
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 02 April 2019
Sec. Aquatic Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fphys.2019.00331
Long non-coding RNAs (lncRNA) have emerged as important regulators of lipid metabolism and have been shown to play multifaceted roles in controlling transcriptional gene regulation, but very little relevant information has been available in fish, especially in non-model fish species. With a feeding trial on a typical marine teleost tongue sole C. semilaevis followed by transcriptomic analysis, the present study investigated the possible involvement of lncRNA in hepatic mRNA expression in response to different levels of dietary DHA and EPA, which are two most important fatty acids for marine fish. An 80-day feeding trial was conducted in a flow-through seawater system, and in this trial three experimental diets differing basically in DHA/EPA ratio, i.e., 0.61 (D/E-0.61), 1.46 (D/E-1.46), and 2.75 (D/E-2.75), were randomly assigned to 9 tanks of experimental fish. A total of 124.04 G high quality genome-wide clean data about coding and non-coding transcripts was obtained in the analysis of hepatic transcriptome. Compared to diet D/E-0.61, D/E-1.46 up-regulated expression of 178 lncRNAs and 2629 mRNAs, and down-regulated that of 47 lncRNAs and 3059 mRNAs, while D/E-2.75 resulted in much less change in gene expression. The co-expression and co-localization analysis of differentially expressed (DE) lncRNA and mRNA among dietary groups were then conducted. The co-expressed DE lncRNA and mRNA were primarily enriched in GO terms such as Metabolic process, Intracellular organelle, Catalytic activity, and Oxidoreductase activity, as well as in KEGG pathways such as Ribosome and Oxidative phosphorylation. Overlap of co-expression and co-localization analysis, i.e., lncRNA–mRNA matches “XR_523541.1–solute carrier family 16, member 5 (slc16a5)” and “LNC_000285–bromodomain adjacent to zinc finger domain 2A (baz2a),” were observed in all inter-group comparisons, indicating that they might crucially mediate the effects of dietary DHA and EPA on hepatic gene expression in tongue sole. In conclusion, this was the first time in marine teleost to investigate the possible lncRNA–mRNA interactions in response to dietary fatty acids. The results provided novel knowledge of lncRNAs in non-model marine teleost, and will serve as important resources for future studies that further investigate the roles of lncRNAs in lipid metabolism of marine teleost.
Long non-coding RNAs (lncRNAs) are a class of RNA molecules with more than 200 bases that function as RNAs with little or no protein-coding capacity (Spizzo et al., 2012). LncRNAs represent a new frontier in molecular biology. More and more studies have demonstrated that lncRNAs play critical roles in various biological processes, including chromatin modification, regulation of transcription, influence of nuclear architecture and regulation of gene expression at post-transcriptional and post-translational levels, and also interact with DNA, RNA and proteins during these processes (Zhu et al., 2013; Kornfeld and Brüning, 2014; Gardini and Shiekhattar, 2015; Lopez-Pajares, 2016; Smekalova et al., 2016; Delás and Hannon, 2017; Long et al., 2017).
LncRNAs have emerged as important regulators of lipid metabolism, and have been shown to influence lipid homeostasis by controlling lipid metabolism in the liver and by regulating adipogenesis (Chen, 2016). In humans and other mammals, loss- and gain-of-function analysis of identified lncRNAs showed that lncRNAs are important regulators of a series of lipid metabolism-related processes such as plasma triglyceride accumulation (Cui et al., 2015; Li et al., 2015), cholesterol transportation, apolipoprotein A1 (APOA1) expression (Halley et al., 2014; Hu et al., 2014), adipose proliferation (Xu et al., 2010; Liu et al., 2014a,b), brown adipose tissue activation (Alvarez-Dominguez et al., 2015), and adipogenesis (Cooper et al., 2014; Divoux et al., 2014; Zhao et al., 2014; Gernapudi et al., 2015; Xiao et al., 2015).
In fish, however, little information has been available either about the identification and characterization of lncRNAs or about the roles of lncRNAs in lipid metabolism. Limited studies on lncRNAs in fish have been restricted to model fish species, and most of the studies focused on developmental biology (Kaushik et al., 2013; Liu et al., 2013; Haque et al., 2014; Dhiman et al., 2015; Al-Tobasei et al., 2016; Wang et al., 2016, 2017; Sarangdhar et al., 2017; Hu et al., 2018). Therefore, following our previous studies on lipid/fatty acid nutrition of marine fish (Zuo et al., 2012a,b; Xu et al., 2016, 2017), the present study was aimed at investigating the potential involvement of lncRNAs in effects of dietary fatty acids on a non-model marine teleost tongue sole Cynoglossus semilaevis, which is also an important aquaculture species. A feeding trial was conducted in this study, followed by hepatic transcriptome analysis.
Docosahexaenoic acid (DHA, 22:6n-3) and eicosapentaenoic acid (EPA, 20:5n-3) are the most important essential fatty acids for marine fish. Previous studies have widely demonstrated the critical roles of DHA and EPA in a series of physiological functions of marine fish such as functions of visual and neural systems (Bell et al., 1995; Furuita and Takeuchi, 1998; Ishizaki et al., 2000, 2001; Benítez-Santana et al., 2007; Noffs et al., 2009), bone development (Gapasin and Duray, 2001; Roo et al., 2009), pigmentation (Villalta et al., 2008; Vizcaíno-Ochoa et al., 2010), stress resistance (Kanazawa, 1997; Liu et al., 2002), immune response (Zuo et al., 2012b; Xu et al., 2016), and reproduction (Wilson, 2009; Xu et al., 2017). However, more fundamental mechanisms involved are still elusive. How DHA and EPA regulate physiological processes at transcriptional and post-transcriptional levels needs to be elucidated. Moreover, compared to total DHA+EPA contents, the effects of DHA/EPA ratio were interesting as well but relatively less studied (Sargent et al., 1999; Kim et al., 2002; Lee et al., 2003; Wu et al., 2003; Skalli and Robin, 2004; Hamre and Harboe, 2008; Wilson, 2009; Lund and Steenfeldt, 2011; ØStbye et al., 2011; Tocher, 2015). Our previous studies with marine species such as large yellow croaker Larmichthys crocea (Zuo et al., 2012a), Japanese seabass Lateolabrax japonicus (Xu et al., 2016) and tongue sole (Xu et al., 2017) have shown the significantly different roles of DHA and EPA in some physiological processes such as non-specific immune response and gonadal steroidogenesis. In the present study, taking advantage of completely sequenced genome data on tongue sole C. semilaevis, a genome-wide hepatic transcriptome analysis was conducted with tongue sole following an 80-day feeding trial with diets containing different DHA/EPA ratios. Potential lncRNA–mRNA interactions were thereafter analyzed based on the co-expression and co-localization analysis of differentially expressed lncRNAs and mRNAs among dietary groups. The results will provide novel knowledge about lncRNAs in non-model marine fish, and will serve as important resources for future studies that further investigate the roles of lncRNAs in lipid metabolism of marine teleost.
The experimental diets, experimental fish and feeding procedure has been described in a previous study of ours (Xu et al., 2018). Briefly, different levels of EPA enriched oil (containing 11.2% DHA and 52.0% EPA; in the form of triglyceride; Xi'an Renbang Biological Science and Technology Co., Ltd., Xi'an, China) and DHA enriched oil (containing 69.5% DHA and 6.6% EPA; in the form of triglyceride; Xi'an Renbang Biological Science and Technology Co., Ltd., Xi'an, China) were supplemented to the basal diet to obtain different DHA/EPA ratios, 0.61, 1.46, and 2.75, and the corresponding diets were designated as D/E-0.61, D/E-1.46, and D/E-2.75, respectively (Tables 1,2).
The feeding trial was conducted in a flow-through seawater system. Juvenile tongue sole C. semilaevis with an average initial weight of 23.40 ± 0.45 g were used in the feeding trial. Each diet was randomly assigned to triplicate tanks of 30 fish each. Fish were hand-fed to apparent satiation twice daily. The feeding trial lasted for 80 days. At the end of the feeding trial, after anesthetized with eugenol, liver samples from 6 fish each tank were collected, frozen with liquid nitrogen and stored at −80°C prior to analysis. The feeding trial was carried out in accordance with the recommendations of Guidelines on Management of Experimental Animals, Animal Care and Use Committee of Yellow Sea Fisheries Research Institute. All sampling protocols, as well as fish rearing practices, were reviewed and approved by the Animal Care and Use Committee of Yellow Sea Fisheries Research Institute.
Total RNA in liver samples was isolated using RNAiso Plus (TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China). RNA degradation and contamination were monitored on 1% agarose gels. No genomic DNA contamination was observed. RNA purity was checked using the Nano Photometer® spectrophotometer (IMPLEN, Westlake Village, CA, USA). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Flurometer (Life Technologies, Carlsbad, CA, USA). RNA integrity was assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA).
