- 1State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Agriculture, University of Swabi, Khyber Pakhtunkhwa, Pakistan
- 3Canterbury Agriculture and Science Centre, AgResearch, Lincoln, New Zealand
Diapause hormone (DH) neuropeptides in insects are produced by the genes belonging to pban/capa family. Previous studies show that DH contains a conserved sequence of WFGPRXa that plays vital role in diapause regulation of some Lepidopteran species. However, the function of DH in other species is still unknown. In order to expand our understanding of DH function in diapause induction, Lom-pban, Lom-capa, and five candidates DH precursor genes (Lom-dh1, Lom-dh2, Lom-dh3, Lom-dh4, Lom-dh5) of Locusta migratoria L. were subsequently cloned. We identified Lom-dh1 to Lom-dh5 as novel genes that encoded five types (type I–V) of 44 tandem repeats of DH-like neuropeptides, which might promote egg diapause of L. migratoria. To test this hypothesis, we identified four types of eight new neuropeptides encoded by Lom-dh using liquid chromatography–tandem mass spectrometry from the central neuron system of L. migratoria under both short (10:14 L:D) and long (16:8 L:D) photoperiods. Later on, we synthesized four type I DH-like neuropeptides, LDH1, SDH1, LDH2, and SDH2, encoded by Lom-dh2/Lom-dh3 and injected them into fifth instar female locusts. Egg diapause incidences were observed after female oviposition. The four DH-like neuropeptides significantly increased the incidence of egg diapause under the short photoperiod, but the response was absent under the long photoperiod. Injection of dsLom-dh into female adults of L. migratoria under the short photoperiod could inhibit egg diapause, with no response under the long photoperiod. This study identified a new member of pban/capa family being the second example beside Bombyx mori, where the DH showed significant role on maternal induction of diapause.
Introduction
The PRXamide family of peptides plays essential roles in insect metabolism, including mating, development, diapause, muscle contraction, and induction of cuticular melanization (Jurenka and Nusawardani, 2011). Leucopyrokinin, the first PRXamide family member, was identified in the cockroach Leucophaea maderae (Holman et al., 1986). Since then, other members of PRXamide family, including CAPA peptides, pyrokinins (PK), ecdysis-triggering hormone, periviscerokinins (PVKs), tryptopyrokinins (TPKs), diapause hormones (DHs), and pheromone biosynthesis-activating neuropeptides (PBANs), have also been identified (Holman et al., 1986; Veenstra, 2014; Jurenka, 2015). Usually, pban family genes are firstly translated into a large inactive protein precursor, and then cleaved by endonuclease and amidation to produce active neuropeptides (Camargo et al., 2012). CAPA and PBAN are two of the most important multifunctional proteins and their gene sequences and structures have been thoroughly studied to reveal their roles in insect development and diapause regulation (Nagasawa et al., 1994; Loi and Tublitz, 2004; Uehara et al., 2011). Most CAPA/PBAN families share the common sequence “FXPRLamide.” The last amino acid of “FXPRLamide” is variable, including L, N, V, and I (Coast and Schooley, 2011; Veenstra, 2014; Jurenka, 2015). Insect CAPA/PBAN peptides are encoded by capa and pban gene. There are three PK peptides (PK-1, PK-2, PK-3), one DH-like peptide (DH-2), and one PBAN peptide (PBAN), expressed by the pban gene in almost all insects (Jurenka and Nusawardani, 2011). The DH-2 peptide is usually characterized by a GMWFGPRLamide ending, with two CAPA peptides (CAPA-1, CAPA-2) and one DH-like peptide (DH-1) expressed by the capa gene. The DH-1 sequence found in the capa gene of insects is also highly conserved with a consensus sequence of GMWFGPRLamide (Jurenka, 2015). Notable exceptions include Anopheles gambiae and L. migratoria for which the consensus sequences are AMWFGPRLamide and PLWFGPRVamide, respectively (Predel and Wegener, 2006). Hence, DH-1 and DH-2 are mostly encoded by a consensus sequence of GMWFGPRLamide. The DH-1 (from CAPA) and DH-2 (from PBAN) of the red flour beetle (Tribolium castaneum) have the same consensus sequence, but the strength of binding with its receptors is totally different. The binding affinities between DHs and DH receptors indicated that DH-1 is more selective for its respective authentic receptors than DH-2 (Jiang et al., 2014). This difference in binding affinity may cause DH-1 and DH-2 species to perform variably (Altstein et al., 2013). There has been functional divergence in DH-1 and DH-2 with DH-2 from PBAN linked to diapause regulation in Lepidoptera, but there is no evidence indicating that DH-1 from CAPA has a similar role (Xu et al., 1995; Xu and Denlinger, 2004; Predel and Neupert, 2007; Schmitt et al., 2015). Evidence indicates that the function of DH-1 and DH-2 have diverged in the PRXamide family (Jiang et al., 2014).
Diapause hormone can inhibit synthesis of cGMP, where the reduction of cGMP leads to increased trehalase activity of ovaries to enhance or stimulate the incorporation of glucose into oocytes through the catabolism of hemolymph trehalose in B. mori (Yamashita, 1981; Ikeda et al., 1993). However, how the environmental signals induced insect diapause by DH is still unknown. L. migratoria is an important insect pest in many parts of Africa, Asia, and Australia (Turkez et al., 2014) with facultative embryonic egg diapause. Unlike most insects, diapause induction of locusts is a trans-generation process, but similar to silkworm B. mori. Changes due to environment in the maternal parent transfer diapause factor to the offspring eggs (Hakomori and Tanaka, 1992; Tanaka, 1992; Tanaka, 1994). The offspring eggs of locusts need to be induced under a low temperature until it ceased development in late anatrepsis stage before embryo entered the diapause (Wardhaugh, 2010). Our previous work has established the conditions for diapause induction in L. migratoria. A temperature ranging from 27 to 30°C can induce diapause in L. migratoria under short photoperiod. Our previous study also demonstrated that insect hormone biosynthesis, the insulin signaling pathway and the peroxisome proliferator-activated receptor (PPAR) signaling pathway were involved in diapause regulation of the locust eggs (Tu et al., 2015; Hao et al., 2017). Meanwhile, the low level of ecdysteroids in both maternal and offspring eggs was observed in the diapause process, whereas the high quantity of ecdysteroids terminated the diapause in L. migratoria (Tawfik et al., 2002). Previous studies on L. migratoria have identified 14 neuropeptides (pyrokinin), two genes in the CAPA/PBAN family, and predicted four TPKs (Clynen et al., 2003; Coast and Schooley, 2011; Veenstra, 2014; Hou et al., 2015). The C-terminal consensus sequences in parts of these neuropeptides and genes composed of WFGPRXa (X = V, I, or Y) are similar to those in Lepidoptera DH-2 (Altstein et al., 2013; Zhang et al., 2015). The locust pyrokinin neuropeptides elicit significant diapause-inducing activity in the lepidopteran silkworm (Nachman et al., 1993). We believe that some of these DH-like neuropeptides may be involved in induction of L. migratoria embryonic diapause (Tanaka and Zhu, 2008; Tu et al., 2015). The functional genes or neuropeptides belonging to CAPA or PBAN in locusts have not yet been studied. Even it is unknown whether gene function divergence is occurring in locusts? Hence, in this study, we cloned the entire pban/capa family genes of L. migratoria. We identified novel Lom-dh genes and DH-like neuropeptides, and even applied injection bioassay and RNAi assay to understand their functions in insect diapause regulation.
Results
Prediction and Cloning of Pyrokinin Family Genes in L. migratoria
To obtain the L. migratoria capa/pban gene sequences, we first aligned other insect taxa CAPA/PBAN family protein precursors with the L. migratoria genome and found 28 target contigs (Supplementary Table S1). Among these, only five contigs, including AVCP010882506 (TPK1), AVCP010410303 (TPK2), AVCP010159529 (TPK3), AVCP010159522 (TPK4), and AVCP010410310 (TPK5), contained the conserved amino acid sequence FGPRV/I/Y of CAPA/PBAN, whereas the other 22 contigs were non-target sequences (Supplementary Table S1). We then aligned other insect taxa CAPA/PBAN family protein precursors once again with the L. migratoria transcriptome (accession number: GCGJ00000000) in NCBI, and found two new target transcripts, including gb| GCGJ01004763.1| (Lom-capa) and gb| GCGJ01044456.1| (Lom-pban) (Supplementary Table S1). We aligned these two new transcripts with the L. migratoria genome, and six other contigs, including the capa gene of gb| AVCP011143372.1|, gb| AVCP011143377.1|, gb| AVCP011143364.1|, gb| AVCP011143367.1|, and the pban gene of gb| AVCP011098992.1|, gb| AVCP011098990.1| (Supplementary Table S1). Finally, we aligned 14 pyrokinin family peptides with the L. migratoria genome and found a contig of gb| AVCP010267064.1| (Lom-PK-2) in NCBI (Supplementary Table S1). In total, 12 contigs related to CAPA/PBAN in L. migratoria genome were obtained. Among them, genome sequences of TPK 1, 3, 4, 5, and mRNA sequence of Lom-capa, Lom-pban had complete coding sequences (CDS), TPK 2 lacked the 5′-end, whereas Lom-PK-2 lacked both a 5′- and 3′-end (Supplementary Figure S1). CDS analysis indicated TPK4 and TPK5 had high identity, in the similar manner as that of TPK2 and TPK3.
Then, Lom-TPK1 (789 bp), Lom-TPK2 (1512 bp), Lom-TPK3 (1767 bp), Lom-TPK4 (732 bp), and Lom-TPK5 (1092 bp) were cloned from the L. migratoria genome DNA and central neural system cDNA, respectively. Lom-pban (498 bp, GenBank accession number: MG517529) and Lom-capa (846 bp, GenBank accession number: MG517530) were cloned from central neural system cDNA of L. migratoria.
Phylogenetic and Codon Bias Analysis of L. migratoria Pyrokinin Family Genes
To compare the phylogenetic position of these neuropeptide precursors, we downloaded all insect CAPA/PBAN precursors into NCBI and together with Lom-CAPA, Lom-PBAN, Lom-TPK1, Lom-TPK2, Lom-TPK3, Lom-TPK4, and Lom-TPK5 were used to construct phylogenetic trees. Previous research showed that the length of CAPA/PBAN family precursors differed. Within the same insect order, they were conserved in similar positions (Supplementary Figure S2), whereas in different orders, they had only the consensus conserved sequence of FXPRL/V/I. Thus, the clustering results were unable to illustrate the phylogenetic position of all insects CAPA/PBAN. The pyrokinin precursors of Hymenoptera, Lepidoptera, Diptera, and L. migratoria, however, were well clustered into CAPA and PBAN (Supplementary Figure S2). Thus, L. migratoria contains both CAPA and PBAN. Lom-TPK1, 2, 3, 4, and 5 were clustered into CAPA (Supplementary Figure S2). However, Lom-TPK1, 2, 3, 4, and 5 are clustered together on a unified branch, and are not clustered with Lom-CAPA.
