- 1Branch for Bioresources, Fraunhofer Institute for Molecular Biology and Applied Ecology, Giessen, Germany
- 2Entomology Department, Max Planck Institute for Chemical Ecology, Jena, Germany
- 3Institute for Insect Biotechnology, Justus-Liebig University of Giessen, Giessen, Germany
Aphids are economically important pest insects that damage plants by phloem feeding and the transmission of plant viruses. Their ability to feed exclusively on nutritionally poor phloem sap is dependent on the obligatory symbiotic bacterium Buchnera aphidicola, but additional facultative symbionts may also be present, a common example of which is Serratia symbiotica. Many Serratia species secrete extracellular enzymes, so we hypothesised that S. symbiotica may produce proteases that help aphids to feed on plants. Molecular analysis, including fluorescence in situ hybridization (FISH), revealed that S. symbiotica colonises the gut, salivary glands and mouthparts (including the stylet) of the pea aphid Acyrthosiphon pisum, providing a mechanism to transfer the symbiont into host plants. S. symbiotica was also detected in plant tissues wounded by the penetrating stylet and was transferred to naïve aphids feeding on plants containing this symbiont. The maintenance of S. symbiotica by repeated transmission via plants may explain the high frequency of this symbiont in aphid populations. Proteomic analysis of the supernatant from a related but cultivable S. symbiotica strain cultured in liquid medium revealed the presence of known and novel proteases including metalloproteases. The corresponding transcripts encoding these S. symbiotica enzymes were detected in A. pisum and in plants carrying the symbiont, although the mRNA was much more abundant in the aphids. Our data suggest that enzymes from S. symbiotica may facilitate the digestion of plant proteins, thereby helping to suppress plant defense, and that the symbionts are important mediators of aphid–plant interactions.
Introduction
Aphids are major crop pests, causing both direct feeding damage and the transmission of important plant viruses (Van Emden and Harrington, 2017). The pea aphid (Acyrthosiphon pisum Harris) is a model for the analysis of symbiosis, and its genome sequence was the first to be published among hemipteran insects (Consortium, 2010; Oliver et al., 2014). These species have specialised mouthparts, including a stylet that penetrates plant tissues such as sieve tubes in order to withdraw the phloem sap (Powell et al., 2006). The adaptation of aphids to this exclusive diet is facilitated by the obligatory bacterial symbiont Buchnera aphidicola, which compensates for the lack of nutrients by providing essential amino acids (Hansen and Moran, 2011). Aphids may also carry a variety of facultative bacterial symbionts (e.g., Serratia symbiotica, Hamiltonella defensa, and Regiella insecticola) that act as mutualists or parasites depending on the context of the environmental interactions (Oliver et al., 2010, 2014).
Facultative symbionts are found in multiple aphid tissues (including the haemolymph, gut, and reproductive system), and are sometimes co-localised with B. aphidicola within specialised structures known as bacteriomes (Moran et al., 2005; Skaljac et al., 2018). Most symbiotic bacteria (obligatory and facultative) are maternally inherited, whereas the extracellular and scattered localization of facultative symbionts facilitates their horizontal transfer, promoting rapid spreading to new hosts (Russell et al., 2003; Chiel et al., 2009; Oliver et al., 2010). Many studies have revealed phylogenetically closely related symbionts in evolutionarily distant hosts, suggesting that bacteria are horizontally transmitted between diverse insect species (Moran et al., 2005, 2008; Ahmed et al., 2013; Skaljac et al., 2017). The complex horizontal transmission routes include shared plants and parasitoids, resulting in the acquisition of novel ecological traits by the host (Russell et al., 2003; Chiel et al., 2009; Caspi-Fluger et al., 2012; Gehrer and Vorburger, 2012; Gonella et al., 2015; Chrostek et al., 2017).
The genus Serratia has spread to diverse habitats and the species in this genus have evolved multiple ecological functions (Petersen and Tisa, 2013). Whereas S. symbiotica is one of the most common facultative symbionts of aphids (Manzano-Marín et al., 2012), other Serratia species are pathogens associated with humans, insects, nematodes, and plants (Petersen and Tisa, 2013). The ubiquity of the genus is correlated with its ability to produce a large number of extracellular proteins (e.g., proteases, lipases, DNAses, and chitinases) that enable the species to thrive within or in close contact with many hosts (Petersen and Tisa, 2014). There are several classes of bacterial proteases, the most common of which is the metalloproteases (Miyoshi, 2013), and their major physiological role is to degrade environmental proteins for bacterial heterotrophic nutrition (Wu and Chen, 2011).
Although S. symbiotica is predominantly a mutualist, it acts as a facultative and protective symbiont in A. pisum and the black bean aphid (Aphis fabae Scopoli), but it has established co-obligate (nutritional) associations with aphids of the Lachninae subfamily and B. aphidicola (Manzano-Marin and Latorre, 2016). S. symbiotica provides many benefits but it also imposes costs on A. pisum by inhibiting reproduction, development and survival (Laughton et al., 2014; Skaljac et al., 2018). Insects must control their symbiont population in order to ensure the success of both partners, and this is frequently associated with trade-offs between investment in life-history traits and the regulation of symbionts (Login et al., 2011; Laughton et al., 2014).
The vast majority of bacterial symbionts have proven difficult to cultivate in the laboratory due to their lifestyle, gene loss, and dependence on host metabolites (Dale and Moran, 2006; Stewart, 2012). However, several cultivable strains of S. symbiotica have recently been isolated from A. fabae and the sage aphid (A. passeriniana Del Guercio; Sabri et al., 2011; Foray et al., 2014; Grigorescu et al., 2018). These strains are transitional forms between free-living and host-dependent symbiotic bacteria and they provide unique opportunities to study different multi-trophic interactions, such as the tritrophic relationship between symbionts, insects and plants (Foray et al., 2014; Renoz et al., 2017).
Bacterial symbionts frequently play a key role in plant–insect interactions, with important implications for plant defence and plant utilisation by insects (Frago et al., 2012; Sugio et al., 2015; Chrostek et al., 2017). Although the diversity of insect symbionts associated with plants has been investigated in detail, the role of symbiotic bacteria in such interactions is unclear. For example, Rickettsia spp. and Wolbachia spp. infect the sweet potato whitefly (Bemisia tabaci Gennadius) and are horizontally transmitted via the host plant to uninfected peers or even different species (Caspi-Fluger et al., 2012; Li S.J. et al., 2017; Li Y.H. et al., 2017). Furthermore, Cardinium spp. are transferred between different phloem-feeding insects via plants carrying the symbiont (Gonella et al., 2015). A common factor in many of these studies is that bacterial symbionts are found in different insect organs, including the salivary glands and stylet, enabling insect hosts to inoculate plant tissues with symbionts. Furthermore, Wolbachia spp. and Rickettsia spp. associated with B. tabaci are viable and persist in reservoir plants for an extended duration, suggesting potential interactions with the plant, such as nutrient uptake (Caspi-Fluger et al., 2012; Chrostek et al., 2017; Li S.J. et al., 2017; Li Y.H. et al., 2017).
Bacterial symbionts are known to help their insect hosts overcome plant defense and adapt to host plants. As a defence mechanism, plants frequently produce inhibitors to destroy proteases secreted by herbivorous insects, thus stopping them from digesting plant proteins (Hansen and Moran, 2014; Sugio et al., 2015; Wielkopolan and Obrepalska-Steplowska, 2016). In turn, insects may produce new protease isoforms that are resistant to plant inhibitors, or they may produce proteases at a higher rate (Wielkopolan and Obrepalska-Steplowska, 2016). Remarkably, gut bacteria in the Western corn rootworm (Diabrotica virgifera virgifera LeConte) and the velvet bean caterpillar (Anticarsia gemmatalis Hübner) produce additional proteases that help the insects to overcome the protease inhibitors produced by plants (Sugio et al., 2015).
Aphids inject infested plants with saliva containing proteases that digest phloem sap proteins, and these enzymes can be inhibited by the broad-spectrum metalloprotease inhibitor EDTA (Furch et al., 2015). Given that Serratia spp. are known to secrete a variety of extracellular enzymes (Hase and Finkelstein, 1993; Renoz et al., 2017), we hypothesise that S. symbiotica proteases may help aphids to exploit plants more efficiently by digesting plant proteins. We therefore investigated the localization of S. symbiotica in aphid mouthparts and wounded plants, analysed the proteome of S. symbiotica cultured in liquid medium to identify secreted proteases, and determined whether the transcripts encoding these enzymes are present in the aphids and also their host plants.