A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations and all samples had RIN values > 8. Six individual samples from the same experimental tank were pooled in equal amounts to obtain a pooling sample for this replicate tank. Nine pooling samples were then used to prepare 9 separate Illumina sequencing libraries (three biological replicates for each dietary group).
After ribosomal RNA was removed with Epicentre Ribo-ZeroTM rRNA Removal Kit (Epicentre, Madison, WI, USA), and rRNA free residues were cleaned up by ethanol precipitation, sequencing libraries were generated using the rRNA-depleted RNA by NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's instructions. Briefly, fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H−). Second strand cDNA synthesis was subsequently performed using DNA Polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3′ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of 150 ~200 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter, Beverly, MA, USA). Then 3 μl USER Enzyme (NEB, Ipswich, MA, USA) was added to size-selected and adaptor-ligated cDNA at 37°C for 15 min followed by 5 min of 95°C treatment before the PCR process. PCR was then carried out with Phusion High-Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified with AMPure XP system (Beckman Coulter, Beverly, USA), and the library quality was assessed on Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, USA).
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer's instructions. After cluster generation, the libraries were sequenced on an Illumina Hiseq2500 platform (Illumina, Inc., San Diego, CA, USA) and 150 bp paired-end reads were generated.
Raw data (raw reads) in fastq format were firstly processed through in-house perl scripts. Clean data (clean reads) were obtained by removing reads containing adapter or poly-N, as well as low quality reads from raw data. At the same time, Q20, Q30, and GC content of the clean data were calculated. All the downstream analysis were based on clean data with high quality. The paired-end clean reads were then aligned to the reference genome (RefSeq assembly accession: GCA_000523025.1; https://www.ncbi.nlm.nih.gov/genome/?term=Cynoglossus%20semilaevis) using TopHat v2.0.9. The mapped reads of each sample were assembled by both Scripture (beta2) (Guttman et al., 2010) and Cufflinks (v2.1.1) (Trapnell et al., 2010) in a reference-based approach. Cuffdiff (v2.1.1) was also used to calculate FPKMs (Fragments Per Kilo-base of exon per Million fragments mapped) of both lncRNAs and coding genes in each sample. FPKMs were calculated based on the length of the fragments and reads count mapped to this fragment. Gene FPKMs were computed by summing the FPKMs of transcripts in each gene group. Cuffdiff provides statistical routines for determining differential expression in digital transcript or gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted with the Benjamini and Hochberg's approach for controlling the false discovery rate (FDR). Transcripts or genes with an adjusted P-value (P-adj) < 0.05 were assigned as differentially expressed (DE) between experimental groups (three biological replicates for each group).
Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) or lncRNA target genes was implemented by the GOseq R package, and GO terms with corrected P < 0.05 were considered significantly enriched by DEGs. KOBAS software was used to test the statistical enrichment of differentially expressed genes or lncRNA target genes in KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways (http://www.genome.jp/kegg/).
To validate the Illumina sequencing data, 10 mRNA and ten lncRNA, which were selected from the most potential “lncRNA-mRNA” interactions based on the co-expression and co-localization analysis of DElncRNAs and DEmRNA among dietary groups, were tested for qRT-PCR analysis, using the same RNA samples for the transcriptome profiling. Specific primers were designed based on the data from GenBank (Table 3), and beta-2-microglobulin (β-2-m) was used as the reference gene according to our previous screening. The real-time PCR was carried out with SYBR Green Real-time PCR Master Mix (TaKaRa Biotechnology (Dalian) Co., Ltd., Dalian, China) in a quantitative thermal cycler (Mastercycler eprealplex, Eppendorf, German). The amplification efficiency for all primers, which was estimated by standard curves based on a 6-step 10-fold dilution series of target template, was within 95 ~105%, and the coefficients of linear regression (R2) were more than 0.99. The detailed program was similar with Xu et al. (2014). The mRNA expression levels were studied by qRT-PCR method: 2−ΔΔCT (Livak and Schmittgen, 2001). The qRT-PCR data were subjected to one-way analysis of variance (ANOVA) in SPSS 16.0 (SPSS Inc., Chicago, USA) for Windows. Differences between means were tested by Tukey's multiple range test. The level of significance was chosen at P < 0.05, and the results were presented as means of triplicate tanks ± standard errors.
Detailed information about methods and materials used in the current study are available from the corresponding author on reasonable request.
In the transcriptomic analysis, three pooled liver RNA samples were prepared for each dietary group (D/E-0.61, D/E-1.46, and D/E-2.75). Nine cDNA libraries were then constructed to perform Illumina sequencing. A total of 278,269,246, 269,325,362, and 279,307,386 clean reads were obtained for groups D/E-0.61, D/E-1.46, and D/E-2.75 respectively, giving rise to total clean bases of 41.74, 40.40 and 41.90 G, respectively (Supplementary Table 1). The average Q20 and Q30 (the sequencing error rate at 1 and 0.1% respectively) of the experimental samples was 96.17 and 90.17% respectively, indicating the high accuracy of the sequencing processes. Raw reads were deposited at the National Center for Biotechnology Information (NCBI)'s Sequence Read Archive under Accession No. SRP127310 (D1E2, D3E2, and D3E1 in the archived data represents groups D/E-0.61, D/E-1.46, and D/E-2.75 respectively). The reads were mapped on the genome of C. semilaevis and the average mapping rates of groups D/E-0.61, D/E-1.46, and D/E-2.75 was 77.62, 76.09, and 72.43%, respectively (Supplementary Table 2). The classification analysis of the reads showed that the total reals were comprised of (average): exon, 0.07%; mRNA, 67.72%; misc_RNA, 0.03%; ncRNA, 0.30%; tRNA, 1.19%; and others, 30.69% (Supplementary Table 3). Regarding the comparison among dietary groups, compared to groups D/E-1.46 and D/E-2.75, group D/E-0.61 had higher proportion of mRNA (70.21% vs. 66.52% and 66.43%), misc_RNA (0.04% vs. 0.02% and 0.02%), and ncRNA (0.41% vs. 0.27% and 0.22%). This indicated that dietary DHA/EPA ratio might have significant effects on transcription of different types of RNA.
Large-scale analysis of the mammalian transcriptome have shown that the number and types of lncRNAs far exceed those of protein-coding mRNAs, while a small proportion of lncRNAs have been reported to have biological functions (Carninci et al., 2005; Birney et al., 2007; Carninci and Hayashizaki, 2007; Kapranov et al., 2007). In fish, however, very little information has been available about the identification and function prediction of lncRNAs. Only limited studies have been published in model fish such as zebrafish Danio rerio and rainbow trout Oncorhynchus mykiss (Kaushik et al., 2013; Liu et al., 2013; Haque et al., 2014; Dhiman et al., 2015; Al-Tobasei et al., 2016; Wang et al., 2016). Moreover, previous studies in both mammals and zebrafish have shown that lncRNAs are less conserved than protein-coding genes (Guttman et al., 2010; Cabili et al., 2011; Ulitsky et al., 2011; Derrien et al., 2012). Therefore, specific investigation is needed to identify and characterize the lncRNA profile in a certain fish species.
In the present study with tongue sole, based on the transcript assembly results, lncRNA was filtered following five steps: (1) number of exon (≥2); (2) length of transcript (>200bp); (3) comparison with annotated transcript (Cuffcompare software): transcripts overlapped with annotated coding exon were eliminated, and transcripts overlapped with annotated lncRNAs were regarded as annotated lncRNA; (4) expression level (Cuffquant software, FPKM≥0.5); (5) prediction of coding potential: transcripts containing no coding potential predicted by any mainstream software (CPC, CNCI, and PFAM) was recognized as novel lncRNAs (see Supplementary Figure 1 for the results), and transcripts with coding potential predicted by at least one software were designated as Transcripts of Uncertain Coding Potential (TUCP). The filtration results following these steps were presented in Figure 1.
Finally, 789 annotated lncRNAs and 638 novel lncRNAs were obtained. These lncRNAs contained 69.9% lincRNA (long intergenic non-coding RNA), 30.1% antisense_lncRNA, but no intronic_lncRNA. Either annotated or novel lncRNAs were shorter and had less exons than mRNA (Figures 2, 3). Also, the average expression level of lncRNA was lower compared to mRNA (Figure 4). These characteristics of tongue sole lncRNAs were similar to those found in other species (Guttman et al., 2010; Cabili et al., 2011; Young and Ponting, 2013; Liang et al., 2015; Núñez-Acuña et al., 2017).
