 10.1523/jneurosci.07-05-01558.1987
Muraro, N. I., and Ceriani, M. F. (2015). Acetylcholine from visual circuits modulates the activity of arousal neurons in Drosophila. J. Neurosci. 35, 16315–16327. doi: 10.1523/JNEUROSCI.1571-15.2015
Nagy, D., Cusumano, P., Andreatta, G., Anduaga, A. M., Hermann-Luibl, C., Reinhard, N., et al. (2019). Peptidergic signalling from clock neurons regulates reproductive dormancy in drosophila melanogaster. PLoS Genet. 15:e1008158. doi: 10.1371/journal.pgen.1008158
Nässel, D. R. (1999). Histamine in the brain of insects: a review. Microsc. Res. Tech. 44, 121–136. doi: 10.1002/(sici)1097-0029(19990115/01)44:2/3<121::aid-jemt6>3.0.co;2-f
Nässel, D. R., Enell, L. E., Santos, J. G., Wegener, C., and Johard, H. A. D. (2008). A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 9:90. doi: 10.1186/1471-2202-9-90
Ni, J. D., Baik, L. S., Holmes, T. C., and Montell, C. (2017). A rhodopsin in the brain functions in circadian photoentrainment in Drosophila. Nature 545, 340–344. doi: 10.1038/nature22325
Ni, J. D., Gurav, A. S., Liu, W., Ogunmowo, T. H., Hackbart, H., Elsheikh, A., et al. (2019). Differential regulation of the drosophila sleep homeostat by circadian and arousal inputs. eLife 8:e40487. doi: 10.7554/eLife.40487
Nitabach, M. N., Sheeba, V., Vera, D. A., Blau, J., and Holmes, T. C. (2005). Membrane electrical excitability is necessary for the free-running larval Drosophila circadian clock. J. Neurobiol. 62, 1–13. doi: 10.1002/neu.20053
Nitz, D. A., Van Swinderen, B., Tononi, G., and Greenspan, R. J. (2002). Electrophysiological correlates of rest and activity in Drosophila melanogaster. Curr. Biol. 12, 1934–1940. doi: 10.1016/s0960-9822(02)01300-3
Niu, Y., Liu, Z., Nian, X., Xu, X., and Zhang, Y. (2019). miR-210 controls the evening phase of circadian locomotor rhythms through repression of Fasciclin 2. PLoS Genet. 15:e1007655. doi: 10.1371/journal.pgen.1007655
Oh, Y., Jang, D., Sonn, J. Y., and Choe, J. (2013). Histamine-HisCl1 receptor axis regulates wake-promoting signals in Drosophila melanogaster. PLoS One 8:e68269. doi: 10.1371/journal.pone.0068269
Omoto, J. J., Kele??, M. F., Nguyen, B. C. M., Bolanos, C., Lovick, J. K., Frye, M. A., et al. (2017). Visual input to the drosophila central complex by developmentally and functionally distinct neuronal populations. Curr. Biol. 27, 1098–1110. doi: 10.1016/j.cub.2017.02.063
O’Tousa, J. E., Baehr, W., Martin, R. L., Hirsh, J., Pak, W. L., and Applebury, M. L. (1985). The Drosophila ninaE gene encodes an opsin. Cell 40, 839–850. doi: 10.1016/0092-8674(85)90343-5
Palgi, M., Greco, D., Lindström, R., Auvinen, P., and Heino, T. I. (2012). Gene expression analysis of Drosophilaa Manf mutants reveals perturbations in membrane traffic and major metabolic changes. BMC Genomics 13:134. doi: 10.1186/1471-2164-13-134
Palgi, M., Lindström, R., Peränen, J., Piepponen, T. P., Saarma, M., and Heino, T. I. (2009). Evidence that DmMANF is an invertebrate neurotrophic factor supporting dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 2429–2434. doi: 10.1073/pnas.0810996106
Pantazis, A., Segaran, A., Liu, C. H., Nikolaev, A., Rister, J., Thum, A. S., et al. (2008). Distinct roles for two histamine receptors (hclA and hclB) at the Drosophila photoreceptor synapse. J. Neurosci. 28, 7250–7259. doi: 10.1523/JNEUROSCI.1654-08.2008
Papatsenko, D., Sheng, G., and Desplan, C. (1997). A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development 124, 1665–1673.