Materials and Methods
Aphids and Bacterial Symbionts
Maintenance of Aphids and Detection of Symbionts
Parthenogenetic A. pisum clone LL01 was reared under controlled conditions on the host plant Vicia faba var. minor as previously described (Luna-Ramirez et al., 2017; Will et al., 2017). The LL01 clone was obtained from Dr. Torsten Will (Justus-Liebig University, Giessen, Germany) and has been used in our research since 2009. We have previously shown that every individual carries B. aphidicola and S. symbiotica (Luna-Ramirez et al., 2017; Skaljac et al., 2018). A previously established, Serratia-free A. pisum line was used as a control, whereas the original (infected) aphid line is described hereafter as Serratia-positive (Skaljac et al., 2018). The infection status of these aphid lines was regularly checked to detect any potential contamination, especially the presence of S. symbiotica in the Serratia-free line.
We detected S. symbiotica in aphids and plants by extracting total DNA from Serratia-positive or Serratia-free aphids and V. faba tissues using the CTAB method (Luna-Ramirez et al., 2017). We then used Serratia-specific primers to detect S. symbiotica 16S rDNA in the aphids and V. faba plants by PCR (Supplementary Table S1). Amplicons were eluted using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Düren, Germany), and sequenced for verification on a 3730xl DNA analyzer (Macrogen Europe, Amsterdam, Netherlands). The resulting sequences were screened against the NCBI nr database using BLAST. The nucleotide sequences of the S. symbiotica 16S rDNA identified in this study were deposited in GenBank under accession numbers MH447605–MH447629 (whole aphid body), MH447630 (aphid gut), and MH447631–MH447632 (V. faba carrying S. symbiotica).
Proteomic analysis was carried out using the cultivable S. symbiotica strain CWBI-2.3 (DSM no. 23270), originally isolated from A. fabae. This strain was obtained from the Leibniz Institute DSMZ (Braunschweig, Germany) and was cultivated as recommended by the supplier. Briefly, the strain was grown in 535 liquid medium at 28°C overnight in a shaking incubator at 200 rpm. Cells were harvested by centrifugation at 453 × g for 30 min at 10°C, and the supernatant was stored at -80°C.
Quantification and Visualisation of S. symbiotica in A. pisum and Its Host Plants
At least three biological replicates of 30 adult A. pisum (10 days old) from Serratia-positive and Serratia-free aphid lines were released into Petri dishes containing V. faba discs (2 cm diameter) on 1% agar. After 2 days, aphids were collected in groups of 10 and stored in absolute ethanol at -20°C. Small strips of V. faba disc (2 cm × 3 mm) were cut from each replicate immediately after feeding and also 5 and 10 days post-feeding. All insect and plant samples were surface sterilised as previously described (Grigorescu et al., 2018) before DNA or further RNA extraction to ensure that S. symbiotica cells and gene expression represented bacteria present inside the tissues.
The abundance of S. symbiotica in the A. pisum and V. faba samples was determined by quantitative PCR (qPCR) as previously described with modifications (Luna-Ramirez et al., 2017). Briefly, genomic DNA was extracted using the CTAB method and a 133-bp fragment of the S. symbiotica dnaK gene (Supplementary Table S1) was amplified using the StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA, United States). The 10-μL reaction mixture comprised 2 μL of DNA template (50 ng/μL), 10 μM of each specific primer and 5 μL of Power SYBR Green PCR Master Mix (Applied Biosystems). For each sample, three independent reactions were carried out for each primer pair. The relative abundance of the dnaK gene in the Serratia-positive and Serratia-free aphid lines was determined after normalisation to the ribosomal protein L32 (rpl32) reference gene in aphids (Pfaffl, 2001). Furthermore, the relative abundance of S. symbiotica in V. faba plants exposed to the two aphid lines was determined after normalisation to the V. faba actin reference gene (Supplementary Table S1). Significant differences in abundance were confirmed using Student’s t-test in IBM SPSS v23 (Armonk, New York, NY, United States), with statistical significance defined as p < 0.05.
We visualised S. symbiotica by fluorescence in situ hybridization (FISH) in dissected mouthparts, salivary glands and guts of adult aphids as we previously described (Luna-Ramirez et al., 2017). In addition, hand-cut longitudinal stem sections of V. faba seedlings that were highly infested with aphids for at least 10 days were analysed by FISH as previously reported (Ghanim et al., 2009). Negative controls consisted of uninfected samples and no-probe staining (Supplementary Figures S1, S2 and Supplementary Table S2). The primers and probe used for the quantification and visualisation of S. symbiotica are listed in Supplementary Table S1.
Horizontal Transmission of S. symbiotica Between A. pisum Individuals via Host Plants
To determine whether S. symbiotica detected in V. faba plants can be acquired by Serratia-free aphids, 30 aphids (10 days old) from the Serratia-positive line were fed on V. faba discs in five replicates for 2 days and then removed (Supplementary Figure S4). Meanwhile, 30 age-synchronised aphids (2 days old) from the Serratia-free line were released onto each V. faba disc previously occupied by the Serratia-positive aphids (Supplementary Figure S3). The Serratia-free aphids were allowed to feed for 3 days before transfer to a cage containing non-infested V. faba plants. These aphids are described hereafter as Serratia-reinfected and were kept in the rearing cage for the next 2 months to ensure the bacterial symbiont could spread among the aphid population.
The V. faba discs, mothers from both aphid lines and their randomly selected offspring were tested by PCR for the presence of S. symbiotica (Figure 1). Two months after infection, at least 30 Serratia-reinfected aphids were individually tested by PCR to confirm the transmission of S. symbiotica (Figure 1 and Supplementary Table S3). The nucleotide sequences of S. symbiotica 16S rDNA identified in this study were deposited in GenBank under accession numbers MK424314–MK424325 for the Serratia-reinfected aphids. The three aphid lines were strictly separated to prevent contamination. However, to avoid false positive transmission results due to potential contamination with the symbiont, we also included a negative control comprising Serratia-free aphids as both donors and recipients (Supplementary Table S3).
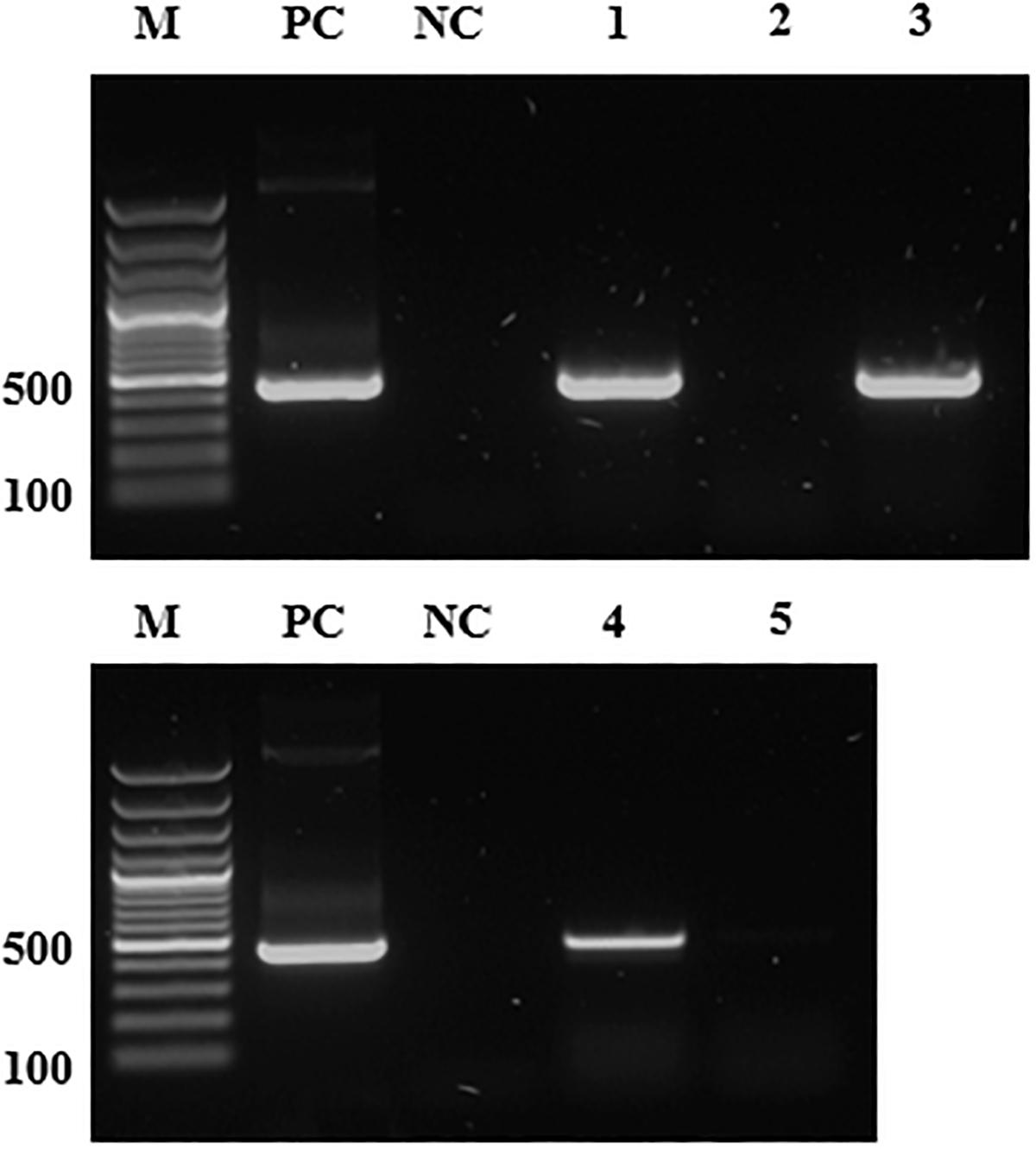
Figure 1. The detection of S. symbiotica genomic DNA by PCR. M, DNA marker (size in base pairs); PC, positive control (pGEM T-Easy vector with S. symbiotica 16S rDNA); NC, negative control (distilled water); lane 1, Serratia-positive aphids; lane 2, Serratia-free aphids; lane 3, Serratia-reinfected aphids (2 months after infection event); lane 4, V. faba plant infested with Serratia-positive aphids; lane 5, V. faba plant infested with Serratia-free aphids. The Serratia specific primers used for PCR are listed in Supplementary Table S1. Amplicon size ∼480 bp.