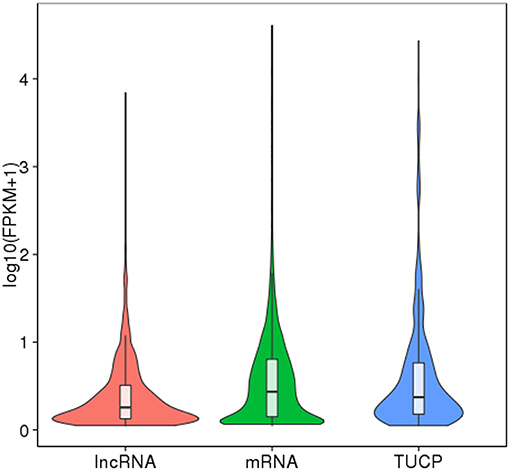
Figure 4. FPKM distribution of transcripts. The boxplot inside shows the FPKM distribution. The five characteristic values indicate maximum, upper quartile, mid-value, lower quartile, and minimum, respectively. The violin diagram shows the FPKM density distribution. The width of violin diagram represents the density of transcript under a certain expression level.
Non-coding RNAs have emerged as important regulators of cellular and systemic lipid metabolism (Chen, 2016; Zhou et al., 2016). In particular, the enigmatic class of long non-coding RNAs have been shown to play multifaceted roles in controlling transcriptional and posttranscriptional gene regulation (Kornfeld and Brüning, 2014; Gardini and Shiekhattar, 2015; Lopez-Pajares, 2016; Delás and Hannon, 2017; Long et al., 2017; Mathy and Chen, 2017).
In the present study, the experimental diets significantly affected the expression levels of both lncRNAs and mRNAs (Table 4). The most significant difference existed between groups D/E-0.61 and D/E-1.46. Compared to group D/E-0.61, D/E-1.46 significantly (adjusted P < 0.05) up-regulated expression of 178 lncRNA and 2629 mRNA, and down-regulated that of 47 lncRNA and 3059 mRNA. However, compared to group D/E-0.61, D/E-2.75 only significantly up-regulated expression of 78 lncRNA and 1925 mRNA, and down-regulated that of 42 lncRNA and 2015 mRNA. Generally, this result indicated that dietary DHA/EPA ratio regulated gene transcription in tongue sole in a dose-dependent manner. This was in accordance with the effects of dietary DHA/EPA ratio on growth performances and other physiological status such as lipid accumulation (Xu et al., 2018). As reported previously, compared to group D/E-0.61, D/E-1.46 but not D/E-2.75 significantly increased the growth rate and whole-body lipid content of juvenile tongue sole. In addition, despite the large number of differentially expressed (DE) genes among dietary groups, only 24 DElncRNA (about 16%) and 638 (about 16%) DEmRNA were overlapped among the three inter-group comparisons, D/E-1.46 vs. D/E-0.61, D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75 (Figure 5). This indicated that each DHA/EPA ratio exerted a different regulation of the liver transcriptome. Moreover, the amount of DElncRNA was in proportion to that of DEmRNA among the inter-group comparisons. This suggested that the transcriptional activity of coding and non-coding RNAs in tongue sole liver was co-expressed in response to dietary DHA/EPA ratio, which was to be evidenced by the co-expression analysis described below.
To confirm the DEG results from the transcriptomic assay, 10 mRNA and 10 lncRNA, which were selected from most potential “lncRNA-mRNA” interactions based on the co-expression and co-localization analysis of DElncRNA and DEmRNA described below, were tested for quantitative RT-PCR analysis. The results showed that transcription of 18 out of 20 selected genes was in good accordance with the transcriptomic results (Table 5, Figure 6). The validation results confirmed the high accuracy of the transcriptomic results.
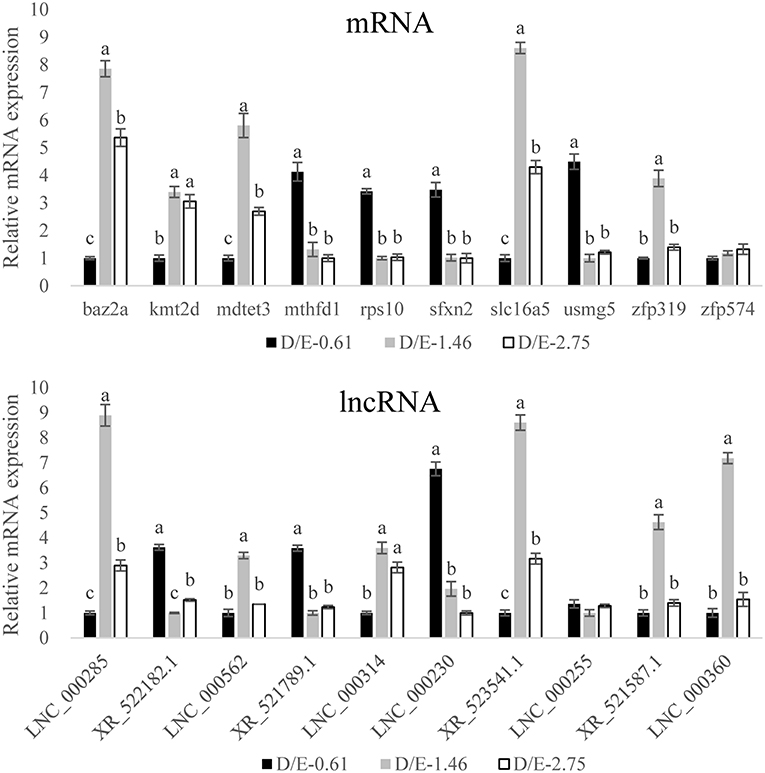
Figure 6. Validation of the transcriptome results by qRT-PCR measurement. The gene levels were expressed relative to β-2-microglobulin. Results are expressed as means ± standard error. For a certain mRNA or lncRNA, bars not sharing same letters denote significant (P < 0.05) difference in gene expression.
In order to estimate the potential lncRNA–mRNA interactions in response to dietary DHA/EPA ratio and consequently to predict the potential roles of the obtained lncRNAs in mRNA expression, co-expression and co-localization relationship of DElncRNA and DEmRNA among dietary groups were analyzed (Table 6). The co-expression analysis showed that 12888, 7659, and 2765 co-expression relationships were observed in inter-group comparisons D/E-1.46 vs. D/E-0.61, D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75, respectively. The significant expression correlations between certain lncRNAs and protein-coding genes have also been observed in a previous study on rainbow trout investigating the intestinal lncRNA and coding RNAs transcription in response to functional diets (Núñez-Acuña et al., 2017). These results highlighted that the regulation of numerous protein-coding genes by dietary nutrients was highly correlated with lncRNAs. The GO enrichment analysis of the co-expression relationships showed that the most enriched GO terms were Metabolic process (including Protein metabolic process, Primary metabolic process, Macromolecule metabolic process, and Organic substance metabolic process), Gene expression, Cellular macromolecule bioynthetic process, Introcellular organelle (including Intracellular non-membrane-bounded organelle), and Oxidoreductase activity (see Figure 7 for D/E-1.46 vs. D/E-0.61, see Supplementary Figures 2A, S3A for D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75, respectively). The KEGG pathway enrichment analysis of the co-expression relationships showed that the most enriched pathways were Ribosome and Oxidative phosphorylation (see Figure 8 for D/E-1.46 vs. D/E-0.61, see Supplementary Figures 2B,S3B for D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75, respectively). These enrichment results were highly similar with the enrichment results for DEmRNA among experimental groups (Supplementary Figures 4,5), indicating the possible wide involvement of lncRNA in the regulation of mRNA expression. On the other hand, these results also indicated the effects of dietary DHA/EPA on a wide range of physiological processes in fish liver, which, however, was not the focus of the present study.
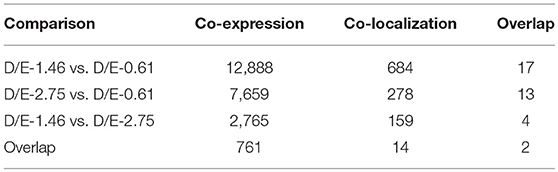
Table 6. Analysis of co-expression and co-localizaiton of differentially expressed lncRNA and mRNA among dietary groups.
