- State Key Laboratory of Wheat and Maize Crop Science, College of Plant Protection, Henan Agricultural University, Zhengzhou, China
Helicoverpa armigera is a universal pest around the world that has been extensively used as a model organism for agricultural pests. Calcineurin (CAN) is an important Ca2+-dependent phosphatase that is participated in various biological pathways. Here, we revealed that CAN inhibition significantly arrested H. armigera larval development by reducing larvae weight, prolonging development time and reducing pupate rates. Furthermore, CAN serves as an immune activator and regulates antimicrobial peptide (AMP; cecropin D, attacin, and gloverin) expression by binding with relish transcript factor in H. armigera. This study provides a potential target to control H. armigera by using synergistic agents for pesticides or plant-mediated RNA interference technology.
Introduction
Calcineurin (CAN) belongs to the protein phosphatase 2B family and acts as a Ca2+-dependent phosphatase. CAN is ubiquitously expressed in most mammalian tissues, and exerts different functions in different tissues (Furman and Norris, 2014). Studies have reported that CAN is predominantly expressed in the brain and plays important roles in various neuronal functions and disorders. This is due to CAN’s vital functions in Ca2+ homeostasis, synaptic plasticity, receptor signaling, and transcription regulation (Jackson et al., 2018). Furthermore, CAN is also found to be expressed in lymphocytes and macrophages, in which CAN has been confirmed to be took part in innate immunity (Bueno et al., 2002; Kang et al., 2007). In addition, In human endothelial cells, CAN functions as a mediator of anaphylaxis (Ballesteros-Martinez et al., 2017). These studies provide key evidence that CAN is a vital signaling molecule that performs a variety of functions in different biological pathways (Liu et al., 1991; Jackson et al., 2018).
In insects, studies on the functions of CAN have mostly been performed in the model organism Drosophila melanogaster. In Drosophila, CAN plays a crucial role in myofilament formation and troponin I isoform transition which affects the development of flight muscles (Gajewski et al., 2006). Interestingly, CAN in Drosophila has also been demonstrated to participate in circadian rhythms regulation (Kweon et al., 2018), axonal transport (Shaw et al., 2013), mitochondrial function (Chang and Min, 2005), olfactory associative learning and long-term memory (Chang et al., 2003), courtship behaviors (Ejima et al., 2004; Sakai and Aigaki, 2010), and female reproductive activities (Ejima et al., 2004; Horner et al., 2006; Takeo et al., 2010). Besides, Drosophila CAN has also been identified to play a role in immune response. When Drosophila were infected with gram-negative bacteria, CAN activity increased (Li and Dijkers, 2015). This promoted the product of relish, a key transcription factor of the immune deficiency (IMD) pathway in the innate immunity, which finally induces antimicrobial gene expression (Dijkers and O’Farrell, 2007; Li and Dijkers, 2015).
Besides Drosophila, relatively few studies have focused on CAN role in other insect species. In the Lepidoptera adult, Bombyx mori, CAN is localized in the cytoplasm of pheromone-producing cells and regulates the production of sex pheromone (Yoshiga et al., 2002). In Helicoverpa armigera, CAN in adults mediates female attraction to males (Zhao et al., 2018). Furthermore, CAN has been confirmed to activate acetyl CoA carboxylase through dephosphorylation, and thus regulate sex pheromone biosynthesis and female mating acceptance in H. armigera (Du et al., 2017; Zhao et al., 2018). While these studies deepen our understanding of CAN function in adults, CAN function in larvae remains elusive and lacking.
Helicoverpa armigera is a universal pest around the world, and many measures have been taken to control this pest, including planting transgenic crops expressing a chemical pesticide, Bacillus thuringiensis (BT) (Downes et al., 2017; Wei et al., 2018). Although these measures work to some extent, they also present many problems, such as insect resistance. In order to find better methods to control H. armigera, it is critical to study the associated genes that involved in certain physiological process, such as genes involved in the immune process. Insect and mammalian immune systems share many similarities (Kaneko et al., 2004; Kaneko and Silverman, 2005). Insects depend solely upon the innate immune responses to survive. In the IMD pathway, the expression of antimicrobial genes is required to respond to gram-negative bacteria, thus making CAN an attractive target to control H. armigera CANs from different species share high amino acid identity, indicating they may share similar functions (Chen and Zhang, 2013; Zhao et al., 2018). Whether the IMD pathway exists in H. armigera larvae has never been addressed.
In this study, CAN from H. armigera (HaCAN) was found to be widely distributed in different developmental stages and different tissues, especially in fourth and fifth instars. Feeding of H. armigera larvae with FK506 significantly affected their development. FK506 treatments also significantly attenuated the expression of antimicrobial peptide (AMP) transcripts (cecropin D, attacin, and gloverin) caused by infection with gram-negative bacteria. For the first time, HaCAN was confirmed to be involved in larval development. HaCAN was also found to interact with relish. The present study reveals a potential target for synergistic agents such as pesticides or plant-mediated RNA interference (RNAi) technology, to control H. armigera.
Materials and Methods
Insect Rearing
Larvae of H. armigera were purchased from the Henan Jiyuan Baiyun Industry Co., Ltd. Larvae were reared on artificial diet at 26 ± 1°C, 60 ± 10% relative humidity (RH), and a photoperiod of 16L:8D h (Du et al., 2017).
Tissue Sampling
Larvae on the second day of every instar (from first to fifth) were collected with three replicates. The collected samples were used for subsequent RNA extraction to analyze the developmental expression profile of HaCAN (GenBank: KR185962.1).
In order to investigate distributions of HaCAN in the different tissues (including midgut, malpighian tubes, salivary glands, fat body, hemocyte, central nervous, and epidermis), the tissues were harvested from more than five larvae on the ice. This served as one biological replicate.
Expression patterns of the endogenous AMPs [cecropin D (GenBank: EU041763.1), attacin (GenBank: AY948540.1), and gloverin (GenBank: KT346373.1)] in the midgut, fat body and epidermis were analyzed by quantitative real-time PCR after tacrolimus (FK506) treatment. Third instar larvae were fed with different doses of FK506 (50, 100, and 200 μM). DMSO (FK506 solvent) treatment was used as control. Midgut, fat body, and epidermis were dissected from 10 larvae and served as one biological replicate. The expression levels of AMPs in larvae were tested 72 h after FK506 (or DMSO) treatments.
Expressions of endogenous AMPs (cecropin D, attacin, and gloverin) in midgut, fat body, and epidermis were analyzed by quantitative real-time PCR after FK506 treatment or Escherichia coli injection. The FK506 inhibitor was dissolved in DMSO. Either 50 μM FK506 or DMSO was fed to third instar larvae. 72 h after FK506 (or DMSO) treatments larvae were then injected with either 3 μL E. coli cells suspended in PBS (4 × 105 cell/μL) or 3 μL PBS. The larvae were incubated for either 1 h or 3 h, after which, the midgut, fat body and epidermis was harvested. All the collected samples were quickly frozen in liquid nitrogen and stored at −80°C for subsequent RNA extraction. Ten larvae were used for one biological replicate.
All the above treatments were carried out with three biological replicates.
