- 1Key Laboratory of Sustainable Forest Ecosystem Management-Ministry of Education, School of Forestry, Northeast Forestry University, Harbin, China
- 2Department of Entomology, Pennsylvania State University, University Park, PA, United States
The ocular albinism type 1 (OA1), a pigment cell-specific integral membrane glycoprotein, is a member of the G-protein-coupled receptor (GPCR) superfamily that binds to heterotrimeric G proteins in mammalian cells. We aimed to characterize the physiological functions an insect OA1 from Lymantria dispar (LdOA1) employs in the regulation of insecticide tolerance. In the present study, we investigated the roles of LdOA1 in response to deltamethrin exposure in both L. dispar and Drosophila melanogaster. LdOA1 was expressed at the lowest level during the 4th instar stage, while LdOA1 was significantly upregulated in the 5th instar and male stages. Knockdown of LdOA1 by injecting dsRNA of LdOA1 into gypsy moth larvae caused a 4.80-fold higher mortality than in control larvae microinjected with dsRNA of GFP under deltamethrin stress. Nine out of 11 L. dispar CYP genes were significantly downregulated under deltamethrin stress in LdOA1 silenced larvae as compared to control larvae. Moreover, the LdOA1 gene was successfully overexpressed in D. melanogaster using transgenic technique. The deltamethrin contact assay showed that the LdOA1 overexpression in flies significantly enhanced the tolerance to deltamethrin compared to the control flies. Furthermore, the downstream Drosophila CYP genes were upregulated in the LdOA1 overexpression flies, suggesting LdOA1 may play a master switch role in P450-mediated metabolic detoxification. This study is the first report of an insect OA1 gene regulating insecticide tolerance and potentially playing a role in the regulation of downstream cytochrome P450 expression. These results contribute to the future development of novel insecticides targeting insect GPCRs.
Introduction
The Asian gypsy moth, Lymantria dispar (Lepidotera: Erebidae), is a key forest pest mainly present in Asia and Europe. As a polyphagous herbivore, L. dispar can feed on at least 500 species of plants in over 100 botanical families, particularly on oak, poplar, and birch (Roy et al., 1995; Lazarevic et al., 1998). Outbreaks of L. dispar lead to serious damage of forests, orchards, and landscaping. In Northern and Eastern China, the Asian gypsy moth has defoliated about two million acres of forest each year over the past 20 years (Sun et al., 2014). Since the last century, the Asian gypsy moth has also been observed in North America and has presented a huge threat to North American forests once the population became established (Roy et al., 1995; United States Department of Agriculture [USDA], 2016). Compared to the subspecies of gypsy moth known in Europe and North America, the “Asian form” gypsy moth has active flying females and a much broader host range (Roy et al., 1995). To manage gypsy moths, a variety of strategies have been used, including chemical control, release of natural enemies (Coccygomimus disparis), biological control (Bacillus thuringiensis), and host plant transgenic engineering (transgenic poplar expressing fusion protein genes of the spider insecticidal peptide and Bt-toxin C-peptide) (Hou et al., 2009; Cao et al., 2010; Li, 2012). Among these strategies, spraying insecticides (e.g., organophosphates, pyrethroids, and carbamates) has remained the most rapid and effective control method in China so far (Ni et al., 2009; Li, 2012). However, it has become increasingly important to develop new insecticides with safe and novel modes of action due to the growing challenges associated with the development of insecticide resistance, off-target effects, and environmental contamination (Audsley and Down, 2015).
G-protein-coupled receptors (GPCRs) comprise one of the largest and most diverse family of membrane proteins, which transduce extracellular signals into cellular responses to hormones, neurotransmitters, and environmental stimulants (Hill et al., 2002; Rosenbaum et al., 2009). The ligands of GPCRs are extremely diverse, including chromophores, neuropeptides and hormones, acetylcholine, biogenic amines, nucleotides and nucleosides, lipids and eicosanoids, olfactory and taste substances, and so on (Broeck, 2001; Klabunde and Hessler, 2002). Ligand binding triggers GPCRs conformational changes, activating complex cytosolic signaling networks and causing intracellular responses. Therefore, GPCRs play various important roles in modulating sense of vision, smell and taste, immune system and autonomic nervous system, regulating reproduction, behavior, osmoregulation, growth, and development, and they draw much attention for drug discovery (Klabunde and Hessler, 2002; Garland, 2013). As a result, GPCRs are targets for many of the best-selling drugs and roughly 30–50% of all medicines in the pharmaceutical market (Klabunde and Hessler, 2002; Filmore, 2004; Garland, 2013).
Insect GPCRs have been recognized as highly attractive targets for new generation insecticide discovery due to their critical functions in insect reproduction, development, behavior, and metabolism (Broeck, 2001; Bai et al., 2011; Caers et al., 2012; Meyer et al., 2012; Audsley and Down, 2015; Bai and Palli, 2016; Hill et al., 2018). Inhibition or overstimulation of insect GPCRs may cause the death of a pest or disrupt its normal physiological processes and reduce pest populations (Mitri et al., 2009; Bai et al., 2011; Caers et al., 2012). Ocular albinism type 1 protein (OA1; GPR143) is a pigment cell-specific glycoprotein with the characteristic seven transmembrane structural features, and amino acid sequence conservation consistent with the GPCR family of membrane proteins. In humans and mice, OA1 mutations have been documented to generate a decrease or deficiency in melanin biogenesis, which often results in ocular albinism (d′Addio et al., 2000). The ocular albinism disease causes severe reductions in visual acuity via an increase of retinal pigment loss by the OA1 mutation (Schiaffino, 2010). Insect melanin plays a key role in pigment synthesis. For example, pigmentation is one of the most variable traits in the genus Drosophila (Wittkopp et al., 2003). However, the functions and mechanisms of insect OA1 in response to insecticide stresses remain unclear. Our previous study reported the transcriptional expression patterns of LdOA1 under deltamethrin, carbaryl, and omethoate stresses (Cao et al., 2014). Here, we further investigated whether the LdOA1 is associated with tolerance to insecticide stresses and involved in the regulation of downstream detoxification genes (e.g., cytochrome P450s) in response to insecticide stresses.
Materials and Methods
Insects
Lymantria dispar eggs and the artificial diet for larvae were purchased from the Research Institute of Forest Ecology, Environment and Protection, Chinese Academy of Forestry (Beijing, China). The L. dispar line purchased had been maintained in the lab for >5 years without exposure to pesticides. Rearing of L. dispar was modified from the protocol of Cao et al. (2015). In brief, gypsy moth larvae were kept in transparent plastic bottles (250 mL) with artificial diets at 25 ± 1°C and 16:8 h light:dark photoperiod. The humidity of the rearing bottles and artificial diets was maintained with botanical sponges soaked with water. Healthy gypsy moth larvae of 3rd instar larvae were used for bioassays, dsRNA injection, and RNA extractions.
