- 1Ubiquitin Project, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan
- 2Graduate School of Life Science, Picobiology Institute, University of Hyogo, Kobe, Japan
- 3Laboratory of Protein Metabolism, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan
F-box proteins, the substrate recognition subunits of SKP1–CUL1–F-box protein (SCF) E3 ubiquitin ligase complexes, play crucial roles in various cellular events mediated by ubiquitination. Several sugar-recognizing F-box proteins exist in both mammalian and plant cells. Although glycoproteins generally reside outside of cells, or in organelles of the secretory pathway, these lectin-type F-box proteins reside in the nucleocytoplasmic compartment. Mammalian sugar-recognizing F-box proteins commonly bind to the innermost position of N-glycans through a unique small hydrophobic pocket in their loops. Two cytosolic F-box proteins, Fbs1 and Fbs2, recognize high-mannose glycans synthesized in the ER, and SCFFbs1 and SCFFbs2 ubiquitinate excess unassembled or misfolded glycoproteins in the ERAD pathway by recognizing the innermost glycans, which serve as signals for aberrant proteins. On the other hand, endomembrane-bound Fbs3 recognizes complex glycans as well as high-mannose glycans, and SCFFbs3 ubiquitinates exposed glycoproteins in damaged lysosomes fated for elimination by selective autophagy. Plants express stress-inducible lectin-type F-box proteins recognizing a wider range of N- and O-glycans, suggesting that the roles of mammalian and plant lectin-type F-box proteins have diverged over the course of evolution to recognize species-specific targets with distinct functions. These sugar-recognizing F-box proteins interpret glycans in the cytosol as markers of unwanted proteins and organelles, and degrade them via the proteasome or autophagy.
Introduction
Ubiquitination occurs in a temporally and spatially specific manner. E3 ubiquitin ligases control ubiquitination by recognizing specific motifs, such as post-translational modifications induced by cell-signaling events or exposed elements that are normally hidden within proteins (Ravid and Hochstrasser, 2008). Cullin-RING E3 ligases (CRLs) are the largest family of E3 enzymes in all eukaryotes (Petroski and Deshaies, 2005). The best characterized CRLs are SCF complexes. Each SCF complex consists of four subunits: a scaffold protein CUL1, a RING protein RBX1, an adaptor protein SKP1, and one of many F-box proteins, which are responsible for substrate recognition. Each F-box proteins consists of an F-box domain, which binds to SKP1, and a divergent carboxy-terminal substrate-binding domain (Zheng et al., 2002). Mammalian F-box proteins have been grouped into three subfamilies according to their substrate-binding domains (Jin et al., 2004): the FBXW and FBXL families possess WD40 repeats and leucine-rich repeats (LRRs) in their binding domains, respectively, whereas the FBXO family does not have any characteristic structural domain(s). The varieties of SCF complexes differ considerably among eukaryotes. For example, there are 22, 72, and 698 F-box proteins in yeast (Saccharomyces cerevisiae), human, plant (Arabidopsis thaliana), respectively (Hua et al., 2011; Lee et al., 2011; Finley et al., 2012). Furthermore, CUL1, RBX1, and SKP1 are invariable components in the SCF complex in yeasts and metazoans, but the Arabidopsis genome encodes 19 SKP1-like proteins (ASK1-19), and some F-box proteins probably interact with several ASK proteins, yielding more diverse SCF complexes in plants (Farras et al., 2001; Gagne et al., 2002; Kuroda et al., 2012).
F-box proteins discriminate among free metabolites and various post-translational modifications in order to correctly ubiquitinate and degrade substrates in cells. For example, the growth-regulating plant hormones auxin/indole-3-acetic acid and jasmonates bind to transport inhibitor response 1 (TIR1) and coronatine-insensitive-1 (COI-1), respectively, to form part of an enlarged protein-binding interface that allows high-affinity interaction with their specific substrate hormone repressors (Tan et al., 2007; Sheard et al., 2010). As a common mechanism in all eukaryotes, many cell-cycle–related F-box proteins recognize phosphorylation in a specific motif in their corresponding substrates (Ang and Wade Harper, 2005; Randle and Laman, 2016). In addition to phosphorylation, other posttranslational modifications are also necessary for ubiquitination of some SCF complex substrates. In mammals, for example, SCFFBXO22-KDM4A and SCFFBXL17 target methylated p53 and acetylated PRMT1, respectively (Johmura et al., 2016; Lai et al., 2017). In addition, glycosylation is recognized by some F-box proteins in both mammals and plants. In contrast to other posttranslational modifications, the sugar chains of glycoproteins exhibit structural complexity and diversity.
Protein glycosylation occurs in the endoplasmic reticulum (ER) and Golgi, and glycoproteins reside within the lumen of secretory pathway organelles or outside the cell. Because they are separated by an endomembrane or the plasma membrane, sugar chains are normally not accessible to the ubiquitination machinery in the cytosol or nucleus. However, there are several opportunities for glycoproteins to appear in the cytosol. The first possibility is the ER-associated degradation (ERAD) pathway, in which unfolded proteins and orphan subunits are degraded by the proteasome after retrograde transport from the ER to the cytosol (Vembar and Brodsky, 2008). In this case, the N-glycan structures of glycoproteins emerging in the cytosol are high-mannose glycans that are modified by ER-resident enzymes. On the other hand, compounds including silica, monosodium urate, and protein amyloids, which are endocytosed from the extracellular milieu, can injure endosomes and lysosomes, causing glycoproteins modified with complex- or hybrid-type glycans to be leaked from these organelles to the cytosol. Furthermore, some specific-glycans on the surfaces of viruses and bacterial toxins that invaded cells via the retro-grade transport pathway may appear in the cytosol. Therefore, sugar chains appearing in the cytosol serve as ubiquitination signal for unwanted proteins and organelles (Yoshida and Tanaka, 2018). In this review, we focus on the substrate recognition mechanisms of sugar-recognizing F-box proteins. We will discuss the differences and similarities in the substrate recognition modes of lectin-type F-box proteins between plants and mammals, from the standpoint of their physiological roles.