Quantitative Real-Time PCR Analysis of HaCAN Expression
Total RNA was extracted from each sample prepared above with TRIzol reagent (Invitrogen, Carlsbad, CA, United States) according to the manufacturer’s instructions. All RNA samples were analyzed by UV spectrophotometry (NanoDrop; Thermo Scientific). A260/A280 ratio of all RNA samples were >1.8. All RNA samples were run on 1% agarose gel to ensure the quality of RNA (data not shown). After that, 1 μg of each DNase I-treated RNA sample was reversely transcribed into first strand cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Daliang, China). The above first-strand cDNA was then used as the template for qRT-PCR. The primers used for qRT-PCR analysis are listed in Table 1. Amplification efficiency of each primer pair was examined on an Applied Biosystems 7500 Fast Real-Time PCR system and calculated using the formula, PCR efficiency = 10–1/slope − 1 (Bustin et al., 2009). All the primer pairs had an amplification efficiency between 97 and 98% (Table 1). QRT-PCR analysis of HaCAN and 18S (internal reference gene; Zhang S. et al., 2015) were performed individually in a 12 μL reaction system containing 6 μL 2 × SYBR® Premix Ex TaqTM II (Eurogentec, Fremont, CA, United States), 5 μM gene-specific forward primer and reverse primer (0.5 μL each), 1 μL template cDNA, and 4 μL nuclease-free water using an Applied Biosystems 7500 Fast Real-Time PCR system. Thermocycler conditions consisted of an initial denaturation for 3 min at 95°C, followed by 35 cycles of denaturation at 95°C for 15 s and annealing/extension at 58°C for 30 s. A melting curve was generated after the termination of PCR cycles to ensure free of junk products. For each gene in each treatment, at least three biological replicates were tested with three technical replicates. The mRNA expression was quantified using the comparative cross threshold method (CT, the PCR cycle number that crosses the signal threshold) (Livak and Schmittgen, 2001). The CT result of the 18S gene was subtracted from the CT result of the target gene to obtain a ΔCT. Normalized fold changes in target gene mRNA expression were expressed as 2–ΔΔCT, where ΔΔCT is equal to ΔCT treated sample – substracted from the ΔCT control (Schmittgen and Livak, 2008).
The expression levels of cecropin D, attacin, and gloverin in H. armigera larvae were analyzed using qRT-PCR method as the above mentioned after treatments with FK506, E. coli, FK506 + E. coli. The primers of cecropin D, attacin, and gloverin are listed in the Table 1.
Effects on H. armigera Development After Feeding With FK506
Tacrolimus (FK506) is a special CAN inhibitor, which inhibits CAN phosphatase activity and affects the insect immunity (Liu et al., 1991, 1992; Dijkers and O’Farrell, 2007). Three concentrations of FK506 (50, 100, and 200 μM) were used to evaluate its effect on the development of H. armigera larvae (DMSO, FK506 solvent, was set as a control). Third instar larvae, 12 h before molting, were transferred onto the artificial diet that was mixed with either FK506 or DMSO. For each treatment, five replicates (30 third instar larvae per replicates × 5 replicates = 150 larvae total per concentration for each treatment) were employed.
In these experiments, the weights of the treated insects were recorded every other day. After 7 days, dead insects were counted to calculate the mortalities. The days, when the treated larvae began to pupate, were record as the larvae’s development time. Successfully pupated insects were counted to calculate pupation rates. For the healthy pupae, the numbers of successfully emerged adults were counted to calculate the adult emergence rate.
Protein–Protein Interactions Between HaCAN and Harelish
A full open reading frame of Harelish (GenBank: KR185962.1) and HaCAN were PCR-amplified using the gene-specific primers pDHB1-N-Harelish (F and R) and pPR3-N-CAN (F and R) from relish-pGEMT and CAN-pGEMT (Table 1) (Du et al., 2017), respectively. The PCR product of Harelish was cloned into bait vector pDHB1-N (Dualsystems Biotech), and HaCAN was cloned into pPR3-N vector (Dualsystems Biotech).
Several colonies of NMY51 (Dualsystems Biotech) were picked from YPAD plates and incubated in 3 mL YPAD media at 30°C, while shaking at 250 rpm for 8–12 h. Upon the culture reaching an OD600 of 0.15–0.3 (about 16–20 h), the culture was centrifuged at 700 × g for 5 min, the supernatant was discarded and pellet was resuspended in 100 mL YPAD. The bacterium solution was centrifuged at 700 × g for 5 min and washed again using 30 mL ddH2O. The collected bacteria were then resuspended in 600 μL 1.1 × TE/LiAc solution [1.1 × TE/LiAc (10 mL): 1.1 mL 1 M LiAc, 1.1 mL 10 × TE Buffer, 8.8 mL ddH2O].
A 5 μL carrier DNA (10 μg/mL) were boiled for 5 min and added into a 1.5 mL eppendorf tube. 500 μL PEG/LiAc solution [1 × PEG/LiAc (10 mL): 8 mL 50% PEG3350, 1 mL 10 × TE Buffer, 1 mL 1 M LiAc] was added with 50 μL NMY51 cells to the carrier DNA tube then mixed with the corresponding plasmids. Four treatments were prepared including 100 ng pDHB1-largeT and 100 ng pDSL-p53, 100 ng pDHB1-relish and 100 ng pOst1-NubI, 100 ng pDHB1-relish and 100 ng pPR3-N, and 100 ng pDHB1-relish and 100 ng pPR3-N-CAN. Each treatment was incubated at 30°C with shaking every 10 min. 20 μL DMSO was added to each tube and the tubes were transferred to a 42°C water bath and incubated for 15 min while, vortexing every 5 min. The solutions were then centrifuged at 700 × g for 5 min and the cell pellets were dissolved in 1 mL 0.9% NaCl. 100 μL suspension was taken from each transformation and plated onto SD-Leucine-Tryptophan plate (DDO; Clontech), and SD-Leucine-Tryptophan-Histidine-Adenine plates (QDO; Clontech). All plates were incubated for 5 days at 30°C (Yu et al., 2015).
Five days after the transformation of cDNA library, nine white colonies from the QDO plates were transferred into 1.5 mL eppendorf tubes containing 500 μL 0.9% NaCl. 0.9% NaCl was used to normalize the OD600 of each treatment to the same value. 1 μL of transformed solution was added onto SD/-LT/X (SD-Leucine-Tryptophan + 40 mg/L X-α-Gal media) (X-α-Gal soluted in N, N-dimethylformamide), SD/-LTH/X (SD-Leucine-Tryptophan-Histidine + 40 mg/L X-α-Gal media), SD/-LTHA/X (SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media), SD/-LTHA/X/10 mM3-AT (SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media + 3 mM 3-AT) and SD/-LTHA/X/10 mM 3-AT (SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media + 10 mM 3-AT) plates. These plates were used to determine the threshold of selection which was increased by either removal of essential amino acids or by using 3-AT, a competitive inhibitor of the his gene product and the highest selection stress used in these four plates (Fetchko and Stagljar, 2004).
Statistical Analysis
Significant differences in the relative expression levels of HaCAN in different instars and different tissues were compared using Tukey’s test with P < 0.05 [analysis of variance (ANOVA) and DPS7.05]. Significant differences in the relative expression levels of cecropin D, attacin, and gloverin in different treatments were compared using Tukey’s test with P < 0.05 (ANOVA and DPS7.05).
Results
Developmental Expression Pattern of HaCAN in Larvae
To investigate the developmental expression pattern of HaCAN in different instars (first-, second-, third-, fourth-, and fifth-instar larvae), qRT-PCR was employed and results revealed that HaCAN was ubiquitously expressed in all of the examined stages. The expression level of HaCAN reached its peak at the first-instar larvae, followed by fifth-instar larvae (Figure 1A).