RNA Extraction, cDNA Synthesis, and qRT-PCR Analysis
Total RNA was extracted from insect samples with Trizol reagent (Invitrogen®) following the manufacturer’s protocol. The total RNA was treated with DNase I (Ambion Inc., Austin, TX, United States) for contaminating genomic DNA (gDNA) removal. Approximately 0.5 μg of DNase I-treated RNA was reverse transcribed to cDNA using 1 μM of oligo (dT) primer in a 10 μL reaction. Synthesized cDNA was diluted with sterile water to 100 μL, and the solution was used as templates for qRT-PCR in an MX3000P machine (StrataGene, Agilent, CA, United States). Actin (MK926773), EF1α (MK926771), and TUB (MK926772) genes were used as reference genes for gypsy moths and RpL32 and ATP Binding Protein (ABP) genes were chosen as internal controls for Drosophila (Willis et al., 2010). The reaction mixture (20 μL) contained 10 μL of SYBR Green real-time PCR master mix (Toyobo, Osaka, Japan), 0.5 μM each of forward and reverse primers, and 2 μL of cDNA template (equivalent to 100 ng of total RNA). DNA amplification was conducted using the following cycling parameters: 94°C for 30 s followed by 45 cycles of 94°C for 12 s, 60°C for 30 s, 72°C for 40 s, and 1 s at 82°C for plate reading. A melting curve generated for each sample at the end of each run was used to assess the purity of the amplified products. Expression levels were calculated using the 2–ΔΔCT method (Pfaffl et al., 2002). qRT-PCR was performed with three independent biological repeats using 20 Drosophila or gypsy moth larvae for each replicate to ensure reproducibility.
RNA Interference (RNAi) Bioassay in L. dispar Larvae
The function of the LdOA1 gene was investigated using the RNA interference (RNAi) technique. Approximately 200–650 bp fragments based on the full length of the LdOA1 gene and a green fluorescent protein gene (GFP-pMW1650, a generous gift from Professor Nannan Liu, Auburn University, Alabama, United States) were used to generate cDNA of the L. dispar larvae and the pMW1650 plasmid as templates, respectively. The specific primers were designed with the T7 promoter (Supplementary Table S1). dsRNA was synthesized in vitro using the MEGAscript T7 High Yield Transcription kit (Ambion) following the manufacturers’ protocol. dsRNA was purified with phenol/chloroform followed by ethanol precipitation. dsRNA was diluted with nuclease-free water, and a 1∼2 μg/μL dsRNA solution (0.5∼1 μL) was microinjected into the penultimate posterior abdominal section of each 3rd instar gypsy moth larvae using an injection needle (MICROLITERTM #65 with 33-gauge needle, Hamilton Co., Reno, NV, United States) under CO2 gas exposure. Control gypsy moth larvae were microinjected with the GFP dsRNA. Injected gypsy moth larvae were removed to recover for 2 h at room temperature and then were reared on an artificial diet under a 16:8 light:dark photoperiod at 25 ± 1°C. After 48 h, 72 h, 96 h, and 120 h, the L. dispar larvae were selected to measure effects of dsRNA microinjection on LdOA1 transcript levels by qRT-PCR.
Deltamethrin Challenge in L. dispar Larvae
To study the effects of deltamethrin on cytochrome P450 genes in L. dispar larvae microinjected with dsRNAs of LdOA1 or GFP (dsLdOA1 or dsGFP), LC20 value after 24 h of deltamethrin (15 mg/L) exposure was used as the treatment concentration (Sun et al., 2016). 15 mg/L of deltamethrin was added to the artificial diet and fed to the 3rd instar larvae at 120 h after microinjection with dsLdOA1 and dsGFP. 20 healthy larvae were collected for each replicate at 24 h and stored at -80°C for RNA extraction. Each treatment was repeated three times. The cumulative mortality of larvae in both control (dsGFP) and treatment (dsLdOA1) was recorded 5 days after the deltamethrin challenge. The transcript levels of L. dispar CYP genes were measured using qRT-PCR. The primers are listed in Supplementary Table S1.
Construction of UAS-LdOA1 Transgenic Drosophila
The Drosophila expressing LdOA1 gene was generated with the GAL4/UAS system following the protocol of Riveron et al. (2014). Briefly, the full-length LdOA1 was amplified from cDNA using RT-PCR with the primers containing restriction sites for EcoRI and XhoI listed in Supplementary Table S1 and ligated into pMD18-T vector. After digesting plasmids LdOA1/pMD18-T and pUAST-attB with EcoRI and XhoI, the LdOA1 gene was subsequently cloned into the pUAST-attB vector to obtain a recombinant plasmid, pUAST-attB-LdOA1. Using PhiC31 system (Markstein et al., 2008), the recombinant plasmid pUAST-attB-LdOA1 was microinjected into the embryos of D. melanogaster strain carrying docking site on chromosome 3 [UAS-phi2b2a; VK5] to generate the transformant line UAS-LdOA1. To ubiquitously express LdOA1 in Drosophila, UAS-LdOA1 flies were crossed with a ubiquitous tissue driver Act5C-Gal4. UAS-LdOA1 crossing with w1118 was used as the control. To confirm the expression of LdOA1 in the transformant Drosophila, the total RNA and DNA were extracted from three pools of ten flies of each strain. The LdOA1 gene was amplified by RT-PCR or PCR.
Drosophila Contact Bioassay and Deltamethrin Challenge
The contact bioassay was used to measure the susceptibility of Drosophila to deltamethrin. 200 μL of acetone was used as a control, and different concentrations of deltamethrin diluted in acetone were coated on the inside of glass scintillation vials (40 mL) by rolling the vial until the acetone evaporated following the protocol described in our previous study (Zhu et al., 2010). The small diet (approximately 1 cm × 1 cm) was placed into vials plugged with cotton balls. Ten 1–3 day(s) old posteclosion Drosophila adults (sex ratio = 1:1) were placed into each vial, and the mortality was scored after 24 h of treatment. Each concentration was repeated three times independently. Drosophila were reared under a 16:8 light:dark photoperiod at 25 ± 1°C. The medium lethal concentration (LC50) and 20% lethal concentration (LC20) values at 24 h obtained from strains (Act5 > UAS-LdOA1 and UAS-LdOA1 > w1118) were calculated by POLO software (LeOra Software Inc., Petaluma, CA, United States). Each treatment was repeated three times independently. The transcriptional expression levels of Drosophila cytochrome P450 genes in UAS-LdOA1 > w1118 and Act5 > UAS-LdOA1 strains were measured by qRT-PCR. Before the bioassay, 20 control (UAS-LdOA1 > w1118) and 20 LdOA1 overexpression (Act5 > UAS-LdOA1) flies (sex ratio = 1:1) were selected to measure the weight. Three replicates were performed. The average weight of each fly in both groups was calculated and compared.
Statistical Analysis
The data are represented in figures as means ± SD. The difference between two treatments was compared by Student’s t-test (two-tailed paired t-test). The difference among multiple samples was calculated by one-way analysis of variance (ANOVA) followed by Student-Newman-Keuls multiple comparisons test using a statistical software package (GraphPad InStat version 3.05). The level of significance was set at P < 0.05.
Results
Developmental Expression Profile of the LdOA1 Gene
The transcriptional expression levels of the LdOA1 gene were examined in different developmental stages of the Asian gypsy moth, including eggs, larvae (1st to 6th instars), pupae, male and female adults. The expression of the LdOA1 gene varies among different developmental stages. Compared with other stages, the expression of LdOA1 in the 4th instar larva exhibited the lowest level. Whereas, the expression level of LdOA1 in the male was the maximum (Figure 1).