Mechanism of N-Glycan Recognition by Sugar-Recognizing F-Box Proteins
N-Glycan Recognition by Mammalian Sugar-Recognizing F-Box Proteins
F-box protein recognizing sugar chain 1 (Fbs1), the first ubiquitin ligase component identified as a sugar-recognizing F-box protein, was purified from mouse brain lysate based on its affinity for an N-glycoprotein (Yoshida et al., 2002). Of the 72 human F-box proteins, only three, Fbs1/FBXO2, Fbs2/FBXO6, and Fbs3/FBXO27, have the ability to bind glycoproteins containing high-mannose glycans, which are synthesized in the ER (Yoshida et al., 2003, 2011).
These F-box proteins recognize Man3GlcNAc2 core in N-glycans, but exhibit diverse binding to various glycan structures (Glenn et al., 2008). Structural analysis reveals that the overall architecture of Fbs1 consists of the F-box domain, a linker domain, and a substrate-binding domain (Figure 1A). The substrate-binding domain of Fbs1 is composed of a 10-stranded β-sandwich with an α-helix; it binds Man3GlcNAc2 through a small hydrophobic pocket in the loops located at the top of the β-sandwich, which protrudes toward E2 (Figure 1B). Man3GlcNAc2 interacts with Fbs1 through hydrogen bonds and/or hydrophobic interactions (Figure 1C; Mizushima et al., 2004, 2007). The core regions of glycans in native glycoproteins are shielded by the amino acid residues surrounding the glycosylation site, but are exposed upon denaturation. Indeed, Fbs1 and Fbs2 prefer to interact with denatured glycoproteins; thus, exposure of the innermost position of N-glycans upon glycoprotein denaturation serves as a signal of misfolding (Yoshida et al., 2005, 2007; Mallinger et al., 2012). Interestingly, a cytosolic N-Glycanase 1 (NGLY1) also recognizes the same position of glycoproteins and is involved in deglycosylation of various substrates prior to their proteasome-mediated degradation via the ERAD pathway (Yamaguchi et al., 2007; Suzuki, 2015). Therefore, ERAD substrates emerging in the cytosol might be denatured.
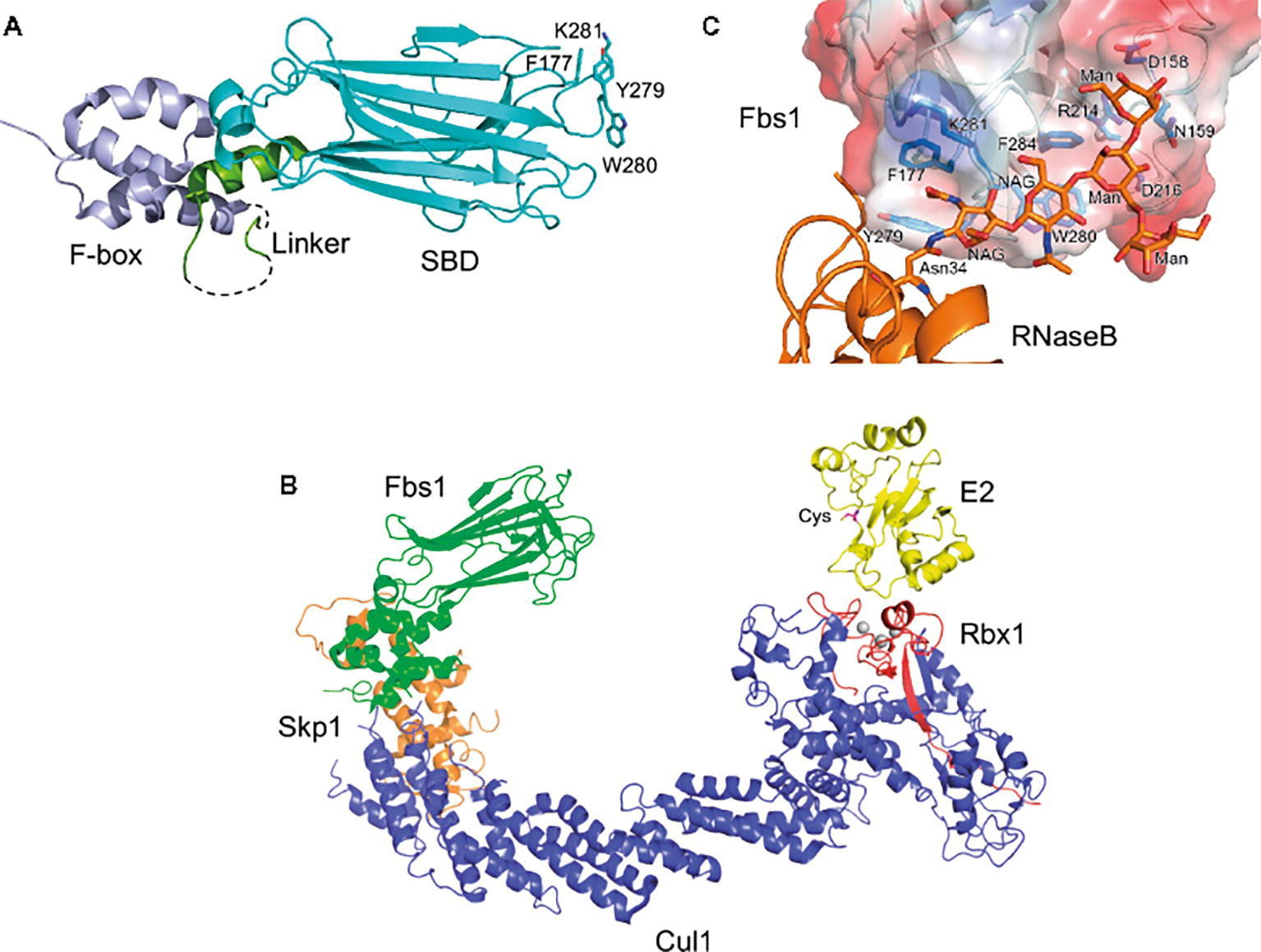
Figure 1. Structure of Fbs1. (A) Overall structure of Fbs1. The F-box domain (F-box), linker, and substrate-binding domain (SBD) are shown in violet, green, and cyan, respectively. Dotted lines represent disordered regions. GlcNAc2-binding residues are depicted as stick models. (B) Model of the SCFFbs1 complex bound to E2. Fbs1, Cul1, Rbx1, Skp1, and E2 are colored green, blue, red, orange, and yellow, respectively. A model of SCFFbs1 was constructed by superposition of the Skp1 subunits from the Skp1–Fbs1, and Skp1–Cul1–Rbx1 structures (Zheng et al., 2002) (PDB ID code 1LDK); the RING-finger domains derived from Rbx1; the c-Cbl subunit of c-Cbl–UbcH7 (Zheng et al., 2000) (PDB ID code 1FBV); and the E2 subunits of c-Cbl-UbcH7. (C) Surface representation of the substrate-binding site of the Fbs1 SBD bound to Man3GlcNAc2 of RNase B. The surface is colored according to the electrostatic potential of the residues (blue, positive; red, negative). Bound RNase B and Man3GlcNAc2 are orange, and the residues involved in the substrate binding are blue.
