- 1Dipartimento di Farmacia e Biotecnologie, Alma Mater Studiorum Università di Bologna, Bologna, Italy
- 2CNR Istituto di Genetica Molecolare, Unità di Bologna, Bologna, Italy
- 3IRCCS, Istituto Ortopedico Rizzoli, Bologna, Italy
- 4Department of Genetics, University of Cambridge, Cambridge, United Kingdom
- 5Cambridge Systems Biology Centre, University of Cambridge, Cambridge, United Kingdom
Introduction
The von-Hippel Lindau (VHL) disease is a hereditary genetic disorder that predisposes to the onset of several highly vascularized benign and malignant tumors, developing with elevate frequency in the central nervous system and kidneys. The most-aggressive VHL tumor is ccRCC, the clear-cell renal cell carcinoma, affecting the kidney. VHL disease etiology can be attributed to the inheritance of a VHL loss-of-function allele, typically a deletion (Gnarra et al., 1994; Herman et al., 1994); this facilitates the somatic inactivation of the other allele (through amorphic mutations or gene silencing through promoter methylation), leading to the onset of the tumorous phenotype (Latif et al., 1993). This reveals the haploinsufficient behavior of the VHL gene.
The high vascularization of VHL tumors can be explained considering that human VHL protein is the substrate-binding subunit of an E3 ubiquitin ligase (Lonergan et al., 1998; Iwai et al., 1999; Kamura et al., 1999) involved in the poly-ubiquitination of HIF-1α transcription factor. This post-translational modification leads HIF-1α to proteosomal degradation (Maxwell et al., 1999). Loss of VHL function causes the stabilization of HIF-1α, triggering cellular response and adaptation to hypoxic conditions (expression of genes involved in glycolysis, angiogenesis and erythropoiesis) (Bader and Hsu, 2012). While this represents the canonical function of VHL, other HIF-1α-independent function of VHL have been identified, thanks to the contribution of model organisms (Hsu, 2012). Indeed, VHL gene function is conserved and also Drosophila has a VHL homolog, the dVHL gene (Adryan et al., 2000; Aso et al., 2000). dVHL is involved in the development of Drosophila vascular system (Adryan et al., 2000; Hsouna et al., 2010) and in morphogenesis of follicular epithelium of the egg chamber (Duchi et al., 2010). Interestingly, some VHL functions are mediated by Awd, an endocytic mediator whose human orthologs are NME1/2 metastasis suppressors (Rosengard et al., 1989). Awd is broadly required during Drosophila development since it is involved in epithelial morphogenesis (Nallamothu et al., 2008; Woolworth et al., 2009; Ignesti et al., 2014) and required for maintaining genomic stability (Romani et al., 2017). Moreover, Awd is also present into the extracellular fluids of Drosophila larvae (Romani et al., 2016, 2018).
In Drosophila, two pairs of monolayered epithelial Malpighian tubules, each composed of 100-150 cells, absolve to osmoregulation and excretion functions (Denholm and Skaer, 2009). Transcriptomic analysis of Malpighian tubules revealed that among genes that are here enriched there are homologs of human genes implicated into renal pathologies (Wang et al., 2004). This justifies the use of Drosophila Malpighian tubules as model system to gain insights into pathophysiology of human kidneys (Dow and Romero, 2010; Miller et al., 2013).
The dVHL1.1 allele is a loss of function mutation of the dVHL locus (Duchi et al., 2010; Hsouna et al., 2010). dVHL1.1/+ flies mimic the genetic condition of VHL patients. We carried out a genome-wide gene expression profiling of whole Malpighian tubules dissected from Drosophila females both heterozygous for the dVHL1.1 mutation and with two wild type copies of the dVHL gene. The comparison of differentially expressed genes in the two genetic backgrounds potentially allows to identify genes that are sensible to dVHL functional copy number. Quality control assessments of the data were performed and results obtained from the differential expression analysis were confirmed by qRT-PCR. With this approach we aimed to provide a well-controlled dataset for a better understanding of the VHL disease. Indeed, even if further molecular and functional characterization are needed, human homologs of the differentially expressed genes, if existing, could have a role in the somatic inactivation of the wild type copy of VHL and/or into the very first phase of cancer onset.
Materials and Methods
Drosophila Stocks and Genotypes
Drosophila flies were raised at 25°C on a standard cornmeal/yeast/agar culture medium. We used y1,w67c23 flies as wild type stock. The dVHL1.1 null mutation has been previously characterized (Duchi et al., 2010; Hsouna et al., 2010).