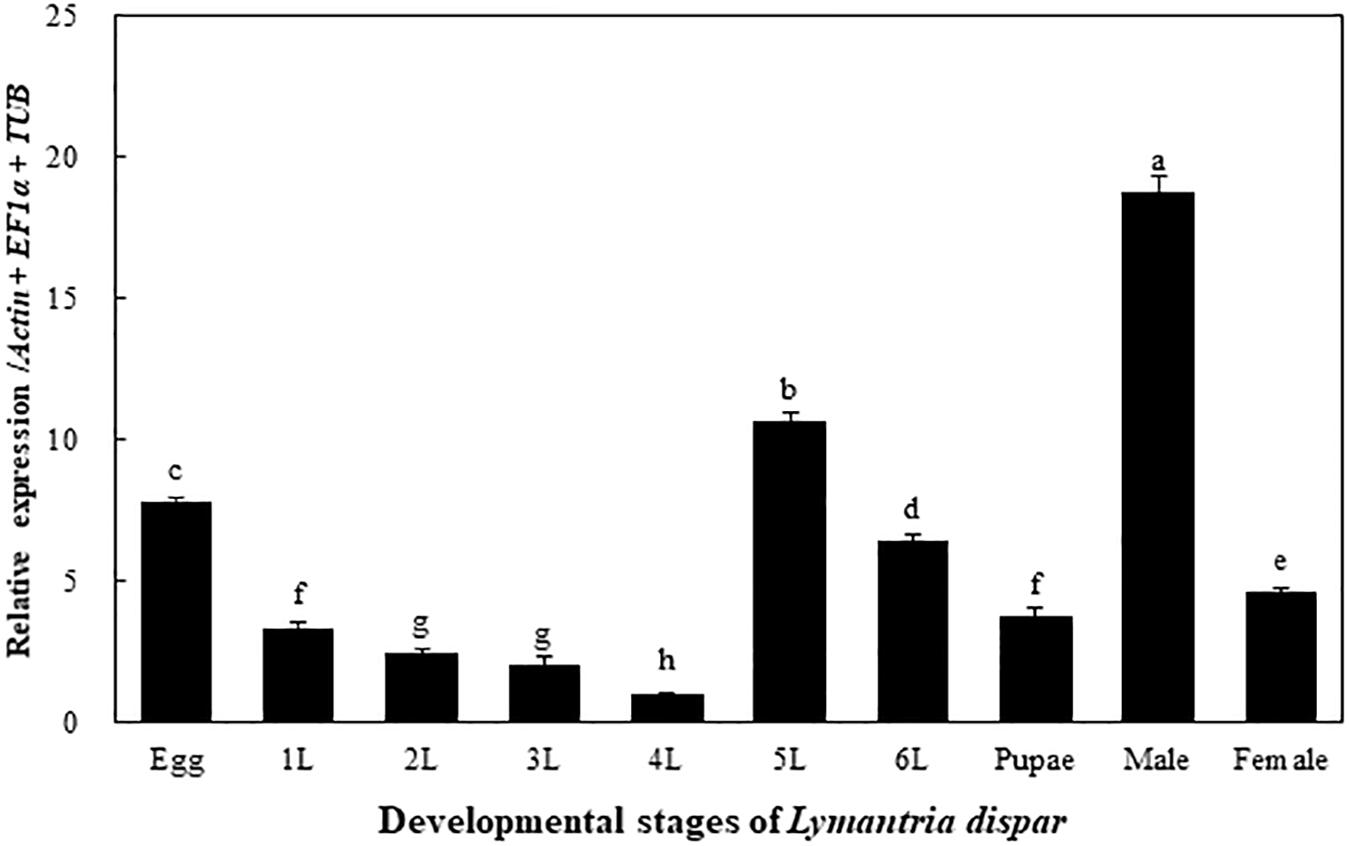
Figure 1. Transcriptional expression of the LdOA1 gene during different developmental stages of L. dispar. 1 to 6 L represent 1st to 6th instar L. dispar larvae. Data were normalized to three most stable reference genes Actin, EF1α, and TUB. The 2–ΔΔCT method was used to calculate relative expression levels. The expression was shown as mean ± SD from three independent assays. Different lowercase letters above bars indicate significant difference among developmental stages according to ANOVA followed by Student-Newman-Keuls multiple comparisons test (P < 0.05).
LdOA1 Silencing and Its Response to Deltamethrin Challenge
To investigate the function of LdOA1 in response to insecticide challenging, we first employed RNA interference (RNAi) technology to knock down the expression of LdOA1 in gypsy moth by injecting dsRNA into the penultimate posterior abdominal section of the 3rd instar larva (Figure 2A). RNAi results showed that the mRNA level of LdOA1 in the 3rd instar larvae microinjected with dsLdOA1 was significantly lower than that in the larvae microinjected with dsGFP (Figure 2B). At 120 h after injection of dsLdOA1, the LdOA1 mRNA level decreased by 70% (Figure 2B).
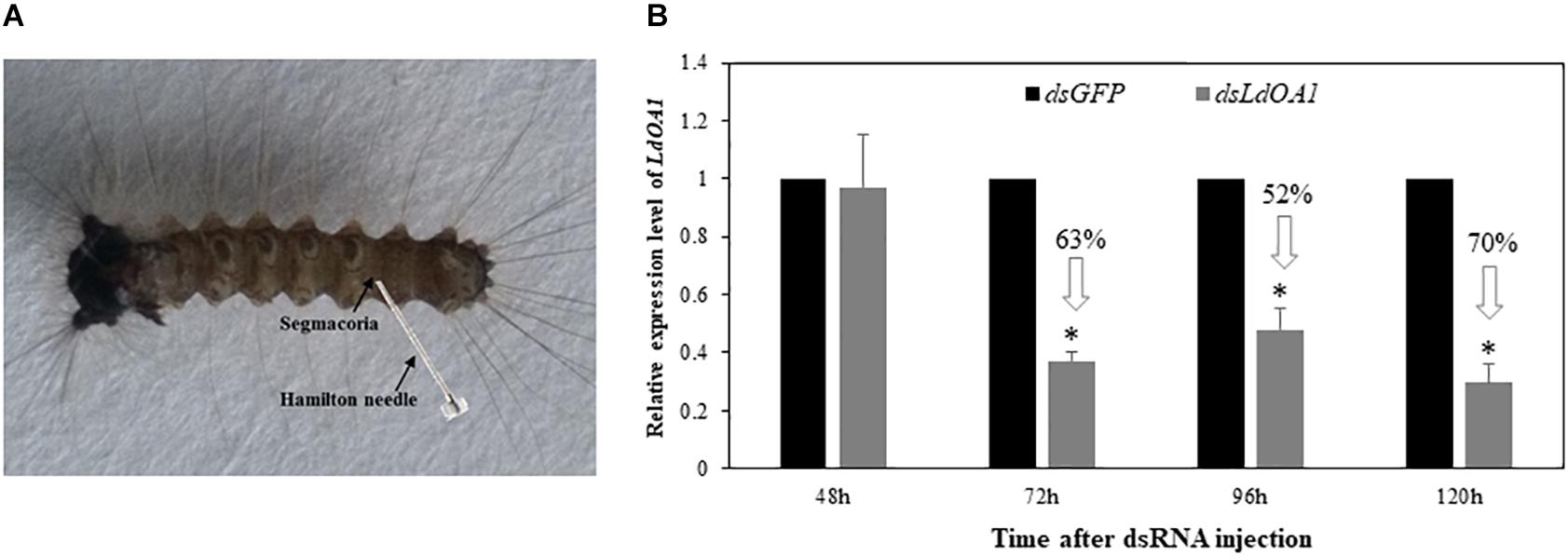
Figure 2. Effect of RNAi on LdOA1 gene expression in 3rd instar L. dispar larvae after dsRNA injection. (A) 3rd instar L. dispar larva receiving dsRNA injection via the segmacorla. (B) LdOA1 transcript levels were determined by qRT-PCR for green fluorescent protein dsRNA-injected (dsGFP) and LdOA1 dsRNA-injected (dsLdOA1) 3rd instar L. dispar larvae. Data were normalized to three most stable reference genes Actin, EF1α, and TUB. The 2–ΔΔCT method was used to calculate relative expression levels. The gene expression was shown as mean ± SD from three independent assays. Asterisk (*) indicates significant difference according to Student’s t-test (P < 0.05).