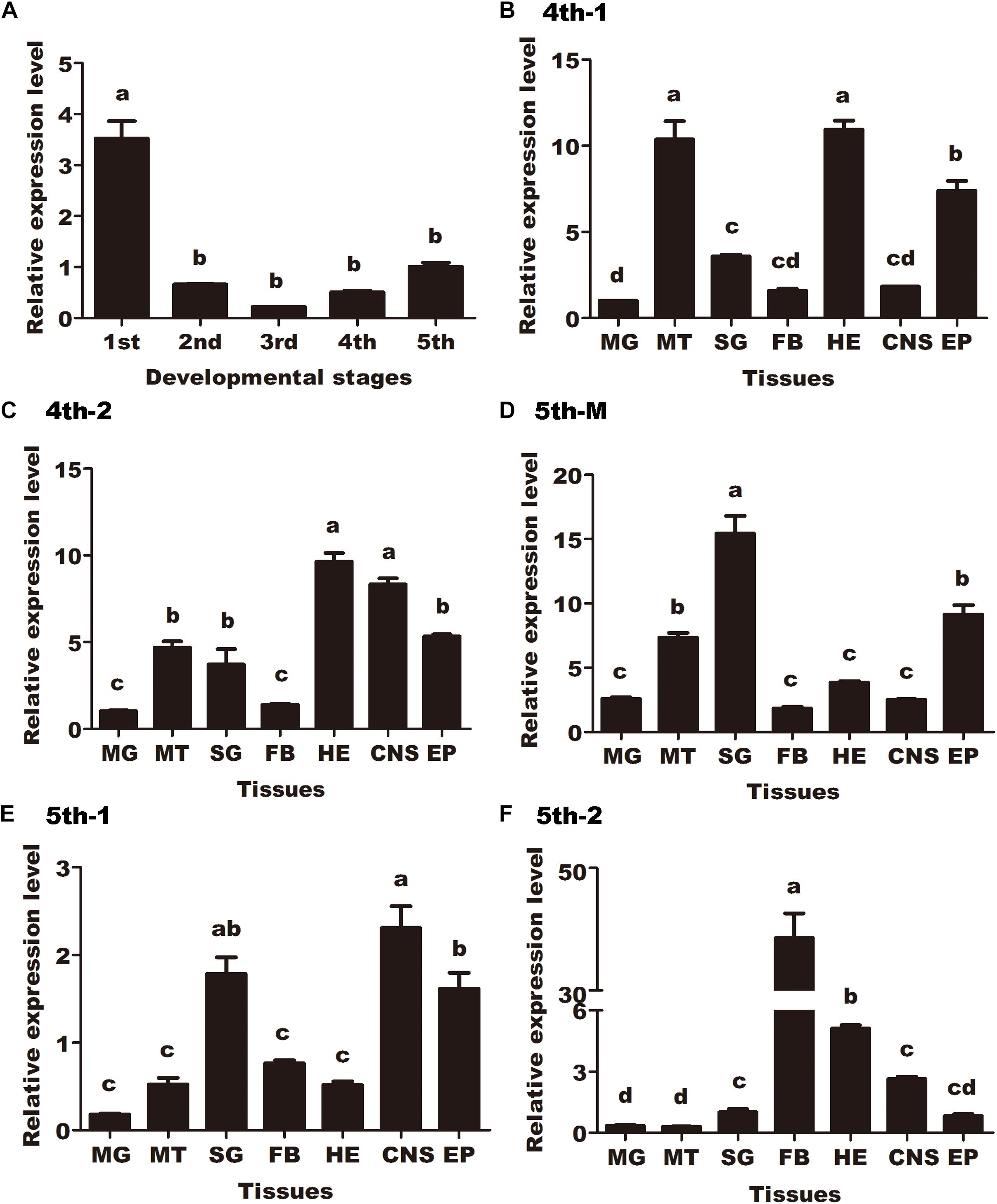
Figure 1. Developmental expression pattern and spatial distributions of Helicoverpa armigera calcineurin A (HaCNA) in larvae as determined by quantitative real-time PCR. (A) Relative expression levels of HaCNA in first-instar larvae (1st), second-instar larvae (2nd), third-instar larvae (3rd), fourth-instar larvae (4th), and fifth-instar larvae (5th). (B) Relative expression levels of HaCNA in different tissues (MG, midgut; MT, malpighian tubes; SG, salivary glands; FB, fat body; HE, hemocyte; CNS, central nervous; EP, epidermis) in the first day of 4th larvae. (C) Relative expression levels of HaCNA in different tissues in the second day of 4th larvae. (D) Relative expression levels of HaCNA in different tissues in fifth instar molting stage. (E) Relative expression levels of HaCNA in different tissues in the first day of 5th larvae. (F) Relative expression levels of HaCNA in different tissues in the second day of 5th larvae. Statistically significant differences for experimental comparisons are indicated by different small letters [analysis of variance (ANOVA) and Tukey’s test, P < 0.05; DPS7.05]. Values shown are means and standard errors.
The spatial distributions of HaCAN were investigated in different tissues (midgut, malpighian tubes, salivary glands, fat body, hemocytes, central nervous system, and epidermis) during different developmental stages (the first and second day of fourth instar larvae, molting day, and the first and second day of fifth-instar larvae). The distribution patterns of HaCAN in different tissues varied (Figures 1B–F), which generally exhibited higher expression levels in the epidermis during the first and second day in fourth instar larvae, the molting day and the first day in fifth instar larvae. Interestingly, HaCAN transcriptional level showed a sudden increase on the second day in fifth instar larvae (Figures 1B–F).
Effect of CAN Inhibitor on H. armigera Development
Four days of FK506 treatment caused significant decreases in body weight compared with controls (DMSO treatment). However, no significant differences were observed between the different FK506 concentrations used (Figure 2A).
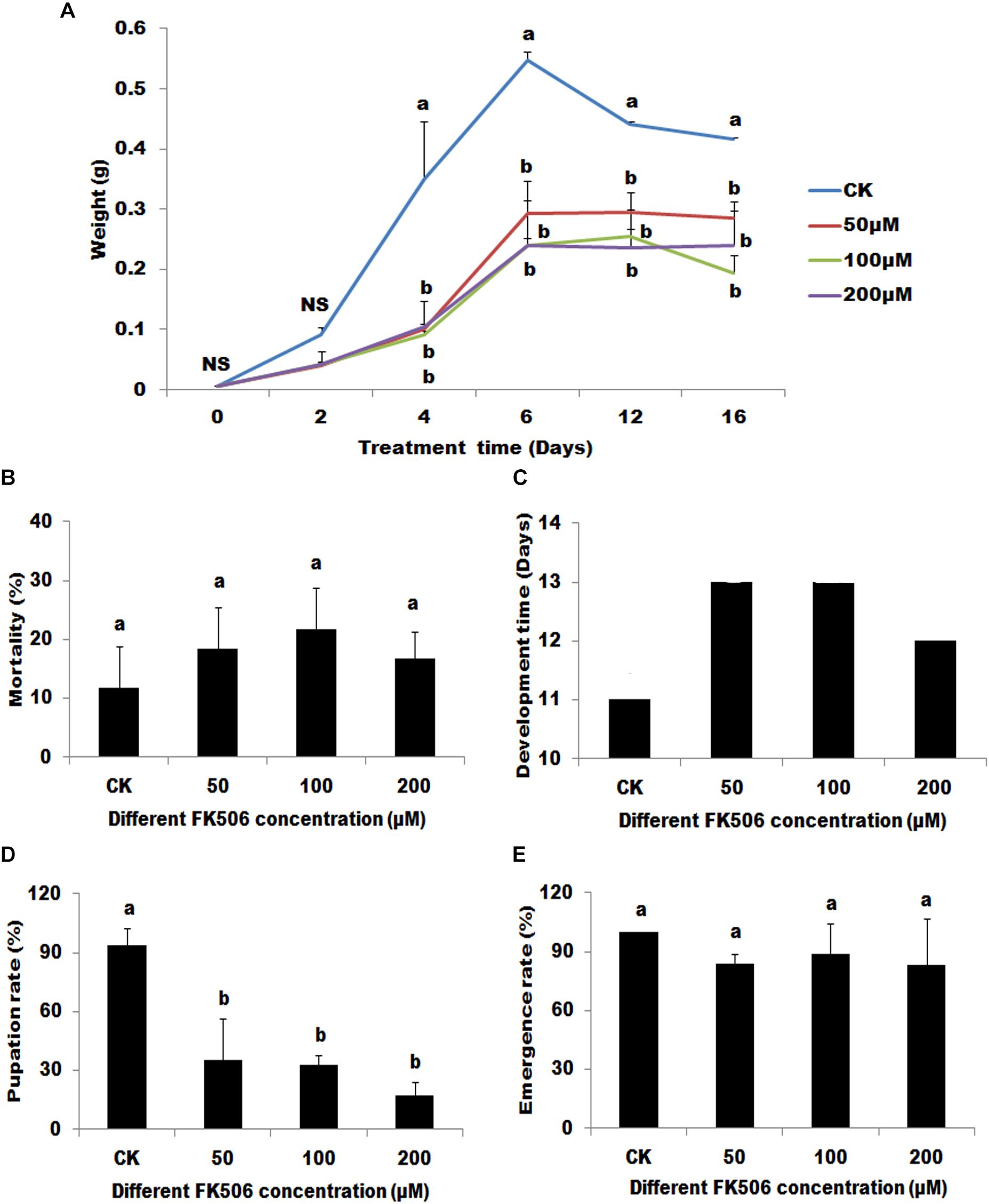
Figure 2. Effects of the calcineurin (CN) inhibitor (FK506) on the Helicoverpa armigera. The larvae were treated begin at the first day of 3rd. (A) The effect of FK506 on the larvae weight. Larvae were fed with different FK506 concentrations (50, 100, and 200 μM) (DMSO as buffer control). (B) The mortality of larvae 7 days after different treatments. (C) The developmental time of the different treatments from the first day of 3rd instar larvae to began to pupate. (D) The pupation rate of the different treatments. (E) The emergence rate of the successfully pupate insects under the different treatments. Statistically significant differences for experimental comparisons are indicated by different small letters [analysis of variance (ANOVA) and Tukey’s test, P < 0.05; DPS7.05]. Values shown are means and standard errors.
