
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 25 July 2019
Sec. Aquatic Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.00952
Tolls/Toll-like receptors (TLRs) play a key role in innate immunity by detecting the invading microbes and subsequently activating downstream signaling cascades. In this study, two new molluscan Toll members (designed as HcToll6 and HcToll7) were identified from triangle-shell pearl mussel (Hyriopsis cumingii). The obtained HcToll6 full-length cDNA was 3207 bp consisting of a 2223 bp open reading frame (ORF) that encoded a peptide of 740 amino acids. HcToll7 cDNA is a 3216 bp molecule that contains an ORF of 2139 bp encoding a protein of 712 amino acids. The deduced HcToll6 and HcToll7 proteins share two common structures: extracellular leucine-rich repeat (LRR) domains and intracellular Toll/interleukin-1 receptor (TIR) domain. Quantitative real-time PCR results showed that HcToll6 and HcToll7 were mainly expressed in the hepatopancreas and the gills, and they responded rapidly to bacterial stimulation. RNA interference by dsRNA results revealed that HcToll6 and HcToll7 RNAi strongly decreased the expression of lysozyme (HcLyso) and defensin (HcDef) in the gills of RNAi-treated mussels with Vibrio parahaemolyticus challenge. As a pattern recognition receptor, the prokaryotic expressed the recombinant LRR domains of HcToll6 and HcToll7 (rHcToll6-LRR and rHcToll7-LRR) could bind to Gram-positive and Gram-negative bacteria and had a strong tendency to recognize lipopolysaccharide (LPS) and peptidoglycan (PNG). rHcToll6-LRR and rHcToll7-LRR exhibited a significant in vitro bactericidal activity against V. parahaemolyticus and Staphylococcus aureus. These findings provide useful information to characterize Tolls in mussels.
Lacking a true adaptive immune system, invertebrates against infectious agents mainly rely on innate immunity (Loker et al., 2004). Innate immune response is the main line of defense, triggering various humoral and cellular activities through signal transduction pathways to defense against microbial infections (Akira et al., 2006). Toll, immune deficiency (IMD), and Janus family tyrosine kinase and signal transducer and activator of transcription (JAK/STAT) pathways are regarded as the three main pathways regulating the immune response of invertebrates (Li and Xiang, 2013). During pathogen invasion, conserved pathogen-associated molecular patters (PAMPs), such as lipopolysaccharide (LPS), peptidoglycan (PNG), lipoteichoic acid, and double-stranded viral RNA, can be discriminated by a wide range of pattern recognition receptors (PRRs) (Medzhitov and Janeway, 2000; Janeway and Medzhitov, 2002; Leulier and Lemaitre, 2008). An important class of PRRs is the Toll receptor superfamily, which includes vertebrate Toll-like receptors (TLRs) and invertebrate Tolls (Takeda and Akira, 2004; Hopkins and Sriskandan, 2005). When triggered by pathogens, the Toll pathway, such as in insects, is induced to activate Spätzle (Yuan et al., 2017). Afterward, Spätzle binds to Toll receptor and mediates downstream molecules containing Myeloid differentiation factor 88 (MyD88), Tube, Pelle, tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6), Cactus, and Dorsal (Anderson, 2000; Silverman and Maniatis, 2001). These cascades ultimately activate NF-κB transcription factors, which regulate the synthesis of specific target genes, such as antibacterial peptide genes (AMPs) (Lemaitre and Hoffmann, 2007).
Toll receptor (dToll) is originally identified in Drosophila melanogaster and then considered as a core component of the anti-Gram-positive bacterial and anti-fungal response in Drosophila signaling pathway (Lemaitre et al., 1996; Kawai and Akira, 2006). At present, 9 Tolls (dToll1-9) in Drosophila (Bilak et al., 2003; Valanne et al., 2011) and 13 members of TLRs (TLR1-13) in mammals (Roach et al., 2005; Akira et al., 2006) have been identified. Tolls and their homolog TLRs are evolutionarily conserved and have crucial roles in the recognition of microbial pathogens (Beutler, 2004). They are a family of transmembrane proteins (type I protein) characterized by a transmembrane region, extracellular leucine-rich repeat (LRR) domains, and an intracellular Toll/interleukin-1 receptor (TIR) domain (Funami et al., 2004; Coscia et al., 2011). Upon the recognition of ligands by LRR domains, the cytoplasmic TIR domain interacts with downstream TIR-containing adaptor proteins, which activate the TLR-mediated signaling cascade (Akira et al., 2006; Valanne et al., 2011). A large number of Tolls/TLRs from various species are involved in the immune responses of hosts to infectious pathogens. For instance, a TLR (CfTLR) from Zhikong scallop Chlamys farreri can activate diverse downstream reactions, including antibacterial activity, antioxidant response, and apoptosis against Vibrio anguillarum (Wang et al., 2011). Four TLR homologs (CgTLRs) have been identified in Crassostrea gigas, and they have a consistent challenge response to multiple PAMPs and are involved in hemocyte activation and TNF induction induced by bacterial infection (Zhang et al., 2013). The expression of CnTLR-1 increases after Vibrio parahaemolyticus, LPS, and Poly I:C challenge and acts as an important factor for an immune defense against pathogens in noble scallop (Chlamys nobilis) (Lu et al., 2016).
Triangle-shell pearl mussel (Hyriopsis cumingii) has been widely cultured for freshwater pearl production in China (Li et al., 2005). The frequent outbreak of diseases causes high mortality and large economic losses to mussel aquaculture. Understanding the native response against infectious diseases is crucial for the sustainable production of this species. In previous studies, seven Tolls, namely, HcToll1 (Ren et al., 2013), HcToll2-1 (Ren et al., 2014), HcTollI, HcToll2-2 (Cao et al., 2016), HcToll3 (Zhang et al., 2017), HcToll4, and HcToll5 (Huang Y. et al., 2018), have been cloned and identified in H. cumingii, and they play crucial roles in bacterial and viral infections. However, the significance of mussel Tolls to immune responses at a protein level has been rarely discussed, and we have yet to fully elucidate the molecular mechanisms involved in how this pathway regulates immune responses. Here, two new Toll family members, designated as HcToll6 and HcToll7, were identified in H. cumingii; their expression in tissues and changes after bacterial infection were examined. The functions of HcToll6 and HcToll7 were also analyzed in antibacterial immunity. The results of this study offer helpful information to characterize Tolls in mussels.
A total of 100 adult H. cumingii with a full shell length of 6–7 cm were obtained from a farm of Wuhu City, China. All the mussels were cultured in our laboratory aeration tank with recirculating water at 25°C for 2 weeks before any experiments.
For the bacterial challenge, 50 μL of activated Staphylococcus aureus (3 × 107 cells) or V. parahaemolyticus (3 × 107 cells) were injected into the adductor muscles of H. cumingii (each group contained 20 mussels). Of these mussels, 20 were similarly injected with 1 mL of PBS (140 mM NaCl; 3 mM KCl; 8 mM Na2HPO4; 1.5 mM KH2PO4, pH 7.4) to serve as controls. The gills and the hepatopancreas of five mussels from each group were collected at 2, 6, 12, and 24 h post-injection and used for the isolation of total RNA. Tissues (hemocytes, hepatopancreas, gills, and mantle) were also collected from untreated mussels for tissue distribution studies.
The cDNA fragments amplified by four specific primers (HcToll6-RNAi-F: 5′-GCGTAATACGACTCACTATAGGCCCGGGCGTGGCAAGATG-3′ and HcToll6-RNAi-R: 5′-GCGTAATACGACTCACTATAGGCAGAGACTCGAGGTATGG-3′; HcToll7-RNAi-F: 5′-GCGTAATACGACTCACTATAGGGGAGAGAACTTGGACAGG-3′ and HcToll7-RNAi-R: 5′-GCGTAATACGACTCACTATAGGGGGCATTCAAAGGCAGACG-3′) linked to the T7 promoter were used as templates for the synthesis of double-stranded RNA (dsRNA) by using T7 RNA polymerase (Fermentas, Waltham, MA, United States) in accordance with previously described methods (Wang et al., 2012). The fragment of green fluorescent protein (GFP) was also obtained with the primers GFP-RNAi-F: 5′-GCGTAATACGACTCACTATAGGTGGTCCCAATTCTCGTGGAAC-3′ and GFP-RNAi-R: 5′-GCGTAATACGACTCACTATAGGCTTGAAGTTGACCTTGATGCC-3′ to synthesize the control dsRNA. Each mussel of the experimental groups (HcToll6-RNAi, HcToll7-RNAi) and the control group (GFP-RNAi) were coinjected with 15 μg of corresponding dsRNA and 50 μL of V. parahaemolyticus (3 × 107 cells). At the same time, V. parahaemolyticus (3 × 107 cells) alone was injected into the mussels as control. After 12 h post-injection, 15 μg of dsHcToll6, dsHcToll7, or dsGFP was injected into the same mussel. The gills of the mussel in each group were collected separately for RNA extraction 24 h after the last injection. The experiments were biologically repeated three times.
Total RNA was extracted from the same tissues by using an RNApure high-purity total RNA rapid extraction kit (Spin-Column; BioTeke, Beijing, China) in accordance with the manufacturer’s protocol. The quality and concentration were checked through agarose gel electrophoresis and NanoDrop 2000 (Beijing GenoStar Biotech Co., Ltd., Beijing, China). First-strand cDNA synthesis for quantitative real-time PCR (qPCR) analysis was performed using a PrimeScript® first-strand cDNA synthesis kit (Takara, Japan) to transcribe poly (A) mRNA with the Oligo-d(T) Primer and random 6-mer primers. The total RNA obtained from the hepatopancreas was also reverse transcribed via a Clontech SMARTerTM RACE cDNA amplification kit (Clontech, Palo Alto, CA, United States) with 5′-CDS Primer A and SMARTerIIA oligos for 5′-RACE Ready cDNA, and 3′-CDS Primer A for 3′-RACE-Ready cDNA. Manufacturer-recommended reaction conditions were used.
