- 1Department of Gastroenterology, Huadong Hospital, Fudan University, Shanghai, China
- 2Department of Geriatrics, Shanghai Key Laboratory of Clinical Geriatric Medicine, Huadong Hospital, Fudan University, Shanghai, China
Non-alcoholic fatty liver disease (NAFLD) is a common disease, which is characterized by the accumulation of triglycerides in the hepatocytes without excess alcohol intake. Circadian rhythms can participate in lipid, glucose, and cholesterol metabolism and are closely related to metabolism seen in this disease. Circadian clock genes can modulate liver lipid metabolism. Desynchrony of circadian rhythms and the influences imparted by external environmental stimuli can increase morbidity. By contrast, synchronizing circadian rhythms can help to alleviate the metabolic disturbance seen in NAFLD. In this review, we have discussed the current research connections that exist between the circadian clock and the metabolism of NAFLD, and we have specifically focused on the key circadian clock genes, Bmal1, Clock, Rev-Erbs, Rors, Pers, Crys, Nocturnin, and DECs.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a form of triglyceride (TG) accumulation at or exceeding 5% of the liver weight without excess alcohol intake (Andronescu et al., 2018). The histological classification distinguishes a range of conditions within NAFLD that vary from hepatic steatosis to non-alcoholic steatohepatitis (NASH), which might evolve to many subsequent conditions that include fibrosis, cirrhosis, liver failure, and hepatocellular carcinoma (Ipsen et al., 2018).
The global prevalence of NAFLD has dramatically increased during the past three decades, as a result of a global epidemic in the incidence of metabolic disorders. The current prevalence rates of NAFLD vary from 17 to 51% in western countries and 25% in Asian countries (Stern and Castera, 2017; Wong et al., 2018). In China, the prevalence varies from 15 to 30%.
In our study, we have revealed that the prevalence of NAFLD in employees of the city of Shanghai, China was 38.17%; a rate that was much higher than we had previously appreciated (Hu et al., 2012; Sherif et al., 2016). Furthermore, it was reported that the prevalence of NAFLD in children is increasing, and although putative mechanisms have received broad discussion, biological reasons accounting for the prevalence and contributions to NAFLD remain an enigma (Friedman et al., 2018).
Multiple factors lead to abnormal accumulation of triglycerides (TGs) in hepatocytes, among which, insulin resistance (IR) is the most essential pathogenesis. The role of insulin in inhibiting the decomposition of fat cells is weakened where IR is seen, resulting in lipolysis from the adipose tissue, and increased uptake of free fatty acids into the liver. At the same time, the utilization of TGs by the liver is inhibited, which provokes lipid deposition in the liver (Asrih and Jornayvaz, 2015). Fat accumulation in the liver can be traced by an increased incidence of de novo lipogenesis. In addition, mitochondrial dysfunction could impair fatty acid beta-oxidation and cause lipid accumulation, which usually precedes NAFLD. Excessive TG is transported out of the cell by binding to intra-hepatic synthesis of VLDL (Ipsen et al., 2018). With impaired fatty acid beta-oxidation or TG transport, the capacity of the liver to clear lipids efficiently is reduced, which might ultimately lead to the development of NAFLD (Figure 1).
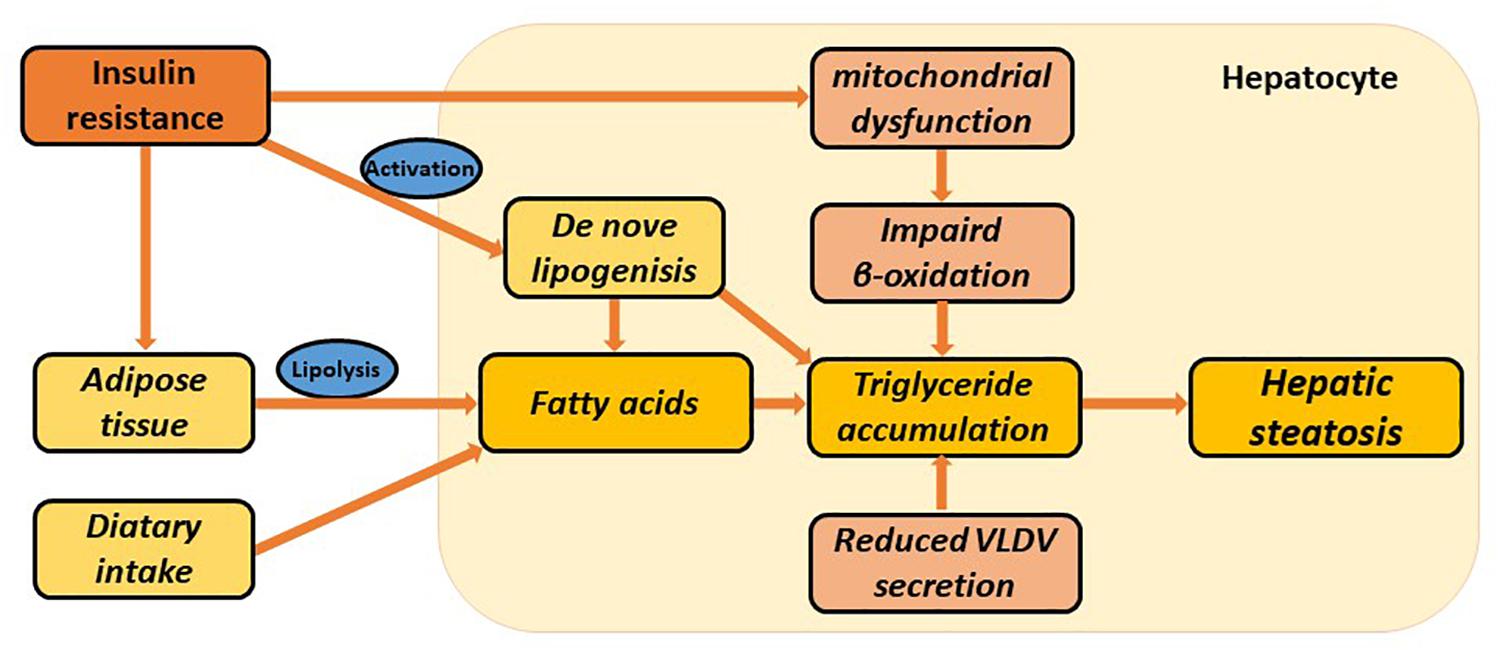
Figure 1. Multiple metabolic disorders involved in the pathogenesis of hepatic steatosis. Hepatic steatosis can be stimulated via increased adipose tissue lipolysis, increased de novo lipogenesis, increased dietary fatty acid uptake, impaired β-oxidation, and reduced VLDL synthesis and secretion. Insulin resistance is particularly involved in several metabolic pathways. These all lead to hepatic triglyceride accumulation and ultimately NAFLD.
Circadian rhythms in mammals are hard-wired biological systems that follow the 24-h cycle of the day, which serve to precisely regulate many of the major physiological activities, including sleep/wake cycles, feeding/fasting cycles, endocrine rhythms and of course metabolic rhythms (Bass and Takahashi, 2010). Studies provide a strong link between circadian rhythm disruption and the onset of a variety of human diseases. Desynchrony of circadian rhythms and the external environment, such as shift work, chronic jet lag, intentional sleep restriction, deprivation and night eating can markedly contribute to increased morbidity. Shift workers for example, exhibit a higher prevalence of obesity and associated disorders, such as NAFLD (Johnston, 2014). By contrast, restoration of normal circadian rhythms can improve overall health and alleviate the observed morbidity (Oosterman et al., 2015). The circadian rhythm system thus plays a key role in human physiology and disease systems.
A connection between the circadian rhythm and NAFLD has relatively recently been proposed (Gnocchi et al., 2015). Circadian rhythms are closely related to metabolic diseases and can participate in lipid, glucose, and cholesterol metabolism. Resynchronizing circadian rhythms can help alleviate this observed metabolic disturbance (Mazzoccoli et al., 2018).