Although both Fbs1 and Fbs2 preferentially bind high-mannose glycans, Fbs3 binds to glycoproteins modified with complex-type glycans, such as transferrin and LAMP2, as well as high-mannose glycans (Glenn et al., 2008; Yoshida et al., 2017). However, the structural information of Fbs3 is not yet available until now, and the mode of recognition by Fbs3 remains to be elucidated.
Mammalian Fbs1-Related F-Box Proteins
Fbs1/FBXO2 exhibits high sequence similarity with other F-box proteins (Winston et al., 1999; Ilyin et al., 2002). In phylogenetic analysis, these proteins cluster into two groups: one group contains Fbs1/FBXO2, Fbs2/FBXO6, and FBXO44a, and the other contains FBXO17 and Fbs3/FBXO27 (Figure 2A). The genes that encode the proteins each group are arranged in tandem with very short intergenic regions, but the two groups map to different chromosomes in both human and mouse (Ilyin et al., 2002; Yoshida and Tanaka, 2010). These F-box proteins contain a highly homologous F-box domain and a substrate-binding domain (SBD) (Figure 2B), but the long N-terminal region and C-terminal tail are unique to Fbs1 and Fbs2, respectively. The SBD of FBXO44 has 68% identity with the corresponding region of Fbs2; the residues necessary for binding to N-glycans are conserved, and its overall structure is similar to Fbs1 and Fbs2. Nonetheless, FBXO44 has no detectable sugar-binding activity (Glenn et al., 2008; Yoshida et al., 2011). The crystal structure of FBXO44–Skp1 revealed that FBXO44 has different hydrogen bond networks than the four loops from Fbs1 and Fbs2, preventing the formation of the sugar-binding pocket (Kumanomidou et al., 2015; Nishio et al., 2016). Thus far, from a structural perspective, it has been challenging to identify factors that determine the substrate specificity of these isoforms.
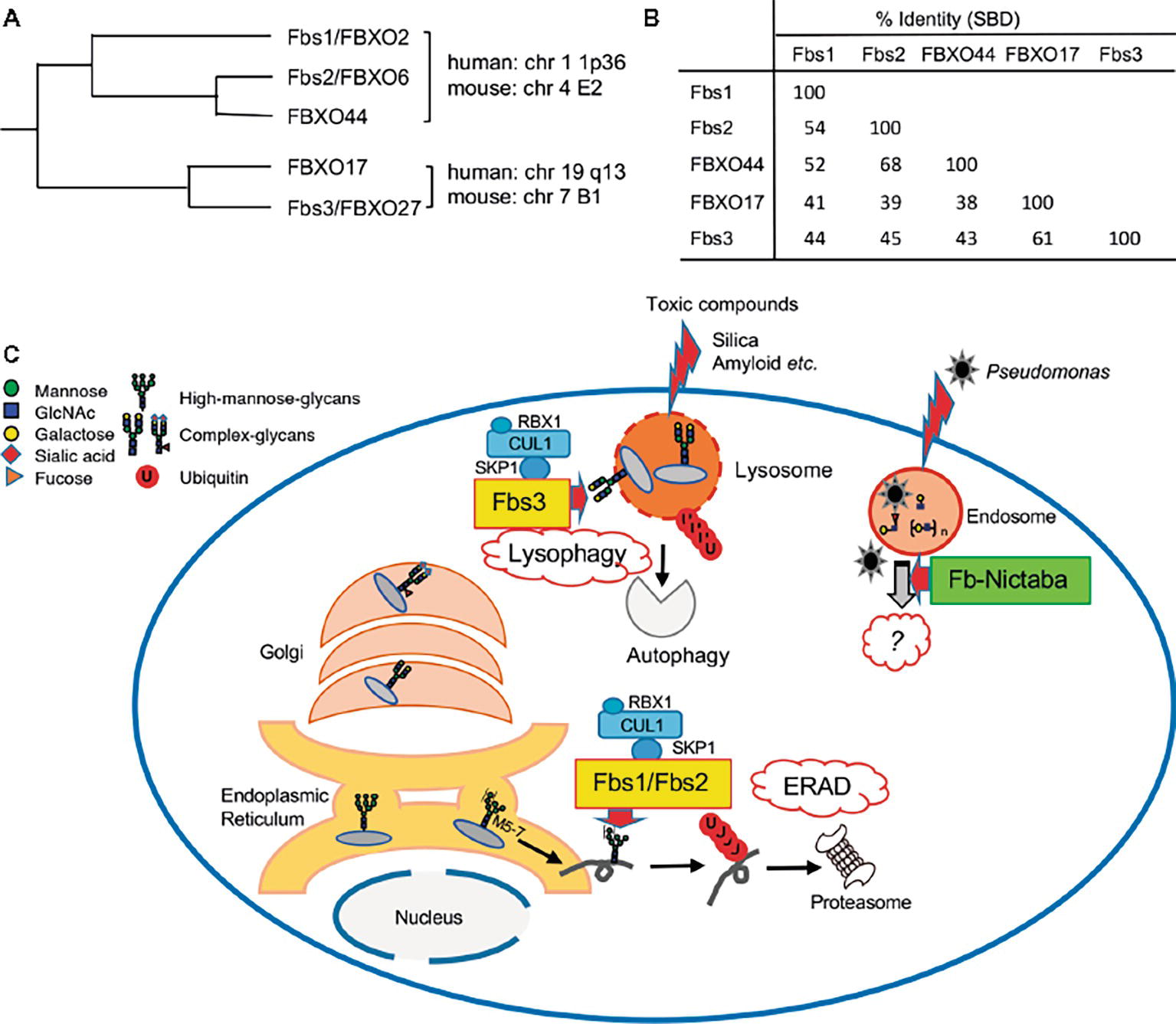
Figure 2. Roles of sugar-recognizing F-box proteins in the cytosol. (A) Phylogenetic tree of Fbs1 homologs. Genomic locations of these genes on human or mouse chromosomes are also shown. (B) Percentage identities of substrate-binding domain (SBD) of Fbs1 homologs. (C) Overview of the functions of sugar-recognizing F-box proteins. Three mammalian F-box proteins (Fbs1, Fbs2, and Fbs3; yellow-colored) form SCF complexes and ubiquitinate glycoproteins in the cytosol, followed by proteasomal or autophagic degradation to maintain cellular homeostasis. They mainly recognize the innermost Man3GlcNAc2 structure in high-mannose glycans, which are attached in the ER, as a signal of misfolded glycoproteins, and act on excess unassembled subunits or misfolded glycoproteins via the ERAD pathway. In contrast to Fbs1 and Fbs2, Fbs3 can bind to complex-type glycans, is targeted to endomembranes via N-myristoylation, and accumulates on organelles such as lysosomes and endosomes ruptured by toxic compounds. SCFFbs3 ubiquitinates exposed the lysosomal glycoprotein LAMP2 to induce autophagy. Dozens of F-box/Nictaba proteins (green) are expressed in plants, but their functions have not been elucidated. These proteins recognize varieties of glycan structure in both N- and O-glycans, and their expression is induced by certain environmental stresses. The Arabidopsis F-box/Nictaba protein functions in defense against pathogens such as Pseudomonas syringae.
Plant Lectin-Type F-Box Proteins