The partial cDNA sequences of HcToll6 and HcToll7 were retrieved from our previous hepatopancreas transcriptome database (unpublished). In this experiment, 3′ and 5′ RACE were performed by using a Clontech Advantage® 2 PCR kit from Takara (Japan) with two pairs of specific primers (HcToll6-F:5′-TGGATAAACGATGTGCTAAGCGACCCC-3′, HcToll6-R: 5′-CTGTGAAGACCACGCAAATAGAGACGGAAC-3′; HcToll7-F: 5′-GAGAGAACTTGGACAGGAAAGGGGGCT-3′, HcToll7-R: 5′-CGTATCGGCAGGTCGCCAAGGGTAAC-3′) to obtain the full-length cDNAs of HcToll6 and HcToll7. The amplification products were purified using a DNA gel extraction kit (Shanghai Generay Biotech Co., Ltd., Shanghai, China), inserted into the pEasy-T3 vector, and transformed into Escherichia coli Trans1-T1 cells (TransGen Biotech, Beijing, China). The putative clones were identified by PCR with M13F and M13R primers. The selected clones were sequenced by a commercial company (Springen, China).
The BLAST algorithm1 was used to analyze the nucleotide and protein sequence homologs. The amino acid sequences of HcToll6 and HcToll7 were translated using ExPASy2. The signal peptide was predicted using SignalP 4.1 Server3, and the domain organization was identified by SMART4. The ExPASy Compute pI/Mw tool5 was used to predict the molecular weight and isoelectric point of HcToll6 and HcToll7. Multiple amino acid sequence alignments were generated with DNAMAN and GENDOC by using the ClustalX 2.06. The phylogenetic relationships among the Toll homologs were determined by constructing a phylogenetic tree via the neighbor-joining (NJ)-embedded algorithm with MEGA 7 and the bootstrapping of 1000 replicates (Kumar et al., 2016).
qPCR was conducted in accordance with previously described methods (Huang et al., 2013) to examine the expression profiles of HcToll6 and HcToll7 transcripts in various samples. The gene specific primers for HcToll6 (HcToll6-RT-F: 5′-AAGTGTTATCTGTTCCGTCTCT-3′ and HcToll6-RT-R: 5′-GTGTATGTTAGTGGGTGGTATCT-3′) and HcToll7 (HcToll7-RT-F: 5′-AGCGAAGAAGAGAATGGG-3′ and HcToll7-RT-R: 5′-AAAGGCAGACGATAGATAGG-3′) were designed using Premier version 5.0. After HcToll6 and HcToll7 were knocked down, two immune-related gene expression levels were analyzed with the primers HcLyso-RT-F: 5′-CTTCTTTCTTGTTGGTCTGC-3′ and HcLyso-RT-R: 5′-CTGGTAGTAGCCACAGGACA-3′; HcDef-RT-F: 5′-GGTGTCGTCTATCTTGCTTC-3′ and HcDef-RT-R: 5′-AGGTTATTTGGTCATCTATTTTG-3′. β-actin was also amplified as a reference gene by using the primers β-actin-RT-F: 5′-GTGGCTACTCCTTCACAACC-3′ and β-actin-RT-R: 5′-GAAGCTAGGCTGGAACAAGG-3′. The data were calculated by the 2-Δ ΔCt methods (Livak and Schmittgen, 2001) and were subjected to statistical analysis. An unpaired sample t-test was conducted, and differences were considered significant at P < 0.05 and extremely significant at P < 0.01.
cDNA fragments encoding the LRR domains of HcToll6 (480 amino acids) and HcToll7 (446 amino acids) were amplified from the hepatopancreas cDNA by using the primers HcToll6-LRR-ex-F: 5′-GGATCCCCAGGAATTCCCACATGTCCAGCGAACTGCA-3′ and HcToll6-LRR-ex-R: 5′-GATGCGGCCGCTCGAGTTAGGTTGGTTTGGCATCTTTCAA-3′; HcToll7-LRR-ex-F: 5′-GGTGATCCCCAGGAATTCCCCCATCACTGATTCGATTTCT-3′ and HcToll7-LRR-ex-R: 5′-GATGCGGCCGCTCGAGTTATCTGCAATCTTCATCCGTTGG-3′. EcoRI and XhoI restriction sites were inserted at the beginning and end of the DNA fragments. After digestion occurred, the PCR products were cloned into the pGEX-6p-2 vector. Recombinant plasmids were transformed into E. coli BL21 (DE3) cells (TransGen Biotech, China), which were then cultured in Luria–Bertani (LB) medium (100 μg/mL ampicillin) at 37°C and 200 rpm. When OD600 of the culture reached 0.6, isopropyl-β-D-thiogalactoside (IPTG; 0.5 mM) was added. After 5 h of culture at 28°C and 200 rpm, the cells were collected by centrifugation at 6000 rpm for 10 min. They were then resuspended in PBS containing 0.1% Triton X-100. After cell sonication and centrifugation were accomplished, the proteins were purified using Glutathione Sepharose 4B chromatography (Gen-Script, United States) in accordance with the manufacturer’s instructions. The purified proteins were analyzed by 12.5% SDS-PAGE and stained with Coomassie brilliant blue G250.
Seven bacterial species, namely, S. aureus, Micrococcus luteus, Bacillus subtilis, Bacillus thuringiensis, V. parahaemolyticus, Aeromonas hydrophila, and E. coli, were used to measure the binding activities of rHcToll6-LRR and rHcToll7-LRR by using a binding assay described previously (Huang et al., 2014). After the bacteria were cultivated, the microorganisms were collected by centrifugation at 6000 rpm for 5 min. Microbial pellets were washed three times with tris buffered saline (TBS; 20 mM Tris–HCl, 150 mM NaCl, pH 7.4) and resuspended in TBS. Each microorganism (approximately 2 × 108 cells) was incubated in 100 μg of rHcToll6-LRR or rHcToll7-LRR by gentle rotation for 60 min at 37°C. After centrifugation at 6000 rpm for 5 min, the harvested cells were rinsed three times with TBS. All of the fractions were analyzed through 12.5% SDS-PAGE, transferred onto a nitrocellulose membrane, and revealed through Western blot analysis by using an anti-GST rabbit antibody (TransGen, Beijing, China).
Enzyme-linked immunosorbent assay (ELISA) was conducted on the basis of a previous study (Huang et al., 2014) to detect the direct binding of rHcToll6-LRR and rHcToll7-LRR to LPS (E. coli serotype 055: B5) and PGN (M. luteus) (Sigma, St. Louis, MO, United States). LPS and PGN were initially dissolved in sterile water at a concentration of 80 μg/mL. Each well of the 96-well plate was then coated with 50 μL of sugar solution. The plate was then incubated overnight at 37°C and heated for 30 min at 60°C. Each well was blocked with 200 μL of bovine serum albumin (BSA; 1 mg/mL) at 37°C for 2 h and then washed with TBS four times. The purified proteins with gradient dilution (0.78, 1.56, 3.125, 6.25, 12.5, and 25 μg/mL in TBS containing 0.1 mg/mL BSA) were added to the wells and incubated at 37°C for 3 h. The wells were washed four times. Afterward, 100 μL of rabbit monoclonal anti-GST antibody (1/2000 diluted in 0.1 mg/mL BSA) was then added and incubated at 37°C for 1 h. Each well was washed four times and then incubated with 100 μL of peroxidase-conjugated goat anti-rabbit IgG (1/5000 diluted) at 37°C for 1 h. The plate was washed four times with TBS and developed with 0.01% 3,3′,5,5′-tetramethylbenzidine (Sigma). Approximately 2 M H2SO4 was used to stop the reaction. Absorbance was read and noted at 450 nm by using a plate reader (BioTek Instruments, Winooski, VT, United States). The assays were performed in triplicate.
The inhibitory effects of rHcToll6-LRR and rHcToll7-LRR on S. aureus and V. parahaemolyticus were tested as described in our previous report with slight modifications (Huang Y. et al., 2016). In this experiment, 50 μL of bacteria was transferred to 5 mL of LB broth, and rHcToll6-LRR and rHcToll7-LRR were separately added at a final concentration of 100 μg/mL. TBS and GST-tag protein served as a blank group and a negative control group, respectively. Each sample was incubated at 37°C with shaking at 200 rpm, and bacterial growth was monitored by checking OD600 every 1 h.
The complete cDNA sequences of HcToll6 and HcToll7 were obtained by using RACE technology. The full-length HcToll6 cDNA was 3207 bp, and it consisted of a 2223 bp open reading frame (ORF) that encoded a peptide of 740 amino acids, a 52 bp 5′ untranslated region (UTR), and a 932 bp 3′ UTR with a potential polyadenylation signal (AATAAA) and a poly (A) tail (Supplementary Figure S1A). SignalP analysis showed that HcToll6 had a signal peptide with 24 amino acids (aa) at the N-terminal. SMART analysis predicted that the deduced HcToll6 protein contained 12 LRR residues, including nine 21–30 aa LRRs, a 32 aa LRR N-terminal domain (LRR-NT), a 24 aa LRR typical (most populated) subfamily domain (LRR-TYP), and a 55 aa LRR C-terminal domain (LRR-CT) in the extracellular domain, a 23 aa transmembrane domain, and a 141 aa intracellular TIR domain (Figure 1). The molecular mass of the mature protein was 86.1 kDa, and the estimated isoelectric point was 7.04. HcToll7 cDNA was 3216 bp, and it was composed of an 84 bp 5′ UTR, a 993 bp 3′ UTR with poly (A), and a 2139 bp ORF encoding a 712 aa HcToll7 protein with a deduced molecular mass of 82.1 kDa and a theoretical pI of 8.69 (Supplementary Figure S1B). Similar to HcToll6 domain organization, HcToll7 contains a 31 aa signal peptide, eight 20–24 aa LRR domains, a 58 aa LRR-CT domain, a 20 aa transmembrane domain, and a 142 aa TIR domain (Figure 1).