In this review, we will discuss current research connections between the circadian clock and the metabolism seen in NAFLD, with a particular focus on key circadian clock genes.
The Molecular Network of Circadian Clock
The molecular circadian clock network is composed of four transcriptional-translational feedback loops. The core loop, which includes Clock (circadian locomotor output cycles kaput) and Bmal1 (brain and muscle-ARNT-like 1, also known as ARNTL1), generates the autonomous circadian rhythm (Yoo et al., 2017) (Figure 2). Clock and Bmal1 heterodimerize and induce the transcription of clock-controlled genes (CCGs) in a process that involves binding with E-box (5′-CACGTG-3′). The heterodimers also direct the transcription of their functional repressors that include period (Per1, 2, and 3) and cryptochrome (Cry1 and 2), with the ultimate formation of a self-regulated loop (Gerber et al., 2015). When the protein expression levels of Pers and Crys achieve a high level, they dimerize and translocate to the nucleus to repress Clock: Bmal1-mediated transcription. Pers and Crys undergo post-translational modifications that induce their degradation in biological readiness for a new circadian cycle. In the second loop, Clock: Bmal1 heterodimers regulate the transcription of the DNA-binding orphan nuclear receptor reverse erythroblastosis virus-α/β (Rev-Erb-α/β, Rev-Erbs), and the retinoid-related orphan receptor-α, -β, and -γ (Ror-α, -β, -γ, and Rors). Rors activate, while Rev-Erbs repress, the transcriptional expression of Bmal1 (Orozco-Solis and Sassone-Corsi, 2014). The Rev-Erbα/b proteins, whose levels increase during the day, bind specific responsive promoter elements (RRE) and inhibit Bmal1 transcription (Guillaumond et al., 2005). At night, Rev-Erbα protein levels are low, allowing for the transcription of Bmal1. Recently, the expression of DECs, which include DEC1 (Bhlhe40/Stra13/Sharp2) and DEC2 (Bhlhe41/Sharp1) have been found to function as clock genes that can form the third autonomous feedback loop. DECs can repress their own transcription by binding to Bmal1 or by binding to E-box sites by competing with Clock: Bmal1 (Sato et al., 2018). DECs can also repress the transcription of Per1 and DBP (albumin d-element binding protein) (Kawamoto et al., 2004). Furthermore, DBP and E4BP4 can bind D-box, activate and suppress transcription to stabilize and fine-tune the Per/Cry feedback loop, and thus promote the formation of the fourth loop (Yamajuku et al., 2011). In the liver, the sex of animals is a pivotal regulator of circadian clock genes that include Rev-Erbα, Rorγ, and Cry1 (Doi, 2012).
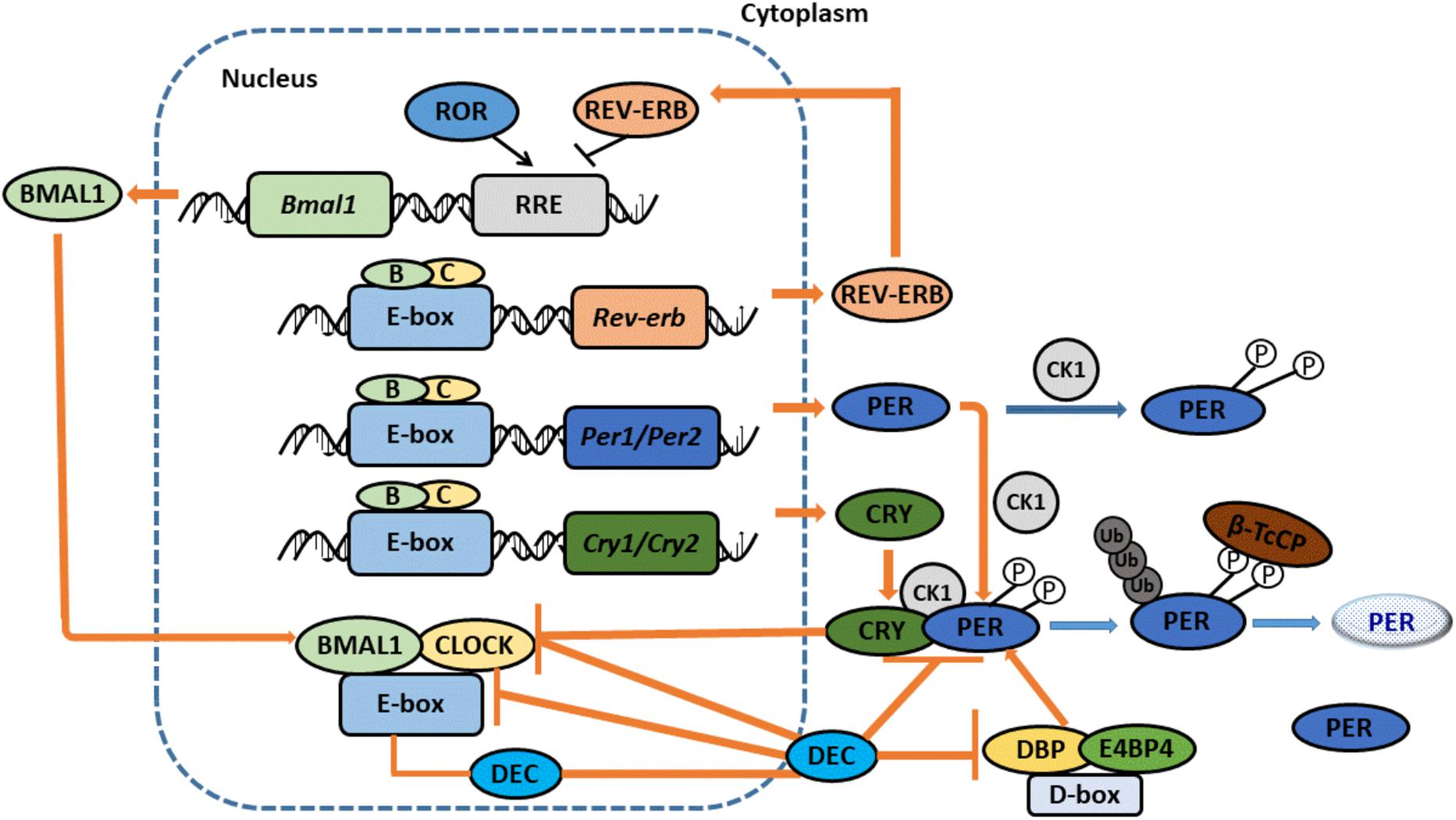
Figure 2. The Molecular Network of Circadian clock. CLOCK and BMAL1 dimerize to stimulate the expression of CCGs with E-box promoter. CLOCK: BMAL1 also activate the expression of the PERS and CRYS. When PERs and CRYs protein levels become high, they dimerize and translocate to the nucleus to repress CLOCK: BMAL1-mediated transcription. PERs and CRYs undergo post-translational modifications that induce their degradation, prepared a new circadian cycle. Proteins REV-ERBα/b, whose levels increase during the day, bind specific responsive promoter elements (RRE) and inhibit BMAL1 transcription. At night, REV-ERBα proteins levels are low, allowing BMAL1’s transcription. DECs also can form an additional loop. DECs repress their own transcription by directly binding to BMAL1 and/or E-box sites. DECs can also repress the transcription of PER1 and DBP. Furthermore, DBP and E4BP4 can bind D-box, activate and suppress transcription to stabilize and fine-tune the PER/CRY feedback loop, and thus promote the formation of the fourth loop. Adopted with modification from Masao Doi.
Circadian Clock Genes and Metabolism in the Setting of Nafld
NAFLD is essentially an imbalance of lipogenesis between free fatty acid (FFA) supply, formation, utilization, and disposal. The circadian clock is also closely associated with metabolism. Circadian clock genes tightly modulate the metabolism of liver lipids, and disruption of the biological circadian rhythms induces lipid accumulation in the liver (Mazzoccoli et al., 2014; Shi et al., 2014). However, the exact mechanisms remain obscure and unresolved at this time. Targeting the clock genes in the mouse model will benefit our improved understanding of how circadian rhythms interact with a variety of metabolic disorders (Sahar and Sassone-Corsi, 2012) (Table 1).