Codon bias analysis showed that the CAI of Lom-pban and Lom-capa are 0.02–0.03; the CBI of Lom-pban and Lom-capa are about 0.5. The FOP of Lom-pban and Lom-capa are about 0.7 and GC3s of Lom-pban and Lom-capa are about 0.9. CAI of Lom-TPK1, 2, 3, 4, and 5 are 0.06–0.09; CBI of Lom-TPK1, 2, 3, 4, and 5 are −0.2 – 0.1; FOP of Lom-TPK1, 2, 3, 4, and 5 are 0.2–0.4; and GC3s of Lom-TPK1, 2, 3, 4, and 5 are about 0.3–0.6. CAI of the reference gene actin, gapdh, and ef are 0.1–0.2; CBI of actin, gapdh, and ef are about 0.0; FOP of actin, gapdh, and ef are about 0.4; and GC3s of actin, gapdh, and ef are 0.4–0.5. CAI, CBI, FOP, and GC3s values of Lom-pban and Lom-capa are significantly different from reference genes and Lom-TPKs. However, there is no significant difference between CBI, FOP, and GC3s values of Lom-TPKs and the reference genes (Supplementary Figure S3). Thus, we considered that Lom-TPK1, 2, 3, 4, and 5 are novel types of genes from Lom-capa and Lom-pban and then we renamed Lom-TPK1, 2, 3, 4, and 5 as Lom-dh1, 2, 3, 4, and 5 (GenBank accession number: MG517531, MG517532, MG517533, MG517534, MG517535), respectively.
Lom-CAPA/PBAN/DH Structure Analysis
We used ORF Predictor to identify the signal peptide and endoproteolytic cleavage site of CAPA/PBAN neuropeptide precursors. This indicated that the new cloned Lom-dh1 to Lom-dh5 genes had no intron or any other alternative splicing transcripts (Figure 1). Furthermore, potential signal peptides of Lom-DH1, 2, 3, 4, and 5 were predicted and demonstrated in Figure 1. All potential neuropeptides could be processed at possible endoproteolytic cleavage sites GR, GRR, KR (Figure 1). Gene structure analysis showed that Lom-DH2 contained eight DH-like neuropeptides, whereas Lom-DH3 contained 11 DH-like neuropeptides. Lom-DH1 contained five DH-like neuropeptides, Lom-DH5 contained 14 DH-like neuropeptides, and Lom-DH4 contained six DH-like neuropeptides (Figure 1). According to the C-terminus consensus sequences, 44 DH-like neuropeptides, encoding by Lom-DH1 to Lom-DH5, can be classified into five types (Table 1). We used Weblogo to build a sequence logo based on consensus sequences of the 13 C-terminal amino acids of all 44 DH-like neuropeptides. The results indicated that two of the consensus amino acid sequences, including LWFGPRI and LWFGPRV appeared most frequently (Supplementary Figure S4). These two amino acid sequences were clustered on the reported L. migratoria neuropeptides. Results indicated they belonged to DH-1 part of the capa gene, the same as Lom-PVK-3 (Figure 2A).

Figure 1. Lom-DH1 to Lom-DH5 protein sequences. Amino acid sequences in black box indicate signal peptide, gray amino acid sequences indicate endoproteolytic cleavage sites, and black amino acid sequences indicate short neuropeptides.
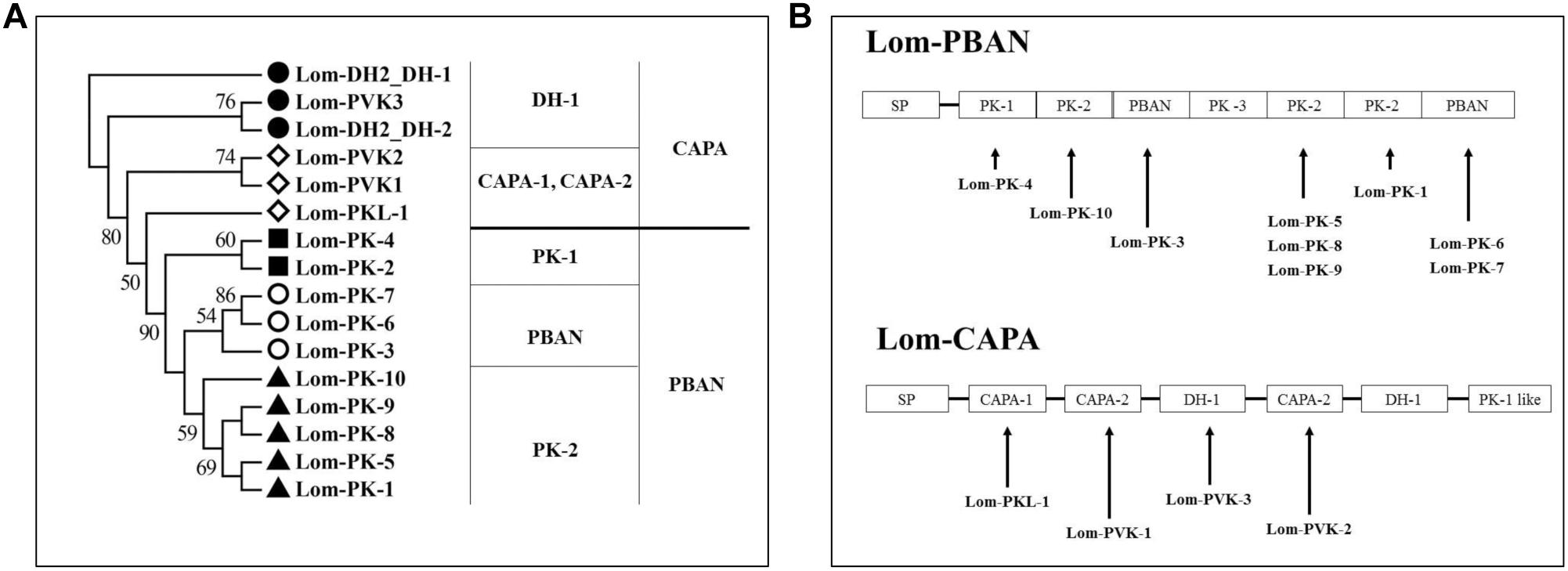
Figure 2. Neuropeptides encoded by capa/phan in Locusta migratoria. (A) Phylogenetic analysis of pyrokinin consensus sequences. (B) Genes of coding neuropeptide; solid line indicates the location of the neuropeptides in the capa/pban genes.
We aligned the Lom-capa and Lom-pban genes to the neuropeptides previously found in L. migratoria and found that 14 neuropeptides, except for Lom-PK-2, were encoded by the Lom-capa or Lom-pban genes (Table 2). The gene structure of Lom-capa was SP_CAPA-1_CAPA-2_DH-1_CAPA-2_DH-1_PK-1 with the segments of CAPA-2_DH-1 repeated two times (Figure 2B). Although the gene structure of Lom-pban was SP_PK-1_PK-2_PBAN_PK-3_PK-2_PK-2_PBAN, there was no DH, with PK-2_PBAN segments repeated twice (Figure 2B). The amino acid sequence of PK-3 was DPPVDGPLVWLPLQVSPRL in the pban gene, but there was no neuropeptide corresponding to PK-3 indicated for L. migratoria. The absence of a PK-3 neuropeptide, even though sequences were present in the pban gene, is potentially caused by a mutation from GR to AR at the endoproteolytic cleavage site, which prevents peptide expression. Thus, the Lom-capa and Lom-pban genes are unique. The former could be defined as the traditional capa gene, and the latter could be defined as the traditional pban gene when compared with other insect taxa. Most importantly, the capa gene, including DH-1, may be involved in locust diapause regulation, whereas the pban gene plays no role in this process.
Novel Pyrokinin Family Neuropeptides Identified by LC-MS/MS
These results demonstrate that most of the 14 PK neuropeptides obtained by previous MS are encoded by Lom-pban and Lom-capa (Figure 2B). We performed LC-MS/MS to identify the neuropeptides from the central neuron system of L. migratoria under short and long photoperiods. We identified all of the 14 PK neuropeptides in previous studies, except Lom-PK-2, Lom-PK-6, and Lom-PK-7. There were minute differences between the 14 PK and our newly identified peptides. The N-terminal of the newly identified Lom-PK-10, Lom-PKL-1, and Lom-PVK-2 is longer than the previously identified ones (Table 2). The terminal amino acid of Lom-PKL is I rather than L (Table 2). Similarly, the fourth amino acid of Lom-PVK-2 is Q rather than E (Table 2). In addition, we identified nine new neuropeptides encoded by Lom-dh1 to Lom-dh5 genes and named them Lom-PK-11, Lom-DH-1, Lom-DH-2, Lom-DH-3, Lom-DH-4, Lom-DH-5, Lom-DH-6, Lom-DH-7, and Lom-DH-8. Lom-DH-1, Lom-DH-2, Lom-DH-3, and Lom-DH-4 were type I DH-like neuropeptides. Lom-DH-5 and Lom-DH-6 were type II DH-like neuropeptides. Lom-DH-7 was a type IV DH-like neuropeptide. Lom-DH-8 was a type V DH-like neuropeptide (Table 2).
DH Function Identified by Injection Assay
To confirm locust DH function, LDH1 (MHSKNVVVQRC IQTGFSDGGSIRNVPETSLWFGPRIa), SDH1 (LWFGPRIa), LDH2 (SNPETNLWFGPRVa) and SDH2 (LWFGPRVa) as shown in Figure 1 were synthesized and injected into fifth instar females independently. All of the treated females turned to adults’ within 72 h of neuropeptides injection. After attaining the adulthood, the insects were reared for additional 14 days afterward they reached to maturity and began eggs laying. The control treatment consisted of fifth instar females injected with distilled water. Under an L:D 10:14 photoperiod and constant temperature of 28°C, we induced diapause in 6.8–24.7% of the eggs oviposited by females treated with LDH1, SDH1, LDH2, and SDH2. This allowed us to test whether DH enhanced or inhibited diapause. Results showed that SDH1 induced diapause in eggs under L:D 10:14 at a constant temperature of 28°C, with 68.1% of the eggs entering diapause after injection of SDH1 at 0.2 nmol/individual. This was significantly higher than the diapause incidence after injection of LDH1 at 0.2 nmol/individual and control (F = 9.344, P = 0.001) (Figure 3A). In the control, 19.1% (range from 10.1 to 24.7%) of the eggs entered diapause (Figures 3A,B). However, no eggs were induced to enter diapause by injection of LDH1, SDH1, LDH2, and SDH2 among the eggs laid and incubated under long photoperiod at 28°C, even though 2 nmol/individual was injected (Figure 3D).