Malpighian Tubules RNA Extraction
Fifty Drosophila females of the appropriate genotype (dVHL1.1/+ or wild type flies) were transferred every day into vials with fresh yeasted food for 5 days. Malpighian tubules were then dissected and 400 μl of TRIzol were added. Homogenization was performed keeping samples on ice. 10 μg of linear polyacrylamide were added before centrifuging at 16,000 g (10 min). 80 μl of chloroform were added to supernatant. Sample was vortexed for 60 s and then centrifuged at 16,000 g (15 min). The upper phase was transferred to a new RNase-free tube. 0.8 volumes of isopropanol were added. RNA was then precipitated for 1 h at −20 μC and pelleted by centrifugation at 16,000 g (30 min). Pellet was then washed with 500 μl of 70% ethanol and centrifuged at 16,000g (5 min). Ethanol was then removed and pellet re-suspended in 15 μl of DEPC water. RNA concentration and purity was assessed through NanoDrop spectrophotometer.
cDNA Generation, Amplification, and Labeling
Four biological replicates were performed. Five Hundred nana gram of RNA were amplified using the SMARTer™ PCR cDNA synthesis kit (Clontech) following manufacture's instruction. Amplified cDNA was then labeled by using the Klenow labeling of double stranded DNA protocol. The number of cycles required to obtain products in exponential phase was determined by performing a PCR using the Advantage® II PCR kit (Clontech) and following manufacture's protocol (5′ PCR Primer II used: 5′-AAGCAGTGGTATCAACGCAGAGT-3′). DNA was purified using QIAquick PCR purification columns. Nine nano gram of cDNA were labeled through incorporation of dCTP conjugated with Cy3 or Cy5 dyes using the BioPrime DNA Labeling System and following manufacture's protocol. Cy3 and Cy5 labeled sample and control pairs were combined in 1.5 ml tubes. The volume was reduced to 25–30 μl in a SpeedVac concentrator before proceeding with Sephadex G50 purification (two per sample), assembled following manufacture's instruction. Sample volumes was reduced to 2–5 μl using a SpeedVac. Finally, 2 μl of 10 mg/ml sonicated salmon sperm DNA were added with 140 μl of hybridization buffer. Samples were then boiled at 100°C (2 min), centrifuged at 16,000 × g (1 min) and then hybridized on slides.
FL003 Array Hybridization
The FlyChip in-house printed FL003 gene expression arrays on FMB PowerMatrix slides using the Genetix Qarray2 (producing 82 arrays per run), consisting of 14,444 transcript-specific 70-mer oligonucleotides were used (GEO accession GPL14121). Four biological replicates were performed including 2 dye swaps. Blocking of slides was performed (as per FMB protocol) by incubating slides for 30 min in 0.1% BSA, 0.2% SDS, 2x SSC (300 mM NaCl, 30 mM Na citrate, pH 7), followed by three washes in clean water. One hundred and thirty five microliters of samples were hybridized for 16 h at 51°C with agitation using the GeneTac Hybridisation station. Slides were then washed with pre-warmed (55°C) wash solution 1 (0.2 × SSC; 0.2% SDS) for 20 min with gentle agitation, followed by 3 washes for 1 min in warm solution 2 (0.2 × SSC), avoiding light exposure, rinsed with MilliQ water at room temperature and finally dried in a centrifuge at 96 × g (5 min).
Data Acquisition and Processing
Slides were scanned using an Axon GenePix 4000B scanner at optimal PMT gain. Manual spot-finding was operated through Dapple (Buhler et al., 2000). Raw data was imported into limma (Bioconductor R package, R version 3.1.0) and Variance stabilizing normalization (vsn) was applied (Huber et al., 2002). Significance analysis was performed using the empirical Bayes method within limma. Due to the low number of significant genes (n = 8 at fdr <= 0.05) thresholds were relaxed to include genes with average M value <-0.5 or >0.5 (M-value is the log2 of the ratio of sample vs. control intensities), and p-value <0.1 (187 genes).