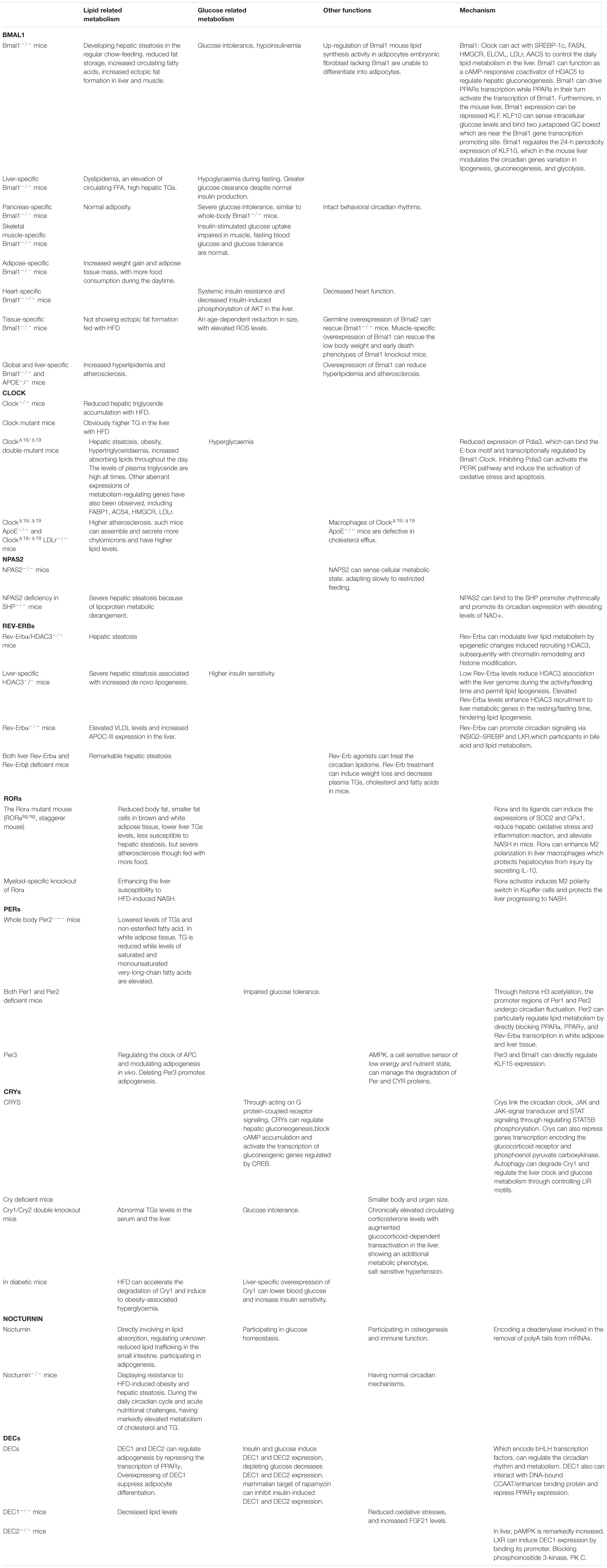
Bmal1
Bmal1 plays an important role in the modulation of fat storage, utilization and adipocyte differentiation. Up-regulation of Bmal1 increases lipid synthesis activity in adipocytes (Tong and Yin, 2013). Bmal1-/- knock-out mice show glucose intolerance, hypoinsulinemia, reduced fat storage, increased circulating fatty acids, increased ectopic fat formation in the liver and muscle, and hepatic steatosis even with regular chow-feeding (McDearmon et al., 2006; Landgraf et al., 2017). Liver-specific Bmal1-/- knock-out mice exhibit hypoglycemia during fasting, and greater glucose clearance despite normal insulin production and dyslipidemia, including high circulating FFA levels and high hepatic TGs (Pan et al., 2016). Pancreas-specific Bmal1-/- knockout mice show severe glucose intolerance, despite intact behavioral circadian rhythms and normal adiposity. Skeletal muscle-specific Bmal1-/- knockout mice show impaired insulin-stimulated glucose uptake in the muscles, while fasting blood glucose and glucose tolerance activities appear normal. Adipose-specific Bmal1-/- knockout mice show increased weight gain and adipose tissue mass, which might be due to increased food consumption during the daytime hours. Heart-specific Bmal1-/- knockout mice show decreased heart function, systemic insulin resistance and decreased insulin-induced AKT phosphorylation in the liver (Nakao et al., 2018). By contrast, global and liver-specific Bmal1-/- knockout and APOE-/- knockout mice show increased hyperlipidemia and atherosclerosis. Hepatic overexpression of Bmal1 in liver-specific Bmal1-/- knockout and APOE-/- knockout mice can reduce hyperlipidemia and atherosclerosis. Mouse embryonic fibroblasts lacking Bmal1 cannot differentiate into adipocytes (Grechez-Cassiau et al., 2008). Furthermore, tissue-specific (i.e., liver, skeletal muscle, fat, bone, spleen, kidney, testis, heart, and lung) Bmal1-/- knockout mice show an age-dependent reduction in size, and do so consistently with elevated levels of ROS (Nakahata et al., 2018). Germline overexpression of Bmal2 can rescue Bmal1-/- knockout mice (Reinke and Asher, 2016). Muscle-specific overexpression of Bmal1 can rescue low body weight and early death phenotypes that are seen in Bmal1 knockout mice (Gooley, 2016).
Bmal1: Clock cooperates with SREBP-1c, and downstream genes like fatty acid synthase (FASN), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), and fatty acid elongase family members (ELOVL), the low-density lipoprotein receptor (LDLr), and acetoacetyl-CoA synthetase (AACS) to modulate daily lipid metabolism in the liver (Friedrichs et al., 2018). Bmal1 can function as a cAMP-responsive coactivator of HDAC5 to regulate hepatic gluconeogenesis (Li et al., 2018). Bmal1 can drive PPARs transcription, while in turn, PPARs can activate the transcription of Bmal1, which drives PPARα and activates PPARα targeted gene expression. PPARα can directly bind PPAR response elements (PPRE), which are located upstream of the transcription initiation site in the Bmal1 promoter where they serve to regulate Bmal1 expression (Matsusue et al., 2014). PPARγ positively acts on Bmal1 expression by binding the same PPRE site (Pettersson-Klein et al., 2018). Further, in the mouse liver, Bmal1 expression is transcriptionally repressed by the member of three-zinc finger family of Kruppel-like transcription factors (KLF). KLF10 (also known as TGFb inducible early gene-1, or TIEG1) can sense intracellular glucose levels and bind two juxtaposed GC boxes that spatially located near the Bmal1 gene transcription promotor site. Bmal1 regulates the 24-h periodic expression of KLF10, which in the mouse liver, modulates variations in circadian gene expression in lipogenesis, gluconeogenesis, and glycolysis. Reciprocal control between PPARα/Bmal1, PPARγ/Bmal1 and KLF10/Bmal1 provides fine metabolic regulation in accord with the circadian rhythms system (Guillaumond et al., 2010).
Interestingly, sex dimorphism can modulate the acrophase of hepatic Bmal1, and can do so because of differential androgenic and estrogenic hormonal circadian regulation. The daily expression of hepatic Bmal1 in lean male mice (LM) is similar to that found in other studies. Acrophase occurs during the transition from dark to light cycles in LM, while in lean female mice (LF), acrophase is found during the transition from light to dark (Pérez-Mendoza et al., 2018). Increased lipogenic gene expression is observed in obese males with reduced levels of hepatic Bmal1 are found at the end of the light phase, while the same observations were found in Bmal1-/- knockout mice (Soeda et al., 2017).