Although seven days of FK506 treatments did not lead to significant changes in larvae mortalities, FK506 treatments significantly arrested the development of the larvae (Figures 2B,C). Pupation was delayed by 1-2 days and the pupation rates were significantly decreased (35.00, 32.47, and 17.36% after treatments with 50, 100, and 200 μM FK506, respectively, versus 93.75% after DMSO treatment) (Figures 2C,D). However, FK506 treatments exhibited no effect on the emergence rate of pupae (Figure 2E).
Protein–Protein Interactions Between HaCAN and Harelish
To test yeast two-hybrid system, pDHB1-LargeT and pDSL-p53 were transformed as a positive control and were able to see growth on the selection plates, which showed that the yeast two-hybrid system works well (Figure 3). The transformation of pDHB1-N-relish and pOst1-NubI grew well on SD-Leucine-Tryptophan (DDO; Clontech) and SD-Leucine-Tryptophan-Histidine-Adenine (QDO; Clontech) plates, respectively. Moreover, the transformation of pBT3-N-relish & pOst1-NubI turned blue in the β-galactosidase assay on SD/-LT/X, SD/-LTH/X, SD/-LTHA/X, SD/-LTHA/X/3mM 3-AT and SD/-LTHA/X/10 mM 3-AT plates, respectively (Figure 3). These experiments demonstrated that pBT3-N-relish plasmids worked well. The vector pPR3-N expresses the N-terminal half of ubiquitin (NubG) and transformation of pBT3-N-relish and pPR3-N displayed less growth on normal selection plate while there were no clonies on QDO plates. There were also no blue colonies present in the β-galactosidase assay on SD/-LT/X, SD/-LTH/X, SD/-LTHA/X, SD/-LTHA/X/10 mM 3-AT plates (Figures 1, 2). These results showed that the clones of pDHB1-N-relish and pPR3-N on QDO plates were false positives, and that the split-ubiquitin membrane yeast two-hybrid system can be used to identify interactions between HaCAN and Harelish. The transformation of pDHB1-N-relish & pPR3-N-CAN grew well on both DDO plates and QDO plates. Moreover, the colonies resulting from this transformation turned blue in a β-galactosidase assay on SD/-LT/X, SD/-LTH/X, SD/-LTHA/X, SD/-LTHA/X/3 mM 3-AT and SD/-LTHA/X/10 mM 3-AT plates, indicating that HaCAN and Harelish interact directly (Figure 3).
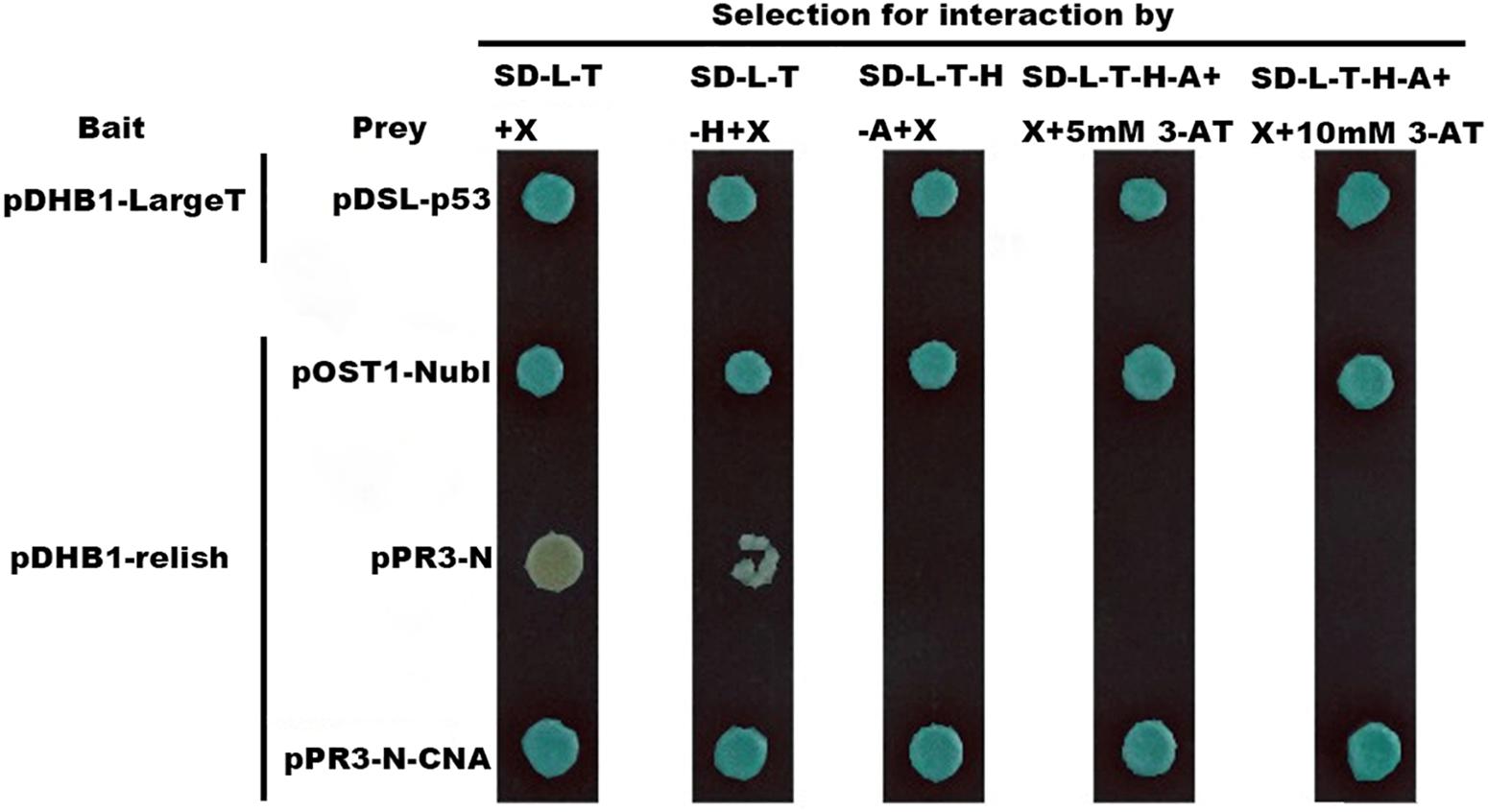
Figure 3. The interactions between HaCNA and Harelish based on yeast two-hybrid system. pDHB1 vector serves as the bait which is fused with the C-terminal half of ubiquitin (Cub) and an artificial transcription factor (LexA-VP6), while in the vector of pPR3-N are fused with the N-terminal half of ubiquitin (NubG, replacement of Ile-13 of wildtype NubI by glycine decreases the affinity between Nub and Cub.). The treatment of pDHB1-LargeT & pDSL-p53 was positive control to show this yeast two-hybrid system works well. SD-L-T + X: SD-Leucine-Tryptophan + 40 mg/L X-α-Gal media (X-α-Gal soluted in N,N-dimethylformamide); SD-L-T-H + X: SD-Leucine-Tryptophan-Histidine + 40 mg/L X-α-Gal media; SD-L-T-H-A + X: SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media; SD-L-T-H-A + X + 5 mM 3-AT: SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media + 5 mM 3-AT; SD-L-T-H-A + X + 10 mM 3-AT: SD-Leucine-Tryptophan-Histidine-Adenine + 40 mg/L X-α-Gal media + 10 mM 3-AT.