Quantitative RT-PCR Analysis
Three biological replicates of Malpighian tubules dissected from 30 females were analyzed. Malpighian tubule total RNA was extracted in TRI Reagent (Sigma-Aldrich) and treated with TURBO DNase (Ambion). RNA was reverse transcribed using the high-capacity RNA-to-cDNA kit (Applied Biosystems) according to the manufacture's protocol. Quantitative real-time PCRs were performed in fast 48-well reaction plates (Applied Biosystems) and analyzed by StepOnePlus real-time PCR system (Applied Biosystems) according to the manufacturer's procedure. For each sample, at least two technical replicates were performed. Primers were designed using Primer 3 software (Untergasser et al., 2012). Parameters for primer design were a length of 18-27 bases, a melting temperature between 57.0-63.0°C, and a GC content from 20-80%. dGrip75 and CG31955 primers were designed in different exons. Expression of target genes was normalized to the widely used reference gene Rp49. The qRT-PCR primers used are listed in Table S1. For each gene of interest, fold changes in expression levels were evaluated by using the ΔΔCt method. The mean fold change and SD were calculated. p-value was calculated using a one-tail t-test analysis on three biological replicates. Dissociation curve analysis was performed to confirm the presence of a single specific product.
Differentially Expressed Genes in dVHL1.1/+ Malpighian Tubules
By using the statistical parameters reported in the material and methods section we recovered 331 hit genes whose expression significantly differ between dVHL1.1/+ and wild type tubules (Figure 1A). One hundred and eighteen are upregulated (red dots) while 321 are downregulated (blue dots). The majority of genes are not significantly differentially expressed (black dots), as expected. By looking at M values of genes in each of the 4 slides we also highlighted those of them for which single absolute M values are higher than 0.5 in at least 3 slides (Figure 1B). This should outline genes for which the alteration in gene expression is reliable (based on alteration reproducibility). Finally, we merged the data in Figures 1A,B and found 187 genes for which the absolute value of average M is higher than 0.5, p-value is lower than 0.1 and in at least 3 out of 4 slides the single absolute M-values are higher than 0.5 (Figure 1C).
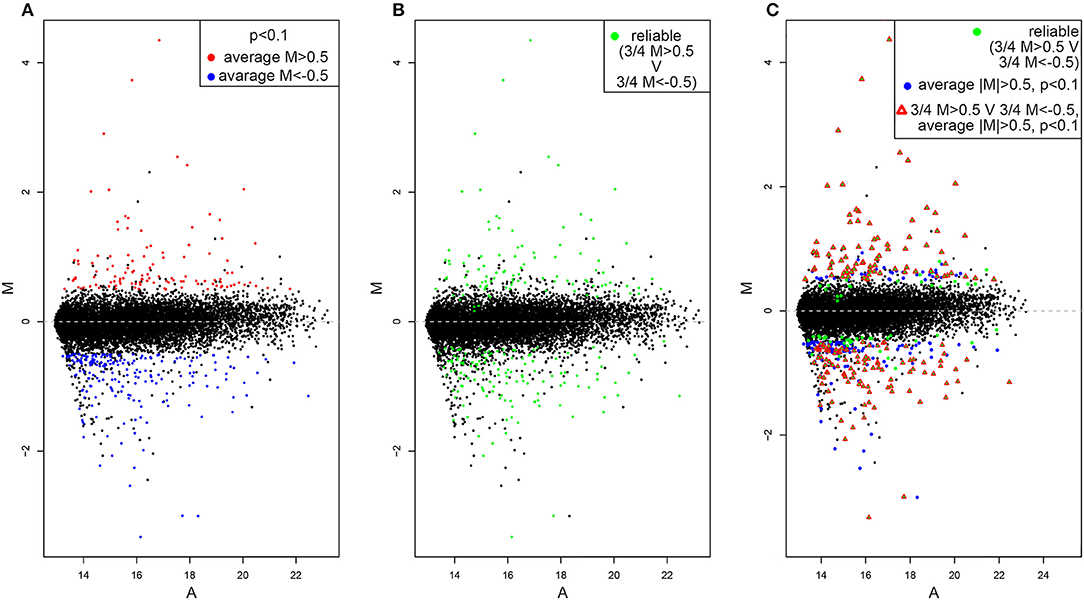
Figure 1. MA-plots of differentially expressed genes in dVHL1.1/+ Malpighian tubules. (A) Upregulated genes (average M > 0.5, p < 0.1) are shown in red while downregulated genes (average M<-0.5, p < 0.1) are shown in blue. (B) Genes for which the alteration in gene expression (M > 0.5 or M<-0.5) was recovered in 3 out of 4 slides are shown in green. (C) Genes that satisfy both the cut-off conditions in (A,B) are shown with red triangle.