Clock
Similarly, Clock is closely associated with NAFLD. Clock-/- knockout mice display a reduced hepatic triglyceride accumulation under HFD conditions. Clock mutant mice with HFD show a clearly increased TG in the liver. Contrary to observations seen in wild-type mice, ClockΔ19/Δ19 double-mutant mice, are characterized by hepatic steatosis, obesity, hypertriglyceridemia and hyperglycemia, and show increased absorption of lipids throughout the day. The levels of plasma triglyceride in ClockΔ19/Δ19 double-mutant mice do not show circadian rhythms and are high at all times of the day. Other aberrant expressions of metabolism-regulating genes have also been observed, including fatty acid binding protein1 (FABP1), acyl-CoA synthetase 4 (ACS4), HMGCR, and of the low-density lipoprotein receptor (LDLr) (Pérez-Mendoza et al., 2014). ClockΔ19/Δ19 double-mutant mice show reduced expression of Pdia3, which can bind the E-box motif, which is transcriptionally regulated by Bmal1: Clock. Inhibiting Pdia3 can activate the PERK-mediated signaling pathway and induces activation of oxidative stress and apoptosis. In addition, the PERK signaling pathway, appears to be the key pathway that is predominantly regulated by Clock.
ClockΔ19/Δ19 ApoE-/- knockout and ClockΔ19/Δ19 LDLr-/- knockout mice show higher levels of incidences of atherosclerosis. Physiologic studies indicate that such mice assemble and secrete more chylomicrons and have higher lipid levels. Moreover, macrophages of ClockΔ19/Δ1 ApoE-/- knockout mice are defective in cholesterol efflux (Sookoian et al., 2008).
Neuronal PAS domain-containing protein 2 (Npas2), paralogous to Clock, is mainly expressed in the brain with lower levels in the peripheral tissues, while Clock is mainly expressed in the peripheral tissues (Reick et al., 2001). Naps2 can also sense the cellular metabolic state. Most likely due to the function of Clock in SCN neurons, loss of Npas2 does not affect feeding patterns or weight gain. Npas2-/- knockout mice adapt slowly to restricted feeding (Schleicher et al., 2015). The orphan nuclear receptor small heterodimer partner (SHP) can inhibit Npas2 gene transcription and promoter activity, while Npas2 can bind to the SHP promoter rhythmically and promote its circadian expression while elevating levels of NAD+. Npas2 deficiency in SHP-/- knockout mice can lead to severe hepatic steatosis because of disrupted lipoprotein metabolism (Lee et al., 2015).
Rev-Erbs and Rors
Rev-Erbα and its close homolog Rev-Erbβ (also known as nuclear receptor subfamily1, group D, member 1; NR1D1) are heme-dependent transcriptional repressors, whereas Ror -α, -β, and -γ (Rors) are transcriptional activators. Rev-Erbs and Rors can recognize the same DNA binding sites (Ror response elements, RREs) and perform opposing regulatory biological functions (Hu et al., 2012).
Rev-Erbs
Rev-Erbα can epigenetically modulate liver lipid metabolism that subsequently recruits histone deacetylase 3 (HDAC3), and induces chromatin remodeling and histone modification. Rev-Erbα/HDAC3-/- knockout mice display hepatic steatosis. Liver-specific HDAC3-/- knockout mice show severe hepatic steatosis that is associated with increased de novo lipogenesis and higher insulin sensitivity. Low Rev-Erbα levels reduce the capacity for HDAC3 to associate with the liver genome during the time of activity/feeding, which permits lipid lipogenesis. By contrast, elevated Rev-Erbα levels enhance HDAC3 recruitment to liver metabolic genes during the resting/fasting time, hindering lipid lipogenesis. Rev-Erbα can promote circadian signaling via the INSIG2–SREBP pathway and the liver X receptor (LXR), which participates in bile acid and lipid metabolism. Rev-Erbα-/- knockout mice show elevated VLDL levels and increased APOC-III expression in the liver. Both liver Rev-Erbα and Rev-Erbβ deficient mice display remarkable hepatic steatosis (Fathi Dizaji, 2018). Rev-Erb agonists have been used to treat the circadian lipidome. Beyond altering circadian behavioral patterns and clock gene expression, Rev-Erb treatment can induce weight loss and decrease the levels of plasma TGs, cholesterol and fatty acids in mice (Liu et al., 2008).
Rors
Rorα and its ligands can induce the expression of SOD2 and GPx1, reduce hepatic oxidative stress and inflammatory reactions, and can alleviate NASH in mice (Monnier et al., 2018). The Rorα mutant mouse (Rorαsg/sg; i.e., the staggerer mouse), though fed with increased quantities of food, has reduced levels of body fat, visibly smaller fat cells in the brown and white adipose tissues, and lower liver TGs levels, as well as decreased susceptibility to hepatic steatosis, contrasted by evidence of severe atherosclerosis (Solt et al., 2012). Moreover, a prior published study found that Rorα can enhance M2 polarization in liver macrophages, which protects hepatocytes from injury by secreting the anti-inflammatory and immuno-modulatory cytokine IL-10. Myeloid-specific knockout of Rorα enhances liver susceptibility to HFD-induced NASH. In addition, the Rorα activator induces an M2 polarity switch in Kuepfer cells and protects the liver progressing to NASH (Han et al., 2014, 2017).
Pers
Through histone H3 acetylation, the promoter regions of Per1 and Per2 undergo circadian fluctuation. Beyond binding core clock genes, Per2 can particularly regulate lipid metabolism by directly blocking PPARα, PPARγ, and Rev-Erbα transcription in white adipose and liver tissue. Whole body Per2-/- knockout mice showed decreased levels of TGs and non-esterified fatty acids. Compared to wild-type mice, white adipose tissue, and TG is reduced, while levels of saturated and mono-unsaturated very-long-chain fatty acids are elevated in Per2-/- knockout mice (Grimaldi et al., 2010). AMPK, which is a cell sensitive sensor of a low energetic and nutritional state, manages the degradation of expressed Per and CYR proteins. Both Per1 and Per2 deficient mice show impaired glucose tolerance. Per3 can regulate the clock of adipocyte precursor cells (APC), and can modulate adipogenesis in vivo. In addition, both Per3 and Bmal1 can directly regulate KLF15 expression. By contrast, deleting Per3 expression can promote adipogenesis.
Crys
Crys link the circadian clock, Janus kinase (JAK) and JAK-signal transducer and activator of transcription (STAT) signaling by regulating STAT5B phosphorylation. By their action on G protein-coupled receptor signaling, Crys can regulate hepatic gluconeogenesis, block cAMP accumulation and activate the transcription of gluconeogenic genes that are regulated by CREB (Zhang et al., 2010). Crys can also repress gene transcription that encode the glucocorticoid receptor and phosphoenol pyruvate carboxykinase. Glucocorticoids induce Per2 expression and affect glucose metabolism under hyperglycemic conditions. In addition, Cry deficient mice have smaller body and organ sizes. Cry1/Cry2 double knockout mice show abnormal TGs levels in the serum and liver, glucose intolerance and chronically elevated circulating corticosterone levels with augmented glucocorticoid-dependent transactivation in the liver. Furthermore, Cry1/Cry2 double knockout mice display an additional metabolic phenotype of salt-sensitive hypertension (Chaudhari et al., 2017). In diabetic mice, liver-specific overexpression of Cry1 can lower blood glucose and increase insulin sensitivity. HFD can accelerate the degradation of Cry1 and induce obesity-associated hyperglycemia. Autophagic pathways can degrade Cry1 and regulate the liver clock and glucose metabolism by modulating the expression of light chain3 (LC3)-interacting region (LIR) motifs (Toledo et al., 2018).
Nocturnin
Nocturnin (also known as Noct, formerly known as Ccrn4l) displays a vigorous circadian rhythm at both the mRNA and protein levels. Nocturnin encodes a deadenylase that is involved in the removal of polyA tails from mRNAs (Green et al., 2007). Nocturnin is not only directly involve in lipid absorption but also appears to be important in regulating unknown reduced lipid trafficking in the small intestine. The exact mechanism responsible for Nocturnin-mediated promotion of lipid secretion remains unknown. Nocturnin has also been confirmed to participate in adipogenesis, glucose homeostasis, osteogenesis and immune functions. Nocturnin in Per2-/- knockout mice have normal circadian mechanisms, although they display resistance to HFD-induced obesity and hepatic steatosis, which indicates that Nocturnin is downstream of the core circadian clock (Stubblefield et al., 2018). During the daily circadian cycle and acute nutritional challenges, Nocturnin in Per2-/- knockout mice have markedly elevated metabolism of cholesterol and TG (Stubblefield et al., 2012).