These Fbs orthologues are encoded in various vertebrate genomes, but other lectin-type F-box proteins are found in plants. High levels of secretory lectins accumulate in plant seeds and vegetative storage tissues, but plants also synthesize small amounts of nucleocytoplasmic lectins in response to specific stress factors and changing environmental conditions. Among nucleocytoplasmic lectins, chimeric proteins that contain F-box domain with Jacalin-related lectins or Nictaba-like lectins were found (Lannoo and Van Damme, 2010). Nictaba, an inducible lectin found in Nicotiana tabacum leaves treated with jasmonates (Chen et al., 2002), recognizes Man3GlcNAc2 core as well as human Fbs family members (Lannoo et al., 2006). However, the three-dimensional conformations and sugar-recognition modes of Nictaba and mammalian Fbs1 differ considerably (Mizushima et al., 2004; Schouppe et al., 2010). Arabidopsis and crops such as soybean express dozens of F-box/Nictaba proteins (Delporte et al., 2015; Van Holle et al., 2017), but to date only an Arabidopsis F-box/Nictaba protein, At2g02360, has been characterized. The expression of At2g02360 is up-regulated after treatment with salicylic acid, heat stress, or infection with Pseudomonas syringae. Plants overexpressing At2g02360 exhibit milder disease symptoms after infection of pathogens, but the molecular mechanisms involved in acquisition of pathogen resistance remain to be elucidated (Stefanowicz et al., 2016). Although At2g02360 shares the highest sequence similarity (64%) to tobacco Nictaba among Arabidopsis F-box/Nictaba proteins, it binds to N- and O-glycans with Galβ1-3GlcNAc and Galβ1-4GlcNAc and poly-N-acetyllactosamine, but not to Man3GlcNAc2 core (Stefanowicz et al., 2012). It is possible that other F-box/Nictaba proteins have different sugar-binding specificities, but the roles of mammalian and plant lectin-type F-box proteins seem to have diverged substantially over the course of to recognize species-specific targets with distinct functions (Figure 2C).
Physiological Roles of Mammalian Sugar-Recognizing F-Box Proteins
As described above, Fbs1 and Fbs2 specifically recognize high-mannose glycans, and in particular the innermost structure. Therefore, SCFFbs1 and SCFFbs2 can ubiquitinate excess unassembled or misfolded glycoproteins in the ERAD pathway by recognizing the innermost glycans as signals for aberrant proteins (Figure 2B). On the other hand, Fbs3, which can also interact with glycoproteins modified with complex-type glycans present in organelles downstream of the Golgi or on the cell surface, contributes to maintenance of organelle homeostasis (Figure 2C).
SCFFbs1 and SCFFbs2 are the E3 enzymes responsible for degradation of integrin β1 that is expressed in excess over integrin α chains, or ERAD substrates such as TCRα, asialoglycoprotein receptor H2a, and CFTRΔ508 (Yoshida et al., 2002, 2003; Groisman et al., 2011; Chen et al., 2016; Ramachandran et al., 2016). The ERAD pathway is a ubiquitous protein quality control system in all eukaryotes, and ERAD substrates are generally ubiquitinated by ER membrane-embedded E3s, such as Hrd1 and gp78 (Vembar and Brodsky, 2008). However, Fbs proteins are encoded only in vertebrate genomes. In particular, Fbs1 expression is restricted to a subset of tissues, and therefore its role in in quality control may be tissue-specific.
Roles of Fbs1 in the Brain and Inner Ear
Originally named neural F-box protein 42 kDa (NFB42) and organ of Corti protein 1 (OCP1), Fbs1 is expressed at high levels in the brain and rodent inner ear (Erhardt et al., 1998; Henzl et al., 2001). Therefore, Fbs1 may function in quality control specifically in the nervous system and inner ear, rather than in the general ERAD system.