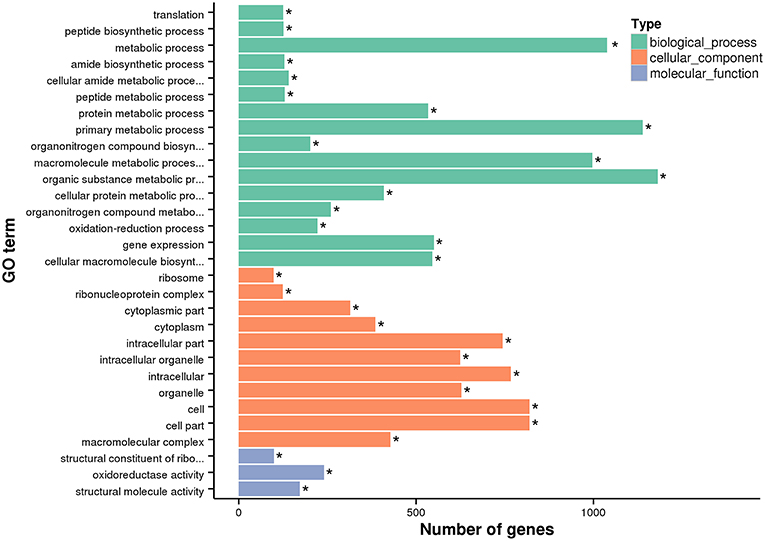
Figure 7. GO enrichment analysis for differentially expressed mRNA in lncRNA-mRNA co-expression analysis for D/E-1.46 vs. D/E-0.61. * denotes significant enrichment (Padj < 0.05, Padj is the adjusted P-value). The GO terms without full title: (1) Cellular amide metabolic process; (2) Organonitrogen compound biosynthetic process; (3) Macromolecule metabolic process; (4) Organic substance metabolic process; (5) Cellular protein metabolic process; (6) Organonitrogen compound metabolic process; (7) Cellular macromolecule biosynthetic process; (8) Structural constituent of ribosome.
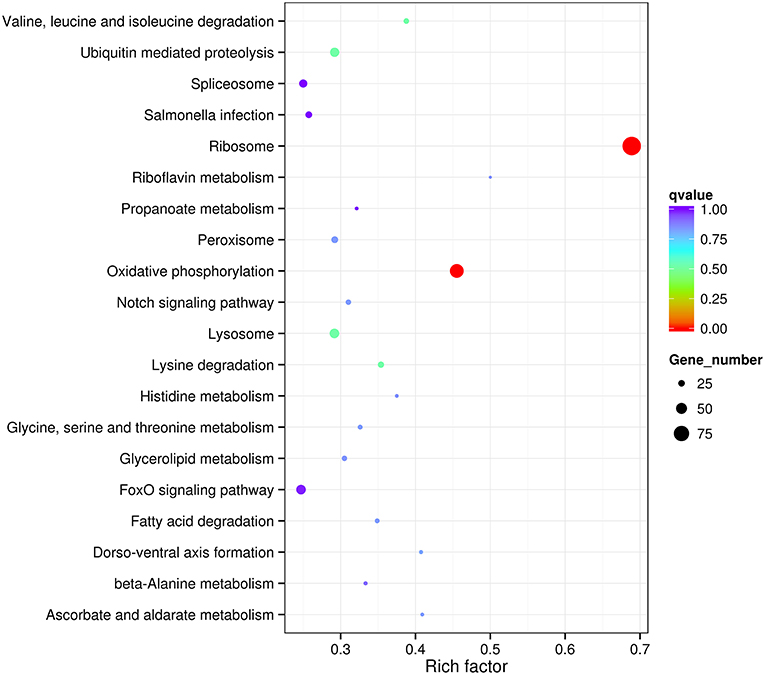
Figure 8. Statistics of KEGG pathway enrichment for differentially expressed mRNA in lncRNA-mRNA co-expression analysis for D/E-1.46 vs. D/E-0.61. Rich factor is the ratio of number of differentially expressed genes in a certain pathway to number of all annotated genes in this pathway. qvalue is corrected P-value by multiple hypothesis test. q-value < 0.05 denotes significant differences.
As some lncRNAs could regulate the expression of coding genes near genomic regions, the co-localization of DElncRNA and DEmRNA among dietary groups were also analyzed. In total, 684, 278, and 159 co-localization relationships were observed in comparisons D/E-1.46 vs. D/E-0.61, D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75, respectively. The co-localization relationships were primarily enriched in GO terms such as RNA metabolic process, Membrane-bounded organelle, Nucleus, RNA polymerase, and Nucleoside-triphosphatase regulator activity, as well as in KEGG pathways such as Ribosome and Glycerolipid metabolism (Supplementary Figures 6–8). Compared to the co-expression relationships, the co-localization relationships were more related to gene expression regulation, indicating the distance from target mRNA probably influenced the transcription-regulating effects of lncRNAs.
Considering that concurrent existence of co-expression and co-localization relationships of a certain lncRNA-mRNA match must increase the possibility of lncRNA-mRNA interaction, the overlap of co-expression and co-localization relationships of DElncRNA and DEmRNA was analyzed (Table 7). A total of 17, 13, and 4 overlapped lncRNA-mRNA matches was obtained for D/E-1.46 vs. D/E-0.61, D/E-2.75 vs. D/E-0.61, and D/E-1.46 vs. D/E-2.75, respectively. Two overlapped matches, “XR_523541.1–solute carrier family 16, member 5 (slc16a5)” and “LNC_000285–bromodomain adjacent to zinc finger domain 2A (baz2a)” were observed in all the three inter-group comparisons, indicating that they might play crucial roles in the effects of dietary DHA and EPA on hepatic gene transcription in tongue sole.
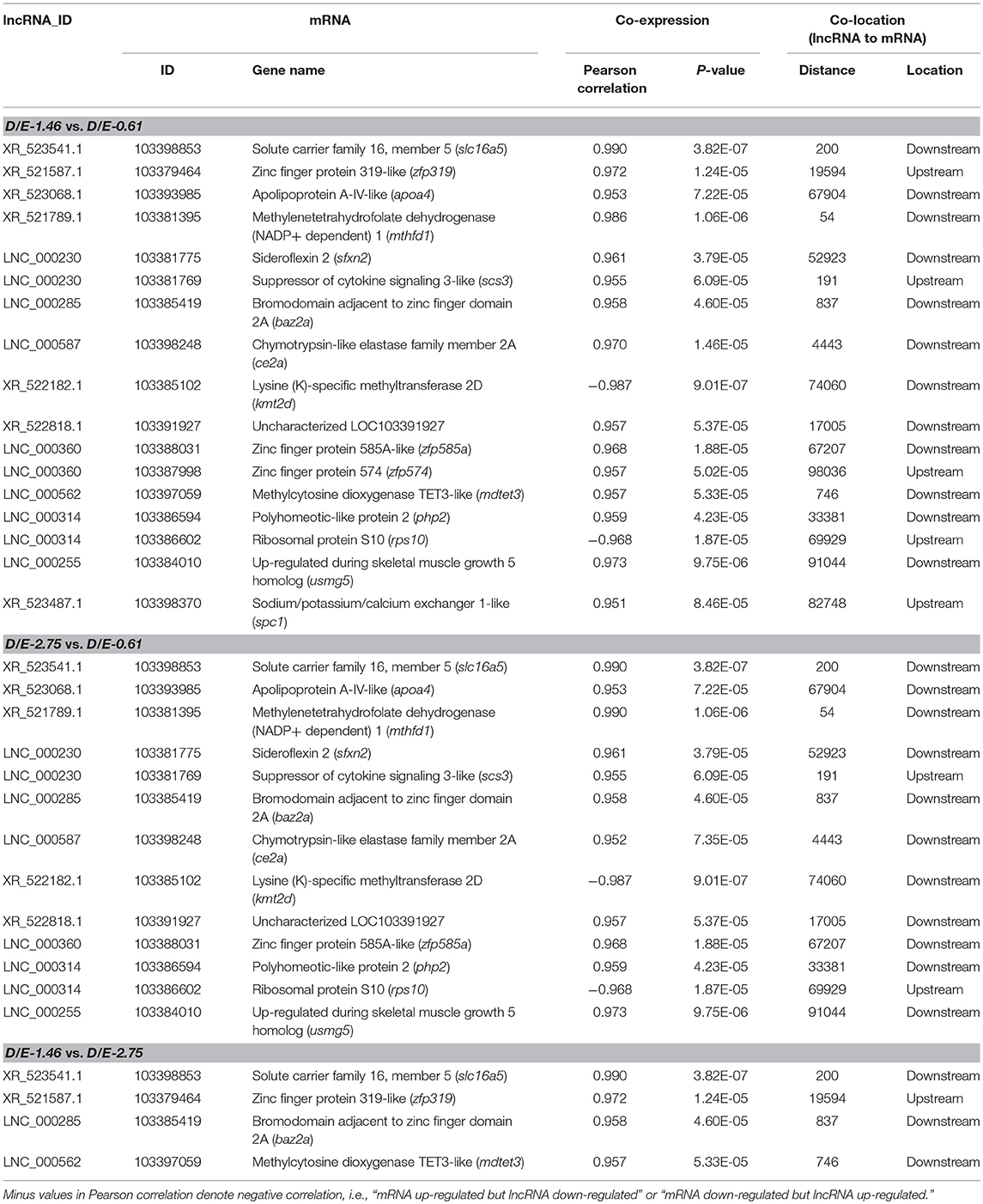
Table 7. Overlap of co-expression and co-localization relationships between differentially expressed lncRNAs and mRNAs among dietary groups.
