- 1Laboratory of Epigenetics, ICS Maugeri S.p.A., Pavia, Italy
- 2Molecular Cardiology Laboratory, IRCCS Policlinico San Donato, Milan, Italy
- 3Institute of Cell Biology and Neurobiology, National Research Council, Università Cattolica di Roma, Rome, Italy
Non-coding RNAs are pivotal for many cellular functions, such as splicing, gene regulation, chromosome structure, and hormone-like activity. Here, we will report about the biology and the general molecular mechanisms associated with long non-coding RNAs (lncRNAs), a class of >200 nucleotides-long ribonucleic acid sequences, and their role in chronic non-transmissible diseases. In particular, we will summarize knowledge about some of the best-characterized lncRNAs, such as H19 and MALAT1, and how they regulate carbohydrate and lipid metabolism as well as protein synthesis and degradation. Evidence is discussed about how lncRNAs expression might affect cellular and organismal metabolism and whether their modulation could provide ground for the development of innovative treatments.
Introduction
Non-coding RNAs (ncRNAs) are a class of ribonucleic acid sequences which do not carry any information for protein translation, at least not for long peptides, but are demonstrably involved in gene and protein regulation (Monticelli et al., 2005), RNA splicing (Kishore and Stamm, 2006), chromosome structure (Park et al., 2002) or, traveling through circulating blood, they play some hormone-like activities (Knoll et al., 2015). Their length principally classifies ncRNAs. Conventionally, 200 nucleotides (nts) are the cut-off length determining to which group they belong to: long non-coding RNAs (lncRNAs) are longer than 200 nts, while small ncRNAs (sncRNA) are much shorter (Pedersen et al., 2006). Interestingly, although not widely used, an alternate classification has been proposed based on ncRNAs biological function, such as gene silencing or transcriptional activation, which might result much more informative and helpful for translational and clinical investigations (Amaral et al., 2011).
Regarding lncRNAs, they include: imprinted lncRNAs, which control expression of imprinted genes (Sleutels et al., 2002); disease-associated lncRNAs which are particularly abundant in pathophysiological conditions (Matouk et al., 2007; Yoshimizu et al., 2008); pathogen-induced lncRNAs which are produced and modulated by exogenous microorganisms (Zhou et al., 2016); bifunctional RNAs that can have more than one role and can also be translated into proteins (Hashimoto et al., 2009); miRNA sponges that interfere with the inhibiting activity of miRNAs (Chen L. et al., 2018; Zhao et al., 2018). Despite a growing body of knowledge about lncRNA regulation and function, it must be said that most lncRNAs identified so far are yet “unknown” being physiologically transcribed but with a mostly undetermined function.
Evidence supports the presence of circular RNAs (circRNAs) which are 500 nucleotides on average (Salzman et al., 2012). Many processes can produce circRNAs; an example is the co-transcription of pre-mRNAs (Zhang et al., 2016) or post-transcriptional back splicing in which the exon excised contains lariat that is not connected to pre-mRNA (Suzuki et al., 2006; Barrett et al., 2015). These differences in circRNA biogenesis might be significant for their biological and pathophysiological properties. Co-transcribed circRNAs, modulate linear mRNA directly in cis (Ashwal-Fluss et al., 2014), while post-transcriptionally generated circRNAs do not (Barrett and Salzman, 2016).
Another class of lncRNAs is the large intergenic ncRNAs (lincRNAs) which, at least in part, controls gene expression recruiting enzymes involved in histone modifications (Khalil et al., 2009). It has been shown that dysregulations of lincRNAs are associated with epigenetic modifications occurring in cancer progression and metastasis (Gupta et al., 2010).
However, lncRNAs share many of their regulatory features with normal coding messenger RNAs (mRNA). Similar to mRNAs lncRNAs are transcribed by RNA Polymerase type II, indeed lncRNA promoters are enriched in histone H3 lysine-4 tri-methylated (H3K4me3) while, along the gene body, tri-methylation is enriched on histone H3 lysine-36 (H3K36me3) residues, a feature that allows RNA Polymerase II to start transcription (Guttman et al., 2009). Besides, lncRNAs undergo post-transcriptional modifications such as splicing, polyadenylation and 5′-capping (Sas-Chen et al., 2017) and their 3′UTR is organized similarly to that of mRNAs (Niazi and Valadkhan, 2012). Nevertheless, lncRNAs have also unique traits: (i) the nucleus is the subcellular compartment in which they are particularly abundant (Cabili et al., 2015); (ii) surprisingly enough, they may have small open reading frames (ORFs) which are sometimes converted in very short peptides of still unknown function (Ma et al., 2013; Aboudehen et al., 2018); (iii) although their abundance is physiologically lower than that of mRNAs, lncRNAs are more cell- or tissue-specific than coding mRNAs (Zimmer et al., 1994).
This review is focused on the pathophysiological relevance of lncRNA associated to, cancer and cardiovascular diseases with special attention paid to those processes arising from dysmetabolic conditions. Also, particular emphasis is given to those lncRNAs for which potential therapeutic and diagnostic applications have been envisaged. A broad description of ncRNA role across a larger number of non-communicable diseases has been provided elsewhere (Ma et al., 2013; Greco et al., 2015; Barrett and Salzman, 2016; Viereck and Thum, 2017; Rafiee et al., 2018) and is beyond the scope of this article.
LncRNAs and Non-Communicable Diseases: An Overview
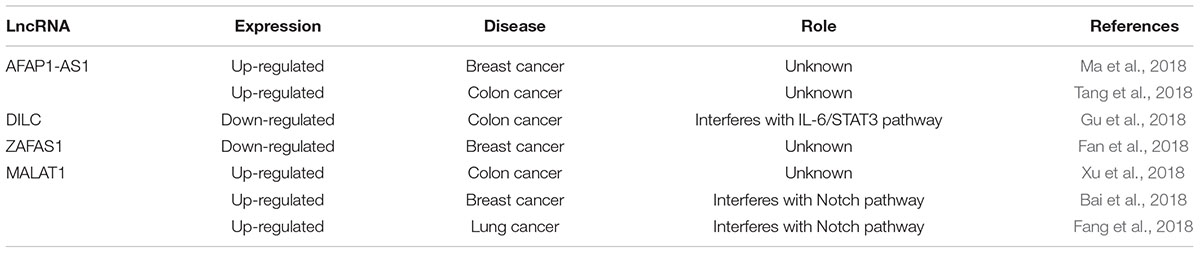
Cancer is probably the most common non-communicable disease in which important prognostic/therapeutic applications are envisaged for lncRNAs. An example of this perspective is given by the actin filament associated protein one antisense RNA1 (AFAP1-AS1) which is up-regulated in breast cancer. Its expression levels correlate well with poor prognosis (Ma et al., 2018). Similarly, high levels of AFAP1-AS1, in colon cancer, have been associated with malignancy (Tang et al., 2018), suggesting an oncogenic role for this lncRNA although the molecular mechanism associated with this property is unclear. On the opposite, the lncRNA-downregulated in liver cancer stem cells (DILC) is down-regulated in colorectal cancer where it has a tumor suppressor function. DILC interferes with the IL-6/STAT3 pathway inhibiting cell growth and metastasis (Gu et al., 2018). Another lncRNA, which has an anticancer function, is named zinc finger antisense 1 (ZFAS1). ZFAS1 is down-regulated in breast cancer, and increasing ZFAS1 level may cause cell cycle arrest and apoptosis (Fan et al., 2018). MALAT1 is probably the best-characterized lncRNAs in cancer biology. In fact, in colorectal cancer high levels of MALAT1 have been associated with increased proliferation and migration (Xu et al., 2018), whereas in oesophageal cancer, it promotes epithelial-to-mesenchymal transition through the modulation of Notch pathway (Chen M. et al., 2018). In addition, high levels of MALAT1 confer resistance to pharmacological treatment, such as cisplatin in lung cancer (Fang et al., 2018). Finally, in ovarian cancer MALAT1 interferes with the Notch pathway conferring chemo-resistance (Bai et al., 2018) (see Table 1).
Nowadays, obstacles in treating cancer via modulation of lncRNA expression apparently arise from cancer heterogeneity and genomic instability. It remains, however, an exciting challenge. A more extensive overview of the role of lncRNAs in cancer is out of the scope of this article while it has been the object of more extensive discussion in other recent reviews (Rafiee et al., 2018; Sanchez Calle et al., 2018).
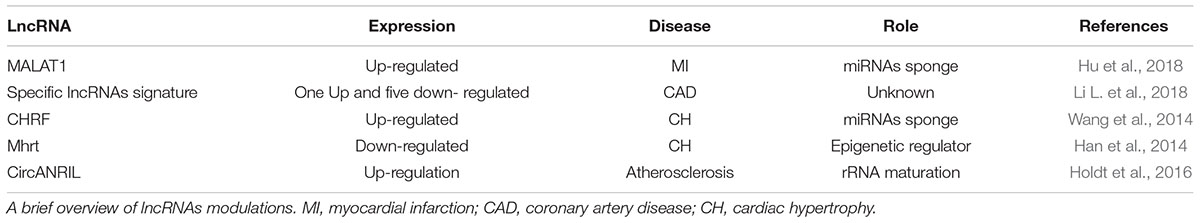
Although probably less characterized than in cancer, the RNA level of different classes of ncRNAs changes significantly in CVD, including miRNAs, circRNAs, and lncRNAs (Viereck and Thum, 2017). Interestingly, a specific lncRNAs signature, composed of six lncRNAs: uc010yfd.1, RNA147299| p0403_imsncRNA819, ENST00000444488.1, ASO3973, ENST00000602558.1, and ENST00000561165.1, has been recently identified in myocardial infarction (MI) and proposed as a sensitive biomarker of coronary disease (Li L. et al., 2018). However, in this pathophysiological condition, also the cancer-associated lncRNA MALAT1 has been proposed to be of potential relevance, being highly expressed in the peripheral blood of infarcted mice (Hu et al., 2018) as a possible consequence of endogenous activation of the hypoxia pathway (Choudhry and Mole, 2016). MALAT1 might act as a molecular sponge for microRNAs, including miR-320, promoting apoptosis in infarcted cardiomyocytes (Hu et al., 2018). Cardiac hypertrophy is another situation in which lncRNAs may play a role. High levels of the cardiac-hypertrophy-related factor (CHRF) lncRNA have been found associated with this pathophysiological condition. This lncRNA sequesters miR-489, known to inhibit the mRNA encoding for the myeloid differentiation primary response gene product 88 (Myd88), increasing cardiomyocyte volume (Wang et al., 2014). On the opposite, the low levels of myosin heavy chain (MHC) associated RNA transcript lncRNAs (Mhrt) have been found associated with cardiac hypertrophy. Indeed, Mhrt binds Brg1, an ATP-dependent chromatin remodeling factor, inhibiting Brg1/DNA interaction (Han et al., 2014). As a result, pro-hypertrophic genes are not efficiently transcribed (Han et al., 2014).