NMDA receptors play crucial roles in neuronal development and information storage in the brain, and SCFFbs1 controls the abundance and localization of their specific subunits, GluN1 and GluN2A (Kato et al., 2005; Atkin et al., 2015). Furthermore, Fbs1 attenuates amyloid-β (Αβ) production through ubiquitination of β-secretase (BACE1) and amyloid precursor protein (APP), and the expression of Fbs1 decreases in the brains of Alzheimer’s disease (AD) patients and Tg2576 mice, a well-characterized model of AD (Gong et al., 2010; Atkin et al., 2014). In primary neurons derived from Tg2576 mice, overexpression of Fbs1 promotes the degradation of BACE1, which is essential for Αβ generation, thereby decreasing the Aβ level (Gong et al., 2010). In addition, the total amount of amyloid precursor protein (APP) in the brain of Fbs1-KO mouse is increased but decreased on the surface of cells in hippocampal neurons, indicating that Fbs1 regulates APP protein levels and processing (Atkin et al., 2014). Interestingly, PGC-1α, a transcriptional coactivator involved in control of energy metabolism, regulates Fbs1 expression, and nicotinamide riboside upregulates BACE1 degradation through enhancing PGC-1α expression (Gong et al., 2010, 2013). Compounds such as nicotinamide that stimulate Fbs1 expression/activity may represent candidate therapeutic agents for AD.
Furthermore, Fbs1-knockout mice exhibit age-related hearing loss with cochlear degeneration and high cochlear levels of the inner-ear gap-junction protein GJB2 (Nelson et al., 2007), which is a multi-pass membrane protein that lacks glycans but nonetheless interacts with Fbs1 (Henzl et al., 2004). Fbs1 is an unusually abundant inner ear protein, and exists as a heterodimer with Skp1 but not as a component of the SCF complex, suggesting that its function in inner-ear homeostasis is distinct from that of the conventional SCF complex (Atkin et al., 2015).
Although the expression of Fbs1 is restricted to specific organs under normal condition, recent studies show that expression of Fbs1, like plant Nictaba, is up-regulated in response to some stressors. In the livers of mice with high-fat diet-induced obesity, Fbs1 expression is induced by the IKKβ/NF-κΒ pathway, and SCFFbs1 disrupts glucose homeostasis via degradation of the insulin receptor (Liu et al., 2017). Infection with Epstein–Barr virus (EBV) also stimulates Fbs1 expression, and induced SCFFbs1 ubiquitinates and degrades EBV glycoprotein B, thereby decreasing the infectivity of progeny viruses (Zhang et al., 2018). Thus, SCFFbs1 may function in the unusual ERAD system that is induced under certain stresses. The expression of Fbs3, like that of Fbs1, is restricted to specific organs, including the brain. Its levels are very low, but it may be induced by as-yet-undetermined stimuli.
Roles of Fbs3
Fbs3 has a unique endomembrane localization due to N-myristoylation. This endomembrane localization, together with its glycoprotein-binding activity, is essential for effective recruitment to damaged organelles. Fbs3 accumulates within ruptured lysosomes or endosomes by treated with the lysosomal damage reagent L-leucyl-l-leucine methyl ester (LLOMe), crystalline silica, or latex beads coated with transfection reagents, whereas neither Fbs1 or Fbs2 behaves in this manner (Yoshida et al., 2017). Lysosomes are specialized organelle that contain a variety of digestive enzymes and play a crucial role in autophagy, but damaged lysosomes are themselves eliminated by a special form of autophagy known as lysophagy. Like mitophagy, a well-characterized form of selective autophagy, ubiquitination is prerequisite for lysophagy, but the ubiquitination substrates and molecular mechanisms underlying lysophagy induction have not been elucidated (Maejima et al., 2013). Following lysosomal damage, Fbs3 quickly moves to ruptured lysosomes by detecting exposed glycoproteins, and SCFFbs3 ubiquitinates several lysosomal glycoproteins. Among the lysosomal glycoproteins, ubiquitination of LAMP2 is especially important for recruitment of components of the autophagic machinery, such as p62 and LC3 (Yoshida et al., 2017). Although SCFFbs3 recognizes and ubiquitinates exposed glycoproteins that are normally sequestered in lysosomes following lysosomal damage, resulting in induction of lysophagy, other glycan-recognition systems are also involved in the autophagy-mediated response to damaged endomembranes.
Galectins are β-galactoside–binding lectins, and mostly reside in the cytosol and nucleus. Galectin-8 accumulates on damaged bacteria-containing vesicles and binds directly to NDP52, an autophagy receptor, thereby triggering a specific form of autophagy called xenophagy and restricting the growth of Salmonella in cells (Thurston et al., 2012; Li et al., 2013). Galectin-3 also accumulates on damaged organelles, such as phagosomes ruptured by infecting Mycobacterium and damaged lysosomes, and interacts with TRIM16, a RING-type ubiquitin ligase. TRIM16 further interacts with the key autophagy regulator ULK1, Beclin1, and ATG16L1, and induces autophagy following their ubiquitination (Chauhan et al., 2016). Thus, several cytosolic glycan-monitoring systems collaborate with the proteasome and autophagy to maintain cellular homeostasis.
Conclusions and Perspectives
Here we have summarized our current knowledge of the sugar-recognition modes and physiological roles of lectin-type F-box proteins. These proteins recognize cytosolic sugar chains, which are normally present in organelles or extracellularly, as aberrant or harmful signals that trigger ubiquitination, leading to alleviation of deleterious cellular states and maintenance of homeostasis. Mammalian sugar-recognizing F-box proteins commonly bind to the innermost position of N-glycans, and cytosolic NGLY1 removes this sugar degron. Mutations in NGLY1 cause an inherited disorder of the ERAD pathway (Enns et al., 2014), and NGLY1 knockout mice are embryonic lethal (Fujihira et al., 2017). Why would F-box proteins fail to rescue the lethality of NGLY1 deficiency, which is caused by excessive glycoprotein in the cytosol? As with Fbs3, the functions of Fbs1 and Fbs2 may be distinct from the ERAD pathway. For instance, Fbs3 accumulates in ruptured lysosomes and preferentially ubiquitinates LAMP2, which plays a crucial role for lysophagy. Thus, identification of the intrinsic substrates for Fbs1 and Fbs2 is essential for understanding their physiological relevance in maintaining cellular homeostasis. Research of these glycoprotein-related F-box proteins knockout mice in models of disease, including AD, would be also useful for assessing their physiological and pathophysiological roles.