Phylogenetic Analysis of S. symbiotica
A phylogenetic tree was constructed using MEGA v7.0 (Kumar et al., 2016). DNA sequence similarities among Serratia species were investigated using the BLAST search tool1. ClustalW was used for multiple sequence alignments with default parameters. The phylogenetic tree was constructed using the maximum-likelihood method with a Tamura-Nei distance matrix. Bootstrap analysis of 1000 replicates was used to deduce confidence levels. The phylogenetic tree was displayed, manipulated and annotated using iTOL v4.2 (Letunic and Bork, 2016).
Proteomic Analysis of S. symbiotica CWBI-2.3 Culture Medium and Identification of Genes Encoding Proteolytic Enzymes in Aphids and Plants
Liquid Chromatography–Mass Spectrometry (LC-MS)
The concentrated supernatant of S. symbiotica CWBI-2.3 cells in 535 medium was fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in 16.5% tricine gradient gels (BioRad, Munich, Germany). The protein bands were stained with Coomassie Brilliant Blue and excised from the gel matrix for tryptic digestion as previously described (Shevchenko et al., 2006). For LC-MS analysis, samples were reconstituted in 50 μL aqueous 1% formic acid and 1 μL of the peptide mixture was injected into a UPLC M-class system (Waters, Eschborn, Germany) coupled online to a Synapt G2-si mass spectrometer equipped with a T-WAVE-IMS device (Waters). Data were acquired in data-dependent acquisition (DDA) and data-independent acquisition (DIA) modes, the latter described as enhanced MSE. DIA analysis was supported by ion mobility separation, i.e., high-definition enhanced MSE (HDMSE) analysis (Distler et al., 2016).
Data Processing and Protein Identification
DDA raw data were first searched against a small database containing common contaminants to remove them (ProteinLynx Global Server v2.5.2, Waters). Remaining spectra were interpreted de novo to yield peptide sequences and used as queries for homology-based searching with MS-BLAST (Shevchenko et al., 2001) installed on a local server. MS-BLAST searches were performed against the NCBI nr database and a refined S. symbiotica database generated by the in silico translation of predicted S. symbiotica genes. In parallel, MS/MS spectra were searched against the NCBI nr database combined with the refined S. symbiotica database using MASCOT v2.5.1. HDMSE data were searched against the refined S. symbiotica protein database and a database containing common contaminants (human keratins and trypsin).
Identification and Expression Analysis of S. symbiotica Protease Genes in Aphids and Plants
Proteolytic enzymes detected in the supernatant of the S. symbiotica CWBI-2.3 strain (Supplementary Table S4) allowed the analysis of the corresponding genes in S. symbiotica infecting A. pisum and its infested host plants. Complementary DNA (cDNA) sequences for most of the S. symbiotica proteases were identified using the Ensembl Bacteria browser2 or NCBI databases3. Gene-specific PCR primers were designed using Primer3 v4.1.04 to amplify specific regions of the transcribed cDNAs (Koressaar and Remm, 2007; Supplementary Table S1).
Total RNA was extracted from the previously described samples, i.e., aphids from Serratia-positive and Serratia-free lines, V. faba containing or lacking the symbiont, and S. symbiotica CWBI-2.3, using the Direct-zol RNA MiniPrep Plus Kit (Zymo Research, Freiburg, Germany). RNA (100 ng) was transcribed using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, Dreieich, Germany) to obtain first-strand cDNA. Amplicons from V. faba samples infested with Serratia-positive aphids were re-amplified because the quantity was low, and were cloned (Supplementary Figures S5, S6) before sequencing together with amplicons from the Serratia-positive aphids and the supernatant of S. symbiotica CWBI-2.3. Cloning and sequencing were carried out as previously described (Skaljac et al., 2018). Accession numbers for the S. symbiotica protease genes are listed in Table 1. The sequences were used to design qRT-PCR primers (Supplementary Table S1) in PrimerQuest (Integrated DNA Technologies, Coralville, IA, United States5). Control samples (Serratia-free aphids and their host plants, as well as non-infested V. faba plants), were negative for the expression of S. symbiotica protease genes. S. symbiotica CWBI-2.3 cDNA was used as a positive control (Supplementary Figure S5).

Table 1. Overview of the genes encoding proteolytic enzymes with associated GenBank accession numbers from S. symbiotica expressed in A. pisum and its host plant V. faba (for additional explanations, see Results section “Proteolytic enzymes associated with S. symbiotica”).
The S. symbiotica genes previously shown to be expressed in V. faba carrying S. symbiotica (DegQ, HtpX, YfgC, SohB, and PepA) were chosen for further expression analysis by qRT-PCR because they may be important for tritrophic interactions between symbionts, insects and plants (Table 1). The expression of the five selected genes in Serratia-free and Serratia-positive aphids was evaluated by qRT-PCR after normalisation to the expression level of the rpl32 reference gene (Pfaffl, 2001). For each sample, three independent reactions were carried out for each primer pair. The qPCR protocol described above was modified so that the cDNA template was diluted 1:2 with RNase-free water before qRT-PCR (2 μL in a total volume of 10 μL). The relevant target genes and primers are listed in Table 1 and Supplementary Table S1. Data were analysed as described above.
Results
S. symbiotica in A. pisum and Its Host Plants
Detection and Visualisation of S. symbiotica
Polymerase chain reaction analysis showed that S. symbiotica was present in every individual of the Serratia-positive line, in multiple tissues including the salivary glands and gut (Supplementary Table S2) confirming findings from our previous study (Skaljac et al., 2018). We found no evidence of the symbiont in the Serratia-free line over many generations of rearing under laboratory conditions (Figure 1). Furthermore, the same PCR also showed that S. symbiotica was present in V. faba plants infested with Serratia-positive aphids, whereas no symbionts were detected in the plants exposed to the Serratia-free aphid line (Figure 1).
Fluorescence in situ hybridization analysis with a probe specific for S. symbiotica was used to confirm the PCR data (Supplementary Table S2) and to reveal the distribution of S. symbiotica within aphid and V. faba tissues. The S. symbiotica signal was abundant in the aphid gut (Figures 2C,D), but also in salivary glands and associated mouthparts (stylet, mandibles, labrum, food, and salivary canal) (Figures 2A–D). At this resolution, we were unable to determine whether S. symbiotica was present in one or both canals, but in either case our results indicated its route into aphids with the phloem sap or outward with the saliva. We also observed S. symbiotica cells in V. faba tissues wounded by the penetrating stylet (Figures 2E,F). The symbiont was not detected in non-infested host plants or those infested with the Serratia-free line.