Parisky, K. M., Agosto, J., Pulver, S. R., Shang, Y., Kuklin, E., Hodge, J. J. L., et al. (2008). PDF Cells Are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron 60, 672–682. doi: 10.1016/j.neuron.2008.10.042
Parisky, K. M., Agosto Rivera, J. L., Donelson, N. C., Kotecha, S., and Griffith, L. C. (2016). Reorganization of sleep by temperature in drosophila requires light, the Homeostat, and the circadian clock. Curr. Biol. 26, 882–892. doi: 10.1016/j.cub.2016.02.011
Perez, S. E., and Steller, H. (1996). Migration of glial cells into retinal axon target field in Drosophila melanogaster. J. Neurobiol. 30, 359–373. doi: 10.1002/(sici)1097-4695(199607)30:3<359::aid-neu5>3.0.co;2-3
Peschel, N., Chen, K. F., Szabo, G., and Stanewsky, R. (2009). Light-Dependent Interactions between the Drosophila Circadian Clock Factors Cryptochrome. Jetlag, and Timeless. Curr. Biol. 19, 241–247. doi: 10.1016/j.cub.2008.12.042
Picot, M., Cusumano, P., Klarsfeld, A., Ueda, R., and Rouyer, F. (2007). Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol. 5:2513–2521. doi: 10.1371/journal.pbio.0050315
Pimentel, D., Donlea, J. M., Talbot, C. B., Song, S. M., Thurston, A. J. F., and Miesenböck, G. (2016). Operation of a homeostatic sleep switch. Nature 536, 333–337. doi: 10.1038/nature19055
Pitman, J. L., McGill, J. J., Keegan, K. P., and Allada, R. (2006). A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756. doi: 10.1038/nature04739
Plaza, S., Chanut-Delalande, H., Fernandes, I., Wassarman, P. M., and Payre, F. (2010). From A to Z: apical structures and zona pellucida-domain proteins. Trends Cell Biol. 20, 524–532. doi: 10.1016/j.tcb.2010.06.002
Poeck, B., Triphan, T., Neuser, K., and Strauss, R. (2008). Locomotor control by the central complex in Drosophila - An analysis of the tay bridge mutant. Dev. Neurobiol. 68, 1046–1058. doi: 10.1002/dneu.20643
Pollack, I., and Hofbauer, A. (1991). Histamine-like immunoreactivity in the visual system and brain of Drosophila melanogaster. Cell Tissue Res. 266, 391–398. doi: 10.1007/BF00318195
Pollock, J. A., and Benzer, S. (1988). Transcript localization of four opsin genes in the three visual organs of Drosophila. RH2 is ocellus specific. Nature 333, 779–782. doi: 10.1038/333779a0
Potdar, S., and Sheeba, V. (2018). Wakefulness is promoted during day time by PDFR signalling to dopaminergic neurons in Drosophila melanogaster. eNeuro 5:ENEURO.129-18.2018. doi: 10.1523/ENEURO.0129-18.2018
Qian, Y., Cao, Y., Deng, B., Yang, G., Li, J., Xu, R., et al. (2017). Sleep homeostasis regulated by 5HT2b receptor in a small subset of neurons in the dorsal fan-shaped body of drosophila. eLife 6:e26519. doi: 10.7554/eLife.26519
Raccuglia, D., Huang, S., Ender, A., Heim, M. M., Laber, D., Suárez-Grimalt, R., et al. (2019). Network-specific synchronization of electrical slow-wave oscillations regulates sleep drive in Drosophila. Curr. Biol. 29, 3611–3621.e3. doi: 10.1016/j.cub.2019.08.070
Raghu, S. V., and Borst, A. (2011). Candidate glutamatergic neurons in the visual system of drosophila. PLoS One 6:e19472. doi: 10.1371/journal.pone.0019472
Rahman, M., Ham, H., Liu, X., Sugiura, Y., Orth, K., and Krämer, H. (2012). Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 15, 871–875. doi: 10.1038/nn.3102