In comparison with mammals, plants have more sugar-recognizing F-box proteins with diverse substrate specificities, but their functions have not been elucidated. The ability of these proteins to form SCF complexes remain to be determined. Among 22 yeast F-box proteins, some function in complexes that lack CUL1, suggesting that not all F-box proteins in plants must form SCF complexes. Future studies should seek to determine their sugar-binding specificities and endogenous interacting proteins, substrates, and components of the complex. The elucidation of the molecular mechanisms underlying induction of sugar-recognizing F-box proteins and promotion of SCF complex formation by various stimuli, as well as detailed analyses of their substrate recognition modes in both plants and mammals, will be crucial to understanding the functions of F-box proteins and cytosolic sugar chains.
Data Availability
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
YY and TM wrote the manuscript with supervision from KT. All authors critically appraised the final version of the paper.
Funding
This work was supported by JSPS KAKENHI Grant Number JP16K07705 (to YY) and JP2600014 (to KT): JST CREST Grant Number JPMJCR18H3 (to YY), and the Takeda Science Foundation (KT).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ang, X. L., and Wade Harper, J. (2005). SCF-mediated protein degradation and cell cycle control. Oncogene 24, 2860–2870. doi: 10.1038/sj.onc.1208614
Atkin, G., Hunt, J., Minakawa, E., Sharkey, L., Tipper, N., Tennant, W., et al. (2014). F-box only protein 2 (Fbxo2) regulates amyloid precursor protein levels and processing. J. Biol. Chem. 289, 7038–7048. doi: 10.1074/jbc.M113.515056
Atkin, G., Moore, S., Lu, Y., Nelson, R. F., Tipper, N., Rajpal, G., et al. (2015). Loss of F-box only protein 2 (Fbxo2) disrupts levels and localization of select NMDA receptor subunits, and promotes aberrant synaptic connectivity. J. Neurosci. 35, 6165–6178. doi: 10.1523/JNEUROSCI.3013-14.2015
Chauhan, S., Kumar, S., Jain, A., Ponpuak, M., Mudd, M. H., Kimura, T., et al. (2016). TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev. Cell 39, 13–27. doi: 10.1016/j.devcel.2016.08.003
Chen, X., Duan, L. H., Luo, P. C., Hu, G., Yu, X., Liu, J., et al. (2016). FBXO6-mediated ubiquitination and degradation of Ero1L inhibits endoplasmic reticulum stress-induced apoptosis. Cell. Physiol. Biochem. 39, 2501–2508. doi: 10.1159/000452517
Chen, Y., Peumans, W. J., Hause, B., Bras, J., Kumar, M., Proost, P., et al. (2002). Jasmonic acid methyl ester induces the synthesis of a cytoplasmic/nuclear chito-oligosaccharide binding lectin in tobacco leaves. FASEB J. 16, 905–907. doi: 10.1096/fj.01-0598fje
Delporte, A., Van Holle, S., Lannoo, N., and Van Damme, E. J. (2015). The tobacco lectin, prototype of the family of Nictaba-related proteins. Curr. Protein Pept. Sci. 16, 5–16. doi: 10.2174/1389203716666150213154107
Enns, G. M., Shashi, V., Bainbridge, M., Gambello, M. J., Zahir, F. R., Bast, T., et al. (2014). Mutations in NGLY1 cause an inherited disorder of the endoplasmic reticulum-associated degradation pathway. Genet. Med. 16, 751–758. doi: 10.1038/gim.2014.22
Erhardt, J. A., Hynicka, W., Dibenedetto, A., Shen, N., Stone, N., Paulson, H., et al. (1998). A novel F box protein, NFB42, is highly enriched in neurons and induces growth arrest. J. Biol. Chem. 273, 35222–35227. doi: 10.1074/jbc.273.52.35222
Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., et al. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756. doi: 10.1093/emboj/20.11.2742
Finley, D., Ulrich, H. D., Sommer, T., and Kaiser, P. (2012). The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360. doi: 10.1534/genetics.112.140467
Fujihira, H., Masahara-Negishi, Y., Tamura, M., Huang, C., Harada, Y., Wakana, S., et al. (2017). Lethality of mice bearing a knockout of the Ngly1-gene is partially rescued by the additional deletion of the Engase gene. PLoS Genet. 13:e1006696. doi: 10.1371/journal.pgen.1006696
Gagne, J. M., Downes, B. P., Shiu, S. H., Durski, A. M., and Vierstra, R. D. (2002). The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11519–11524. doi: 10.1073/pnas.162339999
Glenn, K. A., Nelson, R. F., Wen, H. M., Mallinger, A. J., and Paulson, H. L. (2008). Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 283, 12717–12729. doi: 10.1074/jbc.M709508200
Gong, B., Chen, F., Pan, Y., Arrieta-Cruz, I., Yoshida, Y., Haroutunian, V., et al. (2010). SCFFbx2-E3-ligase-mediated degradation of BACE1 attenuates Alzheimer’s disease amyloidosis and improves synaptic function. Aging Cell 9, 1018–1031. doi: 10.1111/j.1474-9726.2010.00632.x
Gong, B., Pan, Y., Vempati, P., Zhao, W., Knable, L., Ho, L., et al. (2013). Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol. Aging 34, 1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005
Groisman, B., Shenkman, M., Ron, E., and Lederkremer, G. Z. (2011). Mannose trimming is required for delivery of a glycoprotein from EDEM1 to XTP3-B and to late endoplasmic reticulum-associated degradation steps. J. Biol. Chem. 286, 1292–1300. doi: 10.1074/jbc.M110.154849
Henzl, M. T., O’Neal, J., Killick, R., Thalmann, I., and Thalmann, R. (2001). OCP1, an F-box protein, co-localizes with OCP2/SKP1 in the cochlear epithelial gap junction region. Hear. Res. 157, 100–111. doi: 10.1016/S0378-5955(01)00285-4
Henzl, M. T., Thalmann, I., Larson, J. D., Ignatova, E. G., and Thalmann, R. (2004). The cochlear F-box protein OCP1 associates with OCP2 and connexin 26. Hear. Res. 191, 101–109. doi: 10.1016/j.heares.2004.01.005