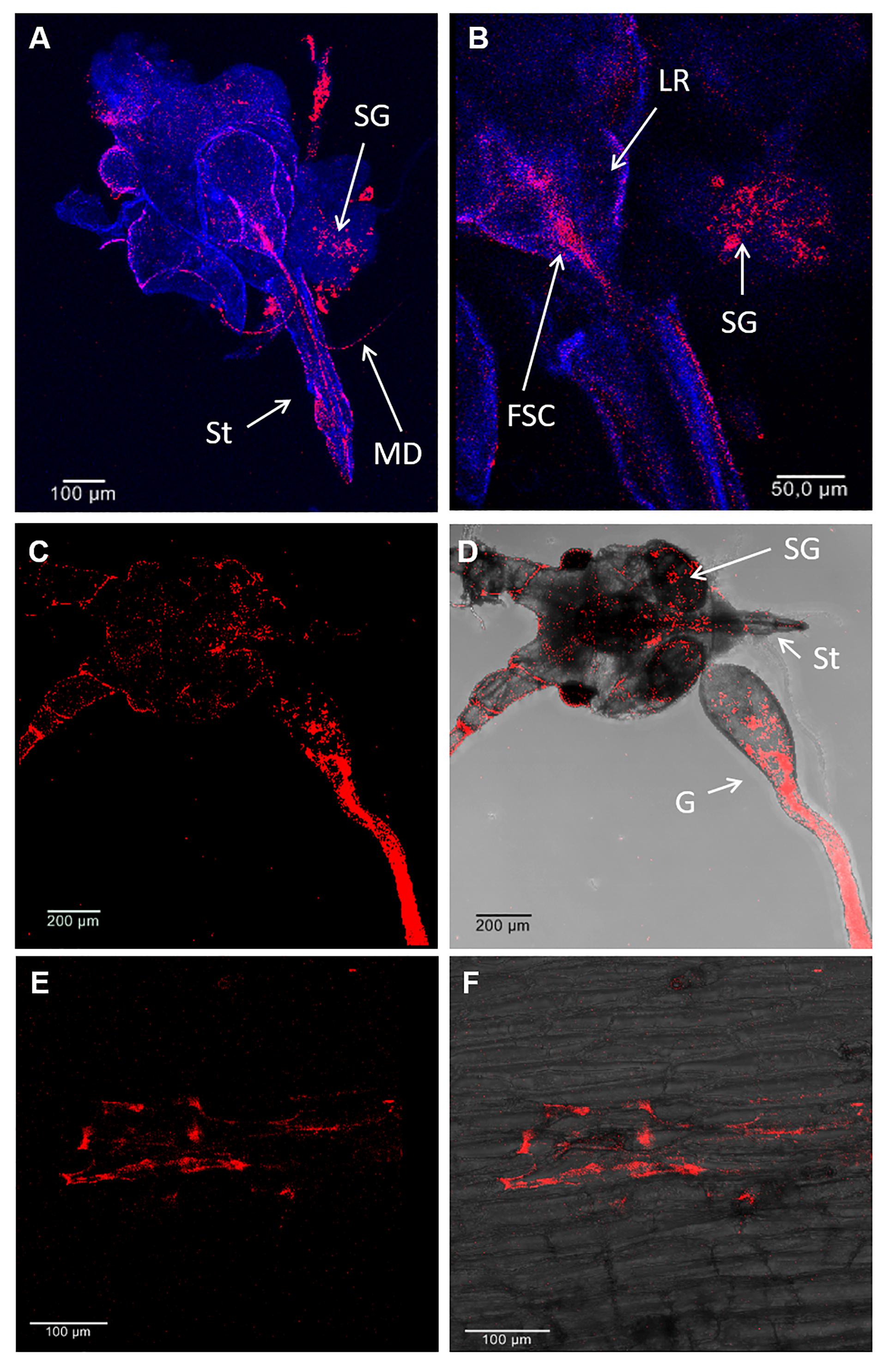
Figure 2. Localization of S. symbiotica by fluorescence in situ hybridization (FISH) in A. pisum mouthparts and V. faba tissues. Detection of S. symbiotica (red) in the head (mouthparts, salivary glands and gut) of a 10-day-old adult aphids (A–D) and V. faba longitudinal stem sections under dark field (E) and bright field (F) imaging. Nuclei were counterstained with DAPI (dark blue). Abbreviations: MD, mandible; SG, salivary gland; St, stylet; LR, labrum; FSC, food and salivary canal; G, gut.
Quantification by qPCR revealed that S. symbiotica was remarkably abundant in Serratia-positive aphids (Supplementary Table S5 and Figure 3A). Furthermore, we detected large numbers of S. symbiotica in V. faba plants after exposure to aphids from the Serratia-positive line for 2 days. When the aphids were removed from the host plants, the numbers of S. symbiotica fell progressively at the subsequent testing points, 5 and 10 days post-feeding (Figure 3B and Supplementary Table S5). However, S. symbiotica was still significantly more abundant in these plants, even 10 days post-feeding, compared to plants exposed to aphids from the Serratia-free line (Figure 3B and Supplementary Table S5).
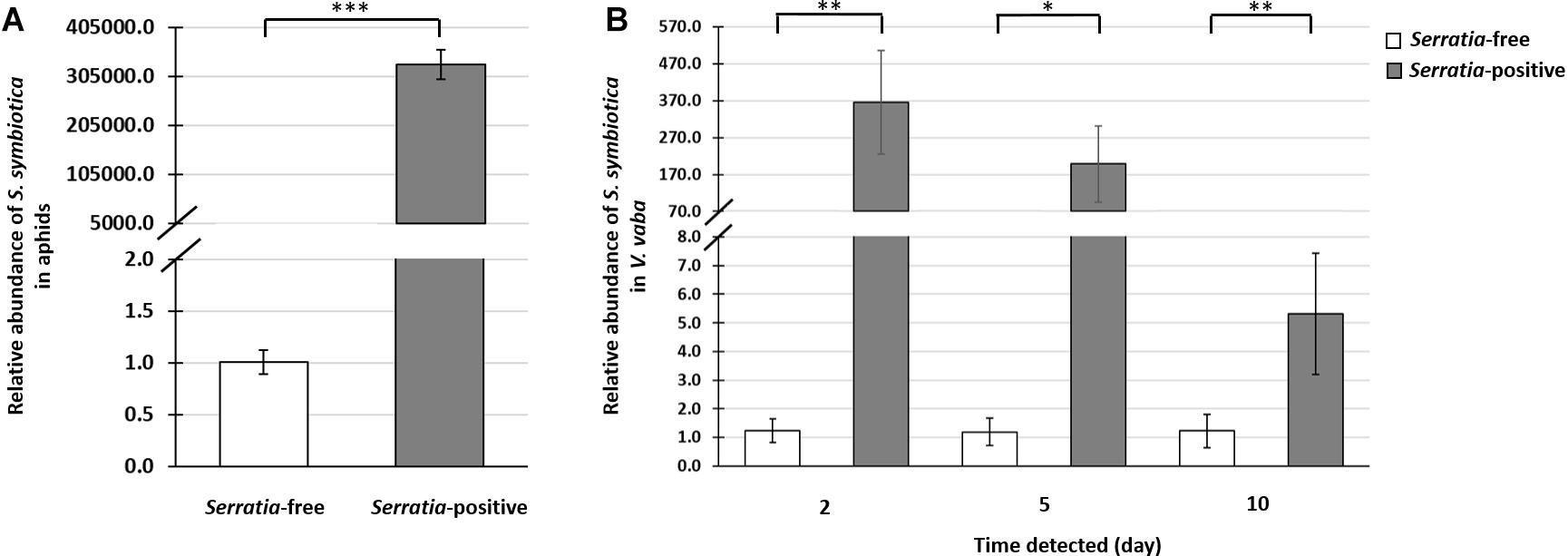
Figure 3. Quantitative PCR analysis of S. symbiotica in A. pisum and V. faba. Data show the relative abundance of the S. symbiotica dnaK gene compared to the rpl32 reference gene in aphids and the actin reference gene in plants. This was used to determine the abundance of S. symbiotica in the Serratia-positive and Serratia-free aphid lines (A), and in V. faba leaves after exposure to each aphid line, after retention times of 2, 5, and 10 days (B). Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Phylogenetic Placement of S. symbiotica
Our phylogenetic analysis of S. symbiotica incorporated 28 partial 16S rDNA sequences derived from the analysis of A. pisum and V. faba specimens. These sequences were compared with reference sequences from GenBank. S. symbiotica from the aphids and V. faba plants in this study clustered together with S. symbiotica CWBI-2.3 isolated from A. fabae, but also with most of the S. symbiotica sequences identified in other clones of A. pisum (Supplementary Figure S4).