In most of the cases, the molecular mechanisms associated with lncRNAs in cardiovascular accidents, heart failure, atherosclerosis, and ischemia is still unknown (Thum and Condorelli, 2015; Chatterjee et al., 2017). It must be said, however, that the multi-layered regulation of lncRNAs expression and function does not facilitate gathering mechanistic insights. For example, the long non-coding/circRNA named ANRIL has a role in CVD and diabetes. In this case, disease conditions have been associated with some single-nucleotide polymorphisms (SNPs) present in its genomic sequence at 9p21 region which is close to CDKN2A and CDKN2B tumor suppressor genes (Helgadottir et al., 2007; Wei et al., 2015). Specifically, in patients with MI, it has been found a significant presence of ANRIL genomic locus variants such as rs10757278 (Helgadottir et al., 2007), rs10965215 and rs10738605 (Cheng et al., 2017), highlighting the concept that MI predisposition might be associated with the genetic variation of an epigenetic non-coding modulator. Moreover, with this evidence, in diabetic patients, other ANRIL SNPs have been involved in cardiovascular illnesses such as coronary artery disease (CAD). T2D patients that carry SNPs rs2891168 have a higher risk to develop CAD compared to controls (Broadbent et al., 2008). Consistently, rs10757274 and rs1333042 SNPs are common in Iranians which suffers from CAD (Mafi Golchin et al., 2017). Also, increased levels of ANRIL have been observed in left ventricle biopsies derived from ischemic heart failure patients (Greco et al., 2016). However, ANRIL exists also in circular form (circANRIL). Recently, circANRIL has been found involved in atherosclerosis because of its function as ribosomal RNA maturation modulator (Holdt et al., 2016). In fact, in vascular smooth muscle cells and macrophages, circANRIL binds pescadillo homolog 1 (PES1), a 60S-preribosomal assembly factor (Holdt et al., 2016). As a result, cells undergo growth arrest and apoptosis a condition that might reduce atherosclerotic plaques formation by culling hyper-proliferating cell types in atherosclerotic plaques (Holdt et al., 2016). Indeed, carriers of a coronary artery disease-protective haplotype displayed increased expression of circANRIL. Importantly, the effect of circANRIL was inverse to that of its linear counterpart (linANRIL), which is downregulated in patients carrying the 9p21 protective genotype (Holdt et al., 2013). Accordingly, the highest circANRIL/linANRIL ratio was found in patients free of coronary artery disease.
Additional data obtained in a rat model of atherosclerosis stress the importance of the techniques used to modulate circANRIL in vivo and of the targeted cells (endothelial cells vs. smooth muscle and macrophages). Indeed, in this rat model, reduced circANRIL in vascular endothelial cells has been correlated with decreased inflammatory factors expression, reduced coronary atherosclerosis, and apoptosis, as well as lower endothelial damage (Song et al., 2017). The lncRNA H19 has been demonstrated to exacerbate cardiac hypertrophy (Liu L. et al., 2016) and atherosclerosis (Pan, 2017; Zhang et al., 2017; Bitarafan et al., 2019). In mouse model has been shown that miR-675, a target of H19, down-regulates CAMKIIδ expression (Liu L. et al., 2016) which is a kinase that induces cardiac hypertrophy (Anderson et al., 2011). The overexpression of H19 leads the transduction of CAMKIIδ increasing cardiomyocyte volume (Liu L. et al., 2016). High levels of H19 have been found in serum patients with atherosclerosis (Pan, 2017), indeed, in mouse vascular smooth muscle cells, upon up-regulation of H19, low level of caspase 3 has been found while proliferation is improved probably due to the activation of MAP-ERK pathway linked to inflammation (Pan, 2017). Associated to H19, in atherosclerotic patients high plasma levels of long intergenic ncRNA predicting cardiac remodeling (LIPCAR) has been observed (Zhang et al., 2017) suggesting these two lncRNAs as novel biomarkers of CVD. However, LIPCAR’s role in the development of the plaque remains unclear. Moreover, the up-regulation of H19 have been reported in peripheral blood mononuclear cells (PBMCs) of coronary artery disease patients but the role of this lncRNA and the molecular mechanisms altered in these cells are still unknown (Bitarafan et al., 2019) (see Table 2).
LncRNAs and Carbohydrate Metabolism
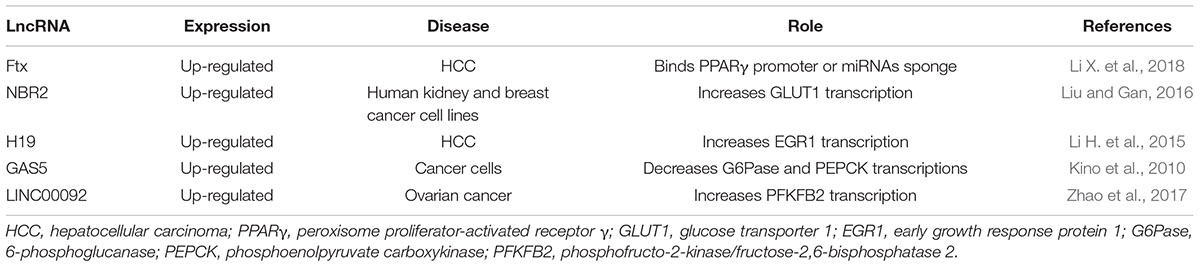
Most of our knowledge about the role of lncRNAs in carbohydrate control comes from studies aimed at understanding cancer energy production. Alteration of carbohydrate metabolism, in fact, is frequent in cancer because transformed cells have an enhanced glucose uptake and predominantly utilize glycolysis in order to provide to their energy needs and growth requirements, a phenomenon notoriously known as Warburg effect (Warburg et al., 1927; Bose and Le, 2018). LncRNA Ftx has been described as a regulator of peroxisome proliferator-activated receptor γ (PPARγ) pathway in hepatocellular carcinoma (HCC) (Li X. et al., 2018). Ftx is involved in lipid (Tsukahara et al., 2017) and carbohydrate metabolism (Janani and Ranjitha Kumari, 2015) and, in HCC, the level of lncRNA Ftx is significantly higher than healthy controls, enhancing glucose consumption, cell proliferation, migration and invasion (Li X. et al., 2018). Recently, analysis of lncRNA Ftx sequence through bioinformatics tools revealed that Ftx overlaps by 118 nucleotides to PPARγ gene and promoter region, suggesting that lncRNA Ftx might bind directly to PPARγ promoter enhancing transcription or acting as a PPARγ miRNA sponge (Li X. et al., 2018). Indeed, in HCC, the PPARγ pathway is more active than in non-transformed cells (or in normal tissue) (Li X. et al., 2018). This evidence suggests a mechanism of action in which a lncRNA can influence gene transcription by direct binding to a promoter region. Thus, targeting PPARγ-dependent pathway by lncRNA Ftx silencing may provide a novel approach for HCC therapy.
NBR2, a lncRNA up-regulated in cancer, is known to activate AMPK in human kidney and breast cancer cell lines, where the uptake of glucose increases in association to the activation of GLUT1 transcription (Liu X. et al., 2016). Consequently, in those cells, high intracellular glucose levels accumulate, providing a reservoir for the energy and biomass necessities of the growing tumor (Liu and Gan, 2016). In this study, in cells treated with phenformin, a drug known to up-regulate transcription of the GLUT family, NBR2 has been observed to be significantly increased. Interestingly, down-regulating NBR2, only GLUT1 resulted down-regulated, suggesting specificity for NBR2 toward a specific GLUT family member (Liu and Gan, 2016). This finding suggests that NBR2 is a phenformin-induced lncRNA that could be proposed as a target for novel anti-tumor drug design and development (Liu and Gan, 2016).
LncRNAs can modulate glycolytic enzymes such as pyruvate kinase isoform M2 (PKM2) which is highly expressed in cancer cells. In fact, in hepatocarcinoma, lncRNA H19 has been found significantly overexpressed (Li H. et al., 2015) a phenomenon paralleled by the up-regulation of PKM2 via transcriptional activation of the early growth response protein 1 (EGR1) (Li H. et al., 2015). Specifically, miR-675, embedded in the first exon of H19, is a repressor of HP1α that regulates EGR1, allowing the transcription of PKM2 (Li H. et al., 2015). Interestingly, PKM2 seems not only involved in glucose metabolism, but it exists into the nucleus where acts as a kinase for some transcription factors, regulating the expression of proteins involved in hypoxia resistance, proliferation, glucose uptake, and mitosis progression (Gao et al., 2012). These findings suggest that PKM2 level could be modulated through H19 lncRNA which modulation might represent a potential new anticancer strategy. In the diabetic mouse model, a recent study demonstrates the involvement of H19 in hepatocytes where it acts on FoxO1 regulation (Goyal et al., 2019). The scientists observed that in hepatic cells, FoxO1 promoter is occupied by p53 instead of H19 binding proteins which have a binding site of FoxO1 promoter sequence (Goyal et al., 2019). P53 interaction on promoter triggers the transcription of FoxO1, and as a result the activation of gluconeogenesis pathway has been reported (Goyal et al., 2019).
GAS5 is another essential tumor-associated lncRNA that, in consequence of its repression activity on the adrenocorticotropic hormone receptor, inhibits the expression of 6-phosphoglucanase (G6Pase) and phosphoenolpyruvate carboxykinase (PEPCK), resulting in a reduction of glucose storage and enhancement of glucose catabolism in cancer cells (Kino et al., 2010).
In ovarian cancer, LINC00092 lncRNA regulates phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 (PFKFB2), enhancing its expression (Zhao et al., 2017). LINC00092 expression is modulated in the response to CXCL14, a chemokine secreted by cancer-associated fibroblasts (CAFs) linked to metastasis (Zhao et al., 2017). The knockdown of LINC00092 decreases PFKFB2 expression and glucose consumption, significantly reducing metastasis. Also, the PFKFB2-induced glycolytic cancer cell phenotype seems critical for the maintenance of CXCL14 secretion from CAFs, suggesting for LINC00092 a regulatory feedback loop between cancer cells and CAFs that is important for the maintenance of the tumor microenvironment (Zhao et al., 2017).
Of particular interest for this review, in heart and skeletal muscle, the lncRNA LIN00116 has been found to encode a transmembrane microprotein, named mitoregulin, that localizes into the inner mitochondrial membrane. In consequence of the unusual coding activity of LIN00116, respiration during glucose degradation is improved, and ROS formation reduced (Stein et al., 2018) (see Table 3 and Figure 1).