Redlin, U. (2001). Neural basis and biological function of masking by light in mammals: suppression of melatonin and locomotor activity. Chronobiol. Int. 18, 737–758. doi: 10.1081/CBI-100107511
Richardt, A., Kemme, T., Wagner, S., Schwarzer, D., Marahiel, M. A., and Hovemann, B. T. (2003). Ebony, a novel nonribosomal peptide synthetase for β-alanine conjugation with biogenic amines in Drosophila. J. Biol. Chem. 278, 41160–41166. doi: 10.1074/jbc.M304303200
Richardt, A., Rybak, J., Störtkuhl, K. F., Meinertzhagen, I. A., and Hovemann, B. T. (2002). Ebony protein in the Drosophila nervous system: optic neuropile expression in glial cells. J. Comp. Neurol. 452, 93–102. doi: 10.1002/cne.10360
Rieger, D., Fraunholz, C., Popp, J., Bichler, D., Dittmann, R., and Helfrich-Förster, C. (2007). The fruit fly Drosophila melanogaster favors dim light and times its activity peaks to early dawn and late dusk. J. Biol. Rhythms 22, 387–399. doi: 10.1177/0748730407306198
Rieger, D., Stanewsky, R., and Helfrich-Förster, C. (2003). Cryptochrome, compound eyes, hofbauer-buchner eyelets, and ocelli play different roles in the entrainment and masking pathway of the locomotor activity rhythm in the fruit fly Drosophila Melanogaster. J. Biol. Rhythms 18, 377–391. doi: 10.1177/0748730403256997
Rister, J., Pauls, D., Schnell, B., Ting, C. Y., Lee, C. H., Sinakevitch, I., et al. (2007). Dissection of the peripheral motion channel in the visual system of Drosophila melanogaster. Neuron 56, 155–170. doi: 10.1016/j.neuron.2007.09.014
Rivera-Alba, M., Vitaladevuni, S. N., Mischenko, Y., Lu, Z., Takemura, S. Y., Scheffer, L., et al. (2011). Wiring economy and volume exclusion determine neuronal placement in the Drosophila brain. Curr. Biol. 21, 2000–2005. doi: 10.1016/j.cub.2011.10.022
Robinson, J. E., Paluch, J., Dickman, D. K., and Joiner, W. J. (2016). ADAR-mediated RNA editing suppresses sleep by acting as a brake on glutamatergic synaptic plasticity. Nat. Commun. 7:10512. doi: 10.1038/ncomms10512
Rosato, E., Codd, V., Mazzotta, G., Piccin, A., Zordan, M., Costa, R., et al. (2001). Light-dependent interaction between Drosophila CRY and the clock protein PER mediated by the carboxy terminus of CRY. Curr. Biol. 11, 909–917. doi: 10.1016/S0960-9822(01)00259-7
Saint-Charles, A., Michard-Vanhée, C., Alejevski, F., Chélot, E., Boivin, A., and Rouyer, F. (2016). Four of the six Drosophila rhodopsin-expressing photoreceptors can mediate circadian entrainment in low light. J. Comp. Neurol. 524, 2828–2844. doi: 10.1002/cne.23994
Salcedo, E., Huber, A., Henrich, S., Chadwell, L. V., Chou, W. H., Paulsen, R., et al. (1999). Blue- and green-absorbing visual pigments of Drosophila: ectopic expression and physiological characterization of the R8 photoreceptor cell- specific Rh5 and Rh6 rhodopsins. J. Neurosci. 19, 10716–10726. doi: 10.1523/jneurosci.19-24-10716.1999
Schlichting, M., Grebler, R., Peschel, N., Yoshii, T., and Helfrich-Förster, C. (2014). Moonlight detection by drosophila’s endogenous clock depends on multiple photopigments in the compound eyes. J. Biol. Rhythms 29, 75–86. doi: 10.1177/0748730413520428