Decs
DECs, which encode the bHLH transcription factors, can regulate the circadian rhythm and metabolism. DEC1 in Per2-/- knockout mice show decreased lipid levels, reduced oxidative stress, and increased fibroblast growth factor 21 (FGF21) expression levels (Fujita et al., 2016). In DEC2 in Per2-/- knockout mice livers, phosphorylation of AMP-activated protein kinase (pAMPK) is remarkably increased. Insulin and glucose can induce DEC1 and DEC2 expression, while under conditions of depleted glucose, the expression of both DEC1 and DEC2 is decreased (Sato et al., 2016). Liver X receptor (LXR) can induce DEC1 expression by binding its promoter. Blocking phosphoinositide 3-kinase, protein kinase C, or the mammalian target of rapamycin can also inhibit insulin-induced DEC1 and DEC2 expression. Both DEC1 and DEC2 can regulate adipogenesis by repressing the transcription of PPARγ. Furthermore, over-expressing DEC1 suppresses adipocyte differentiation. It has also been found that DEC1 can interact with the DNA-bound CCAAT/enhancer binding protein and repress PPARγ expression (Park and Park, 2012).
The Circadian Clock Genes and the Progress of Nafld
The classic “two-hit hypothesis” proposes that the progression of NAFLD is initiated by hepatic fat accumulation (the first hit), and subsequent hits by a combination of oxidative stress, cytokines, bacterial endotoxin or stress at the level of the endoplasmic reticulum (ER) (the second hit). Recently, the coordinated interactions of autophagy and the host gut-microbiota has been identified as also representing additional biological insults (Nseir et al., 2014). We have already discussed above circadian clock genes that can induce hepatic steatosis. Moreover, studies have shown that circadian clock genes induce the progression of NAFLD (Froy, 2017).
Two investigations have suggested a protective effect of PER2 protein expression in acute liver injury and fibrosis. In the carbon tetrachloride-induced hepatitis model, Per2-/- knockout mice progressed to a more severe form of hepatic fibrosis with hepatic stellate cell activation (Chen et al., 2010). In the cholestatic hepatitis model, lack of PER2 expression could also cause more severe fibrotic injury and the accumulation of extra cellular matrix (Chen et al., 2013).
In the context of a progression of liver tumorigenesis, growing evidence suggests that circadian clock genes play key roles in cell cycle regulation, checkpoint determination, genomic stability, and DNA repair. In Cry1/2 double knockout and Clock mutant mice models, Wee1, which monitors cell-cycle progression from the G2 to M transition, exhibits a disrupted circadian expression (Stevenson, 2017). NPAS2 can be overexpressed in hepatocellular carcinoma (HCC) and induces cell survival by promoting cell proliferation and inhibiting mitochondria-related intrinsic apoptosis both in vitro and in vivo. Transcriptionally upregulating CDC25A phosphatase can stimulate NPAS2 expression. Moreover, Bmal1 can heterodimerize with NPAS2, bind to the E-box element in the promoter region of CDC25A and participate in NPAS2-mediated tumor cell survival in HCC (Yuan et al., 2017). Per-/- or Cry-/- knockout mice can form fewer but larger HCCs that are first detected at the age of 50 weeks, while Albcre; Bmal1fl/fl mice form increased numbers but smaller HCCs that are first detected at the age of 70 weeks. Chronic jet lag can increase both the numbers and size of HCCs in Per-/- or Cry-/- knockout mice and the size of HCCs in Albcre; Bmal1fl/fl mice. Both sexes of mutant mice have an increased risk of HCC, while males show a comparatively greater risk of developing HCC than do females (Kettner et al., 2016).
The Circadian Clock Genes and Nafld in Humans
Genomic variants in circadian clock genes are closely associated with hepatic steatosis, and predispose to NAFLD development. In humans, the clinical conditions of obesity, NAFLD and metabolic syndrome are associated with polymorphisms present in the Clock gene. Clock gene variants that include rs11932595 and rs6843722 show a close connection to NAFLD. A remarkable association is also found between the clinical or histologic spectrum of NAFLD and the presence of rs1554483, rs6843722, and rs6850524, and between the fibrosis score and the presence of rs1554483, rs6843722, and rs4864548.
Haplotypic association analyses show that Clock gene variant haplotype frequencies in NAFLD are quite different from those in controls (Sookoian et al., 2007). Clock rs3749474 is associated with total energy intake that might also be influenced by specific cytokine [e.g., monocyte chemotactic protein 1(MCP1), IL-6 and adiponectin] alterations (Ando et al., 2009). The Clock 3111T/C single-nucleotide polymorphism in women correlates with being overweight, the presence of circadian abnormalities and is characterized by “evening-type” subjects. Recognizing Clock type genotypes can help manage the causal roots of the metabolic problem (Bandín et al., 2013).
The Bmal2 rs7958822 genotype shows a significant association with type 2 diabetes (T2DM) among obese Japanese individuals (Yamaguchi et al., 2015). The mRNA expression patterns of Bmal1, Per1, Per2, and Per3 are 24 h rhythmic and lower expression levels in the peripheral leucocytes of T2DM patients, while the transcriptional expression patterns are inversely correlated with HbA1c levels. Clock gene (Per2, Bmal1, and Cry1) expression patterns are sex dependent in human adipose tissues derived from morbidly obese subjects. In addition, the three clock genes are remarkably and negatively associated with the level of total cholesterol and low density lipoprotein (LDL) levels. Per2 expression in the visceral depot is inversely associated with waist circumference (Gomez-Abellan et al., 2008; Garaulet et al., 2010). The methylation status of CpG sites located in clock genes (Clock, Bmal1, and Per2) is associated with obesity, metabolic syndrome and weight loss. The differential methylation of a variety of CpG sites in Clock and Per2 indicates the success of weight-loss, especially in the context of Clock CPGs 5–6 (Milagro et al., 2012). The methylation of several CpGs in the PER3 is indicating the development of childhood obesity (Samblas et al., 2018).
In the Nocturnin gene, rs9684900 is closely related to the body mass index (BMI) among a study of 1, 510 non-diabetic Chinese subjects in Taiwan. Nocturnin mRNA levels in human abdominal adipose tissues are also elevated in obese as compared with non-obese subjects (Chang et al., 2013). Here, we only list a few closely related circadian clock genes, and indeed, we recognize that there are many other genes related to lipid metabolism, which are also controlled by circadian oscillators, which can react to the molecular networks of the circadian clock (Li et al., 2017).
In addition to circadian gene polymorphisms, feeding/fasting cycles, feeding time, sleep deprivation, and sleep quality all have prominent effects on circadian clocks. They can disrupt the circadian rhythms and interfere the metabolism. The optimal feeding time and the optimal sleep duration may decrease the risk of metabolism syndrome and NAFLD (Shetty et al., 2018).
Conclusion
During the past several decades, an understanding of the circadian clock and metabolism has made significant advances. Moreover, environments that include cycles of rest and activity, feeding/fasting times and social stressors, have tremendous impacts on human physiology and metabolism (Angelousi et al., 2018). NAFLD, which is recognized as a lipid metabolic disease is closely connected with the circadian clock. Indeed, an increasing number of circadian rhythm studies have provided important insights that have enabled correlating expression of the circadian clock gene with metabolism in NAFLD within the holistic understanding of the involved molecular mechanisms. However, research investigators in the field of circadian metabolism are only beginning to understand the systems and mechanism, and clearly require additional experimentation to further broaden our comprehension of lipid metabolism in the liver (Wright et al., 2013; Sun et al., 2015). Clinical studies of circadian clock genes remain scarce in NAFLD patients. Hence, new insights that target key circadian clock genes with the intention of treating NAFLD may provide more effective strategies, pharmacological approaches, and improved guidance for specific nutrient components in the human diet.