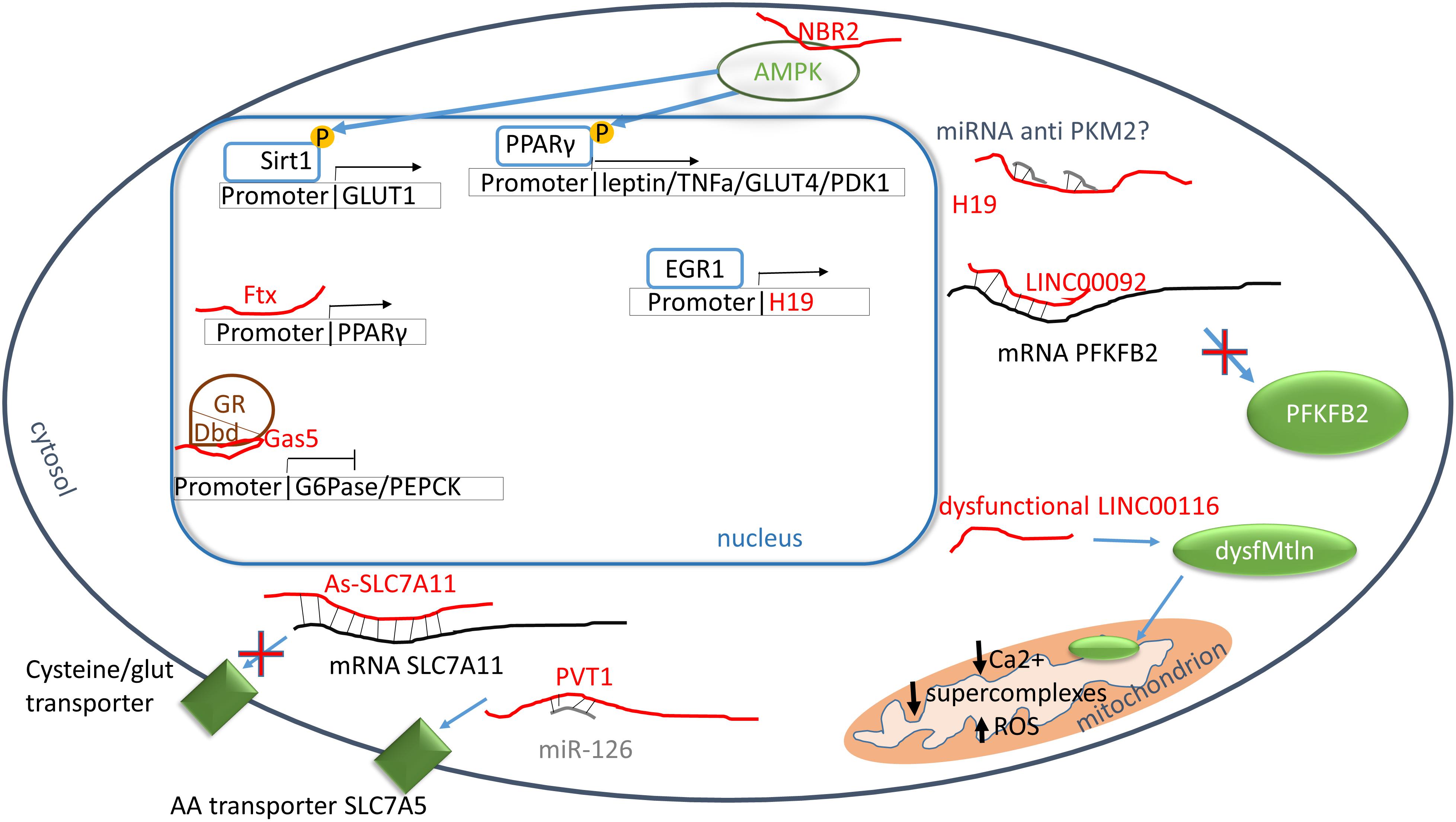
Figure 1. Carbohydrate and protein metabolisms. Red: lncRNAs; green: proteins; blue: transcriptor factors; brown: nuclear receptors. PPARγ promoter binds directly Ftx increasing the transcription. PPARγ is phosphorylated through AMPK pathway and it enhances the transcription of leptin, tumor necrosis factor α (TNFα), glucose channel 4 (GLUT4) and pyruvate dehydrogenase kinase 1 (PDK1). In turn, AMPK activity is enhanced by the binding to NBR2. H19 influences the transcription of pyruvate kinase M2 probably by the inhibition of PKM2 miRNAs, however, more studies are necessary to elucidate the molecular mechanism. The glucocorticoid receptor (GR) is blocked by Gas5 which interacts to GR DNA binding domain resulting in an inhibition of 6-phosphoglucanase (G6Pase) and phosphoenolpyruvate carboxykinase (PFKFB2). LINC00092 stabilizes phosphofructo-2-kinase/fructose-2,6-phosphatase 2 (PFKFB2) mRNA allowing its synthesis. LINC00116 encodes the microprotein mitoregulin (Mtln) which acts into mitochondrial inner membrane increasing the formation of supercomplexes, calcium intake and reducing ROS formation. The alteration of LINC00116 generate dysfunctional of low levels of Mtln. As-SLC7A11 binds SLC7A11 mRNA inhibiting the translation of cysteine/glutamine transporter, meanwhile plasmacytoma variant translocation 1 (PVT1) enhance the synthesis of AA transporter through the binding of miR-126 which is a suppressor of SCL7A5 mRNA.
LncRNAs and Their Role in Amino Acid Metabolism
A recent study demonstrates the regulation of cysteine/glutamate transporter by a lncRNA named As-SLC7A11, which is markedly reduced in epithelial ovarian cancer (Yuan et al., 2017). As a consequence of As-SLC7A11 overexpression, epithelial ovarian cancer cells were not able to use glutamate as AAs precursor or an energy source, inhibiting tumor growth (Yuan et al., 2017). Another lncRNA, the plasmacytoma variant translocation 1 (PVT1) can regulate the intake of AAs through the modulation of the miR-126/SLC7A5 axis (Li H. et al., 2018). In lung cancer, PVT1 level is increased, stimulating the uptake of AAs in cancer cells (Li H. et al., 2018). Moreover, PVT1 is induced by oxidative stress in endothelial cells, with a p53-dependent manner. Accordingly, PVT is also induced in senescent endothelial cells and decreased in PBMCs of long-living individuals (Fuschi et al., 2017) (see Table 4 and Figure 1).
LncRNAs and Their Role in Lipid Metabolism
Alteration in blood lipid level is one of the most relevant risk factor in cardiovascular disease (CVD). It is well known in fact that high amount of LDL cholesterol and apolipoprotein B are associated with atherosclerotic plaques formation (Linton et al., 2000). The dangerousness of the plaque is correlated to the ease of forming vessel obstruction that cause hypoxia possibly leading to necrosis in downstream tissues causing MI or stroke (Linton et al., 2000). To prevent plaque formation, patients are often treated with drugs that inhibit the synthesis of cholesterol such as statins or more recently PCSK9 inhibitors. Cholesterol, fatty acids, and phospholipids are among the most abundant cellular lipids and their synthesis and uptake are strongly regulated, in physiological conditions, by transcription factors named sterol regulatory element binding proteins (SREBPs) (Parraga et al., 1998; Horton, 2002) which exist in three different isoforms: SREBP-1a; SREBP-1c and SREBP2 (Horton, 2002). H19 is one of the lncRNAs that modulates hepatic lipids homeostasis. It directly interacts with the polypyrimidine tract-binding protein 1 (PTBP1) enhancing its binding activity to SREBP1 that, in the presence of a high fat/high sucrose diet, translocates into the nucleus (Liu et al., 2018). Moreover, PTBP1 increases SREBP1 mRNA stability while stimulating SREBP1 cleavage (Liu et al., 2018). As a result, in hepatocytes the nuclear accumulation of SREBP1, upon H19 overexpression, seems mimicking the consequences of a fed state (Liu et al., 2018).
Other study revealed a modulation of SREBP-1c levels through the action of the lncRNA HCV regulated 1 (lncHR1) (Li D. et al., 2018). Here, in a mouse model of triglyceride accumulation, high levels of lncHR1 decreased phosphorylation and activation of the PDK1/ATK/FoxO1 pathway, inhibiting SREBP-1c expression (Li D. et al., 2018). As for many other lncRNAs, the molecular mechanism by which lncHR1 regulates SREBP-1c remains to be clarified.
MALAT1 itself can regulate lipid metabolism. In hepatic cells, recent studies demonstrated its involvement in the regulation of SREBP-1c level (Yan et al., 2016). In liver and HepG2 cells, in fact, after palmitate uptake, MALAT1 expression increase enhancing the nuclear localization of SREBP-1c that transcribes genes involved in lipid metabolism such as stearoyl CoA desaturase 1, fatty acid synthase, acetyl-CoA carboxylase 1 and ATP citrate synthase (Yan et al., 2016). Mechanistically, MALAT1 directly binds SREBP-1c, which is stabilized (Yan et al., 2016). As a result, in hepatocytes, the presence of a sufficient amount of MALAT1 decreases lipid accumulation (Yan et al., 2016).
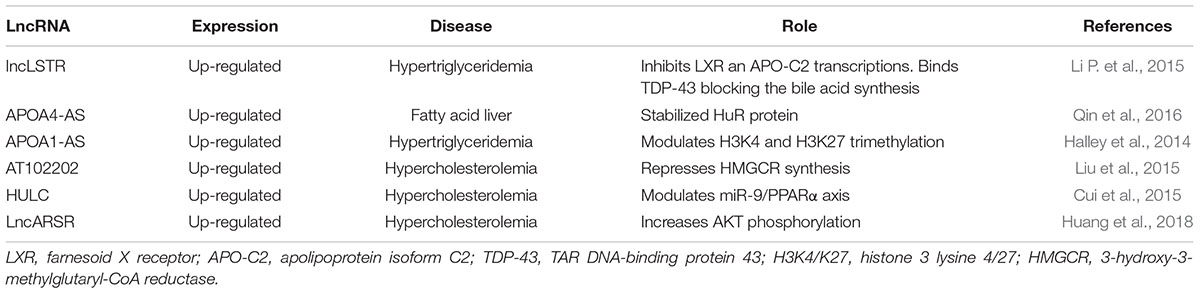
Triglycerides metabolism needs to be strictly regulated in order to avoid the onset of metabolic syndrome and CVD. The liver-specific triglyceride regulator lncRNA Lancaster (lncLSTR) has been demonstrated to regulate triglyceride plasma levels through apolipoprotein C2 (APO-C2) and lipoprotein lipase expression (Li P. et al., 2015). Specifically, lncLSTR inhibits the farnesoid X receptor (LXR) mRNA that synthesizes for a nuclear receptor activated by bile acids and regulates APO-C2 transcription (Li P. et al., 2015). In addition, the presence of a molecular complex has been observed between lncLSTR and the TAR DNA-binding protein 43 (TDP-43) that regulates the expression of sterol 12 α hydroxylase, a key enzyme in the bile acid synthesis (Li P. et al., 2015). As a result, lncLSTR depletion improves triglycerides plasma clearance, and it could be considered as a potential new target for hypertriglyceridemia treatment.