Solute carrier family 16 (SLC16) is a proton-linked monocarboxylate transporter (MCT), which comprises 14 members (Halestrap and Meredith, 2004), and mainly catalyzes the rapid transport of many monocarboxylates across the plasma membrane. While function of some members of this family such as SLC16A1 (also known as MCT1), SLC16A7 (MCT2), SLC16A8 (MCT3), and SLC16A3 (MCT4) have been elucidated (Bröer et al., 1997, 1998; Lin et al., 1998; Grollman et al., 2000; Manning Fox et al., 2000), functions of SLC16A5 (MCT6) are still unknown. Considering the possible roles of SLC16A5 in transmembrane transport of lactate, pyruvate, and ketone bodies, the significant difference in hepatic transcription of slc16a5 among groups with different DHA/EPA ratios indicated that DHA and EPA may have different efficiency and priority in energy supply in tongue sole. The co-expression and co-localization relationship of XR_523541.1 and slc16a5 suggested that the lncRNA XR_523541.1 may be involved in the regulation of slc16a5 transcription by dietary DHA/EPA. Precise functions of both XR_523541.1 and slc16a5 need to be elucidated by future studies.
Baz2a is an essential component of nucleolar remodeling complex, a complex that mediates silencing of a fraction of rDNA by recruiting histone-modifying enzymes and DNA methyltransferases, leading to heterochromatin formation and transcriptional silencing (Gu et al., 2015). The co-expression and co-localization relationship of “LNC_000285–baz2a” indicated that the lncRNA LNC_000285 probably mediated the regulatory effects of dietary DHA/EPA on gene expression through promoting heterochromatin formation. Inducing heterochromatin formation and suppressing mobile elements were major mechanisms of transcription-regulating effects of non-coding RNAs (Cusanelli and Chartrand, 2015; Meller et al., 2015; Acharya et al., 2017; da Rocha and Heard, 2017; Oliva-Rico and Herrera, 2017). In human pancreatic ductal adenocarcinoma, the lncRNA HOTAIR contributed in the silence of MicroRNA-34a by enchancer of zeste homolog 2 through induction of heterochromatin formation (Li et al., 2016). Microsatellite repeat DXZ4-associated long non-coding RNAs also had developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat (Figueroa et al., 2015). Other evidence showed that lncRNA maturation to initiate heterochromatin formation in the nucleolus is required in exit from pluripotency in embryonic stem cells (Savić et al., 2014).
Besides “LNC_000285–baz2a,” the overlapped matches “LNC_000562–methylcytosine dioxygenase TET3-like (mdtet3)” and “LNC_000314–polyhomeotic-like protein 2 (php2)” also provided potential evidence for the roles of lncRNAs in chromatin remodeling. Mdtet3 catalyzes DNA demethylation and thus plays important roles in epigenetic chromatin reprogramming and maintenance of genome stability (Xu et al., 2012; Deplus et al., 2013; Jiang et al., 2017). Php2 is a component of a Polycomb group (PcG) multiprotein PRC1-like complex, which is required to maintain the transcriptionally repressive state of many genes, and acts via chromatin remodeling and modification of histones (Isono et al., 2005).
The overlapped match “XR_522182.1–lysine (K)-specific methyltransferase 2D (kmt2d)” was observed in inter-group comparisons D/E-2.75vs. D/E-0.61 and D/E-1.46vs. D/E-0.61. As a histone methyltransferase, Kmt2d methylates 'Lys-4' of histone H3 into H3K4me, which represents a specific tag for epigenetic transcriptional activation (Cho et al., 2007; Lan et al., 2007). This result indicated the potential roles of lncRNA in epigenetic transcriptional regulation.
Other mechanisms of transcription-regulating effects of lncRNAs were also indicated from the present results. For example, several overlapped “lncRNA–zinc finger protein” matches, such as “XR_521587.1–zinc finger protein 319-like (zfp319),” “LNC_000360–zinc finger protein 574 (zfp574),” and “LNC_000360–zinc finger protein 585A-like (zfp585a),” were observed in the co-expression and co-localization analysis of DElncRNAs and DEmRNAs. Interacting with these zinc finger proteins may be a possible way of lncRNA functioning on gene expression.
Interactions of other lncRNAs with some mRNAs observed in overlapped “lncRNA–mRNA” matches of the current analysis have been reported in human and terrestrial animal studies. For example, the lncRNA–mRNA match “XR_523068.1–apolipoprotein A-IV (apoa4)” was overlapped in the co-expression and co-localization analysis in the present study. A study on mice has shown that an anti-sense lncRNA (APOA4-AS) concordantly and specifically regulated apoa4 expression both in vitro and in vivo with the involvement of the mRNA stabilizing protein HuR (Qin et al., 2016). The alignment analysis did not find homology between mice APOA4-AS and tongue sole XR_523068.1. Moreover, mice APOA4-AS was transcribed from the opposite strand of apoa4 locus (+ strand), and partially overlapped with apoa4 exon 3 and 3'-UTR, while tongue sole XR_523068.1 was far away from tongue sole apoa4 (67904 bp downstream). In mice, gene expression of another apolipoprotein APOC2 was also reported to be regulated by the lncRNA lncLSTR (Li et al., 2015). These results indicated that lncRNAs may play certain roles in gene expression of apolipoproteins, which are key components of lipoproteins and play important roles in lipid transportation. In addition, the mRNA expression of apoa4 in response to dietary DHA/EPA ratio was negatively correlated with the hepatic lipid content of the experimental fish (Xu et al., 2018). The lower hepatic apoa4 expression in group D/E-1.46 may reduce the transportation of lipid out of liver, and thus contribute to higher hepatic lipid accumulation in this group.
Interactions between lncRNAs and suppressor of cytokine signaling 3-like (scs3) or methylenetetrahydrofolate dehydrogenase (NADP+ dependent) (mthfd) have also been observed in human studies. A recent study on postmenopausal osteoporosis observed a probable interaction between lncRNA LINC00963 and scs3 based on analysis of DElncRNA–DEmRNA co-expression network (Fei et al., 2018). However, different from the present study which showed that lncRNA LNC_000230 was 191 bp upstream relative to co-expressed scs3, in that study LINC00963 and scs3 were located in different chromosomes. Regarding mthfd, a study with MCF-7 breast cancer cells showed that a lncRNA, taurine-upregulated gene 1 (tug1), positively regulated the gene expression of mthfd2 (Zhao and Ren, 2016). Tug1 and mthfd2 were also located in different chromosomes, while in the present study, lncRNA XR_521789.1 was 54 bp downstream relative to co-expressed mthfd1.
Other overlapped “lncRNA–mRNA” matches in the co-express and co-localization analysis, such as “LNC_000587 - chymotrypsin-like elastase family member 2A (ce2a),” “LNC_000230–sideroflexin 2 (sfxn2),” “XR_523487.1–sodium/potassium/calcium exchanger 1-like (spc1),” “LNC_000314–ribosomal protein S10 (rps10),”and “LNC_000255–up-regulated during skeletal muscle growth 5 homolog (usmg5),”were novel findings of the present study. No similar or relevant lncRNA-mRNA interactions have been reported in other studies. These findings provided useful sources for further investigation on lncRNA–mRNA interactions, especially regarding the effects of dietary fatty acids.
In conclusion, this was the first time in marine teleost to investigate the possible lncRNA-mRNA interactions in response to dietary fatty acids. The results provided novel knowledge of lncRNAs in non-model marine teleost, and will serve as important resources for future studies that further investigate the roles of lncRNAs in lipid metabolism of marine teleost. Potential lncRNA–mRNA interactions “XR_523541.1–slc16a5” and “LNC_000285–baz2a” may play important roles in effects of dietary DHA/EPA on hepatic gene transcription in tongue sole.
DE, differentially expressed; DEG: differentially expressed gene; slc16a5, solute carrier family 16, member 5; baz2a: bromodomain adjacent to zinc finger domain 2a; mct, monocarboxylate transporter; mdtet3, methylcytosine dioxygenase tet3-like; php2, polyhomeotic-like protein 2; kmt2d, lysine (k)-specific methyltransferase 2d; zfp, zinc finger protein; apo, apolipoprotein; scs3, suppressor of cytokine signaling 3-like; mthfd1, methylenetetrahydrofolate dehydrogenase (nadp+ dependent) 1; ce2a, chymotrypsin-like elastase family member 2a, sfxn2; sideroflexin 2; spc1, sodium/potassium/calcium exchanger 1-like; rps10, ribosomal protein s10; usmg5, up-regulated during skeletal muscle growth 5 homolog.
The raw reads obtained from the transcriptomic analysis have been deposited at the National Center for Biotechnology Information (NCBI)'s Sequence Read Archive under Accession No. SRP127310. Other raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
HX and ML designed the study and wrote the manuscript. LC conducted the feeding trial. HX, LC, and BS did the transcriptomic analysis. LC and YW did the qRT-PCR study. All authors read and approved the final version of the manuscript.