Horizontal Transmission of S. symbiotica in Aphids via Host Plants
The detection of S. symbiotica in the mouthparts of Serratia-positive aphids and wounded plant tissues exposed to these aphids led us to investigate whether this symbiont was transmitted to naïve aphids after feeding on V. faba plants containing the bacteria. When V. faba discs were exposed to Serratia-positive aphids for 2 days, the bacterial symbiont was detected by PCR in all plant samples (Figure 1). Sequences from S. symbiotica detected in the plant were identical to those in the Serratia-positive aphids (Supplementary Figure S4). Releasing Serratia-free aphids to feed on plant discs carrying the symbiont for 3 days enabled the transmission of the symbiont to naïve aphids. This was confirmed by PCR analysis and sequencing 2 months after the infection event (Figure 1 and Supplementary Table S3). The incubation period of 2 months enabled S. symbiotica to spread among all formerly Serratia-free aphids, thus increasing the likelihood of inducing the previously observed biological effects and fitness costs (Skaljac et al., 2018). We did not detect S. symbiotica following the exposure of V. faba to Serratia-free aphids (Figure 1). During our experiments, no symptoms of bacterial disease were observed in V. faba infested with Serratia-positive aphids, indicating that the symbiont is not phytopathogenic in nature.
Proteolytic Enzymes Associated With S. symbiotica
Identification of Proteolytic Enzymes Released by S. symbiotica CWBI-2.3
Sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis of the S. symbiotica CWBI-2.3 culture supernatant revealed a remarkable number of potentially secreted proteins (Supplementary Figure S7). In total, 246 different extracellular proteins were identified by LC-MS/MS and characterised, representing numerous categories of biological processes (Supplementary Table S6). Among these proteins, we identified 15 enzymes with predicted proteolytic activity, including metalloproteases (Supplementary Table S4). These enzymes potentially facilitate the degradation of host plant proteins as their annotations suggest6,7,8. In total, nine S. symbiotica proteases with complete genomic information were included for further analysis (Table 1): the serine endopeptidases DegP and DegQ, the putative IgA-specific Zn-dependent serine endopeptidase HtpX, the putative M48 family peptidase YfgC, the putative peptidase SohB, peptidase D (PepD), aminopeptidase A (PepA) and aminopeptidase N (PepN).
S. symbiotica Genes Encoding Proteolytic Enzymes in A. pisum and Its Host Plants
Having identified nine S. symbiotica CWBI-2.3 extracellular proteases for further analysis, we tested different aphid and plant samples for the presence of the corresponding transcripts. The DegP, DeqQ, HtpX, YfgC, SohB, PepD, PepA, and PepN transcripts were detected in Serratia-positive aphids (Table 1). Furthermore, the DegQ, HtpX, YfgC, SohB, and PepA transcripts were also present (albeit at much lower levels) in plants previously exposed to the Serratia-positive aphids (Table 1 and Supplementary Figure S5). The DegQ, HtpX, YfgC, SohB, and PepA transcripts representing serine endopeptidases and metallopeptidases were selected for further qRT-PCR analysis because they may be relevant in the context of aphid–plant interactions. Quantitative RT-PCR analysis revealed that these five genes were more strongly expressed in Serratia-positive aphids than Serratia-free aphids (Supplementary Table S5 and Figure 4). The same transcripts were below the level of detection in V. faba tissues previously infested with Serratia-positive aphids (Supplementary Figure S5).
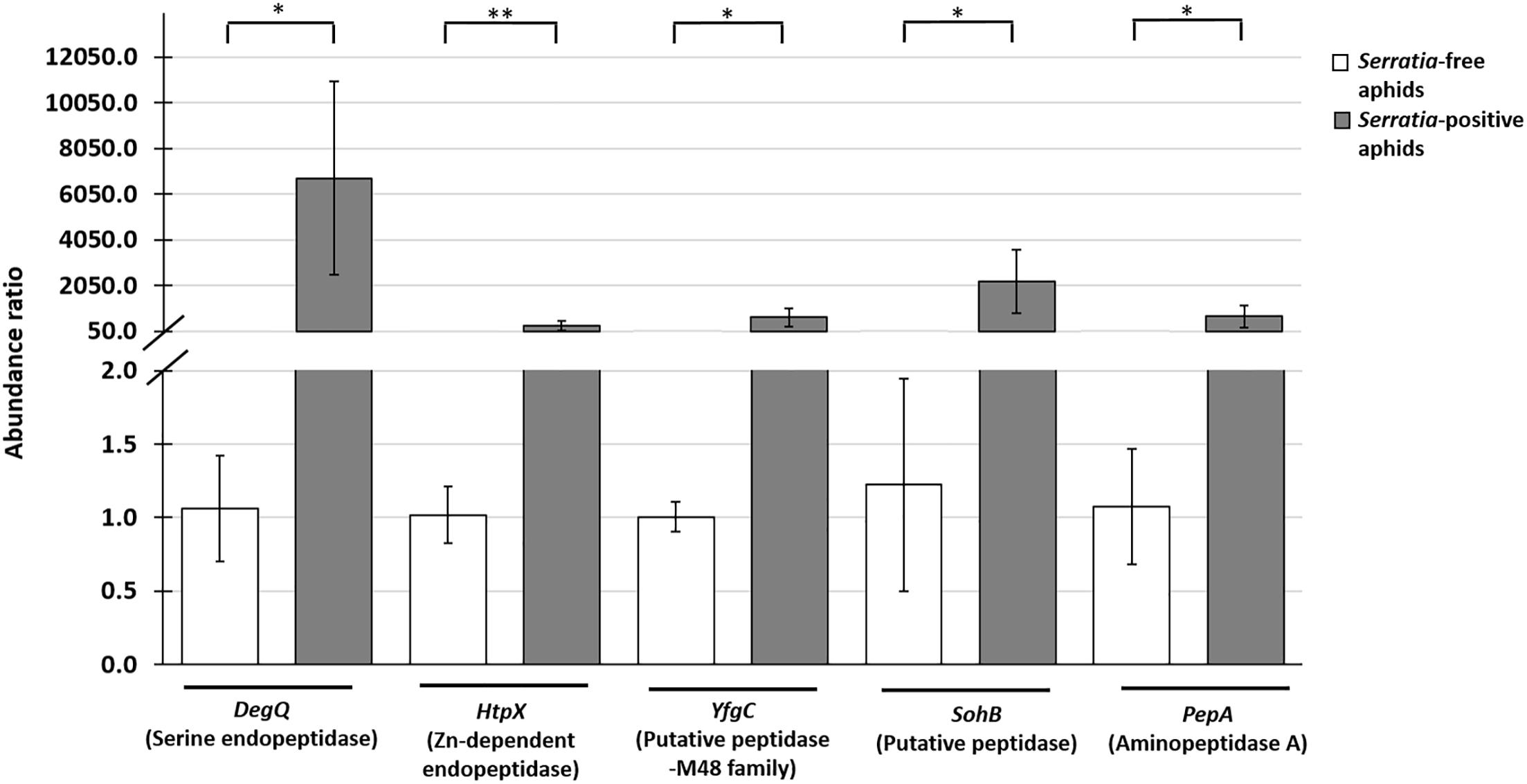
Figure 4. Quantitative RT-PCR analysis showing the relative expression of five S. symbiotica genes (DegQ, HtpX, YfgC, SohB, and PepA) encoding proteolytic enzymes associated with the host plant (Table 1) in Serratia-positive and Serratia-free aphids. The expression data were normalised to the aphid reference gene rpl32. Statistical significance is indicated as follows: ∗p < 0.05, ∗∗p < 0.01.
Discussion
Previous studies have shown that S. symbiotica colonises several A. pisum tissues, specifically the bacteriocytes, gut and haemolymph (Moran et al., 2005; Sabri et al., 2013; Luna-Ramirez et al., 2017; Skaljac et al., 2018). The experiments described here allow us to expand that distribution to include the aphid salivary glands and associated mouthparts (Figures 2A–D). Furthermore, S. symbiotica was detected in the stylet and in wounded plant tissues, providing strong evidence that aphids inoculate host plants with their bacterial symbionts (Figures 2E,F). In agreement with our data, recent studies of bacterial symbionts (e.g., Rickettsia spp., Wolbachia spp., and Cardinium spp.) associated with herbivorous insects (e.g., B. tabaci or Scaphoideus titanus Ball) reported that bacteria found in the feeding apparatus and gut were also observed in the host plants (Skaljac et al., 2010; Brumin et al., 2012; Caspi-Fluger et al., 2012; Chrostek et al., 2017; Li S.J. et al., 2017; Li Y.H. et al., 2017). The localization of cultivable strains of S. symbiotica (e.g., CWBI-2.3) associated mainly with Aphis species is currently thought to be limited to the gut, with no cells detected in the haemolymph (Pons et al., 2019). S. symbiotica CWBI-2.3 is able to colonise the entire A. pisum gut within just a few days after artificial infection via a specialised diet, without triggering an immune response or affecting survival (Renoz et al., 2015). It would be interesting to determine whether non-cultivable S. symbiotica strains are localised differently in the A. pisum as previously shown for Rickettsia spp. in B. tabaci (Gottlieb et al., 2008; Caspi-Fluger et al., 2011). We detected S. symbiotica in many A. pisum tissues (Figure 2D), including the bacteriome and ovarioles, whereas a more restricted distribution was reported in earlier studies (Moran et al., 2005; Luna-Ramirez et al., 2017).