HaCAN Regulates AMPs Expression
To investigate the regulation of AMPs expression by HaCAN, the CAN inhibitor FK506 was used. The results demonstrated that HaCAN inhibition with different FK506 resulted in a significant decrease in the expression levels of several AMPs, including cecropin D, attacin, and gloverin in the midgut, fat body and epidermis of larvae (Figure 4).
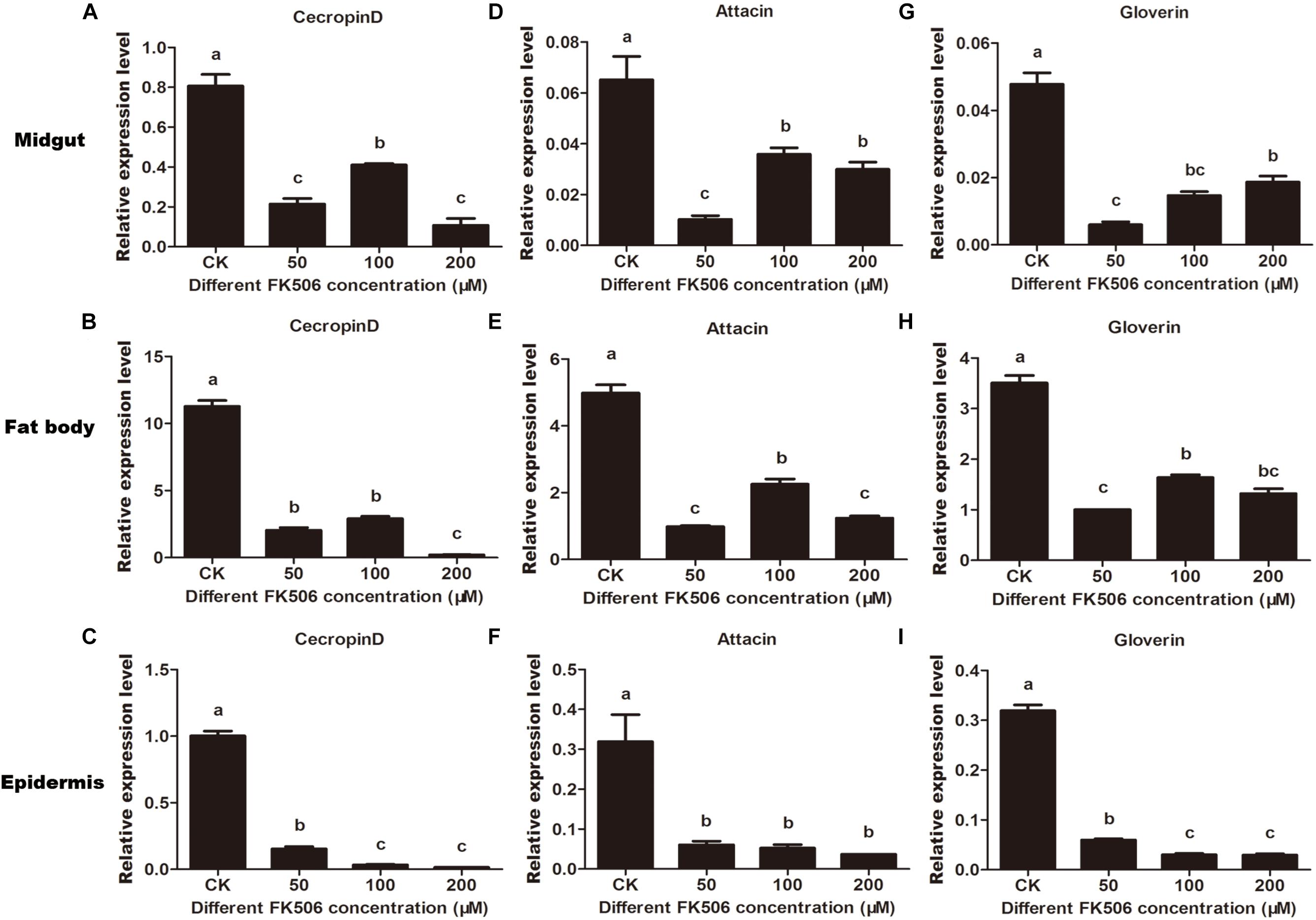
Figure 4. Expression levels of endogenous antimicrobial peptides (cecropin D, attacin, and gloverin) in midgut, fat body and epidermis after feeding FK506 (0, 50, 100, and 200 μM) were analyzed by quantitative real-time PCR. (A–C) Represent the relative expression levels of cecropin D in the migut, fat body, and epidermis, respectively. (D–F) Represent the relative expression levels of attacin in the migut, fat body and epidermis, respectively. (G–I) Represent the relative expression levels of gloverin in the migut, fat body, and epidermis, respectively. Values shown are means and standard errors. Statistically significant differences for experimental comparisons are indicated by different small letters [analysis of variance (ANOVA) and Tukey’s test, P < 0.05; DPS7.05].
In order to further study the HaCAN role on AMP expressions, gram-negative E. coli was injected into larvae fed with 50 μM FK506 for two days prior. A 1 h treatment with E. coli treatment caused significant increases in AMPs’ expressions in the fat body and midgut tissues. Interestingly, the application of FK506 significantly counteracted E. coli–induced AMP expression in the fat body and midgut tissues (Figure 5). While 1 h-E. coli treatment can significantly induced cecropin D expression in the epidermis, FK506 treatment did not significantly hinder this expression (Figure 5). Interestingly, 1 h treatments both E. coli and FK506 had no effect on gloverin expression in the epidermis (Figure 5). Unlike cecropin D and gloverin, the expression pattern of attacin in the epidermis was similar to that the expression pattern seen in the midgut and fat body (Figure 5).
