- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Fujian Provincial Key Laboratory of Insect Ecology, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
The immune system of animals, including insects, is the vital factor to maintain the symbiotic interactions between animals and their associated microbes. However, the effects of gut microbiota on insect immunity remain mostly elusive. Red palm weevil (RPW), Rhynchophorus ferrugineus Olivier, is a destructive pest of palm trees worldwide, which has forged alliances with its gut microbiota. Here, we found that the aposymbiotic insects succumbed at a significantly faster rate than conventionally reared (CR) ones upon bacterial infection. Physiological assays confirmed that CR insects had stronger antimicrobial activity and higher phenoloxidase activity in contrast to germfree (GF) ones, indicating that the systemic immune responses of GF individuals were compromised markedly. Interestingly, under the bacterial challenge conditions, the reassociation of gut microbiota with GF insects could enhance their survival rate by rescuing their immunocompetence. Furthermore, comparative transcriptome analysis uncovered that 35 immune-related genes, including pathogen recognition receptors, effectors and immune signaling pathway, were significantly downregulated in GF insects as compared to CR ones. Collectively, our findings corrobate that intestinal commensal bacteria have profound immunostimulatory effects on RPW larvae. Therefore, knowledge on the effects of gut microbiota on RPW immune defenses may contribute to of set up efficient control strategies of this pest.
Introduction
As in vertebrates, diverse groups of microbes, being generally known as gut microbiota, also colonize insect guts. Gut microbiota has been attested to profoundly affect host physiological traits including nutrition provision (Wong et al., 2014; Janssen and Kersten, 2015), growth and development (Blatch et al., 2010; Wong et al., 2014), mating and foraging behavior (Ami et al., 2010), and gut homeostasis (Engel and Moran, 2013; Douglas, 2015). Considering both pathogenic and beneficial bacterial species possess the conserved immune elicitors, it is the dilemma that insect hosts should preserve the beneficial microbes while ward off pathogenic intruders by mounting effective, yet not damaging defense responses. Accumulating evidence has indicated that some physical barriers, such as peritrophic matrix and mucus layer (Martens et al., 2018), and gut immunity play crucial roles in maintaining the homeostasis of gut microbiota (Lee et al., 2017; Dawadi et al., 2018). For example, gut epithelial cells can secrete reactive oxygen species (ROS) and antimicrobial peptides to regulate the intestinal microbiota in Drosophila melanogaster (Ha et al., 2005; Ryu et al., 2008; Lee et al., 2017). To our knowledge, an increasing number of observations have shown that endosymbiotic bacteria, such as Wolbachia and Wigglesworthia in tsetse flies and Buchnera in aphids, can confer resistance to host against the natural enemies (Oliver et al., 2007; Weiss et al., 2012; Johnson, 2015). Unfortunately, the effects of gut microbiota on the development and proper presence of insect immunity and the mechanisms underlying gut microbiota-induced impacts on host immunity are far from being well understood, especially for the invasive insect pests.
Red palm weevil (RPW), Rhynchophorus ferrugineus Olivier, is a destructive pest for palm trees worldwide (Ju et al., 2011; Wang et al., 2015). In China, it has killed almost 20,000 coconut palms in the area of over 10,000 km2 as an invasive pest since 1997 (Li et al., 2009; Ju et al., 2011; Ge et al., 2015). RPW complete its development from the egg to newborn adult in the trunk of palm trees and the weevil larva is the major infestation stage which feed on tender tissues from the apical growing point of palm trunk (Butera et al., 2012; Shi et al., 2014). Because RPW always live inside palm trunks except for copulation and oviposition, it is usually difficult to find RPW adults and larvae, let alone trying to kill them with chemical pesticides. Consequently, it is urgent to develop integrated pest management tactics based on pheromone traps and biological agents. However, current evidences have indicated that the control efficiency of biological agents is always limited. It is widely acknowledged that insects solely rely on a suite of innate immune responses, comprising cellular and humoral defense mechanisms, to fight pathogenic microbes (Shi et al., 2014). Presently, RPW has been found to live in the symbiotic associations with gut microbiota which is involved in its nutrition metabolism (Muhammad et al., 2017; Prosper et al., 2019). To our knowledge, it is still unknown about if and how commensal microbiota is involved in the proper immune function of insects outside dipteran insects. Because insects often combat pathogens by triggering their innate immune responses, investigations on the intimate relationships between RPW immunity and its gut microbiota are very promising via providing new insights to improve and even drive the development of novel control tactics.
In this study, the influence of commensal microbiota on RPW immunity was determined by comparing the immunocompetence and survival ability of GF and CR RPW larvae upon pathogenic bacterial challenge. Furthermore, transplanting gut microbiota to GF larvae by introducing gut homogenate from CR ones on diet was executed to evaluate whether the compromised immunity phenotypes in GF larvae could be restored. Finally, comparative transcriptome analysis on the immunity of RPW CR and GF larvae was applied to uncover the possible molecular mechanisms behind intestinal bacteria-induced maturation of host immunity. The data we presented here reinforce the understanding of how commensal microbiota affect RPW immunity and set the groundwork for further investigation on the interplay of RPW gut symbionts with the host immune system.
Materials and Methods
Insect Rearing and Generation of Germfree Individuals
Red palm weevil insects were maintained in our laboratory at 27 ± 1°C and 75 ± 5% relative humidity. Larvae were reared in a photoperiod of 24 h dark on artificial diet (Pu et al., 2017), while adults were maintained in climatic chambers as described previously (Muhammad et al., 2017). Treatment cohorts of RPW in this study were designated in four groups: (I) Conventionally reared (CR) larvae contained their inherent gut microbiota; (II) Germfree (GF) ones, being completely devoid of gut microbiota, were generated with the following steps: RPW eggs were washed in 10% sodium hypochlorite solution (NaClO) for 3–5 min, rinsed in 75% ethanol two times to remove the bacteria on the egg surface, and then rinsed with sterilized distilled water two times to exclude the potential effect of sodium hypochlorite and ethanol on the following experiments. The neonatal larvae were fed on the sterile diet (date palm tissue 8.0 g, sucrose 8.0 g, agar 6.0 g, casein 8.0 g, corn flour 10.0 g, yeast extract 12.0 g, avicel 5.0 g, ascorbic acid as vitamin C 1.0 g, potassium sorbate 0.4 g, sodium p-hydroxybenzoate 0.2 g, cholestrol 0.3 g, choline chloride 0.25 g, inositol 0.02 g, and 220 ml distilled water) with antibiotic cocktail (Kanamycin, Tetracycline, Gentamycin, and Erythromycin, the final concentration of 600 mg/L) in the laminar hood (Heal Force safe-1200LC, China). (III) GF + BasSup larvae were established by feeding the gut homogenate from CR larvae to GF individuals (Supplementary Figure S1), and (IV) CR Dech larvae, being from dechorionated eggs and reared on sterile food without antibiotics, contained impaired (≈80% less) gut microbiota (Supplementary Figure S1). GF larvae were always maintained on the food added with antibiotics, but CR Dech. and CR feed on the diet without antibiotics. The 3rd instar GF + BacSup. larvae were provided with food without antibiotics until they were used in the experiments. The procedures on how to generate and maintain these GF and GF + BasSup larvae have been described in detail by Prosper et al. (2019).
Bacteria Preparation for Infection Assays
Serratia marcescens strain RPWL1 (Accession No. MF185369) was previously isolated and identified from RPW gut (Pu and Hou, 2016; Muhammad et al., 2017) and eGFP-tagged Escherichia coli was prepared in LB (Luria-Bertani) medium at least 2 days prior to infection according to the steps of Dawadi et al. (2018). The OD600 (optical density) value of S. marcescens and E. coli solution was 1.89 and 1.00, respectively. All assays were repeated at least three times.
Determination of Phenoloxidase (PO) Activity
To measure the PO response of symbiotic and aposymbiotic individuals and the effect of bacterial infection on the PO activity, the symbiotic (harboring gut microbiota, i.e., CR and GF BacSup.) and aposymbiotic (GF) fourth instar larvae were challenged with S. marcescens. Systemic infection was established by injecting 3 μl (OD600 = 1.89) S. marcescens solution directly into the hemocoel using a 10 μl glass syringe (Hamilton). Parallel controls were established by injecting the same volume of sterile PBS. Before being injected, larvae from different cohorts were chilled on ice for 5–7 min to be immobilized and then their body surface were sterilized in 75% ethanol and washed with sterile water. Six hours post infection, the hemolymph was collected from the larvae as described previously (Muhammad et al., 2017). Hemolymph samples were centrifuged for 5 min at 8000 g (4°C) and then the supernatant was collected into a new tube. The PO activity was assayed using SpectraMax (Molecular Devices, United States) spectrometer according to the methods described by Meng et al. (2016).
The Assays on the Ability of Bacteria Clearance in Hemolymph
To investigate the antimicrobial activity of symbiotic and aposymbiotic RPW individuals, the 4th instar larvae were challenged with 10 μl eGFP-tagged E. coli solution as described above. The same volume of sterile PBS was injected into the same instar larvae as control. Bacterial load in hemolymph was assayed at 6 hpi (hour post-infection) and the hemolymph samples were collected with the above procedures. 100 μl hemolymph samples were spread on the LB agar plates with ampicillin (100 μg/ml) which were incubated for 18 h at 37°C. The CFUs were counted by visualizing the plates under Leica MZFII Stereo Fluorescence Microscope (Leica Microsystems, Germany). Three larvae were used in this assay from each cohort and each treatment was replicated for four times.
Septic Infection and Survival Assay
To determine whether gut microbiota benefit host survival, the symbiotic and aposymbiotic fourth instar larvae were challenged by S. marcescens as described in PO activity assays. The injected larvae were maintained in a separate growth chamber and the number of live or dead larvae was counted every 12 h to monitor insect’s survival. This experiment was repeated three times and 10 larvae were treated in each group of every replicate.
