- 1Department of Basic Science, College of Dental Sciences, University of Ha'il, Ha'il, Saudi Arabia
- 2The University College of Medical Sciences and GTB, Guru Teg Bahadur Hospital (University of Delhi), New Delhi, India
- 3Research and Scientific Studies Unit, College of Nursing & Allied Health Sciences, Jazan University, Jazan, Saudi Arabia
- 4Department of Bioscience & Bioinformatics, Khallikote University, Berhampur, India
- 5Department of Biotechnology, Institute of Engineering and Technology, Lucknow, India
- 6Department of Laboratory Medicine, Faculty of Applied Medical Sciences, Albaha University, Albaha, Saudi Arabia
Angiotensin-converting enzyme (ACE) gene is indispensable for endothelial control and vascular tone regulatory systems, usually affected in Systemic Lupus Erythematosus (SLE). ACE insertion/deletion (I/D) polymorphism may influence the progress of SLE. Earlier studies have investigated this association without any consistency in results. We performed this meta-analysis to evaluate the precise association between ACE I/D polymorphism and SLE susceptibility. The relevant studies were searched until December, 2017 using Medline (PubMed), Google-Scholar and EMBASE search engines. Twenty-five published studies involving 3,308 cases and 4,235 controls were included in this meta-analysis. Statistically significant increased risk was found for allelic (D vs. I: p = 0.007; OR = 1.202, 95% CI = 1.052–1.374), homozygous (DD vs. II: p = 0.025; OR = 1.347, 95% CI = 1.038–1.748), dominant (DD+ID vs. II: p = 0.002; OR = 1.195, 95% CI = 1.070–1.334), and recessive (DD vs. ID+II: p = 0.023; OR = 1.338, 95% CI = 1.042–1.718) genetic models. Subgroup analysis stratified by Asian ethnicity revealed significant risk of SLE in allelic (D vs. I: p = 0.045; OR = 1.238, 95% CI = 1.005–1.525) and marginal risk in dominant (DD+ID vs. II: p = 0.056; OR = 1.192, 95% CI = 0.995–1.428) models; whereas, no association was observed for Caucasian and African population. Publication bias was absent. In conclusion, ACE I/D polymorphism has significant role in overall SLE risk and it can be exploited as a prognostic marker for early SLE predisposition.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease often involving multiple organ inflammation. The clinical consequences of SLE are extremely heterogeneous and generally characterized by pathogenic autoantibody formation against the host's nuclear antigens, immune complex deposition, and end-organ damage (Kyttaris et al., 2006). The precise etiology of SLE is still ambiguous. Earlier studies have proposed that a complex interaction of factors involving gene and environment causes genetic alterations thereby play a key role in the development of SLE in genetically susceptible individuals (Tsao, 2003). In spite of noteworthy advances in understanding the pathogenic role of this disease, diagnosis, prognosis, and therapeutic puzzles are still incomplete. A number of genetic loci have recently been highlighted to be having association with susceptibility to SLE by genome-wide association studies (GWAS), gene association studies, and current advanced single nucleotide polymorphism (SNP) studies (Harley et al., 2006; Gregersen and Olsson, 2009; Frangou et al., 2013). These data clearly highlight the complexity of the genetic interactions involved in SLE progression, suggesting the use of SNPs as promising future biomarkers for assessing genetic background of individuals for prognosis of SLE.
The angiotensin-converting enzyme (ACE, kininase II, EC3.74.15.1) is a zinc metalloproteinase converting angiotensin I (Ang I) into angiotensin II (Ang II), an octapeptide acting as a potent vasopressor and stimulator of aldosterone. Ang II also acts as a growth factor, particularly in kidneys, inducing remodeling of tissue, and fibrosis. Contraction of smooth muscle cells and their proliferation is also known to be induced by Ang II, including adhesion of monocytes and platelets (Morrissey and Klahr, 1998; Kasal and Schiffrin, 2012). ACE gene consists of 26 exons and 25 introns amassing to nearly 24 kb in size and is situated on the long arm of the chromosome 17. Many polymorphic residues have also been identified in ACE; including a widely studied insertion (I) or deletion (D) of a 287-bp fragment on intron 16 (Rigat et al., 1990), causing three possible genotypes—II, ID, and DD. The genotype DD carrying individuals have 2-fold (higher) levels of ACE in serum when compared to the individuals with II genotype. Whereas, ID heterozygotes show a moderate activity while homozygote for I allele reveals the least ACE activity (Sayed-tabatabaei et al., 2006). Therefore, it is possible that this I/D polymorphism may be involved in vascular immunity and SLE pathogenesis. Considering its important role in SLE development, several case-control studies have recently examined the effect of ACE I/D polymorphism on the risk of SLE in different populations. However, due to lack of consistency, their results remained inconclusive (Guan et al., 1997; Sato et al., 1998; Tassiulas et al., 1998; Akai et al., 1999; Pullmann et al., 1999; Molad et al., 2000; Kaufman et al., 2001; Prkacin et al., 2001; Uhm et al., 2002; Douglas et al., 2004; Shin, 2004; El-Shafeey et al., 2005; Saeed et al., 2005; Sprovieri and Sens, 2005; Al-Awadhi et al., 2007; Rabbani et al., 2008; Hussain et al., 2010; Abbas et al., 2012; Gong et al., 2012; Lian et al., 2012; Salimi et al., 2012; Topete-Reyes et al., 2013; Negi et al., 2015; Pradhan et al., 2015; Pitipakorn et al., 2016). The inconsistency in their results may be attributed to their probable small sample sizes with petite statistical power. Studying the association of different polymorphisms with complex diseases always requires large sample size as is recommended in a recent study (Burton et al., 2009).
A meta-analysis, as a statistical tool, can overcome these limitations of single studies with small sample sizes. It combines multiple studies on the same alleles of genes to enhance the statistical power of the analysis and derive more precise and reliable results of the genetic effects. Therefore, we performed this meta-analysis by pooling all the eligible published studies to determine a comprehensive picture of the above said genetic association and understand the role of ACE gene polymorphism as a genetic marker for SLE progression. To maintain the overall quality of this study, we assessed the selected studies on Newcastle Ottawa Scale (NOS) for their quality score. Trial Sequential Analysis (TSA) was used to minimize type-I statistical errors like publication bias and random errors, caused generally by sparse data, in order to quantify the statistical reliability of the included data in the meta-analysis with statistical significance threshold.
Materials and Methods
Search Strategy and Eligibility of the Relevant Studies
A thorough search of studies on the ACE I/D gene polymorphism and its association with SLE susceptibility was conducted in PubMed (Medline), Google Scholar and EMBASE. An update limit of December 2017 was applied. The search terms were as follows: “ACE” “angiotensin converting enzyme” OR “ACE” (polymorphism OR mutation OR variant) in combination with “systemic lupus erythematosus” OR “SLE” (susceptibility OR risk). The focus of the search was human studies only. The titles and abstracts of the all the searched articles were read for initial evaluation, and only studies fulfilling the eligibility criteria were retrieved and used in this meta-analysis. The reference lists of the selected studies was also inspected manually for other pertinent articles.
Study Selection Criteria
The following criteria was used for selection of studies for this meta-analysis: (a) the study must be an evaluation of the association between ACE I/D gene polymorphism and SLE susceptibility, (b) the study must have a case-control design, (c) the study must have enrolled well diagnosed SLE patients and normal healthy controls, (d) the study must have genotypic frequency data available for both patients and controls, (e) the study must be published in English. In case the data for same patient population was reported in more than one publication, the most recent and complete publication was considered for this meta-analysis. The exclusion criteria for the studies were: studies with overlapping data; studies reporting data for patient population only; studies where genotypic frequency data was not available; and review articles. All information regarding selection of the studies is depicted as PRISMA 2009 Flow-Diagram (Figure 1).
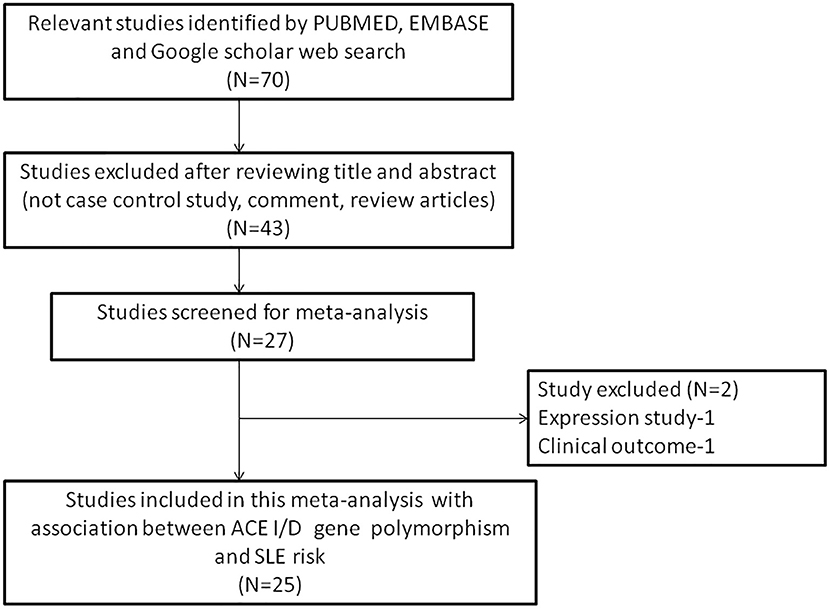
Figure 1. PRISMA 2009 Flow-diagram showing the identification and selection process (inclusion/exclusion) of the pertinent studies for the present meta-analysis.