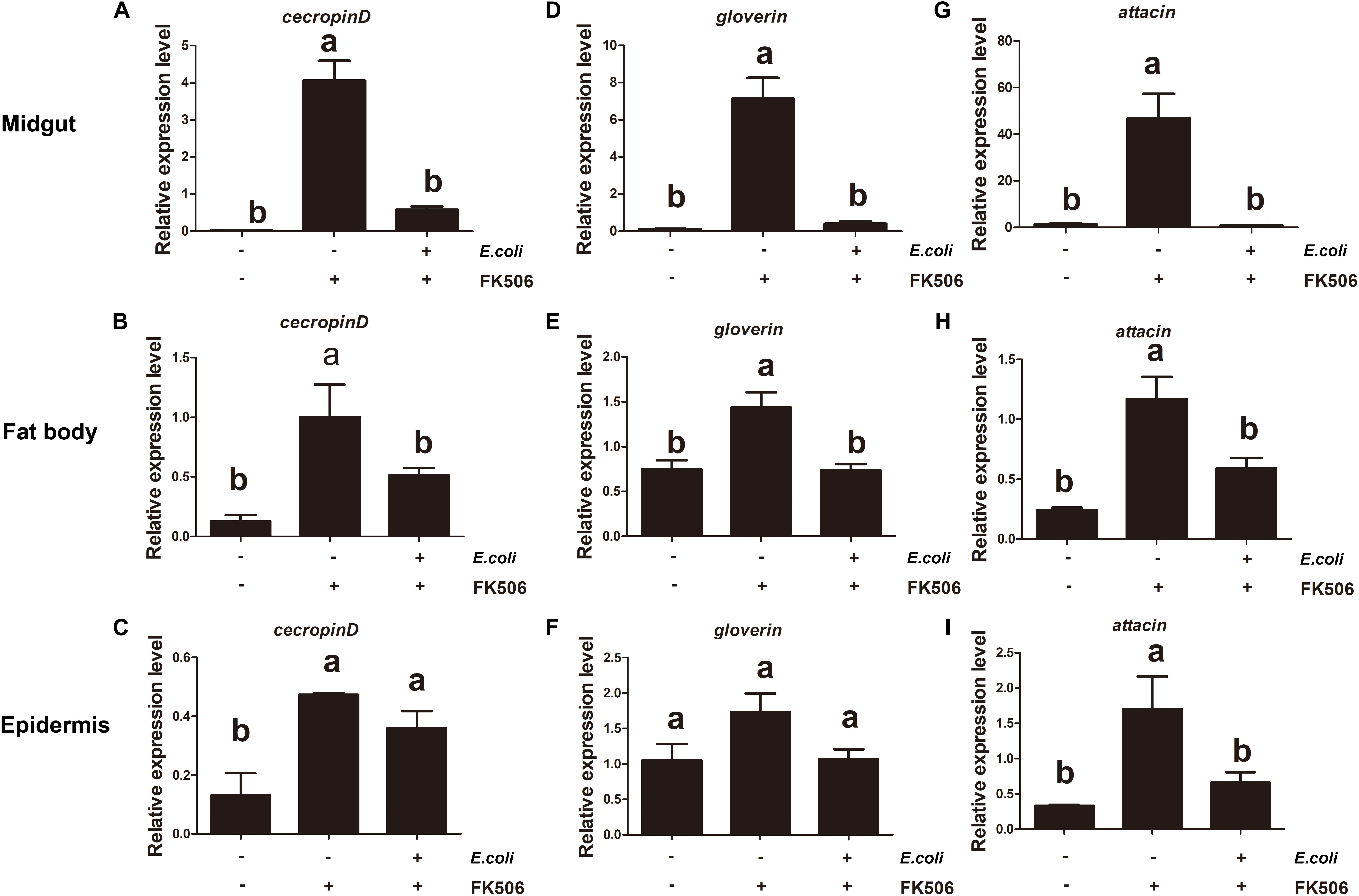
Figure 5. Expression levels of endogenous antimicrobial peptides (cecropin D, attacin, and gloverin) in the midgut, fat body, and epidermis in response to Escherichia coli infection and E. coli + FK506 in fourth instar H. armigera after 1 h treatment. (A–C) Represent the relative expression levels of cecropin D in the migut, fat body, and epidermis, respectively. (D–F) Represent the relative expression levels of attacin in the migut, fat body, and epidermis, respectively. (G–I) Represent the relative expression levels of gloverin in the migut, fat body, and epidermis, respectively. Values shown are means and standard errors. Statistically significant differences for experimental comparisons are indicated by different small letters [analysis of variance (ANOVA) and Tukey’s test, P < 0.05; DPS7.05].
Longer E. coli (for 3 h) treatments resulted in significant increases in the expression levels of attacin and gloverin in the midgut, fat body and epidermis. The E. coli induced increase of these transcripts (attacin and gloverin) was inhibited by FK506 (Figure 6). Although cecropin D expression increases with E. coli stimulation, it was not suppressed by FK506 in the midgut, fat body or epidermis (Figure 6).
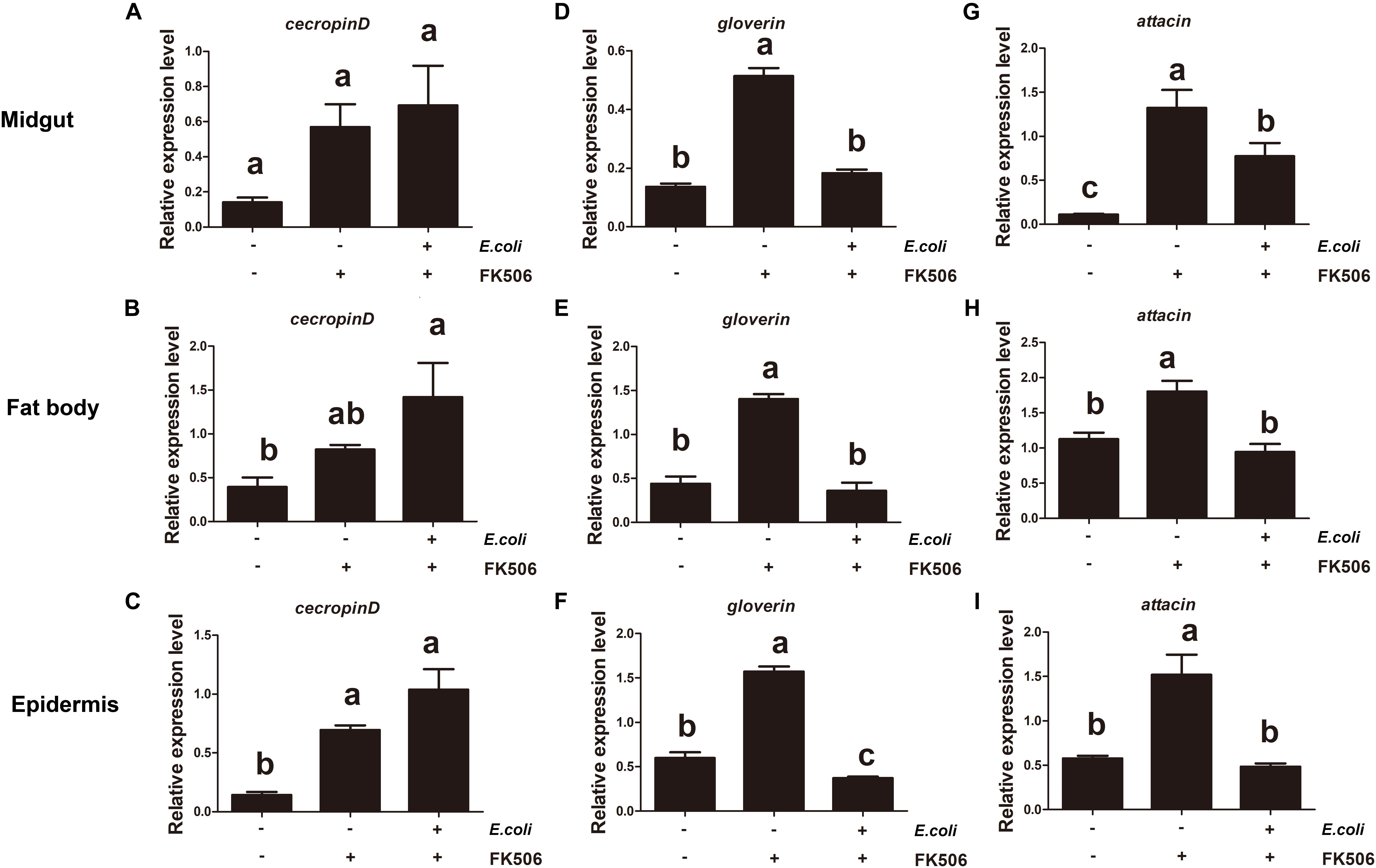
Figure 6. Expression levels of endogenous antimicrobial peptides (cecropin D, attacin, and gloverin) in the midgut, fat body, and epidermis in response to Escherichia coli infection and E. coli + FK506 in fourth instar H. armigera after 3 h treatment. (A–C) Represent the relative expression levels of cecropin D in the migut, fat body, and epidermis, respectively. (D–F) Represent the relative expression levels of attacin in the migut, fat body, and epidermis, respectively. (G–I) Represent the relative expression levels of gloverin in the migut, fat body, and epidermis, respectively. Values shown are means and standard errors. Statistically significant differences for experimental comparisons are indicated by different small letters [analysis of variance (ANOVA) and Tukey’s test, P < 0.05; DPS7.05].
Discussion
Many important life processes in the cell depends on protein phosphorylation/dephosphorylation, which plays an important role in many signal transduction pathways (Patarca, 1996; Lapied et al., 2009). HaCAN is a critical dephosphorylase and its ubiquitous expression in different instars and tissues indicates that HaCAN participates in many biological pathways (Figure 2). The importance of CAN has been reported in other species including mammals and Drosophila (Dijkers and O’Farrell, 2007; Jackson et al., 2018; Kweon et al., 2018). CAN shares a high amino acid identity cross different species, which indicates that CAN may have similar functions in these different species (Chen and Zhang, 2013; Zhao et al., 2018).