Physiologically, levels of plasmatic lipids are regulated by apolipoproteins (APOs). It has been demonstrated that two antisense transcript lncRNAs, APOA1-AS and APOA4-AS, regulate APOA1 and APOA4 expression respectively (Halley et al., 2014; Qin et al., 2016). More in detail, APOA1-AS can modulate histone three lysine 4 (H3K4) and lysine 27 (H3K27) trimethylation (H3K4me3; H3K27me3); indeed, lncRNA silencing enhances H3K4me3 while it reduces H3K27me3 at APO gene cluster (Halley et al., 2014). Consequently, APOA1-AS knockdown increases APOs expression, enhancing lipid clearance (Halley et al., 2014). In fatty liver disease, levels of both APOA4 and APOA4-AS have been found elevated. The antisense lncRNA interacts with a stabilizing protein named HuR to stabilize APOA4 mRNA, (Qin et al., 2016). In an obese mouse model, the silencing of APOA4-AS has been associated with a reduction of cholesterol and triglycerides plasma level, suggesting this lncRNA as a potential target for new anti-hyperlipidemia drugs (Qin et al., 2016).
AT102202 is another lncRNA, which can modulate plasmatic cholesterol level inhibiting its synthesis in hepatocytes. Recently, 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the rate-limiting enzyme for cholesterol synthesis, has been found down-regulated by increasing levels of AT102202 (Liu et al., 2015). However, the molecular mechanism by which HMGCR is repressed remains under investigation. Remarkably, the uptake of plasmatic cholesterol it is mediated by the LDL receptor (LDLR) which is over-expressed by lncRNA RP1-13D10.2, determining a stimulation of LDL uptake and a decrease of plasma cholesterol (Mitchel et al., 2016). Indeed, RP1-13D10.2 expression increases in response to statin treatment, while in the absence of statin as well as after SREBP2 silencing, this lncRNA decreases, suggesting that RP1-13D10.2 transcription could be under control of sterol response element binding protein 2 (Mitchel et al., 2016). The molecular mechanisms involved in RP1-13D10.2 regulation of LDLR transcription are at present unknown.
Alteration of cholesterol metabolism is commonly found in hepatoma cells (Cui et al., 2015). Herein, the lncRNA HULC increases PPARα expression which enhances transcription of acyl-CoA synthetase long-chain family members 1 (ACSL1) (Cui et al., 2015), the enzyme appointed to long-chain fatty acid synthesis. To increase this effect, HULC down-regulates miR-9 which targets 3′UTR of PPARα transcript (Cui et al., 2015). In this context, HULC levels seem controlled by a positive feedback. Indeed, cholesterol stimulates the retinoic acid X-receptor alpha (RXRA) which recognizes and positively regulates HULC promoter (Cui et al., 2015). In a hepatoma mouse model, beneficial effects have been demonstrated by inhibition of the HULC/miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC loop suggesting this feedback mechanism as a new potential therapeutic target (Cui et al., 2015). Moreover, in hepatocytes of hypercholesterolemia patients, high levels of lncRNA Activated in RCC (renal cell carcinoma) with Sunitib Resistance (lncARSR) have been found (Huang et al., 2018) where lncARSR increases the phosphorylation of AKT enhancing SREBP2 transcription (Huang et al., 2018). As a result, genes involved in cholesterol biosynthesis, such as enzymes involved in squalene synthesis including HMG-CoA reductase and HMG-CoA synthase, are up-regulated, while Cyp7a1, a limiting enzyme that converts cholesterol in bile acids, decreases (Huang et al., 2018). Also, lncARSR can also modulate SREBP-1c promoting hepatic lipogenesis. Indeed, downregulating lncARSR ameliorates lipid uptake into liver suggesting this as a new potential target for steatosis treatment (Zhang M. et al., 2018) (see Table 5 and Figure 2).
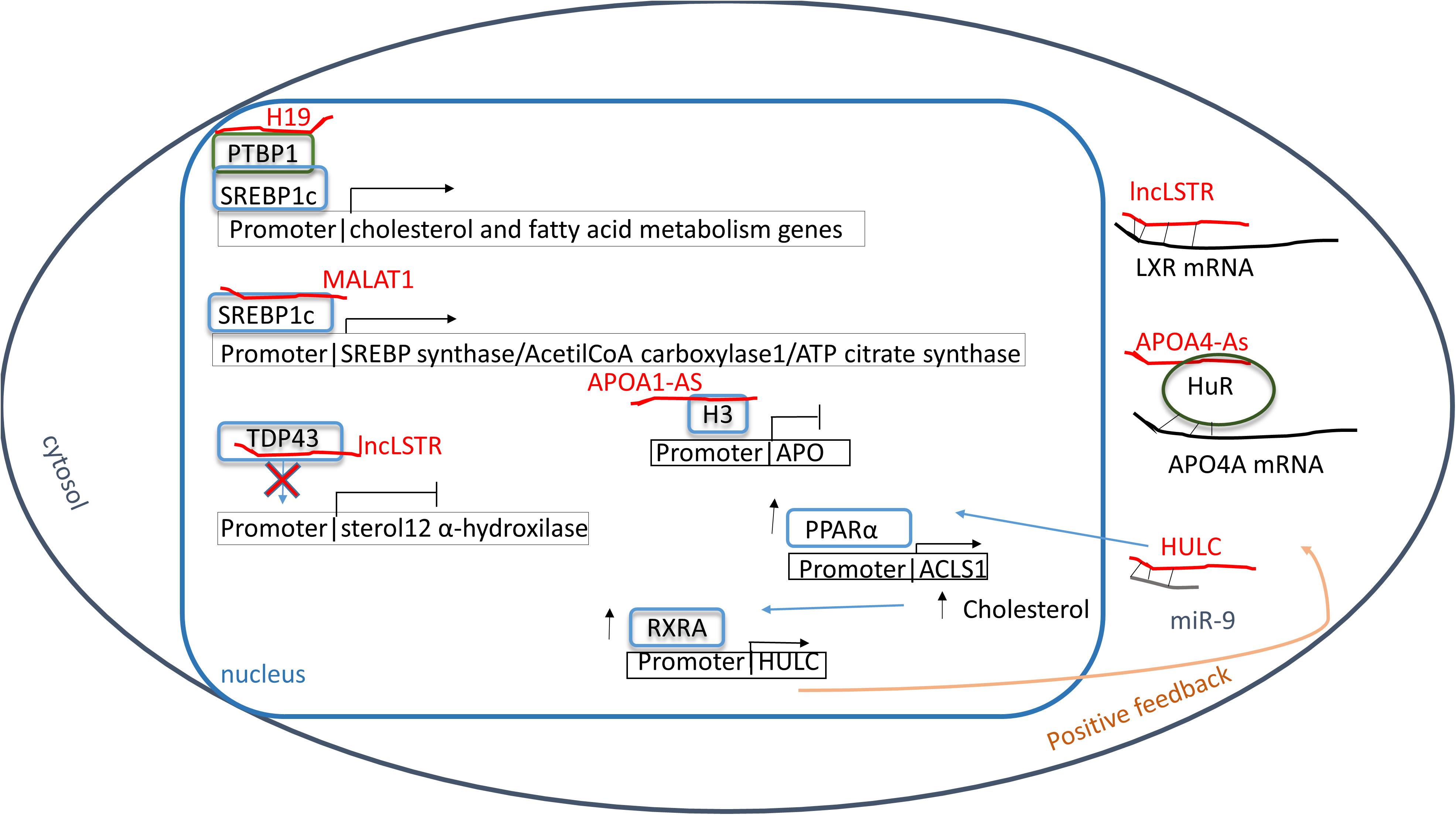
Figure 2. Lipid metabolism. Red: lncRNA; green: proteins; blue: transcriptional factors; gray: miRNAs. Sterol regulatory element binding protein 1c (SREBP1c) can be modulated in different ways: directly binding MALAT1 or by poly-pyrimidine tract-binding protein 1 (PTBP1) that, in turn, binds H19. As result genes involved in lipid anabolism are expressed. The long non-coding RNA lncLSTR inhibits TAR DNA-binding protein 43 (TDP-43) blocking the transcription of sterol 12 α hydroxylase and increasing the hyperlipidemia. Moreover, the hyperlipidemia is enhanced by the inhibition of APO synthesis through the chromatin condensation at APO promoters by the hypomethylation condition of histone 3 at lysine 4 and 27 due to APOA1-As bound. In addition, farnesoid X receptor (LXR) and APOA4 mRNAs are inhibited by lncLSTR and HuR protein helped by APOA4-As respectively reducing the clearance of plasmatic fatty acids. In hypercholesteremia a positive feedback has been shown in which HULC enhances PPARα synthesis. One effect is the increasing of retinoic acid X-receptor alpha (RXRA) level which recognizes and positively regulates HULC promoter.
LncRNAs and Their Role in Chronic Dysmetabolic Conditions
According to the World Health Organization (WHO), in 2016 type 2 diabetes (T2D) has been the most frequently reported metabolic disease in developed countries, affecting about 8.2% of the adult population. T2D is a multifactorial disorder characterized, among other aspects, by high blood glucose and lipid levels (hyperglycemia and hyperlipidemia) in association with insulin resistance and atherosclerosis (Bornfeldt and Tabas, 2011). All these factors contribute to a condition of chronic inflammation leading to micro/macro-vascular alterations often linked to ischemia, chronic ulcers, atherosclerotic plaque formation, cardiomyopathy, amputations, and renal failure (Olefsky and Glass, 2010). Multiple epigenetic mechanisms are involved in the onset of such diabetic complications. Acetylation of histone H3 lysines 9, 14 and that of histone H4 lysine 5 (H3K9, H3K14, and H4K5) and methylation of histone H4 lysine 4 (H3K4) enriched in chromatin regions encoding for inflammatory genes correlated with the exacerbation of T2D (Reddy et al., 2015). Indeed, it has been demonstrated that in cardiac mesenchymal T2D cells the epigenetic drug pentadecylidenemalonate 1b (SPV106), an activator of histone acetylases, can restore the proliferation and differentiation potential lost in cells of cardiac origin isolated from T2D patient (Vecellio et al., 2014). Additional work reported about the presence of specific epigenetic modifications, including the increase in H3K27 tri-methylation and DNA methylation/hydroxymethylation in the chromatin and genomic DNA of cardiac fibroblasts isolated from T2D patients undergoing major cardiac surgery. In this model, a global increase of cytosine methylation has been observed in those genes involved in transcription, metabolism and glucose uptake (Spallotta et al., 2018). To revert this condition, the addition of alpha-ketoglutarate seems beneficial through its interaction with DNA demethylases (Spallotta et al., 2018).