Schlichting, M., Menegazzi, P., Lelito, K. R., Yao, Z., Buhl, E., Benetta, E. D., et al. (2016). A neural network underlying circadian entrainment and photoperiodic adjustment of sleep and activity in Drosophila. J. Neurosci. 36, 9084–9096. doi: 10.1523/JNEUROSCI.0992-16.2016
Schlichting, M., Menegazzi, P., Rosbash, M., and Helfrich-Förster, C. (2019a). A distinct visual pathway mediates high-intensity light adaptation of the circadian clock in drosophila. J. Neurosci. 39, 1621–1630. doi: 10.1523/JNEUROSCI.1497-18.2018
Schlichting, M., Rieger, D., Cusumano, P., Grebler, R., Costa, R., Mazzotta, G. M., et al. (2018). Cryptochrome interacts with actin and enhances eye-mediated light sensitivity of the circadian clock in Drosophila melanogaster. Front. Mol. Neurosci. 11:238. doi: 10.3389/fnmol.2018.00238
Schlichting, M., Weidner, P., Diaz, M., Menegazzi, P., Dalla Benetta, E., Helfrich-Förster, C., et al. (2019b). Light-mediated circuit switching in the drosophila neuronal clock network. Current Biology 29, 3266–3276.e3. doi: 10.1016/j.cub.2019.08.033
Schnaitmann, C., Haikala, V., Abraham, E., Oberhauser, V., Thestrup, T., Griesbeck, O., et al. (2018). Color processing in the early visual system of Drosophila. Cell 172, 318–330.e18. doi: 10.1016/j.cell.2017.12.018
Schubert, F. K., Hagedorn, N., Yoshii, T., Helfrich-Förster, C., and Rieger, D. (2018). Neuroanatomical details of the lateral neurons of Drosophila melanogaster support their functional role in the circadian system. J. Comp. Neurol. 526, 1209–1231. doi: 10.1002/cne.24406
Scott, K., Becker, A., Sun, Y., Hardy, R., and Zuker, C. (1995). Gqα protein function in vivo: genetic dissection of its role in photoreceptor cell physiology. Neuron 15, 919–927. doi: 10.1016/0896-6273(95)90182-5
Seelig, J. D., and Jayaraman, V. (2013). Feature detection and orientation tuning in the Drosophila central complex. Nature 503, 262–266. doi: 10.1038/nature12601
Sengupta, S., Crowe, L. B., You, S., Roberts, M. A., and Jackson, F. R. (2019). A secreted Ig-Domain protein required in both astrocytes and neurons for regulation of Drosophila night sleep. Curr. Biol. 25, 2547–2554.e2. doi: 10.1016/j.cub.2019.06.055
Senthilan, P. R., Grebler, R., Reinhard, N., Rieger, D., and Helfrich-Förster, C. (2019). Role of rhodopsins as circadian photoreceptors in the drosophila melanogaster. Biology 8:6. doi: 10.3390/biology8010006
Senthilan, P. R., and Helfrich-Förster, C. (2016). Rhodopsin 7-The unusual Rhodopsin in Drosophila. PeerJ 4:e2427. doi: 10.7717/peerj.2427
Shafer, O. T., Kim, D. J., Dunbar-Yaffe, R., Nikolaev, V. O., Lohse, M. J., and Taghert, P. H. (2008). Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of drosophila revealed by real-time cyclic AMP imaging. Neuron 58, 223–237. doi: 10.1016/j.neuron.2008.02.018
Shang, Y., Donelson, N. C., Vecsey, C. G., Guo, F., Rosbash, M., and Griffith, L. C. (2013). Short neuropeptide F is a sleep-promoting inhibitory modulator. Neuron 80, 171–183. doi: 10.1016/j.neuron.2013.07.029
Shang, Y., Griffith, L. C., and Rosbash, M. (2008). Light-arousal and circadian photoreception circuits intersect at the large PDF cells of the Drosophila brain. Proc. Natl. Acad. Sci. U.S.A. 105, 19587–19594. doi: 10.1073/pnas.0809577105