Extraction of the Data From Selected Studies
Two investigators (RKM & SAD), independently, extracted and summarized the data from each retrieved study by following the standard procedure. The data collection form was designed and used to collect the data to ensure accuracy while following stringent inclusion/exclusion criteria as described above. The attributes extracted and summarized from the selected articles were: first author's name; publication year; origin country; total number of patients and controls included; study type; association status; genotyping method used; and genotype frequency of patients and controls. Discrepancy, if observed, in the data collected from the selected studies by the two investigators was settled by open discussion in presence of SH (an adjudicator) in order to reach a final consensus.
Newcastle-Ottawa Scale (NOS) Criteria for Quality Assessment
The NOS criteria (Stang, 2010) was used to assess the methodological quality of the selected studies. This was again done separately by two independent investigators (RKM & SAD). Three major aspects are included in the NOS criteria—(i) selection of subjects (0–4 points); (ii) comparability of subjects (0–2 points); (iii) clinical outcomes (0–3 points). Studies getting 5 or more than 5 points or stars were considered as of moderate to high or good quality (Hu et al., 2015). The disagreement, if any, in assigning the points or stars to the study, by the two investigators, was resolved in consultation with the adjudicator SH.
Statistical Analysis
The statistical analyses was performed using Comprehensive Meta-Analysis (CMA) V2 software program (Biostat, USA). The p < 0.05 was considered as statistically significant. All the p-values were two sided. Crude odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were computed to assess the association intensity between the ACE I/D gene polymorphism and SLE susceptibility. The allele contrast, log-additive, dominant, and recessive model pooled ORs were estimated (Woolf, 1955). Chi-square based Q-test was used to perform heterogeneity assumption across the eligible studies (Wu and Li, 1999), and heterogeneity was considered significant when p < 0.05. A fixed effect model was used when p > 0.05 (Mantel and Haenszel, 1959); and a random effect model was used when p < 0.05 (DerSimonian and Laird, 1986). For efficient testing of the heterogeneity, I2 statistics was also employed (Higgins et al., 2003). Chi-square test was used to measure Hardy-Weinberg equilibrium (HWE) in the control population. Egger's linear regression test was used to estimate funnel plot asymmetry, a type of linear regression approach on the natural logarithm ORs scale. The t-test was used to determine the significance of the intercept; statistically significant publication bias was indicated by p < 0.05 (Egger et al., 1997).
Trial Sequential Analysis (TSA)
In an attempt to include all the eligible trials in this meta-analysis, and to minimize the systematic errors (bias) or random errors by chance, we used a novel statistical TSA tool from Copenhagen Trial Unit, Center for Clinical Intervention Research, Denmark. TSA estimates the required information size and adjusts statistical significance thresholds, and also estimates the power of conclusion (Wetterslev et al., 2008; Brok et al., 2009; Turner et al., 2013). TSA indicates no requirement of further trials if the Z curve crosses the monitoring boundary before the required information size is reached; however, if it does not cross the boundary, further trials becomes necessary. The TSA software program, version 0.9 (http://www.ctu.dk/tsa/) was used for TSA analysis.
Cochran-Armitage (CA) Statistics
We determined Cochran-Armitage (CA) statistics for each of the (29) cases considered in our study. CA trend test is the most popular association analysis for determining genetic associations (Sasieni, 1997).
CA trend test statistic (T) is given by the following equation
Where
For a codominant or additive model, M1 = 0; M2 = 1;M3 = 2.
For a Dominant Model, M1 = 0; M2 = 1; M3 = 1.
For a Recessive model, M1 = 0; M2 = 1; M3 = 2.
Here, Ny is the total number of controls, Nx is the total number of cases, Xi and Yi represents the number of cases and controls for each of the three (II, ID, and DD) genotypes.
Results
Literature Search and Meta-Analysis Databases
During the study search the full-texts of all the articles deemed potentially eligible were retrieved. The study eligibility for inclusion was determined by reviewing the full text of all the articles by the first investigator. The second investigator randomly selected 10% of the articles and reviewed them by the same procedure, independently. A complete agreement was observed between the two investigators regarding inclusion and exclusion of the studies. Following the identification of the final set of the eligible articles, one investigator extracted the relevant data from all the studies, and the other investigator independently re-extracted the data from all the included studies to cross-check this step. Table 1, 2 depict the main characteristics and genotype distribution along with minor allele frequency (MAF) in subjects of all the 25 studies included in this meta-analysis, respectively. In NOS analysis for quality score, more than 95% of all the included studies scored 5 stars or more, except the study of Hussain et al. (2010) which scored only 3 stars. This study was included because it possessed all the basic needful information for its consideration in the analysis and its 3 stars suggested a moderate to good quality (Table 3). The sequential process for identification of the eligible studies for the present meta-analysis followed the pre-set inclusion and exclusion criteria shown in Figure 1 (PRISMA 2009 Flow Diagram).
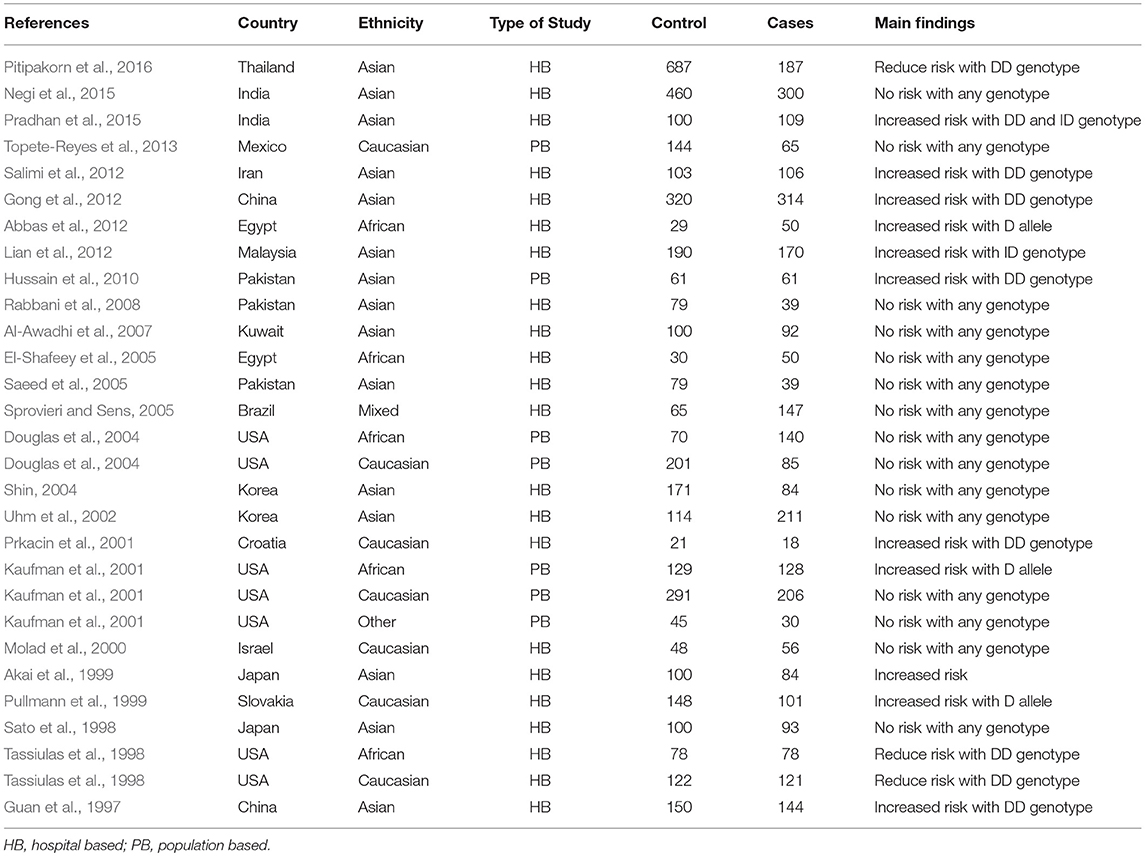
Table 1. Main characteristics of all the studies of ACE I/D polymorphism and SLE risk included in this meta-analysis.
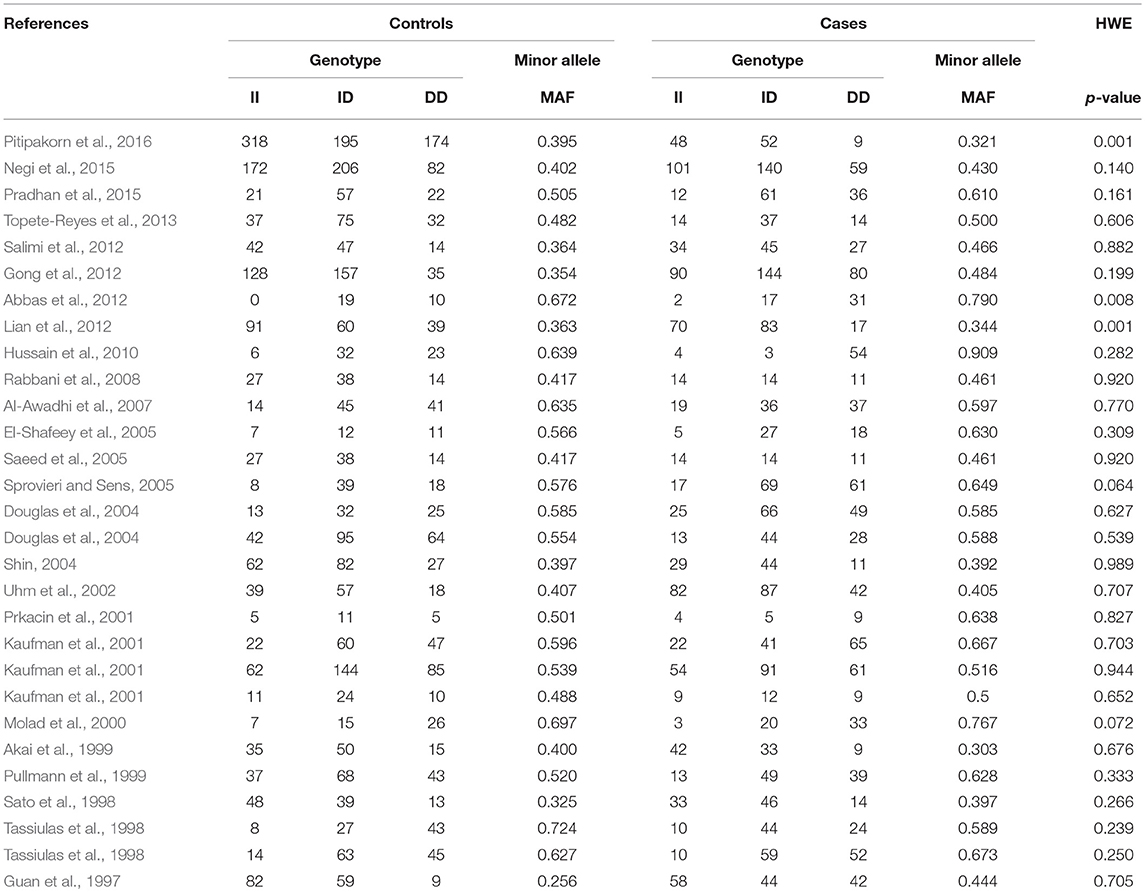
Table 2. Genotypic distribution of ACE I/D gene polymorphism in studies included in this meta-analysis.