The multiple sequence alignment analysis of HcToll6 and HcToll7 showed that they had different amino acid compositions (Supplementary Figure S2). BLAST analysis indicated that the deduced amino acid sequences of HcToll6 and HcToll7 were similar to the amino acid sequences of the Tolls from Mytilus galloprovincialis (XP_021343306.1, 28, 31% similarity), Crassostrea virginica (XP_022327704.1, 28, 31% similarity), Argopecten irradians (AVP74315.1, 26, 31% similarity), and Lingula anatina (XP_013415421.1, 25, 28% similarity). A phylogenetic tree was constructed on the basis of the homologous analysis of 9 Tolls from H. cumingii and 31 Tolls from other species. The relationship showed that the Tolls of H. cumingii were clustered independently, and HcToll6 and HcToll7 had a relatively close evolutionary position with the reported Tolls from M. galloprovincialis, C. virginica, A. irradians, and L. anatina (Figure 2).
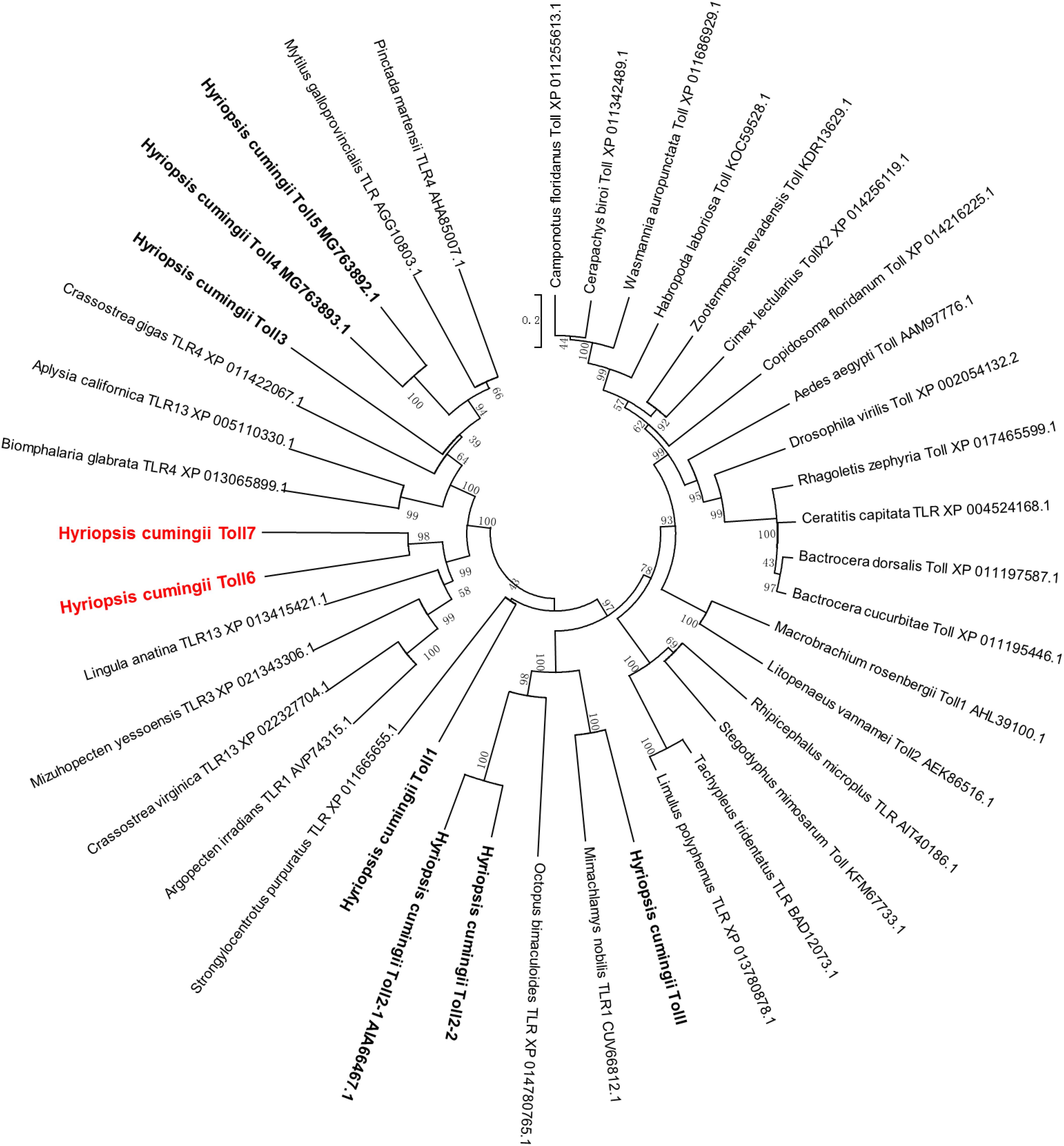
Figure 2. Phylogenetic analysis of HcToll6 and HcToll7 with other Tolls or TLRs. The repeatability of the results was checked by performing 1000 bootstraps on the NJ trees. The scale bar indicates a branch length of 0.2.
The tissue distributions of HcToll6 and HcToll7 were investigated through qPCR. HcToll6 and HcToll7 transcriptions were constitutively expressed in the tested tissues, including hemocytes, hepatopancreas, gills, and mantle, when their expression levels were normalized to the housekeeping gene β-actin. HcToll6 and HcToll7 mRNAs were highly expressed in the hepatopancreas, moderately expressed in the gills, and weakly expressed in the hemocytes and the mantle (Figure 3).
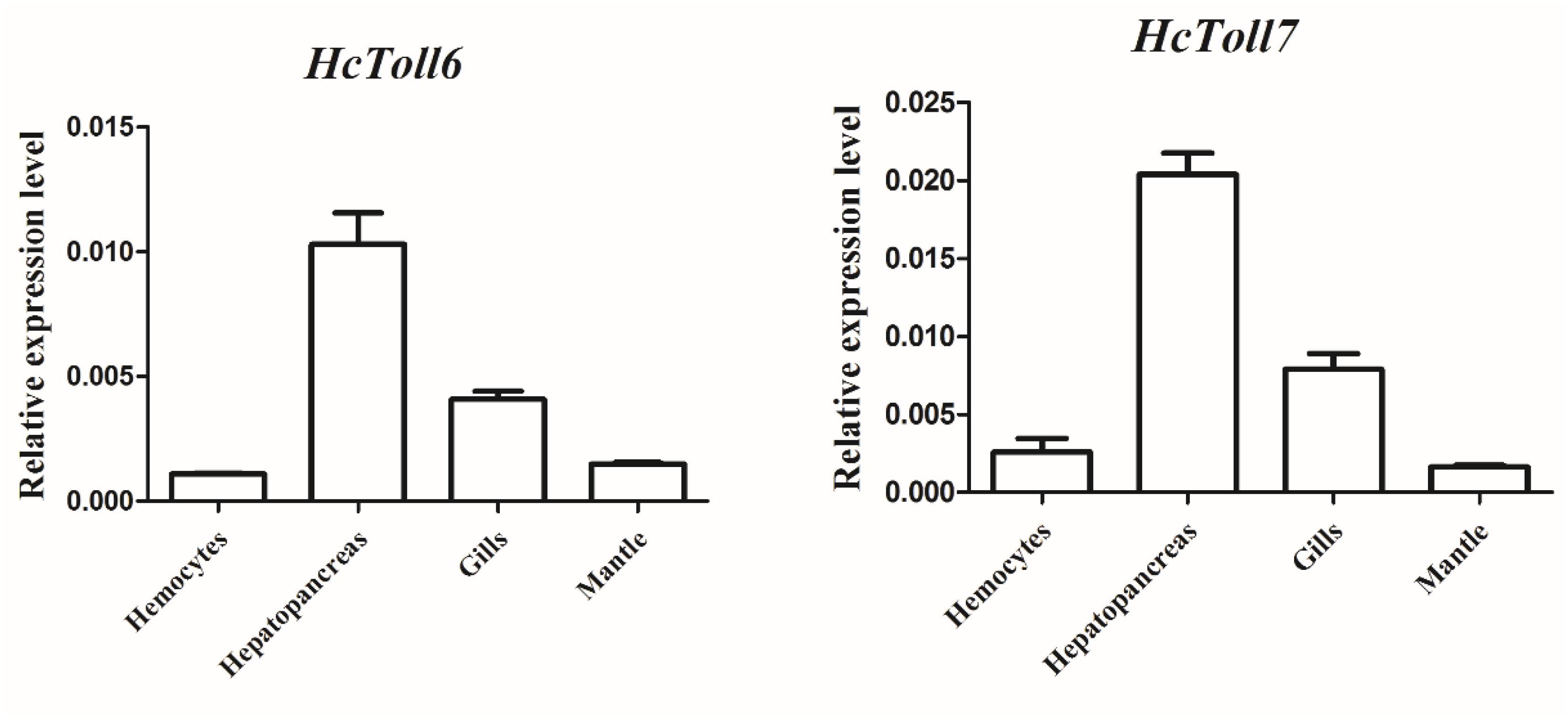
Figure 3. Expression profiles of HcToll6 and HcToll7 in various tissues. The 2-Δ ΔCt method was utilized in the analysis of the expression patterns of HcToll6 and HcToll7 with β-actin as the reference gene. Error bars represent SD.