Gypsy moth larvae with silenced LdOA1 or GFP were then treated with the sublethal dose of deltamethrin (LC20 = 15 mg/L). Under the stress of deltamethrin, larvae treated with dsLdOA1 showed a 4.80-fold greater mortality than the larvae treated with dsGFP (Figure 3A).
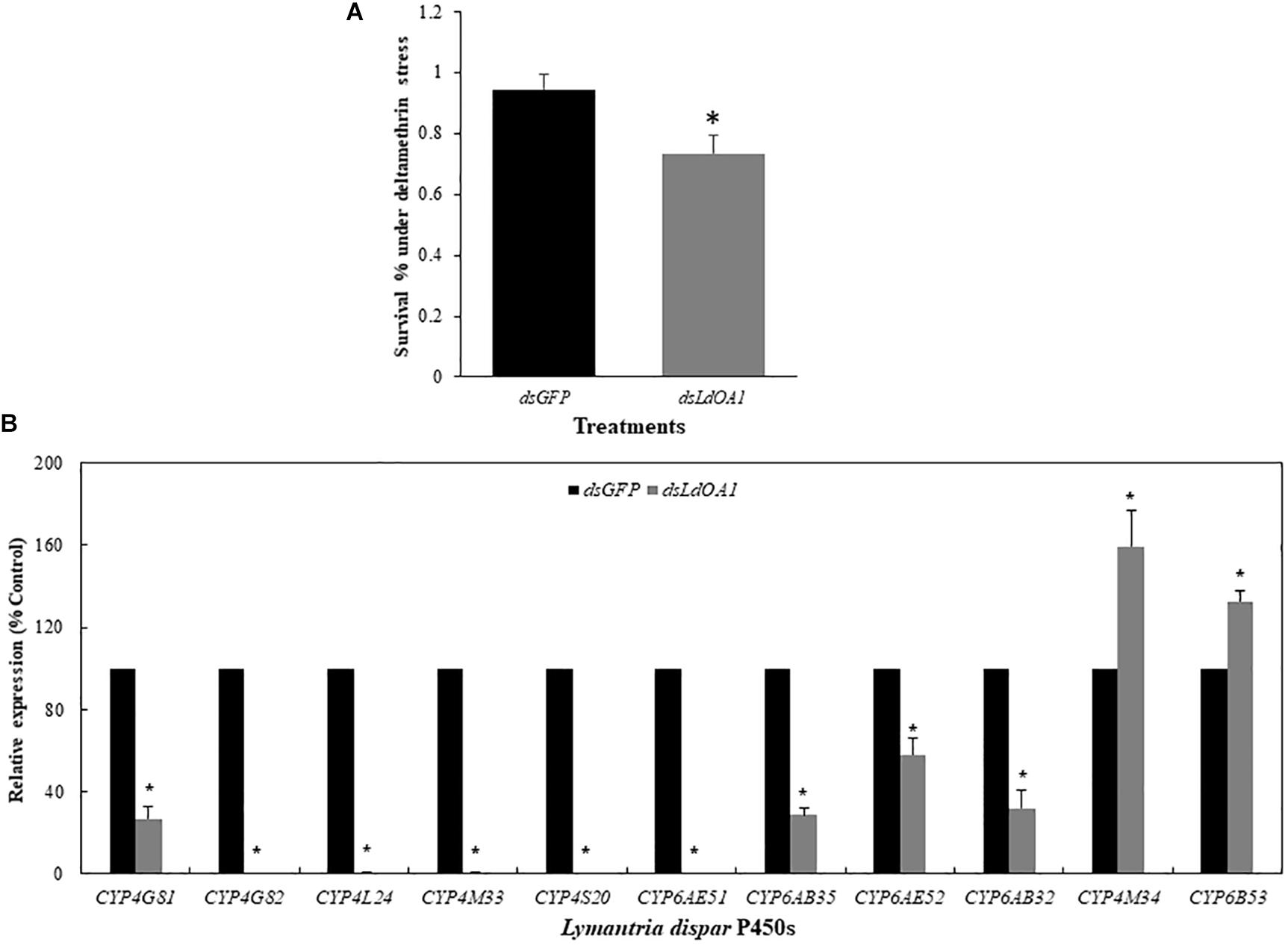
Figure 3. Effects of sublethal deltamethrin (24 h LC20 = 15 mg/L) on 3rd instar L. dispar larvae survival (A) and CYP gene expressions (B) 120 h after injecting dsRNA of green fluorescent protein (GFP, control) or dsRNA of LdOA1. The cumulative mortality of larvae in both control and treatment was recorded 5 days after the deltamethrin challenge. Steady-state CYP transcript levels were determined by qRT-PCR for GFP dsRNA-injected (dsGFP) and LdOA1 dsRNA-injected (dsLdOA1) 3rd instar L. dispar larvae. Data were normalized to Actin, EF1α, and TUB internal controls, and 2–ΔΔCT method was used to obtain relative expression levels, expressed as mean ± SD from assays performed in triplicate. Asterisk (*) indicates significant differences among different treatments at the same gene by Student’s t-test (P < 0.05).
Eleven cytochrome P450 genes were selected from the transcriptome of gypsy moth and our previous studies (Sun et al., 2014; Cao et al., 2015) to detect the effects of LdOA1 RNAi and deltamethrin challenge on the expression of down-stream cytochrome P450 genes (Figure 3B). Except for CYP4M34 and CYP6B53, the expression of nine cytochrome P450 genes was significantly decreased under deltamethrin stress in LdOA1 RNAi larva compared to the control larva (Figure 3B). These results showed that the LdOA1 silencing decreased the tolerance to deltamethrin in L. dispar larva and suppressed cytochrome P450 gene expression under the stress of deltamethrin.
Drosophila With Ectopically Expressed LdOA1 Shows Enhanced Tolerance to Deltamethrin
To further investigate the function of LdOA1 in the regulation of insecticide stress tolerance, the LdOA1 was ectopically expressed in Drosophila using GAL4/UAS system. After confirming the expression of LdOA1 in F1 progeny by RT-PCR and PCR (Supplementary Figure S1), the results of deltamethrin toxicity assay were shown in Table 1. The LC50 values of deltamethrin for the control flies (UAS-LdOA1 > w1118) and LdOA1 overexpressed flies (Act5 > UAS-LdOA1) were 3.97 and 8.03 mg/L, respectively (Table 1), revealing that there was a 2.02-fold higher tolerance to deltamethrin in the flies with ectopically overexpressed LdOA1 gene. The 95% confidence intervals (95% CI) were not overlapped between the LdOA1 overexpression flies and control flies, suggesting the enhancement of deltamethrin tolerance was significant (Table 1).
Expression of Cytochrome P450 Genes in LdOA1-Expressing Drosophila
To investigate the regulation of LdOA1 on Drosophila cytochrome P450 gene expression in LdOA1-expressing Drosophila, the expressions of seven Drosophila cytochrome P450 genes were examined and compared in LdOA1-expressing Drosophila and control flies (Figure 4). Among the seven P450 genes, five of them, including Cyp4ac3, Cyp6a2, Cyp6a9, Cyp6g1, and Cyp6w1, were upregulated in the LdOA1-expressing Drosophila compared to the control flies (Figure 4). The expression levels of the remaining two Drosophila CYP genes (Cyp4e2 and Cyp6a8) exhibited no difference between LdOA1 overexpression files and control files (Figure 4).