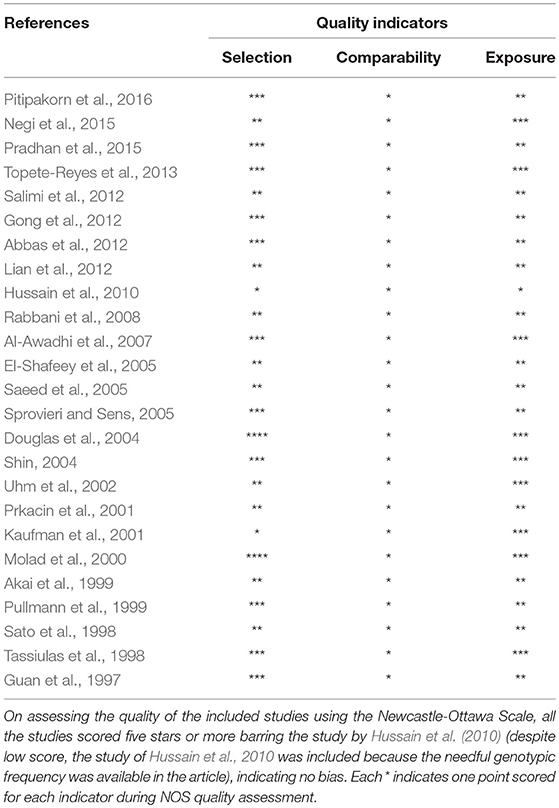
Table 3. Quality assessment conducted according to the Newcastle-Ottawa Scale for all the studies included in the present meta-analysis.
Publication Bias Diagnosis
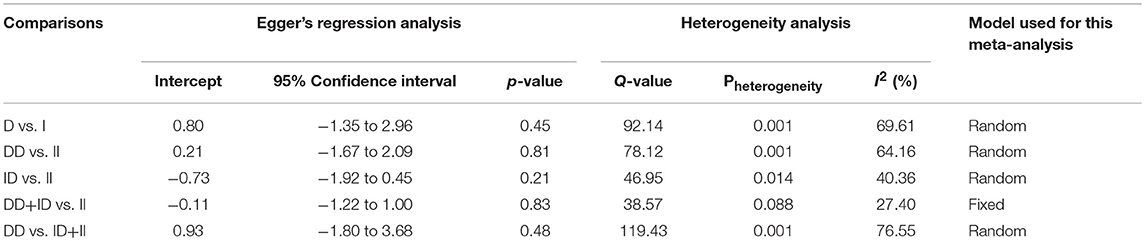
Both, funnel plot asymmetry and Egger's regression statistics were employed to evaluate the publication bias. A p < 0.05 was fixed for the significant publication bias in the present meta-analysis. All the comparison genetic models showed absence of publication bias (p > 0.05) (Table 4, Figure SI1).
Heterogeneity Evaluation
Heterogeneity was tested by Q-test and I2 statistics among the included studies. Four genetic models showed heterogeneity, so the data was synthesized by applying the random effect model (Table 4).
Association of ACE I/D Polymorphism and Overall SLE Susceptibility
When pooled together, the number of cases accumulated to 3,308 and controls to 4,235 from all the included studies. The pooled subjects were examined for the precise association between ACE I/D polymorphism and overall SLE susceptibility. Overall, the pooled analysis suggests significant increased risk between ACE I/D gene polymorphism and overall SLE risk in four genetic models, i.e., allelic (D vs. I: p = 0.007; OR = 1.202, 95% CI = 1.052–1.374), homozygous (DD vs. II: p = 0.025; OR = 1.347, 95% CI = 1.038–1.748), dominant (DD+ID vs. II: p = 0.002; OR = 1.195, 95% CI = 1.070–1.334) and recessive (DD vs. ID+II: p = 0.023; OR = 1.338, 95% CI = 1.042–1.718) (Figure 2). Whereas, heterozygous (ID vs. II: p = 0.250; OR = 1.103, 95% CI = 0.934–1.302) genetic model did not show any SLE risk (Figure 2).
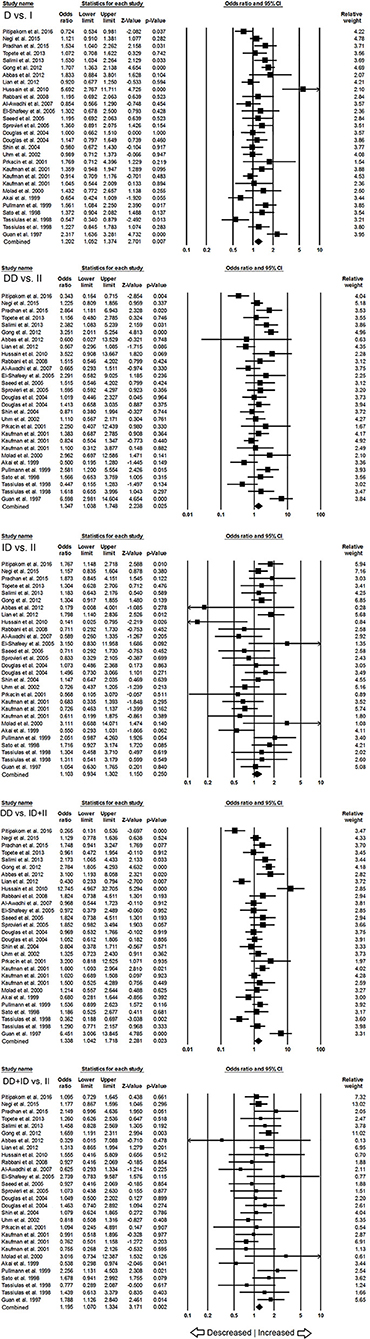
Figure 2. Forest plot of ORs with 95% CI of SLE risk associated with the ACE I/D gene polymorphism for the overall population. Black square represents the value of OR and the size of the square indicates the inverse proportion relative to its variance. Horizontal line is the 95% CI of OR.
Association of ACE I/D Polymorphism and SLE Susceptibility in Asian Population
In subgroup analysis of Asian ethnic population, 15 studies involving a total of 2,083 confirmed SLE cases and 2,844 controls were considered for this analysis. Heterogeneity was observed in four genetic models (Figure SI2), so the random effect models were applied to generate ORs and 95% CIs for the synthesis (Table 5). We observed increased risk of SLE development in allelic (D vs. I: p = 0.045; OR = 1.238, 95% CI = 1.005–1.525) genetic model. Similarly, dominant (DD+ID vs. II: p = 0.056; OR = 1.192, 95% CI = 0.995–1.428) genetic model also showed marginal significant risk. But, other genetic models, i.e., homozygous (DD vs. II: p = 0.141; OR = 1.366, 95% CI = 0.902–2.070), heterozygous (ID vs. II: p = 0.418; OR = 1.094, 95% CI = 0.880–1.360) and recessive (DD vs. ID+II: p = 0.101; OR = 1.437, 95% CI = 0.931–2.217) genetic models did not show any increased or decreased risk of SLE (Figure 3).
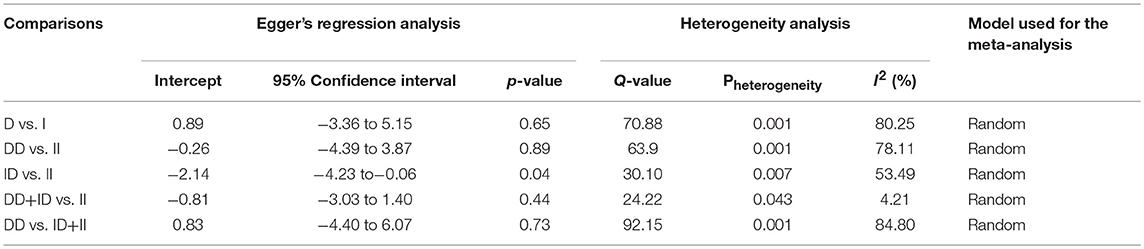
Table 5. Statistics to test publication bias and heterogeneity in the present meta-analysis: Asian population.