After treatment with S. aureus, the HcToll6 transcript level in the gills was obviously downregulated at 12 h and then gradually upregulated at 24 h (Figure 4A), whereas HcToll7 significantly decreased at 2 h and kept a relatively high level from 6 h to 24 h (Figure 5A). HcToll6 and HcToll7 transcription levels in the hepatopancreas were upregulated gradually from 2 h, reached a peak at 12 h, and subsequently remained relatively high until 24 h (Figures 4B, 5B). After the V. parahaemolyticus challenge, the mRNA expression of HcToll6 in the gills slowly decreased at 2 h, gradually increased, and reached a maximum level at 24 h (Figure 4C). HcToll7 expression increased at 2 h, peaked at 12 h, and decreased at 24 h (Figure 5C). In the hepatopancreas, HcToll6 expression reached its first peak at 2 h and decreased from 6 h to 12 h. The expression level reached the second peak at 24 h (Figure 4D). HcToll7 expression increased continuously from 2 h to 12 h and peaked at 24 h (Figure 5D). No changes in the HcToll6 and HcToll7 expression levels of the PBS-injected group were detected for 24 h.
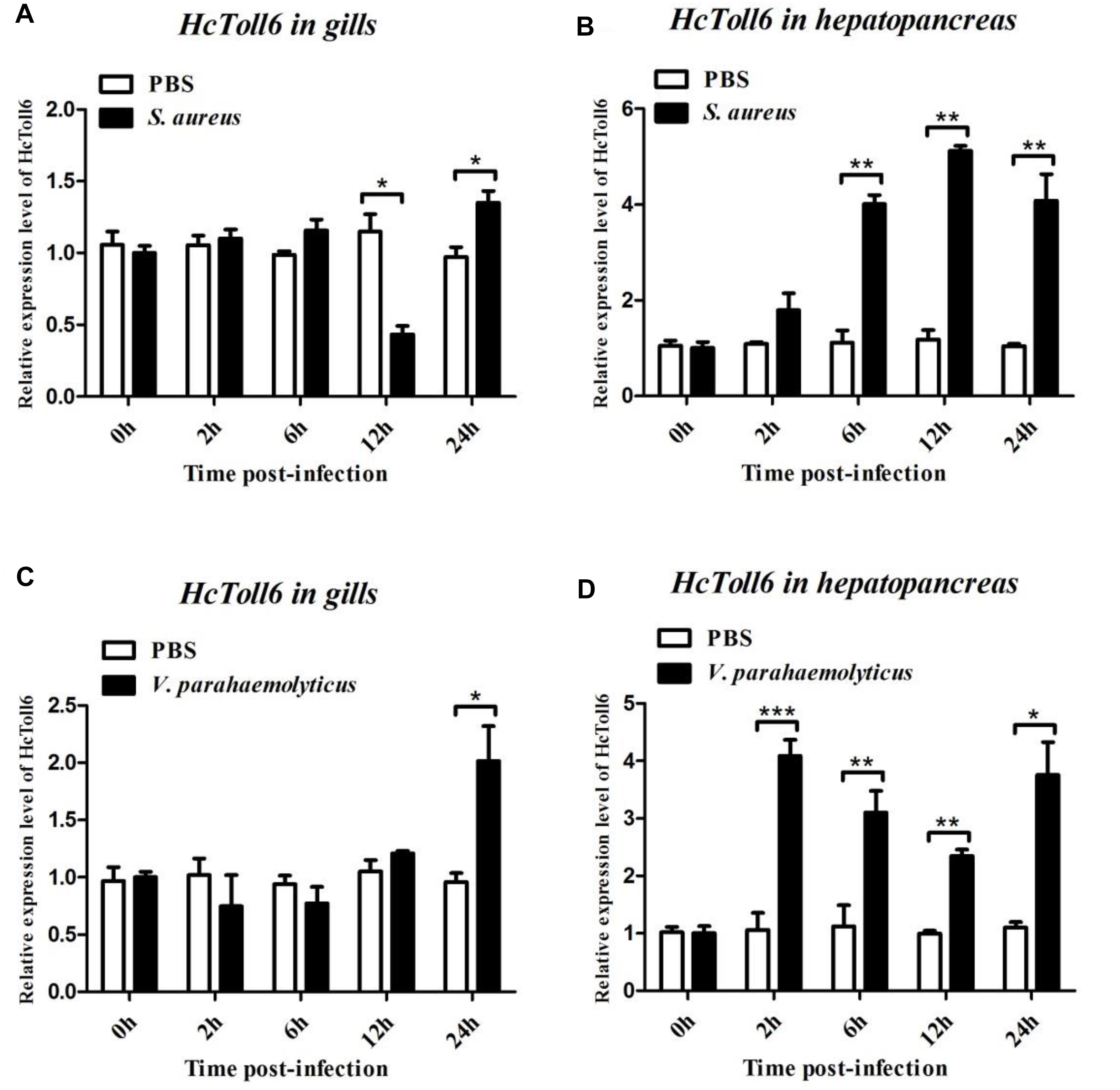
Figure 4. Time-course expression analysis of HcToll6 gene in the gills (A,C) and hepatopancreas (B,D) at 0, 2, 6, 12, and 24 h after injection with S. aureus and V. parahaemolyticus. Expression level was determined by qPCR and normalized to β-actin in triplicate. Statistically significant differences were analyzed by t-test and indicated by asterisks (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
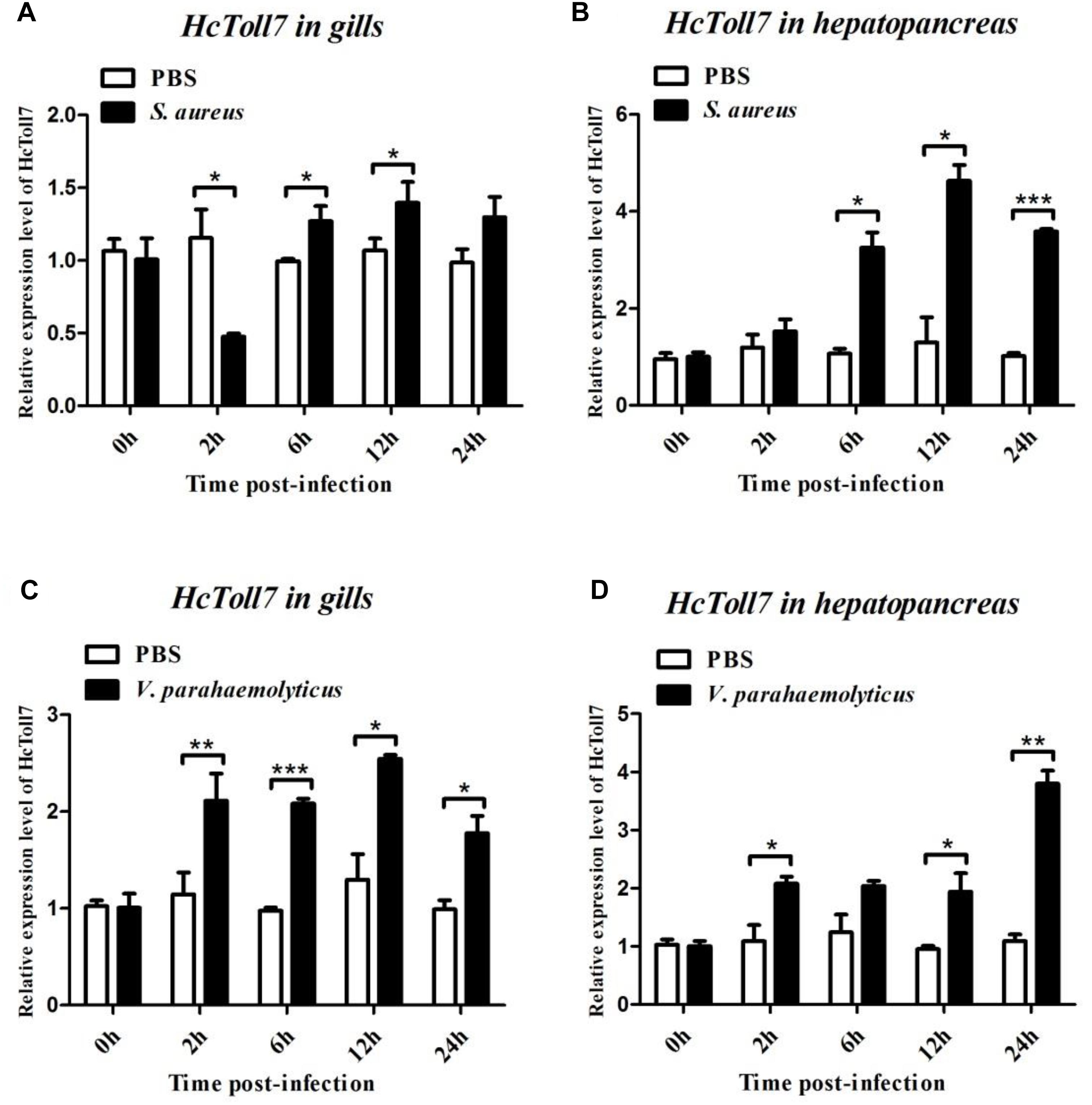
Figure 5. Time-course expression analysis of HcToll7 gene in the gills (A,C) and hepatopancreas (B,D) at 0, 2, 6, 12, and 24 h after injection with S. aureus and V. parahaemolyticus. Expression level was determined by qPCR and normalized to β-actin in triplicate. Statistically significant differences were analyzed by t-test and indicated by asterisks (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
HcToll6 and HcToll7 in H. cumingii were knockdown by dsHcToll6 and dsHcToll7, respectively, to investigate the contribution of HcToll6 and HcToll7 in mussel immunity. In Figures 6A,D, the transcriptional levels of HcToll6 and HcToll7 in the gills were suppressed after dsHcToll6 and dsHcToll7 injection relative to those of the normal group, indicating that silencing was gene specific. The mRNA expression levels of lysozyme (HcLyso) and defensin (HcDef) in the gills were highly induced after stimulation with V. parahaemolyticus. The transcripts of these genes were considerably downregulated in the gills of HcToll6-silenced and HcToll7-silenced mussels challenged with live bacteria in comparison with those in the dsGFP and V. parahaemolyticus only injection groups (Figures 6B,C,E,F). These results implied that HcToll6 and HcToll7 might play a positive role in the regulation of AMPs.