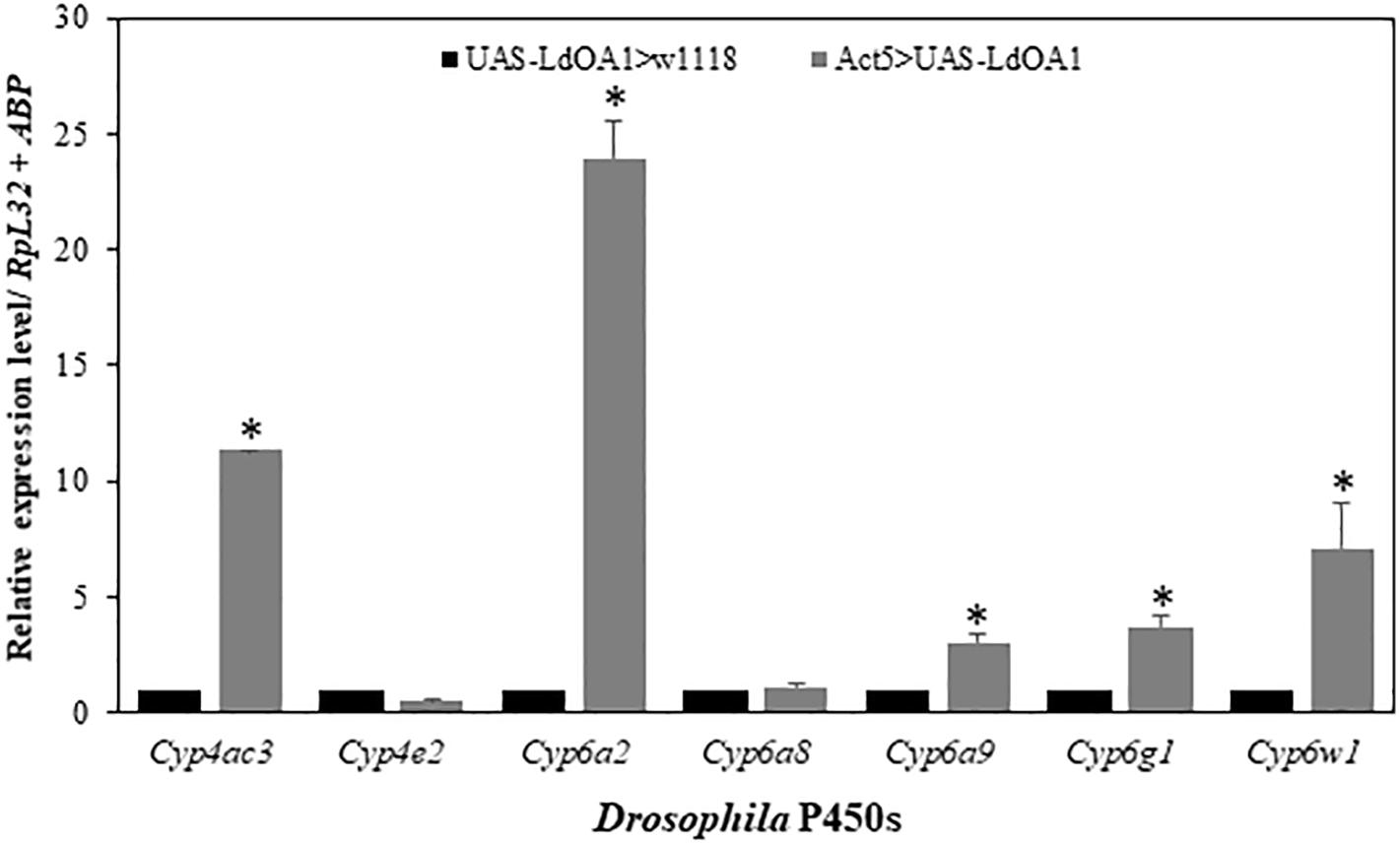
Figure 4. The relative expression levels of Drosophila CYP genes in the LdOA1 overexpression flies (Act5 > UAS-LdOA1) comparing to the control flies (UAS-LdOA1 > w1118). Data were normalized to two most stable reference genes RpL32 and ABP. The 2–ΔΔCT method was used to calculate relative expression levels. The gene expression was shown as mean ± SD from three independent assays. * above bars indicate significant difference according to Student’s t-test (P < 0.05).
Discussion
LdOA1 GPCR is primarily associated with intracellular late endosome/lysosome and melanosomes, with its N-terminus located toward the lumen of the organelle and its C-terminus toward the cytoplasm (d′Addio et al., 2000). The OA1 has been observed to provide the cell with a mechanism to sense melanosome maturation and regulate organelle biogenesis and homeostasis (Raposo and Marks, 2007; Bai et al., 2014). In humans and mice, the OA1 genes encode for pigment cell-specific integral membrane glycoproteins that contain several putative transmembrane domains, and consist of 404 and 405 amino acids, respectively. The OA1 gene is exclusively expressed in the pigment cells of the skin and eye (Bassi et al., 1996; Schiaffino et al., 1996, 1999). To date, the first component of the signaling cascade triggered by OA1 has been identified as Gαi3 (Young et al., 2008, 2011, 2013). However, identity of its downstream targets and the cause of macromelanosome production remain unknown. Recently, Cao et al. (2014) reported that the expression of LdOA1 in 3rd instar L. dispar larvae was significantly inhibited by deltamethrin, carbaryl and omethoate stresses during the 72 h period of exposure. Sun et al. (2016) had provided additional confirmation that knockdown of the LdOA1 gene not only increased the mortality of 3rd instar L. dispar larvae but also decreased their body weight, relative consumption rate (RCR), and approximate digestibility (AD) based on the nutrition utilization index. In the present study, we investigated the roles of LdOA1 in the regulation of deltamethrin tolerance and cytochrome P450 expression in both L. dispar and D. melanogaster. Our study showed that RNAi of LdOA1 in L. dispar larvae led to significantly decreased deltamethrin tolerance (Figure 3A). In contrast, the overexpression of LdOA1 in Drosophila resulted in enhanced tolerance to deltamethrin stress over control flies (Table 1). These results suggest that LdOA1 plays an important role in the regulation of deltamethrin tolerance. Similarly, in Culex pipiens pallens, several GPCRs have shown the functions associated with deltamethrin resistance (Hu et al., 2007; Sun et al., 2012; Zhou et al., 2018). In C. quinquefasciatus, four GPCR-related genes are involved in the regulation of permethrin resistance (Liu et al., 2007; Li et al., 2015). In Tribolium castaneum, RNAi of a GPCR Latrophilin decreases larval tolerance to carbamates and organophosphates, suggesting the role of Latrophilin in insecticide tolerance (Gao et al., 2018). These evidence indicate that GPCR regulatory pathways are a common mechanism in the regulation of insecticide tolerance or resistance (Liu, 2015).