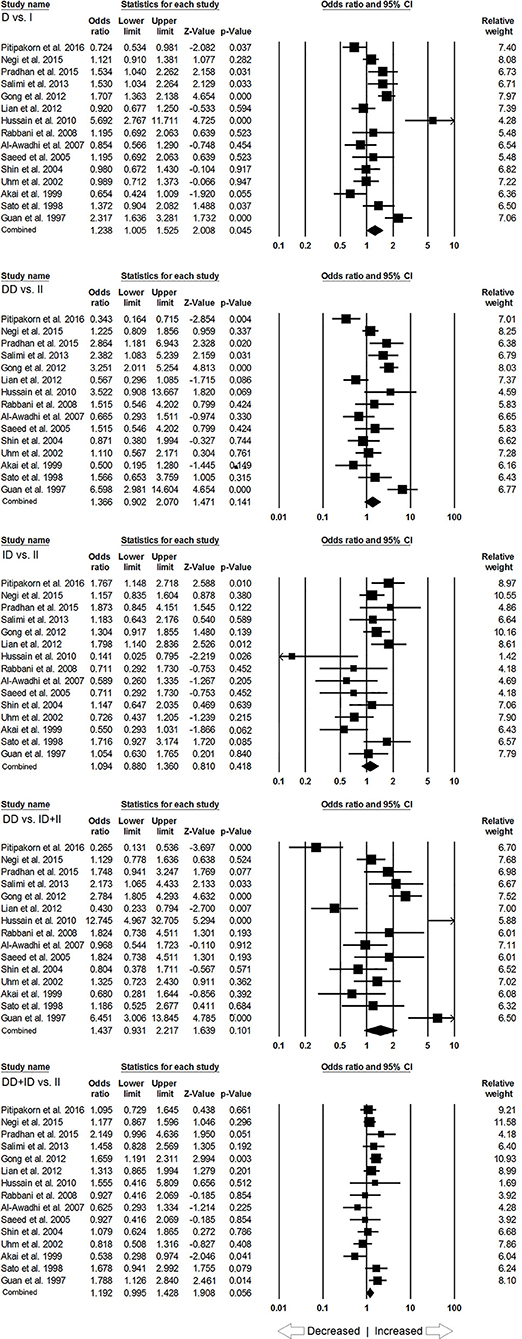
Figure 3. Forest plot of ORs with 95% CI of SLE risk associated with the ACE I/D gene polymorphism for the Asian subgroup population. Black square represents the value of OR and the size of the square indicates the inverse proportion relative to its variance. Horizontal line is the 95% CI of OR.
Association of ACE I/D Polymorphism and SLE Susceptibility in Caucasian Population
In case of subgroup analysis of Caucasian ethnicity population, 7 studies comprising of 652 confirmed SLE cases and 975 controls were included. No heterogeneity was observed in all the genetic models (Figure SI3), so the fixed effect models were applied to generate ORs and 95% CIs (Table 6). After the synthesis, we found that all the genotypic models were not significantly associated with SLE risk, i.e., allelic (D vs. I: p = 0.066; OR = 1.146, 95% CI = 0.991–1.324), homozygous (DD vs. II: p = 0.077; OR = 1.314, 95% CI = 0.971–1.779), heterozygous (ID vs. II: p = 0.355; OR = 1.142, 95% CI = 0.862–1.511), dominant (DD+ID vs. II: p = 0.189; OR = 1.194, 95% CI = 0.917–1.555) and recessive (DD vs. ID+II: p = 0.125; OR = 1.185, 95% CI = 0.954–1.471) genetic models (Figure 4).
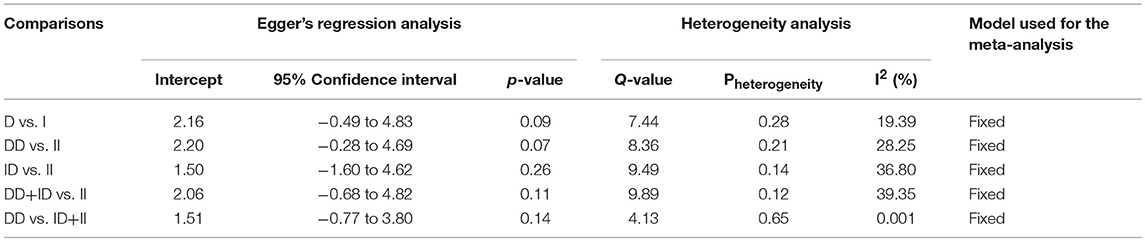
Table 6. Statistics to test publication bias and heterogeneity in the present meta-analysis: Caucasian population.
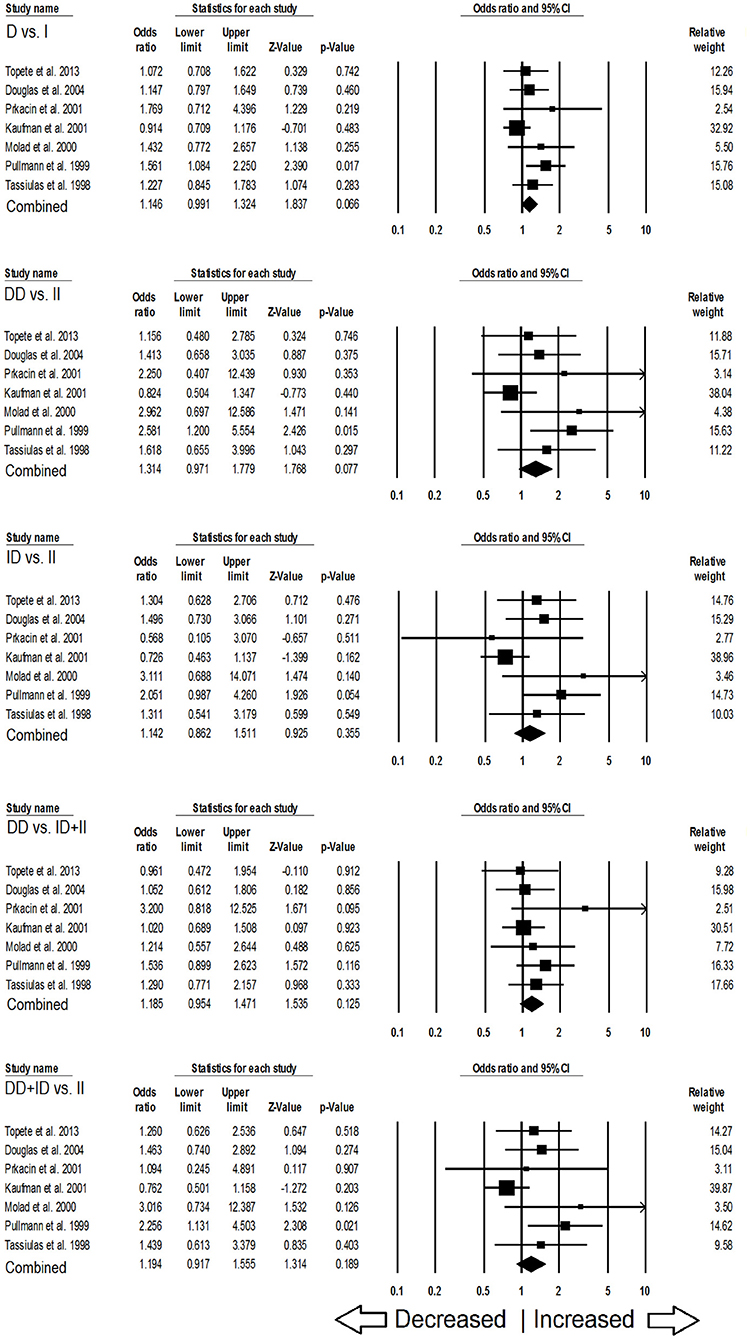
Figure 4. Forest plot of ORs with 95% CI of SLE risk associated with the ACE I/D gene polymorphism for the Caucasian subgroup population. Black square represents the value of OR and the size of the square indicates the inverse proportion relative to its variance. Horizontal line is the 95% CI of OR.
Association of ACE I/D Polymorphism and SLE Susceptibility in African Population
Likewise, in case of subgroup analysis of African ethnicity population, 5 studies with a total number of 446 confirmed SLE cases and 336 controls were included for the pooled analysis. We observed heterogeneity in two genetic models (Figure SI4), so the random effect models were applied to generate ORs and 95% CIs (Table 7). Also, we found that all the genotypic models were not significantly associated with increased or decreased risk of SLE, i.e., allelic (D vs. I: p = 0.683; OR = 1.083, 95% CI = 0.738–1.591), homozygous (DD vs. II: p = 0.757; OR = 1.073, 95% CI = 0.687–1.675), heterozygous (ID vs. II: p = 0.952; OR = 1.014, 95% CI = 0.652–1.575), dominant (DD+ID vs. II: p = 0.791; OR = 1.057, 95% CI = 0.702–1.592) and recessive (DD vs. ID+II: p = 0.766; OR = 1.110, 95% CI = 0.559–2.204) genetic models (Figure 5).
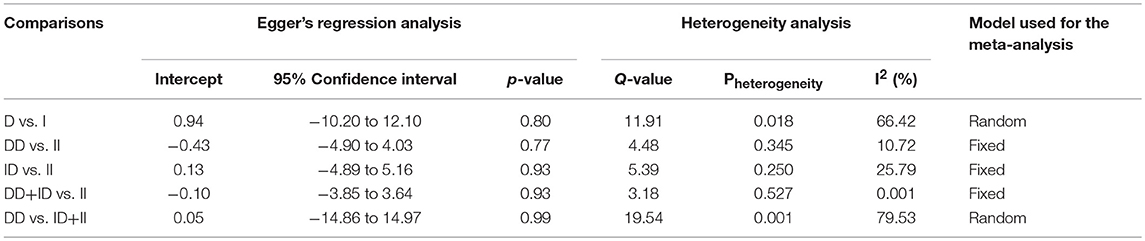
Table 7. Statistics to test publication bias and heterogeneity in the present meta-analysis: African population.