In Israeli populations of B. tabaci, Rickettsia spp. displayed a “scattered” distribution, in which the symbiont was present in the haemocoel, excluding the bacteriocytes, or a “confined” distribution, in which it was restricted to bacteriocytes (Caspi-Fluger et al., 2011). In contrast, we previously reported that Rickettsia spp. are distributed in all B. tabaci tissues, including both the haemocoel and bacteriocytes (Skaljac et al., 2010). The Rickettsia strains with different localization patterns often featured identical sequences, suggesting they are closely related (Caspi-Fluger et al., 2011). However, even the same symbionts can show different localization patterns and fulfil diverse functions in their insect hosts, depending on the environmental conditions (Gottlieb et al., 2008; Caspi-Fluger et al., 2011).
Our results revealed the remarkable abundance of S. symbiotica in V. faba plants after only 2 days of exposure to Serratia-positive aphids (Figure 3B). When the aphids were removed from the feeding site, the S. symbiotica load decreased over the subsequent 10 days (Supplementary Table S5). A similar decline in the number of whitefly-associated Rickettsia spp. was reported in cotton leaves (Li Y.H. et al., 2017), suggesting that the production of chemical defence compounds in plants may correlate with the decline of symbionts in plant tissues. In addition to the retention time of S. symbiotica in V. faba, the viability of symbionts in plant tissues is another key requirement for successful interactions with either the plant or naïve insects (Chrostek et al., 2017). The detection of S. symbiotica mRNAs in V. faba tissues revealed that the symbiont remains alive and transcriptionally active in the plant (Table 1). This was previously shown in the Rickettsia and Wolbachia symbionts of B. tabaci (Caspi-Fluger et al., 2012; Li S.J. et al., 2017; Li Y.H. et al., 2017). Future studies should include experiments to determine whether S. symbiotica is able to multiply in the host plants as previously described for phytopathogenic S. marcescens (Petersen and Tisa, 2013).
The transmission of symbionts via host plants can have a significant impact on the ecology and evolution on both the symbiont and its insect host (Chrostek et al., 2017). For instance, Rickettsia spp. has rapidly spread among populations of B. tabaci across the southwestern United States, significantly affecting life-history traits by accelerating development, promoting survival into adulthood, and encouraging the production of more offspring (Himler et al., 2011). At the same time, the transmission of Rickettsia spp. via plants may have favoured the rapid spreading of this symbiont among populations of B. tabaci (Caspi-Fluger et al., 2012). Symbionts help herbivorous insects to utilise plants (e.g., the gut bacteria in D. virgifera virgifera), whereas other bacteria have evolved from arthropod symbionts into insect-vectored plant pathogens (e.g., Arsenophonus spp.; Sugio et al., 2015; Chrostek et al., 2017). This shows the complexity of the interactions between insects, their symbionts and plants in response to different selection pressures (Shah and Walling, 2017).
We investigated the possibility that S. symbiotica was transmitted to uninfected aphids via the host plant, as previously shown for other insect–symbiont systems (Chrostek et al., 2017). Accordingly, we found that when V. faba plants containing S. symbiotica were fed to uninfected aphids, the plants acted as reservoirs for the efficient transmission of symbionts, resulting in the reinfection of all exposed individuals (Figure 1 and Supplementary Table S3). Several studies have indicated that symbionts of herbivorous insects can be transmitted via honeydew (Darby and Douglas, 2003; Chrostek et al., 2017; Pons et al., 2019). We previously detected S. symbiotica in the honeydew of Serratia-positive A. pisum, so this transmission route cannot be ruled out in natural environments (Skaljac et al., 2018). The transmission route of cultivable S. symbiotica strains (e.g., CWBI-2.3) is unknown in Aphis species, but this study provides important clues to support the plant reservoir hypothesis. Bacterial symbionts are transmitted maternally with high fidelity. We previously detected S. symbiotica in the bacteriomes and ovarioles of A. pisum suggesting that this symbiont probably spreads via both horizontal and vertical transmission (Luna-Ramirez et al., 2017).
Given that S. symbiotica is one of the most common symbionts of aphids and that Serratia species can secrete extracellular enzymes to fulfil their roles in diverse ecological niches, we propose that some of the proteins secreted by S. symbiotica (especially proteolytic enzymes) might help the aphids to exploit their host plants more efficiently (Manzano-Marín et al., 2012; Petersen and Tisa, 2013; Sugio et al., 2015; Renoz et al., 2017). In order to test this hypothesis, we used the cultivable S. symbiotica strain CWBI-2.3 to identify extracellular proteases and investigate the abundance of the corresponding transcripts in aphids and V. faba plants. Our proteomic analysis of the S. symbiotica CWBI-2.3 culture supernatant revealed a diverse spectrum of secreted proteins, in agreement with the recently published membrane and cytosolic proteome of this species (Renoz et al., 2017; Supplementary Tables S4, S6). Our study has expanded the spectrum of S. symbiotica proteolytic enzymes (Renoz et al., 2017) to include serine endopeptidases (DegP and DegQ), M48 family metallopeptidases (HtpX and YfgC), aminopeptidases (PepA and PepN) and the other peptidases listed in Supplementary Table S4. Proteases are well-known virulence factors in pathogenic Serratia species (Petersen and Tisa, 2014) and they play important roles in the degradation of tissues that allow Serratia spp. to survive and proliferate within the host (Matsumoto, 2004).
The proteomic analysis of candidate S. symbiotica proteases in host plant tissues is not feasible due to the competition from endogenous plant proteins, so we focused on the highly sensitive detection of the corresponding transcripts. Most of the S. symbiotica CWBI-2.3 genes encoding proteases in the culture medium were also detected in both Serratia-positive aphids and in plants containing symbiont cells (Table 1). The S. symbiotica protease genes identified in V. faba were strongly expressed in Serratia-positive aphids (Figure 4 and Supplementary Table S5), suggesting that S. symbiotica may indeed help aphids to digest phloem sap proteins and potentially to resist protease inhibitors (Zhu-Salzman and Zeng, 2015). Several studies have highlighted the importance of symbiotic bacteria in the ability of insects to exploit host plants more efficiently by suppressing plant defence mechanisms and/or by expanding the host plant range. For example, this has been shown for B. tabaci and its symbiont H. defensa, and in the Colorado potato beetle (Leptinotarsa decemlineata Say) and its symbionts representing the bacterial genera Stenotrophomonas, Pseudomonas, and Enterobacter (Frago et al., 2012; Su et al., 2015; Sugio et al., 2015; Chung et al., 2017).
In this study, transcripts encoding candidate proteases were present at very low levels in plants previously infested with Serratia-positive aphids (Supplementary Figure S5). This suggests that the detection of transcripts in V. faba is most likely associated with the presence of the symbiont (Table 1). On the other hand, the abundance of S. symbiotica in aphid tissues (Figures 2A–D, 3A) together with the strong expression of protease genes associated with Serratia-positive aphids (Figure 4) suggest that the proteases may be active in the aphid gut and salivary glands but not necessarily in the host plant. These assumptions are supported by previous studies showing that plant-derived protease inhibitors inactivate digestive enzymes in the insect gut, preventing the digestion and absorption of nutrients (Ryan, 1990; Hansen and Moran, 2014). Therefore, S. symbiotica proteases are more likely to fulfil their role in the aphid gut (or salivary glands) rather than the host plants.
In summary, we investigated the localization of S. symbiotica in aphid mouthparts and host plant tissues and confirmed the transmission of this symbiont via plants, potentially explaining its high frequency among aphid populations. We expanded the repertoire of proteolytic enzymes produced by S. symbiotica in liquid medium and confirmed the strong expression of the corresponding genes in aphids and their weaker expression in infested host plants. We conclude that plants serve as reservoirs for the transmission of protease-secreting bacterial symbionts among aphids, suggesting that such symbionts could be important mediators of aphid–plant interactions. Investigating the precise nature of the symbiotic relationship described in this study will help to determine whether S. symbiotica uses proteases to spread among insect hosts, while in return enabling the insect to exploit plants more efficiently by the suppression of protease inhibitors.