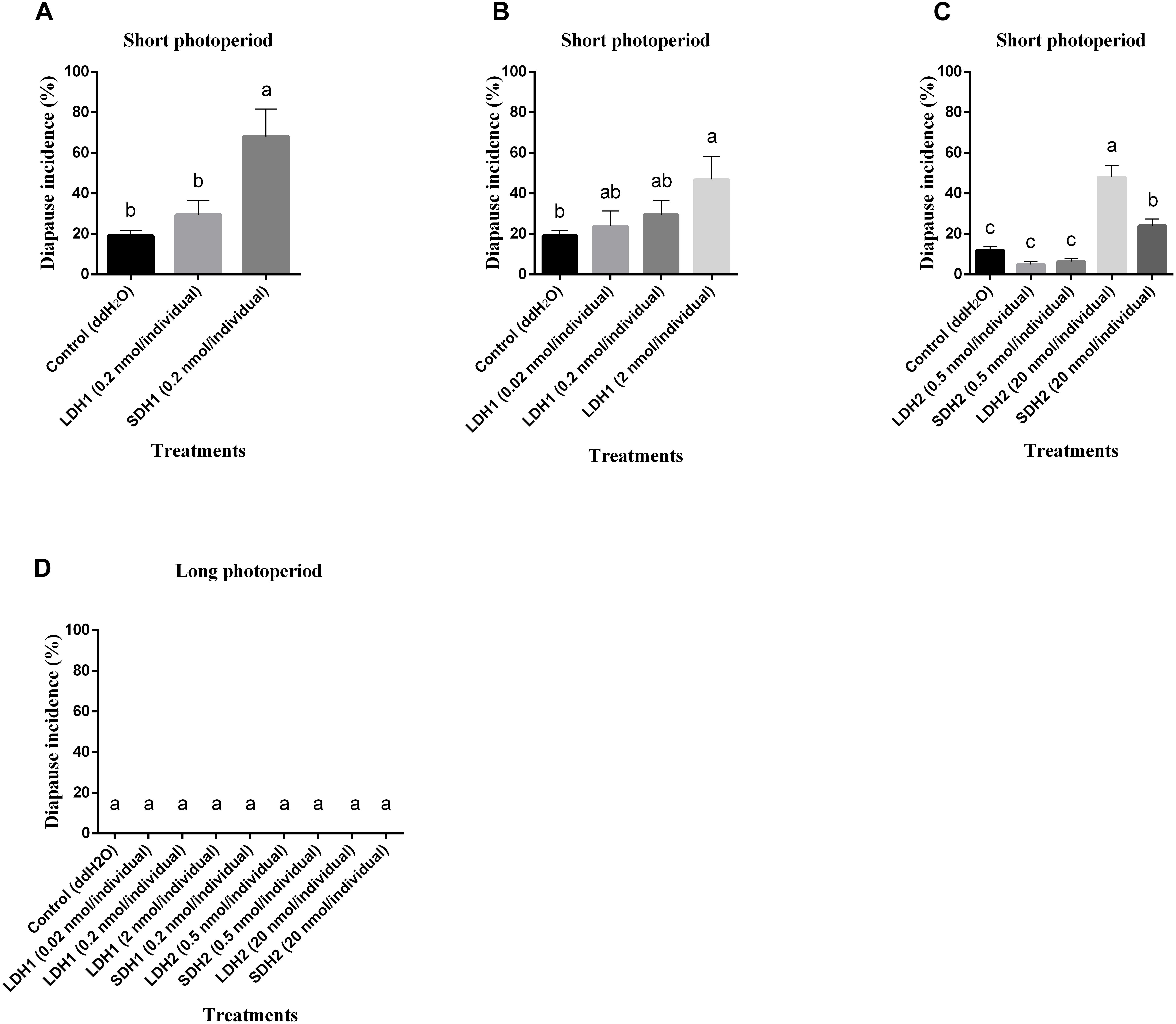
Figure 3. Diapause incidence of L. migratoria eggs after treatment of female parent with selected doses of LDH1 (MHSKNVVVQRCIQTGFSDGGSIRNVPETSLWF GPRIa), SDH1 (LWFGPRIa), LDH2 (SNPETNLWFGPRVa) and SDH2 (LWFGPRVa). (A) Injection of 0.2 nmol/individual of LDH1 or SDH1 at 10:14 L:D and 28°C. (B) Injection of 0.02, 0.2, and 2 nmol/individual LDH1 under 10:14 L:D and 28°C. (C) Injection of 0.5 and 20 nmol/individual of LDH2 or SDH2 under 10:14 L:D and 28°C. (D) Injection of 0.02, 0.2, and 2 nmol/individual LDH1, 0.2 nmol/individual of SDH1, 0.5, and 20 nmol/individual of LDH2 or SDH2 at 16:8 L:D and 28°C. Control treatment was injection of distilled water (control). The lowercase a, b, and c indicated that statistically significant difference of diapause incidence was considered on an error probability of p < 0.05 by one-way ANOVA using Duncan’s multiple-range test.
There was a dose-related effect on diapause induction. Injection of LDH1 at 2 nmol/individual produced 47.0% diapause eggs compared with 23.8% and 30.0% when the dose was 0.02 and 0.2 nmol/individual, respectively (F = 2.407, P = 0.105) (Figure 3B). Under short photoperiod (L:D 10:14) and at a constant temperature of 28°C, injection of 20 nmol/individual of both LDH2 and SDH2 significantly induced diapause (F = 30.197, P = 0.001), but at a lower dose of 0.5 nmol/individual, there was no significant difference between the LDH2, SDH2 treatments and the control group (Figure 3C). No eggs entered diapause when LDH2 and SDH2 were injected into females held under long photoperiod (L:D 16:8) and at a constant temperature of 28°C (Figure 3D).
SDH1 and LDH1 could promote diapause at 0.2 and 2 nmol/individual, respectively. Meanwhile, the dose of 20 nmol/individual promoted diapause in LDH2 and SDH2 treatments. The results also indicated that these synthetic DH peptides could effectively promote locust diapause at a specific dose rate.
Lom-dh Genes Function Identified by RNAi
Expression levels of Lom-dh1, Lom-dh2/3, and Lom-dh4/5 were significantly higher (53.1, 10.6, and 13.5 folds) in maternal locust under short photoperiod as compared to long photoperiod (Figure 4). This demonstrated that the Lom-dh genes might have the function of diapause induction. To verify the function of all Lom-dh genes, dsLom-dh1, dsLom-dh2/3, dsLom-dh4/5, and dsLom-dhall were injected into adult female L. migratoria maintained under L:D10:14 and L:D16:8 photoperiods and at a constant temperature of 28°C. Because Lom-dh2 and Lom-dh3 shared a common region, hence the Lom-dh2 and Lom-dh3 were knocked down simultaneously by the same dsRNA (dsLom-dh2/3). Similarly, the Lom-dh4 and Lom-dh5 were knocked down by the dsRNA (dsLom-dh4/5). dsLom-dhall, which was used to knock down all Lom-dh at once, was a mixture of dsLom-dh1, dsLom-dh2/3, and dsLom-dh4/5. Then we performed RT-PCR to check RNAi efficiency. Relative mRNA levels of dsLom-dh1, dsLom-dh2/3, dsLom-dh4/5, and dsLom-dhall treatments were reduced significantly compared with the control and dsGFP treatments under both L:D 10:14 and L:D 16:8 photoperiods (Figure 5). Females were maintained under these conditions until they reached maturity and commenced eggs laying. Diapause incidences in the control group and dsGFP treatment ranged from 76.23 to 85.59% under L:D 10:14 and at a temperature of 28°C. Meanwhile diapause incidences were 70.34% in dsLom-dh1 treatment, 35.84% in dsLom-dh2/3 treatment, 30.27% in the dsLom-dh4/5 treatment, and 42.59% in the dsLom-dhall treatment at L:D 10:14, 28°C. Compared with the control and the dsGFP treatments, egg diapause incidences were reduced significantly in the dsLom-dh1, dsLom-dh2/3, dsLom-dh4/5, and dsLom-dhall treatments (F = 117.891, P = 0.001) (Figure 6A). No eggs entered diapause resulting from injection of dsLom-dh1, dsLom-dh2/3, and dsLom-dh4/5 where the eggs were laid and incubated under long photoperiod at 28°C (Figure 6B). Results revealed that diapause incidences were significantly down regulated after the knock down of Lom-dh genes independently or jointly. This indicated that all Lom-dh genes could induce diapause of L. migratoria.