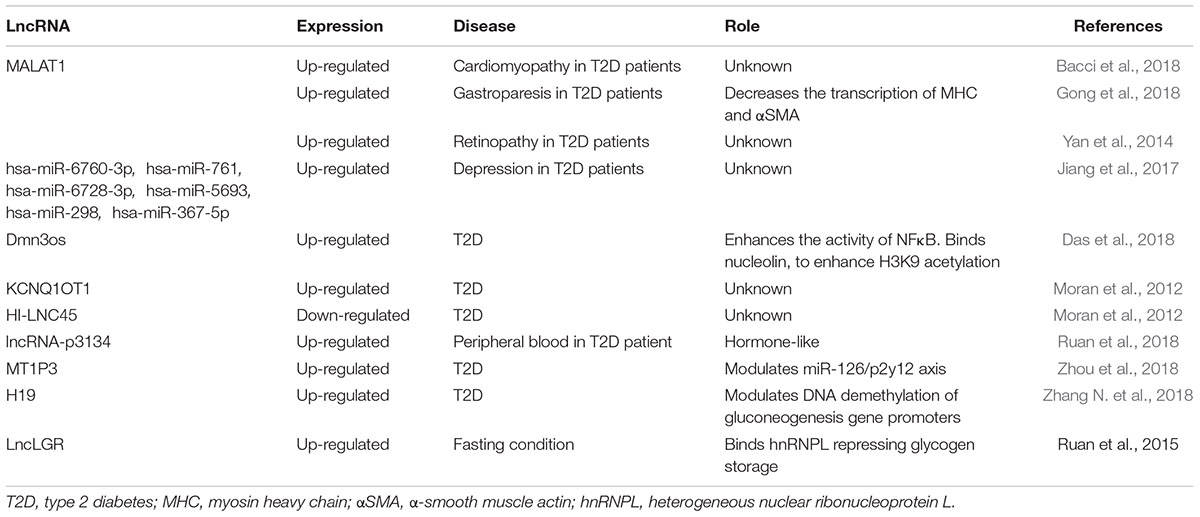
Many lncRNAs have been associated to T2D and diabetes complications such as MALAT1, that correlates to cardiomyopathy (Bacci et al., 2018), or circRNAs targeting hsa-miR-6760-3p, hsa-miR-761, hsa-miR-6728-3p, hsa-miR-5693, hsa-miR-298, and hsa-miR-367-5p which have been correlated to depression in T2D patient (Jiang et al., 2017). However, another lncRNA, the dynamin 3 opposite strand (Dnm3os) has been recently proposed as important in T2D (Das et al., 2018). Specifically, in T2D macrophages, Dnm3os, which can bind and enhance the activity of NFκB transcription factor into the nucleus, is significantly up-regulated as compared to normal controls. As a result, Dnm3os increases the NFκB activity enhancing inflammation, phagocytosis, immune response, cell activation, chemotaxis, and response to wounding (Das et al., 2018). Moreover, Dnm3os has been found interacting with several other critical intracellular targets including nucleolin, the actin-related protein 3, ILF-2, the RNA-binding protein Ralv short isoform, as well as the heterogeneous nuclear ribonucleoprotein U like protein 2, and the variant ribonucleoprotein isoform A1 (Das et al., 2018). Interesting, nucleolin regulates chromatin structure, and the synergic effect of Dnm3os and nucleolin is increasing H3K9 acetylation at IL-6 promoter with a consequent higher inflammatory response (Das et al., 2018).
Another study indicated, in pancreatic β cells of T2D patients, the dysregulation of two lncRNAs: KQT-like subfamily member one opposite strand/antisense transcript 1 (KCNQ1OT1) and HI-LNC45 (Moran et al., 2012). The authors reported that KCNQ1OT1, abundant in cardiac tissue (Vausort et al., 2014), was found over-expressed in diabetic pancreatic cells while HI-LNC45 was significantly lower than in healthy controls (Moran et al., 2012). Despite this evidence, their functional role in diabetes remains unclear.
In addition to its role in cancer and CVD, MALAT1 is also significantly up-regulated in diabetic gastroparesis (DGP) (Gong et al., 2018), which is a clinically relevant complication of diabetes mellitus characterized by bloating, nausea and vomiting (Kashyap and Farrugia, 2010). In DGP rat models and gastric smooth muscle cells isolated from DGP patients, the accumulation of MALAT1 has been associated with the decrease of MHC and alpha-smooth muscle actin (αSMA), which are proteins involved in contractile and adhesion functions (Gong et al., 2018), resulting in a chronic smooth muscle cell enlargement which contributes to symptoms. This study shows that high glucose increases MALAT1 level and that after MALAT1 silencing contractile markers are re-expressed reducing DGP exacerbation at least in diabetic mice (Gong et al., 2018). Although in this condition, MALAT1-dependent mechanism of action has not been elucidated yet, these findings suggest MALAT1 as a potential target for the development of new DGP therapeutic strategies.
Diabetic cardiomyopathy is a critical complication of T2D (Jia et al., 2018). Interestingly, upon prolonged exposure to elevated blood glucose, high level of MALAT1 has been found in mouse cardiomyocytes (Bacci et al., 2018). Sildenafil, the pharmacological inhibitor of phosphodiesterase 5 (PD5), which increases endogenously cardiac and vascular levels of nitric oxide, normalized MALAT1 expression with beneficial effects in an experimental mouse model of diabetic cardiomyopathy (Bacci et al., 2018).
Remarkably, lncRNAs can also act as hormone-like molecules. In fact, fluctuating circulating levels have been detected body fluids such as peripheral blood (Shen et al., 2017) and urine (Terracciano et al., 2017) and lncRNA-p3134 secreted by defective β-cells and circulating in exosomal vesicles, has been found highly expressed in T2D patients’ plasma samples (Ruan et al., 2018) where lncRNA-p3134 expression levels increased proportionally to blood glucose (Ruan et al., 2018). Interestingly, this lncRNA has been shown to increase insulin synthesis in healthy β cells, suggesting a role as an epigenetic regulatory element. Indeed, normal β cells reacted to lncRNA-p3134 reducing caspase 3 and 9 and Bcl2 levels, thus protecting themselves from glucotoxicity (Ruan et al., 2018). Inhibiting PI3K/AKT axis, the protective effect of lncRNA-p3134 was significantly attenuated (Ruan et al., 2018). Despite these findings, the correlation between lncRNA-p3134 regulation/function and insulin signaling remains mostly unclear.
In T2D patients, many epigenetic alterations have been found in non-pancreatic cells. A recent study reported that lncRNA metallothionein one pseudogene 3 (MT1P3) is up-regulated in T2D megakaryocytes (Zhou et al., 2018). MT1P3 regulates the expression of the p2y12 receptor by sponging out miR-126 that targets p2y12 mRNA (Zhou et al., 2018). Also, MT1P3 enhances the p2y12 presence in platelet membranes increasing the normal activity of the receptor (Zhou et al., 2018). P2y12 is a G-coupled receptor, and it is activated by ADP stimulating platelets aggregation (Hu et al., 2017). Clinically, in T2D patients that frequently fail to respond to antiplatelet therapy (Samos et al., 2016), the alteration of the MT1P3 epigenetic pathway seems associated with risk of microcirculation complications and thrombotic microangiopathies higher than in the non-diabetic population (Ding et al., 2017). In this condition, a myocardial re-infarction can occur after an episode of in-stent thrombosis while standard clopidogrel treatment results ineffective (Samos et al., 2014).
One of the mechanisms of action accredited to lncRNAs is binding and regulating transcription factors. An example in this direction is given by lncRNA Meg3 which modulates the musculoaponeurotic fibrosarcoma protein A (MafA) in mouse β cells through interaction with EZH2 (Wang N et al., 2018), one of the methyl-transferases of Polycomb complex (Cao et al., 2002). EZH2 represses MafA inhibitors possibly present in specific genomic regions, allowing the transcription factor to be active (Wang N et al., 2018). In fact, in diabetic mice, Meg3 down-regulation is paralleled by a decrease of MafA with the consequent reduction in insulin synthesis and secretion (Zhang et al., 2005).
In diabetic retinopathy, a severe and common complication of diabetes, aberrant expression of several lncRNAs has been frequently reported (Yan et al., 2014). Here, those lncRNAs involved in MAPK and chemokine signaling pathways, axon guidance, complement, and coagulation cascades and pyruvate metabolism were found up-regulated (Yan et al., 2014). All these alterations are known associated with cellular stress response, DNA repair, epithelium and tube development and lens formation (Yan et al., 2014). Specifically, it has been discovered that MALAT1 is upregulated in retina being present in the aqueous humor of diabetic mice and patients. This result suggests for MALAT1 potential application as a new biomarker of retinopathy (Yan et al., 2014).
Novel findings indicate that lncRNA H19 is highly expressed in diabetic liver (Zhang N. et al., 2018). Here, H19 has been found enriched into the nucleolus and able to regulate gluconeogenesis via DNA methylation control at the level of transcription factor hepatocyte nuclear factor 4 alpha (HNF4A) which transcribes glucose-6-phosphatase (G6PC) and phosphoenolpyruvate carboxykinase 1 (PCK1) (Zhang N. et al., 2018). In hyperglycemia, high levels of H19 in the liver contribute to HNF4A promoter demethylation which results repressed at transcriptional level inhibiting G6PC and PCK1 synthesis essential for glycogen formation and storage (Zhang N. et al., 2018). As a consequence, H19 overexpression promotes hepatic glucose production (Zhang N. et al., 2018) and insulin resistance (Gabory et al., 2010; Zhang N. et al., 2018). Moreover, it has been demonstrated that in hepatic cells H19 binds a set of DNA-binding proteins associated to FoxO1 promoter that inhibit FoxO1 transcription resulting in a downregulation of gluconeogenesis pathway in healthy cells (Goyal et al., 2019). To simulate the diabetic conditions, H19 has been silenced in mouse hepatocytes and as results on FoxO1 promoter p53 has been found associated activating the cascade that exacerbates in insulin resistance (Goyal et al., 2019).
During fasting, the hepatic glucokinase (CGK), another enzyme involved in glycogen synthesis and storage, is physiologically downregulated (Massa et al., 2011). This phenomenon occurs at, least partially, in consequence of the up-regulation of lncRNA liver CGK repressor (lncLGR) (Ruan et al., 2016). It has been demonstrated that during fasting, lncLGR is overexpressed binding to heterogeneous nuclear ribonucleoprotein L (hnRNPL) which is recruited to CGK gene promoter repressing its transcription (Ruan et al., 2016). As a consequence, glycogen is not stored remaining accessible for glucose synthesis (Ruan et al., 2016).
As in CVD, a SNP in ANRIL gene has been found to correlate with T2D exacerbation. Patients who carry rs10811661 (Broadbent et al., 2008) or rs564398 (Kong et al., 2018) have more risk to develop diabetes. Especially, rs564398 causes T2D by the reduction of b-cell proliferation (Kong et al., 2018). All these findings in ANRIL locus suggest that a lncRNA polymorphism can be a new risk factor of non-transmissible diseases. An exciting discovery would be if the lncRNA and its variants could be detected in peripheral blood and used as a diagnostic biomarker.
Another condition, in which the metabolism results altered, is obesity that is defined through the valuation of body mass index (BMI, kg/m2). Conventionally, the cut off between overweight and obese is a BMI higher than 30. The clinical picture of obesity is very complex due to the condition, called metabolic syndrome, in which insulin resistance, glucose intolerance, hypertension, and dyslipidemia coexist (Reaven, 2005).