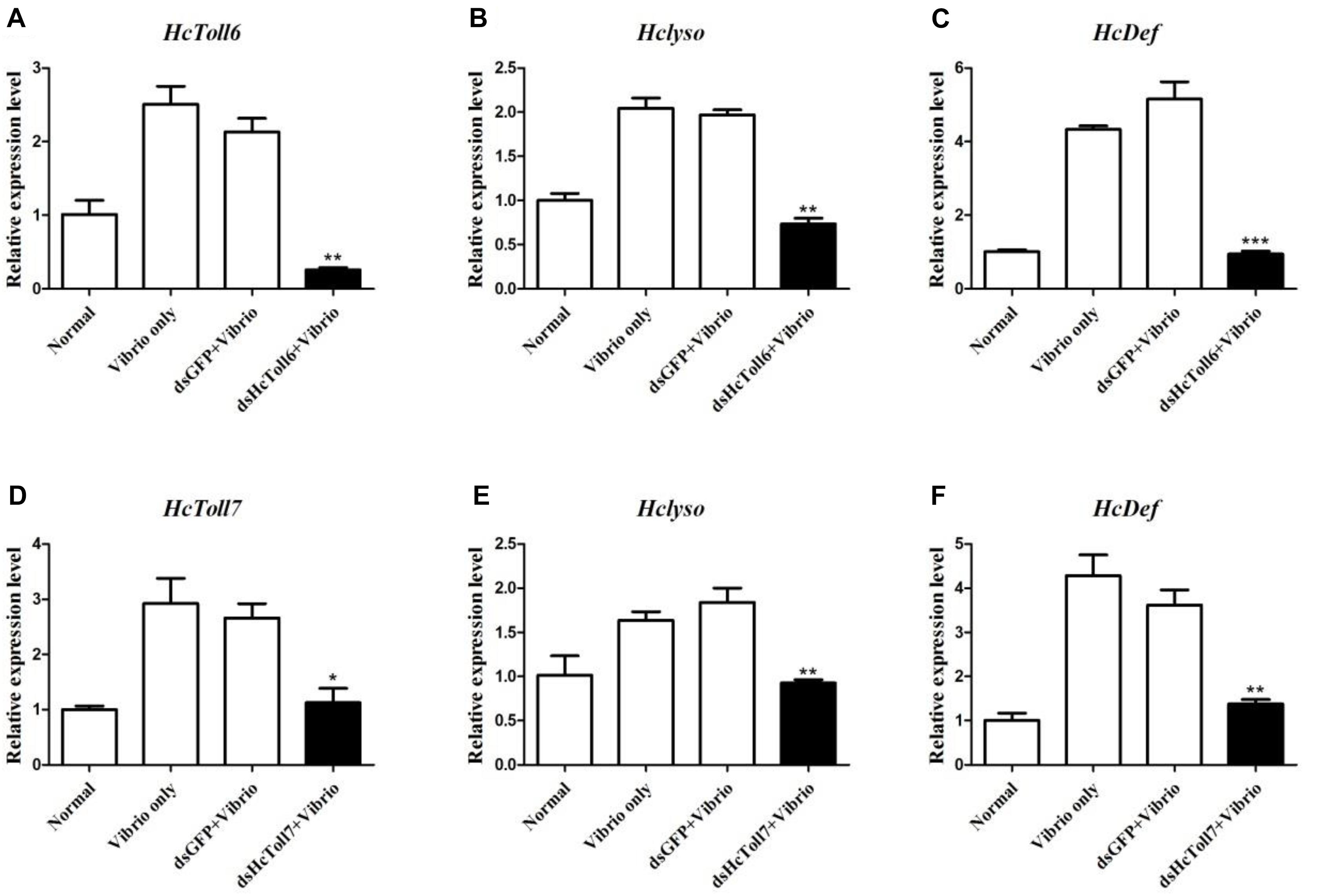
Figure 6. AMP expression patterns in HcToll6 (A–C) and HcToll7 (D–F) knockdown H. cumingii after V. parahaemolyticus infection. The mussels were divided into four groups (Normal, V. parahaemolyticus only, dsGFP and V. parahaemolyticus, dsHcToll6/dsHcToll7, and V. parahaemolyticus). The expression profiles of AMPs (HcLyso and HcDef) in the gills at 36 h after the first injection was investigated by qPCR. All data were obtained from at least three parallel experiments and were expressed as mean ± SD values; asterisks indicate significant differences (∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001).
The recombinant plasmids of pGEX-6p-2-HcToll6-LRR and pGEX-6p-2-HcToll7-LRR were expressed in the prokaryotic expression system after transformation into E. coli BL21 (DE3). After IPTG induction, SDS-PAGE analysis revealed that two proteins with approximate molecular weights of 81.7 and 77.7 kDa were induced (Figures 7A,B, lanes 2), which exactly matched the predicted molecular mass deduced from HcToll6 and HcToll7. Two purified products after Glutathione Sepharose 4B chromatography purification had only one band of approximately 81.7 and 77.7 kDa, which were the same size as the expected recombinant HcToll6-LRR (55.7 kDa) and HcToll7-LRR (51.7 kDa) with a 26 kDa vector GST-tag fragment (Figures 7A,B, lanes 3).
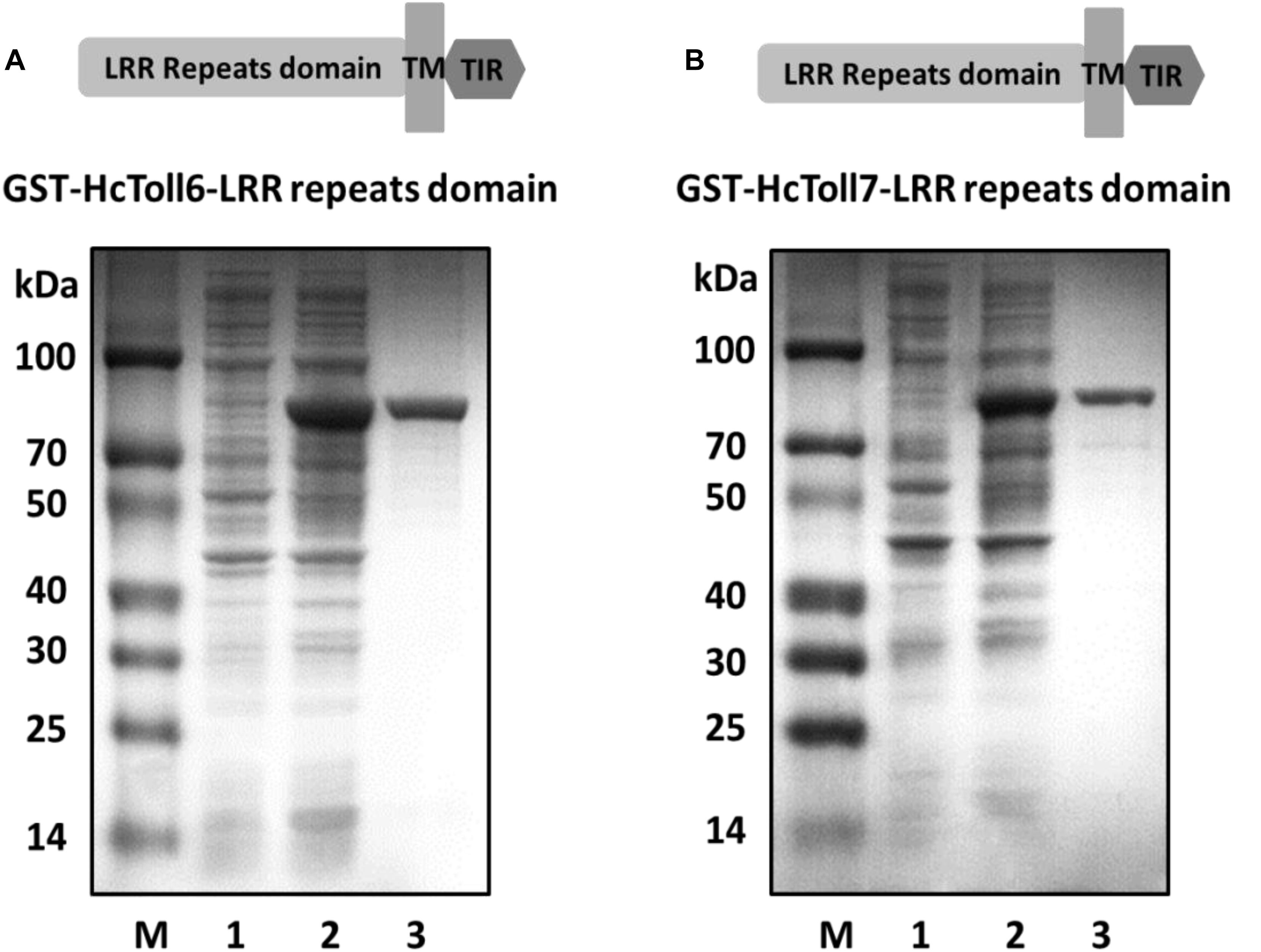
Figure 7. Pattern of recombinant HcToll6-LRR (A) and HcToll7-LRR (B) analyzed by 12.5% SDS-PAGE. Lane M, molecular weight markers; lane 1, total proteins of plasmids without induction; lane 2, total proteins of plasmids with IPTG induction; lane 3, purified rHcToll6-LRR/rHcToll7-LRR.