Transcriptome Analysis on Gut Immunity of CR and GF Larvae
To detect the microbiota-induced changes in gut immunity, comparative RNA-Seq analysis on the guts of the fourth instar CR and GF RPW larvae was applied. Whole guts were dissected in sterile conditions for RNA extraction as described previously (Muhammad et al., 2017). Separate libraries, being constructed from total RNA samples of CR and GF groups, were sequenced on Illumina HiSeqTM 4000 system (Guangzhou Gene Denovo Biotechnology Co., Ltd., China). de novo assembly of transcriptome was performed with Trinity software package. For functional annotation, the unigenes were searched against four databases (Nr, Swissport, KEGG, and COG/KOG) using BLASTx program1 with an E-value of 10–5 cut off threshold. Gene Ontology (GO) analysis was completed with Blast2GO software according to the GO association with Nr annotation results of unigenes. Differential expression analysis between the gut transcriptome of CR and GF insects was executed using a rigorous algorithm. Based on the IDs of previously identified immune genes, putative immunity-related genes from RPW gut transcriptome were searched manually against the immune genes of insects such as Tribolium castaneum (Zou et al., 2007), Locusta migratoria manilensis (Zhang et al., 2015), and Ostrinia furnacalis (Liu et al., 2014). The accession number of our sequence data is SAMN11959955-11959960.
The Impact of Gut Bacteria on the Transcript Abundance of Immunity-Related Genes Validated by RT-qPCR
To validate the intestinal-induced expression level of some important immune genes, 15 genes from four functional categories, i.e., pathogen recognition receptors, signal modulation, signal transduction and immune effectors, were selected for transcript quantification. Among these genes, PGRPs (PGRP-LB, PGRP-LC, PGRP-LE, and PGRP-S1), Relish, Defensin, Coleoptericin, Attacin, and Cecropin have been cloned and verified by us. while Beta-glucosidase, C-type lectin (CTL), EF-hand domain containing protein, Serine protease like protein, C-type lysozyme, and Cathepsin were determined according to the previously published data (Hussain et al., 2016). Three individual guts were pooled as a replicate and TRIzol Reagent (Invitrogen, United States) were used to extract total RNA from these gut samples following the manufacturer’s protocol. Each treatment was independently repeated with at least three times. RNA integrity, quality, and concentrations were assessed by Nanodrop (Thermo scientific 2000) and electrophoresis on 1% agarose gel. Total RNA samples were then stored at −80°C until further use. TransScript® All-in-One First Strand cDNA Synthesis Kit (Cat# AT341) was used for synthesizing cDNA. cDNA was serially diluted (5, 25,125, 625, and 3125 fold) to generate standard curves for the target genes to determine primers efficiency and optimal template concentration. RT-qPCR analysis was conducted to determine the fold changes of genes. The qPCR reaction system (20 μl) comprised 10 μl FastStart Universal SYBR Green Master Mix (ROX, Roche) (Life Technologies, United States), 1 μl cDNA, and 3 μM of each primer. Primers used in this study are listed in Table 1. Reactions were performed in triplicate in 96 well plates on a 7500 Fast Real Time PCR system (Applied Biosystem, Life Technologies) with the following thermal program: denaturation at 95°C for 10 min, 40 cycles of 95°C for 15 s, and amplification at 60°C for 60 s and with a dissociation step.
Expression Analysis of Immunity-Related Genes in Fat Body in Symbiotic and Aposymbiotic Individuals After Bacterial Challenge
To investigate the effects of gut microbiota on the systemic immune response in symbiotic and aposymbiotic individuals, they were challenged with S. marcescens as mentioned above. The fold changes of the immunity-related genes, containing RfPGRP-LB, RfPGRP-LC, RfPGRP-LE, RfPGRP-S1, RfRelish, RfDefensin, RfColeoptericin, RfAttacin, and RfCecropin, were determined by RT-qPCR. 6 h after bacterial challenge, the specimens were dissected for collecting fat body that was subsequently processed for RNA extraction. RNA extraction, cDNA synthesis and RT-qPCR analysis assays were performed as described above.
Statistical Analysis
One-way ANOVA or Independent-Sample t-test was used to detect the statistical differences. All statistical analyses were finished using IBM SPSS Statistics (V. 22.0) and the significance was determined at P < 0.05. Kaplan Meir log-rank test was performed to determine if significance between survival rate of symbiotic and aposymbiotic individuals after bacterial challenge.
Results
The Commensal Microbiota Deprival Compromised the Immunocompetence and Survival Rate of RPW Larvae
Significant differences were detected in PO activity of RPW larvae from four treated groups without infection (ANOVA: F(3,16) = 4.54, P < 0.05) or following the bacterial challenge (ANOVA: F(3,16) = 7.19, P = 0.003). As shown in Figure 1, PO activity of CR individuals was markedly higher than that of GF ones. To further uncover the effects of gut microbiota on the antimicrobial activity of this pest, the ability of RPW larvae to clear the injected bacteria from its hemocoel was measured. 1337.45 ± 478.07 CFU of eGFP-tagged E. coli were harvested from the hemolymph of GF insects, which was ten folds greater than that of CR ones. However, less CFU of eGFP-tagged E. coli were found in the symbiotic insects (CR and GF BacSup) when compared to GF ones, indicating that they had higher pathogen clearance ability than aposymbiotic (GF) or depleted (CR Dech.) symbiotic insects (ANOVA: F(3,12) = 22.10, P < 0.001). Interestingly, no statistical significance was detected in the CFU number between CR and GF BacSup groups, suggesting that the reassociation of gut bacteria with GF larvae could rescue their pathogen clearance ability significantly (Figure 2). After being challenged by S. marcescens, GF larvae died at a significantly higher rate as compared to CR ones (Kaplan Meier Survival analysis, P < 0.05, Figure 3). The corrected mortality of GF BacSup was 62.22 ± 3.84% which was lower than that of GF ones (83.33 ± 15.27%). Collectively, these findings support that gut microbiota of RPW can stimulate the immune system against septic pathogen infection. No obvious differences were determined in the PO, antimicrobial activity and survival percentage of CR and GF Bacsup insects, indicating that introducing commensal bacteria into the gut of GF insects could improve their immunocompetence significantly.
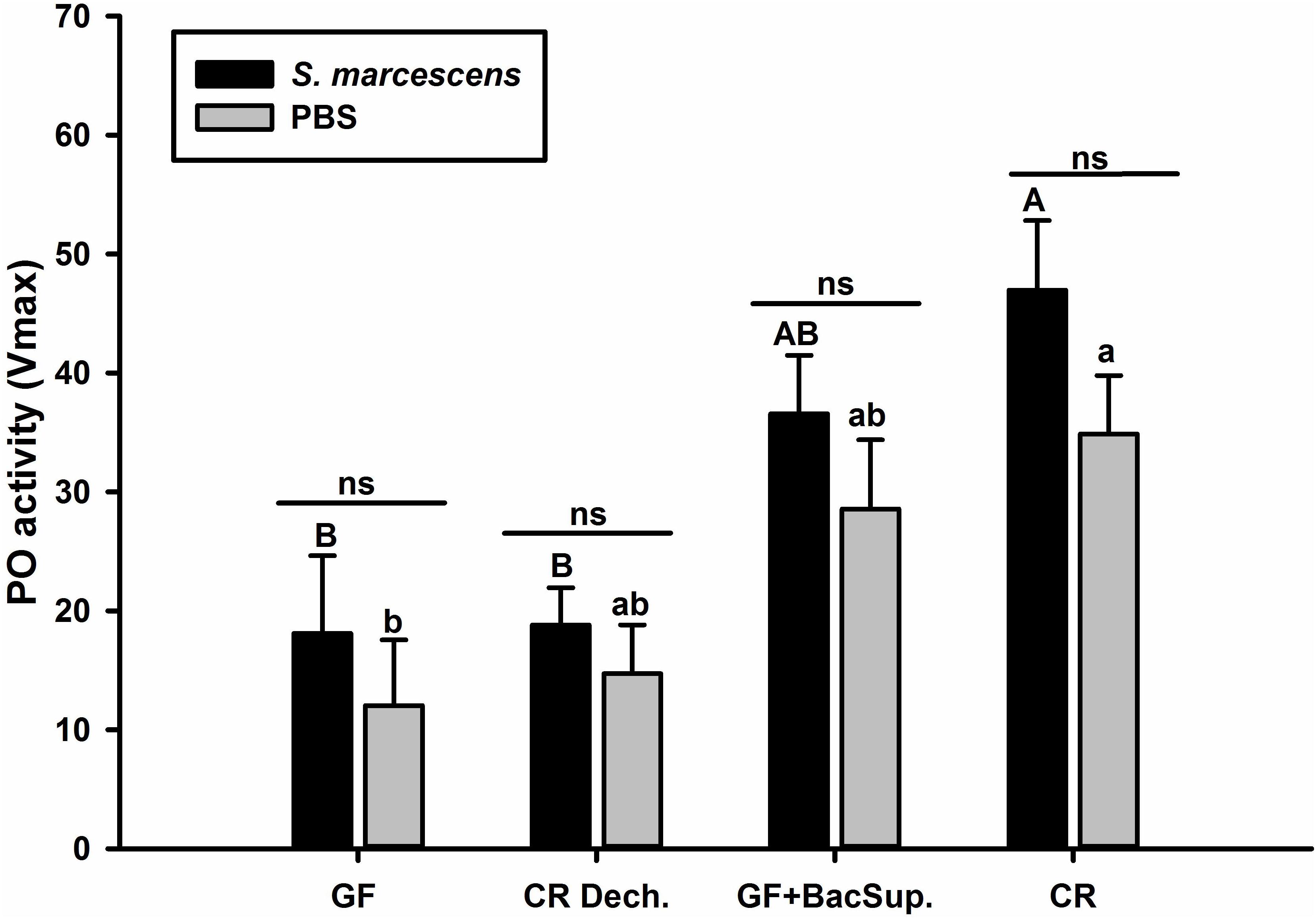
Figure 1. The Phenoloxidase (PO) activity of symbiotic (CR Dech., GF BacSup., and CR) and aposymbiotic (GF) RPW larvae. Uppercase letters indicate the statistical differences of PO activity across different PBS injection groups, whereas the lowercase letters represent the significance of PO activity across different bacterial infection groups using post hoc multiple comparisons Tukey’s HSD (Honest Significant Difference, P < 0.05). Pairwise comparison for each cohort was performed between infected and control counterparts with independent t-test to reveal the effect of bacteria challenge on PO activity. Error bars represent the standard deviation of five independent replicates (ns, P > 0.05).