Moreover, the obesity is a risk factor for many diseases such as CVD, non-alcoholic steatohepatitis (NASH), polycystic ovarian syndrome (PCOS), obstructive sleep apnea (OSA), and cancers (Einhorn et al., 2003). Recently, in obese mouse models, it has been discovered a lncRNA (lncSHGL) which is significantly downregulated in hepatocytes while in obese human a homologous (B4GALT1-AS1) results decreased in hepatocytes too (Wang J. et al., 2018). This study reveals the physiological lncSHGL ability to recruits heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) that increases the efficiency of the translation of calmodulin (CaM) mRNA without altering the transcription (Wang J. et al., 2018). As a result, in healthy hepatocytes, PI3K/Akt pathway is activated while mTOR/SREPB pathway is inhibited and hepatic gluconeogenesis and lipogenesis are suppressed in an insulin-independent way (Wang J. et al., 2018). In conclusion, low levels of B4GALT1-AS1 in obese patients cause a continuous activation of the lipid synthesis determining their accumulation (Wang J. et al., 2018) (see Table 6).
Conclusion
Recent discoveries in epigenetics enlightened important functional and pathophysiological roles for lncRNAs. Dysregulation of this component of our transcriptome, in fact, has been found in pathological condition such as cancer (Terracciano et al., 2017; Chen M. et al., 2018; Ma et al., 2018; Tang et al., 2018), CVD (Thum and Condorelli, 2015; Mitchel et al., 2016; Viereck and Thum, 2017), and dysmetabolic conditions including obesity and T2D (Moran et al., 2012; Yan et al., 2016; Das et al., 2018). Although lncRNAs molecular mechanism of action is far to be clarified, it seems clear that they can act as miRNA sponges (Chen L. et al., 2018; Zhao et al., 2018) or directly binding regulatory proteins such as transcription factors, nuclear hormone receptors (Aiello et al., 2016) or epigenetic enzymes (Khalil et al., 2009), thus controlling gene expression and protein function. Remarkably, lncRNAs have also a pivotal role in carbohydrate and lipid metabolism, and their alteration greatly influences cellular homeostasis (Liu et al., 2018) contributing, for instance, to insulin-resistance (Yan et al., 2016) or formation of atherosclerotic plaques (Huang et al., 2018). In light of this evidence, although our mechanistic knowledge is still limited, lncRNAs seem to play an enormously crucial biological role regulating cellular metabolism at multiple levels, in physiological as well as pathophysiological conditions. This fact is unquestionable and may open essential avenues for the future development of novel therapeutic strategies in chronic diseases.
Author Contributions
All authors have contributed to the conception and design of the article and revision. AM and CG wrote the text. AF and FM provided the suggestions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
FM was supported by Ministero della Salute (Ricerca Corrente, 5X1000 and RF-2011-02347907). AF was supported by National Research Council (CNR)-IBCN c/o Università Cattolica di Roma. CG and AM are supported by Ricerca Corrente Ministero della Salute.
References
Aboudehen, K., Farahani, S., Kanchwala, M., Chan, S. C., Avdulov, S., Mickelson, A., et al. (2018). Long noncoding RNA Hoxb3os is dysregulated in autosomal dominant polycystic kidney disease and regulates mTOR signaling. J. Biol. Chem. 293, 9388–9398. doi: 10.1074/jbc.RA118.001723
Aiello, A., Bacci, L., Re, A., Ripoli, C., Pierconti, F., Pinto, F., et al. (2016). MALAT1 and HOTAIR long Non-coding RNAs play opposite role in estrogen-mediated transcriptional regulation in prostate cancer cells. Sci. Rep. 6:38414. doi: 10.1038/srep38414
Amaral, P. P., Clark, M. B., Gascoigne, D. K., Dinger, M. E., and Mattick, J. S. (2011). lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 39, D146–D151. doi: 10.1093/nar/gkq1138
Anderson, M. E., Brown, J. H., and Bers, D. M. (2011). CaMKII in myocardial hypertrophy and heart failure. J. Mol. Cell. Cardiol. 51, 468–473. doi: 10.1016/j.yjmcc.2011.01.012
Ashwal-Fluss, R., Meyer, M., Pamudurti, N. R., Ivanov, A., Bartok, O., Hanan, M., et al. (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66. doi: 10.1016/j.molcel.2014.08.019
Bacci, L., Barbati, S. A., Colussi, C., Aiello, A., Isidori, A. M., Grassi, C., et al. (2018). Sildenafil normalizes MALAT1 level in diabetic cardiomyopathy. Endocrine 62, 259–262. doi: 10.1007/s12020-018-1599-z
Bai, L., Wang, A., Zhang, Y., Xu, X., and Zhang, X. (2018). Knockdown of MALAT1 enhances chemosensitivity of ovarian cancer cells to cisplatin through inhibiting the Notch1 signaling pathway. Exp. Cell Res. 366, 161–171. doi: 10.1016/j.yexcr.2018.03.014
Barrett, S. P., and Salzman, J. (2016). Circular RNAs: analysis, expression and potential functions. Development 143, 1838–1847. doi: 10.1242/dev.128074
Barrett, S. P., Wang, P. L., and Salzman, J. (2015). Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4:e07540. doi: 10.7554/eLife.07540
Bitarafan, S., Yari, M., Broumand, M. A., Ghaderian, S. M. H., Rahimi, M., Mirfakhraie, R., et al. (2019). Association of increased levels of lncRNA H19 in PBMCs with risk of coronary artery disease. Cell J. 20, 564–568. doi: 10.22074/cellj.2019.5544
Bornfeldt, K. E., and Tabas, I. (2011). Insulin resistance, hyperglycemia, and atherosclerosis. Cell Metab. 14, 575–585. doi: 10.1016/j.cmet.2011.07.015
Bose, S., and Le, A. (2018). Glucose metabolism in cancer. Adv. Exp. Med. Biol. 1063, 3–12. doi: 10.1007/978-3-319-77736-8_1
Broadbent, H. M., Peden, J. F., Lorkowski, S., Goel, A., Ongen, H., Green, F., et al. (2008). Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 17, 806–814. doi: 10.1093/hmg/ddm352
Cabili, M. N., Dunagin, M. C., Mcclanahan, P. D., Biaesch, A., Padovan-Merhar, O., Regev, A., et al. (2015). Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16:20. doi: 10.1186/s13059-015-0586-4
Cao, R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., et al. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039–1043. doi: 10.1126/science.1076997
Chatterjee, S., Bar, C., and Thum, T. (2017). Linc-ing the noncoding genome to heart function: beating hypertrophy. Trends Mol. Med. 23, 577–579. doi: 10.1016/j.molmed.2017.05.007
Chen, L., Hu, N., Wang, C., Zhao, H., and Gu, Y. (2018). Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle 17, 319–329. doi: 10.1080/15384101.2017.1407893
Chen, M., Xia, Z., Chen, C., Hu, W., and Yuan, Y. (2018). LncRNA MALAT1 promotes epithelial-to-mesenchymal transition of esophageal cancer through Ezh2-Notch1 signaling pathway. Anticancer Drugs 29, 767–773. doi: 10.1097/CAD.0000000000000645
Cheng, J., Cai, M. Y., Chen, Y. N., Li, Z. C., Tang, S. S., Yang, X. L., et al. (2017). Variants in ANRIL gene correlated with its expression contribute to myocardial infarction risk. Oncotarget 8, 12607–12619. doi: 10.18632/oncotarget.14721
Choudhry, H., and Mole, D. R. (2016). Hypoxic regulation of the noncoding genome and NEAT1. Brief. Funct. Genomics 15, 174–185. doi: 10.1093/bfgp/elv050
Cui, M., Xiao, Z., Wang, Y., Zheng, M., Song, T., Cai, X., et al. (2015). Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857. doi: 10.1158/0008-5472.CAN-14-1192
Das, S., Reddy, M. A., Senapati, P., Stapleton, K., Lanting, L., Wang, M., et al. (2018). Diabetes mellitus-induced long noncoding RNA Dnm3os regulates macrophage functions and inflammation via nuclear mechanisms. Arterioscler. Thromb. Vasc. Biol. 38, 1806–1820. doi: 10.1161/ATVBAHA.117.310663
Ding, Y., Sun, X., and Shan, P. F. (2017). MicroRNAs and cardiovascular disease in diabetes mellitus. Biomed Res. Int. 2017:4080364. doi: 10.1155/2017/4080364
Einhorn, D., Reaven, G. M., Cobin, R. H., Ford, E., Ganda, O. P., Handelsman, Y., et al. (2003). American College of Endocrinology position statement on the insulin resistance syndrome. Endocr. Pract. 9, 237–252.