Shang, Y., Haynes, P., Pírez, N., Harrington, K. I., Guo, F., Pollack, J., et al. (2011). Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat. Neurosci. 14, 889–895. doi: 10.1038/nn.2860
Shaw, P. J., Cirelli, C., Greenspan, R. J., Tononi, G., Campbell, S. S., Tobler, I., et al. (2000). Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837. doi: 10.1126/science.287.5459.1834
Shaw, P. J., Tortoni, G., Greenspan, R. J., and Robinson, D. F. (2002). Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287–291. doi: 10.1038/417287a
Sheeba, V., Fogle, K. J., Kaneko, M., Rashid, S., Chou, Y. T., Sharma, V. K., et al. (2008a). Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 18, 1537–1545. doi: 10.1016/j.cub.2008.08.033
Sheeba, V., Gu, H., Sharma, V. K., O’Dowd, D. K., and Holmes, T. C. (2008b). Circadian- and light-dependent regulation of resting membrane potential and spontaneous action potential firing of Drosophila circadian pacemaker neurons. J. Neurophysiol. 99, 976–988. doi: 10.1152/jn.00930.2007
Shimada, N., Inami, S., Sato, S., Kitamoto, T., and Sakai, T. (2016). Modulation of light-driven arousal by LIM-homeodomain transcription factor Apterous in large PDF-positive lateral neurons of the Drosophila brain. Sci. Rep. 6:37255. doi: 10.1038/srep37255
Shinomiya, K., Huang, G., Lu, Z., Parag, T., Xu, C. S., Aniceto, R., et al. (2019). Comparisons between the ON- and OFF-edge motion pathways in the Drosophila brain. eLife 8:e40025. doi: 10.7554/eLife.40025
Shinomiya, K., Karuppudurai, T., Lin, T. Y., Lu, Z., Lee, C. H., and Meinertzhagen, I. A. (2014). Candidate neural substrates for off-edge motion detection in drosophila. Curr. Biol. 24, 1062–1070. doi: 10.1016/j.cub.2014.03.051
Sinakevitch-Pean, I., Geffard, M., and Plotnikova, S. I. (2001). Localization of glutamate in the nervous system of the fly Drosophila melanogaster: an immunocytochemical study. J. Evol. Biochem. Physiol. 37, 83–88. doi: 10.1023/A:1017574120553
Sitaraman, D., Aso, Y., Jin, X., Chen, N., Felix, M., Rubin, G. M., et al. (2015a). Propagation of homeostatic sleep signals by segregated synaptic microcircuits of the Drosophila mushroom body. Curr. Biol. 25, 2915–2927. doi: 10.1016/j.cub.2015.09.017
Sitaraman, D., Aso, Y., Rubin, G. M., and Nitabach, M. N. (2015b). Control of sleep by dopaminergic inputs to the drosophila mushroom body. Front. Neural Circuits 9:73. doi: 10.3389/fncir.2015.00073
Sprecher, S. G., and Desplan, C. (2008). Switch of rhodopsin expression in terminally differentiated Drosophila sensory neurons. Nature 454, 533–537. doi: 10.1038/nature07062
Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S. A., et al. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. doi: 10.1016/s0092-8674(00)81638-4
Stark, W. S., and Carlson, S. D. (1986). Ultrastructure of capitate projections in the optic neuropil of Diptera. Cell Tissue Res. 246, 481–486. doi: 10.1007/BF00215187
Stenesen, D., Moehlman, A. T., and Krämer, H. (2015). The carcinine transporter CarT is required in Drosophila photoreceptor neurons to sustain histamine recycling. eLife 4:e10972. doi: 10.7554/eLife.10972