Hua, Z., Zou, C., Shiu, S. H., and Vierstra, R. D. (2011). Phylogenetic comparison of F-Box (FBX) gene superfamily within the plant kingdom reveals divergent evolutionary histories indicative of genomic drift. PLoS One 6:e16219. doi: 10.1371/journal.pone.0016219
Ilyin, G. P., Serandour, A. L., Pigeon, C., Rialland, M., Glaise, D., and Guguen-Guillouzo, C. (2002). A new subfamily of structurally related human F-box proteins. Gene 296, 11–20. doi: 10.1016/S0378-1119(02)00867-3
Jin, J., Cardozo, T., Lovering, R. C., Elledge, S. J., Pagano, M., and Harper, J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580. doi: 10.1101/gad.1255304
Johmura, Y., Sun, J., Kitagawa, K., Nakanishi, K., Kuno, T., Naiki-Ito, A., et al. (2016). SCF(Fbxo22)-KDM4A targets methylated p53 for degradation and regulates senescence. Nat. Commun. 7:10574. doi: 10.1038/ncomms10574
Kato, A., Rouach, N., Nicoll, R. A., and and Bredt, D. S. (2005). Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc. Natl. Acad. Sci. USA 102, 5600–5605. doi: 10.1073/pnas.0501769102
Kumanomidou, T., Nishio, K., Takagi, K., Nakagawa, T., Suzuki, A., Yamane, T., et al. (2015). The structural differences between a glycoprotein specific F-Box protein Fbs1 and its homologous protein FBG3. PLoS One 10:e0140366. doi: 10.1371/journal.pone.0140366
Kuroda, H., Yanagawa, Y., Takahashi, N., Horii, Y., and Matsui, M. (2012). A comprehensive analysis of interaction and localization of Arabidopsis SKP1-like (ASK) and F-box (FBX) proteins. PLoS One 7:e50009. doi: 10.1371/journal.pone.0050009
Lai, Y., Li, J., Li, X., and Zou, C. (2017). Lipopolysaccharide modulates p300 and Sirt1 to promote PRMT1 stability via an SCF(Fbxl17)-recognized acetyldegron. J. Cell Sci. 130, 3578–3587. doi: 10.1242/jcs.206904
Lannoo, N., Peumans, W. J., Pamel, E. V., Alvarez, R., Xiong, T. C., Hause, G., et al. (2006). Localization and in vitro binding studies suggest that the cytoplasmic/nuclear tobacco lectin can interact in situ with high-mannose and complex N-glycans. FEBS Lett. 580, 6329–6337. doi: 10.1016/j.febslet.2006.10.044
Lannoo, N., and Van Damme, E. J. (2010). Nucleocytoplasmic plant lectins. Biochim. Biophys. Acta 1800, 190–201. doi: 10.1016/j.bbagen.2009.07.021
Lee, J. E., Sweredoski, M. J., Graham, R. L., Kolawa, N. J., Smith, G. T., Hess, S., et al. (2011). The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell. Proteomics 10:M110.006460. doi: 10.1074/mcp.M110.006460
Li, S., Wandel, M. P., Li, F., Liu, Z., He, C., Wu, J., et al. (2013). Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy. Sci. Signal. 6:ra9. doi: 10.1126/scisignal.2003730
Liu, B., Lu, H., Li, D., Xiong, X., Gao, L., Wu, Z., et al. (2017). Aberrant expression of FBXO2 disrupts glucose homeostasis through ubiquitin-mediated degradation of insulin receptor in obese mice. Diabetes 66, 689–698. doi: 10.2337/db16-1104
Maejima, I., Takahashi, A., Omori, H., Kimura, T., Takabatake, Y., Saitoh, T., et al. (2013). Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 32, 2336–2347. doi: 10.1038/emboj.2013.171
Mallinger, A., Wen, H. M., Dankle, G. M., and Glenn, K. A. (2012). Using a ubiquitin ligase as an unfolded protein sensor. Biochem. Biophys. Res. Commun. 418, 44–48. doi: 10.1016/j.bbrc.2011.12.109
Mizushima, T., Hirao, T., Yoshida, Y., Lee, S. J., Chiba, T., Iwai, K., et al. (2004). Structural basis of sugar-recognizing ubiquitin ligase. Nat. Struct. Mol. Biol. 11, 365–370. doi: 10.1038/nsmb732
Mizushima, T., Yoshida, Y., Kumanomidou, T., Hasegawa, Y., Suzuki, A., Yamane, T., et al. (2007). Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc. Natl. Acad. Sci. USA 104, 5777–5781.
Nelson, R. F., Glenn, K. A., Zhang, Y., Wen, H., Knutson, T., Gouvion, C. M., et al. (2007). Selective cochlear degeneration in mice lacking the F-box protein, Fbx2, a glycoprotein-specific ubiquitin ligase subunit. J. Neurosci. 27, 5163–5171. doi: 10.1523/JNEUROSCI.0206-07.2007
Nishio, K., Yoshida, Y., Tanaka, K., and Mizushima, T. (2016). Structural analysis of a function-associated loop mutant of the substrate-recognition domain of Fbs1 ubiquitin ligase. Acta. Crystallogr. F Struct. Biol. Commun. 72, 619–626. doi: 10.1107/S2053230X16011018
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. doi: 10.1038/nrm1547
Ramachandran, S., Osterhaus, S. R., Parekh, K. R., Jacobi, A. M., Behlke, M. A., and McCray, P. B. Jr. (2016). SYVN1, NEDD8, and FBXO2 proteins regulate DeltaF508 cystic fibrosis transmembrane conductance regulator (CFTR) ubiquitin-mediated proteasomal degradation. J. Biol. Chem. 291, 25489–25504. doi: 10.1074/jbc.M116.754283
Randle, S. J., and Laman, H. (2016). F-box protein interactions with the hallmark pathways in cancer. Semin. Cancer Biol. 36, 3–17. doi: 10.1016/j.semcancer.2015.09.013
Ravid, T., and Hochstrasser, M. (2008). Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9, 679–690. doi: 10.1038/nrm2468
Schouppe, D., Rouge, P., Lasanajak, Y., Barre, A., Smith, D. F., Proost, P., et al. (2010). Mutational analysis of the carbohydrate binding activity of the tobacco lectin. Glycoconj. J. 27, 613–623. doi: 10.1007/s10719-010-9305-2
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Stefanowicz, K., Lannoo, N., Proost, P., and Van Damme, E. J. (2012). Arabidopsis F-box protein containing a Nictaba-related lectin domain interacts with N-acetyllactosamine structures. FEBS Open Bio. 2, 151–158. doi: 10.1016/j.fob.2012.06.002
Stefanowicz, K., Lannoo, N., Zhao, Y., Eggermont, L., Van Hove, J., Al Atalah, B., et al. (2015). Glycan-binding F-box protein from Arabidopsis thaliana protects plants from Pseudomonas syringae infection. BMC Plant Biol. 16:213.