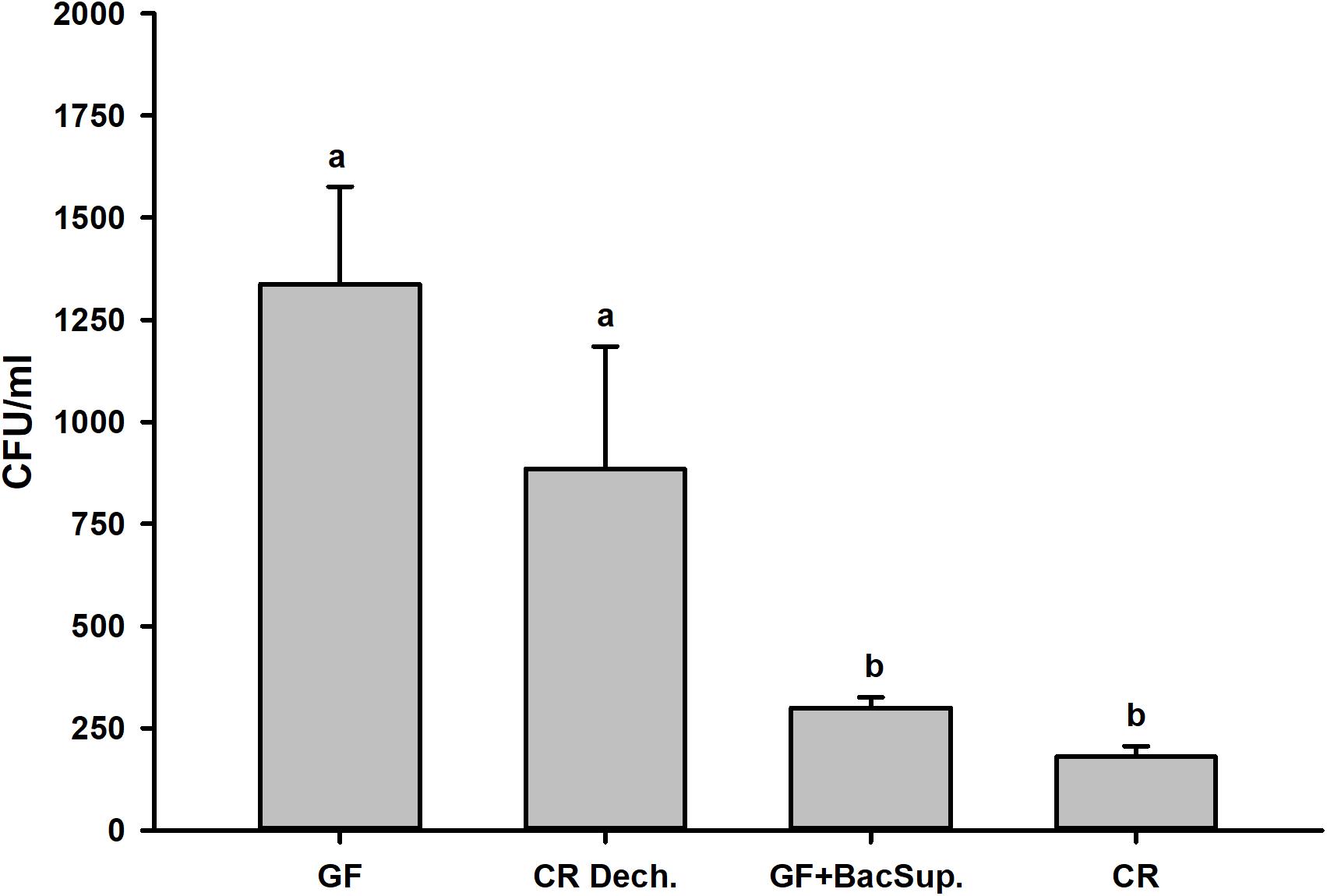
Figure 2. Bacterial clearing ability of the symbiotic (CR Dech., GF BacSup., and CR) and aposymbiotic (GF) RPW larvae.
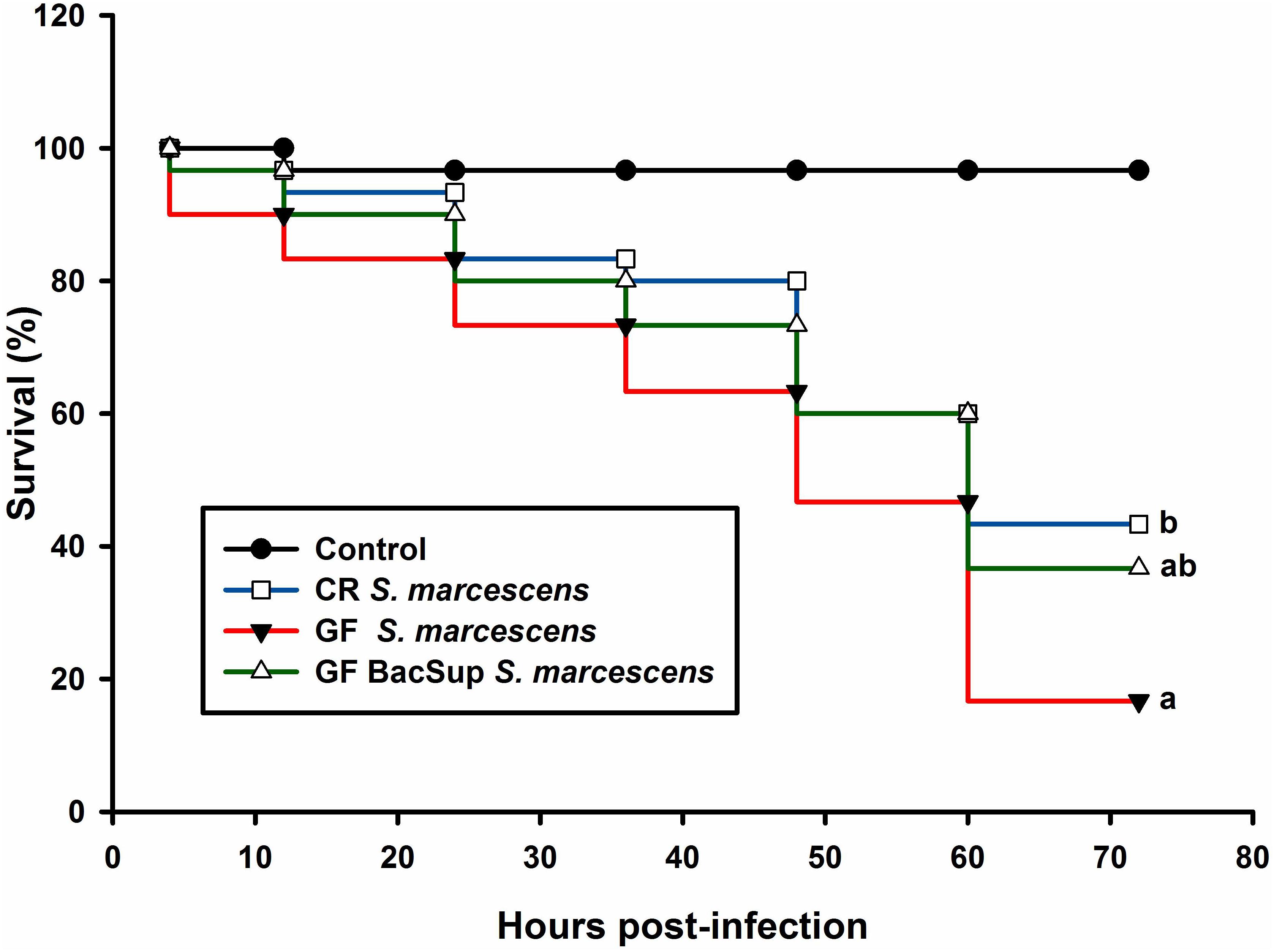
Figure 3. Red palm weevil survival rate of RPW larvae after Serratia marcescens infection. The control (black line) insects were injected with sterile PBS. The symbiotic (CR, blue line and GF BacSup., green line) and aposymbiotic (GF, red line) individuals were injected with Serratia marcescens. Different alphabetic letters represent the significance (Kaplan Meier Survival analysis, Log rank test P < 0.05).