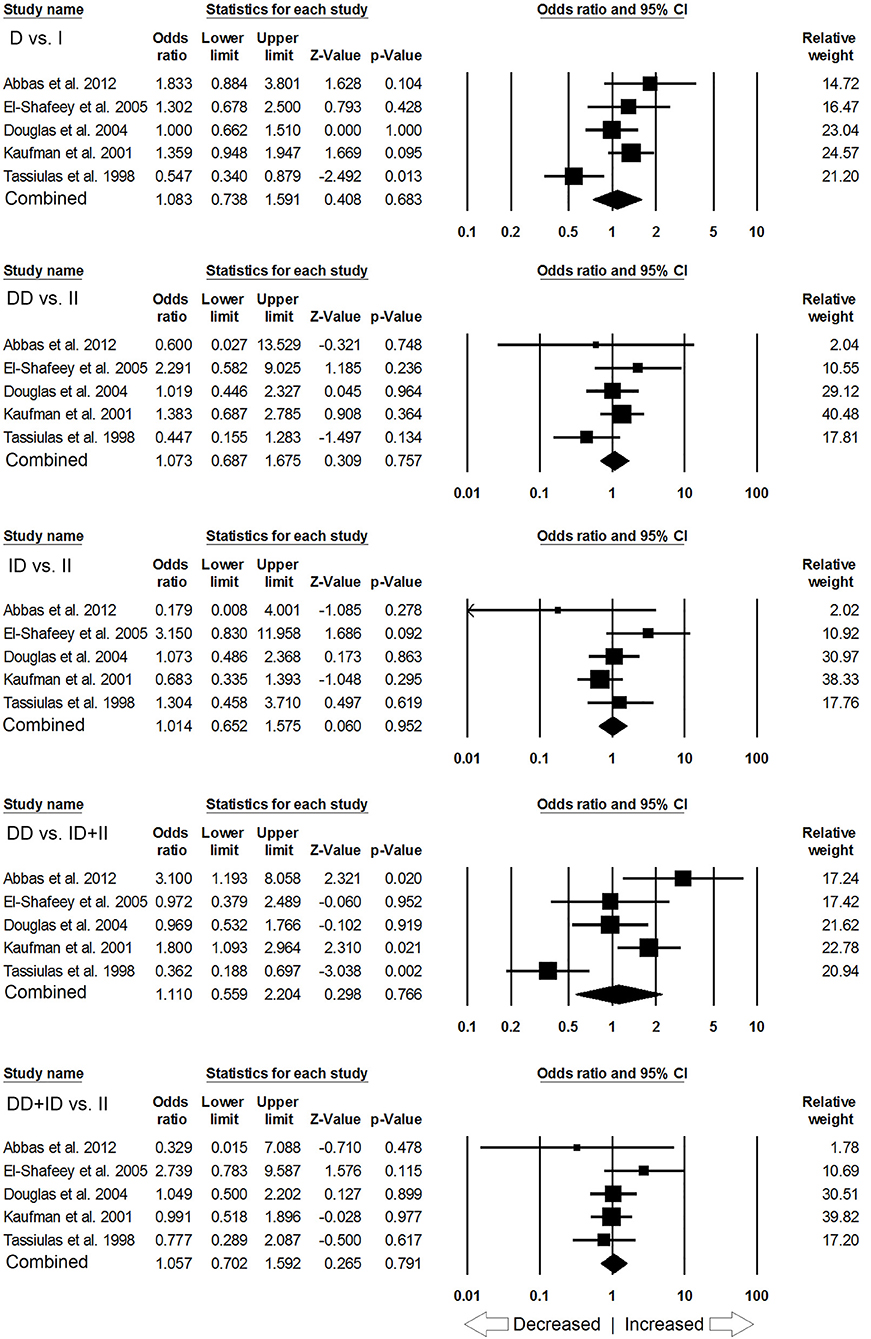
Figure 5. Forest plot of ORs with 95% CI of SLE risk associated with the ACE I/D gene polymorphism for the African subgroup population. Black square represents the value of OR and the size of the square indicates the inverse proportion relative to its variance. Horizontal line is the 95% CI of OR.
Sensitivity Analysis
To appraise the effect of an individual study on the overall SLE risk, we performed leave-one-out sensitivity analysis and recomputed the pooled ORs. The estimated pooled ORs calculated after excluding a single study did not show any differences from the primary values. This suggests that the results of sensitivity analysis were stable for overall SLE risk (Figure SI5). Moreover, the estimated pooled ORs also did not show any change in the subgroup analyses (for Asian, Caucasian, and African ethnicities), which suggested that the results of subgroup analyses were robust (Figure SI6–SI8, respectively).
Trial Sequential Analysis (TSA) of ACE I/D Gene Polymorphism With SLE Risk
Our TSA analysis depicted that cumulative Z curve crossed the trial monitoring boundary before required information size (6762 subjects) was reached. The dominant model was taken as an example in the TSA analysis, which indicated that ACE I/D gene polymorphism is associated with SLE risk and hence no further trials are required (Figure 6A). Subgroup analysis based on ethnicity revealed that Z curve did not cross the trial monitoring boundary before required information size was reached, suggesting insufficiency of cumulative evidence, therefore demanding further trials (Figures 6B–D).
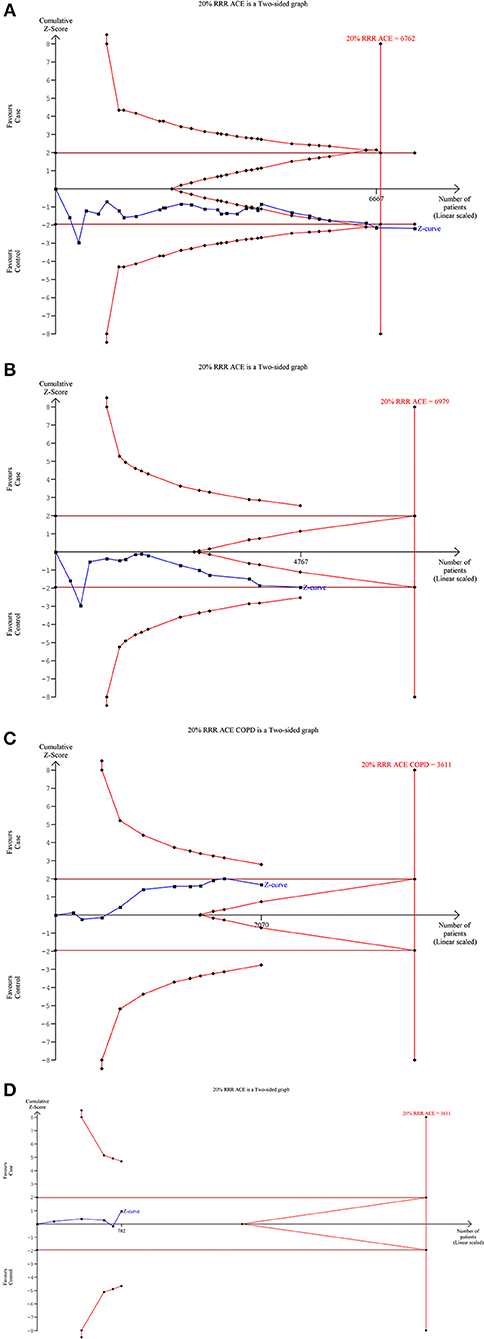
Figure 6. Trial sequence analysis of all the studies on ACE I/D gene polymorphism and SLE risk based on dominant genetic model: (A) Overall, (B) Asian, (C) Caucasian, and (D) African.