Our study also reveals that LdOA1 may play a master switch role for P450-mediated metabolic detoxification. Under deltamethrin stress, the expressions of nine L. dispar P450 genes (CYP4G81, CYP4G82, CYP4L24, CYP4M33, CYP4S20, CYP6AE51, CYP6AB35, CYP6AE52, and CYP6AB32) in gypsy moth larva with LdOA1 silencing was significantly decreased compared to the control larva (Figure 3B). In LdOA1 overexpression Drosophila, five Drosophila P450 genes (Cyp4ac3, Cyp6a2, Cyp6a9, Cyp6g1, and Cyp6w1) were significantly upregulated compared to control flies (Figure 4). Some of these genes (e.g., Cyp6a2, Cyp6g1, and Cyp6w1) have been shown to confer insecticide resistance or link with xenobiotic metabolism in Drosophila populations (Daborn et al., 2002; Battlay et al., 2016; Denecke et al., 2017; Seong et al., 2018; Huang et al., 2019). Our study is consistent with other insect GPCR studies. For example, in C. pipiens pallens, the expressions of CYP6A1 and opsin were upregulated in mosquito cells transfected with arrestin GPCR and suppressed after arrestin silencing (Sun et al., 2012). Knockdown of a rhodopsin-like GPCR gene or its downstream effectors from permethrin resistant C. quinquefasciatus reduced the expression of four permethrin-resistance related P450 genes (Li et al., 2014, 2015; Li and Liu, 2017, 2018). A recent study showed that knockdown of an opsin GPCR, NYD-OP7, from permethrin resistant C. pipiens pallens strain led to reduced expression of five P450 genes from seven genes tested (Zhou et al., 2018).
Cytochrome P450s are extremely important detoxification enzymes and play vital roles in xenobiotic adaptation (Li et al., 2007; Liu and Zhu, 2011; Feyereisen, 2012; Zhu et al., 2016; Zhang et al., 2018). Cytochrome P450-mediated detoxification is a universal mechanism contributing resistance to various pesticides and nature-derived toxins, regardless of the mode of action (Scott et al., 1998; Feyereisen, 2012; Zhu et al., 2013). Therefore, understanding the genetic machinery underpinning regulation of P450-mediated xenobiotic detoxification will facilitate the development of new pest control strategies (Misra et al., 2011; Zhu et al., 2014). Insect nuclear receptors play important roles in the regulation of xenobiotic genes induction (Fahrbach et al., 2012). Recent studies illustrated that an evolutionary conserved Cap’n’collar isoform C (CncC)/Keap1 pathway plays a role in the xenobiotic responses in Drosophila as well as agricultural and urban pests (Misra et al., 2011, 2013; Kalsi and Palli, 2017a, b). Here, we identified a GPCR gene from the Asian gypsy moth, LdOA1, that was involved in the regulation of deltamethrin tolerance potentially through regulating downstream cytochrome P450 genes. However, further experiments are required to investigate whether LdOA1-mediated regulatory pathway directly regulates the expression of cytochrome P450 genes or not. Insect OA1 can serve as a very promising target for more effective pest control. In general, insect GPCRs may therefore be a valuable target for the development of new insecticides.
Data Availability
The datasets generated for this study can be found in the NCBI, Actin (MK926773), EF1α (MK926771), and TUB (MK926772).
Author Contributions
CC, FZ, and ZW conceived and designed the experimental study. LS, PL, CZ, and HD conducted the experiments. CC, FZ, and TM analyzed the data and drafted the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31570642), the Fundamental Research Funds for the Central Universities (Grant No. 2572016DA02), a faculty Start-Up fund from the Pennsylvania State University, and the USDA National Institute of Food and Federal Appropriations under Project #PEN04609 and Accession #1010058.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Lihua Jin (Northeast Forestry University) for kindly providing Drosophila strains and Dr. Hua Bai (Iowa State University) for his guidance on transgenic Drosophila experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00766/full#supplementary-material
FIGURE S1 | The expression of LdOA1 gene was shown in lanes 1 and 2 (Act5 > UAS-LdOA1) but not in lanes 3 and 4 (UAS-LdOA1 > w1118) on 1% agarose gel.
TABLE S1 | Primers used for dsRNA synthesis, transformant Drosophila and qRT-PCR.
References
Audsley, N., and Down, R. E. (2015). G protein coupled receptors as targets for next generation pesticides. Insect Biochem. Mol. Biol. 67, 27–37. doi: 10.1016/j.ibmb.2015.07.014
Bai, H., and Palli, S. R. (2016). Identification of G protein-coupled receptors required for vitellogenin uptake into the oocytes of the red flour beetle, Tribolium castaneum. Sci. Rep. 6:27648. doi: 10.1038/srep27648
Bai, H., Zhu, F., Shah, K., and Palli, S. R. (2011). Large-scale RNAi screen of G protein-coupled receptors involved in larval growth, molting and metamorphosis in the red flour beetle. BMC Genomics 12:388. doi: 10.1186/1471-2164-12-388
Bai, J., Xie, X., Lei, Y., An, G., He, L., and Lv, X. (2014). Ocular albinism type 1-induced melanoma cell migration is mediated through the RAS/RAF/MEK/ERK signaling pathway. Mol. Med. Rep. 10, 491–495. doi: 10.3892/mmr.2014.2154
Bassi, M. T., Incerti, B., Easty, D. J., Sviderskaya, E. V., and Ballabio, A. (1996). Cloning of the murine homolog of the ocular albinism type 1 (OA1) gene: sequence, genomic structure, and expression analysis in pigment cells. Genome Res. 6, 880–885. doi: 10.1101/gr.6.9.880
Battlay, P., Schmidt, J. M., Fournier-Level, A., and Robin, C. (2016). Genomic and transcriptomic associations identify a new insecticide resistance phenotype for the selective sweep at the Cyp6g1 locus of Drosophila melanogaster. G3 6, 2573–2581. doi: 10.1534/g3.116.031054
Broeck, J. V. (2001). Insect G protein-coupled receptors and signal transduction. Arch. Insect Biochem Physiol. 48, 1–12. doi: 10.1002/arch.1054
Caers, J., Verlinden, H., Zels, S., Vandersmissen, H. P., Vuerinckx, K., and Schoofs, L. (2012). More than two decades of research on insect neuropeptide GPCRs: an overview. Front. Endocrinol. 3:151. doi: 10.3389/fendo.2012.00151
Cao, C., Sun, L., Wen, R., Dou, X., and Wang, Z. (2014). Cloning and analysis of LdOA1 in Lymantria dispar and its response to the stress of three kinds of insecticides. Scientia Silvae Sinicae 50, 102–107.
Cao, C., Sun, L., Wen, R., Shang, Q., Ma, L., and Wang, Z. (2015). Characterization of the transcriptome of the asian gypsy moth Lymantria dispar identifies numerous transcripts associated with insecticide resistance. Pest. Biochem.Physiol. 119, 54–61. doi: 10.1016/j.pestbp.2015.02.005
Cao, C. W., Liu, G. F., Wang, Z. Y., Yan, S. C., Ma, L., and Yang, C. P. (2010). Response of the gypsy moth, Lymantria dispar to transgenic poplar, Populus simonii x P. nigra, expressing fusion protein gene of the spider insecticidal peptide and Bt-toxin C-peptide. J. Insect Sci. 10:200. doi: 10.1673/031.010.20001
Daborn, P. J., Yen, J. L., Bogwitz, M. R., Le Goff, G., Feil, E., Jeffers, S., et al. (2002). A single p450 allele associated with insecticide resistance in Drosophila. Science 297, 2253–2256. doi: 10.1126/science.1074170
d′Addio, M., Pizzigoni, A., Bassi, M. T., Baschirotto, C., Valetti, C., Incerti, B., et al. (2000). Defective intracellular transport and processing of OA1 is a major cause of ocular albinism type 1. Hum. Mol. Genet. 9, 3011–3018. doi: 10.1093/hmg/9.20.3011
Denecke, S., Fusetto, R., Martelli, F., Giang, A., Battlay, P., Fournier-Level, A., et al. (2017). Multiple P450s and variation in neuronal genes underpins the response to the insecticide imidacloprid in a population of Drosophila melanogaster. Sci. Rep. 7:11338. doi: 10.1038/s41598-017-11092-5
Fahrbach, S. E., Smagghe, G., and Velarde, R. A. (2012). Insect nuclear receptors. Annu. Rev. Entomol. 57, 83–106. doi: 10.1146/annurev-ento-120710-100607
Feyereisen, R. (2012). “Insect CYP genes and P450 enzymes,” in Insect Molecular Biology and Biochemistry, ed. L. I. Gilbert (Cambridge, MA: Elisevier Academic Press), 236–316. doi: 10.1016/b978-0-12-384747-8.10008-x
Gao, S., Xiong, W., Wei, L., Liu, J., Liu, X., Xie, J., et al. (2018). Transcriptome profiling analysis reveals the role of latrophilin in controlling development, reproduction and insecticide susceptibility in Tribolium castaneum. Genetica 146, 287–302. doi: 10.1007/s10709-018-0020-4
Garland, S. L. (2013). Are GPCRs still a source of new targets? J. Biomol. Screen 18, 947–966. doi: 10.1177/1087057113498418
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., et al. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178.