As an initial step to analyze the differentially expressed genes we performed quantitative real time PCR (qRT-PCR) experiments and we analyzed the transcript levels of two genes that we are interested on studying, dGrip75 and CG31955; rp49 was used as internal reference gene (Figure 2B). The qRT-PCR experiments confirmed that, in dVHL1.1/+ Malpighian tubules, both genes are downregulated, as expected by microarray results (Figure 2A). dGrip75 encodes a γ-tubulin which takes part in the assembly of the γ-tubulin ring complex (γTuRC), located at the centrosomes, at the base of a microtubule. γTuRC has a ring-shaped structure that serves as a template for a microtubule and allows the controlled polymerization of tubulin dimers (Oegema et al., 1999; Moritz et al., 2000). This protein attracted our attention since we have already demonstrated that dVHL is essential in follicle cells via stabilizing microtubules (Duchi et al., 2010). CG31955 encodes a protein with unknown molecular function. An interesting microarray study of Andrew (Chung et al., 2011) showed that CG31955 is downregulated in trachealess (trh) mutant embryos. Trh is the master regulator of trachea development, the Drosophila branched and tubular system which is responsible for transport of oxygen and other gases. Earlier analysis on dVHL highlighted its requirement in this tubular organ: heterozygous and homozygous dVHL1.1 embryos display altered tracheal system (Hsouna et al., 2010).
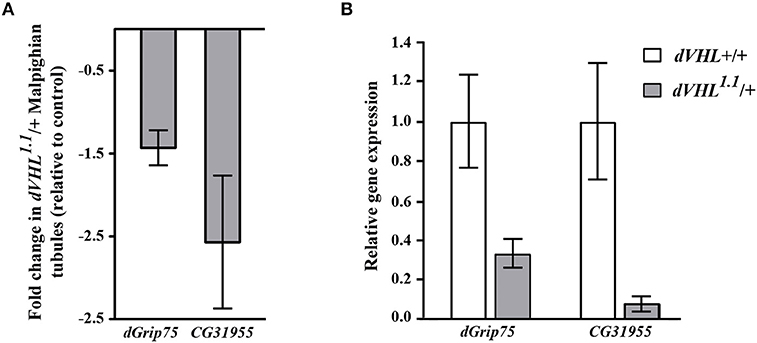
Figure 2. Validation and classification of differentially expressed genes in dVHL1.1/+ Malpighian tubules. (A) Fold change in transcription levels of dGrip75 and CG31955 genes in the microarray analysis. (B) qRT-PCR analysis of dGrip75 and CG31955 transcript levels in individuals of the reported genotypes. 3 biological replicates of tubules from 5-days old dVHL1.1/+ and wild type females were analyzed. Graphs represent mean ± SD; p = 0.0152 (dGrip75) and p = 0.0153 (CG31955).
We screened our candidate list with FlyMine (Lyne et al., 2007) tool for gene ontology (GO) enrichment in biological processes (p < 0.1, Bonferroni test) and we found enrichment in regulation of phosphoprotein phosphatase activity [GO:0043666, p-value = 0.05] and regulation of protein serine/threonine phosphatase activity [GO:0080163, p = 0.07].
Transcriptome alterations in human morphologically normal cells heterozygous for a VHL mutation (derived from VHL patients) have also been analyzed (Peri et al., 2016). A comparison between Drosophila and human datasets could recover strong hits, whose molecular dissection may be performed using Drosophila as a model system.
The limiting-most aspect of this study is intrinsic to omics approaches: functional analyses of candidates are needed to genetically dissect gene functions and pathways, confirming their role into the VHL pathogenesis.
Data Availability
The datasets generated for this study can be found in the GEO data repository (http://www.ncbi.nlm.nih.gov/geo/) under the accession identification number GSE124152.
Author Contributions
MI cultured flies and performed the experiments. BF assisted the microarray experiments. DA performed the qPCR analysis. MI, GG, and VC conceived the experiments and wrote the article.