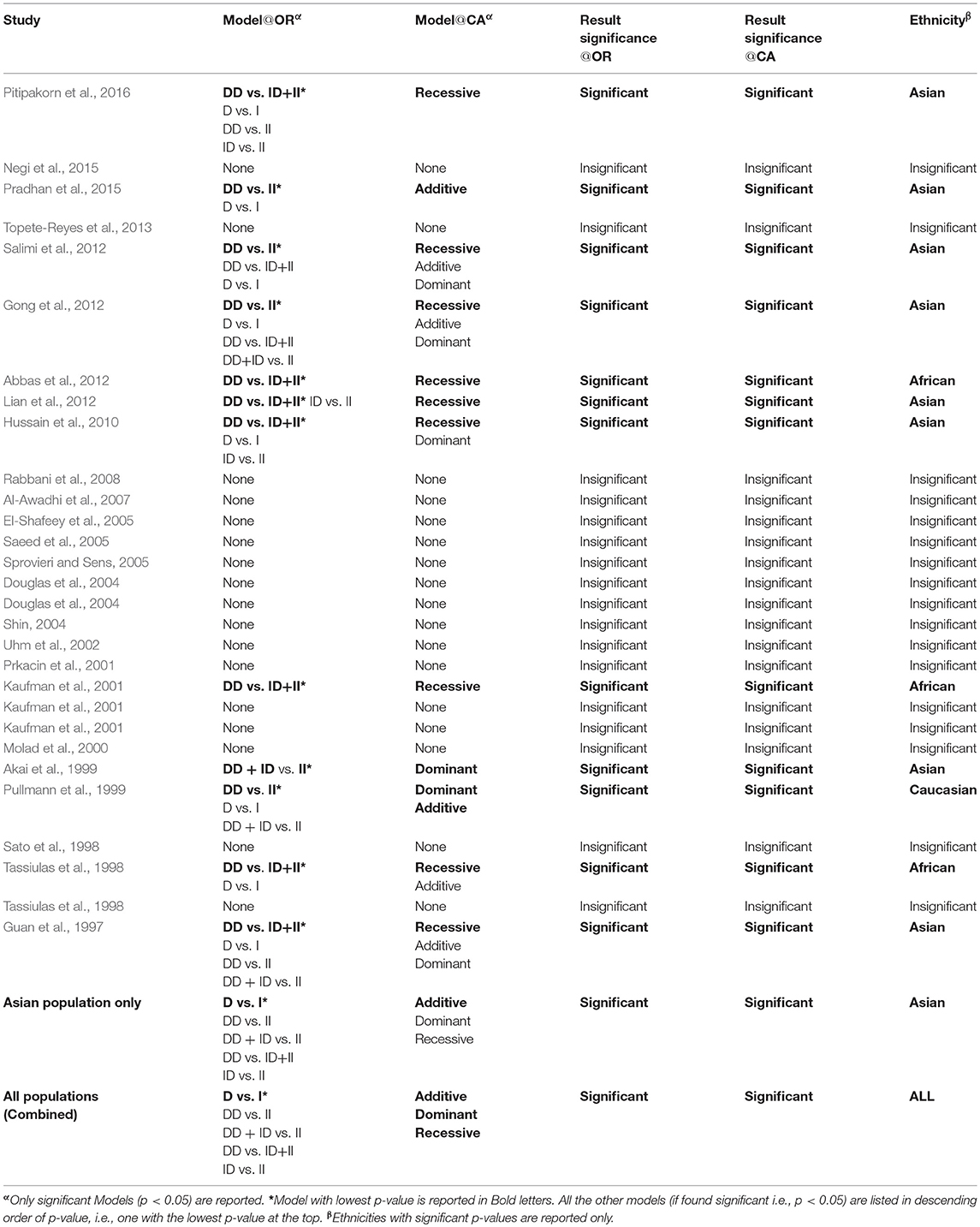
Cochran-Armitage (CA) and Log Odds Ratio (OR)
A comparison of the results obtained from OR and CA trend test is shown in Table 8. It can be observed that significant association was found mostly in the case of Asian population (eight cases out of total twelve significant associations observed as per CA trend test, Table 8). CA results confirm that more than 53% of Asian association studies have a significant association of I/D polymorphism to SLE. For African Population three out of the total five studies considered in this meta-analysis were found to have significant recessive model association to SLE susceptibility; however more studies are required to state a clear association. The Caucasian ethnicity appears to have no significant association of I/D polymorphism to SLE susceptibility (only one out of seven studies considered in this meta-analysis was found to have a significant dominant association). Table 8 reports the Models, which were found significant as per the log odds ratio and CA trend test. It is evident that in all the cases considered the results from CA and Log odds ratio (OR) are same (column 4 and 5, Table 8). It can be observed that the results from the log odds ratio exactly follow the findings from the CA trend test (detailed p-values can be referred from the attached Excel Sheet: Supplementary Material).
Discussion
Earlier studies have reported that individual's SLE susceptibility often present a variety of symptoms that pose difficulty in the disease diagnosis by the physicians, leading to a consequent delay in the onset of the treatment. Host factors, including genetic polymorphisms implicated in autoimmune diseases, might have interpreted this divergence. Thus, there is an urgent need of the identification of the genetic biomarkers that are responsible for the onset and progress of SLE. Nowadays, the current research focus is based on the role of genetic susceptibility in autoimmune disease progression or development.
Renin—angiotensin system (RAS) regulates the arterial blood pressure at both levels—systemic and tissue, and contributes to the immunological responses arising during various phases of nephropathy evolution (Egido, 1996). The major cause of late stage morbidity and mortality in SLE patients is ischemic heart disease (IHD) (Urowitz et al., 1976; Starfelt et al., 1992). It looks evident that ACE plays an important role in SLE etiology by affecting the immune responses and vascular changes. Angiotensin II converted by ACE is a potent pro-inflammatory modulator for immune responses in the renal tissues and shows robust potential in mediating the development and progression of renal disease during SLE (Suzuki et al., 2000; Taal et al., 2000). As the ACE gene has different SNP sites, and research studies have shown that SNPs are capable of changing the structure of the genome and influence protein expression and function that leads to increased risk of different autoimmune diseases, for e.g., SLE. It is possible that Ins/Del (I/D) genotype/allele might confer susceptibility in SLE patients. Individual studies generally have low statistical power to detect the risk, owing to their small sample sizes. Therefore, it is more judicious to estimate the precise relationship of ACE I/D gene polymorphism to understand the contribution of this polymorphism in overall SLE risk.
Meta-analysis, as a statistical strategy, is capable of reducing the pernicious effect of the stochastic processes on studies reporting the false-positive and false-negative associations by pooling the sample size from similar studies. Using the meta-analysis strategy, we in this study pooled the data from all the 25 eligible case-control studies and observed that the individuals carrying D allele are at the higher risk of developing SLE when compared to individuals carrying the wild I allele. This supportive evidence (i.e., the results of meta-analysis) was further confirmed by the TSA and comparison with CA trend test statistics, which authenticated the association of ACE I/D polymorphism with an increased SLE risk. These results confirm that ACE I/D genetic variant may interfere with its expression level and plays a pivotal role in the progression of SLE. Earlier studies have reported that circulating ACE levels vary greatly between individuals and is extremely determined genetically. ACE I/D polymorphism could alter the circulation of ACE levels. It has been found that individuals carrying DD genotype had 2-folds higher circulating ACE levels and lowest among the II genotype carrying individuals (Cambien et al., 1988; Alhenc-Gelas et al., 1991).
Above studies have reported higher serum ACE levels in SLE patients. However, the interruption of the renin angiotensin with ACE inhibitors or angiotensin receptor blockers (ARBs) is recommended as first line adjuvant therapy for the patients with lupus nephritis for proteinuria (Bertsias et al., 2012). Earlier studies on lupus patients and experimental studies on lupus-prone mice suggested that renin angiotensin system inhibition reduces glomerular injury and proteinuria along with transforming growth factor beta, a major mediator of renal fibrosis (De Albuquerque et al., 2004; Tselios et al., 2014). Animal studies have also enlightened the inflammatory components of renin angiotensin and the potential benefits of angiotensin blockade in reducing or eliminating the inflammation in lupus nephritis (Teplitsky et al., 2006). Lupus nephritis patients not using ACE inhibitor have shown association with increased carotid atherosclerosis (Ravenell et al., 2012). Earlier studies have shown that ACE inhibitor delays the occurrence of renal involvement and stabilizes the disease activity in SLE patients (Duran-Barragan et al., 2008). Therefore, these agents may be used in clinical improvement of arterial hypertension and proteinuria in SLE patients.
The present meta-analysis gives a preliminary overview of the involvement of ACE I/D gene polymorphism in SLE etiology and sheds valuable insight on its pathogenesis. Therefore, a better understanding of ACE related genetic, epigenetic, environmental, and clinical factors may add to the effective prevention methods for SLE treatment. The defects in the immune-surveillance complex pathway may shed light on new therapeutic targets for SLE.
During the subgroup analysis of ACE I/D polymorphism and SLE risk in each ethnic group, the pooled analysis demonstrated that ACE I/D polymorphism is significantly associated with SLE risk in Asian population. However, this polymorphism has no role of SLE risk in Caucasian and African population. Since the overall number of studies in non-Asian population is less, it may be possible that the ethnic subgroup analyses of Caucasian and African population may have delineated ambiguous outcomes. Hence, larger studies with bigger sample size from Caucasian and African population are warranted to explore the precise association in this subgroup ethnicity analysis.
The polygenic nature of SLE etiology, and diversity in role of ACE I/D gene polymorphism in developing SLE risk renders implication of a single gene variant for risk of developing this complex autoimmune disease due to clinical heterogeneity and acquired genetic alterations in SLE.
In comparison with the previously published meta-analyses (Zhou et al., 2012; Lee et al., 2013), our study included all the eligible studies which were missed somehow in the previous meta-analyses and also added some new eligible studies providing the largest sample size for ACE I/D polymorphism and SLE risk. Some advantageous features of the present meta-analysis are application of NOS scale for stringent quality assessment where majority of studies showed good quality in terms of sample size, genotypes, and inclusion of patients and healthy controls; adoption of strict search and pre-set selection procedure for the inclusion of the studies; recruitment of more studies (missed in earlier meta-analyses) for increased statistical power and robust conclusion; well exploration of the methodological issues generally occurring in pooled analysis (for e.g., publication bias and sensitivity) which further confirmed the reliability and validity of the present study; and reduction of type I error rate by Trial sequential analysis.
Despite the obvious strengths of this meta-analysis (large sample size and TSA implementation), this study also suffers with several limitations, which must be stated and addressed in the future studies. First, the result of overall population may be biased, perhaps owing to slight over-representation of Asian population. Second, the result of subgroup analysis especially for Caucasian and African, it may be possible that the observed result could be partly due to modest number of studies included. Third, there was a significant heterogeneity in some of the pooled analysis, which may affect the meta-analysis outcome. Fourth, unadjusted estimates form the basis of this meta-analysis and studies published in English language only were included. Unpublished data and ongoing studies were not searched and included. Further, studies reporting negative findings are less likely to be published, hence causing publication bias thereby bringing increase of the associations. Fifth, due to lack of sufficient data, we were not able to study lupus nephritis individually in relationship with ACE allele in this meta-analysis. Sixth, we failed to study the gender associations due to limited data. We hope that peer or future researchers will attempt to study the association between SLE susceptibility and gender specification genotype of ACE I/D in the near future.
In conclusion, the pooled results of independent association studies by meta-analysis confirmed statistically significant association between ACE I/D gene polymorphism and SLE susceptibility. Individuals carrying allele “D” of the ACE I/D polymorphism had greater risk of SLE. The findings will advance our understanding of the role of ACE I/D genetic variant and assist in identifying the “at-risk” individuals. Our results also provide a solid foundation for future genetics studies to focus on ACE related phenotypes and integrative network modules analysis to clarify the potential role of ACE genetic variants in SLE risk. Moreover, more meta-analysis with larger sample size should also be encouraged in future to understand the molecular mechanism of ACE gene with SLE development and to verify the current findings reported in this manuscript.