This work was supported by National Natural Science Foundation of China (31772862), National Key R&D Program of China (2018YFD0900400), Central Public-interest Scientific Institution Basal Research Fund, CAFS (2018HY-ZD0505), and China Agriculture Research System (CARS-47-G15).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2019.00331/full#supplementary-material
Acharya, S., Hartmann, M., and Erhardt, S. (2017). Chromatin-associated noncoding RNAs in development and inheritance. WIREs RNA 8:e1435. doi: 10.1002/wrna.1435
Al-Tobasei, R., Paneru, B., and Salem, M. (2016). Genome-wide discovery of long non-coding RNAs in rainbow trout. PLoS ONE 11:e0148940. doi: 10.1371/journal.pone.0148940
Alvarez-Dominguez, J. R., Bai, Z., Xu, D., Yuan, B., Lo, K. A., Yoon, M. J., et al. (2015). De Novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metab. 21, 764–776. doi: 10.1016/j.cmet.2015.04.003
Bell, M. V., Batty, R. S., Dick, J. R., Fretwell, K., Navarro, J. C., and Sargent, J. R. (1995). Dietary deficiency of docosahexaenoic acid impairs vision at lowlight intensities in juvenile herring (Clupea harengus L.). Lipids 30, 443–449. doi: 10.1007/BF02536303
Benítez-Santana, T., Masuda, R., Carrillo, E. J., Ganuza, E., Valencia, A., Hernández-Cruz, C. M., et al. (2007). Dietary n-3 HUFA deficiency induces a reduced visual response in gilthead seabream Sparus aurata larvae. Aquaculture 264, 408–417. doi: 10.1016/j.aquaculture.2006.10.024
Birney, E., Stamatoyannopoulos, J. A., Dutta, A., Guig,ó, R., Gingeras, T. R., Margulies, E. H., et al. (2007). Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature 447, 799–816. doi: 10.1038/nature05874
Bröer, S., Rahman, B., Pellegri, G., Pellerin, L., Martin, J. L., Verleysdonk, S., et al. (1997). Comparison of lactate transport in astroglial cells and monocarboxylate transporter 1 (MCT 1) expressing Xenopus laevis oocytes. Expression of two different monocarboxylate transporters in astroglial cells and neurons. J. Biol. Chem. 272, 30096–30102. doi: 10.1074/jbc.272.48.30096
Bröer, S., Schneider, H. P., Bröer, A., Rahman, B., Hamprecht, B., and Deitmer, J. W. (1998). Characterization of the monocarboxylate transporter 1 expressed in Xenopus laevis oocytes by changes in cytosolic pH. Biochem. J. 333, 167–174.
Cabili, M. N., Trapnell, C., Goff, L., Koziol, M., Tazon-Vega, B., Regev, A., et al. (2011). Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927. doi: 10.1101/gad.17446611
Carninci, P., and Hayashizaki, Y. (2007). Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 17, 139–144. doi: 10.1016/j.gde.2007.02.008
Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M. C., Maeda, N., et al. (2005). The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. doi: 10.1126/science.1112014
Chen, Z. (2016). Progress and prospects of long noncoding RNAs in lipid homeostasis. Mol. Metab. 5, 164–170. doi: 10.1016/j.molmet.2015.12.003
Cho, Y. W., Hong, T., Hong, S., Guo, H., Yu, H., Kim, D., et al. (2007). PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282, 20395–20406. doi: 10.1074/jbc.M701574200
Cooper, D. R., Carter, G., Li, P., Patel, R., Watson, J. E., and Patel, N. A. (2014). Long non-coding RNA NEAT1 associates with SRp40 to temporally regulate PPARgamma2 splicing during adipogenesis in 3T3-L1 cells. Genes 5, 1050–1063. doi: 10.3390/genes5041050
Cui, M., Xiao, Z., Wang, Y., Zheng, M., Song, T., Cai, X., et al. (2015). Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through a miR-9-mediated RXRA signaling pathway. Cancer Res. 75, 846–857. doi: 10.1158/0008-5472.CAN-14-1192
Cusanelli, E., and Chartrand, P. (2015). Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front. Genet. 6:143. doi: 10.3389/fgene.2015.00143
da Rocha, S. T., and Heard, E. (2017). Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat. Struct. Mol. Biol. 24, 197–204. doi: 10.1038/nsmb.3370
Delás, M. J., and Hannon, G. J. (2017). LncRNA in development and disease: from functions to mechanisms. Open Biol. 7, 170–121. doi: 10.1098/rsob.170121
Deplus, R., Delatte, B., Schwinn, M. K., Defrance, M., Méndez, J., Murphy, N., et al. (2013). TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655. doi: 10.1038/emboj.2012.357
Derrien, T., Johnson, R., Bussotti, G., Tanzer, A., Djebali, S., Tilgner, H., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. doi: 10.1101/gr.132159.111
Dhiman, H., Kapoor, S., Sivadas, A., Sivasubbu, S., and Scaria, V. (2015). zflncRNApedia: a comprehensive online resource for zebrafish long non-coding RNAs. PLoS ONE 10:e0129997. doi: 10.1371/journal.pone.0129997
Divoux, A., Karastergiou, K., Xie, H., Guo, W., Perera, R. J., Fried, S. K., et al. (2014). Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity 22, 1781–1785. doi: 10.1002/oby.20793
Fei, Q., Bai, X., Lin, J., Meng, H., Yang, Y., and Guo, A. (2018). Identification of aberrantly expressed long non-coding RNAs in postmenopausal osteoporosis. Int. J. Mol. Med. 41, 3537–3550.doi: 10.3892/org/ijmm.2018.3575
Figueroa, D. M., Darrow, E. M., and Chadwick, B. P. (2015). Two novel DXZ4-associated long noncoding RNAs show developmental changes in expression coincident with heterochromatin formation at the human (Homo sapiens) macrosatellite repeat. Chrom. Res. 23, 733–752. doi: 10.1007/s10577-015-9479-3
Furuita, H., and Takeuchi, T. (1998). Effects of eicosapentaenoic and docosahexaenoic acids on growth, survival and brain development of larval Japanese flounder Paralichthys olivaceus. Aquaculture 161, 269–279. doi: 10.1016/S0044-8486(97)00275-5
Gapasin, R. S. J., and Duray, M. N. (2001) Effects of DHA-enriched live food on growth, survival incidence of opercular deformities in milkfish (Chanos chanos). Aquaculture 193, 49–63. doi: 10.1016/S0044-8486(00)00469-5
Gardini, A., and Shiekhattar, R. (2015). The many faces of long noncoding RNAs. FEBS J. 282, 1647–1657. doi: 10.1111/febs.13101
Gernapudi, R., Wolfson, B., Zhang, Y., Yao, Y., Yang, P., Asahara, H., et al. (2015). miR-140 promotes expression of long non-coding RNA NEAT1 in adipogenesis. Mol. Cell. Biol. 36, 30–38. doi: 10.1128/MCB.00702-15
Grollman, E. F., Philp, N. J., McPhie, P., Ward, R. D., and Sauer, B. (2000). Determination of transport kinetics of chick MCT3 monocarboxylate transporter from retinal pigment epithelium by expression in genetically modified yeast. Biochemistry 39, 9351–9357. doi: 10.1021/bi000464+
Gu, L., Frommel, S. C., Oakes, C. C., Simon, R., Grupp, K., Gerig, C. Y., et al. (2015). BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nat. Genet. 47, 22–30. doi: 10.1038/ng.3165