There may be ecological and genomic differences between the two S. symbiotica strains used in this study, and accordingly some of the symbiotic proteases originating from the uncultivable strain may have been overlooked. Therefore, future studies should investigate extracellular proteases originating from different S. symbiotica strains released under diverse ecological conditions (e.g., exposure to a range of host plants). Furthermore, it would be interesting to determine the precise functions of the proteases listed in Table 1 to see whether any of them are specifically involved in the suppression of plant defences, the digestion of plant proteins or the proliferation of the symbiont. It would also be valuable to compare defence mechanisms in plants attacked by Serratia-positive and Serratia-free aphids because this symbiont may have the potential to evolve into a plant pathogen that uses aphids as vectors.
Author Contributions
MS, HV, NW, and SM contributed to the study design, carried out the molecular laboratory work, analysed the data, and drafted the manuscript. AV conceived, designed, and coordinated the study, and helped draft the manuscript. All authors agreed to be accountable for the content of the article and give approval for its publication.
Funding
This study was financially supported by the Hessen State Ministry of Higher Education, Research and the Arts (HMWK) via the LOEWE Research Center “Insect Biotechnology and Bioresources.”
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge Jens Grotmann, Phillipp Kirfel, Tobias Kessel, Maximilian Seip and Katja Michaelis from Fraunhofer IME (Giessen, Germany), and Sebastian Beer from University of Applied Sciences Mittelhessen, Institute of Bioprocess Engineering and Pharmaceutical Technology (Giessen, Germany) for their valuable help in this study. We thank Dr. Richard M. Twyman for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00438/full#supplementary-material
Footnotes
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://bacteria.ensembl.org/index.html
- ^ https://www.ncbi.nlm.nih.gov/
- ^ http://primer3.ut.ee/
- ^ http://eu.idtdna.com/PrimerQuest
- ^ https://www.uniprot.org
- ^ https://www.ebi.ac.uk/interpro/
- ^ https://www.ncbi.nlm.nih.gov
References
Ahmed, M. Z., De Barro, P. J., Ren, S. X., Greeff, J. M., and Qiu, B. L. (2013). Evidence for horizontal transmission of secondary endosymbionts in the Bemisia tabaci cryptic species complex. PLoS One 8:e53084. doi: 10.1371/journal.pone.0053084
Brumin, M., Levy, M., and Ghanim, M. (2012). Transovarial transmission of Rickettsia spp. and organ-specific infection of the whitefly Bemisia tabaci. Appl. Environ. Microbiol. 78, 5565–5574. doi: 10.1128/AEM.01184-12
Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Katzir, N., Portnoy, V., Belausov, E., et al. (2012). Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc. Biol. Sci. 279, 1791–1796. doi: 10.1098/rspb.2011.2095
Caspi-Fluger, A., Inbar, M., Mozes-Daube, N., Mouton, L., Hunter, M. S., and Zchori-Fein, E. (2011). Rickettsia ‘in’ and ‘out’: two different localization patterns of a bacterial symbiont in the same insect species. PLoS One 6:e21096. doi: 10.1371/journal.pone.0021096
Chiel, E., Inbar, M., Gottlieb, Y., Kelly, S. E., Asplen, M. K., Hunter, M. S., et al. (2009). Almost there: transmission routes of bacterial symbionts between trophic levels. PLoS One 4:e4767. doi: 10.1371/journal.pone.0004767
Chrostek, E., Pelz-Stelinski, K., Hurst, G. D. D., and Hughes, G. L. (2017). Horizontal transmission of intracellular insect symbionts via plants. Front. Microbiol. 8:2237. doi: 10.3389/fmicb.2017.02237
Chung, S. H., Scully, E. D., Peiffer, M., Geib, S. M., Rosa, C., Hoover, K., et al. (2017). Host plant species determines symbiotic bacterial community mediating suppression of plant defenses. Sci. Rep. 7:39690. doi: 10.1038/srep39690
Consortium, I. A. G. (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
Dale, C., and Moran, N. A. (2006). Molecular interactions between bacterial symbionts and their hosts. Cell 126, 453–465. doi: 10.1016/j.cell.2006.07.014
Darby, A. C., and Douglas, A. E. (2003). Elucidation of the transmission patterns of an insect-borne bacterium. Appl. Environ. Microbiol. 69, 4403–4407. doi: 10.1128/AEM.69.8.4403-4407.2003
Distler, U., Kuharev, J., Navarro, P., and Tenzer, S. (2016). Label-free quantification in ion mobility-enhanced data-independent acquisition proteomics. Nat. Protoc. 11, 795–812. doi: 10.1038/nprot.2016.042
Foray, V., Grigorescu, A. S., Sabri, A., Haubruge, E., Lognay, G., Francis, F., et al. (2014). Whole-genome sequence of serratia symbiotica strain CWBI-2.3T, a free-living symbiont of the black bean aphid Aphis fabae. Genome Announc. 2:e00767-14. doi: 10.1128/genomeA.00767-14
Frago, E., Dicke, M., and Godfray, H. C. (2012). Insect symbionts as hidden players in insect-plant interactions. Trends Ecol. Evol. 27, 705–711. doi: 10.1016/j.tree.2012.08.013
Furch, A. C., van Bel, A. J., and Will, T. (2015). Aphid salivary proteases are capable of degrading sieve-tube proteins. J. Exp. Bot. 66, 533–539. doi: 10.1093/jxb/eru487
Gehrer, L., and Vorburger, C. (2012). Parasitoids as vectors of facultative bacterial endosymbionts in aphids. Biol. Lett. 8:613. doi: 10.1098/rsbl.2012.0144
Ghanim, M., Brumin, M., and Popovski, S. (2009). A simple, rapid and inexpensive method for localization of tomato yellow leaf curl virus and Potato leafroll virus in plant and insect vectors. J. Virol. Methods 159, 311–314. doi: 10.1016/j.jviromet.2009.04.017
Gonella, E., Pajoro, M., Marzorati, M., Crotti, E., Mandrioli, M., Pontini, M., et al. (2015). Plant-mediated interspecific horizontal transmission of an intracellular symbiont in insects. Sci. Rep. 5:15811. doi: 10.1038/srep15811
Gottlieb, Y., Ghanim, M., Gueguen, G., Kontsedalov, S., Vavre, F., Fleury, F., et al. (2008). Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 22, 2591–2599. doi: 10.1096/fj.07-101162
Grigorescu, A. S., Renoz, F., Sabri, A., Foray, V., Hance, T., and Thonart, P. (2018). Accessing the hidden microbial diversity of aphids: an illustration of how culture-dependent methods can be used to decipher the insect microbiota. Microb. Ecol. 75, 1035–1048. doi: 10.1007/s00248-017-1092-x
Hansen, A. K., and Moran, N. A. (2011). Aphid genome expression reveals host–symbiont cooperation in the production of amino acids. Proc. Natl. Acad. Sci. U.S.A. 108, 2849–2854. doi: 10.1073/pnas.1013465108
Hansen, A. K., and Moran, N. A. (2014). The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol. Ecol. 23, 1473–1496. doi: 10.1111/mec.12421
Hase, C. C., and Finkelstein, R. A. (1993). Bacterial extracellular zinc-containing metalloproteases. Microbiol. Rev. 57, 823–837.