Funding
The authors acknowledge funding from the University of Bologna (RFO 2014) to GG and VC.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
A special thank goes to Dr. Boris Adryan, for hosting MI in his laboratory in Cambridge (UK) and for his invaluable help in designing the experimental plan. We thank Patrizia Romani for her precious suggestion on interpreting data. MI has been awarded a PhD fellowship from University of Bologna and MIUR Fondo Giovani and a Marco Polo fellowship for the research period in Cambridge, UK.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00619/full#supplementary-material
References
Adryan, B., Decker, H.J., Papas, T.S., and Hsu, T. (2000). Tracheal development and the von Hippel-Lindau tumor suppressor homolog in Drosophila. Oncogene 19, 2803–2811. doi: 10.1038/sj.onc.1203611
Aso, T., Yamazaki, K., Aigaki, T., and Kitajima, S. (2000). Drosophila von Hippel-Lindau tumor suppressor complex possesses E3 ubiquitin ligase activity. Biochem. Biophys. Res. Commun. 276, 355–361. doi: 10.1006/bbrc.2000.3451
Bader, H.L., and Hsu, T. (2012). Systemic VHL gene functions and the VHL disease. FEBS Lett. 586, 1562–1569. doi: 10.1016/j.febslet.2012.04.032
Buhler, J., Ideker, T., and Haynor, D. (2000). Dapple: Improved Techniques for Finding Spots on DNA Microarrays. UW CSE Technical Report UWTR 2000-08-05.
Chung, S., Chavez, C., and Andrew, D.J. (2011). Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev. Biol. 360, 160–172. doi: 10.1016/j.ydbio.2011.09.014
Denholm, B., and Skaer, H. (2009). Bringing together components of the fly renal system. Curr Opin. Genet. Dev. 19, 526–532. doi: 10.1016/j.gde.2009.08.006
Dow, J.A., and Romero, M.F. (2010). Drosophila provides rapid modeling of renal development, function, and disease. Am. J. Physiol. Renal. Physiol. 299, F1237–1244. doi: 10.1152/ajprenal.00521.2010
Duchi, S., Fagnocchi, L., Cavaliere, V., Hsouna, A., Gargiulo, G., and Hsu, T. (2010). Drosophila VHL tumor-suppressor gene regulates epithelial morphogenesis by promoting microtubule and aPKC stability. Development 137, 1493–1503. doi: 10.1242/dev.042804
Gnarra, J.R., Tory, K., Weng, Y., Schmidt, L., Wei, M.H., and Li, H., et al. (1994). Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat. Genet. 7, 85–90. doi: 10.1038/ng0594-85
Herman, J.G., Latif, F., Weng, Y., Lerman, M.I., Zbar, B., and Liu, S., et al. (1994). Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc. Natl. Acad. Sci. U.S.A. 91, 9700–9704.
Hsouna, A., Nallamothu, G., Kose, N., Guinea, M., Dammai, V., and Hsu, T. (2010). Drosophila von Hippel-Lindau tumor suppressor gene function in epithelial tubule morphogenesis. Mol. Cell. Biol. 30, 3779–3794. doi: 10.1128/MCB.01578-09
Hsu, T. (2012). Complex cellular functions of the von Hippel-Lindau tumor suppressor gene: insights from model organisms. Oncogene 31, 2247–2257. doi: 10.1038/onc.2011.442
Huber, W., von Heydebreck, A., Sultmann, H., Poustka, A., and Vingron, M. (2002). Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18 (Suppl. 1), S96–S104. doi: 10.1093/bioinformatics/18.suppl_1.S96
Ignesti, M., Barraco, M., Nallamothu, G., Woolworth, J.A., Duchi, S., and Gargiulo, G., et al. (2014). Notch signaling during development requires the function of awd, the Drosophila homolog of human metastasis suppressor gene Nm23. BMC Biol. 12:12. doi: 10.1186/1741-7007-12-12
Iwai, K., Yamanaka, K., Kamura, T., Minato, N., Conaway, R.C., and Conaway, J.W., et al. (1999). Identification of the von Hippel-lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. U.S.A. 96, 12436–12441.
Kamura, T., Conrad, M.N., Yan, Q., Conaway, R.C., and Conaway, J.W. (1999). The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes. Dev. 13, 2928–2933.
Latif, F., Duh, F.M., Gnarra, J., Tory, K., Kuzmin, I., and Yao, M., et al. (1993). von Hippel-Lindau syndrome: cloning and identification of the plasma membrane Ca(++)-transporting ATPase isoform 2 gene that resides in the von Hippel-Lindau gene region. Cancer Res. 53, 861–867.