The Induced Higher Expression Level of the Immunity-Related Genes by the Intestinal Bacteria in RPW Larvae
Two cDNA libraries, corresponding to CR and GF groups, were generated and the Illumina HiSeqTM 4000 system yielded an average of 54,820,970 and 57,311,099 clean reads (total reads 336,396,210), respectively. For in-depth elucidation on the interactions between RPW intestine immunity and its gut microbiota, more than 550 immunity-related genes were determined in the gut transcriptome datasets. These immunity-related genes were classified into the following functional categories such as immune recognition, signal transduction and signal modulation (signaling pathways), immune effectors and others (Supplementary Table S1). To determine the effect of gut commensal microbiota on the expression level of immunity-related genes in this pest, pairwise comparisons were performed between the two libraries to reveal the differentially expressed genes (DEGs). Totally, 1,211 unigenes were found to be differentially expressed between CR and GF insects, of which 1,095 unigenes were downregulated and 116 unigenes were upregulated (Figures 4A,B). Interestingly, 35 immunity-related unigenes were found to be differentially expressed in the guts of CR and GF insects. Significant lower expression level of these unigenes was detected in GF insects, indicating the strong association between gut microbiota and RPW immune system. For instance, a number of immune recognition receptors, including 1 C-type lectin and 1 Galectin unigene, were found to be significantly decreased (FDR value < 0.05) in GF insects (Figure 5A). The expression level of these unigenes, being involved in signal modulation and signal transduction group, such as Imd, Toll-like receptor, TNF receptor, Tubulin, Rapamycin, Son of Sevenless, MAPK, Ras-related protein, Serine protease, Calmodulin, and EF-hand domain containing protein (total 23 signaling genes), were also drastically impaired in GF individuals (Figure 5B). Some other immune related genes, including Caspases and Dicer, also presented in GF insects with significantly decreased transcript abundance (Figure 5A). Moreover, the abundance of effector genes, including C-type lysozyme and antimicrobial peptides, were reduced by removing intestinal microbiota from RPW larvae. To confirm these results from RNA sequencing, the transcripts abundance of 15 immune genes was quantified by RT-qPCR. As expected, these genes were expressed in GF insects with significantly lower transcript level as compared to CR ones. For example, the expression level of pathogen recognition receptors, such as RfPGRP-LB, RfPGRP-LE, RfBeta-glucosidase, and RfC-type lectin, was significantly decreased in GF insects. Both Relish and EF-hand domain containing protein genes that mediate the immune signal transduction were significantly compromised by the absence of gut microbiota. Additionally, the immune effector genes, such as RfDefensin, RfAttacin, RfColeoptericin, RfCecropin, RfC-type lysozyme, and RfCathepsin, exhibited in GF insects with strikingly lower transcript abundance (Figure 6). Taken together, these findings demonstrate that gut microbiota can stimulate the expression of immune related genes in the gut of RPW to affect host physiology.
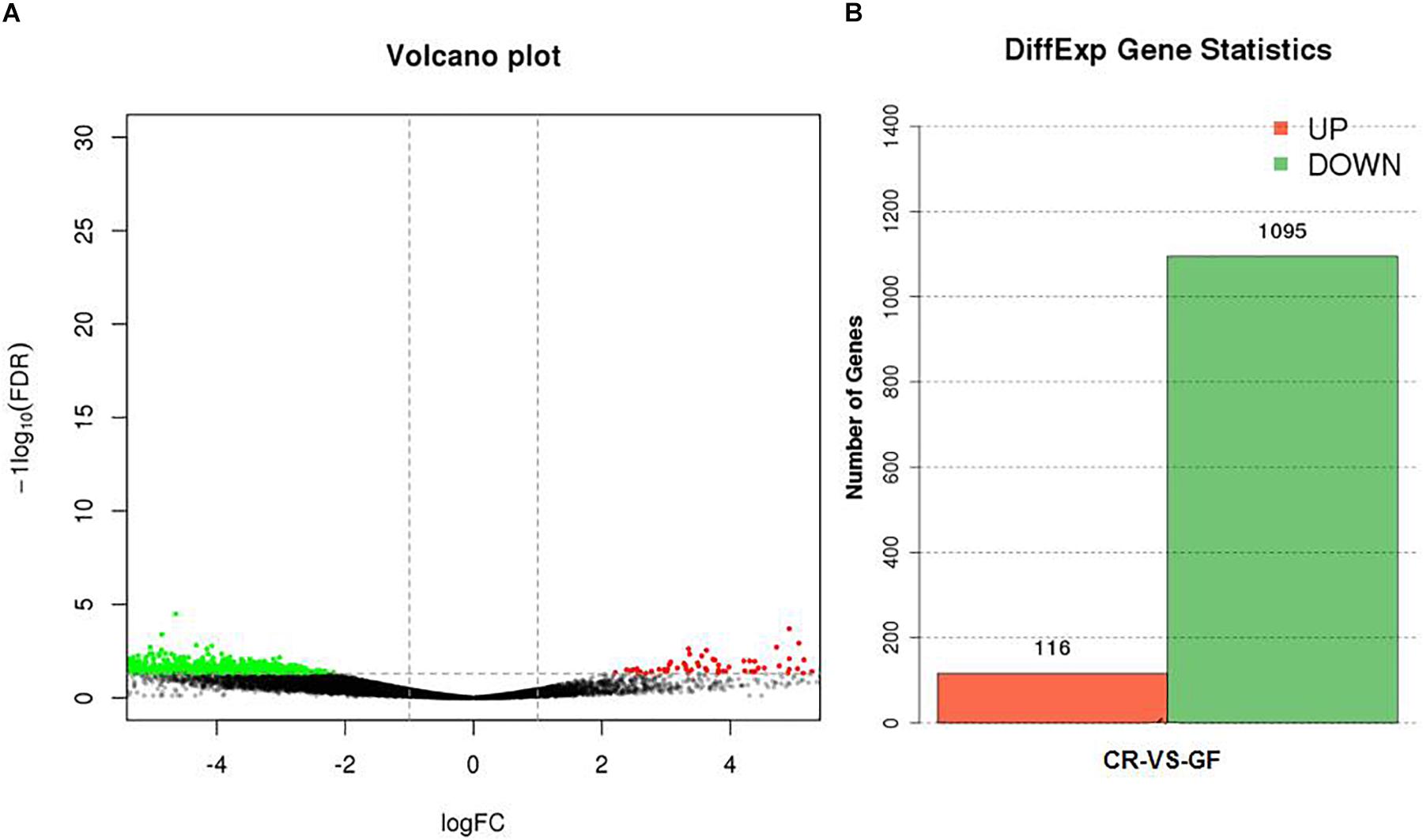
Figure 4. The effect of gut microbiota on the expression level of unigenes in the guts of RPW larvae (A) and the number of genes with significant differences in transcript abundance between CR and GF guts (B). Green means downregulation, red means upregulation and black means no significance.
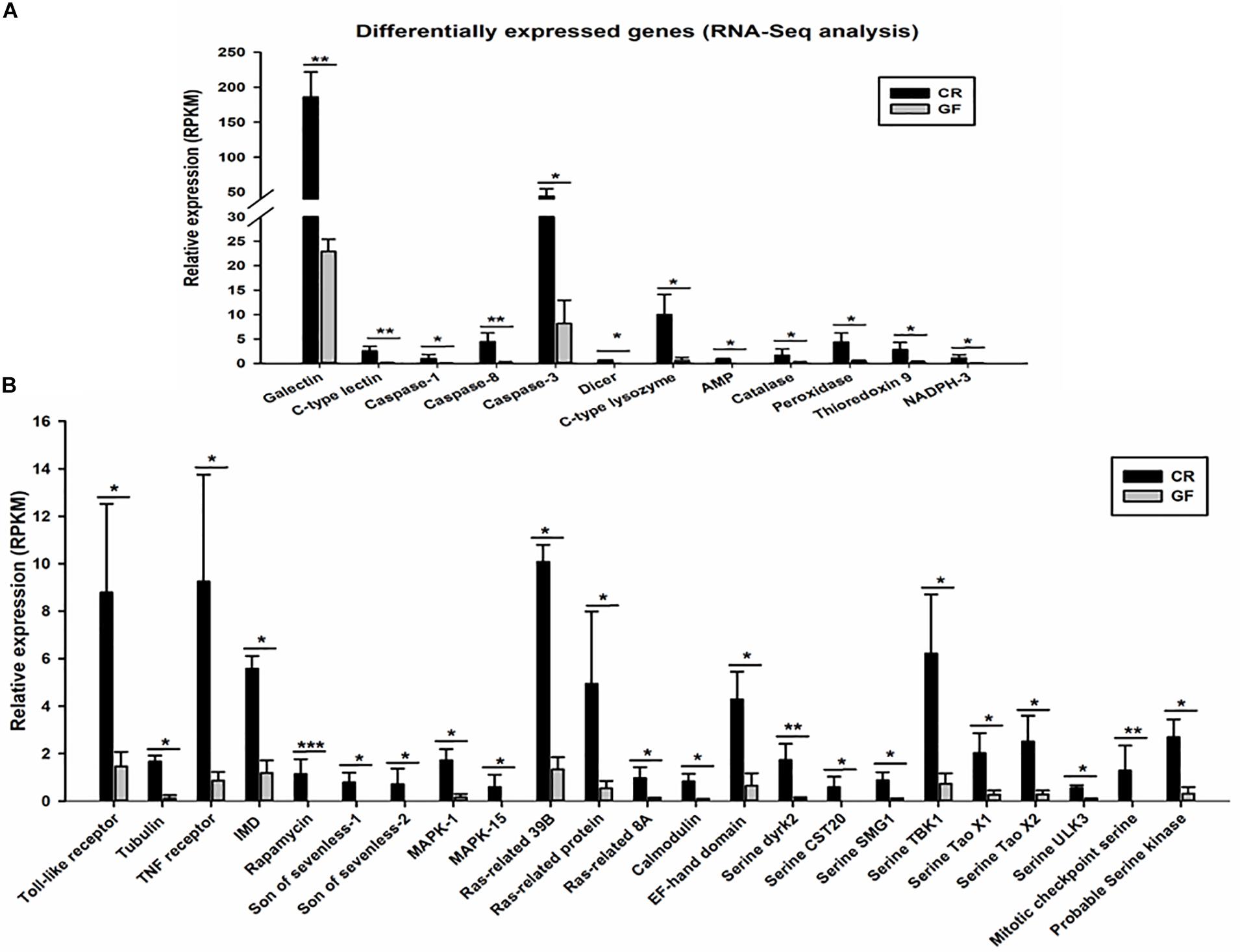