Fan, S., Fan, C., Liu, N., Huang, K., Fang, X., and Wang, K. (2018). Downregulation of the long non-coding RNA ZFAS1 is associated with cell proliferation, migration and invasion in breast cancer. Mol. Med. Rep. 17, 6405–6412. doi: 10.3892/mmr.2018.8707
Fang, Z., Chen, W., Yuan, Z., Liu, X., and Jiang, H. (2018). LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. Biomed. Pharmacother. 101, 536–542. doi: 10.1016/j.biopha.2018.02.130
Fuschi, P., Maimone, B., Gaetano, C., and Martelli, F. (2017). Noncoding RNAs in the vascular system response to oxidative stress. Antioxid. Redox Signal. 30, 992–1010. doi: 10.1089/ars.2017.7229
Gabory, A., Jammes, H., and Dandolo, L. (2010). The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays 32, 473–480. doi: 10.1002/bies.200900170
Gao, X., Wang, H., Yang, J. J., Liu, X., and Liu, Z. R. (2012). Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609. doi: 10.1016/j.molcel.2012.01.001
Gong, Y., Zhu, Y., Zhu, B., Si, X., Heng, D., Tang, Y., et al. (2018). LncRNA MALAT1 is up-regulated in diabetic gastroparesis and involved in high-glucose-induced cellular processes in human gastric smooth muscle cells. Biochem. Biophys. Res. Commun. 496, 401–406. doi: 10.1016/j.bbrc.2018.01.038
Goyal, N., Tiwary, S., Kesharwani, D., and Datta, M. (2019). Long non-coding RNA H19 inhibition promotes hyperglycemia in mice by upregulating hepatic FoxO1 levels and promoting gluconeogenesis. J. Mol. Med. 97, 115–126. doi: 10.1007/s00109-018-1718-6
Greco, S., Gorospe, M., and Martelli, F. (2015). Noncoding RNA in age-related cardiovascular diseases. J. Mol. Cell. Cardiol. 83, 142–155. doi: 10.1016/j.yjmcc.2015.01.011
Greco, S., Zaccagnini, G., Perfetti, A., Fuschi, P., Valaperta, R., Voellenkle, C., et al. (2016). Long noncoding RNA dysregulation in ischemic heart failure. J. Transl. Med. 14:183. doi: 10.1186/s12967-016-0926-5
Gu, L. Q., Xing, X. L., Cai, H., Si, A. F., Hu, X. R., Ma, Q. Y., et al. (2018). Long non-coding RNA DILC suppresses cell proliferation and metastasis in colorectal cancer. Gene 666, 18–26. doi: 10.1016/j.gene.2018.03.100
Gupta, R. A., Shah, N., Wang, K. C., Kim, J., Horlings, H. M., Wong, D. J., et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. doi: 10.1038/nature08975
Guttman, M., Amit, I., Garber, M., French, C., Lin, M. F., Feldser, D., et al. (2009). Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227. doi: 10.1038/nature07672
Halley, P., Kadakkuzha, B. M., Faghihi, M. A., Magistri, M., Zeier, Z., Khorkova, O., et al. (2014). Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 6, 222–230. doi: 10.1016/j.celrep.2013.12.015
Han, P., Li, W., Lin, C. H., Yang, J., Shang, C., Nuernberg, S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. doi: 10.1038/nature13596
Hashimoto, K., Ishida, E., Matsumoto, S., Shibusawa, N., Okada, S., Monden, T., et al. (2009). A liver X receptor (LXR)-beta alternative splicing variant (LXRBSV) acts as an RNA co-activator of LXR-beta. Biochem. Biophys. Res. Commun. 390, 1260–1265. doi: 10.1016/j.bbrc.2009.10.132
Helgadottir, A., Thorleifsson, G., Manolescu, A., Gretarsdottir, S., Blondal, T., Jonasdottir, A., et al. (2007). A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493. doi: 10.1126/science.1142842
Holdt, L. M., Hoffmann, S., Sass, K., Langenberger, D., Scholz, M., Krohn, K., et al. (2013). Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 9:e1003588. doi: 10.1371/journal.pgen.1003588
Holdt, L. M., Stahringer, A., Sass, K., Pichler, G., Kulak, N. A., Wilfert, W., et al. (2016). Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7:12429. doi: 10.1038/ncomms12429
Horton, J. D. (2002). Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem. Soc. Trans. 30, 1091–1095. doi: 10.1042/bst0301091
Hu, H., Wu, J., Li, D., Zhou, J., Yu, H., and Ma, L. (2018). Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed. Pharmacother. 106, 738–746. doi: 10.1016/j.biopha.2018.06.122
Hu, L., Chang, L., Zhang, Y., Zhai, L., Zhang, S., Qi, Z., et al. (2017). Platelets express activated P2Y12 receptor in patients with diabetes mellitus. Circulation 136, 817–833. doi: 10.1161/CIRCULATIONAHA.116.026995
Huang, J., Chen, S., Cai, D., Bian, D., and Wang, F. (2018). Long noncoding RNA lncARSR promotes hepatic cholesterol biosynthesis via modulating Akt/SREBP-2/HMGCR pathway. Life Sci. 203, 48–53. doi: 10.1016/j.lfs.2018.04.028
Janani, C., and Ranjitha Kumari, B. D. (2015). PPAR gamma gene–a review. Diabetes Metab. Syndr. 9, 46–50. doi: 10.1016/j.dsx.2014.09.015
Jia, G., Hill, M. A., and Sowers, J. R. (2018). Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circ. Res. 122, 624–638. doi: 10.1161/CIRCRESAHA.117.311586
Jiang, G., Ma, Y., An, T., Pan, Y., Mo, F., Zhao, D., et al. (2017). Relationships of circular RNA with diabetes and depression. Sci. Rep. 7:7285. doi: 10.1038/s41598-017-07931-0
Kashyap, P., and Farrugia, G. (2010). Diabetic gastroparesis: what we have learned and had to unlearn in the past 5 years. Gut 59, 1716–1726. doi: 10.1136/gut.2009.199703
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Kino, T., Hurt, D. E., Ichijo, T., Nader, N., and Chrousos, G. P. (2010). Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 3:ra8. doi: 10.1126/scisignal.2000568
Kishore, S., and Stamm, S. (2006). Regulation of alternative splicing by snoRNAs. Cold Spring Harb. Symp. Quant. Biol. 71, 329–334. doi: 10.1101/sqb.2006.71.024
Knoll, M., Lodish, H. F., and Sun, L. (2015). Long non-coding RNAs as regulators of the endocrine system. Nat. Rev. Endocrinol. 11, 151–160. doi: 10.1038/nrendo.2014.229
Kong, Y., Sharma, R. B., Ly, S., Stamateris, R. E., Jesdale, W. M., and Alonso, L. C. (2018). CDKN2A/B T2D genome-wide association study risk SNPs impact locus gene expression and proliferation in human islets. Diabetes 67, 872–884. doi: 10.2337/db17-1055
Li, D., Guo, L., Deng, B., Li, M., Yang, T., Yang, F., et al. (2018). Long noncoding RNA HR1 participates in the expression of SREBP1c through phosphorylation of the PDK1/AKT/FoxO1 pathway. Mol. Med. Rep. 18, 2850–2856. doi: 10.3892/mmr.2018.9278
Li, H., Chen, S., Liu, J., Guo, X., Xiang, X., Dong, T., et al. (2018). Long non-coding RNA PVT1-5 promotes cell proliferation by regulating miR-126/SLC7A5 axis in lung cancer. Biochem. Biophys. Res. Commun. 495, 2350–2355. doi: 10.1016/j.bbrc.2017.12.114
Li, L., Wang, L., Li, H., Han, X., Chen, S., Yang, B., et al. (2018). Characterization of LncRNA expression profile and identification of novel LncRNA biomarkers to diagnose coronary artery disease. Atherosclerosis 275, 359–367. doi: 10.1016/j.atherosclerosis.2018.06.866
Li, X., Zhao, Q., Qi, J., Wang, W., Zhang, D., Li, Z., et al. (2018). lncRNA Ftx promotes aerobic glycolysis and tumor progression through the PPARgamma pathway in hepatocellular carcinoma. Int. J. Oncol. 53, 551–566. doi: 10.3892/ijo.2018.4418
Li, H., Li, J., Jia, S., Wu, M., An, J., Zheng, Q., et al. (2015). miR675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget 6, 31958–31984. doi: 10.18632/oncotarget.5579
Li, P., Ruan, X., Yang, L., Kiesewetter, K., Zhao, Y., Luo, H., et al. (2015). A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 21, 455–467. doi: 10.1016/j.cmet.2015.02.004
Linton, M. R. F., Yancey, P. G., Davies, S. S., Jerome, W. G., Linton, E. F., Song, W. L., et al. (2000). “The role of lipids and lipoproteins in atherosclerosis,” in Endotext, eds K. R. Feingold, B. Anawalt, A. Boyce, G. Chrousos, K. Dungan, A. Grossman, et al. (South Dartmouth, MA: MDText.com, Inc).
Liu, C., Yang, Z., Wu, J., Zhang, L., Lee, S., Shin, D. J., et al. (2018). Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 67, 1768–1783. doi: 10.1002/hep.29654
Liu, G., Zheng, X., Xu, Y., Lu, J., Chen, J., and Huang, X. (2015). Long non-coding RNAs expression profile in HepG2 cells reveals the potential role of long non-coding RNAs in the cholesterol metabolism. Chin. Med. J. 128, 91–97. doi: 10.4103/0366-6999.147824
Liu, L., An, X., Li, Z., Song, Y., Li, L., Zuo, S., et al. (2016). The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 111, 56–65. doi: 10.1093/cvr/cvw078
Liu, X., Xiao, Z. D., Han, L., Zhang, J., Lee, S. W., Wang, W., et al. (2016). LncRNA NBR2 engages a metabolic checkpoint by regulating AMPK under energy stress. Nat. Cell Biol. 18, 431–442. doi: 10.1038/ncb3328
Liu, X., and Gan, B. (2016). lncRNA NBR2 modulates cancer cell sensitivity to phenformin through GLUT1. Cell Cycle 15, 3471–3481. doi: 10.1080/15384101.2016.1249545
Ma, D., Chen, C., Wu, J., Wang, H., and Wu, D. (2018). Up-regulated lncRNA AFAP1-AS1 indicates a poor prognosis and promotes carcinogenesis of breast cancer. Breast Cancer 26, 74–83. doi: 10.1007/s12282-018-0891-3
Ma, L., Bajic, V. B., and Zhang, Z. (2013). On the classification of long non-coding RNAs. RNA Biol. 10, 925–933. doi: 10.4161/rna.24604
Mafi Golchin, M., Ghaderian, S. M. H., Akhavan-Niaki, H., Jalalian, R., Heidari, L., and Salami, S. A. (2017). Analysis of two CDKN2B-AS polymorphisms in relation to coronary artery disease patients in north of Iran. Int. J. Mol. Cell. Med. 6, 31–37.
Massa, M. L., Gagliardino, J. J., and Francini, F. (2011). Liver glucokinase: an overview on the regulatory mechanisms of its activity. IUBMB Life 63, 1–6. doi: 10.1002/iub.411
Matouk, I. J., Degroot, N., Mezan, S., Ayesh, S., Abu-Lail, R., Hochberg, A., et al. (2007). The H19 non-coding RNA is essential for human tumor growth. PLoS One 2:e845. doi: 10.1371/journal.pone.0000845
Mitchel, K., Theusch, E., Cubitt, C., Dose, A. C., Stevens, K., Naidoo, D., et al. (2016). RP1-13D10.2 is a novel modulator of statin-induced changes in cholesterol. Circ. Cardiovasc. Genet. 9, 223–230. doi: 10.1161/CIRCGENETICS.115.001274
Monticelli, S., Ansel, K. M., Lee, D. U., and Rao, A. (2005). Regulation of gene expression in mast cells: micro-rNA expression and chromatin structural analysis of cytokine genes. Novartis Found. Symp. 271, 179–187; discussion 187–190, 198–199. doi: 10.1002/9780470033449.ch14
Moran, I., Akerman, I., Van De Bunt, M., Xie, R., Benazra, M., Nammo, T., et al. (2012). Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16, 435–448. doi: 10.1016/j.cmet.2012.08.010
Niazi, F., and Valadkhan, S. (2012). Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3′ UTRs. RNA 18, 825–843. doi: 10.1261/rna.029520.111
Olefsky, J. M., and Glass, C. K. (2010). Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246. doi: 10.1146/annurev-physiol-021909-135846
Pan, J. X. (2017). LncRNA H19 promotes atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 21, 322–328.
Park, Y., Kelley, R. L., Oh, H., Kuroda, M. I., and Meller, V. H. (2002). Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298, 1620–1623. doi: 10.1126/science.1076686
Parraga, A., Bellsolell, L., Ferre-D’amare, A. R., and Burley, S. K. (1998). Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure 6, 661–672. doi: 10.1016/S0969-2126(98)00067-7
Pedersen, J. S., Bejerano, G., Siepel, A., Rosenbloom, K., Lindblad-Toh, K., Lander, E. S., et al. (2006). Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2:e33. doi: 10.1371/journal.pcbi.0020033
Qin, W., Li, X., Xie, L., Li, S., Liu, J., Jia, L., et al. (2016). A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 44, 6423–6433. doi: 10.1093/nar/gkw341
Rafiee, A., Riazi-Rad, F., Havaskary, M., and Nuri, F. (2018). Long noncoding RNAs: regulation, function and cancer. Biotechnol. Genet. Eng. Rev. 34, 153–180. doi: 10.1080/02648725.2018.1471566
Reaven, G. M. (2005). Insulin resistance, the insulin resistance syndrome, and cardiovascular disease. Panminerva Med. 47, 201–210.