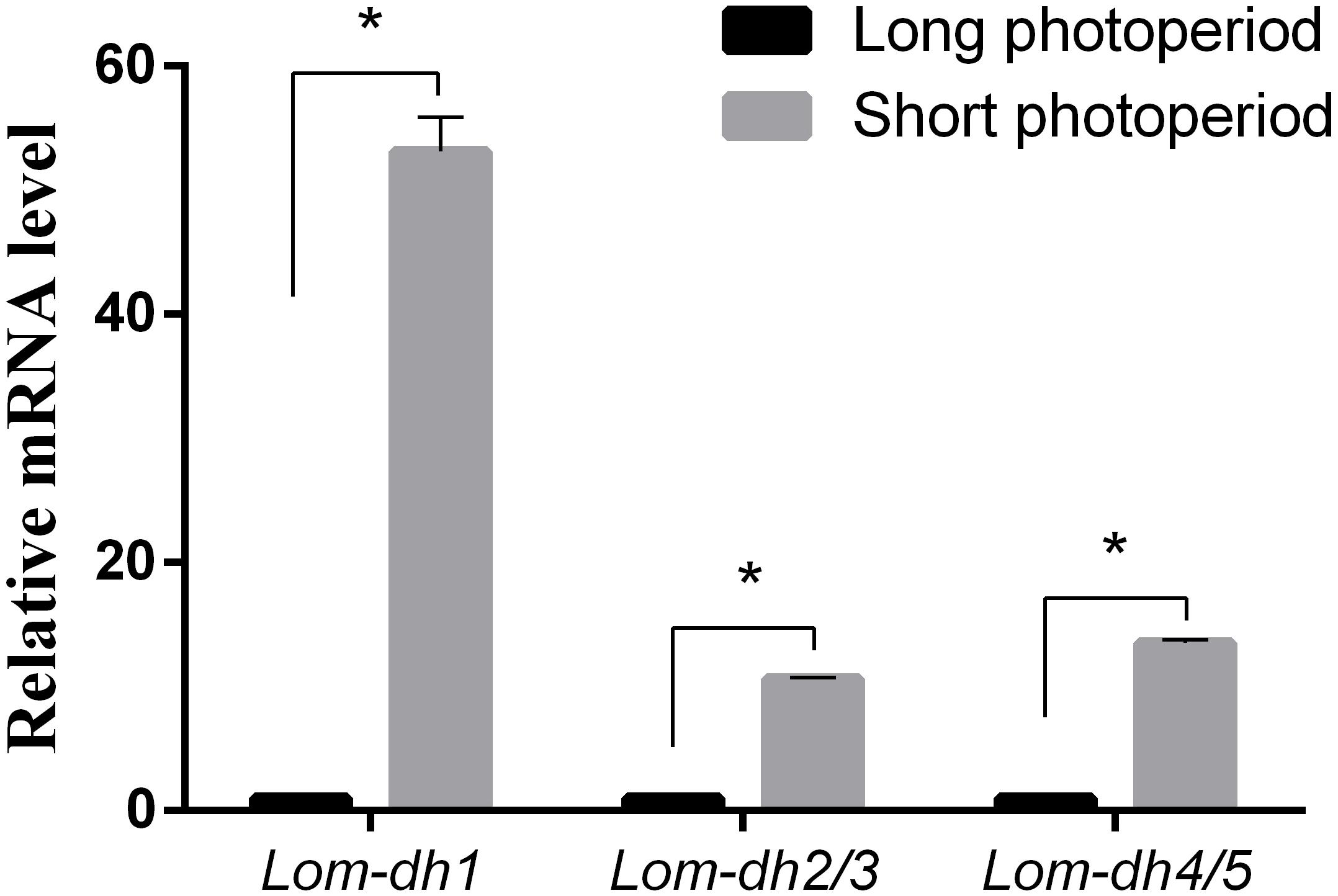
Figure 4. Relative mRNA level of Lom-dh genes detect in L. migratoria under long and short photoperiod. *Indicates an error probability of p < 0.05 by Student’s t-test.
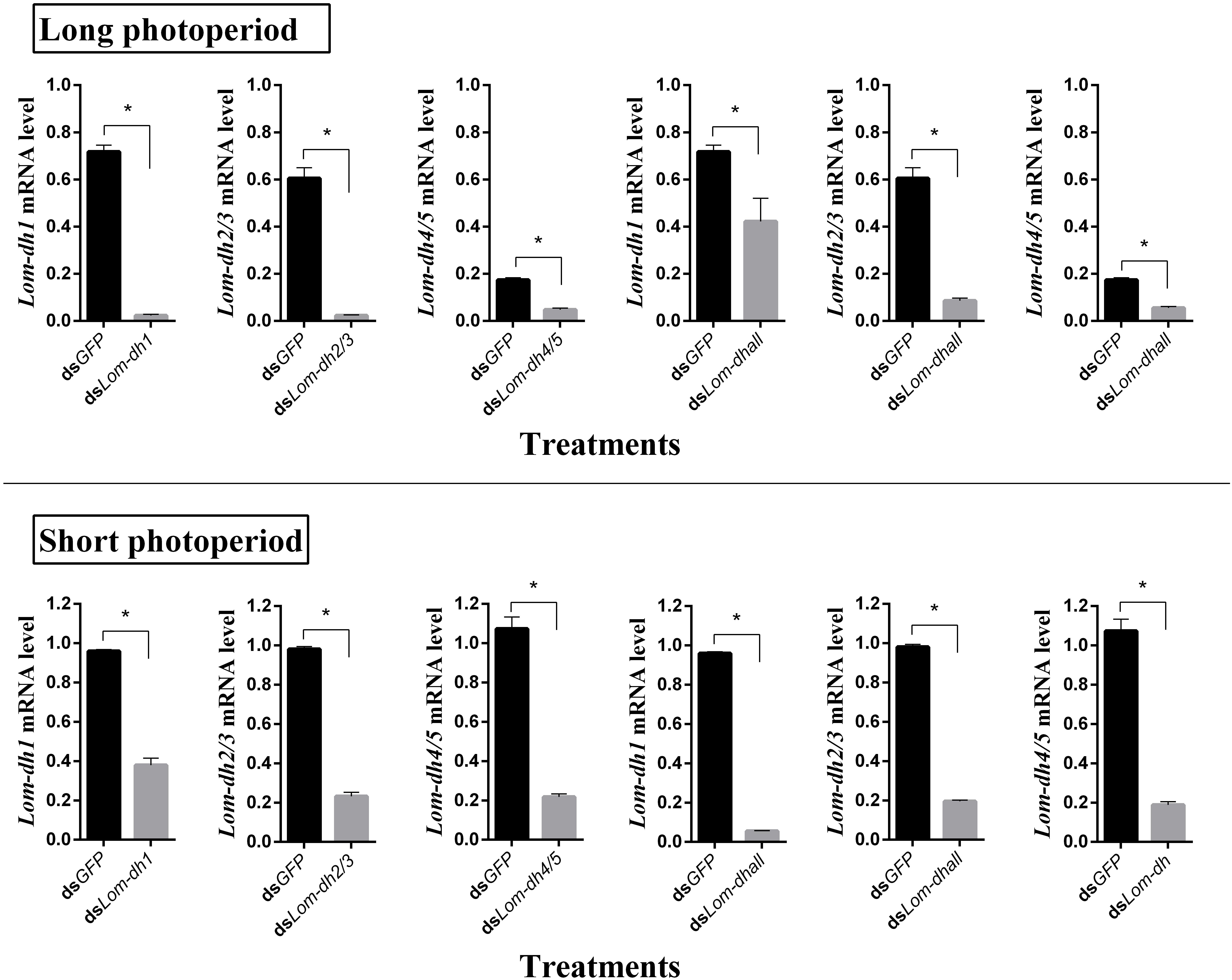
Figure 5. RNAi efficiency of Lom-dh genes detect in L. migratoria under long and short photoperiod. *Indicates an error probability of p < 0.05 by Student’s t-test.
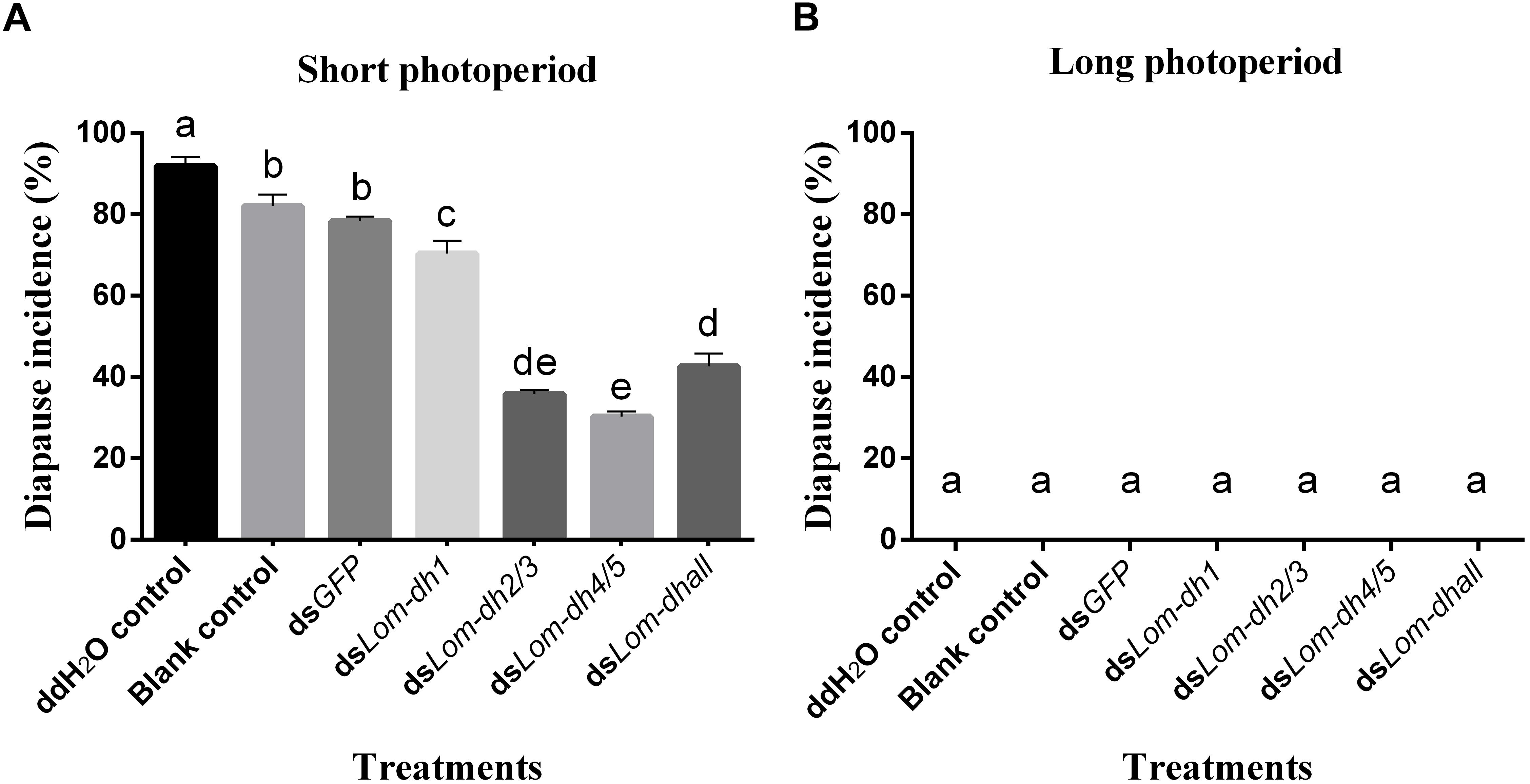
Figure 6. (A) Diapause incidence after injecting dsLom-dh to RNAi Lom-dh genes of L. migratoria under short photoperiod. (B) Diapause incidence after injecting dsLom-dh to RNAi Lom-dh genes of L. migratoria under long photoperiod. The lowercase a, b, c, d, and e indicated that statistically significant difference of diapause incidence was considered on an error probability of p < 0.05 by one-way ANOVA using Duncan’s multiple-range test.