Stoleru, D., Peng, Y., Agosto, J., and Rosbash, M. (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. doi: 10.1038/nature02926
Stratoulias, V., and Heino, T. I. (2015). Analysis of the conserved neurotrophic factor MANF in the Drosophila adult brain. Gene Exp. Patt. 18, 8–15. doi: 10.1016/j.gep.2015.04.002
Strausfeld, N. J. (1971). The organization of the insect visual system (Light microscopy) - I. Projections and arrangements of neurons in the lamina ganglionaris of Diptera. Z. Zellforsch. Mikrosk. Anat. 121, 377–441. doi: 10.1007/BF00337640
Stuart, A. E., Borycz, J., and Meinertzhagen, I. A. (2007). The dynamics of signalling at the histaminergic photoreceptor synapse of arthropods. Prog. Neurobiol. 82, 202–207. doi: 10.1016/j.pneurobio.2007.03.006
Suver, M. P., Huda, A., Iwasaki, N., Safarik, S., and Dickinson, M. H. (2016). An array of descending visual interneurons encoding self-motion in Drosophila. J. Neurosci. 36, 11768–11780. doi: 10.1523/JNEUROSCI.2277-16.2016
Takemura, S. Y., Karuppudurai, T., Ting, C. Y., Lu, Z., Lee, C. H., and Meinertzhagen, I. A. (2011). Cholinergic circuits integrate neighboring visual signals in a drosophila motion detection pathway. Curr. Biol. 21, 2077–2084. doi: 10.1016/j.cub.2011.10.053
Tan, L., Zhang, K. X., Pecot, M. Y., Nagarkar-Jaiswal, S., Lee, P. T., Takemura, S. Y., et al. (2015). Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163, 1756–1769. doi: 10.1016/j.cell.2015.11.021
Technau, G. M. (2007). Fiber number in the mushroom bodies of adult Drosophila melanogaster depends on age, sex and experience. J. Neurogenet. 21, 183–196. doi: 10.1080/01677060701695359
Thakkar, M. M. (2011). Histamine in the regulation of wakefulness. Sleep Med. Rev. 15, 65–74. doi: 10.1016/j.smrv.2010.06.004
Tomita, J., Ueno, T., Mitsuyoshi, M., Kume, S., and Kume, K. (2015). The NMDA Receptor promotes sleep in the fruit fly, Drosophila melanogaster. PLoS One 10:e0128101. doi: 10.1371/journal.pone.0128101
Tracy, T. E., and Gan, L. (2018). Tau-mediated synaptic and neuronal dysfunction in neurodegenerative disease. Curr. Opin. Neurobiol. 51, 134–138. doi: 10.1016/j.conb.2018.04.027
Triphan, T., Poeck, B., Neuser, K., and Strauss, R. (2010). Visual targeting of motor actions in climbing Drosophila. Curr. Biol. 20, 663–668. doi: 10.1016/j.cub.2010.02.055
Ueno, T., Tomita, J., Tanimoto, H., Endo, K., Ito, K., Kume, S., et al. (2012). Identification of a dopamine pathway that regulates sleep and arousal in Drosophila. Nat. Neurosci. 15, 1516–1523. doi: 10.1038/nn.3238
van Alphen, B., Yap, M. H. W., Kirszenblat, L., Kottler, B., and van Swinderen, B. (2013). A dynamic deep sleep stage in Drosophila. J. Neurosci. 33, 6917–6927. doi: 10.1523/JNEUROSCI.0061-13.2013
Van Meyel, D. J., O’Keefe, D. D., Jurata, L. W., Thor, S., Gill, G. N., and Thomas, J. B. (1999). Chip and Apterous physically interact to form a functional complex during Drosophila development. Mol. Cell 4, 259–265. doi: 10.1016/s1097-2765(00)80373-1
Vanin, S., Bhutani, S., Montelli, S., Menegazzi, P., Green, E. W., Pegoraro, M., et al. (2012). Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 474, 371–375. doi: 10.1038/nature10991