Author Contributions
SK, SD, RM, MW, AJ, AP, ML BM, NA, and SH conceived and designed the study and experiments. SD, SK, RM, MW, AP, NA, and SH performed the experiments. SD, SK, RM, AJ, AP, ML, BM, and NA analyzed the data. AJ, MW, AP, BM, ML, NA, SD, and SH contributed reagents, materials, analysis tools. SD, SK, RM, AP, and SH wrote the paper. All authors reviewed and approved the manuscript.
Acknowledgments
We are grateful to the Deanship of Scientific Research, Jazan University, Jazan-45142, Saudi Arabia, for providing access to the Saudi Digital Library for this research study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2018.01793/full#supplementary-material
References
Abbas, D., Ezzat, Y., Hamdy, E., and Gamil, M. (2012). Angiotensin-converting enzyme (ACE) serum levels and gene polymorphism in Egyptian patients with systemic lupus erythematosus. Lupus 21, 103–110. doi: 10.1177/0961203311418268
Akai, Y., Sato, H., Iwano, M., Kurumatani, N., Kurioka, H., Kubo, A., et al. (1999). Association of an insertion polymorphism of angiotensin-converting enzyme gene with the activity of lupus nephritis. Clin. Nephrol. 51, 141–146.
Al-Awadhi, A. M., Haider, M. Z., Hasan, E. A., Botaiban, F., Al-Herz, A., et al. (2007). Angiotensin-converting enzyme gene polymorphism in Kuwaiti patients with systemic lupus erythematosus. Clin. Exp. Rheumatol. 25, 437–442.
Alhenc-Gelas, F., Richard, J. L., Courbon, D., Warnet, J. M., and Corvol, P. (1991). Distribution of plasma angiotensin I-converting enzyme levels in healthy men: relationship to environmental and hormonal parameters. J. Lab Clin. Med. 117, 33–39.
Bertsias, G. K., Tektonidou, M., Amoura, Z., Aringer, M., Bajema, I., Berden, J. H., et al. (2012). Joint European league against rheumatism and european renal association–european dialysis and transplant association (EULAR/ERA-EDTA) recommendations for the management of adult and paediatric lupus nephritis. Ann. Rheum. Dis. 71, 1771–1782. doi: 10.1136/annrheumdis-2012-201940
Brok, J., Thorlund, K., Wetterslev, J., and Gluud, C. (2009). Apparently conclusive meta-analyses may be inconclusive–Trial sequential analysis adjustment of random error risk due to repetitive testing of accumulating data in apparently conclusive neonatal meta-analyses. Int. J. Epidemiol. 38, 287–298. doi: 10.1093/ije/dyn188
Burton, P. R., Hansell, A. L., Fortier, I., Manolio, T. A., Khoury, M. J., Little, J., et al. (2009). Size matters: just how big is BIG?: quantifying realistic sample size requirements for human genome epidemiology. Int. J. Epidemiol. 38, 263–273. doi: 10.1093/ije/dyn147
Cambien, F., Alhenc-Gelas, F., Herbeth, B., Andre, J. L., Rakotovao, R., Gonzales, M. F., et al. (1988). Familial resemblance of plasma angiotensin converting enzyme levels: the Nancy Study. Am. J. Hum. Genet. 43, 774–780.
De Albuquerque, D. A., Saxena, V., Adams, D. E., Boivin, G. P., Brunner, H. I., Witte, D. P., et al. (2004). An ACE inhibitor reduces Th2 cytokines and TGF-b1 and TGF-b2 isoforms in murine lupus nephritis. Kidney Int. 65, 846–859. doi: 10.1111/j.1523-1755.2004.00462.x
DerSimonian, R., and Laird, N. (1986). Meta-analysis in clinical trials. Control Clin. Trials 7, 177–188. doi: 10.1016/0197-2456(86)90046-2
Douglas, G., Reilly, C., Dooley, M. A., Page, G., Cooper, G., and Gilkeson, G. (2004). Angiotensin-converting enzyme (insertion/deletion) and endothelial nitric oxide synthase polymorphisms in patients with systemic lupus erythematosus. J. Rheumatol. 31, 1756–1762.
Duran-Barragan, S., McGwin, G. Jr., Vila, L. M., Reveille, J. D., and Alarcon, G. S. (2008). Angiotensin converting enzyme inhibitors delay the occurrence of renal involvement and are associated with a decreased risk of disease activity in patients with systemic lupus erythematosus–results from LUMINA (LIX): a multiethnic US cohort. Rheumatology 47, 1093–1096. doi: 10.1093/rheumatology/ken208
Egger, M., Davey Smith, G., Schneider, M., and Minder, C. (1997). Bias in meta-analysis detected by a simple, graphical test. BMJ 7109, 629–634. doi: 10.1136/bmj.315.7109.629
Egido, J. (1996). Vasoactive hormones and renal sclerosis. Kidney Int. 49, 578–597. doi: 10.1038/ki.1996.82
El-Shafeey, M. M., El-Shayeb, M., Othman, E., and Elfawy, N. (2005). Angiotensin converting enzyme gene polymorphism in egyptian patients with systemic lupus erythematosus. Egyptian J. Hospital Med. 21, 131–138.
Frangou, E. A., Bertsias, G. K., and Boumpas, D. T. (2013). Gene expression and regulation in systemic lupus erythematosus. Eur. J. Clin. Invest 43, 1084–1096. doi: 10.1111/eci.12130
Gong, A. M, Li, X. Y, Wang, Y. Q., Yan, H. X., Xu, Z. X., Zhao, F., et al. (2012). Association study of ACE polymorphisms and systemic lupus erythematosus in northern chinese han population. Mol. Biol. Rep. 39, 9485–9491. doi: 10.1007/s11033-012-1813-7
Gregersen, P. K., and Olsson, L. M. (2009). Recent advances in the genetics of autoimmune disease. Annu. Rev. Immunol. 27, 363–391. doi: 10.1146/annurev.immunol.021908.132653
Guan, T., Liu, Z., and Chen, Z. (1997). Angiotensin-converting enzyme gene polymorphism and the clinical pathological features and progression in lupus nephritis. Zhonghua Nei Ke Za Zhi 36, 461–464.
Harley, J. B., Kelly, J. A., and Kaufman, K. M. (2006). Unraveling the genetics of systemic lupus erythematosus. Springer Semi. Immunopathol. 28, 119–130. doi: 10.1007/s00281-006-0040-5
Higgins, J. P., Thompson, S. G., Deeks, J. J., and Altman, D. G. (2003). Measuring inconsistency in meta-analyses. BMJ 7414, 557–560. doi: 10.1136/bmj.327.7414.557
Hu, P., Huang, M.-Y., Hu, X.-Y., Xiang, M.-X., Liu, X-,., et al. (2015). Meta-analysis of C242T polymorphism in CYBA genes: risk of acute coronary syndrome is lower in Asians but not in Caucasians. J. Zhejiang Univ. Sci. B. 16, 370–379. doi: 10.1631/jzus.B1400241
Hussain, N., Jaffery, G., Hasnain, S., and Sabri, A. N. (2010). Angiotensin-converting enzyme gene I/D polymorphism in Pakistani systemic lupus erythematosus patients. African J. Biotechnol. 9, 8134–8138. doi: 10.5897/AJB09.087
Kasal, D. A., and Schiffrin, E. L. (2012). Angiotensin II, aldosterone, and anti-inflammatory lymphocytes: interplay and therapeutic opportunities. Int. J. Hypertens. 2012:829786. doi: 10.1155/2012/829786
Kaufman, K. M., Kelly, J., Gray-McGuire, C., Asundi, N., Yu, H., Reid, J., et al. (2001). Linkage analysis of angiotensin-converting enzyme (ACE) insertion/deletion polymorphism and systemic lupus erythematosus. Mol. Cell Endocrinol. 177, 81–85. doi: 10.1016/S0303-7207(01)00424-5
Kyttaris, V. C., Krishnan, S., and Tsokos, G. C. (2006). Systems biology in systemic lupus erythematosus: integrating genes, biology and immune function. Autoimmunity 39, 705–709. doi: 10.1080/08916930601061363
Lee, Y. H., Choi, S. J., Ji, J. D., and Song, G. G. (2013). Association between the angiotensin-converting enzyme insertion/deletion polymorphism and susceptibility to systemic lupus erythematosus: a meta-analysis. J. Renin. Angiotensin. Aldosterone Syst. 14, 248–254. doi: 10.1177/1470320312459979
Lian, L. H., Lau, T. P., Ching, A. S., and Chua, K. H. (2012). Angiotensin-converting enzyme gene I/D dimorphism does not play a major role in the susceptibility of Malaysian systemic lupus erythematosus patients. Genet. Mol. Res. 11, 863–871. doi: 10.4238/2012.April.10.2
Mantel, N., and Haenszel, W. (1959). Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 4, 719–748.