Discussion
Novel DH Neuropeptides Encoded by Novel Lom-dh Genes
Most TPK genes of Orthoptera occur in the capa/pban gene family (Veenstra, 2014). We cloned the entire capa/pban family genes of L. migratoria in our study. Similar to other species, Lom-pban and Lom-capa had introns. Unlike Lom-pban and Lom-capa, there were no introns in TPKs (Lom-TPK1, 2, 3, 4, and 5). Phylogenetic analysis showed that the TPKs in L. migratoria were similar to capa, but not identical. Codon bias analysis also identified a significant difference between capa/pban and TPKs. Thus, we renamed TPK (Lom-TPK1, 2, 3, 4, and 5) as Lom-dh (Lom-dh1, 2, 3, 4, and 5) in L. migratoria to better understand their function and avoid classification confusion. Lom-dh is a new type of DH precursor gene in addition to Lom-capa and Lom-pban.
The pban/capa family is involved in insect growth and development. Diapause is either induced or inhibited by the same gene as seen in some species of Lepidoptera, but their different segments may play different roles (Hasegawa, 1957; Jurenka, 2015; Zhang et al., 2015). For example, the PBAN segment of the pban gene regulates growth and development and the DH-2 segment regulates diapause. In the capa gene, CAPA-1 and the CAPA-2 segment regulate growth and development (Zhang et al., 2015). In L. migratoria, however, no DH-2 segment was found in the Lom-PBAN, CAPA-1_CAPA-2_DH-1 segments contained in Lom-CAPA. Moreover, when we compared all neuropeptides of Lom-PBAN, Lom-CAPA, and Lom-DH1-5 with former studies (Clynen and Schoofs, 2009), we found the former Lom-PVK3 (DH-1 from Lom-CAPA) and four types of eight novel DH neuropeptides (Lom-DH-1 to Lom-DH-8) encoded by the five novel Lom-dh genes.
Most reported CAPA or PBAN precursors in Diptera, Hymenoptera, and Lepidoptera also contained the DH segment (Jurenka, 2015; Zhang et al., 2015). L. migratoria PBAN had no DH segment, similar to Rhodnius prolixus Stål (Hemiptera: Reduviidae), Lygus hesperus Knight (Hemiptera: Miridae), and Drosophila melanogaster Meigen (Diptera: Drosophilidae) (Supplementary Table S2), but while CAPA was found to include a DH segment, it was dissimilar to that found in Solenopsis invicta Buren (Hymenoptera: Formicidae) and Apis mellifera L. (Hymenoptera: Apidae) (Supplementary Table S2) as reported by Rahman et al. (2009) and Veenstra (2010). Moreover, Lom-CAPA contained CAPA-1, CAPA-2, and DH-1 segments along with a PK segment in the C-terminal. These results are similar to Aedes aegypti L. (Diptera: Culicidae), Tribolium castaneum Herbst (Coleoptera: Tenebrionidae), Athalia rosae L. (Hymenoptera: Tenthredinidae) (Supplementary Table S2). In L. migratoria, we found three types of pyrokinins, including Lom-CAPA, Lom-PBAN, and Lom-DH. Teleogryllus commodus Walker (Orthoptera: Gryllidae), Zootermopsis nevadensis Hagen (Isoptera: Termopsidae), and Blattella germanica L. (Blattodea: Blattellidae) also has novel dh genes with eight, six, and 23 DH peptide copies respectively (Supplementary Table S2). On the basis of these data, the CAPA and PBAN of insects have a fixed structure, but one of them may lose the DH segment, and the novel dh genes that we have found in this study contained multiple DH copies (Supplementary Table S2). As the DH functions were not properly documented in previous studies, we considered that at least one of the capa, pban, or dh genes was involved with the DH function. This was vital for insect physiological metabolism. The duplicate dh genes and multiple copies of DH in the DH precursors in Orthoptera may be involved with diapause regulation (Supplementary Table S2). The differentiation of Lom-dh with tandem DHs permits the locust to exploit its environment to maximize offspring survival. L. migratoria, along with termites and cockroaches are important pests. Understanding the function of novel dh genes in the physiological metabolism of these taxa might be helpful for developing new pest control methods.
Novel Lom-dh Genes Can Encode Multiple DH Neuropeptides to Induce Locust Egg Diapause
We found that type I DH-like neuropeptides from novel Lom-dh genes can induce locust diapause (Table 1). Previous studies on diapause in the silkworm B. mori showed that, depending on the neuropeptide, doses ranging from 1 pmol to 20 nmol per individual induced embryonic diapause (Sato et al., 1993). Moreover, the locust pyrokinin Lom-PK is threefold more active than native Bom-DH as a diapause induction agent at the dose of 20–4,000 pmol/individual (Nachman et al., 1993). Injection of 200 pmol SDH1 induced locust diapause at a rate higher than that found when locusts were injected with 2,000 pmol LDH1. This result may relate to the N-terminal hydrophobic structure, as LDH1 contains LWFGPRI (Zhang et al., 2015). For both LDH2 and SDH2, diapause induction occurred only at doses of at least 104 pmol. To increase the induction of diapause using LDH1 and SDH1 beyond 50%, the concentration of doses needed to be increased. Neuropeptides working in combination with receptors are concentration dependent, with the relationship showing evidence of coevolution (Park et al., 2002; Grimmelikhuijzen and Hauser, 2012). Thus, the DH receptor recognition of DH 1 and DH 2 may be caused by the difference in the last amino acid between LWFGPRV and LWFGPRI. This study may aid in the development of other DH analogs and antagonists that could disrupt insect diapause and be used as pest management tools. Insect CAPA neuropeptides may be associated with cold tolerance, but no previous data have indicated that it was involved in locust diapause (Xu and Denlinger, 2003; Terhzaz et al., 2015). DH associated with the pban gene is known to be involved in insect diapause regulation (Jurenka and Nusawardani, 2011). No DH produced by the pban gene in some diapausing insects, such as L. migratoria; this suggests that the DH, in these species, may be derived from capa or dh genes.
To verify the effect of Lom-dh genes on locust diapause at the mRNA level, we injected dsLom-dh1, dsLom-dh2/3, dsLom-dh4/5, and dsLom-dhall to implement RNA interference on all Lom-dh genes. Because of the high similarity between Lom-dh2 and Lom-dh3, Lom-dh4 and Lom-dh5, the same dsRNA (dsLom-dh2/3, dsLom-dh4/5) were used to implement RNA interference on all of them. Moreover, dsLom-dhall, which is composed of equimolar amounts of dsLom-dh1, dsLom-dh2/3, and dsLom-dh4/5, was used to implement RNAi on all Lom-dh genes simultaneously. The interference efficiency test showed that relative mRNA levels were significantly decreased after injection of dsLom-dhall under both long and short photoperiods. This result proved that five different Lom-dh genes are susceptible to RNA interference by simultaneous injection of three dsRNAs in L. migratoria. The results demonstrate that the diapause incidences of locust eggs are significantly (p < 0.05) decreased by RNA interference of Lom-dh1 to Lom-dh5 genes of adult females under short photoperiod. Among these, the dsLom-dh4/5 treatment was the most effective. We did not observe any effects by injecting any dsLom-dh under a long photoperiod. Results showed that the expression level of Lom-dh genes were higher under short photoperiod compared to long photoperiod (Figure 4). These findings are consistent with the results obtained from RNAi.
DH Peptides and Lom-dh Only Have Function Under Short Photoperiod
As early as 1957, the existence of a DH from B. mori maternal subesophageal ganglions (SG) was suggested to act on the developing ovaries via the hemolymph, thereby causing the entry of eggs into diapause (Hasegawa, 1957). Interestingly, previous research found that DH was not capable of inducing pupal diapause in the Helicoverpa/Heliothis complex, but instead it effectively terminates pupal diapause, a surprising contrast to the results noted in B. mori (Xu and Denlinger, 2003; Zhang et al., 2004; Zhao et al., 2004). In previous studies, the functions of DH on diapause regulation haven’t been found in any other insect orders except Lepidoptera. Our work is the first evidence that diapause of L. migratoria, an Orthoptera insect, can be induced by DH. Our results are consistent with the observations on B. mori, but are in contrast to the results with the Helicoverpa/Heliothis complex. Diapause induction of both L. migratoria and B. mori is a trans-generation process. In the Helicoverpa/Heliothis complex, diapause occurs at a fixed developmental stage (pupal stage). This may be the probable reason to explain why DH functions are so different in diapause regulating. However, we cannot confirm why the sequences of these DHs are almost similar, yet their functions in diapause regulation are opposite. The specific mechanism of variable function of DH still remains an interesting and hot topic for future study.
To investigate how photoperiod and DH affect L. migratoria diapause, two important pathways of diapause regulation, i.e., insulin signaling pathway and MAPK signaling pathway, have been described below. Circadian clock oscillations function in the insect brain or peripheral nerve and likely contribute to photoperiodism (Ito et al., 2008; Ikeno et al., 2010). The circadian genes are linked to insulin signaling, which is the most important regulator of normal growth in insects and leads to the production of juvenile hormone through the PI3-kinase/Akt pathway. Then, forkhead box protein O (FOXO) is phosphorylated and it blocks the translocation protein into the nucleus under long-day photoperiod (Lee et al., 2001; Jünger et al., 2003; Sim and Denlinger, 2008, 2013). Suppression of insulin signaling causes the FOXO, translocated into the nucleus, to initiate transcription of downstream genes and induce diapause in D. melanogaster, C. pipiens, and C. elegans under short photoperiod (Gerisch et al., 2001; Williams et al., 2006; Sim and Denlinger, 2008). Previous study in silkworm B. mori revealed that DH activated the DH receptor signals to MAPK signaling pathway through Gq-PLC-PKC-dependent cascade (Jiang et al., 2016). In this study, the DH peptides and dsRNA (dsLom-dh) that were injected, at various concentrations, did not affect L. migratoria diapause incidence under long photoperiod. This suggests that there may be a cross talk between insulin signaling pathway and DH activated MAPK signaling pathway in diapause induction of L. migratoria under long photoperiod where the insulin signaling dominated the diapause regulation and subsequently, the DH activated MAPK signaling was blocked. On the contrary, insulin signaling pathway was inhibited under short photoperiod, thus, the DH activated MAPK signaling pathway takes over, leading to diapause induction. Hence, we suggested that the DH function may be inhibited by insulin signaling pathway under long photoperiod.
Lepidoptera studies have demonstrated that PBAN, subesophageal ganglion neuropeptide (SGNP), and other pyrokinin family neuropeptides have functions similar to DH, but they are less effective than DH in diapause termination in Helicoverpa armigera and Helicoverpa assulta (Zhang et al., 2004; Zhao et al., 2004). All of these pyrokinin family neuropeptides have consensus sequences and could competitively bind to the same GPCR. In addition to encoding I–V type DH-like neuropeptides, the Lom-dh genes can also encode other neuropeptides, such as KGLVASARVa, which may competitively bind DH receptors. Many neuropeptides may exist in L. migratoria to compete with DH-like neuropeptides for DH receptors under long-day photoperiod. Therefore, maybe not all of the DH-like neuropeptides play as an active role in regulating diapause. Our study verified the function of only type I DH. The functions of II–V type DH and other neuropeptides encoded by Lom-dh genes in diapause regulation will require further study.