Figure 5. Expression profile of differentially expressed immune related genes (35 genes). RNA-Seq analysis identified immune recognition, immune effectors, and other immune genes (A) and immune signaling genes (B) significantly downregulated in the guts of GF RPW larvae. The False Discovery Rate (FDR) value was set at P < 0.05 and twofold change only for the unigene to be considered as differently expressed genes (DEGs). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
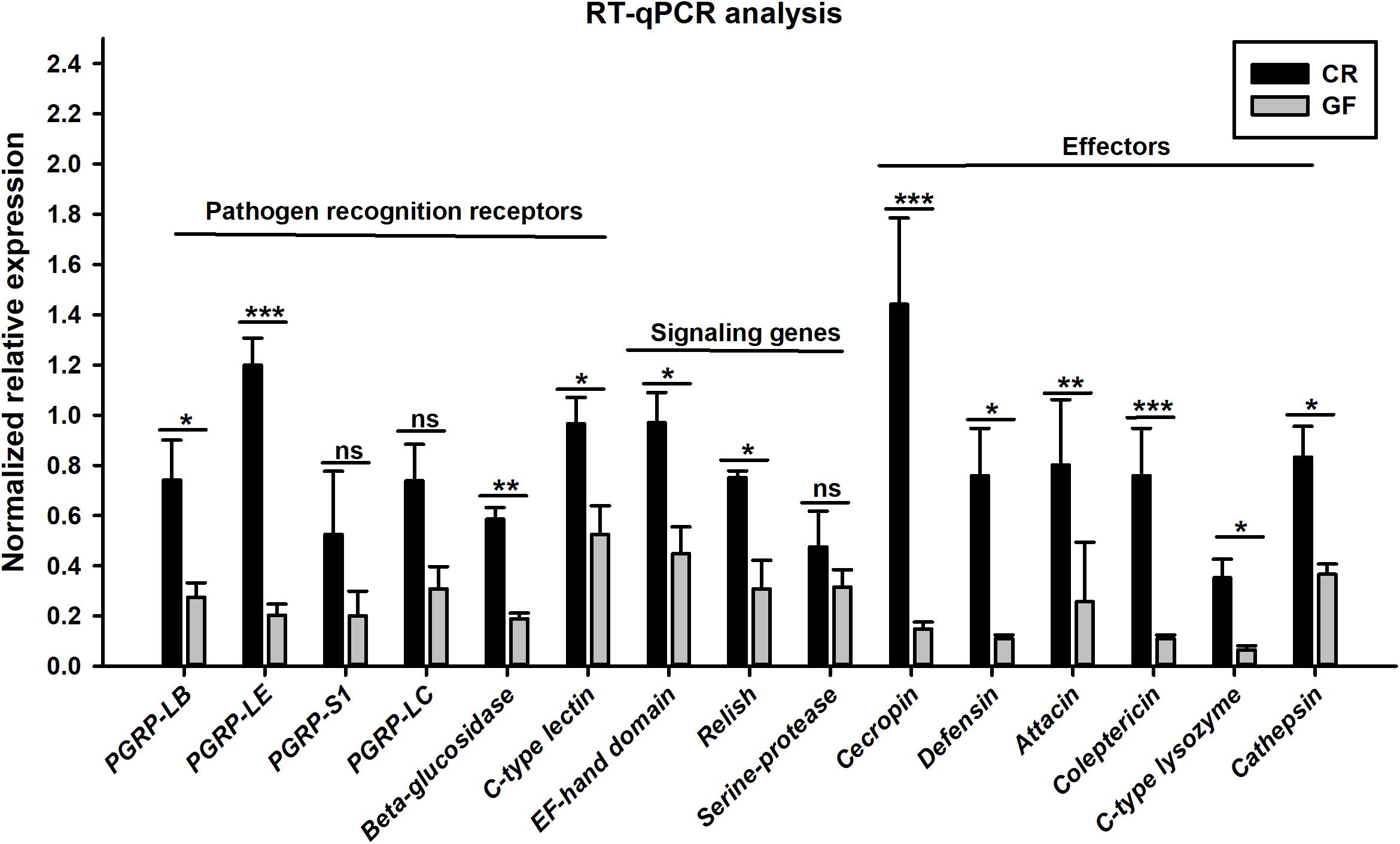
Figure 6. RT-qPCR verification on the transcript abundance of immunity-related genes in the guts of CR and GF RPW larvae. Asterisks indicate statistical difference (P < 0.05) between the CR and GF individuals (t-test: ns, P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). The fold changes of genes were calculated by normalizing to that of the reference gene, Rfβ-actin, with the 2–ΔΔCt method. Error bars represent the standard deviation of three independent biological replicates.
The Symbiotic Insects Exhibit Stronger Magnitude in the Expression Level of Immunity-Related Genes in Fat Body
Gut microbiota has been shown to enhance the immunocompetence and survival rate of RPW larvae under the challenge of S. marcescens. However, the mechanisms underlying the enhancement of gut commensal bacteria on RPW systemic immunity were still unknown. Following the infection, the expression profiles of some important immune genes in fat body, including four PGRP genes (RfPGRP-LB, RfPGRP-LC, RfPGRP-LE, and RfPGRP-S1), RfRelish, and four AMP genes (RfDefensin, RfColeoptericin, RfAttacin, and RfCecropin), were quantified by RT-qPCR. As expected, the GF individuals had decreased expression level of all tested immune genes in both bacterial challenged and non-infected cohorts (Figure 7). Nevertheless, gut bacterial complementation to GF hosts (GF BacSup) exhibited markedly elevated transcript abundance of these tested genes in contrast to GF ones. These results suggested that gut microbiota could enhance the expression level of these PGRP genes in RPW fat body. The challenge of S. marcescens triggered the drastically stronger expression level of four PGRP genes in CR and GF BacSup individuals, indicating that the magnitude of induced expression level of these PGRP genes was strongly dependent on the presence of gut microbiota in RPW. Yet this was not the case for that of RfPGRP-LE, RfPGRP-S1, and RfPGRP-LC in GF individuals. For RfRelish, bacterial infection significantly induced its expression level in fat body of RPW larvae in all groups (Figure 8). As expected, RfDefensin and RfColeoptericin were drastically induced in fat body of aposymbiotic and symbiotic insects by bacterial challenge (Figures 9A,B). But RfAttacin was found to be only upregulated upon bacterial infection in CR individuals (Figure 9C) and only GF individuals exhibited significantly elevated expression level of RfCecropin (Figure 9D). Taken together, these observations revealed that the presence of gut microbiota could drastically enhance the transcript abundance of immunity-related genes in fat body of RPW larvae and then improve their systemic immunocompetence to provide protection against pathogen.
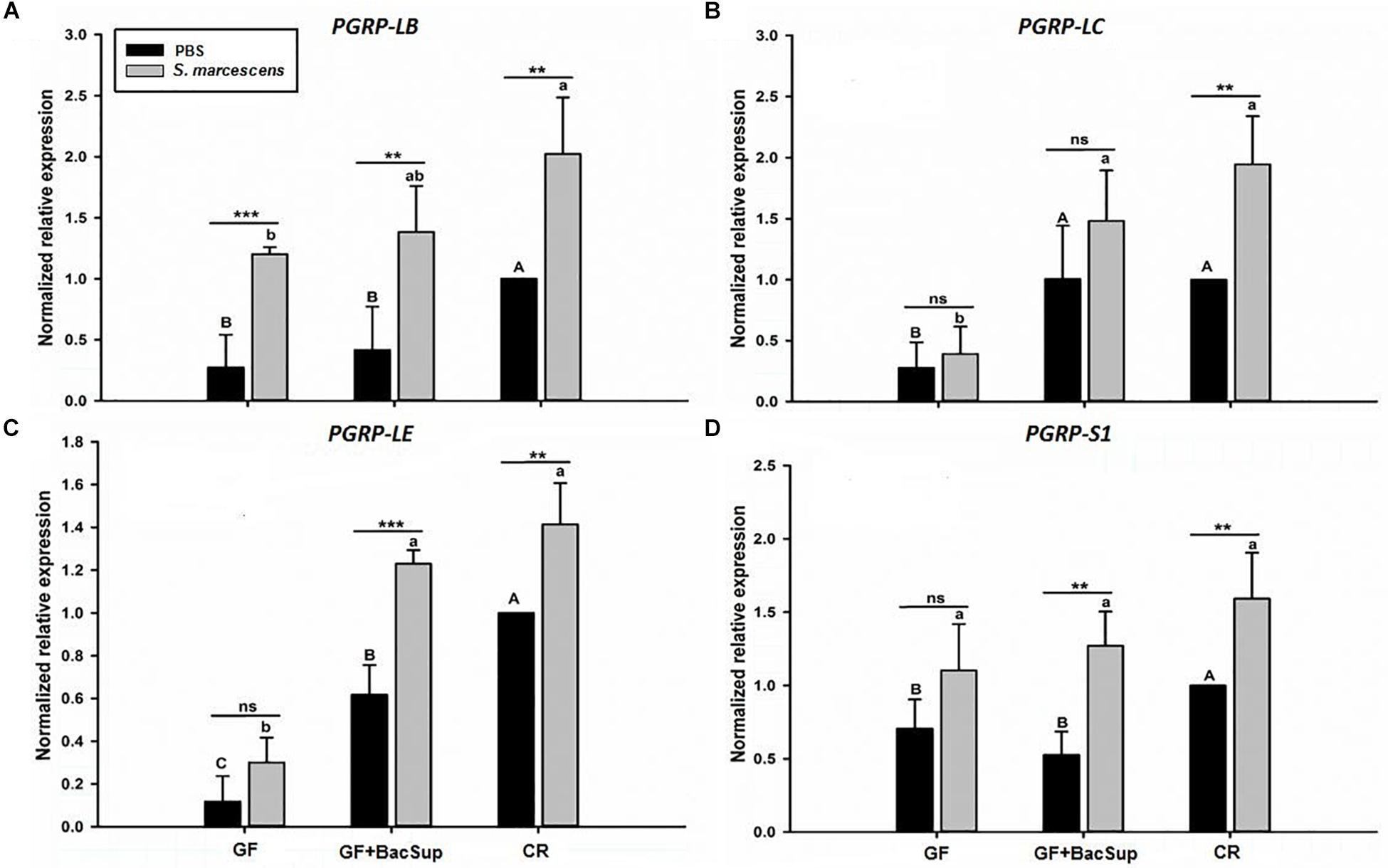
Figure 7. The effect of bacterial challenge on the expression level of four peptidoglycan recognition protein genes in fat body of symbiotic (GF BacSup and CR) and aposymbiotic (GF) RPW larvae. The transcript of PGRP-LB (A), PGRP-LC (B), PGRP-LE (C), and PGRP-S1 (D) are evaluated by RT-qPCR in fat body 6 h post-infection with Serratia marcescens or PBS as control. One-way ANOVA was conducted to evaluate the statistical differences across the groups using Tukey’s HSD post hoc multiple comparison. Uppercase letters indicate the statistical differences of control groups, whereas the lowercase letters represent bacterial infection across the different groups. “∗” presents the significance within each cohort (t-test: ns, P > 0.05, P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Error bars represent the standard deviation of four independent replicates.