Author Contributions
DS designed the research hypotheses and objectives and wrote the manuscript. JC and JY performed detailed searches of the literature. JW and YH analyzed the data. GZ programmed and coordinated the writing. ZB critically analyzed and revised the manuscript.
Funding
This study was generously supported by grants obtained from Shanghai Municipal Commission of Health and Family Planning; Key developing discipline (contract award number: 2015ZB0501).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ando, H., Takamura, T., Matsuzawa-Nagata, N., Shima, K. R., Eto, T., Misu, H., et al. (2009). Clock gene expression in peripheral leucocytes of patients with type 2 diabetes. Diabetologia 2, 329–335. doi: 10.1007/s00125-008-1194-6
Andronescu, C. I., Purcarea, M. R., and Babes, P. A. (2018). Nonalcoholic fatty liver disease: epidemiology, pathogenesis and therapeutic implications. J. Med. Life 11, 20–23.
Angelousi, A., Kassi, E., Nasiri-Ansari, N., Weickert, M. O., Randeva, H., and Kaltsas, G. (2018). Clock genes alterations and endocrine disorders. Eur. J. Clin. Invest. 48:e12927. doi: 10.1111/eci.12927
Asrih, M., and Jornayvaz, F. R. (2015). Metabolic syndrome and nonalcoholic fatty liver disease: is insulin resistance the link? Mol. Cell. Endocrinol. 418, 55–65. doi: 10.1016/j.mce.2015.02.018
Bandín, C., Martinez-Nicolas, A., Ordovás, J. M., Ros Lucas, J. A., Castell, P., Silvente, T., et al. (2013). Differences in circadian rhythmicity in Clock 3111T/C genetic variants in moderate obese women as assessed by thermometry, actimetry and body position. Int. J. Obes. 37, 1044–1050. doi: 10.1038/ijo.2012.180
Bass, J., and Takahashi, J. S. (2010). Circadian integration of metabolism and energetics. Science 330, 1349–1354. doi: 10.1126/science.1195027
Chang, Y. C., Chiu, Y. F., Liu, P. H., Hee, S. W., Chang, T. J., Jiang, Y. D., et al. (2013). Genetic variation in the NOC gene is associated with body mass index in Chinese subjects. PLoS One 26:e69622. doi: 10.1371/journal.pone.0069622
Chaudhari, A., Gupta, R., Patel, S., Velingkaar, N., and Kondratov, R. (2017). Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol. Biol. Cell 15, 834–842. doi: 10.1091/mbc.E16-08-0624
Chen, P., Han, Z., Yang, P., Zhu, L., Hua, Z., and Zhang, J. (2010). Loss of clock gene mPer2 promotes liver fibrosis induced by carbon tetrachloride. Hepatol. Res. 40, 1117–1127. doi: 10.1111/j.1872-034X.2010.00695.x
Chen, P., Kakan, X., Wang, S., Dong, W., Jia, A., Cai, C., et al. (2013). Deletion of clock gene Per2 exacerbates cholestatic liver injury and fibrosis in mice. Exp. Toxicol. Pathol. 65, 427–432. doi: 10.1016/j.etp.2011.12.007
Doi, M. (2012). Circadian clock-deficient mice as a tool for exploring disease etiology. Biol. Pharm. Bull. 35, 1385–1391. doi: 10.1248/bpb.b12-00364
Fathi Dizaji, B. (2018). The investigations of genetic determinants of the metabolic syndrome. Diabetes Metab. Syndr. 12, 783–789. doi: 10.1016/j.dsx.2018.04.009
Friedman, S. L., Neuschwander-Tetri, B. A., Rinella, M., and Sanyal, A. J. (2018). Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 24, 908–922. doi: 10.1038/s41591-018-0104-9
Friedrichs, M., Kolbe, I., Seemann, J., Tsang, A. H., Cherradi, L., Klein, J., et al. (2018). Circadian clock rhythms in different adipose tissue model systems. Chronobiol. Int. 1, 1–10. doi: 10.1080/07420528.2018.1494603
Froy, O. (2017). Circadian rhythms, nutrition and implications for longevity in urban environments. Proc. Nutr. Soc. 25, 1–7. doi: 10.1017/S0029665117003962
Fujita, Y., Makishima, M., and Bhawal, U. K. (2016). Differentiated embryo chondrocyte 1 (DEC1) is a novel negative regulator of hepatic fibroblast growth factor 21 (FGF21) in aging mice. Biochem. Biophys. Res. Commun. 469, 477–482. doi: 10.1016/j.bbrc.2015.12.045
Garaulet, M., Lee, Y. C., Shen, J., Parnell, L. D., Arnett, D. K., Tsai, M. Y., et al. (2010). Genetic variants in human Clock associate with total energy intake and cytokine sleep factors in overweight subjects (GOLDN population). Eur. J. Hum. Genet. 3, 364–369. doi: 10.1038/ejhg.2009.176
Gerber, A., Saini, C., Curie, T., Emmenegger, Y., Rando, G., Gosselin, P., et al. (2015). The systemic control of circadian gene expression. Diabetes. Obes. Metab. 17(Suppl. 1), 23–32. doi: 10.1111/dom.12512
Gnocchi, D., Pedrelli, M., Hurt-Camejo, E., and Parini, P. (2015). Lipids around the clock: focus on circadian rhythms and lipid metabolism. Biology 4, 104–132. doi: 10.3390/biology4010104
Gomez-Abellan, P., Hernandez-Morante, J. J., Lujan, J. A., Madrid, J. A., and Garaulet, M. (2008). Clock genes are implicated in the human metabolic syndrome. Int. J. Obes. 1, 121–128. doi: 10.1038/sj.ijo.0803689
Gooley, J. J. (2016). Circadian regulation of lipid metabolism. Proc. Nutr. Soc. 75, 440–450. doi: 10.1017/S0029665116000288
Grechez-Cassiau, A., Rayet, B., Guillaumond, F., Teboul, M., and Delaunay, F. (2008). The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283, 4535–4542. doi: 10.1074/jbc.M705576200
Green, C. B., Douris, N., Kojima, S., Strayer, C. A., Fogerty, J., Lourim, D., et al. (2007). Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 104, 9888–9893. doi: 10.1073/pnas.0702448104
Grimaldi, B., Bellet, M. M., Katada, S., Astarita, G., Hirayama, J., Amin, R. H., et al. (2010). PER2 controls lipid metabolism by direct regulation of PPARg. Cell Metab. 12, 509–520. doi: 10.1016/j.cmet.2010.10.005
Guillaumond, F., Dardente, H., Giguère, V., and Cermakian, I. (2005). Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20, 391–403. doi: 10.1177/0748730405277232
Guillaumond, F., Gréchez-Cassiau, A., Subramaniam, M., Brangolo, S., Peteri-Brünback, B., Staels, B., et al. (2010). Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol. Cell. Biol. 30, 3059–3070. doi: 10.1128/MCB.01141-09
Han, Y. H., Kim, H. J., Kim, E. J., Kim, K. S., Hong, S., Park, H. G., et al. (2014). RORα decreases oxidative stress through the induction of SOD2 and GPx1 expression and thereby protects against nonalcoholic steatohepatitis in mice. Antioxid. Redox Signal. 20, 2083–2094. doi: 10.1089/ars.2013.5655
Han, Y. H., Kim, H. J., Na, H., Nam, M. W., Kim, J. Y., Kim, J. S., et al. (2017). RORα Induces KLF4-mediated M2 polarization in the liver macrophages that protect against nonalcoholic steatohepatitis. Cell Rep. 5, 124–135. doi: 10.1016/j.celrep.2017.06.017