Conclusion
The diapause hormone, only encoded by capa or pban, and its role in insect diapause regulation, is well researched. In this study, a new capa/pban family member, dh is reported for the first time in L. migratoria. We also identified novel DH neuropeptides. Characters of dh genes are as follows: (1) there is no intron in dh genes. (2) It contains tandem DH-like neuropeptide repeats. (3) Compared to capa and pban, these dh genes have codon bias and they belong to a new type of genes in phylogeny. Finally, the diapause induction function of dh genes was identified by RNAi technology. Moreover, functions of DH neuropeptides encoded by these dh genes were also identified by injection bioassay. Our study shows that multiple DH neuropeptides regulate locust diapause. Diapause regulation by DH in locusts maybe more likely a quantitative character rather than a qualitative character. Our study provides a new perspective for insect diapause regulation. However, how Locusta and other species evolved this new type of genes and their importance to insect physiology still needs to be explored.
Materials and Methods
Insects
The L. migratoria colony studied was maintained at the Institute of Plant Protection, CAAS laboratory for 8 years. Third instars nymphs were collected from the rearing cages and transferred to 20 cm × 20 cm × 28 cm mesh cages in artificial climate chambers (PRX-250B-30, Haishu Saifu Experimental Instrument Factory, Ningbo, China) and reared to adults at 16:8 L:D photoperiod, 60% RH, 28°C conditions to produce non-diapause eggs and at 10:14 L:D photoperiod, 60% RH, 28°C conditions to induce diapause eggs.
Eggs were laid in sand beneath the cages and were collected at an interval of 2 days. Eggs were then transferred into plastic Petri dishes (90 mm × 50 mm) and incubated on vermiculite at 28°C, 60% RH and a 10:14 L:D photoperiod. Nymphs were fed glasshouse grown wheat plants at 16:8 L:D photoperiod, 60% RH and 28°C with food replaced two to three times on daily basis until they reached the adult stage.
To investigate the basis for DH precursor sequences and function, we dissected five adult females from each photoperiod treatment to remove the brain ganglia and nerve cord. The tissues from each locust in each treatment were transferred to 1.5 ml Eppendorf tubes before being snap-frozen in liquid nitrogen and stored at ‑80°C for subsequent gene amplification (see below).
pban/capa Family Gene Sequences Analysis
The insect CAPA/PBAN protein precursor sequences were downloaded from the National Center for Biotechnology Information (NCBI) protein database1. Lom-PK peptides were downloaded from NCBI and merged together as a hypothetical precursor. The L. migratoria whole genome sequences contigs were downloaded in fasta from NCBI Whole Genome Shotgun website2 and analyzed by local BLAST on a desktop PC.
Locusta migratoria genome sequences were used to build searchable BLAST databases3 using ncbi-blast-2.2.30+. Later on, the CAPA/PBAN proteins were aligned to translate locust genome (Camacho et al., 2009). Alignments longer than 100 bp were retained as candidate sequences for identifying the target genes (Supplementary Figure S5).
DNA and cDNA Cloning of the Target Genes
Frozen tissue collected from adult locusts was macerated in 1 × TE buffer before extraction. Genome DNA was extracted using a TaKaRa MiniBEST Universal Genomic DNA Extraction Kit and diluted by ddH2O at a 1:10 ratio. Total RNA was extracted using Invitrogen TRIzol and purified between 1.9 and 2.2 (A260/A280). cDNA was reverse-transcribed using M-MLV reverse transcriptase and recombinant RNase inhibitor (TaKaRa) and diluted by ddH2O at a 1:4 ratio. All DNA and cDNA were prepared for gene amplification. Gene-specific primers were designed according to the predicted pban/capa gene sequences and synthesized by SANGON (Beijing, China). Specific primers were designed to obtain the fragment of the target genes using polymerase chain reaction (PCR; Supplementary Table S3). PCR procedures were performed as follows: 95°C 3 min; 36 cycles of 95°C 30 s; 58°C 30 s; 72°C 90 s; and 72°C 10 min. Bands of interest were gel purified and cloned using the pGEM T-Easy Vector System (Promega) per the manufacturer’s instructions, and then sequenced by Invitrogen, Shanghai, China (Li et al., 2013).
Phylogenetic Analysis of CAPA/PBAN Precursors in Insecta
The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The optimal tree with the sum of branch lengths = 29.94781313 is shown (Supplementary Figure S2). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown next to the branches (Felsenstein, 1985) (Supplementary Figure S2). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965) and are in units of the number of amino acid substitutions per site. The analysis involved 94 amino acid sequences. We removed all ambiguous positions for each sequence pair. The final data-set had a total of 737 positions. Evolutionary analyses were conducted in MEGA6 (Tamura et al., 2013).
Codon Bias Analysis
Codon bias analysis was calculated using CodonW, version 1.4.2 (Peden, 2000). Codon Adaptation Index (CAI) (Sharp and Li, 1987), Frequency of Optimal codons (FOP) (Ikemura, 1981), and Codon Bias Index (CBI) (Bennetzen and Hall, 1982). The G+C content of the third position of synonymous codons (GC3s) was used to evaluate codon bias.
Lom-capa/pban/dh Genes Structure Analysis
For subsequent sequence analysis, the open reading frame (ORF) for Lom-capa/pban genes in L. migratoria genomic DNA was predicted using the website4. Signal peptides were predicted using the hidden Markov model of SignalP4.15 (Veenstra, 2000). Putative cleavage sites in the precursors were analyzed as described by Veenstra (2000) or using web-based NeuroPred program6.
We conducted evolutionary analyses of pyrokinin consensus sequences in MEGA6 (Tamura et al., 2013). The evolutionary history was inferred using the maximum likelihood method based on the Poisson correction model (Zuckerkandl and Pauling, 1965). The tree with the highest log likelihood (−57000.9915) is shown (Figure 2). The percentage of trees in which the associated taxa clustered together is shown next to the branches (Figure 2). Initial tree(s) for the heuristic search were obtained by applying the Neighbor-Joining method to a matrix of pairwise distances estimated using a JTT model. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 16 amino acid sequences. There were a total of seven positions in the final data set.
Liquid Chromatography–Tandem Mass Spectrometry Analysis and Neuropeptide Identification
The central neuron systems of L. migratoria under both long (L:D 16:8) and short (L:D 10:14) photoperiods at a constant temperature of 28°C were homogenized and neuropeptides were extracted at 4°C using a 90:9:1 solution of methanol, H2O, and acetic acid to extract the neuropeptides. The homogenate was then centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected and vacuum-dried using a SpeedVac system (RVC 2–18, Marin Christ, Osterod, Germany) to obtain a neuropeptide sample. Samples were stored at ‑80°C before liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis.
Neuropeptide pellets were re-dissolved in 0.1% formic acid in distilled water, and the final peptide concentration was quantified using a Bradford assay. LC-MS/MS analysis was performed on the Easy-nLC 1000 (Thermo Fisher Scientific, Bremen, Germany) coupled with the LTQ-Orbitrap Elite (Thermo Fisher Scientific) hybrid mass spectrometer with three replicates. Samples were loaded onto a 2 cm long, 100 μm inner diameter fused silica trap column containing 5.0 μm Aqua C18 beads (Thermo Fisher Scientific) for 2 min in buffer A (0.1% formic acid) at a flow rate of 5 μL/min before analytical separation. Peptides were separated on a column packed with 2 μm C18 (100 Å, 75 μm × 50 cm, Thermo Fisher Scientific) at a flow rate of 350 nL/min using the following gradients: from 3 to 8% buffer B in 5 min, from 8 to 20% buffer B in 80 min, from 20 to 30% buffer B in 20 min, from 30 to 90% buffer B in 5 min, and 90% buffer B in 10 min. In data-dependent acquisition mode (range from 300 to 1800 m/z with a resolution of 70,000 at 400 m/z), the 10 most abundant precursor ions with charge states greater than +1 were fragmented and previously acquired precursor ions (repeat count 1, repeat duration: 30 s; exclusion duration 45 s) were dynamically excluded. MS/MS spectra were acquired in higher energy collisional dissociation (HCD) mode with a resolution of 17,500 at 400 m/z and started from 100 m/z using a normalized energy of 30. The MS/MS data were acquired in raw files using Xcalibur software (version 2.2, Thermo Fisher Scientific).
The extracted MS/MS spectra were searched against a composite database containing 3,090 protein sequences of L. migratoria using in-house PEAKS software (version 7.0, Bioinformatics Solutions, Waterloo, ON, Canada). The following modifications were applied: C-terminal amidation (A, −0.98) and pyroglutamination from Q (P, −17.03). The other parameters used were the following: parent ion mass tolerance, 15.0 ppm; fragment ion mass tolerance, 0.05 Da; enzyme, none; and maximum allowed variable post-translational modification per peptide, 2. A fusion target-decoy approach was used for the estimation of the false discovery rate and controlled at ≤1.0% (−10 log P ≥ 20.0) at both protein and peptide levels. Neuropeptide identifications were used only if at least two spectra were identified in one sample.
Bioassay of Neuropeptides Activity
Based on the Lom-DH2 amino acid sequence, four neuropeptides were synthesized (SANGON, Shanghai, China). The neuropeptides were designated as follows: LDH1: MHSKNVVVQRCIQTGFSDGGSIRNVPETSLWFGPRIa, purity > 93.06%; SDH1: LWFGPRIa, purity > 96.27%; LDH2: SNPETNLWFGPRVa, purity > 99.11%; SDH2: LWFGPRVa, purity > 99.10%. For neuropeptide activity bioassays, solutions (10 μl/individual) containing different doses (20–20,000 pmol/individual) of the four neuropeptides (LDH1, SDH1, LDH2, and SDH2) were micro-injected [microprocessor Controller PAX100-3, microapplicator (Burkard, United Kingdom)] into the interstitial membrane between the second and third abdominal segments of the insects with a microapplicator. We randomly removed a total of 100 fifth instar females from the four cages in each photoperiod treatment and injected them with one of the four neuropeptides to provide 25 individuals per treatment. The selected fifth instar nymphs were almost at the verge of adulthood. Another 25 females taken from each photoperiod treatment were injected with distilled water as the controls.