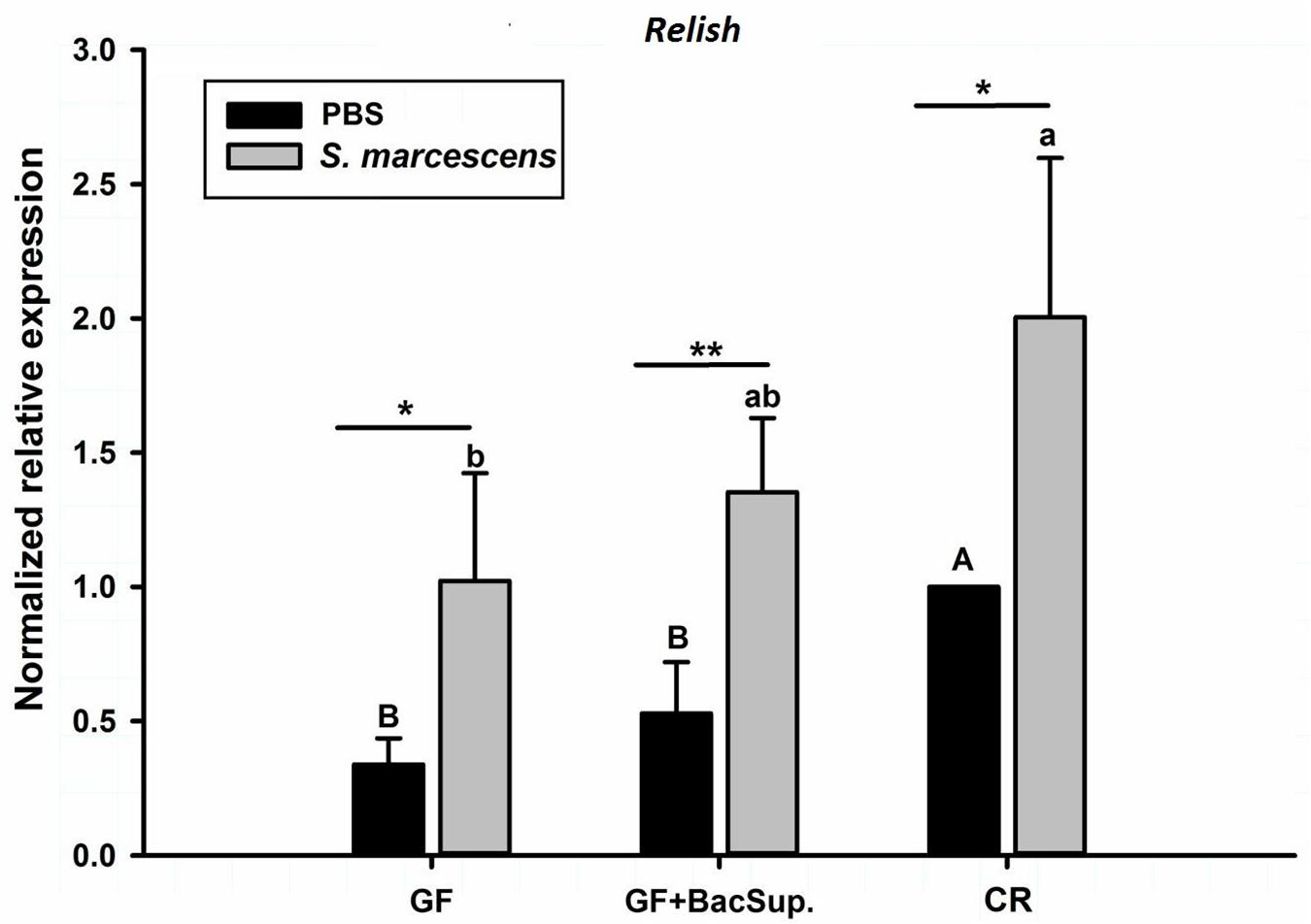
Figure 8. The effect of bacterial challenge on the expression level of Relish in the fat body of symbiotic (GF BacSup and CR) and aposymbiotic (GF) RPW larvae. Uppercase and lowercase letters indicate the statistical differences across control and bacteria challenge groups, respectively. “∗” presents the significance within each cohort (t-test: ∗P < 0.05, ∗∗P < 0.01). Error bars represent the standard deviation of four independent replicates.
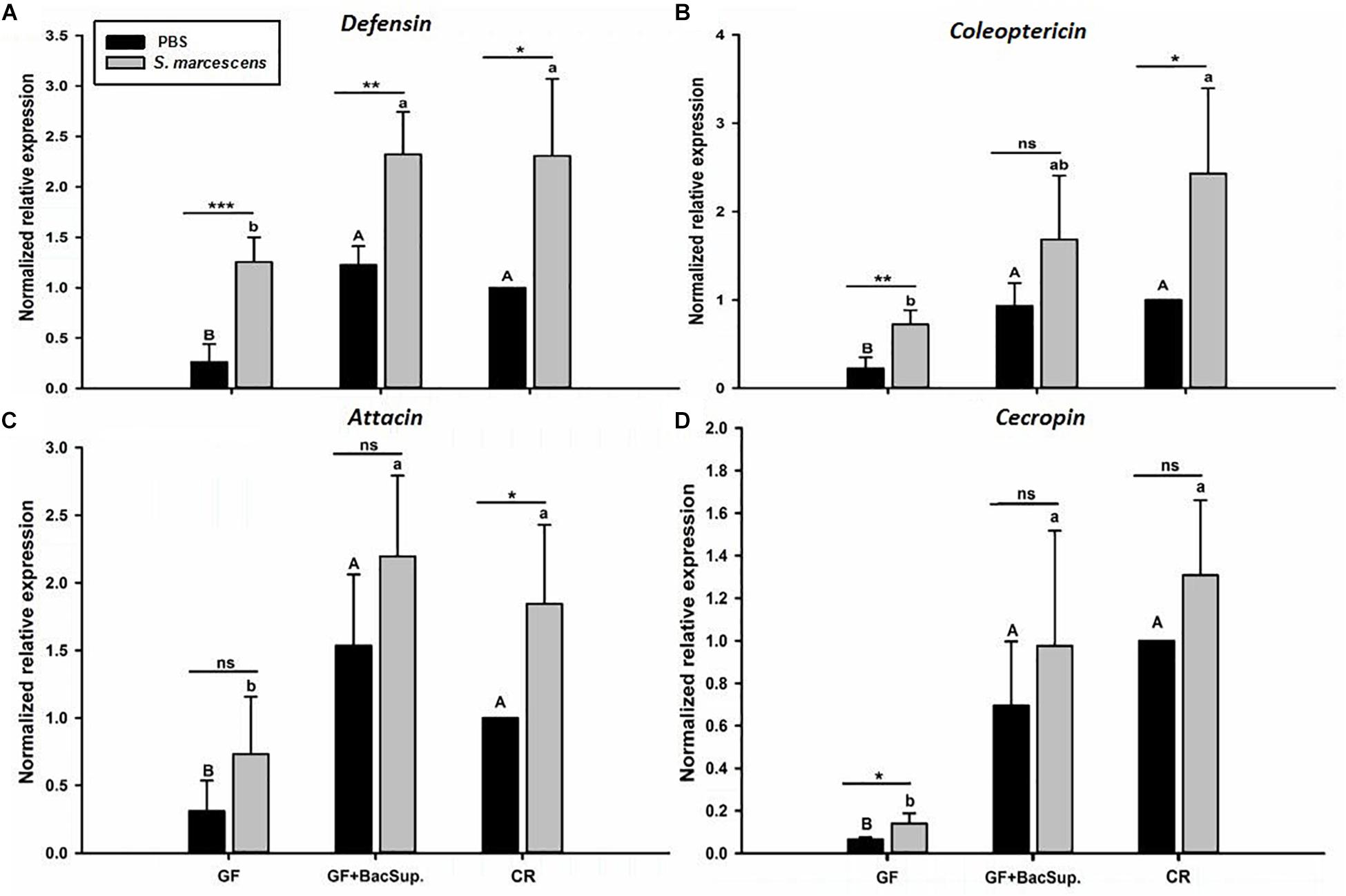
Figure 9. The effect of bacterial challenge on the expression level of antimicrobial peptide genes in fat body of symbiotic (GF BacSup and CKR) and aposymbiotic (GF) RPW larvae. Uppercase and lowercase letters indicate the statistical differences across control and bacteria challenge groups, respectively. “∗” presents the significance within each cohort (t-test: ns, P > 0.05, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001). Error bars represent the standard deviation of four independent replicates. The data was log10 transformed to meet the normality assumptions. The transcript copies of Defensin (A), Coleoptericin (B), Attacin (C) and Cecropin (D) were determined by RT-qPCR.