An in vitro binding assay was conducted to test the binding of rHcToll6-LRR and rHcToll7-LRR to microorganisms. Purified rHcToll6-LRR and rHcToll7-LRR could bind to Gram-positive bacteria (S. aureus, M. luteus, B. subtilis, and B. thuringiensis) and Gram-negative bacteria (V. parahaemolyticus, A. hydrophila, and E. coli) at different band intensities. Figure 8 shows that rHcToll6-LRR could bind more tightly to S. aureus than to others. With the same protein contents, rHcToll7-LRR seemed to bind to most tested bacteria more potently than rHcToll6-LRR (Figure 8). In Figure 9, the direct binding specificity of rHcToll6-LRR and rHcToll7-LRR to sugars was tested through ELISA. Purified rHcToll6-LRR and rHcToll7-LRR could bind to LPS and PGN in a dose-dependent manner. The binding activity of rHcToll6-LRR to LPS was higher than that of PGN, whereas rHcToll7-LRR exhibited a similar binding affinity to LPS and PGN. They could bind to a broad spectrum of microorganisms and polysaccharides, although they had distinct specificities.
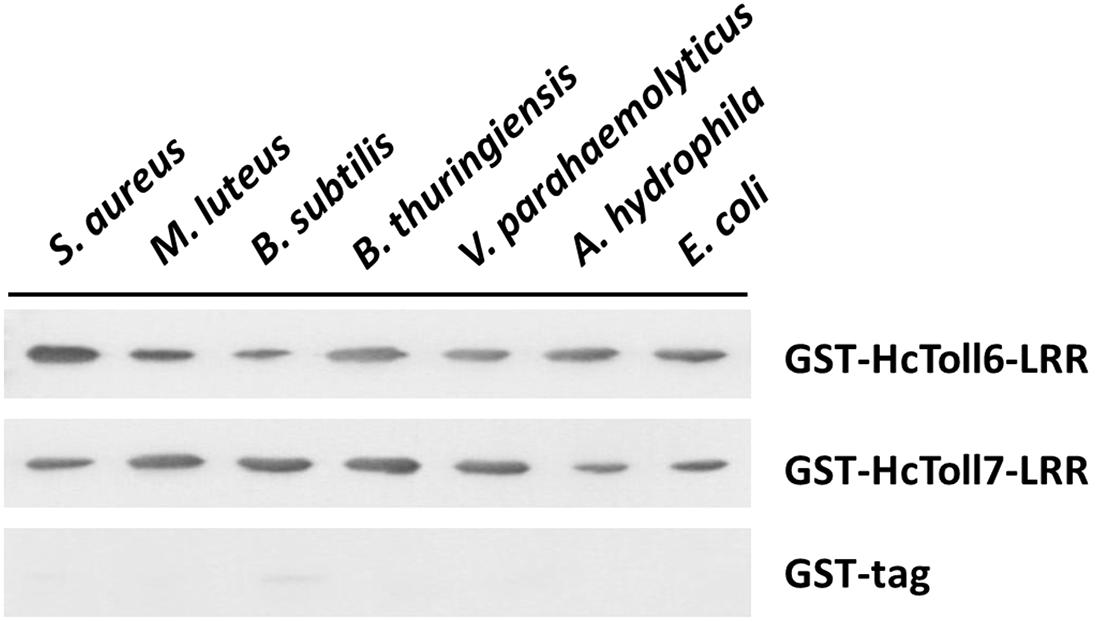
Figure 8. Binding of rHcToll6-LRR and rHcToll7-LRR toward microorganisms (Gram-positive bacteria S. aureus, M. luteus, B. subtilis, B. thuringiensis and Gram-negative bacteria V. parahaemolyticus, A. hydrophila, E. coli) were detected by Western blot analysis.
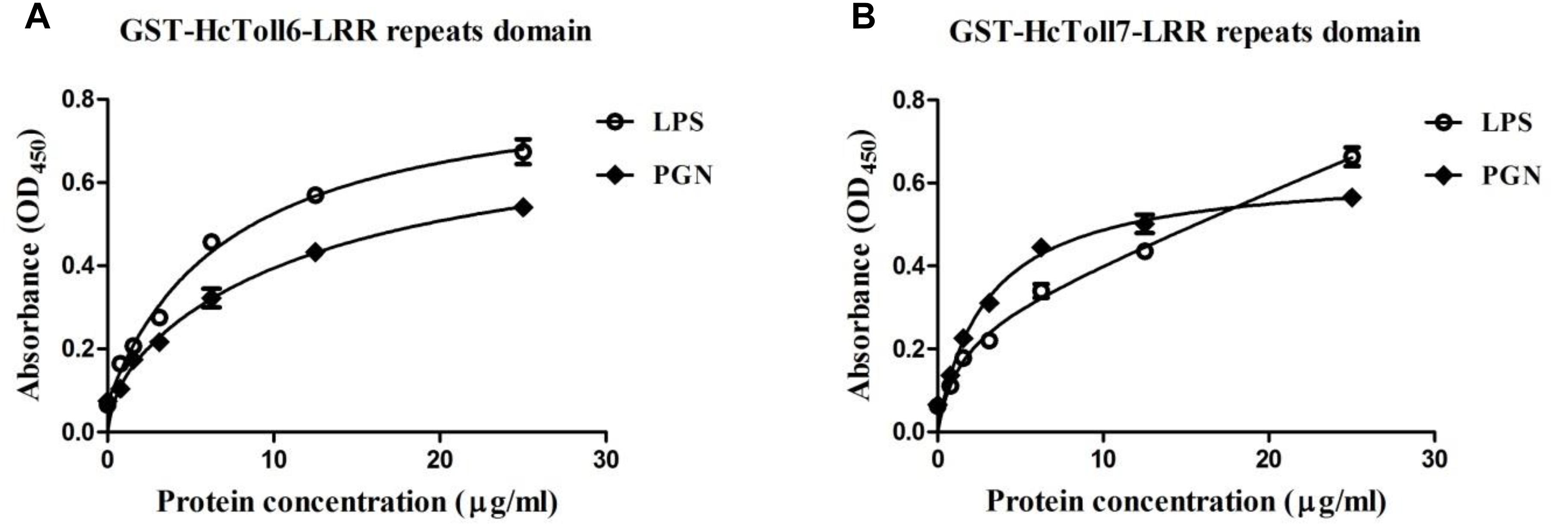
Figure 9. Binding of rHcToll6-LRR (A) and rHcToll7-LRR (B) to polysaccharides determined by ELISA. ELISA was used to quantify the binding of purified rHcToll6-LRR/rHcToll7-LRR to immobilized LPS and PGN.
rHcToll6-LRR and rHcToll7-LRR (100 μg/mL; TBS as the control) were used to examine their inhibitory effects on the growth of S. aureus and V. parahaemolyticus at different times and to determine the antimicrobial activity. The results showed that the growth of S. aureus and V. parahaemolyticus incubated with rHcToll6-LRR and rHcToll7-LRR for 8 h was inhibited significantly compared with that of the control group (Figure 10). No obvious inhibitory activity was observed against the bacteria with GST-tag protein(data were not shown).