Himler, A. G., Bergen, J. E., Kozuch, A., Kelly, S. E., Tabashnik, B. E., Chiel, E., et al. (2011). Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332:254. doi: 10.1126/science.1199410
Koressaar, T., and Remm, M. (2007). Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–1291. doi: 10.1093/bioinformatics/btm091
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laughton, A. M., Fan, M. H., and Gerardo, N. M. (2014). The combined effects of bacterial symbionts and aging on life history traits in the pea aphid, Acyrthosiphon pisum. Appl. Environ. Microbiol. 80, 470–477. doi: 10.1128/AEM.02657-13
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, S. J., Ahmed, M. Z., Lv, N., Shi, P. Q., Wang, X. M., Huang, J. L., et al. (2017). Plant-mediated horizontal transmission of Wolbachia between whiteflies. ISME J. 11, 1019–1028. doi: 10.1038/ismej.2016.164
Li, Y. H., Ahmed, M. Z., Li, S. J., Lv, N., Shi, P. Q., Chen, X. S., et al. (2017). Plant-mediated horizontal transmission of Rickettsia endosymbiont between different whitefly species. FEMS Microbiol. Ecol. 93:fix138. doi: 10.1093/femsec/fix138
Login, F. H., Balmand, S., Vallier, A., Vigneron, A., Rochat, D., Heddi, A., et al. (2011). Antimicrobial peptides keep insect endosymbionts under control. Science 334, 362–365. doi: 10.1126/science.1209728
Luna-Ramirez, K., Skaljac, M., Grotmann, J., Kirfel, P., and Vilcinskas, A. (2017). Orally delivered scorpion antimicrobial peptides exhibit activity against pea aphid (Acyrthosiphon pisum) and its bacterial symbionts. Toxins 9:E261. doi: 10.3390/toxins9090261
Manzano-Marín, A., Lamelas, A., Moya, A., and Latorre, A. (2012). Comparative genomics of Serratia spp.: two paths towards endosymbiotic life. PLoS One 7:e47274. doi: 10.1371/journal.pone.0047274
Manzano-Marin, A., and Latorre, A. (2016). Snapshots of a shrinking partner: genome reduction in Serratia symbiotica. Sci. Rep. 6:32590. doi: 10.1038/srep32590
Matsumoto, K. (2004). Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 385, 1007–1016. doi: 10.1515/BC.2004.131
Miyoshi, S. I. (2013). Extracellular proteolytic enzymes produced by human pathogenic vibrio species. Front. Microbiol. 4:339. doi: 10.3389/fmicb.2013.00339
Moran, N. A., McCutcheon, J. P., and Nakabachi, A. (2008). Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42, 165–190. doi: 10.1146/annurev.genet.41.110306.130119
Moran, N. A., Russell, J. A., Koga, R., and Fukatsu, T. (2005). Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl. Environ. Microbiol. 71, 3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005
Oliver, K. M., Degnan, P. H., Burke, G. R., and Moran, N. A. (2010). Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu. Rev. Entomol. 55, 247–266. doi: 10.1146/annurev-ento-112408-085305
Oliver, K. M., Smith, A. H., and Russell, J. A. (2014). Defensive symbiosis in the real world – advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct. Ecol. 28, 341–355. doi: 10.1111/1365-2435.12133
Petersen, L. M., and Tisa, L. S. (2013). Friend or foe? A review of the mechanisms that drive Serratia towards diverse lifestyles. Can. J. Microbiol. 59, 627–640. doi: 10.1139/cjm-2013-0343
Petersen, L. M., and Tisa, L. S. (2014). Molecular characterization of protease activity in Serratia sp. strain SCBI and its importance in cytotoxicity and virulence. J. Bacteriol. 196, 3923–3936. doi: 10.1128/JB.01908-14
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Pons, I., Renoz, F., Noël, C., and Hance, T. (2019). New insights into the nature of symbiotic associations in aphids: infection process, biological effects and transmission mode of cultivable Serratia symbiotica bacteria. Appl. Environ. Microbiol. doi: 10.1128/AEM.02445-18
Powell, G., Tosh, C. R., and Hardie, J. (2006). Host plant selection by aphids: behavioral, evolutionary, and applied perspectives. Annu. Rev. Entomol. 51, 309–330. doi: 10.1146/annurev.ento.51.110104.151107
Renoz, F., Champagne, A., Degand, H., Morsomme, P., Foray, V., Hance, T., et al. (2017). Toward a better understanding of the mechanisms of symbiosis: a comprehensive proteome map a nascent insect symbiont. PeerJ Preprints 5:e3291. doi: 10.7717/peerj.3291
Renoz, F., Noël, C., Errachid, A., Foray, V., and Hance, T. (2015). Infection dynamic of symbiotic bacteria in the pea aphid Acyrthosiphon pisum gut and host immune response at the early steps in the infection process. PLoS One 10:e0122099. doi: 10.1371/journal.pone.0122099
Russell, J. A., Latorre, A., Sabater-Munoz, B., Moya, A., and Moran, N. A. (2003). Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol. Ecol. 12, 1061–1075. doi: 10.1046/j.1365-294X.2003.01780.x
Ryan, C. A. (1990). Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Ann. Rev. Pathol. 28, 425–449. doi: 10.1146/annurev.py.28.090190.002233
Sabri, A., Leroy, P., Haubruge, E., Hance, T., Frere, I., Destain, J., et al. (2011). Isolation, pure culture and characterization of Serratia symbiotica sp. nov., the R-type of secondary endosymbiont of the black bean aphid Aphis fabae. Int. J. Syst. Evol. Microbiol. 61(Pt 9), 2081–2088. doi: 10.1099/ijs.0.024133-0
Sabri, A., Vandermoten, S., Leroy, P. D., Haubruge, E., Hance, T., Thonart, P., et al. (2013). Proteomic investigation of aphid honeydew reveals an unexpected diversity of proteins. PLoS One 8:e74656. doi: 10.1371/journal.pone.0074656
Shah, J., and Walling, L. (2017). Editorial: advances in plant-hemipteran interactions. Front. Plant Sci. 8:1652. doi: 10.3389/fpls.2017.01652
Shevchenko, A., Sunyaev, S., Loboda, A., Shevchenko, A., Bork, P., Ens, W., et al. (2001). Charting the proteomes of organisms with unsequenced genomes by MALDI-quadrupole time-of-flight mass spectrometry and BLAST homology searching. Anal. Chem. 73, 1917–1926. doi: 10.1021/ac0013709
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V., and Mann, M. (2006). In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860. doi: 10.1038/nprot.2006.468
Skaljac, M., Kanakala, S., Zanic, K., Puizina, J., Pleic, I. L., and Ghanim, M. (2017). Diversity and phylogenetic analyses of bacterial symbionts in three whitefly species from Southeast Europe. Insects 8:E113. doi: 10.3390/insects8040113
Skaljac, M., Kirfel, P., Grotmann, J., and Vilcinskas, A. (2018). Fitness costs of infection with Serratia symbiotica are associated with greater susceptibility to insecticides in the pea aphid Acyrthosiphon pisum. Pest. Manag. Sci. 74, 1829–1836. doi: 10.1002/ps.4881
Skaljac, M., Zanic, K., Ban, S. G., Kontsedalov, S., and Ghanim, M. (2010). Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol. 10:142. doi: 10.1186/1471-2180-10-142
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/JB.00345-12
Su, Q., Oliver, K. M., Xie, W., Wu, Q., Wang, S., and Zhang, Y. (2015). The whitefly-associated facultative symbiont Hamiltonella defensa suppresses induced plant defences in tomato. Funct. Ecol. 29, 1007–1018. doi: 10.1111/1365-2435.12405
Sugio, A., Dubreuil, G., Giron, D., and Simon, J. C. (2015). Plant-insect interactions under bacterial influence: ecological implications and underlying mechanisms. J. Exp. Bot. 66, 467–478. doi: 10.1093/jxb/eru435
Van Emden, H. F., and Harrington, R. (2017). Aphids as Crop Pests. Wallingford: CABI. doi: 10.1079/9781780647098.0000
Wielkopolan, B., and Obrepalska-Steplowska, A. (2016). Three-way interaction among plants, bacteria, and coleopteran insects. Planta 244, 313–332. doi: 10.1007/s00425-016-2543-1
Will, T., Schmidtberg, H., Skaljac, M., and Vilcinskas, A. (2017). Heat shock protein 83 plays pleiotropic roles in embryogenesis, longevity, and fecundity of the pea aphid Acyrthosiphon pisum. Dev. Genes Evol. 227, 1–9. doi: 10.1007/s00427-016-0564-1
Wu, J.-W., and Chen, X. L. (2011). Extracellular metalloproteases from bacteria. Appl. Microbiol. Biotechnol. 92:253. doi: 10.1007/s00253-011-3532-8
Keywords: symbiosis, extracellular proteases, phloem sap, Serratia symbiotica, Vicia faba
Citation: Skaljac M, Vogel H, Wielsch N, Mihajlovic S and Vilcinskas A (2019) Transmission of a Protease-Secreting Bacterial Symbiont Among Pea Aphids via Host Plants. Front. Physiol. 10:438. doi: 10.3389/fphys.2019.00438
Received: 12 November 2018; Accepted: 01 April 2019;
Published: 17 April 2019.
Edited by:
Patrizia Falabella, University of Basilicata, ItalyReviewed by:
Clare L. Casteel, University of California, Davis, United StatesMarylène Poirié, Université Côte d’Azur, France
Copyright © 2019 Skaljac, Vogel, Wielsch, Mihajlovic and Vilcinskas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andreas Vilcinskas, QW5kcmVhcy5WaWxjaW5za2FzQGFncmFyLnVuaS1naWVzc2VuLmRl
 Marisa Skaljac
Marisa Skaljac Heiko Vogel
Heiko Vogel Natalie Wielsch
Natalie Wielsch Sanja Mihajlovic
Sanja Mihajlovic Andreas Vilcinskas
Andreas Vilcinskas