Following treatment, locusts were moved to new mesh cages (20 cm × 20 cm × 28 cm) and provided with bouquets of glasshouse grown wheat. At the same time, 25 adult males were introduced to the cages before being returned to the allocated photoperiod treatments from which they had been collected. The floor of the cages was covered by a 2 cm layer of sieved sterilized sand, with the cage conditions maintained until egg laying commenced. Generally, the time lapse between injection of DH and egg laying was 2–3 weeks. Once oviposition was observed, eggs were collected, using a camel paint brush, after every 48 h for 10 days and transferred to Petri dishes lined with moist filter paper, before transfer to 28°C and 60% RH to slow development. Eggs were kept at 28°C for 30 days until eclosion of first instar nymphs ended. To account for non-viable eggs, remnant unhatched eggs were kept at 4°C for 60 days to provide time to break diapause after which they were incubated at 28°C for 30 days and any further first instar emergence was recorded. The diapause incidence was calculated as following: Diapause incidence (%) = H2/(H1+H2), where H1 are individuals emerging within 30 days at 28°C and H2 individuals emerging following chilling at 4°C for 60 days.
RNA Interference Treatment by Double-Stranded RNA Injection and Real-Time PCR
The dsRNA was generated by in vitro transcription using the T7 RiboMAX system (Promega) according to the manufacturer’s protocol. Templates for in vitro transcription reactions were prepared by PCR amplification from plasmid DNA of the cDNA clone of target genes using the primer pairs with a T7 polymerase promoter sequence at 5′-end (Supplementary Table S3). A total of 10 μl of dsRNAs (1 μg/μl) for the target genes, GFP controls, or water controls were injected into the ventral area between second and third abdominal segments of female adults within 3 days after the nymph to adult molt. The dsLom-dhall was composed of dsLom-dh1, dsLom-dh2/3, and dsLom-dh4/5 with a mole ratio of 1:1:1. For each gene, 75 nymphs were injected and divided into three groups. The effects of RNAi on the mRNA levels were investigated by quantitative real-time PCR (qRT-PCR) at 48 h after injection. To monitor the transcript levels of the target genes, total RNA was extracted from whole bodies. For each target gene, three individuals from each group were used for RNA extraction. The diapause incidence was observed according to previous section of bioassay of neuropeptides activity. Each treatment included 25 female adults that were injected and their eggs were divided into three groups. The effect of RNAi on mRNA levels and diapause incidence was monitored as described earlier.
We used the same methods of RNA extraction and reverse-transcription as described earlier. To quantify the gene expression, the 2–ΔΔCt method was used, with β-actin as the positive control and for normalizing the data. The specific primers used were for qRT-PCR (Supplementary Table S3). We performed RT-PCR with the SYBR Premix ExTaq™ (TaKaRa, Dalian, China) on the ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, United States). The β-actin gene was used as an internal control. The reaction was performed using the following conditions: denaturation at 95 ration 60 s, followed by 40 cycles of amplification (95°C, 15 s; 60°C, 60 s). Melting curve analysis was performed to confirm the specificity of amplification. Each plate was repeated three times in independent runs for all reference and selected genes.
Statistical Analysis
We compared the differences between treatments either by Student’s t-test or by one-way analysis of variance (ANOVA) followed by a Tukey’s test for multiple comparisons. We considered differences significant at P < 0.05 and reported values as mean ± SE. We analyzed data using the SPSS software (version 15.0; SPSS Inc., Chicago, IL, United States).
Author Contributions
ZZ and KH conceived and designed the study. KH performed the experiments, data collection and analysis, and drafted the manuscript. HU, XT, and MM reviewed and edited the manuscript. All authors approved the final manuscript.
Funding
This research was funded by the China Agriculture Research System (CARS-34-07B), China, and the funding body had not involved in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript. We also thank Shan Xue for editing figures and tables.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00767/full#supplementary-material
FIGURE S1 | The seven predicted PBAN, CAPA, tryptopyrokinin precursors, and one pyrokinin amino acid sequences in Locusta migratoria L. Blue sequences indicate signal peptide, green sequences indicate endoproteolytic cleavage sites, and yellow sequences indicates short neuropeptides.
FIGURE S2 | Phylogenetic analysis of CAPA/PBAN precursors in Insecta. Circles represent Hymenoptera, squares represent Lepidoptera, upward triangles represent Diptera, diamonds represent CAPA and PBAN of Locust migratoria, and downward triangles represent TPK of Locusta migratoria.
FIGURE S3 | Codon bias analysis of L. migratoria capa, pban and TPK genes. The lowercase a, b, and c indicated that statistically significant difference of different genes was considered on an error probability of p < 0.05 by one-way ANOVA using Duncan’s multiple-range test.
FIGURE S4 | The sequence logo was created using Weblogo (Crooks et al., 2004).
FIGURE S5 | R mapping result of PBAN/PK precursors based on blasting Locusta migratoria L. genome. The horizontal coordinate indicates genome files, whereas the vertical coordinate indicates mapped amino acid numbers; light blue to dark blue indicates e-value expression level.
TABLE S1 | Candidate pban/capa family gene sequences of the L. migratoria genome contigs.
TABLE S2 | PBAN, CAPA, and DH precursors of insects. The black box indicates consensus sequences, the green amino acid sequence indicates endoproteolytic cleavage sites, and the blue amino acid indicates DH neuropeptides.
TABLE S3 | Specific primers of this chapter.
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/protein/
- ^ http://www.ncbi.nlm.nih.gov/Traces/wgs/?val=AVCP01#contigs
- ^ ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/
- ^ https://omictools.com/genscan-tool
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://stagbeetle.animal.uiuc.edu/cgi-bin/neuropred.py
References
Altstein, M., Hariton, A., and Nachman, R. J. (2013). “FXPRLamide (pyrokinin/PBAN) family,” in Handbook of Biologically Active Peptides, ed. A. J. Kastin (Amsterdam: Elsevier), 255–266. doi: 10.1016/b978-0-12-385095-9.00037-3
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Camargo, A. C., Fernandes, B. L., Cruz, L., and Ferro, E. S. (2012). Bioactive Peptides Produced by Limited Proteolysis. Colloquium Series on Neuropeptides. San Rafael, CA: Morgan & Claypool Life Sciences, 1–92.
Clynen, E., Baggerman, G., Huybrechts, J., Bosch, L. V., De Loof, A., and Schoofs, L. (2003). Peptidomics of the locust corpora allata: identification of novel pyrokinins (-FXPRLamides). Peptides 24, 1493–1500. doi: 10.1016/j.peptides.2003.10.006
Clynen, E., and Schoofs, L. (2009). Peptidomic survey of the locust neuroendocrine system. Insect Biochem. Mol. Biol. 39, 491–507. doi: 10.1016/j.ibmb.2009.06.001
Coast, G. M., and Schooley, D. A. (2011). Toward a consensus nomenclature for insect neuropeptides and peptide hormones. Peptides 32, 620–631. doi: 10.1016/j.peptides.2010.11.006
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x
Gerisch, B., Weitzel, C., Kober-Eisermann, C., Rottiers, V., and Antebi, A. (2001). A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev. Cell 1, 841–851. doi: 10.1016/S1534-5807(01)00085-5
Grimmelikhuijzen, C. J., and Hauser, F. (2012). Mini-review: the evolution of neuropeptide signaling. Regul. Peptides 177, S6–S9. doi: 10.1016/j.regpep.2012.05.001
Hakomori, T., and Tanaka, S. (1992). Genetic control of diapause and other developmental traits in Japanese strains of the migratory locust, (Locusta migratoria L). univoltine vs bivoltine. Jpn. J. Entomol. 60, 319–328.