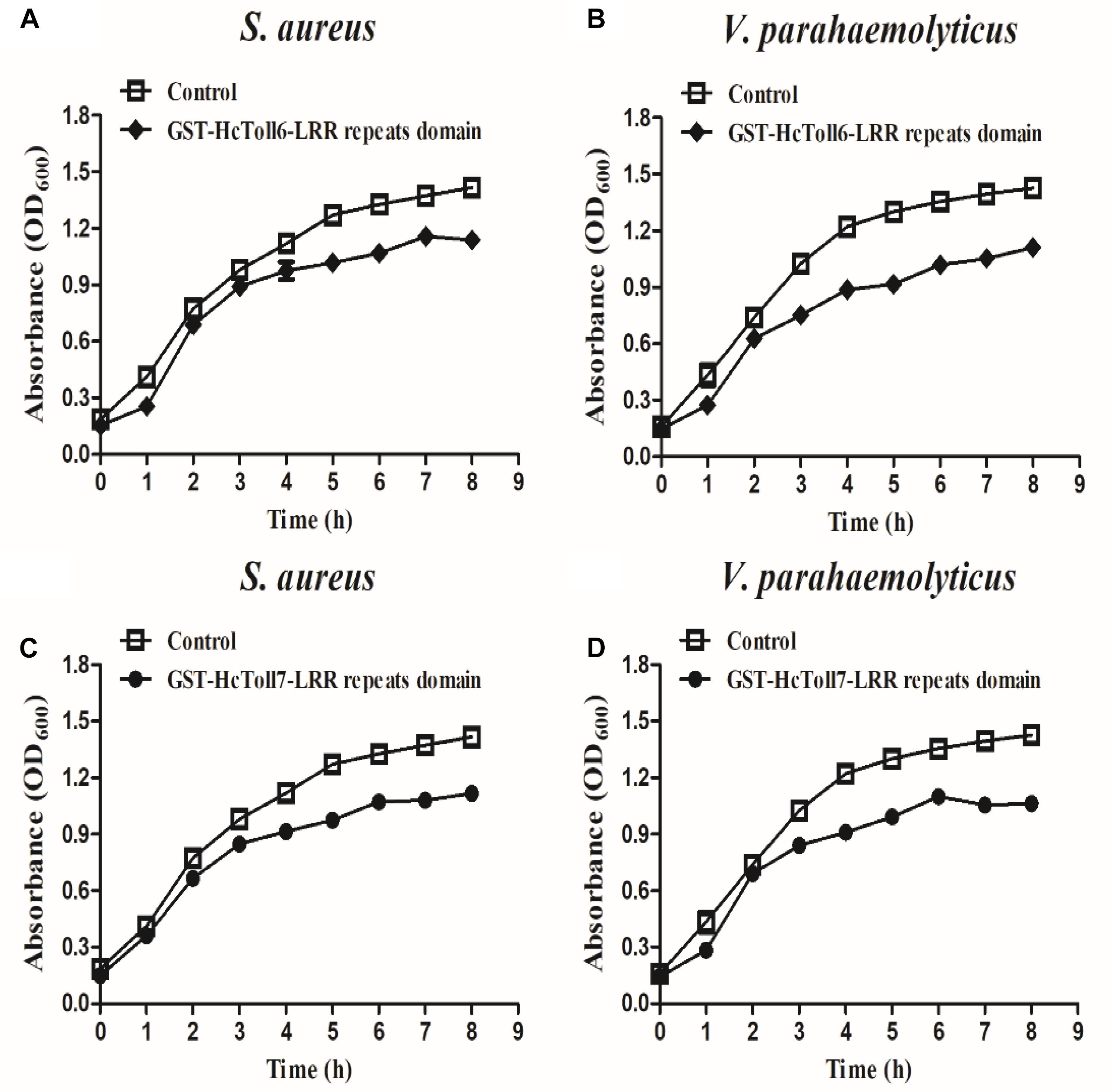
Figure 10. Bacterial growth inhibitory activities of rHcToll6-LRR and rHcToll7-LRR. Antimicrobial activities against cultured S. aureus (A,B) and V. parahaemolyticus (C,D) are shown by the inhibition growth curves. Bacteria were mixed with 100 μg/mL purified rHcToll6-LRR/rHcToll7-LRR (TBS as the control), and OD600 values were measured every 1 h. Data represent mean ± SD values from three independent replications. Error bars represent SD.
Tolls/TLRs play crucial roles in innate immunity by detecting invading microbes and thereby relaying downstream signaling cascades to activate the synthesis of AMPs and eliminate pathogens (Medzhitov, 2001). Recently, a large number of Tolls have been found in mollusks and confirmed to be involved in a range of innate immune responses. Our study was an attempt to elucidate the effect of Tolls in the immune response of mussels to bacterial infections.
In the present study, two Tolls, possessing 2223 and 2193 bp ORFs, were identified in the triangle sail mussel H. cumingii and named HcToll6 and HcToll7. The deduced HcToll6 and HcToll7 proteins share two common structures: extracellular LRR domains and intracellular TIR domain. In contrast to the highly conserved TIR domain, the LRR domains show distinct divergence in sequence and are characterized by tandem copies of LRR motifs (Matsushima et al., 2007). BLAST and multiple sequence alignment analysis of proteins indicated that these two amino acid sequences were not conserved. Moreover, in the phylogenetic tree, HcToll6 and HcToll7 were closely matched to bivalve Tolls (MgToll, CvToll, AiToll, and LaToll) but were not clustered with other known H. cumingii Tolls. These findings indicated the novelty of HcToll6 and HcToll7 in H. cumingii.
In mussels, HcToll6 and HcToll7 were predominantly expressed in immune tissues, such as the hepatopancreas and gills. Gills are water-air-exchange tissues that are frequently exposed to external environments and in direct contact with pathogens (Huang X. et al., 2016). Similar to the fat body of insects and liver of mammals, the hepatopancreas is vital for the immune system and homeostasis maintenance (Jiravanichpaisal et al., 2006). A diverse set of tissues expressing HcToll6 and HcToll7 implies that these genes are biologically important. The mussels were infected with S. aureus and V. parahaemolyticus to explore the expression of HcToll6 and HcToll7 after bacteria challenge. qPCR confirmed that HcToll6 and HcToll7 mRNAs in hepatopancreas and gills were remarkably upregulated when stimulated with bacteria. This result is consistent with many previous reports (Ren et al., 2013; Zhang et al., 2017; Huang Y. et al., 2018). After treatment with different bacteria, the transcript levels of HcToll6 and HcToll7 in different tissues obviously varied. This finding indicated that the Toll functions are diverse, and that Toll exerts different immune effects in response to different stimuli. In invertebrates, the innate immune system recognizes Gram-positive bacteria and fungi mainly through the Toll signaling pathway, and the identification of Gram-negative bacteria mainly depends on the IMD pathway (Parker et al., 2001). In our study, Gram-positive and Gram-negative bacteria could regulate HcToll6 and HcToll7 expression and activate the Toll pathway. This result indicated that different immune regulatory mechanisms might be linked.
Toll was originally identified as an essential molecule for embryonic dorsal/ventral patterning in Drosophila and confirmed to be responsible for the modulation of a large number of AMPs (Valanne et al., 2011). In vitro dsRNA was synthesized and used to silence the expression of HcToll6 and HcToll7 and to examine the role of HcToll6 and HcToll7 in mediating the expression of AMPs. RNA interference (RNAi) involving dsRNA to silence genes has been applied to studies on mussel immunity (Huang D. et al., 2018; Zhao et al., 2018). The successful knockdown of HcToll6/HcToll7 with a unique dsHcToll6/dsHcToll7 provided a practical way of demonstrating their potential function in the innate immune system of mussels. The data showed that HcToll6 and HcToll7 RNAi strongly decreased the expression of HcLyso and HcDef in the gills of RNAi-treated mussels with V. parahaemolyticus challenge and the GFP RNAi group showed no change in gene expression compared with V. parahaemolyticus only group. Lysozymes are the first-line defensive enzymes to resist various invading pathogens and play key roles in the natural defense of most living organisms (D€uring et al., 1999). Defensins are AMPs that serve as a component of the immune defense system of many organisms and possess remarkable microbicidal activities (Wang et al., 2014). We speculated that HcToll6 and HcToll7 were involved in H. cumingii immune response challenged with V. parahaemolyticus and was related to the activation of HcLyso and HcDef expression. Nonetheless, the exact mechanism is unclear and needs an in-depth research.
Next, we found that the prokaryotic expressed recombinant LRR domains of HcToll6 and HcToll7 (rHcToll6-LRR and rHcToll7-LRR) have the ability to binding a wide range of four Gram-positive bacteria and three Gram-negative bacteria. To further explore the possible recognition mechanism, two common PAMPs were selected to detect the binding affinity of rHcToll6-LRR and rHcToll7-LRR, which showed similar affinity to LPS and PGN. The binding property of rHcToll6-LRR and rHcToll7-LRR were quite different from TLRs in mammals and Drosophila but was similar to those of rCgTLR6 or rMjToll1-3, which also had a wide spectrum of binding activities towards various bacteria and LPS and PGN (Wang et al., 2016; Sun et al., 2017). In view of the lack of adaptive immunity, invertebrates were suspected to harbor more PRRs by gene expansion as a compensation to identify diverse pathogens (Wang et al., 2016). More interestingly, rHcToll6-LRR and rHcToll7-LRR exhibited significant bactericidal activity against V. parahaemolyticus and S. aureus in vitro, which confirmed the significant role of Tolls in eliminating invading non-self. Based on these results, we draw a conclusion that HcToll6 and HcToll7 could play a primary role in anti-bacterial defense of H. cumingii. Our study provides more evidence and novel understanding to the potential functions of Tolls in the innate immunity of mussels.
We declare that appropriate ethical approval and licenses were obtained during our research.
YH carried out the experiments and contributed reagents and materials. YH, GSZ, and QR designed the experiments, analyzed the data, and wrote the manuscript. All authors gave final approval for publication.
This study was supported by the China Postdoctoral Science Foundation (2019M651666), the Natural Science Foundation of Jiangsu Province (BK20180501 and BK20171474), the Fundamental Research Funds for the Central Universities (2018B03214), the National Natural Science Foundation of China (Grant No. 31572647), the Natural Science Fund of Colleges and Universities in Jiangsu Province (14KJA240002), the Natural Science Foundation of Shandong Province of China (ZR2019QC002), the Heze University doctoral fund (XY18BS08), and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00952/full#supplementary-material
Akira, S., Uematsu, S., and Takeuchi, O. (2006). Pathogen recognition and innate immunity. Cell 124, 783–801. doi: 10.1016/j.cell.2006.02.015
Anderson, K. V. (2000). Toll signaling pathways in the innate immune response. Curr. Opin. Immunol. 12, 13–19. doi: 10.1016/s0952-7915(99)00045-x
Beutler, B. (2004). Innate immunity: an overview. Mol. Immunol. 40, 845–859. doi: 10.1016/j.molimm.2003.10.005
Bilak, H., Tauszig-Delamasure, S., and Imler, J. L. (2003). Toll and toll-like receptors in Drosophila. Biochem. Soc. Trans. 31, 648–651.
Cao, J., Chen, Y., Jin, M., and Ren, Q. (2016). Enhanced antimicrobial peptide-induced activity in the mollusc Toll-2 family through evolution via tandem Toll/interleukin-1 receptor. R. Soc. Open Sci. 3:160123. doi: 10.1098/rsos.160123
Coscia, M., Giacomelli, S., and Oreste, U. (2011). Toll-like receptors: an overview from invertebrates to vertebrates. Invertebr. Surviv. J. 8, 210–226.