Reddy, M. A., Zhang, E., and Natarajan, R. (2015). Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia 58, 443–455. doi: 10.1007/s00125-014-3462-y
Ruan, X., Li, P., Cangelosi, A., Yang, L., and Cao, H. (2016). A Long non-coding RNA, lncLGR, regulates hepatic glucokinase expression and glycogen storage during fasting. Cell Rep. 14, 1867–1875. doi: 10.1016/j.celrep.2016.01.062
Ruan, Y., Lin, N., Ma, Q., Chen, R., Zhang, Z., Wen, W., et al. (2018). Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet beta-cell function. Cell. Physiol. Biochem. 46, 335–350. doi: 10.1159/000488434
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N., and Brown, P. O. (2012). Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7:e30733. doi: 10.1371/journal.pone.0030733
Samos, M., Fedor, M., Kovar, F., Mokan, M., Bolek, T., Galajda, P., et al. (2016). Type 2 diabetes and ADP receptor blocker therapy. J. Diabetes Res. 2016:6760710. doi: 10.1155/2016/6760710
Samos, M., Simonova, R., Kovar, F., Duraj, L., Fedorova, J., Galajda, P., et al. (2014). Clopidogrel resistance in diabetic patient with acute myocardial infarction due to stent thrombosis. Am. J. Emerg. Med. 32, 461–465. doi: 10.1016/j.ajem.2014.01.006
Sanchez Calle, A., Kawamura, Y., Yamamoto, Y., Takeshita, F., and Ochiya, T. (2018). Emerging roles of long non-coding RNA in cancer. Cancer Sci. 109, 2093–2100. doi: 10.1111/cas.13642
Sas-Chen, A., Srivastava, S., and Yarden, Y. (2017). The short and the long: non-coding RNAs and growth factors in cancer progression. Biochem. Soc. Trans. 45, 51–64. doi: 10.1042/BST20160131
Shen, X., Zhang, Y., Wu, X., Guo, Y., Shi, W., Qi, J., et al. (2017). Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer Biomark. 18, 257–263. doi: 10.3233/CBM-160158
Sleutels, F., Zwart, R., and Barlow, D. P. (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. doi: 10.1038/415810a
Song, C. L., Wang, J. P., Xue, X., Liu, N., Zhang, X. H., Zhao, Z., et al. (2017). Effect of circular ANRIL on the inflammatory response of vascular endothelial cells in a rat model of coronary atherosclerosis. Cell. Physiol. Biochem. 42, 1202–1212. doi: 10.1159/000478918
Spallotta, F., Cencioni, C., Atlante, S., Garella, D., Cocco, M., Mori, M., et al. (2018). Stable oxidative cytosine modifications accumulate in cardiac mesenchymal cells from Type2 diabetes patients: rescue by alpha-ketoglutarate and TET-TDG functional reactivation. Circ. Res. 122, 31–46. doi: 10.1161/CIRCRESAHA.117.311300
Stein, C. S., Jadiya, P., Zhang, X., Mclendon, J. M., Abouassaly, G. M., Witmer, N. H., et al. (2018). Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 23, 3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002
Suzuki, H., Zuo, Y., Wang, J., Zhang, M. Q., Malhotra, A., and Mayeda, A. (2006). Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34:e63. doi: 10.1093/nar/gkl151
Tang, J., Zhong, G., Wu, J., Chen, H., and Jia, Y. (2018). Long noncoding RNA AFAP1-AS1 facilitates tumor growth through enhancer of zeste homolog 2 in colorectal cancer. Am. J. Cancer Res. 8, 892–902.
Terracciano, D., Ferro, M., Terreri, S., Lucarelli, G., D’elia, C., Musi, G., et al. (2017). Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl. Res. 184, 108–117. doi: 10.1016/j.trsl.2017.03.005
Thum, T., and Condorelli, G. (2015). Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 116, 751–762. doi: 10.1161/CIRCRESAHA.116.303549
Tsukahara, T., Matsuda, Y., and Haniu, H. (2017). Lysophospholipid-related diseases and PPARgamma signaling pathway. Int. J. Mol. Sci. 18:E2730. doi: 10.3390/ijms18122730
Vausort, M., Wagner, D. R., and Devaux, Y. (2014). Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 115, 668–677. doi: 10.1161/CIRCRESAHA.115.303836
Vecellio, M., Spallotta, F., Nanni, S., Colussi, C., Cencioni, C., Derlet, A., et al. (2014). The histone acetylase activator pentadecylidenemalonate 1b rescues proliferation and differentiation in the human cardiac mesenchymal cells of type 2 diabetic patients. Diabetes 63, 2132–2147. doi: 10.2337/db13-0731
Viereck, J., and Thum, T. (2017). Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circ. Res. 120, 381–399. doi: 10.1161/CIRCRESAHA.116.308434
Wang, J., Yang, W., Chen, Z., Chen, J., Meng, Y., Feng, B., et al. (2018). Long noncoding RNA lncSHGL recruits hnRNPA1 to suppress hepatic gluconeogenesis and lipogenesis. Diabetes 67, 581–593. doi: 10.2337/db17-0799
Wang, N., Zhu, Y., Xie, M., Wang, L., Jin, F., Li, Y., et al. (2018). Long noncoding RNA Meg3 regulates mafa expression in mouse beta cells by inactivating Rad21, Smc3 or Sin3alpha. Cell Physiol. Biochem. 45, 2031–2043. doi: 10.1159/000487983
Wang, K., Liu, F., Zhou, L. Y., Long, B., Yuan, S. M., Wang, Y., et al. (2014). The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 114, 1377–1388. doi: 10.1161/CIRCRESAHA.114.302476
Warburg, O., Wind, F., and Negelein, E. (1927). The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530. doi: 10.1085/jgp.8.6.519
Wei, F., Cai, C., Feng, S., Lv, J., Li, S., Chang, B., et al. (2015). TOX and CDKN2A/B gene polymorphisms are associated with type 2 diabetes in Han Chinese. Sci. Rep. 5:11900. doi: 10.1038/srep11900
Xu, Y., Zhang, X., Hu, X., Zhou, W., Zhang, P., Zhang, J., et al. (2018). The effects of lncRNA MALAT1 on proliferation, invasion and migration in colorectal cancer through regulating SOX9. Mol. Med. 24:52. doi: 10.1186/s10020-018-0050-5
Yan, B., Tao, Z. F., Li, X. M., Zhang, H., Yao, J., and Jiang, Q. (2014). Aberrant expression of long noncoding RNAs in early diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 55, 941–951. doi: 10.1167/iovs.13-13221
Yan, C., Chen, J., and Chen, N. (2016). Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 6:22640. doi: 10.1038/srep22640
Yoshimizu, T., Miroglio, A., Ripoche, M. A., Gabory, A., Vernucci, M., Riccio, A., et al. (2008). The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. U.S.A. 105, 12417–12422. doi: 10.1073/pnas.0801540105
Yuan, J., Liu, Z., and Song, R. (2017). Antisense lncRNA As-SLC7A11 suppresses epithelial ovarian cancer progression mainly by targeting SLC7A11. Pharmazie 72, 402–407. doi: 10.1691/ph.2017.7449
Zhang, C., Moriguchi, T., Kajihara, M., Esaki, R., Harada, A., Shimohata, H., et al. (2005). MafA is a key regulator of glucose-stimulated insulin secretion. Mol. Cell Biol. 25, 4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005
Zhang, M., Chi, X., Qu, N., and Wang, C. (2018). Long noncoding RNA lncARSR promotes hepatic lipogenesis via Akt/SREBP-1c pathway and contributes to the pathogenesis of nonalcoholic steatohepatitis. Biochem. Biophys. Res. Commun. 499, 66–70. doi: 10.1016/j.bbrc.2018.03.127
Zhang, N., Geng, T., Wang, Z., Zhang, R., Cao, T., Camporez, J. P., et al. (2018). Elevated hepatic expression of H19 long noncoding RNA contributes to diabetic hyperglycemia. JCI Insight 3:120304. doi: 10.1172/jci.insight.120304
Zhang, Y., Xue, W., Li, X., Zhang, J., Chen, S., Zhang, J. L., et al. (2016). The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624. doi: 10.1016/j.celrep.2016.03.058
Zhang, Z., Gao, W., Long, Q. Q., Zhang, J., Li, Y. F., Liu, D. C., et al. (2017). Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 7:7491. doi: 10.1038/s41598-017-07611-z
Zhao, L., Ji, G., Le, X., Wang, C., Xu, L., Feng, M., et al. (2017). Long noncoding RNA LINC00092 acts in cancer-associated fibroblasts to drive glycolysis and progression of ovarian cancer. Cancer Res. 77, 1369–1382. doi: 10.1158/0008-5472.CAN-16-1615
Zhao, Y., Zhao, J., Guo, X., She, J., and Liu, Y. (2018). Long non-coding RNA PVT1, a molecular sponge for miR-149, contributes aberrant metabolic dysfunction and inflammation in IL-1beta-simulated osteoarthritic chondrocytes. Biosci. Rep. 38:BSR20180576. doi: 10.1042/BSR20180576
Zhou, M., Gao, M., Luo, Y., Gui, R., and Ji, H. (2018). Long non-coding RNA metallothionein 1 pseudogene 3 promotes p2y12 expression by sponging miR-126 to activate platelet in diabetic animal model. Platelets doi: 10.1080/09537104.2018.1457781 [Epub ahead of print].
Zhou, X., Chen, H., Zhu, L., Hao, B., Zhang, W., Hua, J., et al. (2016). Helicobacter pylori infection related long noncoding RNA (lncRNA) AF147447 inhibits gastric cancer proliferation and invasion by targeting MUC2 and up-regulating miR-34c. Oncotarget 7, 82770–82782. doi: 10.18632/oncotarget.13165
Keywords: lncRNA, lipids, carbohydrates, amino acids, metabolism, cancer, diabetes, cardiovascular
Citation: Mongelli A, Martelli F, Farsetti A and Gaetano C (2019) The Dark That Matters: Long Non-coding RNAs as Master Regulators of Cellular Metabolism in Non-communicable Diseases. Front. Physiol. 10:369. doi: 10.3389/fphys.2019.00369
Received: 24 January 2019; Accepted: 18 March 2019;
Published: 22 May 2019.
Edited by:
Nada A. Abumrad, Washington University in St. Louis, United StatesReviewed by:
Angelo Baldassare Cefalù, University of Palermo, ItalyAndrew Carley, Wexner Medical Center, The Ohio State University, United States
Copyright © 2019 Mongelli, Martelli, Farsetti and Gaetano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonella Farsetti, YW50b25lbGxhLmZhcnNldHRpQGNuci5pdA==
Carlo Gaetano, Y2FybG8uZ2FldGFub0BpY3NtYXVnZXJpLml0
 Alessia Mongelli
Alessia Mongelli Fabio Martelli
Fabio Martelli Antonella Farsetti3*
Antonella Farsetti3*