Hao, K., Jie, W., Xiong-Bing, T. U., Whitman, D. W., and Zhang, Z. H. (2017). Transcriptomic and proteomic analysis of locusta migratoria eggs at different embryonic stages: comparison for diapause and nondiapause regimes. J. Integr. Agric. 16, 1777–1788. doi: 10.1016/S2095-3119(16)61529-0
Hasegawa, K. (1957). The diapause hormone of the silkworm, Bombyx mori. Nature 179:1300. doi: 10.1038/1791300b0
Holman, G. M., Cook, B. J., and Nachman, R. J. (1986). Primary structure and synthesis of a blocked myotropic neuropeptide isolated from the cockroach, leucophaea maderae. comparative biochemistry and physiology. Comp. Pharmacol. Toxicol. 85, 219–224. doi: 10.1016/0742-8413(86)90077-0
Hou, L., Jiang, F., Yang, P., Wang, X., and Kang, L. (2015). Molecular characterization and expression profiles of neuropeptide precursors in the migratory locust. Insect Biochem. Mol. Biol. 63, 63–71. doi: 10.1016/j.ibmb.2015.05.014
Ikeda, M., Su, Z. H., Saito, H., Imai, K., Sato, Y., Isobe, M., et al. (1993). Induction of embryonic diapause and stimulation of ovary trehalase activity in the silkworm, Bombyx mori, by synthetic diapause hormone. J. Insect Physiol. 39, 889–895. doi: 10.1016/0022-1910(93)90122-8
Ikemura, T. (1981). Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151, 389–409. doi: 10.1016/0022-2836(81)90003-6
Ikeno, T., Tanaka, S. I., Numata, H., and Goto, S. G. (2010). Photoperiodic diapause under the control of circadian clock genes in an insect. BMC Biol. 8:116. doi: 10.1186/1741-7007-8-116
Ito, C., Goto, S. G., Shiga, S., Tomioka, K., and Numata, H. (2008). Peripheral circadian clock for the cuticle deposition rhythm in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 105, 8446–8451. doi: 10.1073/pnas.0800145105
Jiang, H., Wei, Z., Nachman, R. J., Adams, M. E., and Park, Y. (2014). Functional phylogenetics reveals contributions of pleiotropic peptide action to ligand-receptor coevolution. Sci. Rep. 4:6800. doi: 10.1038/srep06800
Jiang, X., Yang, J., Shen, Z., Chen, Y., Shi, L., and Zhou, N. (2016). Agonist-mediated activation of Bombyx mori diapause hormone receptor signals to extracellular signal-regulated kinases 1 and 2 through Gq-PLC-PKC-dependent cascade. Insect Biochem. Mol. Biol. 75, 78–88. doi: 10.1016/j.ibmb.2016.06.005
Jünger, M. A., Rintelen, F., Stocker, H., Wasserman, J. D., Végh, M., Radimerski, T., et al. (2003). The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2:20. doi: 10.1186/1475-4924-2-20
Jurenka, R. (2015). The PRXamide neuropeptide signalling system: conserved in animals. Adv. Insect Physiol. 49, 123–170. doi: 10.1016/bs.aiip.2015.07.001
Jurenka, R., and Nusawardani, T. (2011). The pyrokinin/pheromone biosynthesis-activating neuropeptide (PBAN) family of peptides and their receptors in Insecta: evolutionary trace indicates potential receptor ligand-binding domains. Insect Mol. Biol. 20, 323–334. doi: 10.1111/j.1365-2583.2010.01065.x
Lee, R. Y., Hench, J., and Ruvkun, G. (2001). Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 11, 1950–1957. doi: 10.1016/S0960-9822(01)00595-4
Li, J., Zhou, J., Sun, R., Zhang, H., Zong, S., Luo, Y., et al. (2013). cDNA cloning and sequence determination of the pheromone biosynthesis activating neuropeptide from the seabuckthorn carpenterworm, Holcocerus hippophaecolus (Lepidoptera: Cossidae). Arch. Insect. Biochem. Physiol. 82, 183–195. doi: 10.1002/arch.21084
Loi, P. K., and Tublitz, N. J. (2004). Sequence and expression of the CAPA/CAP2b gene in the tobacco hawkmoth, Manduca sexta. J. Exp. Biol. 207, 3681–3691. doi: 10.1242/jeb.01186
Nachman, R. J., Holman, G. M., Schoofs, L., and Yamashita, O. (1993). Silkworm diapause induction activity of myotropic pyrokinin (fxprlamide) insect neuropeptides. Peptides 14, 1043–1048. doi: 10.1016/0196-9781(93)90084-T
Nagasawa, H., Kuniyoshi, H., Arima, R., Kawano, T., Ando, T., and Suzuki, A. (1994). Structure and activity of Bombyx PBAN. Arch. Insect Biochem. Physiol. 25, 261–270. doi: 10.1002/arch.940250403
Park, Y., Kim, Y. J., and Adams, M. E. (2002). Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc. Natl. Acad. Sci. U.S.A. 99, 11423–11428. doi: 10.1073/pnas.162276199
Predel, R., and Neupert, S. (2007). Social behavior and the evolution of neuropeptide genes: lessons from the honeybee genome. BioEssays 29, 416–421. doi: 10.1002/bies.20571
Predel, R., and Wegener, C. (2006). Biology of the CAPA peptides in insects. Cell. Mol. Life Sci. 63, 2477–2490. doi: 10.1007/s00018-006-6187-3
Rahman, M. M., Fromm, B., Neupert, S., Kreusch, S., and Predel, R. (2009). Extended FMRFamides in dipteran insects: conservative expression in the neuroendocrine system is accompanied by rapid sequence evolution. Gen. Comp. Endocrinol. 162, 52–58. doi: 10.1016/j.ygcen.2008.08.006
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sato, Y., Oguchi, M., Menjo, N., Imai, K., Saito, H., Ikeda, M., et al. (1993). Precursor polyprotein for multiple neuropeptides secreted from the suboesophageal ganglion of the silkworm Bombyx mori: characterization of the cDNA encoding the diapause hormone precursor and identification of additional peptides. Proc. Natl. Acad. Sci. U.S.A. 90, 3251–3255. doi: 10.1073/pnas.90.8.3251
Schmitt, F., Vanselow, J. T., Schlosser, A., Kahnt, J., Rössler, W., and Wegener, C. (2015). Neuropeptidomics of the carpenter ant Camponotus floridanus. J. Proteome Res. 14, 1504–1514. doi: 10.1021/pr5011636
Sharp, P. M., and Li, W. H. (1987). The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15, 1281–1295. doi: 10.1093/nar/15.3.1281
Sim, C., and Denlinger, D. L. (2008). Insulin signaling and FOXO regulate the overwintering diapause of the mosquito Culex pipiens. Proc. Natl. Acad. Sci. U.S.A. 105, 6777–6781. doi: 10.1073/pnas.0802067105
Sim, C., and Denlinger, D. L. (2013). Insulin signaling and the regulation of insect diapause. Front. Physiol. 4:189. doi: 10.3389/fphys.2013.00189
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tanaka, H. (1994). Embryonic diapause and life cycle in the migratory locust, Locusta migratoria L. (Orthoptera: Acrididae), in Kyoto. Appl. Entomol. Zool. 29, 179–191. doi: 10.1303/aez.29.179
Tanaka, S. (1992). The significance of embryonic diapause in a Japanese strain of the migratory locust, Locusta migratoria (Orthoptera, Acrididae). Jpn. J. Entomol. 60, 503–520.
Tanaka, S., and Zhu, D. H. (2008). Geographic variation in embryonic diapause, cold-hardiness and life cycles in the migratory locust Locusta migratoria (Orthoptera: Acrididae) in China. Entomol. Sci. 11, 327–339. doi: 10.1111/j.1479-8298.2008.00284.x
Tawfik, A. I., Tanaka, Y., and Tanaka, S. (2002). Possible involvement of ecdysteroids in embryonic diapause of Locusta migratoria. J. Insect Physiol. 48, 743–749. doi: 10.1016/S0022-1910(02)00099-9
Terhzaz, S., Teets, N. M., Cabrero, P., Henderson, L., Ritchie, M. G., Nachman, R. J., et al. (2015). Insect capa neuropeptides impact desiccation and cold tolerance. Proc. Natl. Acad. Sci. U.S.A. 112, 2882–2887. doi: 10.1073/pnas.1501518112
Tu, X., Wang, J., Hao, K., Whitman, D. W., Fan, Y., Cao, G., et al. (2015). Transcriptomic and proteomic analysis of pre-diapause and non-diapause eggs of migratory locust, Locusta migratoria L. (Orthoptera: Acridoidea). Sci. Rep. 5:11402. doi: 10.1038/srep11402
Turkez, H., Ýncekara, Ü, Güner, A., Aydın, E., Dirican, E., and Togar, B. (2014). The cytogenetic effects of the aqueous extracts of migratory locust (Locusta migratoria L.) in vitro. Toxicol. Industr. Health 30, 233–237. doi: 10.1177/0748233712452610
Uehara, H., Senoh, Y., Yoneda, K., Kato, Y., and Shiomi, K. (2011). An FXPRLamide neuropeptide induces seasonal reproductive polyphenism underlying a life-history tradeoff in the tussock moth. PLoS One 6:e24213. doi: 10.1371/journal.pone.0024213
Veenstra, J. A. (2000). Mono-and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. Physiol. 43, 49–63. doi: 10.1002/(sici)1520-6327(200002)43:2<49::aid-arch1>3.0.co;2-m
Veenstra, J. A. (2010). Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen. Comp. Endocrinol. 167, 86–103. doi: 10.1016/j.ygcen.2010.02.010
Veenstra, J. A. (2014). The contribution of the genomes of a termite and a locust to our understanding of insect neuropeptides and neurohormones. Front. Physiol. 5:454. doi: 10.3389/fphys.2014.00454
Wardhaugh, K. G. (2010). The effects of temperature and moisture on the inception of diapause in eggs of the Australian plague locust, Chortoicetes terminifera walker (Orthoptera: Acrididae). Aus. J. Ecol. 5, 187–191. doi: 10.1111/j.1442-9993.1980.tb01241.x
Williams, K. D., Busto, M., Suster, M. L., So, A. K. C., Ben-Shahar, Y., Leevers, S. J., et al. (2006). Natural variation in Drosophila melanogaster diapause due to the insulin-regulated PI3-kinase. Proc. Natl. Acad. Sci. U.S.A. 103, 15911–15915. doi: 10.1073/pnas.0604592103
Xu, W. H., and Denlinger, D. L. (2003). Molecular characterization of prothoracicotropic hormone and diapause hormone in Heliothis virescens during diapause, and a new role for diapause hormone. Insect Mol. Biol. 12, 509–516. doi: 10.1046/j.1365-2583.2003.00437.x
Xu, W. H., and Denlinger, D. L. (2004). Identification of a cDNA encoding DH, PBAN and other FXPRL neuropeptides from the tobacco hornworm, Manduca sexta, and expression associated with pupal diapause. Peptides 25, 1099–1106. doi: 10.1016/j.peptides.2004.03.021
Xu, W. H., Sato, Y., Ikeda, M., and Yamashita, O. (1995). Molecular characterization of the gene encoding the precursor protein of diapause hormone and pheromone biosynthesis activating neuropeptide (DH-PBAN) of the silkworm, Bombyx mori and its distribution in some insects. Biochim. Biophys. Acta 1261, 83–89. doi: 10.1016/0167-4781(94)00238-X
Yamashita, O. (1981). Hormonal and metabolic control of egg diapause of the silkworm, Bombyx mori (Lepidoptera: Bombycidae). Entomol. Gen. 7, 195–211.
Zhang, Q., Nachman, R. J., and Denlinger, D. L. (2015). Diapause hormone in the Helicoverpa/Heliothis complex: a review of gene expression, peptide structure and activity, analog and antagonist development, and the receptor. Peptides 72, 196–201. doi: 10.1016/j.peptides.2015.05.005
Zhang, T. Y., Sun, J. S., Zhang, L. B., Shen, J. L., and Xu, W. H. (2004). Cloning and expression of the cdna encoding the fxprl family of peptides and a functional analysis of their effect on breaking pupal diapause in Helicoverpa armigera. J. Insect Physiol. 50, 25–33. doi: 10.1016/j.jinsphys.2003.09.006
Zhao, J. Y., Xu, W. H., and Kang, L. (2004). Functional analysis of the SGNP I in the pupal diapause of the oriental tobacco budworm, Helicoverpa assulta (Lepidoptera: Noctuidae). Regul. Peptides 118, 25–31. doi: 10.1016/j.regpep.2003.10.024
Keywords: diapause hormone, diapause induction, tandem repeat, RNAi, neuropeptide
Citation: Hao K, Tu X, Ullah H, McNeill MR and Zhang Z (2019) Novel Lom-dh Genes Play Potential Role in Promoting Egg Diapause of Locusta migratoria L. Front. Physiol. 10:767. doi: 10.3389/fphys.2019.00767
Received: 29 January 2019; Accepted: 03 June 2019;
Published: 18 June 2019.
Edited by:
Fernando Ariel Genta, Oswaldo Cruz Foundation (Fiocruz), BrazilReviewed by:
Wen Liu, Huazhong Agricultural University, ChinaDavid L. Denlinger, The Ohio State University, United States
Copyright © 2019 Hao, Tu, Ullah, McNeill and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zehua Zhang, emhhbmd6ZWh1YUBjYWFzLmNu
†These authors have contributed equally to this work
 Kun Hao
Kun Hao Xiongbing Tu1†
Xiongbing Tu1† Hidayat Ullah
Hidayat Ullah Mark Richard McNeill
Mark Richard McNeill Zehua Zhang
Zehua Zhang