Suzuki, T. The cytoplasmic peptide: N-glycanase (Ngly1)-basic science encounters a human genetic disorder. J. Biochem. 157, 23–34. doi: 10.1093/jb/mvu068
Tan, X., Calderon-Villalobos, L. I., Sharon, M., Zheng, C., Robinson, C. V., Estelle, M., et al. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446, 640–645. doi: 10.1038/nature05731
Thurston, T. L., Wandel, M. P., Von Muhlinen, N., Foeglein, A., and Randow, F. (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418. doi: 10.1038/nature10744
Van Holle, S., Rouge, P., and Van Damme, E. J. M. (2017). Evolution and structural diversification of Nictaba-like lectin genes in food crops with a focus on soybean (Glycine max). Ann. Bot. 119, 901–914. doi: 10.1093/aob/mcw259
Vembar, S. S., and Brodsky, J. L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957. doi: 10.1038/nrm2546
Winston, J. T., Koepp, D. M., Zhu, C., Elledge, S. J., and Harper, J. W. (1999). A family of mammalian F-box proteins. Curr. Biol. 9, 1180–1182. doi: 10.1016/S0960-9822(00)80021-4
Yamaguchi, Y., Hirao, T., Sakata, E., Kamiya, Y., Kurimoto, E., Yoshida, Y., et al. (2007). Fbs1 protects the malfolded glycoproteins from the attack of peptide: N-glycanase. Biochem. Biophys. Res. Commun. 362, 712–716. doi: 10.1016/j.bbrc.2007.08.056
Yoshida, Y., Adachi, E., Fukiya, K., Iwai, K., and Tanaka, K. (2005). Glycoprotein-specific ubiquitin ligases recognize N-glycans in unfolded substrates. EMBO Rep. 6, 239–244. doi: 10.1038/sj.embor.7400351
Yoshida, Y., Chiba, T., Tokunaga, F., Kawasaki, H., Iwai, K., Suzuki, T., et al. (2002). E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442. doi: 10.1038/nature00890
Yoshida, Y., Murakami, A., Iwai, K., and Tanaka, K. (2007). A neural-specific F-box protein Fbs1 functions as a chaperone suppressing glycoprotein aggregation. J. Biol. Chem. 282, 7137–7144. doi: 10.1074/jbc.M611168200
Yoshida, Y., Murakami, A., and Tanaka, K. (2011). Skp1 stabilizes the conformation of F-box proteins. Biochem. Biophys. Res. Commun. 410, 24–28. doi: 10.1016/j.bbrc.2011.05.098
Yoshida, Y., and Tanaka, K. (2010). Lectin-like ERAD players in ER and cytosol. Biochim. Biophys. Acta 1800, 172–180. doi: 10.1016/j.bbagen.2009.07.029
Yoshida, Y., and Tanaka, K. (2018). Cytosolic N-glycans: triggers for ubiquitination directing proteasomal and autophagic degradation: molecular systems for monitoring cytosolic N-glycans as signals for unwanted proteins and organelles. BioEssays 40, 1700215 (1-9). doi: 10.1002/bies.201700215
Yoshida, Y., Tokunaga, F., Chiba, T., Iwai, K., Tanaka, K., and Tai, T. (2003). Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 278, 43877–43884. doi: 10.1074/jbc.M304157200
Yoshida, Y., Yasuda, S., Fujita, T., Hamasaki, M., Murakami, A., Kawawaki, J., et al. (2017). Ubiquitination of exposed glycoproteins by SCFFBXO27 directs damaged lysosomes for autophagy. Proc. Natl. Acad. Sci. USA 114, 8574–8579. doi: 10.1073/pnas.1702615114
Zhang, H. J., Tian, J., Qi, X. K., Xiang, T., He, G. P., Zhang, H., et al. (2018). Epstein-Barr virus activates F-box protein FBXO2 to limit viral infectivity by targeting glycoprotein B for degradation. PLoS Pathog. 14:e1007208. doi: 10.1371/journal.ppat.1007208
Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., et al. (2002). Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416, 703–709. doi: 10.1038/416703a
Keywords: E3 ubiquitin ligase, ERAD, F-box protein, glycoprotein, N-glycan, SCF complex, sugar chain
Citation: Yoshida Y, Mizushima T and Tanaka K (2019) Sugar-Recognizing Ubiquitin Ligases: Action Mechanisms and Physiology. Front. Physiol. 10:104. doi: 10.3389/fphys.2019.00104
Edited by:
Victor M. Bolanos-Garcia, Oxford Brookes University, United KingdomReviewed by:
Sumit Sahni, University of Sydney, AustraliaPaul Kenneth Witting, University of Sydney, Australia
Copyright © 2019 Yoshida, Mizushima and Tanaka. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keiji Tanaka, dGFuYWthLWtqQGlnYWt1a2VuLm9yLmpw
 Yukiko Yoshida
Yukiko Yoshida Tsunehiro Mizushima
Tsunehiro Mizushima Keiji Tanaka
Keiji Tanaka