FK506 is a specific inhibitor of CAN that significantly inhibit CAN dephosphatase activity (Aramburu et al., 2000). In the present study, FK506 was employed to investigate the role of CAN on H. armigera development. FK506 H. armigera insect development by reducing the weight of larvae, prolonging development time, and reducing pupate rates (Figure 2). These results indicate that HaCAN plays an important role in the development of H. armigera larvae. Although treatments with different FK506 concentrations for 7 days exhibited no significant differences in mortality, FK506 treatment clearly affected insect survival. The present study also revealed that HaCAN can regulate AMP’s expression, however, the decrease in HaCAN-regulated AMPs expression alone did not explain the large changes in developmental delay and pupate rate reduction. Thus, the functions of HaCAN needed to be further investigated.
In order to survive, insects must defend themselves against various pathogens. Drosophila employ two pathways (Toll and IMD) against pathogens (Kaneko and Silverman, 2005). Gram-negative bacteria infection promotes the activation of the relish transcription factor through the IMD pathway in Drosophila (Dijkers and O’Farrell, 2007). The expressions of the AMPs attacin, cecropin, and gloverin depend on relish expression in insects (Meng et al., 1999; Xu et al., 2012; Yang et al., 2015). Our results demonstrate that E. coli can induce the expression of these AMPs via the IMD pathway. However, E. coli induced AMP expression can be neutralized by FK506, which indicating that CAN acts as an up-stream regulator of AMP expression. AMP expression depends on the relish transcription factor and our studies confirmed that HaCAN interacts with Harelish (Figure 3). These results provide evidence for the first time that HaCAN can activate relish through the IMD pathway to regulate the expression of cecropin D, attacin, and gloverin under gram-negative bacteria infection. Similar results were also found in Drosophila, in which CAN acted on relish during infection (Li and Dijkers, 2015).
Drosophila utilizes two distinct pathways to express AMP genes (Kaneko and Silverman, 2005). Infection with gram-negative bacteria promotes the activation of the relish transcription factor through the IMD pathway in Drosophila (Dijkers and O’Farrell, 2007). The Toll pathway provides defense against gram-positive bacteria or fungal infection. Activation of the Toll pathway leads to expression of Dorsal and Dif transcription factors, which in turn up-regulate expression of AMP transcripts (Lemaitre et al., 1996, 1997). Our results show that the expression of cecropin D is regulated in part by the IMD pathway, which suggest that HaCAN may participate in the Toll pathway or another immune mechanism. These pathways contribute to the expression of different AMPs by activating different downstream transcription factors (Zhang et al., 2012; Zeng et al., 2019). It may be interesting to find other transcription factors that interact with HaCAN, and how these transcription factors function within immune response in H. armigera. The role of HaCAN in the immune response must also be further investigated in future studies.
Inhibition of HaCAN activity by FK506 significantly affects the development of cotton bollworms, indicating that FK506 may be used as asynergistic agent for pesticides. Moreover, plant-mediated RNAi shows great potential in crop protection. This technology relies on plants to stably express double-stranded RNAs that target essential genes in pest insects (Mamta and Rajam, 2017; Zhang et al., 2017). RNAi-mediated silencing of insect genes took part in various physiological processes were found to be detrimental to the growth, development and survival of these pest insects (Xiong et al., 2013; Zhang J. et al., 2015; Chikate et al., 2016). HaCAN inhibition significantly affects the development of insects (approximately 70% mortality rate with 50 μM FK506 in H. armigera) (Figure 1). It has also been reported that HaCAN can regulate sex pheromone production and change mating behavior (Zhao et al., 2018). An interesting next step will be to study RNAi of HaCAN, as a technology to control H. armigera.
Data Availability Statement
All datasets generated for this study are included in the manuscript/supplementary files.
Author Contributions
JW, MD, and SA conceived and designed the experiments, wrote the manuscript, and shared the microscopic observations and writing responsibilities. LL, SYao, SY, and SZ performed the experiments. JW, MD, LL, SYao, and SA analyzed the data. All authors have read and approved the manuscript for publication.
Funding
This study was supported by the National Natural Science Foundation of China (Grants 31802016 and 31601904), the Educational Commission of Henan Province of China (Grant 18A210003), the Young Talents Promotion Project in Henan Province (Grant 2019HYTP014), and the Youth Innovation Foundation of the Henan Agricultural University (Grant KJCX2018A14).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aramburu, J., Rao, A., and Klee, C. B. (2000). Calcineurin: from structure to function. Curr. Top. Cell. Regul. 36, 237–295. doi: 10.1016/s0070-2137(01)80011-x
Ballesteros-Martinez, C., Mendez-Barbero, N., Montalvo-Yuste, A., Jensen, B. M., Gomez-Cardenosa, A., Klitfod, L., et al. (2017). Endothelial regulator of calcineurin 1 promotes barrier integrity and modulates histamine-induced barrier dysfunction in anaphylaxis. Front. Immunol. 8:1323. doi: 10.3389/fimmu.2017.01323
Bueno, O. F., Brandt, E. B., Rothenberg, M. E., and Molkentin, J. D. (2002). Defective T cell development and function in calcineurin A -deficient mice. Proc. Natl. Acad. Sci. U.S.A. 99, 9398–9403. doi: 10.1073/pnas.152665399
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. doi: 10.1373/clinchem.2008.112797
Chang, K. T., and Min, K.-T. J. (2005). Drosophila melanogaster homolog of down syndrome critical region 1 is critical for mitochondrial function. Nat. Neurosci. 8, 1577–1585. doi: 10.1038/nn1564
Chang, K. T., Shi, Y.-J., and Min, K.-T. J. (2003). The Drosophila homolog of down’s syndrome critical region 1 gene regulates learning: implications for mental retardation. Proc. Natl. Acad. Sci. U.S.A. 100, 15794–15799. doi: 10.1073/pnas.2536696100
Chen, X. E., and Zhang, Y. J. (2013). Molecular cloning and characterization of the calcineurin subunit A from Plutella xylostella. Int. J. Mol. Sci. 14, 20692–20703. doi: 10.3390/ijms141020692
Chikate, Y. R., Dawkar, V. V., Barbole, R. S., Tilak, P. V., Gupta, V. S., and Giri, A. P. (2016). RNAi of selected candidate genes interrupts growth and development of Helicoverpa armigera. Pestic. Biochem. Physiol. 133, 44–51. doi: 10.1016/j.pestbp.2016.03.006
Dijkers, P. F., and O’Farrell, P. H. (2007). Drosophila calcineurin promotes induction of innate immune responses. Curr. Biol. 17, 2087–2093. doi: 10.1016/j.cub.2007.11.001
Downes, S., Kriticos, D., Parry, H., Paull, C., Schellhorn, N., and Zalucki, M. P. (2017). A perspective on management of Helicoverpa armigera: transgenic Bt cotton, IPM, and landscapes. Pest Manag. Sci. 73, 485–492. doi: 10.1002/ps.4461
Du, M., Liu, X., Ma, N., Liu, X., Wei, J., Yin, X., et al. (2017). Calcineurin-mediated dephosphorylation of acetyl-coA carboxylase is required for pheromone biosynthesis activating neuropeptide (PBAN)-induced sex pheromone biosynthesis in Helicoverpa armigera. Mol. cell. Proteomics 16, 2138–2152. doi: 10.1074/mcp.RA117.000065
Ejima, A., Tsuda, M., Takeo, S., Ishii, K., Matsuo, T., and Aigaki, K. (2004). Expression level of sarah, a homolog of DSCR1, is critical for ovulation and female courtship behavior in Drosophila melanogaster. Genetics 168, 2077–2087. doi: 10.1534/genetics.104.029934
Fetchko, M., and Stagljar, I. (2004). Application of the split-ubiquitin membrane yeast two-hybrid system to investigate membrane protein interactions. Methods 32, 349–362. doi: 10.1016/j.ymeth.2003.10.010
Furman, J. L., and Norris, C. M. (2014). Calcineurin and glial signaling: neuroinflammation and beyond. J. Neuroinflammation 11:158. doi: 10.1186/s12974-014-0158-7
Gajewski, K. M., Wang, J., and Schulz, R. A. (2006). Calcineurin function is required for myofilament formation and troponin I isoform transition in Drosophila indirect flight muscle. Dev. Biol. 289, 17–29. doi: 10.1016/j.ydbio.2005.09.039
Horner, V. L., Czank, A., Jang, J. K., Singh, N., Williams, B. C., Puro, J., et al. (2006). The Drosophila calcipressin sarah is required for several aspects of egg activation. Curr. Biol. 16, 1441–1446. doi: 10.1016/j.cub.2006.06.024
Jackson, S., Pallab, B., Kiran, K., Anupom, B., Deepaneeta, S., Harpreet, K., et al. (2018). A friend or foe: calcineurin across the gamut of neurological disorders. ACS Cent. Sci. 4, 805–819. doi: 10.1021/acscentsci.8b00230
Kaneko, T., Goldman, W. E., Mellroth, P., Steiner, H., Fukase, K., Kusumoto, S., et al. (2004). Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20, 637–649. doi: 10.1016/s1074-7613(04)00104-9
Kaneko, T., and Silverman, N. J. (2005). Bacterial recognition and signalling by the Drosophila IMD pathway. Cell. Microbiol. 7, 461–469. doi: 10.1111/j.1462-5822.2005.00504.x
Kang, Y. J., Kusler, B., Otsuka, M., Hughes, M., Suzuki, N., Suzuki, S., et al. (2007). Calcineurin negatively regulates TLR-mediated activation pathways. J. Immunol. 179, 4598–4607. doi: 10.4049/jimmunol.179.7.4598
Kweon, S. H., Lee, J., Lim, C., and Choe, J. (2018). High-amplitude circadian rhythms in Drosophila driven by calcineurin-mediated post-translational control of sarah. Genetics 209, 815–828. doi: 10.1534/genetics.118.300808
Lapied, B., Pennetier, C., Apaire-Marchais, V., Licznar, P., and Corbel, V. (2009). Innovative applications for insect viruses: towards insecticide sensitization. Trends Biotechnol. 27, 190–198. doi: 10.1016/j.tibtech.2008.12.005
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., and Hoffmann, J. A. (1996). The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 14614–14619.