Lonergan, K.M., Iliopoulos, O., Ohh, M., Kamura, T., Conaway, R.C., and Conaway, J.W., et al. (1998). Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol. Cell. Biol. 18, 732–741.
Lyne, R., Smith, R., Rutherford, K., Wakeling, M., Varley, A., and Guillier, F., et al. (2007). FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome. Biol. 8, R129. doi: 10.1186/gb-2007-8-7-r129
Maxwell, P.H., Wiesener, M.S., Chang, G.W., Clifford, S.C., Vaux, E.C., and Cockman, M.E., et al. (1999). The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399, 271–275. doi: 10.1038/20459
Miller, J., Chi, T., Kapahi, P., Kahn, A.J., Kim, M.S., and Hirata, T., et al. (2013). Drosophila melanogaster as an emerging translational model of human nephrolithiasis. J. Urol. 190, 1648–1656. doi: 10.1016/j.juro.2013.03.010
Moritz, M., Braunfeld, M.B., Guenebaut, V., Heuser, J., and Agard, D.A. (2000). Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365–370. doi: 10.1038/35014058
Nallamothu, G., Woolworth, J.A., Dammai, V., and Hsu, T. (2008). Awd, the homolog of metastasis suppressor gene Nm23, regulates Drosophila epithelial cell invasion. Mol. Cell. Biol. 28, 1964–1973. doi: 10.1128/MCB.01743-07
Oegema, K., Wiese, C., Martin, O.C., Milligan, R.A., Iwamatsu, A., and Mitchison, T.J., et al. (1999). Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733.
Peri, S., Caretti, E., Tricarico, R., Devarajan, K., Cheung, M., and Sementino, E., et al. (2016). Haploinsufficiency in tumor predisposition syndromes: altered genomic transcription in morphologically normal cells heterozygous for VHL or TSC mutation. Oncotarget 8, 17628–17642. doi: 10.18632/oncotarget.12192
Romani, P., Duchi, S., Gargiulo, G., and Cavaliere, V. (2017). Evidence for a novel function of Awd in maintenance of genomic stability. Sci. Rep. 7:16820. doi: 10.1038/s41598-017-17217-0
Romani, P., Ignesti, M., Gargiulo, G., Hsu, T., and Cavaliere, V. (2018). Extracellular NME proteins: a player or a bystander? Lab Invest. 98, 248–257. doi: 10.1038/labinvest.2017.102
Romani, P., Papi, A., Ignesti, M., Soccolini, G., Hsu, T., and Gargiulo, G., et al. (2016). Dynamin controls extracellular level of Awd/Nme1 metastasis suppressor protein. Naunyn. Schmiedebergs Arch. Pharmacol. 389, 1171–1182. doi: 10.1007/s00210-016-1268-9
Rosengard, A.M., Krutzsch, H.C., Shearn, A., Biggs, J.R., Barker, E., and Margulies, I.M., et al. (1989). Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature 342, 177–180. doi: 10.1038/342177a0
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B.C., and Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Wang, J., Kean, L., Yang, J., Allan, A.K., Davies, S.A., and Herzyk, P., et al. (2004). Function-informed transcriptome analysis of Drosophila renal tubule. Genome. Biol. 5:R69. doi: 10.1186/gb-2004-5-9-r69
Keywords: microarray analysis, Drosophila VHL, renal carcinoma, kidney, sensitized genetic background
Citation: Ignesti M, Andrenacci D, Fischer B, Cavaliere V and Gargiulo G (2019) Comparative Expression Profiling of Wild Type Drosophila Malpighian Tubules and von Hippel-Lindau Haploinsufficient Mutant. Front. Physiol. 10:619. doi: 10.3389/fphys.2019.00619
Received: 28 December 2018; Accepted: 02 May 2019;
Published: 21 May 2019.
Edited by:
Cristiano De Pitta, University of Padova, ItalyCopyright © 2019 Ignesti, Andrenacci, Fischer, Cavaliere and Gargiulo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marilena Ignesti, bWFyaWxlbmEuaWduZXN0aUB1bmliby5pdA==
Giuseppe Gargiulo, Z2l1c2VwcGUuZ2FyZ2l1bG9AdW5pYm8uaXQ=
 Marilena Ignesti
Marilena Ignesti Davide Andrenacci
Davide Andrenacci Bettina Fischer4,5
Bettina Fischer4,5 Valeria Cavaliere
Valeria Cavaliere Giuseppe Gargiulo
Giuseppe Gargiulo