Guttman, M., Garber, M., Levin, J. Z., Donaghey, J., Robinson, J., Adiconis, X., et al. (2010). Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multiexonic structure of lincRNAs. Nat. Biotechnol. 28, 503–510. doi: 10.1038/nbt.1633
Halestrap, A. P., and Meredith, D. (2004). The SLC16 gene family-from monocarboxylate transporters (MCTs) to aromatic amino acid transporters and beyond. Pfluegers Arch. 447, 619–628. doi: 10.1007/s00424-003-1067-2
Halley, P., Kadakkuzha, B. M., Faghihi, M. A., Magistri, M., Zeier, Z., Khorkova, O., et al. (2014). Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 6, 222–230. doi: 10.1016/j.celrep.2013.12.015
Hamre, K., and Harboe, T. (2008). Artemia enriched with high n-3 HUFA may give a large improvement in performance of Atlantic halibut (Hippoglossus hippoglossus L.) larvae. Aquaculture 277, 239–243. doi: 10.1016/j.aquaculture.2008.02.028
Haque, S., Kaushik, K., Leonard, V. E., Kapoor, S., Sivadas, A., Joshi, A., et al. (2014). Short stories on zebrafish long noncoding RNAs. Zebrafish 11, 499–508. doi: 10.1089/zeb.2014.0994
Hu, H., Tao, B., Chen, J., Zhu, Z., and Hu, W. (2018). Fam60al as a novel factor involved in reprogramming of somatic cell nuclear transfer in zebrafish (Danio rerio). Int. J. Biol. Sci. 14, 78–86. doi: 10.7150/ijbs.22426
Hu, Y. W., Yang, J. Y., Ma, X., Chen, Z. P., Hu, Y. R., Zhao, J. Y., et al. (2014). A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J. Lipid Res. 55, 681–697. doi: 10.1194/jlr.M044669
Ishizaki, Y., Masuda, R., Uematsu, K., Shimizu, K., Arimoto, M., and Takeuchi, T. (2001). The effect of dietary docosahexaenoic acid on schooling behaviour and brain development in larval yellowtail. J. Fish Biol. 58, 1691–1703. doi: 10.1111/j.1095-8649.2001.tb02323.x
Ishizaki, Y., Uematsu, K., and Takeuchi, T. (2000). Preliminary study of the effect of dietary docosahexaenoic acid on the volumetric growth of the brain in larval yellowtail. Fish. Sci. 66, 611–613. doi: 10.1046/j.1444-2906.2000.00096.x
Isono, K., Fujimura, Y., Shinga, J., Yamaki, M. O., Wang, J., Takihara, Y., et al. (2005). Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol. Cell. Biol. 25, 6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005
Jiang, D., Wei, S., Chen, F., Zhang, Y., and Li, J. (2017). TET3-mediated DNA oxidation promotes ATR-dependent DNA damage response. EMBO Rep. 18, 781–796. doi: 10.15252/embr.201643179
Kanazawa, A. (1997). Effects of docosahexaenoic acid and phospholipids on stress tolerance of fish. Aquaculture 155, 129–134. doi: 10.1016/S0044-8486(97)00123-3
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi: 10.1126/science.1138341
Kaushik, K., Leonard, V. E., Shamsudheen, K. V., Lalwani, M. K., Jalali, S., Patowary, A., et al. (2013). Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS ONE 8, e83616. doi: 10.1371/journal.pone.0083616
Kim, K. D., Lee, S. M., Park, H. G., Bai, S. C., and Lee, Y. H. (2002). Essentiality of dietary n-3 highly unsaturated fatty acids in juvenile Japanese flounder Paralichthys olivaceus. J. World Aquacult. Soc. 33, 432–440. doi: 10.1111/j.1749-7345.2002.tb00022.x
Kornfeld, J. W., and Brüning, J. C. (2014). Regulation of metabolism by long, non-coding RNAs. Front. Genet. 5:57. doi: 10.3389/fgene.2014.00057
Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., et al. (2007). A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449, 689–694. doi: 10.1038/nature06192
Lee, S. M., Lee, J. H., and Kim, K. D. (2003). Effect of dietary essential fatty acids on growth, body composition and blood chemistry of juvenile starry flounder (Platichthys stellatus). Aquaculture 225, 269–281. doi: 10.1016/S0044-8486(03)00295-3
Li, C. H., Xiao, Z., Tong, J. H., To, K. F., Fang, X., Cheng, A. S., et al. (2016). EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int. J.Cancer 140, 120–129. doi: 10.1002/ijc.30414
Li, P., Ruan, X., Yang, L., Kiesewetter, K., Zhao, Y., Luo, H., et al. (2015). A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 21, 455–467. doi: 10.1016/j.cmet.2015.02.004
Liang, L. X., Ai, L. Y., Qian, J., Fang, J. Y., and Xu, J. (2015). Long noncoding RNA expression profiles in gut tissues constitutemolecular signatures that reflect the types of microbes. Sci. Rep. 5:11763. doi: 10.1038/srep11763
Lin, R. Y., Vera, J. C., Chaganti, R. S., and Golde, D. W. (1998). Human monocarboxylate transporter 2 (MCT2) is a high affinity pyruvate transporter. J. Biol. Chem. 273, 28959–28965. doi: 10.1074/jbc.273.44.28959
Liu, J., Caballero, M. J., Izquierdo, M., Ali, T. E-S., Hernández-Cruz, C. M., Valencia, A., et al. (2002). Necessity of dietary lecithin and eicosapentaenoic acid for growth, survival, stress resistance and lipoprotein formation in gilthead sea bream Sparus aurata. Fish. Sci. 68, 1165–1172. doi: 10.1046/j.1444-2906.2002.00551.x
Liu, S., Sheng, L., Miao, H., Saunders, T. L., MacDougald, O. A., Koenig, R. J., et al. (2014a). SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J. Biol. Chem. 289, 13000–13009. doi: 10.1074/jbc.M114.564658
Liu, S., Xu, R., Gerin, I., Cawthorn, W. P., Macdougald, O. A., Chen, X. W., et al. (2014b). SRA regulates adipogenesis by modulating p38/JNK phosphorylation and stimulating insulin receptor gene expression and downstream signaling. PLoS ONE 9:e95416. doi: 10.1371/journal.pone.0095416
Liu, Y., Luo, D., Zhao, H., Zhu, Z., Hu, W., and Cheng, C. H. K. (2013). Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS ONE 8:e76387. doi: 10.1371/journal.pone.0076387
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of Relative Gene Expression Data Using Real–Time Quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Long, Y., Wang, X., Youmans, D. T., and Cech, T. R. (2017). How do lncRNAs regulate transcription? Sci. Adv. 3:eaao2110. doi: 10.1126/sciadv.aao2110
Lopez-Pajares, V. (2016). Long non-coding RNA regulation of gene expression during differentiation. Pflugers Arch. 468, 971–981. doi: 10.1007/s00424-016-1809-6
Lund, I., and Steenfeldt, S. J. (2011). The effects of dietary long-chain essential fatty acids on growth and stress tolerance in pikeperch larvae (Sander lucioperca L.). Aquacult. Nutr. 17, 191–199. doi: 10.1111/j.1365-2095.2009.00724.x
Manning Fox, J. E., Meredith, D., and Halestrap, A. P. (2000). Characterisation of human monocarboxylate transporter 4 substantiates its role in lactic acid efflux from skeletal muscle. J. Physiol. 529, 285–293. doi: 10.1111/j.1469-7793.2000.00285.x
Mathy, N. W., and Chen, X. M. (2017). Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J. Biol. Chem. 292, 12375–12382. doi: 10.1074/jbc.R116.760884
Meller, V. H., Joshi, S. S., and Nikita, D. (2015). Modulation of Chromatin by Noncoding RNA. Annu. Rev. Genet. 49, 673–695. doi: 10.1146/annurev-genet-112414-055205.