Lemaitre, B., Reichhart, J.-M., and Hoffmann, J. A. (1997). Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. U.S.A. 94, 14614–14619. doi: 10.1073/pnas.94.26.14614
Li, Y. X., and Dijkers, P. F. (2015). Specific calcineurin isoforms are involved in Drosophila Toll immune signaling. J. Immunol. 194, 168–176. doi: 10.4049/jimmunol.1401080
Liu, J., Albers, M. W., Wandless, T. J., Luan, S., Alberg, D. G., Belshaw, P. J., et al. (1992). Inhibition of T cell signaling by immunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31, 3896–3901. doi: 10.1021/bi00131a002
Liu, J., Farmer, J. D., Lane, W. S., Friedman, J., and Schreiber, S. L. (1991). Calcineurin is a common target of Cyclophilin-Cyclosporin A and FKBP-FK506 complexes. Cell 66, 807–815. doi: 10.1016/0092-8674(91)90124-h
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mamta, B., and Rajam, M. V. (2017). RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. doi: 10.1007/s12298-017-0443-x
Meng, X., Khanuja, B. S., and Ip, Y. T. (1999). Toll receptor-mediated Drosophila immune response requires Dif, an NF- κB factor. Genes Dev. 13, 792–797. doi: 10.1101/gad.13.7.792
Patarca, R. J. (1996). Protein phosphorylation and dephosphorylation in physiologic and oncologic processes. Crit. Rev. Oncog. 7, 343–432. doi: 10.1615/critrevoncog.v7.i5-6.20
Sakai, T., and Aigaki, T. J. (2010). The Drosophila calcineurin regulator, Sarah, is involved in male courtship. Neuroreport 21, 985–988. doi: 10.1097/WNR.0b013e32833eaade
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by th comparative CT method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Shaw, J. L., Chang, K. T., and Lu, B. (2013). Nebula/DSCR1 upregulation delays neurodegeneration and protects against APP-Induced axonal transport defects by restoring calcineurin and GSK-3β signaling. PLoS Genet. 9:e1003792. doi: 10.1371/journal.pgen.1003792
Takeo, S., Hawley, R. S., and Aigaki, T. (2010). Calcineurin and its regulation by Sra/RCAN is required for completion of meiosis in Drosophila. Dev. Biol. 344, 957–967. doi: 10.1016/j.ydbio.2010.06.011
Wei, J., Zhang, L., Yang, S., Xie, B., An, S., and Liang, G. (2018). Assessment of the lethal and sublethal effects by spinetoram on cotton bollworm. PLoS One 13:e0204154. doi: 10.1371/journal.pone.0204154
Xiong, Y., Zeng, H., Zhang, Y., Xu, D., and Qiu, D. (2013). Silencing the HaHR3 gene by transgenic plant-mediated RNAi to disrupt Helicoverpa armigera development. Int. J. Biol. Sci. 9, 370–381. doi: 10.7150/ijbs.5929
Xu, X., Zhong, X., Yi, H., and Yu, X. (2012). Manduca sexta gloverin binds microbial components and is active against Bacteria and Fungi. Dev. Comp. Immunol. 38, 275–284. doi: 10.1016/j.dci.2012.06.012
Yang, J., Wang, X., Tang, S., Shen, Z., and Wu, J. (2015). Peptidoglycan recognition protein S2 from silkworm integument: characterization, microbe-Induced expression, and involvement in the immune-deficiency pathway. J. Insect Sci. 15:20. doi: 10.1093/jisesa/iev007
Yoshiga, T., Yokoyama, N., Imai, N., Ohnishi, A., Moto, K., and Matsumoto, S. (2002). cDNA cloning of calcineurin heterosubunits from the pheromone gland of the silkmoth, Bombyx mori. Insect Biochem. Mol. Biol. 32, 477–486. doi: 10.1016/s0965-1748(01)00125-4
Yu, Y., Li, Y., and Zhang, Y. (2015). Screening of APP interaction proteins by DUALmembrane yeast two-hybrid system. Int. J. Exp. Pathol. 8, 2802–2808.
Zeng, L., Li, Z., and Liu, L. (2019). Research progress in the immunity of insects and the immune mechanisms of five important invasive insects. J. Plant Prot. 46, 6–16.
Zhang, J., Khan, S. A., Hasse, C., Ruf, S., Heckel, D. G., and Bock, R. (2015). Pest control. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347, 991–994. doi: 10.1126/science.1261680
Zhang, S., An, S., Li, Z., Wu, F., Yang, Q., Liu, Y., et al. (2015). Identification and validation of reference genes for normalization of gene expression analysis using qRT-PCR in Helicoverpa armigera (Lepidoptera: Noctuidae). Gene 555, 393–402. doi: 10.1016/j.gene.2014.11.038
Zhang, J., Khan, S. A., and Heckel, D. G. (2017). Next-generation insect-resistant plants RNAi-mediated crop protection. Trends Biotechnol. 8, 871–882. doi: 10.1016/j.tibtech.2017.04.009
Zhang, M., Chu, Y., Zhao, Z., and An, C. (2012). Progress in the molecular mechanisms of the innate immune responses in insects. Acta Entomologica Sinica 55, 1221–1229.
Keywords: calcineurin, AMP, IMD pathway, relish, Helicoverpa armigera
Citation: Wei J, Li L, Yao S, Yang S, Zhou S, Liu X, Du M and An S (2019) Calcineurin-Modulated Antimicrobial Peptide Expression Is Required for the Development of Helicoverpa armigera. Front. Physiol. 10:1312. doi: 10.3389/fphys.2019.01312
Received: 13 August 2019; Accepted: 30 September 2019;
Published: 17 October 2019.
Edited by:
Arash Zibaee, University of Guilan, IranReviewed by:
Mauro Mandrioli, University of Modena and Reggio Emilia, ItalyHao Guo, Chinese Academy of Sciences, China
Copyright © 2019 Wei, Li, Yao, Yang, Zhou, Liu, Du and An. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengfang Du, ZHVtZW5nZmFuZ0AxNjMuY29t
†These authors have contributed equally to this work
 Jizhen Wei
Jizhen Wei Linhong Li†
Linhong Li† Xiaoguang Liu
Xiaoguang Liu Shiheng An
Shiheng An