Hill, C. A., Sharan, S., and Watts, V. J. (2018). Genomics, GPCRs and new targets for the control of insect pests and vectors. Curr. Opin. Insect Sci. 30, 99–106. doi: 10.1016/j.cois.2018.08.010
Hou, Y. Q., Nan, N., and Li, Z. Y. (2009). Research advances of Lymantria dispar. Hebei J. Forest. Orch. Res. 24, 439–444.
Hu, X. B., Sun, Y., Wang, W. J., Yang, M. X., Sun, L. X., Tan, W. B., et al. (2007). Cloning and characterization of NYD-OP7, a novel deltamethrin resistance associated gene from Culex pipiens pallens. Pest Biochem. Physiol. 88, 82–91. doi: 10.1016/j.pestbp.2006.09.004
Huang, K., Chen, W., Zhu, F., Li, P. W. L., Kapahi, P., and Bai, H. (2019). RaboTag translatomic profiling of Drosophila oenocytes under aging and induced oxidative stress. BMC Genomics 20:50. doi: 10.1186/s12864-018-5404-4
Kalsi, M., and Palli, S. R. (2017a). Cap n collar transcription factor regulates multiple genes coding for proteins involved in insecticide detoxification in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. Biol. 90, 43–52. doi: 10.1016/j.ibmb.2017.09.009
Kalsi, M., and Palli, S. R. (2017b). Transcription factor cap n collar C regulates multiple cytochrome P450 genes conferring adaptation to potato plant allelochemicals and resistance to imidacloprid in Leptinotarsa decemlineata (Say). Insect Biochem. Mol. Biol. 83, 1–12. doi: 10.1016/j.ibmb.2017.02.002
Klabunde, T., and Hessler, G. (2002). Drug design strategies for targeting G-protein-coupled receptors. Chembiochem 3, 928–944. doi: 10.1002/1439-7633(20021004)3:10<928::aid-cbic928>3.0.co;2-5
Lazarevic, J., Peric-mataruga, V., Ivanovic, J., and Andjelkovic, M. (1998). Host plant effects on the genetic variation and correlations in the individual performance of the gypsy moth. Funct. Ecol. 12, 141–148. doi: 10.1046/j.1365-2435.1998.00166.x
Li, S. (2012). Research advances of Lymantria dispar control technology. Shandong Forest Sci. Technol. 4, 104–107.
Li, T., Cao, C., Yang, T., Zhang, L., He, L., Xi, Z., et al. (2015). A G-protein-coupled receptor regulation pathway in cytochrome P450-mediated permethrin-resistance in mosquitoes, Culex quinquefasciatus. Sci. Rep. 5:17772. doi: 10.1038/srep17772
Li, T., Liu, L., Zhang, L., and Liu, N. (2014). Role of G-protein-coupled receptor-related genes in insecticide resistance of the mosquito, Culex quinquefasciatus. Sci. Rep. 4:6474. doi: 10.1038/srep06474
Li, T., and Liu, N. (2017). Regulation of P450-mediated permethrin resistance in Culex quinquefasciatus by the GPCR/Galphas/AC/cAMP/PKA signaling cascade. Biochem. Biophys. Rep. 12, 12–19. doi: 10.1016/j.bbrep.2017.08.010
Li, T., and Liu, N. (2018). The function of G-protein-coupled receptor-regulatory cascade in southern house mosquitoes (Diptera: Culicidae). J. Med. Entomol. 55, 862–870. doi: 10.1093/jme/tjy022
Li, X., Schuler, M. A., and Berenbaum, M. R. (2007). Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu. Rev. Entomol. 52, 231–253. doi: 10.1146/annurev.ento.51.110104.151104
Liu, N. (2015). Insecticide resistance in mosquitoes: impact, mechanisms, and research directions. Annu. Rev. Entomol. 60, 537–559. doi: 10.1146/annurev-ento-010814-020828
Liu, N., Liu, H., Zhu, F., and Zhang, L. (2007). Differential expression of genes in pyrethroid resistant and susceptible mosquitoes, Culex quinquefasciatus (S.). Gene 394, 61–68. doi: 10.1016/j.gene.2007.01.032
Liu, N., and Zhu, F. (2011). “House fly cytochrome P450s: their role in insecticide resistance and strategies in the isolation and characterization,” in Recent Advances in Entomological Research, eds T. Liu and L. Kang (Berlin: Springer), 246–257. doi: 10.1007/978-3-642-17815-3_14
Markstein, M., Pitsouli, C., Villalta, C., Celniker, S. E., and Perrimon, N. (2008). Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483. doi: 10.1038/ng.101
Meyer, J. M., Ejendal, K. F. K., Avramova, L. V., Garland-Kuntz, E. E., Giraldo-Calderón, G. I., Brust, T. F., et al. (2012). A “Genome-to-Lead” approach for insecticide discovery: pharmacological characterization and screening of Aedes aegypti D(1)-like dopamine receptors. PLoS Negl. Trop. Dis. 6:e1478. doi: 10.1371/journal.pntd.0001478
Misra, J. R., Horner, M. A., Lam, G., and Thummel, C. S. (2011). Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 25, 1796–1806. doi: 10.1101/gad.17280911
Misra, J. R., Lam, G., and Thummel, C. S. (2013). Constitutive activation of the Nrf2/Keap1 pathway in insecticide-resistant strains of Drosophila. Insect Biochem. Mol. Biol. 43, 1116–1124. doi: 10.1016/j.ibmb.2013.09.005
Mitri, C., Soustelle, L., Framery, B., Bockaert, J., Parmentier, M. L., and Grau, Y. (2009). Plant insecticide L-canavanine repels Drosophila via the insect orphan GPCR DmX. PLoS Biol. 7:e1000147. doi: 10.1371/journal.pbio.1000147
Ni, M., Cai, N. S., Zhang, P., Tian, Y. L., and Xu, T. J. (2009). Toxicity test of different kinds of insecticides to Lymantria dispar and Apocheima cinerarius. J. Northeast Forest Univ. 37, 121–122.
Pfaffl, M. W., Horgan, G. W., and Dempfle, L. (2002). Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids. Res. 30:e36.