Discussion
It has been previously demonstrated that RPW harbors a complex gut microbiota which is involved in the nutrition metabolism (Muhammad et al., 2017; Prosper et al., 2019). We have also verified that the immune system of this pest play the pivotal role in maintaining and modulating the homeostasis of gut commensal microbiota (Dawadi et al., 2018; Xiao et al., 2019). However, the impact of gut microbiota on the immunity of this pest remains elusive. In the present study, it has been determined that the deprival of intestinal microbiota drastically impaired the pathogen clearance and survival ability of RPW larvae after being challenged by E. coli and S. marcescens, respectively. Furthermore, RNA sequencing and RT-qPCR analysis verified that the presence of gut microbiota strongly upregulated the immunity-related genes of RPW larvae, containing pathogen recognition receptors, nuclear factor κB Relish and antimicrobial peptides, as compared to GF ones. These findings provide the experimental evidence to support that colonization of gut commensal microbiota has the stimulatory effects on the immune system of RPW larvae and enhance the immunocompetence of host. Several studies have revealed that gut microbiome can inhibit the development of Plasmodium and other human pathogens in mosquito by upregulating some important immune genes such as cecropins, defensin, and gambicin (Gonzalez-Ceron et al., 2003; Cirimotich et al., 2011; Ricci et al., 2012). Interestingly, Futo et al. (2016) observed that T. castaneum larvae, with significantly lowered microbial load, exhibited decreased survival upon secondary challenge of Bacillus thuringiensis, indicating that gut microbiota plays a crucial role in oral immune priming. Recently, it has been shown that the immune system of honey bee Apis mellifera can be stimulated by its native gut microbiota (Kwong et al., 2017). Specifically, Frischella perrara, a gut bacterium of honey bee, has been verified to cause a strong immune activation and influence gut immunity and homeostasis in the pylorus (Emery et al., 2017). Therefore, our findings, being consistent with these previous reports, highlight that gut commensal bacteria are essential for the development and maintenance of a healthy immune system in insects.
This study uncovered that the presence of gut bacteria could significantly improve the transcript abundance of several pattern recognition receptors (PRRs) and antimicrobial peptides in fat body. Furthermore, RPW larvae without gut bacteria exhibited the lower ability to clear pathogenic bacteria in hemolymph, impaired PO activity and died at faster rate after microbial challenge. It is well known that insect immunity is often mounted through recognizing invaded pathogens by PRRs and then immune signals are transduced via NF-κB pathway to induce the secretion of effectors (Lemaitre and Hoffmann, 2007; Hetru and Hoffmann, 2009). In this context, our data suggest that gut microbiota can enhance the immunity of RPW larvae via the upregulation of immune genes to stimulate antimicrobial peptide secretion. Notably, our previous investigation has determined that enzymatic PGRP-LB acts as a negative regulator on intestinal immunity to maintain the homeostasis of RPW gut microbiota (Dawadi et al., 2018). Our analysis found RPW PGRP-LB gene was significantly upregulated by the colonization of gut microbiota, suggesting that gut commensal bacteria are also involved in the modulation of RPW immunohomeostasis by elevating the expression level of some immunity negative regulators to avoid over-activation of mucosal immunity. RPW gut is colonized by multiple bacterial species which exert different roles on host fitness (Muhammad et al., 2017; Prosper et al., 2019). Recent studies on the immunomodulatory impacts of commensal bacteria on mammals have identified that these effects are specific for individual bacterium, groups of bacteria or specific components of the microbiota (Hooper et al., 2012; Ivanov and Honda, 2012). In this way, RPW gut bacterial composition may influence the type and robustness of host immune responses. Here we just determined the stimulatory effects of intestinal microbiota on RPW immunity by removing the entire commensal microbiota. Therefore, the possible specific roles of individual bacterium or groups of bacteria on the immune system of RPW deserve further investigation through making the gnotobiotic RPW larvae.
In the present investigation, we also provided the evidence to show that gut microbiota do not only affect gut immunity but also mediate the systemic immune response of RPW larvae. To our best knowledge, it is still unknown on the mechanisms underlying gut commensal bacteria modulate the immune system of insect hosts although the immunostimulatory effects of gut microbiome have been determined. Mounting evidence uncovered that gut microbiome can produce numerous bioactive compounds, containing short chain fatty acids (SCFAs), choline metabolites and lipids, that are important for host physiology (Nicholson et al., 2012; Lee and Hase, 2014; Qin and Wade, 2017). In particular, recent reports on mammals have pointed out that SCFAs have important metabolic functions and are crucial for intestinal health (Canfora et al., 2015). For instance, the SCFAs, containing butyrate, propionate, and acetate, promote intestinal epithelial barrier function and regulate the host mucosal immune system via promoting some specific histone post-translational modifications (Vinolo et al., 2011; Krautkramer et al., 2017; Fellows et al., 2018; Schulthess et al., 2019). In D. melanogaster, it has been found that axenic flies have altered insulin signaling and lipid metabolism, and this can be reversed by the provision of a gut microbial metabolite, acetate (Shin et al., 2011; Hang et al., 2014). More importantly, SCFAs can affect mammal cellular and modulate immune responses, in part through affecting gene expression and the epigenome by inhibiting histone deacetylases (Koh et al., 2016; Krautkramer et al., 2016). These reports imply that gut microbiota-derived metabolites act as critical messengers in the crosstalk between microbes and hosts. Consequently, the stimulatory effects of commensal bacteria on the systemic immune responses of RPW larvae might be executed by their fermentative metabolites. The diversity and composition of gut microbiota in insect pests have been studied for over a decade, but the causal relationship among microbe-derived gut metabolites, host signaling pathways and host physiology are still poorly understood outside D. melanogaster. It is well known that intestine commensal bacteria can release peptidoglycan and uracil to induce the expression of AMPs and ROS (Paredes et al., 2011; Lee and Brey, 2013; Dawadi et al., 2018; Xiao et al., 2019). Therefore, another possibility is that gut bacteria-derived peptidoglycan can pass through gut barrier and enter the hemolymph acting as a ligand for NF-κB activation in fat body. Therefore, the exact mechanisms behind commensal bacteria affect the immunity of RPW larvae need extensive dissections by detecting the behavior of microbe-derived metabolites on host physiology.
In conclusion, our results demonstrated that the presence of gut microbiota could drastically improve the survival rate of bacteria challenged RPW larvae via upregulating the important immune genes to enhance host immunocompetence, suggesting that gut bacteria have strong stimulatory effects on the immune system of RPW larvae. Consequently, we found that this pest relies on its intestinal bacteria for its proper immune functions. Because the innate immune responses are the sole way for insect pests to fight off pathogens, harnessing the ability of intestinal microbiota to affect host immunity can be considered as a very promising pest management tactic. On the other hand, our study set a firm foundation to deeply dissect the interplay between RPW and its gut microbiota.
Data Availability Statement
The datasets (PRJNA546480) for this study can be accessed from NCBI SRA (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA546480) under the accession number (SRR9637668–SRR9637673).
Author Contributions
ZS conceived and designed the research. AM, PH, and TJ completed the experiments. AM and ZS analyzed the data. ZS, AM, and YH wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31470656), the National Key Research and Development Project of China (2017YFC1200605), and the Natural Science Foundation of Fujian Province (2018J01705).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.01303/full#supplementary-material
FIGURE S1 | The number of RPW gut bacterial colony-forming uints (CFUs) from four designated groups.
Footnotes
References
Ami, E. B., Yuval, B., and Jurkevitch, E. (2010). Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 4, 28–37. doi: 10.1038/ismej.2009.82
Blatch, S. A., Meyer, K. W., and Harrison, J. F. (2010). Effects of dietary folic acid level and symbiotic folate production on fitness and development in the fruit fly Drosophila melanogaster. Fly 4, 312–319. doi: 10.4161/fly.4.4.13258
Butera, G., Ferraro, C., Colazza, S., Alonzo, G., and Quatrini, P. (2012). The culturable bacterial community of frass produced by larvae of Rhynchophorus ferrugineus olivier (Coleoptera: Curculionidae) in the Canary island date palm. Lett. Appl. Microbiol. 54, 530–536. doi: 10.1111/j.1472-765X.2012.03238.x
Canfora, E. E., Jocken, J. W., and Blaak, E. E. (2015). Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 11, 577–591. doi: 10.1038/nrendo.2015.128
Cirimotich, C. M., Dong, Y., Clayton, A. M., Sandiford, S. L., Souza-Neto, J. A., Mulenga, M., et al. (2011). Natural microbe-mediated refractoriness to Plasmodium infection in Anopheles gambiae. Science 332, 855–858. doi: 10.1126/science.1201618
Dawadi, B., Wang, X., Xiao, R., Muhammad, A., Hou, Y., and Shi, Z. (2018). PGRP-LB homolog acts as a negative modulator of immunity in maintaining the gut-microbe symbiosis of red palm weevil, Rhynchophorus ferrugineus Olivier. Dev. Comp. Immunol. 86, 65–77. doi: 10.1016/j.dci.2018.04.021
Douglas, A. E. (2015). Multiorganismal insects: diversity and function of resident microorganisms. Annu. Rev. Entomol. 60, 17–34. doi: 10.1146/annurev-ento-010814-020822
Emery, O., Schmidt, K., and Engel, P. (2017). Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 26, 2576–2590. doi: 10.1111/mec.14058
Engel, P., and Moran, N. A. (2013). The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735. doi: 10.1111/1574-6976.12025
Fellows, R., Denizot, J., Stellato, C., Cuomo, A., Jain, P., Stoyanova, E., et al. (2018). Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat. Commun. 9:105. doi: 10.1038/s41467-017-02651-5
Futo, M., Armitage, S. A., and Kurtz, J. (2016). Microbiota plays a role in oral immune priming in Tribolium castaneum. Front. Microbiol. 6:1383. doi: 10.3389/fmicb.2015.01383
Ge, X., He, S., Wang, T., Yan, W., and Zong, S. (2015). Potential distribution predicted for Rhynchophorus ferrugineus in China under different climate warming scenarios. PLoS One 10:e0141111. doi: 10.1371/journal.pone.0141111
Gonzalez-Ceron, L., Santillan, F., Rodriguez, M. H., Mendez, D., and Hernandez-Avila, J. E. (2003). Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J. Med. Entomol. 40, 371–374. doi: 10.1603/0022-2585-40.3.371
Ha, E.-M., Oh, C.-T., Bae, Y. S., and Lee, W.-J. (2005). A direct role for dual oxidase in Drosophila gut immunity. Science 310, 847–850. doi: 10.1126/science.1117311
Hang, S., Purdy, A., Robins, W., Wang, Z., Mandal, M., Chang, S., et al. (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. doi: 10.1016/j.chom.2014.10.006
Hetru, C., and Hoffmann, J. A. (2009). NF-κB in the immune response of Drosophila. CSH Perspect. Biol. 1:a000232. doi: 10.1101/cshperspect.a000232
Hooper, L. V., Littman, D. R., and Macpherson, A. J. (2012). Interactions between the microbiota and the immune system. Science 336, 1268–1274. doi: 10.1126/science.1223490
Hussain, A., Rizwan-ul-Haq, M., Al-Ayedh, H., and AlJabr, A. M. (2016). Susceptibility and immune defence mechanisms of Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) against entomopathogenic fungal infections. Int. J. Mol. Sci. 17:1518. doi: 10.3390/ijms17091518
Ivanov, I. I., and Honda, K. (2012). Intestinal commensal microbes as immune modulators. Cell Host Microbe 12, 496–508. doi: 10.1016/j.chom.2012.09.009
Janssen, A. W., and Kersten, S. (2015). The role of the gut microbiota in metabolic health. FASEB J. 29, 3111–3123. doi: 10.1096/fj.14-269514
Johnson, K. N. (2015). Bacteria and antiviral immunity in insects. Curr. Opin. Insect. Sci. 8, 97–103. doi: 10.1016/j.cois.2015.01.008
Ju, R. T., Wang, F., Wan, F. H., and Li, B. (2011). Effect of host plants on development and reproduction of Rhynchophorus ferrugineus (Olivier)(Coleoptera: Curculionidae). J. Pest Sci. 84, 33–39. doi: 10.1007/s10340-010-0323-4
Koh, A., De Vadder, F., Kovatcheva-Datchary, P., and Bäckhed, F. (2016). From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345. doi: 10.1016/j.cell.2016.05.041
Krautkramer, K. A., Kreznar, J. H., Romano, K. A., Vivas, E. I., Barrett-Wilt, G. A., Rabaglia, M. E., et al. (2016). Diet-microbiota interactions mediate global epigenetic programming in multiple tissues. Mol. Cell 64, 982–992. doi: 10.1016/j.molcel.2016.10.025
Krautkramer, K. A., Rey, F. E., and Denu, J. M. (2017). Chemical signaling between gut microbiota and host chromatin: what is your gut really saying? J. Biol. Chem. 292, 8582–8593. doi: 10.1074/jbc.R116.761577
Kwong, W. K., Mancenido, A. L., and Moran, N. A. (2017). Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 4:170003. doi: 10.1098/rsos.170003
Lee, J.-H., Lee, K.-A., and Lee, W.-J. (2017). Microbiota, Gut Physiology, and Insect Immunity, Vol. 52. Amsterdam: Elsevier, 111–138.