Noffs, M. D., Martino, R. C., Trugo, L. C., Urbinati, E. C., Fernandes, J. B. K., and Takahashi, L. S. (2009). Dietary fish oil replacement with lard and soybean oil affects triacylglycerol and phospholipid muscle and liver docosahexaenoic acid content but not in the brain and eyes of surubim juveniles Pseudoplatystoma sp. Fish Physiol. Biochem. 35, 399–412. doi: 10.1007/s10695-008-9264-8
Núñez-Acuña, G., Détrée, C., Gallardoescárate, C., and Gonçalves, A. T. (2017). Functional diets modulate lncRNA-coding RNAs and gene interactions in the intestine of rainbow trout Oncorhynchus mykiss. Mar. Biotechnol. 19, 287–300. doi: 10.1007/s10126-017-9750-z
Oliva-Rico, D., and Herrera, L. A. (2017). Regulated expression of the lncRNA TERRA and its impact on telomere biology. Mech. Ageing Dev. 167, 16–23. doi: 10.1016/j.mad.2017.09.001
ØStbye, T-K., Kjær, M. A., Rørå, A. M. B, Torstensen, B., and Ruyter, B. (2011). High n-3 HUFA levels in the diet of Atlantic salmon affect muscle and mitochondrial membrane lipids and their susceptibility to oxidative stress. Aquacult. Nutr. 17, 177–190. doi: 10.1111/j.1365-2095.2009.00721.x
Qin, W., Li, X., Xie, L., Li, S., Liu, J., Jia, L., et al. (2016). A long non-coding RNA, APOA4-AS, regulates APOA4 expression depending on HuR in mice. Nucleic Acids Res. 44, 6423–6433. doi: 10.1093/nar/gkw341
Roo, F. J., Hernández-Cruz, C. M., Socorro, J. A., Fernández-Palacios, H., Montero, D., and Izquierdo, M. S. (2009). Effect of DHA content in rotifers on the occurrence of skeletal deformities in red porgy Pagrus pagrus (Linnaeus, 1758). Aquaculture 287, 84–93. doi: 10.1016/j.aquaculture.2008.10.010
Sarangdhar, M. A., Chaubey, D., Bhatt, A., Km, M., Kumar, M., Ranjan, S., et al. (2017). A novel long non-coding RNA, durga modulates dendrite density and expression of kalirin in zebrafish. Front. Mol. Neurosci. 10:95. doi: 10.3389/fnmol.2017.00095
Sargent, J. R., Bell, J. G., Mcevoy, L., Tocher, D., and Estevez, A. (1999). Recent developments in the essential fatty acid nutrition of fish. Aquaculture 177, 191–199. doi: 10.1016/S0044-8486(99)00083-6
Savić, N., Bär, D., Leone, S., Frommel, S. C., Weber, F. A., Vollenweider, E., et al. (2014). lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell. 15, 720–734. doi: 10.1016/j.stem.2014.10.005
Skalli, A., and Robin, J. H. (2004). Requirement of n-3 long chain polyunsaturated fatty acids for European sea bass (Dicentrarchus labrax) juveniles: growth and fatty acid composition. Aquaculture 240, 399–415. doi: 10.1016/j.aquaculture.2004.06.036
Smekalova, E. M., Kotelevtsev, Y. V., Leboeuf, D., Shcherbinina, E. Y., Fefilova, A. S., Zatsepin, T. S., et al. (2016). LncRNA in the liver: prospects for fundamental research and therapy by RNA interference. Biochimie 131, 159–172. doi: 10.1016/j.biochi.2016.06.007
Spizzo, R., Almeida, M. I., Colombatti, A., and Calin, G. A. (2012). Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31, 4577–4587. doi: 10.1038/onc.2011.621
Tocher, D. R. (2015). Omega-3 long-chain polyunsaturated fatty acids and aquaculture in perspective. Aquaculture 449, 94–107. doi: 10.1016/j.aquaculture.2015.01.010
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H., and Bartel, D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 147, 1537–1550. doi: 10.1016/j.cell.2011.11.055
Villalta, M., Estévez, A., Bransden, M. P., and Bell, J. G. (2008). Effects of dietary eicosapentaenoic acid on gowth, survival, pigmentation and fatty acid composition in Senegal sole (Solea senegalensis) larvae during Artemia feeding period. Aquacult. Nutr. 14, 232–241. doi: 10.1111/j.1365-2095.2007.00522.x
Vizcaíno-Ochoa, V., Lazo, J. P., Barón–Sevilla, B., and Drawbridge, M. A. (2010). The effect of dietary docosahexaenoic acid (DHA) on growth, survival and pigmentation of California halibut Paralichthys californicus larvae (Ayres, 1810). Aquaculture 302, 228–234. doi: 10.1016/j.aquaculture.2010.02.022
Wang, J., Fu, L., Koganti, P. P., Wang, L., Hand, J. M., Ma, H., et al. (2016). Identification and functional prediction of large intergenic noncoding RNAs (lincRNAs) in rainbow trout (Oncorhynchus mykiss). Mar. Biotechnol. 18, 271–282. doi: 10.1007/s10126-016-9689-5
Wang, X., Lin, J., Li, F., Zhang, C., Li, J., Wang, C., et al. (2017). Screening and functional identification of lncRNAs under β-diketone antibiotic exposure to zebrafish (Danio rerio) using high-throughput sequencing. Aquat. Toxicol. 182, 214–225. doi: 10.1016/j.aquatox
Wilson, R. (2009). Dietary effects of n-3 highly unsaturated fatty acid levels on egg and larval quality, and the fatty acid composition of the eggs of Chilean flounder Paralichthys adspersus broodstock. Aquac. Res. 40, 1400–1409. doi: 10.1111/j.1365-2109.2009.02238.x
Wu, F. C., Ting, Y. Y., and Chen, H. Y. (2003). Dietary docosahexaenoic acid is more optimal than eicosapentaenoic acid affecting the level of cellular defence responses of the juvenile grouper Epinephelus malabaricus. Fish Shellfish Immunol. 14, 223–238. doi: 10.1006/fsim.2002.0433
Xiao, T., Liu, L., Li, H., Sun, Y., Luo, H., Li, T., et al. (2015). Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPalpha. Stem Cell Rep. 5, 856–865. doi: 10.1016/j.stemcr.2015.09.007
Xu, B., Gerin, I., Miao, H., Vu-Phan, D., Johnson, C. N., Xu, R., et al. (2010). Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS ONE 5:e14199. doi: 10.1371/journal.pone.0014199
Xu, H., Cao, L., Wei, Y., Zhang, Y., and Liang, M. (2017). Effects of different dietary DHA:EPA ratios on gonadal steroidogenesis in the marine teleost, tongue sole (Cynoglossus semilaevis) Br. J. Nutr. 118, 179–188. doi: 10.1017/S0007114517001891
Xu, H., Cao, L., Wei, Y., Zhang, Y., and Liang, M. (2018). Lipid contents in farmed fish are influenced by dietary DHA/EPA ratio: a study with the marine flatfish, tongue sole (Cynoglossus semilaevis). Aquaculture 485, 183–190. doi: 10.1016/j.aquaculture.2017.11.054
Xu, H., Dong, X., Ai, Q., Mai, K., Xu, W., Zhang, Y., et al. (2014). Regulation of tissue LC-PUFA contents, Δ6 fatty acyl desaturase (FADS2) gene expression and the methylation of the putative FADS2 gene promoter by different dietary fatty acid profiles in Japanese seabass (Lateolabrax japonicus). PLoS ONE 9:e87726. doi: 10.1371/journal.pone.0087726
Xu, H., Wang, J., Mai, K., Xu, W., Zhang, W., Zhang, Y., et al. (2016). Dietary docosahexaenoic acid to eicosapentaenoic acid (DHA/EPA) ratio influenced growth performance, immune response, stress resistance and tissue fatty acid composition of juvenile Japanese seabass, Lateolabrax japonicus (cuvier). Aquac. Res. 47, 741–757. doi: 10.1111/are.12532
Xu, Y., Xu, C., Kato, A., Tempel, W., Abreu, J. G., Bian, C., et al. (2012). Tet3 CXXC domain and dioxygenase activity cooperatively regulate key genes for Xenopus eye and neural development. Cell 151, 1200–1213. doi: 10.1016/j.cell.2012.11.014
Young, R. S., and Ponting, C. P. (2013). Identification and function of long noncoding RNAs. Essays Biochem. 54, 113–126. doi: 10.1042/bse0540113
Zhao, X-B., and Ren, G-S. (2016). LncRNA taurine-upregulated gene 1 promotes cell proliferation by inhibiting microRNA-9 in MCF-7 cells. J. Breast Cancer 19, 349–357. doi: 10.4048/jbc.2016.19.4.349
Zhao, X. Y., Li, S., Wang, G. X., Yu, Q., and Lin, J. D. (2014). A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 55, 372–382. doi: 10.1016/j.molcel.2014.06.004
Zhou, T., Ding, J. W., Wang, X. A., and Zheng, X. X. (2016). Long noncoding RNAs and atherosclerosis. Atherosclerosis 248, 51–61. doi: 10.1016/j.atherosclerosis.2016.02.025
Zhu, J., Fu, H., Wu, Y., and Zheng, X. (2013). Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 56, 876–885. doi: 10.1007/s11427-013-4553-6
Zuo, R., Ai, Q., Mai, K., Xu, W., Wang, J., Xu, H., et al. (2012a). Effects of dietary docosahexaenoic to eicosapentaenoic acid ratio (DHA/EPA) on growth, nonspecific immunity, expression of some immune related genes and disease resistance of large yellow croaker (Larmichthys crocea) following natural infestation of parasites (Cryptocaryon irritans). Aquaculture 334-337, 101–109. doi: 10.1016/j.aquaculture.2011.12.045
Zuo, R., Ai, Q., Mai, K., Xu, W., Wang, J., Xu, H., et al. (2012b). Effects of dietary n-3 highly unsaturated fatty acids on growth, nonspecific immunity, expression of some immune related genes and disease resistance of large yellow croaker (Larmichthys crocea) following natural infestation of parasites (Cryptocaryon irritans). Fish Shellfish Immunol. 32, 249–258. doi: 10.1016/j.fsi.2011.11.005
Keywords: Cynoglossus semilaevis, diet, DHA/EPA, hepatic transcriptome, lncRNA
Citation: Xu H, Cao L, Sun B, Wei Y and Liang M (2019) Transcriptomic Analysis of Potential “lncRNA–mRNA” Interactions in Liver of the Marine Teleost Cynoglossus semilaevis Fed Diets With Different DHA/EPA Ratios. Front. Physiol. 10:331. doi: 10.3389/fphys.2019.00331
Received: 30 July 2018; Accepted: 11 March 2019;
Published: 02 April 2019.
Edited by:
Youji Wang, Shanghai Ocean University, ChinaReviewed by:
Albert Caballero Solares, Memorial University of Newfoundland, CanadaCopyright © 2019 Xu, Cao, Sun, Wei and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengqing Liang, bGlhbmdtcUB5c2ZyaS5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.