VanVickle-Chavez, S. J., and Van Gelder, R. N. (2007). Action spectrum of Drosophila cryptochrome. J. Biol. Chem. 282, 10561–10566. doi: 10.1074/jbc.M609314200
Veleri, S., Rieger, D., Helfrich-Förster, C., and Stanewsky, R. (2007). Hofbauer-Buchner eyelet affects circadian photosensitivity and coordinates TIM and PER expression in Drosophila clock neurons. J. Biol. Rhythms 22, 29–42. doi: 10.1177/0748730406295754
Vinayak, P., Coupar, J., Hughes, S. E., Fozdar, P., Kilby, J., Garren, E., et al. (2013). Exquisite Light Sensitivity of Drosophila melanogaster Cryptochrome. PLoS Genet. 9:e1003615. doi: 10.1371/journal.pgen.1003615
Wagner, S., Heseding, C., Szlachta, K., True, J. R., Prinz, H., and Hovemann, B. T. (2007). Drosophila photoreceptors express cysteine peptidase Tan. J. Comp. Neurol. 500, 601–611. doi: 10.1002/cne.21138
Walkowicz, L., Kijak, E., Krzeptowski, W., Górska-Andrzejak, J., Stratoulias, V., Woznicka, O., et al. (2017). Downregulation of DmMANF in glial cells results in neurodegeneration and affects sleep and lifespan in Drosophila melanogaster. Front. Neurosci. 11:610. doi: 10.3389/fnins.2017.00610
Wang, T., and Montell, C. (2007). Phototransduction and retinal degeneration in Drosophila. Pflugers Arch. 454, 821–847. doi: 10.1007/s00424-007-0251-1
Wang, T., Xu, H., Oberwinkler, J., Gu, Y., Hardie, R. C., and Montell, C. (2005). Light activation, adaptation, and cell survival functions of the Na +/Ca2+ exchanger CalX. Neuron 45, 367–378. doi: 10.1016/j.neuron.2004.12.046
Wegener, C., Hamasaka, Y., and Nässel, D. R. (2004). Acetylcholine Increases Intracellular Ca2+ Via Nicotinic Receptors in Cultured PDF-Containing Clock Neurons of Drosophila. J. Neurophysiol. 91, 912–923. doi: 10.1152/jn.00678.2003
Weigelt, C. M., Hahn, O., Arlt, K., Gruhn, M., Jahn, A. J., Eßer, J., et al. (2019). Loss of miR-210 leads to progressive retinal degeneration in Drosophila melanogaster. Sci. Alliance 2:e20180049. doi: 10.26508/lsa.201800149
Witte, I., Kreienkamp, H. J., Gewecke, M., and Roeder, T. (2002). Putative histamine-gated chloride channel subunits of the insect visual system and thoracic ganglion. J. Neurochem. 84, 504–514. doi: 10.1046/j.1471-4159.2002.01076.x
Xu, Y., An, F., Borycz, J. A., Borycz, J., Meinertzhagen, I. A., and Wang, T. (2015). Histamine recycling is mediated by CarT, a Carcinine transporter in drosophila photoreceptors. PLoS Genet. 11:1005764. doi: 10.1371/journal.pgen.1005764
Yamaguchi, S., Wolf, R., Desplan, C., and Heisenberg, M. (2008). Motion vision is independent of color in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 105, 4910–4915. doi: 10.1073/pnas.0711484105
Yao, Z., and Shafer, O. T. (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. doi: 10.1126/science.1251285
Yasuyama, K., and Meinertzhagen, I. A. (1999). Extraretinal photoreceptors at the compound eye’s posterior margin in Drosophila melanogaster. J. Comp. Neurol. 412, 193–202. doi: 10.1002/(sici)1096-9861(19990920)412:2<193::aid-cne1>3.0.co;2-0
Yasuyama, K., and Salvaterra, P. M. (1999). Localization of choline acetyltransferase-expressing neurons in Drosophila nervous system. Microsc. Res. Tech. 45, 65–79. doi: 10.1002/(sici)1097-0029(19990415)45:2<65::aid-jemt2>3.0.co;2-0