Lee, W. J., and Brey, P. T. (2013). How microbiomes influence metazoan development: insights from history and Drosophila modeling of gut-microbe interactions. Annu. Rev. Cell Dev. Biol. 29, 571–592. doi: 10.1146/annurev-cellbio-101512-122333
Lee, W. J., and Hase, K. (2014). Gut microbiota–generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424. doi: 10.1038/nchembio.1535
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Li, Y., Zhu, Z. R., Ju, R. T., and Wang, L. S. (2009). The red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae), newly reported from Zhejiang, China and update of geographical distribution. Fla. Entomol. 92, 386–387. doi: 10.1653/024.092.0229
Liu, Y., Shen, D., Zhou, F., Wang, G., and An, C. (2014). Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS One 9:e86436. doi: 10.1371/journal.pone.0086436
Martens, E. C., Neumann, M., and Desai, M. S. (2018). Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 16, 457–470. doi: 10.1038/s41579-018-0036-x
Meng, E., Tang, B., Hou, Y., Chen, X., Chen, J., and Yu, X. (2016). Altered immune function of Octodonta nipae (Maulik) to its pupal endoparasitoid, Tetrastichus brontispae Ferrière. Biochem. Mol. Biol. 198, 100–109. doi: 10.1016/j.cbpb.2016.04.001
Muhammad, A., Fang, Y., Hou, Y., and Shi, Z. (2017). The gut entomotype of red palm weevil Rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae) and their effect on host nutrition metabolism. Front. Microbiol. 8:e2291. doi: 10.3389/fmicb.2017.02291
Nicholson, J. K., Holmes, E., Kinross, J., Burcelin, R., Gibson, G., Jia, W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336, 1262–1267. doi: 10.1126/science.1223813
Oliver, K. M., Campos, J., Moran, N. A., and Hunter, M. S. (2007). Population dynamics of defensive symbionts in aphids. Proc. R. Soc. Lond. Biol. 275, 293–299. doi: 10.1098/rspb.2007.1192
Paredes, J. C., Welchman, D. P., Poidevein, M., and Lemaitre, B. (2011). Negative regulation by amidase PGRPs shapes the Drosophila antibacterial response and protects the fly from innocuous infection. Immunity 35, 770–779. doi: 10.1016/j.immuni.2011.09.018
Prosper, H., Muhammad, A., Ji, T., Xiao, R., Yin, X., Hou, Y., et al. (2019). The promoting effect of gut microbiota on growth and development of red palm weevil, Rhynchophorus ferrugineus (Olivier) (Coleoptera: Dryophthoridae) by modulating its nutritional metabolism. Front. Microbiol. 10:1212. doi: 10.3389/fmicb.2019.01212
Pu, Y., and Hou, Y. (2016). Isolation and identification of bacterial strains with insecticidal activities from Rhynchophorus ferrugineus Olivier (Coleoptera: Curculionidae). J. Appl. Entomol. 140, 617–626. doi: 10.1111/jen.12293
Pu, Y., Ma, T., Hou, Y., and Sun, M. (2017). An entomopathogenic bacterium strain, Bacillus thuringiensis, as a biological control agent against the red palm weevil, Rhynchophorus ferrugineus (Coleoptera: Curculionidae). Pest. Manag. Sci. 73, 1494–1502. doi: 10.1002/ps.4485
Qin, Y., and Wade, P. A. (2017). Crosstalk between the microbiome and epigenome: messages from bugs. J. Biochem. 163, 105–112. doi: 10.1093/jb/mvx080
Ricci, I., Damiani, C., Capone, A., DeFreece, C., Rossi, P., and Favia, G. (2012). Mosquito/microbiota interactions: from complex relationships to biotechnological perspectives. Curr. Opin. Microbiol. 15, 278–284. doi: 10.1016/j.mib.2012.03.004
Ryu, J.-H., Kim, S.-H., Lee, H.-Y., Bai, J. Y., Nam, Y.-D., Bae, J.-W., et al. (2008). Innate immune homeostasis by the homeobox gene caudal and commensal-gut mutualism in Drosophila. Science 319, 777–782. doi: 10.1126/science.1149357
Schulthess, J., Pandey, S., Capitani, M., Rue-Albrecht, K. C., Arnold, I., Franchini, F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 1–14. doi: 10.1016/j.immuni.2018.12.018
Shi, Z., Lin, Y., and Hou, Y. (2014). Mother-derived trans-generational immune priming in the red palm weevil, Rhynchophorus ferrugineus Olivier (Coleoptera, Dryophthoridae). Bull. Entomol. Res. 104, 742–750. doi: 10.1017/S0007485314000583
Shin, S. C., Kim, S.-H., You, H., Kim, B., Kim, A. C., Lee, K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. doi: 10.1126/science.1212782
Vinolo, M. A., Rodrigues, H. G., Nachbar, R. T., and Curi, R. (2011). Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. doi: 10.3390/nu3100858
Wang, G. H., Zhang, X., Hou, Y. M., and Tang, B. Z. (2015). Analysis of the population genetic structure of Rhynchophorus ferrugineus in Fujian, China, revealed by microsatellite loci and mitochondrial COI sequences. Entomol. Exp. Appl. 155, 28–38. doi: 10.1111/eea.12282
Weiss, B. L., Maltz, M., and Aksoy, S. (2012). Obligate symbionts activate immune system development in the tsetse fly. J. Immunol. 188, 3395–3403. doi: 10.4049/jimmunol.1103691
Wong, A. C., Dobson, A. J., and Douglas, A. E. (2014). Gut microbiota dictates the metabolic response of Drosophila to diet. J. Exp. Biol. 217, 1894–1901. doi: 10.1242/jeb.101725
Xiao, R., Wang, X., Xie, E., Ji, T., Li, X., Muhammad, A., et al. (2019). An IMD-like pathway mediates the intestinal immunity to modulate the homeostasis of gut microbiota in Rhychophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Dev. Comp. Immunol. 97, 20–27. doi: 10.1016/j.dci.2019.03.013
Zhang, W., Chen, J., Keyhani, N. O., Zhang, Z., Li, S., and Xia, Y. (2015). Comparative transcriptomic analysis of immune responses of the migratory locust, Locusta migratoria, to challenge by the fungal insect pathogen, Metarhizium acridum. BMC Genomics 16:e867. doi: 10.1186/s12864-015-2089-9
Keywords: gut microbiota, Rhynchophorus ferrugineus, insect immunity, invasive biology, symbiosis
Citation: Muhammad A, Habineza P, Ji T, Hou Y and Shi Z (2019) Intestinal Microbiota Confer Protection by Priming the Immune System of Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Dryophthoridae). Front. Physiol. 10:1303. doi: 10.3389/fphys.2019.01303
Received: 14 May 2019; Accepted: 27 September 2019;
Published: 16 October 2019.
Edited by:
Senthil-Nathan Sengottayan, Manonmaniam Sundaranar University, IndiaReviewed by:
Cecilia Stahl Vieira, Oswaldo Cruz Foundation (Fiocruz), BrazilJean-Michel Drezen, Université de Tours, France
Copyright © 2019 Muhammad, Habineza, Ji, Hou and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youming Hou, ymhou@fafu.edu.cn; Zhanghong Shi, shizh@fafu.edu.cn
 Abrar Muhammad
Abrar Muhammad Prosper Habineza
Prosper Habineza Tianliang Ji
Tianliang Ji Youming Hou
Youming Hou Zhanghong Shi
Zhanghong Shi