D€uring, K., Porsch, P., Mahn, A., Brinkmann, O., and Gieffers, W. (1999). The non-enzymatic microbicidal activity of lysozymes. FEBS Lett. 449, 93–100. doi: 10.1016/s0014-5793(99)00405-6
Funami, K., Matsumoto, M., Oshiumi, H., Akazawa, T., Yamamoto, A., and Seya, T. (2004). The cytoplasmic ’linker region’ in Toll-like receptor 3 controls receptor localization and signaling. Int. Immunol. 16, 1143–1154. doi: 10.1093/intimm/dxh115
Hopkins, P. A., and Sriskandan, S. (2005). Mammalian Toll-like receptors: to immunity and beyond. Clin. Exp. Immunol. 140, 395–407. doi: 10.1111/j.1365-2249.2005.02801.x
Huang, D., Bai, Z., Shen, J., Zhao, L., and Li, J. (2018). Identification of tumor necrosis factor receptor-associated factor 6 in the pearl mussel Hyriopsis cumingii and its involvement in innate immunity and pearl sac formation. Fish Shellfish Immunol. 80, 335–347. doi: 10.1016/j.fsi.2018.06.035
Huang, Y., Han, K., and Ren, Q. (2018). Cloning and analysis of gene expression of two toll receptors in freshwater pearl mussel Hyriopsis cumingii. Front. Physiol. 9:133. doi: 10.3389/fphys.2018.00133
Huang, X., Li, W., Jin, M., Ma, F. T., Huang, Y., and Shi, Y. R. (2016). Single CRD containing lectin from Macrobrachium rosenbergii (MrLec) participates in innate immunity against pathogen infections. Fish Shellfish Immunol. 51, 282–290. doi: 10.1016/j.fsi.2016.01.043
Huang, Y., Wang, W., and Ren, Q. (2016). Function of gC1qR in innate immunity of Chinese mitten crab, Eriocheir sinensis. Dev. Comp. Immunol. 61, 34–41. doi: 10.1016/j.dci.2016.03.016
Huang, Y., An, L., Hui, K. M., Ren, Q., and Wang, W. (2014). An LDLa domain-containing C-type lectin is involved in the innate immunity of Eriocheir sinensis. Dev. Comp. Immunol. 42, 333–344. doi: 10.1016/j.dci.2013.10.004
Huang, Y., Li, Y. R., An, L., Hui, K. M., Ren, Q., and Wang, W. (2013). Cloning and characterization of a clip domain serine protease and its homolog (masquerade) from Eriocheir sinensis. Fish Shellfish Immunol. 35, 1155–1162. doi: 10.1016/j.fsi.2013.07.025
Janeway, C. A. Jr., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216.
Jiravanichpaisal, P., Lee, B. L., and S€oderh€all, K. (2006). Cell-mediated immunity in arthropods: hematopoiesis, coagulation and opsonization. Immunobiology 211, 213–236. doi: 10.1016/j.imbio.2005.10.015
Kawai, T., and Akira, S. (2006). Innate immune recognition of viral infection. Nat. Immunol. 7, 131–137. doi: 10.1038/ni1303
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary Genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lemaitre, B., and Hoffmann, J. (2007). The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. doi: 10.1146/annurev.immunol.25.022106.141615
Lemaitre, B., Nicolas, E., Michaut, L., Reichhart, J. M., and Hoffmann, J. A. (1996). The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983. doi: 10.1016/s0092-8674(00)80172-5
Leulier, F., and Lemaitre, B. (2008). Toll-like receptors taking an evolutionary approach. Nat. Rev. Genet. 9, 165–178. doi: 10.1038/nrg2303
Li, F., and Xiang, J. (2013). Signaling pathways regulating innate immune responses in shrimp. Fish Shellfish Immunol. 34, 973–980. doi: 10.1016/j.fsi.2012.08.023
Li, J. L., Qian, R. H., Bao, B. L., Wang, G. L., and Qi, N. D. (2005). RAPD analysis on genetic diversity among the stocks of Hyriopsis cumingii from the five large lakes of China. J. Shanghai Fish. Univ. 14, 1–5.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2 (–ΔΔC(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loker, E. S., Adema, C. M., Zhang, S. M., and Kepler, T. B. (2004). Invertebrate immune systems-not homogeneous, not simple, not well understood. Immunol. Rev. 198, 10–24. doi: 10.1111/j.0105-2896.2004.0117.x
Lu, Y., Zheng, H., Zhang, H., Yang, J., and Wang, Q. (2016). Cloning and differential expression of a novel toll-like receptor gene in noble scallop Chlamys nobilis with different total carotenoid content. Fish Shellfish Immunol. 56, 229–238. doi: 10.1016/j.fsi.2016.07.007
Matsushima, N., Tanaka, T., Enkhbayar, P., Mikami, T., Taga, M., Yamada, K., et al. (2007). Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics 8:124. doi: 10.1186/1471-2164-8-124
Medzhitov, R., and Janeway, C. A. Jr. (2000). Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173, 89–97. doi: 10.1034/j.1600-065x.2000.917309.x
Parker, J. S., Mizuguchi, K., and Gay, N. J. (2001). A family of proteins related to Spätzle, the toll receptor ligand, are encoded in the Drosophila genome. Proteins 45, 71–80. doi: 10.1002/prot.1125
Ren, Q., Lan, J. F., Zhong, X., Song, X. J., Ma, F., Hui, K. M., et al. (2014). A novel Toll like receptor with two TIR domains (HcToll-2) is involved in regulation of antimicrobial peptide gene expression of Hyriopsis cumingii. Dev. Comp. Immunol. 45, 198–208. doi: 10.1016/j.dci.2014.02.020
Ren, Q., Zhong, X., Yin, S. W., Hao, F. Y., Hui, K. M., Zhang, Z., et al. (2013). The first Toll receptor from the triangle-shell pearl mussel Hyriopsis cumingii. Fish Shellfish Immunol. 34, 1287–1293. doi: 10.1016/j.fsi.2013.02.014
Roach, J. C., Glusman, G., Rowen, L., Kaur, A., Purcell, M. K., and Smith, K. D. (2005). The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. U.S.A. 102, 9577–9582.
Silverman, N., and Maniatis, T. (2001). NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15, 2321–2342. doi: 10.1101/gad.909001
Sun, J. J., Xu, S., He, Z. H., Shi, X. Z., Zhao, X. F., and Wang, J. X. (2017). Activation of toll pathway is different between kuruma shrimp and Drosophila. Front. Immunol. 8:1151. doi: 10.3389/fimmu.2017.01151
Takeda, K., and Akira, S. (2004). TLR signaling pathways. Semin. Immunol. 16, 3–9. doi: 10.1016/j.smim.2003.10.003
Valanne, S., Wang, J. H., and Ramet, M. (2011). The Drosophila toll signaling pathway. J. Immunol. 186, 649–656. doi: 10.4049/jimmunol.1002302
Wang, G. L., Xia, X. L., Li, X. L., Dong, S. J., and Li, J. L. (2014). Molecular characterization and expression patterns of the big defensin gene in freshwater mussel (Hyriopsis cumingii). Genet. Mol. Res. 13, 704–715. doi: 10.4238/2014.January.29.1
Wang, M. Q., Yang, J. L., Zhou, Z., Qiu, L. M., Wang, L. L., and Zhang, H. (2011). A primitive toll-like receptor signaling pathway in mollusk Zhikong scallop Chlamys farreri. Dev. Comp. Immunol. 35, 511–520. doi: 10.1016/j.dci.2010.12.005
Wang, S., Chen, A. J., Shi, L. J., Zhao, X. F., and Wang, J. X. (2012). TRBP and eIF6 homologue in Marsupenaeus japonicus play crucial roles in antiviral response. PLoS One 7:e30057. doi: 10.1371/journal.pone.0030057
Wang, W., Zhang, T., Wang, L., Xu, J., Li, M., and Zhang, A. (2016). A new non-phagocytic TLR6 with broad recognition ligands from Pacific oyster Crassostrea gigas. Dev. Comp. Immunol. 65, 182–190. doi: 10.1016/j.dci.2016.07.010
Yuan, K., Yuan, F. H., Weng, S. P., He, J. G., and Chen, Y. H. (2017). Identification and functional characterization of a novel Spätzle gene in Litopenaeus vannamei. Dev. Comp. Immunol. 68, 46–57. doi: 10.1016/j.dci.2016.11.016
Zhang, H. W., Huang, Y., Man, X., Wang, Y., Hui, K. M., Yin, S. W., et al. (2017). HcToll3 was involved in anti-Vibrio defense in freshwater pearl mussel, Hyriopsis cumingii. Fish Shellfish Immunol. 63, 189–195. doi: 10.1016/j.fsi.2017.02.015
Zhang, Y., He, X., Yu, F., Xiang, Z., Li, J., Thorpe, K. L., et al. (2013). Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PLoS One 8:e76464. doi: 10.1371/journal.pone.0076464
Keywords: Hyriopsis cumingii, innate immunity, Toll receptors, RNAi, antibacterial response
Citation: Huang Y, Zhang G and Ren Q (2019) Molecular Characterization of Two Toll Receptors in Hyriopsis cumingii and Their Potential Roles in Antibacterial Response. Front. Physiol. 10:952. doi: 10.3389/fphys.2019.00952
Received: 16 October 2018; Accepted: 09 July 2019;
Published: 25 July 2019.
Edited by:
Anna Di Cosmo, University of Naples Federico II, ItalyCopyright © 2019 Huang, Zhang and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guosong Zhang, aHp4eXpnc0AxNjMuY29t; Qian Ren, cmVucWlhbjA0MDJAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.