Hu, X., Huang, Y., Bao, Z., Wang, Y., Shi, D., Liu, F., et al. (2012). Prevalence and factors associated with nonalcoholic fatty liver disease in shanghai work-units. BMC Gastroenterol. 12:123. doi: 10.1186/1471-230X-12-123
Ipsen, D. H., Lykkesfeldt, J., and Tveden-Nyborg, P. (2018). Molecular mechanisms of hepatic lipid accumulation in non-alcoholic fatty liver disease. Cell. Mol. Life Sci. 75, 3313–3327. doi: 10.1007/s00018-018-2860-6
Johnston, J. D. (2014). Physiological links between circadian rhythms, metabolism and nutrition. Exp. Physiol. 99, 1133–1137. doi: 10.1113/expphysiol.2014.078295
Kawamoto, T., Noshiro, M., Sato, F., Maemura, K., Takeda, N., Nagai, R., et al. (2004). A novel autofeedback loop of Dec1 transcription involved in circadian rhythm regulation. Biochem. Biophys. Res. Commun. 313, 117–124. doi: 10.1016/j.bbrc.2003.11.099
Kettner, N. M., Voicu, H., Finegold, M. J., Coarfa, C., Sreekumar, A., Putluri, N., et al. (2016). Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 12, 909–924. doi: 10.1016/j.ccell.2016.10.007
Landgraf, D., Neumann, A. M., and Oster, H. (2017). Circadian clock-gastrointestinal peptide interaction in peripheral tissues and the brain. Best Pract. Res. Clin. Endocrinol. Metab. 31, 561–571. doi: 10.1016/j.beem.2017.10.007
Lee, S. M., Zhang, Y., Tsuchiya, H., Smalling, R., Jetten, A. M., and Wang, L. (2015). Small heterodimer partner/neuronal PAS domain protein 2 axis regulates the oscillation of liver lipid metabolism. Hepatology 61, 497–505. doi: 10.1002/hep.27437
Li, J., Lv, S., Qiu, X., Yu, J., Jiang, J., Jin, Y., et al. (2018). BMAL1 functions as a cAMP-responsive coactivator of HDAC5 to regulate hepatic gluconeogenesis. Protein Cell 9, 976–980. doi: 10.1007/s13238-018-0514-y
Li, S., Dou, X., Ning, H., Song, Q., Wei, W., Zhang, X., et al. (2017). Sirtuin 3 acts as a negative regulator of autophagy dictating hepatocyte susceptibility to lipotoxicity. Hepatology 66, 936–952. doi: 10.1002/hep.29229
Liu, A. C., Tran, H. G., Zhang, E. E., Priest, A. A., Welsh, D. K., and Kay, S. A. (2008). Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4:e1000023. doi: 10.1371/journal.pgen.1000023
Matsusue, K., Aibara, D., Hayafuchi, R., Matsuo, K., Takiguchi, S., Gonzalez, F. J., et al. (2014). Hepatic PPARγ and LXRα independently regulate lipid accumulation in the livers of genetically obese mice. FEBS Lett. 588, 2277–2281. doi: 10.1016/j.febslet.2014.05.012
Mazzoccoli, G., De Cosmo, S., and Mazza, T. (2018). The biological clock: a pivotal hub in non-alcoholic fatty liver disease pathogenesis. Front. Physiol. 15:193. doi: 10.3389/fphys.2018.00193
Mazzoccoli, G., Vinciguerra, M., Oben, J., Tarquini, R., and De Cosmo, S. (2014). Non-alcoholic fatty liver disease: the role of nuclear receptors and circadian rhythmicity. Liver Int. 34, 1133–1152. doi: 10.1111/liv.12534
McDearmon, E. L., Patel, K. N., Ko, C. H., Walisser, J. A., Schook, A. C., Chong, J. L., et al. (2006). Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science 24, 1304–1308. doi: 10.1126/science.1132430
Milagro, F. I., Gomez-Abellan, P., Campion, J., Martinez, J. A., Ordovas, J. M., and Garaulet, M. (2012). CLOCK, PER2 and BMAL1 DNA methylation: association with obesity and metabolic syndrome characteristics and monounsaturated fat intake. Chronobiol. Int. 9, 1180–1194. doi: 10.3109/07420528.2012.719967
Monnier, C., Auclair, M., Le Cam, G., Garcia, M. P., and Antoine, B. (2018). The nuclear retinoid-related orphan receptor RORα controls circadian thermogenic programming in white fat depots. Physiol. Rep. 6:13678. doi: 10.14814/phy2.13678
Nakahata, Y., Yasukawa, S., Khaidizar, F. D., Shimba, S., Matsui, T., and Bessho, Y. (2018). Bmal1-deficient mouse fibroblast cells do not provide premature cellular senescence in vitro. Chronobiol. Int. 35, 730–738. doi: 10.1080/07420528.2018.1430038
Nakao, T., Kohsaka, A., Otsuka, T., Thein, Z. L., Le, H. T., Waki, H., et al. (2018). Impact of heart-specific disruption of the circadian clock on systemic glucose metabolism in mice. Chronobiol. Int. 35, 499–510. doi: 10.1080/07420528.2017.1415922
Nseir, W., Hellou, E., and Assy, N. (2014). Role of diet and lifestyle changes in nonalcoholic fatty liver disease. World J. Gastroenterol. 28, 9338–9344. doi: 10.3748/wjg.v20.i28.9338
Oosterman, J. E., Kalsbeek, A., la Fleur, S. E., and Belsham, D. D. (2015). Impact of nutrients on circadian rhythmicity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 308, R337–R350. doi: 10.1152/ajpregu.00322.2014
Orozco-Solis, R., and Sassone-Corsi, P. (2014). Epigenetic control and the circadian clock: linking metabolism to neuronal responses. Neuroscience 264, 76–87. doi: 10.1016/j.neuroscience.2014.01.043
Pan, X., Bradfield, C. A., and Hussain, M. M. (2016). Global and hepatocyte-specific ablation of BMAL1 induces hyperlipidaemia and enhances atherosclerosis. Nat. Commun. 10:13011. doi: 10.1038/ncomms13011
Park, Y. K., and Park, H. (2012). Differentiated embryo chondrocyte 1 (DEC1) represses PPARgamma2 gene through interacting with CCAAT/enhancer binding protein beta (C/EBPbeta) Mol. Cells. 33, 575–581. doi: 10.1007/s10059-012-0002-9
Pérez-Mendoza, M., Rivera-Zavala, J. B., and Díaz-Muñoz, M. (2014). Daytime restricted feeding modifies the daily variations of liver gluconeogenesis: adaptations in biochemical and endocrine regulators. Chronobiol. Int. 31, 815–828. doi: 10.3109/07420528.2014.908898
Pérez-Mendoza, M., Rivera-Zavala, J. B., Rodríguez-Guadarrama, A. H., Montoya-Gomez, L. M., Carmona-Castro, A., Díaz-Muñoz, M., et al. (2018). Daily cycle in hepatic lipid metabolism in obese mice, Neotomodon alstoni: sex difference. Chronobiol. Int. 35, 643–657. doi: 10.1080/07420528.2018.1424178
Pettersson-Klein, A. T., Izadi, M., Ferreira, D. M. S., Cervenka, I., Correia, J. C., Martinez-Redondo, V., et al. (2018). Small molecule PGC-1α1 protein stabilizers induce adipocyte Ucp1 expression and uncoupled mitochondrial respiration. Mol. Metab. 9, 28–42. doi: 10.1016/j.molmet.2018.01.017
Reick, M., Garcia, J. A., Dudley, C., and McKnight, S. L. (2001). NPAS2: an analog of clock operative in the mammalian forebrain. Science 20, 506–509. doi: 10.1126/science.1060699
Reinke, H., and Asher, G. (2016). Circadian clock control of liver metabolic functions. Gastroenterology 150, 574–580. doi: 10.1053/j.gastro.2015.11.043
Sahar, S., and Sassone-Corsi, P. (2012). Regulation of metabolism: the circadian clock dictates the time. Trends Endocrinol. Metab. 23, 1–8. doi: 10.1016/j.tem.2011.10.005