Molad, Y., Gal, E., Magal, N., Sulkes, J., Mukamel, M., Weinberger, A., et al. (2000). Renal outcome and vascular morbidity in systemic lupus erythematosus (SLE): lack of association with the angiotensin-converting enzyme gene polymorphism. Semin. Arthritis Rheum. 30, 132–137. doi: 10.1053/sarh.2000.8365
Morrissey, J. J., and Klahr, S. (1998). Differential effects of ACE and AT1 receptor inhibition on chemoattractant and adhesion molecule synthesis. Am. J. Physiol. 274, F580–6. doi: 10.1152/ajprenal.1998.274.3.F580
Negi, V. S., Devaraju, P., and Gulati, R. (2015). Angiotensin-converting enzyme (ACE) gene insertion/deletion polymorphism is not a risk factor for hypertension in SLE nephritis. Clin. Rheumatol. 34, 1545–1549. doi: 10.1007/s10067-015-2954-6
Pitipakorn, U., Suwannalai, P., Trachoo, O., Rattanasiri, S., Chitphuk, S., Ngamjanyaporn, P., et al. (2016). Angiotensin-converting enzyme gene polymorphism in Thai patients with systemic lupus erythematosus. Int. J. Rheum. Dis. 19, 693–699. doi: 10.1111/1756-185X.12609
Pradhan, V., Kemp, E. H., Nadkar, M., Rajadhyaksha, A., Lokhandwala, K., Patwardhan, M., et al. (2015). Association between the angiotensin converting enzyme gene insertion/deletion polymorphism and susceptibility to systemic lupus erythematosus in an Indian population. Scand. J. Rheumatol. 44, 425–427. doi: 10.3109/03009742.2015.1022214
Prkacin, I., Novak, B., Sertić, J., and Mrzljak, A. (2001). Angiotensin-converting enzyme gene polymorphism in patients with systemic lupus. Acta Med. Croatica 55, 73–76.
Pullmann, R. Jr., Lukác, J., Skerenová, M., Rovensky, J., Melus, V., et al. (1999). Association between systemic lupus erythematosus and insertion/deletion polymorphism of the angiotensin converting enzyme (ACE) gene. Clin. Exp. Rheumatol. 17, 593–596.
Rabbani, M. A., Mahmood, M. S., Mekan, S. F., and Frossard, P. M. (2008). Association of angiotensin-converting enzyme gene dimorphisms with severity of lupus disease. Saudi J. Kidney Dis. Transpl. 19, 761–766.
Ravenell, R. L., Kamen, D. L., Spence, J. D., Hollis, B. W., Fleury, T. J., Janech, M. G., et al. (2012). Premature atherosclerosis is associated with hypovitaminosis D and angiotensin converting enzyme inhibitor non-use in lupus patients. Am. J. Med. Sci. 344, 268–273. doi: 10.1097/MAJ.0b013e31823fa7d9
Rigat, B., Hubert, C., Alhencgelas, F., Cambien, F., Corvol, P., and Soubrier, F. (1990). An insertion deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J. Clin. Invest. 86, 1343–1346. doi: 10.1172/JCI114844
Saeed, M., Mekan, S. F., Rabbani, M. A., Arain, F. M., Arif, M., and Shaharyar, S. (2005). Angiotensin converting enzyme (ACE) gene polymorphisms and lupus disease severity: a promising link. Ann. Rheum. Dis. 64, 164–165. doi: 10.1136/ard.2004.020834
Salimi, S., Naghavi, A., Zakeri, Z., Nabizadeh, S., Mashhadi, F. F., and Sandoughi, M. (2012). Association of DD genotype of insertion/deletion polymorphism of angiotensin converting enzyme gene with systemic lupus erythematosus and lupus nephropathy. Zahedan J. Res. Med. Sci. 15, 69–73.
Sasieni, P. D. (1997). From genotypes to genes: doubling the sample size. Biometrics 53, 1253–1261. doi: 10.2307/2533494
Sato, H., Akai, Y., Iwano, M., Kurumatani, N., Kurioka, H., Kubo, A., et al. (1998). Association of an insertion polymorphism of angiotensin-converting enzyme gene with the activity of systemic lupus erythematosus. Lupus 7, 530–534. doi: 10.1191/096120398678920622
Sayed-tabatabaei, F. A., Oostra, B. A., Isaacs, A., Witteman, J. C., et al. (2006). ACE polymorphisms. Circ. Res. 98, 1123–1133. doi: 10.1161/01.RES.0000223145.74217.e7
Shin, K. S. (2004). Association of Angiotensin-converting enzyme gene polymorphism with the disease activity of systemic lupus erythematosus in Korean children. Korean J. Pediatr. 47, 672–676. doi: 10.1038/srep31243
Sprovieri, S. R., and Sens, Y. A. (2005). Polymorphisms of the renin-angiotensin system genes in Brazilian patients with lupus nephropathy. Lupus 14, 356–362 doi: 10.1191/0961203305lu2093oa
Stang, A. (2010). Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta- analyses. Eur. J. Epidemiol. 25, 603–605. doi: 10.1007/s10654-010-9491-z
Starfelt, G., Eskilsson, J., Nived, O., Truedsson, L., and Valind, S. (1992). Cardiovascular disease in systemic lupus erythematosus: a study of 75 patients from a defined population. Medicine 71, 216–223. doi: 10.1097/00005792-199207000-00004
Suzuki, Y., Ruiz-Ortega, M., and Egido, J. (2000). Angiotensin II: a double-edged sword in inflammation. J. Nephrol. 13, S101–S110.
Taal, M. W., Omer, S. A., Nadim, M. K., and Mackenzie, H. S. (2000). Cellular and molecular mediators in common pathway mechanisms of chronic renal disease progression. Curr. Opin. Nephrol. Hypertens. 9, 323–331. doi: 10.1097/00041552-200007000-00001
Tassiulas, I. O., Aksentijevich, I., Salmon, J. E., Kim, Y., Yarboro, C. H., Vaughan, E. M., et al. (1998). Angiotensin I converting enzyme gene polymorphisms in systemic lupus erythematosus: decreased prevalence of DD genotype in African American patients. Clin. Nephrol. 50, 8–13.
Teplitsky, V., Shoenfeld, Y., and Tanay, A. (2006). The renin-angiotensin system in lupus: Physiology, enes and practice, in animals and humans. Lupus 15, 319–325. doi: 10.1191/0961203306lu2306rr
Topete-Reyes, J. F., Soto-Vargas, J., Morán-Moguel, M. C., Dávalos-Rodríguez, I. P, Chávez-González, E. L., García-de la Torre, I., et al. (2013). Insertion/deletion polymorphism of the angiotensin-converting enzyme gene in lupus nephritis among Mexicans. Immunopharmacol. Immunotoxicol. 35, 174–180. doi: 10.3109/08923973.2012.739175
Tsao, B.P. (2003). The genetics of human systematic lupus erytrhematosus. Trends Immunol. 24, 595–602. doi: 10.1016/j.it.2003.09.006
Tselios, K., Koumaras, C., Urowitz, M. B., and Gladman, D. D. (2014). Do current arterial hypertension treatment guidelines apply to systemic lupus erythematosus patients? A critical appraisal. Semin Arthritis Rheum. 4, 521–525. doi: 10.1016/j.semarthrit.2013.07.007
Turner, R. M., Bird, S. M., and Higgins, J. P. (2013). The impact of study size on meta-analyses: examination of underpowered studies in Cochrane reviews. PLoS ONE 8:e59202. doi: 10.1371/journal.pone.0059202
Uhm, W. S., Lee, H. S., Chung, Y. H., Kim, T. H., Bae, S. C., Joo, K., et al. (2002). Angiotensin-converting enzyme gene polymorphism and vascular manifestations in Korean patients with SLE. Lupus 11, 227–233. doi: 10.1191/0961203302lu174oa
Urowitz, M. B., Bookman, A. A. M., Koehler, B. E., Gordon, D. A., Smythe, H. A., and Ogryzlo, M. A. (1976). The bimodal mortality pattern of systemic lupus erythematosus. Am. J. Med. 60, 221–225. doi: 10.1016/0002-9343(76)90431-9
Wetterslev, J., Thorlund, K., Brok, J., Gluud, C., et al. (2008). Trial sequential analysis may establish when firm evidence is reached in cumulative meta-analysis. J. Clin. Epidemiol. 61, 64–75. doi: 10.1016/j.jclinepi.2007.03.013
Woolf, B. (1955). On estimating the relation between blood group and disease. Ann. Hum. Genet. 19, 251–253. doi: 10.1111/j.1469-1809.1955.tb01348.x
Wu, R., and Li, B. (1999). A multiplicative-epistatic model for analyzing interspecific differences in outcrossing species. Biometrics 2, 355–365.
Zhou, T. B., Liu, Y. G., Lin, N., Qin, Y. H., Huang, K., Shao, M., et al. (2012). Relationship between angiotensin-converting enzyme insertion/deletion gene polymorphism and systemic lupus erythematosus/lupus nephritis: a systematic review and meta-analysis. J. Rheumatol. 39, 686–693. doi: 10.3899/jrheum.110863
Keywords: genetic variants, polymorphism, ACE gene, meta-analysis, genotypic risk, SLE
Citation: Khan S, Dar SA, Mandal RK, Jawed A, Wahid M, Panda AK, Lohani M, Mishra BN, Akhter N and Haque S (2018) Angiotensin-Converting Enzyme Gene I/D Polymorphism Is Associated With Systemic Lupus Erythematosus Susceptibility: An Updated Meta-Analysis and Trial Sequential Analysis. Front. Physiol. 9:1793. doi: 10.3389/fphys.2018.01793
Received: 27 March 2018; Accepted: 28 November 2018;
Published: 17 December 2018.
Edited by:
Joseph L. Greenstein, Johns Hopkins University, United StatesReviewed by:
Kumari Sonal Choudhary, University of California, San Diego, United StatesAna Cristina Simões E. Silva, Universidade Federal de Minas Gerais, Brazil
Andreas Kronbichler, Innsbruck Medical University, Austria
Copyright © 2018 Khan, Dar, Mandal, Jawed, Wahid, Panda, Lohani, Mishra, Akhter and Haque. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shafiul Haque, c2hhZml1bC5oYXF1ZUBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Saif Khan
Saif Khan Sajad A. Dar
Sajad A. Dar Raju K. Mandal
Raju K. Mandal Arshad Jawed
Arshad Jawed Mohd Wahid
Mohd Wahid Aditya K. Panda
Aditya K. Panda Mohtashim Lohani
Mohtashim Lohani Naseem Akhter
Naseem Akhter Shafiul Haque
Shafiul Haque