Yi, W., Zhang, Y., Tian, Y., Guo, J., Li, Y., and Guo, A. (2013). A subset of cholinergic mushroom body neurons requires go signalling to regulate sleep in Drosophila. Sleep 36, 1809–1821. doi: 10.5665/sleep.3206
Yoshii, T., Todo, T., Wülbeck, C., Stanewsky, R., and Helfrich-Förster, C. (2008). Cryptochrome is present in the compound eyes and a subset of Drosophila’s clock neurons. J. Comp. Neurol. 508, 952–966. doi: 10.1002/cne.21702
Yuan, Q., Joiner, W. J., and Sehgal, A. (2006). a sleep-promoting role for the drosophila serotonin receptor 1A. Curr. Biol. 16, 1051–1062. doi: 10.1016/j.cub.2006.04.032
Yuan, Q., Lin, F., Zheng, X., and Sehgal, A. (2005). Serotonin modulates circadian entrainment in Drosophila. Neuron 47, 115–127. doi: 10.1016/j.neuron.2005.05.027
Zerr, D. M., Hall, J. C., Rosbash, M., and Siwicki, K. K. (1990). Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J. Neurosci. 10, 2749–2762. doi: 10.1523/jneurosci.10-08-02749.1990
Zhang, L., Lear, B. C., Seluzicki, A., and Allada, R. (2009). The CRYPTOCHROME photoreceptor gates PDF neuropeptide signalling to set circadian network hierarchy in Drosophila. Curr. Biol. 19, 2050–2055. doi: 10.1016/j.cub.2009.10.058
Zhang, Y., Liu, Y., Bilodeau-Wentworth, D., Hardin, P. E., and Emery, P. (2010). Light and temperature control the contribution of specific DN1 neurons to Drosophila circadian behaviour. Curr. Biol. 20, 600–605. doi: 10.1016/j.cub.2010.02.044
Zheng, L., de Polavieja, G. G., Wolfram, V., Asyali, M. H., Hardie, R. C., and Juusola, M. (2006). Feedback network controls photoreceptor output at the layer of first visual synapses in Drosophila. J. Gen. Physiol. 127, 495–510. doi: 10.1085/jgp.200509470
Zheng, Y., Hirschberg, B., Yuan, J., Wang, A. P., Hunt, D. C., Ludmerer, S. W., et al. (2002). Identification of two novel Drosophila melanogaster histamine-gated chloride channel subunits expressed in the eye. J. Biol. Chem. 277, 2000–2005. doi: 10.1074/jbc.M107635200
Zimmerman, J. E., Chan, M. T., Lenz, O. T., Keenan, B. T., Maislin, G., and Pack, A. I. (2017). Glutamate Is a Wake-Active Neurotransmitter in Drosophila melanogaster. Sleep 40:zsw046. doi: 10.1093/sleep/zsw046
Zuker, C. S., Cowman, A. F., and Rubin, G. M. (1985). Isolation and structure of a rhodopsin gene from D. melanogaster. Cell 40, 851–858. doi: 10.1016/0092-8674(85)90344-7
Keywords: Drosophila, wake-sleep pattern, light, photoreception, neurotransmitters
Citation: Mazzotta GM, Damulewicz M and Cusumano P (2020) Better Sleep at Night: How Light Influences Sleep in Drosophila. Front. Physiol. 11:997. doi: 10.3389/fphys.2020.00997
Received: 28 May 2020; Accepted: 22 July 2020;
Published: 04 September 2020.
Edited by:
Charalambos P. Kyriacou, University of Leicester, United KingdomReviewed by:
Ko-Fan Chen, University of Leicester, United KingdomStephane Dissel, University of Missouri–Kansas City, United States
Copyright © 2020 Mazzotta, Damulewicz and Cusumano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella M. Mazzotta, Z2FicmllbGxhLm1henpvdHRhQHVuaXBkLml0; Paola Cusumano, cGFvbGEuY3VzdW1hbm9AdW5pcGQuaXQ=
 Gabriella M. Mazzotta
Gabriella M. Mazzotta Milena Damulewicz
Milena Damulewicz Paola Cusumano
Paola Cusumano