Samblas, M., Milagro, F. I., Mansego, M. L., Marti, A., Martinez, J. A., and GENOI Members (2018). PTPRS and PER3 methylation levels are associated with childhood obesity: results from a genome-wide methylation analysis. Pediatr. Obes. 13, 149–158. doi: 10.1111/ijpo.12224
Sato, F., Kohsaka, A., Bhawal, U. K., and Muragaki, Y. (2018). Potential roles of Dec and Bmal1 genes in interconnecting circadian clock and energy metabolism. Int. J. Mol. Sci. 19:E781. doi: 10.3390/ijms19030781
Sato, F., Muragaki, Y., Kawamoto, T., Fujimoto, K., Kato, Y., and Zhang, Y. (2016). Rhythmic expression of DEC2 protein in vitro and in vivo. Biomed. Rep. 4, 704–710. doi: 10.3892/br.2016.656
Schleicher, J., Tokarski, C., Marbach, E., Matz-Soja, M., Zellmer, S., Gebhardt, R., et al. (2015). Zonation of hepatic fatty acid metabolism - The diversity of it regulation and the benefit of modeling. Biochim. Biophys. Acta 1851, 641–656. doi: 10.1016/j.bbalip.2015.02.004
Sherif, Z. A., Saeed, A., Ghavimi, S., Nouraie, S.-M., Laiyemo, A. O., Brim, H., et al. (2016). Global epidemiology of non-alcoholic fatty liver disease and perspectives on US minority populations. Dig. Dis. Sci. 61, 1214–1225. doi: 10.1007/s10620-016-4143-0
Shetty, A., Hsu, J. W., Manka, P. P., and Syn, W. K. (2018). Role of the circadian clock in the metabolic syndrome and nonalcoholic fatty liver disease. Dig. Dis. Sci. 63, 3187–3206. doi: 10.1007/s10620-018-5242-x
Shi, D., Zhan, X., Yu, X., Jia, M., Zhang, Y., Yao, J., et al. (2014). Inhibiting CB1 receptors improves lipogenesis in an in vitro non-alcoholic fatty liver disease model. Lipids Health Dis. 13, 173–179. doi: 10.1186/1476-511X-13-173
Soeda, J., Cordero, P., and Li, J. (2017). Hepatic rhythmicity of endoplasmic reticulum stress is disrupted in perinatal and adult mice models of high-fat diet-induced obesity. Int. J. Food Sci. Nutr. 68, 455–466. doi: 10.1080/09637486.2016.1261086
Solt, L. A., Wang, Y., Banerjee, S., Hughes, T., Kojetin, D. J., Lundasen, T., et al. (2012). Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 29, 62–68. doi: 10.1038/nature11030
Sookoian, S., Castano, G., Gemma, C., Gianotti, T. F., and Pirola, C. J. (2007). Common genetic variations in Clock transcription factor are associated with nonalcoholic fatty liver disease. World J. Gastroenterol. 13, 4242–4248. doi: 10.3748/wjg.v13.i31.4242
Sookoian, S., Gemma, C., Gianotti, T. F., Burgueño, A., Castaño, G., and Pirola, C. J. (2008). Genetic variants of clock transcription factor are associated with individual susceptibility to obesity. Am. J. Clin. Nutr. 87, 1606–1615. doi: 10.1093/ajcn/87.6.1606
Stern, C., and Castera, L. (2017). Non-invasive diagnosis of hepatic steatosis. Hepatol. Int. 11, 70–78. doi: 10.1007/s12072-016-9772-z
Stevenson, T. J. (2017). Epigenetic regulation of biological rhythms: an evolutionary ancient molecular timer. Trends Genet. 34, 90–100. doi: 10.1016/j.tig.2017.11.003
Stubblefield, J. J., Gao, P., Kilaru, G., Mukadam, B., Terrien, J., and Green, C. B. (2018). Temporal control of metabolic amplitude by nocturnin. Cell Rep. 30, 1225–1235. doi: 10.1016/j.celrep.2018.01.011
Stubblefield, J. J., Terrien, J., and Green, C. B. (2012). Nocturnin: at the crossroads of clocks and metabolism. Trends Endocrinol. Metab. 23, 326–333. doi: 10.1016/j.tem.2012.03.007
Sun, C., Liu, X., Yi, Z., Xiao, X., Yang, M., Hu, G., et al. (2015). Genome-wide analysis of long noncoding RNA expression profiles in patients with non-alcoholic fatty liver disease. IUBMB Life 67, 847–852. doi: 10.1002/iub.1442
Toledo, M., Batista-Gonzalez, A., Merheb, E., Aoun, M. L., Tarabra, E., Feng, D., et al. (2018). Autophagy Regulates the Liver Clock and Glucose Metabolism by Degrading CRY1. Cell Metab. 11, S1550–S4131. doi: 10.1016/j.cmet.2018.05.023 [Epub ahead of print].
Tong, X., and Yin, L. (2013). Circadian rhythms in liver physiology and liver diseases. Compr. Physiol. 3, 917–940. doi: 10.1002/cphy.c120017
Wong, V. W., Chan, W. K., Chitturi, S., Chawla, Y., Dan, Y. Y., Duseja, A., et al. (2018). Asia-pacific working party on non-alcoholic fatty liver disease guidelines 2017-Part 1: definition, risk factors and assessment. Gastroenterol. Hepatol. 33, 70–85. doi: 10.1111/jgh.13857
Wright, K. P. Jr., McHill, A. W., Birks, B. R., Griffin, B. R., Rusterholz, T., and Chinoy, E. D. (2013). Entrainment of the human circadian clock to the natural light-dark cycle. Curr. Biol. 23, 1554–1558. doi: 10.1016/j.cub.2013.06.039
Yamaguchi, M., Uemura, H., Arisawa, K., Katsuura-Kamano, S., Hamajima, N., Hishida, A., et al. (2015). Association between brain-muscle-ARNT-like protein-2 (BMAL2) gene polymorphism and type 2 diabetes mellitus in obese Japanese individuals: a cross-sectional analysis of the Japan multi- institutional collaborative cohort study. Diabetes Res. Clin. Pract. 3, 301–308. doi: 10.1016/j.diabres.2015.10.009
Yamajuku, D., Shibata, Y., Kitazawa, M., Katakura, T., Urata, H., Kojima, T., et al. (2011). Cellular DBP and E4BP4 proteins are critical for determining the period length of the circadian oscillator. FEBS Lett. 21, 2217–2222. doi: 10.1016/j.febslet.2011.05.038
Yoo, S. H., Kojima, S., Shimomura, K., Koike, N., Buhr, E. D., Furukawa, T., et al. (2017). Period2 3’-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. Proc. Natl. Acad. Sci. U.S.A. 17, E8855–E8864. doi: 10.1073/pnas.1706611114
Yuan, P., Li, J., Zhou, F., Huang, Q., Zhang, J., Guo, X., et al. (2017). NPAS2 promotes cell survival of hepatocellular carcinoma by transactivating CDC25A. Cell Death Dis. 8:e2704. doi: 10.1038/cddis.2017.131
Keywords: circadian rhythm, circadian clock, non-alcoholic fatty liver disease (NAFLD), circadian clock gene, metabolism
Citation: Shi D, Chen J, Wang J, Yao J, Huang Y, Zhang G and Bao Z (2019) Circadian Clock Genes in the Metabolism of Non-alcoholic Fatty Liver Disease. Front. Physiol. 10:423. doi: 10.3389/fphys.2019.00423
Received: 12 January 2019; Accepted: 27 March 2019;
Published: 08 May 2019.
Edited by:
Sara Montagnese, University of Padova, ItalyReviewed by:
Elizabeth Anne Schroder, University of Kentucky, United StatesHenrik Oster, Universität zu Lübeck, Germany
Copyright © 2019 Shi, Chen, Wang, Yao, Huang, Zhang and Bao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Bao, eGlueWk4NjgxQHNpbmEuY29t
 Dongmei Shi
Dongmei Shi Jie Chen
Jie Chen Jiaofeng Wang
Jiaofeng Wang Jianfeng Yao
Jianfeng Yao Yiqin Huang
Yiqin Huang Gansheng Zhang
Gansheng Zhang Zhijun Bao1,2*
Zhijun Bao1,2*