Raposo, G., and Marks, M. S. (2007). Melanosomes-dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Bio. 8, 786–797. doi: 10.1038/nrm2258
Riveron, J. M., Yunta, C., Ibrahim, S. S., Djouaka, R., Irving, H., Menze, B. D., et al. (2014). A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 15:R27. doi: 10.1186/gb-2014-15-2-r27
Rosenbaum, D. M., Rasmussen, S. G., and Kobilka, B. K. (2009). The structure and function of G-protein-coupled receptors. Nature 459, 356–363. doi: 10.1038/nature08144
Roy, A. S., Mcnamara, D. G., and Smith, I. M. (1995). Situation of Lymantria dispar in Europe. EPPO Bull. 25, 611–616. doi: 10.1111/j.1365-2338.1995.tb01112.x
Schiaffino, M. V. (2010). Signaling pathways in melanosome biogenesis and pathology. Int. J. Biochem. Cell Biol. 42, 1094–1104. doi: 10.1016/j.biocel.2010.03.023
Schiaffino, M. V., Baschirotto, C., Pellegrini, G., Montalti, S., Tacchetti, C., De Luca, M., et al. (1996). The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc. Natl. Acad. Sci. U.S.A. 93, 9055–9060. doi: 10.1073/pnas.93.17.9055
Schiaffino, M. V., d’Addio, M., Alloni, A., Baschirotto, C., Valetti, C., Cortese, K., et al. (1999). Ocular albinism: evidence for a defect in an intracellular signal transduction system. Nat. Genet 23, 108–112. doi: 10.1038/12715
Scott, J. G., Liu, N., and Wen, Z. (1998). Insect cytochromes P450: diversity, insecticide resistance and tolerance to plant toxins. Comp. Biochem. Physiol. C Pharmacol. Toxicol. Endocrinol. 121, 147–155. doi: 10.1016/s0742-8413(98)10035-x
Seong, K. M., Coates, B. S., Berenbaum, M. R., Clark, J. M., and Pittendrigh, B. R. (2018). Comparative CYP-omic analysis between the DDT-susceptible and -resistant Drosophila melanogaster strains 91-C and 91-R. Pest Manag. Sci. 74, 2530–2543. doi: 10.1002/ps.4936
Sun, L., Wang, Z., Wu, H., Liu, P., Zou, C., Xue, X., et al. (2016). Role of ocular albinism type 1 (OA1) GPCR in Asian gypsy moth development and transcriptional expression of heat-shock protein genes. Pest Biochem. Physiol. 126, 35–41. doi: 10.1016/j.pestbp.2015.07.004
Sun, L., Wang, Z., Zou, C., and Cao, C. (2014). Transcription profiling of 12 asian gypsy moth (Lymantria dispar) cytochrome P450 genes in response to insecticides. Arch. Insect Biochem. Physiol. 85, 181–194. doi: 10.1002/arch.21152
Sun, Y., Zou, P., Yu, X. Y., Chen, C., Yu, J., Shi, L. N., et al. (2012). Functional characterization of an arrestin gene on insecticide resistance of Culex pipiens pallens. Parasit. Vectors 5:134. doi: 10.1186/1756-3305-5-134
United States Department of Agriculture [USDA] (2016). Pest Alert-Asian Gypsy Moth. Available at: https://www.aphis.usda.gov/publications/plant_health/content/printable_version/fs_phasiangm.pdf (accessed June 12, 2019).
Willis, D. K., Wang, J., and Lindholm, J. R. (2010). Orth A and Goodman WG, Microarray analysis of juvenile hormone response in Drosophila melanogaster S2 cells. J. Insect Sci. 10:66. doi: 10.1673/031.010.6601
Wittkopp, P. J., Williams, B. L., Selegue, J. E., and Carroll, S. B. (2003). Drosophila pigmentation evolution: divergent genotypes underlying convergent phenotypes. Proc. Natl. Acad. Sci. U.S.A. 100, 1808–1813. doi: 10.1073/pnas.0336368100
Young, A., Jiang, M., Wang, Y., Ahmedli, N. B., Ramirez, J., Reese, B. E., et al. (2011). Specific interaction of Galphai3 with the Oa1 G-protein coupled receptor controls the size and density of melanosomes in retinal pigment epithelium. PLoS One 6:e24376. doi: 10.1371/journal.pone.0024376
Young, A., Powelson, E. B., Whitney, I. E., Raven, M. A., Nusinowitz, S., Jiang, M., et al. (2008). Involvement of OA1, an intracellular GPCR, and G alpha i3, its binding protein, in melanosomal biogenesis and optic pathway formation. Invest. Ophthalmol. Vis. Sci. 49, 3245–3252. doi: 10.1167/iovs.08-1806
Young, A., Wang, Y., Ahmedli, N. B., Jiang, M., and Farber, D. B. (2013). A constitutively active Galphai3 protein corrects the abnormal retinal pigment epithelium phenotype of Oa1-/- mice. PLoS One 8:e76240. doi: 10.1371/journal.pone.0076240
Zhang, X., Kang, X., Wu, H., Silver, K., Zhang, J., Ma, E., et al. (2018). Transcriptome-wide survey, gene expression profiling and exogenous chemical-induced transcriptional responses of cytochrome P450 superfamily genes in migratory locust (Locusta migratoria). Insect Biochem. Mol. Biol. 100, 66–77. doi: 10.1016/j.ibmb.2018.06.006
Zhou, D., Duan, B., Xu, Y., Ma, L., Shen, B., Sun, Y., et al. (2018). NYD-OP7/PLC regulatory signaling pathway regulates deltamethrin resistance in Culex pipiens pallens (Diptera: Culicidae). Parasit. Vectors 11:419. doi: 10.1186/s13071-018-3011-5
Zhu, F., Cui, Y., Lavine, L. C., and Walsh, D. B. (2014). “Application of RNAi toward insecticide resistance management,” in Short Views on Insect Biochemistry and Molecular Biology, eds R. Chandrasekar, B. K. Tyagi, Z. Gui, and G. R. Reeck (Manhattan: Academic Publisher), 595–619.
Zhu, F., Gujar, H., Gordon, J. R., Haynes, K. F., Potter, M. F., and Palli, S. R. (2013). Bed bugs evolved unique adaptive strategy to resist pyrethroid insecticides. Sci. Rep. 3:1456. doi: 10.1038/srep01456
Zhu, F., Moural, T. W., Nelson, D. R., and Palli, S. R. (2016). A specialist herbivore pest adaptation to xenobiotics through up-regulation of multiple Cytochrome P450s. Sci. Rep. 6:20421. doi: 10.1038/srep20421
Keywords: G-protein-coupled receptor, Asian gypsy moth, deltamethrin stress, cytochrome P450, physiological function
Citation: Sun L, Liu P, Zhang C, Du H, Wang Z, Moural TW, Zhu F and Cao C (2019) Ocular Albinism Type 1 Regulates Deltamethrin Tolerance in Lymantria dispar and Drosophila melanogaster. Front. Physiol. 10:766. doi: 10.3389/fphys.2019.00766
Received: 26 March 2019; Accepted: 31 May 2019;
Published: 19 June 2019.
Edited by:
Jin-Jun Wang, Southwest University, ChinaReviewed by:
Guo-Qing Li, Nanjing Agricultural University, ChinaAram Megighian, University of Padua, Italy
Copyright © 2019 Sun, Liu, Zhang, Du, Wang, Moural, Zhu and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fang Zhu, ZnV6NTlAcHN1LmVkdQ==; Chuanwang Cao, Y2h1YW53YW5nY2FvQG5lZnUuZWR1LmNu
 Lili Sun1
Lili Sun1 Fang Zhu
Fang Zhu Chuanwang Cao
Chuanwang Cao